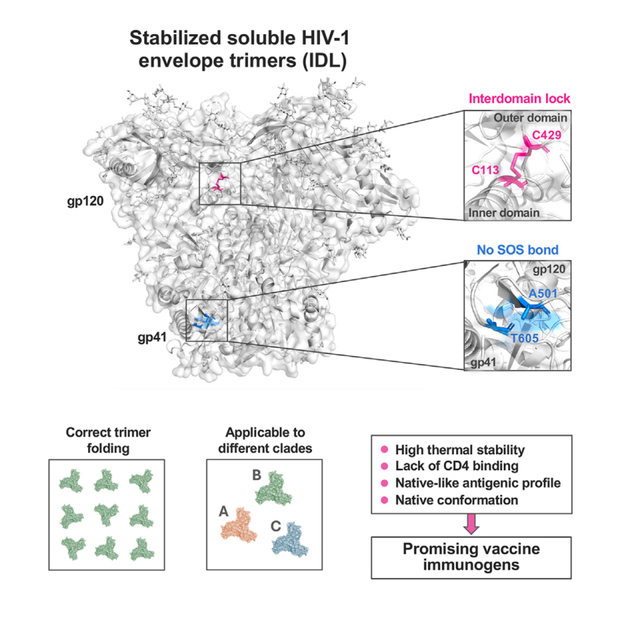
SUMMARY
Soluble HIV-1 envelope (Env) trimers may serve as effective vaccine immunogens. The widely utilized SOSIP trimers have been paramount for structural studies, but the disulfide bond they feature between gp120 and gp41 constrains intersubunit mobility and may alter antigenicity. Here, we report an alternative strategy to generate stabilized soluble Env trimers free of covalent gp120-gp41 bonds. Stabilization was achieved by introducing an intrasubunit disulfide bond between the inner and outer domains of gp120, defined as inter-domain lock (IDL). Correctly folded IDL trimers displaying a native-like antigenic profile were produced for HIV-1 Envs of different clades. Importantly, the IDL design abrogated CD4 binding while not affecting recognition by potent neutralizing antibodies to the CD4-binding site. By cryoelectron microscopy, IDL trimers were shown to adopt a closed prefusion configuration, while single-molecule fluorescence resonance energy transfer documented a high prevalence of native-like conformation. Thus, IDL trimers may be promising candidates as vaccine immunogens.
In brief
Stabilized soluble HIV-1 envelope trimers may be effective vaccine components. Zhang et al. design stabilized soluble trimers without covalent bonds between gp120 and gp41 such as the SOS bridge, a constraint that may alter structure and antigenicity. Given their native-like conformation and lack of CD4 binding, these trimers may have vaccine immunogen potential.
Graphical abstract
INTRODUCTION
The HIV-1 envelope (Env) trimer is the sole target for neutralizing antibodies (NAbs) and hence a major focus for the development of a protective vaccine. While early structural studies have provided important preliminary data on the conformation of the HIV-1 Env glycoprotein,1–3 a major breakthrough for the study of the Env structure came with the design of stabilized soluble trimers, denominated SOSIP trimers, that was achieved with the introduction of a covalent disulfide bond between residues 501 in gp120 and 605 in gp41 (defined SOS), along with an isoleucine-to-proline substitution at residue 559 (defined IP).4,5 A large number of high-resolution structures of SOSIP trimers, mostly in complex with different broadly NAbs (bNAbs), have been determined by X-ray crystallography or cryoelectron microscopy (cryo-EM).6–13 Subsequently, various modifications in the SOSIP design have been proposed with the aim of improving the trimer stability, immunogenicity, and biological features, including: (1) the introduction of hydrophobic mutations to stabilize the core of the trimer14,15; (2) the generation of non-cleavable trimers to replace the cleavage site with a flexible linker16,17; (3) the transplantation of key residues or domains from the more stable BG505 trimer to other strains18; (4) the introduction of new disulfide bonds to lock the trimer in a closed, CD4-unresponsive conformation19–21; (5) the replacement of the I559P mutation with an alternative double-proline stabilization in gp4122; and (6) the use of nanoparticle platforms for a more efficient presentation to the immune system.23 Besides their importance for structural studies, SOSIP trimers have extensively been tested as immunogens in preclinical studies and shown to be highly immunogenic in different animal models.14,21,24–26
Although SOSIP trimers adopt a native-like configuration, some authors have raised concerns about the impact of the sequence modifications, especially the SOS bond that constrains specific regions of gp120 and gp41 into a fixed position. Both the SOS and IP modifications individually were shown to reduce the cell fusion and virus-entry functions of the trimer and to induce subtle alterations in the Env conformation, glycosylation, and antigenicity.27–30 Other authors have counterargued that the SOS and IP modifications only affect the yield of native-like trimers, but not their structural, antigenic, and functional properties.31 Additional concerns came from single-molecule fluorescence resonance energy transfer (smFRET) studies, which revealed that SOSIP trimers predominantly occupy downstream conformations (states 2 and 3), rather than the bona fide pretriggered conformation (state 1) that is prevalent among virion-associated, membrane-bound trimers.32,33 Nevertheless, cryo-EM structures of full-length HIV-1 Env trimers lacking the SOSIP modifications have demonstrated a remarkable similarity with the SOSIP structure.34–36 Furthermore, studies of SOSIP trimers by double electron-electron resonance have suggested a similarity with low-resolution cryo-EM conformations of virion-bound Env spikes.37 The reasons for these discrepant results remain to be elucidated.
In the present study, we describe an alternative strategy to stabilize soluble HIV-1 Env trimers that does not require the introduction of a covalent bond between gp120 and gp41. Previously, we reported the stabilization of the HIV-1 Env trimer in a native-like conformation by introducing an interdomain lock (IDL) within the gp120 subunit via a neo-disulfide bond bridging aspartic acid 113 in the inner domain (C1) with either arginine 429 or lysine 433 in the outer domain (C4), two positions located at close distance in all high-resolution structures of the Env trimer.21 This modification was successfully introduced in both membrane-anchored gp160 and SOSIP trimers derived from different HIV-1 isolates.21 Here, we report that the IDL modification, in the absence of covalent gp120-gp41 bonds such as the SOS, is sufficient to stabilize Env trimers in a soluble form that adopts a native-like conformation. This SOS-free design was successfully applied to HIV-1 Envs from different clades. The IDL design has the additional advantage of abrogating the ability to bind CD4, which may increase vaccine effectiveness by reducing CD4-binding site (CD4-BS) occupancy and preventing CD4-induced conformational changes that compromise the native antigenic profile of the trimer. Studies using smFRET confirmed that IDL-stabilized trimers have an increased prevalence of state 1, which is diminished upon reintroduction of the SOS constraint. Together, these features make IDL trimers promising candidates as HIV-1 vaccine immunogens.
RESULTS
Design of stabilized trimers lacking the SOS constraint
To generate stabilized soluble HIV-1 Env trimers lacking the SOS constraint, we explored the possibility of replacing it with an interdomain disulfide bond that we previously reported to increase the trimer stability and to abrogate binding to CD4.21 Thus, we removed the intersubunit disulfide bond between positions 501 in gp120 and 605 in gp41 and introduced an intrasubunit disulfide bond in gp120, between amino acid (aa) 113 in the inner domain (C1) and aa 429 in the outer domain (C4); a spacer glycine residue was added on each side of aa 429 to reduce the distance from aa 113 (Figure 1A). The IP substitution (I559P), which is critical to prevent gp41 transitioning to the post-fusion state, was maintained, as were the insertion of 6 arginine residues within the gp120-gp41 cleavage site and the C-terminal truncation of gp41 at aa 664 (Figure 1B). The SOS-free soluble trimers were designated IDL trimers. The IDL design was initially applied to the reference clade A Env BG505. The BG505 IDL trimer was efficiently expressed in 293 freestyle (FS) cells and, by size-exclusion chromatography (SEC), appeared as a well-defined peak, clearly distinct from the monomer peak (Figure S1A). The purified BG505 IDL trimer was completely cleaved and, due to the absence of covalent gp120-gp41 bonds, migrated as two separate bands by SDS-PAGE under both reducing and non-reducing conditions, whereas the BG505 SOSIP.664 trimer appeared as a single band under non-reducing conditions and as two bands under reducing conditions (Figure S1B). In addition, we produced a BG505 double mutant containing both the SOSIP (501C–605C) and the IDL (113C-429GCG) disulfide bonds within the same molecule (BG505 SOSIP + IDL). The double-mutant trimer was efficiently expressed in 293FS cells, appeared as a well-defined peak by SEC, and showed the expected migration pattern by SDS-PAGE (Figures S1A and S1B).
Figure 1. Design of stabilizing mutations in SOSIP and IDL trimers.
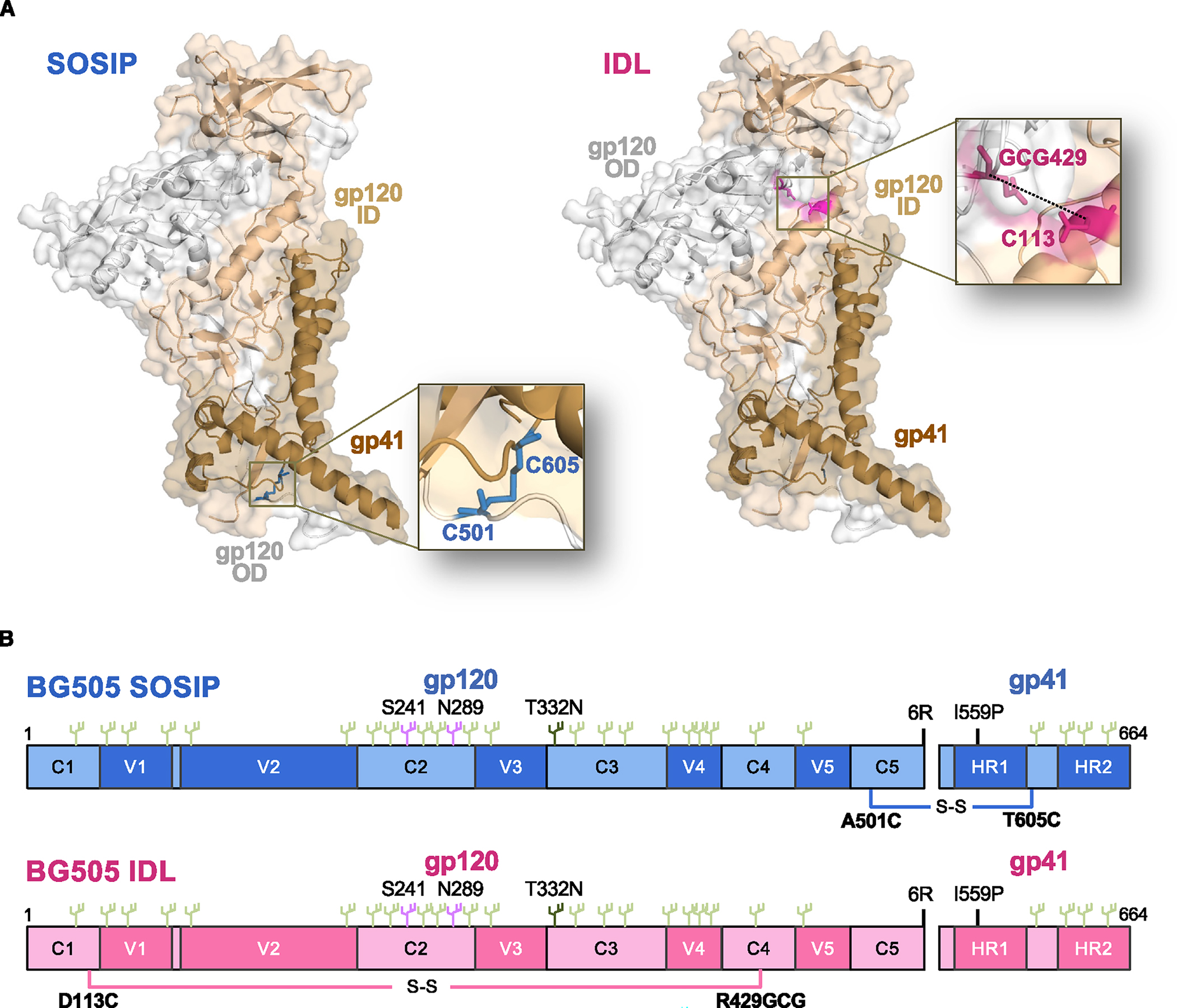
(A) Representation of the stabilizing disulfide bonds in SOSIP trimers (intersubunit bond between residues 501 in gp120 and 605 in gp41) and IDL trimers (in-trasubunit bond between residues 113 and 429, in the inner and outer domains, respectively, of the same gp120 subunit) highlighted on individual gp120/gp41 protomers.
