Abstract
Eukaryotic organisms are composed of different cell types with defined shapes and functions. Specific cell types are produced by the process of cell differentiation, which is regulated by signal transduction pathways. Signaling pathways regulate cell differentiation by sensing cues and controlling the expression of target genes whose products generate cell types with specific attributes. In studying how cells differentiate, fungi have proved valuable models because of their ease of genetic manipulation and striking cell morphologies. Many fungal species undergo filamentous growth—a specialized growth pattern where cells produce elongated tube-like projections. Filamentous growth promotes expansion into new environments, including invasion into plant and animal hosts by fungal pathogens. The same signaling pathways that regulate filamentous growth in fungi also control cell differentiation throughout eukaryotes and include highly conserved mitogen-activated protein kinase (MAPK) pathways, which is the focus of this review. In many fungal species, mucin-type sensors regulate MAPK pathways to control filamentous growth in response to diverse stimuli. Once activated, MAPK pathways reorganize cell polarity, induce changes in cell adhesion, and promote the secretion of degradative enzymes that mediate access to new environments. However, MAPK pathway regulation is complicated because related pathways can share components with each other yet induce unique responses (i.e. signal specificity). In addition, MAPK pathways function in highly integrated networks with other regulatory pathways (i.e. signal integration). Here, we discuss signal specificity and integration in several yeast models (mainly Saccharomyces cerevisiae and Candida albicans) by focusing on the filamentation MAPK pathway. Because of the strong evolutionary ties between species, a deeper understanding of the regulation of filamentous growth in established models and increasingly diverse fungal species can reveal fundamentally new mechanisms underlying eukaryotic cell differentiation.
Keywords: mucin, Rho GTPase, MAPK pathway, signaling specificity, signal integration, comparative evolutionary biology, filamentous growth, pseudohyphae, invasive growth, hyphae, fungal pathogens, pectinases
Introduction
Cell differentiation is a fundamental process underlying the extraordinary biological diversity in nature. Starting with genetically identical progenitor cells, cells can differentiate into a vast array of cell types with different shapes and sizes that are suited for specific functions. These specialized cells are highly organized in multicellular organisms through the process of development (Glover 2000; Basson 2012; Cook and Genever 2013; Brunet and King 2017). Cell differentiation can also occur in single-celled organisms to promote their survival in specific environments, such as in response to stress, and as cells go through their life cycle. Because cells can differentiate into different cell types, a central question is to understand how cell differentiation is regulated. Because the misregulation of cell differentiation is commonly linked to developmental problems and disease, addressing this question is important to understanding human health and organismal development.
During the process of development, cells differentiate in a predefined program. This generally occurs in response to signals (i.e. stimuli) transmitted by signal transduction pathways. Signaling pathways that regulate differentiation can be activated by the detection of extracellular stimuli through receptors or sensors at the cell surface. Once activated, these sensors control effector relay molecules and second messengers, which govern the activity of transcription factors. Transcription factors activated by signaling pathways regulate the expression of target genes whose products function to produce specialized cell types (Kramer 2016; Martinez-Soto and Ruiz-Herrera 2017). Some of these gene products generate specialized cell types by reconfiguring cell polarity or the composition of proteins at the cell surface.
One way that signaling pathways have been studied is by reductionist approaches with genetically tractable model organisms. One valuable group of models has been fungi, which continue to provide valuable insights into the fundamental processes that generate cell-type diversity. The general and distinct biology of each fungal species, including aspects of cell differentiation, come from their evolution in different niches and environments. These niches are diverse as some fungi are free-living saprotrophs (decomposers), others are pathogens (that cause infections), and yet others are symbionts (mutualist and commensal organisms). Originally nonpathogenic models were commonly studied; however, as genetic tools have improved, fungal pathogens have become important models as well. Additionally, cell differentiation in fungi can occur throughout the life cycle, such as during sexual reproduction where diploid cells undergo sporulation to produce haploid progeny (Neiman 2011; Su et al. 2012; Wendland 2020), and haploid cells can mate to form diploids (Bölker and Kahmann 1993; Bardwell 2005; Du and Yang Zhu 2021; Sieber et al. 2023). Many fungal species also undergo filamentous growth (Riquelme 2013; Kiss et al. 2019)—a specialized cell type where elongated cells expand into new environments. Because filamentous growth is morphologically striking and important for disease, it has taken center stage in the studies of fungal differentiation.
How cells make decisions about whether to undergo filamentous growth and how they accomplish this morphological switch is the focus of this review. We will introduce filamentous growth and discuss how it is used as a foraging strategy in various fungal species. We will also discuss one type of pathway that controls filamentous growth, the evolutionarily conserved mitogen-activated protein kinase (MAPK) pathway. Much has been learned about how MAPK pathways are activated, including by mucin-type signaling proteins. We will also explore how different MAPK pathways send specific signals through shared proteins that function in many pathways. In addition to signal specificity, we will discuss how MAPK and other pathways function in networks to integrate a response to multiple signals. Because these pathways control cell differentiation, we will also describe how some of the key target proteins of these pathways produce the filamentous cell type.
Filamentous growth is a conserved cell type across fungal species
At a cellular level, most fungal species exist in one or more different cell types. For most fungi, cells spend at least a portion of their life cycle forming filaments (Fig. 1; Chen et al. 2020; Kumar 2021). These filaments vary widely in appearance depending on the species and life stage and can be categorized in different patterns (e.g. true hyphae or false hyphae). In true hyphae, cells do not fully separate by cytokinesis leaving cytoplasmic connections between cells (Silva et al. 2004; Mouriño-Pérez 2013; Roberson 2020). In some species, there is no separation of cellular compartments (Fig. 1, coenocytic hyphae), while in other species, the formation of septa allows migration of some materials between cells (Fig. 1, septate hyphae). In false hyphae, also called pseudohyphae, cells fully separate by cytokinesis and do not share a cytoplasmic connection. In this state, cells are held together by cell adhesion molecules to form chains of cells (Fig. 1, pseudohyphae). All these patterns are grouped together under the umbrella terms filamentous or filamentous-form growth.
Fig. 1.
Different growth patterns in fungi. Many fungal species undergo filamentous growth, including hyphal and pseudohyphal growth. Some species, such as yeasts, can grow as individual cells and form filaments under the right conditions (arrows).
Filamentous growth is the normal, or default growth form, for many fungal species such as Ashbya gossypii (Schmitz and Philippsen 2011) and Neurospora crassa (Patel and Free 2019). These models have been useful for studying many aspects of filamentous growth including cytoplasmic streaming and nuclear migration, as well as the regulation of the fungal cell wall. Other species include yeasts, which are a large subcategory of single-celled fungi distributed across 2 major subphyla (ascomycetes and basidiomycetes; Shen et al. 2018). In yeasts, filamentous growth is an inducible growth form. For example, in nutrient replete environments yeast cells reproduce by budding, where cells are ovoid and fully separate (Fig. 1, yeast-form growth). However, in response to changes in the environment such as nutrient limitation, yeast cells can also undergo filamentous growth (Fig. 1, arrows; Chen et al. 2020; Kumar 2021). Interestingly, some yeasts (such as the saprotroph Saccharomyces cerevisiae and human pathogen Nakaseomyces glabratus formerly Candida glabrata) only make pseudohyphae (Gimeno et al. 1992; Csank and Haynes 2000), while other yeasts [such as the plant pathogen Ustilago maydis (corn smut)] make septate hyphae (Brefort et al. 2009). The most common human pathogen Candida albicans can exist in multiple filamentous types at its physiological temperature, 37°C (Sudbery 2011; Chen et al. 2020). Contrastingly, another category of human pathogens known as the dimorphic fungi (Histoplasma capsulatum, Blastomyces dermatitidis, and Coccidioides posadasii) exist as yeast-form at physiological temperature (37°C) and filamentous molds at lower temperatures. Therefore, although there is quite a variety in filamentous growth patterns, most species of fungi can undergo filamentous growth during at least part of their life cycle.
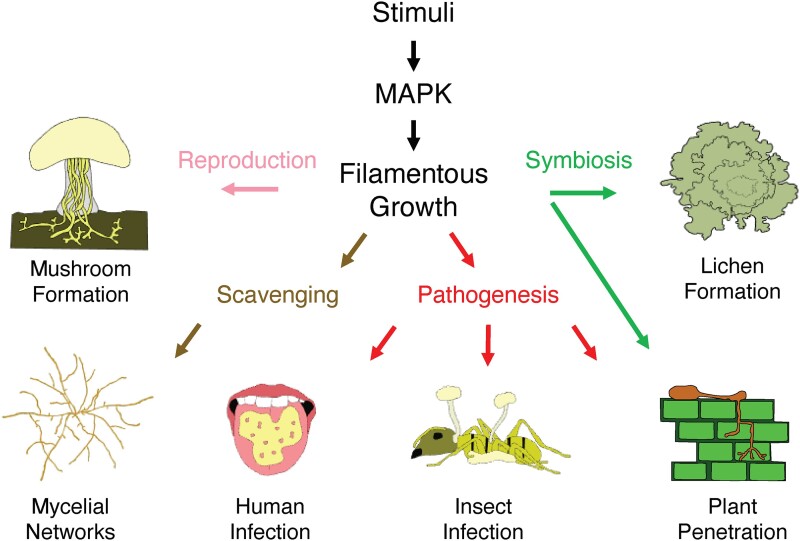
Filamentous growth makes fungi excellent scavengers because filaments grow outwards to allow the expansion of nonmotile cells into new environments. Filaments can also play important roles in sexual reproduction. For example, in some species of multicellular fungi, filaments can switch from a branching pattern to forming parallel arrangements to form reproductive structures, such as mushrooms, baskets, and cushions (Fig. 2, Reproduction, Mushroom Formation; Chiu and Moore 1996; Money 2016). Filamentous growth also generates branching mycelial networks in multicellular species (Fig. 2; Mycelial Networks; Heaton et al. 2012; Fricker et al. 2017), which are important for obtaining and transporting nutrients over large distances and in heterogenous environments. In fact, the largest organism in the world is composed of a single mycelial network, identified as the honey mushroom Armillaria ostoyae, which covers 9 km2 in the Malheur National Forest in Oregon.
Fig. 2.
The breadth of filamentous growth regulation by MAPK pathways. Many types of stimuli trigger MAPK pathways to regulate filamentous growth. Filamentous growth promotes multicellular reproductive structures, as seen in mushrooms, nutrient scavenging in mycelial networks, pathogenesis (in animals such as humans, insects, and plants), and even symbioses with other species, including plants and algae in lichens.
Filaments can also penetrate into substrates when invading new territories, by a process known as invasive growth. For symbiotic mycorrhizal fungi, invasive cells establish mutualistic connections with plant roots to exchange nutrients between the plant and the fungal mycelial networks. This has the effect of greatly expanding the plant's root network to benefit both species, providing sugars to the fungus and micronutrients and water to the plant (Dighton 2009; Bonfante and Genre 2010; Figueiredo et al. 2021). Similarly, in lichen-forming fungi, filamentous growth is important to establish symbiotic connections with algae (Fig. 2; symbiosis, lichen formation; Wang, Li et al. 2023). Although some species of pathogens do not undergo filamentous growth in the host, many human (Brand 2012; Chen et al. 2020), insect (Islam et al. 2021; Zhang, Meng, et al. 2021) and plant pathogens (Kahmann and Kämper 2004; Haueisen and Stukenbrock 2016) form filaments that penetrate and weave their way through the host to extract nutrients and complete their life cycles (Fig. 2, pathogenesis).
The interactions between yeasts and multicellular organisms have become a fertile area for new discoveries since invertebrates such as the roundworm Caenorhabditis elegans (Mylonakis et al. 2007; Pukkila-Worley et al. 2011) and larvae of the wax moth Galleria mellonella (Trevijano-Contador and Zaragoza 2018) have been developed into infection models. These models are surprisingly relevant to mammalian infection and have allowed the genetic dissection of the fungal and host pathways that control colonization and infection (Phadke et al. 2018). For example, multispecies interactions in C. elegans showed that probiotic yeasts (and their secondary metabolites) can inhibit infection by pathogens (Peleg et al. 2008; Kunyeit et al. 2019; Kunyeit et al. 2021). Similarly, the human oral cavity is a complex habitat where microbes display complex interactions and show striking spatial heterogeneity (Montelongo-Jauregui et al. 2019; Banerjee et al. 2024; Sulyanto et al. 2024). Yeasts have evolved specific strategies to colonize the host—and the host has developed responses to limit fungal invasion. Pathogens produce toxins that damage mammalian cells, such as the peptide candidalysin from C. albicans (Moyes et al. 2016; Verma et al. 2017), and humans produce antimicrobial compounds, such as the salivary histatin proteins (Puri and Edgerton 2014) that interfere with fungal colonization by disrupting membrane integrity. Recently, candidalysin has been shown to have a major role in establishing C. albicans colonization amongst the complex bacterial microbiota in the mammalian gut (Liang et al. 2024). Further studies of the regulation of fungal responses in these systems have the potential to reveal how interactions between yeasts, other microorganisms, and the host impacts coevolution and virulence. For many of the above examples, a common regulatory element of filamentous growth and fungal behavioral responses is the MAPK pathway (Fig. 2, MAPK). The MAPK pathway is a signal transduction pathway that responds to diverse stimuli to regulate filamentation responses in many contexts.
Filamentous growth has been studied in many different species, and this review will focus on a few model systems. One is the Baker's yeast S. cerevisiae, which has proved a useful genetically amenable model to study the regulatory pathways that control the response. Yeast gives us an ever-expanding genetic, molecular, and functional genomics toolkit that is readily available (Mortimer and Johnston 1986; Roman 1986; Botstein et al. 1997; Borneman et al. 2006; Replansky et al. 2008; Dowell et al. 2010; Botstein and Fink 2011; Ryan et al. 2012). Even though S. cerevisiae has long been considered mostly a tool to study cell biology, yeast research is now capitalizing on the growing knowledge of the organism's ecology and evolution (Hittinger 2013; Goddard and Greig 2015; Liti 2015; Duan et al. 2018; Peter et al. 2018; Bai et al. 2022; De Chiara et al. 2022; O'Donnell et al. 2023; Peris et al. 2023) to better contextualize differentiation events and the signaling pathways that control them. Furthermore, although S. cerevisiae is a free-living organism not widely considered a pathogen, strains have been cultured from immunocompromised individuals, which has allowed exploration of this facet of yeast biology as well (Wheeler et al. 2003; Granek et al. 2013; Raghavan et al. 2019; Ekdahl et al. 2023).
Another important model is C. albicans (Nobile and Mitchell 2006; Noble et al. 2010; Nobile and Johnson 2015; Basso et al. 2019; Chen et al. 2020), a commensal yeast found in most healthy individuals that can become deadly when the immune system is compromised. Several plant pathogens have also been studied, which allows comparison of the similarities and differences between plant and animal colonization (Lanver et al. 2010, 2014; Liu et al. 2011; Perez-Nadales and Di Pietro 2015; Qin et al. 2021; Wang et al. 2021). As a result of evolution, most fungal species share conserved regulatory pathways that govern the filamentous growth response making model systems highly relevant. Interestingly, although the components of these pathways are conserved, what the pathways sense varies based on the specific cues found in species-specific environments. In addition, depending on the species, the same regulatory pathways can induce quite different responses. By exploring strategies that related organisms use to solve similar biological problems, new insights have emerged (Johnson 2017; Wolfe and Butler 2022). For example, S. cerevisiae has undergone a whole-genome duplication (Wolfe and Shields 1997) in which diversification of gene function has occurred through subfunctionalization for some regulatory processes (Hickman et al. 2011), whereas C. albicans maintains a single regulator for some of these similar processes. Because of the strong evolutionary ties between species, lessons learned by studying filamentous growth in 1 species benefits the overall knowledge of the principles underlying developmental cell-type specification in eukaryotes.
Regulation of filamentous growth by a conserved MAPK pathway
Many fungal species regulate cell differentiation through homologous signaling pathways (Ryan et al. 2012; Kiss et al. 2019). One of the main pathways, which regulates filamentous growth, is called the filamentous growth or filamentation MAPK (fMAPK) pathway (Cek pathway in C. albicans; Liu et al. 1993; Roberts and Fink 1994; Kumar 2021). MAPK pathways regulate filamentous growth in saprotrophs (Madhani et al. 1997; Pandey et al. 2004; Xu et al. 2016), human pathogens (Chen et al. 2020), plant pathogens (Xu 2000; Cho et al. 2007), insect pathogens (Jin et al. 2014; Zhao et al. 2023), and lichen-forming fungi (Wang, Li et al. 2023). MAPK pathways also govern cell differentiation and stress responses in mammals and control critical processes including cell proliferation, cell migration, cell survival, and apoptosis (Seger and Krebs 1995; Roberts et al. 2000; Zhang and Liu 2002; Krens et al. 2006; Shaul and Seger 2007; Taj et al. 2010). As we will see below, key regulatory features of the fMAPK pathway apply throughout fungal species (including pathogens) and to eukaryotes in general. Fungal-specific regulatory elements in turn lend themselves to strategies aimed at curbing virulence in fungal pathogens.
A kinase module controlled by the Rho GTPase Cdc42 regulates multiple MAPK pathways: the puzzle of specificity
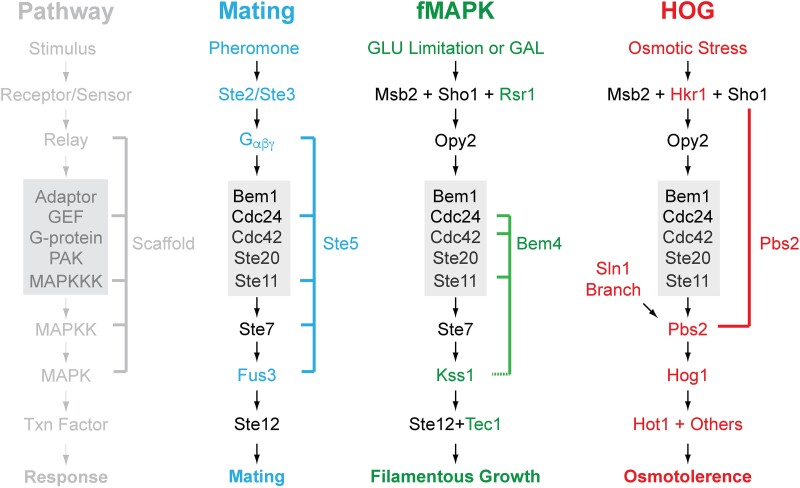
A defining feature of MAPK pathways is they are composed of multiple kinases that phosphorylate and activate each other in a tandem series (Seger and Krebs 1995). The MAP kinase (MAPK, Kss1 for the fMAPK pathway) is phosphorylated and activated by the MAP kinase kinase (MAPKK, Ste7p), which in turn is phosphorylated and activated by a MAP kinase kinase kinase (MAPKKK, Ste11p) (Fig. 3, Cook et al. 1996, 1997; Bardwell, Cook, Voora, et al. 1998; Lee and Elion 1999; Shock et al. 2009). Ste11p is itself activated by phosphorylation by a member of the p21-activated kinases (PAKs), called Ste20p. Ste20p is activated by binding to the ubiquitous Rho-type GTPase Cdc42p. Cdc42p is a member of the monomeric small GTPase superfamily of proteins (Johnson and Pringle 1990; Johnson 1999; Pruyne and Bretscher 2000) and is highly conserved across eukaryotes sharing 81% protein sequence identity between yeast and humans. As for many members of the GTPase family, in its active or guanosine triphosphate (GTP)-bound conformation, Cdc42p binds effector proteins including Ste20p (Peter et al. 1996; Lee and Elion 1999; Tatebayashi et al. 2006) and other proteins involved in polarity organization (Gic1p, Gic2p, and another PAK, Cla4p). Specifically, Cdc42p is activated by the guanine-nucleotide exchange factor (GEF, Cdc24p), which promotes the exchange of GTP for guanosine diphosphate. GTP-bound Cdc42p binds to and relieves autoinhibition of Ste20p's Cdc42p-binding or CRIB domain, which stands for Cdc42/Rac interactive binding motif, allowing the PAK to phosphorylate and activate Ste11p (van Drogen et al. 2000). Additionally, 14-3-3 proteins (Bmh1p and Bmh2p; Roberts et al. 1997) and an adaptor protein (Bem1p; Basu et al. 2020) facilitate activation of Ste20p. Therefore, a signaling cascade composed of a Cdc42p module and 4 kinases in a tandem series represents the core of the fMAPK pathway (Fig. 3, green).
Fig. 3.
Three yeast MAPK pathways share components. The MAPK pathway that regulates filamentous growth (center) shares components with the mating and/or HOG pathways. Box, proteins utilized by all 3 pathways. Scaffolds are shown to the right of each pathway, and a key is shown at left. Not all proteins are shown.
Signaling pathways are not separate entities from each other. Most pathways share common factors, even in the same cell type. In S. cerevisiae, the fMAPK pathway shares components with at least 2 other MAPK pathways: The mating and high osmolarity glycerol (HOG) pathways (Fig. 3; Roberts and Fink 1994; Posas and Saito 1997). The mating (or pheromone response, blue) pathway allows haploid cells to recognize each other by secreted peptide pheromones that leads to cell fusion and the formation of diploids (Herskowitz 1995). The HOG pathway (red) regulates tolerance to osmotic and other stresses (Saito and Tatebayashi 2004; de Nadal and Posas 2022) and modulates filamentous growth (Shock et al. 2009; Yang et al. 2009; Adhikari and Cullen 2014) through 2 redundant branches (Ste11p and Sln1p). Although each pathway senses a different stimulus and induces a distinct set of targets genes, they all utilize the same core module (Fig. 3, gray box). Moreover, any 2 pathways also utilize some of the same components. For example, the mating and fMAPK pathways both require the same MAPKK, Ste7p. How the same core module induces different responses to different stimuli is a longstanding question that extends to signaling modules in general. Studies of MAPK pathways in yeast have revealed fundamental mechanisms that cells have developed to keep their signals straight.
Below, we point out features of the fMAPK pathway in reference to the other pathways. We compare the receptors/sensors between the pathways that contribute to specificity, including the mating G-protein-coupled receptors (GPCRs) and the mucin sensors of the fMAPK and HOG pathways. We discuss how scaffolds maintain insulation between pathways, and the cross talk that occurs when specificity is lost. Finally, we show how different target genes are induced by shared transcription factors. In this way, as we learn about the fMAPK pathway, we will also learn how specificity is maintained between MAPK pathways and address open questions in this area.
A mucin sensor at the head of the fMAPK pathway
Signaling pathways can be activated by many different types of receptors. The sensor for the fMAPK pathway is a member of the mucin family of proteins (Fig. 4, Msb2p; Cullen et al. 2004). Mucins differ from the more common GPCRs, whose ligands are well defined (Lemaire et al. 2004; Xue et al. 2008), structures resolved (Velazhahan et al. 2021, 2022), and connections to effector proteins firmly established (Lee and Dohlman 2008). Mucins by comparison are broadly similar to a diverse group of glycoprotein sensors, such as Notch (Zhou et al. 2022), cadherins (Fulford and McNeill 2020), and dystroglycan (Endo 2015). As a group, these proteins are interesting because of their mechanosensory properties, the varied effects of glycosylation and posttranslational processing on signaling, and the challenges associated with studying large glycoproteins by biochemical and structural methodologies (Pei and Grishin 2017).
