Abstract
In standard neutralization (STAN), virus and antibody are reacted together before inoculation of target cells, and inhibition of almost any of the processes concerned in the early interaction of virus and cell, including inhibition of virus attachment to cell receptors, can be the cause of neutralization by a particular monoclonal antibody (MAb). To simplify the interpretation of antibody action, we carried out a study of postattachment neutralization (PAN), where virus is allowed to attach to target cells before neutralizing antibody is introduced. We used influenza virus A/PR/8/34 (H1N1) and monoclonal immunoglobulin G (IgG) molecules and their Fabs specific to antigenic sites Sb (tip), Ca2 (loop), and Cb (hinge) of the hemagglutinin 1 (HA1) protein. All IgGs and Fabs gave PAN, although with reduced efficiency compared with STAN. Thus, bivalent binding of antibody was not essential for PAN. By definition, none of these MAbs gave PAN by inhibiting virus attachment, and they did not elute attached virus from the target cell or inhibit endocytosis of virus. However, virus-cell fusion, as demonstrated by R18 fluorescence dequenching or hemolysis of red blood cells, was inhibited in direct proportion to neutralization and in a dose-dependent manner and was thus likely to be responsible for the observed neutralization. However, to get PAN, it was necessary to inhibit the activation of the prefusion intermediate, the earliest known form on the fusion pathway that is created when virus is incubated at pH 5 and 4°C. PAN antibodies may act by binding HA trimers in contact with the cell and/or trimers in the immediate vicinity of the virus-cell contact point and so inhibit the recruitment of additional receptor-HA complexes.
Conventional or standard neutralization (STAN) takes place when antibodies bind to virus free in solution. The resulting loss of infectivity can occur by a variety of mechanisms, broadly by aggregation of virions, inhibition of attachment of virus to cell receptors on the target cell, inhibition of virus internalization, inhibition of the entry of the viral genome and associated proteins into the cell, or inhibition of a postentry event (15). However, some antibodies can also neutralize virus that has already attached to the cell, a process called postattachment neutralization (PAN). PAN is of interest since, by definition, it cannot take place by aggregating free virus or by inhibiting virus attachment to the target cell, but it has not been extensively studied and there is little information about the mechanisms of PAN. There have been no reports on the mediation of PAN by Fab fragments.
PAN has been reported for a number of different virus systems, and it is evident that not all monoclonal antibodies (MAbs) that mediate STAN can also mediate PAN. Examples of PAN have been reported with enterovirus 71 (27), poliovirus (54), Venezuelan equine encephalitis virus (42), rotavirus (43), influenza A virus (25), respiratory syncytial virus (34), Newcastle disease virus (44), transmissible gastroenteritis virus (52), vesicular stomatitis virus (7), rabies virus (14), adenovirus (58), human cytomegalovirus (33), African swine fever virus (20), and human immunodeficiency virus type 1 (HIV-1) (1, 3, 4, 11, 26, 29, 31, 37). Sensitivity to PAN has been used as a means of demonstrating the kinetics of virus endocytosis or of virus genome entry into the cell of viruses that fuse with the plasma membrane at neutral pH. The window available for PAN is generally short, on the order of a few minutes, for viruses being endocytosed (see, e.g., reference 44), or quite long (over 60 min) for viruses fusing with the plasma membrane at pH 7 (34). However, the situation is probably more complicated. For example, with HIV-1 the occurrence of PAN was dependent on the epitope, with one MAb giving PAN for up to 90 min of incubation at 37°C (and probably until fusion had been completed) while another MAb gave PAN for only 15 min at 37°C and yet another gave no PAN at all even with infection at 4°C when virus remained at the cell surface (3, 4). Others reported similar data with a different MAb (29). The abbreviation or absence of PAN may reflect the disappearance of epitopes, following changes in conformation of the envelope protein that take place when virus binds to cell receptors and the fusion mechanism is slowly activated (46, 47).
The only report of influenza virus PAN known to the authors showed that virus attached to cells at 3°C was completely susceptible to PAN by polyclonal antiserum to whole virus but at 37°C rapidly became resistant to PAN (25). The mechanism of influenza virus PAN has never been addressed, although there are limited studies of other systems. PAN of rabies virus showed that neutralizing MAbs caused a threefold increase in the release of cell-bound virus (14). The remaining virus was endocytosed but was not infectious, leading to the presumption that PAN was mediated by a postinternalization mechanism. A MAb to the VP8* protein of the rhesus monkey rotavirus also mediated PAN by releasing virus from target cells (43). Vesicular stomatitis virus was susceptible to PAN by a G protein-specific antiserum, but this did not elute virus from the cell. Rather, PAN inhibited the fusion of viral and cell membranes in endosomes, probably by decreasing the sensitivity of G protein to the low-pH-induced conformational changes, although the effect was marginal (7). African swine fever virus was susceptible to PAN by convalescent-phase serum, which prevented internalization of virus by the target cell (20). With poliovirus, only 6 of 19 MAbs that gave standard neutralization mediated PAN, and these did so by preventing uncoating and the conversion of native 135S particles to 80S particles (54). It was concluded that that bivalent binding of MAbs to virions was essential for PAN.
In this work we studied the PAN of a human influenza A virus, mediated by three high-affinity HA1-specific immunoglobulin G (IgG) molecules (H9, H36, and H37) and their Fabs. H36 IgG and Fab and H9 IgG and Fab had similar affinities, suggesting that the IgGs bound monovalently. H37 probably bound bivalently. All IgGs mediated STAN by a combination of inhibition of virus-cell fusion and inhibition of virus attachment to cells. H36 and H37 Fabs mediated STAN solely by inhibiting attachment, while H9 Fabs mediated STAN solely by inhibiting fusion (17, 18). Here we found that all the IgGs and Fabs mediated PAN and that they all acted by inhibiting an early event in the viral low-pH-dependent fusion process. The mechanism(s) of PAN and the differences that we observed between PAN and STAN are discussed.
MATERIALS AND METHODS
Virus.
Influenza virus A/Puerto Rico/8/34 (Mount Sinai strain; H1N1; PR8) was grown in the allantoic cavities of 10-day-old embryonated chicken eggs (Poynden Egg Farm, Goss Oaks, United Kingdom) for 48 h at 33°C. Virus was purified by differential centrifugation and banded on a 10 to 45% (wt/vol) sucrose gradient at 60,000 × g for 90 min and a 20 to 70% (wt/vol) sucrose equilibrium gradient at 60,000 × g for 16 h. Virus was stored at −70°C.
Hemagglutination and the hemagglutination inhibition assay.
Virus was double diluted in phosphate-buffered saline (PBS) in a round-bottom 96-well plate (Greiner, Stonehouse, United Kingdom) and detected by agglutination of 0.13% adult chicken red blood cells (RBCs; Serotech, Kidlington, United Kingdom). The 50% agglutination point (1 HAU) was estimated by interpolation between complete agglutination and no agglutination, and the titer was expressed as the reciprocal of the dilution in HAU per milliliter. For hemagglutination inhibition, antibodies were doubly diluted and mixed with 4 HAU per well for 1 h at 20°C. Uninhibited virus was detected by agglutination of RBCs as above, and the reciprocal of the dilution giving 50% inhibition (1 HIU) was taken as the end point.
Cells.
Madin-Darby canine kidney (MDCK) cells (kindly provided by Wendy Barclay) were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco BRL Life Technologies, Paisley, United Kingdom)–4 mM glutamine (Gibco)–5% (vol/vol) heat-inactivated fetal calf serum (HIFCS; Gibco)–50 μg of gentamicin (Gibco) per ml. Mouse hybridomas were grown in RPMI 1640 (Gibco)–10% HIFCS–2 mM glutamine–50 μg of gentamicin per ml.
Antibodies.
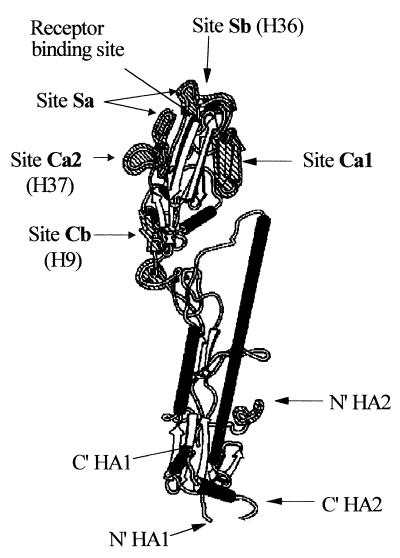
The PR8 HA1-specific MAbs H36-4.5-2 (antigenic site Sb or B of H3, IgG2a), H37-45-5R3 (site Ca2/A, IgG3), and H9-D3-4R2 (site Cb/E, IgG3) (10, 50) were used for PAN. Reverse transcription-PCR and sequencing of neutralization escape mutants showed that there were inferred single-residue changes in the expected antigenic sites. For H36 IgG these were mainly 152D→K, for H37 IgG they were mainly 140K→D, and for H9 it was 75S→P (A. C. Marriott, C. Parry, and N. J. Dimmock, unpublished data). Another HA1-specific MAb used was H37-66-1 (site Sb, IgA). In addition, we used the following HA1-specific MAbs that are specific to low-pH-induced epitopes: Y8-10C2 (site Sa), H2-4C2 (site Cb), H3-4C5 (site Ca2), H18-S13 (site Cb), and H17-L2, which recognizes an epitope in site Ca2 that is lost at low pH (59). All these MAbs were kindly supplied by Walter Gerhard. The NP-specific MAb HB-67 (IgG1) was supplied by Robert G. Webster. MAbs were purified by affinity chromatography on a protein A-Sepharose column (Sigma) as described previously (17).
