Abstract
BamA is the central component of the essential β-barrel assembly machine (BAM), a conserved multi-subunit complex that dynamically inserts and folds β-barrel proteins into the outer membrane of Gram-negative bacteria. Despite recent advances in our mechanistic and structural understanding of BamA, there are few potent and selective tool molecules that can bind to and modulate BamA activity. Here, we explored in vitro selection methods and different BamA/BAM protein formulations to discover peptide macrocycles that kill Escherichia coli by targeting extreme conformational states of BamA. Our studies show that Peptide Targeting BamA-1 (PTB1) targets an extracellular divalent cation-dependent binding site and locks BamA into a closed lateral gate conformation. By contrast, PTB2 targets a luminal binding site and traps BamA into an open lateral gate conformation. Our results will inform future antibiotic discovery efforts targeting BamA and provide a template to prospectively discover modulators of other dynamic integral membrane proteins.
Subject terms: Microbiology, Structural biology
BamA carries out the essential process of folding outer membrane β-barrels in Gram-negative bacteria and is a potential antibiotic target. Here, the authors discover macrocyclic peptide inhibitors that trap BamA in distinct structural conformations.
Introduction
The outer membrane of Gram-negative bacteria is an essential and unique structure that serves as a permeability barrier to cytotoxic molecules and antibiotics1. Integral outer membrane proteins (OMPs) display a characteristic β-barrel structure and perform essential functions, supporting metabolism, transport, adhesion, and virulence2. Canonical bacterial OMPs are composed of 8–26 antiparallel β-strands which form a stable β-barrel structure where the first and last strands are arranged as an antiparallel seam3. Hydrogen bonds between adjacent β-strands confer the high stability and heat-modifiable properties that are hallmarks of β-barrel OMPs3,4. These OMPs typically possess short periplasmic turns and long extracellular loops, where the latter perform key roles in substrate recognition, adhesion, and catalysis4. In contrast to the Sec-mediated folding of integral inner membrane proteins, the folding of OMPs into the outer membrane occurs in the absence of a traditional energy source, such as ATP or the proton motive force.
OMP folding into the bacterial outer membrane is catalyzed by the β-barrel assembly machine (BAM), which is comprised of BamA, an essential and conserved integral OMP, and four periplasmic lipoproteins, BamBCDE, that support BamA function5,6. Escherichia coli BamA is comprised of five N-terminal periplasmic polypeptide transport-associated (POTRA) domains and a C-terminal 16-stranded β-barrel where the first (β1) and last (β16) β-strands form a lateral gate. Structural and mechanistic studies have implicated large-scale conformational changes at the lateral gate as being important for BAM function where the complex has been proposed to cycle between extreme closed and open states7–14. The β-signal, a conserved C-terminal sequence motif present in β-barrel OMPs, is thought to directly engage the BamA lateral gate15. One favored model posits that BAM inserts and folds nascent OMP substrates through β-strand complementation at the lateral gate of BamA, giving rise to a substrate-BamA complex from which the mature OMP ultimately grows and buds into the outer membrane2. Recent high-resolution structural studies have captured snapshots of OMP substrate folding intermediates trapped at the BAM lateral gate, highlighting the apparent dynamics of this essential membrane protein foldase7,8,10,16. However, which conformational states of BAM are permissive or suitable for inhibitor discovery remains unknown, in part, because it is technically challenging to prospectively interrogate different conformational states of dynamic integral membrane protein drug targets.
The BAM complex has been proposed to be an antibacterial target17–22. An antibody BamA inhibitor, MAB1, provided early validation for bactericidal BAM inhibition but suffered from an inability to access BamA within the context of an intact bacterial LPS layer18. A small molecule BamA inhibitor (MRL494) and a chimeric macrocyclic peptide-polymyxin derivative targeting the BAM complex have been described, but their properties have not yet been optimized19,21. The discovery of two natural products, darobactin and dynobactin, have shown broad Gram-negative antibacterial activity by targeting the closed lateral gate of BamA17,23,24, but these suffer from challenges associated with the optimization of natural products into manufacturable drug-like molecules. Here, to advance and diversify the availability of BAM inhibitors, we screen a massive macrocycle peptide library with the intention to interrogate different conformations of BamA and identify two inhibitor series with previously undescribed binding sites and distinct structural mechanisms of action.
Results
Rationale for Prospective Inhibitor Discovery
To our knowledge, beyond engineering disulfide crosslinks, there is no facile way to present distinct conformational states of BamA or the BAM complex in the context of affinity selection experiments. However, existing experimental structures of isolated BamA have revealed a closed lateral gate13,25, whereas the majority of experimental structures of the intact BAM complex (i.e. BamABCDE) present an open lateral gate7,9,11,13,14,26. We, therefore, purified isolated BamA and the intact BAM complex of E. coli to pursue in vitro selection experiments against a large mRNA-display library of 10–14 amino acid macrocyclic peptides consisting of both natural and non-canonical amino acids27 (Supplementary Fig. 1a). Furthermore, we intentionally prepared isolated BamA and the BAM complex in different detergent and non-detergent matrices to account for the possibility that the membrane mimetic environment will alter the conformational state(s) and/or dynamics of BamA/BAM8,9,28 presented in the context of these selection experiments. Specifically, we prepared these protein reagents in the short-chain and long-chain detergents octyl-glucoside, dodecyl-maltoside, dimethyl-dodecylamine oxide, and dodecyl-phosphocholine, as well as in amphipol PMAL-C8 and nanodiscs utilizing a POPE:POPG:POPC lipid mixture. In all selection experiments for BamA, we also intentionally used a construct lacking the POTRA1 and POTRA2 domains in order to minimize the possibility of identifying macrocycles that bind to these periplasmic domains. Sequence analysis after the selections revealed several peptide families that were confirmed to bind to BamA or the BAM complex by surface plasmon resonance (SPR) (Supplementary Fig. 1b, c). Identified macrocycles were screened in a minimal inhibitory concentration (MIC) assay where two distinct peptide scaffolds were discovered to inhibit the growth of wild-type E. coli (Supplementary Fig. 1b). We undertook further characterization of Peptide Targeting BamA-1 (PTB1) and Peptide Targeting BamA-2 (PTB2) to understand their mechanisms of action. Of note, both PTB1 and PTB2 were identified from the short-chain octyl-glucoside detergent-based selection conditions, perhaps suggesting that some feature of this matrix is a suitable mimic of the E. coli outer membrane in the context of the utilized selection technology.
PTB1 is a multi-species BamA inhibitor
Identified using isolated E. coli BamA as bait, the 12-amino acid macrocycle PTB1 was found to have an MIC of 42 µg/mL (25 µM) against a wild-type E. coli strain (Fig. 1A). In line with the sequence conservation of bamA, PTB1 exhibited MICs against wild-type Klebsiella pneumoniae and Enterobacter cloacae, ranging from 42 to 168 µg/mL (25–100 µM) (Fig. 1A). We did not observe MICs against the non-fermenting Gram-negative pathogens Pseudomonas aeruginosa and Acinetobacter baumannii (Fig. 1A), consistent with the more divergent bamA sequences of these species. Activity was not observed against Staphylococcus aureus, which is a Gram-positive bacterium that lacks BamA and an outer membrane (Fig. 1A).
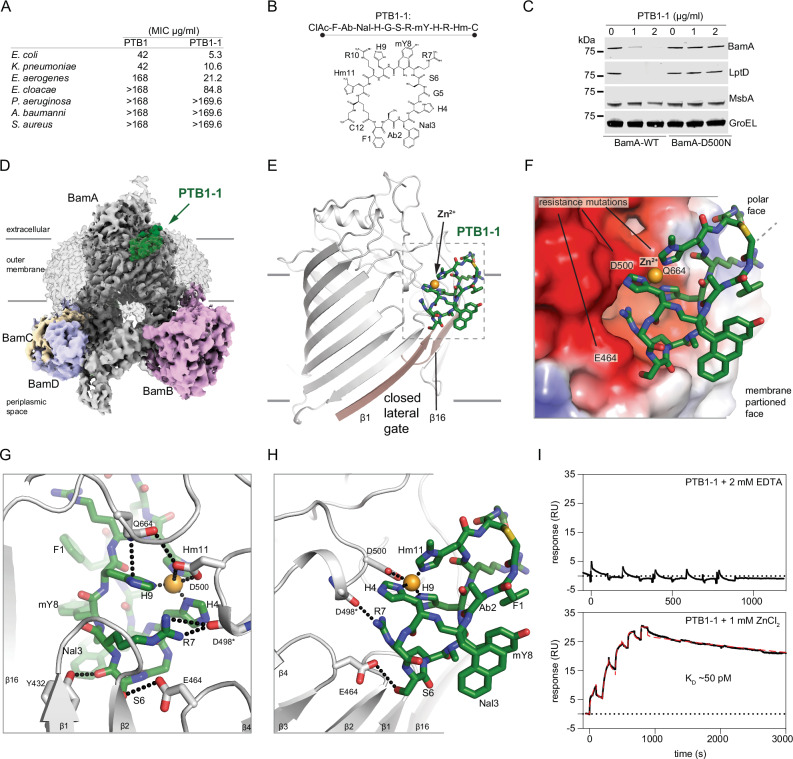
Fig. 1. Activity and cryo-EM structure of the closed-state PTB1-1–BAM complex.
A Minimal inhibitory concentrations (MICs) for BamA-binding macrocycles PTB1 and PTB1-1 on indicated bacterial species. MIC assays were performed in triplicate with mean values reported. The molecular weights of PTB1 and PTB1-1 are 1678.88 and 1705.90 g/mol, respectively. B Amino acid sequence and schematic of BamA-binding PTB1-1 macrocycle. CIAc-F (N-α-chloroacetyl-l-phenylalanine), Ab (l-α-aminobutanoic acid), Nal (β-(1-naphthyl)-l-alanine), H (l-histidine), G (glycine), S (l-serine), R (l-arginine), mY (N-α-methyl-L-tyrosine), Hm (3-methyl-l-histidine), C (l-cysteine). C Western blot of E. coli wild-type (BamA-WT) and a representative PTB1-1-resistant strain (BamA-D500N) for select outer membrane (BamA and LptD), inner membrane (MsbA), and cytoplasmic (GroEL) proteins under increasing concentrations of PTB1-1. Dashes represent location of 75 kDa molecular weight marker on each blot. This is representative of two replicates and the full blots are provided in Source Data. D Cryo-EM map of the PTB1-1–BAM–MAB2 Fab complex. Lower contours of the map that include detergent micelle and MAB2 Fab are shown in transparency. BAM subunits are labeled and BamA is shown in gray, BamB in pink, BamC in wheat, BamD in light blue, and BamE is not visible in the presented orientation. PTB1-1 is shown in green. Approximate outer membrane boundaries are indicated. E Overall view of the PTB1-1–BamA complex. PTB1-1 is shown in green, lateral gate β-strands are shown in tan, and assigned Zn2+ cation is shown as an orange sphere. Approximate outer membrane boundaries are indicated. F Electrostatics of the PTB1-1 binding site on BamA. Select positions where substitutions lead to PTB1-1 resistance are labeled and the assigned Zn2+ cation is shown as an orange sphere. Red, blue, and white represents negative, positive, and neutral charge, respectively. G and H Close-in views of the PTP1-1 interactions with BamA. BamA is shown in gray and PTB1-1 is shown in green, with direct bonding interactions indicated by dotted lines. I Duplicate SPR sensorgram of PTB1-1 interaction with surface-bound BamA in the presence of 2 mM EDTA (top) or 1 mM ZnCl2 (bottom) where the latter is fit using a two-state model, KD = (kd1/kd2)*(kd2/(ka2 + kd2)) to calculate the KD = 50 ± 2 pM in the presence of ZnCl2 (model curve is displayed as a dashed red line). No binding was observed in the presence of EDTA.
