Abstract
Since 2019, SARS-CoV-2 has undergone mutations, resulting in pandemic and epidemic waves. The SARS-CoV-2 spike protein, crucial for cellular entry, binds to the ACE2 receptor exclusively when its receptor-binding domain (RBD) adopts the up-conformation. However, whether ACE2 also interacts with the RBD in the down-conformation to facilitate the conformational shift to RBD-up remains unclear. Herein, we present the structures of the BA.2.86 and the JN.1 spike proteins bound to ACE2. Notably, we successfully observed the ACE2-bound down-RBD, indicating an intermediate structure before the RBD-up conformation. The wider and mobile angle of RBDs in the up-state provides space for ACE2 to interact with the down-RBD, facilitating the transition to the RBD-up state. The K356T, but not N354-linked glycan, contributes to both of infectivity and neutralizing-antibody evasion in BA.2.86. These structural insights the spike-protein dynamics would help understand the mechanisms underlying SARS-CoV-2 infection and its neutralization.
Subject terms: Virus structures, Cryoelectron microscopy, SARS-CoV-2
JN.1 lineage is a globally dominant variant of SARS-CoV-2. Here, the authors examine structural insights into the BA.2.86 and JN.1 spike dynamics to help understand the mechanisms underlying SARS-CoV-2 infection and its neutralizing-antibody evasion.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19), a respiratory infectious disease that led to a global pandemic in 2020. According to data retrieved from Nextstrain (https://nextstrain.org/ncov/gisaid/global/6m) regarding the global status of SARS-CoV-2 evolution, the BA.2.86 variant of SARS-CoV-2 was first identified in July 2023. Notably, BA.2.86 acquired over 30 amino acid substitutions in the spike (S) protein compared to those present in the previously predominant lineage XBB and its parent BA.2, raising concerns about potential immune evasion. Acknowledging its heightened mutational load, the World Health Organization promptly designated BA.2.86 as a variant under monitoring on August 17, 20231. By December 2023, BA.2.86 had become globally prevalent, with over 7500 reported sequences on GISAID. Furthermore, JN.1, a descendant of BA.2.86, with an amino acid substitution in the S-protein, was also spreading rapidly, and, a month later was designated as a variant of interest with BA.2.86 by the WHO2,3. To date, the BA.2.86 lineage, including JN.1, has been detected in over 30 countries. The emergence of BA.2.86 coincides with a surge in cases across several countries. Therefore, studying the structure of the BA.2.86-S-protein, along with the reported virological characteristics of BA.2.864–8, may contribute to a better understanding of this virus.
The S-protein of SARS-CoV-2 exists as a trimer with a pivotal role in viral entry into target cells. During this cellular entry process, the S-protein—comprising the S1 and S2 subunits—initially binds to receptors such as angiotensin-converting enzyme 2 (ACE2), neuropilin-1, and TMEM106B9–12. Among these receptors, ACE2 is the most established receptor associated with pathophysiology13,14. The ACE2 receptor is also utilized by SARS-CoV, the etiologic pathogen of an epidemic that primarily impacted Asia in 2003, and coronavirus NL63, which causes common cold symptoms15. The S1 subunit of the S-protein contains a receptor-binding domain (RBD), which is the primary target of neutralizing antibodies. Meanwhile, the S2 subunit mediates membrane fusion between the viral envelope and the target cell membrane, facilitating the delivery of the viral genome into the host cell cytoplasm. To bind to the ACE2 receptor, the RBD of the S-protein must be in the up conformation16. However, the full-length structure of the S-protein, including the transmembrane domain, reportedly adopts an all-down conformation of the RBD17. Structures of the S-protein of the recent Omicron lineages, predominantly the ectodomain, also reportedly exhibit the all-down conformational state of RBDs18–20, suggesting the requirement for a passive trigger to transition the RBD to the up-conformation. Therefore, the structure of the SARS-CoV-2-S undergoes sequential conformational transitions, progressing from an all-RBD down state to one RBD-up, followed by two RBD-up, and ultimately reaching the three RBD-up states upon ACE2 binding. To activate its fusion ability, the S-protein is cleaved into S1 and S2 subunits by the host protease furin during viral budding21, and the S2 subunit is further cleaved by host proteases, such as transmembrane protease, serine 2 (TMPRSS2), or cathepsin L, during viral entry22. However, an in-depth exploration of ACE2 binding to both up- and down-RBD conformations in the SARS-CoV-2 spike protein is lacking. Consequently, the understanding of critical conformational shifts essential for cellular entry and neutralizing-antibody evasion, particularly in the context of the BA.2.86 variant, remains limited.
In this work, we perform the single-particle analyses of the BA.2.86 S-protein alone and the BA.2.86 and JN.1 S-proteins in complex with the ACE2 receptor using cryo-electron microscopy (cryo-EM). The primary objective is to gain insights into the structural dynamics of ACE2-receptor recognition and neutralizing-antibody evasion in SARS-CoV-2 variants. Additionally, we monitor interactions between ACE2 and RBDs to identify any potential intermediate structural conformations. We believe that our findings can contribute to an enhanced understanding of the mechanism underlying SARS-CoV-2 infection and aid in the development of effective neutralization strategies.
Results
Evolutionary and epidemic features of BA.2.86 lineages
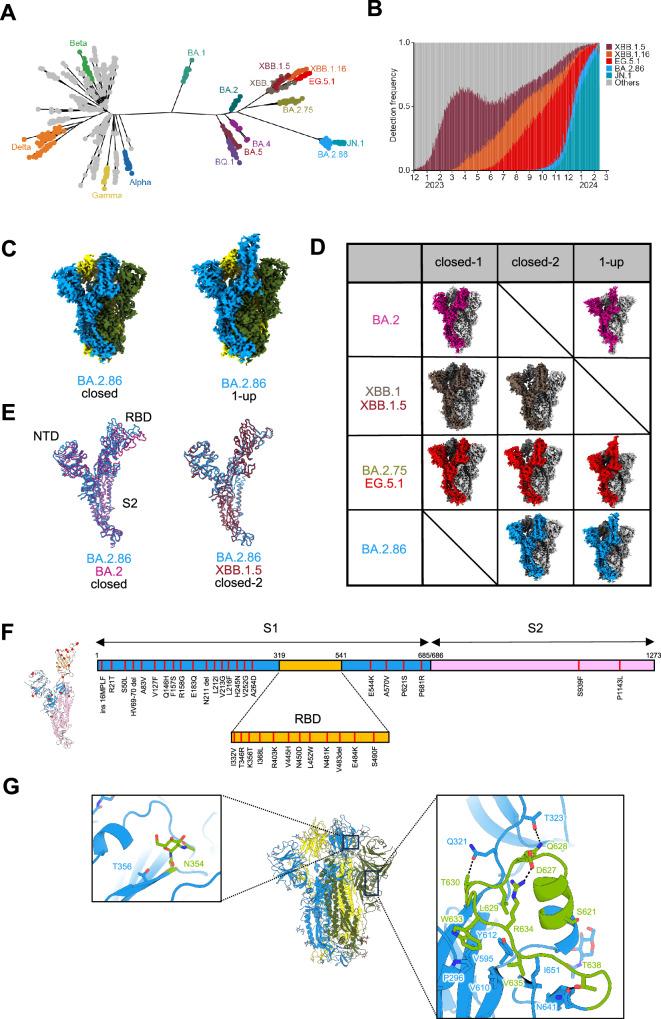
The BA.2.86 lineage, including JN.1, significantly diverged from other SARS-CoV-2 variants, including XBB (Fig. 1A). The BA.2.86 lineage, particularly JN.1, demonstrated rapid global spread, effectively out-competing XBB and establishing itself as the dominant lineage as of February 2024 (Fig. 1B). These evolutionary and epidemic characteristics of BA.2.86 highlight the critical need for its detailed characterization, including the determination of its S-protein structure.
Fig. 1. Evolutionary and epidemic features of BA.2.86 and cryo-EM maps of BA.2.86-S-protein.
A Maximum likelihood tree depicting SARS-CoV-2 evolution. B Detection frequency plot of XB.1.5, XBB.1.6, EG.5.1, BA.2.86, JN.1, and other variants. C Cryo-EM maps of the BA.2.86-S-protein trimer, closed-2 state (left) and one-RBD-up state (right); protomers are sky blue, yellow, and dark olive green. D Cryo-EM maps obtained for each Omicron subvariants. Protomers for BA.2, XBB.1/XBB.1.5, BA.2.75/EG.5.1, and BA.2.86 are shown in dark green, brown, red, and sky blue, respectively. Other protomers are shown in dark gray and light gray. E Superposition of the main-chain structure as protomers of the BA.2.86-S closed-2 state (sky blue), BA.2-S closed-1 state (deep pink), and XBB.1.5-S closed-2 state (raspberry), respectively. F The position of amino-acid substitutions in the BA.2.86-S-protein compared to the XBB.1.5-S-protein. The BA.2.86-S protomer structure (left) and its sequence schematic (right) are shown as the S1 subunit (sky blue), the S2 subunit (pink), and the RBD (bright yellow). G Structure of BA.2.86-S-protein trimer (same colors as in C); close-up view presents the N354-linked glycan (left) and residues 621–640 (right), showing interactions with surrounding residues and glycosylation sites as sticks (leaf green). Dashed lines represent hydrogen bonds.
