ABSTRACT
Thirty years ago, DNA sequences were obtained from an extinct Aotearoa New Zealand animal for the first time. Since then, ancient DNA research has provided many – often unexpected – insights into the origins of New Zealand’s terrestrial and marine vertebrate fauna. Because recent human activities in New Zealand have caused the decline or extinction of many endemic plant, bird, reptile, and marine mammal species, ancient DNA has been instrumental in reconstructing their identities and origins. However, most ancient DNA studies focusing on New Zealand species have been restricted to vertebrates, with small sample sizes, and/or relatively few genetic markers. This has limited their power to infer fine-scale biogeographic patterns, including (pre)historic distributions and range-shifts driven by past climate and environmental change. Recently, ‘next-generation’ methodological and technological advances have broadened the range of hypotheses that can feasibly be tested with ancient DNA. These advances represent an exciting opportunity for further exploring New Zealand biogeography using ancient DNA, but their promise has not yet been fully realised. In this review, we summarise the last 30 years of ancient DNA research into New Zealand faunal biogeography and highlight key objectives, challenges, and possibilities for the next 30 years and beyond.
KEYWORDS: Aotearoa, dispersal, Gondwana, island, palaeogenomics, phylogenetics, phylogeography, speciation, vicariance, Zealandia
Introduction
The first people to arrive in Aotearoa New Zealand (∼1280 A.D.; Wilmshurst et al. 2008) encountered a unique community of animal species characterised by a complete lack of terrestrial non-volant mammals, an unusual preponderance of flightless birds (Figures 1A–B and 2A–C), and living representatives of lineages that are otherwise entirely extinct or unknown from other parts of the world (Worthy and Holdaway 2002; Tennyson and Martinson 2006). New Zealand has subsequently been a puzzle for biogeographers, who study how historical processes contributed to the modern distribution of biodiversity (e.g. Fleming 1962; Craw 1989; Bellamy 1990; Cooper & Millener 1993; McGlone 2005). Where did these species come from? When, and by what means, did their ancestors arrive? While comparative anatomy formed the basis of most early biogeographical research (e.g. Owen 1879), genetics has increasingly become a powerful tool for reconstructing the relationships between organisms (e.g. Cole and Wood 2018) and inferring their likely geographical origins. However, standard genetic techniques examining only living populations using recently collected samples are often unsuitable for testing biogeographic hypotheses concerning New Zealand, due to the drastic population declines and species extinctions that followed human settlement. Inferences made solely from the analysis of modern DNA may be misleading for species that have been reduced to relict distributions, or for taxa where many closely related species are now extinct, as the DNA in contemporary populations is unlikely to preserve their complete evolutionary history. The finding that authentic genetic material – ancient DNA – could be isolated from the remains of old biological material began a new ‘palaeogenetic’ chapter in the biogeographic study of New Zealand’s fauna.
Figure 1.
Ancient DNA has illuminated the previously hidden biogeographic history of many of New Zealand’s extinct birds including A, moa such as moa mōmona eastern moa Emeus crassus (Verry, Mitchell, et al. 2022); B, adzebill (Aptornis spp. [Boast et al. 2019]; pictured with tuatara Sphenodon punctatus – bottom left); C, raptors such as pouākai Haast’s eagle (Aquila moorei; Knapp et al. 2019); D, whēkau laughing owl (Ninox albifacies; Wood et al. 2017); E, New Zealand raven (Corvus moriorum [Scofield et al. 2017] – foreground; pictured with rāpoka Phocarctos sea lions, terns, and tōrea tai oystercatcher Haematopus chathamensis – background); F, Chatham Island duck (Anas chathamica; Mitchell, Wood, et al. 2014); G, ruru hinapō New Zealand owlet-nightjar (Aegotheles novazelandiae; Dumbacher et al. 2003); H, piopio (Turnagra spp. [Gibb et al. 2015] – foreground; pictured with kakariki kowhai yellow-crowned parakeet Cyanoramphus auriceps – background); I, wrens such as Lyall’s wren (Traversia lyalli [Mitchell et al. 2016] – foreground; pictured with tītī wainui fairy prion Pachyptila turtur – background); and J, huia (Heteralocha acutirostris; Dussex et al. 2019). Artwork by Paul Martinson from Tennyson and Martinson (2006) © Te Papa CC-BY_NC_ND 4.0.
Figure 2.
Palaeogenetics has highlighted the impacts of past climate change and human impact on the distribution of New Zealand’s living species including A, kiwi (e.g. roroa great spotted kiwi Apteryx maxima; Shepherd et al. 2012); B, parrots like kākāpo (Strigops habroptila; Dussex et al. 2021); C, penguins such as hoiho yellow-eyed penguin (Megadyptes antipodes antipodes; Boessenkool et al. 2009); D, shags including matapo Otago shag (Leucocarbo chalconotus; Rawlence et al. 2015); E, swans (Cygnus spp.; Rawlence et al. 2017); F, pinnipeds such as rāpoka Phocarctos sea lions (Collins et al. 2014); and G, herpetofauna including Duvaucel’s gecko (Hoplodactylus duvaucelii; Scarsbrook et al. 2022).
In 1992, ancient DNA was recovered from an extinct New Zealand species for the first time: Cooper et al. (1992) amplified short mitochondrial DNA (mtDNA) fragments from the bones and mummified tissue of four different species of moa (Aves: Dinornithiformes; Figure 1A). Shortly thereafter, ancient DNA was isolated from other extinct New Zealand taxa including adzebill (Figure 1B; Houde et al. 1997), rails (Trewick 1997), and ruru hinapō owlet nightjars (Figure 1G; Dumbacher et al. 2003). Since Cooper et al.’s (1992) initial publication, at least 100 papers have used ancient DNA to examine the evolution of New Zealand’s unique flora and fauna (as of 08/06/2022; collated via a Google Scholar search utilising the terms ‘ancient DNA’ and ‘New Zealand’ from 1992 onwards). These papers have investigated a range of topics, including the phylogenetics and phylogeography of both extinct and extant species (e.g. Bunce et al. 2009; Scarsbrook et al. 2022), palaeoecological interactions (e.g. Boast et al. 2018), and the effects of humans (both Polynesian/Māori and European) and environmental change on species and populations (e.g. Rawlence et al. 2012; Rawlence, Perry, et al. 2015).
Although the general use of ancient DNA to study New Zealand’s biodiversity has recently been reviewed by Cole and Wood (2018), here we specifically focus on the contribution of palaeogenetics to our understanding of the biogeography of New Zealand’s fauna. We examine how the past 30 years of ancient DNA research have provided novel insights into the origins of New Zealand’s animals and how species distributions have changed through time. Finally, we conclude by highlighting remaining gaps in our understanding and identifying opportunities for the application of new palaeogenetic techniques to study the biogeography of New Zealand.
Global origins of New Zealand’s faunal biodiversity
New Zealand occupies an important place in biogeographical debates due to its unique geological history, having separated from the remnants of the Gondwanan supercontinent ∼82-55 million years ago (Mya) and since remaining isolated from other large landmasses by hundreds of kilometres of open ocean (Figure 3A; Schellart et al. 2006). This geological isolation once led many biogeographers to assume that the biodiversity of present-day New Zealand was primarily composed of the descendants of lineages that were already present when the landmass separated from Gondwana (i.e. vicariant relicts) – a hypothesis termed ‘Moa’s Ark’ (Bellamy 1990). However, the subsequent observation of close links between flora and fauna from Australia and New Zealand, and other areas of the world (e.g. Fleming 1962), led other researchers to suppose that substantial overwater dispersal had contributed to the modern biodiversity of New Zealand, perhaps more appropriately making it the ‘flypaper of the Pacific’ (McGlone 2005). Palaeogenetics has played a role in these discussions, most visibly by revising our understanding of the biogeographical history of the palaeognathous birds (previously referred to as ‘ratites’) – today represented by the ostrich, rheas, tinamous, emu, cassowary, and kiwi, but also including the extinct moa from New Zealand and the elephant birds of Madagascar (see Allentoft and Rawlence 2012). These birds were long used as a textbook example of Gondwanan vicariance (e.g. Cracraft 1974; Bourdon et al. 2009); that is, their speciation and modern distribution was thought to have been driven primarily by the geographical isolation of non-volant ancestors as the various continental fragments of Gondwana separated over the past 200 million years. Therefore, the flightless ratites were presumed to comprise a monophyletic group, with South America’s volant tinamous sitting sister to the ratite clade (e.g. Bourdon et al. 2009).
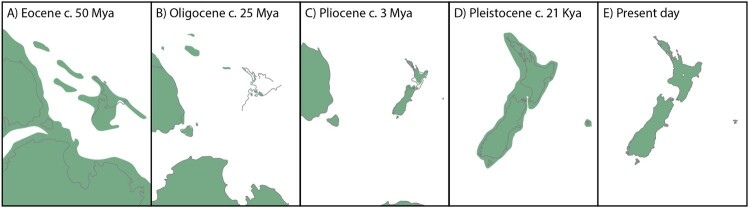
Figure 3.
Maps illustrating the evolution of New Zealand’s landscape and its position relative to Australia, Tasmania, and Antarctica throughout the Cenozoic. Green shading indicates terrestrial areas; grey lines show modern-day coastlines. A, Zealandia breaks from Gondwana 82–50 Mya; B, the Oligocene marine transgression submerges most of the continent c. 25–23 Mya; C, the North and South Islands are separated by marine seaways, and the Chatham Islands form c. 5–3 Mya; D, land bridges during the Last Glacial Maximum (29–19 Kya) connect the three main islands; E, presently, the North and South Islands are separated by Cook Strait.
As expected under the Moa’s Ark hypothesis, the two groups of palaeognathous birds endemic to New Zealand – kiwi (Apteryx spp.; Figure 2A) and moa (Dinornithiformes; Figure 1A) – appeared to be each other’s closest relatives on the basis of morphological comparisons (e.g. Cracraft 1974). However, shortly after the advent of ancient DNA techniques, kiwi were shown to be closer relatives to the emu and cassowary than to moa (Cooper et al. 1992). Subsequent palaeogenetic research utilising mitochondrial genomes and nuclear single nucleotide polymorphisms clarified that the volant South American tinamous were the closest relatives of the moa (Phillips et al. 2010; Baker et al. 2014), while the closest relatives of kiwi were the extinct Madagascan elephant birds (Mitchell, Llamas, et al. 2014; Grealy et al. 2017; Yonezawa et al. 2017). These palaeogenetic data suggested not only that New Zealand’s palaeognathous birds descend from two separate dispersal events, but each group also evolved to become flightless independently – likely after flying to New Zealand after its separation from Gondwana, as suggested by post-Gondwanan genetic estimates for the origin of the kiwi and moa lineages (Cooper et al. 2001; Haddrath and Baker 2001; Phillips et al. 2010; Baker et al. 2014; Mitchell, Llamas, et al. 2014; Grealy et al. 2017). Recent phylogenetic analyses combining palaeognath morphology and DNA have lent support to these evolutionary hypotheses (e.g. Mitchell, Llamas, et al. 2014; McInerney et al. 2019).
