Abstract
It was recently reported that the human endogenous retrovirus HTDV/HERV-K encodes the regulatory protein Rec (formerly designated Corf), which is functionally equivalent to the nuclear export adapter proteins Rev of human immunodeficiency virus and Rex of human T-cell leukemia virus. We have demonstrated that the Rec protein interacts with a characteristic 429-nucleotide RNA element, the Rec-responsive element (RcRE), present in the 3′ long terminal repeat of HTDV/HERV-K transcripts. In analogy to the Rev and Rex proteins, which have distinct RNA binding sites in their responsive elements, we have proposed that Rec may also have a defined binding site in the RcRE. In this report, we demonstrate that not every HTDV/HERV-K copy present in the human genome contains an active RcRE, and we characterize mutations that abrogate Rec function. In addition, we demonstrate that Rec function requires binding to a complex, folded RNA structure rather than binding to a discrete specific binding site, in contrast to Rev and Rex and their homologous responsive elements. We define four stem-loop structures in the RcRE that are essential for Rec function. Finally, we demonstrate that both Rev and Rex can mediate nuclear export through the RcRE but that their binding sites are different from each other and from that of Rec.
HTDV/HERV-K is a human endogenous retrovirus present in about 50 copies in the human genome. In addition, about 10,000 solitary long terminal repeats (LTRs) exist. HTDV/HERV-K elements are genetic footprints of germ line infection with an exogenous predecessor almost 30 million years ago. Like all other retroviruses, HTDV/HERV-K needs to export partially spliced and unspliced mRNAs from the cell nucleus during its life cycle. Retroviruses have developed different systems to circumvent the splicing machinery of the cell by enhanced nuclear export of viral mRNA. Simply structured retroviruses, such as the D-type viruses, contain a constitutive transport element (4, 8), an RNA element that directly interacts with the cellular TAP protein (12), an export receptor involved in the nuclear export of cellular mRNA molecules. Recently, it was reported that Rous sarcoma virus, which contains an element like the constitutive transport element, uses a cellular export factor distinct from TAP (26).
Complex viruses, such as human immunodeficiency virus (HIV) and human T-cell leukemia virus, contain genes encoding a regulatory protein (Rev in HIV and Rex in human T-cell leukemia virus) translated from completely spliced mRNAs that are exported from the nucleus by the regular mRNA export pathway (for reviews, see references 5, 25, and 29). Rev and Rex shuttle into the nucleus and bind as multimers to their responsive elements (Rev-responsive element [RRE] and Rex-responsive element [RxRE]) present in the respective primary viral transcripts. In a second step, they recruit the cellular export receptor exportin 1 (10, 11, 24) to mediate the export of unspliced and incompletely spliced viral transcripts, thus competing with the splicing machinery.
The RRE and RxRE sequences form highly structured RNA elements with several stem-loop structures. Within these tightly folded structures, Rev and Rex bind to defined sites located in a specific stem with an internal bulge (1, 2, 3, 9, 14).
Recently, it was reported that HTDV/HERV-K also encodes a regulatory protein which is a functional homologue of Rev and Rex (18, 21, 31). This protein was previously termed Corf. The fact, however, that the Corf export domain cannot functionally replace the Rev export domain and in many respects rather resembles Rex (unpublished data) prompted us to rename it Rec (regulator of expression encoded by corf). Rec binds to an RNA element, the Rec-responsive element (RcRE), in the 3′ LTR of HTDV/HERV-K transcripts. The RcRE is located in U3 and R and is approximately 430 nucleotides (nt) long (21). In this report, we demonstrate not only that Rec and Rev can use the RcRE, as previously reported (21), but also that Rex exports transcripts encoding the RcRE. Like the RRE and the RxRE, the RcRE seems to be a highly structured RNA element with several stem-loop structures (21, 32). However, little is known about the sequences and structural requirements which enable binding of the Rec protein to the RcRE. Further, it is unknown whether all HTDV/HERV-K copies encode an active RcRE.
Here, we demonstrate that a subset of these elements contains six clusters of mutations within the RcRE region compared to the biologically active RcRE sequence obtained from clone pcK30 (21). We show that the mutated sequences do not support Rec- and Rex-mediated RNA export, whereas Rev is inactive only with some of these sequences. By analyzing hybrid RcRE (HyRcRE) sequences consisting of active and inactive parts, we demonstrate that two of the six clusters abrogate Rec binding and function when mutated, while the others do not affect the functionality of the element. Rev and Rex function is inhibited or even enhanced by different sets of mutations. Computational RNA folding analyses predict that inactivating mutations evoke a very different structure for the RcRE, indicating strict conformational constraints for function. Deletion of single stem-loop structures present in the active folding structure of the RcRE enabled us to identify four stem-loop structures that are essential for Rec function and two stem-loop structures that influence Rev function on the RcRE, while Rex, in contrast, was only moderately influenced by these deletions.
MATERIALS AND METHODS
Plasmid constructions.
Plasmids pHIVgagRRE (pNLcgagA2 in references 6 and 13), pHIVgagRcRE, pREC (formerly named pcORF), pREV (pBsrev in references 7 and 23), and p(−) have been described previously (21, 22). Plasmids pHIVgagRcRELTR3, pHIVgagRcRELTR10, pHIVgagRcRELTR21, pHIVgagRcRELTR23, pHIVgagRcRELTR26, and pHIVgagRcRELTR18 were generated by PCR amplification of different RcRE sequences from several HERV-K LTRs with the same primers as those used to amplify the original RcRE from the pcK30 LTR (21) and replacement of the RRE in pHIVgagRRE with the amplified RcRE sequences by use of SacII and XhoI restriction sites. The resulting RcRE sequences are depicted in Fig. 1. Plasmid pHIVgagRxRE was cloned by PCR amplification of the RxRE from clone pDM128/CMV/RxRE (15) and replacement of the RRE by the RxRE. Plasmids pHIVgagHyRcRE0 to pHIVgagHyRcRE12 were generated by assembly PCR to fuse fragments of the pcK30 LTR to LTR21 and to insert specific clusters of mutations from the pcK30 LTR into the background of LTR21 (see Fig. 3A). The vectors with internal deletions were again cloned by assembly PCR to delete the following nucleotides after ensuring that the predicted RNA secondary structure of the remaining part of the RcRE was not affected by the deletions: pHIVgagdel1, nt 125 to 179; pHIVgagdel2, nt 184 to 233; pHIVgagdel3, nt 235 to 252; pHIVgagdel4, nt 100 to 118; pHIVgagdel5, nt 74 to 91; pHIVgagdel6, nt 272 to 286; pHIVgagdel7, nt 303 to 339; pHIVgagdel11, nt 167 and 168 as well as 174 and 175; pHIVgagdel12, nt 211 and 212; pHIVgagdel13, nt 194 to 197; and pHIVgagdel14, nt 220 and 223. Accession numbers are as follows: RcRE, part of X82272; and Rec, X82271. Plasmid pcRex (27) was termed pREX. A 32P cycle sequencing kit (Amersham) was used to confirm the introduced mutations or deletions. Plasmids were produced in DH5α cells (Gibco BRL). Plasmid DNA was prepared with an endotoxin-free plasmid preparation kit (Qiagen).