Structures of the BA.2.86 S-protein
To gain structural insights into the evolution of the BA.2.86 S-protein, we performed cryo-EM analysis of the BA.2.86 S ectodomain (Fig. 1C, Supplementary Fig. 1A, B, and Supplementary Table 1). We previously reported closed-1 and closed-2 conformational states in the S-protein structures of SARS-CoV-2 variants that emerged after BA.2.75 (BA.2.75, XBB.1, XBB.1.5, and EG.5.1)18–20,23 (Fig. 1D). Notably, closed-2 typically has a more open RBD structure than that of closed-1; however, their S2 subunit is relatively identical. However, structural analysis in this study revealed only the closed-2 state and not the closed-1 state in the BA.2.86 S-protein. Meanwhile, the S-protein structure of BA.2 has only been resolved in the closed-1 state24,25, whereas that of BA.2.86 has only been observed in the closed-2 state, resulting in poor overlap of the main chains of BA.2 and BA.2.86 S-proteins (Fig. 1E). In contrast, the main chains of the closed-2 states of the S-proteins in the BA.2.75, XBB.1, XBB.1.5, and EG.5.1 variants overlapped well with that of the BA.2.86 S-protein (Fig. 1E). One RBD-up state, frequently observed in the S-protein structures of other SARS-CoV-2 variants, was rarely observed in the XBB.1 and XBB.1.5 S-proteins. However, the one RBD-up state was observed again in EG.5.1 and in BA.2.86 (Fig. 1D). The spike protein of BA.2.86 acquired 39 amino acid substitutions (37 in the S1 subunit, 12 in the RBD, and 2 in the S2 subunit) compared with that in the amino acid sequence of the XBB.1.5 S-protein (Fig. 1F); however, the overall structure did not significantly differ from that of the closed-2 state of the previous variants (Fig. 1E). Hence, the survival strategy of SARS-CoV-2 involves the gradual modification of its S-protein structure or conformation, facilitating evolutionary adaptation without undue stress.
Subsequently, we focused on three specific areas where significant structural alterations were observed in BA.2.86 S compared with those in XBB.1.5, given that both S-proteins can adopt a closed-2 state. First, a consensus sequence of N-linked glycosylation was obtained by K356T substitution in the RBD, and the presence of an N-linked glycan structure at N354 was confirmed in this study (Fig. 1G and Supplementary Fig. 1C). Second, the structure corresponding to amino acid residues 621–640 surrounding the furin-cleavage site was observed in the BA.2.86 S-protein (Fig. 1G and Supplementary Fig. 1C); this region was not visible in S-proteins of other variants, except for the S-protein of alpha26. This observation is likely attributed to the influence of P621S, which marks the starting point for structural visibility. Third, a large EM density was observed above V445H at the apex of the closed-2 state of the BA.2.86 S-protein, suggesting an interaction between H445 and this unidentified EM density (Supplementary Fig. 2). Analysis of raw EM images indicated that the formation of a dimer-of-trimer with head-to-head orientation, as reported for the Kappa S-protein27, is unlikely. The disappearance of the unidentified EM density during cryo-EM analysis in EDTA (1 mM)-containing phosphate-buffered saline (PBS) suggests that His accumulation at the S-protein trimer apex may cause metal ion coordination, leading to nonspecific interactions with substances in the solution.
Structures of the ACE2-bound BA.2.86 S-protein
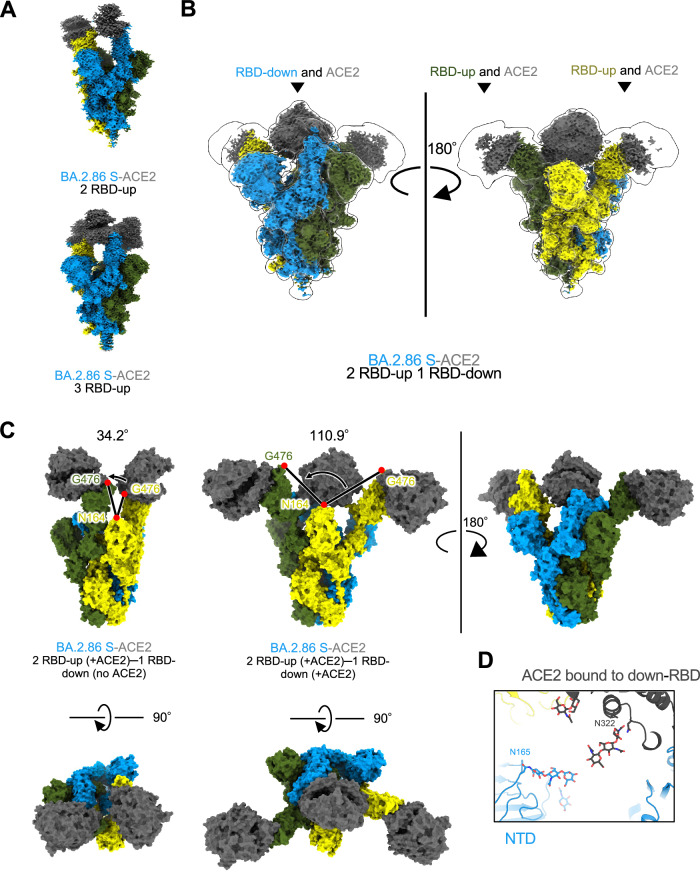
To elucidate the molecular mechanism of ACE2 receptor recognition by BA.2.86 S-protein, we performed cryo-EM analysis of the BA.2.86 S–ACE2 complex (Fig. 2A, B, Supplementary Fig. 3, and Supplementary Table 1). In SARS-CoV-2 variants that emerged after Omicron, most ACE2-bound S-protein structures exhibited one-RBD-up and/or two-RBD-ups. However, in the BA.2.86 S−ACE2 complex, the presence of one-RBD-up was not observed, whereas conformations with two- and three-RBD-ups were evident (Fig. 2A). Notably, the ACE2 receptor was bound to all three RBDs, including two-RBD-ups and one-RBD-down, which is an unprecedented observation. This structure may represent an intermediate state wherein the two-RBD-up transitions to three-RBD-up. The two-RBD-up and one-RBD-down conformation (two-RBD-up–one-RBD-downthree-ACE2) were not modeled owing to unclear EM density during conventional refinement (Fig. 2B and Supplementary Fig. 3, and Supplementary Table 1).
Fig. 2. Cryo-EM maps of BA.2.86-S-protein bound to ACE2.
A–C Cryo-EM maps of BA.2.86-S-protein bound to human ACE2 (S; same colors as Fig. 1C, ACE2; dark gray). A Two-RBD-up state (top), and three-RBD-up state (bottom). B Two-RBD-up−one-RBD-downthree-ACE2 state. C Comparison of the angles formed by the three residues: N164 of the NTD and two G476 of the up-RBD for two-RBD-uptwo-ACE2 (34.2°, left) and two-RBD-up−one-RBD-downthree-ACE2 (110.9°, right), respectively. D Close-up view of the two-RBD-up−one-RBD-downthree-ACE2 state. The N165-linked glycan in the NTD and the N322-linked glycan in ACE2 are situated close to each other.
Consequently, we employed 3D Flexible Refinement (3DFlex)28 to assess the flexibility of the two up-RBDs, revealing significant swaying motion (Fig. 2C and Supplementary Fig. 4A–C, and Supplementary Movie 1). Compared with the two RBD-up bound to ACE2 (two-RBD-uptwo-ACE2; one RBD is down state and not bound to ACE2) (Fig. 2C left top), the two up-RBDs in the two-RBD-up–one-RBD-downthree-ACE2 exhibited increased mobility and wider angles (Fig. 2C right). The angles formed by the three points, N164 of the N-terminal domain (NTD) and G476 of two up-RBDs, were different at 34.2° and 110.9° in the two-RBD-uptwo-ACE2 and two-RBD-up–one-RBD-downthree-ACE2, respectively (Fig. 2C). This heightened mobility of the two up-RBDs in the two-RBD-up–one-RBD-downthree-ACE2 enables ACE2 access. Moreover, binding to a down-RBD may contribute to the down-to-up transition of this domain. In contrast, the RBD-down−ACE2 complex remained stable, allowing for model building. Apart from the RBD–ACE2 interface in the RBD-down−ACE2 complex, we observed a weak stacking interaction between the NTD N165-linked glycan and ACE2 N322-linked glycan, alternated to avoid steric hindrance, which may contribute to the RBD-down−ACE2 stabilization (Fig. 2D).
For the two-RBD-up–one-RBD-downthree-ACE2 conformation, we assume that the up-RBDs become very flexible/mobile in exchange for the stabilization of the down-RBD, and that the balance between the up and down conformations has led to this non-canonical conformation. To test this possibility, the BA.2.86 S-ACE2 complex was treated at a higher temperature of 42 °C and subjected to cryo-EM analysis (Supplementary Fig. 5). The two-RBD-up–one-RBD-downthree-ACE2 conformation was observed in BA.2.86 S−ACE2 treated at 37 °C for 1 hour, but not in that treated at 42 °C for 1 hour. These results may suggest that an intermediate structure, two-RBD-up–one-RBD-downthree-ACE2, has been thermally eliminated, which indicates instability as an S-trimer bound to ACE2. However, the canonical two-RBD-uptwo-ACE2 structure was retained under this condition. Unexpectedly, another non-canonical conformation of one-highly-open RBD and one-partially-open RBD was observed in the BA.2.86 S−ACE2 complex treated at 42 °C, in addition to the canonical two-RBD-uptwo-ACE2 (Supplementary Fig. 5).
