Abstract
Background
Progression of chronic lung disease may lead to the requirement for lung transplant (LTx). Despite improvements in short-term survival after LTx, chronic lung allograft dysfunction (CLAD) remains a critical challenge for long-term survival. This study investigates the molecular and microbial relationships between underlying lung disease and the development of CLAD in bronchoalveolar lavage fluid (BALF) from subjects post-LTx, which is crucial for tailoring treatment strategies specific to allograft dysfunctions.
Methods
Paired 16S rRNA gene amplicon sequencing and untargeted LC–MS/MS metabolomics were performed on 856 BALF samples collected over 10 years from LTx recipients (n = 195) with alpha-1-antitrypsin disease (AATD, n = 23), cystic fibrosis (CF, n = 47), chronic obstructive pulmonary disease (COPD, n = 78), or pulmonary fibrosis (PF, n = 47). Data were analyzed using random forest (RF) machine learning and multivariate statistics for associations with underlying disease and CLAD development.
Results
The BALF microbiome and metabolome after LTx differed significantly according to the underlying disease state (PERMANOVA, p = 0.001), with CF and AATD demonstrating distinct microbiome and metabolome profiles, respectively. Uniqueness in CF was mainly driven by Pseudomonas abundance and its metabolites, whereas AATD had elevated levels of phenylalanine and a lack of shared metabolites with the other underlying diseases. BALF microbiome and metabolome composition were also distinct between those who did or did not develop CLAD during the sample collection period (PERMANOVA, p = 0.001). An increase in the average abundance of Veillonella (AATD, COPD) and Streptococcus (CF, PF) was associated with CLAD development, and decreases in the abundance of phenylalanine-derivative alkaloids (CF, COPD) and glycerophosphorylcholines (CF, COPD, PF) were signatures of the CLAD metabolome. Although the relative abundance of Pseudomonas was not associated with CLAD, the abundance of its virulence metabolites, including siderophores, quorum-sensing quinolones, and phenazines, were elevated in those with CF who developed CLAD. There was a positive correlation between the abundance of these molecules and the abundance of Pseudomonas in the microbiome, but there was no correlation between their abundance and the time in which BALF samples were collected post-LTx.
Conclusions
The BALF microbiome and metabolome after LTx are particularly distinct in those with underlying CF and AATD. These data reflect those who developed CLAD, with increased virulence metabolite production from Pseudomonas, an aspect of CF CLAD cases. These findings shed light on disease-specific microbial and metabolic signatures in LTx recipients, offering valuable insights into the underlying causes of allograft rejection.
Video Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s40168-024-01893-y.
Keywords: Microbiome, Metabolome, Cystic fibrosis, Bronchioalveolar lavage fluids, Chronic lung allograft dysfunction, Lung diseases, Pseudomonas aeruginosa virulence factors
Introduction
Lung transplantation (LTx) is a therapeutic option for patients who develop progressive and severe chronic lung diseases [1]. However, chronic lung allograft dysfunction (CLAD), which is an overarching malady characterized by the chronic and progressive decline in the function of the lung allograft, remains a major barrier to long-term LTx patient survival. CLAD affects 50% of patients at 5 years post-transplantation, requiring extensive treatment and management [2, 3]. Bronchiolitis obliterans syndrome (BOS), the canonical phenotypic manifestation of allograft dysfunction [4, 5], is used as a scaled measure of disease progression (i.e., CLAD stage). BOS severity scores were recently updated [6] to better reflect the stages of severity (0–4) experienced by LTx recipients, including non-obstructive or mixed phenotypes. These CLAD stages can occur in all post-LTx recipients, regardless of different underlying morbidities such as alpha-1-antitrypsin deficiency disease (AATD), cystic fibrosis (CF), chronic obstructive pulmonary disease (COPD), and pulmonary fibrosis (PF) [7–10].
An important aspect of CLAD and long-term survival of patients post-LTx is lung infection. As such, the lung microbiome plays an important role in the health and disease states of LTx recipients, with compelling evidence suggesting its role in modulating immune responses, influencing the risk of allograft dysfunction, and impacting pulmonary function. Post-transplant changes in microbiome composition, coupled with acute and chronic inflammation, can affect disease outcomes and the success of lung transplantation [11–13]. While the microbiome has been investigated in CLAD for patterns with disease development, the metabolome, which constitutes the collective molecular crosstalk between microbes and immune cells, is comparatively unexplored. There is a need to better understand the microbiome and metabolome interactions that are associated with the many clinical manifestations and outcomes of CLAD. However, it remains challenging to navigate the diverse chemical data generated in untargeted metabolomics studies because (i) a large proportion of detectable metabolites are of unknown structure, and (ii) it is difficult to identify their source as either host or microbially derived [14–18]. Recent advances in bioinformatic analyses of 16S rRNA gene microbiome and mass spectrometry data have enabled a more comprehensive interpretation of biological information contained within these highly technical and large datasets [15, 17, 19].
Bronchoalveolar lavage fluid (BALF) is a valuable sample type for studying the lung microbiome and metabolome of the lung allograft due to the direct proximity of sample origin to the site of stress and/or injury in the lung [20]. Investigation of BALF allows for monitoring immunologic changes, detection of early signs of rejection/infection, identification of biomarkers of chronic rejection, and more generally understanding the mechanisms of lung injury [21, 22]. Metabolites detected within BALF can indicate physiological processes of disease progression, microbial lung burden, and possibly, allograft rejection [19, 23, 24]. There are few studies of BALF metabolomes, and those that are available describe the presence of microbial metabolites, xenobiotics, and a myriad of pharmaceuticals [19, 25–27].
Here, we applied a multi-omics-based approach to 856 BALF samples collected longitudinally from 195 LTx recipients to determine whether post-transplant BALF microbiome and metabolome reflected the underlying disease diagnosis and the development of CLAD. Our data offer valuable insights into disease progression post-LTx, pointing to new lines of exploration that could help define the mechanistic basis of allograft dysfunction.
Materials and methods
Sample collection, study subjects, and CLAD staging
Subjects with AATD, CF, COPD, and PF who underwent lung transplantation at the University of Minnesota between 2002 and 2012 consented to have a portion of their post-LTx BALF used for research. BALF was collected as a routine procedure via orotracheal or nasotracheal bronchoscopy per transplant protocol or if undergoing diagnostic BALF collection due to clinical indications, such as new radiographical changes, new respiratory symptoms, or a decrease in forced expiratory volume in one second (FEV1) [28]. Each bronchoscopy consisted of 100–1200 mL sterile saline instilled into a subsegmental location after advancement and occlusion of the airway lumen. BALF was separated into ~ 1.5 mL aliquots and frozen at − 80 °C for further analysis (see supplementary methods).
