Abstract
Early identification of plant fungal diseases is critical for timely treatment, which can prevent significant agricultural losses. While molecular analysis offers high accuracy, it is often expensive and time-consuming. In contrast, image processing combined with machine learning provides a rapid and cost-effective alternative for disease diagnosis. This study presents a novel approach for detecting four common fungal diseases in tomatoes, Botrytis cinerea, Fusarium oxysporum, Alternaria alternata, and Alternaria solani, using both RGB (visible) and hyperspectral (400–950 nm) imaging of plant leaves over the first 11 days post-infection. Data sets were generated from leaf samples, and a range of statistical, texture, and shape features were extracted to train machine learning models. The spectral signatures of each disease were also developed for improved classification. The random forest model achieved the highest accuracy, with classification rates for RGB images of 65%, 71%, 75%, 77%, 83%, and 87% on days 1, 3, 5, 7, 9, and 11, respectively. For hyperspectral images, the classification accuracy increased from 86% on day 1 to 98% by day 11. Two- and three-dimensional spectral analyses clearly differentiated healthy plants from infected ones as early as day 3 for Botrytis cinerea. The Laplacian score method further highlighted key texture features, such as energy at 550 and 841 nm, entropy at 600 nm, correlation at 746 nm, and standard deviation at 905 nm, that contributed most significantly to disease detection. The method developed in this study offers a valuable and efficient tool for accelerating plant disease diagnosis and classification, providing a practical alternative to molecular techniques. .
Keywords: Classification, Feature selection, Image processing, Plant disease, Random forest, Spectral signature
Highlights
-
•
Identification of spectral signature of four tomato fungal diseases, namely: Alternaria alternata, Alternaria solani, Botrytis cinerea, and Fusarium oxysporum.
-
•
Diagnosis and classification of common tomato fungal diseases using RGB and hyperspectral image processing.
-
•
Introduction of important and effective features in RGB and hyperspectral images for diagnosing plant diseases.
-
•
Early detction of plant disease using hyperspectral image processing.
1. Introduction
Diseases in crop plants are the main reason for reducing the quantity and quality of agricultural production. Although symptoms of plant diseases might be evident in various parts of plants, the leaves are usually used to diagnose diseases. Diagnosing plant diseases with the naked eye requires human experts. This method can be accompanied by remarkable error, and a large group of specialists is required to constantly scout agricultural fields [1]. Although direct detection at the molecular level can accurately classify plant diseases, it is time-consuming and costly. In comparison, indirect detection based on machine vision is more attractive in practice due to its non-invasive characteristics and the ability to identify plant diseases through various parameters such as color, morphology, and temperature changes. Automatic disease diagnosis and monitoring can facilitate timely disease control. This can lead to an increased product yield, improved product quality, and reduced pesticide application.
Machine vision and image processing have been extensively used to identify and classify plant diseases based on their symptoms [2,3]. However, this method can only detect disease at late disease development stages, when it might be late for controlling the spread of disease in the entire field. Technological advances have created opportunities for non-destructive and early identification of plant diseases through spectroscopy. Hyperspectral imaging has grown significantly in the last two decades for non-destructive investigation of biotic and abiotic stresses in crops at different spatial and temporal scales [[4], [5], [6]]. Hyperspectral imaging uses a combination of spectroscopy and imaging techniques to obtain detailed information about the full spectral wavelength of a sample for each pixel in the image. This is achieved by taking photographs at various wavelengths and combining them to create a three-dimensional hyperspectral cube. Disease infections can change the biophysical and biochemical characteristics of plants, resulting in structural, spatial, and physiological changes. These alterations can affect the spectral characteristics of plants, which can be identified by hyperspectral imaging platforms.
Close-range remote sensing techniques such as spectral, multispectral, and hyperspectral imaging can help diagnose plant diseases effectively. Hyperspectral imaging is a powerful technology that can capture high spatial and spectral resolutions, offering a superior advantage over RGB and multispectral imaging [7,[8], [9]]. It can provide hundreds of images covering wavelengths in the visible and near-infrared range. The most effective, i.e., sensitive, wavelengths can be identified by combining hyperspectral imaging with feature selection algorithms. This allows to obtain an extensive range of features, including spectral reflectance, vegetation indices, shape, descriptive statistics, and texture information. These features facilitate a more comprehensive understanding of plant diseases, ultimately aiding in better management practices. Much research has been conducted regarding the use of color and hyperspectral images in the diagnosis and classification of plant diseases [10].
Yuan et al. [11] investigated the anthracnose disease in tea plants using hyperspectral imaging. By analyzing the images in the spectral range of 400–1000 nm, they identified wavelengths equal to 542, 686, and 754 nm as the sensitive spectra in detecting the tea anthracnose disease. They also developed an approach for classifying images containing various backgrounds and showed that the approach was not affected by the background of leaves. The accuracy of the disease diagnosis for the classification at the pixel and leaf levels was 94 % and 98 %, respectively. Abdulridha et al. (2020) used hyperspectral imaging in the range of 380–1020 nm and radial basis function (RBF) machine learning (ML) to develop a technique for detecting powdery mildew in squash at different stages of disease development. A total of 29 vegetation indices were used to diagnose diseased and healthy leaves, and the classification accuracy was reported as 89 %. Fernández et al. (2021) investigated the spectral changes caused by powdery mildew in cucumber. They used hyperspectral imaging in the range of 400–900 nm, and developed a principal component analysis (PCA) model to determine the sensitive wavelengths in disease diagnosis. They used two spectral variables, the red-well point and the red-edge inflection point, to extract features. Furthermore, support vector machine (SVM) with linear kernel was used to classify diseased and healthy leaves. The accuracy of the algorithm six days after the disease transmission was 89 %. Zhang et al. [12] investigated the spectral reflectance of rice blast disease using a hyperspectral camera in the range of 400–1000 nm. SVM was used to classify diseased and healthy leaves with classification accuracy of 92.92 %. Wu et al. [13] diagnosed gray mold disease in strawberries using hyperspectral images. The texture and vegetation characteristics were extracted from images at 400–1000 nm from healthy and 24-h infected leaves. Feature selection was performed using ReliefF, while extreme learning machine (ELM), SVM, and k-nearest neighbors (k-NN) were used for classification, the accuracy of which were 93.33, 93.33, and 96.67 %, respectively.
Assessment of plant diseases using sensors involves three stages: diagnosis, identification, and quantification. Sensors should be sensitive to the effects of fungal colonization of plant tissue during pathogen development. Hyperspectral imaging has made it possible to assign pixel-wise spectral signatures to assess small-scale changes typical for the early stages of plant diseases. This information is instrumental in understanding the fundamental processes of plant biology and fungal pathogenesis. Significant changes in the spectral signature of host tissue can be detected during early pre-symptom periods, providing the detection of plant diseases before the emergence of visible symptoms. The literature review shows the limited work on the simultaneous diagnosis of several plant fungal diseases in different stages of the spread in RGB and hyperspectral images. In the meantime, the simultaneous detection of diseases with similar symptoms has attracted the attention of researchers since such tasks are difficult for farmers using their naked eyes during the diagnosis. Several common tomato fungal diseases exhibit similar symptoms on plants. Some of these fungal pathogens include Alternaria alternata, Alternaria solani, Botrytis cinerea, and Fusarium oxysporum.
A. alternata is a fungus that infects only certain cultivars of tomato plants, causing a condition known as Alternaria tomato stem canker. The fungus lives in seeds and seedlings and can spread through spores in air or landing on plants. It is often characterized by a canker that forms on the stem, which is the main symptom of the infection. In severe cases, the cankers enlarge and merge, causing leaf burn. The symptoms start with yellowing and browning of the lower leaves and then on the tips of the leaves and along the leaf petiole margins. This progression continues until the entire leaves are diseased and fall off. A. alternata is infectious and can easily spread to other tomato plants, causing significant economic losses to farmers [14].
Early blight is a common fungal disease caused by A. solani. The penetration of the disease agent into the plants inside the tank causes the tissues around the collar to turn black. As these spots spread to the top of the stem, the seedlings inside the tank are destroyed. The wounds on the stem are irregular, black, and concave and are formed near the junction with the stem. The severity of disease symptoms is more visible in the lower leaves of the plant. These concentric spots are initially dark green and gradually turn brown or black. Leaf spots are usually round or angular and cover the entire width of the leaf when the right environmental conditions are provided. Around the leaf spots, a wide halo with a bright yellow color can be seen. Symptoms on the fruit are also seen as dry and sunken spots and sometimes with black mold [15].
Gray mold, caused by B. cinerea, infects tomato plants. In this type of disease, during the initial days, small brown spots and burnt water appear on the parts of the plant that are in contact with the soil or those that are damaged. Then, gray to brown mold forms on these spots. Eventually, mold grows on young fruits and pods, giving them a fluffy appearance. Sometimes, the symptoms of this disease appear during fruit storage. In both cases, the damage to the product is very high [16].
Fusarium wilt, caused by F. oxysporum, is one of the important tomato diseases. The symptoms of this disease are wilting, yellowing of the lower leaves, browning of the veins, and slow growth. Seedlings that are attacked by this fungus in the early stages of growth die suddenly. Symptoms of the disease can be seen on whole plants in the form of yellowing of the leaves. As the disease continues, the entire leaf blade turns yellow and remains suspended on the stem. The symptoms of the disease on the infected roots are also in the form of dark-colored spots of two to 3 cm. Usually, this disease causes paleness, yellowness, and dead plant tissue from one side. In some cases, with a transverse cut of the stem, very dark vascular tissues can be seen [17].
It is essential to determine the spectral signature and effective features capable of diagnosing the diseases to provide a reliable disease detection technique. Robust feature selection algorithms can identify these effective features. This study aimed at detecting four common fungal diseases of tomatoes on days 1–11 after the disease transmission using RGB and hyperspectral imaging. The diagnosis performance of each type of image, i.e., RGB and hyperspectral, was evaluated in the early, intermediate, and late stages of the disease development, and the spectral signature of each disease was obtained. Then, the most effective features extracted from the images as well as the most effective spectral bands extracted for the diagnosis of each disease were identified.
