Abstract
Primary biliary cholangitis (PBC) is a chronic autoimmune liver disease characterized by multilineage immune dysregulation, which subsequently causes inflammation, fibrosis, and even cirrhosis of liver. Due to the limitation of traditional assays, the local hepatic immunopathogenesis of PBC has not been fully characterized. Here, we utilize single-cell RNA sequencing technology to depict the immune cell landscape and decipher the molecular mechanisms of PBC patients. We reveal that cholangiocytes and hepatic stellate cells are involved in liver inflammation and fibrosis. Moreover, Kupffer cells show increased levels of inflammatory factors and decreased scavenger function related genes, while T cells exhibit enhanced levels of inflammatory factors and reduced cytotoxicity related genes. Interestingly, we identify a liver-resident Th1-like population with JAK-STAT activation in the livers of both PBC patients and murine PBC model. Finally, blocking the JAK-STAT pathway alleviates the liver inflammation and eliminates the liver-resident Th1-like cells in the murine PBC model. In conclusion, our comprehensive single-cell transcriptome profiling expands the understanding of pathological mechanisms of PBC and provides potential targets for the treatment of PBC in patients.
Subject terms: Systems analysis, Autoimmunity, Primary biliary cirrhosis, Chronic inflammation
Primary biliary cholangitis is a chronic autoimmune disease critically linked to immunological dysregulation but the local immune-pathogenesis is poorly understood. Here the authors present single cell transcriptomic characterisation of primary biliary cholangitis and implicates Th1 like cells in a murine model.
Introduction
Primary biliary cholangitis (PBC) is a chronic auto-immune liver disease characterized by destructive lymphocytic cholangitis, the presence of specific serum anti-mitochondrial antibodies (AMAs), and female predominance, which may subsequently result in inflammation, fibrosis, and even cirrhosis of the liver1. Every year, at least 100,000 individuals worldwide are diagnosed of PBC, and at least one in 1000 women over the age of 40 years have PBC2. Up to 40% of patients with PBC have an incomplete response to the main treatment ursodeoxycholic acid (UDCA)3. Because of the distinctive autoimmune disease features, biologically based therapies targeting immune injury have been developed. However, their effectiveness vastly varies and none has entered routine clinical use4–7. Hence, further understanding of the underlying mechanisms of the immunological pathogenesis and progression of PBC are urgently needed.
Dysregulation of multiplex immune cells is a pivotal feature of PBC. Specifically, antigen-presenting cells (APCs), such as dendritic cells (DCs), present bile duct-derived autoantigens and trigger the ensuing autoreactive immune response of CD4+ and CD8+ T lymphocytes by specifically targeting the E2 subunit of the pyruvate dehydrogenase complex located on the mitochondrial inner membrane1,8,9. AMAs produced by plasma cells recognize immunogenic complexes within apoptotic bleb of biliary epithelial cells (BECs, also called cholangiocytes). Those AMA–apotope complexes further stimulate hepatic macrophage and amplify the local inflammatory signaling, which eventually lead to bile duct damage10. Moreover, stimulated cholangiocytes express various chemokines with different functions. For instance, fractalkine recruits CX3CR1-positive mononuclear cells like CD4+ and CD8+ T cells and monocytes to amplify the local inflammatory signals11. Other cell types, including natural killer (NK) cells, natural killer T (NKT) cells, mucosal-associated invariant T (MAIT) cells and hepatic stellate cells (HSCs), also foster the local inflammatory microenvironment, which contribute to progressive ductopenia, cholestasis, and biliary fibrosis12,13.
An important obstacle for the mechanistic understanding of certain clinical diseases may be the unsatisfactory research depth due to previously available methods, especially in cell clustering and analysis. Particularly, transcriptome data from bulk tissue are not sufficient to provide more detailed information on specific cell types. Therefore, more thorough and in-depth (cell and molecular levels) data acquisition and mining methods are warranted and have been under prompt development in recent years. Single-cell RNA sequencing (scRNA-seq) is a powerful tool to investigate the intercellular transcriptomic heterogeneity and has been successfully used to delineate the pathological changes of immune cells in autoimmune liver diseases such as primary sclerosing cholangitis (PSC), autoimmune hepatitis (AIH) and PBC14–18. However, the single-cell transcriptomic features of PBC, especially the characteristics of the immune microenvironment, still require further exploration.
In this study, we applied scRNA-seq to depict the immunological landscape of liver tissues and peripheral blood mononuclear cells (PBMCs) of individuals with and without PBC. We revealed the pathological signatures and established the cellular communication network of immune and non-immune cells in PBC. Importantly, we identified a liver-resident and pro-inflammatory Th1-like cell population that was expanded in the liver, but not PMBCs, of PBC patients. We further validated our findings by using scRNA-seq analysis of 2OA-BSA immunized murine experimental cholangitis and identified the similar liver-resident Th1-like cells. Importantly, the JAK/STAT pathway was activated in both human and mouse Th1-like cells and blocking the JAK/STAT pathway in the murine model ameliorated liver inflammation and repressed Th1-like cells. Our findings expand our understanding of pathological mechanisms of PBC and provide potential therapeutic targets for PBC therapy.
Results
Single-cell transcriptome profiles of liver and PBMC in PBC patients
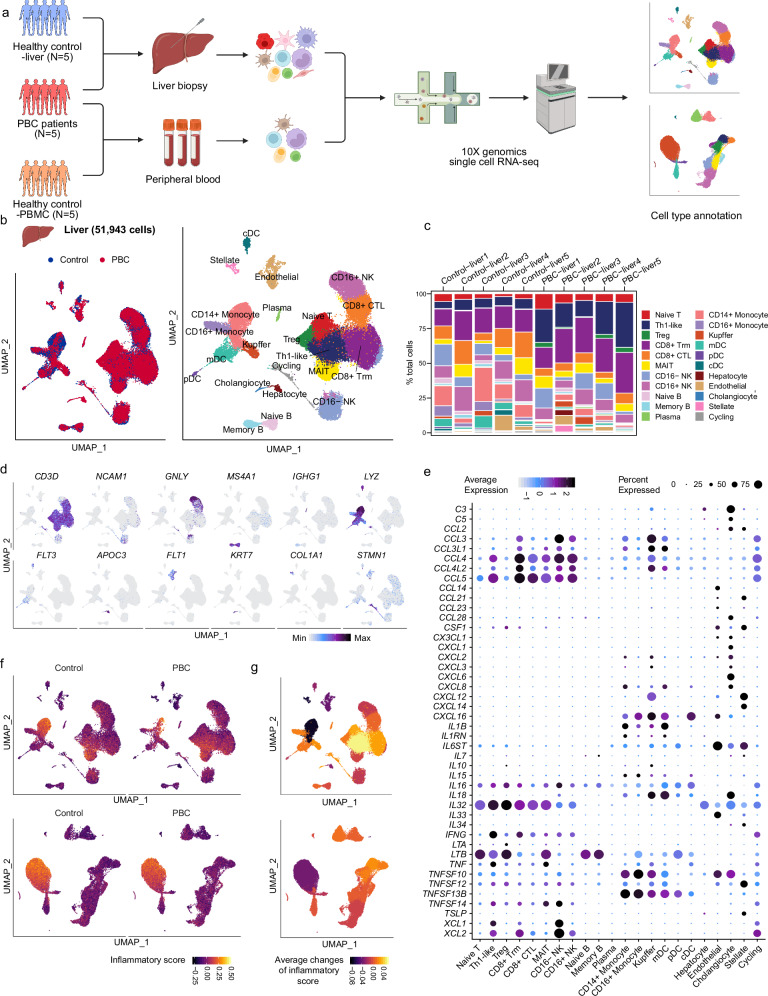
PBC, a chronic autoimmune disorder, manifests with the infiltration of immune cells in the portal tracts, thereby triggering an inflammatory response (Supplementary Fig. 1a). In order to elucidate the characteristics of these infiltrating immune cells, we performed scRNA-seq of liver tissues and PBMC from 5 PBC patients (UDCA treatment-naïve, Nakanuma Stage I~II19), 5 hepatic hemangioma patients (liver healthy control) and 5 healthy donors (PBMC healthy control) (Fig. 1a and Supplementary Data 1). After quality control, we obtained 51,943 and 47,644 single-cell transcriptomes of human liver and PBMC, respectively (Fig. 1b, Supplementary Figs. 1b–e and 2a). After dimension reduction and clustering, liver and PBMC scRNA-seq data were partitioned into 22 and 18 subpopulations, respectively, according to known marker genes (Fig. 1b, c, Supplementary Figs. 2a–d, 3 and Supplementary Data 2, 3). Cell subsets of the liver included Naïve T (CD3D+, TCF7+), Th1-like (CD4+, IFNG+), Treg (CD4+, FOXP3+), CD8+ Trm (CD8B+, GZMK+, CXCR6+), CD8+ CTL (CD8B+, GZMB+), MAIT (SLC4A10+), CD16- NK (NCAM1+, FCGR3A–), CD16+ NK (NCAM1+, FCGR3A+), Naive B (MS4A1+, IGHD+), Memory B (MS4A1+, IGHD–, IGHG1low), Plasma (IGHG1high), CD14+ Monocyte (CD14+, LYZ+), CD16+ Monocyte (FCGR3A+ LYZ+), Kupffer (CD5L+), mDC (FCGR1A+, CD1C+), pDC (IL13RA+), cDC (CLEC9A+), Hepatocyte (PCK1+, BCHE+), Endothelial (FCN2+), Cholangiocyte (KRT7+, EPCAM+), Stellate (ACTA2+, COL1A1+), and Cycling (STMN1+). Cell subsets of PBMC included Progenitor (CD34+), Naive CD4+ T (CD4+, CCR7+), Memory CD4+ T (CD4+, IL7R+), Treg (CD4+, FOXP3+), Naive CD8+ T (CD8B+, CCR7+), Memory CD8+ T (CD8B+, GZMK+), Effector CD8+ T (CD8B+, GZMB+), NK (NCAM1+, KLRD1+), Cycling T (CD3D+, MKI67+), γδT (CD3D+, TRDC+), Naive B (MS4A1+, IGHD+), Memory B (MS4A1+, IGHD–), Plasma (IGHA1+), CD14+ Monocyte (CD14+, LYZ+), CD16+ Monocyte (FCGR3A+ LYZ+), mDC (LYZ+, FLT3+), pDC (IL3RA+), and Platelet (PPBP+). Surprisingly, alterations in the distribution of immune cell subsets in PBC compared to the control group did not exhibit consistent patterns between the liver and PBMC (Fig. 1d, Supplementary Fig. 2e–g and Supplementary Data 4). In the liver of PBC patients, Th1-like and Treg cells were increased, while CD16+ monocytes were decreased. However, similar differences were not identified in PBMC. We also conducted a cell-cell communication analysis to explore the cellular interaction patterns in liver of PBC patients and observed several important markers and cell-cell interaction patterns (Supplementary Fig. 4).
Fig. 1. Single-cell transcriptome profiles of liver and PBMC in PBC patients.
a Schematic of scRNA-seq assay of liver and PBMC from PBC patients and control individuals. Figure 1a created in BioRender. Li, J. (2022) BioRender.com/u80n181. b Uniform Manifold Approximation and Projection (UMAP) showing the distribution of liver cells from PBC or control (left) and the annotated cell population (right). c Bar plots showing the cell ratio of liver cell subsets in each sample. d UMAP showing the expression of marker genes of major cell types in the liver data. e Bubble plot showing the expression of cytokines in each cell subgroup. The size of bubbles indicates the proportion of cells within a cell population that express a given gene. The color represented the average expression of each gene. f UMAP showing the inflammatory score of each cell in the liver (top graphs) and PBMC (bottom graphs) data. g UMAP showing average changes of inflammatory score of each cell subset. Significance was determined by Wilcoxon test.
In order to identify the inflammation factors in each cell subset, we examined the expression of known cytokine and inflammation-related genes within each cell subset (Fig. 1e). Thereafter, these gene sets were used to establish the cytokine score and the inflammatory score to assess the overall cytokine releasing and inflammation level of each cell (Fig. 1f, g, Supplementary Fig. 2h, i and Supplementary Fig. 5). The inflammatory score and cytokine score of multiple cell subsets in the liver, especially Th1-like and CD8+ Trm cells, was elevated in patients with PBC (Fig. 1g, Supplementary Fig. 2i and Supplementary Fig. 5a, b). However, immune cells in PBMCs of PBC patients exhibited only mild inflammation changes (Fig. 1g, Supplementary Figs. 2i and 5c, d). Together, these results suggest that these PBC patients showed increased inflammation in the liver, whereas not in the peripheral blood. Therefore, we focused on investigating the pathological cellular features of the hepatic cells in PBC.