Inclusion criteria were ≥ 18 years of age; diagnosis of AATD, CF, COPD, or PF; and receipt of a bilateral or single lung transplant, while the exclusion criteria were the inability to provide consent and/or tolerate the BALF procedure. Clinical characteristics of subjects in this study, including disease progression scores and samples collected for each, are displayed in both Table 1 and Table S1. BOS-grade was recorded based on changes in FEV1, initially determined using definitions of the International Society for Heart and Lung Transplantation (ISHLT) [29], and updated here to reflect current CLAD definitions [6] (Table S1). The final CLAD stage of each subject was known and recorded at the end of the sample collection period. This allowed us to compare the longitudinal multi-omics data between those subjects who never developed any CLAD stage during the study to those who developed CLAD falling within stages 1 to 4. Notably, CLAD stage diagnosis at the time of sample collection was known for only a subset of subjects, and therefore, was not analyzed for this study. This study did not differentiate between restrictive and mixed phenotypes which would require full lung function testing including total lung capacity (plethysmography) and chest computed tomography (CT) imaging. This protocol was approved by the University of Minnesota IRB (STUDY00004547).
Table 1.
Clinical and demographic characteristics of subjects (total n = 195) and BALF samples (total n = 856) collected post-LTx grouped by underlying disease—alpha-1 antitrypsin deficiency (AATD), cystic fibrosis (CF), chronic obstructive pulmonary disease (COPD), and pulmonary fibrosis (PF). Data include details on BALF sampling, microbiology related to Pseudomonas spp., age at lung transplant (LTx), sex ratio (female/male), and the average number of BALF samples collected per subject. Data are presented for subjects who did or did not develop CLAD
| Clinical and demographic characteristics | AATD | CF | COPD | PF |
|---|---|---|---|---|
| Subjects | 23 | 47 | 78 | 47 |
| Sex ratio (F/M) | 0.64 (9/14) | 0.95 (23/24) | 1.22 (43/35) | 0.42 (14/33) |
| Mean age at LTx (range) | 55 (42–55) | 36 (20–54) | 59 (44–74) | 58 (39–68) |
| Mean FEV1 (range) | 64 (18–96) | 69 (17–100) | 65 (23–100) | 68 (26–99) |
| Subjects with Pseudomonas spp. (%) | 11 (50%) | 32 (70%) | 7 (5%) | 4 (9%) |
| BALF samples | 114 | 170 | 314 | 258 |
| Mean BALF sample collected per subject | 4.96 | 3.62 | 4.02 | 5.50 |
| CLAD ( −) subjects | 6 | 21 | 20 | 15 |
| CLAD ( +) subjects (SD of CLAD Stage) | 17 (1.13) | 26 (1.30) | 58 (0.96) | 32 (0.93) |
DNA extraction and microbiome sequencing in BALF
16S rRNA gene sequence data were generated from bacterial genomic DNA extracted from BALF samples using the Qiagen DNeasy PowerSoil Pro kit. Sequencing of the V4 region was performed using Illumina MiSeq TruSeq 2 × 300 paired-end technology on the Illumina MiSeq platform at the University of Minnesota Genomics Center. Sequence analysis was performed in R as previously described [30]. Cutadapt/2.6 [31] was used to remove primer sequences, with size filtering set to 215 bp at minimum and 285 bp at maximum. Bacterial relative abundance was determined using DADA2 [32]. Reads were dereplicated, paired ends merged, and chimeric reads removed using default options. Genus-level taxonomy was assigned using the Ribosomal Database Project (RDP) Bayesian classifier and SILVA-132 taxonomy training set [33, 34]. R version 4. 2.1 and its packages random forest (version 4.7.1.1), vegan (version 2.6.4), and ggplot2 (version 3.5.0) were also used for these analyses [35–37] (see supplementary methods).
Organic extraction and LC–MS/MS analysis
Methanolic extracts of 856 BALF samples (50:50 v/v) were analyzed on a Thermo QExactive™ mass spectrometer coupled to a Vanquish ultra-high-performance liquid chromatography (UHPLC) system (ThermoFisher) (see supplementary methods). Subsequently, feature-based molecular networking (FBMN) was performed through the Global Natural Produces Social Molecular Networking (GNPS) web tool with a parent and fragment mass ion tolerance of 0.02 Da, a cosine score of 0.65, and a minimum matched peaks minimum of 4 [38–40]. The FBMN job is publicly available at https://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=756456ac794d48b0bba80dbe28e0de66, and raw data files are available in the MassIVE (massive.ucsd.edu) data repository as MSV000085760. Data curation consisted of removing metabolites detected in blanks and removing known pharmaceuticals and their related nodes in the molecular network (see supplementary methods, Fig. S1, and Table S2). The metabolome of the BALF samples was independently analyzed using CANOPUS through SIRIUS to determine in silico chemical classifications among the underlying diseases [41]. Presence/absence frequency and rank abundance of all molecular features were calculated after creating a cutoff by summing the relative abundances of features above 10e5 abundance in samples obtained from subjects with AATD, CF, COPD, or PF.
Statistical analysis
Statistical significance between both microbiome and metabolome data and categorial variables such as underlying disease type and the CLAD development for each subject was assessed through permutational multivariate analysis of variance (PERMANOVA) testing and random forests (RF) machine learning classification analysis [35]. Bray–Curtis dissimilarity matrices were calculated and visualized using principal coordinate analysis (PCoA). PERMANOVA tests were performed for underlying lung diseases and CLAD development (as a categorical variable) with subject-source as an interacting factor to account for variation in the number of samples per subject. Post hoc Dunn’s tests among disease types were performed with R packages Devtools and Vegan (pairwise-adonis) [36, 42]. Variable importance plots from the microbiome and metabolome RF classifications were used to identify variables best contributing to the classifications tested (CLAD or underlying disease). Ranked variables of interest were then tested with a Wilcoxon rank-sum test for significance. Pearson correlations were applied to determine relationships between Pseudomonas-derived metabolites, time, and the average relative abundance of the genus Pseudomonas spp.
Results
Sample collection and clinical design
The two primary objectives for this study were to (i) determine if the microbiome and metabolome of BALF collected after LTx were associated with the underlying pre-diagnosed lung disease, and (ii) identify if the data were different between those who developed CLAD and those who did not across and within underlying disease types. The dataset was composed of longitudinal BALF samples (n = 856) collected over 10 years from 195 subjects with one of four underlying conditions prior to transplant; AATD (n = 23), CF (n = 47), COPD (n = 78), and PF (n = 47). Clinical parameters and patient demographics are presented in Table 1. The final CLAD stage of all subjects was known, even when this measure was recorded after BALF collection had ceased. Thus, the data variation for both the microbiome and metabolome was tested against the development of CLAD (CLAD or no-CLAD (collectively including stages 1–4)) [6].
