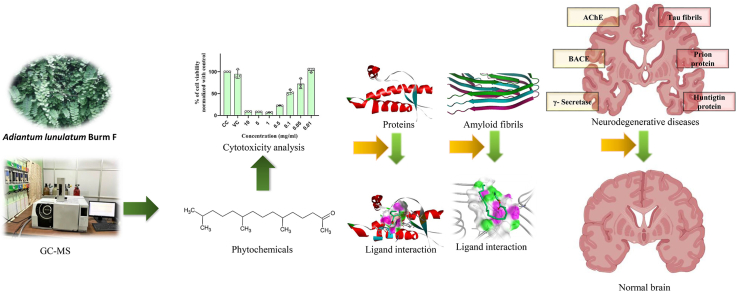
Abstract
In neurodegenerative diseases, amyloid formation by some proteins cause neuronal damage and loss. To prevent this neuronal damage and loss certain pharmaceuticals are available. Many of these pharmaceuticals act on the neurodegenerative disease symptoms but not on the root cause. This study helps to detect more effective agents which directly act on the root cause and reduce the risk of neurodegenerative diseases. To identify new anti-amyloid agents, the folk medicinally important plant Adiantum lunulatum was collected, authenticated, dried, extracted with ethanol and analyzed by GC-MS method. The screening of the identified phytochemicals was done using the webservers swissADME and ProTox-II. In-vitro MTT assay using Neuro-2a cell lines was carried out to determine the cytotoxicity of the extract. The interactions of these phytochemicals with the amyloid forming peptides and proteins were predicted using the molecular docking tools such as AutoDock Vina and BIOVIA discovery studio visualizer 2020. Through GC-MS analysis, 18 different volatile phytochemicals were identified from the ethanol extract. From this, 7 phytochemicals were selected based on the computational non-toxicity prediction. In-vitro cytotoxicity analysis of the ethanol extract using Neuro-2a cell lines detected the IC50 value of 0.09 mg/ml. Of these, the phytochemical P1 (trans, trans-9, 12-Octadecadienoic acid, propyl ester) interacts with tau, and huntingtin proteins, P2 (2-Pentadecanone, 6, 10, 14-trimethyl-) interacts with prion protein. The phytochemicals P1, P3 (Ethyl oleate), P4 (Octadecanoic acid, ethyl ester), and P5 (Phytol) interact with acetylcholinesterase. P2, P4, P5 and P6 (Henicosanal), interact with BACE-1. The phytochemical P3 interacts with γ- Secretase. The interaction of P2 and P5 with BACE-1 and P3 with γ- Secretase show better inhibition in inhibitory constant (Ki) analysis. These phytochemicals have been predicted to show significant potential against the formation or breakdown of peptide/protein amyloids, and further in-vitro studies are necessary to develop them into anti-amyloid agents.
Keywords: β-sheet, Amyloid, Phytochemical, Computation, Neurodegenerative
Graphical abstract
1. Introduction
The neurodegenerative diseases are the major prevalent diseases affecting approximately 50 million people across the world. Geriatric population are more susceptible for neurodegenerative diseases such as Alzheimer's, Parkinson's, Huntington and Prion diseases. These neurodegenerative illnesses are caused by the accumulation of the Aβ peptide [1], α-synuclein [2], huntingtin [3] and prion protein's [4] amyloid fibrils. The self-assembly of these proteins are initiated by the transformation of α-helical rich native form to the β-sheet rich fibrillar form resulting in to the disease condition. The self-assembly is triggered by conditions such as pH, temperature, concentration or chemical agents. For example, β-sheet rich tightly packed α-synuclein fibrils are formed with increase in the ionic strength [5].
In traditional medicine, many plant preparations are useful for the treatment of neurological diseases. One of the important plant type used in these medicinal preparations is Adiantum spp which has effective role in the prevention of neuronal damages and neuroprotective effect, for example, Adiantum capillus-veneris linn plant extracts protect rat brain from inflammation caused by carbendazim pesticide intoxication [6]. It has been reported in a research article published by Abdulqadir et al., 2018 that the methanol and ethyl acetate extracts of Adiantum capillus-veneris linn has the potential to inhibit the enzyme acetylcholinesterase [7]. This enzyme is involved in the cleavage of amyloid precursor protein (APP) producing amyloid β peptide which self assembles to form amyloid fibrils causing Alzheimer's disease. The phytochemicals from methanol and ethyl acetate extracts of Adiantum capillus-veneris linn is responsible for this inhibitory activity. The medicinal plant Adiantum lunulatum belongs to the same species and was expected to contain similar anti-amyloid agents. We have been searching for potential anti-amyloid phytochemicals from a new source and hence we selected this medicinally important plant in this study. Studies on the plant Adiantum lunulatum detected that it contains many classes of pharmacologically important phytochemicals, for example, terpenoids. In folk medicine, Adiantum lunulatum decoction, infusion and juices containing these phytochemicals are used for the treatment of various diseases [[8], [9], [10], [11]]. These phytochemical's detection is commonly carried out by using analytical techniques like GC-MS, LC-MS etc. Computational methods help in the pharmacokinetics prediction to detect the efficacy of these phytochemicals as a medicine [12].
Certain classes of phytochemicals are increasingly effective in the prevention of the self-assembly of amyloidogenic peptides and proteins, phenolics and terpenoids are important among them. For example, through non-covalent interactions, curcumin and its derivatives disrupt or stop the formation of amyloid fibrils of Aβ peptide [13,14]. Curcumin and its analogs also have high anti-amyloid activity against, tau [15], α-synuclein [16] and prion proteins [17]. In this study, we have explored the presence of some phytochemicals from the medicinal plant Adiantum lunulatum, their pharmacological behavior and anti-amyloid potential using various in-vitro and in-silico methods.
2. Materials and methods
2.1. Chemicals, instruments, softwares and webservers
The solvent ethanol for the extraction was obtained from Merk Life Sciences Pvt. Ltd Mumbai. MTT and media supplements were obtained from Himedia (Thane, India). Rotary vacuum evaporator for concentrating the extract was from Heldoph instruments, Germany. Characterization of different compounds present in the extract was analyzed using GC-MS from Shimadzu, Kyoto, Japan. Softwares for molecular docking studies include AutoDock Vina [18] and BIOVIA discovery studio visualizer 2020 (http://discover.3ds.com/discovery-studio-visualizer). Webserver used for the active site prediction was Computed Atlas of Surface Topography of Proteins (Castp) (http://sts.bioe.uic.edu) [19] SwissADME (http://www.swissadme.ch/), and ProTox-II (http://tox.charite.de/protox_II) webservers were used for the prediction of biochemical activities [20].
2.2. Plant materials and extraction
The plant was identified as Adiantum lunulatum Burm F after being validated at Kerala University's Department of Botany Herbarium as a voucher specimen with the number KUBH 11089. The aerial plant components were washed, dried, powdered and macerated in ethanol for eight days, which was filtered and centrifuged at 8000 rpm for 8 min at room temperature. The supernatant was collected, concentrated using a rotary vacuum evaporator, and then used for further studies.
2.3. GC-MS analysis of the extract
GC-MS analysis was carried out using Shimadzu QP2020 GC-MS instrument containing SH-Rxi-5Sil MS capillary column of 30 m × 0.2 mm ID x 0.25 μm df thickness. The carrier gas used was helium at a constant flow rate of 1.20 ml/min. 1 μl (Split ratio 10:1) of the concentrated extract was injected using the injector maintained at 3000C. The column oven temperature was raised to 280 °C at a heating rate of 10 °C per minute and sample was injected 10 min after attaining the target temperature. The ionization chamber was kept at 220 °C and ionized the vaporized sample using EI method. The ions were detected using TCD detector from 3.2 to 34 min. Phytochemicals were identified by comparative analysis of retention time, peak height, and area with the help of WILEY NIST/EPA/NIH Mass spectral library 2017.
2.4. Prediction of toxicity and biochemical activity
The webserver SwissADME (http://www.swissadme.ch/, [21]) was used to predict the critical biochemical activities of the selected phytochemicals based on Lipinski rules. According to this rule, compounds donating less than 5 hydrogen bonds, accepting less than ten hydrogen bonds, having molecular weight below 500 Da and log P below 5 and have absorption or permeation potential [22] may act as drugs. The toxicological profile was predicted using ProTox-II (http://tox.charite.de/protox_II) webserver [20].
2.5. In-vitro analysis of the ethanol extract cytotoxicity on Neuro-2a cell lines
The mouse neuroblastoma cell line Neuro-2a has been acquired from the National Center for Cell Science, Pune. The Neuro-2a cells were cultured in Dulbecco's Modified Eagle Medium containing sodium pyruvate and sodium bicarbonate supplemented with high glucose (4.5 g/L), L-glutamine (0.30 g/L), 1 % 100X Antibiotic Antimycotic solution (10,000 U Penicillin, 10 mg Streptomycin and 25 μg Amphotericin B per ml in 0.9 % normal saline), and 10 % fetal bovine serum (FBS). Cell monolayers were grown in a humidified incubator with 95 % air and 5 % CO2 at 37 °C.
2.5.1. 3-(4, 5-Dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay
Cells were seeded in NGM for 24 h by using 96-well plates to achieve 60–70 % confluency. After 24 h, the cells were differentiated by serum deprivation in high-glucose DMEM for 48 h. Then the cultured Neuro-2a cell lines were treated with varying concentrations of ethanol extract (0.001–10 mg/ml) for 24 h, MTT (0.5 mg/ml) was applied to cells in serum-free media at 37 °C for 1 h and 5 % CO2 to metabolize the yellow MTT to purple formazan. After that, it was dissolved in DMSO, and a microplate spectrophotometer was used to determine the optical density at 570 nm (Thermo Scientific™ Varioskan LUX Multimode Microplate Reader). The viability of the cells at different treatments were determined by percentage of control cell viability [23]. The IC50 value was determined by using the GraphPad Prism version 9.0.0 software.
