Abstract
Treatment of Limbal Stem Cell Deficiency (LSCD), based on autologous transplantation of the patient’s stem cells, is one of the few medical stem cell therapies approved by the European Medicines Agency (EMA). It relies on isolating and culturing in vivo Limbal Epithelial Stem Cells (LESC) and then populating them on the fibrin substrate, creating a scaffold for corneal epithelial regeneration. Such a solution is then implanted into the patient’s eye. The epithelial cell culture process is specific, and its results strongly depend on the initial cell seeding density. Achieving control of the density and repeatability of the process is a desirable aim and can contribute to the success of the therapy. The study aimed to test bioprinting as a potential technique to increase the control over LESCs seeding on a scaffold and improve process reproducibility. Cells were applied to 0.5 mm thick, flat, transparent fibrin substrates using extrusion bioprinting; the control was the traditional manual application of cells using a pipette. The use of 3D printer enabled uniform coverage of the scaffold surface, and LESCs density in printed lines was close to the targeted value. Moreover, printed cells had higher cell viability than those seeded traditionally (91.1 ± 8.2% vs 82.6 ± 12.8%). The growth rate of the epithelium was higher in bioprinted samples. In both methods, the epithelium had favorable phenotypic features (p63 + and CK14 +). 3D printing constitutes a promising approach in LSCD therapy. It provides favorable conditions for LESCs growth and process reproducibility. Its application may lead to reduced cell requirements, thereby to using fewer cells on lower passages, which will contribute to preserving LESCs proliferative potential.
Keywords: LESC, Fibrin, Cell culture, LSCD, Bioprinting, Epithelium, Ocular regeneration
Subject terms: Preclinical research, Stem-cell research
Introduction
Limbal stem cell deficiency (LSCD) is a rare corneal disorder caused by dysfunction and/or insufficient amount of corneal limbal stem cells, affecting 1–5/10 000 people worldwide. It can be genetic, idiopathic, or acquired—certain conditions, i.e., ocular surface disorders, such as Stevens-Johnson syndrome, ocular cicatricial pemphigoid, and chemical or thermal burns, can contribute to developing LSCD and, consequently, to impaired vision or blindness. Efficient and widely available treatment options for LSCD are limited and challenging. Corneal transplantation remains the golden standard of treatment. However, the number of donors, compared to the needs, is insufficient. Moreover, some patients have contraindications for full-thickness keratoplasty1,2.
Transplanting a cultured corneal epithelial cell equivalent containing potential adult stem cells and corneal epithelium constitutes today an alternative enabling the restoration of the ocular surface3. The most commonly performed procedure in unilateral LSCD is the conjunctival limbal autograft (CLAU), in which an opposing pair of two clock-hour-long conjunctival-limbal biopsies are harvested from the healthy eye to be grafted to the affected eye. This technique, however, is associated with the risk of inducing iatrogenic LSCD in the healthy donor eye4,5. Therefore, other therapeutic methods have been introduced. One is cultivated limbal epithelial transplant (CLET)—a technique in which a small biopsy (approximately 2–3 mm2) is harvested in a healthy eye. Cells are then expanded ex vivo on a human amniotic membrane or a fibrin carrier scaffold and grafted onto the affected eye. CLET provides a long-term solution to LSCDs by transplanting a self-renewing limbal stem cell population that can maintain a clear corneal epithelium6,7. This method, however, also has its limitations.
The finding that human limbal cell cultures contain holoclones8 led to the first therapeutic use of such cultures propagated with a feeder layer in the regeneration of corneal epithelium9. Similar therapy, with some modifications, was successfully reproduced by Ivan Schwab in the USA10, Ray Tsai in Taiwan11, and later by V Sangwan in India12. In the above-mentioned treatment, the culture protocol required the use of lethally irradiated 3T3 feeder cells in co-culture with the LESCs.
In recent years, numerous protocols for culturing LESCs have been developed; they differ significantly from each other. They assume either the use of limbal explants (explant technique) or single cell culture (suspension technique), the presence or absence of feeder-layer (FL) usage (e.g. murine 3T3- J2 FL), as well as the application of different LESCs markers, and various culture media compositions8,13–15. Standard complex culturing media therefore contain fetal bovine serum (FBS) in addition to various growth hormones and cholera toxin16. Moreover, isolated LESCs are seeded on different carriers for cell expansion and transplantation in each method. The most frequently used scaffolds are human amniotic membrane (AM) and fibrin matrix gels17,18. What is common for all those protocols is that LESCs are applied using traditional—manual, cell seeding method, i.e., pipetting.
Fibrin is considered a versatile substrate for cell growth, with great potential to be used in tissue regeneration and wound healing19. Next to the amniotic membrane, it is the most frequently used material in CLET procedures, and fibrin glue has been used to secure the amniotic membrane in those therapies20. Fibrin can also be used as a temporary basal membrane for the epithelium to grow on its surface. It is also considered far superior in enhancing the epithelial cells’ survival, growth, and migration compared to collagen I and puramatrix21. Due to its ability to mimic extracellular matrix (ECM), fibrin is considered today the most optimal material for stem cell seeding: it forms a favorable ground for cell implantation and a supportive environment for engraftment or accelerating stem cell differentiation. Thanks to its unique features, it reduces the number of stem cells required for final tissue reconstruction and stimulates stem cell self-renewal22. Moreover, use of the fibrin may also result in obtaining holoclone formation on the scaffold23.
The density of cell deposition on the materials to be implanted is essential due to the paracrine exchange of chemical signals between epithelial cells and stem cells. Too high cell density in the preparation may result in a limited growth surface for the transplanted cells. In turn, too low cell density may lead to loss of cell–cell or cell-ECM adhesion-related survival signals and, as a consequence, to cell death called anoikis, form of apoptosis22. The mechanisms responsible for acute donor cell death after transplantation are complex and result from many factors, including disruption of interaction between cells and loss of survival signals from matrix attachment. Stem cells require a strictly controlled environment to maintain high viability throughout the process, until transplantation22. The use of a 3D printer can provide such accurate monitoring of the cell seeding and growing procedure24.
Cell density can be controlled by extrusion volumes. Moreover, a typical printing pattern consisting of lines separated by empty spaces can result in an epithelial regeneration model, allowing for faster in vitro expansion and reducing the number of cells required. Pramotton et al.25 tested this approach using polydimethylsiloxane (PDMS) magnetic stencil. In their research, seeding epithelial cells at high density in a confined channel led to three times faster complete epithelization of the target surface. However, to the best of our knowledge this approach has not been applied to LESCs yet, and 3D printing of the epithelium has so far been carried out only in such a way that the cells are distributed evenly over the entire surface26,27.
We assume that simple extrusion-based 3D printing of cell suspension combined with well-defined fibrin material (with flat surface) can constitute a faster and more efficient method to obtain epithelium scaffolds for cornea regeneration. In our opinion numerous advantages of this method may contribute to a better standardization of the process, and its wider implementation. However, the 3D printer in this case is used to obtain a two-dimensional pattern, analogous to the pattern obtained by Pramotton, and given the relatively simple structure of the epithelium, the multilayer structure (that could be obtained by 3D printer) is not required.
Printing cells, especially epithelial ones, at the appropriate seeding density provides an optimal growth niche for cells transplanted on fibrin. Such a microenvironment creates a favorable condition for cells to communicate—exchange signals with stem cells to induce the cellular activity, and avoid anoikis process. Such effect cannot be achieved to such an extent by traditional method of cell seeding—pipetting.
The primary objective of this study was to compare LESC cultivation at in vitro conditions using bioprinting and pipetting (droplet seeding) method. LESCs were isolated from porcine eyes, and after the in vitro propagation process, they were seeded on fibrin either by bioprinting or manually, by droplet method. First, the extrusion-based printing process was characterized in terms of (1) initial cell viability during printing, (2) cell density control, (3) printed line diameter, (4) cell distribution on material, (5) print reproducibility and (6) cells viability on the material. The results were compared to those obtained through droplet seeding. Then, flat fibrin scaffolds were seeded with cells using 3D printing and droplet method. The cell coverage area was analyzed for different initial seeding densities over time to assess the rate of epithelialization. Additionally, morphology and phenotype differences were analyzed for both methods to confirm LESCs presence on the fibrin scaffold.
In our opinion, using 3D cell printing to prepare scaffolds in the Good Manufacturing Practice (GMP) regime could significantly improve the quality of Advanced Therapy Medicinal Products (ATMP)28 and hence, increase their presence in the clinical trials. Until now, the cell banks’ quality control of ATMPs preparations has concentrated mostly on the type and number of cells, their origin and viability. Since the density of epithelial cell seeding largely determines the transplantation’s success, optimizing this parameter in scaffold preparation is crucial.
The 3D printing method guarantees a highly repeatable process: not only does it allow to seed cells in a controlled, very precise manner, maintaining appropriate even distances between the patterns, but it also enables the achievement of repeatability of appropriate growth niches for transplanted cells. Such features can contribute to replacing the traditional method of pipetting cell suspensions in the production of cell grafts (ATMP).