BALF multi-omics data variation based on the underlying lung disease pre-transplant
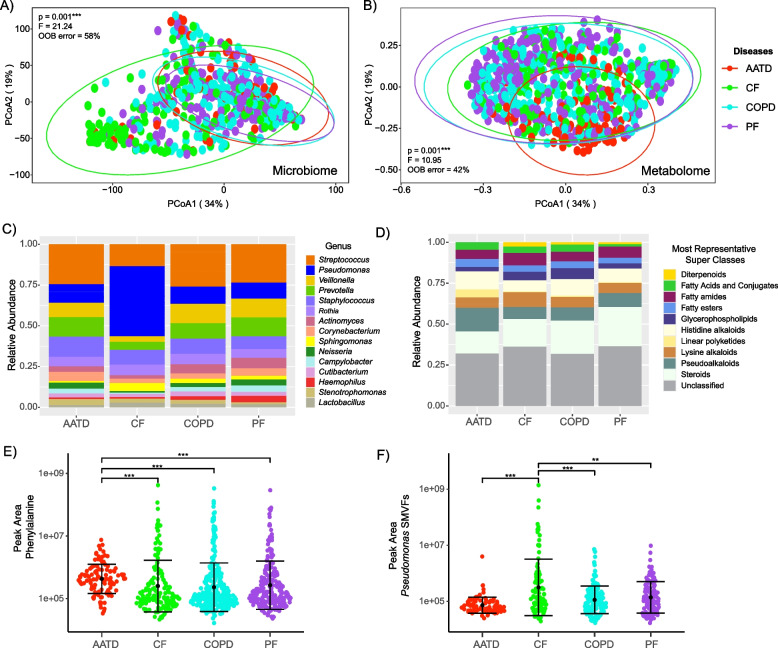
Principal coordinate analysis (PCoA) ordination was used to visualize the microbial and metabolomic variability among the entire dataset by pre-transplant disease diagnosis (Fig. 1). Significant differences in the microbiome composition were observed among the underlying diseases (PERMANOVA F = 21.14, p = 0.001) with CF distinctly clustering apart from AATD, COPD, and PF (Fig. 1A). Pairwise comparisons within disease types verified distinct differences, with only AATD and PF not having significantly different microbiomes from one another (p > 0.005). Differences were also observed in the metabolome by underlying disease (PERMANOVA F = 10.95, p = 0.001), with AATD exhibiting distinct clustering from CF, COPD, and PF (Fig. 1B, Table S3). Significant differences in the metabolomic composition of BALF from subjects with AATD compared to CF (p = 0.0296), COPD (p = 0.0166), and PF (p = 0.0025) were also observed, although the overall metabolome structure was not significantly different among CF, COPD, and PF (Table 2). This analysis signifies that the BALF microbiome and metabolome were different based on the underlying disease of the subject even after LTx with CF and AATD being especially different from the others. These findings demonstrate the clinical relevance of disease-specific molecular signatures post-LTx and underscore the importance of potential therapeutic approaches targeting microbial and metabolomic profiles.
Fig. 1.
Lung allograft molecular profiles vary by pre-LTx condition. A PCoA plot of a Bray–Curtis distance matrix of lung allograft BALF microbiome and B metabolomic profiles based on underlying diseases diagnosed pre-LTx. Ellipses are drawn around the underlying diseases to represent the 95% confidence intervals of the group centroids. C, D Bar plots representing the microbiome and metabolome’s relative abundance among underlying diseases. E, F Sina plots reflecting the difference in the abundance of phenylalanine in AATD and Pseudomonas-derived small molecules virulence factors (SMVFs) in CF. Samples were colored by pre-LTx disease and demonstrated significant differences among disease types. PERMANOVA, random forest out-of-bag (OOB) error, and Wilcoxon test with the significance values > 0.05 (**) and 0.001 (***) are also displayed
Table 2.
Post hoc Dunn test pairwise comparisons of BALF microbiome and metabolome among different diseases (AATD, CF, COPD, and PF). Z-scores quantify the magnitude of difference between underlying diseases
| Pairwise comparisons | Microbiome | Metabolome | ||
|---|---|---|---|---|
| Z-statistic | p-value | Z-statistic | p-value | |
| AATD and CF | 6.2757 | < 0.001 | − 2.5810 | 0.0296 |
| AATD and COPD | 2.5902 | 0.0288 | − 2.7741 | 0.0166 |
| AATD and PF | 0.3868 | > 0.050 | 3.3429 | 0.0025 |
| CF and COPD | − 5.0040 | < 0.001 | 0.1017 | > 0.050 |
| CF and PF | − 7.2503 | < 0.001 | − 0.6433 | > 0.050 |
| COPD and PF | − 2.8529 | 0.0130 | − 0.8660 | > 0.050 |
Microbial and metabolite features driving underlying disease variation in multi-omics data
Analysis of the microbiome composition of BALF revealed a predominance of Streptococcus, Pseudomonas, Veillonella, Prevotella, and Staphylococcus spp. Notably, Pseudomonas exhibited a significantly higher relative abundance in subjects with CF compared to those with AATD (p = 7.40E-13), COPD (p < 0.001), and PF (p < 0.001) and a decrease in Streptococcus, Veillonella, Prevotella, and Actinomyces (Fig. 1C, Fig. S2). Other notable taxonomic differences were higher Staphylococcus and Prevotella abundance in AATD, and higher Haemophilus in PF (Fig. S2).
For the metabolome, in silico molecular classification analysis conducted using CANOPUS cheminformatics software enabled the determination of the molecular family composition of BALF classifying at the level of annotation to superclass [41]. This analysis showed that the BALF metabolome predominantly consisted of steroids, alkaloids, glycerophospholipids, fatty acyls and conjugates, fatty esters, and some other families (Fig. 1D). Molecular family differences across diseases were less notable than in the microbiome, but fatty acyls and conjugates, pseudoalkaloids, and steroid molecular families were significantly different among underlying diseases (Kruskal–Wallis p = 0.03, p = 0.0016, and p = 0.0065, respectively, Fig. S3). As noted in the PCoA plot, AATD stood out as being unique with higher histidine alkaloids, linear polyketides, and lower steroids than the other groups (Fig. S3). Further exploration of the RF variable importance plots for specific molecules contributing to underlying disease uniqueness showed that phenylalanine was more abundant in BALF samples from subjects with underlying AATD (pairwise p-value; CF p = 2.90E-11, COPD p = 4.70E-14, and PF p = 3.10E-13, Fig. 1E). Likewise, our analysis revealed other molecular features, such as elevated fatty acids and glycerophospholipids in CF, COPD, and PF (Fig. S4). We note, however, that a substantial portion of the BALF metabolome (regardless of disease state) remains unclassified, and these molecular family classifications are quite broad (Fig. 1D). Therefore, we compared the metabolome structure across diseases in a more general manner (presence/absence) to determine the degree of chemical sharing. Although most metabolites were shared among the diseases, AATD had fewer shared molecules and overall total metabolites, which were also less abundant compared to the other three disease types, as revealed by the metabolome rank abundance curves (Fig. S5).
