Abstract
The 192-kb linear DNA genome of vaccinia virus has covalently closed hairpin termini that are extremely AT rich and contain 12 extrahelical bases. Vaccinia virus telomeres have previously been implicated in the initiation of viral genome replication; therefore, we sought to determine whether the telomeres form specific protein-DNA complexes. Using an electrophoretic mobility shift assay, we found that extracts prepared from virions and from the cytoplasm of infected cells contain telomere binding activity. Four shifted complexes were detected using hairpin probes representing the viral termini, two of which represent an interaction with the “flip” isoform and two with the “flop” isoform. All of the specificity for protein binding lies within the terminal 65-bp hairpin sequence. Viral hairpins lacking extrahelical bases cannot form the shifted complexes, suggesting that DNA structure is crucial for complex formation. Using an affinity purification protocol, we purified the proteins responsible for hairpin-protein complex formation. The vaccinia virus I1 protein was identified as being necessary and sufficient for the formation of the upper doublet of shifted complexes, and the vaccinia virus I6 protein was shown to form the lower doublet of shifted complexes. Competition and challenge experiments confirmed that the previously uncharacterized I6 protein binds tightly and with great specificity to the hairpin form of the viral telomeric sequence. Incubation of viral hairpins with extracts from infected cells also generates a smaller DNA fragment that is likely to reflect specific nicking at the apex of the hairpin; we show that the vaccinia virus K4 protein is necessary and sufficient for this reaction. We hypothesize that these telomere binding proteins may play a role in the initiation of vaccinia virus genome replication and/or genome encapsidation.
Poxviruses replicate entirely in the cytoplasm of eukaryotic cells (19). Because of this unusual compartmentalization, poxviruses have evolved to encode a repertoire of proteins required to replicate their DNA genome independently of the nuclear environment, including a DNA polymerase, processivity factor, DNA ligase, uracil DNA glycosylase, ribonucleotide reductase, thymidine kinase, topoisomerase, DNA-independent dNTPase, and protein kinase (reviewed in reference 29). Poxviruses contain a double-stranded linear DNA genome ranging from 160 to 300 kb; the vaccinia virus genome is 192 kb in length and is unusually rich in adenine and thymidine residues (67%) (9). The ends of the viral genome (telomeres) contain a hairpin turnaround that links both DNA strands; thus, one can imagine the genome as one continuous polynucleotide strand (1, 2).
Elucidation of the mechanism by which poxviruses replicate their genome has been challenging, but a working model that incorporates the available experimental evidence has been proposed (1, 28, 29). Initiation of DNA synthesis is believed to occur with the introduction of a nick near one or both of the genomic termini; this nick would expose a 3′ hydroxyl that could serve as a primer terminus and allow the viral DNA polymerase to initiate DNA synthesis. Indeed, experiments performed by Pogo et al. allowed them to infer that the vaccinia virus genome becomes nicked after uncoating (20) and that [3H]thymidine is first incorporated into nascent sequences copied from the terminal 200 bp of the viral genome (21, 22). The newly synthesized strand, representing the viral hairpin, would fold back on itself, allowing replication of the entire genome through strand displacement synthesis. This model would predict the formation of genome concatemers; indeed concatemers have been detected in virally infected cells (16), and the sequences required for resolution of genome concatemers into monomers have been well characterized (6, 17). Formation of more complex, branched intermediates as a result of recombinational priming has also been proposed (5, 18).
More recent evidence for the central role of the viral telomeres in the initiation of genome replication emerged from our previous analyses of minichromosome replication (7). Minichromosomes retaining the secondary structure of the viral genome (covalently closed hairpin termini) were constructed by flanking a plasmid stuffer sequence with viral telomeres of various lengths. These minichromosomes were transfected into virally infected cells, and the efficiency with which they replicated was determined. The terminal 150 to 200 bp of the viral telomere were found to be necessary and sufficient for optimal replication efficiency of the minichromosome templates. These 150 to 200 bp included the terminal hairpin and an adjacent 87 bp of unique sequence.
The terminal hairpin has a unique and intriguing sequence and structure. Within viral genomes, these 104 nucleotides (nt) are found in two isoforms, termed flip and flop (1). These isoforms are inverted and complementary with respect to one another, and, although of the same length, they migrate with different electrophoretic mobilities when resolved on a nondenaturing gel (2). Four nucleotides comprise the apex of the hairpin turnaround. Of the remaining 100 nt, 88 are proposed to fully base pair to form the most distal portion of the double-stranded linear genome; the remaining 12 nt are interspersed throughout this region and, lacking a complement, are considered to be extrahelical. Ten of these extrahelical nucleotides are positioned on one strand, and two are on the complementary strand. Extrahelical bases have been identified in the telomeres of all poxvirus genomes, although their position and number vary.
Since the viral telomeres are likely to play an important role in the initiation of genome replication, we hypothesized that they would form specific protein-DNA complexes with virally encoded proteins. In this report, we indeed show that vaccinia virus encodes telomere binding proteins and that this telomere-protein interaction is dependent on the presence of extrahelical bases. By affinity purification, we identify the vaccinia virus I1 and I6 proteins as being responsible for this telomere binding activity. We also show that when telomeres are incubated with extracts from virally infected cells, a smaller DNA fragment is generated that corresponds to a nick at the apex of the hairpin turnaround. We identify the vaccinia virus K4 protein as being responsible for this nicking activity. The implications of these results will be discussed.
MATERIALS AND METHODS
Materials.
Restriction endonucleases, the Klenow fragment of Escherichia coli DNA polymerase, T4 DNA ligase, and DNA molecular weight standards were purchased from New England Biolabs, Inc. (Beverly, Mass.) or Boehringer Mannheim Biochemicals (Indianapolis, Ind.) and used according to the instructions provided by the manufacturer. [32P]-labeled nucleotide triphosphates were acquired from Dupont Life Sciences (Boston, Mass.). Cytosine-d-arabinofuranoside (araC) was obtained from Sigma Chemical Company (St. Louis, Mo.); poly(dI-dC) was obtained from Amersham Pharmacia Biotech (Piscataway, N.J.). Formamide was acquired from Fluka (Milwaukee, Wis.). Oligonucleotides were obtained from IDT (Coralville, Iowa).
Cells and virus.
Monolayer cultures of mouse L cells were maintained in Dulbecco modified Eagle medium (GIBCO-BRL, Gaithersburg, Md.) containing 5% fetal calf serum. Wild-type (wt) vaccinia virus (WR strain), vΔK4 (kindly provided prior to publication by Michael Merchlinsky, U.S. Department of Agriculture, Bethesda, Md.) (D. Eckert, O. Williams, and M. Merchlinsky, unpublished data), vLacI (13), and vindI1 (10) were amplified in monolayer cultures of BSC40 primate cells or suspension cultures of mouse L cells. Viral stocks were prepared from cytoplasmic lysates of infected cells by ultracentrifugation through 36% sucrose. Where indicated, cytosine arabinoside (araC) was added to cells at a final concentration of 20 μM.
Preparation of cytoplasmic extracts.
L cells were infected with wt virus at a multiplicity of infection (MOI) of 10 and harvested at 24 h postinfection (hpi) unless otherwise indicated. Cells were harvested with a rubber policeman and centrifuged at 834 × g for 5 min at 4°C. Cells were washed in 5 volumes of isotonic buffer (10 mM Tris-HCl [pH 7.9], 150 mM NaCl, 5 mM EDTA) and collected by sedimentation. Cells were resuspended in hypotonic lysis buffer (10 mM Tris-HCl [pH 7.9], 10 mM KCl, 5 mM EDTA) and incubated on ice for 10 min prior to disruption with a Dounce homogenizer. Nuclei were removed by sedimentation at 834 × g for 10 min at 4°C. The supernatant was removed, adjusted to 10% glycerol, and assayed for protein concentration by the Bradford assay (Bio-Rad, Hercules, Calif.).
Preparation of virion extracts.
Virions were purified by banding on a 25 to 40% sucrose gradient and then retrieved by ultracentrifugation (13). Virions were then solubilized by resuspension in a solution containing 20 mM Tris (pH 7.8), 20 mM dithiothreitol (DTT), 50 mM KCl, 0.04 mM EDTA, and 0.2% sodium deoxycholate and incubated on ice for 1 h. After sedimentation at 14,000 × g for 30 min at 4°C, the supernatant was collected, passed through a 23-gauge needle to shear viral DNA, and adjusted to 10% glycerol. The soluble virion extract was then applied to a DEAE-cellulose column to remove the viral DNA. The column was equilibrated and developed in a solution containing 100 mM Tris (pH 8.0), 10 mM DTT, 250 mM KCl, 0.2 mM EDTA, and 10% glycerol. Fractions eluting from the column were collected, and their protein concentration was determined by the Bradford assay; peak fractions were pooled.
EMSA.
The standard 30-μl reaction mixture contained 10 mM Tris-HCl (pH 7.4), 100 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, 10% glycerol, 5 μg of bovine serum albumin, 104 cpm of radiolabeled probe, 2.2 μg of poly(dI-dC), and 2 μg of protein extract unless otherwise indicated. Competitor DNA was added to a 100-fold molar excess prior to the addition of protein extract, unless otherwise indicated. Electrophoretic mobility shift assay (EMSA) reactions were incubated at 30°C for 15 min and then applied directly to a nondenaturing gel containing 6% acrylamide, 0.16% bisacrylamide, 2.5% glycerol, and 1× Tris-glycine buffer (50 mM Tris, 380 mM glycine, 2 mM EDTA). Electrophoresis was performed in 1× Tris-glycine buffer at 11.5 V/cm for 5 to 5.5 h at 4°C. Gels were dried and exposed for autoradiography on film or a phosphor screen; data were acquired on a Storm PhosphoImager (Molecular Dynamics, Sunnyvale, Calif.) and quantitated using ImageQuant software (Molecular Dynamics).
Preparation of DNA used in EMSA. (i) Viral hairpins and extended duplexes.
The plasmids pHS, p150, and p65+tet have been described previously (7). Briefly, pHS contains an ∼400-bp palindromic insert derived from the HinfI concatemer junction fragment of vaccinia virus replication intermediates. This insert contains the extended hairpin sequences flanked on each side by an 87-bp unique region and the first of the tandem repeats found within the telomeres of the viral genome. The palindromic insert within p150 contains the extended hairpin sequences and the flanking 87-bp unique region. In addition, this plasmid contains a 6-bp EcoRI site inserted at the central axis of the palindrome. The p65+tet plasmid contains an ∼400-bp chimeric palindrome in which the central 130-bp region of the concatameric junction is flanked on each side by 130 bp derived from the bacterial tetracycline resistance gene. For all of these plasmid inserts, the palindromes are imperfect in the central region that encodes the extended duplex form of the flip and flop hairpins of mature viral genomes. (The pSV9 plasmid used as the templates for the construction of these plasmids was kindly provided by M. Merchlinsky [16]; pHS, p150, and p65+tet were generated by Du and Traktman (7)).
