Abstract
Introduction
The translation of gene expression profiles of SCLC to clinical testing remains relatively unexplored. In this study, gene expression variations in SCLC were evaluated to identify potential biomarkers.
Methods
RNA expression profiling was performed on 44 tumor samples from 35 patients diagnosed with SCLC using the clinically validated RNA Salah Targeted Expression Panel (RNA STEP). RNA sequencing (RNA-Seq) and immunohistochemistry were performed on two different SCLC cohorts, and correlation analyses were performed for the ASCL1, NEUROD1, POU2F3, and YAP1 genes and their corresponding proteins. RNA STEP and RNA-Seq results were evaluated for gene expression profiles and heterogeneity between SCLC primary and metastatic sites. RNA STEP gene expression profiles of independent SCLC samples (n = 35) were compared with lung adenocarcinoma (n = 160) and squamous cell carcinoma results (n = 25).
Results
The RNA STEP results were highly correlated with RNA-Seq and immunohistochemistry results. The dominant transcription regulator by RNA STEP was ASCL1 in 74.2% of the samples, NEUROD1 in 20%, and POU2F3 in 2.9%. The ASCL1, NEUROD1, and POU2F3 gene expression profiles were heterogeneous between primary and metastatic sites. SCLCs displayed markedly high expression for targetable genes DLL3, EZH2, TERT, and RET. SCLCs were found to have relatively colder immune profiles than lung adenocarcinomas and squamous cell carcinomas, characterized by lower expression of HLA genes, immune cell, and immune checkpoint genes, except the LAG3 gene.
Conclusions
Clinical-grade SCLC RNA expression profiling has value for SCLC subtyping, design of clinical trials, and identification of patients for trials and potential targeted therapy.
Keywords: Small cell lung cancer, RNA expression, Biomarker, Clinical testing, Transcriptomics
Introduction
SCLC is a dismal malignancy compromising approximately 14% of all lung cancers (National Comprehensive Cancer Network v.2.24-November 21, 2023). Treating SCLC presents a formidable challenge for oncologists, and the outcomes of treatment remain unsatisfactory. Elucidation of predictive biomarkers, development of better methods for selecting immunotherapy-sensitive populations,1,2 and improvement in therapeutic strategies are needed. In select instances, molecular profiling is currently considered for patients with extensive-stage (ES) SCLC who fall into rare categories, such as those who have minimal tobacco exposure (never smoked to less than 10 cigarettes/d). In addition, molecular profiling may be considered in cases where there is a diagnostic challenge, because additional molecular information may influence the treatment approach (National Comprehensive Cancer Network v.2.24-November 21, 2023).
Developing a deeper understanding of SCLC biology through genomic characterization offers the potential for critically needed advancement in the care and treatment of patients with SCLC. SCLC is characterized by inactivating mutations in the RB1 and TP53 tumor-suppressor genes with these mutations also associated with increased risk for SCLC transformation when identified in EGFR-mutant lung cancers. The neuroendocrine differentiation associated with SCLC is regulated by INSM1 which in turn is regulated by the Notch1-Hes1 signaling pathway.3 Early studies focusing on proteogenomic characterization defined four SCLC molecular subtypes based on high protein expression of the following markers: ASCL1 (SCLC-A), NEUROD1 (SCLC-N), POU2F3 (SCLC-P), and YAP1 (SCLC-Y).4,5 Later studies confirmed SCLC-A, SCLC-N, and SCLC-P as three distinct subtypes but did not validate the SCLC-Y as a distinct subtype.6, 7, 8 These distinct subtypes correlate with therapeutic responsiveness, differences in genetic alterations, and prognosis.4,9,10 In addition, some studies have presented compelling evidence for the existence of an SCLC inflamed subtype (SCLC-I) with lower expression of the ASCL1, NEUROD1, and YAP1 genes, characterized by an inflamed gene signature and mesenchymal features.10 Although these foundational studies have made strides in the genomic characterization of SCLC, there remains an unmet need for the identification of biomarkers and new therapy targets for SCLC.
In this study, the primary objective was to explore SCLC gene expression variations with a clinically validated 204 gene expression panel, the RNA Salah Targeted Expression Panel (RNA STEP), to identify subtypes and potential diagnostic, prognostic, and therapeutic biomarkers.
Materials and Methods
Samples
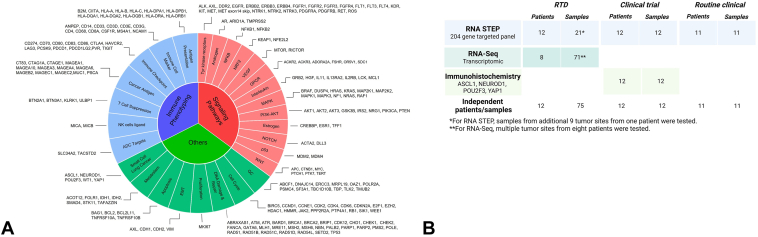
Between November 2022 and November 2023, a total of 589 samples from solid tumors underwent RNA STEP testing (Fig. 1A), including samples from patients with a diagnosis of SCLC (n = 35), lung adenocarcinoma (LUAD, n = 160), and lung squamous cell carcinoma (SqCC, n = 25). This project to study deidentified RNA STEP results from routine clinical care was reviewed by the Advarra Institutional Review Board (IRB, IRB#00000971, MCC 23158) to help ensure that the rights and welfare of research participants were protected. The IRB granted a full waiver of the Health Insurance Portability and Accountability Act (HIPAA) authorization due to the impracticality of gaining retrospective consent from the many patients associated with the clinical results and the low ethical risk due to the deidentified nature of this study. Of the 35 SCLC samples, 11 were collected per routine clinical care. The remaining 24 were obtained through two IRB-approved research protocols: MCC19163 (n = 12), a vaccine-based clinical trial, and MCC18843 (n = 12), the rapid tissue donation (RTD) autopsy research protocol (Table 1, Fig. 1B). The vaccine-based clinical trial was registered on the ClinicalTrials.gov website (NCT00617409). All patients provided a written informed consent, and the treatment protocol was approved by University of South Florida Institutional Review Board. For the RTD study, in all cases where it was feasible, discussion of the study and informed consent was provided during clinical care.11 In addition, to evaluate for intermetastatic heterogeneity, nine additional samples from different tumor sites of one RTD donor (#32) were tested with RNA STEP.
Figure 1.
(A) RNA STEP. Panel targets include 204 genes. (B) Overview of the samples and methods included in the study. RNA STEP, RNA Salah Targeted Expression Panel.
Table 1.
