Summary
Background
While many patients seem to recover from SARS-CoV-2 infections, many patients report experiencing SARS-CoV-2 symptoms for weeks or months after their acute COVID-19 ends, even developing new symptoms weeks after infection. These long-term effects are called post-acute sequelae of SARS-CoV-2 (PASC) or, more commonly, Long COVID. The overall prevalence of Long COVID is currently unknown, and tools are needed to help identify patients at risk for developing long COVID.
Methods
A working group of the Rapid Acceleration of Diagnostics-radical (RADx-rad) program, comprised of individuals from various NIH institutes and centers, in collaboration with REsearching COVID to Enhance Recovery (RECOVER) developed and organized the Long COVID Computational Challenge (L3C), a community challenge aimed at incentivizing the broader scientific community to develop interpretable and accurate methods for identifying patients at risk of developing Long COVID. From August 2022 to December 2022, participants developed Long COVID risk prediction algorithms using the National COVID Cohort Collaborative (N3C) data enclave, a harmonized data repository from over 75 healthcare institutions from across the United States (U.S.).
Findings
Over the course of the challenge, 74 teams designed and built 35 Long COVID prediction models using the N3C data enclave. The top 10 teams all scored above a 0.80 Area Under the Receiver Operator Curve (AUROC) with the highest scoring model achieving a mean AUROC of 0.895. Included in the top submission was a visualization dashboard that built timelines for each patient, updating the risk of a patient developing Long COVID in response to clinical events.
Interpretation
As a result of L3C, federal reviewers identified multiple machine learning models that can be used to identify patients at risk for developing Long COVID. Many of the teams used approaches in their submissions which can be applied to future clinical prediction questions.
Funding
Research reported in this RADx® Rad publication was supported by the National Institutes of Health. Timothy Bergquist, Johanna Loomba, and Emily Pfaff were supported by Axle Subcontract: NCATS-STSS-P00438.
Keywords: Long COVID, PASC, Machine learning, COVID-19, Evaluation, Community challenge
Research in context.
Evidence before this study
Post-acute sequelae of SARS-CoV-2 (PASC), commonly known as Long COVID, is a collection of symptoms that often lingers after the acute phase of a SARS-CoV-2 infection. At the time of the Long COVID Computational challenge, some studies has used the National COVID Cohort Collaborative (N3C) to develop machine learning models for identifying patients with Long COVID and for phenotyping patients with Long COVID, however, no prediction models had been developed to predict whether a patient would develop Long COVID using the data available in N3C. Additionally, most machine learning community challenges focus on quantitative evaluation metrics such as the F1 statistic and Area Under the Receiver Operator Curve as their primary ranking metric. While these are important, focus on quantitative metrics often results in challenge submissions that are uninterpretable and unwieldy in their size. Clinical prediction models need to be interpretable and portable.
Added value of this study
The Long COVID Computational Challenge brought together a large and diverse group of researchers to develop Long COVID risk prediction models on the N3C data repository. The challenge evaluation process incorporated both qualitative and quantitative evaluation metrics to identify a clinically useful, translational, and interpretable model likely to be implemented into a clinical setting.
Implications of all the available evidence
The Long COVID Computational Challenge incentivized the development of accurate, generalizable, and interpretable Long COVID risk prediction models that have the potential of being refined, validated, and implemented into a clinic setting.
Introduction
Long COVID
Studies have shown that recovery from SARS-CoV-2, the virus that causes COVID-19, can vary from person to person. Many patients seem to recover from COVID-19 quickly and completely. However, others report experiencing COVID-19 symptoms that last for weeks or months or developing new symptoms weeks after infection. These long-term effects are called post-acute sequelae of SARS-CoV-2 (PASC) or, more commonly, Long COVID. The overall prevalence of Long COVID is currently unknown, but there is growing evidence that more than a quarter of COVID-19 survivors experience at least one symptom of Long COVID after recovery of the acute illness.1 Research is ongoing to understand prevalence, duration, and clinical outcomes of Long COVID. Symptoms of fatigue, brain fog, shortness of breath, and cardiac damage, among others, have been observed in patients who had only mild initial disease.2, 3, 4 Real world data, which includes electronic health record (EHR) data, is a valuable resource to examine the heterogeneous, multi-system presentation of Long COVID at scale.5,6
The breadth and complexity of data created in today’s health care encounters require advanced analytics to extract meaning from longitudinal data on symptoms, laboratory results, social determinants of health (SDoH), viral variants, electronic health records (EHR), and other relevant data types. Advanced development of software tools and computing capacity has allowed artificial intelligence (AI)/machine learning (ML) approaches to leverage large observational datasets to identify patterns then enable prediction of outcomes at the patient level. These tools have been employed to better characterize the complex and diverse features of Long COVID and can also be used to identify risk factors for Long COVID.5
The Long COVID Computational Challenge
Community challenges are crowd-sourcing exercises where third parties elicit computational solutions from the broader research community for specific research questions. The solicited solutions are compared to each other by a third party and evaluated against a hidden gold standard answer set. The use of a third party helps avoid the self-assessment bias when evaluating machine learning models.7 Community challenges follow a well-established framework to address biomedical research questions. Previous successful scientific community challenges include the DREAM Challenges8,9 the Critical Assessments,10, 11, 12, 13 and the Pediatric COVID-19 Data Challenge.14
The Rapid Acceleration of Diagnostics-radical (RADx-rad) Executive Committee and REsearching COVID to Enhance Recovery (RECOVER) leadership in collaboration with the working group co-chairs sponsored and organized the Long COVID Computational Challenge (L3C). The primary objective of L3C was to focus on the development of prognostic models that serve as open-source tools for using structured medical records to identify which patients infected with SARS-CoV-2 have a high likelihood of developing Long COVID. The challenge was implemented by Sage Bionetworks in the National COVID Cohort Collaborative (N3C) Data Enclave.15,16 Prior to the L3C, no prediction model had been created to predict which patients were at risk for developing Long COVID using N3C data.
Methods
Data in the N3C data enclave
N3C has spearheaded the collection and harmonization of a large EHR data repository that represents over 7 million patients who tested positive for COVID-19 and over 11 million demographically matched patients who tested negative for COVID-19 from 76 Health Care Organizations (HCOs) across the U.S. The N3C Enclave includes demographics as well as longitudinal coded medical data derived from these patients’ EHRs from January of 2018 to the present. The N3C data repository is updated on a weekly basis with updated data from contributing sites as well as new data from newly onboarded contributing sites. The reported gender of the patients is mostly self-reported by the patients, but may vary by HCO.