Interactions between the BA.2.86 S-protein RBD and ACE2
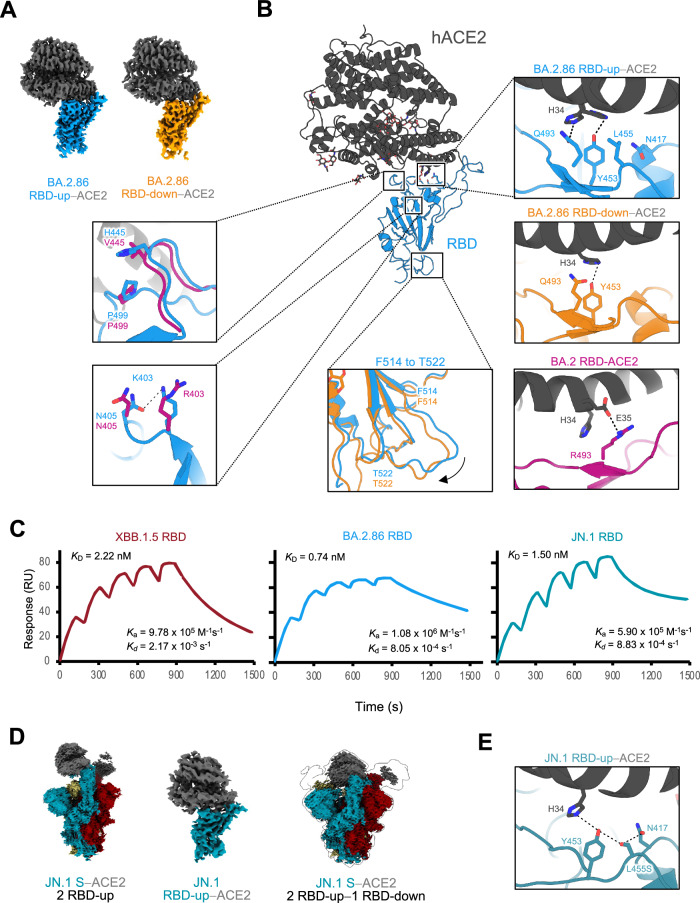
Notably, we observed a complex with ACE2 bound to the BA.2.86 S RBD-down conformation. Hence, we compared two structures in which ACE2 was bound to BA.2.86 S RBD-up or RBD-down. Local refinement was performed on both the RBD-up–ACE2 and RBD-down–ACE2 complexes to comprehensively examine the interactions between ACE2 and RBD-up or RBD-down, and the structures were reconstructed at resolutions of 3.00 Å and 3.05 Å, respectively (Fig. 3A, B, Supplementary Fig. 3, and Supplementary Table 1). No significant differences were detected in the overall structures of either ACE2–RBD; however, slight variations were observed at the interface between ACE2 and the up- or down-RBD (Fig. 3B and Supplementary Fig. 4D, E). First, the main chain from F514 to T522 was shifted outwardly in the up-RBD compared with that in the down-RBD (Fig. 3B). The F514 to T522 loop in the down-RBD was located close to the NTD of the adjacent protomer. In contrast, the loop in the up-RBD was exposed toward the solvent (Fig. 3B and Supplementary Fig. 4D, E). Second, ACE2 H34 interacted bidirectionally with RBD Y453 and Q493 in the RBD-up–ACE2, whereas ACE2 H34 exhibited a unidirectional interaction with RBD Y453 in the RBD-down−ACE2.
Fig. 3. Structures of RBD-ACE2 complexes in the BA.2.86- and JN.1-S- proteins, and binding affinities to ACE2 in the XBB.1.5, BA.2.86, and JN.1.
A Cryo-EM maps of the RBD–ACE2 interface in the RBD-up (sky blue)−ACE2 (dark gray), and RBD-down (orange)−ACE2. B Structure of BA.2.86 S RBD-ACE2 complex (same colors as in A). Close-up views represent residues involved in the corresponding interaction of the BA.2.86 RBD-up–ACE2 complex structure, which differs from the BA.2 or BA.2.86 RBD-down–ACE2 complex structure (BA.2; PDB, 8DM6; BA.2 RBD, deep pink; ACE2, dark gray, BA.2.86 RBD-down; same color as A), are shown. Dashed lines represent hydrogen bonds. C Sensorgrams of SPR analysis evaluating the binding affinities of ACE2 for BA.2.86 S-RBD (sky blue), JN.1 S-RBD (blue-green), and XBB.1.5 S- RBD (raspberry). D Cryo-EM maps of JN.1-S-protein bound to human ACE2 (S; blue-green, raspberry and khaki, ACE2; dark gray). The two-RBD-uptwo-ACE2 state (left), the RBD-ACE2 interface in the RBD-up−ACE2 (middle), and the two-RBD-up−one-RBD-downthree-ACE2 state (right). E Close-up view of the JN.1 S–ACE2 interface (JN.1 S; blue-green, ACE2; dark gray). Dashed lines represent hydrogen bonds.
Subsequently, to comprehend the altered interactions of SARS-CoV-2 with ACE2 caused by SARS-CoV-2 evolution, we compared the RBD–ACE2 interface interactions between BA.2 and BA.2.86. A notable disparity among the variants was primarily observed in the interaction between RBD R/Q493 and ACE2 H34 (Fig. 3B and Supplementary Fig. 4D). In variants predating Omicron, represented by the ancestral strain, the amino acid residue 493 of the S-proteins was Gln. However, the Q493R substitution occurred in the S-proteins of BA.1 and BA.2, leading to the loss of interaction with ACE2 H34 and an alternative interaction with ACE2 E3529,30 (Fig. 3B). Subsequently, in BA.4/5 and subsequent variants, it reverted to Q493; although variations exist in the interactions of each variant with ACE2, the substitution did not occur until BA.2.86 (Fig. 3B). We observed that V445H, a unique characteristic of the BA.2.86 S structure, formed a novel stacking intramolecular interaction with P499 in the RBD (Fig. 3B and Supplementary Fig. 4D). Furthermore, BA.2.86 RBD R403K formed a novel intramolecular interaction with RBD N405 (Fig. 3B and Supplementary Fig. 4D). R403K reportedly enhances the membrane fusion ability4, likely owing to the influence of the altered interaction.
We also compared the binding affinity of ACE2 to the S-RBD of JN.1, which has recently been observed to be prevalent along with BA.2.86, with that of BA.2,86. However, similar to a previous report3, the ACE2-binding affinity of JN. 1 was lower than that of BA.2.86 and higher than that of XBB.1.5 (Fig. 3C). This suggests that the ACE2-binding ability of JN.1 did not influence its prevalence.
Structures of the ACE2-bound JN.1 S-protein
To determine the structural basis for the differences in ACE2 recognition between the S-proteins of BA.2.86 and JN.1, we performed cryo-EM analysis of JN.1-S–ACE2 complex (Fig. 3D, Supplementary Fig. 6, and Supplementary Table 1). In the BA.2.86 S–ACE2 complex, two- and three-RBD-ups were observed; however, only the two-RBD-uptwo-ACE2 structure was found in the JN.1-S–ACE2 complex. Notably, the structure of ACE2 binding to all RBDs in the two-RBD-up–one-RBD-down, which is a characteristic in the BA.2.86 S–ACE2 complex, was observed in JN.1 as well. Similar to the two up-RBDs in the two-RBD-up–one-RBD-downthree-ACE2 conformation of the BA.2.86 S, those of JN.1 also exhibit increased mobility and wider angles (termed highly-open RBDs). ACE2 bound to the highly-open RBDs in the JN.1 S was partially observed, like BA.2.86, owing to the unclear EM density. Accordingly, the highly-open RBDs in the S-proteins may be a shared property in the BA.2.86 lineage, which includes the JN.1 subvariant.
To gain structural insights into the reduced ACE2-binding affinity in the JN.1 S, we performed local refinement on the RBD-up–ACE2 complex, and the structure was reconstructed at resolutions of 4.30 Å (Fig. 3D, Supplementary Fig. 6, and Supplementary Table 1). In the JN.1 S, the L455S substitution occurred, which did not happen in BA.2.86. Neither L455 in the BA.2.86 S nor S455 in the JN.1 S interacts with ACE2; however, the adjacent RBD Y453 forms a hydrogen bond with ACE2 H34 in both S-proteins of BA.2.86 and JN.1 (Fig. 3B, E). S455 in the JN.1 S also forms a hydrogen bond with the adjacent RBD N417. However, the interaction around L455S could not explain the reduced ACE2 affinity in the JN.1 S. Therefore, based on the determined structures, we calculated the shape complementarity (Sc) between RBD and ACE2 in the BA.2.86 and the JN.1. The Sc (0.42) of JN.1 S-RBD–ACE2 was lower than that (0.52) of BA.2.86. The binding free energy (dG= –17.88) of JN.1 S-RBD-ACE2 was increased compared to that (dG= –24.30) of BA.2.86. These values indicate that JN.1 S is less favorable for binding to ACE2 compared to BA.2.86.
Impact of amino acid substitutions resulting in BA.2.86 on cellular entry and neutralization
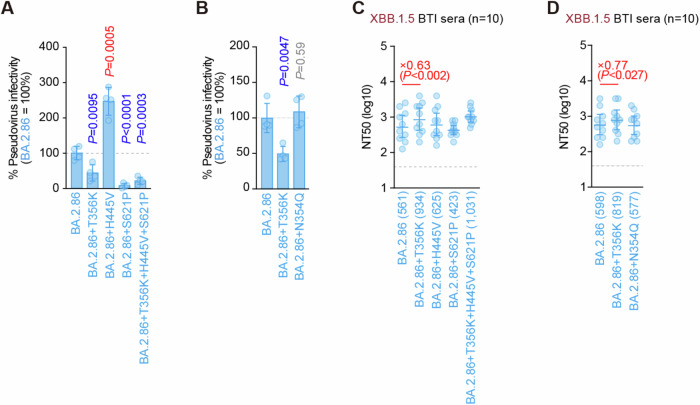
Structural analysis revealed that the three amino acid substitutions, K356T, V445H, and P621S, characteristic of the BA.2.86-S structure, were especially responsible for the structural alterations compared to that in XBB.1.5. Therefore, the impact of these amino acid substitutions on infectivity and neutralization was evaluated in vitro (Fig. 4A–D). First, we introduced single amino acid substitutions reverting to the BA.2 type into the pseudotyped virus bearing the BA.2.86-S- protein and compared its entry efficiency into HOS-ACE2/TMPRSS2 cells. Viruses bearing BA.2.86-S T356K or S621P exhibited decreased infectivity compared with that bearing BA.2.86-S WT (Fig. 4A). Conversely, the virus bearing BA.2.86-S H445V showed increased infectivity compared with that bearing BA.2.86-S WT (Fig. 4A). These findings suggest that K356T and P621S contribute to the increased infectivity of BA.2.86; however, V445H has a negative impact.
Fig. 4. Effects of amino acid substitution on BA.2.86 infectivity and immune evasion.