(B) Schematic representation of SOSIP and IDL trimers from the BG505 Env (clade A) showing the respective disulfide bonds and the most salient sequence features.
Biophysical features of the BG505 IDL trimer
To evaluate the characteristics of the IDL trimers, we first analyzed BG505 IDL trimers by negative-staining electron microscopy (NSEM). The trimers appeared as correctly folded and symmetrical (Figure 2A). To assess the degree of structural stability conferred by the interdomain disulfide bond, we measured the thermal stability of homologous BG505 IDL and SOSIP trimers. Differential scanning calorimetry (DSC) analysis documented a high level of thermal stability for the BG505 IDL trimer, slightly higher than that of the homologous SOSIP trimer (midpoint of thermal denaturation [Tm] = 67.53°C vs. 66.25°C), associated with a wider transition at the half height of the peak (ΔT1/2 5.42 vs. 3.75) (Figure 2B). When SOSIP and IDL stabilizing bonds were both introduced in the same trimer, the thermal stability was further improved (Tm = 69.38°C) (Figure 2B).
Figure 2. Biophysical and immunological features of BG505 SOSIP and IDL trimers.
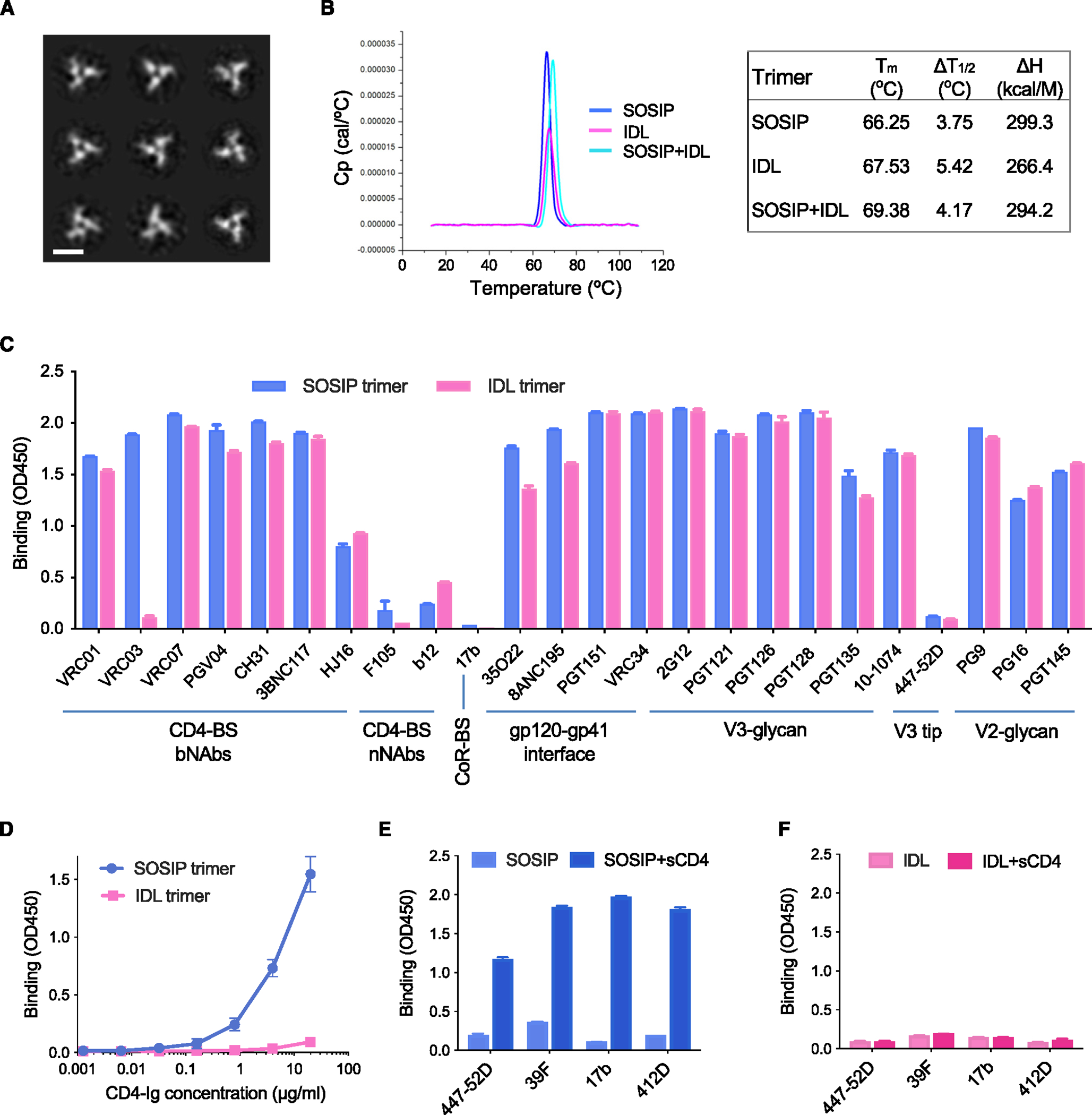
(A) NSEM analysis of BG505 IDL trimers.
(B) DSC measurements of BG505 SOSIP trimers, IDL trimers, and SOSIP-IDL (double mutant) trimers. Calorimetric parameters are presented in the table at the right of the figure.
(C) Binding of bNAbs and weakly/nNAbs antibodies to BG505 SOSIP and IDL trimers, as determined by immunoassays. Antibody target regions are specified on the bottom.
(D) Dose-response binding of soluble CD4 (CD4-Ig) to BG505 SOSIP and IDL trimers, as determined by immunoassays.
(E) Binding of antibodies to CD4-induced epitopes to the BG505 SOSIP trimer before and after treatment with soluble CD4, as determined by immunoassays.
(F) Binding of antibodies to CD4-induced epitopes to the BG505 IDL trimer before and after treatment with soluble CD4, as determined by immunoassays. The ELISA data represent the mean ± SEM of 2 technical replicates.
See also Figure S1.
The BG505 IDL trimer displays a native-like antigenic profile
Analysis of antibody binding by immunoassays demonstrated that the BG505 IDL trimer displays a native-like antigenic profile similar to that of its homologous SOSIP trimer (Figure 2C). Specifically, the IDL trimer showed high levels of binding to bNAbs directed against different antigenic supersites, including the CD4-BS, the gp120/gp41 interface (IF), and the V2-glycan and the V3-glycan regions; in contrast, there was limited or no binding to weakly/non-neutralizing antibodies (nNAbs) directed against the CD4-BS (i.e., F105, b12), the V3-loop tip (i.e., 447–52D), and the co-receptor-BS (CoR-BS) (i.e., 17b) (Figure 2C). The only discrepant results were obtained with VRC03, an antibody that mimics the quaternary interaction of CD4 with two neighboring protomers,38,39 the binding of which was abrogated by the IDL.21
The BG505 IDL trimer lacks CD4-binding capacity
While the BG505 SOSIP trimer showed the expected dose-dependent binding to CD4, the ability of the BG505 IDL trimer to bind CD4 was nearly abrogated, as documented by immunoassays using immunoglobulin (Ig)-chimeric soluble CD4 (CD4-Ig) (Figure 2D). This finding is in agreement with previous results obtained with membrane-bound trimers bearing the 113–429 or 113–431 disulfide bond, which prevents the accommodation of CD4 into the interprotomer groove, presumably by fixing the distance between neighboring protomers.21 Accordingly, treatment with soluble CD4 did not induce any detectable conformational changes in the BG505 IDL trimer, in contrast to the homologous SOSIP trimer, as revealed by reactivity with Abs to CD4-induced epitopes in the V3-loop tip (i.e., 447–52D, 39F) or the CoR-BS (i.e., 17b, 412D) (Figure 2E). This property of the IDL trimer may be advantageous for an HIV-1 Env immunogen to preserve the native-like configuration in the presence of CD4-expressing cells in vivo.
Design of IDL trimers for HIV-1 envelopes of different clades
To evaluate whether the IDL design could be applied to different HIV-1 Envs, we designed IDL trimers based on Envs from different strains and clades. Thus, we produced three IDL trimers from clade B Envs (i.e., JR-FL, AD8, and WITO), and one based on a clade C Env (i.e., DU422). In all of these trimers, the second cysteine was inserted at the alternative position 432 in C4, instead of 429, to facilitate the bond formation with aa 113 in C1. All the trimers showed a well-defined peak by SEC analysis, generally more distinct from that of the monomeric form compared to the homologous SOSIP trimers, especially for the WITO and AD8 Envs (Figure S2).
NSEM analysis showed that all four IDL trimers generated correctly folded trimeric structures (Figures 3A–3D). The results of immunoassays documented that the four IDL trimers adopted a native-like antigenic configuration, as demonstrated by high levels of binding to bNAbs along with limited binding to nNAbs (Figures 3A–3D). Of note, the nNAb 17b, directed to the CoR-BS, was reactive with three of the SOSIP trimers (WITO, AD8, and DU422.1), but was completely negative on the respective IDL trimers. These findings suggest that for these three Envs, the SOSIP design was unable to fully stabilize the soluble trimer, while, in contrast, the IDL design yielded soluble trimers with a tightly closed configuration. Another major difference between IDL and SOSIP trimers was the ability to bind CD4, which was abrogated for all four IDL trimers, while, in contrast, their SOSIP counterparts displayed high levels of CD4 binding. These results provide evidence that the IDL design can be applied successfully to multiple Envs derived from different clades, resulting in well-folded trimeric structures displaying a native-like, closed antigenic configuration and lacking the ability to bind CD4.
Figure 3. Structural and immunological features of IDL trimers from different HIV-1 clades.
