Abstract
Malate dehydrogenase (MDH) is a ubiquitous and central enzyme in cellular metabolism, found in all kingdoms of life, where it plays vital roles in the cytoplasm and various organelles. It catalyzes the reversible NAD+-dependent reduction of L-malate to oxaloacetate. This review describes the reaction mechanism for MDH and the effects of mutations in and around the active site on catalytic activity and substrate specificity, with a particular focus on the loop that encloses the active site after the substrates have bound. While MDH exhibits selectivity for its preferred substrates, mutations can alter the specificity of MDH for each cosubstrate. The kinetic characteristics and similarities of a variety of MDH isozymes are summarized, and they illustrate that the KM values are consistent with the relative concentrations of the substrates in cells. As a result of its existence in different cellular environments, MDH properties vary, making it an attractive model enzyme for studying enzyme activity and structure under different conditions.
Keywords: enzyme kinetics, enzyme specificity, malate dehydrogenase, reaction mechanism
Introduction
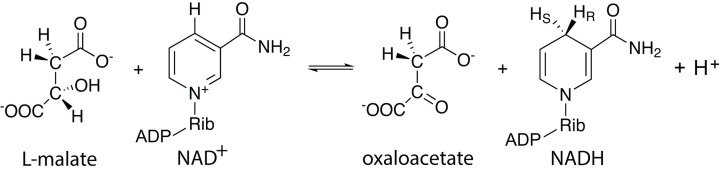
Malate dehydrogenase (MDH, EC 1.1.1.37) catalyzes the reversible conversion of L-malate ((S)-malate) to oxaloacetate, accompanied by the conversion of NAD+ to NADH + H+, with the addition of the pro-R hydrogen to NAD+ (Figure 1) [1,2]. This enzyme is highly conserved across all kingdoms of life. Eukaryotes contain cytosolic MDH as well as isozymes that are specifically transported to organelles, including mitochondria, chloroplasts, peroxisomes, and glyoxysomes. Although the size of MDH varies among organisms and organelles, MDH is approximately 36 kDa and most commonly functions as a dimer. For NADH produced by cytosolic MDH, reducing equivalents in the form of L-malate allows for the net import of NADH into the mitochondria via the L-malate-aspartate shuttle to drive the production of ATP. Mitochondrial MDH catalyzes an essential step in producing oxaloacetate in the citric acid cycle, in addition to NADH for making ATP.
Figure 1. Overall reaction catalyzed by malate dehydrogenase.
The illustrated reaction involves the conversion of L-malate to oxaloacetate, accompanied by the reduction of NAD+ to NADH + H+. Rib: Ribose, HS: pro-S hydrogen, HR: pro-R hydrogen.
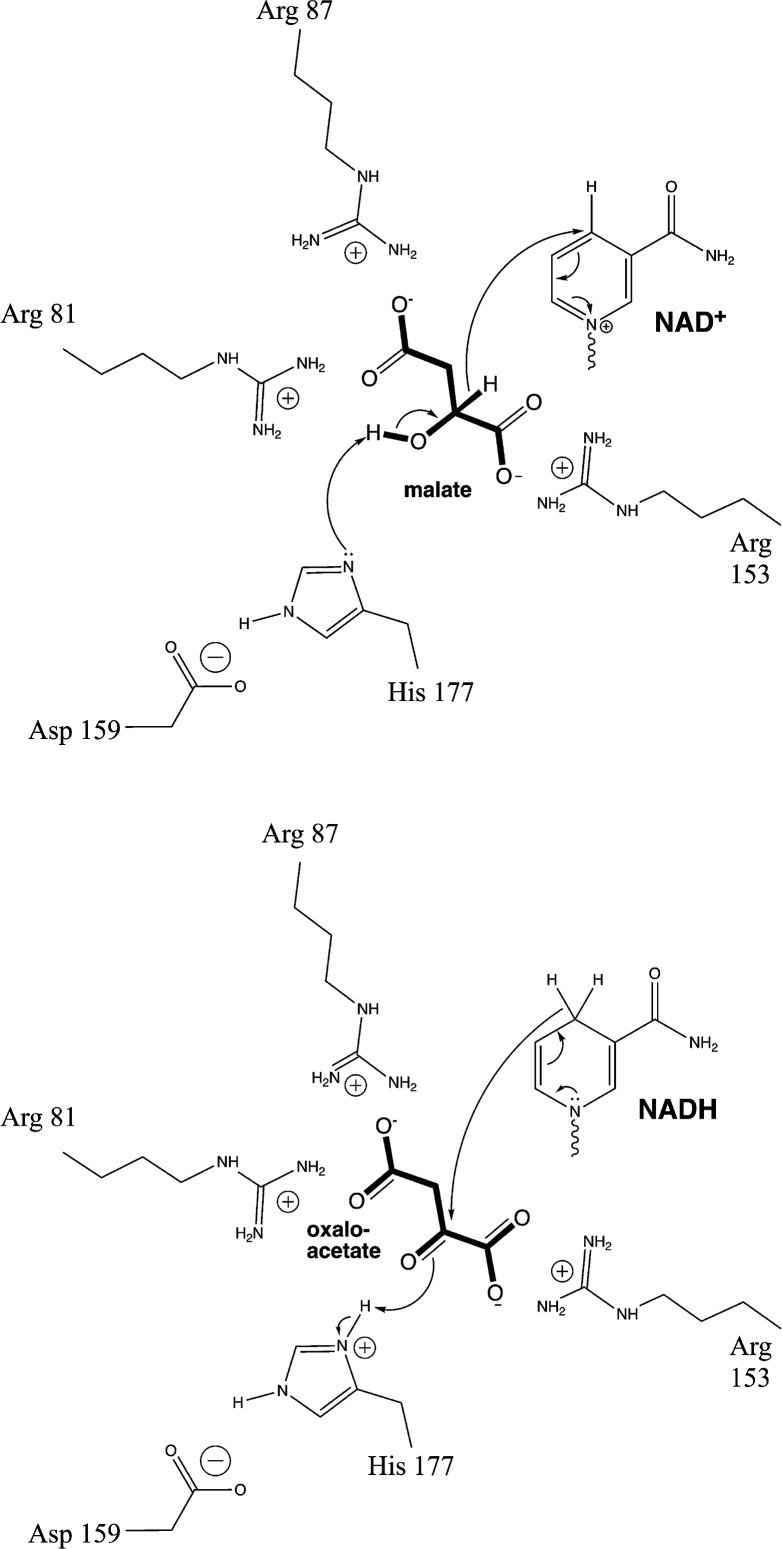
The active site of MDH features a crucial His 177-Asp 150 pair [3] (this review uses the amino acid numbering for Escherichia coli MDH, Uniprot P61889). His 177 forms a hydrogen bond to the hydroxyl group of L-malate, facilitating its conversion to oxaloacetate by removing the hydroxyl proton (Figure 2). L-Malate is oriented in the active site by forming salt bridges between its C1 and C4 carboxylate and Arg 153 and Arg 81, respectively. As described below in more detail, the two substrates bind, and then a flexible loop bearing Arg 81 and a third essential Arg residue, Arg 87, encloses the active site before the reaction proceeds.
