Abstract
The ability to monitor vaccine-elicited CD8+ cytotoxic T-lymphocyte (CTL) responses in simian immunodeficiency virus (SIV)- and simian-human immunodeficiency virus (SHIV)-infected rhesus monkeys has been limited by our knowledge of viral epitopes predictably presented to those lymphocytes by common rhesus monkey MHC class I alleles. We now define an SIV and SHIV Nef CTL epitope (YTSGPGIRY) that is presented to CD8+ T lymphocytes by the common rhesus monkey MHC class I molecule Mamu-A*02. All seven infected Mamu-A*02+ monkeys evaluated demonstrated this response, and peptide-stimulated interferon gamma Elispot assays indicated that the response represents a large proportion of the entire CD8+ T-lymphocyte SIV- or SHIV-specific immune response of these animals. Knowledge of this epitope and MHC class I allele substantially increases the number of available rhesus monkeys that can be used for testing prototype HIV vaccines in this important animal model.
With compelling evidence that a virus-specific cytotoxic T-lymphocyte (CTL) response is of central importance in containing the spread of human immunodeficiency virus type 1 (HIV-1) in infected individuals (12, 13, 18, 23), there is a growing appreciation that an effective HIV-1 vaccine should elicit a high-frequency virus-specific CD8+ CTL population (14). Nonhuman primate models are playing a major role in the preclinical development of HIV-1 vaccine strategies. In particular, simian immunodeficiency virus (SIV)- and simian-human immunodeficiency virus (SHIV)-infected Indian-origin rhesus monkeys have become the most commonly used nonhuman primate models for assessing the immunogenicity and protective efficacy of prototype HIV-1 vaccines (3, 6, 9). It is therefore critical that vaccine-elicited SIV- and SHIV-specific CTL responses can be assessed in Indian-origin rhesus monkeys with quantitative precision and ease.
The detection of SIV- and SHIV-specific CTL responses in rhesus monkeys has been facilitated by our understanding of a dominant SIV Gag CTL epitope referred to as p11Cc-m or p11C (CTPYDINQM) presented to CD8+ T lymphocyte populations by the common Indian-origin rhesus monkey MHC class I allele Mamu-A*01 (2, 15). Appropriately vaccinated and infected Mamu-A*01+ rhesus monkeys universally develop a high-frequency CTL response to this 9-amino-acid fragment of SIV Gag (7, 19, 20). The use of tetramer, Elispot interferon gamma production and cytolytic assays based on an understanding of this peptide and MHC class I allele have allowed the assessment of CTL in this model system with a remarkable degree of reproducibility and quantitative precision. The obvious limitation of this technical approach has, however, been the fact that only approximately 20% of all Indian-origin rhesus monkeys express the Mamu-A*01 allele (11). Thus, it remains extremely difficult to quantitate SIV- and SHIV-specific CTL in the majority of available experimental animals. It would clearly be of enormous benefit to have knowledge of further CTL epitopes of SIV and/or SHIV that are reliably presented to CD8+ T lymphocytes by other common rhesus monkey MHC class I alleles. Such an understanding would allow the use of a larger proportion of available rhesus monkeys for studies in which the quantitation of SIV- and SHIV-specific CTL is important.
To explore the possibility that further predictable CTL epitopes might exist in this animal model, we confirmed the common expression of another MHC class I allele in Indian-origin rhesus monkeys. We then assessed SIV- and SHIV-infected rhesus monkeys for the presence of viral epitopes presented to CD8+ T lymphocytes by this allele. In so doing we defined a novel Nef CTL epitope uniformly presented to immune cells by the MHC class I molecule Mamu-A*02.
MATERIALS AND METHODS
Animals.
All animals used in this study were Indian-origin rhesus monkeys (Macaca mulatta). The monkeys had been shown to express Mamu-A*02 by PCR typing, as described below. Monkeys 19196, 483, and 833 also expressed Mamu-A*01. Monkeys 19196, 24195, and 16097 were chronically infected with SIVmac251. Monkeys 26795, 14282, 18324, 19242, 483, and 833 were chronically infected with SHIV-89.6 or SHIV-89.6P (10, 17). Rhesus monkeys used in this study were maintained in accordance with the guidelines of the Institutional Animal Care and Use Committee for Harvard Medical School and the Guide for the Care and Use of Laboratory Animals [Institute of Laboratory Animal Resources (U.S.) Committee on Care and Use of Laboratory Animals, 1996].
Cell lines and peptides.
B-lymphoblastoid cell lines (B-LCL) were prepared from peripheral blood mononuclear cells as previously described (16, 25). Briefly, whole blood from rhesus monkeys was subjected to Ficoll-diatrizoate density gradient centrifugation, and cells were seeded into a 24-well plate at a density of 5 × 105 per well. Cells were plated in the presence of supernatant from a cell line productively infected with the baboon herpesvirus Herpesvirus papio, and fresh medium was added every 2 to 3 days until transformed foci appeared (approximately 2 to 3 weeks).
Stably transfected cell lines expressing the Mamu-A*02 molecule were prepared as described previously (24). Briefly, HMy2.C1R cells expressing human β2m (21, 22) were transfected by electroporation using a Bio-Rad Gene Pulser II (0.25 kV, 950 μF) with plasmid pKG5-Mamu-A*02 (24) linearized at a unique ScaI site. G418 selection was begun on day 5 posttransfection at 0.8 mg/ml and was increased to 1.6 mg/ml on day 19. Cultures were screened for the presence of surface Mamu-A*02 by flow cytometry of cells labeled with the mouse anti-human immunoglobulin (Ig) monoclonal antibody W6/32 (American Type Culture Collection, 1:500 dilution). W6/32 binds to a conserved region of MHC class I α-chains. Labeled cells were stained with a fluorescein isothiocyanate-labeled goat anti-mouse Ig antibody. Two high-expressing cell populations were selected and cloned at limiting dilution.
