Abstract
Partial E1 envelope glycoprotein gene sequences and complete structural polyprotein sequences were used to compare divergence and construct phylogenetic trees for the genus Alphavirus. Tree topologies indicated that the mosquito-borne alphaviruses could have arisen in either the Old or the New World, with at least two transoceanic introductions to account for their current distribution. The time frame for alphavirus diversification could not be estimated because maximum-likelihood analyses indicated that the nucleotide substitution rate varies considerably across sites within the genome. While most trees showed evolutionary relationships consistent with current antigenic complexes and species, several changes to the current classification are proposed. The recently identified fish alphaviruses salmon pancreas disease virus and sleeping disease virus appear to be variants or subtypes of a new alphavirus species. Southern elephant seal virus is also a new alphavirus distantly related to all of the others analyzed. Tonate virus and Venezuelan equine encephalitis virus strain 78V3531 also appear to be distinct alphavirus species based on genetic, antigenic, and ecological criteria. Trocara virus, isolated from mosquitoes in Brazil and Peru, also represents a new species and probably a new alphavirus complex.
The family Togaviridae is comprised of two genera, Alphavirus and Rubivirus (77). The genus Alphavirus contains at least 24 species (77) that can be classified antigenically into seven complexes (4) (Table 1). As a genus, the alphaviruses are widely distributed throughout the world, inhabiting all of the continents except Antarctica. The geographic distributions of individual species are restricted because of specific ecological conditions and reservoir host and vector restrictions (22, 77).
TABLE 1.
Alphaviruses studieda
| Antigenic complex and speciesb | Strain or subtype | Origin | Source | Distribution | Passage historyc | Reference(s) | GenBank accession no. |
|---|---|---|---|---|---|---|---|
| BF, BFV | BH2193 | Australia, 1974 | C. annulirostris | Australia | sm-2, v-2 | 34 | U73745 |
| EEEV | |||||||
| EEEV1 | 82V2137 | Florida, 1982 | Mosquito | North America, Mexico, Caribbean | NRd | 79 | U01034 |
| EEEV2 | BeAn5122 | Brazil, 1956 | Monkey | Central South America | sm-8 | 2 | AF159559 |
| EEEV3 | 435731 | Panama 1986 | Horse | Central South America | v-2 | 2 | AF159560 |
| EEEV4 | 436087 | Brazil 1985 | Culex sp. | Brazil | sm-1, v-1 | 2 | AF159561 |
| MID, MIDV | SA AR749 | South Africa | A. caballus | Africa | sm-12, BHK-1 | AF398374 | |
| NDU, NDUV | SAAr221D | South Africa, 1959 | A. circumuteolus | Africa | sm-9, BHK-1 | AF398375 | |
| SF | |||||||
| BEBV | MM2354 | Malaysia | Culiseta (Lophoceraomyia) sp. | Malaysia | sm-8, BHK-1 | AF398376 | |
| CHIKV | Ross | Tanganyika, 1953 | Human | Africa, Asia | sm-175, v-1, BHK-1 | 48 | AF192905 |
| GETV | MM2021 | Malaysia, 1955 | C. gelidus | Asia, Oceania | sm-5, BHK-1 | AF398377 | |
| MAYV | TRVL4675 | Trinidad, 1954 | Human serum | South America, Trinidad | sm-13, BHK-1 | AF398378 | |
| ONNV | Gulu | Uganda, 1959 | Human | Africa | sm-1, v-3, BHK-1 | 35 | M33999 |
| RRV | T48 | Australia, 1959 | Mosquito | Oceania, Australia | v-2 | 11 | M20162 |
| RRV (SAGV) | 31407 | Japan, 1956 | Culex sp | Asia, Japan Africa | p-14 | ||
| SFV | NRd | NR | 14, 15 | X04129 | |||
| SFV | Me Tri | Vietnam, 1971 | C. tritaeniorhynchus | Vietnam | AF398380 | ||
| UNAV | CoAr43332 | Colombia, 1964 | Psorophora ferox | South America, Panama | AF398381 | ||
| VEE | |||||||
| VEEV (IAB) | Trinidad donkey | Trinidad, 1943 | Donkey | Central, North, South America | gp-1, v-6, BHK-1 | 25 | L01442 |
| VEEV (IC) | P676 | Venezuela, 1963 | A. triannulatus | South America | sm-1, v-7 | 28 | L04653 |
| VEEV (ID) | 3880 | Panama, 1961 | Human | Central, South America | sm-8, v-1 | 28 | L00930 |
| VEEV (IE) | 68U201 | Guatemala, 1968 | Hamster | Central America, Mexico | sm-1 | 42 | U34999 |
| 78V3531 (VEE-IF) | 78V3531 | Brazil, 1978 | Culex sp. | South America | dec-1, sm-3, v-1 | AF398382 | |
| EVEV (VEE-II) | Fe3-7c | Florida, 1963 | Mosquito | Florida | sm-6, v-1, BHK-1 | 58 | AF075251 |
| MUCV (VEE-IIIA) | BeAn8 | Brazil, 1954 | Monkey | South America, Trinidad | p-8 | AF398383 | |
| MUCV (Tonate virus; VEE-IIIB) | CaAn410d | French Guiana, 1973 | Bird | French Guiana | sm-3, cec-2 | AF398384 | |
| MUCV (Bijou Bridge virus; VEE-IIIB) | Bijou Bridge | Colorado, 1974 | Oeciacus vicarius | Western North America | v-6, cec-2 | AF398385 | |
| 71D1252V (VEE-IIIC) | 71D1252 | Peru, 1971 | Mosquito | Peru | sm-1 | AF398386 | |
| PIXV (VEE-IV) | BeAr35645 | Brazil, 1961 | Mosquito | South America | sm-4 | 27 | AF075256 |
| CABV (VEE-V) | CaAr508 | French Guiana, 1968 | C. portesi | French Guiana | sm-5, BHK-1 | 27 | AF398387 |
| Ag80V (VEE-VI) | Ag80-663V | Argentina, 1980 | C. (Melanoconion) delpontel | Argentina | p-4, sm-2 | 27 | AF398388 |
| WEE | |||||||
| AURAV | BeAr10315 | Brazil, 1959 | Culex (Melanoconion) sp. | South America | p-6, sm-1 | 51 | AF126284 |
| (Sindbis-like virus) | |||||||
| SINV | AR339 | Egypt, 1953 | C. univittatus | Africa, Eurasia, Australia | sm-14, v-1 | 50, 69 | V00073 |
| SINV (Babanki virus) | DAK ArY251 | Cameroon, 1969 | Mansonia africana | Africa | sm-3, BHK-1 | AF398391 | |
| SINV (Kyzylagach virus) | Leiv65A | Azerbaijan, 1969 | C. modestus | Azerbaijan, China | p-8, sm-1, BHK-1 | AF398392 | |
| SINV (Ockelbo virus) | Edsbyn 82-5 | Sweden, 1982 | Culiseta sp. | Europe | p-?, BHK-1 | 56 | M69205 |
| WHAV | M78 | New Zealand, 1962 | C. pervigilans | New Zealand | p-9, sm-1, BHK-1 | AF398394 | |
| WEEV (recombinants) | |||||||
| FMV | 73V1570 | Colorado, 1973 | Passer domesticus | Western North America | nc-1, de-1, sm-1 | AF398389 | |
| FMV (Buggy Creek virus) | 81V1822 | Oklahoma, 1981 | Oeciacus vicarius | Western North America | de-2 | AF398390 | |
| HJV | 64A-1519 | Florida, 1964 | Horse | Eastern North America | p-4, sm-1 | 8 | U52586 |
| WEEV | BFS1703 | California, 1953 | C. tarsalis | South, North America | sm-1, C6/36-1 | 19 | J03854 |
| WEEV | Ag80-646 | Argentina, 1980 | Canoconion (Melocossa) | Argentina | v-2, sm-1, BHK-1 | 6 | AF398393 |
| Unclassified | |||||||
| TROCV | BeAr422431 | Brazil, 1984 | A. serratus | South America | sm-1, BHK-1 | 65 | AF252264 |
| SDV | NR | France, 2000 | Oncorhynchus mykiss | Europe | CHSE-214-? | 69 | AJ238578 |
| SPDV | F93125 | United Kingdom | Salmo salar | Europe, North America | CHSE-214-? | 81 | AJ012631 |
| SESV | NR | Australia | Lepidophthirus macrorhini | Australia | BHK-1 | 32 | EMBL DS44746 |
Species classification according to the most recent report of the ICTV (77); antigenic complexes, subtypes, and varieties according to the SIRACA (4).
Subtypes are shown in parentheses. Virus abbreviations: MUCV, Mucambo virus; TONV, Tonate virus; PIXV, pixuna virus; CABV, Cabassou virus; FMV, Fort Morgan virus; HJV, Highlands virus; WHATV, Whataroa virus; SINV, Sindbis virus; KZLV, Kyzylagach virus; MIDV, Middelburg virus; MAYV, Mayaro virus; SFV, Semliki Forest virus; RRV, Ross River virus; CHIKV, chikungunya virus; ONNV, o'nyong nyong virus; BFV, Barmah Forest virus; SAGV, Sagiama virus; GETV, getah virus; NDUV, Ndumu virus; BEBV, Bebaru virus; TROCV, Trocara virus.
Cell types: DEC, duck embryo cells; SM, suckling mouse; V, Vero; p, passage in unknown cell type; CEC, chicken enbryo cells; gp, guinea pig; BHK, baby hamster kidney; C6/36, A. albopictus larvae; CHSE/214, Chinook salmon embryo cells; NC, newly hatched chicken.
NR, not reported.
Members of the genus Alphavirus are typically maintained in natural cycles involving transmission by an arthropod vector among susceptible vertebrate hosts (60). Virus-host interactions may be highly specific, and sometimes only a single mosquito species is utilized as the principal vector, as has been reported for many Venezuelan equine encephalitis (VEE) complex viruses (74). These specific virus-vector interactions may limit the distribution of many alphaviruses. Possible exceptions to the presumption that all alphaviruses have an arthropod host are the newly identified salmonid viruses salmon pancreas disease virus (SPDV) (81) and sleeping disease virus (SDV) (69). These viruses have been isolated only from diseased Atlantic salmon and rainbow trout, respectively, and are not known to have arthropod vectors. It has been postulated that the sea louse, Lepeophtheirus salmonis, may play a role in the transmission of SPDV, but no evidence to support this hypothesis has been generated. Parasitic lice have been implicated in the transmission of the newly discovered southern elephant seal alphavirus (SESV) from the coast of Australia. SESV has been grouped genetically with the Semliki Forest virus complex (32).
The members of the genus Alphavirus cause a wide range of diseases in humans and animals. Many Old World viruses, including the Ross River, Barmah Forest, Mayaro, o'nyong-nyong, chikungunya, and Sindbis viruses, cause an arthralgia syndrome (47, 52), while encephalitis is caused by VEEV, eastern equine encephalitis virus (EEEV), and western equine encephalitis virus (WEEV) in the New World. In addition to causing febrile illness in equines, pigs, and calves, Getah virus has been reported to potentially induce abortion or stillbirth in pregnant sows (20, 44). Highlands J virus causes dramatic decreases in egg production and mortality in domestic birds (13, 70). Seroprevalence data on many of the remaining alphaviruses indicate that they infect people and/or domestic animals but have unknown clinical manifestations or cause only a mild febrile illness (1, 29–31, 41, 63, 65). Interestingly, alphaviruses causing similar disease symptoms are maintained under diverse ecological conditions and can have a widespread distribution. For example, Mayaro virus is limited geographically to Latin America (46, 64) while o'nyong-nyong virus has never been identified outside of Africa (21, 33, 48). These two viruses cause almost identical clinical signs and symptoms. This unusual epidemiological pattern seen among the various alphaviruses presents some intriguing questions regarding evolutionary relationships of the members of the Alphavirus genus, including the origins of the genus and subsequent geographic expansion of the genus and species.
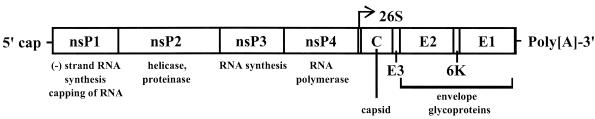
The alphaviruses are small, spherical, enveloped viruses with a genome consisting of a single strand of positive-sense RNA (22, 55, 60). The nonstructural protein genes are encoded in the 5′ two-thirds of the genome, while the structural proteins are translated from a subgenomic mRNA colinear with the 3′ one-third of the genome (Fig. 1). Replication occurs within the cytoplasm, and virions mature by budding through the plasma membrane, where virus-encoded surface glycoproteins E2 and E1 are assimilated. These two glycoproteins are the targets of numerous serologic reactions and tests (e.g., neutralization and hemagglutination inhibition); the alphaviruses show various degrees of antigenic cross-reactivity in these reactions, forming the basis for the seven antigenic complexes, 24 species, and many subtypes and varieties of alphaviruses defined previously (4, 23, 62). The E2 protein is the site of most neutralizing epitopes, while the E1 protein contains more conserved, cross-reactive epitopes.