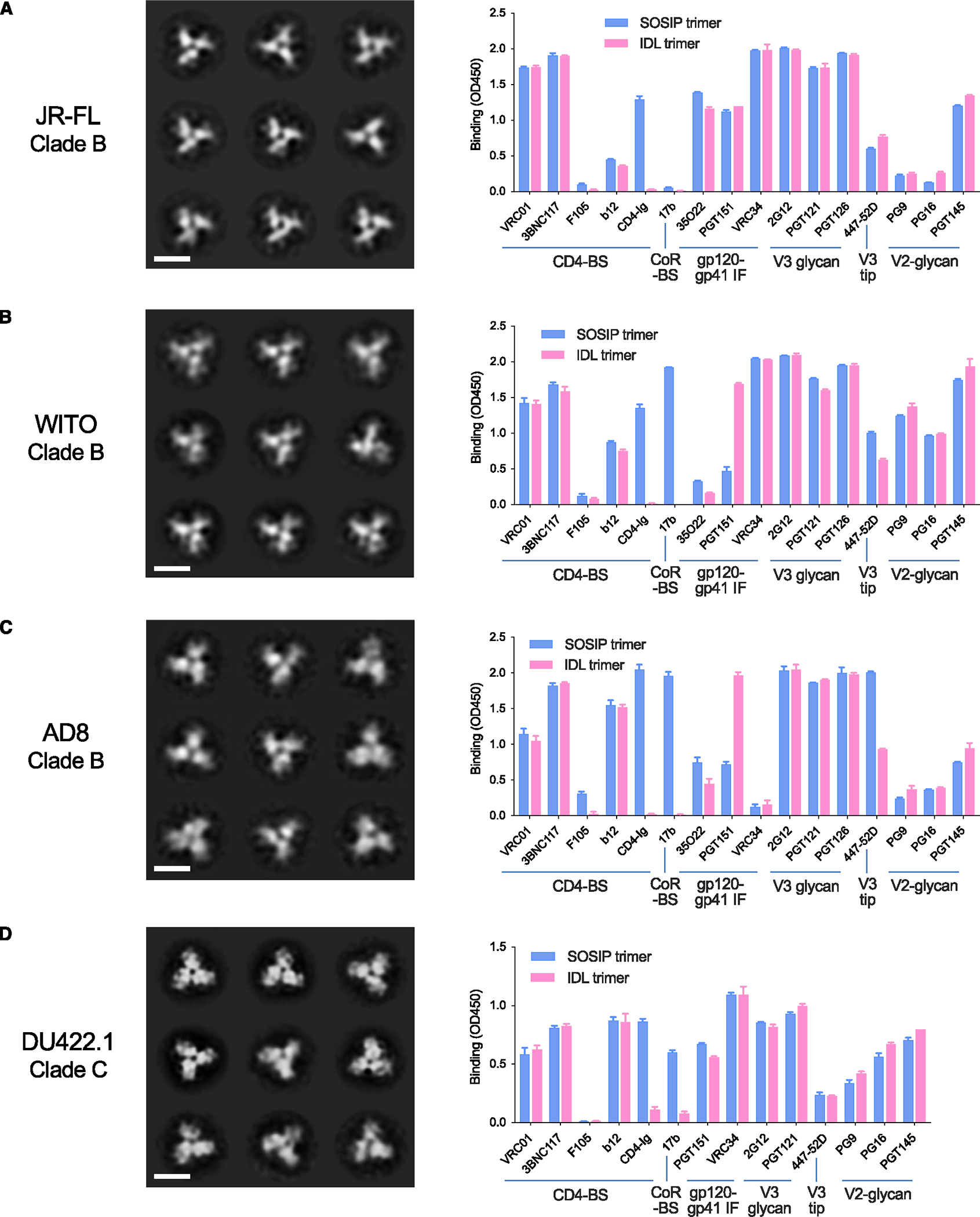
(A) Structure of JR-FL IDL trimers (clade B) by NSEM and comparative binding of homologous IDL and SOSIP trimers to bNAbs and nNAbs by immunoassays.
(B) Structure of WITO IDL (clade B) trimers by NSEM and comparative binding of homologous IDL and SOSIP trimers to bNAbs and nNAbs by immunoassays.
(C) Structure of AD8 IDL trimers (clade B) by NSEM and comparative binding of homologous IDL and SOSIP trimers to bNAbs and nNAbs by immunoassays.
(D) Structure of DU422 IDL trimers (clade C) by NSEM and comparative binding of homologous IDL and SOSIP trimers to bNAbs and nNAbs by immunoassays. The ELISA data represent the mean ± SEM of 2 technical replicates.
See also Figures S2 and S3.
To verify the stability of IDL trimers over time, we incubated homologous IDL and SOSIP trimers derived from the BG505 and JR-FL strains in the cold (4°C) for 14 days resuspended in sterile PBS at the same stock concentration and tested their reactivity with a panel of bNAbs and nNAbs. No significant changes in antibody reactivity were observed for the paired BG505-based trimers, in comparison with freshly thawed aliquots of the same trimer stocks; in contrast, the JR-FL SOSIP trimer exhibited a moderate increase in reactivity with nNAbs to the CD4-BS (i.e., F105, b12), the CoR-BS (i.e., 17b, 412D) and the V3-loop tip (i.e., 447–52D), while the homologous IDL trimer showed only a slight increase in b12 and 447–52D reactivity; for all the other Abs, the reactivity was unchanged (Figure S3). These results demonstrate a fundamentally good stability after prolonged incubation in the cold for both IDL and SOSIP trimers, although for some HIV-1 Envs IDL trimers may be more stable than SOSIP trimers.
Cryo-EM structure of IDL trimers from different HIV-1 clades
To investigate the structural features of IDL trimers, we first obtained a high-resolution structure of the BG505 IDL trimer (clade A) by cryo-EM. For this purpose, we prepared complexes with the antigen-binding fragment (Fab) of two bNAbs, 3BNC117 (anti-CD4-BS), and 10–1074 (anti-V3 glycan), resulting in complexes where each trimer was bound by 6 Fabs (Figures 4A, 4B, and S4; Table S1). From 50,380 particles analyzed, we produced a three-dimensional (3D) reconstruction of the complex (Figures 4A, 4B, and S4; Table S1). The final resolution of the reconstruction using C3 symmetry and non-uniform refinement was 3.3 Å (Figure S4D) using a Fourier shell correlation cutoff of 0.14. The density of the region encompassing the engineered disulfide bond was better than 3 Å (Figure S4E). The high quality of the map allowed for a precise positioning of the 113C-429GCG disulfide bond at the inner-outer domain IF within each protomer (Figures 4C and 4D). The overall structure of the BG505 IDL trimer was similar to the structure of the BG505 SOSIP.664 trimer, with a Cα root-mean-square deviation (RMSD) of 0.72 Å (PDB: 5V8M; Figure 4E).40 We observed a clear density for the two residues that in SOSIP trimers are involved in the SOS bond formation (aa 501 and 605), which were positioned 6.1 Å apart in the BG505 IDL structure, as measured by their Cα atoms (Figure 4F). The lack of a covalent bond between these two residues confirmed that the SOS bond is not necessary to stabilize the HIV-1 Env trimer in a soluble, native-like form. Conversely, the IDL design indicates that intrinsic stabilization of the gp120 subunit is sufficient for the trimer to adopt and maintain a closed prefusion conformation.
Figure 4. Cryo-EM structure of the HIV-1 BG505 IDL Env trimer in complex with Fabs 3BNC117 and 10–1074.
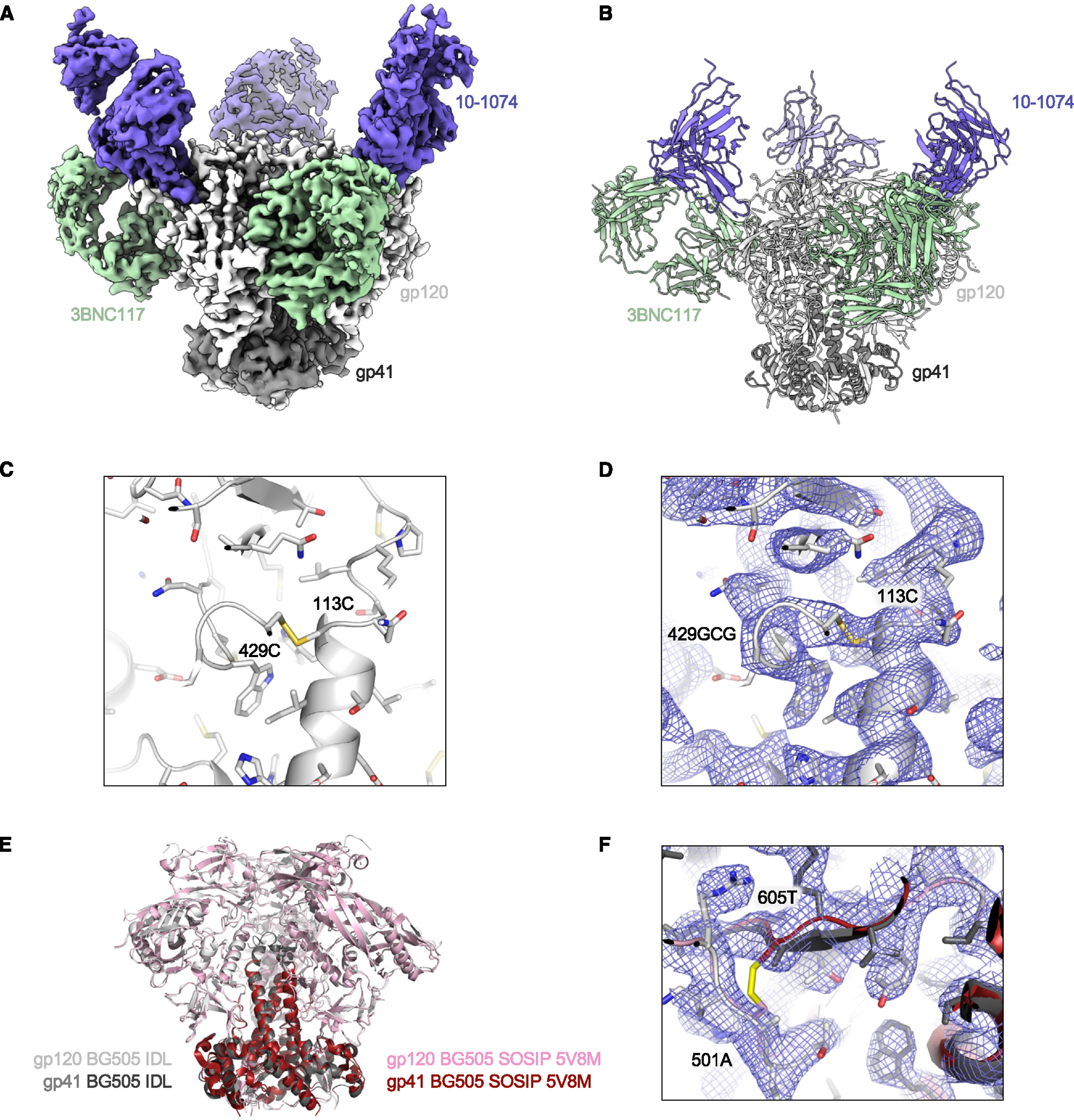
(A) Cryo-EM density is shown for HIV-1 Env BG505 IDL bound to 3BNC117 and 10–1074 Fabs.
(B) Illustration of the trimer structure.
(C) Illustration and stick representation of the engineered disulfide bond.
(D) Volume density is shown in blue mesh and clearly resolved around the disulfide linkage shown in (C).
(E) Overall alignment of gp120 and gp41 IDL (gray and black) compared to SOSIP (pink and maroon, PDB: 5V8M). Illustrations of Env trimers are shown with bound Abs hidden.
(F) The region of disulfide stabilization in SOSIP trimers is shown contrasted with the structure we present colored as in (E), with density as blue mesh.
See also Figures S4 and S7 and Table S1.
Next, we obtained cryo-EM structures for two additional IDL trimers of a different clade (JR-FL and WITO, both clade B). For the JR-FL IDL trimer, we prepared complexes with the Fab fragment of the anti-V3-glycan bNAb PGT122. The complexes showed 3 Fabs bound to one trimer (Figures 5A, 5B, and S5; Table S1). From 31,576 particles analyzed, we produced a 3D reconstruction of the PGT122-Env complex (Figures 5B and S5; Table S1) with a nominal resolution of 4.1 Å (Figures S5D and S5E) using nonuniform refinement and C1 symmetry. The density for the 113C-432GCG bond connecting the inner and outer domains of gp120 was clearly visible within each protomer. The overall structure of the JR-FL IDL trimer was similar to the structure of the JR-FL SOSIP.664 trimer (PDB: 5FYK; Figure 5E), with an RMSD of 1.5 Å across 441 Cα atoms. Residues 501 and 605 were ~6.5 Å apart, as measured from the Cα atoms (Figure 5F). Interestingly, the most prominent particle class of the JR-FL trimers adopted a C1 symmetry, and this asymmetry was not due to variation in the Abs or glycosylation, but rather to slight variations among the three protomers assembling in the trimer. This finding suggests that after removing the non-natural SOS constraint, IDL trimers retain a certain level of conformational heterogeneity among the protomers.
