Abstract
India is experiencing a rapid spread of human immunodeficiency virus type 1 (HIV-1), primarily through heterosexual transmission of subtype C viruses. To delineate the molecular features of HIV-1 circulating in India, we sequenced the V3-V4 region of viral env from 21 individuals attending an HIV clinic in Calcutta, the most populous city in the eastern part of the country, and analyzed these and the other Indian sequences in the HIV database. Twenty individuals were infected with viruses having a subtype C env, and one had viruses with a subtype A env. Analyses of 192 subtype C sequences that included one sequence for each subject from this study and from the HIV database revealed that almost all sequences from India, along with a small number from other countries, form a phylogenetically distinct lineage within subtype C, which we designate CIN. Overall, CIN lineage sequences were more closely related to each other (level of diversity, 10.2%) than to subtype C sequences from Botswana, Burundi, South Africa, Tanzania, and Zimbabwe (range, 15.3 to 20.7%). Of the three positions identified as signature amino acid substitution sites for CIN sequences (K340E, K350A, and G429E), 56% of the CIN sequences contained all three amino acids while 87% of the sequences contained at least two of these substitutions. Among the non-CIN sequences, all three amino acids were present in 2%, while 22% contained two or more of these amino acids. These results suggest that much of the current Indian epidemic is descended from a single introduction into the country. Identification of conserved signature amino acid positions could assist epidemiologic tracking and has implications for the development of a vaccine against subtype C HIV-1 in India.
Human immunodeficiency virus type 1 (HIV-1) infection has been reported in more than 173 countries worldwide (45). Prior to worldwide spread, HIV-1 infections were mainly found in North America, western Europe, and sub-Saharan Africa. While HIV-1 infection appears to have been introduced into India in the mid-1980s, high rates of seroprevalence, especially among commercial sex workers (30; R. C. Bollinger, S. Mehendale, R. Gangakhedkar, T. Quinn, M. Bentley, R. Brookmeyer, D. Gadkari, A. Risbud, A. Divekar, M. Shephard, S. Thilakavathi, and J. Rodrigues, Conf. Adv. AIDS Vaccine Dev., p. 221, 1996), have been documented. If the current trends continue, by one estimate, India may have the highest number of HIV-1 infections of any country by the end of this decade (8, 9, 30).
Genetic analyses of HIV-1 sequences circulating in India have been limited. Initial reports indicated that viruses from India were more closely related to those identified in South Africa than to those in North America or central Africa (16). Subsequent studies have shown that subtype C HIV-1 predominates in India (11, 15, 19, 43, 44), with a small fraction of infections caused by HIV-1 subtypes A and B (3, 19, 44). Genetic characterization of HIV-1 in India has involved mainly the northern, western, and southern parts of the country (11, 19, 29, 33, 43, 44), whereas no information from the eastern part is available. Based on genetic relatedness in heteroduplex mobility assays, Delwart et al. suggested a recent introduction of HIV-1 subtype C in India from one or a set of similar founder strains (15). Similarly, based on viral sequence diversity estimates, Grez et al. suggested the spread of both HIV-1 and HIV-2 from recent ancestors (18). A subsequent study of eight virus isolates from Pune in the southern and New Delhi in the northern part of India found increased levels of genetic heterogeneity between strains (43).
The present study was undertaken to characterize HIV-1 from the eastern part of India and to identify molecular sequence features that distinguish variants circulating in India from those present in other parts of the world. We sampled HIV-1 sequences from individuals attending an HIV clinic in Calcutta, the most populous city in India and located in the eastern part of the country. We sought to identify the molecular features unique to subtype C HIV-1 circulating in India by analyzing 192 env sequences, including subtype C sequences from 20 individuals in this study as well as 172 sequences available in GenBank. We identified a monophyletic lineage of subtype C sequences circulating in India, designated here as CIN, and signature amino acids in the Env associated with these sequences.
Blood samples were obtained in 1999 from 21 subjects recruited from an HIV clinic at the Tropical School of Medicine at Calcutta, India, as part of the Fogarty International Collaborative Research on AIDS. The clinical and transmission information pertaining to each of the 21 individuals is provided in Table 1. Most acquired HIV-1 infection through heterosexual contact and had exposure to multiple sex partners. HIV-1 infection was determined by an enzyme-linked immunosorbent assay (Organon Teknika, Durham, N.C.) and confirmed by Western blotting using a whole HIV-1 lysate (Dupont, Wilmington, Del.). Cellular DNA was isolated from 0.5 to 3.0 ml of whole blood by the PureGene DNA isolation kit (Gentra System, Minneapolis, Minn.). The C2-V5 region of the viral envelope gene was amplified by a nested PCR as previously described (15, 26), using multiple serial dilutions of cellular DNA with primers ED31/BH2 and ES7/ES8 (or DR7/DR8 [26]) in the first and second rounds of PCR, respectively. Multiple HIV-1-negative controls were included in each amplification experiment to identify carryover PCR contamination. PCR products were either directly sequenced or cloned into the pGEM-T vector (Promega, Madison, Wis.) and