Abstract
An immediate-early (IE) gene of human herpesvirus 6 (HHV-6), U95, has similarity at the amino acid level to the murine cytomegalovirus (MCMV) IE2 gene and is related to the human cytomegalovirus (HCMV) US22 gene family. Sequence analyses of U95 cDNA clones revealed that the transcription start site was located about 1.6 kbp upstream of the putative initiating ATG and that the transcript consisted of two exons. A single intron extended from nucleotides 142589 to 144229, which contained ORF U94. A protein with a molecular mass of about 120 kDa was translated from this cDNA clone in an in vitro transcription-translation assay. The transcription start site was found to be 220 bp downstream of the R3 region by primer extension analysis. HHV-6 has three repetitive elements, R1, R2, and R3, in or near the IE-A locus. R3 is composed of 24 copies of a 104- to 107-bp sequence element, which contains multiple putative binding sites for cellular transcription factors such as AP2 and NF-κB, and its biological significance has yet to be elucidated. The region between −710 and +46 relative to the transcription start site of U95 was analyzed in this study. Deletion from −710 to −396, corresponding to three copies of an R3 unit, decreased the promoter activity by 15-fold, and coexpression of IκBα(S32A/S36A) repressed it to almost the same level. Electrophoretic mobility shift assays showed that NF-κB family members p50 and c-Rel bound to NF-κB sites derived from the R3 region. These results demonstrate that R3 strongly enhances the U95 promoter activity and that NF-κB and binding sites for NF-κB in the R3 region play an important role in its activation. Because U95 promoter activity correlated with the number of R3 units, which each contained an NF-κB site, the repetitive organization of R3 is important for regulating U95 transcription.
Human herpesvirus 6 (HHV-6) was first isolated in 1986 from the peripheral blood of patients with lymphoproliferative disorders (42) and AIDS (25; R. S. Tedder, M. Briggs, C. H. Cameron, R. Horess, D. Robertson, and H. Whittle, Letter, Lancet ii:390–392, 1987). The virus was subsequently shown to be ubiquitous in healthy adults, with seropositivity in excess of 90% (39). HHV-6 predominantly infects and replicates in CD4+ lymphocytes (30, 48) and may establish latency in the monocyte/macrophage lineage (26). HHV-6 isolates are segregated into two closely related variants, A (HHV-6A) and B (HHV-6B), based on molecular and biological criteria (1, 3, 9, 18, 45, 53, 54). HHV-6B is the causative agent of exanthem subitum (roseola) (55), a common childhood disease, whereas the pathological role of HHV-6A remains to be determined. Their genomes are double-stranded DNA of approximately 160 kbp, consisting of a unique long region of 140 kbp flanked by 10-kbp direct repeats, and there is 90% homology between the variants. The complete genome sequences of HHV-6A strain U1102 and HHV-6B strains HST and Z29 were determined recently (14, 19, 22).
HHVs are divided into three subgroups, alpha, beta, and gamma, originally based on a diverse collection of in vivo and in vitro biological properties (32, 33, 41). HHV-6 is classified as a member of the betaherpesviruses, represented by human cytomegalovirus (HCMV) as well as HHV-7. This classification was made on the basis of the evolutionary divergence of its genome sequence from other subgroups (6, 14, 15, 19, 22, 29, 37). The betaherpesviruses have extensive domains of similar genetic organization, the conserved herpesvirus gene blocks, in the unique region of their genome, and they include a number of gene families that are characteristic of this subgroup (10). These include the US22, G-protein-coupled receptor, and immunoglobulin gene families. The US22 gene family is the most extensive family found in betaherpesviruses but is absent in the alpha- and gammaherpesviruses. HHV-6 encodes 11 members of the US22 family, DR1, DR2, DR6, DR7, U2, U3, U7, U8, U16, U25, and U95, which are related to the 12 members of this family found in HCMV (19, 36, 51). Some members of this family are spliced and expressed as immediate-early (IE) proteins (28, 38) and are likely to be transcriptional activators (12, 16, 46, 49). Murine cytomegalovirus (MCMV) IE2 has all of these characteristics (7, 27, 34). Since HHV-6 U95 is the positional homolog of MCMV IE2 and has amino acid similarity, it has been expected to be expressed as an IE gene (19).
Recently, using DNA microarrays and Northern blot analyses, we showed that U95 is indeed expressed at the IE stage of infection (unpublished data). The transcription of the HHV-6 genes, like other herpesviruses, generally follows a typical cascade. While some of the regulatory mechanisms were studied for early (E) genes, e.g., U27, U38 (2, 50), and U41 (unpublished data), the regulation of the IE genes has not been elucidated yet. To understand the transcription mechanism of the IE genes, we focused on U95, which is conserved in HHV-6, HHV-7, and MCMV but not in the alpha- and gammaherpesviruses.
In this paper, we present the structure of the U95 transcript. We show that it consists of two exons and that the transcription start site is located 220 bp downstream of the R3 region. HHV-6 has three major repetitive elements, R1, R2, and R3, in or near the IE-A locus (14, 19, 22), and their biological functions remain unclear. R3 has been predicted to regulate the expression of major IE (MIE) genes from the IE-A locus, because R3 has multiple putative binding sites for cellular transcription factors and is located upstream of this locus (14, 31, 51), but no evidence currently exists to support this prediction. Here we demonstrate that HHV-6B R3 plays an important role in regulating the expression of IE gene U95.
MATERIALS AND METHODS
Cells and virus.
Umbilical cord blood mononuclear cells (CBMCs) were separated on a Ficoll-Conray gradient and cultured in RPMI 1640 medium containing 10% fetal calf serum (FCS) and 5 μg of phytohemagglutinin per ml. At 2 or 3 days later, the CBMCs were infected with HHV-6B strain HST, which was isolated from a patient with exanthem subitum. When more than 80% of the cells showed cytopathic effects, the cell culture was frozen and thawed twice and centrifuged at 1,500 × g for 10 min at 4°C. The supernatant was stored at −80°C as a cell-free virus stock. The MT-4 cell line is derived from a human T cell transformed by human T-cell leukemia virus type 1 and was maintained in RPMI 1640 medium supplemented with 10% FCS.
RNA isolation and RT-PCR assay.
Cycloheximide (CHX) and phosphonoformic acid (PFA) were used for protein and viral DNA synthesis inhibition, respectively. PHA-stimulated CBMCs were infected with strain HST as described above and cultured for 24 h in the presence of 50 μg CHX per ml or 200 μg of PFA per ml. Total RNA was extracted from the infected cells as described previously (23).
For reverse transcriptase PCR (RT-PCR), reverse transcription reactions for cDNA synthesis of portions of HHV-6 IE1, DNA polymerase (Pol), glycoprotein H (gH), U95, and cellular elongation factor 1α (EF-1α) were performed at 42°C in a 20-μl solution containing 50 mM Tris-HCl (pH 8.3), 50 mM KCl, 10 mM MgCl2, 3 mM dithiothreitol, 20 U of RAV2 RT (Takara Shuzo), 1 μg of total RNA, and 0.4 μg of oligo(dT). PCR was carried out with EX Taq DNA polymerase. The following pair of primers was used to generate a 396-bp fragment of U95: U95-F1 (5′ TAATATGATCAATCCCATCAAAC 3′) and U95-R1 (5′ GGATTAGGGGGGTTCCGTTTCAGA 3′). The sequences of the other primer sets, used to amplify IE1, Pol, gH, and EF-1α, are described elsewhere (23).
Cloning of U95 cDNAs.
An oligo(dT)-primed cDNA library (35) was prepared from MT-4 cells infected with HHV-6B HST (32) and used to screen for a full-length cDNA clone of U95 mRNA. A synthetic oligonucleotide, U95-S (5′ CTGTCACAATGCAATCTC 3′), which was designed to hybridize to the 5′ region of the U95 open reading frame (ORF), was 5′-end labeled with [γ-32P]ATP. The screening method was described by Sambrook et al. (43). The clones obtained in the first round of screening were checked for the existence of the 3′ end of the U95 ORF by PCR using the following set of primers: U95-F.chip (5′ CTGTGTGAAAATAAATGGGTGCT 3′) and U95-R.chip (5′ CCAATTCAGGATTGCAGATATGT 3′). The cDNAs containing the 3′ end of the U95 ORF were subjected to a second round of screening to isolate them as single clones.
DNA sequencing.