Figure 5. Cryo-EM structure of the HIV-1 JR-FL IDL Env trimer in complex with Fab PGT122.
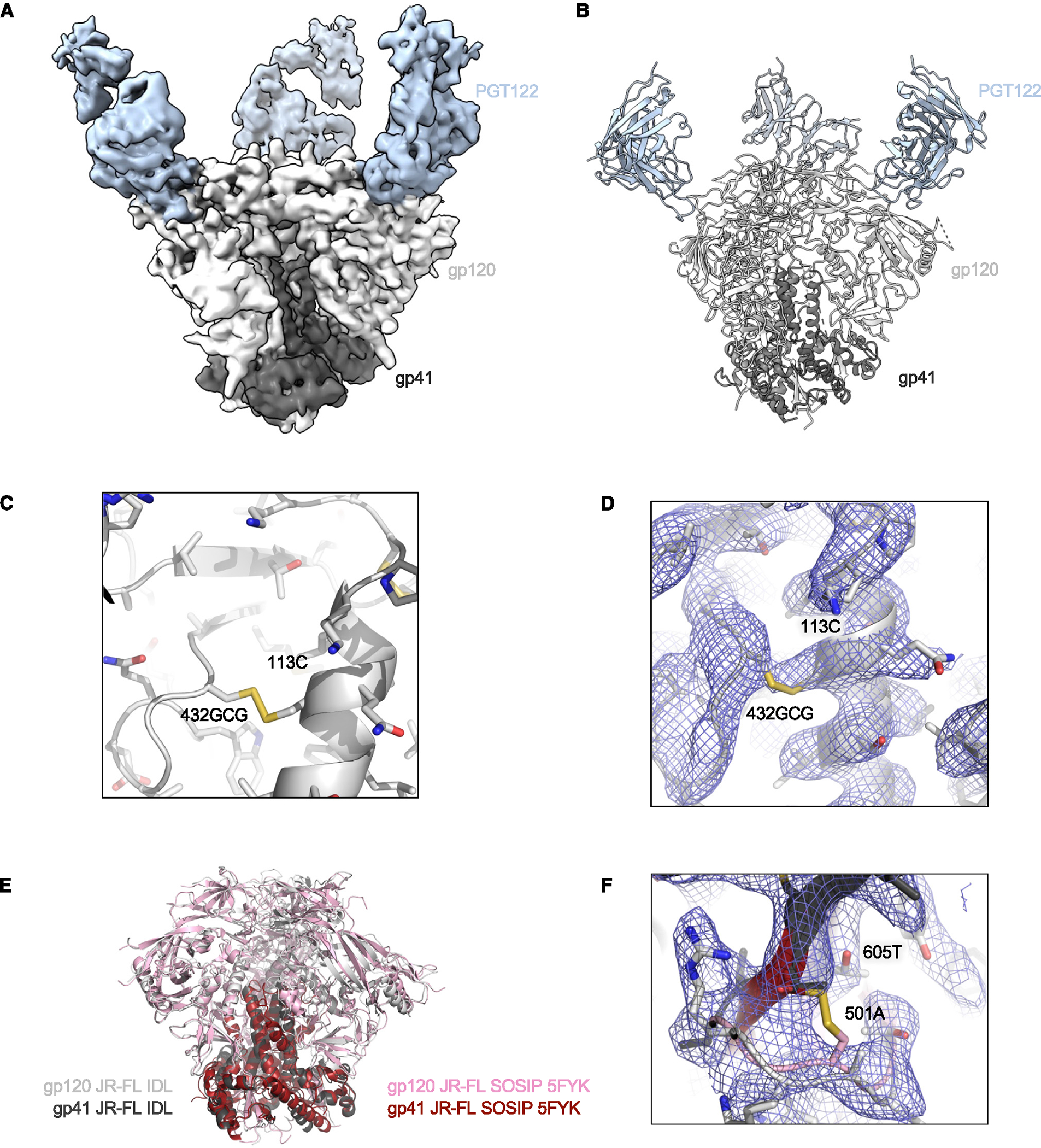
(A) Cryo-EM sensity is shown for HIV-1 Env JR-FL IDL bound to PGT122 Fab.
(B) Illustration of the trimer structure.
(C) Illustration and stick representation of the engineered disulfide bond.
(D) Volume density is shown in blue mesh and clearly resolved around the disulfide linkage shown in (C).
(E) Overall alignment of gp120 and gp41 IDL (gray and black) compared to SOSIP (pink and maroon, PDB: 5FYK). Illustrations of Env trimers are shown with bound Abs hidden.
(F) The region of disulfide stabilization in SOSIP trimers is shown contrasted with the structure we present colored as in (E), with density as blue mesh.
See also Figures S5 and S7 and Table S1.
The cryo-EM structure of the WITO IDL trimer was also determined in complex with the PGT122 Fab (Figures 6A, 6B, and S6; Table S1). Despite a relatively low number of particles for this complex (n = 12,615), the structure was reliably defined. The complexes showed 3 Fabs bound per trimer using C1 symmetry in the initial refinement and C3 symmetry for the final non-uniform refinement. Due to the lower resolution (4.5 Å), the density for the 113C-432GCG bond was less defined than in the other two structures, but the cysteine residues were correctly oriented and at the appropriate distance to establish the disulfide bond (Figures 6C and 6D). Although, for the WITO Env, no SOSIP trimer structure has been solved, comparisons with the BG505 SOSIP trimer showed an RMSD of 1.4 Å (Figure 6E). Residues 501 and 605 were positioned ~6.5 Å apart, as measured from the Cα atoms (Figure 6F).
Figure 6. Cryo-EM structure of the HIV-1 WITO IDL Env trimer in complex with the Fab PGT122.
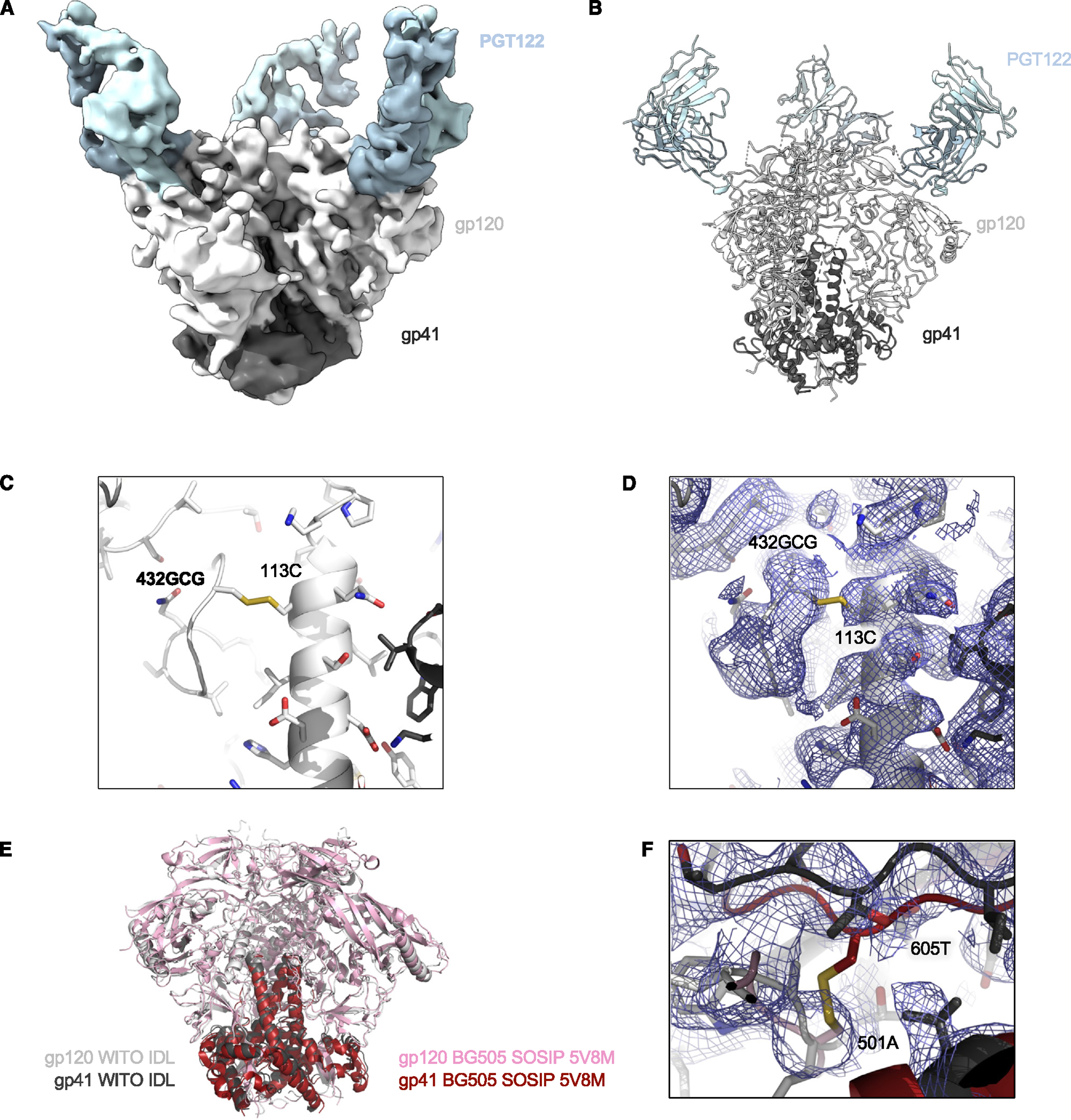
(A) Cryo-EM density is shown for HIV-1 Env WITO IDL bound to PGT122 Fab.
(B) Illustration of the trimer structure.
(C) Illustration and stick representation of the engineered disulfide bond.
(D) Volume density is shown in blue mesh and clearly resolved around the disulfide linkage shown in (C).
(E) Overall alignment of gp120 and gp41 IDL (gray and black) compared to BG505 SOSIP (pink and maroon, PDB: 5V8M). Illustrations of Env trimers are shown with bound Abs hidden.
(F) The region of disulfide stabilization in SOSIP trimers is shown contrasted with the structure we present colored as in (E), with density as blue mesh.
See also Figures S6 and S7 and Table S1.
In summary, cryo-EM analysis confirmed that the IDL design resulted in correctly folded trimers that adopt a stable, native-like structure. The distance between residues 501 and 605 does not differ from the distance that separates these residues in SOSIP trimers, where they are covalently linked (Figure S7B). The relative distance of each protomer to another also resembled that observed for SOSIP trimers, with the exception of the WITO IDL trimer, which is one of the most compact trimers thus far reported, as measured by the distance between residues 501 in different protomers (Figure S7C). Likewise, the RMSDs for both gp120 and gp41 of IDL protomers falls within the standard range of deposited SOSIP trimers (Figures S7D and S7E). Overall, the cryo-EM structures of three IDL trimers confirmed that the IDL design, despite the lack of covalent bonds linking gp120 to gp41, effectively promotes the formation of stable soluble trimers. The additional flexibility conferred by the lack of covalent gp120-gp41 constraints may foster a greater HIV-1 Env conformation diversity, allowing for the exploration of symmetric and asymmetric protomer conformations that are likely to better reflect the native conformation of membrane-bound trimers.