If neither kiwi nor moa – both enduring and popular symbols of the supposedly ‘ancient’ New Zealand faunal assemblage – are true vicariant Gondwanan lineages, then which other elements of the fauna represent more recent arrivals? After rafting away from Gondwana, the crust underlying New Zealand stretched, thinned, and gradually sank, resulting in a near-total marine inundation, often referred to as the ‘Oligocene drowning’ (Figure 3B; Cooper and Cooper 1995). One extreme hypothesis suggests that practically all New Zealand species descend from lineages that arrived following this inundation, which reached its maximum extent around 23 Mya (Campbell and Landis 2001; Waters and Craw 2006; Landis et al. 2008). This hypothesis has proven challenging to test. While fossils from the 16–19 Mya St Bathans (Worthy et al. 2007) and the 23 Mya Foulden Maar (Lee et al. 2016) fossil localities provide a window into biodiversity during and shortly after the marine inundation, no fossil sites are known from the period immediately beforehand with which to draw a comparison. Here genetic data are once again useful, as molecular phylogenies can be calibrated using dated fossils to infer the timing of divergence events. Although most present-day New Zealand faunal lineages do appear to have arrived after (see Wallis and Jorge 2018 for a review of molecular divergence estimates for New Zealand biota), at least some lineages seem to pre-date the marine inundation (e.g. tuatara, Gemmell et al. 2020; pepeketua Leiopelma frogs, Carr et al. 2015; ngaokeoke peripatus, Allwood et al. 2010; Zopheridae beetles, Buckley et al. 2020). However, timescales estimated from genetic data are sensitive to model misspecification, which can lead to imprecise or biased results, and failure to sample the most relevant sister taxa can obscure a lineage’s true age and origin (e.g. Shepherd and Lambert 2007) – these results should thus be treated with caution and interpreted as part of a broader body of evidence (McCulloch and Waters 2019).
Palaeogenetics has been useful for addressing some of these issues. By allowing members of extinct lineages to be included in phylogenetic analyses, the sister-taxa of living organisms can be more confidently determined. For example, palaeogenetic data revealed the whēkau laughing owl (‘Sceloglaux’ albifacies; Figure 1D) to be a member of the Australasian/Oceanian genus Ninox, suggesting an Australian origin for this species (Wood et al. 2017). The inclusion of extinct species can also improve the estimation of molecular divergence dates, with palaeogenetic data suggesting that the ancestors of the extinct piopios (New Zealand ‘thrushes’; Turnagra spp.; Figure 1H) and the upokororo New Zealand grayling (Prototroctes oxyrhynchus) almost certainly arrived after the Oligocene drowning (Johansson et al. 2011; Zuccon and Ericson 2012; Gibb et al. 2015; Scarsbrook et al. In review). Among the post-inundation fossils identified from St Bathans are representatives of two endemic New Zealand bird families – Aptornithidae (Worthy et al. 2011) and Acanthisittidae (Worthy et al. 2010). Aptornitihidae are now extinct, but at the time of human arrival in New Zealand the family was represented by two species of adzebill (Aptornis spp.; Figure 1B). Palaeogenetic data demonstrated that the adzebill lineage diverged from its nearest relatives elsewhere in the world 26–54 Mya (Boast et al. 2019), suggesting that they could plausibly represent pre-inundation arrivals in New Zealand. However, the two adzebill species only diverged around 1 Mya, likely as a result of the closure and opening up of Plio-Pleistocene marine straits in central New Zealand (e.g. Manawatu and Cook Straits; Figure 3C), leaving open the possibility that their common ancestor arrived soon after the marine inundation subsided. Likewise, comparisons of palaeogenetic data from three extinct species of acanthisittid wren (e.g. Figure 1I) to those of the two living species – the tītitipounamu rifleman (Acanthisitta chloris) and pīwauwau rock wren (Xenicus gilviventris) – were equivocal on the timing of arrival of this group in New Zealand (in relation to the Oligocene drowning) with different divergence estimates spanning the period of marine inundation (Mitchell et al. 2016). Thus, while palaeogenetic data can provide clarity in some cases, they are not the panacea for all biogeographical puzzles.
Geological events throughout the Miocene and Plio-Pleistocene continued to shape the distribution of New Zealand’s biodiversity. Tectonic activity along the Alpine Fault starting 23–25 Mya began a series of mountain-building periods that gradually lifted many parts of present-day New Zealand out of the water (Kamp 1986; King 2000). This activity was both creative and destructive, at once creating new landscapes and initiating intense volcanic activity that continues to this day. The Southern Alps grew from 8.5 Mya, fragmenting the landscape and isolating the lineages inhabiting it (Tippett and Kamp 1993; Bunce et al. 2009). Evidence for the biological impact of this orogeny can be observed in palaeogenetic data from moa, which underwent speciation during this time that appears closely linked to the development of mountainous landscapes in the South Island and the creation of new habitats (Baker et al. 2005; Bunce et al. 2009). In addition, new opportunities for speciation arose as offshore islands such as the Rēkohu/Chatham Islands surfaced 2–3 Mya (Figure 3C; Campbell et al. 1988, 2008). Extinct species of duck (Pachyanas chathamica, now Anas chathamica; Figure 1F; Mitchell, Wood, et al. 2014), raven (Corvus moriorum; Figure 1E; Scofield et al. 2017), kākā (Nestor chathamensis; Wood et al. 2014), penguin (Eudyptes warhami; Cole et al. 2019), and petrel (Pterodroma imberi; Tennyson et al. 2015) endemic to the Chatham Islands have been described based on ancient DNA sequences and morphological characters – all apparently descended, with the possible exception of E. warhami, from taxa that dispersed from the main islands of New Zealand in the last three million years.
Emergent mountain ranges and offshore islands were not the sole source of new environments; as Pliocene cooling trends culminated in the first glaciations of the Pleistocene ∼2.5 Mya, the mountainous landscape became intermittently covered in glacial ice, and previously forested areas turned to tussock or shrubland (Carter 2005; Heenan and McGlone 2013). The dramatic changes in vegetative cover resulted in ecological shifts as new avian species began to arrive, taking advantage of newly-opening niches and diverging from their overseas relatives (Rawlence, Scofield, et al. 2019). Palaeogenetic data have demonstrated that the ancestors of the extinct pouakai Haast’s eagle (Aquila moorei; Figure 1C; Bunce et al. 2005; Knapp et al. 2019), kērangi Eyles’ harrier (Circus teauteensis; Knapp et al. 2019), and New Zealand raven (Corvus moriorum; Figure 1E; Scofield et al. 2017) arrived during this time, as did the ancestors of the takahē (Porphyrio hochstetteri; Garcia-R et al. 2014; Garcia-R & Trewick 2015), and possibly other avian groups (e.g. pōuwa/matapu Chatham Island/ New Zealand swan, Cygnus sumnerensis and kakī, Himantopus novaezelandiae; see Rawlence, Scofield, et al. 2019). The source of these Pleistocene lineages appears to have been Australia, likely as a result of prevailing westerly winds, relatively close proximity across the Tasman Sea, and newly available niche space. In contrast, the geographical origin of earlier arrivals to New Zealand appears to be more diverse – for example, palaeogenetic data have revealed possible ancestral links to Madagascar in the kiwi (Mitchell, Llamas, et al. 2014), adzebill (Boast et al. 2019), and New Zealand Anas teals (including the extinct Chatham Island duck, A. chathamica; Mitchell, Wood, et al. 2014). Unexpected affiliations to geographically distant overseas taxa highlight the need to look further afield to better understand the biogeography of New Zealand. Furthermore, these far-flung links emphasise the remarkable capacity of species to disperse (particularly flighted taxa), and are a testament to the invaluable role of palaeogenetic data in piecing together the complexity of the historical processes that shaped New Zealand’s present-day biodiversity.
Phylogeographic impacts of the Pleistocene ice ages
Beyond the global origin of New Zealand’s biodiversity, palaeogenetic data have also begun to reveal finer-scale biogeographic patterns, such as those driven by the repeated glacial-interglacial cycles of the Pleistocene (2.58 Mya–11.65 thousand years ago, Kya). Environmental changes in New Zealand were largely characterised by glaciation of the Southern Alps throughout the South Island and the expansion of open shrublands and grasslands across both main islands during cold glacial periods, with the widespread (re)establishment of open and closed-canopy forests during warm interglacials (Newnham et al. 2013). In the North Island, periodic volcanism has been hypothesised to have generated phylogeographic structure (e.g. moa ruarangi Mantell’s moa, Pachyornis geranoides; Bunce et al. 2009). New Zealand thus presents a useful system to study the impacts of Pleistocene climate change, because humans arrived only in the late thirteenth century at a time of relative climatic stability (Wanner et al. 2008; Wilmshurst et al. 2008; Waters et al. 2017), in contrast to continental ecosystems where anthropogenic impacts often overlap with major climatic/environmental shifts (e.g. Lorenzen et al. 2011; Metcalf et al. 2016; Hocknull et al. 2020).
Phylogeographic studies using modern DNA have revealed the impacts of Pleistocene environmental change on some of New Zealand’s birds (e.g. Goldberg et al. 2011; Dussex et al. 2014; Weston and Robertson 2015), insects (e.g. Trewick 2001; McCulloch et al. 2010; King et al. 2020), and plants (e.g. Shepherd et al. 2007, 2017; Rawlence et al. 2021), with a recent synthesis suggesting that periods of Pleistocene glaciation promoted the structuring and diversification of New Zealand’s alpine biota (Wallis et al. 2016). Speciation/phylogeographic structure potentially driven by glaciation typically manifests in New Zealand as a north–south pattern across the Southern Alps (e.g. McCulloch et al. 2010; Weston and Robertson 2015), although it has also been implicated in the formation of more extensive phylogeographic structure involving multiple barriers (e.g. Weir et al. 2016; King et al. 2020). Relatively few studies focusing on New Zealand have taken advantage of the temporal resolution provided by ancient DNA, which can provide direct insights into the response of species and populations to environmental stressors by analysing genetic data spanning the event itself (e.g. the Pleistocene-Holocene transition 11.6 Kya or the Last Glacial Maximum (LGM) 19–29 Kya; Newnham et al. 2013). Additionally, most ancient DNA studies of New Zealand taxa only incorporate data from Holocene-aged material (see Table S1 in Grealy, Rawlence et al. (2017) for examples from birds) as Pleistocene-aged remains are less common and often have poorer morphological and biomolecular preservation due to their greater age and less favourable taphonomic conditions.
