Abstract
Background
One of the most prevalent cancers worldwide is HCC, which has put patient health at risk. Increasing evidence indicated that messenger RNAs (mRNAs) played significant roles in modulating tumorigenesis. It has been established that Gli1 acts as an oncogene in a number of malignancies. However, more research was necessary to understand the Gli1 regulation mechanism in HCC.
Methods
Microarray technology was used to evaluate the expression of mRNAs. RT-qPCR was utilized to evaluate Gli1 and miR-875-5p expression. To investigate the role of Gli1, tests using CCK-8, EdU, transwell, immunofluorescence, and Western blot analysis was performed. RIP, RNA pull down, and luciferase reporter assays were employed to verify the interaction between Gli1 and miR-875-5p.
Results
In tissues and cells of HCC, Gli1 expression appeared to be upregulated, especially in metastatic samples and advanced stages of the disease. A worse outcome was predicted by elevated Gli1 expression. Additionally, in HCC, Gli1 inhibition impeded the growth, migration, and development of the EMT. Since miR-875-5p was shown to have a molecular target in Gli1, miR-875-5p mediated the negative regulation of Gli1. In HCC tissues, its expression pattern was less prominent. In HCC tissues, there was an inverse relationship between Gli1 expression and miR-875-5p expression. Overexpressing Gli1 helped to partially counteract the suppression of HCC migration, proliferation, and EMT formation by miR-875-5p overexpression.
Conclusions
MiR-875-5p in HCC suppresses tumors by downregulating Gli1, which supplies a novel treatment for HCC patients.
Keywords: miR-875-5p, Gli1, HCC, Hedgehog signaling pathway
1. Introduction
Liver cancer is ranked among the top ten malignant tumors by the World Health Organization [1]. Notably, in countries in Asia and Africa, the most frequent kind of the liver cancer and the main drive of death from cancer is HCC. Surgical treatment and chemotherapy are widely applied to treat HCC patients [2,3], but their effects were considerably discouraged due to high recurrences after treatment [4]. Consequently, in order to improve the prognosis for HCC, it is imperative to look into HCC's regulatory framework.
Messenger RNA (mRNA) is a kind of RNA molecule that has been testified to profoundly participate in the development of diverse cancers [[5], [6], [7]]. Glioma-associated oncogene 1 (Gli1) was reported to promote tumor malignant behaviors. For instance, Gli1 modulates MGMT expression in human glioblastoma [8]. Gli1 has been demonstrated to function as a promoter in a number of cancers, including HCC, yet the specific method by which it regulates HCC remains mostly unclear.
MiRNAs belong to the non-coding RNA family with 20–24 nucleotides [9]. They are increasingly reported to regulate the progression of tumorigenesis by targeting and down-regulating certain disease-related mRNAs [[10], [11], [12]]. For instance, miR-1253 dampens the progression of prostate cancer via targeting EZH2 to deplete its expression [13].MiR-143 inhibits HMGB1 expression, which in turn controls the growth of bladder cancer [14]According to recent research, miR-875-5p functions as SATB2's lung cancer sponge [15]. Additionally, it has been demonstrated that miR-875-5p functions as an inhibitor of tumor growth in the progression of colorectal cancer (CRC) [16]. However, it is unknown how miR-875-5p of HCC functions biologically.
This study set out to look into how the miR-875-5p/Gli1 axis functions in HCC. Our investigation revealed that miR-875-5p effectively targets Gli1 and thereby influences the progression of HCC.
2. Materials and methods
2.1. Tissue samples
At Central Hospital Affiliated to Jiangnan University, 67 patients with HCC provided samples of their disease, as well as 67 samples of nearby noncancerous tissues. All of the collected specimens were kept in storage at −80 °C and frozen in liquid nitrogen right away. Before the procedure, none of these patients had undergone any additional anticancer therapy. Patients signed informed consent forms before to surgery, and the Central Hospital Affiliated to Jiangnan University's Ethics Committee approved this investigation (No. 20200027).
2.2. Cell transfection and cell lines
The Chinese Academy of Science Cell Bank contributed the HCC cell lines and the human normal liver cells L02, which included HCCLM3 SMMC-7721 HepG2 and MHCC97H. All cells were cultured with 10 % fetal bovine serum (FBS) in DMEM medium (Gibbon, USA). The cell line was cultured at 37 °C in a humid atmosphere with 5 % CO2. MiR-875-5p mimics were obtained from Life Technologies (Shanghai, China) and utilized to overexpress miR-875-5p. Additionally, using sh-NC as the negative control, the Gli1 expression was knocked down using the sh-Gli1, sh-Gli1#2, and sh-Gli1#3 vectors. To create pcDNA3.1/Gli1, the entire Gli1 gene was cloned into a pcDNA3.1 vector, overexpressing Gli1 expression while leaving empty pcDNA3.1 as the control. Sangon Inc. (Shanghai, China) supplied the plasmids of sh-Gli1#1, sh-Gli1#2, and sh-Gli1#3, while Sigma Aldrich (Shanghai, China) supplied the pcDNA3.1 plasmids. With the use of Lipofectamine 2000 (Invitrogen, Carlsbad, CA), these vectors were all used to transfect cells.