IDL trimers display a high prevalence of native-like conformation
Although the results of structural analysis revealed a fundamental homology between the structures of IDL and SOSIP trimers, IDL trimers are characterized by the inability to bind CD4, suggesting that they explore a different conformational landscape than the SOSIP trimers. This prompted us to investigate the conformational dynamics of IDL trimers by smFRET. Thus, we introduced site-specific enzymatic sites in the variable regions V1 and V4 to enable the insertion of donor and acceptor fluorophores into the BG505 IDL and the double-mutant (BG505 SOSIP + IDL) trimers.32,33 We generated soluble trimers that predominantly carry labeling tags only in a single protomer. Following purification, soluble trimers were immobilized on quartz slides and imaged by total internal reflection fluorescence microscopy. Parameters and statistics of the three-state Gaussian model are presented in Table S2. Analysis by smFRET demonstrated that the BG505 IDL-stabilized trimer has an increased prevalence of state 1 conformation, which is characteristic of the native trimers displayed on the surface of viruses, and is rarely, if at all, sampled by SOSIP trimers (Figure 7A). Of note, reintroduction of the SOS constraint into the IDL trimer (BG505 SOSIP + IDL double mutant) markedly reduced access to state 1 (Figure 7B), further corroborating the rigidifying effect of the SOS bond on the trimer structure. These results illustrate the potential advantage of generating HIV-1 Env trimers free of the SOS mutations.
Figure 7. The BG505 IDL trimer displays a prevalent native-like conformation.
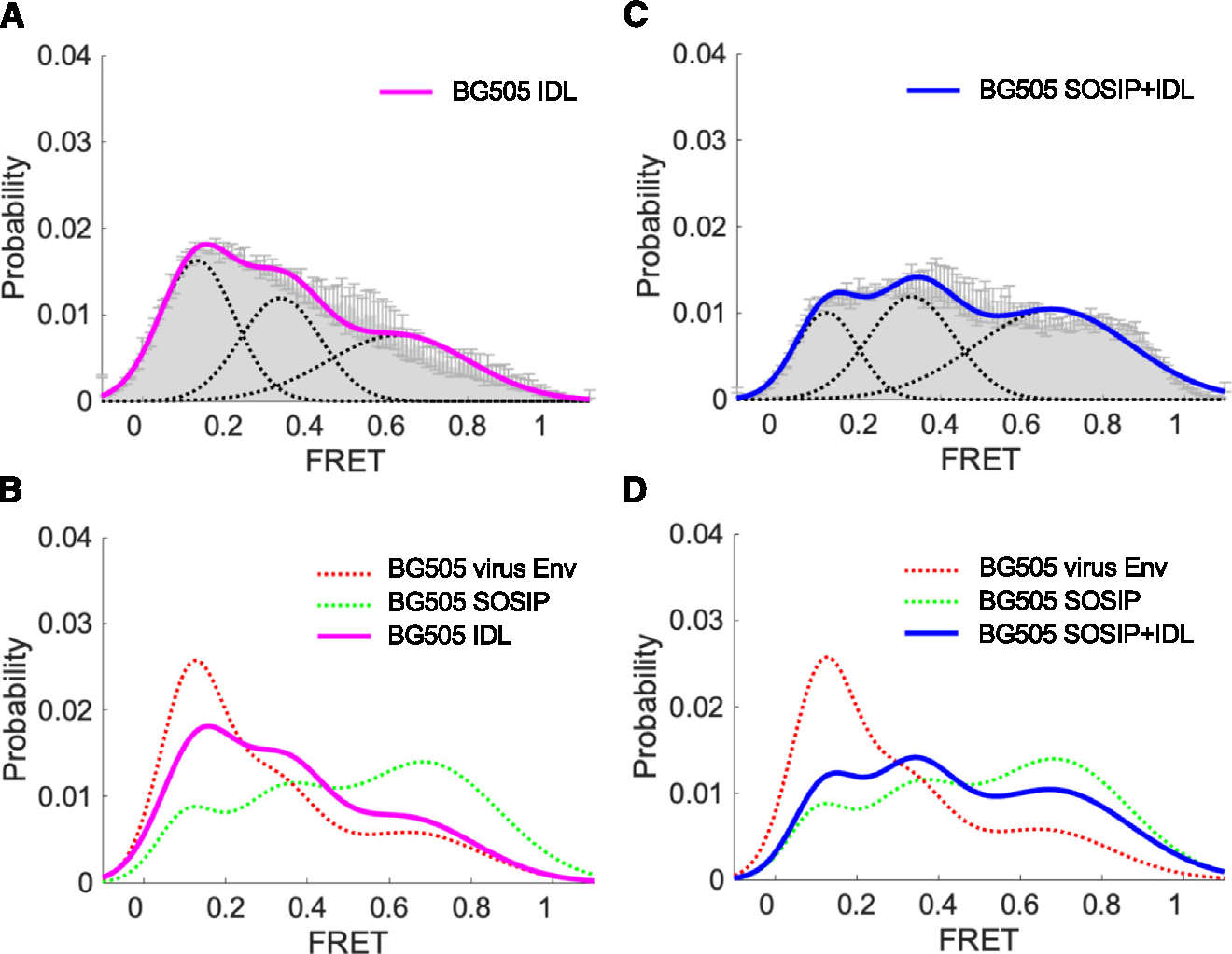
(A) Three-state Gaussian-distributed FRET population histogram (mean ± SD) of the BG505 IDL trimer, reflecting the conformational distribution sampled by the trimers.
(B) Conformational landscape of the BG505 IDL Env trimer (A) overlaid with those of virus-associated, full-length BG505 Env and BG505 SOSIP trimer.33
(C) Conformational distribution of the double mutant BG505 SOSIP + IDL Env trimer as in (A).
(D) Conformational landscape of the BG505 SOSIP + IDL SOSIP (C) overlaid with those of virus-associated, full-length BG505 Env and BG505 SOSIP trimer.33 Parameters and statistics for the smFRET analysis are provided in Table S2.
DISCUSSION
The design of stabilized soluble HIV-1 Env trimers based on the SOSIP modifications has represented a milestone in the field of HIV-1 structural biology leading to fundamental progress in the elucidation of the HIV-1 Env spike conformation.7 However, there are potential limitations with the SOSIP design. Specifically, concerns have been raised about the effect of the SOS modification, a disulfide bond between aa 501 in the gp120 subunit and aa 605 in the gp41 subunit, which appears to constrain the trimer in a rigid configuration that may prevent the adoption of the bona fide native prefusion structure.27,30,32,33 Furthermore, as immunogens, SOSIP trimers did not live up to the initial expectations as they were shown in preclinical models to induce primarily off-target Abs directed to the artificial surface created by gp41 truncation at the trimer base, as well as autologous NAbs that do not qualify as authentic tier 2 neutralizers being directed solely against isolate-specific glycan holes present on the trimer surface.14,24–26 Finally, the original SOSIP design cannot be applied universally to primary HIV-1 strains, as many Envs do not form stable and properly folded trimers after introduction of the SOSIP mutations.22,41
In the present study, we devised an alternative strategy to stabilize HIV-1 Env trimers in a soluble, native-like form, which does not require the introduction of a covalent bond between gp120 and gp41. To achieve this goal, we removed the intersubunit SOS bond and introduced instead an intrasubunit neo-disulfide bond that we previously reported to stabilize the HIV-1 Env trimer in a native-like conformation.21 We found that the IDL modification is sufficient to replace the SOS bond and stabilize soluble trimers from diverse Envs derived from different HIV-1 clades. Such stabilized trimers displayed a high thermal stability and a native-like antigenic profile, with high levels of binding to bNAbs, which was equivalent to or even better than that of their homologous SOSIP counterparts. Although it is unclear, at present, whether the IDL design can be applied to a broader range of Env trimers than the SOSIP design, it is nonetheless important for the field to have multiple options at its disposal for deriving stable soluble trimers, particularly when coping with difficult Envs. In addition, as we demonstrated by combining both the IDL and SOSIP disulfide bonds in the same trimer, more than one stabilizing mutation can simultaneously be used to further improve the stability and manufacturability of soluble trimers. An important added value of the IDL design is that it abrogates binding to the CD4 receptor. This is a desirable feature for an Env-based vaccine immunogen, as CD4 binding results in the occlusion of the CD4-BS, a critical antigenic target for the elicitation of protective Abs by a vaccine. Furthermore, binding to CD4 triggers conformational changes that compromise the native antigenic state of the trimer, and promotes immunogen sequestration on the surface of CD4+ T cells, which lack antigen-presenting functions. Conversely, it is important to emphasize that introduction of the IDL bond does not affect the binding of potent bNAbs targeting the CD4-BS, such as VRC01 and 3BNC117, thereby leaving this crucial antigenic site intact and available for antibody elicitation. Together, these properties make IDL trimers promising candidates as vaccine immunogens.
Structural analysis revealed subtle, yet potentially important differences between IDL and SOSIP trimers. Cryo-EM analysis of three IDL trimers from different clades showed that these trimers can adopt both symmetrical and asymmetrical conformations, as seen particularly with the asymmetry of the JR-FL IDL trimer. While the covalent constraint between gp120 and gp41 that characterizes the SOSIP trimer design appears to rigidify the structure into a strictly symmetrical configuration, the IDL trimer design allows for a greater conformational freedom that may better reflect the innate mobility of the protomers within the context of the full-length, membrane-anchored native trimer. To what extent such innate mobility and the consequent trimer asymmetry affect the Env spike function and epitope accessibility, especially for bNAb epitopes that are the most relevant to vaccine efficacy, remains to be defined. In contrast to the JR-FL IDL trimer, the differences that we observed between the BG505 IDL and SOSIP trimers were minimal, presumably due to the uncommonly compact configuration of this clade A Env, which has certainly facilitated the development of the first stable soluble trimers.4,5 As for the WITO IDL trimer, it displayed a moderate asymmetry, but its most distinctive feature was a remarkably compact assembly of the trimer structure. The biological implications of such compact arrangement remain at present unclear. Another significant difference that we observed between IDL and SOSIP trimers was the prevalence of native-like conformational state occupancy, as determined by smFRET analysis. Thus, we found that IDL trimers display a prevalent state 1 conformational occupancy, which is characteristic of virion-associated native trimers, while state 1 is rarely, if at all, sampled by SOSIP trimers. Of note, reintroduction of the SOS constraint into the IDL trimer (SOSIP + IDL double mutation) drastically reduced the prevalence of state 1 conformation, further corroborating the rigidifying role of the SOS bond on intersubunit mobility within the trimer. These results confirmed the increased conformational freedom of IDL trimers compared to their SOSIP counterparts, which allows them to sample multiple conformational states and, most notably, to adopt a predominant native-like configuration. This feature may constitute an additional advantage for the vaccine-induced elicitation of Abs capable of recognizing with high affinity the native trimer on the surface of infectious virions.
Expanding our knowledge of the native HIV-1 Env structure is key to the elucidation of the structure-function relationships, and thus, to the design of increasingly effective HIV-1 vaccine immunogens. There is no doubt that the wide genetic diversity of HIV-1 is reflected by structural heterogeneity among different strains and clades, but there are inherent limitations to structural studies of diverse Envs since many of them are difficult to stabilize in soluble forms. The alternative strategy described herein offers additional opportunities to derive stable soluble trimers for HIV-1 strains that are not compatible with the SOSIP design. Our data provide evidence that the lack of a covalent bond between gp120 and gp41 allows for a greater intra-molecular mobility within the trimer, particularly in regard to the gp41 subunits, which seems to better reflect the conformational plasticity of the native Env spike. Due to the increased prevalence of native-like conformation and the lack of CD4 binding, IDL trimers may provide a new class of Env-based immunogens that could be included as components of an HIV-1 vaccine. Preclinical studies in relevant animal models will be instrumental in determining the effective value of IDL trimers as vaccine immunogens.