sequenced with the Taq DyeDeoxy terminator cycle sequencer kit (Applied Biosystems Inc., Foster City, Calif.) in a 373 DNA sequencer (Applied Biosystems Inc.). All sequences were subjected to quality control measures to ensure that there were no sample mix-ups or contamination from other sources (23, 25). Sequences corresponding to V3-V4 region were used for the analyses described here. BLAST searches of sequences from each subject identified a best match in the HIV sequence database (21) that was always with another sequence from India. However, each sequence was divergent from those in the database (21) by more than 5%, suggesting an absence of sample mix-ups with previously published sequences. Envelope sequence subtypes were assigned using the genotyping tool (http://www.ncbi.nlm.nih.gov/retroviruses/subtype/subtype.html). Sequences in this study were aligned using CLUSTAL W (41) and manually edited using the Genetic Data Environment program (39). A set of 192 sequences spanning positions 7093 to 7540 of HXB2 included a sequence from each individual in this study and the available subtype C GenBank sequences that span this region. An appropriate evolutionary model for these sequences was selected using the Akaike information criterion (2) as implemented in Modeltest 3.0 (35). Parameters of the chosen model (TVM+I+G) were as follows: equilibrium nucleotide frequencies, fA = 0.4381, fC = 0.1804, fG = 0.1814, fT = 0.2001; proportion of invariable sites, = 0.0499; shape parameter (α) of the 71 distribution reflecting site-to-site rate variability of variable sites, 0.6309; and R matrix values, RA→C = 1.805, RA→G = RC→T = 4.664, RA→T = 0.6892, RC→G = 0.9563, and RG→T = 1. A pairwise distance matrix was calculated based on this model and used in the construction of a neighbor-joining tree in version 4.0b2a of PAUP (40) on a Macintosh G4 computer. To further examine relationships seen in this tree, a subset of subtype C sequences, including all sequences from India as well as reference sequences from the Los Alamos subtype reference alignment (http://hiv-web.lanl.gov/ALIGN_CURRENT/SUBTPE-REF /subtype.html), were selected for a maximum likelihood analysis. Again, an appropriate evolutionary model (TVM+G) for these 60 sequences was selected using the Akaike information criterion. Parameters of this model were as follows: fA = 0.4019 fC = 0.1755, fG = 0.1951, fT = 0.2275; α = 0.4982; RA→C = 3.204, RA→G = RC→T = 7.106, RA→T = 0.7699, RC→G = 1.903, and RG→T = 1.
TABLE 1.
HIV-1-infected subjects evaluated in this study
| Subject | Age (yr) | Sex | Mode of transmission | STD history | No. of partners | Clinical symptoms |
|---|---|---|---|---|---|---|
| 7 | 35 | Male | Heterosexual contact | Yes | >10 | Asymptomatic |
| 10 | 26 | Male | Heterosexual contact | Yes | 10 | Asymptomatic |
| 12 | 32 | Male | Heterosexual contact | No | >10 | Asymptomatic |
| 13 | 36 | Male | Heterosexual contact | No | >10 | Asymptomatic |
| 14 | 22 | Female | Heterosexual contact | No | 1 | Asymptomatic |
| 64 | 20 | Female | Heterosexual contact | Yes | 1 | Genital ulcer |
| 84 | 35 | Male | Heterosexual contact | No | 1 | Genital ulcer |
| 86 | 30 | Male | Intravenous-drug use | Yes | >10 | Genital ulcer |
| 87 | 36 | Male | Blood transfusion | Yes | 1 | Genital ulcer, fever |
| 96 | 40 | Male | Intravenous-drug use | No | 3 | Asymptomatic |
| 97 | 30 | Female | Blood transfusion | No | 1 | Asymptomatic |
| 125 | 33 | Male | Heterosexual contact | No | 4 | Fever, weight loss |
| 221 | 32 | Male | Heterosexual contact | No | 5 | Diarrhea, weight loss |
| 251 | 29 | Male | Heterosexual contact | Yes | 3 | Fever, chest pain, genital ulcer |
| 257 | 34 | Male | Heterosexual contact | No | >10 | Candidiasis |
| 275 | 26 | Male | Heterosexual contact | Yes | 5 | Urethritis, weakness, weight loss |
| 276 | 45 | Male | Heterosexual contact | No | 3 | Fever, diarrhea |
| 277 | 27 | Male | Heterosexual contact | No | 1 | None |
| 293 | 34 | Male | Heterosexual contact | Yes | 1 | Genital ulcer, weakness, weight loss |
| 306 | 32 | Male | Heterosexual contact | No | >10 | Fever, weight loss |
| 321 | 27 | Male | Heterosexual contact | Yes | 7 | Urethritis, weight loss, skin rash |
HIV-1 sequences from Calcutta, India.
We sampled 60 env sequences from 21 individuals and found 20 to be infected with viruses with subtype C env, while one individual (subject 12) was infected with virus bearing a subtype A env. In all but two subjects, sequences from each subject formed monophyletic groups in phylogenetic analysis, supported at about 100% bootstrap levels (data not shown); the exception was two individuals (subjects 13 and 14), whose sequences were highly similar, suggesting epidemiologically linked infections, although no information was available to evaluate this possibility. These findings suggest that majority of HIV-1 isolates circulating in Calcutta possess subtype C env sequences.
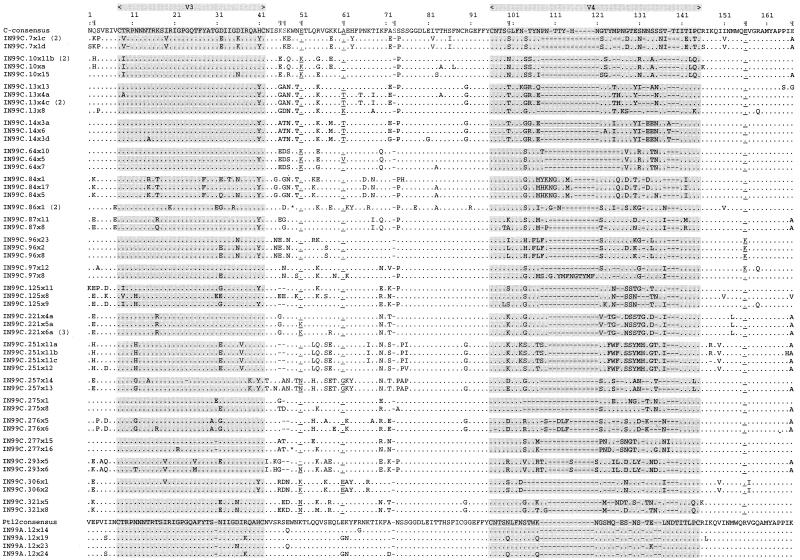
An amino acid alignment representing sequences from each of the 21 individuals is shown in Fig. 1. The V3 loop was conserved in all sequences, the GPGQ motif at the tip of V3 was conserved in all sequences except two, and the conserved dodecapeptide RIGPGQTFYATG (43) (amino acids 20 to 31, corresponding to positions 308 to 321 in HXB2 Env) was found in 13 subjects. The adjacent heptapeptide DIIGDIR (amino acids 32 to 38; positions 322 to 327 in HXB2 Env), often found in other Indian HIV-1 subtype C strains (19, 29), was conserved in nine subjects. The mean viral diversity for nucleic acid sequences present within an individual among the study subjects sampled here was 2.6% and ranged between 0 and 13.6%.