Pseudomonas small molecule virulence factors (SMVFs) were detected in the metabolome after spectral library searching in GNPS, including quinolones, rhamnolipids, phenazines, and the siderophore pyochelin. Due to the importance of this pathogen to airway disease and its enrichment in the microbiome of those with CF after LTx [43], we more closely compared the abundances of these molecules between underlying disease states. These SMVFs were significantly higher in BALF samples from CF subjects who underwent LTx compared to patients with the other diagnoses (Wilcoxon test, p = 0.001; Fig. 1F), matching the Pseudomonas trend in the microbiome data.
In summary, these data show that underlying CF disease is associated with a unique BALF microbiome after LTx, driven predominately by the higher abundance of Pseudomonas. The importance of this organism in CF was verified in the metabolome, where its SMVFs were also enriched. More generally, the metabolome showed that AATD was more distinct than the other underlying diseases, driven by higher amounts of phenylalanine, fewer steroids, and overall, less sharing of metabolites with the others.
Multi-omics variation associated with CLAD development
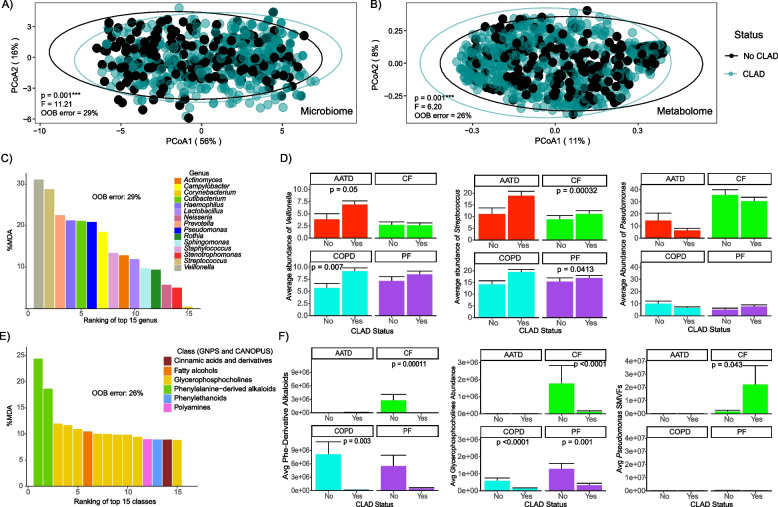
We then compared the microbiome and metabolome composition between those subjects who did not develop CLAD during prospective sample collection to those who developed any CLAD stage (1–4). We observed significant differences in both the microbiome (PERMANOVA, F = 11.21; p = 0.001) and metabolome (PERMANOVA, F = 6.20; p = 0.001) between these two patient cohorts (Fig. 2A, B). As further support for this classification, a supervised random forest analysis of the microbiome data correctly classified 71% of subjects as having CLAD across all underlying diseases and 74% for the BALF metabolome. Therefore, despite variation in the omics data based on the underlying disease, there was still a robust and statistically significant signature for those who did or did not develop CLAD.
Fig. 2.
Lung allograft molecular profiles change according to CLAD onset status and its underlying diseases. A PCoA plot of a Bray–Curtis distance matrix of lung allograft BALF microbiome and B metabolomic profiles based on CLAD diagnosis. C Variable importance plot of the BALF microbiome derived from supervised random forest analysis. D Average abundances of Veillonella, Streptococcus, and Pseudomonas and by CLAD/non-CLAD and underlying disease. E Variable importance plot of the BALF metabolome from supervised random forest analysis. F Average abundances of phenylalanine-derived alkaloids, glycerophosphocholines, and Pseudomonas SMVFs by CLAD status and underlying disease. The top 15 ranked molecular drivers (microbiome and metabolome) the % out-of-bag (OOB) error from RF, Wilcoxon test, and the standard error of the mean (SEM) are shown
Random forest classification was used to rank microbial and metabolite features that best contributed to the correct classifications of CLAD or no-CLAD, and these were tested within each disease group (Fig. 2C, E). There were higher relative abundances of Veillonella and Streptococcus in subjects that developed CLAD, particularly in those with AATD (Wilcoxon test, p = 0.05) and COPD (Wilcoxon test, p = 0.007), while Streptococcus displayed a similar trend in CF (Wilcoxon test, p = 0.00032) and PF (Wilcoxon test, p = 0.0413). As expected, Pseudomonas was the predominant genus among those with CF, though its relative abundance was not significantly different in those who did or did not develop CLAD (Fig. 2D). In addition, we observed a significant increase in Prevotella abundance in subjects with CF and PF that developed CLAD (Wilcoxon test p-values of 0.00032 and 0.0413, respectively) (Fig. S6).
Supervised RF analysis of the metabolome identified two distinct classes of molecules, phenylalanine-derived alkaloids and glycerophosphocholines, as key drivers of changes in the status of CLAD onset (Fig. 2E). We observed a significant decrease in the average abundance of phenylalanine-derived alkaloids in individuals with CF (Wilcoxon test, p = 0.0001) and COPD (Wilcoxon test, p = 0.003) who developed CLAD, mirroring a similar trend for glycerophosphocholines in CF (Wilcoxon test, p < 0.0001), COPD (Wilcoxon test, p < 0.0001), and PF (Wilcoxon test, p = 0.001). Importantly, among subjects with CF, the average abundance of Pseudomonas SMVFs was significantly higher in those who developed CLAD (Wilcoxon test, p = 0.043) (Fig. 2F).