We performed a semi-random selection experiment against purified BamA in the detergent octyl-glucoside to improve the potency of PTB1 and identified PTB1-1 (Fig. 1A, B and Supplementary Fig. 2a). PTB1-1 displayed enhanced MIC activity against wild-type E. coli, K. pneumoniae, E. cloacae, and additionally revealed an MIC against Enterobacter aerogenes (Fig. 1A). Consistent with inhibition of the BAM complex, PTB1-1 reduced OMP levels (Fig. 1C), decreased activity of the OmpT enzyme that requires BAM for folding29, and increased OM permeability as determined by ethidium bromide uptake (Supplementary Fig. 2b, c). PTB1-1 was also found to be bactericidal in a time-kill experiment against E. coli (Supplementary Fig. 2d).
E. coli colonies resistant to PTB1-1 were isolated at a frequency of ~3 × 10−8 and whole genome sequencing revealed mutations restricted to the bamA gene (Supplementary Figs. 2e, 3). Nineteen distinct mutations were identified that led to PTB1-1 resistance, including substitutions of acidic residues within extracellular loops of BamA (Supplementary Figs. 2e, f, and 3). One representative PTB1-1 mutant that was isolated in multiple independent selections, BamA D500N, did not exhibit decreased OMP levels (Fig. 1C) or increased outer membrane permeability in the presence of PTB1-1 (Supplementary Fig. 2c). PTB1 and PTB1-1 do not lyse red blood cells, indicating their activity is specific for Gram-negative bacteria (Supplementary Fig. 2g). Collectively, our data indicate that the PTB1 peptide series targets BamA to antagonize BAM function in the outer membrane.
PTB1-1 targets a closed state of BamA
To define the molecular determinants of the PTB1-1 interaction, we determined a PTB1-1-BAM complex co-structure by cryogenic electron microscopy (cryo-EM) to 3.5 Å resolution in the detergent dodecyl-maltoside (DDM) using a previously characterized non-functional antibody fragment18 as a fiducial marker (Fig. 1D, Supplementary Fig. 4a–i, Supplementary Table 1). In the BAM complex, BamA is observed in a closed lateral gate conformation where PTB1-1 is bound within the extracellular leaflet region of the outer membrane (Fig. 1D, E). PTB1-1 interacts with multiple extracellular loops of BamA that form a composite acidic patch located directly above its closed transmembrane lateral gate (Fig. 1E, F). The PTB1-1 binding site is dramatically remodeled and absent in known open conformations of the BAM complex11,13,14, defining PTB1-1 as a state-dependent antagonist which targets the closed state of BamA. Beyond slight local side-chain adjustments, the closed PTB1-1-bound BAM conformation is essentially unchanged from unbound BamA structures and the periplasmic components observed in BAM complexes, indicating that PTB1-1 binding does not require or induce substantial structural rearrangements in BAM (Supplementary Fig. 4j).
PTB1-1 targets an extracellular binding site
PTB1-1 forms a compacted fold with an amphipathic character where polar side chains are found on one face and hydrophobics on the other (Fig. 1E–H). The cryo-EM map reveals that PTB1-1 is almost completely embedded within the detergent phase, suggesting that it resides within the LPS leaflet of the outer membrane (Fig. 1D). The membrane-exposed face of PTB1-1 is formed by an aromatic cluster of side chains including Phe1, Abu2, Nal3, and N-methyl-Tyr8 (mY8) (Fig. 1F, H). These features indicate that membrane partitioning is likely an important component of the potency and mechanism by which this macrocycle targets BamA30. Notably, the PTB1-1 binding site is distinct in location and nature from previously identified BamA inhibitors MAB1, darobactin, and dynobactin17,24,31.
The polar surface of PTB1-1 makes multipoint contact across extracellular loop 1 (ECL1), ECL2, and ECL3 of BamA (Fig. 1F–H). Ser6 (S6) of PTB1-1 engages the side chain of Tyr432 (ECL1) as well as the acidic side chain of Glu464 (ECL2) and PTB1-1 Arg7 (R7) makes backbone interactions to Asp498 of BamA (ECL3) (Fig. 1G, H). A constellation of three histidine side chains on PTB1-1 (H4, H9, and Hm11) converge towards the acidic side chain of Asp500 (ECL3) where density assigned as a Zn2+ ion (see below) is coordinated directly between the macrocycle and BamA (Fig. 1E–H). We speculate that the bidentate interaction between Gln664 (ECL6) of BamA with Asp500 and PTB1-1 H9 provides a bridging contact that may be required for trapping and stabilizing the lateral gate of BamA in a closed state (Fig. 1G), which would rationalize the observed Gln664His and Gln664Pro resistance mutations (Supplementary Figs. 2e, f, and 3). Mechanistically and structurally, the PTB1-1 macrocycle appears to antagonize the BAM complex by acting as a molecular staple that prevents opening of the lateral gate.
We used native mass spectrometry to confirm that divalent cations can complex with detergent purified BamA (Supplementary Fig. 5a, b). Of the metals tested, only Zn2+, Ni2+, or Cu2+ allowed an interaction between PTB1-1 and BamA, with Zn2+ enabling the most stable complex (Supplementary Fig. 5b). In the presence of Zn2+, PTB1-1 bound to purified BamA with a Kd of ~50 pM as determined by surface plasmon resonance, where divalent cation chelation abolished binding (Fig. 1I). Alanine-scanning substitutions revealed that all three divalent-coordinating histidine side chains of PTB1-1 are essential for BamA binding and antibacterial activity (Fig. 1G, H, Supplementary Fig. 5c). Additionally, PTB1-1 resistance mutations mapped to the intense electronegative surface patch on BamA, including the central Asp500 Zn2+-coordinating side chain (Fig. 1F–H, Supplementary Figs. 2e, 3). Thus, PTB1-1 is a closed-state antagonist that targets a previously unknown extracellular divalent cation site at an electronegative surface that has previously been suggested to be important for BAM complex activity32, raising speculation that metal-binding to E. coli BamA might have physiological implications.
PTB2-1 is an E. coli-specific BamA inhibitor
Selections performed against the intact BAM complex identified PTB2, a 14-amino acid macrocyclic peptide with an MIC of 4 µg/mL (2 µM) against wild-type E. coli (Fig. 2A, B, Supplementary Fig. 1b, c). We performed a semi-random selection experiment against purified BAM complex in octyl-glucoside detergent conditions and identified PTB2-1, a macrocycle that was more selective against E. coli and inactive against all other bacterial species tested (Fig. 2A, Supplementary Fig. 6a). PTB2-1 decreased OMP levels, decreased activity of the OmpT enzyme, increased outer membrane permeability, and was bactericidal as demonstrated in a time-kill experiment (Fig. 2C, Supplementary Fig. 6a–d), consistent with selective inhibition of BamA.
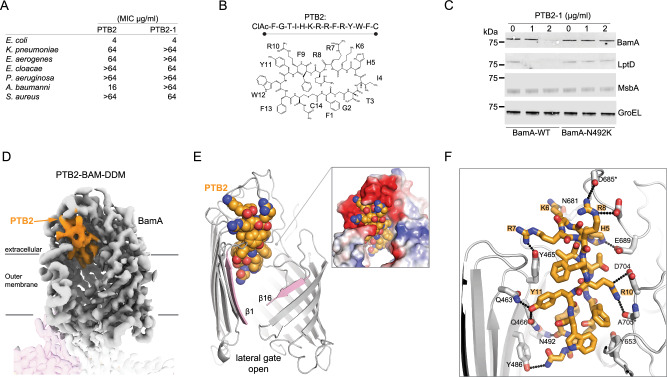
Fig. 2. Activity and cryo-EM structure of the open-state PTB2-BAM-DDM complex.
A Minimal inhibitory concentrations (MICs) for BamA-binding macrocycles of PTB2 and PTB2-1 on indicated bacterial species. MIC assays were performed in triplicate with mean values reported. The molecular weights of PTB2 and PTB2-1 are 2013.33 and 2043.40 g/mol, respectively. B Amino acid sequence and schematic of BamA-binding PTB2 marcocycle. CIAc-F (N-α-chloroacetyl-l-phenylalanine), G (glycine), T (l-threonine), I (l-isoleucine), H (l-histidine), K (l-lysine), R (l-arginine), F (l-phenylalanine), Y (l-tyrosine), W (l-tryptophan), C (l-cysteine). C Western blot of E. coli wild-type (BamA-WT) and a representative PTB2-1-resistant strain (BamA-N492K) for select outer membrane (BamA and LptD), inner membrane (MsbA), and cytoplasmic (GroEL) proteins under increasing concentrations of PTB2-1. Dashes represent the location of the 75 kDa molecular weight marker on each blot. This is representative of 2 replicates, and the full blots are provided in Source Data. D Cryo-EM map of the PTB2–BAM–DDM complex. BamA is shown in gray, BamB in pink, and BamC, BamD, and BamE are present but not labeled/shown for clarity. PTB2 bound in the central lumen is shown in orange. Lines indicate the approximate boundaries of the outer membrane. E Overall view of PTB2 bound to BamA with open lateral gate. PTB2 is shown in orange, and the lateral gate is shown in pink. The electrostatics of the PTB2 binding site are shown (inset) with red, blue, and white representing negative, positive, and neutral charges, respectively. F Close-in view of PTB2 interactions with BamA. BamA is shown in gray and PTB2 is shown in orange, with direct bonding interactions indicated by dotted lines.
Resistance to PTB2-1 was isolated at a frequency of ~1 × 10−7, and sequencing revealed eight discrete on-target mutations, all in bamA, which map to the lumen of the β-barrel (Supplementary Fig. 6e, f). A representative PTB2-1-resistant mutant, BamA N492K, did not display defects in OMP folding or increased outer membrane permeability (Fig. 2C, Supplementary Fig. 6c). The parental PTB2 macrocycle showed cross-resistance to these strains, consistent with the expectation that both macrocycles share a common binding site and determinants in BamA (Supplementary Fig. 6f). PTB2-1 activity was also unaffected by mutations in bamA that led to PTB1 resistance (Supplementary Fig. 6f) and lacked non-specific activity associated with red blood cells lysis (Supplementary Fig. 6g). Overall, the PTB2 macrocycle series represents a distinct class of BamA-targeting antibacterial peptides.
PTB2 is an open-state inhibitor of BamA
To identify the molecular determinants of PTB2 interaction, a co-structure with the BAM complex was determined by cryo-EM to ~3.1 Å resolution in DDM detergent (Supplementary Fig. 7). This revealed that the PTB2 macrocycle binds deep within the central lumen of the BamA β-barrel, where observed contacts rationalize its E. coli-specific activity and luminal-facing resistance mutations (Fig. 2D–F, Supplementary Figs. 6 and 3, Supplementary Table 1). By forming multipoint contacts within the BamA β-barrel, PTB2 traps the lateral gate open in a V-shape conformation that is directly exposed to the outer membrane bilayer (Fig. 2E), identifying PTB2 as an open-state inhibitor of BamA. PTB2 is a compact, rectangular-shaped, amphipathic macrocycle enriched with aromatic residues on one side and basic side chains on the other (Fig. 2E, F). The aromatic face of PTB2 interacts within the central vestibule of BamA, whereas the basic end is intertwined with an acidic outer opening on BamA contributed by ECLs 1–3 (Fig. 2E, F). At the base of the lateral gate, β1 is separated relative to the adjacent periplasmic loop 7 by ~5 Å (Fig. 2E). Overall, the architecture of the complex suggests that PTB2 may gain access to its luminal binding site through the open lateral gate of BamA directly from the outer membrane LPS layer.