2. Material and methods
2.1. Plant material
Tomato plants were cultivated in a research greenhouse at the Gorgan University of Agricultural Sciences and Natural Resources, Iran. Tomato seeds (Solanum lycopersicum Var. 4921) were surface disinfected using sodium hypochlorite 1 % for 60 s. They were then placed in a seedling tray containing coco peat, perlite, sand, field soil, and peat moss with equal proportions. After one month, the seedlings were transferred into 1-kg pots and kept in the greenhouse with 80 % humidity and a temperature of 20 ± 2 °C.
2.2. Sampling and disease isolation from tomato plants
Four tomato fungal pathogens, i.e., A. alternata, A. solani, B. cinerea, and F. oxysporum, which are among the most common and destructive fungal diseases in greenhouse and field cultivation, were investigated in this study. In order to prepare and isolate the disease agents, samples were collected from tomato fields. The samples were then placed separately in paper envelopes and kept at a temperature of 4 °C in the laboratory environment. Using a sterile scalpel, 1 × 1-cm pieces were cut from the infected tissues. The pieces were surface disinfected in 10 % sodium hypochlorite solution for 10 min and immediately washed twice with water. After dewatering with filter paper, the samples were cultured on (potato dextrose agar (PDA) nutrient medium. Then, the plates were incubated for 3–5 days at 25 °C. From the grown colonies that had the characteristics of the disease considered in this study, they were purified by removing single spore or spore chain. The 5-mm pieces of growing colonies were removed and cultured on PDA.
To stimulate the sporulation pattern, the plates were incubated at 24 °C with a light cycle of 8:16 light:dark for 5–7 days. The isolates were identified using the general patterns of sporulation, including the arrangement of spores on the spore carrier, the number of spores in each chain, and the branching pattern of the chains, and the pathogenicity test of the isolates was proved. After 10 days of purification, the surface of Petri dishes was washed with sterile distilled water. Ten microliters of the solution obtained from disease spores was taken by a sampler. The number of disease spores and their concentration were determined using a light microscope with a hemocytometer slide. During the spore counting, the spores attached to the top and right sides of the slides were counted, while the spores located at the bottom and left were not counted. The number of spores allowed was 5 × 106 spores/mL for each disease. Fig. 1 shows the stages of sampling and disease isolation from tomato plants.
Fig. 1.
(A) Collected samples, (B) Washed samples, (C) Sections of disease area, (D) Transferring disease sections to PDA culture medium, (E) Disease growth in PDA culture medium, and (F) Purification of samples.
2.3. Transferring the disease to the plants
In order to transfer the diseases to the tomato plants at the stage of 6–8 leaves, fungal suspensions with a concentration of 5 × 106 spores/mL were sprayed on all the aerial organs of the plants. Also, the control plants were sprayed only with sterile water. For a better transfer of the diseases to the plants, a clean scarper was used to create delicate cuts on the leaves and stems before spraying. A total of 25 pots for each disease as well as 25 pots for the control were considered during the experiments.
2.4. RGB and hyperspectral imaging
Spectral images of healthy and infected leaves were captured daily under laboratory conditions using a spectroscopic camera (Hyspim HSI-Vis_Nir-15fps, Sweden) with a spectral range of 400–950 nm in 1, 3, 5, 7, 9, and 11 days after the disease transfer to the plants. The spectral range between 400 and 950 nm provides sufficient information for the diagnosis and classification of most plant diseases [18]. Plants exhibit unique spectral responses within this range. In particular, changes in the pigmentation and chemical composition of leaves due to disease can lead to distinct spectral patterns in this range that can be identified and classified using hyperspectral imaging [19]. Also, to compare each leaf on various days, RGB images were captured with a mobile phone with a 64-pixel camera (Galaxy A32, Samsung, Korea).
Lighting for both RGB and hyperspectral imaging was provided by two halogen sources installed above the leaf, and a dark background was considered for the samples. To prevent lighting interference, a small dark chamber was prepared where the camera and halogen lamps were placed inside the chamber. The distance between the leaf samples and the camera was 25 cm so the total area of each leaf can be visible in the images captured. To avoid shadows in the images, halogen lamps were installed at an angle of 45° with the horizon. A white paper sample recommended by the manufacturer of the hyperspectral camera was used as a reference to calibrate the reflection of the leaf. The white paper was captured every 10 min to recalibrate the camera because the irradiance of the light source used could change over time. For each pathogen type and healthy plants, 100 hyperspectral and 100 RGB images were taken. Therefore, 500 images per sampling day and 3000 images in total were obtained using the RGB (Fig. 2) and hyperspectral cameras.
Fig. 2.
RGB images of tomato plant leaves on days 1–11 after disease transmission.
2.5. Disease segmentation in RGB images
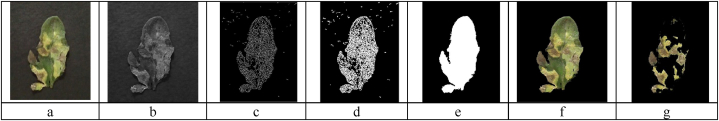
Background removal is a crucial step in extracting image features for ML tasks. It aims to eliminate any potential noise in the obtained image features. This study proposes a combination of pixel clustering and edge detection for automatic background removal. Another critical challenge in natural images is the presence of shadows, as shown in Fig. 3a. To remove the background, first, only the green color band was extracted from the image, as shown in Fig. 3b, and then the Canny algorithm was used for edge detection (Canny, 1986), followed by morphological dilation to fill in the empty spaces in the image (Fig. 3c and d). In the next step, the holes were filled (Fig. 3e) to obtain the image without the presence of a background (Fig. 3f).
Fig. 3.
Background removal and disease segmentation in RGB images, (a) original image, (b) extracting green color band from the image, (c) edge detection, (d) morphological dilation to connect edges, (e) filling in the holes in the edge image, (f) the background removal results, and (g) disease segmentation results. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
K-means clustering was used to segment the diseased and healthy parts of the leaves. This method works based on minimizing the sum of squares of the distance between an object in the image and the corresponding cluster. It includes four steps: (1) picking the center of the K-th cluster either randomly or based on some heuristics; (2) assigning each pixel to a cluster that minimizes the distance between the pixel and the cluster center; (3) computing the cluster centers by averaging all the pixels in the cluster; and finally, (4) repeating steps 2 and 3 until convergence is obtained.
Generally, the selection of the value of K and also the selection of the region of interest (ROI) are made manually, depending on the skill of the user. Sometimes, ROI might not be selected correctly by the user. This means that for each number of images in the database, the ROI number should be manually selected to determine the desired area of the disease. This is very time-consuming and error-prone. Therefore, automatic clustering seems useful for the reliable detection of the disease area in the plant leaves. In this study, the ROI in the K-means clustering (cluster number indicating the disease) was obtained automatically similar to a method introduced by Javidan et al. [[6], [20]] which includes thresholding between the color of the disease area (i.e., symptoms) and the color of the healthy leaf area (Fig. 3g).
2.6. Normalization of hyperspectral images
Multiple factors can impact the performance of ML algorithms, particularly the quality and presentation of the data set. For instance, a data set that contains redundancies, noise, or unreliable data may hinder the ability of the algorithm to extract meaningful insights and provide accurate results. Normalization, a data preprocessing technique, can address some of these issues by transforming the features of a data set to a common scale. By removing potential biases and distortions caused by features of different scales, normalization can improve the performance of ML algorithms. In summary, normalization is a critical step in preparing data sets for ML applications and ensures that the algorithms can discover information and provide acceptable performance.
The first step of hyperspectral image analysis involved smoothing the data using the standard normal transformation to reduce outliers, reconstruct the data into a relational domain, improve data integrity, and provide data smoothing. For this purpose, the probability density function was used (Eq. (1))
| (1) |
where
| (2) |
and μ is the mean and σ is the standard deviation. Inflection points are essential indicators of change within data sets. In healthy data samples, second-order divided differences are used to reveal where these inflection points occur and how they vary across different categories of infected samples. These variations serve as unique identifiers of the samples' signatures, which are presented in this work. The higher-order data functions developed in this work allow for the categorization of plants into their respective classes (healthy and diseased) based on the correlation of these functions at regions of interest in the spectrum. This process reveals the unique features of each sample, providing a precise and accurate assessment of plant health. Fig. 4 shows the spectra before and after normalization.
Fig. 4.
Spectral data before (top) and after (bottom) the normalization process.
2.7. Feature extraction
The similarity of the primary symptoms of the fungal diseases examined in this research made it difficult to visually distinguish the lesions of the diseases. With the process of spreading the disease in the plant, symptoms such as yellowing, withering, and growth disorders appear, and gradually with the provision of suitable conditions and the growth of the disease agents in the plant, the symptoms of the disease spread and cover a larger part of the plant. The effects of damage appear as symptoms of the disease in various degrees according to the plant tolerance level.
