Abstract
Productive infection by adeno-associated virus type 2 (AAV) requires coinfection with a helper virus, e.g., adenovirus or herpesviruses. In the case of adenovirus coinfection, the replication machinery of the host cell performs AAV DNA replication. In contrast, it has been proposed that the herpesvirus replication machinery might replicate AAV DNA. To investigate this question, we have attempted to reconstitute AAV DNA replication in vitro using purified herpes simplex virus type 1 (HSV-1) replication proteins. We show that the HSV-1 UL5, UL8, UL29, UL30, UL42, and UL52 gene products along with the AAV Rep68 protein are sufficient to initiate replication on duplex DNA containing the AAV origins of replication, resulting in products several hundred nucleotides in length. Initiation can occur also on templates containing only a Rep binding site and a terminal resolution site. We further demonstrate that initiation of DNA synthesis can take place with a subset of these factors: Rep68 and the UL29, UL30, and UL42 gene products. Since the HSV polymerase and its accessory factor (the products of the UL30 and UL42 genes) are unable to efficiently perform synthesis by strand displacement, it is likely that in addition to creating a hairpin primer, the AAV Rep protein also acts as a helicase for DNA synthesis. The single-strand DNA binding protein (the UL29 gene product) presumably prevents reannealing of complementary strands. These results suggest that AAV can use the HSV replication apparatus to replicate its DNA. In addition, they may provide a first step for the development of a fully reconstituted AAV replication assay.
Adeno-associated virus type 2 (AAV), a member of the parvovirus family, contains a single-stranded genome of 4,679 bases. The genome of AAV contains two open reading frames, one coding for the replication (Rep) proteins and the other coding for the structural proteins. The Rep proteins are designated Rep78, Rep68, Rep52, and Rep40 according to their apparent molecular weights. They are produced by the use of different transcriptional start sites and splicing patterns. Either Rep68 or Rep78, both of which possess origin binding, helicase, ATPase, and strand- and site-specific nicking activities, is absolutely required for the replication of AAV DNA. The Rep proteins are the only AAV proteins involved in AAV DNA replication, necessitating that most replication functions must be provided by non-AAV proteins. The ends of the AAV genome contain identical origins of DNA replication. Each origin consists of an inverted terminal repeat (ITR) capable of complementary intrastrand base pairing to form a hairpin, thereby providing the replication apparatus with a primer terminus. The hairpin primers can be recreated indefinitely by the process called terminal resolution, which consists of site-specific nicking at the terminal resolution site (TRS), followed by strand displacement synthesis from the nick towards the end of the genome. The newly synthesized double-stranded ITR can then fold back and serve as a primer for synthesis of full-length genomes (reviewed in reference 2). The AAV Rep protein appears to be involved in all of these functions (4, 16, 19, 30, 42).
A curious feature of the biology of AAV is that productive infection generally requires that a helper virus, either adenovirus or a member of the herpesvirus family (reviewed in reference 2), simultaneously or subsequently infect the AAV-infected cell. There are, however, some observations illustrating that AAV may not be completely dependent on a helper virus. It has been shown that limited production of AAV can be achieved upon AAV infection of tissue culture cells that have been treated with DNA-damaging agents such as UV light, hydroxyurea, X rays, and alkylating substances (39, 40, 41). More recently, productive infection by AAV has been observed in epithelial cells maintained in raft culture (25). These cultures were apparently uninfected by any of the known helper viruses. It is also worth noting that AAV DNA can be replicated in extracts from HeLa cells which have not been infected by a helper virus, provided that Rep is supplied (26, 33).
The molecular mechanisms by which helper viruses promote AAV replication vary. The major effect that adenovirus exerts on AAV replication appears to be on gene expression and on promoting the entry of cells into S phase. It has, for example, been demonstrated that the synthesis of the AAV DNA in cells infected with adenovirus is mediated by the cellular replication machinery and not by the adenovirus polymerase (26, 27). One adenovirus protein is apparently directly involved in AAV DNA replication. The adenovirus single-strand DNA binding protein is found in cells at foci of AAV DNA replication and in vitro helps to stabilize single-stranded DNA during DNA synthesis (34, 36).
The role of the herpes simplex virus (HSV) replication machinery in the synthesis of AAV DNA is less clear. It was shown by Handa and Carter (13) that the treatment of HSV- and AAV-coinfected cells with phosphonoacetic acid (PAA), an inhibitor of the HSV polymerase, resulted in a reduction of both HSV and AAV DNA synthesis. This result raised the possibility that synthesis of the AAV DNA might be by the HSV polymerase.
The components of a minimal HSV replication machine (replisome) were identified by Challberg and colleagues (6, 38). They used selective transfection of HSV genes to demonstrate that in cell culture, replication of an HSV origin-containing plasmid could be achieved with only seven HSV genes. These were the genes coding for an origin binding protein (UL9), a single-strand DNA binding protein (UL29), a polymerase (UL30) and its accessory factor (UL42), and a helicase-primase complex (UL5, UL52, and UL8) (reviewed in reference 22).
Transfection of actively dividing HeLa cells with these seven plasmids encoding the HSV type 1 (HSV-1) replication proteins efficiently promoted synthesis of AAV DNA as well as the production of infectious particles (35). The contribution made by the individual replication proteins was also addressed. The HSV-1 origin binding protein, UL9, was not needed. The DNA polymerase, UL30, and its processivity factor, UL42, were also dispensable. In contrast, components of the helicase-primase complex, UL5, UL8, and UL52, as well as the single-strand DNA binding protein, UL29, were required (35). The interpretation of these findings is not straightforward. One would like to imagine that the HSV-1 replisome, which in itself is capable of processive and coupled synthesis of leading and lagging strands, would remain intact also during replication of AAV DNA. However, it is possible that individual components can be utilized for strand displacement synthesis together with AAV Rep and cellular enzymes.
HSV infection typically occurs in nondividing cells, and unlike adenovirus, rather than directing the host cells towards S phase, HSV down regulates host cell functions (28). It would seem that if, in this case, HSV is to serve as a helper for AAV replication, then AAV must be able to either replicate its DNA with the HSV replisome or induce a reversal of the normal HSV down regulation of host cell functions. Therefore, the question of whether in the absence of cellular replication functions AAV can use the HSV replisome to replicate its DNA remains significant.
