Abstract
Regulation of gene expression through histone modifications underlies cell homeostasis and differentiation. Kdm4d and Kdm4dl exhibit a high degree of similarity and demethylate H3K9me3. However, the physiological functions of these proteins remain unclear. In this study, we generated Kdm4dl mutant mice and found that Kdm4dl was dispensable for mouse development. However, through the generation of Kdm4d mutant mice, we unexpectedly found that Kdm4d mutant male mice were subfertile because of impaired sperm motility. The absence of Kdm4d was associated with an altered distribution of H3K9me3 in round spermatids, suggesting that the Kdm4d-mediated adjustment of H3K9me3 levels is required to generate motile sperm. Further analysis revealed that the absence of Kdm4d did not affect the functionality of sperm nuclei in generating offspring. As KDM4D is specifically expressed in the human testes, our results suggest that changes in KDM4D expression or its activity may be a risk factor for human infertility.
Keywords: Kdm4d, Kdm4dl, Male fertility, Mouse
Histone post-translational modifications (PTMs) are regulated by writers and erasers. Writers deposit PTMs, whereas erasers remove PTMs. Although various types of PTMs have been identified on histones, including methylation, acetylation, and phosphorylation, methylation of the N-terminal histone H3 tail, mediated by histone methyltransferases, constitutes an integral part of gene regulation [1, 2]. For example, H3K4me3 is tightly coupled to gene activation, whereas H3K9me3 and H3K27me3 induce the formation of constitutive and facultative heterochromatin, respectively. These histone PTMs are reversible and, therefore, can be dynamically controlled depending on specific cellular contexts, which enables alteration of gene expression patterns during cell differentiation and development.
Histone demethylases can be divided into two classes: the first class consists of flavin adenine dinucleotide (FAD)-dependent lysine-specific demethylases (Lsd or Kdm1 subfamily), and the second class consists of α-ketoglutarate and Fe(II)-dependent oxygenases, Jumonji C (JmjC) domain-containing histone lysine demethylases (Kdm) subfamilies [3,4,5]. The JmjC domain-containing Kdms catalyze demethylation through N-methyl hydroxylation using Fe(II) and α-ketoglutarate as cofactors. These Kdms display distinct specificities for the methylation states and lysine residues of histones. In mice, the JmjC domain-containing Kdm4 subfamily is comprised of Kdm4a, Kdm4b, Kdm4c, Kdm4d, and Kdm4dl. Gene knockout studies in mice have indicated that Kdm4a, Kdm4b, and Kdm4c are required for heart, fetal and mammary gland, and embryonic development, respectively, whereas Kdm4d mutant mice do not display any prominent phenotype [6,7,8,9,10]. A clear structural difference between them is that while the PHD and Tudor domains are present in Kdm4a, Kdm4b, and Kdm4c at their C-terminal regions, Kdm4d and Kdm4dl, which are highly similar in amino acid sequences to each other (71.5% overall similarity), lack these domains and are shorter in protein length (Fig. 1A). In line with these structural differences, Kdm4 subfamily genes show differences in their substrate specificity; Kdm4a, Kdm4b, and Kdm4c, display dual selectivity in demethylating H3K9me2/me3 and H3K36me3, and Kdm4d and Kdm4dl demethylate H3K9me2/me3 and do not recognize H3K36me3. Furthermore, H3K9me3 rather than H3K9me2 is the preferred substrate for Kdm4d and Kdm4dl, indicating that Kdm4d and Kdm4dl are highly specific demethylases targeting H3K9me3 [6, 11,12,13]. These observations suggest that Kdm4d and Kdm4dl play important roles in regulating gene function by demethylating H3K9me3.