FIG. 1.
Alignment of RcRE sequences of different genomic LTRs. Mutational clusters M0 to M5 are depicted in bold letters. cons, consensus.
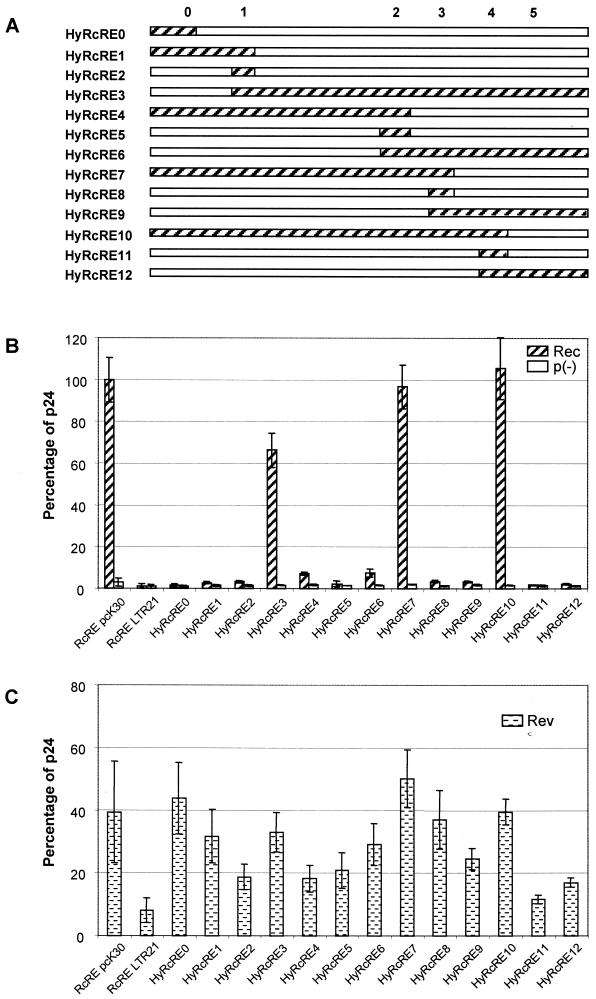
FIG. 3.
Activity of RcRE hybrids containing pcK30 and LTR21 sequences. (A) Schematic representation of hybrid sequences. pcK30 sequences are depicted as hatched bars; LTR21 sequences are depicted as white bars; 0 to 5 indicate the positions of mutations or consensus sequences. (B to D) Determination of the HIV Gag precursor in cell lysates after cotransfection of the indicated pHIVgagHyRcRE vectors into HLtat cells with the effector plasmid pRec or p(−) (B), pRev (C), or pRex (D). Error bars indicate average deviations.
Transfections.
Transfections into HLtat cells (28) were performed with six-well plates (Greiner; 0.5 million cells per well) for HIV p24 quantification; for immunofluorescence studies, the six-well plates contained coverslips. Transfections were carried out with Lipofectamine Plus (Gibco BRL) in accordance with the user manual. Transfectants were analyzed 24 h after transfection. Cell viability was controlled by measuring the total protein content of the cell lysates.
Quantification of HIV p24.
Cells were lysed with 250 μl of cell culture lysis buffer (Promega) per well and used in an HIV AG-1 monoclonal antibody p24 capture assay (Abbott) in accordance with the manufacturer's instructions.
Protein expression and purification.
Escherichia coli BL21(SI) (Invitrogen) was transformed with a pET15 (Novagen) expression plasmid containing the rec sequence. Rec protein expression was induced with 500 mM NaCl for 4 to 5 h. Bacterial pellets were lysed by sonication in 50 mM Capso (Sigma) pH 9.5. Rec protein was purified using SP-Sepharose beads (Amersham).
In vitro transcription and RNA gel shifts.
For in vitro transcription, the RcRE sequences were subcloned into plasmid pBluescript (Stratagene). Linearized plasmids were transcribed with T7 RNA polymerase by use of a Riboprobe in vitro transcription system (Promega) in accordance with the manufacturer's instructions. Probes were labeled with 25 μCi of [α-32P]CTP (Amersham).
RNA-protein binding reactions were performed with 4 × 104 cpm of labeled RNA probe and 10 to 20 ng of Rec protein in 20 μl of buffer containing 50 mM capso-buffer (pH 9.5), 2.5 U of RNasin, 70 mM NaCl, 1 mM MgCl2, 0.5 mM EDTA, 1 mM dithiothreitol, 30 to 50 ng of bovine serum albumin, 4% glycerol, 20 μg of yeast tRNA, and 10 to 20 ng of unrelated RNA transcribed in vitro. Rec was preincubated with the nonspecific RNA competitors for 10 min at room temperature. 32P-labeled probes were denatured at 80°C for 5 min and slowly renatured by cooling to 37°C before addition to the reaction mixture. Incubation was then continued for 15 min at room temperature. For competition experiments, 10 ng of the respective unlabeled RcRE competitor RNA was added to the preincubation mixture. The reaction products were resolved on a native 6% polyacrylamide gel (acrylamide-bisacrylamide, 80:1) containing 2.5% glycerol.
RESULTS
Recently, it has been demonstrated that Rec (formerly Corf) can mediate nuclear export of RNA by binding to its responsive element, RcRE, present in a transcript (21, 31). We mapped the RcRE to a fragment of 429 nt in the U3R region of the 3′ LTR and reported that a smaller element (374 nt) deleted from the 3′ end was partially active (21). Longer deletions at the 3′ end or deletions of about 50 nt at the 5′ end abrogated the function of the RcRE (21), a fact recently confirmed by others (32).
Characteristic clusters of mutations abrogated the RNA export function of RcRE sequences.
To elucidate putative functional domains within the RcRE, we first analyzed whether or not different HTDV/HERV-K LTRs harbor an active RcRE. These sequences, cloned to perform promoter studies (unpublished data), were obtained by PCR using human genomic DNA and primers located in U3 and downstream of U5 to ensure that the sequences were not amplified from solitary LTRs. From the respective RcRE sequences, a consensus sequence was compiled (Fig. 1) that—with the exception of nt 143—was identical to the pcK30 sequence, the RcRE sequence previously used for our studies (21, 22). Comparison of the different RcRE sequences revealed that a subset of them had five or six specific clusters of mutations (M0 to M5) (Fig. 1). Throughout this publication, the mutational clusters are designated Mn, e.g., M1, and the respective consensus sequence at that position is designated Cn, e.g., C1. The RcRE sequence of LTR26 was unique in lacking the M0 mutation. Additionally, most LTR sequences differed from each other by scattered singular point mutations.