Here we have examined replication of AAV DNA in vitro using highly purified HSV-1 replication proteins, AAV Rep68, and double-stranded template DNA containing the AAV origin of DNA replication. Our results demonstrate that AAV Rep68 is required to promote origin-specific initiation of DNA synthesis in the presence of HSV-1 replication proteins. Interestingly, efficient synthesis of AAV DNA is also obtained in the presence of a subset of HSV-1 replication proteins consisting of the single-strand DNA binding protein, UL29, and the HSV-1 DNA polymerase and its accessory factor. The products formed on longer templates, however, were often of less than full length, indicating that DNA synthesis was nonprocessive.
Recombinant AAV is becoming increasingly important as a vector for gene therapy, and HSV-1 is being proposed as a helper virus in vector production (7, 8). An understanding of the mechanisms by which AAV replicates in the presence of a helper virus should be useful in optimizing the efficiency and accuracy of vector production.
MATERIALS AND METHODS
DNA substrates.
The plasmid used for these studies (pAV2DA) is derived from pAV2 and has been described previously (31). pAV2 consists of the entire AAV2 genome inserted into a pBR derivative with BglII linkers (20). The deletion construct pAV2DA(31) was made by digesting pAV2 at the DraIII (AAV nucleotide 235) and ApaI (AAV nucleotide 4045) sites. The construct was treated with T4 polymerase and circularized by religating. Replication substrates were produced by digestion with BglII, which released a duplex complete genome in the case of pAV2 and a duplex minigenome (mAAV) in the case of pAV2DA. pBS-AAV (29) was made by insertion of a double-stranded oligonucleotide equivalent to nucleotides 89 to 133 of AAV; i.e., it contains the Rep binding site (RBS) and the TRS between the XbaI and SalI sites of pBluescript KS(+) plasmid (Stratagene). The plasmid was linearized by digestion at the XmnI site prior to use.
Proteins.
HisRep68 contains six histidine residues fused to the amino-terminal end of the full-length Rep68 protein (29). It was produced in Escherichia coli from a pET 15b vector (New England Biolabs) and purified according to the manufacturer's instructions. The proteins encoded by the HSV UL5, UL8, UL29, UL30, UL42, and UL52 genes were produced from stocks of recombinant Autographa californica nuclear polyhedrosis virus and purified as described previously (9, 10). The purity of each protein was greater than or equal to 95% as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Coomassie blue staining.
DNA replication assay.
Two types of replication assay were performed. For the basic assay (containing no cellular extract), the reaction mixture (15 μl) was 2.7% glycerol; 40 mM HEPES (pH 7.7); 40 mM creatine phosphate (pH 7.7); 7 mM MgCl2; 4 mM ATP; 200 μM each CTP, GTP, and UTP; 100 μM each dATP, dGTP, and dTTP; 10 μM dCTP; 2 mM dithiothreitol (DTT); and 6 mM potassium glutamate. It also contained 2.0 μg of creatine phosphokinase, 10.0 μg of bovine serum albumin, 5 μCi of [α-32P]dCTP (3,000 Ci/mmol; Amersham), and 35 ng of BglII-digested pAV2DA.
Proteins were added to the reaction mixture in the following order: 6,000 fmol of the UL29 protein, 1,400 fmol of the UL8 protein, 1,200 fmol of the UL5-UL52 complex, 750 fmol of the UL30-UL42 complex, and 700 fmol of HisRep68. The UL29 protein was in 10% glycerol–20 mM HEPES (pH 7.6 [NaOH])–0.5 mM EDTA (pH 8.0)–2 mM DTT–300 mM NaCl and contributed a volume of 0.6 μl; the remaining HSV proteins were in 10% glycerol–20 mM HEPES (pH 7.6 [NaOH])–0.5 mM EDTA (pH 8.0)–2 mM DTT–200 mM NaCl and contributed a volume of 2.4 μl to the 15.0-μl total. The reaction mixture was consequently increased by 2% glycerol, 5 mM HEPES, 0.1 mM EDTA, 0.4 mM DTT, and 44 mM NaCl. The reaction mixture was incubated at 37°C for 4 h.
A second replication assay (containing cellular extracts) was performed as described previously (33). The reaction mixture (15 μl) contained 40 mM HEPES (pH 7.7); 40 mM creatine phosphate (pH 7.7); 7 mM MgCl2; 4 mM ATP; 200 μM each CTP, GTP, and UTP; 100 μM each dATP, dGTP, and dTTP; 10 μM dCTP; 2 mM DTT; 6 mM potassium glutamate; 2.0 μg of creatine phosphokinase; approximately 60 μg of HeLa cell extract protein; 0.1 μg of plasmid DNA (BglII-digested pAV2 or pAV2DA); and 100 ng of HisRep68. Reaction mixtures were preincubated at 37°C for 3 h, at which time Rep68, the six HSV proteins, and labeled dCTP (10 μCi of [α-32P]dCTP [3,000 Ci/mmol; Amersham]) were added. Incubations were continued at 37°C for an additional 16 h.
Both the extract-free and the extract-containing assays were terminated by the addition of 50 μl of digestion buffer (20 mM HEPES [pH 7.5], 10 mM KCl, 10 mM EDTA, 1.0% sodium dodecyl sulfate, 50 mM NaCl). Products were passed over a Sephadex 50 spin column and then digested with proteinase K at 1 mg/ml for 2 h at 50°C. Aliquots of the products were separated by electrophoresis on 0.8% agarose gels with Tris-borate-EDTA buffer. The data were analyzed by PhosphorImager (Molecular Dynamics) quantification of dried gels using ImageQuant1.1 software.
Cellular extracts.
Replication extracts from uninfected HeLa cells were prepared as described previously (32); the procedure was a modification of that of Wobbe et al. (37).
RESULTS
DNA synthesis by HSV DNA replication proteins on an AAV substrate is dependent on the AAV Rep protein.