Production of Fabs by digestion of IgG with papain.
IgG2a and IgG3 required different conditions for digestion with immobilized papain, as detailed before (17). Briefly, IgG2a (1 mg) was mixed with 6.2 μl of freshly made 0.01 M cysteine–20 μl EDTA–1 U agarose-immobilized papain (Sigma). This was adjusted to pH 5.5 and shaken for 3 h at 37°C. IgG3 was treated similarly except with 1 M cysteine. Papain was removed by pelleting, the digest was made 5.5 mM with respect to iodoacetamide (Sigma), and the pH was raised to 7.5. Undigested IgG and Fc fragments were removed by repeated passage through protein A-Sepharose. Fabs were dialyzed and concentrated, and their concentration was determined spectrophotometrically (1 optical density [OD] unit = 1.5 mg/ml [24]). All preparations had the expected Mr of approximately 50,000 by polyacrylamide gel electrophoresis (PAGE) and were free of detectable IgG (≤50 ng/ml).
STAN assays.
STAN was measured by plaque assay and by enzyme-linked immunosorbent assay (ELISA). In the plaque assay, virus (120 PFU/100 μl) was mixed with an equal volume of antibody for 1 h at 37°C. MDCK monolayers in six-well plates (Falcon) were rinsed to remove serum and then inoculated with the virus-MAb mix for 20 min at 4°C. Excess virus and antibody were removed by washing three times with cold PBS, and monolayers were overlaid with 0.9% agar (Gibco) in medium 199 containing 0.2% (wt/vol) bovine serum albumin (BSA), 0.01% (wt/vol) DEAE-dextran, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 30 U of TPCK trypsin (Sigma, Poole, United Kingdom) per ml. After incubation for 3 days at 33°C, the monolayers were fixed with formal-saline and stained for plaque counting with toluidine blue (BDH). Neutralization was estimated by taking the percentage of plaques remaining after MAb treatment and subtracting it from 100.
For STAN by ELISA, 20 to 200 HAU of virus was incubated with an equal volume of MAb for 1 h at 37°C and then 100 μl was inoculated onto monolayers in 96-well plates as above. After being washed, the plate was incubated overnight at 37°C in 100 μl of DMEM–1% HIFCS, 4 mM glutamine–20 μg of gentamicin per ml in the absence of trypsin so that virus replicated only within the original infected cell. This maintained the direct response of the neutralization assay. Monolayers were then fixed with 2% paraformaldehyde and blocked with 3% BSA in TBS (0.02 M Tris-HCl, 0.14 M NaCl [pH 7.6]) at 20°C for 90 min. De novo expressed HA on the cell surface was assayed as a measure of virus replication and detected with monoclonal mouse anti-HA IgA (H37-66-1), an anti-IgA–alkaline phosphatase conjugate (Sigma), and dinitrophenyl phosphate (DNPP). The product was read on an optical plate reader (Titertek Multiscan Plus) at 405 nm, where the virus control had an OD of approximately 1.1. When experimental procedures were carried out with 3-cm-diameter dishes, the PAN ELISA was made in the same format to ensure comparability. The procedure was the same as for the 96-well ELISA, except that the volumes were scaled up. The colored product was then transferred to 96-well plates for reading.
PAN assays.
PAN was measured by plaque assay and by ELISA. In the plaque assay, virus was inoculated onto MDCK monolayers in six-well plates as described above at 4°C for 20 min. After washing to remove excess virus, antibody was added for 120 min at 4°C or for 60 min at 37°C. Antibody was then removed by washing, and plaques were allowed to develop under nutrient agar. For neutralization in 96-well plates, a PAN ELISA was used in which monolayers were inoculated with 10 to 100 HAU of virus for 20 min at 4°C. After washing, dilutions of MAb were added as described. Monolayers were then washed and incubated overnight at 37°C as described for STAN.
For determination of the kinetics of PAN, virus was allowed to attach to cells at 4°C as above and incubation was continued at 4 or 37°C. Virus was removed and antibody was added at intervals. This was left on the monolayer for 60 min before being removed by washing. All monolayers were then overlaid and incubated at 37°C for plaques to develop.
Assay for the internalization of virus by cells by ELISA.
The ELISA for virus internalization was carried out as detailed previously (17). Monolayers in 96-well plates were inoculated and PAN was carried out as described above. After being washed, the monolayers were incubated with warm DMEM at 37°C for 30 min to allow the attached virus to be internalized. Noninternalized virus was removed by treating monolayers twice with 0.025 U of Clostridium perfringens neuraminidase at 37°C for 10 min. This removed more than 95% of the attached virus as verified by ELISA, as described previously (17). The monolayers were then permeabilized by freeze-thawing three times and fixed by adding 80 μl of saline followed by 150 μl of methanol at −20°C for 30 min. Virus was detected as virion NP antigen as described above. To show that virus was being internalized by receptor-mediated endocytosis, we used conditions that are known to inhibit this process: either maintaining cells at 4°C (30, 40) or incubating them in hypertonic medium (0.45 M sucrose in medium) at 20°C for 30 min before inoculation of virus. This latter treatment prevents the clathrin lattice formation required for receptor-mediated endocytosis (13, 22). PAN by ELISA was determined at the same time in the same 96-well system.
Assay for virus-cell fusion using virus labeled with R18.
The assay for virus-cell fusion was carried out as detailed before (17). Virus (500 μl of 107 HAU of freshly prepared purified stock) was incubated with 85.5 μM octadecyl rhodamine B chloride (R18; Molecular Probes Europe BV, Leiden, The Netherlands) for 1 h at 20°C in the dark. After centrifugation to remove any precipitate, virus was separated from free R18 by pelleting the virus through 20% sucrose and stored at −70°C. Solubilization of R18-labeled virus in 1% Triton X-100 (BDH) gave a 150-fold increase in fluorescence, indicating that the R18 was self-quenched and incorporated into the virus lipid bilayer. For the assay of fusion or inhibition of fusion, virus (200 HAU in 100 μl) was inoculated onto a 3-cm-diameter monolayer of MDCK cells at 4°C for 25 min. Unattached virus was removed, and antibody was added as before. After being washed, the monolayers were incubated in warm DMEM for 30 min at 37°C. The cells were detached by incubation with cold EDTA for 5 min at 20°C, pelleted, and fixed in cold 2% paraformaldehyde in the dark. Fluorescence was determined, after solubilizing in Triton X-100, with a Luminescence Spectrophotometer LS-5 (Perkin-Elmer Ltd., Beaconsfield, United Kingdom), exciting at 560 nm and emitting at 590 nm, with an emission slit width of 10 nm. To demonstrate that the increase of fluorescent signal was due to virus-cell fusion, cells inoculated with R18-labeled virus were kept at 4°C to inhibit fusion (30, 40) or were treated, prior to infection, with 500 nM bafilomycin A1 from Streptomyces griseus [Calbiochem: Novabiochem (UK) Ltd., Beeston, United Kingdom] at 37°C for 90 min. Bafilomycin is a V-ATPase inhibitor that prevents the acidification of endosomal vesicles and the triggering of HA-mediated fusion (21, 32, 36). The fusion ability of neutralized virus was calculated as a percentage of that achieved by the nonneutralized virus control. To keep the assays comparable, neutralization was determined by PAN ELISA with the same virus-antibody mixtures and in the same 3-cm-diameter dish system as used for the fusion assays.
Inhibition of hemolysis of chicken RBCs by influenza virus.
Hemolysis and hemolysis inhibition assays were adapted from earlier work (45). Virus (10,000 HAU/ml) was incubated with 200 μl of 8% RBCs at 4°C for 30 min on a rotating mixer. The cells were then pelleted to remove unattached virus, washed, and resuspended in 200 μl of diluent or antibody at 37°C for 60 min. After being washed again, RBCs were resuspended in 250 μl of citrate buffer (pH 5) at 37°C for 45 min for hemolysis to take place. The cells were pelleted, and the amount of hemolysis (released hemoglobin) was determined spectrophotometrically at 520 nm. The percent inhibition of hemolysis was calculated by comparing the extent of hemolysis in the virus control to the hemolysis found in the presence of antibody. In other experiments, the effect of neutralization on the hemolysis properties of the prefusion intermediate was investigated. Virus was attached to RBCs as described above and then treated at 4°C in citrate buffer (pH 5) for 30 min to induce the prefusion intermediate. The pH was then adjusted to neutral, and the prefusion intermediate was treated with antibody. Hemolysis was assayed as before.