2.3. Analysis of KEGG pathways and microarrays
Following the manufacturer's recommendations, all of the RNA was taken out from the patient's tissue with HCC, and a RNeasy mini tool (QIAGEN, Germany) was utilized for cleaning. The RNA clarity was measured in great detail using a Qubit Fluorometer (Thermo Fisher Scientific, USA). The analysis revealed the top 200 mRNA expression of HCC. The manufacturer's usual technique was followed to handle the raw data, and KEGG pathway analysis was then used for additional analysis.
2.4. RT-qPCR
Following the directions provided by the maker, Trizol reagent (Invitrogen) was used to extract all of the RNA from the tissues and cells. In order to do reverse transcription, TaqMan® miRNA transformation tool (Applied Biosystems, Foster City, CA, USA) and First Strand Synthesis kit (Invitrogen, Carlsbad, CA, USA) were used to create first strand cDNA. Using gene-specific primers and SYBR® Green qPCR MasterMix (Applied Biosystems, Foster City, CA, USA) in ABI 7500 real-time PCR Systems, the expression of genes was assessed via RT-qPCR. The target gene expression was established using the 2−ΔΔCt technique. As the endogenous normalizers, GAPDH and U6 were employed. The primers were:
Gli1: forward, 5′-TCT GCC CCC ATT GCC CAC TTG-3′and reverse, 5′- TAC ATA GCC CCC AGC CCA TAC CTC-3′;
MiR-875-5p: forward, 5′-CGA ATG GGC CTA AGA TCC CG-3′ and reverse, 5′-GGA GCC CAG CAC TTT GAT CT-3′;
GAPDH: forward, 5′-GAA GGT GAA GGT CGG AGT C-3′ and reverse, 5′-GAA GAT GGT GAT GGG ATT TC-3′;
U6: forward, 5′-ATT GGA ACG ATA CAG AGA AGA TT-3′ and reverse, 5′-GGA ACG CTT CAC GAA TTT G-3′.
2.5. CCK-8 assay
The Cell-Counting Kit 8 (Dojingdo Molecular Technologies, Rockville, Japan) was employed to measure the reproduction of cells. Cell growth was examined 0, 24, 48, 72, and 96 h after transfection. The cells were then cultured for a further 4 h using 10 μl of CCK-8 solution as opposed to the initial medium. Ultimately, a 450 nm wave length was employed in a Bio-Tek, Winooski, Vermont microplate reader to assess the cell growth.
2.6. EdU assay
To evaluate the cell growth, EdU was used. Initially, the cells were cultivated in an EdU mixture for 2 h. After that, the cells were fixed with PBS containing 4 % paraformaldehyde. Have been kept in 70 % ethanol, the prepared cells were colorized employing the Cell-LightTM EdU Apollo®488 In Vitro Imaging Kit (RioBio, China). Fluorescence microscopy was used to monitor the cell development.
2.7. Transwell assay
Serum-free medium was mixed with transfected cells, and the number of 1 × 104 cells was observed in every well within the upper chamber. Concurrently, 10 % FBS-containing DMEM was added to the bottom chamber. For 48 h, cells were grown at 37 °C in an environment with 5 % CO2 and humidity. Then, 4 % alcohol with 0.5 % purple crystals (Sigma-Aldrich, St. Louis, MO, USA), respectively, were employed for coloring and fix cells. An illuminated field microscope (Olympus CKX41, Tokyo, Japan) was utilized to demonstrate the results. A random selection of fields was made in the bottom chamber, and the number of migrating cells in each field was tallied.
2.8. Western blot
The cells were lysed using RIPA buffer, which was supplemented using blockers of polymerase and enzyme (Roche, Shanghai, China). 10 % electrophoresis on a polyacrylamide gel was used to extract protein molecules from the cells. Proteins were placed onto a PVDF membrane following electrophoresis. After coating the membrane with 5 % nonfat milk, primary antibodies were added and it was left to incubate for one night at 4 °C. After that, it was incubated with secondary antibodies at ambient temperatures in the proper manner. Bands of proteins were located using the Thermo Fisher Scientific, Rochester, New York, ECL chemiluminescent Detection System.
2.9. Immunofluorescence assay
After being fixed in 4 % paraformaldehyde for 30 min, HepG2 and SMMC-7721 cells were dissolved in 0.5 % Triton X-100 solution. The serum from goats was used to block cells. Furthermore, the SMMC-7721 and HepG2 cells were treated with secondary antibodies for 1 h at 37 °C after an overnight incubation at 4 °C with the suitable primary antagonists. Lastly, DAPI (Genview Inc., Shanghai, China) was used to stain the SMMC-7721 and HepG2 carcinoma cells, which were then examined under a fluorescence microscope.
2.10. Luciferase reporter assay
Gli1-Mut and Gli1-WT were transfected with pGL3 vectors (Promega, Madison, USA) to achieve the desired result. The synthesised vectors were transmitted together with the miR-875-5p and NC mimics for 48 h in the SMMC-7721 and HepG2 carcinoma cells. With the aid of a luminescence projector test mix (Promega, Madison, WI), the associated luminescence levels were investigated.