To analyze the function of these different RcRE sequences, a well-established test system was used. In transient transfections with a human cell line (HLtat), the amount of HIV Gag expressed from a reporter plasmid depended on the presence of an active responsive element in cis (e.g., RcRE) and a plasmid encoding the viral export adapter (e.g., Rec) in trans. In two previous studies (21, 22), it was demonstrated that Rec-dependent RNA export activity can be measured either by comparing the levels of cytoplasmic and nuclear HIV Gag reporter RNAs containing the RcRE or by measuring the amount of the reporter protein translated from the exported transcripts. Both tests measure nuclear RNA export equally reliably. Throughout this study, nuclear export of HIV Gag transcripts was visualized by immunofluorescence staining of translated HIV Gag (data not shown) and was quantified by measuring the amount of p24 HIV capsid protein in cell lysates. All transfections were repeated six times. Uniformity of transfection efficiencies was monitored by comparing the numbers of cells expressing the respective viral export adapters visualized by immunofluorescence. Within each set of experiments, the HIV Gag expression level mediated by the RcRE sequence of the pcK30 LTR and Rec was set as 100%, and all results obtained are reported as percentages of this value. In addition, all data presented in this publication were confirmed by comparing the levels of cytoplasmic and nuclear reporter transcripts in transfected cells. The level of exported HIV Gag reporter mRNA was always reflected by the level of p24 HIV Gag protein (data not shown).
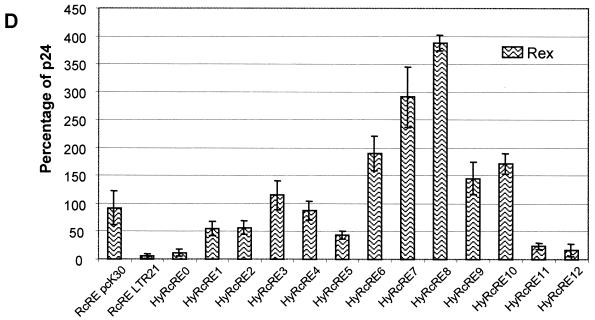
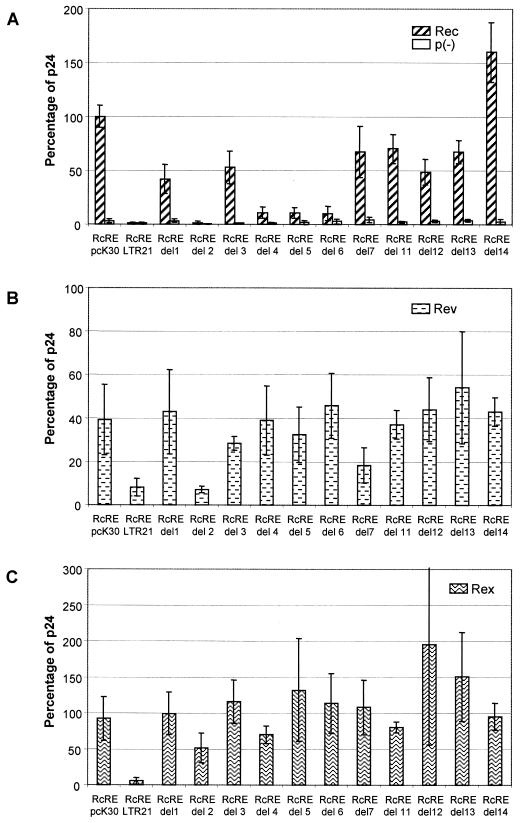
With the different RcRE sequences depicted in Fig. 1, the cotransfection experiments showed that Rec was active on the RcRE sequences present in LTR3, LTR10, and LTR23, with efficiencies comparable to the activation seen with the RcRE from the pcK30 LTR (means of 77% for LTR3 to 144% for LTR23) (Fig. 2A). Interestingly, the RcRE sequences present in LTR18, LTR21, and LTR26 did not support Rec-mediated nuclear export. In these cotransfections, the amount of p24 (1 to 2%) was in the same range as in cotransfections with p(−), a construct lacking the Rec coding sequence (21) (Fig. 2A). The findings were confirmed by immunofluorescence analyses, the results of which were absolutely consistent with the p24 data indicating that the presence of mutational clusters in a subset of RcRE sequences correlated with a lack of export function. In addition, Fig. 2A shows that Rec not only failed to function with the RRE, as described before (21), but also was unable to use the RxRE.
FIG. 2.
Biological activity of RcRE sequences from HTDV/HERV-K genomes. The HIV Gag precursor in cell lysates was determined with an HIV p24 capture assay after cotransfection of the indicated pHIVgagRcRE reporter vectors into HLtat cells with the effector plasmid pRec or p(−) (A), pRev (B), or pRex (C). Expression of pHIVgagRcRE pcK30 in the presence of pRec was set as 100%; all other data are relative to this value. Error bars indicate average deviations.
Rev (Fig. 2B), being active on the RcRE at a reduced level (40% the Rec activity seen with the pcK30 RcRE), as already described (21), was less restrictive than Rec. Within the subset of mutated RcRE sequences, RcRE LTR18 exerted residual export function in cotransfections with Rev (26% the Rec activity seen with the pcK30 RcRE). Only RcRE LTR21 showed significantly reduced export activity (8%), similar to the level reached with the RxRE sequence. RcRE LTR18 differs from RcRE LTR21 by the lack of a nucleotide exchange in clusters M1 (nt 85) and M2 (nt 248), a feature which might explain the less pronounced effect of RcRE LTR18. Surprisingly, RcRE LTR26 was as active in cotransfections with Rev as the pcK30 RcRE (43%). The fact that M1 to M5 did not abrogate Rev function either indicated that C0 might harbor a Rev binding site or indicated that in the presence of C0, a structure was formed that allowed Rev binding.
Rex was nearly as effective as Rec on the pcK30 RcRE (92%), although it was considerably more active with its genuine element, RxRE (2.7 times the Rec activity seen with the pcK30 RcRE) (Fig. 2C). The activity of Rex in cotransfections with the different RcRE sequences displayed a pattern similar to that seen with Rec. The RcRE sequences of LTR3, LTR10, and LTR23 supported export activity, and the RcRE sequences of LTR18, LTR21, and LTR26 did not support export activity. However, from these experiments, it was not possible to identify which mutational cluster abrogated Rec or Rex binding and export activity.
In summary, these results demonstrated that the mutational clusters in LTR21, LTR18, and LTR26 inactivated the RcRE for Rec and Rex and in part for Rev-mediated RNA export function. The most inactive element was LTR21, bearing all point mutations depicted in the mutational clusters in Fig. 1.
RcRE hybrids of pcK30 and LTR21 revealed sequence and structural requirements for Rec, Rev, and Rex functions.