Limitations of the study
An important general limitation inherent to the use of soluble HIV-1 Env trimers as immunogens is the exposure of off-target epitopes, which has significantly hampered the utilization of SOSIP trimers as vaccine components. Indeed, SOSIP trimers were shown to invariably elicit large amounts of nNAbs directed to the trimer base, an artificial surface created by gp41 truncation that is completely hidden in the membrane-anchored Env spike, in addition to autologous NAbs targeting strain-specific glycan holes present on the Env surface.14,24–26 The IDL trimer design reported here does not address these critical issues. However, strategies to overcome these limitations have been devised, including the engineering of new N-linked glycans to cover the trimer base and to fill immunodominant glycan holes.42,43 Such strategies could be applied successfully to IDL trimers. Another limitation of the present study is that it did not evaluate the in vivo immunogenicity of IDL trimers in preclinical models. The results of preclinical studies will be critical to determine whether IDL trimers can be effective components of an HIV-1 vaccine.
STAR★METHODS
RESOURCES AVAILABILITY
Lead contact
Paolo Lusso at plusso@niaid.nih.gov.
Data and code availability
Cryo-EM maps of IDL trimers have been submitted to the PDB database under accession codes 9BEW, 9BF6 and 9BER and to the EMDB database under accession codes 4484, 44482, 44491.
This paper does not report original code;
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
METHOD DETAILS
HIV-1 gp160 mutagenesis
The primers for mutagenizing the HIV-1 Env trimers were designed using the online program provided by Agilent (https://www.agilent.com/store/primerDesignProgram.jsp). The oligos were synthesized by Eurofins Genomics. The gp160 genes from different HIV-1 strain plasmids (BG505 from John P. Moore; JR-FL from the VRC) were mutagenized by site-directed mutagenesis using QuickChange II Site-Directed Mutagenesis Kit (Agilent Technologies). The correct mutagenesis clones were expanded by QIAGEN Plasmid Mega Kit. The others genes were synthesized by Genscript.
Production of HIV-1 envelope trimers
SOSIP.664 and IDL trimers were expressed by co-transfecting the relevant plasmids with a plasmid expressing the cellular protease furin into human embryonic kidney (HEK) 293 free-style (FS) cells. Cell-free supernatants were harvested after 6 days, passed through a 0.22 μm PES filter, and loaded onto a Galanthus nivalis (GNA) lectin column (Vector laboratories). After repeated washing with PBS, bound proteins were eluted with 1 M methyl α-D-mannopyranoside in PBS, followed by dialysis toward PBS. Dialyzed samples were concentrated to 2 mL using Amicon Ultra-15 centrifugal filter units (MWCO 50,000, Millipore) and applied to a PBS pre-equilibrated Superdex 200 SEC column. The peak corresponding to the trimeric form was collected and concentrated, followed by a second round of gel filtration purification and passed through a mAb 447–52D affinity column to remove V3 loop exposed trimer. Finally, purified trimers were concentrated to 1 mg/mL and stored at −80°C.
Differential scanning calorimetry
A high-precision differential scanning VP-DSC microcalorimeter (GE Healthcare/MicroCal) was employed to measure the heat capacity of the trimers. In brief, samples were diluted to 0.3 mg/mL with PBS. Thermal denaturation scans were performed from 30°C to 110°C at a rate of 1°C/min.
Enzyme immunoassays
All the antibodies and HIV-1 Env proteins used in ELISA (Enzyme linked immunosorbent assay) experiments were diluted in 1X casein solution (Vector Laboratories) in PBS. For assessing binding titers of monoclonal antibodies, 96-well ELISA plates (Corning) were coated with 2 μg/mL of Lectin Galanthus nivalis (Sigma) at 4°C overnight. After blocking with 1X casein in PBS, purified trimers (1 μg/mL) were added and incubated for 2 h at RT. After washing, monoclonal HIV antibodies were added individually and incubated for 1 h at RT. After washing, horseradish peroxidase (HRP)-conjugated goat anti human IgG (Jackson ImmunoResearch) was added for 1 h at RT. The reaction was revealed by incubation with 50 μL of substrate (R&D Systems) for 15 min before addition of the stop solution (25 μL). Light absorption was measured at a wavelength of 450 nm by PerkinElmer Enspire plate reader. For experiments of trimer stability, identical stocks of homologous IDL and SOSIP trimers were either kept frozen or incubated for 14 days at 4°C in sterile PBS at the concentration of 1 mg/mL before testing by ELISA.
Fab preparation
Variable regions of the 3BNC117, 10–1074 and PGT122 antibody genes were synthesized (Genscript) and subcloned into the pVRC8400 vector, in which a HRV3C cleavage site was inserted in the heavy-chain hinge region. The heavy and light chain pair was co-transfected in Expi293F cells (Thermo Fisher) using Turbo293 transfection reagent (Speed BioSystems) as described previously.44 On day 6 post transfection, the culture supernatant was harvested and loaded on a protein A column, the column was washed with three column volumes of PBS, and IgG proteins were eluted with a low pH buffer. The eluted IgG proteins were cleaved by HRV3C, and the cleavage mixture was passed through a protein A column to separate the Fab fragments.
Negative-staining electron microscopy
Proteins were diluted to a concentration of about 0.02 mg/mL, adsorbed to freshly glow-discharged carbon-film grids for 15 s, washed with a buffer containing 10 mM HEPES, pH 7 and 150 mM NaCl, and stained with 0.7% uranyl formate. Images were collected semi-automatically with SerialEM45 on an FEI Tecnai T20 electron microscope operating at 200 kV and equipped with a 2k ×2k Eagle CCD camera at a nominal magnification of 100,000 and a pixel size of 0.22 nm. To estimate the fractions of aberrant trimers, particles were picked automatically and manually from micrographs of BG505 SOSIP.664 and IDL trimers using e2boxer from the EMAN2 software package46 and subjected to reference-free two-dimensional classification in EMAN2, SPIDER47 (operation: AP C), and Xmipp.48 The obtained class averages were assigned manually to either the native or aberrant trimer conformation based on expected sharp, symmetrical, propeller-like views typical of correctly folded HIV glycoproteins. The fractions of incorrectly formed trimers were then calculated based on numbers of particles forming the “native” and “aberrant” classes. The three software packages produced similar ratios, which were averaged to produce the final values.
For three-dimensional reconstructions, extended datasets were collected, each containing equal numbers of micrographs obtained at tilt angles of 0°, 15°, 30°, and 45°. Tilted micrographs were divided into two vertical slices each for more accurate contrast transfer function (CTF) correction. The initial datasets for BG505 contained 25,856 and 22,430 particles, respectively. EMAN2 was used to estimate CTF parameters and perform CTF correction by phase flipping. Reference-free 2D class averages were obtained using SPIDER49 by rotationally and translationally aligning all particles in a dataset and performing correspondence analysis and classification. Selected SPIDER class averages were then used to generate initial 3D maps in EMAN2 with C3 symmetry imposed. These maps were filtered to a resolution of 40 Å and used as initial models for three-dimensional reconstruction and refinement using reference projections in SPIDER. The C3 symmetry was used during the reconstruction and refinement. A limit was placed on the number of particles assigned to a single orientation to overcome the strong preferential orientation typical of HIV glycoproteins adsorbed to carbon. A total of 18,142, and 10,370 particles contributed to the final maps of BG505 SOSIP.664 and 113C-429GCG, respectively. The resolutions of the final maps were 17.6 Å and 18.9 Å, respectively, when a Fourier shell correlation (FSC) threshold of 0.5 was used. The maps were visualized using UCSF Chimera.50
To measure the volume of the internal cavity of the trimer glycoproteins, the maps were first low-pass filtered to a resolution of 19 Å, and the map thresholds were chosen such that the three maps had identical volumes as calculated using UCSF Chimera. The contrast of the original maps was then inverted (SPIDER’s operation “NEG”). Each inverted map was loaded in UCSF Chimera along with the original map, and the threshold of the inverted map was adjusted such that the positive density corresponding to the internal cavity slightly exceeded the size of the internal cavity of the original map. The inverted map was then segmented using the watershed algorithm implemented in Segger,51 a component of UCSF Chimera, and the density of the segment(s) corresponding to the internal cavity was extracted. The volume of this density was then measured in UCSF Chimera after adjusting the threshold such that this volume fit exactly in the internal cavity of the original map.
Cryo-EM data collection and processing
Env trimer used in cryo-EM was generated in a 293F cell line as described previously.19 To prepare Env complexes, BG505 IDL, JR-FL IDL, WITO IDL at a final concentration of 2 mg/mL were incubated with 3-fold molar excess of the antibody Fab fragment for 30 min. To prevent aggregation during vitrification, the sample was incubated in 0.085 mM n-Dodecyl β-D-maltoside (DDM). Complexes were vitrified using a Vitrobot Mark IV with a wait time of 30 s and a blot time of 3 s. Data were acquired using the Leginon system52 on a Titan Krios electron microscopes operating at 300kV and fitted with a Gatan K2 Summit direct detection device. The dose was fractionated over 50 raw frames collected over a 10 s exposure time. Individual frames were aligned and dose-weighted with MotionCor2.53 CTF was estimated using the CTFFind4 package.54,55 Particles were picked using DoG Picker within the Appion pipeline.56,57 Particles were extracted using RELION.58 2D classifications, ab initio model generation, and homogeneous nonuniform 3D refinements were carried out in cryoSPARC 2.12.59 The 3D reconstructions were performed using C1 symmetry initially and, after assessing for symmetry, the BG505 and WITO complexes were refined with C3 while JR-FL was refined with C1. Global resolution was calculated in CryoSPARC with an FSC cutoff of 0.143 while the local resolution mapping was determined with a conservative FSC cutoff of 0.5.
Cryo-EM model fitting
For initial fits to the cryo-EM reconstructed maps, we used the coordinates of BG505-SOSIP or JR-FL, PDB IDs: 5V8M, and 5FYK respectively as starting templates. The coordinates were fit to the density more precisely through an iterative process of manual fitting using Coot60 and real space refinement within Phenix,61 Molprobity62 and EMRinger63 were used to check geometry and evaluate structures at each step. Figures were generated in UCSF ChimeraX and PyMOL (https://pymol.org/2/). Local resolution of cryo-EM maps was determined using cryoSPARC.