2.11. RNA pull down assay
miR-875-5p-Wt and miR-875-5p-Mut were transcribed using the Transcript Aid T7 High Yield Extraction Toolkit from ThermoFisher Scientific, USA. Using lysate from cells (SMMC-7721 and HepG2), biotinylated miR-875-5p-Wt, miR-875-5p-Mut, and negative control (Bio-NC) were co-cultivated for 1 h at 4 °C. After purification, RNAs were eluted using elution buffer, and RT-qPCR was used to measure the production of Gli1.
2.12. RIP
The Magna RIP toolkit (Millipore, Billerica, USA) was used to assess the capacity of miR-875-5p to bind with Gli1. Subsequent to the storage of cell lysate in RIP buffer, Ago2 or IgG (negative control) antibody-bearing magnetic beads were added. The beneficial condition was supplied. RT-qPCR was used to determine the comparative levels of Gli1 and miR-875-5p.
2.13. Animal experiment
We bought six-week-old male naked mice from Beijing. China's National Laboratory Animal Center and kept in the animal laboratory using SPF-grade housing. All animal experiments acquired the approval from the Zhengzhou University's Affiliated Cancer Hospital's Animal Research Ethics Committee (Henan Cancer Hospital). In vivo experiment was performed by subcutaneous injection of SMMC-7721 cells into naked mice. Every four days, the malignancy size was determined. Xenograft tumors were carefully excised 28 days after inoculation for weighed measurement after mice were sacrificed.
2.14. Analytical statistics
The SPSS 16.0 software program was used to examine the statistics (SPSS, Chicago, IL). All information was given as a mean ± SD and all the previously described experiments were run in triplicate. One-way ANOVA or the student's t-test were used to assess the distinctions between multiple categories. Making use of the correlation of Spear study, the associations among target genes were investigated. P values were regarded as statistically significant if they were less than 0.05.
3. Results
3.1. In the tissues and cells of hepatocellular carcinoma, Gli1 is extensively expressed
To identify mRNAs involved in HCC, total RNAs were examined with RNA microarray (Arraystar, Rockville, MD), and we found that there were 200 up-regulated mRNAs, as displayed in Fig. 1A. By KEGG pathway analysis, we noticed that Hedgehog pathway had higher enrichment fraction (Fig. 1B). Next, we examined the amounts of shh, Gli1, SOX2, and OCT4 mRNA expression in a single paired HCC tissue. In contrast to corresponding normal liver tissue, the test results showed that one HCC tissue had higher amounts of shh, Gli1, SOX2, c-Myc, Cyclin D1, Cyclin D2, Nanog, and OCT4 mRNA and protein., and Gli1 was most remarkably up-regulated among them (Fig. 1C–D). When comparing HCC tissues to normal human liver tissues, we clearly saw an increase in Gli1 expression (Fig. 1E). Furthermore, we observed an apparent increase in Gli1 expression in advanced stage HCC patients compared to I-II stages (Fig. 1F). In comparison to non-metastatic tissues, metastasis tissues had increased levels of Gli1 expression (Fig. 1G). Furthermore, patients with high Gli1 levels in HCC had a worse prognosis than those with low Gli1 levels, per Kaplan-Meier analysis (Fig. 1H). Fig. 1I, which presents the results of the RT-qPCR assay, indicates that compared to normal liver cells (LO2), cells with HCC (HepG2, SMMC-7721, HCCLM3, MHCC97H) exhibited higher levels of Gli1. According to the aforementioned research, In HCC cell lines and tissues, there is a discernible increased levels of Gli1 phrase, and a negative prognosis is associated with high Gli1 expression.
Fig. 1.
Hepatocellular carcinoma tissues and cells have high expression levels of Gli1, which triggers the Hedgehog signaling pathway.
(A) The microarray was performed to identify mRNAs involved in HCC. (B) KEGG pathway analysis was used to calculate the protein enrichment fraction. (C–D) The mRNA and protein expression levels of shh, Gli1, SOX2, c-Myc, Cyclin D1, Cyclin D2, Nanog, and OCT4 in a single paired HCC tissue were evaluated by RT-qPCR and Western blot. (E–F) Gli1 expression was used by RT-qPCR in the tissue from the liver from both normal and HCC patients, as well as at various tumor lymph node metastasis (TNM) stages. (G) Gli1 expression was found in both metastatic and non-metastasis tissues using RT-qPCR. (H) Using a Kaplan-Meier analysis, the association between Gli1 expression and survival rate was examined. (I) Both normal and HCC liver cells' Gli1 expression was monitored using RT-qPCR. ∗∗P < 0.01 and ∗P < 0.05.