Pseudomonas and its metabolites in BALF samples from subjects with CF after LTx
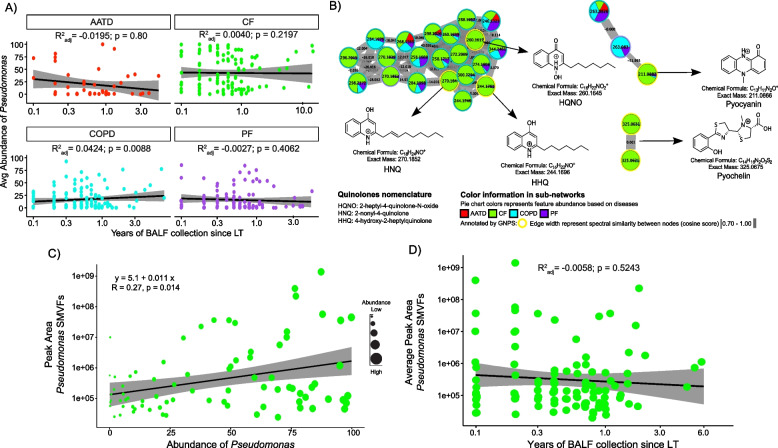
Differentiation of the CF microbiome driven by Pseudomonas and the association of its SMVFs with those who developed CLAD prompted further investigation into the chemical diversity of these compounds, relationships between the two data types, and their dynamics during the time since transplant. We first plotted the average abundance of Pseudomonas over time following LTx and observed that, despite its higher abundance among CF subjects, there were no significant changes over time since LTx for any underlying disease (Fig. 3A). MS/MS molecular networking revealed the presence of quinolones such as 2-heptyl-4-quinolone-N-oxide (HQNO; m/z 260.1645 [M + H]+, C16H22NO2; 10.67 ppm) and 2-nonyl-4-quinolone (HNQ; m/z 270.1852 [M + H]+, C17H24NO; 7.45 ppm), the redox-active virulence factor pyocyanin (m/z 211.0866 [M + H]+, C13H11N2O; 5.64 ppm), and the iron chelating siderophore pyochelin (m/z 325.0675 [M + H]+, C14H16N2O3S2; 10.46 ppm). Molecular network node mapping based on disease showed that the entire molecular family of quinolones and pyochelin was enriched in CF (Fig. 3B). We then tested for a correlation between Pseudomonas relative abundance and its SMVF abundance in the metabolome of the same samples and found a significant positive correlation between them (r = 0.27; p = 0.014) (Fig. 3C). Interestingly, in 60% (n = 46) of subjects with CF, at least one Pseudomonas-derived molecule class was detected in the first BALF sample collected post-transplant (Fig. S7). Detection of these molecules was sporadic overall, but even years after LTx, some subjects still had abundant Pseudomonas SMVF in their BALF metabolome that did not significantly change over time (Fig. 3D).
Fig. 3.
Pseudomonas spp. molecular signatures in BALF samples from CF subjects post-LTx. A Average Pseudomonas abundance among underlying diseases and linear regression over time since LTx. Grey shaded area has 95% confidence interval. B Molecular networks of molecules produced by Pseudomonas spp. identified by GNPS library searching. Each node represents a unique MS/MS spectrum (putative molecule), and connections between nodes are determined by spectral similarity (cosine score) from MS/MS alignment. Pie charts represent total feature abundance colored by the underlying disease. C Linear correlation plot depicting the abundance of SMVFs post-LTx and average abundance of Pseudomonas spp. in CF BALF samples. Dot size denotes Pseudomonas abundance per subject. D Scatter plot of the Pseudomonas SMVFs in CF BALF across years of sampling after LTx and linear regression
Discussion
Clinical outcomes after LTx are idiosyncratic and not easily predictable. To better understand the potential drivers of these outcomes, we applied 16S rRNA gene sequencing and untargeted metabolomics to BALF collected longitudinally from subjects who had undergone LTx for various chronic lung conditions. Based on the structure of our BALF repository, we sought to determine if there was an association between the multi-omics data and underlying disease state and the development of CLAD during the study period. Perhaps the most noteworthy finding was the presence of P. aeruginosa SMVFs in the BALF metabolome, which we investigated further for associations with underlying disease and CLAD.
The post-LTx microbiome differed based on the underlying lung disease, with particularly unique profiles found in those with CF, driven by the higher relative abundance of Pseudomonas. This is not unexpected, as P. aeruginosa is a canonical pathogen of the CF airways and is known to recolonize the allograft after LTx [44, 45]. We note that our data showed the bacterium persists at high relative abundances in many subjects with CF after LTx. The unique aspect of this study was the inclusion of metabolomics data, which identified SMVFs from Pseudomonas as being most abundant in those with underlying CF and higher in those who developed CLAD after LTx. Although some of these metabolites can be produced by other airway pathogens [46], the collective detection of quinolones, rhamnolipids, phenazines, and pyochelin in BALF samples is highly indicative of P. aeruginosa infection and active metabolism in vivo. It is also noteworthy that many SMVFs were detected in the first BALF sample collected after LTx, supporting early colonization of the new allograft by P. aeruginosa immediately after LTx [44]. Although transplant centers perform chest irrigation with antimicrobials, this observation is consistent with the model that the bacterium readily re-colonizes the respiratory tract post-LTx from its reservoir in the upper airways (i.e., paranasal sinuses) [47]. Furthermore, these molecules were also elevated in BALF from those with underlying CF who developed CLAD during the study period versus those who did not. Pseudomonas SMVFs possess cytotoxic properties and can damage host immune and epithelial cells, modulate cytokine production, and interfere with host signaling pathways [48–50], making them of potential interest in the pathology of CLAD or allograft rejection. This opportunistic pathogen regulates the production of various virulence factors through its quorum sensing system, which is mediated through N-acyl-homoserine lactones and alkyl quinolones [51]; only the latter of which were detected in this study. The correlation between the abundance of these molecules and CLAD development indicates they could potentially be explored as biomarkers of bacterial infection in CF post-LTx and as indicators of CLAD progression, though further research and more targeted measurements of their concentrations in BALF will be required. Importantly, they are relatively easy to detect via LC–MS/MS after sample extraction, and this can be done within hours, further supporting their biomarker potential. Though data from this study cannot directly assess this infection reservoir or mechanistic associations with CLAD, the detection of P. aeruginosa SMVFs in BALF so early after LTx is an important finding for understanding disease pathogenesis. It is also important to note that samples from this study were collected before the widespread use of CFTR modulators to treat the underlying cause of CF disease [52]. While data are limited, studies indicate significant benefits from modulator treatment, such as improved lung function, reduced sinus disease, enhanced nutritional status, and lowered infection load in the airways of pwCF. It is not known how these modulators may alter the microbial and metabolite dynamics in pwCF after lung transplant observed here, making a study of the effects of modulators before or after LTx an important area for future research [53, 54].
Metabolomic data identified other aspects of the post-LTx BALF metabolome that were unique based on underlying lung disease, especially those with AATD. The metabolomes of the other chronic lung diseases were also significantly different from one another but more difficult to distinguish overall. The metabolomic uniqueness of AATD was driven by differences in aromatic amino acids (phenylalanine), fewer steroids, an overall lack of shared molecules with the other diseases, and a lower abundance of metabolite features (Figs. S4, S5). We note that all four underlying conditions have unique etiologies, and findings here indicate that, somewhat unexpectedly, the chemical environment of the lung allograft reflects the initial disease of the recipient, particularly in the case of AATD. The high abundance of phenylalanine in allografts from subjects with AATD may reflect increased proteolysis due to increased trypsin-mediated proteolysis, which is a hallmark of this disease [55], though further research will be needed to test this hypothesis.