FIG. 1.
Organization of the Alphavirus genome. Gene products and associated functions are indicated.
Previous studies of the evolutionary relationships among alphaviruses have relied on phylogenetic analyses of either partial or complete sequences from one or more of the seven protein genes (35, 73, 80). Overall, these studies have produced relationships in agreement with the antigenically based approaches used traditionally for alphavirus classification (4, 7, 77). For example, viruses in the VEE (49, 76), EEE (2, 75), and WEE antigenic complexes (80) have each been shown to be monophyletic (WEE complex for the envelope glycoproteins only). Additionally, phylogenetic studies have shown that most of the New World viruses in the WEE antigenic complex (WEEV, Highlands J virus, Fort Morgan virus, and Buggy Creek virus [a variant of Fort Morgan virus]) are descendants of an ancestral alphavirus that resulted from a recombination event; recombination combined the E2 and E1 envelope protein genes from a Sindbis-like virus and the remaining genes from an EEEV-like ancestor (19, 80). The Old World serogroups have been studied in less detail; the chikungunya, o'nyong-nyong, Semliki Forest, and Ross River viruses, belonging to the Semliki Forest virus complex, are monophyletic in some analyses and paraphyletic in others, with Middelburg virus falling into this group in some trees (73, 79).
To provide a more complete understanding of the evolutionary history and mechanisms of emergence of alphaviruses, we conducted a comprehensive examination of the evolution of the genus by sequencing most of the E1 envelope glycoprotein gene for representatives of all alphavirus species (77), as well as major antigenic subtypes and varieties (4). Using phylogenetic methods, these sequences were used to reexamine the evolutionary history and systematics of the genus.
MATERIALS AND METHODS
Virus preparation.
The virus strains used in this study are listed in Table 1. Viruses were diluted and passaged on BHK-21 or Vero 76 cells at a low multiplicity of infection. After approximately 75% of the cells exhibited cytopathic effects, the virus present in the supernatant was concentrated by precipitation with 7% polyethylene glycol and 2.3% NaCl (24). The virus pellet was resuspended in 150 μl of TEN (Tris-EDTA-NaCl) buffer, and 2 ml of Trizol LS (Gibco BRL, Bethesda, Md.) was added in preparation for RNA extraction in accordance with the manufacturer's protocol.
RNA extraction and reverse transcription PCR.
RNA was extracted from one-half of each virus-Trizol suspension in accordance with manufacturer's protocols as described previously (8). cDNAs were synthesized from the RNA by using a poly(T) oligonucleotide primer (T25V-Mlu; 5′-TTACGAATTCACGCGT25V-3′ or T19V). PCR amplification was performed on the first-strand cDNA by using the poly(T) primer and a forward primer designated α10247A (5′-TACCCNTTYATGTGGGG-3′). Thisforward primer anneals to a highly conserved sequence that encodes the putative fusion domain of the E1 protein, and this conservation allowed us to amplify most of the E1 glycoprotein gene from a wide variety of highly divergent alphaviruses. Amplification of the carboxy portion of the E1 gene and the 3′ noncoding region utilized the following parameters: 30 cycles of denaturation at 95°C for 30 s, primer annealing at 49°C for 30 s, and extension at 72°C for 3 min. A 10-min final extension was used to ensure complete product synthesis. For the virus designated Ag80-663, for which the above primer pairs were unsuccessful, the T25V-Mlu primer was used in conjunction with primer E/V7514(+) (5′-ACYCTCTACGGCTRACCTRA-3′) to amplify the entire 26S subgenomic message region. Sequencing was performed on this strain by gene walking using sequentially designed primers (see Table 2).
TABLE 2.
Primers used in reverse transcription PCR and sequencing reactions
| Primera | Sequenced |
|---|---|
| E/V7514(+)b | 5′-ACYCTCTACGGCTRACCTRA-3′ |
| α10247A(+) | 5′-TACCCNTTYATGTGGG-3′ |
| α10552(+) | 5′-CAYGTNCCWTAYACVCAG-3′ |
| α10613(+) | 5′-CCCTAAATACGAAGGCTCC-3′ |
| α10720(+) | 5′-TAACAGCGGGAAATCGTTGC-3′ |
| K10550(+)c | 5′-CACGTTCCGTACACGCAAG-3′ |
| SIN10790(+) | 5′-GTGGAAAAACAACTCAGG-3′ |
| T25V-Mlu(−) | 5′-TTACGAATTCACGCGT25V-3′ |
| T19V(−) | 5′-T19V-3′ |
Each number indicates the position of the 5′ nucleotide of the primer based on the full-length VEEV genome (26).
Primer used to generate a PCR product for the Ag80-663 virus.
Primer used specifically to sequence the Kyzylagach virus.
Y = C or T; R = A or G; N = A, C, G, or T; W = A or T; V = A, C, or G.
Sequencing and genetic analysis.
PCR products ranging in size from 1.1 to 1.8 kb were isolated from 1% agarose gels. The cleaned DNA fragments were either sequenced directly or cloned into pBluescript II SK (Stratagene, La Jolla, Calif.) that had been linearized with SmaI. Restriction enzyme SmaI was included in the ligation reaction to reduce the religation of the vector upon itself. White bacterial colonies were screened for plasmids containing inserts of the correct size. Two selected clones were sequenced by using plasmid-specific T7 promoter and M13 reverse primers. Additional internal sequence was obtained by using virus-specific primers as indicated in Table 2. Sequencing was performed by using an Applied Biosystems (Foster City, Calif.) Prism 377 sequencer and BigDye automated DNA sequencing kit. Deduced amino acid sequences were aligned with those of other alphaviruses sequenced previously (Table 1) by using the PILEUP program in the University of Wisconsin Genetics Computer Group package (10) with manual refinements to preserve codon homology. Pairwise comparisons were performed with PAUP (61) and the GAP program within the Genetics Computer Group package.
Phylogenetic analyses were performed on both the nucleotide and translated amino acid sequences for the E1 gene or complete 26S sequence by using the PAUP program (61). The heuristic algorithm was employed for the maximum-parsimony analysis. The neighbor-joining distance matrix algorithm was used with the Kimura 2 parameter and F84 corrections. Bootstrap resampling to determine confidence values on the groupings within trees was performed with 1,000 replicates (12). For the generation of a maximum-likelihood model for alphavirus sequence evolution, closely related sequences of many different strains of EEEV (2), VEEV (49), WEEV (80), Highlands J virus (8), and chikungunya virus (48) were analyzed to avoid the effects of superimposed nucleotide substitutions. Tree topologies determined previously were used to estimate the transition-transversion ratios and gamma values for the unevenness in the distribution of substitutions across nucleotide sites using the PAUP program (61). These estimates were then used to search for maximum-likelihood trees for the Alphavirus genus.
Nucleotide sequence accession numbers. The GenBank accession numbers for the sequences reported in this paper are AF398374 to AF398393.
RESULTS
E1 3′ NCR amplification and sequencing.
The 3′ noncoding region (NCR) and E1 envelope glycoprotein gene were selected for genetic analyses to take advantage of conserved sequences described previously for primer annealing and PCR amplification (8). The E1 region has also been shown to be phylogenetically informative. Alphavirus cDNAs were synthesized by using an oligo(dT) primer containing a 3′ clamp (T25V-Mlu). By using this primer and a primer from a conserved region of the E1 gene (α10247A), nearly all alphavirus genomes were amplified. The VEE complex virus Ag80-663 (subtype VI) could not be amplified with the α10247A primer, but the entire 26S region of this strain was amplified by using T25V-Mlu and E/V7514(+). This was the only alphavirus that required alternative amplification conditions (see Materials and Methods). An analysis of the genome at the α10247A primer binding site revealed that the primer site was a highly conserved region across the entire Alphavirus genus and was an exact match in strain Ag80-663, making it unclear why this virus was unable to be amplified with this primer.
All of the amplicons generated ranged in size from 1.1 to 1.8 kb (Table 3), depending upon the length of the 3′ NCR. The shortest 3′ NCRs belonged to the 78V3531 and Trocara viruses, and Bebaru virus contained the longest (Table 3). Some of the amplicons (Getah, Una, Babanki, and Trocara viruses) did not contain the conserved alphavirus termini (5′-ATTTTGTTTTTAATATTTC-3′) (45), indicating that perhaps the entire 3′ NCR was not amplified. Trocara virus was unique in that the α10247A primer used in the PCR amplification was present on both ends of the amplicon, suggesting a 3′ NCR of only 34 nucleotides. The use of a longer poly(T) oligonucleotide primer increased the likelihood of obtaining the entire 3′ NCR (T25 compared to T19) but was still unsuccessful in some instances, including that of Trocara virus. However, based on the finding of George and Raju (16) that the classical 19-nt conserved terminal element is not essential for replication or virus maintenance, it is possible that some of these viral sequences that appear to be incomplete (because they lack the entire conserved 3′ terminus) are actually complete. While there was considerable variability in the 3′ NCR sequences, the E1 gene, with the exception of the five or six 3′ terminal codons, was more conserved among all of the alphaviruses. Most viruses were sequenced directly by using the α10247A and α10552(+) primers. However, several required additional primers and the Kyzylagach strain, a subtype of Sindbis virus, was unable to be sequenced with the universal internal primer and required virus-specific primers (Table 2). Occasionally, there were sequence differences between isolates sequenced in our laboratory and those in the GenBank database. The Mucambo (VEE subtype IIIA), Tonate (VEE subtype IIIB), 71D1252 (VEE subtype IIIC), and Ag80-663 (VEE subtype VI) viruses had differences, typically in the third codon and/or synonymous positions, that were most likely due to differences in passage history. The sequence analyses we performed utilized the isolates with the lowest passage histories available, which were generally lower than those of the isolates used to generate sequences already in the GenBank database.
TABLE 3.
Alphavirus E1 3′ NCR amplicon analysis
| Virus | Approx PCR amplicon size (kb) | 3′ NCR size (nt)a |
|---|---|---|
| Middelburg | 1.5 | >329 |
| Ndumu | 1.6 | 537 |
| Bebaru | 1.8 | 595 |
| Getah | 1.4 | >378 |
| Mayaro | 1.3 | 259 |
| Sagiyama | 1.5 | 401 |
| Me Tri | 1.5 | 504 |
| Una | 1.5 | >433 |
| 78V3531 | 1.2 | 82 |
| Mucambo (VEEV-IIIA) | 1.3 | 184 |
| Tonate | 1.3 | 184 |
| Bijou Bridge | 1.4 | 149 |
| 71D1252 | 1.2 | 127 |
| Cabassou | 1.3 | 180 |
| Ag80-663 | 3.9 | 195 |
| Buggy Creek | 1.3 | 200 |
| Babanki | 1.4 | >190 |
| Kyzylagach | 1.4 | 314 |
| Ag80-646 | 1.5 | NAb |
| Trocara | 1.1 | >33 |
Minimum sizes for sequenced isolates that did not contain the 3′ conserved 19 nt are indicated by a greater-than sign.
NA, not available.
Direct comparisons of E1 gene sequences.
To determine the extent of relatedness of all established members of the genus Alphavirus, pairwise comparisons were performed by using the nucleotide and deduced amino acid sequences in the E1 gene coding region (Table 4). The C-terminal 5 to 10 amino acids and their codons were omitted from the analyses because they were highly divergent and could not be aligned reliably (many alignment scores for this fragment did not differ statistically significantly from jumbled alignments). In general, the percentage of sequence divergence correlated inversely with serologic cross-reactivity (4). Viruses within a given antigenic serocomplex were usually genetically more closely related than viruses in different complexes. Those within a given antigenic complex typically had a nucleotide sequence divergence of less than 43% and an amino acid sequence divergence of less than 44%, while interserocomplex comparisons usually exceeded 38 and 40%, respectively. The Middelburg virus complex was the least divergent of the antigenic complexes, with only 33% nucleotide and 31% amino acid sequence divergence compared with some Semliki Forest virus complex viruses, such as Getah virus. In contrast, Trocara virus exhibited considerable sequence divergence versus all other alphaviruses, with at least 43% nucleotide and 47% amino acid sequence divergence. These data support the previous conclusion that Trocara virus probably represents a new antigenic complex in the genus Alphavirus (65).
TABLE 4.