Figure 2. Reaction mechanism for malate dehydrogenase.
The diagram on the top illustrates the mechanism of oxidation of L-malate, while the diagram on the bottom is the reverse, the reduction of oxaloacetate.
Reaction thermodynamics
The equation below represents the reaction catalyzed by MDH, along with the corresponding definition for :
The observed equilibrium constant for MDH greatly favors the formation of L-malate and NAD+. Under near-physiological conditions (38°C, pH 7.0, ionic strength I = 0.25, 1 mM Mg2+), , corresponding to a ΔG = +27 kJ/mol [4]. The endergonic reaction catalyzed by MDH is facilitated within the mitochondrial cellular environment by coupling with downstream metabolic pathways. NADH is utilized in the electron transport chain to produce ATP, and oxaloacetate is a critical intermediate in the citric acid cycle. The enzyme citrate synthase is energetically favored and consumes the produced oxaloacetate, driving the MDH-catalyzed reaction forward. In laboratory settings, the exergonic reverse reaction (conversion of oxaloacetate to L-malate) is often studied.
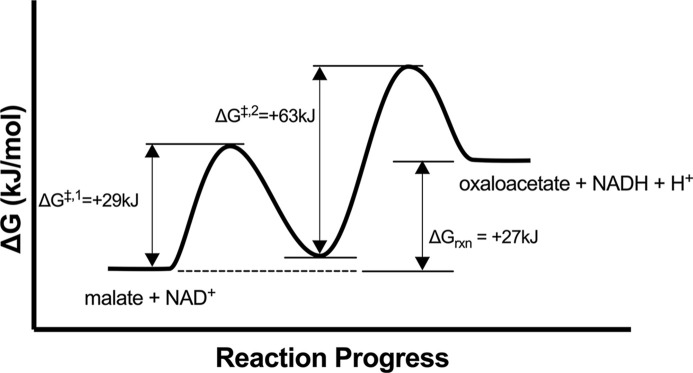
As described below, the L-malate oxidation reaction is more favorable at high pH than the oxaloacetate reduction reaction. Keq increases with rising pH levels, reaching at pH 9 [5]. Despite this increase, the equilibrium still greatly favors oxaloacetate reduction, with the equilibrium percentage of oxaloacetate decreasing slightly from 99.8% at pH 7 to 97.1% at pH 9. Computational modeling of the MDH-catalyzed oxidation of L-malate suggests two distinct steps, each characterized by its own transition states. Initially, His 177 of the E. coli MDH deprotonates the hydroxyl group of L-malate with a calculated activation energy ΔG‡ of +29 kJ/mol (Figure 3). Subsequently, the hydride anion from the C2 of L-malate can transfer to NAD+, representing the rate-limiting catalytic step with a calculated ΔG‡ of +63 kJ/mol [6].
Figure 3. Free energy diagram for MDH.
The reaction profile for conversion of malate to oxaloacetate is shown using thermodynamic data from [4,6].
Exploring the role of the substrate binding loop in enzyme catalysis
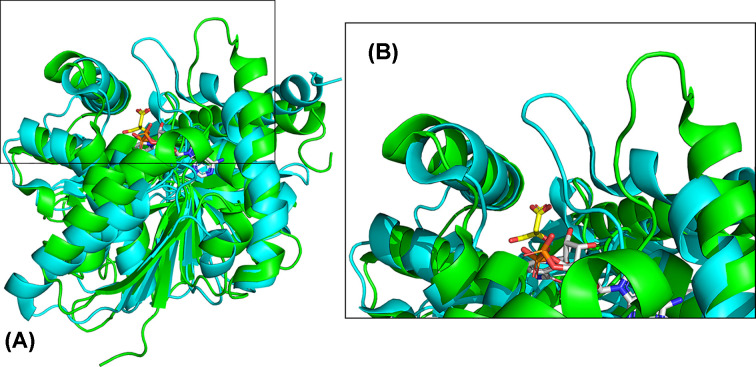
The catalytic dyad and one substrate binding residue, Arg 153, provide initial contacts with L-malate/oxaloacetate, followed by further interactions with a mobile loop between β4 and α4 (residues 76-88), located near the L-malate/oxaloacetate binding site. This loop includes Arg 81 and Arg 87, which interact with the C4 carboxylate and stabilize the anionic transition state, respectively [7]. The structure of MDH from the archaeon Metallosphaera sedula was determined in the presence and absence of NAD+/L-malate [8]. The active site loop is disordered with high crystallographic temperature factors when no substrate is present and becomes more ordered when the substrate binds (Figure 4). In the structure of the enzyme from M. sedula with and without NAD+, the loop is most open in the apoenzyme. When NAD+ binds, the loop partially closes, and then completely closes when L-malate binds. On average, the loop moves 6.8 Å between the most open and most closed forms. Molecular dynamics simulations of Thermus thermophilus MDH revealed increased mobility of the active site loop at higher simulated temperatures, while the mobility of other loops in the enzyme did not change with temperature [9].
Figure 4. Movement of active site loop upon NAD+ and L-malate binding to MDH.