Demographics and Clinical Features of SCLC Cohort Tested by RNA STEP (n = 35)
| Case Number | Age | Sex | Smoking History |
Sample Type |
Therapy-Naive Sample | Tested Tissue |
Tumor Percentage |
|---|---|---|---|---|---|---|---|
| 1 | 79 | F | Former | Clinical | Yes | Liver | 70 |
| 2 | 75 | M | Former | Trial | No | Liver | 15 |
| 3 | 57 | F | Former | RTD | No | Lung | 90 |
| 4 | 59 | M | Current | Clinical | No | Liver | 70 |
| 5 | 47 | M | Former | Trial | No | Adrenal | 90 |
| 6 | 67 | F | Current | RTD | No | Lung | 95 |
| 7 | 77 | M | Former | RTD | No | Lung | 90 |
| 8 | 69 | M | Former | RTD | No | Lung | 85 |
| 9 | 60 | M | Former | Trial | No | Liver | 80 |
| 10 | 59 | M | Former | RTD | No | Liver | 80 |
| 11 | 69 | F | Former | Clinical | No | Liver | 60 |
| 12 | 56 | F | Former | Trial | No | Liver | 90 |
| 13 | 66 | F | Former | Trial | No | Liver | 40 |
| 14 | 54 | F | Former | RTD | No | Lung | 90 |
| 15 | 74 | M | Current | RTD | No | Lymph node | 20 |
| 16 | 59 | F | Former | Trial | No | Lung | 60 |
| 17 | 74 | F | Current | Trial | No | Lymph node | 90 |
| 18 | 66 | F | Former | Trial | No | Adrenal | 70 |
| 19 | 63 | M | Former | RTD | No | Liver | 90 |
| 20 | 66 | F | Current | Clinical | Yes | Lymph node | 30 |
| 21 | 74 | M | Current | Clinical | Yes | Retroperitoneum | 60 |
| 22 | 77 | M | Former | Trial | No | Liver | 60 |
| 23 | 77 | F | Former | Clinical | No | Liver | 50 |
| 24 | 68 | M | Former | Clinical | Yes | Pleural | 95 |
| 25 | 88 | M | Former | RTD | No | Lung | 95 |
| 26 | 78 | M | Former | RTD | No | Lung | 10 |
| 27 | 70 | M | Former | Clinical | No | Lung | 70 |
| 28 | 36 | M | Nonsmoker | Clinical | Yes | Lung | 75 |
| 29 | 72 | F | Nonsmoker | Clinical | No | Lung | 60 |
| 30 | 68 | M | Current | RTD | Yes | Mediastinum | 30 |
| 31 | 59 | F | Former | Trial | No | Liver | 60 |
| 32 | 59 | M | Current | RTD | No | Lung | 90 |
| 33 | 69 | F | Current | Clinical | Yes | Lymph node | 30 |
| 34 | 73 | F | Former | Trial | No | Liver | 70 |
| 35 | 70 | M | Former | Trial | No | Soft tissue | 60 |
Note: All SCLC samples were tested with RNA STEP (N = 35); trial samples were also tested with IHC (N = 12); eight of above 12 RTD samples were also tested with RNA-seq.
F, female; IHC, immunohistochemistry; M, male; RNA-seq, RNA sequencing; RNA STEP, RNA Salah Targeted Expression Panel; RTD, rapid tissue donation.
RNA sequencing (RNA-Seq) was performed on tumor from 71 SCLC samples from eight RTD donors. RNA STEP was performed on 17 of these samples, one sample each from seven RTD donors and 10 samples from one donor (#32). There were 12 samples included in this study from a cohort of 21 patients with SCLC enrolled for a clinical trial with sufficient tissue for both immunohistochemical (IHC) staining and RNA STEP.
RNA STEP
A custom RNA expression panel of 204 genes, RNA STEP, was designed with feedback elicited from Moffitt clinicians about which proteins or biomarkers would be most relevant for future clinical trials. Probes were designed to target the correlative genes to these selected proteins. The RNA STEP assay used the NanoString platform to digitally count the mRNA transcripts in the samples. Each run included a reference universal mRNA control derived from pooled human normal tissues (BioChain Institute, Inc., Newark, CA, Catalog number R4234565) to ensure batch-to-batch consistency and normalize gene signals. Raw data files were processed and normalized using geNorm in the NanoString nSolver 4.0 advanced analysis software. The normalized log2 ratios were computed by subtracting the normalized log2 counts of the universal mRNA control genes from the log2 counts of the individual samples. In the RNA STEP validation, a log2 ratio of more than or equal to 2 was chosen as the criterion to define “high” expression.12 Nevertheless, considering that one SCLC sample had NEUROD1 protein expression by IHC, had a log2 ratio of 1.6 in the RNA STEP analysis, a log2 ratio cutoff of more than or equal to 1 was selected for designating “high” expression in this study. For ASCL1, NEUROD1, POU2F3, and YAP1, the dominant transcription regulator was identified as the gene exhibiting the highest log2 ratio in the sample.
The methodologies for RNA extraction, RNA-Seq, IHC, and statistical analyses are provided in Supplementary Data 1.
Results
SCLC Cohort Demographics and Clinical Features
Of the 35 patients with SCLC included in this study, the mean age was 67 years, 54.2% (19/35) were male, 45.8% (16/35) were female, and 94.3% (33 of 35) had a history of current or former smoking with two patients having a nonsmoking history (Table 1). During clinical care, 94.3% (33/35) patients had undergone systemic therapy, including chemotherapy and immunotherapy, and the remaining 5.7% (2/35) had not received systemic treatment. Of 35 independent samples tested, 20% (7/35) were therapy-naive samples. The distribution of analyzed tissue sites was as follows: 37.1% (13/35) liver, 34.2% (12 of 35) lung, 11.4% (4/35) lymph nodes, 5.7% (2/35) adrenal glands, 2.9% (1/35) pleura, 2.9% (1/35) mediastinum, 2.9% (1/35) retroperitoneum, and 2.9% (1/35) soft tissue. The mean tumor percentage in the analyzed samples was 66.3% (range 10%–95%). In the clinical diagnostic samples, tumors from 94.3% (33/35) patients had positive expression for at least one neuroendocrine marker (synaptophysin, chromogranin A, CD56, INSM1) by IHC. In the remaining two patients (#12 and #34), the IHC results were unavailable for one (#12) and only CD56 IHC was performed in the other (#34) with positivity only in rare tumor cells.
Comparison of RNA STEP Results With RNA-Seq and IHC
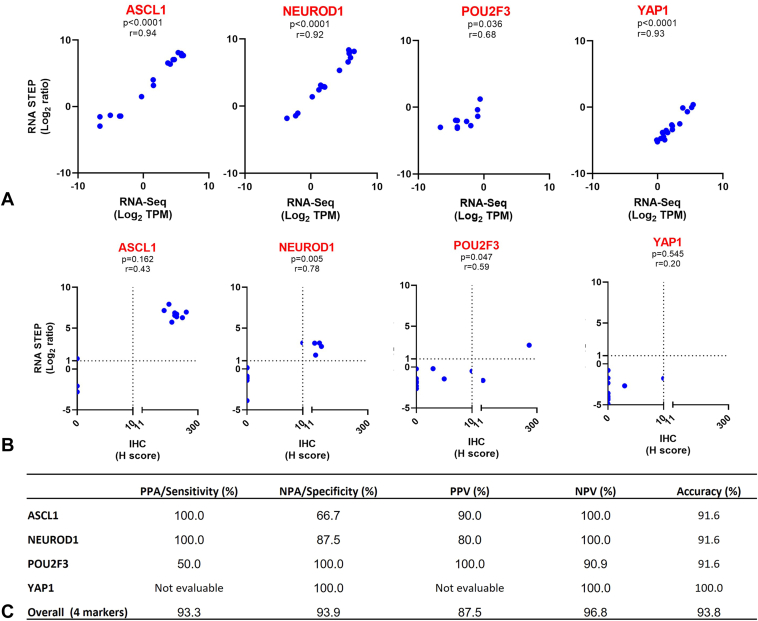
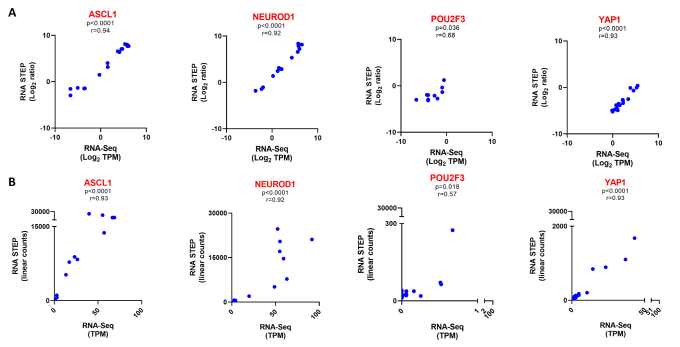
Correlation analyses were conducted to compare RNA STEP versus RNA-Seq results for ASCL1, NEUROD1, POU2F3, and YAP1 gene expression using samples tested with both methods (Supplementary Data 2, 17 samples, eight patients). Comparisons were performed with values converted to a logarithmic scale of 2 (Fig. 2A and Supplementary Data 3A) and with the normalized RNA STEP linear counts versus RNA-Seq TPM (transcripts per million, Supplementary Data 3B). All comparisons for ASCL1, NEUROD1, and YAP1 revealed statistically significant correlations with p values less than 0.0001 and r correlation coefficients more than 0.90. The POU2F3 correlations were also statistically significant, though with weaker correlation values of r equal to 0.68 and p value equal to 0.036 for the normalized logarithmic comparison and r equal to 0.57 and p value equal to 0.018 for the normalized linear count raw data comparison.