Results
Cell isolation and characteristics
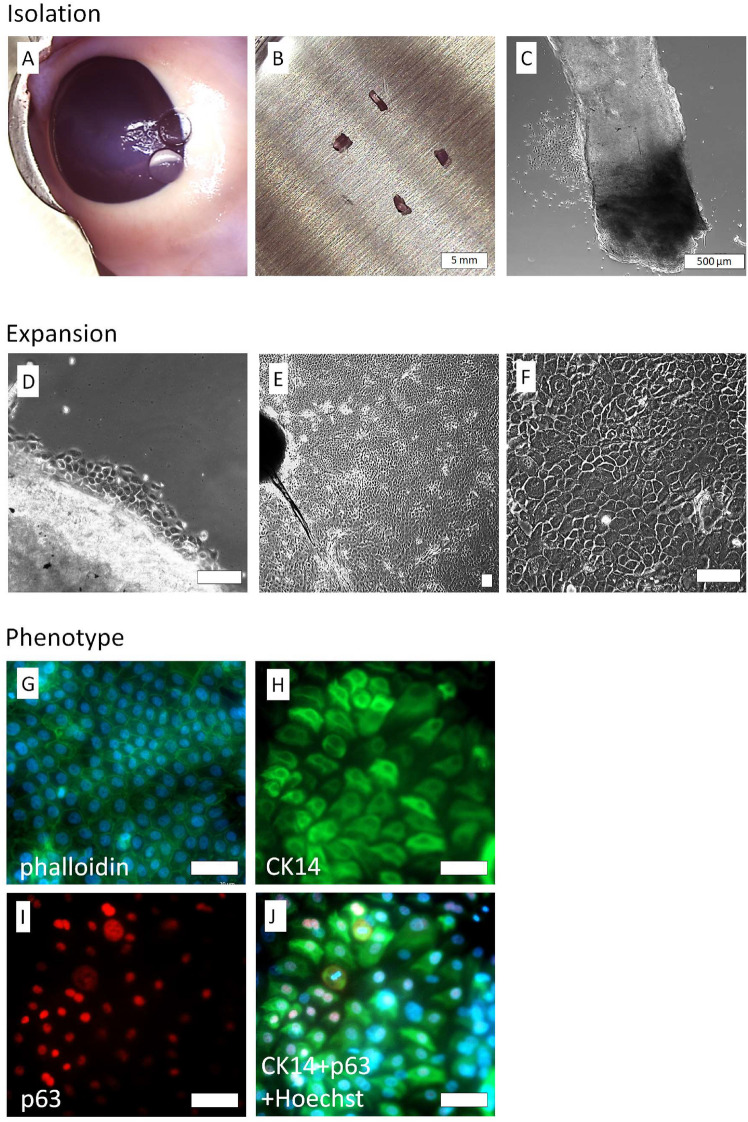
Cell culture containing LESCs was obtained from the explants of the limbal region of porcine eyeballs (Fig. 1A–C). First, those tissues were subjected to enzymatic treatment; after a 3-day culture in CNT medium, the LESCs started to migrate from the explants (Fig. 1D). Their morphology closely resembled limbal epithelial stem cells—small rounded cells with high proliferative potential. Within 7–12 days, most of the surface was covered with cells, although their morphology had become more heterogenic. Also, more giant cells were present (Fig. 1E,F). Fluorescent staining of the cytoskeleton by phalloidin was characteristic of the epithelial cells—most fibers were located in junction areas (Fig. 1G). Also, most cells were CK14 + and p63 + (Fig. 1H–J).
Fig. 1.
LESC cells isolated from porcine eye explants and its characterization. Explants of corneoscleral junction (A, B, C), LESC cells migrated from explants after 3 days (D), confluent culture after 7 days (E, F, scalebar = 100 µm). Cells with characteristic epithelial cytoskeletal structure visualized by phalloidin staining (G). CK14 (H) and p63 (I) antigens are present in high percentage. Additional composite image with Hoechst staining and CK14 and p63 (J) showing that significant fraction of cells are CK14 and p63 positive (scalebar = 200 µm).
Data collected during numerous isolations indicated that the obtained cells had an average viability of 85 ± 0.1% and contained at least 70% of p63 + cells. Cell culture could be carried out until passage P4, although the best results in scaffold preparations were obtained in lower passages. In our experiments with fibrin scaffolds, we used only cells on passage P3.The efficiency of isolation from 1 mm2 explant was approximately 2.6 ± 1.3 × 104 cells on P1, 8.8 ± 4.5 × 104 cells on P2 and 1.1 ± 0.5 × 105 on P3. It should be noted, however, that cell yield was highly determined by individual variability of donors.
Extrusion-based bioprinting process characteristics
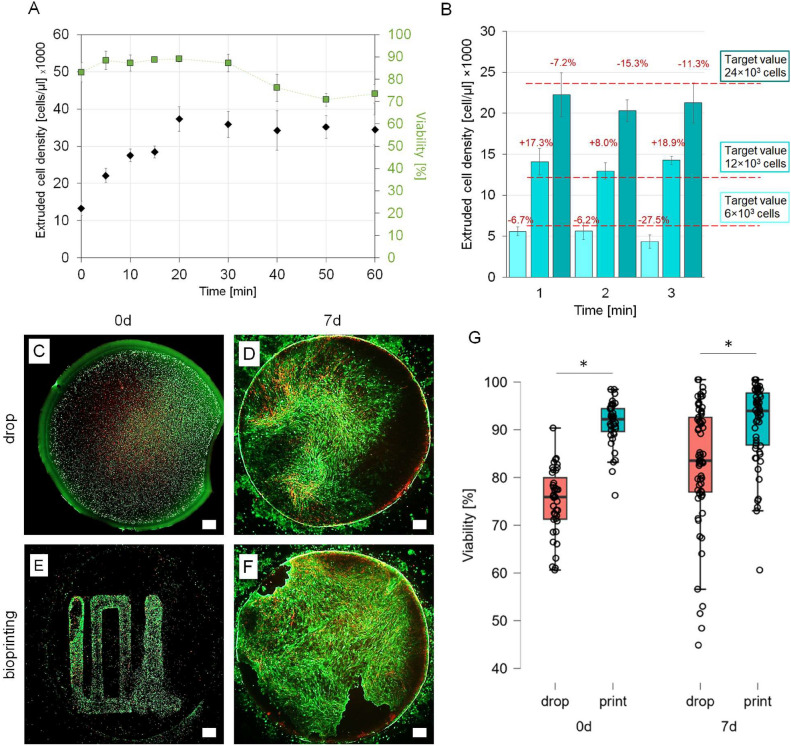
Viability and cell concentration in extruded cell suspension were tested to determine whether the process associated with 3D printing has no harmful effect on the cells. It was also verified whether printing from the cell suspension could be reliable regarding the final cell count in extruded volumes. It was reported that for 100 µl load and 16 000 cell/µl cells sedimented slowly in the printing nozzle, and within 20 min, cell concentration increased almost four times (Fig. 2A). The sedimentation process could be eliminated almost entirely by 1% methyl cellulose (MC); but in this case MC caused deterioration in cell adhesion (S1, Supplementary Data). Performed later experiments with smaller volumes (8 µl) and shorter time (1 min for each print) for cell culture medium without MC demonstrated that extruded cell concentration remained stable (no statistical difference), and obtained results were close to the target values (Fig. 2B). Due to the relatively high stability of the cell suspension in the medium, the use of MC additive was abandoned in the next experiments.
Fig. 2.
Characteristics of the bioprinting process. Cell density and viability of cells extruded from the 3D printer nozzle at selected time points for 60 min (A). Stability of the expected density of extruded cells during the bioprinting process at 1, 2 and 3 min of extrusion (B). Imaging of calcein (green)/PI (red) cell staining for manually seeded (C, D) and bioprinted (E, F) samples on the day of culture initiation and after 7 days of culture. The green color indicates the high cell viability. The red color indicates the dead cells. Graph of cell viability seeded manually and bioprinted at two time points (G). Bioprinted cells were significantly more viable than manually deposited control on the day of culture initiation (*p < 0.001, n = 39) and also after 7 days of culture (*p < 0.001, n = 58). Scalebar = 1 mm.
The viability of cells inside the nozzle was high during the first 30 min of the experiment (Fig. 2A). Printed cells (Fig. 2C,E) had a significantly higher viability than those deposited by pipetting (Fig. 2G), i.e. 91.3 ± 4.6% vs 75 ± 4.6%. In addition, printed cells covered the surface more thoroughly (Fig. 2D,F). After seven days, differences in cell viability between printed and manually pipetted samples were still significant (91.1 ± 8.2% vs 82.6 ± 12.8%, Fig. 2G).
Then, LESCs were printed in an 8 × 8 mm rectilinear dense pattern to cover the entire surface with cells, determine cell distribution in printed area, and compare it with manual drop deposition (Fig. 3). Patterns with 500 cells/mm2 were printed, and the same number of cells as in 0.8 µl volume of printed pattern was placed in drops.
