Abstract
The tegument is an integral and essential structural component of the herpes simplex virus type 1 (HSV-1) virion. The UL37 open reading frame of HSV-1 encodes a 120-kDa virion polypeptide which is a resident of the tegument. To analyze the function of the UL37-encoded polypeptide a null mutation was generated in the gene encoding this protein. In order to propagate this mutant virus, transformed cell lines that express the UL37 gene product in trans were produced. The null mutation was transferred into the virus genome using these complementing cell lines. A mutant virus designated KΔUL37 was isolated based on its ability to form plaques on the complementing cell line but not on nonpermissive (noncomplementing) Vero cells. This virus was unable to grow in Vero cells; therefore, UL37 encodes an essential function of the virus. The mutant virus KΔUL37 produced capsids containing DNA as judged by sedimentation analysis of extracts derived from infected Vero cells. Therefore, the UL37 gene product is not required for DNA cleavage or packaging. The UL37 mutant capsids were tagged with the smallest capsid protein, VP26, fused to green fluorescent protein. This fusion protein decorates the capsid shell and consequently the location of the capsid and the virus particle can be visualized in living cells. Late in infection, KΔUL37 capsids were observed to accumulate at the periphery of the nucleus as judged by the concentration of fluorescence around this organelle. Fluorescence was also observed in the cytoplasm in large puncta. Fluorescence at the plasma membrane, which indicated maturation and egress of virions, was observed in wild-type-infected cells but was absent in KΔUL37-infected cells. Ultrastructural analysis of thin sections of infected cells revealed clusters of DNA-containing capsids in the proximity of the inner nuclear membrane. Occasionally enveloped capsids were observed between the inner and outer nuclear membranes. Clusters of unenveloped capsids were also observed in the cytoplasm of KΔUL37-infected cells. Enveloped virions, which were observed in the cytoplasm of wild-type-infected cells, were never detected in the cytoplasm of KΔUL37-infected cells. Crude cell fractionation of infected cells using detergent lysis demonstrated that two-thirds of the UL37 mutant particles were associated with the nuclear fraction, unlike wild-type particles, which were predominantly in the cytoplasmic fraction. These data suggest that in the absence of UL37, the exit of capsids from the nucleus is slowed. UL37 mutant particles can participate in the initial envelopment at the nuclear membrane, although this process may be impaired in the absence of UL37. Furthermore, the naked capsids deposited in the cytoplasm are unable to progress further in the morphogenesis pathway, which suggests that UL37 is also required for egress and reenvelopment. Therefore, the UL37 gene product plays a key role in the early stages of the maturation pathway that give rise to an infectious virion.
The tegument layer of the herpes simplex virus type 1 (HSV-1) virion is the structure between the DNA-containing capsid and the envelope (34). It is one of the most complex and diverse structures of the virion both in terms of protein composition and the functions encoded by the constituents of this structure. A number of virus-specified polypeptides comprise this structure, including those that function to activate transcription, shut off host protein synthesis, uncoat the virus genome, and phosphorylate virus proteins and others whose functions are still poorly defined (reviewed in references 35 and 44). The tegument displays a duality of functions in virus replication due to the role that the tegument proteins play both at early and late times in infection. The virion proteins incorporated into the tegument structure effectively jump-start the replication cycle. Examples of these proteins include the potent transcriptional activator VP16 (5, 6, 31) and the virion host shutoff (vhs) polypeptide that shuts off host protein synthesis (20, 32). Tegument proteins also function late in infection. This is exemplified by VP16, which is required for virus egress subsequent to exit of these particles from the nucleus (1, 27, 46). It has become increasingly evident that the tegument proteins play a key role in virion morphogenesis.
Tegument proteins occupy approximately one-third of the volume of the virion. A majority of the virion proteins are residents of this structure. Major components of the tegument include VP11/12, VP13/14, VP16, and VP22 (44). VP16 transactivates the immediate-early genes (5, 6, 31), and VP11/12 and VP13/14 function by modulating VP16 activity (23). Although the function of VP22 is unclear, it has the unusual property of cell-to-cell spread in transfected cells (14). Less-abundant components of the tegument include the vhs polypeptide (UL41), the large tegument protein VP1/2 (UL36), and the products of genes UL37, UL17, UL13, UL11, US11, US10, US9, and US3 (reviewed in references 35 and 44). The UL36 gene encodes the largest HSV-1 polypeptide, the tegument protein VP1/2 (26), which is required for uncoating of the viral genome (4, 19). UL36 also encodes a late function during infection, one that is required for virus maturation (13). A null mutant in the UL11 gene product results in a reduction in virus yield but does not abrogate infectivity (3). The UL13 and US3 open reading frames (ORFs) specify protein kinase (7, 16) activity and the US11 gene has been shown to bind RNA and to associate with ribosomes (36). The UL17 gene product is required for cleavage and packaging of viral DNA (38). This catalogue of proteins and some of their identified functions exemplify the enormous variety in the activities of the tegument structure.
The UL37 ORF produces a 120-kDa polypeptide that is expressed late in the infectious cycle (40). This protein is phosphorylated like many of the tegument proteins but the kinase involved may be cellular in origin (2). It is present in virions, specifically in the tegument (25, 39). It was shown that the UL37 polypeptide is retained on single-stranded DNA agarose columns in the presence of ICP8, the major DNA binding protein, suggesting that it may be part of a higher-order complex that associates with DNA (40, 41). The UL37 polypeptide is distributed throughout the infected cell but is predominantly localized to the cytoplasm (24, 25, 39). Recent studies have identified a nuclear export signal which may be responsible for this distribution (45). There is a certain degree of flexibility with regards to the composition of the tegument as has been shown for VP22 (21) and the gene products encoded by UL47 and UL48 (47). However, the copy number of the UL37 polypeptide, which is low in the virion, is invariant (24) and may reflect a difference between this tegument protein and the other abundant proteins in this structure. When this study was initiated, the function of UL37 was unknown. Since it appeared to be associated with a DNA binding protein and because it was a structural protein, it was thought that it may play a role in the association or disassociation of DNA with the capsid structure.
The aim of the research presented in this paper was to analyze the function of the UL37-encoded tegument polypeptide in the virus replication cycle. A null mutation was generated in the gene encoding this polypeptide. Since this gene product was thought to specify an essential function, transformed cell lines were derived that expressed the UL37 gene in trans. These cell lines permitted the isolation of a null mutant in the UL37 gene. The null mutation abrogates virus maturation and the data shown below give new insight into the role of UL37 in the formation of an infectious particle.
MATERIALS AND METHODS
Cells and viruses.
Vero cells and transformed Vero cell lines were grown in minimum essential medium-alpha medium supplemented with 10% fetal calf serum (Life Technologies) and passaged as described by Desai et al. (11). Virus stocks of strain KOS (HSV-1) and the mutant viruses were prepared as previously described (11). Initially 3B13 was used as the permissive cell line for the propagation of the UL37 null mutant, and subsequently BD45 was used and gave yields of approximately 1,000 PFU/cell.
Antibodies.
Rabbit antiserum 780 was a generous gift from Frank Jenkins. This serum was raised against a malE fusion protein expressing the carboxyl one-third of UL37 (2).
Plasmids.
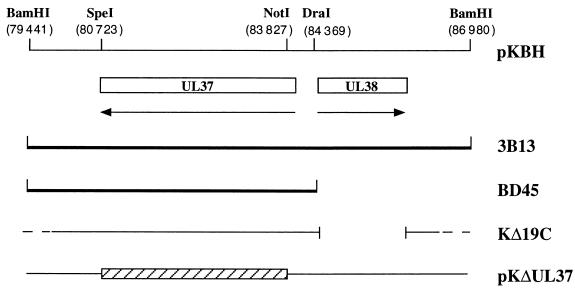
pKBH contains the 7.5-kb BamHI H fragment of the KOS genome (genome nucleotides 79441 to 86980) cloned into pUC19 and contains the ORFs of UL37 and UL38 (Fig. 1) (22). This plasmid was digested with NotI and SpeI, resulting in a deletion of 3.1 kb. The ends were filled in with Klenow and a 14-mer SpeI linker containing translation stop codons in all three reading frames was ligated into the site of the deletion. This plasmid was designated pKΔUL37. pKBD contains the 4.9-kb BamHI-DraI fragment derived from pKBH (Fig. 1) and encodes just the UL37 ORF. Plasmids pKNotI, which encodes the UL26 ORF (10), and pKEL, which contains the EcoRI L fragment of HSV-1 (11), have previously been described.
FIG. 1.
