Abstract
Short bowel syndrome (SBS) leads to severe morbidity and mortality. Intestinal adaptation is crucial in improving outcomes. To understand the human gene pathways associated with adaptation, we perform single-cell transcriptomic analysis of human small intestinal organoids explanted from mice with experimental SBS. We show that transmembrane ion pathways, specifically the transepithelial zinc transport pathway genes SLC39A4 and SLC39A5, are upregulated in SBS. This discovery is corroborated by an external dataset, bulk RT-qPCR, and Western blots. Oral zinc supplementation is shown to improve survival and weight gain of SBS mice and increase the proliferation of intestinal crypt cells in vitro. Finally, we identify the upregulation of SLC39A5 and associated transcription factor KLF5 in biopsied intestinal tissue specimens from patients with SBS. Thus, we identify zinc supplementation as a potential therapy for SBS and describe a xenotransplantation model that provides a platform for discovery in other intestinal diseases.
Subject terms: Intestinal diseases, Experimental models of disease, Animal disease models
SBS leads to severe morbidity and mortality. Here, using a xenotransplantation model, the authors uncover the role of zinc transport pathways in intestinal adaptation in SBS, highlighting the potential of zinc supplementation as a therapeutic approach.
Introduction
Pediatric short bowel syndrome (SBS) is a debilitating condition resulting from substantial loss of functional small intestine1. Despite advancements in patient care, individuals with SBS experience significant morbidity and mortality due to malnutrition and fluid losses, and current treatment strategies are only partly effective2–4. Current approaches to management, such as pharmacologic agents, dietary protocols, surgical procedures, and small bowel transplants, have limitations including variable efficacy, significant side effects, morbidity, cost, and extended hospital stays. In response to significant intestinal loss, the residual intestine undergoes a process of intestinal adaptation, characterized by increases in bowel length, diameter, and villus height5. Importantly, the precise factors that mediate the process of adaptation in SBS remain poorly understood, representing an unmet need in patient care.
Due to the scarcity of large SBS patient tissue repositories, a significant portion of our current knowledge regarding the mechanistic aspects of adaptation has been derived from animal models6–13. Multiple pathways have been implicated in adaptation thus far, including changes in cell proliferation, differentiation, migration, and alterations in intestinal transport, absorption, and secretion. Growth factors including epidermal growth factor (EGF), transforming growth factor-alpha (TGF-α), and insulin-like growth factor (IGF-1), which play roles in modulating epithelial cell proliferation and differentiation are upregulated in SBS14,15. Various transport proteins, such as glucose and sodium-glucose cotransporters, are not only upregulated but also exhibit increased activity, enhancing nutrient and fluid absorption in the intestine7,16,17. Intestinal hormones including glucagon-like peptide-2, ghrelin, and cholecystokinin increase to stimulate villi growth and nutrient absorption. Among these mechanisms, the GLP-2 agonist teduglutide stands out as the sole approved drug for treating SBS. Although many of the molecular regulatory mechanisms remain unknown, several signaling pathways involved in cell proliferation, differentiation, and migration, including the Wnt/β-catenin pathway, Notch pathway, and Hedgehog pathway, have been implicated in SBS18.
To gain a clearer understanding of the most important factors contributing to adaptation in SBS, we employed a model system involving the implantation of human induced pluripotent stem cell (hiPSC)-derived intestinal organoids into immunodeficient Rag1tm1Mom (Rag1 KO) mice subjected to an SBS model that involves more than 75% intestinal resection. This experimental approach allows for determining genetic pathways induced in human tissue in the setting of SBS. These studies revealed an unanticipated role for the transepithelial zinc transport pathway in mediating intestinal adaptation in SBS. Zinc, an essential trace element with critical roles in cellular processes such as growth, division, and differentiation, acts as a component of numerous enzymes and as a co-factor for zinc finger proteins, which interact with RNA and DNA to regulate transcription, DNA repair, cell migration, and protein degradation19. Zinc signaling plays a crucial role in various functions within the body, with particular importance in the gut. Previous studies have demonstrated that zinc deficiency decreases the rate of epithelial cell proliferation, leading to impaired mucosal renewal, increased permeability, and compromised barrier function20–25.
Enterocytes uptake high concentrations of zinc from the gut lumen via the ZIP4 receptor, encoded by SLC39A4. ZIP4 translocates to the apical surface of enterocytes in response to zinc deficiency26,27. Zinc regulates enterocyte function by modulating various signaling pathways25,26,28–32. For example, zinc activates the protein kinase C pathway33, which is involved in cell growth and differentiation. Zinc also plays a role in intestinal immune defense, where its deficiency increases susceptibility to infection and inflammation32. Additionally, zinc is involved in regulating the gut microbiota, with supplementation leading to an increase in beneficial bacteria and a decrease in pathogenic bacteria34. ZIP5, encoded by SLC39A5, is expressed on the basolateral membrane of the intestinal epithelium and plays a role in zinc homeostasis by supplying serum zinc to enterocytes when dietary zinc is low, and in shuttling zinc from the serum into the enterocyte for efflux into the gut lumen when endogenous zinc is high30,35.
In our study, supplementing the diet of SBS mice with zinc resulted in improvement in weight loss and survival. In vitro and in vivo zinc supplementation led to increased proliferation in intestinal crypt cells and remarkable villi elongation in SBS mice compared to controls. These findings provide evidence that zinc supplementation enhances intestinal adaptation, ameliorates weight loss, and improves survival in a mouse model of SBS. Consequently, our study highlights the therapeutic potential of zinc modulation in SBS management.
Results
Establishment of a human–mouse chimera model system to identify pathways that mediate intestinal adaptation in experimental SBS
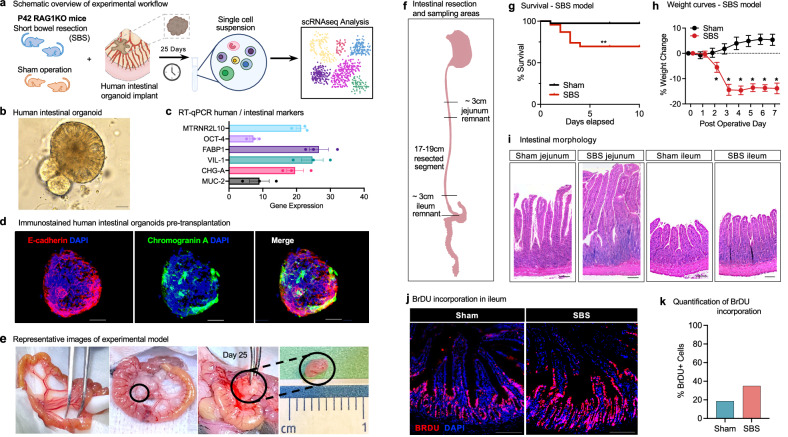
To identify pathways involved in the pathogenesis of SBS and to investigate their potential role in enhancing intestinal adaptation, we established a xenotransplantation human–mouse chimera model using hiPSC-derived intestinal organoids (Fig. 1a). To generate human small intestinal organoids, hiPSCs were first differentiated into endodermal precursors by supplementing the culture medium with Activin A, following established protocols11. The endodermal spheroids were then exposed to Wnt3a, FGF4, and EGF for intestinal lineage differentiation, and then cultured in a 3D matrix with Noggin and R-spondin to form structures resembling small intestinal organoids (Fig. 1b) that consisted of epithelial monolayers surrounding spherical and cystic central lumens36,37. In validation experiments, these human-derived organoids were found to express human-specific genes associated with intestinal stem cells and epithelial differentiation, including MTRN2RNL10 (encoding humanin-like ten peptide, which is specific to human tissue), OCT4 (octamer-binding transcription factor four controlling embryonic development and pluripotency marker), FABP1 (fatty acid binding protein 1), VIL1 (villin 1, encoding an actin-binding protein responsible for intestinal epithelial cell morphology), CHGA (chromogranin A, a neuroendocrine secretory protein produced by enterochromaffin-like cells), and MUC2 (mucin2 produced by goblet cells), as confirmed by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) (Fig. 1c). Immunostaining of the organoids further revealed the presence of epithelial-specific proteins, such as epithelial Cadherin 1 and CHGA (Fig. 1d). Importantly, the organoids were composed not only of epithelial cells, but also included cells of mesenchymal origin, as confirmed by the expression of collagen and matrix-associated proteins revealed by single cell RNA sequencing (scRNAseq) (Table 1). This incorporation of diverse cell types mirrors the complexity of human intestinal tissue in vivo, providing a platform for investigating pathways that may exhibit co-dependency between various cell lineages.
Fig. 1. Development of a human–mouse chimera model system for the identification of pathways that mediate intestinal adaptation in experimental SBS.
a Schematic overview of the experimental workflow for the human small intestinal organoid xenotransplantation SBS model. Six-week-old Rag1 KO C57BL/6 mice underwent experimental SBS (n = 5) or sham surgery (n = 5). Human intestinal organoids, differentiated from iPSCs, were implanted into the mesentery using surgical glue. Xenotransplants were harvested after 25 days for single-cell RNA sequencing analysis. b Bright field microscopy of human small intestinal organoids differentiated from iPSC (Day 42). Magnification ×20. Scale bar 100 μm. c RT-qPCR analysis of intestinal-specific markers in human organoids pre-xenotransplantation, showing increased expression relative to housekeeping gene RPLP0. Data from three independent organoid cultures are presented as mean ± standard error of the mean (SEM). d Immunofluorescence staining of pre-xenotransplantation organoids for epithelial and intestinal markers. Scale bar 100 µm. e Images of mouse intestine pre-resection, during xenotransplantation, and at harvest on Day 25. f Schematic of intestinal resection and sampling areas in the SBS model. 75% of the small bowel was resected, with a jejuno-ileal anastomosis. Sham controls underwent transection and anastomosis without resection. g Survival curves for SBS (n = 23) and sham (n = 40) mice with human xenotransplants. SBS mice had ~70% survival at 10 days vs ~100% for sham (log-rank test, p = 0.0015). h Growth curves showing significant weight loss in SBS mice (n = 6) compared to steady weight gain in sham controls (n = 7). Weight differences were significant at all indicated time points (t-test, *p < 0.05). Data are mean ± SEM. i H&E staining of jejunum and ileum from Rag1 KO mice post-xenotransplant. SBS jejunal villi were significantly lengthened; ileal villi were modestly increased. Scale bar 100 µm. j Immunofluorescence images of BrdU staining in intestinal tissue post-xenotransplant. BrdU incorporation was higher in SBS ileum compared to sham. Scale bar 100 µm. k Quantification of BrdU-positive cells in intestinal crypts showed significantly higher incorporation in SBS compared to sham controls. Source data are provided as a separate file.
Table 1.
List of marker genes for cell cluster identification
| Cluster | Markers |
|---|---|
| Enterocyte 1 | APOE, FABPs, KLF5, ANPEP, VIL1, CHH17 |
| Enterocyte 2 | APOE, FABPs, KLF5, ANPEP, VIL1, CHH17 |
| Stem-like fibroblast | COLs, FBN1, IGFBP6, FSTL1, SPARC, DCN |
| Mature fibroblast | COLs, FBN1, MMP2, IGFBP6, FSTL1, SPARC, DCN |
| Myeloid 1 | CEBPB, TLR2 |
| Myeloid 2 | HLA-DQB1, MRC1, AIF1 |
| Myeloid 3 | CREG1, CTSS, CD9, CAPG |
| Mucin producing | MUC1, MUC5AC |
To evaluate the molecular mechanisms mediating intestinal adaptation in SBS, 5–6-week-old C57BL/6 (C57 black 6) Rag1 knockout (Rag1 KO) mice underwent 75% small bowel resection followed by primary jejunoileal anastomosis38. Briefly, a 1 cm midline abdominal skin incision was made between the inferior rib and the iliac crest, along with a deeper incision in the linea alba to access the peritoneum. The small intestine was eviscerated, and 75% of the small bowel was excised, leaving approximately 3 cm of residual intestine proximally and distally (Fig. 1f). A jejunoileal anastomosis was then performed using interrupted 8–0 silk sutures, which are each 0.04 mm in diameter. Mice subjected to this experimental model exhibited characteristic features observed in human SBS, including reduced survival (Fig. 1g), significant weight loss (Fig. 1h), and evidence of intestinal adaptation such as villi expansion and crypt proliferation (Fig. 1i–k), when compared to sham-treated mice that underwent laparotomy, intestinal transection, and anastomosis alone.
