Abstract
We previously described a two-component protein nanoparticle vaccine platform that displays 60 copies of the SARS-CoV-2 spike protein RBD (RBD-NP). The vaccine, when adjuvanted with AS03, was shown to elicit robust neutralizing antibody and CD4 T cell responses in Phase I/II clinical trials, met its primary co-endpoints in a Phase III trial, and has been licensed by multiple regulatory authorities under the brand name SKYCovioneTM. Here we characterize the biophysical properties, stability, antigenicity, and immunogenicity of RBD-NP immunogens incorporating mutations from the B.1.351 (β) and P.1 (γ) variants of concern (VOCs) that emerged in 2020. We also show that the RBD-NP platform can be adapted to the Omicron strains BA.5 and XBB.1.5. We compare β and γ variant and E484K point mutant nanoparticle immunogens to the nanoparticle displaying the Wu-1 RBD, as well as to soluble prefusion-stabilized (HexaPro) spike trimers harboring VOC-derived mutations. We find the properties of immunogens based on different SARS-CoV-2 variants can differ substantially, which could affect the viability of variant vaccine development. Introducing stabilizing mutations in the linoleic acid binding site of the RBD-NPs resulted in increased physical stability compared to versions lacking the stabilizing mutations without deleteriously affecting immunogenicity. The RBD-NP immunogens and HexaPro trimers, as well as combinations of VOC-based immunogens, elicited comparable levels of neutralizing antibodies against distinct VOCs. Our results demonstrate that RBD-NP-based vaccines can elicit neutralizing antibody responses against SARS-CoV-2 variants and can be rapidly designed and stabilized, demonstrating the potential of two-component RBD-NPs as a platform for the development of broadly protective coronavirus vaccines.
Subject terms: Protein vaccines, Vaccines
Introduction
SARS-CoV-2 has had a major public health impact since its emergence in late 20191. The rapid development of novel vaccines against the ancestral (Wu-1) strain of SARS-CoV-2 proved an effective public health measure in preventing severe disease and death amongst the vaccinated2,3. However, mutations in the spike of emerging variants has dramatically reduced the serum neutralization potency of infected and immunized individuals against new variants and variants of concern (VOCs)4,5. For example, neutralizing antibody levels against the B.1.351 (β) variant, one of the first VOCs, dropped ~13 and ~7-9 fold in pre-infected and twice-immunized individuals respectively when compared to the ancestral strain6. Since then, there has only been an increase in variants originating from the B.1.1.529 lineage, with the EG.5 sublineage having 42 and 24 mutations in the full-length spike and receptor binding domain (RBD), respectively7–9. Although mRNA vaccines based on the full-length, membrane-anchored spike protein have proven amenable to strain updates2,4,10,11, there is little information on the ability of other vaccine platforms to tolerate antigen updates. This topic merits further investigation as it is relevant to updating current vaccines to address emerging variants, the suitability of various vaccine platforms for pandemic preparedness and response, and understanding the interplay between the spike mutational landscape and vaccine-elicited immune responses.
We previously developed a computationally designed receptor binding domain protein nanoparticle vaccine (RBD-NP) that elicited strong protection against the ancestral strain of SARS-CoV-2 in mice and non-human primates and met its Phase III clinical trial endpoints, leading to licensure in South Korea and the UK as well as an Emergency Use Listing from the WHO12–15. Although we and others have begun to explore mosaic RBD-NPs as broadly protective sarbecovirus vaccine candidates16–18, questions remain about how often SARS-CoV-2 vaccines should be updated to keep pace with emerging variants of concern and the amenability of various vaccine platforms to strain updates.
Here we describe variant updated RBD-NP vaccines based on the ancestral (Wu-1), B.1.351(β), and P.1(γ) lineages benchmarked against prefusion-stabilized (HexaPro) spikes19. We find that although the RBD-NP based upon the ancestral lineage (Wu-1-RBD-NP) was stable over 28 days, RBD-NPs displaying wild-type RBDs from these VOCs exhibited much poorer solution properties. Although introducing stabilizing mutations we previously identified in the linoleic acid binding site20 improved stability and immunogenicity in some cases, our results suggest that additional RBD-stabilizing mutations may be necessary to enable reliable strain updates in this vaccine platform.
Results
To gauge the manufacturability and immunogenicity of VOC vaccines based on the RBD-NP platform, we produced RBD-NPs displaying several different RBDs for immunogenicity studies (all amino acid sequences for novel proteins provided in Supplementary Table 1). All RBD-NP vaccines were based on the two-component self-assembling protein nanoparticle I53-50, which consists of 12 pentameric and 20 RBD-bearing trimeric building blocks for a total of 60 RBDs presented on each nanoparticle12,21. As previously described, we expressed and purified the antigen-bearing components in Expi293F cells and the I53-50B.4PT1 pentamer in E. coli prior to assembling the nanoparticle immunogens in vitro16 (Fig. 1A). We generated RBD-NPs displaying RBDs from the Wu-1, B.1.351 (β), and P.1 (γ) (K417N/T, E484K, N501Y) lineages with and without the previously reported “Rpk9” stabilizing mutations to the linoleic binding pocket (Y365F, F392W, V395I)20, as these were the dominant circulating strains with prominent escape mutations prior to the Omicron lineages (Fig. 1B). An RBD-NP vaccine bearing the single point mutation E484K was also created to probe the effects of this mutation on stability, antigenicity, and vaccine-elicited neutralizing antibody titers and breadth. SARS-CoV-2 strains containing this substitution escape protection by several monoclonal antibody therapies such as LY-CoV555, which resulted in discontinuation of their clinical use22.
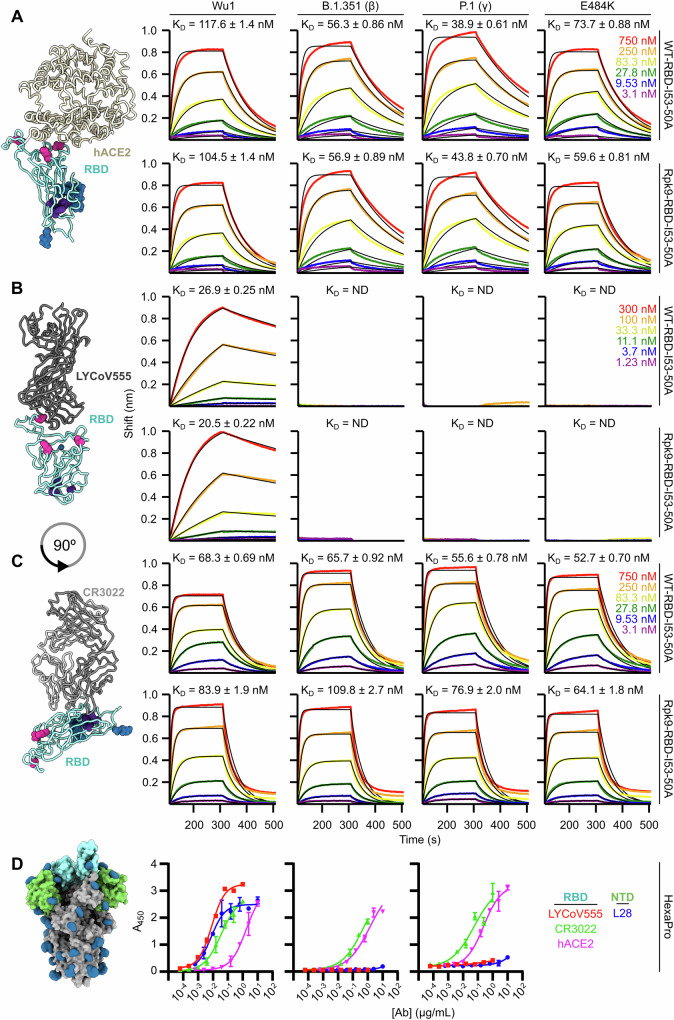
Fig. 1. SARS-CoV-2 variant vaccine design and characterization.
A Graphical representation of the production and characterization of SARS-CoV-2 RBD-NP vaccines, created with BioRender.com. B Molecular surface representation of the SARS-CoV-2 HexaPro trimer in the prefusion-stabilized conformation in gray19 (PDB: 6XKL). N-linked glycans in dark blue. A single RBD is boxed and expanded with bound hACE2 receptor61 (PDB: 6M0J) and CR3022 Fab (PDB: 6W41)23 shown for reference. Pink spheres indicate β and γ RBD mutations (K417N/T, E484K, and N501Y). Purple spheres indicate stabilizing Rpk9 mutations (Y365F, F392W, and V395I). C Representative DLS of 8 monovalent RBD-NPs with and without Rpk9 mutations. D Structural models of 8 monovalent RBD-NPs and 3 prefusion-stabilized HexaPro trimers alongside representative nsEM micrographs of each immunogen. All graphical representations of proteins made using ChimeraX61.
