Abstract
During lytic infections, the virion host shutoff (Vhs) protein (UL41) of herpes simplex virus destabilizes both host and viral mRNAs. By accelerating mRNA decay, it helps determine the levels and kinetics of viral and cellular gene expression. In vivo, Vhs shows a strong preference for mRNAs, as opposed to non-mRNAs, and degrades the 5′ end of mRNAs prior to the 3′ end. In contrast, partially purified Vhs is not restricted to mRNAs and causes cleavage of target RNAs at various sites throughout the molecule. To explain this discrepancy, we searched for cellular proteins that interact with Vhs using the Saccharomyces cerevisiae two-hybrid system. Vhs was found to interact with the human translation initiation factor, eIF4H. This interaction was verified by glutathione S-transferase pull-down experiments and by coimmunoprecipitation of Vhs and epitope-tagged eIF4H from extracts of mammalian cells. The interaction was abolished by several point mutations in Vhs that abrogate its ability to degrade mRNAs in vivo. The results suggest that Vhs is a viral mRNA degradation factor that is targeted to mRNAs, and to regions of translation initiation, through an interaction with eIF4H.
Controls of the rate of mRNA decay play an important role in mammalian gene expression (25, 37). Different mRNAs have characteristic half-lives that range from less than 15 min to more than 10 h. The decay rates of some mRNAs are dependent upon the stage of the cell cycle or are responsive to external stimuli such as exposure of the cells to hormones or growth factors (3, 6). While much has been learned concerning mRNA degradation pathways in yeast (39, 43), considerably less is known about the decay mechanisms in mammalian cells. Although a number of mammalian RNases have been isolated, it is unclear which enzymes are components of the mRNA degradation machinery, how they are targeted to mRNAs, or how their activities are controlled (1, 2). In addition, the regulated degradation of many mRNAs depends upon their being translated; yet the mechanism of this linkage is unclear (14). In this context, the virion host shutoff (Vhs) protein of herpes simplex virus (HSV) provides an attractive model system for studying mechanisms of mammalian mRNA decay.
Vhs is a 58-kDa polypeptide encoded by the viral UL41 open reading frame, which is a minor structural component of HSV virions (20, 33, 40). During lytic infections, copies of the Vhs (UL41) polypeptide, which enter the cell as components of infecting virions, accelerate the degradation of cellular mRNAs, contributing to an overall decrease in host protein synthesis (11, 38). Following the onset of viral transcription, Vhs also accelerates the turnover of viral mRNAs (19, 27, 28, 42). By shortening the half-lives of all mRNAs, it helps redirect the cell from host to viral gene expression, and facilitates the sequential expression of different classes of viral genes (31).
A number of studies suggest that Vhs (UL41) is itself an endonuclease. The UL41 homologues from HSV and other alpha herpesviruses share sequence homologies with a family of mammalian, Saccharomyces cerevisiae, bacterial, and phage nucleases (5, 9; D. N. Everly, Jr., P. Feng, I. S. Mian, and G. S. Read, unpublished data). For several of these homologues, point mutations that alter key conserved residues abrogate the nuclease activity, and mutations that alter the corresponding residues of Vhs inactivate its ability to induce mRNA decay (Everly et al., unpublished). Extracts of detergent-solubilized virions contain an endonuclease activity that is Vhs dependent since it is found in wild-type but not Vhs mutant virions and can be inhibited by Vhs-specific antisera (44). Furthermore, rabbit reticulocyte lysates containing in vitro-translated Vhs contain an endonuclease activity (7, 44). Although these studies are suggestive, the virion preparations that exhibited nuclease activity probably contained a number of contaminating cellular proteins, while the reticulocyte lysates contained the full complement of cytoplasmic proteins. Thus, one cannot exclude the possibility that Vhs activates a cellular endonuclease but is not itself a nuclease.
Although Vhs does not discriminate between mRNAs, it does exhibit two important kinds of selectivity. First, it exhibits a strong preference for mRNAs, as opposed to non-mRNAs. This is true in vivo (27, 28) as well as in in vitro decay reactions containing either cytoplasmic extracts from infected cells (18, 41) or in vitro-translated Vhs (44). Second, recent studies suggest that Vhs does not cleave mRNAs at random sites but may initiate degradation near regions of translation initiation. In infected cells, sequences near the 5′ end of an mRNA are degraded prior to those near the 3′ end of the transcript (17), while in rabbit reticulocyte lysates, in vitro-translated Vhs initially targets sites near the 5′ end of an mRNA (7). In addition, in vitro-translated Vhs preferentially induces cleavage at sites downstream from a picornavirus internal ribosome entry site (IRES) (8, 23). In contrast, the Vhs activity in solubilized virions is not restricted to mRNAs and cleaves target RNAs at multiple sites throughout the molecule (44). These data suggest that, in the absence of key cellular factors, the Vhs-dependent endonuclease is significantly less specific than in intact cells. To explain this discrepancy, we searched for cellular proteins that interact with Vhs using the yeast two-hybrid system and coimmunoprecipitation assays. The results show that Vhs interacts with the cellular translation initiation factor eIF4H and suggest a mechanism for targeting Vhs to mRNAs and to regions of translation initiation, as well as for linking mRNA decay and translation.
MATERIALS AND METHODS
Cells and virus.
Vero cells and HSV type 1 (HSV-1), strain KOS, were grown as described previously (29). The recombinant vaccinia virus vvT7 expresses the T7 RNA polymerase (26) and was obtained from Lindsey Hutt-Fletcher (University of Missouri—Kansas City).
Antibodies.
A polyclonal rabbit antiserum raised against a Vhs-LacZ fusion protein has been described previously (33). Monoclonal antibody that reacts with an epitope of the influenza virus hemagglutinin (HA) was purchased from Covance.
Plasmids. (i) Plasmids containing wild-type and mutant Vhs alleles.
The plasmid pKOSamp contains the Vhs (UL41) open reading frame from wild-type HSV-1 (KOS) cloned into the vector pcDNA1.1 A (Invitrogen) (10). Various mutant Vhs alleles have been constructed by site-directed mutagenesis of pKOSamp. Each of the alleles D34N, D82N, E192Q, D194N, D195N, T211S, T211A, D213N, D215N, and D261N contains a single amino acid change in the Vhs allele of HSV-1 (KOS). The name of each allele includes the number of the residue that is altered, preceded by the wild-type amino acid and followed by the amino acid to which it is changed. For example, in D194N an aspartic acid at residue 194 is changed to asparagine. Each mutation alters an amino acid that is conserved in Vhs and a number of cellular nucleases and that has been shown to be important to the activity of some of those cellular enzymes. Each mutation also abrogates the mRNA degradative activity of the Vhs allele following transfection of mammalian cells. Construction and characterization of these alleles will be described elsewhere (Everly et al., unpublished). Construction of R435H, in which arginine 435 is changed to histidine, has been described previously (10). The mutation in the allele referred to in this work as T214I is that carried by the mutant virus Vhs1. This mutant lacks detectable Vhs activity and has been characterized extensively in previous studies (9, 18, 28, 29, 32). The mutants K(89–489) and K(1–453) encode Vhs polypeptides lacking the first 88 or last 36 amino acids of the wild-type polypeptide, respectively. The structure of each mutant allele was confirmed by DNA sequencing.
