Abstract
The mechanism of human-to-human transmission of the polyomaviruses JC virus (JCV) and BK virus (BKV) has not been firmly established with regard to possible human exposure. JCV and BKV have been found in sewage samples from different geographical areas in Europe, Africa, and the United States, with average concentrations of 102 to 103 JCV particles/ml and 101 to 102 BKV particles/ml. Selected polyomavirus-positive sewage samples were further characterized. The JCV and BKV present in these samples were identified by sequencing of the intergenic region (the region found between the T antigen and VP coding regions) of JCV and the VP1 region of BKV. The regulatory region of the JCV and BKV strains found in sewage samples presented archetypal or archetype-like genetic structures, as described for urine samples. The stability (the time required for a 90% reduction in the virus concentration) of the viral particles in sewage at 20°C was estimated to be 26.7 days for JCV and 53.6 days for BKV. The presence of JCV in 50% of the shellfish samples analyzed confirmed the stability of these viral particles in the environment. BKV and JCV particles were also found to be stable at pH 5; however, treatment at a pH lower than 3 resulted in the detection of free viral DNA. Since most humans are infected with JCV and BKV, these data indicate that the ingestion of contaminated water or food could represent a possible portal of entrance of these viruses or polyomavirus DNA into the human population.
JC virus (JCV) and BK virus (BKV) are nonenveloped, icosahedral viruses with double-stranded, negatively supercoiled, circular DNA genomes of approximately 5.13 kb. The polyomavirus genome consists of an early region, a late region, and a regulatory region (RR) containing promoters, enhancers, and the replication origin. The genome is transcribed bidirectionally from the origin. It codes for the early region proteins (large T and small t antigens) that regulate the transcription of the late region proteins (VP1, VP2, VP3, and agnoprotein). JCV and BKV are associated primarily with progressive multifocal leukoencephalopathy (PML) and hemorrhagic cystitis, respectively, and a role for these viruses in human cancer has been suggested (23). Both viruses are found at high frequencies throughout most human populations, and their pathogenicity, which is associated primarily with immunocompromised states, has attracted more attention due to the immunosuppression associated with AIDS. As determined by seroconversion, primary infection with BKV occurs early in childhood, while JCV infection occurs slightly more toward adolescence (17, 36, 51). Initial infection is not apparent and rarely causes clinical disease, although respiratory symptoms or urinary tract disease is sometimes found in the case of BKV (18, 21, 37). JCV and BKV can be detected in tonsillar tissue (19, 32), and the hypothesis that the respiratory tract is the primary site of viral infection has been suggested. After the initial infection, the virus disseminates and establishes a persistent infection in renal tissue throughout life (27, 46). The presence of JCV DNA in the upper and lower parts of the human gastrointestinal tract and particularly in the mucosa of the human colon and colorectal cancers has been recently described (29, 42).
In a previous study, using nested PCR methods that we developed for studying viruses in wastewater (39, 40), we reported the presence of human polyomavirus JCV and BKV DNAs, but not simian virus 40 DNA, in urban sewage (12). In this study, we report the presence of JCV and BKV in sewage from other geographical areas. In addition, we have detected JCV and BKV in shellfish, have evaluated the stability of JCV and BKV virions in the environment, and have begun to determine the genetic characteristics of the strains excreted by populations in different geographical areas. Human adenoviruses (HAd) were used in this study as a control for human contamination of the samples. Also, shellfish were used as biosensors for contamination studies since they filter large volumes of water, concentrating, especially in the digestive tract, any viruses that are present in the water. We believe that viruses excreted in urine and feces are transmitted through what is known as fecal contamination, which includes viruses excreted in feces and urine. Our findings of high levels of JCV and BKV in most sewage samples and the relative stability of these viruses under environmental conditions suggest that the alimentary tract could be an important point of exposure and transmission of these viruses among humans.
MATERIALS AND METHODS
Viruses.
BKV and JCV positive controls were obtained from the urine of a healthy woman who had been pregnant for 38 weeks. We also used JCV obtained from cerebrospinal fluid (CSF) samples kindly donated by J. L. Pérez, Microbiology Department, Hospital de Bellvitge, Barcelona, Spain. Adenovirus type 2 (prototype) grown on A549 cells and partially purified was used as a positive control.
Sewage samples.
Twenty-four raw sewage samples from different geographical areas were analyzed. Nine samples (BCN17 to BCN25) were collected from October 1998 to June 2000 in the sewers of Barcelona, Spain. Each sample was collected in a sterile 500-ml polyethylene container, kept at 4°C for less than 8 h until the viral particles were concentrated in phosphate-buffered saline (PBS), and stored at −80°C.
Five samples (G1 to G5) were collected in Patras, Greece, during June and July 1999. Four samples (E1 to E4) were collected in Cairo, Egypt, during July 1999, and six samples (W1 to W6) were collected in Washington, D.C., during December 1999. These samples were collected and shipped, frozen, to Spain, where they were concentrated in PBS and stored at −80°C.
Shellfish samples.
Six oyster (Crassostrea gigas) samples and four mussel (Mytilus galloprovincialis) samples were obtained from shellfish-growing in areas with different levels of fecal pollution located in the delta of the Ebro river. Shellfish sample analysis was carried out by following a slightly modified form of the protocol of Pina et al. (39) and Muniain-Mujika et al. (33). The digestive diverticulum was the only part of the animals tested in this analysis. These samples were analyzed for the presence of different human viruses, including: hepatitis A virus, enteroviruses, HAd, JCV, and BKV.
Concentration of viral particles and nucleic acid extraction.
Recovery of viral particles and nucleic acid extraction were carried out as described previously (40). Briefly, 40-ml sewage samples were ultracentrifuged (229,600 × g for 1 h at 4°C) to pellet all of the viral particles together with any suspended material. The sediment was then eluted by mixing it with 4 ml of 0.25 N glycine buffer (pH 9.5) on ice for 30 min, and the suspended solids were separated by centrifugation at 12,000 × g for 15 min after the addition of 4 ml of 2× PBS. Viruses were finally pelleted by ultracentrifugation (229,600 × g for 1 h at 4°C), resuspended in 0.1 ml of 1× PBS, and stored at −80°C.
Viral nucleic acids were extracted by using a procedure that uses guanidinium thiocyanate and adsorption of the nucleic acids to silica particles (13), providing clean nucleic acids for molecular studies.
Enzymatic amplification.
Ten-microliter aliquots of the extracted nucleic acids were used in each test, corresponding to a 4-ml sewage sample, 1 g of shellfish, 1 ml of urine, or 10 μl of CSF, depending on the origin of the samples analyzed. Serial tenfold dilutions were also analyzed in order to carry out a semiquantitative analysis of the samples studied by limiting-dilution experiments. Knowing the sensitivity of the detection procedure applied, we could estimate the concentration of the sewage samples by considering the lower decimal dilution positive for the nested-PCR assay.