To identify potential human genes involved in mediating adaptation in SBS, we implanted human intestinal organoids into the mesentery of mice that underwent either SBS surgery (n = 5) or a sham operation (control group) (n = 5)39. After allowing 3–4 weeks for intestinal adaptation to occur, the human small intestinal organoid implants that had been exposed to the SBS microenvironment were harvested (Fig.1e) and subjected to scRNAseq analysis. Mice in this model tolerated the surgical procedures well, with the survival rate of the sham group implanted with human organoid implants reaching approximately 90%, while the SBS group demonstrated worse outcomes (Fig. 1h). In the experimental group, organoids were implanted into the intestinal mesentery of mice that had undergone resection of 75% of the small bowel followed by intestinal anastomosis, as previously described38. This resection protocol resulted in predictable weight loss of approximately 15% (Fig. 1g) and characteristic intestinal adaptation, consisting of elongation of the intestinal crypts and increased villus height, as confirmed by histological examination of jejunal and ileal sections (Fig. 1h).
Distinct transcriptional signatures are revealed by single-cell RNA sequencing analysis of human intestinal organoid explants in mice with SBS
To understand the mechanisms that mediate intestinal adaptation in SBS for therapeutic use, we next conducted a comprehensive analysis of human small intestinal organoids explanted from SBS mice. Using scRNAseq, we assessed human small intestinal organoid explants harvested from five mice with experimentally induced SBS and five mice subjected to exploratory laparotomy without intestinal resection which served as sham controls. Following the dissociation of freshly harvested implants into single cells, droplet-based scRNAseq was performed. Low-quality cells and sequences derived from contaminating mouse cells were excluded from the analysis, allowing a focus solely on cells of human origin (Supplementary Fig. 1).
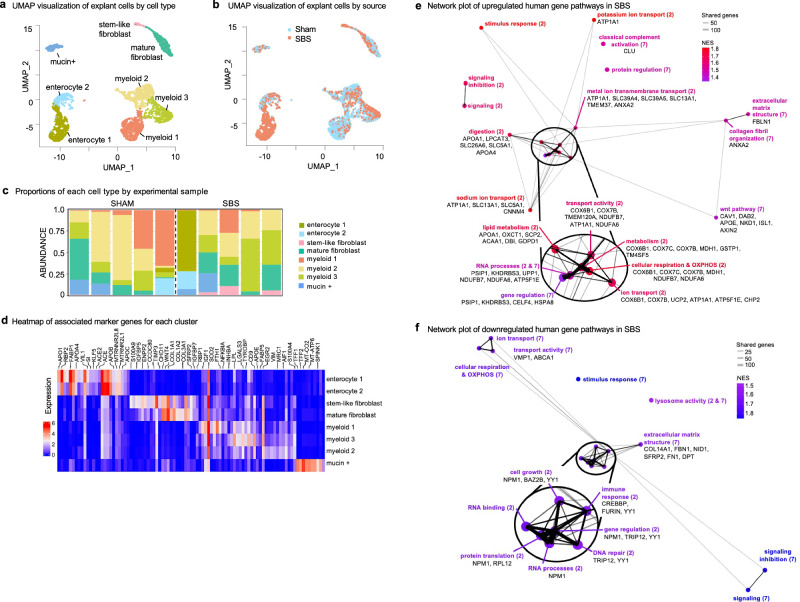
Using this approach, we analyzed an average of 1660 and 1645 genes and 6693 and 6432 transcripts per cell per cell in sham and SBS explants respectively. Utilizing the R package Seurat, clustering of 3160 human intestinal organoid cells from the ten mice generated a detailed map consisting of nine distinct transcriptionally similar subpopulations, visualized as uniform manifold approximation and projection (UMAP) plots (Fig. 2a, b). By examining the expression of known marker genes, these clusters were identified as either epithelial or mesenchymal in origin (Fig. 2c, d). Two populations of epithelial-derived cells were observed, including enterocytes that expressed markers such as VIL1 and FABP1, and one cluster of mucin-producing cells. Also noted was a cluster of mesenchymal-derived cells that exhibited fibroblast-like or myeloid-like characteristics including the expression of COL1A1 and COL1A2. Notably, the enterocyte 1 and stem-like fibroblast clusters were composed entirely of cells derived from SBS mice explants (Fig. 2a, b). Furthermore, clusters dominated by SBS-derived sequences displayed significantly upregulated expression of specific genes, corroborating previous studies investigating the expression of intestinal genes in prior animal studies of SBS6,7,13. Noteworthy examples include retinol-binding protein 2 (RBP2), apolipoprotein a1 (APOA1), and apolipoprotein a4 (APOA4), which are involved in lipid trafficking7,13, solute carrier family 26 (SLC26A6) genes associated with ion transport13, and Wnt signaling pathway gene WNT46.
Fig. 2. Distinct transcriptional signatures are revealed by single-cell RNA sequencing analysis of human intestinal organoid explants harvested from mice with SBS.
a UMAP projection of single-cell data from human small intestine organoid implants in SBS (n = 5) and sham mice (n = 5). Each dot represents a cell, with coordinates determined by transcriptome expression via principal components analysis and subsequent UMAP dimensional reduction. Clusters are color-coded and labeled by cell type. b UMAP projection colored by SBS cells (red) and sham cells (blue). The Enterocyte 1 and stem-like fibroblast clusters are predominantly derived from SBS samples. c Barplot showing the proportions of each cell type identified in scRNA-seq analysis of human intestinal organoid explants from mice subjected to either sham or SBS surgery. Each bar represents a cell type, and the x-axis indicates the experimental group. The height of each bar corresponds to the percentage of cells of that type in the total cell population for that sample. d Heatmap illustrating DEGs in human intestinal organoids derived from sham and SBS mice. The top genes within each cluster and the corresponding marker genes identified by scRNA-seq analysis are displayed. Rows represent individual genes, and columns represent cell clusters or individual marker genes. Color intensity reflects gene expression levels, with red indicating high expression and blue indicating low expression. Horizontal bars above the heatmap denote marker genes for each cell cluster. e Network plot generated from gene set enrichment analysis (GSEA) showing upregulated pathways between enterocyte 1 (95% SBS) and enterocyte 2 (59% SBS) as well as stem-like fibroblast (96% SBS) and mature fibroblast (52% SBS). Node size correlates with the number of genes examined within those gene sets. Edges represent connections between nodes, with thickness proportional to the number of shared genes. The top six genes differentially expressed with statistical significance between the compared cell types and listed in the core enrichment of the GSEA analysis are displayed alongside each pathway. f Network plot of downregulated pathways in SBS, generated from GSEA showing downregulated pathways between Enterocyte 1 (95% SBS) and Enterocyte 2 (59% SBS) as well as stem-like fibroblast (96% SBS) and mature fibroblast (52% SBS). Two-sided Wilcoxon rank sum test; filtering for genes with a Bonferroni-corrected p-value of less than 0.05. Source data are provided.
For a comprehensive analysis of the key pathways involved in the adaptation process of human SBS, we next conducted differential expression analysis and gene set enrichment analysis (GSEA) focusing on intestinal cells. Specifically, we compared cells from each cluster dominated by SBS-induced cells (enterocyte cluster 1 and stem-like fibroblast clusters) with corresponding cells of the same lineage representing the baseline or sham state. Hence cells from enterocyte cluster 1 (95% SBS) were compared to enterocyte 2 (59% SBS) and those from stem-like fibroblast (96% SBS) were compared to mature fibroblast (52% SBS). GSEA using biological process and molecular function databases identified 76 upregulated pathways in SBS, which were further consolidated into 19 related pathways based on shared genes and functions. A network plot was generated to visualize the inter-connectedness of these pathways using the graph 2.1.0 function (Fig. 2e). The network plot revealed an array of upregulated gene pathways in SBS, encompassing processes such as digestion and lipid transport, metabolism and cellular respiration, ion and molecular transport, extracellular matrix organization, RNA processing, and cell signaling. Among the statistically significant differentially expressed genes (DEGs) that contributed to these pathways, several genes were identified, including ECM-associated FBLN1 and ANXA2, cytochrome c oxidase subunit genes (COX6B1, COX7C, COX7B) involved in the electron transport chain, and cell migration-associated genes caveolin CAV1 and disabled-2 DAB2. Mesenchymal cells were proportionally distributed across organoid implants derived from both SBS and sham-treated groups. However, further analysis revealed some differences between the groups. Mesenchymal cells from the SBS group exhibited enhanced expression of genes and pathways crucial for intestinal stem cell support, anti-inflammatory responses, and barrier integrity. Notably, genes such as ANXA1, SOX9, and HIF1A were upregulated in mesenchymal cells extracted from SBS mice post-implantation, suggesting a unique adaptive response in the mesenchymal compartment. Importantly, and what caught our attention, was our observation of the upregulation of multiple ion transport pathways, including zinc transport genes SLC39A4 and SLC39A5 in human intestinal enterocytes explanted from SBS mice. In addition to the upregulated pathways, we analyzed downregulated pathways and associated genes similarly (Fig. 2f). This comprehensive analysis sheds light on the molecular networks driving adaptation in SBS in human tissue and highlights multiple targets for therapeutic intervention and reveals the potential roles of ion transport genes in intestinal adaptation.
Zinc transport genes SLC39A4 and SLC39A5 are upregulated in SBS
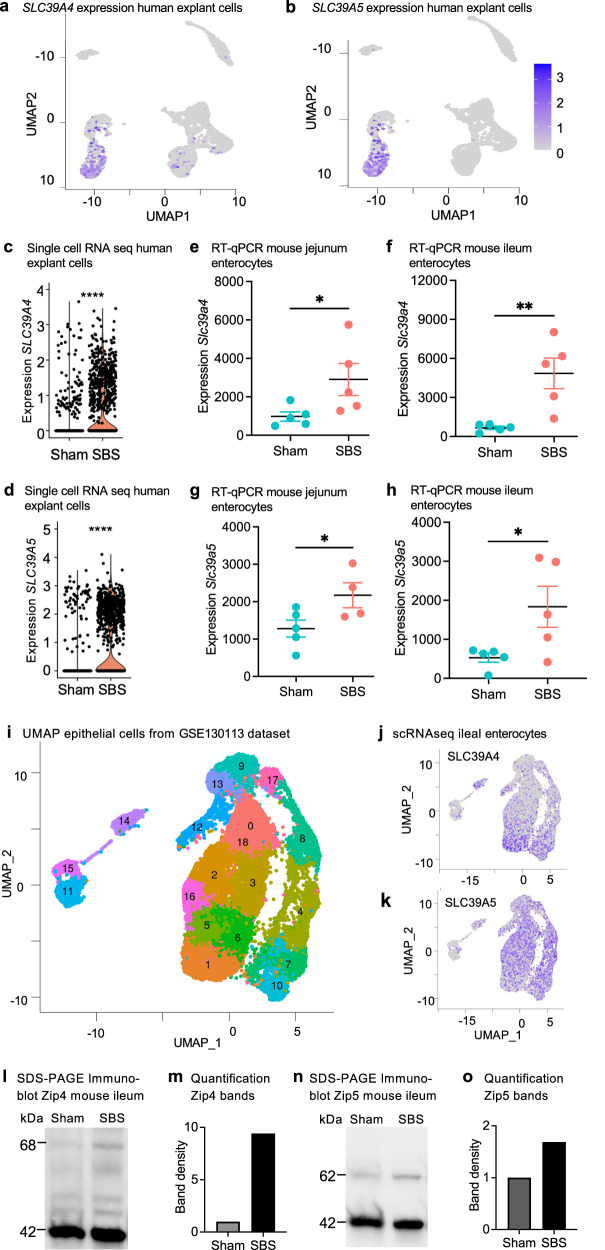
Our analysis of scRNAseq data revealed the upregulation of zinc transport genes SLC39A4 and SLC39A5 in human intestinal organoid explants from SBS mice. These genes were mainly upregulated in the enterocyte compartment, particularly in enterocyte cluster 2 associated with SBS (Fig. 3a, b). 56% of enterocyte cluster 2 expressed high levels of SLC39A4 and 63.2% expressed SLC39A5, compared to 13.41% and 14.92% respectively in cluster 5 enterocyte cells (Fig. 3a, b). Comparing expression levels between cells derived from sham and SBS, we observed increased expression of SLC39A4 and SLC39A5 (p = 2.2 × 10−6) in human organoids in the setting of SBS (Fig. 3c, d).