The sequences of the U95 cDNA clones were determined by the dideoxy-chain termination method using the SequiTherm EXCEL II DNA sequencing kits-LC (Epicentre Technologies). The cycle-sequencing reaction was carried out with 5′-end-labeled primers with fluorescent dye IRD800, pGAD424-F (5′ TGTTTAATACCACTACAATGGATG 3′) and pGAD424-R (5′ TTGAGATGGTGCACGATGCACAG 3′), as specified by the manufacturer, and the products were analyzed on 4% polyacrylamide–7 M urea sequencing gels on a 4000L DNA sequencer (Li-Cor).
Prior to performing the reporter gene assays and electrophoretic mobility shift assays (EMSAs), the DNA sequences of all plasmid constructions and mutations were confirmed as described above with vector-specific universal primers.
In vitro transcription-translation.
The expression vector encoding the U95 protein was prepared as follows. The clone containing full-length U95 cDNA was digested with NotI, because NotI sites were present in adapters that flanked the ends of the cDNAs that were inserted into the vector for the library. The U95 cDNA was excised from the library vector as a NotI fragment and inserted into pcDNA3 1(+) at a NotI site. The orientation of the U95 cDNA was checked by PCR. In vitro transcription-translation reactions were carried out with the TNT T7 quick coupled transcription-translation system (Promega) in the presence of [35S]methionine, as recommended by the manufacturer. The in vitro translation product and the immunoprecipitation products were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (7% polyacrylamide) and treated with En3Hance (NEN). The gel was dried and exposed to a BAS-III imaging plate (Fujifilm). The autoradiogram was imaged and analyzed with a BAS 2000 II Bio-imaging analyzer (Fujix).
Immunoprecipitation.
MT-4 cells (4 × 107) were infected with HST strain and cultured in the presence of 200 μg of PFA per ml for 48 h. Mock- or HST-infected cells were labeled with 100 μCi of [35S]methionine per ml for 3 h, lysed in TNE buffer (10 mM Tris [pH 7.8], 1% NP-40, 0.15 M NaCl, 1 mM EDTA, 2 μg each of leupeptin, aprotinin, and pepstatin per ml), and subjected to immunoprecipitation using rabbit polyclonal antiserum to U95. Cell lysate was preincubated with protein G-Sepharose which had been blocked with 0.1% FCS to remove proteins nonspecifically bound to Sepharose beads. Subsequently the supernatant was incubated at 4°C overnight with anti-U95 antiserum-bound protein G-Sepharose. The Sepharose beads were rinsed five times with TNE buffer and resuspended in sodium dodecyl sulfate sample buffer. The final product was analyzed at the same time as the in vitro translation product.
Primer extension.
Total RNA was extracted from MT-4 cells that were uninfected or infected with HHV-6B, using RNeasy (Qiagen), and the poly(A)+ RNA was isolated using Oligotex-dT30 super (TaKaRa). A 24-mer oligonucleotide, U95-PE (5′ CCAGCTGCGATGTTTTCGTCACAC 3′), was 5′-end labeled with IRD800 (Aloka) and hybridized with 1 μg of poly(A)+ RNA, and the primer was extended with SuperScript II RT (GIBCO-BRL), as specified by the manufacturer for cDNA synthesis. The extension products were concentrated by ethanol precipitation, dissolved in Tris-EDTA (TE), and analyzed on a 6% polyacrylamide–7 M urea sequencing gel using a 4000L DNA sequencer.
Construction of reporter plasmids for the U95 promoter and repetitive R3 unit.
A 756-bp fragment and all deletion mutants of the U95 promoter were generated by PCR with the Expand high-fidelity PCR system (Boehringer Mannheim). Genomic DNA prepared from MT-4 cells infected with HHV-6B was used as the template. The oligonucleotide primer U95P-R (5′ AGTCAAGCTTCTGTACCGCTGTGTCCAAGCTACG 3′) was used for all PCRs to generate the 3′ end of U95 promoter. U95P-F1 (5′ AGTCCTCGAGCTCACCCGCTTGGGTAGGAAAGAC 3′) was used as a 5′ primer to amplify a series of 5′ deletion mutants: U95P-710, U95P-605, U95P-500, U95P-396, and U95P-290. This primer hybridized to the 5′ ends of the five repetitive elements that ended at the above sites (−710, −605, −500, −396, and −290) in the promoter. Oligonucleotides U95P-F2 (5′ AGTCCTCGAGTATATCTCTATCCAATCAGCACTC 3′), U95P-F3 (5′ AGCTCGAGCGAATCAAAAGCCGTGAAGTAG 3′), and U95P-F4 (5′ AGCTCGAGCCTACCACGCCTATTAACTTCAG 3′) were used for the amplification of deletions U95P-186, U95P-102, and U95P-50, respectively. The amplified fragments were digested, subcloned into the pGL3-Basic vector (Promega) at XhoI and HindIII sites, and sequenced.
Fragments of R3 units, 106 bp long and containing NF-κB(R3) (R3-A) and NF-κB(TT) (R3-B), were generated by PCR. The following sets of primers were used to amplify R3-A and R3-B, respectively: the R3-AF (5′ GTAAGCTTGAGGAAAGACCTAAACCCGC 3′) and R3-R (5′ GTAAGCTTCCAAGCGGGTGAGAACCTT 3′) pair and the R3-BF (5′ GTAAGCTTGTAGGAAAGACTTTAACCGC 3′) and R3-R pair. The underlined 6-mers indicate a HindIII site. The amplified fragments were digested with HindIII and subcloned into pBluescript. The copy number of each unit inserted into the vector was confirmed by sequencing. The one to four copies of R3-A and R3-B were cut out with SacI and XhoI from the vector and inserted into the upstream region of the U95 promoter of pU95P-186 at the SacI and XhoI sites.
Transient transfection and luciferase assays.
Plasmid DNA was transfected into 2 × 106 MT-4 cells using Lipofectamine PLUS reagent (GIBCO-BRL), as specified by the manufacturer. For promoter deletion analyses and to assay the effect of R3 unit copy numbers, cells were transfected with 1 μg of each reporter plasmid described above and 5 ng of pRL-SV40. To examine the ability of IκB to inhibit U95 promoter activity, pME-IκBα(S32A/S36A), a generous gift from Junichiro Inoue, was used. This plasmid expresses a mutant IκBα that has serine-to-alanine amino acid substitutions at residues 32 and 36. The substitutions permit the molecule to escape phosphorylation-dependent degradation, resulting in constant suppression of NF-κB. Cells were transfected with 0.5 μg of the promoter deletion constructs (pU95P-710, pU95P-605, pU95P-500, pU95P-396, pU95P-290, or pU95P-186) and pME-IκBα(S32A/S36A) or control vector pEF-BOS with 50 ng of pRL-TK. The control plasmid, pEF-BOS, contains the same EF-1α promoter as pME-1κBα(S32A/S36A) and was kindly provided from Shigekazu Nagata. pRL-SV40 and pRL-TK were used for normalizing variations in transfection efficiency.
Cell lysates were prepared 16 h after transfection using the dual-luciferase assay kit (Promega), and luciferase activities were measured using a Lumat LB9507 photon counter (EG&G Berthold). The luciferase activities were assessed in triplicate samples that were prepared from independently transfected cells, and the mean and standard deviation were calculated to ascertain the statistical significance of the data.
Preparation and labeling of oligonucleotides for EMSAs.
