Summary
Deficits in chemosensory processing are associated with healthy aging, as well as numerous neurodegenerative disorders, including Alzheimer’s disease (AD). The fruit fly, Drosophila melanogaster, is a powerful model for studying chemosensation, aging, and aging-related pathologies, yet the effects of aging and neurodegeneration on taste function remain largely unexplored. Aging impaired response to sugars, but not medium-chain fatty acids that are sensed by a shared population of neurons. Selective expression of the human amyloid beta (Aβ) peptide phenocopied the effects of aging. Functional imaging of gustatory axon terminals revealed reduced response to sugar, but not fatty acids. Axonal innervation of the fly taste center was largely intact in aged flies; however, axonal innervation was reduced upon expression of Aβ. A comparison of transcript expression within the sugar-sensing taste neurons revealed age-related changes in 66 genes. Together, these findings suggest that different mechanisms underly taste deficits in aged and AD model flies.
Subject areas: Biological sciences, Neuroscience, Sensory neuroscience
Graphical abstract
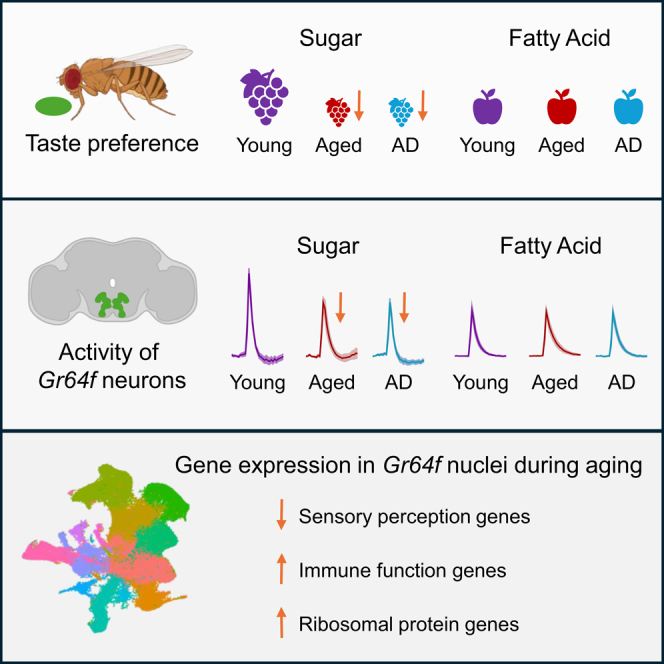
Highlights
-
•
Taste sensitivity to sugars, but not fatty acids diminishes with age
-
•
Taste decline is accelerated in Alzheimer’s model flies
-
•
Aged and Alzheimer’s model flies maintain physiological response to fatty acids
Biological sciences; Neuroscience; Sensory neuroscience
Introduction
Aging is associated with deficits in numerous sensory modalities including taste and smell.1,2,3,4 In humans, several neurodegenerative diseases including Alzheimer’s disease (AD) and Parkinson’s disease (PD) have been shown to disrupt primary sensory cells, though less is known about the physiological and functional effects of healthy aging.3,5 Most neurodegenerative diseases exhibit chemosensory deficits that precede motor or memory deficits, which are hallmarks of PD and AD, respectively.6,7 The taste system provides a particularly tractable system for examining the effects of aging on chemosensory processing. Unlike olfaction, taste is composed of relatively few modalities that elicit reflexive responses. Investigating how taste response changes in aging and AD model animals has the potential to identify the genetic and neural basis contributing to age-related chemosensory decline.
The fruit fly, Drosophila melanogaster, has developed into a powerful model to study taste processing because of its amenability to genetic manipulation and functional parallels in taste processing between flies and mammals.8,9,10 Additionally, the gustatory system of Drosophila is amenable to in vivo Ca2+ imaging and electrophysiology, both of which can be coupled with robust behavioral assays that measure reflexive taste response and food consumption.11 Among the main classes of non-overlapping gustatory neurons are sweet-sensing neurons, which promote feeding and are labeled by expression of the gustatory receptor 64f (Gr64f); these neurons respond to both sugars and fatty acids.12,13,14 The well-defined function and projection of these neurons provide a system to examine how gustatory function changes in normal and pathological aging.
The short lifespan of fruit flies provides a system for examining the genetic basis of age-related deficits in many behavioral and physiological traits. The effects of aging are accelerated in the neurodegenerative disease model Drosophila expressing amyloidogenic forms of human amyloid beta (Aβ) and recapitulate several key features of AD, including Aβ accumulation, age-dependent learning impairment, and neurodegeneration.15,16,17 In a commonly used model, a variant of human Aβ1-42 harboring the mutation E22G, termed the Arctic mutation, induces robust neurodegeneration as well as behavioral deficits.17,18,19,20,21 While most experiments examining the effects of AD variants in flies drive AD-associated variants throughout the brain, targeted expression of disease-causing variants in defined populations of neurons allows for investigation of how aging and Aβ expression impact the connectivity and physiology of individual circuits.
Here, we examine the effects of aging or Arctic-bearing Aβ1-42 expression on gustatory function and behavior. We find that both healthy aging and pathological aging through cell-autonomous expression of Arctic in sweet taste neurons result in deficits in the detection of sugars, but not fatty acids. These tastants are detected by shared neurons but are activated by unique taste receptors and intercellular signaling pathways. Our findings suggest that both aging and Arctic-bearing Aβ1-42 expression disrupt cellular physiology in a modality-specific manner. We also identify many candidate regulators of age-related deficits in taste. Therefore, gustatory neurons provide a model to study the mechanisms underlying aging and the neurodegeneration-related decline of sensory processing.
Results
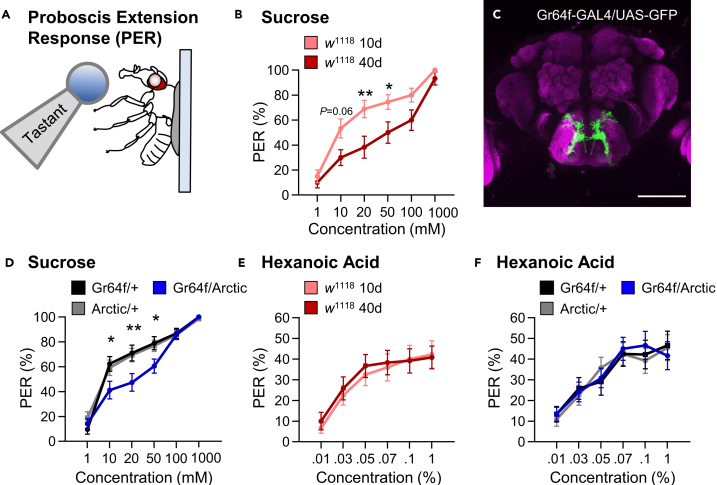
To examine the relationship between aging and taste, we measured the proboscis extension response (PER) against varying concentrations of sucrose in young w1118 10-day-old flies and aged 40-day-old flies. In this assay, a tastant is provided to the proboscis, eliciting a reflexive response independent of post-ingestive feedback (Figure 1A).22 To define response sensitivity, flies were provided with different concentrations of sucrose ranging from 1 mM to 1 M. In 10-day-old flies, the response to these concentrations ranged from 15% to 100%, providing a range to measure taste sensitivity. The sucrose response did not differ at concentrations of 1 mM and 1 M between young and aged flies; however, there was a significant decrease in responsiveness to sucrose in aged flies at intermediate concentrations of 20 and 50 mM (Figure 1B). This decrease in responsiveness to sucrose only occurred after 40 days of age, as there was no significant difference in PER to sucrose between young 10-day-old flies and flies that were 20 or 30 days of age (Figures S1A and S1B). To determine whether this aging-dependent decline in taste sensitivity to sucrose can be generalized to other sweet tastants, we measured PER to different concentrations of fructose. Like our results using sucrose, we found that taste sensitivity to fructose significantly declines at intermediate concentrations (10 and 20 mM; Figure S2A). The finding that sugar taste does not differ at high concentrations suggests that flies are capable of recognizing sugar and performing the motor task of full proboscis extension, even though sensitivity to sweet tastants is diminished in aged animals.
Figure 1.
Taste response to sugar, but not fatty acids, is reduced at intermediate concentrations in aging and AD model flies
(A) Proboscis extension response (PER) was measured in female flies after 24 h of starvation. Either sucrose or hexanoic acid was applied to the fly’s labellum for a maximum of 2 s and then removed to observe proboscis extension reflex.
(B) There is a significant effect of age on PER to sucrose in w1118 flies (two-way ANOVA: F1,278 = 23.77, p < 0.0001; n = 20–30).
(C) Expression pattern of Gr64f is visualized with GFP. Gr64f-expressing neurons project to the subesophageal zone of the brain. Background staining is NC82 antibody (magenta). Scale bar: 100 μm.
(D) There is a significant effect of Arctic expression on PER to sucrose (two-way ANOVA: F2,702 = 7.529, p < 0.0006; n = 28–49).
(E) There is no effect of age on PER to hexanoic acid in w1118 flies (two-way ANOVA: F1,415 = 0.2959, p < 0.5867; n = 20–40).
(F) There is no effect of Arctic expression on PER to hexanoic acid (two-way ANOVA: F2,671 = 0.0396, p < 0.9612; n = 30–40). Data are represented as mean ± SEM. ∗p < 0.05; ∗∗p < 0.01.