smFRET analysis
Sample preparation, data acquisition, and analysis of fluorescently labeled soluble Env trimers for smFRET experiments were similar to those previously described.32,33 Two Env constructs, V1V4-tagged and untagged IDL, were used to express BG505 IDL trimers suitable for smFRET experiments, in which tagged IDL Env (or IDL SOSIP Env) carries two short labeling peptides (Q3: GQQQLG; A1: GDSLDMLEWSLM) in gp120 V1 and V4.32,33 Soluble Env trimers were his-tagged for protein purification and immobilization. Fluorescently labelable soluble Env IDL was generated by co-transfecting 293F cells with a 20:1 ratio of plasmids expressing untagged and V1V4-tagged IDL trimers supplemented by a furin plasmid. Recombinant Env trimers were then purified by affinity chromatography using a Ni-NTA column, followed by further purification by size-exclusion chromatography. The diluted ratio statistically ensured that most of the assembled Env trimers were unlabeled, while among fluorescently labeled Env trimers, the majority were single-protomer labeled.32,33 Purified BG505 IDL trimers were site-specifically labeled with donor and acceptor fluorophores at introduced peptide sites, followed by removing free dye using Zeba spin desalting columns. The donor dye was Cy3B-cadaverine, and the acceptor dye was Cy5-CoA (Lumidyne Technologies). The site-specific labeling chemistry was as previously described.32,33
smFRET experiments of fluorescently labeled Env proteins were performed on a customized prism-based total internal reflection fluorescence (prism-TIRF) microscope, as previously described.32,33 Labeled Env proteins were incubated with anti-6xhis biotin antibodies for 2 h on ice before imaging. Env proteins were further immobilized on passivated streptavidin-coated microfluidic chambers through biotin-streptavidin interactions. This was followed by inflexing an oxygen-scavenger (with triplet quencher mix) imaging buffer into the chambers to remove free Env proteins and prolong image durations. Immobilized Env trimers were excited by the evanescent field generated by the total internal reflection of a 532 nm CW laser (Opus, Laser Quantum). Fluorescence intensities were collected through a 60x Nikon objective, spectrally split by a multi-channel beam splitter MultiCam-LS device (Cairn), and simultaneously recorded by two ORCA-Flash4.0v2 sCMOS cameras (Hamamatsu) for 80 s with 40 ms exposure time. smFRET image stacks were processed by an SPARTAN software package64 and customized MATLAB-based scripts. Fluorescence trajectories (time series) were first extracted from image stacks. Extracted signals were further used to calculate the FRET efficiency trajectories based on FRET = IA/(ID + IA), where ID and IA are the fluorescence intensities of the donor and acceptor, respectively. The smFRET trajectories included for following histogram analysis were those with sufficient signal-to-noise ratio, prolonged FRET lifetime, and FRET-indicating negative correlation between donor and acceptor trajectories. More than one hundred FRET trajectories (130 for BG505 IDL and 165 for BG505 IDL SOSIP) were compiled into respective FRET population histograms. Histograms were then fitted into the sum of previously well-defined three-state Gaussian distributions,32,33 which correspond to three primary conformational states: a pre-sstriggered (~0.1-FRET centered Gaussian), pre-fusion closed (~0.65-FRET centered Gaussian), and the CD4-bound completely open (0.3-FRET centered Gaussian). Parameters and statistics for smFRET analysis are provided in Table S2.
Quantification and statistical analysisCα distances were measured using PyMol Measurement function. The IDL structures were submitted to Dali Server (http://ekhidna2.biocenter.helsinki.fi/dali/).65 Heuristic PBD search and All against all comparison methods were employed to compare new IDL structures against those deposited PBD. The RSMD values were calculated by Dali server. Unrelated, gp120 monomer, or redundant PDB structures were removed manually before statistical analysis. Statistically significant differences were determined by Mann-Whitney U test or paired t test in Prism.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Monoclonal anti-HIV-1 gp120 2G12 | NIH AIDS Reagent Program | Cat# 1476 |
| Monoclonal anti-HIV-1 gp120VRC01 | NIH AIDS Reagent Program | Cat# 12033 |
| Monoclonal anti-HIV-1 gp120 F105 | NIH AIDS Reagent Program | Cat# 857 |
| Monoclonal anti-HIV-1 gp120 10-1074 | NIH AIDS Reagent Program | Cat# 12477 |
| Monoclonal anti-HIV-1 gp120 PGT128 | John R. Mascola, VRC, NIH | N/A |
| Monoclonal anti-HIV-1 gp120 PG9 | John R. Mascola, VRC, NIH | N/A |
| Monoclonal anti-HIV-1 gp120 PG16 | John R. Mascola, VRC, NIH | N/A |
| Monoclonal anti-HIV-1 gp120 35O22 | NIH AIDS Reagent Program | Cat# 12586 |
| Monoclonal anti-HIV-1 gp120 447-52D | NIH AIDS Reagent Program | Cat# 4030 |
| Monoclonal anti-HIV-1 gp120 412d | John R. Mascola, VRC, NIH | N/A |
| Monoclonal anti-HIV-1 gp120 PGV04 | John R. Mascola, VRC, NIH | N/A |
| Monoclonal anti-HIV-1 gp120 CH31 | NIH AIDS Reagent Program | Cat# 12565 |
| Monoclonal anti-HIV-1 gp120 3BNC117 | NIH AIDS Reagent Program | Cat# 12474 |
| Monoclonal anti-HIV-1 gp120 HJ16 | NIH AIDS Reagent Program | Cat# 12138 |
| Monoclonal anti-HIV-1 gp120 b12 | NIH AIDS Reagent Program | Cat# 2640 |
| Monoclonal anti-HIV-1 gp120 17b | NIH AIDS Reagent Program | Cat# 4091 |
| Monoclonal anti-HIV-1 gp120 8ANC15 | John R. Mascola, VRC, NIH | N/A |
| Monoclonal anti-HIV-1 gp120 PGT145 | NIH AIDS Reagent Program | Cat# 12703 |
| Monoclonal anti-HIV-1 gp120VRC03 | John R. Mascola, VRC, NIH | N/A |
| Monoclonal anti-HIV-1 gp120VRC07 | John R. Mascola, VRC, NIH | N/A |
|
| ||
| Bacterial and Virus Strains | ||
|
| ||
| NEB 5-alpha Competent E. coli | New England BioLabs | Cat# C2987H |
|
| ||
| Chemicals, Peptides, and Recombinant Proteins | ||
|
| ||
| sCD4-183aa (2-domain) | NIH AIDS Reagent Program | Cat# 7356 |
| Human soluble CD4 | NIH AIDS Reagent Program | Cat# 4615 |
| Lectin from Galanthus nivalis | Sigma-Aldrich | Cat# L8275-5MG |
| BG505.T332N·SOSIP.664 gp140 trimer | This study | N/A |
| BG505.T332N SOSIP IDL gp140 trimer | This study | N/A |
| JR-FL.SOSIP.664 gp140 trimer | This study | N/A |
| JR-FL IDL gp140 trimer | This study | N/A |
| WITO IDL gp140 trimer | This study | N/A |
| DU422 IDL gp140 trimer | This study | N/A |
| CH0848 IDL gp140 trimer | This study | N/A |
|
| ||
| Critical Commercial Assays | ||
|
| ||
| Luciferase 1000 Assay System | Promega | Cat# E4550 |
| Alliance HIV-1 p24 Antigen ELISA | PerkinElmer | Cat# NEK050B001KT |
| QuickChange II Site-Directed Mutagenesis Kit | Agilent | Cat# 200524 |
| Plasmid Mega Kit (25) | QIAGEN | Cat# 12181 |
| LiFect293 | LifeSCT | Cat# M0002-01 |
|
| ||
| Deposited Data | ||
|
| ||
| PDB | 9BEW | BG505 IDL trimer |
| PDB | 9BER | JR-FL IDL trimer |
| PDB | 9BF6 | WITO IDL trimer |
| EMDB | 44484 | BG505 IDL trimer |
| EMDB | 44482 | JR-FL IDL trimer |
| EMDB | 44491 | WITO IDL trimer |
|
| ||
| Experimental Models: Cell Lines | ||
|
| ||
| Expi293F cells | ThermoFisher Scientific Inc | Cat# A14527 |
| FreeStyle 293-F cells | ThermoFisher Scientific Inc | Cat# R79007 |
|
| ||
| Recombinant DNA | ||
|
| ||
| pVRC8400 vector | https://www.addgene.org | Cat# 63160 |
| pVRC8400 vector-JR-FL SOSIP.664 | This study | N/A |
| pVRC8400 vector-BG505 IDL SOSIP.664 | This study | N/A |
| pVRC8400 vector-WITO IDL SOSIP.664 | This study | N/A |
| pVRC8400 vector-DU422 IDL SOSIP.664 | This study | N/A |
| pVRC8400 vector-CH0848 IDL SOSIP.664 | This study | N/A |
|
| ||
| Software and Algorithms | ||
|
| ||
| FlowJo v.9.9.4 | FlowJo, LLC | https://www.www.flowjo.com; RRID: SCR_008520 |
| QuikChange Primer Design | Agilent | https://www.agilent.com/store/primerDesignProgram.jsp |
| Pymol | The PyMOL Molecular Graphics System, Version 2.0 Schrodinger, LLC. | https://pymol.org/2/ |
| PRISM 8 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| Leginon | Suloway et al., 200552 | https://sbgrid.org/software/titles/leginon |
| CTFFind4 | Rohou and Grigorieff, 201554 | https://grigoriefflab.janelia.org/ctfflnd4 |
| Dog Picker | Voss et al., 200957 | https://omictools.com/dog-picker-tool |
| RELION | Scheres, 201258 | https://www3.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| cryoSPARC | Punjani et al., 201759 | https://cryosparc.com |
| EMRinger | Baradetal., 201563 | https://github.com/fraser-lab/EMRinger |
| Phenix | Adams et al., 200461 | https://sbgrid.org/software/ |
| Coot | Emsley and Cowtan, 200460 | https://sbgrid.org/software/ |
| Leginon | Suloway et al., 200552 | https://sbgrid.org/software/titles/leginon |
Highlights.
Introduction of an interdomain disulfide bond (IDL) stabilizes soluble HIV-1 envelope trimers
IDL trimers do not possess covalent bonds between gp120 and gp41 (such as the SOS)
IDL trimers from different HIV-1 clades display a native-like structure and antigenic profile
IDL trimers bind with high affinity to broad and potent neutralizing antibodies but not to CD4
ACKNOWLEDGMENTS
We thank Susan Zolla-Pazner and James E. Robinson for the gift of anti-Env monoclonal Abs and the AIDS Reagent Program for providing the reagents indicated in the STAR Methods section. This research was supported by the Intramural Programs of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases (NIAID), NIH, by the Vaccine Research Center, an intramural division of the NIAID, NIH, from an R37 grant (AI150560) from NIAID/NIH, to W.M., and from an R01 AI181600 grant from NIAID/NIH to M.L. Some of this work was performed at the Simons Electron Microscopy Center and the National Resource for Automated Molecular Microscopy, located at the New York Structural Biology Center, supported by grants from the Simons Foundation (SF349247) and from the NIH National Institute of General Medical Sciences (GM103310), with additional support from NYSTAR and the New York State Assembly.
Footnotes
DECLARATION OF INTERESTS
P.L. and P.Z. applied for a patent describing HIV-1 envelope trimers stabilized in a native-like, non-CD4-binding conformation (US Provisional Patent Application No. 62/306,006, filed on March 9, 2016, entitled “Recombinant HIV-1 envelope proteins and their use”).
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114518.