2.6. Molecular docking of the phytochemicals with amyloid fibrils
2.6.1. Ligand preparation for docking
The structure coordinates of various ligands such as Neophytadiene (PubChem CID: 10446); 2-Pentadecanone, 6,10,14-trimethyl- (PubChem CID: 10408); Phytol (PubChem CID: 5280435); Octadecanoic acid, ethyl ester (PubChem CID: 8122); trans,trans-9,12-Octadecadienoic acid, propyl ester (PubChem CID: 18432395); Ethyl Oleate (PubChem CID: 5363269); Henicosanal (PubChem CID: 12052911) were obtained from PubChem Database (https://pubchem.ncbi.nlm.nih.gov/) in SDF file format. BIOVIA discovery studio visualizer 2020 (http://discover.3ds.com/discovery-studio-visualizer) and AutoDock Vina were used to prepare ligands for docking [18].
2.6.2. Target (protein) preparation for molecular docking
The initial coordinates of the targets Aβ peptide (PDB ID: 6W0O), huntingtin protein (PDB ID: 6N8C), α-synuclein protein (PDB ID: 6FLT), prion protein (PDB ID: 1QLX), tau protein (PDB ID: 6VH7), acetylcholinesterase (PDB ID: 4M0E), BACE1 (PDB ID: 6EQM), γ-Secretase (PDB ID: 7C9I) fibrils were obtained from the RCSB PDB database (https://www.rcsb.org/structure). The structures were optimized using AutoDock Vina software by removing all water molecules, and adding Kollman charges and nonpolar hydrogens.
2.6.2.1. Active site prediction
Computed Atlas of Surface Topography of Proteins (CASTp) is an online tool for active site prediction which accepts protein fibril structures in PDB format [19]. Active site prediction helped to determine the protein fibril's cavities or possible ligand binding pockets.
2.6.2.2. Grid preparation
The autodock vina used for the preparation of the grid box on each protein fibril. The grid box center coordinate for Aβ peptide grid dimension 90 X 90 × 90 Å as X = 216.050, Y = 215.943, Z = 216.175, huntingtin protein grid dimension 40 X 40 X 40 for as X = 0.002, Y = 0.011, Z = 0.006, α-synuclein grid dimension as 60 X 60 X 60 X = 116.376, Y = 116.378, Z = 92.089 prion grid dimension as 40 X 40 X 40 X = 0.019, Y = 0.099, Z = -0.039, and tau grid dimension 90 X 90 X 90 as X = 190.897, Y = 190.864, Z = 188.078. Acetylcholinesterase with grid dimension 60 X 60 X 60 for as X = −0.727, Y = −51.740, Z = 3.216, BACE-1 with grid dimension 40 X 40 X 40 for as X = 21.651, Y = 75.240, Z = 15.407 and γ –Secretase with grid dimension 60 X 60 X 60 for as X = 173.310, Y = 175.625, Z = 188.081.
2.6.3. Molecular docking
Molecular docking of identified phytochemicals with the fibrils was performed with BIOVIA discovery studio visualizer 2020 (http://disco ver.3ds.com/discovery-studio-visualizer) and AutoDock Vina [18] softwares. The ligand-fibril complex with a Root Mean Square Deviation (RMSD) of 0.00 was chosen as the optimal interaction position from among eight docking ligand poses. The ligand's affinity to protein fibril was determined by the lowest binding energy (kcal/mol), number of hydrogen bonding and hydrophobic interactions. Compounds having well-studied anti-amyloid action were used as the docking standard. The docked complex structures were analyzed using the BIOVIA discovery studio visualizer 2020 (http://disco.3ds.com/discovery-studio-visualizer) software. Confirmation of the ligand protein interaction was carried out by using SwissDock webserver [24].
2.7. Molecular interaction studies by using SwissDock webserver
The interaction of screened phytochemicals with proteins and peptides showing amyloid formation was carried out by using SwissDock webserver. The grid box center coordinate for tau peptide are X = 216.050, Y = 215.943, Z = 216.175 and grid dimension 30 X 30 × 30 Å. The grid box center coordinate for prion protein are X = 0.019, Y = 0.099, Z = -0.039 with grid dimension as 30 X 30 × 30 Å. The grid box center coordinate for huntingtin protein are X = 0.002, Y = 0.011, Z = 0.006 with grid dimension 30 X 30 × 30 Å. Acetylcholinesterase with grid box center coordinates of X = −0.727, Y = −51.740, Z = 3.216 with grid dimension 10 X 10 × 10Å. BACE-1 with grid box center coordinates of X = 21.651, Y = 75.240, Z = 15.407 with grid dimension 30 X 30 × 30 Å. γ –Secretase grid box center coordinates of X = 173.310, Y = 175.625, Z = 188.081 with grid dimension 20 X 20 × 20 Å. Molecular docking was carried out by SwissDock in presence of AutoDock Vina and by Attracting cavities [[24], [25], [26]].
Inhibitory constant Ki was calculated by using the equation,
ΔG -binding Gibbs free energy, R (gas constant) is 1.9872036 cal/mol and T (temperature) is 298.15 K [27].
3. Results
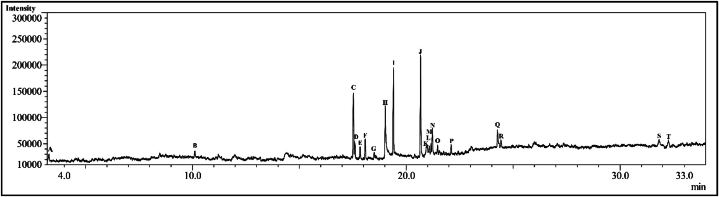
Using GC-MS technique, we have identified 18 phytochemicals from the ethanol extract of Adiantum lunulatum plant (Fig. 1). The name, retention time (RT), molecular weight (MW) and molecular formula of the phytochemicals identified from ethanol extract are given in Table 1.
Fig. 1.
Chromatogram of the ethanol extract.
Table 1.
The name, retention time (RT), molecular weight (MW) and molecular formula of the phytochemicals identified from ethanol extract using GC-MS method.
| Sl. No | Compounds | RT (min) | MW (g/mol) | Molecular Formula |
|---|---|---|---|---|
| 1 | Formamide, N-methoxy- | 3.291 | 75.07 | C2H5NO2 |
| 2 | 2-Methoxy-4-vinylphenol | 10.112 | 150.17 | C9H10O2 |
| 3 | Neophytadiene | 17.527 | 278.5 | C20H38 |
| 4 | 2-Pentadecanone, 6,10,14-trimethyl- | 17.592 | 268.5 | C18H36O |
| 5 | Benzene, (1-methylnonadecyl)- | 18.500 | 358.6 | C26H46 |
| 6 | n-Hexadecanoic acid | 19.017 | 256.42 | C16H32O2 |
| 7 | Hexadecanoic acid, ethyl ester | 19.400 | 284.5 | C18H36O2 |
| 8 | Phytol | 20.667 | 296.5 | C20H40O |
| 9 | 7-Tetradecenal, (Z)- | 20.974 | 210.36 | C14H26O |
| 10 | Carbonic acid, but-3-yn-1-yl hexadecyl ester | 21.076 | 338.52 | C21H38O3 |
| 11 | trans,trans-9,12-Octadecadienoic acid, propyl ester | 21.162 | 322.5 | C21H38O2 |
| 12 | Ethyl Oleate | 21.221 | 310.51 | C20H38O2 |
| 13 | Octadecanoic acid, ethyl ester | 21.464 | 312.53 | C20H40O2 |
| 14 | Oxirane, hexadecyl- | 22.101 | 268.47 | C18H36O |
| 15 | Hexadecanoic acid, 2-hydroxy-1-(hydroxymethyl)ethyl ester | 24.265 | 330.5 | C19H38O4 |
| 16 | Henicosanal | 24.434 | 310.6 | C21H42O |
| 17 | Stigmasta-5,22-dien-3-ol, acetate, (3.beta.,22Z)- | 31.826 | 454.7 | C31H50O2 |
| 18 | Stigmast-5-en-3-ol, oleate | 32.259 | 679.2 | C47H82O2 |
3.1. Ligand screening based on their predicted biochemical properties
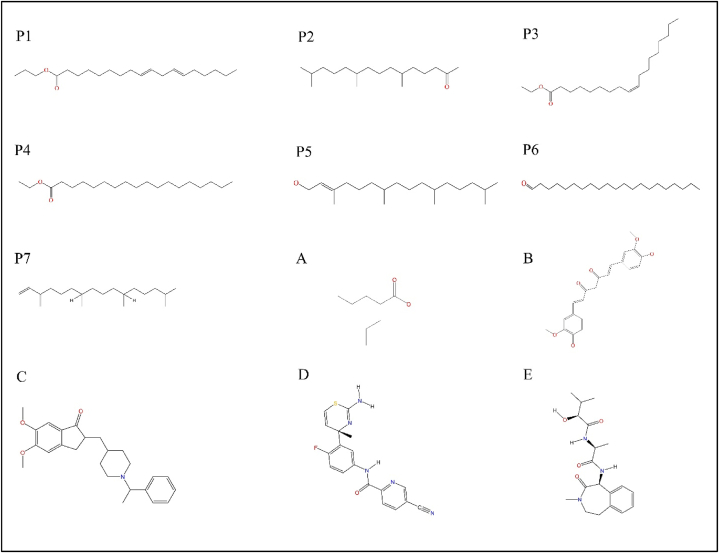
As a first step to identify the most potent anti-amyloid phytochemicals, the biochemical properties of all phytochemicals identified from the ethanol extract were predicted. From these biochemical predictions, 7 phytochemicals lacking cytotoxicity and hepatotoxicity were chosen for molecular docking studies. These include P1 (trans, trans-9, 12-octadecadienoic acid, propyl ester), P2 (2-pentadecanone, 6, 10, 14-trimethyl-), P3 (ethyl oleate), P4 (octadecanoic acid, ethyl ester), P5 (phytol), P6 (henicosanal) and P7 (neophytadiene) (Table 2). The different predicted biochemical properties of these compounds are given in Table 2.