(ii) Plasmids for yeast two-hybrid interactions.
The plasmid pAS2-Vhs contains the UL41 open reading frame of HSV-1 (KOS) inserted between the EcoRI and BamHI sites of pAS2-1 (Clontech). pAS2-Vhs encodes a fusion protein containing the Gal4 DNA binding domain (amino acids 1 through 147) followed by 16 amino acids encoded by the polylinker and the entire 489-amino-acid Vhs polypeptide. pAS2-Vhs was used to screen a commercially available library of HeLa cell cDNAs cloned into pACT2 (Clontech). pACT2-4H and pACT2-4Hi are two plasmids isolated from that library. To confirm the specificity of the two-hybrid interaction, the vectors used to express Vhs and eIF4Hi were switched. The two-hybrid assay then was repeated using pAS2-4Hi, which encodes a fusion protein of the Gal4 DNA binding domain and eIF4Hi, and pACT2-Vhs, which encodes a Gal4 activation domain-Vhs fusion protein.
(iii) Plasmids for GST pull-down experiments.
For GST pull-down experiments, the eIF4H open reading frame was PCR amplified and cloned between the BamHI and SalI sites of pGEX-5-3 (Amersham-Pharmacia) to yield pGST-4H, which encodes a glutathione S-transferase (GST)–eIF4H fusion protein that can be expressed in bacteria. A similar procedure was used to construct pGST-4Hi, which encodes a fusion protein of GST and eIF4Hi.
(iv) Plasmids for transient expression of Vhs and eIF4H.
For expression of an epitope-tagged version of eIF4H in mammalian cells, sequences encoding HA-tagged eIF4H were PCR amplified from pACT2-4H and inserted between the NcoI and SacI sites of pTM1, to yield pTM1-4H. Parallel procedures were used to construct pTM1-4Hi, encoding an HA-tagged eIF4Hi. To express Vhs, the UL41 open reading frame was PCR amplified and inserted between the NcoI and BamHI sites of pTM1, to yield pTM1-Vhs.
Yeast two-hybrid screen.
A search for cellular cDNAs encoding Vhs-interacting proteins was performed using the Matchmaker Two-Hybrid System 2 (Clontech) according to standard techniques (12). Yeast strain Y190, which contains the his3 and lacZ genes driven by Gal4-responsive promoters, was transformed with pAS2-Vhs. Cells receiving the bait plasmid were transformed with plasmids from a commercially available library of HeLa cell cDNAs in the Gal4 activation domain vector pACT2 (Clontech). Cells that grew on synthetic dropout (SD) medium containing 35 mM 3-aminotriazole (3-AT) and lacking tryptophan, leucine, and histidine potentially did so because of activation of the Gal4-responsive his3 gene. These were replica plated onto filters and screened for lacZ expression according to standard protocols (Clontech). cDNA-containing plasmids were isolated from blue colonies, amplified in Escherichia coli strain KC8 (Clontech), and analyzed by restriction enzyme digestion. Selected plasmids were used to retransform yeast strain Y187 along with the pAS2-Vhs. Plasmids that activated lacZ expression in the presence of pAS2-Vhs, but not in the presence of the DNA binding domain vector pAS2-1 or an unrelated bait plasmid containing a cDNA for lamin C (Clontech), were judged to encode potential Vhs-interacting proteins. cDNA sequences were determined and compared to known DNA and protein sequences using the BLAST network service and sequence analysis software of the National Center for Biotechnology Information. Quantitative analyses of LacZ expression in yeast containing selected bait and cDNA-containing plasmids were performed according to published protocols (4).
Recombination-based two-hybrid assay.
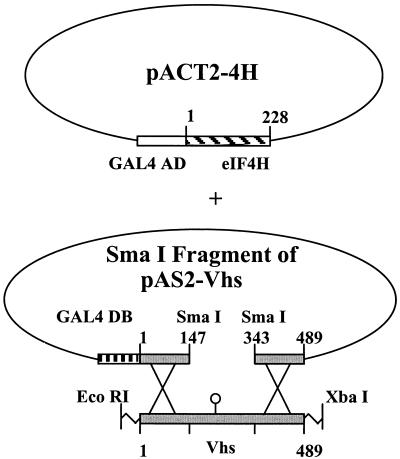
A recombination-based yeast two-hybrid assay was performed as described previously (30). Yeast strain Y190 was transformed with pACT2-4H and the large SmaI fragment of pAS2-Vhs (see Fig. 4). This fragment contains all of the pAS2-Vhs sequences except those encoding Vhs amino acids 148 through 343. Transformations also included an EcoRI-XbaI fragment from another plasmid containing the entire mutant Vhs allele whose binding to eIF4H was to be tested. Transformants were plated on SD medium containing 35 mM 3-AT and lacking tryptophan, leucine, and histidine to select for yeast that grew because transcription of the Gal4-responsive his3 gene had been activated. Within the yeast, recombination between the SmaI fragment of pAS2-Vhs and the fragment containing the mutant Vhs gene resulted in reconstitution of a pAS2-Vhs plasmid containing the Vhs mutation. Transformations that contained an EcoRI-XbaI fragment carrying the wild-type Vhs allele gave rise to 200 to 300 colonies on the selective medium which also expressed the lacZ gene from a Gal4-responsive promoter. In contrast, transformations that contained just pACT2-4H and the large SmaI fragment of pAS2-Vhs, but no EcoRI-XbaI fragment containing a Vhs allele, gave rise to at most 5 to 10 colonies on selective medium. Mutant Vhs alleles that gave rise to a number of colonies comparable to that of wild-type Vhs were judged to encode proteins that retained eIF4H binding activity. In contrast, several mutant alleles gave rise to no more than the background number of colonies and were judged to encode proteins that had lost the ability to interact with eIF4H. All of the mutants gave rise to a number of colonies comparable to that of wild-type Vhs or a number that was indistinguishable from the background.
FIG. 4.
