Abstract
The SNF2-related CBP activator protein, SrCap (pronounced “sir cap”), shares homology with the SNF2/SWI2 protein family. SrCap was cloned through its ability to bind CBP. SrCap can function as a CBP coactivator and can activate transcription in a reporter assay when expressed as a Gal-SrCap fusion protein. A monoclonal antibody raised against the carboxyl terminus of SrCap coimmunoprecipitates CBP/p300, supporting the model that SrCap is a CBP binding protein and that these proteins can be found together in a cellular protein complex. In addition, several cellular proteins are coimmunoprecipitated by the SrCap-specific antibody. Since adenovirus E1A proteins interact with CBP/p300 proteins, we examined what proteins could be copurified in a SrCap-specific coimmunoprecipitation assay from lysates of adenovirus-infected cells. While E1A proteins were not detected in this complex, to our surprise, we observed the presence of an infected-cell-specific band of 72 kDa, which we suspected might be the adenovirus DNA binding protein, DBP. The adenovirus DBP is a multifunctional protein involved in several aspects of the adenovirus life cycle, including an ability to modulate transcription. The identity of DBP was confirmed by DBP-specific Western blot analysis and by reimmunoprecipitating DBP from denatured SrCap-specific protein complexes. Using in vitro-translated DBP and SrCap proteins, we demonstrated that these proteins interact. To determine whether this interaction could affect SrCap-mediated transcription, we tested whether increasing amounts of DBP could modulate the Gal-SrCap transcription activity. We observed that DBP inhibited Gal-SrCap transcription activity in a dose-dependent manner. These data suggest a novel mechanism of adenovirus host cell control by which DBP binds to and inactivates SrCap, a member of the SNF2 chromatin-remodeling protein family.
The adenovirus DNA binding protein, DBP, is well studied for its role in adenovirus replication (3, 7, 12). DBP's replication function can be reconstituted in vitro and involves functions and interactions of at least three viral proteins, including the adenovirus DNA polymerase, precursor terminal protein, and DBP. In vitro replication function can be enhanced by the addition of two cellular transcription factors, nuclear factor I (NFI) and Oct1 (28). However, DBP is also implicated in several other essential functions important in the adenovirus life cycle. These include virion assembly (21), host range determination (1, 9), mRNA stability (5), and transformation (23).
In addition, DBP has roles in transcriptional regulation. DBP can enhance its own expression, and mutant studies demonstrate that only the highly phosphorylated forms of DBP grant this activation (20). DBP can regulate transcription directed by virus promoters (2). In these studies, DBP was shown to enhance transcription from the adenovirus E1A, E2A, and major late promoters and the adeno-associated virus P5 promoter but was also found to slightly inhibit the adenovirus E4 promoter. The mechanistic difference that explains these opposing activities is unknown. DBP is also implicated in enhancing the binding of NFI to its recognition site in the adenovirus replication origin (4, 25). NFI is a member of a family of factors that function in both DNA replication and transcription (14). Another target of DBP-induced transcriptional modulation is the transcription factor USF (upstream stimulatory factor). DBP enhances the binding of USF to its recognition site, resulting in an enhanced stimulation of in vitro transcription by USF (29). The fact that DBP can apparently both activate and inhibit transcription suggests that DBP is functioning through separate mechanisms.
In addition, DBP has been found in a stable cellular protein complex with a molecular mass of more than 650 kDa (24). This complex is devoid of viral replication proteins, suggesting that it is not a viral replication complex. The complex is also devoid of nucleic acids, indicating that its protein-protein associations are nucleic acid independent; nevertheless, this complex has the ability to bind DNA. These data suggest that DBP has as-yet-unidentified cellular protein interactions that may function in the adenovirus life cycle.
The SNF2-related CBP activator protein (SrCap) is a high-molecular-weight protein that was cloned in a yeast two-hybrid assay on the basis that it interacts with amino acids (aa) 227 to 460 of CBP (CREB-binding protein) (13). This region of CBP was shown to be important for CBP to function as a CREB coactivator (27). SrCap is a member of the SNF2 protein family of DNA-dependent ATPases, whose members' functions include chromatin remodeling, DNA repair, and regulation of transcription (26). SrCap can enhance the ability of CBP to activate transcription (13). SrCap and the adenovirus E1A proteins both bind to overlapping binding regions on CBP (16, 27). Using a mammalian two-hybrid reporter assay that functions through the SrCap-CBP interacting domains, we have demonstrated that wild-type E1A proteins, but not a CBP/p300-binding-negative E1A mutant, can inhibit transcription activity in this system, presumably through E1A proteins disrupting the SrCap-CBP interaction (13).
In this study, we used a monoclonal antibody raised against the carboxyl-terminal 239 aa of SrCap to probe for SrCap-associated proteins. This approach identifies an apparent multiprotein complex that includes SrCap and CBP/p300 proteins. To our surprise, when this protein complex was purified from lysates of adenovirus-infected cells, DBP copurified along with this complex. We demonstrate that in vitro-translated DBP and SrCap proteins can interact and that DBP can inhibit SrCap-mediated transcription. These data suggest a novel mechanism of adenovirus DBP-mediated transcriptional control.
MATERIALS AND METHODS
Plasmids.