Figure 2.
Comparison of RNA STEP, RNA-Seq, and IHC results. (A) RNA STEP and RNA-Seq revealed a statistically significant correlation for four transcriptional factors of SCLC (n = 17, p-values all <0.05). Numbers in the x and y axis are illustrated as log2 values of RNA-Seq TPM values and log2 ratio of RNA STEP, respectively. (B) Statistically significant correlations were observed in the comparison of RNA STEP (log2 ratio) and IHC (H score) results for the NEUROD1 and POU2F3 genes (n = 12, p < 0.05). (C) In the comparative analyses of RNA STEP and IHC results, the PPA or sensitivity reached 100% for the ASCL1 and NEUROD1 genes, whereas the NPA or specificity achieved 100% for the POU2F3 and NEUROD1 genes. The YAP1 gene was not assessable for PPA or PPV due to the absence of any positive results. IHC, immunohistochemistry; NPA, negative percent agreement; PPA, positive percent agreement; RNA-Seq, RNA sequencing; RNA STEP, RNA Salah Targeted Expression Panel.
To evaluate whether the RNA STEP results also correlated with protein expression, RNA STEP results were compared with protein expression by IHC for 12 samples (Supplementary Data 4). Despite the low number of samples, a significant correlation was observed between the RNA and protein expression for NEUROD1 (N = 12, r = 0.78, p = 0.005) and POU2F3 genes (r = 0.59, p = 0.047), but no significant correlation was observed for the ASCL1 and YAP1 genes (Fig. 2B). Concordance between the RNA STEP and IHC results was also evaluated using a log2 ratio cutoff of more than or equal to 1 and a H score cutoff of more than 10 to classify as positive for high gene and protein expression, respectively. The positive percent agreements (PPAs or sensitivity) for the ASCL1, NEUROD1, and POU2F3 genes were 100%, 100%, and 50%, respectively. The YAP1 gene could not be evaluated for PPA because there were no positive results by either assay. The negative percent agreements (NPAs or specificity) were 66.7%, 87.5%, 100%, and 100% for the ASCL1, NEUROD1, POU2F3, and YAP1 genes, respectively. Overall, PPA and NPA for the four markers were 93.3% and 93.9%, respectively (Fig. 2C).
Gene Expression Levels
High expression of ASCL1, NEUROD1, and POU2F3 genes was observed in 88.6% (31/35), 57.2% (20/35), and 11.4% (4/35) of the samples with RNA STEP, respectively. None of the samples had high YAP1 gene expression (Fig. 3A and Supplementary Data 5).
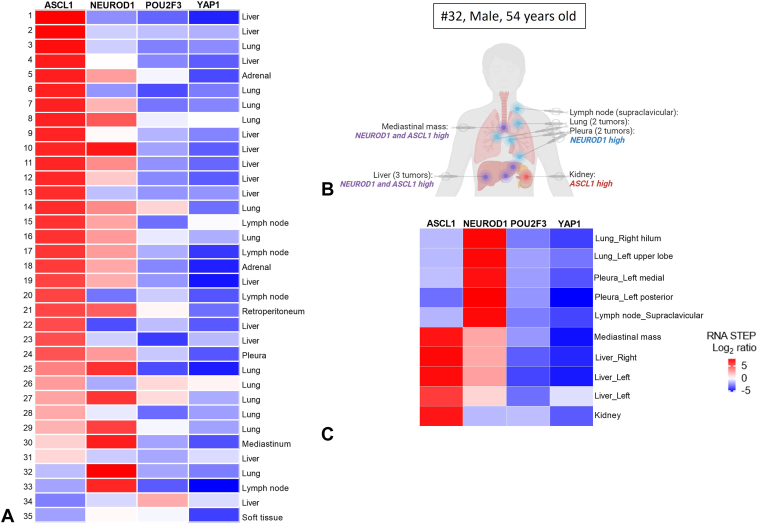
Figure 3.
ASCL1, NEUROD1, POU2F3, and YAP1 gene expression profile of SCLC. (A) RNA expression levels of ASCL1, NEUROD1, POU2F3, and YAP1 genes in the tested 35 tumor samples by RNA STEP. RNA STEP results of samples 1 to 31 and 32 to 33 were ordered by highest to lowest ASCL1 and NEUROD1 results, respectively. (B, C) Comparison of ASCL1, NEUROD1, POU2F3, and YAP1 gene expression levels in primary and metastatic tumors of same patient (#32, 54 y old, male) by RNA STEP. The distinct expression profiles observed in primary and metastatic tissues highlight substantial intrapatient heterogeneity. (B) Visual representation illustrating the gene expression profile of each tumor site. Created using BioRender.com. (C) Gene expression levels assessed by RNA STEP. RNA STEP, RNA Salah Targeted Expression Panel.
In 42.8% (15/35) of the samples, high expression of only one of the transcription regulator genes was observed. In 60% (20/35) of the samples, high expression of two or three genes was observed as follows: 45.7% (16/35) samples with ASCL1 and NEUROD1 high gene expression, 2.9% (1/35) samples with ASCL1 and POU2F3 high gene expression, and 5.7% (2/35) samples with ASCL1, NEUROD1, and POU2F3 high gene expression. One sample did not have high expression of any of these transcription regulation genes (Fig. 3A and Supplementary Data 5).
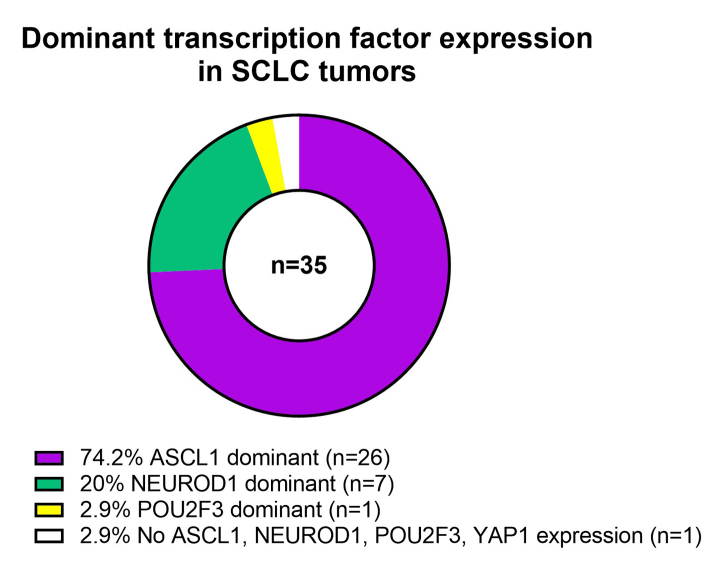
The transcription regulator gene with the most dominant (highest) expression was ASCL1 in 74.2% (26/35), NEUROD1 in 20% (7/35), and POU2F3 in 2.9% (1/35) (Supplementary Data 6). Of note, in the one sample with dominant POU2F3 gene expression (#34), only rare tumor cells had expression of CD56, a neuroendocrine marker, with IHC.