Aiming to analyze the mutational clusters in detail in order to identify a putative Rec, Rev, or Rex binding site, we cloned a set of HyRcRE sequences. They consisted of fusions of the pcK30 LTR and LTR21. A schematic representation of the sequence composition is depicted in Fig. 3A. The positions of mutational clusters (M) respective to the consensus sequence (C) are indicated. All HyRcRE sequences were tested for their ability to mediate the nuclear export of HIV Gag reporter RNA in cotransfections with Rec, Rev, and Rex. The controls mentioned above (visualization of the viral export adapter and the reporter protein by immunofluorescence as well as comparison of cytoplasmic and nuclear RNA levels) were included, but only the data obtained by measuring the level of HIV Gag p24 are shown.
The results obtained in transfections with Rec (Fig. 3B) showed that wild-type Rec activity correlated with a sequence context containing C1, C2, and C3 in hybrids 3, 7, and 10, although the presence of M0 in HyRcRE3 induced a slight reduction in Rec activity (to 66%). Conversely, the presence of M1 in HyRcRE6 and of M3 in HyRcRE4 led to a dramatic reduction in Rec activity (to 7 to 8%). None of the other hybrids supported significant HIV p24 Gag expression. Since the introduction of neither C1 (HyRcRE2) nor C2 (HyRcRE5) nor C3 (HyRcRE8) into the inactive RcRE backbone of LTR21 restored Rec activity, it could be deduced that the entire region between C1 and C3 was essential for Rec function. In further experiments, C1 and C3 were introduced into the inactive RcRE LTR21 backbone (HyRcRE13) or C2 was mutated to M2 in the active pcK30 RcRE backbone (HyRcRE15). In cotransfections with Rec, these two mutants exerted approximately 60% the export activity measured with the pcK30 RcRE (data not shown). In summary, the hybrid analyses revealed that in cotransfections with Rec, cluster M4 or M5 (HyRcRE7 and HyRcRE10) had no influence on RcRE activity, cluster M0 (HyRcRE3 and HyRcRE13) or M2 (HyRcRE13 and HyRcRE15) reduced RcRE activity by 40%, and the presence of both C1 and C3 was essential for function. As it is highly unlikely that a region spanning more than 210 nt represents a single sequence-specific protein binding site, one can conclude that the active hybrids fold into a complex RNA structure which enables Rec binding.
Cotransfections with Rev resulted in a different picture. Rev activity was not completely abrogated in any of the hybrids, although the presence of M0 to M3 seemed to be responsible for the low activity in HyRcRE11 and HyRcRE12. Full activity of Rev correlated with the presence of C3 (HyRcRE3 and HyRcRE6 to HyRcRE10). Notably, C3 alone in the context of M0 to M2 and M4 or M5 (HyRcRE8) rescued Rev activity, indicating that C3 allowed the formation of a Rev binding site in this context. In addition, C0 alone (HyRcRE0) or in combination with C1 (HyRcRE1) also restored Rev activity in an inactive context. These results are consistent with the observation that C0 in RcRE LTR26 was fully functional with Rev. In summary, the data indicated that the requirements for the binding of Rev to the RcRE differed from those of Rec. Rev may have two discrete primary binding sites in the RcRE which may be used independently because either C0 or C3 alone was sufficient to fully restore export activity. These binding sites may be more sequence specific than the Rec binding site because they require smaller parts of the consensus RcRE sequence.
Cotransfections with Rex again showed a different pattern of sequence requirements for function (Fig. 3D). HyRcRE3 and HyRcRE6 to HyRcRE10, which contained C3, were active or even hyperactive compared to the pcK30 RcRE. C3 alone in the context of M0 to M2 and M4 to M5 (HyRcRE8) resulted in dramatic activation (388%), followed by HyRcRE7 (290%), in which M4 and M5 were present. As C3 sequences introduced into RcRE LTR21 even exceeded the activity seen with Rex binding to its genuine element, RxRE (270%) (Fig. 2C), one can conclude that a new, very potent Rex binding site was created in this sequence context. Only for HyRcRE4 did Rex function depend not on C3 but on the presence of C0 to C2, indicating the existence of a second Rex binding site. As neither C0 nor C1 nor C2 alone rescued Rex activity, Rex binding to this site may depend on structural requirements rather than on sequence-specific requirements only.
In summary, these data suggested that rather than a discrete binding site, Rec function required the presence of a large sequence region (C1 to C3) that might fold into a complex structure. Rev function depended on the presence of either C0 or C3, indicating the presence of two independent and discrete binding sites. Rex function depended on the presence either of a putative sequence-specific binding site in C3 or a larger sequence region comprising C0 to C2.
Predicted secondary structures of the RcREpcK30 LTR, RcRELTR21, and the hybrids differed significantly.
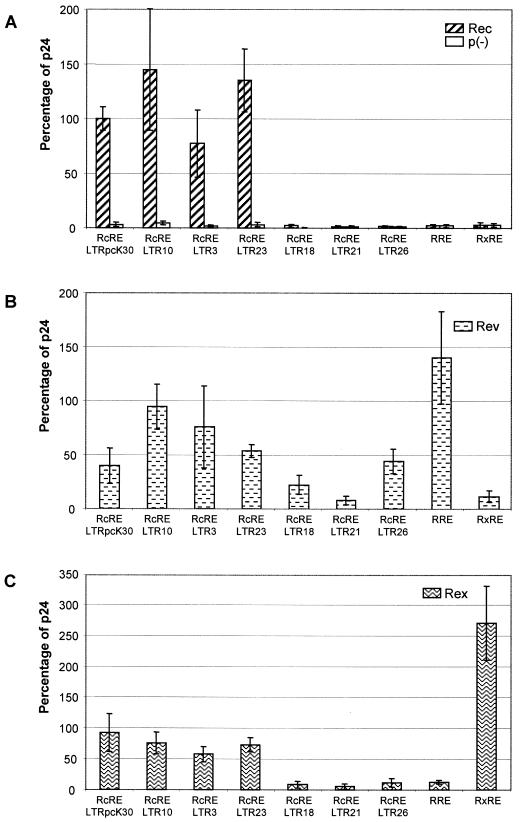
It has been previously shown that Rev and Rex bind to a discrete site in their respective responsive elements that is exposed in a larger, tightly folded region, as predicted by computer analyses (1, 3, 9, 14, 16). Computational approaches have repeatedly allowed the definition of the responsive elements of retroviral export adapters and were recently used to generate a candidate structure for the RcRE (reference 32 and references therein). We applied comparable computer software—the DNASIS RNA folding software (Hitachi) based on algorithms described by Zuker (33)—to compare the predicted structures of the active (pcK30) and inactive (LTR21) RcREs as well as their hybrids. As shown in Fig. 4, the predicted structures of the RcREpcK30 and RcRELTR21 were both rather complex. Due to the six clusters of nucleotide exchanges, they differed significantly, especially in the shape and the arrangement of the stem-loop formations. In both structures, however, several major stem-loop formations (SL1 to SL7) were discerned in the inner core segment from nt 60 to nt 340, including C1 to C4 (M1 to M4, respectively). The 5′ and 3′ ends of the element folded back into a single long stem-loop stretch in the RcREpcK30 and a set of several smaller stem-loop formations in RcRELTR21.