To generate pcDNA-DBP, a DNA fragment was amplified from adenovirus serotype 2 DNA that encoded the DBP reading frame. The PCR primers encoded sequences that contain NheI or BamHI sites and a consensus Kozak sequence, as well as an approximately 20-nucleic-acid sequence 5′ or 3′ of the DBP sequence, respectively. The DNA fragment encoding DBP was cloned into pcDNA3.1(−)MycHis Version C (Invitrogen). The sequence of the DBP coding region was confirmed by DNA sequence analysis, and DBP expression from this plasmid was confirmed by immunoprecipitating DBP from transfected cells using a DBP-specific monoclonal antibody. The pGal-CAT, pGAL-VP16, and pGal-SRCAP plasmids were generated as previously described (13). The SrCap gene's GenBank accession number is AF143946.
Generation of monoclonal antibodies.
A monoclonal antibody (designated 253) was raised against a highly purified His-tagged SrCap fusion protein encoding carboxyl terminal aa 2733 to 2971. In addition, a DBP-specific monoclonal antibody (designated 218.2) against highly purified DBP from adenovirus type 2-infected HeLa cells was isolated. These antibodies were generated by immunizing BALB/c mice and using standard hybridoma development technology to isolate hybridoma cell lines that secrete the SrCap and DBP antibodies (10).
Immunoprecipitations.
A549 cells were metabolically labeled in 3 ml of Met-free, Cys-free Dulbecco's modified Eagle medium containing Tran35S-label (100 μCi/ml; ICN) per 10-cm-diameter plate for 2 h prior to lysis and immunoprecipitated as previously described (18). Typically, nearly confluent A549 cells from a 10-cm-diameter plate were lysed in 1 ml of a buffer containing 0.1% Nonidet P-40, 250 mM sodium chloride, 20 mM sodium phosphate (pH 7.0), 30 mM sodium pyrophosphate, 5 mM EDTA, and 10 mM sodium fluoride supplemented with 5 mM dithiothreitol and protease and phosphatase inhibitors (100 kIU of aprotinin and 1 μg [each] of leupeptin and pepstatin per ml). Lysates were precleared in the presence of 100 μl of a 10% (wt/vol) slurry of Staphylococcus aureus. Lysis using these nonionic detergent lysis conditions tends to preserve protein-protein interactions (18). Proteins were immunoprecipitated using 100 μl of hybridoma supernatant and 100 μl of a 3% (wt/vol) slurry of Sepharose CL-4B–protein A beads. Proteins were resolved on a sodium dodecyl sulfate (SDS)–7.5% polyacrylamide gel, and the dried gel was subjected to autoradiography. The SrCap-specific and M73 (E1A-specific) antibodies are of the same isotype (immunoglobulin G2a). For the denaturation experiment (see Fig. 5B), 35S-labeled SrCap-specific protein complexes were purified on Sepharose CL-4B–protein A beads from lysates of adenovirus-infected cells. A portion of this “native” SrCap immune complex was run in lane 1. The remaining beads containing SrCap-specific complexes were split into two equal portions, resuspended in 50 μl of lysis buffer containing 1% SDS and 10% 2-mercaptoethanol, and boiled for 5 min. Samples were cooled, and 1 ml of lysis buffer was added to each sample. Samples were spun briefly to remove initial beads, and supernatants were transferred to new microcentrifuge tubes. Proteins were reimmunoprecipitated with SrCap-specific or DBP-specific monoclonal antibodies and Sepharose CL-4B–protein A beads. Reimmunoprecipitated proteins were resolved on an SDS-polyacrylamide gel. Adenovirus serotypes 2 and 5 were used in these studies at a concentration of 50 to 100 PFU/cell.
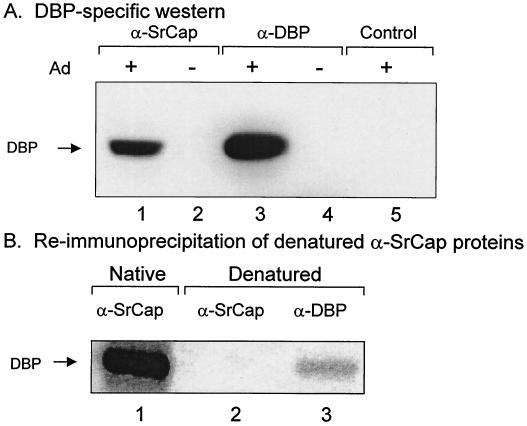
FIG. 5.
The 72-kDa protein present in the SrCap-specific coimmunoprecipitation is identified as the adenovirus DNA binding protein, DBP. (A) Proteins from uninfected (−) or adenovirus-infected (+) A549 cell lysates were immunoprecipitated with the SrCap-specific or DBP-specific monoclonal antibodies and resolved by SDS-PAGE and then transferred to a PVDF Western blot membrane. The membrane was probed with a DBP-specific monoclonal antibody. The position of DBP is indicated to the left. Control antibody was specific for the retinoblastoma gene product. (B) Proteins from uninfected (−) or adenovirus-infected (+) A549 cell lysates were immunoprecipitated with the SrCap-specific or DBP-specific monoclonal antibodies. These immune complexes were denatured (as described in Materials and Methods) and reimmunoprecipitated with a DBP-specific monoclonal antibody. The position of DBP is indicated to the left.
Western blot analysis.