Nonetheless ancient DNA has improved our understanding of the impacts of the Pleistocene on New Zealand’s extinct species, particularly the moa. Bunce et al. (2009) analysed ancient mitochondrial DNA from all nine currently recognised moa species, finding four geographically distinct mitochondrial lineages in the alpine specialist moa pukepuke upland moa (Megalapteryx didinus), and suggested that Pleistocene glaciations led to this deep phylogeographic structure. Similarly, Rawlence et al. (2012) reported phylogeographic structure between northern and southern populations of the moa waewae taumaha heavy-footed moa (Pachyornis elephantopus), likely due to contraction into separate glacial refugia. Palaeogenetic and isotopic data also suggests that moa koukou crested moa (Pachyornis australis), moa nunui South Island giant moa (Dinornis robustus), and heavy-footed moa were able to adapt to changing climates and environmental conditions by tracking spatial changes in their preferred habitat through time (Rawlence et al. 2012; Lomolino et al. 2021). In contrast to the phylogeographic structuring observed in some moa species, other species do not exhibit such structure, but have still been significantly impacted by the Pleistocene glacial-interglacial cycles. Bunce et al. (2009) suggested the open-forest specialist moa mōmona eastern moa (Emeus crassus) suffered a pronounced population bottleneck, based on the comparatively shallow nature of the lineage divergence within this species. Further evidence for a population bottleneck in eastern moa was subsequently obtained from an expanded dataset that included nuclear microsatellites (Allentoft et al. 2014), while evidence from whole mitochondrial genomes provided support for post-LGM expansion of eastern moa from a glacial refugium in the southern South Island (Verry, Mitchell, et al. 2022). Glacial maxima also appear to have provided new opportunities for movement, with moa possibly using land bridges revealed by falling sea levels (Figure 3D) to disperse between the North Island, South Island, and Rakiura/Stewart Island (Bunce et al. 2009; Verry, Schmidt, et al. 2022). These dynamic and species-specific responses of moa to past climatic and environmental change are underpinned by moa biology including habitat preference and diet (e.g. Wood et al. 2020).
Palaeogenetic research has also revealed the impacts of glacial cycles on kiwi (Apteryx spp.), huia (Heteralocha acutirostris; Figure 1J), South Island kōkako (Callaeas cinereus), kākāpō (Strigops habroptila), shearwaters (Puffinus spp.), and Duvaucel’s gecko (Hoplodactylus duvaucelii; Figure 2G). Glaciation appears to have driven diversification in kiwi (Shepherd and Lambert 2008; Shepherd et al. 2012; Weir et al. 2016), shearwaters (Tennyson & Shepherd 2017), and South Island populations of Duvaucel’s gecko (Scarsbrook et al. 2022), while forest-dwelling huia and South Island kōkako experienced population declines during glacial periods (Dussex et al. 2019). Dussex et al. (2018) analysed mitochondrial genomes retrieved from museum skins of the kākāpō and suggested that South Island kākāpō expanded from northern and southern refugia following the end of the LGM (though Mudge et al. 2022 challenged this interpretation). These results emphasise that responses to climatic and environmental change are often species specific, highlighting the need for palaeogenetic data from extinct taxa in order to fully understand how Pleistocene glacial cycles shaped New Zealand’s biodiversity.
Human impacts on biodiversity
The arrival of humans to New Zealand in the late thirteenth century and their subsequent impacts (e.g. Anderson 1989; Tennyson and Martinson 2006; Perry et al. 2014; Rawlence et al. in press) resulted in now well-characterised changes to many plant and animal species – including dramatic population declines, local extirpations, and global extinctions – which are reflected in the changing contents of both fossil and archaeological sites (e.g. Tennyson and Martinson 2006, Rawlence et al. in press). However, palaeogenetic assessment of these contents have revealed previously cryptic responses to anthropogenic impacts, which were undetectable using traditional comparative methods, and has helped quantify the true extent of anthropogenic impacts on New Zealand’s fauna. Prior to the advent of palaeogenetics Western knowledge of human impacts on the biodiversity of New Zealand was limited to identification of widespread avian (and some herpetofaunal) extinctions and range contractions based on morphological analysis of the fossil and archaeological record (e.g. Worthy 1987, 1991; Worthy and Holdaway 2002; Tennyson and Martinson 2006). Combined palaeogenetic and morphological reanalyses have (1) allowed taxonomic identifications based on morphology to be tested, resulting in the recent description of several new extinct species of penguin (Boessenkool et al. 2009; Cole et al. 2019); shag (Figure 2D; Rawlence, Till, et al. 2017), swan (Figure 2E; Rawlence, Kardamaki, et al. 2017), and petrel (Tennyson et al. 2015), and new genetic lineages of rāpoka sea lion (Figure 2F; Collins et al. 2014; Rawlence et al. 2016); and (2) the provenance of historical museum specimens, which often underlie our understanding of the severity of human impact, to be verified (e.g. Boessenkool et al. 2010; Shepherd et al. 2013; Rawlence, Kennedy, et al. 2014; Verry et al. 2019; Mudge et al. 2022; Scofield et al. 2021). The identification of cryptic biodiversity (i.e. previously unrecognised species/lineages and biological turnover events) enables a better understanding of the true extent of human impacts on New Zealand’s biodiversity by establishing a more complete ecological baseline prior to anthropogenically induced extinctions/extirpations.
In all cases, the previously unrecognised extinct species identified using palaeogenetic data were traditionally considered synonymous with their living congeners (which in some cases were thought to be relictual species with formally widespread distributions), as minimal morphological variation made them difficult to diagnose taxonomically without the use of ancient DNA. This was especially true when the age of specimens was not constrained using radiocarbon dating, as for early analyses of penguin and swan remains (e.g. Worthy 1997; Worthy 1998c). For example, the current distribution of hoiho yellow-eyed penguins (Megadyptes antipodes antipodes; Figure 2C) was thought to be a relict of a previously more widespread distribution around mainland New Zealand’s coastline (e.g. Worthy 1997). However, palaeogenetic analyses showed that palaeontological and early archaeological remains belonged to a genetically and morphologically distinct lineage described as the extinct Waitaha penguin (M. a. waitaha) that was formerly widespread across Rakiura/Stewart Island, the South Island, and southern North Island (Boessenkool et al. 2009; Rawlence, Perry, et al. 2015; Rawlence, Tennyson, et al. 2019). Further research by Cole et al. (2019) highlighted the presence of another extinct Megadyptes lineage – Richdale’s penguin (M. a. richdalei) – on the Rēkohu/Chatham Islands. Cryptic species have also been found in crested penguins (Eudyptes spp.) in the New Zealand region, including Warham’s penguin (E. warhami), the bones of which were originally thought to belong to tawaki nana hī erect-crested (E. sclateri) or tawaki Fiordland crested penguin (E. pachyrhynchus).
The discovery of previously unrecognised species in the New Zealand fossil and archaeological record has also revealed several cryptic biological turnover events since human arrival in New Zealand. For example, the simultaneous human-driven extinction of the Waitaha penguin and the prehistoric New Zealand lineage of Phocarctos sea lion by the early-mid 1400s preceded the successful recolonisation of mainland New Zealand by yellow-eyed penguin and the subantarctic sea lion lineage by the late 1400s, which was facilitated by decreased human hunting in southern New Zealand after the extinction of large prey animals like moa, sea lions, and ihupuku southern elephant seals (Rawlence, Perry, et al. 2015; Waters et al. 2017). While these extinctions occurred at a time of relative climatic stability, the recolonisations did not occur until the Little Ice Age (∼1500–1800 CE) – 20–50 years after the extinction event – which allowed these cold-adapted species to colonise New Zealand from their subantarctic homeland; highlighting that climatic conditions need to be favourable for species migration even in the event of available niche space and the absence of competitive exclusion. Other biological turnover events have been documented in Cygnus swans (i.e. extinction of C. sumnerensis and replacement with C. atratus; Rawlence, Kardamaki, et al. 2017) and Eudyptula penguins (replacement of kororā New Zealand little blue populations with Australian populations in coastal Otago; Grosser et al. 2016). While the extinction of Cygnus swans and the pronounced population bottleneck in the lead up to the extinction of little blue penguins in Otago occurred contemporaneously with Megadyptes penguins and Phocarctos sea lions, their recolonisations were offset, not occurring until the 1700–1800s when conditions were favourable for the establishment of populations. Knowledge about these biological turnover events may have important implications for species conservation management: in some cases, recent colonists have been termed ‘native invaders’ or ‘maladapted colonisers’ with questions asked about whether they have adapted to local climatic conditions and the role of ecological surrogates in the modern New Zealand ecosystem (Waters and Grosser 2016).
Many species suffered pronounced range contractions, and in some cases underwent severe population bottlenecks and significant changes in haplotype frequency as populations moved into and out of refugia, as in kekeno fur seals, kākāpō, and kawau tikitiki spotted and matapo Otago shags (Rawlence, Kennedy, et al. 2015; Bergner et al. 2016; Salis et al. 2016; Seersholm et al. 2018; Rawlence, Rayner, et al. 2019). Many of these refugia were in areas where the intensity of human occupation and activity were low after the extinction of much larger prey (e.g. moa, sea lions). In southern New Zealand, Little Ice Age cooling prompted people to move to warmer northerly areas where horticulture of crops from Polynesia was supported (e.g. northern South Island and the North Island). Not all range contractions, however, resulted in a decrease of genetic diversity (e.g. Dussex et al. 2015). While the range of Fiordland crested penguins (E. pachyrhynchus) contracted from the southern North Island, and northern and eastern South Island to Fiordland, core genetic diversity was preserved in this refugium (Cole et al. 2019). Range contractions and population bottlenecks may also have resulted in a decrease of phenotypic variation exhibited by surviving populations compared to prehistoric populations, as observed in Duvaucel’s gecko (Scarsbrook et al. 2021, 2022).