Fig. 3. Zinc transport genes SLC39A4 and SLC39A5 are upregulated in SBS.
UMAP visualization of human small intestinal xenotransplants from five SBS mice and five sham controls. Higher expression of SLC39A4 (a) and SLC39A5 (b) is observed within the enterocyte 1 cluster derived from SBS. Violin plots depict the elevated expression of SLC39A4 (c) and SLC39A5 (d) in human SBS xenotransplants compared to sham. Wilcoxon rank sum test, ****p < 0.0001. RT-qPCR of isolated intestinal epithelial cells of SBS mice (n = 5) shows the increased relative expression of Slc39a4 in the jejunum (e) and ileum (f) and of Slc39a5 expression in SBS jejunum (g) and ileum (h) compared to sham control (n = 5). Mann–Whitney U-test. *p ≤ 0.05; **p ≤ 0.01. Data represent mean ± SEM. i UMAP projection of processed, filtered, and clustered SBS epithelial cell data from the gene expression omnibus GSE130113 generated in Seurat. Feature plots illustrate the expression of j SLC39A4, and k SLC39A5 genes in mouse SBS native epithelial cells in GSE130113. Representative Western blot analysis of Zip4 (l) and Zip5 (m) protein expression in sham and SBS mice. Quantification analysis of Zip4 (n) Zip5 (o) protein band density from blots comparing ileum tissue from sham and SBS mice fed a control diet. Source data are provided as a Source Data file.
We next sought to correlate these observed increases in zinc receptor expression observed in human enterocytes explanted from SBS mice with the expression of genes directly within the intestinal epithelium. To do so, we compared levels of Slc39a4 and Slc39a5 transcripts by RT-qPCR analysis following SBS vs sham surgery on WT C57BL/6 mice. Consistent with our scRNAseq results in the human organoids, we observed increased Slc39a4 transcript levels in SBS jejunum (p = 0.0281), and ileum (p = 0.0038) compared to sham controls (Fig. 3e, f), and elevated numbers of Slc39a5 transcripts in SBS jejunum (p = 0.0280) and SBS ileum (p = 0.0476) compared to sham controls (Fig. 3g, h).
Having confirmed the expression of these key transporter genes in the setting of SBS in both mouse and human tissue, we next sought external corroboration of our findings, and therefore turned to an SBS scRNA-seq dataset unaffiliated with our studies. Analysis of the GSE130113 dataset40, which included epithelial cell samples from mice undergoing 50% proximal small bowel resection compared to sham surgery, replicated the presence of multiple epithelial cell clusters (Fig. 3i) using the Seurat package. Specifically, Slc39a4 exhibited increased expression mainly in mature proximal and mature distal enterocytes (Fig. 3j), while Slc39a5 was upregulated in nearly all enterocyte cell clusters (Fig. 3k). These cell populations were abundant in samples derived from SBS ileal tissue.
We next sought to confirm these findings at the protein level. As shown in Fig. 3l–o, SDS-PAGE immunoblotting of ileal tissue revealed the upregulation of Zip4 and Zip5 proteins in SBS intestinal tissue compared to sham, consistent with the transcript data. Taken together, our findings highlight the upregulation of the zinc transport genes SLC39A4 and SLC39A5 in both human and mouse intestinal tissue from various models of SBS. These findings suggest their potential involvement in the adaptive response to SBS and the modulation of zinc homeostasis in the intestinal epithelium.
Zinc supplementation promotes intestinal stem proliferation in juvenile mouse jejunal enteroids and human small intestinal organoids in vitro
To enhance our understanding of how zinc transport affects intestinal adaptation in SBS, we first conducted in vitro experiments using primary mouse enteroid cultures. Enteroids derived from the jejunal crypt cells of juvenile WT C57BL/6 mice were subjected to varying zinc concentrations, including control culture media that was deficient in zinc, 40 μM zinc acetate supplementation, or 2 μM of the high-affinity zinc chelator N, N, N’, N’-tetrakis (2-pyridinylmethyl)-1,2-ethanediamine (TPEN). As shown in Fig. 4, zinc-treated enteroids exhibited increased growth compared to the control group (average diameter 205.5 ± 9.5 μm, p < 0.0001), while TPEN-treated cells were significantly smaller (Fig. 4a–c, m) (average diameter 70.7 ± 6.5 μm, p = 0.0004). Since enterocyte proliferation is a key component of the adaptation response18, we next assessed proliferation by BrdU labeling. After 8 h, we observed a significant increase in the number of BrdU-positive cells in the zinc-treated enteroids compared to the control group (Fig. 4d–f, m). In contrast, TPEN-induced zinc inhibition led to a reduction in BrdU labeling (Fig. 4e). Immunohistochemistry analysis of the expression of cell replication markers Ki67 and PCNA further confirmed the increased proliferation activity in the zinc-treated cells and reduction in the TPEN-treated cells (Fig. 4g–l, m).
Fig. 4. Zinc supplementation promotes intestinal stem proliferation in juvenile mouse jejunal enteroids and human small intestinal organoids in vitro.
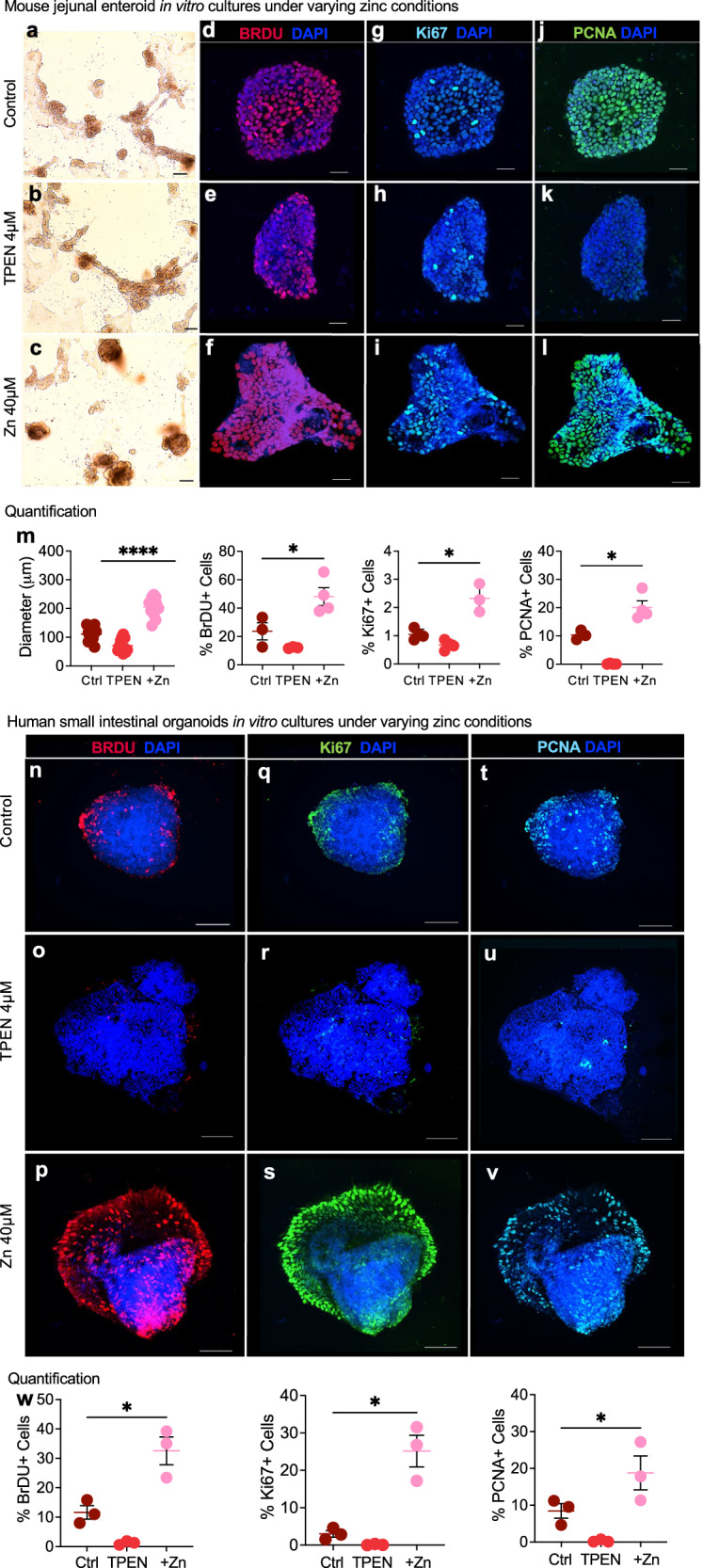
Representative light microscopy images of juvenile WT C57BL/6 mice jejunal enteroids following 24 h of treatment with control a 4 µM TPEN and b 40 µM zinc acetate. c Magnification 10×; scale bar 100 mm. d Representative immunofluorescence images of BRDU staining in jejunal enteroids indicating proliferation after an 8-h pre-treatment with control, d 4 µM TPEN, and e 40 µM zinc acetate. f Magnification 20×; scale bar 100 mm. Representative immunofluorescence images showing Ki67 expression in jejunal enteroids treated with control (g), 4 µM TPEN (h), and 40 µM zinc acetate (i). Magnification 20×; scale bar 100 mm. Representative immunofluorescence images visualizing PCNA expression in jejunal enteroids following treatment with control (j), 4 µM TPEN (k), and 40 µM zinc acetate (l). Magnification 20×; scale bar 100 µm. m Quantification of jejunal enteroid sizes (n = 12 per experimental group) and fluorescence for BrdU, Ki67, and PCNA following 24 h of treatment with control, TPEN, or zinc acetate (n = 3 or 4 per group). Data represent compiled measurements derived from two independent experiments. Enteroid size: t-test. ****p ≤ 0.0001. Quantification of fluorescence: Mann–Whitney U-test, *p ≤ 0.05. Error bars indicate ±SEM. Representative immunofluorescence images of human small intestinal organoids treated with control (n), 4 µM TPEN (o), and 40 µM zinc acetate (p) visualizing BrdU to assess proliferation after 8-h pre-treatment. Magnification 25×; scale bar 50 mm. Representative immunofluorescence images displaying Ki67 expression in human small intestinal organoids treated with control (q), 4 µM TPEN treatment (r), and 40 µM zinc acetate treatment (s). Magnification 25×; scale bar 50 μm. Representative immunofluorescence images showing PCNA expression in human small intestinal organoids treated with control (t), 4 µM TPEN, and (u) 40 μM zinc acetate (v). Magnification 25×; scale bar 50 μm. w Fluorescence quantification of BrdU, Ki67, and PCNA staining following 24 h of treatment with control, TPEN, or zinc acetate. Data are compiled from measurements of three intact stained organoids derived from two independent experiments. Error bars indicate ± SEM. Mann–Whitney U-test, *p ≤ 0.05. Source data are provided as a Source Data file.