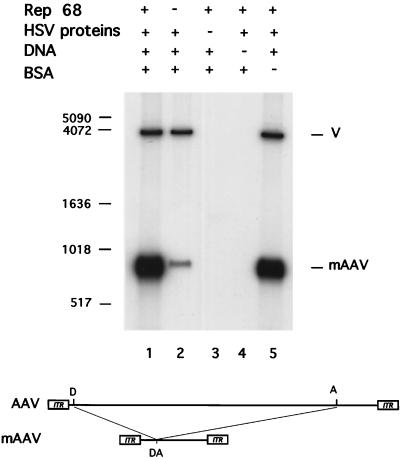
We set out to determine whether purified HSV replication proteins in combination with the AAV Rep protein could initiate DNA synthesis in an AAV origin-dependent manner. We employed a minimal AAV genome, referred to as mAAV, as a substrate. Previous observations have indicated that shorter genomes have a substantial replicative advantage over full-length genomes in uninfected HeLa cell extracts (31). Consequently, we used a linear AAV minigenome which can be excised from the plasmid pAV2DA. The plasmid pAV2DA had been derived from pAV2 by excising the AAV sequences between nucleotides 235 and 4045 (31). To create the substrate for the assay, the AAV sequences are separated from vector sequences by digesting pAV2DA with BglII, thereby producing the double-stranded mAAV genome of 870 nucleotides with intact copies of both ITRs as well as adjacent sequences (Fig. 1). Also present is an equimolar amount of the vector backbone.
FIG. 1.
Incorporation of nucleotides into the AAV minigenome is dependent upon the AAV Rep protein. The assay was performed as described in Materials and Methods except for the omission of selected reagents as indicated. Also shown is a diagram illustrating the derivation of the AAV minigenome from the full-length AAV genome. ITRs are indicated by rectangles. D and A, DraIII and ApaI sites used to generate the mini-AAV construct. Sizes in base pairs are shown on the left. V, vector; BSA, bovine serum albumin
The Rep protein is a His-tagged form of Rep68, produced in and purified from E. coli (29). The six HSV-1 proteins used in this study (the products of the UL5, UL8, UL29, UL30, UL42, and UL52 genes) were all expressed in and purified to near homogeneity from insect cells infected with recombinant baculoviruses. These six HSV proteins plus the product of the UL9 gene had been shown to constitute an HSV replisome (6, 38).
We made the assumption that the AAV Rep68 protein might substitute for the HSV-1 origin binding function and direct the activity of the HSV replication complex to the AAV template. Consequently, we omitted the UL9 gene product. Our results show that the complete system is capable of robust synthesis of mAAV DNA (Fig. 1, lane 1). The vector component, on the other hand, supports only limited synthesis of DNA, suggesting that DNA synthesis was dependent on the AAV origins of DNA replication (Fig. 1, lane 1). The ratio of newly synthesized mAAV DNA to vector DNA was 10:1. Synthesis of mAAV DNA was largely Rep68 dependent (Fig. 1, lane 2). The ratio of newly synthesized mAAV to vector DNA in the absence of Rep68 was 1:5, which corresponds approximately to ratio of the molecular weights of the DNA substrates. The Rep68 protein therefore appears to stimulate synthesis of mAAV approximately 50-fold under these conditions.
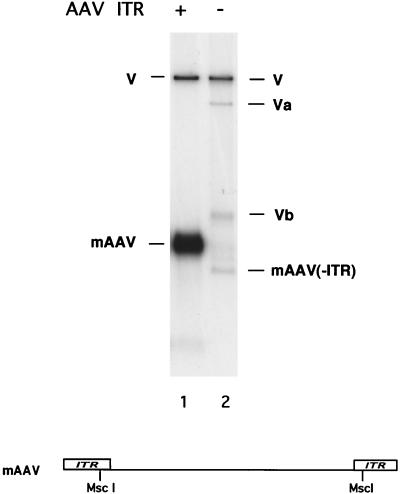
Rep68-dependent DNA synthesis by HSV-1 replication proteins requires the AAV ITRs.
To test if Rep68-dependent DNA synthesis required the AAV ITRs, which contain the origins of AAV DNA replication, we compared replication of intact mAAV to replication of a template from which the ITRs had been removed (Fig. 2). Cleavage of mAAV by MscI removes almost all of both ITRs, except for the D region (the innermost 25 bases of the ITR). As shown above, intact mAAV readily supported DNA synthesis (Fig. 2, lane 1). In contrast, very little DNA synthesis was obtained with the truncated version of mAAV referred to as mAAV(−ITR) (Fig. 2, lane 2). These results demonstrate that the ITRs (i.e., AAV origins) are required for Rep68 to direct the activity of the HSV-1 replication proteins to the mAAV template.
FIG. 2.
Incorporation of nucleotides into the AAV minigenome by Rep-HSV is dependent upon the presence of the AAV ITRs. The assay was performed as described in Materials and Methods except that in lane 2 the ITRs were removed from the substrate by MscI digestion. Va and Vb, vector fragments resulting from MscI digestion at the one MscI site in the vector. Due to an overlapping Dam methylation site, the efficiency of digestion at this site is only 2% (New England Biolabs). A diagram illustrating the portion of the AAV minigenome which is removed by digestion with MscI is also shown. V, vector. The ITRs are indicated by rectangles.
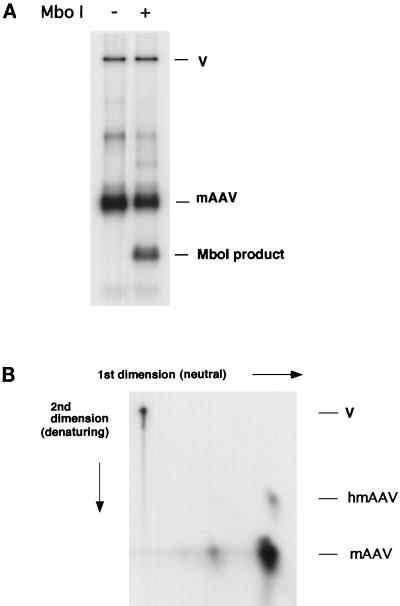
Characterization of replication products.
The replication products of the basic assay were characterized by restriction enzyme analysis as well as two-dimensional gel electrophoresis. First, an MboI digestion of the replication products was performed (Fig. 3A). The mAAV genome contains only one MboI restriction site located precisely at its center. Cleavage at this site implies that the products consist of double-stranded unmethylated DNA, demonstrating that both strands are newly synthesized. Furthermore, successful cleavage by MboI indicates that DNA synthesis must have extended at least through one half of the 840-bp mAAV genome. The average extent of MboI cleavage of the mAAV products from three experiments was 37%. In contrast, fewer than 1% of vector sequences were digested despite the presence of 20 MboI sites.
FIG. 3.