Future directions
From palaeogenetics to palaeogenomics
High-throughput DNA sequencing technologies have significantly increased the amount of sequencing data obtainable from ancient material, ushering in the age of ‘palaeogenomics’ – the analysis of genome-scale data from ancient samples (e.g. Mitchell and Rawlence 2021). Although many ancient DNA studies continue to rely on mitochondrial DNA (e.g. Scarsbrook et al. 2022; Verry, Mitchell, et al. 2022) – due to its wide use in phylogeography and the relative ease with which it can be retrieved from ancient remains – the field has seen a shift towards the sequencing of nuclear DNA via the hybridisation capture of genome-wide single nucleotide polymorphisms (SNPs; e.g. McCormack et al. 2016) or shotgun sequencing of whole nuclear genomes (e.g. Dussex et al. 2019, 2021). Analysis of genome-wide nuclear data can overcome many of the factors that may potentially mislead studies based only on the mitochondrial genome – interspecific hybridisation, substitution saturation, and incomplete lineage sorting (Xia et al. 2003; Funk and Omland 2003; however, see Phillips and Zakaria 2021) – making palaeogenomics a powerful tool for the investigation of past demography, hybridisation, and selection (Shapiro and Hofreiter 2014; Mitchell and Rawlence 2021).
While palaeogenomics is now routinely conducted in other countries, it has yet to be widely applied in New Zealand and thus represents a major opportunity for research over the coming decades (see Mitchell and Rawlence 2021). One recent example demonstrates the power of this approach: genomes from historical museum skins of kākāpō revealed that the modern kākāpō population (founded by individuals from Rakiura/Stewart Island) contains a lower number of deleterious mutations relative to the now extinct mainland population (Dussex et al. 2021), hinting at different population histories. High-throughput sequencing of nuclear DNA from additional species will likely be integral to reconstructing the pre-human phylogeography of many of New Zealand’s unique animals, how they responded to past climate change and human impact (i.e. change in population size, cryptic extinctions, range contractions/expansions, or even biological turnover events), the role of hybridisation and gene flow in their evolution, and their adaptation to past biotic and abiotic factors.
Methodological advancements and new sources of ancient DNA
The majority of palaeogenetic research to date has relied on extracting ancient DNA from individual bones where destructive sampling can be supported (i.e. the bone is big enough to sample without destroying important osteological characters or morphometric landmarks), mostly limiting the range of species that can feasibly be studied to large animals. However, owing to methodological advances, ancient DNA research is now practical for a much wider range of specimens (e.g. insects, feathers, eggshell), broadening the scope of hypotheses that can be tested with palaeogenetics (e.g. King et al. 2009; Rawlence et al. 2009; Huynen et al. 2010; Oskam et al. 2011; Scarsbrook et al. 2022). For example, new ancient DNA extraction techniques have been developed for formalin preserved specimens (Ruane and Austin 2017; Hahn et al. 2021; Straube et al. 2021; Scarsbrook et al. In review), which are common in museum collections, especially for certain taxonomic groups (e.g. fish). In addition, minimally destructive DNA extraction techniques – requiring only temporary immersion of the specimen in a liquid buffer, leaving no discernible morphological damage – have allowed ancient DNA to be extracted from small bones that would otherwise be destroyed by standard sampling techniques (Tennyson et al. 2015; Scarsbrook et al. 2022).
Ancient DNA research within New Zealand has focused almost exclusively on large vertebrates (primarily birds and pinnipeds), with little focus on smaller and/or more cryptic animals (e.g. herpetofauna, invertebrates). This is likely due to the increased availability of vertebrate remains relative to other organisms, as New Zealand contains an exemplary Late Quaternary subfossil record (Holdaway et al. 2001), and the bones of vertebrates are more likely to be preserved in the fossil record than the soft tissues of many invertebrates. Additionally, the bones of large vertebrates are often more amenable to the destructive sampling required for ancient DNA analysis. This taxonomic bias distorts our understanding of New Zealand biogeography, as the evolution of vertebrate species may not necessarily represent the evolution of the biota more generally. However, new ancient DNA methods mean that greater focus can now be placed on taxa from New Zealand that have previously been neglected in palaeogenetic research. Molluscs in particular offer an exciting opportunity to use ancient DNA to understand how ecological communities functioned prior to human arrival, as new DNA extraction methodologies facilitate the retrieval of ancient DNA from shells (Der Sarkissian et al. 2017, 2020).
In the absence of identifiable remains, genetic information – palaeoenvironmental DNA – can instead be retrieved from environmental archives or ichnofossils, simultaneously providing information about past animal, plant, fungal, and microbial diversity. Most palaeoenvironmental DNA research in New Zealand has focused on reconstructing moa diet (e.g. Wood et al. 2008, 2012; Wood, Wilmshurst, Richardson, et al. 2013; Boast et al. 2018), ecological niche partitioning of different moa species (e.g. Wood, Wilmshurst, Richardson, et al. 2013), and parasitology (e.g. Wood, Wilmshurst, Rawlence, et al. 2013; Boast et al. 2018) based on coprolite specimens, with limited research conducted on palaeoenvironmental DNA from sediment cores (Willerslev et al. 2003; Haile et al. 2007; Wilmshurst et al. 2014). Wilmshurst et al. (2014) used palaeoenvironmental DNA to show that the pre-human podocarp dominated forest of the Poor Knights Islands was almost completely replaced by angiosperm-based forest following anthropogenic disturbance. The Lakes 380 project is also extracting bacterial, plant, and animal DNA from lake sediment cores (e.g. Brasell et al. 2021). Using palaeoenvironmental DNA techniques it should be possible to track community level responses to climate and environmental change from the Pleistocene through to the present day (e.g. Seersholm et al. 2020; Dussex et al. 2021), including how humans have influenced the distribution of plants and animals through extinctions and translocations. However, determining the reliability of the palaeoenvironmental DNA record is vitally important (see Rawlence, Lowe, et al. 2014), as palaeoenvironmental DNA has been shown to migrate between sediment layers (Haile et al. 2007). Additionally, the accuracy of species identifications based on palaeoenvironmental DNA relies on access to comprehensive and well-annotated databases (e.g. GenBank), making the public release of palaeoenvironmental DNA data advantageous where possible.
An exciting application of palaeoenvironmental DNA techniques is ‘bulk bone metabarcoding’, which focuses on determining the taxonomic composition of randomised samples of non-diagnostic bone – ‘frag bags’ – from archaeological and palaeontological excavations (e.g. Murray et al. 2013). Not only is bulk bone metabarcoding able to determine the previously unrecognised presence and absence of species in these deposits (including species not usually morphologically preserved; e.g. sharks, rays, freshwater fish, small birds), but quantitative haplotype data can also be obtained, enabling the past phylogeography of species to be reconstructed (Seersholm et al. 2018). Grealy et al. (2015) used this method to document the presence of eight different avian families within a single palaeontological site in Canterbury, while Seersholm et al. (2018), conducted a nationwide bulk bone metabarcoding survey of both palaeontological and archaeological sites, reconstructing the past subsistence strategies of Māori and showing that kākāpō from the North and South Island were genetically distinct and the species underwent two consecutive bottlenecks in response to the arrival of Polynesians and then Europeans (cf. Bergner et al. 2016). Future studies utilising ancient DNA from multiple sources (including palaeoenvironmental DNA and novel substrates) will enable researchers to investigate past evolutionary patterns and processes at the ecosystem/community level (Dussex, Bergfeldt et al. 2021) facilitating a more comprehensive understanding of anthropogenic, climatic, and environmental influences on New Zealand’s biodiversity.
Conclusion
Thirty years of palaeogenetics research have revolutionised our understanding of the origins of New Zealand’s unique fauna and how past processes have shaped their modern distributions. However, as highlighted in this review, ancient DNA is not a panacea – it has its own challenges and limitations. While New Zealand contains a multitude of Holocene-aged terrestrial vertebrate remains, Pleistocene-aged remains are typically rarer (e.g. Worthy 1998a, 1998b) and may be less likely than younger specimens to yield usable ancient DNA sequences, even with modern high-throughput DNA sequencing techniques (Kistler et al. 2017; Verry 2021). In addition, the incompleteness of the fossil record and various taphonomic biases limit the number and type of species available to be studied using ancient DNA. Consequently, ancient DNA is but one tool in the biogeographer’s toolbox, though a tool that – with recent methodological and technological advances – has now truly come of age (Mitchell and Rawlence 2021).
Moving forward, a combination of palaeogenomics (including minimally-destructive sampling) and palaeoenvironmental DNA with morphological analysis (including 3D geometric morphometrics), radiometric dating, stable dietary isotopes, ecological niche modelling, and mātauranga Māori will help to answer a wider variety of outstanding questions relating to New Zealand’s biogeographic past. For example, the pre-human diversity of New Zealand’s invertebrates and plants are almost entirely unexplored using palaeogenetic techniques, but such data may be crucial for fully reconstructing the composition and distribution of vanished ecosystems like the kōwhai (Sophora microphylla) forests that may formerly have dominated Central Otago (Pole 2022). In addition, mysteries still persist even among New Zealand’s comparatively well-studied avifauna: what are the geographical and temporal origins of the New Zealand geese (Cnemiornis spp.) or the mergansers (Mergus spp.) – now completely absent from Australasia/Oceania – that were formerly found in the New Zealand region? More uncertainties surround the responses of New Zealand’s herpetofauna to past anthropogenic activities and pre-human climate change. By integrating modern palaeogenetic techniques with other lines of evidence, we may be able to determine the evolutionary and biogeographic relationships of New Zealand’s enigmatic biodiversity in its entirety – even down to the microbial level – and how the dynamic geological, climatic, and human history of this island archipelago has shaped its inhabitants.
Acknowledgements
Thank you to Graham Wallis and Thomas Buckley for the invitation to write this review, and to Colin Miskelly, Matthias Dehling, Tony Whitehead, and Philip Griffin for permission to use their photos from New Zealand Birds Online.
Funding Statement
This work was supported by the University of Otago and the Royal Society of New Zealand Marsden Fund [grant number 16-UOO-096]. KJM was supported by a Fast Start Grant awarded by the Royal Society of New Zealand Marsden Fund [grant number 20-UOO-130].