E1 envelope glycoprotein gene sequence divergence among alphavirusesa
| Virusb | Amt of sequence divergence from:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FV | EEEV1 | EEEV2 | EEEV3 | EEEV4 | MIDV | NDUV | BEBV | CHIKV | GETV | |
| BFV | 0.46 | 0.47 | 0.49 | 0.48 | 0.42 | 0.48 | 0.40 | 0.49 | 0.44 | |
| EEEV1 | 0.42 | 0.11 | 0.09 | 0.09 | 0.45 | 0.50 | 0.49 | 0.50 | 0.48 | |
| EEEV2 | 0.43 | 0.25 | 0.04 | 0.08 | 0.47 | 0.50 | 0.51 | 0.51 | 0.48 | |
| EEEV3 | 0.43 | 0.23 | 0.18 | 0.06 | 0.47 | 0.50 | 0.51 | 0.51 | 0.48 | |
| EEEV4 | 0.43 | 0.23 | 0.20 | 0.20 | 0.47 | 0.50 | 0.50 | 0.51 | 0.47 | |
| MIDV | 0.41 | 0.42 | 0.42 | 0.43 | 0.43 | 0.46 | 0.31 | 0.42 | 0.31 | |
| NDUV | 0.43 | 0.44 | 0.43 | 0.45 | 0.45 | 0.40 | 0.45 | 0.49 | 0.43 | |
| BEBV | 0.40 | 0.44 | 0.44 | 0.45 | 0.44 | 0.35 | 0.42 | 0.41 | 0.29 | |
| CHIKV | 0.44 | 0.45 | 0.45 | 0.45 | 0.45 | 0.42 | 0.44 | 0.41 | 0.41 | |
| GETV | 0.40 | 0.44 | 0.42 | 0.43 | 0.43 | 0.33 | 0.39 | 0.33 | 0.40 | |
| MAYV | 0.43 | 0.43 | 0.44 | 0.43 | 0.44 | 0.36 | 0.41 | 0.36 | 0.40 | 0.35 |
| ONNV | 0.43 | 0.42 | 0.45 | 0.46 | 0.45 | 0.41 | 0.44 | 0.40 | 0.24 | 0.40 |
| RRV | 0.41 | 0.43 | 0.42 | 0.44 | 0.43 | 0.35 | 0.40 | 0.31 | 0.39 | 0.24 |
| RRV (SAGV) | 0.40 | 0.44 | 0.43 | 0.43 | 0.43 | 0.34 | 0.38 | 0.33 | 0.39 | 0.06 |
| SFV | 0.41 | 0.43 | 0.44 | 0.45 | 0.43 | 0.35 | 0.41 | 0.32 | 0.39 | 0.31 |
| SFV (Me Tri virus) | 0.42 | 0.44 | 0.44 | 0.45 | 0.44 | 0.36 | 0.41 | 0.32 | 0.39 | 0.31 |
| UNAV | 0.42 | 0.42 | 0.44 | 0.43 | 0.43 | 0.36 | 0.42 | 0.32 | 0.40 | 0.32 |
| VEEV (IAB) | 0.46 | 0.38 | 0.38 | 0.38 | 0.38 | 0.46 | 0.46 | 0.46 | 0.45 | 0.43 |
| VEEV (IC) | 0.46 | 0.38 | 0.37 | 0.38 | 0.38 | 0.45 | 0.45 | 0.46 | 0.44 | 0.43 |
| VEEV (ID) | 0.46 | 0.38 | 0.38 | 0.38 | 0.38 | 0.47 | 0.46 | 0.45 | 0.44 | 0.43 |
| VEEV (IE) | 0.46 | 0.37 | 0.39 | 0.39 | 0.39 | 0.47 | 0.46 | 0.46 | 0.46 | 0.45 |
| 78V3531 (VEE-IF) | 0.46 | 0.40 | 0.40 | 0.41 | 0.40 | 0.47 | 0.44 | 0.45 | 0.44 | 0.45 |
| EVEV (VEE-II) | 0.46 | 0.38 | 0.39 | 0.37 | 0.38 | 0.46 | 0.46 | 0.46 | 0.46 | 0.44 |
| MUCV (VEE-IIIA) | 0.43 | 0.38 | 0.37 | 0.38 | 0.37 | 0.46 | 0.46 | 0.47 | 0.46 | 0.44 |
| MUCV (Tonate virus; VEE-IIIB) | 0.43 | 0.37 | 0.39 | 0.40 | 0.39 | 0.47 | 0.47 | 0.45 | 0.46 | 0.43 |
| MUCV (Bijou Bridge virus; VEE-IIIB) | 0.43 | 0.37 | 0.39 | 0.40 | 0.39 | 0.47 | 0.47 | 0.46 | 0.46 | 0.44 |
| 71D1252 (VEE-IIIC) | 0.42 | 0.37 | 0.36 | 0.37 | 0.37 | 0.45 | 0.47 | 0.45 | 0.45 | 0.44 |
| PIXV (VEE-IV) | 0.46 | 0.39 | 0.39 | 0.40 | 0.39 | 0.46 | 0.47 | 0.46 | 0.47 | 0.45 |
| CABV (VEE-V) | 0.45 | 0.37 | 0.37 | 0.38 | 0.36 | 0.46 | 0.47 | 0.46 | 0.46 | 0.42 |
| Ag80V (VEE-VI) | 0.44 | 0.37 | 0.38 | 0.39 | 0.38 | 0.47 | 0.46 | 0.46 | 0.46 | 0.44 |
| AURAV | 0.47 | 0.46 | 0.46 | 0.47 | 0.46 | 0.46 | 0.50 | 0.47 | 0.50 | 0.48 |
| FMV | 0.46 | 0.45 | 0.47 | 0.47 | 0.45 | 0.49 | 0.49 | 0.49 | 0.51 | 0.48 |
| FMV (BCV) | 0.46 | 0.45 | 0.47 | 0.47 | 0.45 | 0.49 | 0.48 | 0.48 | 0.50 | 0.47 |
| HJV | 0.47 | 0.44 | 0.46 | 0.44 | 0.45 | 0.47 | 0.50 | 0.47 | 0.49 | 0.47 |
| SINV | 0.45 | 0.43 | 0.43 | 0.45 | 0.44 | 0.48 | 0.48 | 0.48 | 0.49 | 0.46 |
| SINV (BABV) | 0.45 | 0.43 | 0.44 | 0.43 | 0.44 | 0.46 | 0.49 | 0.49 | 0.48 | 0.45 |
| SINV (KZLV) | 0.46 | 0.44 | 0.44 | 0.44 | 0.43 | 0.48 | 0.50 | 0.50 | 0.49 | 0.47 |
| SINV (OCKV) | 0.45 | 0.43 | 0.44 | 0.44 | 0.44 | 0.47 | 0.49 | 0.49 | 0.49 | 0.45 |
| WEEV | 0.49 | 0.46 | 0.48 | 0.47 | 0.45 | 0.49 | 0.50 | 0.49 | 0.51 | 0.48 |
| WEEV (Ag80–646) | 0.49 | 0.46 | 0.48 | 0.46 | 0.46 | 0.47 | 0.49 | 0.51 | 0.51 | 0.48 |
| WHATV | 0.45 | 0.43 | 0.43 | 0.43 | 0.43 | 0.45 | 0.47 | 0.46 | 0.49 | 0.47 |
| TROCV | 0.49 | 0.44 | 0.45 | 0.44 | 0.45 | 0.48 | 0.48 | 0.47 | 0.46 | 0.45 |
| SDV | 0.51 | 0.50 | 0.50 | 0.53 | 0.52 | 0.51 | 0.51 | 0.50 | 0.50 | 0.51 |
| SPDV | 0.51 | 0.50 | 0.49 | 0.52 | 0.51 | 0.51 | 0.51 | 0.50 | 0.50 | 0.50 |
| Amt of sequence divergence from:
| |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AYV | ONNV | RRV | RRV (SAGV) | SFV | SFV (Me Trivirus) | UNAV | VEEV (IAB) | VEEV (IC) | VEEV (ID) | VEEV (IE) | 78V3531 (VEE-IF) |
| 0.45 | 0.49 | 0.42 | 0.44 | 0.43 | 0.44 | 0.40 | 0.51 | 0.51 | 0.51 | 0.50 | 0.52 |
| 0.48 | 0.49 | 0.47 | 0.47 | 0.46 | 0.47 | 0.48 | 0.37 | 0.37 | 0.37 | 0.36 | 0.40 |
| 0.48 | 0.49 | 0.49 | 0.48 | 0.47 | 0.48 | 0.49 | 0.39 | 0.39 | 0.39 | 0.39 | 0.40 |
| 0.48 | 0.51 | 0.49 | 0.48 | 0.47 | 0.48 | 0.48 | 0.38 | 0.38 | 0.38 | 0.38 | 0.40 |
| 0.48 | 0.49 | 0.47 | 0.47 | 0.47 | 0.48 | 0.49 | 0.37 | 0.37 | 0.37 | 0.37 | 0.39 |
| 0.34 | 0.42 | 0.30 | 0.31 | 0.31 | 0.30 | 0.32 | 0.49 | 0.49 | 0.49 | 0.49 | 0.49 |
| 0.44 | 0.48 | 0.41 | 0.42 | 0.45 | 0.45 | 0.44 | 0.51 | 0.51 | 0.52 | 0.51 | 0.51 |
| 0.32 | 0.40 | 0.27 | 0.28 | 0.25 | 0.25 | 0.28 | 0.49 | 0.49 | 0.50 | 0.50 | 0.50 |
| 0.40 | 0.13 | 0.40 | 0.41 | 0.40 | 0.40 | 0.41 | 0.48 | 0.48 | 0.48 | 0.48 | 0.48 |
| 0.33 | 0.43 | 0.14 | 0.04 | 0.27 | 0.26 | 0.25 | 0.46 | 0.46 | 0.46 | 0.48 | 0.48 |
| 0.40 | 0.30 | 0.32 | 0.30 | 0.31 | 0.27 | 0.47 | 0.47 | 0.47 | 0.47 | 0.47 | |
| 0.38 | 0.40 | 0.42 | 0.40 | 0.40 | 0.41 | 0.47 | 0.47 | 0.47 | 0.46 | 0.46 | |
| 0.33 | 0.38 | 0.13 | 0.22 | 0.22 | 0.23 | 0.46 | 0.46 | 0.46 | 0.47 | 0.48 | |
| 0.34 | 0.40 | 0.25 | 0.25 | 0.25 | 0.26 | 0.46 | 0.46 | 0.46 | 0.48 | 0.49 | |
| 0.34 | 0.39 | 0.30 | 0.31 | 0.02 | 0.26 | 0.48 | 0.48 | 0.48 | 0.49 | 0.49 | |
| 0.34 | 0.39 | 0.30 | 0.30 | 0.01 | 0.26 | 0.49 | 0.49 | 0.49 | 0.50 | 0.50 | |
| 0.33 | 0.40 | 0.32 | 0.32 | 0.31 | 0.32 | 0.47 | 0.47 | 0.47 | 0.48 | 0.48 | |
| 0.44 | 0.45 | 0.45 | 0.44 | 0.47 | 0.48 | 0.46 | 0.00 | 0.06 | 0.07 | 0.17 | |
| 0.45 | 0.45 | 0.44 | 0.43 | 0.47 | 0.47 | 0.46 | 0.03 | 0.06 | 0.07 | 0.47 | |
| 0.45 | 0.44 | 0.44 | 0.43 | 0.47 | 0.47 | 0.45 | 0.05 | 0.05 | 0.07 | 0.18 | |
| 0.44 | 0.46 | 0.46 | 0.45 | 0.45 | 0.46 | 0.46 | 0.21 | 0.20 | 0.20 | 0.18 | |
| 0.46 | 0.45 | 0.46 | 0.45 | 0.46 | 0.46 | 0.46 | 0.27 | 0.27 | 0.27 | 0.27 | |
| 0.45 | 0.45 | 0.44 | 0.44 | 0.47 | 0.47 | 0.45 | 0.10 | 0.10 | 0.10 | 0.20 | 0.27 |
| 0.46 | 0.43 | 0.45 | 0.45 | 0.45 | 0.45 | 0.46 | 0.27 | 0.27 | 0.26 | 0.26 | 0.27 |
| 0.46 | 0.43 | 0.45 | 0.43 | 0.46 | 0.46 | 0.46 | 0.24 | 0.25 | 0.24 | 0.25 | 0.28 |
| 0.46 | 0.43 | 0.46 | 0.43 | 0.46 | 0.46 | 0.47 | 0.24 | 0.25 | 0.24 | 0.25 | 0.28 |
| 0.45 | 0.45 | 0.44 | 0.44 | 0.47 | 0.47 | 0.45 | 0.25 | 0.25 | 0.26 | 0.25 | 0.27 |
| 0.47 | 0.46 | 0.45 | 0.45 | 0.47 | 0.47 | 0.44 | 0.26 | 0.25 | 0.26 | 0.27 | 0.29 |
| 0.46 | 0.46 | 0.44 | 0.43 | 0.46 | 0.46 | 0.46 | 0.25 | 0.25 | 0.25 | 0.26 | 0.29 |
| 0.46 | 0.43 | 0.44 | 0.44 | 0.46 | 0.47 | 0.44 | 0.26 | 0.26 | 0.26 | 0.27 | 0.22 |
| 0.48 | 0.50 | 0.48 | 0.47 | 0.49 | 0.49 | 0.47 | 0.45 | 0.45 | 0.47 | 0.45 | 0.45 |
| 0.48 | 0.51 | 0.47 | 0.48 | 0.49 | 0.49 | 0.49 | 0.47 | 0.47 | 0.47 | 0.46 | 0.45 |
| 0.48 | 0.50 | 0.47 | 0.48 | 0.49 | 0.49 | 0.49 | 0.47 | 0.46 | 0.46 | 0.45 | 0.45 |
| 0.46 | 0.49 | 0.47 | 0.47 | 0.46 | 0.46 | 0.46 | 0.46 | 0.47 | 0.46 | 0.46 | 0.47 |
| 0.47 | 0.48 | 0.46 | 0.45 | 0.47 | 0.47 | 0.47 | 0.44 | 0.44 | 0.44 | 0.43 | 0.45 |
| 0.48 | 0.48 | 0.46 | 0.44 | 0.47 | 0.47 | 0.47 | 0.43 | 0.44 | 0.43 | 0.44 | 0.45 |
| 0.48 | 0.47 | 0.46 | 0.46 | 0.47 | 0.47 | 0.46 | 0.44 | 0.44 | 0.44 | 0.44 | 0.45 |
| 0.47 | 0.48 | 0.45 | 0.44 | 0.46 | 0.47 | 0.47 | 0.44 | 0.44 | 0.43 | 0.44 | 0.46 |
| 0.47 | 0.48 | 0.48 | 0.48 | 0.46 | 0.46 | 0.47 | 0.46 | 0.47 | 0.46 | 0.46 | 0.47 |
| 0.48 | 0.49 | 0.47 | 0.48 | 0.47 | 0.47 | 0.49 | 0.48 | 0.48 | 0.47 | 0.47 | 0.48 |
| 0.46 | 0.49 | 0.47 | 0.47 | 0.46 | 0.46 | 0.46 | 0.43 | 0.42 | 0.42 | 0.45 | 0.44 |
| 0.46 | 0.46 | 0.47 | 0.45 | 0.48 | 0.48 | 0.46 | 0.44 | 0.44 | 0.44 | 0.45 | 0.43 |
| 0.52 | 0.53 | 0.50 | 0.51 | 0.50 | 0.49 | 0.49 | 0.51 | 0.50 | 0.50 | 0.53 | 0.50 |
