Abstract
The viral determinants that underlie human immunodeficiency virus type 1 (HIV-1) neurotropism are unknown, due in part to limited studies on viruses isolated from brain. Previous studies suggest that brain-derived viruses are macrophage tropic (M-tropic) and principally use CCR5 for virus entry. To better understand HIV-1 neurotropism, we isolated primary viruses from autopsy brain, cerebral spinal fluid, blood, spleen, and lymph node samples from AIDS patients with dementia and HIV-1 encephalitis. Isolates were characterized to determine coreceptor usage and replication capacity in peripheral blood mononuclear cells (PBMC), monocyte-derived macrophages (MDM), and microglia. Env V1/V2 and V3 heteroduplex tracking assay and sequence analyses were performed to characterize distinct variants in viral quasispecies. Viruses isolated from brain, which consisted of variants that were distinct from those in lymphoid tissues, used CCR5 (R5), CXCR4 (X4), or both coreceptors (R5X4). Minor usage of CCR2b, CCR3, CCR8, and Apj was also observed. Primary brain and lymphoid isolates that replicated to high levels in MDM showed a similar capacity to replicate in microglia. Six of 11 R5 isolates that replicated efficiently in PBMC could not replicate in MDM or microglia due to a block in virus entry. CD4 overexpression in microglia transduced with retroviral vectors had no effect on the restricted replication of these virus strains. Furthermore, infection of transfected cells expressing different amounts of CD4 or CCR5 with M-tropic and non-M-tropic R5 isolates revealed a similar dependence on CD4 and CCR5 levels for entry, suggesting that the entry block was not due to low levels of either receptor. Studies using TAK-779 and AMD3100 showed that two highly M-tropic isolates entered microglia primarily via CXCR4. These results suggest that HIV-1 tropism for macrophages and microglia is restricted at the entry level by a mechanism independent of coreceptor specificity. These findings provide evidence that M-tropism rather than CCR5 usage predicts HIV-1 neurotropism.
Human immunodeficiency virus type 1 (HIV-1) infects macrophages and microglia in the central nervous system (CNS) and frequently causes dementia and other neurological disorders in AIDS patients (65, 87). CNS infection can cause HIV-1 encephalitis, which is characterized by reactive astrocytes, myelin pallor, microglial nodules, perivascular inflammation, multinucleated giant cells, and neuronal loss. Neuroinvasion by HIV-1 occurs through trafficking of infected monocytes and possibly lymphocytes across the blood-brain barrier (87). Infected macrophages and microglia in the brain represent a significant cellular reservoir for long-term viral persistence (reviewed in references 83 and 93). Other tissues that harbor persistently infected macrophages include lung, lymph node, spleen, and bone marrow. Macrophages are less susceptible to the cytopathic effects of HIV-1 than CD4+ T cells (37, 38, 48, 70), so they may continue to shed virus for the duration of their normal life span. Most drugs used in highly active antiretroviral therapy have relatively poor CNS penetration (83, 97). Therefore, CNS infection is a major barrier to effective antiviral therapy.
The tropism of HIV-1 is determined by the interaction of the HIV-1 envelope glycoprotein with CD4 and a particular coreceptor. Macrophage-tropic (M-tropic) HIV-1 isolates primarily use CCR5 (R5) as a coreceptor (these are referred to as R5 viruses) (2, 12, 17, 26, 27), whereas T-cell line-tropic HIV-1 isolates use CXCR4 (X4) (33). Dualtropic viruses (R5X4) use both coreceptors. A subset of viruses can also use alternative coreceptors, including CCR3, CCR2b, CCR8, Apj, Strl33 (BONZO), Gpr1, Gpr15 (BOB), CX3CR1 (V28), ChemR23, and RDC1 (11–13, 18, 26, 28, 29, 31, 50, 53, 64, 89, 90, 96), but the role of these coreceptors in vivo is unknown. In some patients, disease progression is associated with a general broadening of virus tropism by expansion of coreceptor usage (14). HIV-1 enters the CNS in the early stages of infection. However, it is late in the course of disease progression, when X4 and R5X4 isolates emerge, that neurological symptoms such as dementia typically arise.
CCR5 is the major coreceptor for HIV-1 infection of macrophages and microglia (1, 36, 41, 42, 45, 95). Furthermore, previous studies suggest that CCR5 is the principal coreceptor used by HIV-1 isolated from brain (1, 12, 45, 62, 95, 101). Most laboratory-adapted X4 viruses, such as IIIB and NL4-3, do not replicate efficiently in macrophages and microglia (19, 45, 60, 81, 91, 103, 107). However, macrophages and microglia can support efficient replication by a subset of primary X4 viruses (46, 81, 98, 99, 105). CCR3 is expressed on microglia and may facilitate infection by certain HIV-1 strains (45). Apj, CCR8, Gpr15, and Strl33 can be used by some brain-derived viruses at low efficiency (1, 45, 95), but the role of these coreceptors in mediating infection of macrophages and microglia is unknown.
The genetic evolution of HIV-1 within the brain is distinct from that in lymphoid tissues and other organs (9, 24, 39, 51, 58, 94, 104, 106). Specific sequences within Env, particularly the V3 region, are associated with brain infection (51, 58, 85, 86, 104, 106). Whereas one previous study suggests that some primary HIV-1 isolates show preferential tropism for microglia compared to blood monocyte-derived macrophages (MDM) (103), other studies suggest that the tropisms of HIV-1 isolates for microglia and macrophages are similar (41, 46). Thus, specific determinants that underlie HIV-1 neurotropism remain unresolved.
Relatively few brain-derived HIV-1 isolates from neurologically well-characterized patients are available to study HIV-1 neurotropism (39, 40, 62, 63, 69, 79, 101). To better understand HIV-1 neurotropism, we isolated and characterized primary viruses from autopsy brain, cerebrospinal fluid (CSF), spinal cord, blood, spleen, and lymph node samples from AIDS patients with dementia and HIV-1 encephalitis. We found that CCR5 usage was neither necessary nor sufficient for M tropism. However, M-tropism, irrespective of coreceptor usage, predicted the ability of primary HIV-1 strains from brain and other tissues to replicate in microglia. These findings suggest that M-tropism rather than CCR5 usage predicts HIV-1 neurotropism.
MATERIALS AND METHODS
Subjects.
Autopsy brain, CSF, peripheral blood, lymph node, or spleen samples were collected from 18 patients who died of AIDS and were stored at −80°C. Clinical characteristics (risk factor for acquiring HIV-1 infection, last CD4 cell count, use of antiretroviral therapies, and clinical history of dementia) of these patients are described in Table 1. None had evidence of a CNS opportunistic infection or neoplasm at autopsy except for patient CB3, who was diagnosed with primary CNS lymphoma. Sixteen patients were male and two (UK1 and UK3) were female. Patients MACS1 through MACS12 were participants in the Chicago component of the Multicenter AIDS Cohort Study (MACS). Tissue samples from patients UK1 through UK4 were obtained from the Edinburgh Brain Bank. Patients CB1 and CB3 were treated at Fairfield Hospital, Victoria, Australia. Brain tissue samples were obtained from the frontal lobe. Additional samples from basal ganglia were obtained for the UK patients.
TABLE 1.
Clinical history and neuropathology of study subjectsa
| Patient | Risk factorb | Last CD4 count (cells/μl) | Antiretroviral(s) | Clinical dementia | HIV-1 encephalitis | Giant cellsc | HIV-1 isolation from brain |
|---|---|---|---|---|---|---|---|
| MACS1 | MH | 2 | None | Yes | Severe | +++ | + |
| MACS2 | MH | 52 | AZT | Yes | Moderate | + | + |
| MACS3 | MH | 95 | None | Yes | Moderate | + | + |
| MACS4 | MH | 8 | None | No | None | − | − |
| MACS5 | MH | 14 | AZT | Yes | None | − | − |
| MACS6 | MH | 3 | Indinavir | Yes | Mild | + | − |
| MACS7 | MH | 44 | AZT | No | None | − | − |
| MACS8 | MH | 36 | NAd | No | None | − | − |
| MACS9 | MH | 4 | None | No | None | − | − |
| MACS10 | MH | 70 | None | No | None | − | − |
| MACS11 | MH | 7 | D4Te | No | None | − | − |
| MACS12 | MH | 149 | None | Yes | None | − | − |
| UK1 | IVDU | 87 | ddc (1 mo) | Yes | Moderate | ++ | + |
| UK2 | HET | 297 | None | No | Mild | + | − |
| UK3 | IVDU | 137 | AZT, ddI (6 wk) | Yes | Severe | ++ | − |
| UK4 | MH | 8 | D4T (1 mo) | No | Mild | ++ | − |
| CB1 | MH | 10 | ddI (prior AZT) | Yes | Severe | NA | + |
| CB3 | MH | 5 | ddI (prior AZT and ddC) | Yes | Severe | NA | + |
Tissue samples were collected at autopsy from 18 patients who died of AIDS. The presence of clinical dementia and HIV-1 encephalitis was determined by the clinical history and neuropathological examination at the time of autopsy. Brain tissue samples were collected from frontal lobe (MACS and CB patients), or frontal lobe and basal ganglia (UK patients).
MH male homosexual; IVDU, intravenous drug user; HET, heterosexual transmission; mo, months; wk, weeks; N/A, not available.
Frequency of multinucleated giant cells observed in autopsy brain tissue sections. −, +, ++, and +++, none, occasional, moderate, and high, respectively.
NA, not available.
d4T, stavudine..
Cells.
Peripheral blood mononuclear cells (PBMC) were purified from blood of healthy HIV-1-negative donors by Ficoll-Hypaque density gradient centrifugation, stimulated with 2 μg of phytohemagglutinin (PHA) (Sigma, St. Louis, Mo.) per ml for 3 days, and cultured in RPMI 1640 medium supplemented with 10% (vol/vol) fetal bovine serum (Mediatech, Herndon, Va.), 100 μg of penicillin and streptomycin per ml, and 20 U of interleukin-2 (IL-2) (Boehringer, Mannheim, Germany) per ml. CD8+ T cells were depleted by magnetic separation with anti-CD8-conjugated magnetic beads (Miltenyi Biotech, Auburn, Calif.). MDM were purified from PBMC by plastic adherence and cultured for 5 days in RPMI 1640 medium supplemented with 10% (vol/vol) human AB+ serum (Nabi, Boca Raton, Fla.), 100 μg of penicillin and streptomycin per ml, and 12.5 ng of macrophage colony-stimulating factor (M-CSF) per ml. Primary human fetal brain cultures which contain a mixture of astrocytes, neurons, and microglia were prepared as previously described (81) and cultured in Dulbecco modified Eagle medium supplemented with 10% (vol/vol) bovine calf serum (HyClone, Logan, Utah), 100 μg of penicillin and streptomycin per ml, 2 mM l-glutamine, 1 mM sodium pyruvate, and 5 ng of M-CSF per ml. The protocol for tissue procurement was approved by an institutional review board and in compliance with federal regulations. Cf2-Luc cells (30), derived from the Cf2th canine thymocyte cell line (12), stably express the luciferase gene under the control of the HIV-1 long terminal repeat (LTR). Cf2-Luc cells were cultured in Dulbecco modified Eagle medium supplemented with 10% (vol/vol) fetal bovine serum, 100 μg of penicillin and streptomycin per ml, and 0.7 mg of G418 (Gibco BRL, Gaithersburg, Md.) per ml.
Isolation of HIV-1.
Autopsy brain and lymphoid tissue samples (approximately 2 to 4 mm3) were rinsed three times in 2 ml of RPMI 1640 medium containing 100 μg of penicillin and streptomycin per ml, 5 μg of amphotericin B (Fungizone) per ml, and 60 μg of anti-PPLO (Gibco BRL) per ml and then rinsed three times in the same medium without amphotericin B and anti-PPLO prior to homogenization. The protocol used to isolate HIV-1 from tissue samples minimizes contamination by peripheral blood and has been described elsewhere (62); it was used with the following modifications. Brain and spleen samples were homogenized in 2 ml of RPMI 1640 medium by repeated suction using sterile plastic transfer pipettes. Lymph node and spinal cord samples were processed in 2 ml of RPMI 1640 medium using TenBroeck ground glass tissue grinders (Wheaton, Millville, N.J.). Tissue homogenates or 1 ml of filtered (0.45-μm-pore-size filter) CSF samples were then added to 10 × 106 CD8-depleted PBMC, incubated at 37°C for 1 h, and cultured in 10 ml of growth medium containing 20 U of IL-2 per ml. Fifty percent medium changes were performed twice weekly, with residual tissue debris removed during the first and second medium changes. For virus isolation from PBMC, 2 × 106 cells were added to 5 × 106 PBMC from a normal uninfected donor and cultured as described above. Five million fresh PHA-activated, CD8-depleted PBMC from a different donor were added at every second medium change. For virus isolation using MDM as target cells, tissue homogenates were added to confluent monolayers of MDM in T-25 tissue culture flasks and incubated at 37°C for 3 h in a volume of 2.5 ml. Culture medium containing 10% (vol/vol) human AB+ serum (Nabi) and 12.5 ng of M-CSF per ml was then added to a final volume of 10 ml. Fifty percent medium changes were performed twice weekly. Supernatants were tested for reverse transcriptase (RT) activity using [3H]dTTP incorporation as previously described (81). Supernatants testing positive for RT were filtered through 0.45-μm-pore-size filters and stored at −80°C.
RNA isolation and RT-PCR.