Probes for EMSAs were produced by digesting these plasmids with SalI to release the concatameric junction fragment insert (∼400 bp for pHS and p65+tet, ∼300 bp for p150). The recessed 3′ termini generated by SalI digestion were made flush using the Klenow fragment of E. coli DNA polymerase in the presence of α-32P-labeled nucleoside triphosphates. The reaction was stopped by heating to 68°C for 15 min, and the DNA was resolved on an agarose gel. After staining with ethidium bromide, the fragment was excised, glass purified (30), and eluted in Tris-EDTA. The fragments were either used directly as probes in the EMSA (in the extended duplex form) or converted to the hairpin forms of the viral telomere by heating them to 95°C for 4 min and cooling them immediately in an ice-water bath for 5 min. Probe recovery was quantitated by measuring the absorbance at an optical density of 260 nm (OD260) or by ethidium bromide-UV visualization using an AlphaImager digital gel documentation system (Alpha Innotech, San Leandro, Calif.); specific activity of the radiolabeled probes was determined by Cerenkov counting. When nonradiolabeled extended duplex or hairpin DNAs were prepared for use as competitors in the EMSA reactions, they were prepared in an analogous fashion except for the use of nonradiolabeled deoxynucleoside triphosphates in the Klenow extension reaction.
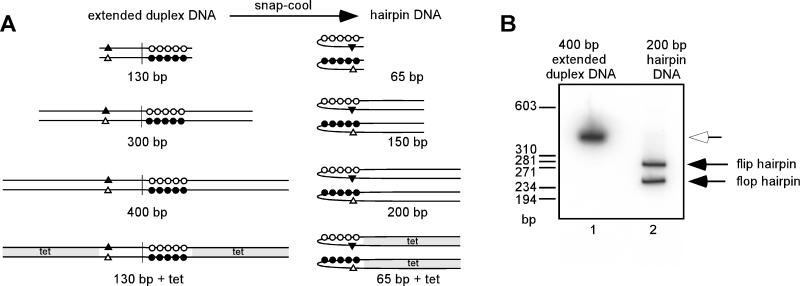
Throughout this report, “200 bp” and “400 bp” represent the hairpin and extended duplex forms, respectively, of the ∼400-bp HinfI concatemer junction fragment released from pHS by restriction enzyme digestion; “150 bp” and “300 bp” represent the hairpin and extended duplex forms, respectively, of the 300-bp insert released from p150 by restriction enzyme digestion; and “65 bp+tet” and “130 bp+tet” represent the hairpin and extended duplex forms, respectively, of the ∼400-bp insert released from p65+tet by restriction enzyme digestion. The extended duplexes described above are completely base paired and linear. Upon heating and snap-cooling, they each generate two hairpin isoforms. These isoforms (flip and flop) are inverted and complementary with respect to one another and contain extrahelical bases. Figure 1A shows a schematic representation of these probes.
FIG. 1.
DNA constructs used in EMSAs. (A) Schematic of DNA probes and competitors. Viral telomere sequences were present in one of two conformations: extended duplex or hairpin form. The extended duplex form represents the concatemer junction fragment found in replication intermediates and is palindromic with the exception of the nucleotides that will eventually become extrahelical. In this schematic, each circle and triangle denotes 2 nt that will eventually become extrahelical, and the vertical line denotes the axis of symmetry. The extended duplex form is completely base paired (open circles anneal with complementary closed circles, and closed triangle anneals with complementary open triangle). To generate viral hairpins, extended duplex DNA was heated to denature the two strands and then cooled rapidly. The two hairpins generated, flip and flop, are incompletely base paired and accurately resemble the telomeres of the viral genome. Nomenclature for the DNA constructs is related to the length of viral sequence. The 130-bp extended duplex is snap-cooled to form 65-bp hairpins. The 300-bp extended duplex is snap-cooled to form 150-bp hairpins. The 400-bp extended duplex is snap-cooled to form 200-bp hairpins. The 130-bp+tet construct contains 130 bp of viral sequence plus nonviral sequences derived from the bacterial tetracycline resistance gene (tet, shaded), such that the extended duplex totals 400 bp in length. The hairpins formed from this extended duplex are 200 bp in length and contain the terminal 65 bp of viral sequence plus 135 bp of bacterial sequence. (B) Nondenaturing gel analysis of viral hairpins. The 400-bp extended duplex was terminally radiolabeled and resolved on a 5% polyacrylamide–TBE gel before and after snap-cooling (lanes 1 and 2, respectively). The white arrow indicates the 400-bp extended duplex form, and the two solid arrows indicate the flip and flop hairpins that are generated. Note that even though each viral hairpin is 200 bp in length, the two isoforms migrate more slowly than expected and possess distinct electrophoretic mobilities. The sizes of the linear DNA markers are indicated at the left in base pairs.
ii. Synthetic hairpin competitors.
A 26-nt palindromic oligonucleotide was snap-cooled and filled in using the Klenow fragment of E. coli DNA polymerase to form a perfectly annealed 13-bp hairpin with four thymidine residues at its apex (7). A 38-nt palindromic oligonucleotide containing a vitamin D response element from the mouse osteopontin gene promoter (DR3′) was snap-cooled to form a perfectly annealed 19-bp hairpin (4).
iii. Oligonucleotide competitors.
Oligonucleotides were annealed in Tris-EDTA containing 100 mM NaCl by heating equimolar amounts to 100°C for 5 min and cooling slowly to room temperature over 2 h. When annealed, T oligo (5′ ATTTCCTTCAGCAGATAGGAACCATACTGATTCACAT) and B oligo (5′ ATGTGAATCAGTATGGTTCCTATCTGCTGAAGGAAAT) formed a 37-bp duplex. T oligo and B + 1 oligo (5′ ATGTGAATCAGTCATGGTTCCTATCTGCTGAAGGAAAT) formed a 37-bp duplex containing one extrahelical base (bold) (+1EHB). T oligo and B + 2 oligo (5′ ATGTGAATCAGTCATGGTGTCCTATCTGCTGAAGGAAAT) formed a 37-bp duplex containing two extrahelical bases spaced 5 nt apart (bold) (+2EHB).
Visualization of flip and flop hairpins by nondenaturing polyacrylamide gel electrophoresis.
The radiolabeled 400-bp extended duplex probe and the 200-bp hairpins generated upon snap-cooling were adjusted to contain 5% glycerol, applied to a 5% polyacrylamide gel cast and run in 1× TBE (100 mM Tris, 83 mM boric acid, 1 mM EDTA) at 12 V/cm for 2.5 h. A lane containing HaeIII-digested phage φX174 DNA markers was visualized by ethidium bromide staining; the remainder of the gel was dried and exposed for autoradiography.
Selective radiolabeling of the flip or flop isoform of the viral hairpins.
To radiolabel only the flip isoform of the viral telomere, p65+tet was digested with Asp718 and radiolabeled with the Klenow subunit of E. coli DNA polymerase I in the presence of [α-P32]dATP, [α-P32]TTP, dGTP, and dCTP. The reaction was terminated by heating to 68°C for 15 min, and the DNA was retrieved by phenol-chloroform extraction and ethanol precipitation. The DNA was then digested with BamHI to release the extended duplex fragment, which was resolved by agarose gel electrophoresis, purified on glass beads, and subjected to heating and snap-cooling to form hairpins. To radiolabel only the flop isoform of the viral telomere, p65+tet was digested with BamHI and then radiolabeled, heat treated, and purified as described above. The DNA was then digested with Asp718 to release the extended duplex fragment, which was gel purified and snap-cooled to form hairpins.
Construction of viral hairpins that lack extrahelical bases.
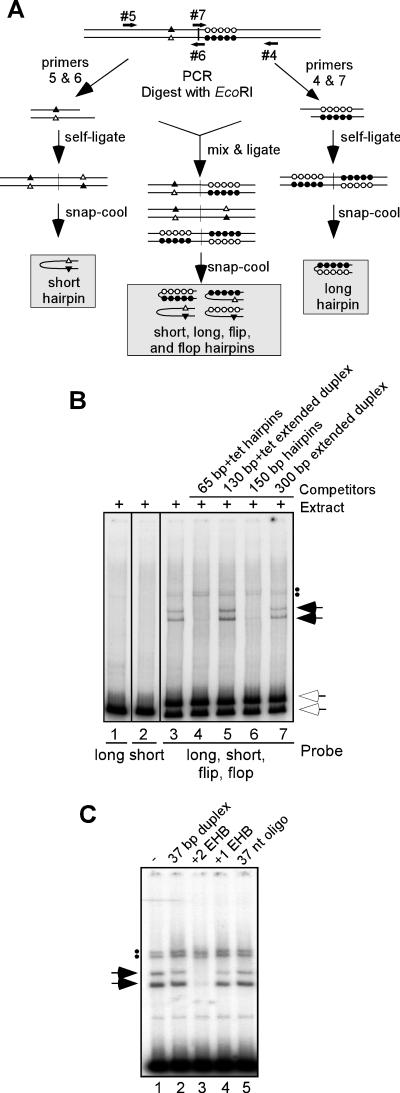
Construction of viral hairpins lacking extrahelical bases relied on PCRs designed to amplify the individual halves of the 300-bp concatameric resolution fragment engineered to contain an EcoRI site at the central axis of the palindrome. Self-ligation of the individual products yielded completely palindromic fragments which, in turn, generated homogeneous and completely base-paired hairpins upon heating and snap-cooling (see Fig. 3A). A PCR was performed using primer #4 (5′ GCGGATCCGTAGACTGTGTATAAAGCGATCG) and primer #7 (5′ GGGAATTCAAGTTAGTAAATTATATATATAAT) to amplify half of the concatemer junction fragment, yielding a 158-bp product. Another PCR was performed using primer #5 (5′ GCGGTACCGTCGACTCTATAAAGCGATCG) and primer #6 (5′ GGGAATTCTAGTTAGATAAATTAATAATATATAAG) to amplify the opposite half of the concatemer junction fragment, yielding a 142-bp product. Primers #6 and #7 contained EcoRI sites (underlined). The above-described PCRs were performed in the presence of [α-P32]dATP and [α-P32]TTP. The radiolabeled PCR products were purified, digested with EcoRI, purified, and incubated at 16°C overnight in the presence of T4 DNA ligase. The 142-bp fragment underwent self-ligation to form completely palindromic 284-bp dimers by virtue of the overhangs generated by EcoRI digestion. Similarly, the 158-bp product yielded 316-bp palindromic dimers upon self-ligation. A ligation reaction containing both the 142- and 158-bp products was also performed. This ligation yielded three products: a 300-bp imperfect palindrome (heterotypic ligation) and the 284- and 316-bp perfect palindromes (homotypic ligations).
FIG. 3.