Characterization of replication products. (A) MboI digestion of the products of an assay performed as described in Materials and Methods. The MboI digestion products are indicated. (B) Two-dimensional gel electrophoresis of the replication products. Products were separated first under neutral conditions (horizontal dimension) and then under alkaline conditions (vertical dimension). The vector (V), AAV minigenome, and hairpinned AAV minigenome (hmAAV) are indicated.
Replication products were also examined by two-dimensional agarose gel electrophoresis (Fig. 3B). The products were first separated under neutral conditions. In the second dimension, electrophoresis was performed under alkaline conditions. We found that incorporation of labeled nucleotides into the mAAV genome results in production of full-length DNA chains that are essentially free of nicks (Fig. 3B). Only a small fraction of the mAAV genomes were in a hairpin configuration, as indicated by slow migration in the second dimension. It is likely that synthesis proceeds through a hairpin intermediate but that hairpinned structures in this assay are efficiently nicked by Rep68 (27, 33). The hairpin molecules remaining have presumably escaped nicking and terminal resolution. A small amount of replication products migrates in the first dimension at a position consistent with their being mAAV dimers. However, their migration during denaturing electrophoresis implies that the molecules are unit-length mAAV genomes, which in the neutral dimension migrate as noncovalently linked tandem molecules. Their presence is suggestive of a terminal resolution step in this assay. It is possible that partial resolution, i.e., Rep-dependent nicking at the TRS followed by partial rather than complete duplication of the hairpin, would give rise to free single-stranded ends derived from the ITR sequences that would permit the formation of noncovalently linked dimers.
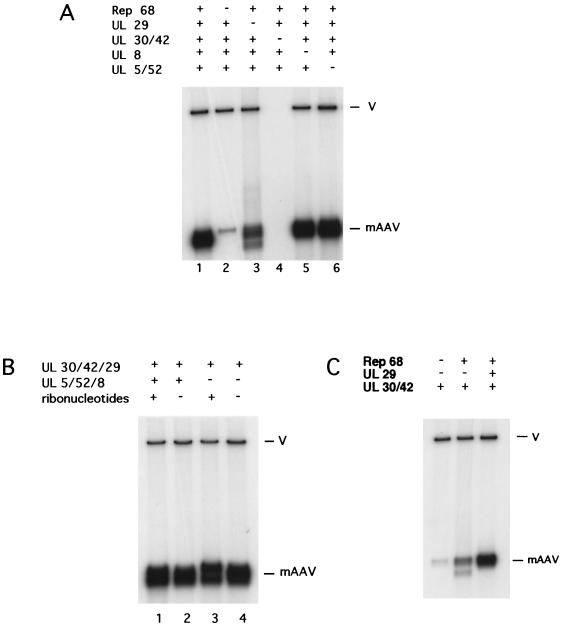
Rep-dependent DNA synthesis can be performed by a subset of HSV-1 replication proteins.
To examine the contribution of the HSV-1 replication proteins to Rep-dependent DNA synthesis, a set of reactions in which individual components were omitted were performed. In the absence of Rep68, no specific synthesis of mAAV was seen (Fig. 4A, lane 2). Omission of the HSV-1 DNA polymerase, UL30, and its accessory protein, UL42, eliminated all DNA synthesis (Fig. 4A, lane 4). Interestingly, the single-strand DNA binding protein ICP8 (encoded by UL29) was required for efficient DNA synthesis (Fig. 4A, lane 3). Without the UL29 product, the incorporation of radioactively labeled nucleotides was reduced by approximately 65%. Moreover, the replication products were more heterogeneous (Fig. 4A, lane 3). The omission of either the UL5-UL52 components of the helicase-primase complex or the UL8 gene product did not reduce total synthesis (Fig. 4A, lanes 5 and 6). Quantification of the replication products revealed that the omission of the UL5 and UL52 proteins reproducibly resulted in a slight increase of about 25% of the total synthesis of DNA. Possibly, the presence of free UL8 protein may counteract the tendency of ICP8 (the UL29 gene product) to inhibit DNA synthesis at high concentrations (11).
FIG. 4.
Synthesis requires only Rep68 and proteins coded by the HSV UL30, UL42, and UL29 genes. (A) The assay was performed as described in Materials and Methods with the omission of selected reagents (designated by their genes) as indicated. (B) The assay was performed as described in Materials and Methods with the omission of UL5, UL8, and UL52 proteins and ribonucleotides as indicated. (C) Assays were performed as described in Materials and Methods except that the protein components are only Rep68 and the UL29 and UL30-U42 proteins as indicated. V, vector.
To investigate further the contributions of the products of the UL5, UL52, and UL8 genes, we tested the effect of removing all three proteins from the assay. In Fig. 4B, a comparison of lanes 1 and 3 shows that Rep68 and the products of the UL30, UL42, and UL29 genes alone result in approximately as much synthesis as Rep68 and the products of all six HSV genes. A comparison of lanes 1 and 2 demonstrates that when the helicase-primase complex is present, the omission of ribonucleotides, which would prevent primase activity, has no significant effect on total incorporation. Taken together these observations suggest that lagging-strand synthesis is not required for production of mAAV DNA. There is also the suggestion that the presence of ribonucleotides can influence the outcome of the synthesis reactions (note the bimodal distribution of products in Fig. 4B, lanes 1 and 3). Perhaps they affect the availability of divalent cations or modify the properties of Rep68.
We also investigated whether the HSV-1 DNA polymerase alone could cooperate with Rep68 and support the production of mAAV DNA. Experiments using Rep68 and the HSV-1 DNA polymerase, consisting of the UL30 and UL42 gene products, were performed in the presence or absence of the single-strand DNA binding protein (the UL29 gene product ICP8). Our results show that in the absence of ICP8 only small amounts of mAAV DNA were produced and that these products were structurally heterogeneous (Fig. 4C, lane 2). However, the results (Fig. 4C) suggest that even in the absence of the other four HSV proteins, the polymerase-accessory protein complex is able to functionally interact with Rep. Nevertheless there might be no direct interaction or contact between the Rep protein and the UL30 and UL42 products. Rep may simply alter the structure of the ITR in a manner that permits some synthesis by the HSV polymerase and its accessory protein.