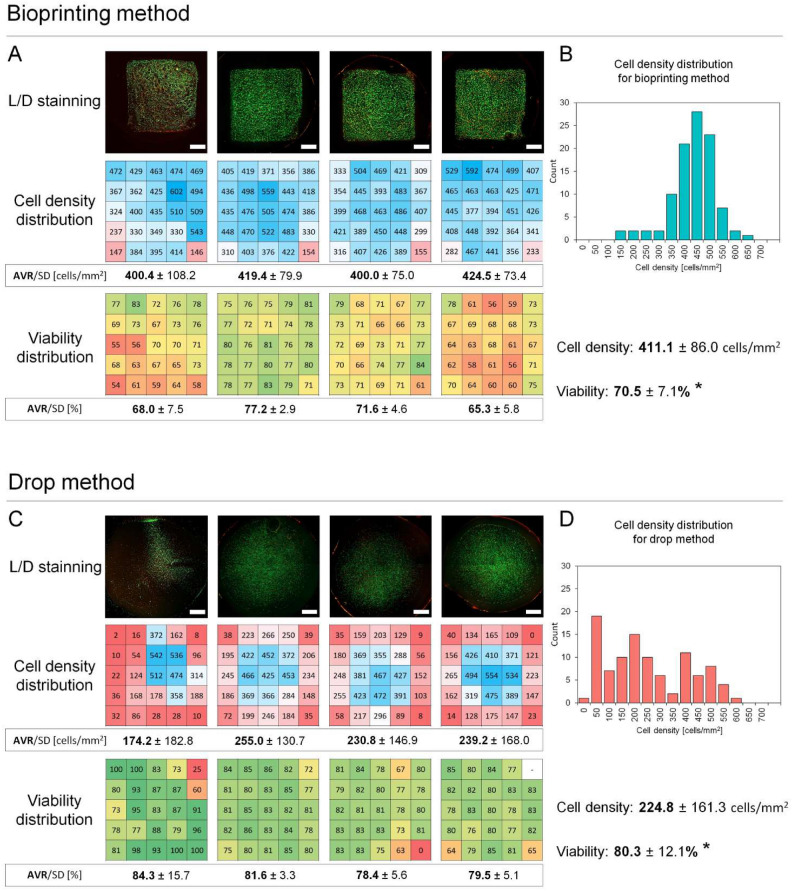
Fig. 3.
Cell density distribution (cell/mm2) and viability (%) for dense bioprinted vs drop deposited samples. Imaging of calcein (green)/PI (red) cell staining, heatmaps of cell density distribution and cell viability after their deposition on fibrin scaffolds by bioprinting (A) and drop method (C). Histograms of cell density distribution for bioprinting (B) and drop method (D). Fibrin surface can covered by cells more homogamically by bioprinting compared to manual drop deposition where cells are mostly located in central area and cell density on fibrin edges is very low. However dense bioprinting cases cell damage which is represented by significant drop in cell viability compared to control (p = 0.0203, n = 4). Scalebar = 2 mm.
Cell density on fibrin has been shown to be more homogenous when cells are deposited by bioprinting compared to manual cell deposition (411.1 ± 86.0 cells/mm2 vs 224.8 ± 161.3 cells/mm2, Fig. 3). For bioprinted samples, the density of cells in most areas ranged from 350–600 cells/mm2 (Fig. 3B). Lower cell counts were noted only on the edges (at the start and stop printing points). For traditionally prepared samples, cell density ranged from 50–550 cells/mm2, and higher cell counts were only noted in the central area (Fig. 3D). In contrast, the edges of counting areas were almost entirely depleted with cells. In this case, the count of cells was nearly twice smaller because cells were deposited outside the counting area (and, frequently, outside the fibrin scaffold). Aggregated data of the distribution of cell counts presented in the histogram showed relatively narrow and unimodal distribution for bioprinted samples and bimodal, wide distribution for manually prepared models (Fig. 3B,D). The repeatability of the average cell density is higher for the printing method than for the manual method. The average cell density for the printing method ranged from 400.4 to 424.5 cells/mm2 (max. 3.3% variation) and the average density for the manual method ranged from 174.2 to 255 cells/mm2 (max. 22.5% variation).
LESCs’ viability 20 min after dense bioprinting was significantly lower than of the manually deposited control (70.5 ± 5.2% vs 80.3 ± 3.6%, p = 0.02, n = 4, Fig. 3A,C).
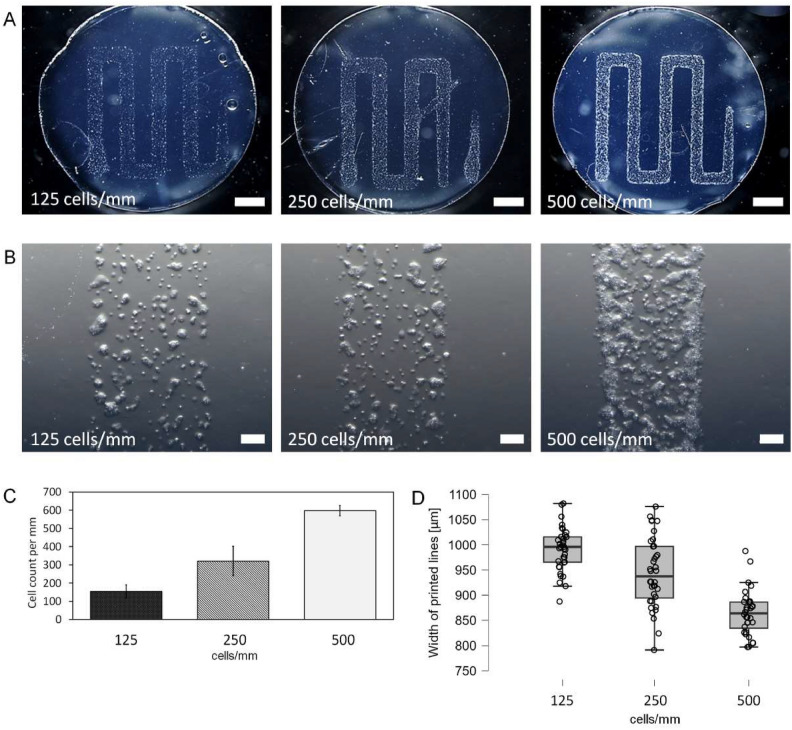
For bioprints with widely spared lines (1.2 mm apart), such parameters as deposited cell density in line and line width were measured. The experiment was performed for three numbers of cells: 6 × 103, 1.2 × 104 and 2.4 × 104, which gives 125, 250 and 500 cells/mm of printed path, respectively (Fig. 4). Cells were not evenly distributed in lines but were forming groups and were located more densely on the edges of the lines (Fig. 4B). Cell count in 1 mm line was slightly higher than intended values for all samples (Fig. 4C). After 15 min most of the cells’ morphology indicated cell adhesion (Fig. 4B).
Fig. 4.
Rectilinear patterns printed with LESC cells on fibrin for tree different cell concentrations. Microphotograph of a fibrin sample with the pattern of cell suspension printed for three different concentrations (A). Microscopic image of the distribution of cells on a printed line section (B). Cell count in each bioprint per mm of printed path (C). Width of printed cell lines for each cell concentration (D). Cell count in each print is close to targeted values of 500, 250 and 125 cells/mm. Cell distribution in line is not homogenic – cells are mostly located in groups on the edges of the lines. Scalebar = 2 mm (A), scalebar = 0.2 mm (B).
Line width was similar within groups and within each sample. Line width variation from the average value was less than 5% for all samples within a group. Individual lines had less than 8.6% variation from average sample line width (Table 1). Line width decreased with increasing cell concentration (Fig. 4D). Differences in line width between 125 vs 250 cells/mm, 125 vs 500 cells/mm, 250 vs 500 cells/mm were statistically significant (Table 1). Concluding, the obtained line width was much higher than printing nozzle diameter of 200 µm.
Table 1.