Both the microbiome and metabolome data are also associated with CLAD development. In terms of the microbiome, RF machine learning identified greater relative abundances of Veillonella (AATD and COPD) and Streptococcus (CF and PF) in those who developed CLAD compared to those who did not. Although both genera are common components of the human oral microbiome, their higher abundance in the airways of those with CLAD may indicate more frequent aspiration from the oral cavity to the allograft. The presence of anaerobic bacteria such as Veillonella and Streptococcus, which have been linked with an increased host inflammatory response, may also reflect a developing dysbiosis in the lung microbiome characteristic of that associated with CF, COPD, and PF [56–58]. Interestingly, we did not find an association between Pseudomonas abundance and CLAD development in any disease subgroup, even though its increased relative abundance in airway fluid LTx has been linked to an increased risk of CLAD or death [59].
Metabolome features associated with CLAD development were primarily phenylalanine-derived alkaloids, glycerophosphocholines, and the Pseudomonas metabolites described above. Higher glycerophosphocholines in those with CLAD is notable because of their presence in pulmonary surfactant comprising about 90% of its composition, which is primarily synthesized by alveolar type 2 cells (AT2Cs). In the context of LTx and allograft dysfunction, understanding the role of surfactants may be important, because acute respiratory distress syndrome (ARDS) is characterized by disturbances in surfactant composition, leading to impaired functionality [60, 61]. The deficiency of surfactant is a common feature of ARDS reviewed in [62], and is also observed in conditions such as pneumonia and sepsis-induced acute lung injury. Additionally, surfactant deficiency and alterations in lipid composition are associated with CF and PF [63, 64]. Future studies focusing on characterizing surfactant lipidome alterations post-LTx could provide valuable insights into disease mechanisms and guide the development of therapeutic strategies.
Some caveats to our study should be considered. Though our sample set is large covering four chronic diseases with broad disease severity (Table S1), the longitudinal nature of the sampling approach was not uniform across subjects (time and number of specimens). This is due to the opportunistic and non-interventional nature of the sampling approach where BALF samples were collected during routine visits and not specifically for this study. Furthermore, we simplified our clinical data on CLAD to compare only those who developed CLAD during any point of the collection period to those who did not. Though clinical information on CLAD severity scores (BOS-grade) was available for some subjects at the time of BALF sampling, these were not consistent across the population, making any multi-omics comparisons to CLAD severity potentially biased and limiting our sample size across the four disease states. Those subjects classified as not having developed CLAD during the sample period may have eventually developed this disease after collections ceased, but this is not known. Thus, we were careful to provide a comparison only to those subjects that developed CLAD during the study period compared to those who did not to minimize these potential confounders, but still provide useful information about the microbiology and biochemistry of this important disease manifestation after LTx. Additionally, an inherent and universal limitation in the field of metabolomics is that a large portion of molecules within BALF samples remain unannotated, including those that contributed to trends seen with CLAD. Low annotation rates in untargeted mass spectrometry studies remain a significant challenge [65], leaving most molecules detected in even the best-studied samples unknown. Though the low annotation rate could be viewed as a limitation, our LC–MS/MS approach removes background contaminants and noise signals, increasing confidence that these molecules reflect the onset of CLAD biology despite their anonymity.
Our study highlights the microbial and metabolite features of lung allografts after LTx. Somewhat unexpectedly, considering the allograft itself is assumed healthy, both datasets demonstrated unique signatures of the underlying disease of the recipient, especially for those with CF and AATD. This indicates that microbiological and metabolic processes from the LTx recipient have a direct impact on the allograft immediately post-LTx and that some of these processes can be associated with the development of CLAD. The role of Pseudomonas in allograft dysfunction and rejection is well established, and this study further implicates this organism and its SMVFs as potentially harmful to those with LTx, especially those with CF. Further work is needed to accurately quantify the microbes and metabolites identified in this study for their potential development as biomarkers of disease after LTx and to better understand the microbiological and immune dynamics that underlie chronic graft rejection.
Supplementary Information
Acknowledgements
We thank Anthony Schilmiller for his constant support during sample processing in the mass spectrometry and metabolomics core laboratory at Michigan State University. We thank the staff at the University of Minnesota Genomics Center for their guidance on sample preparation and sequencing. We thank Bonnie Holme for maintenance of the O’Brien BALF Specimen database. The CF Foundation grant HUNTER18ABO and the National Institute of Allergy and Infectious Diseases (R01AI145925) provided funding.
Authors’ contributions
R.H. and R. Q. conceptualized the study. K. M. performed BALF/CLAD stages tabulation. C. M., T. W., A. G., R. H., and R. Q. performed data acquisition, analysis, and interpretation. C. M., R. Q., and R. H. wrote the original and final versions of the manuscript. K. M. and M. H. edited the manuscript. All authors read and approved the final manuscript.
Funding
This project was funded by the Cystic Fibrosis Foundation (HUNTER18AB0) awarded to Ryan Hunter and the National Institutes of Health (R01AI145925) awarded to Robert Quinn.
Availability of data and materials
No datasets were generated or analysed during the current study.