RNA STEP testing was performed successfully on 10 autopsy tumor samples from multiple tumor sites in one male RTD donor (#32) with SCLC. Tumors from the lungs, pleural tumors, and a supraclavicular lymph node had sole high NEUROD1 expression. Both high NEUROD1 and ASCL1 expression was detected in the liver tumors and a mediastinal mass. By contrast, a more geographically distant kidney tumor had sole high ASCL1 expression (Fig. 3B and C). Analyses of RNA-Seq results (Supplementary Data 7) on this patient revealed a similar pattern of positivity as the RNA STEP results.
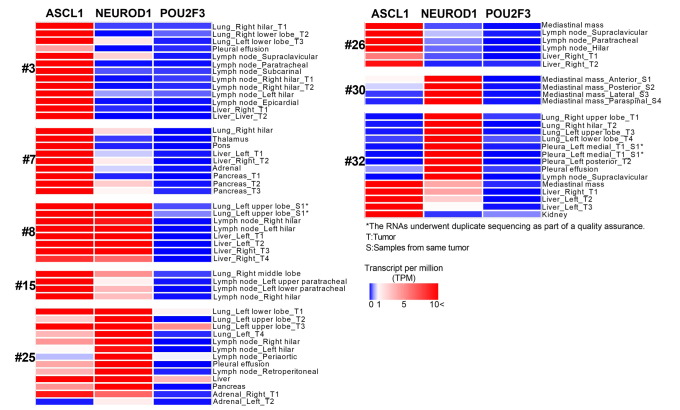
RNA-Seq analyses of multiple tumor sites from eight RTD donors with SCLC highlighted the high prevalence of intermetastatic heterogeneity13 (#3, 7, 25, 32) for transcriptional regulator gene expression (Supplementary Data 7 and 8).
SCLC, LUAD, and SqCC Gene Expression Profiles
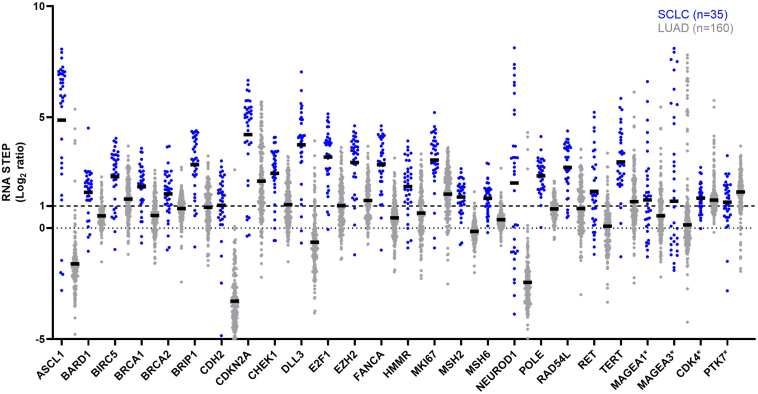
Among the 204 genes analyzed for expression with RNA STEP, 26 genes exhibited a high mean gene expression in the 35 tested SCLC samples (mean log2 ratio ≥ 1) (Fig. 4, Supplementary Data 9). Many of these genes play crucial roles in diverse pathways, including the basic helix-loop-helix family of transcription factors, DNA damage and repair processes, cell cycle regulation, epithelial-mesenchymal transition, NOTCH signaling, WNT signaling, tyrosine kinases, cancer antigens, and a proliferation marker. Within this subset of 26 genes with high general expression in SCLC, 22 genes also had significantly (p < 0.0001) higher relative expression in SCLC compared with LUAD (Fig. 4).
Figure 4.
Gene expression analysis in SCLC and LUAD. RNA STEP revealed high expression of 26 genes in SCLC (mean log2 ratio ≥ 1 by RNA STEP). Among these, 22 genes exhibited significantly higher expression in SCLC compared with LUAD (p < 0.0001). The black bars represent the mean values. ∗No statistically significant difference was detected for the MAGEA1, MAGEA3, CDK4, and PTK74 genes between SCLC and LUAD. LUAD, lung adenocarcinoma; RNA STEP, RNA Salah Targeted Expression Panel.
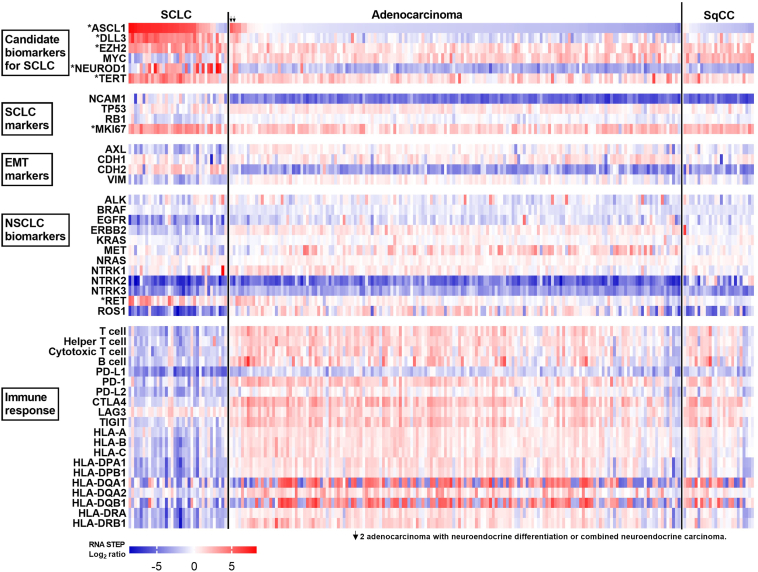
Within the lung cancer cohort of 220 samples, high ASCL1 expression was observed in 88.6% (31/35), 3.7% (6/160), and 0% (0/25) of the SCLC, LUAD, and SqCC samples, respectively. The clinicopathologic features of the six patients with LUAD with high ASCL1 expression were reviewed. Neuroendocrine differentiation or combined neuroendocrine carcinoma was observed in two of the six samples (Fig. 5, arrows). Neuroendocrine differentiation was observed in the same biopsy sample of one patient, and the subsequent resection specimen from the other patient revealed a large cell neuroendocrine carcinoma component. High NEUROD1 expression was found in 57.2% (20/35), 2.5% (4/160), and 4% (1/25) of the SCLC, LUAD, and SqCC samples, respectively. High DLL3 and EZH2 expression was observed in 91.4% (32/35) and 91.4% (32/35) in SCLCs, 11.2% (18/160) and 52.5% (84/160) in LUADs, and 24% (6/25) and 76% (19/25) in SqCCs, respectively (Fig. 5). SCLCs had high TERT expression in 91.4% (32/35) of samples versus 51.2% (82/160) in LUAD and 56% (14/25) in SqCC samples. High CDKN2A expression was observed in 85.8% (30/35) of the SCLC samples, and high MYC expression was found in 25.8% (9/35) of the SCLC samples (Fig. 5).
Figure 5.
Gene expression profile of SCLC (n = 35), adenocarcinoma (n = 160), and SqCC (n = 25). ∗Genes exhibiting elevated expression level in SCLC (mean log2 ratio ≥) and having a statistically significant higher in expression in SCLC compared with LUAD (p < 0.0001). LUAD, lung adenocarcinoma; SqCC, squamous cell carcinoma.