(A) Overall loop movement with open in green (4kde) and closed in cyan (6ihe). (B) Close-up of loop movement. L-Malate is shown in sticks with yellow carbons, and NAD+ is shown in sticks with gray carbons.
The loop conformations of the MDH enzymes from Chlorobium vibrioforme and Chloroflexus aurantiacus, adapted to mesophilic and thermophilic environments, respectively, were studied by molecular dynamics simulations [10]. The MDH from the thermophile exhibited more salt bridges, leading to the stabilization of the active site loop by residues even in the absence of substrate. This suggests that the loop is less mobile in the thermophile.
Effect of loop mutations on MDH activity
MDH displays a high level of substrate discrimination, showing a six order of magnitude difference in the reduction rate of oxaloacetate compared to pyruvate. This specificity arises from charge balancing between substrates and the active site of MDH [11]. The activated complex formed upon oxaloacetate and NADH binding includes a positively charged imidazolium ion acting as a general acid, along with two negative charges contributed by each carboxylate of oxaloacetate, resulting in a net negative charge of -1. When oxaloacetate binds, the two loop Arg residues (81 and 87) are driven into close proximity to each other and the C4 carboxylate of oxaloacetate. As pyruvate lacks this complementary negative charge of oxaloacetate, charge repulsion prevents a catalytically productive conformation, leading to the 106-fold discrimination [11].
Given the critical role of the active site loop in substrate binding, it is noteworthy that Arg 81 emerges as a key determinant for substrate specificity. In contrast, the related lactate dehydrogenase (LDH) has a Gln residue at the corresponding position. LDH has detectable activity with oxaloacetate, while MDH has comparatively little detectable activity with pyruvate. Mutating the Gln of LDH to an Arg at the corresponding position is critical in creating an LDH that is more specific for oxaloacetate than pyruvate [12]. Conversely, mutating this arginine to glutamine (R81Q) in MDH results in higher specificity for pyruvate, but significantly lower activity than wild-type MDH with oxaloacetate. However, this activity can be improved with other loop mutations; notably, the mutation of the nearby Ala 80 to proline results in a four-fold increase in kcat/KM [13]. Due to the critical role of loop residues in conferring specificity and the tendency for enzymes with loop mutations to retain some activity, the loop has become a subject of study in Course-based Undergraduate Research Experiences [14].
Furthermore, other mutations in MDH have been found to impact its activity. For instance, the R102Q mutation induces loop closure in E. coli MDH, enhancing thermal stability [15]. Mutation of the Arg 153 residue that binds to the C1 carboxylate of the substrate to Cys (R153C) results in a mutant with diminished catalytic activity, although KM values for L-malate and oxaloacetate remain unaffected [16]. Some mutations, such as Q11G and N119G in Plasmodium falciparum MDH, lead to the loss of critical hydrogen bonding interactions with NADH and an increased KM for oxaloacetate compared with the wild-type [17].
Human MDH2 shows at least three orders of magnitude greater activity with oxaloacetate compared with the longer diacid substrate, α-ketoglutarate. Despite this reduced activity, MDH2 may contribute to the production of 2-hydroxyglutarate in cancer cells. Notably, as the pH is lowered from 7.4 to 6.8, commonly associated with the hypoxic conditions of cells, the generation of 2-hydroxyglutarate by MDH increases while oxaloacetate reduction decreases over the same pH change [18].
Cosubstrate specificity in MDH
MDH generally prefers NAD+ over NADP+, although NADP+-dependent MDH enzymes do exist in chloroplasts [19]. Cosubstrate specificity is determined by interactions in the β2-loop-α3 region (residues 34-38 in E. coli MDH). The cytoplasmic Thermus thermophilus (UniProt ID P10584, formerly Thermus flavus) MDH efficiently reduces oxaloacetate using both NADH and NADPH, with a 22-fold higher value of kcat/KM with NADH (8.5 × 107 M−1 s−1 and 3.9 × 106 M−1 s−1, respectively), along with a 14-fold lower KM value for NADH [20].
Mutation of the β2-loop-α3 region in T. thermophilus (amino acids 42-48) to the corresponding residues in chloroplastic Zea mays MDH (UniProt ID P15719, amino acids 127-133) leads to a 7-fold increase in efficiency using NADPH as a cosubstrate and a 71-fold decrease when using NADH. Part of the decrease in the KM for NADPH utilization may be due to the mutation of a Gln to Arg causing ionic attraction with the phosphate of NADPH. However, mutation of other residues in the loop improves the relative efficiency of NADPH-dependent reduction of oxaloacetate [20]. In Bacillus subtilis, substituting these same residues converts the wild-type MDH, with a 200-fold preference for NADH, into a mutant enzyme with a 9-fold preference for NADPH. Additionally, this mutant displayed a lower KM value, higher kcat, and higher kcat/KM values for NADPH than wild-type NADPH-dependent MDHs from T. flavus and Corynebacterium glutamicum [21]. The ability to alter the coenzyme specificity of B. subtilis MDH from NADH to NADPH opens up new possibilities for utilizing this enzyme in industrial processes that require NADPH-dependent reactions. Understanding the impact of mutations on protein-ligand binding affinity and kinetics is crucial, and computational methods offer valuable insights into MDH mutational dynamics to predict these effects [22].
The Rossman fold found in MDH is common to many nucleotide-binding enzymes, including S-adenosyl methionine (SAM)-dependent enzymes. Although E. coli MDH has no detectable affinity for SAM, removing three amino acids (ΔAla9, ΔGly10, ΔGln14) from the V/IxGxxGxxG motif found in the β1-loop-α1 region of E. coli MDH to create instead a V/IGxGxG motif, typically found in SAM binding Rossman folds, leads to an MDH that binds SAM (Kd = 13 µM) and has no observable binding to NAD+ [23]. These deletions shorten the α1 helix and flatten the loop, expanding the active site to bind SAM.