Dynamic light scattering (DLS) and negative stain electron microscopy (nsEM) revealed that all eight monovalent RBD-NPs (four variants with and without Rpk9 mutations) were of the expected hydrodynamic diameter and adopted the known morphology of icosahedral I53-50-based immunogens (Fig. 1C, D). As benchmarks, we expressed prefusion-stabilized (HexaPro) spike proteins bearing the same VOC mutations (except for the E484K point mutant) in Expi293F cells, purified them by immobilized metal-ion affinity chromatography (IMAC), and structurally validated them by nsEM (Fig. 1D). These data suggested that the RBD-I53-50-NP and HexaPro immunogen formats are robust to variations in RBD or spike protein sequences.
To understand the effects of VOC and Rpk9 mutations on antigenicity, we used biolayer interferometry (BLI) to measure the binding of the trimeric antigen-bearing components to monomeric hACE2 receptor and two Fabs from antibodies that bind the ancestral Wu-1 RBD, LY-CoV555 and CR302223–25. Since some VOC mutations are in the receptor binding motif, we determined the affinity of each component for hACE2, finding that there were minimal differences between variant-matched WT and Rpk9 antigen-bearing components (Fig. 2A). We then compared binding to LY-CoV555, expecting that the E484K substitution would abrogate binding as previously reported22. As expected, all antigen-bearing components with the E484K substitution failed to bind LY-CoV555 Fab at any concentration tested (1.23–300 nM) (Fig. 2B). Finally, we determined the affinity of each component to CR3022 Fab to understand the potential impact of the nearby Rpk9 mutations on this conserved cryptic epitope23. Our findings indicate that Rpk9 mutations result in a minimal but consistent reduction in CR3022 binding, with no pair of designs showing even a two-fold difference in KD (Fig. 2C).
Fig. 2. Antigenic characterization of variant antigens.
Left, Models of RBD with bound (A) hACE2 receptor, (B) LY-CoV55524 (PDB: 7KMG), and C) CR3022 Fab, depicted as in Fig. 1. Right, Binding of (A) monomeric hACE2, (B) LY-CoV555, and (C) CR3022 Fab to immobilized VOC-RBD-I53-50A trimers. Data are shown in colors and global fits as black lines, with the KD of each interaction indicated. D Left, Molecular surface representation of SARS-CoV-2 HexaPro prefusion-stabilized trimer19 (PDB: 6XKL) with RBDs and NTDs highlighted in light blue and green, respectively. Right, ELISA binding titers to hACE2 receptor, RBD-specific IgG (LY-CoV555 and CR3022), and NTD-specific IgG (S2L28). All graphical representations of proteins made using ChimeraX61. Error bars represent standard error of mean from three replicates.
Antigenic characterization of Wu-1 and VOC HexaPros by ELISA recapitulated the BLI obtained with trimeric RBD-I53-50A components. Both VOC HexaPros lost all binding to LY-CoV555 while maintaining similar antigenic characteristics at the hACE2 and CR3022 binding sites (Fig. 2D). We also used the NTD-directed S2L28 monoclonal antibody26 to evaluate potential differences in antigenicity among the three HexaPro NTDs. Both VOC HexaPros failed to bind S2L28 mAb, indicating antigenic remodeling of this highly variable domain. In summary, all HexaPros and antigen-bearing components, with or without the Rpk9 mutations, were antigenically intact, exhibiting the expected binding profiles.
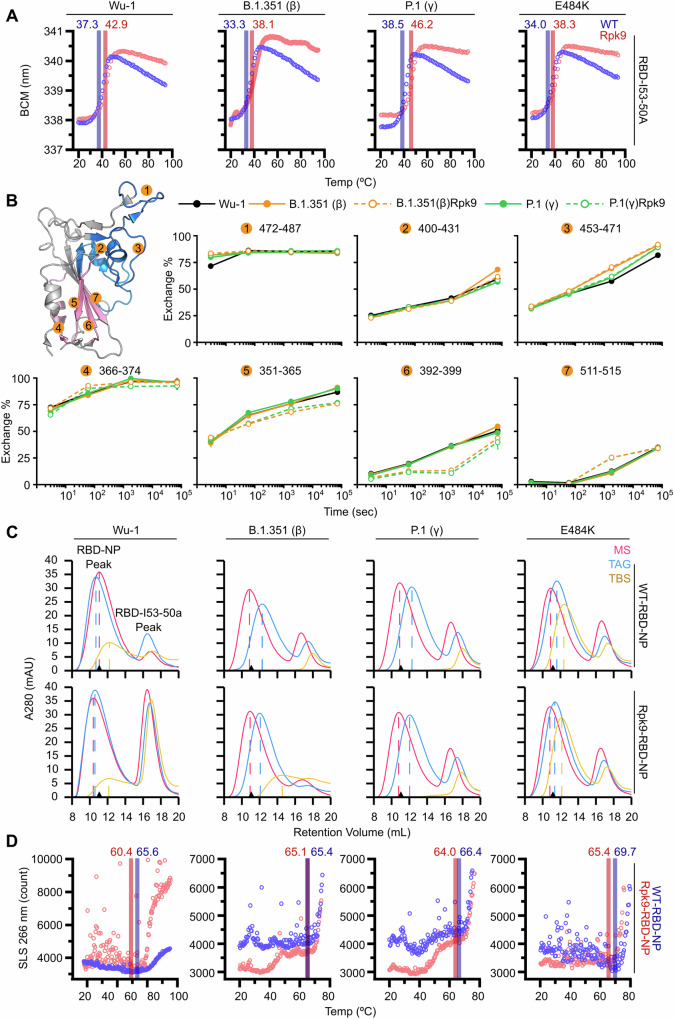
Next, to determine the impact of the Rpk9 mutations on VOC antigen stability, thermal melts and hydrogen/deuterium exchange mass spectrometry (HDX-MS) were performed on the trimeric I53-50A components. Melting temperatures for each VOC antigen-bearing component were obtained by nano differential scanning fluorimetry (nanoDSF), monitoring intrinsic tryptophan fluorescence from 20 °C to 95 °C. The melting temperature (Tm) of each Rpk9-containing component was about 5 °C higher than its strain-matched WT construct, revealing that the Rpk9 mutations increase the thermal stability of VOC RBDs as they did for the ancestral Wu-1 RBD (Fig. 3A). Previously, the Rpk9 mutations were reported to increase the Tm of Wu-1-RBD-I53-50A from ~49 °C to ~54 °C (ref. 20). Here, we observed an increase from ~37 °C to ~43 °C. We attribute the difference to the inclusion of CHAPS, a zwitterionic detergent, in the buffers used in the present study. Nevertheless, the Rpk9 mutations consistently increase Tm by ~5–6 °C in the Wu-1-RBD-I53-50A regardless of the differences in buffer. The Rpk9 mutations also reduced hydrogen/deuterium exchange in the region of the linoleic acid binding site, specifically in peptides spanning residues 351-365 and 392-399 (Fig. 3B). However, there was no difference in exchange between variant-matched RBDs in peptides comprising residues 400-431, 453-471, and 472-487, indicating that—in accordance with our antigenic characterization—the Rpk9 mutations have minimal effect on the local stability of surface neutralizing epitopes.
Fig. 3. Stabilizing effects of Rpk9 mutations in variant RBD components and NPs.
A Representative melting curves of RBD-I53-50A trimers with and without Rpk9 mutations. Melting temperatures (Tm) are indicated. B Hydrogen/deuterium exchange mass spectrometry of selected RBD peptides. Left, RBD structure with observed peptides numbered and regions where the VOC-related and Rpk9 mutations reside represented in blue and pink, respectively. C SEC chromatograms of RBD-NPs in three different buffers. MS: 50 mM Tris pH 7.4, 185 mM NaCl, 100 mM arginine-HCl, 4.5% v/v glycerol, 0.75% w/v CHAPS; TAG: 50 mM Tris pH 8, 150 mM NaCl, 100 mM arginine-HCl, 5% v/v glycerol; TBS: 50 mM Tris pH 8, 150 mM NaCl. Major peak at ~10.5 mL represents assembled NP and minor peak ~17 mL represents excess VOC-RBD-I53-50A. Black triangle on the x axis represents the Wu-1-RBD-NP peak in MS. D Representative aggregation profiles of purified RBD-NPs, with temperature (Tagg) indicated.
We previously showed that the Rpk9 mutations improved the solution properties of the Wu-1-RBD-NP, particularly in excipient-free buffers lacking detergent20. To determine the effects of the Rpk9 mutations in VOC-RBD-NPs, we utilized SEC in decreasingly excipient-rich buffers. We found that Rpk9-RBD-NPs did not show major differences in solution properties compared to the WT-RBD-NPs (Fig. 3C). Unexpectedly, the temperatures at which the Rpk9-RBD-NPs began to aggregate were slightly lower than WT-RBD-NPs (Fig. 3D). This differs from previously published data on Rpk9-Wu-1-RBD-NP20, which is likely explained by the use of different buffers (previously 50 mM Tris pH 8, 150 mM NaCl, 100 mM arginine-HCl, 5% v/v glycerol versus currently 50 mM Tris pH 7.4, 150 mM NaCl, 100 mM arginine-HCl, 5% v/v glycerol, 0.75% w/v CHAPS). Overall, although the Rpk9 mutations led to increases in the stability of components bearing the β and γ RBDs, nanoparticles displaying these RBDs had relatively poor solution properties compared to the ancestral Wu-1 immunogen, even in excipient-rich buffers. Interestingly, NPs displaying the RBD of the more recent OBA.4/5 variants required the Rpk9 mutation for expression and assembly: we were unable to detect expression of the wild-type OBA.4/5 RBD fused to I53-50A. However, RBD-NPs containing the OXBB.1.5 variant RBD could be readily produced in high yield and were well-behaved in solution (Supplementary Fig. 1).