Cholangiocytes and hepatic stellate cells showed increased expression of inflammation and matrix remodeling genes in PBC
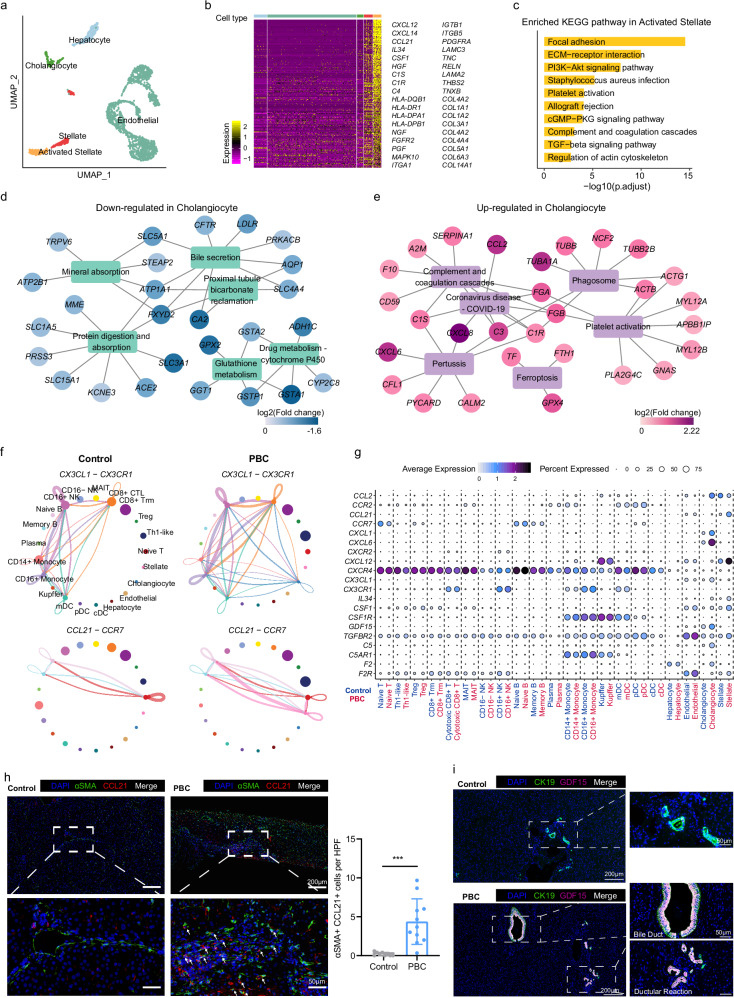
After re-clustering non-immune cells in the liver, we found a cluster of activated stellate cells that predominantly originated from the PBC group, although these cells were primarily from one PBC patient (Fig. 2a, Supplementary Fig. 6a and Supplementary Data 5). The activated stellate cells highly expressed genes related to ECM and cell adhesion, ECM receptor, immune rejection and transforming growth factor-β (TGF-β) pathways (Fig. 2b, c and Supplementary Data 5), suggesting that activated stellate cells might play vital roles in promoting extracellular matrix remodeling and eventual liver fibrosis in PBC.
Fig. 2. Characteristics of hepatic non-immune cells in PBC patients.
a UMAP showing the distribution of non-immune cells. b Heatmap showing the marker genes of activated stellate cells. The color bar at the top of the figure indicates the cell types shown in Fig. 2a. c The enriched KEGG pathway of marker genes of activated stellate cells. P value was estimated in R package clusterProfiler using one-tailed Fisher’s Exact test with Benjamini & Hochberg adjusted. Downregulated (d) or upregulated (e) genes and enriched KEGG pathways in cholangiocytes of PBC patients. f Circle diagram showing the cell-cell interaction mediated by CX3CL1-CX3CR1 and CCL21-CCR7 interactions in PBC and control. g Bubble plot showing the expression of cell communication related genes in each cell subgroup of control and PBC groups. h Representative immunofluorescent images (left) showing the distribution of DAPI (blue), αSMA (green) and CCL21 (red) in the liver biopsy samples (PBC patients n = 11, control patients n = 9) and summary data showing αSMA+ CCL21+cells per HPF in control and PBC liver (right). Each dot represents a biological replicate from different patients. Data are presented as means ± s.d. Significance was determined by two-tailed Student’s t-test, ***P < 0.001. Source data are provided as a Source Data file including exact P values. i Representative immunofluorescent images showing the distribution of DAPI (blue), CK19 (green) and GDF15 (purple) in the liver from PBC (n = 5) and control (n = 5).
Several pathways related to physiological functions of cholangiocytes were downregulated in cholangiocytes of PBC patients, including Bile secretion, Protein digestion and absorption, and Glutathione metabolism (Fig. 2d). “Biliary bicarbonate umbrella” is an alkaline barrier created by biliary secretion of bicarbonate and have protective properties against highly concentrated bile acids (BAs)20. Anion exchange protein 2 (AE2; also known as SLC4A2) is downregulated in cholangiocytes of PBC patients21. Of note, we found that genes essential for bicarbonate secretion and “biliary bicarbonate umbrella” maintenance, such as CFTR, AQP1, CA2, SLC5A1 and SLC4A420,22, were significantly depressed in cholangiocytes of PBC patients (Fig. 2d and Supplementary Data 6), which might be an important reason for injury of cholangiocytes in PBC. Moreover, the cholangiocytes of PBC patients had higher activation of inflammation-related genes (CCL2, CXCL8, C3, FGA) and pathways (Coronavirus disease—COVID-19, Complement and coagulation cascades, Ferroptosis, etc.)20, implying that cholangiocytes turned into senescence and participated in the bile duct inflammatory response (Fig. 2e). Finally, we found that some pro-inflammatory genes were specifically expressed in cholangiocytes (CCL28, CXCL1, CXCL6) and stellate cells (CCL21, CXCL14, TSLP), suggesting that these cells might play special pro-inflammatory roles in PBC (Fig. 1e).
Through cell-cell communication analysis, we found that several pathways related to immune cell migration or activation were enhanced in PBC (Fig. 2f, g and Supplementary Fig. 6b, c), including previously reported CX3CL1-CX3CR1 (cholangiocytes recruit CD8+ CTL and CD16+ NK), CCL2-CCR2 (cholangiocytes recruit monocyte and DC), IL34 and CSF1-CSR1R (cholangiocytes and stellate cells activate myeloid cells), and CXCL12-CXCR4 (cholangiocytes and stellate cells recruit all immune cells). Surprisingly, we found that CCL21, the ligand of CCR7, was specifically activated in stellate cells of PBC patients (Fig. 2f, g and Supplementary Data 5). Immunofluorescence staining results also showed the number of CCL21+αSMA+ stellate cells (activated stellate cells) was significantly elevated in PBC samples compared with normal control (Fig. 2h). These unique findings suggest that naïve T and B cells with high expression of CCR7 might be directly recruited by stellate cells to local inflammatory regions and differentiated respectively into T helper and plasma cells in the inflammatory microenvironment.
Growth differentiation factor 15 (GDF15) belongs to the TGF-β family. Accumulating data emerge that GDF15 is elevated in patients with chronic liver diseases, including PBC, and related to the process of hepatic fibrosis23–25. However, the source of GDF15 was still unclear. Our scRNA-seq data showed that GDF15 was specifically expressed in cholangiocytes and was strikingly elevated in PBC patients (Fig. 2g). Immunofluorescence staining verified that GDF15 was predominantly secreted by CK19+ cholangiocytes in the portal area of PBC patients, while the hyperplastic cholangiocytes of ductular reaction also possessed GDF15 secreting capacity (Fig. 2i), indicating the potential role of GDF15 in the hepatic regeneration and fibrosis. Collectively, these results suggest that senescent cholangiocytes and activated stellate cells played indispensable roles in amplifying inflammatory signals and promoting liver fibrosis in PBC (Supplementary Fig. 6d).
Hepatic Kupffer cells had decreased scavenger and increased inflammatory functions in PBC
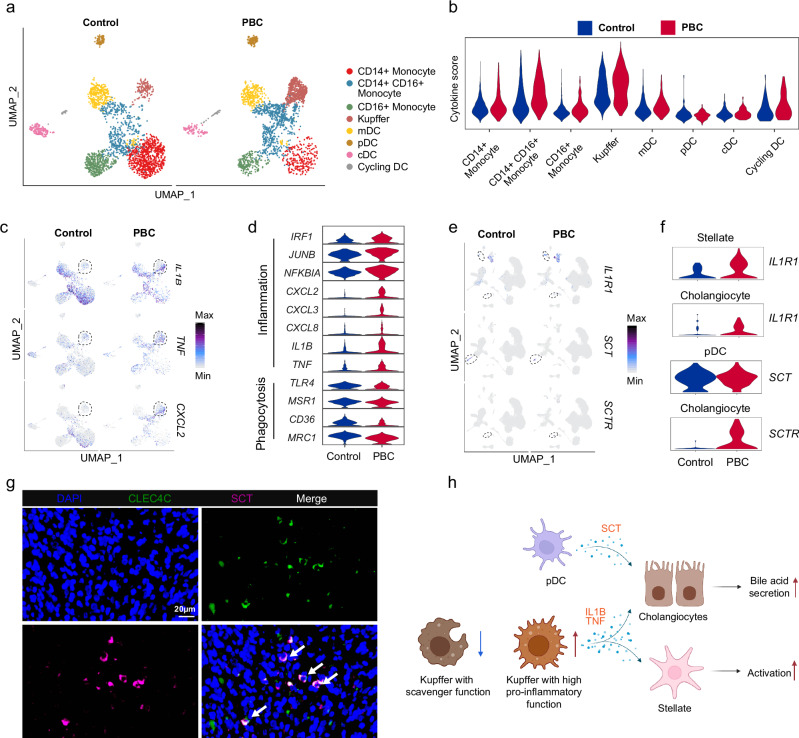
We then sub-grouped myeloid cells and annotated an additional cell subset (CD14+ CD16+ Monocyte) expressing both CD14 and FCGR3A (CD16) that was initially classified as CD14+ Monocyte (Fig. 3a, Supplementary Fig. 7a and Supplementary Data 7). We found that the proportion of CD14+ monocytes were decreased while the proportion of liver resident Kupffer cells were increased among myeloid cells (Supplementary Fig. 7b and Supplementary Data 4), although these Kupffer cells were primarily obtained from two PBC patients. Moreover, the proportion of Kupffer cells in total liver cells was also not decreased like monocytes (Fig. 1c). The bile acid receptor GPBAR1 (TGR5) was mainly expressed in myeloid cells (Supplementary Fig. 7c), suggesting that bile acids may have a specific effect on myeloid cells. Recent research revealed that the expression of inflammatory and phagocytic genes in monocytes was decreased and negatively correlated with the GPBAR1 expression in biliary atresia, suggesting that bile acids may inhibit the function of monocytes26. Thus, we further analyzed the correlation between the GPBAR1 expression and the inflammation score of monocytes and found a negative correlation (Supplementary Fig. 7d). However, there was no correlation between the expression of GPBAR1 and cell cycle genes in monocytes (Supplementary Fig. 7e–g). Moreover, the expression of inflammatory genes of monocytes did not decrease in PBC. These results suggest that bile acids might not be the main factor affecting the number of monocytes. In addition, we found that the expression of CXCR4, a key receptor related to chemotaxis, decreased in monocytes (CD14+ Monocyte: log2FC = 0.508; CD16+ Monocyte: log2FC = 0.596) (Fig. 2g), suggesting that the reduction of monocyte number may not be a result of inhibiting the proliferation by bile acids, but by the decline of monocyte migration. Interestingly, we found that the reduction of the CD16+ monocyte number was closely related to the increase of TBIL and GGT (Supplementary Fig. 7h, i).
Fig. 3. Characteristics of hepatic myeloid cells in PBC patients.
a UMAP showing the distribution of each subset of myeloid cells from the liver of control and PBC groups. b Violin plot showing the cytokine sore of each subset of myeloid cells from the liver of control and PBC groups. Significance was determined by two-tailed Wilcoxon test. c UMAP showing expressions of IL1B, TNF and CXCL2. d Violin plot showing differentially expressed genes related to inflammation and phagocytosis in Kupffer cells. UMAP (e) and violin plot (f) showing the expression of IL1R1, SCT and SCTR in pDC, stellate and cholangiocytes from the liver of control and PBC groups. g Representative immunofluorescent images of DAPI (blue), CLEC4C (green) and SCT (purple) in the liver from PBC (n = 5). h Schematic diagram depicting the signature of pDC and Kupffer cells in PBC. Figure 3h created in BioRender. Li, J. (2022) BioRender.com/a57d541.
Although the number of monocytes was decreased in the liver of PBC patients, the cytokine score and the expression of pro-inflammatory genes such as IL1B and CXCL2 in Kupffer cells were increased, as well as TNF (log2FC = 0.29, P < 0.001 and adjust-P = 1) exhibited an upward trend (Fig. 3b–d and Supplementary Data 8). Furthermore, there was a rising trend in the expression of the IL1B receptor IL1R1 (log2FC = 0.56, P < 0.001 and adjust-P = 1) in stellate cells, indicating an increased IL1 signaling pathway between Kupffer cells and stellate cells in PBC (Fig. 3e, f and Supplementary Fig. 7j). Furthermore, we also observed that the expression of scavenger function related receptors TLR4 (log2FC = 0.21, adjust-P < 0.001), MSR1 (log2FC = 0.26, adjust-P < 0.001), CD36 and MRC1 was decreased in Kupffer cells (Fig. 3d). These results imply that the enhanced proinflammatory roles of Kupffer cells may aggravate the local inflammatory response in the liver, while the inhibition of the scavenger function of Kupffer cells may slow down the clearance of senescent cholangiocytes and activated stellate cells (Fig. 3h).
The secretin (SCT)/secretin receptor (SCTR) axis is the major regulator of ductal bile secretion27. The increased stimulation of the SCT/SCTR signaling was observed in early-stage PBC patients and PBC mouse model28. SCT is primary secreted by epithelial cells, such as intestinal S cells. However, we found that SCT was also expressed in pDC, which was verified by immunofluorescence assay (Fig. 3e–g). Since SCTR was specifically expressed and upregulated in cholangiocytes, it is theoretically plausible that pDC might play a direct regulatory role through governing ductal bile secretion in PBC (Fig. 3e, f, h). To sum up, these data suggest that Kupffer cells in the liver had decreased scavenger and increased inflammatory functions in PBC.