A, B Lentivirus-based pseudovirus assay. HOS-ACE2/TMPRSS2 cells were infected with pseudoviruses bearing each S-protein of BA.2.86 and its derivatives. The amount of input virus was normalized to that of HIV-1 p24 capsid protein. The percent infectivity of BA.2.86 derivatives compared to that of BA.2.86 is shown. The horizontal dashed line indicates the mean value of the percentage infectivity of BA.2.86. Assays were performed in quadruplicate, and a representative result of four independent assays is shown. Data are presented as the mean ± SD. Each dot indicates the result of an individual replicate. Statistically significant differences versus BA.2.86 are determined by two-sided Student’s t tests. The p values for the difference of each infectivity are indicated in A (vs BA.2.86 + T356K, p = 0.0095; vs BA.2.86 + H445V, p = 0.0005; vs BA.2.86 + S621P, p < 0.0001; and vs BA.2.86 + T356K + H445V + S621P, p = 0.0003) and in (B) (vs BA.2.86 + T356K, p = 0.0047 and vs BA.2.86 + N354Q, p = 0.59). Increased and decreased infectivity are shown in red and blue, respectively. C, D Neutralization assay. Assays were performed with pseudoviruses harboring the S-proteins of BA.2.86 and its derivatives. Convalescent sera were used, which were from fully vaccinated individuals who had been infected with XBB.1.5 (four 3-dose vaccinated, three 4-dose vaccinated, two 5-dose vaccinated, and one 6-dose vaccinated; total =10 donors). Assays for each serum sample were performed in quadruplicate to determine the 50% neutralization titer (NT50). Each dot represents one NT50 value, and the geometric mean and 95% confidence interval are shown. Numbers in parentheses indicates the geometric mean of NT50 values. The horizontal dashed line indicates the detection limit (40-fold). Statistically significant differences vs. BA.2.86 were determined by two-sided Wilcoxon signed-rank tests. The p-values less than 0.05 for the difference of each NT50 are indicated in C (vs BA.2.86 + T356K, p = 0.0020; vs BA.2.86 + H445V, p = 0.22; vs BA.2.86 + S621P, p = 0.32; and vs BA.2.86 + T356K + H445V + S621P, p = 0.10) and in D (vs BA.2.86 + T356K, p = 0.027 and vs BA.2.86 + N354Q, p = 0.77). NT50 fold changes compared with BA.2.86 are indicated by X.
Subsequently, we compared neutralizing-antibody evasion using the same mutant pseudotyped viruses with XBB.1.5 breakthrough-infection human sera (Fig. 4C). Only the virus bearing BA.2.86-S T356K exhibited decreased neutralizing-antibody evasion (Fig. 4C). This indicates that the K356T substitution acquired in the BA.2.86-S-protein contributes to enhanced neutralizing-antibody evasion. K356T represents a novel site for acquired N354-linked glycosylation of the S-protein of BA.2.86 (Fig. 1G); therefore this glycosylation might contribute to the heightened neutralizing-antibody evasion ability of BA.2.86. To evaluate the possibility that the novel N354-linked glycosylation might play an important role in neutralizing-antibody evasion, we further compared infectivity and neutralizing-antibody evasion in a similar manner using the pseudotyped virus bearing BA.2.86-S N354Q (Fig. 4B, D). A substitution to Gln, which exhibits the most similar side chain structure to Asn, was employed to evaluate only the effect of the glycosylation defect. Both of infectivity and neutralizing-antibody evasion by pseudotyped virus bearing BA.2.86-S N354Q were comparable to those of BA.2.86-S WT (Fig. 4B, D). Taken together, the K356T acquired in the BA.2.86 S would contribute to the enhancement of infectivity and neutralizing-antibody evasion by itself, rather than through the effect of glycosylation.
Discussion
During the evolution of SARS-CoV-2, the S-protein has undergone recurrent amino acid substitutions. Correspondingly, various S-protein structures have been determined, demonstrating both closed-1 and closed-2 states, as observed in BA.2.75, XBB.1, XBB.1.5, and EG.5.118–20,23; S-proteins adopting the closed-1 state, exemplified by BA.224; and those displaying the closed-2 state, as demonstrated by BA.2.86 in this study, all within the context of RBD all-down states. Note that because the inability to identify a specific state within a cryo-EM dataset can typically be a function of the data processing workflow, these structures are not the only available structure in each variant. Nonetheless, these conformational states of S-proteins appear to be gradually altering with SARS-CoV-2 evolution (Fig. 1D). Although these conformational state variations may influence cell entry and neutralizing-antibody evasion, the impact of specific conformational states remained unclear owing to technical challenges in fixing these states in vitro and in vivo. Despite the BA.2.86-S-protein acquiring 39 amino acid substitutions compared with that in the XBB.1.5-S-protein, its overall structure did not significantly differ from that of the closed-2 states of other variants (Fig. 1D, E). This suggests that the survival strategy of SARS-CoV-2 may not allow amino acid substitutions that would substantially alter the overall structure.
Structural insights and neutralization assays using XBB.1.5 breakthrough-infection human sera revealed that the newly acquired K356T in the BA.2.86 S contributes to its enhanced neutralizing-antibody evasion (Fig. 4C). However, modification of the coupled N354-linked glycan did not contribute to it (Fig. 4D). K356T also contributes to increased infectivity in BA.2.86 (Fig. 4A), rendering it a crucial mutation responsible for its prevalence. The S50L substitution, alongside K356T, facilitates efficient entry into lung cells31 without inducing structural changes. Additionally, P621S allows the amino acid residues 621–640, adjacent to the furin-cleavage site, to be a visible structural region in BA.2.86 (Fig. 1G). The cleavage efficiency of the furin-cleavage site in this region is reportedly higher in BA.2.86 than that in BA.24, suggesting that structural stabilization in this region of the BA.2.86 S-protein may influence furin-cleavage efficiency.
On neutralizing-antibody evasion in BA.2.86, we only examined the amino acid substitutions, K356T, V445H, and P621S, in the BA.2.86 S, where the structural alterations were obvious compared to that in XBB.1.5. Hence, other substitutions acquired by BA.2.86 that are reportedly to be important for changes in neutralizing potential were mapped onto the determined BA.2.86-S structure32–34 (Supplementary Fig. 7A). Notably, that the used sera and the panel of monoclonal antibodies differ from that in our experiments. Based on the mapping, nine substitutions, H245N, K356T, R403K, V445H, L452W, N481K, A484K, E554K, and P621S enhanced antibody evasion potential, and one, I332V, reduced it. To assess the neutralizing-antibody evasion potential in BA.2.86 with a different approach, we employed a computational evaluation of the antibody binding potential to the S-proteins of ancestral strain, XBB.1.5, and BA.2.86. A total of 34 antibodies whose RBD-bound structures are registered in the Protein Data Bank (PDB), which are either therapeutic antibodies (in clinical trials or on the market) or neutralizing antibodies, were utilized (Supplementary Fig. 7B and Supplementary Table 2). The interaction energies against these anti-S antibodies were significantly reduced in the BA.2.86-S-protein compared to those of ancestral strain, but not significantly different from those of XBB.1.5 (Supplementary Fig. 7C). In fact, most of the 39 substitutions occurred in the S1 subunit. However, the substitutions were dispersed throughout the structure, not concentrated in the receptor-binding motif and specific RBD epitopes classified as class 1 to 4 epitopes35 (Fig. 1F and Supplementary Fig. 7D). It implies that BA.2.86 may not escape greatly from neutralizing antibodies at serum levels in vaccinated and XBB lineage-infected individuals.
Upon examining the SARS-CoV-2 S-RBD–ACE2 interface, the most notable difference among variants lies in the interaction centered on RBD R/Q493 and ACE2 H34 or E35 (Fig. 3B). The Q493R substitution, occurring only in BA.1 and BA.2, forms an interaction with ACE2 E3536–38. In other variants, a unique hydrogen-bonding network is formed or lost, including surrounding residues centered on RBD Q493 and ACE2 H34, likely having a significant role in ACE2 binding. In fact, the interaction of ACE2 with the up- or down-RBD in BA.2.86 S exhibited a slight difference in the orientation of H34 in ACE2 (Fig. 3B). Hence, SARS-CoV-2 S-RBD R/Q493 and ACE2 H34 may act as modulators of amino acid substitutions within the RBD when the S-protein of each variant binds to ACE2.
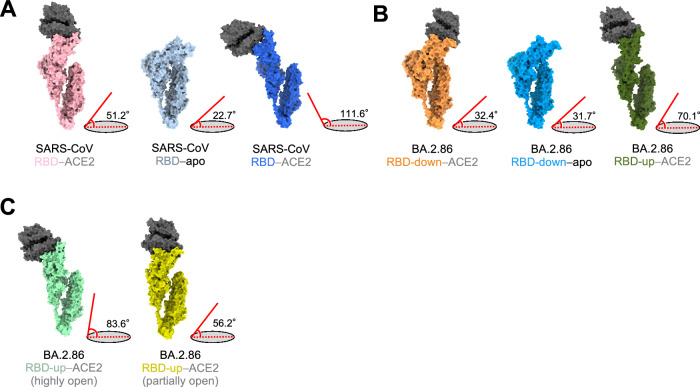
To transition from an all-RBD-down state to one RBD-up, the down-RBD must first adopt the up-conformation and bind to ACE2. Our findings suggest that a similar principle applies to the transition from two-RBD-up to three-RBD-up. However, in the transition from two-RBD-up to three-RBD-up, we found that the two up-RBDs bound to ACE2 exhibit considerable flexibility, resulting in a swaying motion, creating space for an approaching ACE2 receptor. ACE2 binding to the down-RBD then may assist the transition to three-RBD-up (Fig. 2B, C and Supplementary Movie 1 and 2). Indeed, some antibodies reportedly bind to down-RBDs39; our results support the significance of neutralizing antibodies binding to down-RBDs for viral neutralization. Distinct RBD opening angles have also been reported in the cryo-EM structures of SARS-CoV bound to ACE2 under conditions that mimic its entry process40. Of the angles formed between long axes of the SARS-CoV RBD and horizontal plane, the smallest angle is 51.2°. Compared to an angle of 22.7° formed by the SARS-CoV RBD in the closed state, an angle of 51.2° would render a partially-open RBD state (Fig. 5A). In contrast, the angles formed by the down-RBD of the ACE2-bound and apo form in BA.2.86-S-proteins are nearly identical, with values of 32.4° and 31.7°, respectively (Fig. 5B). Accordingly, the structure of BA.2.86-S-RBD-down bound to ACE2 suggests a unique ACE2-S structure. The angle of the highly-open RBD observed when the BA.2.86-ACE2 complex was treated at 42 °C was 83.6°, and the angle of the partially-open RBD was 56.3° (Fig. 5C). The variation in the open angles of RBDs may reflect the nature of variants and the entry process. Furthermore, a unique ACE2-binding mode of the BA.2.86 and JN.1 S-proteins may contribute to neutralizing-antibody evasion because they can bind to ACE2 even though the RBD is down-state and does not require exposure of its RBM that a major target of neutralizing antibodies.