Fig. 4.
The fMAPK pathway. Msb2p, Sho1p, and Opy2p function at the plasma membrane. Msb2p is induced by glucose (GLU) limitation and growth in galactose (GAL), which results in under-glycosylation of the protein and cleavage of an inhibitory extracellular domain by the yapsin protease, Yps1p. Through a partially defined mechanism, Msb2p, Sho1p, and Opy2p regulate cytosolic proteins, including adaptors, Bem1p and Bem4p, the Cdc42p GTPase module, and the MAPK cascade.
Mucins are conserved in fungi, invertebrates, and vertebrates and are highly glycosylated proteins that are secreted to provide a layer of protection, and a layer of lubrication for epithelium tissue in animals. Because of their presence in the gut and oral epithelium, mucins play important roles in the digestive and immune systems (Johansson and Hansson 2016). Mucins that regulate signaling pathways have a single-pass transmembrane helix and cytosolic domain and are called signaling mucins (Corfield 2015). In humans, the signaling mucin MUC1 regulates the RAS-MEK-ERK pathway, which controls key cellular responses including cell proliferation (Theodoropoulos and Carraway 2007) and when misregulated is a main cause of cancer. MUC1 is itself up-regulated in many cancers and is therefore a target for immunotherapies (Singh and Bandyopadhyay 2007; Sousa et al. 2016; Supruniuk and Radziejewska 2021; Qing et al. 2022). One defining feature of mucins is a tandem repeat region rich in serine, threonine, and proline (S/T/P) residues (Desseyn et al. 1998; Cao et al. 2012). Because S/T residues are targets of O-linked glycosylation (in yeast typically poly-mannosylation), mucins can be abundantly glycosylated; so much so that the carbohydrate content outweighs the protein content of the protein. Variation in the number of repeats can occur and have phenotypic consequences. For example, increases in the repeat number of MUC1 is associated with kidney disease (Dvela-Levitt et al. 2019). The roles mucin signalers play in regulatory pathways and their prevalence in disease make them important molecules of interest.
Msb2p shares many defining features of signaling mucins (Cullen et al. 2004). It is a single-pass transmembrane protein that is highly glycosylated, resides at the cell surface, and is required for activity of the fMAPK pathway. Msb2p also contains an S/T/P-rich region of tandem repeats of 17 amino acids in length (Fig. 4, repeats). The S/T/P region is part of a larger portion of the extracellular domain (from residues 100 to 950 amino acids) that inhibits the fMAPK pathway (Vadaie et al. 2008). Moreover, this large portion of the extracellular domain is shed from cells, which is a typical feature of mucins and other signaling glycoproteins. Shedding of the inhibitory domain of Msb2p requires members of a family of glycosylphosphatidylinositol anchored aspartyl proteases called yapsins (Krysan et al. 2005). Processing and release of the inhibitory domain of Msb2p leads to cleavage-dependent activation of the fMAPK pathway. Cleavage-dependent activation is common to the activation of other glycoprotein sensors, including Notch (van Tetering and Vooijs 2011), adhesion GPCRs (Araç et al. 2012), and even several bacterial proteases (Jeong et al. 2020), including one that when shed induces virulence (Dong et al. 2004).
What is Msb2p sensing? An established inducer of filamentous growth is nutrient limitation. The limitation of a preferred carbon source (e.g. glucose) and/or nitrogen induces filamentous growth in S. cerevisiae (Gimeno et al. 1992; Roberts and Fink 1994; Cullen and Sprague 2000), human pathogens (Brown et al. 1999; Csank and Haynes 2000; Biswas and Morschhäuser 2005; Morschhäuser 2011; Ng et al. 2016), and plant pathogens (Banuett and Herskowitz 1994; Van den Ackerveken et al. 1994). Moreover, the expression of genes related to filamentous growth is linked in some cases to glucose repression, where adequate glucose inhibits the expression of the metabolism of other carbon sources (Carlson 1999).
Msb2p may respond to glucose limitation in a noncanonical way. Upon glucose limitation, the glycosylation of Msb2p becomes reduced, presumably because the substrates for its glycosylation are glucose and mannose moieties (Fig. 4, Adhikari, Vadaie et al. 2015). Msb2p is glycosylated in the endoplasmic reticulum (ER), and problems with protein folding/glycosylation in the ER can trigger the unfolded protein response (UPR, Fig. 4), a regulatory pathway in the ER that is responsible for dealing with protein folding stress in eukaryotes including fungi and mammals (Walter and Ron 2011). The UPR induces the levels of yapsin proteases resulting in elevated cleavage of Msb2p (Adhikari, Vadaie et al. 2015). In this way, Msb2p functions as an unconventional glucose sensor that is activated by cleavage when carbon levels become limited (Fig. 4). In support of this idea, problems in protein glycosylation, especially O-linked glycosylation by the mannoyltransferase Pmt4p (Yang et al. 2009), also result in elevated Msb2p-dependent fMAPK pathway activity (Cullen et al. 2004).
In addition to nutrient limitation, recent studies suggest that the fMAPK pathway responds to other stimuli connected to the cell's ability to colonize fruit-based ecological niches (Vandermeulen and Cullen 2023). One stimulus is galactose, a nonpreferred carbon source found in some plant-rich environments (Botha 2011; Gunina and Kuzyakov 2015) and certain fruits (Folsom et al. 1974; Asgar et al. 2003; Qi and Tester 2019). Galactose stimulates the activity of the fMAPK pathway (Karunanithi and Cullen 2012; Adhikari and Cullen 2014) beyond glucose limitation and requires galactose metabolism (Vandermeulen and Cullen 2023). Another stimulus is pectin (Vandermeulen and Cullen 2023), a major component of plant cell walls (JARVIS 1984; Willats et al. 2001; Voragen et al. 2009; Zdunek et al. 2021). Pectin may be sensed by Msb2p at the cell surface directly, as some mammalian mucins expressed in gut tissue interact with pectin (Thirawong et al. 2008; Sriamornsak et al. 2010). Alternatively, pectin molecules are large and may bring Msb2p and other sensors (Sho1p, discussed below) into a complex. Similarly, pectin and galactose induce the secretion of pectolytic enzymes in the absence of glucose in other fungi, such as N. crassa (Polizeli et al. 1991; Crotti et al. 1996; Crotti, Terenzi, Jorge, de Lourdes et al. 1998; Crotti, Terenzi, Jorge, Polizeli 1998).
The third molecule that induces the fMAPK pathway is the metabolic byproduct ethanol (Vandermeulen and Cullen 2023). Ethanol and other alcohols in S. cerevisiae (Dickinson 1996; Lorenz et al. 2000; Chen and Fink 2006; Wuster and Babu 2009) and C. albicans (Chen et al. 2004; Nickerson et al. 2006; Albuquerque and Casadevall 2012) are soluble metabolites and indicators of cell density, which in microbes is known as quorum sensing (Ng and Bassler 2009; Albuquerque and Casadevall 2012; Whiteley et al. 2017; Zhao et al. 2020; Tian et al. 2021). In S. cerevisiae, ethanol can act as an inhibitor of microbial competitors (Piskur et al. 2006; Dashko et al. 2014) and an attractant for insect vectors (Goddard et al. 2010; Stefanini et al. 2012; Palanca et al. 2013; Buser et al. 2014; Liti 2015).
When combined with galactose, ethanol induces the fMAPK pathway to high (i.e. near maximal) levels, suggesting quorum and nutrient-sensing mechanisms can be additive to pathway activity (Vandermeulen and Cullen 2023). Ethanol may be detected through its ability to disrupt the structure of cell membranes (which is where the sensors/receptors are located) or its ability to denature proteins and activate heat shock proteins (HSPs; Toth et al. 2014; Cohen 2018). Alternatively, the ethanol signal may come through a separate signaling pathway that regulates fMAPK pathway activity (RAS-PKA, discussed below) as Ras2p is more critical to fMAPK pathway activity than Msb2p in response to ethanol (Vandermeulen and Cullen 2023). The fMAPK pathway may also sense cell density through cell-to-cell contact, as Msb2p has been proposed to function as a pressure sensor in some contexts (Delarue et al. 2017) although this possibility has not been tested in the context of fMAPK pathway signaling.
Msb2p interacts with the tetraspan protein Sho1 and cysteine-rich protein Opy2
Msb2p functions with 2 other proteins at the plasma membrane (Fig. 4). One is Sho1p (Maeda et al. 1995; O'Rourke and Herskowitz 1998; Cullen et al. 2004), a tetraspan protein important for filamentous growth signaling in fungi (Lambou et al. 2008). Sho1p has 4 transmembrane helices and a cytosolic region containing an SH3 domain. The cytosolic domain of Sho1p functions as an adaptor that scaffolds together multiple proteins that regulate the MAPK pathway, including the PAK Ste20p and MAPKKK Ste11p (Tatebayashi et al. 2006). By comparison, the SH3 domain binds to the MAPKK of the HOG pathway, Pbs2p (Maeda et al. 1995). Loss of Pbs2p results in crosstalk, where salt induces the activation of the fMAPK and mating pathways (O'Rourke and Herskowitz 1998). Msb2p interacts with Sho1p and may activate the protein. The activatable part of Sho1p is connected to an external loop between the second and third transmembrane helices, which when mutated (typically P120L) causes hyperactivity (Tatebayashi et al. 2007; Vadaie et al. 2008). Msb2p and Sho1p also interact with Opy2p, a single-pass transmembrane protein that contains an extracellular region with 8 cysteines that form disulfide bonds in a predicted knot structure (Wu et al. 1999, 2006; Yamamoto et al. 2010). Opy2p by its Ras-associated domain binds to an adaptor protein (Ste50p) that recruits and activates the MAPKKK Ste11p at the plasma membrane (Fig. 4; Wu et al. 1999; Ramezani-Rad 2003). Msb2p, Sho1p, and Opy2p form a complex that assembles in predictable hetero-oligomeric ratios (Tatebayashi et al. 2015). Puzzlingly, each member of the complex is localized and turned over independently of the other members. Msb2p is turned over rapidly by the ubiquitination of lysines in the cytosolic region of the protein (Adhikari, Vadaie et al. 2015), whereas Sho1p and Opy2p are stable proteins turned over at lower rates (Adhikari, Caccamise et al. 2015). It will be interesting to understand how these proteins cooperate in signaling with their unique turnover and localization properties.
Conserved and unique features of mucins in different fungal species
Msb2p, Sho1p, and Opy2p have homologs in other species, although the sensing mechanisms of the complex may be tailored for specific ecological niches. In C. albicans, Msb2p and Sho1p homologs induce filamentous growth by activation of the homologous Cek1p MAPK pathway (Csank et al. 1998; Román et al. 2009). During activation, Msb2p is cleaved by the aspartyl-type protease Sap8p [(Puri et al. 2012) similar to S. cerevisiaeYps1p] and shed into the extracellular matrix. The aspartyl-type proteases are members of large families: there are 5 cell wall anchored yapsins in S. cerevisiae (Krysan et al. 2005), 11 yapsins in C. glabrata (Kaur et al. 2007), 10 secreted aspartyl proteases (8 secreted, and 2 cell-wall anchored) in C. albicans (Monod et al. 1994; Albrecht et al. 2006), and 7 in Candida auris (Kim et al. 2023). The amplification of gene families can produce functional diversity. For example, by regulating the expression, activity, and specificity of these proteases, cells can tailor protease activity and hence penetration into specific environments.
In the extracellular space, the large glycol-domain of Msb2p functions to protect cells against antimicrobial peptides (Szafranski-Schneider et al. 2012). Cleavage of the C. albicansMsb2p occurs in nutrient-limiting environments and elevated temperatures corresponding to host conditions (37°C). Moreover, in this species, Msb2p may specifically play a role in temperature sensing (Saraswat et al. 2016). Interestingly, in another human pathogen, Aspergillus fumigatus, the Msb2p homolog induces a related MAPK pathway controlling cell integrity (Gurgel et al. 2019).
Msb2p and Sho1p homologs also regulate MAPK pathways involved in host recognition, appressorium formation, filamentous growth, and invasion in the plant pathogens. These include U. maydis (Lanver et al. 2010, 2014), Fusarium oxysporum (Perez-Nadales and Di Pietro 2015), Magnaporthe oryzae (Liu et al. 2011), Colletotrichum gloeosporioides (Wang et al. 2021), and Aspergillus flavus (Qin et al. 2021). Host recognition by Msb2p and Sho1p occurs by sensing hydrophobicity and by chemical signals associated with the cuticle of the plant surface. Msb2p regulates a related MAPK pathway (i.e. HOG) in Arthrobotrys oligospora that induces a different type of differentiation program for trap formation by nematode predators (Kuo et al. 2020). Therefore, Msb2p-type mucins have functional homologs in other fungal species, although their sensing mechanisms and effector MAPK pathways differ, perhaps as a result of selective pressure due to diverse ecological niches.
Scaffolds promote specificity between MAPK pathways that share components
How do proteins at the plasma membrane (Msb2p, Sho1p, and Opy2p) connect to and regulate the Cdc42p-dependent MAPK cascade? One clue toward answering this question comes from protein scaffolds. In signaling pathways, scaffolds bind to general and pathway-specific proteins to induce a pathway-specific response (DiRusso et al. 2022). In the mating pathway, the scaffold Ste5p links the GPCR, namely the Gβ protein Ste4p to Ste20p, Ste11p, Ste7p, and the MAPK Fus3p (Fig. 3; Zalatan et al. 2012). In this way, signals emanating at the receptor are selectively and efficiently directed toward a specific MAPK pathway. Furthermore, Ste5p binding to Fus3p catalytically unlocks the protein, enabling it to be phosphorylated and activated by its MAPKK Ste7p (Good et al. 2009; Coyle et al. 2013). As mentioned above, Pbs2p is the MAPKK and scaffold for the HOG pathway that binds to the tetraspan protein Sho1p. Again, this has the effect of linking a protein that operates at the plasma membrane (Sho1p) to a pathway-specific MAPK (Hog1p, by way of Pbs2p, Fig. 3).
For the fMAPK pathway, a different type of scaffold functions specifically in the fMAPK but not mating or HOG pathways, called Bem4p (Figs. 3 and 4; Pitoniak et al. 2015). Bem4p is a Cdc42p-binding protein with homology to SMG GDS-type regulators of GTPases (Brandt et al. 2021). Bem4p is required for activation of the fMAPK pathway but not the mating or HOG pathways. Moreover, Bem4p binding to Cdc42p protects the protein from turnover (González et al. 2023). Specifically, the ubiquitin ligase Rsp5p and HSPs Ssa1p and Ssa2p mediate turnover of GTP-bound Cdc42p, and this effect is countered by Bem4p. Although many GTPases are known to be regulated by turnover, here the turnover of the active or GTP-bound conformation of Cdc42p leads to attenuation of the fMAPK pathway. The current model is that Bem4p in some manner protects Cdc42p to sustain fMAPK pathway activity. Cdc42p protection does not impact the mating pathway because that pathway is gated: Fus3p cannot be phosphorylated by Ste7p without the activation of Ste5p (Good et al. 2009; Coyle et al. 2013). Therefore, Bem4p functions at a distinct point in the pathway to amplify signals generated by activated Cdc42p.
Bem4p not only interacts with Cdc42p but also with its major activator, the GEF Cdc24p. Two other proteins also bind Cdc24p and promote Cdc42p function in the fMAPK pathway (Fig. 4, Rsr1 and Bem1). One of these proteins is the Ras-type GTPase Rsr1p. Interestingly, Rsr1p is itself activated by proteins that control the direction in which cells grow or bud, called bud-site-selection proteins, a mechanism of polarity establishment (Park et al. 1997). In haploid cells, cells grow back toward the mother in an axial pattern when nutrients are plentiful and away from the mother to make pseudohyphae in a distal pattern when nutrients become limiting (Chant and Pringle 1995). Bud-site-selection proteins located at either pole recruit Rsr1p, which directs GEF-dependent activation of Cdc42p at the growth site. Interestingly, Rsr1p contributes to fMAPK pathway activation (Basu et al. 2016), presumably as a way of integrating positional information into the regulation of MAPK pathway signaling.
In addition to Bem4p and Rsr1p, the third protein that binds Cdc24p and regulates Cdc42p is the main polarity scaffold Bem1p. Bem1p regulates Cdc42p in multiple contexts—it is required for symmetry breaking during polarity establishment (Woods and Lew 2019), and it regulates the mating, fMAPK, and HOG pathways (Leeuw et al. 1995; Elion 2000; Tanaka et al. 2014). Nevertheless, a direct comparison shows that different domains of Bem1p play different roles in different pathways (Fig. 4, mark on Bem1; Basu et al. 2020). Similarly, specific residues of the adaptor Ste50p mediate pathway-specific functions (Sharmeen et al. 2019). Therefore, even proteins that are shared between pathways can have pathway-specific determinants. It will be interesting to learn how Msb2p and Sho1p connect to cytosolic signaling proteins (Fig. 4, top question mark), and how Bem4p, Rsr1p, and Bem1p coordinate GEF activation in the specific context of filamentous growth.
Much remains to be learned by additional studies on the fMAPK pathway. One example comes from the fact that 2 mucins regulate MAPK pathways in yeast. One is Msb2p and the other is Hkr1p (Fig. 3). Msb2p regulates the fMAPK pathway, while Msb2p and Hkr1p regulate the HOG pathway (Tanaka et al. 2014). Disentangling specificity between these mucins is an area of intense interest. A second example comes from evidence for a Ste20p-independent branch of the MAPK pathway (Fig. 4, curved arrow, bottom question mark). In the HOG pathway, Ste20p can be bypassed by the addition of salt (Raitt et al. 2000). Similarly, conditions that hyperactivate the fMAPK pathway (galactose with ethanol; Vandermeulen and Cullen 2023) or some alleles that hyperactivate the fMAPK pathway (González et al. 2023), lead to a signal that requires Ste11p but not Ste20p. Future work can clarify this mechanism to understand how it works and why it evolved.
Regulation of transcription factors and gene expression by the fMAPK pathway
In yeast, the MAPK Kss1p functions mainly in the fMAPK pathway, although it can modulate mating in some circumstances (Ma et al. 1995; Cherkasova et al. 1999; Farley et al. 1999). Unlike other MAPKs, Kss1p functions in an inhibitory capacity until it is phosphorylated [Fig. 4, block arrow at Kss1, (Cook et al. 1997; Gustin et al. 1998)]. Relief of the inhibitory role of Kss1p by phosphorylation by Ste7p leads to the activation of several transcription factors. One of these is Ste12p, a homeodomain protein that regulates the fMAPK and mating pathways. Here, the puzzle of specificity surfaces again, this time at the transcription factor level. How is Ste12p directed to a specific gene set? The answer to this question comes from the discovery of a TEA/ATTS family transcription factor, named for the members of the family, Tec1p, AbaA, TEF1/TEAD1, and Scalloped. Tec1p can hetero-dimerize with Ste12p (Fig. 4; Laloux et al. 1990; Madhani and Fink 1997; Borneman et al. 2006; Chou et al. 2006). Ste12p and Tec1p each recognize different DNA-specific binding motifs and are conserved across fungi (Schweizer et al. 2000; Vallim et al. 2000; Wong Sak Hoi and Dumas 2010; León-Ramírez et al. 2022). During mating, Ste12p binds pheromone response elements as a homodimer. Tec1p does not interfere with mating in this setting because Fus3p phosphorylates Tec1p, which targets the protein for ubiquitination and destruction by the 26S proteosome (Bao et al. 2004; Chou et al. 2004). In this way, Ste12p is directed solely to the mating pathway upon exposure to pheromone. In filamentous growth inducing environments, Ste12p forms a Ste12p–Tec1p heterodimer, leading to these transcription factors binding to distinct sites containing TCS elements, which contain adjacent pheromone (Ste12p) and filamentation (Tec1p) binding sites (Zeitlinger et al. 2003; Zhou et al. 2020), to regulate filamentation related target genes (Madhani et al. 1999; Roberts et al. 2000; Heise et al. 2010; Adhikari and Cullen 2014; van der Felden et al. 2014; Chow, Starr, et al. 2019; Zhou et al. 2020).
The transcription factors Ste12p and Tec1p are regulated by transcriptional modulators, Msa1p and Msa2p (van der Felden et al. 2014), and are inhibited by the transcriptional repressors, Dig1p and Dig2p (Cook et al. 1996, 1997; Bardwell, Cook, Zhu-Shimoni et al. 1998). Ste12p and Tec1p also have independent regulatory mechanisms (Kohler et al. 2002; Heise et al. 2010; van der Felden et al. 2014) some of which depend on the environment (Vandermeulen and Cullen 2023), which may be due to their regulation by unique mechanisms. For example, Ste12p can also associate with Mcm1p, an essential transcription factor (Zeitlinger et al. 2003) and is phosphorylated by Cdk8p, a cyclin-dependent kinase (CDK; Nelson et al. 2003). Tec1p can be independently modulated by SUMOylation (Wang et al. 2009). Preferences in binding sites may also influence transcription factor specificity (Dorrity et al. 2018).
Transcriptional targets of the fMAPK pathway control aspects of the filamentation response
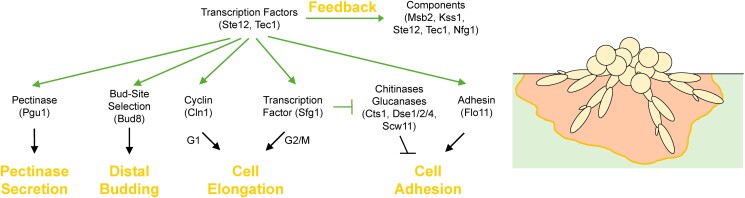
The main function of the transcription factors for the fMAPK pathway is to control the expression of genes whose products mount the filamentous growth response. The specific gene set based on multiple studies contains more than fifty genes that regulate cell differentiation, cell adhesion, and enzyme secretion (Madhani et al. 1999; Zeitlinger et al. 2003; Adhikari and Cullen 2014; Chow, Starr et al. 2019; Vandermeulen and Cullen 2020). In addition, the fMAPK pathway up-regulates several of its own components (e.g. MSB2, KSS1, STE12, and TEC1) to induce positive feedback, presumably to amplify the initial signal. The fMAPK pathway also up-regulates negative regulators of the pathway (i.e. NFG1/YLRO42c, RGS2, RPI1, and TIP1) to modulate pathway activity and filamentous growth (Vandermeulen and Cullen 2020). Collectively, these feedback-control mechanisms allow the cell to precisely set the level of its filamentous growth response.