PTB2 can bind to the open lateral gate
Two additional cryo-EM map features were observed alongside β1 of the open BamA lateral gate nearly 10 Å away from PTB2 located within the lumen (Fig. 3A, Supplementary Fig. 7). Artificial intelligence (AI)-assisted map analysis33 and model building allowed us to assign two additional copies of PTB2 within these extra densities organized parallel to the open lateral gate within the OM (Fig. 3B). The first membrane-embedded macrocycle, PTB2-lg-1 (PTB2-lateral gate-1), is well-defined and forms backbone-mediated hydrogen-bonds along the β1-strand of BamA, as if to mimic engagement of a β-signal motif from a native substrate15 (Fig. 3B). When bound to BAM at the open lateral gate in a transmembrane environment, PTB2-lg-1 is folded into a β-hairpin-like structure (Fig. 3B, C). Remarkably, the local folding of PTB2-lg-1 and PTB2 bound within the lumen (PTB2-lumen) differ substantially. For example, the Phe1-Trp12-Arg10 side chains which form a hydrophobic core in PTB2-lg-1 rearrange, whereby the Phe1 (F1) side chain becomes wedged in-between the Trp12 (W12) indole ring and the aliphatic portion of Arg10 (R10) to form a different hydrophobic core structure in PTB2-lumen (Fig. 3C). The ability of PTB2 to assume at least two distinct folded conformations is likely dictated, in part, by the local environment: being submersed within the lipid bilayer when bound to the lateral gate (PTB2-lg-1) or solvent-exposed when bound within the lumen of BamA (PTB2-lumen). Although its density is less well-defined, a second membrane-embedded macrocycle (PTB2-lg-2) is bound along the edge of PTB2-lg-1 distal to the BamA lateral gate (Fig. 3A, B). PTB2-lg-2 appears to form backbone-mediated interactions with PTB2-lg-1 and inserts into the membrane with an inverted orientation relative to PTB2-lg-1, resulting in an asymmetric PTB2-lg-1/PTB2-lg-2 dimeric structure that tapers into the membrane bilayer (Fig. 3A, B).
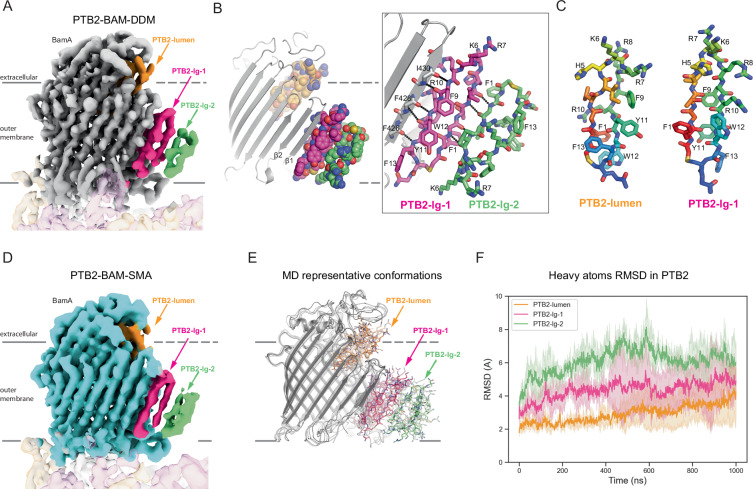
Fig. 3. PTB2 occupies the lateral gate of BamA.
A Cryo-EM map of the PTB2–BAM–DDM complex. PTB2-lumen bound in the BamA β-barrel lumen is shown in orange and the two PTB2 molecules bound at the lateral gate of BamA are shown in pink (PTB2-lg-1) and green (PTB2-lg-2). BamA is shown in gray and the approximate boundaries of the outer membrane are indicated. B Model and interactions between the two PTB2 molecules bound at the lateral gate of BamA. PTB2-lumen bound in the BamA β-barrel lumen (orange) and the two macrocycles bound at the later gate, PTB2-lg-1 (pink) and PTB2-lg-2 (green), are shown. C Comparison of the PTB2 macrocycles bound in the BamA β-barrel lumen (PTB2-lumen) and at the lateral gate of BamA (PTB2-lg-1). D Cryo-EM map of the PTB2–BAM–SMA complex. Three bound PTB2 molecules are shown bound in the BamA β-barrel lumen (PTB2-lumen, orange) and at the BamA lateral gate, PTB2-lg-1 (pink) and PTB2-lg-2 (green), are shown. BamA is shown in cyan and the approximate boundaries of the outer membrane are indicated. E Representative molecular dynamics (MD) conformations of PTM2–BAM–DDM from simulation frames sampled every 250 ns. BamA is shown in gray, PTB2 bound in the BamA β-barrel lumen in orange, PTB2-lg-1 in pink, and PTB2-lg-2 in green. The approximate boundaries of the outer membrane are indicated. MD input–output files in Source Data. F Heavy atoms RMSD of the three PTB2 macrocycles bound to BamA in the PTB2–BAM–DDM complex over time from molecular dynamics (MD) simulations. Data with PTB2-lumen, PTB2-lg-1, and PTB2-lg-2 are colored as shown in Fig. 1E. An average of three independent runs is plotted and MD input–output files are provided in Source Data.
To evaluate the structure of BAM in a native membrane environment, we extracted and purified BAM from E. coli cells using styrene maleic acid (SMA)-copolymer technology (Supplementary Fig. 8a, b). When purified in the presence of PTB2 and visualized to ~3.3 Å resolution (Supplementary Fig. 8c–h, Supplementary Table 1), the PTB2–BAM–SMA complex revealed strong extra map features adjacent to the β1 lateral gate, establishing that PTB2-lg-1 and PTB2-lg-2 can partition into a native outer membrane and interact with BAM (Fig. 3D). Beyond very slight adjustments within the ECL1 and ECL2 loops that occur to accommodate interactions with PTB2, the PTB2–BAM–SMA and PTB2–BAM–DDM complex structures are both highly similar to the apo-BAM-SMA/DDM complex that we have also determined (RMSD < 0.7 Å over all residues; Supplementary Figs. 9a–h and 10a–h, Supplementary Table 1), demonstrating that PTB2 can target and trap an open conformation of the BAM complex that may exist within the native outer membrane.
Molecular dynamics studies of the PTB2–BAM complex revealed that the overall system remained stable over ~1 µs (Fig. 3E, F, Supplementary Fig. 11a–c). The conformations of BamA and PTB2-lumen remained most stable throughout the simulation, whereas slightly larger conformational fluctuations were observed for PTB2-lg-1, while PTB2-lg-2 was the most dynamic within the system (Fig. 3E, F). The hydrogen bond interactions between BamA and PTB2-lg-1 were maintained throughout the entire simulation (Supplementary Fig. 11c), highlighting how non-native ligands might remain stably bound to the open lateral gate of the BAM complex.
Crystallization traps an inverted non-physiological BamA substrate
We determined a ~3 Å co-crystal structure of the isolated BamA β-barrel in complex with PTB2, which revealed PTB2 bound only within the central lumen (PTB2-lumen) (Fig. 4A; Supplementary Table 2, Supplementary Fig. 12). Unexpectedly, we observed a symmetry-related double-barrel structure where a 16-stranded inverted non-substrate BamA (ins-BamA) is bound alongside the BamA β1-strand of the open lateral gate (Fig. 4A; Supplementary Table 2, Supplementary Fig. 12). We recognize that this double-barrel complex is non-physiological and a consequence of crystallization, but nonetheless find that it provides interesting observations about BamA. When in complex with PTB2-lumen and ins-BamA, β1 is separated relative to the adjacent periplasmic turn 7 by ~10 Å at the base of the lateral gate, which is significantly wider than the open lateral gate observed in our PTB2-BAM cryo-EM complex structures (Fig. 2E). This suggests that PTB2 binding does not formally restrict widening of the lateral gate. Examination of PTB2-bound structures does demonstrate that the presence of PTB2-luman would sterically clash with the walls of the β-barrel in the closed conformation of BamA, indicating that PTB2-bound structures are not compatible with substantial closure of the BAM lateral gate. Overall, this structural analysis may suggest a mechanism whereby PTB2 prevents conformations of BAM required for the OMP folding cycle7,10,16.
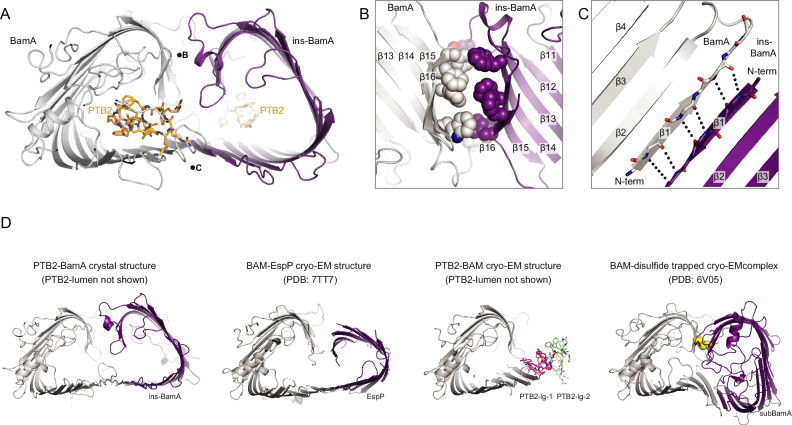
Fig. 4. Crystal structure of the open lateral gate PTB2–BamA complex.
A Overall arrangement of two PTB2–BamA complexes observed within the X-ray crystal lattice. One BamA molecule is named ins-BamA (purple), and the other BamA is shown in gray. The PTB2 macrocycle is shown in orange. Positions of the views shown in panels B and C are noted with the corresponding letter. B Close-in view of the BamA (gray)/ins-BamA (purple) β14–β16/β14–β16 interaction interface. C Close-in view of the BamA (gray)-ins-BamA (purple) β1–β1 interaction interface. D Comparison of the crystal structure of PTB2-BamA (left, BamA in gray and ins-BamA in purple), the cryo-EM structure of BAM–EspP complex8 (middle-left, BamA in gray and EspP in purple), the cryo-EM structure of PTB2-BAM (middle-right, BamA in gray, PTB2-lg-1 shown in pink and PTB2-lg-2 shown in green), and the cryo-EM structure of BAM–disulfide trapped complex7 (right, BamA in gray, subBamA in purple, and disulfide in yellow).
A conglomerate of aromatic and aliphatic side chains forms numerous van der Waals and surface complementarity interactions across β14–β16 between BamA and ins-BamA, highlighting the characteristic aromatic belts found within OMPs3,34 (Fig. 4B). These large interfaces may stabilize the double-barrel complex, suggesting how BamA might chaperone late-stage folding intermediates. Moreover, both ends of the double-barrel structure are engaged in complementary protein–protein interactions (Fig. 4B), which may restrict membrane lipids from entering the central, solvent-accessible lumen during the late stages of folding.
The PTB2-BamA crystal structure reveals extensive backbone-mediated β1–β1 strand interactions between BamA and ins-BamA at the open lateral gate (Fig. 4C). Although the topology of the ins-BamA is inverted with respect to the outer membrane and non-physiological, this observation highlights the ability of the lateral gate to engage diverse β-strand-containing ligands. This finding, together with the visualization of PTB2-lg-1, may suggest avenues for future inhibitor design strategies aimed at targeting the open lateral gate of BAM.