Fig. 5. Comparisons of the angles between RBDs and the horizontal plane.
A–C Protomer of S-proteins in SARS-CoV and SARS-CoV-2 BA.2.86 S─ACE2 complex. Angles between the axis of RBDs in the SARS-CoV-2 BA.2.86 or SARS-CoV and the horizontal plane are shown to the right of each conformation. A Surfaces of SARS-CoV S─ACE2 complex. Left: ACE2-bound conformation 1 (S; light pink). Middle: Unbound-down conformation (S; blue gray). Right: ACE2-bound conformation 3 (S; blue). B Surfaces of BA.2.86 S–ACE2 complex. Left: RBD-down–ACE2 conformation (S; orange). Middle: RBD-down conformation in the apo form (S; sky blue). Right: RBD-up–ACE2 (S; dark olive green). C Surfaces of BA.2.86 S─ACE2 complex treated at 42 °C for 1 h. Left: highly-open RBD conformation (S; mint green). Right: partially-open RBD conformation (S; dark yellow).
Collectively, our findings suggest that comprehensively investigating and elucidating the structural details of the SARS-CoV-2 S-protein, including those from different variants, is crucial to understanding the complex mechanisms underlying SARS-CoV-2 cellular entry. Additionally, they may provide valuable insights for the designing of therapeutics, developing vaccines, and informing public health strategies.
Methods
Ethics statement
All protocols involving specimens from human subjects recruited at Interpark Kuramochi Clinic were reviewed and approved by the Institutional Review Board of Interpark Kuramochi Clinic (approval ID: G2021-004). All human subjects provided written informed consent. All protocols for the use of human specimens were reviewed and approved by the Institutional Review Boards of The Institute of Medical Science, The University of Tokyo (approval IDs: 2021-1-0416 and 2021-18-0617). We have obtained consent to publish information that identifies individuals (including three or more indirect identifiers such as exact age, sex, medical history, vaccination history or medical center of the study participants).
Human serum collection
Convalescent sera were collected from fully vaccinated individuals who had been infected with XBB.1.5 (four 3-dose vaccinated, three 4-dose vaccinated, two 5-dose vaccinated, and one 6-dose vaccinated; the time interval between the last vaccination and infection, 44–435 days; 15–46 days after testing. n = 10 in total; average age: 50.4 years, range: 18–74 years, 30% male). The SARS-CoV-2 variants were identified as previously described2,18,41,42. Sera were inactivated at 56 °C for 30 minutes and stored at –80 °C until use. Details of the convalescent sera are summarized in Supplementary Table 3.
Detection frequency
We calculated the detection frequency of SARS-CoV-2 isolates that circulated from November 1, 2022 to February 19, 2024 with the GISAID viral genomic surveillance data (https://www.gisaid.org; EPI SET ID: EPI_SET_240301rn)43. We excluded the data of the SARS-CoV-2 isolate that (i) lacks collection date and PANGO lineage information; (ii) was retrieved from non-human animals; and (iii) was sampled by quarantine. The lineage of each isolate was reassigned using Nextclade v2.14.044. The Nextclade classification system was used to classify the variants, with 23 A as XBB.1.5; 23B and 23 G as XBB.1.16; 23 F and 23H as EG.5.1; 23I without S: L455S mutation as BA.2.86; 23I with S:L455S mutation as JN.1; and other clades as other. The detection frequency plot was created using ggplot2 v3.4.4.
Phylogenetic tree reconstruction
We obtained genomic sequences and surveillance data of 729 SARS-CoV-2 isolates used to reconstruct the representative SARS-CoV-2 phylogenetic tree in our previous paper4 from the GISAID database. We excluded the data of SARS-CoV-2 isolates that meet the filtering criteria mentioned earlier (see Detection frequency) and whose genomic sequences are no longer than 28,000 base pairs and contain ≥2% of unknown (N) nucleotides. We randomly selected 20 genomic sequences of the SARS-CoV-2 JN.1 variant that were classified based on the Nextclade lineage classification system and satisfy the same filtering criteria. We then pooled the data of the selected JN.1 genomic sequences with those in our previous paper’s dataset. Next, the genomic sequences were aligned to the genomic sequence of the Wuhan-Hu-1 SARS-CoV-2 isolate (NC_045512.2) using the reference-guide multiple pairwise alignment strategy implemented in ViralMSA v1.1.2445. Gaps in the alignment were removed automatically using TrimAl v1.4.rev22 with -gappyout mode46, and the flanking edges at positions 1–292 and 29,588–29,771 were removed manually. The genomic sequence of Wuhan-Hu-1 was subsequently omitted from the alignment. Then, a preliminary phylogenetic tree of SARS-CoV-2 sublineages was reconstructed from the alignment using maximum likelihood-based IQ-TREE v2.2.047. The best-fit nucleotide substitution model was selected automatically using ModelFinder48. Branch support was assessed using ultrafast bootstrap approximation49 with 1000 bootstrap replicates. We identified genomic sequences causing branch length outliers in the preliminary tree using the Rosner test implemented in the EnvStats R package v2.7.050 using R v4.2.2 (https://www.r-project.org/). These genetic sequences were discarded from the final tree reconstruction. The final tree, consisting of 720 SARS-CoV-2 genomic sequences (EPI SET ID: EPI_SET_240301bk), was then reconstructed using the methods described earlier. The tree was visualized using ggtree R package v3.6.251.
Protein expression and purification
S-protein ectodomain (BA.2.86, JN.1), S-protein RBD (BA.2.86, JN.1, XBB.1.5) and human ACE2 were expressed and purified as previously described52. Briefly, pHLsec expression plasmids, encoding the BA.2.86 S-protein ectodomain with six proline substitutions (F817P, A892P, A899P, A942P, K986P, and V987P)53 and deleting the furin-cleavage site (RRAR to GSAG substitution) with a T4-foldon domain, the S-protein RBD (residues 322–536), or soluble human ACE2 (residues 19–617), were constructed. Plasmids expressing the BA.2.86-S ectodomain were generated by DNA synthesis (Eurofins). Plasmids expressing the JN.1-S ectodomain were generated by site-directed overlap extension PCR using pHLsec BA.2.86-S as the template. The resulting PCR fragments, S-protein RBDs and soluble human ACE2, were subcloned into the AgeI-KpnI site of the pHLsec vector using the In-Fusion HD Cloning Kit (Takara, Cat# Z9650N). Nucleotide sequences were determined by DNA sequencing services (Eurofins), and the sequence data were analyzed by SnapGene software v6.1.1 (www.snapgene.com). Details of the primers are summarized in Supplementary Table 4. Each plasmid was transfected into HEK293S GnTI(−) cells, and the expressed proteins in the cell culture supernatant were purified using a cOmplete His-Tag Purification Resin (Roche, Cat# 5893682001) affinity column and either Superose 6 Increase 10/300 GL size-exclusion chromatography (Cytiva, Cat# 29091596) for the BA.2.86-S and JN.1-S ectodomains or Superdex 75 Increase 10/300 GL size-exclusion chromatography (Cytiva, Cat# 29148721) for the S-protein RBD or soluble human ACE2.
Surface plasmon resonance (SPR) analysis
Soluble human ACE2 was covalently immobilized onto a CM5 chip (Cytiva, Cat# 29104988) covalently. A serial dilution of S-protein RBD (BA.2.86, JN.1,and XBB.1.5) ranging in concentrations from 100 to 6.25 nM was prepared in the HBS-EP buffer (Cytiva, Cat# BR100188) and injected on the chip. The response units were recorded in a single cycle mode at 25 °C using the Biacore T200 system. The resulting data were fitted to a 1:1 binding model using Biacore T200 Evaluation software.
Cryo-EM sample preparation and data collection
The BA.2.86 and JN.1 S-protein solutions were incubated at 37 °C for 1 h before cryo-EM grid preparation. An equal volume of PBS with 2 mM EDTA was added to the BA.2.86 S-protein solution (final 1 mM EDTA) to clarify the unknown cryo-EM density. To prepare the BA.2.86 and JN.1 S–ACE2 complex solutions, the purified ACE2 was incubated with BA.2.86- and JN.1-S-proteins at a molar ratio of 1: 3.2 molar ratio (spike: ACE2) at 18 °C for 15 min. Another BA.2.86 S–ACE2 complex solution was incubated at 42 °C for 1 hour to increase the mobility of RBD-ACE2. The samples were then applied to a Quantifoil R2.0/2.0 Cu 300 mesh grid (Quantifoil Micro Tools GmbH), which was freshly glow-discharged for 60 s at 10 mA using PIB-10 (Vacuum Device). The grids were plunged into liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific) with the following settings: temperature 18 °C, humidity 100%, blotting time 5 s, and blotting force 5.