To define the shelf-life stability of the Rpk9-bearing VOC vaccines, we conducted a 28-day stability assessment. A suite of biophysical characterization assays was used to analyze the physical and antigenic integrity of RBD nanoparticle immunogens at four temperatures: <-70, 2–8, 22–27, and 35–40 °C. As previously described, Wu-1-RBD-NP was stable over the 4-week study12, showing changes only at 35–40 °C, where some aggregation was observed at day 28 by nsEM and a slight increase in hydrodynamic diameter by DLS (Fig. 4 and Supplementary File 1). Antigenicity, as noted by hACE2-Fc binding relative to reference samples stored at −70 °C, decreased starting at day 7 only when the sample was incubated at 35–40 °C. At all other temperatures, there were no major differences over the 28-day study in appearance by SDS-PAGE or nsEM, antigenicity, UV/vis absorbance, or hydrodynamic diameter.
Fig. 4. Shelf-life stability of SARS-CoV-2 variant RBD-NPs.
A Summary of SDS-PAGE and nsEM stability data over 4 weeks. N/A, not assessed. Cocktail formulations include Wu-1-RBD-NP formulated with β-RBD-NP, βRpk9-RBD-NP, γ-RBD-NP, or γRpk9-RBD-NP. Detailed RBD-NP construct information can be found in Supplementary Table 2. B Binding to hACE2-Fc over 4 weeks. Immunogens were analyzed for hACE2-Fc binding by BLI after storage at the various temperatures, normalizing against a <-70°C reference sample at each time point. C Summary of UV/vis stability data over 4 weeks, using 320/280 nm absorbance ratio as a measure of particulate scattering. D Hydrodynamic diameter and polydispersity measured by DLS over 4 weeks. Raw data provided in Supplementary File 1.
Consistent with previous reports6, the β-RBD-NP proved the least stable throughout the study, showing degradation at 22–27 and 35–40 °C by SDS-PAGE; aggregation from 2–8 °C up to 35–40 °C by nsEM, UV/vis, and DLS; and a progressive loss of hACE2 binding with increasing temperature. The γ-RBD-NP showed similar trends at mid- to high-temperature, although they were less pronounced. Introduction of the Rpk9 mutations dramatically increased the stability of these immunogens by maintaining the expected hydrodynamic diameters throughout the study duration, although in each case there was still a decrease in antigenicity over time, especially at 35–40 °C.
We also compared the shelf-life stabilities of nanoparticle cocktails in which the Wu-1-RBD-NP was mixed with variant RBD-NPs. When combined with the β-RBD-NP (cWu-1β-RBD-NP), Wu-1-RBD-NP seemed to improve temperature-induced aggregation and loss of antigenicity compared to β-RBD-NP alone. The differences were not as dramatic in the cWu-1γ-RBD-NP compared to monovalent γ-RBD-NP, but generally followed the same pattern. Again, introducing the Rpk9 mutations into the variant RBD-NPs improved immunogen stability in cWu-1-βRpk9-RBD-NP (Wu-1-RBD-NP + βRpk9-RBD-NP) and cWu-1-γRpk9-RBD-NP (Wu-1-RBD-NP + γRpk9-RBD-NP) compared to cWu-1β-RBD-NP and cWu-1γ-RBD-NP, respectively, but did not completely prevent loss of antigenicity at 35–40 °C.
Collectively, these data show that the Rpk9 mutations increased the stability and solution properties of variant RBD nanoparticles. Furthermore, they establish that the Wu-1-RBD-NP has higher physical and antigenic stability than nanoparticles displaying the β and γ variant RBDs, a finding with implications for vaccine updates.
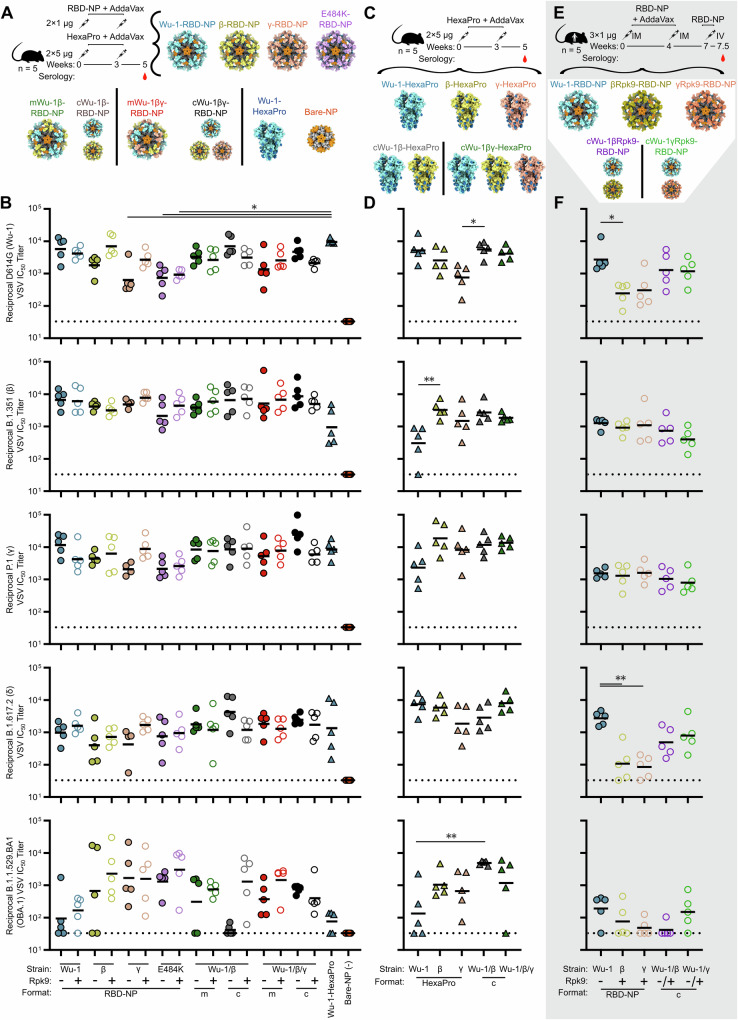
Finally, we immunized mice with 17 different vaccine candidates including monovalent Wu-1, β, γ, and E484K RBD-NPs with and without the Rpk9 mutations; mosaic NPs (mRBD-NPs) co-displaying Wu-1/β or Wu-1/β/γ RBDs on the same nanoparticle; cocktails of NPs (cRBD-NPs) consisting of mixtures of monovalent NPs; and individual or cocktails of prefusion-stabilized (HexaPro) Wu-1, β, and γ spike trimers (Fig. 5A, C and Supplementary File 1). Groups of five BALB/c mice were first immunized at weeks 0 and 3 with 1 μg of RBD-NPs or 5 μg of Wu-1 HexaPro, and serum neutralizing antibody titers were tested against pseudotyped VSV bearing G614, β, γ, B.1.617.2 (δ), or B.1.1.529 (Omicron BA.1) S. All immunogens elicited neutralizing antibodies 2 weeks post-boost against all viruses tested, although there were subsets of mice in several groups that did not yield detectable neutralizing activity against BA.1 (Fig. 5B). A negative control immunogen consisting of bare I53-50 lacking any displayed antigen did not induce any neutralizing activity, as expected. The Rpk9-RBD-NPs tended to elicit comparable or higher neutralizing antibody (nAb) titers than their WT counterparts, although none of the differences were statistically significant. The Wu-1-RBD-NP elicited high nAb titers against all VOC pseudoviruses tested, with geometric mean titers (GMTs) ranging from 1 × 103 to 1.2 × 104, except B.1.1.529 (BA.1; GMT 94.5). Introduction of the stabilizing Rpk9 mutations to the β-RBD-NP improved neutralization of all vaccine-mismatched pseudoviruses tested (Wu-1, γ, δ, and BA.1) by 3.9×, 1.4×, 1.8×, and 3.4× respectively, while maintaining similar nAb titers to the vaccine-matched pseudovirus. The Rpk9 mutations also improved nAb titers elicited by the γ-RBD-NP against Wu-1, β, γ, and δ S by 4.3×, 1.6×, 4.3×, and 4.1×, while GMT nAb titers against BA.1 were relatively unchanged. Interestingly, although the Rpk9 mutations did not consistently improve nAb titers in mice immunized with E484K-RBD-NP, these nanoparticles induced nAb titers against BA.1 14× and 18× higher than the corresponding ancestral (Wu-1) RBD-NPs. The Rpk9 mutations generally did not seem to improve nAb titers in mosaic and cocktail formulations, except against BA.1, where they increased nAb titers by 2.4×, 31×, and 3.9× in the mosaic bivalent, cocktail bivalent, and mosaic trivalent RBD-NP groups, respectively. Regardless of the Rpk9 mutations, NP vaccines comprising multiple VOC RBDs elicited comparable titers to all VOCs, both vaccine-matched (Wu-1, β, or γ) or -mismatched (δ or OBA.1), compared to their monovalent counterparts.