Strategy of the recombination-based yeast two-hybrid assay. Yeast strain Y190 was transformed with pACT2-4H and the large SmaI fragment of pAS2-Vhs. Transformations also included an EcoRI-XbaI fragment from another plasmid containing the entire mutant Vhs allele whose binding to eIF4H was to be tested. Within the yeast, recombination between the SmaI fragment of pAS2-Vhs and the fragment containing the mutant Vhs gene resulted in reconstitution of a pAS2-Vhs plasmid containing the Vhs mutation. Transformants were plated on SD medium containing 35 mM 3-AT and lacking tryptophan, leucine, and histidine to select for yeast that grew because transcription of the Gal4-responsive his3 gene had been activated. Transformations that contained a Vhs allele encoding a protein that bound eIF4H gave rise to 200 to 300 colonies on the selective medium, while control transformations containing just pACT2-4H and the large SmaI fragment of pAS2-Vhs gave rise to at most 5 to 10 colonies.
In vitro transcription and translation.
Vhs protein was produced by coupled in vitro transcription and translation using the TnT T7 Quick Coupled Transcription/Translation System from Promega. Translation reactions were adjusted to 50 mM Tris-HCl (pH 8.0)–100 mM NaCl–1% Nonidet P-40–1 mM EDTA and used in GST pull-down experiments.
Virus infection and preparation of infected cell extracts.
Vero cells were infected with 5 PFU of HSV-1 (KOS)/cell (28). Sixteen hours later, the cells were harvested and lysed by resuspension at a concentration of 107cells/ml in ice-cold lysis buffer (50 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1% NP-40, 1 mM EDTA) containing protease inhibitors (2 μg of aprotinin per ml and 50 μg of phenylmethylsulfonyl fluoride per ml). After 10 min on ice, the nuclei were pelleted, and the supernatant was saved for GST pull-down experiments.
GST pull-down assays.
E. coli, strain BL21, was transformed with plasmids encoding fusion proteins of GST and wild-type or mutant forms of eIF4H. Overnight cultures were induced for 4 h with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), after which the bacteria were harvested, resuspended in phosphate-buffered saline containing 1% (vol/vol) Triton X-100 and lysozyme (0.1 mg/ml), and lysed by sonication. Bacterial extracts were clarified and mixed with a 50% (vol/vol) slurry of glutathione-Sepharose 4B in phosphate-buffered saline (50 μl per 1 ml of lysate). After 15 min on ice, the beads were pelleted, washed, and resuspended in 0.4-ml portions of either cytoplasmic supernatant from HSV-infected cells or rabbit reticulocyte lysates containing in vitro-translated Vhs. Thirty minutes later the beads were pelleted and washed. Complexes of GST-eIF4H bound to Vhs were eluted with a small volume of 10 mM glutathione in 50 mM Tris (pH 8.0) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). [35S]methionine-labeled Vhs from in vitro translation reactions was detected by autoradiography of the dried gel, while unlabeled Vhs from lysates of infected cells was detected by Western blotting (33).
Coprecipitation of Vhs and eIF4H from mammalian cells.
Vero cells were grown to 90% confluence in 100-mm-diameter petri dishes and infected with vaccinia virus vTF7-39 (5 PFU/cell) as described previously (21). Thirty minutes later, they were transfected with 5 μg of pTM1-Vhs and 5 μg of either pTM1-4Hi or a mixture of pTM1-4Hi and pTM1-4H. Twenty hours after transfection, the cells were harvested and lysed, and immunoprecipitates were prepared using either a Vhs-specific polyclonal antiserum or HA-specific monoclonal antibody (33). Precipitated proteins were resolved by SDS-PAGE and analyzed by Western blotting using the reciprocal antibody.
RESULTS
Two-hybrid screen for Vhs-interacting proteins.
To identify cellular proteins that interact with Vhs, the entire Vhs (UL41) open reading frame of HSV-1 was cloned downstream from the Gal4 DNA binding domain and used as bait in a yeast two-hybrid screen of a HeLa cell cDNA library cloned downstream from the Gal4 activation domain. HeLa cells were chosen as the source of cellular cDNAs because they are readily infected with HSV-1 and are very susceptible to Vhs-induced mRNA degradation. Of 1.7 × 107 clones that were screened, 90 were judged to be candidates for cDNAs encoding Vhs-interacting proteins since they induced simultaneous expression of his3 and lacZ genes from Gal4-responsive promoters. Analysis of all 90 cDNAs revealed that they fell into five groups on the basis of their size and restriction patterns. Selected cDNA-containing plasmids were amplified in E. coli and reintroduced into yeast, where, once again, they activated a Gal4-responsive promoter in the presence of a Vhs-containing bait plasmid. Transcription was not activated when the cDNAs were used in combination with the Gal4 DNA binding domain vector lacking Vhs or with an unrelated bait plasmid containing a lamin C cDNA. Interchanging the vectors in which Vhs and the cDNAs were expressed did not affect their ability to activate transcription of a Gal4-responsive promoter, further verifying the specificity of the two-hybrid interaction.
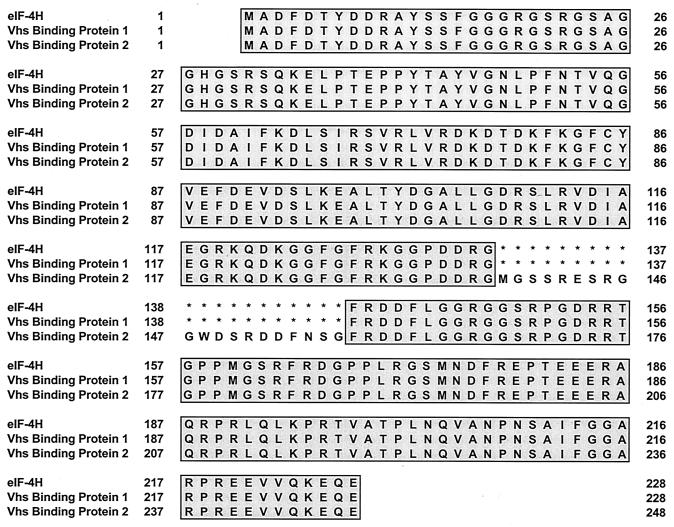
Seven of the cDNAs were sequenced, including representatives of each of the five groups. Each of the seven was predicted to encode one of two related polypeptides (Fig. 1). Differences between the restriction patterns of cDNAs that encoded the same polypeptide, but fell into different groups, could be explained by differences in the size of the cDNA inserts. Four of the cDNAs encoded a protein whose sequence was a perfect match to that of eukaryotic translation initiation factor eIF4H (34, 35). The other three encoded a recently reported isotype of eIF4H, which is produced from an alternatively spliced mRNA and contains an insertion of 20 amino acids after residue 137 (24). This protein was designated eIF4Hi.
FIG. 1.
Sequences of eIF4H and two cellular Vhs binding proteins. The published sequence of eIF4H is shown, along with the deduced sequences of two Vhs binding proteins identified in the yeast two-hybrid screen. Amino acids that are identical to those of eIF4H are shaded grey. Asterisks indicate the absence of an amino acid and were introduced to facilitate alignment of eIF4H, Vhs binding protein 1, and Vhs binding protein 2.