Intrahepatic T cells of PBC patients manifested enhanced pro-inflammatory function and depressed cytotoxicity
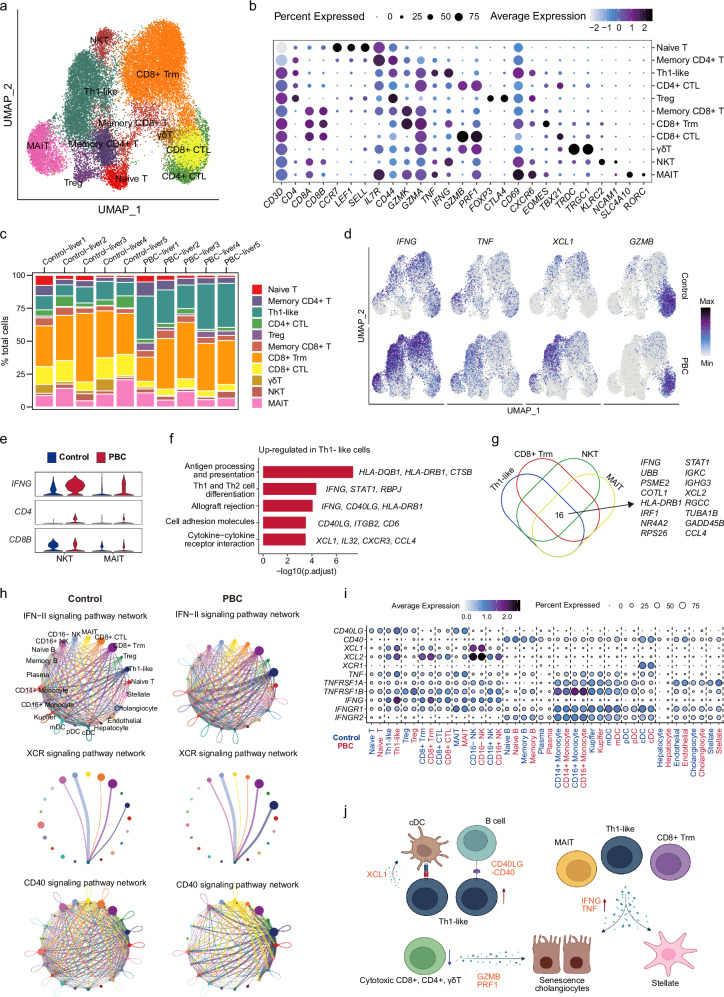
To elucidate the role of intrahepatic T cells in the immunopathology of PBC, we re-clustered and categorized T cells into 11 subpopulations, including Naive T (CCR7+LEF+), Memory CD4+ T (CD4+IL7R+IFNG-GZMB-), Th1-like (CD4+IFNG+TNF+), CD4+ CTL (CD4+GZMB+), Treg (CD4+FOXP3+), Memory CD8+ T (CD8+GZMK+), CD8+ Trm (GZMK+CD69+CXCR6+), CD8+ CTL (CD8+GZMB+), γδT (TRDC+TRGC1+), NKT (KLRC2+NCAM1+), and MAIT (SLC4A10+RORC+) (Fig. 4a, b, Supplementary Fig. 8a and Supplementary Data 9). We noticed that the proportion of Treg cells within the T cells was also increased in the PBC group (Supplementary Fig. 8b and Supplementary Data 4). To verify this result, we performed flow cytometry on liver biopsies of early stage PBC patients and healthy control samples, revealing a significant elevation of both CD25+Foxp3+ and CD25-Foxp3+ Tregs in PBC liver (Supplementary Fig. 8c).
Fig. 4. Characteristics of hepatic T cells in PBC patients.
a UMAP showing the distribution of T cells. b Bubble plot showing the expression of marker genes in T cell subsets. c Bar plots showing the cell ratio of T cell subset in each sample. d UMAP showing the expression of IFNG, TNF, XCL1, GZMB in control and PBC groups. e Violin plot showing the expression of IFNG, CD4, CD8B in NKT and MAIT cells from control and PBC groups. f Bar plot showing the enriched KEGG pathways of un-regulated genes in Th1-like cells from PBC. P value was estimated in R package clusterProfiler using one-tailed Fisher’s Exact test with Benjamini & Hochberg adjusted. g Venn plot showing the genes upregulated in Th1-like, CD8+ Trm, NKT and MAIT cells in PBC compared with the control. h Circle diagrams showing the cell-cell interaction difference of IFN-II, XCR and CD40 signaling pathways between PBC and control. i Bubble plot showing the expression of cell communication related genes in each cell subgroup of control and PBC groups. j Schematic model summarizing the characterization of T cells in liver of PBC patients. Figure 4j created in BioRender. Li, J. (2022) BioRender.com/y68a484.
To gain further insights into the distinctions between Treg cells from PBC patients and control subjects, we identified differentially expressed genes in Treg cells from PBC patients (Supplementary Fig. 8d and Supplementary Data 10). Interestingly, we noticed an upregulation in CTLA4 expression, which is associated with immune suppression, as well as an upregulation in LTB and IL32 expression, which are linked to immune activation. Furthermore, there was an increase in the expression of the transcription factor STAT3, which promotes the differentiation of Tregs into effector T cells, while the expression of FOXP1, which regulates immune suppression functions, was markedly diminished (Supplementary Fig. 8d). The simultaneous activation of these genes with opposing functions implies that Treg cells in PBC patients were in an aberrantly activated state. However, it remains to be determined whether these Treg cells exert inhibitory or pro-inflammatory roles within the immune microenvironment of PBC, necessitating further validation through functional experiments in future investigations.
The proportions of CD4+ CTL, CD8+ CTL and γδT cells with high expression of CX3CR1 and GZMB were decreased, while the proportion of Th1-like cells with strong pro-inflammatory property was significantly increased (Fig. 4c, Supplementary Fig. 8b and Supplementary Data 4). Moreover, the percentages of IFNG+ and CD4+ T cells in total T cells were increased, while the proportions of GZMB+, PRF1+ and CD8+ T cells were decreased among T cells (Fig. 4d and Supplementary Fig. 8e). Our flow cytometry results also showed the reduction of PRF1+ CD8+ T cells in PBC liver biopsy samples compared with healthy controls (Supplementary Fig. 8f), which is consistent with our findings in the scRNA-seq data (Fig. 4c and Supplementary Fig. 8e). Consistently, the cytotoxic function of CX3CR1+ CD8+ T and CX3CR1+ NK cells had been found to be depressed in biliary atresia26. Impaired cytotoxic function might prevent the clearance of senescent cholangiocytes and activated stellate cells, thereby exacerbating the disease.
Furthermore, we observed an increase in the proportion of CD4+ T cells in total T cells and a reduction in the proportion of CD8+ T cells in PBC (Supplementary Fig. 8e). This phenomenon was also observed in NKT and MAIT (Fig. 4e). The skewed ratio of CD4+/CD8+ T cells might be partially caused by the increased IL16 secretion in multiple cell subsets, such as Naive T (log2FC = 0.243, adjusted P = 0.028), Memory CD4+ T (log2FC = 0.232, adjusted P < 0.001), Th1-like (log2FC = 0.191, adjusted P < 0.001), CD4+ CTL (log2FC = 0.218, adjusted P < 0.001), Treg (log2FC = 0.447, adjusted P < 0.001), CD8+ Trm (log2FC = 0.171, adjusted P < 0.01), and MAIT (log2FC = 0.198, adjusted P < 0.001) (Supplementary Fig. 8g). Pro-inflammatory genes (IFNG, CCL4, HLA-DQB1, XCL1, STAT1, etc) involved in the regulation of antigen presentation, Th1 and Th2 cell differentiation, cell adhesion, and cytokine-cytokine receptor interaction were highly expressed in Th1-like cells in PBC (Fig. 4f). Moreover, the expression of many pro-inflammatory genes (IFNG, IRF1, STAT1, CCL4, etc) was also increased in CD8+ Trm, NKT, and MAIT cells (Fig. 4g and Supplementary Data 10).
Cell-cell interaction analysis revealed that IFN-II, XCR, TNF and CD40 signaling pathways were significantly enhanced in Th1-like cells (Fig. 4h and Supplementary Fig. 8h). Since IFNG is considered a driver of cholangitis1, the enhanced IFN-II signaling observed in Th1-like, CD8+ Trm, and MAIT cells may play crucial roles in the development of cholangitis in PBC. In addition, the interaction between XCL1, XCL2 and XCR1 related to T cell activation and antigen presentation in Th1-like cells and cDC cells was significantly enhanced (Fig. 4h, i)29,30. Since XCR was specifically expressed in cDCs, cDCs might play an indispensable role in PBC development by activating Th1-like cells. In order to further validate the results of cell-cell interactions, we performed cell-cell interaction analysis using CellphoneDB and found that the results from CellphoneDB also support our findings (Supplementary Fig. 9).
Overall, our analysis of the T cell compartment revealed that multiple T cell subsets showed enhanced inflammatory response, especially in Th1-like cells (Fig. 4j). Among the cell subsets with increased inflammation, Th1-like cells had the most remarkable changes in the inflammatory score and the cell number, which suggested that Th1-like cells played a dominant pro-inflammatory role in PBC.
Pro-inflammatory Th1-like cells were restricted to liver tissue and correlated with disease severity
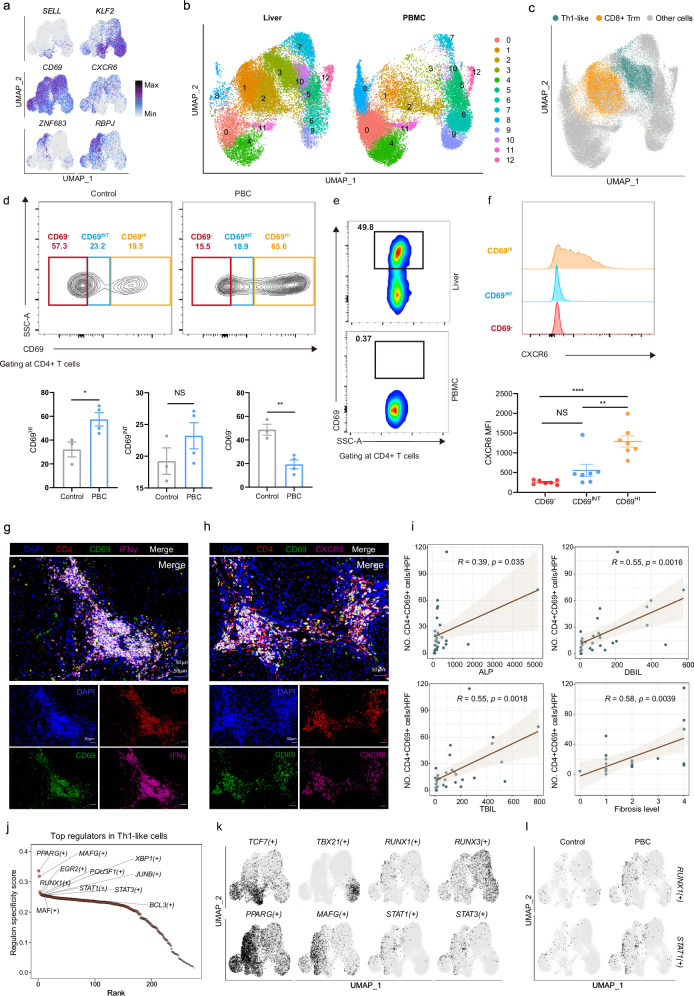
We found that Th1-like cells expressed genes related to tissue residency, such as CXCR6, CD69, ZNF683 (Hobbit), and RBPJ, and barely expressed genes related to T cell homing, like SELL (CD62L) and KLF2 (Fig. 5a and Supplementary Data 9). In order to further clarify the tissue-resident status of the Th1-like cluster, we combined and re-clustered T cells of PBMC and liver. As shown in Fig. 5b, cells of the cluster 3 were specifically derived from the liver and increased in PBC (Supplementary Fig. 8i). The major source of this cluster was from the Th1-like cells (Fig. 5c).
Fig. 5. Characteristics of liver resident Th1-like cells in PBC patients.
a UMAP showing the expression of SELL, KLF2, CD69, CXCR6, ZNF683, and RBPJ in T cells. b UMAP showing the distribution of liver and PBMC T cells. c UMAP showing the distribution of Th1-like and CD8+ Trm cells from liver. d Gating strategy showing CD69-, CD69INT and CD69HI populations. Representative flow cytometry plot for CD4+ T cell distribution in control liver, PBC liver and summary data showing % CD4+ T cells in control and PBC liver (control n = 3, PBC n = 4). Data are presented as means ± s.d. Significance was determined by two-tailed Student’s t-test, *P < 0.05, **P < 0.01. Source data are provided as a Source Data file including exact P values. e Representative flow cytometry plot for CD4+ T cell distribution in Liver, PBMC from same PBC patient. f MFI of CXCR6 expressions in the three populations in control and PBC liver (control n = 3, PBC n = 4). The results are presented as the mean with SD, and P values were calculated using one-way ANOVA **P < 0.01, ****P < 0.0001. g Representative immunofluorescent images (n = 5) of DAPI (blue), CD4 (red), CD69 (green), and IFNγ (purple) in the liver of PBC patients. Source data are provided as a Source Data file including exact P values. h Representative immunofluorescent images (n = 5) of DAPI (blue), CD4 (red), CD69 (green), and CXCR6 (purple) in the liver of PBC patients. i Scatter plot with linear regression line showing the correlation between the percentage of CD4+CD69+ cells in the liver of PBC patients and pathological features (ALP, DBIL, TBIL, Fibrosis level). ALP, alkaline phosphatase. DBIL, direct bilirubin. TBIL, total bilirubin. Grey area represented 95% confidence intervals. n = 32. The line indicates the linear regression fit with the 95% confidence interval as error band. The Pearson correlation analysis was used to assess if there is a significant correlation. j Scatter plot showing the top cell type specific regulons in Th1-like cells. k UMAP showing binarized AUC score of TCF7(+), TBX21(+), RUNX3(+), RUNX1(+), PPARG(+), MAFG(+), STAT1(+), STAT3(+). Black and grey indicate that the activity of a regulon is ‘ON’ and ‘OFF’, respectively. l UMAP showing binarized AUC score of RUNX1(+) and STAT1(+) in control and PBC groups.