Movies were collected on a Krios G4 (Thermo Fisher Scientific) operated at 300 kV with a K3 direct electron detector (Gatan) at a nominal magnification of 130,000 (0.67 per physical pixel) using a GIF-Biocontinuum energy filter (Gatan) with a 20 eV slit width. The movies were collected with a total exposure of 1.5 s and a total dose of 52.45 (BA.2.86 S), 51.16 (BA.2.86 S with 1 mM EDTA), 51.39 (BA.2.86 S–ACE2; dataset 1), 51.1 (BA.2.86 S–ACE2; dataset 2), 51.0 (BA.2.86 S–ACE2 at 42 °C for 1 hour), and 51.2 (JN.1 S–ACE2) e/Å2 over 50 frames. A total of 3,500 (BA.2.86 S), 801 (BA.2.86 S with 1 mM EDTA), 6,000 (BA.2.86 S–ACE2; dataset 1), 6,719 (BA.2.86 S–ACE2; dataset 2), 4,813 (BA.2.86 S–ACE2 at 42 °C for 1 hour), and 9,018 (JN.1 S–ACE2) movies for BA.2.86 S–ACE2 complexes were collected at a nominal defocus range of 0.8 – 1.8 µm using EPU software (Thermo Fisher Scientific).
Cryo-EM image processing
All datasets were processed using cryoSPARC v4.3.154. For BA.2.86 S, movie frames were aligned, dose-weighted, and CTF-estimated using Patch Motion correction and Patch CTF estimation. A total of 1,156,425 particles were blob-picked, and reference-free 2D classification (K = 150, batch = 200, iteration = 30) was performed to remove the junk particles. A total of 465,894 particles were used for ab-initio reconstruction to obtain the initial models. Two rounds of heterogeneous refinement were performed to classify the closed and one-up conformations of the RBD. For the one-up conformation, 3D classification without alignment focused on the up RBD (K = 4, force hard classification, input mode = simple) was performed, and a class that clearly showed up RBD conformation. A final map was reconstructed by non-uniform refinement, and a local refinement focusing on the RBD was carried out to support model building. For the closed conformation, once the particles were aligned and expanded by non-uniform refinement with C3 symmetry, they were further expanded with C3 symmetry. A 3D classification without alignment focused on the down RBD (K = 6, force hard classification, input mode = simple) was performed, which clearly showed down RBD conformation. A final map was reconstructed by non-uniform refinement with C3 symmetry, and a local refinement focusing on the down RBD was carried out to support model building.
For BA.2.86 S under PBS condition with 1 mM EDTA, movie frames were aligned, dose-weighted, and CTF-estimated using Patch Motion correction and Patch CTF estimation. A total of 280,627 particles were blob-picked, and reference-free 2D classification (K = 150, batch = 200, iteration = 30) was performed to remove junk particles. Heterogeneous refinement was performed using EMD-3562320 (SARS-CoV-2 XBB.1 spike closed state) as a reference map, followed by 3D classification without alignment focused on the down RBD (K = 4, force hard classification, input mode = simple). The final map was reconstructed by non-uniform refinement with C3 symmetry.
For the BA.2.86 S–ACE2 complex, movie frames were aligned, dose-weighted, and CTF-estimated using Patch Motion correction and Patch CTF estimation. A total of 1,537,510 (dataset 1) and 1,909,117 (dataset 2) particles were blob-picked, and reference-free 2D classifications (K = 150, batch = 200, iteration = 30) were performed to remove junk particles on each dataset separately. The initial models were reconstructed using 403,943 particles belonging to dataset 1; two rounds of heterogeneous refinement were performed on each dataset to reconstruct the BA.2.86 S–ACE2 complex map using the initial models as references.
For the RBD-up state bound to ACE2, to address the flexibility of the RBD-up and ACE2 interface, once the particles were aligned by non-uniform refinement with C3 symmetry, the particles were expanded with C3 symmetry. 3D classification (K = 6, force hard classification, input mode = simple) focused on the RBD-up; ACE2 interface without alignment was performed to remove those appearing unclear on the RBD-ACE2 map. The duplicated particles were removed, followed by heterogeneous refinement, and non-uniform refinements were performed to obtain maps of the two-up or three-up RBD states bound to ACE2. A local map of the RBD-up and ACE2 interface was obtained by the iterative runs of local refinement and 3D classification without alignment.
For the RBD-down state bound to ACE2, once the particles were aligned by non-uniform refinement followed by 3D classification (K = 4, force hard classification, input mode = simple) focused on RBD-down and ACE2 interface without alignment was performed to obtain the particles that clearly showed the RBD-down and ACE2. An additional 3D classification (K = 4, force hard classification, input mode = simple) was performed, and global and local maps were obtained by non-uniform refinement or local refinement.
To address the flexibility of the ACE2-bound RBD-up state, 3D Variability Analysis, and subsequent 3D Flexible Refinement28 were performed. 3D Variability Analysis (Number of modes: 4, Filter resolution: 10 Å) was performed using the downsampled particles (96 pixels, 4.02 Å/pix). For the 3D Flex training, the four components solved by 3D Variability analysis were used to initialize the latent coordinates, and the particles were downsampled to 128 pixels (3.01 Å/pix). The mesh was segmented into seven subregions corresponding to three RBDs, three NTDs, and an S2 subunit. All RBD and NTD segments were connected to the S2 subunit. After the training was completed, the volume series was generated by 20 frames on each latent coordinate.
For the BA.2.86 S–ACE2 complex incubated at 42 °C for 1 hour, movie frames were aligned, dose-weighted, and CTF-estimated using Patch Motion correction and Patch CTF estimation. A total of 1,541,394 particles were blob-picked, and reference-free 2D classification (K = 150, batch = 200, iteration = 30) was performed to remove junk particles. Heterogeneous refinement was performed using the initial model of BA.2.86 S–ACE2 complex as a reference map. For unclearly RBD-ACE2 state, 3D classification without alignment focused on the down RBD (K = 4, force hard classification, input mode = simple). For the two-RBD-uptwo ACE2, 3D classification without alignment focused on the RBD-up (K = 4, force hard classification, input mode = simple). The final map was reconstructed by non-uniform refinement. For a non-canonical two-up state (one-highly-open RBD and one-partially-open RBD) bound to ACE2, 3D classification without alignment focused on the two-up RBDs (K = 4, force hard classification, input mode = simple). The global map was obtained by non-uniform refinement.
For the JN.1 S–ACE2 complex, movie frames were aligned, dose-weighted, and CTF-estimated using Patch Motion correction and Patch CTF estimation. A total of 1,099,575 particles were blob-picked, and reference-free 2D classifications (K = 150, batch = 200, iteration = 30) were performed to remove junk particles on each dataset separately. Two rounds of heterogeneous refinement were performed using the initial model of BA.2.86 S–ACE2 complex as a reference map. For the two-RBD-uptwo ACE2, 3D classification (K = 4, force hard classification, input mode = simple) focused on the RBD-up and ACE2 interface without alignment was performed to remove the classes showing the unclear RBD-ACE2 map. A local map of the RBD-up and ACE2 interface was obtained by the iterative runs of local refinement and 3D classification without alignment. For the two-RBD-up−one-RBD-downthree ACE2, once the particles were aligned by non-uniform refinement followed by 3D classification (K = 4, force hard classification, input mode = simple) focused on RBD-down and ACE2 interface without alignment was performed to select the particles that clearly showed the RBD-down and ACE2. The global map was obtained by non-uniform refinement.
The reported global resolutions are based on the gold-standard Fourier shell correlation curve (FSC = 0.143) criteria. Local resolutions were calculated using cryoSPARC55. Figures related to data processing and reconstructed maps were prepared using UCSF Chimera (version 1.16)56 and UCSF Chimera X (version 1.4)57.
Cryo-EM model building and analysis
The structures of the SARS-CoV-2 XBB.1 S-protein closed-2 state (PDB: 8IOT19) and/or human ACE2 protein (PDB:7XB038) were fitted to the corresponding maps using UCSF Chimera. Iterative rounds of manual fitting in Coot (version 0.9.8.7)58 and real-space refinement in Phenix (version 1.20.1)59 were performed to improve the non-ideal rotamers, bond angles, and Ramachandran outliers. The final model was validated using MolProbity software60. The structural models shown in the surface, cartoon, and stick presentations in the figures were prepared using the PyMOL Molecular Graphics System, Version 2.5.0 (http://pymol.sourceforge.net).
To calculate the angles of RBD-apo or RBD-ACE2 complexes, the axis of RBDs was generated. Then the angles between horizontal plane and the axis were calculated with UCSF Chimera X angle command.
Docking simulation
Docking simulations between 34 therapeutically relevant antibodies61,62 and RBD variants were performed using Rosetta release 3.1363. Only high-resolution refinement was employed. The amino acid sequences of each variant were obtained from the GISAID database (https://gisaid.org/lineage-comparison/). Initial backbone coordinates were obtained from the structures of each antibody/RBD complex in the PDB. The side chains of variants were modeled using PyMOL 2.5.0 (http://pymol.sourceforge.net), and deletions in the RBDs were modeled using Modeller 10.564 when necessary. In each docking run, we generated 20 docking poses and averaged the binding energies (I_sc) of the top ten poses to evaluate the physicochemical compatibility of each antibody/RBD complex. For group comparisons, statistical tests were performed using the Friedman test followed by a post-hoc Nemenyi test, utilizing Python libraries such as scipy and scikit_posthocs. The level of significance was set at 5%.
Docking simulations between RBDs and human ACE2 were also performed, as described above. In addition to binding energies, shape complementarity (Sc)65 was assessed using InterfaceAnalyzer in Rosetta, based on the top ten docking poses.
Plasmid construction
Plasmids expressing the SARS-CoV-2 spike proteins of BA.2.86, and its derivatives were prepared in our previous studies42,66,67. Plasmids expressing the spike proteins of BA.2.86 derivatives were generated by site-directed overlap extension PCR using pC-SARS2-S BA.2.86 as the template. The resulting PCR fragments were subcloned into the KpnI-NotI site of the pCAGGS vector68 using the In-Fusion HD Cloning Kit (Takara, Cat# Z9650N). Details of the primers are summarized in Supplementary Table 4. Nucleotide sequences were determined by DNA sequencing services (Eurofins), and the sequence data were analyzed by SnapGene software v6.1.1 (www.snapgene.com).