In vitro interaction between Vhs and eIF4H.
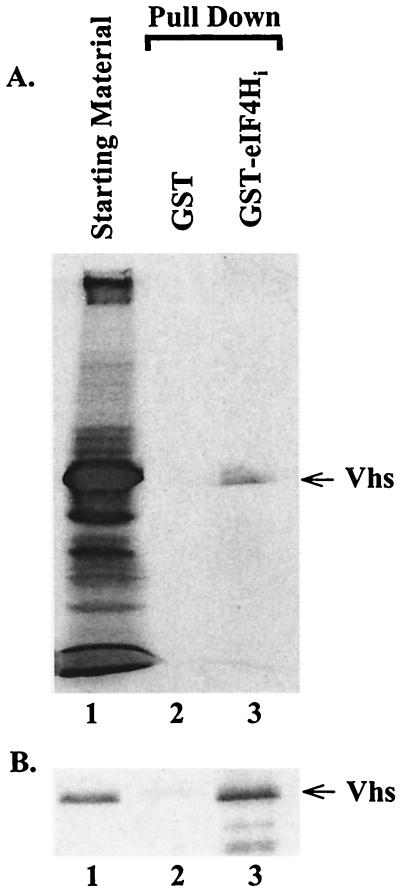
To corroborate the results of the two-hybrid assay, eIF4Hi was cloned into pGEX-5X-3 (Pharmacia) to produce pGST-4Hi, a plasmid encoding a fusion protein of GST and eIF4Hi. GST-eIF4Hi was produced in bacteria, bound to glutathione-Sepharose, and tested for its ability to interact with Vhs in solution. Two sources of Vhs were used: [35S]methionine-labeled Vhs produced by in vitro transcription and translation in rabbit reticulocyte lysates and unlabeled Vhs from lysates of HSV-infected cells. Control reactions assayed the binding of Vhs to GST produced in bacteria that contained the pGEX-5X-3 vector. After incubation with Vhs, the beads were washed and the bound proteins were eluted with 10 mM glutathione. Bound proteins were analyzed by SDS-PAGE and autoradiography to detect 35S-labeled Vhs (Fig. 2A) or by SDS-PAGE and Western blotting to detect Vhs from infected cell lysates (Fig. 2B). Vhs from both sources bound to beads containing GST-eIF4Hi, but not to beads charged with just GST, indicating a specific interaction between Vhs and eIF4Hi.
FIG. 2.
Binding of Vhs to a GST-eIF4Hi fusion protein in vitro. GST or a GST-eIF4Hi was bound to glutathione-Sepharose 4B and incubated with rabbit reticulocyte lysates containing in vitro-translated Vhs (A) or a cytoplasmic extract from HSV-1 infected cells (B). Bound proteins were eluted with 10 mM glutathione and analyzed by SDS -PAGE. In vitro-translated proteins were detected by autoradiography, while Vhs from infected cells was detected by Western blotting. In both panels, lane 1 contains the starting material, lane 2 contains proteins that bound to GST, and lane 3 contains proteins that bound to GST-eIF4Hi.
Coimmunoprecipitation of Vhs and eIF4H.
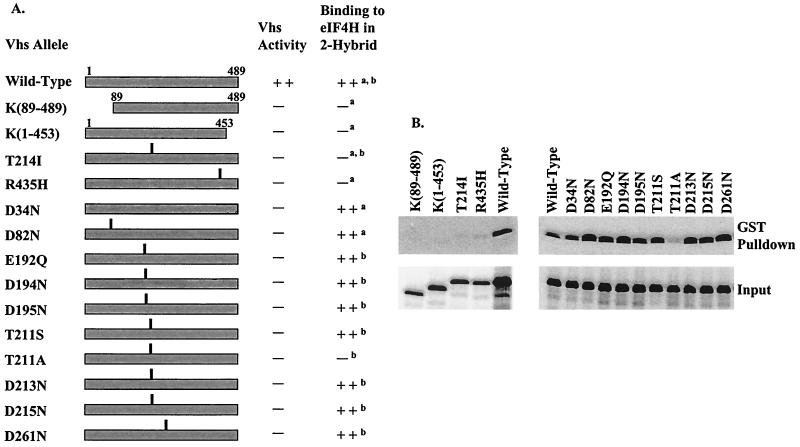
To determine whether Vhs and eIF4H interact in vivo, coimmunoprecipitation experiments were performed in which HA-tagged forms of eIF4H and eIF4Hi were expressed in Vero cells in the presence and absence of Vhs. After lysis of the cells, immunoprecipitates were prepared using either Vhs-specific antisera or an HA-specific monoclonal antibody and examined for the presence of coprecipitated proteins by Western blotting using the reciprocal antibody. Previous studies have shown that small amounts of the Vhs protein are produced in cells transfected with the wild-type UL41 allele, while significantly more of the protein is expressed in cells transfected with inactive Vhs alleles (29). This is presumably due to the fact that active Vhs protein degrades its own mRNA, thereby reducing the amount of the protein that is produced in transfected cells. Consequently, to maximize the amount of the Vhs protein that was produced, we used a Vhs allele (D194N) with a single point mutation that changes an aspartic acid to asparagine at residue 194. Vhs shares a number of highly conserved residues with a family of mammalian, yeast, bacterial, and phage nucleases, among them aspartate 194. For several of these cellular homologues, structural studies and site-directed mutagenesis have implicated aspartate 194 as essential for nuclease activity. Therefore, the D194N allele was constructed in anticipation that it would encode a Vhs protein that lacks nuclease activity and would be unable to induce mRNA decay in transfected cells. Recent studies have proven this to be the case (Everly et al., unpublished). In addition, experiments (see Fig. 5) show that the D194N Vhs protein retains the ability to interact with eIF4H in the yeast two-hybrid system and in GST pull-down assays.
FIG. 5.
eIF4H binding by wild-type and mutant Vhs. (A) The structures of the wild-type Vhs protein and various mutants. For deletion mutants, the Vhs residues included in the mutant protein are indicated. For each point mutant, the location of the altered residue is indicated by the vertical line above the bar representing the protein. The in vivo mRNA-degradative activity of each Vhs protein is shown in the column immediately to the right of the diagram. This was assayed in transfected Vero cells for all of the alleles and during virus infections for wild-type Vhs and the mutant T214I. The right-hand column indicates whether a Vhs protein binds (++) or does not (−) bind eIF4H in the conventional (superscript letter a) or recombination-based (superscript letter b) yeast two-hybrid system. (B) GST pull-down assays. Wild-type and mutant Vhs polypeptides were produced by in vitro translation and analyzed for the ability to bind a GST-eIF4H fusion protein as described for Fig. 2. Material that bound to GST-eIF4H and was eluted with 10 mM glutathione is shown in the upper gels (GST Pulldown). Aliquots of the total in vitro-translated material are shown in the lower gels (Input).