Detection of low-pH-induced conformational changes in the HA by ELISA.
An ELISA plate was coated overnight with 100 HIU of an HA1-specific IgA (H37-66-1) and blocked with 3% BSA in TBS-Tween for 2 h. The IgA was then used to capture virus (100 HAU), which was then treated at pH 5 or 7.5 at 37°C. The pH was then adjusted to 7.5, and the reactivity of virus with various HA1-specific IgGs was determined. In another experiment, virus (200 HAU) was captured on a monolayer of paraformaldehyde-fixed MDCK cells at 4°C. Unattached virus was removed by washing, and bound virus was incubated with Fab at 4 or 37°C for 2 or 1 h, respectively. These temperatures were then maintained for the rest of the experiment. After washing, citrate buffer was added, with half the plate at pH 5 and the other half at pH 7.5 for 30 min. The pH was returned to neutral, and MAbs in 1% BSA in TBS-Tween were added for 1 h at 4°C to detect pH-induced changes in epitopes of HA1. Unbound MAb was removed, and the cells were treated with methanol at −20°C for 20 min and blocked with 3% BSA in TBS-Tween. Bound MAb was detected at room temperature using a rabbit antiserum specific for the Fc region of mouse IgG conjugated with alkaline phosphatase followed by DNPP in diethanolamine buffer.
Detection of low-pH-induced conformational changes in the HA by acquisition of sensitivity to digestion with proteinase K.
Purified virus (100,000 HAU/ml) was incubated with antibody at 37°C for 1 h. The pH of the neutralization mix was then kept at pH 7.5 or lowered to pH 5 by the addition of 0.1 M HCl and incubated at 37°C for 30 min. The pH of the neutralization mix was then restored to neutrality. The change of the HA to the low-pH conformation was detected by incubation with proteinase K (1 μg/ml at 37°C for 30 min). Digestion was stopped by the addition of cracking buffer with dithiothreitol, and the digestion mix was analyzed on a 12% polyacrylamide gel under reducing conditions. The gel was fixed, and proteins were stained with colloidal Coomassie brilliant blue.
Data analysis.
Dose-response curves were calculated by nonlinear regression analysis using Prism Graphpad software. To determine if they were significantly different, we compared them using an unpaired t test, where P < 0.05 is considered significant.
RESULTS
Comparison of PAN and STAN mediated by IgGs.
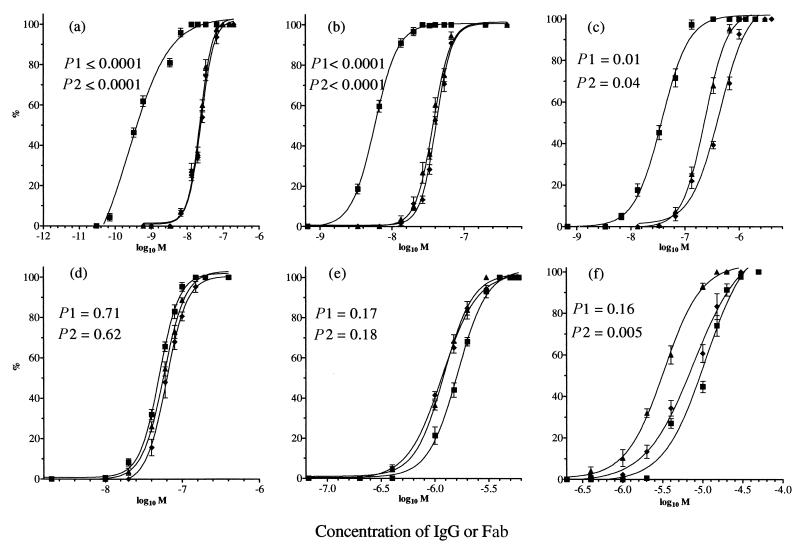
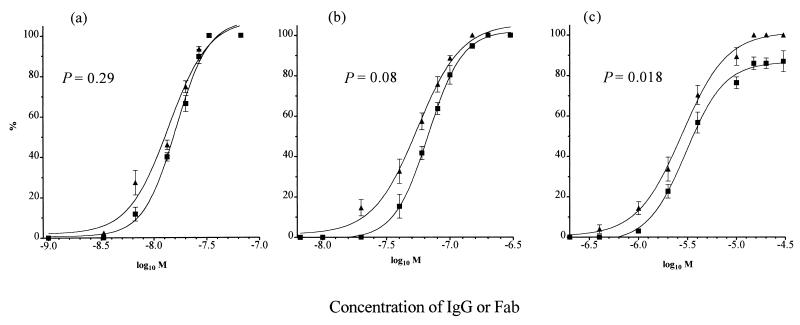
PAN was investigated using three HA1-specific IgGs, H36 (IgG2a, site Sb), H37 (IgG3, site Ca2), and H9 (IgG3, site Cb) (Fig. 1). Virus was allowed to attach to MDCK cell monolayers at 4°C, and then dose-response curves were constructed at 4°C, to give the IgG the most opportunity to act, and at 37°C, to mimic physiological conditions. STAN assays were carried out at the same time, with the same virus and the same batch of monolayers. Figure 2a to c show that all IgGs mediated PAN and that this was essentially the same at both 4 and 37°C. However, PAN by all three MAbs was significantly reduced compared to STAN (P values are given in Fig. 2), with a difference of 37-fold at N50 (the concentration required for 50% neutralization) for H36 but only 8-fold for H37 and H9 (Table 1). The differences were less at N90.
FIG. 1.
Model of an HA1 monomer of PR8 influenza virus showing the positions of the antigenic sites and epitopes of antibodies used in this study (adapted from references 10 and 56).
FIG. 2.
Comparison of the percent PAN and STAN of PR8 virus by plaque assay in MDCK cell monolayers, as a function of IgG or Fab concentration (M). For PAN, virus was allowed to attach to a prechilled monolayer at 4°C and washed, and then antibody was added at 4°C for 120 min or 37°C for 60 min. For STAN, virus-antibody mixtures were incubated together at 20°C and inoculated onto monolayers at 37°C for 60 min. Monolayers were washed to remove virus and antibody before being overlaid with agar. (a and d) H36 IgG (a) and Fab (d); (b and e) H37 IgG (b) and Fab (e); (c and f) H9 IgG (c) and Fab (f). ⧫, PAN at 4°C; ▴, PAN at 37°C; ■, STAN at 37°C. Virus controls gave approximately 50 PFU/plate. Data are the mean of three experiments, and the bars represent the standard error of the mean. Curves were generated by Prism Graphpad software. R was >0.97 for all curves. The value of P1 shows the significance of the difference between the two curves of 4°C PAN and 37°C STAN, and that of P2 shows the same for 37°C PAN and 37°C STAN. These were calculated using an unpaired t test. P < 0.05 is considered significant.
TABLE 1.
Comparison of PAN and STAN at 37°C
| MAb | IgG
|
Fab
|
||
|---|---|---|---|---|
| PAN50/STAN50 | PAN90/STAN90 | PAN50/STAN50 | PAN90/STAN90 | |
| H36 | 36.7 | 10.9 | 1.0 | 1.0 |
| H37 | 8.3 | 5.2 | 1.0 | 1.0 |
| H9 | 7.5 | 7.0 | 0.48 | 0.34 |
Comparison of PAN and STAN mediated by Fabs.
All three Fabs mediated PAN, but the N50 of each was reduced by 3.6- to 18-fold compared with that of IgG (Table 2). PAN curves for H36 and H37 Fabs were the same at 4 and 37°C and, in contrast to the IgGs, did not differ significantly from STAN curves (Fig. 2d and e; Table 1). H9 Fab differed in two aspects: (i) PAN at 37°C was significantly higher than STAN at 37°C, with an N50 and N90 that were two- and threefold higher, respectively (Fig. 2f; Table 1), and (ii) PAN at 37°C was greater than PAN at 4°C. This may be because the H9 epitope is more accessible to Fab at 37°C than at 4°C.
TABLE 2.
Summary of data on PAN of PR8
| MAb | Antigenic site | N50 (nM)
|
Inhibition of internalization | Inhibition of fusion | Inhibition of hemolysis | Inhibition of prefusion intermediate | Inhibition of exposure of low-pH epitopes | Inhibition of gain in protease sensitivity at low pH | |
|---|---|---|---|---|---|---|---|---|---|
| IgG | Fabc | ||||||||
| H36 | Sb | 22 | 78 (3.6) | No | Yes | Yes | Yes | No | No |
| H37 | Ca2 | 58 | 1,060 (18.3) | No | Yes | Yes | Yes | No | NDb |
| H9 | Cb | 340 | 4,400 (12.9) | No | Yesa | Yesa | Yes | No | ND |
H9 Fab inhibited fusion to a plateau of approximately 80%; the others inhibited fusion completely. Columns 4 to 7 summarize the activity of all IgGs and Fabs, and columns 8 and 9 show the activity of the Fabs only.