Schematic representation of the BamHI H region of the KOS genome. The BamHI H region of KOS (7.5 kb) contains genes UL37 and UL38 (22), with the latter being the gene for the capsid protein VP19C. The UL37 and UL38 ORFs specify 1,123 and 465 amino acid residues, respectively. Two cell lines expressing UL37 in trans were constructed. 3B13 and BD45 were transformed with the BamHI H and BamHI-to-DraI fragments, respectively. The virus designated KΔ19C contains a deletion in the UL38 gene which eliminates residues 1 to 415 of VP19C (29). Plasmid pKΔUL37 was constructed by deletion of sequences between the NotI restriction enzyme site at the 5′ end and the SpeI site at the 3′ end of UL37. This resulted in the deletion of residues 86 to 1120. The relevant restriction enzyme sites and the genome nucleotide numbers in parentheses are shown at the top of the figure.
Construction of transformed Vero cell lines.
The procedure of DeLuca et al. (8) was followed for transformation of Vero cells and was also described by Desai et al. (9). Subconfluent monolayers of Vero cells were cotransfected with pSV2-neo (1.0 μg) (43) and a three- or fivefold molar excess of the plasmid encoding the UL37 gene. These same cells were also transformed with pKEL (11) and pKNotI (10). G418-resistant colonies were tested for the ability to support the replication of KUL26ΔZ, a null mutant of the UL26 ORF (10). One such cell line, designated 3B13, was chosen for the isolation of the UL37 null mutant. Subsequently, after the isolation of the UL37 null mutant, cells were similarly transformed with a plasmid (pKBD) encoding the BamHI-to-DraI fragment derived from pKBH (Fig. 1) which encodes just the UL37 ORF. Out of 63 G418-resistant cell lines derived, 8 complemented the growth of the UL37 null mutant. One cell line designated BD45 was used thereafter for propagation of KΔUL37.
Marker transfer and marker rescue.
Marker transfer of the null mutation was essentially carried out as described by Person and Desai (29). Subconfluent monolayers of cells (0.75 × 106) in 60-mm-diameter dishes were cotransfected with 2 μg of linearized plasmid and 5 μg of infected cell DNA. When foci were observed (48 h after transfection), the cell monolayers were harvested, freeze-thawed once, and sonicated, and the total virus progeny was titered. Single plaque isolates were tested for their ability to plate on the noncomplementing Vero cells and the complementing cell lines. Isolates that exhibited the mutant phenotype were plaque purified three times prior to further characterization. A KΔUL37 rescued virus was isolated following cotransfection of BD45 cells with KΔUL37 virus DNA and linearized pKBD. The transfection progeny was screened for the ability to plate on the noncomplementing Vero cell line. One virus, designated KΔUL37R, that displayed this phenotype was purified and characterized.
Southern blot hybridization.
Southern blot analysis was performed as described by Desai et al. (9).
Western blot analysis.
Unlabeled infected cell extracts were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to Immobilon-P membranes (Millipore) in Tris-glycine buffer using a Bio-Rad mini-transblot apparatus. Transfer buffer and procedures were used according to the manufacturer's protocol. The membrane was incubated in 5% bovine serum albumin (BSA) in TN buffer (10 mM Tris [pH 7.4], 150 mM NaCl) overnight at 4°C to block nonspecific reactivity. Filters were then incubated with an appropriate dilution of antibody in 5% BSA–TN for 90 min at room temperature. Following antibody reactivity, the membranes were washed three times for 7 min each at room temperature in TN followed by two washes in TN containing 0.5% NP-40 (10 min each) and finally three washes in TN buffer (7 min each). Antigen detection was performed by incubation with 125I-labeled protein A (NEN-DuPont) diluted 1:1,000 in 5% BSA–TN for 2 h at room temperature. The filters were washed as described above and dried prior to autoradiography.
Sedimentation analysis of capsids.
Sedimentation analysis of capsids from infected cells was performed as described by Desai et al. (9) and Person and Desai (29). All gradients were made using a BioComp Gradient Mate (BioComp). Generally, lysates for sedimentation were prepared from cells (107) in 100-mm-diameter dishes. Infected cells were harvested by scraping into phosphate-buffered saline (PBS), pelleted, washed once in PBS, and repelleted. The cell pellet was resuspended in 2× capsid lysis buffer (CLB; 2% Triton X-100, 2 M NaCl, 10 mM Tris [pH 7.5], and 2 mM EDTA) and the lysate was sonicated prior to sedimentation. This procedure was used to derive the total capsid population from infected cells. Crude fractionation of infected cells was performed by lysis of cells in Triton lysis buffer (2% Triton X-100, 300 mM NaCl, 0.5% deoxycholic acid in PBS). The nuclei were pelleted, lysed in 2× CLB, and sonicated, and the nuclear lysate and the supernatant representing the cytoplasmic fraction were sedimented.
Electron microscopy.
Vero cells (107) in 100-mm-diameter dishes were infected at a multiplicity of infection (MOI) of 10 PFU/cell. The samples were processed for transmission electron microscopy (TEM) as described by Hendricks et al. (18). The cells were fixed for 1 h at room temperature in a solution containing 2.5% glutaraldehyde in 100 mM cacodylate. The cells were then lifted off the dish, pelleted, and subsequently osmicated in Palade's OsO4 (1 h at 4°C). The pellet was then washed three times in 100 mM cacodylate, pH 7.4, treated with 1% tannic acid for 30 min at room temperature, washed three times in double-distilled water, and incubated overnight in Kellenberger's uranyl acetate (18). The pellet was then dehydrated through a graded series of ethanol and embedded in EMBED-812. Sections were cut on a Leica Ultracut UCT ultramicrotome, collected onto 400-mesh nickel grids, poststained in uranyl acetate and lead citrate, and observed in a Philips EM410 TEM.
Confocal and deconvolving microscopy.
Confluent monolayers of cells in eight-well LabTek chamber slides (2.5 × 105 cells per tray) were infected at an MOI of 10 PFU/cell. At various times postinfection the cells were rinsed twice in PBS and overlaid with PBS for microscopy. Confocal analysis was carried out using the Noran “Oz” confocal microscope as described by Desai (13). Deconvolution microscopy was carried out using an Applied Precision Deltavision Imaging System. Stacks were collected and deconvolved, selected images were rendered in three dimensions (3-D) utilizing Deltavision software, and the figure was prepared in Adobe Photoshop.
Data preparation.
For figure preparation, autoradiographs were scanned at 300 dots per inch into Adobe Photoshop. Electron microscope negatives were scanned in the same manner. Confocal images were saved as 8-bit TIFF files and imported into Photoshop for figure presentation.
RESULTS
Isolation of transformed cell lines that express the UL37 ORF.
At the onset of this study it was assumed that the UL37 gene encodes a protein with an essential function. This assumption was based on the inability to isolate a UL37 mutant virus on Vero cells. Therefore, complementing cell lines were needed that would support the replication of viruses specifying mutations in the gene encoding UL37. Since we did not have a mutant virus in the UL37 locus to select for complementing cells, we used an indirect approach to obtain helper cell lines. Previously we had simultaneously transfected cells with plasmids expressing different capsid genes and obtained cells that express all the transfected genes (9, 10, 29). The goal was to select for cells expressing the UL26 gene products and to use these cells to isolate a UL37 null mutant with the hope that these cells would also express UL37. Vero cells were simultaneously cotransfected with pSV2neo (43) and plasmids pKBH, pKNotI (10), and pKEL (11). Plasmid pKBH contains genes UL37 and UL38 (Fig. 1), pKNotI contains the UL26 ORF, and pKEL contains the UL35 ORF (22). A plasmid carrying UL35 was also included because at that time we did not know it was unessential for replication of the virus in cell culture (11). Colonies that were resistant to the drug G418 were harvested and tested for their ability to complement KΔUL26Z, a null mutant virus of UL26 (10). Forty-seven G418-resistant stable transformants were isolated, of which five were able to support the replication of KΔUL26Z. Of these, 3B13 (Fig. 1) gave the highest plating efficiency for KΔUL26Z and was used in the subsequent experiments. Although this indirect approach was not ideal, the goal was to use these cell lines to isolate a UL37 null mutant virus. This mutant could then be used to select for complementing cell lines that were transformed with only the UL37 gene.
Construction and isolation of a null mutant in the UL37 gene.