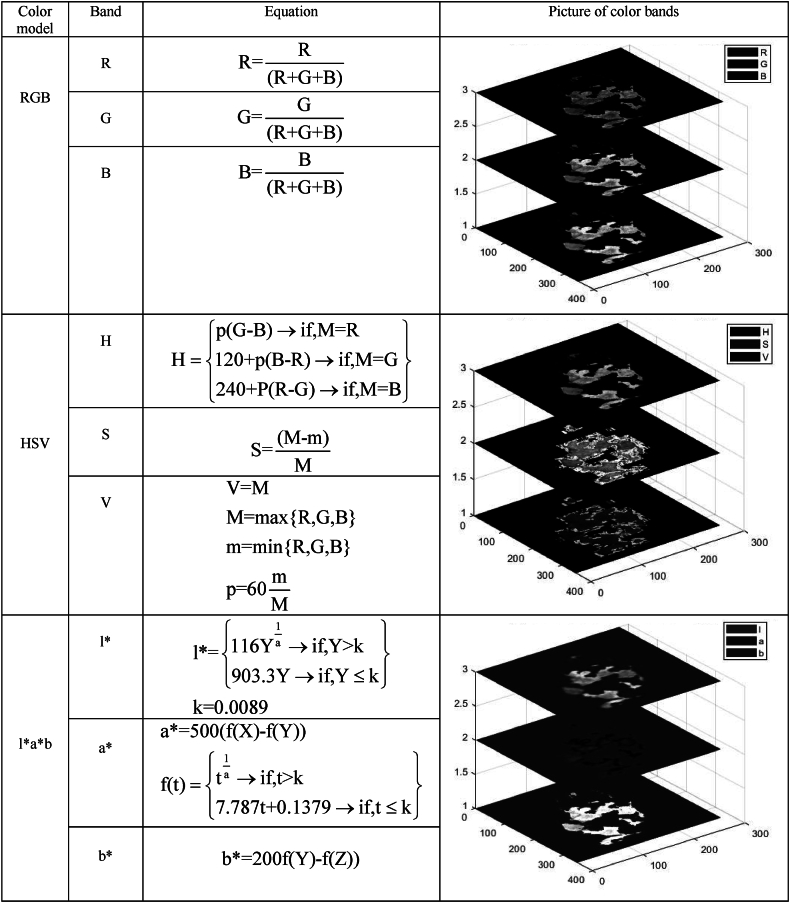
Obtaining the most relevant features from images is crucial for effective disease diagnosis in plants. Plant disease detection and classification based on images is challenging since they are exposed to different lighting conditions, can be affected by noise, and may undergo various transformations, including changes in the angle and height of the camera relative to the image sample [21]. This emphasizes the extraction of scale-invariant features that are not affected by the distance and angle of the camera during image acquisition. Color and texture features can provide us with useful scale-invariant information in plant disease diagnosis [22]. Descriptive color statistical features (mean, maximum, standard deviation, median) and texture features from gray-level co-occurrence matrix (GLCM) (contrast, correlation, energy, homogeneity, mean, standard deviation, entropy, root mean square (RMS), variance, smoothness, kurtosis, skewness, and inventive design method (IDM)) in each spectrum of hyperspectral images and each color band in color models of RGB, HSV, and l∗a∗b for RGB images (Fig. 5), as well as shape features (area, perimeter, number of objects, major and minor axis length of the spots, eccentricity index, circularity, circular index, compression index, and shape factors), were extracted from the images. During extracting texture features from the GLCM, the distance and direction were considered 1 pixel and 0°, respectively, according to the literature [23]. Table 1 shows the features extracted from RGB and hyperspectral images used in this research along with their description.
Fig. 5.
Disease symptoms in RGB, HSV, and l∗a∗b color spaces and the features in each band.
Table 1.
Features extracted from RGB and hyperspectral images for disease diagnosis.
| Feature | Description | Equation | Reference |
|---|---|---|---|
| Descriptive color statistical features (1–36) | |||
| Mean | he average value of the data. | Ashfaq et al. [24] | |
| Maximum | The largest value in the data. | Ashfaq et al. [24] | |
| Standard Deviation | A measure of the spread of the data. | Ashfaq et al. [24] | |
| Median | The number in the middle of the data set when it is sorted in increasing order. | Ashfaq et al. [24] | |
| Textural features (37–154) | |||
| Contrast | This is a measure of the intensity difference between neighboring pixels. | Haralick et al. [25] | |
| Correlation | The degree of linear dependence between neighboring pixels. | Haralick et al. [25] | |
| Energy | The sum of the squares of all the possible patterns of intensity difference between neighboring pixels in all directions. | Haralick et al. [25] | |
| Homogeneity | Measures the uniformity of the neighboring pixels | Haralick et al. [25] | |
| Mean | The average intensity value of neighboring pixels. | Haralick et al. [25] | |
| Standard Deviation | Measures the spread of the intensity values of the neighboring pixels. | Haralick et al. [25] | |
| Entropy | Measure of the amount of information stored in the texture image. | Haralick et al. [25] | |
| RMS | Measures the square root of the average of the sum of the squares of the difference between intensity values of neighboring pixels. | Haralick et al. [25] | |
| Variance | A measure of the spread of the intensity values of the neighboring pixels. | Haralick et al. [25] | |
| Smoothness | Measures the degree to which the neighboring pixels have similar intensity values. | – | Haralick et al. [25] |
| Kurtosis | A measure of the peakedness of the distribution of the intensity values of the neighboring pixels. | Haralick et al. [25] | |
| Skewness | A measure of the asymmetric distribution of the intensity values of the neighboring pixels. | Haralick et al. [25] | |
| IDM | A sophisticated GLCM feature that captures complex texture features in the neighborhood of a pixel. | – | Haralick et al. [25] |
| Shape features (155–166) | |||
| Area | The mean of number of pixels in each region segmented in the image | Vishnoi et al. (2021) | |
| Perimeter | The mean of number of boundary pixels on each region segmented in the image | 2 (length + width) | Vishnoi et al. (2021) |
| Number of objects | Number of disease spots in the leaf image | – | – |
| Major axis length | The distance along the longest axis of an object, in pixels or other appropriate units. | Vishnoi et al. (2021) | |
| Minor axis length | The distance along the shortest axis of an object, in pixels or other appropriate units. | Vishnoi et al. (2021) | |
| eccentricity index | A measurement of the difference between the lengths of the major and minor axis, normalized to range between 0 and 1. | Vishnoi et al. (2021) | |
| Circularity | A ratio of a circle to the bounding box of an object, with a value of 1 indicating a perfect circle and values approaching 0 indicating a more complex or amorphous shape. | Vishnoi et al. (2021) | |
| Circular index | Mean of circularity of each segmented region in the image | Javidan et al. [26] | |
| Compactness index | Mean of circularity of each segmented region in the image | Javidan et al. [26] | |
| Shape factors 1 | Dimensionless numerical values calculated based on at least two simple shape measures, which makes them independent to object orientation, translation, and scale. These features may provide useful information for the morphological description of plants. Primary shape features, namely area, perimeter, and major and minor axis length values of each plant are extracted from binary images | Javidan et al. [26] | |
| Shape factors 2 | Javidan et al. [26] | ||
| Shape factors 3 | Javidan et al. (2024) | ||
2.8. Feature selection
Identifying the most effective features in improving the classification results is one of the main challenges of ML. It is necessary to provide a list of the most influential features extracted for each disease in various spectrum/color bands to be used in the diagnosis of plant diseases. In this situation, the most effective features among descriptive statistical, texture, and shape features can be introduced as a kind of spectral/color signature in recognizing plant diseases.
Many algorithms have been proposed for feature selection, of which the Laplacian score method has shown great performance in problems related to image-based object detection [[27], [28], [29]]. This method, which is used in this work for feature selection, considers the fact that the data of the same class are usually located in the neighborhood of each other in the feature space. The importance (relevance) of a feature can be measured by calculating its power in preserving the locality information of the samples. In this method, samples are first mapped to a nearest-neighbor graph using an arbitrary distance measure. Then, the Laplacian scores are calculated for the features. Features that are more important (more relevant) than other features will have a lower Laplacian score than other features and vice versa. In the end, a clustering method such as K-means is used to determine the best subset of features.
2.9. Classification methods
Classification is one of the main subfields of data mining and ML and is used to learn how to assign a class label to an input sample [30]. In this study, various machines, including SVM, k-NN, decision tree (DT), and random forest (RF) were used to classify RGB and hyperspectral images.
2.10. Performance evaluation criteria
Accuracy (Eq. (3)), precision (Eq. (4)), sensitivity (Eq. (5)), specificity (Eq. (6)), and F-measure (Eq. (7)) were used to investigate the performance of the models
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
where TP, TN, FP, and FN are true positives, true negatives, false positives, and false negatives, respectively.
3. Results and discussion
3.1. Disease classification results in RGB and hyperspectral images
After extracting the descriptive color statistical, texture, and shape features from RGB and hyperspectral images to construct data sets for both groups of images, classification was performed to recognize five classes including four classes for the diseases and one for the healthy leaves. Table 2 shows the accuracy of disease classification by various ML models utilizing RGB and hyperspectral imaging (HSI) data for days 1, 3, 5, 7, 9, and 11. According to the table, the classification results using hyperspectral data are better than those using RGB data in all ML models investigated in this work. The classification performance in all models has increased over time, which is due to the growth and expansion of the diseases throughout the infected plants, increasing the infected area on plant leaves. The disease classification accuracy using SVM, k-NN, DT, and RF models on day 11 for RGB images was 70, 55, 65, and 87 %, respectively. The accuracy for hyperspectral images on day 11 was 91, 82, 87, and 98 %, respectively, showing the promising performance of the RF model in both types of imaging (see Table 3).
Table 2.
Accuracy of the ML models for classification of common tomato fungal diseases.
| Type of imaging | ML models | Accuracy % |
|||||
|---|---|---|---|---|---|---|---|
| Day 1 | Day 3 | Day 5 | Day 7 | Day 9 | Day 11 | ||
| RGB | SVM | 55 | 57 | 61 | 63 | 66 | 70 |
| k-NN | 42 | 45 | 47 | 50 | 52 | 55 | |
| DT | 50 | 53 | 56 | 58 | 61 | 65 | |
| RF | 65 | 71 | 75 | 77 | 83 | 87 | |
| HSI | SVM | 73 | 76 | 78 | 82 | 87 | 91 |
| k-NN | 70 | 73 | 75 | 77 | 79 | 82 | |
| DT | 71 | 76 | 78 | 80 | 84 | 87 | |
| RF | 86 | 90 | 92 | 95 | 97 | 98 | |
Table 3.