Table 2.
The predicted biochemical properties of the phytochemicals, without cytotoxicity and hepatotoxicity, from ethanolic extract of Adiantum Lunulatum.
| Code | Lipinski rule/violation | GI | BBB | A | WS | CT | HT | OT mol/kg | HU | SP (log Kp) |
|---|---|---|---|---|---|---|---|---|---|---|
| P1 | Yes/1 | L | N | N | MS | N | N | 1.588 | 0.102 | −2.77 cm/s |
| P2 | Yes/1 | H | N | N | MS | N | N | 1.532 | 0.244 | −3.00 cm/s |
| P3 | Yes/1 | L | N | N | MS | N | N | 1.663 | 0.07 | −2.49 cm/s |
| P4 | Yes/1 | L | N | N | PS | N | N | 1.68 | 0.13 | −1.84 cm/s |
| P5 | Yes/1 | L | N | N | MS | N | N | 1.607 | 0.05 | −2.29 cm/s |
| P6 | Yes/1 | L | N | N | PS | N | N | 1.615 | −0.134 | −1.26 cm/s |
| P7 | Yes/1 | L | N | N | PS | N | N | 1.473 | 0.272 | −1.17 cm/s |
Phytochemicals P1 to P7 follow the Lipinski rules with single violation MLOGP>4.15. These phytochemicals GI- Gastrointestinal absorption, BBB-Blood brain barrier permeability, A- AMES toxicity, WS- water solubility, CT- Cytotoxicity, HT-Hepatotoxicity, OT-Oral toxicity LD50 [mol/kg], PS- Poorly soluble, HU- human max tolerated dose log mg/kg/day, SP- Skin permeability were reported. Y-Yes, N-no, L-low, H-High. MS- medium solubility.
The phytochemical P1 has low gastrointestinal absorption, no AMES toxicity, moderate water solubility, oral toxicity LD50 1.588 mol/kg, and human max tolerated dose 0.102 log mg/kg/day. The phytochemical P2 has high gastrointestinal absorption, no AMES toxicity, moderate water solubility, oral toxicity LD50 1.532 mol/kg, and human max tolerated dose 0.244 log mg/kg/day. The phytochemical P3 has low gastrointestinal absorption, no AMES toxicity, moderately water soluble, oral toxicity LD50 1.663 mol/kg, and human max tolerated dose 0.07 log mg/kg/day. The phytochemical P4 has low gastrointestinal absorption, no AMES toxicity, less water soluble, oral toxicity LD50 1.68 mol/kg, and human max tolerated dose 0.13 log mg/kg/day. The phytochemical P5 has low gastrointestinal absorption, AMES toxicity, moderate water soluble, oral toxicity LD50 1.607 mol/kg, and human max tolerated dose 0.05 log mg/kg/day. The phytochemical P6 with low gastrointestinal absorption, no AMES toxicity, low water soluble, oral toxicity LD50 1.615 mol/kg, and human max tolerated dose −0.134 log mg/kg/day. The phytochemical P7 has low gastrointestinal absorption, AMES toxicity, less water soluble, oral toxicity LD50 1.473 mol/kg, and human max tolerated dose is 0.272 log mg/kg/day. The phytochemicals P1 to P7 follow the rule of Lipinski's with single violation MLOGP>4.15 and have good drug likeness. Prediction showed that these phytochemicals have low blood brain barrier crossing ability in normal conditions, but in neurodegenerative patients the permeability of blood brain barrier is higher allowing the crossing of these molecules.
3.2. Cytotoxicity of ethanol extract on the Neuro-2a cell lines
The ethanol extract, containing above mentioned phytochemicals, has been used for the determination of cytotoxicity on Neuro-2a cell lines and the metabolic activity was determined using the MTT assay.
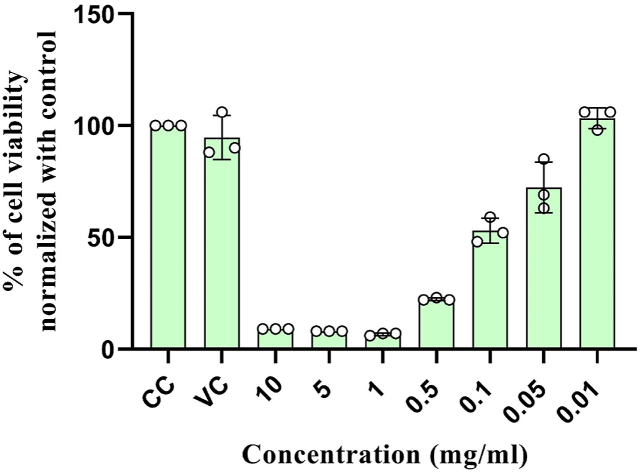
Absorbance of MTT assay was represented in mean ± standard deviation (SD), Control cells were treated with normal growth medium, and DMSO at 10 mg/ml was used as vehicle control. Different concentration of ethanol extract 10, 5, 1 and 0.5 mg/ml show high level of Neuro-2a cytotoxicity. Ethanol extract at concentration 0.1, 0.05, and 0.01 mg/ml is effective in maintaining the normal cell metabolism. The calculated IC50 value of ethanol extract was 0.09 mg/ml and concentration below 0.01 mg/ml was considered as safe (Graph 1) (Table 3) .
Graph 1.
In-vivo cytotoxicity analysis on Neuro-2a cell lines.
Table 3.
In-vivo cytotoxicity analysis on Neuro-2a cell lines.
| Sl no | Concentration of ethanol extract (mg/ml) | Mean ± SD |
|---|---|---|
| 1. | Cell control | 1.65 ± 0.06 |
| 2. | Vehicle control | 1.56 ± 0.11 |
| 3. | 10 mg/ml | 0.15 ± 0.00 |
| 4. | 5 mg/ml | 0.13 ± 0.01 |
| 5. | 1 mg/ml | 0.11 ± 0.00 |
| 6. | 0.5 mg/ml | 0.37 ± 0.01 |
| 7. | 0.1 mg/ml | 0.87 ± 0.07 |
| 8. | 0.05 mg/ml | 1.19 ± 0.20 |
| 9. | 0.01 mg/ml | 1.71 ± 0.13 |
3.3. Molecular docking study of the selected phytochemicals with different protein fibrils
3.3.1. Tau protein
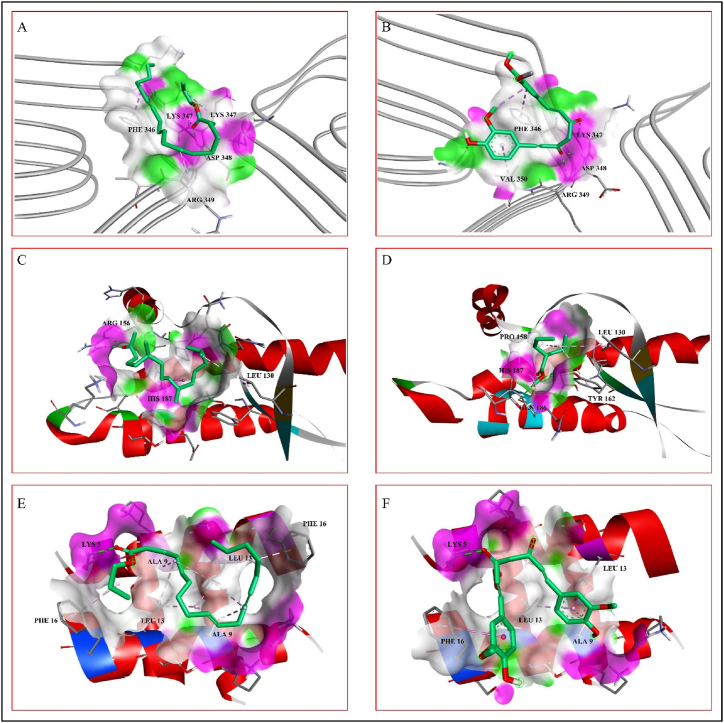
Fibril formation by the self-assembly of tau protein causes Alzheimer's disease. It was reported that the phytochemical curcumin has the potential to destabilize the amyloid fibrils of tau protein [15]. In this docking study, curcumin is selected as a standard for comparing the binding affinities of the selected phytochemicals from Adiantum Lunulatum. The binding affinity of curcumin with tau protein fibril is −4.7 kcal/mol and it forms hydrogen bonds with the A chain amino acid residues LYS347, ASP348, and ARG349 and hydrophobic interactions with the A chain residues PHE346 and VAL350. Out of the above mentioned seven phytochemicals, the phytochemical P1 interacts with tau protein fibrils by forming hydrogen bonds with the A chain residues LYS347 and ASP348, as well as hydrophobic interactions with the A chain residues PHE346, LYS347, and ARG349 with a binding affinity of −2.9 kcal/mol (Table 4). The interactions between the tau protein fibrils and the ligands are shown in Fig. 2A and B. According to the SwissDock prediction the phytochemical P1 has binding affinity of −3.629 kcal/mol, inhibitory constant (Ki) calculated as 2181 μM, attracting cavity (AC) score of −3.658105 and SwissParam score of −4.3364 to tau protein (Table 6).
Table 4.