Viral RNA was extracted from 140 μl of cell-free virus supernatant using a QIAmp viral RNA kit (Qiagen, Valencia, Calif.) according to the manufacturers' protocol. The Env V1/V2 region was reverse transcribed using a modified Titan One Tube RT-PCR System (Roche Molecular Biochemicals). Primers that span the V1/V2 region used for RT-PCR were 5′-TTATGGGATCAAAGCCTAAAGCCATGTGTA-3′ (V1) and 5′-CCTAATTCCATGTGTACATTGTACTGTGCT-3′ (V2). RT-PCR was performed as follows. Twenty-microliter reaction mixtures consisted of 5 μl of viral RNA, 1× RT-PCR buffer, 5 mM dithiothreitol, 1 mM each deoxynucleoside triphosphate, 15 pmol of V2 primer, 10 U of RNase inhibitor (Roche Molecular Biochemicals), and 12 U of avian myeloblastosis virus RT (Roche Molecular Biochemicals). Reaction mixtures were incubated at 42°C for 30 min and then at 99°C for 2 min to inactivate the avian myeloblastosis virus RT. Thirty microliters of PCR mix (1× Titan RT-PCR buffer, 5 mM dithiothreitol, 15 pmol of V1 primer, and 0.5 μl of Titan enzyme mix) was then added to the reverse transcription reaction mixtures. PCR was carried out in a Stratagene 40 Robocycler (1 cycle of 95°C for 2 min 45 s, 55°C for 45 s, and 72°C for 2 min; 40 cycles of 95°C for 45 s, 55°C for 45 s, and 72°C for 2 to 5 min; and 1 cycle of 95°C for 45 s, 55°C for 45 s, and 72°C for 8 min). The Env V3 region was reverse transcribed using primers 5′-GAATCTGTAGAAATTAATTGTACAAGACCC-3′ (V3L4) and 5′-TTTTGCTCTACTAATGTTACAATGTGCTTG-3′ (V3R5) as previously described (76).
V1/V2 and V3 HTA.
The V1/V2 probe was generated by PCR amplification of a 420-bp segment of the HIV-1 JR-FL Env using primers V1 and V2 and was radioactively labeled with α-35S-dATP (K. McGrath and R. Swanstrom, unpublished data). This fragment spans the entire V1/V2 region and part of the C2 region. Labeling was achieved by digestion of the V1/V2 PCR product with NdeI followed by incorporation of α-35S-dATP (50 μCi, 1,250 Ci/mmol; NEN Life Science, Boston, Mass.) in a fill-in reaction with 10 U of the Klenow fragment of DNA polymerase I. Klenow fragment was inactivated by heat and the addition of EDTA. Unincorporated nucleotides were removed using a QIAquick spin column (Qiagen). The V3 probe was generated from a 141-bp segment of the JR-FL Env and radioactively labeled with α-35S-dATP as previously described (77). V1/V2 heteroduplex reaction mixtures consisted of 1× annealing buffer (1 M NaCl, 100 mM Tris-HCl [pH 7.5], 20 mM EDTA), 8 μl of unpurified RT-PCR product, 0.1 μM V1 and V2 primers, and 1 μl of labeled V1/V2 JR-FL probe. V1 and V2 primers were included in the heteroduplex formation reaction mixtures to normalize for PCRs containing excess primer after amplification. The reaction products were denatured at 99°C for 2 min and then allowed to anneal at room temperature for 5 min. The heteroduplexes were separated in 6% polyacrylamide (37.5:1 acrylamide-bisacrylamide) gels in 1× Tris-borate-EDTA buffer. Heteroduplexes were visualized by autoradiography of dried heteroduplex tracking assay (HTA) gels. V3 heteroduplex reactions were performed as previously described (77). The procedure is identical to the protocol described above except for the use of V3L4 primer and V3 probe in heteroduplex reactions and separation of the heteroduplexes in 12% polyacrylamide.
Sequence analysis.
PCR products were sequenced by ABI dye terminator sequencing (Perkin-Elmer Corp.) and analyzed using MacVector 7.0 (Genetics Computer Group, Madison, Wis.).
Coreceptor usage.
To determine coreceptor usage by primary HIV-1 isolates, Cf2-Luc cells were cotransfected with 10 μg of plasmid pcDNA3-CD4 and 20 μg of plasmid pcDNA3 containing CCR2b, CCR3, CCR5, CCR8, CXCR4, CX3CR1, Gpr1, Gpr15, Strl33, or Apj using the calcium phosphate method and infected 48 h later by incubation with 10,000 3H cpm RT units of HIV-1 in the presence of 2 μg of Polybrene (Sigma) per ml. Cf2-Luc cells transfected with pcDNA3-CD4 alone were used as negative controls. After overnight infection, virus was removed and the cells were cultured for an additional 48 h prior to lysis in 200 μl of cell lysis buffer (Promega, Madison, Wis.). Cell lysates were cleared by centrifugation, and assayed for luciferase activity (Promega) according to the manufacturer's protocol.
HIV-1 replication kinetics.
Five million PHA-activated PBMC were infected by incubation with 50,000 3H cpm RT units of virus supernatant in a volume of 2 ml for 3 h at 37°C. Virus was then removed, and PBMC were washed three times with phosphate-buffered saline (PBS) and cultured in medium containing 20 U of IL-2 per ml for 18 days. Fifty percent medium changes were performed twice weekly, and supernatants were tested for HIV-1 by RT assays. MDM were isolated from PBMC by plastic adherence and allowed to mature for 5 days prior to seeding in six-well tissue culture plates at approximately 90% confluence. Virus equivalent to 50,000 3H cpm RT units in a volume of 2 ml was allowed to adsorb to the cell monolayers for 3 h at 37°C. Virus was then removed, and cells were rinsed three times with PBS prior to addition of 2 ml of culture medium. Fifty percent medium changes were performed twice weekly for 28 days, and supernatants were tested for HIV-1 replication by RT assays. Mixed fetal brain cells were seeded into 48-well tissue culture plates coated with poly-l-lysine at a density of 1.6 × 105/well and cultured for 5 days. Microglia were then added at a density of 0.2 × 105/well and cultured further for 2 days. Brain cell cultures were infected by overnight incubation with 10,000 3H cpm RT units of HIV-1 in a volume of 0.5 ml. Virus was removed, and cells were rinsed three times with warm culture medium prior to addition of 0.25 ml of mixed brain cell-conditioned medium. After culture for 4 days, 0.25 ml of fresh culture medium was added and cells were incubated for an additional 24 days. Fifty percent medium changes were performed weekly, and supernatants were tested for HIV-1 replication by p24 antigen enzyme-linked immunosorbent assay (ELISA) (NEN) using the manufacturer's protocol.
Virus inhibition studies.
Monoclonal antibodies against CCR5 and CXCR4 (2D7 and 12G5, respectively) or small-molecule inhibitors of CCR5 and CXCR4 (TAK779 and AMD3100, respectively) were used for virus inhibition studies. Mixed fetal brain cells containing microglia were preincubated with 10 μg of 2D7 or 12G5 antibodies (PharMingen) per ml, 100 nM TAK-779 (4), or 1.2 μM AMD3100 (25, 92) for 1 h prior to infection with HIV-1 isolates containing the same concentration of antibody or inhibitor. Infected cells were cultured for 28 days as described above in the presence of each antibody or inhibitor.
Real-time PCR.
MDM cultured for 5 days (105 cells) were infected by incubation with 20,000 3H cpm RT units of virus supernatant in a volume of 0.5 ml for 18 or 36 h. Virus stocks were treated with 10 U of RNase-free DNase (Promega) in the presence of 10 mM MgCl2 for 1 h at room temperature prior to infection. Mock-infected cells were incubated with 0.5 ml of DNase-treated medium. ADA virus inactivated by incubation at 56°C for 1 h was used as a negative control. At 18 or 36 h after infection, cells were rinsed three times with PBS, removed with PBS containing 2.5 mM EDTA, pelleted, and stored at −80°C.
DNA was obtained from frozen cell pellets following exposure to urea lysis buffer and subsequent phenol-chloroform extraction as previously described (108). Amplification and detection were performed on an Applied Biosystems PRISM 7700 Sequence Detection System using the Taqman reagent kit in 5 mM MgCl2. A 25-μl reaction volume and the standard Taqman amplification cycle were used as recommended by the manufacturer. The oligonucleotides used for detection of early HIV-1 DNA (R-U5 of the LTR) reverse transcripts were as follows. The forward primer SR1 (5′-CAAGTAGTGTGTGCCCGTCTGT-3′) was used at a concentration of 300 nM. This primer corresponds to nucleotides 560 to 581 in the R region of the HIV-1 JR-CSF LTR. The reverse primer used was AA55 (5′-CTGCTAGAGATTTTCCACACTGAC-3′) at 150 nM. This primer binds to the U5 region of the LTR and has been described previously (108). The fluorescent probe ZXF (6FAM 5′-TGTGACTCTGGTAACTAGAGATCCCTCAGACCC-3′ TAMRA), used at 200 nM, corresponds to nucleotides 584 to 616 in the JR-CSF LTR. The oligonucleotides used to detect full-length (LTR-gag) HIV-1 reverse transcripts included the forward primer SR1 and the reverse primer M661 (5′-CCTGCGTCGAGAGAGCTCCTCTGG-3′) at 150 nM. This primer binds to sequences in the beginning of gag and has been previously described (108). The probe for this condition was identical to that used for early HIV-1 reverse transcripts. Thus, the combination of primers SR1 and AA55 amplifies sequences in R-U5 of the LTR and detects early events in reverse transcription. The combination of SR1 and M667 amplifies the LTR-gag junction, which is formed near the completion of the reverse transcription process. The oligonucleotides used to detect the human beta-globin gene were as follows. The forward primer BGF1 (5′-CAACCTCAAACAGACACCATGG-3′) was used at a concentration of 300 nM. This primer corresponds to nucleotides 846 to 866 in the human beta-globin gene. Reverse primer BGR1 (5′-TCCACGTTCACCTTGCCC-3′) was used at 150 nM. BGR1 corresponds to nucleotides 911 to 928 in the human beta-globin sequence. Together these primers amplify a band of 83 nucleotides in length. The fluorescent probe BGX1 (6FAM 5′-CTCCTGAGGAGAAGTCTGCCGTTACTGCC-3′ TAMRA) was used at 200 nM. Probe BGX1 corresponds to nucleotides 877 to 903 in the human beta-globin sequence. Oligonucleotides were purchased from Applied Biosystems and Annovis. All amplifications were performed in parallel with a set of known quantitative standards. The standard curve used to determine HIV DNA levels ranged from 10 to 20,000 copies of cloned HIV DNA (108). The standard curve used to determine levels of beta-globin gene sequences consisted of DNA derived from 10 to 100,000 normal human peripheral blood lymphocytes. Quantitation of HIV-1 sequences was achieved by extrapolation from these standard curves.
Retroviral transduction of microglia.
The envelope-deficient simian-human immunodeficiency virus (SHIV) vectors pSIvec1ΔenvGFP and pSIvec1ΔenvhuCD4, used to transduce microglia with green fluorescent protein (GFP) or human CD4, respectively, have been described previously (6, 49). Viruses pseudotyped with vesicular stomatitis virus G protein were produced by transfection of 293T cells with pSIvec1ΔenvGFP or pSIvec1ΔenvhuCD4 plus pHCMV-G and a Rev-expressing plasmid at a ratio of 20:2:1. Microglia were cultured in six-well plates at a density of 0.5 × 106/well and transduced by overnight infection with 50,000 3H cpm RT units of virus in a volume of 1 ml containing 3 μg of Polybrene per ml. Virus was removed and cells were rinsed twice with medium prior to culturing for an additional 48 h. Transduced microglia were added to wells of 48-well tissue culture plates containing 1.6 × 105 mixed fetal brain cells at a density of 0.2 × 105/well and infected with HIV-1 as described above or analyzed for expression of GFP in cells or CD4 on the cell surface. For analysis of GFP expression, cells were washed twice with PBS, fixed in 400 μl of 4% (wt/vol) paraformaldehyde, and analyzed by flow cytometry. Cell surface CD4 expression was measured by staining with phycoerythrin-conjugated anti-human CD4 (PharMingen) and analysis by flow cytometry.
Nucleotide sequence accession numbers.
The V1/V2 and V3 nucleotide sequences reported here have been assigned GenBank accession no. AF414888 to AF414912.
RESULTS
Isolation of primary HIV-1 viruses.
To generate a panel of primary HIV-1 isolates from brain and other tissues, we obtained autopsy samples of brain, CSF, lymph node, spleen, and blood from 18 AIDS patients (Table 1). Ten patients had HIV-1 dementia, and 10 had HIV-1 encephalitis. Brain isolates were obtained from 6 of 18 frontal lobe samples (33%) (patients MACS1, MACS2, MACS3, UK1, CB1, and CB3) by coculture with CD8-depleted PBMC. All six patients with positive brain isolations had dementia and HIV-1 encephalitis. Two received no antiretroviral therapy, one received zidovudine (AZT) monotherapy, one received dideoxycytosine (ddC) for 1 month, and two received dideoxyinosine (ddI) or ddC as monotherapies after developing resistance to AZT. Virus was not recovered from any of the samples of basal ganglia. Of six patients with positive brain virus isolations, five viruses from blood or lymphoid tissues (83%) and two viruses from CSF (33%) were subsequently isolated. A spinal cord-derived isolate was also obtained from patient CB3. Thus, positive brain isolations were obtained in 6 of 10 patients with HIV-1 encephalitis (60%) compared to 0 of 8 patients without HIV-1 encephalitis (0%), and the frequency of virus isolation from brain and CSF was much lower than from peripheral blood and lymphoid tissues. Virus isolation was also attempted with 16 brain tissue samples (MACS and UK patients), five spleen samples (patient MACS1 and UK patients), and two lymph node samples (patients MACS2 and MACS3) using MDM as target cells. By coculture with MDM, isolates were recovered from brain and spleen of patient MACS1 and from brain of patient MACS3 (data not shown) but not from 20 other tissue samples. Thus, coculture with CD8-depleted PBMC was more sensitive for isolation of HIV-1 from tissue samples than coculture with MDM.