To further validate the residency of Th1-like cells, we performed flow cytometry on liver biopsies from PBC patients and healthy controls, following Zania Stamataki’s strategy31. Our data showed that the proportion of CD69HICD4+ T cells was increased in PBC patients’ liver tissue, compared with healthy control (Fig. 5d). Furthermore, these CD69HICD4+ T cells were not found in the PBMC from the same PBC patient (Fig. 5e) and expressed more CXCR6 than other CD4+ T cells, which is consistent with our scRNA-seq data (Fig. 5f).
By performing immunofluorescence staining on hepatic samples of patients with PBC, we found that liver-infiltrating CD4+ T cells co-expressed the tissue-resident molecules (CD69, CXCR6) and pro-inflammatory cytokines (IFNγ, TNFα) (Fig. 5g, h and Supplementary Fig. 8j). Interestingly, most of these cells located in the portal area, suggesting that stellate cells might recruit and activate these CD4+ cells. The number of CD4+CD69+ cells showed significantly positive correlations with APL, DBIL, TBIL, IgG and the degree of fibrosis (Fig. 5i and Supplementary Fig. 8k). Instead, there was no significant correlation between the number of CD69+CD4+ T cells and GGT, AMA-M2 or the inflammation level (Supplementary Fig. 8k). These results indicated that tissue resident Th1-like cells were involved in the development of PBC and promoted the inflammation and fibrosis of hepatic tissue.
In order to better understand the transcriptional regulatory network in Th1-like cells, we calculated the relative activities of each transcription factor (TF) and its target genes (regulons) in each cell by using SCENIC32. We further calculated the regulon specific score (RSS) to find out the cell type-specific regulators, such as naïve T, cytotoxic T and CD8+ Trm corresponding to the activation of TCF7, TBX21 and RUNX3, respectively (Fig. 5j, k, Supplementary Fig. 8l and Supplementary Data 11). TFs with the most specific activation in Th1-like cells were further enhanced in PBC group, including RUNX1 and STAT1 (Fig. 5l and Supplementary Fig. 8m), implying that the activation of RUNX1 and STAT1 may play an important role in the effector function of Th1-like cells.
To verify whether the results observed in PBC are PBC-specific features, we compared our PBC data with public PSC data14 to compare the differences of immunopathology between these two diseases. After performing quality control on the PSC dataset (Supplementary Fig. 10a), we utilized our annotated T cell dataset as a reference to annotate the T cells from PSC patients. We then projected these PSC T cells onto the UMAP plot of T cells presented in Fig. 4a (Supplementary Fig. 10b). To assess the expression of marker genes, we compared the T cell subpopulations from PBC and PSC. Interestingly, we discovered that these marker genes exhibited similar expression patterns in T cell subpopulations from both PBC and PSC (Supplementary Fig. 10c). Additionally, we observed an increase in the proportion of Th1-like cells and a decrease in cytotoxic T cells (CD4+ CTL, CD8+ CTL) in PSC, which aligns with the results observed in PBC (Supplementary Fig. 10d). In brief, these data suggest that although both PBC and PSC patients exhibit an increase in Th1-like cells.
Pro-inflammatory Th1-like cells expanded in the liver of PBC mice
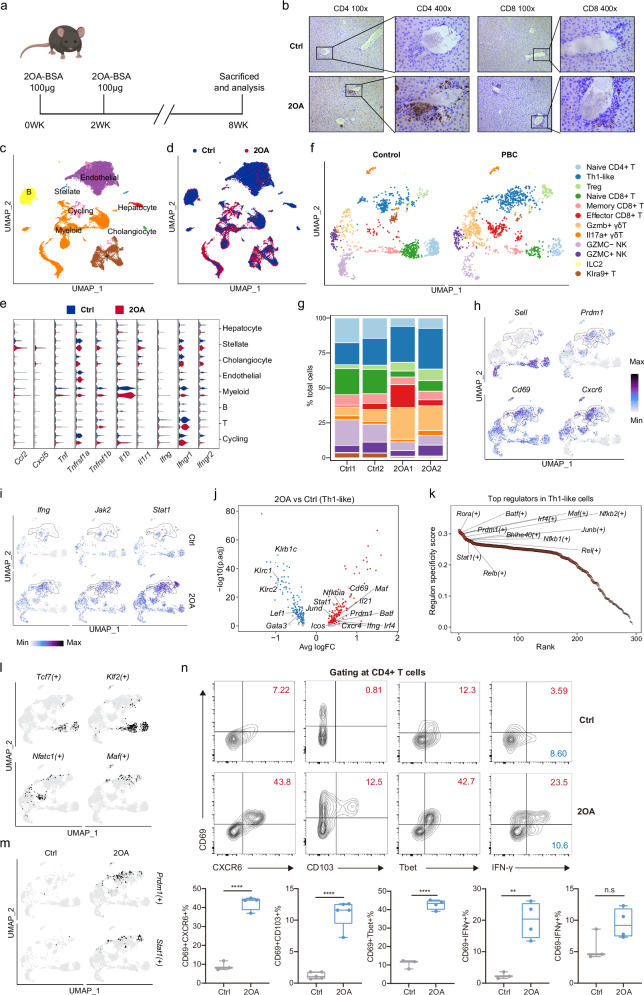
In order to investigate the mechanisms underlying PBC immunopathology, we took advantage of murine model of PBC, in which a typical portal CD4+ and CD8+ T cells infiltrations occur in mice immunized with 2-octynoic acid coupled to bovine serum albumin (2OA-BSA) (Fig. 6a, b). To enable cross-species comparison, we performed scRNA-seq on intrahepatic cells isolated from control mice and mice immunized with 2OA-BSA. After quality control, we obtained high-quality scRNA-seq data of 30,334 cells (Fig. 6c, d and Supplementary Fig. 11a). According to marker genes, cells were broadly categorized into 8 clusters, including hepatocytes (Pck1, Hamp), stellate cells (Acta2, Col1a1), cholangiocytes (Epcam), endothelial cells (Bmp2, clec4g), myeloid cells (Cd14, Lyz2), B cells (Ms4a1, Ighd), T cells (Cd3d) and Cycling cells (Mki67) (Fig. 6c, Supplementary Fig. 11b, c and Supplementary Data 12). The proportions of myeloid and T cells in the 2OA-BSA group were increased, whereas the proportions of non-immune cells such as cholangiocytes and endothelial cells were decreased (Supplementary Fig. 11d and Supplementary Data 4). We also observed expression changes of cellular interaction-related genes and found some of them were consistent with results from PBC patients (Fig. 6e and Supplementary Data 13). Expression levels of chemokines such as Ccl2 and Cxcl5 were significantly increased in stellate cells in the 2OA-BSA group. Myeloid cells showed increased expression of Tnf and Il1b while T cells showed increased expression of Ifng. Moreover, the expression of receptors of above mentioned pro-inflammatory factors showed an increasing trend in cholangiocytes or stellate cells, such as Tnfrsf1a (Stellate cell: log2FC = 0.14, adjust-P = 0.006), Tnfrsf1b (Stellate cell: log2FC = 0.12, adjust-P = 0.02; Cholangiocyte: log2FC = 0.29, P = 0.005, adjust-P = 1), Il1r1 (Stellate cell: log2FC = 0.34, P = 0.002, adjust-P = 1). Collectively, these results suggest that cholangiocytes and stellate cells might be activated by pro-inflammatory factors such as Tnf, Il1b and Ifng in this murine model and produce a variety of chemokines to recruit immune cells to amplify inflammatory signals.
Fig. 6. Identification and characterization of Th1-like cells in the liver of PBC mice.
a Schematic of the PBC mouse model generated using 2OA-BSA. Figure 6a created in BioRender. Gu, T. (2024) BioRender.com/b84j502. b Representative immunohistochemical images showing distributions of CD4+ and CD8+ T cells in the liver from ctr and 2OA-BSA groups. ctrl indicated control group. All experiments are repeated 5 times for each group. c UMAP showing annotated cell subsets. d UMAP showing the distribution of cells from control and 2OA-BSA groups. e Expression of pro-inflammatory related genes in each cell subset from control and 2OA-BSA groups. f UMAP showing the distribution of annotated T cell subsets from control and 2OA-BSA groups. g Bar plots showing the cell ratio of T cell subsets in different samples and groups. The color represents the T cell subpopulation shown in Fig. 6f. h UMAP showing the expression of Sell, Prdm1, Cd69, Cxcr6. i UMAP showing the expression of Ifng, Jak2, Stat1 in control and 2OA-BSA groups. j Scatter plot showing the differentially expressed genes in Th1-like cells from the 2OA-BSA group relative to the control group. Red dots represented upregulated genes. Blue dots represented downregulated genes. The P values were estimated in R package seurat using MAST with Bonferroni test adjusted. k Scatter plot showing the top cell type specific regulons in Th1-like cells. l UMAP showing binarized AUC score of Tcf7(+), Klf2(+), Nfatc1(+), Mafg(+). m UMAP showing binarized AUC score of Prdm1(+) Stat1(+) in control and 2OA-BSA groups. n Flow cytometry analysis (top graphs) and percentages (bottom graphs) of CD69+CXCR6+ cells (n = 3 Ctrl, n = 4 2OA), CD69+CD103+ cells (n = 5 Ctrl, n = 5 2OA), CD69+Tbet+ cells (n = 3 Ctrl, n = 4 2OA), CD69+IFN-γ+ cells (n = 3 Ctrl, n = 4 2OA) and CD69-IFN-γ+ cells (n = 3 Ctrl, n = 4 2OA) among CD4+ T cells from the liver samples of 2OA-BSA and Ctrl group mice. Each dot represents a biological replicate from different mice. Box plots show median (center line), the upper and lower quantiles (box), and the range of the data (whiskers). Data are presented as means ± s.d. Significance was determined by two-tailed Student’s t-test, **P < 0.01, ****P < 0.0001. Source data are provided as a Source Data file including exact P values.
To identify the changes in Kupffer cells in the 2OA-BSA induced PBC murine model, we divided myeloid cells into four subtypes: monocytes, Kupffer cells, DCs, and neutrophils (Supplementary Fig. 11e,f and Supplementary Data 14). In alignment with the scRNA-seq data gathered from PBC patients, we observed an upregulation of Tnf (log2FC = 0.114, adjusted P-value < 0.001) and Cxcl2 (log2FC = 0.239, adjusted P-value < 0.001) in Kupffer cells, which are known to be associated with pro-inflammation (Supplementary Fig. 11g). Conversely, Cd36 and Mrc1, genes related to phagocytosis, showed a downregulated expression. Intriguingly, we noticed a similar alteration in gene expression patterns within monocytes (Supplementary Fig. 11g and Supplementary Data 15).
After re-clustering T cells, we obtained 12 cell subsets, including Naïve CD4+ T (Cd4+, Ccr7+), Th1-like (Cd4+, Ifng, Tnf), Treg (Cd4+Foxp3+), Naïve CD8+ T (Cd8b1+, Ccr7+), Memory CD8+ T (Cd8b1+, Ccr7low), Effector CD8+ T (Cd8b1+, Gzmb+), Gzmb+ γδT (Trdc+, Gzmb+), Il17a+ γδT (Trdc+, Il17a+), Gzmc- NK (Ncr1+, Gzmc-), Gzmc+ NK (Ncr1+, Gzmc+), ILC2 (Kit+, Gata3+, Il13+), Klra9+ T (Cd3d+, Klra9+) (Fig. 6f, Supplementary Fig. 11h and Supplementary Data 16). We also found that a Th1-like population with high Ifng expression had an increasing trend in the 2OA-BSA group (Fig. 6g and Supplementary Data 4) that showed tissue resident characteristics (Sell–, Prdm1+, Cd69+, Cxcr6+) (Fig. 6h). Moreover, pro-inflammatory genes in Th1-like cells, such as Ifng, Il21, Icos, Stat1, Prdm1, were increased in the 2OA-BSA group (Fig. 6i, j and Supplementary Fig. 11i and Supplementary Data 17). The TFs activated in each cell subtype of mouse T cells were identified by SCENIC (Fig. 6k, l, Supplementary Fig. 11j, k and Supplementary Data 18). We found that Prdm1 and Stat1 were specifically activated in mouse Th1-like cells of the 2OA-BSA group (Fig. 6m and Supplementary Fig. 11k), indicating that Th1-like cells had similar transcriptional regulation characteristics in human and rodent livers. Moreover, there was an increased proportion of Treg cells observed in the 2OA group (Fig. 6g). While the levels of Ltb expression did not exhibit a significant increase in Treg cells from the 2OA group, we discovered the decreased expression of Tcf7 and Klf3 associated with T cell homeostasis, and a downward trend in Foxp1 (log2FC = 0.44, P = 0.009, adjust-P = 1) related to immunosuppression. Conversely, we found increased expression of Stat1 and Tnfrsf4, as well as a trend of elevated expression of Stat3 (log2FC = 0.75, P < 0.001, adjust-P = 1) and Ctla4 (log2FC = 0.76, P < 0.001, adjust-P = 1), which aligned with the patterns observed in the scRNA-seq data from PBC patients (Supplementary Fig. 11i and Supplementary Fig. 8d). These findings imply that Treg cells may also exist in an abnormally activated state in the 2OA-BSA murine model.
