Abstract
Rhopalosiphum padi virus (RhPV) is one of several picorna-like viruses that infect insects; sequence analysis has revealed distinct differences between these agents and mammalian picornaviruses. RhPV has a single-stranded positive-sense RNA genome of about 10 kb; unlike the genomes of Picornaviridae, however, this genome contains two long open reading frames (ORFs). ORF1 encodes the virus nonstructural proteins, while the downstream ORF, ORF2, specifies the structural proteins. Both ORFs are preceded by long untranslated regions (UTRs). The intergenic UTR is known to contain an internal ribosome entry site (IRES) which directs non-AUG-initiated translation of ORF2. We have examined the 5′ UTR of RhPV for IRES activity by translating synthetic dicistronic mRNAs containing this sequence in a variety of systems. We now report that the 5′ UTR contains an element which directs internal initiation of protein synthesis from an AUG codon in mammalian, plant, and Drosophila in vitro translation systems. In contrast, the encephalomyocarditis virus IRES functions only in the mammalian system. The RhPV 5′ IRES element has features in common with picornavirus IRES elements, in that no coding sequence is required for IRES function, but also with cellular IRES elements, as deletion analysis indicates that this IRES element does not have sharply defined boundaries.
Rhopalosiphum padi virus (RhPV) is an insect virus with a narrow host range, infecting aphids of the Rhopalosiphum and Schizaphis families (6, 11). Virus infection reduces both the life span and the reproductive capacity of the insects (6). RhPV was initially classified within the Picornaviridae, based largely on its physicochemical properties. However, sequence analysis has prompted a reevaluation of this attribution, and RhPV is now considered to belong to a group of insect viruses (the cricket paralysis-like viruses) with a picornavirus-like capsid structure but a distinct genome organization (15, 16). Other members of this group include Drosophila C virus, Plautia stali intestinal virus (PSIV), and cricket paralysis virus (CrPV) itself. The RNA genome of each of these viruses, including RhPV, encodes two polyproteins in separate open reading frames (ORFs) (15). ORF1 encodes nonstructural proteins that possess sequence similarity to both mammalian picornavirus and plant comovirus proteins. ORF2 encodes the three structural proteins that also show similarity to picornavirus capsid proteins. Both ORFs are preceded by long untranslated regions (UTRs) about 500 nucleotides (nt) long. In contrast, mammalian picornaviruses encode one long polyprotein with the structural proteins at the N-terminal region and the nonstructural proteins at the C terminus. The genome organization of the cricket paralysis-like viruses resembles that of the caliciviruses (23). However, there is no evidence for the production of a subgenomic RNA by these insect viruses, and calicivirus ORFs are not preceded by long UTRs.
It is well established that the initiation of translation on picornavirus RNA occurs by a cap-independent mechanism that is directed by an internal ribosome entry site (IRES) element within the 5′ UTR of the genome (reviewed in references 2 and 3). Picornavirus IRES elements are grouped into two major classes according to their predicted secondary structure and their activity in vitro; there is little sequence identity between the two classes (reviewed in references 2 and 14). One class contains IRES elements from the enteroviruses and rhinoviruses, while the second class contains the cardiovirus and aphthovirus IRES elements. The latter IRES elements function efficiently in the rabbit reticulocyte lysate (RRL) translation system. In contrast, the poliovirus and rhinovirus IRES elements are inefficient in this system until the reaction is supplemented with HeLa cell extracts (5, 8). The hepatitis A virus IRES is distinct from those listed above and forms a minor class on its own; it can function in the RRL system, but its activity is stimulated in this system by liver cell and not HeLa cell extracts (12). These findings highlight the importance of cellular trans-acting factors in the mechanism of IRES action and could provide some explanation for the cellular tropism of picornaviruses. Indeed, it has been demonstrated that the intracellular activities of different picornavirus IRES elements vary in different cell types (4, 18).