FIG. 4.
Predicted folding structures of active and inactive RcREs. (A) pcK30. (B) LTR21. The structures were predicted with DNASIS software using algorithms defined by Zuker (33).
Computational analysis of all the hybrid RNAs led to an interesting observation. Only the three HyRcRE sequences that were functional with Rec had predicted structures which resembled the pcK30 RcRE structure, displaying an inner core with stem-loop formations SL1 to SL7. Table 1 summarizes which stem-loop formations were retained or destroyed by the presence of the different mutational clusters in the hybrids. The free energy calculated by the computer software did not differ significantly between active and inactive elements. Surprisingly, however, all sequences tested, whether they were biologically active or inactive, contained SL2 and SL7. Rec was only active when SL1 to SL7 were all present, but it remained unclear to which of the seven stem-loop formations Rec would bind. Rev and Rex function either correlated with the presence of all seven stem-loop formations or, in some instances, correlated with the presence of SL2, SL3, and SL7. However, SL3 did not fold in HyRcRE1 and HyRcRE0, which efficiently supported Rev-mediated export, or in HyRcRE4, which efficiently supported Rex-mediated export.
TABLE 1.
Compilation of the presence of predicted stem-loop structures in the inner core of the HyRcRE sequences and their functionality
| RcRE | Stem-loop structuresa
|
Functionb
|
Free energy (kcal/mol)c | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SL1 | SL2 | SL3 | SL4 | SL5 | SL6 | SL7 | Rec | Rev | Rex | ||
| pcK30 | + | + | + | + | + | + | + | +++ | +++ | +++ | −227.6 |
| LTR21 | − | + | − | + | (+) | − | [+] | − | ± | − | −217.8 |
| HyRcRE0 | − | + | − | + | (+) | − | [+] | − | +++ | ± | −217.0 |
| HyRcRE1 | − | + | − | − | − | − | [+] | − | +++ | ++ | −222.2 |
| HyRcRE2 | − | + | − | − | − | − | [+] | − | ++ | ++ | −220.2 |
| HyRcRE3 | + | + | + | + | + | + | + | +++ | +++ | +++ | −223.2 |
| HyRcRE4 | − | + | − | − | − | − | [+] | − | ++ | +++ | −225.8 |
| HyRcRE5 | − | + | − | + | (+) | − | [+] | − | ++ | ++ | −222.3 |
| HyRcRE6 | − | + | + | − | − | − | [+] | − | +++ | ++++ | −221.1 |
| HyRcRE7 | + | + | + | + | + | + | [+] | +++ | +++ | +++++ | −227.4 |
| HyRcRE8 | − | + | (+) | − | − | − | [+] | − | +++ | ++++++ | −217.8 |
| HyRcRE9 | − | + | (+) | − | − | − | + | − | +++ | ++++ | −216.0 |
| HyRcRE10 | + | + | + | + | + | + | + | +++ | +++ | ++++ | −223.3 |
| HyRcRE11 | − | + | − | + | (+) | − | + | − | + | + | −213.7 |
| HyRcRE12 | − | + | − | + | (+) | − | + | − | ++ | ± | −213.0 |
+ and −, presence and absence, respectively, of the structure; (+), the structure contains point mutations compared to pcK30; [+], the structure contains point mutations compared to pcK30 and is slightly modified.
++++++, >3.5-fold standard; +++++, 2.5-fold standard; ++++, 1.5-fold standard; +++, standard; ++, 0.5-fold standard; +, 0.25-fold standard; ±, <0.25-fold standard; −, no activity. Activity with RcREpcK30 is defined as standard.
Calculated as DNA energy values.
Deletion of individual predicted stem-loop structures in the RcRE and their influence on Rec, Rev, and Rex function.
To elucidate the putative role of the diverse stem-loop structures in the biological functions of Rec and the other retroviral export adapters, a further set of mutated RcRE sequences was cloned. In each of these constructs, a single stem-loop was deleted (del1 to del7) (Fig. 4A) in a way that did not alter the formation of the predicted remaining stem-loop structures, which was controlled by folding of each of the deletion RcREs (data not shown). In addition to these large deletion mutants, four mutants (del11 to del14) (Fig. 4A) that each eliminated one or two (del11) of the small bulges in SL1 and SL2 were cloned. del11 affected a sequence with homology to the Rex binding site on the RxRE (21).
The results of the functional analyses in cotransfections of these constructs with Rec are shown in Fig. 5A. Again, the usual control experiments (immunofluorescence to monitor transfection efficiencies and comparison of nuclear export at the RNA level) were performed (data not shown). Impressively, deletion of SL2 completely abrogated Rec-mediated HIV Gag export, indicating that this stem-loop was essential for Rec function. The fact, however, that SL2 was present in all RcREs which did not support Rec activity showed that this stem-loop was necessary but not sufficient for an active element. Concomitantly, deletion of SL4, SL5, or SL6 significantly abrogated Rec function, hinting at severe structural constraints for Rec binding. Notably, the correct folding of SL5 and SL6 depended on the presence of C1 to C3 (Table 1). All other deletions, except del14, which had some enhancing effect, reduced Rec function at most by only half, indicating that SL1, SL3, SL7, or one of the bulges in SL1 or SL2 was not essential for Rec function. The data that Rec function was abrogated or highly impaired by deletion of any of the four stem-loop structures mentioned above (SL2, SL4, SL5, and SL6) hinted at the possibility that Rec binding to the RcRE needed a complex, folded structure rather than a discrete binding site.
FIG. 5.
Activity of RcRE deletion mutants of pcK30. The HIV Gag precursor in cell lysates was determined after cotransfection of the indicated pHIVgagdel vectors (for exact positions of deletions, see Materials and Methods) into HLtat cells with the effector plasmid pRec or p(−) (A), pRev (B), or pRex (C). Error bars indicate average deviations.
For Rev function, the hybrid analyses suggested the existence of two independent binding sites, one of which correlated with the presence of C3, which is located partly in SL6. The fact that del6 had no effect on Rev activity (Fig. 5B) can therefore be explained by the presence of C0 in this construct. In cotransfections with Rev, deletion of SL2 also reduced the Rev-mediated export function to the level of RcRE LTR21, indicating that this stem-loop structure was necessary for the recognition of either of the two Rev binding sites. Of all other deletions, only deletion of SL7 reduced Rev function by half.