The luminal binding site is required for PTB2 inhibition
We next considered the contribution of PTB2-lumen and PTB2-lg-1 to the mechanism of BAM inhibition. The following observations collectively point to PTB2-lumen as a critical determinant of PTB2 inhibitory activity. First, Klebsiella and Enterobacter BamA have identical sequences at the lateral gate when compared to E. coli BamA (Supplementary Fig. 3), and yet neither species is inhibited by PTB2 (Fig. 2A). Second, Klebsiella and Enterobacter BamA have mutations within their β-barrel lumen compared to E. coli BamA (Supplementary Fig. 3), suggesting that they are naturally resistant to PTB2-lumen binding. Finally, all PTB2 resistance mutations in E. coli BamA were identified at sites expected to directly disrupt PTB2-lumen binding, but no PTB2 resistance mutations were isolated at the lateral gate.
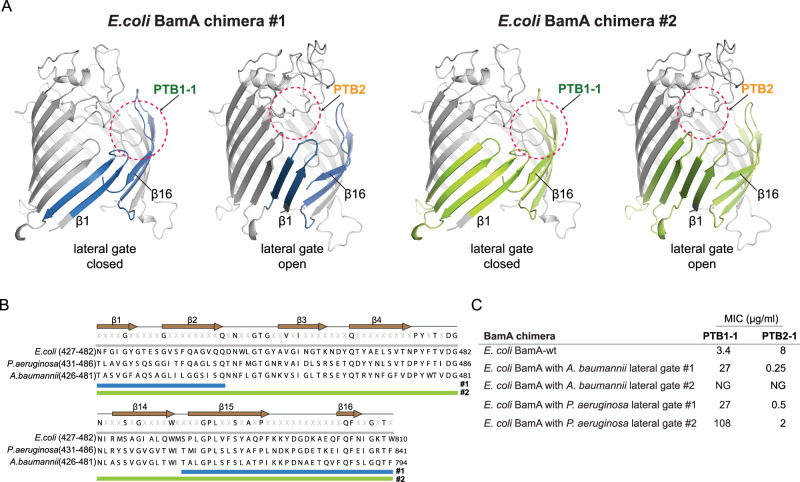
To formally evaluate the role of PTB2-lumen, we generated chimeras between E. coli bamA and bamA from more divergent PTB2-insensitive species (i.e., Acinetobacter and Pseudomonas; Fig. 2A), where significant sequence differences are found at the respective BamA lateral gates (Fig. 5B, Supplementary Fig. 3). We generated two chimeric construct designs, the first (chimera #1) substituted the β1–β2 and β15–β16 hairpin regions of the lateral gate of E. coli BamA for the equivalent reagents in these non-PTB2-sensitive strains; and the second (chimera #2) substituted the β1–β4 and β14–β16 hairpin regions of the lateral gate of E. coli BamA. Importantly, both chimeric construct designs alter the lateral gate region but maintain the residues observed in the cryo-EM structure expected to coordinate the PTB2-lumen macrocycle. The chimera #1 BamA proteins supported the growth of E. coli, as did the Pseudomonas–E. coli chimera #2, demonstrating that these proteins are functionally competent. The Acinetobacter–E. coli chimera #2 failed to support growth, demonstrating that there appear to be sequences within this region of the E. coli BamA protein that are required to support sufficient function.
Fig. 5. E. coli BamA lateral gate chimeras remain sensitive to PTB2 inhibition.
A The lateral gate of the E. coli BamA barrel was substituted with the lateral gate sequence from A. baumannii or P. aeruginosa. As described in the main text, chimera #2 (green, right) has more extensive substitutions compared to chimera #1 (blue, left), and all swapped regions are highlighted in the context of our PTB1-1–BAM structure (left: closed lateral gate) and PTB2-BAM structure (right: open lateral gate), respectively. The approximate binding site location of PTB1-1 and PTB2 are circled with pink dashed lines. B Sequence alignments of BamA from E. coli, P. aeruginosa, and A. baumannii in the regions encompassing chimeras #1 (blue bar) and #2 (green bar) substitutions. C Minimal Inhibitory Concentrations (MICs) for BamA-binding macrocycles of PTB1-1 and PTB2-1 with E. coli strains producing the indicated BamA chimeras. MIC assays were performed in triplicate with mean values reported. Note, the more extensive chimera #2 with the A. baumannii sequence failed to grow under the assay conditions. NG no growth.
We next evaluated the pharmacology of the PTB1-1 and PTB2-1 macrocycles against the chimeric BamA proteins that were able to support E. coli growth. All the chimeric strains become less sensitive to PTB1-1 (Fig. 5C), confirming that our chimeric protein designs significantly alter the BamA closed lateral gate sequence and PTB1-1 binding site. By contrast, all chimeric strains remain sensitive to PTB2-1 growth inhibition (Fig. 5C), consistent with our construct design strategy whereby these chimeric proteins maintain the residues observed in the cryo-EM structure that coordinate the PTB2-lumen macrocycle (Fig. 5A, B). Overall, these results establish that the lumen binding site is required for PTB2 inhibition. However, an inhibitory role for PTB2-lg-1 binding at the lateral gate cannot be formally ruled out.
Discussion
We employed an in vitro macrocycle selection platform using different purified membrane protein constructs presented in distinct membrane mimetic formulations to identify bactericidal peptides that trap the BAM complex in extreme conformational states. These efforts allowed us to identify two series of BamA inhibitors that have distinct binding sites compared to previously structurally characterized BAM antagonists (Supplementary Figs. 13b, 14)17,23,24. Therefore, the logic of our approach may represent a generalizable strategy to present distinct conformational states of integral membrane proteins to enable the prospective in vitro discovery of state-dependent modulators of many important drug targets.
PTB1-1 targets a binding site above the lateral gate where it appears to lock BamA into a closed state (Fig. 1G). This observation suggests that PTB1-1 antagonizes the opening of the lateral gate of BAM, confirming that an open lateral gate is likely required for substrate insertion and folding into the outer membrane7,10,16. The amphipathic nature of PTB1-1 suggests that it may first partition into the outer membrane phase prior to engaging its membrane-proximal extracellular binding site on BamA30. It is also notable that PTB1-1 targets an electronegative surface of BamA in a divalent cation-dependent manner, and that frequent PTB1-resistance mutations occur at this metal coordination site. These key metal-coordinating residues are not present in the extracellular loops of Acinetobacter or Pseudomonas BamA, which rationalizes why PTB1-1 lacks broader spectrum bactericidal activity. The PTB1-1 binding site on BamA is distinct in location and nature from previously identified closed-state inhibitors darobactin, dynobactin, and peptide 3 (Supplementary Figs. 13b,14)17,19,23,24, highlighting the power of our in vitro approach to identify relevant modulatory sites, providing future opportunities for closed-state inhibitor discovery targeting BAM.
The PTB2 macrocycle traps an open lateral gate of the BAM complex by wedging deeply into the vestibule of BamA, defining PTB2 as an open-state inhibitor of BAM with a unique binding site relative to the open-state MAB1 inhibitor31. This result further validates the strength of our discovery approach to identify state-dependent modulators. The amphipathic nature of PTB2 suggests that it may first partition into the outer membrane and access its binding site within the BamA lumen through the open lateral gate directly from the membrane phase30. The membrane-partitioning potential of PTB2 is directly highlighted by the PTB2-lg-1 and PTB2-lg-2 macrocycles, which are observed in a transmembrane orientation alongside the open lateral gate of BAM, embedded within the lipid bilayer. Although the position of PTB2-lg-1 bound at the open BamA lateral gate is highly reminiscent of the binding site targeted by the closed-state BamA inhibitor darobactin17 (Supplementary Fig. 13b), the E. coli-specific spectrum (Fig. 2A), resistance profile (Supplementary Fig. 6e), and results with chimeric BamA strains (Fig. 5C) indicates that PTB2-lumen is critical for antagonizing BAM function. Nevertheless, the experimental observation of diverse occupants bound at the BamA lateral gate, namely PTB2-lg-1 (Fig. 3A, B) ins-BamA (Fig. 4), darobactin17, BamA7, and EspP8, demonstrate the diversity of ligand or substrate coordination that can occur along β-strand 1 of BamA. Beyond implications for β-barrel folding, these observations may provide inspiration for future inhibitor discovery and design considerations targeting the lateral gate of BAM.
PTB2 binding may allow BamA to sample slightly different open conformational states (Supplementary Fig. 13c). If we consider that the BamA–ins-BamA double-barrel complex structure may mimic a late-stage folding intermediate, it is notable that the trajectory of the ins-BamA substrate emanating from the lateral gate is similar to prior BAM complex structures with stalled EspP folding intermediates7,8 (Fig. 4D). It has been proposed that BamA serves to bring the N- and C-terminal β-strands of the folding substrate into proximity during late-stages of folding, where a terminating β-strand complementation event occurs to complete OMP folding and release the mature β-barrel into the outer membrane7,10. Thus, we speculate that PTB2 may prevent the conformations required to achieve this late-stage strand exchange and substrate release by acting as a molecular wedge within BamA. Alternatively, PTB2 might prevent substrate engagement on BAM by steric exclusion (PTB2-lumen) or direct competition (PTB2-lg-1), but determining these potential contributions will require further experimental characterization.
In this study, we performed in vitro selections while intentionally using purified recombinant membrane protein constructs reconstituted into membrane mimetic matrices expected to bias distinct conformational states. This approach allowed us to discover distinct classes of potent and selective macrocycle inhibitors that trap extreme conformational states of BAM and reveal pharmacological concepts for targeting BamA. Notably, and in stark contrast to our findings with MAB118, both the bactericidal PTB1 and PTB2 macrocycle series discovered here can access BamA within the context of an intact LPS-containing outer membrane bilayer. Our structural studies indicate that both PTB1 and PTB2 are amphipathic peptides, a feature that may contribute to their ability to access and engage BAM in the context of the intact bacterial outer membrane bilayer. This may highlight the importance of screening different membrane matrices when pursuing in vitro ligand discovery methods against integral membrane proteins. Overall, in addition to providing tool molecules that may enable future antibiotic discovery targeting BamA, our study provides generalizable concepts to prospectively discover modulators against other dynamic membrane protein targets.
Methods
Expression and purification of BAM, BamA-POTRA3-5, and β-barrel-only of BamA
The E. coli BamABCDE (BAM) expression plasmid was constructed as previously described35. An 8 × histidine tag was inserted at the C terminus of BamE. BAM complex was expressed in E. coli BL21(DE3). Cells were grown in Terrific Broth (TB) and induced with 0.5 mM isopropyl-β-d-1-thiogalactopyranoside at 37 °C when the absorbance of the cell culture at 600 nm reached 0.6. After 4 h of expression, cells were harvested by centrifugation (5422 × g, 20 min). The cell pellet was resuspended (10 mL/g) in lysis buffer A (50 mM Tris, pH 8.0, 300 mM NaCl, 5 mM imidazole, 1 × complete protease inhibitor mixture; Roche), and the resuspended cells were disrupted by passing through a microfluidizer at 10,000 psi twice. The cell lysate was added with 1% dodecyl-β-d-maltopyranoside (DDM; all detergents were purchased from Anatrace) and incubated with stirring overnight at 4 °C. The suspension was ultracentrifuged with a 45Ti rotor (125,171 × g, 1 h, 4 °C). The supernatant was passed over 4 mL Ni-NTA resin (Qiagen) in a disposable column (Bio-Rad Laboratories). Resin was washed with 10CV (column volumes) buffer containing 50 mM Tris, pH 8.0, 300 mM NaCl, 50 mM imidazole, 0.01% DDM and eluted with 5CV buffer consisting of 50 mM Tris, pH 8.0, 300 mM NaCl, 300 mM imidazole, 0.01% DDM. The eluent was purified by a Superdex 200 16/60 column (GE Healthcare) using 50 mM Tris, pH 8.0, 150 mM NaCl, and 0.05% DDM. The yield is around 4 mg/L. For cryoEM sample preparation as described below, the purified BAM complex was further purified over a Superose 6 3.2/300 column (GE Healthcare) using 50 mM Tris, pH 8.0, 150 mM NaCl, and 0.05% DDM.