Amplifications were carried out in a 50-μl reaction mixture containing 10 mM Tris-HCl (pH 8.3 at 25°C), 50 mM KCl, 1.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 2 U of Ampli Taq DNA polymerase (Perkin-Elmer Cetus), and the corresponding primers at the corresponding concentrations (25 μM for all polyomavirus amplifications). In all PCR assays, the first cycle of denaturation was carried out for 4 min at 94°C. The conditions for the 29-cycle amplification were as follows: denaturing at 92°C for 60 s, annealing at the corresponding annealing temperature for 60 s, and extension at 72°C for 75 s. All amplifications were completed with 4 min of extension at 72°C. The PCR amplifications of adenovirus genomes were carried out as previously described (12). JCV genomes were amplified by using EP1A and EP2A as external primers and P1A and P2A (modified from Kunitake et al. [28]) as internal primers and an annealing temperature of 59°C in both PCRs. BKV genomes were amplified by using external primers BK1 and BK2 at an annealing temperature of 46°C and internal primers BK4 and BK6 at an annealing temperature of 50°C. All of the primer sequences used in this study are represented in Table 1. The results were analyzed by agarose gel electrophoresis using ethidium bromide as a stain.
TABLE 1.
Oligonucleotide primers used for PCR amplification and sequencing of human polyomaviruses BKV and JCV
| Virus type (region) | Position | Primer | Sequencea |
|---|---|---|---|
| JCV (IGR) | 2062–2087b | EP1A | 5′-TGAATGTTGGGTTCCTGATCCCACC-3′ |
| JCV (IGR) | 2774–2798 | EP2A | 5′-ACCCATTCTTGACTTTCCTAGAGAG-3′ |
| JCV (IGR) | 2099–2124 | P1Ad | 5′-CAAGATATTTTGGGACACTAACAGG-3′ |
| JCV (IGR) | 2742–2766 | P2Ad | 5′-CCATGTCCAGAGTCTTCTGCTTCAG-3′ |
| JCV (IGR) | 2511–2536 | JCSR | 5′-TGATTACAGCATTTTTGTCTGCAAC-3′ |
| JCV (IGR) | 2364–2388 | JCSL | 5′-GGAAGTCCTTCTGTTAATTAAATCAG-3′ |
| JCV (RR) | 4992–5011 | JR1e | 5′-CCCTATTCAGCACTTTGTCC-3′ |
| JCV (RR) | 428–447 | JR2e | 5′-CAAACCACTGTGTCTCTGTC-3′ |
| JCV (RR) | 5060–5079 | JR3e | 5′-GGGAATTTCCCTGGCCTCCT-3′ |
| JCV (RR) | 298–317 | JR4e | 5′-ACTTTCACAGAAGCCTTACG-3′ |
| BKV (VP1) | 1452–1467c | BK1 | 5′-TATTGCCCCAGGAGGT-3′ |
| BKV (VP1) | 2132–2148 | BK2 | 5′-AACATTTTCCCCTCCTG-3′ |
| BKV (VP1) | 1762–1781 | BK4 | 5′-AGTAGATTTCCACAGGTTAG-3′ |
| BKV (VP1) | 1486–1506 | BK6 | 5′-CCAGGGGCAGCTCCCAAAAAG-3′ |
| BKV (VP1) | 1425–1442 | BK3 | 5′-ACTGTAACACCTGCTCTT-3′ |
| BKV (RR) | 5024–5043 | BR1 | 5′-CCCTGTTWARRACTTTATCC-3′ |
| BKV (RR) | 431–457 | BR2 | 5′-GTAAAGCAGTGGTACTTT-3′ |
| BKV (RR) | 5083–5102 | BR3 | 5′-ATAGTTTTGCTAGGCCTCAG-3′ |
| BKV (RR) | 305–322 | BR4 | 5′-CAACTTTCACTGAAGCTT-3′ |
Quality control of the amplification method.
Standard precautions were applied in all of the manipulations in order to reduce the probability of sample contamination by amplified DNA molecules. All of the samples were analyzed twice in independent experiments, and a negative control was added every two samples.
Characterization of the JCV genomes.
The amplicons obtained from the nested PCR when using primers to amplify the intergenic region (IGR) of nine JCV-positive samples (W1 to W6, E1, E2, and G1) and a positive CSF sample (CSFK) were further characterized by amplifying products of the first-round PCR by using primers P1A-JCSR and P2A-JCSL at an annealing temperature of 55°C. That permitted us to amplify the IGR of JCV in two overlapping fragments. Sequencing of these fragments was carried out by using the same two pairs of primers.
Two JCV-positive samples (W5 and G3) and two samples that tested positive for JCV in a previous study (BCN26 and BCN28) were analyzed by using primers suitable for amplification of the RR. JR1 and JR2 were used in the first-round PCR, and JR3 and JR4 were used in the nested PCR, and in both cases, the annealing temperature was 55°C. These primers have been described by Monaco et al. (32). The amplicons obtained from the nested PCR were further characterized by sequencing with primers JR3 and JR4.
Characterization of BKV genomes.
Two BKV-positive samples (W2 and W5), other samples that had tested positive for BKV in a previous study (U3, BCN26, and BCN28), and a control urine sample (BCNU) were analyzed by using primers suitable for amplification of the RR, i.e., BR1 and BR2 at 44°C for the first PCR and BR3 and BR4 at 46°C for the nested PCR. We further characterized these samples by sequencing amplicons obtained from the nested PCR by using primers BR3 and BR4.
The amplicons obtained from the first-strand PCR using primers BK1 and BK2 of five samples, two positive samples (W3 and BCN17), two other positive samples that tested positive for BKV in a previous study (BCN29, BCN30), and the urine sample used as a control (BCNU), were further amplified in a nested PCR using primers BK2 and BK3 (at an annealing temperature of 46°C), which amplify a VP1 region used for the typing of these viruses (24). The amplicons obtained were sequenced by using BK2 and BK3.
Sequencing of nested-PCR products.
Products obtained from the nested PCR were purified with the QIAquick PCR purification kit (QIAGEN, Inc.). Both strands of the purified DNA amplicons were sequenced with the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction kit with Ampli Taq DNA polymerase FS (Perkin-Elmer, Applied Biosystems) by following the manufacturer's instructions. The conditions for the 25-cycle sequencing amplification were as follows: denaturing at 96°C for 10 s, annealing at 50°C for 5 s, and extension at 60°C for 4 min. The primers used for sequencing were used at a concentration of 2.5 μM. The results were checked by using the ABI PRISM 377 automated sequencer (Perkin-Elmer, Applied Biosystems). The sequences were compared with the GenBank and EMBL databases by using the basic BLAST program of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/BLAST/). Alignments of the sequences were carried out by using the ClustalW program of the European Bioinformatics Institute of the European Molecular Biology Laboratory (http://www.ebi.ac.uk/clustalw/).
Phylogenetic analysis of JCV.
Sequenced regions of JCV were analyzed by using the seqboot and neighbor programs included in the PHYLIP software package (15). The phylogenetic tree constructed by the neighbor-joining (NJ) method and bootstrap analysis was visualized by using the TREEVIEW 1.5 program (38). The JCV isolates used in the phylogenetic analysis are described in Table 2. The NJ method was also applied for analysis of the full sequences of 20 of the strains described in Table 2 (the 20 classified into types and subtypes by Jobes et al. [25]) and the partial 461-nucleotide (nt) sequence used in our analysis.
