Abstract
Skin identity is controlled by intrinsic features of the epidermis and dermis and their interactions. Modifying skin identity has clinical potential, such as the conversion of residual limb and stump (nonvolar) skin of amputees to pressure-responsive palmoplantar (volar) skin to enhance prosthesis use and minimize skin breakdown. Greater keratin 9 (KRT9) expression, higher epidermal thickness, keratinocyte cytoplasmic size, collagen length, and elastin are markers of volar skin and likely contribute to volar skin resiliency. Given fibroblasts’ capacity to modify keratinocyte differentiation, we hypothesized that volar fibroblasts influence these features. Bioprinted skin constructs confirmed the capacity of volar fibroblasts to induce volar keratinocyte features. A clinical trial of healthy volunteers demonstrated that injecting volar fibroblasts into nonvolar skin increased volar features that lasted up to 5 months, highlighting a potential cellular therapy.
INTRODUCTION:
More than 1.7 million people in United States live with limb amputations, often resulting from diabetes, vasculopathy, or trauma. Amputees often experience skin breakdown at the stump site due to friction and pressure from long-term use of prosthetics. The objective of this study was to convert nonvolar skin to pressure-responsive volar (palmoplantar) skin by means of autologous volar fibroblast transplantation, given the ability for fibroblasts to modulate skin function. We used primary cultured volar (sole) or nonvolar (scalp) fibroblasts from healthy volunteers to test for distinctive responses to pressure, capacity for modifying skin identity in bioprints, and potential cell therapy through autologous engraftment of fibroblasts in a clinical trial.
RATIONALE:
Fibroblasts retain positional memory of their origin when expanded in vitro. Also, fibroblasts modify skin function, including the differentiation of the epidermis. Given that volar fibroblasts likely also have distinct responses to pressure, we hypothesized their utility as an ectopic cellular therapy to enhance pressure adaptation in otherwise nonpressure-bearing areas.
RESULTS:
In a two-dimensional culture system, volar fibroblasts showed higher proliferation and migration rates compared with nonvolar fibroblasts. The higher migration rate of volar fibroblasts was characterized by a distinctive slingshot movement not modified by static (continuous) pressure treatment. Dynamic (on and off) pressure treatment highlighted distinct gene expression changes in volar versus nonvolar fibroblasts, including particular HOX genes. Volar fibroblasts in bioprinted skin induced volar features in nonvolar epidermis, such as greater epidermal thickness, larger keratinocyte size, and increased amounts of KRT9 and FGF7 proteins. Similarly to the results observed with bioprinted skin, a clinical trial in which human volunteers received injections of ectopic autologous sole fibroblasts into nonvolar skin confirmed modified volar features in the nonvolar skin. These features included increased skin firmness, greater epidermal thickness, larger keratinocyte size, longer dermal collagen fiber length, increased dermal elastin expression, and elevated expressions of KRT9, KRT17, pEGFR (Y1068), FGFBP1, FGF7, and RBPJ proteins. Single-cell transcriptomic analysis further confirmed gene expression modifications associated with epidermal development, keratinization, epidermal cell differentiation, and cell shape at sole fibroblast injection sites 2 weeks to 17 months after injection. Spatial transcriptomics confirmed single-cell RNA sequencing to detect the stable engraftment of ectopic volar fibroblast population.
CONCLUSION:
These data demonstrate the distinctive properties and therapeutic potential of volar fibroblasts to modify skin identity toward a volar phenotype and greater pressure adaptation. Future work is required to enhance the engraftment rate and degree of phenotype switching with ectopic fibroblasts. ■
Graphical Abstract
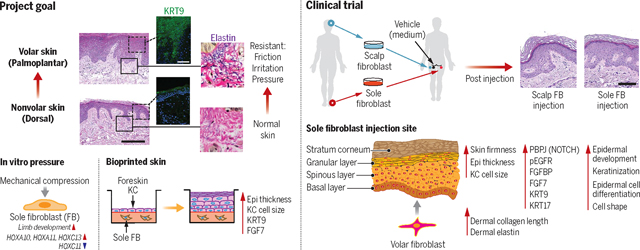
Project goal, pressure response of sole fibroblasts, bioprinted skin, and ectopic autologous cell therapy clinical trial. (Top left) An overview of our project goal. (Bottom left) Pressure-responsive genes in volar fibroblast. (Bottom middle) Morphological and gene expression changes in bioprinted skin with foreskin keratinocytes and sole fibroblasts. (Right) Clinical trial design and morphological and protein expression changes at the sole fibroblast injection site.
Cellular adaptation to pressure at volar (palmoplantar) sites likely began with the first terrestrial tetrapod, but the basic biology and translational potential are understudied. By contrast, the diversity of fibroblast populations common to most skin sites has received more recent attention (1, 2). Building from others’ work (3–7), we hypothesized that palmoplantar (volar) fibroblasts endow some of the pressure adaptation of volar skin and can be used as a cell therapy to prevent ectopic pressure-related skin problems, such as stump-site skin erosions in amputees or decubitus ulcers in the infirm. In the case of amputees, more than 1.7 million (about 1 out of every 200) people in the United States suffer from limb loss, predominantly as a result of diabetes, vasculopathy, or trauma. Despite great strides in prosthetic designs, many amputees self-restrict their activity because of inconvenience, pain, and actual or feared stump dermatoses from prosthesis use (8). Given the paucity of attention to the skin side of the limb-prosthesis interface, we hypothesized that enhancing volar features at the residual limb in those with limb loss might enhance prosthesis use and improve quality of life. Volar fibroblasts, therefore, deserve further investigation as therapeutic candidates.
Volar skin has not been fully characterized, but several features are well appreciated. Foremost, volar skin has a thick epidermis, whose distinct morphological layers contribute to its function (9). Starting superficially, the stratum corneum (SC) is the outermost layer of dead, anucleated keratinocytes that are cross-linked to form a strong, water-impermeable layer. In volar skin, the SC is thicker than that of nonvolar skin—up to nearly six times thicker in the palm compared with the dorsum of the hand (10). Progressing deeper, the layer of living keratinocytes beneath the SC in volar skin is roughly 1.5 times thicker than that of nonvolar skin (11, 12). Dermal thickness is estimated to be about 1.2 to 1.5 times thicker when comparing palm tissue to nonvolar skin with high-frequency ultrasonography (13). Collagen in human volar plantar skin also has a higher frequency of thicker cross-sectional diameter compared with human nonvolar abdominal skin (14). The subcutaneous fat layer beneath the dermis, however, has not been well described in volar skin. All of these features—and recently described cellular gene expression differences (15–19)—likely enhance pressure distribution and cell viability. These adaptations likely accommodate circumstances of extreme pressure (up to several thousand kilopascals), such as during running, which is estimated to produce pressures equivalent to a force of 2.5 times body weight over the area of the foot (20).
As a candidate to promote volar features, fibroblasts generally have dramatic effects on epidermal function, such as the dermal papillae control of hair morphogenesis and cycling (21, 22). Older literature supports this key role of the mesenchyme in modulating epidermal function (23) and even of volar fibroblasts in inducing volar features in the epidermis (7). More recently, anatomically diverse fibroblasts were shown to have retained homeobox signatures that code their positional memory over a large number of in vitro passages (3, 4, 24). These studies also confirmed that volar fibroblasts can induce the gene expression of volar keratin 9 (KRT9) (6, 25). In functional human studies, Yamaguchi et al. (6) showed a model in which epidermis-only grafts spliced onto human palmoplantar dermis led to KRT9-expressing keratinocytes. Although these studies confirmed the logic of using ectopic fibroblasts, they did not characterize pressure responses of volar fibroblasts, their use in human subjects, or single-cell resolution of epidermal-mesenchymal interactions.
In this study, our aim was to test the hypothesis that volar fibroblasts have distinctive properties to adapt to pressure and to modify skin identity in human subjects. To test this, we used in vitro live imaging, mechanical pressure testing, gene expression characterization, and bioprinting. We also conducted a human trial in which we analyzed the behavior of volar fibroblasts at single-cell resolution. Our results demonstrate that volar cells have specific features that are associated with long-term changes in human subjects after injection. This provides a platform for testing future improvements and iterations toward potential human-cell therapies.
Results
Volar and nonvolar fibroblasts respond to pressure differently
Given that pressure is a distinct environmental exposure that native volar fibroblasts routinely experience at magnitudes of up to several thousand kilopascals in the case of jogging (20), we applied 100 kPa of static hydraulic pressure for 3 hours to volar (sole) or nonvolar (scalp) fibroblasts (Fig. 1A). We used scalp fibroblasts as a nonvolar control to avoid dorsalventral and distal-proximal axis confounders. Volar and nonvolar fibroblasts were then assessed for proliferation and migration speed. Ki67 staining showed that volar fibroblasts were significantly more proliferative (P = 0.02) compared with nonvolar fibroblasts. However, the application of pressure did not show differences between volar and nonvolar fibroblasts (Fig. 1B). Although pressure did not alter migration speed, volar fibroblasts showed significantly higher migration speed than nonvolar fibroblasts (P ≤ 0.005 both with and without pressure, Fig. 1C). This was demonstrated by the “slingshot” movement (26) of volar fibroblasts, which refers to intermittent, high-speed migration, as shown in the relatively noncontinuous trace of volar fibroblasts of Fig. 1D and movies S1 to S4. The histogram in Fig. 1D indicates higher frequencies of rapid displacements in volar fibroblasts in comparison with nonvolar fibroblasts. These studies show that even under static pressure, volar fibroblasts maintain phenotypic differences in proliferation and migration.
Fig. 1. Volar fibroblasts showed distinct responses to pressure and induced volar features in 3D-printed skin tissue constructs.
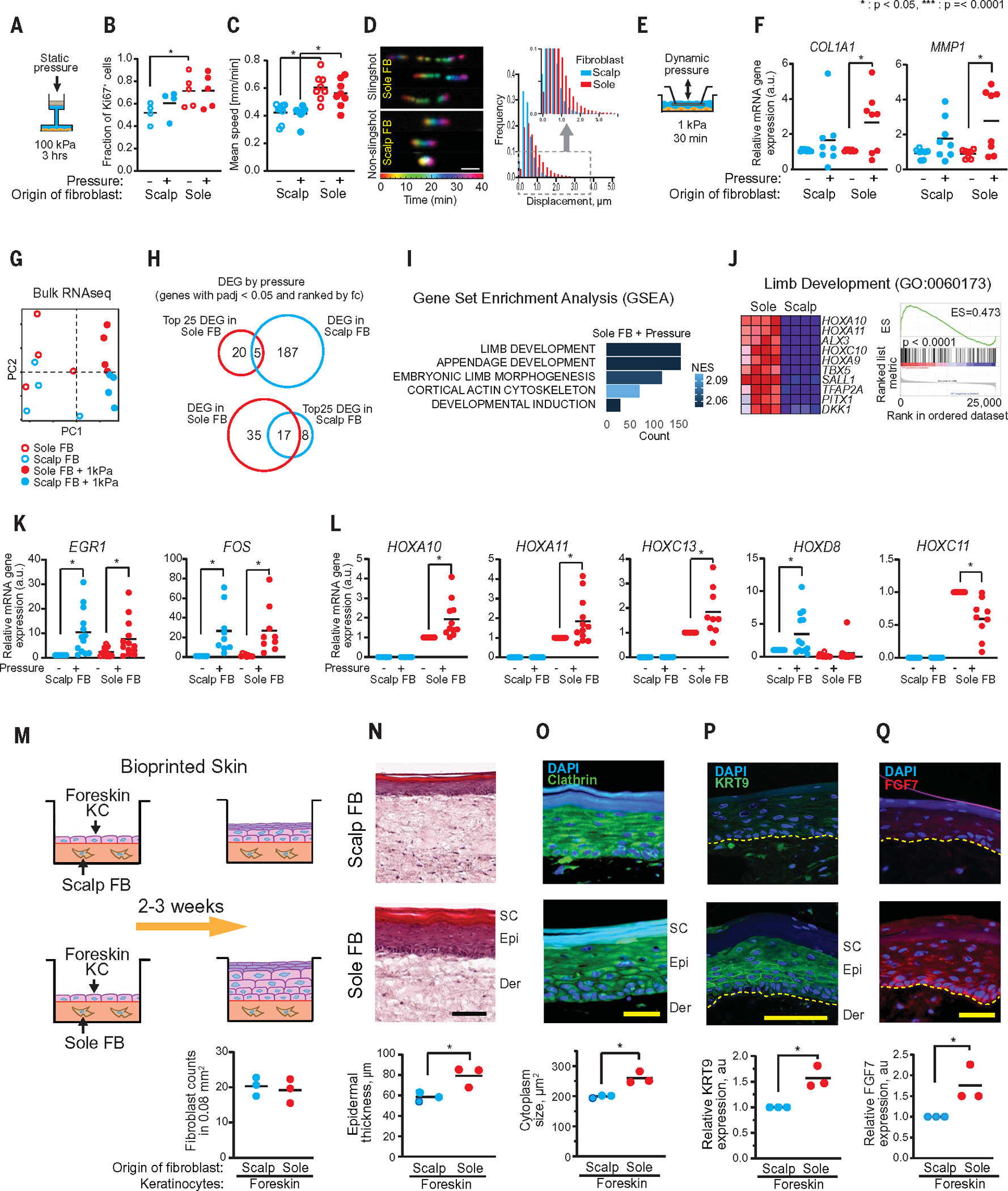
(A) Schematic diagram illustrates an in vitro static continuous pressurizing system that applied 100 kPa of hydraulic pressure to cultured fibroblasts for 3 hours. (B) Proliferation and cell count of Ki67-stained fibroblasts were measured with flow cytometry. Sole fibroblasts show significantly (P = 0.02) increased fraction of Ki67+ compared with scalp fibroblasts (n = 4 distinct subjects with 1 repeat in scalp fibroblasts, n = 5 distinct subjects with 1 repeat in sole fibroblasts). Pressurizing scalp or sole fibroblasts does not show significant changes in Ki67+ fraction. (C) Mean migration speed was measured by microscopy and analyzed with ImageJ. Sole fibroblasts showed significantly higher migration speed compared with scalp fibroblasts both without and with pressure (P = 0.005 and P = 0.004, respectively). No significant difference was observed between pressurized and nonpressurized fibroblasts (n = 4 distinct subjects with 1 repeat in scalp fibroblasts, n = 5 distinct subjects with 1 repeat in sole fibroblasts). (D) Representative images of scalp and sole fibroblast migration tracings and histogram. Noncontinuous trace of sole fibroblasts compared with continuous trace of scalp fibroblasts demonstrates characteristic “slingshot” movement of sole fibroblasts. Sole fibroblasts show higher frequencies at longer displacement distances. Histogram represents 664 nonvolar and 385 volar fibroblasts from a single subject (n = 7 distinct subjects). (E) Schematic diagram illustrates an in vitro dynamic oscillatory pressurizing system that applied 1 kPa in 0.025 Hz (20 s on, 20 s off) intervals to cultured sole or scalp fibroblasts for 30 min. (F) Scatter plots of quantitative PCR (qPCR) data indicate COL1A1 and MMP1 mRNA expression in response to dynamic oscillatory pressure. Only sole fibroblasts significantly increased expression of COL1A1 (P = 0.03) and MMP1 (P = 0.01) after pressure (n = 8 distinct subjects with 3 repeats in scalp and sole fibroblasts). (G) PCA plot from bulk RNA-seq data (n = 3 distinct subjects). (H) Overlap of top 25 up-regulated DEG in sole fibroblasts associated with pressure treatment and full DEG in scalp fibroblasts associated with pressure treatment, or vice versa. (I) Selected GO pathways in GSEA of transcripts modified by pressure in sole fibroblasts (full list is in fig. S3B). (J) Top 10 genes associated with the limb development pathway are shown as a heatmap and enrichment plot. (K) Scatter plots of qPCR data illustrate significant increases in mRNA expression of EGR1 (P = 0.0002 in scalp fibroblast, P = 0.05 in sole fibroblast, n = 13 distinct subjects with 3 repeats) and FOS (P = 0.01 in scalp; P = 0.01 in sole fibroblast, n = 9 distinct subjects with 3 repeats) in both scalp and sole fibroblasts in response to pressure. (L) Scatter plots of qPCR data for HOX genes. HOXA10 (P = 0.009, n = 11 distinct subjects with 3 repeats), HOXA11 (P = 0.026, n = 12 distinct subjects with 3 repeats), and HOXC13 (P = 0.02, n = 9 distinct subjects with 3 repeats) are up-regulated only in pressure-treated sole fibroblasts. HOXD8 is up-regulated only in pressure-treated scalp fibroblasts (P = 0.005, n = 12 distinct subjects with 3 repeats). HOXC11 is down-regulated only in pressure-treated sole fibroblasts (P = 0.004, n = 9 distinct subjects with 3 repeats). (M) Schematic presentation of bioprinted skin using foreskin keratinocytes (KCs) and scalp or sole fibroblasts. Numbers of fibroblasts per 0.08 mm2 were counted (n = 3 distinct subjects with 4 repeats). There were no significant differences between scalp and sole fibroblasts in bioprinted skin. (N) Representative image of hematoxylin and eosin (H&E) stain at 20× magnification. Epidermal thickness was significantly higher in bioprinted skin with sole fibroblasts (P = 0.03, n = 3 distinct subjects with 4 repeats). Scale bar, 100 μm. (O) Representative image of clathrin/DAPI immunostain at 40× magnification. Cytoplasm size in keratinocytes was significantly larger in bioprinted skin with sole fibroblasts (p = 0.05, n = 3 distinct subjects with 4 repeats). Scale bar, 50 μm. (P) Representative image of KRT9/DAPI immunostain at 20× magnification. Epidermal KRT9 expression was significantly greater in bioprinted skin with sole fibroblasts (P = 0.04, n = 3 distinct subjects with 4 repeats). Scale bar, 100 μm. (Q) Representative image of FGF7/DAPI (4′,6-diamidino-2-phenylindole) immunostain at 20× magnification. Epidermal FGF7 expression was significantly greater in bioprinted skin with sole fibroblasts (P = 0.04, n = 3 distinct subjects with 4 repeats). Scale bar, 100 μm. SC, stratum corneum; Epi, epidermis; Der, dermis.