ND, not done.
Values in parentheses are Fab N50/IgG N50 ratios.
Kinetics of PAN mediated by H36 IgG and Fab.
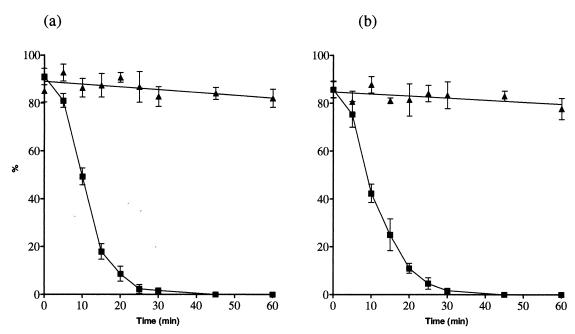
Attachment of virus to cells at 4°C followed by incubation at 4 or 37°C allows the window of time during which PAN can take place to be determined. In the experiment in Fig. 3, virus was allowed to attach and was then incubated at the temperature indicated. Antibody was added at intervals at the times indicated by the experimental points. Figure 3 shows that at 4°C the virus was susceptible to PAN by IgG and Fab for the entire incubation period while at 37°C PAN was reduced by 50% in 10 min and by 90% in 20 min. PAN by an equivalent neutralizing concentration of Fab followed very similar kinetics (Fig. 3b). Similar kinetics were also followed by PAN with H37 and H9 IgGs (data not shown). Such a loss of PAN can mean that the virus has been internalized by the cell, that the epitope is no longer available, or that the pathway through which neutralization is mediated has been closed as a result of virus-cell interactions.
FIG. 3.
Analysis of the kinetics of PAN of PR8 virus by H36 IgG and Fab at 4 and 37°C in MDCK cell monolayers, as a function of antibody concentration (M). PAN is expressed as a percentage. (a) IgG (55 nM); (b) Fab (100 nM). Data were generated from plaque assays and are the mean of two experiments. Bars represent the standard error of the mean. Curves were generated as in Fig. 2. ▴, 4°C; ■, 37°C.
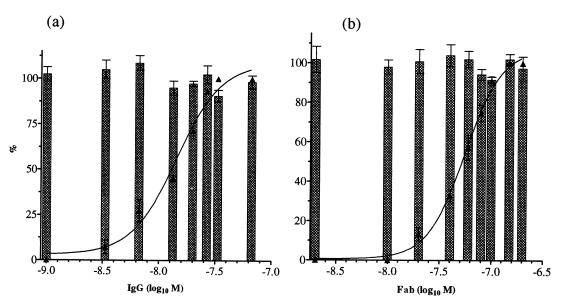
Mechanisms of PAN. (i) IgGs and Fabs did not elute virus from the target cell.
The possibility that PAN resulted from virus being eluted from the cell was investigated by carrying out PAN and then determining how much virus had been internalized by the cell. This was done by determining its acquired resistance to removal from the cell by treatment with bacterial neuraminidase. Figure 4 shows that there was no loss of virus internalization at any of the IgG or Fab concentrations used. Similar data were obtained with H37 and H9 IgGs and their Fabs (data not shown). Controls designed to demonstrate that virus in this system was being internalized by endocytosis were carried out either by keeping cells at 4°C to inhibit the endocytotic process or by treating monolayers with hypertonic sucrose to disperse the clathrin lattice around endocytic vesicles. These were found to result in a loss of ≥90 or ≥79% of virus internalization, respectively (data not shown).
FIG. 4.
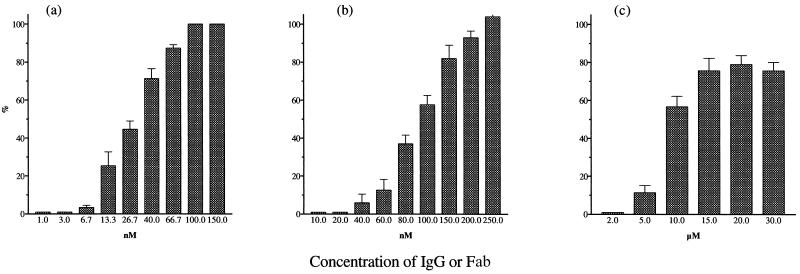
Analysis of the relationship between PAN and internalization of PR8 virus by MDCK cell monolayers as a function of antibody concentration (M). PAN and internalization were assayed in parallel in the same batch of monolayers in 96-well trays, both by ELISA, and are expressed as a percentage. (a) H36 IgG; (b) Fab. ▴, PAN. Columns represent internalization. The internalization virus control gave values of 1.2 to 1.4 OD units, and the PAN virus control gave a value of 1.1 OD units. Data are the mean of three experiments, and the bars represent the standard error of the mean. Curves were generated as in Fig. 2. R > 0.97 for all curves.
(ii) IgGs and Fabs inhibited fusion of virus with the target cell in direct proportion to neutralization.
Virus labeled with the fluorescent dye R18 was allowed to attach to cells, and then PAN was carried out over a range of concentrations with IgG or Fab. Neutralization and virus-cell fusion (by fluorescence dequenching) were then determined using the same batch of cells. Control experiments to demonstrate that R18 dequenching did in fact result from virus-cell fusion were carried out at 4°C or by treating monolayers with bafilomycin to inhibit the protonation of endocytic vesicles. These conditions resulted in fusion activity losses of ≥87% at 4°C and of ≥79% with bafilomycin (data not shown). Figure 5a and b show that the neutralization and virus-cell fusion curves did not differ significantly with either H36 IgG or Fab. Similar data were obtained with H37 and H9 IgG and H37 Fab (not shown). However, with H9 Fab there was a small but significant difference between the amounts of neutralization at N50 and fusion inhibition. In addition, this difference increased with higher concentrations and fusion was never completely inhibited. These differences were found consistently in three separate experiments (Fig. 5c).
FIG. 5.
Analysis of the relationship between PAN and virus-cell fusion of R18-labeled PR8 virus as a function of antibody concentration (M). PAN and fusion are expressed as a percentage. All assays were carried out at the same time in the same batch of 3-cm-diameter monolayers. PAN was measured by ELISA, and fusion was measured by fluorescence dequenching of R18. (a and b) H36 IgG (a) and Fab (b); (c) H9 Fab. ▴, PAN; ■, fusion inhibition. Virus controls gave values of 1.2 to 1.4 OD units. All data are the mean of three experiments, and the bars represent the standard error of the mean. Curves were generated as in Fig. 2. R > 0.97 for all curves. P values were calculated using an unpaired t test, where P < 0.05 is considered significant.
Effect of IgGs and Fabs on RBC hemolysis by attached virus.
Hemolysis has long been used as a model for virus-directed fusion. It is not strictly the same process but is thought to be analogous (23). Here virus was bound to RBCs and incubated with antibody and the pH was lowered to 5 to allow hemolysis to occur. The extent of hemolysis was determined spectrophotmetrically. Data for H36 IgG and Fab show that hemolysis was inhibited up to 100% as the antibody concentration increased (Fig. 6a and b). The experiment was repeated with H37 and H9 IgG and H37 Fab, and similar data (not shown) were obtained. H9 Fab, however, proved unable to inhibit hemolysis beyond 80% (Fig. 6c), recalling the situation found with fusion inhibition above (Fig. 5c).
FIG. 6.
Inhibition of hemolysis of RBCs by preattached PR8 virus by antibody. Hemolysis is presented as a percentage. Virus was incubated with cells at 4°C for 30 min, and antibody was added for 1 h at 37°C. Incubation was then continued at pH 5 and 37°C for 45 min to allow hemolysis to occur. (a and b) H36 IgG (a) and Fab (b); (c) H9 Fab. Cells were pelleted, and released hemoglobin was determined spectrophotometrically at 520 nm. Data are the mean of four experiments, and the bars represent the standard error of the mean.
Further investigation of the involvement of virus-cell fusion in PAN.