Classification results using RGB images and the RF model for each disease type on days 1–11.
|
Day 1= 65 % |
Day 3= 71 % |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AS | BOT | FUS | H | AA | AS | BOT | FUS | H | ||
| TP | 9 | 12 | 14 | 11 | 19 | TP | 9 | 12 | 14 | 11 | 19 |
| FP | 9 | 9 | 12 | 4 | 1 | FP | 9 | 9 | 12 | 3 | 1 |
| FN | 17 | 11 | 3 | 3 | 1 | FN | 17 | 11 | 2 | 3 | 1 |
| TN | 65 | 68 | 71 | 82 | 79 | TN | 64 | 67 | 71 | 82 | 78 |
| Precision | 0.50 | 0.57 | 0.54 | 0.73 | 0.95 | Precision | 0.50 | 0.57 | 0.54 | 0.79 | 0.95 |
| Sensitivity | 0.35 | 0.52 | 0.82 | 0.79 | 0.95 | Sensitivity | 0.35 | 0.52 | 0.88 | 0.79 | 0.95 |
| Specificity | 0.88 | 0.88 | 0.86 | 0.95 | 0.99 | Specificity | 0.88 | 0.88 | 0.86 | 0.96 | 0.99 |
| Accuracy | 0.65 | 0.65 | 0.65 | 0.65 | 0.65 | Accuracy | 0.66 | 0.66 | 0.66 | 0.66 | 0.66 |
| F-Measure | 0.41 | 0.55 | 0.65 | 0.76 | 0.95 | F-Measure | 0.41 | 0.55 | 0.67 | 0.79 | 0.95 |
| Day 5= 75 % | Day 7= 77 % | ||||||||||
| AA |
AS |
BOT |
FUS |
H |
AA |
AS |
BOT |
FUS |
H |
||
| TP | 12 | 12 | 21 | 10 | 20 | TP | 10 | 11 | 15 | 21 | 20 |
| FP | 6 | 8 | 7 | 4 | 2 | FP | 10 | 5 | 4 | 1 | 2 |
| FN | 13 | 6 | 4 | 3 | 1 | FN | 8 | 7 | 3 | 3 | 1 |
| TN | 71 | 76 | 70 | 85 | 79 | TN | 71 | 76 | 77 | 74 | 76 |
| Precision | 0.67 | 0.60 | 0.75 | 0.71 | 0.91 | Precision | 0.5 | 0.69 | 0.79 | 0.95 | 0.91 |
| Sensitivity | 0.48 | 0.67 | 0.84 | 0.77 | 0.95 | Sensitivity | 0.56 | 0.61 | 0.83 | 0.88 | 0.95 |
| Specificity | 0.92 | 0.90 | 0.91 | 0.96 | 0.98 | Specificity | 0.88 | 0.94 | 0.95 | 0.99 | 0.97 |
| Accuracy | 0.74 | 0.74 | 0.74 | 0.74 | 0.74 | Accuracy | 0.78 | 0.78 | 0.78 | 0.78 | 0.78 |
| F-Measure | 0.56 | 0.63 | 0.79 | 0.74 | 0.93 | F-Measure | 0.53 | 0.65 | 0.81 | 0.91 | 0.93 |
| Day 9= 83 % | Day 11= 87 % | ||||||||||
| AA |
AS |
BOT |
FUS |
H |
AA |
AS |
BOT |
FUS |
H |
||
| TP | 13 | 14 | 15 | 19 | 22 | TP | 13 | 20 | 20 | 19 | 15 |
| FP | 9 | 3 | 1 | 1 | 3 | FP | 2 | 6 | 5 | 0 | 0 |
| FN | 4 | 6 | 6 | 1 | 0 | FN | 9 | 4 | 0 | 0 | 0 |
| TN | 74 | 77 | 78 | 79 | 75 | TN | 76 | 70 | 75 | 81 | 85 |
| Precision | 0.59 | 0.82 | 0.94 | 0.95 | 0.88 | Precision | 0.87 | 0.77 | 0.80 | 1.00 | 1.00 |
| Sensitivity | 0.76 | 0.70 | 0.71 | 0.95 | 1.00 | Sensitivity | 0.59 | 0.83 | 1.00 | 1.00 | 1.00 |
| Specificity | 0.89 | 0.96 | 0.99 | 0.99 | 0.96 | Specificity | 0.97 | 0.92 | 0.94 | 1.00 | 1.00 |
| Accuracy | 0.83 | 0.83 | 0.83 | 0.83 | 0.83 | Accuracy | 0.87 | 0.87 | 0.87 | 0.87 | 0.87 |
| F-Measure | 0.67 | 0.76 | 0.81 | 0.95 | 0.94 | F-Measure | 0.70 | 0.80 | 0.89 | 1.00 | 1.00 |
AA: A. alternata, AS: A. solani, BOT: B. cinerea, FUS: F. oxysporum, and H: healthy samples.
To further investigate the performance of the RF model in classifying the diseases, precision, sensitivity, specificity, and F-measure values were calculated for each group of diseases. Using hyperspectral images led to higher classification accuracies; the lowest accuracy was 86 % for day 1, and the highest accuracy was 98 % for day 11. A total of 650 spectral bands in the hyperspectral images improved the performance of the classification algorithm in analyzing more relevant data for more accurate classification. However, more time was required for the classification. On the other hand, the RGB images were analyzed only in nine color bands, and their highest classification accuracy was 87 % on day 11, i.e., when the diseases showed more visible symptoms. Xie et al. [31] worked on eight texture features from hyperspectral images for two diseases in tomato plants, i.e., early blight and late blight, and the classification accuracy in their proposed method was about 78 %. They only extracted texture features and did not examine the results for different days of disease transmission. Also, they did not compare the performance of the classification utilizing RBG and hyperspectral images.
3.2. Spectral signatures of the tomato fungal diseases
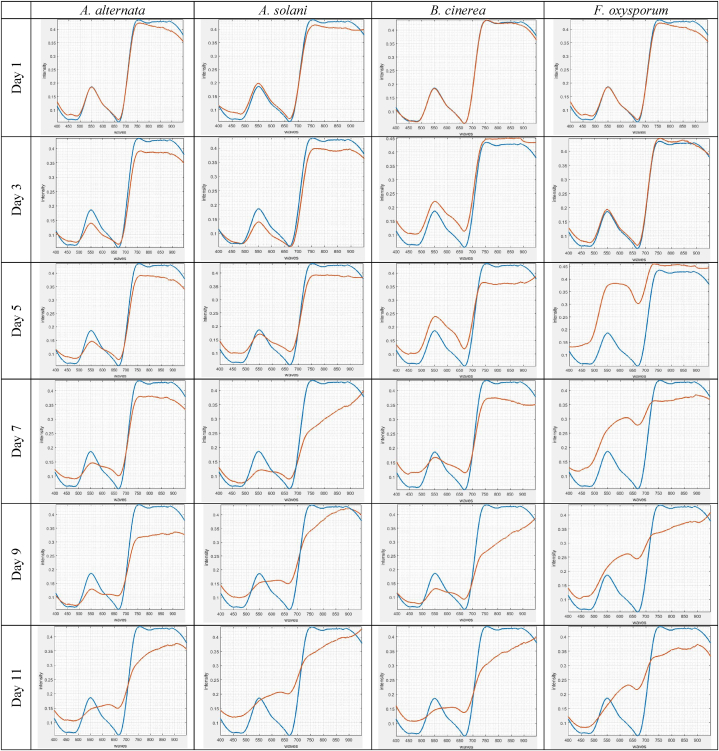
Fig. 6 shows the spectral signatures of the tomato plant diseases investigated in this work on days 1–11 for an arbitrary pixel in the diseased area. The plots are drawn versus the spectral signature of healthy plants. Any change in the spectral response of diseased plants compared to healthy plants can lead to the identification of the disease. This spectral distinction can help diagnose and identify the disease in early stages. Spectral reflectance of healthy leaf tissue from adjacent pixels on a leaf section was completely homogeneous. Minor changes were due to slight heterogeneity of leaf texture, the surface structure of tomato leaves, and its interaction with radiation.
Fig. 6.
Spectral signatures in healthy (blue lines) and diseased (red lines) samples from day 1–11 after the disease transmission. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
According to Fig. 6, on the first day, the ranges (signatures) of all four disease groups and healthy leaves are similar, making it impossible to identify the disease on this day. This can be seen in both the visible and invisible ranges. On day 3, B. cinerea showed clear spectral differences (symptoms) in the spectral range of 530–580 nm (visible range) and 740–900 nm (invisible range), meaning that pixel reflection in this disease is different from the reflection of the healthy pixel.
Over time, spectral changes were observed with the spread of the disease in the leaves of the plant and the establishment of the disease in the healthy area of the leaf, as well as the reaction of the initially infected tissue to dry and necrotic tissue. In F. oxysporum, the difference with the healthy spectrum was observed in the spectral ranges of 500–650 nm and 700–950 nm on day 5. On this day, the response (spectral signature) for A. alternata disease was still similar to the third day. The difference was observed in A. alternata and A. solani on day 7. In the following days, with the further spread of the disease in all four groups, noticeable changes in the spectrum of the disease compared to the healthy spectrum were observed. Therefore, clear differences between healthy samples and B. cinerea, F. oxysporum, A. alternata, and A. solani were found on days 3, 5, 7, and 7 after the disease transmission to the plant, respectively.
Fig. 7 shows the spectral signatures of the studied diseases compared to the signatures of healthy leaves on the first day of exerting clear differences with healthy leaves. As can be seen in the figure, the spectral data for the four groups of diseases are more effective in the ranges of 500–550 nm and 740–950 nm for identifying and diagnosing the disease in the early stages.
Fig. 7.
Spectral signatures for diseases, (A) A. alternata, (B) A. solani, (C) B. cinerea, and (D) F. oxysporum, on the first day of exerting clear differences with healthy leaves.
The average spectral signatures of diseased samples compared to healthy samples are shown in three-dimensional plots in Fig. 8 for an arbitrary pixel row selected from the diseased area. These spectral signatures can be used as a marker for various diseases and help identify the diseases. The spectral reflectance recorded for A. alternata in the range of 550–950 nm was lower than that of the healthy plants, making this range a good indicator for disease identification. In the case of A. solani disease, the spectral reflectance in the range of 550–950 nm showed fewer differences with healthy leaves than the A. alternata disease. The symptoms of B. cinerea caused a change in spectral reflectance in the range of 700–950 nm compared to healthy plants. Finally, the symptoms of F. oxysporum disease, which had a spectral signature more distinct than all other diseases, showed a strong light reflection in the ranges of 500–650 nm and 700–950 nm compared to the healthy plants.