Cell differentiation by polarity and cell cycle target genes
Perhaps the most striking feature of filamentous growth is the ability of cells to elongate and begin moving away from parent cells in search of nutrient-rich environments. The apparent complexity of this growth pattern is caused by induction of a few target genes. The change in polarity or growth direction is particularly clear in haploid cells, which switch from growth toward the mother cell in an axial pattern to growth away from the mother in a distal pattern. Transcriptional induction of the distal pole marker, BUD8 (Fig. 5; Chant and Pringle 1995; Zahner et al. 1996; Taheri et al. 2000; Ni and Snyder 2001; Cullen and Sprague 2002) promotes this outward growth. In addition, reduced levels of an axial-specific protein, Axl1p (Cullen and Sprague 2002) solidifies the distal pattern.
Fig. 5.
Key transcriptional targets of the fMAPK pathway in yeast. Upon activation of the fMAPK pathway, target genes are induced by transcription factors Ste12 and Tec1. Genes encoding pathway components are induced to generate positive (for Msb2, Kss1, Ste12, and Tec1) or negative feedback (Nfg1). Genes encoding proteins that degrade the plant cell wall (Pgu1), alter polarity toward distal growth (Bud8), delay the cell cycle (Cln1 and Sfg1), and promote cell adhesion, by inhibition of cell separation by Sfg1 (Cts1, Dse1/2/4, and Scw11) and the major adhesin flocculin (Flo11) are also induced. At right, diagram of the resulting phenotypic changes that occur during filamentous growth.
The change in the length of the cell is caused by a delay in cell cycle progression. MAPK pathways commonly regulate progression through the cell cycle by affecting cyclins that bind to and activate CDKs. In this case, the fMAPK pathway induces expression of CLN1, which encodes a G1 cyclin (Hadwiger et al. 1989), whose induction leads to a delay in progression through the cell cycle resulting in an elongated cell morphology (Fig. 5; Madhani et al. 1999). Cell cycle extension is also mediated by a second mechanism. The fMAPK pathway induces expression of SFG1, which encodes a transcription factor that contributes to cell elongation (Vandermeulen and Cullen 2020) presumably by causing a delay in the cell cycle at the G2/M phase (White et al. 2009). In other fungi, cell polarization depends on steep phosphatidyl inositol gradients (Vernay et al. 2012), Cdc42 regulation by multiple control proteins (Bassilana et al. 2005; Brand et al. 2014), and the RAM or regulation of Ace2 and morphogenesis pathway (Bharucha et al. 2011; Chadwick et al. 2022).
Cell adhesion
MAPK pathways also up-regulate the expression of genes that control cell adhesion, which is a major aspect of the filamentous growth response. Many fungi have numerous genes that encode cell adhesion molecules that serve different purposes depending on context, which have been extensively reviewed (Hoyer 2001; Bruckner and Mosch 2012; Lipke 2018; Essen et al. 2020). In S. cerevisiae, mechanisms underlying the regulation of cell adhesion have been worked out, which includes the fMAPK pathway-dependent up-regulation of the expression of FLO11 and SFG1 (Fig. 5), 2 genes that encode proteins that regulate separate cell adhesion mechanisms.
A large amount of research has been done on FLO11, which encodes a cell surface mucin-type protein and major cell adhesin involved in filamentous growth in S. cerevisiae (Fig. 5, Lambrechts et al. 1996; Lo and Dranginis 1996, 1998; Guo et al. 2000). For example, extensive structure and function analysis has been performed on Flo11p (Verstrepen et al. 2005; Christiaens et al. 2012; Meem and Cullen 2012; Kraushaar et al. 2015), revealing hydrophobic and homotypic binding sites. Additionally, studies on the regulation of FLO11 gene expression reveal extensive regulation at its unusually large promoter (Rupp et al. 1999; Palecek et al. 2000; Borneman et al. 2006; Barrales et al. 2008; Bumgarner et al. 2012), and evolutionary studies have shown strain and functional variation (Fidalgo et al. 2006; Barua et al. 2016; Brückner et al. 2020). Moreover, Flo11p has been shown to be shed into the extracellular space to attenuate adhesion and contribute to formation of a mucus layer (Karunanithi et al. 2010). Flo11p is also critical for biofilm/mat formation (Reynolds 2018), and the protein can be expressed in a variegated manor among individuals in a population (Halme et al. 2004). Other adhesin genes exist in S. cerevisiae (e.g. FLO1, FLO10, FLO9, FLO5; Guo et al. 2000; Veelders et al. 2010; Bruckner and Mosch 2012; Christiaens et al. 2012); however, they are thought to be silenced at the gene level in some strain backgrounds (Halme et al. 2004) and only play a minor role compared to FLO11. These adhesins are involved in promoting the formation of adhesion clusters in liquid environments (flocs when suspended, flors when on the surface; Lindquist 1952; Mill 1964; Bruckner and Mosch 2012).
Beyond Flo11p, the inhibition of cell-wall-degrading enzymes that regulate cell separation also plays a role in regulating cell adhesion (Vandermeulen and Cullen 2020). A family of cell-wall-degrading enzymes (e.g. glucanases and endo-chitinases; King and Butler 1998; Doolin et al. 2001; Baladrón et al. 2002; Roncero and Sanchez 2010) promote cell separation between mother and daughter cells to reduce adhesion. The fMAPK pathway up-regulates expression of SFG1 (Vandermeulen and Cullen 2020), which encodes a transcription factor that inhibits the expression of cell-wall-degrading enzymes (Fujita et al. 2005; White et al. 2009; Fig. 5). Ace2p is another transcription factor that also inhibits expression of these enzymes (King and Butler 1998; Doolin et al. 2001).
For human pathogens, such as C. albicans, adhesion molecules can play even more critical roles in modulating the strength and specificity of cellular interactions. One key example is the recognition of host cells (Hoyer 2001; Martin et al. 2021). A second is forming fungicidal resistant multicellular biofilms (Chandra et al. 2005; Nobile and Mitchell 2006; Flemming and Wingender 2010; Desai et al. 2014; Silva-Dias et al. 2015) that adhere to other microbes or abiotic surfaces, including plastics found in medical equipment (Kennedy et al. 1989; Pereira et al. 2021; Ponde et al. 2021). Therefore, it is not surprising that there are more cell adhesion molecules in pathogens, and that they are extensively regulated. C. albicans has more than a dozen adhesion molecules in the ALS gene family (Hoyer 2001; Lipke et al. 2012), which have structural homology to the FLO proteins, as well as cell-wall-degrading enzymes that include an ACE2 homolog (Kelly et al. 2004). Many of the adhesion molecules in C. albicans create strong adhesive forces to form biofilms through amyloid-type aggregation (Garcia et al. 2011; Lipke et al. 2018). Different combinations of adhesins show different properties, suggestive of an adhesion code, which could promote redundancy or phenotypic plasticity depending on the environment (Rosiana et al. 2021). Future research may allow analysis of the function of each adhesin, which at present is hampered by the presumed redundancy in these protein families.
For plant-associated fungi (Tucker and Talbot 2001; Taylor et al. 2022) or insect pathogens (Shang et al. 2024), adhesion molecules play critical roles on the host surface. This includes specific cell adhesion molecules found on the adhesive structure called the appressorium (Mendgen et al. 1996; Tucker and Talbot 2001; Ryder and Talbot 2015; Chethana et al. 2021), a specialized structure that generates turgor pressure for breaking through the cuticle of the plant surface for plant pathogens (Kahmann and Kämper 2004), the insect cuticle for insect pathogens (Ortiz-Urquiza and Keyhani 2013), or the plant root for symbiotic mycorrhizal fungi (Demoor et al. 2019). Recently, the insect pathogen Metarhizium robertsii was found to contain a histone lysine methyltransferase called Ash1p, which is up-regulated upon exposure to the insect cuticle to promote the production of peroxisomes that promote lipid hydrolysis (through Pex16p) to produce large amounts of glycerol for turgor generation (Wang, Lai et al. 2023). Additional types of adhesion molecules have been discovered in the plant-associated fungus U. maydis where a novel adhesin promotes hyphal aggregation in plant tumors (Lep1p; Fukada et al. 2021). Likewise, proteins similar to FAS-domain containing fasciclins have been found in U. maydis (Mueller et al. 2008) and the Shiitake mushroom forming fungus Lentinula edodes (Miyazaki et al. 2007).
Additional roles for cell adhesion molecules outside of filamentous growth include aiding the formation of traps in predatory nematode-trapping fungi (Jiang et al. 2017; Lipke 2018), promoting adherence of sexual partners (i.e. agglutination) before the fusion of cells (de Nobel et al. 1995; Zhao et al. 2001; Muller et al. 2003), and acting as a mechanism to recognize “same” individuals or individuals of different species to promote kin selection (Dranginis et al. 2007; Smukalla et al. 2008; Shinn-Thomas and Mohler 2011; Kraushaar et al. 2015; Brückner et al. 2020). Overall, fungi contain numerous mechanisms to regulate cell adhesion with a repertoire of different adhesion strategies that allows for context-specific cell adhesion regulation and nuanced responses when invading new environments and performing other critical tasks.
Secreted enzymes
In addition to the morphological and adhesive changes that occur during filamentous growth, many fungi also secrete molecules into the extracellular milieu (Fig. 5). Broadly speaking, these molecules act to break down extracellular materials and facilitate entry into new environments. Fungi can secrete proteases, like the yapsins and Saps mentioned above, that break down host tissues for human pathogens (Hube 1996; Naglik et al. 2004; Kaur et al. 2007; Tobouti et al. 2016; Safiya et al. 2023), cuticle-degrading enzymes that break down the insect exoskeleton in insect pathogens (Charnley and St. Leger 1991; Zhang, Meng, et al. 2021), and pectinases and cellulases that break down pectin and cellulose, respectively, in the plant cell wall for plant pathogens and saprotrophic fungi (Collmer and Keen 1986; Jayani et al. 2005; Benz et al. 2014; Panchapakesan and Shankar 2016; Lange et al. 2019; Berbee et al. 2020).
In S. cerevisiae, the fMAPK pathway up-regulates expression of the PGU1 gene that encodes a secreted endo-polygalacturnase (Blanco et al. 1998), which breaks down pectin in the environment (Madhani et al. 1999; Gognies et al. 2001, 2006; Gognies and Belarbi 2002) including in the rind of citrus fruit (Vandermeulen and Cullen 2023). It is likely that the fMAPK pathway evolved to regulate pectinase production as the pathway also senses pectin in the external environment as discussed above. The fMAPK pathway also up-regulates the secreted enzyme encoded by SUC2, which is an invertase for hydrolyzing sucrose (Carlson et al. 1981) and contributes to social behaviors (Greig and Travisano 2004; Maclean and Brandon 2008; Koschwanez et al. 2011), such as by the formation of invasive aggregates that allow cells to invade in large groups by making gouges into surfaces (Chow, Dionne et al. 2019).
From pathways to networks: the integration of signals by multiple pathways
Besides the fMAPK pathway, other pathways sense and integrate signals into the filamentous growth response. In pathogens such as C. albicans (Kadosh and Johnson 2005; Hall et al. 2010; Shapiro and Cowen 2010; Du et al. 2012; Su et al. 2018; Pentland et al. 2021) and Na. glabratus (C. glabrata; Sasani et al. 2016; Hassan et al. 2021), these signals include serum, human body temperature (37°C), and elevated CO2 concentrations. The host immune system can also suppress filamentous growth in pathogens (Gow et al. 2011; Kavanaugh et al. 2014; Vila et al. 2017; Takagi et al. 2022). In plant pathogens, signals include the plant surface (Lanver et al. 2010; Liu et al. 2011; Leroch et al. 2015; Perez-Nadales and Di Pietro 2015; Wang et al. 2021), the plant cell wall (Polizeli et al. 1991; Gognies et al. 2001; Gognies and Belarbi 2002; Vandermeulen and Cullen 2023), and plant hormones, including indoleacetic acid (Prusty et al. 2004) and ethylene (Kolattukudy et al. 1995).
These signals are sensed by a variety of signaling pathways that can be as important for filamentous growth as the fMAPK pathway. One of these is the Ras-cAMP-regulated Protein Kinase A (RAS-PKA) pathway (Kronstad et al. 1998; Borges-Walmsley and Walmsley 2000; Fortwendel 2015; Kayikci and Magwene 2018; Dautt-Castro et al. 2021; Hong et al. 2024). The RAS-PKA pathway induces filamentous growth in response to nitrogen- and/or glucose-limited environments (Gimeno et al. 1992; Pan and Heitman 1999; Hogan and Sundstrom 2009; Cullen and Sprague 2012). The RAS-PKA pathway also regulates cell growth and metabolism (Morishita et al. 1995; Schmelzle et al. 2004; Kunkel et al. 2019) and depending on the species can play roles in other differentiation-type responses. These include responses central to the life cycle, such as sporulation and mating (Fillinger et al. 2002; Lee and Kronstad 2002; McDonald et al. 2009; Fortwendel 2015; Wendland 2020), and appressorium formation (Zhou et al. 2014; Li et al. 2017; Zhu et al. 2017; Qu et al. 2021). A long history of evolutionary conservation links mating, sporulation, and filamentous growth to nutrient starvation in many species, which may explain the requirement for a master nutrient-sensing pathway (RAS) pathway in overseeing these responses.
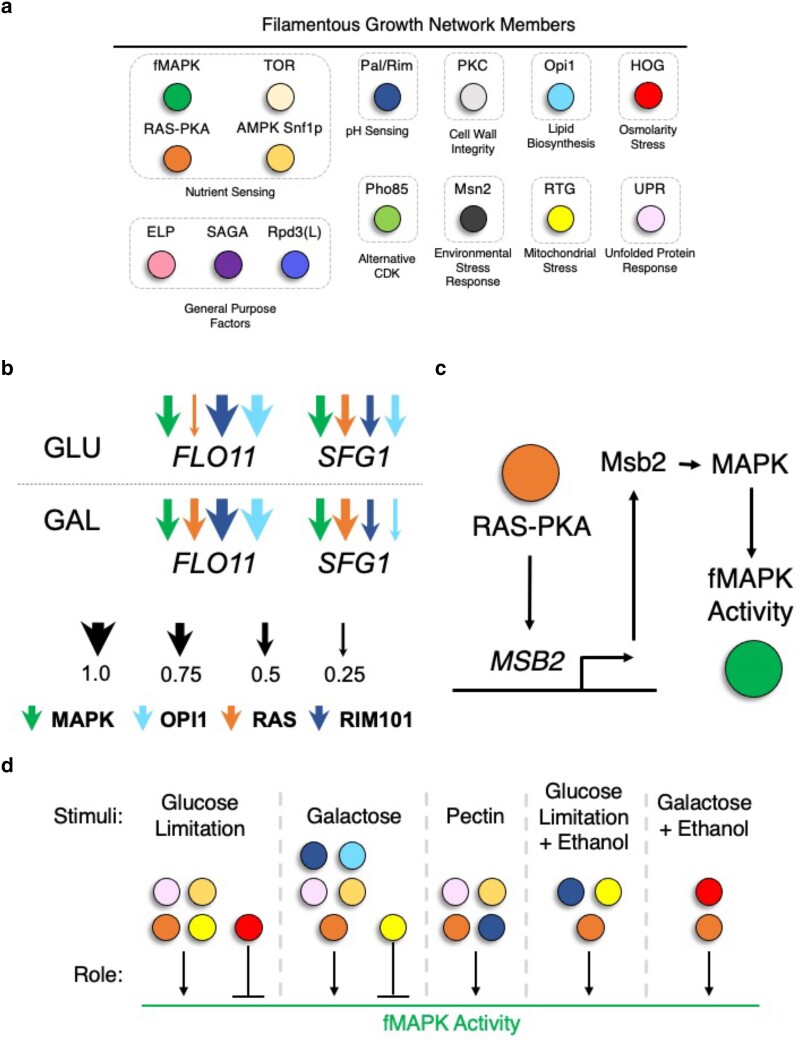
Many other regulatory proteins and pathways beyond the fMAPK and RAS-PKA pathways have been identified by comprehensive genetic screens in S. cerevisiae (Fig. 6a; Jin et al. 2008; Xu et al. 2010; Ryan et al. 2012; Kiss et al. 2019). These include the Pal/Rim pathway, which positively regulates filamentous growth, pathogenicity, and stress tolerance across fungi in response to pH changes through a related group of transcription factors (e.g. PacC in Aspergillus nidulans and Rim101p in S. cerevisiae and C. albicans; Li and Mitchell 1997; Peñalva Miguel and Arst Herbert 2002; Davis 2003; Lamb and Mitchell 2003; Barrales et al. 2008; Mira et al. 2009; Selvig and Alspaugh 2011; Du and Huang 2016; Li et al. 2022; Vandermeulen and Cullen 2022). Neutral to alkaline pH stimulates filamentous growth in S. cerevisiae (Li and Mitchell 1997; Davis, Edwards, et al. 2000; Davis, Wilson et al. 2000; Lamb and Mitchell 2003; Barrales et al. 2008; Mira et al. 2009) and C. albicans (Nobile et al. 2008; Du and Huang 2016), whereas an acidic pH stimulates filamentous growth in U. maydis (Mayorga and Gold 1999; Martínez-Espinoza et al. 2004), likely because auxins produced by the fungus cause a reduction in pH in plant tissue (Guevara-Lara et al. 2000; Martínez-Espinoza et al. 2004).
Fig. 6.
The filamentation regulatory network in S. cerevisiae. a) Regulatory pathways and protein complexes that impact filamentous growth, b) Many of these pathways regulate each other's target genes, c) One pathway can regulate the levels of another pathway's components (cross-pathway feedback), d) Different pathways regulate the activity of the fMAPK pathway under different conditions. The network in C. albicans is even more extensive.
Other pathways include the UPR, as mentioned above; the retrograde (RTG) pathway, a nuclear pathway that responds to mitochondrial stress (Bui and Labedzka-Dmoch 2024) and filamentous growth (Aun et al. 2013; Gonzalez et al. 2017; Rollenhagen et al. 2020); the AMP-dependent protein kinase (AMPK) Snf1p, which regulates glucose repression (Carlson 1999; Simpson-Lavy and Kupiec 2023) and filamentous growth (Cullen and Sprague 2000); the target of rapamycin (TOR) pathway, a conserved pathway in eukaryotes that regulates cell metabolism, survival, ribosome biogenesis, and growth (Laplante and Sabtini 2009; Laplante and Sabatini 2012; Gutiérrez-Santiago and Navarro 2023; Wang, Zheng et al. 2023) and controls many aspects of filamentous growth (Cutler et al. 2001); the cell wall integrity protein kinase C pathway (Levin 2005; Birkaya et al. 2009; Zhang, Wang et al. 2021; Yoshimi et al. 2022); a transcriptional regulator of phospholipid biogenesis, Opi1p (Reynolds 2006; Chen et al. 2015; Vandermeulen and Cullen 2022); the alternative CDK Pho85p (Carroll and O'Shea 2002; Huang et al. 2007; Lee et al. 2007; Chavel et al. 2014; Vandermeulen and Cullen 2022); and the general yeast stress response transcription factor Msn2p (Estruch and Carlson 1993; Martínez-Pastor et al. 1996; Gasch et al. 2000). Some pathways, such as the RTG, TOR, and Pho85p have been shown to exhibit conditional role reversals in S. cerevisiae in that they switch between positive and negative regulators depending on the environment (Vandermeulen and Cullen 2022).
Additionally, several general-purpose protein complexes also play a role in filamentous growth (Fig. 6a; Abdullah and Cullen 2009; Chavel et al. 2014; Chow, Starr et al. 2019; Vandermeulen and Cullen 2022). These factors include the chromatin-remodeling complex Rpd3p(L) (Kurdistani and Grunstein 2003); the elongator complex (ELP; Fellows et al. 2000; Krogan and Greenblatt 2001; Dong et al. 2015); and the chromatin-remodeling complex and transcription coactivator Spt-Ada-Gcn5 acetyltransferase (SAGA; Roberts and Winston 1997; Kohler et al. 2010; Hirsch et al. 2015). How these general-purpose factors function during filamentous growth to promote the response is not clear.
Complex networks like the one described above exist in many eukaryotes and appear to be the norm. The filamentation regulatory network in C. albicans is even more complicated than in S. cerevisiae and in some cases is wired differently. For example, in S. cerevisiae, glucose limitation is an inducer of filamentous/invasive growth through AMPK Snf1p (Cullen and Sprague 2000). Snf1p regulates filamentous growth by relieving the inhibitory effects of the transcriptional repressor Nrg1p (Kuchin et al. 2002), which is a negative regulator of glucose-repressed genes (Zhou and Winston 2001). In C. albicans, nutrient deprivation is also a trigger of filamentous growth (Huang 2012; Chow et al. 2021). However, in C. albicans, Snf1p is an essential protein (Petter et al. 1997). As in S. cerevisiae, Nrg1p also negatively regulates the filamentation response in C. albicans (Braun et al. 2001; Murad et al. 2001), although not by Snf1p. Rather, in C. albicans, Nrg1p is inhibited in a sequential manner by the RAS-PKA pathway and reduced TOR activity (Lu et al. 2011). Therefore, the RAS-PKA and TOR pathways are equally important for signaling and filamentation responses in C. albicans but in some ways work through different regulatory mechanisms (Shapiro et al. 2009; Inglis and Sherlock 2013; Huang et al. 2019; Qi et al. 2022). This is just 1 example showing how the same type of protein can be regulated by different upstream pathways in different organisms to achieve the same outcome. Another example can be seen when exploring how the RAS-PKA pathway controls pH-dependent responses in C. albicans (Hollomon et al. 2016), whereas pH-dependent responses are more exclusive to the Pal/Rim pathway in S. cerevisiae.
How do multiple pathways control a biological response in a coordinated manner? For filamentous growth, 1 way is that regulatory pathways coregulate shared target genes. For example, numerous transcription factors from different signaling pathways converge to regulate the same gene (e.g. FLO11; Rupp et al. 1999; Borneman et al. 2006, 2007). The fMAPK pathway, RAS-PKA pathway, SAGA complex, Pal/Rim pathway, RTG pathway, Opi1p transcriptional regulator, ELP complex, and chromatin-remodeling complex Rpd3(L) also regulate the expression of a subset of the same genes that induce filamentous growth (Chavel et al. 2014; Chow, Starr et al. 2019). Additionally, the RAS-PKA and TOR pathways regulate the same genes based on different temporal contexts after glucose induction (Kunkel et al. 2019; Plank 2022). Recently, studying the up-regulation of 2 different target genes that regulate cell adhesion (FLO11 or SFG1) has revealed insight into how an integrated signaling network can generate phenotypic plasticity based on the environment. This is achieved because the expression of FLO11 and SFG1 are both conditionally and differentially regulated by several different signaling pathways (e.g. RAS-PKA, Pal/Rim, Opi1p, fMAPK, Fig. 6b;Vandermeulen and Cullen 2022). The fact that numerous pathways can fine-tune the level of gene expression of filamentous targets allows for a highly nuanced and coordinated response.