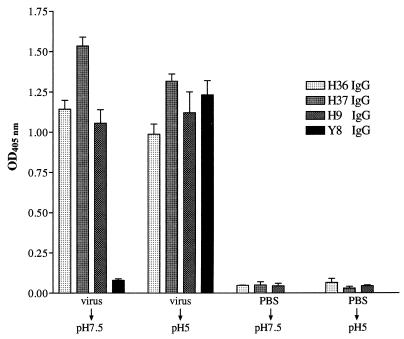
We have shown above that all antibodies used in this study inhibited virus-mediated fusion, a complex, multistage process that is activated by exposure to low pH. The details of the fusion process are incompletely understood, but the current view is that HA2 is metastable and undergoes major conformational changes at low pH that jackknife its N-terminal fusion peptide into proximity with the target cell membrane (9). These conformational changes can also be detected by the exposure of new epitopes and the acquired sensitivity of the HA to protease digestion. The following experiments were designed to determine which part of this process our antibodies inhibited. However, we first needed to determine if our antibodies bound to the low-pH form of the virus. Figure 7 shows that MAb Y8, specific for a low-pH-induced epitope, bound poorly to virus kept at pH 7.5 but 16-fold better to virus exposed to pH 5. However H9, H36, and H37 bound equally well to both forms of virus, showing that their epitopes were not pH sensitive.
FIG. 7.
H9, H36, H37, and Y8 IgGs bound to virus that had been treated at pH 5. Virus was captured by HA-specific IgA and treated at pH 5 or 7.5 at 37°C as shown. The pH was then adjusted to 7.5, and the virus was incubated with various IgGs. MAb Y8 is specific for a low-pH-induced epitope in the Sa site of HA1. Binding of antibodies was determined by ELISA. PBS controls show the level of nonspecific binding of IgGs. Data are the mean of four experiments, and the bars represent the standard error of the mean.
(i) IgGs and Fabs inhibit formation of the prefusion intermediate.
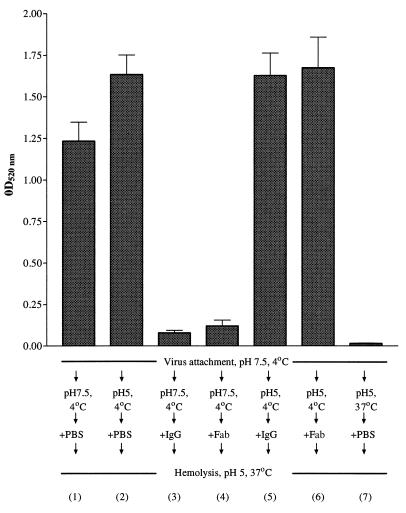
The prefusion intermediate is a form of the HA that is induced when PR8 virus is exposed to pH 5 at 4°C (35, 51, 53), and here we determined the effect of our antibodies (H9, H36, and H37) on the formation of the prefusion intermediate and its function once it had been formed. In the following experiment, virus was prebound to chicken RBCs and then exposed to various permutations of low pH and antibody. Figure 8 shows that cell-bound virus exposed to pH 5 at 37°C (column 1) lysed RBCs as expected and that cell-bound virus exposed to pH 5 at 4°C (to form the prefusion intermediate [column 2]) gave about 25% more fusion. When cell-bound virus was incubated with H36 IgG or Fab before the pH was lowered to 5 (columns 3 and 4), hemolysis was inhibited. However, when cell-bound virus was incubated at pH 5 and 4°C to form the prefusion intermediate, before adding IgG or Fab (columns 5 and 6), no inhibition of hemolysis was seen. As expected, the control of exposing virus to pH 5 at 37°C before mixing with cells gave no hemolysis (column 7). The experiments were repeated with H37 and H9 IgGs and Fabs and gave similar results (data not shown). Thus, to inhibit hemolysis, antibodies had to be present prior to the induction of the prefusion intermediate (pH 5 and 4°C). This suggested that the antibodies inhibited a very early stage in the fusion process.
FIG. 8.
Hemolysis of chicken RBCs by the prefusion intermediate of PR8 and its inhibition by neutralizing amounts of H36 IgG and Fab. Virus (10,000 HAU/ml) was mixed with H36 IgG (67 nM) or H36 Fab (200 nM). These concentrations of antibody gave 90% PAN of the virus used here in MDCK cells. Virus was attached to RBCs at 4°C for 30 min at pH 7.5 and then treated at pH 5 and 4°C for 30 min to activate the prefusion intermediate or at pH 7.5. Virus in column 7 was treated at pH 5 and 37°C before being mixed with RBCs. PBS or H36 IgG or Fab was then added at 37°C for 60 min, and mixtures were incubated for 45 min at pH 5 and 37°C for hemolysis to occur. Released hemoglobin was determined spectrophotometrically at 520 nm. Data are the mean of three experiments, and the bars represent the standard error of the mean. Numbers in parentheses refer to each experimental procedure.
(ii) Fabs do not inhibit exposure of epitopes specific for the low-pH form of the HA.
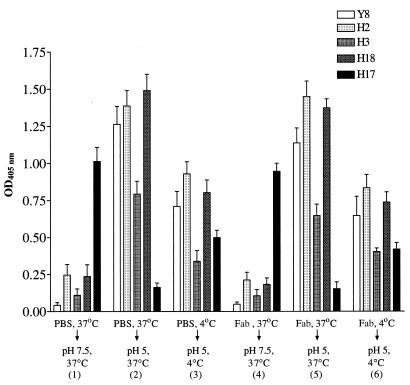
We investigated whether our Fabs inhibited the low-pH-mediated conformational changes in the HA. To detect these conformational changes, we employed a panel of MAbs that recognize pH-dependent epitopes. Virus was first allowed to attach to MDCK monolayers, which had been fixed so that any conformational changes that were due to the combined effects of binding to cell receptors and low pH might be detected, without virus being internalized. After the virus had attached to cells, the monolayers were incubated with H36, H37, or H9 Fabs or PBS and then exposed to pH 5 to induce conformational changes in the HA. These were detected by the binding of IgGs that recognize low-pH-sensitive epitopes, and this was demonstrated using an Fc-specific ELISA. (Therefore we were unable to investigate the effects of IgGs using this protocol.) Data for H36 Fab are shown in Fig. 9. In the absence of any Fab (Fig. 9, panels 1 and 2), pH 5 incubation at 37°C gave a 4.3-fold decrease in the binding of H17, a MAb that recognizes only the pH-neutral form of the HA, and increased the binding of the four MAbs that were specific for the pH 5 form of the HA. Quantitation (summarized in Table 3) showed that incubation at pH 5 and 4°C (panel 3) was about 50% as effective at losing MAb H17 reactivity and exposing low-pH epitopes as was incubation at pH 5 and 37°C. Incubation of cell-bound virus with 200 nM H36 Fab (enough to cause >99% PAN), before incubation at pH 5 or 7.5 (panels 4 to 6), did not alter the binding of the panel of MAbs in any way. Repeating the experiment with H37 Fab and H9 Fab gave similar results (data not shown). Thus, all Fabs at a concentration sufficient to cause >99% PAN had no effect on the low-pH-induced conformational changes of the HA detected by low-pH-specific MAbs.
FIG. 9.
PAN amounts of H36 Fab fail to inhibit the appearance of low pH-induced conformational changes in the HA1 of PR8 virus bound to fixed MDCK cells monolayers. Virus (200 HAU) was allowed to attach to cells at 4°C and then incubated with 200 nM H36 Fab at 4 or 37°C as indicated. The Fab gave >99% PAN of the virus used here in MDCK cells. After washing, virus-cell complexes were incubated at pH 5 or 7.5 and at 4 or 37°C for 30 min as shown and then probed with IgGs (as shown) specific for the pH-sensitive epitopes of the HA. Binding of antibodies was determined by ELISA. Data are the mean of three experiments, and the bars represent the standard error of the mean. Numbers in parentheses refer to each experimental procedure.
TABLE 3.
Comparison of the binding of MAbs to pH-sensitive epitopes in the prefusion intermediate (pH 5 and 4°C) or the densensitized (pH 5 and 37°C) form of the HA of PR8 virus to the pH 7.5 forma
| MAbb | Binding to pH 5, 4°C virus/pH 7.5 virus | Binding to pH 5, 37°C virus/pH 7.5 virus |
|---|---|---|
| Y8 | 14.0 | 25.0 |
| H2 | 3.4 | 5.4 |
| H3 | 2.0 | 5.4 |
| H18 | 3.2 | 6.0 |
| H17 | 0.5 | 0.16 |
Data from Fig. 9.
The epitopes of MAbs Y8, H2, H3, and H18 are exposed, and the epitope of H17 is reduced at low pH.
(iii) Fab H36 does not inhibit induction of the low pH-induced protease-sensitive form of the HA.
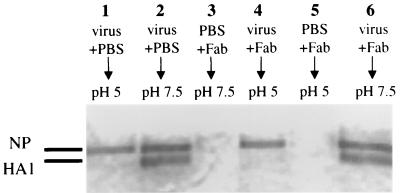
Another manifestation of exposure of virus to low pH is the acquired sensitivity of the HA to cleavage by proteinase K. Virus was incubated with H36 Fab and then adjusted to pH 5 or 7.5 at 37°C for 30 min. The pH was returned to 7.5, and virus was incubated with 1 μg of proteinase K per ml at 37°C for 30 min. PAGE analysis showed that HA1 of the virus maintained at pH 7.5 ran in its expected position (Fig. 10, lanes 2 and 6), but HA1 of the virus that was incubated at pH 5 without antibody was degraded (lane 1). Incubation with H36 Fab did not prevent digestion (lane 4). By staining, there was a molar excess of Fab over HA (data not shown). The internal proteins NP (Fig. 10) and M1 (not shown) were unaffected by any of the incubation conditions, as expected. Thus, we see that H36 Fab did not prevent the low-pH-mediated conformational changes in the HA that are detected by acquisition of protease sensitivity.