The intergenic regions (IGRs) of both PSIV and RhPV have recently been shown to contain IRES elements. These direct the translation of the second ORFs, and the initiation of translation on both virus IGRs occurs at non-AUG start codons: on PSIV RNA, a CAA codon is used (21), while a CCU codon is probably used on RhPV RNA (7). Both the IGR and the 5′ UTR of CrPV have very recently been shown to contain IRES elements (25). In common with the situation for the related PSIV and RhPV, the initiation of CrPV ORF2 takes place at a non-AUG codon (CCU) (25). Both the 5′ UTR and the IGR sequences of CrPV were reported to function as IRES elements in insect cells and in the RRL in vitro translation system. Furthermore, the IGR IRES but not the 5′ UTR IRES of CrPV functioned in the wheat germ in vitro translation system (25).
The 5′ UTR of RhPV has many features in common with the 5′ ends of mammalian picornavirus RNAs. The RhPV 5′ terminus is uncapped and is predicted to form extensive secondary structure (15). The 5′ UTR of RhPV is predicted to be 580 nt long and is highly A and U rich. It has been suggested, but not proven, that the initiation of protein synthesis occurs at the third AUG (15). The RhPV 5′ UTR contains two AUG codons upstream of ORF1 but out of frame with the coding sequence of this gene; both are followed quickly by termination codons and are therefore unlikely to be used. However, there is another AUG codon, 6 nt downstream of the proposed ORF1 initiation site, which could also function as an initiation codon. Many of these features are also found in picornavirus IRES elements and prompted us to examine the 5′ UTR of RhPV for the presence of an IRES element which could direct the cap-independent initiation of translation of ORF1. To date, most IRES elements isolated from virus or mammalian mRNAs have been shown to be functional only in mammalian cells and translation systems derived from them. Here, we report that the 5′ UTR of RhPV contains an IRES element which functions in the RRL, wheat germ, and Drosophila in vitro translation systems.
MATERIALS AND METHODS
Plasmids.
DNA preparations and manipulations were performed using standard methods as described by Sambrook et al. (20) or by manufacturers of the reagents used. cDNA containing the 5′ UTR and ORF1 sequences of RhPV (15) was kindly donated by L. Domier (University of California). As the hepatitis C virus IRES element has been shown to require some coding sequence for efficient function (17), a sequence from the 5′ end of ORF1 was included in some of the constructs. For these, the 5′ UTR (nt 1 to 579) and the first 15 codons of the ORF1 sequence were amplified by PCR from RhPV cDNA using forward primer RFOR1 (Table 1) and reverse primer RREV624 (Table 1), each containing a BamHI site. The PCR product was ligated into pGEMTeasy (Promega), and the structures of the created plasmids were verified. The RhPV cDNA (624 nt) was then released by BamHI digestion and inserted in both orientations at the unique BamHI site of the dicistronic plasmid pGEM-CAT/LUC (24), between the two reporter genes (Fig. 1). The plasmid containing the RhPV 5′ UTR in the sense (genomic) orientation was designated pGEM-CAT/RhPVs/LUC (abbreviated RhPVs), and that containing this element in the antisense orientation was called pGEM-CAT/RhPVas/LUC (abbreviated RhPVas). The structures of the plasmid constructs were verified by restriction enzyme digestion and sequencing. The dicistronic plasmid pGEM-CAT/EMC/LUC (abbreviated EMC) containing the encephalomyocarditis virus (EMCV) IRES element has been described previously (24). The same 5′ portion of the RhPV sequence was also inserted at the unique BamHI site of a second dicistronic vector, pGUS/RXB/HOOK (19), between the β-glucuronidase (GUS) and HOOK reporter genes.
TABLE 1.
Sequences of oligonucleotides used to create RhPV 5′ UTR cDNA fragmentsa
| Primer name | Primer sequence |
|---|---|
| RFOR1 | 5′ ATAGGATCCGATAAAAGAACCTATCACACCG |
| RREV624 | 5′ TATGGATCCTGCGTTGAACTGACTTTGGT |
| RREV579 | 5′ ACGGATCCTATAAATAGATAAAG |
| RREV463 | 5′ ACGGATCCATATACAGAAGATAT |
| RREV374 | 5′ ACGGATCCTTGTTACGCAACTAG |
| RREV588 | 5′ ACGGATCCCGTAGACTATATAAA |
| RFOR100 | 5′ ACGGATCCATACGATATACTTAT |
BamHI sites, highlighted in bold, were added to enable cloning into the pGEM-CAT/LUC vector as described in the text.