Fibroblasts are highly mechanoresponsive, being capable of differentiating in response to stretching and compressive forces (27–29). To mimic native volar fibroblasts’ exposure to dynamic environmental pressure (such as from walking), we next used an on/off oscillatory pressure model, applying 1 kPa onto in vitro cultured sole or scalp fibroblasts at 0.025 Hz (20 s on, 20 s off) for 30 min (Fig. 1E and movie S5). In response to mechanical stresses, fibroblasts express ECM- and inflammation-related genes (29). Two well-studied ECM-related genes are COL1A1, which increases after stretching, and MMP-1, which increases following pressurization (27, 28). We measured mRNA levels of COL1A1 and MMP-1 as indicators of fibroblast response to pressure. Sole fibroblasts showed a significant increase in COL1A1 mRNA expression after dynamic oscillatory pressure treatment (P = 0.03), but scalp fibroblasts did not (Fig. 1F). Similarly, MMP1 mRNA expression significantly increased in sole fibroblasts after pressure treatment (P = 0.01) but not in scalp fibroblasts (Fig. 1F). We also constructed three-dimensional (3D) collagen matrices with scalp or sole fibroblasts and measured collagen I and elastin protein levels (fig. S1A), finding no differences between them (fig. S1B). We attempted to apply pressure to 3D constructs, but the constructs did not withstand the applied forces (fig. S1C).
We explored the mechanical related proteins Yes-associated protein (YAP) and focal adhesion kinase (FAK). YAP regulates cell proliferation, regeneration, and stem cell renewal (30) through nuclear translocation (31). FAK, a tyrosine kinase, regulates cell adhesion and motility, with its activity proportional to applied tension forces (32). In our dynamic oscillatory pressure model, gene expression of FAK and YAP did not show significant changes in response to the pressure in either volar or nonvolar fibroblasts (fig. S1D). Total protein expression of FAK and YAP also did not significantly change in either volar or nonvolar fibroblasts (fig. S1, E to H). However, volar fibroblasts showed higher rates of YAP nuclear translocation at baseline, and application of pressure increased YAP nuclear translocation in nonvolar fibroblasts but decreased nuclear translocation in volar fibroblasts (fig. S1, G and I).
To test whether the above changes were generalizable across the transcriptome, we plotted bulk RNA sequencing (bulk RNA-seq) results in a principal components analysis (PCA). These results confirm that volar and nonvolar fibroblasts are different both at baseline and in response to pressure (Fig. 1G). To further explore gene expression differences, we separately identified differentially expressed genes (DEG) by fold change after pressure in either sole or scalp fibroblasts (listed in table S1 and volcano plots in fig. S2). To quantify the difference in pressure response between sole and scalp fibroblasts, we measured the overlap between these two lists. To avoid minor differences in fold change and a direct comparison of the top 25 genes ranked by fold change, we determined whether the top 25 genes induced by pressure were at all present in any transcript differentially affected by pressure (Padj < 0.05) from the opposite cell type. The top 25 DEG induced by pressure in sole fibroblasts had little overlap with DEG affected by pressure in scalp fibroblasts (Fig. 1H, top Venn diagram); by contrast, the top 25 DEG induced by pressure in scalp fibroblasts had much higher overlap with DEG modified by pressure in sole fibroblasts (Fig. 1H, bottom Venn diagram). The genes represented in the Venn diagram are listed in table S2. This result highlights a particular pressure-responsive gene program for volar fibroblasts. We similarly analyzed the data by gene set enrichment analysis (GSEA) (33), and sole fibroblasts demonstrated categories of limb development (Fig. 1, I and J) that were not detected in scalp fibroblasts (figs. S3 and S4).
To explore the responses of volar fibroblasts as demonstrated by RNA sequencing (fig. S2 to S4), we verified notable transcripts modified by pressure in each cell type by reverse transcription polymerase chain reaction (RT-PCR). FOS and EGR1 were commonly up-regulated after pressure treatment in both the scalp and sole fibroblasts (Fig. 1K, fig. S2A, and table S1). However, we found divergent responses to pressure in HOX genes. Several genes in this family (HOXA10, HOXA11, and HOXC13) were up-regulated with pressure treatment in sole fibroblasts, but they were absent in scalp fibroblasts (Fig. 1L, fig. S2B, and table S3). Analogously, HOXD8 was up-regulated with pressure only in the scalp fibroblasts. By contrast, HOXC11 was down-regulated with pressure only in the sole fibroblasts. These results demonstrate the effect of pressure on the differentiation state of volar versus nonvolar fibroblasts.
Bioprinted volar fibroblasts induce histological changes in nonvolar keratinocytes
Engineered skin equivalents incorporating volar fibroblasts were compared with control samples to investigate the mesenchymal-epithelial communication. Dermal constructs were bioprinted with patient fibroblasts taken from either the sole or scalp, whereas the epidermis was sourced from foreskin keratinocytes in both cases. We allowed 2 to 3 weeks for keratinocytes proliferation and differentiation before bioprint viability loss (Fig. 1M). Fibroblast counts showed no significant difference between scalp and sole fibroblasts in bioprinted skin (Fig. 1M). Next, histologic endpoints of epidermal thickness, cytoplasm size, and expressions of KRT9 and FGF7 were evaluated. Compared with scalp fibroblasts, sole fibroblasts induced a significantly thicker epidermis (P = 0.03, Fig. 1N), a significant increase in cytoplasmic area of keratinocytes (P = 0.05, Fig. 1O), and higher epidermal expression of KRT9 and FGF7 (P = 0.04 for both, Fig. 1, P and Q). Collectively, the bioprinted skin constructs highlight the capacity for mesenchyme to define epidermal features and therefore the potential use of volar fibroblasts to induce volar skin features.
In a human trial, ectopic volar fibroblasts induced histologic changes in nonvolar skin
To explore the therapeutic potential of volar fibroblasts to protect against ectopic pressure in patients, we conducted a phase 1 trial, injecting ectopic volar fibroblasts into nonvolar skin in healthy volunteers (inclusion and exclusion criteria for study participants are listed in table S4). As controls, vehicle and nonvolar (scalp) fibroblasts were injected into the left thigh, separated by at least 5 cm, and volar fibroblasts were injected into the right thigh (Fig. 2A and fig. S5A). After 2 or 5 months, injection site tissues were excised for testing and to remove grafted cells. Variables tested included total cell dose, delivery schedule, and site preparation with superficial wounding; results were either aggregated to ignore these modifications (Fig. 2) or unaggregated (fig. S5, B to E). Because unaggregated subgroups testing different doses, schedules, and site preparation with wounding were smaller in size, individual treatments were mostly not significantly different in measured endpoints (fig. S5, B to E). However, divided dosing appeared to increase efficacy in a previous animal model (34) and showed significant changes with multiple injections (P < 0.05, fig. S5, B to E), and therefore the most common treatment regimen was an injection of a total of 10 million fibroblasts, but divided into three doses delivered on Monday, Wednesday, and Friday (MWF injections).
Fig. 2. In a human trial, ectopic volar fibroblasts induced histologic changes in nonvolar skin.
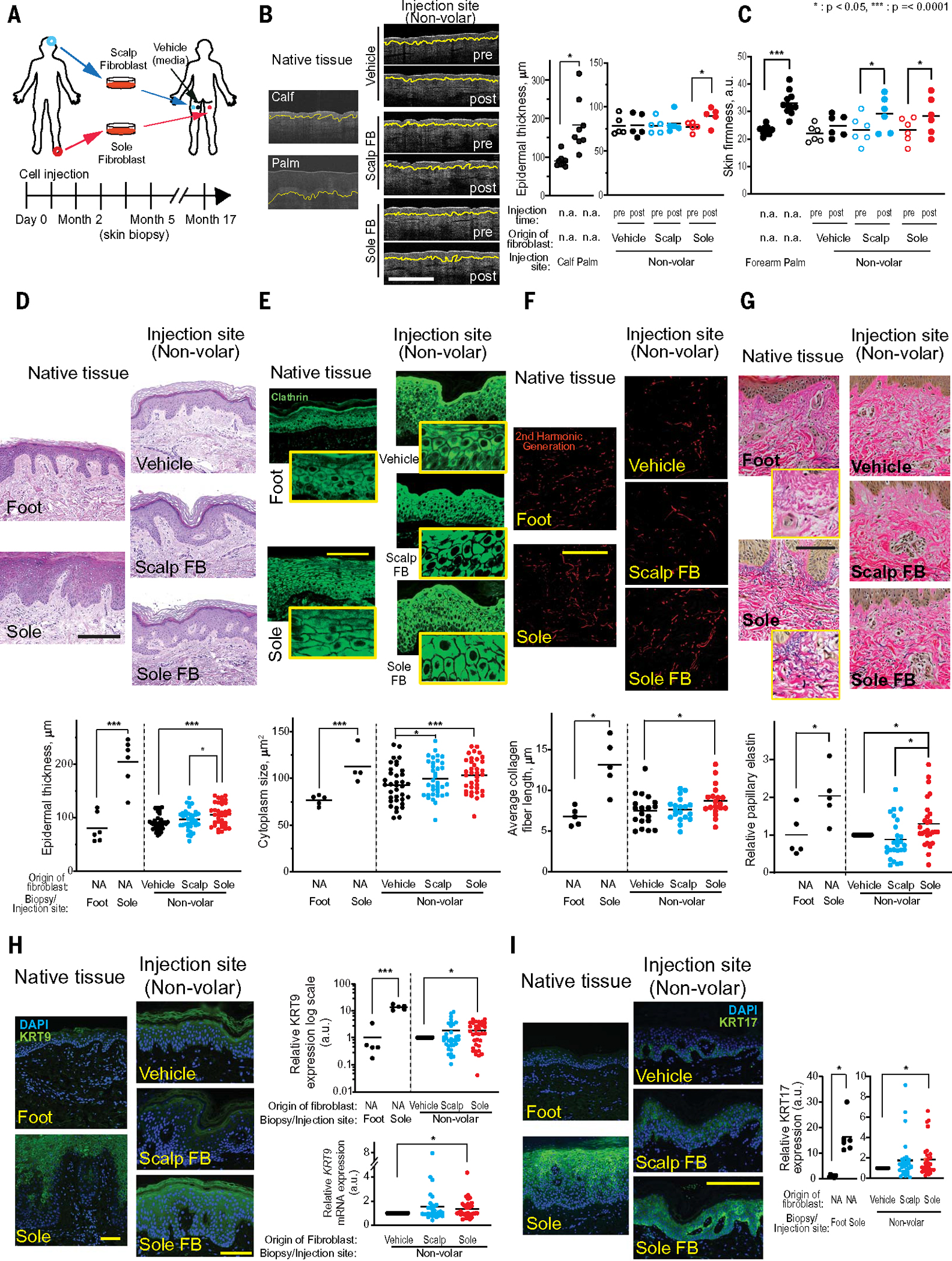
(A) Schematic presentation of experimental design for human clinical trial. (B) Representative images from OCT. Yellow line indicates dermal-epidermal junction (DEJ). Pre- and post-fibroblast injection time points showed that native volar (palm) epidermis was significantly thicker than native nonvolar (calf) epidermis (P = 0.002, n = 8 distinct subjects with 4 readings). The epidermis of sole fibroblast–injected sites (Sole FB-post) was significantly thicker compared to preinjection (Sole FB-pre, P = 0.009) or post-vehicle–injected control sites (Vehicle, P = 0.042, n = 5 distinct subjects with 4 readings). Scale bar, 500 μm. (C) Skin firmness measurement by durometer. Native volar (palm) skin was significantly firmer than native nonvolar (forearm) epidermis (P < 0.0001, n = 10 distinct subjects with 4 readings). Skin at scalp fibroblast–injected and sole fibroblast–injected sites was firmer than at preinjection (P = 0.007 and P = 0.015, respectively, n = 6 distinctive subjects with 4 readings). (D) Representative images of H&E stain from native and injection sites. Histological imaging of epidermal thickness in excised native tissue and injected sites. Native volar (sole) epidermis was significantly thicker than nonvolar (foot) epidermis (P < 0.0001, n = 6 distinct subjects with 1 repeat). Epidermal thickness in the Sole FB–injected sites was significantly thicker than that in the Scalp FB–injected sites (P = 0.04) or Vehicle-injected sites (P = 0.0001, n = 32 distinct subjects with 1 repeat). Scale bar, 100 μm. (E) Representative images of clathrin stain. Fluorescent imaging of clathrin protein to determine cytoplasm size in keratinocytes of native tissue and injected sites. Cytoplasm was significantly larger in native volar tissue compared to native nonvolar tissue (P < 0.0001, n = 5 distinct subjects with 1 repeat). Cytoplasm size in Sole FB– or Scalp FB–injected sites was larger compared to Vehicle-injected sites (P = 0.0001 and 0.006, respectively, n = 32 distinct subjects with 1 repeat). Scale bar, 100 μm. (F) Representative images of collagen fibers by second harmonic generation (SHG). Collagen fibers in dermis of native tissue and injected sites were detected by SHG. Collagen fiber lengths were measured according to collagen fibril detection. Fibers were significantly longer in native volar tissue compared to native nonvolar tissue (P = 0.01, n = 5 distinct subjects with 1 repeat). Fibers in the Sole FB–injected sites were significantly larger compared with Vehicle-injected sites (P = 0.04, n = 18 distinct subjects with 1 repeat). Scale bar, 100 μm. (G) Representative images of elastin stain. Native volar region (sole) had more elastin than native nonvolar regions (foot) (P = 0.04, n = 5 distinct subjects with 1 repeat). Injection of volar fibroblast in a nonvolar site led to increased elastin compared with similar sites’ vehicular injections (P = 0.05) and nonvolar injections (P = 0.002, n = 27 distinct subjects with 1 repeat). Scale bar, 100 μm. (H) Protein and mRNA expression of epidermal KRT9. Representative images are protein KRT9 stain. Scale bar, 100 μm. Epidermal KRT9 expression in native volar tissue was higher compared with native non-volar tissue (P < 0.0001, n = 5 distinct subjects with 1 repeat). Epidermal protein and mRNA KRT9 expression in the Sole FB–injected sites (n = 31 distinct subjects with 2 repeats) was significantly higher than in Vehicle-injected sites (P = 0.004 and 0.04, respectively). (I) KRT17 in native tissue and injected sites. KRT17 expression in native volar tissue was significantly higher than that in nonvolar tissue (P = 0.0003, n = 5 distinct subjects with 1 repeat). KRT17 expression in the Sole FB–injected sites was significantly higher compared with Vehicle-injected sites (P = 0.02, n = 31 distinct subjects with 1 repeat). Scale bar, 100 μm.