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Allentoft ME, Heller R, Oskam CL, Lorenzen ED, Hale ML, Gilbert MTP, Jacomb C, Holdaway RN, Bunce M.. 2014. Extinct New Zealand megafauna were not in decline before human colonization. Proceedings of the National Academy of Sciences. 111(13):4922–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allentoft ME, Rawlence NJ.. 2012. Moa’s Ark or volant ghosts of Gondwana? Insights from nineteen years of ancient DNA research on the extinct moa (Aves: Dinornithiformes) of New Zealand. Annals of Anatomy-Anatomischer Anzeiger. 194(1):36–51. [DOI] [PubMed] [Google Scholar]
- Allwood J, Gleeson D, Mayer G, Daniels S, Beggs JR, Buckley TR.. 2010. Support for vicariant origins of the New Zealand Onychophora. Journal of Biogeography. 37(4):669–681. [Google Scholar]
- Anderson A. 1989. Mechanics of overkill in the extinction of New Zealand moas. Journal of Archaeological Science. 16(2):137–151. [Google Scholar]
- Baker AJ, Haddrath O, McPherson JD, Cloutier A.. 2014. Genomic support for a moa–tinamou clade and adaptive morphological convergence in flightless ratites. Molecular Biology and Evolution. 31(7):1686–1696. [DOI] [PubMed] [Google Scholar]
- Baker AJ, Huynen LJ, Haddrath O, Millar CD, Lambert DM.. 2005. Reconstructing the tempo and mode of evolution in an extinct clade of birds with ancient DNA: the giant moas of New Zealand. Proceedings of the National Academy of Sciences. 102(23):8257–8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellamy D. 1990. Moa’s Ark: The Voyage of New Zealand. Auckland: Viking. [Google Scholar]
- Bergner LM, Dussex N, Jamieson IG, Robertson BC.. 2016. European colonization, not Polynesian arrival, impacted population size and genetic diversity in the critically endangered New Zealand kākāpō. Journal of Heredity. 107(7):593–602. [DOI] [PubMed] [Google Scholar]
- Boast AP, Chapman B, Herrera MB, Worthy TH, Scofield RP, Tennyson AJD, Houde P, Bunce M, Cooper A, Mitchell KJ.. 2019. Mitochondrial genomes from New Zealand’s extinct adzebills (Aves: Aptornithidae: Aptornis) support a sister-taxon relationship with the Afro-Madagascan Sarothruridae. Diversity. 11:24. [Google Scholar]
- Boast AP, Weyrich LS, Wood JR, Metcalf JL, Knight R, Cooper A.. 2018. Coprolites reveal ecological interactions lost with the extinction of New Zealand birds. Proceedings of the National Academy of Sciences. 115(7):1546–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boessenkool S, Austin JJ, Worthy TH, Scofield P, Cooper A, Seddon PJ, Waters JM.. 2009. Relict or colonizer? Extinction and range expansion of penguins in southern New Zealand. Proceedings of the Royal Society B: Biological Sciences. 276(1658):815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boessenkool S, Star B, Seddon PJ, Waters JM.. 2010. Temporal genetic samples indicate small effective population size of the endangered yellow-eyed penguin. Conservation Genetics. 11(2):539–546. [Google Scholar]
- Bourdon E, De Ricqles A, Cubo J.. 2009. A new Transantarctic relationship: morphological evidence for a Rheidae-Dromaiidae-Casuariidae clade (Aves, Palaeognathae, Ratitae). Zoological Journal of the Linnean Society. 156(3):641–663. [Google Scholar]
- Brasell KA, Howarth J, Pearman JK, Fitzsimons SJ, Zaiko A, Pochon X, Vandergoes MJ, Simon KS, Wood SA.. 2021. Lake microbial communities are not resistant or resilient to repeated large-scale natural pulse disturbances. Molecular Ecology. 30(20):5137–5150. [DOI] [PubMed] [Google Scholar]
- Buckley TR, Lord NP, Ramon-Laca A, Allwood JS, Leschen RAB.. 2020. Multiple lineages of hyper-diverse Zopheridae beetles survived the New Zealand Oligocene Drowning. Journal of Biogeography. 47(4):927–940. [Google Scholar]
- Bunce M, Szulkin M, Lerner HRL, Barnes I, Shapiro B, Cooper A, Holdaway RN.. 2005. Ancient DNA provides new insights into the evolutionary history of New Zealand’s extinct giant eagle. PLoS Biology. 3(1):e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunce M, Worthy TH, Phillips MJ, Holdaway RN, Willerslev E, Haile J, Shapiro B, Scofield RP, Drummond A, Kamp PJJ, et al. . 2009. The evolutionary history of the extinct ratite moa and New Zealand Neogene paleogeography. Proceedings of the National Academy of Sciences. 106(49):20646–20651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell HJ, Andrews PB, Beu AG, Edwards AR, Hornibrook N, Laird MG, Maxwell PA, Watters WA.. 1988. Cretaceous-Cenozoic lithostratigraphy of the Chatham Islands. Journal of the Royal Society of New Zealand. 18(3):285–308. [Google Scholar]
- Campbell HJ, Begg J, Beu A, Carter B, Curtis N, Davies G, Emberson R, Given D, Goldberg J, Holt K, et al. . 2008. Geological considerations relating to the Chatham Islands, mainland New Zealand, and the history of New Zealand terrestrial life. Geological Society of New Zealand Miscellaneous Publication. 126.
- Campbell HJ, Landis CA.. 2001. New Zealand awash. New Zealand Geographic. 51:6–7. [Google Scholar]
- Carr LM, McLenachan PA, Waddell PJ, Gemmell NJ, Penny D.. 2015. Analyses of the mitochondrial genome of Leiopelma hochstetteri argues against the full drowning of New Zealand. Journal of Biogeography. 42(6):1066–1076. [Google Scholar]
- Carter RM. 2005. A New Zealand climatic template back to c. 3.9 Ma: ODP site 1119, Canterbury Bight, south-west Pacific Ocean, and its relationship to onland successions. Journal of the Royal Society of New Zealand. 35(1–2):9–42. [Google Scholar]
- Cole TL, Ksepka DT, Mitchell KJ, Tennyson AJD, Thomas DB, Pan H, Zhang G, Rawlence NJ, Wood JR, Bover P, et al. . 2019. Mitogenomes uncover extinct penguin taxa and reveal island formation as a key driver of speciation. Molecular Biology and Evolution. 36(4):784–797. [DOI] [PubMed] [Google Scholar]
- Cole TL, Wood JR.. 2018. The ancient DNA revolution: the latest era in unearthing New Zealand’s faunal history. New Zealand Journal of Zoology. 45(2):91–120. [Google Scholar]
- Collins CJ, Rawlence NJ, Prost S, Anderson CN, Knapp M, Scofield RP, Robertson BC, Smith I, Matisoo-Smith EA, Chilvers BL, et al. . 2014. Extinction and recolonization of coastal megafauna following human arrival in New Zealand. Proceedings of the Royal Society B: Biological Sciences. 281(1786):20140097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper A, Cooper RA.. 1995. The Oligocene bottleneck and New Zealand biota: genetic record of a past environmental crisis. Proceedings of the Royal Society of London. Series B: Biological Sciences. 261(1362):293–302. [DOI] [PubMed] [Google Scholar]
- Cooper A, Lalueza-Fox C, Anderson S, Rambaut A, Austin J, Ward R.. 2001. Complete mitochondrial genome sequences of two extinct moas clarify ratite evolution. Nature. 409(6821):704–707. [DOI] [PubMed] [Google Scholar]
- Cooper A, Mourer-Chauviré C, Chambers GK, von Haeseler A, Wilson AC, Pääbo S.. 1992. Independent origins of New Zealand moas and kiwis. Proceedings of the National Academy of Sciences. 89(18):8741–8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper RA, Millener PR.. 1993. The New Zealand biota: historical background and new research. Trends in Ecology & Evolution. 8(12):429–433. [DOI] [PubMed] [Google Scholar]
- Cracraft J. 1974. Phylogeny and evolution of the ratite birds. Ibis. 116(4):494–521. [Google Scholar]
- Craw R. 1989. New Zealand biogeography: a panbiogeographic approach. New Zealand Journal of Zoology. 16(4):527–547. [Google Scholar]
- Der Sarkissian C, Pichereau V, Dupont C, Ilsøe PC, Perrigault M, Butler P, Chauvaud L, Eiríksson J, Scourse J, Paillard C, et al. . 2017. Ancient DNA analysis identifies marine mollusc shells as new metagenomic archives of the past. Molecular Ecology Resources. 17(5):835–853. [DOI] [PubMed] [Google Scholar]
- Dumbacher JP, Pratt TK, Fleischer RC.. 2003. Phylogeny of the owlet-nightjars (Aves: Aegothelidae) based on mitochondrial DNA sequence. Molecular Phylogenetics and Evolution. 29(3):540–549. [DOI] [PubMed] [Google Scholar]
- Dussex N, Bergfeldt N, de Anca Prado V, Dehasque M, Diez-del-Molino D, Ersmark E, Kanellidou F, Larsson P, Lemez S, Lord Eet al. . 2021. Integrating multi-taxon palaeogenomes and sedimentary ancient DNA to study past ecosystem dynamics. Proceedings of the Royal Society B: Biological Sciences. 288(1957):20211252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussex N, Rawlence NJ, Robertson BC.. 2015. Ancient and contemporary DNA reveal a pre-human decline but no population bottleneck associated with recent human persecution in the kea (Nestor notabilis). PLoS ONE. 10(2):e0118522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussex N, van der Valk T, Morales HE, Wheat CW, Díez-del-Molino D, von Seth J, Foster Y, Kutschera VE, Guschanski K, Rhie A, et al. . 2021. Population genomics of the critically endangered kākāpō. Cell Genomics. 1(1):100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussex N, von Seth J, Knapp M, Kardailsky O, Robertson BC, Dalén L.. 2019. Complete genomes of two extinct New Zealand passerines show responses to climate fluctuations but no evidence for genomic erosion prior to extinction. Biology Letters. 15(9):20190491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussex N, von Seth J, Robertson BC, Dalén L.. 2018. Full mitogenomes in the critically endangered kākāpō reveal major post-glacial and anthropogenic effects on neutral genetic diversity. Genes. 9(4):220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dussex N, Wegmann D, Robertson BC.. 2014. Postglacial expansion and not human influence best explains the population structure in the endangered kea (Nestor notabilis). Molecular Ecology. 23(9):2193–2209. [DOI] [PubMed] [Google Scholar]
- Fleming CA. 1962. New Zealand biogeography—A palaeontologist’s approach. Tuatara. 10(2):53–107. [Google Scholar]