TABLE 2.
JCV types used for phylogenetic analysisa
| Strain(s) | Origin (ethnicity)b | GenBank accession no(s). | Reference | Type |
|---|---|---|---|---|
| Mad-1 | United States (C) | J0227 | 16 | 1 |
| 124 | United States (C) | AF015526 | 4 | 1 |
| 123 | United States (C) | AF015227 | 4 | 1 |
| Tokyo-1 | Japan | AF030085 | 7 | 2A |
| 224 | United States (H) | AF015529 | 7 | 2A |
| 225 | United States (NA) | AF015530 | 7 | 2A |
| 226 | United States (NA) | AF015531 | 7 | 2A |
| 223 | United States (AA) | AF015532 | 7 | 2B |
| 227 | United States (C) | AF015533 | 7 | 2B |
| GS/K | Germany | AF004349 | 30 | 2B |
| GS/B | Germany | AF004350 | 30 | 2B |
| 228 | United States (NA) | AF015534 | 7 | 2C |
| 229 | United States (C) | AF015535 | 7 | 2C |
| 230 | United States (AA) | AF015536 | 7 | 2C |
| 308 | Tanzania | U73500 | 2 | 3 |
| 312 | United States (AA) | U73502 | 2 | 3 |
| 311 | United States (AA) | U73501 | 2 | 3 |
| 402 | United States (C) | AF015528 | 4 | 4 |
| 601 | United States (AA) | AF015537 | 6 | 6 |
| Tai-3 | Taiwan | U61771 | 35 | 7 |
| BCNU-2, -16, -8, -15 | Spain | AF119345–49 | 12 | |
| U3 | Sweden | AF119350 | 12 | |
| F2 | France | AF119351 | 12 | |
| P3, P1 | South Africa | AF119352–53 | 12 | |
| CSFB, -E, -J | Spain | AF119354–56 | 12 | |
| CSFK | Spain | AF304389 | This paper | |
| G1 | Greece | AF303943 | This paper | |
| E1–2 | Egypt | AF303944–45 | This paper | |
| W1–3/W4–6 | United States | AF303946–48/AF304386–88 | This paper |
Strains and genomes used to construct the phylogentic tree are represented. The geographic origins of the strains are shown. Some data are from the work of Jobes et al. (25).
C, Caucasian; H, Hispanic; NA, Native American; AA, African American.
Stability of human polyomavirus and HAd particles in sewage.
One liter each of three sewage samples (BCN23, BCN24, and BCN25) was kept in a sterile beaker in a heater at 20°C. One liter of PBS spiked with 84 ml of urine from a healthy woman pregnant for 38 weeks that was positive for both JCV and BKV was used as a positive control. Aliquots of 40 ml of each sample were analyzed within a 5-month period for the presence of JCV and BKV on days 0, 3, 10, 17, 30, 45, 62, 78, 92, 120, and 150. The presence of HAd was also analyzed on days 30, 60, and 92. The presence of viral particles was evaluated, and in order to find out if the viral particles had remained intact, viral concentrates were treated with DNase before nucleic acid extraction. One aliquot of each sample was treated with DNase with the appropriate buffer in order to destroy free DNA, and the other aliquot was treated with DNase buffer but no DNase for analysis of total DNA under equivalent conditions. Twenty-five microliters of DNase buffer containing Tris-HCl (pH 7.5) at 100 mM, MgCl2 at 20 mM, bovine serum albumin at 100 μg/ml, 25 μl of viral particles, and 1 μl of DNase I (Amersham Pharmacia Biotech Inc.) at 10,000 U/ml (DNase treatment) or 1 μl of sterile water was mixed before nucleic acid extraction.
In order to obtain the estimated t90 and t99 (times required for 90 and 99% reductions of the viral concentration), we computed a linear regression model with the logarithm of the estimated concentration of viral particles detected by nested PCR expressed as PCR genomic equivalents. More precisely, the model is log yt = at + log y0, where yt is the mean of the three observed values of y at time t, y0 is the value of the PCR genomic equivalents at time zero, and a is the slope of the regression line. The log-transform is suggested by the proportional relationship between the mean and the standard deviation of the PCR genomic equivalents at every time and is strongly supported by inspection of the residuals of the model. However, the usual inferences in linear regression (i.e., signification test of correlation, confidence intervals, etc.) are not applicable because of the known dependence of the observed values of the PCR genomic equivalents in the stability assays. For our purposes, the model attempts to give an approximate (descriptive) approach to t90 and t99. Inverting the equation described above, estimates of t90 and t99 are computed by substituting for yt 10 and 1%, respectively, of the observed value of yt at time zero (t0).
Stability of JCV and BKV particles at acidic pHs.
A sewage sample at pH 7 to 7.5 was divided into three aliquots, and their pHs were adjusted to 1, 3, and 5 with 35% HCl. The samples were magnetically stirred at room temperature. After 30 min, the pH was neutralized with 1 M NaOH. The samples were then concentrated and treated with DNase or left untreated before nucleic acid extraction. This assay was repeated while keeping the samples at a low pH for 1.30 h.
Nucleotide sequence accession numbers.
The JCV IGR sequences reported in this paper have been deposited in the GenBank database under accession no. AF303943 to AF303948 and AF304386 to AF304389. The JCV RR sequences obtained in this study are identical to those described by Bofill-Mas et al. (12) (accession no. AF120242; archetypal JCV RR sequence obtained from sewage samples). The BKV RR sequences analyzed have been deposited under accession no. AF356528 to AF356531, and the BKV VP1 region sequences have been deposited under accession no. AF356534 to AF356538.
RESULTS
Human polyomaviruses in sewage and shellfish samples.
Viruses excreted by people in their feces or urine are found in urban sewage and seawater with fecal contamination. Since shellfish filter large volumes of seawater, viruses that are present in water accumulate and remain in shellfish digestive tissues. Thus, analysis of the viruses present in urban sewage and shellfish samples will give information about the excretion patterns of polyomaviruses in specific communities. The sensitivity of the nested PCR assay was tested by following the procedure applied in a previous study (11, 12). The observed sensitivity of the nested PCR was 5 JCV genome copies and 50 BKV genome copies. A sensitivity of 1 to 10 HAd particles has been reported previously (40).
The results obtained in the limiting-dilution experiments when analyzing sewage samples collected in the United States, Egypt, Greece, and Spain are shown in Table 3 and reflect a very high level of excretion of JCV, BKV, and HAd. All of the samples from these areas were positive for HAd, JCV, and BKV.
TABLE 3.
