Abstract
Despite widespread application during the coronavirus disease-19 pandemic, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection using patient-performed rapid antigen tests (RATs) is limited, especially regarding the Delta and Omicron variants. Therefore, in this study, we evaluated the performance of RATs in identifying Delta and Omicron infections in self-test settings. In this multicenter clinical performance study conducted in Korea between November 2021 and February 2022, we included participants without prior diagnostic device experience. Using 2 RAT types, we compared the results with real-time reverse transcriptase-polymerase chain reaction testing, focusing on clinical sensitivity and specificity. Reverse transcriptase-polymerase chain reaction helped confirm 77 SARS-CoV-2 infections among 280 participants. RATs exhibited high positive agreement for Omicron detection but lower rates for Delta, especially among partially vaccinated individuals. This study provides direct evidence that RATs, originally developed for ancestral strains of SARS-CoV-2, effectively detect major variants such as Delta and Omicron in real patient/clinical settings. By confirming variant presence through sequencing, our research offers significant and novel insights into the performance of RATs, particularly in the context of breakthrough infections postvaccination, with precise data on vaccination status and timing obtained from government records.
Keywords: Delta variant, Omicron variant, patient-performed testing, rapid antigen tests, SARS-CoV-2
1. Introduction
Rapid antigen tests (RATs) were widely adopted during the coronavirus disease-19 (COVID-19) pandemic, leading to the rapid diagnosis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Laboratory studies have demonstrated the ability of RATs to effectively detect emerging variants, such as Omicron.[1,2] Furthermore, as field-based validations,[3,4] clinical studies have confirmed the ability of RATs to identify Omicron-infected individuals, including those in a community walk-up setting where trained technicians performed specimen collection and testing.[5] However, the diagnostic accuracy of RAT in at-home or self-test settings remains understudied. Therefore, we aimed to determine the performance of RATs in detecting the Delta and Omicron variants in self-test clinical settings.
2. Methods
This sponsor-initiated, multicenter, prospective clinical performance study was conducted in Korea between November 2021 and February 2022. Participants were recruited from 2 university hospitals and a community treatment center (CTC) affiliated to and run by one of the hospitals, including individuals who visited for COVID-19 symptoms or testing, or were hospitalized or quarantined in the CTC due to COVID-19, or needed a COVID-19 screening test prior to hospital admission or for other procedural reasons. Upon consenting to participate in the study, the participants self-performed 2 RATs on anterior nasal swabs (described in detail below), and on the same day, healthcare providers collected nasopharyngeal swab samples for a real-time reverse transcriptase-polymerase chain reaction (rRT-PCR). The results of the rRT-PCR were not disclosed to the participants, as they were reported much later than the RAT results. For symptomatic participants, COVID-19-related symptoms and date of onset were scrutinized and documented.
We included participants aged >2 years who had no experience or training in testing with in vitro diagnostic devices, had not recently participated in similar studies or other diagnostic device trials, and provided informed consent. Participants included both symptomatic and asymptomatic individuals to ensure a mix of cases. Individuals with nasal injuries or bleeding, visual impairments affecting their ability to read the test results, or involvement in other diagnostic device studies were excluded. For positive participants, additional criteria were as follows: (1) symptomatic individuals were included if symptoms began within 7 days prior to enrollment; (2) asymptomatic individuals were included if recruitment date was within 7 days from initial positive diagnosis; (3) individuals receiving antiviral treatment for COVID-19 were excluded.
Since the first confirmed case of COVID-19 in South Korea on January 20, 2020, the disease was designated and managed as a Class 1 infectious disease under the Infectious Disease Control and Prevention Act. All confirmed cases, no matter how mildly symptomatic or even asymptomatic, were legally required to be isolated at the time of this study, with most patients being admitted to hospitals or CTCs.[6] Although RATs were approved by the Korea Ministry of Food and Drug Safety as in vitro diagnostic kits in April 2021, they were not recognized as confirmatory diagnostic tests at that time. Therefore, most hospitals and public health centers relied on the highly accurate rRT-PCR for diagnosis before and during our study period.[6] Therefore, positive participants cannot help but be aware of their positive status before entering the study, as it was required to have already been confirmed by rRT-PCR at a different facility (mostly public health centers) prior to admission to the study sites. However, healthcare providers collected nasopharyngeal swabs for an additional rRT-PCR test on the same day when RATs were performed; it was against this additional rRT-PCR that the performance of the RATs was evaluated, and the rRT-PCR results were not disclosed to the participants before they reported the RAT results to a physician.