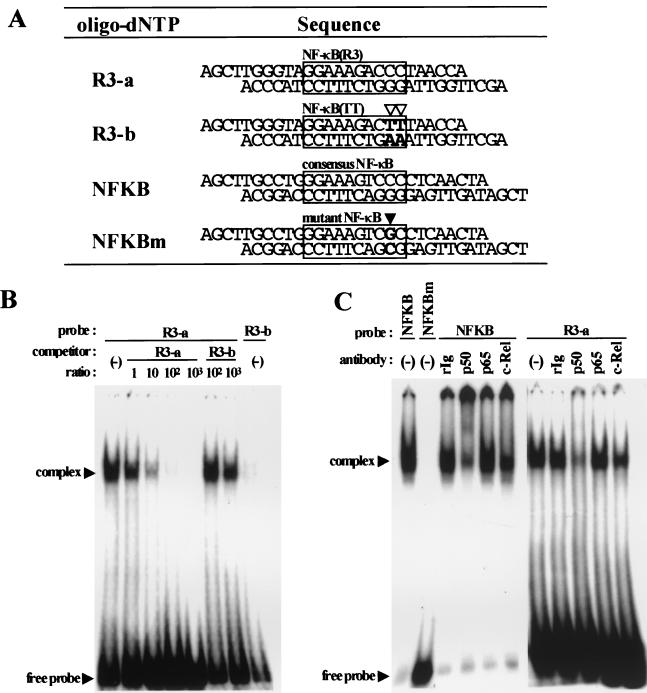
Double-stranded oligonucleotides for EMSAs were designed to contain BamHI and HindIII sites at their ends. They were prepared by annealing a pair of synthetic oligonucleotides, which were then subcloned into the pUC19 vector. The plasmids containing the double-stranded oligonucleotides were double digested with BamHI and HindIII, and the target fragments were separated by polyacrylamide gel electrophoresis (12% polyacrylamide) and purified as described elsewhere (4). A 1 pmol portion of double-stranded oligonucleotides was labeled with [α-32P]dCTP (Amersham Pharmacia) using the Klenow fragment (Takara Shuzo) as specified by the manufacturer and purified with Quick spin column G-50 Sephadex (Boehringer Mannheim). The sequences of the oligonucleotides used in this study are described in Fig. 5A.
FIG. 5.
EMSAs. (A) Double-stranded oligonucleotide sequences used in EMSAs. R3-a contains the R3-derived NF-κB sequence [termed NF-κB(R3)], and R3-b contains the mutant NF-κB sequence [termed NF-κB(TT)]. Putative PEA3 motif (AGGAAA) is conserved in either oligonucleotide. NFKB and NFKBm contain consensus or its mutant motifs for NF-κB, respectively. Each NF-κB motif is boxed, and mutated bases are indicated by open and solid triangles in R3-b and NFKBm, respectively. (B) Cold competition assays. The indicated molar excess of unlabeled competitor was added to the preincubation reaction mixture, and then 32P-labeled probes were added. (C) Supershift analyses of NF-κB family bound to NFKB and R3-a probes with specific antibodies to p50, p65, and c-Rel. Nuclear extracts were preincubated with each antibody, and then 32P-labeled probes were added. NFKBm was used to identify the specific complex.
EMSAs.
Nuclear extracts were prepared from 5 × 108 MT-4 cells by the method of Dignam et al. (13). Gel shift reactions were performed in a total volume of 24 μl as follows. Nuclear extract (8 μg) was preincubated for 5 min at room temperature with 3 μg of poly(dI-dC) in a binding buffer containing 20 mM HEPES (pH 7.9), 1 mM MgCl2, 10% glycerol, 5 mM dithiothreitol, 0.7 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, and 2 μg each of leupeptin, aprotinin, and pepstatin per ml. Subsequently, 0.1 ng (2 × 104 cpm) of the labeled probe was added to the mixture, which was then incubated for an additional 30 min at room temperature. The complexes were resolved at room temperature for 1 h at 180 V on a 4.5% nondenaturing acrylamide gel containing 2% glycerol and 1X TBE (45 mM Tris [pH 8.3], 45 mM boric acid, 1 mM EDTA). Competition experiments were performed by supplementing the reaction mixtures with 0.1, 1, 10, or 100 ng (∼1, 10, 100, or 1,000-fold molar excess, respectively) of unlabeled competitor probe.
For the supershift experiments, nuclear extracts were preincubated for 1 h at 4°C in the presence of 2 μg of the indicated antibody (Upstate Biotechnology), poly(dI-dC), and binding buffer. The labeled probe was then added, and the mixture was incubated at room temperature for 30 min before being separated on a gel. The gels were dried at 80°C for 1 h on a decompression dryer and exposed to BioMax MS film (Kodak) with intensifying screens at −70°C.
RESULTS
U95 mRNA is expressed in the IE stage in vivo.
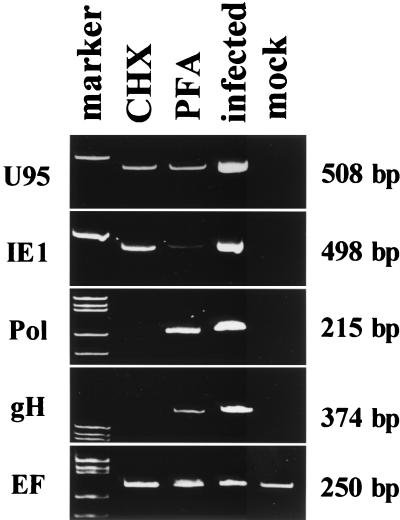
U95 is a member of the HCMV US22 gene family and a positional homolog of the MCMV IE2 gene, to which it also has a partial similarity in its amino acid sequence (19). On the basis of analogy to MCMV IE2, we expected U95 to be an IE gene. Recently, we used DNA microarrays and Northern blot analyses to show that the U95 transcript is indeed expressed with IE kinetics in MT-4 cells infected with HHV-6B (unpublished data). To further confirm whether U95 was transcribed in the IE stage in primary cells, such as CBMCs infected with HHV-6B, we performed an RT-PCR analysis in the presence of the protein synthesis inhibitor CHX. The U95 mRNA was detected in the presence of CHX, as was the IE1 mRNA; in contrast, the Pol and gH mRNAs were not detected under these conditions (Fig. 1, lane CHX). The Pol and gH mRNAs were classed as early genes because the viral DNA synthesis inhibitor PFA did not block their expression (Fig. 1, lane PFA). None of these four viral mRNAs were detected in mock-infected cells. mRNA for a cellular gene, EF-1α, was detected in both CHX-treated and untreated cells, as well as in mock-infected cells. These results showed that the U95 mRNA was transcribed without de novo protein synthesis, indicating that U95 is an IE gene.
FIG. 1.
RT-PCR analyses for the transcripts of U95, IE1, Pol, gH, and EF 1α in HST-infected CBMCs treated with CHX and PFA. Total RNAs prepared from mock-infected and HST-infected cells, which had been treated with CHX for 24 h or PFA for 24 h or left untreated, were used. The U95 band was detectable in the presence of CHX and PFA, as well as the IE1 band, while the bands of Pol and gH could not be detected in the presence of CHX. EF1α, the cellular endogenous gene, was transcribed under all conditions.
The U95 transcript is spliced and composed of two exons.
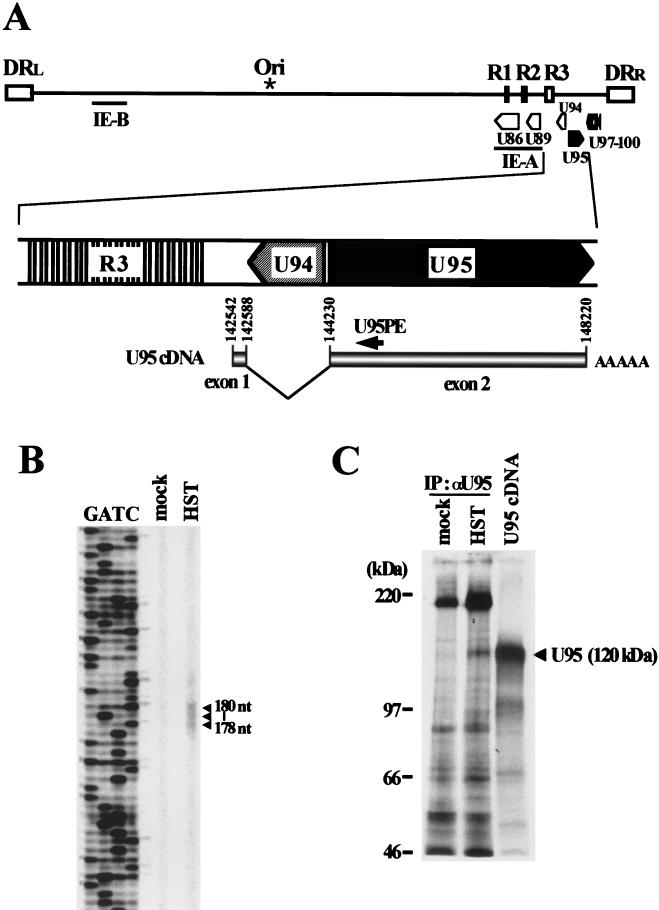
To identify the structure of the transcripts encoded by U95, we determined the nucleotide sequences of four cDNA clones isolated from the cDNA library. Sequencing analysis revealed that the U95 transcripts were composed of two exons, as shown in Fig. 2A. The 5′ end of the cDNAs was at or near nucleotide (nt) 142559, which is located between the R3 region and ORF U94. A 1.6-kb intron containing U94 in an inverted orientation was spliced out between a splice donor site at nt 142588 and a splice acceptor site at nt 144230. The 3′ poly(A) tail began at nt 148220, with a polyadenylation signal at nt 148199. All four clones had the same splicing sites and 3′ end. The translation initiation site was located at the start site of the second exon. To determine the transcription start site, we performed primer extension analysis.