FIG. 1.
Structure of the RhPV genome and plasmids used in this study. Various fragments of the 5′ end of the RhPV genome were amplified by PCR using primers containing BamHI sites, digested, and inserted between the CAT and LUC ORFs (at the unique BamHI site) in plasmid pGEM-CAT/LUC as described in Materials and Methods. Nucleotide numbers corresponding to the fragments are shown. AUG codons within the RhPV 5′ UTR are indicated by arrowheads. The IGR of RhPV is also indicated.
Three truncated versions of the RhPVs plasmid containing 3′-end deletions were constructed. RhPVΔ1 was created by PCR amplification using primers RFOR1 and RREV579. The primer sequences are shown in Table 1, and the 5′ UTR fragments created are illustrated in Fig. 1. The RhPVΔ1 fragment contains the probable complete 5′ UTR (nt 1 to 579) but terminates immediately upstream of the predicted initiation codon at nt 580. RhPVΔ2 contains nt 1 to 463 and was created using primers RFOR1 and RREV463. RhPVΔ3 contains nt 1 to 374 and was amplified using primers RFOR1 and RREV374. Finally, a single 5′-end deletion containing nt 100 to 588 (RhPVΔ4) was constructed using primers RF0R100 and RREV588. All fragments were inserted into the pGEM-CAT/LUC vector as described above. s
Coupled transcription-translation reactions.
The dicistronic plasmids (2 μg) were assayed in an RRL coupled transcription-translation system (TNT Quick system; Promega) or the wheat germ-based coupled TNT system (Promega) essentially as described by the manufacturer. [35S]methionine-labeled products were analyzed on sodium dodecyl sulfate (SDS)-polyacrylamide gels, and dried gels were exposed to Fuji X-ray film. Luciferase (LUC) activity was measured using a LUC assay kit (Promega) and a Bio-orbit luminometer.
In vitro transcription reactions.
The pGEM-CAT/LUC-based plasmids were linearized with XhoI, and transcripts were made using T7 RNA polymerase (Epicentre). For the Drosophila extract translation reactions, capped RNA transcripts were prepared using a CapScribe system (Boehringer Mannheim).
Northern blot analysis.
RNA transcript size and integrity were analyzed by Northern blot analysis using a probe specific for the LUC sequence. A HindIII-SacI fragment from pLUC (Promega) was labeled with [α-32P]dCTP using Ready-To-Go labeling beads (Amersham Pharmacia Biotech) and purified on a ProbeQuant G50 purification column (Amersham Pharmacia Biotech). A control transcript corresponding to the LUC sequence was made from XhoI-linearized pGEM-LUC (Promega) using SP6 RNA polymerase (Promega). Transcripts were analyzed by agarose gel electrophoresis, transferred to a nylon membrane (Boehringer Mannheim), and probed with the LUC probe. Membranes were exposed to film and visualized by autoradiography.
In vitro translation reactions.
RRL (25 μl; Promega) was programmed with 25 ng of RNA as described in the manufacturer's protocol. Samples were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and autoradiography. Alternatively, aliquots were assayed for LUC activity as described above. Drosophila embryo extracts (12 μl; kind gifts from Fatima Gebauer, European Molecular Biology Laboratory) were programmed with 15 ng of capped RNA transcripts as described previously (10) but without spermidine and dithiothreitol. Samples were assayed for LUC activity as described above.
RESULTS
The 5′ UTR of RhPV contains an efficient IRES.
A dicistronic reporter plasmid (RhPVs) was constructed in which the 5′ terminus of the RhPV genome (all 579 bases of the 5′ UTR plus the first 15 codons of RhPV ORF1) was inserted between the coding sequences for chloramphenicol acetyltransferase (CAT) and LUC (Fig. 1). A second dicistronic construct was prepared which contained the identical RhPV sequence but inserted in the opposite (antisense) orientation (RhPVas). When the RNA transcripts were translated, the presence of an active IRES element led to the expression of the second ORF (LUC), whereas cap-dependent translation was monitored by CAT expression. The activities of each plasmid were assessed initially with a coupled transcription-translation system based on RRL. Plasmid pGEM-CAT/EMC/LUC (EMC), containing the well-characterized EMCV IRES, was used as a positive control, and plasmid pGEM-CAT/LUC, which lacks any IRES sequences, was used as a negative control.