Extrahelical bases are required for recognition by telomere binding proteins. (A) Strategy for synthesis of viral hairpins lacking extrahelical bases. PCR was performed to amplify the halves of the concatemer junction. Primers 5 and 6 amplified the half giving rise to the two extrahelical bases (triangles), and primers 4 and 7 amplified the half giving rise to the 10 extrahelical bases (circles). PCR was performed in the presence of [α-P32]dATP and [α-P32]dTTP. PCR products of 150 bp in length were gel purified and digested with EcoRI (primers 6 and 7 contained an EcoRI site). These products were then self-ligated to create completely palindromic fragments that, when snap-cooled, gave hairpins that were completely base paired: short form (open triangle annealing with closed triangle) and long form (closed circles annealing with open circles). When the two PCR products were mixed, ligated, and snap-cooled, a mix of four types of hairpins was formed: the flip and flop isoforms that contain extrahelical bases as well as the long and short forms that lack extrahelical bases. (B) DNA-protein complex formation requires the presence of extrahelical bases. Hairpins made using the strategy described for panel A were utilized as probes in the standard EMSA: lane 1, long probe; lane 2, short probe; lanes 3 to 7, mix of short, long, flip, and flop probes. Competitor DNA in either the hairpin (lanes 4 and 6) or extended duplex (lanes 5 and 7) form was added in 100-fold molar excess to the mixed probe prior to the addition of cytoplasmic extract harvested at 24 hpi (compare lane 3 with lanes 4 to 7). The upper white arrow indicates the migration of the free flip and flop hairpins, while the lower white arrow indicates the migration of the free long and short hairpins. The solid arrows and dots indicate the lower and upper doublets of shifted complexes formed with the mixed probe, respectively. No shifted complexes were formed on either the long or short perfect hairpins that lack extrahelical bases. (C) Oligonucleotide duplexes containing two extrahelical bases compete for the telomere binding activity. EMSA reactions were performed using 1.5 fmol of 65-bp+tet probe. Competitor DNA (67 pmol) was added to EMSA reactions prior to the addition of cytoplasmic extract. Lane 1, no competitor; lane 2, a completely base paired 37-mer duplex; lane 3, a 37-bp duplex with two extrahelical bases (+2 EHB); lane 4, a 37-bp duplex with one extrahelical base (+1 EHB); lane 5, single-stranded 37-nt oligonucleotide. The solid arrows and dots indicate the lower and upper doublets of shifted complexes, respectively. Only the +2 EHB oligonucleotide competes for complex formation.
The ligated products were resolved from the unligated input DNA on an 8% polyacrylamide TBE gel; the DNA was visualized by ethidium bromide staining and UV illumination, excised, and eluted from the gel as described previously (24). The linear double-stranded products were then heated and snap-cooled to form hairpins. The 284-bp palindromic product formed a 142-bp hairpin (termed short) that is completely base paired (the two normally extrahelical bases now have complements). The 316-bp palindromic product formed a 158-bp hairpin (termed long) that is also completely base paired (the 10 extrahelical bases now have complements). Heating and snap-cooling of the ligation performed with both the 142- and 158-bp products yielded four hairpins: the long and short forms generated by self-ligation as well as the flip and flop forms that are normally present in the viral genomes.
Denaturing gel electrophoresis of viral hairpins.
Reactions prepared for EMSAs were incubated for 30 min at 30°C. Reactions were then treated with 10 μg of proteinase K for an additional 30 min at 30°C. An equal volume of 2× sample buffer (96% formamide, 20 mM EDTA, xylene cyanol, and bromophenol blue) was added to each reaction. Samples were heated to 95°C for 3 min and then applied to a 5% acrylamide–0.25% bisacrylamide gel containing 16 M formamide and cast and run in 20 mM KH2PO4 (pH 7.4)–1 mM EDTA (8). Electrophoresis was performed at 14 V/cm until the bromophenol blue had migrated off the gel. The gel was fixed in 5% methanol–5% acetic acid for 45 min, dried, and exposed for autoradiography.
Affinity purification of telomere binding proteins using biotinylated hairpins conjugated to streptavidin-coated magnetic beads.
pHS was digested with SalI to release the 400-bp junction fragment, which was then terminally biotinylated using the Klenow fragment of E. coli DNA polymerase I in the presence of 20 μM biotin-11-dUTP. The 400-bp fragment was resolved on an agarose gel, purified on glass beads, and snap-cooled to form 200-bp hairpins. Biotinylated hairpins were conjugated to Dynabeads M-280 streptavidin-coated magnetic beads (Dynal, Oslo, Norway) in 1× B&W buffer (5 mM Tris-HCl [pH 7.4], 1 M NaCl, 0.5 mM EDTA) for 30 min at room temperature with occasional mixing. DNA was bound at a ratio of 40 pmol of 200-bp DNA per mg of beads, according to the manufacturer's instructions. The amount of biotinylated DNA conjugated to the beads was monitored by measuring the OD260 of DNA before and after incubation with beads. Magnetic beads were pulled down using a Dynal EC-1 magnet.
Hairpin-conjugated beads were washed in 1× gel shift buffer (10 mM Tris-HCl [pH 7.4], 0.2 mM EDTA, 0.5 mM DTT) containing 150 mM KCl. Typical affinity purification reactions were performed by incubating 1.5 mg of hairpin-conjugated beads in a reaction containing 1× gel shift buffer, infected cytoplasmic L cell extract (from 500 μg to 2 mg), 150 mM KCl, 10% glycerol, 100 μg of poly(dI-dC), and protease inhibitors (1 μg of leupeptin/μl, 1 μg of pepstatin/μl, 1 mM phenylmethylsulfonyl fluoride) for 30 min at 30°C. Beads were washed with solutions of increasing ionic strength, and proteins were eluted at room temperature in 1× gel shift buffer containing 10% glycerol and protease inhibitors. Fractions were analyzed by EMSA and stored at −80°C.
Identification of proteins by MALDI-TOF mass spectrometry.
Fractions from affinity purifications were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by silver staining. Bands of interest were excised and analyzed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry and post-source decay fragment analysis by John Leszyk at the Laboratory for Protein Microsequencing and Proteomic Mass Spectrometry, University of Massachusetts Medical School, Shrewsbury, Mass.
Cloning of the vaccinia virus I6 gene.
The I6 gene was amplified by PCR from genomic viral DNA (WR strain) using an upstream primer (5′ CCATCTCCATATGAATAACTTTGTTAAAAC) and downstream primer (5′ CCGGATCCTCAAAGAATATGTGACAAAG). The resulting 1,155-bp fragment was digested with NdeI (underlined) and BamHI (bold) and ligated into pET14b plasmid DNA (26) (Clontech, Palo Alto, Calif.) that had been previously digested with NdeI and BamHI and treated with calf intestine alkaline phosphatase. This placed the I6 gene in frame with an amino-terminal hexahistidine tag and under the control of a T7 RNA polymerase promoter.
Cloning of the vaccinia virus K4 gene.
The K4 gene was amplified by PCR from genomic viral DNA (WR strain) using an upstream primer (5′ CACCATGGCATATGAATCCGGATAATAC) and downstream primer (5′ CCGGATCCTTATTCAAGAGAATATT). The resulting 1,275-bp fragment was digested with NdeI (underlined) and BamHI (bold) and ligated into pET16b plasmid DNA that had been previously digested with NdeI and BamHI and treated with calf intestine alkaline phosphatase. This placed the K4 gene in frame with an amino-terminal decahistidine tag and under the control of a T7 RNA polymerase promoter.
Expression and purification of recombinant HisI6 protein.
BL21(DE3) E. coli cells transformed with pET14b-HisI6 were grown in tryptone-phosphate medium supplemented with 50 μg of ampicillin/ml to an OD550 of 0.5 to 0.6. Cells were then treated with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and 2% ethanol. After 20 min on ice, cultures were incubated at 20°C for 36 h with agitation (250 rpm). Cells were harvested by sedimentation and lysed by incubation in 1× binding buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 10% glycerol, 5 mM imidazole) on ice for 30 min. Cells were disrupted using a French press, and the lysate was clarified by sedimentation at 14,400 × g for 20 min at 4°C. The supernatant was filtered through a 0.45 μM syringe filter and applied to a Ni-nitrilotriacetic acid agarose column (Qiagen). The column was washed with binding buffer, and proteins were eluted in binding buffer supplemented with increasing concentrations of imidazole.
Peak fractions were pooled, diluted to 250 mM NaCl, and applied to a heparin-agarose column (Sigma) equilibrated in a solution containing 20 mM Tris (pH 7.4), 250 mM NaCl, 1 mM EDTA, 1 mM DTT, and 10% glycerol. The column was washed with the same buffer, and proteins were eluted with increasing concentrations of NaCl. Fractions were analyzed by SDS-PAGE and visualized by silver staining.
Expression and purification of recombinant HisK4 protein.
HMS174 E. coli cells transformed with pET16b-HisK4 were grown in Luria-Bertani medium supplemented with 50 μg of ampicillin/ml and 0.2% maltose to an OD550 of 0.5 to 0.6. Cells were then infected with the lambda bacteriophage CE6 (which encodes the T7 RNA polymerase) (26). After 20 min of adsorption at room temperature without shaking, cultures were maintained at 37°C for 30 min with agitation (250 rpm) and then shifted to 25°C with agitation for 3 h. Cells were harvested by sedimentation and lysed by incubation in 1× binding buffer (20 mM Tris-HCl [pH 7.9], 500 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 10% glycerol, 5 mM imidazole) containing 2 mg of lysozyme/ml on ice for 30 min. Cells were disrupted using a French press, and the lysate was clarified by sedimentation at 14,400 × g for 20 min at 4°C. The supernatant was filtered through a 0.45-μM-pore-size syringe filter and applied to a Ni-nitrilotriacetic acid agarose column (Qiagen). The column was washed with binding buffer, and proteins were eluted in binding buffer containing increasing concentrations of imidazole. Fractions were analyzed by SDS-PAGE and visualized by silver staining. Peak fractions were pooled, concentrated using a Centricon-10 spin concentrator (Millipore, Bedford, Mass.), and desalted by dilution and reconcentration in 20 mM Tris-HCl [pH 7.9], 100 mM NaCl, 1 mM EDTA, and 10% glycerol.
Immunoblot analysis.
After resolution by SDS-PAGE, proteins were transferred electrophoretically to nitrocellulose filters in Tris-glycine buffer (27). Filters were probed with either INDIA His-probe-horseradish peroxidase (Pierce, Rockford, Ill.) or with an anti-I1 primary antiserum (10) and a horseradish peroxidase-conjugated secondary antiserum. Proteins were then detected by chemiluminescence (Pierce).
Computer analysis.
Autoradiographs were scanned with a SAPHIR scanner (Linotype-Hell Co., Hauppauge, N.Y.) and adjusted with Adobe Photoshop software (Adobe Systems Inc., San Jose, Calif.). Labeling of figures was performed using Canvas software (Deneba Systems, Miami, Fla.). Data were plotted using SigmaPlot software (SSPS, Chicago, Ill.)
Primers.
The sequences for all PCR primers used for cloning were based on the vaccinia virus genomic sequence, Copenhagen strain, GenBank accession number NC_001559.
RESULTS
Our laboratory has previously developed a minichromosome replication assay designed to determine which cis-acting sequences within the vaccinia virus genome are necessary for DNA replication. We observed that telomeres containing the terminal 200 bp of the vaccinia virus genome are sufficient to confer optimal replication efficiency on minichromosome templates when transfected into virally infected cells (7). These termini are comprised, primarily, of a 52-bp AT-rich hairpin and a flanking region of 87 bp. The hairpin has interesting structural characteristics, including the distal turnaround and the presence of extrahelical bases (10 on one strand, 2 on the other); moreover, within the viral genome, the hairpin is found in two isoforms that are inverted and complementary with respect to one another [termed flip and flop] (1). The 87-bp sequence is less unusual in nature, but it is indispensable for minichromosome replication. Based on our assumption that the hairpin and/or the 87-bp sequence might recruit key proteins to the telomeres during genome replication, we initiated a search for such telomere-protein complexes.
Preparation of DNA probes for use in EMSA.