The addition of ICP8 resulted in a significant stimulation of DNA synthesis and homogeneous products (Fig. 4C, lane 3). It seems likely that Rep68 creates primer termini by promoting strand separation of the ends of the DNA template and the formation of hairpin structures (42). In addition, Rep68 may also act as the DNA helicase, providing the DNA polymerase with access to single-stranded DNA. The UL29 protein, ICP8, would be expected to prevent reannealing of complementary single strands. It is also possible that high concentrations of ICP8 might promote passive unwinding of duplex DNA. It is somewhat surprising that this substrate with BglII ends can serve as a template. The BglII ends would seem to render the 3′ ends of a hairpin structure unpaired. The ability of the substrate to replicate might be due to the fact that the 3′ end of a BglII site contains only one base, an A.
Initiation of replication from a substrate containing internal AAV RBS and TRS.
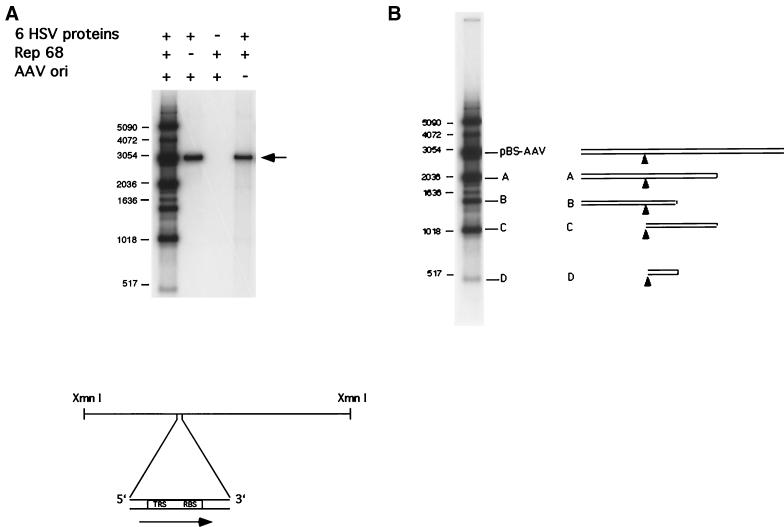
The experiments described above do not show that HSV-1-mediated replication can proceed from a single-stranded nick introduced by Rep68 at the TRS. This must happen during the terminal resolution stage of authentic DNA replication. To determine whether replication from the Rep-induced nick is possible, we employed a substrate, pBS-AAV, described previously (29). This substrate contains the AAV RBS and TRS inserted into the polylinker of the plasmid pBluescript KS(+). The plasmids pKS(+) and pBS-AAV were linearized at the XmnI site. The AAV RBS and TRS were thus located approximately 1,000 and 2,000 nucleotides from the ends of the template molecules (Fig. 5A). Rep68 greatly stimulated synthesis by the HSV replisome (Fig. 5A, lane 1). The increase in DNA synthesis of molecules greater than 500 nucleotides in length was approximately 35-fold in comparison to the assay in which Rep68 was omitted, as determined by PhosphorImager analysis. DNA synthesis was dependent on the AAV RBS and TRS sequences (Fig. 5A, lane 4). Our minimal system consisting of Rep68 and the HSV UL30, UL42, and UL29 proteins gave similar results (data not shown).
FIG. 5.
Replication assay on the minimal AAV origin. The substrate was a KS(+) plasmid with a copy of the AAV minimal origin (i.e., an RBS and an TRS) inserted into the polylinker, pBS-AAV. [As a control pKS(+) without an insert was used.] Plasmids were linearized at the XmnI site prior to use. (A) The arrow in the upper panel indicates the position of full-length product. Sizes are shown in base pairs at left. The lower panel is a diagram of the origin-containing substrate with the relative positions of the RBS and TRS illustrated. The arrow in the lower panel indicates the expected point of initiation and direction of replication. (B) Diagram illustrating the structures of various fold-back replication structures. Shown to the right are the structures of the substrate species, pBS-AAV, and fold-back or hairpinned bands designated A to D. Black triangles designate locations of RBS-TRS.
An analysis of the products by restriction enzyme digestion, in particular with DpnI and MboI, was consistent with initiation of synthesis in the region of the AAV origin and extension in the expected direction by single-strand displacement (data not shown). Products which migrate more rapidly than the starting substrate during gel electrophoresis appear to result from displacement of newly synthesized strands from the template at certain sequences followed by fold-back synthesis. The products of fold-back synthesis are illustrated in Fig. 5B. Products migrating more slowly than the starting substrate are branched structures, formed by stalling of the replication complex. These results suggest that the DNA replication in this assay is not fully processive and that substantial displacement of the replication complex is occurring at specific positions. Although there is a deficiency in processivity, the results from this substrate and the mAAV substrate show that the HSV-1 replisome is capable of cooperating with the AAV Rep protein to mediate the initiation of DNA synthesis from an AAV origin.
Effects of cellular extracts on Rep-dependent DNA synthesis by the HSV-1 replisome.
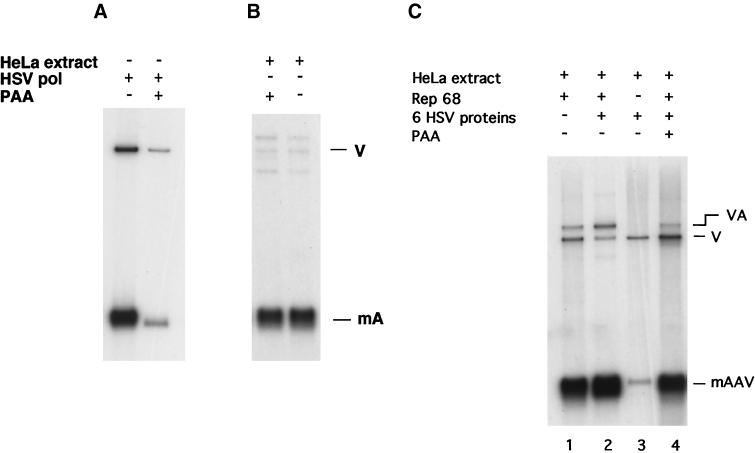
A question that arises is whether the HSV replication complex can initiate replication at the AAV origin in cells. We have made a first attempt at answering this question by employing a cell-free replication assay that has been used previously to study AAV DNA replication. In that assay a duplex AAV genome and the Rep68 protein are added to an extract made from rapidly growing uninfected HeLa cells. Using the cell extract assay, a limited replication of the full-length genome by Rep68 and cellular replication factors has been observed (33). Consequently, a problem with the use of the cell extract assay to measure the activity of the HSV complex is simultaneous replication of the AAV constructs by cellular replication proteins. This is especially the case with the minigenome, which replicates quite well in cellular extracts (31). To distinguish replication by the HSV polymerase from replication by the cellular polymerase, we have used PAA, which specifically inhibits the HSV polymerase (24). Figure 6A shows PAA inhibition of replication of the AAV minigenome by the six HSV proteins in the absence of extract. Figure 6B shows that in a cellular extract in the absence of the HSV replication complex, the addition of PAA has no effect on the replication of the AAV minigenome, demonstrating that PAA does not inhibit the cell extract polymerase that replicates AAV DNA.