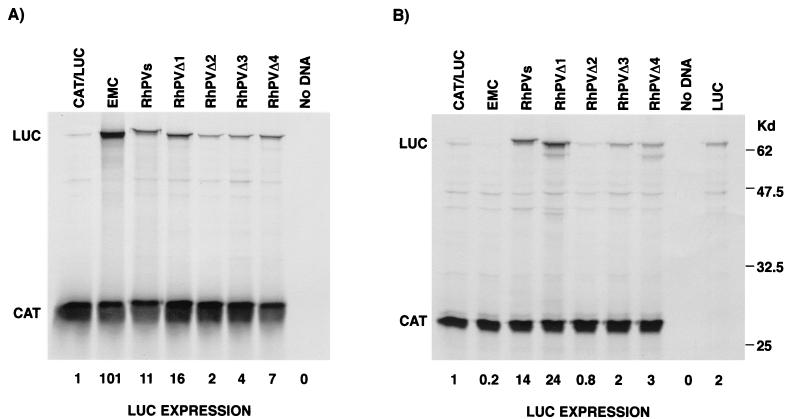
All plasmids induced efficient expression of CAT (Fig. 2A). Reactions containing the RhPVs and EMC constructs also produced high levels of LUC expression (Fig. 2A). LUC enzyme activity from plasmid RhPVs was measured at about 20 to 30% that observed with the EMCV IRES (data not shown). Little LUC expression was detected from plasmid RhPVas or from pGEM-CAT/LUC, which contains no IRES element (Fig. 2A). The ability of the RhPV sequence to promote internal initiation was also tested in a different context using a GUS-HOOK dicistronic construct as described previously (19). With this construct, too, efficient expression of the second ORF was achieved only when the RhPV sequence was inserted between the GUS and HOOK ORFs in the sense orientation (data not shown).
FIG. 2.
The RhPV 5′ UTR displays IRES activity in vitro. Plasmids encoding dicistronic mRNAs containing the indicated virus sequences were analyzed using in vitro transcription-translation systems or in vitro-derived RNA transcripts were analyzed in the RRL translation system as described in Materials and Methods. Samples were analyzed by SDS-PAGE and autoradiography. (A) Transcription-translation in RRL system. (B) Translation in RRL system with in vitro-derived transcripts. (C) Transcription-translation in wheat germ translation system. IRES-containing plasmids are indicated by the name of the IRES insert. Results shown are representative of three separate experiments.
To confirm the IRES activity of the RhPV 5′ UTR, the RhPV sequence was also tested for IRES activity in the RRL in vitro translation system programmed with in vitro-derived transcripts (Fig. 2B). The RhPV sequence in RhPVs directed translation of the LUC sequence, consistent with the result obtained in the TNT system (Fig. 2A). Thus, we conclude that the RhPV 5′ UTR contains an IRES element that is active in the RRL system.
The RhPV IRES functions efficiently in the wheat germ translation system.
RhPV is believed to make use of plants only as passive vehicles for the transmission of infection to other aphids, but the virus genome does have some similarity to that of the comoviruses, which actively replicate in plants (11). Thus, it was possible that the reported lack of RhPV replication in plant cells might be due to a failure of IRES function in this environment. We therefore examined the ability of the RhPV IRES to direct translation initiation in the wheat germ translation system. Reporter plasmids EMC, RhPVs, and RhPVas were analyzed in a wheat germ-based coupled T7 transcription-translation system. As expected, efficient expression of CAT was observed for all plasmids. The EMCV IRES was totally inactive in this plant system and showed less LUC expression than the pGEM-CAT/LUC control, lacking any IRES element. However, LUC was very efficiently expressed from the RhPVs construct (17-fold above that in the control), and once again this expression was abrogated when the 5′ UTR was present in the antisense form (Fig. 2C). The CrPV IGR has also recently been reported to function in the wheat germ translation system (25). However, our data contrast with the inactivity of the CrPV 5′ UTR in the wheat germ translation system reported by these authors.