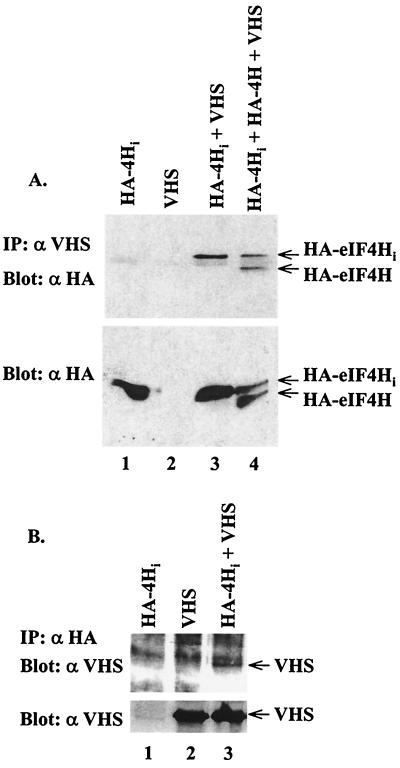
Readily detectable amounts of HA-tagged eIF4H and eIF4Hi were produced in the presence and absence of Vhs, as shown by Western blots of unfractionated lysates from transfected cells (Fig. 3A, bottom panel). However, they were precipitated by Vhs-specific antiserum only from lysates of cells that also expressed Vhs (Fig. 3A, top panel; lanes 1 and 3). A faint band was observed in all four lanes that migrates slightly faster than eIF4Hi and more slowly than eIF4H (Fig. 3A, top panel). This band was judged to be due to nonspecific interaction of a cellular protein with the anti-HA monoclonal antibody on Western blots. Transfection of cells with a mixture of plasmids encoding Vhs, eIF4H, and eIF4Hi resulted in the precipitation of approximately equal amounts of eIF4H and eIF4Hi, suggesting that the 20-amino-acid insertion that is present in eIF4Hi had little effect upon its ability to interact with Vhs.
FIG. 3.
Coimmunoprecipitation of Vhs and epitope-tagged eIF4H from mammalian cells. (A and B) Vero cells were infected with the recombinant vaccinia virus vTF7-39 and transfected with plasmids expressing HA-tagged eIF4Hi (lanes 1); Vhs (lanes 2); HA-tagged eIF4Hi and Vhs (lanes 3); or HA-tagged eIF4Hi, HA-tagged eIF4H, and Vhs (lane 4, panel A). Cell extracts were prepared and immunoprecipitated (upper gels) using Vhs-specific antiserum (A) or HA-specific monoclonal antibody (B). Immunoprecipitates (IP) were analyzed for coprecipitated proteins by Western blotting (upper gels) using the reciprocal antibody—either the HA-specific monoclonal antibody (A) or Vhs-specific antiserum (B). To check for protein expression, an aliquot of the cell lysate was analyzed directly by Western blotting (lower gels) using HA-specific monoclonal antibody (A) or Vhs-specific antiserum (B).
When the immunoprecipitates were prepared using anti-HA monoclonal antibody and probed on Western blots with anti-Vhs antiserum, a substantially higher background was observed on all lanes of the gel (Fig. 3B, upper panel). Nevertheless, Vhs was observed only in precipitates from cells that also expressed HA-tagged eIF4Hi (Fig. 3B, upper panel; compare lanes 2 and 3). Taken together, the results indicate that Vhs interacts with eIF4H and eIF4Hi in mammalian cells.
Some inactive Vhs mutants fail to bind eIF4H.
If this interaction between Vhs and eIF4H is important for Vhs activity, one would expect that some mutant Vhs polypeptides, which do not induce mRNA decay, would also fail to bind eIF4H. To test this prediction, we screened a panel of Vhs mutants for the ability to interact with eIF4H using the conventional two-hybrid assay or a recombination-based two-hybrid system as diagrammed in Fig. 4. Each of the mutants is unable to stimulate mRNA decay, either during a virus infection or in transfected cells (10, 29, 32; Everly et al., unpublished). The mutant proteins were also tested for binding of GST-eIF4H in a GST pull-down assay. All of the assays yielded similar results (Fig. 5).
Removing 88 amino acids from the amino terminus [K(89–489)] or 36 amino acids from the carboxyl terminus [K(1–453)] abolished the ability of Vhs to interact with eIF4H either in the two-hybrid system or in the GST pull-down assay (Fig. 5). Whether this was because the domain of Vhs that interacts with eIF4H is formed by folding together of the two termini of the protein or simply because these deletions cause misfolding of the protein is unknown. However, a Vhs polypeptide containing only amino acids 1 through 382 of the wild-type protein retains enough of its normal interactions with other proteins to be packaged into virions (29).
We also examined a collection of Vhs point mutants (Fig. 5). Two of these, T214I and R435H, are spontaneous mutants isolated on the basis of their inability to induce mRNA decay. Both mutants were significantly compromised with regard to their ability to bind eIF4H. T214I is the mutation carried by the mutant virus Vhs1 (28, 32). The mutant polypeptide is incorporated into virions (33) and induces residual nuclease activity in in vitro translation reactions (23), indicating that it retains at least some of its normal activities. Ten other mutants (D34N, D82N, E192Q, D194N, D195N, T211S, T211A, D213N, D215N, and D261N) were constructed by site-directed mutagenesis. These mutants were designed to alter key residues that are conserved between Vhs and a family of cellular nucleases, with the anticipation that the changes would abolish the nuclease activity of Vhs (Everly et al., unpublished). Nine of the mutants lack the ability to induce mRNA decay yet are still able to bind eIF4H. One of the mutants, T211A, does not stimulate mRNA degradation and does not bind eIF4H. Interestingly, a more conservative change at the same residue (T211S) results in an inactive Vhs polypeptide that still binds eIF4H. The observation that three Vhs point mutants which lack mRNA-degradative activity fail to bind eIF4H strongly suggests that an interaction between the two proteins is important for Vhs activity.
eIF4H mutants that fail to bind Vhs.