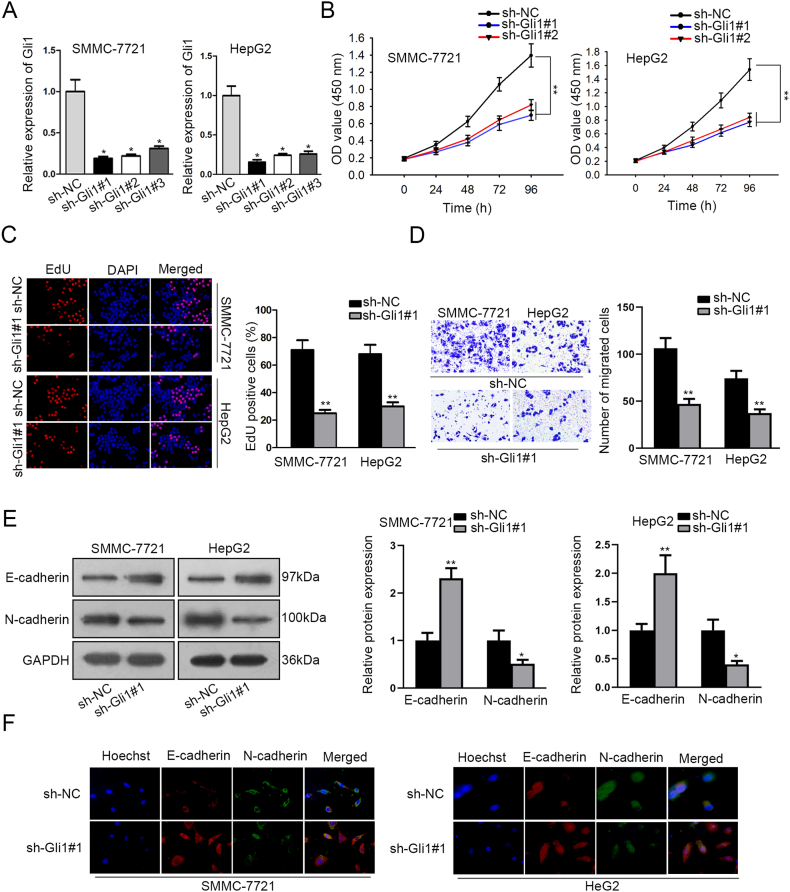
3.2. In HCC cells, Gli1 knockdown inhibits the process of proliferation, migration, and EMT
By transfecting sh-Gli1#1, sh-Gli1#2, and sh-Gli1#3 and using sh-NC as a negative control, Gli1 was knocked down in order to examine its possible biological function in HCC carcinogenesis. Using the RT-qPCR method, the knockdown efficiency of sh-Gli1#1/2/3 in the HepG2 and SMMC-7721 cells was evaluated. When compared to the negative control, SMMC-7721 and HepG2 cells that were implanted using sh-Gli1#1/2/3 revealed a discernible decrease in Gli1 expression, as shown in Fig. 2A. Therefore, for the subsequent loss-of-function tests, sh-Gli1#1 and sh-Gli1#2 were used. As depicted in Fig. 2B, Gli1 deficiency attenuated cell vitality. The EdU assay also showed that fewer EdU-positive cells were present after Gli1 knockdown (Fig. 2C). Additionally, the transwell assay verified that Gli1 silencing reduced the quantity of migrating HCC cells (Fig. 2D). To ascertain if Gli1 aided in the development of EMT, the Western blot test was employed. The findings demonstrated that in SMMC-7721 and HepG2 cells, Gli1 deletion increased the expression of E-cadherin while decreasing the expression of N-cadherin (Fig. 2E). According to the immunofluorescence assay results, suppression of Gli1 lowered N-cadherin expression while increasing E-expression cadherin (Fig. 2F). Also, our findings suggested that Gli1 downregulation inhibits HCC cell motility, proliferation, and EMT.
Fig. 2.
In HCC cells, Gli1 knockdown restricts the process of proliferation, EMT and migration
(A)The efficiency of sh-Gli1#1/2/3 and Gli1 expression in SMMC-7721 and HepG2 cells transfected with sh-Gli1#1 or sh-Gli1#2 was assessed using RT-qPCR. (B–C) The CCK-8 and EdU assays were used to measure the proliferation of cells. (D) The transwell assay was used to determine movement of cells. (E–F) Western blot and immunofluorescence assays were utilized to evaluate the expression of proteins associated with EMT. ∗∗P < 0.01 and ∗P < 0.05.