The RhPV IRES functions in Drosophila extracts.
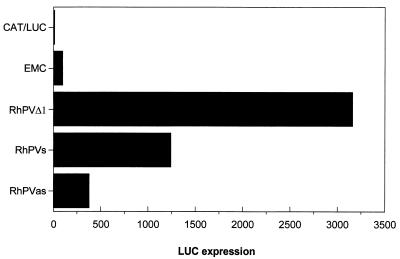
RhPV infects only a narrow range of aphid species; host cell-dependent restriction of IRES function could be a possible contributor to the determination of host range, and we therefore examined the ability of the RhPV IRES to function in a Drosophila-based in vitro translation system (10). Capped transcripts were made in vitro using T7 RNA polymerase and translated in Drosophila lysates. On analysis of LUC expression, the EMCV IRES was found to be very inefficient in this system (Fig. 3). In contrast, the RhPV 5′ UTR directed LUC expression nearly 30-fold above the background expression obtained from the negative control plasmid (pGEM-CAT/LUC) and the EMCV IRES (Fig. 3). It should be noted that the level of LUC expression was about 10-fold lower in the Drosophila system than in the RRL translation system (the Drosophila extract is not treated with nuclease and hence contains cellular mRNAs).
FIG. 3.
The RhPV 5′ UTR displays IRES activity in a Drosophila translation system. Drosophila translation extracts were programmed with RNA transcripts of the form CAT-IRES-LUC. LUC activities were measured using a Promega LUC assay kit as described in Materials and Methods. LUC activity observed from the EMCV IRES was set at 100. IRES-containing plasmids are indicated by the name of the IRES insert. Similar results were obtained in two separate experiments.
Integrity of RNA transcripts containing the RhPV 5′ UTR.
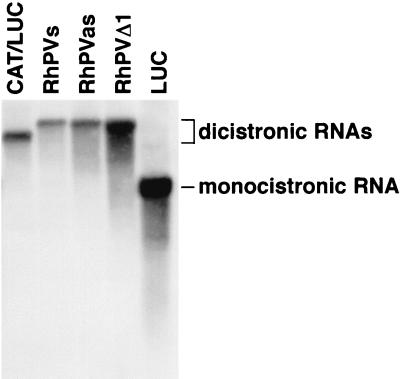
In order to confirm the size and integrity of the dicistronic RNA transcripts generated by T7 RNA polymerase, these RNAs were analyzed by electrophoresis on an agarose gel, transferred to a nylon membrane, and probed with a 32P-labeled probe specific for the LUC sequence. A single species of RNA of the expected size was detected in each instance (Fig. 4), indicating that the RhPV sequence did not contain a cryptic T7 promoter or induce RNA cleavage which could have generated monocistronic LUC transcripts.
FIG. 4.
Integrity of RNA transcripts containing the RhPV sequence. In vitro-derived RNA transcripts were analyzed by agarose gel electrophoresis, transferred to a nylon membrane, and probed with a 32P-labeled probe specific for the LUC sequence. An autoradiograph is shown. The IRES-containing dicistronic transcripts are referred to by the name of the IRES. Note the smaller product obtained from monocistronic pGEM-LUC (lane LUC).
Mapping the 5′ and 3′ boundaries of the RhPV IRES.