In Drosophila, PER involves the activation of a sensory circuit that includes gustatory neurons, interneurons, motor neurons, and musculature.11,23,24 In flies, pan-neuronal expression of the Aβ (Aβ1-42) variant Arctic significantly increases Aβ1-42 mRNA expression in the head and accumulation of the Aβ1-42 protein concentration over time scales with activity impairment and survivorship.25 Further, broad expression of this transgene recapitulates many aspects of premature aging including disrupted sleep, memory impairment, and premature death.17,18,19,20,21,26,27 To determine whether dysfunction in the sensory component of the taste circuit alone is sufficient to drive this decline in taste sensitivity, we expressed Arctic exclusively in sweet taste neurons labeled by the Gr64f receptor (Figure 1C). We tested 20-day-old flies because this is prior to age-related loss of taste in controls (Figure S1), allowing for the identification of accelerated loss of taste. PER was significantly diminished in 20-day-old flies expressing Arctic in sweet taste neurons (Gr64f-GAL4 > UAS-Arctic) compared to controls harboring Gr64f-GAL4 or UAS-Arctic alone (Figure 1D). Similarly, a decline in taste sensitivity in flies that express Arctic in Gr64f neurons was also observed in response to fructose (Figure S2B). Further, there was no effect on the expression of the non-toxic 40-amino acid form of Aβ (Aβ1-40) to sucrose presentation (Figure S2C), suggesting that the results observed with Arctic are related to the pathogenicity of Aβ1-42.28,29 Therefore, Arctic expression induces cell-autonomous deficits in sweet taste response, phenocopying the effects of aging and supporting the notion that reduced sensitivity of taste neurons underlies aging-related reductions in PER.
The Gr64f-expressing taste neurons detect numerous appetitive tastants, including sugars and fatty acids.30,31,32 While these neurons are activated by both sugars and fatty acids, the response to tastants is conferred by distinct signaling pathways with sugar response dependent on the Gsα subunit,33,34 while fatty acid response is dependent on parallel signaling pathways that include phospholipase C (PLC) and ionotropic receptor 56d (Ir56d).30;31;35 To determine whether aging impairs response to both tastants, we compared the response of young and aged flies to hexanoic acid, a medium-chain fatty acid that robustly activates Gr64f neurons (Figure 1E).31,32 Across six different concentrations ranging from 0.01% to 1%, there was no difference in response to hexanoic acid. Higher concentrations of this tastant are aversive; therefore, they were not tested in this assay.30 Together, these findings suggest that fatty acid taste is not impaired in aged animals.
To determine whether Arctic expression in Gr64f neurons impairs fatty acid taste, we measured taste response to hexanoic acid in 20-day-old flies expressing UAS-Arctic in taste neurons (Figure 1F). There was no difference in PER between control flies harboring Gr64f and experimental flies expressing Arctic in Gr64f neurons (Gr64f-GAL4 > UAS-Arctic). We also measured PER to different concentrations of octanoic acid, a second fatty acid that is also activated by Gr64f neurons.32 There was no significant difference in taste response to octanoic acid between young and aged flies or flies expressing Arctic in Gr64f neurons (Figures S2D and S2E). Similarly, expression of the non-toxic Aβ1-40 did not affect taste response to hexanoic acid (Figure S2F). Therefore, both natural aging and taste neuron-specific expression of Arctic impairs response to sugars without affecting fatty acid taste response.
To explore the possibility that other appetitive taste modalities are affected by aging, we measured taste response to sour (citric acid and acetic acid) and protein (phenylalanine and serine), which activate nonoverlapping populations of neurons with Gr64f.36,37 We observed no significant difference in taste response to sour tastants between 10-day-old and 40-day-old flies (Figures S2G and S2H). In contrast, taste response to amino acids were significantly reduced in aged 40-day-old flies when compared to young 10-day-old flies (Figures S2I and S2J), suggesting the modality-specific effects of aging on taste response is not specific to sugars.
We next sought to determine whether the observed differences in aging and AD model flies are sex specific. We found that the sensitivity to sucrose, but not hexanoic acid, is reduced in 40-day-old male flies compared to their 10-day-old counterparts (Figures S3A and S3B). Similarly, expression of Arctic in Gr64f neurons in 20-day-old male flies impairs sensitivity to sucrose, but not fatty acids (Figures S3C and S3D). Therefore, both aging and Arctic expression selectively impact sucrose response in male and female flies.
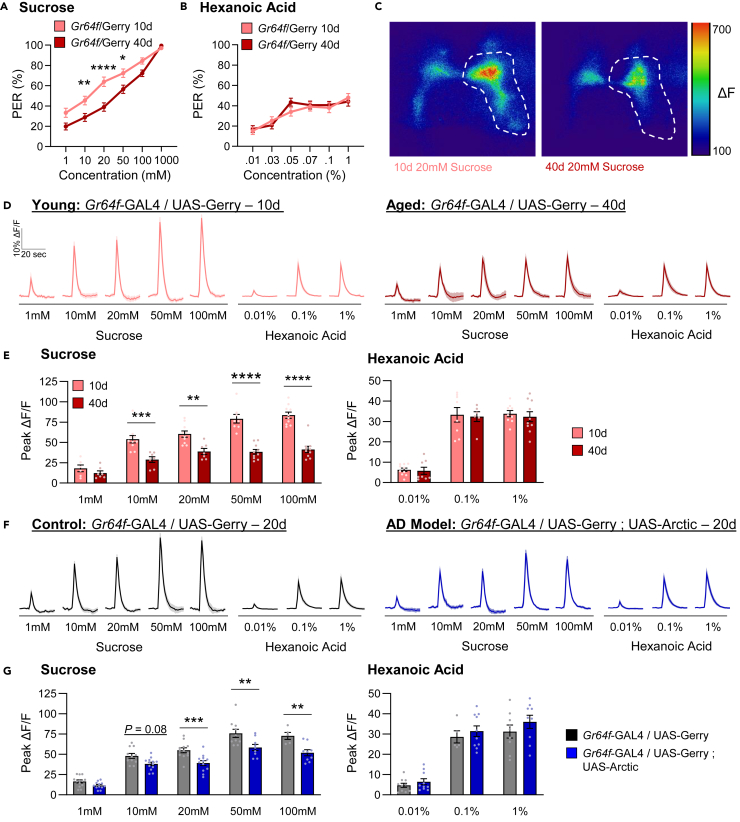
It is possible that the decline in taste response is due to neurodegeneration of the taste neurons, loss of connectivity with taste neuron targets, or reduced responsiveness of taste neurons. Given that the decline in taste response is modality-specific, we hypothesize that neural responsiveness to sugars, but not to fatty acids change with age. To test this hypothesis, we first measured neural activity of Gr64f neurons to tastant presentation. To directly assess the effects of aging on neural activity, we expressed the genetically encoded Ca2+ sensor GCaMP6s-mCherry (UAS-Gerry) in Gr64f neurons. Quantifying the ratio of stable mCherry signal to Ca2+ induced GCaMP6s provides a ratiometric quantification of evoked activity.38,39,40 Similar to w1118 flies, taste response to sucrose declines during aging in this genetic background (Gr64f-GAL4 > UAS-Gerry), while taste response to hexanoic acid remained unchanged (Figures 2A and 2B). Using these flies, we applied an in vivo preparation to record responsiveness of projections in the subesophageal zone (SEZ) when tastants were applied directly to the proboscis (Figure 2C).31 At concentrations ranging from 10 to 100 mM, the responses of 40-day-old flies to sucrose were significantly reduced compared to 10-day-old flies (Figures 2D and 2E). To determine if aging also impacts fatty acid taste response, we compared the response in the SEZ to hexanoic acid in 10-day-old and 40-day-old flies (Figure 2D). Given the lack of statistically significant differences in taste preference behavior to hexanoic acid, we choose to limit the number of concentrations tested to 0.01%, 0.1%, and 1%. Across all three concentrations, there were no differences in Ca2+ response between 10-day-old and 40-day-old flies (Figure 2E). Therefore, consistent with our behavioral results, aging diminishes taste neuron response to sucrose, but not hexanoic acid presentation.
Figure 2.
Neuronal activity in Gr64f neurons is reduced during aging and in an AD model upon sucrose but not hexanoic acid presentation
(A) There is a significant effect of age on PER to sucrose in Gr64f/Gerry flies (two-way ANOVA: F1,625 = 40.54, p < 0.0001; n = 47–60).
(B) There is no effect of age on PER to hexanoic acid in Gr64f/Gerry flies (two-way ANOVA: F1,684 = 0.2826, p = 0.5952; n = 57–59).
(C) Representative pseudocolor images of GCaMP/mCherry fluorescence in Gr64f neurons in response to 20mM sucrose in 10 (left) and 40-day-old flies (right). The peak in UAS-GCaMP6 fluorescence is shown. Scale bar: 50 μm.
(D) The average ratio of GCaMP/mCherry of Gr64f neurons in response to each tastant and concentration in 10-day-old and 40-day-old flies. The shaded region of each trace indicates ±SEM.
(E) Average peak change in the ratio of GCaMP/mCherry for data shown in (D). There is a significant effect of age on neuronal response to sucrose (two-way ANOVA: F1,75 = 112.1, p < 0.0001; n = 6–10), but not to hexanoic acid (two-way ANOVA: F1,45 = 0.2846, p < 0.5963; n = 6–10).
(F) The average ratio of GCaMP/mCherry of Gr64f neurons in response to each tastant and concentration in control and AD model flies.
(G) Average peak change in the ratio of GCaMP/mCherry for data shown in (F). There is a significant effect of Arctic expression on neuronal response to sucrose (two-way ANOVA: F1,96 = 47.80, p < 0.0001; N = 5–13), but not to hexanoic acid (two-way ANOVA: F1,47 = 2.218, p < 0.1431; n = 5–10). Data are represented as mean ± SEM. ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
To determine if the effects of aging are shared in flies with cell-autonomous expression of Arctic in taste neurons, we expressed UAS-Gerry in taste neurons and measured neural activity to sucrose and hexanoic acid presentation in 20-day-old flies (Figure 2F). Consistent with the results comparing 10-day-old and 40-day-old flies, the response to sucrose was reduced at intermediate (20 mM) concentrations in experimental flies expressing Arctic (Gr64f > UAS-Arctic) compared to age-matched flies harboring Gr64f-GAL4 alone (Figure 2G). We also observed a significant reduction in responsiveness to sucrose at high (100 mM) concentrations (Figure 2G), despite no change in taste preference at this concentration (Figure 1D). It is possible this is because the activity of Gr64f neurons in Arctic-expressing flies is above the threshold of detection of sucrose at this concentration. There were no significant differences in response to hexanoic acid at any of the concentrations tested (Figure 2G). Therefore, selective expression of Arctic in sweet-sensing taste neurons has a selective effect on lowering sugar sensitivity, similar to the effects of aging on taste response. Taken together, the reduced behavioral response to sucrose in both aged and Arctic flies may be attributed to the observed deficit in Gr64f neurons.