Interactions of selected phytochemicals with neurodegenerative proteins and fibrils.
| Protein/peptide | Ligands | BA (kcal/mol) | Ki (μM) | Hydrogen bonding | Hydrophobic interactions |
|---|---|---|---|---|---|
| Tau peptide | P1 | −2.9 | 7468 | A:LYS347, A:ASP348 | A:LYS347, A:ARG349, A:PHE346 |
| Prion protein | P2 | −3.8 | 1634 | A:ARG156,A:ARG156 | A:LEU130, A:HIS187 |
| Huntingtin protein | P1 | −4.0 | 1166 | D:LYS5, D:LYS5 | C:ALA9, C:ALA9, C:LEU13, D:ALA9, D:LEU13, C:LEU13, D:LEU13, D:LEU13, C:PHE16, D:PHE16 |
| Standards | |||||
| Tau peptide | SD1 | −4.7 | 357 | A:LYS347, A:ASP348, A:ARG349, |
A:PHE346, A:VAL350 |
| Prion protein | SD2 | −3.8 | 1634 | A:TYR162, A:HIS187, A:GLN186, |
A:PRO158, A:LEU130, A:TYR162 |
| Huntingtin protein | SD1 | −5.1 | 182 | D:LYS5, D:LYS5 | D:LEU13, C:PHE16, C:ALA9, C:LEU13 |
BA-binding affinity. Curcumin SD1, Valproic acid –SD2.
Fig. 2.
Phytochemical interaction with the peptide/protein fibrils of neurodegenerative diseases. A- Tau protein interaction of trans,trans-9,12-Octadecadienoic acid, propyl ester. B- Tau protein interaction of standard curcumin. C- Prion protein interaction of 2-Pentadecanone, 6,10,14-trimethyl- D- Prion protein interaction of valproic acid. E- Huntingtin protein interaction of trans,trans-9,12-Octadecadienoic acid, propyl ester. F- Huntingtin protein interaction of standard curcumin.
Table 6.
SwissDock prediction of the phytochemical interaction with anti-amyloid proteins. SwissParam used to detect the interactions through the AC score and SwissParam score.
| Protein/Peptides | Ligands | BA (Kcal/mol) | Ki (μM) | AC score | SwissParam score (Kcal/mol) |
|---|---|---|---|---|---|
| Tau | P1 | −3.629 | 2181 | −3.658105 | −4.3364 |
| Prion | P2 | −5.013 | 211 | −31.797964 | −6.2641 |
| Huntingtin | P1 | −4.051 | 1069 | −22.283152 | −5.0610 |
| ACE | P1 | −5.27 | 137 | −44.568258 | −7.3496 |
| ACE | P3 | −5.205 | 152 | −53.131895 | −6.9867 |
| ACE | P4 | −5.079 | 188 | −57.341387 | −6.8157 |
| ACE | P5 | −4.968 | 227 | −39.488361 | −6.9718 |
| BACE 1 | P2 | −5.338 | 122 | −36.198006 | −7.4459 |
| BACE 1 | P4 | −4.285 | 720 | −57.705446 | −7.0953 |
| BACE 1 | P5 | −5.553 | 85 | −35.430713 | −6.7800 |
| BACE 1 | P6 | −4.401 | 592 | −50.277182 | −6.8360 |
| γ-Secretase | P3 | −4.683 | 368 | −32.107416 | −5.6657 |
BA-binding affinity, AC- Attracting cavity,ACE- Acetylcholinesterase, BACE 1- β-site amyloid precursor protein (APP) cleaving enzyme-1.
3.3.2. Prion protein
Conversion of prion protein from native to scrapie form leads to self-assembly causing prion diseases. According to previous reports valproic acid is effective against the self-assembly of prion proteins [28]. In this study, valproic acid is used as standard with a binding affinity of −3.8 kcal/mol forming hydrogen bonds with the residues TYR162, GLN186 and HIS187 and hydrophobic interactions with the residues LEU130, PRO158 and TYR162. In a similar fashion, the phytochemical P2 interacts with the prion protein with a binding affinity of −3.8 kcal/mol forming hydrogen bonds with the residue ARG156, and hydrophobic interactions with the residues LEU130 and HIS187 (Table .4). The interactions between the prion protein fibrils and the ligands are shown in Fig. 2C and D. According to SwissDock prediction P2 has binding affinity of −5.013 kcal/mol with inhibitory constant (Ki) calculated as 211 μM, attracting cavity (AC) score of −31.797964 and SwissParam score of −6.2641 to prion protein.
3.3.3. Huntingtin protein
Huntingtin protein form amyloids by self-assembly which cause Huntington diseases. To prevent this self-assembly of huntingtin protein, phytochemicals like curcumin are effective [29]. Curcumin interacts with huntingtin protein form two hydrogen bonds with the D chain residue LYS5, and one hydrogen bond with the D chain residue LEU13 as well as hydrophobic interactions with the C chain residues ALA9, LEU13 and PHE16 with a binding affinity of −5.1 kcal/mol. Similarly, the phytochemical P1 interacts with huntingtin protein forming hydrogen bond with the D chain residue LYS5 and hydrophobic interactions with the C chain residues ALA9, LEU13, PHE16, and D chain residues ALA9, LEU13, and PHE16 with a binding affinity of −4.0 kcal/mol (Table 4). The interactions between the huntingtin protein and the ligands are shown in Fig. 2E and F. According to SwissDock prediction detected that phytochemical P1 has binding affinity of −4.051 kcal/mol with inhibitory constant (Ki) calculated as 1069 μM, attracting cavity (AC) score of −22.283152 and SwissParam score of −5.0610 to huntingtin protein.
3.4. Molecular docking study of the selected phytochemicals with the acetylcholine sterase, BACE-1 and γ –secretase proteins
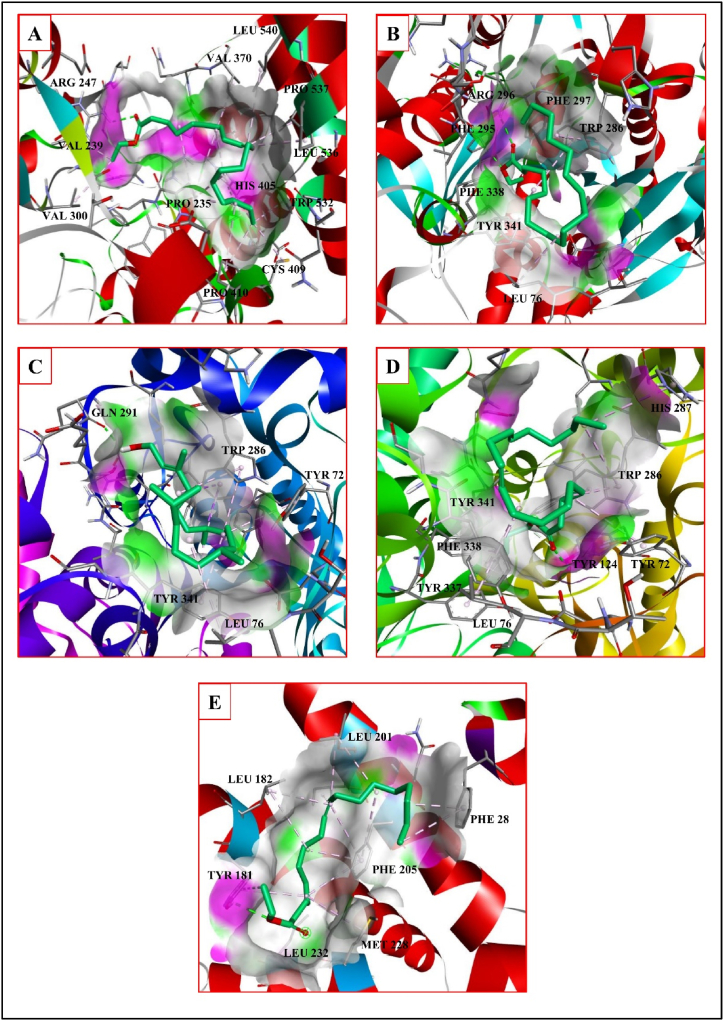
Acetylcholinesterase inhibitors help to alleviate inflammatory response, apoptosis, oxidative stress and protein aggregation symptoms of neurodegenerative disorders [30]. For example, the compound donepezil lessens the brain pathology by reducing the neuronal inflammation and oxidative stress, by recovering the mitochondrial function and preventing the Aβ fibril accumulation [31]. Donepezil forms hydrogen bonds with the A chain residues TYR337 and PHE338 and hydrophobic interactions with the A chain residues TYR72 TRP286, and TYR341, with a binding affinity of −8.9 kcal/mol. The phytochemical P1 interacts with acetylcholinesterase with a binding affinity of −5.0 kcal/mol, forming a hydrogen bond with the A chain residue TYR124 and hydrophobic contacts with the A chain residues TYR72, LEU76, TRP286, HIS287, TYR341 TYR337, and PHE338 (Fig. 3D). The phytochemical P3 interacts with a binding affinity of −5.2 kcal/mol through forming hydrogen bonds with the A chain residue ARG247 and hydrophobic interactions with the A chain residues PRO235, VAL370, PRO410, PRO537, LEU536, LEU540, CYS409, PRO410, VAL239, VAL300, HIS405, and TRP532 (Fig. 3A). The phytochemical P4 interacts with a binding affinity of −5.2 kcal/mol forming hydrogen bonds with the A chain residues PHE 295 and ARG 296 as well as hydrophobic interactions with the A chain residues LEU76, TRP286, PHE 297, TYR 341 and PHE 338 (Fig. 3B). The phytochemical P5 interacts with a binding affinity of −5.1 kcal/mol forming hydrogen bonds with the B chain residue GLN291 and hydrophobic interactions with the B chain residues TYR72, LEU76, TRP286 and TYR341 (Fig. 3C) (Table 5.). Interactions of Acetylcholinesterase with the standard are shown in Fig. 5A. According to SwissDock prediction phytochemical P1 has binding affinity of −5.27 kcal/mol with inhibitory constant (Ki) of 137 μM, attracting cavity score of (AC) score of −44.568258 and SwissParam score of −7.3496. Phytochemical P3 has binding affinity of −5.205 kcal/mol with inhibitory constant (Ki) calculated as 152 μM, attracting cavity (AC) score of −53.131895 and SwissParam score of −6.9867. Phytochemical P4 has binding affinity of −5.079 kcal/mol with inhibitory constant (Ki) of 188 μM, attracting cavity (AC) score of −57.341387 and SwissParam score of −6.8157. Phytochemical P5 has binding affinity of −4.968 kcal/mol with inhibitory constant (Ki) of 227 μM, attracting cavity score of (AC) score of −39.488361 and SwissParam score of −6.9718 to enzyme Acetylcholinesterase (Table 6).