Width of bioprinted lines for samples printed with three cell concentrations. Kruskal–Wallis test 125 vs 250 cells/mm #p = 0.0186, 125 vs 500 cells/mm ##p < 0.001, 250 vs 500 cells/mm ###p < 0.001.
| Sample | 125 cells/mm | 250 cells/mm | 500 cells/mm | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample number | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 |
| Line number | |||||||||
| I | 1031,4 | 1081,6 | 975,0 | 972,2 | 1027,3 | 997,1 | 871,4 | 966,9 | 886,2 |
| 886,8 | 1015,3 | 964,1 | 918,6 | 948,0 | 979,7 | 878,4 | 925,0 | 856,1 | |
| 974,5 | 980,6 | 1015,2 | 918,4 | 926,6 | 951,4 | 894,3 | 987,6 | 844,9 | |
| II | 965,2 | 992,7 | 937,6 | 887,7 | 896,4 | 1048,6 | 886,5 | 875,0 | 816,3 |
| 956,0 | 1001,4 | 1039,2 | 853,4 | 1007,2 | 1047,5 | 886,3 | 873,9 | 845,8 | |
| 989,9 | 1018,5 | 1014,9 | 902,0 | 1010,8 | 1056,2 | 861,5 | 866,2 | 825,9 | |
| III | 1008,9 | 1032,9 | 991,1 | 912,5 | 996,6 | 966,7 | 918,6 | 887,4 | 856,1 |
| 998,4 | 936,4 | 955,0 | 926,6 | 955,4 | 1076,0 | 854,1 | 906,5 | 856,1 | |
| 1002,9 | 966,9 | 995,4 | 864,6 | 925,8 | 976,9 | 822,2 | 846,0 | 876,7 | |
| IV | 917,3 | 1012,2 | 924,3 | 876,6 | 948,3 | 1047,3 | 797,5 | 837,1 | 797,8 |
| 1024,0 | 1055,5 | 941,4 | 870,2 | 874,5 | 916,7 | 886,0 | 805,1 | 825,7 | |
| 995,3 | 1079,3 | 1005,6 | 888,2 | 823,9 | 791,6 | 878,3 | 825,8 | 805,7 | |
| Average for lines | |||||||||
| I | 964,2 | 1025,8 | 984,8 | 936,4 | 967,3 | 976,1 | 881,4 | 959,8 | 862,4 |
| II | 970,4 | 1004,2 | 997,2 | 881,0 | 971,5 | 1050,8 | 878,1 | 871,7 | 829,3 |
| III | 1003,4 | 978,7 | 980,5 | 901,3 | 959,2 | 1006,5 | 865,0 | 880,0 | 863,0 |
| IV | 978,9 | 1049,0 | 957,1 | 878,3 | 882,3 | 918,5 | 853,9 | 822,7 | 809,7 |
| Average for samples | |||||||||
| I–IV | 979,2 | 1014,4 | 979,9 | 899,3 | 945,1 | 988,0 | 869,6 | 883,5 | 841,1 |
| ± 40,9 | ± 41,7 | ± 34,5 | ± 31,5 | ± 58,1 | ± 75,8 | ± 31,3 | ± 52,9 | ± 26,2 | |
| Total average | 991.2 ± 42.5 #,## | 944.1 ± 68.4 #,### | 864.8 ± 42.4 ##,### | ||||||
| Max. spread (in group) | 1.2% | 2.3% | 1.1% | 4.7% | 0.1% | 4.6% | 0.6% | 2.2% | 2.7% |
| Max. spread (in sample) | 2.5% | 3.5% | 2.3% | 4.1% | 6.6% | 7.0% | 1.8% | 8.6% | 3.7% |
The initial retention of the cells on the fibrin and the further cell spreading over the surface for different conditions were assessed (Fig. S2, Supplementary Data). A concentrated drop of cell suspension with a concentration similar to that of bioprinting was placed on a fibrin surface to simulate this process and determine proper fibrin preparation (dehydration level) and the first incubation time before adding the medium. Even after successful printing, we observed that cells did not retain on a surface; an added medium washed them out. On the other hand, prolonged incubation, meant to give more time for cell adhesion, has often led to over-drying and death of cells. 5 min of dehydration and 1 h of incubation caused minimal cell retention. In this case, longer incubation (1.5 and 2 h) reversed this undesirable phenomenon. For more dehydrated (10 min) fibrin, 2 h incubation led to over-drying and loss of proliferative potential; however, cells retain on a surface and proliferate properly for shorter (1 and 1.5 h) incubations. For most dehydrated fibrines (15 min), only short incubation led to the preservation of the proliferative potential of LESCs.
Scaffolds characteristics—viability, cell coverage, density, phenotyping
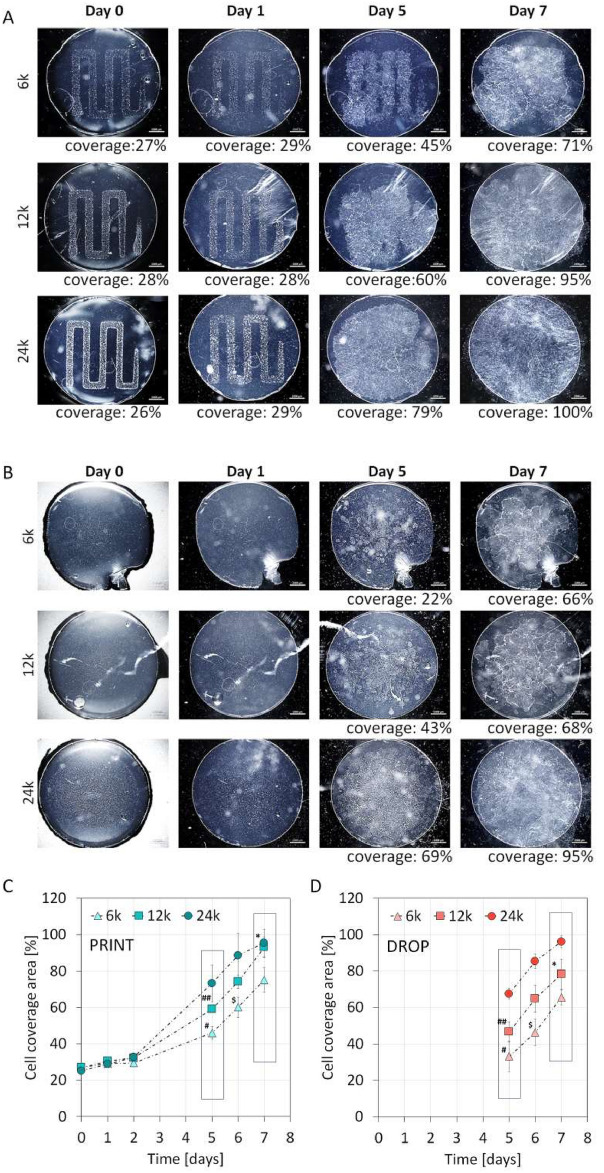
Cell cultures of LESC on fibrin, which were applied using traditional and bioprinting methods, are shown in Fig. 5A,B. LESCs were applied in the same number for each method – 6 × 103, 1.2 × 104 and 2.4 × 104 and cell morphology and growth area were analyzed over seven days. Results showed that cell spreading (Fig. 5C,D) was more efficient for bioprinting when fewer cells were applied (6 × 103 and 1.2 × 104). Furthermore, the 1.2 × 104 sample printing method resulted in almost full coverage after seven days, while for the 1.2 × 104 manually seeded sample, only 60% coverage was achieved at that time (significantly different, p = 0.0243). Similarly, cell coverage after five days for 6 × 103 and 1.2 × 104 was more advanced for the printing method (p = 0.0403, p = 0.0373, respectively). Interestingly, higher initial cell numbers of 2.4 × 104 led to similar results for manual and bioprinting methods.
Fig. 5.
Epithelium cell growth on a fibrin surface over 7 day culture for bioprinted samples as well as for manually dispensed cells (a, b). Cell spreading is more efficient for bioprinting method when smaller number of cells is applied (6 × 103 and 1.2 × 104, #p = 0.0403, ##p = 0.0373, $p = 0.0251, *p = 0.0243, n = 3). Higher initial cell number (2.4 × 104) led to similar results for both manual and bioprinting methods (not significant)(c, d).
Changes in cell morphology were also visible. Cells applied manually on the surface in lower numbers grew in separate spots – clones (Fig. 5B, 6k, day 7). For the bioprinting method, clones were also visible, but in this case, the surface was mostly covered with a dense, confluent layer of cells (Fig. 5A, 6k, day 7).