Demographic analysis depicts the subjects’ ethnicities, genders, and ages (Table 1). Minor side effects, such as bruising, tenderness, thickening, scaling, peeling, itching, and hyperpigmentation, are listed in table S5. The highest dose—a single injection of 30 million fibroblasts—led to occasional hyperpigmentation, greatest at the volar fibroblast injection sites (fig. S6A). Histological analysis revealed that high-dose ectopic fibroblasts induced epidermal downgrowths of benign-appearing keratinocytes without nuclear atypia. This histology is reminiscent of benign dermatofibromas (35, 36) characterized by keratinocyte overgrowth (fig. S6B). These epidermal downgrowths were not seen in other injection regimens.
Table 1.
Demographic analysis.
| Characteristic | |
|---|---|
| Total subject, n | 39 |
| Age, mean (years) | 35.9 |
| (range) | (19 to 63) |
| White | 21 |
| African American | 7 |
Self-reported
The effect of injected volar fibroblasts on nonvolar skin in human subjects was measured using optical coherence tomography (OCT) for epidermal thickness and a durometer for skin firmness. In all measures, we compared injection sites with native volar versus nonvolar skin. OCT images showed that native volar skin in the palm had a significantly thicker epidermis than native nonvolar skin on the calf (P = 0.002, Fig. 2B). In a recapitulation of this volar feature, the injection of sole fibroblasts into nonvolar skin (designated “Sole FB” in figures) significantly increased epidermal thickness (P = 0.009), but this was not evident after either vehicle or scalp fibroblast injections (designated “Scalp FB” in figures) (Fig. 2B). Skin firmness measurements revealed that normal or native volar skin of the palm was significantly firmer than nonvolar skin of the forearm (P < 0.0001, Fig. 2C). Whereas vehicle injection (designated “Vehicle” in figures) skin did not show significant increases in skin firmness, injection of either sole or scalp fibroblasts did significantly increase skin firmness (P = 0.007 and 0.015, respectively, Fig. 2C).
Histological analysis of excised skin samples confirmed noninvasive measurements above and in vitro bioprint experiment results. Epithelial thickness in histologic samples matched OCT results. Native volar skin of the sole was significantly thicker than nonvolar skin of the foot (P < 0.0001), and epidermal thickness significantly increased in nonvolar skin after volar fibroblast injection (P = 0.0001, Fig. 2D). Next, we analyzed cytoplasmic size. We found that a larger cytoplasmic area is a feature of native sole volar keratinocytes compared with those of the native nonvolar dorsal foot (P < 0.0001, Fig. 2E). Similarly, the cytoplasmic size of nonvolar keratinocytes significantly increased after injection of volar fibroblasts (P = 0.0001, Fig. 2E). We also characterized dermal changes after cellular injection. Native volar skin of the sole had significantly longer average collagen fibers than nonvolar skin of the dorsal foot (P = 0.01, Fig. 2F). As for the epidermal end points, we found that volar fibroblast therapy partially induced this dermal volar feature as well. The average collagen fiber length was significantly greater in nonvolar skin injected with volar fibroblasts compared with that found in skin injected with vehicle (P = 0.04). Lastly, we investigated changes in dermal elastin. To obtain consistent results for elastin quantification from a large number of slides, we applied artificial intelligence (AI) machine learning to detect elastin fibers using intersection over union (IoU) (fig. S7). In the papillary dermis, native volar (sole) regions had more elastin than native nonvolar (dorsal foot) regions (P = 0.04, Fig. 2G). Volar fibroblasts also enhanced this volar feature, with more elastin detected in the sole fibroblast injection sites compared with the vehicle (P = 0.05) or scalp fibroblast injection sites (P = 0.002, Fig. 2G).
Next, we measured whether volar fibroblasts up-regulate proteins elevated in volar skin (25). Immunohistochemistry confirmed that KRT9 and KRT17 are elevated in native volar skin (Fig. 2, H and I). Subsequent to volar fibroblast injections in nonvolar skin, KRT9 (P = 0.004, Fig. 2H) and KRT17 (P = 0.02, Fig. 2I) expression was significantly increased as compared with vehicle injection. There was no significant increase of these proteins seen after scalp fibroblast injection.
Native volar skin of sole showed significantly thicker stratified corneum compared with nonvolar skin of the dorsal foot (P < 0.0001, fig. S8A). However, sole fibroblast injection induced a modest increase in stratified corneum thickness at 2 months after injection (fig. S8, A and B). These results demonstrate the partial induction of gross histological and select markers of volar skin after volar fibroblast injection into nonvolar skin. Next, we attempted to characterize the global gene expression changes after ectopic-cell injection.
RNA sequencing defines tissue changes after ectopic fibroblast injection
To better characterize the tissue changes after ectopic-cell injections in humans, we performed bulk RNA-seq and single-cell RNA sequencing (scRNA-seq) on excised tissue from time points 2 weeks to 17 months after injection. We first compared single-dose injection of 10 million fibroblasts versus three fractionated injections (total of 10 million fibroblasts) over the first week (MWF injections), maintaining a consistent total fibroblast dosage in both cases at the 5-month postinjection time point using bulk RNA-seq (fig. S9A). Previous mouse model studies support divided dosing for long-term engraftment, perhaps owing to enhanced graft survival (34). Five months after injection in three subjects per regimen, bulk RNA-seq of epidermis revealed distinct gene changes with both single and fractionated injections. Compared with the vehicle injection sites, 109 and 108 (scalp fibroblast injection sites) and 118 and 128 (sole fibroblast injection sites) transcripts were significantly up- or down-regulated in single and fractionated injections, respectively (Padj < 0.05; fig. S9, B and C, and tables S6 and S7). Gene ontology (GO) analysis of these transcripts showed differences in how scalp and sole fibroblasts affect ectopic skin (fig. S9, B and C). Commonly modified transcripts with scalp fibroblasts included DLG2 (up-regulated), RAI14 (up-regulated), WWP2 (up-regulated), and FAM13A (up in 1× and down in 3×). For volar fibroblast injections, commonly modified transcripts included ITPRIP (up-regulated) and ZNF562 (down-regulated) as shown in fig. S9D. GSEA analysis revealed the top 100 distinctively expressed genes (fig. S10) and GO categories (fig. S11), demonstrating that fractionated sole fibroblast dosing produced a greater diversity of GO categories, including cornified envelope and desmosome, compared with single dosing or scalp fibroblast injections (fig. S11). The enrichment scores for the cornified envelope and desmosome categories were higher for sole fibroblasts than for scalp fibroblasts with fractionated dosing (fig. S12). These results suggest that divided volar fibroblast dosing is more effective in modifying epidermal gene expression in nonvolar epidermis.
To explore this mechanism, we performed divided dosing injections of fibroblasts or vehicle followed by whole-skin scRNA-seq at 2-week, 2-month, 5-month, and 17-month post-injection time points. A total of 79,735 preprocessed cells from all samples and time points were preprocessed, with each cluster identified using Seurat (37). Clustering of integrated human skin cells in uniform manifold approximation and projection (UMAP) at each post-injection time point resulted in distinct grouping of keratinocytes, fibroblasts, immune cells, endothelial cells, and melanocytes (Fig. 3A, figs. S13 to S16, and tables S8 to S11). As expected, at 2 weeks after injection, there was an increased fibroblast population at the volar fibroblast injection sites compared with vehicle injection (fig. S13B). This was not observed at the scalp fibroblast injection sites. By 2 months after injection, fibroblast proportions were stabilized and were no longer elevated in any injection site (figs. S14 to S16).
Fig. 3. scRNAseq of human skin after ectopic-cell injection demonstrates dynamic tissue changes.
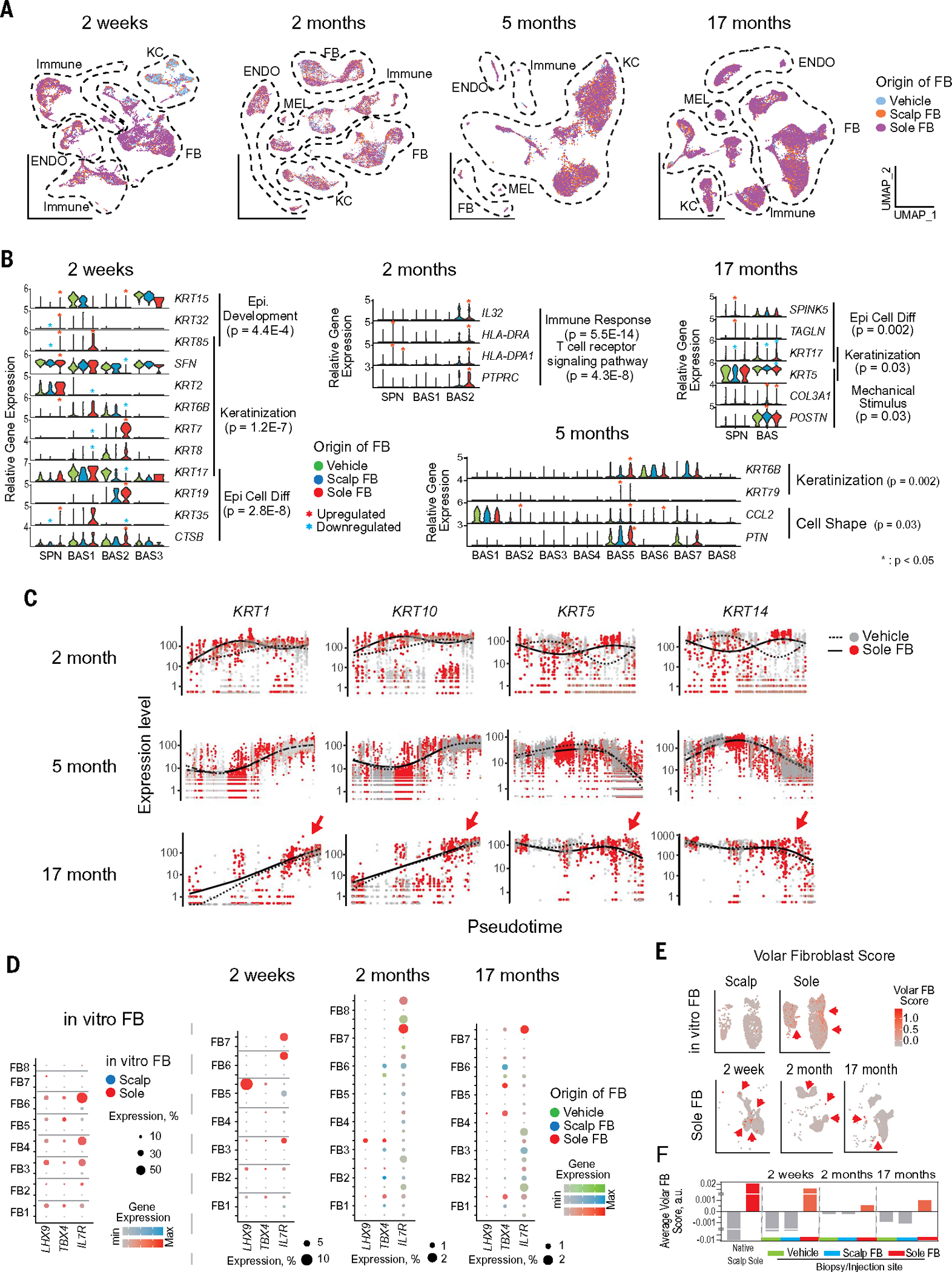
(A) Scatter plots show UMAP cell clusters at 2-week, 2-month, 5-month, and 17-month postinjection time points. (B) Violin plots of relative gene expression from differentially expressed genes (DEG) in spinous (SPN) and basal (BAS) keratinocyte clusters from injected sites. Each violin plot represents DEG from 2-week, 2-month, 5-month, and 17-month time points. DEG with Padj < 0.05 were further analyzed by DAVID analysis and selected for presentation. (C) Keratinocyte pseudotime single-cell trajectories based on KRT1, 10, 5, and 14 gene expressions from Sole FB–injected sites (red; solid line) and Vehicle injected sites (gray; dotted line). KRT1/10 are expressed higher in the earlier pseudotimes, whereas KRT5/14 are expressed higher in later pseudotimes after sole fibroblast injection. (D) Dot plot of volar genes LHX9/TBX4/IL7R expressed in cultured native preinjection fibroblasts (left) versus fibroblast clusters from injected sites (right). LHX9/TBX4/IL7R are expressed in preinjection native sole fibroblasts but not scalp fibroblasts. These genes were also detected in the skin sites with volar fibroblast injection but decreased in abundance with time. (E) UMAP plots of volar fibroblast–specific gene expression score (LHX9/TBX4/IL7R) in native scalp and sole fibroblasts (top) and Sole FB–injected sites with various time points (bottom). Volar fibroblast scores were calculated using volar-specific genes in Fig. 3D. (F) Average volar fibroblast scores in fibroblast clusters from vehicle, Scalp FB, and Sole FB injection sites at different postinjection time points were calculated.
