Abstract
Earlier studies have shown that cdc2 kinase is activated during herpes simplex virus 1 infection and that its activity is enhanced late in infection even though the levels of cyclin A and B are decreased below levels of detection. Furthermore, activation of cdc2 requires the presence of infected cell protein no. 22 and the UL13 protein kinase, the same gene products required for optimal expression of a subset of late genes exemplified by US11, UL38, and UL41. The possibility that the activation of cdc2 and expression of this subset may be connected emerged from the observation that dominant negative cdc2 specifically blocked the expression of US11 protein in cells infected and expressing dominant negative cdc2. Here we report that in the course of searching for a putative cognate partner for cdc2 that may have replaced cyclins A and B, we noted that the DNA polymerase processivity factor encoded by the UL42 gene contains a degenerate cyclin box and has been reported to be structurally related to proliferating cell nuclear antigen, which also binds cdk2. Consistent with this finding, we report that (i) UL42 is able to physically interact with cdc2 at both the amino-terminal and carboxyl-terminal domains, (ii) the carboxyl-terminal domain of UL42 can be phosphorylated by cdc2, (iii) immunoprecipitates obtained with anti UL42 antibody contained a roscovitine-sensitive kinase activity, (iv) kinase activity associated with UL42 could be immunodepleted by antibody to cdc2, and (v) UL42 transfected into cells associates with a nocodazole-enhanced kinase. We conclude that UL42 can associate with cdc2 and that the kinase activity has the characteristic traits of cdc2 kinase.
In this report we show that the mitotic cyclin-dependent kinase interacts with the herpes simplex virus 1 (HSV-1) protein product of the UL42 open reading frame. The viral protein functions as a DNA polymerase-associated processivity factor. We have identified this protein as a potential virally encoded partner for cdc2 on the basis of its cyclin-like characteristics, and in this report we show that it interacts physically with cdc2. Relevant to this report are the following.
(i) The roots of this investigation rest on the observation that infected-cell protein 0 (ICP0), a promiscuous transactivator encoded by the α0 gene, binds to and stabilizes cyclin D3 (13, 31). In the ensuing investigation it became apparent that the stabilization of cyclin D3 was not associated with the transition from G1 to S phase of the cell cycle. Specifically, cdk2 was inactive and members of the E2F family of proteins required for transcriptional activation of the S phase genes were either sequestered in the cytoplasm or rendered inactive (2, 8, 23, 32). It also became apparent that while HSV stabilized cyclin D3, at least two other herpesviruses, herpessaimiri virus and human herpesvirus 8, encoded cyclin D homologs (17, 21). The obvious conclusion was that herpesviruses require D cyclins although, at least in the case of HSV, for other purposes than those used by the host cell. Ultimately, cyclin D3 was shown to play a role in the translocation of ICP0 from the nucleus to the cytoplasm in HSV-1-infected cells (32).
(ii) The evolving studies on the interaction of cyclin D3 with viral proteins led us to investigate the mitotic cyclins and their kinase. In these studies we found that while cyclins A and B turned over by 8 h after infection, their partner, cdc2, was stabilized and actively phosphorylated histone H1 (1). Moreover, the stabilization of cdc2 required the expression of two viral genes, a regulatory protein, ICP22, encoded by the α22 gene and a viral protein kinase encoded by the UL13 open reading frame.
(iii) The requirement for ICP22 and UL13 protein kinase for the stabilization of cdc2 was investigated in two series of experiments. First, HSV-1 genes form several groups whose expression is coordinately regulated and sequentially ordered (26). The expression of α genes, the first set to be expressed, does not require prior synthesis of viral proteins. The expression of β genes requires α gene products but does not require viral DNA synthesis. The γ1 genes are expressed in the absence of viral DNA synthesis, but their expression is significantly enhanced by the onset of synthesis of viral DNA. Lastly, the γ2 genes require viral DNA for their expression. Of the γ2 genes, a small subset exemplified by US11, UL38, and UL41, require the presence of ICP22 and UL13 protein kinase, the same proteins required for the stabilization of cdc2 (22, 25, 28). To test the connection, cells were transfected with a dominant negative form of cdc2 (cdc-dn) and then infected with wild-type HSV-1. The results were that infected cells expressing cdc2-dn also expressed representative α, β, and γ1 proteins but not the US11 protein (3).
The second series of experiments centered on the pathway by which cdc2 becomes activated. cdc2, like other cyclin-dependent kinases, is present throughout the cell cycle, but its activity is tightly regulated (14). In the case of cdc2, the kinase activity is turned off by phosphorylation by wee-1 and myt-1 and activated by dephosphorylation by activated (phosphorylated) cdc25C and, at a subsequent stage, by phosphorylation by cyclin-dependent activating kinase. These studies indicated that in wild-type virus-infected cells wee-1 is modified and cdc25C is hyperphosphorylated (1). The posttranslational modification of cdc25C phosphatase required the presence of ICP22 and UL13 protein kinase.
These studies led to the conclusion that cdc2 is specifically activated by virally induced modification of cdc2 regulators.
(iv) In uninfected cells, activated cdc2 forms a heterodimer with its cognate partner, cyclin A or cyclin B. Inasmuch as cyclins A and B are no longer detectable at the time after infection when cdc2 is fully active, it could be expected that cdc2 acquired a new cognate partner. The members of the cyclin family members have a characteristic cyclin box motif described as a 32-amino-acid pattern that is also shared by the cyclin homologs encoded by herpesvirus saimiri, human herpesvirus 8, and Autographa californica nucleopolyhedrovirus (6, 17, 21). The consensus phosphorylation site of activated cdc2 is S/T-P-X-K/R/H (11, 18). Earlier we have shown that the sequence encoded by exon 2 of ICP0 and ICP4 containing the consensus site are indeed phosphorylated by cdc2, and other herpesvirus proteins carrying this consensus sequence have been shown to serve as substrates of cdc2 (3, 4). However, these substrates do not appear to have the properties of a putative cognate partner inasmuch as they do not have cyclin box motif. In an attempt to determine whether any HSV protein matched the predicted structure of a cyclin, the HSV-1 open reading frames were scanned for the presence of this motif. The results were that although no HSV-1 protein contained a perfect cyclin box consensus pattern, a degenerate version of the consensus pattern was identified in the UL42 open reading frame. Starting at amino acid 51, the sequence of UL42 is RTSLLDSLLVMGDRGILIHNTIFGEQVF-LPLEH, which resembles a cyclin box motif. The hypothesis that UL42 protein could serve as a partner for cdc2 emerged from the report that the crystal structure of UL42 (34) contains elements similar to those of proliferating-cell nuclear antigen (PCNA). PCNA has been previously reported to interact with cdk2 (16). The objectives of this study were to test this hypothesis. We report that UL42 is able to physically associate with cdc2. The carboxy half of UL42 can be phosphorylated by cdc2 kinase. Immunoprecipitation with UL42 antibody pulls down a kinase activity with properties similar to those of cdc2 in that it is inhibited by roscovitine, immunodepleted by cdc2 antibody, and enhanced by nocodazole.