Synthetic peptides were obtained from Quality Controlled Biochemicals (Hopkinton, Mass.), a division of Biosource.
PCR typing of Mamu-A*02+ rhesus monkeys.
DNA was extracted from rhesus monkey peripheral blood lymphocytes (PBL) and amplified using allele-specific primers (11). DNA from monkeys that tested positive for the presence of the Mamu-A*02 allele by PCR was sequenced to confirm the identification. For screening, EDTA-preserved whole blood from monkeys was subjected to Ficoll-diatrizoate density gradient centrifugation to isolate leukocytes, and the washed cell pellets were resuspended in 200 μl of phosphate-buffered saline (PBS). DNA extraction was then carried out using a QIAmp blood kit (Qiagen Inc., Chatsworth, Calif.). PCR was performed on 500 ng of extracted DNA in a 50-μl reaction consisting of 60 mM Tris-HCl (pH 8.5), 15 mM ammonium sulfate, 1.5 mM MgCl2, 1 mM deoxyribonucleoside triphosphates (0.25 mM each), 1.5 U of AmpliTaq polymerase (Perkin-Elmer), and allele-specific primers. The Mamu-A*02-specific primers A*02/F (5′-GTG GGT GGA GCA GGA GGG TCC A-3′) and A*02/R3 (5′-CAG CAC CTC AGG GTG GCC TCT-3′) were each used at a concentration of 5 μg/ml. Primers 5′MDRB (5′-GCC TCG AGT GTC CCC CCA GCA CGT TTC-3′) and 3′MDRB (5′-GCA AGC TTT CAC CTC GCC GCT G-3′) were each used at a concentration of 1.5 μg/ml. The latter primer pair are specific for a conserved MHC class II sequence (based on the macaque homologue of HLA-DRB3) and was included in the PCR as an internal control. PCR was carried out using a Perkin-Elmer GeneAmp 9600 thermocycler (Perkin-Elmer Inc., Norwalk, Conn.). Samples were denatured for 2 min at 94°C and then cycled at 94°C for 30 s, 67°C for 30 s, and 72°C for 90 s, a total of 30 times. Potential Mamu-A*02-positive monkeys were identified by the presence of two bands on 1% agarose gels: the expected 260-bp MDRB product and a 1.3-kb Mamu-A*02-specific product. Sequencing of samples identified by PCR screening was carried out at a central core sequencing facility on an ABI-373 stretch DNA sequencing machine, using ABI AmpliTaq FS dye terminator chemistry (Perkin-Elmer). The following sequencing primers were used: A*02/F4 (5′-TGG GAC CGG GAG ACA CGG AA-3′), A*02-Int2/F (5′-CGG TTT CAT TTT CAG TTG-3′), A*02Int3/R (5′-GAT TTT ATC CTT AAT TGT GTC-3′), A*02Int2/R (5′-CAA CTG AAA ATG AAA CCG-3′), and A*02/R4 (5′-CCC TCC AGG TAG GTT CTG TG-3′).
CTL assays.
Lymphocytes were isolated from approximately 10 ml of monkey peripheral blood by Ficoll-diatrizoate gradient centrifugation. Cultures were started with 4 × 106 to 5 × 106 PBL per ml in RPMI-1640 containing 12% heat-inactivated fetal calf serum and antibiotics. Peptides were added at various concentrations. On day 3 of culture, 20 U of recombinant human interleukin-2 per ml was added. On days 11 to 14 of culture, the lymphocytes were centrifuged over a Ficoll gradient and assessed as effector cells in a standard 4-h 51Cr release CTL assay. Target cells were B-LCL made from Mamu-A*02+ monkeys or were Mamu-A*02 transfectants of the MHC class I-deficient human cell line C1R that had been incubated for 1.5 h with peptide at various concentrations, as well as 75 to 100 μCi of sodium [51Cr]chromate. After being washed, 104 target cells per well were added to 96-well U-bottomed plates. Effectors were added at various effector-to-target cell (E:T) ratios in a final volume of 200 μl of RPMI 1640 with 12% heat-inactivated fetal calf serum and incubated for 4 h at 37°C. Then 50 μl of the supernatant was added to 200 μl of scintillation fluid, and the mixture was analyzed in a 1450 Microbeta liquid scintillation counter. Specific lysis was calculated as [(experimental release − spontaneous release)/(maximal release − spontaneous release)] × 100.
Elispot analysis.
EDTA-preserved whole blood from rhesus monkeys was subjected to Ficoll-diatrizoate density gradient centrifugation to isolate leukocytes, and cells were then seeded into 96-well plates (Millipore MultiScreen) that had been coated the night before with a mouse anti-human interferon gamma antibody. Plates were then washed three times with PBST (0.25% Tween 20 in Dulbecco's phosphate-buffered saline [Gibco-BRL]). Plates were next blocked for 2 h at 37°C with 5% fetal calf serum (FCS) in Dulbecco's phosphate-buffered saline and then washed three times with PBST and once with RPMI 1640 medium supplemented with 10% FCS prior to seeding. Cells were seeded in RPMI with 10% FCS at a density of 2 × 105 cells per well in the presence of peptide (8 μg/ml) or medium alone, in triplicate. Plates were then incubated for 18 h at 37°C in a 5% CO2 atmosphere. Plates were washed nine times with PBST and once with water and then incubated in the presence of biotinylated rabbit polyclonal anti-human interferon gamma antiserum (Biosource) for 2 h at room temperature. Plates were then washed again and incubated with streptavidin-alkaline phosphatase (Southern Biotechnology, Birmingham, Ala.) for 2 h at room temperature. After a final rinse, a chromogenic substrate was added (1-Step nitro blue tetrazolium/5-bromo-4-chloro-3-indolylphosphate; Pierce, Rockford, Ill.). After 10 to 15 min, plates were washed thoroughly with water and air dried. Spots were counted on an imaging system put together by Hitech Instruments, Inc., a division of Olympus Scientific Instruments (Edgemont, Pa.) using Image-Pro Plus image-processing software.