3.3. In HCC cells, the target gene for miR-875-5p is Gli1
Next, we looked into Gli1's regulatory mechanism in HCC. Numerous investigations suggested that miRNAs could alter the expression of target genes [17,18], so we hypothesize that Gli1 might induce cellular effects by modulation of downstream target inside of HCCs. Http://www.targetscan.org prediction results indicated the presence of Gli1 locations for binding in miR-875-5p. (Fig. 3A). MiR-875-5p was chosen because in various malignancies, it has been reported to block the invasion and EMT process [15,19]. Using the following tests, the link and correlation between Gli1 and miR-875-5p were verified. Using RT-qPCR to detect the overexpression efficiency of miR-875-5p (Fig. 3B). The luciferase experiment's outcomes demonstrated that the transfected HepG2 carcinoma cells with miR-875-5p and SMMC-7721 copies had significantly lower pGL3-Gli1-WT luciferase activity. On the other hand, luciferase activity in the pGL3-Gli1-Mut groups did not significantly change (Fig. 3C), indicating that Gli1 could connect with miR-875-5p. Additionally, the Bio-miR-875-5p-WT group clearly had higher Gli1 expression based on the RNA pulldown test (Fig. 3D). Moreover, the RIP experiment showed that Gli1 and miR-875-5p may co-immunoprecipitate with the Ago2 antibody instead of the IgG antibody (Fig. 3E). Using multiplex RT-qPCR analysis and the Western blot test, to further support miR-875-5p′s regulatory role on Gli1, the impacts of miR-875-5p expression of Gli1 mRNA and expression of proteins were examined. Its findings demonstrated that transfecting SMMC-7721 and HepG2 cells using miR-875-5p imitators reduced Gli1's protein and mRNA (Fig. 3F). Furthermore, we also validated the negative regulation of miR-875-5p on Gli1 based on additional analyses. Using RT-qPCR, we discovered that the expression of miR-875-5p is significantly lower in HCC tissues compared to normal liver tissues (Fig. 3G). And, through Spearman's correlation analysis and Kaplan-Meier analysis, we found that miR-875-5p is negatively correlated with Gli1, and the survival rate in the low miR-875-5p level group is significantly lower than that in the high-level group (Fig. 3H–I). MiR-875-5p mimics were incorporated into HepG2 and SMMC-7721 cells. Examination of Western blot revealed that the levels of protein expression for Gli1, SOX2, c-Myc, Cyclin D1, Cyclin D2, and OCT4 were much lower in cells that had miR-875-5p mimics transfected (Fig. 3J). Next, we found that Gli1 knockdown had no obvious effect on miR-875-5p expression (Fig. S1A). To learn more about the role that miR-875-5p plays in HCC, we employed gain-of-function experiments. Operational studies showed that dramatically decreased by miR-875-5p overexpression cells' capacity to proliferate (Figs. S1B–C) and move (Fig. S1D). The suppression of EMT progression by overexpressing miR-875-5p was further verified by Western blot results, which showed a reduction in N-cadherin and an elevation in E-cadherin levels (Fig. S1E). In conclusion, these results demonstrated that the miR-875-5p can target and interact with Gli1, as well as having a detrimental effect on Gli1 expression in HCC.
Fig. 3.
In HCC cells, miR-875-5p target gene is Gli1.
(A) To find purportedly binding sites between miR-875-5p and Gli1, Targetscan was employed. (B) RT-qPCR was used to detect the overexpression efficiency of miR-875-5p. (C–E) Luciferase reporter assay, RNA pull down and RIP confirmed the interaction between Gli1 and miR-875-5p. (F) RT-qPCR and Western blot were used to assess Gli1 mRNA and protein expression following the overexpression of miR-875-5p in certain HCC cells. (G) RT-qPCR was used to detect the expression level of miR-875-5p in normal liver tissues and HCC tissues. (H) The Spearman's correlation analysis of Gli1 and miR-875-5p. (I) Kaplan-Meier analysis demonstrated the correlation among miR-875-5p expression and survival time. (J) Gli1, SOX2, c-Myc, Cyclin D1, Cyclin D2, and OCT4 protein expression in SMMC-7721 and HepG2 cells implanted with either NC or miR-875-5p mimics was evaluated by Western blot. ∗∗P < 0.01 and ∗P < 0.05.
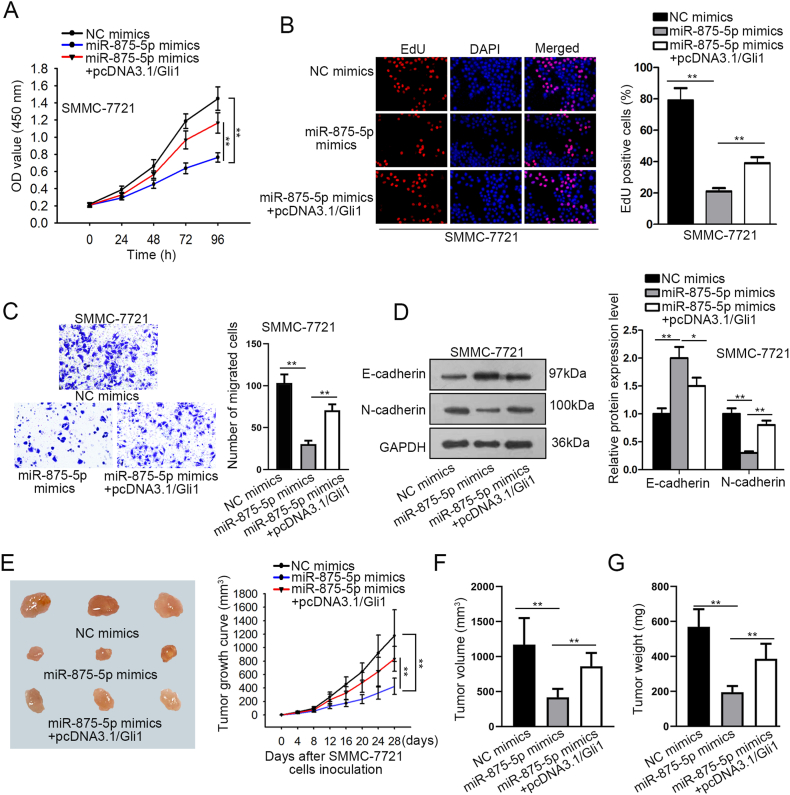
3.4. Gli1 overexpression reduces the restraint effect of miR-875-5p overexpression on HCC migration, proliferation, the EMT process