| 0.52 | 0.52 | 0.49 | 0.50 | 0.50 | 0.50 | 0.49 | 0.50 | 0.50 | 0.50 | 0.52 | 0.49 |
| Amt of sequence divergence from:
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| EVEV (VEEV-II) | MUCV (VEEV-IIIA) | MUCV (Tonate virus; VEEV-IIIB) | MUCV (Bijou Bridge virus; VEEV-IIIB) | 71D1252 (VEEV-IIIC) | PIXV (VEEV-IV) | CABV (VEEV-V) | Ag80 (VEEV-VI) | AURAV | FMV | FMV (BCV) |
| 0.51 | 0.50 | 0.49 | 0.49 | 0.50 | 0.50 | 0.50 | 0.50 | 0.54 | 0.54 | 0.54 |
| 0.37 | 0.37 | 0.36 | 0.37 | 0.36 | 0.39 | 0.38 | 0.39 | 0.50 | 0.49 | 0.50 |
| 0.39 | 0.38 | 0.38 | 0.38 | 0.37 | 0.41 | 0.40 | 0.40 | 0.53 | 0.52 | 0.52 |
| 0.37 | 0.37 | 0.37 | 0.38 | 0.36 | 0.40 | 0.39 | 0.39 | 0.53 | 0.52 | 0.51 |
| 0.37 | 0.37 | 0.37 | 0.37 | 0.36 | 0.40 | 0.38 | 0.39 | 0.51 | 0.51 | 0.51 |
| 0.48 | 0.48 | 0.47 | 0.48 | 0.49 | 0.49 | 0.49 | 0.48 | 0.52 | 0.54 | 0.54 |
| 0.51 | 0.53 | 0.52 | 0.53 | 0.53 | 0.51 | 0.52 | 0.51 | 0.58 | 0.55 | 0.55 |
| 0.49 | 0.51 | 0.49 | 0.50 | 0.49 | 0.49 | 0.49 | 0.49 | 0.53 | 0.54 | 0.53 |
| 0.48 | 0.51 | 0.50 | 0.51 | 0.49 | 0.49 | 0.48 | 0.48 | 0.54 | 0.57 | 0.56 |
| 0.46 | 0.48 | 0.47 | 0.48 | 0.49 | 0.48 | 0.45 | 0.47 | 0.55 | 0.53 | 0.53 |
| 0.47 | 0.49 | 0.49 | 0.50 | 0.49 | 0.47 | 0.49 | 0.47 | 0.52 | 0.53 | 0.53 |
| 0.47 | 0.49 | 0.47 | 0.48 | 0.47 | 0.48 | 0.48 | 0.46 | 0.54 | 0.57 | 0.56 |
| 0.46 | 0.47 | 0.47 | 0.48 | 0.47 | 0.48 | 0.46 | 0.47 | 0.53 | 0.51 | 0.51 |
| 0.46 | 0.49 | 0.47 | 0.48 | 0.47 | 0.48 | 0.45 | 0.47 | 0.55 | 0.53 | 0.53 |
| 0.48 | 0.49 | 0.49 | 0.50 | 0.49 | 0.50 | 0.49 | 0.49 | 0.54 | 0.55 | 0.55 |
| 0.49 | 0.49 | 0.49 | 0.50 | 0.49 | 0.50 | 0.50 | 0.50 | 0.54 | 0.55 | 0.54 |
| 0.47 | 0.48 | 0.48 | 0.49 | 0.50 | 0.47 | 0.46 | 0.46 | 0.50 | 0.53 | 0.53 |
| 0.03 | 0.14 | 0.14 | 0.14 | 0.16 | 0.18 | 0.14 | 0.17 | 0.50 | 0.49 | 0.49 |
| 0.03 | 0.14 | 0.14 | 0.14 | 0.16 | 0.18 | 0.14 | 0.17 | 0.50 | 0.49 | 0.49 |
| 0.03 | 0.14 | 0.14 | 0.15 | 0.16 | 0.19 | 0.14 | 0.17 | 0.50 | 0.49 | 0.50 |
| 0.07 | 0.14 | 0.14 | 0.15 | 0.15 | 0.19 | 0.16 | 0.16 | 0.50 | 0.49 | 0.49 |
| 0.18 | 0.19 | 0.18 | 0.18 | 0.19 | 0.19 | 0.20 | 0.11 | 0.50 | 0.51 | 0.51 |
| 0.14 | 0.14 | 0.15 | 0.17 | 0.19 | 0.15 | 0.17 | 0.49 | 0.49 | 0.49 | |
| 0.27 | 0.07 | 0.08 | 0.10 | 0.20 | 0.17 | 0.19 | 0.48 | 0.48 | 0.48 | |
| 0.25 | 0.16 | 0.02 | 0.07 | 0.19 | 0.15 | 0.17 | 0.49 | 0.48 | 0.48 | |
| 0.26 | 0.16 | 0.01 | 0.08 | 0.20 | 0.16 | 0.17 | 0.49 | 0.48 | 0.48 | |
| 0.26 | 0.21 | 0.21 | 0.21 | 0.19 | 0.18 | 0.18 | 0.48 | 0.48 | 0.49 | |
| 0.26 | 0.27 | 0.27 | 0.27 | 0.27 | 0.18 | 0.19 | 0.48 | 0.49 | 0.49 | |
| 0.26 | 0.25 | 0.25 | 0.25 | 0.28 | 0.29 | 0.19 | 0.50 | 0.51 | 0.51 | |
| 0.27 | 0.26 | 0.27 | 0.26 | 0.26 | 0.28 | 0.27 | 0.51 | 0.49 | 0.49 | |
| 0.46 | 0.45 | 0.44 | 0.45 | 0.45 | 0.45 | 0.45 | 0.45 | 0.40 | 0.39 | |
| 0.47 | 0.45 | 0.46 | 0.46 | 0.45 | 0.47 | 0.46 | 0.47 | 0.38 | 0.03 | |
| 0.46 | 0.44 | 0.45 | 0.45 | 0.45 | 0.47 | 0.45 | 0.46 | 0.39 | 0.05 | |
| 0.47 | 0.46 | 0.45 | 0.45 | 0.46 | 0.48 | 0.45 | 0.46 | 0.41 | 0.32 | 0.31 |
| 0.44 | 0.42 | 0.43 | 0.43 | 0.42 | 0.45 | 0.42 | 0.44 | 0.38 | 0.32 | 0.33 |
| 0.44 | 0.43 | 0.44 | 0.44 | 0.42 | 0.45 | 0.42 | 0.44 | 0.37 | 0.32 | 0.32 |
| 0.44 | 0.42 | 0.43 | 0.43 | 0.43 | 0.44 | 0.43 | 0.43 | 0.38 | 0.32 | 0.32 |
| 0.44 | 0.43 | 0.44 | 0.44 | 0.43 | 0.45 | 0.42 | 0.44 | 0.38 | 0.32 | 0.32 |
| 0.46 | 0.46 | 0.45 | 0.46 | 0.46 | 0.46 | 0.46 | 0.46 | 0.41 | 0.30 | 0.31 |
| 0.48 | 0.46 | 0.46 | 0.46 | 0.47 | 0.46 | 0.47 | 0.47 | 0.42 | 0.32 | 0.33 |
| 0.44 | 0.44 | 0.44 | 0.44 | 0.43 | 0.44 | 0.43 | 0.44 | 0.38 | 0.33 | 0.33 |
| 0.45 | 0.44 | 0.45 | 0.45 | 0.43 | 0.45 | 0.43 | 0.43 | 0.46 | 0.48 | 0.47 |
| 0.52 | 0.49 | 0.49 | 0.49 | 0.51 | 0.51 | 0.50 | 0.50 | 0.51 | 0.54 | 0.54 |
| 0.52 | 0.50 | 0.49 | 0.49 | 0.51 | 0.50 | 0.49 | 0.50 | 0.50 | 0.53 | 0.54 |
| Amt of sequence divergence from:
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| JV | SINV | SINV (BABV) | SINV (KZLY) | SINV (OCKY) | WEEV | WEEV (Ag80-646) | WHATV | TROCV | SDV | SPDV |
| 0.56 | 0.53 | 0.54 | 0.54 | 0.54 | 0.56 | 0.57 | 0.54 | 0.55 | 0.63 | 0.62 |
| 0.51 | 0.48 | 0.49 | 0.48 | 0.48 | 0.50 | 0.51 | 0.47 | 0.50 | 0.61 | 0.60 |
| 0.50 | 0.51 | 0.51 | 0.50 | 0.51 | 0.52 | 0.52 | 0.48 | 0.51 | 0.61 | 0.59 |
| 0.49 | 0.50 | 0.51 | 0.49 | 0.50 | 0.51 | 0.52 | 0.49 | 0.51 | 0.62 | 0.61 |
| 0.50 | 0.49 | 0.50 | 0.49 | 0.49 | 0.51 | 0.51 | 0.48 | 0.51 | 0.62 | 0.61 |
| 0.53 | 0.53 | 0.54 | 0.54 | 0.53 | 0.53 | 0.53 | 0.51 | 0.51 | 0.64 | 0.64 |
| 0.58 | 0.56 | 0.56 | 0.57 | 0.56 | 0.56 | 0.57 | 0.54 | 0.54 | 0.65 | 0.64 |
| 0.55 | 0.55 | 0.56 | 0.56 | 0.55 | 0.55 | 0.56 | 0.54 | 0.49 | 0.65 | 0.64 |
| 0.58 | 0.55 | 0.55 | 0.55 | 0.55 | 0.59 | 0.59 | 0.55 | 0.49 | 0.62 | 0.62 |
| 0.54 | 0.53 | 0.53 | 0.54 | 0.53 | 0.54 | 0.55 | 0.54 | 0.49 | 0.66 | 0.66 |
| 0.54 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.54 | 0.52 | 0.51 | 0.62 | 0.62 |
| 0.58 | 0.55 | 0.56 | 0.56 | 0.55 | 0.58 | 0.58 | 0.54 | 0.48 | 0.61 | 0.61 |
| 0.51 | 0.50 | 0.51 | 0.51 | 0.50 | 0.53 | 0.53 | 0.51 | 0.49 | 0.62 | 0.62 |
| 0.54 | 0.52 | 0.53 | 0.53 | 0.52 | 0.54 | 0.55 | 0.53 | 0.49 | 0.65 | 0.66 |
| 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.54 | 0.52 | 0.51 | 0.63 | 0.63 |
| 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.53 | 0.54 | 0.52 | 0.52 | 0.63 | 0.63 |
| 0.54 | 0.53 | 0.54 | 0.53 | 0.53 | 0.54 | 0.55 | 0.51 | 0.49 | 0.62 | 0.62 |
| 0.50 | 0.50 | 0.50 | 0.51 | 0.50 | 0.51 | 0.52 | 0.49 | 0.48 | 0.62 | 0.62 |
| 0.50 | 0.50 | 0.50 | 0.51 | 0.50 | 0.51 | 0.52 | 0.49 | 0.48 | 0.62 | 0.62 |
| 0.50 | 0.50 | 0.50 | 0.51 | 0.50 | 0.51 | 0.52 | 0.50 | 0.48 | 0.62 | 0.62 |
| 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.50 | 0.51 | 0.49 | 0.48 | 0.62 | 0.62 |
| 0.52 | 0.51 | 0.52 | 0.51 | 0.51 | 0.52 | 0.53 | 0.50 | 0.47 | 0.60 | 0.59 |
| 0.50 | 0.49 | 0.50 | 0.50 | 0.50 | 0.51 | 0.52 | 0.49 | 0.48 | 0.62 | 0.61 |
| 0.50 | 0.49 | 0.49 | 0.49 | 0.49 | 0.51 | 0.51 | 0.49 | 0.49 | 0.62 | 0.61 |
| 0.50 | 0.49 | 0.49 | 0.50 | 0.49 | 0.51 | 0.51 | 0.49 | 0.49 | 0.61 | 0.61 |
| 0.50 | 0.49 | 0.49 | 0.50 | 0.50 | 0.51 | 0.51 | 0.50 | 0.50 | 0.61 | 0.61 |
| 0.50 | 0.49 | 0.49 | 0.49 | 0.49 | 0.51 | 0.52 | 0.49 | 0.49 | 0.61 | 0.60 |
| 0.51 | 0.49 | 0.50 | 0.50 | 0.49 | 0.51 | 0.52 | 0.50 | 0.47 | 0.61 | 0.61 |
| 0.51 | 0.49 | 0.49 | 0.50 | 0.49 | 0.52 | 0.54 | 0.49 | 0.47 | 0.62 | 0.62 |
| 0.51 | 0.49 | 0.50 | 0.50 | 0.50 | 0.51 | 0.53 | 0.50 | 0.48 | 0.61 | 0.61 |
| 0.43 | 0.39 | 0.40 | 0.41 | 0.39 | 0.42 | 0.43 | 0.39 | 0.50 | 0.60 | 0.59 |
| 0.24 | 0.26 | 0.26 | 0.27 | 0.26 | 0.22 | 0.25 | 0.28 | 0.50 | 0.64 | 0.64 |
| 0.24 | 0.26 | 0.26 | 0.26 | 0.26 | 0.23 | 0.56 | 0.28 | 0.49 | 0.64 | 0.64 |
| 0.25 | 0.25 | 0.27 | 0.25 | 0.10 | 0.12 | 0.28 | 0.51 | 0.65 | 0.65 | |
| 0.34 | 0.02 | 0.06 | 0.01 | 0.23 | 0.25 | 0.19 | 0.53 | 0.62 | 0.61 | |
| 0.32 | 0.05 | 0.06 | 0.01 | 0.24 | 0.25 | 0.20 | 0.53 | 0.62 | 0.62 | |
| 0.34 | 0.18 | 0.18 | 0.05 | 0.25 | 0.26 | 0.20 | 0.52 | 0.63 | 0.62 | |
| 0.32 | 0.03 | 0.03 | 0.17 | 0.24 | 0.25 | 0.19 | 0.53 | 0.62 | 0.61 | |
| 0.23 | 0.30 | 0.30 | 0.31 | 0.30 | 0.04 | 0.27 | 0.52 | 0.65 | 0.65 | |
| 0.24 | 0.33 | 0.33 | 0.33 | 0.32 | 0.11 | 0.27 | 0.53 | 0.65 | 0.65 | |
| 0.33 | 0.28 | 0.27 | 0.30 | 0.27 | 0.34 | 0.34 | 0.50 | 0.61 | 0.60 | |
| 0.45 | 0.46 | 0.46 | 0.47 | 0.46 | 0.47 | 0.46 | 0.45 | 0.65 | 0.64 | |
| 0.54 | 0.52 | 0.52 | 0.52 | 0.52 | 0.55 | 0.54 | 0.52 | 0.53 | 0.02 | |
| 0.55 | 0.51 | 0.50 | 0.52 | 0.51 | 0.55 | 0.54 | 0.52 | 0.53 | 0.05 | |
The upper diagonal indicates amino acid sequence divergence; the lower diagonal indicates uncorrected nucleotide sequence divergence.
Abbreviations are found in Table 1, footnote b. BCV, Buggy Creek virus; BABV, Babanki virus; KZLV, Kyzylagach virus; OCKV, Ockelbo virus.