Fig. 3.
Phytochemical interaction with the proteins of associated neurodegenerative diseases. A- Acetylcholinesterase interaction of ethyl oleate. B- Acetylcholinesterase interaction of octadecanoic acid, ethyl ester. C- Acetylcholinesterase interaction of phytol. D- Acetylcholinesterase interaction of trans, trans-9,12-Octadecadienoic acid, propyl ester. E- γ-Secretase interaction of ethyl oleate.
Table 5.
Interactions of phytochemicals with Acetylcholinesterase, BACE-1 and γ –Secretase proteins.
| Ligand | BA (kcal/mol) | Ki (μM) | Hydrogen bonding | Hydrophobic interactions |
|---|---|---|---|---|
| Acetylcholinesterase | ||||
| P1 | −5.0 | 215 | A:TYR124, A:TYR124 | A:LEU76, A:TYR72, A:TRP286, A:TRP286, A:TRP286, A:HIS287, A:TYR337, A:PHE338, A:TYR341, A:TYR341 |
| P3 | −5.2 | 154 | A:ARG247 | A:PRO235, A:PRO235, A:PRO235, A:PRO235, A:VAL370, A:PRO410, A:PRO537, A:LEU536, A:LEU540, A:CYS409, A:PRO410, A:VAL239, A:VAL300, A:HIS405, A:HIS405, A:HIS405, A:TRP532 |
| P4 | −5.2 | 154 | A:PHE295, A:ARG296 | A:LEU76, A:TRP286, A:TRP286, A:TRP286, A:PHE297, A:PHE338, A:TYR341 |
| P5 | −5.1 | 182 | B:GLN291 | B:TRP286, B:TRP286, B:LEU76, B:TYR72, B:TRP286, B:TRP286, B:TRP286, B:TYR341, B:TYR341 |
| BACE-1 | ||||
| P2 | −5.7 | 66 | A:THR231 | A:LEU30, A:TYR71, A:TRP115, A:TRP115 |
| P4 | −5.2 | 154 | A:ASP228 | A:TYR71, A:TYR71, A:PHE108, A:TYR198 |
| P5 | −6.0 | 40 | A:GLY230 | A:TYR71, A:ILE118, A:TYR71, A:PHE108 |
| P6 | −5.2 | 154 | A:ASP32 | A:VAL332, A:TYR71, A:TYR71, A:PHE108, A:TRP115 |
| γ- Secretase | ||||
| P3 |
−6.0 |
40 |
B:TYR181 |
B:MET228, B:LEU232, B:LEU182, B:LEU182, B:LEU201, B:ILE202, B:LEU201, B:TYR181, B:TYR181, B:PHE205, B:PHE205, B:PHE205, B:PHE205, D:PHE28, D:PHE28. |
|
Standards | ||||
| Acetylcholinesterase | ||||
| SD3 | −8.9 | 0.3 | A:PHE338, A:TYR337 | A:TYR341, A:TRP286, A:TRP286, A:TYR341, A:TYR72, A:TYR72, A:TRP286 |
| BACE-1 | ||||
| SD4 | −4.2 | 984 | A:ASP32, A:GLY34 | A:TYR71 [o], A:TYR71, A:PHE108 |
| γ- Secretase | ||||
| SD5 | −7.6 | 3 | B:ASN204, B:LEU201 | B:LEU182 |
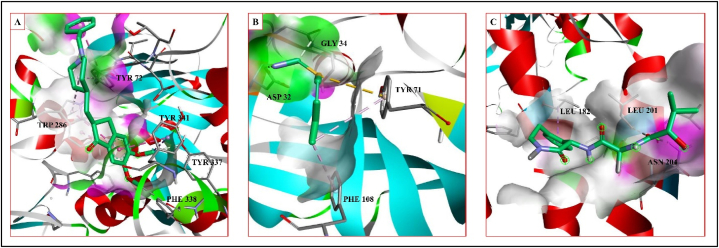
BA-binding affinity, [o]- other interaction. Donepezil –SD3, Atabecestat SD4, Semagacestat-SD5.
Fig. 5.
Interaction of standard with the proteins of associated neurodegenerative diseases. A- Acetylcholineesterase interaction of donepezil. B- BACE-1 interaction of atabecestat. C- γ-Secretase interaction of semagacestat.
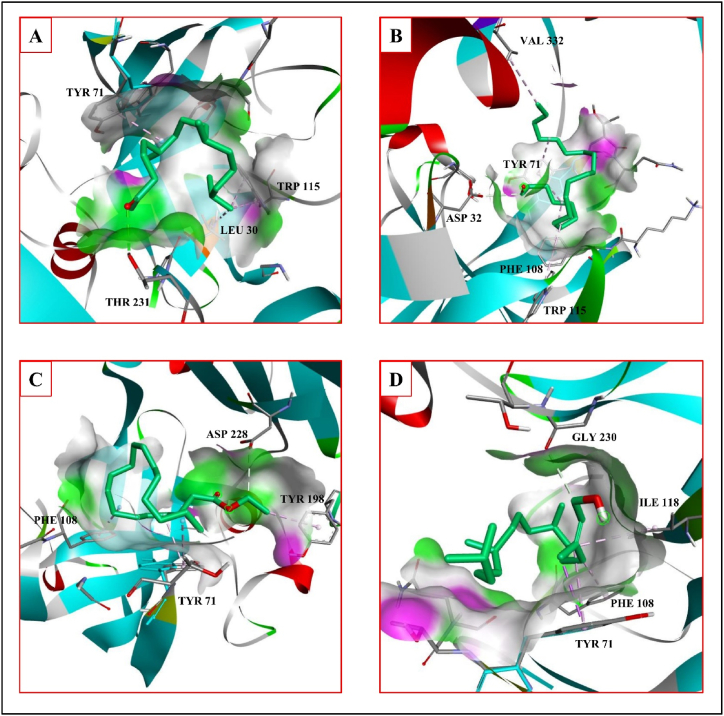
β-site amyloid precursor protein (APP) cleaving enzyme-1 (BACE-1) induces the Aβ protein generation. BACE-1 inhibitor has the ability to prevent the Aβ formation, for example, atabecestat reduces the formation of the Aβ amyloid fibrils [32]. Atabecestat form hydrogen bonds with the A chain residues ASP32 and GLY34, and hydrophobic interactions with A chain residues TYR71, PHE108 and PHE108 with a binding affinity of −4.2 kcal/mol. Phytochemical P2 interacts with BACE-1 with a binding affinity of −5.7 kcal/mol forming hydrogen bonds with the A chain residue THR231, and hydrophobic interactions with the A chain residues LEU30, TYR71 and TRP115 (Fig. 4A). The phytochemical P4 interacts with a binding affinity of −5.2 kcal/mol forming hydrogen bond with the A chain residue ASP228 and hydrophobic interactions with the A chain residues TYR71, PHE108 and TYR198 (Fig. 4C). The phytochemical P5 interacts with a binding affinity of −6.0 kcal/mol forming hydrogen bond with the A chain residue GLY230 and hydrophobic interactions with A chain residues TYR71, PHE108 and ILE118 (Fig. 4D). The phytochemical P6 form hydrogen bond with the A chain residue ASP32 and hydrophobic interactions with the A chain residues VAL332, TYR71, PHE108 and TRP115 with a binding affinity of −5.2 kcal/mol (Fig. 4B) (Table 5). The interactions of the BACE-1 with the standard are shown in Fig. 5B. According to SwissDock prediction phytochemical P2 has binding affinity of −5.338 kcal/mol with inhibitory constant (Ki) of 122 μM, attracting cavity score of AC score of −36.198006 and SwissParam score of −7.4459. Phytochemical P6 has binding affinity of −4.401 kcal/mol with inhibitory constant (Ki) of 592 μM, attracting cavity AC score of −50.277182 and SwissParam score of −6.8360. Phytochemical P4 has binding affinity of −4.285 kcal/mol with inhibitory constant (Ki) of 720 μM, attracting cavity (AC) score of −57.705446 and SwissParam score of −7.0953. Phytochemical P5 has binding affinity of −5.553 kcal/mol with inhibitory constant (Ki) of 85 μM, attracting cavity (AC) score of −35.430713 and SwissParam score of −6.7800 to the enzyme BACE-1 (Table 6).
Fig. 4.
Phytochemical interaction with the proteins associated neurodegenerative diseases. A- BACE-1 interaction of 2-Pentadecanone, 6,10,14-trimethyl- B- BACE-1 interaction of henicosanal. C- BACE-1 interaction of octadecanoic acid, ethyl ester. D- BACE-1 interaction of phytol.