Encouraged by our initial findings, we expanded our in vitro studies to human intestinal organoids derived from iPSCs. These organoids were cultured in a control medium, treated with 4 μM TPEN, or supplemented with 40 μM zinc acetate. In zinc-treated organoids, we observed a robust increase in the expression of migration and proliferation markers, including BRDU, Ki67, and PCNA (Fig. 4p–v, w). Conversely, TPEN treatment led to a near-complete suppression of these markers (Fig. 4o–u, w). Organoids treated with sodium acetate, serving as controls for acetate effects, exhibited similar expression levels of the proliferation markers BRDU and TPEN as those observed in the control medium. However, PCNA expression was found to be decreased in these control organoids (Supplementary Fig. 2a–d)
Zinc supplementation in murine SBS enhances intestinal proliferation and adaptation in vivo
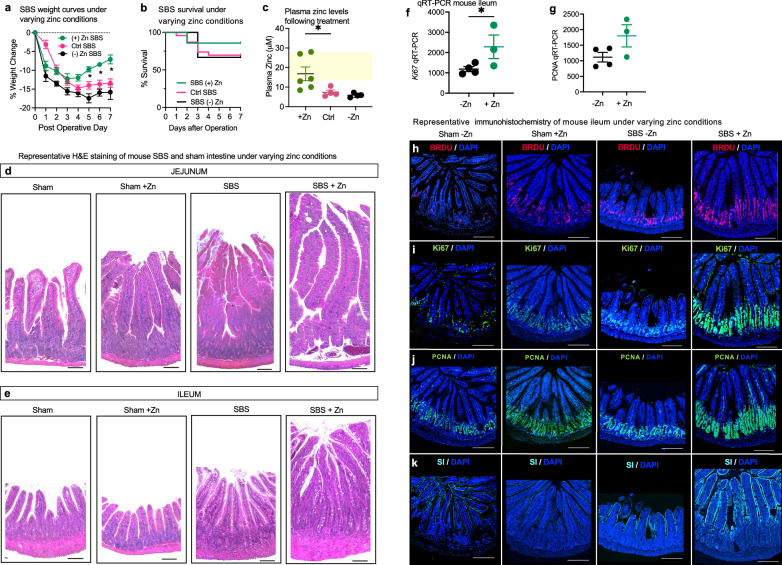
Given our findings of upregulated SLC39A4 and SLC39A5 in human small intestinal organoid and mouse intestinal tissue in the setting of SBS, as well as in vitro evidence of zinc-induced proliferation in enteroids, we next sought to evaluate whether enteral zinc supplementation could enhance intestinal adaptation in SBS. To do so, we next conducted a randomized study in WT C57BL/6 mice, where zinc was administered in the liquid diet at a dose of 65 mg/kg/day, while other groups received a standard liquid diet (13.5 mg/kg/day) or a zinc-depleted diet (estimated 0 mg/kg/day). A separate group of control mice was administered sodium acetate at a dosage of 80 mg/kg/day. This was implemented to specifically control for the effects of acetate, isolating its impact from other variables in the study. Experimental SBS was then induced, and zinc supplementation or deficiency was maintained throughout the postoperative period. Functional adaptation was indirectly assessed by monitoring changes in body weight during the post-operative period. Strikingly, we discovered that SBS mice receiving zinc supplementation exhibited significant weight gain compared to those on standard or zinc-depleted diets (Fig. 5a). By day 7, mice on the standard diet lost 13.38 ± 1.11%, whereas mice in the zinc supplementation group had lost approximately 7.1 ± 1.19% of their original weight (p = 0.0284). Furthermore, while differences in the survival curves did not reach statistical significance, zinc supplementation showed a trend toward improved survival rates in SBS mice compared to control SBS mice at day 7, with survival rates of 85.7% vs 66.67%, respectively (Fig. 5b). Plasma zinc levels in the majority of the zinc-supplemented SBS mice were found to be within the normal to supratherapeutic range29 (Fig. 5c). In contrast, mice on the standard and zinc-depleted diets exhibited low plasma zinc levels. Additionally, growth and survival curves for mice treated with sodium acetate did not differ from those on the standard diet, as shown in Supplementary Fig. 2e–g.
Fig. 5. Zinc supplementation in murine SBS enhances intestinal proliferation and adaptation in vivo.
a Growth curves for SBS mice treated with control (Ctrl SBS) (n = 10), zinc supplementation ((+) Zn SBS) (n-6), and zinc-depleted diet ((−) Zn SBS) (n = 6) over 7 days. Significant weight change is observed in (+) Zn SBS compared to Ctrl SBS from Day 5 onward. Data points are mean ± SEM. t-test, *p < 0.05 at indicated time points. b Survival curves comparing the three groups within the 7-day SBS model. c Plasma zinc levels measured by colorimetric assay (Abcam). Zinc-supplemented mice (n = 6) achieved plasma zinc levels within or above the normal range (12–25 µmol/l, yellow-shaded area). SBS mice on control (n = 4) and zinc-depleted diets (n = 4) did not reach this range. Data are mean ± SEM. t-test, p = 0.0336. H&E staining of the jejunum (d) and ileum (e) tissue following the 7-day SBS model. Mice were fed either a control or a zinc-supplemented diet. Magnification: 20x; scale bar: 100 µm. RT-qPCR analysis of ileal tissue from −Zn SBS mice (n = 4) vs +Zn SBS mice (n = 3), showing ~ 2-fold increase in Ki67 expression (f, p < 0.05) and a trend towards increased PCNA expression (g) in the zinc-supplemented group (p = 0.0540). Data are mean ± SEM. Representative immunofluorescence images of intestinal tissue from SBS mice following 10 days of post-operative treatment with zinc depleted or zinc-supplemented diet. h BRDU staining 24 h after gavage; Ki67 expression (i), PCNA expression (j), and SI expression (k) under the same conditions. Magnification 10×; scale bar 100 µm. Source data are provided as a Source Data file.
Based upon these findings, we assessed directly whether zinc supplementation could alter intestinal adaptation to SBS, and thus advance the potential clinical relevance of these findings. We assessed structural adaptation histologically by measuring villus height and crypt depth and compared these parameters across the various treatment groups. As shown in Fig. 5d, e, morphometric analysis of hematoxylin-and-eosin (H&E) stained sections revealed a significant increase in average villus height in control SBS villi (649.6 ± 16.28 μm, jejunum), compared with sham mice (521.4 ± 23.34 μm, jejunum), that was significantly increased for SBS villi in the setting of zinc treatment (881 ± 24.77 μm, jejunum). Differences in villus length between the sham and SBS groups were consistent across all intestinal sections, irrespective of whether mice were fed a standard diet or treated with sodium acetate (Fig. 1i and Supplementary Fig. 2h–j).
We directly evaluated functional adaptation by performing qRT-PCR and immunohistological analyses on tissue samples from this study. This assessment included the use of antibodies targeting proliferative markers BRDU, Ki67, and PCNA, as well as measurements of sucrase-isomaltase (SI) enzyme levels. Our findings indicated a 2-fold increase in Ki67 expression (Fig. 5f), and although there was a trend towards higher PCNA levels, these changes did not reach statistical significance (Fig. 5g). Immunohistological analysis further revealed that zinc depletion was associated with reduced expression of the proliferation markers BRDU, Ki67, and PCNA in sham mice while zinc supplementation diminished SI staining. Notably, SBS mice receiving 65 mg/kg/day of zinc compared to those on a zinc-depleted diet showed increased expression of all analyzed proliferation markers, as well as SI (Fig. 5h–k). These findings reveal that zinc supplementation can enhance intestinal adaptation, leading us to evaluate the clinical relevance of these findings in humans with SBS.
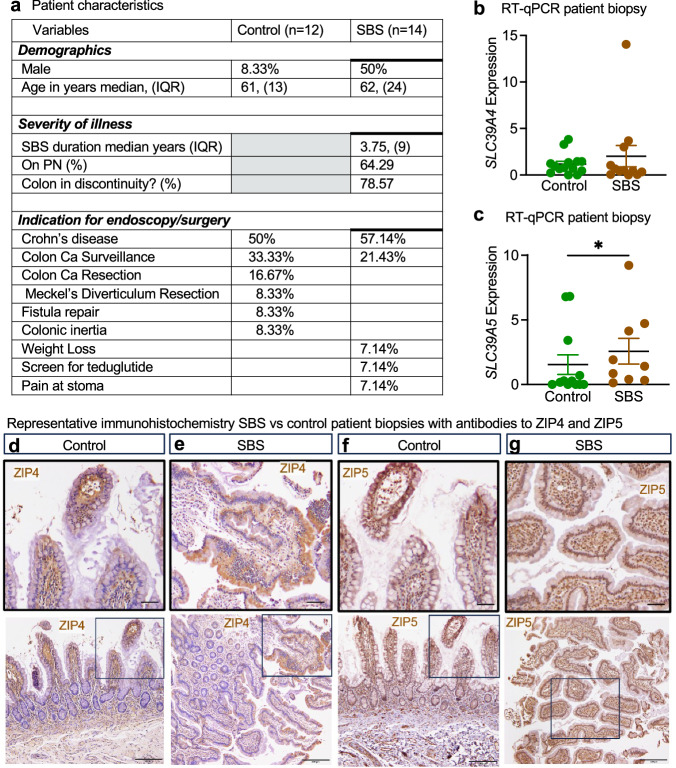
Intestinal tissue from SBS patients shows increased expression and protein translocation of ZIP4 and ZIP5 to putative sites
To confirm the clinical relevance of zinc transport in human SBS, we next conducted a retrospective analysis of intestinal biopsies from 26 patients who had undergone endoscopic or surgical procedures between 2008 and 2020. Among these patients, 14 were diagnosed with clinical SBS, with approximately 65% dependent on parenteral nutrition. The average duration of SBS among these individuals was 3.75 years (Fig. 6a). We assessed the expression levels of SLC39A4 and SLC39A5 in their intestinal tissue using qRT-PCR and immunohistochemistry. qRT-PCR analysis revealed significantly higher levels of SLC39A5 expression in SBS patients compared to the control group (p = 0.0346) (Fig. 6c). Of note, the expression of the counterregulatory transporter SLC39A4 was unchanged (Fig. 6b). Given the critical role of translational and post-translational regulation in the function of zinc transport proteins22,41–43, we further examined the protein expression of ZIP4 and ZIP5 in the clinical biopsies using immunohistochemistry. Specifically, ZIP4 was found to be localized at the cell surface of enterocytes in SBS patients, while in control tissue, ZIP4 remained within the cytoplasm (Fig. 6d, e). Similarly, ZIP5 was primarily cytoplasmic in control patients but exhibited strong localization to the epithelial basement membrane in SBS patients (Fig. 6f, g). The observed distinctive pattern of localization for primary zinc transepithelial receptors implies an avid enterocytic appetite for zinc in the setting of SBS, underlining its pivotal role in fostering intestinal adaptation.