FIG. 2.
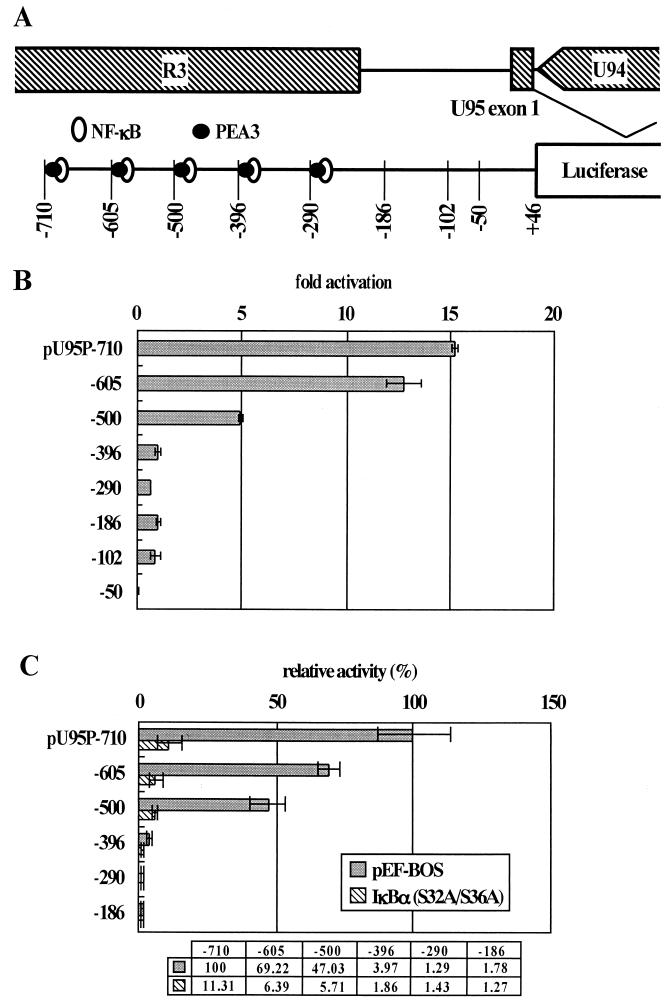
Analyses of U95 cDNA. (A) Relative genomic locations of the U95 ORF and the structure of the U95 transcript. The transcription start site (nt 142572) was determined by primer extension using the IRD800-labeled U95PE primer. All four cDNA clones had the same splice donor and acceptor sites and 3′ ends. (B) Primer extension analysis. mRNAs extracted from HST-infected and uninfected MT-4 cells at 72 h. postinfection were used. Sequencing ladders for sizing were generated from pcDNA3.1(+)-cU95 using the same primer. (C) In vitro transcription-translation and immunoprecipitation of U95. In vitro transcription-translation from pcDNA3.1(+)-cU95 was performed by using TNT T7 quick coupled transcription-translation system. Endogenous U95 protein was immunoprecipitated from HST-infected and 200-μg-of-PFA-per-ml-treated MT-4 cells at 48 h postinfection by using rabbit polyclonal antiserum to U95 as described in Materials and Methods.
As shown in Fig. 2B, three bands that differed from one another in length by 1 base were detected in the mRNAs from MT-4 cells infected with HST but not from uninfected cells. By comparison with a sequencing ladder, the three primer extension products were determined to be 178, 179, and 180 bases long, indicating that the start site was located about 16 bp upstream of the 5′ end of the cDNA clones. We assumed the longest to be a fully extended product and determined the transcription start site to be at nt 142542.
To assess whether the cDNA encoded full-length U95, we performed in vitro transcription-translation analysis. The translation product had a molecular mass of about 120 kDa, which is slightly smaller than the predicted size of 133 kDa. Therefore, we determined the endogenous size of U95 protein expressed in HST-infected MT-4 cells by immunoprecipitation using rabbit polyclonal antiserum to U95. As shown in Fig. 2C, the immunoprecipitated U95 protein showed the same electrophoretic mobility as that of the in vitro translation product, indicating that the molecular mass of 120 kDa represents the intact U95 protein. These data demonstrated that the cDNAs cloned in this study encoded the full-length U95 protein.
R3 strongly enhances U95 promoter activity.
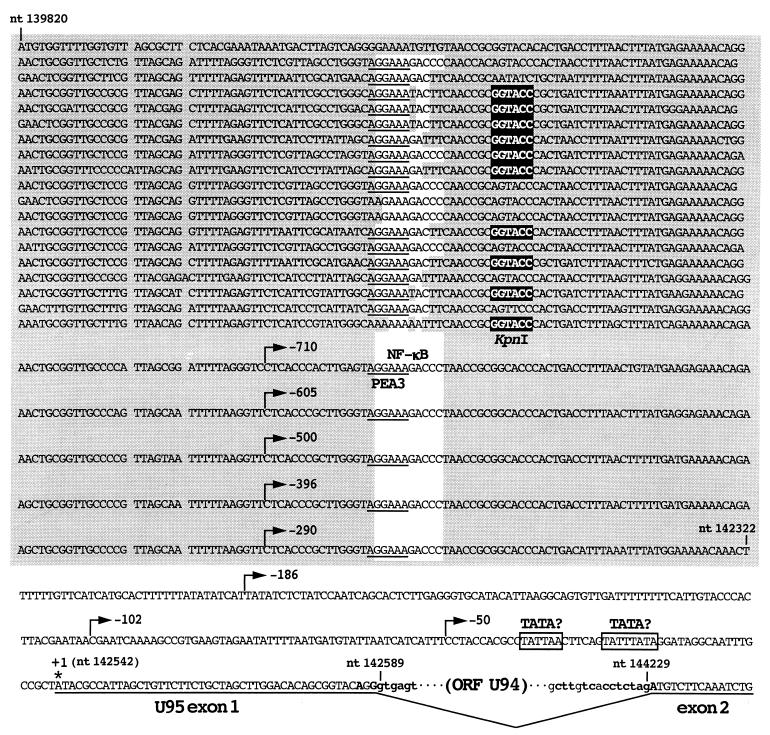
The R3 region is one of the major repetitive elements included in the unique region of the HHV-6 genome and is located upstream of the IE-A locus. It is composed of 24 copies of tandemly repeated 104- to 107-bp units, contains 10 recognition sites for KpnI, and has abundant putative binding sites for cellular transcription factors (Fig. 3) (14, 22). It has been speculated that R3 might function as an enhancer of MIE genes (14, 31, 51). Our data showing that the transcription start site of U95 was located 220 bp downstream of the R3 region raised the intriguing possibility that R3 might directly enhance the promoter activity of U95.
FIG. 3.
Nucleotide sequence of the R3 region and the U95 promoter of HHV-6B strain HST. Numbers with “nt” indicate genome coordinates of HST, and numbers by the arrows indicate the 5′ deletion points of the U95 promoter constructs relative to the transcription start site (+1 asterisk). The R3 region sequence is shaded, and 9 putative binding sites for NF-κB (white background), 20 sites for PEA3 (underlined), and 10 KpnI recognition sites (white letters in black box) are indicated. Potential TATA motifs, which lie at positions −27 to −20 and −39 to −34 relative to the transcription start site, are boxed. Lowercase letters indicate spliced-out region, and boldface letters indicate consensus splice donor and acceptor sequences.