MATERIALS AND METHODS
Cells and viruses.
HEp-2 cells were initially obtained from the American Type Culture Collection and maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum (NBCS). HSV-1(F) is the prototype HSV-1 wild-type strain used in this laboratory (9).
Plasmids.
The UL42 gene was cloned into PGEX4T-1 and pCDNA3.1(+) and cdc2-dn was cloned into PGEX4T-1 by PCR. The oligonucleotides listed below containing terminal EcoRI or XhoI restrictions sites were used in PCR to generate full-length UL42 or N- and C-terminal portions of UL42 from the BamHI I fragment of HSV-1(F) within pRB130. Full-length cdc2-dn was generated from a plasmid kindly provided by Sander van den Heuvel (30). The oligonucleotides used for this purpose were oligonucleotide 1 (5′ CC GAA TTC ATG ACG GAT TCC CCT GGC GG), oligonucleotide 2 (5′ CCG CTC GAG G TCA GGG GAA TCC AAA ACC), oligonucleotide 3 (5′ CC GAA TTC ATG GTG TCG TCC AGC ACC AGC), oligonucleotide 4 (5′ CCG CTC GAG G TCA CTT GGT GAG CGC GTT G), oligonucleotide 5 (5′ CC GAA TTC ATG GAA GAT TAT ACC AAA ATA G), and oligonucleotide 6 (5′ CCG CTC GAG G CTA CAT CTT CTT AAT CTG ATT G). Full-length UL42 (amino acids 1 to 488) was generated with oligonucleotides 1 and 2, N-terminal UL42 (amino acids 1 to 244) was generated with oligonucleotides 1 and 4, and C-terminal UL42 (amino acids 226 to 488) was generated with oligonucleotides 2 and 3. The PCR products were ligated into EcoRI-XhoI-digested pGEX4T-1 or pCDNA3.1(+). Plasmids with UL42 inserts were sequence verified at the ligation juncture. Full-length cdc2-dn was generated by PCR with oligonucleotides 5 and 6. The PCR product and PGEX4T-1 were digested with EcoRI-XhoI and ligated. The sequence of plasmid PGEX4T-1/cdc2-dn was verified.
Production and purification of GST fusion proteins.
Glutathione S-transferase (GST) fusion proteins containing GST alone or GST fused to full-length UL42, N-terminal UL42, C-terminal UL42, or full-length cdc2-dn were produced as previously described (3). Briefly, Escherichia coli BL21 cells were transformed with the above-mentioned GST-encoding plasmids (PGEX4T-1 based). Fusion protein production was induced with 100 μM IPTG (isopropyl-β-d-thiogalactopyranoside). Bacteria were lysed, GST fusion proteins were absorbed to glutathione-agarose beads (Sigma), and fusion proteins were eluted with 10 mM glutathione in 50 mM Tris (pH 8.0). The eluted protein solution was dialyzed against phosphate-buffered saline. Protein production was assessed by polyacrylamide gel electrophoresis followed by Coomassie brilliant blue staining, and protein concentrations were measured by the Bradford assay (Bio-Rad).
In vitro transcription-translation.
In vitro-coupled transcription-translation of UL42 was carried out with the TNT T7 Quick Coupled Transcription/Translation System as described by the manufacturer (Promega). Briefly, 40 μl of reticulocyte lysate master mix was combined with 25 μCi of [35S]methionine and 1 μg of plasmid DNA [pCDNA3.1(+) with full-length, amino-terminal, or carboxyl-terminal UL42]. The mixture was reacted at 30°C for 90 min. An aliquot was taken out, and the remainder of the reaction mixture was subjected to GST pull down as follows: 300 μl of IP buffer (20 mM Tris [pH 8.0], 1 mM EDTA, 0.5% NP-40, 200 mM NaCl, 0.1 mM sodium orthovanadate, 10 mM NaF, 2 mM dithiothreitol [DTT], 100 μg each of phenylmethylsulfonyl fluoride and tolylsulfonyl phenylalanyl chloromethyl ketone per ml, 2 μg each of aprotonin and leupeptin per ml) was added to the reaction mixture, and samples were processed as described below for the GST pull down assay. Blots were analyzed by autoradiography and PhosphorImager (Storm 860; Molecular Dynamics) analysis.
GST pull down assay.
HEp-2 cells were lysed in high-salt lysis buffer (20 mM Tris [pH 8.0], 1 mM EDTA, 0.5% NP-40, 400 mM NaCl, 0.1 mM sodium orthovanadate, 10 mM NaF, 2 mM dithiothreitol, 100 μg each of phenylmethylsulfonyl fluoride and tolylsulfonyl phenylalanyl chloromethyl ketone per ml, 2 μg each of aprotonin and leupeptin per ml) on ice for 1 h. Insoluble material was pelleted by centrifugation. The supernatant was brought up in an equal volume of high-salt lysis buffer without NaCl (final concentration, 200 mM NaCl). Samples were precleared with 50 μl of a 50% slurry of glutathione beads for 2 h at 4°C. The precleared supernatant was incubated for 3 h at 4°C with either 2, 5, or 10 μl of a 50% slurry of glutathione beads bound to GST alone or GST fused to the above-mentioned UL42 protein constructs. The beads were pelleted by centrifugation and washed three times with IP buffer. Then 40 μl of SDS gel-loading buffer (2% sodium dodecyl sulfate [SDS], 5% β-mercaptoethanol, 50 mM Tris [pH 6.8], 2.75% sucrose) was added to the beads. The samples were heated to 95°C for 5 min, resolved by polyacrylamide gel electrophoresis, transferred to nitrocellulose membrane, and immunoblotted with antibody to cdc2 (SC-54; Santa Cruz Biotechnology). The blots were developed by peroxidase-conjugated secondary antibody and reacted with chemoluminescent substrate (Super Signal; Pierce).