V1V2 and V3 HTA and sequence analysis.
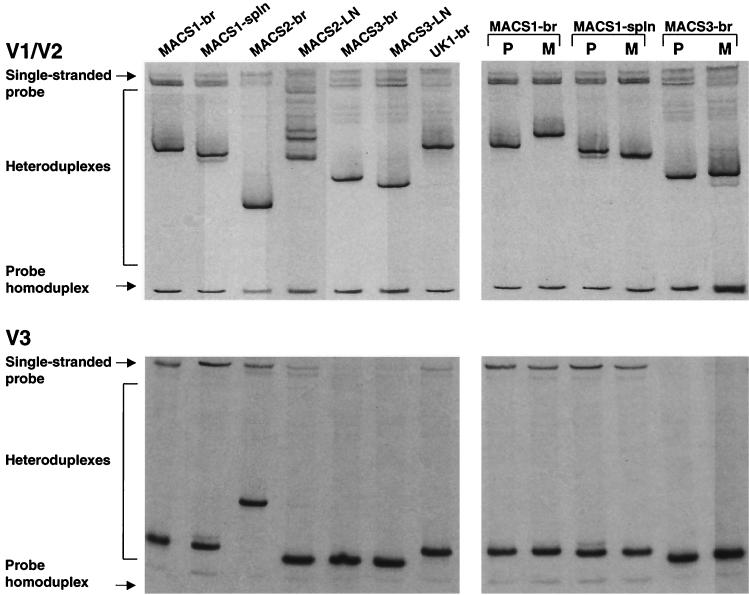
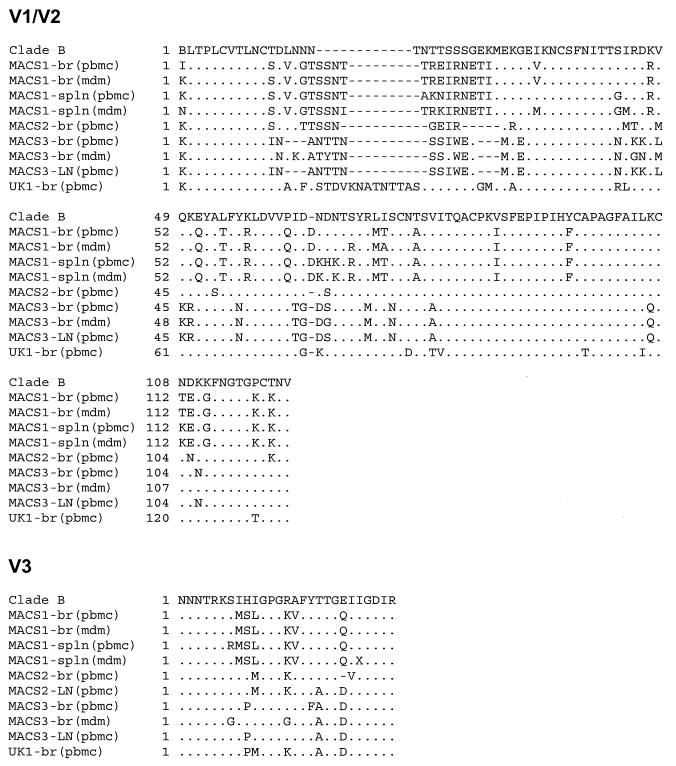
To determine whether distinct viral variants are present in the brain isolates compared to lymphoid tissue isolates, Env V1/V2 and V3 HTA and sequence analyses were performed using RT-PCR products generated from viral RNA (Fig. 1, left panels). The V1/V2 and V3 heteroduplex patterns consisted of one major band for each isolate except the MACS2 lymph node isolate (MACS2-LN), which showed a V1/V2 heteroduplex pattern consisting of several major bands. V1/V2 HTA analysis demonstrated distinct major variants in each of the four brain isolates from different patients and in brain compared to spleen or lymph node isolates from the same patient. For two patients, additional V1/V2 variants not detected in brain isolates were detected in the corresponding spleen (MACS1) and lymph node (MACS2) isolates. The V3 heteroduplex patterns also demonstrated distinct variants in each of the four brain isolates. The major V3 heteroduplexes detected in brain and lymphoid tissue isolates migrated similarly for two patients (MACS1 and MACS3) and differently for one patient (MACS2). A minor V3 variant not detected in the brain isolate from patient MACS1 was detected in the corresponding spleen isolate. The differences in V1/V2 and V3 heteroduplex formation were consistent with amino acid sequence changes detected between brain and lymphoid isolates from the same patient (Fig. 2). The distinct mobility of V1/V2 heteroduplexes for the MACS3 brain isolate (MACS3-br) and MACS3-LN despite identical amino acid sequences most likely resulted from several silent changes in the V1/V2 nucleotide sequence (GenBank accession numbers AF414888 to AF414912). The significant shift of the V3 heteroduplex for MACS2-br is consistent with a single V3 codon deletion. Thus, distinct HIV-1 variants were detected in brain isolates from different patients and in brain and lymphoid tissue isolates from the same patient.
FIG. 1.
V1/V2 and V3 HTA analysis. HIV-1 Env V1/V2 and V3 regions were amplified by RT-PCR of viral RNA and subjected to HTA analysis as described in Materials and Methods. (Left panels) V1/V2 and V3 heteroduplex patterns of HIV-1 isolated from brain and lymphoid tissue by coculture with CD8-depleted PBMC. (Right panels) V1/V2 and V3 heteroduplex patterns of HIV-1 isolated from the same tissues by coculture with CD8-depleted PBMC (lanes P) or MDM (lanes M). Virus isolates were from brain (br), lymph node (LN), or spleen (spln) tissues.
FIG. 2.
V1/V2 and V3 amino acid sequence analysis. The amino acid sequences were obtained from RT-PCR-amplified HIV-1 Env V1/V2 and V3 regions as described in Materials and Methods. V1/V2 and V3 alignments are compared to the clade B consensus sequence. Dots indicate residues identical to the clade B consensus, dashes indicate gaps, and X indicates uncertainty at the nucleotide level.
We also compared the V1/V2 and V3 heteroduplex patterns and amino acid sequences between isolates recovered by coculture with CD8-depleted PBMC versus MDM (Fig. 1, right panels). V1/V2 HTA analysis of isolates recovered from brain and spleen of patient MACS1 and brain of patient MACS3 demonstrated distinct major variants in PBMC- compared to MDM-derived isolates. The major V1/V2 variant detected in the MDM-derived MACS1 spleen isolate (MACS1-spln) was detected as a minor variant in the PBMC-derived isolate. An additional minor variant not present in the PBMC-derived MACS3-br isolate was detected in the MDM-derived isolate. V3 HTA analysis demonstrated similar (MACS1-br and MACS1-spln) or distinct (MACS3-br) major variants in PBMC- compared to MDM-derived isolates. A minor variant not detected in the MDM-derived MACS1-spln isolate was present in the PBMC-derived isolate, consistent with the V1/V2 heteroduplex patterns. The differences in V1/V2 and V3 heteroduplex formation between PBMC- and MDM-derived isolates were consistent with amino acid sequence changes detected in the same isolates (Fig. 2). These results demonstrate that coculture of brain or spleen tissue with CD8-depleted PBMC or MDM results in a cell-dependent selection of distinct viral variants.
Coreceptor usage.
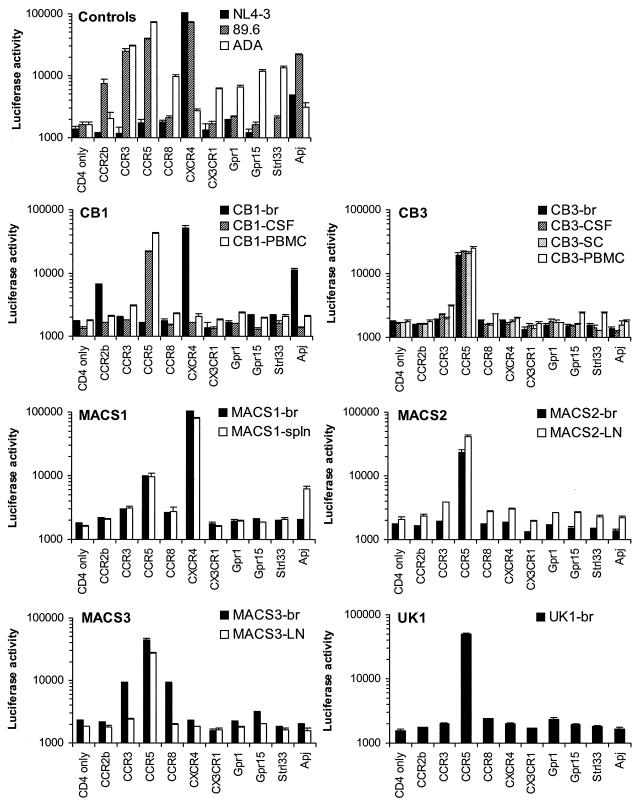
To determine whether primary HIV-1 brain isolates from patients with HIV-1 dementia exhibit particular patterns of chemokine coreceptor usage, viruses isolated by coculture with CD8-depleted PBMC were characterized for the ability to use CCR5, CXCR4, or alternative coreceptors for virus entry using Cf2-Luc cells (Fig. 3). The X4, R5, and R5X4 strains NL4-3, ADA, and 89.6, respectively, were used as positive controls. NL4-3 used CXCR4 and Apj; ADA used CCR3, CCR5, CCR8, CX3CR1, Strl33, Gpr15, Gpr1, and Apj; and 89.6 used CCR2b, CCR3, CCR5, CXCR4, and Apj as coreceptors for virus entry, as described in previous studies (11, 12, 17, 18, 26–29, 31, 64, 89). The use of Gpr1 by ADA was not detected in most of these studies but has occasionally been observed at low levels, particularly in fusion assays (28, 81). These differences may reflect the particular assay system used. Brain-derived HIV-1 isolates used CCR5 (four of six isolates), CXCR4 (one of six isolates), or both CXCR4 and CCR5 (one of six isolates) as principal coreceptors for virus entry. Minor usage of CCR2b, CCR3, CCR8, or Apj was demonstrated for two brain isolates. The pattern of coreceptor usage was similar between isolates from brain and other tissue compartments for three patients (CB3, MACS1, and MACS2) but was different for two patients (CB1 and MACS3). Thus, primary brain HIV-1 isolates from patients with dementia and HIV-1 encephalitis displayed diverse patterns of coreceptor usage and were not restricted to the use of CCR5 for virus entry.
FIG. 3.
Coreceptor usage by primary HIV-1 isolates. Cf2-Luc cells were transfected with pcDNA3-CD4 alone or cotransfected with pcDNA3-CD4 and pcDNA3 expressing CCR2b, CCR3, CCR5, CCR8, CXCR4, CX3CR1, Gpr1, Gpr15, Strl33, or Apj and infected with equivalent amounts of each HIV-1 isolate as described in Materials and Methods. Viruses were isolated from brain (br), CSF, PBMC, lymph node (LN), spleen (spln), and spinal cord (SC) by coculture with CD8-depleted PBMC. Cell lysates were prepared at 48 h postinfection and assayed for luciferase activity. Data are expressed as means from duplicate infections. Error bars represent standard deviations. Similar results were obtained in two independent experiments.
We next determined whether the pattern of coreceptor usage differed between HIV-1 isolates recovered by coculture with CD8-depleted PBMC versus MDM as target cells (Table 2). The levels of CCR5-mediated virus entry by the MACS1-br, MACS1-spln, and MACS3-br viruses isolated using MDM were lower than, equivalent to, or higher than, respectively, those by viruses isolated from the same tissues using CD8-depleted PBMC. In contrast, the levels of CXCR4-mediated virus entry by the MACS1-br and MACS1-spln viruses isolated using MDM were much lower than those by viruses isolated from the same tissues using PBMC. In contrast to viruses isolated using PBMC, usage of CCR3 (MACS3-br), CCR8 (MACS3-br), and Apj (MACS1-spln) was not detected for viruses isolated using MDM. These findings indicate that the pattern of coreceptor usage by viruses isolated using CD8-depleted PBMC was more diverse than that by viruses isolated using MDM. Furthermore, coculture with CD8-depleted PBMC allowed recovery of virus from more tissue samples. Therefore, viruses isolated by coculture with CD8-depleted PBMC were used for all subsequent experiments.
TABLE 2.
Coreceptor usage by HIV-1 viruses isolated using CD8-depleted PBMC versus MDM as target cells
| Virus | Target cells used for isolation | Coreceptor usagea
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | CCR2b | CCR3 | CCR5 | CCR8 | CXCR4 | CX3CR1 | Gpr1 | Gpr15 | Strl33 | Apj | ||
| MACS1-br | MDM | − | − | − | ++ | − | + | − | − | − | − | − |
| PBMC | − | − | − | +++ | − | +++ | − | − | − | − | − | |
| MACS1-spin | MDM | − | − | − | +++ | − | + | − | − | − | − | − |
| PBMC | − | − | − | +++ | − | +++ | − | − | − | − | + | |
| MACS3-br | MDM | − | − | − | +++ | − | − | − | − | − | − | − |
| PBMC | − | − | + | ++ | + | − | − | − | +/− | − | − | |
The pattern of coreceptor usage of viruses isolated using CD8-depleted PBMC or MDM as target cells was determined in Cf2-Luc cells, as described for Fig. 3. Control cells were transfected with CD4 alone. Entry levels were scored as +++ (>50,000 luciferase activity units), ++ (between 30,000 and 50,000 luciferase activity units), + (between 10,000 and 30,000 luciferase activity units), +/− (between 5,000 and 10,000 luciferase activity units), or − (<5,000 luciferase activity units).