E. coli BamA-POTRA3-5 (residues 173−810), with an N-terminal AviTag followed by an 8 × histidine tag, was cloned into the pET-52a vector. The expression plasmid was transformed with E. coli BL21(DE3). Cells were grown in TB-auto induction media (17 °C, 72 h). Harvested cells were resuspended in the lysis buffer B (50 mM Tris, pH 8.0, 300 mM NaCl, 1 × complete protease inhibitor mixture; Roche) and were disrupted by passing through a microfluidizer at 10,000 psi twice. The lysate was centrifuged (18,668 × g, 30 min, 4 °C). The pellet was cleaned twice with lysis buffer B and collected by centrifugation. The isolated inclusion body was resuspended in 50 mM Tris pH 8, 6 M Guanidine hydrochloride, and 1 mM Tris (2-carboxyethyl) phosphine hydrochloride (TCEP) and stirred at 4 °C overnight. The denatured sample solution was cleared by centrifugation in a 45 Ti rotor (125,171 × g, 1 h, 4 °C). Clarified supernatant was diluted into the refolding buffer consisting of 50 mM Tris pH 8, 200 mM NaCl, 0.3% Anzergent 3-12 to a final protein concentration of about 0.02 mg/mL, while the refolding solution was stirred at room temperature. Refolded BamA-POTRA3-5 was passed over Ni-NTA resin (Qiagen) in a disposable column (Bio-Rad Laboratories). The resin was washed with buffer containing 50 mM Tris, pH 8.0, 300 mM NaCl, 25 mM imidazole, 0.3% Anzergent 3-12 and eluted with buffer consisting of 50 mM Tris, pH 8.0, 300 mM NaCl, 300 mM imidazole, 0.3% Anzergent 3-12. The eluent was applied onto a Superdex 200 16/60 column (GE Healthcare) in a buffer consisting of 25 mM Tris, pH 8.0, 150 mM NaCl, and 1.5% octyl-β-d-glucopyranoside (OG). Purified BamA-POTRA3-5 was used in mRNA-display selections.
β-barrel domain of E. coli BamA (residues 426–810) was cloned into the pET-52b vector without signal peptide and affinity tags. The isolation, solubilization, and denaturation of the inclusion body were the same as BamA-POTRA3-5. The refolded β-barrel domain of BamA was applied to a Mono Q column (GE Healthcare) which was equilibrated with 50 mM Tris pH 8, 0.3% Anzergent 3-12. After washing with 10 column volumes of 50 mM Tris pH 8, 0.6% tetraethylene glycol monooctyl ether (C8E4), the protein was eluted with a buffer consisting of 25 mM Tris pH 8.0, 0.6% C8E4, 1 M NaCl. The peak fractions were concentrated and applied onto a Superdex 200 16/60 column (GE Healthcare) in 25 mM Tris pH 8.0, 150 mM NaCl, 0.6% C8E4.
Purification of PTB2–BAM complex in native nanodiscs
During the whole purification process, 2 µM PTB2 was added. The cell pellet of BAM complex, which was expressed in E. coli was resuspended in the lysis buffer (50 mM Tris, pH 8.0, 500 mM NaCl, 5 mM Imidazole, 10% glycerol and 1 × complete protease inhibitor mixture; Roche). The resuspended cells were lysed with an ultrasonicator (BRANSON Digital Sonifier) for 4 min (1 s ON/6 s OFF). The lysate was added with a final 2.5% (w/v) SMALP-200 (POLYSCOPE). After mixing on a Vortex Mixer at room temperature for 1 h, the mixture was homogenized using a dounce homogenizer and mixed for another 1 h at room temperature. The protein solution was ultracentrifuged (125,171 × g, 1 h, 4 °C), and the supernatant was diluted 1:10 with the lysis buffer. The diluted protein solution was incubated with the cobalt resin (Takara Bio) at 4 °C overnight. The protein–resin solution was passed through a disposable column (Bio-Rad Laboratories). The resin was washed with 10 CV washing buffer (50 mM Tris, pH 8.0, 500 mM NaCl, 20 mM Imidazole) and eluted with 10CV elution buffer (50 mM Tris, pH 8.0, 500 mM NaCl, 300 mM Imidazole). The eluted protein was concentrated and applied to a Superose 6 3.2/300 column (GE Healthcare) using 50 mM Tris, pH 8.0, and 150 mM NaCl. The peak fractions were used for cryoEM sample preparation as below. The apo-BAM–SMA complex was purified exactly as described above, except PTB2 was omitted from all steps.
Biotinylation of BAM, BamA-POTRA3-5, and β-barrel-only of BamA
For specific labeling of BAM, an AviTag following an 8 × histidine tag was inserted into the C-terminal of BamE. The biotinylation of BAM and BamA-POTRA3-5 (residues 173–810) was performed using a BirA-500 biotin–protein ligase reaction kit (Avidity LLC) according to the manufacturer’s protocol. The chemical biotinylation of the untagged β-barrel domain of BamA (residues 426–810) was accomplished using EZ-Link™ NHS-PEG4-Biotin (Thermo Fisher Scientific) as described in the manufacturer’s protocol. After the reaction, labeled proteins were further polished by a Superdex 200 10/300 column (GE Healthcare) in a buffer consisting of 25 mM Tris, pH 8.0, 150 mM NaCl, and 1.5% OG to remove excess BirA enzyme or non-reacted NHS-PEG4-Biotin.
Macrocyclic peptide library design
A thioether-macrocyclic peptide library was constructed by using N-chroloacetyl l-phenylalanine (ClAc-F) as an initiator in a genetically reprogrammed in vitro translation system36,37. The two genetic codes, NNW and NNU, were designed. The NNW code contains all 18 natural amino acids except for Met and Cys. The NNU code contains four N-methyl amino acids: N-methyl-l-phenylalanine (MeF), N-methyl-l-glycine (MeG), N-methyl-l-norleucine (MeNle), and N-methyl-l-alanine (MeA) in addition to 11 natural amino acids (Ser, Tyr, Trp, Leu, Pro, His, Arg, Asn, Val, Asp, and Gly). The two mRNA libraries, referred to as NNW or NNU library, were designed to have an AUG (ClAc-F) initiator codon followed by 8–11 NNW or NNU codons, which code random amino acid residues, followed by a fixed UGG codon that assigns Cys and a sequence coding a G4S2 peptide linker. After in vitro translation, a thioether bond formed spontaneously between the N-terminal ClAc group of the initiator l-phenylalanine residue and the sulfhydryl group of a downstream cysteine residue to generate the macrocyclic peptides.
Semi-random libraries for PTB1 and PTB2 were designed such that the parental amino acids at each position can be changed to ones with similar side chain properties. The codon table for PTB1 semi-random library contains 18 non-canonical amino acids (F4C, Nal1, Bph, W5C, F3F, MeHis, 1 Pa, H3M, Abu/Ab, Aib, d-Ala, Ahp, MeTyr/mY, MeTrp, MePhe, MeSer, MeNle, and MeGly) in addition to 12 natural amino acids (Phe, Trp, Tyr, Lys, Arg, His, Val, Ile, Ser, Gly, Glu, and Asp); the codon table for PTB2 to contain 15 non-canonical (F4C, Nal1, Bph, W5C, F3F, MeHis, 1 Pa, H3M, Abu/Ab, Aib, Ahp, MePhe, MeSer, MeNle, and MeGly) in addition to 11 natural amino acids (Phe, Trp, Tyr, Lys, Arg, His, Val, Ile, Ser, Gly, and Thr). The two mRNA libraries were designed to have a ClAc-F initiator codon followed by a semi-randomized region, a Cys residue, and a G4S2 linker.
Selection of BamA and BAM-binding molecules
Affinity selection of macrocyclic peptides binding to BamA or BAM was performed using E. coli BamA-biotin or BAM-biotin solubilized in 1.5% n-Octyl-β-d-glucopyranoside (OG, Anatrace). Briefly, the 10 mM mRNA library (containing 1014 unique peptides) was hybridized with a peptide-linker (11 mM) at RT for 3 min. The mRNA library was translated at 37 °C for 30 min in the reprogrammed in vitro translation system to generate the peptide–mRNA fusion library27,37. Each reaction contained 2 mM mRNA–peptide-linker conjugate, 12.5 mM initiator tRNA (tRNAfMet aminoacylated with ClAc-l-Phe), and 25 mM of each elongator tRNA aminoacylated with the specified non-canonical/canonical amino acids. In the first round of selections, translation was performed at a 100 mL scale. After the translation, the reaction was quenched with 17 mM EDTA. The product was subsequently reverse-transcribed using RNase H minus reverse transcriptase (Promega) at 42 °C for 30 min, and buffer was exchanged for HBS-OG buffer: 25 mM HEPES–NaOH (pH 7.4), 150 mM NaCl, 1.5% OG. For affinity selection, the peptide-mRNA/cDNA solution was incubated with 250 nM biotinylated E. coli BamA or BAM for 60 min at 4 °C and the streptavidin-coated beads (Dynabeads M-280 Streptavidin, Thermo) were further added and incubated for 10 min to isolate binders. The beads were washed three times with cold HBS-OG buffer, the cDNA was eluted from the beads by heating for 5 min at 95 °C, and fractional recovery from the affinity selection step was assessed by quantitative PCR using Sybr Green I on a LightCycler thermal cycler (Roche). After eight rounds of affinity maturation, two additional rounds of off-rate selections were performed by increasing the wash stringency before elution to identify high-affinity binders. Sequencing of the final enriched cDNA was carried out using a MiSeq next-generation sequencer (Illumina).
Binding ELISA
Biotinylated BamA or BAM was immobilized on streptavidin-coated plate (Nunc) by incubating 60 nM of protein solutions for 0.5 h at room temperature. After washing the plate, 1 mL of in vitro translated FLAG-tagged peptides were incubated with 50 mL HBS-OG in the plate for 1 h. After washing by HBS-OG (300 mL, 3 times), the plate was incubated with anti-Flag-HRP antibody (Monoclonal ANTI-FLAG® M2-Peroxidase (HRP) antibody produced in mouse, Sigma) for 0.5 h. Color development was achieved by adding TMB substrate (Sera Care, USA), and the reaction was stopped by adding an equal volume of TMB stop solution (Sera Care, USA). Absorbances were recorded at OD 450 using the microplate reader EnSpire (Perkin).
Synthesis of PTB1, PTB2 and derivatives
Thioether macrocyclic peptides were synthesized using standard Fmoc solid phase peptide synthesis (SPPS). Following the coupling of all amino acids, the deprotected N-terminus was chloroacetylated on-resin followed by global deprotection using a trifluoroacetic acid (TFA) deprotection cocktail. The peptides were then precipitated from the deprotection solution by adding over 10-fold excess diethyl ether. Crude peptide pellets were then dissolved and re-pelleted 3 times using diethyl ether. After the final wash, the pellet was left to dry, and then the pellet was resuspended in DMSO, followed by the addition of triethylamine for intramolecular cyclization via the formation of a thioether bond between the thiol of the cysteine and N-terminal chloroacetyl group. Upon completion of cyclization, the reaction was quenched with AcOH, and the cyclic peptide was purified using standard reverse-phase HPLC methods. The molecular masses were confirmed by single quadrupole LC/MS (LCMS-2020 systems, Shimadzu).