Fig. 6. Intestinal tissue from SBS patients shows increased expression and protein translocation of ZIP4 and ZIP5 to putative sites.
a Table describes a total of 26 patients that were included in this study. b, c RT-qPCR of tissue derived from human intestinal biopsies. Gene expression levels were normalized by SI expression to estimate expression levels in enterocytes. SLC39A4 (b) is expressed at similar levels in control (n = 14) compared to SBS (n = 12). Approximately 1.5-fold higher SLC39A5 (c) expression in SBS (n = 9) compared to control (n = 12). Data points represent means, and bars represent ±SEM. Mann–Whitney U-test, p = 0.0346. Representative immunostaining of ZIP4 in control (d) and SBS (e) human intestinal tissue biopsies; staining of ZIP5 in control (f) and SBS (g) biopsy tissue demonstrate changes in the localization of zinc transport proteins in response to SBS. Tissue from over 25 biopsies from participants across both the SBS group and the control group were analyzed. Magnification 20×, upper panel scale bar 50 μm; lower panel scale bar 200 μm. Source data are provided as a Source Data file.
Gene regulatory network analysis uncovers KLF5 as a key regulator of ZIP4 expression in SBS
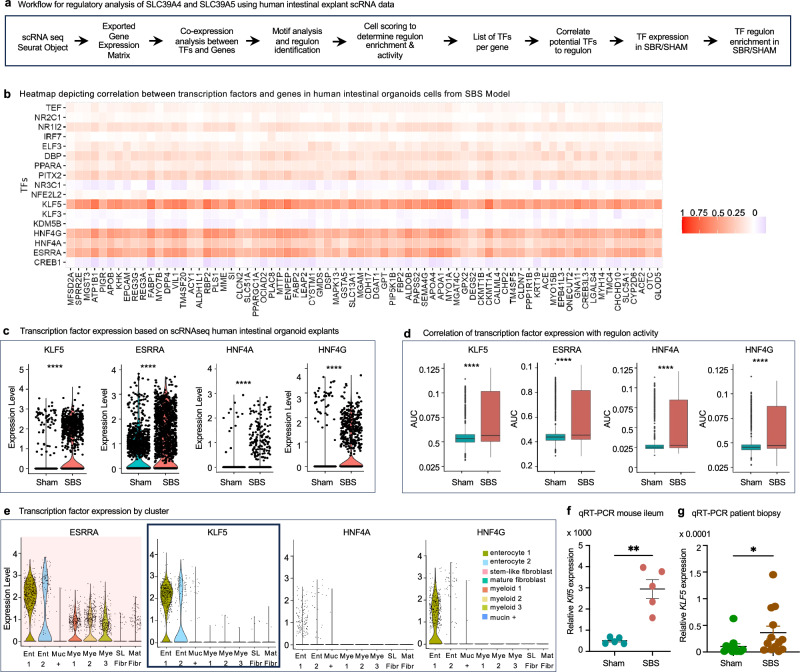
To identify the key TF governing the expression of ZIP4 and ZIP5 in SBS, we employed a combination of bioinformatics analyses and experimental validation. We determined the transcriptional regulatory network governing the expression of ZIP4 and ZIP5 in SBS, by employing the pySCENIC tool to infer transcription factor activity based on gene expression patterns and known transcription factor binding site information (Fig. 7a). Through this analysis, we identified 17 candidate TF that potentially upregulate the regulon encompassing SLC39A4 and SLC39A5.
Fig. 7. Gene regulatory network analysis uncovers KLF5 as a key regulator of ZIP4 expression in SBS.
a Workflow for regulatory analysis of SLC39A4 and SLC39A5. TF was identified using pySCENIC and analyzed for best fit. b Heatmap showing the correlation between TF and gene expression in human intestinal organoid explants from scRNA-seq data. Columns represent genes, and rows represent TFs, with color intensity indicating correlation (red for positive, blue for negative). KLF5, HNF4A, HNF4G, and ESRRA were identified as potential regulators due to their strong correlation with SLC39A4 and SLC39A5 expression. c Violin plots of TF expression in sham and SBS human intestinal explant cells. All four TFs (KLF5, HNF4A, HNF4G, and ESRRA) show significantly higher expression in SBS compared to sham. Wilcoxon rank sum test, ****p < 0.0001. d AUC scores indicate increased regulon activity for each TF in SBS vs sham, suggesting regulon enrichment in cells expressing these TFs. Box plots display AUC score distributions, with the median indicated by a central line, bounds of the box corresponding to the lower (Q1) and upper (Q3) quartiles, and whiskers extending to 1.5 times the interquartile range. Wilcoxon rank sum test, ****p < 0.0001. Data are from 3090 cells (1729 from five SBS mice, 1361 from five sham mice). e Violin plots showing TF expression across cell clusters from human small intestinal explant scRNA-seq analysis, highlighting KLF5 specificity within the regulon to enterocyte cells. f qRT-PCR reveals increased Klf5 expression in ileal enterocyte cells from SBS mice (n = 5) vs sham controls (n = 5), p = 0.004. g Increased Klf5 expression in ileum tissue from SBS patients (n = 13) compared to controls (n = 12) by qRT-PCR, p = 0.0173. Data are mean ± SEM. Mann–Whitney U-test, *p ≤ 0.05; **p ≤ 0.01. Source data is provided as a Source Data file.
To narrow down these TF, we identified genes with the highest co-expression to SLC39A4 and SLC39A5, creating a potential regulon. We then correlated the expression of all these genes to all 17 TF and found that four TF stood out as having a strong correlation across this potential regulon: KLF5, ESSRA, HNF4A, and HNF4G (Fig. 7b). To further examine these potential regulators, we looked at their expression in SBS compared to sham-derived cells and found that all four had significantly higher expression in SBS cells (Fig. 7c).
We next evaluated the area under the curve (AUC) scores generated by pySCENIC, which are used as a metric of regulon enrichment. This value also indicates that the regulons of these four TF are significantly enriched in SBS compared to sham (Fig. 7d). Since the expression of SLC39A4 and SLC39A5 are specific to enterocytes, we checked our pool of potential regulators for this pattern. Among these candidate TF, it became evident that ESSRA was not exclusively expressed in enterocyte cells; its expression was also observed in various other clusters.
When considering the remaining candidates, KLF5 had the highest expression across enterocytes, suggesting it is the primary regulator responsible for the transcription of SLC39A4, SLC39A5, and their associated genes within the identified regulon in enterocyte cells (Fig. 7e). Finally, we validated the expression and activity of Klf5 in our experimental model. Our analysis revealed a noteworthy upregulation of Klf5 in native mouse SBS enterocytes in comparison to sham controls (Fig. 7f). Most remarkably, though overall levels were low compared to housekeeping genes, KLF5 expression was statistically higher in tissue obtained from SBS patient biopsies compared to control patient intestinal tissue (Fig. 7g).
Discussion
SBS poses a significant clinical challenge, with the goal of treatment being to restore enteral autonomy and reduce dependence on parenteral nutrition. To develop additional therapies and improve patient outcomes, we sought to deepen our understanding of the mechanisms underlying adaptation. We employed an organoid xenotransplantation model of SBS in mice to map the human gene pathways involved. Our analysis revealed significant upregulation of transepithelial zinc transport genes SLC39A4 and SLC39A5 in human organoids implanted in SBS mice.
Despite the known biological and clinical manifestations associated with zinc deficiency, our study is the first to demonstrate a direct correlation between impaired zinc homeostasis and intestinal adaptation. Zinc needs in humans vary by age and physiological status, with daily recommendations ranging from 2 mg for infants to 11 mg for adult males44. In healthy individuals, zinc absorption primarily occurs in the proximal small intestine27,45. Several factors can inhibit this process, including phytates, dietary fiber, excessive calcium and iron levels, oxalates, polyphenols, certain medications, and excessive alcohol consumption22,46,47. These inhibitors can bind to zinc, form insoluble complexes, or compete for absorption, reducing zinc’s bioavailability48. The body stores zinc mainly in skeletal muscle and bone, with additional storage in the liver, skin, kidneys, and prostate49. Whole-body zinc content remains stable over a wide range of dietary zinc concentrations due to efficient homeostatic mechanisms. Excess endogenous zinc is secreted into the intestine and excreted in feces50.
In our study, when examining clinical SBS samples from patients, approximately 65% of whom were reliant on TPN, we observed the localization of ZIP4 at the luminal border. This observation indicates the presence of ongoing zinc deficiency. Additionally, SLC39A5 showed elevated expression, and its corresponding protein was abundantly present, localized to the epithelial basement of SBS enterocytes, suggesting increased transport of zinc from the systemic circulation to the enterocytes. These findings overall point towards the ramping up of redundant mechanisms for transporting luminal and systemic zinc to SBS enterocyte cells. Consequently, it is conceivable that SBS patients may require supratherapeutic zinc supplementation to achieve the levels required to facilitate adaptation effectively. To address this hypothesis, a targeted clinical study is warranted. Furthermore, exploring agents capable of modulating the zinc transport proteins may be essential for potential combination therapy with zinc supplementation.
Our findings also highlight the involvement of four TF—KLF5, HNF4A, HNF4G, and ESSRA—in the regulation of ZIP4 and ZIP5 expression in SBS. While the initial three TF are known for their roles in intestinal epithelial cell differentiation and intestinal development, ESRRA, recognized for its involvement in mitochondrial function and energy metabolism51, added a novel dimension to our understanding. These identified TF displayed co-expression with, and co-regulation of previously upregulated genes linked to SBS, such as RPB2, APOA4, SI, VIL1, and CREB3L37,8,40,52. Remarkably, a previous single-cell RNA sequencing study in mouse cells underscored the increased expression of KLF5 in intestinal epithelial cells post-small bowel resection, highlighting its pivotal role in cellular reprogramming during intestinal adaptation40.
This study addresses the existing knowledge gaps in SBS research by utilizing an innovative approach. Given the scarcity of large SBS patient tissue repositories, we employed an organoid xenotransplantation model in mice to map the human genes and pathways involved in SBS-associated adaptation. Through this approach, we identified that zinc supplementation could represent a novel therapeutic strategy for SBS. The significance of our study lies in its translational potential. By using human tissue-derived data, we provide findings that are readily translatable to the clinical setting. While we have confirmed differences in zinc transporter expression and localization in intestinal tissue from SBS patients compared to controls, our findings will be strengthened by future clinical trials to evaluate the effects of zinc supplementation, ZIP4, and ZIP5 modulators in SBS patients. These endeavors will provide additional data to support the role of zinc homeostasis in improving outcomes in SBS.
An important consideration in our study is that the hiPSC-derived organoids were not in direct contact with the intestinal lumen of the mouse. This isolation from the lumen suggests that the observed changes in the hiPSC-derived intestinal organoids are likely driven by circulating or humoral factors derived from the mesenteric circulation of the host. Cytokines, growth factors, and hormones significantly influence the behavior and differentiation of intestinal stem cells. However, our data may not have captured all the physiological and cellular effects induced by direct exposure to intestinal contents, including nutrients, microbiota, and luminal secretions, along with their associated mechanisms.
Nevertheless, we have developed a discovery platform that holds promise for new avenues for research and the development of innovative therapeutics not only for SBS but also for other intestinal absorptive disorders. Moreover, this platform will contribute to elucidating cellular processes governing human intestinal stem cell function.