Isolation of CD8+ lymphocytes.
Cultured lymphocytes were centrifuged over a Ficoll-diatrizoate gradient, washed, resuspended at 108 per ml in PBS with phycoerythrin-labeled anti-CD8 antibody, and incubated at 4°C for 10 min. They were washed and resuspended in the same volume of PBS containing magnetic beads linked to antiphycoerythrin (Miltenyi Biotec Inc., Auburn, Calif.) and incubated at 4°C for 15 min. They were washed and resuspended in 0.5 ml of PBS and separated using the AutoMACS from Miltenyi. Aliquots of CD8+ lymphocyte-enriched and CD8+ lymphocyte-depleted cells were assessed on a FACScaliber, using four-color staining for expression of CD3, CD8α, CD8αβ, and CD4.
RESULTS
To initiate these studies, a cohort of 100 Indian-origin rhesus monkeys were screened for their expression of the relatively common MHC class I allele Mamu-A*02 (24). This group of monkeys comprised animals from five distinct colonies. Screening was done by PCR amplification of cDNA derived from PBL of these animals (Fig. 1). Positive signals were detected in samples from 18 of these 100 monkeys, indicating that 18% of the animals expressed the Mamu-A*02 allele. DNA sequencing of the entire α2 domain coding region as well as 122 nucleotides of the region encoding the α1 domain of the Mamu-A*02 molecule was carried out on the PCR-amplified products from all of the putative Mamu-A*02+ rhesus monkeys. Fifteen animals had sequences identical to the previously published cDNA sequence for this allele. Three monkeys had a single nucleotide change resulting in a predicted amino acid substitution of threonine for alanine at residue 85 (5) of the protein product. In the experiments described in the present report, lymphocytes were evaluated from monkeys expressing both Mamu-A*02 sequences. cDNAs extracted from PBL from a second cohort of 123 Indian-origin rhesus monkeys were similarly screened, and 35 were found to a have a PCR-amplified Mamu-A*02 product, indicating an allele frequency of 28% in this population. The PCR-amplified products from the cDNAs of this cohort of monkeys were not sequenced. The high frequency of this MHC class I allele among captive Indian-origin rhesus monkeys suggested that Mamu-A*02 would be a useful molecule for further studies.
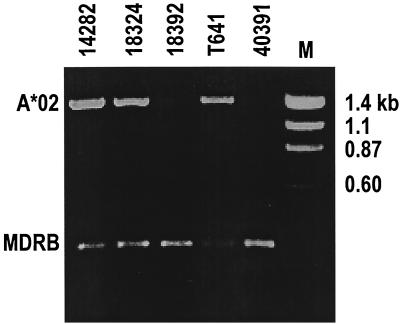
FIG. 1.
PCR typing of Mamu-A*02+ rhesus monkeys. DNA from PBL of five representative monkeys was amplified using allele-specific primers as described in Materials and Methods. Samples were run on a 1% agarose gel. MDRB represents the position of a 260-bp internal-control amplification product expected to be present in all samples tested. A*02 indicates the position of the 1.3-kb Mamu-A*02-specific amplification product. Based on the data shown, monkeys 14282, 18324, and T641 were tentatively identified as Mamu-A*02+, pending sequence confirmation.
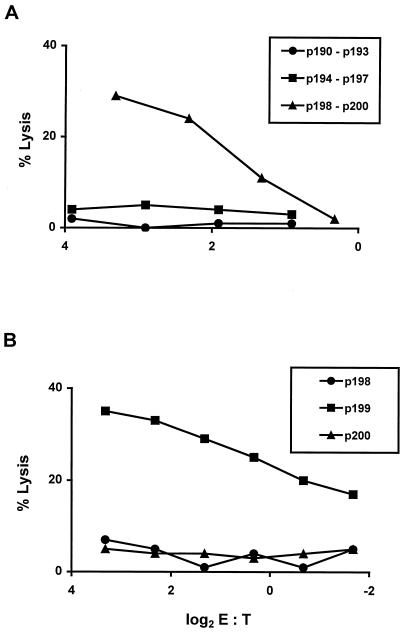
We next sought to determine whether a Mamu-A*02-presented Nef CTL epitope might be defined. To initiate this evaluation, PBL from a Mamu-A*02+ Mamu-A*01− SIVmac-infected rhesus monkey were stimulated in vitro with distinct pools of 25-amino-acid peptides that spanned SIVmac239 Nef, overlapping each other by 8-amino-acids. These peptide-stimulated PBL were then assessed for their ability to lyse Mamu-A*02+ B-LCL pulsed with the same pool of peptides that had been used to stimulate them (Fig. 2A). Target cell lysis was detected by the PBL population stimulated with the peptide pool p198 to p200. To determine which of the peptides in that pool contained the epitope recognized by the effector cells, the same effectors were assessed for the ability to lyse Mamu-A*02+ B-LCL pulsed with the individual peptides (Fig. 2B). Target cells were sensitized for lysis by p199, suggesting that a CTL epitope existed within that 25-amino-acid peptide fragment.
FIG. 2.
SIV Nef peptide-specific cytolytic activity of PBL from an SIV-infected rhesus monkey is localized to the 25-amino-acid peptide p199. (A) PBL from monkey 24195 (an SIVmac251-infected Mamu-A*01− Mamu-A*02+ rhesus monkey) were stimulated in vitro with a mix of the overlapping SIV Nef peptides p190 to p193, p194 to p197, or p198 to p200, as indicated, at 20 μg/ml for each peptide. On day 11 of culture, the lymphocytes were assessed as effector cells in a standard 4-h 51Cr release CTL assay. Target cells were B-LCL made from monkey 16097 (Mamu-A*01− Mamu-A*02+) that had been incubated for 1.5 h in the indicated mix of peptides at 20 μg/ml per peptide. (B) PBL from monkey 24195 were stimulated in vitro with a mix of the overlapping SIV Nef peptides p198, p199, and p200 at 20 μg/ml each. On day 13 of culture, the lymphocytes were assessed as effector CTL as described above, using B-LCL from monkey 16097 that had been incubated overnight with 20 μg of the indicated peptide per ml.