We chose to use an EMSA to investigate whether probes representing the viral telomeres did indeed form stable and specific interactions with viral and/or cellular proteins. To prepare probes that accurately represented the viral telomeres, we utilize a cloned version of the vaccinia virus concatameric junction fragment. This was accomplished by digesting the plasmid pHS with SalI to release the 400-bp insert derived from the vaccinia virus concatameric junction fragment; the recessed 3′ termini of this fragment were then filled in with radiolabeled deoxynucleoside triphosphates. This extended duplex form was heated to dissociate the strands and then snap-cooled immediately in an ice-water bath, causing each strand to fold back on itself to form self-annealed hairpins. Because the concatameric junction fragment is actually an imperfect palindrome, two different hairpins are formed, one in the flip conformation and the other in the flop conformation (see Fig. 1A for a schematic explanation). These hairpins accurately represent the sequences and structures that exist at the termini of the vaccinia virus genome.
To confirm the formation of the hairpin isoforms, the radiolabeled concatameric junction fragment was analyzed by nondenaturing gel electrophoresis before and after snap-cooling. Figure 1B shows that before snap-cooling, the concatameric junction fragment migrates at approximately 400 bp (lane 1). After snap-cooling, the two dissociated strands (each 400 nt) have formed distinct 200-bp hairpins (lane 2). Note that even though the flip and flop hairpins contain the same number of nucleotides, they migrate with different electrophoretic mobilities, as previously observed (2). These hairpins possess the extrahelical bases found in the viral telomeres, and it is presumably these structural features that are responsible for the distinct electrophoretic mobilities of the two isoforms.
Viral proteins form four complexes with telomeric hairpins.
Radiolabeled hairpins containing the terminal 200 bp of the vaccinia virus genome were incubated with cytoplasmic extracts and analyzed by nondenaturing gel electrophoresis to monitor DNA-protein complex formation. While a cytoplasmic extract from uninfected cells did not alter the migration of the hairpin probes, a cytoplasmic extract prepared from virally infected cells harvested at 24 hpi caused the formation of four shifted complexes: a lower doublet and an upper doublet (Fig. 2A, compare lanes 1 and 2). It should be emphasized that these reactions were performed in the presence of an excess of poly(dI-dC) and were not stabilized by the addition of fixative. To determine the specificity of the telomere binding activity, a variety of competition experiments were performed. The added 100-fold molar excess of nonviral hairpins (13- and 19-bp) did not compete for complex formation, as evidenced by the unchanged intensity of the shifted doublets (lanes 3 and 4). These data, along with additional data presented later in this report, suggest that recognition of the telomere probe is not due to a generic affinity for the turnaround feature of the hairpins.
FIG. 2.
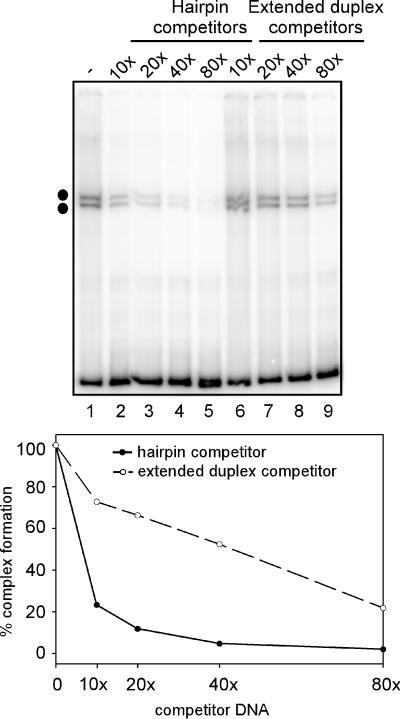
Viral proteins form four specific complexes with telomeric hairpins. (A) All the specificity for binding lies within the terminal 65-bp telomeric sequence. EMSA reactions were performed using the 200-bp radiolabeled hairpin probe derived from the vaccinia virus telomere and cytoplasmic extracts prepared from uninfected cells (UE, lane 1) or infected cells harvested at 24 hpi (IE, lanes 2 to 12). Reactions were incubated for 15 min at 30°C and resolved on a nondenaturing 6% polyacrylamide Tris-glycine gel. Competitor DNA in either the hairpin (lanes 3 to 8) or extended duplex (lanes 9 to 12) conformation was added in 100-fold molar excess prior to addition of cytoplasmic extract, as indicated above each lane. The two arrows indicate specific DNA-protein complexes that form on viral hairpins exclusively in the presence of virally infected cell extracts and can be competed away by viral hairpins (lanes 5 to 8) but not by nonviral hairpins (lanes 3 and 4) or the extended duplex form of the telomeric sequences (lanes 9 to 12). The two dots indicate specific DNA-protein complexes formed exclusively in the presence of virally infected cell extracts that are not subjected to competition by the addition of any DNA competitors. (B) Hairpin conformation is required for DNA-protein complex formation. EMSA reactions were performed using radiolabeled DNA corresponding to hairpin or extended duplex forms of the viral telomere and cytoplasmic extracts harvested from infected cells at 24 hpi. The following probes were used: lanes 1 to 3, 150-bp hairpins; lanes 4 to 6, 200-bp hairpins; lanes 7 and 8, 65-bp+tet hairpins; lanes 9 to 11, 300-bp extended duplex; lanes 12 to 14, 400-bp extended duplex; lanes 15 to 17, 130-bp+tet extended duplex. Competitor DNA in either the hairpin (lanes 2, 3, 5, 6, and 8) or extended duplex (lanes 10, 11, 13, 14, 16, and 17) conformation was added in 100-fold molar excess prior to the addition of cytoplasmic extract, as indicated by the angled text above each lane. The two arrows and two dots indicate four specific DNA-protein complexes that form on viral hairpin probes but not extended duplex probes.
To define which regions of the viral telomere were responsible for binding, competitions were also performed using hairpins containing either the terminal 200, 150, or 65 bp of viral sequences (hairpins illustrated in Fig. 1A; data shown in Fig. 2A, lanes 5 to 8). All of these unlabeled viral hairpins served as competitors for the lower doublet of shifted complexes, as evidenced by the disappearance of these bands. Interestingly, when the extended duplex forms of these hairpins (which contain the same primary sequence but a different secondary structure) were tested for their ability to compete in the EMSA reactions, they were found to be inert (Fig. 2A, lanes 9 to 12), suggesting that the proteins that interact with the viral telomeres are likely to recognize some or all of the unique structural features of the hairpin isoforms.
These data were confirmed in additional experiments in which each of the hairpin and extended duplex forms were radiolabeled and used as probes in EMSA reactions. Protein complexes formed on both the 150-bp and 65-bp+tet viral hairpins, yielding four shifted complexes that were comparable to those seen with the 200-bp hairpin (Fig. 2B, lanes 1, 4, and 7); none of the extended duplexes were competent to form shifted complexes (Fig. 2B, lanes 9 to 17). As predicted from the data shown in Fig. 2A, unlabeled versions of all of the viral hairpins could compete for the formation of the lower doublet of shifted complexes (Fig. 2B, lanes 2, 3, 5, 6, and 8). Taken together, these data indicate that the terminal 65 bp of the viral telomeres contain all of the structural and sequence features that are recognized by the telomere binding protein(s) responsible for the formation of the upper and lower doublets of shifted complexes.
Interestingly, the upper doublet of shifted complexes could not be competed away in any of these experiments. Nevertheless, formation of this doublet of shifted complexes appeared to be specific, since it required the use of hairpin probes rather than extended duplex probes and was generated by extracts prepared from infected, but not uninfected, cells.
Extrahelical bases are required for telomere binding activity.
The data shown above indicated that the hairpin form, but not the extended duplex form, of the telomeric sequences formed specific and stable complexes with proteins found exclusively within infected cells. One key difference between these two forms of DNA is the presence of extrahelical bases: extended duplex DNA is completely base paired, while viral hairpins are incompletely base paired. To determine whether the extrahelical bases that are so characteristic of the poxvirus hairpin telomeres are required for telomere binding activity, novel viral hairpins were constructed that were completely base paired (see Fig. 3A for a schematic representation). Briefly, PCR was performed using the 300-bp concatemer junction fragment as a template and primer sets that separately amplified the individual “halves” of the concatameric junction fragment—the short and long halves that would normally contribute the 2 or 10 extrahelical bases to the final hairpin form, respectively. Each of the two PCR products was digested with EcoRI and self-ligated to generate a fully palindromic duplex containing an EcoRI site at the axis of symmetry.
When these palindromic products were snap-cooled, each formed a fully self-complementary hairpin. Within the long hairpin, the 10 bases that are normally extrahelical now could anneal to their complements, as did the two normally extrahelical bases in the short hairpin (Fig. 3A). These radiolabeled hairpins were then used as probes in EMSA reactions. The long and short forms of the hairpins, which are essentially 150-bp perfectly base paired hairpins, did not generate any shifted complexes in EMSA reactions (Fig. 3B, lanes 1 and 2). These hairpins were also unable to serve as competitors for the telomere binding activity (data not shown).
When a mixed ligation reaction was performed using both the long and short EcoRI-digested PCR products, three possible ligation products were formed: long-to-long, short-to-short, and short-to-long. When snap-cooled, four different types of hairpins were formed: the fully duplexed short and long forms and the extrahelical base-containing flip and flop forms. These flip and flop hairpins, in contrast to the long and short, formed the characteristic upper and lower doublets of shifted complexes (lane 3). Detection of these complexes was, as usual, abrogated by the inclusion of hairpin but not extended duplex competitors (lanes 4 to 7). These data provide strong evidence that the extrahelical bases are indeed essential for the specific recognition of the telomeres by proteins present within infected cells. Furthermore, the fact that the perfectly base paired hairpin probes did not form shifted complexes also reinforces our previous observations that the telomere binding proteins do not have nonspecific hairpin binding activity.
To further analyze the necessity and sufficiency of extrahelical bases for complex formation, competitions were performed using oligonucleotide heteroduplexes, some of which contained extrahelical bases. While a perfectly base-paired 37-mer duplex was not able to compete for the formation of the lower doublet of shifted complexes (Fig. 3C, lane 2), a 37-mer duplex containing two extrahelical bases, spaced 5 nt apart, competed strongly when present in vast excess (lane 3). This competition did not occur when the 37-mer contained only one extrahelical base (lane 4) or when only a single-stranded oligonucleotide was used (lane 5). The sequences for these oligos, which did not contain any homology to the AT-rich viral hairpins, were taken from a previous report in which EMSA reactions were used to study the binding specificity of the mismatch repair proteins MSH2 and MSH6 (14). These data strongly reinforce the conclusion that the extrahelical features of the viral hairpins are crucial for recognition by the telomere binding proteins and that multiple extrahelical nucleotides are probably required. None of the oligonucleotide competitors were able to interfere with upper-doublet complex formation under these conditions.
Different isoforms of the viral telomeres form electrophoretically distinct complexes.
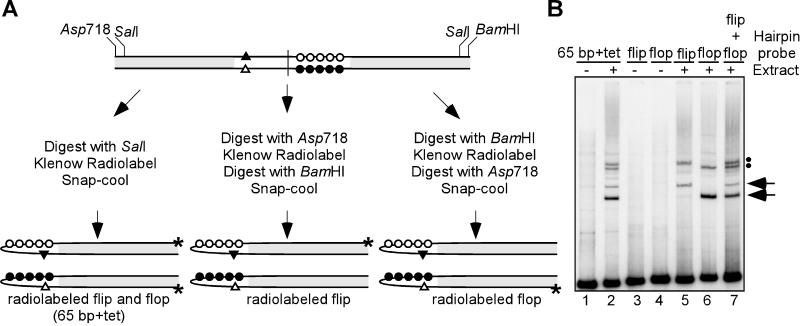
As mentioned above, vaccinia virus telomeres exist in two isoforms (flip and flop) that are inverted and complementary with respect to one another. As shown in Fig. 1B, the flip and flop isoforms migrate with different electrophoretic mobilities under certain nondenaturing conditions despite being identical in length. We therefore wished to test the possibility that the doublets of shifted complexes seen reproducibly in our EMSA reactions reflect the formation of distinct complexes on the flip and flop isoforms. We were able to selectively radiolabel the individual isoforms by virtue of unique restriction enzyme sites present in the p65+tet plasmid (see Fig. 4A for a schematic representation).