Summary of results obtained by analysis of 52 sewage samples for the presence of human polyomaviruses
| Area (no. of samples) | No. positive/total (no. of viral particles/ml)
|
||
|---|---|---|---|
| HAd | JCV | BKV | |
| Barcelona, Spain (16a + 9) | 24/25 (101–102) | 23/24 (101–102) | 20/25 (1–101) |
| Nancy, France (4)a | 4/4 (101–102) | 4/4 (102–103) | 4/4 (1–101) |
| Umeå, Sweden (4)a | 4/4 (1–101) | 4/4 (102–103) | 4/4 (1–101) |
| Pretoria, South Africa (4)a | 4/4 (102–103) | 4/4 (101–102) | 4/4 (101–102) |
| Cairo, Egypt (4) | 4/4 (102–103) | 4/4 (102–103) | 4/4 (101–102) |
| Patras, Greece (5) | 5/5 (102–103) | 5/5 (102–103) | 4/4 (102–103) |
| Washington, D.C. (6) | 6/6 (102–103) | 6/6 (103–104) | 6/6 (102–103) |
| Total (52) | 51/52 (102–103) | 50/51 (102–103) | 46/51 (101–102) |
Samples analyzed in a previous study (12).
Table 3 also reflects results obtained in a previous study with samples collected in South Africa, France, Sweden, and Spain (12). The total percentage of positive samples and estimated mean values of HAd, JCV, and BKV are represented.
Of the 10 shellfish samples analyzed, 8 were positive for at least one of the human viruses analyzed (HAd or JCV) and the average Escherichia coli level of these samples was 40 bacteria/100 g. These data indicates that these samples had been in contact with fecal contamination of human origin. Seven samples were positive for HAd, with a mean concentration of 1 to 10 viral particles/g. Six samples were positive for hepatitis A virus, with mean concentrations of 10 viral particles/g. Five samples were positive for JCV at concentrations between 1 and 10 viral particles/g. All of the samples tested negative for BKV.
Characterization of the IGR of JCV.
The IGR of JCV has been studied largely as a tool with which to trace human migrations, since it presents differences corresponding to the different geographic origins of the populations that excrete the viruses.
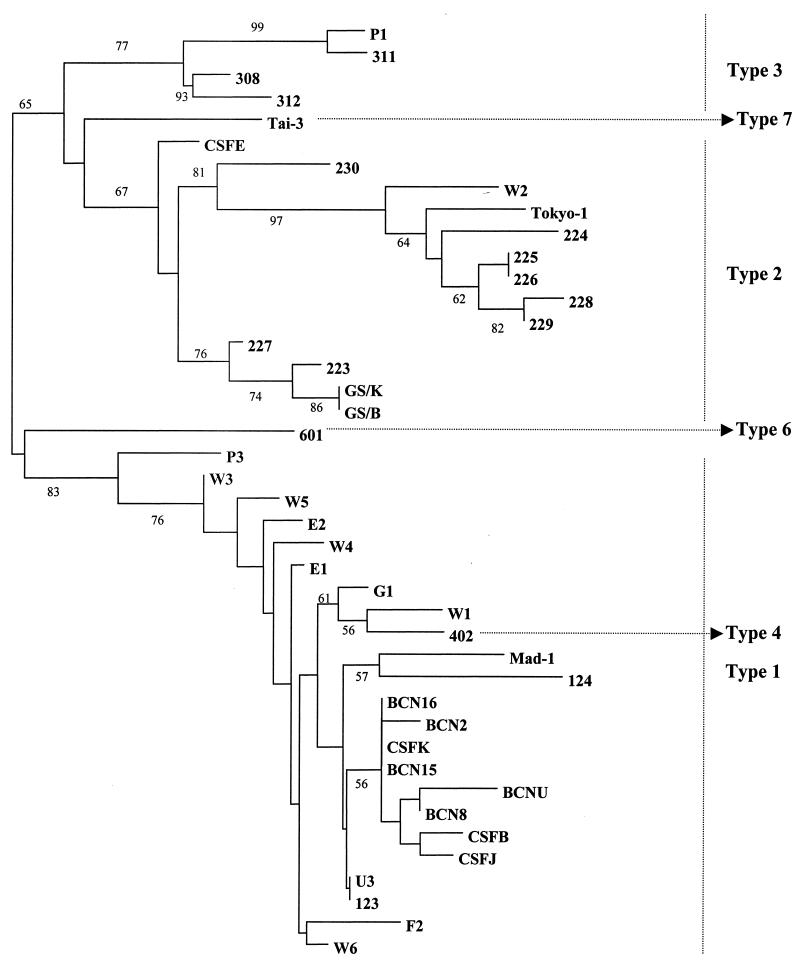
Sequencing of the IGR of JCV classifies the JCV genomes into at least seven types and a larger number of subtypes (25). Type 1 is found in Europeans, types 2 and 7 are found in Asians, and types 3 and 6 are found in Africans (3, 14, 20); type 6 is found in western and central Africa but not in eastern Africa (3, 14, 20). Types 1, 2, and 3 present two or more subtypes (A, B, C… ). The inhabitants of America present genotypes characteristic of other continents. African Americans present mainly type 3, while European Americans present type 1 and Native Americans present type 2A, which is typical of their northeastern Asian origins (8).
We sequenced 461 nt of the IGR of JCV-positive samples. The consistency of the phylogenetic analysis using the 461-bp sequence of the IGR was evaluated by comparing trees obtained by using this 461-bp sequence with phylogenetic trees obtained by using the full-length JCV sequences described in the GenBank and EMBL data banks. The two types of trees showed significant similarity (data not shown).
The sequences analyzed confirmed the specificity of the nested PCR amplification, since all viral sequences were identified as JCV when specific primers were used. These results also confirmed the absence of cross-contamination, since all of the viral sequences analyzed were different from the positive controls used in the assays.
The phylogenetic tree obtained by the NJ method is shown in Fig. 1. The strains used to construct this tree are represented in Table 2.
FIG. 1.
NJ tree constructed to represent phylogenetic relationships among 42 JCV sequences (see Table 3) for nt 2177 to 2637 of the IGR. The bootstrap confidence levels obtained for 100 replicates are shown (only significant values are indicated).
Characterization of the RRs of JCV genomes.
In order to check whether or not the genomes detected presented the archetypal transcriptional control regions (TCRs) excreted by individuals in urine or the rearranged TCRs typically found in the CSF of PML patients, we analyzed the RR sequences from four JCV-positive samples and compared these sequences with sequences previously obtained. All of the sequences obtained from sewage samples and urine samples were identical in these 187 nt and were also identical to the archetypal consensus sequence.
Characterization of BKV genomes.