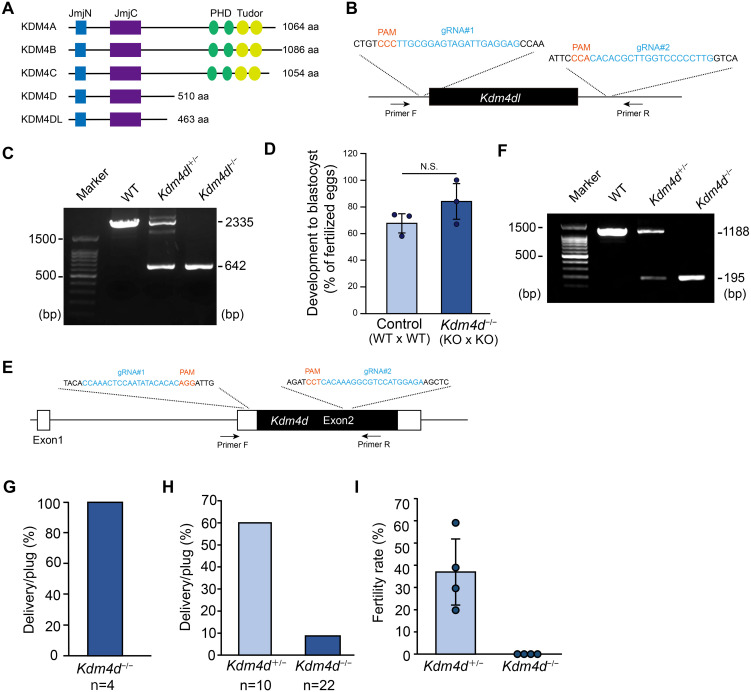
Fig. 1.
Kdm4dl is dispensable for development while Kdm4d is required for male fertility. (A) An illustration showing the protein structure of the Kdm4 family genes. N-terminal JmjN and JmjC domains as well as C-terminal PHD and Tudor domains are indicated. aa, amino acids. (B) Schematic diagram of Kdm4dl gene knockout by CRISPR-Cas9 system; gRNA#1 and gRNA#2 were used to induce genomic deletion. Primers indicated were used for genotyping. (C) Representative genotyping PCR results of Kdm4dl mutant mice. (D) Developmental efficiency to the blastocyst stage. In vitro fertilization was performed with mice with indicated genotypes (n = 3). Mean ± SD are indicated. (E) Schematic diagram of Kdm4d gene knockout by CRISPR-Cas9 system as in (A). (F) Representative genotyping PCR results of Kdm4d mutant mice. (G) Delivery rate of Kdm4d−/− female mice. Kdm4d−/− female mice were crossed with wild-type males; n indicates the number of plugs examined. Four independent female mice were used. (H) Delivery rate of Kdm4d mutant male mice. Kdm4d mutant male mice were crossed with wild-type females; n indicates the number of plugs examined. Five heterozygous and six homozygous male mice were used. (I) Fertility rate upon in vitro fertilization. Wild-type eggs and sperms from Kdm4d mutant mice were used for IVF and pronuclear formation was examined. Mean ± SD are indicated. Four independent male mice were used for each experimental group.
Kdm4d is specifically expressed in the testis [6]. Kdm4d knockout (KO) mice were generated as described previously to investigate their physiological roles. Kdm4d KO mice showed a prominent increase in H3K9me3 levels in round spermatids, indicating that Kdm4d was responsible for H3K9me3 turnover in round spermatids. However, this study found that fertility was not affected in this Kdm4d KO mouse line. In contrast to Kdm4d, Kdm4dl is expressed in mice upon zygotic genome activation (ZGA) after fertilization [14]. KDM4E, the human homolog of Kdm4dl, is also activated during ZGA in human embryos [15], suggesting that Kdm4dl plays an important role in early embryonic development. However, whether zygotic expression of Kdm4dl is required for development remains unclear.
Materials and Methods
Animals
Mice aged 8–10 weeks of age were used in this study. All animal experiments were approved by the Animal Experiments Committee of the University of Yamanashi (A4-1) and were performed according to the guidelines for animal experiments at the University of Yamanashi. The mice were housed in cages under specific pathogen-free conditions and had free access to water and food.
Generation of Kdm4dl and Kdm4d knockout mice
The crRNAs (Integrated DNA Technologies, Coralville, Iowa, USA) were annealed with tracrRNA and Cas9 protein (TAKARA BIO, Shiga, Japan) was added to the annealed sgRNA solution. The resulting CRISPR-Cas9 complex solution, containing 1 μM annealed sgRNA and 300 nM Cas9 protein, was injected into the cytoplasm of the fertilized eggs. Kdm4dl and Kdm4d mutant mice were generated from the C57BL/6J and C57BL/6N, respectively. The sgRNA target sequences were as follows.