The construction of a null mutation in the UL37 gene and transfer of the mutation to the KOS genome were carried out using the 3B13 transformed cell line as host. The BamHI H fragment of HSV-1 strain KOS (pKBH) was used for this construction (Fig. 1). Most of the UL37 ORF was deleted by digestion of pKBH with NotI and SpeI followed by religation. A SpeI linker containing translation termination signals in all three reading frames was inserted at the site of this deletion. In addition to polypeptide termination after amino acid residue 86, residues 86 to 1120 were deleted by this procedure, and this plasmid was designated pKΔUL37. A virus designated KΔ19C (Fig. 1) was isolated in our laboratory and specifies a null mutation in the UL38 ORF (29). UL38 encodes the essential capsid protein VP19C (29, 32). This virus was propagated on cell line C32, a Vero cell line transformed with the gene that encodes UL38 but not UL37 (29). Although 3B13 cells were transformed with the BamHI H DNA fragment, which encodes UL37 and UL38, they did not support the growth of the VP19C null mutant. Preliminary experiments indicated that these cells might express UL37.
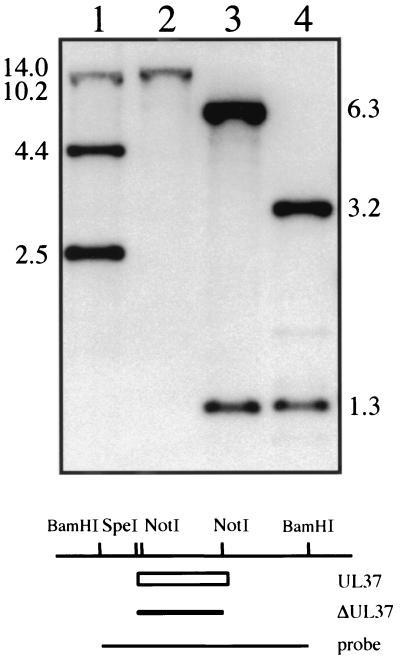
In order to recombine the UL37 null mutation into the virus genome, a marker rescue, marker transfer experiment was performed. The goal was to use plasmid pKΔUL37 in cotransfection assays with KΔ19C viral DNA to rescue the deletion in UL38 (VP19C) and at the same time to transfer the UL37 mutation into the virus (Fig. 1). Such viruses would be identified by plating on 3B13 cells, which support the growth of wild-type viruses or those containing a null mutation in UL37, but not in KΔ19C. Cells were cotransfected with linearized pKΔUL37 and viral DNA derived from KΔ19C. The viruses that could arise from this transfection are the original parent, KΔ19C; wild-type virus by rescue of the UL38 deletion but not transfer of the UL37 mutation; a double mutant recombinant of both ΔUL37 and ΔUL38; and the desired UL37 mutant. Only the wild-type virus and the UL37 null mutant could replicate on 3B13 cells. Progeny virus from the transfection was harvested and assayed for the ability to form plaques on 3B13 cells. Those plaques that arose on this cell line were then tested for the ability to plate on Vero and C32 cells. Approximately 50% of the isolates grew only on 3B13 monolayers and not on Vero or C32 cells. Therefore, in these viruses the deletion in KΔ19C was rescued and at the same time the mutation in UL37 was transferred into virus. One such virus was purified further and was designated KΔUL37. Since this virus was unable to replicate on Vero cells, UL37 specifies an essential function. To confirm the introduction of the plasmid-specified mutation into the virus, the genome of KΔUL37 was examined by Southern blot hybridization. Small batches of infected cell DNA were prepared and digested with restriction enzymes and the resulting fragments were analyzed by blot hybridization (Fig. 2). The probe (BamHI H) hybridized to a 2.5-kb NotI fragment within the UL37 gene and to 10.2- and 4.4-kb fragments 5′ and 3′, respectively, of this NotI fragment (lane 1). In KΔUL37 DNA, hybridization was only observed to a 14-kb fragment (lane 2) due to the deletion of the two NotI sites in BamHI H (Fig. 1). For KOS DNA digested with BamHI and SpeI (lane 3), the probe hybridized to 6.3- and 1.3-kb fragments. In KΔUL37 DNA (lane 4) the 6.3-kb fragment was reduced in size to 3.2 kb due to the deletion in UL37 followed by the addition of a SpeI linker. Therefore, KΔUL37 contains the specified deletion in the UL37 ORF.
FIG. 2.
Southern blot analysis of the KΔUL37 genome. Two micrograms of KOS (lanes 1 and 3) and KΔUL37 (lanes 2 and 4) viral DNA was digested with NotI (lanes 1 and 2) or BamHI and SpeI (lanes 3 and 4) and resolved by agarose gel electrophoresis prior to analysis by Southern blot hybridization. Filters were probed with a 32P-labeled DNA probe corresponding to the BamHI H fragment. The size of hybridized fragments in kilobases is indicated at the sides of the gel. The schematic at the bottom of the figure shows the BamHI H region of HSV-1. The open box depicts the UL37 ORF, the filled box depicts the UL37 deletion, and the line at the bottom depicts the probe used for hybridization. Relevant restriction enzyme sites are shown at the top.
Isolation of a complementing cell line that expresses only the UL37 gene.
The 3B13 cell line was transformed with multiple ORFs and was useful for the initial isolation of the UL37 null mutant; however, the presence of multiple genes in this cell line may obscure mutations that are not in the UL37 gene. Therefore, cell lines were generated that express only the UL37 ORF. Vero cells were transformed with a plasmid that contains the BamHI-DraI fragment derived from pKBH (Fig. 1). This fragment encodes just the UL37 ORF (22). Transformed colonies were isolated and screened for their ability to support the replication of the UL37 null mutant. Out of 63 G418-resistant cell lines isolated, 8 were able to support the replication of KΔUL37. One cell line designated BD45 was chosen for further analysis. Single-step growth experiments of the UL37 null mutant were performed in Vero, 3B13, and BD45 cells. The mean burst size calculated from a number of experiments in BD45 cells was 1,400 PFU/cell, whereas the burst size in 3B13 cells was consistently 10-fold less. The burst size of KOS in Vero cells was 1,000 PFU/cell. The ability to propagate the UL37 mutant in a cell line that expressed only UL37 provided strong genetic evidence that the mutation resides in the UL37 locus. A marker-rescued virus was constructed to further ensure that the UL37 mutant phenotype was due solely to the absence of the mutated gene. This virus was isolated following cotransfection of BD45 cells with KΔUL37 viral DNA and a plasmid encoding the wild-type copy of the UL37 gene (pKBD). The transfection progeny was tested for the ability to replicate on noncomplementing cells and plaques were detected on these cells indicative of successful rescue of the UL37 mutation. One isolate was further purified and designated KΔUL37R. This virus gave a mean burst size of 1,000 PFU/cell when grown in Vero cells compared to wild-type virus (KOS), which gave a burst size of 1,110 PFU/cell. This revertant virus behaved similarly to wild-type virus as judged by its replication in Vero cells. Therefore, the UL37 mutation was rescued by DNA sequences encoding the UL37 gene.
The UL37 polypeptide was not detected in Vero cells infected with KΔUL37.
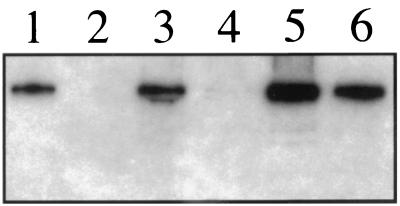
Analysis of [35S]methionine-labeled infected cell lysates using SDS-PAGE showed that the UL37 mutant accumulates the full spectrum of infected cell polypeptides upon infection of Vero cells (data not shown). In order to confirm the absence of the UL37 polypeptide in Vero cells infected with KΔUL37, Western blot assays were carried out on infected cell extracts using antisera raised to the carboxy one-third of the protein (2) (Fig. 3). Radioactivity corresponding to the polypeptide encoded by the UL37 gene product was observed in lysates prepared from KOS-infected Vero cells (lane 1). This polypeptide was not detected in extracts derived from KΔUL37-infected Vero cells (lane 2). The UL37 polypeptide was observed in extracts derived from BD45 cells infected with KΔUL37 (lane 6). However, the UL37 protein was barely detectable using this assay in KΔUL37-infected 3B13 cells (lane 4). The reduced accumulation of the UL37 polypeptide in 3B13 cells most likely accounts for the poor yield of KΔUL37 obtained in these cells.
FIG. 3.
Western blot analysis of KΔUL37-infected cells. Vero (lanes 1 and 2), 3B13 (lanes 3 and 4), and BD45 (lanes 5 and 6) cells were infected at an MOI of 10 PFU/cell with KOS (lanes 1, 3, and 5) or KΔUL37 (lanes 2, 4, and 6). Infected cells were harvested 24 h after infection and protein extracts were prepared. Total infected cell polypeptides were resolved by SDS-PAGE (12% acrylamide) and transferred onto a membrane. The proteins on the filter were incubated with UL37 antisera (708; see reference 2) and hybridization was monitored by 125I-labeled protein A.