A second way that signaling pathways control filamentous growth in a coordinated manner is through major transcription factors regulating each other's gene expression (Borneman et al. 2006). In many cases, this can create a type of cross feedback and has been shown to amplify and in some cases modulate transcription factor activity. Similarly, the transcription factor of 1 pathway has been found at the promoter of a gene encoding the components of another pathway, referred to as cross-pathway feedback. For example, the RAS-PKA pathway up-regulates MSB2 gene expression (Fig. 6c; Chow, Starr et al. 2019).
A third way is that pathways can regulate each other's activities (Mosch et al. 1996; Reinders et al. 1998; Chavel et al. 2010; Brückner et al. 2011). For example, the kinases of 1 pathway can regulate the activity and localization of the kinases in another (Bharucha et al. 2008). A classic example discovered in yeast is the fact that the RAS-PKA pathway can regulate fMAPK pathway activity by Ras2p (Mosch et al. 1996, 1999). It has since been shown that more than 5 pathways/protein complexes regulate the activity of the fMAPK pathway (Chavel et al. 2010). However, different pathways regulate fMAPK pathway activity depending on which stimuli are present and some even show opposing effects (i.e. role reversals) across environments (Fig. 6d; Vandermeulen and Cullen 2022, 2023). Therefore, a highly integrated network of protein complexes and signaling pathways regulate filamentous growth. This allows for the integration of multiple signals and coordination of cellular responses, including perhaps to modulate phenotypic plasticity.
In C. albicans, these combinatorial control mechanisms are evident during biofilm growth, which is regulated by a huge transcriptional network overseeing more than 1,000 genes (Nobile et al. 2012). These genes are coregulated by a core group of 9 transcription factors, the promoters of which are directly bound by most of the other regulators in a an intricate spiderweb of regulatory interactions that may allow integrated control by multiple environmental stimuli. Mammalian differentiation pathways similarly operate in vast uncharted networks. Addressing network-related questions in a simple model is relevant given the relatively small number of proteins and interactions compared to metazoans. Deciphering how signals become integrated into a cohesive response is critical to comprehensively understand the regulation of cell differentiation.
Conclusions and future directions
Much of the vast diversity in nature can be explained by the specialization of cells into differentiated cell types that perform highly specific functions. Progress toward understanding cell-type specialization has come in part from studying fungal model systems. In a handful of yeast models, genetic and molecular tools have been developed that have elucidated the molecular basis of the striking morphological diversity observed in nature. Remarkably, a few common pathways, such as MAPK pathways, control aspects of cell differentiation across many fungal species, and this logic extends to eukaryotes in general. Therefore, a satisfying feature of studying signaling pathways in fungal models is that molecular insights can be broadly applied across eukaryotes to understand how cell differentiation is regulated. Likewise, fungal-specific elements identified in these models provide potential road maps toward curbing fungal pathogenesis in plants and animals, including emerging fungal pathogens, such as C. auris (Yue et al. 2018; Egger et al. 2022).
Many questions remain to be answered surrounding MAPK pathway regulation during filamentous growth. It remains unclear what mucin sensors are sensing and how different mucins control different pathways. In addition, the way that mucins regulate the Cdc42p module is an open question. How the MAPK pathway operates in different modes (Ste20p dependent and Ste20p independent) in different contexts is not well understood. Equally fascinating are questions surrounding the broader network. How do signaling pathways regulate each other's activities? Is the network static, or does it change in different settings to modulate the filamentation response? Given that genetic screens have identified hundreds of proteins that impact filamentous growth, understanding the global picture of the response will be a major undertaking. New model systems, now approachable by genetic tools, such as clustered regularly interspaced short palindromic repeats or CRISPR may provide new insights into longstanding questions surrounding pathogenesis, symbiosis, and multispecies interactions. It will be interesting to “listen in” on the conversations that fungal cells have with their hosts and neighboring microbes at the molecular level. Future work aimed at addressing these important questions is highly related to human health from the perspective of fungal pathogenesis. Future work can also reveal fundamental insights into the regulation of eukaryotic cell differentiation.
Supplementary Material
Acknowledgments
Apologies to labs whose work was not cited due to page limitations. Thanks to laboratory members for reading the manuscript.
Contributor Information
Matthew D Vandermeulen, Department of Biological Sciences, University at Buffalo, Buffalo, NY 14260-1300, USA.
Michael C Lorenz, Department of Microbiology and Molecular Genetics, University of Texas McGovern Medical School, Houston, TX 77030, USA.
Paul J Cullen, Department of Biological Sciences, University at Buffalo, Buffalo, NY 14260-1300, USA.
Data availability
Genes and proteins are linked to Saccharomyces Genome Database (SGD) https://www.yeastgenome.org/.
Funding
The work was supported by grants from the National Institutes of Health to P.J.C. (R01GM098629) and M.C.L. (R01AI143304).
Literature cited
- Abdullah U, Cullen PJ. 2009. The tRNA modification complex elongator regulates the Cdc42-dependent mitogen-activated protein kinase pathway that controls filamentous growth in yeast. Eukaryot Cell. 8(9):1362–1372. doi: 10.1128/EC.00015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari H, Caccamise LM, Pande T, Cullen PJ. 2015. Comparative analysis of transmembrane regulators of the filamentous growth mitogen-activated protein kinase pathway uncovers functional and regulatory differences. Eukaryotic Cell. 14(9):868–883. doi: 10.1128/EC.00085-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari H, Cullen PJ. 2014. Metabolic respiration induces AMPK- and Ire1p-dependent activation of the p38-type HOG MAPK pathway. PLoS Genet. 10(10):e1004734. doi: 10.1371/journal.pgen.1004734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari H, Vadaie N, Chow J, Caccamise LM, Chavel CA, Li B, Bowitch A, Stefan CJ, Cullen PJ. 2015. Role of the unfolded protein response in regulating the mucin-dependent filamentous-growth mitogen-activated protein kinase pathway. Mol Cell Biol. 35(8):1414–1432. doi: 10.1128/MCB.01501-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht A, Felk A, Pichova I, Naglik JR, Schaller M, de Groot P, MacCallum D, Odds FC, Schäfer W, Klis F, et al. 2006. Glycosylphosphatidylinositol-anchored proteases of Candida albicans target proteins necessary for both cellular processes and host-pathogen interactions. J Biol Chem. 281(2):688–694. doi: 10.1074/jbc.M509297200. [DOI] [PubMed] [Google Scholar]
- Albuquerque P, Casadevall A. 2012. Quorum sensing in fungi–a review. Med Mycol. 50(4):337–345. doi: 10.3109/13693786.2011.652201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araç D, Boucard AA, Bolliger MF, Nguyen J, Soltis SM, Südhof TC, Brunger AT. 2012. A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. EMBO J. 31(6):1364–1378. doi: 10.1038/emboj.2012.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asgar MA, Yamauchi R, Kato K. 2003. Modification of pectin in Japanese persimmon fruit during the sun-drying process. Food Chem. 81(4):555–560. doi: 10.1016/S0308-8146(02)00489-2. [DOI] [Google Scholar]
- Aun A, Tamm T, Sedman J. 2013. Dysfunctional mitochondria modulate cAMP-PKA signaling and filamentous and invasive growth of Saccharomyces cerevisiae. Genetics. 193(2):467–481. doi: 10.1534/genetics.112.147389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai FY, Han DY, Duan SF, Wang QM. 2022. The ecology and evolution of the Baker's yeast Saccharomyces cerevisiae. Genes (Basel). 13(2):230. doi: 10.3390/genes13020230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baladrón V, Ufano S, Dueñas E, Martín-Cuadrado AB, del Rey F, Vázquez de Aldana CR. 2002. Eng1p, an endo-1,3-beta-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot Cell. 1(5):774–786. doi: 10.1128/EC.1.5.774-786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A, Kang C-Y, An M, Koff BB, Sunder S, Kumar A, Tenuta LMA, Stockbridge RB. 2024. Fluoride export is required for the competitive fitness of pathogenic microorganisms in dental biofilm models. mBio. 15(5):e0018424. doi: 10.1128/mbio.00184-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuett F, Herskowitz I. 1994. Morphological transitions in the life cycle of Ustilago maydis and their genetic control by the a and b loci. Exp Mycol. 18(3):247–266. doi: 10.1006/emyc.1994.1024. [DOI] [Google Scholar]
- Bao MZ, Schwartz MA, Cantin GT, Yates JR III, Madhani HD. 2004. Pheromone-dependent destruction of the Tec1 transcription factor is required for MAP kinase signaling specificity in yeast. Cell. 119(7):991–1000. doi: 10.1016/j.cell.2004.11.052. [DOI] [PubMed] [Google Scholar]
- Bardwell L. 2005. A walk-through of the yeast mating pheromone response pathway. Peptides. 26(2):339–350. doi: 10.1016/j.peptides.2004.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell L, Cook JG, Voora D, Baggott DM, Martinez AR, Thorner J. 1998. Repression of yeast Ste12 transcription factor by direct binding of unphosphorylated Kss1 MAPK and its regulation by the Ste7 MEK. Genes Dev. 12(18):2887–2898. doi: 10.1101/gad.12.18.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell L, Cook JG, Zhu-Shimoni JX, Voora D, Thorner J. 1998. Differential regulation of transcription: repression by unactivated mitogen-activated protein kinase Kss1 requires the Dig1 and Dig2 proteins. Proc Natl Acad Sci U S A. 95(26):15400–15405. doi: 10.1073/pnas.95.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrales RR, Jimenez J, Ibeas JI. 2008. Identification of novel activation mechanisms for FLO11 regulation in Saccharomyces cerevisiae. Genetics. 178(1):145–156. doi: 10.1534/genetics.107.081315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barua S, Li L, Lipke PN, Dranginis AM. 2016. Molecular basis for strain variation in the Saccharomyces cerevisiae Adhesin Flo11p. mSphere. 1(4):24. doi: 10.1128/mSphere.00129-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassilana M, Hopkins J, Arkowitz RA. 2005. Regulation of the Cdc42/Cdc24 GTPase module during Candida albicans hyphal growth. Eukaryot Cell. 4(3):588–603. doi: 10.1128/EC.4.3.588-603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso V, d'Enfert C, Znaidi S, Bachellier-Bassi S. 2019. From genes to networks: the regulatory circuitry controlling Candida albicans morphogenesis. Curr Top Microbiol Immunol. 422:61–99. doi: 10.1007/82_2018_144. [DOI] [PubMed] [Google Scholar]
- Basson MA. 2012. Signaling in cell differentiation and morphogenesis. Cold Spring Harb Perspect Biol. 4(6):a008151. doi: 10.1101/cshperspect.a008151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, González B, Li B, Kimble G, Kozminski KG, Cullen PJ. 2020. Functions for Cdc42p BEM adaptors in regulating a differentiation-type MAP kinase pathway. Mol Biol Cell. 31(6):491–510. doi: 10.1091/mbc.E19-08-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu S, Vadaie N, Prabhakar A, Li B, Adhikari H, Pitoniak A, Chow J, Chavel CA, Cullen PJ. 2016. Spatial landmarks regulate a Cdc42-dependent MAPK pathway to control differentiation and the response to positional compromise. Proc Natl Acad Sci U S A. 113(14):E2019–E2028. doi: 10.1073/pnas.1522679113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz JP, Chau BH, Zheng D, Bauer S, Glass NL, Somerville CR. 2014. A comparative systems analysis of polysaccharide-elicited responses in Neurospora crassa reveals carbon source-specific cellular adaptations. Mol Microbiol. 91(2):275–299. doi: 10.1111/mmi.12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbee ML, Strullu-Derrien C, Delaux P-M, Strother PK, Kenrick P, Selosse M-A, Taylor JW. 2020. Genomic and fossil windows into the secret lives of the most ancient fungi. Nat Rev Microbiol. 18(12):717–730. doi: 10.1038/s41579-020-0426-8. [DOI] [PubMed] [Google Scholar]
- Bharucha N, Chabrier-Rosello Y, Xu T, Johnson C, Sobczynski S, Song Q, Dobry CJ, Eckwahl MJ, Anderson CP, Benjamin AJ, et al. 2011. A large-scale complex haploinsufficiency-based genetic interaction screen in Candida albicans: analysis of the RAM network during morphogenesis. PLoS Genet. 7(4):e1002058. doi: 10.1371/journal.pgen.1002058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharucha N, Ma J, Dobry CJ, Lawson SK, Yang Z, Kumar A. 2008. Analysis of the yeast kinome reveals a network of regulated protein localization during filamentous growth. Mol Biol Cell. 19(7):2708–2717. doi: 10.1091/mbc.e07-11-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkaya B, Maddi A, Joshi J, Free SJ, Cullen PJ. 2009. Role of the cell wall integrity and filamentous growth mitogen-activated protein kinase pathways in cell wall remodeling during filamentous growth. Eukaryot Cell. 8(8):1118–1133. doi: 10.1128/EC.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas K, Morschhäuser J. 2005. The Mep2p ammonium permease controls nitrogen starvation-induced filamentous growth in Candida albicans. Mol Microbiol. 56(3):649–669. doi: 10.1111/j.1365-2958.2005.04576.x. [DOI] [PubMed] [Google Scholar]
- Blanco P, Sieiro C, Reboredo NM, Villa TG. 1998. Cloning, molecular characterization, and expression of an endo-polygalacturonase-encoding gene from Saccharomyces cerevisiae IM1-8b. FEMS Microbiol Lett. 164(2):249–255. doi: 10.1111/j.1574-6968.1998.tb13094.x. [DOI] [PubMed] [Google Scholar]
- Bölker M, Kahmann R. 1993. Sexual pheromones and mating responses in fungi. Plant Cell. 5(10):1461–1469. doi: 10.1105/tpc.5.10.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfante P, Genre A. 2010. Mechanisms underlying beneficial plant–fungus interactions in mycorrhizal symbiosis. Nat Commun. 1(1):48. doi: 10.1038/ncomms1046. [DOI] [PubMed] [Google Scholar]
- Borges-Walmsley MI, Walmsley AR. 2000. cAMP signalling in pathogenic fungi: control of dimorphic switching and pathogenicity. Trends Microbiol. 8(3):133–141. doi: 10.1016/S0966-842X(00)01698-X. [DOI] [PubMed] [Google Scholar]
- Borneman AR, Leigh-Bell JA, Yu H, Bertone P, Gerstein M, Snyder M. 2006. Target hub proteins serve as master regulators of development in yeast. Genes Dev. 20(4):435–448. doi: 10.1101/gad.1389306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borneman AR, Zhang ZD, Rozowsky J, Seringhaus MR, Gerstein M, Snyder M. 2007. Transcription factor binding site identification in yeast: a comparison of high-density oligonucleotide and PCR-based microarray platforms. Funct Integr Genomics. 7(4):335–345. doi: 10.1007/s10142-007-0054-7. [DOI] [PubMed] [Google Scholar]
- Botha A. 2011. The importance and ecology of yeasts in soil. Soil Biol Biochem. 43(1):1–8. doi: 10.1016/j.soilbio.2010.10.001. [DOI] [Google Scholar]
- Botstein D, Chervitz SA, Cherry M. 1997. Yeast as a model organism. Science. 277(5330):1259–1260. doi: 10.1126/science.277.5330.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D, Fink GR. 2011. Yeast: an experimental organism for 21st Century biology. Genetics. 189(3):695–704. doi: 10.1534/genetics.111.130765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. 2012. Hyphal growth in human fungal pathogens and its role in virulence. Int J Microbiol. 2012:517529. doi: 10.1155/2012/517529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AC, Morrison E, Milne S, Gonia S, Gale CA, Gow NAR. 2014. Cdc42 GTPase dynamics control directional growth responses. Proc Natl Acad Sci U S A. 111(2):811–816. doi: 10.1073/pnas.1307264111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt AC, Koehn OJ, Williams CL. 2021. SmgGDS: an emerging master regulator of prenylation and trafficking by small GTPases in the Ras and Rho families. Front Mol Biosci. 8:685135. doi: 10.3389/fmolb.2021.685135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun BR, Kadosh D, Johnson AD. 2001. NRG1, a repressor of filamentous growth in C. albicans, is down-regulated during filament induction. EMBO J. 20(17):4753–4761. doi: 10.1093/emboj/20.17.4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brefort T, Doehlemann G, Mendoza-Mendoza A, Reissmann S, Djamei A, Kahmann R. 2009. Ustilago maydis as a pathogen. Annu Rev Phytopathol. 47(1):423–445. doi: 10.1146/annurev-phyto-080508-081923. [DOI] [PubMed] [Google Scholar]
- Brown DH Jr, Giusani AD, Chen X, Kumamoto CA. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol Microbiol. 34(4):651–662. doi: 10.1046/j.1365-2958.1999.01619.x. [DOI] [PubMed] [Google Scholar]
- Brückner S, Kern S, Birke R, Saugar I, Ulrich HD, Mösch H-U. 2011. The TEA transcription factor Tec1 links TOR and MAPK pathways to coordinate yeast development. Genetics. 189(2):479–494. doi: 10.1534/genetics.111.133629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner S, Mosch HU. 2012. Choosing the right lifestyle: adhesion and development in Saccharomyces cerevisiae. FEMS Microbiol Rev. 36(1):25–58. doi: 10.1111/j.1574-6976.2011.00275.x. [DOI] [PubMed] [Google Scholar]
- Brückner S, Schubert R, Kraushaar T, Hartmann R, Hoffmann D, Jelli E, Drescher K, Müller DJ, Oliver Essen L, Mösch H-U. 2020. Kin discrimination in social yeast is mediated by cell surface receptors of the Flo11 adhesin family. Elife. 9:e55587. doi: 10.7554/eLife.55587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet T, King N. 2017. The origin of animal multicellularity and cell differentiation. Dev Cell. 43(2):124–140. doi: 10.1016/j.devcel.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui THD, Labedzka-Dmoch K. 2024. RetroGREAT signaling: the lessons we learn from yeast. IUBMB Life. 76(1):26–37. doi: 10.1002/iub.2775. [DOI] [PubMed] [Google Scholar]
- Bumgarner SL, Neuert G, Voight BF, Symbor-Nagrabska A, Grisafi P, van Oudenaarden A, Fink GR. 2012. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Mol Cell. 45(4):470–482. doi: 10.1016/j.molcel.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buser CC, Newcomb RD, Gaskett AC, Goddard MR. 2014. Niche construction initiates the evolution of mutualistic interactions. Ecol Lett. 17(10):1257–1264. doi: 10.1111/ele.12331. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang TT, DeMaria G, Sheehan JK, Kesimer M. 2012. Mapping the protein domain structures of the respiratory mucins: a mucin proteome coverage study. J Proteome Res. 11(8):4013–4023. doi: 10.1021/pr300058z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M. 1999. Glucose repression in yeast. Curr Opin Microbiol. 2(2):202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- Carlson M, Osmond BC, Botstein D. 1981. Mutants of yeast defective in sucrose utilization. Genetics. 98(1):25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll AS, O'Shea EK. 2002. Pho85 and signaling environmental conditions. Trends Biochem Sci. 27(2):87–93. doi: 10.1016/S0968-0004(01)02040-0. [DOI] [PubMed] [Google Scholar]
- Chadwick BJ, Pham T, Xie X, Ristow LC, Krysan DJ, Lin X. 2022. The RAM signaling pathway links morphology, thermotolerance, and CO2 tolerance in the global fungal pathogen Cryptococcus neoformans. Elife. 11:e82563. doi: 10.7554/eLife.82563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra J, Zhou G, Ghannoum MA. 2005. Fungal biofilms and antimycotics. Curr Drug Targets. 6(8):887–894. doi: 10.2174/138945005774912762. [DOI] [PubMed] [Google Scholar]
- Chant J, Pringle JR. 1995. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J Cell Biol. 129(3):751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnley AK, St. Leger RJ. 1991. The role of cuticle-degrading enzymes in fungal pathogenesis in insects. In: Cole GT, Hoch HC, editors. The Fungal Spore and Disease Initiation in Plants and Animals. Boston (MA): Springer. p. 267–286. [Google Scholar]
- Chavel CA, Caccamise LM, Li B, Cullen PJ. 2014. Global regulation of a differentiation MAPK pathway in yeast. Genetics. 198(3):1309–1328. doi: 10.1534/genetics.114.168252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavel CA, Dionne HM, Birkaya B, Joshi J, Cullen PJ. 2010. Multiple signals converge on a differentiation MAPK pathway. PLoS Genet. 6(3):e1000883. doi: 10.1371/journal.pgen.1000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Fink GR. 2006. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 20(9):1150–1161. doi: 10.1101/gad.1411806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Fujita M, Feng Q, Clardy J, Fink GR. 2004. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc Natl Acad Sci U S A. 101(14):5048–5052. doi: 10.1073/pnas.0401416101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zhou X, Ren B, Cheng L. 2020. The regulation of hyphae growth in Candida albicans. Virulence. 11(1):337–348. doi: 10.1080/21505594.2020.1748930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-L, de Bernardis F, Yu S-J, Sandini S, Kauffman S, Tams RN, Bethea E, Reynolds TB. 2015. Candida albicans OPI1 regulates filamentous growth and virulence in vaginal infections, but not inositol biosynthesis. PLoS One. 10(1):e0116974. doi: 10.1371/journal.pone.0116974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkasova V, Lyons DM, Elion EA. 1999. Fus3p and Kss1p control G1 arrest in Saccharomyces cerevisiae through a balance of distinct arrest and proliferative functions that operate in parallel with Far1p. Genetics. 151(3):989–1004. doi: 10.1093/genetics/151.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chethana KWT, Jayawardena RS, Chen Y-J, Konta S, Tibpromma S, Abeywickrama PD, Gomdola D, Balasuriya A, Xu J, Lumyong S, et al. 2021. Diversity and function of appressoria. Pathogens. 10(6):746. doi: 10.3390/pathogens10060746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S-W, Moore D. 1996. Patterns in Fungal Development. Cambridge, England; New York: Cambridge University Press. [Google Scholar]