FIG. 4.
Different isoforms of the viral telomeres form electrophoretically distinct complexes. (A) Strategy for selective labeling of the flip or flop viral hairpins. To label the individual isoforms of the viral hairpins, p65+tet was digested with either Asp718 (to label flip) or BamHI (to label flop) prior to the 3′-labeling reaction and subdigestion with the other enzyme. Initial digestion of p65+tet with SalI allows for simultaneous labeling of both isoforms. (B) EMSA using radiolabeled flip and flop hairpins. Hairpins prepared using the above strategy were used as probes in EMSA reactions: lanes 1 and 2, simultaneously labeled 65-bp+tet hairpins; lanes 3 and 5, radiolabeled flip hairpin; lanes 4 and 6, radiolabeled flop hairpin; lane 7, a mixture of radiolabeled flip and flop hairpins. EMSA reactions were performed in the presence (lanes 2, 5, 6, and 7) or absence (lanes 1, 3, and 4) of cytoplasmic extract prepared from infected cells at 24 hpi. The solid arrows and dots indicate the lower and upper doublets of shifted complexes, respectively.
When only the flip isoform of the viral telomere was radiolabeled, two DNA-protein complexes representing the upper band of each doublet were observed (Fig. 4B, lane 5). In contrast, when only the flop isoform was radiolabeled, two DNA-protein complexes representing the lower band of each doublet were observed (lane 6). Combining the two probes in one reaction restored the upper doublet/lower doublet pattern seen when both isoforms were radiolabeled simultaneously (compare lanes 2 and 7). These data indicated that the flip and flop isoforms each participate in the formation of two protein-DNA complexes and that the electrophoretic mobilities of these complexes are isoform specific.
Telomere binding activity accumulates after the onset of viral DNA replication and is encapsidated in virions.
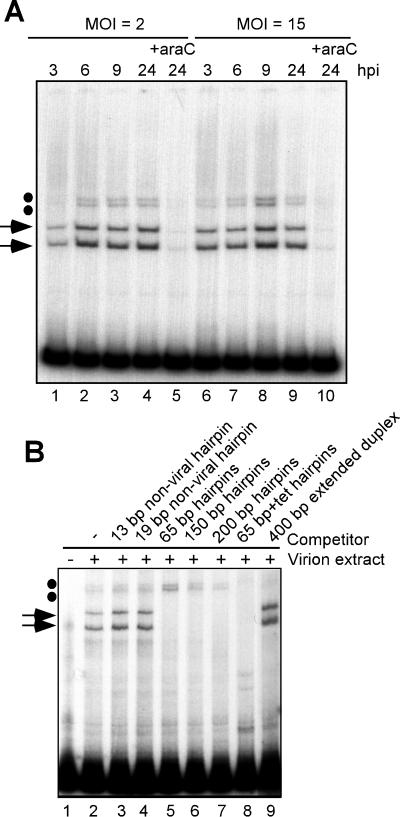
Up to this point, EMSA experiments were performed using a cytoplasmic extract harvested at 24 h postinfection. To begin to understand the possible functions of these telomere binding activities in vivo, cytoplasmic extracts were harvested at various times after infection and assayed by EMSA (Fig. 5A). Telomere binding activity was first detected at 3 hpi and was present throughout the viral life cycle in cells infected at either low or high MOI (lanes 1 to 4 and 6 to 9). Cells infected in the presence of araC, an inhibitor of viral DNA replication and hence of the subsequent stages of intermediate and late gene expression, did not accumulate telomere binding activity (lanes 6 and 10). This suggests that one or more of the proteins required for telomere binding activity is expressed as an intermediate or late gene product, or, alternatively, that the activity of these proteins is manifested only after the onset of DNA replication, possibly due to posttranslational modification.
FIG. 5.
Telomere binding activity accumulates after the onset of DNA replication and is encapsidated in virions. (A) Time course of telomere binding activity. Cytoplasmic extracts were prepared at various hpi from cells infected at an MOI of 2 or 15 and analyzed by EMSA using the 65-bp+tet hairpin probe. Lanes 1 to 4, extracts harvested at 3, 6, 9, or 24 hpi from cells infected at an MOI of 2; lanes 6 to 9, extracts harvested at 3, 6, 9, or 24 hpi from cells infected at an MOI of 15. For lanes 5 and 10, cells were treated with 20 μM araC throughout the infection so as to block DNA replication and hence intermediate and late gene expression. (B) EMSA using virion extracts. A soluble extract of purified wt virions was applied to a DEAE-cellulose column to remove viral DNA. The protein extract was then analyzed by EMSA using the 150-bp viral hairpin probe (compare lanes 1 and 2). Competitor DNA in either the hairpin (lanes 3 to 8) or extended duplex (lane 9) conformation was added in 100-fold molar excess prior to addition of virion extract. Competitors were as follows: lane 3, 13-bp nonviral synthetic hairpin; lane 4, 19-bp nonviral synthetic hairpin; lanes 5 to 8, 65-bp, 150-bp, 200-bp and 65-bp+tet viral hairpins, respectively; lane 9, 400-bp extended duplex. The arrows and dots indicate the lower and upper doublets of shifted complexes, respectively.
We postulated that since the telomere binding activity was present at late times of infection, it might also be encapsidated in virions. We performed EMSA reactions using a soluble protein extract prepared from purified, wt virions (Fig. 5B). Indeed, telomere binding activity was detected in virions, with the appearance of the characteristic upper doublet and lower doublet of shifted complexes (lane 2). The encapsidated telomere binding activity retained the binding specificity described above for the cytoplasmic activity, since viral hairpins (lanes 5 to 8), but not nonviral hairpins (lanes 3 and 4) or extended duplexes (lane 9), were able to compete for the formation of the lower doublet of shifted complexes.
The vaccinia virus I1 protein is responsible for the formation of the upper doublet of shifted complexes.
As a prelude to addressing the function(s) of the telomere binding proteins, we initiated efforts to purify them using an affinity purification protocol. Viral hairpins were terminally biotinylated and conjugated to streptavidin-coated magnetic beads. Our approach was to isolate specific hairpin-protein complexes from a cytoplasmic extract in a single chromatographic step by including an excess of soluble poly(dI-dC) in the reactions to eliminate the retrieval of nonspecific DNA-binding proteins.
The hairpin beads were incubated with a cytoplasmic extract harvested at 24 hpi, under conditions similar to those used in EMSA reactions and in the presence of poly(dI-dC). Beads containing hairpin-protein complexes were retrieved using a magnet, washed, and developed with the stepwise application of buffers containing increasing concentrations of KCl. Elutions from the hairpin beads were analyzed by both EMSA and SDS-PAGE (Fig. 6A and B, respectively). Telomere binding activity was present in the initial cytoplasmic extract, as shown by EMSA (Fig. 6A, lane 1), and remained associated with the hairpin beads until the application of the 500 mM KCl (upper and lower doublets, lane 6) and 1,000 mM KCl washes (lower doublet, lane 7).
FIG. 6.
The vaccinia virus I1 protein binds telomeres and is responsible for the upper doublet of shifted complexes. (A and B) Identification of I1 by affinity purification. The 200-bp viral hairpins were biotinylated and conjugated to streptavidin-coated magnetic beads. Hairpin beads were then incubated with an infected cell extract using conditions similar to those used in EMSA reactions, including the presence of poly(dI-dC) and 150 mM KCl. Beads were collected using a magnet and developed with buffer containing increasing concentrations of KCl. Washes were assayed for telomere binding activity by EMSA using the 65-bp+tet hairpin probe (A) and analyzed in parallel by SDS-PAGE and silver staining (B). Lanes 1, cytoplasmic extract before incubation with beads; lanes 2, cytoplasmic extract after incubation with beads (flow through); lanes 3 and 4, 150 mM KCl washes; lanes 5, 250 mM KCl wash; lanes 6, 500 mM KCl wash; lanes 7, 1,000 mM KCl wash. The 35-kDa band in the 500 mM KCl wash (panel B, lane 6, lower gray arrow) was excised and identified as the vaccinia virus I1 protein by mass spectroscopy (see the text). Protein standards are shown at the right with their molecular masses indicated in kilodaltons. (C) The vaccinia virus I1 protein is necessary and sufficient for complex formation. Cytoplasmic extracts from uninfected cells (lane 1) or infected cells harvested at 24 hpi (lane 2 to 4) were analyzed by EMSA using the 65-bp+tet hairpin probe (upper panel) and by immunoblot analysis using a polyclonal anti-I1 serum (lower panel). Cells were infected with the following: lane 2, wt virus (wtVV); lane 3, vLacI (a virus expressing the lac repressor protein); lane 4, vindI1 in the absence of IPTG. In lane 5, 320 ng of His-tagged recombinant I1 protein (HisI1) was used in the EMSA reaction (upper panel) and immunoblot analysis (lower panel). Dots and black arrows in panels A and C indicate the upper and lower doublets of shifted complexes, respectively; gray arrows in panel B indicate the 35- and 40-kDa proteins discussed in the text. Protein standards are shown at the right with their molecular masses indicated in kilodaltons.
Analysis of the elutions by SDS-PAGE and silver staining revealed that a prominent protein with a molecular mass of 35 kDa was present only in the 500 mM KCl fraction (Fig. 6B, lane 6), while a protein of 40 kDa appeared to be restricted to the 1,000 mM KCl fraction (lane 7). As controls, similar pull-down experiments were performed with extracts prepared from uninfected cells or with empty beads. It was clear from these control pull-down experiments that the 35- and 40-kDa proteins were specific to infected cell extracts and that their retention and purification required the presence of the viral hairpins on the magnetic beads (data not shown).
The 35-kDa protein was excised from the gel and submitted for analysis by mass spectrometry. MALDI-TOF mass analysis of the trypsinized protein identified three peptide sequences that matched amino acids 2AEFEDQLVFNSISAR16, 166DESIQLDEK174, and 236IASILSLETVK246 of the vaccinia virus I1 protein, respectively. We had previously identified I1 as a 35-kDa viral protein that is expressed at late times during infection, encapsidated within the virion core, and essential for the assembly of mature virions (10). Recombinant I1 protein had been shown to bind single-stranded and double-stranded DNA in an EMSA.
Results of the affinity purification experiment shown in Fig. 6 indicated that I1 was the predominant protein in the 500 mM KCl fraction that was competent for upper-doublet complex formation. Two approaches were taken to test whether the I1 protein was directly responsible for the formation of these shifted complexes. First, the radiolabeled hairpin probe was incubated in EMSA reactions containing a cytoplasmic extract prepared from cells infected with vindI1 (a virus in which the I1 gene is under the control of the lac operator/repressor) (10) in the absence of IPTG. Indeed, when expression of the I1 protein was repressed, the extract no longer formed the upper doublet of telomere-protein complexes (Fig. 6, panel C, top, lane 4). Immunoblot analysis of these extracts confirmed that they did not contain detectable I1 protein (Panel C, bottom, lane 4). This result indicated that the I1 protein is necessary for the formation of the upper doublet of shifted complexes.