The reference standard used was rRT-PCR. Nasopharyngeal swabs were used for rRT-PCR testing and also variant typing. Two kits were used for rRT-PCR testing according to the study sites: either the PowerChek SARS-CoV-2 Real-time PCR kit (Kogene Biotech, Seoul, South Korea) or the STANDARD M nCoV Real-time Detection kit (SD Biosensor, Suwon, South Korea), with cycle threshold (Ct) values below 38 and 36, respectively, considered positive. The variant type was determined using Sanger sequencing of the receptor binding domain hotspot.[7]
The index tests were 2 COVID-19 antigen test kits: the Panbio COVID-19 Antigen self-test (Abbott Rapid Diagnostics Jena GmbH, Jena, Germany) and the STANDARD Q COVID-19 Ag Home test (SD Biosensor, Suwon, Republic of Korea). According to the manufacturer’s instructions, participants self-collected nasal swabs and immediately extracted and reacted them with the test devices for themselves. They performed the tests at home or, in case of hospitalized or CTC-quarantined participants, at their facilities. The order of the test kits was alternated among participants to ensure balanced evaluation of both kits; after recording the results of the first test, participants immediately (<20 minutes from the first test) collected another swab to conduct the second RAT. Participants also self-interpreted their test results according to the instructions, sending photographs of the test devices along with their own interpretation to a physician via mobile phone text message as soon as possible. There were no cases where the photos were discrepant with the interpretation. The researchers provided no feedback to the participants on RAT results submitted via text message.
The sample size was determined based on the clinical sensitivity and specificity goals set by the Ministry of Food and Drug Safety in Korea. The authors targeted a clinical sensitivity of at least 90% and a specificity of 99%, with respective 95% confidence interval (CI) lower limits of 80% and 97%. Incorporating a 10% margin to account for potential dropout rates, the estimated sample size requirements were 78 positive and 214 negative samples.
Clinical sensitivity (positive percent agreement, PPA) and specificity were evaluated against the rRT-PCR results. The 95% CIs were calculated using the Clopper–Pearson method. Indeterminate results were reevaluated, and participants were retested if necessary. Persistently indeterminate results were excluded from the final analysis.
The Institutional Review Boards of Pusan National University Hospital and Gyeongsang National University Changwon Hospital (approval numbers: 2110-026-108 and 2021-11-003, respectively) approved this sponsor-initiated, multicenter clinical performance study performed in Korea between November 2021 and February 2022. The study was conducted in compliance with the ethical standards of the Declaration of Helsinki. All participants provided written informed consent.
3. Results
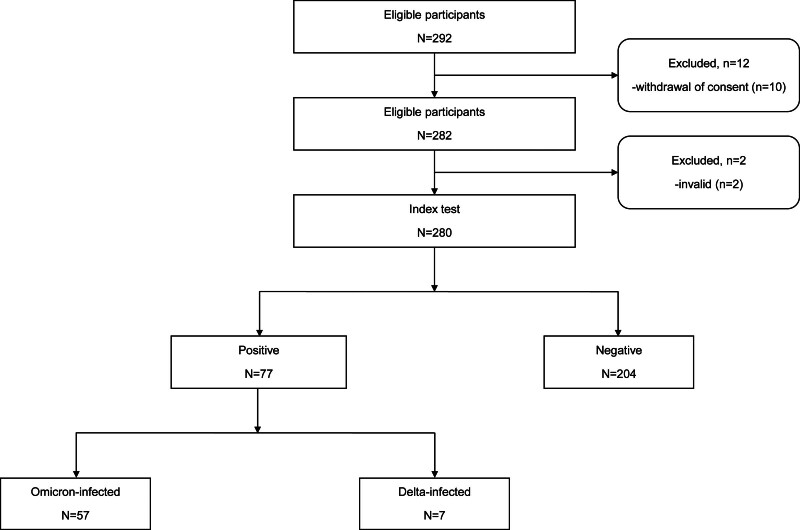
The study initially identified 292 eligible participants (Fig. 1). Of these, 10 participants were excluded due to withdrawal of consent, and 2 participants were excluded as invalid due to technical issues, resulting in 280 participants completing the index test. Among the 280 enrolled participants, rRT-PCR confirmed that 77 (27.5%) were SARS-CoV-2-infected (Table 1). Both the RATs showed a specificity of 100.0% (95% CI: 98.2–100.0%).