Attempts to delineate the 5′ and 3′ boundaries of the RhPV 5′ UTR IRES were made by construction of truncated versions in which sequences were removed from either end of the insert contained in the RhPVs construct. Truncated versions of RhPVs were generated by PCR and cloned into the pGEM-CAT/LUC dicistronic vector as described above (Fig. 1). Each plasmid was tested in the RRL and wheat germ-based coupled transcription-translation systems. Plasmid RhPVΔ1 efficiently expressed both CAT and LUC (Fig. 2B and 5A); deletion of the coding sequence from the RhPVs construct had no negative effect on the amount of LUC expressed in the RRL system. Note the faster migration of the LUC protein produced from this construct than of the fusion protein generated by the inclusion of 15 amino acids of the virus coding sequence in the RhPVs construct (Fig. 5A). Quantitation of LUC expression by phosphorimager analysis showed that expression from the RhPVΔ1 construct was greater than that from the RhPVs construct. Similarly, the activity of the RhPV IRES measured by protein synthesis in the wheat germ translation system and in the Drosophila translation system was not inhibited by deletion of these codons (Fig. 3 and 5B). A modest increase in LUC expression was observed by the removal of the virus coding sequence from the constructs (1.5-fold increase; Fig. 5B). Thus, the RhPV IRES does not extend into the virus coding sequence. Deletion of the 3′ end of the RhPV 5′ UTR (removal of nt 464 to 579) generated construct RhPVΔ2. This deletion significantly reduced the ability of the 5′ UTR to direct internal initiation in both translation systems (Fig. 5), and LUC expression fell to about 13% of that directed by the full-length 5′ UTR in RhPVΔ1. Intriguingly, removal of a further 89 nt from the 3′ end (construct RhPVΔ3; removal of nt 375 to 579) partially restored IRES activity (25% of that seen with RhPVΔ1). Deletion of the first 99 bases from the 5′ end of RhPVs (RhPVΔ4) had less of an effect on LUC activity (44% less activity than that seen with RhPVΔ1 in the RRL system) in both coupled transcription-translation systems, indicating that these 5′ sequences may be less critical for IRES function (Fig. 5).
FIG. 5.
Delimitation of the RhPV 5′ UTR sequences required for IRES activity in vitro. Dicistronic plasmids containing the RhPV 5′ UTR and truncated versions of this sequence were analyzed in the RRL (A) and wheat germ (B) coupled transcription-translation systems as described in Materials and Methods. Samples were analyzed by SDS-PAGE and autoradiography. LUC expression was also measured using a LUC assay kit, and values are calculated relative to the activity obtained with no IRES element (plasmid pGEM-CAT/LUC) (lane CAT/LUC). Similar results were obtained in two separate experiments. In panel B, a monocistronic LUC plasmid was also included as a positive control (lane LUC).
DISCUSSION
The results presented here demonstrate that the 5′ UTR of RhPV contains an IRES element. This IRES functions efficiently in the RRL in vitro translation system, albeit displaying less activity than the well-studied EMCV IRES. However, in addition, the RhPV 5′ UTR IRES functions well in other systems in which the EMCV IRES is essentially inactive. The RhPV IRES stimulated LUC expression in a Drosophila translation system about 30-fold more than the pGEM-CAT/LUC vector alone. The lack of EMCV IRES activity observed with this system is in line with data from Finkelstein et al. (9), who found that the EMCV IRES was inefficient in directing internal initiation in a range of different insect cells. The data obtained with the RhPV 5′ IRES in the Drosophila system are consistent with data recently reported for the CrPV IRES elements (25). IRES activity was reported for both the 5′ UTR and the IGR of CrPV in the RRL system and insect cells, although the IGR demonstrated higher activity than the 5′ UTR. However, Wilson et al. (25) reported that the 5′ UTR of CrPV was inactive in wheat germ extracts, although the CrPV IGR was active in this system. In contrast, the RhPV 5′ IRES was shown here (Fig. 2 and 5) to function very efficiently in the wheat germ translation system. Clearly, functional differences exist between the two virus genomes, and the RhPV 5′ UTR and IGR IRES elements need to be compared in different translation systems.
Although RhPV displays physicochemical characteristics similar to those of the mammalian picornaviruses, it is clearly distinguished from these viruses by the differences in its genome organization. However, both RhPV and the mammalian picornaviruses make use of internal ribosome entry in the initiation of translation. The 5′ UTRs of mammalian picornaviruses (600 to 1,300 nt) are generally longer than that of RhPV (580 nt); however, only about 450 nt of picornavirus RNA is required for IRES function, and deletions of this region from either end result in a complete loss of activity (1, 24). The 5′ UTR of RhPV also does not have a polypyrimidine tract found near the 3′ end of all picornavirus IRES elements (reviewed in reference 3).