Declarations
Ethics approval and consent to participate
This study adhered to the principles outlined in the Declaration of Helsinki. Ethical approval and written informed consent for subject participation were performed under the approved protocol at the University of Minnesota Internal Review Boards (STUDY00004547).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ryan C. Hunter, Email: rhunter2@buffalo.edu
Robert A. Quinn, Email: quinnrob@msu.edu
References
- 1.Afonso JE, Werebe E de C, Carraro RM, Teixeira RH de OB, Fernandes LM, Abdalla LG, et al. Lung transplantation. Einstein. 2015;13:297–304. [DOI] [PMC free article] [PubMed]
- 2.Yusen RD. Technology and outcomes assessment in lung transplantation. Proc Am Thorac Soc. 2009;6:128–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khush KK, Cherikh WS, Chambers DC, Goldfarb S, Hayes D, Kucheryavaya AY, et al. The international thoracic organ transplant registry of the international society for heart and lung transplantation: thirty-fifth adult heart transplantation report-2018; focus theme: multiorgan transplantation. J Heart Lung Transplant Off Publ Int Soc Heart Transplant. 2018;37:1155–68. [DOI] [PubMed] [Google Scholar]
- 4.Tissot A, Danger R, Claustre J, Magnan A, Brouard S. Early identification of chronic lung allograft dysfunction: the need of biomarkers. Front Immunol. 2019;10:1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DerHovanessian A, Wallace WD, Lynch JP, Belperio JA, Weigt SS. Chronic lung allograft dysfunction: evolving concepts and therapies. Semin Respir Crit Care Med. 2018;39:155–71. [DOI] [PubMed] [Google Scholar]
- 6.Verleden GM, Glanville AR, Lease ED, Fisher AJ, Calabrese F, Corris PA, et al. Chronic lung allograft dysfunction: definition, diagnostic criteria, and approaches to treatment-a consensus report from the Pulmonary Council of the ISHLT. J Heart Lung Transplant Off Publ Int Soc Heart Transplant. 2019;38:493–503. [DOI] [PubMed] [Google Scholar]
- 7.Conrad A, Janciauskiene S, Köhnlein T, Fuge J, Ivanyi P, Tudorache I, et al. Impact of alpha 1-antitrypsin deficiency and prior augmentation therapy on patients’ survival after lung transplantation. Eur Respir J. 2017;50:1700962. [DOI] [PubMed] [Google Scholar]
- 8.Kneidinger N, Milger K, Janitza S, Ceelen F, Leuschner G, Dinkel J, et al. Lung volumes predict survival in patients with chronic lung allograft dysfunction. Eur Respir J. 2017;49:1601315. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez IE, Heinzelmann K, Verleden S, Eickelberg O. Characteristic patterns in the fibrotic lung Comparing idiopathic pulmonary fibrosis with chronic lung allograft dysfunction. Ann Am Thorac Soc. 2015;12(Suppl 1):S34-41. [DOI] [PubMed] [Google Scholar]
- 10.Blatter J, Sweet S. Lung transplantation in cystic fibrosis: trends and controversies. Pediatr Allergy Immunol Pulmonol. 2015;28:237–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cribbs SK, Beck JM. Microbiome in the pathogenesis of cystic fibrosis and lung transplant-related disease. Transl Res J Lab Clin Med. 2017;179:84–96. [DOI] [PubMed] [Google Scholar]
- 12.McGinniss JE, Whiteside SA, Simon-Soro A, Diamond JM, Christie JD, Bushman FD, et al. The lung microbiome in lung transplantation. J Heart Lung Transplant Off Publ Int Soc Heart Transplant. 2021;40:733–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borewicz K, Pragman AA, Kim HB, Hertz M, Wendt C, Isaacson RE. Longitudinal analysis of the lung microbiome in lung transplantation. FEMS Microbiol Lett. 2013;339:57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Da Silva RR, Lopes NP, Silva DB. Chapter 3. Metabolomics. In: Peporine Lopes N, Roberto Da Silva R, editors. Chem Biol. Cambridge: Royal Society of Chemistry; 2017. p. 57–81.
- 15.Raghuvanshi R, Vasco K, Vázquez-Baeza Y, Jiang L, Morton JT, Li D, et al. High-resolution longitudinal dynamics of the cystic fibrosis sputum microbiome and metabolome through antibiotic therapy. mSystems. 2020;5:e00292–20. [DOI] [PMC free article] [PubMed]
- 16.Singh S, Natalini JG, Segal LN. Lung microbial-host interface through the lens of multi-omics. Mucosal Immunol. 2022;15:837–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin C, Guzior DV, Gonzalez CT, Okros M, Mielke J, Padillo L, et al. Longitudinal microbial and molecular dynamics in the cystic fibrosis lung after Elexacaftor–Tezacaftor–Ivacaftor therapy. Respir Res. 2023;24:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garg N, Kapono C, Lim YW, Koyama N, Vermeij MJA, Conrad D, et al. Mass spectral similarity for untargeted metabolomics data analysis of complex mixtures. Int J Mass Spectrom. 2015;377:719–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watzenboeck ML, Gorki A-D, Quattrone F, Gawish R, Schwarz S, Lambers C, et al. Multi-omics profiling predicts allograft function after lung transplantation. Eur Respir J. 2022;59:2003292. [DOI] [PubMed] [Google Scholar]
- 20.Walmsley S, Cruickshank-Quinn C, Quinn K, Zhang X, Petrache I, Bowler RP, et al. A prototypic small molecule database for bronchoalveolar lavage-based metabolomics. Sci Data. 2018;5: 180060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, Wroblewski M, Hertz MI, Wendt CH, Cervenka TM, Nelsestuen GL. Analysis of chronic lung transplant rejection by MALDI-TOF profiles of bronchoalveolar lavage fluid. Proteomics. 2006;6:1001–10. [DOI] [PubMed] [Google Scholar]
- 22.Reynaud-Gaubert M, Thomas P, Gregoire R, Badier M, Cau P, Sampol J, et al. Clinical utility of bronchoalveolar lavage cell phenotype analyses in the postoperative monitoring of lung transplant recipients. Eur J Cardio-Thorac Surg Off J Eur Assoc Cardio-Thorac Surg. 2002;21:60–6. [DOI] [PubMed] [Google Scholar]
- 23.Das S, Bernasconi E, Koutsokera A, Wurlod D-A, Tripathi V, Bonilla-Rosso G, et al. A prevalent and culturable microbiota links ecological balance to clinical stability of the human lung after transplantation. Nat Commun. 2021;12:2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Combs MP, Wheeler DS, Luth JE, Falkowski NR, Walker NM, Erb-Downward JR, et al. Lung microbiota predict chronic rejection in healthy lung transplant recipients: a prospective cohort study. Lancet Respir Med. 2021;9:601–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans CR, Karnovsky A, Kovach MA, Standiford TJ, Burant CF, Stringer KA. Untargeted LC-MS metabolomics of bronchoalveolar lavage fluid differentiates acute respiratory distress syndrome from health. J Proteome Res. 2014;13:640–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cribbs SK, Park Y, Guidot DM, Martin GS, Brown LA, Lennox J, et al. Metabolomics of bronchoalveolar lavage differentiate healthy HIV-1-infected subjects from controls. AIDS Res Hum Retroviruses. 2014;30:579–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walter S, Gudowius P, Bosshammer J, Romling U, Weissbrodt H, Schurmann W, et al. Epidemiology of chronic Pseudomonas aeruginosa infections in the airways of lung transplant recipients with cystic fibrosis. Thorax. 1997;52:318–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gimino VJ, Lande JD, Berryman TR, King RA, Hertz MI. Gene expression profiling of bronchoalveolar lavage cells in acute lung rejection. Am J Respir Crit Care Med. 2003;168:1237–42. [DOI] [PubMed] [Google Scholar]
- 29.Estenne M, Hertz MI. Bronchiolitis obliterans after human lung transplantation. Am J Respir Crit Care Med. 2002;166:440–4. [DOI] [PubMed] [Google Scholar]
- 30.Lucas SK, Villarreal AR, Ahmad MM, Itabiyi A, Feddema E, Boyer HC, et al. Anaerobic microbiota derived from the upper airways impact Staphylococcus aureus physiology. Infect Immun. 2021;89: e0015321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–2. [Google Scholar]
- 32.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Breiman L. Random Forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- 36.Oksanen J, Simpson GL, Blanchet FG, Kindt R, Legendre P, Minchin PR, et al. vegan: community ecology package. 2001. p. 2.6–6.1. Available from: https://CRAN.R-project.org/package=vegan.