Kdm4d #1: CCAAACTCCAATATACACAC
Kdm4d #2: TCTCCATGGACGCCTTTGTG
Kdm4dl #1: CAAGGGGGACCAAGCGTGTG
Kdm4dl #2: CTCCTCAATCTACTCCGCAA
In vitro fertilization (IVF) and embryo transfer
Female mice were superovulated by injecting 5 IU of pregnant mature serum gonadotropin (PMSG, ASKA Pharmaceutical, Tokyo, Japan), followed by an injection of 5 IU of human chorionic gonadotropin (hCG, ASKA Pharmaceutical) 46–48 h later. Sperm were collected from the cauda epididymis, placed in 0.2 ml human tubal fluid (HTF) medium (in-house), and incubated for 1 h for capacitation. Cumulus-oocyte complexes were collected 14–16 h after hCG injection and used for insemination with capacitated sperms. Five to six hours after insemination, embryos that formed two pronuclei were considered fertilized eggs. For embryo transfer, 2-cell stage embryos were transferred into the oviducts of pseudopregnant ICR mice.
Analyses of sperm motility
Capacitated sperms were prepared as described above, placed on the Sperm Motility Analysis System Animal (DITECT, Tokyo, Japan), and analyzed using Computer-Aided Sperm Analysis. More than 400 sperms were recorded for each mouse. The straight-line velocity, ‘curvilinear velocity’, ‘average path velocity,’ and ‘beating cross frequency’ were recorded.
Intracytoplasmic sperm injection (ICSI)
ICSI was performed as previously described [16]. Briefly, cumulus-oocyte complexes were collected in 0.1 ml of HTF medium prepared in-house, and cumulus were removed by 0.1% hyaluronidase (Sigma-Aldrich, St. Louis, MO, USA) treatment. After washing with HEPES-buffered CZB (H-CZB) medium (in-house), the oocytes were placed in H-CZB for micromanipulation. The tail of the sperm was removed using several piezo pulses with a piezo drive micromanipulator (Prime Tech, Tokyo, Japan) and the head of the sperm was injected into eggs from wild-type C57BL/6N females. Pronuclear formation was verified 5–6 h after intracytoplasmic sperm injection (ICSI). Zygotes were cultured in CZB medium until further use.
IVF with zona-free eggs
IVF with zona-free eggs was performed as described in the IVF section, except that the zona pellucida was removed prior to insemination. To remove the zona pellucida from the eggs, Acidic Tyrode’s solution (Sigma-Aldrich) was added for 1 min, and the resulting zona-free oocytes were placed in HTF medium. Capacitated sperm from Kdm4d knockout mice were used for IVF.
Immunohistochemistry
For immunohistochemistry of the testis sections, fresh testes were embedded in OCT compound (Sakura Finetek, Tokyo, Japan) and sectioned at a thickness of 12 μm with a cryostat (Thermo Fisher Scientific, Waltham, MA, USA). The sections were treated with 4% PFA in PBS for 10 min at room temperature and incubated in a blocking buffer (3% bovine serum albumin in PBS with 0.1% Triton X-100) for 30 min. The sections were then incubated with primary antibodies diluted in blocking buffer at 4°C, overnight. The primary antibodies used were rabbit polyclonal anti-H3K9me3 (1:500; Active Motif, Carlsbad, CA, USA: 39161) and rabbit monoclonal H3K27me3 antibodies (1:500; CST, Danvers, MA, USA: 9733). The secondary antibody used was donkey anti-rabbit IgG conjugated to Alexa Fluor 555 (1:1000; Thermo Fisher Scientific, A31572). Slides were mounted using VECTASHIELD (Vector Laboratories, Burlingame, CA, USA) containing DAPI. Images were captured using an FV1200 confocal microscope (Olympus, Tokyo, Japan). The relative fluorescence intensity was analyzed using the plot profile function of the ImageJ software.
Hematoxylin and eosin staining
Testes were fixed with 4% paraformaldehyde (PFA) in PBS overnight, incubated sequentially in 10% sucrose in PBS for 2 h, 20% sucrose in PBS for 2 h, and then 30% sucrose in PBS overnight at 4°C, and embedded in OCT compound. Sections were prepared and stained with hematoxylin and eosin. Sections were observed under a BX51 microscope (Olympus).
Statistical analysis
Statistical analyses were performed using the R (http://www.r-project.org) or Excel software (Microsoft, Redmond, WA, USA).