Second, to test whether the I1 protein interacts directly with viral telomeres, an EMSA reaction was performed in which the radiolabeled hairpin probe was incubated with recombinant, His-tagged I1 protein (HisI1) (Fig. 6C, top, lane 5) (10). Recombinant I1 protein formed a doublet of shifted complexes that exhibited a slightly slower electrophoretic mobility than the typical upper doublet seen with wt-infected cell extracts. This retardation is most likely due to the presence of the hexahistidine tag on the recombinant I1 protein, which adds ∼2 kDa to the I1 protein and causes it to migrate more slowly than authentic I1 (Fig. 6C, bottom, lane 5). From these cumulative data, we conclude that the vaccinia virus I1 protein forms a stable and specific complex with viral hairpins in the presence of poly(dI-dC) and that the I1 protein is both necessary and sufficient for formation of the upper doublet of shifted complexes.
In the EMSA reactions performed up to this point, we were never able to inhibit formation of the upper doublet of radiolabeled, shifted complexes by addition of competitor DNA. A typical EMSA reaction contains approximately 1,000 fmol of poly(dI-dC3000) and 1 fmol of radiolabeled hairpin probe. Since I1 has been previously shown to bind DNA nonspecifically, it is likely to bind both the hairpin probe and the vast excess of poly(dI-dC) present in these EMSA reactions. When nonradiolabeled competitor hairpins are added (at approximately 100 fmol per reaction), this competitor might drive I1 protein off of poly(dI-dC) and thus not appear to compete for complex formation on the radiolabeled hairpin probe.
EMSA reactions were therefore performed using purified HisI1 protein in the absence of poly(dI-dC). HisI1 formed the upper doublet of shifted complexes when incubated with radiolabeled viral hairpins (Fig. 7, upper panel, lane 1). Under these conditions, nonradiolabeled viral hairpins did indeed compete for complex formation in a concentration-dependent manner (lanes 2 to 5). To determine whether I1 discriminated between viral hairpins and the extended duplex form of the same sequence, the extended duplex form was also tested for its ability to compete for upper doublet formation (Fig. 7, lanes 6 to 9). Viral hairpins were a better competitor than the extended duplex DNA; these data are represented graphically in the lower panel. Whereas a 10-fold molar excess of viral hairpin DNA was able to reduce the yield of complex to 20% of that seen in the absence of competitor, an 80-fold molar excess of extended duplex DNA was required to achieve a comparable level of complex disruption (compare lanes 9 and 2). Thus, HisI1, while able to bind both forms of DNA, has a higher affinity for the hairpin form that resembles the viral telomere.
FIG. 7.
Analysis of I1-hairpin complex formation. EMSA reactions were performed using 30 fmol of recombinant HisI1 protein and 1.3 fmol of radiolabeled 200-bp viral hairpins in the absence of poly(dI-dC) (upper panel). Competitor DNA in either the hairpin (lanes 2 to 5) or extended duplex (lanes 6 to 9) conformation was added to reactions prior to the addition of HisI1, in the molar excess indicated above each lane. The two dots indicate the DNA-protein complexes that form on viral hairpins in the presence of the I1 protein. The levels of I1-hairpin complexes observed in each lane were quantitated using a Storm Phosphorimager; a graphic representation is shown in the lower panel. The level of complex formed in the absence of competitor (lane 1) was set to 100%, and the remaining data were normalized relative to this level.
The vaccinia virus I6 protein is responsible for the formation of the lower doublet of shifted complexes.
Our initial affinity purification experiment (Fig. 6) did not yield sufficient levels of the 40-kDa protein found in the 1,000 mM KCl elution for us to determine its identity. We repeated the purification with some modifications to optimize the recovery of the protein(s) responsible for lower doublet formation. Basically, we used extracts prepared from cells infected with vindI1 in the absence of IPTG; these extracts lack I1 and form only the lower doublet of complexes. In addition, we used more protein and modified the elution protocol slightly. Fractions from this modified affinity purification were analyzed by both EMSA and SDS-PAGE (Fig. 8A and B, respectively). The lower doublet of telomere binding activity was present in the input cytoplasmic extract as shown by EMSA (Fig. 8A, lane 1), remained associated with the hairpin beads through the application of buffer containing 350 mM KCl (lanes 3 to 6), and was eluted in the 1,000 mM KCl wash (lane 7).
FIG. 8.
The vaccinia virus I6 protein binds telomeres and is responsible for the lower doublet of shifted complexes. (A and B) Identification of I6 by affinity purification. The 200-bp viral hairpins were biotinlyated and conjugated to streptavidin-coated magnetic beads. Hairpin-conjugated beads were then incubated with a cytoplasmic extract prepared from cells infected with vindI1 in the absence of IPTG; the conditions used were similar to those used in EMSA, including the presence of poly(dI-dC) and 150 mM KCl. Beads were collected using a magnet and developed with buffer containing increasing concentrations of KCl. Washes were assayed for telomere binding activity by EMSA using the 65-bp+tet hairpin probe (A) and analyzed in parallel by SDS-PAGE and silver staining (B). Lanes 1, cytoplasmic extract before incubation with beads; lanes 2, cytoplasmic extract after incubation with beads (flow through); lanes 3 and 4, 150 mM KCl washes; lanes 5, 250 mM KCl wash; lanes 6, 350 mM KCl wash; lanes 7, 1,000 mM KCl wash. The 40-kDa band in the 1,000 mM KCl wash (panel B, lane 7) was excised and identified as the vaccinia virus I6 protein by mass spectroscopy (see the text). Protein standards are shown at the right with their molecular masses indicated in kilodaltons. (C) Expression and purification of HisI6. The vaccinia virus I6 gene was cloned into pET14b and expressed in E. coli with an N′-terminal hexahistidine tag. Extracts were applied to a nickel-agarose column, and the HisI6 fraction was further purified on a heparin-agarose column developed with increasing concentrations of NaCl. Peak elutions from the nickel-agarose column (Fraction I, lane 1) and heparin-agarose column (Fraction II, lanes 2 and 3) were analyzed by SDS-PAGE and visualized by silver staining (lanes 1 and 2) or immunoblot analysis using a probe reactive with the polyhistidine tag (lane 3). The final preparation (Fraction II) contained a single 40-kDa species. (D) HisI6 forms the lower doublet of shifted complexes. EMSA reactions were performed using the 65 bp+tet hairpin probe and a cytoplasmic extract prepared from uninfected cells (lane 1) or infected cells harvested at 24 hpi (lane 2). For lanes 3 and 4, EMSA reactions were performed using increasing amounts of the HisI6 protein (Fraction II, 150 and 300 ng, respectively). In panels A and D, the black arrows indicate the lower doublet of shifted complexes; in panel B, the gray arrow indicates the 40-kDa protein chosen for analysis.
As shown by SDS-PAGE analysis (Fig. 8B), the use of more protein in these pull-down reactions increased the background of nonspecific proteins seen in all the elution fractions. Nevertheless, it was apparent that the 1,000 mM KCl fraction contained a major protein species with a molecular mass of approximately 40 kDa (lane 7). This band, which corresponded to that seen in our earlier experiment, was carefully excised from the gel and submitted for analysis by mass spectrometry. Following trypsin digestion, MALDI-TOF mass analysis identified six peptide sequences that matched amino acids 114HFNSSLISIK124, 262TFTPLNASPYIPK274, 276IVSLLDLPSNVEIK289, 294GGVDFITHINNK305, 321NSTFSGTFIK330, and 346SSFPVPTIK354 of the vaccinia virus I6 protein.
There are no previous publications concerning the vaccinia virus I6 gene, which is predicted to encode a protein with a molecular mass of 43.5 kDa, nor are any potential functions suggested by sequence analysis. In order to test if the vaccinia virus I6 protein interacts directly with viral telomeres, the I6 gene was first amplified by PCR from the vaccinia virus genome and cloned into a pET expression vector, such that it would be expressed as a fusion protein containing a hexahistidine tag at the amino terminus. The HisI6 protein was bacterially expressed and purified on a nickel-agarose column (Fig. 8C, lane 1). Peak elutions were then applied to a heparin-agarose column to further purify the HisI6 protein (lane 2). A 40-kDa band was recovered, and its identity as the HisI6 protein was confirmed by showing its reactivity with a probe specific for the histidine tag (lane 3).
When recombinant HisI6 protein was incubated with viral hairpins in EMSA reactions, two shifted complexes were formed in a dose-dependent manner; these comigrated with the lower doublet of shifted complexes seen with cytoplasmic extracts of wt-infected cells (Fig. 8D, lanes 3 and 4 versus lane 2). We thus concluded that the vaccinia virus I6 protein interacts directly with hairpins derived from the viral telomeres and is responsible for the lower doublet of telomere binding activity.
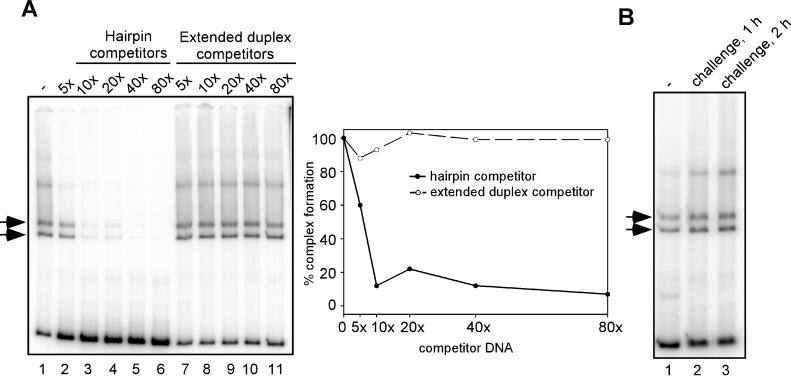
As an initial investigation into the binding specificity of I6, we examined whether the purified I6 protein displayed a binding preference for viral hairpins. As described above for HisI1, EMSA reactions were performed in which HisI6 protein was added to hairpin probes that had been mixed with competitor DNA in either the hairpin or extended duplex conformation (Fig. 9). Whereas competitor DNA in the hairpin conformation was able to inhibit complex formation by 90% at a 10-fold molar excess (Fig. 9A, lanes 2 to 6), extended duplex DNA did not perturb complex formation even when added at a 100-fold molar excess (lanes 7 to 11); a graphic representation of the data is shown at the right. This competition experiment provides a clear demonstration of the specificity with which HisI6 discriminated between the hairpin and extended duplex forms of the viral telomeric DNA. We next performed a challenge experiment to evaluate the stability of the HisI6-hairpin interaction. As before, EMSA reactions were performed using HisI6 and the radiolabeled hairpin probe; reactions were then either analyzed directly or challenged with a 20-fold molar excess of nonradiolabeled hairpin DNA. Figure 9B shows the results of the 1- and 2-h challenge experiments: the intensity of the radiolabeled shifted complex remained unchanged (compare lane 1 with lanes 2 and 3). These data indicate that even when challenged for 2 h, the bound HisI6 protein did not disassociate from the complex and redistribute on the available DNA. Taken together, these data provide compelling evidence that the vaccinia virus I6 protein binds to telomeric hairpin probes with a high degree of specificity and stability.