FIG. 10.
H36 Fab fails to inhibit the acquisition of protease sensitivity of PR8 HA at pH 5. Virus (100,000 HAU) was incubated with 2.5 μM H36 Fab at 37°C for 1 h. The Fab gave 90% STAN of the virus used here in MDCK cells. The mixture was then held at pH 5 or 7.5 at 37°C for 30 min as shown. After the pH was restored to 7.5, all mixtures were reacted with proteinase K (1 μg/ml) at 37°C for 30 min to determine if conformational changes to the HA had taken place. Lanes 3 and 5 contain Fab and no virus. Virus not incubated with protease was indistinguishable from virus in lane 2 (data not shown). Proteins were analyzed by PAGE under reducing conditions and stained with Coomassie blue.
DISCUSSION
Comparison of PAN and STAN.
IgGs H36 (IgG2a to HA1 site Sb/B), H37 (IgG3 to site Ca2/A), and H9 (IgG3 to site Cb/E) mediated STAN by a combination of inhibition of virus-cell fusion and inhibition of virus attachment to cells; H36 and H37 Fabs inhibited attachment only, while H9 Fab inhibited fusion only (17, 18). Here we found that all IgGs and Fabs mediated PAN (Fig. 2). Thus PAN of influenza virus was independent of antibody isotype, antigenic site, and size and valency of the ligand, although the efficiency of PAN may be affected by any one of these parameters; it also contrasts with the PAN of poliovirus, since that virus does not undergo PAN with Fabs (54). It is of interest that the efficiency of PAN (H36 > H37 > H9) reflects the efficiency of STAN (17), which implies that the antibodies have an inherent neutralization efficiency, possibly due to their ability to inhibit HA-cell or HA-HA interactions. The efficiency of IgG-mediated PAN was lower (8- to 37-fold at N50) than that of IgG-mediated STAN (Table 1), depending on the antibody. PAN mediated by Fabs was less efficient (3.6- to 18-fold) than PAN mediated by IgG (Table 2). Affinity was not an issue here, at least for H36 and H9 since their IgGs and Fabs had very similar values (17, 18). However, it is of interest that Fabs effected PAN and STAN with a similar efficiency (Table 1), even though Fabs H36 and H37 mediated PAN (by inhibition of fusion) and STAN (by inhibition of virus attachment to target cells) (17) by different mechanisms. It may be that the relatively small size of the Fabs facilitated access to their epitopes and permitted the observed equality of PAN and STAN.
Why is IgG-PAN less efficient than IgG-STAN?
PAN requires virus to be attached and exposed on the surface of the target cell and is limited by the rate of virus internalization. At 4°C there was negligible internalization over 60 min, and PAN was effective throughout this period (Fig. 3). However at 37°C, PAN decreased rapidly, and after 10 min approximately 50% of virus was refractory to PAN. This requirement to act rapidly may also explain why PAN needs a higher concentration of antibody than STAN does.
An explanation for the difference in efficiency of STAN and PAN may simply be that the processes operate by different mechanisms. All three IgGs used here display a complex mechanism of STAN, in which both inhibition of virus-cell fusion (predominantly) and inhibition of virus-cell attachment act to reduce infectivity (17, 18). In contrast, PAN takes place solely by inhibition of fusion. In STAN, the two neutralization mechanisms may act cumulatively or synergistically, and this may increase neutralization efficiency.
A second explanation centers on the attachment of influenza virus to N-acetylneuraminic acid (NANA) residues on the cell surface. Individually, this is a low-affinity reaction that is strengthened by the recruitment of multiple NANA molecules (19). After attachment, the part of the virus surface that is in contact with the cell is likely to be less readily available to antibody than is the rest of the surface of the bound or free virus particles. If PAN was mediated by antibody binding to the exposed part of the attached virus, one would expect PAN and STAN to have a similar efficiency. However, STAN is the more efficient, suggesting that PAN antibodies bind to HA trimers in contact with the cell and block fusion (see below) and/or bind trimers in the immediate vicinity and inhibit the recruitment of additional receptor-HA complexes necessary for the fusion process. Such HA trimers may bind antibody less readily because they are less accessible to antibody per se or because receptor binding alters the conformation of the HA. The latter seems unlikely because there would have to be an effect on epitopes in three discrete antigenic sites (Sb/B, Ca2/A and Cb/E).
Mechanism of PAN.
By definition, PAN is mediated by antibody that is added after virus has attached to the target cell, and it acts after attachment. Since antibodies did not mediate PAN by eluting virus from the cell and did not inhibit internalization of virus by the target cell (Fig. 4), they evidently inhibited a postinternalization event. Indeed, we demonstrated (Fig. 5) that they inhibited virus-cell fusion in proportion to the loss of infectivity (except with high concentrations of H9 Fab) and inhibited hemolysis of red blood cells (Fig. 6). Thus, the inhibitory mechanism of PAN is centered on the fusion process. However, this is complex, and we do not know if the six IgGs and Fabs used here inhibited the same part of the fusion process. If they did not, this would help to explain the different efficiencies of PAN by the IgGs and the Fabs. Any unaccounted-for infectivity (as with high concentrations of H9 Fab) might result from inhibition of a postfusion event, as described previously for influenza A virus (2, 16, 38, 39, 41), but further investigation is required to establish this.
Prefusion intermediate and PAN.
Virus-cell fusion is a complex process in which exposure to low pH triggers a cascade of conformational changes of the HA trimers. Recent characterization of some of these conformational changes provided us with the opportunity to begin to investigate and pin down the specific stage(s) of the fusion process that our PAN antibodies were inhibiting. The pH 5 structure, which is characterized by dissociation of the HA1 globular domains and extensive rearrangements of HA2 including exposure of the normally hidden hydrophobic fusion region, was originally proposed as the fusion-active conformation (9, 57), but this is now regarded as an inactive end product, with fusion being mediated by transition from a transient conformational or prefusion intermediate to the extensively dissociated structure (51, 55). In other words the HA moves from the tense (T) state in the virion to the relaxed (R) or intermediate state to the densensitized (D) or pH 5 state (28). The R state retains most of the morphology of the T spike but is less well defined (28, 49) and can be obtained by incubation at pH 5 and 4°C (51, 53). The prefusion intermediate is not fusion active, although it releases at least some of its complement of HA2 fusion peptides, as shown by a gain in hydrophobicity and its precipitation with anti-fusion peptide antibody. There are also small conformational changes in either the globular HA1 heads or the association of the individual HAs that constitute the trimer, as these had increased interaction with MAbs specific for the low pH form of HA1 (35, 53).
In agreement with others, our prefusion intermediate was fusion inactive and failed to hemolyze RBC during a prolonged (4 h) incubation at 4°C (51, 53). However, our prefusion intermediate (data not shown) and that of others (48) was converted to fusion activity at pH 7.5 and 37°C, whereas others found it necessary to use pH 5 and 37°C (35). IgGs and Fabs that mediated PAN inhibited hemolysis of RBCs by previously bound virus only if added before the virus had been converted into the prefusion intermediate (Fig. 8). To further characterize the prefusion intermediate generated in our hands, virus attached to fixed MDCK cells was incubated with neutralizing Fab and converted into the prefusion intermediate by incubation at pH 5 and 4°C and then probed with a panel of conformation-specific MAbs (Fig. 9). Attachment to cells (without internalization) was used so that we would detect any additional changes to the HA that resulted from the interaction of virus with its receptors and antibody to the same virus particle. Binding of the prefusion intermediate to MAb H17, specific for the neutral pH form of the HA, was reduced twofold, compared with a sixfold reduction seen after the virus had been incubated at pH 5 and 37°C. Binding to the prefusion intermediate of four other MAbs that were specific for epitopes induced at low pH was increased across the board compared to the binding to HA trimer at neutral pH. However, this increase in binding was again approximately twofold lower than that seen with the pH 5 and 37°C structure. Therefore, on both counts, the HA1 globular head or the association of HA in the trimers underwent conformational changes at low pH that were less extensive at 4°C than at 37°C. Electron microscopy also showed no detectable dissociation of the globular heads in the prefusion intermediate (28, 49).
To determine if Fabs that gave PAN would inhibit the pH 5 conformational changes detected with MAbs to pH-sensitive epitopes, we attached virus to the fixed MDCK cells described above and reacted this with a concentration of neutralizing Fabs that gave 99% PAN. None of these pH 5 conformational changes was inhibited (Fig. 9). Finally, we showed that the acquisition of protease sensitivity by HA at pH 5 and 37°C was unaffected by preincubation with H36 Fab (Fig. 10). Thus, for PAN it was necessary for antibodies to bind their epitopes prior to initiation of one of the earliest known points in the fusion process. However, this did not prevent the majority of HA trimers from going through their low-pH conformational transition.