The Long COVID Computational Challenge question
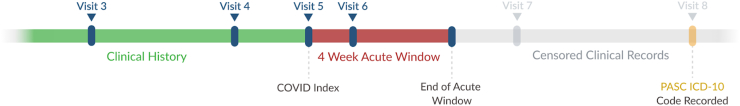
From August 25, 2022 to December 18, 2022 the NIH ran the Long COVID Computational Challenge. The L3C organizers asked participants to use data available in the enclave to predict the risk of a patient developing Long COVID at least 4 weeks after their initial positive SARS-CoV-2 test result (defined as a documented positive result from a qualitative rt-PCR or Antigen lab test within 7 days of a visit). Long COVID cases were selected using the ICD-10-CM U09.9 diagnosis code (“Post-COVID-19 condition, unspecified”) and two time references in a patient's clinical record: their COVID index date and their 4 week acute window. The COVID index date is the date of the patient's first lab-based record of SARS-CoV-2 infection. The 4 week acute window is the 4 weeks after their COVID index date. U09.9 is the diagnosis code made available to US physicians in October of 2021 to code Long COVID in a patient’s EHR. In this challenge, Long COVID was defined as a “True Positive” case as any patient who had a U09.9 diagnosis code entered in their clinical record after the 4 week acute window (Fig. 1). This acute period was necessary to include since the severity and length of a COVID infection can vary from patient to patient and was based on the CDC definition of Long COVID with a potential start of symptoms beginning 4 weeks after the initial COVID infection.2
Fig. 1.
Censoring protocol of patient records. All records available in the N3C enclave prior to (clinical history) and within 4 weeks after (4 week acute window) the COVID index date were available for use by the models. All records after the 4 week acute window were removed from the training and testing datasets. ICD-10-CM code U09.9 that occurred after the 4 week acute window indicated a patient with Long COVID.
Statistics
L3C challenge datasets
Inclusion criteria and sampling protocols
The L3C patient selection process involved identifying patients who tested positive for SARS-CoV-2 and then narrowing to a set of Long COVID cases and matched controls. Possible patients without Long COVID (patients without U09.9 recorded in the EHR) were used as controls and selected from the patients who tested positive for SARS-CoV-2 who had at least one visit after the 4 week acute window and who had no record of U09.9 diagnosis. Control patients were randomly selected at a 1:4 ratio of patients with Long COVID: patients without Long COVID. For this sampling, patients with a U09.9 ICD9 code within 4 weeks of their COVID index were included in the patients with Long COVID count and ratio. To control for data availability differences across patients, patients were matched on the distribution of visits prior to their COVID index visit, meaning that the proportion of patients in both the patients with Long COVID and patients without Long COVID cohorts who had one visit, two visits, three visits, etc. prior to their COVID index date was the same. Patients were not matched on their COVID index date, but solely on their visit counts prior to their COVID index date.
Challenge training and testing datasets
During the challenge, two datasets were used to evaluate model performance: the Hold Out Dataset 1 and the Two Site Dataset 2. Each of these datasets were split into a training and a testing set. For all datasets, all clinical records after the end of the 4 week acute period in each patient clinical record were removed, as participants were asked to build models to predict the chance of Long COVID given only data during and before the 4 week acute period (Fig. 1).
The Hold Out Training Dataset 1 was available to participants during the model development phase (Supplemental Fig 1). This dataset provided a traditional random sample of the available data and contained 9031 cases, 46,226 matched controls, plus 2415 patients who had a U09.9 code in their clinical record after the COVID index date but during the 4 week acute period (These patients are not considered cases, but were provided for teams’ use in exploratory analysis.) The Hold Out Testing Dataset 1 contained 2257 cases and 11,557 controls and was hidden from participants during the challenge but was used to calculate one of the AUROC scores (Table 1).
Table 1.
Demographic breakdown from the datasets used in the L3C.
| Demographic | Hold Out Dataset 1 |
Two Site Dataset 2 |
||||||
|---|---|---|---|---|---|---|---|---|
| Training |
Testing |
Training |
Testing |
|||||
| No PASC | PASC | No PASC | PASC | No PASC | PASC | No PASC | PASC | |
| Age | ||||||||
| 0–17 | 6907 | 276 | 1205 | 71 | 14,489 | 573 | 1571 | 79 |
| 18–34 | 12,876 | 1152 | 1862 | 294 | 24,606 | 2165 | 3904 | 395 |
| 35–59 | 17,100 | 4270 | 2914 | 1041 | 32,028 | 8366 | 4747 | 1445 |
| 60+ | 9937 | 3080 | 1890 | 763 | 19,373 | 5994 | 2530 | 1256 |
| Unknown | 1821 | 253 | 485 | 57 | 4475 | 285 | 108 | 40 |
| Ethnicity | ||||||||
| Not Hispanic or Latino | 37,872 | 7444 | 6585 | 1865 | 73,539 | 15,027 | 9939 | 2684 |
| Hispanic or Latino | 4731 | 852 | 1046 | 220 | 11,674 | 1806 | 1241 | 273 |
| Unknown | 6038 | 735 | 725 | 141 | 9758 | 550 | 1680 | 258 |
| Gender | ||||||||
| Female | 28,243 | 5749 | 5048 | 1428 | 53,768 | 11,266 | 6915 | 2002 |
| Male | 20,350 | 3282 | 3306 | 798 | 41,117 | 6116 | 5942 | 1213 |
| Race | ||||||||
| White | 27,788 | 6302 | 5913 | 1517 | 61,231 | 12,818 | 9791 | 2621 |
| Black | 10,305 | 1618 | 1232 | 432 | 16,334 | 2854 | 507 | 122 |
| Other | 9477 | 949 | 990 | 233 | 14,663 | 1371 | 2531 | 464 |
| Asian | 1071 | 162 | 221 | 44 | 2743 | 340 | 31 | <20 |
| Severity | ||||||||
| Hospitalization | 6679 | 4478 | 1604 | 1124 | 16,337 | 7795 | 1771 | 1641 |
| No Hospitalization | 41,962 | 4553 | 6752 | 1102 | 78,634 | 9588 | 11,089 | 1575 |
The testing data represents the data that was held out from the N3C data that was available at the time of the start of the challenge. The two site validation data represents data from the two sites that were split off from the data after the latest N3C data had been included. Counts of patients who had an unknown gender were not included due to the small counts in compliance with the N3C publication policy. Minor skews (±5) in the counts of the Age, Gender, Ethnicity, and Race of the Two Site Dataset 2 Testing PASC column have been implemented to mask the true count of the category with a <20 count. Patient’s with hospitalization include patients that have either an inpatient visit or emergency room visit during the 4 week acute window.
Over the course of the challenge, new data accumulated within the N3C enclave that was not included in either the training or testing Hold Out Dataset 1. We took this accumulated new data, combined it with the full Hold Out Dataset 1, and resplit the data into the Two Site Training Dataset 2 and the Two Site Testing Dataset 2. The Two Site Testing Dataset 2 represented data from two HCOs that were excluded from the Two Site Training Dataset 2. The purpose of this dataset was to assess generalizability of the models to health systems not represented in the training data. The Two Site Training Dataset 2 represented 17,383 cases, 89,992 controls, and 4979 patients who had a U09.9 code in their record during the 4 week acute period. The Two Site Testing Dataset 2 represented 3219 cases and 12,860 controls (Table 1).
Evaluation process
The evaluation process was carried out in two phases: the quantitative evaluation and the qualitative evaluation. In order to qualify for the quantitative evaluation, L3C teams were required to submit their models by the challenge deadline (December 18, 2022) along with a short manuscript describing their methods, rationale, and preliminary results. All evaluation metrics were generated by the L3C organizers independent of the participating teams. Qualification for the qualitative phase required ranking among the top 10 submissions during the quantitative phase.