To examine this possibility, a 756-bp fragment that extended from −710 to +46 relative to the transcription start site of U95 was subcloned into the pGL3 Basic vector (Promega). The resultant plasmid, pU95P-710, contained a 491-bp region of R3. The plasmid was transfected into MT-4 cells, and the luciferase activity was measured. As shown in Fig. 4B, pU95P-710 had more than 15-fold-greater activity than did a 232-bp fragment (pU95P-186) that extended from −186 to +50 relative to the transcription start site upstream of the luciferase gene and contained no more of the R3 region. The activity of pU95P-710 was almost two-thirds that of the MIE promoter of the IE-A locus (data not shown). These findings indicated that the upstream region has strong promoter activity. To determine whether cis-acting elements were essential for the promoter activity, a series of 5′ deletion mutants were constructed and transfected into MT-4 cells. The results, shown in Fig. 4B, indicated that most of the activity of the U95 promoter is attributable to the region upstream of nt −396. This region lies within R3, as shown in Fig. 4A. Detailed analysis of the deletion mutants revealed that four regions (−710 to −605, −605 to −500, −500 to −396, and −102 to −50) were responsible for the promoter activity. Three of the four cis elements lie within the R3 region, and more than 90% of the activity of the U95 promoter is attributable to R3, indicating the significance of the R3 region in regulating the transcription of U95.
FIG. 4.
(A) Schematic organization of the U95 promoter. Putative binding sites for NF-κB and PEA3 and potential TATA motifs are shown. Numbers indicate 5′ deletion points relative to the transcription start site. (B) Effect of the indicated 5′ deletions on the U95 promoter activity. Mean fold activities relative to that of pU95P-186-transfected cells and standard deviations for three independent experiments performed in triplicate are plotted. (C) Effect of coexpression of 1κBα(S32A/S36A) on the U95 promoter activity. The luciferase reporter plasmids (0.5 μg) were cotransfected with effector plasmid pME-1κBα(S32A/S36A) (0.5 μg) or control plasmid pEF-BOS (0.5 μg). The mean percent activities relative to 100% for cells cotransfected with pU95P-710 and control plasmid are indicated below the panel. The error bars plot the standard deviation for three independent experiments, each done in triplicate.
NF-κB plays a major role in U95 promoter activation.
The three regions in R3 that significantly affected promoter activity corresponded to repeated units with almost the same nucleotide sequence. Each unit contains a binding site for NF-κB that overlaps with a site for polyomavirus enhancer A binding protein 3 (PEA3) (14). PEA3 is required for efficient early-gene transcription of polyomavirus and adenovirus. NF-κB is involved in gene regulation of other herpesviruses, such as herpes simplex virus type 1 (40), Epstein-Barr virus (47), and HCMV (8, 11, 44, 52). As a first step in examining whether NF-κB was involved in gene activation of U95, we used an NF-κB-specific inhibitor, IκB. IκB masks the nuclear localization signal of NF-κB by complexing with it, thereby trapping NF-κB in the cytoplasm and inhibiting its function of transcriptional activation (17). We used pME-IκBα(S32A/S36A), a construct that expresses mutant IκBα possessing serine-to-alanine substitutions at residues 32 and 36, which allow the mutant to escape phosphorylation-dependent degradation, resulting in constant inhibition of NF-κB. pME-IκBα(S32A/S36A) and each of the series of reporter plasmids were cotransfected into MT-4 cells, and the luciferase activities were measured. The first three constructs, pU95P-710, pU95P-605, and pU95P-500 contained three, two, and one of the three cis elements of the R3 region described above, respectively. As shown in Fig. 4C, IκB suppressed the luciferase activities of pU95P-710 and pU95P-605 to 11 and 6%, respectively, of the levels seen using a control vector. In addition, IκB reduced the activity of pU95P-500 to the level seen with pU95P-396 alone. IκB had almost no effect on pU95P-396, pU95P-290, and pU95P-186, indicating that the cis element from −102 to 50 was not controlled by NF-κB. These results indicated that the activating region upstream of −396 in R3 was controlled by NF-κB.
p50 and c-Rel bind to the NF-κB sites in the R3 region.
The above data suggest that the binding sites for NF-κB in the R3 region function as cis-acting elements of the U95 promoter. R3 units contain potential binding sites for NF-κB in 9 of the 24 repeats [GGAAAGACCC, referred to as NF-κB(R3) (see below)]. Five of these nine sites are concentrated upstream of the U95 transcription start site (Fig. 3 and 4A). Of 24 repeats 8 encompass mutated NF-κB binding sites that have base substitutions from CC to TT at the 3′ end of the motif [GGAAAGACTT, referred to as NF-κB(TT) (see below)], as shown in Fig. 3. To determine whether cellular transcription factors bind to these sites in the R3 region, we performed EMSAs using double-stranded oligonucleotides, R3-a and R3-b, which contained NF-κB(R3) and NF-κB(TT), respectively (Fig. 5A). These oligonucleotides were labeled with [α-32P]dCTP and used as probes. As shown in Fig. 5B, R3-a formed a DNA-protein complex but R3-b did not. A 1- to 1,000-fold molar excess of unlabeled R3-a was sufficient to compete for the complexed protein, but a 100-fold molar excess of R3-b was not. A 1,000-fold molar excess of cold R3-b showed a slight competitive effect with R3-a, but the competition level was almost the same as that seen with an equimolar excess of R3-a, suggesting that this competition was nonspecific. These data indicated that the complex formation is specific for R3-a, i.e., for NF-κB(R3). All the R3 repeat units contain a potential binding site for a transcription factor, PEA3 [AGGAA(A/G)], as shown in Fig. 3. Although R3-a and R3-b also contain this site, R3-b exhibited no complex formation and unlabeled R3-b did not compete with R3-a for the complex. These data indicated that there was no protein in the nuclear extract prepared from MT-4 cells that binds to the PEA3 site, implying that the PEA3 site is not functional in MT-4 cells.
To determine whether NF-κB family members are involved in the complex formation described above, we performed supershift analysis. In this experiment, we used two additional probes, NFKB and NFKBm (Fig. 5A). These probes contain consensus and mutated NF-κB-binding sites, respectively, and were used as positive and negative controls. As shown in Fig. 5C, NFKB formed a DNA-protein complex in the nuclear extract of MT-4 cells but NFKBm did not. Antibodies to p50, p65, or c-Rel were incubated with nuclear extracts prepared from MT-4 cells, prior to the addition of probes. The complex formation with NFKB was partially inhibited by the anti-p50 and anti-c-Rel antibodies. Control rabbit immunoglobulin and anti-p65 had no effect on the complex, indicating that this inhibition was antibody specific. These data demonstrated that MT-4 cells contain p50 and c-Rel, which could bind to a consensus NF-κB site. When we used R3-a as a probe, antibodies against p50 and c-Rel, but not control rabbit immunoglobulin or anti-p65, partially blocked the complex formation. These data demonstrated that the NF-κB-binding sites observed in the R3 region are bound by NF-κB family members p50 and c-Rel.
Repetitive effect of R3 units on U95 promoter activity.
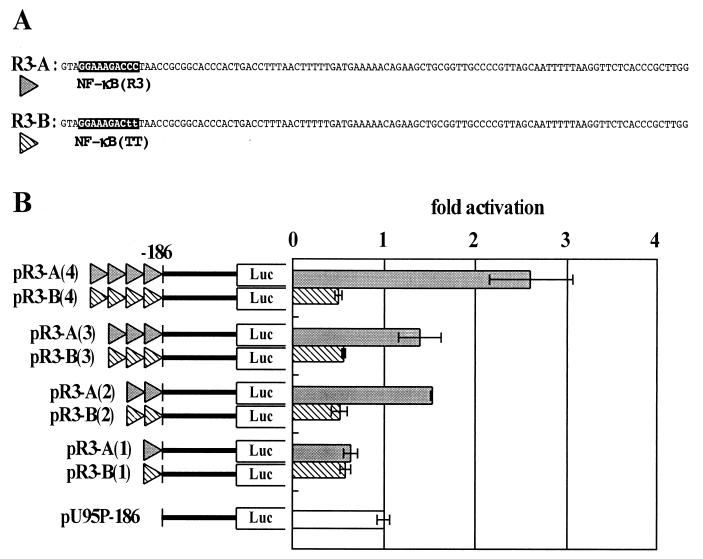
The results of the U95 promoter deletion assays implied that most U95 promoter activity depended on the copy number of the NF-κB sites (Fig. 4A and B) and that the NF-κB(TT) site did not affect the promoter activity. We next investigated whether the repeat structure of R3 was important for the regulation of the U95 promoter. As discussed above, the R3 region consists mainly of two kinds of units, with respect to the NF-κB sites, which we termed R3-A and R3-B (Fig. 6A). R3-A and R3-B encompass NF-κB(R3) and NF-κB(TT), respectively. As shown in Fig. 3, R3-A and related sequences are tandemly arranged in five repeats upstream of nt −186. R3-B and related sequences are found upstream of nt −710.
