Abstract
Coexpression of bovine papillomavirus L1 with L2 mutants lacking either eight N-terminal or nine C-terminal amino acids that encode positively charged domains resulted in wild-type levels of viral genome encapsidation. Despite wild-type binding to the cell surface, the resulting virions were noninfectious. An L2 mutant encoding a scrambled version of the nine C-terminal residues restored infectivity, in contrast to an L2 mutant encoding a scrambled version of the N-terminal residues.
The papillomavirus capsid comprises two genetically unrelated viral proteins called L1 and L2 (2) surrounding an ∼8-kb circular DNA genome bound to histones. L1 is able to self-assemble into virus-like particles (VLPs), morphologically and immunologically very similar to virions (5), that are both necessary and sufficient for binding to the cell surface (14). When coexpressed with L1, L2 is incorporated into VLPs in a similar ratio to virions purified from warts (6, 26). L1 expressed in trans in mammalian cells harboring episomal DNA forms virions (19), although L2 dramatically (>50-fold) (23) increases the efficiency of DNA encapsidation (12, 16, 24). Using a series of L2 deletion mutants, we have recently identified two domains in bovine papillomavirus type 1 (BPV1) L2 that participate in the interaction with L1 (11). One maps to residues 129 to 246, near the middle of L2, while the other involves residues 384 to 460, near the C terminus of L2. Each L1 interaction domain of L2 was found to also affect the efficiency of viral genome encapsidation.
The positively charged residues at the N and C termini of human papillomavirus type 6 (HPV6) L2 function as redundant nuclear localization signals (17); furthermore, the residues at the N terminus of HPV16 L2, but not those at the C terminus, bind to DNA in a sequence-independent manner in vitro (25). BPV1 L2 is 469 amino acids long, with groups of positively charged residues at the extreme N terminus and the extreme C terminus. With the goal of directly assessing the role of the positively charged termini in the generation of infectious virions, we constructed a BPV1 L2 with a deletion of either amino acids 2 to 9 (L2Δ2–9) or the final nine amino acids (L2Δ461–469). We also constructed a mutant with a deletion of internal L2 residues 422 to 431 (L2Δ422–431) to serve as a control. These deletion mutants were cloned into vector pSFV-1, from which recombinant defective Semliki Forest viruses (SFVs) encoding these deletion mutants were generated (12). The activities of the mutants were then tested in several L2-dependent assays, including one for infectivity.
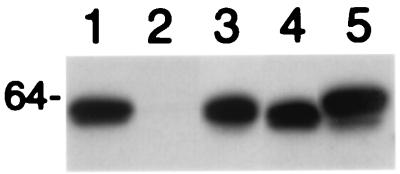
To examine L1-L2 interactions, BHK21 cells were coinfected with recombinant SFVs that express L1 and each mutant L2 (12). Cell lysates were prepared 24 h postinfection, and sequential immunoprecipitations were performed initially with preimmune rabbit serum, then rabbit anti-BPV1 L1 VLP (5), and finally rabbit anti-BPV1 L2–6His (15). The presence of L2 in immunoprecipitates was detected by Western blotting with monoclonal antibody C6 (7) (Fig. 1). When expressed alone, L2 was not immunoprecipitated by rabbit anti-BPV1 L1 VLP (Fig. 1, lane 2). L2 was absent from preimmune rabbit serum immunoprecipitates, thus demonstrating specificity (not shown). The three L2 deletion mutants were immunoprecipitated with anti-L1 as for wild-type L2 (Fig. 1), demonstrating equivalent association with L1 for L2, L2Δ2–9, L2Δ461–469 and L2Δ422–431. Furthermore, equivalent quantities of L2 were detected upon subsequent immunoprecipitation with antiserum to six-histidine-tagged full-length L2 (L2–6His), confirming equivalent expression in each sample (not shown).
FIG. 1.