- Cho Y, Cramer RA, Kim K-H, Davis J, Mitchell TK, Figuli P, Pryor BM, Lemasters E, Lawrence CB. 2007. The Fus3/Kss1 MAP kinase homolog Amk1 regulates the expression of genes encoding hydrolytic enzymes in Alternaria brassicicola. Fungal Genet Biol. 44(6):543–553. doi: 10.1016/j.fgb.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Chou S, Huang L, Liu H. 2004. Fus3-regulated Tec1 degradation through SCFCdc4 determines MAPK signaling specificity during mating in yeast. Cell. 119(7):981–990. doi: 10.1016/j.cell.2004.11.053. [DOI] [PubMed] [Google Scholar]
- Chou S, Lane S, Liu H. 2006. Regulation of mating and filamentation genes by two distinct Ste12 complexes in Saccharomyces cerevisiae. Mol Cell Biol. 26(13):4794–4805. doi: 10.1128/MCB.02053-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow EWL, Pang LM, Wang Y. 2021. From Jekyll to Hyde: the yeast–hyphal transition of Candida albicans. Pathogens. 10(7):859. doi: 10.3390/pathogens10070859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J, Dionne HM, Prabhakar A, Mehrotra A, Somboonthum J, Gonzalez B, Edgerton M, Cullen PJ. 2019. Aggregate filamentous growth responses in yeast. mSphere. 4(2):e00702–e00718. doi: 10.1128/mSphere.00702-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow J, Starr I, Jamalzadeh S, Muniz O, Kumar A, Gokcumen O, Ferkey DM, Cullen PJ. 2019. Filamentation regulatory pathways control adhesion-dependent surface responses in yeast. Genetics. 212(3):667–690. doi: 10.1534/genetics.119.302004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiaens JF, Van Mulders SE, Duitama J, Brown CA, Ghequire MG, De Meester L, Michiels J, Wenseleers T, Voordeckers K, Verstrepen KJ. 2012. Functional divergence of gene duplicates through ectopic recombination. EMBO Rep. 13(12):1145–1151. doi: 10.1038/embor.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen BE. 2018. Membrane thickness as a key factor contributing to the activation of osmosensors and essential ras signaling pathways. Front Cell Dev Biol. 6:76. doi: 10.3389/fcell.2018.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collmer AC, Keen NT. 1986. The role of pectic enzymes in plant pathogenesis. Annu Rev Phytopathol. 24(1):383–409. doi: 10.1146/annurev.py.24.090186.002123. [DOI] [Google Scholar]
- Cook D, Genever P. 2013. Regulation of mesenchymal stem cell differentiation. In: Hime G, Abud H, editors. Transcriptional and Translational Regulation of Stem Cells. Dordrecht: Springer. p. 213–229. [DOI] [PubMed] [Google Scholar]
- Cook JG, Bardwell L, Kron SJ, Thorner J. 1996. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 10(22):2831–2848. doi: 10.1101/gad.10.22.2831. [DOI] [PubMed] [Google Scholar]
- Cook JG, Bardwell L, Thorner J. 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature. 390(6655):85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- Corfield AP. 2015. Mucins: a biologically relevant glycan barrier in mucosal protection. Biochim Biophys Acta. 1850(1):236–252. doi: 10.1016/j.bbagen.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Coyle SM, Flores J, Lim WA. 2013. Exploitation of latent allostery enables the evolution of new modes of MAP kinase regulation. Cell. 154(4):875–887. doi: 10.1016/j.cell.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotti LB, Jorge JA, Terenzi HF, Polizeli MLTM. 1996. Purification and characterization of galactose-induced pectinases from the exo-1 mutant strain of Neurospora crassa. In: Visser J, Voragen AGJ, editors. Progress in Biotechnology. Vol. 14. Amsterdam: Elsevier. p. 787–792. doi: 10.1016/S0921-0423(96)80317-0. [DOI] [Google Scholar]
- Crotti LB, Terenzi HF, Jorge JA, de Lourdes M, Polizeli ML. 1998. Regulation of pectic enzymes from the exo-1 mutant strain of Neurospora crassa: effects of glucose, galactose, and galacturonic acid. J Basic Microbiol. 38(3):181–188. doi:. [DOI] [PubMed] [Google Scholar]
- Crotti LB, Terenzi HF, Jorge JA, Polizeli MLTM. 1998. Characterization of galactose-induced extracellular and intracellular pectolytic activities from the exo-1 mutant strain of Neurospora crassa. J Ind Microbiol Biotech. 20(3–4):238–243. doi: 10.1038/sj.jim.2900519. [DOI] [Google Scholar]
- Csank C, Haynes K. 2000. Candida glabrata displays pseudohyphal growth. FEMS Microbiol Lett. 189(1):115–120. doi: 10.1111/j.1574-6968.2000.tb09216.x. [DOI] [PubMed] [Google Scholar]
- Csank C, Schröppel K, Leberer E, Harcus D, Mohamed O, Meloche S, Thomas DY, Whiteway M. 1998. Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun. 66(6):2713–2721. doi: 10.1128/IAI.66.6.2713-2721.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Sabbagh W, Graham E, Irick MM, van Olden EK, Neal C, Delrow J, Bardwell L, Sprague GF. 2004. A signaling mucin at the head of the Cdc42- and MAPK-dependent filamentous growth pathway in yeast. Genes Dev. 18(14):1695–1708. doi: 10.1101/gad.1178604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF Jr. 2000. Glucose depletion causes haploid invasive growth in yeast. Proc Natl Acad Sci U S A. 97(25):13619–13624. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF Jr. 2002. The roles of bud-site-selection proteins during haploid invasive growth in yeast. Mol Biol Cell. 13(9):2990–3004. doi: 10.1091/mbc.e02-03-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Sprague GF Jr. 2012. The regulation of filamentous growth in yeast. Genetics. 190(1):23–49. doi: 10.1534/genetics.111.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler NS, Pan X, Heitman J, Cardenas ME. 2001. The TOR signal transduction cascade controls cellular differentiation in response to nutrients. Mol Biol Cell. 12(12):4103–4113. doi: 10.1091/mbc.12.12.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashko S, Zhou N, Compagno C, Piškur J. 2014. Why, when, and how did yeast evolve alcoholic fermentation? FEMS Yeast Res. 14(6):826–832. doi: 10.1111/1567-1364.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dautt-Castro M, Rosendo-Vargas M, Casas-Flores S. 2021. The small GTPases in fungal signaling conservation and function. Cells. 10(5):1039. doi: 10.3390/cells10051039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D. 2003. Adaptation to environmental pH in Candida albicans and its relation to pathogenesis. Curr Genet. 44(1):1–7. doi: 10.1007/s00294-003-0415-2. [DOI] [PubMed] [Google Scholar]
- Davis D, Edwards JE Jr, Mitchell AP, Ibrahim AS. 2000. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect Immun. 68(10):5953–5959. doi: 10.1128/IAI.68.10.5953-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D, Wilson RB, Mitchell AP. 2000. RIM101-dependent and-independent pathways govern pH responses in Candida albicans. Mol Cell Biol. 20(3):971–978. doi: 10.1128/MCB.20.3.971-978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Chiara M, Barré BP, Persson K, Irizar A, Vischioni C, Khaiwal S, Stenberg S, Amadi OC, Žun G, Doberšek K, et al. 2022. Domestication reprogrammed the budding yeast life cycle. Nat Ecol Evol. 6(4):448–460. doi: 10.1038/s41559-022-01671-9. [DOI] [PubMed] [Google Scholar]
- Delarue M, Poterewicz G, Hoxha O, Choi J, Yoo W, Kayser J, Holt L, Hallatschek O. 2017. SCWISh network is essential for survival under mechanical pressure. Proc Natl Acad Sci U S A. 114(51):13465–13470. doi: 10.1073/pnas.1711204114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demoor A, Silar P, Brun S. 2019. Appressorium: the breakthrough in Dikarya. J Fungi (Basel). 5(3):72. doi: 10.3390/jof5030072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E, Posas F. 2022. The HOG pathway and the regulation of osmoadaptive responses in yeast. FEMS Yeast Res. 22(1):foac013. doi: 10.1093/femsyr/foac013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nobel H, Pike J, Lipke PN, Kurjan J. 1995. Genetics of a-agglutunin function in Saccharomyces cerevisiae. Mol Gen Genet. 247(4):409–415. doi: 10.1007/BF00293141. [DOI] [PubMed] [Google Scholar]
- Desai JV, Mitchell AP, Andes DR. 2014. Fungal biofilms, drug resistance, and recurrent infection. Cold Spring Harb Perspect Med. 4(10):a019729. doi: 10.1101/cshperspect.a019729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desseyn JL, Buisine MP, Porchet N, Aubert JP, Laine A. 1998. Genomic organization of the human mucin gene MUC5B. cDNA and genomic sequences upstream of the large central exon. J Biol Chem. 273(46):30157–30164. doi: 10.1074/jbc.273.46.30157. [DOI] [PubMed] [Google Scholar]
- Dickinson JR. 1996. Fuse1 alcohols induce hyphal-like extensions and pseudohyphal formationin yeasts. Microbiology. 142(6):1391–1397. doi: 10.1099/13500872-142-6-1391. [DOI] [PubMed] [Google Scholar]
- Dighton J. 2009. Mycorrhizae. In: Schaechter M, editor. Encyclopedia of Microbiology. 3rd ed. Oxford: Academic Press. p. 153–162. [Google Scholar]
- DiRusso CJ, Dashtiahangar M, Gilmore TD. 2022. Scaffold proteins as dynamic integrators of biological processes. J Biol Chem. 298(12):102628. doi: 10.1016/j.jbc.2022.102628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Lin Z, Diao W, Li D, Chu X, Wang Z, Zhou H, Xie Z, Shen Y, Long J. 2015. The Elp2 subunit is essential for elongator complex assembly and functional regulation. Structure. 23(6):1078–1086. doi: 10.1016/j.str.2015.03.018. [DOI] [PubMed] [Google Scholar]
- Dong F, Pirbhai M, Zhong Y, Zhong G. 2004. Cleavage-dependent activation of a chlamydia-secreted protease. Mol Microbiol. 52(5):1487–1494. doi: 10.1111/j.1365-2958.2004.04072.x. [DOI] [PubMed] [Google Scholar]
- Doolin MT, Johnson AL, Johnston LH, Butler G. 2001. Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol Microbiol. 40(2):422–432. doi: 10.1046/j.1365-2958.2001.02388.x. [DOI] [PubMed] [Google Scholar]
- Dorrity MW, Cuperus JT, Carlisle JA, Fields S, Queitsch C. 2018. Preferences in a trait decision determined by transcription factor variants. Proc Natl Acad Sci U S A. 115(34):E7997–E8006. doi: 10.1073/pnas.1805882115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell RD, Ryan O, Jansen A, Cheung D, Agarwala S, Danford T, Bernstein DA, Rolfe PA, Heisler LE, Chin B, et al. 2010. Genotype to phenotype: a complex problem. Science. 328(5977):469. doi: 10.1126/science.1189015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. 2007. A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol Mol Biol Rev. 71(2):282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Guan G, Xie J, Cottier F, Sun Y, Jia W, Mühlschlegel FA, Huang G. 2012. The transcription factor Flo8 mediates CO2 sensing in the human fungal pathogen Candida albicans. Mol Biol Cell. 23(14):2692–2701. doi: 10.1091/mbc.e12-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Huang G. 2016. Environmental pH adaption and morphological transitions in Candida albicans. Curr Genet. 62(2):283–286. doi: 10.1007/s00294-015-0540-8. [DOI] [PubMed] [Google Scholar]
- Du X-H, Yang Zhu L. 2021. Mating systems in true morels (Morchella). Microbiol Mol Biol Rev. 85(3):e0022020. doi: 10.1128/mmbr.00220-00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S-F, Han P-J, Wang Q-M, Liu W-Q, Shi J-Y, Li K, Zhang X-L, Bai F-Y. 2018. The origin and adaptive evolution of domesticated populations of yeast from Far East Asia. Nat Commun. 9(1):2690. doi: 10.1038/s41467-018-05106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvela-Levitt M, Kost-Alimova M, Emani M, Kohnert E, Thompson R, Sidhom E-H, Rivadeneira A, Sahakian N, Roignot J, Papagregoriou G, et al. 2019. Small molecule targets TMED9 and promotes lysosomal degradation to reverse proteinopathy. Cell. 178(3):521–535.e23. doi: 10.1016/j.cell.2019.07.002. [DOI] [PubMed] [Google Scholar]
- Egger NB, Kainz K, Schulze A, Bauer MA, Madeo F, Carmona-Gutierrez D. 2022. The rise of Candida auris: from unique traits to co-infection potential. Microb Cell. 9(8):141–144. doi: 10.15698/mic2022.08.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekdahl LI, Salcedo JA, Dungan MM, Mason DV, Myagmarsuren D, Murphy HA. 2023. Selection on plastic adherence leads to hyper-multicellular strains and incidental virulence in the budding yeast. Elife. 12:e81056. doi: 10.7554/eLife.81056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion EA. 2000. Pheromone response, mating and cell biology. Curr Opin Microbiol. 3(6):573–581. doi: 10.1016/S1369-5274(00)00143-0. [DOI] [PubMed] [Google Scholar]
- Endo T. 2015. Glycobiology of α-dystroglycan and muscular dystrophy. J Biochem. 157(1):1–12. doi: 10.1093/jb/mvu066. [DOI] [PubMed] [Google Scholar]
- Essen L-O, Vogt MS, Mösch H-U. 2020. Diversity of GPI-anchored fungal adhesins. Biol Chem. 401(12):1389–1405. doi: 10.1515/hsz-2020-0199. [DOI] [PubMed] [Google Scholar]
- Estruch F, Carlson M. 1993. Two homologous zinc finger genes identified by multicopy suppression in a SNF1 protein kinase mutant of Saccharomyces cerevisiae. Mol Cell Biol. 13(7):3872–3881. doi: 10.1128/mcb.13.7.3872-3881.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley FW, Satterberg B, Goldsmith EJ, Elion EA. 1999. Relative dependence of different outputs of the Saccharomyces cerevisiae pheromone response pathway on the MAP kinase Fus3p. Genetics. 151(4):1425–1444. doi: 10.1093/genetics/151.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fellows J, Erdjument-Bromage H, Tempst P, Svejstrup JQ. 2000. The Elp2 subunit of elongator and elongating RNA polymerase II holoenzyme is a WD40 repeat protein. J Biol Chem. 275(17):12896–12899. doi: 10.1074/jbc.275.17.12896. [DOI] [PubMed] [Google Scholar]
- Fidalgo M, Barrales RR, Ibeas JI, Jimenez J. 2006. Adaptive evolution by mutations in the FLO11 gene. Proc Natl Acad Sci U S A. 103(30):11228–11233. doi: 10.1073/pnas.0601713103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo AF, Boy J, Guggenberger G. 2021. Common mycorrhizae network: a review of the theories and mechanisms behind underground interactions. Front Fungal Biol. 2:735299. doi: 10.3389/ffunb.2021.735299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillinger S, Chaveroche M-K, Shimizu K, Keller N, D’Enfert C. 2002. cAMP and ras signalling independently control spore germination in the filamentous fungus Aspergillus nidulans. Mol Microbiol. 44(4):1001–1016. doi: 10.1046/j.1365-2958.2002.02933.x. [DOI] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol. 8(9):623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Folsom BL, Wagner GH, Scrivner CL. 1974. Comparison of soil carbohydrate in several prairie and forest soils by gas-liquid chromatography. Soil Sci Soc Am J. 38(2):305–309. doi: 10.2136/sssaj1974.03615995003800020027x. [DOI] [Google Scholar]
- Fortwendel JR. 2015. Orchestration of morphogenesis in filamentous fungi: conserved roles for Ras signaling networks. Fungal Biol Rev. 29(2):54–62. doi: 10.1016/j.fbr.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker MD, Heaton LLM, Jones NS, Boddy L. 2017. The mycelium as a network. In: Heitman J, Howlett BJ, Crous PW, Stukenbrock EH, James TY, Gow NAR, editors. The Fungal Kingdom. Washington (DC): American Society of Microbiology. p. 335–367. [Google Scholar]
- Fujita A, Hiroko T, Hiroko F, Oka C. 2005. Enhancement of superficial pseudohyphal growth by overexpression of the SFG1 gene in yeast Saccharomyces cerevisiae. Gene. 363:97–104. doi: 10.1016/j.gene.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Fukada F, Rössel N, Münch K, Glatter T, Kahmann R. 2021. A small Ustilago maydis effector acts as a novel adhesin for hyphal aggregation in plant tumors. New Phytol. 231(1):416–431. doi: 10.1111/nph.17389. [DOI] [PubMed] [Google Scholar]
- Fulford AD, McNeill H. 2020. Fat/Dachsous family cadherins in cell and tissue organisation. Curr Opin Cell Biol. 62:96–103. doi: 10.1016/j.ceb.2019.10.006. [DOI] [PubMed] [Google Scholar]
- Garcia MC, Lee JT, Ramsook CB, Alsteens D, Dufrêne YF, Lipke PN. 2011. A role for amyloid in cell aggregation and biofilm formation. PLoS One. 6(3):e17632. doi: 10.1371/journal.pone.0017632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 11(12):4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno CJ, Ljungdahl PO, Styles CA, Fink GR. 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 68(6):1077–1090. doi: 10.1016/0092-8674(92)90079-R. [DOI] [PubMed] [Google Scholar]
- Glover BJ. 2000. Differentiation in plant epidermal cells. J Exp Bot. 51(344):497–505. doi: 10.1093/jexbot/51.344.497. [DOI] [PubMed] [Google Scholar]
- Goddard MR, Anfang N, Tang R, Gardner RC, Jun C. 2010. A distinct population of Saccharomyces cerevisiae in New Zealand: evidence for local dispersal by insects and human-aided global dispersal in oak barrels. Environ Microbiol. 12(1):63–73. doi: 10.1111/j.1462-2920.2009.02035.x. [DOI] [PubMed] [Google Scholar]
- Goddard MR, Greig D. 2015. Saccharomyces cerevisiae: a nomadic yeast with no niche? FEMS Yeast Res. 15(3):fov009. doi: 10.1093/femsyr/fov009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gognies S, Barka EA, Gainvors-Claisse A, Belarbi A. 2006. Interactions between yeasts and grapevines: filamentous growth, endopolygalacturonase and phytopathogenicity of colonizing yeasts. Microb Ecol. 51(1):109–116. doi: 10.1007/s00248-005-0098-y. [DOI] [PubMed] [Google Scholar]
- Gognies S, Belarbi A. 2002. Endopolygalacturonase of Saccharomyces cerevisiae: involvement in pseudohyphae development of haploids and in pathogenicity on Vitis vinifera. Plant Sci. 163(4):759–769. doi: 10.1016/S0168-9452(02)00211-X. [DOI] [Google Scholar]
- Gognies S, Simon G, Belarbi A. 2001. Regulation of the expression of endopolygalacturonase gene PGU1 in Saccharomyces. Yeast. 18(5):423–432. doi: 10.1002/yea.693. [DOI] [PubMed] [Google Scholar]
- Gonzalez B, Mas A, Beltran G, Cullen PJ, Torija MJ. 2017. Role of mitochondrial retrograde pathway in regulating ethanol-inducible filamentous growth in yeast. Front Physiol. 8:148. doi: 10.3389/fphys.2017.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González B, Mirzaei M, Basu S, Pujari AN, Vandermeulen MD, Prabhakar A, Cullen PJ. 2023. Turnover and bypass of p21-activated kinase during Cdc42-dependent MAPK signaling in yeast. J Biol Chem. 299(11):105297. doi: 10.1016/j.jbc.2023.105297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good M, Tang G, Singleton J, Reményi A, Lim WA. 2009. The Ste5 scaffold directs mating signaling by catalytically unlocking the Fus3 MAP kinase for activation. Cell. 136(6):1085–1097. doi: 10.1016/j.cell.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow NAR, van de Veerdonk FL, Brown AJP, Netea MG. 2011. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol. 10(2):112–122. doi: 10.1038/nrmicro2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granek JA, Murray D, Kayrkci O, Magwene PM. 2013. The genetic architecture of biofilm formation in a clinical isolate of Saccharomyces cerevisiae. Genetics. 193(2):587–600. doi: 10.1534/genetics.112.142067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greig D, Travisano M. 2004. The Prisoner's Dilemma and polymorphism in yeast SUC genes. Proc Biol Sci. 271(suppl_3):S25–S26. doi: 10.1098/rsbl.2003.0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara-Lara F, Valverde ME, Paredes-López O. 2000. Is pathogenicity of Ustilago maydis (huitlacoche) strains on maize related to in vitro production of indole-3-acetic acid? World J Microbiol Biotechnol. 16(5):481–490. doi: 10.1023/A:1008906013524. [DOI] [Google Scholar]
- Gunina A, Kuzyakov Y. 2015. Sugars in soil and sweets for microorganisms: review of origin, content, composition and fate. Soil Biol Biochem. 90:87–100. doi: 10.1016/j.soilbio.2015.07.021. [DOI] [Google Scholar]
- Guo B, Styles CA, Feng Q, Fink GR. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc Natl Acad Sci U S A. 97(22):12158–12163. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurgel ILdS, Jorge KTdOS, Malacco NLSdO, Souza JAM, Rocha MC, Fernandes MF, Martins FRB, Malavazi I, Teixeira MM, Soriani FM. 2019. The Aspergillus fumigatus mucin MsbA regulates the cell wall integrity pathway and controls recognition of the fungus by the immune system. mSphere. 4(3):e00350–e00319. doi: 10.1128/mSphere.00350-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin MC, Albertyn J, Alexander M, Davenport K. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 62(4):1264–1300. doi: 10.1128/MMBR.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez-Santiago F, Navarro F. 2023. Transcription by the three RNA polymerases under the control of the TOR signaling pathway in Saccharomyces cerevisiae. Biomolecules. 13(4):642. doi: 10.3390/biom13040642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadwiger JA, Wittenberg C, Richardson HE, de Barros Lopes M, Reed SI. 1989. A family of cyclin homologs that control the G1 phase in yeast. Proc Natl Acad Sci U S A. 86(16):6255–6259. doi: 10.1073/pnas.86.16.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall RA, De Sordi L, Maccallum DM, Topal H, Eaton R, Bloor JW, Robinson GK, Levin LR, Buck J, Wang Y, et al. 2010. CO(2) acts as a signalling molecule in populations of the fungal pathogen Candida albicans. PLoS Pathog. 6(11):e1001193. doi: 10.1371/journal.ppat.1001193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme A, Bumgarner S, Styles C, Fink GR. 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 116(3):405–415. doi: 10.1016/S0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]
- Hassan Y, Chew SY, Than LTL. 2021. Candida glabrata: pathogenicity and resistance mechanisms for adaptation and survival. J Fungi (Basel). 7(8):667. doi: 10.3390/jof7080667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haueisen J, Stukenbrock EH. 2016. Life cycle specialization of filamentous pathogens—colonization and reproduction in plant tissues. Curr Opin Microbiol. 32:31–37. doi: 10.1016/j.mib.2016.04.015. [DOI] [PubMed] [Google Scholar]
- Heaton L, Obara B, Grau V, Jones N, Nakagaki T, Boddy L, Fricker MD. 2012. Analysis of fungal networks. Fungal Biol Rev. 26(1):12–29. doi: 10.1016/j.fbr.2012.02.001. [DOI] [Google Scholar]