FIG. 1.
Deduced amino acid sequences of partial HIV-1 env sequences obtained from the 21 subjects in this study. Sequences from 20 subjects harboring subtype C were aligned with the subtype C consensus sequence. Subtype A sequences from subject 12 were aligned with a consensus sequence derived from the four sequences sampled from this individual. IN99C and IN99A in the names indicate the year of sampling and subtype assignment. Numbers in parentheses indicate the number of sequences with identical amino acid sequences. The regions corresponding to V3 and V4 in the envelope protein are highlighted. The nine amino acid positions identified to be particularly discriminatory for subtype CIN sequences (Table 2) are indicated (¶). In addition, the amino acids at positions 51, 61, and 156 (corresponding to positions 340, 350, and 429, respectively) that were conserved in more than 70% of the CIN sequences are underlined. Within the alignment, dots indicate identity with the consensus sequence, dashes indicate deletions, and asterisks indicate stop codons.
A switch in virus phenotype from R5 (non-syncytium inducing on MT-2 cells with CCR5 coreceptor usage) into X4 (syncytium inducing on MT-2 cells along with the utilization of the CXCR4 molecule as a coreceptor) is associated with accelerated disease progression in HIV-1 Env subtypes B, D, and E (5–7, 17, 38). Consistent with previous reports indicating a low prevalence of X4 viruses among subtype C viruses (12, 34, 42), none of the sequences analyzed in this study were found to have basic amino acids at V3 loop positions 11, 24, and 25 (positions 18, 31, and 32 in Fig. 1), previously shown to be linked to a switch to the X4 phenotype (13, 14).
Geographic structure in CIN sequences.
When the sequences from this study were compared to GenBank sequences in a BLAST search, the best matches and nearly all of the high-scoring matches were also from India. These results prompted us to test for the presence of geographic structure in sequences sampled within India as well in subtype C sequences sampled from South Africa, Botswana, South Africa, Burundi, Tanzania, and Zimbabwe. We used the Slatkin-Maddison method, previously adapted to test for tissue-compartmental structure of HIV (4, 36). We counted the number of changes (or steps) from one locale (country or city) to another in an observed phylogram and compared this number to those seen for 10,000 randomly constructed trees using MacClade version 3.08 (28). We inferred that there is significant geographic structure if fewer changes are seen in the observed tree than in 95% of the random trees. We sought evidence of geographic structure at three levels: (i) among all the 23 countries for which sequences were available, (ii) between the six countries (India, South Africa, Botswana, Burundi, Tanzania, and Zimbabwe) from which eight or more sequences in the region examined were available, and (iii) among cities within India. Amino acid signature sequences were identified using VESPA (20, 22).
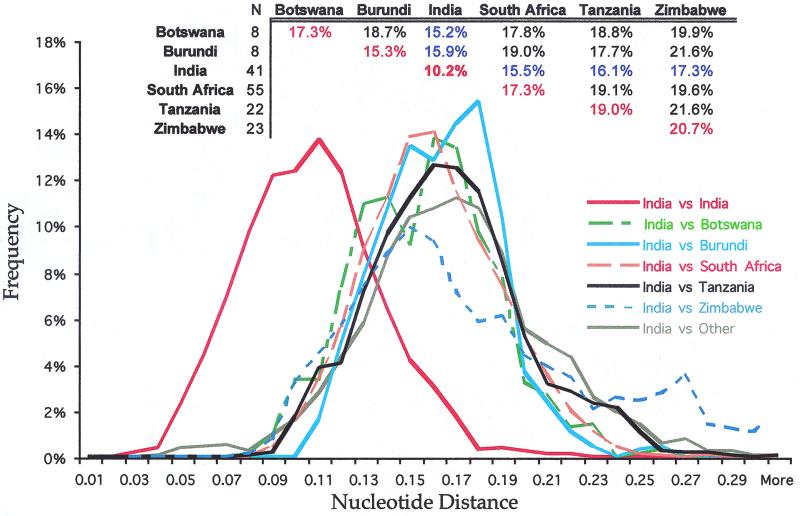
We compared subtype C sequences from Botswana, Burundi, India, South Africa, Tanzania, and Zimbabwe to evaluate levels of viral diversity within each country. Sequences sampled within India exhibited a lower level of diversity (10.2%) than those from other countries, which ranged from 15% in Burundi to 20% in Zimbabwe (Fig. 2, inset). Indian sequences differed from sequences from other countries by an average of 14 to 17%, closer than all other between-country comparisons. In view of the small numbers of sequences involved in these comparisons, the statistical significance of this observation remains to be confirmed.
FIG. 2.