Methods
This study was conducted in compliance with all relevant ethical regulations. All animal work followed a protocol approved by the Johns Hopkins University Institutional Animal Care and Use Committee (protocol MO20M276). All animal protocols were meticulously reviewed and approved by Johns Hopkins University and all practices adhered strictly to the principles outlined in ‘The Guide for the Care and Use of Laboratory Animals’ and the Public Health Service Policy on Humane Care and Use of Laboratory Animals. The Institutional Review Board of Washington University School of Medicine approved the use of human tissue in this study (IRB Number 201504100), and informed written consent was obtained from all participants.
hiPSC culture conditions and human intestinal organoid differentiation
Human iPSC (hiPSC) cell line ASE-9209 was obtained from Applied Stem Cell (Milpitas CA). The cell line was generated from normal human dermal fibroblasts of a 47-year-old female using episomal reprogramming methods. The hiPSC were cultured under feeder-free conditions using StemFlex™ Medium (ThermoFisher Scientific) on human embryonic stem cell qualified Matrigel (Corning) coated tissue culture plates. Passaging was performed using 0.5 M EDTA in PBS (ThermoFisher Scientific).
Differentiation was carried out as previously described36,37. hIPSC grown to 70–80% confluence were plated to 100000 cells/well. Cells were then treated with 100 ng/ml Activin A (R&D Systems) for 3 days in RPMI supplemented with 2 mM l-glutamine, and penicillin-streptomycin with increasing concentration of dFBS daily. This resulted in the generation of a definitive endoderm monolayer, which was further cultured supplemented with 500 ng/ml FGF-4 (Prepotech) and 3 μM CHIR99201(Sigma) to generate midgut spheroids. The culture medium was changed daily for 4 days. Spheroids were then collected, and resuspended into warm Matrigel which was left to solidify at 37 °C. Matrigel cultures were then maintained in Advanced DMEM/F-12 supplemented with B27, N2, 15 mM HEPES, 2 mM l-glutamine, and penicillin-streptomycin, all from ThermoFisher Scientific, for 14 days. Mature organoids were then selected and replated in fresh Matrigel and growth media consisting of the components described above and supplemented with 100 ng/ml EGF (Sigma).
mRNA isolation and quantitative PCR
mRNA was isolated using the RNeasy mini kit (Qiagen) and complementary DNA was synthesized from 0.5 μg RNA using QuantiTect Reverse Transcription kit (Qiagen) per the standard protocol. Quantification was performed using iTaq™ universal SYBR® Green supermix (Bio-Rad) on the CFX96 Real-Time System (Bio-Rad). CFX Manager 3.1 software (BioRad) was used for data collection and analysis. Relative mRNA expression levels were normalized to the expression of endogenous housekeeping gene ribosomal protein lateral stalk subunit P0 (Rplp0). The primers used are listed in Table 2.
Table 2.
PCR primers
| Gene | Species | Forward primer sequence | Reverse primer sequence |
|---|---|---|---|
| MTRNR2L10 | Human | CTCCGCAAATTTTACCCCGC | GCGGCCATTGAACGTATGTC |
| OCT-4 | Human | GAGTGAGAGGCAACCTGGAG | GAACCACACTCGGACCACAT |
| FABP1 | Human | GCTGGAGACAATGACAGGGG | TATGTCGCCGTTGAGTTCGG |
| VIL1 | Human | CTTCCAGAAGTGGACAGCGT | GCATGGATGTGGCATCGAAC |
| CHGA | Human | AAGGTGATGAAGTGCGTCCTGGAA | AGCAGATTCTGGTGTCGCAGGATA |
| MUC2 | Human | TAGTGGAGATTGTGCCGCTGAAGT | AGAGCCCATCGAAGGTGACAAAGT |
| SLC39A4 | Human | GTTGGAGTCAGCGAGGAGAG | CGGGTCCCGTACTTTCAACA |
| SLC39A5 | Human | GACCACTCATTGGCTGACCA | GAGTTCCTAGTCCAGCAGCC |
| Slc39a4 | Mouse | ATATCTCTGCAGCTGGCACC | GTAGTTCCGGGGTCTCCTCT |
| Slc39a5 | Mouse | GGATGGTCCTCCTGGGAGAT | GTGGCAGAAGACTGCTAGGG |
| Klf5 | Mouse | CGACGTATCCACTTCTGCGA | CTTCTCGCCCGTATGAGTCC |
Immunofluorescence
For immunofluorescence staining, mature human organoids differentiated from IPSC were cultured in chamber slides precoated with laminin mouse protein (ThermoFisher). Organoids were washed with PBS and fixed with 4% PFA for 30 min then rinsed and permeabilized with 0.1% Triton-X 100 for 30 min. Tissue was probed with primary antibodies overnight at 4 °C, and subsequently with secondary antibody and 4′,6-diamidino-2-phenylindole (DAPI, Biolegend) for 1 h at room temperature, and then mounted in Gelvatol mounting media (Sigma-Aldrich) for imaging. Imaging was performed using NIS-Elements AR v4.10.01 software on a Nikon Eclipse Ti Confocal microscope. Antibodies used were as follows: goat anti-e-cadherin (R&D Biosystems AF748 1:200), rabbit anti-chromogranin-A (Abcam ab15160 1:200), and rhodamine phalloidin (Sigma R415 1:200). Secondaries were Alexa Fluor® 680 donkey Anti-Goat IgG (H + L) antibody (Life Technologies A-21084 1:1000) and Alexa Fluor® 488 Donkey Anti-Rabbit IgG (H + L) antibody (Life Technologies A-21206 1:1000).
Animal studies
We employed a preclinical experimental model that closely mimics SBS in mice, and we correlated our findings with observations made in patients and in resected intestinal specimens from patients diagnosed with SBS. To conduct these experiments, mice used in the study were specifically bred in-house to create various mutant mouse lines. WT (stock no: 000664) and Rag1 KO (stock no: 002216) C57BL/6 mice were obtained from the Jackson Laboratory. In experiments involving genetically modified mice, all mice underwent a minimum of eight backcrosses with C57BL/6. Mice were housed in a controlled environment with a 12-h light/dark cycle. The ambient temperature was maintained at 22 ± 2 °C, and the relative humidity was kept at 50 ± 10%. Mice had ad libitum access to liquid feed throughout the study.
Xenotransplantation SBS model
We created a novel xenotransplantation model of human SBS based on a 50–75% resection model described by our lab and others. Anesthesia with 90 mg/kg ketamine and 10 mg/kg xylazine was administered on 6-week-old C57BL/6 Rag1 KO mice of both sexes, following which the skin and linea alba were incised to allow for evisceration of the small bowel. For SBS mice, 75% of the small bowel was then resected beginning approximately 3–4 cm from the ligament of Treitz to 4 cm from the ileocecal valve. The remnant small intestinal segments were anastomosed using 12–15 interrupted 8–0 nylon sutures (Ethicon). Sham mice underwent transection of the terminal ileum about 4 cm from the ileocecal valve and subsequent anastomosis similarly. Three microliter of octyl/butyl cyanoacrylate adhesive glue (GLUture, Abbott Laboratories) was then placed on the small bowel mesenteric tissue, and human small intestinal organoids attached to a bed of Matrigel were harvested from in vitro culture, dropped into the glue, and allowed to dry for 10 min. The small intestine was then returned to the abdominal cavity and the abdomen was closed in two layers with 3–0 and 4–0 Vicryl (Ethicon) sutures following 2 ml of normal saline administered intraperitoneally. A rodent liquid diet (LBS Biotechnology F1259SP) was provided ad libitum in the peri-operative period. Mice received a 2 ml subcutaneous infusion of normal saline per day through postoperative day 3. Weights were followed for 25–28 days after surgery, and mice were sacrificed to harvest xenotransplants. All surgical procedures were performed with the aid of an operative microscope.
Tissue processing, morphometric analysis, and immunohistochemistry
To assess for adaptation in native mouse intestines, we harvested jejunum and ileum tissue for fixation in 4% PFA for 24 h. Fixed tissues were embedded in paraffin, cut into 5–10 μm sections, and stained with hematoxylin-and-eosin using standard protocols. Imaging was performed using light microscopy (Leica). At least 25 appropriately oriented and intact villi were measured per mouse to assess adaptation. Paraffin sections were processed in parallel for immunofluorescence staining. Briefly, 5-10um jejunum and ileum sections were rehydrated and heated in 10 mmol/l citric acid buffer at a pH of 6 for antigen retrieval. Tissue was permeabilized with 0.1% Tween-20 and stained with rat anti-BRDU (Novus NB500-169/ab6326 1:200) goat SI Antibody (Santa Cruz sc-27603 1:200), rabbit Anti-Ki67 antibody (Abcam ab-a5580 1:200), or mouse anti PCNA (Santa Cruz sc-56 1:200) overnight at 4 °C. Sections was then stained with DAPI (Biolegend 1:1000), Alexa Fluor® 680 donkey anti-goat IgG (H + L) antibody (Life Technologies A-21084 1:1000), Alexa Fluor® 594 donkey anti-rat IgG (H + L) antibody (Life Technologies A-21209 1:1000), Alexa Fluor® 488 donkey anti-rabbit IgG (H + L) antibody (Life Technologies A-21206 1:1000) and Alexa Fluor® 488 donkey anti-mouse IgG (H + L) antibody (Life Technologies A-21202 1:1000) for 1 h at room temperature and mounted in Gelvatol mounting media (Sigma-Aldrich) for imaging.
scRNAseq library preparation
scRNAseq was performed on human intestinal organoid implants from five SBS mice and five sham control mice. Xenotransplants were harvested 3–4 weeks following surgery and immediately dissociated into single-cell suspension by using Accumax (Sigma). A single-cell library was constructed by the JHU (Johns Hopkins University) School of Medicine Sequencing Core. Cell counts and viability were determined using the Cell Countess II with Trypan Blue. A maximum volume of 43.3 μl/sample was used for processing to target up to 10,000 cells. Cells were combined with RT reagents and loaded onto 10× Next GEM Chip G along with 3’ v3.1 gel beads. The NextGEM protocol was run on the 10× Chromium Controller to create GEMs, composed of a single cell, gel bead with a unique barcode and UMI primer, and RT reagents. 100 ul of emulsion was retrieved from the chip and incubated (45 min at 53 °C, 5 min at 85 °C, cool to 4 °C), generating barcoded cDNA from each cell. The GEMs were broken using Recovery Agent and cDNA was cleaned, following the manufacturer’s instructions using MyOne SILANE beads. cDNA was amplified for 11 cycles (3 min @ 98 °C, 11 cycles: 15 s @ 98 °C, 20 s @ 63 °C, 1 min @ 72 °C; 1 min @ 72 °C, cool to 4 °C). Samples were cleaned using 0.6× SPRIselect beads. QC was completed using Qubit and Bioanalyzer to determine size and concentrations. Ten microliters of amplified cDNA were carried into library prep. Fragmentation, end repair, and A-tailing were performed (5 min @ 32 °C, 30 min @ 65 °C, cool to 4 °C), and samples were cleaned up using double-sided size selection (0.6× and 0.8×) with SPRIselect beads. Adapter ligation (15 min @ 20 °C, cool to 4 °C), 0.8× cleanup, and amplification are performed, with PCR using unique i7 index sequences. Libraries undergo a final cleanup using double-sided size selection (0.6× and 0.8×) with SPRIselect beads. Library QC is performed using Qubit, Bioanalyzer, and KAPA library quantification qPCR kit. Libraries are sequenced on the Illumina NovaSeq 6000 using v1.5 kits, targeting 50 K reads/cell, at read lengths of 28 (R1), 8 (i7), 91 (R2). Demultiplexing and FASTQ generation are completed using Illumina’s BaseSpace software.
scRNAseq analysis
UMI count matrices were generated by aligning fastq files to GRCh38 human genome reference assembly with the 10x Genomics Cell Ranger 6.1.1 count function53. Preprocessing and QC filtering were performed with the R package Seurat 4.254. First, the count matrices were converted to a Seurat object with the Read10X function. Cells with less than 300 genes, more than 4000 genes, or over 25% mitochondrial expression were filtered out to reduce levels of dead cells and doublets. After filtering, there were 3160 cells expressing 36,601 genes. Next data was normalized, and the top 2000 variable genes were identified with the FindVariableGenes function. This function uses the variance stabilizing transformation method of identifying top variable genes with a regularized negative binomial regression model.