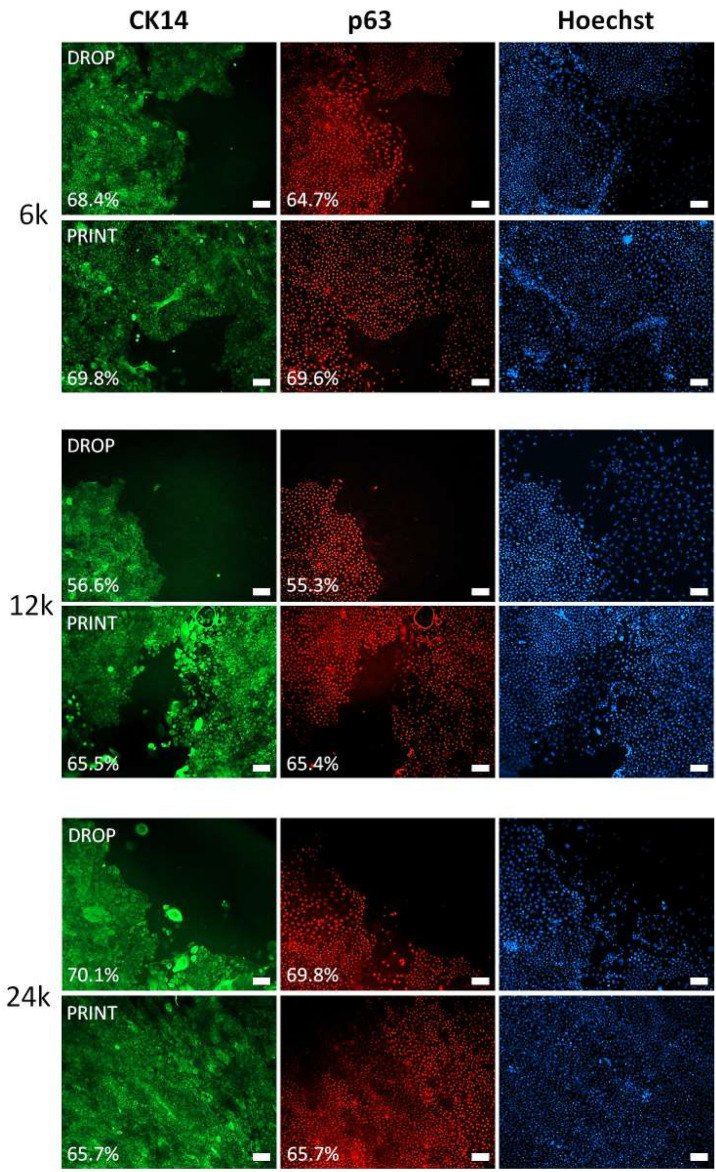
Phenotypes for previously described groups at day seven were analyzed by p63 and CK14 immunostaining (Fig. 6A). Most of the cells were p63 and CK14 positive; however, edges of expanding epithelia in many cases revealed the presence of fully differentiated keratinocytes or fibroblasts—characterized by the absence of p63 and CK14 antigens. Those cells usually were separated and did not form a confluent cell layer. They were often observed in 6 × 103 and 1.2 × 104 drop samples, as there were more areas with less confluency. However, statistical analysis of p63 and CK14 percentages did not show significant differences between manual and printing techniques (Fig. S3 D, E, Supplementary Data). Several p63 and CK14 positive cells for each sample were presented (Fig. S3 A, B, Supplementary Data). The samples produced by the printing method had a narrower cell number value distribution of p63 and CK14 than those produced by pipetting. Only for 2.4 × 104 cells seeded manually were no visible changes in p63 and CK14 cell number value distribution. Changes in p63 + and CK14 + cell number distribution were mainly caused by the fact that 6 × 103 and 1.2 × 104 samples were less thoroughly covered by cells at day seven than their printed counterparts. Concluding, the total number of cells on scaffolds was significantly higher for the printing method, as shown by quantitative data from Hoechst staining (Fig. S3 C, Supplementary Data).
Fig. 6.
CK14 and p63 positive cells on a fibrin surface after 7 day culture for bioprinted samples as well as for manual method. High percentage of CK14 + and p63 + was noted in all groups. Images from immunostaining show mostly areas partially covered by cell to illustrate morphological differences of expanding epithelia. Differentiated (CK14- and p63-) cells are more abundant on the edges of samples produced by drop method. Scalebar = 100 µm.
Discussion
Cell isolation and characterization
The LESCs needed for the bioprinting assessment were obtained by a mechano-enzymatic method involving partial digestion of the scrap and then culture and migration of the cells. The culture allowed the efficient isolation of cells in the amount needed to populate fibrin scaffolds. Their characteristics indicated epithelial cells, which was confirmed by the presence of p63 and CK14. These antigens frequently appear in publications describing the LESCs characteristics29,30. However, there is no clear statement regarding which antigens are specific for corneal epithelial stem cells. LESCs do not express CK3 or CK12 but have been reported to express CK19, CK1431,32, vimentin33, and ABCG231. The TP63 gene generates six isoforms34. Ocular keratinocytes may contain all the ΔN isoforms, but ΔNp63α (referred to as p63) is by far the most abundant14,35. It is present in the limbus but not in the uninjured central cornea29,36, and is expressed in holoclones but not in paraclones35.
In our experiment, the cells had the p63 antigen in a significant percentage (> 80%), similar to what was previously presented in literature30,37,38. It needs to be clarified what proportion of the cells are stem cells with the highest proliferative potential and how many are TAC (Transient Amplifying Cells) cells. Holoclones and meroclones have the highest proliferative potential (100 divisions and 40 divisions, respectively) and are characterized by the p63 antigen. Terminally differentiated cells with low proliferative potential forming paraclones are not p63 positive29.
The proliferative potential of the cells has a significant impact on their ability to form epithelium on a scaffold (irrespective of the cell application method) and the success of future cell therapy. It has been reported that > 3% levels of p63 + correlate with a 78.1% treatment success rate15. However, it is worth noting that individual variability also plays an important role.
In our study, porcine limbal cells were used. The porcine eye shows a strong resemblance to the human ocular surface, and is characterized by the presence of limbal interpalisade crypt structures and similar functionality to the human epithelial tissue. Seyed-Safi et al. demonstrated the presence of p63 + /CK15 + /CK3- cells in the basal layer of the porcine limbal epithelium, indicating a similar phenotype of the porcine and human limbal epithelium and concluding that porcine tissue is a useful model for studying limbal stem cell behavior39–41.
Bioprinting process
Flat fibrines, obtained by casting between two defined and flat surfaces, allowed on printing cells using extrusion without hydrogel or other thickener. The proposed approach enabled the application of cells on the surface in a controlled manner, either as a compact patch or as separated lines. It also allowed cell density and viability monitoring, provided, that specific conditions are met. One of them is the flatness of the fibrin. As the printer nozzle moves very close to the material, small changes in the thickness of the hydrogel can contribute to breaks in the strand due to approaching either too close or too far. Coming too close may result in rubbing cells against the surface and their mechanical damage. In our study, observed decreased cell viability in printed compact patches was most likely due to the nozzle snagging on already applied cells. In comparison, viability was not reduced when the lines were printed separately.
Interestingly, the extrusion of the cell suspension from the nozzle did not result in a cell viability loss, as it in the case of hydrogels42 and the formation of high shear forces in the nozzle. In our experiment, a low-viscosity suspension was extruded at relatively low rates. We found that the viability of the cells may also be affected by holding them in the nozzle at a relatively high concentration before the printing process. However, it has been shown that even 30 min of holding the cells in the printer nozzle did not result in viability loss. More problematic was the sedimentation of the cells, which occurred quite rapidly and could lead to a four times increase in the suspension density. This phenomenon, however, did not significantly affect cell density obtained for small suspension volumes and an instantaneous printing process (up to 3 min).
The handling of concentrated cell suspensions also turned out challenging. It is likely that either too high or too low measured cell density values may have been caused by the difficulty of preparing a cell suspension.
The lower cell viability for the manual method may have been due to the initial strong concentration of cells in the middle part of the sample, which may have limited the ability to adhere, as well as affecting the availability of oxygen and nutrients. Also, the rate at which the cell suspension was extruded from the pipette tip was higher than with the bioprinter (this for concentrated cell suspensions may have exerted higher shear forces).
The printing process went well, and the printed lines were repeatable with only minor variability—value was less than 5% for all samples within a group. The thickness of the printed lines was much greater than the thickness of the 200 µm nozzle due to the process of droplet adhesion onto the hydrophilic material. It is known that for materials of higher surface energy compared to bioink surface tension the bioink will spread43. It is likely that for 500 cells/mm samples—the effect of cell clustering at the edge was dictated by snatching cells by the medium flow moving away from the nozzle. Cell density, in this case, had affected line thickness. For higher cell densities, the lines were thinner due to the higher viscosity of the suspension and the lower medium content. In a more viscous suspension, flow on a fibrin surface was suppressed.
The bioprinting process has been shown to allow much greater control over the density of cells on the surface than a manual application of cell suspension droplets. In the latter case, the cells spread throughout the fibrin surface, but a high concentration remained only in the center. Occasionally, cell suspension was spilled from the surface, and many cells dropped outside the sample. In the case of bioprinting, it was possible to apply the cells as a compact patch over a limited area. The cell density in this area remained even, and the fluctuations were much smaller than those of the droplet method. In the case of the droplet method, the edges of the material had a cell density more than 10 × lower than the center. On the other hand, the density distribution was relatively narrow for bioprinting, and the cells were also concentrated at the edges. Average cell density for bioprinting method was also more repeatable with lower variability than that for manual method (respectively 3.3% vs 22.5% maximal variation from the group average).
Controlling cell seeding density in such a manner has a crucial impact on maintaining epithelial cells’ correct phenotype and growth potential. For this type of cells, growth at low densities is not favorable. In this case, it was possible to simulate a growth niche. Limbal cells needed to be cultivated in sufficient seeding concentrations to obtain enough paracrine signals from one another to survive in serum-free conditions or a medium with low calcium concentration44. In our case, FBS was present in the culture, but no feeder layer or amniotic membrane was used. So far, to the best of our knowledge, there has been no report of culturing cornea epithelia on fibrin without a feeder layer. Cell density, proper preparation of the surface, and proper seeding process might have been crucial in this case. However, as experiments have shown, it is even more advantageous to use separate lines in which the cells are compacted, as total surface coverage leads to decreased viability due to probable mechanical damage. This approach is similar to the wound healing model presented by Pramotton25. This method provides local densification of cells in separated lines. After confluency is reached within these lines, epithelium spreads toward unseeded areas. This mechanism resembles physiological conditions when the epithelium is partially damaged.