Quantitative model ranking
The Area Under the Receiver Operator Curve (AUROC)17 was calculated for each model on both the Hold Out Testing Dataset 1 and Two Site Testing Dataset 2. Models were ranked against each other by calculating the mean of these two AUROCs and ranking on that mean. Prior to testing with Two Site Testing Dataset 2, the models were re-trained on the Two Site Training Dataset 2 by the L3C organizers.
Qualitative model ranking
Qualifying models from the quantitative phase moved to the qualitative evaluation. Models were subjectively scored by a review panel of federal employees, using metrics designed to judge model clinical utility and reproducibility. The clinical utility metrics included lead time, interpretability, transferability, and translational feasibility. The metrics included in reproducibility were technical reproducibility, prediction reproducibility, and documentation. Additional information on the qualitative evaluation metrics can be found in the Supplemental materials (Supplemental Description, Qualitative Review). The final rankings were dictated by the qualitative metrics.
Post-challenge analysis of highest scoring model
After the top performers were announced, level 3 (limited dataset) was accessed for a final model evaluation in order to enable real world date filtering and also to take advantage of the data that had accumulated since the start of the challenge. The L3C organizers created a third dataset using the limited data from the N3C data enclave which includes non-shifted dates, in order to filter the training and testing cohorts to patients who have clinical records after October 2021, the month in which the U09.9 diagnosis code was first introduced. The inclusion criteria represented all patients who had their COVID index date after October 1, 2021, who had a least one visit within one year after their acute COVID window, and who was a patient at an HCO with at least 1% of the patients in the data enclave from that site being patients with Long COVID. The Limited Testing Dataset 3 represented the largest available HCO from this dataset, and the Limited Training Dataset 3 included the remaining HCOs. Models were re-trained on the Limited Training Dataset 3 and scored using the Limited Testing Dataset 3.
The World Health Organization’s (WHO) current definition of Long COVID is persistent or recurrent COVID-19 symptoms 3 months after the acute COVID-19 phase.18 In order to evaluate whether the highest scoring model could potentially transfer to this alternative definition, we evaluated the highest scoring model using a dataset derived from the Hold Out Testing Dataset 1, where patients who received their U09.9 ICD-10 code 90 days after their initial COVID diagnosis were treated as true positives and patients who never had a U09.9 ICD-10 code in their record were treated as the negatives. We did not re-train the models but used the models trained on the Hold Out Training Dataset 1.
Ethics
The N3C data transfer to NCATS is performed under a Johns Hopkins University Reliance Protocol #IRB00249128 or individual site agreements with NIH. The N3C Data Enclave is managed under the authority of the NIH; information can be found at https://ncats.nih.gov/n3c/resources. The analyses described in this publication were conducted with data or tools accessed through the NCATS N3C Data Enclave https://covid.cd2h.org. The data in this challenge was used under the Data Use Request DUR-E6CBB51.
Role of funders
Research reported in this RADx® Rad publication was supported by the National Institutes of Health. The funders of this study were involved in the study design and model evaluation. The funders were not involved in the analysis and interpretation of results or writing of this manuscript. Timothy Bergquist, Johanna Loomba, and Emily Pfaff were supported by Axle Subcontract: NCATS-STSS-P00438. The N3C Publication committee confirmed that this manuscript is in accordance with N3C data use and attribution policies (MSID:1526.544); however, this content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the N3C program. The analyses described in this publication were conducted with data or tools accessed through the NCATS N3C Data Enclave https://covid.cd2h.org and N3C Attribution & Publication Policy v 1.2-2020-08-25b supported by NCATS U24 TR002306, Axle Informatics Subcontract: NCATS-P00438-B.
Results
Participants were asked to use data available in the N3C enclave to predict the risk of a patient developing Long COVID at least 4 weeks after their initial COVID positive test result. Over the course of the challenge, 74 teams designed and built their Long COVID prediction models in the N3C Enclave, resulting in 35 final submissions.
Highest performing models
Overall, the top 10 models scored above 0.8 mean AUROC on both testing datasets (Table 2). The most common method used was XGBoost, followed by LightGBM and other ensemble methods. Features selected by the highest scoring models included patient demographics, visit data (outpatient, ED, and inpatient encounters), measurements (e.g., temperature, BMI, and lab results), conditions (diagnoses and symptoms), drugs (e.g., vaccine records, COVID treatments, medications for comorbid conditions), procedures (e.g., ventilation), and observations (e.g., smoking status). While many of the teams limited the number of features in their models, the largest model included 6000 features while the smallest model contained 131 features with an option for a 36-feature model. Each of the top 10 models was scored by a panel of federal reviewers, with each reviewer scoring their assigned models on a scale from 0 to 10. The highest scoring submission, from Team Convalesco, received a qualitative score of 8.29 while the lowest scoring model scored a 4.69 (Table 2).
Table 2.
Model scores across testing datasets including AUROCs achieved and qualitative evaluation scores.
| Team name | Team institution(s) | Hold out AUROC (95% CI) | Two site AUROC (95% CI) | AUROC difference | Mean AUROC | Qualitative score |
|---|---|---|---|---|---|---|
| Convalesco | University of Chicago | 0.879 (0.873, 0.884) | 0.911 (0.907, 0.915) | 0.032 | 0.895 | 8.29 |
| GAIL | Geisinger | 0.889 (0.884, 0.894) | 0.805 (0.799, 0.812) | −0.084 | 0.847 | 7.52 |
| UC Berkeley Center for Targeted Machine Learning | UC Berkeley | 0.864 (0.858, 0.874) | 0.859 (0.854, 0.865) | −0.005 | 0.862 | 7.39 |
| UW-Madison-BMI | University of Wisconsin–Madison | 0.886 (0.88, 0.893) | 0.841 (0.835, 0.846) | −0.045 | 0.864 | 6.84 |
| Ruvos | Ruvos | 0.851 (0.832, 0.844) | 0.838 (0.832, 0.844) | −0.013 | 0.844 | 6.77 |
| Anonymous Group 1 | 0.884 (0.877, 0.891) | 0.835 (0.829, 0.841) | −0.05 | 0.86 | 5.78 | |
| Anonymous Group 2 | 0.853 (0.846, 0.86) | 0.824 (0.816, 0.83) | −0.029 | 0.839 | 5.57 | |
| Penn | Penn | 0.889 (0.883, 0.895) | 0.841 (0.834, 0.847) | −0.048 | 0.865 | 5.37 |
| Anonymous Group 4 | 0.905 (0.9, 0.91) | 0.836 (0.83, 0.841) | −0.07 | 0.87 | 4.8 | |
| Anonymous Group 5 | 0.837 (0.832, 0.846) | 0.836 (0.83, 0.842) | −0.001 | 0.836 | 4.69 |
Models not explicitly named have been masked as anonymous groups. The final rankings were based on the qualitative scores which combined aspects of reproducibility, interpretability, and translational feasibility (See Supplemental materials for more information).