FIG. 6.
The HSV replication complex functions in the context of a HeLa cell extract. (A) Replication of the AAV minigenome with Rep and the six HSV proteins in the presence and absence of PAA (20 μg/ml), an inhibitor of the HSV polymerase (pol), performed as described in Materials and Methods. (B) Replication of the AAV minigenome with Rep, HeLa extract, and the UL29 protein. Shown are assays with and without PAA. (C) Replication of the AAV minigenome in a HeLa cell extract with and without the added HSV replication complex. PAA was added to lane 4. The relative incorporations into the AAV minigenomes as determined by phosphorimager analysis are as follows: lane 1, 1.00; lane 2, 1.42; lane 3, not determined; lane 4, 1.01. The band designated VA is produced by a ligation activity present in the extract which religates some of the AAV minigenome and vector fragments. This species contains an AAV origin and therefore will show incorporation in the presence of Rep.
We modified the cell extract assay by adding the six HSV replication proteins just prior to the addition of Rep68. Figure 6C shows the results of this assay. With the addition of the HSV replication complex to the HeLa cell extract (lane 2), there is a slight increase in replication of the mAAV. Upon the addition of PAA (lane 4), replication is reduced to the level of that for extract unsupplemented with HSV proteins. These results suggest that with the AAV minigenome, the products of an assay with the HSV proteins and Rep68 are apparently the same in both the absence and the presence of extract from uninfected HeLa cells.
DISCUSSION
Here we have presented results demonstrating the initiation of DNA synthesis in the presence of AAV Rep68 and the products of the HSV-1 UL5, UL8, UL52, UL29, UL30, and UL42 genes that is dependent on the AAV origins of DNA replication. Efficient DNA replication was also seen with a minimal system consisting of Rep68 and the products of the UL30, UL42, and UL29 genes. Surprisingly, while we observed efficient initiation on the duplex AAV minigenome with Rep and the six HSV proteins, we were unable to achieve initiation on the duplex full-length AAV genome with the same seven proteins. In our assays Rep68 apparently creates primer termini either by helicase action at the ends of the double-stranded template molecules or by site-specific endonucleolytic cleavage. In addition, Rep68 most likely acts as the DNA helicase facilitating strand displacement. The HSV-1 DNA polymerase-UL42 complex is able to efficiently utilize primers created by Rep68 but needs the single-strand DNA binding protein (the UL29 gene product) to synthesize long stretches of DNA.
In the case of longer substrates, e.g., pBS-AAV, the products of DNA synthesis were heterogeneous. Molecules of full length were seen, but most products were of less than full length, formed apparently by displacement from the template and fold-back synthesis (Fig. 5B). The reason for this lack of full-length synthesis is presently unknown but may reflect a requirement for a different stoichiometry of replication factors. It has been shown previously that the activity of an HSV replication complex is very sensitive to the ratios of different factors and Mg2+ concentration (11, 12). An alternative possibility is a requirement for an as-yet-unknown cellular factor.
The results shown in Fig. 6 suggest that initiation mediated by the HSV polymerase occurs in the presence of cellular proteins. Therefore, unless processes specific to the intact cell prevent initiation (e.g., the sequestering of AAV DNA and HSV replication proteins in separate nuclear compartments), it is likely that this initiation occurs in the AAV- and HSV-coinfected cell.
Initiation of replication occurred both on a complete double-stranded AAV ITR and on a linear molecule containing a minimal origin, i.e., an RBS and a TRS, located 1,000 nucleotides from the closer end. The latter substrate serves to model the mechanism of initiation that must occur on the ITR during terminal resolution and also that which is predicted to occur on the integrated AAV genome upon its rescue from latency.
Previously, transfection experiments have been used to determine which HSV genes were required for a helper effect. Weindler and Heilbronn showed incontrovertibly that a helper effect could be supplied by a subset of the HSV replication genes, namely, the helicase-primase genes UL5, UL8, and UL52 and the gene for the single-strand DNA binding protein, UL29 (35). The absence of the UL30 and UL42 genes is somewhat surprising. Weindler and Heilbronn (35) suggested that cellular DNA polymerases were replicating AAV DNA in concert with the HSV single-strand DNA binding protein.
There are several previous examples of the HSV replication complex being affected by the AAV Rep protein, namely, Rep inhibition of HSV DNA replication and HSV-induced gene amplification (1, 15, 17). There are also examples of HSV replication proteins interacting with non-HSV replication factors. Blumel and Matz (3) and Heilbronn and zur Hausen (14) showed that in nonpermissive hamster cells infected with HSV, replication of simian virus 40 DNA became possible. Lee et al. (21) demonstrated an interaction between the HSV origin binding protein and the cellular DNA polymerase alpha-primase, leading those authors to suggest that cellular replication enzymes might be involved in HSV DNA replication. Taken together, these experiments suggest that the possibilities for cooperation between HSV and non-HSV replication machineries may be rather complex. By analogy, it may be that in the AAV- and HSV-coinfected cell, replication of AAV DNA might be by both cellular and HSV factors acting in concert.
In one model for a helper effect, herpesviruses might help to create subnuclear compartments, perhaps related to the viral prereplicative foci and replication compartments, to facilitate AAV replication. Interestingly, transfection of cells with a mixture of expression plasmids for the UL5, UL8, UL52, and UL9 genes has been shown to localize the single-strand DNA binding protein (ICP8) to punctate sites in the nucleus (23). It would be of great interest to learn whether replication of AAV DNA might be associated with such foci. We can only demonstrate that ICP8, the UL29 gene product, has a direct stimulatory effect on AAV DNA synthesis. However, there is a well-documented functional interaction between the UL29 and UL8 gene products (10, 11). It is therefore not unlikely that proper functioning of the UL29 protein during AAV replication in vivo might require assistance from the helicase-primase complex. The use of mutant genes encoding functionally impaired HSV-1 replication proteins that retain their ability to localize appropriately in the nucleus might help determine whether the enzymatic activities of the replication proteins are needed for the helper effect.