Figure 1.
Study participant flow diagram. This flow diagram outlines the selection process of study participants. A total of 292 participants were initially identified as eligible for the study. After excluding 12 participants (10 withdrew consent and 2 provided invalid test results), 280 participants were included in the index test. Among these, 77 participants tested positive for SARS-CoV-2 via rRT-PCR, while 204 tested negative. Of the positive cases, 57 were infected with the Omicron variant, and 7 with the Delta variant. The settings for testing included both home environments and facilities for those who were hospitalized or quarantined in COVID-19 treatment centers. COVID-19 = coronavirus disease-19, rRT-PCR = real-time reverse transcriptase-polymerase chain reaction, SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2.
Table 1.
Characteristics of rRT-PCR-positive participants.
| rRT-PCR-positive (N = 77) | |
|---|---|
| Age, mean (SD), years | 39.0 (14.0) |
| Male sex, No. (%) | 28 (36%) |
| Asymptomatic, No. (%) | 26 (34%) |
| Days since symptom onset,* mean (range) | 3.3 (2–6) |
| Vaccine status,† No. (%) | |
| Not vaccinated | 6 (8%) |
| Fully vaccinated‡ | 36 (51%) |
| Not fully vaccinated | 29 (41%) |
| Ct value, mean (range) | |
| E gene | 19.99 (12.81–36.41) |
| ORF1ab gene | 19.01 (12.71–36.14) |
| Variant type, No. (%) | |
| Delta | 7 (9%) |
| Omicron | 57 (74%) |
| Unevaluable | 13 (17%) |
Ct = cycle threshold, rRT-PCR = real-time reverse transcriptase-polymerase chain reaction, SD = standard deviation.
Analyzed for symptomatic participants only (N = 51).
Six participants had missing data.
Defined as persons who received all recommended doses of vaccines at the time of initial diagnosis; participants were considered fully vaccinated 2 weeks after receiving a third (booster) dose or 2 weeks after but not exceeding 90 days of receiving their primary series.
The mean age of the rRT-PCR-positive participants was 39 ± 14 years, and 36% were male. Among the positive cases, 34% (26/77) were asymptomatic, with a mean of 3.3 days since symptom onset in the symptomatic group (range 2–6 days, ≤4 days accounting for 78%). The vaccination details were as follows: 8% (6/77) were not vaccinated, 51% (36/77) were fully vaccinated, and 41% (29/77) were not fully vaccinated. Fully vaccinated individuals were defined as those who had received all recommended doses at the time of initial diagnosis, that is, who were ≥2 weeks post a third (booster) dose or 14 to 90 days from their primary vaccination series. Of the rRT-PCR-positive cases, 74% (57/77) were infected with the Omicron variant (including 49 with BA.1 and 8 with BA.2 subvariants) and 9% (7/77) with the Delta variant. Eleven cases were not subjected to variant typing, and sequencing failed in 2 cases.