FIG. 9.
Analysis of I6-hairpin complex formation. (A) HisI6 binds to viral hairpins but not to extended duplex DNA. EMSA reactions were performed using 30 fmol of recombinant HisI6 protein and 1.3 fmol of radiolabeled 200-bp viral hairpins in the absence of poly(dI-dC). Competitor DNA in either the hairpin (lanes 2 to 6) or extended duplex (lanes 7 to 11) conformation was added to reactions prior to addition of HisI6, in the molar excess indicated above each lane. The two arrows indicate the DNA-protein complexes that form on viral hairpins in the presence of the I6 protein. The levels of I6-hairpin complexes observed in each lane were quantitated using a Storm Phosphorimager; a graphic representation is shown in the right panel. The level of complex formed in the absence of competitor (lane 1) was set to 100%, and the remaining data were normalized relative to this level. (B) HisI6 forms highly stable complexes with viral hairpins. EMSA reactions were performed using 30 fmol of recombinant HisI6 protein and 1.3 fmol of radiolabeled 200-bp viral hairpins in the absence of poly(dI-dC). For lanes 2 and 3, HisI6 was allowed to bind to the hairpin probe for 15 min, at which point a 20-fold molar excess of nonradiolabeled, hairpin competitor DNA was added to the EMSA reaction for 1 and 2 h, respectively.
The vaccinia virus K4 protein nicks viral hairpins.
Nicking of the viral telomeres has been proposed to occur during the initiation of genome replication and the resolution of concatemeric intermediates to mature, monomer genomes. Thus, it seemed feasible that the viral hairpins were becoming cleaved during their incubation with the cytoplasmic extracts from infected cells and that the complexes observed by EMSA might reflect such a covalent modification of the hairpin probes. To address this possibility, EMSA reactions were performed as usual and then applied to formamide-containing polyacrylamide gels after treatment of the samples with proteinase K. Formamide-acrylamide gels are highly denaturing and provide accurate molecular weight estimates for fragments that contain high levels of secondary structure, such as hairpins (8). The efficacy of this approach is demonstrated in Fig. 10A, in which the 200-bp hairpin probe is shown to migrate with an apparent size of 400 nt, indicating that it is fully unfolded (lane 1).
FIG. 10.
The vaccinia virus K4 protein nicks viral hairpins. (A) Viral hairpins become nicked by a viral protein. EMSA reactions were performed using either 65 bp+tet hairpin probes (lanes 1 and 2) or the 130 bp+tet extended duplex probe (lanes 3 and 4). Reactions were incubated for 30 min at 30°C, treated with proteinase K for an additional 30 min, and applied to a denaturing 5% polyacrylamide–16 M formamide gel. Lanes 1 and 3, cytoplasmic extract prepared from uninfected cells; lanes 2 and 4, cytoplasmic extract prepared from wt-infected cells at 24 hpi. The open arrow indicates the full-length probe (∼400 nt), while the solid arrow indicates the nicked radiolabeled product (∼220 nt). The electrophoretic profile of the DNA markers is shown at the left, with the fragment sizes indicated in nucleotides. (B) Expression and purification of HisK4. The vaccinia virus K4 gene was cloned into pET16b and expressed in E. coli with an N′-terminal decahistidine tag. The HisK4 protein was enriched by chromatography on a nickel-agarose column; peak fractions were pooled, concentrated, and analyzed by SDS-PAGE and silver staining (lane 1) and immunoblot analysis with a probe reactive with the polyhistidine tag (lane 2). The enriched fraction contains the full-length 50-kDa HisK4 protein along with a number of other his-tagged species. Protein standards are shown at the left, with their molecular masses indicated in kilodaltons. (C) K4 is necessary and sufficient for the nicking of viral hairpins. EMSA reactions were performed using the 65 bp+tet hairpin probe, digested with proteinase K, and analyzed by formamide-denaturing gel electrophoresis. Reactions were performed using cytoplasmic extracts prepared from uninfected cells (lane 1) or from wt-infected (lane 2) or vΔK4-infected (lane 5) cells harvested at 24 hpi or using 300 ng of HisK4 protein prior to (lane 3) or after (lane 4) desalting. The open arrow indicates the full-length probe, while the solid and gray arrows indicate nicked radiolabeled products. The electrophoretic profile of the DNA markers is shown at the left, with the fragment sizes indicated in nucleotides.
While incubation of the probe with a cytoplasmic extract prepared from uninfected cells did not alter its integrity (Fig. 10A, lane 1), incubation with an extract prepared from virally infected cells harvested at 24 hpi generated a novel DNA band approximately half the size of the viral hairpin (lane 2). Although it is formally possible that this band represents exonucleolytic trimming of the probe to a discrete size, it is most likely that the new band arises from the introduction of a nick at or near the apex of the hairpin turnaround. Nicking did not occur when the probe was in the extended duplex conformation (lane 4), reinforcing the importance of probe structure in these reactions. These data suggest that viral hairpins are being recognized and cleaved by a virally encoded nuclease during EMSA reactions. When EMSA reactions were performed with purified HisI1 or HisI6 protein and then analyzed on formamide-acrylamide gels, it was clear that neither protein was able to generate the nicked fragment (data not shown).
The nicking activity, however, was reminiscent of past reports of a vaccinia virus-encoded nicking-joining (NJ) enzyme that cleaves extruded cruciform DNA at its apex (15). The protein responsible for this activity has been recently identified as the product of the vaccinia virus K4 gene (M. Merchlinsky, personal communication). Two approaches were taken by us to test directly whether the K4 protein was responsible for generating the faster-migrating band observed in EMSA reactions analyzed on formamide-acrylamide gels. We first tested whether the purified HisK4 protein could nick viral hairpins. HisK4 protein expressed in E. coli was partially purified on a nickel-agarose column (Fig. 10B, lane 1). This preparation contained a 50-kDa band that corresponded to the predicted molecular mass of the K4 protein as well as a number of other species shown to react with a probe specific for the histidine tag (lane 2). These smaller proteins are likely to represent premature termination products or proteolytic fragments of HisK4.
When EMSA reactions were performed with this partially purified preparation of recombinant HisK4 and analyzed on a formamide-acrylamide gel, a 220-nt band identical to the one generated by infected cell extracts was seen (Fig. 10C, compare lane 3 with lane 2). When the K4 preparation was desalted such that its final NaCl concentration was reduced from 500 to 100 mM, an equivalent amount of K4 protein was shown to generate an increased amount of the 220-nt product and a novel species that migrated with an apparent size of approximately 170 nt (lane 4). We believe that K4 is primarily nicking the viral hairpins at their apex in our assay; however, nicking at other sites can occur under conditions of reduced ionic strength.
The K4 open reading frame was recently reported to be nonessential for normal viral propagation; a virus in which the K4 gene is deleted (vΔK4) is viable in tissue culture (M. Merchlinsky, personal communication). To confirm that the K4 protein was responsible for nicking the viral hairpins, EMSA reactions were performed using cytoplasmic extracts prepared from cells infected with vΔK4. Extracts lacking K4 were unable to nick viral hairpins (Fig. 10C, lane 5). Thus, the K4 protein is necessary and sufficient for the hairpin nicking seen in our reactions. As expected, the absence of K4 did not impair the ability of these extracts to form the upper and lower doublets of shifted complexes in EMSA reactions (data not shown).
DISCUSSION
This report demonstrates that vaccinia virus-infected cells and vaccinia virus virions contain several proteins that form specific interactions with hairpin probes representing the telomeres of the viral genome. The stability and specificity of the complexes seen is underscored by the fact that their formation was analyzed in the absence of fixative and in the presence of a vast excess of the nonspecific competitor poly(dI-dC). Two doublets of shifted complexes (upper and lower) were visualized reproducibly; each hairpin isoform (flip and flop) generated one member of each doublet. The association of the hairpin DNA with the telomere binding proteins must exaggerate the inherent difference in the electrophoretic mobilities of the two isoforms that can be seen during nondenaturing gel electrophoresis.
Our previous data have shown that the terminal 200 bp of the viral telomeres are necessary and sufficient to confer optimal replication efficiency on minichromosome templates (7). Here we show that the terminal 65 bp, which are not sufficient for minichromosome replication, do contain all of the specificity required for formation of the protein-DNA complexes studied in this report. These 65 bp have several interesting structural features, including the hairpin turnaround and the characteristic extrahelical bases. The importance of these structural features was corroborated by our demonstration that the extended duplex form of the probe could not participate in, or disrupt, complex formation. Moreover, after using a PCR-ligation strategy to construct perfectly base paired hairpins that otherwise resembled the viral telomeres, we demonstrated that the extrahelical bases are indeed necessary for recognition by the telomere binding proteins.
To further investigate the role of extrahelical bases, we performed competition experiments using oligonucleotides containing extrahelical bases. These oligonucleotides had no sequence similarity to the viral telomeres; in fact, their sequence was taken from published studies on proteins that participate in mismatch repair by binding to extrahelical bases (14). A vast excess of a 37-bp duplex that contained two extrahelical bases spaced 5 nt apart was able to compete for the formation of the lower doublet of shifted complexes, while the same duplex containing 1 or 0 extrahelical bases could not. The dependence of the competition ability on the presence of extrahelical bases was reproducible; however, it must be mentioned that competition with the duplex that contained two extrahelical bases was 2 orders of magnitude less efficient than that obtained with the viral telomeres (40,000-fold versus 10-fold for complete disruption of complex formation). Whether the dramatically higher affinity of the telomere binding proteins for the viral hairpins is due to the increased number of extrahelical bases (12 versus 2), the hairpin turnaround, the AT richness of the viral sequences, or specific sequence motifs is worthy of further study. Nevertheless, the telomere binding activity was completely abrogated by a duplex containing two extrahelical bases and was completely unaffected by a homologous duplex that was completely base paired. In conjunction with our finding that the telomere binding proteins do not interact with perfectly base-paired viral hairpins, these data strengthen the conclusion that the extrahelical bases within the viral telomeres are recognizable features that are likely to play an important function in some facet of genome utilization.
It is interesting to speculate about what role the extrahelical bases might play in marking viral telomeres. They could serve to distinguish the telomeres of mature viral genomes from the homologous sequences found at the junction of replication intermediates. They could, in an analogous way to the insertion/deletion lesions that recruit the mismatch repair machinery of prokaryotes and eukaryotes, serve as specific binding sites for a complex of proteins. In the case of the DNA repair machinery, this complex contains the mismatch-binding protein MutS (or its eukaryotic homologs, MSH2, -3, and -6) and the MutH endonuclease (11). In the case of vaccinia virus, recognition by the telomere binding proteins discussed in this report might serve to recruit an endonuclease to the telomeres that might provide the nick required to initiate viral DNA replication. With this in mind, our future studies will include the generation of minichromosomes that contain perfectly base-paired telomeres that lack the extrahelical bases: these may have lost their competence as replication templates in vivo.
The apparent specificity and stability of the interactions between the telomere binding proteins and the hairpin probe allowed us to develop an effective affinity purification protocol. In a single chromatographic step, we could identify the two key protein species which directed the formation of the upper and lower doublets of protein-DNA complexes. These were shown to be the products of the I1 and I6 genes, respectively. The vaccinia virus I1 protein is expressed during the late stage of viral gene expression and encapsidated at approximately 700 copies per virion (10). These data support our observation that telomere binding activity accumulates after the onset of DNA replication and is present in virions. Although the I1 protein has been shown to bind DNA irrespective of sequence or conformation, our studies show that it binds preferentially to viral hairpins containing extrahelical bases. Whether the affinity of I1 for the telomeric sequence and/or structure contributes to its role in virion maturation remains to be determined in future studies.