MDH kinetic parameters
Steady-state enzyme kinetics data are available for both the forward and reverse reactions of many MDH isozymes from various organisms (Table 1). Older experiments often used racemic mixtures of L- and D-malate, and the table indicates when the L enantiomer was utilized. The typical reaction conditions for assays with oxaloacetate included 30–100 mM Tris or phosphate buffer, pH 7–8, with 0–100 mM KCl or NaCl at 30–37°C. As described in this review, enzyme assays using L-malate as a substrate were frequently conducted at a higher pH. For most enzymes, the KM for oxaloacetate is lower than for L-malate, and the KM for NADH is lower than for NAD+, which is reasonable given the lower cellular concentrations of oxaloacetate and NADH compared to L-malate and NAD+, respectively [24]. The kcat values for L-malate oxidation were much smaller than for oxaloacetate reduction, with MDH from P. falciparum exhibiting the highest kcat for L-malate oxidation [25].
Table 1. Kinetic constants for a variety of purified MDH isozymes.
| Organism | UniProtKB unique entry | Substrate | KM (μM) | kcat (s−1) | kcat/KM (M−1s−1) | Citation |
|---|---|---|---|---|---|---|
| H. sapiens (cyto*) | P40925-1 | OAA | 33.4 ± 0.2 | 200 ± 20 | 6 × 106 | [26] |
| OAA | 58 ± 12 | [27] | ||||
| NADH | 14.3 ± 1.5 | N.D. | ||||
| L-Malate | 660 ± 230 | |||||
| NAD+ | 39 ± 14 | |||||
| S. scrofa (cyto) | P11708 | OAA | 30 | 475 | 1.36 × 107 | [28–30] |
| OAA | 35 ± 2 | |||||
| OAA | 8.3 ± 0.6 | |||||
| NADH | 21 ± 1 | |||||
| S. scrofa (mito) | P00346 | OAA | 18, 35 | N.D. | N.D. | [30–32] |
| NADH | 38, 15 | |||||
| L-Malate | 370, 1100 | |||||
| NAD+ | 230, 480 | |||||
| C. vulgaris (mito) | P17783 | OAA | 150 | N.D. | N.D. | [33] |
| NADH | 110 | |||||
| L-Malate | 3690 | |||||
| NAD+ | 460 | |||||
| C. vulgaris (glyox) | P19446 | OAA | 180 | N.D. | N.D. | [33] |
| NADH | 130 | |||||
| L-Malate | 7180 | |||||
| NAD+ | 460 | |||||
| A. thaliana (cyto) | P93819 | OAA | 238 ± 21 | 608 ± 22 | 2.6 × 106 | [34] |
| NADH | 72 ± 7 | 677 ± 24 | 9.0 × 106 | |||
| A. thaliana (chloro) | Q9SN86 | OAA | 324 ± 109 | 9.9 ± 1.4 | 3.1 × 104 | [35] |
| NADH | 431 ± 28 | 13 ± 9 | 3.2 × 104 | |||
| C. elegans (cyto) | Q9UAV5 | OAA | 54 ± 4 | [36] | ||
| NADH | 61 ± 6 | 350 ± 21 | 5.7 × 106 | |||
| C. elegans (mito) | O02640 | OAA | 52 ± 4 | [36] | ||
| NADH | 107 ± 5 | 460 ± 23 | 4.3 × 106 | |||
| P. falciparum | Q6VVP7 | OAA | 30 ± 1 | 960 ± 70 | 3.2 × 107 | [25] |
| NADH | 36 ± 2 | 950 ± 80 | 2.6 × 107 | |||
| L-Malate | 1350 ± 24 | 250 ± 19 | 1.8 × 105 | |||
| NAD+ | 152 ± 13 | 150 ± 20 | 9.8 × 105 | |||
| E. coli | P61889 | OAA | 40-50 | 900-930 | 2.3 × 107 | [37,38] |
| NADH | 61 ± 2 | |||||
| L-Malate | 2600 ± 200 | 21 | 8.1 × 103 | |||
| NAD+ | 260 ± 30 | |||||
| T. thermophilus | P10584 | OAA | 9.8 | N.D. | N.D. | [39] |
| NADH | 6 | |||||
| L-Malate | 26 | |||||
| NAD+ | 19 | |||||
| S. coelicolor | Q9K3J3 | OAA | 189 | 1870 | 1 × 104 | [40] |
| NADH | 83 | 542 | 6.56 × 103 | |||
| L-Malate | 494 | 4.71 | 9.53 | |||
| NAD+ | 150 | 3.66 | 24.4 | |||
| H. marismortui | Q07841 | OAA | 130 | N.D. | N.D. | [41] |
| NADH | 12 |
chloro, chloroplastic; cyto, cytosolic; glyox, glyoxysomal; mito, mitochondrial; N.D., not done in the published article; OAA, oxaloacetate.
Kinetic diversity across MDH isoforms in cellular organelles
When kinetic constants were available for both the cytoplasmic and mitochondrial isozymes in the same organism, the KM values for oxaloacetate were often similar. However, the KM for NADH varied, with higher values observed for the mitochondrial isozyme in most cases, as seen in Caenorhabditis elegans [36]. This observation is consistent with the higher NADH concentration found in the mitochondria (reviewed in [42]). The chloroplastic MDH from Arabidopsis thaliana also had a higher KMfor NADH compared to the cytosolic enzyme [34,35]. The KM values for NADH from different steady-state kinetics experiments with Sus scrofa isoforms were more varied [30,32]. Detailed time-course measurements and kinetic models for both cytosolic and mitochondrial S. scrofa MDH isoforms predicted higher activity for cytosolic MDH at lower NADH concentrations [43], suggesting that the cytosolic MDH also has a lower KM for NADH. This comprehensive understanding of MDH kinetics across diverse cellular contexts underscores the adaptability and crucial role of MDH in cellular metabolism.
MDH kinetics and environmental factors
MDH follows an ordered Bi-Bi kinetic mechanism characterized by the sequential binding of NAD+ and then L-malate, followed by the release of oxaloacetate and then NADH [5,43–46]. The kinetic evidence supports a compulsory order reaction mechanism, and some studies suggest that an enzyme isomerization is involved [47–49]. Lodola et al. found that dimeric MDH has two independent active sites that can function simultaneously [50].