Fig. 8.
Three-dimensional spectral signatures for diseases for an arbitrary pixel row selected from the diseased area, (A) A. alternata, (B) A. solani, (C) B. cinerea, (D) F. oxysporum, and (E) healthy samples. Both raw (right) and preprocessed (left) data are shown in the figure.
Research confirms that plant-pathogen interactions induce unique spectral signatures due to the involvement of different chemicals [32]. Hyperspectral imaging technology allows for the analysis of the interaction between plants and light, including transmission, absorption, and reflection [33]. However, limitations in existing transmittance sensors hinder the measurement of both reflectance and transmittance [34]. Different substances, including chlorophyll, exhibit specific absorption and reflectance values in specific wavebands, leading to unique spectral features [35]. Reflectance peaks near 550 nm and 700 nm correspond to the strong reflection region of chlorophyll, and the red edge phenomenon results from the low red reflectance and high internal leaf scattering of chlorophyll [36]. Moisture absorption troughs near 1470 nm and 1940 nm and low reflectance in the range of 1300–2500 nm indicate plant moisture content and CO2 emissions [37,38]. Hyperspectral imaging of biological processes during plant-pathogen interactions enables disease detection using hyperspectral imaging technology [39,40]. The spatial resolution of hyperspectral cameras can be improved using a hyperspectral microscope to study diseases at smaller scales [41]. Changes in reflectance can be attributed to damage to the chemical components of the leaf surface, necrotic tissue, or typical fungal structures, resulting in chlorosis [42,43]. The accuracy and efficiency of disease detection and classification using hyperspectral imaging technology can positively affect plant management and crop yields (Behmann et al., 2015) (see Table 4).
Table 4.
Classification results using hyperspectral images and the RF model for each disease type on days 1–11.
|
Day 1= 86 % |
Day 3= 90 % |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AS | BOT | FUS | H | AA | AS | BOT | FUS | H | ||
| TP | 15 | 14 | 15 | 20 | 22 | TP | 17 | 17 | 20 | 16 | 20 |
| FP | 9 | 1 | 3 | 1 | 0 | FP | 3 | 4 | 2 | 1 | 0 |
| FN | 2 | 6 | 6 | 0 | 0 | FN | 3 | 4 | 0 | 3 | 0 |
| TN | 74 | 79 | 76 | 79 | 78 | TN | 77 | 75 | 78 | 80 | 80 |
| Precision | 0.63 | 0.93 | 0.83 | 0.95 | 1.00 | Precision | 0.85 | 0.81 | 0.91 | 0.94 | 1.00 |
| Sensitivity | 0.88 | 0.70 | 0.71 | 1.00 | 1.00 | Sensitivity | 0.85 | 0.81 | 1.00 | 0.84 | 1.00 |
| Specificity | 0.89 | 0.99 | 0.96 | 0.99 | 1.00 | Specificity | 0.96 | 0.95 | 0.98 | 0.99 | 1.00 |
| Accuracy | 0.86 | 0.86 | 0.86 | 0.86 | 0.86 | Accuracy | 0.90 | 0.90 | 0.90 | 0.90 | 0.90 |
| F-Measure | 0.73 | 0.80 | 0.77 | 0.98 | 1.00 | F-Measure | 0.85 | 0.81 | 0.95 | 0.89 | 1.00 |
| Day 5= 92 % | Day 7= 95 % | ||||||||||
| AA |
AS |
BOT |
FUS |
H |
AA |
AS |
BOT |
FUS |
H |
||
| TP | 19 | 24 | 15 | 19 | 15 | TP | 18 | 17 | 19 | 19 | 22 |
| FP | 4 | 1 | 1 | 2 | 0 | FP | 1 | 2 | 0 | 2 | 0 |
| FN | 3 | 0 | 5 | 0 | 0 | FN | 3 | 0 | 1 | 1 | 0 |
| TN | 74 | 75 | 79 | 79 | 85 | TN | 78 | 81 | 80 | 78 | 78 |
| Precision | 0.83 | 0.96 | 0.94 | 0.90 | 1.00 | Precision | 0.95 | 0.89 | 1.00 | 0.90 | 1.00 |
| Sensitivity | 0.86 | 1.00 | 0.75 | 1.00 | 1.00 | Sensitivity | 0.86 | 1.00 | 0.95 | 0.95 | 1.00 |
| Specificity | 0.95 | 0.99 | 0.99 | 0.98 | 1.00 | Specificity | 0.99 | 0.98 | 1.00 | 0.98 | 1.00 |
| Accuracy | 0.92 | 0.92 | 0.92 | 0.92 | 0.92 | Accuracy | 0.95 | 0.95 | 0.95 | 0.95 | 0.95 |
| F-Measure | 0.84 | 0.98 | 0.83 | 0.95 | 1.00 | F-Measure | 0.90 | 0.94 | 0.97 | 0.93 | 1.00 |
| Day 9= 97 % | Day 11= 98 % | ||||||||||
| AA |
AS |
BOT |
FUS |
H |
AA |
AS |
BOT |
FUS |
H |
||
| TP | 16 | 19 | 21 | 19 | 22 | TP | 22 | 23 | 19 | 19 | 15 |
| FP | 1 | 0 | 2 | 0 | 0 | FP | 0 | 1 | 0 | 1 | 0 |
| FN | 1 | 1 | 0 | 1 | 0 | FN | 0 | 1 | 1 | 0 | 0 |
| TN | 82 | 80 | 77 | 80 | 78 | TN | 78 | 75 | 80 | 80 | 85 |
| Precision | 0.94 | 1.00 | 0.91 | 1.00 | 1.00 | Precision | 1.00 | 0.96 | 1.00 | 0.95 | 1.00 |
| Sensitivity | 0.94 | 0.95 | 1.00 | 0.95 | 1.00 | Sensitivity | 1.00 | 0.96 | 0.95 | 1.00 | 1.00 |
| Specificity | 0.99 | 1.00 | 0.97 | 1.00 | 1.00 | Specificity | 1.00 | 0.99 | 1.00 | 0.99 | 1.00 |
| Accuracy | 0.97 | 0.97 | 0.97 | 0.97 | 0.97 | Accuracy | 0.98 | 0.98 | 0.98 | 0.98 | 0.98 |
| F-Measure | 0.94 | 0.97 | 0.95 | 0.97 | 1.00 | F-Measure | 1.00 | 0.96 | 0.97 | 0.97 | 1.00 |
AA: A. alternata, AS: A. solani, BOT: B. cinerea, FUS: F. oxysporum, and H: healthy samples.
3.3. Feature selection results
The effective features and bands in two groups of RGB and hyperspectral images are shown in Table 5. Among the top ten selected features, the texture features had the most impact, followed by the descriptive color statistical and shape features. As can be seen in the images of disease symptoms, texture and color characteristics are clearly effective in diagnosing the study diseases. According to Table 5, energy, entropy, and standard deviation are among the most essential features during the disease diagnosis. Energy is a useful feature in plant disease detection and classification because it measures the intensity of the spectral signal at different frequencies. Changes in the texture and structure of plant leaves, stems, or roots caused by diseases can result in changes in the energy distribution of spectral data. High energy values can indicate the presence of healthy vegetation, while lower energy values may correspond to diseased or damaged vegetation [44]. Entropy is also a useful feature in plant disease detection because it measures the randomness or disorder in a set of data, in this case, the spectral data of plant leaves, stems, or roots. Higher entropy values may correspond to areas of variability in plant coloration, caused by a variety of disease-related changes. Standard deviation is also an essential feature in plant disease detection because it measures the spread of data around its mean value. Higher standard deviation values may indicate greater variability in the spectral data, implying the presence of disease-related changes. Chen et al. (2022) proposed a method to detect bacterial wilt disease of tomatoes on leaf and stem. They used a combination of genetic algorithms and SVM and showed that disease diagnosis on the stem with an accuracy of 92.6 % is better than the diagnosis on the leaf with an accuracy of 90.7 %. In previous studies using hyperspectral images for crop disease monitoring, spectral and textural features were used simultaneously to develop a reliable disease detection model [[45], [46], [47], [48]].
Table 5.
Effective features and bands in identifying and classifying diseases in RGB and hyperspectral images.
| HSI |
RGB |
||||||
|---|---|---|---|---|---|---|---|
| Rank | Features Type | Feature Selected | Band (nm) | Rank | Features Type | Feature Selected | Band |
| 1 | Texture | Energy | 550 | 1 | Texture | Entropy | l |
| 2 | Texture | Entropy | 600 | 2 | Descriptive statistics | Median | G |
| 3 | Descriptive statistics | Max | 480 | 3 | Texture | Standard Deviation | R |
| 4 | Descriptive statistics | mean | 663 | 4 | Descriptive statistics | mean | l |
| 5 | Texture | Correlation | 746 | 5 | Texture | Energy | R |
| 6 | Texture | Standard Deviation | 886 | 6 | Shape | Perimeter | – |
| 7 | Descriptive statistics | Median | 850 | 7 | Texture | Median | b |
| 8 | Descriptive statistics | Median | 460 | 8 | Texture | Skewness | H |
| 9 | Texture | Energy | 841 | 9 | Descriptive statistics | mean | G |
| 10 | Texture | Standard Deviation | 905 | 10 | Shape | Area | – |
Median, maximum, and mean are commonly used color features in RGB and hyperspectral image analysis. The median is a robust measure of the central tendency of a sample or population. It is often preferred over the mean for its stability to outliers and less sensitivity to the presence of extreme values. In plant disease detection using hyperspectral imaging data, the median can provide a robust measure of the average gray-level value of healthy vegetation. The median is also useful in identifying changes in the spectral properties or gray-level composition of diseased vegetation by identifying areas with lower median values. Maximum is the largest value in a sample or data set. In plant disease detection using hyperspectral imaging data, the maximum can help identify areas of intense or highly variable vegetation. Mean is the arithmetic average, being the representative measure of the central tendency of a sample or population. Mean can be useful in identifying changes in the spectral properties or gray-level composition of diseased vegetation.