The replication properties of KΔUL37.
The virus isolated above and designated KΔUL37 was unable to form plaques on noncomplementing Vero cells. This indicated that UL37 specifies an essential function. To quantitate this growth defect the plating efficiency of KΔUL37 and also its growth properties were examined. The plaquing efficiency of wild-type virus KOS, KΔUL37, and the revertant virus KΔUL37R was examined on Vero and BD45 cells (Table 1). The plating efficiency of KOS and KΔUL37R was similar on both Vero and BD45 cells. The stock of KΔUL37 used in this assay gave a titer of 1010 PFU/ml on BD45 cells. When the mutant was plated on Vero cells at a concentration of 1.0 PFU/cell (106 PFU), there was complete lysis of the cells. Dilution to 0.1 PFU/cell did not reveal any plaques on Vero cells; therefore, the titer on Vero cells could not be determined but was judged to be less than 105.
TABLE 1.
Plating efficiency of wild-type HSV-1 strain KOS, KΔUL37, and KΔUL37R on Vero and BD45 cells
| Virus | Plating efficiency (PFU/ml) on:
|
|
|---|---|---|
| Vero cells | BD45 cells | |
| KOS | 1.1 × 1010 | 8.9 × 109 |
| KΔUL37 | <105 | 1.33 × 1010 |
| KΔUL37R | 5.4 × 109 | 4.6 × 109 |
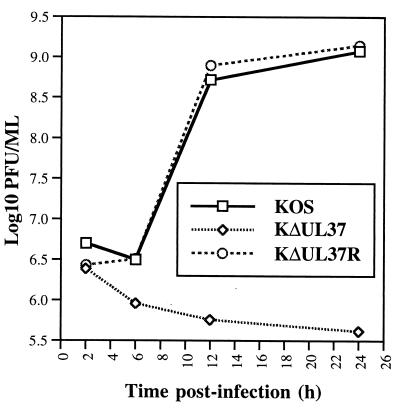
Single-step growth assays were performed to determine the growth properties of the UL37 mutant in Vero cells. Replicate Vero cell monolayers were infected with KOS, KΔUL37, and KΔUL37R at an MOI of 10 PFU/cell and the virus yield at different times postinfection was determined (Fig. 4). Both KOS and KΔUL37R replicate to appreciable levels in Vero cells. At the end of a 24-h growth cycle both viruses gave a burst size of approximately 1,000 PFU/cell. As expected, the UL37 mutant did not grow in Vero cells. Infection of the complementing cells (BD45) with KΔUL37 gave a yield of 1,400 PFU/cell following a single cycle of growth. Thus, UL37 is essential for virus replication.
FIG. 4.
Single-step growth of KΔUL37. Vero cells in 35-mm-diameter dishes (106 cells) were infected with KOS, KΔUL37, and KΔUL37R at an MOI of 10 PFU/cell. At various times postinfection the cultures were harvested and the virus yield was determined by titration on BD45 cells.
Capsid formation in KΔUL37-infected cells.
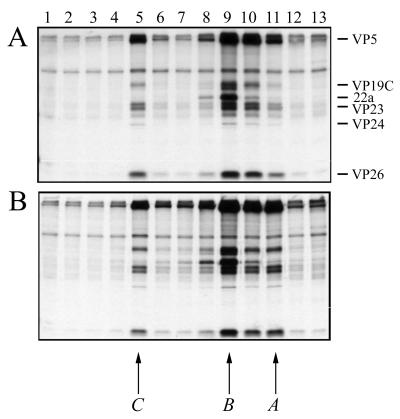
Previously, Southern blot analysis of KΔUL37-infected cell DNA revealed the presence of genome termini indicative of cleavage of viral DNA into genome-length molecules (data not shown). The detection of cleaved DNA suggested that DNA-filled capsids may be present in KΔUL37-infected cells. To confirm this and to study the assembly process, capsids were analyzed by sedimentation of radiolabeled nuclear extracts through sucrose gradients. The result of this experiment is shown in Fig. 5. Three peaks of radioactivity were detected for KOS (panel A) nuclear extracts and they correspond to DNA-filled C capsids (fraction 5), scaffold-filled B capsids (fraction 9), and empty A capsids (fraction 11). A, B, and C capsids all contain the shell proteins (VP5, VP19C, VP23, and VP26) and the protease VP24. The scaffold protein 22a was detected only in B capsids, as expected. Capsids were detected in KΔUL37 (panel B) extracts and were of similar composition and sedimentation profile to wild-type capsids. Therefore, capsid assembly and viral DNA packaging into capsids were detected in Vero cells infected with the UL37 null mutant.
FIG. 5.
Capsid formation in KΔUL37-infected cells. Vero cell monolayers (107 cells in 100-mm-diameter dishes) were infected with KOS (A) and KΔUL37 (B) at an MOI of 10 PFU/cell and labeled with [35S]methionine from 8 to 24 h postinfection. Nuclear extracts were prepared and layered onto 20 to 50% sucrose gradients. Fractions collected after sedimentation were analyzed by SDS-PAGE (17% acrylamide). Direction of sedimentation was from right to left. The positions of capsid proteins are indicated on the right of the figure for KOS. The positions at which A, B, and C capsids sediment are indicated below the figure.
Observation of the cellular localization of KΔUL37 capsids using a GFP tag.
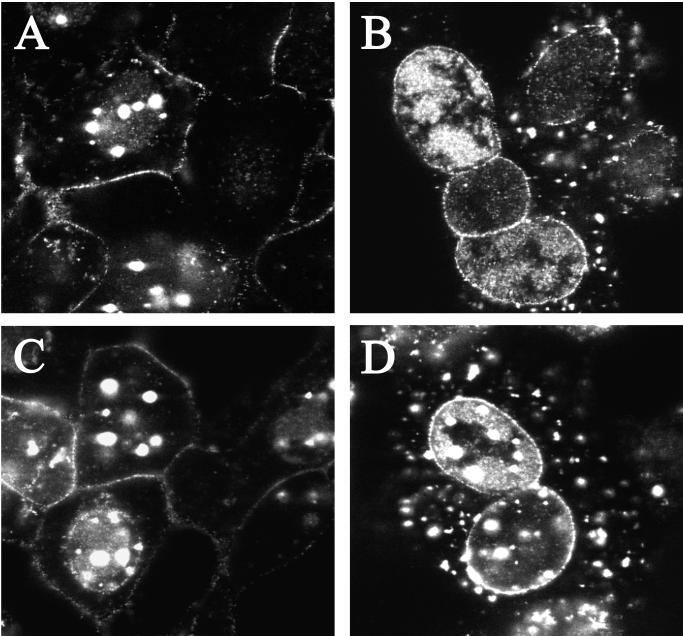
In order to visualize the fate of the UL37 mutant particles in living infected cells a green fluorescent protein (GFP) tag was used. The tag used in this case was a fusion between the smallest capsid protein of HSV-1, VP26, and GFP. K26GFP is a virus that expresses this VP26-GFP fusion protein (12). This virus replicates with the same growth properties as wild-type virus. The fusion protein decorates the capsid surface, and consequently the virus capsid and the mature virion are tagged with a fluorescent tag that can be visualized in living cells using light microscopy. The VP26-GFP marker was crossed into the genome of the UL37 mutant by coinfecting cells (BD45) with K26GFP and the null mutant virus. Individual plaques were scored with the fluorescence microscope and for a host-range phenotype. Viruses were isolated that gave rise to fluorescent plaques on the complementing cell line but not on Vero cells. The UL37 null mutant virus containing the VP26-GFP marker was designated KΔUL37-GFP. Capsids isolated from cells infected with this virus contained the VP26-GFP protein (data not shown), and therefore the absence of UL37 did not alter the ability of the GFP fusion to bind to capsids. Cells were infected with the GFP-tagged viruses and imaged using the confocal microscope (Fig. 6). During the course of the wild-type infection the fluorescence visualized followed the typical course predictive of the virus replication cycle. Thus, early in infection fluorescence was predominantly nuclear, indicative of capsids being assembled (data not shown). This was also seen in the UL37 null mutant-infected cells. As time progressed fluorescence was detected at the plasma membrane and began to accumulate here, indicative of wild-type virus that had matured and translocated to the cell surface (panels A and C).
FIG. 6.
Analysis in living cells of the replication of GFP-tagged KΔUL37. Cells were infected with K26GFP (A and C) and KΔUL37-GFP (B and D) at an MOI of 10 PFU/cell. Live cells were visualized in a confocal microscope at 16 (A and B) and 18 (C and D) h after infection. Magnification, ×100.