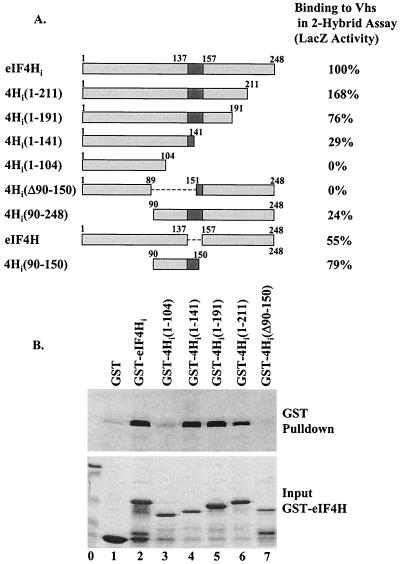
To investigate the domain of eIF4H required for Vhs binding, a series of eIF4H deletion mutants were tested for their ability to interact with Vhs in the yeast two-hybrid and GST pull-down assays (Fig. 6). Shortening eIF4H from the carboxyl terminus to 191 amino acids reduced its ability to bind Vhs only marginally. Further shortening to 141 amino acids reduced binding to Vhs somewhat; although a strong interaction still remained. However, truncation of eIF4H to 104 amino acids abolished detectable binding to Vhs, suggesting that residues between amino acids 105 and 141 are important for the interaction. Consistent with this, deletion of amino acids 90 to 150 abolished the interaction. Deletion of amino acids 1 through 89 reduced binding to Vhs only a few fold, indicating that the first 89 amino acids of eIF4H do not contribute significantly to the interaction. Finally, a fragment containing amino acids 90 through 150 of eIF4Hi resulted in almost as strong a two-hybrid interaction as did full-length eIF4Hi. Amino acids 138 through 150 of this fragment are present in eIF4Hi but not in eIF4H. Since Vhs interacts with both eIF4H and eIF4Hi, a fragment containing amino acids 90 through 137 of eIF4H is sufficient to allow strong interaction with Vhs.
FIG. 6.
Vhs binding by eIF4H mutants. (A) The structures of eIF4H mutants are shown, with residues shared by eIF4H and eIF4Hi shaded light grey and those unique to eIF4Hi shaded dark grey. At the right is the relative binding of each eIF4H protein to Vhs in the conventional yeast two-hybrid assay. Binding is expressed as the amount of lacZ activity resulting from the activation of a GAL4-dependent lacZ gene, where the activity induced by eIF4Hi is defined as 100%. (B) Wild-type Vhs was produced by in vitro translation and analyzed for the ability to bind various fusion proteins containing GST fused to mutant forms of eIF4H. Vhs bound by the various GST-eIF4H polypeptides and eluted with 10 mM glutathione is shown in the upper gel (GST Pulldown). A Coomassie-stained gel of the mutant GST-eIF4H proteins is shown below (Input).
DISCUSSION
These studies demonstrate that the Vhs protein of HSV interacts with the cellular translation initiation factor eIF4H in vitro, in yeast, and in mammalian cells. The biologic importance of this interaction is suggested by the observation that several point mutations in Vhs, which abrogate its ability to induce mRNA decay, also abolish its ability to bind eIF4H.
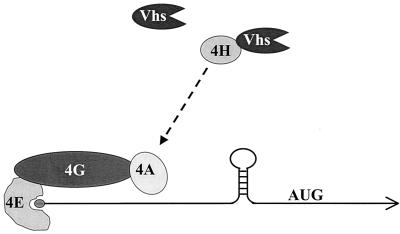
Although in vivo Vhs is targeted to mRNAs and perhaps to regions of translation initiation, the Vhs-dependent nuclease that is observed in extracts of partially purified virions is much less specific (44). Our observation that Vhs binds eIF4H suggests a mechanism for targeting the Vhs activity. eIF4H shares a region of sequence homology with eIF4B and appears to be functionally similar in that both proteins stimulate the RNA helicase of eIF4A, possibly by increasing its processivity (34–36). Along with eIF4E and eIF4G, eIF4A is a component of the tripartite cap binding complex eIF4F (13). Thus, eIF4H appears to act at an early stage of cap-dependent translation initiation to help unwind mRNA secondary structure and facilitate scanning by the small ribosomal subunit (15). The data suggest a model in which binding to eIF4H somehow targets Vhs to mRNAs, as opposed to non-mRNAs, and to regions of translation initiation (Fig. 7).
FIG. 7.
Model for Vhs targeting. eIF4H (4H) stimulates the RNA helicase and RNA-dependent ATPase activities of eIF4A (4A). eIF4A, in turn, is a component of the tripartite cap-binding complex eIF4F. The dashed arrow depicts the functional, and perhaps physical, interaction of eIF4H with eIF4A. Vhs is targeted to mRNAs, as opposed to non-mRNAs, and to regions of translation initiation through its interaction with eIF4H.
While this model is attractive in outline, many of its details remain to be developed and tested. Although the model is based upon known functions of eIF4H and other cellular translation factors, it remains to be proven that binding to eIF4H actually leads to the localization of Vhs near regions of translation initiation. An implication of the model is that targeting of Vhs may be dependent not only on its interaction with eIF4H but also on a series of protein-protein and protein-RNA interactions involving other translation factors. Thus, Vhs binds eIF4H, which in turn interacts functionally, and perhaps physically, with eIF4A. eIF4A binds eIF4G, which interacts with eIF4E, which binds the mRNA cap. Whether all or only some of these interactions are required for efficient targeting of Vhs, or whether other protein-protein or protein-RNA interactions are also required, remains to be determined. In addition, it is interesting to speculate whether the Vhs-eIF4H interaction inhibits the normal activity of eIF4H. If this is the case, Vhs may affect gene expression both by degrading mRNA and by inhibiting the rate of translation initiation.
These results also are significant because of the light they shed upon mechanisms of eukaryotic mRNA decay in general. In eukaryotes, the regulated degradation of many mRNAs is dependent upon their being translated (14, 39). The observation that a viral mRNA degradation protein binds a cellular translation initiation factor provides a novel example of one way in which this linkage between mRNA decay and translation can be accomplished.
Although it is clear that Vhs induces mRNA turnover, it is uncertain whether it is itself a nuclease or, instead, somehow activates a cellular enzyme. Data suggesting that Vhs is an RNase include the observation that the Vhs homologues of alpha herpesviruses share sequence homologies and a number of key conserved residues with a family of mammalian, yeast, bacterial, and phage nucleases. For several of these nucleases, point mutations that alter key conserved residues abrogate the nuclease activity, and mutations that alter the corresponding residues of Vhs inactivate its ability to induce mRNA decay (Everly et al., unpublished). While these results are suggestive, the Vhs protein has not been purified and definitively proven to have nuclease activity. However, recent progress has been made in this area by taking advantage of the interaction between Vhs and eIF4H. In our laboratory, initial attempts to express Vhs in bacteria resulted in the production of Vhs protein that was insoluble, except in buffers containing high concentrations of guanidine hydrochloride or urea. However, coexpression of Vhs and a GST-eIF4H fusion protein resulted in the formation of soluble complexes of Vhs–GST-eIF4H that could be isolated by binding to glutathione-Sepharose and subsequent ion exchange chromatography. Complexes containing the wild-type Vhs protein were found to have RNase activity, while complexes containing either of two mutant forms of Vhs, which lack mRNA-degradative activity in mammalian cells but still bind eIF4H, lacked detectable nuclease activity (Everly et al., unpublished). The results indicate that Vhs indeed is an RNase, either by itself or as a complex with eIF4H. Interestingly, Lu and coworkers recently reported that extracts from yeast that expressed wild-type Vhs lacked detectable RNase activity but that RNase activity was observed after the extracts were supplemented with rabbit reticulocyte lysates (22). This observation is consistent with the possibility that one or more mammalian factors, such as eIF4H, are required to activate the nuclease activity of Vhs. However, a number of alternative interpretations exist. Additional characterization of purified Vhs and Vhs-eIF4H complex are required.