We queried how fibroblast injection into human subjects modifies gene expression. GO analysis with DEG (Padj < 0.05) demonstrated multiple specific categories after 2 weeks (Fig. 3B). Particularly, genes associated with epidermal development, keratinization, and epidermal cell differentiation were modified. For example, in the epidermal development, volar fibroblasts up-regulated KRT15, KRT32, and KRT85 in both spinous (SPN) and multiple basal (BAS) keratinocyte populations, but nonvolar fibroblasts down-regulated KRT32 and KRT85 in the spinous keratinocyte population. Similarly, in the epithelial cell differentiation, volar fibroblasts up-regulated KRT19, KRT35, and CTSB in SPN, BAS1, and BAS2 keratinocyte populations. However nonvolar fibroblasts down-regulated KRT35 in SPN keratinocyte population (full list in table S12). At 2 months after injection, only immune response–related GO categories were detected. Up-regulated genes at the sole fibroblast injection sites included IL32, HLA-DRA, and PTPRC, and at the scalp fibroblast injection sites, HLA-DRA was up-regulated, perhaps during immune clearance of excess fibroblasts (full list in table S13). At 5 months after fibroblast injection, we detected keratinization and cell-shape categories. The keratinization category included an up-regulation of KRT6B—a known cause of volar keratodermas (38)—in the volar fibroblast injection sites (full list in table S14). At the latest time point, 17 months after injection, genes associated with keratinization and epithelial differentiation were still detected (full list in table S15). Detailed analyses of individual time points are presented in figs. S13G to S16G. The overall mRNA levels of KRT9 at injection sites were not strongly induced by engrafted fibroblasts, but KRT9 detection persisted throughout the time points (fig. S17A). For immune cells, increased mast cells were detected in fibroblast injection sites at 2 and 17 months after injection (fig. S17B). This result suggests that mast cells might be involved in an ectopic fibroblast clearance process. Although no global cell-cycle changes were seen in aggregated cells (fig. S18A), specific patterns emerged in basal versus spinous keratinocytes after fibroblast cell therapy (fig. S18, B and C). Generally, injected fibroblasts decreased the proportion of basal keratinocytes in the G1 phase, with an increase in S/G2M fraction at most time points. By contrast, spinous keratinocytes showed an early increase in G1 fraction and a decrease in G2M/S fraction. This effect was more pronounced at 2 weeks after injection with volar fibroblasts and is consistent with recently described cell-cycle differences in native volar versus nonvolar skin (17). Taken together, these results highlight a persistent effect of injected fibroblasts on altered keratinocyte differentiation after injection.
Injected fibroblast modification of keratinocyte single-cell trajectories in pseudotime
Given the above modification of keratinization GO categories after fibroblast injection, we examined individual keratin transcript changes in pseudotime trajectories with or without injected fibroblasts. Using Seurat keratinocyte clusters, we plotted gene expression kinetics in UMAP by Monocle 3 (Fig. 3C and figs. S19 to S21). Postinjection samples generally showed increased spinous keratinocyte markers (KRT1/10) and decreased basal keratinocyte markers (KRT5/14) over the pseudotime (figs. S19 to S21). To examine fibroblast injection effect on pseudotime progression, we overlapped pseudotime data from the vehicle injection and volar fibroblast injection sites (Fig. 3C). At 2 months after injection, volar fibroblasts (solid line) led to a higher level of differentiated keratins (KRT1/10) and lower basal keratins (KRT5/14) at early pseudotimes, with higher basal keratins at later pseudotime points compared with vehicle (dotted line). At subsequent time points, trajectories were similar, but different cell densities at particular pseudotimes appeared suggestive of a similar pattern. In particular, at 17 months after injection, the sole fibroblast injection sites showed a higher cell density at the latest pseudotime with higher levels of both KRT1/10 and KRT5/14. Together, these results further suggest how ectopic fibroblasts modify keratinocyte differentiation in human subjects.
Ectopic fibroblast persistence
The evidence of altered epidermal gene expression by injected fibroblasts motivated a search to identify ectopic fibroblasts in scRNA-seq data. We first sought to identify volar fibroblast markers and performed scRNA-seq data analysis on cultured scalp and sole fibroblasts before injection (fig. S22, A to C). Cell-cycle analysis (fig. S22B, bottom) confirmed the greater proliferation rate of volar fibroblasts seen in Fig. 1A. DAVID (the database for annotation, visualization and integrated discovery) analysis with DEG showed hair follicle morphogenesis and cell morphogenesis (fig. S22D). Genes associated with hair follicle morphogenesis (FGF7 and GFBP5) were up-regulated in scalp fibroblasts, and genes associated with cell morphogenesis (NRG1 and IL7R) were up-regulated in sole fibroblasts (full list in table S16). Combining these results with previous microarray data (25) on volar versus nonvolar skin tissues (table S17), we identified three candidate transcripts up-regulated in sole fibroblasts: LHX9, TBX4, and IL7R (fig. S22E). These were up-regulated to some degree in each in vitro sole fibroblast subpopulation (Fig. 3D). We next measured these markers in human subjects after cellular injection. Although technical limitations prevented analyzing the 5-month postinjection data owing to low fibroblast capture (fig. S15B), we detected an enrichment of these markers in fibroblast populations from skin after volar fibroblast injection at all other time points. However, this enrichment degree was modest. After volar fibroblast injection, the percentage of fibroblasts expressing volar markers peaked at 10% at 2 weeks after injection but decreased to about 1% by 17 months after injection (Fig. 3D). We then calculated the volar fibroblast score (39) using the LHX9, TBX4, and IL7R genes. Native volar fibroblasts showed high volar fibroblast scored cells (Fig. 3E, top). At different time points, sole fibroblast injection sites showed high volar fibroblast scored cells (Fig. 3E, bottom, and fig. S22F). Volar fibroblast–injected sites showed the positive average volar fibroblast scores, whereas nonvolar fibroblast–injected sites showed negative scores (Fig. 3F). This suggests a modest persistence of ectopic fibroblasts.
Cell-cell communications at various postinjection time points
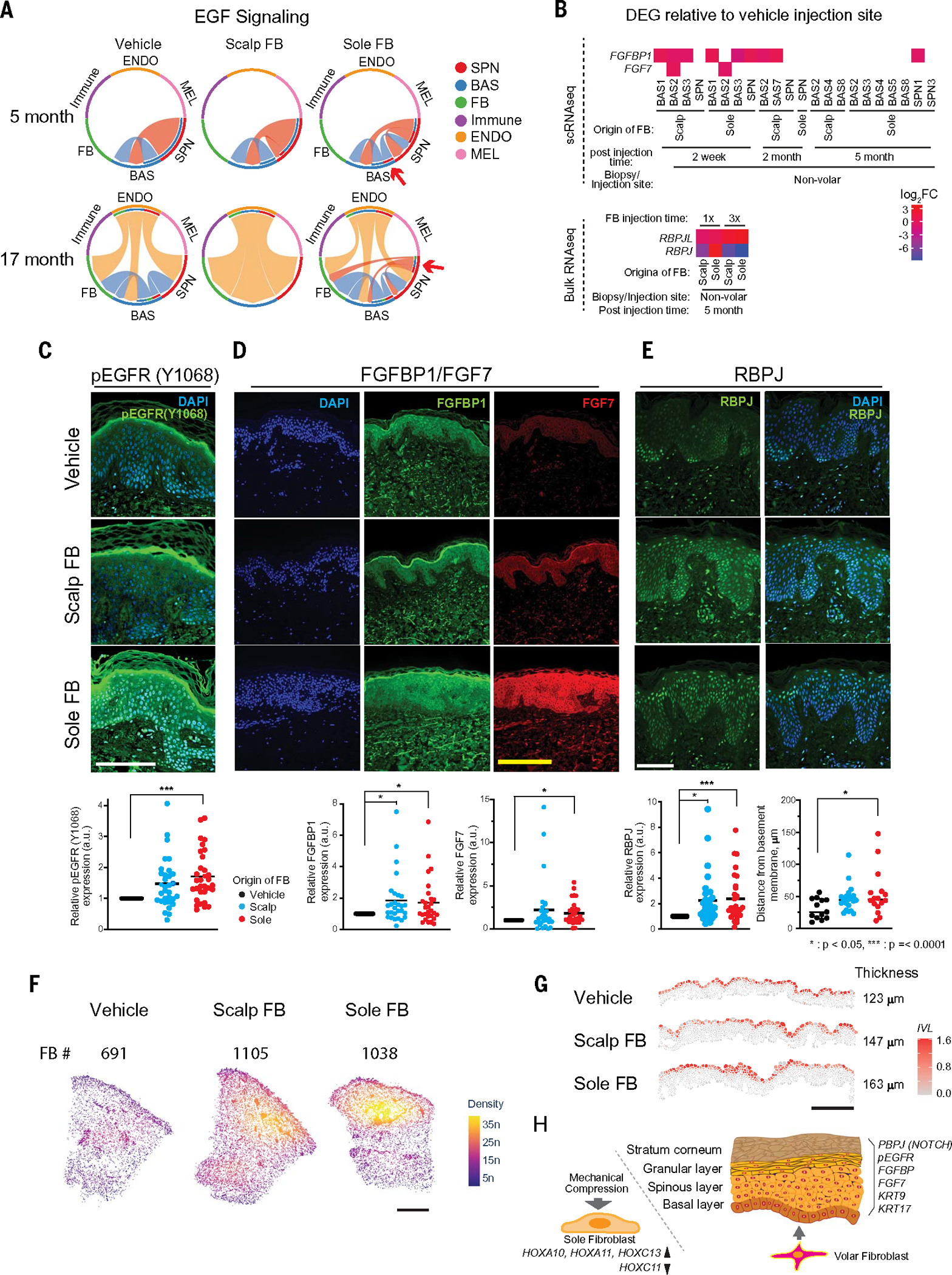
The effect of injected fibroblasts on epidermal function implies that cell-cell communication mediates tissue changes. To investigate this, we analyzed cell-cell interactions of scRNA-seq data using CellChat (40). The interactions were visualized in heatmaps that show matched ligand-receptor expression between cell types (fig. S23). Regardless of cell injection, the greatest interactions were between fibroblasts as senders and basal keratinocytes as recipients. No global, high-magnitude changes in communication were observed when comparing vehicle-injected sites with volar or nonvolar fibroblast–injected sites, although subtle differences existed (fig. S23). Instead, we focused on pathways mediating mesenchymal-epithelial crosstalk in volar biology (17). CellChat circle plots of EGF signaling showed sender-receptor relationships in injection sites at 5- and 17-month postinjection time points (Fig. 4A, full time points in fig. S24). At 2 weeks, epidermal growth factor (EGF) signaling from immune cell senders dominated in scalp fibroblast or sole fibroblast injection sites, indicating immune responses to transplanted cells compared with vehicle injection sites (fig. S24A). At 2 months, interactions were similar across injection sites. However, at 5 months, cross-talk between BAS and SPN cells were observed in the sole fibroblast injection sites, persisting at 17 months (Fig. 4A). The EGF signaling heatmap revealed elevated SPN-SPN communication in the sole fibroblast injection sites at both the 5- and 17-month postinjection time points, compared with the vehicle or scalp fibroblast injection sites (fig. S24B). scRNA-seq also showed up-regulated amphiregulin (AREG)—another EGF receptor ligand—in SPN (sole fibroblast injection sites) at 17 months (table S15). These results suggest a possible role for EGF in mediating some effects of ectopic volar fibroblasts.
Fig. 4. Cell-to-cell signaling through EGF, FGF, and NOTCH pathways was modified by ectopic fibroblasts.