Coimmunoprecipitation of BPV1 L2 deletion mutants with L1. BHK21 cells were coinfected with recombinant SFV expressing BPV1 L1 and L2 deletion mutants, and 24 h postinfection, the cells were harvested and lysed. Immunopreciptation was performed with preimmune serum (not shown) and then rabbit anti-BPV1 L1 VLP (lanes 1 to 5). L2 was detected by Western blotting with monoclonal antibody C6 in anti-L1 VLP immunoprecipitates from lysates containing L1 (lanes 1 and 3 to 5) and full-length L2 (lanes 1 and 2), L2Δ2–9 (lane 3), L2Δ422–431 (lane 4), and L2Δ461–469 (lane 5).
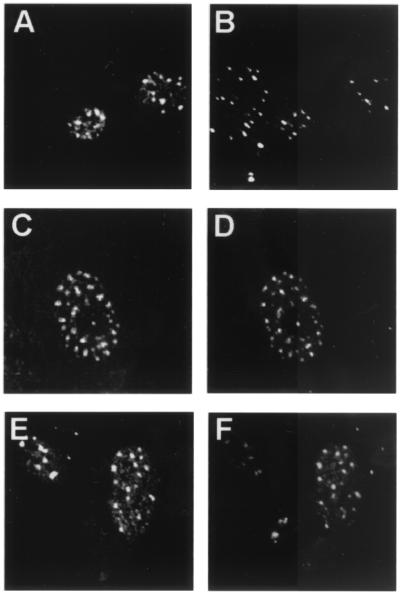
Before examining their infectivity, we sought to verify that the mutants behaved similarly to the wild type in several biochemical assays. We previously demonstrated that BPV1 L2 traffics to the subnuclear domains termed promonocytic leukemia protein (PML)-oncogenic domains (PODs), also referred to as ND10 (1). L2 can also induce the POD localization of BPV1 L1 and E2, two proteins that exhibit a diffuse nuclear pattern in the absence of L2. To analyze their subcellular localization, the BPV1 L2 N-terminal and C-terminal deletion mutants were expressed in BPHE-1 cells by infection with recombinant SFV. The BPHE-1 line was clonally derived from BPV1-infected primary hamster embryo cells; it contains 50 to 200 BPV1 episomes per cell, but fails to express detectable levels of the capsid proteins (1, 12, 22). The BPV1 L2 deletion mutant proteins were immunohistochemically stained with rabbit antiserum to 6His-L2 and visualized by confocal fluorescence microscopy, as described previously (1). L2Δ2–9 and L2Δ461–469 accumulated in the nucleus and associated with PODs (as assessed by colocalization with anti-PML monoclonal antibody 5E10 staining) as for wild-type L2 (Fig. 2). Colocalization of deletion mutant and wild-type L2 with anti-E2 monoclonal antibody B202 or with anti-L1 monoclonal antibody 5B6 staining was also assessed. The L2 deletion mutants were found to retain the ability to localize E2 and L1 to PODs (data not shown) as described for wild-type L2 (1).
FIG. 2.
Colocalization of BPV1 L2 deletion mutants with PML. BPHE-1 cells were infected with the L2-SFV recombinants. The cells were fixed, and double-staining immunolocalization against the L2 proteins and PML was performed. Infection of cells with full-length L2 is shown in panels A and B, infection with L2Δ2–9 is shown in panels C and D, and infection with L2Δ461–469 is shown in panels E and F. The same field is shown in each pair of panels. Anti-L2 staining is shown in panels A, C, and E, and anti-PML staining is shown in panels B, D, and F.