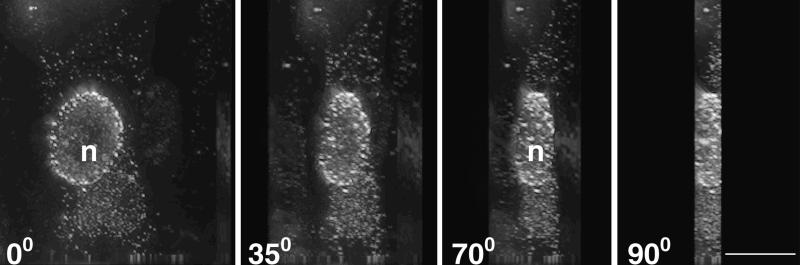
In the UL37 null mutant-infected cells (Fig. 6B and D), the most striking phenotype clearly observed in the majority of the cells was the punctate fluorescence around the nuclear periphery. Accumulation at the nuclear periphery was also seen occasionally in wild-type-infected cells (see cell in panel C). To analyze this phenotype further, deconvolving microscopy was used on cells similarly infected with this mutant. Optical sections (0.2 μm thick) were collected of infected cells. These stacks were deconvolved and 3-D rendered utilizing the Deltavision 3-D restoration software (API, Seattle, Wash.), thereby allowing for a more detailed visualization of the GFP-labeled capsids within the cell. The fluorescence observed was punctate and regularly distributed about the periphery of the nucleus, essentially forming discrete clusters of variable dimensions (Fig. 7, 0° to 90° views). The side view (90°) demonstrates clearly the large quantities of fluorescence emanating from the nucleus in contrast to that detected in the cytoplasm. These data show that GFP-labeled UL37 mutant capsids are restricted to clusters and/or aggregates in the nucleus even at this late stage of the infectious cycle. The other striking observation in the mutant-infected cells was the absence of plasma membrane fluorescence (Fig. 6B and D and Fig. 7). This would indicate that egress of UL37 mutant particles to the cell surface was disrupted. Cytoplasmic fluorescence was also present in the UL37 mutant-infected cells (Fig. 6D). This fluorescence was contained in larger puncta, indicating aggregates or clusters of particles. The cytoplasmic fluorescence did not translocate to the cell surface even late in the replication cycle. The pattern of fluorescence observed in complementing BD45 cells infected with KΔUL37 was similar to that seen in wild-type-infected cells. Thus, the mutant phenotype can be rescued in the complementing cell line. In the absence of the UL37-encoded function, fluorescence at the plasma membrane indicative of mature viruses was never observed. Fluorescence accumulates around the nucleus; therefore, virus egress from the nucleus to the cell surface was severely impaired.
FIG. 7.
KΔUL37-GFP-infected Vero cells analyzed using deconvolving microscopy. Cells were infected as described in the legend for Fig. 6 for 15 h and fixed with 5% formaldehyde in PBS containing 2% sucrose. Images were collected on a Zeiss axiovert light microscope equipped with a Deltavision 3-D Restoration system. The images were deconvolved and 3-D rendered, and optical planes of sections are presented from 0° to 90°. Bar = 10 μm. n, nucleus.
Ultrastructural analysis of KΔUL37-infected cells.
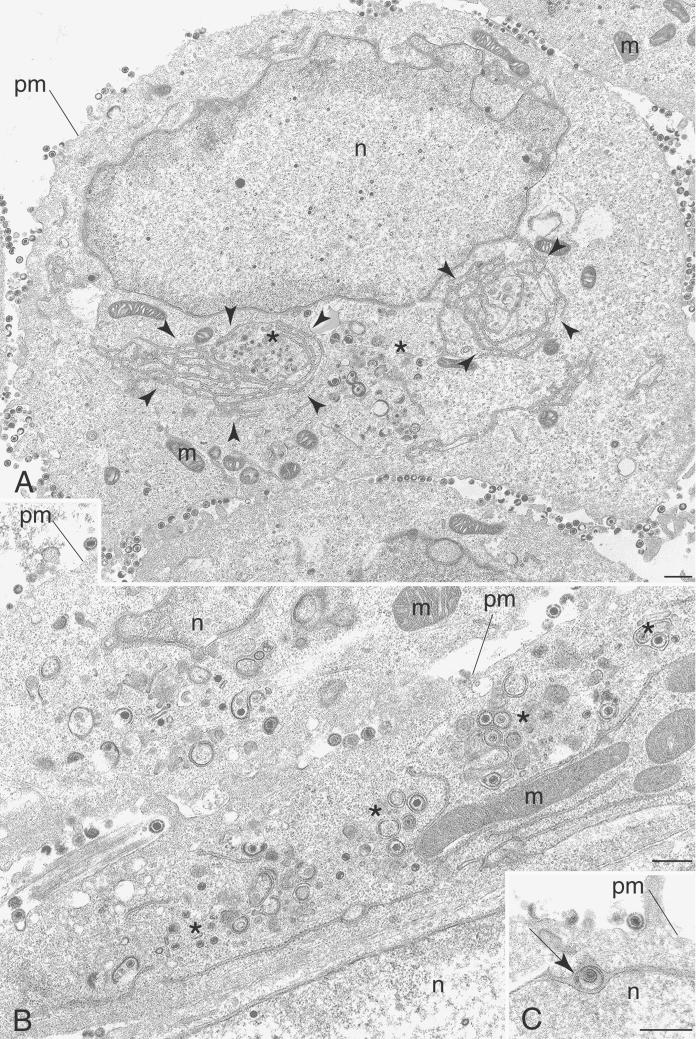
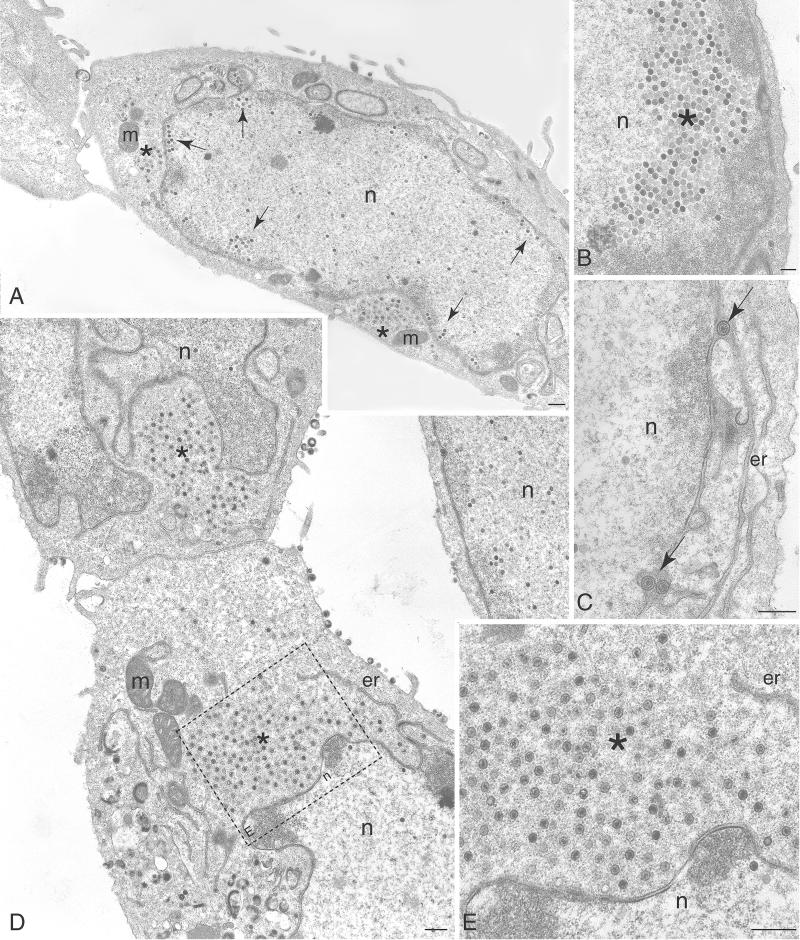
Sedimentation analysis of infected cell lysates revealed the presence of a diffuse light-scattering band corresponding to virions in gradients of KOS and KΔUL37R extracts. This was absent in gradients of KΔUL37 extracts, in which a very faint light-scattering band at the position where C capsids sediment was observed (data not shown). Additionally, light microscopy of GFP-tagged particles revealed the absence of fluorescence at the plasma membrane. We therefore undertook an ultrastructural analysis of cells infected with the UL37 null mutant in order to elucidate the nature of the mutant particles detected in the sucrose gradients and to visualize their fate in the infected cell. Vero cell monolayers were infected with either KOS (Fig. 8) or KΔUL37 (Fig. 9) and the cells were fixed 16 h after infection. Thin sections were prepared, stained, and examined by TEM. In KOS-infected cells enveloped virions were evident in between the nuclear membranes (Fig. 8C, see arrow), in the cytoplasm (Fig. 8A and B), and in the extracellular space (Fig. 8A), which is indicative of a productive infection. Also present in the cytoplasm were naked capsids (Fig. 8B, see asterisk). These could represent capsids that have exited the nucleus and are in the process of translocating to a cytoplasmic site for reenvelopment. In addition, capsids that appear to be in the process of envelopment (Fig. 8B, see asterisk) were detected. At low magnification, the endoplasmic reticulum (ER) was seen to be hypertrophied in both wild-type- and mutant-infected cells (Fig. 8A, see arrowheads). This amplification of the ER is commonly observed late in infection and is typical of HSV-infected cells (28, 34). In some cases capsids are enclosed by these membranes (Fig. 8A, see asterisk). Several electron micrographs of cells infected with KΔUL37 were examined, some of which are shown in Fig. 9. DNA-containing capsids were observed in three locations of the cell. There were groups of capsids close to the inner nuclear membrane (Fig. 9A and B), there were enveloped capsids between the inner and outer nuclear membranes (Fig. 9C, see arrows), and there were clusters of unenveloped capsids in the cytoplasm (Fig. 9A, D, and E, see asterisks). These data provide a clear picture of the fate of the UL37 mutant capsids. Examination of the cytoplasm of these infected cells did not reveal any enveloped particles. In the image of the whole cell (Fig. 9A), the phenotype of the UL37 mutant is clear. Capsids cluster close to the inner nuclear membrane (marked by arrows) and clusters of unenveloped capsids are deposited in the cytoplasm (marked by asterisks) close to the nucleus. In order to quantitate the numbers of nuclear capsids versus cytoplasmic particles and capsids, 20 cells were examined for each virus and the locations and numbers of capsids were enumerated. The data from this analysis are shown in Table 2. The total numbers of both wild-type and mutant particles were pooled for this analysis. The numbers of wild-type particles enumerated were lower than those of the mutant due to the productive nature of the infection and the egress of virus into the extracellular medium. In wild-type-infected cells the distribution of the particles, which included both enveloped capsids and naked capsids, was similar in the different cell compartments. The quantitation of the mutant particles shows that in these infected cells there is an accumulation of particles in the nucleus; 58% of the capsids are still in the nucleus at this late stage of infection. Of these nuclear capsids close to half (22%) were in the proximity of the nuclear envelope. Thus, in the absence of UL37, capsids appear to accumulate in the nucleus. Some capsids do undergo envelopment and are then deposited in the cytoplasm. The cytoplasmic capsids, however, do not mature into enveloped virions. These data together with the fluorescence results suggest that the UL37 mutant particles are impeded in their exit from the nucleus and subsequent maturation into virions is abrogated.