FIG. 6.
Effect of repetition of the R3 unit on U95 promoter activity. (A) Schematic representation of PCR-generated R3 units containing NF-κB(R3) or NF-κB(TT) (termed R3-A and R3-B, respectively). (B) The left map represents the copy numbers of each R3 unit inserted immediately upstream of U95P-186. MT-4 cells were transfected with each construct and harvested 16 h later, and luciferase activities were measured. The right panel indicates the mean fold activities relative to that of pU95P-186-transfected cells in three independent experiments. The luciferase activities were normalized as described in Materials and Methods, and error bars plot the standard deviation for each set of triplicate samples.
To assess the effect of their repetition on U95 promoter activity, R3-A and R3-B were generated by PCR and inserted upstream of nt −186 as a single copy or up to four tandem repeats, as shown in Fig. 6B. One copy of each unit slightly suppressed the luciferase activity of pU95P-186, which did not contain an R3 unit. Two copies of R3-A enhanced the promoter activity 1.5-fold compared with pU95P-186, but three copies showed no further enhancement. Four copies increased the promoter activity 2.5-fold compared with pU95P-186. There was, however, a significant difference in activities between pU95P-605 and pR3-A(4) relative to pU95P-186 although both promoters contain the same four copies of NF-κB sites. The activity of pU95P-605 was increased about 12.5-fold compared with that of pU95P-186, whereas that of pR3-A(4) was increased only 2.5-fold. This discrepancy could be explained by the sequence differences between two promoters. The R3 units of pU95P-605 have sequence variation, but those of pR3-A have an identical sequence. Besides, HindIII sites were introduced in the promoter sequence of pR3-A series to repeat the R3-A units. These differences might contribute to the level of promoter activity in a significant way. As expected, R3-B repetition had no effect on the promoter activity. These data demonstrate that the repeat structure of R3 indeed plays a critical role in the regulation of the U95 promoter.
DISCUSSION
HHV-6 has an ORF, U95, that has sequence similarity to MCMV IE2 and belongs to the US22 gene family that was originally identified in HCMV (19). We recently showed that U95 is expressed at the IE stage of infection. Like other herpesviruses, the transcription of the HHV-6 genes follows a cascading pattern that involves an orderly regulation of viral genes, starting with the IE genes and followed by the early and finally the late families of genes, during productive infection. Most of the regulatory mechanisms for this cascade remain to be determined, and particularly little is known about the regulation of the IE genes. To elucidate the mechanism of IE gene expression, we focused on U95 and aimed to determine the transcription factors and cis-acting elements that function at the IE stage of infection.
ORF U95 is located at the right end of the unique region of the HHV-6 genome, near the R3 region, on the other side of R3 from the IE-A locus. It is transcribed in the direction opposite to that of the MIEs (Fig. 2). The ORFs encoding an adeno-associated virus type 2 rep homolog (U94) and glycoprotein 82/105 (U97-100) lie on the complementary strand on the left and right sides of U95, respectively. Analyses of cDNA clones encoding the U95 protein revealed that the transcript originated 220 bp downstream of the R3 region and consisted of two exons. Exon 1 was 47 bases long and was separated from exon 2 by a 1.8-kb intron that contained a complementary strand of ORF U94.
To determine the promoter region of U95, we carried out primer extension analysis. We detected three bands that differed from one another in length by 1 base. The most probable explanation for these three bands is the cap effect, that is, the incomplete termination of reverse transcription at the methylated base next to the cap site, which results in extension products that are 1 to 2 bases shorter than full length (21). Secondary structure or degradation of a template RNA will also cause the incomplete termination of reverse transcription but is unlikely to cause differences of a single base. We concluded that the two shorter bands were generated by the cap effect and that the longest represented the true full-length extension product, and we used its sequence to determine the transcription start site.
We found two putative TATA boxes at −20 to −27 (TATTTATA) and at −34 to −39 (TATTAA) relative to the determined start site (Fig. 3). One of the two is expected to be functional.
Luciferase assays of a series of 5′ deletion mutants revealed that more than 90% of the U95 promoter activity was attributable to an approximately 490-bp region of R3 (Fig. 4B). The promoter region that extended from −710 to +46 was necessary and sufficient for full activity of the U95 promoter, because its activity was the same as that of a 3-kbp region upstream of the transcription start site of U95 that included the entire R3 region (data not shown). Therefore, these results indicated that a portion of the R3 region is involved in the activation of the U95 promoter.
We determined that NF-κB played an important role in the activation of the U95 promoter, because its activity was suppressed by IκBα(S32A/S36A) (Fig. 4C). To identify the members of the NF-κB family involved in the transcriptional regulation, we performed EMSAs using specific antibodies to p50, p65, and c-Rel. The DNA-protein complex formation was specifically inhibited by the addition of both anti-p50 and anti-c-Rel antibodies, suggesting that p50 and c-Rel formed heterodimers and bound to NF-κB sites in the R3 region (Fig. 5C). We did not investigate the other members of the NF-κB family, such as p52 and RelB, because we could detect only a complex in which p50 and c-Rel were included. When antibodies to these molecules were added to the nuclear extracts, complex formation was prevented but no supershifted band was observed when using both probes containing a consensus NF-κB site or an R3-derived NF-κB site (NFKB and R3-a, respectively). This phenomenon was probably due to the characteristics of these antibodies. Partial disappearance of the complex was also likely to be due to the affinity, purity, and concentration of the antibodies used in this study.
The free NFKB probe was almost depleted by incubation with nuclear extract, while the free R3-a probe remained abundant, suggesting that NF-κB binds to the consensus motif with much higher affinity than to the NF-κB(R3) site. Comparison between the sequences of the consensus NF-κB (GGAAAGtCCC) and NF-κB(R3) (GGAAAGaCCC) sites revealed a single-base substitution (indicated by a lowercase letter), which probably caused the difference in the binding affinity of NF-κB. The low activities in the luciferase assays using reporter plasmids containing one or two NF-κB(R3) sites are probably also attributable to the low affinity of NF-κB for these sites.
In HCMV, NF-κB is reported to activate the MIE promoter (MIEP) (11, 44) and the US3 promoter (8, 52). As with the U95 promoter, these promoter regions contain repeated sequences that bear NF-κB sites. While the HHV-6 U95 promoter has five tandemly repeated 106-bp units that each contain an NF-κB site, the HCMV MIEP has four 18-bp units that each contain an NF-κB site scattered throughout approximately 400 bp of the enhancer region and the US3 promoter also has five tandemly repeated 18-bp units that each contain an NF-κB site. The basic organization of the repetitive elements is, however, different in each of these promoters, and there are no conserved motifs other than the NF-κB sites. In addition, the NF-κB sequences in the MIEP enhancer (CGGGGACTTTCC) and US3 promoter [GGAAAGT(C/A)CC] are either identical to the consensus NF-κB site used in the EMSAs or more similar to the consensus than to the NF-κB(R3) of the U95 promoter. We observed that the promoter activity of the HCMV MIE gene was higher than that of U95. This observation might be partially explained by the difference in the affinities of NF-κB to the cis elements of these two genes.
Many groups have reported that the Ets family physically interacts with NF-κB and synergistically enhances some cellular and viral gene promoter activities, e.g., IL-2Ra, IL-12 p40, and the human immunodeficiency virus enhancer (5, 20, 24). In the R3 region, putative binding sites for PEA3, a member of the Ets family, overlapped with NF-κB sites, suggesting that PEA3 might interact with NF-κB. However, the EMSAs showed that no protein bound to the PEA3 sites. Therefore, we excluded the possibility that PEA3 was involved in the activation of the U95 promoter. Thus, these data indicated that NF-κB independently enhanced the U95 promoter activity.
The biological function of R3 has been unknown, although it has been suggested that R3 might play a role in the transcriptional regulation of the IE-A locus, because it contains multiple putative binding sites for cellular transcription factors such as NF-κB and AP2 (14, 31, 51). Furthermore, it was difficult to predict the regulatory role of R3 in U95 transcription because the 1.8-kbp region containing ORF U94 lies between R3 and ORF U95. Therefore, it was unexpected that the transcription start site of U95 was located 220 bp downstream of R3. Fortunately, however, this observation provided information that was extremely useful in predicting the role of R3 in U95 transcription.