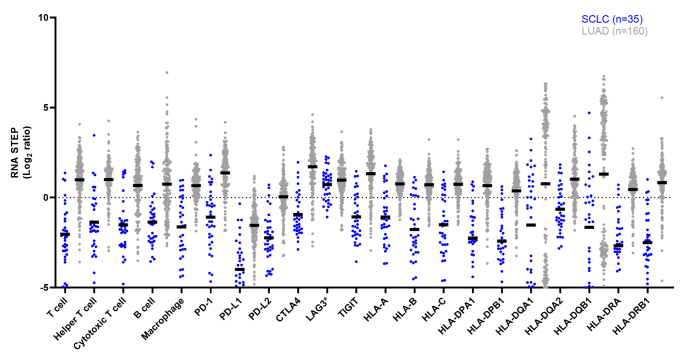
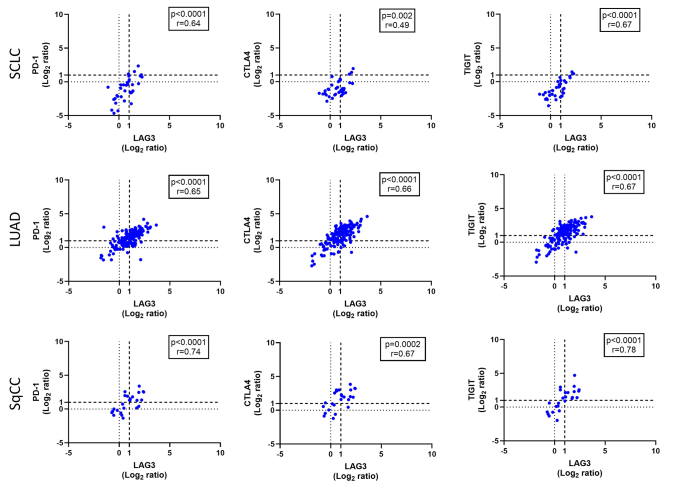
Tyrosine kinase genes associated with targeted therapy in NSCLC, such as EGFR, ERBB2, MET, and ROS1, had lower expression in SCLC than LUAD (Supplementary Data 9). In contrast, high RET expression was observed in 60% (21/35) of SCLC versus 16.9% (27/160) of LUAD and 4% (1/25) of SqCC samples. In addition, 93 genes had a mean log2 ratio less than 0 in SCLC and exhibited a significant decrease (p < 0.0001) in SCLC compared with LUAD (Supplementary Data 10). The SCLC cohort had generally lower expression of genes associated with T cells, helper T cells, cytotoxic T cells, B cells, and the HLA genes than the LUAD cohort, reflecting a generally “colder” immune microenvironment. Immune checkpoint markers, except LAG3, were also lower in the SCLC versus LUAD samples, suggesting a lower prevalence of immune cells and lower HLA antigen presentation in SCLC. Of 14 SCLC cases with high LAG3 expression, none had concurrently high programmed death-ligand 1 (PD-L1 protein, CD274 gene) expression, three had high programmed cell death protein-1 (PD-1 protein, PDCD1 gene) expression, and three had high CTLA4 expression, including one SCLC case with high LAG3, PDCD1, and CTLA4 expression (Supplementary Data 11). The mean LAG3 gene expression was similar between SCLC (mean log2 ratio = 0.72) and LUAD (mean log2 ratio = 0.97), despite the lower expression of immune cell markers in SCLC. The prevalence of samples with high LAG3 expression was also similar between SCLC (40.0%, 14/35) and LUAD (51.2%, 82/160). In SCLC, LUAD, and SqCC, LAG3 had a positive significant correlation with PDCD1, CTLA4, and TIGIT, despite gene expression values for PDCD1, CTLA4, and TIGIT being generally low in SCLC (all p < 0.05, Supplementary Data 12).
Discussion
Therapeutic advancements for SCLC have historically fallen behind those for NSCLC. Nonetheless, in recent years, substantial strides have been made in understanding the molecular aspects of SCLC biology. Comprehensive genomic and transcriptomic analyses by tissue-based and blood-based testing have revealed molecular subtypes within SCLC categorized based on the gene expression levels of the following four transcriptional regulators: ASCL1, NEUROD1, POU2F3, and YAP1. Blood, unlike SCLC tissue, is easily available, and blood-based methods can rapidly provide molecular information from circulating biomarkers and cell free DNA/cell free RNA, making them ideal for ongoing monitoring.14,15 Nevertheless, blood-based testing may have limitations in sensitivity and specificity, especially when detecting low-abundance targets or in early stage disease. Future studies should aim to validate and compare blood- and tissue-based methods in lung cancer to fully establish their comparative effectiveness. In this study, the primary objective was to analyze gene expression variations, including ASCL1, NEUROD1, POU2F3, and YAP1, in SCLC with a clinical tissue-based targeted RNA expression panel, RNA STEP. Another aim was to gain a better understanding of the gene expression landscape of SCLC, especially genes associated with potential as diagnostic, prognostic, and therapeutic biomarkers.
RNA STEP was previously validated for clinical use with testing of more than 100 clinical samples. This study also aimed to evaluate the assay’s correlation for SCLC transcription regulator genes as compared with RNA-Seq and IHC results from the same samples. RNA STEP exhibited statistically significant correlations with RNA-Seq for ASCL1, NEUROD1, and YAP1 (all p < 0.0001), and, though weaker, also for POU2F3 (p = 0.036). RNA STEP and IHC results revealed 93.8% overall accuracy for all four markers. These findings underscore the ability of RNA STEP to provide accurate expression levels for these transcriptional regulators with the caveat that the sample size for these comparisons was small.
After assuring that the RNA STEP assay results were similar to RNA-Seq and IHC results, the prevalence of high ASCL1, NEUROD1, POU2F3, and YAP1 gene expression in 35 SCLC samples with RNA STEP data was explored. High expression of the ASCL1, NEUROD1, and POU2F3 genes was detected in 88.6%, 57.2%, and 11.4% of the SCLC samples, respectively. Consistent with previous studies,16 none of the SCLC samples had high YAP1 expression in our cohort. The dominant transcription regulator was ASCL1 in 74.2%, NEUROD1 in 20%, and POU2F3 in 2.9% of the samples (Supplementary Data 6). The average ASCL1 gene expression was approximately fivefold higher in SCLC samples relatively to the pooled control sample (Supplementary Data 9). This observation aligns with the established role of the ASCL1 transcription factor as the master regulator in SCLC. The one sample with dominant POU2F3 gene expression had neuroendocrine marker expression in only a few tumor cells by IHC consistent with a previous report describing the lower neuroendocrine character of tumors with high POU2F3 levels.17 Of note, 54.2% of the samples had high expression of more than one transcription regulator gene. In the initial studies that delineated molecular subtypes, the occurrence of double/triple expression of transcription regulators was not reported. This may be attributed to the utilization of experimental models (human cell line models, genetically engineered animal models, and patient-derived xenografts) in these studies rather than human tumor samples which have a heterogeneous nature.18 In experimental models, there could be a clonal selection bias favoring one transcription regulator and suppressing others in the emerging monoclonal population, such that expression of only one marker is detected as opposed to the double/triple marker expression that may have been present in the native human samples.19
Furthermore, 50% of SCLCs may undergo subtype switching or exhibit a loss of expression of the ASCL1 and NEUROD1 transcription factors as the disease progresses.20,21 Rapid autopsies provide primary and metastatic samples to enable studies of both intratumoral and intermetastatic heterogeneity, including molecular changes during disease progression.22 In our study, a comparative analysis of RNA STEP results by testing primary and metastatic tumors from 12 RTD donors was conducted. Notably, for donor #32, lung and pleural tumors, along with a lymph node, displayed high gene expression of NEUROD1 and low expression of ASCL1. Conversely, the mediastinal mass, liver, and kidney samples exhibited higher ASCL1 than NEUROD1 gene expression (Fig. 3B and C). In a previous study, loss of ASCL1 expression was documented during progression20; conversely, our study reveals the loss of NEUROD1 expression in distant metastatic sites.