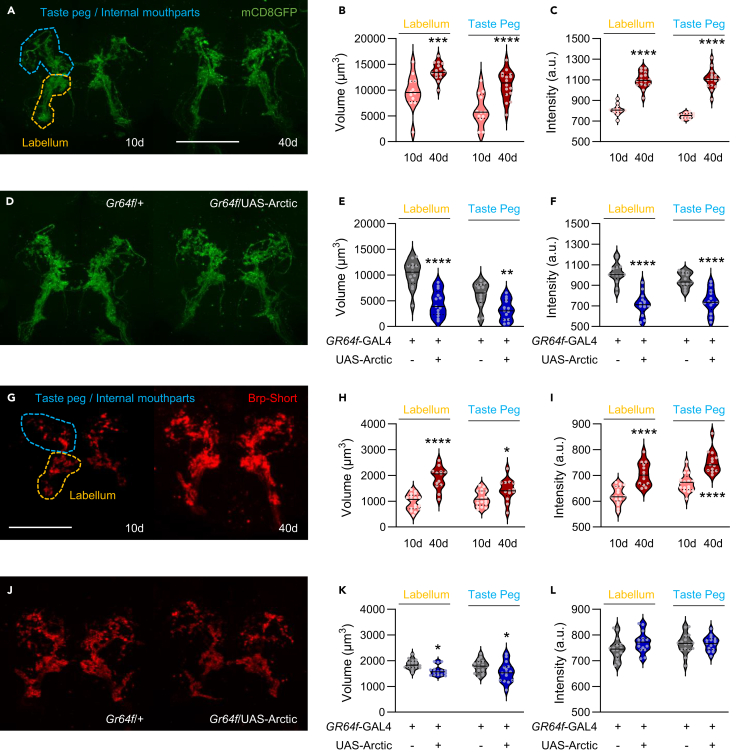
It is also possible that reductions in synaptic innervation of the suboesophageal zone contribute to age-dependent loss of sucrose responsiveness. To explore this possibility, we directly measured innervation of the SEZ Gr64f neurons. We expressed the membrane-bound marker mCD8GFP in Gr64f neurons and quantified immunofluorescence from sensory terminals that innervate the SEZ at days 10 and 40 (Figure 3A). The Gr64f neurons project to distinct regions of the SEZ, with neurons from the labellum projecting to the ventral region of the SEZ, while neurons from the taste pegs and internal mouthparts primarily project to the dorsal region of the SEZ.38,39,40 The volume and intensity within these two regions were increased in both labellum and taste peg regions (Figures 3B and 3C). This increase in intensity may be attributable to either the accumulation of membrane-bound GFP at the synapse during aging, or to a strengthening of connections. Together, these results suggest that the reduced PER in response to sucrose is not due to large-scale neurodegeneration or loss of SEZ innervation.
Figure 3.
Aging and Arctic expression have differential effects on the axon terminals and active zone scaffold of Gr64f neurons
(A) Expression pattern of Gr64f in 10-day-old and 40-day-old flies is visualized with GFP (green). ROIs indicating the labellar (yellow) and the taste peg/internal mouthpart projections (blue) are shown. Scale bar: 50 μm.
(B) There is a significant effect of age on Gr64f neurite volume (two-way repeated measures ANOVA: F1,26 = 23.29, p < 0.0001; n = 12–16).
(C) There is a significant effect of age on Gr64f neurite intensity (two-way repeated measures ANOVA: F1,26 = 167.5, p < 0.0001; n = 12–16).
(D) Expression pattern of Gr64f in control and AD model flies is visualized with GFP (green).
(E) There is a significant effect of Arctic expression on Gr64f neurite volume (two-way repeated measures ANOVA: F1,24 = 22.98, p < 0.0001; n = 13).
(F) There is a significant effect of Arctic expression on Gr64f neurite intensity (two-way repeated measures ANOVA: F1,24 = 45.01, p < 0.0001; n = 13).
(G) The active zone of Gr64f neurons in young and aged flies is visualized with Brp-Short (red). ROIs indicating the labellar (yellow) and the taste peg/internal mouthpart projections (blue) are shown. Scale bar: 50 μm.
(H) There is a significant effect of age on Brp volume (two-way repeated measures ANOVA: F1,28 = 26.18, p < 0.0001; n = 13–17).
(I) There is a significant effect of age on Brp intensity (two-way repeated measures ANOVA: F1,28 = 43.09, p < 0.0001; n = 13–17).
(J) The active zone of Gr64f neurons in control and AD model flies is visualized with Brp-Short (red).
(K) There is a significant effect of Arctic expression on Brp volume (two-way repeated measures ANOVA: F1,29 = 8.244, p < 0.0076; n = 15–16).
(L) There is no effect of Arctic expression on Brp intensity (two-way repeated measures ANOVA: F1,29 = 1.706, p < 0.2018; n = 15–16). The median (solid line) as well as 25th and 75th percentiles (dotted lines) are shown. a.u. = arbitrary units. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
We also sought to directly compare the effects of cell-autonomous Arctic expression on innervation of Gr64f axon terminals. A comparison between Gr64f controls and flies expressing Arctic in Gr64f neurons (Gr64f-GAL4 > UAS-Arctic) revealed a significant reduction in both volume and intensity in projections from the labellum as well as from the taste pegs and internal mouthparts of 20-day-old flies (Figures 3D–3F). Therefore, reduced synaptic innervation of axon terminals that project from the labellum may contribute to the reduced response to sucrose in flies that express Arctic in Gr64f neurons. Together, these findings raise the possibility that different mechanisms underlie natural aging- and Arctic-dependent decline in taste response to sugar.
It is possible that the change in signal intensity of Gr64f neurons with age and Arctic expression is driven by changes in the active zone scaffold, as it has been previously shown that aging substantially increases levels of the active zone component Bruchpilot (Brp).41,42,43,44 To label the active zones of Gr64f neurons, we expressed UAS-Brp-short,45 a nonfunctional Brp that localizes to endogenous Brp sites. We quantified Brp at Gr64f terminals of 10-day-old and 40-day-old flies (Figure 3G). Both the volume and intensity of synaptic Brp levels in the labellar and taste peg regions were significantly elevated in aged flies, suggesting that the loss of synaptic proteins does not account for the reduced response to sucrose (Figures 3H and 3I). To determine whether these aging-dependent changes to the SEZ are similar in AD model flies, we next compared the effects of cell-autonomous Arctic expression on Brp levels in Gr64f neurons (Figure 3J). We found that Arctic expression in Gr64f neurons (Gr64f-GAL4 > UAS-Arctic, UAS-Brp-short) significantly decreases volume, but not signal intensity in 20-day-old flies (Figures 3K and 3L). These findings suggest that aging and cell-autonomous Arctic expression have differential effects on innervation of the SEZ and assembly of the active zone in Gr64f neurons, raising the possibility that different mechanisms regulate the decline in taste response to sugar in aging and AD model flies.
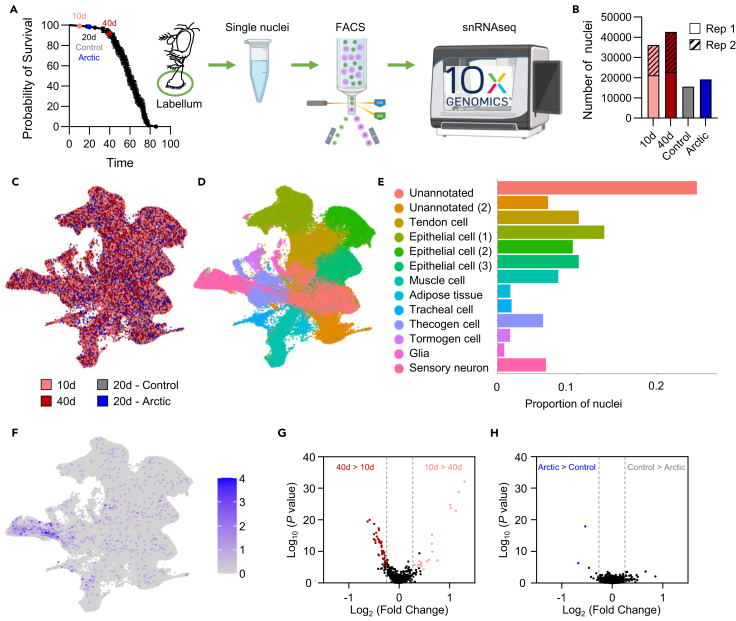
To identify genes associated with aging and Arctic-expression in chemosensory neurons, we performed 10× Genomics-based single-nucleus RNA sequencing (snRNA-seq)46 on the labellar tissue of young (10-day-old) and aged (40-day-old) flies as well as 20-day-old control flies and 20-day-old flies expressing the Arctic transgene in sweet taste neurons (Figure 4A). This approach allowed us to examine the transcriptomes of individual sweet taste neurons and identify changes in gene expression associated with aging. Two independent rounds of sequencing were performed for each young and aged condition (with ∼400 labellum per replicate, while only one round of sequencing was performed for the control and Arctic samples. After filtering and correcting for batch effects, there were a total of 36,194 young nuclei and 42,639 aged nuclei in the pooled replicates, while the control and Arctic samples contained 15,581 and 19,179 nuclei, respectively (Figures 4B and 4C). For the aging comparison, the pooled samples from young nuclei had a median of 558 genes and 1,116 Unique Molecular Identifiers (UMIs) per cell, while the pooled samples from aged nuclei had a median of 557 genes and 1,080 UMIs. For the AD comparison, the control sample had a median of 595 genes and 1,267 UMIs per cell, while the Arctic sample had a median of 637 genes and 1,266 UMIs. The detected numbers of expressed genes and UMIs were largely consistent between all samples (Figure S4).
Figure 4.
snRNA-sequencing of the labellum during aging and in AD model flies
(A) Schematic of snRNA-sequencing workflow. The labellum of adult flies were dissected and dissociated to obtain single nuclei for 10× Genomics snRNA-seq. Error bars indicate ±SEM. FACS: florescence-activated cell sorting.