FIG. 8.
Wild-type-infected Vero cells exhibit normal distribution of virus particles by conventional TEM. (A) Particles were variously observed at the plasma membrane (pm), in the nucleus (n), in hypertrophied ER nests (arrowheads), and generally throughout the cytoplasm (asterisks). (B) Higher magnification of wild-type-infected cell shows normal distribution of enveloped and naked capsids. (C) Wild-type enveloped virus was seen within the nuclear envelope (arrow). m, mitochondria. Bars = 0.5 μm.
FIG. 9.
Capsids of the UL37 null mutant are seen to accumulate in the nucleus and cytoplasm of infected Vero cells. (A and B) UL37 null mutant capsids accumulate in clusters (arrows) within the nucleus, and in many cases the accumulation is quite large (asterisk), generally in close proximity to the nuclear envelope. (C) UL37 mutant enveloped capsids are seen in the perinuclear ER cisternae (arrows). (D) UL37 mutant capsids accumulate extensively in the cytoplasm (asterisks). (E) High magnification of boxed area in panel D showing extensive cytoplasmic accumulation of UL37 mutant capsids (asterisk). er, endoplasmic reticulum; m, mitochondria; n, nucleus. Bars = 1.0 μm (A), 0.5 μm (B and C), 1.0 μm (D), and 0.5 μm (E).
TABLE 2.
Distribution of HSV particles (capsids and virions) in wild-type- and KΔUL37-infected Vero cells observed by TEM
| Virusa | Distribution of HSV particles [no. of particles (%)]
|
|||
|---|---|---|---|---|
| Extracellular | Cytoplasm | Nucleus | Nuclear peripheryb | |
| KOS | 271 (6.9) | 85 (2.2) | 297 (7.5) | 67 (1.7) |
| KΔUL37 | 155 (3.9)c | 842 (21.4) | 1,402 (35.7) | 817 (22.1) |
The total number of particles counted for both wild-type and mutant was 3,936 (n = 20 cells for each virus).
Particles within 2 μm of the nuclear periphery.
The particles of KΔUL37 observed outside the cell were most likely light particles.
Association of the KΔUL37 capsids with the nuclear fraction of infected cells.
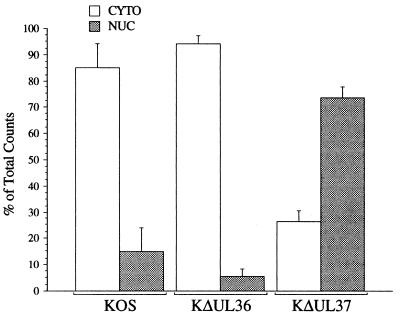
The ultrastructural study revealed accumulation of KΔUL37 C capsids close to the inner nuclear membrane. Unenveloped capsids were observed in the cytoplasm. GFP-tagged particles were also seen to accumulate at the periphery of the nucleus as judged by fluorescence microscopy. Although cytoplasmic fluorescence was detected, no particles were transported to the cell surface. Previously it had been observed that a virus specifying a null mutant in another tegument protein encoded by UL36 was also defective for maturation (13). The UL36 mutant particles, unlike those of the UL37 mutant, were not restricted to the nucleus but were distributed throughout the cytoplasm. Another noticeable difference between these two viruses was observed when infected cell extracts were subjected to sedimentation in sucrose gradients. The quantity of DNA-filled C capsids as judged by the incorporation of [3H]thymidine was much greater for the UL36 mutant than that for the UL37 mutant. Furthermore, when extracts were treated with detergent the yields of C capsids for the UL37 mutant were increased significantly. One possible explanation for these observations was that the UL37 mutant particles were tightly associated with the nucleus and are liberated when the cells are treated with agents that lyse or disrupt nuclei. In order to quantitate the association of KΔUL37 capsids with the nucleus, infected cells were fractionated and the levels of DNA-filled capsids associated with the nucleus and cytoplasm were determined by sedimentation assays in sucrose gradients. The quantities of DNA-filled capsids were monitored by including [3H]thymidine during the infection. Infected cells were fractionated into crude nuclear and cytoplasmic fractions by treatment with PBS containing 2% Triton X-100, 300 mM NaCl, and 0.5% deoxycholic acid. The supernatant and the pellet were both subjected to sedimentation in sucrose gradients. The pellet was lysed in 2× CLB (see Materials and Methods) followed by sonication. This buffer contained 2% Triton X-100 and a high salt concentration, and this treatment solubilized and disrupted all membranes, thus liberating capsids. This buffer also solubilizes virion envelopes, thus converting KOS virion particles into C capsids. Following sedimentation of the lysates the light-scattering bands of the capsid particles were visualized and the radioactivity present in the peak fractions which contained C capsids was determined for each cellular fraction and illustrated in Fig. 10. Data are plotted as a percentage of the total counts present in either the cytoplasmic or nuclear fraction. For both the UL36 null mutant and KOS gradients the majority of the radioactivity was present in the cytoplasmic fraction. This was not the case for the gradients of the UL37 null mutant lysates, in which 75% of the radioactivity representing the DNA-filled capsids was present in the nuclear fraction. The confluence of data obtained from the ultrastructural, light microscopy, and fractionation analyses therefore shows that late in infection the maturation and processing of the KΔUL37 DNA-containing capsids were substantially slowed kinetically, resulting in significant accumulations of capsids in the nucleus and unenveloped capsids in the cytoplasm.
FIG. 10.