Summary.
Under conditions of PAN, all our IgGs and Fabs (with the exception of H9 Fab at high concentration) inhibited fusion in direct proportion to loss of infectivity. To do this, they have to bind virus before activation of the prefusion intermediate. There are several ways in which fusion might be inhibited. One is for HA1-specific antibody to cross-link the HA1 trimer, so that the HA2 fusion peptide cannot be liberated. However, since monovalent Fabs mediate PAN, any cross-linking would have to be within a single antibody footprint. In addition, our Fabs were directed against three discrete antigenic sites, and each would have to have the type of structure that can be cross-linked by a single footprint, and this seems unlikely. A more persuasive possibility flows from the fact that H36 IgG and Fab and H9 IgG and Fab have nearly identical affinities (17, 18) and yet the IgGs were more efficient at PAN than their Fabs were. In addition, H36 IgG and Fab both bind virus monovalently (S. Hardy and N. J. Dimmock, unpublished data), so that valency of binding is not at issue here. The main remaining difference is the threefold-larger mass of the IgG, suggesting that steric hindrance may be the important factor, with PAN being mediated by antibody that binds either HA trimers in contact with cell receptors or HA trimers in the immediate vicinity of the HA-cell receptor contact zone. The lower efficiency of PAN by Fabs would be explained if steric hindrance required more of the smaller Fab molecules bound at the neutralization site than of bound IgG molecules. Steric hindrance may prevent released fusion peptides from inserting into the membrane or may prevent the subsequent interaction of trimers that form the fusion pore. It is thought that fewer than 6 of the 700 or so HA trimers are needed for fusion (5, 6, 8, 12). Although we have not investigated the occupancy of the HA trimers by antibody, the amount of Fab used gave 99% PAN and was probably sufficient to bind to the majority of trimers. Even so, conformational changes in the majority of HAs were not inhibited by the Fab. Thus, PAN may result from inhibition of conformational changes in the small number of trimers actually responsible for fusion, but this was undetectable against the high background of low-pH-activated trimers. Trimers that mediate PAN may be distinguished from those that do not by their reaction with the cell receptor, with formation of an antibody-HA-receptor complex being a key event. Alternatively, and more simply, it may be that proximity of a trimer to the HA-receptor complex is what distinguishes a PAN-active trimer from the others.
ACKNOWLEDGMENTS
M.J.E. was supported by a studentship from the BBSRC.
We thank Walter Gerhard (The Wistar Institute, Philadelphia, Pa.) and Robert G. Webster (St. Jude Children's Research Hospital, Memphis, Tenn.) for their generous gifts of hybridomas and MAbs, Wendy Barclay (University of Reading, Reading, United Kingdom) for the MDCK cells, and Lesley McLain for her input into the manuscript.
REFERENCES
- 1.Arendrup M, Sönnerborg A, Svennerholm B, Åkerblom L, Nielsen C, Clausen H, Olofsson S, Nielsen J O, Hansen J-E S. Neutralizing antibody response during human immunodeficiency virus type 1 infection: type and group specificity and viral escape. J Gen Virol. 1993;74:855–863. doi: 10.1099/0022-1317-74-5-855. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong S J, Dimmock N J. Neutralization of influenza virus by low concentrations of HA-specific polymeric IgA inhibits viral fusion activity but activation of the ribonucleoprotein is also inhibited. J Virol. 1992;66:3823–3832. doi: 10.1128/jvi.66.6.3823-3832.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong S J, Dimmock N J. Varying temperature-dependence of post-attachment neutralization of human immunodeficiency virus type 1 by monoclonal antibodies to gp120: identification of a very early fusion-independent event as a neutralization target. J Gen Virol. 1996;77:1397–1402. doi: 10.1099/0022-1317-77-7-1397. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong S J, McInerney T L, McLain L, Wahren B, Hinkula J, Levi M, Dimmock N J. Two neutralizing anti-V3 monoclonal antibodies act by affecting different functions of human immunodeficiency virus type 1. J Gen Virol. 1996;77:2931–2941. doi: 10.1099/0022-1317-77-12-2931. [DOI] [PubMed] [Google Scholar]
- 5.Bentz J. Minimal aggregate size and minimal fusion unit for the first fusion pore of influenza hemagglutinin-mediated membrane fusion. Biophys J. 2000;78:227–245. doi: 10.1016/S0006-3495(00)76587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentz J, Ellens H, Alford D. Architecture of the influenza hemagglutinin fusion site. In: Bentz J, editor. Viral fusion mechanisms. Boca Raton, Fla: CRC Press, Inc.; 1993. pp. 163–199. [Google Scholar]
- 7.Blumenthal R, Bali-Puri A, Walter A, Covell D, Eidelman O. pH-dependent fusion of vesicular stomatitis virus with Vero cells. J Biol Chem. 1987;262:13614–13619. [PubMed] [Google Scholar]
- 8.Blumenthal R, Sarkar D P, Durell S, Howard D E, Morris S J. Dilation of the influenza hemagglutinin fusion pore revealed by the kinetics of individual cell-cell fusion events. J Cell Biol. 1996;135:63–71. doi: 10.1083/jcb.135.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature (London) 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 10.Caton A J, Brownlee G G, Yewdell J W, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 11.Cleveland S M, Jones T D, Dimmock N J. Properties of a neutralizing antibody that recognises a conformational form of epitope ERDRD in the C-terminal tail of human immunodeficiency virus type 1. J Gen Virol. 2000;81:1251–1260. doi: 10.1099/0022-1317-81-5-1251. [DOI] [PubMed] [Google Scholar]
- 12.Danieli T, Pelletier S, Henis Y I, White J M. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daukas G, Zigmond S H. Inhibition of receptor-mediated but not fluid-phase endocytosis in polymorphonuclear leukocytes. J Cell Biol. 1985;101:1673–1679. doi: 10.1083/jcb.101.5.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietzschold B, Tollis M, Lafon M, Wunner W H, Koprowski H. Mechanisms of rabies virus neutralization by glycoprotein specific monoclonal antibodies. Virology. 1987;161:29–36. doi: 10.1016/0042-6822(87)90167-x. [DOI] [PubMed] [Google Scholar]
- 15.Dimmock N J. Neutralization of animal viruses. Curr Top Microbiol Immunol. 1993;183:1–149. doi: 10.1007/978-3-642-77849-0. [DOI] [PubMed] [Google Scholar]
- 16.Dimmock N J, Taylor H P, Carver A S. Interaction of neutralized influenza virus avian and mammalian cells. In: Compans R W, Bishop D H L, editors. Segmented negative strand viruses. London, United Kingdom: Academic Press, Ltd.; 1984. pp. 355–359. [Google Scholar]
- 17.Edwards M J, Dimmock N J. Two influenza A virus haemagglutinin-specific Fabs neutralize by inhibiting virus attachment to target cells, while neutralization by their IgGs is complex and occurs through fusion-inhibition and attachment-inhibition simultaneously. Virology. 2000;278:423–435. doi: 10.1006/viro.2000.0631. [DOI] [PubMed] [Google Scholar]
- 18.Edwards M J, Dimmock N J. A haemagglutinin (HA1)-specific Fab neutralizes influenza A virus by inhibiting fusion of virus to target cells. J Gen Virol. 2001;82:1387–1395. doi: 10.1099/0022-1317-82-6-1387. [DOI] [PubMed] [Google Scholar]
- 19.Glick G D, Knowles J R. Molecular recognition of bivalent sialosides by influenza virus. J Am Chem Soc. 1991;113:4701–4703. [Google Scholar]
- 20.Gómez-Puertas P, Rodríguez F, Oviedo J M, Ramiro-Ibáñez F, Ruiz-Gonzalvo F, Alonso C, Escribano J M. Neutralizing antibodies to different proteins of African swine fever virus inhibit both virus attachment and internalization. J Virol. 1996;70:5689–5694. doi: 10.1128/jvi.70.8.5689-5694.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guinea R, Carrasco L. Requirement for vacuolar proton-ATPase activity during entry of influenza virus into cells. J Virol. 1995;69:2306–2312. doi: 10.1128/jvi.69.4.2306-2312.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heuser J E, Anderson R G W. Hypertonic media inhibit receptor-mediated endocytosis by blocking clathrin-coated pit formation. J Cell Biol. 1989;108:389–400. doi: 10.1083/jcb.108.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang R T C, Rott R, Klenk H D. Influenza viruses cause hemolysis and fusion of cells. Virology. 1981;110:243–247. doi: 10.1016/0042-6822(81)90030-1. [DOI] [PubMed] [Google Scholar]