The PPA between rRT-PCR and RATs by symptomaticity, days since symptom onset, viral load, and vaccination status is shown in Table 2. The Panbio kit detected infection in 97.4% (75/77, 95% CI: 90.9–99.7%) and the STANDARD Q kit detected infection in 94.8% (73/77, 95% CI: 87.2–98.6%) of individuals who tested positive using the rRT-PCR. Among symptomatic cases, both the Panbio and the STANDARD Q detected 98.0% (50/51, 95% CI: 89.6–100%). For asymptomatic cases, the Panbio detected 96.2% (25/26, 95% CI: 80.4–99.9%), whereas the STANDARD Q detected 88.5% (23/26, 95% CI: 69.8–97.6%). In those tested 2 to 4 days after symptom onset, both the Panbio and the STANDARD Q detected 97.5% (39/40, 95% CI: 86.8–99.9%) of infections. In those tested 5 to 6 days after symptom onset, both tests detected 100% (11/11, 95% CI: 71.5–100%) of infections. Regarding vaccine status, among the non-vaccinated group, both the Panbio and the STANDARD Q detected 100% (6/6, 95% CI: 54.1–100%). In the fully vaccinated group, the Panbio detected 97.2% (35/36, 95% CI: 85.5–99.9%), and the STANDARD Q test detected 91.7% (33/36, 95% CI: 77.5–98.2%) of infections. In the not-fully vaccinated group, both the Panbio and the STANDARD Q detected 96.6% (28/29, 95% CI: 82.2–99.9%) of infections. In those infected with the Delta variant, both the Panbio and the STANDARD Q detected 85.7% (6/7, 95% CI: 42.1–99.6%) of infections. In those infected with the Omicron variant, the Panbio test detected 98.2% (56/57, 95% CI: 90.6–100%), and the STANDARD Q test detected 94.7% (54/57, 95% CI: 85.4–98.9%) of infections.
Table 2.
Comparison of positive percentage agreement between rRT-PCR and rapid antigen tests across variants by symptomaticity, days since symptom onset, and vaccination status.
| No. | Panbio COVID-19 antigen self-test | STANDARD Q COVID-19 Ag home test | |||
|---|---|---|---|---|---|
| TP/(TP + FN) | PPA (95% CI)* | TP/(TP + FN) | PPA (95% CI)* | ||
| All rRT-PCR-positive cases | 77 | 75/77 | 97.4 (90.9–99.7) | 73/77 | 94.8 (87.2–98.6) |
| Symptomaticity | |||||
| Asymptomatic | 26 | 25/26 | 96.2 (80.4–99.9) | 23/26 | 88.5 (69.8–97.6) |
| Symptomatic | 51 | 50/51 | 98.0 (89.6–100) | 50/51 | 98.0 (89.6–100) |
| Days since symptom onset | |||||
| 2–4 | 40 | 39/40 | 97.5 (86.8–99.9) | 39/40 | 97.5 (86.8–99.9) |
| 5–6 | 11 | 11/11 | 100 (71.5–100) | 11/11 | 100 (71.5–100) |
| Vaccine status† | |||||
| Not vaccinated | 6 | 6/6 | 100 (54.1–100) | 6/6 | 100 (54.1–100) |
| Fully vaccinated‡ | 36 | 35/36 | 97.2 (85.5–99.9) | 33/36 | 91.7 (77.5–98.2) |
| Not fully vaccinated | 29 | 28/29 | 96.6 (82.2–99.9) | 28/29 | 96.6 (82.2–99.9) |
| Delta variant | 7 | 6/7 | 85.7 (42.1–99.6) | 6/7 | 85.7 (42.1–99.6) |
| Symptomaticity | |||||
| Asymptomatic | 0 | NC | NC | NC | NC |
| Symptomatic | 7 | 6/7 | 85.7 (42.1–99.6) | 6/7 | 85.7 (42.1–99.6) |
| Days since symptom onset | |||||
| 2–4 | 7 | 6/7 | 85.7 (42.1–99.6) | 6/7 | 85.7 (42.1–99.6) |
| 5–6 | 0 | NC | NC | NC | NC |
| Vaccine status | |||||
| Not vaccinated | 2 | 2/2 | 100 (15.8–100) | 2/2 | 100 (15.8–100) |
| Fully vaccinated‡ | 2 | 2/2 | 100 (15.8–100) | 2/2 | 100 (15.8–100) |
| Not fully vaccinated | 3 | 2/3 | 66.7 (9.4–0.99.2) | 2/3 | 66.7 (9.4–99.2) |
| Omicron variant | 57 | 56/57 | 98.2 (90.6–100) | 54/57 | 94.7 (85.4–98.9) |
| Symptomaticity | |||||
| Asymptomatic | 22 | 21/22 | 95.5 (77.2–99.9) | 19/22 | 86.4 (65.1–97.1) |
| Symptomatic | 35 | 35/35 | 100 (90.0–100) | 35/35 | 100 (90.0–100) |
| Days since symptom onset | |||||
| 2–4 | 26 | 26/26 | 100 (86.8–100) | 26/26 | 100 (86.8–10) |
| 5–6 | 9 | 9/9 | 100 (66.4–100) | 9/9 | 100 (66.4–100) |
| Vaccine status§ | |||||
| Not vaccinated | 3 | 3/3 | 100 (29.2–100) | 3/3 | 100 (29.2–100) |
| Fully vaccinated‡ | 31 | 30/31 | 96.8 (83.3–99.9) | 28/31 | 90.3 (74.2–98.0) |
| Not fully vaccinated | 19 | 19/19 | 100 (82.3–100) | 19/19 | 100 (82.3–100) |
CI = confidence interval; COVID-19, coronavirus disease-19, Ct = cycle threshold, FN = false negative, NC = not calculable, PPA = positive percentage agreement, rRT-PCR = real-time reverse transcriptase-polymerase chain reaction, TP = true positive.