Immunoprecipitation.
HEp-2 cells were infected with 10 PFU of HSV-1(F) per cell. The cells were harvested 12 h after infection and lysed in high-salt lysis buffer. Samples were kept on ice for 1 h, and insoluble material was pelleted by centrifugation. The supernatant was brought up in an equal volume of lysis buffer without NaCl (final concentration, 200 mM NaCl). Lysates were precleared with preimmune serum for 2 h, and 50 μl of a 50% slurry of protein A-conjugated to agarose was added. Samples were centrifuged, and the supernatant was transferred to new tubes. Antibody to UL42 was added to the samples, and immunocomplexes were collected by the addition of 40 μl of 50% protein– A slurry (29).
Immunoprecipitation-linked in vitro kinase assay.
Mock- or HSV-1-infected HEp-2 cells were harvested and immunoprecipitated with antibodies to cdc2 or UL42 as above. Immunocomplexes were collected with 20 μl of a 50% protein A slurry. The samples were washed twice with IP buffer, twice with low-salt buffer (20 mM Tris [pH 8.0], 1 mM EDTA, 0.5% NP-40, 1 mM NaCl, 2 mM dithiothreitol), and twice with incomplete kinase buffer (50 mM Tris [pH 7.4], 10 mM MgCl2, 5 mM dithiothreitol). The beads were then resuspended in 20 μl of incomplete kinase buffer with 0, 2, 10, or 40 μM roscovitine (Calbiochem); all samples contained 0.4% dimethyl sulfoxide. The samples were incubated for 5 min at 30°C, and 20 μl of complete kinase buffer (50 mM Tris [pH 7.4], 10 mM MgCl2, 5 mM dithiothreitol, 10 μM ATP, 20 μCi of [γ-32P]ATP, 2 μg of histone H1) was added, with final concentrations of roscovitine of 0, 1, 5, or 20 μM. Samples were incubated for an additional 20 min at 30°C. Reactions were terminated by addition of SDS gel-loading buffer and heated to 95°C for 5 min. The samples were resolved by polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and analyzed by autoradiography. Quantification of 32P phosphorylation of the substrates was done with the aid of a PhosphorImager (Storm 860).
Purified cdc2 Kinase Assay.
GST fusion proteins were reacted with purified recombinant cdc2 kinase (New England Biolabs). A 25 U portion of of cdc2 kinase was combined with either 2 μl of a 50% slurry of glutathione beads attached to GST or GST-UL42 fusion proteins in 30 μl of reaction buffer supplemented with 100 μM ATP and 20 μCi of [γ-32P]ATP. The samples were reacted at 30°C for 30 min, and the reactions were terminated by the addition of SDS gel-loading buffer and heated to 95°C for 5 min. The samples were resolved by polyacrylamide gel electrophoresis, transferred to nitrocellulose membrane, and analyzed by autoradiography. Quantification of 32P phosphorylation of the substrates was done with the aid of a PhosphorImager (Storm 860).
Immunodepletion kinase assay.
HSV-1(F)-infected HEp-2 cell lysates were harvested, lysed, and precleared as above. Samples were then immunodepleted of cdc2 or UL42 by incubation with the respective antibody for 2 h at 4°C, 50 μl of a 50% protein A slurry was added for 1 h at 4°C, and the samples were pelleted by centrifugation. The supernatant was then transferred to a new tube, antibody to cdc2 or UL42 was added for 4 h at 4°C, and immunocomplexes were collected by the addition of 20 ml of a 50% protein A slurry for 1 h at 4°C. Samples were washed as above (i.e., twice in IP buffer, twice in low-salt buffer, and twice in incomplete kinase buffer) and incubated in 40 μl of complete kinase buffer for 20 min at 30°C. The samples were then processed as above by bis-polyacrylamide gel electrophoresis.
Transient expression following plasmid transfection.
pCDNA3.1(+) encoding full-length UL42 was transfected into cells using Lipofectamine Plus (Gibco BRL). HEp-2 cells were seeded in T-25-cm2 flasks the day prior to transfection in DMEM containing 10% NBCS. The following day, 2 μg of plasmid was mixed with 10 μl of Plus reagent in 375 μl of DMEM (no serum and no antibiotics) for 15 min. Then 375 μl of DMEM containing 15 μl of Lipofectamine was added to the plasmid DNA. The mixture was incubated for an additional 15 min and added to HEp-2 cells in 2 ml of DMEM without serum or antibiotics. At 4 h later, 2.8 ml of DMEM (20% NBCS and 2× antibiotics) was added to the cells. The cells were harvested as above in high-salt lysis buffer 36 h after transfection. For nocodazole-treated samples, 24 h after transfection the cell culture medium was replaced with medium containing 5 μg of nocodazole (Sigma). per ml. Nocodazole-treated flasks were harvested 12 h after the addition of nocodazole. Kinase assays were performed as above.
RESULTS
HSV-1 UL42 and cdc2 physically interact.
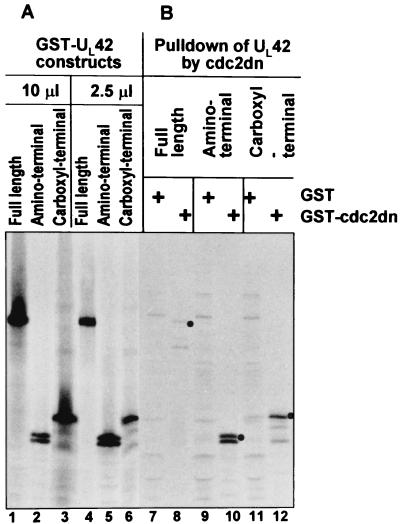
Two studies were done to determine if UL42 and cdc2 associate with each other. In the first study, pcDNA3.1(+) containing full-length, N-terminal, or C-terminal UL42 was in vitro transcribed and translated in the presence of [35S]methionine. The products of the in vitro transcription-translation of the UL42-encoding plasmids are shown in Fig. 1 (lanes 1 to 6). GST or GST-cdc2-dn fusion protein were generated in E. coli and captured on glutathione-agarose beads. GST or GST-cdc2-dn was then mixed and allowed to react with the radiolabeled in vitro-translated UL42 to determine if GST–cdc2-dn could specifically pull down UL42 (Fig. 1). GST–cdc2-dn pulled down full-length UL42 whereas GST did not (lanes 7 and 8). Next, we determined whether either the amino terminus or carboxyl terminus of UL42 interacted with cdc2. Interestingly, polypeptides containing both termini of the full-length UL42 protein were also specifically pulled down by GST–cdc2-dn but not by GST (lanes 9 to 12).