Proteins were resolved by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride (PVDF) membrane in transfer buffer (25 mM Tris and 0.2 M glycine in 5% [vol/vol] methanol) in a Bio-Rad Trans Blot apparatus according to the manufacturer's recommendation. The membrane was blocked in phosphate-buffered saline (PBS) containing 5% nonfat dry milk for 1 h, followed by incubation in PBS containing primary antibody for 1 h. The membrane was washed three times with PBS containing 0.1% Tween 20 (PBS-T) over a 30-min period. The membrane was then incubated with a horseradish peroxidase-conjugated goat anti-mouse secondary antibody (catalog no. NA931; Amersham Pharmacia) for 1 h. The membrane was washed again three times with PBS-T over a 30-min period. Proteins were visualized using an ECL Western blotting detection kit (Amersham) and film.
In vitro translation.
Indicated plasmids containing the T7 promoter were transcribed and translated in reticulocyte lysate (TNT T7 coupled system; Promega) containing [35S]methionine for 1 h. Reticulocyte lysate containing translated protein(s) was immunoprecipitated with SrCap-specific or DBP-specific monoclonal antibodies. Immunocomplexes were washed extensively with lysis buffer, and the purified proteins were resolved by SDS-PAGE. Labeled proteins were visualized by autoradiography.
CAT reporter assays.
Transfections were performed as previously described (13). Cells were plated at 2.5 × 105/well in six-well plates 18 h prior to transfection. Each transfection utilized 200 ng of pGal-CAT as reporter plasmid and the indicated plasmids. The LipofectAMINE transfection method was performed according to the directions of the manufacturer (Life Technologies, Inc.). Cells were harvested 48 h after transfection and assayed for chloramphenicol acetyltransferase (CAT) activity using the phase-extraction method.
SrCap expression in baculovirus.
Baculovirus expressing a histidine-tagged SrCap protein (aa 1 to 2971) was generated using the BAC-to-BAC Baculovirus Expression System (BRL) following the manufacturer's recommendations.
RESULTS
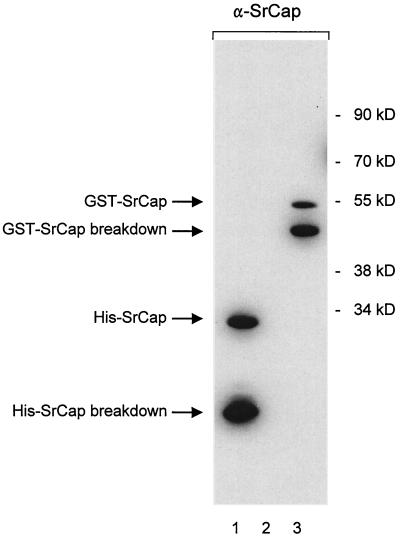
To characterize the SrCap protein, we have generated a monoclonal antibody that specifically recognizes the carboxyl terminus of SrCap. This was demonstrated by Western blot analysis using this antibody to probe a membrane containing a His-tagged SrCap carboxyl-terminal 239-aa fusion protein (Fig. 1, lane 1). This His-tagged SrCap fusion protein was the antigen used to induce the immune response in the mouse. To exclude the possibility that the antibody recognized the histidine portion of the immunogen, we ran bacterial lysate from nontransformed cells (lane 2) or from cells expressing a glutathione S-transferase (GST)–SrCap fusion protein containing the same carboxyl-terminal amino acids as before (lane 3). These results indicate that the monoclonal antibody recognizes SrCap carboxyl-terminal sequences and that it does not recognize the His portion of the antigen or show any aberrant specificities to bacterial proteins.
FIG. 1.
Characterization of the SrCap-specific monoclonal antibody that was raised against the 239-aa carboxyl terminus of the SrCap protein. Either purified His-tagged SrCap carboxyl terminus (lane 1), total cell protein from bacterial lysates (lane 2), or bacterial lysates containing a GST-tagged SrCap fusion protein (lane 3) were resolved by SDS–12% PAGE and transferred to a Western blot membrane. Both His- and GST-tagged SrCap proteins contained the carboxyl-terminal 239 aa of SrCap. This membrane was probed with the SrCap-specific antibody. Positions of His-GST fusion proteins and molecular mass markers are indicated on the left and right, respectively.
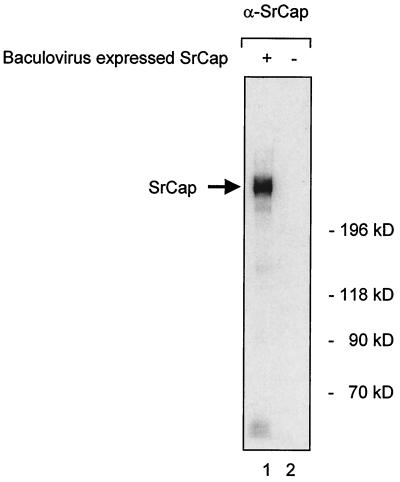
We tested the specificity of the antibody for endogenous SrCap protein in eukaryotic cells by running 200 μg of total cell protein from Sf9 insect cells that were uninfected or infected with baculovirus expressing the complete SrCap gene on a low-concentration SDS-PAGE gel. The separated proteins were transferred to a membrane and probed by Western blotting with the SrCap-specific antibody. A single prominent protein of the predicted molecular mass (ca. 315 kDa) was detected only in the lane containing total cell protein from Sf9 cells that express the SrCap gene (Fig. 2, lane 1); this result indicates that the full-length SrCap protein can be recognized in eukaryotic cells, and the antibody does not show specificity for any other proteins in these cells.
FIG. 2.