Fig. 5. Variant RBD-NP vaccines elicit high neutralizing Ab titers to SARS-CoV-2 variants in mice.
A, C Immunization scheme of VOC-RBD-NP, VOC-HexaPro, and the NP scaffold (Bare-NP) vaccines in Balb/c mice. Structural models of VOC-RBD-NPs, VOC-HexaPro, and Bare-NP vaccines. B, D Two weeks post boost neutralizing Ab titers for monovalent (RBD-NP), mosaics (m), and cocktails (c) of VOC with and without Rpk9 (+/−) mutations, Bare-NP, WT-VOC-HexaPro, and cocktails (c) of WT-VOC-HexaPro against various VOC pseudoviruses. Background neutralization is denoted by a dotted line. E Immunization scheme of VOC-RBD-NP vaccines in Darwin mice. Structural models of VOC-RBD-NP vaccines. F 0.5 week post IV boost neutralizing Ab titers for VOC-RBD-NP vaccines against various VOC pseudoviruses. Cocktail formulations including Wu-1-RBD-NP were formulated with βRpk9-RBD-NP or γRpk9-RBD-NP. All graphical representations of proteins made using ChimeraX61. Detailed RBD-NP immunogen information can be found in Supplementary Table 2. Kruskal–Wallis tests were performed to compare two groups to determine whether they were statistically different. Significance is indicated with stars: ∗p < 0.05; ∗∗p < 0.01; and non-significant differences are not shown.
To understand if the inclusion of the N-terminal domain (NTD) and S2 subunit in immunogens improves elicitation of vaccine-mismatched nAbs, we immunized BALB/c mice with 5 μg of VOC-HexaPro trimers and cocktails thereof (Fig. 5C). Although there is some evidence suggesting that incorporation of the NTD can elicit cross-reactive Abs, a minority of nAbs are NTD-directed27–30. Mice vaccinated with Wu-1-HexaPro, β-HexaPro, and γ-HexaPro elicited high levels of neutralizing antibodies to their vaccine-matched pseudoviruses, however those immunized with Wu-1-HexaPro experienced substantial drops in vaccine-mismatched nAb titers compared to the other HexaPro immunogens, except for δ neutralization (Fig. 5D). Both cocktail formulations of VOC-HexaPros elicited high levels of nAbs across all VOC tested, including the highly divergent δ and BA.1 strains. We conclude that HexaPro immunogen formulations including the β and γ lineages provide increased levels of neutralizing antibodies compared to those containing solely Wu-1-HexaPro in heterologous pseudovirus neutralization.
Based on the stability data and immunogenicity data from the two previous studies, we selected five RBD-NP vaccine candidates for evaluation in transgenic human Ig locus mice (Darwin mice) (Fig. 5E). Wu-1-RBD-NP elicited significantly higher and comparable G614 neutralization compared to VOC-Rpk9-RBD-NPs and cocktails of Wu-1- and VOC-Rpk9-RBD-NPs, respectively (Fig. 5F). Against both β and γ pseudoviruses, the Wu-1-RBD-NP, VOC-Rpk9-RBD-NPs, and cocktail immunogens generated equivalent nAb titers. Surprisingly, in Darwin mice immunization with Wu-1-RBD-NP yielded the highest nAb titers against δ and BA.1. Darwin mice have a human Ig repertoire31 and therefore may generate human-like polyclonal responses against the RBD. Both human and primate polyclonal responses have previously been reported to hyperfocus on position 484 in the RBD, which may explain some of the distinct sensitivities of neutralization breadth in BALB/c mice compared to Darwin mice16,32,33. Additionally, Darwin mice were bred from C57BL/6 and 129S7 mice, and the former have previously been shown to generate lower binding and neutralizing antibody titers than BALB/c mice in response to immunization2. In summary, we conclude that Wu-1-RBD-NP elicits the most consistent levels of vaccine-matched and -mismatched neutralizing antibodies in humanized mice, while c/mRBD-NP and HexaPro formulations containing antigens from multiple VOCs maintain the most consistent neutralizing antibody levels in BALB/c mice.
Discussion
RBD-I53-50 nanoparticles have proven to be robust and versatile immunogens for coronavirus vaccines. The Wu-1-RBD-NP originally described in 2020 was shown to elicit high levels of neutralizing antibodies in Phase III clinical trials14 and has been licensed for use in multiple countries, establishing computationally designed protein nanoparticle vaccines as a commercial vaccine platform. Despite the inclusion of only the RBD, the performance of RBD-NP vaccines has proven similar to vaccines that also include the NTD and the conserved S2 fusion machinery, including against vaccine-mismatched variants. For example, non-human primates boosted with the βRpk9-RBD-NP described here 6 months after a primary immunization series of either the Wu-1-RBD-NP or Wu-1 HexaPro were similarly protected against SARS-CoV-2 Omicron BA.1 challenge 6 months later34. Furthermore, monoclonal antibodies isolated from both sets of animals showed similar potency and breadth against a diverse panel of sarbecoviruses35. Additionally, administration of a third dose of the licensed RBD-NP vaccine SKYCovione™ in humans 7 months after the primary series increased neutralizing antibody titers against Omicron BA.1 75-fold, reaching levels 25-fold higher than peak titers after the primary series14. Similar results against Omicron BA.1 were observed after a third dose of mRNA-127336. These data, together with the production of RBD-NPs displaying various VOC (this work) and sarbecovirus16 RBDs, highlight RBD-I53-50 nanoparticles as a platform for coronavirus vaccine development.
However, our data reinforce that the properties of immunogens based on different SARS-CoV-2 variants can differ substantially and could affect the viability of variant vaccine development. For example, the poor stability and solution properties of the recombinant β spike protein were noted soon after the emergence of B.1.35137, and we observed similar effects in the β- and γ-RBD-NPs. Both of these RBD-NPs exhibited poor expression yields and a marked tendency to aggregate compared to the Wu-1-RBD-NP. Introducing the stabilizing Rpk9 mutations improved expression and the solution properties of these RBD-NPs somewhat, but did not fully rescue them. The OBA.4/5-RBD-NP could only be produced when the Rpk9 mutations were added, as expression was undetectable without them. By contrast, the OXBB.1.5-RBD-NP could be readily produced in high yield and was well-behaved in solution. These differences between individual variants and the inability of the Rpk9 mutations to ubiquitously improve VOC-RBD stability is a limitation of the current work. Identifying additional stabilizing mutations that are portable across sarbecovirus RBDs would help ensure that effective vaccines could be rapidly produced for any new variant38,39. Understanding how mutations that arise in new variants affect pre-existing immunity has also proven helpful for informing when vaccine strain updates are needed40–43.
Since the emergence of the virus, SARS-CoV-2 vaccine development efforts worldwide have shown that the time required to manufacture protein-based vaccines presents a challenge for updating vaccines in response to ongoing antigenic evolution. By contrast, the significantly shorter timelines for mRNA vaccine manufacture have facilitated the development of updated vaccines44–46. Approaches that combine the potency of protein nanoparticle vaccines with the manufacturing speed of mRNA, such as mRNA vaccines that encode secreted protein nanoparticle immunogens47, could become a powerful platform for vaccine development against emerging and evolving infectious diseases. Alternatively, broadly protective protein nanoparticle vaccines that induce immunity against multiple related viruses may preclude the need to frequently update vaccines. Several multivalent RBD nanoparticle vaccines have yielded promising preclinical data and are advancing toward clinical trials16–18. These vaccines utilize the mosaic nanoparticle display technology that was originally developed for influenza vaccines48–50 and is currently being evaluated clinically (NCT04896086 and NCT05968989). We note that the multivalent RBD-I53-50 nanoparticle vaccine currently in preclinical development extends the work presented here by displaying stabilized RBD antigens from several different sarbecoviruses51. The variable behavior we observed in the present study from protein-based vaccines derived from different SARS-CoV-2 isolates highlights the need for technologies such as mosaic nanoparticle display and mRNA-launched nanoparticles that may facilitate the rapid development of protective vaccines for antigenically variable families of viruses.