Top performing model
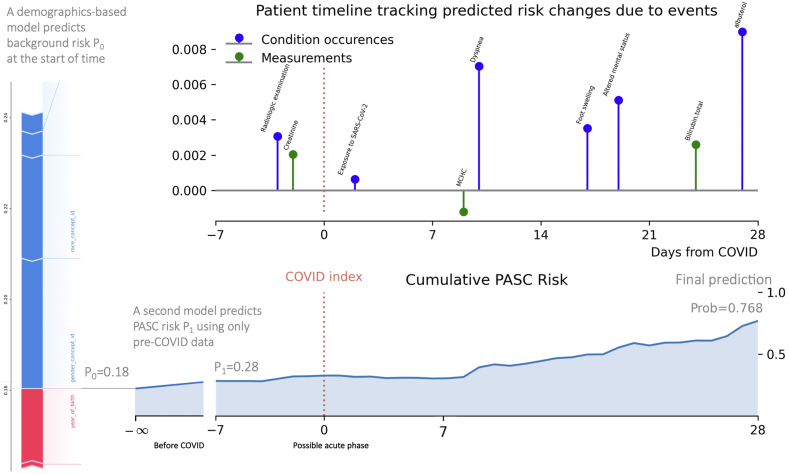
The top performing model, submitted by team Convalesco based out of the University of Chicago, achieved an AUROC of 0.87 on the Hold Out Testing Dataset 1 (Fig. 2d), and increased in performance to 0.91 on Two Site Testing Dataset 2 (the only team in the top 10 to improve) (Fig. 2e). In addition to having the highest mean AUROC, Convalesco also had the highest qualitative score from the federal reviewers. Using a weighted average of 3 LightGBM models and 1 XGBoost model to build a lightweight, portable model, their final submission used 131 total features (100 temporal features and 31 static demographic features) and included a cumulative-risk-over-time visualization dashboard for model interpretation (Fig. 3). Their submission built patient timelines, updating the risk of a patient developing Long COVID in response to new events entering the record. Clinical features for this model included conditions from the acute phase that are sometimes associated with PASC later (such as fatigue, pain, weakness, and dyspnea) as well as prior viral exposure of any type, bloodwork, oxygen saturation, and certain drug exposures during the acute phase. In addition to the larger model of 131 features, Convalesco also built a model with 36 temporal features that achieved a 0.795 AUROC on the Hold Out Testing Dataset 1 (Fig. 2d) and an AUROC of 0.908 on the Two Site Testing Dataset 2 (Fig. 2e) and that improved in calibration between the two datasets (Fig. 2a and b). In addition to the challenge evaluation, Convalesco’s model was retrained using the Limited Dataset 3. Convalesco’s main model improved to an AUC of 0.940, with their 36-feature model improving to an AUC of 0.938 (Fig. 2f). The calibration curves on all the models were good with Brier Scores19 of ∼0.02 (Fig. 2c). The brief project report submitted during the challenge can be found in the Supplemental materials. A table with the 36 temporal features used can be found in Table 2 of the supplemental challenge write up. Convalesco’s model continued to perform well on the test data using the WHO’s 3-month Long COVID definition, with the main model achieving an AUROC of 0.880 and the 36 feature model scoring a 0.800 AUROC, both without re-training.
Fig. 2.
Performance metrics for Convalesco’s highest scoring submission. The calibration curves and area under the receiver operator curves from Convalesco’s highest scoring submission. Each sub-graph shows individual model performances from Convalesco’s submission. The “Main Model” is the model that was evaluated and scored for the L3C evaluation. Model 100 includes only 100 temporal features, Model 36 includes just the top 36 temporal features, and Model Z includes the same 100 temporal features but excludes racial information and data contributor identifiers. (a) The calibration curves from the model on the Hold Out Testing dataset. (b) The calibration curves from the model on the Two Site Testing dataset. (c) The calibration curves from the model on the level 3 post-challenge Limited Testing dataset. (d) The receiver operator curves from the model on the Hold Out Testing dataset. (e) The receiver operator curves from the model on the Two Site Testing dataset. (f) The receiver operator curves from the model on the level 3 post-challenge Limited Testing dataset. While the model wasn’t well calibrated to the Hold Out testing dataset, the model generalized well to two out of sample datasets from separate data contributing partners and improved further after re-training and evaluation on the level 3 limited dataset.
Fig. 3.
Interpretability dashboard from Convalesco’s submission. The chart represents a prototype patient risk timeline. The top graph shows the single-event contributions toward the predicted PASC risk at Day 28. The risk change was calculated based on the difference between the final prediction and the hypothetical risk using all data except one event. Only a subsample of events are shown. The bottom chart shows the day-by-day predictions of cumulative risk based on events prior to the day.
2nd highest performing model
The 2nd highest performing model, submitted by team GAIL (Geisinger AI Laboratory based out of Geisinger Health System) was a portable, efficient, and accurate model using fewer features than the competition, resulting in a translational Long COVID prediction clinical decision support tool that included design plans for EHR-ready summary visualizations for clinician interpretation. A description of their methods is available in the Supplemental materials as well as their preprint manuscript.20
3rd highest performing model
The 3rd placed model, submitted by the UC Berkeley Center for Targeted Machine Learning team built a clinical prediction model that was a weighted combination of many smaller prediction models (this combined model is known as an ensemble or a Super Learner). The model used various aspects of a patient’s health such as cardiovascular health, respiratory health, history of hospital use, and age to predict the patient’s risk for developing PASC/Long COVID. A description of their methods is available in the Supplemental materials as well as their manuscript.21
Honorable mentions
While the honorable mention teams did not have a top three ranking, their approaches stood out during the qualitative evaluation as unique approaches worth highlighting. Team UW-Madison-BMI (UW-Madison Department of Biostatistics & Medical Informatics (BMI)) built a Long COVID prediction model by looking at high-level clinical concepts in a patient’s clinical history to evaluate their risk of developing Long COVID. Team Penn took a unique approach to this challenge and developed a Long COVID prediction model that looked at both static clinically relevant data points as well as dynamically selected data points. This grounded their model in clinical relevance but allowed it to adapt to future changes in new data. Team Ruvos developed a prediction model that used broad categories of disease to predict a patient’s risk of developing Long COVID. Their model was highly generalizable to new EHR data. Each team’s brief project report submitted during the challenge are found in the Supplemental materials.
Discussion
Community challenges can serve to coalesce research communities from an array of scientific domains to catalyze innovation around research questions of interest. The L3C brought together a group of researchers from clinical and machine learning domains from academia and private institutions to build clinical prediction models on a large, national data repository. Prior to this challenge, no risk prediction model for developing Long COVID had been built using the N3C data enclave. L3C incentivized 225 researchers comprising 74 research teams to build 35 Long COVID risk prediction models. The community challenge showed that a wide variety of approaches to the data and models can be applied with similar results. The top teams were set apart from the others primarily by their clinical relevance, their generalizability, and their model’s interpretability. Our multi-faceted quantitative model evaluation provided a robust approach to identifying the top ten models. Furthermore, our use of qualitative metrics, particularly interpretability and translational feasibility, incentivized the development of models with creative interpretability dashboards and thoughtful feature selection.