Calculated using the Clopper–Pearson method.
Six participants had missing data.
Defined as persons who received all recommended doses of vaccines at the time of testing; participants were considered fully vaccinated 2 weeks after receiving a third (booster) dose or 2 weeks after but not exceeding 90 days of receiving their primary series.
Four participants had missing data.
Participants with a Ct value ≤20 showed a 100% detection rate (52/52, 95% CI: 93.2–100%). In those with Ct values between 20 and 30, the detection rate was 95.7% (22/23, 95% CI: 78.1–99.9%). In those with Ct values >30 the detection rate was 50.0% (1/2, 95% CI: 13.0–98.7%).
4. Discussion
Although at-home or patient-performed RAT kits are designed for use by the general population, self-testing by untrained individuals is typically prone to errors. The sensitivity of RATs may be lower than that of the tests performed under strictly controlled conditions due to inappropriate sampling, contamination, and interpretation. Our findings demonstrated mostly consistent results between laboratory tests and participant-performed RATs, implying the practical effectiveness of these RATs when used in real-world settings. Additionally, our study showed that Panbio and STANDARD Q conducted in a self-test setting can detect Delta- and Omicron-infected individuals, which is consistent with the findings of previous laboratory and clinical studies.[1,2,4,5] Both RATs were effective in detecting Omicron-infected individuals who were vaccinated and/or asymptomatic.
A study by Soni et al[8] examined the performance of RATs in symptomatic and asymptomatic individuals, providing a crucial benchmark for our analysis. In their study involving 154 PCR-positive patients, RATs detected SARS-CoV-2 with a sensitivity of 62.7% in 97 asymptomatic individuals and 93.4% in 57 symptomatic individuals. This was in contrast with our findings of no apparent difference according to symptomaticity, with a sensitivity of 98.0% (50/51) in symptomatic individuals and 96.2% (25/26) in asymptomatic individuals.
One potential explanation for these differences may be differences in the viral load. In the study by Soni et al,[8] over 75% of the asymptomatic individuals had a Ct value over 30, indicative of a low viral load, whereas in those with Ct values under 30, the difference in sensitivity between symptomatic and asymptomatic individuals was smaller. In our study, only 2 participants had Ct values below 30, and the majority had Ct values below 20, indicating high viral loads. The high proportion of participants with high viral loads in our cohort likely contributed to the minimal difference in RAT performance between asymptomatic and symptomatic individuals. These findings suggest that while RATs generally perform better in individuals with higher viral loads, the minimal discrepancy in sensitivity between asymptomatic and symptomatic cases in our study highlights the potential of RATs to reliably detect COVID-19 across different clinical presentations, provided the viral load is sufficiently high.
Venekamp et al[9] also found low sensitivity for 3 commonly used RATs in asymptomatic individuals during the Omicron period, with sensitivity ranging from 20.9% to 27.5%. After applying a high viral load cutoff, sensitivities improved, highlighting the importance of viral load in RAT performance. Additionally, a systematic review by Anand et al[10] emphasized the variability in RAT performance across different settings, with better outcomes observed in symptomatic individuals with higher average viral loads than in asymptomatic individuals. Winnett et al[11] further demonstrated that daily nasal RATs missed a substantial portion of individuals infected with the Omicron variant. They found that combining nasal and throat swabs significantly improved detection rates possibly due to the higher viral loads in throat swabs, which suggests that multispecimen approaches might be necessary to enhance RAT performance in detecting infectious individuals. Collectively, these studies show that RAT performance improves with higher viral loads and may require additional strategies, such as repeated testing and multispecimen approaches, for enhanced diagnostic accuracy.