Cell culture
The Lenti-X 293 T cell line (Takara, Cat# 632180) and HOS-ACE2/TMPRSS2 cells (gifted by Dr. Kenzo Tokunaga), a derivative of HOS cells (a human osteosarcoma cell line; ATCC CRL-1543) stably expressing human ACE2 and TMPRSS269,70, were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Wako, Cat# 044- 29765) containing 10% fetal bovine serum (Sigma-Aldrich Cat# 172012-500 ML), 100 units of penicillin and 100 µg/mL streptomycin (Sigma-Aldrich, Cat# P4333-100ML).
Pseudovirus preparation
Pseudoviruses were prepared as previously described42,66,71. Briefly, lentivirus (HIV-1)-based, luciferase-expressing reporter viruses were pseudotyped with the SARS-CoV-2 S. One day prior to transfection, the LentiX-293T cells were seeded at a density of 2 × 106 cells. Subsequently, they were cotransfected with 1 μg of psPAX2-IN/HiBiT (a packaging plasmid encoding the HiBiT-tag-fused integrase70), 1 μg pWPI-Luc2 (a reporter plasmid encoding a firefly luciferase gene70) and 500 ng of plasmids expressing parental S or its derivatives using TransIT-293 transfection reagent (Mirus, Cat# MIR2704) according to the manufacturer’s protocol. Two days post-transfection, the culture supernatants were harvested and filtered. The amount of produced pseudovirus particles was quantified using the HiBiT assay and the Nano Glo HiBiT lytic detection system (Promega, Cat# N3040) as previously described70. In this system, HiBiT peptide is produced with HIV-1 integrase and forms NanoLuc luciferase with LgBiT, which is supplemented with substrates. In each pseudovirus particle, the detected HiBiT value is correlated with the amount of the pseudovirus capsid protein, HIV-1 p24 protein70. Therefore, we calculated the amount of HIV-1 p24 capsid protein based on the HiBiT value measured, according to a previously described method70. To measure viral infectivity, the same amount of pseudovirus normalized to the HIV-1 p24 capsid protein was inoculated into HOS-ACE2/TMPRSS2 cells. At two days postinfection, the infected cells were lysed with a Bright-Glo luciferase assay system (Promega, Cat# E2620), and the luminescent signal produced by the firefly luciferase reaction was measured using a GloMax explorer multimode microplate reader 3500 (Promega). The pseudoviruses were stored at –80 °C until use.
Neutralization assay
Neutralization assays were performed as previously described2,42,66,67 with some modifications. The assays were mainly conducted by a semi-automated high-throughput method using Fluent780 (Tecan)72. The SARS-CoV-2 spike pseudoviruses (counting ~100,000 relative light units) and serially diluted (40-fold to 29,160-fold dilution at the final concentration) heat-inactivated sera were manually prepared in a 2-ml 96-well plate (Greiner, Cat# 780271) and in 96-well microplates (ThermoFisher Scientific, Cat# 168136), respectively. The pseudoviruses were dispensed and mixed with the sera in 384-well plates (ThermoFisher Scientific, Cat# 164610) on Fluent780 (Tecan). Pseudoviruses without sera were included as controls. After incubation at 37 °C for 1 hour, HOS-ACE2/TMPRSS2 cells (3000 cells/30 μL) were added to the 20 μL mixture of pseudovirus and serum in a 384-well white plate on the device. Two days post- infection, the infected cells were lysed with a Bright-Glo luciferase assay system (Promega, Cat# E2620) on Fluent780 (Tecan), and the luminescent signal was measured and processed using an Infinite200 and a Magellan (TECAN). The assay of each serum sample was performed in quadruplicate, and the 50% neutralization titer (NT50) was calculated using Prism 9 (GraphPad Software).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Source data
Acknowledgements
We would like to thank all members of the Japanese Consortium on Structural Virology (JX-Vir). The supercomputing resource in this study was provided by the Human Genome Center at the Institute of Medical Science, The University of Tokyo. This study was supported in part by AMED ASPIRE (JP24jf0126002, to T. Hashiguchi, and K. S.); AMED SCARDA Kyoto University Immunomonitoring Center (KIC) (JP223fa627009, to T. Hashiguchi); AMED SCARDA Japan Initiative for World-leading Vaccine Research and Development Centers UTOPIA (JP223fa627001, to K. S.), AMED SCARDA Program on R&D of new generation vaccine including new modality application (JP223fa727002, to K. S., and JP243fa727002, to Y. T.); AMED SCARDA Hokkaido University Institute for Vaccine Research and Development (HU-IVReD) (JP223fa627005, to K. M. and JP243fa627005, to Y. T.); AMED BINDS (JP17am0101093 and JP22ama121037, to K. M.); AMED CREST (JP21fk0108463 and JP22gm1810004, to K. M. and JP23gm1810004 to D. K. and Y. T.); AMED (JP20ae0101047 to K. M. and JP23wm0325047 to D. K., T. Hashiguhi, and Y. T.); JSPS KAKENHI Fund for the Promotion of Joint International Research (International Leading Research) (JP23K20041 to K. T. K., and K. S.); The Scientific Research on Innovative Areas and International Group from the MEXT/JSPS KAKENHI (JP20H05873 to K. M.); JSPS Core-to-Core Program (A. Advanced Research Networks) (JPJSCCA20240006, to T. Hashiguchi); JST CREST (JPMJCR20H4, to K. S. and JPMJCR20H8, to T. Hashiguchi); The Cooperative Research Program (Joint Usage/Research Center program) of Institute for Life and Medical Sciences, Kyoto University (to K. S. and K. M.); Hokkaido University Biosurface project (to K. M.); The Naito Foundation (to T. Hashiguchi); Takeda Science Foundation (to K. M.).
Author contributions
A. P., K. O., and J. I. performed phylogenetic and bioinformatics analyses. H. Y., Y. Anraku, K. T. K., Y. N. N., Y. Atarashi, T. Hemmi, D. K., Y. T., S. K., J. S., H. S., K. M., and T. Hashiguchi performed structural and protein-science analyses. Y. K. and K. S. performed cell culture experiments. J. I. performed statistical analyses. H. Y., Y. Anraku, J. I., K. S., and T. Hashiguchi designed the experiments and interpreted the results. H. Y., K. T. K., J. I. and T. Hashiguchi wrote the original manuscript. All authors reviewed and approved the final manuscript for publication.
Peer review
Peer review information
Nature Communications thanks George Gao and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The GISAID datasets used in this study are available from the GISAID database (https://www.gisaid.org; EPI_SET_240301rn and EPI_SET_240301bk). The supplemental tables for the GISAID datasets are available in the GitHub repository (https://github.com/TheSatoLab/BA.2.86_RBD). The atomic coordinates and cryo-EM maps of the structures of BA.2.86 S-protein closed state (PDB: 8WXL, EMD-37910), RBD 1-up state (PDB: 8XUX, EMD-38459), RBD 2-up state in complex with ACE2 (PDB: 8XUY, EMD-38686), RBD 3-up state in complex with ACE2 (PDB: 8XVM, EMD-38690), RBD 2-up and 1-down state in complex with ACE2 (PDB: 8XUZ, EMD-38687), up-RBD and ACE2 interface (PDB: 8XV0, EMD-38688), and down-RBD and ACE2 interface (PDB: 8XV1, EMD-38689), treated at 42 °C (EMD-60905[https://www.ebi.ac.uk/emdb/EMD-60905]) and JN.1-S-protein RBD 2-up state in complex with ACE2 (EMD-60904[https://www.ebi.ac.uk/emdb/EMD-60904]), RBD 2-up and 1-down state in complex with ACE2 (EMD-60906 [https://www.ebi.ac.uk/emdb/EMD-60906]) and up-RBD and ACE2 interface (PDB: 9IU1, EMD-60886) generated in this study have been deposited in the Protein Data Bank (www.rcsb.org), and Electron Microscopy Data Bank (www.ebi.ac.uk/emdb/). Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Kei Sato, Email: keisato@g.ecc.u-tokyo.ac.jp.
Takao Hashiguchi, Email: hashiguchi.takao.1a@kyoto-u.ac.jp.
The Genotype to Phenotype Japan (G2P-Japan) Consortium:
Arnon Plianchaisuk, Keita Matsuno, Naganori Nao, Hirofumi Sawa, Keita Mizuma, Jingshu Li, Izumi Kida, Yume Mimura, Yuma Ohari, Shinya Tanaka, Masumi Tsuda, Lei Wang, Yoshikata Oda, Zannatul Ferdous, Kenji Shishido, Hiromi Mohri, Miki Iida, Takasuke Fukuhara, Tomokazu Tamura, Rigel Suzuki, Saori Suzuki, Shuhei Tsujino, Hayato Ito, Naoko Misawa, Ziyi Guo, Alfredo A. Hinay, Jr., Kaoru Usui, Wilaiporn Saikruang, Spyridon Lytras, Keiya Uriu, Ryo Yoshimura, Shusuke Kawakubo, Luca Nishumura, Yusuke Kosugi, Shigeru Fujita, Jarel Elgin M.Tolentino, Luo Chen, Lin Pan, Wenye Li, Maximilian Stanley Yo, Kio Horinaka, Mai Suganami, Mika Chiba, Kyoko Yasuda, Keiko Iida, Adam Patrick Strange, Naomi Ohsumi, Shiho Tanaka, Eiko Ogawa, Tsuki Fukuda, Rina Osujo, Kazuhisa Yoshimura, Kenji Sadamas, Mami Nagashima, Hiroyuki Asakura, Isao Yoshida, So Nakagawa, Kazuo Takayama, Rina Hashimoto, Sayaka Deguchi, Yukio Watanabe, Yoshitaka Nakata, Hiroki Futatsusako, Ayaka Sakamoto, Naoko Yasuhara, Tateki Suzuki, Yukari Nakajima, Takashi Irie, Ryoko Kawabata, Kaori Sasaki-Tabata, Terumasa Ikeda, Hesham Nasser, Ryo Shimizu, M. S. T. Monira Begum, Michael Jonathan, Yuka Mugita, Sharee Leong, Otowa Takahashi, Takamasa Ueno, Chihiro Motozono, Mako Toyoda, Akatsuki Saito, Anon Kosaka, Miki Kawano, Natsumi Matsubara, Tomoko Nishiuchi, Jiri Zahradnik, Prokopios Andrikopoulos, Miguel Padilla-Blanco, and Aditi Konar
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52808-2.