REFERENCES
- 1.Zhu P, Liu J, Bess J Jr., Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, and Roux KH (2006). Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441, 847–852. 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 2.Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, and Hendrickson WA (1998). Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659. 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, and Subramaniam S (2008). Molecular architecture of native HIV-1 gp120 trimers. Nature 455, 109–113. 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, and Moore JP (2000). A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74, 627–643. 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, et al. (2013). A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 9, e1003618. 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. (2013). Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490. 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. (2013). Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477–1483. 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pancera M, Zhou T, Druz A, Georgiev IS, Soto C, Gorman J, Huang J, Acharya P, Chuang GY, Ofek G, et al. (2014). Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature 514, 455–461. 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scharf L, Wang H, Gao H, Chen S, McDowall AW, and Bjorkman PJ (2015). Broadly Neutralizing Antibody 8ANC195 Recognizes Closed and Open States of HIV-1 Env. Cell 162, 1379–1390. 10.1016/j.cell.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart-Jones GB, Soto C, Lemmin T, Chuang GY, Druz A, Kong R, Thomas PV, Wagh K, Zhou T, Behrens AJ, et al. (2016). Trimeric HIV-1-Env Structures Define Glycan Shields from Clades A, B, and G. Cell 165, 813–826. 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ozorowski G, Pallesen J, de Val N, Lyumkis D, Cottrell CA, Torres JL, Copps J, Stanfield RL, Cupo A, Pugach P, et al. (2017). Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 547, 360–363. 10.1038/nature23010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schommers P, Gruell H, Abernathy ME, Tran MK, Dingens AS, Gristick HB, Barnes CO, Schoofs T, Schlotz M, Vanshylla K, et al. (2020). Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody. Cell 180, 471–489. 10.1016/j.cell.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roark RS, Li H, Williams WB, Chug H, Mason RD, Gorman J, Wang S, Lee FH, Rando J, Bonsignori M, et al. (2021). Recapitulation of HIV-1 Env-antibody coevolution in macaques leading to neutralization breadth. Science 371, eabd2638. 10.1126/science.abd2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Taeye SW, Ozorowski G, Torrents de la Pena, A., Guttman, M., Julien, J.P., van den Kerkhof, T.L., Burger, J.A., Pritchard, L.K., Pugach, P., Yasmeen, A., et al. (2015). Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes. Cell 163, 1702–1715. 10.1016/j.cell.2015.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chuang GY, Geng H, Pancera M, Xu K, Cheng C, Acharya P, Chambers M, Druz A, Tsybovsky Y, Wanninger TG, et al. (2017). Structure-Based Design of a Soluble Prefusion-Closed HIV-1 Env Trimer with Reduced CD4 Affinity and Improved Immunogenicity. J. Virol. 91, e02268–16. 10.1128/JVI.02268-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma SK, de Val N, Bale S, Guenaga J, Tran K, Feng Y, Dubrovskaya V, Ward AB, and Wyatt RT (2015). Cleavage-independent HIV-1 Env trimers engineered as soluble native spike mimetics for vaccine design. Cell Rep. 11, 539–550. 10.1016/j.celrep.2015.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong L, He L, de Val N, Vora N, Morris CD, Azadnia P, Sok D, Zhou B, Burton DR, Ward AB, et al. (2016). Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability. Nat. Commun. 7, 12040. 10.1038/ncomms12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rawi R, Rutten L, Lai YT, Olia AS, Blokland S, Juraszek J, Shen CH, Tsybovsky Y, Verardi R, Yang Y, et al. (2020). Automated Design by Structure-Based Stabilization and Consensus Repair to Achieve Prefusion-Closed Envelope Trimers in a Wide Variety of HIV Strains. Cell Rep. 33, 108432. 10.1016/j.celrep.2020.108432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwon YD, Pancera M, Acharya P, Georgiev IS, Crooks ET, Gorman J, Joyce MG, Guttman M, Ma X, Narpala S, et al. (2015). Crystal structure, conformational fixation and entry-related interactions of mature ligand-free HIV-1 Env. Nat. Struct. Mol. Biol. 22, 522–531. 10.1038/nsmb.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de la Pena AT, Julien JP, de Taeye SW, Garces F, Guttman M, Ozorowski G, Pritchard LK, Behrens AJ, Go EP, Burger JA, et al. (2017). Improving the Immunogenicity of Native-like HIV-1 Envelope Trimers by Hyperstabilization. Cell Rep. 20, 1805–1817. 10.1016/j.celrep.2017.07.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang P, Gorman J, Geng H, Liu Q, Lin Y, Tsybovsky Y, Go EP, Dey B, Andine T, Kwon A, et al. (2018). Interdomain Stabilization Impairs CD4 Binding and Improves Immunogenicity of the HIV-1 Envelope Trimer. Cell Host Microbe 23, 832–844. 10.1016/j.chom.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wrapp D, Mu Z, Thakur B, Janowska K, Ajayi O, Barr M, Parks R, Mansouri K, Edwards RJ, Hahn BH, et al. (2023). Structure-Based Stabilization of SOSIP Env Enhances Recombinant Ectodomain Durability and Yield. J. Virol. 97, e0167322. 10.1128/jvi.01673-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sliepen K, Han BW, Bontjer I, Mooij P, Garces F, Behrens AJ, Rantalainen K, Kumar S, Sarkar A, Brouwer PJM, et al. (2019). Structure and immunogenicity of a stabilized HIV-1 envelope trimer based on a group-M consensus sequence. Nat. Commun. 10, 2355. 10.1038/s41467-019-10262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, et al. (2015). HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349, aac4223. 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, Carnathan DG, Chandrashekar A, Cirelli KM, Cottrell CA, et al. (2017). Elicitation of Robust Tier 2 Neutralizing Antibody Responses in Nonhuman Primates by HIV Envelope Trimer Immunization Using Optimized Approaches. Immunity 46, 1073–1088. 10.1016/j.immuni.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pauthner MG, Nkolola JP, Havenar-Daughton C, Murrell B, Reiss SM, Bastidas R, Prevost J, Nedellec R, von Bredow B, Abbink P, et al. (2019). Vaccine-Induced Protection from Homologous Tier 2 SHIV Challenge in Nonhuman Primates Depends on Serum-Neutralizing Antibody Titers. Immunity 50, 241–252. 10.1016/j.immuni.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alsahafi N, Debbeche O, Sodroski J, and Finzi A (2015). Effects of the I559P gp41 change on the conformation and function of the human immunodeficiency virus (HIV-1) membrane envelope glycoprotein trimer. PLoS One 10, e0122111. 10.1371/journal.pone.0122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alsahafi N, Anand SP, Castillo-Menendez L, Verly MM, Medjahed H, Prevost J, Herschhorn A, Richard J, Schon A, Melillo B, et al. (2018). SOSIP Changes Affect Human Immunodeficiency Virus Type 1 Envelope Glycoprotein Conformation and CD4 Engagement. J. Virol. 92, e01080–18. 10.1128/JVI.01080-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen HT, Alsahafi N, Finzi A, and Sodroski JG (2019). Effects of the SOS (A501C/T605C) and DS (I201C/A433C) Disulfide Bonds on HIV-1 Membrane Envelope Glycoprotein Conformation and Function. J. Virol. 93, e00304–19. 10.1128/JVI.00304-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tong T, D’Addabbo A, Xu J, Chawla H, Nguyen A, Ochoa P, Crispin M, and Binley JM (2023). Impact of stabilizing mutations on the antigenic profile and glycosylation of membrane-expressed HIV-1 envelope glycoprotein. PLoS Pathog. 19, e1011452. 10.1371/journal.ppat.1011452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ringe RP, Colin P, Torres JL, Yasmeen A, Lee WH, Cupo A, Ward AB, Klasse PJ, and Moore JP (2019). SOS and IP Modifications Predominantly Affect the Yield but Not Other Properties of SOSIP.664 HIV-1 Env Glycoprotein Trimers. J. Virol. 94, e01521–19. 10.1128/JVI.01521-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB 3rd, Kwong PD, et al. (2014). Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 346, 759–763. 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu M, Ma X, Castillo-Menendez LR, Gorman J, Alsahafi N, Ermel U, Terry DS, Chambers M, Peng D, Zhang B, et al. (2019). Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 568, 415–419. 10.1038/s41586-019-1101-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JH, Ozorowski G, and Ward AB (2016). Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351, 1043–1048. 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Torrents de la Peña A., Rantalainen K., Cottrell CA., Allen JD., van Gils MJ., Torres JL., Crispin M., Sanders RW., and Ward AB. (2019). Similarities and differences between native HIV-1 envelope glycoprotein trimers and stabilized soluble trimer mimetics. PLoS Pathog. 15, e1007920. 10.1371/journal.ppat.1007920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pan J, Peng H, Chen B, and Harrison SC (2020). Cryo-EM Structure of Full-length HIV-1 Env Bound With the Fab of Antibody PG16. J. Mol. Biol. 432, 1158–1168. 10.1016/j.jmb.2019.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stadtmueller BM, Bridges MD, Dam KM, Lerch MT, Huey-Tubman KE, Hubbell WL, and Bjorkman PJ (2018). DEER Spectroscopy Measurements Reveal Multiple Conformations of HIV-1 SOSIP Envelopes that Show Similarities with Envelopes on Native Virions. Immunity 49, 235–246. 10.1016/j.immuni.2018.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Q, Acharya P, Dolan MA, Zhang P, Guzzo C, Lu J, Kwon A, Gururani D, Miao H, Bylund T, et al. (2017). Quaternary contact in the initial interaction of CD4 with the HIV-1 envelope trimer. Nat. Struct. Mol. Biol. 24, 370–378. 10.1038/nsmb.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Q, Lai YT, Zhang P, Louder MK, Pegu A, Rawi R, Asokan M, Chen X, Shen CH, Chuang GY, et al. (2019). Improvement of antibody functionality by structure-guided paratope engraftment. Nat. Commun. 10, 721. 10.1038/s41467-019-08658-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JH, Andrabi R, Su CY, Yasmeen A, Julien JP, Kong L, Wu NC, McBride R, Sok D, Pauthner M, et al. (2017). A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic beta-Hairpin Structure. Immunity 46, 690–702. 10.1016/j.immuni.2017.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rutten L, Lai YT, Blokland S, Truan D, Bisschop IJM, Strokappe NM, Koornneef A, van Manen D, Chuang GY, Farney SK, et al. (2018). A Universal Approach to Optimize the Folding and Stability of Prefusion-Closed HIV-1 Envelope Trimers. Cell Rep. 23, 584–595. 10.1016/j.celrep.2018.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Derking R, and Sanders RW (2021). Structure-guided envelope trimer design in HIV-1 vaccine development: a narrative review. J. Int. AIDS Soc. 24, e25797. 10.1002/jia2.25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olia AS, Cheng C, Zhou T, Biju A, Harris DR, Changela A, Duan H, Ivleva VB, Kong WP, Ou L, et al. (2023). Soluble prefusion-closed HIV-envelope trimers with glycan-covered bases. iScience 26, 107403. 10.1016/j.isci.2023.107403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwon YD, Chuang GY, Zhang B, Bailer RT, Doria-Rose NA, Gindin TS, Lin B, Louder MK, McKee K, O’Dell S, et al. (2018). Surface-Matrix Screening Identifies Semi-specific Interactions that Improve Potency of a Near Pan-reactive HIV-1-Neutralizing Antibody. Cell Rep. 22, 1798–1809. 10.1016/j.celrep.2018.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mastronarde DN (2005). Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51. 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 46.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, and Ludtke SJ (2007). EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46. 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, and Leith A (1996). SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199. 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 48.Scheres SH, Nunez-Ramirez R, Sorzano CO, Carazo JM, and Marabini R (2008). Image processing for electron microscopy single-particle analysis using XMIPP. Nat. Protoc. 3, 977–990. 10.1038/nprot.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, and Frank J (2008). SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 3, 1941–1974. 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, and Ferrin TE (2004). UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 51.Pintilie GD, Zhang J, Goddard TD, Chiu W, and Gossard DC (2010). Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 170, 427–438. 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, and Carragher B (2005). Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60. 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 53.Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, and Agard DA (2017). MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332. 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rohou A, and Grigorieff N (2015). CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221. 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang K (2016). Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12. 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. (2009). Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102. 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voss NR, Yoshioka CK, Radermacher M, Potter CS, and Carragher B (2009). DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213. 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Scheres SH (2012). RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530. 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Punjani A, Rubinstein JL, Fleet DJ, and Brubaker MA (2017). cryo-SPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296. 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 60.Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 61.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davis IW, Murray LW, Richardson JS, and Richardson DC (2004). MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615–W619. 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barad BA, Echols N, Wang RY, Cheng Y, DiMaio F, Adams PD, and Fraser JS (2015). EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy. Nat. Methods 12, 943–946. 10.1038/nmeth.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Juette MF, Terry DS, Wasserman MR, Altman RB, Zhou Z, Zhao H, and Blanchard SC (2016). Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat. Methods 13, 341–344. 10.1038/nmeth.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holm L, Laiho A, Toronen P, and Salgado M (2023). DALI shines a light on remote homologs: One hundred discoveries. Protein Sci. 32, e4519. 10.1002/pro.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps of IDL trimers have been submitted to the PDB database under accession codes 9BEW, 9BF6 and 9BER and to the EMDB database under accession codes 4484, 44482, 44491.
This paper does not report original code;
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