DNA distances between subtype C sequences sampled from various countries. The inset shows the mean DNA distances for comparison of sequences sampled within and between each of the six countries where eight or more sequences were available for comparison. The solid red line in the plot depicts the distribution of DNA distances when sequences sampled within India were compared to each other. Other lines illustrate the distribution of pairwise DNA distances when sequences from India were compared to sequences from each of the other countries.
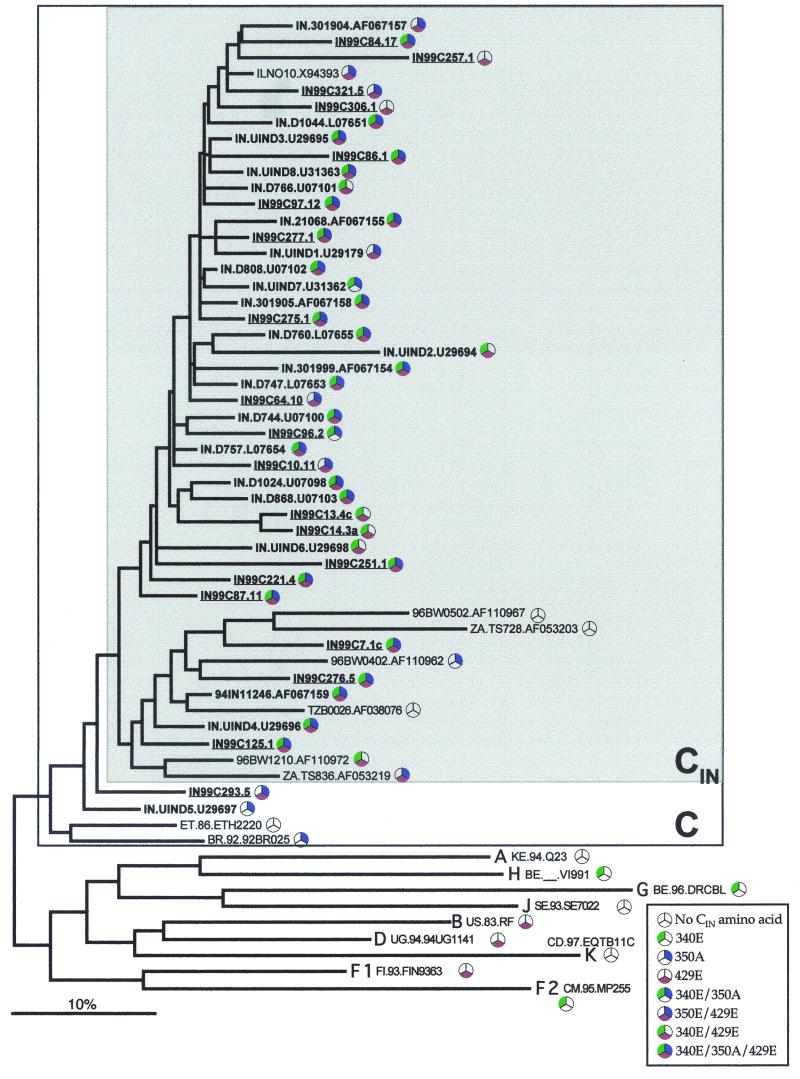
In an assessment of phylogenetic relationships among the 192 known subtype C sequences from 23 countries (Fig. 3), an overall star-like phylogeny was observed, although several clusters were also evident. While no clusters including more than several sequences had substantial bootstrap support (but see below), sequences from India generally clustered together more than sequences from other countries. The sequences were tested for the presence of geographic distribution at three levels. Geographic clustering over the 23 countries was highly significant (74 steps observed; P < 0.0001), indicating a country-dependent distribution of sequences. Among six countries with sufficient sequences (eight or more) to test for geographic structure on a country-by-country basis (Botswana, Burundi, India, South Africa, Tanzania, and Zimbabwe), the Slatkin-Maddison test (28) showed that sequences from India, South Africa, and Zimbabwe had geographic structure with a probability significantly greater than random expectations (9, 25, and 14 changes from one country to another in the reconstructed neighbor-joining trees, respectively; P < 0.0001 for each comparison). However, unlike sequences from South Africa and Zimbabwe, which were scattered in numerous lineages, almost all sequences from India formed a monophyletic lineage that we designate here as CIN (Fig. 3). To test for the presence of geographic clustering within the Indian subcontinent, we examined all 42 available sequences, of which 20 were from Calcutta in the east (this study), 8 were from Bombay in the west, 5 were from Pune in the south, and one was from Goa in the southwest; the geographic origin of 8 sequences was unknown. We observed 12 geographic switches on the maximum-likelihood tree, a figure that is within what might be frequently observed when examining a set of 10,000 random trees (P = 0.3372). Thus, no significant geographic clustering of sequences was found in different regions within India.
FIG. 3.
Phylogenetic relationships among subtype C HIV-1 env sequences sampled from different countries. Neighbor-joining analysis using 192 sequences encoding V3-V4 region was implemented using the TVM+I+G evolutionary model as described in the text.
We next performed a maximum-likelihood phylogenetic analysis on a subset of subtype C sequences that included all Indian sequences plus those that were closely related in the neighbor-joining analysis and the subtype reference sequences (http://hiv-web.lanl.gov/ALIGN_CURRENT/SUBTPE-REF /subtype.html) (Fig. 4). Consistent with neighbor-joining analysis, most subtype C sequences from India formed a strong monophyletic group that contained just one sequence from Israel from an unpublished study (GenBank accession no. X94393) (Gehring et al., unpublished data). A few Indian sequences also clustered in a second lineage with a small number of sequences from Botswana, South Africa, and Tanzania in another lineage. When complete gp160 subtype C sequences were examined (data not shown), sequences from India clustered with a 92% bootstrap support. These included the 94IN11246 sequence in the second lineage, while the African gp160 sequences represented in this lineage were not found in the Indian cluster. The shaded box representing CIN sequences in Fig. 4 was observed in several high-likelihood trees and included all the CIN sequences seen in these trees.