(A) Circle plots by CellChat show sender-receptor relationships in EGF signaling pathways at 5-month and 17-month postinjection time points. The signal sender is located on the outer circle, and the recipient is located on the inner circle. Shading visualizes the pathway connections. SPN, spinous; BAS, basal; ENDO, endothelial; MEL, melanocyte. (B) FGF pathway (top, FGF7/FGFBP1) and NOTCH pathway (bottom, RBPJL/RBPJ) expression in scRNA-seq analysis (top) and bulk RNA-seq analysis (bottom). DEG with P < 0.05 were selected for presentation. (C) Representative images of pEGFR-Y1068 stain, showing pEGFR-Y1068 protein expression in injected sites. Expression was significantly higher in Sole FB–injected sites compared with Vehicle-injected sites (P = 0.0001, n = 32 distinct subjects with 1 repeat). Scale bar, 100 μm. (D) Representative images of FGFBP1 and FGF7 stains. FGFBP1 and FGF7 protein expression in injected sites. Sole FB–injected sites exhibited greater expression of FGFBP1 (P = 0.04) and FGF7 (P = 0 0.007) compared with Vehicle-injected sites. The Scalp FB–injected sites also exhibited greater FGFBP1 expression compared with Vehicle-injected sites (P = 0.024, n = 28 distinct subjects with 1 repeat). Scale bar, 100 μm. (E) Representative images of RBPJ stain. RBPJ (RBPSUH) protein expression in injected sites. RBPJ was expressed at significantly higher levels in the Sole FB– or Scalp FB–injected sites as compared with Vehicle-injected sites (P = 0.001 and 0.007, respectively, n = 29 distinct subjects with 1 repeat). Scale bar, 100 μm. Distance of RBPJ (RBPSUH)–positive keratinocytes from the basement membrane was measured. RBPJ-positive keratinocytes in Sole FB–injected sites were located significantly farther from the basement membrane compared with the keratinocytes in the Vehicle-injected sites (P = 0.03, n = 20 distinct subjects with 1 repeat). (F) Fibroblast density by Xenium in situ spatial transcriptomics at the 2-month postinjection time point. Increased fibroblast densities in the papillary dermis were detected at Scalp and Sole FB injection sites. Fibroblast cell numbers are indicated above the Xenium spatial plot. Scale bar, 1 mm. (G) Epidermal IVL expression at the 2-month postinjection time point as shown in (F). Higher levels of IVL expression across multiple keratinocyte layers were detected at the Sole FB sites. The IVL expressions in the entire epidermis are depicted in fig. S26F. Scale bar, 250 μm. (H) Schematic presentation of volar fibroblast response to pressure and modification of epidermal differentiation.
We next queried other ligands involved in epithelial-mesenchymal cross-talk. Also known as keratinocyte growth factor (KGF), fibroblast growth factor 7 (FGF7) is highly important for epithelial cell proliferation, differentiation (41), and organ and limb development (42, 43). Compared with the vehicle injection sites, FGF7 and FGFBP1 were up-regulated in keratinocytes from the sole or scalp fibroblast injection sites by scRNA-seq (P < 0.05, Fig. 4B). Similarly, NOTCH signaling, important for epidermal differentiation, was analyzed. Bulk RNA-seq revealed that RBPJL and RBPJ expression in the sole or scalp fibroblast injection sites were significantly modified compared with the vehicle injection sites (P < 0.05, Fig. 4B). These results suggest that, in addition to the EGF pathway, FGF and NOTCH signaling also contributed to the effects of ectopic fibroblasts on tissue.
Up-regulated proteins in p-EGFR, FGF, and NOTCH pathways in keratinocytes exposed to ectopic sole fibroblasts
We sought to verify the above RNA-seq data. We used immunofluorescence to measure candidate cell-signaling proteins in skin from our human subjects after ectopic fibroblast injection. Activation of EGF receptor (EGFR) by ligands initiates dimerization and phosphorylation (p-EGFR) at Y1068, Y1173, Y1211, or Y1289, which we tested using specific antibodies that recognize these phosphorylated residues. We found that EGFR phosphorylation at Y1068 was significantly up-regulated in keratinocytes 5 months after sole fibroblast injection (P = 0.0001, Fig. 4C), which is consistent with the EGF signaling pathway in RNA analysis. Next, we measured the protein expression of FGF7 and of three FGF7-associated proteins—FGFBP1, FGFBP2, and FGFR. FGF7 and FGFBP1 were elevated in the sole fibroblast injection sites, and FGFBP1 was also elevated in the scalp fibroblast injection sites (Fig. 4D), which is consistent with RNA analysis. Lastly, we measured the nuclear expression of RBPJ protein, a NOTCH signaling transcription factor involved in epidermal differentiation (44). The overall nuclear RBPJ content was high in both volar and nonvolar fibroblast injection sites (P = 0.001 and 0.007, respectively) compared with the vehicle injection sites (Fig. 4E). Moreover, in volar fibroblast injection sites, the average distances from the basement membrane in nuclear RBPJ-positive keratinocytes were located significantly farther from the basement membrane than in vehicle injection sites (P = 0.03, Fig. 4E). This indicates that injected ectopic volar fibroblasts promote RBPJ expression in more suprabasal keratinocytes in nonvolar epidermis, which is reminiscent of the altered differentiation in native volar epidermis. These results suggest that ectopic volar fibroblasts alter epidermal biology through EGF, FGF, and NOTCH signaling pathways.
Spatial transcriptomics detects increased fibroblast numbers and epidermal differentiation at injection sites
In a final assay for tissue changes after ectopic cell injection, we measured the positional transcriptome at 2 months after injection using the human multitissue and cancer panel from 10x Xenium, supplemented with a custom-designed panel (table S18). We applied scSorter to annotate cell types on the basis of selected markers that are analogous to scRNA marker genes (table S19). Each cell type is visualized in a UMAP plot and a spatial cluster plot (fig. S25). Although ectopic fibroblasts retaining a volar signature were few in scRNA-seq (Fig. 3D), we observed a 50% increase in fibroblasts numbers at injection sites (Fig. 4F and Table 2), with highest density for the ectopic fibroblasts in the papillary dermis. Unsupervised clustering with Seurat identified cell types in heatmap and UMAP plots (fig. S26, A and B). Top genes for cluster identification are listed in table S20. Cell types, identified according to gene expression and spatial location, were depicted in a spatial cluster plot (fig. S26C). For cross-referencing, we measured other genes from the references. We found that APCDD1, confirmed as a papillary fibroblast enriched marker (16), was highly present in papillary regions in Xenium data throughout all injection sites (fig. S26D). As shown in fig. S26E, we confirmed that the SPN scores (KRT1/10) were elevated in the spinous keratinocyte layers, whereas the BAS scores (KRT5/14) were elevated in the basal layers. Consistent with our previous analyses, the layer of suprabasal keratinocytes expressing the IVL was expanded at the sole fibroblast injection sites (Fig. 4G shows zoomed-in images; overall images are provided in fig. S26F). Next, we integrated scRNA and Xenium data from the 2-month postinjection time point. Similar cluster distributions on UMAPs are shown in fig. S27A. The top genes (table S21) from integrated data revealed that FB7 and FB8 clusters expressed volar TBX4 and HOX genes (HOXA10 and HOXA11). Spatial plots showed a higher presence of the volar specific FB8 on the sole fibroblast injection sites compared with the vehicle or scalp fibroblast injection sites (fig. S27B). Therefore, spatial transcriptomic data support the presence of engrafted ectopic fibroblasts at injection sites, with a stronger effect on expanding the epidermal layer by ectopic sole fibroblasts.
Table 2.
Cell numbers per mm2.
| Injection site | Fibroblast | Keratinocyte | Endothelial | Immune | Unknown | Total cells |
|---|---|---|---|---|---|---|
| Vehicle | 691 | 486 | 87 | 246 | 38 | 1548 |
| Scalp FB | 1105 | 417 | 97 | 151 | 13 | 1783 |
| Sole FB | 1038 | 570 | 142 | 261 | 36 | 2048 |
Discussion
Cell therapy holds great promise in medicine, given the capacity of cells to act therapeutically but also with sensing capacities and curative potential. Hematopoietic cell therapy, in routine use for decades, has led to innovations such as engineered chimeric antigen receptor T (CAR-T) cells for cancers (45). However, cell therapy in solid organs has been more challenging owing to the lack of models available to refine concepts and approaches. We propose to capitalize on the accessibility and properties of volar fibroblasts to test for their ability to modify skin differentiation and eventually to be a therapy for patients experiencing ectopic pressure in skin areas—such as amputees with distal limb skin problems, for example. Although larger tissue structural and macroscopic adaptations at focal pressure-bearing areas have been well documented, cellular adaptations at these areas are less studied. In this study, we began such an analysis by comparing some of the fundamental differences of volar fibroblasts in response to pressure and subsequently by testing them in human subjects to assess how they impact tissue function. Our results support the therapeutic potential of these cells and the need to optimize a therapy for dermatoses associated with ectopic pressure such as decubitus ulcers.
Our exploration of cell therapy with volar fibroblasts builds on previous efforts. The instructive capacity of the dermis to regulate epidermal function was identified as early as the 1960s (7, 23), with fibroblasts likely mediating this effect (5, 6). What enables the therapeutic use of site-specific fibroblasts is that their positional memory encoded by HOX genes (such as HOXA13, HOXB, and HOXD) is retained in vitro through many passages by stable epigenetics (4, 24). Consistent with these findings, we have shown that volar fibroblasts retain their migration patterns, gene expression, and response to pressure. In bioprinted skin tissue, they modify epidermal morphology and gene expression. These findings support the idea that ectopic volar fibroblasts in humans modify tissue features such as skin firmness, epidermal thickness, and gene expression via EGF, FGF, and NOTCH signaling pathways. The schematic presentation in Fig. 4H summarizes the pressure response of volar fibroblast and expression changes in epidermal differentiation by volar fibroblasts.
Recent articles describing scRNA-seq of human native volar and nonvolar tissue provide an important template to interpret our work (15, 17). Wiedemann et al. (17) conducted pairwise comparisons (palmoplantar versus nonpalmoplantar) of scRNA-seq data and revealed cornification and keratinization pathways in native sole skin. In this work, we found overlapping pathways induced by ectopic volar fibroblasts in a nonvolar site through bulk and scRNA-seq analysis. Wiedemann et al. also identified gene clusters particular to the volar skin and highlighted sole skin–specific high expression of stress keratins (KRT6, KRT16, and KRT17), GJB2/6, GRHL3, FOSL1, and SOX9, as well as volar fibroblast– or nonvolar fibroblast–specific genes from bulk and scRNA-seq (17). Merleev et al. identified up-regulated lipid-associated metabolic genes, including PPTLC1/2 and CERS4/6 in palm samples (15). Ascensión et al. also identified human dermal fibroblast subpopulations and categorized these in three groups that are based on specific gene clusters (18). Philippeos et al. (16) confirmed the elevation of several papillary enriched markers, including APCDD1 and CD39 (ENTPD1). APCDD1, the papillary fibroblast positional marker gene (16, 18), correlated with our spatial transcriptomic data. However, we did not observe a strong correlation between the volar-specific gene sets from the references (15, 17) and our spatial transcriptomic data, particularly at the volar fibroblast–injected sites; indeed, using our volar gene score, we identified only 1% of fibroblasts that retain a volar gene signature 2 months after volar fibroblast injection. This is in sharp contrast to the 50% increase in fibroblast numbers in ectopic fibroblast injection sites at 2 months after injection. This finding strongly implies that rather than a lack of engraftment, which is widely considered the main barrier to cellular therapy, local cellular reprogramming and co-optation of ectopic fibroblasts to a host genetic expression program is more likely to limit cellular therapy efficacy. Although ectopic fibroblast therapy does adjust tissue identity, it does not induce a large-scale and complete identity conversion in keratinocytes. This is consistent, for example, with the modest increases in end points from our study, such as SC thickness or KRT9 protein and mRNA. These results highlight the clear differences between fully native volar skin and a heterogeneous mix of native nonvolar skin cells with a subpopulation of ectopic volar fibroblasts.
The above findings raise the important question of why—despite finding many modulations of epidermal function by ectopic volar fibroblasts—our human trial results are not a full phenotypic conversion to volar skin features. Similarly, injection of scalp fibroblasts did not lead to visibly modified hair growth at the injected sites. One likely explanation is that although fibroblasts do modify epidermal function, intrinsic epigenetic differences also likely control it; a second is that our human trial on normal subjects did not expose skin to pressure and that volar fibroblasts might more powerfully modulate tissue function in this context. A related explanation in the case of scalp fibroblasts is that specific fibroblast subpopulations such as dermal papillae and dermal sheath cells are required to modify hair function. A final reason is found in the low percentage of fibroblasts that retained volar markers at the latest time point tested, 17 months after injection. One factor contributing to this result is that we did not clear native fibroblasts with chemotherapy, as is done, for example, in the context of hematopoietic transplants, although we did briefly try local wounding for this purpose. This question will require further investigation. Similarly, low engraftment rates might result from active clearance of ectopic cells. Defining mechanisms of ectopic fibroblast sensing and clearance is therefore an exciting future line of investigation and perhaps even relevant to metastasis clearance. One promising result is that mast cells were elevated in fibroblast-injected areas at 2- and 17-month postinjection time points, implicating a mast cell–driven process as one candidate for ectopic-cell clearance. Defining the divergent extracellular signals that lead to ectopic-cell clearance will likely improve cell therapy for many solid organs. However, perhaps more important will be defining the mechanisms by which host cells reprogram ectopic cells to a host tissue identity; the large increase of fibroblast numbers as detected in spatial transcriptomics contrasts with the low number of fibroblasts that retain a volar identity after volar fibroblast injection as detected by scRNA-seq. What are the extracellular cues that reprogram volar fibroblasts to lose this volar gene signature? The available datasets, accessibility of skin, low morbidity of this intervention, and relative feasibility of our protocol will hopefully create platforms for other investigators to address these questions.
Another important question our work raises is whether particular fibroblast subpopulations would enrich the effects seen. For the sake of easier clinical testing, we opted to use a generalized fibroblast population from volar skin that we characterized by scRNA-seq to define its heterogeneity. One obstacle to studying human fibroblast subpopulations is that—although dermal subpopulations have been carefully dissected in mice (1) and there is growing research in human fibroblast subpopulations (15–18)—a comparatively robust effort has not been made with human fibroblast populations, particularly regarding their ability to influence skin identity. Our work provides a template that future researchers can use to test individual human fibroblasts for their capacity to modify skin function in the bioprint assay and correct this discrepancy.
Further work should also investigate some gene hits from our work, such as ITPRIP and ZNF562, the shared transcripts altered with both single and divided dosing of volar fibroblasts to skin. Similarly, an investigation of whether mismatched epidermal-mesenchymal signaling leads to decreased ectopic volar fibroblast survival also deserves study.
Our work demonstrates distinctive properties and therapeutic potential of volar fibroblasts through in vitro, bioprinted, and human in vivo studies. We currently have full regulatory approval and are enrolling amputees in a phase 2 study (NCT03947450) to further explore a future therapy for treating skin conditions of inappropriate pressure. This and future work defining methods of optimization in phase 1 trials could provide foundational insights to optimize cellular engraftment and tissue modification.
Materials and methods
Human fibroblast study
All study procedures took place under the Johns Hopkins Institutional Review Board (NA_0006868) with FDA Investigational New Drug Application approval from the Center for Biologics Evaluation and Research (IND 15658). Participants were enrolled at the Johns Hopkins Department of Dermatology, Baltimore, Maryland. The study lasted for up to 36 months and consisted of no more than 14 visits.