FIG. 1.
Autoradiographic image of in vitro-transcribed/translated UL42 followed by GST-cdc2dn pull down. Full-length (FL) or amino (N′) or carboxyl (C′) halves of UL42 protein were in vitro transcribed and translated (lanes 1 to 6). The in vitro-translated UL42 proteins were then reacted with GST or GST–cdc2-dn for pull down assays (lanes 7 to 12), electrophoretically separated, and developed by autoradiography.
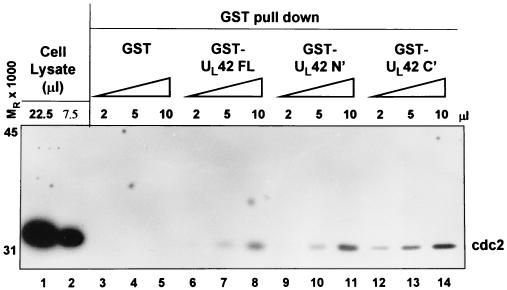
The above study was done with a dominant negative mutant of cdc2. The dominant negative mutant has a single-amino-acid substitution of Asp146Asn, such that it cannot coordinate ATP to the catalytic domain of cdc2 (30). To determine if UL42 could interact with wild-type cdc2, the following study was done. The above UL42 constructs (FL, N′, or C′) were produced as GST fusion proteins in E. coli and collected on glutathione-agarose beads. HEp-2 whole-cell lysates were then incubated with 2, 5, or 10 μl of glutathione-agarose beads bound to GST or GST fused to full-length, amino-terminal, or carboxyl-terminal UL42. Following extensive washing, the beads were solubilized and the proteins bound to the beads were subjected to electrophoresis in denaturing gels, transferred to a nitrocellulose sheet and reacted with antibody against cdc2. As shown in Fig. 2 GST-UL42 pulled down cdc2 from whole-cell lysates in a concentration-dependent manner (Figure 2, lanes 3 to 8). GST fused to the amino or carboxyl terminus of UL42 also pulled down cdc2 from cell lysates (lanes 9 to 14), whereas GST did not. cdc2 from whole-cell lysates is shown in lanes 1 and 2.
FIG. 2.
cdc2 immunoblot of HEp-2 cell lysates reacted with GST or GST fusion proteins expressing full-length (FL) or amino (N′) or carboxyl (C′) halves of UL42 protein. Immunoreactivity of cdc2 from whole-cell lysates is shown in lanes 1 and 2. Whole-cell lysates were then incubated with 2, 5, or 10 μl of GST (lanes 3 to 5), GST-UL42 FL (lanes 6 to 8), GST-UL42 N′ (lanes 9 to 11), or GST-UL42 C′ (lanes 12 to 14), and GST beads were collected, electrophoretically separated, and immunoblotted for cdc2.
We conclude from these studies that cdc2 and UL42 can physically associate with each other and that the sites of interaction reside at both the amino and carboxyl termini of UL42.
The carboxy terminus of UL42 is phosphorylated by cdc2 kinase.
Since both the amino and carboxy termini of UL42 could interact with cdc2, we next determined if UL42 could be phosphorylated by cdc2 kinase. cdc2 is a proline-directed serine/threonine kinase, and its substrate is located 3 amino acids downstream from a basic amino acid (11, 18). The minimally defined consensus phosphorylation site for cdc2 kinase is S/T-P-x-K/R/H. UL42 does not have a serine or threonine that fits the consensus phosphorylation site for cdc2 kinase. However, UL42 does have a putative cdc2 phosphorylation site that resembles that found on cyclins. In in vitro kinase assays, cdc2 can phosphorylate the cyclin associated with it (15). This site is loosely S/T-P-x-P. Amino acids 294 to 297 of UL42 are T-P-V-P.
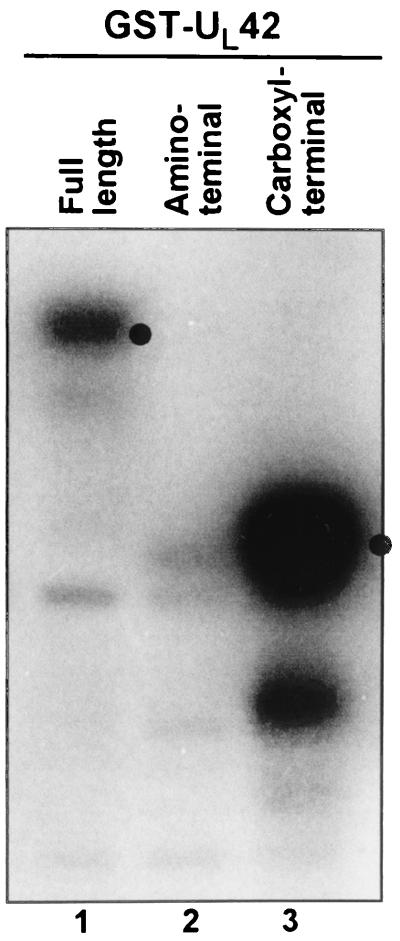
GST fusion proteins of full-length (1 to 488), amino-terminal (1 to 244), or carboxyl-terminal (226 to 488) UL42 were generated and used as substrate for the purified cdc2 kinase as described in Materials and Methods. As shown in Fig. 3, GST fused to full-length UL42 or to the carboxyl-terminal domain was phosphorylated by cdc2 whereas the amino-terminal domain of UL42 was not phosphorylated. These results are consistent with the prediction of a cyclin-like cdc2 phosphorylation site in the carboxyl-terminal domain of UL42.
FIG. 3.