Generally, MDH activity has an optimal temperature range between 30 and 40°C. Deviations from this range are somewhat related to the organism’s physiological temperature range. Thermostability also varies among MDH isoforms, with cytosolic isoforms being more stable than mitochondrial isoforms, such as in C. elegans [36], S. scrofa [28,51,52], and Sphyraena idiastes [30]. However, these enzymes from eukaryotic species are notably less stable than MDH from the mesophile Streptomyces coelicolor and the thermophilic bacterium T. thermophilus. S. coelicolor MDH remains active at 50°C but rapidly inactivates above this temperature, while T. thermophilus MDH retains activity at 90°C for 60 min [20,39,40,51]. A larger number of salt bridges between subunits contributes to thermal stability [28,53]. However, thermal stability is a complex trait influenced by multiple factors, including electrostatic interactions, hydrophobic interactions, hydrogen bonding, and overall protein folding [28,53–55].
Optimal enzyme activity is achieved when MDH reduces oxaloacetate near pH 8, and activity is generally high from pH ∼7–8.5 [5,21,36,56]. Interestingly, the optimal pH for some thermostable MDHs varies with temperature [39,40]. The L-malate oxidation rate is higher at pH > 8 [25,39,43,56]. The pH-dependency of MDH kinetic behavior involves additional ionic states and still follows the ordered Bi-Bi mechanism but is modified to account for enzyme protonation states [5,43,57]. The mitochondrial porcine MDH binds a proton on a His residue upon binding to NADH [58], and a His is deprotonated before L-malate binds [1]. This is consistent with the active site base, His 177, having the appropriate protonation state in each catalytic direction. Also, the interaction of NAD+ with MDH was dependent on the protonation of two unidentified residues, with pKa values of ∼6 and 7–8 [5,43].
MDH regulation
Excessive oxaloacetate inhibits cytoplasmic and mitochondrial MDH, but at concentrations too high to influence enzyme activity in the cell [29,59]. Interestingly, oxaloacetate may also inhibit NAD+ binding, potentially with greater potency than its impact on NADH [47]. L-malate activates MDH at a site that is different from the active site [31,46]. Additionally, citrate is an allosteric regulator of MDH, and the influence of citrate on MDH activity is intricate and multifaceted. Citrate-activated mitochondrial MDH (mMDH) promotes the oxidation of L-malate [48], and this may be another mechanism to promote this energetically unfavorable reaction. However, citrate inhibited oxaloacetate reduction by mMDH and in both directions of the reaction by cytoplasmic MDH (cMDH) [48]. Interestingly, it was later found that citrate only activated mMDH when L-malate and NAD+ concentrations were elevated (2.5–10 mM and 1–5 mM, respectively), while citrate inhibited L-malate oxidation at low concentrations of L-malate or NAD+ [49]. All three effectors (oxaloacetate, L-malate, and citrate) bind to the same putative allosteric site [48].
Human mMDH is an allosteric enzyme that interacts cooperatively with L-malate, with tetramers showing higher activity than dimers [60]. When using NADH as a cofactor, human mMDH is activated by fumarate that binds at the dimer interface at a site ∼30 Å from the active site [60]. The enzyme is also inhibited by ATP, which binds to a site at the tetramer interface (also called the exosite) where ATP/ADP and NAD+/NADH can bind. The binding of ATP leads to the dimer being more stable than the tetramer, causing inhibition [61]. ADP has a similar but much weaker effect. The binding of NAD+ to the tetramer interface leads to the tetramer being more stable and increased activity. Other mammalian MDH enzymes (cMDH and mMDH-NADP+) have not been shown to be similarly regulated.
Summary
In vivo, the reaction catalyzed by malate dehydrogenase mainly proceeds in the direction of L-malate oxidation, but in the lab, it is easier to study the reaction in the spontaneous reverse direction.
MDH is present in all known organisms, and there are different isozymes in organelles.
Multiple investigations of the flexible active site loop and other residues have shown that MDH is a good model system for investigating the basis of substrate specificity.
Regulation of MDH is complex, and the enzyme is affected allosterically by several molecules.
There is little published work on human MDH, which may be a fruitful area of future study.
Abbreviations
- ΔG‡
activation energy
- A. thaliana
Arabidopsis thaliana
- ATP
adenosine triphosphate
- C. elegans
Caenorhabditis elegans
- C. vulgaris
Citrullus vulgaris
- chloro
chloroplastic
- cMDH
cytoplasmic malate dehydrogenase
- cyto
cytoplasmic
- E. coli
Escherichia coli
- glyox
glyoxosomal
- H. marismortui
Haloarcula marismortui
- H. sapiens
Homo sapiens
- kDa
kilodalton
- M. sedula
Metallosphaera sedula
- MDH
malate dehydrogenase
- mito
mitochondrial
- mMDH
mitochondrial malate dehydrogenase
- NAD+
nicotinamide adenine dinucleotide
- NADH
dihydronicotinamide adenine dinucleotide
- NADP+
nicotinamide adenine dinucleotide phosphate
- NADPH
dihydronicotinamide adenine dinucleotide phosphate
- OAA
oxaloacetate
- P. falciparum
Plasmodium falciparum
- S. coelicolor
Streptomyces coelicolor
- S. scrofa
Sus scrofa
- SAM
S-adenosyl methionine
- T. thermophilus
Thermus thermophilus (formerly Thermus flavus)