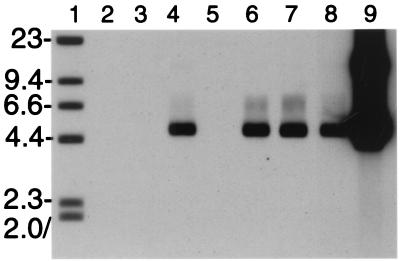
The capacity of the three L2 deletion mutants to encapsidate the viral genome was determined. BPHE-1 cells were coinfected with recombinant SFV expressing BPV1 L1 and mutant L2. Thirty hours postinfection, the cells were harvested and lysed. The capsid proteins were immunoprecipitated with rabbit anti-BPV1 VLP, and the immunoprecipitates were treated with DNase I to eliminate nonencapsulated genomes. Crude extract of bovine wart was used as a positive control. The amount of DNase I-resistant BPV1 genome present in the immunoprecipitates was assessed by Southern blotting (Fig. 3). Consistent with similar papillomavirus assembly systems (16, 23), DNase I-resistant BPV genome, migrating as supercoiled DNA, was readily recovered from immunoprecipitates of BPHE-1 cells expressing BPV1 L1 together with BPV1 L2, although not with HPV16 L2 (Fig. 3). Wild-type levels of genome encapsidation were observed when BPV1 L1 was coexpressed with each of the three deletions of L2. As negative controls, no DNase I-resistant BPV genome was observed in immunoprecipitates from BPHE-1 cells expressing either L1 or L2 alone (Fig. 3, lanes 2 and 3), although on extended exposure, a weak band could be seen for L1 only (at least 50-fold lower than for L1 and L2 together), but not L2 (data not shown).
FIG. 3.
Encapsidation of the BPV1 genome by L1 and L2 deletion mutants in BPHE-1 cells. BPHE-1 cells, which harbor 50 to 200 BPV1 episomes per cell, were coinfected with recombinant SFV expressing BPV1 L1 and L2 deletion mutants, and 30 h postinfection, the cells were harvested and lysed. By using rabbit anti-BPV1 L1 VLP, L1 was immunoprecipitated from lysates containing L1 (lanes 2 and 4 to 8) and full-length L2 (lanes 3 and 4), full-length HPV16 L2 (lane 5), L2Δ2–9 (lane 6), L2Δ461–469 (lane 7), and L2Δ422–431 (lane 8) or from crude extract of bovine wart (lane 9). The immunoprecipitates were treated with DNase I for 1 h and washed. Undigested DNA present in the immunoprecipitates was recovered and separated on a 0.8% agarose gel with a biotinylated HindIII digest of λ (lane 1). The presence of viral DNA was detected by Southern blotting with a biotinylated probe corresponding to the EcoRI-BamHI large fragment of the BPV1 genome.
Having found that the mutants behaved as did the wild type with regard to localization to PODs, recruitment of L1 and E2, and viral genome encapsidation, the ability of each L2 deletion mutant to produce infectious BPV was also determined. Each L2 mutant was coexpressed with wild-type BPV1 L1 in BPHE-1 cells with recombinant SFV (12), the BPHE-1 cells were harvested and lysed 30 h postinfection, and the resulting BPV1 infectivity in the extracts was assayed by using in vitro focal transformation of C127C mouse fibroblasts (3). The focus-forming activity resulting from BPHE-1 extracts from cells coexpressing wild-type L1 with the L2Δ422–431 mutant was similar to that obtained from extracts that coexpressed L1 with wild-type L2 (Table 1). This result indicates that infectivity can be retained despite a 10-amino-acid deletion in at least one region of L2. In contrast to L2Δ422–431, neither L2Δ2–9 nor L2Δ461–469 generated infectious BPV1 when coexpressed with L1 in BPHE-1 cells (Table 1). These results indicate that coexpression of L1 with L2Δ2–9 or L2Δ461–469 in BPHE-1 cells has generated defective virions, thus implying a new role for L2 in viral infection.
TABLE 1.
Generation of infectious BPV1 by coexpression of BPV1 L1 and L2 deletion mutants in BPHE-1 cellsa
| Capsid protein(s) overexpressed in BPHE-1 cells | Mean no. of foci | SD |
|---|---|---|
| L1 | 0 | 0 |
| L2 | 0 | 0 |
| L1 + L2 | 81 | 17.6 |
| L1 + L2Δ2–9 | 0 | 0 |
| L1 + L2Δ461–469 | 0 | 0 |
| L1 + L2Δ422–431 | 86.3 | 15.3 |
| L1 + L2mix2–9 | 0.3 | 0.6 |
| L1 + L2mix461–469 | 80.3 | 10.5 |
BPHE-1 cells were coinfected with recombinant SFV expressing BPV1 L1 or L2 alone, or BPV1 L1 was coexpressed with L2, L2Δ2–9, L2Δ461–469, L2mix2–9, L2mix461–469, or L2Δ422–431, and 30 h postinfection, the cells were harvested and lysed in 1 ml of phosphate-buffered saline by sonication. Monolayers of C127C cells were infected by addition of lysates for 1 h at 37°C, washed, maintained in culture for 3 weeks, and stained. The mean number and standard deviation (SD) of foci per plate were calculated from three independent experiments.