FIG. 4.
Maximum likelihood (TVM+G evolutionary model) phylogram of all CIN lineage sequences along with other sequences sampled from India and reference sequences for other subtypes. CIN lineage sequences identified in Fig. 3 are shown within the gray box. CIN lineage sequences clustered into two lineages, one containing only sequences from India (except one from Israel in an unpublished study; ILNO10.X94393) and another containing a small number of sequences from African countries. Sequences from India are in bold, and those isolated in this study are underlined. Sequence identifiers show the two-letter ISO 3166 country codes (http://www.din.de/gremien/nas/nabd/iso3166ma/codlstp1/en_listp1.html) and the year of isolation, when available. The log likelihood score for the phylogram was −5654.97269.
Signature amino acids in CIN sequences.
We next assessed whether subtype C sequences from India had amino acid substitutions characteristic of their origin. Using VESPA (20, 22), we found that the CIN lineage consensus sequence differed at nine amino acids from that of other subtype C sequences (Table 2). Eight of these amino acids were outside the variable regions, while one at 415G was within the V4 region. Based on an abundance of at least 70%, we identified K340E, K350A, and G429E as signature amino acid substitutions characteristic of the CIN lineage. Fifty-six percent of CIN sequences contained all three of the signature amino acids (340E, 350A, and 429E), compared to 2% of the non-CIN sequences. Similarly, 87% of the CIN sequences contained two or more of the CIN signature amino acid residues, compared to 22% in the non-CIN sequences. Differences in the representation of each of these three amino acids, singly and in combination, between CIN and non-CIN sequences were significant (P < 0.001, chi-square test).
TABLE 2.
Amino acids characteristic of CIN lineage and their potential evolutionary and structural significance
| Subtype | n | Prevalence of CIN amino acid at Env positiona:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 290 [T] | 335 [R]b | 336 [A]b | 340 [N]b | 350 [R] | 363 [K] | 415 [Q] | 429 [K]cd | 440 [S]be | ||
| CIN | 46 | 0.58 (Q) | 0.44 (K) | 0.36 (D) | 0.73 (E) | 0.78 (A) | 0.60 (S) | 0.44 (G) | 0.84 (E) | 0.6 (E) |
| Cf | 146 | 0.65 (E) | 0.30 (E) | 0.15 (E) | 0.46 (K) | 0.30 (K) | 0.59 (P) | 0.32 (K) | 0.46 (G) | 0.44 (A) |
| A | 177 | 0.18 | 0.01 | 0.00 | ||||||
| B | 234 | 0.06 | 0.00 | 0.65 | ||||||
| D | 104 | 0.02 | 0.00 | 0.35 | ||||||
| E | 91 | 0.66 | 0.00 | 0.08 | ||||||
| F | 25 | 0.04 | 0.00 | 0.20 | ||||||
| G | 91 | 0.60 | 0.03 | 0.00 | ||||||
| H | 13 | 0.15 | 0.00 | 0.00 | ||||||
| J | 2 | 0.00 | 0.00 | 0.00 | ||||||
| K | 6 | 0.00 | 0.00 | 0.17 | ||||||
| Group O | 12 | 0.42 | 0.00 | 0.00 | ||||||
| CRF-AGg | 54 | 0.09 | 0.00 | 0.00 | ||||||
Positions and letters in brackets correspond to the HXB2 prototype Env sequence (GenBank accession no. K03455). Values are prevalences of the CIN amino acid in the indicated subtype Env sequence (except for data for subtype C; see below). In an alignment of 192 sequences, the consensus in CIN differed from the remaining C sequences at the nine positions shown. Using a cutoff of 70%, three positions (in bold) appear to have a signature amino acid corresponding to CIN lineage sequences. All available sequences were used for all subtypes except subtype B.
Site has significantly predominant nonsynonymous substitutions indicative of positive selection acting on the site (46).
Site has significantly predominant synonymous substitutions indicative of negative selection acting on the site (46).
Site has been shown to interact with CD4 in the gp120 crystal structure (24).
Site has been shown to interact with CCR5 in the gp120 crystal structure (37).
Letters in parentheses are the residues most prevalent in non-CIN subtype C sequences, and values in this row are prevalences of these amino acids.
Circulating recombinant form comprising subtypes A and G.
More striking was the representation of 340E, 350A, and 429E in the Indian sequences within the CIN lineage (Fig. 4). Of the 39 Indian sequences within the CIN lineage, 26 (67%) had all three signature amino acid residues, 38 (97%) had at least two, and one (2.6%) had one. Of the seven non-Indian sequences within the CIN lineage, three had none of these residues, while three sequences (one each from Botswana, South Africa, and Israel) contained two of them, and one sequence from Botswana contained just 350E. Similar patterns were evident when the presence of these amino acids was identified on the neighbor-joining tree with all the 192 sequences examined in this study (data not shown).
To evaluate uniqueness of the three signature amino acids from the CIN lineage in other subtypes, we determined their prevalence in a data set of sequences from other subtypes from the Los Alamos database (Table 2). 340E was present in a high proportion of Env sequences from subtypes G (60%) and E (66%) and group O (42%), as well as in lesser proportions of sequences from subtype A (18%) and H (15%). 429E was also found in a substantial proportion of sequences from subtypes B (65%) and D (35%) and in a smaller proportion of sequences from subtypes F (20%) and K (17%). In contrast, 350A was observed at very low frequencies in all non-subtype C sequences.