Next, the data was scaled, and dimensional reduction was performed with principal component analysis. After using an elbow plot to visualize the percentage of variance explained by each component, the top seven principal components were selected for UMAP reduction, the resulting dimension plot had the cells sorted into nine distinct clusters based on transcriptome expression. The FindAllMarkers function was used to determine marker genes for each cluster. To identify the cell type of each cluster, its DEGs were compared with Pearson and Spearman correlation to published single-cell data sets with identified cell types55. Cluster cell type predictions were further verified through the expression of marker genes (Table 1). The R package ComplexHeatmap 2.14.0 was used to generate a heatmap of the top DEGs and marker genes of interest to show the contrast in gene expression between clusters.
GSEA for clusters 2 vs 5 and clusters 7 vs 4 was performed with the R package clusterProfiler 4.6.0 function gseGO56. GSEA was run twice for each comparison, once with biological process annotations and again against molecular function annotations. Get sets for comparison had to have at least ten genes and be tested against a maximum of 500 annotated genes. p-values were adjusted with Benjamini–Hochberg multiple testing correction. Results were combined into one large data frame and limited to an adjusted p-value under 0.05. Separate upregulated and downregulated pathways were then created by separating positive and negative NES values. Pathways with similar gene expression were combined into more general pathway terms. Values of these similar G O (GO) were averaged. We then created a network plot with the network function of ggraph 2.1.0. Additionally, we compared core enrichment genes to DEGs between cluster 2 vs 5 and cluster 7 vs 4 to determine which gene expression levels are simultaneously significantly changed between treatments and are primary contributors to pathway enrichment. These genes with the highest fold change were added to the plot.
Primary juvenile mouse small intestinal enteroid and human small intestinal organoid culture
Intestinal crypts of C57BL/6 mice under 3 weeks of age were isolated and cultured as previously described57. Briefly, intestinal crypts were isolated and plated following resuspension in Matrigel (BD Biosciences). Culture medium (advanced Dulbecco’s modified Eagle medium/F12 supplemented with penicillin/streptomycin, 10 mmol/l HEPES, Glutamax, 1× N2, 1× B27 (Thermo Fisher Scientific), and 1 mmol/l N-acetylcysteine (Sigma) was supplemented with the following growth factors; EGF at 50 ng/ml (Life Technologies), Noggin at 100 ng/ml (PeproTech) and R-spondin1 at 500 ng/ml (R and D Biosystems). The culture medium was changed every 3–4 days. Human small intestinal organoids derived from iPSCs were treated in a similar fashion.
To characterize the proliferative effects of zinc, cultured small intestinal enteroids, and organoids were treated with the control medium described above, supplemented with 40 μM sodium acetate, 4 μM TPEN, or 40 μM zinc acetate (Sigma) for 24 h. BRDU was added for 8 h. The enteroids were then washed with PBS, fixed with 4% PFA, and permeabilized with 0.1% Triton-X 100. Primary antibodies at 1:500 dilution were added overnight at 4 °C, and subsequently secondary antibody and DAPI (Biolegend) for 1 h at room temperature. Slides were mounted with Gelvatol (Sigma-Aldrich) for imaging. Imaging was performed using NIS-Elements AR v4.10.01 software on a Nikon Eclipse Ti Confocal microscope. was used to take images for IF staining.
Antibodies used were as follows: rabbit anti-Ki67 (Abcam ab15580-100 1:200), rat anti-BRDU (Novus NB500-169/ab6326 1:200), and mouse anti PCNA (Santa Cruz sc-56 1:200). Secondaries were Alexa Fluor® 680 donkey anti-goat IgG (H + L) antibody (Life Technologies A-21084 1:1000), Alexa Fluor® 594 donkey anti-rat IgG (H + L) antibody (Life Technologies A-21209 1:1000) and Alexa Fluor® 488 donkey anti-rabbit IgG (H + L) antibody (Life Technologies A-212061:1000). ImageJ software was used to determine the pixel intensity of the regions of interest, normalized to DAPI fluorescence of each enteroid under study.
Rescue zinc therapy for SBS WT mice
6-week-old WT C57BL/6 mice were randomized to receive a standard liquid diet (LBS Biotechnology F1259SP) ad libitum, with dietary supplementation of up to 65 mg/kg/day of zinc acetate (Sigma), or a specially formulated zinc-free liquid diet prepared with albumin (LBS Biotechnology F10353SP) 3 days prior to operation. Mice in each group underwent 75% small intestinal resection as described in the xenotransplantation model. No xenotransplantation was performed. Mice were kept on specified diets for their designated groups through postoperative day 7, after which they were sacrificed. Intestinal tissue was collected for Western blotting and histological analysis as described above.
Western blotting
Jejunal tissue harvested from zinc and control-treated SBS and sham mice were processed into lysates and subjected to western blotting as previously described using antibodies to ZIP4 (20 625-1-AP Proteintech 1:500), ZIP5 (OAPB01216 Aviva Systems 1:1000), and beta-actin (mouse anti-beta actin—HRP A00730-200 Genentech; 1:1000 dilution).
Patient population
Patients diagnosed with SBS (n = 14) were recruited from the Gastroenterology Clinic at Washington University in St. Louis School of Medicine and Barnes-Jewish Hospital. SBS was defined as a residual small bowel length of no more than 200 cm, with or without part or all of the colon in continuity.
All patients were allowed to consume oral enteral diets, with some receiving intravenous (parenteral) nutrition support along with routine IV micronutrient supplementation, including zinc. Those without parenteral nutrition were prescribed a once-daily multivitamin, and some required oral zinc supplementation. Control subjects (n = 12) were recruited from the Center for Advanced Medicine Endoscopy Center and the general surgical/colorectal surgical service. Control patients underwent routine endoscopic colon cancer screening or polyp surveillance, confirmed to have normal small intestines during endoscopy. Ileal biopsies were obtained during colonoscopy or from surgical resection specimens, during which areas of normal ileum were biopsied at the margins. Biopsies from patients with SBS were performed between 2016 and 2019, while those from control subjects spanned from 2008 to 2020. Detailed patient demographics are available in Fig. 6a.
Immunohistochemistry
Human tissue biopsies were processed in formalin, dehydrated with 70% ethanol, and embedded in paraffin. 5μm sections were cut, rehydrated, and subjected to antigen retrieval in a pressure cooker for 3 min at 18PSI at 99 °C. After blocking with 0.3% peroxidase (HX0635-3, MilliporeSigma) and blocking buffer (MilliporeSigma), specimens were incubated at 4 °C overnight with human anti-ZIP4 (1:200 20625-1-AP, Proteintech) or human anti-ZIP5 (1:50, OAPB01216, Aviva). The bound antibodies were detected using HRP-Polymer (Biocare Medical) and developed using DAB (Betazid DAB, Biocare Medical). Detection was performed using Sections that were counterstained with hematoxylin and visualized under an Olympus BX43 microscope (Olympus Life Science).
mRNA isolation and quantitative RT-PCR
RNA extraction was carried out using TRIzol reagent (Invitrogen, Thermo Fisher Scientific). cDNA was synthesized from 1 μg/μl of RNA using the SuperScript II RT cDNA synthesis kit (Invitrogen, Thermo Fisher Scientific). The cDNA was then amplified using the PCR 2720 Thermal Cycler (Applied Biosystems, Thermo Fisher Scientific). Gene expression analysis was conducted on the StepOnePlus system using Fast SYBR Green Master Mix (Applied Biosystems, Thermo Fisher Scientific). Primer sequences are provided in Table 2. Cycle threshold values were normalized to villin levels, and fold induction was calculated using the ΔΔCt method.
Transcription factor analysis
Regulatory analysis of SLC39A4 and SLC39A5 in human intestinal organoids was performed with pySCENIC 0.12.158. The Seurat object containing scRNA seq data was converted to a gene expression matrix with Seurat’s GetAssayData function. This assay was then loaded into a loom file with SCopeLoomR 0.13.0. This loom file was used as input for pySCENIC and the pySCENIC output was analyzed in SCope online59. This provided a list of 17 potential TF for both SLC39A4 and SLC39A5. These TF were further investigated. First, genes that are highly co-expressed with SLC39A4 and SLC39A5 were identified through Spearman correlation in base R. The correlation matrix showed that SLC39A4 and SLC39A5 had similar top genes, with genes showing a higher correlation in SLC39A5 than SLC39A4. Genes with a correlation over 0.5 to SLC39A5 expression were kept as co-expressed genes. The expression profiles of these genes were then correlated to the transcription of all 17 TF with rcorr from the package Hmisc 5.0–160. Of these, four TF had a higher correlation to the co-expressed genes, and additionally, all correlations for these TF were statistically significant.
We then examined the expression patterns of these four TF of interest in SBS vs SHAM. Violin plots were generated with Seurat comparing SLC39A4 and SLC39A5 to transcription factor expression in SBS and SHAM. Additionally, the output loom file from pySCENIC was loaded into R with ScopeLoomR. AUC scores for each regulon were loaded into a matrix from the loom filed using get_regulons_AUC from SCopeLoomR and getAUC from AUCell 1.20.258. Generated boxplots for regulations of interest to determine if their activity is influenced by SBS.
Statistical analysis and reproducibility
All experiments were performed on at least three independent samples and yielded consistent results. Statistical analyses were performed using Prism 9.0 (GraphPad Software, San Diego, CA). Differences between groups were evaluated using a two-tailed unpaired t-test for all parametric variables, and the Mann–Whitney U-test for non-parametric data. Single-cell gene expression analysis utilized two-tailed non-parametric Wilcoxon rank sum tests. Values of p < 0.05 were considered significant. Data are presented as mean ± SEM, unless otherwise noted. Sample sizes (n) are indicated in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
Dr. Tyler J. Creamer and Linda Orzolek of the Johns Hopkins Single Cell & Transcriptomics Core. D.J.H. is supported by National Institutes of Health grants R35GM141956. M.E.S.S., C.L. and D.S. have been supported by T32DK007713.
Author contributions
The project was conceived by D.J.H., M.E.S.S., and C.P.S. M.E.S.S., C.P.S., and D.J.H. designed the experiments. M.E.S.S. and D.J.H. wrote the manuscript. M.E.S., H.M., C.C., and C.P.S. prepared the figures. M.E.S. performed all in vitro and in vivo experiments with support from P.L., C.C., J. Du, J. Ding, M.M., C.L., K.T., S.S., D.S., and Z.R. H.M. and R.A. performed single-cell RNA sequencing data analysis. W.F. and M.W. provided support for histology. S.W. performed mouse qRT PCR. C.P.S. performed mouse qRT PCR analysis. D.C.R. and V.G. performed patient tissue qRT PCR, IHC experiments, and associated analyses. T.P. assisted with animal work. M.E.S.S., C.P.S., D.C.R, S.A., and D.J.H. performed a critical review of the data and manuscript.
Peer review
Peer review information
Nature Communications thanks Mikko Pakarinen and Kristen Seiler for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw and processed single-cell RNA sequencing data generated in this study have been deposited in the NCBI Gene Expression Omnibus database under accession code GSE271065 Source data are provided with this paper. The graph matrices generated in this study are provided in the Supplementary Information/Source Data file. The human single-cell RNA sequencing data used in this study for verification was downloaded from the NCBI Gene Expression Omnibus database under accession code GSE130113. Source data are provided in this paper.
Code availability
Code for the analysis is available on GitHub at 10.5281/zenodo.13323988
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chhinder P. Sodhi, Email: csodhi1@jhmi.edu
David J. Hackam, Email: dhackam1@jhmi.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52216-6.