In this study, we found that the R3 region of HHV-6B strongly enhanced the promoter activity of IE gene U95, which lies on the side of R3 that is opposite the IE-A locus, via five tandemly repeated NF-κB/Rel-binding sites. Because the genomic DNA sequence of HHV-6A has significant similarity to that of HHV-6B (14, 22), ORF U95 of HHV-6A is probably expressed similarly to ORF U95 of HHV-6B. However, some characteristics of the R3 region are slightly different between HHV-6B and HHV-6A. In HHV-6B, there are some minor variations within the sequence of each R3 unit and the NF-κB sites are conserved in only 9 of the 24 units. The existence of the five tandem units that contain an NF-κB site in the portion of R3 closest to U95 is probably necessary to maintain the activity of the U95 promoter. HHV-6B R3 is a relatively heterogeneous collection of various units, and its effects on the transcription of U95 are affected by the proximity of its various regions to the coding sequence. In HHV-6A, in contrast, the DNA sequences are highly conserved among the R3 units. NF-κB sites are conserved in all the units but one, and R3 is a homogeneous collection of nearly identical units and would not be expected to exhibit polarity with respect to its effects on transcription. If NF-κB binds equally well to all of the conserved motifs in the R3 region, it is possible that HHV-6A R3 is involved in the transcriptional regulation of the IE-A locus as well as of U95, while there is little or no possibility that the HHV-6B R3 region could do so, as judged by the polarity of the NF-κB motifs. If the effect of the R3 region on the IE-A locus is in fact different among variants of HHV-6, it will be of interest to investigate whether R3 affects different biological properties as well. These possibilities remain to be elucidated in future investigations.
ACKNOWLEDGMENTS
We thank J. Inoue for his generous gift of the pME-IκBα(S32A/S36A) plasmid and S. Nagata for kindly providing the pEF-BOS plasmid.
This study was partly supported by a grant-in-aid for general scientific research from the Ministry of Education, Science and Culture of Japan.
REFERENCES
- 1.Ablashi D V, Balachandran N, Josephs S F, Hung C L, Krueger G R, Kramarsky B, Salahuddin S Z, Gallo R C. Genomic polymorphism, growth properties, and immunologic variations in human herpesvirus-6 isolates. Virology. 1991;184:545–552. doi: 10.1016/0042-6822(91)90424-a. [DOI] [PubMed] [Google Scholar]
- 2.Agulnick A D, Thompson J R, Ricciardi R P. An ATF/CREB site is the major regulatory element in the human herpesvirus 6 DNA polymerase promoter. J Virol. 1994;68:2970–2977. doi: 10.1128/jvi.68.5.2970-2977.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aubin J T, Collandre H, Candotti D, Ingrand D, Rouzioux C, Burgard M, Richard S, Huraux J M, Agut H. Several groups among human herpesvirus 6 strains can be distinguished by Southern blotting and polymerase chain reaction. J Clin Microbiol. 1991;29:367–372. doi: 10.1128/jcm.29.2.367-372.1991. . (Erratum, 30:2524, 1992.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. Vol. 1. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 5.Bassuk A G, Anandappa R T, Leiden J M. Physical interactions between Ets and NF-κB/NFAT proteins play an important role in their cooperative activation of the human immunodeficiency virus enhancer in T cells. J Virol. 1997;71:3563–3573. doi: 10.1128/jvi.71.5.3563-3573.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berneman Z N, Ablashi D V, Li G, Eger-Fletcher M, Reitz M S, Jr, Hung C L, Brus I, Komaroff A L, Gallo R C. Human herpesvirus 7 is a T-lymphotropic virus and is related to, but significantly different from, human herpesvirus 6 and human cytomegalovirus. Proc Natl Acad Sci USA. 1992;89:10552–10556. doi: 10.1073/pnas.89.21.10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cardin R D, Abenes G B, Stoddart C A, Mocarski E S. Murine cytomegalovirus IE2, an activator of gene expression, is dispensable for growth and latency in mice. Virology. 1995;209:236–241. doi: 10.1006/viro.1995.1249. [DOI] [PubMed] [Google Scholar]
- 8.Chan Y J, Tseng W P, Hayward G S. Two distinct upstream regulatory domains containing multicopy cellular transcription factor binding sites provide basal repression and inducible enhancer characteristics to the immediate-early IES (US3) promoter from human cytomegalovirus. J Virol. 1996;70:5312–5328. doi: 10.1128/jvi.70.8.5312-5328.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandran B, Tirawatnapong S, Pfeiffer B, Ablashi D V. Antigenic relationships among human herpesvirus-6 isolates. J Med Virol. 1992;37:247–254. doi: 10.1002/jmv.1890370403. [DOI] [PubMed] [Google Scholar]
- 10.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison C A, Kouzarides T, Martignetti J A, et al. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 11.Cherrington J M, Mocarski E S. Human cytomegalovirus ie1 transactivates the alpha promoter-enhancer via an 18-base-pair repeat element. J Virol. 1989;63:1435–1440. doi: 10.1128/jvi.63.3.1435-1440.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colberg-Poley A M, Santomenna L D, Harlow P P, Benfield P A, Tenney D J. Human cytomegalovirus US3 and UL36–38 immediate-early proteins regulate gene expression. J Virol. 1992;66:95–105. doi: 10.1128/jvi.66.1.95-105.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dominguez G, Dambaugh T R, Stamey F R, Dewhurst S, Inoue N, Pellett P E. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J Virol. 1999;73:8040–8052. doi: 10.1128/jvi.73.10.8040-8052.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Efstathiou S, Lawrence G L, Brown C M, Barrell B G. Identification of homologues to the human cytomegalovirus US22 gene family in human herpesvirus 6. J Gen Virol. 1992;73:1661–1671. doi: 10.1099/0022-1317-73-7-1661. [DOI] [PubMed] [Google Scholar]
- 16.Geng Y Q, Chandran B, Josephs S F, Wood C. Identification and characterization of a human herpesvirus 6 gene segment that trans activates the human immunodeficiency virus type 1 promoter. J Virol. 1992;66:1564–1570. doi: 10.1128/jvi.66.3.1564-1570.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh S, May M J, Kopp E B. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 18.Gompels U A, Carrigan D R, Carss A L, Arno J. Two groups of human herpesvirus 6 identified by sequence analyses of laboratory strains and variants from Hodgkin's lymphoma and bone marrow transplant patients. J Gen Virol. 1993;74:613–622. doi: 10.1099/0022-1317-74-4-613. [DOI] [PubMed] [Google Scholar]
- 19.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 20.Gri G, Savio D, Trinchieri G, Ma X. Synergistic regulation of the human interleukin-12 p40 promoter by NF-κB and Ets transcription factors in Epstein-Barr virus-transformed B cells and macrophages. J Biol Chem. 1998;273:6431–6438. doi: 10.1074/jbc.273.11.6431. [DOI] [PubMed] [Google Scholar]
- 21.Hames B D, Higgins S J. Gene transcription. New York, N.Y: Oxford University Press; 1993. [Google Scholar]
- 22.Isegawa Y, Mukai T, Nakano K, Kagawa M, Chen J, Mori Y, Sunagawa T, Kawanishi K, Sashihara J, Hata A, Zou P, Kosuge H, Yamanishi K. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J Virol. 1999;73:8053–8063. doi: 10.1128/jvi.73.10.8053-8063.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isegawa Y, Ping Z, Nakano K, Sugimoto N, Yamanishi K. Human herpesvirus 6 open reading frame U12 encodes a functional beta-chemokine receptor. J Virol. 1998;72:6104–6112. doi: 10.1128/jvi.72.7.6104-6112.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.John S, Reeves R B, Lin J X, Child R, Leiden J M, Thompson C B, Leonard W J. Regulation of cell-type-specific interleukin-2 receptor alpha-chain gene expression: potential role of physical interactions between Elf-1, HMG-I(Y), and NF-κB family proteins. Mol Cell Biol. 1995;15:1786–1796. doi: 10.1128/mcb.15.3.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Josephs S F, Salahuddin S Z, Ablashi D V, Schachter F, Wong-Staal F, Gallo R C. Genomic analysis of the human B-lymphotropic virus (HBLV) Science. 1986;234:601–603. doi: 10.1126/science.3020691. [DOI] [PubMed] [Google Scholar]