We also examined the frequency of CIN lineage signature amino acids over time using sequences previously reported from India. When sampling time for the sequences was not provided, the year of publication was considered for such analyses. All the sequences from the years 1991 (18) and 1993 (16), contained CIN signature amino acids 340E and 429E, while 83% of the sequences from 1991 contained 350A. Subsequently, 350A and 429E were found among 65 to 98% of the sequences in the years 1994 (43), 1995 (19), 1999 (this study), and 2000 (32). Amino acid 340E was present in about 60% of the sequences in the years 1994, 1995, and 1999 but in only 14% of the 36 sequences from the year 2000 in the one report (32).
This is the first report describing sequences sampled from the eastern part of India (Calcutta). Our analysis of sequences from this study as well as that reported in earlier studies indicates that the viral heterogeneity among sequences sampled from Calcutta appears to be representative of the entire pool of viruses reported from India. The robustness of our findings stem from analyses of 192 sequences from 23 countries, while the presence of similar monophyletic structure for sequences from India was previously reported from analysis of full genomes from Botswana (n = 23) and India (n = 5) (31). We have also observed a similar monophyletic lineage with more than 90% bootstrap support for full-length gp160 sequences from different parts of India, but the numbers of available full-length gp160 sequences are very small (data not shown). The results and analyses presented in this study are consistent with a strong founder effect for HIV-1 infections in the Indian subcontinent (15, 18). Our results suggest a lack of new introductions into India or, at a minimum, a lack of substantial spread of newly introduced subtype C variants in the populations examined to date. This finding is relevant to strain choice in the development of a targeted HIV-1 vaccine for India.
Signature amino acid sites identified in this study may have evolutionary, structural, and viral phenotypic significance. For instance, of the nine sites differentially conserved in CIN lineage sequences (Table 2), four were proposed (46) to be positively selected, while amino acid site 429 was suggested to be negatively selected. Position 429 is also involved in making contact with CD4, while position 440 has been shown to make contact with CCR5 (24, 37). Yamaguchi-Kabata and Gojobori (46) suggested that since the main chain at position 429 interacts with CD4, the side chain residue may change without altering its binding with CD4. Although position 440 is in the C4 region, Carrillo and Ratner (10) have shown that changes at this site are necessary for X4 viruses to infect T cells. As illustrated in Table 2, amino acids at 340 and 429 that are unique to the CIN lineage within subtype C also appear to be conserved in some non-C HIV-1 subtypes. These findings imply that in addition to being the potential sequelae of a founder effect, the signature amino acid substitutions in CIN lineage may also be bound by structure-function constraints.
More HIV-1-infected individuals are infected with subtype C viruses than with any other subtype. These infections are predominantly found in the underdeveloped parts of the world, including India, sub-Saharan Africa, Brazil, and China. India is expected to have the greatest number of HIV-1-infected individuals in the near future (8). Since no medical preventative or therapeutic options are currently available in India, it is necessary to characterize the molecular epidemiologic features of virus that are circulating in India and to use this information in the development of vaccines appropriate for the Indian subcontinent. This study presents a first step in this direction by identifying molecular features unique to subtype C viruses in India. Such an approach may have applications in other epidemics: for example, a genetic cluster has been reported for HIV-1 subtype C sequences circulating in Ethiopia (1).
The epidemiologic importance of subtype A HIV-1 infections in India needs to be defined in more detail. Cassol et al. (11) reported subtype A viruses in Indian HIV-1 sequences isolated as early as 1992 in 2 of 27 individuals. Maitra et al. (29) reported two subtype A infections among 13 individuals, and we found one subtype A Env infection among 21 individuals in Calcutta. It remains to be seen whether subtype A virus sequences in India exhibit a founder effect, but there is no evidence that the frequency of subtype A viruses is approaching the level of subtype C viruses in India. Nevertheless, the role of subtype A viruses could become important in view of the documented spread of recombinant progeny between subtype A and C viruses (27).
Nucleotide sequence accession number.
Sequences obtained in this study have been deposited in GenBank under accession numbers AF392555 to AF392614.
Acknowledgments
We thank Surya Ghosh for clinical assistance and Judy Malenka for secretarial assistance as well as the participants of the study at the Calcutta School of Tropical Medicine, India.
This work was supported by AIDS-FIRCA grant R03 TH00971, a Center for AIDS Research grant to the University of Washington (AI27757), and the Boeing Foundation.