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Bernstein L.H. and Everse J. (1978) Studies on the mechanism of the malate dehydrogenase reaction. J. Biol. Chem. 253, 8702–8707 10.1016/S0021-9258(17)34234-5 [DOI] [PubMed] [Google Scholar]
- 2.Oppenheimer N.J. (1986) The stereospecificity of oxidation of alpha-[4R-2H]NADH by dehydrogenases. J. Biol. Chem. 261, 12209–12212 10.1016/S0021-9258(18)67225-4 [DOI] [PubMed] [Google Scholar]
- 3.Birktoft J.J. and Banaszak L.J. (1983) The presence of a histidine-aspartic acid pair in the active site of 2-hydroxyacid dehydrogenases. X-ray refinement of cytoplasmic malate dehydrogenase. J. Biol. Chem. 258, 472–482 10.1016/S0021-9258(18)33280-0 [DOI] [PubMed] [Google Scholar]
- 4.Guynn R.W., Gelberg H.J. and Veech R.L. (1973) Equilibrium constants of the malate dehydrogenase, citrate synthase, citrate lyase, and acetyl coenzyme A hydrolysis reactions under physiological conditions. J. Biol. Chem., Elsevier 248, 6957–6965 10.1016/S0021-9258(19)43346-2 [DOI] [PubMed] [Google Scholar]
- 5.Dasika S.K., Vinnakota K.C. and Beard D.A. (2015) Determination of the catalytic mechanism for mitochondrial malate dehydrogenase. Biophys. J., Elsevier 108, 408–419 10.1016/j.bpj.2014.11.3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunningham M.A., Ho L.L., Nguyen D.T., Gillilan R.E. and Bash P.A. (1997) Simulation of the enzyme reaction mechanism of malate dehydrogenase. Biochemistry 36, 4800–4816 10.1021/bi962734n [DOI] [PubMed] [Google Scholar]
- 7.Hall M.D. and Banaszak L.J. (1993) Crystal structure of a ternary complex of Escherichia coli malate dehydrogenase citrate and NAD at 1.9 Å resolution. J. Mol. Biol. 232, 213–222 10.1006/jmbi.1993.1377 [DOI] [PubMed] [Google Scholar]
- 8.Lee D., Hong J. and Kim K.-J. (2019) Crystal structure and biochemical characterization of malate dehydrogenase from Metallosphaera sedula. Biochem. Biophys. Res. Commun. 509, 833–838 10.1016/j.bbrc.2019.01.018 [DOI] [PubMed] [Google Scholar]
- 9.Hung C.-H., Hwang T.-S., Chang Y.-Y., Luo H.-R., Wu S.-P. and Hsu C.-H. (2013) Crystal structures and molecular dynamics simulations of thermophilic malate dehydrogenase reveal critical loop motion for co-substrate binding. PLoS ONE 8, e83091 10.1371/journal.pone.0083091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalimeri M., Girard E., Madern D. and Sterpone F. (2014) Interface matters: the stiffness route to stability of a thermophilic tetrameric malate dehydrogenase. PLoS ONE 9, e113895, Public Library of Science 10.1371/journal.pone.0113895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman A.D., Cortés A., Dafforn T.R., Clarke A.R. and Brady R.L. (1999) Structural basis of substrate specificity in malate dehydrogenases: crystal structure of a ternary complex of porcine cytoplasmic malate dehydrogenase, alpha-ketomalonate and tetrahydoNAD. J. Mol. Biol. 285, 703–712 10.1006/jmbi.1998.2357 [DOI] [PubMed] [Google Scholar]
- 12.Wilks H.M., Hart K.W., Feeney R., Dunn C.R., Muirhead H., Chia W.N.et al. (1988) A specific, highly active malate dehydrogenase by redesign of a lactate dehydrogenase framework. Science 242, 1541–1544 10.1126/science.3201242 [DOI] [PubMed] [Google Scholar]
- 13.Boernke W.E., Millard C.S., Stevens P.W., Kakar S.N., Stevens F.J. and Donnelly M.I. (1995) Stringency of substrate-specificity of Escherichia coli malate-dehydrogenase. Arch. Bioch. Biophys. 322, 43–52 10.1006/abbi.1995.1434 [DOI] [PubMed] [Google Scholar]
- 14.Provost J.J. (2022) Increasing access for biochemistry research in undergraduate education: The malate dehydrogenase CURE community. J. Biol. Chem. 298, 102298 10.1016/j.jbc.2022.102298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goward C.R., Miller J., Nicholls D.J., Irons L.I., Scawen M.D., O'brien R.et al. (1994) A single amino acid mutation enhances the thermal stability of Escherichia coli malate dehydrogenase. Eur. J. Biochem. 224, 249–255 10.1111/j.1432-1033.1994.tb20018.x [DOI] [PubMed] [Google Scholar]
- 16.Wright S.K. and Viola R.E. (2001) Alteration of the specificity of malate dehydrogenase by chemical modulation of an active site arginine. J. Biol. Chem. 276, 31151–31155 10.1074/jbc.M100892200 [DOI] [PubMed] [Google Scholar]
- 17.Pradhan A., Tripathi A.K., Desai P.V., Mukherjee P.K., Avery M.A., Walker L.A.et al. (2009) Structure and function of Plasmodium falciparum malate dehydrogenase: Role of critical amino acids in co-substrate binding pocket. Biochimie 91, 1509–1517 10.1016/j.biochi.2009.09.005 [DOI] [PubMed] [Google Scholar]
- 18.Nadtochiy S.M., Schafer X., Fu D., Nehrke K., Munger J. and Brookes P.S. (2016) Acidic pH is a metabolic switch for 2-hydroxyglutarate generation and signaling. J. Biol. Chem. 291, 20188–20197 10.1074/jbc.M116.738799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferte N., Jacquot J.-P. and Meunier J.-C. (1986) Structural, immunological and kinetic comparisons of NADP-dependent malate dehydrogenases from spinach (C3) and corn (C4) chloroplasts. Eur. J. Biochem. 154, 587–595 10.1111/j.1432-1033.1986.tb09439.x [DOI] [PubMed] [Google Scholar]
- 20.Nishiyama M., Birktoft J.J. and Beppu T. (1993) Alteration of coenzyme specificity of malate dehydrogenase from Thermus flavus by site-directed mutagenesis. J. Biol. Chem. 268, 4656–4660 10.1016/S0021-9258(18)53446-3 [DOI] [PubMed] [Google Scholar]