Studies on diagnosing and classifying plant diseases have focused more on texture features than other features ([49]; Zhang et al. [50]. Pourreza et al. [51] investigated hyperspectral images for strawberry diseases. They selected texture features from images at the wavelength of 591 nm. The researchers believe that using only one image at one wavelength to extract texture features may not be appropriate, as some useful texture features may be present at other wavelengths [50]. Therefore, all wavelengths should be examined and feature selection algorithms should be used to select the best ones [31].
The shape features are rarely used in plant disease detection studies since they are not scale-invariant features. In a fixed lighting and image acquisition situation, perimeter and area are useful features in hyperspectral image analysis for plant disease detection since they can provide qualitative information about the size, shape, and extent of the affected area of the plant, which can be useful in identifying and differentiating between healthy vegetation and vegetation with disease-related changes.
3.4. Practical applications of the results
This study emphasizes the superiority of hyperspectral data of the plant leaves compared to their RGB data in plant disease diagnosis. Using hyperspectral imaging, it was found that three days after the transmission of fungal diseases to tomato plants was a reliable stage to identify the type and severity of the diseases, with an accuracy of 92 %. The detection accuracy on day 3 was 71 % when using RGB images for disease identification. Besides, the most differences among the diseases at various stages of growth were obtained at the ranges of 500–550 nm and 740–950 nm. This means that constructing a hand-held optical device equipped with several light-emitting diodes/photodiodes that emit/receive light in the range of visible 500–550 nm and near-infrared 740–950 nm can provide the necessary data to identify major tomato fungal diseases with a promising performance. In this condition, a low-cost portable (or UAV-based) device can also be used for the detection tasks instead of transferring the leaf samples to a laboratory for wide-range hyperspectral imaging using costly equipment. Moreover, instead of implementing a real-time machine-learning decision-making unit in the device that increases the detection's costs, the spectral data obtained by the sensor can be transferred to a server equipped with a high-performance processor through an Internet of Things (IoT) platform, and the results of disease identification can be available to the user in near real-time.
3.5. Discussion
Changes in plant pigments during disease can occur due to a decrease in photosynthesis and alterations in plant biochemical processes. During disease development, leaves may undergo necrosis, causing the loss of chlorophyll pigmentation, leading to a decrease in the green color [52]. Pigments are chemical compounds, including chlorophyll, carotenoids, and anthocyanins, being essential for photosynthesis and plant coloration. Pigments can be altered by pathogens affecting the host plant's physiological, biochemical, and metabolic properties, altering optical features [53,54]. It should be noted that the spectral properties of leaves are not static, but rather change over time with various factors such as plant growth, senescence, decay, and disease development [55]. Despite the potential of hyperspectral imaging for plant disease diagnosis, the complex nature of the hyperspectral data and the need for efficient processing methods are significant challenges to maximizing its capabilities for identifying and managing plant diseases [56].
Image data acquisition with high precision and resolution allows the sensor to capture detailed information from the electromagnetic spectrum. However, the large number of variables (i.e., features, wavelengths) measured leads to high-dimensional data that increases the complexity of processing to produce relevant information [57]. In addition, the spectral data assessed in near-contiguous variables likely exhibits similar or overlapping information, leading to potential data redundancy and increased complexity in the analysis interpretation and the chance of overfitting occurrence [58]. As a result, feature selection methods have been developed to identify the most relevant and distinctive spectral features without sacrificing important information. In recent years, the demand for ML-enabled spectral sensing systems to identify plant diseases has increased. These systems often rely on ML models for processing the hyperspectral data, with the goal of maximizing the accuracy of disease identification [59]. However, the complexity of these models and the need for efficient processing methodologies to produce accurate and interpretable results are significant challenges to unlocking the full potential of hyperspectral imaging for improved plant disease diagnosis and management [60]. Table 6 compares several key studies on the diagnosis and classification of tomato diseases, including the present study.
Table 6.
Comparison of several studies on tomato disease diagnosis using image processing.
| Diseases/pathogen | Wavelength Range | Feature Extraction | Classifier | Accuracy (%) | References |
|---|---|---|---|---|---|
| B. cinerea | 380–1023 nm | Mean reflectance values of the region of interest (ROI) | k-Nearest Neighbors (k-NN) | 61 | Xie et al. [61] |
| Late blight, target spots, and bacterial spots | 400–2400 nm | Vegetation Indices | k-Nearest Neighbors (k-NN) | Asymptomatic = 63 Early Stage = 74 Late Stage = 77 |
Lu et al. [62] |
| Spotted wilt virus | 395–1005 nm | – | OR-AC-GAN | 96 | Wang et al. [33] |
| Yellow leaf curl, bacterial spot and target spot | 380–1020 nm | Vegetation Indices | Stepwise Discriminant Analysis (STDA) Radial Basis Function (RBF) |
95 83 |
Abdulridha et al. (2020) |
| Tomato chlorosis virus | 310–1100 nm | – | XY-fusion network (XY-F) Multilayer Perceptron with Automated Relevance Determination (MLP–ARD) |
88 92 |
Morellos et al. [63] |
| Leaf mildew | 900–1700 nm | Texture + color + shape | backpropagation neural network (BPNN) + Genetic Algorithm (GA) | 97 | Zhang et al. [50] |
| A. alternata, A. solani, B. cinerea, and F. oxysporum | 400–950 nm for hyperspectral Imaging RGB, HSV, and l∗a∗b for RGB Images |
Descriptive statistics, Texture, and Shape |
Random forest and Random forest + Laplacian Score | For RGB images: Asymptomatic = 65 Early Stage = 75 Late Stage = 87 For hyperspectral images: Asymptomatic = 86 Early Stage = 92 Late Stage = 98 |
This Study |
The most remarkable challenge in past research is the lack of large data sets to be used in various deep learning methods such as convolutional neural networks [33]. Deep learning methods are usually considered black box methods, meaning that the features extracted in a network cannot be categorized as shape/color/texture features [6]. Therefore, the correct extraction of the main features such as texture, color, and shape from the images of diseased leaves to train the ML methods is a more effective approach when the data set is not large [50].
The classification of diseases with almost identical symptoms is challenging. To address this challenge, the results obtained by the feature selection algorithm showed that groups of different features such as texture, color, and shapes have an essential role in classification. Another challenges are not diagnosing the disease in the early days and the limited spectrum of color cameras. Using hyperspectral cameras helps users to detect diseases in the early stages and prevent their spread in the field. Plant disease detection based on only RGB color images is still not accurate and robust especially when the number of diseases with similar symptoms to detect increases [21].
Hyperspectral cameras are rather expensive and require an expert to operate them, while a cellphone is sufficient to take RGB images. Although ML-based models have shown significant performance in some applications [64,65], they can still require extensive training data and well-defined algorithms to accurately diagnose plant diseases. Additionally, using such methods requires the availability of appropriate hardware, software, and technical expertise, which might be limiting for some users [66]. There are limitations associated with the use of plant disease diagnosis technology by RGB and hyperspectral image processing, especially related to the collection and management of large amounts of quality data required to train ML models. Despite these limitations, digital image analysis is still an emerging and promising field in plant disease diagnosis and assessment, and its accuracy and efficiency are expected to be improved with further research and development.
4. Conclusion
A method for early identification of four common tomato diseases has been presented based on their spectral signature and morphological characteristics. For this purpose, hyperspectral and RGB images were used to classify the diseases from the first to the eleventh day after the transmission of the disease. A data set was prepared based on RGB and hyperspectral images collected on days 1, 3, 5, 7, 9, and 11 after disease transmission. The spectral signatures of the diseases were recognized using the Laplacian score feature selection algorithm, while a reliable disease detection was obtained in the early stages of disease development using the RF model. The results showed that the proposed ML-based method can identify B. cinerea on day 3 after disease transmission, F. oxysporum on day 5, and A. alternata and A. solani on day 7. The disease classification accuracy using RGB images on days 1, 3, 5, 7, 9, and 11 was 65, 71, 75, 77, 83 and 87 %, respectively, while using hyperspectral images, the accuracy was 86, 90, 92, 95, 97 and 98 %, respectively. The findings of this research show that compared to RGB visible image processing, hyperspectral imaging has a reliable performance in classifying tomato fungal diseases in early disease development stages.
Funding
(Not applicable)
Data availability statement
(The data supporting the results of this study are available from the first author upon reasonable request)
CRediT authorship contribution statement
Seyed Mohamad Javidan: Writing – review & editing, Writing – original draft, Validation, Software, Methodology, Investigation. Ahmad Banakar: Supervision. Keyvan Asefpour Vakilian: Writing – review & editing, Writing – original draft, Methodology. Yiannis Ampatzidis: Writing – review & editing, Writing – original draft, Methodology. Kamran Rahnama: Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Contributor Information
Seyed Mohamad Javidan, Email: Mohamad.javidan@modares.ac.ir.
Ahmad Banakar, Email: ah_banakar@modares.ac.ir.
Keyvan Asefpour Vakilian, Email: keyvan.asefpour@gau.ac.ir.
Yiannis Ampatzidis, Email: i.ampatzidis@ufl.edu.
Kamran Rahnama, Email: kamranrahnama1995@gmail.com.