The highest ranked model, Convalesco, designed a portable, generalizable model that improved in performance when evaluated against the Two Site Testing Dataset 2. Due to the heterogeneity of coding patterns across health systems, it is difficult to design a model that does not overfit to local patterns, but is generalizable to data partners not included in the training data. This model was one of only two models to improve on the external data partners, and had the largest improvement of any model submitted. Although fluctuations in scores are typical across training sets, the fact that the score did not worsen, suggests that Convalesco’s initial high performance was not related to overfitting. The team demonstrated that model performance was not significantly compromised when some features of primary importance were removed, further demonstrating the resilience of the model. Convalesco’s model performed exceptionally well using just 36 temporal features and a third version of their model, Model Z, which removed HCO specific information and included additional censoring protocols to further refine the training data, generalized well to all the testing datasets (Fig. 2). Additionally, each of their models performed well on the WHO’s 3-month definition of Long COVID, indicating that these models are not reliant on a 4-week post acute COVID definition and could be adapted to alternative definitions.
Of the 36 temporal features, some of the most important features are related to hospital utilization such as “Outpatient visit” and “Hospitalization”; however, other important features, such as glucose in serum, could be of interest for follow-up investigation (Supplemental materials, Top Performer Model write up, Table 2). There is literature suggesting that acute COVID-19 can induce persistent hyperglycemia22, 23, 24 so it is possible that Convalesco’s model identified biologically relevant lab values that could be used to better understand long COVID; however, further validation is required.
One aspect of Convalesco’s model that stood out during the qualitative evaluation was the potential transferability of their models to a new health system. When implementing a model into clinical care, each feature must be mapped to an existing datastream within the health system. As models grow, it becomes less likely that each new variable will have a corresponding variable available in the new system. Convalesco’s model being the smallest submission while maintaining the highest accuracy should serve as a lesson for future clinical prediction model development. Increasing the size of the feature space is not a guarantee of improved model performance, but will usually decrease translational feasibility to a new health system.
A second key characteristic was the interpretable charts for showcasing patient-level longitudinal risk factors that informed a patient’s predicted risk score (Fig. 3). These patient-level timelines of clinical events showed the change in the predicted risk of developing Long COVID caused by individual clinical events. Interpretability methods that not only highlight important variables in a patient record but also highlights specific events that contribute to health risks increases the model’s clinical utility.
In order to be translated into clinical use, each model will need to have their variables mapped to the equivalent data variables available at the implementing site. Once this is complete, the model will need to be fine-tuned (given additional training) on data from the implementing site and then prospectively evaluated on new incoming patients to ensure that the model can maintain accuracy while in use in a live clinical scenario. Finally, the model will need to be incorporated into clinical workflows such that the right information is getting to a clinician at the right time. Convalesco’s model shows unique promise in being able to succeed through each of these implementation steps since it uses a small feature space, shows good performance on multiple datasets, and incorporates an easily understandable visualization dashboard.
The techniques used in the top performing submissions are likely generalizable to other clinical prediction problems and the code for all highlighted models have been made available under an open source license (See Data Sharing). These models could be repurposed and adapted to address future clinical prediction problems. Convalesco’s visualization technique is not Long COVID specific and could be generalized to other clinical prediction problems or incorporated into clinical decision support tools. While the resulting models demonstrated the feasibility of machine learning based prognostics for Long COVID, further development and validation of these models is needed by the broader scientific community with more diverse datasets.
Common pitfalls of solutions submitted to L3C
Many of the teams incorporated questionable features into their models that were not generalizable outside of the N3C enclave or to a live EHR system. Of note, many models used the masked ID of the patient’s health care system as a variable. This feature has known predictive properties within the N3C Enclave for predicting who might be labeled with Long COVID based on local coding practices. It may also cause the model to assign higher probability of Long COVID to patients at sites that see and code more patients with Long COVID. This variable does not capture underlying biological mechanisms, is not meaningful to personalized prediction models based on patient features rather than site features, and would not transfer well to an external clinic.
A few teams identified proxies or synonyms of the Long COVID U09.9 ICD-10-CM code (e.g., “Post-viral syndrome,” “Sequelae of infectious disease”, “Long COVID specialty clinic visit”) that appeared in the patient chart during the acute phase, and used those in their model. While, in many cases, these features had outsized importance within the models, and improved their AUROC, these features would not be helpful in a clinical setting as the goal was to predict who is at risk for Long COVID, not who already has Long COVID.
Caveats and limitations
This challenge was run on a large, centralized collection of EHR data collected and harmonized from 76 contributing data partners. The limitations of this challenge include any and all limitations from using structured data (such as ICD mapped codes) from the EHR as the data source, including: (1) varying charting practices across clinicians, clinics, and HCOs, (2) non-representative patient population in terms of demographics, health, and social determinants of health, (3) incomplete health data capture on any given patient at the participating sites, meaning that some facts are not captured and drug or condition start dates noted in the record may be later than true start dates, (4) incomplete or lack of data capture from unstructured data sources, (5) incomplete or lack of data capturing environmental exposures or other co-exposures, (6) and varying coding practices across HCOs and over time (e.g., ICD code U09.9 was not universally adopted nor was the code in use prior to October 2021).
Additional limitations should be noted that are related to all research involving harmonized, multi-institutional EHR data: (1) the N3C sample is large, but is still a subset of HCOs in the US; (2) the lack of raw notes data excludes the use of Long COVID symptoms (e.g., post-exertional malaise) that may only be recorded in clinical notes; and (3) some data is lost when mapping from local EHRs to local data warehouses from which data extracts are shared. Also note that for regulatory reasons, this challenge used the deidentified N3C data where each patient record is date shifted by a random number of days, prohibiting use of real world dates in these predictive models.
Lastly, it is essential that we recognize the time biases related to the evolving nature of the medical community's understanding of Long COVID, coding practices, and patient care seeking behaviors related to this set of conditions. While this challenge illuminated the strengths and weaknesses of predictive models of Long COVID, it is essential to continue to retrain, evaluate and refine these models over time. Predictive models and computable phenotypes leveraging real world electronic health data can only be evaluated using patients who had access to care and who were also diagnosed by a provider. Therefore we must be cautious to not apply them in a way that would reinforce implicit provider biases.
Community challenges serve as catalysts for driving research and building new communities of researchers from diverse backgrounds to tackle open questions and develop innovative solutions. The Long COVID Computational Challenge resulted in 35 Long COVID risk prediction models with the top performing team producing a light weight, accurate model that incorporated a patient-level timeline visualization dashboard highlighting clinical events that were contributing to the increasing or decreasing risk of a patient developing Long COVID. Our approach of integrating qualitative scoring metrics into the machine learning evaluation process incentivized the development of an interpretable and portable Long COVID risk prediction model. Similarly, our use of a multi-healthcare organization data repository like N3C and multi-faceted quantitative evaluations produced an accurate and generalizable model that has the potential of being refined, validated, and implemented into a clinic setting. The top performing methods are generalizable to other clinical prediction problems, and as a result of this challenge, have been made publicly available to serve as templates for researchers using standardized electronic health record data to build clinical prediction models.