Replication in PBMC.
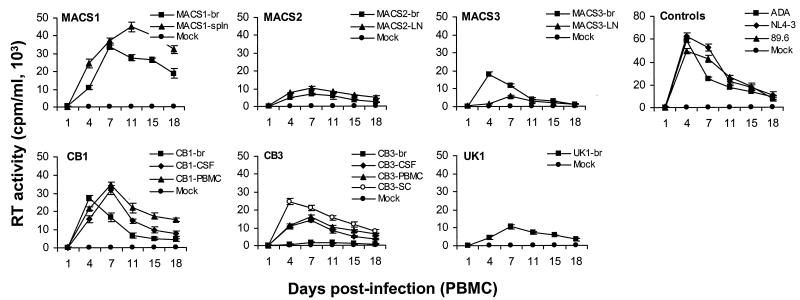
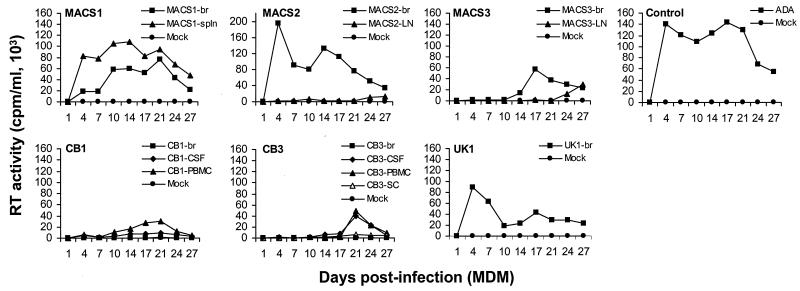
We examined the capacity of primary brain HIV-1 isolates to replicate in PBMC compared to viruses isolated from CSF, spinal cord, or lymphoid tissue from the same patient (Fig. 4). The ADA, NL4-3, and 89.6 positive control viruses replicated to high levels. The brain isolates replicated to variable levels. Two of six (MACS1-br and CB1-br) reached peak levels of replication similar to those of the control viruses, three (MACS2-br, MACS3-br, and UK1-br) replicated at moderate levels, and one (CB3-br) replicated at low levels. The viruses derived from CSF, spinal cord, and lymphoid tissue also replicated to variable levels. Three of eight viruses (CB1-CSF, CB1-PBMC, and MACS1-spln) replicated at high levels, four viruses (CB3-CSF, CB3-PBMC, the spinal cord isolate from patient CB3 [CB3-SC], and MACS2-LN) replicated at moderate levels, and one virus (MACS3-LN) replicated at low levels. Replication capacity in PBMC was similar between viruses isolated from brain and other tissue compartments for three patients (CB1, MACS1, and MACS2) but was different for two patients (MACS3 and CB3). The brain-derived isolate from patient MACS3 reached higher peak levels of replication than viruses isolated from lymph node, whereas the virus isolated from brain from patient CB3 replicated at lower levels than virus isolated from CSF, PBMC, or spinal cord. Thus, the replication capacity of brain-derived isolates in PBMC was diverse.
FIG. 4.
Replication kinetics in PBMC. PBMC were infected with equivalent amounts of each virus, as described in Materials and Methods, and cultured for 18 days. Virus production in culture supernatants was measured by RT assays. Values shown are means from duplicate infections. Error bars represent standard deviations. Results are representative of two independent experiments using cells obtained from different donors.
Replication in MDM.
HIV-1 isolates from brain are typically M-tropic (36–38, 62, 63, 79). To determine the relationship between coreceptor usage and M-tropism, replication kinetics were analyzed in MDM (Fig. 5). The ADA strain, used as a positive control, replicated to high levels. Four of six viruses isolated from brain and five of eight viruses isolated from CSF or lymphoid tissue were replication competent in MDM. However, only 2 of 11 R5 viruses (MACS2-br and UK1-br) and 2 of 2 R5X4 viruses (MACS1-br and MACS1-spln) replicated to high levels. In contrast, nine R5 viruses replicated at low levels or showed no evidence of productive infection. The X4 CB1-br isolate also showed no evidence of productive infection. These results indicate that only a subset of primary R5 isolates that replicate in PBMC can productively infect MDM, suggesting that CCR5 usage is not sufficient for M-tropism.
FIG. 5.
Replication kinetics in MDM. MDM were infected with equivalent amounts of each virus, as described in Materials and Methods, and cultured for 27 days. Virus production in culture supernatants was measured by RT assays. Results are representative of two independent experiments using cells obtained from different donors.
Replication in microglia.
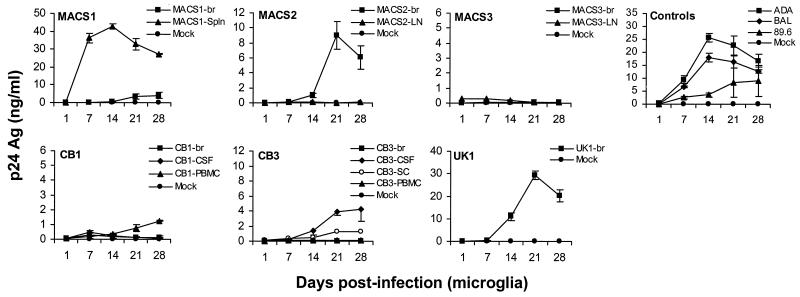
To determine the relationship between coreceptor usage, M-tropism, and neurotropism, we analyzed replication kinetics in microglia (Fig. 6). The R5 ADA and BAL control viruses replicated at high levels, whereas the R5X4 89.6 virus replicated at moderate levels. The capacity of primary HIV-1 viruses to replicate in microglia was generally similar to that demonstrated in MDM (Table 3). However, three isolates (CB3-PBMC, MACS1-br, and MACS3-br) replicated at lower levels in microglia than in MDM. Two of 11 R5 isolates (MACS2-br and UK1-br) replicated to high levels, three (CB1-PBMC, CB3-CSF, and CB3-SC) replicated to moderate or low levels, and six (CB1-CSF, CB3-br, CB3-PBMC, MACS2-LN, MACS3-br, and MACS3-LN) showed no evidence of productive infection. Both R5X4 viruses (MACS1-br and MACS1-spln) replicated in microglia, but in contrast to replication in MDM, only MACS1-spln replicated to high levels. All three viruses that replicated to high levels in microglia (MACS1-spln, MACS2-br, and UK1-br,) replicated to high levels in MDM. Surprisingly, two of these viruses (MACS2-br and UK1-br) replicated inefficiently in PBMC (Fig. 4 and Table 3). Together, these data indicate that M-tropism rather than CCR5 usage predicts the ability of HIV-1 isolates to replicate in microglia. Thus, CCR5 usage is not sufficient for microglia tropism.
FIG. 6.
Replication kinetics in microglia. Mixed brain cell cultures containing microglia were infected with equivalent amounts of each virus as described in Materials and Methods and cultured for 28 days. HIV-1 production in culture supernatants was measured by p24 antigen (Ag) ELISA (NEN). Data are represented as means from duplicate infections. Error bars represent standard deviations. Results are representative of three independent experiments using cells obtained from different donors.
TABLE 3.
Summary of viral phenotypesa
| Virus | Coreceptor usageb | Replication capacityc in:
|
||
|---|---|---|---|---|
| PBMC | Macrophages | Microglia | ||
| MACS1-br | CXCR4, CCR5 | +++ | ++ | + |
| MACS1-spin | CXCR4, CCR5 (Apj) | +++ | +++ | +++ |
| MACS2-br | CCR5 | + | +++ | ++ |
| MACS2-LN | CCR5 | + | +/− | − |
| MACS3-br | CCR5, (CCR3, CCR8) | ++ | + | − |
| MACS3-LN | CCR5 | +/− | +/− | − |
| UK1-br | CCR5 | + | ++ | +++ |
| CB1-br | CXCR4, (CCR2b, Apj) | +++ | − | − |
| CB1-CSF | CCR5 | +++ | − | − |
| CB1-PBMC | CCR5 | +++ | +/− | +/− |
| CB3-br | CCR5 | +/− | − | − |
| CB3-CSF | CCR5 | ++ | + | + |
| CB3-PBMC | CCR5 | ++ | + | − |
| CB3-SC | CCR5 | ++ | − | +/− |
Viruses isolated by coculture with CD8-depleted PBMC were characterized for coreceptor usage and the ability to replicate in PBMC, MDM, and microglia.
Minor usage of coreceptors by HIV-1 isolates, where entry levels were <10-fold above background, is shown in parentheses.
For replication in PBMC, virus levels were scored as +++ (>25,000 cpm/ml), ++ (between 10,000 and 25,000 cpm/ml), + (between 5,000 and 10,000 cpm/ml), +/− (between 1000 and 5000 cpm/ml), or − (<1000 cpm/ml). For replication in MDM, virus levels were scored as +++ (>100,000 cpm/ml), ++ (between 50,000 and 100,000 cpm/ml), + (between 10,000 and 50,000 cpm/ml), +/− (between 5000 and 10,000 cpm/ml), or − (<5000 cpm/ml). For replication in microglia, virus levels were scored as +++ (>25 ng/ml p24), ++ (between 10 and 25 ng/ml p24), + (between 5 and 10 ng/ml p24), +/− (between 0.5 and 5 ng/ml p24), or − (<0.5 ng/ml p24).
Sensitivity to CCR5 and CXCR4 inhibitors in microglia.
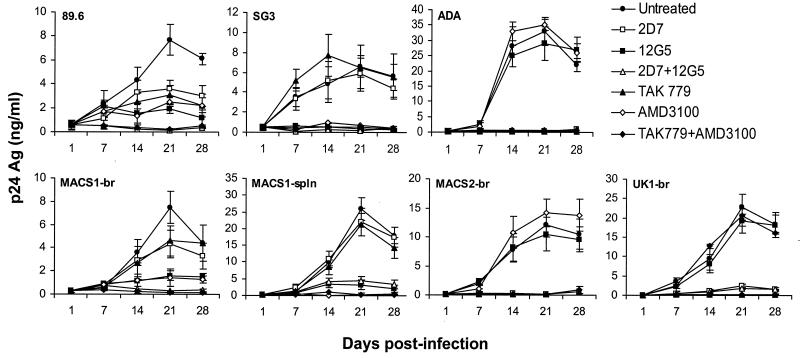
The sensitivity of primary M-tropic HIV-1 viruses to coreceptor-targeted inhibitors was tested in microglia by treating primary brain cultures with monoclonal antibodies against CCR5 or CXCR4 (2D7 and 12G5, respectively) or with small-molecule inhibitors of CCR5 or CXCR4 (TAK779 and AMD3100, respectively) (4, 25, 92) (Fig. 7). The R5 ADA, R5X4 89.6, and X4 SG3 (43) isolates were used as positive controls. 2D7 or TAK779 completely abolished infection of microglia by ADA and reduced infection by 89.6 by approximately 50%. 12G5 or AMD3100 completely abolished infection by SG3 and reduced infection by 89.6 by approximately 70%. Combinations of 2D7 and 12G5 or TAK779 and AMD3100 completely abolished infection by 89.6. 2D7 or TAK779 completely abolished infection by the R5 viruses MACS2-br and UK1-br but had no effect (MACS1-spln) or a minimal effect (MACS1-br) on the R5X4 viruses MACS1-br and MACS1-spln. In contrast, 12G5 or AMD3100 completely inhibited infection by MACS1-br and MACS1-spln but had no effect on the R5 viruses, indicating that these primary R5X4 viruses principally use CXCR4 for entry in microglia. Combinations of 2D7 and 12G5 or TAK779 and AMD3100 completely abolished infection by all primary M-tropic viruses tested, indicating that coreceptors other than CCR5 and CXCR4 are not used by these isolates for infection of microglia. The results further suggest that infection of microglia is highly sensitive to inhibition by coreceptor-targeted inhibitors.
FIG. 7.
Inhibition of HIV-1 infection in microglia by CCR5 and CXCR4 inhibitors. Mixed brain cell cultures containing microglia were treated with monoclonal antibody 2D7 (10 μg/ml), 12G5 (10 μg/ml), or both or treated with TAK779 (100 nM), AMD3100 (1.2 μM), or both for 1 h prior to infection. Untreated cells contained no antibody or inhibitor. Cells were infected with neurotropic primary isolates MACS1-br (R5X4), MACS1-spln (R5X4), MACS2-br (R5), and UK1-br (R5) or with control viruses 89.6 (R5X4), SG3 (X4), and ADA (R5) individually in the presence of each antibody or inhibitor as described in Materials and Methods and cultured for 28 days. HIV-1 production in culture supernatants was measured by p24 antigen (Ag) ELISA (NEN). Data are represented as means from duplicate infections. Error bars represent standard deviations. Results are representative of two independent experiments using cells obtained from different donors. The in vitro coreceptor usage phenotype for each virus determined with transfected Cf2-Luc cells (Fig. 3) is shown in parentheses.
Analysis of HIV-1 entry in macrophages by real-time PCR.