Methods
Cell lines
Expi293F cells are derived from the HEK293F cell line (Life Technologies). Expi293F cells were grown in Expi293 Expression Medium (Life Technologies), cultured at 36.5 °C with 8% CO2 and shaking at 150 rpm. HEK293T/17 is a female human embryonic kidney cell line (ATCC). The HEK-ACE2 adherent cell line was obtained through BEI Resources, NIAID, NIH: Human Embryonic Kidney Cells (HEK293T) Expressing Human Angiotensin-Converting Enzyme 2, HEK293T-hACE2 Cell Line, NR-52511. The VeroE6-TMPRSS2 cell line is an African Green monkey kidney cell line expressing TMPRSS2 (ref. 52). All adherent cells were cultured at 37 °C with 5% CO2 in flasks with DMEM + 10% FBS (Hyclone) + 1% penicillin-streptomycin. Cell lines other than Expi293F were not tested for mycoplasma contamination nor authenticated.
Mice
Four week-old female BALB/c mice (order code 047, BALB/cAnNHsd strain) were obtained from Envigo, and maintained in a specific pathogen-free facility at the University of Washington, Seattle, WA, accredited by the American Association for the Accreditation of Laboratory Animal Care International (AAALAC). Animal procedures were performed under the approvals of the Institutional Animal Care and Use Committee (IACUC) of University of Washington, Seattle, WA. Kymab, a Sanofi Company’s proprietary IntelliSelect™ transgenic mouse platform, known as Darwin, has complete human antibody loci with a non-rearranged human antibody variable and constant germline repertoire. Consequently, the antibodies produced by these mice are fully human.
Plasmid construction
The Wu-1-SARS-CoV-2, B.1.351(β)-SARS-CoV-2, P.1(γ)-SARS-CoV-2, and E484K RBDs (N-RFPN…KKST-C) were genetically fused to the N terminus of the trimeric I53-50A nanoparticle component using a 16-residue glycine and serine linker. The RBD-16GS-I53-50A fusions were cloned into pCMV/R using the Xba1 and AvrII restriction sites using Gibson assembly or by GenScript. All RBD-bearing components contained an N-terminal mu-phosphatase signal peptide and a C-terminal octa-histidine tag. Rpk9 mutants were designed the same way except with mutations at Y365F, F392W, V395I. The Wu-1, B.1.351(β), and P.1(γ)-SARS-CoV-2 S-6P ectodomain trimers were synthesized by GenScript into pCMV/R with an N-terminal BM40 signal peptide and a C-terminal T4 fibritin foldon trimerization domain and octa-histidine tag. The constructs contained the HexaPro mutations (proline substitutions at residues F817P, A892P, A899P, A942P, K986P, and V987P) and 682SGAG685 substitution at the furin cleavage site.
Transient transfection
Proteins were produced using endotoxin-free DNA in Expi293F cells grown in suspension using Expi293F expression medium (Life Technologies) at 33 °C, 70% humidity, 8% CO2 rotating at 150 rpm. The cultures were transfected using PEI-MAX (Polyscience) with cells grown to a density of 3.0 million cells per mL and cultivated for 3 days. Supernatants were clarified by centrifugation (5 min at 4000 rcf), addition of PDADMAC solution to a final concentration of 0.0375% (Sigma Aldrich, #409014), and a second centrifugation (5 min at 4000 rcf).
Microbial plasmid construction, protein expression and purification of I53-50B.4PT1
I53-50B.4PT1 plasmid was synthesized by GenScript in pET29b between the NdeI and XhoI restriction sites with a double-stop codon just before the C-terminal polyhistidine tag. Protein was expressed in Lemo21(DE3) cells (NEB) in LB (10 g Tryptone, 5 g Yeast Extract, 10 g NaCl) grown in a 10 L BioFlo 320 Fermenter (Eppendorf). At inoculation, impeller speed was set to 225 rpm, SPLM set to 5 with O2 supplementation as part of the dissolved-oxygen aeration cascade, and the temperature set to 37 °C. At the onset of a DO spike (OD ~ 12), the culture was fed with a bolus addition of 100 mL of 100% glycerol and induced with 1 mM IPTG. During this time, the culture temperature was reduced to 18 °C, and O2 supplementation was ceased, with expression continuing until an OD ~ 20. The culture was harvested by centrifugation and the pellets were resuspended in PBS, homogenized, and then lysed by microfluidization using a Microfluidics M110P at 18,000 psi using 3 discrete passes. Following sample clarification by centrifugation (24,000 g for 30 min), the supernatant was discarded, and protein was extracted from the inclusion bodies. First, the pellet was washed with PBS supplemented with 0.1% Triton X-100, pH 8.0. After this initial wash and sample clarification by centrifugation, the pellet was washed with PBS supplemented with 1 M NaCl, pH 8.0. Following the second wash, the protein was extracted from the pellet into PBS supplemented with 2 M urea and 0.75% CHAPS (3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate), pH 8.0. Following extraction, the sample was applied to a DEAE Sepharose FF column (Cytiva) on an ӒKTA Avant150 FPLC system (Cytiva). After sample binding, the column was washed with 5 column volumes (CV) of PBS supplemented with 0.1% Triton X-100, pH 8.0, followed by a 5 CV wash with PBS supplemented with 0.75% CHAPs, pH 8.0 in series. The protein was eluted with 3 CV of PBS supplemented with 500 mM NaCl, pH 8.0. After purification, fractions were pooled and concentrated in 10 K molecular weight cutoff (MWCO) centrifugal filters (Millipore), sterile-filtered (0.22 μm), and tested to confirm low endotoxin levels before use in nanoparticle assembly.
Protein purification
Proteins containing His tags were purified from clarified supernatants via a batch bind method where each clarified supernatant was supplemented with 1 M Tris-HCl pH 8.0 to a final concentration of 45 mM and 5 M NaCl to a final concentration of ∼310 mM. Talon cobalt affinity resin (Takara) (for S-6P) or Nickel Sepharose Excel resin (Cytiva) (for VOC-RBD-I53-50A) were added to the treated supernatants and allowed to incubate for 15 min with gentle shaking. Resin was collected using vacuum filtration with a 0.2 μm filter and transferred to a gravity column. The resin was washed with 20 mM Tris pH 8.0, 300 mM NaCl, and the protein was eluted with 3 CV of 20 mM Tris pH 8.0, 300 mM NaCl, 300 mM imidazole. The batch bind process was then repeated and the first and second elutions combined. SDS-PAGE was used to assess purity. VOC-RBD-I53-50A fusion protein IMAC elutions were concentrated to >1 mg/mL and subjected to three rounds of dialysis into 50 mM Tris pH 7.4, 185 mM NaCl, 100 mM L-arginine, 4.5% glycerol, and 0.75% w/v CHAPS (MAGiC Sauce, MS or Mixture of Arginine, Glycerol, in CHAPS) in a hydrated 10 K MWCO dialysis cassette (Thermo Scientific). VOC-HexaPro elutions were concentrated to >0.5 mg/mL and subjected to three rounds of dialysis into 50 mM Tris pH 8, 185 mM NaCl, 0.25% w/v L-histidine, 5% v/v glycerol in a hydrated 10 K MWCO dialysis cassette (Thermo Scientific). Clarified supernatants of cells expressing monoclonal antibodies and human ACE2-Fc were purified using a MabSelect PrismA 2.6 × 5 cm column (Cytiva) on an ӒKTA Avant150 FPLC (Cytiva). Bound antibodies were washed with five CV of 20 mM NaPO4, 150 mM NaCl pH 7.2, then five CV of 20 mM NaPO4, 1 M NaCl, pH 7.4 and eluted with three CV of 100 mM glycine at pH 3.0. The eluate was neutralized with 2 M Trizma base to 50 mM final concentration. SDS-PAGE was used to assess purity.
In vitro nanoparticle assembly and purification
Total protein concentration of purified individual nanoparticle components was determined by measuring absorbance at 280 nm using a UV/vis spectrophotometer (Agilent Cary 8454) and calculated extinction coefficients. The assembly steps were performed at room temperature with addition in the following order: VOC-RBD-I53-50A trimeric fusion protein, followed by additional buffer (50 mM Tris pH 7.4, 185 mM NaCl, 100 mM L-arginine, 4.5% glycerol, and 0.75% w/v CHAPS) as needed to achieve desired final concentration, and finally I53-50B.4PT1 pentameric component (in 50 mM Tris pH 8, 500 mM NaCl, 0.75% w/v CHAPS), with a molar ratio of VOC-RBD-I53-50A:I53-50B.4PT1 of 1.1:1. All VOC-RBD-NP in vitro assemblies were incubated briefly at room temperature before subsequent purification by SEC in order to remove residual unassembled VOC-RBD-I53-50A component. A Superose 6 Increase 10/300 GL column was used for nanoparticle purification. Assembled nanoparticles were purified in 50 mM Tris pH 7.4, 185 mM NaCl, 100 mM Arginine, 4.5% v/v glycerol, and 0.75% w/v CHAPS, and elute at ∼11 mL on the Superose 6 column. Assembled nanoparticles were sterile-filtered (0.22 μm) immediately prior to column application and following pooling of fractions.