Results
Kdm4dl is dispensable for mouse development
We first examined the role of Kdm4dl, which is activated zygotically after fertilization. A previous report indicated that the depletion of Kdm4dl through the injection of Cas9-sgRNA complex into mouse zygotes resulted in failure in development to the blastocyst stage (approximately 60% of embryos were reported to be abnormal at the blastocyst stage) [14]. Thus, this result suggests that the Kdm4dl-mediated regulation of histone modifications may be important for early development. Thus, we created a Kdm4dl KO mouse line by deleting the entire Kdm4dl locus using CRISPR-Cas9. We confirmed that 1,693 bp deletion occurred at the Kdm4dl gene locus (Figs. 1B and C; Supplementary Fig. 1), and heterozygous Kdm4dl (Kdm4dl+/−) females and males were crossed to examine the viability or lethality of Kdm4dl null (Kdm4dl−/−) mice. In contrast to the previous observation [14], Kdm4dl−/− mice were born at an expected mendelian ratio (+/+: +/−: −/− = 20:44:22) and grew normally. In addition, in vitro fertilization using Kdm4dl−/− oocytes and Kdm4dl−/− sperm generated blastocysts normally (Fig. 1D). Thus, we conclude that Kdm4dl is dispensable for mouse development.
Kdm4d knockout male mice are subfertile
We suspect that Kdm4d, which possesses amino acid sequences highly similar to those of Kdm4dl, may compensate for the absence of Kdm4dl. To test this hypothesis, it was necessary to create double-knockout mice for Kdm4d and Kdm4dl. However, we considered it important to examine the phenotype of Kdm4d single-KO mice before creating and analyzing these mice, although a previous report indicated that Kdm4d KO mice show normal viability and fertility [6]. Therefore, we created Kdm4d KO mice using CRISPR-Cas9, as was done for the Kdm4dl mice (Fig. 1E). We deleted a large part of the exon 2 of Kdm4d (993 bp deletion), in which all the coding region consisted of 1,533 bp, suggesting that our Kdm4d−/− mice are null for Kdm4d function (Figs. 1E and F; Supplementary Fig. 2). Crosses between heterozygous females and males indicated that Kdm4d−/− mice were born nearly at a mendelian ratio (+/+: +/−: −/− = 18:46:23). We then crossed Kdm4d−/− mice with wild-type mice to verify their fertility. Hereafter, we used Kdm4d+/− mice as a control. We found that Kdm4d−/− females are fertile (Fig. 1G). However, Kdm4d−/− males displayed limited ability to give rise to offspring (Fig. 1H). In vitro fertilization using wild-type eggs and Kdm4d+/− or Kdm4d−/− mouse sperm indicated that Kdm4d−/− mouse sperm have severe defects in fertilizing with wild-type eggs (Fig. 1I). This result was unanticipated because Kdm4d−/− males were not completely sterile. We believe that the in vitro environment is not optimal compared to the in vivo environment and that the sperm phenotype is exacerbated in vitro.
Sperm motility is affected in Kdm4d knockout mice
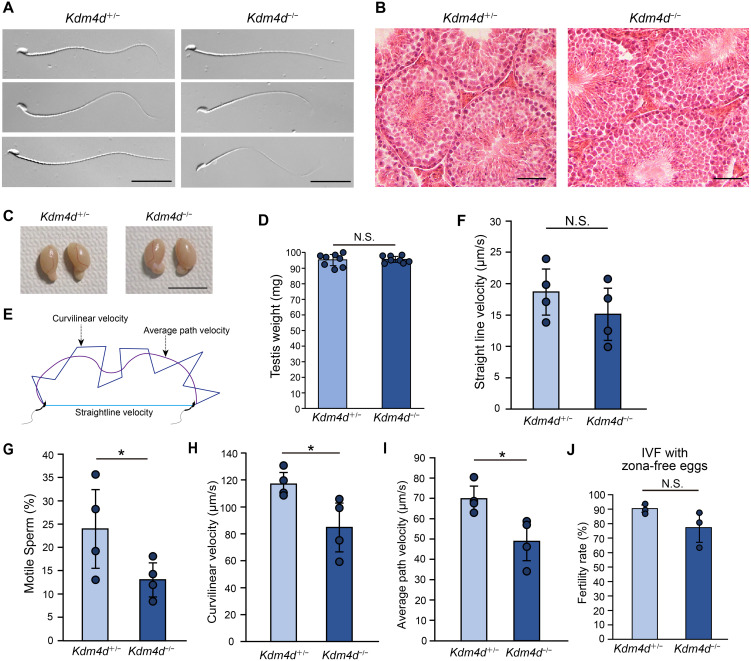
The reason for the subfertility seen in Kdm4d−/− males was further investigated. Sperm morphology in Kdm4d−/− mice were virtually normal (Fig. 2A), and we did not detect histological abnormalities in the testis (Fig. 2B). Consistently, the weight of testis was unchanged in Kdm4d−/− mice (Figs. 2C and D). During in vitro fertilization, we noticed that the motility of Kdm4d−/− mouse sperm appeared to be affected. Quantitative analyses revealed that the fraction of motile sperm was reduced, and sperm movement was significantly slower in Kdm4d−/− mice compared to the control (Figs. 2E, F, G, H, and I). We speculated that sperm motility is required to penetrate the zona pellucida and subsequently fertilize eggs. As expected, removal of the zona pellucida followed by in vitro fertilization could rescue the fertility defects observed in Kdm4d−/− mouse sperm (Fig. 2J). These results suggest that impaired sperm motility underlies the subfertility of Kdm4d−/− male mice.