In cotransfections with Rex (Fig. 5C), none of the deletions abrogated Rex function, although deletion of SL2 again had the most intense inactivating effect and deletion of SL4 evoked a slight reduction of function. Deletions del12, del13, and del14 did not significantly alter Rex export function, whereas del11, which would affect a putative Rex binding sequence (21), had a very minor reducing effect.
In summary, these results demonstrated that binding of the three retroviral export proteins required different structural properties in the RcRE. Rec function depended on the presence of at least four stem-loop structures (SL2, SL4, SL5 and SL6), while Rev was influenced by SL2 and partially by SL7 and Rex did not absolutely depend on any of the predicted stem-loop structures.
In vitro binding studies.
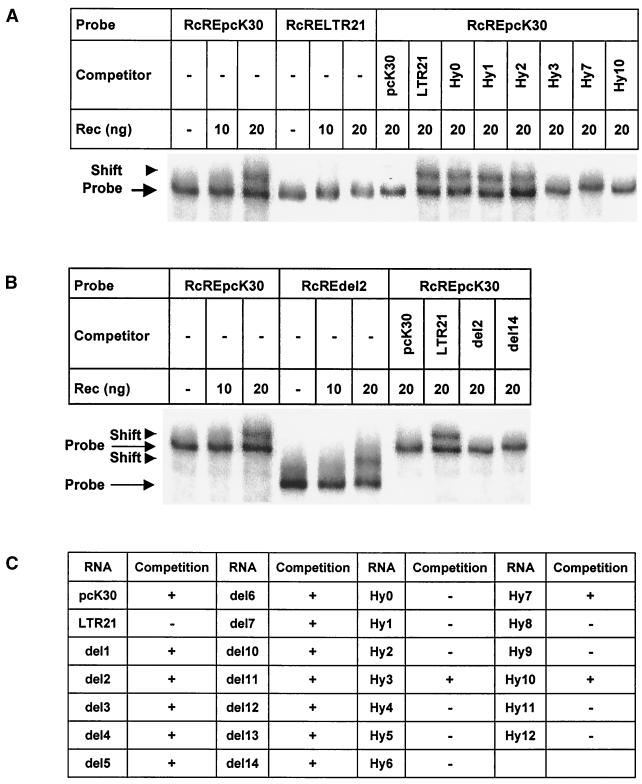
To confirm that the in vivo function of Rec was mediated by direct binding of Rec to the RcRE and to elucidate the sequence and/or structural requirements for this binding, we analyzed the ability of purified recombinant Rec protein to bind to the different RcRE RNAs transcribed in vitro in RNA gel shift assays (Fig. 6). Rec specifically bound to the biologically active pcK30 RcRE RNA, and no binding to biologically inactive RcRE LTR21 was observed (Fig. 6A). Binding of Rec to the HyRcRE sequences was measured as the ability of excess unlabeled HyRcRE RNA transcripts to compete for binding of Rec to a 32P-labeled pcK30 RcRE RNA probe. The results are summarized in Fig. 6A and C. Only the biologically active RcRE transcripts (HyRcRE3, HyRcRE7, and HyRcRE10 as well as pcK30) were able to compete for Rec binding to the labeled pcK30 RcRE RNA probe. These in vitro data confirmed the in vivo data obtained with the LTR and HyRcRE sequences and demonstrated that direct binding of Rec to these elements was a prerequisite for the biological activity measured, that is, for the RcRE-dependent nuclear export of HIV Gag reporter transcripts.
FIG. 6.
In vitro binding of recombinant purified Rec to RcRE sequences transcribed in vitro. RNA gel shift assays were performed as described in Materials and Methods. (A) Titration of Rec on biologically active pcK30 RcRE and on inactive RcRE LTR21 (six lanes at left). Competition for Rec binding to pcK30 RcRE with active (pcK30, Hy3, Hy7, and Hy10) and inactive (LTR21, Hy0, Hy1, and Hy2) RcREs (eight lanes at right). (B) Titration of Rec on active pcK30 RcRE and on inactive RcREdel2 (six lanes at left). Competition for Rec binding to pcK30 RcRE with active (pcK30 and del14) and inactive (LTR21 and del2) RcREs (four lanes at right). (C) Summary of the competition experiments.
When the ability of Rec to bind to the RcRE deletion mutants was analyzed using the same in vitro binding and competition assay (Fig. 6B and C), the binding of Rec to all deletion mutants was observed; this result indicated that the deletion of individual stem-loop structures did not abrogate Rec binding, although the deletion of stem-loop structures SL2, SL4, SL5, and SL6 significantly abrogated Rec function in vivo (Fig. 5A). The results showed that Rec could bind to more than one contact point within the RcRE and that no single sequence-specific binding site exists.
DISCUSSION
In recent years, it has become evident that many proteins are involved in RNA processing and export from the nucleus. Among the first factors identified in this scenario were the retroviral export adapters Rev and Rex, which mediate export of viral transcripts by binding to their respective responsive elements, RRE and RxRE, as multimers and by subsequent recruitment of cellular export receptor exportin 1 to the export complex (5, 20, 25, 29). Although the retroviral export adapters Rev and Rex bind to discrete RNA sequences, these sequences are located in stem-loop structures with internal bulges in the context of a larger RNA segment which is tightly folded into a very complex specific structure and which is essential for function (1, 2, 3, 9, 14). The human endogenous retrovirus HTDV/HERV-K encodes a functional equivalent to Rev and Rex. Its responsive element, RcRE, is located in the U3R region of 3′ LTRs (21). However, the sequence and structural requirements for Rec binding to the RcRE were not yet elucidated, although a recent publication, in which Rec binding to the RcRE was analyzed using deletion mutants, gave some insights (32). Yang et al. (32) applied a wide variety of different experimental test systems (in vitro binding and yeast two- and three-hybrid systems, two rather different tests for biological activity in mammalian cells); however, these systems sometimes produced contradicting results. We therefore mainly relied on testing for biological activity, that is, for export activity from the cell nucleus. We used a mammalian transient transfection assay (21) in which the export activity of the retroviral export adapters was measured with an RcRE-dependent HIV Gag reporter construct that did not contain splice sites (6, 13). The assay was controlled by monitoring transfection efficiencies and by confirming that the level of reporter protein measured reflected the level of reporter RNA exported from the nucleus. In previous publications, this test system and these controls were instrumental in defining the RcRE and in identifying relevant domains in the Rec protein (21, 22). In this publication, data obtained with these controls are mentioned but not shown.
Since more than 30 copies of HTDV/HERV-K elements are present in the human genome, we analyzed the ability of diverse RcRE sequences to support Rec-mediated RNA export. Not surprisingly, some RcRE sequences were inactive. HTDV/HERV-K arose from germ line infection of Old World monkey predecessors with an exogenous retrovirus 30 million years ago. Vertical transmission of retroviruses no longer depends on the presence of replication-competent genomes and, since retroviruses may have a pathogenic potential, selection against intact genomes could be expected (17, 19). Indeed, most human endogenous retrovirus sequences known so far are rather defective, and none of them is infectious. In contrast, it is quite surprising that HTDV/HERV-K elements have retained open reading frames for the expression of viral genes (19) and that this expression is regulated by a viral export adapter and a responsive element. The conservation of these regulatory sequences is rather fascinating, because their presence could influence the regulation of cellular genes. It will be interesting to identify the exact positions of active RcREs in the genome and neighboring genes.