- 37.Wickham H, Chang W, Henry L, Pedersen TL, Takahashi K, Wilke C, et al. ggplot2: create elegant data visualisations using the grammar of graphics. 2024. Available from: https://cran.r-project.org/web/packages/ggplot2/index.html.
- 38.Nothias L-F, Petras D, Schmid R, Dührkop K, Rainer J, Sarvepalli A, et al. Feature-based molecular networking in the GNPS analysis environment. Nat Methods. 2020;17:905–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pluskal T, Castillo S, Villar-Briones A, Orešič M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34:828–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dührkop K, Nothias L-F, Fleischauer M, Reher R, Ludwig M, Hoffmann MA, et al. Systematic classification of unknown metabolites using high-resolution fragmentation mass spectra. Nat Biotechnol. 2021;39:462–71. [DOI] [PubMed] [Google Scholar]
- 42.Wickham H, Hester J, Chang W, Bryan J, RStudio. devtools: tools to make developing R packages easier. 2022. Available from: https://cran.r-project.org/web/packages/devtools/index.html.
- 43.Thornton CS, Acosta N, Surette MG, Parkins MD. Exploring the cystic fibrosis lung microbiome: making the most of a sticky situation. J Pediatr Infect Dis Soc. 2022;11:S13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dugger DT, Fung M, Zlock L, Caldera S, Sharp L, Hays SR, et al. Cystic fibrosis lung transplant recipients have suppressed airway interferon responses during Pseudomonas infection. Cell Rep Med. 2020;1: 100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holm AE, Schultz HHL, Johansen HK, Pressler T, Lund TK, Iversen M, et al. Bacterial re-colonization occurs early after lung transplantation in cystic fibrosis patients. J Clin Med. 2021;10:1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diggle SP, Lumjiaktase P, Dipilato F, Winzer K, Kunakorn M, Barrett DA, et al. Functional genetic analysis reveals a 2-Alkyl-4-quinolone signaling system in the human pathogen Burkholderia pseudomallei and related bacteria. Chem Biol. 2006;13:701–10. [DOI] [PubMed] [Google Scholar]
- 47.Syed SA, Whelan FJ, Waddell B, Rabin HR, Parkins MD, Surette MG. Reemergence of lower-airway microbiota in lung transplant patients with cystic fibrosis. Ann Am Thorac Soc. 2016;13:2132–42. [DOI] [PubMed] [Google Scholar]
- 48.Savchenko V, Szamosvári D, Bao Y, Pignitter M, Böttcher T. Biosynthetic flexibility of Pseudomonas aeruginosa leads to hydroxylated 2-alkylquinolones with proinflammatory host response. Commun Chem. 2023;6:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McClure CD, Schiller NL. Effects of Pseudomonas aeruginosa rhamnolipids on human monocyte-derived macrophages. J Leukoc Biol. 1992;51:97–102. [DOI] [PubMed] [Google Scholar]
- 50.Zulianello L, Canard C, Köhler T, Caille D, Lacroix J-S, Meda P. Rhamnolipids are virulence factors that promote early infiltration of primary human airway epithelia by Pseudomonas aeruginosa. Infect Immun. 2006;74:3134–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Migiyama Y, Kaneko Y, Yanagihara K, Morohoshi T, Morinaga Y, Nakamura S, et al. Efficacy of AiiM, an N-acylhomoserine lactonase, against Pseudomonas aeruginosa in a mouse model of acute pneumonia. Antimicrob Agents Chemother. 2013;57:3653–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Caverly LJ, Riquelme SA, Hisert KB. The impact of highly effective modulator therapy on cystic fibrosis microbiology and inflammation. Clin Chest Med. 2022;43:647–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Middleton PG, Mall MA, Dřevínek P, Lands LC, McKone EF, Polineni D, et al. Elexacaftor-tezacaftor-ivacaftor for cystic fibrosis with a single Phe508del allele. N Engl J Med. 2019;381:1809–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nichols DP, Morgan SJ, Skalland M, Vo AT, Van Dalfsen JM, Singh SB, et al. Pharmacologic improvement of CFTR function rapidly decreases sputum pathogen density, but lung infections generally persist. J Clin Invest. 2023;133: e167957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Quinn RA, Adem S, Mills RH, Comstock W, DeRight GL, Humphrey G, et al. Neutrophilic proteolysis in the cystic fibrosis lung correlates with a pathogenic microbiome. Microbiome. 2019;7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Natalini JG, Singh S, Segal LN. The dynamic lung microbiome in health and disease. Nat Rev Microbiol. 2023;21:222–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caverly LJ, Huang YJ, Sze MA. Past, present, and future research on the lung microbiome in inflammatory airway disease. Chest. 2019;156:376–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Son J-H, Kim JH, Chang HS, Park J-S, Park C-S. Relationship of microbial profile with airway immune response in eosinophilic or neutrophilic inflammation of asthmatics. Allergy Asthma Immunol Res. 2020;12:412–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Combs MP, Wheeler DS, Luth J, Falkowski NR, Chanderraj R, Walker NM, et al. Increasing relative abundance of Pseudomonads predicts chronic rejection after lung transplant. J Heart Lung Transplant. 2020;39:S65. [Google Scholar]
- 60.Günther A, Siebert C, Schmidt R, Ziegler S, Grimminger F, Yabut M, et al. Surfactant alterations in severe pneumonia, acute respiratory distress syndrome, and cardiogenic lung edema. Am J Respir Crit Care Med. 1996;153:176–84. [DOI] [PubMed] [Google Scholar]
- 61.Schmidt R, Meier U, Yabut-Perez M, Walmrath D, Grimminger F, Seeger W, et al. Alteration of fatty acid profiles in different pulmonary surfactant phospholipids in acute respiratory distress syndrome and severe pneumonia. Am J Respir Crit Care Med. 2001;163:95–100. [DOI] [PubMed] [Google Scholar]
- 62.Lewis JF, Jobe AH. Surfactant and the adult respiratory distress syndrome. Am Rev Respir Dis. 1993;147:218–33. [DOI] [PubMed] [Google Scholar]
- 63.Gunasekara L, Al-Saiedy M, Green F, Pratt R, Bjornson C, Yang A, et al. Pulmonary surfactant dysfunction in pediatric cystic fibrosis: mechanisms and reversal with a lipid-sequestering drug. J Cyst Fibros. 2017;16:565–72. [DOI] [PubMed] [Google Scholar]
- 64.Günther A, Schmidt R, Nix F, Yabut-Perez M, Guth C, Rosseau S, et al. Surfactant abnormalities in idiopathic pulmonary fibrosis, hypersensitivity pneumonitis and sarcoidosis. Eur Respir J. 1999;14:565–73. [DOI] [PubMed] [Google Scholar]
- 65.da Silva RR, Dorrestein PC, Quinn RA. Illuminating the dark matter in metabolomics. Proc Natl Acad Sci U S A. 2015;112:12549–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analysed during the current study.