References
- 1.Duggan, C. P. & Jaksic, T. Pediatric intestinal failure. N. Engl. J. Med.377, 666–675 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Hess, R. A., Welch, K. B., Brown, P. I. & Teitelbaum, D. H. Survival outcomes of pediatric intestinal failure patients: analysis of factors contributing to improved survival over the past two decades. J. Surg. Res.170, 27–31 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Modi, B. P. et al. Improved survival in a multidisciplinary short bowel syndrome program. J. Pediatr. Surg.43, 20–24 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amin, S. C., Pappas, C., Iyengar, H. & Maheshwari, A. Short bowel syndrome in the NICU. Clin. Perinatol.40, 53–68 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gentilini, M. V., Rumbo, M. & Gondolesi, G. E. Organoid transplantation: new avenues to treat short bowel syndrome. Transplantation105, 2130–2131 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Gazit, V. A. et al. Stem cell and niche regulation in human short bowel syndrome. JCI Insight5, e137905 (2020). [DOI] [PMC free article] [PubMed]
- 7.Wang, L., Tang, Y., Rubin, D. C. & Levin, M. S. Chronically administered retinoic acid has trophic effects in the rat small intestine and promotes adaptation in a resection model of short bowel syndrome. Am. J. Physiol. Gastrointest. Liver Physiol.292, G1559–G1569 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Wang, J. L., Swartz-Basile, D. A., Rubin, D. C. & Levin, M. S. Retinoic acid stimulates early cellular proliferation in the adapting remnant rat small intestine after partial resection. J. Nutr.127, 1297–1303 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Rowland, K. J. et al. Up-regulation of hypoxia-inducible factor 1 alpha and hemodynamic responses following massive small bowel resection. J. Pediatr. Surg.48, 1330–1339 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schall, K. A. et al. Adult zebrafish intestine resection: a novel model of short bowel syndrome, adaptation, and intestinal stem cell regeneration. Am. J. Physiol. Gastrointest. Liver Physiol.309, G135–G145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schall, K. A. et al. Short bowel syndrome results in increased gene expression associated with proliferation, inflammation, bile acid synthesis and immune system activation: RNA sequencing a zebrafish SBS model. BMC Genomics18, 23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courtney, C. M. et al. Small bowel resection increases paracellular gut barrier permeability via alterations of tight junction complexes mediated by intestinal TLR4. J. Surg. Res.258, 73–81 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugimoto, S. et al. An organoid-based organ-repurposing approach to treat short bowel syndrome. Nature592, 99–104 (2021). [DOI] [PubMed] [Google Scholar]
- 14.McMellen, M. E., Wakeman, D., Longshore, S. W., McDuffie, L. A. & Warner, B. W. Growth factors: possible roles for clinical management of the short bowel syndrome. Semin Pediatr. Surg.19, 35–43 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warner, B. W. The pathogenesis of resection-associated intestinal adaptation. Cell Mol. Gastroenterol. Hepatol.2, 429–438 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whang, E. E. et al. Enterocyte functional adaptation following intestinal resection. J. Surg. Res.60, 370–374 (1996). [DOI] [PubMed] [Google Scholar]
- 17.Hines, O. J. et al. Adaptation of the Na+/glucose cotransporter following intestinal resection. J. Surg. Res.57, 22–27 (1994). [DOI] [PubMed] [Google Scholar]
- 18.Rubin, D. C. & Levin, M. S. Mechanisms of intestinal adaptation. Best. Pr. Res. Clin. Gastroenterol.30, 237–248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hara, T., Yoshigai, E., Ohashi, T. & Fukada, T. Zinc transporters as potential therapeutic targets: an updated review. J. Pharm. Sci.148, 221–228 (2022). [DOI] [PubMed] [Google Scholar]
- 20.Finamore, A., Massimi, M., Conti Devirgiliis, L. & Mengheri, E. Zinc deficiency induces membrane barrier damage and increases neutrophil transmigration in Caco-2 cells. J. Nutr.138, 1664–1670 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Hoque, K. M. & Binder, H. J. Zinc in the treatment of acute diarrhea: current status and assessment. Gastroenterology130, 2201–2205 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Zhong, W., McClain, C. J., Cave, M., Kang, Y. J. & Zhou, Z. The role of zinc deficiency in alcohol-induced intestinal barrier dysfunction. Am. J. Physiol. Gastrointest. Liver Physiol.298, G625–G633 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhong, W. et al. Paneth cell dysfunction mediates alcohol-related steatohepatitis through promoting bacterial translocation in mice: role of zinc deficiency. Hepatology71, 1575–1591 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Queiroz, C. A. et al. Zinc treatment ameliorates diarrhea and intestinal inflammation in undernourished rats. BMC Gastroenterol.14, 136 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyoshi, Y., Tanabe, S. & Suzuki, T. Cellular zinc is required for intestinal epithelial barrier maintenance via the regulation of claudin-3 and occludin expression. Am. J. Physiol. Gastrointest. Liver Physiol.311, G105–G116 (2016). [DOI] [PubMed] [Google Scholar]
- 26.Küry, S. et al. Identification of SLC39A4, a gene involved in acrodermatitis enteropathica. Nat. Genet.31, 239–240 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Hennigar, S. R., Olson, C. I., Kelley, A. M. & McClung, J. P. Slc39a4 in the small intestine predicts zinc absorption and utilization: a comprehensive analysis of zinc transporter expression in response to diets of varied zinc content in young mice. J. Nutr. Biochem.101, 108927 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Ziegler, T. R., Evans, M. E., Fernández-Estívariz, C. & Jones, D. P. Trophic and cytoprotective nutrition for intestinal adaptation, mucosal repair, and barrier function. Annu Rev. Nutr.23, 229–261 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Hambidge, M. Biomarkers of trace mineral intake and status. J. Nutr.133, 948S–955S (2003). [DOI] [PubMed] [Google Scholar]
- 30.Dufner-Beattie, J., Kuo, Y. M., Gitschier, J. & Andrews, G. K. The adaptive response to dietary zinc in mice involves the differential cellular localization and zinc regulation of the zinc transporters ZIP4 and ZIP5. J. Biol. Chem.279, 49082–49090 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Liuzzi, J. P. & Cousins, R. J. Mammalian zinc transporters. Annu Rev. Nutr.24, 151–172 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Wessels, I., Maywald, M. & Rink, L. Zinc as a gatekeeper of immune function. Nutrients9, 1286 (2017). [DOI] [PMC free article] [PubMed]
- 33.Shao, Y. et al. Zinc enhances intestinal epithelial barrier function through the PI3K/AKT/mTOR signaling pathway in Caco-2 cells. J. Nutr. Biochem.43, 18–26 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Knoell, D. L. et al. Zinc deficiency increases organ damage and mortality in a murine model of polymicrobial sepsis. Crit. Care Med.37, 1380–1388 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, F., Kim, B. E., Petris, M. J. & Eide, D. J. The mammalian Zip5 protein is a zinc transporter that localizes to the basolateral surface of polarized cells. J. Biol. Chem.279, 51433–51441 (2004). [DOI] [PubMed] [Google Scholar]
- 36.McCracken, K. W., Howell, J. C., Wells, J. M. & Spence, J. R. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat. Protoc.6, 1920–1928 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spence, J. R. et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature470, 105–109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, A. et al. A central role for lipocalin-2 in the adaptation to short-bowel syndrome through down-regulation of IL22 in mice. Cell Mol. Gastroenterol. Hepatol.10, 309–326 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cortez, A. R. et al. Transplantation of human intestinal organoids into the mouse mesentery: a more physiologic and anatomic engraftment site. Surgery164, 643–650 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seiler, K. M. et al. Single-cell analysis reveals regional reprogramming during adaptation to massive small bowel resection in mice. Cell Mol. Gastroenterol. Hepatol.8, 407–426 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao, X., Kim, B. E., Wang, F., Eide, D. J. & Petris, M. J. A histidine-rich cluster mediates the ubiquitination and degradation of the human zinc transporter, hZIP4, and protects against zinc cytotoxicity. J. Biol. Chem.282, 6992–7000 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Weaver, B. P., Dufner-Beattie, J., Kambe, T. & Andrews, G. K. Novel zinc-responsive post-transcriptional mechanisms reciprocally regulate expression of the mouse Slc39a4 and Slc39a5 zinc transporters (Zip4 and Zip5). Biol. Chem.388, 1301–1312 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andrews, G. K. Regulation and function of Zip4, the acrodermatitis enteropathica gene. Biochem Soc. Trans.36, 1242–1246 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trumbo, P., Yates, A. A., Schlicker, S. & Poos, M. Dietary reference intakes: vitamin A, vitamin K, arsenic, boron, chromium, copper, iodine, iron, manganese, molybdenum, nickel, silicon, vanadium, and zinc. J. Am. Diet. Assoc.101, 294–301 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Lee, H. H., Prasad, A. S., Brewer, G. J. & Owyang, C. Zinc absorption in human small intestine. Am. J. Physiol.256, G87–G91 (1989). [DOI] [PubMed] [Google Scholar]
- 46.Bakaloudi, D. R. et al. Intake and adequacy of the vegan diet. A systematic review of the evidence. Clin. Nutr.40, 3503–3521 (2021). [DOI] [PubMed] [Google Scholar]
- 47.McClain, C., Vatsalya, V. & Cave, M. Role of zinc in the development/progression of alcoholic liver disease. Curr. Treat. Options Gastroenterol.15, 285–295 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suliburska, J. et al. Diuretics, Ca-antagonists, and angiotensin-converting enzyme inhibitors affect zinc status in hypertensive patients on monotherapy: a randomized trial. Nutrients10, 1284 (2018). [DOI] [PMC free article] [PubMed]
- 49.King, J. C., Shames, D. M. & Woodhouse, L. R. Zinc homeostasis in humans. J. Nutr.130, 1360S–1366S (2000). [DOI] [PubMed] [Google Scholar]
- 50.Liuzzi, J. P., Bobo, J. A., Lichten, L. A., Samuelson, D. A. & Cousins, R. J. Responsive transporter genes within the murine intestinal-pancreatic axis form a basis of zinc homeostasis. Proc. Natl. Acad. Sci. USA101, 14355–14360 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tran, A. et al. Estrogen-related receptor alpha (ERRα) is a key regulator of intestinal homeostasis and protects against colitis. Sci. Rep.11, 15073 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hebiguchi, T. et al. Massive bowel resection upregulates the intestinal mRNA expression levels of cellular retinol-binding protein II and apolipoprotein A-IV and alters the intestinal vitamin A status in rats. Int J. Mol. Med.35, 724–730 (2015). [DOI] [PubMed] [Google Scholar]
- 53.Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun.8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell184, 3573–3587.e3529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fawkner-Corbett, D. et al. Spatiotemporal analysis of human intestinal development at single-cell resolution. Cell184, 810–826.e823 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation2, 100141 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology141, 1762–1772 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davie, K. et al. A single-cell transcriptome atlas of the aging drosophila brain. Cell174, 982–998.e920 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frank, E. H. Hmisc: Harrell miscellaneous. R Package version 5.0-1. https://CRAN.R-project.org/package=Hmisc (2023).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and processed single-cell RNA sequencing data generated in this study have been deposited in the NCBI Gene Expression Omnibus database under accession code GSE271065 Source data are provided with this paper. The graph matrices generated in this study are provided in the Supplementary Information/Source Data file. The human single-cell RNA sequencing data used in this study for verification was downloaded from the NCBI Gene Expression Omnibus database under accession code GSE130113. Source data are provided in this paper.
Code for the analysis is available on GitHub at 10.5281/zenodo.13323988