γ-Secretase increases the Aβ formation by removing the trans-membrane domain of amyloid β-protein precursor (APP). Inhibitors of the γ-Secretase, for example, semagacestat, are effective in preventing the Aβ protein formation [33]. In-silico study showed that the γ –secretase inhibitor semagacestat forms hydrogen bonds with the B chain residues ASN204 and LEU201, and hydrophobic interaction with the B chain residue LEU182 with a binding affinity of −7.6 kcal/mol. The phytochemical P3 interacts with a binding affinity of −6.0 kcal/mol forming hydrogen bond with the B chain residue TYR 181 and hydrophobic interactions with the B chain residues TYR18, LEU182, LEU201, ILE202, PHE205 MET228, LEU232, and D chain PHE28 (Table 5.). The interactions of the γ-Secretase with the ligands are shown in Fig. 3F and with the standard are shown in Fig. 5C. According to SwissDock prediction the phytochemical P3 has binding affinity of −4.683 kcal/mol with inhibitory constant (Ki) of 368 μM, attracting cavity (AC) score of −32.107416 and SwissParam score of −5.6657 to the enzyme γ-Secretase (Table 6).
4. Discussion
Amyloid formation, caused by the self-assembly of amino acids, peptides, and proteins, is the root cause of neurodegenerative diseases. Although there is no perfect cure for amyloidosis, targeted drug treatment can alleviate symptoms and reduce further amyloid fibril formation. Majority of currently used drugs function indirectly to slow disease development via. specific receptor molecules/enzymes. These enzyme inhibitors, include acetylcholinesterase inhibitors, glutamate inhibitors, α-secretase modulators, and β-site amyloid precursor protein cleaving enzyme (BACE) inhibitors [34]. Phytochemicals have been effective in inhibiting or destabilizing the production of amino acid, peptide, and protein amyloids e.g scyllo-inositol is effective against the Aβ, α-synuclein and huntingtin protein amyloid formation. These phytochemicals may be useful for the treatment of amyloid diseases [35]. The treatment by using these phytochemicals has several advantages over currently used neurodegenerative medicines, due to its low cost, efficacy, low toxicity, and the capacity to diminish amyloid-related pathology.
The plant Adiantum lunulatum was used in the traditional medicine to treat various diseases like fever, asthma, cough, dermatological problems, wounds [36] and injury [37]. These medicinal effects of Adiantum lunulatum preparations are due to the presence of certain phytochemicals. Our study detected the presence of 18 different phytochemicals in Adiantum lunulatum ethanol extract. Out of these, 7 phytochemicals are important due to their less cytotoxicity (Table 2, Fig. 6). Some phytochemicals like P2 were previously identified from the Adiantum flabellulatum plant [38]. It has been reported that the phytochemical P7 from Turbinaria ornata has antioxidant activity by reducing the production of nitric oxide by decreasing the nitric oxide synthase activity and has effective anti-inflammatory activity [39]. 9, 12-Octadecadienoic acid (Z,Z)-is a conjugated isomer of the linoleic acid which reduces the Aβ production by increasing the level of β-site APP-cleaving enzyme 1 (BACE1) and APP (amyloid precursor protein) level in early endosome but the site of APP cleavage is in late endosome where the level of these protein's colocalization is reduced. This results in to the decrease in the APP cleavage and Aβ production [40]. Acute and chronic neurodegenerative disorders result into glutamate excitotoxicity caused by the N-methyl-d-aspartate (NMDA) receptor. Studies reported that at a concentration of 500 μM conjugated linoleic acid prevent the neuronal cell death by glutamate excitotoxicity [41]. The phytochemicals 2, 4 di-tert-butylphenol and Quinoline 1, 2 dihydro 2, 2, 4 trimethyl from Adiantum lunulatum have the ability to cross blood brain barrier and have no cytotoxicity [42].
Fig. 6.
Chemical structure of different phytochemicals isolated from the ethanol extract and standards used for molecular docking analysis. P1- trans,trans-9,12-Octadecadienoic acid, propyl ester, P2-2-Pentadecanone, 6,10,14-trimethyl-, P3- Ethyl Oleate, P4- Octadecanoic acid, ethyl ester, P5- Phytol, P6- Henicosanal, P7- Neophytadiene, A- Valproic acid, B- curcumin, C- Donepezil, D- Atabecestat, E- Semagacestat.
In this study we propose some phytochemicals as new probable anti-amyloid agents. Based on the lack of cytotoxicity we selected 7 phytochemicals from the ethanol extract of Adiantum lunulatum. To understand the anti-amyloid potential of these selected phytochemicals we have used computer aided molecular docking methods. The binding affinities of these phytochemicals on the neurodegenerative proteins and fibrils were compared with the standard anti-amyloid agents. Structures of these phytochemicals and standard anti-amyloid agents are given in the Fig. 6. We found that these phytochemicals from the Adiantum lunulatum have identical binding affinity (binding energy and intermolecular interactions) to the targets tau protein, prion protein and huntingtin Protein. These phytochemicals show better interaction with amyloid forming enzymes such as Acetylcholinesterase, β-site amyloid precursor protein (APP) cleaving enzyme-1 (BACE-1), γ-Secretase as that of the standard anti-amyloid agents. For example, phytochemical P1 interacts with tau protein through hydrogen bond and hydrophobic interactions. These interactions are identical to that of the standard curcumin with PHE346, LYS347, and ASP348 residues. The interaction of P2 with prion protein is identical to that of the standard valproic acid with the amino-acid residues HIS187 and LEU130. P1 shows interactions with huntingtin protein and these interactions are similar to that of the curcumin with the residues LYS5, ALA9, LEU13, and PHE16.
Acetylcholinesterase is mainly found in the neuromuscular junctions and cholinergic brain synapses. It has role in the termination of impulse transmission at cholinergic synapses by hydrolysis of neurotransmitter Acetylcholine to acetate and choline. Acetylcholinesterase inhibitors prevent the hydrolysis of Acetylcholine. Acetylcholinesterase inhibitors belong to two groups, irreversible and reversible. Loss of cholinergic neurons in the brain and decreased level of ACh is associated with the Alzheimer's disease [43]. Studies reported that the acetylcholinesterase promotes the formation of the amyloid beta peptide fragments and enhances the aggregation of the Aβ (12–28), Aβ (25–35) and Aβ 1–40 peptides [44,45]. Many pharmaceuticals have inhibitory role against the Acetylcholinesterase activity. Some of them are rivastigmine, donepezil, galantamine, huperzine A, Phenserine and Poiphen. Some examples of natural compounds with this property includes Limonoids and Mangiferin [30]. In our study P1, P3, P4, and P5 interact with Acetylcholinesterase by forming hydrogen bond and hydrophobic interactions. The amino acids of Acetylcholinesterase where these phytochemicals bind are identical to that of the standard donepezil (PHE338, TYR337, TYR341, TRP286, TYR341 and TYR72). It was reported that the active site of Acetylcholinesterase contains the amino-acids TYR72, TYR124, TRP286, TYR337, TYR341, PHE338 which are involved in the interaction with the inhibitors too [46].
β-site amyloid precursor protein (APP) cleaving enzyme-1 (BACE-1) is involved in the β-amyloid (Aβ) peptide generation. In a study using mice, targeted deletion of the BACE-1 in microgelia reduces the Aβ amyloid peptide formation. Deletion of the BACE-1 enhances the lysosomal proteases which enhances the Aβ degradation and hence protect from the Alzheimer's diseases [47]. Few examples of BACE-1 inhibitors are Verubecestat, Lanabecestat, Atabecestat, Elenbecestat, 1,3-Thiazine, 1,3-Oxazine, 1,4-Oxazine, Thiomorpholinedioxide, Thiadiazinanedioxide, Piperazine, Tetrahydropyridine-2-amines, Pyrrolidine and 1,3-Oxazoline [48]. The phytochemicals P2, P4, P5 and P6 studied here have hydrogen and hydrophobic interactions with BACE-1. These interactions are identical to that of the standard atabecestat interactions with the amino acids ASP32, TYR71 and PHE108.
γ-Secretase perform the removal of the trans-membrane domain from the amyloid β-protein precursor (APP) to form the amyloid β-protein (Aβ). Therefore inhibitors of the γ-Secretase are effective in preventing the Aβ protein formation. These inhibitors show abnormalities in gastrointestinal tract, by inhibiting the notch signaling, and also in thymus and spleen [49]. Some examples of the γ-Secretase inhibitors at pre-clinical development are AL101, Avagacestat, GSI-IX, begacestat and semagacestat [50]. Studies reported that some phytochemicals inhibit γ –Secretase, for example, rutin binds to the catalytic site of γ-Secretase and prevent its activity [51]. The phytochemicals P3 show hydrogen bond and hydrophobic interactions with γ–Secretase. These interactions with the amino-acids B: LEU182 and B: LEU201 are identical to the interaction of the standard semagacestat.
In this study, using computer aided docking, we found that six phytochemicals (P1 to P6) identified from the ethanol extract of Adiantum lunulatum have comparable affinity towards the neurodegenerative proteins and fibrils with that of the known anti-amyloid agents. Analysis of inhibitory constant (Ki) based on AutoDock Vina program binding affinity detected that the phytochemicals P1and P2 has less interaction with the tau, prion and huntingtin proteins. Inhibitory constant (Ki) shows significant interaction potential of P2 and P5 with the BACE1 enzyme. P3 shows good interaction potential with the γ-Secretase with Ki value of 40 μM. SwissDock webserver based Inhibitory constant (Ki) calculation detected that phytochemicals P1, P3, P4 and P5 have good interaction with enzyme Acetylcholinesterase. The phytochemical P2 has good Ki value with Prion protein and enzyme BACE1. Phytochemical P5 also shows good Ki value with the enzyme BACE1. Another important criteria to be satisfied by these phytochemicals for acting as an anti-amyloid agent is their Blood Brain Barrier crossing potential. Neurodegenerative diseases cause damage to the blood brain barrier which result in to an increased entry of the compounds to the brain [52,53]. Studies reported that the nano-encapsulated compounds cross the blood brain barrier in patients with neurodegenerative diseases. For example, phytol loaded poly (lactic-co-glycolic acid) nanoparticle effective in crossing the blood brain barrier and effective in treatment of Alzheimer's diseases [54]. So, the newly identified anti-amyloid phytochemicals may have the ability to cross the blood brain barrier which makes them more probable anti-amyloid agents.