(B) Number of nuclei collected from each sample.
(C) Uniform Manifold Approximation and Projection (UMAP) visualization.
(D) UMAP visualization representing unique clusters. Each color and dot on the plot represent a unique cluster and nucleus, respectively.
(E) The proportion of nuclei for each broad cell cluster shown in (D).
(F) UMAP visualization representing the average expression of Gr64f+ nuclei.
(G) Volcano plot depicting differentially expressed genes within the Gr64f+ nuclei between 10-day-old and 40-day-old flies.
(H) Volcano plot depicting differentially expressed genes within the Gr64f+ nuclei between control and Arctic-expressing flies. Genotypes: 10d: w1118; 40d: w1118; control: Gr64f-Gal4/+; Arctic: Gr64f-GAL4/UAS-Arctic.
To identify which types of nuclei are present among the samples, we performed clustering analysis and identified 13 distinct clusters, including a sensory neuron cluster (Figures 4D and 4E, bright pink). These clusters were identified based on known marker genes for each of these tissue types. In the case of sensory neurons, the top three marker genes include paralytic (para), long non-coding RNA:noe (lncRNA:noe), and pumilio (pum; Figure S5A). Additional markers of this cluster include neural-specific genes, such as Synaptobrevin (nSyb), Bruchpilot (Brp), Synaptotagmin 1 (Syt1), and cadherin-N (CadN), as well as numerous odorant-binding proteins (OBPs) (Table S1). We next sought to examine the effect of aging on gene expression within the sensory neuron cluster. To identify genes that change expression between the young and aged conditions, we performed differential gene expression analysis (Figure S5B; Table S2). We identified 91 differentially expressed genes (DEGs), with 65 genes upregulated and 26 genes downregulated in the aged condition. We next assessed whether there was statistical enrichment of Gene Ontology (GO) terms between these groups (Figure S5C; Table S3). We found significant upregulation of genes associated with translation and ribosome biogenesis, while significant downregulation was observed for genes involved in nervous system process and the perception of chemical stimuli. Overall, these findings suggest increased translation as a possible compensatory mechanism for reduced chemosensory expression.
Gene expression analysis of individual nuclei provides an opportunity to investigate the effect of aging and AD on gene expression, specifically within Gr64f+ sweet taste neurons. We identified 404 and 360 nuclei that express Gr64f in the young and aged pooled replicates, while 163 and 139 Gr64f+ nuclei were identified in the control and Arctic samples, respectively. (Figure 4F). To identify transcriptomic differences between the young and aged Gr64f+ nuclei, we performed differential gene expression analysis and identified 66 DEGs with 49 genes upregulated and 17 genes downregulated in the aged condition (Figure 4G; Table S4). There was substantial overlap between the Gr64f+ DEGs and those identified in the sensory cluster. We found that 63 of these genes overlap, suggesting that the effects of aging on sensory neurons may be generalizable to other taste neurons. These include the upregulated gene activity-regulated cytoskeleton-associated protein 1 (Arc1) that has retrovirus function and is involved in transsynaptic communication,47 as well as multiple genes involved in the innate immune response, e.g., Listericin and virus-induced RNA 1.48,49,50 We also identified upregulation of 38 ribosomal proteins. Further, numerous OBPs were found to be downregulated with age, raising the possibility that they are directly involved in diminished taste. We next assessed whether there was statistical enrichment of GO terms among these DEGs and found significant downregulation of genes involved in perception of chemical stimuli, while those associated with translation and ribosome biogenesis were significantly upregulated (Figure S6A; Table S5). These findings further support the notion that changes in Gr64f+ nuclei may be generalizable to other sensory nuclei.
To identify genes associated with Arctic-expression within Gr64f+ sweet taste neurons, we next performed differential gene expression analysis of Gr64f+ control and Arctic-expressing nuclei. We identified only 2 DEGs, both of which were upregulated in the Arctic condition (Figure 4H; Table S4). These genes included one ribosomal RNA (mt:lrRNA) and one long non-coding RNA (CR40469), neither of which overlap with Gr64f+ aging DEGs. The ribosomal RNA, mt:lrRNA is a structural component of the mitochondrial ribosome, and upregulation of mitochondrial RNA has been observed in humans with AD.51 Overall, these findings suggest that the molecular signatures of aging are not clearly present in flies expressing Arctic in Gr64f+ neurons.
Different signaling pathways are responsive to fatty acids and sugars, with sugars activating the cAMP signaling pathway33 and fatty acids activating the PLC signaling pathway.35 To determine whether aging or Arctic expression affects the expression of genes associated with either sugar or fatty acid taste perception, we examined genes whose expression has been previously implicated in either of these pathways (Figures S6B–S6E).30,31,32,33,34,52,53 However, there were no significant differences in expression observed in the examined genes with respect to either of these pathways. Given the low detection rate of these genes, we cannot rule out the possibility that aging affects the expression of other aspects of these pathways.
Discussion
Our findings reveal that aging in Drosophila is associated with reduced sensitivity to sugar taste. There is an abundance of evidence that both olfactory and taste sensitivity is impaired during aging in humans and rodents.54,55,56 In humans, the reduced sensitivity to tastants appears to occur across taste modalities, suggesting a generalized effect of normal aging on taste.4 However, studies in mice reveal selective reductions in some modalities, but not others, for example, taste sensitivity loss appears to be selective to sugar, but not other tastants including salt, acid, and bitter substances.56 Therefore, our findings that age-related taste loss in Drosophila occurs in response to sugars but not fatty acids reveals that the effects of modality-specific taste loss in aged animals are present across both mammalian and non-mammalian species.
In humans, chemosensory deficits are present in many neurodegenerative diseases.57,58,59 For example, a decline in olfactory function is reported to precede other neurological deficits in both AD and PD,60,61 suggesting that olfactory receptor neurons are highly sensitive to neurodegenerative processes. Additionally, taste perception is disrupted, and taste bud number is reported to be reduced in AD model mice, though other studies in different AD mice models have found no difference in taste.62,63 However, studies in both humans and rodent models have not specifically examined whether the effects on taste are due to the primary sensory cells themselves, or to downstream factors. We expressed the AD E693G variant of Aβ, Arctic, selectively in taste neurons to determine whether cell-autonomous expression of this variant phenocopies the taste deficits observed during natural aging. Behaviorally and physiologically, these flies phenocopied naturally aged flies, supporting the notion that changes within the primary taste neurons underlie age-related loss of sugar taste in flies, rather than downstream processing or motor mechanisms. In agreement with these findings, aging in mammals directly impacts taste receptor cells.56
We find that both natural aging and expression of Arctic in taste neurons lead to impaired sucrose response without affecting fatty acid taste. Sugars and fatty acids respond by different intercellular signaling pathways and receptors, with sugars activating Gsα33 and fatty acids activating a complex set of intercellular signaling molecules including norpA/PLC.35 These findings raise the possibility that Gsα is more sensitive to the effects of aging, while PLC is more resilient. Consistent with this notion, aged mice display a reduction in the sugar receptor TAS1R, but not PLC.56 While these findings raise the possibility that cAMP signaling is selectively reduced in Gr64f neurons, we did not observe significant expression differences in genes associated with sugar or fatty acid taste. Future studies investigating how specific components of these signaling pathways are modulated at the protein level may provide additional insight into the mechanisms by which taste sensitivity changes across the lifespan. Further, the use of genetically encoded reporters, including the cAMP reporters cAMPr and ePAC, provide the opportunity to directly test the hypothesis that age selectively impacts signaling pathway(s) associated with sugar sensing.64,65
A growing body of literature supports the notion that flies expressing human AD pathology-associated variants recapitulate many of the phenotypes associated with the disorder. For example, expression of Aβ variants, including Arctic, result in reductions in sleep,17,66 memory,67,68 and longevity.20,21,69 Most studies to date have used broad expression of Aβ, either in all neurons or large neural structures within the brain. Therefore, little is known about the cell-autonomous impacts of Aβ expression on Drosophila behavior. Our finding that the expression of Arctic in sweet taste neurons impairs sugar taste response provides an additional application to study the cell type-specific effects of human Aβ variants on neuronal function and physiology. We find that selective expression of Arctic in taste neurons results in a decrease in SEZ innervation. We reported these results by monitoring mCD8GFP, whose axonal transport is mediated by an actin- and vesicle-based trafficking system that regulates delivery of synaptic components to target synapses.70 This is consistent with our findings in which delivery of brp to synaptic terminals in the SEZ is impaired in flies expressing Arctic in Gr64f neurons. In humans, Alzheimer’s is associated with synaptopathy, and synapse loss or reductions have been reported in numerous brain regions including the hippocampus.71,72 While anatomical synaptic loss has not been documented in mouse models of AD, hippocampal physiology supports the loss of presynaptic signaling,73 consistent with the results that we observed. Therefore, it is surprising that these flies maintain their response to fatty acids. It is possible that either the subset of neurons that are sensitive to fatty acids are resilient to the effects of Arctic expression, or that only a subset of Gr64f neurons are required for fatty acid taste.
Here, we identify several candidate genes that are differentially expressed in Gr64f+ neurons between young and aged flies. For example, multiple OBPs are downregulated in aging flies, and while these genes are broadly known to modulate olfaction, they are also expressed in the taste sensilla of insects.74 While less is known about the regulation of taste by OBPs, one of the proposed functions of OPBs in the taste system is to modulate the clearance of bitter compounds. We find that obp18a is downregulated in Gr64f+ neurons of aged flies. A previous study demonstrated that downregulation of obp18a is associated with decreased consumption of bitter tastants.75 Although gustatory OPBs have been previously mapped to thecogen support cells,69 it is possible they serve diverse functions across multiple cell types. Alternatively, it is possible that OBPs interact directly with sweet taste neurons to modulate their activity, as is the case with obp49, which has been shown to be required for suppression of sweet tastants upon exposure to bitter substances.76 Medium-chain fatty acids activate both bitter and sweet gustatory receptor neurons, raising the possibility that the age-dependent shift in OBP expression contributes to the decrease in sugar sensitivity with age.32 Further study of how obp18a and other OBP family genes contribute to sugar taste during aging may inform our understanding of why there is a reduction in sugar, but not fatty acid taste.