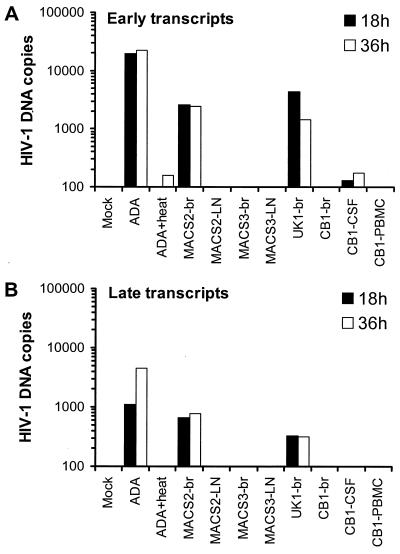
To determine whether the inability of certain R5 isolates to replicate in MDM and microglia was due to a block at the entry or postentry level, real-time PCR was used to quantify HIV-1 DNA sequences that appear early and late in the reverse transcription process (Fig. 8). To detect early events in reverse transcription, primers that span the first region of the viral genome involved in reverse transcription, the R-U5 region of the LTR, were used. To detect late stages of reverse transcription, primers that span the LTR-gag junction were used (see Materials and Methods). MDM infected with ADA or heat-inactivated ADA served as positive and negative controls, respectively. Viruses capable of productively infecting MDM produced high levels of fully reverse-transcribed viral DNA. In contrast, viruses that did not productively infect these cells showed no evidence of early or late reverse transcription products at 18 h postinfection.
FIG. 8.
Real-time PCR analysis of early (A) and late (B) viral transcripts in MDM. MDM were infected with equivalent amounts of DNase-treated virus stocks and analyzed by real-time PCR at 18 or 36 h after infection as described in Materials and Methods. Mock-infected cells were incubated with DNase-treated medium. ADA was heat inactivated by incubation at 56°C for 1 h. The results are expressed as copies of HIV-1 DNA per 10,000 copies of beta-globin. The data shown are representative of two independent experiments using cells obtained from different donors.
To exclude the possibility that this block to reverse transcription was due to slow kinetics of the reverse transcription process in macrophage lineage cells (80), we similarly assessed the levels of viral DNA at 36 h postinfection. No additional DNA sequences in cells originally negative for viral DNA at 18 h were seen. Thus, there is a complete absence of reverse transcription products in these cells. Similar results were obtained in mixed brain cultures, but the levels of reverse transcription products were 10- to 20-fold lower than those in MDM due to the low percentage of microglia in these cultures (data not shown). These experiments demonstrate that a subset of primary R5 isolates (hereafter termed non-M-tropic R5 isolates) cannot replicate in MDM and microglia due to a block in virus entry or an early step prior to reverse transcription.
Effect of CD4 and CCR5 levels on infection by primary R5 isolates.
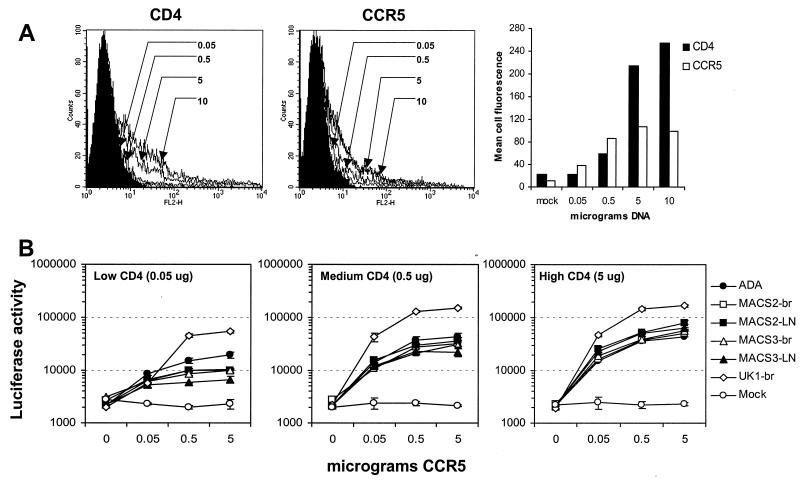
The expression of CD4 on MDM and microglia is significantly lower than that on primary T cells (61, 82). Furthermore, at low concentrations of CD4, relatively high levels of CCR5 are required for entry by some primary R5 viruses (84). Therefore, we investigated whether M-tropic R5 viruses could utilize lower levels of CD4 and/or CCR5 than non-M-tropic R5 viruses. Cf2-Luc cells were transfected with different amounts of CD4- and CCR5-expressing plasmid, ranging from 0.05 to 10 μg, and analyzed by flow cytometry to measure the relative levels of cell surface expression of each receptor (Fig. 9A). A close relationship was found between the levels of CD4 and CCR5 expression and the amount of CD4- and CCR5-expressing plasmid used for transfection. However, this relationship was linear only for transfections of 0.05 to 5 μg of either plasmid. At low, medium, or high levels of CD4 (0.05, 0.5, and 5 μg of CD4-expressing plasmid, respectively), all R5 isolates except UK1-br utilized low, medium, or high levels of CCR5 (0.05, 0.5, and 5 μg of CCR5-expressing plasmid, respectively) with similar efficiency (Fig. 9B). UK1-br entered cells more efficiently than the other viruses, even in cells expressing low levels of CD4 or CCR5, suggesting that UK1-br has reduced dependence on CD4 and CCR5. These findings demonstrate a similar dependence on CD4 and CCR5 levels for entry between M-tropic (e.g., ADA and MACS2-br) and non-M-tropic (e.g., MACS2-LN, MACS3-br, and MACS3-LN) R5 viruses. Therefore, the entry block to infection of MDM and microglia by non-M-tropic R5 viruses is not due to an intrinsic inability to use low levels of CD4 or CCR5 for virus entry.
FIG. 9.
Effect of CD4 and CCR5 levels on infection by R5 viruses. (A) Cf2-Luc cells were transfected with 0.05, 0.5, 5, or 10 μg of CD4- or CCR5-expressing plasmid and analyzed for surface expression of CD4 or CCR5 by flow cytometry. The total amount of DNA in each transfection was adjusted to 10 μg with pCDNA3. (B) Cotransfected Cf2-Luc cells expressing low, medium, or high levels of CD4 (0.05, 0.5, and 5 μg of CD4-expressing plasmid, respectively) and low, medium, or high levels of CCR5 (0.05, 0.5, and 5 μg of CCR5-expressing plasmid, respectively) were infected with equivalent amounts of M-tropic (ADA, MACS2-br, and UK1-br) or non-M-tropic (MACS2-LN, MACS3-br, and MACS3-LN) R5 virus as described in Materials and Methods. Cell lysates were prepared at 48 h postinfection and assayed for luciferase activity. Data are expressed as means from duplicate infections. Error bars represent standard deviations. Similar results were obtained in two independent experiments.
Effect of CD4 overexpression on HIV-1 replication in microglia.
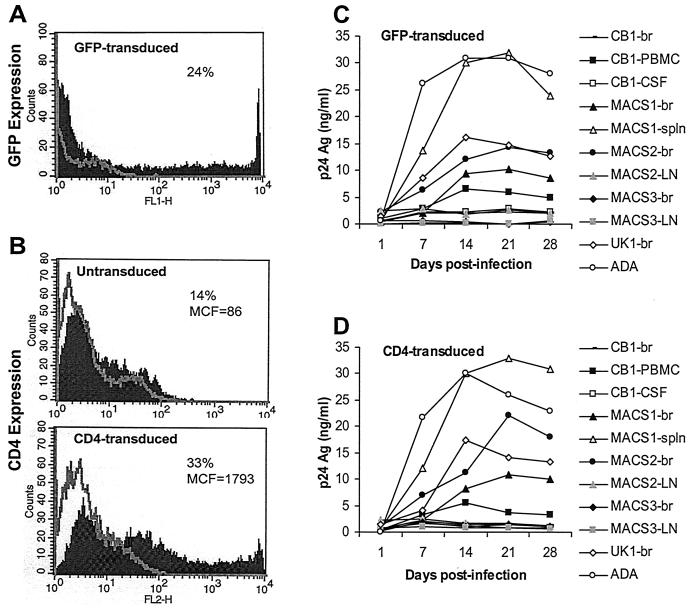
Recent studies have shown that overexpression of CD4 on rhesus macrophages can rescue productive infection by M-tropic HIV-1 or T-tropic simian immunodeficiency virus (SIV) strains (6, 73). To determine whether low levels of CD4 expression on microglia account for the entry block of non-M-tropic R5 viruses, microglia were transduced with the SHIV vector pSIvec1ΔenvhuCD4 to overexpress CD4 and then infected with HIV-1 isolates. To control for the effect of SHIV vector elements on HIV-1 replication, control cultures were similarly transduced with pSIvec1ΔenvGFP. Twenty-four percent of microglia were effectively transduced, as determined by GFP expression (Fig. 10A). Transduction of cells by CD4 resulted in a greater-than-2-fold increase in the number of CD4-positive cells and a 20-fold increase in the mean fluorescence intensity of CD4 (Fig. 10B). The levels of CCR5 expressed on untransduced and CD4-transduced microglia were similar (data not shown). CD4 overexpression was not able to restore virus infection by primary non-M-tropic R5 viruses (e.g., MACS2-LN, MACS3-br, MACS3-LN, and CB1-CSF) and did not enhance replication of M-tropic viruses (e.g., ADA and MACS1-spln) (Fig. 10C and D). Replication kinetics in GFP- and CD4-transduced microglia were similar to those in untransduced cells (data not shown). These results suggest that the inability of non-M-tropic R5 isolates to replicate in microglia is not due to insufficient cell surface expression of CD4.
FIG. 10.
Effect of high CD4 expression on virus replication in microglia. Microglia were untreated or transduced with retroviral vectors to express GFP (A) or CD4 (B). Microglia expressing high levels of CD4 (D) or GFP as a control (C) were infected with equivalent amounts of each HIV-1 isolate as described in Materials and Methods and cultured for 28 days. Virus production in culture supernatants was measured by p24 antigen (Ag) ELISA (NEN). Results are representative of those from two independent experiments with cells obtained from different donors. Percentages represent the proportion of cells expressing GFP or CD4. MCF, mean cell fluorescence.
DISCUSSION
In this study, we isolated and characterized primary viruses from CNS, peripheral blood, and lymphoid tissue from six AIDS patients with dementia and HIV-1 encephalitis. Brain-derived viruses, which consisted of variants that were distinct from those in lymphoid tissues, exhibited diverse patterns of coreceptor usage and used CCR5 (four of six isolates), CXCR4 (one of six isolates), or both CCR5 and CXCR4 (one of six isolates) as primary coreceptors for virus entry. Additional minor usage of CCR3, CCR2b, Apj, or CCR8 was demonstrated for two of six viruses isolated from brain. Previous studies have suggested that brain-derived HIV-1 viruses principally use CCR5 for virus entry (1, 41, 42, 45, 62, 95, 101). However, relatively few brain-derived viruses have been isolated and characterized. Our studies show that CXCR4 can mediate efficient virus entry into microglia and suggest that CXCR4 usage by viruses in brain may be more prevalent than originally thought. Moreover, we demonstrate an association between the abilities of primary viruses to replicate in MDM and microglia irrespective of coreceptor usage. In other studies, we cloned full-length HIV-1 env genes directly from brain and showed that a subset could use both CCR5 and CXCR4 for virus entry (A. Ohagen, A. Devitt, K. J. Kunstman, P. R. Gorry, P. Rose, B. Korber, J. Taylor, R. Levy, R. Murphy, S. Wolinsky, and D. Gabuzda, unpublished data). These findings suggest that M-tropism rather than CCR5 usage predicts HIV-1 neurotropism. Previous studies frequently defined HIV-1 viruses in the brain as M-tropic based on genetic analysis of gp120 Env sequences in the V3 region (20, 39, 51, 58, 85, 88). However, recent studies suggest that the V3 net charge does not predict coreceptor usage or M-tropism (62, 100; Ohagen et al., unpublished data). Although the V3 sequences of the R5X4 viruses MACS1-br and MACS1-spln and the R5 viruses MACS2-LN, MACS3-br, MACS3-LN, and UK1-br contain key amino acids previously shown to distinguish syncytium-inducing and non-syncytium-inducing viruses, respectively (Fig. 2) (71), the highly M-tropic R5 virus MACS2-br does not contain distinguishing sequence changes. Together, these findings indicate that determinants of M-tropism are more complex than coreceptor usage or predictions based on V3 sequences. When the full spectrum of determinants that underlie M-tropism is taken into account, M-tropism of HIV-1 or SIV may indeed predict neurotropism in vivo.
M-tropic HIV-1 viruses typically utilize CCR5 for entry in primary CD4+ cells. However, our studies showed that CCR5 usage is neither necessary nor sufficient for M-tropism. We found that 6 of 11 primary R5 isolates could not replicate in MDM and microglia. Furthermore, inhibition of CXCR4 by 12G5 or AMD3100 abolished virus replication in microglia by the highly M-tropic R5X4 isolates MACS1-br and MACS1-spln, indicating that these isolates entered cells primarily via CXCR4. These findings are consistent with previous studies that failed to establish a strict correlation between CCR5 usage and M-tropism (10, 15, 21, 52, 55) and showed a lack of M- tropism by some R5 HIV-1 clones obtained from brain, lymph node, spleen, and lung tissue (22). Thus, the terms M-tropism and CCR5 usage cannot be used interchangeably.