Autoradiographic image of UL42 phosphorylation by cdc2 kinase. Full-length (lane 1), N-terminal (lane 2), C-terminal (lane 3) UL42 GST fusion proteins were incubated with purified cdc2 kinase in the presence of [γ-32P]ATP, separated by gel electrophoresis, and developed by autoradiography. Dots indicate the molecular weight of the respective GST fusion proteins.
Anti-UL42 antibody precipitates a roscovitine-sensitive kinase activity from HSV-1-infected cells.
The purpose of the series of experiments in this section was to determine whether anti-UL42 antibody precipitated a kinase from infected cells and whether this kinase had the characteristics of cdc2.
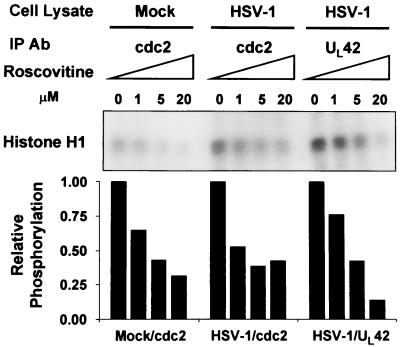
Roscovitine has been reported to be a highly selective inhibitor of cdc2, cdk2, and cdk5 cyclin-dependent kinases (19). Mock- or HSV-1-infected cell lysates were reacted with antibodies to either cdc2 or UL42. The immunoprecipitates were mock treated or reacted with roscovitine and then assayed for their ability to phosphorylate histone H1. To determine the sensitivity of cdc2 kinase to roscovitine, the cdc2 immunoprecipitated from uninfected HEp-2 lysates served as a positive control. The results shown in Fig. 4 were as follows. Both the anti-cdc2 and the anti-UL42 antibodies precipitated a kinase activity that phosphorylated histone H1. The precipitated kinase activities were sensitive to roscovitine (50% inhibitory concentration between 1 and 5 μM). The sensitivity of the kinase activities precipitated from infected cells was similar to that of the kinase activity precipitated by the cdc2 antibody from mock-infected cells. Moreover, as we have previously shown, the cdc2 immunoprecipitated with the anti-cdc2 antibody from HSV-1-infected cell lysates showed elevated kinase activity compared to that from uninfected cell lysates (1). The UL42 antibody did not immunoprecipitate appreciable histone H1 kinase activity from uninfected cells (data not shown).
FIG. 4.
Autoradiographic image of roscovitine-sensitive histone H1 phosphorylation by cdc2 or UL42 immunocomplexes. Mock- or HSV-1-infected HEp-2 cell lysates were subjected to immunoprecipitation (IP) by antibodies (Ab) to cdc2 or UL42. Immunocomplexes were preincubated with 0, 1, 5, or 20 μM roscovitine and subjected to in vitro kinase assays using histone H1 as the substrate. Phosphorylated histone 1 from the reactions was resolved by polyacrylamide gel electrophoresis, and the blots were developed by autoradiography. Histone phosphorylation was quantified by PhosphorImager analysis. Samples not treated roscovitine were assigned a value of 1.
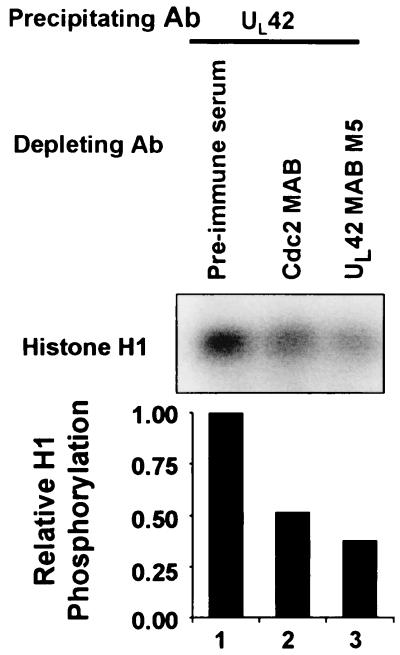
The kinase activity coprecipitated by the anti- UL42 antibody can be immunodepleted by the anti-cdc2 antibody.
To determine if cdc2 was immunoprecipitated with UL42 antibody, the following experiment was done. Lysates from HSV-1-infected HEp-2 cell lysates were reacted with preimmune serum, cdc2 antibody, or UL42 antibody. The immune complexes were then removed, and the immunodepleted lysates were reacted with antibody to UL42. The immune complexes were then harvested and assessed for their ability to phosphorylate histone H1 kinase. The results indicate that the anti-UL42 antibody depleted 63% of the UL42 associated histone H1 kinase activity compared to lysates immunodepleted with pre-immune sera (Fig. 5, lanes 1 and 3) and that lysates immunodepleted with antibody to cdc2 resulted in a 48% reduction of UL42-associated histone H1 kinase activity compared to that of control lysates immunodepleted with preimmune sera (lanes 1 and 2).
FIG. 5.
Autoradiographic image of histone H1 kinase activity associated with UL42 immunocomplexes following immunodepletion. HSV-1-infected HEp2 cell lysates were immunodepleted with preimmune serum (lane 1), antibody (Ab) to cdc2 (lane 2), or antibody to UL42 (lane 3). The immunodepleted lysates were then immunprecipitated with antibody to UL42. Immunocomplexes were assayed for histone H1 kinase activity. Phosphorylated histone H1 was resolved by polyacrylamide gel electrophoresis, and the blots were developed by autoradiography. Histone phosphorylation was quantified by PhosphorImager analysis. The sample immunodepleted with preimmune serume was given a value of 1.
The results therefore indicate that the histone H1 kinase activity associated with UL42 antibody could be attenuated by immunodepleting the infected-cell lysate with cdc2 antibody.
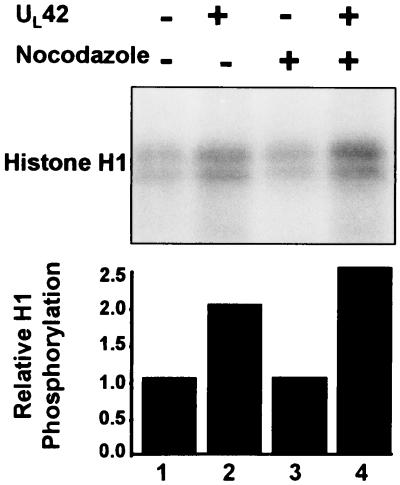
UL42 made in cells transfected with a plasmid encoding the protein is associated with histone H1 kinase activity.