RNA-Seq results provided additional support for the presence of transcription regulator intermetastatic heterogeneity (Supplementary Data 8). The heterogeneity of transcription regulator gene expression by tumor site suggests that a uniform treatment strategy based on transcription regulator subtype evaluation of one tumor site may not be optimal. For example, if a trial was designed to exclude patients with low ASCL1 expression and the lung from the patient associated with #32 was tested, this patient would be excluded from the trial, despite having high ASCL1 expression in the untested metastatic sites. Rather than designing trials based on transcription regulator subtypes, a more effective strategy might be to consider the most common general characteristics of SCLC and design trials with minimal exclusion criteria. The trials can aim to identify which patients might need a different therapeutic strategy. These results also provide evidence for the concept that different metastases may require different types of treatment, which is consistent with the common observation that some lesions regress or remain stable during therapy, but others progress.
RNA STEP results also provided a glimpse into the molecular landscape of SCLC. Of 204 genes covered by RNA STEP, 26 genes had high expression in more than half of the SCLC cases with 22 significantly higher expression in SCLC than LUAD (p < 0.0001). These genes included the major transcriptional regulators, ASCL1 and NEUROD1, and DLL3, which is regulated by ASCL1 (Fig. 4). The DLL3 protein is highly expressed on the cell surface of SCLC and plays a pivotal role as a negative regulator of NOTCH signaling.19,23 As of September 8, 2023, there are 327 clinical trials including patients with SCLC on the clinicaltrials.gov website.1 Of these, 22 specifically focus on DLL3, and within this subset, six are actively recruiting. The DeLLphi-301 trial in patients with ES SCLC reported an overall response rate of 40% to DLL3-targeted therapy tarlatamab-dlle (Imdelltra, Amgen, Inc.) leading to accelerated approval by the United States Food and Drug Administration (FDA) on May 16, 2024, which was biomarker agnostic.24 With RNA STEP, high DLL3 expression was detected in 91.4% of SCLC samples, irrespective of ASCL1 expression. Although some studies indicate higher DLL3 expression in mainly the ASCL1-high group, others, including our study, also had high DLL3 expression in NEUROD1-expressing SCLCs.19 The high prevalence of high DLL3 gene expression in SCLC by RNA STEP corresponds with the high response rate to DLL3-targeted therapy in this patient population.
As a member of the PRC2 family, EZH2 serves as a transcription factor and activates trimethylation of H3K27me3, which alters the expression of downstream target genes resulting in cell proliferation, apoptosis, and senescence.25 Activation of the EZH2 gene is associated with metastasis and reduced therapy response in lung cancer.26,27 Thus, EZH2 might be a therapeutic target to consider for patients with SCLC. A subset of 25.8% of the SCLC samples in this study had high MYC expression. The MYC family genes—MYC, MYCL, and MYCN—are recognized as oncogenic drivers and are considered potential biomarkers for SCLC.28 A trial targeting MYC could benefit from considering high MYC expression as an inclusion criteria; however, additional arms or sites might be needed to assure accrual if a similar cutoff for high expression is used. Comparative analysis of gene expression levels for key genes in SCLC versus LUAD revealed a similar prevalence of high MYC expression in SCLC and LUAD.
Comparative analyses of SCLC with LUAD and SqCC revealed a higher frequency of high RET expression in SCLC (60%) than LUAD (16.9%) or SqCC (4%), consistent with the findings of a study29 that reported 80% of SCLC cases had strong RET positivity by IHC. The RET gene encodes a transmembrane receptor, and the activation of this receptor triggers multiple oncogenic pathways. In 2022, FDA approved RET inhibitors for patients with RET fusion-positive solid cancers.30 In this study, 10 SCLC samples received previous clinical testing for RET fusions with clinical next-generation sequencing (Moffitt STAR, Illumina TSO500 platform). Although none of these samples had a RET fusion, six of 10 had elevated RET expression level by RNA STEP. One study reported that chromatin structure and promoter hypomethylation affect RET expression in cancer cells.31,32 As such, the high RET expression in our SCLC cohort might be associated with either copy number gains or epigenetic mechanisms. RET amplifications have been reported in many tumor types, including lung cancer.33 Platt et al34 reported that incidence of RET amplification is higher than rearrangements in NSCLC (2.8% versus 0.7%). Nevertheless, RET gene alterations have not been well studied in SCLC, and the clinical significance of RET copy number changes and their correlation with increased RET protein expression have not been well characterized yet.33
Other therapeutic targets for NSCLC, such as the ALK, BRAF, and EGFR genes, had lower expression in SCLC. In our cohort, high TERT expression was detected in 91.4% of SCLCs suggesting its potential as a therapeutic candidate for the treatment of SCLC. The TERT gene plays a key role in carcinogenesis by coding a protein which can prevent progressive shortening of telomeres by the reverse transcriptase activity.35 The results of this study are consistent with a previous study that reported elevated TERT expression level in SCLC, particularly in SCLCs that have undergone transformation from LUAD.36 To the best of our knowledge, the number of studies describing the high prevalence of TERT expression is limited. One study describes how abnormal methylation of TERT promotor leads to higher TERT expression which enhances the progression and radiotherapy resistance in SCLC.37 Another study suggests that TERT inhibitors (NU-1) can promote antitumor immunity after radiation.38
The survival of cancer cells and the effectiveness of therapy are heavily affected by the immune microenvironment. Nevertheless, our understanding of the immune microenvironment in SCLC and reliable biomarkers for predicting response to immunotherapy in SCLC remain elusive.39 Previous studies have found an association between survival postimmunotherapy with the infiltration of cytotoxic cells, coupled with high MHC-I expression in pretreatment samples from patients with SCLC.40 In accordance with other studies,41,42 our study revealed an immune cold profile in SCLC with low expression of markers for T lymphocytes, including CD4+ helper T cells and CD8+ cytotoxic T cells, immune checkpoint proteins (except the LAG3 gene), and HLA genes. Although expression of genes associated with PD-1 and CTLA4 was generally lower than LAG3 gene expression, their expression still correlated with LAG3 expression. It may be that the lower overall prevalence of immune cells in SCLC causes a parallel lower level of immune checkpoint expression, though with continued expression of LAG3, PD-1, and CTLA4 on specific immune cells. We recognize that this RNA expression study provides only a glimpse of the immune cold environment of SCLC. Future studies, such as with special technologies like multiplex immunofluorescence, are needed to better elucidate the mechanisms that cause the low immunogenicity in SCLC. Mechanistic possibilities for the immune cold environment include impaired crosstalk between T cells and conventional-type dendritic cells and low expression of HLA genes.
The LAG3 protein, encoded by the LAG3 gene, inhibits T cell responses by binding to stable peptide-MHC class II and is a promising target for immunotherapy.43,44 In our cohort, 40% of SCLCs (14 of 35) had high LAG3 expression. This finding aligns with analysis of a public data set,45 which described elevated LAG3 expression level in SCLC compared with normal lung tissue. These results underscore the potential of LAG3 as a candidate biomarker in SCLC, supporting the importance of clinical trials to study the potential for LAG3 checkpoint inhibition (NCT03219268, NCT03365791, NCT03538028). Of note, inhibition of LAG3 may be necessary, but insufficient alone, to ignite immune activation against the tumor, especially with the low expression of HLA genes in SCLC. A therapeutic strategy that does not require HLA antigen presentation may be needed for immune activation. Anti-LAG3 plus a bispecific T cell engager targeting an SCLC surface protein might activate T cells without relying on HLA antigen presentation.46
Conclusion
The complementary addition of clinical RNA expression profiling in SCLC to established diagnostic techniques provides support for molecular diagnostic classification. The implementation of RNA expression profiling of SCLC clinical samples harnesses the opportunity to inform clinical trial design and translational biomarker studies. The RNA STEP has an extraction-free, simple, and rapid workflow of 3 days which works well with FFPE samples that often have degraded and low RNA yield and offers potential for a comprehensive perspective on the molecular distinctions within tumors. This panel may unveil novel biomarkers, identify prospects for clinical trials, and present therapeutic opportunities for the management of SCLC.