Complex formation of BAM-MAB2-Fab
Preparation of Fab from MAB2 (a non-functional anti-BamA antibody18), was prepared by standard protocols. Recombinant E. coli BAM was incubated with a molar excess of MAB2 Fab on ice for 1 h. The formed complex was purified over a Superose 6 10/300 GL column (GE Healthcare) equilibrated in 50 mM Tris pH 8.0, 150 mM NaCl, and 0.05% DDM. The peak fractions were collected, concentrated, and further purified over a Superose 6 3.2/300 column (GE Healthcare) using 50 mM Tris, pH 8.0, 150 mM NaCl, and 0.05% DDM. The peak fractions were used for complex formation with PTB1 for cryo-EM sample preparation, as described below.
EM sample preparation and imaging
For the apo structure of BamABCDE (BAM) complex prepared in DDM detergent (apo BAM-DDM), Au substrate Quantifoil (Quantifoil GMBH) cryo-EM grids with hole diameter/spacing of 0.6/1.0 µm with 25 nm-thick Au foil were incubated with a thiol reactive, self-assembling reaction mixture of 4 mM monothiolalkane(C11)PEG6-OH (11-mercaptoundecyl) hexaethyleneglycol (SPT-0011P6, SensoPath Technologies, Inc., Bozeman, MT) to improve sample behavior38. Grids were incubated with this self-assembled monolayer (SAM) solution for 24 h. Prior to grid freezing, grids were removed from the SAM solution and rinsed with EtOH. 3 µL of BAM complex (4-5 mg/mL) was applied to grids. Grids were then blotted for 3 s and plunged into liquid ethane, using the Leica Microsystems automatic plunge freezer (EM GP2, Leica Microsystems, Buffalo Grove, IL). For the PTB1-1–BAM structure, the MAB2 antibody fragment, which has no functional impact on BAM activity18 or PTB1 series pharmacology (and was included in our initial experiments as a fiducial marker), was included, and the BAM–MAB2 Fab complex (4–5 mg/mL) was incubated with PTB1-1 macrocycle at 1:2 molar ratio on ice for 30 min. The sample was applied to Ultrafoil R0.6/1.0 (300 mesh) cryo-EM grids (Quantifoil GMBH) which have been plasma cleaned using the Solarus plasma cleaner (Gatan, Pleasanton, CA) and plunge frozen as above. For the PTB2-BAM-DDM structure, apo BAM-DDM (4–5 mg/mL) was incubated with PTB2 macrocycle at a 1:2 molar ratio on ice for 1 h. The sample was applied to grids as above and plunge frozen. For apo BAM-SMA and PTB2-BAM-SMA structures, the sample of apo BAM-SMA or BAM-SMA co-purified with PTB2 was diluted as a final concentration of 1–4 mg/mL. The sample was applied to Ultrafoil R1.2/1.3 (300 mesh) cryo-EM grids (Quantifoil GMBH), previously glow discharged in Solarus plasma cleaner (Gatan, Pleasanton, CA). The grids were plunge-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher).
Apo BAM-DDM and PTB2-BAM-SMA were collected using SerialEM39 on a Titan Krios (Thermo Fisher Scientific, Waltham, MA) operated at 300 keV and equipped with a K3 direct electron detector with BioQuantum energy filter or a Falcon 4 with a Selectris, respectively. PTB1-1-BAM and PTB2-BAM-DDM were collected on a Titan Krios (Thermo Fisher Scientific, Waltham, MA) operated at 300 keV and equipped with a K2 direct electron detector. The apo Bam-SMA sample was collected on a Glacios electron microscope (Thermo Fisher Scientific, Waltham, MA) operated at 200 keV and equipped with a K2 direct electron detector camera (Gatan Inc., Pleasanton, CA). Full data collection parameters for each sample are shown in Supplementary Table 1.
Cryo-EM data processing
Image processing was performed using cisTEM40, RELION41, and cryoSPARC42. For PTB1-1-BAM, cisTEM was first used for motion correction of all raw images, CTF estimation as well as particle picking. For cisTEM processing, images were resampled by Fourier cropping from an original pixel size of 0.849–1.2 Å. 1,360,976 particles from 18,078 micrographs which were collected from grid 1 were subsequently put through multiple rounds of reference-free 2D classification. An ab initio 3D reference model was generated within cisTEM, followed by auto-refine. The resulting 3D map was used for the following 3D classifications in cisTEM. The particle projections from 1 out of 6 classes were subjected to further auto refinement. Subsequently, 18,078 micrographs collected from grid1 together with 20,078 micrographs collected from grid 2 were reprocessed and picked using Warp43. The selected 929,167 particles by Warp were subjected to two rounds of 2D classifications in RELION. After removing particles in the junk classes, the remaining 717,112 particles were used for the following 3D Classification in RELION using 3D reconstruction obtained from cisTEM refinements. A refined set of 124,444 particles from 1 out of 7 classes was imported to cryoSPARC and further refined using non-uniform refinement44.
Apo BAM-DDM, PTB2-BAM-DDM, Apo-BAM-SMA, and PTB2-BAM-SMA were processed using the same workflow. CryoSPARC was first used for motion correction of all raw movies, CTF estimation, and particle picking. 1162k/493k/2889k/2002k particles were picked from 9684/17,756/3596/18,319 micrographs, respectively. Instead of performing 2D classification, all sorting was done in 3D by generating 5 ab initio references and using them for supervised 3D classification. This was repeated 4 times (3 times in the case of Apo-BAM-DDM) to obtain a homogenous set of particles. In the first sorting iteration for PTB2-BAM-SMA, the ab initio algorithm did not produce a reference matching the complex of interest due to a particularly heterogenous initial particle set, and we chose to use a similar reference from PTB2-BAM-DDM as a 6th reference for 3D classification. Maps were low-pass filtered to 40 Å before each step to avoid high-resolution reference bias. Particles from each homogenous set were refined globally using non-uniform refinement in cryoSPARC. Finally, focused refinement on 3 separate parts of each complex was performed to better account for flexibility, and the resulting maps were recombined to produce the final maps. Global resolution of the composite maps was estimated using gold-standard Fourier shell correlation (FSC = 0.143), and local resolution was determined using windowed FSC calculation in RELION.
Both apo BAM-SMA and PTB2-BAM-SMA were also processed in a similar way. cryoSPARC’s Live processing was first used for motion correction, CTF estimation, and particle picking. As for apo BAM-SMA, 1,683,000 particles were picked from 3958 micrographs. And for PTB2-BAM-SMA, 1,091,000 particles were picked from 18,319 micrographs. The particles were then used for ab initio map reconstruction with 5 classes. The resulting maps were merged with previously refined 3D classes from apo BAM-DDM to perform 3D classification, and particles from the best resulting 3D class were selected. This procedure was repeated 3 times. Particles from the best 3D class of the final classification round were then used for 3D refinement.
To analyze PTB2 binding in all PTB2-containing complexes, the intra-membrane region was refined locally. Local 3D refinement was also used for flexible regions, and the results were merged to obtain a composite map for each complex. Global resolution of the composite maps was estimated using gold-standard Fourier shell correlation (FSC = 0.143), and local resolution was determined using windowed FSC calculation in RELION.
Cryo-EM model building
The BAM structure (PDB: 5D0O) with a closed lateral gate was docked as a rigid body into the PTB1-1-BAM-MAB2 Fab cryo-EM map using UCSF Chimera45. The PTB1-1 macrocycle model was built using Coot46. Multiple rounds of model rebuilding or optimization were performed in Coot42 with iterative rounds of real-space refinement performed using Phenix47.
For the apo BAM–DDM and PTB2–BAM–DDM complex, the BAM structure (PDB: 5lJO) was used as a template and docked as a rigid body into apo BAM or the PTB2–BAM map. The PTB2 (lumen) macrocycle model was built using Coot46. The PTB2-lg-1 and PTB2-lg-2 macrocycle models were first predicted by ModelAngelo33 and further built manually in Coot. Iterative rounds of real-space refinement in Phenix48 and model building in Coot were performed. Apo BAM–SMA and PTB2–BAM–SMA complexes were built as described above by using apo BAM–DDM and PTB2–BAM–DDM models, respectively. All final models were validated by MolProbity49. Structural comparisons and figures were made with UCSF Chimera45, UCSF ChimeraX50, and PyMOL (The PyMOL Molecular Graphics System, Version 2.4.1 Schrödinger, LLC.). Structure refinement statistics are provided in Supplementary Table 1.
Crystallization, data collection and structure determination of PTB2-BamA
Complex was formed by mixing 20 mg/mL of β-barrel domain of BamA with PTB2 macrocycle at 1:2 molar ratio. The crystals were grown by hanging-drop vapor diffusion at 18 °C in 100 mM Tris–HCl, pH 8.0, 100 mM NaCl, 325 mM sodium acetate, and 21% PEG 400. Crystals were harvested by quick transfer directly into a cryoprotectant solution containing 20% glycerol and flash-cooled in liquid nitrogen. Diffraction data were collected at the Advanced Photon Source Northeastern Collaborative Access Team (APS NECAT) beamline 24-ID-C. Following extensive optimization and crystal screening efforts, X-ray diffraction data from our single best crystal was highly anisotropic (a* = 4.05 Å, b* = 2.70 Å, c* = 2.85 Å); data were integrated and scaled using autoPROC51 including anisotropy correction performed using STARANISO52 (http://staraniso.globalphasing.org/cgi-bin/staraniso.cgi). Initially, the structure was determined by molecular replacement in PHENIX47 using BamA from PDB 5LJO14 as the search model, finding four molecules in the asymmetric unit, with clear unassigned extra electron density in both Fo−Fc and 2Fo−Fc maps representing the unmodeled but bound luminal PTB2. When the PTB2-BAM cryo-EM structure became available (described above), this model was subsequently used as the starting template for molecular replacement due to the high quality of the available cryo-EM maps and resulting PTB2-BamA model. Manual adjustments to the resulting model were performed in Coot46, and iterative rounds of crystallographic refinement using Phenix47 and model building in Coot46 were guided by inspection of omit maps, where strict geometry and secondary structure restraints were applied throughout refinement. Despite efforts to assess non-crystallographic symmetry, potential crystal lattice pathologies, and alternative space groups, crystallographic refinement did not progress beyond Rwork/Rfree 32.5/36.7, potentially due to the anisotropy of the data set; but we did not truncate the resolution of data during refinement. The geometry of the final PTB2-BamA/subPTB2-BamA model was assessed using MolProbity44. Structural figures were prepared with the PyMol software (The PyMOL Molecular Graphics System v.1.8, Schrödinger, LLC., 2015). The data collection and refinement statistics are provided in Supplementary Table 2.
Molecular dynamics simulations system setup, protocol, and analysis
The cryo-EM structure PTB2-BAM complex was first prepared for simulation using the 2021-3 release of Maestro (Schrodinger) (Schrödinger Release 2021-3). The BamA POTRA domains were truncated at residue 422 to reduce the system size. Protein Preparation Wizard53 was used to cap the N- and C-termini with acetyl and N-methyl amide groups, respectively. Protonation states for His, Glu, Asp and conformational flips of His, Asn, Gln side chains were optimized using PROPKA54 at pH 7.4. Restrained minimization was carried out using the OPLS4 force field55 with heavy atoms converging to a root mean square deviation (RMSD) of 0.3 Å. The prepared system was finally embedded into a pre-equilibrated DMPC membrane (with an initial orientation based on the OPM database), solvated with SPC waters and Na+ Cl− ions were added to a 0.15 mM concentration.