Participants were recruited through online advertisements and fliers within the community. After interested individuals contacted the research team, intake visits were scheduled and screening for inclusion/exclusion criteria was conducted to ensure their eligibility for participation (table S4). After informed consent was provided, bloodwork was completed to further assess for viral infections that could place health care workers at risk or compromise cell expansion. The volunteer and research team then established a study timeline: an initial biopsy, injections, monitoring visits, and graft extraction. The experimental details are provided in subsequent sections.
All study activities were approved and overseen by the Johns Hopkins Institutional Review Board, and all patients provided written informed consent. Study participants were provided monetary compensation at key milestone visits (baseline biopsy, injection, graft removal, and final evaluation) for their time as determined by the study team and Johns Hopkins Institutional Review Board.
Volar and nonvolar skin sample collection, fibroblast isolation, and expansion
Skin sites were identified for collection of volar (palmar hand or the medial heel of the plantar foot) and nonvolar (bilateral postauricular scalp) tissue. Prior to the excision, 1% lidocaine with epinephrine (1:100,000) was administered for local anesthesia. Full-thickness punch biopsies (4 to 6 mm in diameter and extending up to the fascia depth) were then obtained using sterile procedures as approved by the Johns Hopkins Institutional Review Board (NA_0006868), with at most four tissue samples obtained for each skin type. Wounds were sutured, and the standard of care was provided. Sutures were removed at a later time, once the wound healed sufficiently and prior to the start of injections. The tissue samples were placed in sterile tubes containing Dulbecco’s phosphate buffered saline (DPBS, Gibco, 14190) supplemented with penicillin (100 units/ml), streptomycin (100 μg/ml), and amphotericin B (0.25 μg/ml). The tubes were labeled with subject initials, their distinct participant ID, the date and time, and the skin type (volar or nonvolar). Upon immediate subsequent delivery to the Johns Hopkins Cellular Therapy Laboratory, the samples were either processed for fibroblast isolation and expansion or placed in a 2° to 8°C refrigerator for up to 24 hours.
The obtained biopsy underwent two washes in conical tubes with 70% isopropyl alcohol, followed by two additional washes in alternating solutions of DPBS containing 100 units/ml of penicillin, 100 mg/ml of streptomycin, and 0.25 mg/ml of amphotericin B (Sigma-Aldrich, A5955). After washing, each sample was transferred to a conical tube containing 10 ml of digestion medium. Digestion medium was prepared by diluting 600 μl of dispase (Corning, 354235) in 15 ml of fibroblast growth medium (FGM-2, Lonza, CC-3132). Tubes containing the tissue and dispase were incubated in a 2° to 8°C refrigerator for 12 to 24 hours. After digestion, each biopsy sample was aseptically removed from the conical tube and placed in a sterile 10 cm2 culture dish and allowed to remain undisturbed for 2 to 5 min. The dermis and epidermis were then peeled apart using sterile forceps.
The dermal samples were transferred to 6-well plates (Corning Costar Cell Bind, 3335) where they air dried for 30 to 45 min and became firmly adhered to the surface of the well. 2 to 4 mL of FGM-2 was then added, and the plates were incubated in a 37°C, CO2 humidified incubator. As fibroblasts migrated from the dermal tissue and attached to the well, the medium in the wells was changed every 2 to 4 days. After 10 to 20 days of culture, the adherent cells were detached using TrypLE Select (Gibco, 12563), an animal-free, recombinant enzyme for cell detachment. Once the fibroblasts had expanded beyond the threshold necessary for required dose and quality control testing, the cell suspension was cryopreserved in a vapor phase nitrogen freezer in freezing medium containing human serum albumin, hetastarch, and dimethyl sulfoxide (DMSO) (2% human serum albumin, 5% DMSO, 6% hetastarch in sterile saline) at a defined fibroblast cell concentration.
Tattoo application for better visualization of injection site
To help visualize the injection/monitoring site consistently between visits, the skin on participants was marked with tattoo ink on the day of their initial biopsy. Specifically, ink (< 1.25 ml, Black Tattoo ink, Vanarsdale Innovative Products, Inc. #VIP-1PI) was administered using the needle of a 25-gauge 5/8-in. syringe, without the need for the barrel or plunger. Relying on capillary action alone, the shaft of the needle was loaded with ink up to past the bevel. The needle was then inserted into the target skin site, with the bevel facing up and at an angle of 30° to the skin. Once the bevel was completely submerged, the site was marked due to ink leaving the lumen of the needle. The needle was then removed.
This procedure was performed on the participant on the day of the initial biopsy to allow for the ink to fully integrate and for inflammation in the skin to resolve before the first cellular injection. The application of the tattoo was completed with the guidance of Phuoc Tran, MD, PhD, and Valerie Briner, Assistant Chief Therapist for Radiation Oncology at the Johns Hopkins Hospital. These tattoos were temporary and were removed at the time of graft excision near the end of the study.
Autologous fibroblast injections and sample collection from the injected site
Once the cell count of autologous fibroblasts isolated and expanded from the biopsy was sufficient, the volunteer was contacted and scheduled for injections. Three locations were chosen for the three treatment groups: vehicle, volar fibroblast, and nonvolar fibroblast injections. These three locations were distributed among the participant’s left and right posterior thighs, with at least 5 cm of separation between the center of each injection site to ensure there was no overlap (fig. S5A). The thigh was chosen because of the large distances between injection sites, and because in some cases thigh skin is used as a skin flap in amputees and would be a potential future site of therapeutic volar fibroblast injection in amputees. The isolated and expanded fibroblasts were collected from the Johns Hopkins Cell Therapy Core and used within 2 hours of their retrieval. Freezing medium with fibroblasts suspended or not (the freezing vehicle) was thawed and loaded into syringes using a blunt tip needle. This needle was then replaced with a 25-gauge 5/8-in. needle for injection. Prior to treatment delivery, 0.5 ml of 1% lidocaine with epinephrine (1:100,000) was administered to the subject for local anesthesia. The injection was then performed into site marked by tattoo ink, as described above.
The dosage per injection was varied (10 million cells for one injection in one visit, 30 million cells for one injection in one visit, 10 million cells for one injection in one visit following with immediate superficial wound using curettes, 10 million cells for one injection, 1 month post superficial wound using curettes, 1 million cells and two times of DMSO injections in three visits, 3.33 million for three injections in three visits, fig. S5B). An equivalent volume of medium was used for the vehicle injection. When we tested curetting, the skin was scraped with a curette to remove the epidermis and very superficial dermis prior to cell injection. The injection took place at a rate no greater than 1 ml/minute, targeted below the epidermisdermal junction or sub-epidermally. This resulted in a wheal of about 3- to 4-mm diameter on the subject’s thigh. The injection site was then bandaged, and after 20 min, vital signs were recorded. Prior to discharge, participants were instructed to keep the injection site covered with a padded dressing for 24 to 48 hours and to avoid manipulating the site, such as by scratching or applying ice or other topical remedies. The participant returned for at most six monitoring visits for the research team to evaluate the injection site superficially.
At the conclusion of the study, tissue was excised from the injection site using a biopsy bunch (6 mm in diameter, up to fascia depth) after administration of local anesthetic as described for the initial biopsy. The sample was then processed immediately or stored in a 2° to 8°C refrigerator for up to 24 hours. Half of the sample was processed for histology and the other half for RNA extraction, or the whole sample was used for scRNA-seq. The participants returned at a later time for suture removal and for a study closeout visit.
Skin firmness readings
Skin firmness readings were obtained under approval of the Johns Hopkins Institutional Review Board (NA_0006868) using a noninvasive handheld OO scale Model DD-4 Digital Durometer made by Rex Gauge (Buffalo Grove, IL). The instrument was calibrated as per the manufacturer’s standards. Measurements were taken at six anatomical sites on the left and right posterior thighs of subjects across multiple time points spanning up to 22 weeks. They were obtained with subjects lying prone and lower legs raised passively. All measurements were taken at an angle perpendicular to the skin surface, which was parallel to the floor. Five readings were obtained per site, and mean values were calculated.
Skin thickness measurement by OCT
Epidermal thickness measurements were obtained under Hopkins IRB (NA_0006868) using a handheld, noninvasive Ganymede Series Spectral Domain Optical Coherence Tomography (OCT) system (Model GAN610C1) with Z-spacer manufactured by Thorlabs, Inc (Newton, NJ). Tattoo markings performed at baseline visits were used to identify three anatomical sites on participants’ bilateral posterior thighs. With subjects lying prone, 16 OCT images were collected per site per visit in 2D acquisition mode at four axial planes (90°, 0°, 45°, and −45°) across multiple time points spanning up to 22 weeks. Dataset Parameters were: refractive index: 1.45; size: x: 3000, z: 939; FOV: x: 6.00, z: 1.79; pixel size: x: 2.00, z: 1.90; fixed pixel size; center: x: 0, y: 0; angle: (see above); # of frames: 1; speed/sensitivity: 100 kHz A-scan rate; a-scan averaging: 3; b-scan averaging: 1. Images were analyzed using ImageJ, and mean values were calculated.
Static osmotic pressure on in vitro cultured fibroblasts
Human sole and scalp fibroblasts acquired from healthy volunteers were cultured with FGM-2 at 37°C and 5% CO2 until over 90% confluency was achieved. For the static osmotic pressure treatment, fibroblasts were plated into 24-well plates. 1% PEG 300 (Sigma-Aldrich, 91462) in FGM-2 (v/v) was used to apply 100 kPa osmotic pressure on fibroblasts in a sealed system. For tracking of cell migration, Hoechst 33342 (Thermo Scientific, 62249) was added to the culture medium to achieve a final concentration of 2 μM, and cells were incubated for 30 min before imaging. A stage-top incubator maintained the environment at 37°C, 5% CO2, and 35% humidity throughout the entire imaging session. Fluorescent images of cell nuclei were acquired at 2-min intervals for 4 hours with a 5x objective. Tracking was performed in ImageJ (46) with TrackMate (v7.7.2) (47). For Ki67 staining, cells were grown to 50% confluency before osmotic pressure treatment was applied for 1 hour. Cells were then fixed with 4% PFA for 10 min, permeabilized with 0.1% Triton X-100 for 10 min, and blocked with 2% BSA + 0.1% Tween-20 solution for 30 min. Mouse-anti-human Ki67 antibody (eBioscience, 14–5699-82) was diluted 1:500 in 1% BSA and cells were incubated in this antibody solution at 4°C overnight. Goat-anti-mouse IgG DyLight 594 (Abcam, ab96873) was diluted 1:1000 in 1% BSA and cells were incubated with these secondary antibodies for 2 hours at room temperature. Each well was washed three times with PBS between steps. Fluorescent images were acquired with a 10x objective. Images were acquired using a Leica SP8 microscope.
Dynamic oscillatory pressure treatment on in vitro cultured fibroblasts
Human sole and scalp fibroblasts from healthy volunteers were plated into 6-well plates and fibroblasts were cultured in FGM-2 at 37°C and 5% CO2 until reaching over 90% confluency. Experimental groups were subjected to pressure treatment of 1 kPa applied 20 s on and 20 s off, over 30 min using our in vitro dynamic oscillatory pressurization system (movie S5). Higher than 1 kPa or longer than 30 min of dynamic pressure treatment in open system caused cell death (data not shown). For RNA extraction, immediately post-pressure, FGM-2 was aspirated, cells were washed with PBS, and then cells were harvested by adding 350 μl lysis buffer (1:10 β-mercaptoethanol to RLT buffer) directly to the 6-well plate and cell lysate was stored in a −20°C freezer. Control groups followed a similar protocol, except the cells were directly harvested after reaching confluency and did not undergo pressurization. For immunofluorescence staining, cells were fixed in 4% PFA and permeabilized in 0.5% Triton X-100, followed by blocking with 3% BSA. Cells were then incubated with FAK primary antibody conjugated with Alexa Fluor 488 (Invitrogen, ZF002) or YAP primary antibody (Cell Signaling, 14074), followed by secondary antibody conjugated with Alexa Fluor 488 (Invitrogen, A11034).
3D culture of fibroblasts in collagen gel and immunofluorescent staining of collagen 1 and elastin
The collagen gel was prepared at a concentration of 2.0 mg/ml using the 3D Collagen I Matrix Kit (rat tail Collagen I, ScienCell, 8688), and following the manufacturer’s instructions, the indicated distilled water volume was substituted by the same volume of DMEM (Gibco, 11965–092). Either human sole or scalp fibroblasts (2.0 × 105) were mixed with 1 μl of collagen gel. For 3D cell culture, 100 μl of collagen gel and cell mixture was added as a single drop in the center of the chamber in two-chamber cell culture slides (Celltreat, 229162). The cell culture slides were incubated at 37°C and 5% CO2 for 1 hour according to the manufacturer’s instructions to let the collagen gel polymerize. After 1 hour of polymerization, 1 ml of FBM-2 (Lonza, CC-3132) was added to each chamber. The cell culture slides were incubated at 37°C and 5% CO2 for 48 hours. Fixation and immunofluorescence staining of 3D blocks of collagen and cell mixture were performed based on previous research (48). Briefly, 3D blocks were fixed in PFA for 1 hour. The cells in 3D block were permeabilized with 0.5% Triton X-100 for 10 min. After blocking nonspecific antibody binding using 10% fetal bovine serum (FBS, Sigma, F4135), 3D blocks were permeabilized with 0.05% Tween/PBS for 5 min then incubated with antibodies specific for collagen 1 (Abcam, ab138492) and elastin conjugated with Alexa Fluor 680 (Santa Cruz Biotechnology, sc-58756). The nuclei were visualized with DAPI solution (Thermo Scientific, 62248) staining. Mounting solution (ProLong Glass Antifade Mountant, Invitrogen, P36980) was added to the top of each 3D collagen block and a coverslip was used to press the 3D block and cover the slide.
3D skin bioprinting
Previously expanded human sole and scalp fibroblasts from normal healthy volunteers and foreskin epithelial keratinocytes (KC) (Sciencell, 2100) were harvested for printing at 60 to 80% confluency. Skin constructs were created using the method previously described (49) with the following modifications. Dermal layers were fabricated as described, using a hydrogel comprised of gelatin (Sigma G1890, final concentration of 0.045 mg/ml), collagen (Corning 354249, final concentration of 4 mg/ml), and fibrinogen (Sigma F3879, final concentration of 7.7 mg/ml). Rather than apply KC immediately, the constructs were incubated overnight prior to manual KC application (150k cells/cm2), which eliminated the necessity of basal laminin application.