To validate our findings in the liver resident Th1-like cells, we performed flow cytometry on murine intrahepatic cells. Importantly, FACS analysis revealed that the proportion of hepatic CD69+CXCR6+ CD4+ T cells was substantially increased in 2OA-BSA mice compared to control mice (Fig. 6n and Supplementary Fig. 12a). Moreover, frequencies of CD4+CD69+Tbet+ and CD4+CD69+IFNγ+ T cells were increased in the liver of 2OA-BSA mice (Fig. 6n). However, the proportion of CD4+CD69-IFNγ+ T cells did not change significantly. Strikingly, CD4+CD69+ T cells of 2OA-BSA mice manifested a higher level of IFNγ compared to CD4+CD69- T cells of 2OA-BSA mice and CD69+CD4+ T cells from control mice (Fig. 6n and Supplementary Fig. 12b), suggesting that CD4+ T cells with the tissue-resident surface molecule signature expanded in the liver of 2OA-BSA mice and those cells manifested distinct Th1-like characteristics and proinflammatory ability by secreting IFNγ. We further examined hepatic CD4+ T cells using a FACS analysis to check the expression of various cell surface molecules, along with CD69. FACS analysis of expressions of CD69, CD44, and CD62L revealed that the majority of CD69+CD62L– CD4+ T cells expressed a high level of CD44 and that the frequency of CD69+CD62L–CD44+ CD4+ Trm cells was significantly increased in 2OA-BSA mice at the expense of CD69–CD62L+CD44– CD4+ naïve T (Tn) cells (Supplementary Fig. 12c). Expansion of the CD69+CD62L– cell population was also observed in the CD8+ T cell subset (Supplementary Fig. 13a, b). Overall, these data indicate that mouse cells similar to Th1-like cells that we identified in PBC patients also participated in the cholangitis development in this murine model and might overlap with formerly reported tissue resident memory T cells.
Blocking the JAK/STAT pathway relieved bile duct damage of PBC mice and ameliorated liver resident Th1-like cells
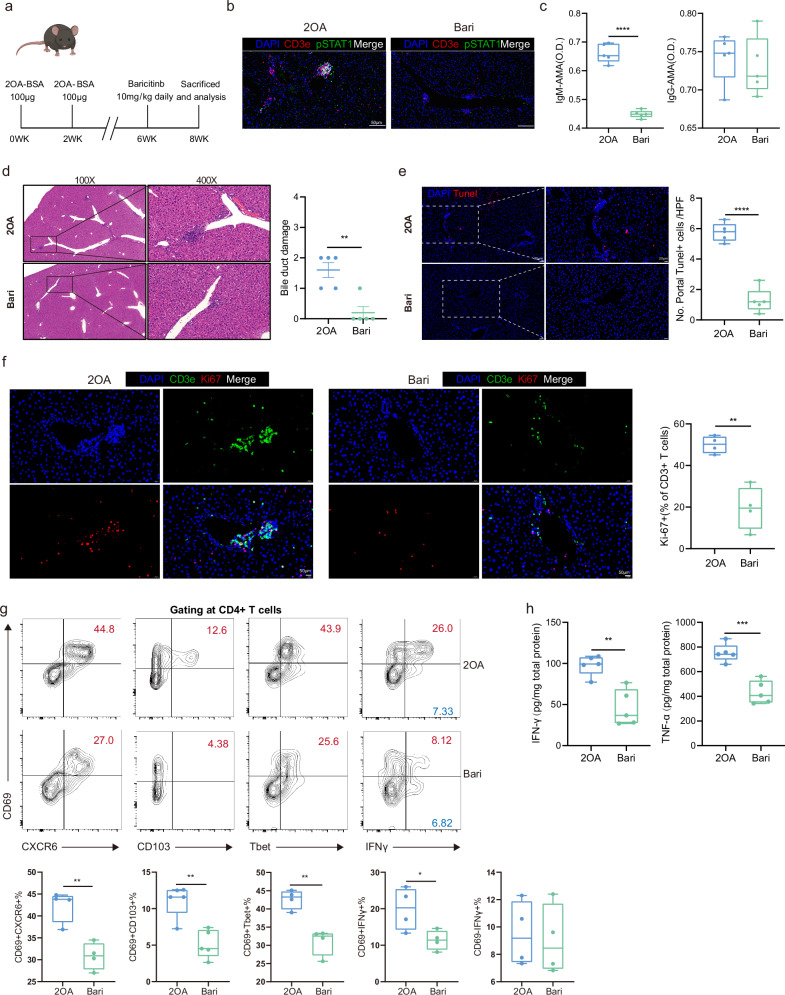
Based on our scRNA-seq analysis of pathogenic liver resident Th1-like cells and the potential involvement of the JAK/STAT pathway we identified in both human PBC patients and a rodent PBC model, we hypothesized that baricitinib, a JAK1/JAK2 inhibitor approved for the treatment of rheumatoid arthritis, might block the immunological disorder caused by Th1-like cells. After a two-week administration of baricitinib, pSTAT1+ levels in hepatic CD3e+ T cells were decreased (Fig. 7a, b). Serum IgM-AMAs were significantly decreased in baricitinib treated group (Fig. 7c). The infiltration of inflammatory cells in the portal area was significantly reduced and the necrosis of cholangiocytes was relieved, as shown by HE staining and counting of portal TUNEL+ cells (Fig. 7d, e and Supplementary Fig. 12d). We examined whether JAK-STAT blockade affected the expansion of tissue-infiltrating T cells. About 50% of hepatic CD3+ T cells expressed the proliferation marker Ki-67 in the 2OA-BSA group, indicating the contribution of clonal expansion to the inflammatory progression (Fig. 7f). The baricitinib treatment significantly restrained the T cell expansion (Fig. 7f). Local protein levels of IFNγ, TNFα were also significantly decreased in baricitinib treated group (Fig. 7h).
Fig. 7. Effects of baricitinb on cholangitis in PBC mice.
a Schematic of baricitinb treatment in 2OA-BSA induced PBC mouse model. Figure 7a created in BioRender. Gu, T. (2024) BioRender.com/b84j502. b Representative immunofluorescent images (n = 5) of DAPI (blue), CD3e (red) and pSTAT1 (green) in the liver of 2OA-BSA and Bari (baricitinb treatment group) mice. c Box plots showing concentrations of IgM-AMA and IgG-AMA in the blood from 2OA-BSA (n = 5) and Bari (Baricitinb treatment group) mice (n = 5). Each dot represents a biological replicate from different mice. Data are presented as means ± s.d. Significance was determined by two-tailed Student’s t-test, ****P < 0.0001. d Representative hematoxylin-eosin (H&E) staining images showing bile duct damage in 2OA-BSA and Bari mice (left) and box plot showing the difference of bile duct damage between 2OA-BSA (n = 5) and Bari (n = 5) mice (right). Each dot represents a biological replicate from different mice. Data are presented as means ± s.d. Significance was determined by two-tailed Student’s t-test, **P < 0.01. Source data are provided as a Source Data file including exact P values. e Representative immunofluorescent images of DAPI (blue) and TUNEL (red) in the liver of 2OA-BSA and Bari mice (left) and boxplot showing the difference of TUNEL+ cell number between 2OA-BSA (n = 5) and Bari (n = 5) mice liver samples (right). Data are presented as means ± s.d. Significance was determined by two-tailed Student’s t-test,****P < 0.0001.f Representative immunofluorescent images (n = 4) of DAPI (blue), CD3e (green) and Ki67(red) in the liver of 2OA-BSA (left) and Bari (middle) mice and boxplot showing the proportion of Ki67+ cells in CD3e+ T cells from the liver samples of 2OA-BSA (n = 4) and Bari (n = 4) mice (right). Data are presented as means ± s.d. Significance was determined by two-tailed Student’s t-test, **p < 0.01. g Representative flow cytometry analysis images (top graphs) and percentages (bottom graphs) of CD69+CXCR6+ cells (n = 4 2OA, n = 4 Bari), CD69+CD103+ cells (n = 5 2OA, n = 5 Bari), CD69+Tbet+ cells (n = 4 2OA, n = 4 Bari), CD69+IFN-γ+ cells (n = 4 2OA, n = 4 Bari) and CD69–IFN-γ+ cells (n = 4 2OA, n = 4 Bari) among CD4+ T cells from the liver of 2OA-BSA and Bari mice. The samples of 2OA-BSA are the same samples shown in Fig. 6n. Each dot represents a biological replicate from different mice. Data are presented as means ± s.d. Significance was determined by two-tailed Student’s t-test, *P < 0.05, **P < 0.01. h Boxplots showing concentrations of IFN-γ and TNFα in the liver samples of 2OA-BSA (n = 5) and Bari (n = 5) mice. Each dot represents a biological replicate from different mice. Data are presented as means ± s.d. Significance was determined by two-tailed Student’s t-test, **P < 0.01, ***P < 0.001. In c, e and f–h, box plots show median (center line), the upper and lower quantiles (box), and the range of the data (whiskers). Source data are provided as a Source Data file including exact P values.
Compared with the untreated group, the proportions of CD4+CD69+CXCR6+, CD4+CD69+CD103+, CD4+CD69+T-bet+ and CD4+CD69+IFNγ+ T cell populations decreased significantly, whereas the proportion of CD4+CD69–IFN+ T cells was similar in the baricitinib treated group (Fig. 7g). These results suggest that the alleviation of hepatic inflammation was linked with the declined occurrence of IFNγ-secreting Th1-like cells and such therapeutic effects were more likely rendered by the CD69+ liver-resident group. Moreover, the proportion of Trm cells in the baricitinib-treated group was also significantly decreased, with a major elevation in the proportion of Tn cells and relatively slight elevation in the proportion of Tcm cells (Supplementary Fig. 12e). Cytometric analysis of CD8+ Trm revealed an insignificant effect on IFNγ-secreting CD8+ Trm cluster, which might contribute to the difference in the sensitivity to the 2-week baricitinib treatment between CD8+ and CD4+ T cells (Supplementary Fig. 13a, b). Conclusively, these data suggest that baricitinib could alleviate mouse cholangitis in the model immunized by 2OA-BSA.
Discussion
Currently, the comprehensive and finely-tuned immunological interactions between non-parenchymal cells and immune cells in the PBC inflammatory microenvironment have not been fully understood. Here, we took the advantage of scRNA-seq to investigate the pathological characteristics of PBC at single cell level. Our study illustrated mechanisms of several different cell types in early-stage PBC pathogenesis. For instance, cholangiocytes and HSCs showed pro-inflammatory and pro-fibrogenic roles with enhanced cell-type specific chemokines and fibrotic signaling, while Kupffer cells manifested reinforced pro-inflammatory roles but reduced scavenger function-related genes. Interestingly, cytotoxic T cells were decreased while pro-inflammatory T cells were increased in PBC. Together, our single-cell transcriptomics-based approach provided a powerful resource for investigating the pathological mechanism of early-stage PBC.
Recently, studies have utilized scRNA-seq to explore the aberrant transcriptional signatures in the livers of PBC patients and mouse models of PBC16–18. Ref. 17, conducted scRNA-seq on an experimental cholangitis mouse model, indicating that the dnTGF-βRII model is primarily driven by CD8 T cells, which may differ from the local immune environment in early-stage PBC. Recently, ref. 18, performed scRNA-seq on five early-stage PBC patients, focusing on the role of DUOX2+ACE2+ small cholangiocytes and B cell function. They found the DUOX2+ACE2+ small cholangiocytes specifically diminished in patients with PBC. Our data also demonstrate that the expression levels of DUOX2 and ACE2 are significantly reduced in cholangiocytes from the PBC cohort (Supplementary Data 6). Our findings provide further evidence supporting the hypothesis that the DUOX2+ACE2+ cholangiocyte subpopulation may represent the specific cellular targets in PBC. Additionally, ref. 16, analyzed PBC data from cirrhotic explanted advanced-stage PBC tissue sections, comparing them to healthy control and PSC. Interestingly, they also observed an increased proportion of activated HSCs in both PBC and PSC samples, with elevated expression of CCL21. The spatial transcriptomics and immunofluorescence data revealed co-localization of stellate cells with CD4+ T cells in PSC samples. However, their data indicated that IFNG was predominantly upregulated in CD8+ T and NK cells, which might be attributed to the differences in disease progression of PBC.
Although not belonging to immune cells, cholangiocytes, and HSCs have been considered to participate in the inflammatory signaling cascade of PBC1, but the detailed mechanisms are elusive. In this study, we dissected a chemotactic interactome among cholangiocytes, HSCs, and intra-scar inflammatory cells, reinforcing that these cells play indispensable roles in the inflammatory signaling network in the liver. Additionally, “Biliary bicarbonate umbrella” maintenance is important for protecting bile duct cells from bile acid damage20,22. Former study revealed the deficiency of bicarbonate transporter AE2 in PBC13, which may exert its effect by breaking pH homeostasis of bile duct. Here, we found that several other anion exchangers were downregulated in the liver of PBC patients, which provides candidate molecules for “Biliary bicarbonate umbrella” maintenance. However, further mechanistic study is necessary to define key up-stream regulators for the downregulation of bicarbonate-releasing related anion exchangers, which may generate therapeutic targets.