The SrCap-specific antibody recognizes full-length SrCap protein. Total cell protein from SF9 cells that were mock infected (lane 2) or infected with baculovirus that expresses the complete SrCap gene (lane 1) were resolved by SDS–5% PAGE and transferred to a Western blot membrane. This membrane was probed with the SrCap-specific antibody. Positions of baculovirus-expressed full-length SrCap and molecular weight markers are indicated.
The SrCap-specific antibody coimmunoprecipitates a series of cellular proteins.
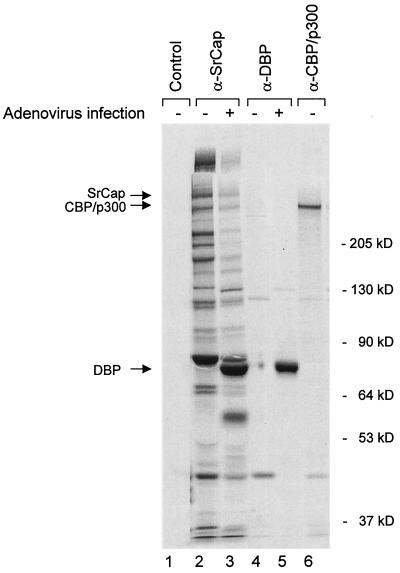
Since adenovirus E1A proteins bind to and regulate the SrCap partner, CBP/p300, we were interested in knowing the effect of adenovirus infection on endogenous SrCap complexes. To determine what proteins are immunoprecipitated by the SrCap-specific antibody in the presence or absence of adenovirus infection, proteins from 35S-labeled lysates from uninfected or adenovirus serotype 2-infected human A549 cells were harvested at 20 h after infection and immunoprecipitated with the SrCap-specific or control antibodies. The SrCap-specific antibody coimmunoprecipitated a series of cellular proteins from uninfected cell lysates (Fig. 3, lane 2), consistent with the expectation from its association with p300 that it is involved in multicomponent protein complexes. This complex set of proteins was also observed in coimmunoprecipitations from HeLa, 293, COS, Vero, Chinese hamster ovary (CHO), and NIH 3T3 cells (data not shown).
FIG. 3.
Several proteins are coimmunoprecipitated with the SrCap-specific monoclonal antibody. Proteins from 35S-labeled uninfected (−) or adenovirus-infected (+) A549 cell lysates were immunoprecipitated with control (E1A-specific antibody M73), SrCap-specific, DBP-specific, or CBP/p300-specific (NM11) monoclonal antibodies and resolved by SDS–7.5% PAGE, as indicated. A549 cells were infected with adenovirus serotype 2 for 20 h prior to lysis. Positions of known proteins are shown to the left. Positions of molecular mass markers are shown to the right. In addition to the molecular mass markers we used the actual molecular mass of CBP/p300 (ca. 265 kDa) to calculate the expected migration position of SrCap (ca. 315 kDa).
As expected, the immune complex included a protein of the expected size of the SrCap protein (predicted unmodified size of 315 kDa) as indicated in Fig. 3. Since SrCap was cloned on the basis of its interaction with CBP, we also wished to know whether any of the SrCap-associated proteins in the immune complex correspond to the CBP/p300 proteins. As shown, there is a 300-kDa protein band present in the SrCap coimmunoprecipitation that comigrated with CBP/p300 proteins purified by direct immunoprecipitation with the p300/CBP-specific monoclonal antibody NM11 (6) (Fig. 3, lane 6). We verified that the 300-kDa protein in the SrCap immunoprecipitation is CBP/p300 by Western blot analysis (described below).
In infected cells, the SrCap-specific antibody coimmunoprecipitated a similar series of proteins, but at least two infection-specific bands were apparent in the complex, migrating at 72 and 55 kDa (Fig. 3, lane 3). These apparent viral proteins were also observed in SrCap-specific coimmunoprecipitations from lysates of adenovirus serotype 5-infected cells (data not shown).
Since DBP is well expressed at 20 h after infection, we suspected that DBP might be the 72-kDa virus-specific band in the complex. We tested this possibility by comparing SrCap-specific and DBP-specific immunoprecipitations. The 72-kDa virus-specific band seen in the SrCap immune complexes precisely comigrates with adenovirus 72-kDa DNA binding protein purified with DBP-specific antibodies (Fig. 3, compare lanes 3 and 5, respectively).
It has previously been reported that DNA binding proteins can coprecipitate nonspecifically due to contaminating DNA present in the cell lysate. This nonspecific association can be disrupted by the simple addition of micrococcal nuclease or ethidium bromide to the cell lysate (17). Since DBP has DNA binding activity and SrCap, as a member of the SNF2 protein family, has a putative DNA binding activity, we tested whether they associate through a “bridge” of potential contaminating DNA present in the cell lysate. Treatment of SrCap-specific protein complexes with ethidium bromide or micrococcal nuclease did not disrupt any protein members from this complex, including the virus-specific bands, suggesting that the SrCap protein associations are DNA independent (data not shown).
Labeling of most of the host cell proteins visible in the lysate from infected cells is less efficient than that in uninfected cells. This is typical of adenovirus-infected cells. We have demonstrated in time-course-of-infection experiments that the overall decrease in radioactivity incorporated into each of the cellular proteins (Fig. 3, compare lanes 2 and 3) correlates with the onset of adenovirus-induced host cell shutoff that begins at the late phase of the virus life cycle (data not shown).
CBP/p300 is present in SrCap-specific immunoprecipitations.