To illustrate how miR-875-5p suppresses tumor growth in the HCC cells by controlling Gli1, CCK-8, and EdU assays were employed to measure proliferation potential. The results of the experiment verified that HCC cell growth is inhibited in cells that had miR-875-5p mimics transfected. However, the detrimental impact of the miR-875-5p mimics on HCC cell increase was reversed using co-transfection of pcDNA3.1/Gli1 (Fig. 4A–B). Additionally, the overexpression of Gli1 enhanced the movement capacity of cells in HCC, which was previously suppressed via miR-875-5p overexpression (Fig. 4C). Gli1 up-regulation offset the miR-875-5p inhibitory effects overexpression on EMT progression, as Western blot assay showed (Fig. 4D). According to every lab result mentioned above, the negative effects of miR-875-5p overexpression on cell in HCC proliferation, migration, and EMT development are offset by Gli1 overexpression. We ran in vivo tests to support this conclusion even more. We found that the miR-875-5p mimics could slow down tumor growth and shrink tumor volume and weight, but these effects were counteracted by overexpressing Gli1 (Fig. 4E–G). These findings provided strong evidence for the miR-875-5p/Gli1 regulatory axis in impacting HCC development.
Fig. 4.
The suppression of miR-875-5p on HCC proliferation, EMT and migration process is rescued by Gli1.
(A–B) Tests for proliferation of cells were conducted using CCK-8 and EdU assays. (C) The analysis of cell migration was done using the transwell assay. (D) To examine the proteins connected to the EMT process, a Western blot was done. (E–G). Subcutaneous injection of SMMC-7721 cells into nude mice was used for in vivo investigations. Comparisons and analyses were conducted between the growth curve, volume of xenograft tumors, and weight for NC mimics, miR-875-5p mimics, and miR-875-5p mimics + pcDNA3.1/Gli1 group. ∗∗P < 0.01 and ∗P < 0.05.
4. Discussion
High incidence and a dismal prognosis are the hallmarks of HCC, an etiology that contributes significantly to global mortality [1]. Prevenient investigations of molecular mechanism in cancers have shed some light on the target-directed treatment strategy [[20], [21], [22]]. Nevertheless, current study on molecular performance is just a tip of iceberg in the anti-cancer research field of HCC. Messenger RNA has been proved could be translated into protein to affect the tumorigenesis and progression in a variety of cancers. Experiment-based mechanical exploration has uncovered that mRNA could be extensively regulated by upstream miRNAs, and even lncRNAs. For example, MTDH found that lncRNA SNHG1 up-regulated a miR-145-5p target into hasten the proliferation of lung cancer mobile that are not small cell [6]. The effect of repression of miR-375 on YAP1 was offset by lncRNA MLK7-AS1, thus promoting ovarian cancer development [5]. As a member of Gli family, Gli1 functions as a transcription factor to exert its impacts on Hedgehog pathway [23,24]. Accumulating reports revealed that Gli1 elicits its vital role on several cancers, including colorectal, breast, and stomach cancers [[25], [26], [27]]. Moreover, Gli1 was reported to accelerate colorectal cancer metastasis by activating EMT [28]. Nevertheless, whether Gli1 regulate EMT progression in HCC remains elusive. In our research, the expression of Gli1 is markedly high in tumor sample, indicating its association with metastasis in HCC. It was well-documented that EMT progression is the hallmark of metastasis [29]. EMT reactivation during cancer progression could aggravate the metastatic phenotype. E-cadherin is the marker of epithelial phenotype, while N-cadherin is the marker of mesenchymal phenotype. We further find that Gli1 knockdown inhibits the progression of EMT by regulating the levels of the E− and N-cadherin proteins. As GLi1 is the crucial protein associated with hedgehog signaling pathway, these findings revealed that hedgehog signaling pathway was involved in EMT progression. This result is consistent with the hedgehog signaling pathway's promotion of EMT progression in cervical cancer [30]. Moreover, knockdown of Gli1 inhibits HCC proliferation ability. This repressing effect of Gli1 knockdown on proliferation accorded with the oncogenic properties of Gli1 in gastric cancer [31].
MiRNAs have been shown to control gene expression, which in turn affects a number of cellular processes in cancer, including migration, apoptosis, and proliferation. For example, the hepatocellular carcinoma cells' ability to proliferate is regulated by the LINC01287-87/miR-298/STAT3 axis [32]. MiR-486-5p exert tumor suppressor role via negatively regulating Sirt1 in suppressing liver cancer cell proliferation [33]. Of note, miR-875-5p was reported to suppress the development of colorectal carcinoma [19]. Nevertheless, It's unclear how miR-875-5p functions in HCC. In our exploration, according to the prediction of Targetscan, miR-875-5p might harbor binding sites for Gli1. We then showed that miR-875-5p adversely impacted Gli1 additionally that it might interact with miR-875-5p. In rescue experiments, Gli1 amplification undid the suppressive effects of miR-875-5p on proliferation., migration, and the EMT process.