- Heise B, van der Felden J, Kern S, Malcher M, Brückner S, Mösch H-U. 2010. The TEA transcription factor Tec1 confers promoter-specific gene regulation by Ste12-dependent and -independent mechanisms. Eukaryot Cell. 9(4):514–531. doi: 10.1128/EC.00251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. 1995. MAP kinase pathways in yeast: for mating and more. Cell. 80(2):187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Hickman MA, Froyd CA, Rusche LN. 2011. Reinventing heterochromatin in budding yeasts: Sir2 and the origin recognition complex take center stage. Eukaryot Cell. 10(9):1183–1192. doi: 10.1128/EC.05123-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch CL, Coban Akdemir Z, Wang L, Jayakumaran G, Trcka D, Weiss A, Hernandez JJ, Pan Q, Han H, Xu X, et al. 2015. Myc and SAGA rewire an alternative splicing network during early somatic cell reprogramming. Genes Dev. 29(8):803–816. doi: 10.1101/gad.255109.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger CT. 2013. Saccharomyces diversity and evolution: a budding model genus. Trends Genet. 29(5):309–317. doi: 10.1016/j.tig.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Hogan DA, Sundstrom P. 2009. The Ras/cAMP/PKA signaling pathway and virulence in Candida albicans. Future Microbiol. 4(10):1263–1270. doi: 10.2217/fmb.09.106. [DOI] [PubMed] [Google Scholar]
- Hollomon JM, Grahl N, Willger SD, Koeppen K, Hogan DA. 2016. Global role of cyclic AMP signaling in pH-dependent responses in Candida albicans. mSphere. 1(6):e00283–e00216. doi: 10.1128/mSphere.00283-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Shang J, Sun Y, Tang G, Wang C. 2024. Fungal infection of insects: molecular insights and prospects. Trends Microbiol. 32(3):302–316. doi: 10.1016/j.tim.2023.09.005. [DOI] [PubMed] [Google Scholar]
- Hoyer LL. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9(4):176–180. doi: 10.1016/S0966-842X(01)01984-9. [DOI] [PubMed] [Google Scholar]
- Huang D, Friesen H, Andrews B. 2007. Pho85, a multifunctional cyclin-dependent protein kinase in budding yeast. Mol Microbiol. 66(2):303–314. doi: 10.1111/j.1365-2958.2007.05914.x. [DOI] [PubMed] [Google Scholar]
- Huang G. 2012. Regulation of phenotypic transitions in the fungal pathogen Candida albicans. Virulence. 3(3):251–261. doi: 10.4161/viru.20010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Huang Q, Wei Y, Wang Y, Du H. 2019. Multiple roles and diverse regulation of the Ras/cAMP/protein kinase A pathway in Candida albicans. Mol Microbiol. 111(1):6–16. doi: 10.1111/mmi.14148. [DOI] [PubMed] [Google Scholar]
- Hube B. 1996. Candida albicans secreted aspartyl proteinases. Curr Top Med Mycol. 7:55–69. Available from https://pubmed.ncbi.nlm.nih.gov/9504059/. [PubMed] [Google Scholar]
- Inglis DO, Sherlock G. 2013. Ras signaling gets fine-tuned: regulation of multiple pathogenic traits of Candida albicans. Eukaryot Cell. 12(10):1316–1325. doi: 10.1128/EC.00094-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam W, Adnan M, Shabbir A, Naveed H, Abubakar YS, Qasim M, Tayyab M, Noman A, Nisar MS, Khan KA, et al. 2021. Insect-fungal-interactions: a detailed review on entomopathogenic fungi pathogenicity to combat insect pests. Microb Pathog. 159:105122. doi: 10.1016/j.micpath.2021.105122. [DOI] [PubMed] [Google Scholar]
- Jarvis MC. 1984. Structure and properties of pectin gels in plant cell walls. Plant Cell Environ. 7(3):153–164. doi: 10.1111/1365-3040.ep11614586. [DOI] [Google Scholar]
- Jayani RS, Saxena S, Gupta R. 2005. Microbial pectinolytic enzymes: a review. Process Biochem. 40(9):2931–2944. doi: 10.1016/j.procbio.2005.03.026. [DOI] [Google Scholar]
- Jeong S, Ahn J, Kwon AR, Ha NC. 2020. Cleavage-dependent activation of ATP-dependent protease HslUV from Staphylococcus aureus. Mol Cells. 43(8):694–704. doi: 10.14348/molcells.2020.0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Xiang M, Liu X. 2017. Nematode-trapping fungi. Microbiol Spectr. 5(1):10–1128. doi: 10.1128/microbiolspec.FUNK-0022-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin K, Han L, Xia Y. 2014. Mamk1, a FUS3/KSS1-type mitogen-activated protein kinase gene, is required for appressorium formation, and insect cuticle penetration of the entomopathogenic fungus Metarhizium acridum. J Invertebr Pathol. 115:68–75. doi: 10.1016/j.jip.2013.10.014. [DOI] [PubMed] [Google Scholar]
- Jin R, Dobry CJ, McCown PJ, Kumar A. 2008. Large-scale analysis of yeast filamentous growth by systematic gene disruption and overexpression. Mol Biol Cell. 19(1):284–296. doi: 10.1091/mbc.e07-05-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Hansson GC. 2016. Immunological aspects of intestinal mucus and mucins. Nat Rev Immunol. 16(10):639–649. doi: 10.1038/nri.2016.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD. 2017. The rewiring of transcription circuits in evolution. Curr Opin Genet Dev. 47:121–127. doi: 10.1016/j.gde.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DI. 1999. Cdc42: an essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol Mol Biol Rev. 63(1):54–105. doi: 10.1128/MMBR.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DI, Pringle JR. 1990. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 111(1):143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh D, Johnson AD. 2005. Induction of the Candida albicans filamentous growth program by relief of transcriptional repression: a genome-wide analysis. Mol Biol Cell. 16(6):2903–2912. doi: 10.1091/mbc.e05-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahmann R, Kämper J. 2004. Ustilago maydis: how its biology relates to pathogenic development. New Phytol. 164(1):31–42. doi: 10.1111/j.1469-8137.2004.01156.x. [DOI] [PubMed] [Google Scholar]
- Karunanithi S, Cullen PJ. 2012. The filamentous growth MAPK pathway responds to glucose starvation through the Mig1/2 transcriptional repressors in Saccharomyces cerevisiae. Genetics. 192(3):869–887. doi: 10.1534/genetics.112.142661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunanithi S, Vadaie N, Chavel CA, Birkaya B, Joshi J, Grell L, Cullen PJ. 2010. Shedding of the mucin-like flocculin Flo11p reveals a new aspect of fungal adhesion regulation. Curr Biol. 20(15):1389–1395. doi: 10.1016/j.cub.2010.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur R, Ma B, Cormack BP. 2007. A family of glycosylphosphatidylinositol-linked aspartyl proteases is required for virulence of Candida glabrata. Proc Natl Acad Sci U S A. 104(18):7628–7633. doi: 10.1073/pnas.0611195104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh NL, Zhang AQ, Nobile CJ, Johnson AD, Ribbeck K. 2014. Mucins suppress virulence traits of Candida albicans. mBio. 5(6):e01911. doi: 10.1128/mBio.01911-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayikci O, Magwene PM. 2018. Divergent roles for cAMP-PKA signaling in the regulation of filamentous growth in Saccharomyces cerevisiae and Saccharomyces bayanus. G3 (Bethesda). 8(11):3529–3538. doi: 10.1534/g3.118.200413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MT, MacCallum DM, Clancy SD, Odds FC, Brown AJP, Butler G. 2004. The Candida albicans CaACE2 gene affects morphogenesis, adherence and virulence. Mol Microbiol. 53(3):969–983. doi: 10.1111/j.1365-2958.2004.04185.x. [DOI] [PubMed] [Google Scholar]
- Kennedy MJ, Rogers AL, Yancey RJ Jr. 1989. Environmental alteration and phenotypic regulation of Candida albicans adhesion to plastic. Infect Immun. 57(12):3876–3881. doi: 10.1128/iai.57.12.3876-3881.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Lee KT, Bahn YS. 2023. Secreted aspartyl protease 3 regulated by the Ras/cAMP/PKA pathway promotes the virulence of Candida auris. Front Cell Infect Microbiol. 13:1257897. doi: 10.3389/fcimb.2023.1257897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King L, Butler G. 1998. Ace2p, a regulator of CTS1 (chitinase) expression, affects pseudohyphal production in Saccharomyces cerevisiae. Curr Genet. 34(3):183–191. doi: 10.1007/s002940050384. [DOI] [PubMed] [Google Scholar]
- Kiss E, Hegedüs B, Virágh M, Varga T, Merényi Z, Kószó T, Bálint B, Prasanna AN, Krizsán K, Kocsubé S, et al. 2019. Comparative genomics reveals the origin of fungal hyphae and multicellularity. Nat Commun. 10(1):4080. doi: 10.1038/s41467-019-12085-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A, Zimmerman E, Schneider M, Hurt E, Zheng N. 2010. Structural basis for assembly and activation of the heterotetrameric SAGA histone H2B deubiquitinase module. Cell. 141(4):606–617. doi: 10.1016/j.cell.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler T, Wesche S, Taheri N, Braus GH, Mosch HU. 2002. Dual role of the Saccharomyces cerevisiae TEA/ATTS family transcription factor Tec1p in regulation of gene expression and cellular development. Eukaryot Cell. 1(5):673–686. doi: 10.1128/EC.1.5.673-686.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolattukudy PE, Rogers LM, Li D, Hwang CS, Flaishman MA. 1995. Surface signaling in pathogenesis. Proc Natl Acad Sci U S A. 92(10):4080–4087. doi: 10.1073/pnas.92.10.4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschwanez JH, Foster KR, Murray AW. 2011. Sucrose utilization in budding yeast as a model for the origin of undifferentiated multicellularity. PLoS Biol. 9(8):e1001122. doi: 10.1371/journal.pbio.1001122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer IM. 2016. Signal Transduction. London: Elsevier/Academic Press. [Google Scholar]
- Kraushaar T, Brückner S, Veelders M, Rhinow D, Schreiner F, Birke R, Pagenstecher A, Mösch H-U, Essen L-O. 2015. Interactions by the fungal Flo11 adhesin depend on a fibronectin type III-like adhesin domain girdled by aromatic bands. Structure. 23(6):1005–1017. doi: 10.1016/j.str.2015.03.021. [DOI] [PubMed] [Google Scholar]
- Krens SF, Spaink HP, Snaar-Jagalska BE. 2006. Functions of the MAPK family in vertebrate-development. FEBS Lett. 580(21):4984–4990. doi: 10.1016/j.febslet.2006.08.025. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Greenblatt JF. 2001. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol Cell Biol. 21(23):8203–8212. doi: 10.1128/MCB.21.23.8203-8212.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad J, De Maria A, Funnell D, Laidlaw RD, Lee N, de Sá Mário M, Ramesh M. 1998. Signaling via cAMP in fungi: interconnections with mitogen-activated protein kinase pathways. Arch Microbiol. 170(6):395–404. doi: 10.1007/s002030050659. [DOI] [PubMed] [Google Scholar]
- Krysan DJ, Ting EL, Abeijon C, Kroos L, Fuller RS. 2005. Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryot Cell. 4(8):1364–1374. doi: 10.1128/EC.4.8.1364-1374.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchin S, Vyas VK, Carlson M. 2002. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Mol Cell Biol. 22(12):3994–4000. doi: 10.1128/MCB.22.12.3994-4000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. 2021. The complex genetic basis and multilayered regulatory control of yeast pseudohyphal growth. Annu Rev Genet. 55(1):1–21. doi: 10.1146/annurev-genet-071719-020249. [DOI] [PubMed] [Google Scholar]
- Kunkel J, Luo X, Capaldi AP. 2019. Integrated TORC1 and PKA signaling control the temporal activation of glucose-induced gene expression in yeast. Nat Commun. 10(1):3558. doi: 10.1038/s41467-019-11540-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunyeit L, Kurrey NK, Anu-Appaiah KA, Rao RP. 2019. Probiotic yeasts inhibit virulence of non-albicans Candida species. mBio. 10(5):e02307–e02319. doi: 10.1128/mbio.02307-02319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunyeit L, Kurrey NK, Anu-Appaiah KA, Rao RP. 2021. Secondary metabolites from food-derived yeasts inhibit virulence of Candida albicans. mBio. 12(4):e0189121. doi: 10.1128/mBio.01891-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C-Y, Chen S-A, Hsueh Y-P. 2020. The high osmolarity glycerol (HOG) pathway functions in osmosensing, trap morphogenesis and conidiation of the nematode-trapping fungus Arthrobotrys oligospora. J Fungi (Basel). 6(4):91. doi: 10.3390/jof6040191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani SK, Grunstein M. 2003. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol. 4(4):276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- Laloux I, Dubois E, Dewerchin M, Jacobs E. 1990. TEC1, a gene involved in the activation of Ty1 and Ty1-mediated gene expression in Saccharomyces cerevisiae: cloning and molecular analysis. Mol Cell Biol. 10(7):3541–3550. doi: 10.1128/mcb.10.7.3541-3550.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb TM, Mitchell AP. 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol Cell Biol. 23(2):677–686. doi: 10.1128/MCB.23.2.677-686.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambou K, Tharreau D, Kohler A, Sirven C, Marguerettaz M, Barbisan C, Sexton AC, Kellner EM, Martin F, Howlett BJ, et al. 2008. Fungi have three tetraspanin families with distinct functions. BMC Genomics. 9(1):63. doi: 10.1186/1471-2164-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts MG, Bauer FF, Marmur J, Pretorius IS. 1996. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc Natl Acad Sci U S A. 93(16):8419–8424. doi: 10.1073/pnas.93.16.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange L, Pilgaard B, Herbst F-A, Busk PK, Gleason F, Pedersen AG. 2019. Origin of fungal biomass degrading enzymes: evolution, diversity and function of enzymes of early lineage fungi. Fungal Biol Rev. 33(1):82–97. doi: 10.1016/j.fbr.2018.09.001. [DOI] [Google Scholar]
- Lanver D, Berndt P, Tollot M, Naik V, Vranes M, Warmann T, Münch K, Rössel N, Kahmann R. 2014. Plant surface cues prime Ustilago maydis for biotrophic development. PLoS Pathog. 10(7):e1004272. doi: 10.1371/journal.ppat.1004272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanver D, Mendoza-Mendoza A, Brachmann A, Kahmann R. 2010. Sho1 and Msb2-related proteins regulate appressorium development in the smut fungus Ustilago maydis. Plant Cell. 22(6):2085–2101. doi: 10.1105/tpc.109.073734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. 2012. mTOR signaling in growth control and disease. Cell. 149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante M, Sabtini DM. 2009. mTOR signaling at a glance. J Cell Sci. 122(20):3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BN, Elion EA. 1999. The MAPKKK Ste11 regulates vegetative growth through a kinase cascade of shared signaling components. Proc Natl Acad Sci U S A. 96(22):12679–12684. doi: 10.1073/pnas.96.22.12679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Dohlman HG. 2008. Coactivation of G protein signaling by cell-surface receptors and an intracellular exchange factor. Curr Biol. 18(3):211–215. doi: 10.1016/j.cub.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Kronstad J. 2002. Ras2 controls morphogenesis, pheromone response, and pathogenicity in the fungal pathogen Ustilago maydis. Eukaryotic Cell. 1(6):954–966. doi: 10.1128/EC.1.6.954-966.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Mulugu S, York JD, O'Shea EK. 2007. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 316(5821):109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuw T, Fourest-Lieuvin A, Wu C, Chenevert J, Clark K, Whiteway M, Thomas DY, Leberer E. 1995. Pheromone response in yeast: association of Bem1p with proteins of the MAP kinase cascade and actin. Science. 270(5239):1210–1213. doi: 10.1126/science.270.5239.1210. [DOI] [PubMed] [Google Scholar]
- Lemaire K, Van de Velde S, Van Dijck P, Thevelein JM. 2004. Glucose and sucrose act as agonist and mannose as antagonist ligands of the G protein-coupled receptor Gpr1 in the yeast Saccharomyces cerevisiae. Mol Cell. 16(2):293–299. doi: 10.1016/j.molcel.2004.10.004. [DOI] [PubMed] [Google Scholar]
- León-Ramírez CG, Sánchez-Arreguin JA, Cabrera-Ponce JL, Martínez-Soto D, Ortiz-Castellanos ML, Aréchiga-Carvajal ET, Salazar-Chávez MF, Sánchez-Segura L, Ruiz-Herrera J. 2022. Tec1, a member of the TEA transcription factors family, is involved in virulence and basidiocarp development in Ustilago maydis. Int Microbiol. 25(1):17–26. doi: 10.1007/s10123-021-00188-8. [DOI] [PubMed] [Google Scholar]
- Leroch M, Mueller N, Hinsenkamp I, Hahn M. 2015. The signalling mucin Msb2 regulates surface sensing and host penetration via BMP1 MAP kinase signalling in Botrytis cinerea. Mol Plant Pathol. 16(8):787–798. doi: 10.1111/mpp.12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DE. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 69(2):262–291. doi: 10.1128/MMBR.69.2.262-291.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Chen Y, Tian S. 2022. Function of pH-dependent transcription factor PacC in regulating development, pathogenicity, and mycotoxin biosynthesis of phytopathogenic fungi. FEBS J. 289(7):1723–1730. doi: 10.1111/febs.15808. [DOI] [PubMed] [Google Scholar]
- Li W, Mitchell AP. 1997. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics. 145(1):63–73. doi: 10.1093/genetics/145.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang X, Hu S, Liu H, Xu J-R. 2017. PKA activity is essential for relieving the suppression of hyphal growth and appressorium formation by MoSfl1 in Magnaporthe oryzae. PLoS Genet. 13(8):e1006954. doi: 10.1371/journal.pgen.1006954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S-H, Sircaik S, Dainis J, Kakade P, Penumutchu S, McDonough LD, Chen Y-H, Frazer C, Schille TB, Allert S, et al. 2024. The hyphal-specific toxin candidalysin promotes fungal gut commensalism. Nature. 627(8004):620–627. doi: 10.1038/s41586-024-07142-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist W. 1952. Cell surface constituents and yeast flocculation. Nature. 170(4326):544–545. doi: 10.1038/170544a0. [DOI] [PubMed] [Google Scholar]
- Lipke PN. 2018. What we do not know about fungal cell adhesion molecules. J Fungi. 4(2):59. doi: 10.3390/jof4020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipke PN, Garcia MC, Alsteens D, Ramsook CB, Klotz SA, Dufrêne YF. 2012. Strengthening relationships: amyloids create adhesion nanodomains in yeasts. Trends Microbiol. 20(2):59–65. doi: 10.1016/j.tim.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipke PN, Klotz SA, Dufrene YF, Jackson DN, Garcia-Sherman MC. 2018. Amyloid-like β-aggregates as force-sensitive switches in fungal biofilms and infections. Microbiol Mol Biol Rev. 82(1):e00035–e00017. doi: 10.1128/MMBR.00035-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G. 2015. The fascinating and secret wild life of the budding yeast S. cerevisiae. Elife. 4:e05835. doi: 10.7554/eLife.05835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Styles CA, Fink GR. 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 262(5140):1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- Liu W, Zhou X, Li G, Li L, Kong L, Wang C, Zhang H, Xu J-R. 2011. Multiple plant surface signals are sensed by different mechanisms in the rice blast fungus for appressorium formation. PLoS Pathog. 7(1):e1001261. doi: 10.1371/journal.ppat.1001261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Dranginis AM. 1996. FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. J Bacteriol. 178(24):7144–7151. doi: 10.1128/jb.178.24.7144-7151.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo WS, Dranginis AM. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol Biol Cell. 9(1):161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MC, Cutler NS, Heitman J. 2000. Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol Biol Cell. 11(1):183–199. doi: 10.1091/mbc.11.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Su C, Wang A, Liu H. 2011. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS Biol. 9(7):e1001105. doi: 10.1371/journal.pbio.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Cook JG, Thorner J. 1995. Phosphorylation and localization of Kss1, a MAP kinase of the Saccharomyces cerevisiae pheromone response pathway. Mol Biol Cell. 6(7):889–909. doi: 10.1091/mbc.6.7.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean CR, Brandon C. 2008. Stable public goods cooperation and dynamic social interactions in yeast. J Evol Biol. 21(6):1836–1843. doi: 10.1111/j.1420-9101.2008.01579.x. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Fink GR. 1997. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 275(5304):1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Galitski T, Lander ES, Fink GR. 1999. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc Natl Acad Sci U S A. 96(22):12530–12535. doi: 10.1073/pnas.96.22.12530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Styles CA, Fink GR. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell. 91(5):673–684. doi: 10.1016/S0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- Maeda T, Takekawa M, Saito H. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science. 269(5223):554–558. doi: 10.1126/science.7624781. [DOI] [PubMed] [Google Scholar]
- Martin H, Kavanagh K, Velasco-Torrijos T. 2021. Targeting adhesion in fungal pathogen Candida albicans. Future Med Chem. 13(3):313–334. doi: 10.4155/fmc-2020-0052. [DOI] [PubMed] [Google Scholar]
- Martínez-Espinoza AD, Ruiz-Herrera J, León-Ramírez CG, Gold SE. 2004. MAP kinase and cAMP signaling pathways modulate the pH-induced yeast-to-mycelium dimorphic transition in the corn smut fungus Ustilago maydis. Curr Microbiol. 49(4):274–281. doi: 10.1007/s00284-004-4315-6. [DOI] [PubMed] [Google Scholar]
- Martínez-Pastor MT, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, Estruch F. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15(9):2227–2235. doi: 10.1002/j.1460-2075.1996.tb00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Soto D, Ruiz-Herrera J. 2017. Functional analysis of the MAPK pathways in fungi. Rev Iberoam Micol. 34(4):192–202. doi: 10.1016/j.riam.2017.02.006. [DOI] [PubMed] [Google Scholar]
- Mayorga ME, Gold SE. 1999. A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol Microbiol. 34(3):485–497. doi: 10.1046/j.1365-2958.1999.01610.x. [DOI] [PubMed] [Google Scholar]
- McDonald CM, Wagner M, Dunham MJ, Shin ME, Ahmed NT, Winter E. 2009. The Ras/cAMP pathway and the CDK-like kinase Ime2 regulate the MAPK Smk1 and spore morphogenesis in Saccharomyces cerevisiae. Genetics. 181(2):511–523. doi: 10.1534/genetics.108.098434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meem MH, Cullen PJ. 2012. The impact of protein glycosylation on Flo11-dependent adherence in Saccharomyces cerevisiae. FEMS Yeast Res. 12(7):809–818. doi: 10.1111/j.1567-1364.2012.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendgen K, Hahn M, Deising H. 1996. Morphogenesis and mechanisms of penetration by plant pathogenic fungi. Annu Rev Phytopathol. 34(1):367–386. doi: 10.1146/annurev.phyto.34.1.367. [DOI] [PubMed] [Google Scholar]
- Mill PJ. 1964. The nature of the interactions between flocculent cells in the flocculation of Saccharomyces cerevisiae. J Gen Microbiol. 35(1):61–68. doi: 10.1099/00221287-35-1-61. [DOI] [PubMed] [Google Scholar]