Within antigenic complexes, different Alphavirus species generally showed at least 21% nucleotide and 8% amino acid sequence divergence. One exception was Everglades virus (EVEV), which differed from some strains of VEEV by only 10% at the nucleotide level and 3% at the amino acid level. Me Tri virus, which is associated with central nervous system disease in Vietnamese children and is considered a new alphavirus on the basis of antigenic tests (18), differed by only 2% in its nucleotide sequence and 1% in its amino acid sequence from Semliki Forest virus. Different subtypes of a given alphavirus had as little as 3% nucleotide and 1% amino acid sequence difference (Sindbis virus) and as much as 25 and 13%, respectively (Ross River and subtype Sagiyama viruses). The maximum divergence between subtypes was found in VEE subtype IF (78V3531), which differed from all other VEE complex viruses by at least 22% at the nucleotide sequence level and 19% at the amino acid sequence level. As in previous analyses (49), this virus grouped with VEE complex subtype VI (with the Ag80-663 strain, a species considered distinct from VEEV) but was still quite distantly related.
The sequences of the two fish viruses were the most distinct genetically, with at least 49% nucleotide and 59% amino acid sequence divergence versus all other alphaviruses. SDV and SPDV had only 5% nucleotide and 2% amino acid sequence differences, values comparable to those of subtypes of other alphaviruses.
Phylogenetic analysis of E1 gene sequences.
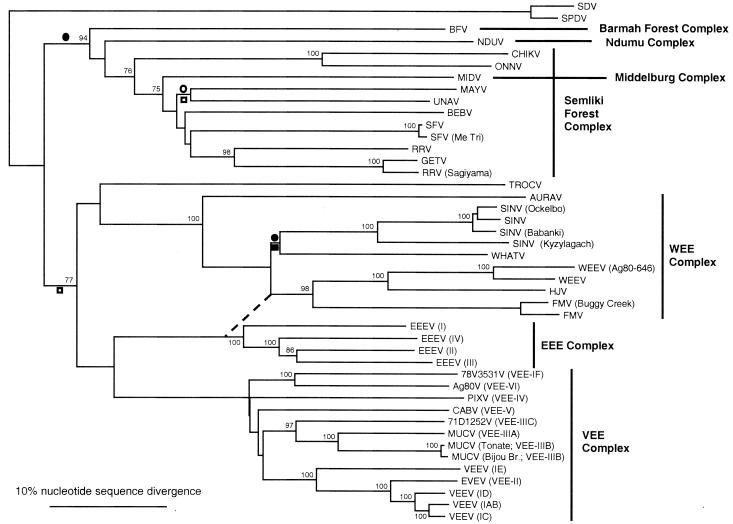
Initially, phylogenetic analyses were performed on the E1 gene region by using the maximum-parsimony and neighbor-joining methods. These methods produced trees with similar topologies, differing primarily in the relationships among some serocomplexes and within the Semliki Forest complex. Viruses with inconsistent placement included the Barmah Forest, Middelburg, Mayaro, Una, and Trocara viruses. Neighbor joining grouped the Barmah Forest and Ndumu viruses at the base of the Semliki Forest clade and placed Middelburg virus within the Semliki Forest group. In neighbor-joining trees, Trocara virus was basal to the WEE complex, which grouped with the EEEV-VEEV clade (Fig. 2). Maximum parsimony placed Middelburg virus outside of the Semliki Forest virus clade without transversion weighting and placed Trocara virus at the base of a nonfish alphavirus clade (not shown). The placement of the Cabassou and Pixuna viruses within the VEE complex was also inconsistent when different methods were used. In general, analyses using amino acid sequences generated results similar to those described above, with diminished resolution within some terminal groupings due to loss of informative, synonymous nucleotides. When all of the methods were used, midpoint rooting placed the fish virus clade at the base of the alphavirus tree, indicating that these viruses probably diverged from the mosquito-borne alphaviruses very early in the evolution of the genus.
FIG. 2.
Phylogenetic tree of all Alphavirus species, and selected subtypes and variants, generated from partial E1 envelope glycoprotein gene sequences by using the neighbor-joining program with the F84 distance formula (61). Virus abbreviations are found in Table 1, footnote 6. Numbers refer to bootstrap values for clades defined by the adjacent node. The topology of Cabassou virus (CABV) and Pixuna virus (PIXV) within the VEEV complex was constrained based on the more robust results of the complete structural polyprotein sequence analysis (see Fig. 3), and this topology was not significantly less likely based on maximum-likelihood analyses with the E1 gene sequences. The open circle adjacent to a branch indicates hypothetical Old to New World introduction, and the closed circle indicates New to Old World introduction, assuming a New World origin; the open square indicates Old to New World introduction, and the closed square indicates New to Old World introduction, assuming an Old World origin of the nonfish Alphavirus clade.
In an attempt to resolve the topological discrepancies described above, the maximum-likelihood method was used, based on a sequence evolution model derived from previously published detailed analyses of many strains from a given Alphavirus species or complex (2, 8, 43, 48, 49). Maximum-likelihood analysis of these data sets provided a mean estimate of 4.5 for the Ti/Tv ratio and a mean gamma value of 0.24. When these values were used, the topologies generated by the maximum-parsimony and neighbor-joining methods were evaluated by using a Kishino-Hasegawa likelihood test (61). The topology generated by the neighbor-joining method, shown in Fig. 2, was significantly more likely (maximum likelihood, P < 0.03) than the topologies generated by the maximum-parsimony method.
Phylogenetic analyses of complete structural gene sequences.
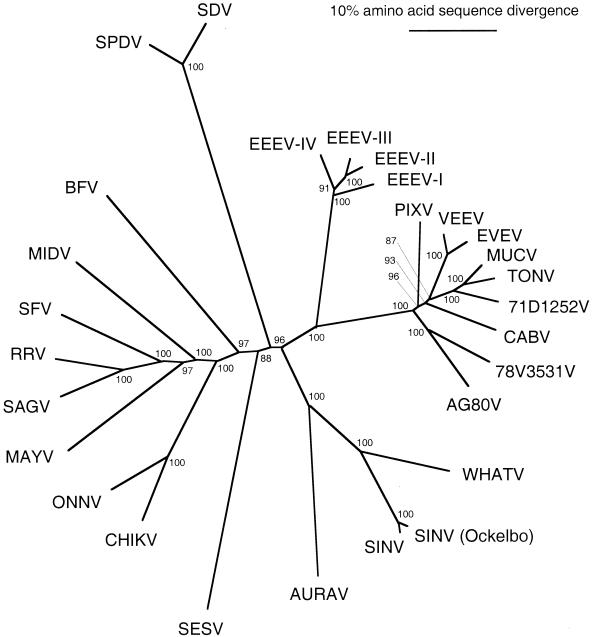
In an attempt to resolve further some of the discrepancies in the tree topologies generated from partial E1 protein gene sequences, we analyzed the complete nonstructural and structural protein gene sequences available for all alphavirus species by using the methods described above. Included in this analysis was the partial structural polyprotein sequence of SESV (32). Individual genes were also analyzed, and no evidence of recombination (topology differences supported by bootstrap values of 80% or greater), aside from the recombinant WEEV-Highlands J virus-Fort Morgan virus group described previously (19, 80), was detected. Structural polyprotein gene trees were consistently more robust than those constructed from nonstructural genes and also included more alphavirus representatives. Therefore, we focused on the structural polyprotein gene analyses.
Trees generated by using both the maximum-parsimony and neighbor-joining methods had identical topologies, except for the placement of Middelburg virus, which fell within the Semliki Forest complex when the neighbor-joining method was used and was basal to the Semliki Forest virus complex when the maximum-parsimony method was used. The neighbor-joining tree generated by using amino acid sequences, which had higher bootstrap values than all others, is shown in Fig. 3. Because this tree had robust groupings for the VEE complex, we applied the VEE topology to the partial E1 protein gene sequence analysis (Fig. 2) and compared the maximum-likelihood values generated for both the E1 and structural polyprotein topologies. The likelihood ratios indicated that the neighbor-joining topology generated by using structural polyprotein sequences was as likely as the original topology generated with E1 nucleotide sequences. The original E1 topology, which placed Cabassou virus at the base of the VEE clade, with Pixuna virus a sister group to VEEV and EVEV, was not significantly more likely when our sequence evolution model was used (P > 0.3). Therefore, we believe that Fig. 2 represents the most accurate topology available for the genus Alphavirus. The fish viruses were even more clearly the outliers in the complete structural polyprotein analyses than in the trees generated from partial E1 sequences, providing stronger evidence that they would represent the basal clade in a rooted tree, as indicated in the midpoint rooted trees (Fig. 2). SESV also appeared to be quite distinct genetically from all of the mosquito-borne alphaviruses, with an amino acid sequence divergence level equivalent to that of a distinct antigenic complex. However, the distance of the SESV branch could be somewhat misleading if the missing regions (part of the capsid and E1 protein sequences) are less divergent than the included sequence regions (E3, E2, and 6K).
FIG. 3.