The vaccinia virus I6 protein was found to be responsible for the formation of the lower doublet of shifted complexes. I6 appears to bind the telomeric hairpins with great specificity and stability, as shown by our competition and challenge experiments. There are no previous reports concerning the I6 protein, and analysis of its sequence does not provide any clues as to how it recognizes DNA or what the consequences of DNA binding might be. The vaccinia virus I6 protein is conserved among poxviruses, and a high degree of sequence identity can be found even between vaccinia virus and the distantly related molluscum contagiosum virus (34% identity, 59% similarity). Our functional studies suggest that I6 will be encapsidated and is likely to be expressed as a late protein or to become activated by posttranslational modifications that depend upon late proteins. Since the DNA sequence surrounding the translational start of the I6 open reading frame does not contain the TAAATG motif that is characteristic of late genes and the coding sequence does not contain the T5NT motif that would signal transcriptional termination during early gene expression, a definitive assessment of when I6 is expressed awaits future analysis.
Our EMSA data indicate that binding of the I1 and I6 proteins to the telomeric probes is mutually exclusive. When both proteins are present, no higher-order complexes are seen that are distinct from those generated by each protein alone. It may be that binding by one of the proteins to the telomeres predominates in vivo or that their independent binding to different telomeres participates in differential marking of the two ends of the viral genome.
At the present time, we have no data which address the functional significance of I6 as a telomere binding protein. Furthermore, the inducible recombinant vindI1, although effective in uncovering a key role for I1 in virion morphogenesis, cannot be used to address all of the possible functions of I1. Since viral stocks must be generated from infections carried out in the presence of IPTG and hence of I1 expression, the vindI1 virions encapsidate a large amount of I1, which will be present during the next round of infection. Therefore, if I1 were to play an important role in DNA replication, the encapsidated I1 would probably mask this requirement. Thus, the potential role of I1 and/or I6 in DNA replication remains an open question. Initially we felt that our observation that telomere binding activity accumulated after the onset of viral genome replication appeared to discount such a role. However, the fact that the telomere binding proteins are encapsidated allows one to propose that they mark the telomeres as sites of replication initiation and can become activated during the next round of infection and/or serve to recruit binding partners or enzymes that drive the initiation of replication. In this sense, they might be analogous to the origin recognition complex (ORC) in eukaryotic cells, which is constitutively bound to replication origins and serves as a landing pad for key components of the replication machinery that join during the G1 phase of the cell (25). Alternatively, the telomere binding proteins might not participate in DNA replication at all. They might be involved in marking mature genomes for encapsidation into virions (6). I1 and/or I6 could recognize and bind to mature genomes by virtue of the extrahelical bases and escort the DNA into immature virions as part of a process that is known to involve the A32 ATPase (3). Further analysis of the I6 gene is under way to determine if it has an essential role in viral replication.
In the course of characterizing proteins that interacted with vaccinia virus telomeres, we also observed that viral hairpins were becoming covalently modified after incubation with a cytoplasmic extract prepared from infected cells at 24 hpi. This modification appeared to occur at or near the apex of the hairpin. This activity was reminiscent of the encapsidated, 50-kDa NJ enzyme (12, 23). This enzyme was reported to nick the apex of extruded cruciforms in an ATP-independent, divalent cation-independent manner (15) and to have a limited ability to join the nicked ends. The NJ enzyme was recently identified as the product of the K4 gene (Eckert et al., unpublished). By preparing and analyzing recombinant K4 protein and by utilizing extracts of cells infected with the deletion mutant vΔK4 (Eckert et al., unpublished), we could show that the K4 protein was necessary and sufficient for the hairpin cleavage that we had been seeing. The role of the K4 gene in the viral life cycle remains unknown. It appears to be nonessential for growth of vaccinia virus in tissue culture, and its specificity in vitro for the apices of hairpin DNA is not compatible with a role in the resolution of concatemeric intermediates to monomeric genomes. Although some of the other enzymes with known roles in viral DNA metabolism are dispensable in tissue culture (DNA ligase, ribonucleotide reductase, thymidine kinase), their functions are not dispensable and are instead supplied by the corresponding cellular enzymes. Not enough is known about the K4 enzyme to propose a similar interpretation. The NJ enzyme has been shown to leave a free 3′-phosphate group after cleaving DNA, which could not serve as a primer terminus for a replicative DNA polymerase. Since K4 has been shown to be encapsidated, it might instead have a role in modulating the torsional stress of viral DNA during genome packaging or during the transcription of early genes which occurs within the core during the next round of infection.
In sum, these studies are the first to define the interactions of the unique poxviral telomeres with specific proteins. We have identified I1 and I6 as telomere binding proteins and demonstrated that their recognition of telomeric DNA requires the extrahelical bases that are such striking characteristics of the poxviral telomeres. Understanding how these proteins recognize these unique DNA elements, and what role(s) they play in the viral life cycle, will be of significant interest. The telomeres are known to be the major cis-acting elements that drive DNA replication and are likely to be involved in genome encapsidation. Therefore, these studies should provide a new and intriguing perspective for dissecting how the viral genome is faithfully replicated and accurately disseminated.
ACKNOWLEDGMENTS
This work was supported by a grant to P.T. from the National Institutes of Health (R01 AI21758).
We thank Nancy Klemperer and Neal Lue for helpful discussions and express our appreciation to Nancy Klemperer and Jeremy Ward for providing the recombinant I1 protein and to Michael Merchlinsky for generously providing the vΔK4 virus prior to publication.
REFERENCES
- 1.Baroudy B M, Venkatesan S, Moss B. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell. 1982;28:315–324. doi: 10.1016/0092-8674(82)90349-x. [DOI] [PubMed] [Google Scholar]
- 2.Baroudy B M, Venkatesan S, Moss B. Structure and replication of vaccinia virus telomeres. Cold Spring Harbor Symp Quant Biol. 1983;47(Pt. 2):723–729. doi: 10.1101/sqb.1983.047.01.083. [DOI] [PubMed] [Google Scholar]
- 3.Cassetti M C, Merchlinsky M, Wolffe E J, Weisberg A S, Moss B. DNA packaging mutant: repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J Virol. 1998;72:5769–5780. doi: 10.1128/jvi.72.7.5769-5780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheskis B, Freedman L P. Ligand modulates the conversion of DNA-bound vitamin D3 receptor (VDR) homodimers into VDR-retinoid X receptor heterodimers. Mol Cell Biol. 1994;14:3329–3338. doi: 10.1128/mcb.14.5.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeLange A M. Identification of temperature-sensitive mutants of vaccinia virus that are defective in conversion of concatemeric replicative intermediates to the mature linear DNA genome. J Virol. 1989;63:2437–2444. doi: 10.1128/jvi.63.6.2437-2444.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeLange A M, McFadden G. The role of telomeres in poxvirus DNA replication. Curr Top Microbiol Immunol. 1990;163:71–92. doi: 10.1007/978-3-642-75605-4_3. [DOI] [PubMed] [Google Scholar]
- 7.Du S, Traktman P. Vaccinia virus DNA replication: two hundred base pairs of telomeric sequence confer optimal replication efficiency on minichromosome templates. Proc Natl Acad Sci USA. 1996;93:9693–9698. doi: 10.1073/pnas.93.18.9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank R, Muller D, Wolff C. Identification and suppression of secondary structures formed from deoxy-oligonucleotides during electrophoresis in denaturing polyacrylamide-gels. Nucleic Acids Res. 1981;9:4967–4979. doi: 10.1093/nar/9.19.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–263. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 10.Klemperer N, Ward J, Evans E, Traktman P. The vaccinia virus I1 protein is essential for the assembly of mature virions. J Virol. 1997;71:9285–9294. doi: 10.1128/jvi.71.12.9285-9294.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolodner R D, Marsischky G T. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 12.Lakritz N, Foglesong P D, Reddy M, Baum S, Hurwitz J, Bauer W R. A vaccinia virus DNase preparation which cross-links superhelical DNA. J Virol. 1985;53:935–943. doi: 10.1128/jvi.53.3.935-943.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu K, Lemon B, Traktman P. The dual-specificity phosphatase encoded by vaccinia virus, VH1, is essential for viral transcription in vivo and in vitro. J Virol. 1995;69:7823–7834. doi: 10.1128/jvi.69.12.7823-7834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsischky G T, Kolodner R D. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 complex and mispaired bases in DNA. J Biol Chem. 1999;274:26668–26682. doi: 10.1074/jbc.274.38.26668. [DOI] [PubMed] [Google Scholar]
- 15.Merchlinsky M, Garon C F, Moss B. Molecular cloning and sequence of the concatemer junction from vaccinia virus replicative DNA. Viral nuclease cleavage sites in cruciform structures. J Mol Biol. 1988;199:399–413. doi: 10.1016/0022-2836(88)90613-4. [DOI] [PubMed] [Google Scholar]
- 16.Merchlinsky M, Moss B. Resolution of linear minichromosomes with hairpin ends from circular plasmids containing vaccinia virus concatemer junctions. Cell. 1986;45:879–884. doi: 10.1016/0092-8674(86)90562-3. [DOI] [PubMed] [Google Scholar]
- 17.Merchlinsky M, Moss B. Nucleotide sequence required for resolution of the concatemer junction of vaccinia virus DNA. J Virol. 1989;63:4354–4361. doi: 10.1128/jvi.63.10.4354-4361.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merchlinsky M, Moss B. Resolution of vaccinia virus DNA concatemer junctions requires late-gene expression. J Virol. 1989;63:1595–1603. doi: 10.1128/jvi.63.4.1595-1603.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moss B. Poxviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2637–2671. [Google Scholar]
- 20.Pogo B G. Changes in parental vaccinia virus DNA after viral penetration into cells. Virology. 1980;101:520–524. doi: 10.1016/0042-6822(80)90466-3. [DOI] [PubMed] [Google Scholar]
- 21.Pogo B G, Berkowitz E M, Dales S. Investigation of vaccinia virus DNA replication employing a conditional lethal mutant defective in DNA. Virology. 1984;132:436–444. doi: 10.1016/0042-6822(84)90048-5. [DOI] [PubMed] [Google Scholar]
- 22.Pogo B G, O'Shea M, Freimuth P. Initiation and termination of vaccinia virus DNA replication. Virology. 1981;108:241–248. doi: 10.1016/0042-6822(81)90543-2. [DOI] [PubMed] [Google Scholar]
- 23.Reddy M K, Bauer W R. Activation of the vaccinia virus nicking-joining enzyme by trypsinization. J Biol Chem. 1989;264:443–449. [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 25.Stillman B. Cell cycle control of DNA replication. Science. 1996;274:1659–1664. doi: 10.1126/science.274.5293.1659. [DOI] [PubMed] [Google Scholar]
- 26.Studier F W, Rosenberg A L, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 27.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Traktman P. Molecular genetic analysis and biochemical analysis of poxvirus DNA replication. Semin Virol. 1991;2:291–304. [Google Scholar]
- 29.Traktman P. Poxvirus DNA replication. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 775–798. [Google Scholar]
- 30.Vogelstein B, Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci USA. 1979;76:615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]