- Funk DJ, Omland KE.. 2003. Species-level paraphyly and polyphyly: frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annual Review of Ecology, Evolution, and Systematics. 34:397–423. [Google Scholar]
- Garcia-R JC, Gibb GC, Trewick SA.. 2014. Deep global evolutionary radiation in birds: diversification and trait evolution in the cosmopolitan bird family Rallidae. Molecular Phylogenetics and Evolution. 81:96–108. [DOI] [PubMed] [Google Scholar]
- Garcia-R JC, Trewick SA.. 2015. Dispersal and speciation in purple swamphens (Rallidae: Porphyrio). The Auk. 132(1):140–155. [Google Scholar]
- Gemmell NJ, Rutherford K, Prost S, Tollis M, Winter D, Macey JR, Adelson DL, Suh A, Bertozzi T, Grau JH, et al. . 2020. The tuatara genome reveals ancient features of amniote evolution. Nature. 584:403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb GC, England R, Hartig G, McLenachan PA, Taylor Smith BL, McComish BJ, Cooper A, Penny D.. 2015. New Zealand passerines help clarify the diversification of major songbird lineages during the Oligocene. Genome Biology and Evolution. 7(11):2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J, Trewick SA, Powlesland RG.. 2011. Population structure and biogeography of Hemiphaga pigeons (Aves: Columbidae) on islands in the New Zealand region. Journal of Biogeography. 38(2):285–298. [Google Scholar]
- Grealy A, Phillips M, Miller G, Gilbert MTP, Rouillard J-M, Lambert D, Bunce M, Haile J.. 2017. Eggshell palaeogenomics: Palaeognath evolutionary history revealed through ancient nuclear and mitochondrial DNA from Madagascan elephant bird (Aepyornis sp.) eggshell. Molecular Phylogenetics and Evolution. 109:151–163. [DOI] [PubMed] [Google Scholar]
- Grealy A, Rawlence NJ, Bunce M.. 2017. Time to spread your wings: a review of the avian ancient DNA field. Genes. 8(7):184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grealy AC, McDowell MC, Scofield P, Murray DC, Fusco DA, Haile J, Prideaux GJ, Bunce M.. 2015. A critical evaluation of how ancient DNA bulk bone metabarcoding complements traditional morphological analysis of fossil assemblages. Quaternary Science Reviews. 128:37–47. [Google Scholar]
- Grosser S, Rawlence NJ, Anderson CN, Smith IW, Scofield RP, Waters JM.. 2016. Invader or resident? Ancient-DNA reveals rapid species turnover in New Zealand little penguins. Proceedings of the Royal Society B: Biological Sciences. 283(1824):20152879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrath O, Baker AJ.. 2001. Complete mitochondrial DNA geonome sequences of extinct birds: ratite phylogenetics and the vicariance biogeography hypothesis. Proceedings of the Royal Society of London. Series B: Biological Sciences. 268(1470):939–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn EE, Alexander MR, Grealy A, Stiller J, Gardiner DM, Holleley CE.. 2021. Unlocking inaccessible historical genomes preserved in formalin. Molecular Ecology Resources. In Press. doi: 10.1111/1755-0998.13505 [DOI] [PubMed] [Google Scholar]
- Haile J, Holdaway R, Oliver K, Bunce M, Gilbert MTP, Nielsen R, Munch K, Ho SYW, Shapiro B, Willerslev E.. 2007. Ancient DNA chronology within sediment deposits: are paleobiological reconstructions possible and is DNA leaching a factor? Molecular Biology and Evolution. 24(4):982–989. [DOI] [PubMed] [Google Scholar]
- Heenan PB, McGlone MS.. 2013. Evolution of New Zealand alpine and open-habitat plant species during the late Cenozoic. New Zealand Journal of Ecology. 37(1):105–113. [Google Scholar]
- Hocknull SA, Lewis R, Arnold LJ, Pietsch T, Joannes-Boyau R, Price GJ, Moss P, Wood R, Dosseto A, Louys J, et al. . 2020. Extinction of eastern Sahul megafauna coincides with sustained environmental deterioration. Nature Communications. 11(1):2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdaway RN, Worthy TH, Tennyson AJD.. 2001. A working list of breeding bird species of the New Zealand region at first human contact. New Zealand Journal of Zoology. 28(2):119–187. [Google Scholar]
- Houde P, Cooper A, Leslie E, Strand AE, Montaño GA.. 1997. Chapter 5 – phylogeny and evolution of 12S rDNA in Gruiformes (Aves). In: Mindell DP, editor. Avian Molecular Evolution and Systematics. San Diego: Academic Press; p. 121–158. [Google Scholar]
- Huynen L, Gill BJ, Millar CD, Lambert DM.. 2010. Ancient DNA reveals extreme egg morphology and nesting behavior in New Zealand’s extinct moa. Proceedings of the National Academy of Sciences. 107(37):16201–16206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson US, Pasquet E, Irestedt M.. 2011. The New Zealand thrush: an extinct oriole. PLoS ONE. 6(9):e24317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp PJJ. 1986. The mid-Cenozoic Challenger Rift System of western New Zealand and its implications for the age of Alpine Fault inception. Geological Society of America Bulletin. 97(3):255–281. [Google Scholar]
- King GA, Gilbert MTP, Willerslev E, Collins MJ, Kenward H.. 2009. Recovery of DNA from archaeological insect remains: first results, problems and potential. Journal of Archaeological Science. 36(5):1179–1183. [Google Scholar]
- King KJ, Lewis DM, Waters JM, Wallis GP.. 2020. Persisting in a glaciated landscape: Pleistocene microrefugia evidenced by the tree wētā Hemideina maori in central South Island, New Zealand. Journal of Biogeography. 47(11):2518–2531. [Google Scholar]
- King PR. 2000. Tectonic reconstructions of New Zealand: 40 Ma to the present. New Zealand Journal of Geology and Geophysics. 43(4):611–638. [Google Scholar]
- Kistler L, Ware R, Smith O, Collins M, Allaby RG.. 2017. A new model for ancient DNA decay based on paleogenomic meta-analysis. Nucleic Acids Research. 45(11):6310–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp M, Thomas JE, Haile J, Prost S, Ho SYW, Dussex N, Cameron-Christie S, Kardailsky O, Barnett R, Bunce M, et al. . 2019. Mitogenomic evidence of close relationships between New Zealand’s extinct giant raptors and small-sized Australian sister-taxa. Molecular Phylogenetics and Evolution. 134:122–128. [DOI] [PubMed] [Google Scholar]
- Landis CA, Campbell HJ, Begg JG, Mildenhall DC, Paterson AM, Trewick SA.. 2008. The Waipounamu Erosion Surface: questioning the antiquity of the New Zealand land surface and terrestrial fauna and flora. Geological Magazine. 145(2):173–197. [Google Scholar]
- Lee DE, Kaulfuss U, Conran JG, Bannister JM, Lindqvist JK.. 2016. Biodiversity and palaeoecology of Foulden Maar: an early Miocene Konservat-Lagerstätte deposit in southern New Zealand. Alcheringa: An Australasian Journal of Palaeontology. 40(4):525–541. [Google Scholar]
- Lomolino MV, Tomlinson S, Wood JR, Wilmshurst JM, Fordham DA.. 2021. Geographic and ecological segregation in an extinct guild of flightless birds: New Zealand’s moa. Frontiers of Biogeography. 13(4):e53416. [Google Scholar]
- Lorenzen ED, Nogues-Bravo D, Orlando L, Weinstock J, Binladen J, Marske KA, Ugan A, Borregaard MK, Gilbert MT, Nielsen R, et al. . 2011. Species-specific responses of Late Quaternary megafauna to climate and humans. Nature. 479(7373):359–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack JE, Tsai WLE, Faircloth BC.. 2016. Sequence capture of ultraconserved elements from bird museum specimens. Molecular Ecology Resources. 16(5):1189–1203. [DOI] [PubMed] [Google Scholar]
- McCulloch GA, Wallis GP, Waters JM.. 2010. Onset of glaciation drove simultaneous vicariant isolation of alpine insects in New Zealand. Evolution. 64(7):2033–2043. [DOI] [PubMed] [Google Scholar]
- McCulloch GA, Waters JM.. 2019. Phylogenetic divergence of island biotas: molecular dates, extinction, and “relict” lineages. Molecular Ecology. 28(19):4354–4362. [DOI] [PubMed] [Google Scholar]
- McGlone MS. 2005. Goodbye Gondwana. Journal of Biogeography. 32(5):739–740. [Google Scholar]
- McInerney PL, Lee MSY, Clement AM, Worthy TH.. 2019. The phylogenetic significance of the morphology of the syrinx, hyoid and larynx, of the southern cassowary, Casuarius casuarius (Aves, Palaeognathae). BMC Evolutionary Ecology. 19:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf JL, Turney C, Barnett R, Martin F, Bray SC, Vilstrup JT, Orlando L, Salas-Gismondi R, Loponte D, Medina M, et al. . 2016. Synergistic roles of climate warming and human occupation in Patagonian megafaunal extinctions during the Last Deglaciation. Science Advances. 2(6):e1501682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KJ, Llamas B, Soubrier J, Rawlence NJ, Worthy TH, Wood J, Lee MSY, Cooper A.. 2014. Ancient DNA reveals elephant birds and kiwi are sister taxa and clarifies ratite bird evolution. Science. 344(6186):898–900. [DOI] [PubMed] [Google Scholar]
- Mitchell KJ, Rawlence NJ.. 2021. Examining natural history through the lens of palaeogenomics. Trends in Ecology & Evolution. 36(3):258–267. [DOI] [PubMed] [Google Scholar]
- Mitchell KJ, Wood JR, Llamas B, McLenachan PA, Kardailsky O, Scofield RP, Worthy TH, Cooper A.. 2016. Ancient mitochondrial genomes clarify the evolutionary history of New Zealand’s enigmatic acanthisittid wrens. Molecular Phylogenetics and Evolution. 102:295–304. [DOI] [PubMed] [Google Scholar]
- Mitchell KJ, Wood JR, Scofield RP, Llamas B, Cooper A.. 2014. Ancient mitochondrial genome reveals unsuspected taxonomic affinity of the extinct Chatham duck (Pachyanas chathamica) and resolves divergence times for New Zealand and sub-Antarctic brown teals. Molecular Phylogenetics and Evolution. 70:420–428. [DOI] [PubMed] [Google Scholar]
- Mudge C, Gray LJ, Austin JJ.. 2022. Using mitochondrial DNA to identify the provenance of 19th century kākāpō skins held in Australia’s oldest natural history collection, the Macleay. Emu - Austral Ornithology. 122(1):16–26. [Google Scholar]