In our study, a significant challenge requiring optimization was the retention of cells on the fibrin after the printing process. The question was which degree of fibrin hydration was optimal for cell adhesion and what the length of incubation should before adding the medium. We found that cells adhered weakly to hydrated fibrin and were flushed when the medium was added. Dehydrated fibrin favored cell adhesion; however, the incubation time needed to be shortened as it caused cells to become too dry and lose viability. The experiment showed that for printing accurately (in a small, medium volume), it is necessary to strictly control the humidity parameters as it can significantly impact on the success of the entire scaffold preparation process.
Scaffolds characteristics
The process of epithelium growth on the scaffold differed between the printing and the traditional method. This phenomenon was associated with the way the cells were applied. Cells applied in drops via pipetting were distributed throughout the fibrin, reaching the highest density of cells in the middle. In the center, the densest layer of epithelium (sample 6 k, Fig. 6) was formed, and it gradually moved towards the edges of the fibrin. When lines of cells were printed, the confluent layer formed within these lines in the first few days of the culture. After that, gradual overgrowth proceeded, but the path for the epithelium to spread was shorter than in the traditional method.
Moreover, in the case of printing, shortly after confluence was reached, the epithelium started growing as one front from every side of the printed pattern. Such a phenomenon is analogous to the wound healing model. Under such conditions, through the proximity of other cells on the other side of the gap, a migration of cell accelerates45. This mechanism translated into an acceleration in the rate of surface overgrowth, although, in our case, the overgrowth was not as significant as previously described in the literature for HaCat cells25. This mechanism is illustrated by the fact that, for the 6 k sample, the expansion of cells leading to the fusion of gaps was significantly faster than the expansion leading into areas at the periphery of the sample (see sample 6 k, day 5, Fig. 5). The bioprinting method allowed faster surface overgrowth only for samples with a relatively low seeding density of 6 and 12 k. For a density of 24 k, the printing produced results comparable to those of the traditional method. This fact may indicate a specific threshold of the cell density on the fibrin necessary for a proper overgrowth process.
The phenotype of the cells after the overgrowth period showed no significant differences in the levels of p63 and CK14 antigens for either application method. In all cases, the percentage of p63 + and CK14 + cells were similar. The approximately 80% p63 + cells for cultures on fibrin was very similar to that of the p63 + percentage on culture plates, indicating that the proliferative potential of cells plated on fibrin was preserved. Those observations were similar to previously reported30,37,38. Comparing the 3D printing with the traditional method in terms of phenotype showed only minor differences. Scaffolds obtained by the 3D printing were characterized by greater p63 and CK14 cell percentage homogeneity and p63 and CK14 total number (S3, Supplementary Data). Samples from the traditional method were characterized by a favorable phenotype in the center, where the culture density was high. However, the edges were dominated by larger cells or cells with unfavorable fibroblast-like morphology and phenotype (S4, Supplementary Data).
The use of low cell counts, condensed in lines resembling a wound model, could be a good approach from the point of view of the lower need for cells required for therapy. It is also possible to use a smaller number of cells at a lower passage but with a higher proliferative potential. It has been shown that as the passage number increases, the dividing potential of the cells decreases, and the success of the therapy is closely correlated with the presence of stem cells (p63 +)8.
The feeder layers promote niche regulation and stemness of cultivated cells. Though no adverse reactions have been reported in the use of 3T3 feeder layers in large case series46,47, avoiding xenogeneic material may reduce the risk of animal-derived infection and potential graft rejection. Moreover, clinical studies using non-xenogeneic reagents are also currently being conducted20,37,48.
According to the authors, using the 3D cell printing on scaffolds in the GMP standard system will increase the quality of ATMPs preparations in human trials. It will ensure the repeatability in obtaining appropriate growth niches for transplanted cells. New developments in additive manufacturing and regenerative medicine have the potential to radically disrupt the traditional pipelines of therapy development and medical device manufacture. These technologies constitute a challenge for regulators as the traditional regulatory frameworks are designed for mass-manufactured therapies rather than personalized solutions. Moreover, 3D bioprinting technologies present another dimension of complexity by including living cells in the manufacturing process. Until now, cell banks’ quality control of ATMP preparations has concentrated mainly on the type and number of cells, their origin and viability. Regulators (EMA/FDA) typically require testing cell phenotypes to confirm purity, potency, and identity in product lot release specifications. Since the density of epithelial cell seeding determines significantly the success of transplantation, it is also necessary to focus on this parameter. The use of 3D cell printing, with its potential to seed cells in a controlled, highly repeatable manner, could contribute to replacing the traditional method of pipetting cell suspensions in the production of cell grafts (ATMP). In Europe, for example, the ATMP Regulation includes a ‘hospital exemption’ for non-routine, custom-made products that are not intended to be marketed. EU (EMA) regulation allows the application of regulatory exemptions to advanced medicinal products, including cell-therapy medicinal products, that 3D bioprinting may be covered by (and thus be exempt)49.
Conclusion
The research demonstrated the feasibility of printing cells on a fibrin layer as a reproducible and high-quality method for obtaining scaffolds for corneal regeneration. Compared to the traditional manual technique, bioprinting yielded a more reproducible cell distribution which resulted in higher viability and accelerated surface overgrowth. The phenotype of cells on the bioprinted scaffolds was similar to traditional scaffolds; however, the number of cells was significantly higher. Due to the reproducible nature of the culture and the more homogeneous scaffold parameters obtained, this method could be more effective in LSCD treatment.
Materials and methods
Fibrin scaffold preparation
Fibrin hydrogels were prepared by modifying commercially available fibrin glue (Tisseel Lyo, Baxter). For the final mixture, 900 µl of fibrinogen dissolved in PBS with aprotinin, 50 µl of thrombin dissolved in PBS and 50 µl of CaCl2 solution (WarChem) were used. The final ingredient concentration was: 3.6% fibrinogen, 2.5 IU/ml thrombin, 1000 KIU/ml aprotinin and 5 mM CaCl2.
Fibrin scaffolds were formed into 0.6 mm thick rectangular surfaces by pouring the final mixture into a mold consisting of two parallel flat Teflon surfaces separated by a 0.6 mm thick spacer and incubated at room temperature for 30 min for fibrin gelation. Then, fibrin hydrogels were washed twice in PBS, dried at room temperature for 15 min transferred to an airtight container, and stored for at least 3 days at 4 °C. Before cell seeding, small circular fibrin scaffolds were prepared by cutting rectangular fibrin hydrogels using a 14 mm diameter punch.
The above-described fibrin forming process, using two parallel surfaces, enabled the production of fibrin scaffolds of the desired thickness and flatness, which was necessary for the correct cells to be 3D printed on the surface.
Cell isolation
LESCs were obtained from pig’s eyes taken from a local slaughterhouse. First, the eyes were rinsed in PBS solution (Corning), sterilized in 5% iodopovidone (EGIS Pharmaceuticals PLC, 10%, diluted in PBS) and stored in CnT-XP3 solution (CELLnTEC) with 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco) in 4 °C. The corneal limbal tissue was punched out of the pig’s eyes using a 3 mm biopsy punch and then cut into pieces with a scalpel. The obtained tissue fragments were treated with 100 U/ml collagenase II (17,101,015, Gibco) for two hours, then rinsed with PBS and placed in CnT-PR-HC (CELLnTEC) medium with 5% FBS (fetal bovine serum, 16,140,071, Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin mixture in standard conditions (37 °C) in a humidified atmosphere of 5% CO2 and 95% air. Cell migration from explants was observed under an inverted phase-contrast microscope (Axio Vert.A1, Zeiss, Germany).
Cell culture
After 6–10 days, the number of migrated cells allowed to carry out the first passage. Cells with explants were rinsed with PBS, detached with TrypLE™ Express Enzyme (Gibco) solution and centrifuged at 230 × g for 3 min. Cells were counted and seeded on φ100 plates pre-coated with a mixture of 0.01 mg/ml fibronectin (F2006, Sigma-Aldrich), 0.03 mg/ml bovine collagen (PureCol, 5006, Sigma-Aldrich) and 0.01 mg/ml bovine serum albumin (A8022, Sigma-Aldrich). Cells were routinely cultured in CnT-PR-HC medium supplemented with 5% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin in a humidified atmosphere containing 5% CO2 and 95% air at 37 °C. The culture medium was changed every 2–3 days. Cell morphology was controlled using a phase-contrast microscope (PrimoVert, Zeis, Germany). Cultures with small cell sizes and high nucleus-to-cytoplasm ratios were selected for further experiments.