- 21.Ge Y.-D., Guo Y.-T., Jiang L.-L., Wang H.-H., Hou S.-L. and Su F.-Z. (2023) Enzymatic characterization and coenzyme specificity conversion of a novel dimeric malate dehydrogenase from Bacillus subtilis. Protein J. 42, 14–23 10.1007/s10930-022-10087-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D.D., Ou-Yang L., Xie H., Zhu M. and Yan H. (2020) Predicting the impacts of mutations on protein-ligand binding affinity based on molecular dynamics simulations and machine learning methods. Comput. Struct. Biotech. J. 18, 439–454 10.1016/j.csbj.2020.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toledo-Patiño S., Pascarelli S., Uechi G. and Laurino P. (2022) Insertions and deletions mediated functional divergence of Rossmann fold enzymes. Proc. Natl. Acad. Sci. U.S.A. 119, e2207965119 10.1073/pnas.2207965119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Opie L.H. and Owen P. (1975) Effects of increased mechanical work by isolated perfused rat heart during production or uptake of ketone bodies. Assessment of mitochondrial oxidized to reduced free nicotinamide-adenine dinucleotide ratios and oxaloacetate concentrations. Biochem. J. 148, 403–415 10.1042/bj1480403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tripathi A.K., Desai P.V., Pradhan A., Khan S.I., Avery M.A., Walker L.A.et al. (2004) An alpha-proteobacterial type malate dehydrogenase may complement LDH function in Plasmodium falciparum. Cloning and biochemical characterization of the enzyme. Eur. J. Biochem. 271, 3488–3502 10.1111/j.1432-1033.2004.04281.x [DOI] [PubMed] [Google Scholar]
- 26.Cornely K. and Stack T. (2024) hMDH1 oxaloacetate kinetic data. Mendeley Data V1, 10.17632/63myn5n4c8.1 [DOI] [Google Scholar]
- 27.Crow K.E., Braggins T.J. and Hardman M.J. (1983) Human liver cytosolic malate dehydrogenase: purification, kinetic properties, and role in ethanol metabolism. Arch. Biochem. Biophys. 225, 621–629 10.1016/0003-9861(83)90073-5 [DOI] [PubMed] [Google Scholar]
- 28.Trejo F., Gelpí J.Ll., Ferrer A., Boronat A., Busquets M. and Cortés A. (2001) Contribution of engineered electrostatic interactions to the stability of cytosolic malate dehydrogenase. Protein Eng. Des. Sel. 14, 911–917 10.1093/protein/14.11.911 [DOI] [PubMed] [Google Scholar]
- 29.Bernstein L.H., Grisham M.B., Cole K.D. and Everse J. (1978) Substrate inhibition of the mitochondrial and cytoplasmic malate dehydrogenases. J. Biol. Chem. 253, 8697–8701 10.1016/S0021-9258(17)34233-3 [DOI] [PubMed] [Google Scholar]
- 30.Lin J.-J., Yang T.-H., Wahlstrand B.D., Fields P.A. and Somero G.N. (2002) Phylogenetic relationships and biochemical properties of the duplicated cytosolic and mitochondrial isoforms of malate dehydrogenase from a teleost fish, Sphyraena idiastes. J. Mol. Evol. 54, 107–117 10.1007/s00239-001-0023-z [DOI] [PubMed] [Google Scholar]
- 31.Telegdi M., Wolfe D.V. and Wolfe R.G. (1973) Malate Dehydrogenase. XII. Initial rate kinetic studies of substrate activation of porcine mitochondrial enzyme by malate. J. Biol. Chem. 248, 6484–6489 10.1016/S0021-9258(19)43471-6 [DOI] [PubMed] [Google Scholar]
- 32.DuVal G., Swaisgood H.E. and Horton H.R. (1985) Some kinetic characteristics of immobilized protomers and native dimers of mitochondrial malate dehydrogenase: an examination of the enzyme mechanism. Biochemistry 24, 2067–2072 10.1021/bi00329a039 [DOI] [PubMed] [Google Scholar]
- 33.Walk R.-A. and Hock B. (1977) Glyoxysomal and mitochondrial malate dehydrogenase of watermelon (Citrullus vulgaris) cotyledons: II. Kinetic properties of the purified isoenzymes. Planta 136, 221–228 10.1007/BF00385988 [DOI] [PubMed] [Google Scholar]
- 34.Huang J., Niazi A.K., Young D., Rosado L.A., Vertommen D., Bodra N.et al. (2018) Self-protection of cytosolic malate dehydrogenase against oxidative stress in Arabidopsis. J. Exp. Bot. 69, 3491–3505 10.1093/jxb/erx396 [DOI] [PubMed] [Google Scholar]
- 35.An Y., Cao Y. and Xu Y. (2016) Purification and characterization of the plastid-localized NAD-dependent malate dehydrogenase from Arabidopsis thaliana. Biotechnol. Appl. Biochem. 63, 490–496 10.1002/bab.1406 [DOI] [PubMed] [Google Scholar]
- 36.Thomas M.J., Cassidy E.R., Robinson D.S. and Walstrom K.M. (2022) Kinetic characterization and thermostability of C. elegans cytoplasmic and mitochondrial malate dehydrogenases. BBA-Proteins Proteom 1870, 140722 10.1016/j.bbapap.2021.140722 [DOI] [PubMed] [Google Scholar]
- 37.Yin Y. and Kirsch J.F. (2007) Identification of functional paralog shift mutations: conversion of Escherichia coli malate dehydrogenase to a lactate dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 104, 17353–17357 10.1073/pnas.0708265104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muslin E.H., Li D., Stevens F.J., Donnelly M., Schiffer M. and Anderson L.E. (1995) Engineering a domain-locking disulfide into a bacterial malate dehydrogenase produces a redox-sensitive enzyme. Biophys. J. 68, 2218–2223 10.1016/S0006-3495(95)80430-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iijima S., Saiki T. and Beppu T. (1980) Physicochemical and catalytic properties of thermostable malate dehydrogenase from an extreme thermophile Thermus Flavus AT-62. Biochim. Biophys. Acta 613, 1–9 10.1016/0005-2744(80)90185-0 [DOI] [PubMed] [Google Scholar]