Unrooted phylogenetic tree of Alphavirus species generated from complete structural polyprotein amino acid sequences by using the neighbor-joining program (61). Virus abbreviations are found in Table 1, footnote 6. Numbers refer to bootstrap values for clades defined by the indicated node.
DISCUSSION
Evolutionary origin of the alphaviruses.
Previous analyses of Alphavirus evolution suggested that the genus originated in the New World from an insect-borne plant virus (35, 73, 79). The present analyses are also consistent with this hypothesis. Excluding the fish and seal viruses, a New World origin would require at least three transoceanic introductions between the hemispheres: (i) transport of the ancestor of the Barmah Forest-Ndumu-Middelburg-Semliki Forest virus complexes from the New World to the Old World, (ii) transport of the ancestor of the Sindbis and Whataroa viruses to the Old World, and (iii) transport of the ancestor of the Mayaro and Una viruses from the Old World to the New World (Fig. 2). However, an Old World origin is also consistent with three transoceanic introductions between the hemispheres: (i) transport of the ancestor of the Trocara virus-WEE-EEE-VEE complexes from the Old World to the New World; (ii) transport of the ancestor of the Sindbis and Whataroa viruses to the Old World, and (iii) transport of the ancestor of the Mayaro and Una viruses from the Old World to the New World (Fig. 2). These equally parsimonious scenarios do not favor either hypothesis over the other. An ancestral alphavirus presumably adapted to fish in the distant past to form the SDV-SPDV lineage. The possible transmission of SESV by insects (lice) strengthens the hypothesis that alphaviruses arose as insect-borne or insect viruses.
Previous estimates placed the origin of the alphaviruses several thousand years ago (73, 79). However, the methods employed previously relied on the assumption of an equal rate of substitutions across nucleotide or amino acid positions in the alphavirus genome. Our data clearly indicate that this assumption is invalid; all estimates of the uniformity of nucleotide changes across sites are far from uniform, with an average gamma value of only 0.24 for those viruses examined in detail (range, 0.05 to 0.31). This nonuniformity in nucleotide substitutions across sites, combined with the saturation of nucleotide changes in many positions, indicates that estimates on the order of thousands of years ago for the alphavirus ancestor are far too recent. An accurate time estimate for the alphavirus progenitor may be impossible due to these factors. Another example of the problems with estimating internal branch lengths is illustrated by our analysis of the recombination event between EEEV- and Sindbis virus-like ancestors leading to the WEEV Fort Morgan virus-Highlands J virus group (19, 80). The interior branch lengths produced with most of the phylogenetic methods yielded different horizontal positions for the internal branches shown previously to represent the recombinant ancestors (80) (Fig. 2). The fact that these ancestors did not occur at the same horizontal position (the dashed line in Fig. 2 cannot be drawn vertically) indicates error in the internal branch lengths of either the EEE or the WEE complex clade or both clades.
Because homologous sequences for the structural proteins cannot be identified in viruses outside the genus Alphavirus, even in rubella virus, which comprises the other genus (Rubivirus) in the family Togaviridae, our trees could not be rooted by using an outgroup. If midpoint rooting is used, the fish viruses are consistently placed at the base of our alphavirus trees; this rooting relies on the assumption of a constant rate of evolution across different lineages in the tree. The WEE complex recombination example described above implies that this assumption is not completely correct and suggests that an unrooted tree is the most accurate representation of the genus at this time.
Mechanisms of Alphavirus radiation.
Previous studies of Alphavirus diversification have emphasized host switching events and geographic introductions in the evolution of the genus (2, 48, 49, 73, 80). Examination of the complete Alphavirus phylogeny confirms the importance of these mechanisms. The Alphavirus phylogenies also show numerous examples of host switching events, such as the presumed introduction of EEEV into North America, accompanied by switching from Culex to Culiseta mosquito vectors (73). EVEV was presumably introduced into Florida from Central or South America and adapted to Culex cedecei, which occurs only in North America. Chikungunya virus is believed to have originated in East Africa in a nonhuman primate-sylvatic Aedes mosquito transmission cycle and later was introduced into Asia along with the urban vector Aedes aegypti (48). O'nyong-nyong virus is believed to have evolved from a chikungunya-like virus that adapted to Anopheles mosquito vectors, a unique trait among alphaviruses (48).
The diversity exhibited by alphavirus groups may be influenced strongly by host mobility. Viruses that utilize reservoir hosts with limited mobility, such as small mammals, tend to be quite diverse and have nonoverlapping distributions. The best examples are the VEE complex viruses, which use primarily rodent hosts and Culex (Melanoconion) mosquito vectors with a limited flight range. VEE complex viruses occur nearly throughout the neotropics and subtropics, but the distributions of the various subtypes are discrete, for the most part. A similar epidemiological phenomenon is seen among the isolates of Ross River virus from Australia (37). Viruses that use birds as their reservoir hosts, such as Sindbis virus, EEEV, and WEEV in North America, are less diverse, and each variant or topotype tends to occupy a greater geographic range (37, 54). Host mobility presumably limits virus diversity by preventing geographic isolation and allopatric divergence and by favoring competitive exclusion of closely related viruses that are mixed over large geographic ranges.
Alphavirus systematics.
Initially, Alphavirus classification was defined by the Subcommittee on Interrelationships Among Catalogued Arboviruses (SIRACA) of the American Committee on Arthropod-Borne Viruses, which relied completely on antigenic cross-reactivity in tests such as hemagglutination inhibition, complement fixation, and neutralization (4, 7). These criteria identified seven antigenic complexes of alphaviruses that contained members displaying greater cross-reactivity to each other than to members of other complexes. Different Alphavirus species were defined as viruses with fourfold or greater differences in cross-reactivity in both directions (one virus reacted against antibody from a second, and the second virus reacted against antibody produced against the first) compared to homologous (a given virus reacted against antibody produced against itself) antibody-antigen reactions. Subtypes were considered viruses with fourfold differences in one direction only, while antigenic varieties were distinguishable only with special tests like hemagglutination inhibition or monoclonal antibody assays (4, 7).
The International Committee on the Taxonomy of Viruses (ICTV) has established taxonomic criteria for alphaviruses and has limited its classification to species within the genus (no complexes or subtypes are defined). Currently, the ICTV defines a virus species as a “polythetic class of viruses that constitute a replicating lineage and occupy a particular ecological niche” (67, 68). This definition includes additional criteria in comparison to the SIRACA classification, but this leads to more subjective interpretation in some cases. For example, EVEV is currently considered a species distinct from VEEV (77) (Table 1), although the SIRACA classification includes it as a subtype of VEEV (4). Phylogenetic studies examining VEEV subtype I viruses in greater detail have shown clearly that EVEV falls within the VEEV subtype IAB/C/D clade (49, 53). A completely natural classification would not include this kind of a paraphyletic taxon and would consider EVEV a variant of VEEV, along with all of the subtype I strains except 78V3531 (Fig. 2). However, EVEV clearly constitutes a replicating lineage (it occurs only in Florida and is genetically distinct based on this distribution) and occupies a particular ecological niche (for example, it uses a mosquito vector different from those of all other VEE complex viruses). Also, EVEV has not been associated with the emergence of epidemics and epizootics like the subtype ID and IE viruses (74). Synonymizing EVEV with VEEV has been previously proposed (27, 39); although justified in many theoretical respects, this would have important practical implications due to biological safety recommendations (66). An additional example of the difficulties in virus classification and taxonomy is the original classification of Barmah Forest virus in the family Bunyaviridae based on antigenic criteria (9, 38). However, subsequent genetic characterization revealed it to be a member of the Alphavirus genus based on virion structure, mode of replication, and nucleic acid and protein sequences.
Despite the fundamental differences between the antigenic and polythetic species definitions, the systematics of the alphaviruses developed on antigenic grounds alone (4) agree remarkably well with those of the ICTV (77). The more detailed nature of the SIRACA classification of antigenic subtypes can lead to minor genetic changes that have a dramatic affect on antigenicity and thus the rapid appearance of new taxa. An example is an antigenic subtype of EEEV isolated from a human in Mississippi in 1983 (5). Although this strain met antigenic criteria as a subtype, genetic analyses demonstrated that minor genetic changes resulted in the addition of an N-linked glycosylation site in the E2 protein (78). Although there was no evidence that this genotype persisted beyond 1983, these kinds of antigenic changes could be epidemiologically important. Another example is VEEV, where only one or two amino acid substitutions in the E2 envelope glycoprotein can result in the generation of subtype IC equine-virulent strains from enzootic, equine-avirulent subtype ID progenitors (72). These changes may have dramatic effects on pathogenicity and host range, leading to epizootics. A completely natural classification would not distinguish these subtypes because they are paraphyletic and the epizootic viruses do not appear to constitute ongoing lineages. However, subtyping of VEEV is extremely important for public health purposes and classifications must balance theoretical and practical considerations.
Alphavirus complexes.
The seven antigenic complexes of alphaviruses (4) appear to accurately reflect clades of viruses that share medically important characteristics. For example, members of the EEE and VEE complexes share encephalitic potential in equines and humans, while the Semliki Forest virus complex viruses generally produce an arthralgic syndrome. The grouping of Barmah Forest virus with the Semliki Forest virus complex viruses is consistent with their sharing this pathogenic trait. The WEE complex includes viruses that produce both arthralgic (Sindbis virus-like clade) and encephalitic (WEEV and Highlands J virus) syndromes. WEEV and Highlands J virus are descendants of a recombinant alphavirus, and their encephalitic potential presumably reflects the genetic contribution (nonstructural proteins, capsid, and 3′ NCR) of the EEEV-like ancestor rather than the Sindbis virus-like glycoprotein genes (19, 80). The only inconsistency of the established Alphavirus complexes with evolutionary relationships is Middelburg virus, which is classified as a separate antigenic complex based on antigenic relationships (4). While there are very few isolates available and the epidemiologic patterns of the virus are unknown, Middelburg virus may be a member of the Semliki Forest virus complex clade (Fig. 2 and 3).
Interestingly, serological characterizations may provide some insight into the relationships of the various complex clades. For example, when monoclonal antibodies generated against the Semliki Forest Virus nucleocapsid are used in antibody capture assays, they cross-react with members of the Semliki Forest virus, WEEV, EEEV, Middelburg virus, and Ndumu virus complexes but do not cross-react at all with VEEV or Barmah Forest virus (17). As the nucleocapsid is one of the more conserved virion proteins, this may reflect some ancient relationships among the alphaviruses.
Recommended revisions of the genus Alphavirus.
Although our phylogenetic data generally supported the current Alphavirus classification, several discrepancies were noted. (i) Virus strain 78V3531 (VEE subtype IF according to SIRACA) is quite distinct phylogenetically from VEEV, and its closest relative is Ag80-646 (Ag80V). Although its transmission cycle has not been characterized and its niche cannot therefore be evaluated, this virus probably warrants species designation based on the clear distinction of its genetic lineage, its isolation in a part of Brazil not known to be inhabited by other VEEV complex alphaviruses, and its antigenic distinction (3). Unlike VEEV, it is also avirulent for adult mice and is not associated with VEEV outbreaks. (ii) Tonate virus, a member of VEEV complex subtype III, is quite distinct from the other members of subtype III, with at least 16% nucleotide and 7% amino acid sequence divergence (Table 4). In addition to their antigenic differences, the Tonate and Mucambo viruses apparently use different reservoir hosts (birds and small mammals, respectively) (71). They should probably be considered distinct species. The Bijou Bridge strain from western North America, also a bird virus and apparently transmitted by nest bugs (40), is appropriately considered a strain of Tonate virus due to its genetic similarity and similar niche. (iii) Although its transmission cycle remains obscure, Trocara virus also appears to be a new Alphavirus species based on genetic distinctions from all other species (65). The antigenic comparisons suggesting that Trocara virus represents a new antigenic complex are not as comprehensive as our sequence comparisons, and cross-reactions with members of several Alphavirus serocomplexes were very weak. (iv) Me Tri virus, originally reported to be a new Alphavirus based on antigenic criteria, is genetically very close to Semliki Forest virus and does not appear to constitute a separate lineage (although lineage is a rather arbitrary term); its genetic distance from Semliki Forest virus is similar to the distances among other Alphavirus subtypes or strains, and it should probably be considered a subtype or strain of Semliki Forest virus. (v) Sagiyama virus, considered by SIRACA to be a subtype of Getah virus, along with Ross River virus and Bebaru virus (4), and considered a subtype of Ross River virus in the most recent ICTV classification (77), is much more closely related to Getah virus than to Ross River or Bebaru virus. Based on our genetic data alone, the Ross River, Bebaru, and Getah viruses should be retained as distinct Alphavirus species but, as suggested by Shirako and Yamaguchi (57), Sagiyama virus should be considered a subtype of Getah virus. (vi) Kyzylagach virus, which was originally isolated in Azerbaijan and was recently identified in China (36), appears to be one of the most distinct subtypes of Sindbis virus yet identified. The genetic data indicate that it could be classified as either a subtype of Sindbis virus or a distinct species (18% divergence at the nucleotide level and 6 to 8% divergence at the amino acid level). Additionally, of all of the viruses analyzed in this study, this is the only virus that could not be sequenced with the degenerate alphavirus sequencing primers; it required species-specific primers. Because SIN viruses are usually transmitted among avian hosts and maintain a high degree of genetic homogeneity, the fact that Kyzylagach virus exists in a lineage so independent from all other SIN viruses suggests that it could be classified as a distinct species. (vii) SDV and SPDV, although not yet compared antigenically to the alphaviruses (69, 81), also appear to represent a distinct complex based on their sequence divergence. They clearly occupy dramatically different niches and genetic lineages from all remaining alphaviruses, indicating that they are not variants of an established species. However, the very small amount of sequence divergence between the two fish viruses suggests that SDV is really a strain or subtype of the novel Alphavirus species SPDV. (viii) SESV also represents a new Alphavirus species, as reported previously (32). It appears to be quite distinct genetically from all of the mosquito-borne alphaviruses, with the amino acid sequence divergence level of a distinct antigenic complex.