Cells on fibrin scaffolds were cultured in a growth medium consisting of a 1:1 mixture of CnT-PR-HC and Dulbecco’s Modified Eagle’s Medium (DMEM, 11,966–025, Gibco) supplemented with 5% FBS, 0.5 mM sodium pyruvate (25-000CIR, Corning), 0.5% (v/v) amino acid solution (non-essential amino acids, 11,140–035, Gibco), 100 U/ml penicillin, 100 μg/ml streptomycin and the addition of 150 KIU/ml aprotinin (from Tisseel Lyo, Baxter). The culture medium was changed every 2–3 days.
Determining the parameters of scaffold preparation for the cell seeding
The optimal drying time of the fibrin scaffold before seeding cells and the adjustment of the appropriate cell attachment time before adding the culture medium were analyzed. For this purpose, the fibrin scaffolds were air-dried in a laminar chamber for 5, 10, and 15 min before cell seeding. Cells were then manually seeded in a very small volume (0.2 μl) of concentrated LESCs suspension in the center of a fibrin scaffold (φ14 mm) placed on φ60 mm dishes. Samples were then incubated without adding medium for 15 min in the cell incubator; and next 30 μl of the medium was added to the edge of the scaffold. Samples were then incubated for another 1, 1.5, or 2 h before adding the medium’s total volume (2.5 ml). The surface of the fibrin scaffold was covered with cells, and the morphology of cells on fibrin was analyzed over 12 days.
Cell culture on scaffolds obtained by traditional method
Fibrin scaffolds were placed in φ 60 mm dishes and incubated in CnT-PR-HC medium in 5% CO2, at 37 °C for 60 min. LESCs were then manually seeded at densities of 6 × 103, 1.2 × 104 and 2.4 × 104 in a 30 µl drop covering the whole fibrin surface using an automated pipette and placed in the incubator for 90 min to attach the cells to the fibrin surface. After this time, 2 ml of CnT/DMEM medium was added. The culture was maintained for 7 days. The medium was changed every 2–3 days.
Cell culture on scaffolds obtained by bioprinting
Printing of LESC was performed using a 200 µl microsyringe (1725 TLX, Hamilton, USA) with a conical tip, mounted in the Syringe Pump Printhead (no. 20855) of the BioX bioprinter (Cellink, Sweden). Printing on the fibrin surface was performed with a φ0.2 mm nozzle, with a speed of 10 mm/s, and an extrusion speed of 17.2 nl/mm at room temperature.
Three printing patterns were used for cell printing: (1) for the viability test, the first pattern created in the shape of a square with 3 parallel lines inside the square with a total print length of 64 mm, (2) for cell distribution test, the second pattern in the shape of a square with 9 parallel lines inside the square with a total length of 112 mm, (3) for long-term culture, third pattern, having no outer edges, only 5 parallel lines spaced 1.8 mm with total print length of 48 mm.
The cell-printed fibrin scaffolds were incubated for 15 min to ensure appropriate cell attachment to the fibrin surface. Then, 30 µl of CnT/DMEM medium was added around the scaffold for another 60 min. After this time, cell cultures on scaffolds were supplemented with 2 ml of medium. The culture was carried out for 7 days. The medium was changed every 2–3 days.
Stability of cell number during bioprinting
The number of cells extruded during the bioprinting process was analyzed at intervals (1, 2, 3 min or 5, 10, 15, 20, 30, 40, 50 and 60 min) to check whether the given number of cells (6 × 103, 1.2 × 104 or 2.4 × 104 cells) is actually extruded in the total print volume and whether it changes over time. Cells were extruded into the Eppendorf tube, diluted by medium, mixed with trypan blue and counted in Bürker counting chamber. Three separate counts were made for each time point.
Cell viability after bioprinting
The viability of printed cells was evaluated by fluorescent staining with Live/Dead solution containing 1% calcein AM (17,783, Sigma-Aldrich) and 1% propidium iodide (P4864, Sigma-Aldrich) in CnT-PR-HC medium. Fibrin scaffolds were incubated with Live/Dead solution for 10 min before cell printing; then cells were printed on scaffolds according to pattern 1 (extrusion rate: 500 cells/mm) and visualized after 15 min with a fluorescent microscope (AxioZoom, Zeiss, Germany) equipped with Zeiss HXP 200 C Fluorescence Light Source. The control sample cells were manually applied to the fibrin surface in a 30 µl drop. After 7 days, the viability of the culture was analyzed by 15 min staining with a Live/Dead solution. Live and dead cells were counted in each of the 1 mm2 spots for four samples.
Cell distribution on fibrin scaffolds
To determine cell distribution on fibrin scaffolds, cells were printed (extrusion rate of 250 cells/mm) according to pattern 2 (dense printing) and visualized by Live/Dead staining. The control samples constituted cells manually applied to the fibrin surface in a 30 µl drop. The number of live and dead cells was counted in 25 areas (1 mm2) arranged in a 5 × 5 rectangular grid for each sample. Collected data was presented as maps representing each sample’s cell distribution and viability distribution and as histograms for printing and control groups.
Characteristic of printed lines
Cells were printed at densities of 6 × 103, 1.2 × 104, or 2.4 × 104 per sample according to pattern and counted to check whether the number of cells in the printed lines was consistent with theoretical assumptions. Counts from each of the four samples collected from 1 mm samples of the printed line were averaged and represented as a bar chart.
The line thickness of every print for three different cell concentrations (6 × 103, 1.2 × 104, or 2.4 × 104 cells) was measured. Each printed line was measured in 3 points. Collected data was represented as table.
Variation in group was presented as the ratio of the difference between the average line width of sample and the average line width for the group to the average line width for the group expressed as a percentage. Variation in sample was presented as the ratio of the difference between the longest line width of sample and the average line width of sample to the average line width of sample expressed as a percentage.
Characterization of cell cultures spread on fibrin
The cell spreading on the fibrin scaffolds (n = 3) printed with 125, 250, and 500 cells/mm cell concentration and corresponding control cultures obtained by manual seeding were analyzed by evaluating the fibrin surface covered by the cells. The stage of fibrin coverage by the cells was visualized using a microscope with oblique contrast illumination (AxioZoom, Zeiss, Germany) for 7 or 12 days. The area of the fibrin scaffold covered by the cells was manually marked using a graphics program (Zen Blue, Zeiss + ImageJ). Then, the number of pixels and the percentage of coverage relative to the total fibrin scaffold size was determined. In the case of samples with cells seeded manually, it was not possible to determine the degree of coverage during the first 2–4 days due to the spread of single cells across the fibrin surface.
Cell phenotyping on fibrin scaffolds
Immunofluorescent staining was performed to confirm the stem cell-associated markers p63 and CK14 expression. Cultured cells on fibrin were fixed with 4% paraformaldehyde (252,549, Sigma-Aldrich) for 5 min, washed twice with 0.1% Triton X-100 (T8787, Sigma-Aldrich), and permeabilized for 15 min. Then cells were washed and blocked with 5% Hores Serum (H1138, Sigma-Aldrich) in 0.1% Triton X-100 for 30 min at room temperature or 4 °C overnight. Next, the samples were incubated with primary antibodies including PE-conjugated rabbit monoclonal anti-human p63-α (#56,687, Cell Signaling Technology, diluted 1:100) and FITC-conjugated mouse monoclonal anti-human CK14 (LS-C535410, LsBio, diluted 1:100) at 4 °C overnight. Then, the samples were washed and incubated with secondary antibodies, including Alexa Fluor 568-conjugated goat anti-rabbit IgG (A11001, Invitrogen, diluted 1:5000) and Alexa Fluor 488-conjugated goat anti-mouse IgG (A11036, Invitrogen, diluted 1:2500) at RT for 3 h. To visualize the cytoskeleton of the cells, fixed cells were stained with phalloidin (A12379, Invitrogen) for 40 min. After washing, Hoechst solution (1 ul/ml) (H1399, Invitrogen) was used for nuclei staining. The stained cells were observed on a fluorescent microscope (AxioZoom, AxioObserver, Zeiss). The p63α -positive cells were identified by the red fluorescent signals in the cell nuclei, and the CK14-positive cells were identified by the green fluorescent signals in the cell cytoplasm.
For each sample, 100 fields of 100 × 100 µm evenly distributed throughout the sample area were generated using the software (ZenBlue). Then, the p63-positive cells, CK14-positive cells, and the total number of cells in Hoechst staining from each field were counted.
Statistical analysis
Statistical analysis was performed using the Statistica (v14.0 StatSoft). Results were presented as mean ± SD or/and median with interquartile range. To check normality, the Shapiro–Wilk test was performed. The Mann–Whitney rank sum test or Student’s t-test was used to evaluate the significance of the two groups. In order to compare more than two groups, in the case of normal distribution of results, one-way ANOVA with Tukey’s or Newman-Keuls post hoc test was performed; on the contrary, the nonparametric Kruskal–Wallis test was used. P < 0.05 was considered statistically significant.
Supplementary Information
Author contributions
KP, KJ conducted the primary experiment, analysed and interpreted the data, and played a significant role in writing the manuscript. RD contribute by moulding fibrin material for experiments. AK, JZD, RB, DD, EW consulted research concept, provided supervision, consulted results and reviewed the final version of the manuscript. All authors carefully reviewed and approved the final manuscript.