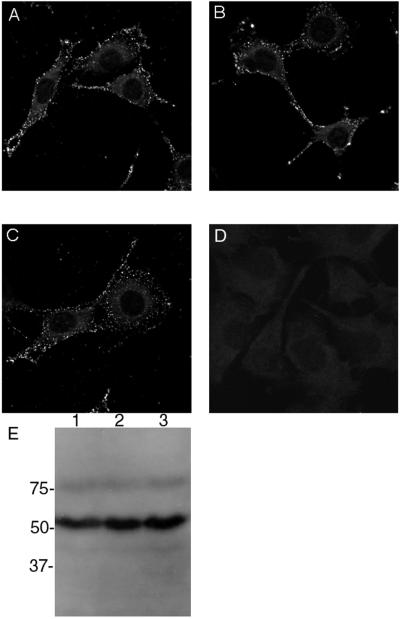
One possible explanation for the lack of infectivity by L2Δ2–9 or L2Δ461–469 could be that the presence of mutant L2 in virions might have interfered with the efficiency of virion binding to cells. This possibility has become more plausible, following the recent demonstration that L2 alone is able to bind to cell surfaces in a sequence-dependent manner (4), although other studies have suggested that L2 may function in a postbinding step (9, 13–15, 20). To examine whether L2Δ2–9 or L2Δ461–469 in virions might interfere with binding to cell surfaces, L1 was coexpressed with either L2, L2Δ2–9, or L2Δ461–469 in BPHE-1 cells. The resultant virions were separated from pentamers and empty VLPs by rate zonal centrifugation through a 20 to 40% (wt/vol) sucrose gradient for 90 min at 160,000 x g at 4°C. Equivalent quantities of virions or buffer alone were incubated with mouse C127C cells for 1 h at 4°C. The cells were washed, and bound virions were detected by indirect immunofluorescence with monoclonal antibody 5B6 to L1. Levels of binding of L1/L2, L1/L2Δ2–9, and L1/L2Δ461–469 virions to C127C cells were equivalent (Fig. 4A, B, and C, respectively), each forming a speckled pattern on the cell surface that was absent from control cells (Fig. 4D). The quality and quantity of the L1/L2, L1/L2Δ2–9, and L1/L2Δ461–469 virion preparations were very similar when compared by Western blot analysis with rabbit antiserum to BPV1 L1/L2 VLPs (Fig. 4E). Therefore, incorporation of the L2 deletion mutants into virions did not prevent them from binding to cell surfaces, implying that the deletion mutants did not induce misfolding of the virions and that the L2 termini function during infection after virion binding to cell surfaces.
FIG. 4.
Binding of BPV1 virions containing L2 deletion mutants to the cell surface. L1 was coexpressed with recombinant SFV with either wild-type L2, L2Δ2–9, or L2Δ461–49 in BPHE-1 cells. Upon cell lysis by sonication, virions were separated from pentamers and empty VLPs by rate zonal centrifugation through a 20 to 40% (wt/vol) sucrose gradient for 90 min at 160,000 × g at 4°C. L1/L2 (A and E, lane 1), L1/L2Δ2–9 (B and E, lane 2), and L1/L2Δ461–49 (C and E, lane 3) virion preparations and buffer alone (D) were incubated at equivalent concentrations for 1 h at 4°C with mouse C127C cells. The cells were washed, and bound virions were detected by indirect immunofluorescence with monoclonal antibody 5B6 to L1. The quality and quantity of virion preparations was estimated by Western blot analysis with rabbit antiserum to BPV1 L1/L2 VLPs (E). In panel E, molecular masses are given to the left in kilodaltons.