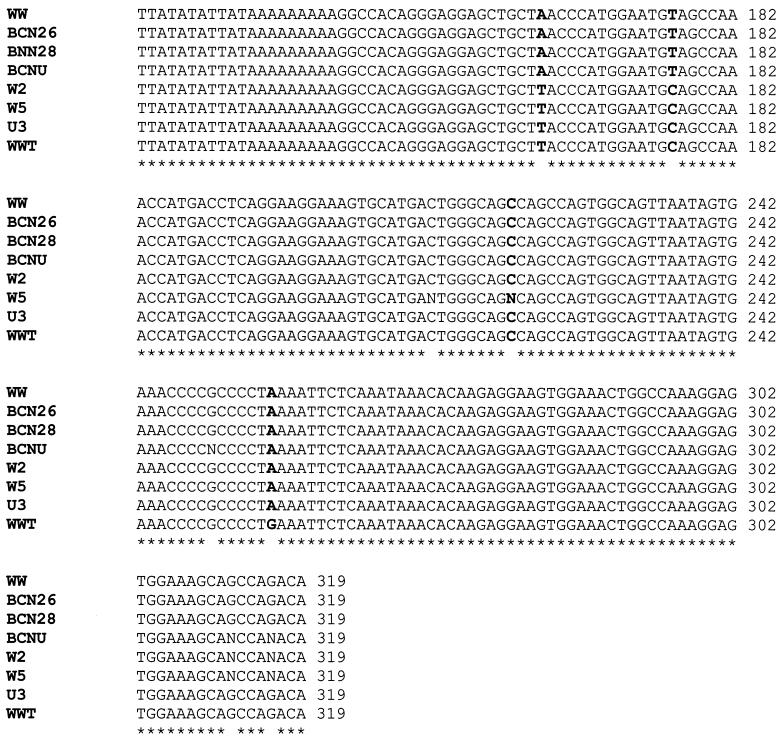
It has been reported that BKV transcriptional RRs also fall into two groups, archetypal and rearranged, depending on their sequence. We sequenced the RRs of some samples positive for BKV and a urine sample used as a control. BKV studied in sewage and urine presented archetypal (WW) or archetype-like (WW-T) structures of the RR, as previously described (43, 49). Alignments of these sequences are shown in Fig. 2. We could establish two groups of archetypal regions that differed in 2 of 180 nt. Samples from the United States and Sweden were almost identical (with the exception of some undetermined nucleotides) and differed by 1 nt from BKWW-T (nt 256, numbering of the Gardner strain). Sewage samples tested from Barcelona and the urine sample used as a control (from a Spanish individual) were identical in the identified nucleotides (three nucleotides in the urine sample could not be defined) and identical to BKWW. The relevant consensus elements, such as protein binding sites and regulatory elements, that have been described by Markowitz et al. (31) were conserved in the genomes analyzed. Differences between the two groups were restricted to nucleotides that present polymorphisms (nt 163 and 176).
FIG. 2.
Annealing of the RRs of some of the BKV-positive samples sequenced (Gardner numbering). Asterisks represent similarity between nucleotides. Bold nucleotides represent sites of diversity in the BKV genome. Archetypal (WW) and archetype-like (WW-T) sequences were also included in the annealing.
To begin typing the BKV present in these sewage samples, we sequenced the VP1 region of five BKV-positive samples. Of the five samples sequenced, four presented the characteristics defined by Jin et al. (24) for BKV type 1, the type most frequently detected in human populations. One of the samples collected in Barcelona (BCN29) seemed to be identical to type 2, the second BKV type most frequent in human populations, with the exception of one nucleotide that corresponded to type 1, 3, or 4 but not type 2. Alignments of the sequences detected are presented in Fig. 2. We also observed some indeterminate nucleotides in the polymorphic sites.
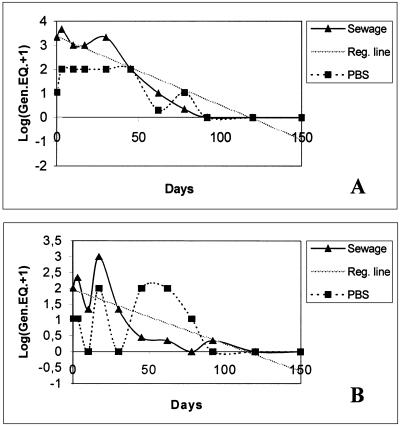
Stability of human polyomaviruses and HAd particles in sewage.
There is no previous information on the behavior of human polyomaviruses in the environment. Data on the stability of these viral particles in the environment could contribute to a better evaluation of their mechanism of transmission. By analyzing the three samples of raw sewage from Barcelona (BCN23, BCN24, and BCN25) and the control (PBS spiked with urine positive for both BKV and JCV), which were all kept at 20°C for 150 days, we found that both BKV and JCV were detected until day 92. The viruses detected during this time appeared to represent intact virions, since treatment with DNase prior to nucleic acid extraction did not eliminate the viral genomes (Fig. 3). The presence of HAd DNA was also analyzed at days 30, 45, and 92 by using DNase. HAd viral particles were detected at days 30 and 45, but disrupted (DNase-sensitive) virions were obtained at day 92.
FIG. 3.
Annealing of the VP1 regions of some of the BKV-positive samples sequenced. Asterisks represent similarity between nucleotides. Bold nucleotides represent sites used for BKV typing.
In evaluating the stability of JCV in sewage at 20°C, we estimated a t90 of 26.7 days and a t99 of 61.5 days. For BKV, the estimated t90 was 53.6 days and the estimated t99 was 96.8 days. These data are approximate and were calculated in accordance with the regression line obtained (Fig. 4).
FIG. 4.
Stability of JCV (A) and BKV (B) in sewage samples. The regression (Reg.) line, the transformed average number of genome equivalents (Gen.EQ+1) detected by nested PCR in the three samples (sewage), and the number of genome equivalents of a spiked PBS control are represented.
Stability of JCV and BKV particles at acidic pH.
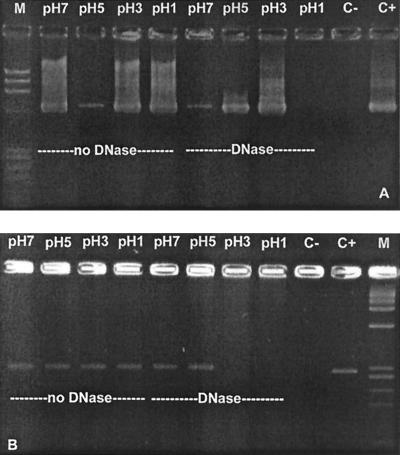
Data on the resistance of polyomaviruses to acidic pH could be useful when considering the gastrointestinal tract as a possible route of transmission of these viruses. Viruses detected in sewage may be ingested through contaminated water or food. After analyzing human polyomaviruses present in a sewage sample treated at pHs 1, 3, 5, and 7 for 30 min, we could detect the DNAs of both JCV and BKV at all of these pHs, including pH 1. When we used DNase before nucleic acid extraction, we found that BKV DNA detected at pHs 1 and 3 was free DNA, implying that the BKV particles were disrupted at these pHs. Free JCV DNA was also found after 90 min at pH 3. However, after 30 min at pH 3, the viral particles appeared to be intact in half of the assays carried out (Fig. 5).
FIG. 5.
Electrophoretic gel showing the bands obtained by analysis of a sewage sample containing JCV (A) and BKV (B) after exposure to different pHs for 30 min. The pH assayed and also the absence or presence of DNase treatment are indicated. Lanes M contained molecular size standards. C−, negative control; C+, positive control.
DISCUSSION
All of the populations in the different geographical areas that we sampled excreted a high number of human polyomaviruses. These high levels of human polyomaviruses in urban sewage probably came from urine, since JCV has been detected in 20 to 80% of adult urine samples, depending on age (26) and ethnic group (8). Our finding of a higher concentration of JCV than BKV in sewage agrees with a study on the genomic sequences of JCV and BKV in 176 urine samples by Shah et al. (47). In that study, 37% JCV and 5% BKV viruria was observed and immunosuppression was associated with a higher frequency of BKV viruria.