UV/vis
UV/vis absorbance was measured using an Agilent Technologies Cary 8454. Samples were applied to a 10 mm, 50 μL quartz cell (Starna Cells, Inc.) and absorbance was measured from 180 to 1000 nm. Net absorbance at 280 nm, obtained from measurement and single reference wavelength baseline subtraction, was used with calculated extinction coefficients and molecular weights to obtain protein concentration. The ratio of absorbance at 320/280 nm was used to determine relative aggregation levels in real-time stability study samples. Samples were diluted with respective purification/instrument blanking buffers to obtain an absorbance between 0.1 and 1.0. All data produced from the UV/vis instrument was processed in the 845x UV/visible System software.
Endotoxin measurements
Endotoxin levels in protein samples were measured using the EndoSafe Nexgen-MCS System (Charles River). Samples were diluted 1:100 in Endotoxin-free LAL reagent water, and applied into wells of an EndoSafe LAL reagent cartridge. Charles River EndoScan-V software was used to analyze endotoxin content, automatically back-calculating for the dilution factor. Endotoxin values were reported as EU/mL which were then converted to EU/mg based on UV/vis measurements. Our threshold for samples suitable for immunization was <50 EU/mg.
Dynamic light scattering
Dynamic light scattering (DLS) was used to measure hydrodynamic diameter (Dh) and % Polydispersity (%Pd) of RBD-NPs, mRBD-NPs, and cRBD-NPs on an UNcle Nano-DSF (UNchained Laboratories). Sample was applied to a 8.8 µL quartz capillary cassette (UNi, UNchained Laboratories) and measured with 10 acquisitions of 5 s each, using auto-attenuation of the laser. Increased viscosity due to 4.5% v/v glycerol in the RBD nanoparticle buffer was accounted for by the UNcle Client software in Dh measurements.
Negative stain electron microscopy
VOC-RBD-NP and VOC-HexaPro vaccines were first diluted to 75 µg/mL or 30 µg/mL in 50 mM Tris pH 7.4, 185 mM NaCl, 100 mM L-arginine, 4.5% v/v glycerol, 0.75% w/v CHAPS or 50 mM Tris pH 8, 185 mM NaCl, 0.25% w/v L-histidine, 5% v/v glycerol prior to application of 3 µL of sample onto freshly glow-discharged 300-mesh copper grids. Sample was incubated on the grid for 1 min before 6 µL of 0.75% w/v uranyl formate stain was applied to the grid. Stain was blotted off with filter paper, then the grids were dipped into another 6 µL of stain and then repeated once more. Finally, the stain was blotted away and the grids were allowed to dry for 1 min. Prepared grids were imaged in a Talos model L120C electron microscope at 57,000× or 92,000×.
hACE2-Fc, CR3022, and LYCoV555 IgG digestion
hACE2-Fc was digested with thrombin protease (Sigma Aldrich) in the presence of 2.5 mM CaCl2 at a 1:300 w/w thrombin:protein ratio. The reaction was incubated at ambient temperature for 16–18 h with gentle rocking. Following incubation, the reaction mixture was concentrated using Ultracel 10 K MWCO centrifugal filters (Millipore Amicon Ultra) and sterile filtered (0.22 μM). Cleaved hACE2 monomer was separated from uncleaved hACE2-Fc and the cleaved Fc regions using Protein A purification (see Protein purification above) on a HiScreen MabSelect SuRe column (Cytiva) using an ӒKTA avant 25 FPLC (Cytiva). Cleaved hACE2 monomer was collected in the flowthrough, sterile-filtered (0.22 μm), and quantified by UV/vis.
LysC (New England BioLabs) was diluted to 10 ng/μL in 10 mM Tris pH 8 and added to CR3022 or LYCoV555 IgG at 1:2000 w/w LysC:IgG and subsequently incubated for 18 h at 37 °C with orbital shaking at 230 rpm. The cleavage reaction was concentrated using Ultracel 10 K MWCO centrifugal filters (Millipore Amicon Ultra) and sterile filtered (0.22 μM). Cleaved CR3022 or LYCoV555 mAb was separated from uncleaved CR3022 or LYCoV555 IgG and the Fc portion of cleaved IgG, using Protein A purification as described above. Cleaved CR3022 or LYCoV555 was collected in the flowthrough, sterile-filtered (0.22 μm), and quantified by UV/vis.
Bio-layer interferometry (affinity determination)
Affinity assays were performed and analyzed using BLI on an Octet Red 96 System (Pall Forté Bio/Sartorius) at ambient temperature with shaking at 1000 rpm. VOC-RBD-I53-50A trimeric components were diluted to 40 μg/mL in Kinetics buffer (1× HEPES-EP+ (Pall Forté Bio), 0.05% nonfat milk, and 0.02% sodium azide). Monomeric hACE2, CR3022, and LYCoV555 Fab were diluted to 750 or 300 nM in Kinetics buffer and serially diluted three-fold for a final concentration of 3.1 or 1.23 nM. Reagents were applied to a black 96-well Greiner Bio-one microplate at 200 μL per well as described below. VOC-RBD-I53-50A components were immobilized onto Anti-Penta-HIS (HIS1K) biosensors per manufacturer instructions (Forté Bio) except using the following sensor incubation times. HIS1K biosensors were hydrated in water for 10 min, and were then equilibrated in Kinetics buffer for 60 s. The HIS1K tips were loaded with diluted trimeric VOC-RBD-I53-50A component or monomeric RBD for 150 s and washed with Kinetics buffer for 60 s. The association step was performed by dipping the HIS1K biosensors with immobilized immunogen into diluted hACE2 monomer, CR3022, or LYCoV555 Fab for 200 s, then dissociation was measured by inserting the biosensors back into Kinetics buffer for 200 s. The data were baseline subtracted and the plots fitted using the Pall FortéBio/Sartorius analysis software (version 12.0). Plots in Fig. 2 show the association and dissociation steps.
ELISA (antigenicity)
For anti-HexaPro ELISA, 50 μL of 2 μg/mL HexaPro S was plated onto 384-well Nunc Maxisorp (ThermoFisher) plates in PBS and sealed overnight at RT. The next day, the plates were slapped dry and blocked with Casein (ThermoFisher) for 1 h at 37 °C. Plates were washed 4× in Tris-Buffered Saline Tween (TBST) using a plate washer (BioTek) and 1:5 serial dilutions of mAb were made in 40 μL TBST and incubated at 37 °C for 1 h. Plates were washed 4× with TBST on the BioTek plate washer, then anti-human (Invitrogen) horseradish peroxidase-conjugated antibodies were diluted 1:5,000 and 40 μL added to each well and incubated at 37 °C for 1 h. Plates were washed 4× in TBST on the BioTek plate washer and 40 μL of room temperature TMB (SeraCare) was added to every well for ∼5 min at room temperature. The reaction was quenched with the addition of 40 μL of 1 N HCl. Plates were immediately read at 450 nm on a BioTek plate reader and data plotted and fit in Prism (GraphPad) using nonlinear regression sigmoidal, 4PL, X is log(concentration) to determine EC50 values from curve fits. ELISA experiments were performed three separate times and representative experiments are shown.
Hydrogen/Deuterium-exchange mass spectrometry
3 μg of VOC-RBD-I53-50A and VOC-Rpk9-RBD-I53-50A mutants were H/D exchanged in the deuteration buffer (pH* 7.5, 85% D2O, Cambridge Isotope Laboratories, Inc.) at 23 °C for 3, 60, 1800, and 72000 s, respectively. H/D exchanged samples were immediately mixed with an equal volume of ice-cold quench buffer (8 M urea, 200 mM tris(2-chloroethyl) phosphate (TCEP), 0.2% formic acid (FA)) to a final pH 2.5 and flash frozen in liquid nitrogen. Samples were analyzed by LC-MS on a Waters Synapt G2-Si mass spectrometer using a custom-built cooling box to maintain sample digestion and injection at 0 °C. Quenched samples were loaded over an immobilized pepsin column (2.1 × 50 mm) with a 200 μL/min flow of loading buffer (0.1% trifluoroacetic acid (TFA), 2% acetonitrile (ACN)). Peptides were trapped on a Waters CSH C18 trap cartridge (2.1 × 5 mm) and resolved over a Waters CSH C18 1 × 100 mm 1.7 μm column with a linear gradient from 3% to 40% B over 18 min (A: 2% ACN, 0.1% FA, 0.025% TFA; B: 100% ACN, 0.1% FA, flow rate of 40 μL/min). A series of washes steps were performed between all samples to minimize carryover53. The total deuterated control was made by collecting a pepsin digest elution, drying by speed-vacuum, and incubating in deuteration buffer for 1 h at 85 °C. Peptides were identified and updated based on our previously examined lists20,54 and validated using DriftScope™ (Waters). Deuterium uptake analysis was performed with HDExaminer (Sierra Analytics) and HX-Express v255,56. Peaks were identified from the peptide spectra with binomial fitting applied. The deuterium uptake level was normalized relative to total deuterated control.