At present it is unclear how many times the Vhs nuclease cleaves an mRNA. Cleavage at a single site near the 5′ end would be sufficient to inhibit further cap-dependent translation, while cellular 5′-to-3′ and 3′-to-5′ exonucleases exist which, in theory, could degrade the resulting fragments. Alternatively, Vhs may cleave mRNAs multiple times at sites that are progressively closer to the 3′ end. Data from Elgadi and coworkers suggest that this may occur in rabbit reticulocyte lysates containing in vitro-translated Vhs (7). Whether it occurs in vivo remains to be determined.
The present results suggest obvious models for the role of eIF4H in Vhs-mediated degradation of mRNAs that are undergoing cap-dependent translation. However, Vhs also has been shown to induce endonuclease cleavage of mRNAs downstream from a picornavirus IRES in vitro (8, 23). Whether eIF4H is required for Vhs-directed decay of IRES-containing mRNAs is unclear. At present, it is unknown whether eIF4H is required for IRES-dependent translation. However, with the notable exception of eIF4E, many of the initiation factors that are required for cap-dependent translation also are required for initiation from a picornavirus IRES (16). If eIF4H is involved in initiation from picornavirus IRESs, it may play a similar role in the Vhs-mediated degradation of these mRNAs and those undergoing cap-dependent translation. However, Lu and coworkers recently reported that a Vhs polypeptide containing the T214I point mutation induces a residual amount of IRES-directed endonuclease activity in the rabbit reticulocyte in vitro degradation system (23). Interestingly, this mutation greatly diminishes binding of Vhs to eIF4H (Fig. 5). One possibility is that the T214I polypeptide retains a residual amount of eIF4H binding activity, which is sufficient for the residual amount of IRES-directed cleavage that is observed in vitro. Alternatively, Vhs may recognize IRES elements directly or through a cellular factor other than eIF4H. These and other questions are under investigation.
ACKNOWLEDGMENTS
We thank Stan Person for helpful discussions concerning the yeast two-hybrid system. Lindsey Hutt-Fletcher provided plasmids and virus, as well as invaluable and friendly advice concerning the expression of epitope-tagged proteins in mammalian cells. We are indebted to Kelley Thomas and Krys Morris at the UMKC Molecular Biology Core Facility for sequencing mutant Vhs and eIF4H alleles. Finally, we thank Marino Martinez-Carrion for inspirational leadership.
This work was supported by grant AI21501 from the National Institute of Allergy and Infectious Diseases and by a grant from the University of Missouri Research Board.
REFERENCES
- 1.Chernokalskaya E, Dubell A N, Cunningham K S, Hanson M N, Dompenciel R E, Schoenberg D R. A polysomal ribonuclease involved in the destabilization of albumin mRNA is a novel member of the peroxidase gene family. RNA Publ RNA Soc. 1998;4:1537–1548. doi: 10.1017/s1355838298980451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cunningham K S, Dodson R E, Nagel M A, Shapiro D J, Schoenberg D R. Vigilin binding selectively inhibits cleavage of the vitellogenin mRNA 3′-untranslated region by the mRNA endonuclease polysomal ribonuclease 1. Proc Natl Acad Sci USA. 2000;97:12498–12502. doi: 10.1073/pnas.220425497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cunningham K S, Hanson M N, Schoenberg D R. Polysomal ribonuclease 1 exists in a latent form on polysomes prior to estrogen activation of mRNA decay. Nucleic Acids Res. 2001;29:1156–1162. doi: 10.1093/nar/29.5.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desai P, Person S. Molecular interactions between the HSV-1 capsid proteins as measured by the yeast two-hybrid system. Virology. 1996;220:516–521. doi: 10.1006/viro.1996.0341. [DOI] [PubMed] [Google Scholar]
- 5.Doherty A J, Serpell L C, Pointing C P. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dompenciel R E, Garnepudi V R, Schoenberg D R. Purification and characterization of an estrogen-regulated Xenopus liver polysomal nuclease involved in the selective destabilization of albumin mRNA. J Biol Chem. 1995;270:6108–6118. doi: 10.1074/jbc.270.11.6108. [DOI] [PubMed] [Google Scholar]
- 7.Elgadi M M, Hayes C E, Smiley J R. The herpes simplex virus vhs protein induces endoribonucleolytic cleavage of target RNAs in cell extracts. J Virol. 1999;73:7153–7164. doi: 10.1128/jvi.73.9.7153-7164.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elgadi M M, Smiley J R. Picornavirus internal ribosome entry site elements target RNA cleavage events induced by the herpes simplex virus virion host shutoff protein. J Virol. 1999;73:9222–9231. doi: 10.1128/jvi.73.11.9222-9231.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Everly D N, Jr, Read G S. Mutational analysis of the virion host shutoff gene (UL41) of herpes simplex virus (HSV): characterization of HSV type 1 (HSV-1)/HSV-2 chimeras. J Virol. 1997;71:7157–7166. doi: 10.1128/jvi.71.10.7157-7166.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everly D N, Jr, Read G S. Site-directed mutagenesis of the virion host shutoff gene (UL41) of herpes simplex virus (HSV): analysis of functional differences between HSV type 1 (HSV-1) and HSV-2 alleles. J Virol. 1999;73:9117–9129. doi: 10.1128/jvi.73.11.9117-9129.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenwick M L, McMenamin M M. Early virion-associated suppression of cellular protein synthesis by herpes simplex virus is accompanied by inactivation of mRNA. J Gen Virol. 1984;65:1225–1228. doi: 10.1099/0022-1317-65-7-1225. [DOI] [PubMed] [Google Scholar]
- 12.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–247. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 13.Gingras A C, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 14.Grosset C, Chen C Y, Xu N, Sonenberg N, Jacquemin-Sablon H, Shyu A B. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell. 2000;103:29–40. doi: 10.1016/s0092-8674(00)00102-1. [DOI] [PubMed] [Google Scholar]
- 15.Hershey J W B, Merrick W C. Pathway and mechanism of initiation of protein synthesis. In: Sonenberg N, Hershey J W B, Mathews M B, editors. Translational control of gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 33–89. [Google Scholar]
- 16.Jackson R J. Translation initiation on picornavirus RNA. In: Sonenberg N, Hershey J W B, Mathews M B, editors. Translational control of gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 869–900. [Google Scholar]