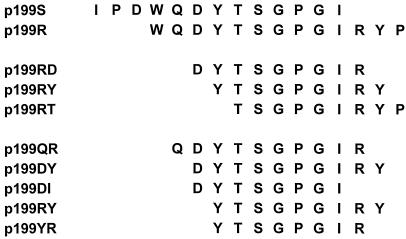
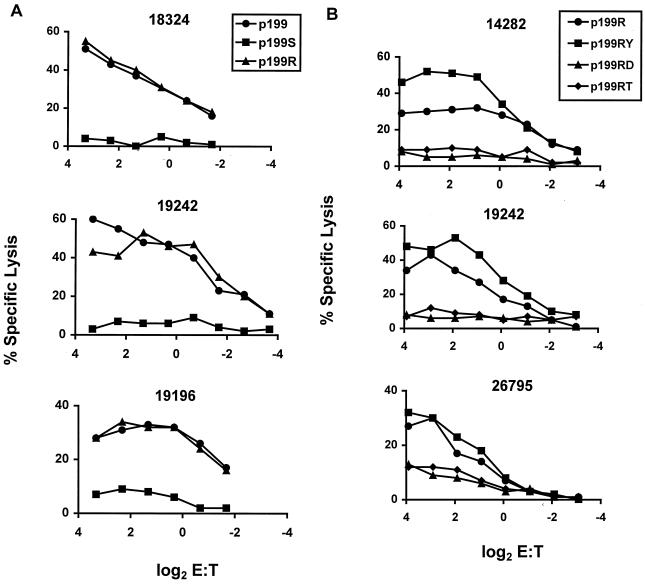
Studies were then done to map the precise CTL epitope contained within the 25-amino-acid p199 sequence. PBL from three Mamu-A*02+ SIVmac- or SHIV-infected rhesus monkeys were stimulated with p199 and assessed for their ability to lyse a single Mamu-A*02+ B-LCL pulsed with p199 or two 13-amino-acid fragments of p199, p199S and p199R (Fig. 3 and 4A). The p199-stimulated PBL populations lysed Mamu-A*02+ target cells pulsed with the 25-amino-acid p199 and p199R, suggesting that the epitope was contained within that 13-amino-acid sequence. To define with greater precision the minimal epitope recognized by these effector T lymphocytes, PBL from three Mamu-A*02+ SHIV-infected rhesus monkeys were stimulated in vitro with the 13-amino-acid peptide p199R and assessed for their ability to lyse a Mamu-A*02+ target cell that was pulsed with p199R or three 9-amino-acid peptides that comprised fragments of the p199R sequence (Fig. 3 and 4B). The peptide-stimulated effector cells lysed the target cells pulsed with p199R and with the 9-amino-acid p199RY fragment. This observation suggested that p199RY may be the optimal epitope recognized by these effector T cells.
FIG. 3.
Sequences of SIV Nef p199-derived peptides.
FIG. 4.
SIV Nef peptide-specific cytolytic activity of PBL from infected Mamu-A*02+ rhesus monkeys is localized to the 9-amino-acid peptide p199RY. (A) PBL from 18324, 19242 (two SHIV-infected Mamu-A*01− Mamu-A*02+ rhesus monkeys) and 19196 (an SIVmac251-infected Mamu-A*01+ Mamu-A*02+ rhesus monkey) were stimulated in vitro with p 199 (20 μg/ml). On day 12 of culture, the lymphocytes were assessed as effector cells in a standard 4-h 51Cr release CTL assay. Target cells were B-LCL made from monkey 19196 (used in experiments with effectors from monkey 19242 or monkey 19196) or monkey 24195 (Mamu-A*01− Mamu-A*02+; used in experiments with effectors from 18324) that had been incubated overnight with p199 (20 μg/ml) or either of the 13-amino-acid peptides p199R or p199S. (B) PBL from monkeys 26795, 14282, and 19242 (three SHIV-infected Mamu-A*01− Mamu-A*02+ rhesus monkeys) were stimulated in vitro with 5 μg of the SIV Nef 13-amino-acid peptide p199R per ml. On days 11 and 12 of culture, the lymphocytes were assessed as effector cells in a standard 4-h 51Cr release CTL assay. Target cells were B-LCL made from monkey 24195 that had been incubated overnight with 5 μg/ml of either SIV Nef p199R or one of the SIV Nef 9-amino-acid peptides p199RD, p199RY, or p199RT.