The primers applied in this study have proven to be specific, sensitive, and able to detect a wide variety of JCV strains. Sequences obtained from the IGRs of 22 JCV genomes were analyzed in phylogenetic studies, and the clusters obtained were used for classification of the viral genomes. The amplified sequences of 461 nt include the 3′ end of VP1, a noncoding short region of about 69 nt, and the 3′ end of the large T antigen. Hatwell et al. (22) found that this region encloses a large number of sequence differences between strains and that these sequences are useful for comparing JCV genomes of diverse origins. Most of the samples studied presented sequences closely related to European types. Only one genome detected in sewage was found to be closely related to Asian strains, i.e., sample W2, from sewage collected in the Washington, D.C., area. This isolate was classified as type 2A, typical of the northeastern Asian origin of Native Americans. A sequence detected in sewage from South Africa was the only one classified as JCV type 3. Samples G1 and W1, collected in Greece and the United States, respectively, were closely related to strain 402, the prototype of previously described type 4, which is closely related to type 1. The genotypes studied appear to be some of the most abundant genotypes in the population. However, a study of the diversity of types excreted in a specific population would require multiple samples of sewage and cloning and sequencing of the diverse genomes that could be amplified.
The JCV strains obtained around the world fall into distinct types and have been described as markers that may be useful in tracing ancient and modern human migrations (1, 2, 9, 48, 52).
Like JCV, BKV also appears to have two different RR structures, archetypal and rearranged (34, 43, 44). When BKV samples were analyzed to determine whether they contained archetypal regions, we found two different types of archetypal regions similar to those defined as BKWW (archetypal) and BKWW-T (archetypal-like) by Sundsfjord et al. (49) among naturally occurring BKV strains.
The BKV genomes from a few samples were typed by sequencing of the VP1 region (24), and with one exception, all of them were found to be of genomic subtype I, which has been described as the BKV type most frequently detected in the human population.
The high prevalence of human polyomaviruses observed in urban sewage suggests that humans are being exposed at a high frequency to JCV and BKV through the digestive tract when ingesting water and foods, such as vegetables or shellfish, previously exposed to fecal contamination. Passage of the contaminated food through the digestive tract could produce infection in the intestinal epithelium or lymphoid cells if JCV and BKV were as resistant to the proteolytic enzymes of the digestive tract as are other nonenveloped viruses that are excreted. The alimentary tract was proposed by Sundsfjord et al. (50) as a portal of infection by JCV and BKV. After studying nasopharyngeal aspirates in children with respiratory infections and saliva from immunodeficient and immunocompetent adult patients, only BKV DNA (but no infectious BKV) was detected in 2 of 201 nasopharyngeal aspirates. However, other routes of infection by these viruses have not been ruled out, since tonsil tissue has also been suggested as a possible site of initial JCV infection (32). This possibility may also be considered as potentially complementary to entry at the intestinal level because swallowed material passes the mouth and nasopharynx and viruses transmitted through the oral-fecal route are often able to multiply in the throat. Human polyomaviruses are relatively stable at acidic pH, and when ingested with food, viruses may be protected, as has been shown for other intestinal pathogens. Gastric secretion is known to be about pH 1, although the total pH of the stomach is highly variable, depending on the ingested material and the stimuli influencing the secretions. The contents of the stomach always have a pH higher than 4 when they pass into the duodenum. We have observed intact JCV particles after treatment at pH 5 for 90 min and also after treatment at pH 3 for 30 min. The stability of these viral particles was confirmed by treatment with DNase. Viruses could enter humans through the digestive tract by infecting intestinal epithelia, by pinocytic mechanisms in the cells, or through M cells in Peyer's patches. Although archetypal JCV is highly difficult to grown in cell cultures, further studies are currently under way to evaluate the potential infectivity of the excreted polyomaviruses in the gastrointestinal tract.
Moreover, there are other aspects that need to be considered. It has been shown that foreign DNA ingested with food is not completely degraded in the gastrointestinal tract in an animal model and a study by Schubbert et al. (45) suggests that transport of foreign DNA through the intestinal wall and Peyer's patches to peripheral blood leukocytes and into several organs can occur. According to this information, free polyomavirus DNA, either completely or partially digested, could also enter the human body through the gastrointestinal tract. JCV DNA has been described in the epithelium of the upper and lower parts of the gastrointestinal tract (42), and although it has been not proven, this could be a direct consequence of the frequent ingestion of polyomaviruses and the intake of viral particles or viral DNA by the oral route.
JCV strains fall into two groups, designated archetypal and rearranged, based on the structure of their TCRs. The TCR of archetypal JCV contains a single copy of the promoter and enhancer, while the rearranged strains contain deletions and duplications in this region (5, 10, 52, 53). The archetypal strains can be isolated from normal individuals and immunocompromised patients and are believed to be the viruses that spread throughout the population and establish persistent infections; however, this has not been firmly established. The rearranged strains found in PML patients appear to be derived from the archetypal strains which can be isolated from the same individuals (53). All of the JCV genomes amplified from the sewage samples and the urine sample contained identical archetypal structures in the RR. In contrast, the CSF samples from PML patients used as a positive control in these experiments (CSFE and CSFB) presented rearranged genomes (12) with duplicated regions, including the binding sites for a variety of transcription factors. Recently, Ricciardiello et al. (41) have described the presence of Mad-1 strains in the human colon. It is necessary to analyze the source of these strains, which may have originated from viremia associated with reactivation of persistent infections, from infection by the small percentage of excreted strains containing tandem repeats sporadically identified in urine (6), or from rearrangements in the RR of archetypal strains.
Analysis of the polyomaviruses excreted by the human population by studying the viral nucleic acids extracted from urban sewage provides information on the status of these viruses in the environment, the potential transmissibility of polyomaviruses and polyomavirus genes, and the characteristics and diversity of the viral strains excreted by humans in different geographic locations.
ACKNOWLEDGMENTS
This work was supported by the Center for Biologics Evaluation and Research, FDA. Sílvia Bofill-Mas is a fellow of the Generalitat de Catalunya.
We thank the Serveis Científico-Tècnics of the University of Barcelona for sequencing of PCR products. We also thank A. Vantarakis (University of Patras, Patras, Greece) and A. Shoaeb (Environmental Virology Laboratory, National Research Center, Cairo, Egypt) for collaboration in obtaining the samples in their areas. We especially thank Rosa Bufías for providing excellent technical assistance. We gratefully acknowledge A. M. Lewis (Office of Vaccine Research and Review, CBER, FDA) for providing samples from the United States and a very useful consultation.