It was also possible that the absence of charge resulting from deletion of the amino acids at the N or C terminus might account for the loss of infectivity. To examine this possibility, two additional mutants were generated. One encoded the N-terminal amino acids in scrambled form (L2mix2–9, which replaces MSARKRVKR with MRVKSKRAR). The other encoded the 9 C-terminal amino acids in scrambled form (L2mix461–469, which replaces RKRKKRKHA with KRKHRARKK). When L2mix461–469 was coexpressed with L1 in BPHE-1, the resulting extract was found to be as infectious as wild-type L2 (Table 1). Thus, compared with the noninfectious L2Δ461–469 mutant, restoration of the positive charge at the L2 C terminus, even in scrambled form, was able to restore infectivity. Since L2Δ422–431 retains the ability to form infectious virions, the length of L2 does not appear to be critical, but further testing of neutral or acid substitutions in residues 461 to 469 is warranted to confirm the significance of the positive charge in infectivity. In contrast, the L2mix2–9 mutant, when coexpressed with L1 in BPHE-1, did not produce infectious virus. This negative result suggests that the extreme N terminus makes a sequence-specific contribution to infectivity.
In addition to its relationship to virion assembly (11, 12, 24), L2 in virions has been shown to participate in pseudoviral infectivity in systems in which pseudovirions are formed in vitro with either L1 alone or the combination of L1 and L2, with naked reporter DNA that is either encapsidated or bound to the capsid surface (4, 9, 18, 21). L2 is not absolutely necessary for infectivity in these systems, but its presence enhances the efficiency of infection. The presence of L2 enhances the infectivity of HPV33 pseudovirion generated in COS-7 cells by ∼10-fold (19). When an L2 protein in which residues 108 to 111 have been mutated is substituted for wild-type L2 in HPV16 L1/L2 pseudovirions, their infectivity approximates that of pseudovirions with only L1 (4). This suggests that the effect of L2 is mediated by sequences near the middle of L2 that are exposed at the capsid surface (4).
We have in the current report identified a function for L2 in infectivity that also appears to be independent of the ability of L2 to participate in virion assembly. The infectivity function was revealed by using two L2 deletion mutants, one lacking amino acids 2 through 9 (L2Δ2–9) and the other lacking the final nine C-terminal amino acids (L2Δ461–469). Both mutants behaved as the wild type for each assay related to virion assembly, including the genome encapsidation assay, but both mutants were deficient for infectivity. Since both mutant proteins retain the exposed L2 amino acids identified by Kawana et al. (4) as required for the L2-dependent infectivity of the pseudovirions and both mutant proteins appear to be incorporated into virions with the same efficiency as wild-type L2, it is likely that the L2 infectivity function described here is distinct.
Since both termini of L2 contain several positively charged amino acids, we assessed the possible role of the charge, rather than sequence, by reconstituting the charge at each terminus of L2 with a scrambled version of the authentic peptide. Infectivity was restored with the scrambled C-terminal mutant L2mix461–469, while the scrambled N-terminal mutant, L2mix2–9, remained deficient for infectivity. Thus, at the C terminus, where 8 of the 9 amino acids are charged, a positive charge seems to be the main determinant of infectivity. In contrast, sequence specificity at the N terminus, where only 5 of the 8 amino acids are charged, appears to be critical for infectivity.
We speculate that the function we have identified for L2 operates after the initial binding of virions to the cell surface. This inference is consistent with earlier studies, suggesting that L1 is both necessary and sufficient for virion binding to cell surfaces (9, 13, 14, 20) and that L2-specific antibodies can neutralize infection without inhibiting such binding (14, 15). Since L2 alone can bind and enter cells (4), L2 may bind a secondary receptor. L2 also interacts in vitro through positively charged side chains with DNA independently of nucleotide sequence (8, 25), and L2 is associated with episomes in vivo (16). We therefore speculate that L2 may function in the uncoating of virions and delivery of the viral genome to the nucleus, via a combination of the putative L2 receptor and the positively charged L2 termini bound to the viral genome (4, 25). During infection by the papovavirus simian virus 40, cytoplasmic or nuclear microinjection of antibody to the minor capsid protein VP3 can prevent infection (10), suggesting that this minor capsid protein targeted to the nucleus could play a role in infection similar to that postulated for L2.