Inactive and active RcRE sequences differed by five or six clusters of mutations (M0 to M5) (Fig. 1 and 2A). Rec activity was entirely restricted to its own responsive element, and Rec was unable to use the RRE, as already reported (21), or the RxRE (Fig. 2A). Rev and Rex, however, could use RcRE sequences to mediate HIV Gag RNA export, as reported before (21), although they were less effective than in cotransfections with their genuine responsive elements (Fig. 2B and C). Interestingly, the presence of mutational clusters M0 to M5 led to a reduction in Rex-mediated export activity similar to that observed with Rec (Fig. 1 and 2C), while Rev activity was undisturbed by the presence of M1 to M5 in RcRE LTR26 (Fig. 1 and 2B).
Hybrid analyses revealed that Rec function depended on the presence of a rather large sequence comprising C1 to C3 (Fig. 3A and B). Mutations in C1 or C3 led to biologically inactive RcRE sequences which were not able to bind Rec (Fig. 3B and Fig. 6A and C). The hybrid mutants also demonstrated that the presence of either cluster M0 or cluster M2 led to a slight reduction in Rec-mediated export activity and that clusters M4 and M5 had no effect.
Hybrid analyses suggested the existence of two Rev binding sites, as the introduction of either C0 or C3 fully restored Rev-mediated export activity in an inactive backbone. None of these sites showed sequence homology to the Rev binding site in the RRE. Overall, however, none of the mutations introduced into the active RNA backbone had a severe abrogating effect on Rev function.
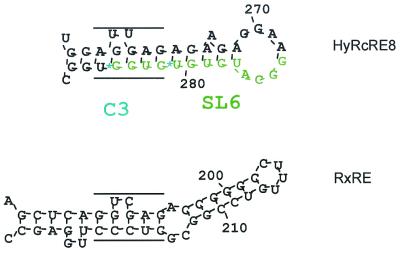
Interestingly, when C3 was present in the otherwise inactive RcRE LTR21 backbone (HyRcRE8) (Fig. 3A and D), Rex export function considerably exceeded that observed with the RxRE. With the RNA folding prediction algorithm, HyRcRE8 displayed a unique stem-loop which folded only in this hybrid and contained a sequence with 75% homology to the experimentally defined Rex binding site in the RxRE (Fig. 7). The observation that Rex was also active when C0, C1, and C2 were present in HyRcRE4 hinted at a second Rex interaction site. Again, similar to Rev function, Rex function was not completely abrogated by any of the hybrids. The observations that Rev and Rex probably had two independent binding sites and that the most efficient mutant probably created a new sequence-specific Rex binding site in an otherwise inactive structural context might indicate that only Rec was completely adapted to its genuine responsive element and that optimized structures could be generated for Rex and probably also for Rev. One could even imagine that the RxRE evolved from the ancient RcREs present in the human genome.
FIG. 7.
Predicted folding structures of single stem-loop formations in HyRcRE8 and RxRE. The structures were predicted with DNASIS, the RxRE secondary structure being identical to that described by Van Brussel et al. (30). The hypothetical Rex binding site in HyRcRE8 and the defined Rex binding site in the RxRE are indicated by the parallel lines.
Computational analyses predicted considerably divergent secondary structures for the active and inactive RcRE sequences as well as for the hybrid sequences (Table 1 and Fig. 4). The main differences affected an inner core segment comprising SL1 to SL7. Deletion of single stem-loop structures revealed that SL2, SL4, SL5, and SL6 were all important for in vivo Rec function (Fig. 5A), although RcRE deletion mutants missing only one of these four stem-loop structures still bound Rec in vitro (Fig. 6D). Several explanations can be given for this controversial result. Formation of a biologically active export complex may depend on interaction of Rec with more than a single contact point within a complex, folded RNA structure, and deletion of one contact point may not abrogate in vitro binding. Alternatively, more than one Rec molecule may be necessary to form an active export complex in vivo, and individual molecules of this oligomeric complex may bind to different but adjacent stem-loop structures. From in vivo competition experiments with Rec mutants, we could estimate that oligomeric complexes were formed containing either dimers or tetramers (22; unpublished data). In addition, cellular export factors may influence the formation, stability, affinity, and kinetics of active export complexes in vivo. Most likely the Rec-RcRE complexes formed in vitro did not completely resemble the functional export complexes formed in vivo.
Some of these results were also obtained in a recent study (32). The authors used an RNA folding algorithm that produced a slightly different active structure than the one presented here, a result which may also have been caused by the fact that the sequence depicted by Yang et al. (32) had eight nucleotide exchanges and one insertion compared to pcK30. Nevertheless, the stem-loop formations identified in their structures (SLIIa to SLIIf and SLIII) resembled SL1 to SL7. The main difference came from the fact that their stem-loop formations were arranged around a large central loop, whereas SL1 to SL7 formed a more complex core in which loops of SL3, SL4, SL5, and SL6 clustered in a center surrounded by SL1, SL2, and SL7. Yang et al. (32) identified two Rec binding sites with a test system in which a Tat-Rec fusion protein activated an HIV LTR that carried either SLIIb (equivalent to SL4) or SLIIf (equivalent to SL6) instead of the Tat-responsive element. SLIIb would be influenced by M1, and SLIIf might be influenced by M3. This information implies that if two Rec binding sites truly exist, either C1 or C3 alone should have rescued Rec activity, a result not obtained in our hybrid analysis. Unfortunately, the authors did not include SLIIc (equivalent to SL1), SLIId (almost identical to SL2), SLIIe (almost identical to SL3), SLIIa (equivalent to SL5), or SLIII (very similar to SL7) in their assay. In the study of Yang et al. (32), all seven stem-loop deletion mutants completely lacked biological activity, whereas our data showed that deletion of SL1, SL3, and SL7 reduced but did not completely abrogate Rec-mediated export activity (Fig. 5A). This major discrepancy may be explained either by differences in the extent of the sequence deleted or by different properties of the test systems used. In summary, the results reported by Yang et al. (32) do not contradict the results presented here.
The data presented in this publication, especially the hybrid analysis, revealed that the biological activity of Rec correlated with a large sequence in the RcRE containing clusters C1 and C3. Introduction of a single C cluster into an inactive backbone did not restore Rec activity. The biological activity of the hybrids also correlated with the in vitro RNA binding data. Rec in vivo function depended further on the presence of four putative stem-loop structures that were predicted by computational analysis only when the active sequence (C1 to C3) was present in the RcRE. A reasonable interpretation of the data is the hypothesis that Rec function required interaction with a complex, folded RNA structure within RcRE rather than binding to a discrete specific binding site.