Fig. 2.
Kdm4d mutant mice display impaired sperm motility. (A) Morphology of sperm from Kdm4d mutant mice. Scale bar, 30 μm. (B) HE-staining of testis sections. Scale bar, 50 μm. (C) Morphology of testis from Kdm4d mutant mice. Scale bar, 1 cm. (D) Weight of testis from Kdm4d mutant mice. Mean ± SD are indicated. N.S., not significant (Student’s t-test). Four independent male mice were used for each experimental group. (E) Schematic representation of sperm motility analysis. (F–I) Sperm motility indices. Straightline velocity (F), fraction of motile sperm (G), curvilinear velocity (H), and average path velocity (I) are shown. Mean ± SD are indicated. * P < 0.05 (Student’s t-test). Four independent male mice were used for each experimental group. (J) Fertility rate upon in vitro fertilization. Data are shown as in Fig. 1H except that zona-free eggs were used in this assay. N.S., not significant (Student’s t-test). Three independent male mice were used for each experimental group.
H3K9me3 distribution in round spermatids is altered in Kdm4d knockout mice
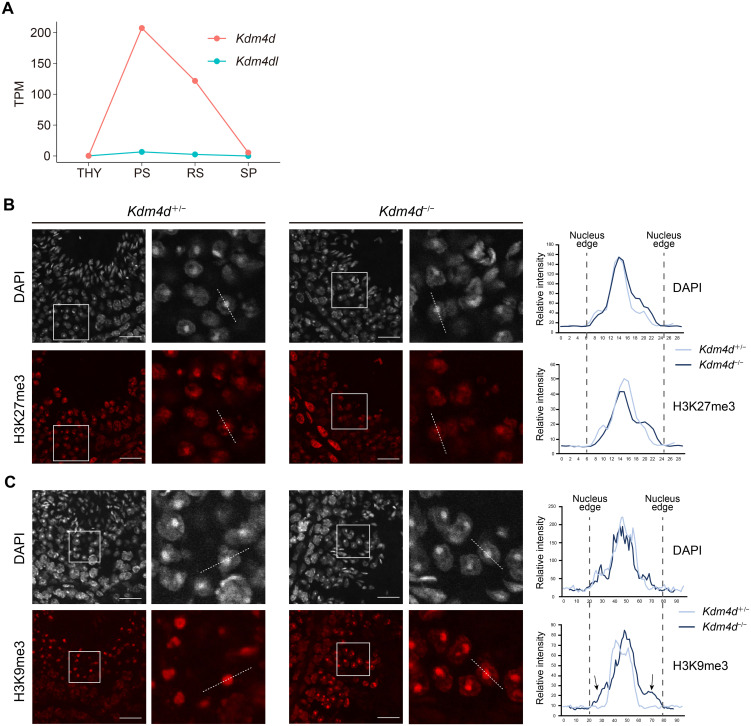
Kdm4d targets H3K9me3 for demethylation. Kdm4d is required to adjust H3K9me3 levels in round spermatids [6]. Analysis of publicly available RNA-seq data during spermatogenesis confirmed the highly upregulated expression of Kdm4d in round spermatids (Fig. 3A). We then performed immunofluorescence staining of the testis sections using an H3K9me3 antibody. We also performed immunofluorescent staining for H3K27me3, a repressive histone marker. While H3K27me3 distribution was unchanged (Fig. 3B), we observed an altered distribution of H3K9me3 in round spermatids (Fig. 3C). In the control, H3K9me3 was confined to the chromocenter located around the center of the nuclei as a single DAPI spot and was not detectable in the nuclear region surrounding the chromocenters. On the contrary, in Kdm4d−/− mice, H3K9me3 was readily detectable at the nuclear region surrounding the chromocenters while H3K9me3 enrichment at the chromocenters was still observed. As a result, the nuclei edges of round spermatids were visible with H3K9me3 staining in Kdm4d−/− mice (Fig. 3C). These results suggest that Kdm4d-mediated H3K9me3 regulation in round spermatids is required for the generation of motile sperms.