References
- 1.Mohammad Zamani D., Javidan S.M., Zand M., Rasouli M. Detection of cucumber fruit on plant image using artificial neural network. Journal of Agricultural Machinery. 2023;13(1):27–39. doi: 10.22067/jam.2022.73827.1077. [DOI] [Google Scholar]
- 2.Cruz A., Ampatzidis Y., Pierro R., Materazzi A., Panattoni A., De Bellis L., Luvisi A. Detection of grapevine yellows symptoms in Vitis vinifera L. with artificial intelligence. Comput. Electron. Agric. 2019;157:63–76. doi: 10.1016/j.compag.2018.12.028. Elsevier BV. [DOI] [Google Scholar]
- 3.Momeny M., Jahanbakhshi A., Neshat A.A., Hadipour-Rokni R., Zhang Y.-D., Ampatzidis Y. Detection of citrus black spot disease and ripeness level in orange fruit using learning-to-augment incorporated deep networks. Ecol. Inf. 2022;71 doi: 10.1016/j.ecoinf.2022.101829. Elsevier BV. [DOI] [Google Scholar]
- 4.Abdulridha J., Ampatzidis Y., Qureshi J., Roberts P. Identification and classification of downy mildew severity stages in watermelon utilizing aerial and ground remote sensing and machine learning. Front. Plant Sci. 2022;13 doi: 10.3389/fpls.2022.791018. Frontiers Media SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hariharan J., Ampatzidis Y., Abdulridha J., Batuman O. An AI-based spectral data analysis process for recognizing unique plant biomarkers and disease features. Comput. Electron. Agric. 2023;204 doi: 10.1016/j.compag.2022.107574. Elsevier BV. [DOI] [Google Scholar]
- 6.Javidan S.M., Banakar A., Asefpour Vakilian K., Ampatzidis Y. Diagnosis of grape leaf diseases using automatic K-means clustering and machine learning. Smart Agricultural Technology. 2023;3 doi: 10.1016/j.atech.2022.100081. Elsevier BV. [DOI] [Google Scholar]
- 7.Hariharan J., Fuller J., Ampatzidis Y., Abdulridha J., Lerwill A. Finite difference analysis and bivariate correlation of hyperspectral data for detecting laurel wilt disease and nutritional deficiency in avocado. Rem. Sens. 2019;11(15):1748. doi: 10.3390/rs11151748. MDPI AG. [DOI] [Google Scholar]
- 8.Luvisi A., Ampatzidis Y., De Bellis L. Plant pathology and information technology: opportunity for management of disease outbreak and applications in regulation frameworks. Sustainability. 2016;8(8):831. doi: 10.3390/su8080831. MDPI AG. [DOI] [Google Scholar]
- 9.Javidan S.M. vol. 4. Skeena Publishers; 2023. Identifying plant pests and diseases with artificial intelligence: a short comment. (International Journal on Engineering Technologies and Informatics). 4. [DOI] [Google Scholar]
- 10.Ampatzidis Y., De Bellis L., Luvisi A. iPathology: robotic applications and management of plants and plant diseases. Sustainability. 2017;9(6):1010. doi: 10.3390/su9061010. MDPI AG. [DOI] [Google Scholar]
- 11.Yuan L., Yan P., Han W., Huang Y., Wang B., Zhang J., Zhang H., Bao Z. Detection of anthracnose in tea plants based on hyperspectral imaging. Comput. Electron. Agric. 2019;167 doi: 10.1016/j.compag.2019.105039. Elsevier BV. [DOI] [Google Scholar]
- 12.Zhang G., Xu T., Tian Y. Hyperspectral imaging-based classification of rice leaf blast severity over multiple growth stages. Research Square Platform LLC. 2022 doi: 10.21203/rs.3.rs-1756611/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu G., Fang Y., Jiang Q., Cui M., Li N., Ou Y., Diao Z., Zhang B. vol. 204. 2023. Early identification of strawberry leaves disease utilizing hyperspectral imaging combing with spectral features, multiple vegetation indices and textural features. (Computers and Electronics in Agriculture). Elsevier BV. [DOI] [Google Scholar]
- 14.Panthee D.R., Pandey A., Paudel R. Multiple foliar fungal disease management in tomatoes: a comprehensive approach. Int. J. Plant Biol. 2024;15(1):69–93. doi: 10.3390/ijpb15010007. MDPI AG. [DOI] [Google Scholar]
- 15.Li Q., Feng Y., Li J., Hai Y., Si L., Tan C., Peng J., Hu Z., Li Z., Li C., Hao D., Tang W. Multi-omics approaches to understand pathogenicity during potato early blight disease caused by Alternaria solani. Front. Microbiol. 2024;15 doi: 10.3389/fmicb.2024.1357579. Frontiers Media SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ismail A.M., Mosa M.A., El-Ganainy S.M. Chitosan-decorated copper oxide nanocomposite: investigation of its antifungal activity against tomato gray mold caused by Botrytis cinerea. Polymers. 2023;15(5):1099. doi: 10.3390/polym15051099. MDPI AG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panno S., Davino S., Caruso A.G., Bertacca S., Crnogorac A., Mandić A., Noris E., Matić S. A review of the most common and economically important diseases that undermine the cultivation of tomato crop in the mediterranean basin. Agronomy. 2021;11(11):2188. doi: 10.3390/agronomy11112188. MDPI AG. [DOI] [Google Scholar]
- 18.Terentev A., Dolzhenko V., Fedotov A., Eremenko D. Current state of hyperspectral remote sensing for early plant disease detection: a review. Sensors. 2022;22(3):757. doi: 10.3390/s22030757. MDPI AG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Golhani K., Balasundram S.K., Vadamalai G., Pradhan B. A review of neural networks in plant disease detection using hyperspectral data. Information Processing in Agriculture. 2018;5(3):354–371. doi: 10.1016/j.inpa.2018.05.002. Elsevier BV. [DOI] [Google Scholar]
- 20.Javidan S.M., Banakar A., Vakilian K.A., Ampatzidis Y. Journal. Vol. 116. Wiley; 2023. Tomato leaf diseases classification using image processing and weighted ensemble learning. In Agronomy; pp. 1029–1049. Issue 3. [DOI] [Google Scholar]
- 21.Javidan S.M., Banakar A., Rahnama K., Vakilian K.A., Ampatzidis Y. Feature engineering to identify plant diseases using image processing and artificial intelligence: a comprehensive review. Smart Agricultural Technology. 2024;8 doi: 10.1016/j.atech.2024.100480. Elsevier BV. [DOI] [Google Scholar]
- 22.Javidan S.M., Banakar A., Vakilian K.A., Ampatzidis Y., Rahnama K. In Multimedia Tools and Applications. Springer Science and Business Media LLC; 2024. Diagnosing the spores of tomato fungal diseases using microscopic image processing and machine learning. [DOI] [Google Scholar]
- 23.Asefpour Vakilian K., Massah J. A farmer-assistant robot for nitrogen fertilizing management of greenhouse crops. Comput. Electron. Agric. 2017;139:153–163. doi: 10.1016/j.compag.2017.05.012. Elsevier BV. [DOI] [Google Scholar]
- 24.Ashfaq M., Minallah N., Ullah Z., Ahmad A.M., Saeed A., Hafeez A. Performance analysis of low-level and high-level intuitive features for melanoma detection. Electronics. 2019;8(6):672. doi: 10.3390/electronics8060672. [DOI] [Google Scholar]
- 25.Haralick R.M., Shanmugam K., Dinstein I. On some quickly computable features for texture. Symposium of Computer Image Processing and Recognition, University of Missouri, Columbia. 1972;2(2) 12-2. [Google Scholar]
- 26.Javidan S.M., Ampatzidis Y., Asefpor Vakilian K., Mohammadzamani D. 2024 10th International Conference on Artificial Intelligence and Robotics (QICAR) IEEE; 2024. A novel approach for automated strawberry fruit varieties classification using image processing and machine learning. [DOI] [Google Scholar]
- 27.Doan N.-Q., Azzag H., Lebbah M. 2018 International Joint Conference on Neural Networks (IJCNN). 2018 International Joint Conference on Neural Networks (IJCNN) IEEE; 2018. Hierarchical Laplacian score for unsupervised feature selection. [DOI] [Google Scholar]
- 28.Javidan S.M., Banakar A., Asefpour Vakilian K., Ampatzidis Y. 2022 8th Iranian Conference on Signal Processing and Intelligent Systems (ICSPIS) IEEE; 2022. A feature selection method using slime mould optimization algorithm in order to diagnose plant leaf diseases; pp. 1–5. [DOI] [Google Scholar]
- 29.Sarkar C., Gupta D., Gupta U., Hazarika B.B. vol. 145. 2023. Leaf disease detection using machine learning and deep learning: review and challenges. (Applied Soft Computing). Elsevier BV. [DOI] [Google Scholar]
- 30.Asefpour Vakilian K., Massah J. An apple grading system according to European fruit quality standards using Gabor filter and artificial neural networks. Scientific Study & Research. Chemistry & Chemical Engineering, Biotechnology, Food Industry. 2016;17(1):75–85. University of Bacău. [Google Scholar]
- 31.Xie C., Shao Y., Li X., He Y. vol. 5. Springer Science and Business Media LLC; 2015. Detection of early blight and late blight diseases on tomato leaves using hyperspectral imaging. (Scientific Reports). 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan L., Li H., Li C., Wang A., Yang Y., Wang P. Hyperspectral sensing of plant diseases: principle and methods. Agronomy. 2022;12(6):1451. doi: 10.3390/agronomy12061451. MDPI AG. [DOI] [Google Scholar]
- 33.Wang D., Vinson R., Holmes M., Seibel G., Bechar A., Nof S., Tao Y. Early detection of tomato spotted wilt virus by hyperspectral imaging and outlier removal auxiliary classifier generative adversarial nets (OR-AC-GAN) Sci. Rep. 2019;9(1) doi: 10.1038/s41598-019-40066-y. Springer Science and Business Media LLC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hennessy A., Clarke K., Lewis M. Hyperspectral classification of plants: a review of waveband selection generalisability. Rem. Sens. 2020;12(1):113. doi: 10.3390/rs12010113. MDPI AG. [DOI] [Google Scholar]