5. Conclusion
Amyloid formation by the self-assembly of amino acids, peptides and proteins is the important underlying cause of neurodegenerative diseases. The search for new the anti-amyloid agents screened many compounds of synthetic and natural origin. The screening of more effective compounds with less toxicity ends up with phytochemicals. The plants belonging to Adiantum species are commonly used in folk medicine to treat different diseases. We used Adiantum lunulatum extract to detect different volatile bioactive phytochemicals with probable anti-amyloid properties. Initially, we identified these compounds from the extract using GC-MS analysis method. The identified phytochemicals belong to different classes and some of the compounds belonging to these phytochemical classes were reported to have anti-amyloid activity, for example, curcumin from curcuma spp has significant anti-amylodogenic potential. Seven phytochemicals identified from Adiantum lunulatum extract were predicted to have no cytotoxicity and some of them have anti-amyloid potential using in-silico methods. Of this some phytochemicals like P1, P2, P3, P4 and P5 have good inhibitory potential to interact with enzymes involved in the formation of amyloid β fibrils. Further confirmation of the anti-amyloid potential of these compounds by using in-vitro and in-vivo methods will lead to the discovery of new more potential anti-amyloid agents.
Funding
This work is done by using the Junior Research Fellowship to Jenat P J in the year 2022 by Mahatma Gandhi University and also by using the CSIR Junior Research Fellowship of Ann Liya Sajan and Ajmal Jalal.
Data availability statement
Data will be made available on request.
CRediT authorship contribution statement
Jenat Pazheparambil Jerom: Writing – original draft, Visualization, Software, Investigation, Formal analysis, Conceptualization. Ajmal Jalal: Visualization, Validation. Ann Liya Sajan: Validation. Reshma Soman: Visualization. Raveendran Harikumaran Nair: Supervision, Project administration. Sunilkumar Puthenpurackal Narayanan: Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We thank Mahatma Gandhi University, Kottayam for the providing the lab facility for the completion of the work.
Contributor Information
Raveendran Harikumaran Nair, Email: harinair@mgu.ac.in.
Sunilkumar Puthenpurackal Narayanan, Email: sunilkumarpn@mgu.ac.in.
References
- 1.Banerjee S., Hashemi M., Zagorski K., Lyubchenko Y.L. Interaction of Aβ42 with membranes triggers the self-assembly into oligomers. Int. J. Mol. Sci. 2020;21:1129. doi: 10.3390/ijms21031129. PMID: 32046252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hardenberg M.C., Sinnige T., Casford S., Dada S.T., Poudel C., Robinson E.A., Fuxreiter M., Kaminksi C.F., Kaminski Schierle G.S., Nollen E.A.A., Dobson C.M., Vendruscolo M. Observation of an α-synuclein liquid droplet state and its maturation into Lewy body-like assemblies. J. Mol. Cell Biol. 2021;13:282–294. doi: 10.1093/jmcb/mjaa075. PMID: 33386842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scherzinger E., Sittler A., Schweiger K., Heiser V., Lurz R., Hasenbank R., Bates G.P., Lehrach H., Wanker E.E. Self-assembly of polyglutamine-containing huntingtin fragments into amyloid-like fibrils: implications for Huntington's disease pathology. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4604–4609. doi: 10.1073/pnas.96.8.4604. PMID: 10200309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narayanan S.P., Nair D.G., Schaal D., Barbosa de Aguiar M., Wenzel S., Kremer W., Schwarzinger S., Kalbitzer H.R. Structural transitions in full-length human prion protein detected by xenon as probe and spin labeling of the N-terminal domain. Sci. Rep. 2016;6 doi: 10.1038/srep28419. PMID: 27341298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roeters S.J., Iyer A., Pletikapić G., Kogan V., Subramaniam V., Woutersen S. Evidence for intramolecular antiparallel beta-sheet structure in alpha-synuclein fibrils from a combination of two-dimensional infrared spectroscopy and atomic force microscopy. Sci. Rep. 2017;7 doi: 10.1038/srep41051. PMID: 28112214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Madboli A.E.A., Seif M.M. Adiantum capillus-veneris Linn protects female reproductive system against carbendazim toxicity in rats: immunohistochemical, histopathological, and pathophysiological studies. Environ. Sci. Pollut. Res. Int. 2021;28:19768–19782. doi: 10.1007/s11356-020-11279-w. PMID: 33405113. [DOI] [PubMed] [Google Scholar]
- 7.Ako A., Yavuz C.S., Gokhan Z. Phenolic compounds, antioxidant properties and enzyme inhibition ability of Adiantum capillus veneris L. Linked to Alzheimer's disease, diabetes mellitus and Skin disorders. Curr. Org. Chem. 2018;22:1697–1703. doi: 10.2174/1385272822666180711145256. [DOI] [Google Scholar]
- 8.Brahmachari G., Chatterjee D. Triterpenes from Adiantum lunulactum. Fitoterapia. 2002;73:363–368. doi: 10.1016/s0367-326x(02)00119-3. PMID: 12165329. [DOI] [PubMed] [Google Scholar]
- 9.Reddy V.L., Ravikanth V., Rao T.P., Diwan P.V., Venkateswarlu Y. A new triterpenoid from the fern Adiantum lunulatum and evaluation of antibacterial activity. Phytochemistry. 2001;56:173–175. doi: 10.1016/s0031-9422(00)00334-4. PMID: 11219810. [DOI] [PubMed] [Google Scholar]
- 10.Mithraja M.J., Antonisamy J.M., Mahesh M., Paul Z.M., Jeeva S. Inter-specific variation studies on the phyto-constituents of Christella and Adiantum using phytochemical methods. Asian Pac. J. Trop. Biomed. 2012;2:S40–S45. doi: 10.1016/S2221-1691(12)60127-0. [DOI] [Google Scholar]
- 11.Ali M.S., Amin M.R., Kamal C.M., Hossain M.A. In-vitro antioxidant, cytotoxic, thrombolytic activities and phytochemical evaluation of methanol extract of the A. philippense L. leaves. Asian Pac. J. Trop. Biomed. 2013;3:464–469. doi: 10.1016/S2221-1691(13)60097-0. PMID: 23730559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu F., Zhou Y., Li L., Shen X., Chen G., Wang X., Liang X., Tan M., Huang Z. Computational approaches in preclinical studies on drug discovery and development. Front. Chem. 2020;8:726. doi: 10.3389/fchem.2020.00726. PMID: 33062633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kocaadam B., Şanlier N. Curcumin, an active component of turmeric (Curcuma longa), and its effects on health. Crit. Rev. Food Sci. Nutr. 2017;57:2889–2895. doi: 10.1080/10408398.2015.1077195. PMID: 26528921. [DOI] [PubMed] [Google Scholar]
- 14.Ono K., Hasegawa K., Naiki H., Yamada M. Curcumin has potent anti-amyloidogenic effects for Alzheimer's beta-amyloid fibrils in vitro. J. Neurosci. Res. 2004;75:742–750. doi: 10.1002/jnr.20025. PMID: 14994335. [DOI] [PubMed] [Google Scholar]
- 15.Rane J.S., Bhaumik P., Panda D. Curcumin inhibits tau aggregation and disintegrates preformed tau filaments in vitro. J. Alzheimers. 2017;60:999–1014. doi: 10.3233/JAD-170351. PMID: 28984591. [DOI] [PubMed] [Google Scholar]
- 16.Pandey N., Strider J., Nolan W.C., Yan S.X., Galvin J.E. Curcumin inhibits aggregation of alpha-synuclein. Acta Neuropathol. 2008;115:479–489. doi: 10.1007/s00401-007-0332-4. PMID: 18189141. [DOI] [PubMed] [Google Scholar]
- 17.Lin C.F., Yu K.H., Jheng C.P., Chung R., Lee C.I. Curcumin reduces amyloid fibrillation of prion protein and decreases reactive oxidative stress. Pathogens. 2013;2:506–519. doi: 10.3390/pathogens2030506. PMID: 25437204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. PMID: 19499576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tian W., Chen C., Lei X., Zhao J., Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:W363–W367. doi: 10.1093/nar/gky473. PMID: 29860391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banerjee P., Eckert A.O., Schrey A.K., Preissner R. ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018;46:W257–W263. doi: 10.1093/nar/gky318. PMID: 29718510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7 doi: 10.1038/srep42717. PMID: 28256516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. PMID: 11259830. [DOI] [PubMed] [Google Scholar]
- 23.Datta A., Park J.E., Li X., Zhang H., Ho Z.S., Heese K., Lim S.K., Tam J.P., Sze S.K. Phenotyping of an in vitro model of ischemic penumbra by iTRAQ-based shotgun quantitative proteomics. J. Proteome Res. 2010;9:472–484. doi: 10.1021/pr900829h. PMID: 19916522. [DOI] [PubMed] [Google Scholar]