Accumulating evidences show that, as a part of immune sensing network, heterogeneous tissue-resident T cells monitor local environmental changes across the human body33. The activation of those cells upon recognizing innocuous antigens or self-antigens can cause severe damage to multi organs, indicating its pivotal role in the autoimmunity at different anatomical sites34–37. Several recent studies have identified the pro-inflammatory role of hepatic resident T cell subsets in autoimmune liver disease. For example, a significant increase of IFNγ and GZMB-secreting CD103+ CD8+ Trm cells was reported to positively correlate with the disease severity in AIH patients38. The up-to date data showed that the number of those cells at baseline was also linked with responsiveness to UDCA in PBC39. Very recently, Zania Stamataki et al., have demonstrated that high CD69 expression marks a CD4+ T cell population capable of long-term residence within the liver and robust production of Th1 cytokine responses31. Here, we firstly utilized scRNA-seq and reported a dramatic expansion of pathogenic Th1-like population with high expression of CD69, CXCR6 and ZNF683 (Hobbit)40–43 that were related to tissue residency and differentiation. This cell cluster secreted IFNγ which was identified as a critical cytokine for the pathogenesis of PBC and its overexpression can cause bile duct injury in mice, similar to in PBC patients44,45. Besides, IL12-JAK-STAT4 pathway orchestrated Th1 activation and drove inflammation in human body46. Several susceptibility loci related to this pathway have been revealed by genome-wide association studies (GWAS) of PBC cohorts, which support the importance of Th1 activation in PBC progression46,47. Our results demonstrated that elevated frequencies of hepatic CXCR6+CD69+IFNγ+ Th1-like cells can be found in PBC patients with both early and advanced stages of liver fibrosis and the level of CD69+CD4+ T cells positively correlated with clinical biochemical markers (ALP, TBIL, DBIL and IgG) in PBC. Recently, A previously uncharacterized population of liver-resident naive-like CD4+ T cells were identified to contribute to the pathogenesis of PSC by single-cell RNA sequencing14. By re-analyzing their PSC data, we found an increase in the proportion of Th1-like cells, which is consistent with the trend we observed in PBC. While, Th1-like cells in PBC patients seems like have stronger pro-inflammatory characteristics. Data from our murine PBC model further supported the destructive ability of Th1-like cells with typical tissue-resident characterization by secreting abundant IFNγ. Regarding the devasting role of IFNγ and the skewed CD4/CD8 ratio, we assume that the prominent priming of liver resident Th1-like cells shouldn’t be neglected in the immunopathology of PBC. Together, our results suggest that the liver-resident Th1-like cells could be an important driver of PBC and depletion of Th1-like cells may be beneficial to improving cholangitis.
For PBC therapy, since about 40% of patients do not show satisfactory response to UDCA, these patients need additional treatments to reach therapeutic goals3 and to prevent their PBC from progressing into hepatic fibrosis or even cirrhosis48,49. Because PBC is an autoimmune liver disease, traditional immunomodulatory drugs such as methotrexate, colchicine and budesonide have been tested to treat “UDCA non-responders”. However, several clinical trials failed to provide sufficient evidence to support the usage of those immunosuppressive agents for treating PBC4–7. By identifying cell-type specific regulators, we found that the JAK/STAT pathway of the liver-resident Th1-like cells was activated in both PBC patients and the 2OA-BSA murine PBC model. JAK inhibitors have been used in the treatment of multiple autoimmune inflammatory diseases50. Here we demonstrated that a two-week administration of the JAK inhibitor baricitinb into the 2OA-BSA cholangitis mouse model resulted in obvious histological remission with lower hepatic cytokine levels and reduced number of liver-resident Th1-like cells. These data suggest that baricitinib could be applied to repress liver-resident Th1-like cells in future therapies of PBC. Further clinical trials are urgently needed to investigate the safety and efficacy of baricitinb and other JAK inhibitors to treat UDCA non-responders PBC patients.
Due to limitations in our sample size, we have made certain assumptions about intriguing findings without experimental validation. These include the correlation between monocyte proportions and TBIL levels, alterations in the pro-inflammatory and phagocytic functions of Kupffer cells, changes in the number of cytotoxic T cells, and their association with disease progression. Here, we reported an elevation of Tregs in both early-stage PBC patients and 2OA -BSA murine model. Former studies also have found that the number of FOXP3+ Tregs was increased in the inflammation sites of an organ-specific autoimmune disease, but did not change in the peripheral blood51. However, due to the restriction of Tregs’ number acquired via liver biopsies, we are currently unable to verify whether these Treg cells undergo pro-inflammatory functional changes. In the future, collecting a larger sample size and different stages of PBC samples will enable us to conduct further pathological correlation analysis and functional experiments, providing deeper insights into the pathogenesis mechanisms of PBC.
In summary, our study provides the transcriptome atlas of the liver and PBMC of PBC patients at the single-cell level and improves the understanding of the molecular mechanisms underlying the occurrence and development of PBC. Moreover, we have identified a liver-resident Th1-like cell population in PBC, which provides a potential therapeutic target. Finally, the potential therapeutic effect of baricitinb in the PBC animal model provides supports for further clinical studies for its expanded indication.
Methods
Study subjects
Local approval for procuring human liver tissue and blood samples for scRNA-seq and histological analysis was obtained from the First Affiliated Hospital of Zhejiang University Ethics Committee, following principles of the Declaration of Helsinki (Ethics approval number: 2021-39). All subjects provided written informed consent.
For scRNA-seq assay, healthy background non-lesion liver tissue (n = 5) was obtained intraoperatively from patients undergoing surgical liver resection for hepatic hemangioma at the Hepatobiliary and Pancreatic Unit, Department of Clinical Surgery, the First Affiliated Hospital of Zhejiang University. Patients with a known history of chronic liver disease, abnormal liver function tests or those who had received systemic chemotherapy within the last four months were excluded from this cohort.
PBC liver tissue (n = 5) was obtained from patients (UDCA treatment-naïve, AMA-M2 positive, ANA negative, GP210 negative) undergoing diagnostic liver biopsy at the Department of infectious diseases, the First Affiliated Hospital of Zhejiang University. Blood from PBC patients was obtained before the diagnostic liver biopsy. All patients met the standard diagnostic criteria of PBC, as described elsewhere49. Scheuer scoring system was used to evaluate fibrosis and inflammation of liver biopsies52, Nakanuma staging system was used to determine the PBC stage19.
Blood from healthy control donors (n = 5) were obtained from medical examination center, the First Affiliated Hospital of Zhejiang University. The clinical characteristics of 15 individuals are noted in Supplementary Data 1. For immunohistochemistry study, liver samples from patients with PBC were derived from ultrasound-guided needle liver biopsies and liver biopsies of liver transplantation recipients; normal liver tissues collected from 5 patients with hepatic hemangioma were used as the normal control group. The clinical characteristics of PBC biopsies used in correlation analysis of CD69+CD4+ T cells and clinical statistics were ALP (n = 30, 486.4 ± 937.5 μ/L), GGT (n = 30, 368.1 ± 302.5 μ/L), TBIL (n = 30, 170.2 ± 195.8 μmol/L), DBIL (n = 30, 133.0 ± 154.5 μmol/L), IgG (n = 24, 1453.8 ± 611.4 mg/dL), AMA-M2(n = 26, 2.3 ± 1.6), fibrosis level (n = 23, 2 ± 1.2), and inflammation level (n = 23, 2.6 ± 0.6).
For flowcytometry 3 healthy background non-lesion liver tissue and 4 PBC liver tissue were acquired the same way as aforementioned. The clinical characteristics of heathy control were ALT (n = 3, 20 ± 8.5 μ/L), AST (n = 3, 21.7 ± 6.7 μ/L), ALP (n = 3, 75.7 ± 34.3 μ/L), GGT (n = 3, 32.3 ± 32.9 μ/L), TBIL (n = 3, 5.6 ± 2.1 μmol/L), DBIL (n = 3, 2.5 ± 0.90 μmol/L). 4 PBC liver tissue was obtained from patients (UDCA treatment-naïve, AMA-M2 positive, ANA negative, GP210 negative) were acquired the same way as aforementioned. The clinical characteristics of PBC were ALT (n = 4, 47.8 ± 32.8 μ/L), AST (n = 4, 45 ± 23,2 μ/L), ALP (n = 4, 171.5 ± 53.8 μ/L), GGT (n = 4, 107.8 ± 113.4 μ/L), TBIL (n = 4, 14.2 ± 7.7 μmol/L), DBIL (n = 4, 5.5 ± 1.80 μmol/L).
To validate the activation of HSCs in PBC, 9 healthy background non-lesion liver tissue and 11 PBC liver tissue were acquired the same way as aforementioned. The clinical characteristics of heathy control were ALP (n = 9, 68.4 ± 17.0 μ/L), GGT (n = 9, 16.4 ± 5.2 μ/L), TBIL (n = 9, 10.0 ± 4.7 μmol/L), DBIL (n = 9,3.7 ± 1.7 μmol/L), IgG (n = 1,1837.0 mg/dL), AMA-M2(n = 0), fibrosis level (n = 0), and inflammation level (n = 0). 11 PBC liver tissue was obtained from patients (UDCA treatment-naïve, AMA-M2 positive, ANA negative, GP210 negative) were acquired. The clinical characteristics of PBC were ALP (n = 11, 295.3 ± 201.2 μ/L), GGT (n = 11, 208.6 ± 162.3 μ/L), TBIL (n = 11, 12.0 ± 5.6 μmol/L), DBIL (n = 11, 7.1 ± 4.6 μmol/L), IgG (n = 10, 1516.6 ± 592.3 mg/dL), AMA-M2 (n = 11, 2.0 ± 1.3), fibrosis level (n = 11,1.0 ± 0.0), and inflammation level (n = 11, 1.5 ± 0.7).
The upper limit of normal biochemical features is listed as below. ULN of ALT = 50 μ/L, ULN of AST = 40 μ/L, ULN of ALP = 125 μ/L, GGT = 60 μ/L, ULN of TBIL = 26 μmol/L, ULN of DBIL = 8 μmol/L, ULN of IgM = 220 mg/dL, ULN of IgG = 1740 mg/dL.
Human liver tissue dissociation and PBMC isolation
Human liver tissue was obtained from liver fine needle aspiration biopsy of patients with PBC. Control human liver tissue was the non-involved surrounding tissue obtained from patients undergoing partial hepatectomy for hepatic hemangioma. Samples were collected from the First Affiliated Hospital of Zhejiang University. The freshly human livers were kept on ice in RIPM1640 + 10%FBS solution. Single-cell suspension was obtained by grinding the tissue and sequentially digested with 37 °C pre-warmed solution containing 10 mM HEPES (Sigma) and 400 μg/mL Liberase TM (Roche). Samples were enzymatically dissociated for 10 min at 37 °C in a water bath, then put on ice with EDTA addition to a final concentration of 10 mM to stop Liberase digestion. After centrifuging at 800 × g for 5 min, the supernatant was decanted, and the pellet was resuspended in ACK lysis buffer to lyse the red blood cells. Cells were washed twice in PBS + 2% FBS and filtered through 40 μm mesh. PBMCs were isolated from blood using human lymphocyte separation medium. The samples collected at different time points were frozen and stored in liquid nitrogen, awaiting their use for scRNA-seq experiments.
Multiplex immunochemistry staining, immunohistochemistry and TUNEL staining
Opal Polaris 7-Color Manual IHC Kit (NEL861001KT) was used for multiplex Immunohistochemistry staining. Multiplex immunochemistry staining was performed following the manufacture’s protocol with minor modification. Liver sections were dewaxed and rehydrated. After appropriate antigen retrieval, slides were blocked with blocking buffer for 10 min at room temperature (RT). For multiplex immunohistochemistry staining of CD69+CXCR6+CD4+ T cells, the primary antibody, anti-CD69 (ab233396, Abcam) was incubated at RT for 2 h. The slides were washed 3 × 2 min in TBST at RT with agitation, followed by incubation in Opal Polymer HRP Rb for 10 min at RT, Rinsing and washing with TBST again. After Drained off excess wash buffer, each slide was pipetted with 300 μL of Opal520 (Green) working solution and incubated at RT for 10 min. Washed and rinsed the slides in AR buffer. Microwaved the slides filled with AR buffer to strip the primary-secondary-HRP complex. Repeated the procedure to introduce the anti-CXCR6 (ab273116, Abcam) primary antibody and the Opal690 (Violet). Then introduced the anti-CD4 (14-2444-82, Invitrogen) primary antibody and Opal570 (Red). Finally applied the DAPI Working Solution. Slides were imaged using Olympus VS200 and BX63. Other antibodies used for human multiplex immunohistochemistry staining included: anti-IFNγ (ab231036, Abcam), anti-TNFα (ab270264, Abcam), anti-CK19 (ab7754, Abcam), anti-GDF15 (ab206414, Abcam), antiCCL21 (ab23116, Abcam), anti-αSMA (ab124964, Abcam), anti-CLEC4C (ab239078, Abcam), anti-Secretin (bs-0088R, Bioss). Antibodies used for the murine multiplex immunochemistry staining included: Ki67(ab15580, Abcam), anti-Phospho-Stat1 Ser727 (8826, CST), CD3 (ab16669, Abcam). Anti-CD4 (ab183685, Abcam), anti-CD8 (ab217344, Abcam) was used for murine 3,3’-Diaminobenzidine (DAB) IHC. In Situ, Cell Death Detection Kit, POD (11684817910, Roche) was used for TUNEL staining following the manufacture’s protocol. For quantification, five representative high-power fields were selected and single and/or double-positive cells were counted using ImageJ (National Institutes of Health).