We quantified early and late viral transcripts by real-time PCR and determined that non-M-tropic R5 isolates were unable to replicate in MDM and microglia due to a block in an early step prior to reverse transcription, presumably at the level of virus entry. Entry and postentry restrictions to replication in MDM have been described for some R5 and X4 HIV-1 viruses. Several studies have demonstrated an entry block in MDM for some R5 viruses (34, 52, 62, 74, 75). Our findings suggest that a similar block to virus entry exists in microglia. Arthos et al. (3) showed that a subset of R5 viruses can enter MDM and synthesize early viral transcripts but are blocked at the postentry level due to an inability to induce signaling via CCR5. Postentry blocks have also been demonstrated for some laboratory-adapted X4 HIV-1 and SIV strains, possibly occurring during or after nuclear translocation of viral DNA (56, 91). In contrast to these studies, our findings suggest that non-M-tropic R5 isolates may be restricted by entry rather than postentry blocks. However, the possibility of a defect in early postpenetration steps prior to reverse transcription (i.e., uncoating) cannot be excluded, and further studies are required to determine whether the restriction is at the entry or postentry step.
Recent studies demonstrated that low levels of CD4 expressed on macrophages from rhesus macaques account for the lack of infection by M-tropic HIV-1 or T-tropic SIV strains (6, 73). Based on these and other studies (19, 60, 84), it has been proposed that low levels of CD4 expression restrict entry in MDM and microglia for some HIV-1 strains. However, we found that CD4 overexpression in microglia was not sufficient to rescue infection by non-M-tropic primary R5 isolates. Furthermore, M-tropic and non-M-tropic R5 viruses showed a similar dependence on CD4 and CCR5 levels for entry in transfected Cf2-Luc cells. Interestingly, we found that UK1-br, a virus isolated from the brain of an intravenous drug user, showed an increased capacity to infect cells expressing low levels of CD4 or CCR5 compared to other primary isolates. Further studies are required to determine whether this results from higher affinity for these receptors, increased exposure of the CCR5 binding site, or yet another mechanism (23). The Env amino acid sequence of UK1-br revealed loss of a potential N-linked glycosylation site at asparagine 76 in the V1/V2 stem region, which corresponds to position 197 in HXB2 (Fig. 2). The elimination of a glycosylation site at this position is sufficient for CD4-independent infection by HIV-1 ADA (57). Studies to determine whether this amino acid change enhances CCR5 affinity and/or reduces CD4 dependence of UK1-br are in progress. Together, these results suggest that the inability of non-M-tropic R5 isolates to replicate in MDM and microglia is not due to insufficient levels of CD4 or CCR5 or to an intrinsic inability to interact efficiently with these receptors. The mechanisms that underlie the restriction to MDM and microglia infection remain to be determined, but possibilities include differences in CCR5 conformation (23, 47) and/or posttranslational modifications such as sulfation (32) or O-linked glycosylation (8, 32, 35) that exist between macrophages, resting T cells, and activated T cells. Other cell-specific factors (23), as well as viral factors such as Nef, could also influence M tropism (5, 72).
We demonstrated an association between the abilities of primary HIV-1 viruses to replicate in MDM and microglia. However, a few viruses replicated slightly (CB3-PBMC and MACS3-br) or moderately (MACS1-br) better in MDM than in microglia (Table 3). This discrepancy most likely reflects the greater numbers and high purity of MDM used as target cells compared to the microglia in mixed brain cell cultures. Other explanations include possible differences in native CD4 and/or coreceptor density or conformation (reference 23 and references therein). Our results are consistent with studies that demonstrated similar tropism of HIV-1 strains for replication in MDM and microglia (41, 46), but they contrast with a study that showed an increased ability of some viruses to replicate in microglia compared to MDM (103). One factor that may explain these discrepancies is the use of different cell culture systems. Our studies and those by Ghorpade et al. (41) used fetal microglia cultured under similar conditions with medium containing M-CSF, whereas Strizki et al. (103) used adult microglia cultured in medium containing giant cell tumor supernatant. Further studies that directly examine HIV-1 replication in fetal versus adult microglia using similar culture conditions may help to elucidate any intrinsic differences in susceptibility to HIV-1 infection that may exist between these two tissue sources. One brain isolate (CB1-br) that principally used CXCR4 for entry (Fig. 3) could not replicate in MDM or microglia (Fig. 5 and 6), thus bringing its neurotropism into question. This isolate might be a minor X4 variant that is not representative of the predominant viral quasispecies, possibly a blood contaminant.
We isolated virus from 6 of 18 autopsy brain tissue samples from patients with AIDS. All patients with positive isolations of virus from brain had dementia and were diagnosed with HIV-1 encephalitis at autopsy. In contrast, brain-derived viruses were not recovered from patients without HIV-1 encephalitis. These findings suggest that a higher local burden of virus in the brain (44) increased the chance of successful virus isolation. Another factor that probably enhanced the success of our virus isolations was the lack of antiviral therapies or use of single agents. Consistent with previous studies (37, 39, 69, 78), the frequency of successful virus isolation from brain was much lower than that of isolation from peripheral blood or lymphoid tissue. Presumably, this discrepancy is due to higher viral loads in peripheral blood and lymphoid compartments compared to brain (7, 24, 44). Sampling different regions of brain might increase the frequency of virus isolation and/or recovery of additional viral variants (101).
V1/V2 and V3 HTA and sequence analyses of viruses isolated from brain revealed major viral species that were distinct from those in viruses isolated from spleen or lymph node of the same patients. The V1/V2 probe was more sensitive than the V3 probe for detecting distinct viral variants, most likely due to greater variability of V1/V2 sequences compared to V3 sequences. Clustered mutations, deletions, or insertions are necessary for changes in heteroduplex mobility. The predicted V1/V2 and V3 amino acid sequences of brain and lymph node isolates from patient MACS3 were identical, despite a shift in mobility of the major V1/V2 heteroduplexes. The difference in V1/V2 heteroduplex mobility for these two isolates most likely resulted from several silent changes in the V1/V2 nucleotide sequences (GenBank accession numbers AF414888 to AF414912). These results demonstrate distinct viral variants in brain compared to lymphoid tissues from the same patients, consistent with previous studies (9, 24, 39, 51, 58, 94, 104, 106), and provide further evidence for tissue-specific compartmentalization of HIV-1.
We found a higher prevalence of brain-derived viruses that used CXCR4 as a coreceptor for entry than in previous studies (1, 37, 39, 95). One difference between our study and previous reports that may explain this finding is the use of CD8-depleted PBMC rather than MDM to isolate viruses. We used this method because it has been shown to be the most sensitive method for virus isolation from lymphocytes and monocytes (16, 59, 102). As expected, we found that coculture with CD8-depleted PBMC was more sensitive for isolation of HIV-1 from brain and lymphoid tissue samples than coculture with MDM. Chemokine receptor expression on PBMC is more heterogeneous than that on MDM. In addition, CD4 expression is higher on PBMC than on MDM (61, 82). Therefore, PBMC would be expected to be more permissive than MDM when used for isolation of primary HIV-1 viruses and thereby would allow the recovery of more diverse strains. Indeed, we found that compared to CD8-depleted PBMC, MDM exerted a selection bias that favored isolation of R5 HIV-1 strains and restricted recovery of variants that used CXCR4 or alternative coreceptors (Table 2). Consistent with this finding, V1/V2 and V3 HTA and sequence analysis of viruses isolated from the same tissues by both methods revealed a cell-dependent selection of distinct viral species.
Our studies suggest that M-tropism, irrespective of coreceptor usage, predicts HIV-1 neurotropism. Consistent with this model, a SHIV (chimeric SIV-HIV) variant that is neurotropic and neurovirulent in rhesus macaques in vivo contains the env gene from the T-cell line-tropic HIV-1 IIIB strain that was adapted for growth in MDM and uses only CXCR4 for virus entry (66). Furthermore, recent studies using SCID mice inoculated intracerebrally with HIV-1-infected MDM demonstrated a relationship between neuropathological changes and the level of virus replication in MDM (A. Nukuna, H. E. Gendelman, J. Limoges, L. Poluektova, J. Rasmussen, A. Ghorpade, and Y. Persidsky, submitted for publication). Neurotropic HIV-1 or SIV isolates are not necessarily neurovirulent (54, 67, 68, 86). In vitro studies have shown that X4 viruses induce neuronal apoptosis more frequently than R5 viruses (81, 109, 110). Similarly, X4 viruses are generally more cytopathic than R5 viruses for cells of the immune system. High-level replication of X4 viruses in MDM and microglia may represent a pathogenic phenotype in the CNS that contributes to neurodegenerative mechanisms in HIV-1 dementia. Understanding the role of strain variability in HIV-1 neurotropism and neurovirulence may advance the development of therapeutics to inhibit CNS infection and prevent neurologic injury in AIDS patients.
ACKNOWLEDGMENTS
We thank H. Naif for assistance with HIV-1 isolation protocols; A. Mehle for sequence analysis; A. Ohagen, J. Wang, M. Farzan, J. Sodroski, S. Gartner, and C. Wood for helpful discussions; and F. Brannan for management of the Edinburgh HIV Brain Bank. We are also grateful to J. Sodroski and B. Etemad-Moghadam for providing Cf2-Luc cells, J. Sodroski for providing SHIV vectors, J. Moore for providing TAK-779, and J. Sodroski, R. Doms, and S. Peiper for coreceptor plasmids.
This work was supported by NIH grants NS37277 and NS35734 to D.G., AI36059 and AI36554 to J.A.Z., AI44667 to R.S., and DA13127 to J.E.B. We also acknowledge support from the Multicenter AIDS Cohort Study (U01 AI35039), a NIDA supplement, and the G. Harold and Leila Mathers Charitable Foundation. Core facilities were supported by Center for AIDS Research grants (AI28691, AI28697, HD37260, and CA79458) and DFCI/Harvard Center for Cancer Research grants. The Edinburgh Brain Bank is supported by UKMRC SPG9708080. K.R. was supported in part by NIH training grant T32-AI07419. K.C. was supported in part by the Robert Wood Johnson Foundation. D.G. and J.A.Z. are Elizabeth Glaser Scientists supported by the Pediatric AIDS Foundation.