We also identify numerous genes that are upregulated with aging. The finding that the immediate-early gene Arc1 is upregulated in Gr64f+ neurons of aged flies is of particular interest. Arc1 has complex functions in Drosophila including the mediation of transsynaptic RNA trafficking.47 It is possible that either upregulation of Arc1 reflects an overall reduction in Gr64f neuron activity, or that there is altered communication between gustatory receptor neurons and their targets in the SEZ. Together, the 66 genes that are differentially expressed provide the basis for future screening and analysis of age-dependent changes in taste neurons. We have also performed snRNA-seq in 20-day-old flies expressing a human AD variant. We see few DEGs within this cluster. It is possible that the study is underpowered, or the 20 days time point is too early to reveal transcriptional dysregulation. Nevertheless, these findings suggest the behavioral and physiological reductions in taste sensitivity in aged and AD model flies derive from independent mechanisms.
Limitations of the study
While we showed that sugar, but not fatty acid taste preference declines during aging and in an AD fly model, a more thorough investigation of additional taste modalities beyond those measured in this study is needed to inform our understanding of which taste modalities are affected by aging and in models of neurodegenerative disease. This study utilized the PER assay to quantify taste preference behavior. However, this approach is limited to appetitive tastants, and additional assay systems will be required to identify changes in sensitivity to aversive taste stimuli. Lastly, while our snRNA-seq analysis identified several candidate genes associated with aging and AD in sweet-taste neurons, this approach resulted in low read depth per nuclei. Subsequent validation of these candidate genes will be needed to confirm their role in mediating changes in taste preference during aging and in an AD fly model.
Taken together, we have found that aging is associated with reduced sensitivity to sugars but not fatty acids. These findings suggest that the intracellular signaling properties associated with sugar and fatty acid taste are differentially impacted during aging. While expression of an Aβ1-42 variant results in similar behavioral and physiological phenotypes as naturally aged flies, the effects on the connectivity and transcriptional regulation of Gr64f neurons differs. Together, these findings establish the taste system as a model to investigate age- and neurodegenerative disease-associated reductions in sensory function.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Elizabeth B. Brown (email: EBBrown@bio.fsu.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
Raw behavioral and imaging data reported in this study are available in this paper’s supplemental information. snRNA-seq data have been deposited onto GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
All original code is available in this paper’s supplemental information.
-
•
Any additional information required to reanalyze the data reported in this paper will be available from the lead contact upon request.
Acknowledgments
We would like to thank members of the Keene and Dahanukar labs for technical assistance and helpful discussion. This work was supported by the National Institutes of Health grants R01NS085252 to A.C.K., R01DC017390 to A.C.K. and A.D., as well as R00AG071833 to E.B.B. This work was also supported by the NIH T32 grant DC000044 and the National Science Foundation REU grant DBI-1852175.
Author contributions
Conceptualization, E.B.B. and A.C.K.; methodology, E.B.B., E.L., and A.M.-P.; investigation, E.B.B., E.L., R.R., Z.P., A.M.-P., and S.M.; writing – original draft, E.B.B., E.L., and A.C.K.; writing – review & editing, E.B.B., E.L., A.M.-P., A.D., and A.C.K.; funding acquisition, E.B.B., A.D., and A.C.K.; resources, E.B.B. and A.C.K.; supervision, E.B.B. and A.C.K.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-bruchpilot (nc82-s) | Developmental Studies Hybridoma Bank | nc82 RRID: AB_2314866 |
| Donkey anti-mouse Alexa Fluor 647 | Invitrogen | A-31571 |
| Chemicals, peptides, and recombinant proteins | ||
| Bloomington Formulation Nutri-fly food | Genesee Scientific | 66–113 |
| Hexanoic Acid | Sigma Aldrich | 21530 |
| Octanoic Acid | Sigma Aldrich | O3907 |
| L-serine | Sigma Aldrich | 84959 |
| L-phenylalanine | Sigma Aldrich | P5482 |
| Citric Acid | Sigma Aldrich | 251275 |
| Acetic Acid | Sigma Aldrich | 695092 |
| Sucrose | Fisher Scientific | FS S5-500 |
| Fructose | Fisher Scientific | 57-48-7 |
| PBS | Fisher Scientific | BP243820 |
| Paraformaldehyde | Sigma Aldrich | 158127 |
| Triton X-100 | Sigma Aldrich | T8787 |
| VECTASHIELD Antifade Mounting Medium | Vector Labs | H1000 |
| NaCl | Sigma Aldrich | S9888 |
| KCl | Sigma Aldrich | P9541 |
| MgCl2 | Sigma Aldrich | 208337 |
| CaCl2 | Sigma Aldrich | C4901 |
| HEPES-NaOH | Fisher Scientific | 50-197-1 |
| Deposited data | ||
| Raw behavioral and imaging data | This paper | Data S1 |
| Raw and analyzed sequence data | This paper | GSE273715 |
| The script used for Seurat analysis | This paper | Data S2 |
| Experimental models: organisms/strains | ||
| w1118 fly strain | Bloomington Drosophila Stock Center (BDSC) | 5905 |
| Gr64f-GAL4 | BDSC | 57668 |
| UAS-mCD8:GFP | BDSC | 32186 |
| UAS-Aβ1-40 | BDSC | 64215 |
| UAS-Arctic | Crowther et al.21 | – |
| UAS-GCaMP-R | Cho et al.40 | – |
| UAS-Brp-Short | Mosca and Luo45 | – |
| Software and algorithms | ||
| ImageJ/FIJI | Schindelin et al.77 | v1.54j |
| Nikon AR Image Analysis software | Nikon | v5.20.02 |
| Microsoft Office 365 | Microsoft | v2406 |
| Prism | Graphpad software | v10.2.3 |
| Python | https://www.anaconda.com/products/individual | v3.8.0 |
| R | https://www.r-project.org/ | v4.3 |
| RStudio | https://www.rstudio.com/products/rstudio/download/ | v3.6.0 |
| Cell Ranger | 10 Genomics | v6.0 |
| Seurat | Hao et al.78 | v4.3.0 |
| Cell Marker Enrichment tool (DRscDB) | Hu et al.79 | – |
| clusterProfiler | Wu et al.80 | v4.8.3 |
| Other | ||
| Incubators | Percival | I36-VL |
| Laser-scanning confocal microscope | Nikon | A1R |
| Micromanipulator | Narishige International USA | MM-3 |
| Magnetic Stand | Narishige International USA | GJ-8 |
| Capillary Tubes | World Precision Instruments | 1B120F-4 |
| Dental Glue | Ivoclar Vivadent | 595953WW |
Experimental model and subject details
Fly stocks and maintenance
Flies were grown and maintained on standard food media (Bloomington Recipe; Genesee Scientific, San Diego, CA). Flies were housed in incubators (Powers Scientific; Warminster, PA, USA) on a 12:12 LD cycle at 25°C with a humidity level of 55–65%. The following fly strains were ordered from the Bloomington Stock Center: w1118 (#5905; Levis et al.81), Gr64f-GAL4 (#57668; Dahanukar et al.14), UAS-mCD8GFP (#32186; Pfeiffer et al.82), UAS-Aβ1-40 (#64215; Jonson et al.29). UAS-Arctic was provided by Matt Kayser and was previously described in Crowther et al.,21 UAS-GCaMP-R was provided by Greg Macleod and was previously described in Cho et al.,40 and UAS-Brp-Short was provided by Tim Mosca and was previously described in Mosca and Luo.45 All flies were backcrossed to the w1118 genetic background for a minimum of 6 generations prior to testing. For aging experiments, w1118 flies were isolated 1 day post-eclosion, sorted by sex into vials of ∼30 flies, and then transferred to new vials every other day until they reached the indicated age for each experiment.
Method details
Proboscis extension response
Female flies were starved for 24 h prior to each experiment and then PER was measured as previously described Masek and Keene11 and Brown et al.83 In experiments using males, flies were starved for 18 h prior to each experiment. Briefly, flies were anesthetized with CO2 and then glued to a microscope slide (#12-550-15; Fisher Scientific) so that their head and proboscis were free to move while their thorax and abdomen remained restrained. After a 60 min acclimation period in a humidified box, flies were presented with water and allowed to drink freely until satiated. Flies that did not stop responding to water within 5 min were discarded. A wick made of Kimwipe (#06–666; Fisher Scientific) was placed partially inside a capillary tube (#1B120F-4; World Precision Instruments; Sarasota, FL) and then saturated with tastant. The saturated wick was then manually applied to the tip of the proboscis for 1–2 s and proboscis extension reflex was monitored. Only full extensions were counted as a positive response. Each tastant was presented a total of three times, with 1 min between each presentation. Both sugars and fatty acids were dissolved in water and tested at the indicated concentration. PER was calculated as the percentage of proboscis extensions divided by the total number of tastant presentations. For example, a fly that extends its proboscis twice out of the three presentations will have a PER response of 66%. Experiments were run ∼3 times per week until completion. Genotype and tastant presentation were randomized to ensure data reproducibility.
Immunohistochemistry
For innervation and active zone measurements, the brains of female flies were dissected in ice-cold phosphate buffered saline (PBS) (#BP243820; Fisher Scientific) and fixed in 4% formaldehyde, PBS, and 0.5% Triton X- for 30 min at room temperature. Brains were rinsed 3 with PBS and 0.5% Triton X-(PBST) for 10 min at room temperature and then mounted in Vectashield (VECTOR Laboratories; Burlingame, CA). For the representative image of Gr64f expression, brains were dissected in PBS and then fixed for 30 min at room temperature. Brains were rinsed 3 with PBST for 10 min then incubated overnight at 4°C. The next day, brains were incubated in primary antibody (1:20 mouse nc82; Iowa Hybridoma Bank; The Developmental Studies Hybridoma Bank; Iowa City, Iowa, USA) diluted in 0.5% PBST at 4°C for 48 h. Next, the brains were rinsed 3 with PBST for 10 min at room temperature and placed in secondary antibody (1:400 donkey anti-mouse Alexa Fluor 647; #A-31571; ThermoFisher Scientific; Waltham, Massachusetts, USA) for 90 min at room temperature. The brains were again rinsed 3 with PBST for 10 min at room temperature and then mounted in Vectashield. All brains were imaged in 1 μm z-plane interval sections on a Nikon A1R confocal microscope (Nikon; Tokyo, Japan) using a 20X oil immersion objective.