Together, this study shows that the miR-875-5p targets Gli1 expression and adversely modifies it to reduce HCC carcinogenesis and therefore deactivate the hedgehog signaling chain. This study provided compelling support for the potential target value of miR-875-5p for future HCC therapeutic approaches. However, the clinical significance of miR-875-5p still needs to be further validated by more clinical trials. Besides, we didn't probe into the potential upstream regulatory molecules of miR-875-5p/Gli1 in our research, which is another deficiency of our research. In our upcoming investigation, we will investigate the putative upstream regulatory molecules of miR-875-5p/Gli1.
Ethics statement
This research approved by Central Hospital Affiliated to Jiangnan University's Ethics Committee (No. 20200027).
Data availability statement
Research data can be obtained from the corresponding author if necessary.
CRediT authorship contribution statement
Qi Zhang: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Investigation, Formal analysis, Data curation. Miao Yu: Writing – review & editing, Writing – original draft, Visualization, Supervision, Investigation. Leilei Yang: Writing – review & editing, Writing – original draft, Validation, Supervision, Formal analysis. Defeng Sun: Writing – review & editing, Writing – original draft, Project administration, Conceptualization.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e37771.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
figs1.
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA A Cancer J. Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Hartke J., Johnson M., Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin. Diagn. Pathol. 2017;34(2):153–159. doi: 10.1053/j.semdp.2016.12.011. [DOI] [PubMed] [Google Scholar]
- 3.Grandhi M.S., Kim A.K., Ronnekleiv-Kelly S.M., Kamel I.R., Ghasebeh M.A., Pawlik T.M. Hepatocellular carcinoma: from diagnosis to treatment. Surgical oncology. 2016;25(2):74–85. doi: 10.1016/j.suronc.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 4.Zhao Q.J., Zhang J., Xu L., Liu F.F. Identification of a five-long non-coding RNA signature to improve the prognosis prediction for patients with hepatocellular carcinoma. World J. Gastroenterol. 2018;24(30):3426–3439. doi: 10.3748/wjg.v24.i30.3426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan H., Li H., Li P., et al. Long noncoding RNA MLK7-AS1 promotes ovarian cancer cells progression by modulating miR-375/YAP1 axis. J. Exp. Clin. Cancer Res. : CR. 2018;37(1):237. doi: 10.1186/s13046-018-0910-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Lu Q., Shan S., Li Y., Zhu D., Jin W., Ren T. Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. Faseb. J. : official publication of the Federation of American Societies for Experimental Biology. 2018;32(7):3957–3967. doi: 10.1096/fj.201701237RR. [DOI] [PubMed] [Google Scholar]
- 7.Yan S., Tang Z., Chen K., et al. Long noncoding RNA MIR31HG inhibits hepatocellular carcinoma proliferation and metastasis by sponging microRNA-575 to modulate ST7L expression. J. Exp. Clin. Cancer Res. : CR. 2018;37(1):214. doi: 10.1186/s13046-018-0853-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Wang K., Chen D., Qian Z., Cui D., Gao L., Lou M. Hedgehog/Gli1 signaling pathway regulates MGMT expression and chemoresistance to temozolomide in human glioblastoma. Cancer Cell Int. 2017;17:117. doi: 10.1186/s12935-017-0491-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Wang K., Jin J., Ma T., Zhai H. MiR-139-5p inhibits the tumorigenesis and progression of oral squamous carcinoma cells by targeting HOXA9. J. Cell Mol. Med. 2017;21(12):3730–3740. doi: 10.1111/jcmm.13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y.W., Xia R., Lu K., et al. LincRNAFEZF1-AS1 represses p21 expression to promote gastric cancer proliferation through LSD1-Mediated H3K4me2 demethylation. Mol. Cancer. 2017;16(1):39. doi: 10.1186/s12943-017-0588-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai F., Wu Y., Lu Y., An C., Zheng X., Dai L., Guo Y., Zhang L., Li H., Xu W., Gao W. Crosstalk between RNA m6A Modification and non-coding RNA contributes to cancer growth and progression. Mol. Ther. Nucleic Acids. 2020 Aug 8;22:62–71. doi: 10.1016/j.omtn.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y., Gu M., Liu C., et al. Long noncoding RNA FOXC2-AS1 facilitates the proliferation and progression of prostate cancer via targeting miR-1253/EZH2. Gene. 2019;686:37–42. doi: 10.1016/j.gene.2018.10.085. [DOI] [PubMed] [Google Scholar]
- 14.Luo J., Chen J., Li H., et al. LncRNA UCA1 promotes the invasion and EMT of bladder cancer cells by regulating the miR-143/HMGB1 pathway. Oncol. Lett. 2017;14(5):5556–5562. doi: 10.3892/ol.2017.6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J., Lu Y., Ding H., et al. The miR-875-5p inhibits SATB2 to promote the invasion of lung cancer cells. Gene. 2018;644:13–19. doi: 10.1016/j.gene.2017.11.066. [DOI] [PubMed] [Google Scholar]