REFERENCES
- 1.Albright A V, Shieh J T C, Itoh T, Lee B, Pleasure D, O'Connor M J, Doms R W, Gonzalez-Scarano F. Microglia express CCR5, CXCR4, and CCR3, but of these, CCR5 is the principal coreceptor for human immunodeficiency virus type 1 dementia isolates. J Virol. 1999;73:205–213. doi: 10.1128/jvi.73.1.205-213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Arthos J, Rubbert A, Rabin R L, Cicala C, Machado E, Wildt K, Hanbach M, Steenbeke T D, Swofford R, Farber J M, Fauci A S. CCR5 signal transduction in macrophages by human immunodeficiency virus and simian immunodeficiency virus envelopes. J Virol. 2000;74:6418–6424. doi: 10.1128/jvi.74.14.6418-6424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, Meguro K, Fujino M. A small molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc Natl Acad Sci USA. 1999;96:5698–5703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balliet J W, Kolson D L, Eiger G, Kim F M, McGann K A, Srinivasan A, Collman R. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: mutational analysis of a primary HIV-1 isolate. Virology. 1994;200:623–631. doi: 10.1006/viro.1994.1225. [DOI] [PubMed] [Google Scholar]
- 6.Bannert N, Schenten D, Craig S, Sodroski J. The level of CD4 expression limits infection of primary rhesus monkey macrophages by a T-tropic simian immunodeficiency virus and macrophagetropic human immunodeficiency viruses. J Virol. 2000;74:10984–10993. doi: 10.1128/jvi.74.23.10984-10993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell J E, Busuttil A, Ironside J W, Rebus S, Donaldson Y K, Simmonds P, Peutherer J F. Human immunodeficiency virus and the brain: investigation of virus load and neuropathologic changes in pre-AIDS subjects. J Infect Dis. 1993;168:818–824. doi: 10.1093/infdis/168.4.818. [DOI] [PubMed] [Google Scholar]
- 8.Carlsson S R, Sasaki H, Fukuda M. Structural variations of O-linked oligosaccharides present in leukosialin isolated from erythroid, myeloid, and T-lymphoid cells. J Biol Chem. 1986;261:12787–12795. [PubMed] [Google Scholar]
- 9.Chang J, Jozwiak R, Wang B, Ng T, Ge Y C, Bolton W, Dwyer D E, Randle C, Osborn R, Cunningham A C, Saksena N D. Unique HIV type 1 V3 region sequences derived from six different regions of brain: region-specific evolution within host-determined quasispecies. AIDS Res Hum Retroviruses. 1998;14:25–30. doi: 10.1089/aid.1998.14.25. [DOI] [PubMed] [Google Scholar]
- 10.Cheng-Mayer C, Liu R, Landau N R, Stamatatos L. Macrophage tropism of human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. J Virol. 1997;71:1657–1661. doi: 10.1128/jvi.71.2.1657-1661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choe H, Farzan M, Konkel M, Martin K, Sun Y, Marcon L, Cayabyab M, Berman M, Dorf M E, Gerard N, Gerard C, Sodroski J. The orphan seven-transmembrane receptor apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J Virol. 1998;72:6113–6118. doi: 10.1128/jvi.72.7.6113-6118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 13.Combadiere C, Salzwedel K, Smith E D, Tiffany H L, Berger E A, Murphy P M. Identification of CX3CR1: a chemotactic receptor for the human CX3C chemokine fractalkine and a fusion coreceptor for HIV-1. J Biol Chem. 1998;273:23799–23804. doi: 10.1074/jbc.273.37.23799. [DOI] [PubMed] [Google Scholar]
- 14.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham A L, Li S, Juarez J, Lynch G, Alali M, Naif H. The level of HIV infection of macrophages is determined by interaction of viral and host cell genotypes. J Leukoc Biol. 2000;68:311–317. [PubMed] [Google Scholar]
- 16.Crowe S M, Sonza S. HIV-1 can be recovered from a variety of cells including peripheral blood monocytes of patients receiving highly active antiretroviral therapy: a further obstacle to eradication. J Leukoc Biol. 2000;68:345–350. [PubMed] [Google Scholar]
- 17.Deng H K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 18.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 19.Dimitrov D S, Norwood D, Stantchev T S, Feng Y, Xiao X, Broder C C. A mechanism of resistance to HIV-1 entry: inefficient interactions of CXCR4 with CD4 and gp120 in macrophages. Virology. 1999;259:1–6. doi: 10.1006/viro.1999.9747. [DOI] [PubMed] [Google Scholar]
- 20.Di Stefano M, Wilt S, Gray F, Dubois-Dalcq M, Chiodi F. HIV type 1 V3 sequences and the development of dementia during AIDS. AIDS Res Hum Retroviruses. 1996;12:471–476. doi: 10.1089/aid.1996.12.471. [DOI] [PubMed] [Google Scholar]
- 21.Dittmar M T, McKnight A, Simmons G, Clapham P R, Weiss R A, Simmonds P. HIV-1 tropism and co-receptor use. Nature. 1997;385:495–496. doi: 10.1038/385495a0. [DOI] [PubMed] [Google Scholar]
- 22.Dittmar M T, Simmons G, Donaldson Y, Simmons P, Clapham P R, Schulz T F, Weiss R A. Biological characterization of human immunodeficiency virus type 1 clones derived from different organs of an AIDS patient by long-range PCR. J Virol. 1997;71:5140–5147. doi: 10.1128/jvi.71.7.5140-5147.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Doms R W. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology. 2000;276:229–237. doi: 10.1006/viro.2000.0612. [DOI] [PubMed] [Google Scholar]
- 24.Donaldson Y K, Bell J E, Holmes E C, Hughes E S, Brown H K, Simmonds P. In vivo distribution and cytopathology of variants of human immunodeficiency virus type 1 showing restricted sequence variability in the V3 loop. J Virol. 1994;68:5991–6005. doi: 10.1128/jvi.68.9.5991-6005.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donzella G A, Schols D, Lin S W, Este J A, Nagashima K A, Maddon P J, Allaway G P, Sakmar T P, Henson G, De Clercq E, Moore J P. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat Med. 1998;4:72–77. doi: 10.1038/nm0198-072. [DOI] [PubMed] [Google Scholar]
- 26.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3 and CKR-2b as fusion cofactors. Cell. 1996;85:149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 27.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 28.Edinger A L, Hoffman T L, Sharron M, Lee B, O'Dowd B, Doms R W. Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology. 1998;249:367–378. doi: 10.1006/viro.1998.9306. [DOI] [PubMed] [Google Scholar]
- 29.Edinger A L, Hoffman T L, Sharron M, Lee B, Yi Y, Choe W, Kolson D L, Mitrovic B, Zhou Y, Faulds D, Collman R G, Hesselgesser J, Horuk R, Doms R W. An orphan seven-transmembrane domain receptor expressed widely in the brain functions as a coreceptor for human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol. 1998;72:7934–7940. doi: 10.1128/jvi.72.10.7934-7940.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Etemad-Moghadam B, Sun Y, Nicholson E K, Fernandes M, Liou K, Gomila R, Lee J, Sodroski J. Envelope glycoprotein determinants of increased fusogenicity in a pathogenic simian-human immunodeficiency virus (SHIV-KB9) passaged in vivo. J Virol. 2000;74:4433–4440. doi: 10.1128/jvi.74.9.4433-4440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard N, Gerard C, Sodroski J. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard N P, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 33.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 34.Fouchier R A M, Brouwer M, Kootstra N A, Huisman H G, Schuitemaker H. HIV-1 macrophage tropism is determined at multiple levels of the viral replication cycle. J Clin Investig. 1994;94:1806–1814. doi: 10.1172/JCI117529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fukuda M, Carlsson S R, Klock J C, Dell A. Structures of O-linked oligosaccharides isolated from normal granulocytes, chronic myelogenous leukemia cells, and acute myelogenous leukemia cells. J Biol Chem. 1986;261:12796–12806. [PubMed] [Google Scholar]
- 36.Gabuzda D, Wang J. Chemokine receptors and virus entry in the central nervous system. J Neurovirol. 1999;5:643–658. doi: 10.3109/13550289909021293. [DOI] [PubMed] [Google Scholar]
- 37.Gartner S, Markovits P, Markovits D M, Betts R F, Popovic M. Virus isolation from and identification of HTLV-III/LAV-producing cells in brain tissue from a patient with AIDS. JAMA. 1986;256:2365–2371. [PubMed] [Google Scholar]
- 38.Gartner S, Markovits P, Markovits D M, Kaplan M H, Gallo R C, Popovic M. The role of mononuclear phagocytes in HTLV-III LAV infection. Science. 1986;233:214–218. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 39.Gartner S, McDonald R A, Hunter E A, Bouwman F, Liu Y, Popovic M. Gp120 sequence variation in brain and in T-lymphocyte human immunodeficiency virus type 1 isolates. J Hum Virol. 1997;1:3–18. [PubMed] [Google Scholar]
- 40.Gartner S, Popovic M. Macrophage tropism of HIV-1. AIDS Res Hum Retroviruses. 1990;6:1017–1021. doi: 10.1089/aid.1990.6.1017. [DOI] [PubMed] [Google Scholar]
- 41.Ghorpade A, Nukuna A, Che M, Haggerty S, Persidsky Y, Carter E, Carhart L, Shafer L, Gendelman H E. Human immunodeficiency virus neurotropism: an analysis of viral replication and cytopathicity for divergent strains in monocytes and microglia. J Virol. 1998;72:3340–3350. doi: 10.1128/jvi.72.4.3340-3350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghorpade A, Xia M E, Hyman B T, Persidsky Y, Nukuna A, Bock P, Che M, Limoges J, Gendelman H E, Mackay C R. Role of the β-chemokine receptors CCR3 and CCR5 in human immunodeficiency virus type 1 infection of monocytes and microglia. J Virol. 1998;72:3351–3361. doi: 10.1128/jvi.72.4.3351-3361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh S K, Fultz P N, Keddie E, Saag M S, Sharp P M, Hahn B H, Shaw G M. A molecular clone of HIV-1 tropic and cytopathic for human and chimpanzee lymphocytes. Virology. 1993;194:858–864. doi: 10.1006/viro.1993.1331. [DOI] [PubMed] [Google Scholar]
- 44.Gosztonyi G, Artigas J, Lamperth L, Webster H D. Human immunodeficiency virus (HIV) distribution in HIV encephalitis: study of 19 cases with combined use of in situ hybridization and immunocytochemistry. J Neuropathol Exp Neurol. 1994;53:521–534. doi: 10.1097/00005072-199409000-00012. [DOI] [PubMed] [Google Scholar]
- 45.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Buscigilo J, Yang X, Hofmann W, Newman W, MacKay C R, Sodroski J, Gabuzda D. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 46.Hibbitts S, Reeves J D, Simmons G, Gray P W, Epstein L G, Schols D, De Clercq E, Wells T N C, Proudfoot A E I, Clapham P R. Coreceptor ligand inhibition of fetal brain cell infection by HIV Type 1. AIDS Res Hum Retroviruses. 1999;15:989–1000. doi: 10.1089/088922299310502. [DOI] [PubMed] [Google Scholar]
- 47.Hill C M, Kwon D, Jones M, Davis C B, Marmon S, Daugherty B L, DeMartino J A, Springer M S, Unutmaz D, Littman D R. The amino terminus of human CCR5 is required for its function as a receptor for diverse human and simian immunodeficiency virus envelope glycoproteins. Virology. 1998;248:357–371. doi: 10.1006/viro.1998.9283. [DOI] [PubMed] [Google Scholar]
- 48.Ho D D, Rota T R, Hirsch M S. Infection of monocyte/macrophages by human T lymphotropic virus type III. J Clin Investig. 1986;77:1712–1715. doi: 10.1172/JCI112491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoffman W, Schubert D, LaBonte J, Munson L, Gibson S, Scammell J, Ferringo P, Sodroski J. Species-specific, postentry barriers to primate immunodeficiency virus infection. J Virol. 1999;73:10020–10028. doi: 10.1128/jvi.73.12.10020-10028.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horuk R, Hesselgesser J, Zhou Y, Faulds D, Halks-Millert M, Harvey S, Taub D, Samson M, Parmentier M, Rucker J, Doranz B J, Doms R W. The CC chemokine I-309 inhibits CCR8-dependent infection by diverse HIV-1 strains. J Biol Chem. 1998;273:386–391. doi: 10.1074/jbc.273.1.386. [DOI] [PubMed] [Google Scholar]
- 51.Hughes E S, Bell J E, Simmons P. Investigation of the dynamics of the spread of human immunodeficiency virus to brain and other tissues by evolutionary analysis of sequences from the p17gag and env genes. J Virol. 1997;71:1272–1280. doi: 10.1128/jvi.71.2.1272-1280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hung C S, Pontow S, Ratner L. Relationship between productive HIV-1 infection of macrophages and CCR5 utilization. Virology. 1999;264:278–288. doi: 10.1006/viro.1999.0013. [DOI] [PubMed] [Google Scholar]
- 53.Jinno A, Shimizu N, Soda Y, Haraguchi Y, Kitamura T, Hoshino H. Identification of the chemokine receptor TER1/CCR8 expressed in brain-derived cells and T cells as a new coreceptor for HIV-1 infection. Biochem Biophys Res Commun. 1998;243:497–502. doi: 10.1006/bbrc.1998.8130. [DOI] [PubMed] [Google Scholar]
- 54.Joag S V, Stephens E B, Gailbreath D, Zhu W, Li Z, Foresman L, Zhao L J, Pinson D M, Narayan O. Simian immunodeficiency virus SIVmac chimeric virus whose env gene was derived from SIV-encephalitic brain is macrophage tropic but not neurovirulent. J Virol. 1995;69:1367–1369. doi: 10.1128/jvi.69.2.1367-1369.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kato K, Saito H, Takebe Y. Role of naturally occurring basic amino acid substitutions in the human immunodeficiency virus type 1 subtype E envelope V3 loop on viral coreceptor usage and cell tropism. J Virol. 1999;73:5520–5526. doi: 10.1128/jvi.73.7.5520-5526.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim S S, You X J, Harmon M-E, Overbaugh J, Fan H. Use of helper-free replication-defective simian immunodeficiency virus-based vectors to study macrophage and T tropism: evidence for distinct levels of restriction in primary macrophages and a T-cell line. J Virol. 2001;75:2288–2300. doi: 10.1128/JVI.75.5.2288-2300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J Virol. 2001;75:3435–3443. doi: 10.1128/JVI.75.7.3435-3443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Korber B T M, Kunstman K J, Patterson B K, Furtado M, McEvilly M M, Levy R, Wolinsky S M. Genetic differences between blood- and brain-derived viral sequences from human immunodeficiency virus type 1-infected patients: evidence of conserved elements in the V3 region of the envelope protein of brain-derived sequences. J Virol. 1994;68:7467–7481. doi: 10.1128/jvi.68.11.7467-7481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lafeuillade A, Poggi C, Chadapaud S, Hittinger G, Chouraqui M, Pisapia M, Delbeke E. Pilot study of a combination of highly active antiretroviral therapy and cytokines to induce HIV-1 remission. J Acquir Immune Defic Syndr. 2001;26:44–55. doi: 10.1097/00126334-200101010-00006. [DOI] [PubMed] [Google Scholar]