Our experiments to define the RhPV 5′ IRES more closely have shown that this IRES does not require the virus coding sequence for optimal activity. Deletion of the coding sequence (construct RhPVΔ1) moderately enhanced IRES activity. The lack of a requirement for the coding sequence is reminiscent of the picornaviruses but differentiates the RhPV IRES from that of hepatitis C virus, where about 30 nt of the coding sequence is apparently required for optimal activity (17). In addition, construct RhPVΔ1 also lacked the initiator AUG codon at position 588, and this construct was the most efficient in directing internal initiation. This finding supports the suggestion of Moon et al. (15) that AUG 580 is the functional initiation codon; however, it does not exclude occasional use of AUG 588.
The RhPV IRES continued to function, albeit at a reduced efficiency, when almost 100 bases were removed from either end of the 5′ UTR. Deletion of 200 nt from the 3′ end of the RhPV 5′ UTR had less effect on IRES activity than deletion of only 100 nt from the same end of the UTR. This was a surprising result, and more work is required to fully explain this finding; however, it may indicate that the boundaries of the 5′ IRES are not sharply defined, a property that is shared with certain IRES elements of cellular origin. For example, a variety of fragments of the immunoglobulin heavy-chain binding protein IRES element are known to retain IRES function (26), but this and certain other cellular IRES elements (e.g., c-myc) do not function in the RRL system or when expressed in the cytoplasm of mammalian cells (13, 22). Studies on the IGR IRES of RhPV (7) showed that deletion of sequences from the 3′ end of the 186-nt IRES inhibited IRES activity in vitro, although no quantitation of this effect was reported. However, the RhPV 5′ UTR IRES is clearly different from the IGR IRES, which initiates at a non-AUG codon. Folding analysis suggests that the IGR IRES elements from both RhPV and PSIV should form a pseudoknot at the 3′ end (7, 21). This feature is thought to be crucial in directing non-AUG initiation of translation. Clearly, further comparison of the 5′ UTR and IGR IRES elements of these viruses is required to determine the structural features responsible for these functional properties.
The ability of the RhPV 5′ UTR to function not only in the RRL system, but also in both plant and insect translation systems suggests potential utility of this IRES in both insect and plant expression systems.
ACKNOWLEDGMENTS
This work was funded by the Royal Society (support given to L.O.R.) and the BBSRC (support given to L.O.R. and G.J.B.). A BBSRC studentship award given to K.E.W. is also gratefully acknowledged.
REFERENCES
- 1.Belsham G J, Brangwyn J K. A region of the 5′ noncoding region of foot-and-mouth disease virus RNA directs efficient internal initiation of protein synthesis within cells: involvement with the role of L protease in translational control. J Virol. 1990;64:5389–5395. doi: 10.1128/jvi.64.11.5389-5395.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belsham G J, Jackson R J. Translation initiation on picornavirus RNA. In: Sonenberg N, Hershey J W B, Mathews M B, editors. Translational control of gene expression. Monograph 39. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 869–900. [Google Scholar]
- 3.Belsham G J, Sonenberg N. RNA-protein interactions in picornavirus translational regulation. Microbiol Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borman A M, Kirchweger R, Ziegler E, Rhoads R E, Skern T, Kean K M. eIF4G and its proteolytic cleavage products: effect on initiation of protein synthesis from capped, uncapped, and IRES-containing mRNAs. RNA. 1997;3:186–196. [PMC free article] [PubMed] [Google Scholar]
- 5.Brown B A, Ehrenfeld E. Translation of poliovirus RNA in vitro: changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology. 1979;97:396–405. doi: 10.1016/0042-6822(79)90350-7. [DOI] [PubMed] [Google Scholar]
- 6.D'Arcy C J, Burnett P A, Hewings A D. Detection, biological effects, and transmission of a virus from the aphid Rhopalosiphum padi. Virology. 1981;114:268–272. doi: 10.1016/0042-6822(81)90275-0. [DOI] [PubMed] [Google Scholar]