ACKNOWLEDGMENTS
We thank Robert Tesh, Robert Shope, and Hilda Guzman for providing some of the alphaviruses used in our analyses.
A.M.P. was supported by the James W. McLaughlin Fellowship Fund and NIH T32 Training Grant on Emerging and Reemerging Infectious Diseases AI-07536. A.C.B. was supported by a James L. McLaughlin Infection and Immunity Fellowship and NIH Emerging Tropical Diseases T32 training grant AI-107526. This research was supported by National Institutes of Health grants AI-10984 to Robert Tesh, AI-39800 to S.C.W., and AI-10793 to J.H.S. and E.G.S.
REFERENCES
- 1.Bianchi T I, Aviles G, Monath T P, Sabattini M S. Western equine encephalomyelitis: virulence markers and their epidemiological significance. Am J Trop Med Hyg. 1993;49:322–328. doi: 10.4269/ajtmh.1993.49.322. [DOI] [PubMed] [Google Scholar]
- 2.Brault A C, Powers A M, Chavez C L, Lopez R N, Cachon M F, Gutierrez L F, Kang W, Tesh R B, Shope R E, Weaver S C. Genetic and antigenic diversity among eastern equine encephalitis viruses from North, Central, and South America. Am J Trop Med Hyg. 1999;61:579–586. doi: 10.4269/ajtmh.1999.61.579. [DOI] [PubMed] [Google Scholar]
- 3.Calisher C, Kinney R, de Souza Lopes O, Trent D, Monath T, Francy D. Identification of a new Venezuelan equine encephalitis virus from Brazil. Am J Trop Med Hyg. 1982;31:1260–1272. doi: 10.4269/ajtmh.1982.31.1260. [DOI] [PubMed] [Google Scholar]
- 4.Calisher C H, Karabatsos N. Arbovirus serogroups: definition and geographic distribution. In: Monath T P, editor. The arboviruses: epidemiology and ecology. I. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 19–57. [Google Scholar]
- 5.Calisher C H, Karabatsos N, Foster J P, Pallansch M, Roehrig J T. Identification of an antigenic subtype of eastern equine encephalitis virus isolated from a human. J Clin Microbiol. 1990;28:373–374. doi: 10.1128/jcm.28.2.373-374.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calisher C H, Monath T P, Mitchell C J, Sabattini M S, Cropp C B, Kerschner J, Hunt A R, Lazuick J S. Arbovirus investigations in Argentina, 1977–1980. III. Identification and characterization of viruses isolated, including new subtypes of western and Venezuelan equine encephalitis viruses and four new bunyaviruses (Las Maloyas, Resistencia, Barranqueras, and Antequera) Am J Trop Med Hyg. 1985;34:956–965. [PubMed] [Google Scholar]
- 7.Calisher C H, Shope R E, Brandt W, Casals J, Karabatsos N, Murphy F A, Tesh R B, Wiebe M E. Proposed antigenic classification of registered arboviruses. Intervirology. 1980;14:229–232. doi: 10.1159/000149190. [DOI] [PubMed] [Google Scholar]
- 8.Cilnis M J, Kang W, Weaver S C. Genetic conservation of Highlands J viruses. Virology. 1996;218:343–351. doi: 10.1006/viro.1996.0203. [DOI] [PubMed] [Google Scholar]
- 9.Dalgarno L, Short N J, Hardy C M, Bell J R, Strauss J H, Marshall I D. Characterization of Barmah forest virus: an alphavirus with some unusual properties. Virology. 1984;133:416–426. doi: 10.1016/0042-6822(84)90407-0. [DOI] [PubMed] [Google Scholar]
- 10.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faragher S G, Meek A D J, Rice C M, Dalgarno L. Genome sequences of a mouse-avirulent and a mouse-virulent strain of Ross River virus. Virology. 1988;163:509–526. doi: 10.1016/0042-6822(88)90292-9. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 13.Ficken M D, Wages D P, Guy J S, Quinn J A, Emory W H. High mortality of domestic turkeys associated with Highlands J virus and eastern equine encephalitis virus infections. Avian Dis. 1993;37:585–590. [PubMed] [Google Scholar]
- 14.Garoff H, Frischauf A M, Simons K, Lehrach H, Delius H. The capsid protein of Semliki Forest virus has clusters of basic amino acids and prolines in its amino-terminal region. Proc Natl Acad Sci USA. 1980;77:6376–6380. doi: 10.1073/pnas.77.11.6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garoff H, Frischauf A M, Simons K, Lehrach H, Delius H. Nucleotide sequence of cDNA coding for Semliki Forest virus membrane glycoproteins. Nature. 1980;288:236–241. doi: 10.1038/288236a0. [DOI] [PubMed] [Google Scholar]
- 16.George J, Raju R. Alphavirus RNA genome repair and evolution: molecular characterization of infectious Sindbis virus isolates lacking a known conserved motif at the 3′ end of the genome. J Virol. 2000;74:9776–9785. doi: 10.1128/jvi.74.20.9776-9785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greiser-Wilke I, Moenning V, Kaaden O R, Figueiredo L T. Most alphaviruses share a conserved epitopic region on their nucleocapsid protein. J Gen Virol. 1989;70:743–748. doi: 10.1099/0022-1317-70-3-743. [DOI] [PubMed] [Google Scholar]
- 18.Ha D Q, Calisher C H, Tien P H, Karabatsos N, Gubler D J. Isolation of a newly recognized alphavirus from mosquitoes in Vietnam and evidence for human infection and disease. Am J Trop Med Hyg. 1995;53:100–104. [PubMed] [Google Scholar]
- 19.Hahn C S, Lustig S, Strauss E G, Strauss J H. Western equine encephalitis virus is a recombinant virus. Proc Natl Acad Sci USA. 1988;85:5997–6001. doi: 10.1073/pnas.85.16.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hohdatsu T, Ide S, Yamagishi H, Eiguchi Y, Nagano H, Maehara N, Tanaka Y, Fujisaki Y, Yago K, Taguchi K, et al. Enzyme-linked immunosorbent assay for the serological survey of Getah virus in pigs. Nippon Juigaku Zasshi. 1990;52:835–837. doi: 10.1292/jvms1939.52.835. [DOI] [PubMed] [Google Scholar]
- 21.Johnson B K. O'nyong-nyong virus disease. In: Monath T P, editor. The arboviruses: epidemiology and ecology. III. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 217–223. [Google Scholar]
- 22.Johnston R E, Peters C J. Alphaviruses. In: Fields B N, Knipe D M, Howley P M, editors. Virology. 3rd edition. New York, N.Y: Lippincott-Raven; 1996. pp. 843–898. [Google Scholar]
- 23.Karabatsos N. Antigenic relationships of group A arboviruses by plaque reduction neutralization testing. Am J Trop Med Hyg. 1975;24:527–532. doi: 10.4269/ajtmh.1975.24.527. [DOI] [PubMed] [Google Scholar]
- 24.Killington R A, Stokes A, Hierholzer J C. Virus purification. In: Mahy B W J, Kangro H O, editors. Virology methods manual. San Diego, Calif: Academic Press, Inc.; 1996. pp. 71–89. [Google Scholar]
- 25.Kinney R M, Johnson B J, Brown V L, Trent D W. Nucleotide sequence of the 26 S mRNA of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and deduced sequence of the encoded structural proteins. Virology. 1986;152:400–413. doi: 10.1016/0042-6822(86)90142-x. [DOI] [PubMed] [Google Scholar]
- 26.Kinney R M, Johnson B J, Welch J B, Tsuchiya K R, Trent D W. The full-length nucleotide sequences of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and its attenuated vaccine derivative, strain TC-83. Virology. 1989;170:19–30. doi: 10.1016/0042-6822(89)90347-4. [DOI] [PubMed] [Google Scholar]
- 27.Kinney R M, Pfeffer M, Tsuchiya K R, Chang G J, Roehrig J T. Nucleotide sequences of the 26S mRNAs of the viruses defining the Venezuelan equine encephalitis antigenic complex. Am J Trop Med Hyg. 1998;59:952–964. doi: 10.4269/ajtmh.1998.59.952. [DOI] [PubMed] [Google Scholar]
- 28.Kinney R M, Tsuchiya K R, Sneider J M, Trent D W. Genetic evidence that epizootic Venezuelan equine encephalitis (VEE) viruses may have evolved from enzootic VEE subtype I-D virus. Virology. 1992;191:569–580. doi: 10.1016/0042-6822(92)90232-e. [DOI] [PubMed] [Google Scholar]
- 29.Kumanomido T, Kamada M, Wada R, Kenemaru T, Sugiura T, Akiyama Y. Pathogenicity for horses of original Sagiyama virus, a member of the Getah virus group. Vet Microbiol. 1988;17:367–373. doi: 10.1016/0378-1135(88)90050-8. [DOI] [PubMed] [Google Scholar]
- 30.Kumanomido T, Wada R, Kanemaru T, Kamada M, Akiyama Y, Matumoto M. Transplacental infection in mice inoculated with Getah virus. Vet Microbiol. 1988;16:129–136. doi: 10.1016/0378-1135(88)90037-5. [DOI] [PubMed] [Google Scholar]
- 31.Kumanomido T, Wada R, Kanemaru T, Kamada M, Hirasawa K, Akiyama Y. Clinical and virological observations on swine experimentally infected with Getah virus. Vet Microbiol. 1988;16:295–301. doi: 10.1016/0378-1135(88)90033-8. [DOI] [PubMed] [Google Scholar]
- 32.La Linn M, Gardner J, Warrilow D, Darnell G A, McMahon C R, Field I, Hyatt A D, Slade R W, Suhrbier A. Arbovirus of marine mammals: a new alphavirus isolated from the elephant seal louse, Lepidophthirus macrorhini. J Virol. 2001;75:4103–4109. doi: 10.1128/JVI.75.9.4103-4109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lanciotti R S, Ludwig M L, Rwaguma E B, Lutwama J J, Kram T M, Karabatsos N, Cropp B C, Miller B R. Emergence of epidemic O'nyong-nyong fever in Uganda after a 35-year absence: genetic characterization of the virus. Virology. 1998;252:258–268. doi: 10.1006/viro.1998.9437. [DOI] [PubMed] [Google Scholar]
- 34.Lee E, Stocks C, Lobigs P, Hislop A, Straub J, Marshall I, Weir R, Dalgarno L. Nucleotide sequence of the Barmah Forest virus genome. Virology. 1997;227:509–514. doi: 10.1006/viro.1996.8343. [DOI] [PubMed] [Google Scholar]
- 35.Levinson R S, Strauss J H, Strauss E G. Complete sequence of the genomic RNA of o'nyong-nyong virus and its use in the construction of alphavirus phylogenetic trees. Virology. 1990;175:110–123. doi: 10.1016/0042-6822(90)90191-s. [DOI] [PubMed] [Google Scholar]
- 36.Liang G D, Li L, Zhou G L, Fu S H, Li Q P, Li F S, He H H, Jin Q, He Y, Chen B Q, Hou Y D. Isolation and complete nucleotide sequence of a Chinese Sindbis-like virus. J Gen Virol. 2000;81:1347–1351. doi: 10.1099/0022-1317-81-5-1347. [DOI] [PubMed] [Google Scholar]
- 37.Mackenzie J S, Smith D W. Mosquito-borne viruses and epidemic polyarthritis. Med J Aust. 1996;164:90–93. doi: 10.5694/j.1326-5377.1996.tb101357.x. [DOI] [PubMed] [Google Scholar]
- 38.Marshall I D, Woodroofe G M, Hirsch S. Viruses recovered from mosquitoes and wildlife serum collected in the Murray Valley of South-eastern Australia, February 1974, during an epidemic of encephalitis. Aust J Exp Biol Med Sci. 1982;60:457–470. doi: 10.1038/icb.1982.51. [DOI] [PubMed] [Google Scholar]
- 39.Meissner J D, Huang C Y, Pfeffer M, Kinney R M. Sequencing of prototype viruses in the Venezuelan equine encephalitis antigenic complex. Virus Res. 1999;64:43–59. doi: 10.1016/S0168-1702(99)00078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monath T P, Lazuick J S, Cropp C B, Rush W A, Calisher C H, Kinney R M, Trent D W, Kemp G E, Bowen G S, Francy D B. Recovery of Tonate virus (“Bijou Bridge” strain), a member of the Venezuelan equine encephalomyelitis virus complex, from cliff swallow nest bugs (Oeciacus vicarius) and nestling birds in North America. Am J Trop Med Hyg. 1980;29:969–983. doi: 10.4269/ajtmh.1980.29.969. [DOI] [PubMed] [Google Scholar]