In vivo calcium imaging
Flies expressing UAS-GCaMP-R (GCaMP6.0 and mCherry) in Gr64f neurons were starved for 24 h prior to imaging, as previously described.Brown et al.32 Flies were anesthetized on ice and then restrained inside of a cut 200 μL pipette tip so that their head and proboscis were accessible, while their body and tarsi remained restrained. The proboscis was manually extended and then a small amount of dental glue (#595953WW; Ivoclar Vivadent Inc.; Amherst, NY) was applied between the labium and the side of the pipette tip, ensuring the same position throughout the experiment. Next, both antennae were removed. A small hole was cut into a 1 cm2 piece of aluminum foil and then fixed to the fly using dental glue, creating a sealed window of cuticle exposed. Artificial hemolymph (140 mM NaCl, 2 mM KCl, 4.5 mM MgCl2, 1.5 mM CaCl2, and 5 mM HEPES-NaOH with pH = 7.1) was applied to the window and then the cuticle and connective tissue were dissected to expose the SEZ. Mounted flies were placed on a Nikon A1R confocal microscope and then imaged using a 20 water-dipping objective lens. The pinhole was opened to allow a thicker optical section to be monitored. Gr64f neurons were simultaneously excited with wavelengths of 488 nm (FITC) and 561 nm (TRITC). All recordings were taken at 4 Hz with 256 resolution. Like PER, tastants were applied to the proboscis for 1–2 s with a wick, which was operated using a micromanipulator (Narishige International USA, Inc.; Amityville, NY). Experiments were run ∼3 times per week until completion.
Single-nucleus RNA sequencing
Tissue collection, nuclei isolation, library preparation, and sequencing
Fly labellums were dissected at 10 and 40 days (for aging samples) or at 20 days (for control and AD model samples) and then placed into 1.5 mL RNase-free Eppendorf tubes, flash-frozen in liquid nitrogen, and then stored at −80°C. Approximately ∼400 labellum were collected for each treatment and stored on dry ice. Nuclei isolation and snRNAseq were performed by Singulomics Corporation (Singulomics.com; New York). To isolate nuclei, tissue was homogenized and lysed with Triton X-100 in RNase-free water. The nuclei were then purified, centrifuged, resuspended in PBS with RNase Inhibitor, and diluted to 700 nuclei/μL. Standardized 10 capture and library preparation were performed using the 10 Genomics Chromium Next GEM 3′ Single Cell Reagent kit v3.1 (10 Genomics; Pleasanton, CA). The libraries were then sequenced with Illumina NovaSeq 6000 (Illumina; San Diego, CA). The snRNA-seq raw sequencing files were processed with CellRanger 6.0 (10 Genomics; Pleasanton, CA). The sequencing reads were mapped to the Drosophila reference genome (Flybase r6.54).
Data normalization and integration
All analysis of snRNA-seq data was performed in R (v4.3) using Seurat (v4.3.0; Hao et al.78). Unless otherwise noted, default parameters were used for all operations. Each sample was independently normalized using the SCTransform function (v2).84,85 Following normalization, integration features were selected using the SelectIntegrationFeatures function, with nFeatures = 3000. Normalized data was prepared for integration with the PrepSCTIntegration function, using the features identified in the previous step. Integration anchors were identified using the FindIntegrationAnchors function, with normalization.method = “SCT”. Finally, the data was integrated with the IntegrateData command, using the previously identified anchors.
Cell type clustering and dimensionality reduction
Following integration, Principal Component Analysis was performed using the RunPCA function, with npcs = 30. The resulting PCs were used to identify clusters, with a resolution of 0.3, resulting in 13 total clusters. The integrated data was prepared for marker identification by running the PrepSCTFindMarkers function, and then markers were identified with the FindAllMarkers command, using the MAST statistical test,86 which has been found to perform well on single-cell datasets.87 The list of marker genes was used to annotate cluster identity. First, we compared the marker genes for each cluster to previously generated datasets in Drosophila, using the Cell Marker Enrichment tool (DRscDB; https://www.flyrnai.org/tools/single_cell/web/enrichment 79). Wherever possible, we refined our labels by comparing marker genes with published literature marking known cell types.88
Differential gene expression (differentially expressed gene) aging analysis
To identify genes in Gr64f+ nuclei that are differentially expressed in aged flies, we applied the FindMarkers function on these cells, again using the MAST statistical test, and set group.by = ’age’. Genes with an adjusted p value less than 0.05 and an average Log2 Fold Change greater than 0.25 or less than −0.25 were considered differentially expressed and used for downstream analysis.
Gene Ontology analysis
GO pathway analyses were conducted in R (v.4.3.0), using the clusterProfiler package (v4.8.3; Wu et al.80). For gene set enrichment analyses, a log2FC value was calculated between the young (10days) and aged (40days) groups for each gene expressed in the cell population of interest. The sorted log2FC list was passed to the gseGO function (with options: ont = ‘BP’, keyType = ‘SYMBOL’, minGSSize = 3, maxGSSize = 800, OrgDb = org.Dm.e.g.,.db, pvalueCutoff = 0.01). The results of this test were passed to the dotplot function for visualization.
Quantification and statistical analyses
Quantification of volume and fluorescence intensity
Quantification of mCD8GFP or Brp-short was performed by generating a sum intensity projection of the Gr64f neurons. A threshold of 491 (low) to 65535 (high) was applied to all images prior to quantification. One region of interest (ROI) was drawn around the dorsal taste peg and internal mouthpart projections while a second ROI was drawn around the ventral labellar projections. For volume measurements, the sum of the total number of fluorescent pixels in each slice was used. For intensity measurements, the mean of the intensity averaged from each slice was used. Images are presented as the z stack projection through the entire brain and processed using Fiji.77
Quantification of calcium activity
To quantify UAS-GCaMP-R activity, regions of interest were first drawn manually around the Gr64f projections of one hemisphere, alternating between flies. For each frame, the mean fluorescence intensity for FITC and TRITC was subtracted from background mean fluorescence intensity. Then, the fluorescence ratio of GCaMP6.0 to mCherry was calculated. Next, baseline fluorescence was calculated as the average fluorescence ratio of the first 5 frames, 10 s prior to tastant application. For each frame, the % change in fluorescence (%ΔF/F) was then calculated as: [(peak fluorescence ratio - baseline fluorescence ratio)/baseline fluorescence ratio] ∗ 100. Average fluorescence traces were created by taking the average and standard error of %ΔF/F for each recording of a specific tastant.
Statistical analysis
All measurements are presented as bar graphs or line graphs showing mean ± standard error. Measurements of PER were not normally distributed; therefore, the non-parametric restricted maximum likelihood estimation was used. For all other measurements, either a t-test or a two-way analysis of variance (ANOVA) was used. Post hoc analyses were performed using Sidak’s multiple comparisons test. Statistical analyses and data presentation were performed using InStat software (GraphPad Software 8.0; San Diego, CA). Sample sizes for behavioral and functional imaging experiments are consistent with previous studies.32 Generally, ∼30–50 flies were used for each experimental or control group for behavioral experiments and ∼10 flies per group for imaging experiments.
Published: September 10, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110919.
Contributor Information
Elizabeth B. Brown, Email: ebbrown@bio.fsu.edu.
Alex C. Keene, Email: akeene@bio.tamu.edu.