CRediT Authorship Contribution Statement
Hilal Ozakinci: Conceptualization, investigation, sample collection, methodology, project supervision, pathology review, data analysis, writing-original draft, review and editing.
Aileen Y. Alontaga: Methodology-RNA STEP, writing-review and editing.
Pedro Cano: Developed the informatics pipeline of RNA STEP.
John M. Koomen: Supervision, Writing-Review and editing.
Bradford A. Perez: Methodology-RNA-Seq, Supervision, Data Acquisition, Writing-review and editing.
Amer A. Beg: Data analysis, supervision, writing-review and editing.
Alberto A. Chiappori: Funding Acquisition, Conceptualization, Writing-Review and editing.
Eric B. Haura: Project conceptualization, Supervision, Funding Acquisition, Writing-Review and editing.
Theresa A. Boyle: Project conceptualization, funding acquisition, methodology-RNA STEP, data analysis, supervision, writing-original draft, review and editing.
All authors read and approved the final manuscript.
Study Approval
This work has been carried out in accordance with The Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans. Tissue samples were procured in line with WHO Guiding Principles on Human Cell, Tissue and Organ Transplantation. Clinical trial and rapid tissue donation were approved by the Institutional Review Board (Advarra, Columbia, MD, Pro00014653 and Pro00030829, respectively). RNA STEP analyses were conducted with a study protocol exemption (MCCC23158, Pro00076403).
Research Data/Data Availability
The data that support the findings of this study are available from the corresponding authors on reasonable request.
Submission Declaration and Verification
This work is not under consideration for publication elsewhere.
Disclosure
Dr. Chiappori received funding from Bristol-Myers Squibb for the clinical trial (MCC19163). Dr. Boyle and Dr. Koomen declare grants/contracts with Bristol-Myers Squibb unrelated to this research. Dr. Perez declares receiving grants from Bristol-Myers Squibb; providing consulting services for AstraZeneca, Bristol-Myers Squibb, G1 Therapeutics, and Novocure; and having board membership in Out of Zion. Dr. Haura declares providing consulting services for Kanaph Therapeutics and ORI Capital II; receiving research funding from Revolution Medicines; and providing advisory services for RevMed and Janssen, all unrelated to this research. The remaining authors declare no conflict of interest.
Acknowledgments
The RNA STEP validation and testing was generously funded by the Salah Foundation. The rapid tissue donation project was funded by Bristol-Myers Squibb and the Moffitt Lung Cancer Center of Excellence. The clinical trial (MCC 19163) was funded by Bristol Myers Squibb, and the biomarker research for this manuscript was funded by the Jennifer Stevens Lung Cancer Research Fund, Thoracic Oncology, and Moffitt Cancer Center. The Tissue Core, Molecular Genomics Core, and Biostatistics and Bioinformatics Shared Resource are funded in part by the National Cancer Institute Cancer Center Support grant, which confers Moffitt’s status as a National Cancer Institute–designated Comprehensive Cancer Center (NCI P30-CA076292).
We extend our heartfelt appreciation to the patients and their loved ones who graciously provided consent for participation in the thoracic rapid tissue donation program and to the participants of the clinical trial who contributed samples which supported this research.
Footnotes
Cite this article as: Ozakinci H, Alontaga AY, Cano P, et al. Unveiling the molecular features of SCLC with a clinical RNA expression panel. JTO Clin Res Rep. 2024;5:100723.
Note: To access the supplementary material accompanying this article, visit the online version of the JTO Clinical and Research Reports at www.jtocrr.org and at https://doi.org/10.1016/j.jtocrr.2024.100723.
Supplementary Data
Supplementary Data 10.
Supplementary Data 12.
Supplementary Data 3.
Supplementary Data 6.
Supplementary Data 7.
References
- 1.College of American Pathologists. The CAP cancer protocols. https://www.cap.org/protocols-and-guidelines/cancer-reporting-tools/cancer-protocol-templates. Accessed July 2024.
- 2.Cedrés S., Ponce-Aix S., Pardo-Aranda N., et al. Analysis of expression of PTEN/PI3K pathway and programmed cell death ligand 1 (PD-L1) in malignant pleural mesothelioma (MPM) Lung Cancer. 2016;96:1–6. doi: 10.1016/j.lungcan.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Fujino K., Motooka Y., Hassan W.A., et al. Insulinoma-associated Protein 1 is a crucial regulator of neuroendocrine differentiation in lung cancer. Am J Pathol. 2015;185:3164–3177. doi: 10.1016/j.ajpath.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 4.Rudin C.M., Poirier J.T., Byers L.A., et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer. 2019;19:289–297. doi: 10.1038/s41568-019-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qu S., Fetsch P., Thomas A., et al. Molecular subtypes of primary SCLC tumors and their associations with neuroendocrine and therapeutic markers. J Thorac Oncol. 2022;17:141–153. doi: 10.1016/j.jtho.2021.08.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ng J., Cai L., Girard L., et al. Molecular and pathologic characterization of YAP1-expressing small cell lung cancer cell lines leads to reclassification as SMARCA4-deficient malignancies. Clin Cancer Res. 2024;30:1846–1858. doi: 10.1158/1078-0432.CCR-23-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caeser R., Egger J.V., Chavan S., et al. Genomic and transcriptomic analysis of a library of small cell lung cancer patient-derived xenografts. Nat Commun. 2022;13:2144. doi: 10.1038/s41467-022-29794-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hwang S., Hong T.H., Kim H.K., et al. Whole-section landscape analysis of molecular subtypes in curatively resected small cell lung cancer: clinicopathologic features and prognostic significance. Mod Pathol. 2023;36 doi: 10.1016/j.modpat.2023.100184. [DOI] [PubMed] [Google Scholar]
- 9.Schwendenwein A., Megyesfalvi Z., Barany N., et al. Molecular profiles of small cell lung cancer subtypes: therapeutic implications. Mol Ther Oncolytics. 2021;20:470–483. doi: 10.1016/j.omto.2021.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gay C.M., Stewart C.A., Park E.M., et al. Patterns of transcription factor programs and immune pathway activation define four major subtypes of SCLC with distinct therapeutic vulnerabilities. Cancer Cell. 2021;39:346–360.e7. doi: 10.1016/j.ccell.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyle T.A., Quinn G.P., Schabath M.B., et al. A community-based lung cancer rapid tissue donation protocol provides high-quality drug-resistant specimens for proteogenomic analyses. Cancer Med. 2020;9:225–237. doi: 10.1002/cam4.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alontaga A.Y., Cano P., Ozakinci H., et al. Implementation of a high-accuracy targeted gene expression panel for clinical care. J Mol Diagn. 2024;26:685–699. doi: 10.1016/j.jmoldx.2024.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.J Saller J., Boyle T.A. Molecular pathology of lung cancer. Cold Spring Harb Perspect Med. 2022;12 doi: 10.1101/cshperspect.a037812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muscarella L.A., Mazza T., Fabrizio F.P., et al. Neuroendocrine-related circulating transcripts in small-cell lung cancers: detection methods and future perspectives. Cancers (Basel) 2021;13:1339. doi: 10.3390/cancers13061339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiatt J.B., Doebley A.L., Arnold H.U., et al. Molecular phenotyping of small cell lung cancer using targeted cfDNA profiling of transcriptional regulatory regions. Sci Adv. 2024;10 doi: 10.1126/sciadv.adk2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Megyesfalvi Z., Barany N., Lantos A., et al. Expression patterns and prognostic relevance of subtype-specific transcription factors in surgically resected small-cell lung cancer: an international multicenter study. J Pathol. 2022;257:674–686. doi: 10.1002/path.5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poirier J.T., George J., Owonikoko T.K., et al. New approaches to SCLC therapy: from the laboratory to the clinic. J Thorac Oncol. 2020;15:520–540. doi: 10.1016/j.jtho.2020.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horvath L., Lang C., Boettiger K., Aigner C., Dome B., Megyesfalvi Z. Potential subtype-specific therapeutic approaches in small cell lung cancer. Curr Opin Oncol. 2024;36:51–56. doi: 10.1097/CCO.0000000000001005. [DOI] [PubMed] [Google Scholar]