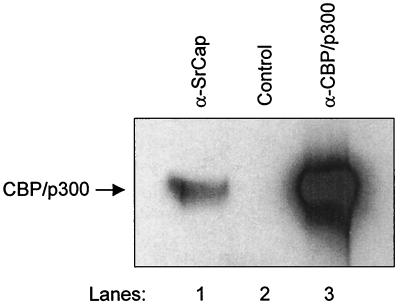
To determine whether the 300-kDa protein immunoprecipitated by the SrCap-specific antibody is CBP/p300, a PVDF Western blot membrane containing SrCap-specific or CBP/p300-specific immunoprecipitated proteins was probed with the CBP/p300-specific monoclonal antibody, NM11 (6). As shown in Fig. 4, CBP/p300 was clearly detected in the SrCap-specific immunoprecipitation, and it comigrated with CBP/p300 immunoprecipitated directly by NM11 (compare lanes 1 and 3). This result indicates that members of the CBP/p300 protein family are present in these complexes. No 300-kDa signal was detected in the control immunoprecipitation (lane 2).
FIG. 4.
CBP/p300 is present in the SrCap-specific monoclonal antibody coimmunoprecipitation. Purified proteins from either the SrCap-specific, no-antibody control, or CBP/p300-specific (NM11) immunoprecipitations from HeLa cell lysates were resolved by SDS-PAGE and transferred to a PVDF Western blot membrane, as indicated. After blocking, the membrane was probed with NM11. Bound antibody was detected by ECL. The position of CBP/p300 is indicated to the left.
The adenovirus DNA binding protein is present in SrCap-specific immunoprecipitations.
Since an apparent adenovirus-encoded 72-kDa protein is observed in the SrCap-specific coimmunoprecipitation, we tested whether this protein is DBP by DBP-specific Western blot analysis of the SrCap protein complex (Fig. 5A) or by immunoprecipitating DBP directly from denatured 35S-labeled SrCap protein complex (Fig. 5B). As shown in Fig. 5A, DBP was obtained only from SrCap protein complexes coimmunoprecipitated from lysates of adenovirus-infected cells and not from SrCap protein complexes isolated from uninfected cells (lane 2). The detected DBP protein in lane 1 comigrated with the DBP that was directly immunoprecipitated with a DBP-specific monoclonal antibody (lane 3). DBP was not observed in a DBP-specific immunoprecipitation from uninfected cells (lane 4) or a pRB control immunoprecipitation from adenovirus-infected cells (lane 5). As shown in Fig. 5B, putative DBP was immunoprecipitated from a 35S-labeled SrCap protein complex isolated from lysates of adenovirus-infected cells (lane 3); this DBP comigrated with DBP isolated with the SrCap protein complex (lane 1). This protein is not observed in a control SrCap-specific immunoprecipitation (lane 2). In addition, we have performed V8 protease digestions of the 72-kDa SrCap-specific protein and DBP protein isolated by DBP-specific immunoprecipitation and have observed identical proteolytic peptide profiles (data not shown). We conclude from these data that the 72-kDa protein observed in the SrCap-specific coimmunoprecipitations from lysates of adenovirus-infected cells is DBP.
DBP can interact directly with the SrCap protein in vitro
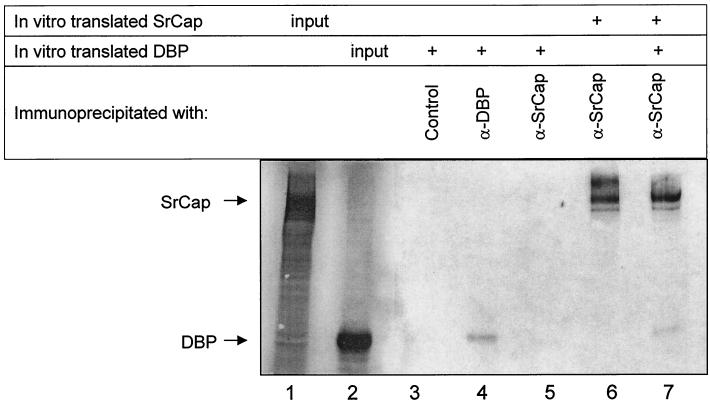
To determine whether DBP can directly bind to the SrCap protein, we translated these proteins in vitro and tested whether SrCap-specific antibody could coimmunoprecipitate DBP through a protein-protein association with SrCap. Due to difficulties with translating full-length SrCap (2,971 aa), we used a construct that encodes the carboxyl-terminal half of the SrCap protein (aa 1275 to 2971). This region of SrCap has the ability to mediate transcription in the absence of its intact ATPase domain (13). The in vitro translation products of the SrCap and DBP cDNAs are shown in Fig. 6, lanes 1 and 2, respectively. Portions of these in vitro translation products were immunoprecipitated with the indicated antibodies. The SrCap-specific or pRB control antibody was not able to immunoprecipitate in vitro-translated DBP (lanes 3 and 5, respectively), indicating that the SrCap-specific antibody does not bind directly to DBP and that DBP does not nonspecifically come down in these reactions. The DBP-specific and SrCap-specific antibodies were able to immunoprecipitate a portion of the in vitro-translated DBP and SrCap proteins, respectively (lanes 4 and 6). When the SrCap-specific antibody was used to immunoprecipitate mixed portions of both in vitro-translated DBP and SrCap proteins, DBP was detected, suggesting that DBP can interact directly with the SrCap protein (lane 7). This result also tentatively maps a DBP binding region within SrCap aa 1275 to 2971. We have attempted the converse experiment without observing SrCap binding to DBP in a DBP-specific immunoprecipitation. This is possibly due to competition of our DBP-specific antibodies and the SrCap product for the same region of DBP. This DBP-SrCap interaction does not rule out the possibility that DBP binds to other regions of SrCap or has other protein-protein associations in the putative SrCap-specific protein complex.