- 16.El Bezawy R., Cominetti D., Fenderico N., et al. miR-875-5p counteracts epithelial-to-mesenchymal transition and enhances radiation response in prostate cancer through repression of the EGFR-ZEB1 axis. Cancer Lett. 2017;395:53–62. doi: 10.1016/j.canlet.2017.02.033. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., Chen L., Wu Z., et al. miR-124-3p functions as a tumor suppressor in breast cancer by targeting CBL. BMC Cancer. 2016;16(1):826. doi: 10.1186/s12885-016-2862-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu J., Lin H., Li G., et al. The miR-367-3p increases sorafenib chemotherapy efficacy to suppress hepatocellular carcinoma metastasis through altering the androgen receptor signals. EBioMedicine. 2016;12:55–67. doi: 10.1016/j.ebiom.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang T., Cai X., Li Q., et al. Hsa-miR-875-5p exerts tumor suppressor function through down-regulation of EGFR in colorectal carcinoma (CRC) Oncotarget. 2016;7(27):42225–42240. doi: 10.18632/oncotarget.9944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L., Wang J., Cao Y., et al. Molecular evidence for better efficacy of hypocrellin A and oleanolic acid combination in suppression of HCC growth. Eur. J. Pharmacol. 2019;842:281–290. doi: 10.1016/j.ejphar.2018.10.042. [DOI] [PubMed] [Google Scholar]
- 21.Zhao X., Liu Y., Yu S. Long noncoding RNA AWPPH promotes hepatocellular carcinoma progression through YBX1 and serves as a prognostic biomarker. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2017;1863(7):1805–1816. doi: 10.1016/j.bbadis.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 22.Ding H., Liu J., Liu B., Zeng Y., Chen P., Su Y. Long noncoding RNA PVT1 inhibits interferon-alpha mediated therapy for hepatocellular carcinoma cells by interacting with signal transducer and activator of transcription 1. Biochem. Biophys. Res. Commun. 2018;500(4):973–980. doi: 10.1016/j.bbrc.2018.04.219. [DOI] [PubMed] [Google Scholar]
- 23.Briscoe J., Therond P.P. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 2013;14(7):416–429. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 24.Cohen M.M. Hedgehog signaling update. Am. J. Med. Genet. 2010;152a(8):1875–1914. doi: 10.1002/ajmg.a.32909. [DOI] [PubMed] [Google Scholar]
- 25.Yu B., Gu D., Zhang X., Li J., Liu B., Xie J. GLI1-mediated regulation of side population is responsible for drug resistance in gastric cancer. Oncotarget. 2017;8(16):27412–27427. doi: 10.18632/oncotarget.16174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni W., Yang Z., Qi W., Cui C., Cui Y., Xuan Y. Gli1 is a potential cancer stem cell marker and predicts poor prognosis in ductal breast carcinoma. Hum. Pathol. 2017;69:38–45. doi: 10.1016/j.humpath.2017.08.038. [DOI] [PubMed] [Google Scholar]
- 27.Wang X., Yao Y., Zhu X. The influence of aberrant expression of GLI1/p-S6K on colorectal cancer. Biochem. Biophys. Res. Commun. 2018;503(4):3198–3204. doi: 10.1016/j.bbrc.2018.08.124. [DOI] [PubMed] [Google Scholar]
- 28.Zhang C., Wang Y., Feng Y., et al. Gli1 promotes colorectal cancer metastasis in a Foxm1-dependent manner by activating EMT and PI3K-AKT signaling. Oncotarget. 2016;7(52):86134–86147. doi: 10.18632/oncotarget.13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chaffer C.L., San Juan BP., Lim E., Weinberg R.A. EMT, cell plasticity and metastasis. Cancer Metastasis Rev. 2016;35(4):645–654. doi: 10.1007/s10555-016-9648-7. [DOI] [PubMed] [Google Scholar]
- 30.Sharma A., De R., Javed S., Srinivasan R., Pal A., Bhattacharyya S. Sonic hedgehog pathway activation regulates cervical cancer stem cell characteristics during epithelial to mesenchymal transition. J. Cell. Physiol. 2019 doi: 10.1002/jcp.28231. [DOI] [PubMed] [Google Scholar]
- 31.Yan W., Deng Y., Zhang Y., et al. DZIP1 promotes proliferation, migration, and invasion of oral squamous carcinoma through the GLI1/3 pathway. Translational oncology. 2019;12(11):1504–1515. doi: 10.1016/j.tranon.2019.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mo Y., He L., Lai Z., et al. LINC01287/miR-298/STAT3 feedback loop regulates growth and the epithelial-to-mesenchymal transition phenotype in hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. : CR. 2018;37(1):149. doi: 10.1186/s13046-018-0831-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan X., Liu X., Wang Z., et al. MicroRNA4865p functions as a tumor suppressor of proliferation and cancer stemlike cell properties by targeting Sirt1 in liver cancer. Oncol. Rep. 2019;41(3):1938–1948. doi: 10.3892/or.2018.6930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Research data can be obtained from the corresponding author if necessary.