To facilitate fibrinogen–fibrin conversion, 300 μl KC medium (Lonza 00192060) containing thrombin (5 NIH unit/ml; Sigma T6884) was placed in each well, beneath the transwell insert. After 1.5 hours incubation at RT, the constructs were submerged in epidermalization medium (containing Aprotinin 0.025 mg/ml, Sigma A4529) and incubated for 1 week with medium changes every 2 to 3 days. Tissues were then transferred to deep-well plates (Greiner 665110) in cornification medium (4 ml/well) and incubated for another week. After 2 to 3 weeks, tissues were fixed in 4% paraformaldehyde and embedded in OCT for hematoxylin and eosin (H&E) staining or immunohistochemistry (IHC).
Histology and immunofluorescence staining
The skin biopsies were cut in half, and one half was preserved in RNAlater (Invitrogen, AM7020) for RNA extraction and the other half was utilized for tissue slide preparation. Tissues were rinsed with DPBS and stored in 4% paraformaldehyde at 4°C overnight for fixation. The samples were then embedded in paraffin at the Johns Hopkins Core Facility. Tissue slides were cut into 5-μm thick slices and stained with hematoxylin and eosin (H&E). For immunofluorescence staining, tissues were deparaffinized, rehydrated, and then treated with antigen retrieval solution (Invitrogen, 00–4955-58) at 120°C for 7 min using a decloaking chamber (Biocare medical, DC2002). After blocking nonspecific antibody binding using 3% bovine serum albumin (BSA, Fisher, BP9703), tissue slides were incubated with antibodies specific for KRT9 (affinity purified from rabbit serum with antigen KRT9 aa451-aa476 by Thermo Scientific), clathrin conjugated with Alexa 488 (Invitrogen, MA1065A488), p-EGFR (Y1068) (cell signal, 3777), RBPSUH for RBPJ (cell signal, 5313) following with secondary antibody conjugated with Alexa 488 targeting rabbit IgG (Invitrogen, A11034). The nuclei were visualized with diaminofluorescein (DAPI, Thermo Scientific, 62248) staining. After mounting tissue by Prolong Glass Antifade Mountant (Invitrogen), P36980 microscopic images were acquired. For immunostaining of 3D skin bioprinting slides, slides were incubated at 45°C for 30 min. Subsequently, the process continued with blocking, following the same steps as for FFPE slides.
Microscopic images, image analysis, and statistical analysis
Microscopic images were obtained by Leica DM4000 B LED microscope (Leica) with Leica Application Suite X (LAS X, Leica), Olympus VS200 slide scanner (Evident) or Zeiss 710NLO-Meta (Zeiss) for second harmonic generation (SHG). All image reading and grading was performed blinded. For YAP nuclear translocations, approximately 5000 cells were selected for each group, and the degree of overlap between nucleus and YAP protein for each cell was calculated using VS200. For 3D culture microscopic images, z-stacked fluorescent images were obtained with total of 41 z-planes and 1.18-μm spacing. Collagen fibrils were detected by SHG and collagen fiber lengths were measured based on collagen fibril detection. For the analysis, 20 volar or nonvolar fibroblasts per biological sample were randomly selected. Intensity of signal from single cells was averaged from seven consecutive z-stacked images. Images from OCT and microscopy were analyzed using ImageJ (46) for epidermal thickness, cytoplasmic size, and collagen fiber length. Nuclear RBPJ expression was identified by DAPI colocalization using CellProfiler (50). Distances from RBPJ positive cells to the dermis-epidermis junction (DEJ) were measured using ImageJ. Data collected from image analyses were statistically analyzed by using one-sample t-tests or one-way ANOVA tests, followed by Tukey’s multiple comparisons test using GraphPad Prism (v9.1.2, GraphPad Software, https://www.graphpad.com/).
Elastin measurement using AI deep learning
Elastin area measurements were done using Olympus VS200 (Evident). An artificial intelligence (AI) deep learning was trained using the intersection over union (IoU) method to identify elastin filaments and calculate their area. The thickness of elastin filaments varies greatly by location, papillary and reticular elastin. For accurate identification of all filament types, the model was trained to identify two subtypes of elastin: reticular (approx. greater than 2 μm in width) and papillary elastin (approx. less than 2 μm in width). Learning iterations were performed until IoU value was close to 1. Individual images were uploaded to VS200 and all elastin filaments with their corresponding areas were identified by the AI. Detection of elastin was confirmed by visual screening. Since elastin level in papillary area shows big differences between native volar (sole) and nonvolar (foot) skin, papillary area in injection sites was chosen as the focus for analysis using the deep learning model to determine relative elastin density. The papillary dermis was determined to be close to 200 μm deep on average. A 200 μm by 250 μm region of interest (ROI) was thus created to cover a portion of the papillary dermis. For each image, three ROIs in the papillary area were created and average values (elastin expression/0.05 mm2) were calculated.
RNA isolation and bulk RNA-seq
The skin biopsy sample was first incubated in 0.4% dispase II (Sigma, D4693) in a PBS solution at 4°C overnight. This resulted in the separation of the epidermal and dermal layers, thereby allowing for separate processing of the two layers and subsequently higher epidermal RNA concentrations for retrieval and analysis. After separation, the samples were stored in RNAlater (Invitrogen, AM7020) at −80°C until later processing. The tissues were then homogenized and RNA was extracted and purified using the RNeasy kit (Qiagen, 74106). During this purification process, DNase (Qiagen, 79254) was used to remove any contaminating DNA. The final concentration and purity of the RNA samples were measured using the NanoDrop 2000C spectrometer (Thermo Scientific). Because the 6-mm biopsy sample of the dermis had a low RNA concentration, only the total RNA from the epidermis was processed for RNA-seq. A total of 500 ng RNA sample isolated from epidermis was prepared for bulk RNA-seq. cDNA and library were prepared, and integrity was screened by Agilent 2100 bioanalyzer (Agilent) and sequencing was done by Illumina HiSeq2500. Data was deposited in the GEO database [GSE231998].
scRNA-seq from cultured cells or skin biopsy samples
Single-cell transcriptome 3′ gene sequencing from autologous fibroblasts and injection site biopsies was performed. Autologous fibroblasts from the scalp and sole were cultured in FGM-2 media. Fibroblasts were trypsinized using 0.05% TrypLE Select and washed with 0.04 (w/v) % BSA (Fisher Scientific, BP9703) in PBS. A total of 3 × 106 scalp or sole autologous fibroblasts was prepared in suspension by filtering through a 40-μm strainer. Skin biopsies from the 5-month time point were rinsed with DPBS and cut finely with scalpels. Small pieces of tissue were incubated with 1 ml of 0.4 mg/ml of Liberase TL (Roche, 05401020001) in DMEM with 100 units/ml of penicillin, 100 mg/ml of streptomycin and washed with PBS then incubated with 0.05% TrypLE Select. Cell suspension was washed with 10% FBS in DMEM with 100 units/ml of penicillin, 100 mg/ml of treptomycin then incubated with 100 units/ml of DNAse I (New England BioLabs, M0303). Then we used whole skin dissociation kit (human, Miltenyi Biotec, 130–101-540) for increased yield. Skin biopsies from other time points (2-week, 2-month, and 17-month) were processed using this kit according to the manufacturer’s protocol. Briefly, the skin biopsy was cut into small pieces with scalpels and transferred into a gentleMACS C tube (Milltenyi Biotec, 130096334) with enzyme mix then incubated for 3 hours at 37°C. Cells were extracted from the tissue using gentleMACS Dissociator (Miltenyi). In both methods, the cell suspension was prepared by filtering through a 70-μm strainer and was washed with 0.04% BSA in PBS.
To achieve live single-cell suspension, Propidium Iodide (BioLegend, 421301) negative and Vybrant DyeCycle Violet (Invitrogen, V35003) positive single cells were sorted using either a MoFlo Legacy or MoFlo XDP sorter (Beckman Coulter, Miami, Texas). The single-cell library was prepared using the 10x Genomics Chromium Single Cell Gene Expression platform (10x Genomics) and sequenced by Illumina NovaSeq 6000 (Illumina) by the Genetic Resources Facility at Johns Hopkins School of Medicine. The resulting raw base call (BCL) files were processed through the Cell Ranger Pipeline for FASTQ files, alignment, filtering, barcode counting, and UMI counting.
Spatial transcriptomics analysis of skin biopsy samples
The skin biopsies from a single subject at 2-month post-injection time point were rinsed with DPBS and stored in 4% paraformaldehyde at 4°C overnight for fixation. Tissue samples were prepared further following to manufacturer’s protocol (10x, CG000578 Xenium In Situ for FFPE Tissue Preparation Guide) in Hopkins histology core facility. Briefly, the samples were embedded in paraffin and then, tissues were cut into 5-μm thick slices and three samples from vehicle, scalp fibroblast, or sole fibroblast injection sites were placed on single Xenium slide (10x, PN 300941). Xenium slide with sections was stored in a desiccator at room temperature before scanning. The Xenium slide was processed according to manufacturer’s protocol (10x, CG000582 Xenium In Situ Gene Expression) at the Hopkins core facility. Processed slide was scanned by Xenium Analyzer (10x, PN-1000481) and data were processed through standard 10x Xenium workflow according to manufacturer’s protocol (10x, CG000584 Xenium Analyzer) to generate cell id by gene and transcript by location coordinate. For the detection, predesigned panel from Xenium (350 genes on human multitissue and cancer panel) and custom design panel (35 genes) (table S18) were used.
Bioinformatics data analysis
Paired-end reads from bulk RNA-seq were aligned to the human reference genome, hg38 (USCS genome website) by using STAR in the RSEM Ubuntu package. Differential gene expression was analyzed by DESeq2 with lower than 0.05 of adjusted P value (Padj). GO analysis was performed using web-based DAVID bioinformatics (https://david.ncifcrf.gov/) (51). These samples were then combined into one object. GO terms with differential enrichment were selected and made into a separate object. Normalized bulk RNA-seq data by DESeq2 was further analyzed by GSEA algorithm (33). Ontology gene sets (C5) were used for GSEA and GO pathways.
Downstream bioinformatic analysis of scRNA-seq was performed by using the Seurat package (ver. 3.0) in R (37). Standard preprocessing workflow was performed to filter out low count cells, multiplets, and dead cells. A total of 100,398 cells were processed for quality control analysis: 15,606 cells (4437 native scalp fibroblasts, 11169 native sole fibroblasts), 14,461 cells [4243 vehicle injection, 3770 scalp fibroblast injection, and 6448 sole fibroblast injection site cells] from 2 weeks, 27,179 cells (8576 vehicle injection, 9106 scalp fibroblast injection, and 9497 sole fibroblast injection site cells) from 2 months, 13,050 cells (4057 vehicle injection, 4239 scalp fibroblast injection, and 4304 sole fibroblast injection site cells) from 5 months, and 30,102 cells (7962 vehicle injection, 10507 scalp fibroblast injection, and 11633 sole fibroblast injection site cells) from 17 months. Cells with between 200 and 5000 feature counts and <10% mitochondrial counts were selected to generate Seurat objects of expression matrix. Preprocessing resulted in 10,938 cells (2012 scalp fibroblasts, 8926 sole fibroblasts) from the native site, 10,916 cells (2904 vehicle injection, 2688 scalp fibroblast injection, and 5324 sole fibroblast injection site cells) from 2 weeks, 22,404 cells (6851 vehicle injection, 7598 scalp fibroblast injection, and 7955 sole fibroblast injection site cells) from 2 months, 11,021 cells (3484 vehicle injection, 3651 scalp fibroblast injection, and 3886 sole fibroblast injection site cells) from 5 months, and 24,456 cells (6356 vehicle injection, 8207 scalp fibroblast injection, and 9893 sole fibroblast injection site cells) from 17 months. Distinct Seurat objects from vehicle, scalp, and sole injected site samples were merged into a single Seurat object to perform normalization, find variable features, integration to find distinct clusters. Individual clusters were identified based on their gene expression profile (52, 53). Markers for the specific cell types were KRT1/10 (spinous keratinocyte, SPN), KRT5/14 (basal keratinocyte, BAS), LUM/BGN/COL1A1 (fibroblast, FB), CD3E/CD69 (T cell), TRDC (γδT cell), CD14 (macrophage, MAC), EREG/S100A9 (dendritic cell, DEN), CD1C (Langerhans, LAN), MS4A2 (MAST), PECAM1 (endothelial cell, ENDO), and PMEL (melanocyte, MEL). Differentially expressed genes in distinct clusters of sole fibroblast injection or scalp fibroblast injection site samples compared to identical clusters in vehicle site samples were identified by default Wilcox method in the Seurat package with lower than 0.05 of adjusted P value (Padj) were selected for GO analysis. Genes in each GO pathway were selected by using DAVID bioinformatics. Trapnell et al. showed that single-cell trajectory analysis enables the recovery of gene expression kinetics in cellular processes including differentiation (54). For pseudotime analysis, the Seurat object of keratinocyte subsets was reclustered after which pseudotime and lineage analysis were performed by Monocle3 (55, 56). Briefly, single-cell trajectory analysis uses a sequence of gene expression changes and determines the position of the cells in the trajectory. Root cells were identified by the user based on high expression of KRT5/14. Normalized count data and cell group information from the Seurat object metadata were extracted and the CellChat R package was used for cell-cell communication (http://cellchat.org) (40). To identify the origin of autologous fibroblasts among fibroblasts from the injection sites, only the fibroblast subset was extracted and analyzed. Data were deposited in the GEO database [GSE231998].
For spatial transcriptomic data, Xenium performs cell segmentation for single-cell level spatial analysis. Briefly, Xenium employs DAPI staining to recognize single cells, and cell boundaries are inferred by expanding the 2D masks of nuclei morphologies. The given cell IDs were used for the further single-cell level analysis. The transcript counts per cell were scaled to 10,000 and log-transformed using a natural logarithm. Initially, scSorter was used to annotate cell types based on known marker genes (57). The cell-type annotations were superimposed on the UMAP dimension reduction plot to visually validate the accuracy of cell-type classification. To investigate changes in cell-type composition at the injection sites, the number of each cell type in the slices were calculated. For comparative analysis, section areas were approximately calculated using a Delaunay diagram with SciPy packages (58). Delaunay diagram transformed the transcripts with coordinates into concatenated triangles. After filtering out outlier transcripts with an arbitrary distance threshold, the total areas of the triangles were computed to approximate the areas of the slices. The cellular composition of each slice is determined by dividing the number of cells for each cell type by the total areas of the slices. For further investigation of the fibroblast densities in each slice, kernel density estimation was specifically applied to fibroblast cells to estimate the local fibroblast densities. Other cell types were excluded to prevent potential confounding effects on the density estimation at the injection sites. Secondly, Seurat in R package was used for unsupervised clustering. Cell types were identified by marker genes which were used in our scRNA-seq analysis. For epidermal differentiation analysis, epidermal layer composing granular (GRN), spinous (SPN) and basal (BAS) keratinocytes was isolated. Differentiation marker genes, including IVL, KRT1, KRT10, KRT5, and KRT14 were used for further analysis. The SPN score, the module score of IVL, KRT1 and KRT10 expression, showed high expressions in the supra layer. The BAS score, the module score of KRT5 and KRT14 expressions, was predominantly localized to the basal layer in the epidermis. We integrated Xenium and scRNA datasets using Seurat to explore overlap between two datasets. The integrated Seurat object was then processed for normalization and to find variable features, and individual clusters were identified based on their gene expression profile similar to scRNA datasets.