- 41.Monath T P, Sabattini M S, Pauli R, Daffner J F, Mitchell C J, Bowen G S, Cropp C B. Arbovirus investigations in Argentina, 1977–1980. IV. Serologic surveys and sentinel equine program. Am J Trop Med Hyg. 1985;34:966–675. [PubMed] [Google Scholar]
- 42.Oberste M S, Parker M D, Smith J F. Complete sequence of Venezuelan equine encephalitis virus subtype IE reveals conserved and hypervariable domains within the C terminus of nsP3. Virology. 1996;219:314–320. doi: 10.1006/viro.1996.0254. [DOI] [PubMed] [Google Scholar]
- 43.Oberste M S, Schmura S M, Weaver S C, Smith J F. Geographic distribution of Venezuelan equine encephalitis virus subtype IE genotypes in Central America and Mexico. Am J Trop Med Hyg. 1999;60:630–634. doi: 10.4269/ajtmh.1999.60.630. [DOI] [PubMed] [Google Scholar]
- 44.Peiris J S, Amerasinghe P H, Amerasinghe F P, Calisher C H, Perera L P, Arunagiri C K, Munasingha N B, Karunaratne S H. Viruses isolated from mosquitoes collected in Sri Lanka. Am J Trop Med Hyg. 1994;51:154–161. doi: 10.4269/ajtmh.1994.51.154. [DOI] [PubMed] [Google Scholar]
- 45.Pfeffer M, Kinney R M, Kaaden O R. The alphavirus 3′-nontranslated region: size heterogeneity and arrangement of repeated sequence elements. Virology. 1998;240:100–108. doi: 10.1006/viro.1997.8907. [DOI] [PubMed] [Google Scholar]
- 46.Pinheiro F P, LeDuc J W. Mayaro virus disease. In: Monath T P, editor. The arboviruses: epidemiology and ecology. Boca Raton, Fla: CRC Press, Inc.; 1988. p. 137. [Google Scholar]
- 47.Poidinger M, Roy S, Hall R A, Turley P J, Scherret J H, Lindsay M D, Broom A K, Mackenzie J S. Genetic stability among temporally and geographically diverse isolates of Barmah Forest virus. Am J Trop Med Hyg. 1997;57:230–234. doi: 10.4269/ajtmh.1997.57.230. [DOI] [PubMed] [Google Scholar]
- 48.Powers A M, Brault A C, Tesh R B, Weaver S C. Re-emergence of chikungunya and o'nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J Gen Virol. 2000;81:471–479. doi: 10.1099/0022-1317-81-2-471. [DOI] [PubMed] [Google Scholar]
- 49.Powers A M, Oberste M S, Brault A C, Rico-Hesse R, Schmura S M, Smith J F, Kang W, Sweeney W P, Weaver S C. Repeated emergence of epidemic/epizootic Venezuelan equine encephalitis from a single genotype of enzootic subtype ID virus. J Virol. 1997;71:6697–6705. doi: 10.1128/jvi.71.9.6697-6705.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rice C M, Strauss J H. Nucleotide sequence of the 26S mRNA of Sindbis virus and deduced sequence of the encoded virus structural proteins. Proc Natl Acad Sci USA. 1981;78:2062–2066. doi: 10.1073/pnas.78.4.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rumenapf T, Strauss E G, Strauss J H. Aura virus is a New World representative of Sindbis-like viruses. Virology. 1995;208:621–633. doi: 10.1006/viro.1995.1193. [DOI] [PubMed] [Google Scholar]
- 52.Russell R C. Vectors vs. humans in Australia—who is on top down under? An update on vector-borne disease and research on vectors in Australia. J Vect Ecol. 1998;23:1–46. [PubMed] [Google Scholar]
- 53.Salas R A, Garcia C Z, Liria J, Barrera R, Navarro J C, Medina G, Vasquez C, Fernandez Z, Weaver S C. Ecological studies of enzootic Venezuelan equine encephalitis in north-central Venezuela, 1997–1998. Am J Trop Med Hyg. 2001;64:84–92. doi: 10.4269/ajtmh.2001.64.84. [DOI] [PubMed] [Google Scholar]
- 54.Sammels L M, Lindsay M D, Poidinger M, Coelen R J, Mackenzie J S. Geographic distribution and evolution of Sindbis virus in Australia. J Gen Virol. 1999;80:739–748. doi: 10.1099/0022-1317-80-3-739. [DOI] [PubMed] [Google Scholar]
- 55.Schlesinger S, Schlesinger M J. Togaviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields' virology. 3rd Edition. New York, N.Y: Raven Press; 1996. pp. 825–842. [Google Scholar]
- 56.Shirako Y, Niklasson B, Dalrymple J M, Strauss E G, Strauss J H. Structure of the Ockelbo virus genome and its relationship to other Sindbis viruses. Virology. 1991;182:753–764. doi: 10.1016/0042-6822(91)90616-j. [DOI] [PubMed] [Google Scholar]
- 57.Shirako Y, Yamaguchi Y. Genome structure of Sagiyama virus and its relatedness to other alphaviruses. J Gen Virol. 2000;81:1353–1360. doi: 10.1099/0022-1317-81-5-1353. [DOI] [PubMed] [Google Scholar]
- 58.Sneider J M, Kinney R M, Tsuchiya K R, Trent D W. Molecular evidence that epizootic Venezuelan equine encephalitis (VEE) I-AB viruses are not evolutionary derivatives of enzootic VEE subtype I-E or II viruses. J Gen Virol. 1993;74:519–523. doi: 10.1099/0022-1317-74-3-519. [DOI] [PubMed] [Google Scholar]
- 59.Strauss E G, Rice C M, Strauss J H. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology. 1984;133:92–110. doi: 10.1016/0042-6822(84)90428-8. [DOI] [PubMed] [Google Scholar]
- 60.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swofford D L. PAUP∗. Phylogenetic analysis using parsimony (∗ and other methods). Version 4. Sunderland, Mass: Sinauer Associates; 1998. [Google Scholar]
- 62.Tesh R B. Arthritides caused by mosquito-borne viruses. Ann Rev Med. 1982;33:31–40. doi: 10.1146/annurev.me.33.020182.000335. [DOI] [PubMed] [Google Scholar]
- 63.Tesh R B, Gajdusek D C, Garruto R M. The distribution and prevalence of group A arbovirus neutralizing antibodies among human populations in Southeast Asia and the Pacific Islands. Am J Trop Med Hyg. 1975;25:664. doi: 10.4269/ajtmh.1975.24.664. [DOI] [PubMed] [Google Scholar]
- 64.Tesh R B, Watts D M, Russell K L, Damodaran C, Calampa C, Cabezas C, Ramirez G, Vasquez B, Hayes C G, Rossi C A, Powers A M, Hice C L, Chandler L J, Cropp B C, Karabatsos N, Roehrig J T, Gubler D J. Mayaro virus disease: an emerging mosquito-borne zoonosis in tropical South America. Clin Infect Dis. 1999;28:67–73. doi: 10.1086/515070. [DOI] [PubMed] [Google Scholar]
- 65.Travassos da Rosa A P, Turell M J, Watts D M, Powers A M, Vasconcelos P F, Jones J W, Klein T A, Dohm D J, Shope R E, Degallier N, Popov V L, Russell K L, Weaver S C, Guzman H, Calampa C, Brault A C, Lemon A P, Tesh R B. Trocara virus: a newly recognized alphavirus (Togaviridae) isolated from mosquitoes in the Amazon basin. Am J Trop Med Hyg. 2001;64:93–97. doi: 10.4269/ajtmh.2001.64.93. [DOI] [PubMed] [Google Scholar]
- 66.U.S. Department of Health and Human Services. Biosafety in microbiological and biomedical laboratories. 4th edition. Washington, D.C.: U.S. Government Printing Office; 1999. [Google Scholar]
- 67.Van Regenmortel M H. Virus species, a much overlooked but essential concept in virus classification. Intervirology. 1990;31:241–254. doi: 10.1159/000150159. [DOI] [PubMed] [Google Scholar]
- 68.Van Regenmortel M H V. Introduction to the species concept in virus taxonomy. In: van Regenmortel M H V, Fauquet C M, Bishop D H L, Carstens E B, Estes M K, Lemon S M, Maniloff J, Mayo M A, McGeogh D J, Pringle C R, Wickner R B, editors. Virus taxonomy. Classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. San Diego, Calif: Academic Press, Inc.; 2000. pp. 3–16. [Google Scholar]
- 69.Villoing S, Bearzotti M, Chilmonczyk S, Castric J, Bremont M. Rainbow trout sleeping disease virus is an atypical alphavirus. J Virol. 2000;74:173–183. doi: 10.1128/jvi.74.1.173-183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wages D P, Ficken M D, Guy J S, Cummings T S, Jennings S R. Egg-production drop in turkeys associated with alphaviruses: eastern equine encephalitis virus and Highlands J virus. Avian Dis. 1993;37:1163–1166. [PubMed] [Google Scholar]
- 71.Walton T E, Grayson M A. Venezuelan equine encephalomyelitis. In: Monath T P, editor. The arboviruses: epidemiology and ecology. IV. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 203–231. [Google Scholar]
- 72.Wang E, Barrera R, Boshell J, Ferro C, Freier J E, Navarro J C, Salas R, Vasquez C, Weaver S C. Genetic and phenotypic changes accompanying the emergence of epizootic subtype IC Venezuelan equine encephalitis viruses from an enzootic subtype ID progenitor. J Virol. 1999;73:4266–4271. doi: 10.1128/jvi.73.5.4266-4271.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weaver S C. Evolution of alphaviruses. In: Gibbs A J, Calisher C H, Garcia-Arenal F, editors. Molecular basis of virus evolution. Cambridge, England: Cambridge University Press; 1995. pp. 501–530. [Google Scholar]
- 74.Weaver S C. Recurrent emergence of Venezuelan equine encephalomyelitis. In: Scheld W M, Hughes J, editors. Emerging infections I. Washington, D.C.: ASM Press; 1998. pp. 27–42. [Google Scholar]
- 75.Weaver S C, Bellew L A, Hagenbaugh A, Mallampalli V, Holland J J, Scott T W. Evolution of alphaviruses in the eastern equine encephalomyelitis complex. J Virol. 1994;68:158–169. doi: 10.1128/jvi.68.1.158-169.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Weaver S C, Bellew L A, Rico-Hesse R. Phylogenetic analysis of alphaviruses in the Venezuelan equine encephalitis complex and identification of the source of epizootic viruses. Virology. 1992;191:282–290. doi: 10.1016/0042-6822(92)90190-z. [DOI] [PubMed] [Google Scholar]
- 77.Weaver S C, Dalgarno L, Frey T K, Huang H V, Kinney R M, Rice C M, Roehrig J T, Shope R E, Strauss E G. Family Togaviridae. In: van Regenmortel M H V, Fauquet C M, Bishop D H L, Carstens E B, Estes M K, Lemon S M, Maniloff J, Mayo M A, McGeogh D J, Pringle C R, Wickner R B, editors. Virus taxonomy. Classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. San Diego, Calif: Academic Press, Inc.; 2000. pp. 879–889. [Google Scholar]
- 78.Weaver S C, Hagenbaugh A, Bellew L A, Calisher C H. Genetic characterization of an antigenic subtype of eastern equine encephalomyelitis virus. Arch Virol. 1992;127:305–314. doi: 10.1007/BF01309592. [DOI] [PubMed] [Google Scholar]
- 79.Weaver S C, Hagenbaugh A, Bellew L A, Netesov S V, Volchkov V E, Chang G-J J, Clarke D K, Gousset L, Scott T W, Trent D W, Holland J J. A comparison of the nucleotide sequences of eastern and western equine encephalomyelitis viruses with those of other alphaviruses and related RNA viruses. Virology. 1993;197:375–390. doi: 10.1006/viro.1993.1599. [DOI] [PubMed] [Google Scholar]
- 80.Weaver S C, Kang W, Shirako Y, Rumenapf T, Strauss E G, Strauss J H. Recombinational history and molecular evolution of western equine encephalomyelitis complex alphaviruses. J Virol. 1997;71:613–623. doi: 10.1128/jvi.71.1.613-623.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weston J H, Welsh M D, McLoughlin M F, Todd D. Salmon pancreas disease virus, an alphavirus infecting farmed Atlantic salmon, Salmo salar L. Virology. 1999;256:188–195. doi: 10.1006/viro.1999.9654. [DOI] [PubMed] [Google Scholar]