Animals, induction of cholangitis and Baricitinib treatment
Adult male C57BL/6JCrl mice aged 8–10 weeks were purchased from Charles River (Shanghai). Mice were housed under specific pathogen-free conditions at the Zhejiang University. All experimental protocols were approved by the First Affiliated Hospital of Zhejiang University Ethics Committee. Two octynoic acid was acquired from Aladin(B301308) and subsequently conjugated to BSA (SRE0096, Sigma-Aldrich) via a refined procedure. Initially, 2OA was solubilized in anhydrous dimethyl ether (506008, Sigma-Aldrich), and NHS (130672, Sigma-Aldrich) was introduced. The mixture was chilled to 0 °C and agitated for 20 min. Subsequently, dicyclohexylcarbodiimide (379115, Sigma-Aldrich) was added, and the reaction mixture was gradually warmed to room temperature overnight. After filtration, the solution underwent roto-evaporation under reduced pressure for concentration, redissolution in ethy lether(1696894, Sigma-Aldrich), and sequential washes with water, 1 M NaHCO3(792519, Sigma-Aldrich), and brine. It was then dried over magnesium sulfate (203726, Sigma-Aldrich), filtered, and concentrated once more. The purified product was isolated via flash chromatography employing a 30% ethyl acetate/hexane(270989/485624, Sigma-Aldrich) mixture.The NHS-activated 2OA was dissolved in DMSO and efficiently coupled to the lysine residues of BSA. The reaction was allowed to proceed for 3 h, followed by dialysis against PBS (P2272, Sigma-Aldrich) to remove excess reagents and byproducts. Murine model of autoimmune cholangitis was established as M. Eric Gershwin reported53, 100 μg 2OA-BSA (in 50 μl PBS) were emulsified with 50 μl of Complete Freund’s Adjuvant (CFA, Sigma-Aldrich) and injected intraperitoneally into 6-week-old female mice. Two days after the initial immunization with 2OA-BSA, mice received 100 ng of pertussis toxin (List Biological Laboratories) in 100 μl PBS by I.P. After two weeks, the mice were re-boosted with 100 μg 2OA-BSA (in 50 μl PBS) emulsified with 50 μl of Incomplete Freund’s Adjuvant (IFA, Sigma-Aldrich) I.P. Mice received, baricitinib (10 mg/kg) in 0.5% methylcellulose, or vehicle only (0.5% methylcellulose), as indicated, once daily per oral gavage 6 weeks after initial 2OA-BSA immunization for 2 weeks. The dosage of baricitinib applied were based on previous works, as exemplified by ref. 54.
Detection of serum anti-PDC-E2 antibodies, hepatic IFNγ and TNFα
Serum titers of IgM and IgG anti-PDC-E2 autoantibodies were measured by ELISA using purified recombinant PDC-E2 to coat the ELISA plates. The serum samples were tested at a dilution of 1:250. The liver tissue was lysed and homogenized. Supernatants of the sample were collected, and the total protein level was quantified by BCA Kit (23227, Thermo). Then the appropriate amount of sample was used for the detection of IFNγ and TNFα level via IFNγ ELISA kit (ELM-IFNg-1, RayBiotech) and TNFα (EKM-TNFa-1, RayBiotech).
Histopathology scoring of H&E
Scoring of murine PBC hepatic inflammation was performed on coded H&E-stained sections of liver using a set of indices by a “blinded” pathologist; these indices quantitated the degree of portal inflammation, liver inflammation, bile duct damage. Each section was scored as either 0 ¼ no significant change, 1 ¼ minimal, 2 ¼ mild, 3 ¼ moderate, and 4 ¼ severe pathology. Details of this scoring system have been described55.
Flow cytometry
Single-cell suspensions were prepared, and surface stained in fluorescence-activated cell sorting (FACS) buffer (phosphate-buffered saline + 1.5% fetal bovine serum) with fluorescence-conjugated antibodies on ice for 20–30 min. For the detection of IFNγ, liver mononuclear cells were incubated with Leukocyte Activation Cocktail, with BD GolgiPlug (550583, BD Biosciences) at 37 °C for 4 h, then incubated with surface antibodies, permeabilized with Cytofix/Cytoperm reagent (562574, BD Biosciences), and then stained with intracellular antibodies. Anti-CD69 antibody was pre-incubated before the procedure of activation to exclude the possibility of activation-induced elevation of CD69. Antibodies used for murine FACS analysis included: BV510 Hamster Anti-Mouse CD45 (563891, BD); BV786 Hamster Anti-Mouse CD3e (564379, BD); BV786 Hamster Anti-Mouse TCRb (742484, BD); BUV395 Rat Anti-Mouse CD4 (563790, BD); APC Rat Anti-Mouse CD8a (553035, BD); BV650 Rat Anti-Mouse CD44 (740455, BD); PE-Cy™7 Rat Anti-Mouse CD62L (560516, BD); FITC Hamster Anti-Mouse CD69 (104505, Biolegend); PE Hamster Anti-Mouse CD103 (121406, Biolegend). For intracellular staining, FITC Rat Anti-Mouse IFN-γ (554411, BD); BV605 Mouse Anti-Mouse Tbet (644817, BD) were used. Antibodies used for human FACS analysis included: APC-H7 Mouse anti-Human CD45(560178,BD); BUV395 Mouse Anti-Human CD3(564001,BD); FITC Mouse Anti-Human TCR αβ(555547,BD); BV480 Mouse Anti-Human CD4(566104,BD); BUV737 Mouse Anti-Human CD25(612806,BD); Alexa Fluor® 647 Mouse anti-Human FoxP3(560045,BD); PE-Cy™7 Mouse Anti-Human CD69 (557745,BD); PE anti-human CD186 (CXCR6) Antibody(356003,Biolegend); Alexa Fluor® 700 Mouse Anti-Human CD8 (557945,BD); PE-CF594 Mouse Anti-Human Perforin(563763;BD). Stained cells were assessed on CytoFLEX LX (Beckman) using FlowJo software (Tree Star, Ashland, OR). Stained cells were assessed on BD Fortessa and CytoFLEX LX(Beckman) using FlowJo software (Tree Star, Ashland, OR).
scRNA-seq assay
Single-cell RNA-seq libraries construction were prepared using the 10× Genomics Chromium Single Cell 3ʹ Reagent Kits v3, according to the manufacturer’s instructions. Briefly, prepared single cell suspension of liver and PBMC was washed twice using PBS + 0.4%BSA. Cells were encapsulated into droplets at a targeted cell recovery of cells. After the reverse transcription step, emulsions were broken and Barcoded cDNA was purified with Dynabeads, followed by PCR amplification. Amplified cDNA was then used for 3′gene expression library construction. For gene expression library construction, 50 ng of amplified cDNA was fragmented and end-repaired, double-size selected with SPRIselect beads, and sequenced on an NovaSeq platform (Illumia) to generate 150 bp paired-end Reads. The library preparations for all scRNA-seq samples in this study were completed within one day. Each channel was used to collect 6000 cells. After data filtering, the human liver data had an average of 5194 cells/sample and a sequencing depth of ~6055 UMI counts/cell; the human PBMC data had an average of 4764 cells/sample and a sequencing depth of ~7664 UMI counts/cell; the mouse data had an average of 5056 cells/sample and a sequencing depth of ~7607 UMI counts/cell.
scRNA-seq data pre-processing and quality control
Cellranger (v3.1) was used to perform scRNA-seq data pre-processing include deduplication, aligning to the human reference genome (GRCh38), and generating a cell × gene raw count matrix composing all samples using default parameters. Raw count matrix was transformed into a Seurat object by R package Seurat (v3.2)56. Genes expressed in less than 3 cells were removed. High-quality cells were retained according to the following criteria: more than 200 expressed genes, less than 20% mitochondrial transcript, UMI number between 2000 and 25,000.
Normalization, dimensionality reduction and clustering
The scRNA-seq count matrix was normalized using NormalizeData function in Seurat with default parameters. The top 4000 variable genes were used to perform principal component analysis (PCA). For cell subset dimensionality reduction, the top 2000 variable genes were used to perform PCA. R package Harmony (v0.99.9)57 was used to remove batch effects with default settings by iteratively corrected PCA embeddings based on top 40 PCA components. Then we used first 40 components resulted in harmony to perform cell clustering and non-linear dimensionality reduction (Uniform Manifold Approximation and Projection, UMAP).
Doublet removal and cell type annotation
After the initial clustering, we found that there were some abnormal cells (doublet cells) expressing multiple cell type specific genes at the same time. So, we used the Scran (v1.12.1) to filter doublet cells based on top 20 marker genes in each cluster58. Moreover, we also removed abnormal cell clusters that simultaneously expressed multiple cell type specific genes. After removing the double cells, we performed dimensionality reduction and re-clustering filtered cells fellow up with the above process. The cell clusters were annotated depending on the expression level of known cell markers. scRNA-seq data was visualized by Seurat utilities and R package ggplot259. UMAP plots are used to compare the differences in cell abundance among different groups, with the cell numbers of each group downsampled to a consistent amount.
Comparison of cell subsets abundance
The NB GLM method with edgeR was used to assess whether there were differences in the abundance of each cell subpopulation among different groups as discribed in “https://bioconductor.org/books/3.13/OSCA.multisample/differential-abundance.html#ref-lun2017testing”. FDR less than 0.05 were considered statistically significant.
Identification and functional analysis of differentially expressed genes
Seurat function FindAllMarkers was used to find the cell type specific genes or differentially expressed genes between PBC and control samples with “logfc.threshold = 0.4, test.use = “MAST”” and “logfc.threshold = 0.3, test.use = “MAST”” respectively. The P values were adjusted by Bonferroni test. Most marker genes and differentially expressed genes are provided in Supplementary Data 2-10,11-17. For genes that may have a close relationship with the progression of PBC disease but have log-transformed fold changes less than 0.3, we have described the fold changes and adjusted P-values of these genes in the results section. R package clusterProfiler (v3.12) was applied to perform pathway enrichment analysis of given gene sets60.
Transcription factor activity analysis
To explore the transcriptional regulatory network in T cells, we applied SCENIC (v1.2.4) to identify the relative activity of transcription factors in each cell32. Briefly, we established the co-expression modules of known human transcription factors and potential target genes. Due to the inherent stochastic nature of constructing the co-expression network, we iteratively built the co-expression network 30 times and calculated the average to obtain more robust results. Motif analysis was used to further screen TFs and their target genes that might have a direct regulatory relationship. Such a TF and its target genes were called a regulon. The AUCell algorithm was used to score each regulon in each cell (AUC score). The higher AUC score represented the higher activation of the corresponding regulon. To better show the difference of activation of each regulon in different cells, we binarized the AUC score matrix using an automatically determined cutoff for each regulon. Moreover, we calculated the regulon specificity score (RSS) to find the TFs specifically activated in each cell subsets.
Acquisition of gene signatures
To obtain the inflammatory score of each cell, we first established an inflammation-related gene set based on the well-known cytokines (Fig. 1i) and inflammation-related genes refers to the gene set previously used to evaluate the inflammatory level of IBD61. The gene set of cytokines included chemokines, interferons, interleukins, TNF family members and other cytokines, such as TSLP, CSF1, CSF2, CSF3, LTA, LTB. Inflammation related genes included TNF, TGFB3, TGFB2, TGFB1, STAT6, STAT4, STAT3, STAT1, S100A9, S100A8, RORC, RORA, RELA, NFKB1, JUN, IL6, IL5, IL4R, IL4, IL2RG, IL23R, IL23A, IL22, IL21R, IL21, IL2, IL1B, IL1A, IL18RAP, IL18R1, IL18, IL17A, IL13, IL12RB2, IL12RB1, IL12B, IL12A, IL10, IFNGR2, IFNGR1, IFNG. Then, the cytokine score and inflammatory score of each cell was calculated using Seurat’s function AddMouduleScore with default parameters. Briefly, the average expression of all genes in the inflammatory gene set was calculated, then spliced the expression matrix into 24 bins (default) according to the average value. 100 genes outside the inflammatory gene set were randomly selected from each bin as the control gene set. The inflammation score was obtained by subtracting the average expression of background genes from the average expression of inflammatory genes. The cell cycle score was calculated in a similar way. The S and G2M phase scores of each cell were produced using CellCycleScoring function in Seurat. Significance of gene signature difference was determined by Wilcoxon test.
Cell-cell communication analysis
We utilized the R package CellChat (v0.5.5)62 and python package CellphoneDB (v3.1)63 for inferring the cell-cell communications based on the expression of known ligand–receptor pairs in different cell types of liver scRNA-seq data.For CellChat, normalized count matrix was transformed into a CellChat object and subset the count matrix by the genes of known ligand–receptor pairs. The communication probability of ligand–receptor pairs and signaling pathways were computed using computeCommunProb and computeCommunProbPathway with default parameters. The circle plots used to show cell-cell interactions were conducted by netVisual_aggregate function. For CellphoneDB, we utilized the normalized count matrix as the input file and processed the data using “cellphonedb method statistical_analysis” with default parameters, obtaining significant interacting receptor-ligand pairs in both the PBC group and the control group. The results of CellphoneDB were visualized in R.
PSC scRNA-seq data processing
The scRNA-seq data of CD3+ T cells in liver samples of PSC patients were downloaded from “https://www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-10143 ”14 The data deduplication, alignment and quality control were performed according to the scRNA-seq analysis pipeline. The filtered high-quality scRNA-seq data were initially clustered and a small number of non-T cells were removed. Our annotated T-cell subtypes were used as a reference dataset to annotate the T cells in PSC using the TransferData function in Seurat. Then, the MapQuery function was used to embed cells into the UMAP structure of our T cell data.