Supplemental information
References
- 1.Doty R.L., Kamath V. The influences of age on olfaction: a review. Front. Psychol. 2014;5:20. doi: 10.3389/fpsyg.2014.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doty R.L. Epidemiology of smell and taste dysfunction. Handb. Clin. Neurol. 2019;164:3–13. doi: 10.1016/B978-0-444-63855-7.00001-0. [DOI] [PubMed] [Google Scholar]
- 3.Kondo K., Kikuta S., Ueha R., Suzukawa K., Yamasoba T. Age-related olfactory dysfunction: epidemiology, pathophysiology, and clinical management. Front. Aging Neurosci. 2020;12:208. doi: 10.3389/fnagi.2020.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Methven L., Allen V.J., Withers C.A., Gosney M.A. Ageing and taste. Proc. Nutr. Soc. 2012;71:556–565. doi: 10.1017/S0029665112000742. [DOI] [PubMed] [Google Scholar]
- 5.Son G., Jahanshahi A., Yoo S.J., Boonstra J.T., Hopkins D.A., Steinbusch H.W.M., Moon C. Olfactory neuropathology in Alzheimer’s disease: a sign of ongoing neurodegeneration. BMB Rep. 2021;54:295–304. doi: 10.5483/BMBRep.2021.54.6.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haehner A., Hummel T., Reichmann H. Olfactory loss in Parkinson’s Disease. Parkinsons Dis. 2011;2011 doi: 10.4061/2011/450939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy C. Olfactory and other sensory impairments in Alzheimer disease. Nat. Rev. Neurol. 2019;15:11–24. doi: 10.1038/s41582-018-0097-5. [DOI] [PubMed] [Google Scholar]
- 8.Thorne N., Chromey C., Bray S., Amrein H. Taste Perception and Coding in Drosophila. Curr Biolo. 2004;14:1065–1079. doi: 10.1016/j.cub.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 9.Vosshall L.B., Stocker R.F. Molecular architecture of smell and taste in Drosophila. Annu. Rev. Neurosci. 2007;30:505–533. doi: 10.1146/annurev.neuro.30.051606.094306. [DOI] [PubMed] [Google Scholar]
- 10.Yarmolinsky D.A., Zuker C.S., Ryba N.J.P. Common Sense about Taste: From Mammals to Insects. Cell. 2009;139:234–244. doi: 10.1016/j.cell.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masek P., Keene A.C. Gustatory processing and taste memory in Drosophila. J. Neurogenet. 2016;30:112–121. doi: 10.1080/01677063.2016.1185104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiao Y., Moon S.J., Wang X., Ren Q., Montell C. Gr64f Is Required in Combination with Other Gustatory Receptors for Sugar Detection in Drosophila. Curr. Biol. 2008;18:1797–1801. doi: 10.1016/j.cub.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slone J., Daniels J., Amrein H. Sugar Receptors in Drosophila. Curr. Biol. 2007;17:1809–1816. doi: 10.1016/j.cub.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dahanukar A., Lei Y.T., Kwon J.Y., Carlson J.R. Two Gr Genes Underlie Sugar Reception in Drosophila. Neuron. 2007;56:503–516. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beharry C., Alaniz M.E., Alonso A.D.C. Expression of Alzheimer-like pathological human tau induces a behavioral motor and olfactory learning deficit in Drosophila melanogaster. J. Alzheimers Dis. 2013;37:539–550. doi: 10.3233/JAD-130617. [DOI] [PubMed] [Google Scholar]
- 16.Dissel S., Angadi V., Kirszenblat L., Suzuki Y., Donlea J., Klose M., Koch Z., English D., Winsky-Sommerer R., van Swinderen B., Shaw P.J. Sleep restores behavioral plasticity to drosophila mutants. Curr. Biol. 2015;25:1270–1281. doi: 10.1016/j.cub.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabuchi M., Lone S.R., Liu S., Liu Q., Zhang J., Spira A.P., Wu M.N. Sleep Interacts with aβ to Modulate Intrinsic Neuronal Excitability. Curr. Biol. 2015;25:702–712. doi: 10.1016/j.cub.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen K.F., Possidente B., Lomas D.A., Crowther D.C. The central molecular clock is robust in the face of behavioural arrhythmia in a Drosophila model of Alzheimer’s disease. Dis. Model. Mech. 2014;7:445–458. doi: 10.1242/dmm.014134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sofola O., Kerr F., Rogers I., Killick R., Augustin H., Gandy C., Allen M.J., Hardy J., Lovestone S., Partridge L. Inhibition of GSK-3 ameliorates Aβ pathology in an adult-onset Drosophila model of Alzheimer’s disease. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iijima K., Chiang H.C., Hearn S.A., Hakker I., Gatt A., Shenton C., Granger L., Leung A., Iijima-Ando K., Zhong Y. Aβ42 Mutants with Different Aggregation Profiles Induce Distinct Pathologies in Drosophila. PLoS One. 2008;3 doi: 10.1371/journal.pone.0001703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crowther D.C., Kinghorn K.J., Miranda E., Page R., Curry J.A., Duthie F.A.I., Gubb D.C., Lomas D.A. Intraneuronal Aβ, non-amyloid aggregates and neurodegeneration in a Drosophila model of Alzheimer’s disease. Neuroscience. 2005;132:123–135. doi: 10.1016/j.neuroscience.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 22.Keene A.C., Masek P. Optogenetic induction of aversive taste memory. Neuroscience. 2012;222:173–180. doi: 10.1016/j.neuroscience.2012.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scott K. Gustatory Processing in Drosophila melanogaster. Annu. Rev. Entomol. 2018;63:15–30. doi: 10.1146/annurev-ento-020117-043331. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y.C.D., Dahanukar A. Recent advances in the genetic basis of taste detection in Drosophila. Cell. Mol. Life Sci. 2020;77:1087–1101. doi: 10.1007/s00018-019-03320-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers I., Kerr F., Martinez P., Hardy J., Lovestone S., Partridge L. Ageing increases vulnerability to aβ42 toxicity in Drosophila. PLoS One. 2012;7 doi: 10.1371/journal.pone.0040569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martín-Peña A., Rincón-Limas D.E., Fernandez-Fúnez P. Engineered Hsp70 chaperones prevent Aβ42-induced memory impairments in a Drosophila model of Alzheimer’s disease. Sci. Rep. 2018;8:9915. doi: 10.1038/s41598-018-28341-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martín-Peña A., Rincón-Limas D.E., Fernandez-Fúnez P. Anti-Aβ single-chain variable fragment antibodies restore memory acquisition in a Drosophila model of Alzheimer’s disease. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-11594-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iijima K., Liu H.P., Chiang A.S., Hearn S.A., Konsolaki M., Zhong Y. Dissecting the pathological effects of human Aβ40 and Aβ42 in Drosophila: A potential model for Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2004;101:6623–6628. doi: 10.1073/pnas.0400895101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jonson M., Pokrzywa M., Starkenberg A., Hammarstrom P., Thor S. Systematic Aβ Analysis in Drosophila Reveals High Toxicity for the 1-42, 3-42 and 11-42 Peptides, and Emphasizes N- and C-Terminal Residues. PLoS One. 2015;10 doi: 10.1371/journal.pone.0133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn J.E., Chen Y., Amrein H. Molecular basis of fatty acid taste in Drosophila. Elife. 2017;6:e30115. doi: 10.7554/eLife.30115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tauber J.M., Brown E.B., Li Y., Yurgel M.E., Masek P., Keene A.C. A subset of sweet-sensing neurons identified by IR56d are necessary and sufficient for fatty acid taste. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1007059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown E.B., Shah K.D., Palermo J., Dey M., Dahanukar A., Keene A.C. Ir56d-dependent fatty acid responses in Drosophila uncover taste discrimination between different classes of fatty acids. Elife. 2021;10 doi: 10.7554/eLife.67878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ueno K., Kohatsu S., Clay C., Forte M., Isono K., Kidokoro Y. Gsα Is Involved in Sugar Perception in Drosophila melanogaster. J. Neurosci. 2006;26:6143–6152. doi: 10.1523/JNEUROSCI.0857-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q.P., Lin Y.Q., Lai M.L., Su Z., Oyston L.J., Clark T., Park S.J., Khuong T.M., Lau M.T., Shenton v., et al. PGCα Controls Sucrose Taste Sensitization in Drosophila. Cell Rep. 2020;31:1. doi: 10.1016/j.celrep.2020.03.044. [DOI] [PubMed] [Google Scholar]
- 35.Kim H., Kim H., Kwon J.Y., Seo J.T., Shin D.M., Moon S.J. Drosophila Gr64e mediates fatty acid sensing via the phospholipase C pathway. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aryal B., Dhakal S., Shrestha B., Lee Y. Molecular and neuronal mechanisms for amino acid taste perception in the Drosophila labellum. Curr. Biol. 2022;32:1376–1386.e4. doi: 10.1016/j.cub.2022.01.060. [DOI] [PubMed] [Google Scholar]
- 37.Mi T., Mack J.O., Lee C.M., Zhang Y.V. Molecular and cellular basis of acid taste sensation in Drosophila. Nat. Commun. 2021;12:3730. doi: 10.1038/s41467-021-23490-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yurgel M.E., Kakad P., Zandawala M., Nässel D.R., Godenschwege T.A., Keene A.C. A single pair of leucokinin neurons are modulated by feeding state and regulate sleep–metabolism interactions. PLoS Biol. 2019;17 doi: 10.1371/journal.pbio.2006409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ugur B., Bao H., Stawarski M., Duraine L.R., Zuo Z., Lin Y.Q., Neely G.G., Macleod G.T., Chapman E.R., Bellen H.J. The Krebs Cycle Enzyme Isocitrate Dehydrogenase 3A Couples Mitochondrial Metabolism to Synaptic Transmission. Cell Rep. 2017;21:3794–3806. doi: 10.1016/j.celrep.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho J.H., Swanson C.J., Chen J., Li A., Lippert L.G., Boye S.E., Rose K., Sivaramakrishnan S., Chuong C.M., Chow R.H. The GCaMP-R Family of Genetically Encoded Ratiometric Calcium Indicators. ACS Chem. Biol. 2017;12:1066–1074. doi: 10.1021/acschembio.6b00883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fouquet W., Owald D., Wichmann C., Mertel S., Depner H., Dyba M., Hallermann S., Kittel R.J., Eimer S., Sigrist S.J. Maturation of active zone assembly by Drosophila Bruchpilot. J. Cell Biol. 2009;186:129–145. doi: 10.1083/jcb.200812150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagh D.A., Rasse T.M., Asan E., Hofbauer A., Schwenkert I., Dürrbeck H., Buchner S., Dabauvalle M.C., Schmidt M., Qin G., et al. Bruchpilot, a Protein with Homology to ELKS/CAST, Is Required for Structural Integrity and Function of Synaptic Active Zones in Drosophila. Neuron. 2006;49:833–844. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 43.Matkovic T., Siebert M., Knoche E., Depner H., Mertel S., Owald D., Schmidt M., Thomas U., Sickmann A., Kamin D., et al. The Bruchpilot cytomatrix determines the size of the readily releasable pool of synaptic vesicles. J. Cell Biol. 2013;202:667–683. doi: 10.1083/jcb.201301072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kittel R.J., Wichmann C., Rasse T.M., Fouquet W., Schmidt M., Schmid A., Wagh D.A., Pawlu C., Kellner R.R., Willig K.I., et al. Bruchpilot Promotes Active Zone Assembly, Ca2+ Channel Clustering, and Vesicle Release. Science. 2006;312:1051–1054. doi: 10.1126/science.1126308. [DOI] [PubMed] [Google Scholar]