Fig. 3.
Distribution of H3K9me3 is altered in Kdm4d mutant mouse round spermatids. (A) Line plots showing Kdm4d (red) and Kdm4dl (blue) expression levels during spermatogenesis based on published RNA-seq data (GSE55060 and DRA000484) [24, 25]. THY, undifferentiated spermatogonia; PS, pachytene spermatocytes; RS, round spermatids; SP, sperm. (B, C) Immunohistochemistry for Kdm4d mutant mouse testis sections. H3K27me3 (B) or H3K9me3 (C) antibody was used. Scale bar, 25 μm. Signal intensity at the indicated dashed lines was measured and indicated in the right panels. Arrows in (C) indicate excessive H3K9me3 signals at the chromatin region surrounding chromocenters.
Generation of offspring through intracytoplasmic injection of Kdm4d knockout mouse sperm
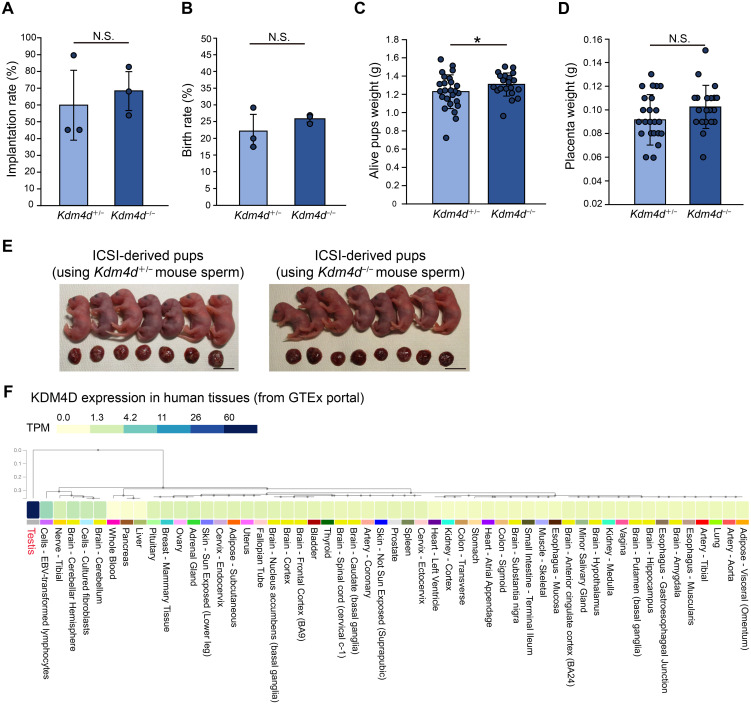
H3K9me3 distribution in round spermatids was altered in Kdm4d−/− mice. Although histones are largely replaced by protamines during spermiogenesis, it has been suggested that the retained histones and histone modifications in the sperm affect development of the next generation [17,18,19]. Therefore, alterations in epigenetic modification patterns occur in Kdm4d−/− mouse sperm, potentially impacting the development of the next generation. To address this possibility as well as the functionality of sperm nuclei of Kdm4d−/− mice, we performed intracytoplasmic sperm injection (ICSI) and transferred ICSI 2-cell embryos to pseudopregnant female mice. We observed no defects in implantation rate, birth rate, and placenta weight whereas a slight increase in pups weight was observed when Kdm4d−/− mouse sperm were used for ICSI for unknown reasons (Figs. 4A, B, C, D and E). Thus, it is likely that Kdm4d-mediated H3K9me3 regulation is required for the generation of functional motile sperms, however is dispensable for the formation of functional sperm nuclei, which permit the development of the next generation.
Fig. 4.
Nuclei of Kdm4d mutant mouse sperm can generate offspring normally. (A–D) Development of ICSI embryos generated by Kdm4d+/− or Kdm4d−/− mouse sperm. Wild-type eggs and Kdm4d+/− or Kdm4d−/− mouse sperm were used for ICSI. Implantation rate (A), birth rate (B), pups weight (C), and placenta weight (D) were analyzed. Mean ± SD are indicated. * P < 0.05 (Student’s t-test). Three independent male mice were used for each experimental group. (E) Pictures showing offspring generated by ICSI. Scale bar, 1 cm. (F) Expression of KDM4D in human tissues. KDM4D is specifically expressed in testis. Data are obtained from the GTEx portal (https://www.gtexportal.org/home/).