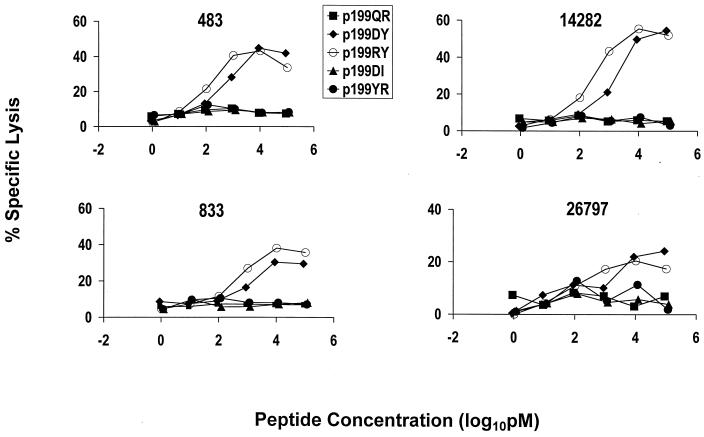
To confirm the fine specificity of these effector T-lymphocyte populations, a further study was done to assess the relative efficiency of target cell sensitization using five peptides, 8 to 10 amino acids in length, that covered this region of SIVmac Nef. PBL from four Mamu-A*02+ monkeys previously infected with SIVmac or SHIV were stimulated in vitro with the 13-amino-acid peptide p199R and assessed for their ability to lyse a Mamu-A*02+ target cell pulsed with limiting concentrations of each of these five peptides (Fig. 3 and 5). Consistently, both the 9-amino-acid peptide p199RY and the 10-amino-acid peptide p199DY were much more efficient than the other peptides at sensitizing target cells. Because p199RY was consistently slightly more efficient than p199DY at target cell sensitization, and because it is the shorter of the two peptides, we assume that the optimal Nef CTL epitope is p199RY, YTSGPGIRY.
FIG. 5.
Fine specificity of SIV Nef peptide-specific cytolytic activity of PBL from infected Mamu-A*02+ rhesus monkeys. PBL from monkeys 483, 833, 14282, and 26795 (four SIV- or SHIV-infected Mamu-A*02+ rhesus monkeys) were stimulated in vitro with the SIV Nef 13-amino-acid peptide p199R (5 μg/ml). On days 13 and 14 of culture, the lymphocytes were assessed as effector cells in a standard 4-h 51Cr release CTL assay. Target cells were B-LCL of either of the Mamu-A*02+ rhesus monkeys (24195 and 19196) that had been incubated with various concentrations of the indicated peptides.
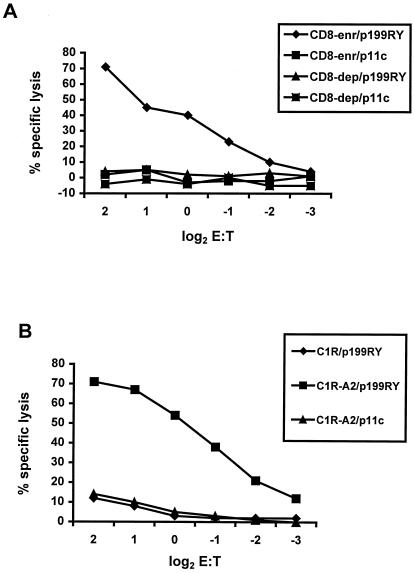
To demonstrate that p199RY is indeed a CTL epitope, experiments were performed to define the phenotype of the effector cells responsible for the recognition of the peptide and the precise MHC class I molecule that binds and presents it to T lymphocytes. PBL from SHIV-infected Mamu-A*02+ rhesus monkeys were stimulated in vitro with p199RY, fractionated into CD8+ lymphocyte-enriched and CD8+ lymphocyte-depleted populations, and assessed as effector cells for the recognition and lysis of p199RY-pulsed, autologous B-LCL (Fig. 6A). Target cell lysis was mediated by the CD8 lymphocyte-enriched but not the CD8 lymphocyte-depleted effector cells. Effector cells generated by p199RY stimulation of PBL from an SHIV-infected Mamu-A*02+ rhesus monkey were then evaluated for their ability to lyse untransfected or Mamu-A*02-transfected C1R cells pulsed with p199RY or the irrelevant control peptide p11C (Fig. 6B). Only the Mamu-A*02-transfected C1R cells pulsed with p199RY were lysed. Thus, the 9-amino-acid peptide p199RY is presented to CD8+ CTL by the rhesus monkey MHC class I molecule Mamu-A*02.
FIG. 6.
SIV Nef peptide-specific cytolytic activity of PBL from an SIV-infected rhesus monkey is mediated by CD8+ T lymphocytes and restricted by Mamu-A*02. (A) PBL from monkey 19242 (a SHIV-infected Mamu-A*01− Mamu-A*02+ rhesus monkey) were stimulated in vitro with 0.1 μg/ml of the SIV Nef 9-amino-acid peptide p199RY. On day 13 of culture, the lymphocytes were sorted for CD8 expression, and then CD8-enriched (97% CD3+ CD8+ CD4−) and CD8-depleted (5% CD3+ CD8+ CD4−) populations were assessed as effector cells in a standard 4-h 51Cr-release CTL assay. Target cells were autologous B-LCL that had been incubated for 1.5 h with 1 μg/ml p199RY or SIV gag p11c, as a negative control. (B) PBL from monkey 26795 (a SHIV-infected Mamu-A*01− Mamu-A*02+ rhesus monkey) were stimulated in vitro with 1 μg/ml p199RY. On day 14 of culture, the lymphocytes were assessed as effector cells in a standard 4-h 51Cr release CTL assay. Target cells were either the MHC class I-deficient human B-LCL C1R or a transfectant expressing Mamu-A*02 that had been incubated for 1.5 h with 1 μg/ml p199RY or SIV gag p11c, as a negative control.
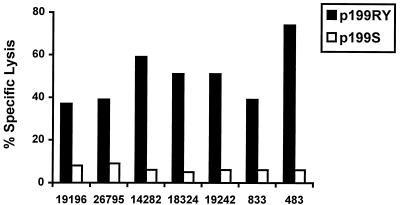
Most viral CTL epitopes that have been defined in outbred populations of nonhuman primates and humans are nondominant (26). Thus, CTL specific for these viral epitopes can be detected in only a subpopulation of infected or vaccinated individuals who express the MHC class I molecule capable of presenting that peptide fragment (7). We reasoned that this newly defined Nef epitope would prove most useful in HIV vaccine development and AIDS pathogenesis studies if it were predictably recognized by CTL in a majority of immune Mamu-A*02+ rhesus monkeys. To determine whether the optimal epitope peptide p199RY is indeed predictably recognized, PBL from seven Mamu-A*02+ rhesus monkeys infected with SIVmac or SHIV were stimulated in vitro with p199RY and assessed for their ability to lyse Mamu-A*02+ B-LCL pulsed with p199RY or a control peptide (p199S). The effector cells generated from all seven of the infected Mamu-A*02+ rhesus monkeys lysed p199RY-pulsed Mamu-A*02+ target cells (Fig. 7).