- Murray DC, Haile J, Dortch J, White NE, Haouchar D, Bellgard MI, Allcock RJ, Prideaux GJ, Bunce M.. 2013. Scrapheap Challenge: a novel bulk-bone metabarcoding method to investigate ancient DNA in faunal assemblages. Scientific Reports. 3:3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newnham R, McGlone M, Moar N, Wilmshurst J, Vandergoes M.. 2013. The vegetation cover of New Zealand at the Last Glacial Maximum. Quaternary Science Reviews. 74:202–214. [Google Scholar]
- Oskam CL, Jacomb C, Allentoft ME, Walter R, Paul Scofield R, Haile J, Holdaway RN, Bunce M.. 2011. Molecular and morphological analyses of avian eggshell excavated from a late thirteenth century earth oven. Journal of Archaeological Science. 38(10):2589–2595. [Google Scholar]
- Owen R. 1879. Memoirs on the extinct wingless birds of New Zealand, with an appendix on those of England, Australia, Newfoundland, Mauritius, and Rodriguez. London: J. Van Voorst. [Google Scholar]
- Perry GLW, Wilmshurst JM, McGlone M.. 2014. Ecology and long-term history of fire in New Zealand. New Zealand Journal of Ecology. 38(2):157–176. [Google Scholar]
- Phillips MJ, Gibb GC, Crimp EA, Penny D.. 2010. Tinamous and moa flock together: mitochondrial genome sequence analysis reveals independent losses of flight among ratites. Systematic Biology. 59(1):90–107. [DOI] [PubMed] [Google Scholar]
- Phillips MJ, Zakaria SS.. 2021. Enhancing mitogenomic phylogeny and resolving the relationships of extinct megafaunal placental mammals. Molecular Phylogenetics and Evolution. 158:107082. [DOI] [PubMed] [Google Scholar]
- Pole M. 2022. A vanished ecosystem: Sophora microphylla (kōwhai) dominated forest recorded in mid-late Holocene rock shelters in Central Otago, New Zealand. Palaeontologia Electronica. 25(1):a1. [Google Scholar]
- Rawlence NJ, Collins CJ, Anderson CNK, Maxwell JJ, Smith IWG, Robertson BC, Knapp M, Horsburgh KA, Stanton J-AL, Scofield RP, et al. . 2016. Human-mediated extirpation of the unique Chatham Islands sea lion and implications for the conservation management of remaining New Zealand sea lion populations. Molecular Ecology. 25(16):3950–3961. [DOI] [PubMed] [Google Scholar]
- Rawlence NJ, Kardamaki A, Easton LJ, Tennyson AJD, Scofield RP, Waters JM.. 2017. Ancient DNA and morphometric analysis reveal extinction and replacement of New Zealand’s unique black swans. Proceedings of the Royal Society B: Biological Sciences. 284(1859):20170876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlence NJ, Kennedy M, Anderson CN, Prost S, Till CE, Smith IW, Scofield RP, Tennyson AJ, Hamel J, Lalas C, et al. . 2015. Geographically contrasting biodiversity reductions in a widespread New Zealand seabird. Molecular Ecology. 24(18):4605–4616. [DOI] [PubMed] [Google Scholar]
- Rawlence NJ, Kennedy M, Waters JM, Scofield RP.. 2014. Morphological and ancient DNA analyses reveal inaccurate labels on two of Buller’s bird specimens. Journal of the Royal Society of New Zealand. 44(4):163–169. [Google Scholar]
- Rawlence NJ, Lowe DJ, Wood JR, Young JM, Churchman GJ, Huang Y-T, Cooper A.. 2014. Using palaeoenvironmental DNA to reconstruct past environments: progress and prospects. Journal of Quaternary Science. 29(7):610–626. [Google Scholar]
- Rawlence NJ, Metcalf JL, Wood JR, Worthy TH, Austin JJ, Cooper A.. 2012. The effect of climate and environmental change on the megafaunal moa of New Zealand in the absence of humans. Quaternary Science Reviews. 50:141–153. [Google Scholar]
- Rawlence NJ, Perry GLW, Smith IWG, Scofield RP, Tennyson AJD, Matisoo-Smith EA, Boessenkool S, Austin JJ, Waters JM.. 2015. Radiocarbon-dating and ancient DNA reveal rapid replacement of extinct prehistoric penguins. Quaternary Science Reviews. 112:59–65. [Google Scholar]
- Rawlence NJ, Potter BCM, Dussex N, Scarsbrook L, Orlovich DA, Waters JM, McGlone M, Knapp M.. 2021. Plio-Pleistocene environmental changes shape present day phylogeography of New Zealand’s southern beeches (Nothofagaceae). New Zealand Journal of Botany. 59(1):55–71. [Google Scholar]
- Rawlence NJ, Rayner MJ, Lovegrove TG, Stoddart D, Vermeulen M, Easton LJ, Tennyson AJD, Scofield RP, Kennedy M, Spencer H, et al. . 2019. Archival DNA reveals cryptic biodiversity within the Spotted Shag (Phalacrocorax punctatus) from New Zealand. The Condor. 121(3):duz029. [Google Scholar]
- Rawlence NJ, Scofield RP, McGlone MS, Knapp M.. 2019. History repeats: large scale synchronous biological turnover in avifauna from the Plio-Pleistocene and Late Holocene of New Zealand. Frontiers in Ecology and Evolution. 7:201900158. [Google Scholar]
- Rawlence NJ, Tennyson AJD, Cole TL, Verry AJF, Scofield RP.. 2019. Evidence for breeding of Megadyptes penguins in the North Island at the time of human arrival. New Zealand Journal of Zoology. 46(2):165–173. [Google Scholar]
- Rawlence NJ, Till CE, Easton LJ, Spencer HG, Schuckard R, Melville DS, Scofield RP, Tennyson AJD, Rayner MJ, Waters JM, et al. . 2017. Speciation, range contraction and extinction in the endemic New Zealand King Shag complex. Molecular Phylogenetics and Evolution. 115:197–209. [DOI] [PubMed] [Google Scholar]
- Rawlence NJ, Verry AJF, Greig K, Maxwell JJ, Shepherd LS, Walter R.. In press. Reconstructing the impact of humans on Aotearoa New Zealand’s biodiversity. In: Decocq G, editor. Historical Ecology. Paris: Springer. [Google Scholar]
- Rawlence NJ, Wood JR, Armstrong KN, Cooper A.. 2009. DNA content and distribution in ancient feathers and potential to reconstruct the plumage of extinct avian taxa. Proceedings of the Royal Society B: Biological Sciences. 276(1672):3395–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruane S, Austin CC.. 2017. Phylogenomics using formalin-fixed and 100+ year-old intractable natural history specimens. Molecular Ecology Resources. 17(5):1003–1008. [DOI] [PubMed] [Google Scholar]
- Salis AT, Easton LJ, Robertson BC, Gemmell N, Smith IWG, Weisler MI, Waters JM, Rawlence NJ.. 2016. Myth or relict: does ancient DNA detect the enigmatic Upland seal? Molecular Phylogenetics and Evolution. 97:101–106. [DOI] [PubMed] [Google Scholar]
- Sarkissian CD, Möller P, Hofman CA, Ilsøe P, Rick TC, Schiøtte T, Sørensen MV, Dalén L, Orlando L.. 2020. Unveiling the ecological applications of ancient DNA from mollusk shells. Frontiers in Ecology and Evolution. 8:37. [Google Scholar]
- Scarsbrook L, Mitchell KJ, McGee MD, Closs GP, Rawlence NJ.. In review. Ancient DNA from the extinct New Zealand grayling (Prototroctes oxyrhynchus) reveals evidence for Miocene marine dispersal. BioRxiv. doi: 10.1101/2022.05.12.491729. [DOI]
- Scarsbrook L, Sherratt E, Hitchmough RA, Rawlence NJ.. 2021. Skeletal variation in extant species enables systematic identification of New Zealand’s large, subfossil diplodactylids. BMC Ecology and Evolution. 21(1):67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarsbrook L, Verry AJF, Walton K, Hitchmough RA, Rawlence NJ.. 2022. Ancient mitochondrial genomes recovered from small-vertebrate bones through minimally destructive DNA extraction: an example using Duvaucel’s gecko. Molecular Ecology. In Press. doi: 10.1111/mec.16434. [DOI] [PubMed] [Google Scholar]
- Schellart WP, Lister GS, Toy VG.. 2006. A Late Cretaceous and Cenozoic reconstruction of the Southwest Pacific region: tectonics controlled by subduction and slab rollback processes. Earth-Science Reviews. 76(3–4):191–233. [Google Scholar]
- Scofield RP, Mitchell KJ, Wood JR, De Pietri VL, Jarvie S, Llamas B, Cooper A.. 2017. The origin and phylogenetic relationships of the New Zealand ravens. Molecular Phylogenetics and Evolution. 106:136–143. [DOI] [PubMed] [Google Scholar]
- Scofield RP, Wood JR, de Nascimento L, Robertson HA, Colbourne RM, De Pietri VL, Innes J, Weir JT.. 2021. Identification of the type locality of the South Island brown kiwi Apteryx australis. Conservation Genetics. 22(4):645–652. [Google Scholar]
- Seersholm FV, Cole TL, Grealy A, Rawlence NJ, Greig K, Knapp M, Stat M, Hansen AJ, Easton LJ, Shepherd L, et al. . 2018. Subsistence practices, past biodiversity, and anthropogenic impacts revealed by New Zealand-wide ancient DNA survey. Proceedings of the National Academy of Sciences. 115(30):7771–7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seersholm FV, Werndly DJ, Grealy A, Johnson T, Keenan Early EM, Lundelius EL, Winsborough B, Farr GE, Toomey R, Hansen AJ, et al. . 2020. Rapid range shifts and megafaunal extinctions associated with late Pleistocene climate change. Nature Communications. 11(1):2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro B, Hofreiter M.. 2014. A paleogenomic perspective on evolution and gene function: new insights from ancient DNA. Science. 343(6169):1236573. [DOI] [PubMed] [Google Scholar]
- Shepherd LD, de Lange PJ, Perrie LR, Heenan PB.. 2017. Chloroplast phylogeography of New Zealand Sophora trees (Fabaceae): extensive hybridization and widespread Last Glacial Maximum survival. Journal of Biogeography. 44(7):1640–1651. [Google Scholar]
- Shepherd LD, Lambert DM.. 2007. The relationships and origins of the New Zealand wattlebirds (Passeriformes, Callaeatidae) from DNA sequence analyses. Molecular Phylogenetics and Evolution. 43(2):480–492. [DOI] [PubMed] [Google Scholar]
- Shepherd LD, Lambert DM.. 2008. Ancient DNA and conservation: lessons from the endangered kiwi of New Zealand. Molecular Ecology. 17(9):2174–2184. [DOI] [PubMed] [Google Scholar]
- Shepherd LD, Perrie LR, Brownsey PJ.. 2007. Fire and ice: volcanic and glacial impacts on the phylogeography of the New Zealand forest fern Asplenium hookerianum. Molecular Ecology. 16(21):4536–4549. [DOI] [PubMed] [Google Scholar]
- Shepherd LD, Tennyson AJD, Lambert DM.. 2013. Using ancient DNA to enhance museum collections: a case study of rare kiwi (Apteryx spp.) specimens. Journal of the Royal Society of New Zealand. 43(3):119–127. [Google Scholar]