- 19.Baine M.K., Hsieh M.S., Lai W.V., et al. SCLC subtypes defined by ASCL1, NEUROD1, POU2F3, and YAP1: a comprehensive immunohistochemical and histopathologic characterization. J Thorac Oncol. 2020;15:1823–1835. doi: 10.1016/j.jtho.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiang C.L., Huang H.C., Luo Y.H., et al. Clinical utility of immunohistochemical subtyping in patients with small cell lung cancer. Lung Cancer. 2024;188 doi: 10.1016/j.lungcan.2024.107473. [DOI] [PubMed] [Google Scholar]
- 21.Lim J.S., Ibaseta A., Fischer M.M., et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature. 2017;545:360–364. doi: 10.1038/nature22323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Megyesfalvi Z., Heeke S., Drapkin B.J., et al. Unfolding the secrets of small cell lung cancer progression: novel approaches and insights through rapid autopsies. Cancer Cell. 2023;41:1535–1540. doi: 10.1016/j.ccell.2023.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borromeo M.D., Savage T.K., Kollipara R.K., et al. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. 2016;16:1259–1272. doi: 10.1016/j.celrep.2016.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahn M.J., Cho B.C., Felip E., et al. Tarlatamab for patients with previously treated small-cell lung cancer. N Engl J Med. 2023;389:2063–2075. doi: 10.1056/NEJMoa2307980. [DOI] [PubMed] [Google Scholar]
- 25.Duan R., Du W., Guo W. EZH2: a novel target for cancer treatment. J Hematol Oncol. 2020;13:104. doi: 10.1186/s13045-020-00937-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Entezari M., Taheriazam A., Paskeh M.D.A., et al. The pharmacological and biological importance of EZH2 signaling in lung cancer. Biomed Pharmacother. 2023;160 doi: 10.1016/j.biopha.2023.114313. [DOI] [PubMed] [Google Scholar]
- 27.Gardner E.E., Lok B.H., Schneeberger V.E., et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 axis. Cancer Cell. 2017;31:286–299. doi: 10.1016/j.ccell.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brägelmann J., Böhm S., Guthrie M.R., Mollaoglu G., Oliver T.G., Sos M.L. Family matters: how MYC family oncogenes impact small cell lung cancer. Cell Cycle. 2017;16:1489–1498. doi: 10.1080/15384101.2017.1339849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dabir S., Babakoohi S., Kluge A., et al. RET mutation and expression in small-cell lung cancer. J Thorac Oncol. 2014;9:1316–1323. doi: 10.1097/JTO.0000000000000234. [DOI] [PubMed] [Google Scholar]
- 30.Duke E.S., Bradford D., Marcovitz M., et al. FDA approval summary: selpercatinib for the treatment of advanced RET fusion-positive solid tumors. Clin Cancer Res. 2023;29:3573–3578. doi: 10.1158/1078-0432.CCR-23-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shakiba E., Movahedi M., Majd A., Hedayati M. Investigating the expression and promoter methylation of RET gene in patients with medullary thyroid cancer with unmutated RET. J Cell Physiol. 2019;234:16304–16311. doi: 10.1002/jcp.28295. [DOI] [PubMed] [Google Scholar]
- 32.Griseri P., Garrone O., Lo Sardo A., et al. Genetic and epigenetic factors affect RET gene expression in breast cancer cell lines and influence survival in patients. Oncotarget. 2016;7:26465–26479. doi: 10.18632/oncotarget.8417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Desilets A., Repetto M., Yang S.R., Sherman E.J., Drilon A. RET-altered cancers-a tumor-agnostic review of biology, diagnosis and targeted therapy activity. Cancers (Basel) 2023;15:4146. doi: 10.3390/cancers15164146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Platt A., Morten J., Ji Q., et al. A retrospective analysis of RET translocation, gene copy number gain and expression in NSCLC patients treated with vandetanib in four randomized Phase III studies. BMC Cancer. 2015;15:171. doi: 10.1186/s12885-015-1146-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dratwa M., Wysoczańska B., Łacina P., Kubik T., Bogunia-Kubik K. TERT-regulation and roles in cancer formation. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.589929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mc Leer A., Foll M., Brevet M., et al. Detection of acquired TERT amplification in addition to predisposing p53 and Rb pathways alterations in EGFR-mutant lung adenocarcinomas transformed into small-cell lung cancers. Lung Cancer. 2022;167:98–106. doi: 10.1016/j.lungcan.2022.01.008. [DOI] [PubMed] [Google Scholar]
- 37.Zhai G., Li J., Zheng J., et al. hTERT promoter methylation promotes small cell lung cancer progression and radiotherapy resistance. J Radiat Res. 2020;61:674–683. doi: 10.1093/jrr/rraa052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., Betori R.C., Pagacz J., et al. Targeting telomerase reverse transcriptase with the covalent inhibitor NU-1 confers immunogenic radiation sensitization. Cell Chem Biol. 2022;29:1517–1531.e7. doi: 10.1016/j.chembiol.2022.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang S., Cheng Y. Immunotherapy for extensive-stage small-cell lung cancer: current landscape and future perspectives. Front Oncol. 2023;13 doi: 10.3389/fonc.2023.1142081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudin C.M., Balli D., Lai W.V., et al. Clinical benefit from immunotherapy in patients with SCLC is associated with tumor capacity for antigen presentation. J Thorac Oncol. 2023;18:1222–1232. doi: 10.1016/j.jtho.2023.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen M., Chen R., Jin Y., et al. Cold and heterogeneous T cell repertoire is associated with copy number aberrations and loss of immune genes in small-cell lung cancer. Nat Commun. 2021;12:6655. doi: 10.1038/s41467-021-26821-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He Y., Rozeboom L., Rivard C.J., et al. MHC class II expression in lung cancer. Lung Cancer. 2017;112:75–80. doi: 10.1016/j.lungcan.2017.07.030. [DOI] [PubMed] [Google Scholar]
- 43.Aggarwal V., Workman C.J., Vignali D.A.A. LAG-3 as the third checkpoint inhibitor. Nat Immunol. 2023;24:1415–1422. doi: 10.1038/s41590-023-01569-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maruhashi T., Sugiura D., Okazaki I.M., et al. Binding of LAG-3 to stable peptide-MHC class II limits T cell function and suppresses autoimmunity and anti-cancer immunity. Immunity. 2022;55:912–924.e8. doi: 10.1016/j.immuni.2022.03.013. [DOI] [PubMed] [Google Scholar]
- 45.Sun H., Dai J., Zhao L., et al. Lymphocyte activation gene-3 is associated with programmed death-ligand 1 and programmed cell death protein 1 in small cell lung cancer. Ann Transl Med. 2021;9:1468. doi: 10.21037/atm-21-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szijj P.A., Gray M.A., Ribi M.K., et al. Chemical generation of checkpoint inhibitory T cell engagers for the treatment of cancer. Nat Chem. 2023;15:1636–1647. doi: 10.1038/s41557-023-01280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.