- 24.Hudson L, Hay F C. Practical immunology. 3rd ed. Oxford, United Kingdom: Blackwell Scientific Publications; 1989. [Google Scholar]
- 25.Ishada N, Ackermann W W. Growth characteristics of influenza virus. Properties of the initial cell-virus complex. J Exp Med. 1956;104:501–505. doi: 10.1084/jem.104.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jackson N A C, Levi M, Wahren B, Dimmock N J. Mechanism of action of a 17 amino acid microantibody specific for the V3 loop that neutralizes free HIV-1 virions. J Gen Virol. 1999;80:225–236. doi: 10.1099/0022-1317-80-1-225. [DOI] [PubMed] [Google Scholar]
- 27.Kjellén L. A hypothesis accounting for the effect of the host cell on neutralization-resistant virus. J Gen Virol. 1985;66:2279–2283. doi: 10.1099/0022-1317-66-10-2279. [DOI] [PubMed] [Google Scholar]
- 28.Korte T, Ludwig K, Booy F P, Blumenthal R, Hermann A. Conformational intermediates and fusion activity of influenza virus hemagglutinin. J Virol. 1999;73:4567–4574. doi: 10.1128/jvi.73.6.4567-4574.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu S, Putney S D, Robinson H L. Human imunodeficiency virus entry into T cells: more rapid escape from an anti-V3 loop than an antireceptor antibody. J Virol. 1992;66:2547–2550. doi: 10.1128/jvi.66.4.2547-2550.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matlin K S, Reggio H, Helenius A, Simons K. Infectious entry pathway of influenza virus in a canine kidney cell line. J Cell Biol. 1981;91:601–613. doi: 10.1083/jcb.91.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nara P L. HIV-neutralization: evidence for rapid, binding/post-binding neutralization from infected humans, chimpanzees and gp120-vaccinated animals. In: Lerner R A, Ginsberg H, Chanock R M, Brown F, editors. Vaccines 89: modern approaches to new vaccines including the prevention of AIDS. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. pp. 137–144. [Google Scholar]
- 32.Ochiai H, Sinya S, Hirabayashi T, Shimizu Y, Terasawa K. Inhibitory effect of bafilomycin A1, a specific inhibitor of vacuolar-type proton pump, on the growth of influenza A and B viruses in MDCK cells. Antiviral Res. 1995;27:425–430. doi: 10.1016/0166-3542(95)00040-s. [DOI] [PubMed] [Google Scholar]
- 33.Ohizumi Y, Suzuki H, Matsumoto Y-I, Mashuo Y, Numazaki Y. Neutralizing mechanisms of two human monoclonal antibodies against human cytomegalovirus glycoprotein 130/55. J Gen Virol. 1992;73:2705–2707. doi: 10.1099/0022-1317-73-10-2705. [DOI] [PubMed] [Google Scholar]
- 34.Osiowy C, Anderson R. Neutralization of repiratory syncytial virus after cell attachment. J Virol. 1995;69:1271–1274. doi: 10.1128/jvi.69.2.1271-1274.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pak C C, Krumbiegel M, Blumenthal R. Intermediates in influenza virus PR/8 hemagglutinin-induced membrane fusion. J Gen Virol. 1994;75:395–399. doi: 10.1099/0022-1317-75-2-395. [DOI] [PubMed] [Google Scholar]
- 36.Palokangas H, Metsikkö K, Väänänen K. Active vacuolar H+ATPase is required for both endocytic and exocytic processes during viral infection of BHK-21 cells. J Biol Chem. 1994;269:17577–17585. [PubMed] [Google Scholar]
- 37.Pelchen-Matthews A, Clapham P, Marsh M. Role of CD4 endocytosis in human immunodeficiency virus. J Virol. 1995;69:8164–8168. doi: 10.1128/jvi.69.12.8164-8168.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Possee R D, Dimmock N J. Neutralization of influenza virus by antibody: attachment, uptake and uncoating of neutralized virus in chick embryo cells. ICN-UCLA Symp Mol Cell Biol. 1981;21:473–480. [Google Scholar]
- 39.Possee R D, Schild G C, Dimmock N J. Studies on the mechanism of neutralization of influenza virus by antibody: evidence that neutralizing antibody (anti-haemagglutinin) inactivates influenza virus by inhibiting virion transcriptase activity. J Gen Virol. 1982;58:373–386. doi: 10.1099/0022-1317-58-2-373. [DOI] [PubMed] [Google Scholar]
- 40.Richman D D, Hostetler K Y, Yazaki P J, Clark S. Fate of influenza A virion proteins after entry into subcellular fractions of LLC cells and the effect of amantadine. Virology. 1986;151:200–210. doi: 10.1016/0042-6822(86)90042-5. [DOI] [PubMed] [Google Scholar]
- 41.Rigg R J, Carver A S, Dimmock N J. IgG-neutralized influenza virus undergoes primary, but not secondary uncoating in vivo. J Gen Virol. 1989;70:2097–2109. doi: 10.1099/0022-1317-70-8-2097. [DOI] [PubMed] [Google Scholar]
- 42.Roehrig J T, Hunt A R, Kinney R M, Mathews J H. In vitro mechanisms of monoclonal antibody neutralization of alphaviruses. Virology. 1988;165:66–73. doi: 10.1016/0042-6822(88)90659-9. [DOI] [PubMed] [Google Scholar]
- 43.Ruggeri F M, Greenberg H B. Antibodies to the trypsin cleavage peptide VP8∗ neutralize rotavirus by inhibiting binding of virions to target cells in culture. J Virol. 1991;65:2211–2219. doi: 10.1128/jvi.65.5.2211-2219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russell P H. Newcastle disease virus: the effect of monoclonal antibody in the overlay on virus penetration and the immunoselection of variants. J Gen Virol. 1984;65:795–798. doi: 10.1099/0022-1317-65-4-795. [DOI] [PubMed] [Google Scholar]
- 45.Sato S B, Kawaski K, Ohnishi S-I. Hemolytic activity of influenza virus hemagglutinin glycoproteins activated in mildly acidic environments. Proc Natl Acad Sci USA. 1983;80:3153–3157. doi: 10.1073/pnas.80.11.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sattentau Q J, Moore J P. Conformational changes induced in the human immunodeficiency virus glycoprotein by soluble CD4. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sattentau Q J, Moore J P, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope protein of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schoch C, Blumenthal R, Clague M J. A long-lived fusion state for influenza virus-erythrocyte complexes committed to fusion at neutral pH. FEBS Lett. 1992;311:221–225. doi: 10.1016/0014-5793(92)81107-w. [DOI] [PubMed] [Google Scholar]
- 49.Shangguan T, Siegel D P, Lear J D, Axelson P H, Alford D, Bentz J. Morphological changes and fusogenic activity of influenza virus hemagglutinin. Biophys J. 1998;74:54–62. doi: 10.1016/S0006-3495(98)77766-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Staudt L M, Gerhard W. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. I. Significant variation in repertoire expression between individual mice. J Exp Med. 1983;157:687–704. doi: 10.1084/jem.157.2.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stegmann T, White J M, Helenius A. Intermediates in influenza induced membrane fusion. EMBO J. 1990;9:4231–4241. doi: 10.1002/j.1460-2075.1990.tb07871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suñé C, Jiménez G, Correa I, Bullido M J, Gebauer F, Smerdou C, Enjuanes L. Mechanisms of transmissible gastroenteritis coronavirus neutralization. Virology. 1990;177:559–569. doi: 10.1016/0042-6822(90)90521-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsurudome N, Glück R, Graf R, Falchetto R, Schaller U, Brunner J. Lipid interactions of the hemagglutinin HA2 NH2-terminal segment during influenza virus-induced membrane fusion. J Biol Chem. 1992;267:20225–20232. [PubMed] [Google Scholar]
- 54.Vrijsen R, Mosser A, Boeyé A. Postadsorbtion neutralization of poliovirus. J Virol. 1993;67:3126–3133. doi: 10.1128/jvi.67.6.3126-3133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White J M, Wilson I A. Anti-peptide antibodies detect steps in a protein conformational change: low pH activation of the influenza virus hemagglutinin. J Cell Biol. 1987;105:2887–2896. doi: 10.1083/jcb.105.6.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiley D C, Wilson I A, Skehel J J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature (London) 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- 57.Wilson L A, Skehel J J, Wiley D C. Structure of the haemagglutinin membrane protein of influenza virus at 3 Å resolution. Nature (London) 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 58.Wohlfart C. Neutralization of adenoviruses: kinetics, stoichiometry and mechanisms. J Virol. 1988;62:2321–2328. doi: 10.1128/jvi.62.7.2321-2328.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yewdell J W, Gerhard W, Bächi T. Monoclonal anti-hemagglutinin antibodies that detect irreversible alterations that coincide with the acid activation of influenza virus A/PR/8/34-mediated hemolysis. J Virol. 1983;48:239–248. doi: 10.1128/jvi.48.1.239-248.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]