The purpose of the series of experiments in this section was to determine whether the association of the UL42 protein with kinase activity for histone H1 required the presence of other viral proteins. HEp-2 cells were transiently transfected with pcDNA3.1 encoding UL42 and were mock treated or incubated in medium containing nocodazole. UL42 protein production from transfection was verified by immunoblotting cell lysates with antibody to UL42 (data not shown). The cells were harvested 36 h after transfection, and the lysates prepared as described in Materials and Methods were reacted with anti UL42 antibody. The precipitates were tested for histone H1 phosphorylation. The results were as follows. (i) The baseline histone H1 phosphorylation obtained by immune complexes obtained from mock-transfected cells was not enhanced by nocodazole treatment of cells and most probably represented nonspecific binding of cellular kinases during immunoprecipitation (Fig. 6, lanes 1 and 3). (ii) The UL42 immunoprecipitate from transfected cells exhibited a twofold increase in relative histone H1 phosphorylation compared to that from nontransfected cells (lanes 1 and 2). (iii) Nocodazole treatment of UL42-transfected cells resulted in a further appreciable enhancement of histone H1 kinase activity (lanes 2 and 4). These results indicate that the association of UL42 with a nocodazole-enhanced cellular kinase activity for histone H1 does not require the presence of other viral gene products.
FIG. 6.
Autoradiographic image of histone H1 kinase activity from UL42 transfected cells. HEp-2 cells were transfected with pCDNA3.1 encoding UL42 (lanes 2 and 4). Cells were also treated with nocodazole following transfection (lanes 3 and 4). Cell lysates were then immunoprecipitated with antibody to UL42 and subjected to histone H1 kinase assays. Phosphorylated histone H1 was electrophoretically separated in a denaturing polyacrylamide gel and subjected to autoradiography. Histone phosphorylation was quantified by PhosphorImager analysis. Untransfected lysate without nocodazole treatment was assigned a value of 1.
DISCUSSION
The studies conducted in the past several years have shown that HSV gene products scavenge the cell for cellular regulatory proteins which may be stabilized and translocated to specific compartments. In some instances, the purpose of the diversion seems clear. For example, ICP34.5 binds and redirects cellular protein phosphatase 1 to dephosphorylate the α subunit of elongation initiator factor 2 that has been phosphorylated by the activated protein kinase R (7, 10). In the case of the association of cyclin D3 and ICP0, the accumulated evidence suggests that the ultimate effect of the new function of cyclin D3 is to expedite the translocation of ICP0 to the cytoplasm (32). In other instances, either the purpose of the diversion or the mechanism by which the diverted cell protein performs its function remain unclear. The objectives of the studies reported here were to solve a mystery related to the stabilization and activation of cdc2 late in infection,
Specifically, earlier studies indicated that HSV-1 was specifically activating cdc2 kinase by specific alterations in protein levels, activity, and posttranslational modifications of both cdc2 and regulators of cdc2 activation (1). Thus, between 8 and 12 h after infection, cdc2 kinase activity was significantly enhanced, the levels of cyclins A and B were dramatically reduced, and cdc25C and wee-1 were posttranslationally modified. HSV-1-induced hyperphosphorylation of cdc25C is especially striking, since it appears to enhance the phosphatase activity that is required to relieve inhibitory phosphorylations within the kinase domain of cdc2. Both the modifications of cdc25C phosphatase and the activation of cdc2 kinase activities require the presence of ICP22 and of the UL13 protein kinase. The problem we wished to address was that cdc2 plays a role in the expression of a subset of late (γ2) proteins exemplified by US11 but that if cdc2 behaved like all other cyclin-dependent kinases, it should have acquired a surrogate cyclin to replace the missing cyclins A and B.
The studies described in this report attempt to reconcile this apparent paradox. Analyses of the open reading frame of HSV-1 genome led to the identification of the UL42 protein as a viral gene product with properties similar to those of bona fide cyclins. In this report we show that cdc2 interacts physically and is phosphorylated by cdc2 in vitro and that a kinase activity characteristic of cdc2 can be precipitated from infected cells or cells transfected with UL42 protein by the anti-UL42 antibody. Curiously, the association of cdc2 with a factor involved in DNA synthesis has a precedent. Thus, UL42 protein is structurally related to PCNA and the association of PCNA with cdk2 has been reported (16, 34).
Irrespective of whether UL42 is the surrogate cyclin partner of cdc2, the interaction of cdc2 with the UL42 DNA synthesis processivity factor may be significant and related to the expression of the subset of γ2 proteins exemplified by US11. As noted above, cdc2 kinase is activated between 8 and 12 h after infection, and functional cdc2 kinase activity was necessary for the accumulation of US11 protein, a member of a unique subset of γ2 proteins that also requires ICP22 and the UL13 protein kinase for maximum accumulation (1, 3, 22, 25, 28). Since γ2 gene expression is intimately linked to viral DNA replication, it could be predicted that a viral protein involved in DNA synthesis facilitated the function of cdc2. UL42, expressed with β kinetics, appears to satisfy this requirement. The kinetics of expression of UL42 coincides with the activation cdc2 kinase in infected cells, and the known function of UL42 is to act as a processivity factor for viral DNA polymerase links it to γ2 gene expression.
The present studies begin to further clarify a pathway for the production of HSV-1 late proteins. A summary model of the results obtained to date is presented in Fig. 7. ICP22 and UL13 are involved in “priming” cdc2 to become active, most probably by modifying the proteins that regulate the function of cdc2. The data presented here indicate that UL42 can associate with cdc2. UL42 appears to have at least two binding sites for cdc2. While both the amino and carboxy halves of UL42 bind cdc2, only the carboxy half is efficiently phosphorylated by cdc2 kinase. This would imply that a portion of the carboxyl-terminal half of UL42 protein binds in proximity to the kinase domain of cdc2. Immunoprecipitation with UL42 antibody also brings down a cdc2-like kinase activity. Taken together, these data indicate that the association of UL42 with cdc2 occurs in the context of a functional cdc2 kinase activity.
FIG. 7.