FIG. 6.
DBP can bind SrCap in vitro. SrCap and DBP proteins were in vitro translated as described in Material and Methods. Roughly 2% of these translation products were run in lanes 1 and 2, respectively. The remaining portions of each translation product were divided equally into the indicated immunoprecipitation reactions. Proteins purified in each of these immunoprecipitations were resolved by SDS-PAGE and were detected by autoradiography. Positions of the SrCap and DBP proteins are indicated to the left. Only alternating lanes of this gel were loaded to avoid any possibility that detected proteins were due to gel loading artifacts.
DBP inhibits SrCap-mediated transcription.
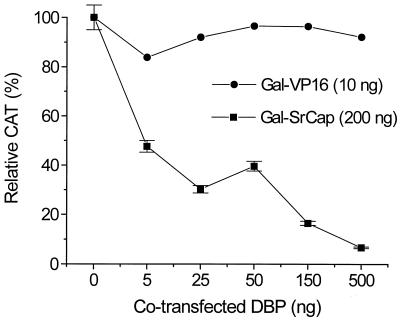
We have previously reported that SrCap, like other members of the SNF2 family, can activate transcription of a CAT reporter plasmid when expressed as a Gal-SrCap (aa 1275 to 2971) fusion protein (13). This is the same region of SrCap that was shown to bind to DBP in Fig. 6. Since DBP is found in a SrCap-specific coimmunoprecipitation and we have demonstrated that DBP and SrCap proteins can interact, we tested whether DBP had an effect on SrCap-mediated transcription. We transfected a reporter plasmid, pGRE-CAT, which contains a promoter with a GAL-responsive element, with plasmids expressing either Gal-VP16 or Gal-SrCap (1275 to 2971) fusion proteins together with increasing amounts (0, 5, 25, 50, 150, or 500 ng) of pcDNA-DBP into CHO cells. CHO cells were used because of their high transfection efficiency. In addition, transfections were normalized to equal picomolar amounts of plasmid DNAs (using decreasing amounts of empty vector control plasmid pcDNA3.1) and also to total amount of DNA (using salmon sperm DNA). The relative amounts of CAT activity are shown in Fig. 7. As shown, increasing amounts of transfected DBP had relatively little effect on Gal-VP16 CAT activity, indicating that DBP did not nonspecifically inhibit transcription in this system. However, Gal-SrCap activity was specifically inhibited by DBP in a dose-dependent manner to less than 10%. DBP inhibition of SrCap-mediated transcription was also observed in transfected HeLa cells (data not shown).
FIG. 7.
DBP inhibits SrCap-mediated transcription in a dose-dependent manner. Regulation of SrCap-mediated transcription by DBP was assessed by transient transcription assay. Chinese hamster ovary (CHO TC9) cells were transfected with the reporter plasmid pGal-CAT (100 ng), either pGal-SrCap (200 ng) or pGal-VP16 (10 ng), and increasing amounts of pcDNA-DBP, as indicated. Transfections were normalized to equal picomolar amounts of plasmid DNAs (using decreasing amounts of empty vector pcDNA) and also to total amount of DNA (using salmon sperm DNA). Values are the means ± standard error (error bars) from two separate experiments in which each point was performed in triplicate. The absolute CAT activities obtained for Gal-VP16 and Gal-SrCap in the absence of pcDNA-DBP (0 ng) are 30,000 and 3,000 cpm, respectively.
DISCUSSION
In this report, we identified a novel cellular target for adenovirus DBP-induced host cell transcription control. We have found that DBP can bind to the SNF2-related CBP-activator protein, SrCap, and this interaction leads to inhibition of SrCap-mediated transcription. SrCap is a member of the SNF2 protein family that was cloned based on its ability to bind to CBP and was subsequently shown to function as an activator of CBP (13). In addition, we have also found that SrCap activates transcription of several promoters, including the phosphoenolpyruvate carboxykinase (PEPCK), somatostatin, enkaphalin, and mouse mammary tumor virus promoters. This activation appears to function through CREB and glucocorticoid receptor-mediated transcription mechanisms (J. Chrivia, unpublished observations) and supports the model that SrCap functions as an activator of CBP.
In uninfected cells we demonstrate that SrCap and a series of cellular proteins are purified by coimmunoprecipitation using a SrCap-specific monoclonal antibody, indicating that SrCap is present in a multicomponent cellular protein complex. This is consistent with the fact that members of the SNF2 protein family exist in multiprotein complexes (11). As expected, CBP and/or p300 proteins are present in SrCap-specific coimmunoprecipitations, consistent with the fact that SrCap was cloned as a CBP-binding protein and that members of the CBP/p300 protein family also interact with several cellular proteins, such as nuclear steroid receptors, basal transcription factors, and RNA polymerase II (8).