Association of KΔUL37 DNA-filled capsids with the nuclear fraction of infected cells. Vero cells (2 × 107 cells in 100-mm-diameter dishes) were infected with KOS, KΔUL37, and KΔUL36 at an MOI of 10 PFU/cell. Infected cells were metabolically labeled with [3H]thymidine from 8 to 24 h after infection. Infected cells were fractioned into crude cytoplasmic (CYTO) and nuclear (NUC) fractions and the lysates were sedimented through 20 to 50% sucrose gradients. The total radioactivity present in the peak fractions, which contained C capsids, from each gradient is plotted as a percentage of the total radioactivity, that is, the sum of the CYTO and NUC radioactivity. Error bars indicate standard deviations of the data.
DISCUSSION
The UL37 gene product is a structural component of the virion particle. Specifically it is a resident of the tegument of the virion (25, 39). The functional role of this gene product in the virus replication cycle was determined by the isolation of a null mutant in the gene encoding UL37. Complementing cell lines were derived that express the UL37 gene product upon infection and these were used to isolate and propagate the null mutant virus. The null mutation in the plasmid DNA encoding UL37 was transferred into the virus genome by homologous recombination using the complementing cell lines. A virus designated KΔUL37 was isolated and purified and was unable to replicate on noncomplementing Vero cells. When the genotype of this virus was examined by Southern blot analysis, it was discovered that it contained the introduced null mutation, thereby establishing for the first time the essential role of the UL37 gene product in the replication of the virus.
Cleavage and packaging of viral DNA occurred normally in Vero cells infected with the UL37 null mutant since both terminal end fragments of the virus genome were detected and C capsids were observed in nuclear extracts. However, these capsids do not mature into enveloped viruses. This was demonstrated by sedimentation analysis of infected cell lysates, which showed that the mutant virus particles have sedimentation properties similar to C capsids. Definitive evidence was provided visually by the ultrastructural analysis of infected cells. This revealed the presence of unenveloped DNA-filled capsids in the cytoplasm. These cytoplasmic capsids were present in clusters and they were never observed adjacent to any membrane structures which would indicate an envelopment process. The only enveloped KΔUL37 virions observed were those between the inner and outer nuclear membranes. Therefore, the absence of UL37 has a profound effect on virus maturation of cytoplasmic capsids.
Recent studies support a mechanism which involves envelopment of the capsid at the inner nuclear membrane followed by de-envelopment at the outer nuclear membrane (references 15, 17 and 42 and references therein). The naked cytosolic capsids are then translocated to a cytoplasmic site for final maturation. Because the UL37 mutant cytoplasmic capsids do not become enveloped, UL37 may be required for translocation of these capsids to a cytoplasmic site for reenvelopment (via a motor, for example) or it may actually be required for directly initiating reenvelopment (via recruitment of membrane or membrane components). Since the mutant capsids were never observed in close proximity to membrane structures (rather, in many cases, the clusters of capsids were proximal to the nucleus) it seems reasonable that a credible role for UL37 would be in the transportation of these capsids to the site for reenvelopment and subsequent maturation.
The trafficking of the UL37 mutant capsids in cells was followed by using the VP26-GFP tag. The UL37 capsids, as assayed by fluorescence, were predominantly restricted to the periphery of the nucleus and to large cytoplasmic clusters. Plasma membrane fluorescence indicative of mature viruses during their egress from the cell was not detected. One of the most dramatic phenotypes of the UL37 mutation was the restriction of the capsids to the nucleus even at late times in infection. This was seen by the accumulation of fluorescence of GFP-tagged capsids at the nuclear periphery, by ultrastructural analysis which showed groups of capsids adjacent to the inner nuclear membrane, and by cell fractionation studies which revealed that two-thirds of the capsids were still associated with the nuclear fraction. This suggests that even though this virus is able to undergo initial envelopment at the nuclear membrane, this envelopment process is somehow slowed or impaired, implicating UL37 in the envelopment process at the nucleus. The exact mechanism by which this protein facilitates envelopment at this site is unclear. Since the UL34-encoded membrane protein is required for initial envelopment at the nuclear membrane (36), it is possible that UL37 interacts with this protein to facilitate the attachment of the capsid to membranes, thus initiating envelopment. In any event the phenotype of the UL37 mutant indicates that it is required for initial envelopment at the inner nuclear membrane and that subsequent to this it may be required for translocating capsids to a cytoplasmic site for final envelopment. Therefore, UL37 appears to be a multifunctional protein required at different stages in the maturation pathway.
The question of where and when the UL37 polypeptide is incorporated into the maturing virion is important in light of the UL37 null mutant phenotype. This would also define at which point in the virus maturation process the function of UL37 is required. Previous studies using antibodies to UL37 in conjunction with immunofluorescence assays showed that fluorescence corresponding to UL37 protein was distributed throughout the cell but that the fluorescence was more abundant in the cytoplasm than in the nucleus (23, 24, 38). Recent studies have shown that there is a nuclear export signal in the UL37 coding region which may be responsible for this asymmetric distribution (44). The results of the null mutant suggest that UL37 is required early in the maturation process, presumably at the nuclear membrane. Therefore, UL37 may be incorporated at or close to the nuclear periphery. Immunoelectron microscopy could reveal where and when the association of UL37 with the maturing virion occurs. One corollary to the phenotype of the UL37 mutation is that the absence of this protein may have a global effect on the functions or localization of other virion structural proteins. The absence of envelopment of cytoplasmic capsids may be due not to the absence of UL37 per se but due to the disruption of complex protein-protein interactions that take place during virion maturation. The human cytomegalovirus homologue of UL37 has been shown to interact with the homologue of UL36 (M. E. Harmon and W. G. Gibson, unpublished data). The absence of UL37 may also affect the integrity and distribution of the host cell secretory components or the cellular cytoskeleton, both of which play an important role in trafficking capsids from the nucleus to the cell surface.
What has become increasingly evident is the importance of the tegument proteins in the maturation process of the enveloped virus. To date three tegument proteins have been shown to have deleterious and lethal effects on the maturation process. These are VP16 (1, 27, 46), VP1/2 (UL36) (13), and now as described in this paper the product of the UL37 gene. The last two proteins are minor components of the tegument and yet they have a dramatic effect on the virus maturation process. The null mutation in the UL36 gene product results in the presence of numerous cytoplasmic capsids that are not enveloped (13). A simple explanation for this phenotype is that they cannot be transported to the correct cellular compartment for final envelopment. The phenotype of the VP16 null mutant is similar in that cytoplasmic naked capsids were observed in infected cells, even though capsids were initially observed to envelop at the nuclear membrane (27). The phenotype of the UL37 mutant suggests that this protein may act before VP16 and UL36 at an earlier stage of virion morphogenesis, namely, for initial envelopment at the inner nuclear membrane, and that subsequent to this it may be required for translocating capsids to the cytoplasmic site for final envelopment.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI33077 from the National Institutes of Health.
We acknowledge discussions of the data with Wade Gibson and members of his laboratory and his continued support. We also acknowledge the kind gift of UL37 antisera 708 from Frank Jenkins.