Contributors
Project administration: TB, JL, EP; data curation: TB, JL, EP; figures: TB, FX, ZZ, YZ; formal analysis: TB, JL, EP; software: FX, ZZ, YZ, EM, BB, GS, TM, GD, AT, ZB, YJ, HL, JC, SS, RP, AM, RP, ML, JC, AH, JG, GC, NV, ZL, YW, AS, JH, ZD, QL, MN, JH, DM, EW, HP, EG, JH, JB; writing - original draft: TB, JL, EP; writing - review & editing: All co-authors.
TB and JL accessed and verified the underlying data. All authors read and approved the final version of the manuscript. Authorship was determined using ICMJE recommendations.
The Long COVID Computational Challenge Participants submitted models to the L3C for evaluation.
The N3C Consortium collected, harmonized, and governed the data used in this study and provided the computational infrastructure for this study.
Data sharing statement
This study used both deidentified and limited data from the National COVID Cohort Collaborative. Interested researchers can request access to N3C at https://ncats.nih.gov/n3c/about/applying-for-access. Code for defining the challenge cohorts, transforming raw OMOP data, and comparing the challenge models for this analysis is available through the NCATS N3C Data Enclave covid.cd2h.org/enclave with access procedures as described above. The project is available under the Data Use Request RP-D5AE34 “NIH Long COVID Computational Challenge (L3C)”. All six of the models discussed in this publication are publicly available on github and can be found at:
Convalesco: https://github.com/levinas/long-covid-prediction.
GAIL: https://github.com/geisinger-ai-lab/gail-l3c.
UC Berkeley Center for Targeted Machine Learning: https://github.com/BerkeleyBiostats/l3c_ctml.
Penn: https://github.com/nvelingker/Penn-Long-COVID-Prediction-Model.
Ruvos: https://github.com/JakeHightower/N3C-Long-Covid-Challenge.
UWisc-BMI: https://github.com/GGGGFan/L3C_Solution_Team_UW_Madison_BMI.
Declaration of interests
Danielle Mowery serves as an unpaid member of the Epic Cosmos Governing Council. Romain Pirracchio received funding from the FDA CERSI grant U01FD005978 and the PCORI grant P0562155 and received a consulting honorarium from Phillips. Martin van der Laan received funding from the NIAID grant 5R01AI074345. Johanna Loomba received contract funding from the NIH RECOVER program. Emily Pfaff received funding from the NIH and PCORI. The views expressed in this manuscript are solely those of the authors and do not necessarily represent those of the National Institutes of Health, the U.S. Department of Health and Human Services or the U.S. government. Qi Long was supported by grants from the NIH.
Acknowledgements
Danielle Mowery was supported by NIH/NCATS UL1TR001878. Qi Long was supported by grants from the NIH. Zachary Butzin-Dozier, John Colford, Haodong Li, Yunwen Ji, Alan Hubbard, Jeremy Coyle, Andrew Mertens, and Seraphina Shi were supported by The Bill & Melinda Gates Foundation global development grant OPP1165144.
We would like to thank the following individuals for their contributions to the conception, development, and evaluation of the L3C:
Jue Chen, Erin Iturriaga, Orlando Lopez, and Ivonne H. Schulman for co-chairing the NIH Long COVID Computational Challenge working group and for spearheading the development and execution of the challenge. Gabriel Anaya, Denis Buxton, Qi Duan, Josh Fessel, Taylor Gilliland, Aminul Islam, Bonnie Joubert, Bill Kapogiannis, Sweta Ladwa, Leonie Misquitta, Asif Rizwan, Rachel Scheinert, Daniel Shaughnessy, Danilo Tagle, Yanli Wang, and Amy Wernimont for supporting the development and execution of the challenge as members of the NIH Long COVID Computational Challenge working group. Jennifer Banarez, Thomas Bennett, Adrienne Lane, and Catherine Perrone, for their administrative and project management support. Maya Choudhury, Saad Ljazouli, and Amin Manna for their technical support and enclave expertise. Jake Albrecht for his administrative support.
We would like to thank The Long COVID Computational Challenge Participants for their participation in the L3C and for their many submissions to the challenge.
The N3C Publication committee confirmed that this manuscript is in accordance with N3C data use and attribution policies (MSID:1526.544); however, this content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the N3C program. The analyses described in this publication were conducted with data or tools accessed through the NCATS N3C Data Enclave https://covid.cd2h.org and N3C Attribution & Publication Policy v 1.2-2020-08-25b supported by NCATS U24 TR002306, Axle Informatics Subcontract: NCATS-P00438-B. This research was possible because of the patients whose information is included within the data and the organizations (https://ncats.nih.gov/n3c/resources/data-contribution/data-transfer-agreement-signatories) and scientists who have contributed to the on-going development of this community resource (https://doi.org/10.1093/jamia/ocaa196).
We gratefully acknowledge the following core contributors to N3C: Meredith Adams, Caleb Alexander, Benjamin Amor, Alfred (Jerrod) Anzalone, Benjamin Bates, Will Beasley, Tellen D. Bennett, Mark M. Bissell, Eilis Boudreau, Samuel Bozzette, Katie Rebecca Bradwell, Carolyn Bramante, Don Brown, Penny Wung Burgoon, John Buse, Tiffany Callahan, Kenrick Dwain Cato, Scott Chapman, Christopher Chute, Jaylyn Clark, Marshall Clark, Will Cooper, Lesley Cottrell, Karen Crowley, Mariam Deacy, Christopher Dillon, David A. Eichmann, Mary Emmett, Rebecca Erwin-Cohen, Patricia A. Francis, Evan French, Rafael Fuentes, Davera Gabriel, Joel Gagnier, Nicole Garbarini, Jin Ge, Kenneth R. Gersing, Andrew T. Girvin, Valery Gordon, Alexis Graves, Justin Guinney, Melissa A. Haendel, Jeremy Richard Harper, J.W. Awori Hayanga, Brian Hendricks, Wenndy Hernandez, Elaine Hill, William Hillegass, Stephanie S. Hong, Dan Housman, Robert Hurley, Jessica Islam, Randeep Jawa, Steve Johnson, Rishi Kamaleswaran, Warren A. Kibbe, Farrukh M Koraishy, Kristin Kostka, Michael G. Kurilla, Adam M. Lee, Harold Lehmann, Hongfang Liu, Johanna Loomba, Charisse Madlock-Brown, Sandeep Mallipattu, Amin Manna, Federico Mariona, Emily Carlson Marti, Greg Martin, Jomol Mathew, Diego Mazzotti, Julie A. McMurry, Hemalkumar Mehta, Sam G. Michael, Robert T. Miller, Leonie Misquitta, Richard A. Moffitt, Michele Morris, Kimberly Murray, Lavance Northington, Shawn T. O'Neil, Amy Olex, Matvey B. Palchuk, Brijesh Patel, Rena Patel, Philip R.O. Payne, Emily R. Pfaff, Jami Pincavitch, Lili Portilla, Fred Prior, Saiju Pyarajan, Lee Allan Pyles, Nabeel Qureshi, Peter Robinson, Joni L. Rutter, Ofer Sadan, Nasia Safdar, Amit Saha, Joel H. Saltz, Joel Saltz, Mary Morrison Saltz, Clare Schmitt, Soko Setoguchi, Noha Sharafeldin, Anjali A. Sharathkumar, Usman Sheikh, Hythem Sidky, George Sokos, Andrew Southerland, Heidi Spratt, Justin Starren, Vignesh Subbian, Christine Suver, Cliff Takemoto, Meredith Temple-O'Connor, Umit Topaloglu, Satyanarayana Vedula, Anita Walden, Kellie M. Walters, Cavin Ward-Caviness, Adam B. Wilcox, Ken Wilkins, Andrew E. Williams, Chunlei Wu, Elizabeth Zampino, Xiaohan Tanner Zhang, Andrea Zhou, and Richard L. Zhu Details of contributions available at covid.cd2h.org/core-contributor.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105333.