FIG. 7.
Seven of seven SIV- or SHIV-infected Mamu-A*02+ rhesus monkeys demonstrate CTL recognition of SIV Nef p199RY (YTSGPGIRY). PBL from seven rhesus monkeys, as indicated, were stimulated in vitro with 5 μg/ml SIV Nef p199RY. On days 13 and 14 of culture, the lymphocytes were assessed as effector cells in a standard 4-h 51Cr release CTL assay. Target cells were B-LCL of monkey 24195 (Mamu-A*01− Mamu-A*02+) that had been incubated overnight in 5 μg/ml of either SIV Nef p199RY or SIV Nef p199S as a negative control. For effectors from monkeys 833 and 483, B-LCL from monkey 19196 (Mamu-A*01+ Mamu-A*02+) were used as targets, treated as above. Effectors were added at a 2:1 E:T ratio.
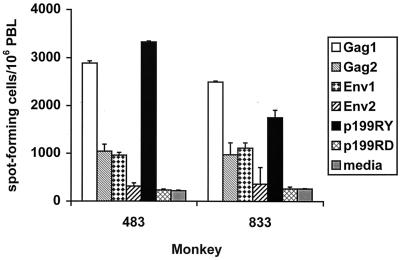
Peptide-stimulated lymphocytes from two infected Mamu-A*01+ and Mamu-A*02+ rhesus monkeys were evaluated for interferon gamma production by Elispot assay to determine whether p199RY is recognized by a sizable proportion of their virus-specific T-cell populations. By doing this study with lymphoctyes from monkeys that are both Mamu-A*01+ and Mamu-A*02+, the magnitude of the CTL populations specific for the dominant Gag p11C epitope and the newly defined Nef epitope could be compared. PBL from SHIV-infected Mamu-A*01+ and Mamu-A*02+ monkeys were stimulated with pools of 85 overlapping 20-amino-acid peptides representing the entire HIV-1 89.6P Env, 50 overlapping 20-amino-acid peptides representing the entire SIVmac 239 Gag, or the single peptide p199RY or p199RD as a control. These peptide-stimulated lymphocytes were then assessed for interferon gamma Elispots. As shown in Fig. 8, a very large proportion of the Elispot response elicited by all the various peptides was generated in response to the single 9-amino-acid Nef peptide p199RY. In fact, the magnitude of the p199RY-specific T-cell response was comparable to the response stimulated by the peptide pool (Gag 1) containing the p11C epitope.
FIG. 8.
The interferon gamma response to p199RY by PBL from Mamu-A*01+ Mamu-A*02+ SHIV-infected rhesus monkeys constitutes a significant portion of the response to all peptides spanning Env and Gag, as measured by Elispot analysis. PBL from 483 and 833 (two SHIV-infected Mamu-A*01+ Mamu-A*02+ rhesus monkeys) were added at 2 × 105 per well to 96-well plates that had been coated with a mouse anti-human interferon gamma antibody. Peptide or medium alone was added in triplicate wells and incubated at 37°C for 18 h. Peptides used were p199RY or p199RD as a negative control at 8 μg/ml; two pools containing a combined total of 50 overlapping 20-mers at 8 μg/ml each, representing the entire SIVmac239 Gag protein; and two pools containing a combined total of 85 overlapping 20-mers at 8 μg/ml each, representing the entire HIV-1 89.6P Env protein. Plates were sequentially washed and incubated with biotinylated polyclonal anti-human interferon gamma, streptavidin-alkaline phosphatase, and chromogenic substrate, as described in Materials and Methods. The number of spot-forming cells per 106 PBL is shown as the mean ± standard deviation of triplicate wells.
DISCUSSION
Dominance is a relative rather than an absolute property of CTL epitopes. For example, while CTL specific for the p54 SIV Env epitope are readily detected in most SIV-infected Mamu-A*01+ rhesus monkeys and might therefore be considered dominant (8), the frequency of the circulating effectors that recognize this epitope in infected monkeys does not appear to be as high as those that are specific for p11C when assessed by tetramer staining (data not shown). Whether p199RY represents a dominant CTL epitope remains an open question. While the peptide Elispot experiment shown in Fig. 8 suggests that the T-cell response to p199RY may represent a sizable proportion of the entire virus-specific T-cell repertoires of these infected monkeys, the study was performed comparing responses to an optimal 9-amino-acid peptide with those to pools of 20-amino-acid peptides. The frequency of CTL in infected and vaccinated Mamu-A*02+ monkeys specific for the p199RY epitope may or may not prove to be as high as those in Mamu-A*01+ monkeys specific for p11C.
Not all SIV or SHIV epitopes that can be recognized by a sizable proportion of the virus-specific CD8+ T-cell population are necessarily useful in monitoring CTL responses in vaccine or pathogenesis studies in rhesus monkeys. A dominant Tat epitope-specific CTL response in Mamu-A*01+ rhesus monkeys that is detected during the initial days following SIV infection has recently been described (1). However, this response has also been shown to disappear in chronically infected monkeys, a phenomenon attributed to viral mutation away from CTL recognition. In that particular case, long-term monitoring of the Tat-specific CTL response would be of little utility. The observation in the present study that the p199RY-specific CTL response is readily detected in a cohort of chronically infected monkeys suggests that viral mutation at this epitope to escape CTL recognition is not occurring.