- 60.Lapham C K, Zaitseva M B, Lee S, Romanstseva T, Golding H. Fusion of monocytes and macrophages with HIV-1 correlates with biochemical properties of CXCR4 and CCR5. Nat Med. 1999;5:303–308. doi: 10.1038/6523. [DOI] [PubMed] [Google Scholar]
- 61.Lewin S R, Sonza S, Irving L B, McDonald C F, Mills J, Crowe S M. Surface CD4 is critical to in vitro HIV infection of human alveolar macrophages. AIDS Res Hum Retroviruses. 1996;12:877–883. doi: 10.1089/aid.1996.12.877. [DOI] [PubMed] [Google Scholar]
- 62.Li S, Juarez J, Alali M, Dwyer D, Collman R, Cunningham A, Naif H M. Persistent CCR5 utilization and enhanced macrophage tropism by primary blood human immunodeficiency virus type 1 isolates from advanced stages of disease and comparison to tissue-derived isolates. J Virol. 1999;73:9741–9755. doi: 10.1128/jvi.73.12.9741-9755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Hui H, Burgess C J, Price R W, Sharp P M, Hahn B H, Shaw G M. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J Virol. 1992;66:6587–6600. doi: 10.1128/jvi.66.11.6587-6600.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao F, Alkhatib G, Peden K W C, Sharma G, Berger E A, Farber J M. STRL33, A novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lipton S A, Gendelman H E. Dementia associated with the acquired immunodeficiency syndrome. N Engl J Med. 1995;332:934–940. doi: 10.1056/NEJM199504063321407. [DOI] [PubMed] [Google Scholar]
- 66.Liu Z Q, Muhkerjee S, Sahni M, McCormick-Davis C, Leung K, Li Z, Gattone V H, Tian C, Doms R W, Hoffman T L, Raghavan R, Narayan O, Stephens E B. Derivation and biological characterization of a molecular clone of SHIVKU-2 that causes AIDS, neurological disease, and renal disease in rhesus macaques. Virology. 1999;260:295–307. doi: 10.1006/viro.1999.9812. [DOI] [PubMed] [Google Scholar]
- 67.Mankowski J L, Flaherty M T, Spelman J P, Hauer D A, Didier P J, Amedee A M, Murphy-Corb M, Kirstein L M, Munoz A, Clements J E, Zink M C. Pathogenesis of simian immunodeficiency virus encephalitis: viral determinants of neurovirulence. J Virol. 1997;71:6055–6060. doi: 10.1128/jvi.71.8.6055-6060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mankowski J L, Spelman J P, Ressetar H G, Strandberg J D, Laterra J, Carter D L, Clements J E, Zink M C. Neurovirulent simian immunodeficiency virus replicates productively in endothelial cells of the central nervous system in vivo and in vitro. J Virol. 1994;68:8202–8208. doi: 10.1128/jvi.68.12.8202-8208.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McGavin C H, Land S A, Sebire K L, Hooker D J, Gurusinghe A D, Birch C J. Syncytium-inducing phenotype and zidovudine susceptibility of HIV-1 isolated from post-mortem tissue. AIDS. 1996;10:47–53. doi: 10.1097/00002030-199601000-00007. [DOI] [PubMed] [Google Scholar]
- 70.Meltzer M S, Nakamura M, Hansen B D, Turpin J A, Kalter D C, Gendelman H E. Macrophages as susceptible targets for HIV infection, persistent viral reservoirs in tissue, and key immunoregulatory cells that control levels of virus replication and extent of disease. AIDS Res Hum Retroviruses. 1990;6:967–971. doi: 10.1089/aid.1990.6.967. [DOI] [PubMed] [Google Scholar]
- 71.Milich L, Margolin B H, Swanstrom R. Patterns of amino acid variability in NSI-like and SI-like V3 sequences and a linked change in the CD4-binding domain of the HIV-1 Env protein. Virology. 1997;239:108–118. doi: 10.1006/viro.1997.8821. [DOI] [PubMed] [Google Scholar]
- 72.Miller M D, Warmerdam M T, Gaston I, Greene W C, Feinberg M B. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J Exp Med. 1994;179:101–131. doi: 10.1084/jem.179.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mori K, Rosenzweig M, Desrosiers R C. Mechanisms for adaptation of simian immunodeficiency virus to replicate in alveolar macrophages. J Virol. 2000;74:10852–10859. doi: 10.1128/jvi.74.22.10852-10859.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Naif H M, Li S, Alali M, Chang J, Mayne C, Sullivan J, Cunningham A L. Definition of the stage of host cell genetic restriction of replication of human immunodeficiency virus type 1 in monocytes and monocyte-derived macrophages using twins. J Virol. 1999;73:4866–4881. doi: 10.1128/jvi.73.6.4866-4881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Naif H M, Li S, Alali M, Sloane A, Wu L, Kelly M, Lynch G, Lloyd A, Cunningham A L. CCR5 expression correlates with susceptibility of maturing monocytes to human immunodeficiency virus type 1 infection. J Virol. 1998;72:830–836. doi: 10.1128/jvi.72.1.830-836.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nelson J A E, Baribaud F, Edwards T, Swanstrom R. Patterns of changes in human immunodeficiency virus type 1 V3 sequence populations late in infection. J Virol. 2000;74:8494–8501. doi: 10.1128/jvi.74.18.8494-8501.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nelson J A E, Fiscus S A, Swanstrom R. Evolutionary variants of the human immunodeficiency virus type 1 V3 region characterized by using a heteroduplex tracking assay. J Virol. 1997;71:8750–8758. doi: 10.1128/jvi.71.11.8750-8758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nyberg M, Suni J, Haltia M. Isolation of human immunodeficiency virus (HIV) at autopsy one to six days postmortem. Am J Clin Pathol. 1990;94:422–425. doi: 10.1093/ajcp/94.4.422. [DOI] [PubMed] [Google Scholar]
- 79.O'Brien W A, Koyanagi Y, Namazie A, Zhao J Q, Diagne A, Idler K, Zack J A, Chen I S. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside of the CD4-binding domain. Nature. 1990;348:69–73. doi: 10.1038/348069a0. [DOI] [PubMed] [Google Scholar]
- 80.O'Brien W A, Namazi A, Kalhor H, Mao S-H, Zack J A, Chen I S Y. Kinetics of human immunodeficiency virus type 1 reverse transcription in blood mononuclear phagocytes are slowed by limitations of nucleoside precursors. J Virol. 1994;68:1258–1263. doi: 10.1128/jvi.68.2.1258-1263.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ohagen A, Ghosh S, He J, Huang K, Chen Y, Yuan M, Osathanondh R, Gartner S, Shi B, Shaw G, Gabuzda D. Apoptosis induced by infection of primary brain cultures with diverse human immunodeficiency virus type 1 isolates: evidence for a role of the envelope. J Virol. 1999;73:897–906. doi: 10.1128/jvi.73.2.897-906.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ometto L, Zanchetta M, Cabrelle A, Esposito G, Mainardi M, Chieco-Bianchi L, De Rossi A. Restriction of HIV type 1 infection in macrophages heterozygous for a deletion in the CC-chemokine receptor 5 gene. AIDS Res Hum Retroviruses. 1999;15:1441–1452. doi: 10.1089/088922299309955. [DOI] [PubMed] [Google Scholar]
- 83.Pierson T, McArthur J, Siliciano R S. Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu Rev Immunol. 2000;18:665–708. doi: 10.1146/annurev.immunol.18.1.665. [DOI] [PubMed] [Google Scholar]
- 84.Platt E J, Wehrly K, Kuhmann S E, Chesebro B, Kabat D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Power C, McArthur J C, Johnson R T, Griffin D E, Glass J D, Perryman S, Chesebro B. Demented and nondemented patients with AIDS differ in brain-derived human immunodeficiency virus type 1 envelope sequences. J Virol. 1994;68:4643–4649. doi: 10.1128/jvi.68.7.4643-4649.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Power C, McArthur J C, Nath A, Wehrly K, Mayne M, Nishio J, Langelier T, Johnson R T, Chesebro B. Neuronal death induced by brain-derived human immunodeficiency virus type 1 envelope genes differs between demented and nondemented AIDS patients. J Virol. 1998;72:9045–9053. doi: 10.1128/jvi.72.11.9045-9053.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Price R W. Neurological complications of HIV infection. Lancet. 1996;348:445–452. doi: 10.1016/S0140-6736(95)11035-6. [DOI] [PubMed] [Google Scholar]
- 88.Reddy R T, Achim C L, Sirko D A, Tehranchi S, Kraus F G, Wong-Staal F, Wiley C A the HIV Neurobehavioral Research Group. Sequence analysis of the V3 loop in brain and spleen of patients with HIV encephalitis. AIDS Res Hum Retroviruses. 1996;12:477–482. doi: 10.1089/aid.1996.12.477. [DOI] [PubMed] [Google Scholar]
- 89.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Samson M, Edinger A L, Stordeur P, Rucker J, Verhasselt V, Sharron M, Govaerts C, Mollereau C, Vassart G, Doms R W, Parmentier M. ChemR23, a putative chemoattractant receptor, is expressed in monocyte-derived dendritic cells and macrophages and is a coreceptor for SIV and some primary HIV-1 strains. Eur J Immunol. 1998;28:1689–1700. doi: 10.1002/(SICI)1521-4141(199805)28:05<1689::AID-IMMU1689>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 91.Schmidtmayerova H, Alfano M, Nuovo G, Bukrinsky M. Human immunodeficiency virus type 1 T-lymphotropic strains enter macrophages via a CD4- and CXCR4-mediated pathway: replication is restricted at a postentry level. J Virol. 1998;72:4633–4642. doi: 10.1128/jvi.72.6.4633-4642.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schols D, Struyf S, Van Damme J, Este J A, Henson G, De Clercq E. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J Exp Med. 1997;186:1383–1388. doi: 10.1084/jem.186.8.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schrager L K, D'Souza M P. Cellular and anatomical reservoirs of HIV-1 in patients receiving potent antiretroviral combination therapy. JAMA. 1998;280:67–71. doi: 10.1001/jama.280.1.67. [DOI] [PubMed] [Google Scholar]
- 94.Shapshak P, Segal D M, Crandall K A, Fujimura R K, Zhang B T, Xin K Q, Okuda K, Petito C K, Eisdorfer C, Goodkin K. Independent evolution of HIV type 1 in different brain regions. AIDS Res Hum Retroviruses. 1999;15:811–820. doi: 10.1089/088922299310719. [DOI] [PubMed] [Google Scholar]
- 95.Shieh J T C, Albright A V, Sharron M, Gartner S, Strizki J, Doms R W, Gonzalez-Scarano F. Chemokine receptor utilization by human immunodeficiency virus type 1 isolates that replicate in microglia. J Virol. 1998;72:4243–4249. doi: 10.1128/jvi.72.5.4243-4249.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shimizu N, Soda Y, Kanbe K, Liu H Y, Jinno A, Kitamura T, Hoshino H. An orphan G protein-coupled receptor, GPR1, acts as a coreceptor to allow replication of human immunodeficiency virus types 1 and 2 in brain-derived cells. J Virol. 1999;73:5231–5239. doi: 10.1128/jvi.73.6.5231-5239.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Siliciano R F. Latency and reservoirs for HIV-1. AIDS. 1999;13(Suppl. A):S49–S58. [PubMed] [Google Scholar]
- 98.Simmons G, Reeves J D, McKnight A, Dejucq N, Hibbitts S, Power C A, Aarons E, Schols D, De Clercq E, Proudfoot A E I, Clapham P R. CXCR4 as a functional coreceptor for human immunodeficiency virus type 1 infection of primary macrophages. J Virol. 1998;72:8453–8457. doi: 10.1128/jvi.72.10.8453-8457.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Simmons G, Wilkinson D, Reeves J D, Dittmar M, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic, and most can use either LESTR or CCR5 as coreceptors. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Singh A, Collman R C. Heterogeneous spectrum of coreceptor usage among variants within a dualtropic human immunodeficiency virus type 1 primary-isolate quasispecies. J Virol. 2000;74:10229–10235. doi: 10.1128/jvi.74.21.10229-10235.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Smit T K, Wang B, Ng T, Osborne R, Brew B, Saksena N K. Varied tropism of HIV-1 isolates derived from different regions of adult brain cortex discriminate between patients with and without AIDS dementia complex (ADC): evidence for neurotropic HIV variants. Virology. 2001;279:509–526. doi: 10.1006/viro.2000.0681. [DOI] [PubMed] [Google Scholar]
- 102.Sonza S, Mutimer H P, Oelrichs R, Jardine D, Harvey K, Dunne A, Purcell D F, Birch C, Crowe S M. Monocytes harbour replication-competent, non-latent HIV-1 in patients on highly active antiretroviral therapy. AIDS. 2001;15:17–22. doi: 10.1097/00002030-200101050-00005. [DOI] [PubMed] [Google Scholar]
- 103.Strizki J M, Albright A V, Sheng H, O'Connor M, Perrin L, Gonzalez-Scarano F. Infection of primary human microglia and monocyte-derived macrophages with human immunodeficiency virus type 1 isolates: evidence of differential tropism. J Virol. 1996;70:7654–7662. doi: 10.1128/jvi.70.11.7654-7662.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.van't Wout A B, Ran L J, Kuiken C L, Kootstra N A, Pais S T, Schuitemaker H. Analysis of the temporal relationship between human immunodeficiency virus type 1 quasispecies in sequential blood samples and various organs obtained at autopsy. J Virol. 1998;72:488–496. doi: 10.1128/jvi.72.1.488-496.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Verani A, Pesenti E, Polo S, Tresoldi E, Scarlatti G, Lusso P, Siccardi A G, Vercelli D. CXCR4 is a functional coreceptor for infection of human macrophages by CXCR4-dependent primary HIV-1 isolates. J Immunol. 1998;161:2084–2088. [PubMed] [Google Scholar]
- 106.Wong J K, Ignacio C C, Torriani F, Havlir D, Fitch N J S, Richman D D. In vivo compartmentalization of human immunodeficiency virus: evidence from the examination of pol sequences from autopsy tissues. J Virol. 1997;71:2059–2071. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yi Y, Rana S, Turner J D, Gaddis N, Collman R G. CXCR-4 is expressed by primary macrophages and supports CCR5-independent infection by dual-tropic but not T-tropic isolates of human immunodeficiency virus type 1. J Virol. 1998;72:772–777. doi: 10.1128/jvi.72.1.772-777.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S Y. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]
- 109.Zheng J, Ghorpade A, Niemann D, Cotter R L, Thylin M R, Epstein L, Swartz J M, Shepard R B, Liu X, Nukuna A, Gendelman H E. Lymphotropic virions affect chemokine receptor-mediated neural signaling and apoptosis: implications for human immunodeficiency virus type 1-associated dementia. J Virol. 1999;73:8256–8267. doi: 10.1128/jvi.73.10.8256-8267.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zheng J, Thylin M R, Ghorpade A, Xiong H, Persidsky Y, Cotter R, Niemann D, Che M, Zeng Y C, Gelbard H A, Shepard R B, Swartz J M, Gendelman H E. Intracellular CXCR4 signaling, neuronal apoptosis and neuropathogenic mechanisms of HIV-1-associated dementia. J Neuroimmunol. 1999;98:185–200. doi: 10.1016/s0165-5728(99)00049-1. [DOI] [PubMed] [Google Scholar]