- 45.Mosca T.J., Luo L. Synaptic organization of the Drosophila antennal lobe and its regulation by the Teneurins. Elife. 2014;3 doi: 10.7554/eLife.03726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng G.X.Y., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J., et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017;8:14049. doi: 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ashley J., Cordy B., Lucia D., Fradkin L.G., Budnik V., Thomson T. Retrovirus-like Gag Protein Arc1 Binds RNA and Traffics across Synaptic Boutons. Cell. 2018;172:262–274.e11. doi: 10.1016/j.cell.2017.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goto A., Yano T., Terashima J., Iwashita S., Oshima Y., Kurata S. Cooperative Regulation of the Induction of the Novel Antibacterial Listericin by Peptidoglycan Recognition Protein LE and the JAK-STAT Pathway. J. Biol. Chem. 2010;285:15731–15738. doi: 10.1074/jbc.M109.082115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kemp C., Mueller S., Goto A., Barbier V., Paro S., Bonnay F., Dostert C., Troxler L., Hetru C., Meignin C., et al. Broad RNA Interference–Mediated Antiviral Immunity and Virus-Specific Inducible Responses in Drosophila. J. Immunol. 2013;190:650–658. doi: 10.4049/jimmunol.1102486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dostert C., Jouanguy E., Irving P., Troxler L., Galiana-Arnoux D., Hetru C., Hoffmann J.A., Imler J.L. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat. Immunol. 2005;6:946–953. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- 51.Kim K.M., Meng Q., Perez de Acha O., Mustapic M., Cheng A., Eren E., Kundu G., Piao Y., Munk R., Wood W.H., 3rd, et al. Mitochondrial RNA in Alzheimer’s Disease circulating extracellular vesicles. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.581882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Masek P., Keene A.C. Drosophila fatty acid taste signals through the PLC pathway in sugar-sensing neurons. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim S.H., Lee Y., Akitake B., Woodward O.M., Guggino W.B., Montell C. Drosophila TRPA1 channel mediates chemical avoidance in gustatory receptor neurons. Proc. Natl. Acad. Sci. 2010;107:8440–8445. doi: 10.1073/pnas.1001425107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schiffman S.S. Effects of aging on the human taste system. Ann. N. Y. Acad. Sci. 2009;1170:725–729. doi: 10.1111/j.1749-6632.2009.03924.x. [DOI] [PubMed] [Google Scholar]
- 55.Tzeng W.Y., Figarella K., Garaschuk O. Olfactory impairment in men and mice related to aging and amyloid-induced pathology. Pflugers Arch. 2021;473:805–821. doi: 10.1007/s00424-021-02527-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shin Y.K., Cong W.N., Cai H., Kim W., Maudsley S., Egan J.M., Martin B. Age-related changes in mouse taste bud morphology, hormone expression, and taste responsivity. J. Gerontol. A Biol. Sci. Med. Sci. 2012;67:336–344. doi: 10.1093/gerona/glr192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Claus L.E., Leland E.M., Tai K.Y., Schlosser R.J., Kamath V., Lane A.P., Rowan N.R. Olfactory loss and beyond: a practical review of chemosensory dysfunction. J. Am. Board Fam. Med. 2022;35:406–419. doi: 10.3122/jabfm.2022.02.210373. [DOI] [PubMed] [Google Scholar]
- 58.Doty R.L., Hawkes C.H. In: Doty R.L., editor. Vol. 164. 2019. Chemosensory Dysfunction in Neurodegenerative Diseases; pp. 325–360. (Handb. Clin. Neurol.). [DOI] [PubMed] [Google Scholar]
- 59.Wilson D.M., Cookson M.R., Van Den Bosch L., Zetterberg H., Holtzman D.M., Dewachter I. Hallmarks of neurodegenerative diseases. Cell. 2023;186:693–714. doi: 10.1016/j.cell.2022.12.032. [DOI] [PubMed] [Google Scholar]
- 60.Müller A., Reichmann H., Livermore A., Hummel T. Olfactory function in idiopathic Parkinson’s disease (IPD): results from cross-sectional studies in IPD patients and long-term follow-up of de-novo IPD patients. J. Neural. Transm. 2002;109:805–811. doi: 10.1007/s007020200067. [DOI] [PubMed] [Google Scholar]
- 61.Tian Q., Bilgel M., Moghekar A.R., Ferrucci L., Resnick S.M. Olfaction, cognitive impairment, and PET biomarkers in community-dwelling older adults. J. Alzheimers Dis. 2022;86:1275–1285. doi: 10.3233/JAD-210636. [DOI] [PubMed] [Google Scholar]
- 62.Ansoleaga B., Garcia-Esparcia P., Llorens F., Moreno J., Aso E., Ferrer I. Dysregulation of brain olfactory and taste receptors in AD, PSP, and CJD and AD-related model. Neuroscience. 2013;248:369–382. doi: 10.1016/j.neuroscience.2013.06.034. [DOI] [PubMed] [Google Scholar]
- 63.Narukawa M., Takahashi S., Saito T., Saido T.C., Misaka T. Analysis of taste sensitivities in App Knock-In Mouse Model of Alzheimer’s Disease. J. Alzheimers Dis. 2020;76:997–1004. doi: 10.3233/JAD-200284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shafer O.T., Kim D.J., Dunbar-Yaffe R., Nikolaev V.O., Lohse M.J., Taghert P.H. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hackley C.R., Mazzoni E.O., Blau J. cAMPr: a single-wavelength fluorescent sensor for cyclic AMP. Sci. Signal. 2018;11 doi: 10.1126/scisignal.aah3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gerstner J.R., Lenz O., Vanderheyden W.M., Chan M.T., Pfeiffenberger C., Pack A.I. Amyloid-β induces sleep fragmentation that is rescued by fatty acid binding proteins in Drosophila. J. Neurosci. Res. 2017;95:1548–1564. doi: 10.1002/jnr.23778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kaldun J.C., Lone S.R., Humbert Camps A.M., Fritsch C., Widmer Y.F., Stein J.V., Tomchik S.M., Sprecher S.G. Dopamine, sleep, and neuronal excitability modulate amyloid-β–mediated forgetting in Drosophila. PLoS Biol. 2021;19 doi: 10.1371/journal.pbio.3001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chiang H.C., Wang L., Xie Z., Yau A., Zhong Y. PI3 kinase signaling is involved in Aβ-induced memory loss in Drosophila. Proc. Natl. Acad. Sci. USA. 2010;107:7060–7065. doi: 10.1073/pnas.0909314107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kerr F., Augustin H., Piper M.D.W., Gandy C., Allen M.J., Lovestone S., Partridge L. Dietary restriction delays aging but not neuronal dysfunction in Drosophila models of Alzheimer’s disease. Neurobiol. Aging. 2011;32:1977–1989. doi: 10.1016/j.neurobiolaging.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martin-Peña A., Ferrus A. CCB is involved in actin-based axonal transport of selected synaptic proteins. J. Neurosci. 2020;40:542–556. doi: 10.1523/JNEUROSCI.0915-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Montero-Crespo M., Domínguez-Álvaro M., Alonso-Nanclares L., DeFelipe J., Blazquez-Llorca L. Three-dimensional analysis of synaptic organization in the hippocampal CA1 field in Alzheimer’s disease. Brain. 2021;144:553–573. doi: 10.1093/brain/awaa406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meftah S., Gan J. Alzheimer’s disease as a synaptopathy: Evidence for dysfunction of synapses during disease progression. Front. Synaptic Neurosci. 2023;15 doi: 10.3389/fnsyn.2023.1129036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arroyo-García L.E., Isla A.G., Andrade-Talavera Y., Balleza-Tapia H., Loera-Valencia R., Alvarez-Jimenez L., Pizzirusso G., Tambaro S., Nilsson P., Fisahn A. Impaired spike-gamma coupling of area CA3 fast-spiking interneurons as the earliest functional impairment in the App NL-GF mouse model of Alzheimer’s disease. Mol. Psychiatry. 2021;26:5557–5567. doi: 10.1038/s41380-021-01257-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ozaki M. Olfactory Concepts of Insect Control - Alternative to Insecticides. Springer; Cham: 2019. Odorant-binding Proteins in Taste System: Putative Roles in Taste Sensation and Behavior; pp. 187–204. [Google Scholar]
- 75.Swarup S., Morozova T.V., Sridhar S., Nokes M., Anholt R.R.H. Modulation of feeding behavior by odorant-binding proteins in Drosophila melanogaster. Chem. Senses. 2014;39:125–132. doi: 10.1093/chemse/bjt061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jeong Y.T., Shim J., Oh S.R., Yoon H.I., Kim C.H., Moon S.J., Montell C. An odorant-binding protein required for suppression of sweet taste by bitter chemicals. Neuron. 2013;79:725–737. doi: 10.1016/j.neuron.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hu Y., Tattikota S.G., Liu Y., Comjean A., Gao Y., Forman C., Kim G., Rodiger J., Papatheodorou I., Dos Santos G., et al. DRscDB: A single-cell RNA-seq resource for data mining and data comparison across species. Comput. Struct. Biotechnol. J. 2021;19:2018–2026. doi: 10.1016/j.csbj.2021.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L.I., et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2 doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Levis R., Hazelrigg T., Rubin G.M. Effects of genomic position on the expression of transduced copies of the white gene of Drosophila. Science. 1985;229:558–561. doi: 10.1126/science.2992080. [DOI] [PubMed] [Google Scholar]
- 82.Pfeiffer B.D., Ngo T.T.B., Hibbard K.L., Murphy C., Jenett A., Truman J.W., Rubin G.M. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brown E., Worden K., Li Y., Masek P., Keene A.C. Measurement of Reflexive Feeding Response in Drosophila. Cold Spring Harb Protoc. 2023;2023 doi: 10.1101/pdb.prot108092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Choudhary S., Satija R. Comparison and evaluation of statistical error models for scRNA-seq. Genome Biol. 2022;23:27. doi: 10.1186/s13059-021-02584-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Finak G., McDavid A., Yajima M., Deng J., Gersuk V., Shalek A.K., Slichter C.K., Miller H.W., McElrath M.J., Prlic M., et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16:278. doi: 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Luecken M.D., Theis F.J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 2019;15 doi: 10.15252/msb.20188746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Prelic S., Pal Mahadevan V., Venkateswaran V., Lavista-Llanos S., Hansson B.S., Wicher D. Functional interaction between Drosophila olfactory sensory neurons and their support cells. Front. Cell. Neurosci. 2021;15 doi: 10.3389/fncel.2021.789086. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw behavioral and imaging data reported in this study are available in this paper’s supplemental information. snRNA-seq data have been deposited onto GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
All original code is available in this paper’s supplemental information.
-
•
Any additional information required to reanalyze the data reported in this paper will be available from the lead contact upon request.