The most likely immediate utility of this newly defined epitope will be in expanding the number of available Indian-origin rhesus monkeys that can be used in studies in which precise quantitation of CTL responses can be performed. For example, in a cohort of 123 rhesus monkeys recently screened for expression of the Mamu-A*01 and Mamu-A*02 alleles, 29 were Mamu-A*01-only positive, 35 were Mamu-A*02-only positive, and 2 were positive for both alleles (data not shown). Thus, 54% of all the monkeys tested expressed at least one of these alleles. This newly defined epitope will clearly prove to be of great importance in monitoring CTL responses specific for SIV or SHIV in rhesus monkeys.
The utility of this defined epitope for AIDS investigators will be determined, in part, by the degree to which this particular Nef sequence is conserved among various SIV and SHIV isolates in common experimental use. In fact, this 9-amino-acid sequence is highly conserved among nef genes encoded by the SIV and SHIV viruses that are employed at this time in nonhuman primate research. SIVmac251, SIVmac239, SIV-SMH4, and SIVmac32H nef genes encode the identical 9-amino-acid sequence used to construct the peptide employed in this study (YTSGPGIRY). All of the SHIV chimeras in use by various investigators incorporate the SIVmac239 nef gene and therefore are predicted to express this epitope. The SIV-MNE sequence has a single isoleucine-to-proline substitution at position 7. SIV-MB670 differs at two amino acids, a serine-to-proline substitution at position 3, and an isoleucine-to-glutamate substitution at position 7.
Extensive studies in numerous laboratories worldwide have led to the definition of no viral epitopes in HIV-1-infected humans that are predictably recognized by CTL from all individuals expressing a shared MHC class I molecule, yet a far less concerted effort has led to the definition of a number of such predictable CTL epitopes in SIVmac-infected Indian-origin rhesus monkeys. While there is no obvious explanation for this disparity, two possibilities bear consideration. First, the variety of MHC class I molecules that can bind viral peptide fragments and present them to CD8+ CTL in rhesus monkeys is greater than in humans. It is thought that rhesus monkeys have at least two A locus homologs and three B locus homologs, whereas humans have only single A and B loci (4). The greater potential number of MHC class I molecules that can present viral peptide fragments to CTL in a single rhesus monkey may affect the likelihood of common epitopes' being selected. Second, the ability to detect CTL responses in an individual is probably diminished if CD4+ T lymphocyte help is absent. It is possible that infections of monkeys with SIVmac and some SHIV isolates causes a less dramatic loss of T helper cells than HIV-1 infections in humans. Thus, consistent CTL responses may be more readily demonstrated in SIVmac-infected monkeys than in HIV-1-infected humans.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI85343 AI20729, and AI28691 from the National Institute for Allergy and Infectious Diseases.
We thank David I. Watkins for the Mamu-A*02 plasmid and Nicole Siciliano for assistance in preparing the manuscript.
REFERENCES
- 1.Allen T M, O'Connor D H, Jing P, Dzuris J L, Mothe B R, Vogel T U, Dunphy E, Liebl M E, Emerson C, Wilson N, Kunstman K J, Wang X, Allison D B, Hughes A L, Desrosiers R C, Altman J D, Wolinsky S M, Sette A, Watkins D I. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature. 2000;407:386–390. doi: 10.1038/35030124. [DOI] [PubMed] [Google Scholar]
- 2.Allen T M, Sidney J, del Guercio M F, Glickman R L, Lensmeyer G L, Wiebe D A, DeMars R, Pauza C D, Johnson R P, Sette A, Watkins D I. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J Immunol. 1998;160:6062–6071. [PubMed] [Google Scholar]
- 3.Barouch D H, Santra S, Schmitz J E, Kuroda M J, Fu T M, Wagner W, Bilska M, Craiu A, Zheng X X, Krivulka G R, Beaudry K, Lifton M A, Nickerson C E, Trigona W L, Punt K, Freed D C, Guan L, Dubey S, Casimiro D, Simon A, Davies M E, Chastain M, Strom T B, Gelman R S, Montefiori D C, Lewis M G, Emini E A, Shriver J W, Letvin N L. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science. 2000;290:486–492. doi: 10.1126/science.290.5491.486. [DOI] [PubMed] [Google Scholar]
- 4.Boyson J E, Shufflebotham C, Cadavid L F, Urvater J A, Knapp L A, Hughes A L, Watkins D I. The MHC class I genes of the rhesus monkey: different evolutionary histories of MHC class I and II genes in primates. J Immunol. 1996;156:4656–4665. [PubMed] [Google Scholar]
- 5.Charini W A, Letvin N L. CTL responses of rhesus monkeys to AIDS viruses: epitopes and their restricting MHC class-I alleles, p. IV-19–IV-24. In: Bette J P M, Korber TM, Brander C, Walker B D, Haynes B F, Koup R, editors. HIV molecular immunology database 1997. Los Alamos, N.Mex: Los Alamos National Laboratory; 1997. [Google Scholar]
- 6.Daniel M D, Kirchhoff F, Czajak S C, Sehgal P K, Desrosiers R C. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science. 1992;258:1938–1941. doi: 10.1126/science.1470917. [DOI] [PubMed] [Google Scholar]