- 35.Nevalainen O., Honkavaara E., Tuominen S., Viljanen N., Hakala T., Yu X., Hyyppä J., Saari H., Pölönen I., Imai N., Tommaselli A. Individual tree detection and classification with UAV-based photogrammetric point clouds and hyperspectral imaging. Rem. Sens. 2017;9(3):185. doi: 10.3390/rs9030185. MDPI AG. [DOI] [Google Scholar]
- 36.Heim R., Jürgens N., Große-Stoltenberg A., Oldeland J. The effect of epidermal structures on leaf spectral signatures of ice plants (Aizoaceae) Rem. Sens. 2015;7(12):16901–16914. doi: 10.3390/rs71215862. MDPI AG. [DOI] [Google Scholar]
- 37.Mutanga O., Van Aardt J., Kumar L. Imaging spectroscopy (hyperspectral remote sensing) in southern Africa: an overview. South Afr. J. Sci. 2009;105(5):193–198. [Google Scholar]
- 38.Lai Y., Zhang J., Song Y., Gong Z. Retrieval and Evaluation of Chlorophyll-a Concentration in reservoirs with main water supply function in Beijing, China, based on landsat satellite images. Int. J. Environ. Res. Publ. Health. 2021;18(9):4419. doi: 10.3390/ijerph18094419. MDPI AG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roth G.A., Tahiliani S., Neu‐Baker N.M., Brenner S.A. Hyperspectral microscopy as an analytical tool for nanomaterials. WIREs Nanomedicine and Nanobiotechnology. 2015;7(4):565–579. doi: 10.1002/wnan.1330. Wiley. [DOI] [PubMed] [Google Scholar]
- 40.Pu H., Lin L., Sun D. Principles of hyperspectral microscope imaging techniques and their applications in food quality and safety detection: a review. Compr. Rev. Food Sci. Food Saf. 2019;18(4):853–866. doi: 10.1111/1541-4337.12432. Wiley. [DOI] [PubMed] [Google Scholar]
- 41.Mahlein A.-K., Rumpf T., Welke P., Dehne H.-W., Plümer L., Steiner U., Oerke E.-C. Development of spectral indices for detecting and identifying plant diseases. Remote Sensing of Environment. 2013;128:21–30. doi: 10.1016/j.rse.2012.09.019. Elsevier BV. [DOI] [Google Scholar]
- 42.Qu J.-H., Sun D.-W., Cheng J.-H., Pu H. Mapping moisture contents in grass carp (Ctenopharyngodon idella) slices under different freeze drying periods by Vis-NIR hyperspectral imaging. LWT. 2017;75:529–536. doi: 10.1016/j.lwt.2016.09.024. Elsevier BV. [DOI] [Google Scholar]
- 43.Oerke E.-C. Remote sensing of diseases. Annu. Rev. Phytopathol. 2020;58(1):225–252. doi: 10.1146/annurev-phyto-010820-012832. Annual Reviews. [DOI] [PubMed] [Google Scholar]
- 44.Asefpour Vakilian K., Massah J. Design, development and performance evaluation of a robot to early detection of nitrogen deficiency in greenhouse cucumber (Cucumis sativus) with machine vision. Int. J. Agric. Res. Rev. 2012;2(4):448–454. [Google Scholar]
- 45.Xiao Y., Dong Y., Huang W., Liu L., Ma H. Wheat fusarium head blight detection using UAV-based spectral and texture features in optimal window size. Rem. Sens. 2021;13(13):2437. doi: 10.3390/rs13132437. MDPI AG. [DOI] [Google Scholar]
- 46.Khan I.H., Liu H., Li W., Cao A., Wang X., Liu H., Cheng T., Tian Y., Zhu Y., Cao W., Yao X. Early detection of powdery mildew disease and accurate quantification of its severity using hyperspectral images in wheat. Rem. Sens. 2021;13(18):3612. doi: 10.3390/rs13183612. MDPI AG. [DOI] [Google Scholar]
- 47.Guo A., Huang W., Dong Y., Ye H., Ma H., Liu B., Wu W., Ren Y., Ruan C., Geng Y. Wheat yellow rust detection using UAV-based hyperspectral technology. Rem. Sens. 2021;13(1):123. doi: 10.3390/rs13010123. MDPI AG. [DOI] [Google Scholar]
- 48.Nguyen C., Sagan V., Skobalski J., Severo J.I. Early detection of wheat yellow rust disease and its impact on terminal yield with multi-spectral UAV-imagery. Rem. Sens. 2023;15(13):3301. doi: 10.3390/rs15133301. MDPI AG. [DOI] [Google Scholar]
- 49.Kamruzzaman M., ElMasry G., Sun D.-W., Allen P. Application of NIR hyperspectral imaging for discrimination of lamb muscles. J. Food Eng. 2011;104(3):332–340. doi: 10.1016/j.jfoodeng.2010.12.024. Elsevier BV. [DOI] [Google Scholar]
- 50.Zhang X., Wang Y., Zhou Z., Zhang Y., Wang X. Detection method for tomato leaf mildew based on hyperspectral fusion terahertz technology. Foods. 2023;12(3):535. doi: 10.3390/foods12030535. MDPI AG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pourreza A., Lee W.S., Ehsani E.R., Etxeberria E. In Transactions of the ASABE. American Society of Agricultural and Biological Engineers (ASABE); 2014. Citrus huanglongbing detection using narrow-band imaging and polarized illumination; pp. 259–272. [DOI] [Google Scholar]
- 52.Trifunović-Momčilov M., Milošević S., Marković M., Đurić M., Jevremović S., Dragićević I.Č., Subotić A.R. Changes in photosynthetic pigments content in non-transformed and AtCKX transgenic centaury (Centaurium erythraea Rafn) shoots grown under salt stress in vitro. Agronomy. 2021;11(10):2056. doi: 10.3390/agronomy11102056. MDPI AG. [DOI] [Google Scholar]
- 53.Wang Y.M., Ostendorf B., Gautam D., Habili N., Pagay V. Plant viral disease detection: from molecular diagnosis to optical sensing technology—a multidisciplinary review. Rem. Sens. 2022;14(7):1542. doi: 10.3390/rs14071542. MDPI AG. [DOI] [Google Scholar]
- 54.Ashikhmin A., Bolshakov M., Pashkovskiy P., Vereshchagin M., Khudyakova A., Shirshikova G., Kozhevnikova A., Kosobryukhov A., Kreslavski V., Kuznetsov V., Allakhverdiev S.I. The adaptive role of carotenoids and anthocyanins in Solanum lycopersicum pigment mutants under high irradiance. Cells. 2023;12(Issue 21):2569. doi: 10.3390/cells12212569. MDPI AG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anderegg J., Yu K., Aasen H., Walter A., Liebisch F., Hund A. Spectral vegetation indices to track senescence dynamics in diverse wheat germplasm. Front. Plant Sci. 2020;10 doi: 10.3389/fpls.2019.01749. Frontiers Media SA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reis Pereira M., Verrelst J., Tosin R., Rivera Caicedo J.P., Tavares F., Neves dos Santos F., Cunha M. Plant disease diagnosis based on hyperspectral sensing: comparative analysis of parametric spectral vegetation indices and nonparametric Gaussian process classification approaches. Agronomy. 2024;14(3):493. doi: 10.3390/agronomy14030493. MDPI AG. [DOI] [Google Scholar]
- 57.Zahra A., Qureshi R., Sajjad M., Sadak F., Nawaz M., Khan H.A., Uzair M. Current advances in imaging spectroscopy and its state-of-the-art applications. Expert Syst. Appl. 2024;238 doi: 10.1016/j.eswa.2023.122172. Elsevier BV. [DOI] [Google Scholar]
- 58.McCraine D., Samiappan S., Kohler L., Sullivan T., Will D.J. Automated hyperspectral feature selection and classification of wildlife using uncrewed aerial vehicles. Rem. Sens. 2024;16(2):406. doi: 10.3390/rs16020406. MDPI AG. [DOI] [Google Scholar]
- 59.Salamai A.A., Ajabnoor N., Khalid W.E., Ali M.M., Murayr A.A. Lesion-aware visual transformer network for Paddy diseases detection in precision agriculture. Eur. J. Agron. 2023;148 doi: 10.1016/j.eja.2023.126884. Elsevier BV. [DOI] [Google Scholar]
- 60.González-Rodríguez V.E., Izquierdo-Bueno I., Cantoral J.M., Carbú M., Garrido C. Artificial intelligence: a promising tool for application in phytopathology. Horticulturae. 2024;10(3):197. doi: 10.3390/horticulturae10030197. MDPI AG. [DOI] [Google Scholar]
- 61.Xie C., Yang C., He Y. Hyperspectral imaging for classification of healthy and gray mold diseased tomato leaves with different infection severities. Comput. Electron. Agric. 2017;135:154–162. doi: 10.1016/j.compag.2016.12.015. Elsevier BV. [DOI] [Google Scholar]
- 62.Lu J., Ehsani R., Shi Y., de Castro A.I., Wang S. vol. 8. Springer Science and Business Media LLC; 2018. Detection of multi-tomato leaf diseases (late blight, target and bacterial spots) in different stages by using a spectral-based sensor. (Scientific Reports). 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morellos A., Tziotzios G., Orfanidou C., Pantazi X.E., Sarantaris C., Maliogka V., Alexandridis T.K., Moshou D. Non-destructive early detection and quantitative severity stage classification of tomato chlorosis virus (ToCV) infection in young tomato plants using Vis–NIR spectroscopy. Rem. Sens. 2020;12(12):1920. doi: 10.3390/rs12121920. MDPI AG. [DOI] [Google Scholar]
- 64.Hashemi A., Asefpour Vakilian K., Khazaei J., Massah J. An artificial neural network modeling for force control system of a robotic pruning machine. J. Inf. Organ. Sci. 2014;38(Issue 1):35–41. University of Zagreb. [Google Scholar]
- 65.Massah J., Asefpour Vakilian K., Torktaz S. Supervised machine learning algorithms can predict penetration resistance in mineral-fertilized soils. Commun. Soil Sci. Plant Anal. 2019;50(17):2169–2177. doi: 10.1080/00103624.2019.1654505. [DOI] [Google Scholar]
- 66.Mahlein A.-K. Plant disease detection by imaging sensors – parallels and specific demands for precision agriculture and plant phenotyping. Plant Dis. 2016;100(2):241–251. doi: 10.1094/pdis-03-15-0340-fe. Scientific Societies. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
(The data supporting the results of this study are available from the first author upon reasonable request)