It should be noted that recent advances in transcription and chromatin remodeling have led to the identification of two distinct families of chromatin remodeling proteins, those that remodel chromatin by targeting and modifying DNA structure in an ATP-dependent mechanism (i.e., SNF2 protein family) and those that remodel chromatin by targeting and modifying proteins that are essential for maintaining chromatin structure, through acetyltransferase activity (i.e., CBP/p300 protein family). It has been proposed that these families act in concert to coordinate remodeling of chromatin (22). The SrCap-CBP/p300 association represents a cellular protein complex that contains members of both protein families. We speculate that the SrCap-CBP/p300 proteins may function synergistically in transcription and chromatin remodeling.
CBP/p300 proteins are key cellular targets of adenovirus growth control of host cells (19). The adenovirus protein E1A has been demonstrated to block the transcriptional activity of several transcription factors which utilize CBP/p300 as a coactivator. E1A binds to at least four distinct regions within CBP/p300: the amino-terminal end, the histone acetyltransferase domain, and two sites in the C-terminal end. Binding of E1A to the amino-terminal end of CBP also blocks the binding of SrCap to this region (13). This result suggested that adenovirus infection might alter the SrCap protein complex.
To our surprise, while E1A proteins were not observed, we found two adenovirus infection-specific proteins associated with the SrCap protein complex that migrated at 55 and 72 kDa. We have identified the 72-kDa SrCap-associated protein as the adenovirus 72-kDa DNA binding protein, DBP. We verified that the 72-kDa protein is DBP by using three separate experimental approaches: DBP was detected in a DBP-specific Western blot analysis of the SrCap complex, DBP was immunoprecipitated from denatured SrCap protein complex by DBP-specific monoclonal antibody, and the presence of DBP was indicated by comparing the partial proteolytic peptide digest pattern of 72-kDa SrCap-associate protein with that of DBP. The lack of a SrCap signal in the DBP-specific immunoprecipitation (Fig. 3, lane 5) is not surprising since only a small percentage of DBP is associated with a large cellular protein complex (24).
The DBP-SrCap complex protein associations were not effected by micrococcal nuclease or ethidium bromide treatment, which digests or disrupts the structure of DNA, respectively. This indicates that although DBP is a DNA binding protein, association with the SrCap protein complex is not through nonspecific binding to DNA. These results are also consistent with previous studies that used these same techniques to demonstrate that DBP exists as part of a high-molecular-weight complex that is free of DNA (24). In addition, our in vitro studies demonstrate that DBP interacts directly with SrCap. They indicate that a least one DBP binding site resides within the C-terminal end of SrCap.
Since the same carboxyl-terminal region of SrCap that was demonstrated to interact with DBP can function as a transcriptional activator when expressed as a Gal-SrCap fusion protein in a CAT reporter assay, we tested whether DBP could affect SrCap-mediated transcription. DBP was found to inhibit SrCap-mediated transcription in a dose-dependent manner, presumably through a direct protein-protein interaction with SrCap. The fact that there was no inhibition of the control Gal-VP16 transcriptional activator, even at the highest concentration of DBP, suggests that inhibition is specific for the presence of SrCap protein sequences in this system and argues against a model in which inhibition is due to a general nonspecific inhibitory effect resulting from DBP's DNA binding activity.
DBP-induced inhibition of a SrCap functional activity may contribute to the fact that DBP is toxic to cells, as demonstrated by the fact that stable DBP cell lines that constitutively express DBP cannot be made. In this model, DBP is inactivating the transcriptional activity and possibly the putative chromatin remodeling activity of SrCap. Since SrCap is an activator of CBP/p300 proteins, which are themselves chromatin remodeling proteins and key cell growth regulators that are implicated in regulating many transcription factors, a DBP-SrCap association could have detrimental effects on cell growth control.
Since the ability of adenovirus to mount a productive viral infection is highly species specific and DBP is implicated in host range determination, the DBP-SrCap interaction may also play a role in the determination of host range. This can be envisioned since mutations in DBP give it the ability to alter the host range of the virus (15). These data make the DBP-SrCap interaction a potentially intriguing mechanism of host cell regulation by DBP.
ACKNOWLEDGMENTS
We thank G. Chinnadurai, E. Moran, and W. Wold for helpful discussions and reading the manuscript.
This work was supported by Public Health Service grant CA-68066 from the National Cancer Institute.