REFERENCES
- 1.Abebe A, Pollakis G, Fontanet A L, Fisseha B, Tegbaru B, Kliphuis A, Tesfaye G, Negassa H, Cornelissen M, Goudsmit J, Renke de Wit T F. Identification of a genetic subcluster of HIV type 1 subtype C (C′) widespread in Ethiopia. AIDS Res Hum Retrovir. 2000;16:1909–1914. doi: 10.1089/08892220050195865. [DOI] [PubMed] [Google Scholar]
- 2.Akaike H. A new look at statistical model identification. IEEE Trans Automatic Control. 1974;19:716–723. [Google Scholar]
- 3.Baskar P V, Ray S C, Rao R, Quinn T C, Hildreth J E, Bollinger R C. Presence in India of HIV type 1 similar to North American strains. AIDS Res Hum Retrovir. 1994;10:1039–1041. doi: 10.1089/aid.1994.10.1039. [DOI] [PubMed] [Google Scholar]
- 4.Beerli P, Grassly N C, Kuhner M K, Nickle D, Pybus O, Rain M, Rambaut A, Rodrigo A G, Wang Y. Population genetics of HIV: parameter estimation using genealogy-based methods. In: Rodrigo A G, Learn G H, editors. Computational and evolutionary analyses of HIV sequences. Boston, Mass: Kluwer Academic Publishers; 2001. pp. 217–258. [Google Scholar]
- 5.Berger E, Doms R, Fenyo E, Korber B, Littman D, Moore J, Sattentau Q, Schuitemaker H, Sodroski J, Weiss R. A new classification for HIV-1. Nature. 1998;391:240. doi: 10.1038/34571. [DOI] [PubMed] [Google Scholar]
- 6.Berger E A. HIV entry and tropism: the chemokine receptor connection. AIDS. 1997;11(Suppl. A):S3–S16. [PubMed] [Google Scholar]
- 7.Bjorndal A, Deng H, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyo E M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bollinger R C, Tripathy S P, Quinn T C. The human immunodeficiency virus epidemic in India. Current magnitude and future projections. Medicine (Baltimore) 1995;74:97–106. doi: 10.1097/00005792-199503000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Brookmeyer R, Quinn T, Shepherd M, Mehendale S, Rodrigues J, Bollinger R. The AIDS epidemic in India: a new method for estimating current human immunodeficiency virus (HIV) incidence rates. Am J Epidemiol. 1995;142:709–713. doi: 10.1093/oxfordjournals.aje.a117700. [DOI] [PubMed] [Google Scholar]
- 10.Carrillo A, Ratner L. Human immunodeficiency virus type 1 tropism for T-lymphoid cell lines: role of the V3 loop and C4 envelope determinants. J Virol. 1996;70:1301–1309. doi: 10.1128/jvi.70.2.1301-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cassol S, Weniger B G, Babu P G, Salminen M O, Zheng X, Htoon M T, Delaney A, O'Shaughnessy M, Ou C Y. Detection of HIV type 1 env subtypes A, B, C, and E in Asia using dried blood spots: a new surveillance tool for molecular epidemiology. AIDS Res Hum Retrovir. 1996;12:1435–1441. doi: 10.1089/aid.1996.12.1435. [DOI] [PubMed] [Google Scholar]
- 12.Cecilia D, Kulkarni S S, Tripathy S P, Gangakhedkar R R, Paranjape R S, Gadkari D A. Absence of coreceptor switch with disease progression in human immunodeficiency virus infections in India. Virology. 2000;271:253–258. doi: 10.1006/viro.2000.0297. [DOI] [PubMed] [Google Scholar]
- 13.de Jong J J, de Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the HIV-1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J Virol. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Jong J J, Goudsmit J, Keulen W, Klaver B, Krone W, Tersmette M, de Ronde A. Human immunodeficiency virus type 1 clones chimeric for the envelope V3 domain differ in syncytium formation and replication capacity. J Virol. 1992;66:757–765. doi: 10.1128/jvi.66.2.757-765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delwart E L, Shpaer E G, McCutchan F E, Louwagie J, Grez M, Rübsamen-Waigmann H, Mullins J I. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science. 1993;262:1257–1261. doi: 10.1126/science.8235655. [DOI] [PubMed] [Google Scholar]
- 16.Dietrich U, Grez M, von Briesen H, Panhans B, Geissendorfer M, Kuhnel H, Maniar J, Mahambre G, Becker W B, Becker M L, et al. HIV-1 strains from India are highly divergent from prototypic African and US/European strains, but are linked to a South African isolate. AIDS. 1993;7:23–27. doi: 10.1097/00002030-199301000-00003. [DOI] [PubMed] [Google Scholar]
- 17.Fouchier R A M, Groenink M, Kootstra N A, Tersmette M, Huisman H G, Miedema F, Schuitemaker H. Phenotype-associated sequence variation in the third variable domain (V3) of the human immunodeficiency virus type 1 gp120 molecule. J Virol. 1992;66:3183–3187. doi: 10.1128/jvi.66.5.3183-3187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grez M, Dietrich U, Balfe P, von Briesen H, Maniar J K, Mahambre G, Delwart E L, Mullins J I, Rübsamen-Waigmann H. Genetic analysis of human immunodeficiency virus type 1 and 2 (HIV-1 and HIV-2) mixed infections in India reveals a recent spread of HIV-1 and HIV-2 from a single ancestor for each of these viruses. J Virol. 1994;68:2161–2168. doi: 10.1128/jvi.68.4.2161-2168.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jameel S, Zafrullah M, Ahmad M, Kapoor G S, Sehgal S. A genetic analysis of HIV-1 from Punjab, India reveals the presence of multiple variants. AIDS. 1995;9:685–690. doi: 10.1097/00002030-199507000-00003. [DOI] [PubMed] [Google Scholar]
- 20.Korber B. HIV sequence signatures and similarities. In: Rodrigo A G, Learn G H, editors. Computational and evolutionary analyses of HIV sequences. Boston, Mass: Kluwer Academic Publishers; 2000. pp. 55–72. [Google Scholar]
- 21.Korber B, Kuiken C, Foley B, Hahn B, McCutchan F, Mellors J W, Sodroski J, editors. Human retroviruses and AIDS 1998. A compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Theoretical Biology and Biophysics, Los Alamos National Laboratory; 1998. [Google Scholar]
- 22.Korber B, Myers G. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res Hum Retrovir. 1992;8:1549–1560. doi: 10.1089/aid.1992.8.1549. [DOI] [PubMed] [Google Scholar]