Discussion
Our results indicate that Kdm4dl is dispensable for mouse development. Although we did not observe any prominent phenotypes, it is possible that Kdm4dl functions redundantly with other Kdm family genes, as KDM4E, a human ortholog of Kdm4dl, is also expressed during early development. Future investigations should address this issue. In this study, we provide data indicating that Kdm4d knockout mice show male subfertility, which is in contrast to a previous study [6]. We believe that these differences could be attributed to differences in the mouse genetic backgrounds used in these studies (C57BL/6 versus C57BL/6;129 mixed background). In humans, KDM4D was specifically expressed in the testes (Fig. 4F). Therefore, our results suggest that inactivation of KDM4D could be a risk factor for male infertility in humans, although further investigation is required to address this possibility.
While we were able to detect the impaired sperm motility of Kdm4d−/− mouse sperm, the mechanism underlying this phenotype is still unclear. We speculate that H3K9me3 removal by Kdm4d in round spermatids permitted the expression of genes required for the acquisition of full sperm motility. This hypothesis is supported by increasing evidence demonstrating the importance of H3K9me3 regulation during spermatogenesis. An early study showed that double-null mice for Suv39h1 and Suv39h2, methyltransferases responsible for H3K9me3, resulted in the absence of sperm owing to failures in early meiotic processes [20]. Similarly, Setdb1, another enzyme that catalyzes H3K9me3, regulates chromosome pairing, meiotic synapsis, and meiotic sex chromosome inactivation (MSCI) during meiosis and ensures male fertility [21, 22]. Moreover, while high levels of H3K9me3 are observed on the X chromosome in spermatocytes and spermatids, possibly due to MSCI, some spermatid-specific genes are reactivated after meiosis, and this reactivation is accompanied by the loss of H3K9me3 enrichment [23]. Thus, in addition to methylation, active demethylation of H3K9me3, in the absence of cell proliferation, appears to contribute to male fertility. Future studies should examine whether, and to what extent, Kdm4d contributes to these processes.
Conflict of interests
The authors declare no competing interests.
Supplementary
Acknowledgments
We thank Satoshi Kishigami (University of Yamanashi) for sharing the SMAS system to analyze sperm motility and the members of the Advanced Biotechnology Center at the University of Yamanashi for assistance with mouse maintenance. This work was supported by grants from the MEXT Grant-in-Aid for Scientific Research on Innovative Areas (JP19H05756 to T.I. and JP19H05750 and JP19H05749 to M.I.), Takeda Science Foundation (T.I.), Mochida Memorial Foundation for Medical and Pharmaceutical Research (T.I.), Naito Foundation (T.I.), and Uehara Memorial Foundation (T.I). Z.X. was supported by a Conohana Fellowship from the University of Yamanashi.
References
- 1.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet 2016; 17: 487–500. [DOI] [PubMed] [Google Scholar]
- 2.Jambhekar A, Dhall A, Shi Y. Roles and regulation of histone methylation in animal development. Nat Rev Mol Cell Biol 2019; 20: 625–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 2006; 7: 715–727. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell 2007; 25: 1–14. [DOI] [PubMed] [Google Scholar]
- 5.Shi YG, Tsukada Y. The discovery of histone demethylases. Cold Spring Harb Perspect Biol 2013; 5: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwamori N, Zhao M, Meistrich ML, Matzuk MM. The testis-enriched histone demethylase, KDM4D, regulates methylation of histone H3 lysine 9 during spermatogenesis in the mouse but is dispensable for fertility. Biol Reprod 2011; 84: 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawazu M, Saso K, Tong KI, McQuire T, Goto K, Son DO, Wakeham A, Miyagishi M, Mak TW, Okada H. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS One 2011; 6: e17830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang QJ, Chen HZ, Wang L, Liu DP, Hill JA, Liu ZP. The histone trimethyllysine demethylase JMJD2A promotes cardiac hypertrophy in response to hypertrophic stimuli in mice. J Clin Invest 2011; 121: 2447–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozaki Y, Fujiwara K, Ikeda M, Ozaki T, Terui T, Soma M, Inazawa J, Nagase H. The oncogenic role of GASC1 in chemically induced mouse skin cancer. Mamm Genome 2015; 26: 591–597. [DOI] [PubMed] [Google Scholar]