Statistics & reproducibility
No statistical method was used to predetermine the sample size. The sample sizes of sequencing were primarily decided based on the availability of samples and previously published research articles of the same type. Detailed protocols for analyzing data and gene expression data are described in the Methods. R was applied for wilcoxon test and correlation analysis with scRNA-seq data. For the experimental data, GraphPad Prism 7 was used to perform statistical analyses and graphics production. All the statistical analyses were performed in two-tailed manner. P values less than 0.05 were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
This work was funded by grants from the National Key R&D Program of China No. 2022YFC2304500 (Y.Y.), 2022YFA1103500 (P.Q.), the National Natural Science Foundation of China No. 82000187 (P.J.), 82222003, 92268117, 82161138028 (P.Q.), 82370570 (X.J.), the Zhejiang Provincial Natural Science Foundation of China Z24H080001(P.Q.) LZ21H030002, Z24H080001 (X.J.), LTGY24H160016 (X.W.), the Key R&D Program of Zhejiang No. 2024SSYS0024 (P.Q.), the Department of Science and Technology of Zhejiang Province No. 2023R01012 (P.Q.), the Fundamental Research Funds for the Central Universities No. 226-2024-00007 (P.Q.) and ZJU-Xiongkai Group Research & Development Center for Medical Device No. ZDXK-2023-00XX (X.J.). We are grateful to Ms. Chao Bi, Ms. Jingyao Chen and Ms. Qiong Huang at the Core Facilities, Zhejiang University School of Medicine for technical assistance in flow cytometry and confocal microscopy. We thank Huiling Zhao (Universal Biotech Company, Shanghai) for the assistance in flow cytometry data analysis. This manuscript was edited at Life Science Editors.
Author contributions
Y.Y., P.Q., and X.J. designed experiments, supervised the whole research process and took responsibility for the work of the study. C.J. collected human samples, performed all murine experiments, and analyzed relative data and performed flow cytometry of human liver samples. P.J., P.Q. analyzed, interpreted, and reviewed the scRNA-seq data. Z.Z. processed samples into single cells for single-cell sequencing. L.Z., J.L., G.Y. offers clinical diagnosis expertise for PBC patients. X.Q., J.C., Y.Z. collected human liver samples and performed flow cytometry of human liver samples. X.W.. provided the pathological score of the PBC samples. W.K., Y.R., M.L., L.X., W.C. collected clinical data of PBC patients. B.W. and J.X. synthesized 2OA-BSA for murine PBC model. C.J., P.J., X.J., Y.Y., P.Q., and Y.H. reviewed the manuscript and took part in the interpretation of data.
Peer review
Peer review information
Nature Communications thanks Vincenzo Ronca and the other, anonymous, reviewers for their contribution to the peer review of this work.
Data availability
All generated human and mouse scRNAseq data in this study have been deposited into Genome Sequence Archive for (GSA) and GSA for Human with accession number HRA008003 and CRA017680. scRNA-seq data of PSC patients14can be download from the European Bioinformatics Institute (EBI) ArrayExpress database under accession number E-MTAB-10143. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ciliang Jin, Penglei Jiang, Zhaoru Zhang.
Contributor Information
Xi Jin, Email: jxfl007@zju.edu.cn.
Pengxu Qian, Email: axu@zju.edu.cn.
Yida Yang, Email: yidayang65@zju.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-53104-9.
References
- 1.Gulamhusein, A. F. & Hirschfield, G. M. Primary biliary cholangitis: pathogenesis and therapeutic opportunities. Nat. Rev. Gastroenterol. Hepatol.17, 93–110 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Boonstra, K., Beuers, U. & Ponsioen, C. Y. Epidemiology of primary sclerosing cholangitis and primary biliary cirrhosis: a systematic review. J. Hepatol.56, 1181–1188 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Shah, R. A. & Kowdley, K. V. Current and potential treatments for primary biliary cholangitis. Lancet Gastroenterol. Hepatol.5, 306–315 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Combes, B. et al. Methotrexate (MTX) plus ursodeoxycholic acid (UDCA) in the treatment of primary biliary cirrhosis. Hepatology42, 1184–1193 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Koch, A., Brahm, J., Antezana, C., Smok, G. & Cumsille, M. A. The combination of ursodeoxycholic acid and methotrexate for primary biliary cirrhosis is not better than ursodeoxycholic acid alone. J. Hepatol.27, 143–149 (1997). [DOI] [PubMed] [Google Scholar]
- 6.Vuoristo, M. et al. A placebo-controlled trial of primary biliary cirrhosis treatment with colchicine and ursodeoxycholic acid. Gastroenterology108, 1470–1478 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Hirschfield, G. M. et al. A placebo-controlled randomised trial of budesonide for PBC following an insufficient response to UDCA. J. Hepatol.74, 321–329 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Kita, H. et al. Identification of HLA-A2-restricted CD8(+) cytotoxic T cell responses in primary biliary cirrhosis: T cell activation is augmented by immune complexes cross-presented by dendritic cells. J. Exp. Med195, 113–123 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akbar, S. M. et al. Peripheral blood T-cell responses to pyruvate dehydrogenase complex in primary biliary cirrhosis: role of antigen-presenting dendritic cells. Eur. J. Clin. Invest.31, 639–646 (2001). [DOI] [PubMed] [Google Scholar]
- 10.Lleo, A. et al. Biliary apotopes and anti-mitochondrial antibodies activate innate immune responses in primary biliary cirrhosis. Hepatology52, 987–998 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isse, K. et al. Fractalkine and CX3CR1 are involved in the recruitment of intraepithelial lymphocytes of intrahepatic bile ducts. Hepatology41, 506–516 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Lleo, A., Leung, P. S. C., Hirschfield, G. M. & Gershwin, E. M. The pathogenesis of primary biliary cholangitis: a comprehensive review. Semin Liver Dis.40, 34–48 (2020). [DOI] [PubMed] [Google Scholar]
- 13.Lleo, A., Wang, G. Q., Gershwin, M. E. & Hirschfield, G. M. Primary biliary cholangitis. Lancet396, 1915–1926 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Poch, T. et al. Single-cell atlas of hepatic T cells reveals expansion of liver-resident naive-like CD4(+) T cells in primary sclerosing cholangitis. J. Hepatol.75, 414–423 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renand, A. et al. Integrative molecular profiling of autoreactive CD4 T cells in autoimmune hepatitis. J. Hepatol.73, 1379–1390 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Andrews, T. S. et al. Single-cell, single-nucleus, and spatial transcriptomics characterization of the immunological landscape in the healthy and PSC human liver. J. Hepatol.80, 730–743 (2024). [DOI] [PubMed] [Google Scholar]
- 17.Han, Y. et al. Single-cell characterization of hepatic CD8(+) T cells in a murine model of primary biliary cholangitis. Front. Immunol.13, 860311 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, X. et al. Unique DUOX2(+)ACE2(+) small cholangiocytes are pathogenic targets for primary biliary cholangitis. Nat. Commun.14, 29 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakanuma, Y. et al. Application of a new histological staging and grading system for primary biliary cirrhosis to liver biopsy specimens: Interobserver agreement. Pathol. Int.60, 167–174 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Banales, J. M. et al. Cholangiocyte pathobiology. Nat. Rev. Gastroenterol. Hepatol.16, 269–281 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medina, J. F., Martínez, A., Vazquez, J. J. & Prieto, J. Decreased anion exchanger 2 immunoreactivity in the liver of patients with primary biliary cirrhosis. Hepatology25, 12–17 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Trampert, D. C., van de Graaf, S. F. J., Jongejan, A., Oude Elferink, R. P. J. & Beuers, U. Hepatobiliary acid-base homeostasis: Insights from analogous secretory epithelia. J. Hepatol.74, 428–441 (2021). [DOI] [PubMed] [Google Scholar]
- 23.Lee, E. S. et al. Growth differentiation factor 15 predicts chronic liver disease severity. Gut Liver11, 276–282 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, Z. et al. Association between circulating growth differentiation factor 15 and cirrhotic primary biliary cholangitis. BioMed. Res. Int.2020, 1–11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koo, B. K. et al. Growth differentiation factor 15 predicts advanced fibrosis in biopsy-proven non-alcoholic fatty liver disease. Liver Int. Off. J. Int. Assoc. Study Liver38, 695–705 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Wang, J. et al. Liver immune profiling reveals pathogenesis and therapeutics for biliary atresia. Cell183, 1867–1883.e1826 (2020). [DOI] [PubMed] [Google Scholar]
- 27.Wu, N. et al. Functional role of the secretin/secretin receptor signaling during cholestatic liver injury. Hepatology72, 2219–2227 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kennedy, L. et al. Secretin/secretin receptor signaling mediates biliary damage and liver fibrosis in early-stage primary biliary cholangitis. FASEB J.33, 10269–10279 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deczkowska, A. et al. XCR1(+) type 1 conventional dendritic cells drive liver pathology in non-alcoholic steatohepatitis. Nat. Med27, 1043–1054 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Tesfaye, D. Y. et al. Targeting Xcr1 on dendritic cells rapidly induce Th1-associated immune responses that contribute to protection against influenza infection. Front. Immunol.13, 752714 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiggins, B. G. et al. The human liver microenvironment shapes the homing and function of CD4(+) T-cell populations. Gut71, 1399–1411 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods14, 1083–1086 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Masopust, D. & Soerens, A. G. Tissue-resident T cells and other resident leukocytes. Annu. Rev. Immunol.37, 521–546 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheuk, S. et al. CD49a expression defines tissue-resident CD8(+) T cells poised for cytotoxic function in human skin. Immunity46, 287–300 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheuk, S. et al. Epidermal Th22 and Tc17 cells form a localized disease memory in clinically healed psoriasis. J. Immunol.192, 3111–3120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kleinschek, M. A. et al. Circulating and gut-resident human Th17 cells express CD161 and promote intestinal inflammation. J. Exp. Med.206, 525–534 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sasaki, K. et al. Relapsing-remitting central nervous system autoimmunity mediated by GFAP-specific CD8 T cells. J. Immunol.192, 3029–3042 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.You, Z. et al. The clinical significance of hepatic CD69(+) CD103(+) CD8(+) resident-memory T cells in autoimmune hepatitis. Hepatology74, 847–863 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Huang, B. et al. NUDT1 promotes the accumulation and longevity of CD103(+) TRM cells in primary biliary cholangitis. J. Hepatol.10.1016/j.jhep.2022.06.014 (2022). [DOI] [PubMed]
- 40.Fernandez-Ruiz, D. et al. Liver-resident memory CD8(+) T cells form a front-line defense against malaria liver-stage infection. Immunity45, 889–902 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Mackay, L. K. et al. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat. Immunol.14, 1294–1301 (2013). [DOI] [PubMed] [Google Scholar]
- 42.Mackay, L. K. et al. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science352, 459–463 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Kumar, B. V. et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep.20, 2921–2934 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueno, K. et al. Integrated GWAS and mRNA microarray analysis identified IFNG and CD40L as the central upstream regulators in primary biliary cholangitis. Hepatol. Commun.4, 724–738 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bae, H. R. et al. Chronic expression of interferon-gamma leads to murine autoimmune cholangitis with a female predominance. Hepatology64, 1189–1201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liaskou, E. et al. Increased sensitivity of Treg cells from patients with PBC to low dose IL-12 drives their differentiation into IFN-gamma secreting cells. J. Autoimmun.94, 143–155 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Cordell, H. J. et al. An international genome-wide meta-analysis of primary biliary cholangitis: Novel risk loci and candidate drugs. J. Hepatol.75, 572–581 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirschfield, G. M. et al. Ustekinumab for patients with primary biliary cholangitis who have an inadequate response to ursodeoxycholic acid: a proof-of-concept study. Hepatology64, 189–199 (2016). [DOI] [PubMed] [Google Scholar]
- 49.European Association for the Study of the Liver. Electronic address, e. e. e. & European Association for the Study of the, L EASL Clinical Practice Guidelines: The diagnosis and management of patients with primary biliary cholangitis. J. Hepatol.67, 145–172 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Winthrop, K. L. The emerging safety profile of JAK inhibitors in rheumatic disease. Nat. Rev. Rheumatol.13, 234–243 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Sakaguchi, S. et al. Regulatory T cells and human disease. Annu. Rev. Immunol.38, 541–566 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Scheuer, P. J. Classification of chronic viral hepatitis: a need for reassessment. J. Hepatol.13, 372–374 (1991). [DOI] [PubMed] [Google Scholar]
- 53.Dhirapong, A. et al. Therapeutic effect of cytotoxic T lymphocyte antigen 4/immunoglobulin on a murine model of primary biliary cirrhosis. Hepatology57, 708–715 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fridman, J. S. et al. Selective inhibition of JAK1 and JAK2 is efficacious in rodent models of arthritis: preclinical characterization of INCB028050. J. Immunol.184, 5298–5307 (2010). [DOI] [PubMed] [Google Scholar]
- 55.Wu, S. J. et al. Innate immunity and primary biliary cirrhosis: activated invariant natural killer T cells exacerbate murine autoimmune cholangitis and fibrosis. Hepatology53, 915–925 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stuart, T. et al. Comprehensive integration of single-cell data. Cell177, 1888–1902.e1821 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korsunsky, I., Millard, N. & Fan, J. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed]
- 58.Lun, A. T., McCarthy, D. J. & Marioni, J. C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Research5, 2122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.H, W. ggplot2: Elegant Graphics for Data Analysis. (Springer-Verlag New York, 2016).
- 60.Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics J. Integr. Biol.16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smillie, C. S. et al. Intra- and inter-cellular rewiring of the human colon during ulcerative colitis. Cell178, 714–730.e722 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin, S. et al. Inference and analysis of cell-cell communication using CellChat. Nat. Commun.12, 1088 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia-Alonso, L. et al. Mapping the temporal and spatial dynamics of the human endometrium in vivo and in vitro. Nat. Genet.53, 1698–1711 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
All generated human and mouse scRNAseq data in this study have been deposited into Genome Sequence Archive for (GSA) and GSA for Human with accession number HRA008003 and CRA017680. scRNA-seq data of PSC patients14can be download from the European Bioinformatics Institute (EBI) ArrayExpress database under accession number E-MTAB-10143. Source data are provided with this paper.