Acknowledgments
We are most grateful to Elliot Androphy and the late Jian Zhou for providing monoclonal antibodies B202 and C6. We also thank Carl Olson for the bovine papillomavirus. We are grateful to Jon Yewdell, Jack Bennink, and the Laboratory of Viral Diseases at the National Institutes of Health (NIH) for the use of their confocal microscope.
This work was supported by NIH intramural funding and grant 1 PO1 AI48203-01 to R.B.S.R., the Richard TeLinde endowment, and the Cancer Research Institute (R.B.S.R.).
REFERENCES
- 1.Day P M, Roden R B S, Lowy D R, Schiller J T. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J Virol. 1998;72:142–150. doi: 10.1128/jvi.72.1.142-150.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doorbar J, Gallimore P H. Identification of proteins encoded by the L1 and L2 open reading frames of human papillomavirus 1a. J Virol. 1987;61:2793–2799. doi: 10.1128/jvi.61.9.2793-2799.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dvoretzky I, Shober R, Chattopadhyay S K, Lowy D R. A quantitative in vitro focus assay for bovine papilloma virus. Virology. 1980;103:369–375. doi: 10.1016/0042-6822(80)90195-6. [DOI] [PubMed] [Google Scholar]
- 4.Kawana Y, Kawana K, Yoshikawa H, Taketani Y, Yoshiike K, Kanda T. Human papillomavirus type 16 minor capsid protein L2 N-terminal region containing a common neutralization epitope binds to the cell surface and enters the cytoplasm. J Virol. 2001;75:2331–2336. doi: 10.1128/JVI.75.5.2331-2336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kirnbauer R, Booy F, Cheng N, Lowy D R, Schiller J T. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci USA. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirnbauer R, Taub J, Greenstone H, Roden R, Dürst M, Gissmann L, Lowy D R, Schiller J T. Efficient self-assembly of human papillomavirus type 16 L1 and L1–L2 into virus-like particles. J Virol. 1993;67:6929–6936. doi: 10.1128/jvi.67.12.6929-6936.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu W J, Gissmann L, Sun X Y, Kanjanahaluethai A, Muller M, Doorbar J, Zhou J. Sequence close to the N-terminus of L2 protein is displayed on the surface of bovine papillomavirus type 1 virions. Virology. 1997;227:474–483. doi: 10.1006/viro.1996.8348. [DOI] [PubMed] [Google Scholar]
- 8.Mallon R G, Wojciechowicz D, Defendi V. DNA-binding activity of papillomavirus proteins. J Virol. 1987;61:1655–1660. doi: 10.1128/jvi.61.5.1655-1660.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller M, Gissmann L, Cristiano R J, Sun X-Y, Frazer I H, Jenson A B, Alonso A, Zentgraf H, Zhou J. Papillomavirus capsid binding and uptake by cells from different tissues and species. J Virol. 1995;69:948–954. doi: 10.1128/jvi.69.2.948-954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakanishi A, Clever J, Yamada M, Li P P, Kasamatsu H. Association with capsid proteins promotes nuclear targeting of simian virus 40 DNA. Proc Natl Acad Sci USA. 1996;93:96–100. doi: 10.1073/pnas.93.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okun M M, Day P M, Greenstone H L, Booy F P, Lowy D R, Schiller J T, Roden R B S. L1 interaction domains of papillomavirus L2 necessary for viral genome encapsidation. J Virol. 2001;75:4332–4342. doi: 10.1128/JVI.75.9.4332-4342.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roden R B S, Greenstone H L, Kirnbauer R, Booy F P, Jessie J, Lowy D R, Schiller J T. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J Virol. 1996;70:5875–5883. doi: 10.1128/jvi.70.9.5875-5883.