Contributor Information
Timothy Bergquist, Email: timothy.bergquist@mssm.edu.
Long COVID Computational Challenge Participants:
Ataes Aggarwal, Joseph Agor, Amera Al-Amery, Oluwatobiloba Aminu, Adit Anand, Corneliu Antonescu, Mehak Arora, Sayed Asaduzzaman, Tanner Asmussen, Mahdi Baghbanzadeh, Frazier Baker, Bridget Bangert, Laila Bekhet, Biplab Bhattacharya, Jenny Blase, Zachary Butzin-Dozier, Brian Caffo, Hao Chang, Zeyuan Chen, Jiandong Chen, Jeffrey Chiang, Peter Cho, Robert Cockrell, Parker Combs, Jeremy Coyle, Ciara Crosby, Zongyu Dai, Ran Dai, Anseh Danesharasteh, Elif Yildirim, Grant Delong, Ryan Demilt, Kaiwen Deng, Sanjoy Dey, Rohan Dhamdhere, Andrew Dickson, Phoebe Dijour, Dong Dinh, Richard Dixon, Albi Domi, Souradeep Dutta, Mirna Elizondo, Zeynep Ertem, Solomon Feuerwerker, Danica Fliss, Jennifer Fowler, Sunyang Fu, Kelly Gardner, Neil Getty, Mohamed Ghalwash, Logan Gloster, Phil Greer, Yuanfang Guan, Colby Ham, Samer Hanoudi, Jeremy Harper, Nathaniel Hendrix, Leeor Hershkovich, Jake Hightower, Junjie Hu, Jiani Huang, Yu Huang, Tongtong Huang, Junguk Hur, Monica Isgut, Hamid Ismail, Grant Izmirlian, Kuk Jang, Christianah Jemiyo, Hayoung Jeong, Yunwen Ji, Xiayan Ji, Ming Jiang, Sihang Jiang, Xiaoqian Jiang, Yuye Jiang, Akin Johnson, Zach Analyst, Saarthak Kapse, Uri Kartoun, Dukka KC, Zahra Fard, Tim Kosfeld, Spencer Krichevsky, Mike Kuo, Dale Larie, Lauren Lederer, Shan Leng, Ziyang Li, Hongyang Li, Haodong Li, Jianfu Li, Tiantian Li, Xinwen Liang, Hengyue Liang, Feifan Liu, Daniel Liu, Gang Luo, Tamanna Munia, Ravi Madduri, Vithal Madhira, Shivali Mani, Farzaneh Mansourifard, Robert Matson, Andrew Mertens, Vangelis Metsis, Pablo Meyer, Catherine Mikhailova, Dante Miller, Christopher Milo, Elliot Mitchell, Gourav Modanwal, Ronald Moore, David Morgenthaler, Rasim Musal, Mayur Naik, Vinit Nalawade, Rohan Narain, Saideep Narendrula, Alena Obiri, Satoshi Okawa, Chima Okechukwu, Toluwanimi Olorunnisola, Tim Ossowski, Harsh Parekh, Jean Park, Saaya Patel, Jason Patterson, Chetan Paul, Le Peng, Diana Perkins, Suresh Pokharel, Dmytro Poplavskiy, Zach Pryor, Sarah Pungitore, Hong Qin, Salahaldeen Rababa, Mahbubur Rahman, Elior Rahmani, Gholamali Rahnavard, Md Raihan, Suraj Rajendran, Sarangan Ravichandran, Chandan Reddy, Abel Reyes, Ali Roghanizad, Sean Rouffa, Xiaoyang Ruan, Arpita Saha, Sahil Sawant, Melody Schiaffino, Diego Seira, Saurav Sengupta, Ruslan Shalaev, Gaurav Shetty, Seraphina Shi, Linh Shinguyen, Karnika Singh, Soumya Sinha, Damien Socia, Halen Stalians, Charalambos Stavropoulos, Jan Strube, Devika Subramanian, Jiehuan Sun, Ju Sun, Chengkun Sun, Prathic Sundararajan, Salmonn Talebi, Edward Tawiah, Jelena Tesic, Mikaela Thiess, Raymond Tian, Luke Torre-Healy; Ming-Tse Tsai, David Tyus, Madhurima Vardhan, Neelay Velingker, Benjamin Walzer, Jacob Walzer, Junda Wang, Lu Wang, Will Wang, Jonathan Wang, Yisen Wang, Chad Weatherly, Fanyou Wu, Yifeng Wu, Yinjun Wu, Fangfang Xia, Hao Yan, Zhichao Yang, Biao Ye, Rui Yin, Changyu Yin, Yun Yoo, Albert You, June Yu, Martin Zanaj, Zachary Zaiman, Kai Zhang, Xiaoyi Zhang, Tianmai Zhang, Zixuan Zhao, Degui Zhi, Yishan Zhong, Huixue Zhou, Andrea Zhou, Yuanda Zhu, Yitan Zhu, and Sophie Zhu
N3C Consortium:
Meredith Adams, Caleb Alexander, Benjamin Amor, Alfred Anzalone, Benjamin Bates, Will Beasley, Tellen Bennett, Mark Bissell, Eilis Boudreau, Samuel Bozzette, Katie Bradwell, Carolyn Bramante, Don Brown, Penny Burgoon, John Buse, Tiffany Callahan, Kenrick Cato, Scott Chapman, Christopher Chute, Jaylyn Clark, Marshall Clark, Will Cooper, Lesley Cottrell, Karen Crowley, Mariam Deacy, Christopher Dillon, David Eichmann, Mary Emmett, Rebecca Erwin-Cohen, Patricia Francis, Evan French, Rafael Fuentes, Davera Gabriel, Joel Gagnier, Nicole Garbarini, Jin Ge, Kenneth Gersing, Andrew Girvin, Valery Gordon, Alexis Graves, Justin Guinney, Melissa Haendel, Jeremy Harper, J.W. Hayanga, Brian Hendricks, Wenndy Hernandez, Elaine Hill, William Hillegass, Stephanie Hong, Dan Housman, Robert Hurley, Jessica Islam, Randeep Jawa, Steve Johnson, Rishi Kamaleswaran, Warren Kibbe, Farrukh Koraishy, Kristin Kostka, Michael Kurilla, Adam Lee, Harold Lehmann, Hongfang Liu, Johanna Loomba, Charisse Madlock-Brown; Sandeep Mallipattu, Amin Manna, Federico Mariona, Emily Marti, Greg Martin, Jomol Mathew, Diego Mazzotti, Julie McMurry, Hemalkumar Mehta, Sam Michael, Robert Miller, Leonie Misquitta, Richard Moffitt, Michele Morris, Kimberly Murray, Lavance Northington, Shawn O’Neil, Amy Olex, Matvey Palchuk, Brijesh Patel, Rena Patel, Philip Payne, Emily Pfaff, Jami Pincavitch, Lili Portilla, Fred Prior, Saiju Pyarajan, Lee Pyles, Nabeel Qureshi, Peter Robinson, Joni Rutter, Ofer Sadan, Nasia Safdar, Amit Saha, Joel Saltz, Mary Saltz, Clare Schmitt, Soko Setoguchi, Noha Sharafeldin, Anjali Sharathkumar, Usman Sheikh, Hythem Sidky, George Sokos, Andrew Southerland, Heidi Spratt, Justin Starren, Vignesh Subbian, Christine Suver, Cliff Takemoto, Meredith Temple-O'Connor, Umit Topaloglu, Satyanarayana Vedula, Anita Walden, Kellie Walters, Cavin Ward-Caviness, Adam Wilcox, Ken Wilkins, Andrew Williams, Chunlei Wu, Elizabeth Zampino, Xiaohan Zhang, Andrea Zhou, and Richard Zhu
Appendix ASupplementary data
References
- 1.National Center for Health Statistics . Long COVID; 2023. U.S. Census Bureau, household pulse survey, 2022–2023.https://www.cdc.gov/nchs/covid19/pulse/long-covid.htm [Google Scholar]