- Mira NP, Lourenco AB, Fernandes AR, Becker JD, Sa-Correia I. 2009. The RIM101 pathway has a role in Saccharomyces cerevisiae adaptive response and resistance to propionic acid and other weak acids. FEMS Yeast Res. 9(2):202–216. doi: 10.1111/j.1567-1364.2008.00473.x. [DOI] [PubMed] [Google Scholar]
- Miyazaki Y, Kaneko S, Sunagawa M, Shishido K, Yamazaki T, Nakamura M, Babasaki K. 2007. The fruiting-specific Le.flp1 gene, encoding a novel fungal fasciclin-like protein, of the basidiomycetous mushroom Lentinula edodes. Curr Genet. 51(6):367–375. doi: 10.1007/s00294-007-0133-2. [DOI] [PubMed] [Google Scholar]
- Money NP. 2016. Fungi: A Very Short Introduction. Oxford: Oxford University Press. Available from https://archive.org/details/fungiveryshortin0000mone. [Google Scholar]
- Monod M, Togni G, Hube B, Sanglard D. 1994. Multiplicity of genes encoding secreted aspartic proteinases in Candida species. Mol Microbiol. 13(2):357–368. doi: 10.1111/j.1365-2958.1994.tb00429.x. [DOI] [PubMed] [Google Scholar]
- Montelongo-Jauregui D, Saville SP, Lopez-Ribot JL. 2019. Contributions of Candida albicans dimorphism, adhesive interactions, and extracellular matrix to the formation of dual-species biofilms with Streptococcus gordonii. mBio. 10(3):e01179–e01119. doi: 10.1128/mBio.01179-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morishita T, Mitsuzawa H, Nakafuku M, Nakamura S, Hattori S, Anraku Y. 1995. Requirement of Saccharomyces cerevisiae Ras for completion of mitosis. Science. 270(5239):1213–1215. doi: 10.1126/science.270.5239.1213. [DOI] [PubMed] [Google Scholar]
- Morschhäuser J. 2011. Nitrogen regulation of morphogenesis and protease secretion in Candida albicans. Int J Med Microbiol. 301(5):390–394. doi: 10.1016/j.ijmm.2011.04.005. [DOI] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. 1986. Genealogy of principal strains of the yeast genetic stock center. Genetics. 113(1):35–43. doi: 10.1093/genetics/113.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosch HU, Kubler E, Krappmann S, Fink GR, Braus GH. 1999. Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol Biol Cell. 10(5):1325–1335. doi: 10.1091/mbc.10.5.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosch HU, Roberts RL, Fink GR. 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 93(11):5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouriño-Pérez RR. 2013. Septum development in filamentous ascomycetes. Fungal Biol Rev. 27(1):1–9. doi: 10.1016/j.fbr.2013.02.002. [DOI] [Google Scholar]
- Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Höfs S, Gratacap RL, Robbins J, Runglall M, et al. 2016. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature. 532(7597):64–68. doi: 10.1038/nature17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller O, Kahmann R, Aguilar G, Trejo-Aguilar B, Wu A, de Vries RP. 2008. The secretome of the maize pathogen Ustilago maydis. Fungal Genet Biol. 45(suppl 1):63–70. doi: 10.1016/j.fgb.2008.03.012. [DOI] [PubMed] [Google Scholar]
- Muller EM, Mackin NA, Erdman SE, Cunningham KW. 2003. Fig 1p facilitates Ca2+ influx and cell fusion during mating of Saccharomyces cerevisiae. J Biol Chem. 278(40):38461–38469. doi: 10.1074/jbc.M304089200. [DOI] [PubMed] [Google Scholar]
- Murad AM, Leng P, Straffon M, Wishart J, Macaskill S, MacCallum D, Schnell N, Talibi D, Marechal D, Tekaia F, et al. 2001. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans. EMBO J. 20(17):4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonakis E, Casadevall A, Ausubel FM. 2007. Exploiting amoeboid and non-vertebrate animal model systems to study the virulence of human pathogenic fungi. PLoS Pathog. 3(7):e101. doi: 10.1371/journal.ppat.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naglik J, Albrecht A, Bader O, Hube B. 2004. Candida albicans proteinases and host/pathogen interactions. Cell Microbiol. 6(10):915–926. doi: 10.1111/j.1462-5822.2004.00439.x. [DOI] [PubMed] [Google Scholar]
- Neiman AM. 2011. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics. 189(3):737–765. doi: 10.1534/genetics.111.127126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C, Goto S, Lund K, Hung W, Sadowski I. 2003. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature. 421(6919):187–190. doi: 10.1038/nature01243. [DOI] [PubMed] [Google Scholar]
- Ng TS, Desa MNM, Sandai D, Chong PP, Than LTL. 2016. Growth, biofilm formation, antifungal susceptibility and oxidative stress resistance of Candida glabrata are affected by different glucose concentrations. Infect Genet Evol. 40:331–338. doi: 10.1016/j.meegid.2015.09.004. [DOI] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu Rev Genet. 43(1):197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Snyder M. 2001. A genomic study of the bipolar bud site selection pattern in Saccharomyces cerevisiae. Mol Biol Cell. 12(7):2147–2170. doi: 10.1091/mbc.12.7.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson KW, Atkin AL, Hornby JM. 2006. Quorum sensing in dimorphic fungi: farnesol and beyond. Appl Environ Microbiol. 72(6):3805–3813. doi: 10.1128/AEM.02765-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 148(1–2):126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Johnson AD. 2015. Candida albicans biofilms and human disease. Annu Rev Microbiol. 69(1):71–92. doi: 10.1146/annurev-micro-091014-104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile CJ, Mitchell AP. 2006. Genetics and genomics of Candida albicans biofilm formation. Cell Microbiol. 8(9):1382–1391. doi: 10.1111/j.1462-5822.2006.00761.x. [DOI] [PubMed] [Google Scholar]
- Nobile CJ, Solis N, Myers CL, Fay AJ, Deneault J-S, Nantel A, Mitchell AP, Filler SG. 2008. Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell Microbiol. 10(11):2180–2196. doi: 10.1111/j.1462-5822.2008.01198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, French S, Kohn LA, Chen V, Johnson AD. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 42(7):590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell S, Yue J-X, Saada OA, Agier N, Caradec C, Cokelaer T, De Chiara M, Delmas S, Dutreux F, Fournier T, et al. 2023. Telomere-to-telomere assemblies of 142 strains characterize the genome structural landscape in Saccharomyces cerevisiae. Nat Genet. 55(8):1390–1399. doi: 10.1038/s41588-023-01459-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke SM, Herskowitz I. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12(18):2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz-Urquiza A, Keyhani NO. 2013. Action on the surface: entomopathogenic fungi versus the insect cuticle. Insects. 4(3):357–374. doi: 10.3390/insects4030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanca L, Gaskett AC, Gunther CS, Newcomb RD, Goddard MR. 2013. Quantifying variation in the ability of yeasts to attract Drosophila melanogaster. PLoS One. 8(9):e75332. doi: 10.1371/journal.pone.0075332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palecek SP, Parikh AS, Kron SJ. 2000. Genetic analysis reveals that FLO11 upregulation and cell polarization independently regulate invasive growth in Saccharomyces cerevisiae. Genetics. 156(3):1005–1023. doi: 10.1093/genetics/156.3.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Heitman J. 1999. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol Cell Biol. 19(7):4874–4887. doi: 10.1128/MCB.19.7.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchapakesan A, Shankar N. 2016. Chapter 2—Fungal cellulases: an overview. In: Gupta VK, editor. New and Future Developments in Microbial Biotechnology and Bioengineering. Amsterdam (The Netherlands): Elsevier. p. 9–18. [Google Scholar]
- Pandey A, Roca MG, Read ND, Glass NL. 2004. Role of a mitogen-activated protein kinase pathway during conidial germination and hyphal fusion in Neurospora crassa. Eukaryotic Cell. 3(2):348–358. doi: 10.1128/EC.3.2.348-358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HO, Bi E, Pringle JR, Herskowitz I. 1997. Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc Natl Acad Sci U S A. 94(9):4463–4468. doi: 10.1073/pnas.94.9.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel PK, Free SJ. 2019. The genetics and biochemistry of cell wall structure and synthesis in Neurospora crassa, a model filamentous fungus. Front Microbiol. 10:2294. doi: 10.3389/fmicb.2019.02294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Grishin NV. 2017. Expansion of divergent SEA domains in cell surface proteins and nucleoporin 54. Protein Sci. 26(3):617–630. doi: 10.1002/pro.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg AY, Tampakakis E, Fuchs BB, Eliopoulos GM, Moellering RC, Mylonakis E. 2008. Prokaryote-eukaryote interactions identified by using Caenorhabditis elegans. Proc Natl Acad Sci U S A. 105(38):14585–14590. doi: 10.1073/pnas.0805048105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalva Miguel A, Arst Herbert N. 2002. Regulation of gene expression by ambient pH in filamentous fungi and yeasts. Microbiol Mol Biol Rev. 66(3):426–446. doi: 10.1128/MMBR.66.3.426-446.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentland DR, Davis J, Mühlschlegel FA, Gourlay CW. 2021. CO2 enhances the formation, nutrient scavenging and drug resistance properties of C. albicans biofilms. NPJ Biofilms Microbiomes. 7(1):67. doi: 10.1038/s41522-021-00238-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira R, dos Santos Fontenelle RO, de Brito EHS, de Morais SM. 2021. Biofilm of Candida albicans: formation, regulation and resistance. J Appl Microbiol. 131(1):11–22. doi: 10.1111/jam.14949. [DOI] [PubMed] [Google Scholar]
- Perez-Nadales E, Di Pietro A. 2015. The transmembrane protein Sho1 cooperates with the mucin Msb2 to regulate invasive growth and plant infection in Fusarium oxysporum. Mol Plant Pathol. 16(6):593–603. doi: 10.1111/mpp.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris D, Ubbelohde EJ, Kuang MC, Kominek J, Langdon QK, Adams M, Koshalek JA, Hulfachor AB, Opulente DA, Hall DJ, et al. 2023. Macroevolutionary diversity of traits and genomes in the model yeast genus Saccharomyces. Nat Commun. 14(1):690. doi: 10.1038/s41467-023-36139-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter J, De Chiara M, Friedrich A, Yue J-X, Pflieger D, Bergström A, Sigwalt A, Barre B, Freel K, Llored A, et al. 2018. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature. 556(7701):339–344. doi: 10.1038/s41586-018-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Neiman AM, Park HO, van Lohuizen M, Herskowitz I. 1996. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 15(24):7046–7059. doi: 10.1002/j.1460-2075.1996.tb01096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petter R, Chang YC, Kwon-Chung KJ. 1997. A gene homologous to Saccharomyces cerevisiae SNF1 appears to be essential for the viability of Candida albicans. Infect Immun. 65(12):4909–4917. doi: 10.1128/iai.65.12.4909-4917.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadke SS, Maclean CJ, Zhao SY, Mueller EA, Michelotti LA, Norman KL, Kumar A, James TY. 2018. Genome-wide screen for Saccharomyces cerevisiae genes contributing to opportunistic pathogenicity in an invertebrate model host. G3 (Bethesda). 8(1):63–78. doi: 10.1534/g3.117.300245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskur J, Rozpedowska E, Polakova S, Merico A, Compagno C. 2006. How did Saccharomyces evolve to become a good brewer? Trends Genet. 22(4):183–186. doi: 10.1016/j.tig.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Pitoniak A, Chavel CA, Chow J, Smith J, Camara D, Karunanithi S, Li B, Wolfe KH, Cullen PJ. 2015. Cdc42p-interacting protein Bem4p regulates the filamentous-growth mitogen-activated protein kinase pathway. Mol Cell Biol. 35(2):417–436. doi: 10.1128/MCB.00850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank M. 2022. Interaction of TOR and PKA Signaling in S. cerevisiae. Biomolecules. 12(2):210. doi: 10.3390/biom12020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polizeli MDLT, Jorge JA, Terenzi HF. 1991. Pectinase production by Neurospora crassa: purification and biochemical characterization of extracellular polygalacturonase activity. Microbiology. 137:1815–1823. doi: 10.1099/00221287-137-8-1815. [DOI] [PubMed] [Google Scholar]
- Ponde NO, Lortal L, Ramage G, Naglik JR, Richardson JP. 2021. Candida albicans biofilms and polymicrobial interactions. Crit Rev Microbiol. 47(1):91–111. doi: 10.1080/1040841X.2020.1843400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Saito H. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science. 276(5319):1702–1705. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- Prusty R, Grisafi P, Fink GR. 2004. The plant hormone indoleacetic acid induces invasive growth in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 101(12):4153–4157. doi: 10.1073/pnas.0400659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A. 2000. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J Cell Sci. 113(3):365–375. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- Pukkila-Worley R, Ausubel FM, Mylonakis E. 2011. Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses. PLoS Pathog. 7(6):e1002074. doi: 10.1371/journal.ppat.1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri S, Edgerton M. 2014. How does it kill? Understanding the candidacidal mechanism of salivary histatin 5. Eukaryot Cell. 13(8):958–964. doi: 10.1128/EC.00095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri S, Kumar R, Chadha S, Tati S, Conti HR, Hube B, Cullen PJ, Edgerton M. 2012. Secreted aspartic protease cleavage of Candida albicans Msb2 activates Cek1 MAPK signaling affecting biofilm formation and oropharyngeal candidiasis. PLoS One. 7(11):e46020. doi: 10.1371/journal.pone.0046020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W, Acosta-Zaldivar M, Flanagan PR, Liu N-N, Jani N, Fierro JF, Andrés MT, Moran GP, Köhler JR. 2022. Stress- and metabolic responses of Candida albicans require Tor1 kinase N-terminal HEAT repeats. PLoS Pathog. 18(6):e1010089. doi: 10.1371/journal.ppat.1010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Tester RF. 2019. Fructose, galactose and glucose—in health and disease. Clin Nutr ESPEN. 33:18–28. doi: 10.1016/j.clnesp.2019.07.004. [DOI] [PubMed] [Google Scholar]
- Qin L, Li D, Zhao J, Yang G, Wang Y, Yang K, Tumukunde E, Wang S, Yuan J. 2021. The membrane mucin Msb2 regulates aflatoxin biosynthesis and pathogenicity in fungus Aspergillus flavus. Microb Biotechnol. 14(2):628–642. doi: 10.1111/1751-7915.13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing L, Li Q, Dong Z. 2022. MUC1: an emerging target in cancer treatment and diagnosis. Bull Cancer. 109(11):1202–1216. doi: 10.1016/j.bulcan.2022.08.001. [DOI] [PubMed] [Google Scholar]
- Qu Y, Wang J, Huang P, Liu X, Lu J, Lin F-C. 2021. Poral2 is involved in appressorium formation and virulence via pmk1 MAPK pathways in the rice blast fungus Pyricularia oryzae. Front Plant Sci. 12:702368. doi: 10.3389/fpls.2021.702368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan V, Aquadro CF, Alani E. 2019. Baker's yeast clinical isolates provide a model for how pathogenic yeasts adapt to stress. Trends Genet. 35(11):804–817. doi: 10.1016/j.tig.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raitt DC, Posas F, Saito H. 2000. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 19(17):4623–4631. doi: 10.1093/emboj/19.17.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramezani-Rad M. 2003. The role of adaptor protein Ste50-dependent regulation of the MAPKKK Ste11 in multiple signalling pathways of yeast. Curr Genet. 43(3):161–170. doi: 10.1007/s00294-003-0383-6. [DOI] [PubMed] [Google Scholar]
- Reinders A, Burckert N, Boller T, Wiemken A, De Virgilio C. 1998. Saccharomyces cerevisiae cAMP-dependent protein kinase controls entry into stationary phase through the Rim15p protein kinase. Genes Dev. 12(18):2943–2955. doi: 10.1101/gad.12.18.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Replansky T, Koufopanou V, Greig D, Bell G. 2008. Saccharomyces sensu stricto as a model system for evolution and ecology. Trends Ecol Evol. 23(9):494–501. doi: 10.1016/j.tree.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Reynolds TB. 2006. The Opi1p transcription factor affects expression of FLO11, mat formation, and invasive growth in Saccharomyces cerevisiae. Eukaryot Cell. 5(8):1266–1275. doi: 10.1128/EC.00022-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds TB. 2018. Going with the flo: the role of Flo11-dependent and independent interactions in yeast mat formation. J Fungi (Basel). 4(4):132. doi: 10.3390/jof4040132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme M. 2013. Tip growth in filamentous fungi: a road trip to the apex. Annu Rev Microbiol. 67(1):587–609. doi: 10.1146/annurev-micro-092412-155652. [DOI] [PubMed] [Google Scholar]
- Roberson RW. 2020. Subcellular structure and behaviour in fungal hyphae. J Microsc. 280(2):75–85. doi: 10.1111/jmi.12945. [DOI] [PubMed] [Google Scholar]
- Roberts RL, Fink GR. 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8(24):2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Roberts CJ, Nelson B, Marton MJ, Stoughton R, Meyer MR, Bennett HA, He YD, Dai H, Walker WL, Hughes TR, et al. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 287(5454):873–880. doi: 10.1126/science.287.5454.873. [DOI] [PubMed] [Google Scholar]
- Roberts RL, Mösch HU, Fink GR. 1997. 14-3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell. 89(7):1055–1065. doi: 10.1016/S0092-8674(00)80293-7. [DOI] [PubMed] [Google Scholar]
- Roberts SM, Winston F. 1997. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 147(2):451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollenhagen C, Mamtani S, Ma D, Dixit R, Eszterhas S, Lee SA. 2020. The role of secretory pathways in Candida albicans pathogenesis. J Fungi. 6(1):26. doi: 10.3390/jof6010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román E, Cottier F, Ernst Joachim F, Pla J. 2009. Msb2 signaling mucin controls activation of cek1 mitogen-activated protein kinase in Candida albicans. Eukaryot Cell. 8(8):1235–1249. doi: 10.1128/EC.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman H. 1986. The early days of yeast genetics: a personal narrative. Annu Rev Genet. 20(1):1–14. doi: 10.1146/annurev.ge.20.120186.000245. [DOI] [PubMed] [Google Scholar]
- Roncero C, Sanchez Y. 2010. Cell separation and the maintenance of cell integrity during cytokinesis in yeast: the assembly of a septum. Yeast. 27(8):521–530. doi: 10.1002/yea.1779. [DOI] [PubMed] [Google Scholar]
- Rosiana S, Zhang L, Kim GH, Revtovich AV, Uthayakumar D, Sukumaran A, Geddes-McAlister J, Kirienko NV, Shapiro RS. 2021. Comprehensive genetic analysis of adhesin proteins and their role in virulence of Candida albicans. Genetics. 217(2):iyab003. doi: 10.1093/genetics/iyab003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp S, Summers E, Lo HJ, Madhani H, Fink G. 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18(5):1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan O, Shapiro RS, Kurat CF, Mayhew D, Baryshnikova A, Chin B, Lin Z-Y, Cox MJ, Vizeacoumar F, Cheung D, et al. 2012. Global gene deletion analysis exploring yeast filamentous growth. Science. 337(6100):1353–1356. doi: 10.1126/science.1224339. [DOI] [PubMed] [Google Scholar]
- Ryder LS, Talbot NJ. 2015. Regulation of appressorium development in pathogenic fungi. Curr Opin Plant Biol. 26:8–13. doi: 10.1016/j.pbi.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safiya SN, Girija ASS, Priyadharsini VJ. 2023. Molecular detection of secreted aspartyl proteinases (Saps) from dental isolates of Candida albicans and targeting with Psidium guajava biocompounds: an in vitro and in silico analysis. Cureus. 15(11):e49143. doi: 10.7759/cureus.49143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Tatebayashi K. 2004. Regulation of the osmoregulatory HOG MAPK cascade in yeast. J Biochem. 136(3):267–272. doi: 10.1093/jb/mvh135. [DOI] [PubMed] [Google Scholar]
- Saraswat D, Kumar R, Pande T, Edgerton M, Cullen PJ. 2016. Signalling mucin Msb2 regulates adaptation to thermal stress in Candida albicans. Mol Microbiol. 100(3):425–441. doi: 10.1111/mmi.13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasani E, Khodavaisy S, Agha Kuchak Afshari S, Darabian S, Aala F, Rezaie S. 2016. Pseudohyphae formation in Candida glabrata due to CO2 exposure. Curr Med Mycol. 2(4):49–52. doi: 10.18869/acadpub.cmm.2.4.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzle T, Beck T, Martin DE, Hall MN. 2004. Activation of the RAS/cyclic AMP pathway suppresses a TOR deficiency in yeast. Mol Cell Biol. 24(1):338–351. doi: 10.1128/MCB.24.1.338-351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz HP, Philippsen P. 2011. Evolution of multinucleated Ashbya gossypii hyphae from a budding yeast-like ancestor. Fungal Biol. 115(6):557–568. doi: 10.1016/j.funbio.2011.02.015. [DOI] [PubMed] [Google Scholar]
- Schweizer A, Rupp S, Taylor BN, Rollinghoff M, Schroppel K. 2000. The TEA/ATTS transcription factor CaTec1p regulates hyphal development and virulence in Candida albicans. Mol Microbiol. 38(3):435–445. doi: 10.1046/j.1365-2958.2000.02132.x. [DOI] [PubMed] [Google Scholar]
- Seger R, Krebs EG. 1995. The MAPK signaling cascade. FASEB J. 9(9):726–735. doi: 10.1096/fasebj.9.9.7601337. [DOI] [PubMed] [Google Scholar]
- Selvig K, Alspaugh JA. 2011. Ph response pathways in fungi: adapting to host-derived and environmental signals. Mycobiology. 39(4):249–256. doi: 10.5941/MYCO.2011.39.4.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Hong S, Wang C. 2024. Fights on the surface prior to fungal invasion of insects. PLoS Pathog. 20(2):e1011994. doi: 10.1371/journal.ppat.1011994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, Cowen LE. 2010. Coupling temperature sensing and development. Virulence. 1(1):45–48. doi: 10.4161/viru.1.1.10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RS, Uppuluri P, Zaas AK, Collins C, Senn H, Perfect JR, Heitman J, Cowen LE. 2009. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr Biol. 19(8):621–629. doi: 10.1016/j.cub.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharmeen N, Sulea T, Whiteway M, Wu C. 2019. The adaptor protein Ste50 directly modulates yeast MAPK signaling specificity through differential connections of its RA domain. Mol Biol Cell. 30(6):794–807. doi: 10.1091/mbc.E18-11-0708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul YD, Seger R. 2007. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim Biophys Acta. 1773(8):1213–1226. doi: 10.1016/j.bbamcr.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Shen X-X, Opulente DA, Kominek J, Zhou X, Steenwyk JL, Buh KV, Haase MAB, Wisecaver JH, Wang M, Doering DT, et al. 2018. Tempo and mode of genome evolution in the budding yeast subphylum. Cell. 175(6):1533–1545.e20. doi: 10.1016/j.cell.2018.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinn-Thomas JH, Mohler WA. 2011. New insights into the mechanisms and roles of cell–cell fusion. Int Rev Cell Mol Biol. 289:149–209. doi: 10.1016/B978-0-12-386039-2.00005-5. [DOI] [PubMed] [Google Scholar]