Nanoscale differential scanning fluorimetry (Component and NP)
nanoDSF was used to measure the relative melting temperature (Tm) and aggregation (Tagg) temperatures of the individual VOC-RBD-I5350As and VOC-RBD-NPs, respectively, on an UNcle Nano-DSF (UNchained Laboratories). VOC-RBD-I5350As and VOC-RBD-NPs were diluted to 0.5 mg/mL in MAGiC Sauce (50 mM Tris pH 7.4, 185 mM NaCl, 100 mM L-arginine, 4.5% glycerol, and 0.75% w/v CHAPS) and 8.8 µL were applied to a 8.8 µL quartz capillary cassette (UNi, UNchained Laboratories). The barycentric mean (BCM) and static light scattering at 488 nm (SLS 488 nm) were taken from 20 °C to 95 °C. Tm and Tagg were approximated by analyzing when the derivative or second derivative of the line fit for BCM or SLS 488 nm, respectively, was approximately equal to zero.
Bio-layer interferometry
Binding of hACE2-Fc to monovalent VOC-RBD-NP, mVOC-RBD-NP, and cVOD-RBD-NP was analyzed for antigenicity experiments and real-time stability studies using an Octet Red 96 System (Pall FortéBio/Sartorius) at ambient temperature with shaking at 1000 rpm. Protein samples were diluted to 100 nM in Kinetics buffer (1 × HEPES-EP+ (Pall Forté Bio), 0.05% nonfat milk, and 0.02% sodium azide). Buffer, receptor, and analyte were then applied to a black 96-well Greiner Bio-one microplate at 200 μL per well. Protein A biosensors (FortéBio/Sartorius) were first hydrated for 10 min in Kinetics buffer, then dipped into hACE2-Fc diluted to 10 μg/mL in Kinetics buffer in the immobilization step. After 150 s, the tips were transferred to Kinetics buffer for 60 s to reach a baseline. The association step was performed by dipping the loaded biosensors into the immunogens for 200 s, and subsequent dissociation was performed by dipping the biosensors back into Kinetics buffer for an additional 200 s. The data were baseline subtracted prior for plotting using the FortéBio analysis software (version 12.0).
BALB/c mice immunizations
At 8 weeks of age, 5 female BALB/c mice per dosing group were vaccinated with a prime immunization, and 3 weeks later mice were boosted with a second vaccination (IACUC protocol 4470.01). Prior to inoculation, immunogen suspensions were gently mixed 1:1 vol/vol with AddaVax adjuvant (Invivogen, San Diego, CA) to reach a final concentration of 0.01 mg/mL antigen and 0.13125% w/v CHAPS. Mice were injected intramuscularly into the gastrocnemius muscle of each hind leg using a 27-gauge needle (BD, San Diego, CA) with 50 μL per injection site (100 μL total) of immunogen under isoflurane anesthesia. Specifically, mice underwent induction of anesthesia in an induction chamber using 3–5% isoflurane with an O2 flow rate of 1 L/min. After assessing depth of anesthesia via toe pinch, mice were transferred from induction chambers into nosecones with the same isoflurane and oxygen parameters for immunization. Euthermia was maintained throughout the procedure using circulating warm water pads. Following immunization, mice recovered from anesthesia in separate recovery chambers. To obtain sera all mice were bled 2 weeks after prime and boost immunizations. Blood was collected via submental venous puncture and rested in 1.5 mL plastic Eppendorf tubes at room temperature for 30 min to allow for coagulation. Serum was separated from red blood cells via centrifugation at 2000 g for 10 min. Complement factors in isolated serum were heat-inactivated via incubation at 56 °C for 60 min. Serum was stored at 4 °C or −80 °C until use. After the post-boost blood collection, mice were euthanized in CO2 chambers with secondary confirmation via cervical dislocation. BALB/c immunogenicity studies were repeated twice.
Darwin mice immunizations
Kymab Darwin mice, equivalent to the Kymouse31 in the IgH locus except the immunoglobulin constant regions are also fully human, and with a fully human IgL kappa loci, were used in this study (a mix of males and females, 10 weeks of age). Five mice per dosing group were vaccinated with a prime immunization and 3 weeks later boosted with a second vaccination. Prior to inoculation, immunogen suspensions were gently mixed 1:1 vol/vol with AddaVax adjuvant (Invivogen) to reach a final concentration of 0.025 mg/mL antigen. Mice were injected intramuscularly into the tibialis muscle of each hind leg using a 30-gauge needle (BD) with 20 μL per injection site (40 μL total) of immunogen under isoflurane anesthesia. A final boost was administered intravenously (50 μL) with no adjuvant at week 7 in order to recruit circulating B cells to the spleen. Mice were sacrificed 5 days later under UK Home Office Schedule 1 (rising concentration of CO2) and spleen, lymph nodes, and bone marrow cryopreserved. Whole blood (0.1 mL) was collected 7.5 weeks after the first immunization. Serum was separated from hematocrit via centrifugation at 2000 g for 10 min. Serum was stored at −20 °C. All mice were maintained and all procedures carried out under United Kingdom Home Office License 70/8718 and with the approval of the Wellcome Trust Sanger Institute Animal Welfare and Ethical Review Body.
Pseudovirus production
SARS-CoV-2 Wu-1 G614 S, B.1.351 S, P1, B.1.617.2 S, and B.1.1.259.BA.1 S pseudotyped VSV viruses were prepared as described previously57–59. Briefly, 293 T cells in DMEM supplemented with 10% FBS, 1% PenStrep seeded in polylysine coated 10-cm dishes were transfected with the plasmid encoding for the corresponding S glycoprotein using lipofectamine 2000 (Life Technologies) following manufacturer’s indications. One day post-transfection, cells were infected with VSV(G∗ΔG-luciferase) and after 2 h were washed five times with warm DMEM before adding medium supplemented with anti-VSV-G antibody (I1- mouse hybridoma supernatant, CRL- 2700, ATCC). Virus pseudotypes were harvested 18–24 h post-inoculation, clarified by centrifugation at 2500 g for 10 min, filtered through a 0.45 μm cutoff membrane, concentrated 10 times with a 30 kDa MWCO Sartorius membrane, aliquoted, and stored at −80 °C prior to use at a 1:25 dilution in DMEM.
Pseudovirus neutralization
HEK293-hACE2 cells60 were cultured in DMEM with 10% FBS (Hyclone) and 1% PenStrep with 8% CO2 in a 37 °C incubator (ThermoFisher). One day prior to infection, 40 μL of poly-lysine (Sigma) was placed into white 96-well plates and incubated with rotation for 5 min. Poly-lysine was removed, plates were dried for 5 min then washed 1× with water prior to plating with ~40,000 cells. The following day, cells were checked to be at 80% confluence. In an empty half-area 96-well plate, a 1:3 serial dilution of sera was made in DMEM and diluted pseudovirus was then added to the serial dilution and incubated at room temperature for 30–60 min. After incubation, the sera-virus mixture was added to the cells and incubated at 37 °C for 2 hours. Following the 2 h incubation, 40 μL of 20% FBS-2% PenStrep DMEM was added for overnight incubation. After 16–20 h 40 μL/well of One-Glo-EX substrate (Promega) was added to the cells and incubated in the dark for 5–10 min prior reading on a BioTek plate reader. Measurements were done in at least duplicate with distinct pseudovirus batches and backbones. Relative luciferase units were plotted and normalized in Prism (GraphPad). Nonlinear regression of log(inhibitor) versus normalized response was used to determine IC50 values from curve fits.
Quantification and statistical analysis
Statistical details of experiments can be found in the figure legends. For mouse experiments, 5 BALB/c animals were used and experiments were completed in at least duplicate. Geometric mean titers were calculated. Kruskal–Wallis tests were performed to compare two groups to determine whether they were statistically different. Significance is indicated with stars: ∗p < 0.05; ∗∗p < 0.01 and non-significant differences are not shown.
Supplementary information
Acknowledgements
We thank Ratika Krishnamurty for excellent program management. This study was supported by the Bill & Melinda Gates Foundation (INV-010680, OPP1126258, and OPP1159947), a generous gift from the Audacious Project, a generous gift from Jodi Green and Mike Halperin, a generous gift from the Hanauer family, the Defense Threat Reduction Agency (HDTRA1-18-1-0001), the National Institute of General Medical Sciences (R01GM120553), the National Institute of Allergy and Infectious Diseases (DP1AI158186, HHSN272201700059C, and P01AI167966), a Pew Biomedical Scholars Award, Investigators in the Pathogenesis of Infectious Disease Awards from the Burroughs Wellcome Fund, and Fast Grants. D.V. is an Investigator of the Howard Hughes Medical Institute and the Hans Neurath Endowed Chair in Biochemistry at the University of Washington.