- 7.Egan M A, Charini W A, Kuroda M J, Schmitz J E, Racz P, Tenner-Racz K, Manson K, Wyand M, Lifton M A, Nickerson C E, Fu T, Shiver J W, Letvin N L. Simian immunodeficiency virus (SIV) Gag DNA-vaccinated rhesus monkeys develop secondary cytotoxic T-lymphocyte responses and control viral replication after pathogenic SIV infection. J Virol. 2000;74:7485–7495. doi: 10.1128/jvi.74.16.7485-7495.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furchner M, Erickson A L, Allen T, Watkins D I, Sette A, Johnson P R, Walker C M. The simian immunodeficiency virus envelope glycoprotein contains two epitopes presented by the Mamu-A*01 class I molecule. J Virol. 1999;73:8035–8039. doi: 10.1128/jvi.73.10.8035-8039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirsch V M, Fuerst T R, Sutter G, Carroll M W, Yang L C, Goldstein S, Piatak M, Elkins W R, Alvord W G, Montefiori D C, Moss B, Lifson J D. Patterns of viral replication correlate with outcome in simian immunodeficiency virus (SIV)-infected macaques: effect of prior immunization with a trivalent SIV vaccine in modified vaccinia virus Ankara. J Virol. 1996;70:3741–3752. doi: 10.1128/jvi.70.6.3741-3752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlsson G B, Halloran M, Li J, Park I W, Gomila R, Reimann K A, Axthelm M K, Iliff S A, Letvin N L, Sodroski J. Characterization of molecularly cloned simian-human immunodeficiency viruses causing rapid CD4+ lymphocyte depletion in rhesus monkeys. J Virol. 1997;71:4218–4225. doi: 10.1128/jvi.71.6.4218-4225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knapp L A, Lehmann E, Piekarczyk M S, Urvater J A, Watkins D I. A high frequency of Mamu-A*01 in the rhesus macaque detected by polymerase chain reaction with sequence-specific primers and direct sequencing. Tissue Antigens. 1997;50:657–661. doi: 10.1111/j.1399-0039.1997.tb02927.x. [DOI] [PubMed] [Google Scholar]
- 12.Koup R A, Safrit J T, Cao Y, Andrews C A, McLeod G, Borkowsky W, Farthing C, Ho D D. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J Virol. 1994;68:4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuroda M J, Schmitz J E, Charini W A, Nickerson C E, Lifton M A, Lord C I, Forman M A, Letvin N L. Emergence of CTL coincides with clearance of virus during primary simian immunodeficiency virus infection in rhesus monkeys. J Immunol. 1999;162:5127–5133. [PubMed] [Google Scholar]
- 14.Letvin N L. Progress in the development of an HIV-1 vaccine. Science. 1998;280:1875–1880. doi: 10.1126/science.280.5371.1875. [DOI] [PubMed] [Google Scholar]
- 15.Miller M D, Yamamoto H, Hughes A L, Watkins D I, Letvin N L. Definition of an epitope and MHC class I molecule recognized by gag-specific cytotoxic T lymphocytes in SIVmac-infected rhesus monkeys. J Immunol. 1991;147:320–329. [PubMed] [Google Scholar]
- 16.Rabin H, Neubauer. Hopkins R H R F, 3rd, Dzhikidze E K, Shevtsova Z V, Lapin B A. Transforming activity and antigenicity of an Epstein-Barr-like virus from lymphoblastoid cell lines of baboons with lymphoid disease. Intervirology. 1977;8:240–249. doi: 10.1159/000148899. [DOI] [PubMed] [Google Scholar]
- 17.Reimann K A, Li J T, Veazey R, Halloran M, Park I W, Karlsson G B, Sodroski J, Letvin N L. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J Virol. 1996;70:6922–6928. doi: 10.1128/jvi.70.10.6922-6928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitz J E, Kuroda M J, Santra S, Sasseville V G, Simon M A, Lifton M A, Racz P, Tenner-Racz K, Dalesandro M, Scallon B J, Ghrayeb J, Forman M A, Montefiori D C, Rieber E P, Letvin N L, Reimann K A. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 19.Seth A, Ourmanov I, Kuroda M J, Schmitz J E, Carroll M W, Wyatt L S, Moss B, Forman M A, Hirsch V M, Letvin N L. Recombinant modified vaccinia virus Ankara-simian immunodeficiency virus gag pol elicits cytotoxic T lymphocytes in rhesus monkeys detected by a major histocompatibility complex class I/peptide tetramer. Proc Natl Acad Sci USA. 1998;95:10112–10116. doi: 10.1073/pnas.95.17.10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen L, Chen Z W, Miller M D, Stallard V, Mazzara G P, Panicali D L, Letvin N L. Recombinant virus vaccine-induced SIV-specific CD8+ cytotoxic T lymphocytes. Science. 1991;252:440–443. doi: 10.1126/science.1708168. [DOI] [PubMed] [Google Scholar]
- 21.Storkus W J, Howell D N, Salter R D, Dawson J R, Cresswell P. NK susceptibility varies inversely with target cell class I HLA antigen expression. J Immunol. 1987;138:1657–1659. [PubMed] [Google Scholar]
- 22.Voss G, Letvin N L. Definition of human immunodeficiency virus type 1 gp120 and gp41 cytotoxic T-lymphocyte epitopes and their restricting major histocompatibility complex class I alleles in simian-human immunodeficiency virus-infected rhesus monkeys. J Virol. 1996;70:7335–7340. doi: 10.1128/jvi.70.10.7335-7340.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker C M, Moody D J, Stites D P, Levy J A. CD8+ lymphocytes can control HIV infection in vitro by suppressing virus replication. Science. 1986;234:1563–1566. doi: 10.1126/science.2431484. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe N, McAdam S N, Boyson J E, Piekarczyk M S, Yasutomi Y, Watkins D I, Letvin N L. A simian immunodeficiency virus envelope V3 cytotoxic T-lymphocyte epitope in rhesus monkeys and its restricting major histocompatibility complex class I molecule Mamu-A*02. J Virol. 1994;68:6690–6696. doi: 10.1128/jvi.68.10.6690-6696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto H, Miller M D, Tsubota H, Watkins D I, Mazzara G P, Stallard V, Panicali D L, Aldovini A, Young R A, Letvin N L. Studies of cloned simian immunodeficiency virus-specific T lymphocytes. gag-specific cytotoxic T lymphocytes exhibit a restricted epitope specificity. J Immunol. 1990;144:3385–3391. [PubMed] [Google Scholar]
- 26.Yewdell J W, Bennink J R. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu Rev Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]