- 2.CDC . Centers for Disease Control and Prevention; 2023. Long COVID or post-COVID conditions.https://www.cdc.gov/coronavirus/2019-ncov/long-term-effects/index.html [Google Scholar]
- 3.Deer R.R., Rock M.A., Vasilevsky N., et al. Characterizing long COVID: deep phenotype of a complex condition. EBioMedicine. 2021;74 doi: 10.1016/j.ebiom.2021.103722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brightling C.E., Evans R.A. Long COVID: which symptoms can be attributed to SARS-CoV-2 infection? Lancet. 2022;400:411–413. doi: 10.1016/S0140-6736(22)01385-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pfaff E.R., Girvin A.T., Bennett T.D., et al. Identifying who has long COVID in the USA: a machine learning approach using N3C data. Lancet Digital Health. 2022;4(7):e532–e541. doi: 10.1016/S2589-7500(22)00048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reese J.T., Blau H., Casiraghi E., et al. Generalisable long COVID subtypes: findings from the NIH N3C and RECOVER programmes. EBioMedicine. 2023;87 doi: 10.1016/j.ebiom.2022.104413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norel R., Rice J.J., Stolovitzky G. The self-assessment trap: can we all be better than average? Mol Syst Biol. 2011;7:537. doi: 10.1038/msb.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saez-Rodriguez J., Costello J.C., Friend S.H., et al. Crowdsourcing biomedical research: leveraging communities as innovation engines. Nat Rev Genet. 2016;17:470–486. doi: 10.1038/nrg.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer P., Saez-Rodriguez J. Advances in systems biology modeling: 10 years of crowdsourcing DREAM challenges. Cell Syst. 2021;12:636–653. doi: 10.1016/j.cels.2021.05.015. [DOI] [PubMed] [Google Scholar]
- 10.Andreoletti G., Pal L.R., Moult J., Brenner S.E. Reports from the fifth edition of CAGI: the critical assessment of genome interpretation. Hum Mutat. 2019;40:1197–1201. doi: 10.1002/humu.23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kryshtafovych A., Schwede T., Topf M., Fidelis K., Moult J. Critical assessment of methods of protein structure prediction (CASP)-Round XIV. Proteins. 2021;89:1607–1617. doi: 10.1002/prot.26237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou N., Jiang Y., Bergquist T.R., et al. The CAFA challenge reports improved protein function prediction and new functional annotations for hundreds of genes through experimental screens. Genome Biol. 2019;20:244. doi: 10.1186/s13059-019-1835-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai B., Li B., Kiga N., et al. Matching phenotypes to whole genomes: lessons learned from four iterations of the personal genome project community challenges. Hum Mutat. 2017;38:1266–1276. doi: 10.1002/humu.23265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergquist T., Wax M., Bennett T.D., et al. A framework for future national pediatric pandemic respiratory disease severity triage: the HHS pediatric COVID-19 data challenge. J Clin Transl Sci. 2023;7:e175. doi: 10.1017/cts.2023.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haendel M.A., Chute C.G., Bennett T.D., et al. The national COVID cohort collaborative (N3C): rationale, design, infrastructure, and deployment. J Am Med Inform Assoc. 2021;28:427–443. doi: 10.1093/jamia/ocaa196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennett T.D., Moffitt R.A., Hajagos J.G., et al. Clinical characterization and prediction of clinical severity of SARS-CoV-2 infection among US adults using data from the US national COVID cohort collaborative. JAMA Netw Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.16901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic test evaluation. Caspian J Intern Med. 2013;4:627–635. [PMC free article] [PubMed] [Google Scholar]
- 18.Post COVID-19 condition (Long COVID) n.d. https://www.who.int/europe/news-room/fact-sheets/item/post-covid-19-condition
- 19.Brier G.W. Verification of forecasts expressed in terms of probability. Mon Weather Rev. 1950;78:1–3. [Google Scholar]
- 20.Bhattacharya B., DeLong G., Mitchell E.G., Munia T.T.K., Shetty G., Tariq A. A Long COVID risk predictor focused on clinical workflow integration. medRxiv. 2023 doi: 10.1101/2023.05.26.23290243. [DOI] [Google Scholar]
- 21.Butzin-Dozier Z., Ji Y., Li H., Coyle J., Shi J., Philips R.V., et al. Predicting long COVID in the national COVID cohort collaborative using super learner: cohort study. JMIR Public Health Surveill. 2023;10(1) doi: 10.2196/53322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Downes J.M., Foster J.A. Prolonged hyperglycemia in three patients with type 2 diabetes after COVID-19 infection: a case series. J Fam Med Prim Care. 2021;10:2041–2043. doi: 10.4103/jfmpc.jfmpc_2271_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goel V., Raizada A., Aggarwal A., et al. Long-term persistence of COVID-induced hyperglycemia: a cohort study. Am J Trop Med Hyg. 2024;110:512–517. doi: 10.4269/ajtmh.22-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Emiroglu C., Dicle M., Yesiloglu C., Gorpelioglu S., Aypak C. Association between newly diagnosed hyperglycemia/diabetes mellitus, atherogenic index of plasma and obesity in post-COVID-19 syndrome patients. Endocrine. 2024;84:470–480. doi: 10.1007/s12020-023-03611-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.