Molecular dynamics simulations were carried out using the Desmond suite (Schrödinger 2021-3)56 with OPLS4 force field. Three independent simulations were performed with each undergoing an equilibration stage followed by a 1 µs production run. The membrane relaxation protocol in the Schrödinger software was used to equilibrate the system. Snapshots from each trajectory were saved every 1000 ps during the production phase of the simulation. We performed analysis on the simulation frames after aligning all frames to the initial structure, using the first β-strand of the BamA β-barrel domain for alignment. RMSD and distances between centroids were analyzed using Simulation Event Analysis and figures were generated with PyMOL (Pymol Schrodinger) and seaborn python package57. Input and output files from molecular dynamics simulations are provided in the Source Data.
Growth conditions
Bacterial cultures were grown at 37 °C in Luria-Bertani (LB, Millers Sigma-Aldrich L3522) broth or agar plates, prepared according to the manufacturer’s instructions. When appropriate, media was supplemented with kanamycin (50 μg/mL), carbenicillin (50 μg/mL), chloramphenicol (12.5 μg/mL), hygromycin (200 μg/mL), gentamicin (10 μg/mL) and arabinose (0.2% vol/vol).
Bacterial strains and plasmids
Bacterial strains and relevant primers are listed in Supplementary Table 3.
OmpT fluorescent peptide-substrate cell-based folding assay
The OmpT assay for monitoring BAM activity was performed as described previously with minor modifications14,29,58,59. Bacterial strains were grown in LB to the early log phase and normalized to OD600 0.2 in the growth medium. A 50 μL solution was prepared as follows: 5 μL of bacteria were added to 45 μL fluorogenic peptide, Abz-Ala-Arg-Arg-Tyr(NO2)-NH2, (Peptide Synthesis) diluted into PBS to a final concentration of 50 μM. The mixture was immediately monitored for fluorescence produced on a Spectramax plate reader for 3 h with readings every 2 min (Ex 325 nm, Em 430 nm). The normalized fluorescence was determined by dividing each measurement by the starting measurement.
Minimum inhibitory concentration (MIC) determination
MICs were determined by performing two-fold serial dilutions of peptides in LB broth to a final volume of 0.1 mL in round-bottom 96-well assay plates (Corning Life Sciences No. 3788). Peptides were initially resuspended in 100% DMSO to 10 mM and subsequently diluted in LB medium to the appropriate concentration. Each well was inoculated with 5 × 105 CFU/mL of the screening stain and incubated at 37 °C without agitation for 18 h. Plates were scored by eye, and the lowest compound concentration preventing visible growth was determined to be the MIC.
Time-kill assay
Log-phase bacterial cells were diluted to 106 CFU/mL and incubated with peptides. Viable bacterial cells were measured by CFU plating onto agar medium at the indicated time points.
Frequency of resistance (FOR)
Log-phase bacterial cells were diluted to 1–5 × 107 CFU/mL in LB broth to a final volume of 0.2 mL containing 2–4 × MIC of each peptide in round-bottom 96-well assay plates (Corning Life Sciences No. 3788). A minimum of 300 wells were monitored for bacterial growth after incubation at 37 °C for 2 days. Selections were performed using ratios of bacterial cells to peptide concentration such that <5% of the wells grew up, minimizing the chance of resistant wells containing more than one resistant isolate. Wells with bacterial growth were streaked for isolation and confirmed to be resistant. The frequency of resistance for each strain and peptide was determined by dividing the number of wells that regrew after 2 days at 37 °C by the initial viable cell count.
Red blood cell (RBC) lysis assay
Human blood samples were collected from volunteers (Genentech Samples for Science Program protocols approved by the Western Institutional Review Board (protocol number CEHS-CP 307.2, IRB tracking number 20080040)). Written informed consent was obtained and no personal or medical history was specified, provided, or collected for volunteers. Macrocycles or 0.5% Triton X-100 (100% lysis control) were diluted in PBS in a 96-well clear round bottom plate at two times the final concentration in 60 μL per well. Whole heparinized human blood was diluted to 4% in PBS and 60 μL added to macrocycle dilutions (final blood concentration 2%). Reactions were incubated statically at 37 °C for 4 h. Cells were pelleted by centrifugation at 600 × g for 3 min and 60 μL of supernatant was removed and read on a SpectraMax M5 plate reader (Molecular Devices).
SDS-PAGE, Western immunoblotting and antibodies
Bacterial cells were diluted to OD600 0.01 in growth media with the peptide of interest. Bacteria and peptides were incubated statically for 3 h at 37 °C and pelleted. Samples were resuspended in 1 × LDS sample buffer (Thermo Fisher Scientific) and boiled for 5 min prior to loading on a 4–12% Bis–Tris SDS–PAGE gel. Proteins were transferred onto cellulose membranes using the iBlot 2 Dry Blotting System (Thermo Fisher Scientific). Membranes were blocked for 1 h in Intercept TBS Blocking Buffer (Li-Cor), washed, then incubated either overnight at 4 °C or room temperature for 1 h with the following primary antibodies: rat anti-BamA MAB2 (1 μg/mL, Genentech18), human anti-LptD 3D11 (1 μg/mL, Genentech60), Rabbit anti-MsbA (50 μg/mL, Genentech61), and rabbit anti-GroEL (1:25,000, Enzo). Appropriate IRDye-linked secondary antibodies (Li-Cor) were diluted 1:10,000 in TBST and incubated with the membrane for 1 h at RT. Blots were developed using the Odyssey imaging system (Li-Cor).
Ethidium bromide accumulation assay
Ethidium bromide accumulation was measured as previously described62. Bacterial strains were grown to log phase, washed in PBS and resuspended to OD600 0.2. An aliquot of 180 μL of cells was added to a 96-well black flat-bottom plate (Costar). Next, 20 μL of ethidium bromide (100 μM) was added to the cells and PBS controls to a final concentration of 10 μM. The plate was incubated at 37 °C for 2 h. Fluorescence was read (Ex 515 nm, Em 600 nm).
Surface plasmon resonance (SPR)
For data shown in Supplementary Fig. 1c, the binding constants between the selected macrocyclic peptides and BamA or BAM were measured by SPR analysis by using a BIAcore T200 instrument (Cytiva) as part of our high-throughput macrocycle discovery approach. The HBS-N buffer (10 mM HEPES (pH 7.4), 150 mM NaCl, Cytiva) containing 1% DMSO and 1.5% OG was used as running buffer. Biotinylated BamA or BAM was immobilized on a sensor chip surface through biotin-streptavidin interactions by using a Biotin CAPture kit (Cytiva). Macrocyclic peptides binding to BamA and BAM were measured by injecting five concentrations of each macrocyclic peptide at a flow rate of 30 mL/min at 25 °C. Binding constants were quantified by a single-cycle kinetics method and all data were fitted to a standard 1:1 binding model in order to monitor binding, but these were not utilized for quantitative binding analyses.
For data shown in Fig. 1I, the BamA β-barrel construct was chemically biotinylated as described above to enable coupling to a streptavidin-coated sensing surface. A series S SA sensor chip was docked in a Biacore T200 and the analysis temperature was set to 20 °C. The system was equilibrated in running buffer (50 mM HEPES, 150 mM NaCl, 1 mM ZnCl2, 1.5% (w/v) BG, pH 7.5) and a peptide dilution series was prepared in running buffer containing 1 mM ZnCl2. A second dilution series was prepared in the same running buffer, but with 2 mM EDTA added in order to strip BamA of bound Zn2+. BamA was captured onto two separate sensing regions giving response values of 740RU and 1200RU, respectively. Each sample was injected at 100 μL/min for 100 s in series and the EDTA-containing peptide samples were analyzed before the free ZnCl2-containing peptide samples. The recorded binding-response curves were imported into Biacore S200 Biaevaluation software (Cytiva Inc., Marlborough, NY, USA), double-referenced and fit to a two-state binding-interaction model where interaction parameters were constrained to global values.
Native mass spectrometry
Ammonium acetate, acetic acid, formic acid, and trifluoroacetic acid were purchased from Sigma-Aldrich (St. Louis, MO). Acetonitrile (ACN) was purchased from Fisher Scientific (Hampton, NH).
BamA at 1 mM in 25 mM Tris, pH 8.0, 100 mM NaCl, 1.5% OG was buffer-exchanged using Micro Bio-SpinTM 6 columns (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The column was first centrifuged at 1500 × g at 4 °C to flush the Tris storage buffer and then rinsed four times in 10 mM ammonium acetate (Sigma) by loading the column with 0.5 mL of buffer and centrifuging for 1 min at 1500 × g and at 4 °C. The collection tube was emptied after each spin. The column was then placed in a fresh collection Eppendorf tube, and 25–50 µg sample was added on the resin at the center of the column. The column was then centrifuged for 10 min at 550 × g and at 4 °C. For the cation titration experiments, copper, zinc, magnesium, calcium, or nickel was added to ammonium acetate buffer exchanged BamA protein in increments from 0 to 100 µM. Following cation addition 1 µM of the binding peptide was added. To deplete metal ions during binding evaluations, 1 mM EDTA was added to reactions prior to buffer exchange or at the last step of the reaction following peptide introduction.
Samples were directly infused into an Exactive Plus Extended Mass Range Orbitrap Mass Spectrometer (Thermo-Fisher Scientific, San Jose, CA, USA)63 via nanospray ionization using a TriversaTM Nanomate (Advion, Inc., Ithaca, NY, USA). The instrument was set in EMR MS mode for intact mass analysis. The complex was analyzed under the following acquisition parameters: capillary temp, 250 °C; S-lens RF level, 200; probe heater temp, 350; scan rage, 1500–10,000 m/z; desolvation, in-source CID 120 ev, CE 0; resolution, 17500 at m/z 200; polarity, positive; microscans, 10; AGC target, 3 × 106; maximum injection time, 50 ms; AGC mode, fixed; averaging, 0; source DC offset, 25 V; injection flatapole DC, 8 V; inter flatapole lens, 7 V; bent flatapole DC, 6 V, transfer multipole DC tune offset, 0 V; C-trap entrance lens tune offset, 0 V; trapping gas pressure setting, 2 source dissociation (SID), 200 V. Mass spectrometric data were analyzed using Protein Deconvolution v4.0 software (Thermo-Fisher Scientific, San Jose, CA, USA). The instrument was mass calibrated as described previously using a solution of Cs64. Mass spectrometry data was deposited to MassIVE as detailed in the Data Availability statement.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank our Genentech and PeptiDream colleagues for their support, including A. Estevez, A. Rohou, Y. Franke, E. Brown, S. Hymowitz and A. Chan. We also thank Wilson Phung for assistance with uploading data.
Author contributions
D.S. performed protein purification for selection, crystallization and cryo-EM samples. K.M.S. performed microbiology and related molecular biology experiments with assistance from H.G., M.K.A. and A.K.M and input from P.A.S. and S.T.R. D.S., D.T., M.J., and C.P.A. determined cryo-EM structures with input from Cl.C., D.S. and J.P. determined the crystal structure. J.G.Q. performed SPR experiments. W.L. and G.H. performed native mass spectrometry experiments with input from W.S. S.S. and C.T. generated key protein expression reagents. Y.Y. and B.D.S. carried out molecular dynamics studies. Hiroshi I., Hiroko I., T.S., and H.Y performed selection assays with input and oversight from J.N., P.C.R., and C.N.C. C.E.H., W.J.F., M-W.T. and N.S. provided input on experiments. P.A.S. performed microbiology experiments that initially characterized PTB1 and PTB2. D.S., C.P.A., Cl. C., S.T.R. and J.P. analyzed X-ray and cryo-EM structures. D.S., S.T.R. and J.P. prepared the manuscript with input and assistance from all authors. C.N.C., S.T.R. and J.P. co-supervised the project and are co-corresponding senior authors.