ACKNOWLEDGMENTS
We especially thank V. Morozov for much helpful advice and J. Blümel, M. Chudy, M. Nübling, B. Schröder, H. Seitz, and H. Willkommen for intense discussions. We also thank J. Hauber (Erlangen, Germany) for generous donation of plasmids pDM128/CMV/RxRE and pcRex as well as B. Felber and G. Pavlakis (Frederick, Md.) for donation of plasmids pNLcgagA2 and pBsrev.
REFERENCES
- 1.Ahmed Y F, Hanly S M, Malim M H, Cullen B R, Greene W C. Structure-function analyses of the HTLV-I Rex and HIV-1 Rev RNA response elements: insights into the mechanism of Rex and Rev action. Genes Dev. 1990;4:1014–1022. doi: 10.1101/gad.4.6.1014. [DOI] [PubMed] [Google Scholar]
- 2.Bartel D P, Zapp M L, Green M R, Szostak J W. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell. 1991;67:529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- 3.Bogerd H P, Tiley L S, Cullen B R. Specific binding of the human T-cell leukemia virus type 1 Rex protein to a short RNA sequence located within the Rex-response element. J Virol. 1992;66:7572–7575. doi: 10.1128/jvi.66.12.7572-7575.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K-T, Rekosh D, Hammarskjöld M-L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cullen B R. HIV-1 auxillary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 6.D'Agostino D M, Felber B K, Harrison J E, Pavlakis G N. The Rev protein of human immunodeficiency virus type 1 promotes polysomal association and translation of gag/pol and vpu/env mRNAs. Mol Cell Biol. 1992;12:1375–1386. doi: 10.1128/mcb.12.3.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Agostino D M, Ciminale V, Pavlakis G N, Chieco-Bianchi L. Intracellular trafficking of the human immunodeficiency virus type 1 Rev protein: involvement of continued rRNA synthesis in nuclear retention. AIDS Res Hum Retrovir. 1995;11:1063–1071. doi: 10.1089/aid.1995.11.1063. [DOI] [PubMed] [Google Scholar]
- 8.Ernst R K, Bray M, Rekosch D, Hammarskjöld M-L. Secondary structure and mutational analysis of Mason-Pfizer monkey virus RNA constitutive transport element. RNA. 1997;3:210–222. [PMC free article] [PubMed] [Google Scholar]
- 9.Felber K B, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis G N. rev protein of human immunodeficiency virus type 1 affects the stability and transport of viral mRNA. Proc Natl Acad Sci USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM 1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 11.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 12.Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 13.Hadzopoulou-Cladaras M, Felber B K, Cladaras C, Athanassopoulos A, Tse A, Pavlakis G N. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J Virol. 1989;63:1265–1274. doi: 10.1128/jvi.63.3.1265-1274.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heaphy S, Dingwall C, Ernberg I, Gait M J, Green S M, Karn J, Lowe A D, Singh M, Skinner M A. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell. 1990;60:685–693. doi: 10.1016/0092-8674(90)90671-z. [DOI] [PubMed] [Google Scholar]
- 15.Heger P, Rosorius O, Koch C, Casari G, Grassmann R, Hauber J. Multimer formation is not essential for nuclear export of human T-cell leukemia virus type 1 Rex trans-activator protein. J Virol. 1998;72:8659–8668. doi: 10.1128/jvi.72.11.8659-8668.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X J, Hope T J, Bond B L, McDonald D, Grahl K, Parslow T G. Minimal Rev-response element for type 1 human immunodeficiency virus. J Virol. 1991;65:2131–2134. doi: 10.1128/jvi.65.4.2131-2134.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Löwer R. The pathogenic potential of endogenous retroviruses: facts and fantasies. Trends Microbiol. 1999;7:350–356. doi: 10.1016/s0966-842x(99)01565-6. [DOI] [PubMed] [Google Scholar]
- 18.Löwer R, Tönjes R R, Korbmacher C, Kurth R, Löwer J. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HDTV/HERV-K. J Virol. 1995;69:141–149. doi: 10.1128/jvi.69.1.141-149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Löwer R, Löwer J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madore S J, Tiley L S, Malim M H, Cullen B R. Sequence requirements for Rev multimerization in vivo. Virology. 1994;202:186–194. doi: 10.1006/viro.1994.1334. [DOI] [PubMed] [Google Scholar]
- 21.Magin C, Löwer R, Löwer J. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J Virol. 1999;73:9496–9507. doi: 10.1128/jvi.73.11.9496-9507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Magin C, Hesse J, Löwer J, Löwer R. Corf, the Rev/Rex homologue of HTDV/HERV-K, encodes an arginine-rich nuclear localization signal that exerts a trans-dominant phenotype when mutated. Virology. 2000;274:11–16. doi: 10.1006/viro.2000.0438. [DOI] [PubMed] [Google Scholar]
- 23.Mermer B, Felber B K, Campbell M, Pavlakis G N. Identification of trans-dominant HIV-1 rev protein mutants by direct transfer of bacterially produced proteins into human cells. Nucleic Acids Res. 1990;18:2037–2044. doi: 10.1093/nar/18.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neville M, Stutz F, Lee L, Davis I L, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 25.Nigg E A. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 26.Paca R E, Ogert R A, Hibbert C S, Izaurralde E, Beemon K L. Rous sarcoma virus DR posttranscriptional elements use a novel RNA export pathway. J Virol. 2000;74:9507–9514. doi: 10.1128/jvi.74.20.9507-9514.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rimsky L, Hauber J, Dukovich M, Malim M H, Langlois A, Cullen B R, Greene W C. Functional replacement of the HIV-1 rev protein by the HTLV-1 rex protein. Nature. 1988;335:738–740. doi: 10.1038/335738a0. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz S, Felber B K, Benko D M, Fenyö E-M, Pavlakis G N. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J Virol. 1990;64:2519–2529. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ullman K S, Powers M A, Forbes D J. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 30.Van Brussel M, Goubau P, Rousseau R, Desmyter J, Vandamme A-M. Complete nucleotide sequence of the new simian T-lymphotrophic virus, STLV-PH969 from a Hamadryas baboon, and unusual features of its long terminal repeat. J Virol. 1997;71:5464–5472. doi: 10.1128/jvi.71.7.5464-5472.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang J, Bogerd H, Peng S, Wiegand H, Truant R, Cullen B R. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc Natl Acad Sci USA. 1999;96:13404–13408. doi: 10.1073/pnas.96.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Bogerd H, Le S-Y, Cullen B R. The human endogenous retrovirus K Rev response element coincides with a predicted RNA folding region. RNA. 2000;6:1–14. doi: 10.1017/s135583820000100x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]