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roden R B S, Hubbert N L, Kirnbauer R, Breitburd F, Lowy D R, Schiller J T. Papillomavirus L1 capsids agglutinate mouse erythrocytes through a proteinaceous receptor. J Virol. 1995;69:5147–5151. doi: 10.1128/jvi.69.8.5147-5151.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roden R B S, Kirnbauer R, Jenson A B, Lowy D R, Schiller J T. Interaction of papillomaviruses with the cell surface. J Virol. 1994;68:7260–7266. doi: 10.1128/jvi.68.11.7260-7266.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roden R B S, Weissinger E M, Henderson D W, Booy F, Kirnbauer R, Mushinski J F, Lowy D R, Schiller J T. Neutralization of bovine papillomavirus by antibodies to L1 and L2 capsid proteins. J Virol. 1994;68:7570–7574. doi: 10.1128/jvi.68.11.7570-7574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stauffer Y, Raj K, Masternak K, Beard P. Infectious human papillomavirus type 18 pseudovirions. J Mol Biol. 1998;283:529–536. doi: 10.1006/jmbi.1998.2113. [DOI] [PubMed] [Google Scholar]
- 17.Sun X Y, Frazer I, Muller M, Gissmann L, Zhou J. Sequences required for the nuclear targeting and accumulation of human papillomavirus type 6B L2 protein. Virology. 1995;213:321–327. doi: 10.1006/viro.1995.0005. [DOI] [PubMed] [Google Scholar]
- 18.Touze A, Coursaget P. In vitro gene transfer using human papillomavirus-like particles. Nucleic Acids Res. 1998;26:1317–1323. doi: 10.1093/nar/26.5.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Unckell F, Streeck R E, Sapp M. Generation and neutralization of pseudovirions of human papillomavirus type 33. J Virol. 1997;71:2934–2939. doi: 10.1128/jvi.71.4.2934-2939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Volpers C, Unckell F, Schirmacher P, Streeck R E, Sapp M. Binding and internalization of human papillomavirus type 33 virus-like particles by eukaryotic cells. J Virol. 1995;69:3258–3264. doi: 10.1128/jvi.69.6.3258-3264.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeager M D, Aste-Amezaga M, Brown D R, Martin M M, Shah M J, Cook J C, Christensen N D, Ackerson C, Lowe R S, Smith J F, Keller P, Jansen K U. Neutralization of human papillomavirus (HPV) pseudovirions: a novel and efficient approach to detect and characterize HPV neutralizing antibodies. Virology. 2000;278:570–577. doi: 10.1006/viro.2000.0674. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y-L, Lewis A, Jr, Wade-Glass M, Schlegel R. Levels of bovine papillomavirus RNA and protein expression correlate with variations in the tumorigenic phenotype of hamster cells. J Virol. 1987;61:2924–2928. doi: 10.1128/jvi.61.9.2924-2928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao K N, Sun X Y, Frazer I H, Zhou J. DNA packaging by L1 and L2 capsid proteins of bovine papillomavirus type 1. Virology. 1998;243:482–491. doi: 10.1006/viro.1998.9091. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J, Stenzel D J, Sun X Y, Frazer I H. Synthesis and assembly of infectious bovine papillomavirus particles in vitro. J Gen Virol. 1993;74:763–768. doi: 10.1099/0022-1317-74-4-763. [DOI] [PubMed] [Google Scholar]
- 25.Zhou J, Sun X-Y, Louis K, Frazer I H. Interaction of human papillomavirus (HPV) type 16 capsid proteins with HPV DNA requires an intact L2 N-terminal sequence. J Virol. 1994;68:619–625. doi: 10.1128/jvi.68.2.619-625.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J, Sun X Y, Stenzel D J, Frazer I H. Expression of vaccinia recombinant HPV 16 L1 and L2 ORF proteins in epithelial cells is sufficient for assembly of HPV virion-like particles. Virology. 1991;185:251–257. doi: 10.1016/0042-6822(91)90772-4. [DOI] [PubMed] [Google Scholar]