Peer review
Peer review information
Nature Communications thanks Haohao Dong and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The 3D cryo-EM map of PTB1-1-BAM, Apo BAM-DDM, PTB2-BAM-DDM, Apo BAM-SMA and PTB2-BAM-SMA have been deposited into the Electron Microscopy Data Bank [https://www.ebi.ac.uk/emdb/] under accession code EMD-45765, EMD-45764, EMD-45767, EMD-45766, and EMD-45768. The coordinates of PTB1-1-BAM, Apo BAM-DDM, PTB2-BAM-DDM, Apo BAM-SMA and PTB2-BAM-SMA have been deposited in the Protein Data Bank [https://www.rcsb.org/] with accession codes 9CNX, 9CNW, 9CNZ, 9CNY, and 9CO0, respectively. The crystal structure of PTB2-BamA has been deposited in the Protein Data Bank (https://www.rcsb.org/) under accession code 9CO2. Mass spectrometry data have been deposited in MassIVE under accession code MSV000095321 [10.25345/C5QV3CF81]. Source data are provided with this paper.
Competing interests
D.S., K.M.S., D.T., Y.Y., C.P.A., M.J., J.G.Q., W.L., G.H., H.S.G., M.K.A., S.S., C.T., C.E.H., W.J.F., M.W.T., N.S., W.S., B.D.S., Cl.C., P.A.S., C.N.C., S.T.R., and J.P. are or were employees of Genentech Inc. Hiroshi I., Hiroko I., T.S., H.Y., J.N., and P.C.R. are employees of PeptiDream Inc. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Dawei Sun, Kelly M. Storek, Dimitry Tegunov.
Contributor Information
Christian N. Cunningham, Email: C-cunningham@peptidream.com
Steven T. Rutherford, Email: rutherford.steven@gene.com
Jian Payandeh, Email: jpayandeh@exelixis.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52512-1.
References
- 1.Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev.67, 593–656 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomasek, D. & Kahne, D. The assembly of β-barrel outer membrane proteins. Curr. Opin. Microbiol.60, 16–23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koebnik, R., Locher, K. P. & Gelder, P. V. Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol.37, 239–253 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Franklin, M. W. & Slusky, J. S. G. Tight turns of outer membrane proteins: an analysis of sequence, structure, and hydrogen bonding. J. Mol. Biol.430, 3251–3265 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu, T. et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell121, 235–245 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Voulhoux, R., Bos, M. P., Geurtsen, J., Mols, M. & Tommassen, J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science299, 262–265 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Tomasek, D. et al. Structure of a nascent membrane protein as it folds on the BAM complex. Nature583, 473–478 (2020). [DOI] [PMC free article] [PubMed]
- 8.Doyle, M. T. et al. Cryo-EM structures reveal multiple stages of bacterial outer membrane protein folding. Cell185, 1143–1156.e13 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiao, L. et al. Structures of the β‐barrel assembly machine recognizing outer membrane protein substrates. FASEB J.35, e21207 (2021). [DOI] [PubMed] [Google Scholar]
- 10.Shen, C. et al. Structural basis of BAM-mediated outer membrane β-barrel protein assembly. Nature617, 185–193 (2023). [DOI] [PubMed]
- 11.Gu, Y. et al. Structural basis of outer membrane protein insertion by the BAM complex. Nature531, 64–69 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Han, L. et al. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat. Struct. Mol. Biol.23, 192–196 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Bakelar, J., Buchanan, S. K. & Noinaj, N. The structure of the β-barrel assembly machinery complex. Science351, 180–186 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iadanza, M. G. et al. Lateral opening in the intact β-barrel assembly machinery captured by cryo-EM. Nat. Commun.7, 12865–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robert, V. et al. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol.4, e377 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Noinaj, N., Kuszak, A. J., Balusek, C., Gumbart, J. C. & Buchanan, S. K. Lateral opening and exit pore formation are required for BamA function. Structure22, 1055–1062 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaur, H. et al. The antibiotic darobactin mimics a β-strand to inhibit outer membrane insertase. Nature593, 125–129 (2021). [DOI] [PubMed]
- 18.Storek, K. M. et al. Monoclonal antibody targeting the β-barrel assembly machine of Escherichia coli is bactericidal. Proc. Natl Acad. Sci. USA115, 3692–3697 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luther, A. et al. Chimeric peptidomimetic antibiotics against Gram-negative bacteria. Nature576, 452–458 (2019). [DOI] [PubMed]
- 20.Kaur, H. et al. Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach. J. Biomol. NMR73, 375–384 (2019). [DOI] [PubMed]
- 21.Hart, E. M. et al. A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier. Proc. Natl Acad. Sci. USA24, 201912345–10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagan, C. L., Wzorek, J. S. & Kahne, D. Inhibition of the β-barrel assembly machine by a peptide that binds BamD. Proc. Natl Acad. Sci. USA112, 2011–2016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imai, Y. et al. A new antibiotic selectively kills Gram-negative pathogens. Nature576, 459–464 (2020). [DOI] [PMC free article] [PubMed]
- 24.Miller, R. D. et al. Computational identification of a systemic antibiotic for Gram-negative bacteria. Nat. Microbiol7, 1661–1672 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong, H. et al. Structural basis for outer membrane lipopolysaccharide insertion. Nature511, 52–56 (2014). [DOI] [PubMed]
- 26.Iadanza, M. G. et al. Distortion of the bilayer and dynamics of the BAM complex in lipid nanodiscs. Commun. Biol.3, 766 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishizawa, T., Kawakami, T., Reid, P. C. & Murakami, H. TRAP display: a high-speed selection method for the generation of functional polypeptides. J. Am. Chem. Soc.135, 5433–5440 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Morgado, L., Zeth, K., Burmann, B. M., Maier, T. & Hiller, S. Characterization of the insertase BamA in three different membrane mimetics by solution NMR spectroscopy. J. Biomol. NMR61, 333–345 (2015). [DOI] [PubMed] [Google Scholar]
- 29.Storek, K. M. et al. The Escherichia coli β-barrel assembly machinery is sensitized to perturbations under high membrane fluidity. J. Bacteriol.201, 593–15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Payandeh, J. & Volgraf, M. Ligand binding at the protein–lipid interface: strategic considerations for drug design. Nat. Rev. Drug Discov.20, 710–722 (2021). [DOI] [PubMed]
- 31.White, P. et al. The role of membrane destabilisation and protein dynamics in BAM catalysed OMP folding. Nat. Commun.12, 4174 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ni, D. et al. Structural and functional analysis of the β-barrel domain of BamA from Escherichia coli. FASEB J.28, 2677–2685 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Jamali, K. et al. Automated model building and protein identification in cryo-EM maps. Nature628, 450–487 (2024). [DOI] [PMC free article] [PubMed]
- 34.Schulz, G. E. The structure of bacterial outer membrane proteins. Biochim. Biophys. Acta BBA-Biomembr.1565, 308–317 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Roman-Hernandez, G., Peterson, J. H. & Bernstein, H. D. Reconstitution of bacterial autotransporter assembly using purified components. eLife3, 711–720 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kashiwagi, K. & Reid, P. C. Rapid Display Method in Translational Synthesis of Peptide. PCT WO2011049157https://patents.google.com/patent/WO2011049157A1/en?oq=WO2011049157 (2010).
- 37.Goto, Y., Katoh, T. & Suga, H. Flexizymes for genetic code reprogramming. Nat. Protoc.6, 779–790 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Meyerson, J. R. et al. Self-assembled monolayers improve protein distribution on holey carbon cryo-EM supports. Sci. Rep.4, 7084 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol.152, 36–51 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Grant, T., Rohou, A. & Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. Elife7, e35383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheres, S. H. W. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol.180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Tegunov, D. & Cramer, P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods16, 1146–1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods17, 1214–1221 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem.25, 1605–1612 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D Biol. Crystallogr.66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr.66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Afonine, P. V. et al. Real‐space refinement in PHENIX for cryo‐EM and crystallography. Acta Crystallogr. Sect. D74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr.66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci.27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. Sect. D Biol. Crystallogr.67, 293–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tickle, I. J. et al. Anisotropy of the Diffraction Limit and Bayesian Estimation of Structure Amplitudes (Global Phasing Ltd., 2020).
- 53.Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R. & Sherman, W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aid. Mol. Des.27, 221–234 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Søndergaard, C. R., Olsson, M. H. M., Rostkowski, M. & Jensen, J. H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theory Comput.7, 2284–2295 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Lu, C. et al. OPLS4: improving force field accuracy on challenging regimes of chemical space. J. Chem. Theory Comput.17, 4291–4300 (2021). [DOI] [PubMed] [Google Scholar]
- 56.Bowers, K. J. et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. In SC ’06: Proc. 2006 ACM IEEE Conference on Supercomputing (chair: Barbara Horner-Miller)10.1145/1188455 (2006).
- 57.Waskom, M. L. Seaborn: statistical data visualization. J. Open Source Softw.6, 3021 (2021). [Google Scholar]
- 58.Kramer, R. A., Zandwijken, D., Egmond, M. R. & Dekker, N. In vitro folding, purification and characterization of Escherichia coli outer membrane protease OmpT. Eur. J. Biochem.267, 885–893 (2000). [DOI] [PubMed] [Google Scholar]
- 59.Hagan, C. L., Kim, S. & Kahne, D. Reconstitution of outer membrane protein assembly from purified components. Science328, 890–892 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Storek, K. M. et al. Massive antibody discovery used to probe structure-function relationships of the essential outer membrane protein LptD. eLife8, 3002 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alexander, M. K. et al. Disrupting Gram-negative bacterial outer membrane biosynthesis through inhibition of the lipopolysaccharide transporter MsbA. Antimicrob. Agents Chemother.62, 2561–15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coldham, N. G., Webber, M., Woodward, M. J. & Piddock, L. J. V. A 96-well plate fluorescence assay for assessment of cellular permeability and active efflux in Salmonella enterica serovar Typhimurium and Escherichia coli. J. Antimicrob. Chemother.65, 1655–1663 (2010). [DOI] [PubMed] [Google Scholar]
- 63.Rose, R. J., Damoc, E., Denisov, E., Makarov, A. & Heck, A. J. R. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat. Methods9, 1084–1086 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Rosati, S., Yang, Y., Barendregt, A. & Heck, A. J. R. Detailed mass analysis of structural heterogeneity in monoclonal antibodies using native mass spectrometry. Nat. Protoc.9, 967–976 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 3D cryo-EM map of PTB1-1-BAM, Apo BAM-DDM, PTB2-BAM-DDM, Apo BAM-SMA and PTB2-BAM-SMA have been deposited into the Electron Microscopy Data Bank [https://www.ebi.ac.uk/emdb/] under accession code EMD-45765, EMD-45764, EMD-45767, EMD-45766, and EMD-45768. The coordinates of PTB1-1-BAM, Apo BAM-DDM, PTB2-BAM-DDM, Apo BAM-SMA and PTB2-BAM-SMA have been deposited in the Protein Data Bank [https://www.rcsb.org/] with accession codes 9CNX, 9CNW, 9CNZ, 9CNY, and 9CO0, respectively. The crystal structure of PTB2-BamA has been deposited in the Protein Data Bank (https://www.rcsb.org/) under accession code 9CO2. Mass spectrometry data have been deposited in MassIVE under accession code MSV000095321 [10.25345/C5QV3CF81]. Source data are provided with this paper.