- 24.Bugnon M., Röhrig U.F., Goullieux M., Perez M.A.S., Daina A., Michielin O., Zoete V. SwissDock 2024: major enhancements for small-molecule docking with attracting cavities and AutoDock vina. Nucleic Acids Res. 2024;52:W324–W332. doi: 10.1093/nar/gkae300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Röhrig U.F., Goullieux M., Bugnon M., Zoete V. Attracting Cavities 2.0: improving the flexibility and robustness for small-molecule docking. J. Chem. Inf. Model. 2023;63:3925–3940. doi: 10.1021/acs.jcim.3c00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zoete V., Schuepbach T., Bovigny C., Chaskar P., Daina A., Röhrig U.F., Michielin O. Attracting cavities for docking. Replacing the rough energy landscape of the protein by a smooth attracting landscape. J. Comput. Chem. 2016;37:437–447. doi: 10.1002/jcc.24249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hussain W., Amir A., Rasool N. Computer-aided study of selective flavonoids against chikungunya virus replication using molecular docking and DFT-based approach. Struct. Chem. 2020;31:1363–1374. doi: 10.1007/s11224-020-01507-x. [DOI] [Google Scholar]
- 28.Legendre C., Casagrande F., Andrieu T., Dormont D., Clayette P. Sodium valproate does not augment Prpsc in murine neuroblastoma cells. Neurotox. Res. 2007;12:205–208. doi: 10.1007/BF03033916. PMID: 17967743. [DOI] [PubMed] [Google Scholar]
- 29.Hickey M.A., Zhu C., Medvedeva V., Lerner R.P., Patassini S., Franich N.R., Maiti P., Frautschy S.A., Zeitlin S., Levine M.S., Chesselet M.F. Improvement of neuropathology and transcriptional deficits in CAG 140 knock-in mice supports a beneficial effect of dietary curcumin in Huntington's disease. Mol. Neurodegener. 2012;7:12. doi: 10.1186/1750-1326-7-12. PMID: 22475209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walczak-Nowicka Ł.J., Herbet M. Acetylcholinesterase inhibitors in the treatment of neurodegenerative diseases and the role of acetylcholinesterase in their pathogenesis. Int. J. Mol. Sci. 2021;22:9290. doi: 10.3390/ijms22179290. PMID: 34502198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ongnok B., Khuanjing T., Chunchai T., Kerdphoo S., Jaiwongkam T., Chattipakorn N., Chattipakorn S.C. Donepezil provides neuroprotective effects against brain injury and Alzheimer's pathology under conditions of cardiac ischemia/reperfusion injury. Biochim. Biophys. Acta, Mol. Basis Dis. 2021;1867 doi: 10.1016/j.bbadis.2020.165975. PMID: 32956775. [DOI] [PubMed] [Google Scholar]
- 32.Novak G., Streffer J.R., Timmers M., Henley D., Brashear H.R., Bogert J., Russu A., Janssens L., Tesseur I., Tritsmans L., Van Nueten L., Engelborghs S. Long-term safety and tolerability of atabecestat (JNJ-54861911), an oral BACE1 inhibitor, in early Alzheimer's disease spectrum patients: a randomized, double-blind, placebo-controlled study and a two-period extension study. Alzheimer's Res. Ther. 2020;12:58. doi: 10.1186/s13195-020-00614-5. PMID: 32410694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Henley D.B., Sundell K.L., Sethuraman G., Dowsett S.A., May P.C. Safety profile of semagacestat, a gamma-secretase inhibitor: IDENTITY trial findings. Curr. Med. Res. Opin. 2014;30:2021–2032. doi: 10.1185/03007995.2014.939167. PMID: 24983746. [DOI] [PubMed] [Google Scholar]
- 34.Alhazmi H.A., Albratty M. An update on the novel and approved drugs for Alzheimer disease. Saudi Pharmaceut. J. 2022;30:1755–1764. doi: 10.1016/j.jsps.2022.10.004. PMID: 36601504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jerom J.P., Madhukumar S., Nair R.H., Narayanan S.P. Anti-amyloid potential of some phytochemicals against Aβ-peptide and α-synuclein, tau, prion, and Huntingtin protein. Drug Discov. Today. 2023;28 doi: 10.1016/j.drudis.2023.103802. PMID: 37858630. [DOI] [PubMed] [Google Scholar]
- 36.Singh H., Husain T., Agnihotri P., Pande P.C., Khatoon S. An ethnobotanical study of medicinal plants used in sacred groves of Kumaon Himalaya, Uttarakhand, India. J. Ethnopharmacol. 2014;154:98–108. doi: 10.1016/j.jep.2014.03.026. PMID: 24685588. [DOI] [PubMed] [Google Scholar]
- 37.Zheng X.L., Xing F.W. Ethnobotanical study on medicinal plants around Mt.Yinggeling, Hainan Island, China. J. Ethnopharmacol. 2009;124:197–210. doi: 10.1016/j.jep.2009.04.042. PMID: 19409476. [DOI] [PubMed] [Google Scholar]
- 38.Kang W.Y., Ji Z.Q., Wang J.M. Composition of the essential oil of Adiantum flabellulatum. Chem. Nat. Compd. 2009;45:575–577. doi: 10.1007/s10600-009-9371-5. [DOI] [Google Scholar]
- 39.Bhardwaj M., Sali V.K., Mani S., Vasanthi H.R. Neophytadiene from Turbinaria ornata suppresses LPS-induced inflammatory response in RAW 264.7 macrophages and sprague dawley rats. Inflammation. 2020;43:937–950. doi: 10.1007/s10753-020-01179-z. PMID: 31981060. [DOI] [PubMed] [Google Scholar]
- 40.Hata S., Kano K., Kikuchi K., Kinoshita S., Sobu Y., Saito H., Saito T., Saido T.C., Sano Y., Taru H., Aoki J., Komano H., Tomita T., Natori S., Suzuki T. Suppression of amyloid-β secretion from neurons by cis-9, trans-11-octadecadienoic acid, an isomer of conjugated linoleic acid. J. Neurochem. 2021;159:603–617. doi: 10.1111/jnc.15490.PMID:34379812. [DOI] [PubMed] [Google Scholar]
- 41.Joo N.E., Park C.S. Inhibition of excitotoxicity in cultured rat cortical neurons by a mixture of conjugated linoleic acid isomers. Pharmacol. Res. 2003;47:305–310. doi: 10.1016/s1043-6618(03)00008-2. PMID: 12644387. [DOI] [PubMed] [Google Scholar]
- 42.Jerom J.P., Nair R.H., Sajan A.L., Manirajan B.A., Mohammed S. GC-MS Screening of Adiantum lunulatum Burm. F Phytochemicals and interaction with COX-2, TRPV1, and TRPC3 proteins-bioinformatics approach. Curr. Bioact. Compd. 2023;19 doi: 10.2174/1573407218666220901114151. [DOI] [Google Scholar]
- 43.Colović M.B., Krstić D.Z., Lazarević-Pašti T.D., Bondžić A.M., Vasić V.M. Acetylcholinesterase inhibitors: pharmacology and toxicology. Curr. Neuropharmacol. 2013;11:315–335. doi: 10.2174/1570159X11311030006. PMID: 24179466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alvarez A., Opazo C., Alarcón R., Garrido J., Inestrosa N.C. Acetylcholinesterase promotes the aggregation of amyloid-beta-peptide fragments by forming a complex with the growing fibrils. J. Mol. Biol. 1997;272:348–361. doi: 10.1006/jmbi.1997.1245.PMID:9325095. [DOI] [PubMed] [Google Scholar]
- 45.Inestrosa N.C., Alvarez A., Pérez C.A., Moreno R.D., Vicente M., Linker C., Casanueva O.I., Soto C., Garrido J. Acetylcholinesterase accelerates assembly of amyloid-beta-peptides into Alzheimer's fibrils: possible role of the peripheral site of the enzyme. Neuron. 1996;16:881–891. doi: 10.1016/s0896-6273(00)80108-7.PMID:8608006. [DOI] [PubMed] [Google Scholar]
- 46.Son M., Park C., Rampogu S., Zeb A., Lee K.W. Discovery of novel acetylcholinesterase inhibitors as potential candidates for the treatment of Alzheimer's disease. Int. J. Mol. Sci. 2019;20:1000. doi: 10.3390/ijms20041000. PMID: 30823604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh N., Das B., Zhou J., Hu X., Yan R. Targeted BACE-1 inhibition in microglia enhances amyloid clearance and improved cognitive performance. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abo3610.PMID:35857844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsiao C.C., Rombouts F., Gijsen H.J.M. New evolutions in the BACE1 inhibitor field from 2014 to 2018. Bioorg. Med. Chem. Lett. 2019;29:761–777. doi: 10.1016/j.bmcl.2018.12.049. PMID: 30709653. [DOI] [PubMed] [Google Scholar]
- 49.Barten D.M., Meredith J.E.Jr., Zaczek R., Houston J.G., Albright C.F. Gamma-secretase inhibitors for Alzheimer's disease: balancing efficacy and toxicity. Drugs R. 2006;7:87–97. doi: 10.2165/00126839-200607020-00003. PMID: 16542055. [DOI] [PubMed] [Google Scholar]
- 50.McCaw T.R., Inga E., Chen H., Jaskula-Sztul R., Dudeja V., Bibb J.A., Ren B., Rose J.B. Gamma secretase inhibitors in cancer: a current perspective on clinical performance. Oncol. 2021;26:e608–e621. doi: 10.1002/onco.13627. PMID: 33284507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh A.K., Shuaib M., Prajapati K.S., Kumar S. Rutin potentially binds the gamma secretase catalytic site, down regulates the notch signaling pathway and reduces sphere formation in colonospheres. Metabolites. 2022;12:926. doi: 10.3390/metabo12100926. PMID: 36295828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carvey P.M., Hendey B., Monahan A.J. The blood-brain barrier in neurodegenerative disease: a rhetorical perspective. J. Neurochem. 2009;111:291–314. doi: 10.1111/j.1471-4159.2009.06319.x. PMID: 19659460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sweeney M.D., Sagare A.P., Zlokovic B.V. Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 2018;14:133–150. doi: 10.1038/nrneurol.2017.188.PMID:29377008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sathya S., Shanmuganathan B., Saranya S., Vaidevi S., Ruckmani K., Pandima Devi K. Phytol-loaded PLGA nanoparticle as a modulator of Alzheimer's toxic Aβ peptide aggregation and fibrillation associated with impaired neuronal cell function. Artif. Cells, Nanomed. Biotechnol. 2018;46:1719–1730. doi: 10.1080/21691401.2017.1391822. PMID: 29069924. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.