Schematic diagram of cdc2 regulation and activity in the uninfected (top) and HSV-1-infected (bottom) cell. In uninfected cells, cdc2 is inactivated by wee-1 and myt-1 kinases and activated by cdc25C phosphatase, cyclin-dependent activating kinase (CAK), and association with cyclin B at the G2/M interphase. HSV-1 infection targets wee-1 and cdc25C through the actions of ICP22 and UL13, and UL42 associates with cdc2. The substrates for cdc2 in infected cells include ICP0, ICP4, and gI. The activation of cdc2 in the infected cell is associated with optimal accumulation of a subset of γ2 proteins exemplified by Us11 protein.
A common theme emerging from the studies on HSV-1 proteins is their multifunctionality. With coding capacity at a premium in viruses, one method to achieve greater versatility is for viral proteins to play multiple roles. The studies presented here begin to assess a novel function for the UL42 viral protein. Studies to date on UL42 have focused on its ability to associate with viral DNA polymerase and act as a processivity factor for DNA replication (26). UL42 is an unorthodox processivity factor since it also binds to DNA. Recently, the viral origin binding protein UL9 was reported to interact with UL42 (20). Interestingly, UL9 appears to be phosphorylated by a cellular kinase (12). Also, other viral proteins associated with DNA replication machinery contain consensus phosphorylation sites for cdc2 kinase, including DNA polymerase (UL30), helicase/primase (UL52), and single-stranded DNA binding protein (ICP8/UL29) (3). Recent studies also indicated that ICP0 continued to undergo phosphorylation between 6 and 10 h after infection and that this phosphorylation was blocked by the cdc2 inhibitor roscovitine (4). Thus, one hypothesis that could be envisioned is that UL42 acts as an adapter or bridge protein to bring in substrates that are subsequently phosphorylated by cdc2 kinase. This scenario is akin to one recently reported for PCNA (16). The interaction of PCNA with cdk2/cyclin A results in the phosphorylation of PCNA-interacting proteins such as replication factor C and DNA ligase I. In this example, cdk2 is also associated with cyclin A, and PCNA serves as a substrate specificity protein.
The intermingling of cell cycle proteins with the virus life cycle appears to be critical for viral replication. Certain members of the herpesvirus family encode functional homologs of cyclins to induce cdk activity. While cyclins tend to have short half-lives and are expressed only during certain phases of the cell cycle, the cyclin-dependent kinase subunits are relatively stable. By encoding the cyclin component, viruses overcome a rate-limiting step in cdk activation. A key question that still remains unresolved is what are the critical substrates for cyclin-dependent kinases activated in the infected cell. Viral proteins from many herpesvirus family members have been reported to be phosphorylated by these kinases (3, 4, 24, 33). HSV-1 infection of roscovitine-treated cells has been reported to grossly interfere with viral replication and gene expression (27). Also, a consensus cdc2 phosphorylation site in a core protein of hepadnavirus has been reported to be involved in capsid assembly and disassembly (5). The identification of key substrates for cyclin-dependent kinases within cells has been a difficult road. Hopefully, the elucidation of these substrates as well as functional consequences in the virally infected cell may also point to important paradigms within the cell.
ACKNOWLEDGMENTS
We thank Stephen J. Kron, Guoying Zhou, and Maria-Teresa Sciortino for invaluable discussions; S. van den Heuvel for providing plasmid encoding cdc2dn; and D. J. Tenney for providing antibodies to UL42.
These studies were aided by grants from the National Cancer Institute (CA87761, CA83939, CA71933, and CA78766), U.S. Public Health Service.
REFERENCES
- 1.Advani S J, Brandimarti R, Weichselbaum R R, Roizman B. The disappearance of cyclins A and B and the increase in activity of the G2/M-phase cellular kinase cdc2 in herpes simplex virus 1-infected cells require expression of the α22/US1.5 and UL13 viral genes. J Virol. 2000;74:8–15. [PMC free article] [PubMed] [Google Scholar]
- 2.Advani S J, Weichselbaum R R, Roizman B. E2F protein are posttranslationally modified concomitantly with a reduction in nuclear binding activity in cells infected with herpes simplex virus 1. J Virol. 2000;74:7842–7850. doi: 10.1128/jvi.74.17.7842-7850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Advani S J, Weichselbaum R R, Roizman B. The role of cdc2 in the expression of herpes simplex virus genes. Proc Natl Acad Sci USA. 2000;97:10996–11001. doi: 10.1073/pnas.200375297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Advani S J, Hagglund R, Weichselbaum R R, Roizman B. Posttranslational processing of infected cell proteins 0 and 4 of herpes simplex virus 1 is sequential and reflects the subcellular compartment in which the proteins localize. J Virol. 2001;75:7904–7912. doi: 10.1128/JVI.75.17.7904-7912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrasa M I, Guo J T, Saputelli J, Mason W S, Seeger C. Does a cdc2 kinase-like recognition motif on the core protein of hepadnavirus regulate assembly and disintegration of capsids? J Virol. 2001;75:2024–2028. doi: 10.1128/JVI.75.4.2024-2028.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belyavskyi M, Braunagel S C, Summers M D. The structural protein ODV-EC27 of Autographa californica nucleopolyhedrovirus is a multifunctional viral cyclin. Proc Natl Acad Sci USA. 1998;95:11205–11210. doi: 10.1073/pnas.95.19.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou J, Chen J J, Gross M, Roizman B. Association of a Mr 90,000 phosphoprotein with protein kinase PKR in cells exhibiting enhanced phosphorylation of translation initiation factor eIF-2 alpha and premature shutoff of protein synthesis after infection with γ134.5 mutants of herpes simplex virus 1. Proc Natl Acad Sci USA. 1995;92:10516–10520. doi: 10.1073/pnas.92.23.10516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehmann G L, McLean T I, Bachenheimer S L. Herpes simplex virus type 1 infection imposes a G1/S block in asynchronously growing cells and prevent G1 entry in quiescent cells. Virology. 2000;267:335–349. doi: 10.1006/viro.1999.0147. [DOI] [PubMed] [Google Scholar]