- 10.Duncan AR, Vitobello A, Collins SC, Vancollie VE, Lelliott CJ, Rodan L, Shi J, Seman AR, Agolini E, Novelli A, Prontera P, Guillen Sacoto MJ, Santiago-Sim T, Trimouille A, Goizet C, Nizon M, Bruel AL, Philippe C, Grant PE, Wojcik MH, Stoler J, Genetti CA, van Dooren MF, Maas SM, Alders M, Faivre L, Sorlin A, Yoon G, Yalcin B, Agrawal PB. Heterozygous variants in KDM4B lead to global developmental delay and neuroanatomical defects. Am J Hum Genet 2020; 107: 1170–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M, Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 2006; 125: 467–481. [DOI] [PubMed] [Google Scholar]
- 12.Shin S, Janknecht R. Diversity within the JMJD2 histone demethylase family. Biochem Biophys Res Commun 2007; 353: 973–977. [DOI] [PubMed] [Google Scholar]
- 13.Hillringhaus L, Yue WW, Rose NR, Ng SS, Gileadi C, Loenarz C, Bello SH, Bray JE, Schofield CJ, Oppermann U. Structural and evolutionary basis for the dual substrate selectivity of human KDM4 histone demethylase family. J Biol Chem 2011; 286: 41616–41625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiao Y, Ren C, Huang S, Yuan J, Liu X, Fan J, Lin J, Wu S, Chen Q, Bo X, Li X, Huang X, Liu Z, Shu W. High-resolution annotation of the mouse preimplantation embryo transcriptome using long-read sequencing. Nat Commun 2020; 11: 2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrickson PG, Doráis JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, Nix DA, Peterson CM, Tapscott SJ, Carrell DT, Cairns BR. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet 2017; 49: 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakamoto M, Ito D, Inoue R, Wakayama S, Kikuchi Y, Yang L, Hayashi E, Emura R, Shiura H, Kohda T, Namekawa SH, Ishiuchi T, Wakayama T, Ooga M. Paternally inherited H3K27me3 affects chromatin accessibility in mouse embryos produced by round spermatid injection. Development 2022; 149: 149. [DOI] [PubMed] [Google Scholar]
- 17.Erkek S, Hisano M, Liang CY, Gill M, Murr R, Dieker J, Schübeler D, van der Vlag J, Stadler MB, Peters AH. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat Struct Mol Biol 2013; 20: 868–875. [DOI] [PubMed] [Google Scholar]
- 18.Siklenka K, Erkek S, Godmann M, Lambrot R, McGraw S, Lafleur C, Cohen T, Xia J, Suderman M, Hallett M, Trasler J, Peters AH, Kimmins S. Disruption of histone methylation in developing sperm impairs offspring health transgenerationally. Science 2015; 350: aab2006. [DOI] [PubMed] [Google Scholar]
- 19.Lismer A, Dumeaux V, Lafleur C, Lambrot R, Brind’Amour J, Lorincz MC, Kimmins S. Histone H3 lysine 4 trimethylation in sperm is transmitted to the embryo and associated with diet-induced phenotypes in the offspring. Dev Cell 2021; 56: 671–686.e6. [DOI] [PubMed] [Google Scholar]
- 20.Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 2001; 107: 323–337. [DOI] [PubMed] [Google Scholar]
- 21.Hirota T, Blakeley P, Sangrithi MN, Mahadevaiah SK, Encheva V, Snijders AP, ElInati E, Ojarikre OA, de Rooij DG, Niakan KK, Turner JMA. SETDB1 links the meiotic DNA damage response to sex chromosome silencing in mice. Dev Cell 2018; 47: 645–659.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng EC, Hsieh CL, Liu N, Wang J, Zhong M, Chen T, Li E, Lin H. The essential function of SETDB1 in homologous chromosome pairing and synapsis during meiosis. Cell Rep 2021; 34: 108575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ernst C, Eling N, Martinez-Jimenez CP, Marioni JC, Odom DT. Staged developmental mapping and X chromosome transcriptional dynamics during mouse spermatogenesis. Nat Commun 2019; 10: 1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, Suzuki Y, Kono T. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet 2012; 8: e1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasegawa K, Sin HS, Maezawa S, Broering TJ, Kartashov AV, Alavattam KG, Ichijima Y, Zhang F, Bacon WC, Greis KD, Andreassen PR, Barski A, Namekawa SH. SCML2 establishes the male germline epigenome through regulation of histone H2A ubiquitination. Dev Cell 2015; 32: 574–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.