In their report, Weindler and Heilbronn noted that the absence of UL30 or UL42 led to somewhat (almost 10-fold) reduced levels of both DNA synthesis and new AAV particles compared to those after transfection of a complete set of HSV replication genes (35). Those authors used dividing tissue culture cells, and it may be that if the HSV polymerase and its accessory factor are present, they make a significant contribution to AAV DNA synthesis even in the presence of a functioning host cell replication apparatus.
However, the cells most commonly infected by HSV, epithelial cells and neurons, are often nondividing and therefore do not have an active replication machinery. (It has recently been suggested that human epidermal cells may also be the natural host cells for AAV [25].) Furthermore unlike adenovirus, HSV apparently shuts down host cell functions upon infection (28). Consequently, the capacity to use the HSV DNA replication machinery may be important for a successful AAV life cycle in vivo. In addition, as noted previously, the HSV life cycle and HSV's possible role as a helper for AAV provide a rationale for a latent phase for AAV in vivo (5). Upon infecting a neuronal cell, HSV commonly ceases replicating and enters a latent state. The accompanying AAV, no longer able to replicate its DNA, might also be induced to enter a latent state, in its case by site-specifically integrating its genome into chromosome 19 (18). Factors resulting from or inducing HSV release from latency might also activate a latent AAV.
The findings in this report suggest the possibility of in vitro AAV DNA synthesis for the purpose of gene therapy experiments. This would be of interest if replication of AAV genomes in vitro could be coupled to assembly of viral capsids and production of infectious virions. The production of less-than-full-length molecules during our in vitro DNA replication assays might, in fact, be in part due to the lack of direct coupling to the later stages of formation of virions. Considering the limited number of gene products involved in AAV DNA replication, it should not be an insurmountable biochemical task to establish a reconstituted system for production of infectious virions.
ACKNOWLEDGMENTS
We thank Nathalie Dutheil for helpful comments.
This work was supported in part by NIH grants DK55609 and DK57746 (to R.M.L.).
REFERENCES
- 1.Bantel-Schaal U, zur Hausen H. Adeno-associated viruses inhibit SV40 DNA amplification and replication of herpes simplex virus in SV40-transformed hamster cells. Virology. 1988;164:64–74. doi: 10.1016/0042-6822(88)90620-4. [DOI] [PubMed] [Google Scholar]
- 2.Berns K I. Parvoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2173–2197. [Google Scholar]
- 3.Blumel J, Matz B. Study on simian virus 40 DNA synthesis in herpes simplex virus-infected cells. Virology. 1996;217:407–412. doi: 10.1006/viro.1996.0132. [DOI] [PubMed] [Google Scholar]
- 4.Brister J R, Muzyczka N. Rep-mediated nicking of the adeno-associated virus origin requires two biochemical activities, DNA helicase activity and transesterification. J Virol. 1999;73:9325–9336. doi: 10.1128/jvi.73.11.9325-9336.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buller R M, Janik J E, Sebring E D, Rose J A. Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. J Virol. 1981;40:241–247. doi: 10.1128/jvi.40.1.241-247.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Challberg M D. A method for identifying the viral genes required for herpesvirus DNA replication. Proc Natl Acad Sci USA. 1986;83:9094–9098. doi: 10.1073/pnas.83.23.9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conway J, Rhys C, Zolotukhin I, Zolotukhin S, Muzyczka N, Hayward G, Byrne B. High-titer recombinant adeno-associated virus production utilizing a recombinant herpes simplex virus type I vector expressing AAV-2 Rep and Cap. Gene Ther. 1999;6:986–993. doi: 10.1038/sj.gt.3300937. [DOI] [PubMed] [Google Scholar]
- 8.Conway J E, Zolotukhin S, Muzyczka N, Hayward G S, Byrne B J. Recombinant adeno-associated virus type 2 replication and packaging is entirely supported by a herpes simplex virus type 1 amplicon expressing Rep and Cap. J Virol. 1997;71:8780–8789. doi: 10.1128/jvi.71.11.8780-8789.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crute J J, Lehman I R. Herpes simplex-1 DNA polymerase. Identification of an intrinsic 5′-3′ exonuclease with ribonuclease H activity. J Biol Chem. 1989;264:19266–19270. [PubMed] [Google Scholar]
- 10.Falkenberg M, Bushnell D A, Elias P, Lehman I R. The UL8 subunit of the heterotrimeric herpes simplex virus type 1 helicase-primase is required for the unwinding of single strand DNA-binding protein (ICP8)-coated DNA substrates. J Biol Chem. 1997;272:22766–22770. doi: 10.1074/jbc.272.36.22766. [DOI] [PubMed] [Google Scholar]
- 11.Falkenberg M, Lehman I R, Elias P. Leading and lagging strand DNA synthesis in vitro by a reconstituted herpes simplex virus type 1 replisome. Proc Natl Acad Sci USA. 2000;97:3896–3900. doi: 10.1073/pnas.97.8.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falkenberg M, Elias P, Lehman I R. The herpes simplex virus type 1 helicase-primase. Analysis of helicase activity. J Biol Chem. 1998;273:32154–32157. doi: 10.1074/jbc.273.48.32154. [DOI] [PubMed] [Google Scholar]
- 13.Handa H, Carter B J. Adeno-associated virus DNA replication complexes in herpes simplex virus or adenovirus-infected cells. J Biol Chem. 1979;254:6603–6610. [PubMed] [Google Scholar]
- 14.Heilbronn R, zur Hausen H. A subset of herpes simplex virus replication genes induces DNA amplification within the host cell genome. J Virol. 1989;63:3683–3692. doi: 10.1128/jvi.63.9.3683-3692.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heilbronn R, Burkle A, Stephan S, zur Hausen H. The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J Virol. 1990;64:3012–3018. doi: 10.1128/jvi.64.6.3012-3018.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Im D S, Muzyczka N. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 17.Kleinschmidt J A, Mohler M, Weindler F W, Heilbronn R. Sequence elements of the adeno-associated virus rep gene required for suppression of herpes-simplex-virus-induced DNA amplification. Virology. 1995;206:254–262. doi: 10.1016/s0042-6822(95)80040-9. [DOI] [PubMed] [Google Scholar]