Funding
Research was a part of project “Scaffold for corneal reconstruction using 3D printing and bio-printing” as part of Priority Axis: I. Modern economy for the measure: 1.2 Research, development and innovation in enterprises of the Regional Operational Program of the Silesian Voivodeship for 2014–2020 (European Regional Development Fund).
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-73383-y.
References
- 1.Hu, J. C. W. & Trief, D. A narrative review of limbal stem cell deficiency & severe ocular surface disease. Ann. Eye Sci. 8, 13–13 (2023). [Google Scholar]
- 2.Limbal stem cell deficiency - ORPHA:171673. https://www.orpha.net/en/disease/detail/171673.
- 3.Yokoo, S., Yamagami, S., Usui, T., Amano, S. & Araie, M. Human corneal epithelial equivalents for ocular surface reconstruction in a complete serum-free culture system without unknown factors. Investig. Opthalmology Vis. Sci.49, 2438 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Holland, E. J. & Schwartz, G. S. The evolution of epithelial transplantation for severe ocular surface disease and a proposed classification system. Cornea. 10.1097/00003226-199611000-00003 (1996). [PubMed] [Google Scholar]
- 5.Lang, S. J., Böhringer, D., Geerling, G. & Reinhard, T. Long-term results of allogenic penetrating limbo-keratoplasty: 20 years of experience. Eye 31, 372–378 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolli, S., Ahmad, S., Lako, M. & Figueiredo, F. Successful clinical implementation of corneal epithelial stem cell therapy for treatment of unilateral limbal stem cell deficiency. Stem Cells 28, 597–610 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Guérin, L.-P. et al. Cultured autologous corneal epithelia for the treatment of unilateral limbal stem cell deficiency: a case series of 15 patients. Biomedicines 10, 1958 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pellegrini, G. et al. Location and clonal analysis of stem cells and their differentiated progeny in the human ocular surface. J. Cell Biol. 145, 769–782 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pellegrini, G. et al. Long-term restoration of damaged corneal surfaces with autologous cultivated corneal epithelium. Lancet 349, 990–993 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Schwab, I. R., Reyes, M. & Isseroff, R. R. Successful transplantation of bioengineered tissue replacements in patients with ocular surface disease. Cornea 19, 421–426 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Tsai, R.J.-F., Li, L.-M. & Chen, J.-K. Reconstruction of damaged corneas by transplantation of autologous limbal epithelial cells. N. Engl. J. Med. 343, 86–93 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Sangwan, V. S., Vemuganti, G. K., Singh, S. & Balasubramanian, D. Successful reconstruction of damaged ocular outer surface in humans using limbal and conjuctival stem cell culture methods. Biosci. Rep. 23, 169–174 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Ramos, T., Scott, D. & Ahmad, S. An update on ocular surface epithelial stem cells: cornea and conjunctiva. Stem Cells Int. 2015, 1–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Di Iorio, E. et al. Techniques for culture and assessment of limbal stem cell grafts. Ocul. Surf. 8, 146–153 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Rama, P. et al. Limbal stem-cell therapy and long-term corneal regeneration. N. Engl. J. Med. 363, 147–155 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Pellegrini, G. & De Luca, M. Eyes on the prize: limbal stem cells and corneal restoration. Cell Stem Cell 15, 121–122 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Pedrotti, E. et al. In vivo confocal microscopy 1 year after autologous cultured limbal stem cell grafts. Ophthalmology 122, 1660–1668 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Rama, P. et al. Autologous fibrin-cultured limbal stem cells permanently restore the corneal surface of patients with total limbal stem cell deficiency. Transplantation 72, 1478–1485 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Ahmed, T. A. E., Dare, E. V. & Hincke, M. Fibrin: a versatile scaffold for tissue engineering applications. Tissue Eng. Part B Rev. 14, 199–215 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Sangwan, V. S. et al. Clinical outcomes of xeno-free autologous cultivated limbal epithelial transplantation: a 10-year study. Br. J. Ophthalmol. 95, 1525–1529 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Forni, M. F. et al. Comparison between different biomaterial scaffolds for limbal-derived stem cells growth and enrichment. Curr. Eye Res. 38, 27–34 (2013). [DOI] [PubMed] [Google Scholar]
- 22.He, N. et al. Extracellular matrix can recover the downregulation of adhesion molecules after cell detachment and enhance endothelial cell engraftment. Sci. Rep. 5, 10902 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright, B., Mi, S. & Connon, C. J. Towards the use of hydrogels in the treatment of limbal stem cell deficiency. Drug Discov. Today 18, 79–86 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeffries, G. D. M. et al. 3D micro-organisation printing of mammalian cells to generate biological tissues. Sci. Rep. 10, 19529 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pramotton, F. M. et al. Optimized topological and topographical expansion of epithelia. ACS Biomater. Sci. Eng. 5, 3922–3934 (2019). [DOI] [PubMed] [Google Scholar]
- 26.Cubo, N., Garcia, M., Del Cañizo, J. F., Velasco, D. & Jorcano, J. L. 3D bioprinting of functional human skin: production and in vivo analysis. Biofabrication 9, 015006 (2016). [DOI] [PubMed] [Google Scholar]
- 27.DenizDerman, I. et al. High-throughput bioprinting of the nasal epithelium using patient-derived nasal epithelial cells. Biofabrication 15, 044103 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pellegrini, G. et al. From discovery to approval of an advanced therapy medicinal product-containing stem cells, in the EU. Regen. Med. 11, 407–420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pellegrini, G. et al. p63 identifies keratinocyte stem cells. Proc. Natl. Acad. Sci. 98, 3156–3161 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharifi, A. M., Darabi, R. & Jadidi, K. Isolation, culture, characterization and optimization of human corneal stem cells.. Biocell Off J. Soc. Latinoam. Microsc. Electron. Al 34, 53–55 (2010). [PubMed] [Google Scholar]
- 31.Chen, Z. et al. Characterization of putative stem cell phenotype in human limbal epithelia. STEM CELLS 22, 355–366 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurpakus, M. A., Maniaci, M. T. & Esco, M. Expression of keratins K12, K4 and K14 during development of ocular surface epithelium. Curr. Eye Res. 13, 805–814 (1994). [DOI] [PubMed] [Google Scholar]
- 33.Schlötzer-Schrehardt, U. & Kruse, F. E. Identification and characterization of limbal stem cells. Exp. Eye Res. 81, 247–264 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Yang, A. et al. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol. Cell 2, 305–316 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Di Iorio, E. et al. Isoforms of ΔNp63 and the migration of ocular limbal cells in human corneal regeneration. Proc. Natl. Acad. Sci. 102, 9523–9528 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barbaro, V. et al. C/EBPδ regulates cell cycle and self-renewal of human limbal stem cells. J. Cell Biol. 177, 1037–1049 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zakaria, N. et al. Results of a phase I/II clinical trial: standardized, non-xenogenic, cultivated limbal stem cell transplantation. J. Transl. Med. 12, 58 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ekpo, P. et al. Characterization of limbal explant sites: optimization of stem cell outgrowth in in vitro culture. PLOS ONE 15, e0233075 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crespo-Moral, M., García-Posadas, L., López-García, A. & Diebold, Y. Histological and immunohistochemical characterization of the porcine ocular surface. PLOS ONE 15, e0227732 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seyed-Safi, A. G. & Daniels, J. T. A validated porcine corneal organ culture model to study the limbal response to corneal epithelial injury. Exp. Eye Res. 197, 108063 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Notara, M., Schrader, S. & Daniels, J. T. The porcine limbal epithelial stem cell niche as a new model for the study of transplanted tissue-engineered human limbal epithelial cells. Tissue Eng. Part A 17, 741–750 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Malekpour, A. & Chen, X. Printability and cell viability in extrusion-based bioprinting from experimental, computational, and machine learning views. J. Funct. Biomater. 13, 40 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Naghieh, S. & Chen, X. Printability–A key issue in extrusion-based bioprinting. J. Pharm. Anal. 11, 564–579 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lužnik, Z. et al. Towards xeno-free cultures of human limbal stem cells for ocular surface reconstruction. Cell Tissue Bank. 18, 461–474 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Farooqui, R. & Fenteany, G. Multiple rows of cells behind an epithelial wound edge extend cryptic lamellipodia to collectively drive cell-sheet movement. J. Cell Sci. 118, 51–63 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Green, H. The birth of therapy with cultured cells. BioEssays 30, 897–903 (2008). [DOI] [PubMed] [Google Scholar]
- 47.De Luca, M., Pellegrini, G. & Green, H. Regeneration of squamous epithelia from stem cells of cultured grafts. Regen. Med. 1, 45–57 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Sejpal, K. et al. Cultivated limbal epithelial transplantation in children with ocular surface burns. JAMA Ophthalmol. 131, 731 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Mladenovska, T., Choong, P. F., Wallace, G. G. & O’Connell, C. D. The regulatory challenge of 3D bioprinting. Regen. Med. 18, 659–674 (2023). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.