In a study by Drain et al,[12] the accuracy of RATs was evaluated at different stages of the COVID-19 pandemic, specifically during the pre-Delta, Delta, and Omicron phases. The PPA for RATs was 90.7% during the Delta phase and 83.6% during the Omicron phase. However, when the Ct value was below 30, indicative of a higher viral load, the PPA for RATs was 100% for both Delta and Omicron variants. In our study, the Panbio COVID-19 Antigen Self-test had a PPA of 98.2% for the Omicron and 85.7% for the Delta variant, whereas the STANDARD Q COVID-19 Ag Home test had a PPA of 94.7% for the Omicron and 85.7% for the Delta variant. These results imply a relatively lower performance of RATs in detecting Delta compared to Omicron in our dataset. Considering that the number of participants confirmed with the Delta variant in our study was relatively small (n = 7), this could potentially account for the observed discrepancy in RAT performance between the 2 variants. This limited sample size might not entirely capture the RATs’ efficacy against the Delta variant, potentially skewing the comparative analysis.
Another study limitation was that the Sanger sequencing of the receptor binding domain hotspot was adopted to determine the variant, which may be more prone to errors compared with whole-genome sequencing. Despite these limitations, our study provides robust evidence supporting the reliability of RATs in real-world settings, particularly for Omicron detection.
In conclusion, the present study provides direct evidence that RATs, originally developed for ancestral strains, are effective in detecting major variants, such as Delta and Omicron, in real patient/clinical settings. The findings are particularly significant for Omicron detection in breakthrough infections postvaccination, where RATs showed high accuracy. The results suggest that RATs can be confidently used in public health measures against COVID-19, facilitating timely and accurate self-diagnosis, which is crucial for controlling the spread of the virus. These study insights are valuable for clinical practice, supporting the continued use and potential expansion of RATs in diverse healthcare settings.
Author contributions
Conceptualization: Jongyoun Yi, Kye-Hyung Kim.
Data curation: Jongyoun Yi, Jongmin Kim, Mee Kyung Ko.
Formal analysis: Jongyoun Yi, Jongmin Kim, Kye-Hyung Kim.
Funding acquisition: Jongyoun Yi, Kye-Hyung Kim.
Investigation: Jongyoun Yi, Mee Kyung Ko, Kye-Hyung Kim.
Methodology: Jongyoun Yi, Kye-Hyung Kim.
Resources: Mee Kyung Ko, Shinwon Lee, Soon Ok Lee, Jeong Eun Lee, Jeongha Mok, Mi-Hyun Kim, Jung Seop Eom, Sunjoo Kim, Kye-Hyung Kim.
Software: Jongmin Kim.
Writing – original draft: Jongmin Kim.
Writing – review & editing: Jongyoun Yi, Jongmin Kim, Mee Kyung Ko, Shinwon Lee, Soon Ok Lee, Jeong Eun Lee, Jeongha Mok, Mi-Hyun Kim, Jung Seop Eom, Sunjoo Kim, Kye-Hyung Kim.
Abbreviations:
- COVID-19
- coronavirus disease-19
- Ct
- cycle threshold
- CTC
- community treatment center
- PPA
- positive percentage agreement
- RATs
- rapid antigen tests
- RBD
- receptor binding domain
- rRT-PCR
- real-time reverse transcriptase-polymerase chain reaction
- SARS-CoV-2
- severe acute respiratory syndrome coronavirus 2
This work was supported by the Abbott Diagnostic Korea Inc (recipient: Jongyoun Yi), the National Research Foundation of Korea (NRF) grant funded by the Korea government (Ministry of Science and ICT) (NRF-2022R1F1A1065194, recipient: Kye-Hyung Kim), and clinical research grant from Pusan National University Hospital in 2022 (recipient: Kye-Hyung Kim).