REFERENCES
- 1.Agostini H T, Jobes D V, Chima S C, Ryschkewitsch C F, Stoner G L. Natural and pathogenic variation in the JC virus genome. Recent Res Dev Virol. 1999;1:683–701. [Google Scholar]
- 2.Agostini H T, Ryschkewitsch C F, Brubaker G R, Shao J, Stoner G L. Five complete genomes of JC virus type 3 from Africans and African Americans. Arch Virol. 1997;142:637–655. doi: 10.1007/s007050050108. [DOI] [PubMed] [Google Scholar]
- 3.Agostini H T, Ryschkewitsch C F, Stoner G L. Genotype profile of human polyomavirus JC excreted in urine of immunocompetent individuals. J Clin Microbiol. 1996;34:159–164. doi: 10.1128/jcm.34.1.159-164.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agostini H T, Ryschkewitsch C F, Stoner G L. JC virus type 1 has multiple subtypes; three new complete genomes. J Gen Virol. 1998;79:801–805. doi: 10.1099/0022-1317-79-4-801. [DOI] [PubMed] [Google Scholar]
- 5.Agostini H T, Ryschkewitsch C F, Stoner G L. Rearrangements of archetypal regulatory regions in JC virus (JCV) genomes from urine. Res Virol. 1998;149:163–170. doi: 10.1016/s0923-2516(98)80034-4. [DOI] [PubMed] [Google Scholar]
- 6.Agostini H T, Ryschkewitsch C F, Stoner G L. Complete genome of a JC virus genotype type 6 from the brain of an African American with progressive multifocal leukoencephalopathy. J Hum Virol. 1998;1:267–272. [PubMed] [Google Scholar]
- 7.Agostini H T, Shishido-Hara Y, Baumhefner R W, Singer E J, Ryschkewitsch C F, Stoner G L. JC virus type 2: definition of subtypes based on DNA sequence analysis of ten complete genomes. J Virol. 1998;79:1143–1151. doi: 10.1099/0022-1317-79-5-1143. [DOI] [PubMed] [Google Scholar]
- 8.Agostini H T, Yanagihara R, Davis V, Ryschkewitsch C F, Stoner G L. Asian genotypes of JC virus in Native Americans and in a Pacific Island population: Markers of viral evolution and human migration. Proc Natl Acad Sci USA. 1997;94:14542–14546. doi: 10.1073/pnas.94.26.14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ault G S, Stoner G L. Two major types of JC virus defined in progressive multifocal leukoencephalopathy brain by early and late coding region DNA sequences. J Gen Virol. 1992;73:2669–2678. doi: 10.1099/0022-1317-73-10-2669. [DOI] [PubMed] [Google Scholar]
- 10.Ault G S, Stoner G L. Human polyomavirus JC promoter/enhancer rearrangement patterns from progressive multifocal leukoencephalopathy brain are unique derivatives of a single archetypal structure. Nucleic Acids Res. 1993;27:517–525. doi: 10.1099/0022-1317-74-8-1499. [DOI] [PubMed] [Google Scholar]
- 11.Bergh O, Børsheim K Y, Bratbak G, Hendal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 12.Bofill-Mas S, Pina S, Girones R. Documenting the epidemiologic patterns of polyomaviruses in human populations by studying their presence in urban sewage. Appl Environ Microbiol. 2000;66:238–245. doi: 10.1128/aem.66.1.238-245.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boom R, Sol C J A, Salimans M M M, Jansen C J P M E, Wertheim-van Dillen, van der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503. doi: 10.1128/jcm.28.3.495-503.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chima S C, Ryschkewitsch C F, Stoner G L. Molecular epidemiology of human polyomavirus JC in the Biaka pygmies and Bantu of Central Africa. Mem Inst Oswaldo Cruz. 1998;93:615–623. doi: 10.1590/s0074-02761998000500010. [DOI] [PubMed] [Google Scholar]
- 15.Felsenstein J. PHYLIP. Phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 16.Frisque R J, Bream G L, Canella M T. Human polyomavirus JC virus genome. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner S D. Prevalence in England of antibody to human polyomavirus (BK) BMJ. 1973;1:77–78. doi: 10.1136/bmj.1.5845.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goudsmit J, Baak M L, Sleterus K W, van der Noordaa J. Human papovavirus isolated from urine of a child with acute tonsillitis. Br Med J. 1981;283:1363–1364. doi: 10.1136/bmj.283.6303.1363-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goudsmit J, Wertheim-van Dillen P, Van Strien A, van der Noordaa J. The role of BK virus in acute respiratory tract disease and the presence of BKV DNA in tonsils. J Med Virol. 1982;10:91–99. doi: 10.1002/jmv.1890100203. [DOI] [PubMed] [Google Scholar]
- 20.Guo J, Kitamura T, Ebiara H, Sugimoto C, Kunitake T, Takehisa J, Na Y Q, Al-Ahdal M N, Hallin A, Kawabe K, Taguchi F, Yogo Y. Geographical distribution of the human polyomavirus JC virus type A and B and isolation of a new type from Ghana. J Gen Virol. 1996;77:919–927. doi: 10.1099/0022-1317-77-5-919. [DOI] [PubMed] [Google Scholar]
- 21.Hashida Y, Gaffney P C, Yunis E J. Acute hemorrhagic cystitis of childhood and papovavirus-like particles. J Pediatr. 1976;89:85–87. doi: 10.1016/s0022-3476(76)80936-5. [DOI] [PubMed] [Google Scholar]
- 22.Hatwell J N, Sharp P M. Evolution of human polyomavirus JC. J Gen Virol. 2000;81:1191–1200. doi: 10.1099/0022-1317-81-5-1191. [DOI] [PubMed] [Google Scholar]
- 23.Imperiale M J. The human polyomaviruses, BKV and JCV: molecular pathogenesis of acute disease and potential role in cancer. Virology. 2000;267:1–7. doi: 10.1006/viro.1999.0092. [DOI] [PubMed] [Google Scholar]
- 24.Jin L, Gibson E, Booth J C, Clewley J P. Genomic typing of BK virus in clinical specimens by direct sequencing of polymerase chain reaction products. J Med Virol. 1993;41:11–17. doi: 10.1002/jmv.1890410104. [DOI] [PubMed] [Google Scholar]
- 25.Jobes D V, Sylvester C C, Ryschkewitsch C F, Stoner G L. Phylogenetic analysis of 22 complete genomes of the human polyomavirus JC virus. J Gen Virol. 1998;79:2491–2498. doi: 10.1099/0022-1317-79-10-2491. [DOI] [PubMed] [Google Scholar]
- 26.Kitamura T, Aso Y, Kuniyoshi N, Hara K, Yogo Y. High incidence of urinary JC virus excretion in nonimmunosuppressed older patients. J Infect Dis. 1990;161:1128–1133. doi: 10.1093/infdis/161.6.1128. [DOI] [PubMed] [Google Scholar]