- 9.Ejercito P, Kieff E D, Roizman B. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 10.He B, Gross M, Roizman B. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1 alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc Natl Acad Sci USA. 1997;94:843–848. doi: 10.1073/pnas.94.3.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes J K, Solomon M J. A predictive scale for evaluating cyclin-dependent kinase substrates. J Biol Chem. 1996;271:25240–25246. doi: 10.1074/jbc.271.41.25240. [DOI] [PubMed] [Google Scholar]
- 12.Isler J A, Schaffer P A. Phosphorylation of the herpes simplex virus 1 origin binding protein. J Virol. 2001;75:628–637. doi: 10.1128/JVI.75.2.628-637.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328–7336. doi: 10.1128/jvi.71.10.7328-7336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King R W, Jackson P K, Kirschner M W. Mitosis in transition. Cell. 1994;79:563–571. doi: 10.1016/0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 15.Kleinberger T, Shenk T. A protein kinase is present in a complex with adenovirus E1A proteins. Proc Natl Acad Sci USA. 1991;88:11143–11147. doi: 10.1073/pnas.88.24.11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koundrioukoff S, Jonsson Z O, Hagan S, de Jong R N, van der Vliet P C, Hottiger M O, Hubscher U. A direct interaction between proliferating cell nuclear antigen (PCNA) and cdk2 targets PCNA-interacting proteins for phosphorylation. J Biol Chem. 2000;275:22882–22887. doi: 10.1074/jbc.M001850200. [DOI] [PubMed] [Google Scholar]
- 17.Li M, Lee H, Yoon D W, Albrecht J C, Fleckenstein B, Neipel F, Jung J U. Kaposi's sarcoma-associated herpesvirus encodes a functional cyclin. J Virol. 1997;71:1984–1991. doi: 10.1128/jvi.71.3.1984-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marin O, Meggio F, Draetta G, Pinna L A. The consensus site for cdc2 kinase and for casein kinase-2 are mutually incompatible. A study with peptides derived from the beta-subunit of casein kinase-2. FEBS Lett. 1992;301:111–114. doi: 10.1016/0014-5793(92)80221-2. [DOI] [PubMed] [Google Scholar]
- 19.Meijer L, Borgne A, Mulner O, Chong J P, Blow J J, Inagaki N, Delcros J G, Moulinoux J P. Biochemical and cellular effects of roscovitine, a potent and selective inhibitor of the cyclin-dependent kinases cdc2, cdk2, and cdk5. Eur J Biochem. 1997;243:527–536. doi: 10.1111/j.1432-1033.1997.t01-2-00527.x. [DOI] [PubMed] [Google Scholar]
- 20.Monahan S J, Grinstead L A, Olivieri W, Parris D S. Interaction between the herpes simplex virus type 1 origin-binding and DNA polymerase accessory proteins. Virology. 1998;241:122–130. doi: 10.1006/viro.1997.8953. [DOI] [PubMed] [Google Scholar]
- 21.Nicholas J, Cameron K R, Honess R W. Herpesvirus saimiri encodes homologues of G protein-coupled receptors and cyclins. Nature. 1992;355:362–365. doi: 10.1038/355362a0. [DOI] [PubMed] [Google Scholar]
- 22.Ogle W O, Roizman B. Functional anatomy of herpes simplex virus 1 overlapping genes encoding infected-cell protein 22 and US1.5 protein. J Virol. 1999;73:4305–4315. doi: 10.1128/jvi.73.5.4305-4315.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olgiate J, Ehmann G L, Vidyarthi S, Hilton M J, Bachenheimer S L. Herpes simplex virus induces intracellular redistribution of E2F4 and accumulation of E2F pocket protein complexes. Virology. 1999;258:257–270. doi: 10.1006/viro.1999.9755. [DOI] [PubMed] [Google Scholar]
- 24.Polson A G, Huang L, Lukac D M, Blethrow J D, Morgan D O, Burlingame A L, Ganem D. Kaposi's sarcoma-associated herpesvirus K-bZIP protein is phosphorylated by cyclin-dependent kinases. J Virol. 2001;75:3175–3184. doi: 10.1128/JVI.75.7.3175-3184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Purves F C, Roizman B. The UL13 gene of herpes simplex virus 1 encodes the function for posttranslational processing associated with the phosphorylation of the regulatory protein α22. Proc Natl Acad Sci USA. 1992;89:7310–7314. doi: 10.1073/pnas.89.16.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2231–2296. [Google Scholar]
- 27.Schang L M, Phillips J, Schaffer P A. Requirement for cellular cyclin-dependent kinases in herpes simplex virus replication and transcription. J Virol. 1998;72:5626–5637. doi: 10.1128/jvi.72.7.5626-5637.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sears A E, Halliburton I W, Meignier B, Silver S, Roizman B. Herpes simplex virus 1 mutant defective in the α22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J Virol. 1985;55:338–346. doi: 10.1128/jvi.55.2.338-346.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheaffer A K, Hurlburt W W, Stevens J T, Bifano M, Hamatake R K, Colonno R J, Tenney D J. Characterization of monoclonal antibodies recognizing amino and carboxy-terminal epitopes of the herpes simplex virus UL42 protein. Virus Res. 1995;38:305–314. doi: 10.1016/0168-1702(95)00047-t. [DOI] [PubMed] [Google Scholar]
- 30.van den Heuvel S, Harlow E. Distinct roles for cyclin-dependent kinases in cell cycle control. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 31.Van Sant C, Kawaguchi Y, Roizman B. A single amino acid substitution in the cyclin D binding domain of the infected cell protein no. 0 abrogates the neuroinvasiveness of herpes simplex virus without affecting its ability to replicate. Proc Natl Acad Sci USA. 1999;96:8184–8189. doi: 10.1073/pnas.96.14.8184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Sant C, Lopez P, Advani S J, Roizman B. Role of cyclin D3 in the biology of herpes simplex virus 1 ICP0. J Virol. 2001;75:1888–1898. doi: 10.1128/JVI.75.4.1888-1898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye M, Duus K M, Peng J, Price D H, Grose C. Varicella-zoster virus Fc receptor component gI is phosphorylated on its endodomain by a cyclin-dependent kinase. J Virol. 1999;73:1320–1330. doi: 10.1128/jvi.73.2.1320-1330.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zuccola H J, Filman D J, Coen D M, Hogle J M. The crystal structure of an unusual processivity factor, herpes simplex UL42, bound to the C terminus of its cognate polymerase. Mol Cell. 2000;5:267–278. doi: 10.1016/s1097-2765(00)80422-0. [DOI] [PubMed] [Google Scholar]