- 17.Karr B M, Read G S. The virion host shutoff function of herpes simplex virus degrades the 5′ end of a target mRNA before the 3′ end. Virology. 1999;264:195–204. doi: 10.1006/viro.1999.9986. [DOI] [PubMed] [Google Scholar]
- 18.Krikorian C R, Read G S. An in vitro mRNA degradation system to study the virion host shutoff function of herpes simplex virus. J Virol. 1991;65:112–122. doi: 10.1128/jvi.65.1.112-122.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwong A D, Frenkel N. Herpes simplex virus-infected cells contain a function(s) that destabilizes both host and viral mRNAs. Proc Natl Acad Sci USA. 1987;84:1926–1930. doi: 10.1073/pnas.84.7.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong A D, Kruper J A, Frenkel N. Herpes simplex virus virion host shutoff function. J Virol. 1988;62:912–921. doi: 10.1128/jvi.62.3.912-921.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Q, Turk S M, Hutt-Fletcher L M. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J Virol. 1995;69:3987–3994. doi: 10.1128/jvi.69.7.3987-3994.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu P, Jones F E, Saffran H A, Smiley J R. Herpes simplex virus virion host shutoff protein requires a mammalian factor for efficient in vitro endoribonuclease activity. J Virol. 2001;75:1172–1185. doi: 10.1128/JVI.75.3.1172-1185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu P, Saffran H A, Smiley J R. The vhs1 mutant form of herpes simplex virus virion host shutoff protein retains significant internal ribosome entry site-directed RNA cleavage activity. J Virol. 2001;75:1072–1076. doi: 10.1128/JVI.75.2.1072-1076.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martindale D W, Wilson M D, Wang D, Burke R D, Chen X, Duronio V, Koop B F. Comparative genomic sequence analysis of the Williams syndrome region (LIMK1-RFC2) of human chromosome 7q11.23. Mamm Genome. 2000;11:890–898. doi: 10.1007/s003350010166. [DOI] [PubMed] [Google Scholar]
- 25.Mitchell P, Tollervey D. mRNA stability in eukaryotes. Curr Opin Genet Dev. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 26.Moss B, Elroy-Stein O, Mizukami T, Alexander W A, Fuerst T R. New mammalian expression vectors. Nature. 1990;348:91–92. doi: 10.1038/348091a0. [DOI] [PubMed] [Google Scholar]
- 27.Oroskar A A, Read G S. A mutant of herpes simplex virus type 1 exhibits increased stability of immediate-early (alpha) mRNAs. J Virol. 1987;61:604–606. doi: 10.1128/jvi.61.2.604-606.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oroskar A A, Read G S. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J Virol. 1989;63:1897–1906. doi: 10.1128/jvi.63.5.1897-1906.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pak A S, Everly D N, Knight K, Read G S. The virion host shutoff protein of herpes simplex virus inhibits reporter gene expression in the absence of other viral gene products. Virology. 1995;211:491–506. doi: 10.1006/viro.1995.1431. [DOI] [PubMed] [Google Scholar]
- 30.Petermann R, Mossier B M, Aryee D N, Kovar H. A recombination based method to rapidly assess specificity of two-hybrid clones in yeast. Nucleic Acids Res. 1998;26:2252–2253. doi: 10.1093/nar/26.9.2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Read G S. Control of mRNA stability during herpes simplex virus infections. In: Harford J B, Morris D R, editors. mRNA metabolism and post-transcriptional gene regulation. New York, N.Y: Wiley-Liss, Inc.; 1997. pp. 311–321. [Google Scholar]
- 32.Read G S, Frenkel N. Herpes simplex virus mutants defective in the virion-associated shutoff of host polypeptide synthesis and exhibiting abnormal synthesis of alpha (immediate-early) polypeptides. J Virol. 1983;46:498–512. doi: 10.1128/jvi.46.2.498-512.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Read G S, Karr B M, Knight K. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J Virol. 1993;67:7149–7160. doi: 10.1128/jvi.67.12.7149-7160.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richter N J, Rogers G W, Ensold J O H, Merrick W C. Further biochemical and kinetic characterization of human eukaryotic initiation factor 4H. J Biol Chem. 1999;274:35415–35424. doi: 10.1074/jbc.274.50.35415. [DOI] [PubMed] [Google Scholar]
- 35.Richter-Cook N J, Dever T E, Ensold J O H, Merrick W C. Purification and characterization of a new eukaryotic protein translation factor—eukaryotic initiation factor 4H. J Biol Chem. 1998;273:7579–7587. doi: 10.1074/jbc.273.13.7579. [DOI] [PubMed] [Google Scholar]
- 36.Rogers G W, Jr, Richter N J, Merrick W C. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 37.Ross J. mRNA stability in mammalian cells. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schek N, Bachenheimer S L. Degradation of cellular mRNAs induced by a virion-associated factor during herpes simplex virus infection of Vero cells. J Virol. 1985;55:601–610. doi: 10.1128/jvi.55.3.601-610.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwartz D C, Parker R. Interaction of mRNA translation and mRNA degradation in Saccharomyces cerevisiae. In: Sonenberg N, Hershey J W B, Mathews M B, editors. Translational control of gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2000. pp. 807–826. [Google Scholar]
- 40.Smibert C A, Johnson D C, Smiley J R. Identification and characterization of the virion-induced host shutoff product of herpes simplex virus gene UL41. J Gen Virol. 1992;73:467–470. doi: 10.1099/0022-1317-73-2-467. [DOI] [PubMed] [Google Scholar]
- 41.Sorenson C M, Hart P A, Ross J. Analysis of herpes simplex virus-induced mRNA destabilizing activity using an in vitro mRNA decay system. Nucleic Acids Res. 1991;19:4459–4465. doi: 10.1093/nar/19.16.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strom T, Frenkel N. Effects of herpes simplex virus on mRNA stability. J Virol. 1987;61:2198–2207. doi: 10.1128/jvi.61.7.2198-2207.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tucker M, Parker R. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu Rev Biochem. 2000;69:571–595. doi: 10.1146/annurev.biochem.69.1.571. [DOI] [PubMed] [Google Scholar]
- 44.Zelus B D, Stewart R S, Ross J. The virion host shutoff protein of herpes simplex virus type 1: messenger ribonucleolytic activity in vitro. J Virol. 1996;70:2411–2419. doi: 10.1128/jvi.70.4.2411-2419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]