- 18.Kotin R M, Siniscalco M, Samulski R J, Zhu X D, Hunter L, Laughlin C A, McLaughlin S, Muzyczka N, Rocchi M, Berns K I. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kyostio S R, Owens R A. Identification of mutant adeno-associated virus Rep proteins which are dominant-negative for DNA helicase activity. Biochem Biophys Res Commun. 1996;220:294–299. doi: 10.1006/bbrc.1996.0399. [DOI] [PubMed] [Google Scholar]
- 20.Laughlin C A, Tratschin J D, Coon H, Carter B J. Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene. 1983;23:65–73. doi: 10.1016/0378-1119(83)90217-2. [DOI] [PubMed] [Google Scholar]
- 21.Lee S S, Dong Q, Wang T S, Lehman I R. Interaction of herpes simplex virus 1 origin-binding protein with DNA polymerase alpha. Proc Natl Acad Sci USA. 1995;92:7882–7886. doi: 10.1073/pnas.92.17.7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehman I R, Boehmer P E. Replication of herpes simplex virus DNA. J Biol Chem. 1999;274:28059–28062. doi: 10.1074/jbc.274.40.28059. [DOI] [PubMed] [Google Scholar]
- 23.Liptak L M, Uprichard S L, Knipe D M. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J Virol. 1996;70:1759–1767. doi: 10.1128/jvi.70.3.1759-1767.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mao J C, Robishaw E E. Mode of inhibition of herpes simplex virus DNA polymerase by phosphonoacetate. Biochemistry. 1975;14:5475–5479. doi: 10.1021/bi00696a015. [DOI] [PubMed] [Google Scholar]
- 25.Meyers C, Mane M, Kokorina N, Alam S, Hermonat P L. Ubiquitous human adeno-associated virus type 2 autonomously replicates in differentiating keratinocytes of a normal skin model. Virology. 2000;272:338–346. doi: 10.1006/viro.2000.0385. [DOI] [PubMed] [Google Scholar]
- 26.Ni T H, McDonald W F, Zolotukhin I, Melendy T, Waga S, Stillman B, Muzyczka N. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J Virol. 1998;72:2777–2787. doi: 10.1128/jvi.72.4.2777-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ni T H, Zhou X, McCarty D M, Zolotukhin I, Muzyczka N. In vitro replication of adeno-associated virus DNA. J Virol. 1994;68:1128–1138. doi: 10.1128/jvi.68.2.1128-1138.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roizman B, Roane P R., Jr Multiplication of herpes simplex virus. II. The relation between protein synthesis and the duplication of viral DNA in infected HEp-2 cells. Virology. 1964;22:262–269. doi: 10.1016/0042-6822(64)90011-x. [DOI] [PubMed] [Google Scholar]
- 29.Smith D H, Ward P, Linden R M. Comparative characterization of Rep proteins from the helper-dependent adeno-associated virus type 2 and the autonomous goose parvovirus. J Virol. 1999;73:2930–2937. doi: 10.1128/jvi.73.4.2930-2937.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snyder R O, Samulski R J, Muzyczka N. In vitro resolution of covalently joined AAV chromosome ends. Cell. 1990;60:105–113. doi: 10.1016/0092-8674(90)90720-y. [DOI] [PubMed] [Google Scholar]
- 31.Ward P, Berns K I. In vitro replication of adeno-associated virus DNA: enhancement by extracts from adenovirus-infected HeLa cells. J Virol. 1996;70:4495–4501. doi: 10.1128/jvi.70.7.4495-4501.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ward P, Berns K I. In vitro rescue of an integrated hybrid adeno-associated virus/simian virus 40 genome. J Mol Biol. 1991;218:791–804. doi: 10.1016/0022-2836(91)90267-a. [DOI] [PubMed] [Google Scholar]
- 33.Ward P, Urcelay E, Kotin R, Safer B, Berns K I. Adeno-associated virus DNA replication in vitro: activation by a maltose binding protein/Rep 68 fusion protein. J Virol. 1994;68:6029–6037. doi: 10.1128/jvi.68.9.6029-6037.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward P, Dean F B, O'Donnell M E, Berns K I. Role of the adenovirus DNA-binding protein in in vitro adeno-associated virus DNA replication. J Virol. 1998;72:420–427. doi: 10.1128/jvi.72.1.420-427.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weindler F W, Heilbronn R. A subset of herpes simplex virus replication genes provides helper functions for productive adeno-associated virus replication. J Virol. 1991;65:2476–2483. doi: 10.1128/jvi.65.5.2476-2483.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weitzman M D, Fisher K J, Wilson J M. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J Virol. 1996;70:1845–1854. doi: 10.1128/jvi.70.3.1845-1854.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wobbe C R, Dean F, Weissbach L, Hurwitz J. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc Natl Acad Sci USA. 1985;82:5710–5714. doi: 10.1073/pnas.82.17.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu C A, Nelson N J, McGeoch D J, Challberg M D. Identification of herpes simplex virus type 1 genes required for origin-dependent DNA synthesis. J Virol. 1988;62:435–443. doi: 10.1128/jvi.62.2.435-443.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yakinoglu A O, Heilbronn R, Burkle A, Schlehofer J R, zur Hausen H. DNA amplification of adeno-associated virus as a response to cellular genotoxic stress. Cancer Res. 1988;48:3123–3129. [PubMed] [Google Scholar]
- 40.Yakobson B, Hrynko T A, Peak M J, Winocour E. Replication of adeno-associated virus in cells irradiated with UV light at 254 nm. J Virol. 1989;63:1023–1030. doi: 10.1128/jvi.63.3.1023-1030.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yakobson B, Koch T, Winocour E. Replication of adeno-associated virus in synchronized cells without the addition of a helper virus. J Virol. 1987;61:972–981. doi: 10.1128/jvi.61.4.972-981.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou X, Zolotukhin I, Im D S, Muzyczka N. Biochemical characterization of adeno-associated virus rep68 DNA helicase and ATPase activities. J Virol. 1999;73:1580–1590. doi: 10.1128/jvi.73.2.1580-1590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]