- Shock TR, Thompson J, Yates JR III, Madhani HD. 2009. Hog1 mitogen-activated protein kinase (MAPK) interrupts signal transduction between the Kss1 MAPK and the Tec1 transcription factor to maintain pathway specificity. Eukaryot Cell. 8(4):606–616. doi: 10.1128/EC.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber B, Coronas-Serna JM, Martin SG. 2023. A focus on yeast mating: from pheromone signaling to cell-cell fusion. Semin Cell Dev Biol. 133:83–95. doi: 10.1016/j.semcdb.2022.02.003. [DOI] [PubMed] [Google Scholar]
- Silva-Dias A, Miranda IM, Branco J, Monteiro-Soares M, Pina-Vaz C, Rodrigues AG. 2015. Adhesion, biofilm formation, cell surface hydrophobicity, and antifungal planktonic susceptibility: relationship among Candida spp. Front Microbiol. 6:205. doi: 10.3389/fmicb.2015.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Gonzales S, Palacios J, Oliver G. 2004. Fungi: the fungal hypha. In: Robinson RK, Batt CA, Patel BD, editors. Encyclopedia of Food Microbiology. San Diego (CA): Academic Press. p. 850–853. Available from https://www.scribd.com/document/388916021/Encyclopedia-of-Food-Microbiology-Richard-K-Ed-Robinson-pdf. [Google Scholar]
- Simpson-Lavy K, Kupiec M. 2023. Glucose inhibits yeast AMPK (Snf1) by three independent mechanisms. Biology (Basel). 12(7):1007. doi: 10.3390/biology12071007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Bandyopadhyay D. 2007. MUC1: a target molecule for cancer therapy. Cancer Biol Ther. 6(4):481–486. doi: 10.4161/cbt.6.4.4201. [DOI] [PubMed] [Google Scholar]
- Smukalla S, Caldara M, Pochet N, Beauvais A, Guadagnini S, Yan C, Vinces MD, Jansen A, Prevost MC, Latgé J-P, et al. 2008. FLO1 is a variable green beard gene that drives biofilm-like cooperation in budding yeast. Cell. 135(4):726–737. doi: 10.1016/j.cell.2008.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa AM, Grandgenett PM, David L, Almeida R, Hollingsworth MA, Santos-Silva F. 2016. Reflections on MUC1 glycoprotein: the hidden potential of isoforms in carcinogenesis. APMIS. 124(11):913–924. doi: 10.1111/apm.12587. [DOI] [PubMed] [Google Scholar]
- Sriamornsak P, Wattanakorn N, Takeuchi H. 2010. Study on the mucoadhesion mechanism of pectin by atomic force microscopy and mucin-particle method. Carbohydr Polym. 79(1):54–59. doi: 10.1016/j.carbpol.2009.07.018. [DOI] [Google Scholar]
- Stefanini I, Dapporto L, Legras J-L, Calabretta A, Di Paola M, De Filippo C, Viola R, Capretti P, Polsinelli M, Turillazzi S, et al. 2012. Role of social wasps in Saccharomyces cerevisiae ecology and evolution. Proc Natl Acad Sci U S A. 109(33):13398–13403. doi: 10.1073/pnas.1208362109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su C, Yu J, Lu Y. 2018. Hyphal development in Candida albicans from different cell states. Curr Genet. 64(6):1239–1243. doi: 10.1007/s00294-018-0845-5. [DOI] [PubMed] [Google Scholar]
- Su Y-Y, Qi Y-L, Cai L. 2012. Induction of sporulation in plant pathogenic fungi. Mycology. 3(3):195–200. doi: 10.1080/21501203.2012.719042. [DOI] [Google Scholar]
- Sudbery PE. 2011. Growth of Candida albicans hyphae. Nat Rev Microbiol. 9(10):737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- Sulyanto RM, Beall CJ, Ha K, Montesano J, Juang J, Dickson JR, Hashmi SB, Bradbury S, Leys EJ, Edgerton M, et al. 2024. Fungi and bacteria occupy distinct spatial niches within carious dentin. PLoS Pathog. 20(5):e1011865. doi: 10.1371/journal.ppat.1011865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supruniuk K, Radziejewska I. 2021. MUC1 is an oncoprotein with a significant role in apoptosis (Review). Int J Oncol. 59(3):68. doi: 10.3892/ijo.2021.5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafranski-Schneider E, Swidergall M, Cottier F, Tielker D, Román E, Pla J, Ernst JF. 2012. Msb2 shedding protects Candida albicans against antimicrobial peptides. PLoS Pathog. 8(2):e1002501. doi: 10.1371/journal.ppat.1002501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri N, Kohler T, Braus GH, Mosch HU. 2000. Asymmetrically localized Bud8p and Bud9p proteins control yeast cell polarity and development. EMBO J. 19(24):6686–6696. doi: 10.1093/emboj/19.24.6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taj G, Agarwal P, Grant M, Kumar A. 2010. MAPK machinery in plants: recognition and response to different stresses through multiple signal transduction pathways. Plant Signal Behav. 5(11):1370–1378. doi: 10.4161/psb.5.11.13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi J, Aoki K, Turner BS, Lamont S, Lehoux S, Kavanaugh N, Gulati M, Valle Arevalo A, Lawrence TJ, Kim CY, et al. 2022. Mucin O-glycans are natural inhibitors of Candida albicans pathogenicity. Nat Chem Biol. 18(7):762–773. doi: 10.1038/s41589-022-01035-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Tatebayashi K, Nishimura A, Yamamoto K, Yang H-Y, Saito H. 2014. Yeast osmosensors Hkr1 and Msb2 activate the Hog1 MAPK cascade by different mechanisms. Sci Signal. 7(314):ra21. doi: 10.1126/scisignal.2004780. [DOI] [PubMed] [Google Scholar]
- Tatebayashi K, Tanaka K, Yang H-Y, Yamamoto K, Matsushita Y, Tomida T, Imai M, Saito H. 2007. Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. EMBO J. 26(15):3521–3533. doi: 10.1038/sj.emboj.7601796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebayashi K, Yamamoto K, Nagoya M, Takayama T, Nishimura A, Sakurai M, Momma T, Saito H. 2015. Osmosensing and scaffolding functions of the oligomeric four-transmembrane domain osmosensor Sho1. Nat Commun. 6(1):6975. doi: 10.1038/ncomms7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatebayashi K, Yamamoto K, Tanaka K, Tomida T, Maruoka T, Kasukawa E, Saito H. 2006. Adaptor functions of Cdc42, Ste50, and Sho1 in the yeast osmoregulatory HOG MAPK pathway. EMBO J. 25(13):3033–3044. doi: 10.1038/sj.emboj.7601192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JT, Harting R, Shalaby S, Kenerley CM, Braus GH, Horwitz BA. 2022. Adhesion as a focus in trichoderma–root interactions. J Fungi. 8(4):372. doi: 10.3390/jof8040372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoropoulos G, Carraway KL. 2007. Molecular signaling in the regulation of mucins. J Cell Biochem. 102(5):1103–1116. doi: 10.1002/jcb.21539. [DOI] [PubMed] [Google Scholar]
- Thirawong N, Kennedy RA, Sriamornsak P. 2008. Viscometric study of pectin–mucin interaction and its mucoadhesive bond strength. Carbohydr Polym. 71(2):170–179. doi: 10.1016/j.carbpol.2007.05.026. [DOI] [Google Scholar]
- Tian X, Ding H, Ke W, Wang L. 2021. Quorum sensing in fungal species. Annu Rev Microbiol. 75(1):449–469. doi: 10.1146/annurev-micro-060321-045510. [DOI] [PubMed] [Google Scholar]
- Tobouti PL, Casaroto AR, de Almeida RSC, de Paula Ramos S, Dionísio TJ, Porto VC, Santos CF, Lara VS. 2016. Expression of secreted aspartyl proteinases in an experimental model of Candida albicans-associated denture stomatitis. J Prosthodont. 25(2):127–134. doi: 10.1111/jopr.12285. [DOI] [PubMed] [Google Scholar]
- Toth ME, Vigh L, Santha M. 2014. Alcohol stress, membranes, and chaperones. Cell Stress Chaperones. 19(3):299–309. doi: 10.1007/s12192-013-0472-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevijano-Contador N, Zaragoza O. 2018. Immune response of Galleria mellonella against human fungal pathogens. J Fungi (Basel). 5(1):3. doi: 10.3390/jof5010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker SL, Talbot NJ. 2001. Surface attachment and pre-penetration stage development by plant pathogenic fungi. Annu Rev Phytopathol. 39(1):385–417. doi: 10.1146/annurev.phyto.39.1.385. [DOI] [PubMed] [Google Scholar]
- Vadaie N, Dionne H, Akajagbor DS, Nickerson SR, Krysan DJ, Cullen PJ. 2008. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J Cell Biol. 181(7):1073–1081. doi: 10.1083/jcb.200704079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallim MA, Miller KY, Miller BL. 2000. Aspergillus SteA (Sterile12-like) is a homeodomain-C2/H2-Zn+2 finger transcription factor required for sexual reproduction. Mol Microbiol. 36(2):290–301. doi: 10.1046/j.1365-2958.2000.01874.x. [DOI] [PubMed] [Google Scholar]
- van den Ackerveken GFJM, Dunn RM, Cozijnsen AJ, Vossen JPMJ, Van den Broek HWJ, De Wit PJGM. 1994. Nitrogen limitation induces expression of the avirulence gene avr9 in the tomato pathogen Cladosporium fulvum. Mol Gen Genet. 243(3):277–285. doi: 10.1007/BF00301063. [DOI] [PubMed] [Google Scholar]
- van der Felden J, Weisser S, Bruckner S, Lenz P, Mosch HU. 2014. The transcription factors Tec1 and Ste12 interact with coregulators Msa1 and Msa2 to activate adhesion and multicellular development. Mol Cell Biol. 34(12):2283–2293. doi: 10.1128/MCB.01599-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermeulen MD, Cullen PJ. 2020. New aspects of invasive growth regulation identified by functional profiling of MAPK pathway targets in Saccharomyces cerevisiae. Genetics. 216(1):95–116. doi: 10.1534/genetics.120.303369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermeulen MD, Cullen PJ. 2022. Gene by environment interactions reveal new regulatory aspects of signaling network plasticity. PLoS Genet. 18(1):e1009988. doi: 10.1371/journal.pgen.1009988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandermeulen MD, Cullen PJ. 2023. Ecological inducers of the yeast filamentous growth pathway reveal environment-dependent roles for pathway components. mSphere. 8(5):e0028423. doi: 10.1128/msphere.00284-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Drogen F, O’Rourke SM, Stucke VM, Jaquenoud M, Neiman AM, Peter M. 2000. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr Biol. 10(11):630–639. doi: 10.1016/S0960-9822(00)00511-X. [DOI] [PubMed] [Google Scholar]
- van Tetering G, Vooijs M. 2011. Proteolytic cleavage of Notch: “HIT and RUN”. Curr Mol Med. 11(4):255–269. doi: 10.2174/156652411795677972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veelders M, Brückner S, Ott D, Unverzagt C, Mösch H-U, Essen L-O. 2010. Structural basis of flocculin-mediated social behavior in yeast. Proc Natl Acad Sci U S A. 107(52):22511–22516. doi: 10.1073/pnas.1013210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazhahan V, Ma N, Pándy-Szekeres G, Kooistra AJ, Lee Y, Gloriam DE, Vaidehi N, Tate CG. 2021. Structure of the class D GPCR Ste2 dimer coupled to two G proteins. Nature. 589(7840):148–153. doi: 10.1038/s41586-020-2994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazhahan V, Ma N, Vaidehi N, Tate CG. 2022. Activation mechanism of the class D fungal GPCR dimer Ste2. Nature. 603(7902):743–748. doi: 10.1038/s41586-022-04498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma AH, Richardson JP, Zhou C, Coleman BM, Moyes DL, Ho J, Huppler AR, Ramani K, McGeachy MJ, Mufazalov IA, et al. 2017. Oral epithelial cells orchestrate innate type 17 responses to Candida albicans through the virulence factor candidalysin. Sci Immunol. 2(17):eaam8834. doi: 10.1126/sciimmunol.aam8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernay A, Schaub S, Guillas I, Bassilana M, Arkowitz RA. 2012. A steep phosphoinositide bis-phosphate gradient forms during fungal filamentous growth. J Cell Biol. 198(4):711–730. doi: 10.1083/jcb.201203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen KJ, Jansen A, Lewitter F, Fink GR. 2005. Intragenic tandem repeats generate functional variability. Nat Genet. 37(9):986–990. doi: 10.1038/ng1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila T, Romo JA, Pierce CG, McHardy SF, Saville SP, Lopez-Ribot JL. 2017. Targeting Candida albicans filamentation for antifungal drug development. Virulence. 8(2):150–158. doi: 10.1080/21505594.2016.1197444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voragen AGJ, Coenen G-J, Verhoef RP, Schols HA. 2009. Pectin, a versatile polysaccharide present in plant cell walls. Struct Chem. 20(2):263–275. doi: 10.1007/s11224-009-9442-z. [DOI] [Google Scholar]
- Walter P, Ron D. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 334(6059):1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wang L, Lai Y, Chen J, Cao X, Zheng W, Dong L, Zheng Y, Li F, Wei G, Wang S. 2023. The ASH1–PEX16 regulatory pathway controls peroxisome biogenesis for appressorium-mediated insect infection by a fungal pathogen. Proc Natl Acad Sci U S A. 120(4):e2217145120. doi: 10.1073/pnas.2217145120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu D, Tian C. 2021. Mucin Msb2 cooperates with the transmembrane protein Sho1 in various plant surface signal sensing and pathogenic processes in the poplar anthracnose fungus Colletotrichum gloeosporioides. Mol Plant Pathol. 22(12):1553–1573. doi: 10.1111/mpp.13126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Abu Irqeba A, Ayalew M, Suntay K. 2009. Sumoylation of transcription factor Tec1 regulates signaling of mitogen-activated protein kinase pathways in yeast. PLoS One. 4(10):e7456. doi: 10.1371/journal.pone.0007456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li R, Wang D, Qian B, Bian Z, Wei J, Wei X, Xu J-R. 2023. Regulation of symbiotic interactions and primitive lichen differentiation by UMP1 MAP kinase in Umbilicaria muhlenbergii. Nat Commun. 14(1):6972. doi: 10.1038/s41467-023-42675-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zheng X, Li G, Wang X. 2023. TORC1 signaling in fungi: from yeasts to filamentous fungi. Microorganisms. 11(1):218. doi: 10.3390/microorganisms11010218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland J. 2020. Sporulation in Ashbya gossypii. J Fungi. 6(3):157. doi: 10.3390/jof6030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler RT, Kupiec M, Magnelli P, Abeijon C, Fink GR. 2003. A Saccharomyces cerevisiae mutant with increased virulence. Proc Natl Acad Sci U S A. 100(5):2766–2770. doi: 10.1073/pnas.0437995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MA, Riles L, Cohen BA. 2009. A systematic screen for transcriptional regulators of the yeast cell cycle. Genetics. 181(2):435–446. doi: 10.1534/genetics.108.098145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M, Diggle SP, Greenberg EP. 2017. Progress in and promise of bacterial quorum sensing research. Nature. 551(7680):313–320. doi: 10.1038/nature24624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willats WGT, McCartney L, Mackie W, Knox JP. 2001. Pectin: cell biology and prospects for functional analysis. Plant Mol Biol. 47(1/2):9–27. doi: 10.1023/A:1010662911148. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Butler G. 2022. Mating-type switching in budding yeasts, from flip/flop inversion to cassette mechanisms. Microbiol Mol Biol Rev. 86(2):e0000721. doi: 10.1128/mmbr.00007-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 387(6634):708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Wong Sak Hoi J, Dumas B. 2010. Ste12 and Ste12-like proteins, fungal transcription factors regulating development and pathogenicity. Eukaryot Cell. 9(4):480–485. doi: 10.1128/EC.00333-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods B, Lew DJ. 2019. Polarity establishment by Cdc42: key roles for positive feedback and differential mobility. Small GTPases. 10(2):130–137. doi: 10.1080/21541248.2016.1275370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Jansen G, Zhang J, Thomas DY, Whiteway M. 2006. Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes Dev. 20(6):734–746. doi: 10.1101/gad.1375706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Leberer E, Thomas DY, Whiteway M. 1999. Functional characterization of the interaction of Ste50p with Ste11p MAPKKK in Saccharomyces cerevisiae. Mol Biol Cell. 10(7):2425–2440. doi: 10.1091/mbc.10.7.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuster A, Babu MM. 2009. Transcriptional control of the quorum sensing response in yeast. Mol Biosyst. 6(1):134–141. doi: 10.1039/B913579K. [DOI] [PubMed] [Google Scholar]
- Xu C, Liu R, Zhang Q, Chen X, Qian Y, Fang W. 2016. The diversification of evolutionarily conserved MAPK cascades correlates with the evolution of fungal species and development of lifestyles. Genome Biol Evol. 9(2):311–322. doi: 10.1093/gbe/evw051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J-R. 2000. MAP kinases in fungal pathogens. Fungal Genet Biol. 31(3):137–152. doi: 10.1006/fgbi.2000.1237. [DOI] [PubMed] [Google Scholar]
- Xu T, Shively CA, Jin R, Eckwahl MJ, Dobry CJ, Song Q, Kumar A. 2010. A profile of differentially abundant proteins at the yeast cell periphery during pseudohyphal growth. J Biol Chem. 285(20):15476–15488. doi: 10.1074/jbc.M110.114926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue C, Hsueh YP, Heitman J. 2008. Magnificent seven: roles of G protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol Rev. 32(6):1010–1032. doi: 10.1111/j.1574-6976.2008.00131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Tatebayashi K, Tanaka K, Saito H. 2010. Dynamic control of yeast MAP kinase network by induced association and dissociation between the Ste50 scaffold and the Opy2 membrane anchor. Mol Cell. 40(1):87–98. doi: 10.1016/j.molcel.2010.09.011. [DOI] [PubMed] [Google Scholar]
- Yang HY, Tatebayashi K, Yamamoto K, Saito H. 2009. Glycosylation defects activate filamentous growth Kss1 MAPK and inhibit osmoregulatory Hog1 MAPK. EMBO J. 28(10):1380–1391. doi: 10.1038/emboj.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi A, Miyazawa K, Kawauchi M, Abe K. 2022. Cell wall integrity and its industrial applications in filamentous fungi. J Fungi. 8(5):435. doi: 10.3390/jof8050435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue H, Bing J, Zheng Q, Zhang Y, Hu T, Du H, Wang H, Huang G. 2018. Filamentation in Candida auris, an emerging fungal pathogen of humans: passage through the mammalian body induces a heritable phenotypic switch. Emerg Microbes Infect. 7(1):188. doi: 10.1038/s41426-018-0187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahner JE, Harkins HA, Pringle JR. 1996. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 16(4):1857–1870. doi: 10.1128/MCB.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan JG, Coyle SM, Rajan S, Sidhu SS, Lim WA. 2012. Conformational control of the Ste5 scaffold protein insulates against MAP kinase misactivation. Science. 337(6099):1218–1222. doi: 10.1126/science.1220683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdunek A, Pieczywek PM, Cybulska J. 2021. The primary, secondary, and structures of higher levels of pectin polysaccharides. Compr Rev Food Sci Food Saf. 20(1):1101–1117. doi: 10.1111/1541-4337.12689. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Simon I, Harbison CT, Hannett NM, Volkert TL, Fink GR, Young RA. 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell. 113(3):395–404. doi: 10.1016/S0092-8674(03)00301-5. [DOI] [PubMed] [Google Scholar]
- Zhang W, Liu HT. 2002. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 12(1):9–18. doi: 10.1038/sj.cr.7290105. [DOI] [PubMed] [Google Scholar]
- Zhang X, Meng Y, Huang Y, Zhang D, Fang W. 2021. A novel cascade allows Metarhizium robertsii to distinguish cuticle and hemocoel microenvironments during infection of insects. PLoS Biol. 19(8):e3001360. doi: 10.1371/journal.pbio.3001360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang Z, Jiang C, Xu J-R. 2021. Regulation of biotic interactions and responses to abiotic stresses by MAP kinase pathways in plant pathogenic fungi. Stress Biol. 1(1):5. doi: 10.1007/s44154-021-00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Shen ZM, Kahn PC, Lipke PN. 2001. Interaction of alpha-agglutinin and a-agglutinin, Saccharomyces cerevisiae sexual cell adhesion molecules. J Bacteriol. 183(9):2874–2880. doi: 10.1128/JB.183.9.2874-2880.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yu Z, Ding T. 2020. Quorum-sensing regulation of antimicrobial resistance in bacteria. Microorganisms. 8(3):425. doi: 10.3390/microorganisms8030425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Jiang Y, Wang H, Lu Z, Huang S, Luo Z, Zhang L, Lv T, Tang X, Zhang Y. 2023. Fus3/Kss1-MAP kinase and Ste12-like control distinct biocontrol-traits besides regulation of insect cuticle penetration via phosphorylation cascade in a filamentous fungal pathogen. Pest Manag Sci. 79(7):2611–2624. doi: 10.1002/ps.7446. [DOI] [PubMed] [Google Scholar]
- Zhou W, Dorrity MW, Bubb KL, Queitsch C, Fields S. 2020. Binding and regulation of transcription by yeast Ste12 variants to drive mating and invasion phenotypes. Genetics. 214(2):397–407. doi: 10.1534/genetics.119.302929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Lin W, Long Y, Yang Y, Zhang H, Wu K, Chu Q. 2022. Notch signaling pathway: architecture, disease, and therapeutics. Signal Transduct Target Ther. 7(1):95. doi: 10.1038/s41392-022-00934-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Winston F. 2001. NRG1 is required for glucose repression of the SUC2 and GAL genes of Saccharomyces cerevisiae. BMC Genet. 2(1):5. doi: 10.1186/1471-2156-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zhao X, Xue C, Dai Y, Xu J-R. 2014. Bypassing both surface attachment and surface recognition requirements for appressorium formation by overactive ras signaling in Magnaporthe oryzae. Mol Plant Microbe Interact. 27(9):996–1004. doi: 10.1094/MPMI-02-14-0052-R. [DOI] [PubMed] [Google Scholar]
- Zhu W, Zhou M, Xiong Z, Peng F, Wei W. 2017. The cAMP-PKA signaling pathway regulates pathogenicity, hyphal growth, appressorial formation, conidiation, and stress tolerance in Colletotrichum higginsianum. Front Microbiol. 8:1416. doi: 10.3389/fmicb.2017.01416. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genes and proteins are linked to Saccharomyces Genome Database (SGD) https://www.yeastgenome.org/.