- Shepherd LD, Worthy TH, Tennyson AJD, Scofield RP, Ramstad KM, Lambert DM.. 2012. Ancient DNA analyses reveal contrasting phylogeographic patterns amongst kiwi (Apteryx spp.) and a recently extinct lineage of spotted kiwi. PLoS ONE. 7(8):e42384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straube N, Lyra ML, Paijmans JLA, Preick M, Basler N, Penner J, Rödel M-O, Westbury MV, Haddad CFB, Barlow A, et al. . 2021. Successful application of ancient DNA extraction and library construction protocols to museum wet collection specimens. Molecular Ecology Resources. 21(7):2299–2315. [DOI] [PubMed] [Google Scholar]
- Tennyson AJD, Cooper JH, Shepherd LD.. 2015. A new species of extinct Pterodroma petrel (procellariiformes: Procellariidae) from the Chatham Islands, New Zealand. Bulletin of the British Ornithologists’ Club. 135(3):267–277. [Google Scholar]
- Tennyson AJD, Martinson P.. 2006. Extinct Birds of New Zealand. Wellington: Te Papa Press. [Google Scholar]
- Tennyson AJD, Shepherd LD.. 2017. DNA reveals the relationships of the extinct Scarlett’s shearwater Puffinus spelaeus (Procellariiformes: Procellariidae). Journal of Ornithology. 158(2):379–384. [Google Scholar]
- Tippett JM, Kamp PJJ.. 1993. The role of faulting in rock uplift in the Southern Alps, New Zealand. New Zealand Journal of Geology and Geophysics. 36(4):497–504. [Google Scholar]
- Trewick SA. 1997. Flightlessness and phylogeny amongst endemic rails (Aves: Rallidae) of the New Zealand region. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences. 352(1352):429–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick SA. 2001. Scree weta phylogeography: surviving glaciation and implications for Pleistocene biogeography in New Zealand. New Zealand Journal of Zoology. 28(3):291–298. [Google Scholar]
- Verry AJF. 2021. Investigating the response of New Zealand avifauna to past climatic and environmental change: insights from ancient DNA [Unpublished PhD Thesis]. University of Otago. [Google Scholar]
- Verry AJF, Mitchell KJ, Rawlence NJ.. 2022. Genetic evidence for post-glacial expansion from a southern refugium in the eastern moa (Emeus crassus). Biology Letters. 18(5):20220013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verry AJF, Scarsbrook L, Scofield RP, Tennyson AJD, Weston KA, Robertson BC, Rawlence NJ.. 2019. Who, where, what, wren? Using ancient DNA to examine the veracity of museum specimen data: a case study of the New Zealand rock wren (Xenicus gilviventris). Frontiers in Ecology and Evolution. 7:496. [Google Scholar]
- Verry AJF, Schmidt M, Rawlence NJ.. 2022. A partial skeleton provides evidence for the former occurrence of moa populations on Rakiura Stewart Island. New Zealand Journal of Ecology. 46(1):3458. [Google Scholar]
- Wallis GP, Jorge F.. 2018. Going under down under? Lineage ages argue for extensive survival of the Oligocene marine transgression on Zealandia. Molecular Ecology. 27(22):4368–4396. [DOI] [PubMed] [Google Scholar]
- Wallis GP, Waters JM, Upton P, Craw D.. 2016. Transverse alpine speciation driven by glaciation. Trends in Ecology & Evolution. 31(12):916–926. [DOI] [PubMed] [Google Scholar]
- Wanner H, Beer J, Bütikofer J, Crowley TJ, Cubasch U, Flückiger J, Goosse H, Grosjean M, Joos F, Kaplan JO, et al. . 2008. Mid- to Late Holocene climate change: an overview. Quaternary Science Reviews. 27(19):1791–1828. [Google Scholar]
- Waters JM, Craw D.. 2006. Goodbye Gondwana? New Zealand biogeography, geology, and the problem of circularity. Systematic Biology. 55(2):351–356. [DOI] [PubMed] [Google Scholar]
- Waters JM, Fraser CI, Maxwell JJ, Rawlence NJ.. 2017. Did interaction between human pressure and Little Ice Age drive biological turnover in New Zealand? Journal of Biogeography. 44(7):1481–1490. [Google Scholar]
- Waters JM, Grosser S.. 2016. Managing shifting species: ancient DNA reveals conservation conundrums in a dynamic world. Bioessays. 38(11):1177–1184. [DOI] [PubMed] [Google Scholar]
- Weir JT, Haddrath O, Robertson HA, Colbourne RM, Baker AJ.. 2016. Explosive ice age diversification of kiwi. Proceedings of the National Academy of Sciences. 113(38):E5580–E5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston KA, Robertson BC.. 2015. Population structure within an alpine archipelago: strong signature of past climate change in the New Zealand rock wren (Xenicus gilviventris). Molecular Ecology. 24(18):4778–4794. [DOI] [PubMed] [Google Scholar]
- Willerslev E, Hansen AJ, Binladen J, Brand TB, Gilbert MT, Shapiro B, Bunce M, Wiuf C, Gilichinsky DA, Cooper A.. 2003. Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science. 300(5620):791–795. [DOI] [PubMed] [Google Scholar]
- Wilmshurst JM, Anderson AJ, Higham TFG, Worthy TH.. 2008. Dating the late prehistoric dispersal of Polynesians to New Zealand using the commensal Pacific rat. Proceedings of the National Academy of Sciences. 105(22):7676–7680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmshurst JM, Moar NT, Wood JR, Bellingham PJ, Findlater AM, Robinson JJ, Stone C.. 2014. Use of pollen and ancient DNA as conservation baselines for offshore islands in New Zealand. Conservation Biology. 28(1):202–212. [DOI] [PubMed] [Google Scholar]
- Wood JR, Mitchell KJ, Scofield RP, De Pietri VL, Rawlence NJ, Cooper A.. 2017. Phylogenetic relationships and terrestrial adaptations of the extinct laughing owl, Sceloglaux albifacies (Aves: Strigidae). Zoological Journal of the Linnean Society. 179(4):907–918. [Google Scholar]
- Wood JR, Mitchell KJ, Scofield RP, Tennyson AJD, Fidler AE, Wilmshurst JM, Llamas B, Cooper A.. 2014. An extinct nestorid parrot (Aves, Psittaciformes, Nestoridae) from the Chatham Islands, New Zealand. Zoological Journal of the Linnean Society. 172(1):185–199. [Google Scholar]
- Wood JR, Rawlence NJ, Rogers GM, Austin JJ, Worthy TH, Cooper A.. 2008. Coprolite deposits reveal the diet and ecology of the extinct New Zealand megaherbivore moa (Aves, Dinornithiformes). Quaternary Science Reviews. 27(27):2593–2602. [Google Scholar]
- Wood JR, Richardson SJ, McGlone M, Wilmshurst JM.. 2020. The diets of moa (Aves: Dinornithiformes). New Zealand Journal of Ecology. 44(1):3397. [Google Scholar]
- Wood JR, Wilmshurst JM, Rawlence NJ, Bonner KI, Worthy TH, Kinsella JM, Cooper A.. 2013. A megafauna’s microfauna: gastrointestinal parasites of New Zealand’s extinct moa (Aves: Dinornithiformes). PLoS ONE. 8(2):e57315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JR, Wilmshurst JM, Richardson SJ, Rawlence NJ, Wagstaff SJ, Worthy TH, Cooper A.. 2013. Resolving lost herbivore community structure using coprolites of four sympatric moa species (Aves: Dinornithiformes). Proceedings of the National Academy of Sciences. 110(42):16910–16915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JR, Wilmshurst JM, Wagstaff SJ, Worthy TH, Rawlence NJ, Cooper A.. 2012. High-resolution coproecology: using coprolites to reconstruct the habits and habitats of New Zealand’s extinct upland moa (Megalapteryx didinus). PLoS ONE. 7(6):e40025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthy T, Tennyson AD, Scofield RP.. 2011. Fossils reveal an early Miocene presence of the aberrant gruiform Aves: Aptornithidae in New Zealand. Journal of Ornithology. 152(3):669–680. [Google Scholar]
- Worthy TH. 1987. Osteology of Leiopelma (Amphibia: Leiopelmatidae) and descriptions of three new subfossil Leiopelma species. Journal of the Royal Society of New Zealand. 17:201–251. [Google Scholar]
- Worthy TH. 1991. Fossil skink bones from Northland, New Zealand, and description of a new species of Cyclodina, Scincidae. Journal of the Royal Society of New Zealand. 21:329–348. [Google Scholar]
- Worthy TH. 1997. The identification of fossil Eudyptes and Megadyptes bones at Marfells Beach, Marlborough, South Island. New Zealand Natural Sciences. 23:71–85. [Google Scholar]
- Worthy TH. 1998a. Quaternary fossil faunas of Otago, South Island, New Zealand. Journal of the Royal Society of New Zealand. 28(3):421–521. [Google Scholar]
- Worthy TH. 1998b. The Quaternary fossil avifauna of Southland, South Island, New Zealand. Journal of the Royal Society of New Zealand. 28(4):537–589. [Google Scholar]
- Worthy TH. 1998c. A remarkable fossil and archaeological avifauna from Marfells Beach, Lake Grassmere, South Island, New Zealand. Records of the Canterbury Museum. 12:79–176. [Google Scholar]
- Worthy TH, Hand SJ, Nguyen JMT, Tennyson AJD, Worthy JP, Scofield RP, Boles WE, Archer M.. 2010. Biogeographical and phylogenetic implications of an early Miocene wren (Aves: Passeriformes: Acanthisittidae) from New Zealand. Journal of Vertebrate Paleontology. 30(2):479–498. [Google Scholar]
- Worthy TH, Holdaway RN.. 2002. The Lost World of the Moa: Prehistoric Life of New Zealand. Christchurch: Canterbury University Press. [Google Scholar]
- Worthy TH, Tennyson AJD, Jones C, McNamara JA, Douglas BJ.. 2007. Miocene waterfowl and other birds from central Otago, New Zealand. Journal of Systematic Palaeontology. 5(1):1–39. [Google Scholar]
- Xia X, Xie Z, Salemi M, Chen L, Wang Y.. 2003. An index of substitution saturation and its application. Molecular Phylogenetics and Evolution. 26(1):1–7. [DOI] [PubMed] [Google Scholar]
- Yonezawa T, Segawa T, Mori H, Campos PF, Hongoh Y, Endo H, Akiyoshi A, Kohno N, Nishida S, Wu J, et al. . 2017. Phylogenomics and morphology of extinct paleognaths reveal the origin and evolution of the ratites. Current Biology. 27(1):68–77. [DOI] [PubMed] [Google Scholar]
- Zuccon D, Ericson PG.. 2012. Molecular and morphological evidences place the extinct New Zealand endemic Turnagra capensis in the Oriolidae. Molecular Phylogenetics and Evolution. 62(1):414–426. [DOI] [PubMed] [Google Scholar]