REFERENCES
- 1.Anderson C W, Hardy M M, Dunn J J, Klessig D F. Independent, spontaneous mutants of adenovirus type 2-simian virus 40 hybrid Ad2+ND3 that grow efficiently in monkey cells possess identical mutations in the adenovirus type 2 DNA-binding protein gene. J Virol. 1983;48:31–39. doi: 10.1128/jvi.48.1.31-39.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang L S, Shenk T. The adenovirus DNA-binding protein stimulates the rate of transcription directed by adenovirus and adeno-associated virus promoters. J Virol. 1990;64:2103–2109. doi: 10.1128/jvi.64.5.2103-2109.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chase J W, Williams K R. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 4.Cleat P H, Hay R T. Co-operative interactions between NFI and the adenovirus DNA binding protein at the adenovirus origin of replication. EMBO J. 1989;8:1841–1848. doi: 10.1002/j.1460-2075.1989.tb03579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cleghon V, Voelkerding K, Morin N, Delsert C, Klessig D F. Isolation and characterization of a viable adenovirus mutant defective in nuclear transport of the DNA-binding protein. J Virol. 1989;63:2289–2299. doi: 10.1128/jvi.63.5.2289-2299.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dallas P B, Yaciuk P, Moran E. Monoclonal antibody NM11 recognizes a C-terminal epitope shared by p300 and CBP. Hybridoma. 1997;16:273–275. doi: 10.1089/hyb.1997.16.273. [DOI] [PubMed] [Google Scholar]
- 7.de Jong R N, van der Vliet P C. Mechanism of DNA replication in eukaryotic cells: cellular host factors stimulating adenovirus DNA replication. Gene. 1999;236:1–12. doi: 10.1016/s0378-1119(99)00249-8. [DOI] [PubMed] [Google Scholar]
- 8.Goodman R H, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 9.Harfst E, Leppard K N. A comparative analysis of the phosphorylation and biochemical properties of wild type and host range variant DNA binding proteins of human adenovirus 5. Virus Genes. 1999;18:97–106. doi: 10.1023/a:1008009630695. [DOI] [PubMed] [Google Scholar]
- 10.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 11.Havas K, Whitehouse I O-H T. ATP-dependent chromatin remodeling activities. Cell Mol Life Sci. 2001;58:673–682. doi: 10.1007/PL00000891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hay R T, Freeman A, Leith I, Monaghan A, Webster A. Molecular interactions during adenovirus DNA replication. Curr Top Microbiol Immunol. 1995;199(Part 2):31–48. doi: 10.1007/978-3-642-79499-5_2. [DOI] [PubMed] [Google Scholar]
- 13.Johnston H, Kneer J, Chackalaparampil I, Yaciuk P, Chrivia J. Identification of a novel SNF2/SWI2 protein family member, SRCAP, which interacts with CREB-binding protein. J Biol Chem. 1999;274:16370–16376. doi: 10.1074/jbc.274.23.16370. [DOI] [PubMed] [Google Scholar]
- 14.Jones K A, Kadonaga J T, Rosenfeld P J, Kelly T J, Tjian R. A cellular DNA-binding protein that activates eukaryotic transcription and DNA replication. Cell. 1987;48:79–89. doi: 10.1016/0092-8674(87)90358-8. [DOI] [PubMed] [Google Scholar]
- 15.Klessig D F, Grodzicker T. Mutations that allow human Ad2 and Ad5 to express late genes in monkey cells map in the viral gene encoding the 72K DNA binding protein. Cell. 1979;17:957–966. doi: 10.1016/0092-8674(79)90335-0. [DOI] [PubMed] [Google Scholar]
- 16.Kurokawa R, Kalafus D, Ogliastro M H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 17.Lai J S, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moran E, Yaciuk P. Immunoprecipitation of E1A-containing protein complexes. In: Wold W S M, editor. Adenovirus methods and protocols. Totowa, N.J: Humana; 1998. pp. 195–202. [Google Scholar]
- 19.Moran E, Zerler B. Interactions between cell growth regulation domains in the products of the adenovirus E1A oncogene. Mol Cell Biol. 1988;8:1756–1764. doi: 10.1128/mcb.8.4.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morin N, Delsert C, Klessig D F. Mutations that affect phosphorylation of the adenovirus DNA-binding protein alter its ability to enhance its own synthesis. J Virol. 1989;63:5228–5237. doi: 10.1128/jvi.63.12.5228-5237.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicolas J C, Sarnow P, Girard M, Levine A J. Host range temperature-conditional mutants in the adenovirus DNA binding protein are defective in the assembly of infectious virus. Virology. 1983;126:228–239. doi: 10.1016/0042-6822(83)90474-9. [DOI] [PubMed] [Google Scholar]
- 22.Pollard K J, Peterson C L. Chromatin remodeling: a marriage between two families? Bioessays. 1998;20:771–780. doi: 10.1002/(SICI)1521-1878(199809)20:9<771::AID-BIES10>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 23.Rice S A, Klessig D F, Williams J. Multiple effects of the 72-kDa, adenovirus-specified DNA binding protein on the efficiency of cellular transformation. Virology. 1987;156:366–376. doi: 10.1016/0042-6822(87)90416-8. [DOI] [PubMed] [Google Scholar]
- 24.Ricigliano J W, Brough D E, Klessig D F. Identification of a high-molecular-weight cellular protein complex containing the adenovirus DNA binding protein. Virology. 1994;202:715–723. doi: 10.1006/viro.1994.1393. [DOI] [PubMed] [Google Scholar]
- 25.Stuiver M H, van der Vliet P C. Adenovirus DNA-binding protein forms a multimeric protein complex with double-stranded DNA and enhances binding of nuclear factor I. J Virol. 1990;64:379–386. doi: 10.1128/jvi.64.1.379-386.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sudarsanam P, Winston F. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 2000;16:345–351. doi: 10.1016/s0168-9525(00)02060-6. [DOI] [PubMed] [Google Scholar]
- 27.Swope D L, Mueller C L, Chrivia J C. CREB-binding protein activates transcription through multiple domains. J Biol Chem. 1996;271:28138–28145. doi: 10.1074/jbc.271.45.28138. [DOI] [PubMed] [Google Scholar]
- 28.van der Vliet P C. Adenovirus DNA replication. Curr Top Microbiol Immunol. 1995;199(Part 2):1–30. doi: 10.1007/978-3-642-79499-5_1. [DOI] [PubMed] [Google Scholar]
- 29.Zijderveld D C, d'Adda di Fagagna F, Giacca M, Timmers H T, van der Vliet P C. Stimulation of the adenovirus major late promoter in vitro by transcription factor USF is enhanced by the adenovirus DNA binding protein. J Virol. 1994;68:8288–8295. doi: 10.1128/jvi.68.12.8288-8295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]