Author contributions
D.V. and N.P.K. contributed equally to the manuscript. Conception of the study: M.C.M., E.K., A.C.W., D.V., and N.P.K. Data acquisition and analysis: M.C.M., E.K., M.J.N., C.C., M.J., K.R.S., C.S., A.P., A.V., D.P., C.S., C.O., J.C.K., M.P., M.M., S.W., B.F., R.R., and D.E. Supervision: L.C., D.C., P.K., K.L., D.V., and N.P.K. Manuscript drafting and figure preparation: M.C.M., E.K., A.C.W., D.V., and N.P.K. Manuscript editing: All authors.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Competing interests
A.C.W., D.E., D.V., and N.P.K. are named as inventors on patent applications filed by the University of Washington based on the studies presented in this paper. The King lab has received unrelated sponsored research agreements from Pfizer and GSK. D.V. is a consultant for and has received an unrelated sponsored research agreement from Vir Biotechnology Inc. P.K. and A.P. were employees and shareholders of Kymab Ltd, now Kymab, a Sanofi Company. The other authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Marcos C. Miranda, Elizabeth Kepl.
Contributor Information
David Veesler, Email: dveesler@uw.edu.
Neil P. King, Email: neilking@uw.edu
Supplementary information
The online version contains supplementary material available at 10.1038/s41541-024-00982-1.
References
- 1.Novel Coronavirus – China. https://www.who.int/emergencies/disease-outbreak-news/item/2020-DON233.
- 2.Corbett, K. S. et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature586, 567–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Doremalen, N. et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature586, 578–582 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang, Z. et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature592, 616–622 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tegally, H. et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature592, 438–443 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Zhou, D. et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell184, 2348–2361.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viana, R. et al. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in Southern Africa. Nature603, 679–686 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu, G. & Moriyama, E. N. 2019nCoVR-A comprehensive genomic resource for SARS-CoV-2 variant surveillance. Innovation (Camb.)2, 100150 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tracking SARS-CoV-2 variants. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants.
- 10.Gagne, M. et al. mRNA-1273 or mRNA-Omicron boost in vaccinated macaques elicits similar B cell expansion, neutralizing responses, and protection from Omicron. Cell185, 1556–1571.e18 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corbett, K. S. et al. Protection against SARS-CoV-2 Beta variant in mRNA-1273 vaccine-boosted nonhuman primates. Science374, 1343–1353 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Walls, A. C. et al. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell183, 1367–1382.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arunachalam, P. S. et al. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature594, 253–258 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Song, J. Y. et al. Immunogenicity and safety of SARS-CoV-2 recombinant protein nanoparticle vaccine GBP510 adjuvanted with AS03: interim results of a randomised, active-controlled, observer-blinded, phase 3 trial. eClinicalMedicine64, 102140 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song, J. Y. et al. Safety and immunogenicity of a SARS-CoV-2 recombinant protein nanoparticle vaccine (GBP510) adjuvanted with AS03: A randomised, placebo-controlled, observer-blinded phase 1/2 trial. EClinicalMedicine51, 101569 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walls, A. C. et al. Elicitation of broadly protective sarbecovirus immunity by receptor-binding domain nanoparticle vaccines. Cell184, 5432–5447.e16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen, A. A. et al. Mosaic RBD nanoparticles protect against challenge by diverse sarbecoviruses in animal models. Science377, eabq0839 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen, A. A. et al. Mosaic nanoparticles elicit cross-reactive immune responses to zoonotic coronaviruses in mice. Science371, 735–741 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsieh, C.-L. et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science369, 1501–1505 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellis, D. et al. Stabilization of the SARS-CoV-2 spike receptor-binding domain using deep mutational scanning and structure-based design. Front. Immunol.12, 710263 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bale, J. B. et al. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science353, 389–394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Starr, T. N., Greaney, A. J., Dingens, A. S. & Bloom, J. D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep. Med.2, 100255 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan, M. et al. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science368, 630–633 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, B. E. et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med.13, eabf1906 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.ter Meulen, J. et al. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med.3, e237 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCallum, M. et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell184, 2332–2347.e16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piccoli, L. et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell183, 1024–1042.e21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCallum, M. et al. Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement. Science375, 864–868 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, L. et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature584, 450–456 (2020). [DOI] [PubMed] [Google Scholar]
- 30.Cerutti, G. et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe29, 819–833.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richardson, E. et al. Characterisation of the immune repertoire of a humanised transgenic mouse through immunophenotyping and high-throughput sequencing. Elife12, e81629 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greaney, A. J. et al. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe29, 463–476.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walls, A. C. et al. Distinct sensitivities to SARS-CoV-2 variants in vaccinated humans and mice. Cell Rep.40, 111299 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arunachalam, P. S. et al. Durable protection against the SARS-CoV-2 Omicron variant is induced by an adjuvanted subunit vaccine. Sci. Transl. Med.14, eabq4130 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng, Y. et al. Broadly neutralizing antibodies against sarbecoviruses generated by immunization of macaques with an AS03-adjuvanted COVID-19 vaccine. Sci. Transl. Med.15, eadg7404 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pajon, R. et al. SARS-CoV-2 Omicron variant neutralization after mRNA-1273 booster vaccination. N. Engl. J. Med.386, 1088–1091 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gobeil, S. M.-C. et al. Effect of natural mutations of SARS-CoV-2 on spike structure, conformation, and antigenicity. Science373, eabi6226 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leonard, A. C. et al. Stabilization of the SARS-CoV-2 receptor binding domain by protein core redesign and deep mutational scanning. Protein Eng. Des. Sel.35, gzac002 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmed, S. et al. A stabilized, monomeric, receptor binding domain elicits high-titer neutralizing antibodies against all SARS-CoV-2 variants of concern. Front. Immunol.12, 765211 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Starr, T. N. et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell182, 1295–1310.e20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Starr, T. N. et al. Deep mutational scans for ACE2 binding, RBD expression, and antibody escape in the SARS-CoV-2 Omicron BA.1 and BA.2 receptor-binding domains. PLoS Pathog.18, e1010951 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao, Y. et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature614, 521–529 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, D. J. et al. Mapping the antigenic and genetic evolution of influenza virus. Science305, 371–376 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Chalkias, S. et al. Original SARS-CoV-2 monovalent and Omicron BA.4/BA.5 bivalent COVID-19 mRNA vaccines: phase 2/3 trial interim results. Nat. Med.29, 2325–2333 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chalkias, S. et al. Three-month antibody persistence of a bivalent Omicron-containing booster vaccine against COVID-19. Nat. Commun.14, 5125 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Usdan, L. et al. A bivalent Omicron-BA.4/BA.5-Adapted BNT162b2 booster in ≥12-year-olds. Clin. Infect. Dis. 10.1093/cid/ciad718 (2023). [DOI] [PMC free article] [PubMed]
- 47.Mu, Z. et al. mRNA-encoded HIV-1 Env trimer ferritin nanoparticles induce monoclonal antibodies that neutralize heterologous HIV-1 isolates in mice. Cell Rep.38, 110514 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanekiyo, M. et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol.20, 362–372 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyoglu-Barnum, S. et al. Quadrivalent influenza nanoparticle vaccines induce broad protection. Nature592, 623–628 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cohen, A. A. et al. Construction, characterization, and immunization of nanoparticles that display a diverse array of influenza HA trimers. PLoS One16, e0247963 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.CEPI. CEPI expands partnership with SK bioscience to develop a ‘variant-proof’ vaccine against SARS-CoV and SARS-CoV-2 variants –. CEPIhttps://cepi.net/news_cepi/cepi-expands-partnership-with-sk-bioscience-to-develop-a-variant-proof-vaccine-against-sars-cov-and-sars-cov-2-variants/ (2021).
- 52.Lempp, F. A. et al. Lectins enhance SARS-CoV-2 infection and influence neutralizing antibodies. Nature10.1038/s41586-021-03925-1 (2021). [DOI] [PubMed]
- 53.Verkerke, H. P. et al. Epitope-independent purification of native-like envelope trimers from diverse HIV-1 isolates. J. Virol.90, 9471–9482 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen, C. et al. hACE2-induced allosteric activation in SARS-CoV versus SARS-CoV-2 spike assemblies revealed by structural dynamics. ACS Infect. Dis.9, 1180–1189 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guttman, M., Weis, D. D., Engen, J. R. & Lee, K. K. Analysis of overlapped and noisy hydrogen/deuterium exchange mass spectra. J. Am. Soc. Mass Spectrom.24, 1906–1912 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weis, D. D., Engen, J. R. & Kass, I. J. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J. Am. Soc. Mass Spectrom.17, 1700–1703 (2006). [DOI] [PubMed] [Google Scholar]
- 57.Sauer, M. M. et al. Structural basis for broad coronavirus neutralization. Nat. Struct. Mol. Biol.28, 478–486 (2021). [DOI] [PubMed] [Google Scholar]
- 58.Walls, A. C. et al. SARS-CoV-2 breakthrough infections elicit potent, broad, and durable neutralizing antibody responses. Cell185, 872–880.e3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCallum, M. et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science374, 1621–1626 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Crawford, K. H. D. et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses12, 513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meng, E. C. et al. UCSF ChimeraX: tools for structure building and analysis. Protein Sci.32, e4792 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.