- 7.Domier L L, McCoppin N K, D'Arcy C J. Sequence requirements for translation initiation of Rhopalosiphum padi virus ORF2. Virology. 2000;268:264–271. doi: 10.1006/viro.2000.0189. [DOI] [PubMed] [Google Scholar]
- 8.Dorner A J, Semler B L, Jackson R J, Hanecak R, Duprey E, Wimmer E. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysate. J Virol. 1984;50:507–514. doi: 10.1128/jvi.50.2.507-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finkelstein Y, Faktor O, Elroy-Stein O, Levi B-Z. The use of bi-cistronic transfer vectors for the baculovirus expression system. J Biotechnol. 1999;75:33–44. doi: 10.1016/s0168-1656(99)00131-5. [DOI] [PubMed] [Google Scholar]
- 10.Gebauer F, Corona D F V, Preiss T, Becker P B, Hentze M W. Translational control of dosage compensation in Drosophila by Sex-lethal: cooperative silencing via the 5′ and 3′ UTRs of msl-2 mRNA is independent of the poly(A) tail. EMBO J. 1999;18:6146–6154. doi: 10.1093/emboj/18.21.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gildow F E, D'Arcy C J. Cytopathology and experimental host range of Rhopalosiphum padi virus, a small isometric RNA virus infecting cereal grain aphids. J Invertebr Pathol. 1990;55:245–257. [Google Scholar]
- 12.Glass M J, Summers D F. Identification of a trans-acting activity from liver that stimulates hepatitis A virus translation in vitro. Virology. 1993;193:1047–1050. doi: 10.1006/viro.1993.1225. [DOI] [PubMed] [Google Scholar]
- 13.Izuka N, Chen C, Yang Q, Johannes G, Sarnow P. Cap-independent translation and internal initiation of translation in eukaryotic cellular mRNAs. Curr Top Microbiol Immunol. 1995;203:155–177. doi: 10.1007/978-3-642-79663-0_8. [DOI] [PubMed] [Google Scholar]
- 14.Jackson R J, Hunt S L, Gibbs C L, Kaminski A. Internal initiation of translation of picornavirus RNAs. Mol Biol Rep. 1994;19:147–159. doi: 10.1007/BF00986957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moon J S, Domier L L, McCoppin N K, D'Arcy C J, Jin H. Nucleotide sequence analysis shows that Rhopalosiphum padi virus is a member of a novel group of insect-infecting RNA viruses. Virology. 1998;243:54–65. doi: 10.1006/viro.1998.9043. [DOI] [PubMed] [Google Scholar]
- 16.Pringle C R. Virus taxonomy—1999. Arch Virol. 1999;144:421–429. doi: 10.1007/s007050050515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reynolds J E, Kaminski A, Kettinen H J, Grace K, Clarke B E, Carroll A R, Rowlands D J, Jackson R J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts L O, Seamons R A, Belsham G J. Recognition of picornavirus internal ribosome entry sites within cells; influence of cellular and viral proteins. RNA. 1998;4:520–529. doi: 10.1017/s1355838298971989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson M E M, Seamons R A, Belsham G J. A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA. 1999;5:1167–1179. doi: 10.1017/s1355838299990301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 21.Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J Virol. 1999;73:1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoneley M, Soubkhankulova T, Le Quesne J P C, Coldwell M, Jopling C L, Belsham G J, Willis A E. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 2000;28:687–694. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thiel H-J, König M. Caliciviruses: an overview. Vet Microbiol. 1999;69:55–62. doi: 10.1016/s0378-1135(99)00088-7. [DOI] [PubMed] [Google Scholar]
- 24.van der Velden A, Kaminski A, Jackson R J, Belsham G J. Defective point mutants of the encephalomyocarditis virus internal ribosome entry site can be complemented in trans. Virology. 1995;214:82–90. doi: 10.1006/viro.1995.9952. [DOI] [PubMed] [Google Scholar]
- 25.Wilson J E, Powell M J, Hoover S E, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol Cell Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Q, Sarnow P. Location of the internal ribosome entry site in the 5′ non-coding region of the immunoglobulin heavy-chain binding protein (BiP) mRNA: evidence for specific RNA-protein interactions. Nucleic Acids Res. 1997;25:2800–2807. doi: 10.1093/nar/25.14.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]