References
- 1.WHO. BA.2.86 Initial Risk Evaluation, 21 November, 2023.) (2023).
- 2.Kaku, Y. et al. Virological characteristics of the SARS-CoV-2 JN.1 variant. Lancet Infect. Dis.24, e82 (2024). [DOI] [PubMed] [Google Scholar]
- 3.Yang, S. et al. Fast evolution of SARS-CoV-2 BA.2.86 to JN.1 under heavy immune pressure. Lancet Infect. Dis.24, e70–e72 (2024). [DOI] [PubMed] [Google Scholar]
- 4.Tamura, T. et al. Virological characteristics of the SARS-CoV-2 BA.2.86 variant. Cell Host Microbe32, 170–180.e112 (2024). [DOI] [PubMed] [Google Scholar]
- 5.Yang, X. et al. Immune escape of BA.2.86 is comparable to XBB subvariants from the plasma of BA.5- and BA.5-XBB-convalescent subpopulations. J. Med Virol.96, e29417 (2024). [DOI] [PubMed] [Google Scholar]
- 6.Willett, B. J. et al. Omicron BA.2.86 cross-neutralising activity in community sera from the UK. Lancet402, 2075–2076 (2023). [DOI] [PubMed] [Google Scholar]
- 7.Qu, P. et al. Immune evasion, infectivity, and fusogenicity of SARS-CoV-2 omicron BA.2.86 and FLip variants. Cell.187, 585–595.e6 (2023). [DOI] [PMC free article] [PubMed]
- 8.Khan, K. et al. Evolution and neutralization escape of the SARS-CoV-2 BA.2.86 subvariant. Nat. Commun.14, 8078 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li, W. et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature426, 450–454 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantuti-Castelvetri, L. et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science370, 856–860 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daly, J. L. et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science370, 861–865 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baggen, J. et al. TMEM106B is a receptor mediating ACE2-independent SARS-CoV-2 cell entry. Cell186, 3427–3442.e3422 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamming, I. et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol.203, 631–637 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell181, 271–280.e278 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann, H. et al. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA102, 7988–7993 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wrapp, D. et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science367, 1260–1263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai, Y. et al. Distinct conformational states of SARS-CoV-2 spike protein. Science369, 1586–1592 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saito, A. et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2.75 variant. Cell Host Microbe30, 1540–1555.e1515 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tamura, T. et al. Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants. Nat. Commun.14, 2800 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamura, T. et al. Virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant. Nat. Commun.15, 1176 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, B. A. et al. Furin cleavage site is key to SARS-CoV-2 pathogenesis. Nature591, 293–299 (2020). [DOI] [PMC free article] [PubMed]
- 22.Jackson, C. B., Farzan, M., Chen, B. & Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol.23, 3–20 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsujino, S. et al. Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant. Microbiol Immunol. 68, 305–330 (2024). [DOI] [PubMed]
- 24.Stalls, V. et al. Cryo-EM structures of SARS-CoV-2 Omicron BA.2 spike. Cell Rep.39, 111009 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao, Y. et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature608, 593–602 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai, Y. et al. Structural basis for enhanced infectivity and immune evasion of SARS-CoV-2 variants. Science373, 642–648 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saville, J. W. et al. Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants. Nat. Commun.13, 742 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Punjani, A. & Fleet, D. J. 3DFlex: determining structure and motion of flexible proteins from cryo-EM. Nat. Methods20, 860–870 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mannar, D. et al. Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding. Cell Rep.37, 110156 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Philip, A. M., Ahmed, W. S. & Biswas, K. H. Reversal of the unique Q493R mutation increases the affinity of Omicron S1-RBD for ACE2. Comput Struct. Biotechnol. J.21, 1966–1977 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, L. et al. SARS-CoV-2 BA.2.86 enters lung cells and evades neutralizing antibodies with high efficiency. Cell187, 596–608.e517 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao, Y. et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature614, 521–529 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, Q. et al. Antigenicity and receptor affinity of SARS-CoV-2 BA.2.86 spike. Nature624, 639–644 (2023). [DOI] [PubMed] [Google Scholar]
- 34.Bdeir, N. et al. Reverse mutational scanning of spike BA.2.86 identifies the epitopes contributing to immune escape from polyclonal sera. Preprint at medRxiv, 10.1101/2024.01.03.23300575 (2024).
- 35.Barnes, C. O. et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature588, 682–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao, Z. et al. Omicron SARS-CoV-2 mutations stabilize spike up-RBD conformation and lead to a non-RBM-binding monoclonal antibody escape. Nat. Commun.13, 4958 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, Y. et al. Structural and biochemical mechanism for increased infectivity and immune evasion of Omicron BA.2 variant compared to BA.1 and their possible mouse origins. Cell Res. 32, 609–620 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, L. et al. Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1. Cell185, 2952–2960.e2910 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen, Y. et al. Broadly neutralizing antibodies to SARS-CoV-2 and other human coronaviruses. Nat. Rev. Immunol.23, 189–199 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song, W., Gui, M., Wang, X. & Xiang, Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog.14, e1007236 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzuki, R. et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature603, 700–705 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uriu, K. et al. Transmissibility, infectivity, and immune evasion of the SARS-CoV-2 BA.2.86 variant. Lancet Infect. Dis.23, e460–e461 (2023). [DOI] [PubMed] [Google Scholar]
- 43.Khare, S. et al. GISAID’s Role in Pandemic Response. China CDC Wkly3, 1049–1051 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aksamentov, I., Roemer, C., Hodcroft, E. B. & Neher, R. A. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw.6, 3773 (2021). [Google Scholar]
- 45.Moshiri, N. ViralMSA: massively scalable reference-guided multiple sequence alignment of viral genomes. Bioinformatics37, 714–716 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minh, B. Q. et al. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol.37, 1530–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods14, 587–589 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol.35, 518–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Millard, S. P. An R Package for Environmental Statistics. (Springer, New York, 2013).
- 51.Yu, G. Using ggtree to visualize data on tree-like structures. Curr. Protoc. Bioinforma.69, e96 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Hashiguchi, T. et al. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat. Struct. Mol. Biol.18, 135–141 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Hsieh, C. L. et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science369, 1501–1505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Cardone, G., Heymann, J. B. & Steven, A. C. One number does not fit all: mapping local variations in resolution in cryo-EM reconstructions. J. Struct. Biol.184, 226–236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput Chem.25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci.27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D. Biol. Crystallogr66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D. Struct. Biol.75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams, C. J. et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci.27, 293–315 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider, C., Raybould, M. I. J. & Deane, C. M. SAbDab in the age of biotherapeutics: Updates including SAbDab-nano, the nanobody structure tracker. Nucleic Acids Res. 50, D1368–d1372 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dunbar, J. et al. SAbDab: the structural antibody database. Nucleic Acids Res. 42, D1140–D1146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leman, J. K. et al. Macromolecular modeling and design in Rosetta: recent methods and frameworks. Nat. Methods17, 665–680 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eswar, N. et al. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinforma. Chapter5, 6 (2006). Unit-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lawrence, M. C. & Colman, P. M. Shape complementarity at protein/protein interfaces. J. Mol. Biol.234, 946–950 (1993). [DOI] [PubMed] [Google Scholar]
- 66.Kaku, Y. et al. Antiviral efficacy of the SARS-CoV-2 XBB breakthrough infection sera against omicron subvariants including EG.5. Lancet Infect. Dis.23, e395–e396 (2023). [DOI] [PubMed] [Google Scholar]
- 67.Kosugi, Y. et al. Characteristics of the SARS-CoV-2 omicron HK.3 variant harbouring the FLip substitution. Lancet Microbe.5, e313 (2024). [DOI] [PubMed]
- 68.Niwa, H., Yamamura, K. & Miyazaki, J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 108, 193–199 (1991). [DOI] [PubMed] [Google Scholar]
- 69.Ferreira, I. et al. SARS-CoV-2 B.1.617 mutations L452R and E484Q are not synergistic for antibody evasion. J. Infect. Dis.224, 989–994 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ozono, S. et al. SARS-CoV-2 D614G spike mutation increases entry efficiency with enhanced ACE2-binding affinity. Nat. Commun.12, 848 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Motozono, C. et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe29, 1124–1136.e1111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garcia-Beltran, W. F. et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell185, 457–466.e454 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The GISAID datasets used in this study are available from the GISAID database (https://www.gisaid.org; EPI_SET_240301rn and EPI_SET_240301bk). The supplemental tables for the GISAID datasets are available in the GitHub repository (https://github.com/TheSatoLab/BA.2.86_RBD). The atomic coordinates and cryo-EM maps of the structures of BA.2.86 S-protein closed state (PDB: 8WXL, EMD-37910), RBD 1-up state (PDB: 8XUX, EMD-38459), RBD 2-up state in complex with ACE2 (PDB: 8XUY, EMD-38686), RBD 3-up state in complex with ACE2 (PDB: 8XVM, EMD-38690), RBD 2-up and 1-down state in complex with ACE2 (PDB: 8XUZ, EMD-38687), up-RBD and ACE2 interface (PDB: 8XV0, EMD-38688), and down-RBD and ACE2 interface (PDB: 8XV1, EMD-38689), treated at 42 °C (EMD-60905[https://www.ebi.ac.uk/emdb/EMD-60905]) and JN.1-S-protein RBD 2-up state in complex with ACE2 (EMD-60904[https://www.ebi.ac.uk/emdb/EMD-60904]), RBD 2-up and 1-down state in complex with ACE2 (EMD-60906 [https://www.ebi.ac.uk/emdb/EMD-60906]) and up-RBD and ACE2 interface (PDB: 9IU1, EMD-60886) generated in this study have been deposited in the Protein Data Bank (www.rcsb.org), and Electron Microscopy Data Bank (www.ebi.ac.uk/emdb/). Source data are provided with this paper.