REFERENCES
- 1.Ace C I, Dalrymple M A, Ramsay F H, Preston V G, Preston C M. Mutational analysis of the herpes simplex virus type 1 trans-inducing factor Vmw65. J Gen Virol. 1988;69:2595–2605. doi: 10.1099/0022-1317-69-10-2595. [DOI] [PubMed] [Google Scholar]
- 2.Albright A G, Jenkins F J. The herpes simplex virus UL37 protein is phosphorylated in infected cells. J Virol. 1993;67:4842–4847. doi: 10.1128/jvi.67.8.4842-4847.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines J D, Roizman B. The UL11 gene of herpes simplex virus type 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J Virol. 1992;66:5168–5174. doi: 10.1128/jvi.66.8.5168-5174.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batterson W, Furlong D, Roizman B. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batterson W, Roizman B. Characterization of the herpes simplex virion-associated factor responsible for the induction of α genes. J Virol. 1983;46:371–377. doi: 10.1128/jvi.46.2.371-377.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell M E M, Palfreyman J W, Preston C M. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for the stimulation of immediate early transcription. J Mol Biol. 1984;180:1–19. doi: 10.1016/0022-2836(84)90427-3. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham C, Davison A J, Dolan A, Frame M C, McGeoch D J, Meredith D M, Moss H W M, Orr A C. The UL13 virion protein of herpes simplex virus type 1 is phosphorylated by a novel virus-induced protein kinase. J Gen Virol. 1992;73:303–311. doi: 10.1099/0022-1317-73-2-303. [DOI] [PubMed] [Google Scholar]
- 8.DeLuca N A, McCarthy A M, Schaffer P A. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;56:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desai P, DeLuca N A, Glorioso J C, Person S. Mutations in herpes simplex virus type 1 genes encoding VP5 and VP23 abrogate capsid formation and cleavage of replicated DNA. J Virol. 1993;67:1357–1364. doi: 10.1128/jvi.67.3.1357-1364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desai P, Watkins S C, Person S. The size and symmetry of B capsids of herpes simplex virus type 1 are determined by the gene products of the UL26 open reading frame. J Virol. 1994;68:5365–5374. doi: 10.1128/jvi.68.9.5365-5374.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai P, DeLuca N A, Person S. Herpes simplex virus type 1 VP26 is not essential for replication in cell culture but influences production of infectious virus in the nervous system of infected mice. Virology. 1998;247:115–124. doi: 10.1006/viro.1998.9230. [DOI] [PubMed] [Google Scholar]
- 12.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol. 1998;72:7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desai P J. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J Virol. 2000;74:11608–11618. doi: 10.1128/jvi.74.24.11608-11618.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliot G, O'Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 15.Enquist L W, Husak P J, Banfield B W, Smith G A. Infection and spread of alphaherpesviruses in the nervous system. Adv Virus Res. 1998;51:237–347. doi: 10.1016/s0065-3527(08)60787-3. [DOI] [PubMed] [Google Scholar]
- 16.Frame M C, Purves F C, McGeoch D J, Marsden H S, Leader D P. Identification of the herpes simplex virus protein kinase as the product of viral gene US3. J Gen Virol. 1987;68:2699–2704. doi: 10.1099/0022-1317-68-10-2699. [DOI] [PubMed] [Google Scholar]
- 17.Granzow H, Klupp B G, Fuchs W, Viets J, Osterrieder N, Mettenleiter T C. Egress of alphaherpesviruses: comparative ultrastructural study. J Virol. 2001;75:3675–3684. doi: 10.1128/JVI.75.8.3675-3684.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendricks L C, McCaffery J M, Palade G E, Farquhar M G. Disruption of ER to Golgi transport leads to the accumulation of large aggregates containing β-COP in pancreatic acinar cells. Mol Biol Cell. 1993;4:413–424. doi: 10.1091/mbc.4.4.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knipe D M, Ruyechan W T, Roizman B. Molecular genetics of herpes simplex virus. VI. Characteriztion of a temperature-sensitive mutant defective in the expression of all early viral gene products. J Virol. 1979;38:539–547. doi: 10.1128/jvi.38.2.539-547.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong A D, Kruper J A, Frenkel N. Herpes simplex virus virion host shutoff function. J Virol. 1988;62:912–921. doi: 10.1128/jvi.62.3.912-921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leslie J, Rixon F J, McLauchlan J. Overexpression of the herpes simplex virus type 1 tegument protein VP22 increases its incorporation into virus particles. Virology. 1996;220:60–68. doi: 10.1006/viro.1996.0286. [DOI] [PubMed] [Google Scholar]
- 22.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 23.McKnight J L C, Pellett P E, Jenkins F J, Roizman B. Characterization and nucleotide sequence of two herpes simplex virus 1 genes whose products modulate α-trans-inducing factor-dependent activation of α genes. J Virol. 1987;61:992–1001. doi: 10.1128/jvi.61.4.992-1001.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLauchlan J. The abundance of the herpes simplex virus type 1 UL37 tegument protein in virus particles is closely controlled. J Gen Virol. 1997;78:189–194. doi: 10.1099/0022-1317-78-1-189. [DOI] [PubMed] [Google Scholar]
- 25.McLauchlan J, Liefkens K, Stow N D. The herpes simplex virus type 1 UL37 gene product is a component of virus particles. J Gen Virol. 1994;75:2047–2052. doi: 10.1099/0022-1317-75-8-2047. [DOI] [PubMed] [Google Scholar]
- 26.McNabb D S, Courtney R J. Analysis of the UL36 open reading frame encoding the large tegument protein (ICP1/2) of herpes simplex virus type 1. J Virol. 1992;66:7581–7584. doi: 10.1128/jvi.66.12.7581-7584.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mossman K L, Sherburne R, Lavery C, Duncan J, Smiley J R. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J Virol. 2000;74:6287–6299. doi: 10.1128/jvi.74.14.6287-6299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nii S, Morgan C, Rose H M. Electron microscopy of herpes simplex virus. II. Sequence of development. J Virol. 1968;2:517–536. doi: 10.1128/jvi.2.5.517-536.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Person S, Desai P. Capsids are formed in a mutant virus blocked at the maturation site of the UL26 and UL26.5 open reading frame of herpes simplex virus type 1 but are not formed in a null mutant of UL38 (VP19C) Virology. 1998;242:193–203. doi: 10.1006/viro.1997.9005. [DOI] [PubMed] [Google Scholar]
- 30.Pertuiset B, Boccara M, Cebrian J, Berthelot N, Chousterman S, Puvion-Dutilleul F, Sisman J, Sheldrick P. Physical mapping and nucleotide sequence of a herpes simplex virus type 1 gene required for capsid assembly. J Virol. 1989;63:2169–2179. doi: 10.1128/jvi.63.5.2169-2179.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Post L E, Mackem S, Roizman B. Regulation of α genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with α gene promoters. Cell. 1981;24:555–565. doi: 10.1016/0092-8674(81)90346-9. [DOI] [PubMed] [Google Scholar]
- 32.Read G S, Frenkel N. Herpes simplex virus mutants defective in the virion shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of α (immediate-early) viral polypeptides. J Virol. 1983;46:498–512. doi: 10.1128/jvi.46.2.498-512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rixon F J, Davison M D, Davison A J. Identification of the genes encoding two capsid proteins of herpes simplex virus type 1 by direct amino acid sequencing. J Gen Virol. 1990;71:1211–1214. doi: 10.1099/0022-1317-71-5-1211. [DOI] [PubMed] [Google Scholar]
- 34.Roizman B, Furlong D. The replication of herpesviruses. In: Fraenkel-Conrat H, Wagner R R, editors. Comprehensive virology. New York, N.Y: Plenum Press; 1974. pp. 11–68. [Google Scholar]
- 35.Roizman B, Sears A. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2231–2295. [Google Scholar]
- 36.Roller R J, Roizman B. The herpes simplex virus 1 RNA binding protein US11 is a virion component and associates with ribosomal 60S subunits. J Virol. 1992;66:3624–3632. doi: 10.1128/jvi.66.6.3624-3632.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roller R J, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J Virol. 2000;74:117–129. doi: 10.1128/jvi.74.1.117-129.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salmon B, Cunningham C, Davison A J, Harris W J, Baines J D. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J Virol. 1998;72:3779–3788. doi: 10.1128/jvi.72.5.3779-3788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitz J B, Albright A G, Kinchington P R, Jenkins F J. The UL37 protein of herpes simplex virus type 1 is associated with the tegument of purified virions. Virology. 1995;206:1055–1065. doi: 10.1006/viro.1995.1028. [DOI] [PubMed] [Google Scholar]
- 40.Shelton L S G, Pensiero M N, Jenkins F J. Identification and characterization of the herpes simplex virus type 1 protein encoded by the UL37 open reading frame. J Virol. 1990;64:6101–6109. doi: 10.1128/jvi.64.12.6101-6109.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shelton L S G, Albright A G, Ruyechan W T, Jenkins F J. Retention of herpes simplex virus type 1 (HSV-1) UL37 protein on single-stranded DNA columns requires the HSV-1 ICP8 protein. J Virol. 1994;68:521–525. doi: 10.1128/jvi.68.1.521-525.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skepper J N, Whiteley A, Browne H, Minson A. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment-deenvelopment-reenvelopment pathway. J Virol. 2001;75:5697–5702. doi: 10.1128/JVI.75.12.5697-5702.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Southern P J, Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under the control of the SV40 early region promoter. J Mol Appl Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- 44.Steven A C, Spear P G. Herpesvirus capsid assembly and envelopment. In: Burnett R, Chiu W, Garcea R, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1996. pp. 312–351. [Google Scholar]
- 45.Watanabe D, Ushijima Y, Goshima F, Takakuwa H, Tomita Y, Nishiyama Y. Identification of nuclear export signal in UL37 protein of herpes simplex virus type 2. Biochem Biophys Res Commun. 2000;276:1248–1254. doi: 10.1006/bbrc.2000.3600. [DOI] [PubMed] [Google Scholar]
- 46.Weinheimer S P, Boyd B A, Durham S K, Resnick J L, O'Boyle D R. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J Virol. 1992;66:258–269. doi: 10.1128/jvi.66.1.258-269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, McKnight J L C. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J Virol. 1993;67:1482–1492. doi: 10.1128/jvi.67.3.1482-1492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]