- 40.Ge Y.-D., Cao Z.-Y., Wang Z.-D., Chen L.-L., Zhu Y.-M. and Zhu G.-P. (2010) Identification and Biochemical Characterization of a Thermostable Malate Dehydrogenase from the Mesophile Streptomyces coelicolor A3(2). Biosci. Biotech. Bioch. 74, 2194–2201 10.1271/bbb.100357 [DOI] [PubMed] [Google Scholar]
- 41.Madern D. and Ebel C. (2007) Influence of an anion-binding site in the stabilization of halophilic malate dehydrogenase from Haloarcula marismortui. Biochimie 89, 981–987 10.1016/j.biochi.2007.03.008 [DOI] [PubMed] [Google Scholar]
- 42.Stein L.R. and Imai S. (2012) The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol. Metab. 23, 420–428 10.1016/j.tem.2012.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dasika S.K., Vinnakota K.C. and Beard D.A. (2015) Characterization of the kinetics of cardiac cytosolic malate dehydrogenase and comparative analysis of cytosolic and mitochondrial isoforms. Biophys. J. 108, 420–430 10.1016/j.bpj.2014.11.3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Silverstein E. and Sulebele G. (1969) Catalytic mechanism of pig heart mitochondrial malate dehydrogenase studied by kinetics at equilibrium. Biochemistry 8, 2543–2550 10.1021/bi00834a042 [DOI] [PubMed] [Google Scholar]
- 45.Harada K. and Wolfe R.G. (1968) Malic dehydrogenase. VII. The catalytic mechanism and possible role of identical protein subunits. J. Biol. Chem. 243, 4131–4137 10.1016/S0021-9258(18)93289-8 [DOI] [PubMed] [Google Scholar]
- 46.Mueggler P.A. and Wolfe R.G. (1978) Malate dehydrogenase. Kinetic studies of substrate activation of supernatant enzyme by L-malate. Biochemistry 17, 4615–4620 10.1021/bi00615a006 [DOI] [PubMed] [Google Scholar]
- 47.Heyde E. and Ainsworth S. (1968) Kinetic studies on the mechanism of the malate dehydrogenase reaction. J. Biol. Chem. 243, 2413–2423 10.1016/S0021-9258(18)93490-3 [DOI] [PubMed] [Google Scholar]
- 48.Mullinax T.R., Mock J.N., McEvily A.J. and Harrison J.H. (1982) Regulation of mitochondrial malate dehydrogenase. Evidence for an allosteric citrate-binding site. J. Biol. Chem. 257, 13233–13239 10.1016/S0021-9258(18)33435-5 [DOI] [PubMed] [Google Scholar]
- 49.Gelpí J.L., Dordal A., Montserrat J., Mazo A. and Cortés A. (1992) Kinetic studies of the regulation of mitochondrial malate dehydrogenase by citrate. Biochem. J. 283, 289–297 10.1042/bj2830289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lodola A., Shore J.D., Parker D.M. and Holbrook J. (1978) Malate dehydrogenase of the cytosol. A kinetic investigation of the reaction mechanism and a comparison with lactate dehydrogenase. Biochem. J. 175, 987–998 10.1042/bj1750987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Duffield M.L., Nicholls D.J., Atkinson T. and Scawen M.D. (1994) An investigation of the thermal stabilities of two malate dehydrogenases by comparison of their three-dimensional structures. J. Mol. Graphics 12, 14–21 10.1016/0263-7855(94)80003-0 [DOI] [PubMed] [Google Scholar]
- 52.Müller J. and Klein C. (1982) Stability of dehydrogenases. III. Malate dehydrogenases. Biochim. Biophys. Acta 707, 133–141 10.1016/0167-4838(82)90406-X [DOI] [PubMed] [Google Scholar]
- 53.Dalhus B., Saarinen M., Sauer U.H., Eklund P., Johansson K., Karlsson A.et al. (2002) Structural basis for thermophilic protein stability: structures of thermophilic and mesophilic malate dehydrogenases. J. Mol. Biol. 318, 707–721 10.1016/S0022-2836(02)00050-5 [DOI] [PubMed] [Google Scholar]
- 54.Brininger C., Spradlin S., Cobani L. and Evilia C. (2018) The more adaptive to change, the more likely you are to survive: Protein adaptation in extremophiles. Seminars Cell Dev. Biol. 84, 158–169 10.1016/j.semcdb.2017.12.016 [DOI] [PubMed] [Google Scholar]
- 55.Goldenzweig A. and Fleishman S.J. (2018) Principles of protein stability and their application in computational design. Ann. Rev. Biochem. 87, 105–129 10.1146/annurev-biochem-062917-012102 [DOI] [PubMed] [Google Scholar]
- 56.Walk R.-A., Michaeli S. and Hock B. (1977) Glyoxysomal and Mitochondrial Malate Dehydrogenase of Watermelon (Citrullus vulgaris) Cotyledons: I. Molecular Properties of the Purified Isoenzymes. Planta, Springer 136, 211–220, 10.1007/BF00385987 [DOI] [PubMed] [Google Scholar]
- 57.Raval D.N. and Wolfe R.G. (1962) Malic dehydrogenase. IV. pH Dependence of the kinetic parameters. Biochemistry 1, 1118–1123 10.1021/bi00912a024 [DOI] [PubMed] [Google Scholar]
- 58.Hodges C.T., Jurgensen S.R. and Harrison J.H. (1980) Investigation of the pH dependence of proton uptake by porcine heart mitochondrial malate dehydrogenase upon binding of NADH. Arch. Biochem. Biophys. 203, 580–586 10.1016/0003-9861(80)90215-5 [DOI] [PubMed] [Google Scholar]
- 59.Raval D.N. and Wolfe R.G. (1963) Malic dehydrogenase. V. Kinetic studies of substrate inhibition by oxalacetate. Biochemistry 2, 220–224 10.1021/bi00902a003 [DOI] [PubMed] [Google Scholar]
- 60.Hung H.-C., Kuo M.-W., Chang G.-G. and Liu G.-Y. (2005) Characterization of the functional role of allosteric site residue Asp102 in the regulatory mechanism of human mitochondrial NAD(P)+-dependent malate dehydrogenase (malic enzyme). Biochem. J. 392, 39–45 10.1042/BJ20050641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsieh J.-Y., Shih W.-T., Kuo Y.-H., Liu G.-Y. and Hung H.-C. (2019) Functional roles of metabolic intermediates in regulating the human mitochondrial NAD(P)+-dependent malic enzyme. Sci. Rep. 9, 9081 10.1038/s41598-019-45282-0 [DOI] [PMC free article] [PubMed] [Google Scholar]