Supplementary Material
ACKNOWLEDGMENTS
We thank the Johns Hopkins Cell Therapy/Bone Marrow Transplant Core, including V. Lemas, J. D. Sproul, R. Edward, and M. Muth. We also thank H. Zhang and J. Margolick from the Flow Cytometry Cell Sorting Core Facility at Bloomberg School of Public Health, Johns Hopkins University, for doing fluorescence-activated cell sorting. We would like to thank the Hopkins Core for 10x scRNA-seq and 10x Xenium in situ, and M. K. Graham for advising 10x single-cell data analysis. The authors are grateful to J. Gillman, L. Rose, J. Ojeda, J. Sgambato, and A. Jaishankar for grant management. The authors would like to also acknowledge the Combat Casualty Care Research Program (CCCRP) and the Extremity Trauma and Amputation Center of Excellence (EACE) team members at Walter Reed National Military Medical Center, Uniformed Services University of the Health Sciences, Brooke Army Medical Center, and Naval Medical Center San Diego for their contributions to, and participation in, the subsequent phase 2 ongoing multisite study.
Funding: This work was funded under DARPA A66001–10-1–4081, AFIRM2 ER-11, and W81XWH-16-C-0167 from the US Department of Defense; philanthropic support including Northrop Grumman Electronic Systems, the Maryland Stem Cell Fund, and R01 AR064297 and AR074846/RMIP UG3AR079376 grants from the NIH to L.A.G. This work also was funded under 21RT0264 - FA9550–21-1–0284 from the Air Force Office of Scientific Research to Y.C. The Flow Cytometry Cell Sorting Core facility was supported by the Johns Hopkins Center for Aids Research (CFAR) grants 5P30AI094189–04 (Chaisson), 1S10OD016315–01, and 1S10RR13777001.
Footnotes
Competing interests: L.A.G. has received grant support paid to his institution, Johns Hopkins University, from Sun Pharma Advanced Research Company (SPARC). This grant is to investigate intellectual property of which Johns Hopkins University is the owner and of which L.A.G. is one of several inventors and for which he is under a licensing agreement with SPARC, and which has resulted in royalty payments to inventors. This grant and royalty payments are not related to the research presented in this manuscript.
REFERENCES AND NOTES
- 1.Driskell RR et al. , Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 504, 277–281 (2013). doi: 10.1038/nature12783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu Z et al. , Anatomically distinct fibroblast subsets determine skin autoimmune patterns. Nature 601, 118–124 (2022). doi: 10.1038/s41586-021-04221-8 [DOI] [PubMed] [Google Scholar]
- 3.Chang HY et al. , Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 99, 12877–12882 (2002). doi: 10.1073/pnas.162488599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinn JL, Bondre C, Gladstone HB, Brown PO, Chang HY, Anatomic demarcation by positional variation in fibroblast gene expression programs. PLOS Genet. 2, e119 (2006). doi: 10.1371/journal.pgen.0020119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rinn JL et al. , A systems biology approach to anatomic diversity of skin. J. Invest. Dermatol. 128, 776–782 (2008). doi: 10.1038/sj.jid.5700986 [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi Y et al. , Regulation of keratin 9 in nonpalmoplantar keratinocytes by palmoplantar fibroblasts through epithelial-mesenchymal interactions. J. Invest. Dermatol. 112, 483–488 (1999). doi: 10.1046/j.1523-1747.1999.00544.x [DOI] [PubMed] [Google Scholar]
- 7.Billingham RE, Silvers WK, Studies on the conservation of epidermal specificies of skin and certain mucosas in adult mammals. J. Exp. Med. 125, 429–446 (1967). doi: 10.1084/jem.125.3.429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang NB, Garza LA, Foote CE, Kang S, Meyerle JH, High prevalence of stump dermatoses 38 years or more after amputation. Arch. Dermatol. 148, 1283–1286 (2012). doi: 10.1001/archdermatol.2012.3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maiti R et al. , Morphological parametric mapping of 21 skin sites throughout the body using optical coherence tomography. J. Mech. Behav. Biomed. Mater. 102, 103501 (2020). doi: 10.1016/j.jmbbm.2019.103501; [DOI] [PubMed] [Google Scholar]
- 10.Egawa M, Hirao T, Takahashi M, In vivo estimation of stratum corneum thickness from water concentration profiles obtained with Raman spectroscopy. Acta Derm. Venereol. 87, 4–8 (2007). doi: 10.2340/00015555-0183 [DOI] [PubMed] [Google Scholar]
- 11.Fruhstorfer H, Abel U, Garthe CD, Knüttel A, Thickness of the stratum corneum of the volar fingertips. Clin. Anat. 13, 429–433 (2000). doi: [DOI] [PubMed] [Google Scholar]
- 12.Sandby-Møller J, Poulsen T, Wulf HC, Epidermal thickness at different body sites: Relationship to age, gender, pigmentation, blood content, skin type and smoking habits. Acta Derm. Venereol. 83, 410–413 (2003). doi: 10.1080/00015550310015419 [DOI] [PubMed] [Google Scholar]
- 13.Firooz A et al. , The influence of gender and age on the thickness and echo-density of skin. Skin Res. Technol. 23, 13–20 (2017). doi: 10.1111/srt.12294 [DOI] [PubMed] [Google Scholar]
- 14.Yasuda M, Yamaguchi S, Miyachi Y, Ishikawa O, Takahashi K, Palmoplantar collagen bundle size correlated with a characteristic spatial expression of decorin and lumican. Exp. Dermatol. 25, 318–320 (2016). doi: 10.1111/exd.12908 [DOI] [PubMed] [Google Scholar]
- 15.Merleev AA et al. , Biogeographic and disease-specific alterations in epidermal lipid composition and single-cell analysis of acral keratinocytes. JCI Insight 7, e159762 (2022). doi: 10.1172/jci.insight.159762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Philippeos C et al. , Spatial and Single-Cell Transcriptional Profiling Identifies Functionally Distinct Human Dermal Fibroblast Subpopulations. J. Invest. Dermatol. 138, 811–825 (2018). doi: 10.1016/j.jid.2018.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wiedemann J et al. , Differential cell composition and split epidermal differentiation in human palm, sole, and hip skin. Cell Rep. 42, 111994 (2023). doi: 10.1016/j.celrep.2023.111994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ascensión AM, Fuertes-Álvarez S, Ibañez-Solé O, Izeta A, Araúzo-Bravo MJ, Human Dermal Fibroblast Subpopulations Are Conserved across Single-Cell RNA Sequencing Studies. J. Invest. Dermatol. 141, 1735–1744.e35 (2021). doi: 10.1016/j.jid.2020.11.028 [DOI] [PubMed] [Google Scholar]
- 19.Xu Y et al. , A single-cell transcriptome atlas profiles early organogenesis in human embryos. Nat. Cell Biol. 25, 604–615 (2023). doi: 10.1038/s41556-023-01108-w [DOI] [PubMed] [Google Scholar]
- 20.Lieberman DE et al. , Foot strike patterns and collision forces in habitually barefoot versus shod runners. Nature 463, 531–535 (2010). doi: 10.1038/nature08723 [DOI] [PubMed] [Google Scholar]
- 21.Morgan BA, The dermal papilla: An instructive niche for epithelial stem and progenitor cells in development and regeneration of the hair follicle. Cold Spring Harb. Perspect. Med. 4, a015180 (2014). doi: 10.1101/cshperspect.a015180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rendl M, Polak L, Fuchs E, BMP signaling in dermal papilla cells is required for their hair follicle-inductive properties. Genes Dev. 22, 543–557 (2008). doi: 10.1101/gad.1614408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Billingham RE, Silvers WK, The origin and conservation of epidermal specificities. N. Engl. J. Med. 268, 539–545 (1963). doi: 10.1056/NEJM196303072681008 [DOI] [PubMed] [Google Scholar]
- 24.Rinn JL et al. , A dermal HOX transcriptional program regulates site-specific epidermal fate. Genes Dev. 22, 303–307 (2008). doi: 10.1101/gad.1610508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim D et al. , To Control Site-Specific Skin Gene Expression, Autocrine Mimics Paracrine Canonical Wnt Signaling and Is Activated Ectopically in Skin Disease. Am. J. Pathol. 186, 1140–1150 (2016). doi: 10.1016/j.ajpath.2015.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang WY, Davidson CD, Lin D, Baker BM, Actomyosin contractility-dependent matrix stretch and recoil induces rapid cell migration. Nat. Commun. 10, 1186 (2019). doi: 10.1038/s41467-019-09121-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hara M, Fujii T, Hashizume R, Nomura Y, Effect of strain on human dermal fibroblasts in a three-dimensional collagen sponge. Cytotechnology 66, 723–728 (2014). doi: 10.1007/s10616-013-9638-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Y, Macarak EJ, Korostoff JM, Howard PS, Compression and tension: Differential effects on matrix accumulation by periodontal ligament fibroblasts in vitro. Connect. Tissue Res. 45, 28–39 (2004). doi: 10.1080/03008200490278124 [DOI] [PubMed] [Google Scholar]
- 29.Wang JH, Thampatty BP, Lin JS, Im HJ, Mechanoregulation of gene expression in fibroblasts. Gene 391, 1–15 (2007). doi: 10.1016/j.gene.2007.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyer K, Lammers NC, Bugaj LJ, Garcia HG, Weiner OD, Optogenetic control of YAP reveals a dynamic communication code for stem cell fate and proliferation. Nat. Commun. 14, 6929 (2023). doi: 10.1038/s41467-023-42643-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franklin JM, Ghosh RP, Shi Q, Reddick MP, Liphardt JT, Concerted localization-resets precede YAP-dependent transcription. Nat. Commun. 11, 4581 (2020). doi: 10.1038/s41467-020-18368-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li X, Combs III JD, Salaita K, Shu X, Polarized focal adhesion kinase activity within a focal adhesion during cell migration. Nat. Chem. Biol. 19, 1458–1468 (2023). doi: 10.1038/s41589-023-01353-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). doi: 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Felfly H, Trudel M, Successful correction of murine sickle cell disease with reduced stem cell requirements reinforced by fractionated marrow infusions. Br. J. Haematol. 148, 646–658 (2010). doi: 10.1111/j.1365-2141.2009.07985.x [DOI] [PubMed] [Google Scholar]
- 35.Alves JV, Matos DM, Barreiros HF, Bártolo EA, Variants of dermatofibroma—A histopathological study. An. Bras. Dermatol. 89, 472–477 (2014). doi: 10.1590/abd1806-4841.20142629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hornick JL, Cutaneous soft tissue tumors: How do we make sense of fibrous and “fibrohistiocytic” tumors with confusing names and similar appearances? Mod. Pathol. 33 (Suppl 1), 56–65 (2020). doi: 10.1038/s41379-019-0388-4 [DOI] [PubMed] [Google Scholar]
- 37.Stuart T et al. , Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21 (2019). doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zieman AG, Coulombe PA, Pathophysiology of pachyonychia congenita-associated palmoplantar keratoderma: New insights into skin epithelial homeostasis and avenues for treatment. Br. J. Dermatol. 182, 564–573 (2020). doi: 10.1111/bjd.18033; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tirosh I et al. , Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016). doi: 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin S et al. , Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 12, 1088 (2021). doi: 10.1038/s41467-021-21246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yun YR et al. , Fibroblast growth factors: Biology, function, and application for tissue regeneration. J. Tissue Eng. 2010, 218142 (2010). doi: 10.4061/2010/218142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yonei-Tamura S et al. , FGF7 and FGF10 directly induce the apical ectodermal ridge in chick embryos. Dev. Biol. 211, 133–143 (1999). doi: 10.1006/dbio.1999.9290 [DOI] [PubMed] [Google Scholar]
- 43.Zinkle A, Mohammadi M, Structural Biology of the FGF7 Subfamily. Front. Genet. 10, 102 (2019). doi: 10.3389/fgene.2019.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moriyama M et al. , Multiple roles of Notch signaling in the regulation of epidermal development. Dev. Cell 14, 594–604 (2008). doi: 10.1016/j.devcel.2008.01.017 [DOI] [PubMed] [Google Scholar]
- 45.Milone MC et al. , Engineering enhanced CAR T-cells for improved cancer therapy. Nat. Cancer 2, 780–793 (2021). doi: 10.1038/s43018-021-00241-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider CA, Rasband WS, Eliceiri KW, NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). doi: 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tinevez JY et al. , TrackMate: An open and extensible platform for single-particle tracking. Methods 115, 80–90 (2017). doi: 10.1016/j.ymeth.2016.09.016 [DOI] [PubMed] [Google Scholar]
- 48.Artym VV, Matsumoto K, Imaging cells in three-dimensional collagen matrix. Curr. Protoc. Cell Biol. 48, 10.18.1–10.18.20 (2010). doi: 10.1002/0471143030.cb1018s48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Derr K et al. , Fully Three-Dimensional Bioprinted Skin Equivalent Constructs with Validated Morphology and Barrier Function. Tissue Eng. Part C Methods 25, 334–343 (2019). doi: 10.1089/ten.tec.2018.0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McQuin C et al. , CellProfiler 3.0: Next-generation image processing for biology. PLOS Biol. 16, e2005970 (2018). doi: 10.1371/journal.pbio.2005970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang DW, Sherman BT, Lempicki RA, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). doi: 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 52.Cheng JB et al. , Transcriptional Programming of Normal and Inflamed Human Epidermis at Single-Cell Resolution. Cell Rep. 25, 871–883 (2018). doi: 10.1016/j.celrep.2018.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang S et al. , Single cell transcriptomics of human epidermis identifies basal stem cell transition states. Nat. Commun. 11, 4239 (2020). doi: 10.1038/s41467-020-18075-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C et al. , The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). doi: 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cao J et al. , The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019). doi: 10.1038/s41586-019-0969-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qiu X et al. , Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315 (2017). doi: 10.1038/nmeth.4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo H, Li J, scSorter: Assigning cells to known cell types according to marker genes. Genome Biol. 22, 69 (2021). doi: 10.1186/s13059-021-02281-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Virtanen P et al. , SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020). doi: 10.1038/s41592-019-0686-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