- 27.Kitamura T, Sugimoto C, Kato A, Ebihara H, Suzuki M, Taguchi F, Kawabe K, Yogo Y. Persistent JC virus (JCV) infection is demonstrated by continuous shedding of the same JCV strains. J Clin Microbiol. 1997;35:1255–1257. doi: 10.1128/jcm.35.5.1255-1257.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kunitake T, Kitamura T, Guo J, Taguchi F, Kawabe K, Yogo Y. Parent-to-child transmission is relatively common in the spread of the human polyomavirus JC virus. J Clin Microbiol. 1995;33:1448–1451. doi: 10.1128/jcm.33.6.1448-1451.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laghi L, Randolph A E, Chauhan D P, Marra G, Major E O, Neel J V, Boland C R. JC virus DNA is present in the mucosa of the human colon and colorectal cancers. Proc Natl Acad Sci USA. 1999;96:7484–7489. doi: 10.1073/pnas.96.13.7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loeber G, Dorries K. DNA rearrangements in organ-specific variants of polyomavirus JC strain GS. J Virol. 1988;62:1730–1735. doi: 10.1128/jvi.62.5.1730-1735.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markowitz R-B, Eaton B A, Kubik M F, Latorra D, McGregor J A, Dynan W S. BK virus and JC virus shed during pregnancy have predominantly archetypal regulatory regions. J Virol. 1991;65:4515–4519. doi: 10.1128/jvi.65.8.4515-4519.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monaco M C G, Jensen P N, Hou J, Durham L C, Major E O. Detection of JC virus DNA in human tonsil tissue: evidence for site of initial viral infection. J Virol. 1998;72:9918–9923. doi: 10.1128/jvi.72.12.9918-9923.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muniain-Mujika I, Girones R, Lucena F. Viral contamination of shellfish: evaluation of methods and analysis of bacteriophages and human viruses. J Virol Methods. 2000;89:109–118. doi: 10.1016/s0166-0934(00)00208-1. [DOI] [PubMed] [Google Scholar]
- 34.Negrini M, Sabbioni S, Arthur R R, Castagnoli A, Barbanti-Brodano G. Prevalence of the archetypal regulatory region and sequence polymorphisms in nonpassaged BK virus variants. J Virol. 1991;65:5092–5095. doi: 10.1128/jvi.65.9.5092-5095.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ou W C, Tsai R T, Wang M, Fung C Y, Hseu T H, Chang D. Genomic cloning and sequence analysis of Taiwan-3 human polyomavirus JC virus. J Formosan Med Assoc. 1997;96:511–516. [PubMed] [Google Scholar]
- 36.Padgett B L, Walker D L. Prevalence of antibodies in human sera against JC virus, an isolate from a case of progressive multifocal leukoencephalopathy. J Infect Dis. 1973;127:467–470. doi: 10.1093/infdis/127.4.467. [DOI] [PubMed] [Google Scholar]
- 37.Padgett B L, Walker D L, Desquitado M M, Kim D U. BK virus and non-haemorrhagic cystitis in a child. Lancet. 1983;1:770. doi: 10.1016/s0140-6736(83)92062-7. [DOI] [PubMed] [Google Scholar]
- 38.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 39.Pina S, Puig M, Lucena F, Jofre J, Girones R. Viral pollution in the environment and shellfish: human adenovirus detection by PCR as an index of human viruses. Appl Environ Microbiol. 1998;64:3376–3382. doi: 10.1128/aem.64.9.3376-3382.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puig M, Jofre J, Lucena F, Allard A, Wadell G, Girones R. Detection of adenoviruses and enteroviruses in polluted waters by nested PCR amplification. Appl Environ Microbiol. 1994;60:2963–2970. doi: 10.1128/aem.60.8.2963-2970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ricciardiello L, Chang D K, Laghi L, Goel A, Chang C L, Boland C R. Mad-1 is the exclusive JC virus strain present in human colon, and its transcriptional control region has a deleted 98-base-pair sequence in colon cancer tissues. J Virol. 2001;75:1996–2001. doi: 10.1128/JVI.75.4.1996-2001.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ricciardiello L, Laghi L, Ramamirtham P, Chang C L, Chang D K, Randolph A E, Boland C R. JC virus DNA sequences are frequently present in human upper and lower gastrointestinal tract. Gastroenterology. 2000;119:1228–1235. doi: 10.1053/gast.2000.19269. [DOI] [PubMed] [Google Scholar]
- 43.Rubinstein R, Pare N, Harley E H. Structure and function of the transcriptional control region of nonpassaged BK virus. J Virol. 1987;61:1747–1750. doi: 10.1128/jvi.61.5.1747-1750.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rubinstein R, Schoonakker B C, Harley E H. Recurring theme of changes in the transcriptional control region of BK virus during adaptation to cell culture. J Virol. 1991;65:1600–1604. doi: 10.1128/jvi.65.3.1600-1604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schubbert R, Renz D, Schmitz B, Doerfler W. Foreign (M13) DNA ingested by mice reaches peripheral leukocytes, spleen, and liver via the intestinal wall mucosa and can be covalently linked to mouse DNA. Proc Natl Acad Sci USA. 1997;94:961–966. doi: 10.1073/pnas.94.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah K V. Polyomaviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Raven Publishers; 1995. pp. 1997–2025. [Google Scholar]
- 47.Shah K V, Daniel R W, Strickler H D, Goedert J J. Investigation of human urine for genomic sequences of the primate polyomaviruses simian virus 40, BK, and JC virus. J Infect Dis. 1997;176:1618–1621. doi: 10.1086/517340. [DOI] [PubMed] [Google Scholar]
- 48.Sugimoto C, Kitamura T, Guo J, Al-Ahdal M N, Shchelkunov S N, Otova B, Ondrejka P, Chollet J Y, El-Safi S, Ettayebi M, Gresenguet G, Kocagoz T, Chaiyarasamee S, Thant K Z, Thein S, Moe K, Kobayashi N, Taguchi F, Yogo Y. Typing of urinary JCV DNA offers a novel means of tracing human migrations. Proc Natl Acad Sci USA. 1997;94:9191–9196. doi: 10.1073/pnas.94.17.9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sundsfjord A, Johansen T, Flaegstad T, Moens U, Villand P, Subramani S, Traavik T. At least two types of control regions can be found among naturally occurring BK virus strains. J Virol. 1990;64:3864–3871. doi: 10.1128/jvi.64.8.3864-3871.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sundfsfjord A, Spein A R, Lucht E, Flaegstad T, Seternes O M, Traavik T. Detection of BK virus DNA in nasopharyngeal aspirates from children with respiratory infections but not in saliva from immunodeficient and immunocompetent adult patients. J Clin Microbiol. 1994;32:1390–1394. doi: 10.1128/jcm.32.5.1390-1394.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taguchi F, Kajioka J, Miyamura T. Prevalence rate and age of acquisition of antibodies against JC virus and BK virus in human sera. Microbiol Immunol. 1982;26:1057–1064. doi: 10.1111/j.1348-0421.1982.tb00254.x. [DOI] [PubMed] [Google Scholar]
- 52.Yogo Y, Kitamura T, Sugimoto C, Hara K, Iida T, Taguchi F, Tajima A, Kawabe K, Aso Y. Sequence rearrangement in JC virus DNAs molecularly cloned from immunosuppressed renal transplant patients. J Virol. 1991;65:2422–2428. doi: 10.1128/jvi.65.5.2422-2428.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yogo Y, Kitamura T, Sugimoto C, Ueki T, Aso Y, Hara K, Taguchi F. Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J Virol. 1990;64:3139–3143. doi: 10.1128/jvi.64.6.3139-3143.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]