The Institutional Review Boards of Pusan National University Hospital and Gyeongsang National University Changwon Hospital (approval numbers: 2110-026-108 and 2021-11-003, respectively) approved this sponsor-initiated, multicenter clinical performance study performed in Korea between November 2021 and February 2022. The study was conducted in compliance with the ethical standards of the Declaration of Helsinki. All participants provided written informed consent.
The authors have no conflicts of interest to disclose.
The datasets generated during and/or analyzed during the current study are not publicly available, but are available from the corresponding author on reasonable request.
How to cite this article: Yi J, Kim J, Ko MK, Lee S, Lee SO, Lee JE, Mok J, Kim M-H, Eom JS, Kim S, Kim K-H. Diagnostic accuracy of rapid antigen tests for detection of Delta and Omicron variants of severe acute respiratory syndrome coronavirus 2: Direct evidence from prospective clinical settings. Medicine 2024;103:40(e39995).
JY and JK contributed equally to this work.
Contributor Information
Jongyoun Yi, Email: socioliberal@yahoo.co.kr.
Jongmin Kim, Email: luckyupper@naver.com.
Mee Kyung Ko, Email: qeqazwsx@hanmail.net.
Shinwon Lee, Email: ebenezere.lee@gmail.com.
Soon Ok Lee, Email: lsook81@hanmail.net.
Jeong Eun Lee, Email: godprayer166@naver.com.
Jeongha Mok, Email: mokgamokga@gmail.com.
Mi-Hyun Kim, Email: lovefull777@naver.com.
Jung Seop Eom, Email: ejspulm@gmail.com.
Sunjoo Kim, Email: sjkim8239@hanmail.net.
References
- [1].Regan J, Flynn JP, Choudhary MC, et al. Detection of the Omicron variant virus with the Abbott BinaxNow SARS-CoV-2 rapid antigen assay. Open Forum Infect Dis. 2022;9:ofac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Stanley S, Hamel DJ, Wolf ID, et al. Limit of detection for rapid antigen testing of the SARS-CoV-2 Omicron and Delta variants of concern using live-virus culture. J Clin Microbiol. 2022;60:e0014022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Coronavirus (COVID-19) Update: December 28, 2021. 2021. https://web.archive.org/web/20211229220840/https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-december-28-2021. Accessed January 23, 2023. [Google Scholar]
- [4].Drain PK. Rapid diagnostic testing for SARS-CoV-2. N Engl J Med. 2022;386:264–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Schrom J, Marquez C, Pilarowski G, et al. Comparison of SARS-CoV-2 reverse transcriptase polymerase chain reaction and BinaxNOW rapid antigen tests at a community site during an Omicron surge: a cross-sectional study. Ann Intern Med. 2022;175:682–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ahn S, Jang J, Park SY, Yang S, Ryu B, Shin E. Outbreak report of COVID-19 during designation of class 1 infectious disease in the Republic of Korea (January 20, 2020 and April 24, 2022). Public Health Wkly Rep. 2022;15:1759–72. [Google Scholar]
- [7].Lee SH. A routine sanger sequencing target specific mutation assay for SARS-CoV-2 variants of concern and interest. Viruses. 2021;13:2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Soni A, Herbert C, Lin H, et al. Performance of rapid antigen tests to detect symptomatic and asymptomatic SARS-CoV-2 infection: a prospective Cohort study. Ann Intern Med. 2023;176:975–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Venekamp RP, Schuit E, Hooft L, et al. Diagnostic accuracy of SARS-CoV-2 rapid antigen self-tests in asymptomatic individuals in the Omicron period: a cross-sectional study. Clin Microbiol Infect. 2023;29:391.e1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Anand A, Vialard F, Esmail A, et al. Self-tests for COVID-19: what is the evidence? A living systematic review and meta-analysis (2020–2023). PLOS Glob Public Health. 2024;4:e0002336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Viloria Winnett A, Akana R, Shelby N, et al. Daily SARS-CoV-2 nasal antigen tests miss infected and presumably infectious people due to viral load differences among specimen types. Microbiol Spectr. 2023;11:e0129523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Drain PK, Bemer M, Morton JF, et al. Accuracy of 2 rapid antigen tests during 3 phases of SARS-CoV-2 variants. JAMA Netw Open. 2022;5:e2228143. [DOI] [PMC free article] [PubMed] [Google Scholar]