- 26.Kondo K, Kondo T, Okuno T, Takahashi M, Yamanishi K. Latent human herpesvirus 6 infection of human monocytes/macrophages. J Gen Virol. 1991;72:1401–1408. doi: 10.1099/0022-1317-72-6-1401. [DOI] [PubMed] [Google Scholar]
- 27.Koszinowski U H, Keil G M, Volkmer H, Fibi M R, Ebeling-Keil A, Munch K. The 89,000-Mr murine cytomegalovirus immediate-early protein activates gene transcription. J Virol. 1986;58:59–66. doi: 10.1128/jvi.58.1.59-66.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouzarides T, Bankier A T, Satchwell S C, Preddy E, Barrell B G. An immediate early gene of human cytomegalovirus encodes a potential membrane glycoprotein. Virology. 1988;165:151–164. doi: 10.1016/0042-6822(88)90668-x. . (Erratum, 167:326–327, 1988.) [DOI] [PubMed] [Google Scholar]
- 29.Lawrence G L, Chee M, Craxton M A, Gompels U A, Honess R W, Barrell B G. Human herpesvirus 6 is closely related to human cytomegalovirus. J Virol. 1990;64:287–299. doi: 10.1128/jvi.64.1.287-299.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lusso P, Markham P D, Tschachler E, di Marzo Veronese F, Salahuddin S Z, Ablashi D V, Pahwa S, Krohn K, Gallo R C. In vitro cellular tropism of human B-lymphotropic virus (human herpesvirus-6) J Exp Med. 1988;167:1659–1670. doi: 10.1084/jem.167.5.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin M E, Nicholas J, Thomson B J, Newman C, Honess R W. Identification of a transactivating function mapping to the putative immediate-early locus of human herpesvirus 6. J Virol. 1991;65:5381–5390. doi: 10.1128/jvi.65.10.5381-5390.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGeoch D J. Evolutionary relationships of virion glycoprotein genes in the S regions of alphaherpesvirus genomes. J Gen Virol. 1990;71:2361–2367. doi: 10.1099/0022-1317-71-10-2361. [DOI] [PubMed] [Google Scholar]
- 33.McGeoch D J. The genomes of the human herpesviruses: contents, relationships, and evolution. Annu Rev Microbiol. 1989;43:235–265. doi: 10.1146/annurev.mi.43.100189.001315. [DOI] [PubMed] [Google Scholar]
- 34.Messerle M, Keil G M, Koszinowski U H. Structure and expression of murine cytomegalovirus immediate-early gene 2. J Virol. 1991;65:1638–1643. doi: 10.1128/jvi.65.3.1638-1643.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mori Y, Yagi H, Shimamoto T, Isegawa Y, Sunagawa T, Inagi R, Kondo K, Tano Y, Yamanishi K. Analysis of human herpesvirus 6 U3 gene, which is a positonal homolog of human cytomegalovirus UL 24 gene. Virology. 1998;249:129–139. doi: 10.1006/viro.1998.9305. [DOI] [PubMed] [Google Scholar]
- 36.Neipel F, Ellinger K, Fleckenstein B. The unique region of the human herpesvirus 6 genome is essentially collinear with the UL segment of human cytomegalovirus. J Gen Virol. 1991;72:2293–2297. doi: 10.1099/0022-1317-72-9-2293. [DOI] [PubMed] [Google Scholar]
- 37.Nicholas J. Determination and analysis of the complete nucleotide sequence of human herpesvirus. J Virol. 1996;70:5975–5989. doi: 10.1128/jvi.70.9.5975-5989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholas J, Martin M E. Nucleotide sequence analysis of a 38.5-kilobase-pair region of the genome of human herpesvirus 6 encoding human cytomegalovirus immediate-early gene homologs and transactivating functions. J Virol. 1994;68:597–610. doi: 10.1128/jvi.68.2.597-610.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okuno T, Takahashi K, Balachandra K, Shiraki K, Yamanishi K, Takahashi M, Baba K. Seroepidemiology of human herpesvirus 6 infection in normal children and adults. J Clin Microbiol. 1989;27:651–653. doi: 10.1128/jcm.27.4.651-653.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel A, Hanson J, McLean T L, Olgiate J, Hilton M, Miller W E, Bachenheimer S L. Herpes simplex type 1 induction of persistent NF-kappa B nuclear translocation increase the efficiency of virus replication. Virology. 1998;247:212–222. doi: 10.1006/viro.1998.9243. [DOI] [PubMed] [Google Scholar]
- 41.Roizmann B, Desrosiers R C, Fleckenstein B, Lopez C, Minson A C, Studdert M J. The family Herpesviridae: an update. The Herpesvirus Study Group of the International Committee on Taxonomy of Viruses. Arch Virol. 1992;123:425–449. doi: 10.1007/BF01317276. [DOI] [PubMed] [Google Scholar]
- 42.Salahuddin S Z, Ablashi D V, Markham P D, Josephs S F, Sturzenegger S, Kaplan M, Halligan G, Biberfeld P, Wong-Staal F, Kramarsky B, et al. Isolation of a new virus, HBLV, in patients with lymphoproliferative disorders. Science. 1986;234:596–601. doi: 10.1126/science.2876520. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Sambucetti L C, Cherrington J M, Wilkinson G W, Mocarski E S. NF-kappa B activation of the cytomegalovirus enhancer is mediated by a viral transactivator and by T cell stimulation. EMBO J. 1989;8:4251–4258. doi: 10.1002/j.1460-2075.1989.tb08610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schirmer E C, Wyatt L S, Yamanishi K, Rodriguez W J, Frenkel N. Differentiation between two distinct classes of viruses now classified as human herpesvirus 6. Proc Natl Acad Sci USA. 1991;88:5922–5926. doi: 10.1073/pnas.88.13.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stasiak P C, Mocarski E S. Transactivation of the cytomegalovirus ICP36 gene promoter requires the alpha gene product TRS1 in addition to IE1 and IE2. J Virol. 1992;66:1050–1058. doi: 10.1128/jvi.66.2.1050-1058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugano N, Chen W, Roberts M L, Cooper N R. Epstein-Barr virus binding to CD21 activates the initial viral promoter via NF-κB induction. J Exp Med. 1997;186:731–737. doi: 10.1084/jem.186.5.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi K, Sonoda S, Higashi K, Kondo T, Takahashi H, Takahashi M, Yamanishi K. Predominant CD4 T-lymphocyte tropism of human herpesvirus 6-related virus. J Virol. 1989;63:3161–3163. doi: 10.1128/jvi.63.7.3161-3163.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson J, Choudhury S, Kashanchi F, Doniger J, Berneman Z, Frenkel N, Rosenthal L J. A transforming fragment within the direct repeat region of human herpesvirus type 6 that transactivates HIV-1. Oncogene. 1994;9:1167–1175. [PubMed] [Google Scholar]
- 50.Thompson J R, Agulnick A D, Ricciardi R P. A novel cis element essential for stimulated transcription of the p41 promoter of human herpesvirus 6. J Virol. 1994;68:4478–4485. doi: 10.1128/jvi.68.7.4478-4485.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomson B J, Honess R W. The right end of the unique region of the genome of human herpesvirus 6U1102 contains a candidate immediate early gene enhancer and a homologue of the human cytomegalovirus US22 gene family. J Gen Virol. 1992;73:1649–1660. doi: 10.1099/0022-1317-73-7-1649. [DOI] [PubMed] [Google Scholar]
- 52.Thrower A R, Bullock G C, Bissell J E, Stinski M F. Regulation of a human cytomegalovirus immediate-early gene (US3) by a silencer-enhancer combination. J Virol. 1996;70:91–100. doi: 10.1128/jvi.70.1.91-100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wyatt L S, Balachandran N, Frenkel N. Variations in the replication and antigenic properties of human herpesvirus 6 strains. J Infect Dis. 1990;162:852–857. doi: 10.1093/infdis/162.4.852. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto T, Mukai T, Kondo K, Yamanishi K. Variation of DNA sequence in immediate-early gene of human herpesvirus 6 and variant identification by PCR. J Clin Microbiol. 1994;32:473–476. doi: 10.1128/jcm.32.2.473-476.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamanishi K, Okuno T, Shiraki K, Takahashi M, Kondo T, Asano Y, Kurata T. Identification of human herpesvirus-6 as a causal agent for exanthem subitum. Lancet. 1988;i:1065–1067. doi: 10.1016/s0140-6736(88)91893-4. [DOI] [PubMed] [Google Scholar]