- 23.Korber B T M, Learn G, Mullins J I, Hahn B H, Wolinsky S. Protecting HIV sequence databases. Nature. 1995;378:242–243. doi: 10.1038/378242a0. [DOI] [PubMed] [Google Scholar]
- 24.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Learn G H, Korber B T M, Foley B, Hahn B H, Wolinsky S M, Mullins J I. Maintaining the integrity of HIV sequence databases. J Virol. 1996;70:5720–5730. doi: 10.1128/jvi.70.8.5720-5730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu S L, Schacker T, Musey L, Shriner D, McElrath M J, Corey L, Mullins J I. Divergent patterns of progression to AIDS after infection from the same source: human immunodeficiency virus type 1 evolution and antiviral responses. J Virol. 1997;71:4284–4295. doi: 10.1128/jvi.71.6.4284-4295.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lole K S, Bollinger R C, Paranjape R S, Gadkari D, Kulkarni S S, Novak N G, Ingersoll R, Sheppard H W, Ray S C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maddison W P, Maddison D R. MacClade—analysis of phylogeny and character evolution, version 3. Sunderland, Mass: Sinauer Associates, Inc.; 1992. [Google Scholar]
- 29.Maitra A, Singh B, Banu S, Deshpande A, Robbins K, Kalish M L, Broor S, Seth P. Subtypes of HIV type 1 circulating in India: partial envelope sequences. AIDS Res Hum Retrovir. 1999;15:941–944. doi: 10.1089/088922299310656. [DOI] [PubMed] [Google Scholar]
- 30.Mehendale S M, Rodrigues J J, Brookmeyer R S, Gangakhedkar R R, Divekar A D, Gokhale M R, Risbud A R, Paranjape R S, Shepherd M E, Rompalo A E, et al. Incidence and predictors of human immunodeficiency virus type 1 seroconversion in patients attending sexually transmitted disease clinics in India. J Infect Dis. 1995;172:1486–1491. doi: 10.1093/infdis/172.6.1486. [DOI] [PubMed] [Google Scholar]
- 31.Novitsky V A, Montano M A, McLane M F, Renjifo B, Vannberg F, Foley B T, Ndung'u T P, Rahman M, Makhema M J, Marlink R, Essex M. Molecular cloning and phylogenetic analysis of human immunodeficiency virus type 1 subtype C: a set of 23 full-length clones from Botswana. J Virol. 1999;73:4427–4432. doi: 10.1128/jvi.73.5.4427-4432.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oelrichs R B, Shrestha I L, Anderson D A, Deacon N J. The explosive human immunodeficiency virus type 1 epidemic among injecting drug users of Kathmandu, Nepal, is caused by a subtype C virus of restricted genetic diversity. J Virol. 2000;74:1149–1157. doi: 10.1128/jvi.74.3.1149-1157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Panda S, Wang G, Sarkar S, Perez C M, Chakraborty S, Agarwal A, Dorman K, Sarkar K, Detels R, Kaplan A H. Characterization of V3 loop of HIV type 1 spreading rapidly among injection drug users of Manipur, India: a molecular epidemiological perspective. AIDS Res Hum Retrovir. 1996;12:1571–1573. doi: 10.1089/aid.1996.12.1571. [DOI] [PubMed] [Google Scholar]
- 34.Peeters M, Vincent R, Perret J L, Lasky M, Patrel D, Liegeois F, Courgnaud V, Seng R, Matton T, Molinier S, Delaporte E. Evidence for differences in MT2 cell tropism according to genetic subtypes of HIV-1: syncytium-inducing variants seem rare among subtype C HIV-1 viruses. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20:115–121. doi: 10.1097/00042560-199902010-00002. [DOI] [PubMed] [Google Scholar]
- 35.Posada D, Crandall K A. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 36.Poss M, Rodrigo A G, Gosink J J, Learn G H, de Vange Panteleeff D, Martin H L, Jr, Bwayo J, Kreiss J K, Overbaugh J. Evolution of envelope sequences from the genital tract and peripheral blood of women infected with clade A human immunodeficiency virus type 1. J Virol. 1998;72:8240–8251. doi: 10.1128/jvi.72.10.8240-8251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 38.Shankarappa R, Margolick J B, Gange S J, Rodrigo A G, Upchurch D, Farzadegan H, Gupta P, Rinaldo C R, Learn G H, He X, Huang X L, Mullins J I. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith S W, Overbeek R, Woese C R, Gilbert W, Gillevet P M. The Genetic Data Environment: An expandable GUI for multiple sequence analysis. Comput Appl Biol Sci. 1994;10:671–675. doi: 10.1093/bioinformatics/10.6.671. [DOI] [PubMed] [Google Scholar]
- 40.Swofford D L. PAUP 4.0: phylogenetic analysis using parsimony (and other methods), 4.0b2a. Sunderland, Mass: Sinauer Associates, Inc.; 1999. [Google Scholar]
- 41.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tien P C, Chiu T, Latif A, Ray S, Batra M, Contag C H, Zejena L, Mbizvo M, Delwart E L, Mullins J I, Katzenstein D A. Primary subtype C HIV-1 infection in Harare, Zimbabwe. J Acquir Immune Defic Syndr Hum Retrovirol. 1999;20:147–153. doi: 10.1097/00042560-199902010-00006. [DOI] [PubMed] [Google Scholar]
- 43.Tripathy S, Renjifo B, Wang W K, McLane M F, Bollinger R, Rodrigues J, Osterman J, Essex M. Envelope glycoprotein 120 sequences of primary HIV type 1 isolates from Pune and New Delhi, India. AIDS Res Hum Retrovir. 1996;12:1199–1202. doi: 10.1089/aid.1996.12.1199. [DOI] [PubMed] [Google Scholar]
- 44.Tsuchie H, Saraswathy T S, Sinniah M, Vijayamalar B, Maniar J K, Monzon O T, Santana R T, Paladin F J, Wasi C, Thongcharoen P, et al. HIV-1 variants in South and South-East Asia. Int J STD AIDS. 1995;6:117–120. doi: 10.1177/095646249500600211. [DOI] [PubMed] [Google Scholar]
- 45.O W H UNAIDS. The global epidemic. UNAIDS report. Geneva, Switzerland: UNAIDS; 1998. [Google Scholar]
- 46.Yamaguchi-Kabata Y, Gojobori T. Reevaluation of amino acid variability of the human immunodeficiency virus type 1 gp120 envelope glycoprotein and prediction of new discontinuous epitopes J. Virol. 2000;74:4335–4350. doi: 10.1128/jvi.74.9.4335-4350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]