Abstract
Background
Lung cancer is one of the major threats to human life worldwide. MiR‐190 has been found to perform essential roles in multiple cancer progression; however, there have been no studies focused on its function and underlying regulatory mechanism in lung cancer.
Method
The miR‐190 expression was detected by real‐time quantitative polymerase chain reaction (RT‐qPCR). The cell functional experiments, including cell counting kit‐8 (CCK‐8), colony formation and transwell assay were conducted in vitro, as well as animal experiments performed in vivo. The regulation and potential binding sites of CBX4 on miR‐190 were predicted by TCGA data set and JASPAR website and verified by ChIP assay and dual‐luciferase reporter assay. The prospects binding site of miR‐190‐3p on CBX4 3′UTR region was predicted by StarBase and verified by dual‐luciferase reporter assay.
Results
MiR‐190 was decreased in lung cancer cells. The overexpression of miR‐190 had no effects on cell proliferation, but significantly inhibited cancer metastasis both in vitro and in vivo. Moreover, miR‐190 expression could be transcriptionally inhibited by CBX4, and CBX4 was the direct target of miR‐190‐3p.
Conclusion
MiR‐190 served as a cancer metastasis inhibitor in lung cancer and formed a regulatory loop with CBX4. These findings provided emerging insights into therapeutic targets and strategies for metastatic lung cancer.
Keywords: CBX4, lung cancer, metastasis, miR‐190
CBX4 could inhibit miR‐190 expression transcriptionally, and CBX4 was a direct target of miR‐190‐3p, and formed a loop in regulating lung cancer metastasis.
INTRODUCTION
Lung cancer is one of the most frequently diagnosed malignancies and cardinal cause for cancer‐related deaths worldwide. 1 , 2 Lung cancer is mainly divided into small cell lung cancer (SCLC) and non‐small cell lung cancer (NSCLC). 3 NSCLC including large cell carcinoma, squamous cell carcinoma and adenocarcinoma, occupy more than 80% of primary lung cancer cases. 4 Among all these pathological characteristics, late diagnosis, unimproved therapeutic tools, relapse and drug resistance, especially cancer metastasis, make the prognosis of lung cancer patients unsatisfactory. 5 , 6 Metastasis is the main reason of cancer death, and despite accumulating advances in understanding lung cancer metastasis, the prognosis for these patients is still unfavorable. 7 , 8 , 9 Thus, further investigation in understanding the mechanisms underlying lung cancer metastasis are essential for improving lung cancer therapy.
MicroRNAs (miRNA) are a kind of small non‐coding RNA molecule, usually constituted by 22 nucleotides. 10 , 11 , 12 Increasing evidence has shown that miRNAs play important roles in lung cancer progression. 13 , 14 Li et al. reported that miR‐1284 could inhibit cell growth and induces apoptosis of lung cancer cells, indicating miR‐1284 may have a key role in lung tumorigenesis. 15 In addition, Chen et al. also demonstrated that miRNA‐148a could suppress migration and invasion of lung cancer by regulating Wnt signaling, and was able to serve as a prognostic factor. 16 Chen et al. found that miR‐182‐5p could promote breast cancer proliferation and invasion both in vivo and in vitro by targeting the CMTM7‐Wnt axis. 17 miR‐190 is located at chromosome 15q22.2, serving as an intron region of TLN2 gene. Previous studies have established the important roles of miR‐190 in multiple cancer types. Some authors believe that miR‐190 serves as a tumor suppressor. Hao et al. reported that miR‐190 attenuates the invasion and migration abilities of hepatocellular carcinoma (HCC) by inhibiting epithelial‐mesenchymal transition (EMT). 18 Our previous study also found that miR‐190 inhibited breast cancer metastasis by regulating TGF‐beta‐induced EMT process. 18 However, other studies consider miR‐190 as a tumor promoting factor. Jia et al. reported that miR‐190 was upregulated in gastric cancer tissues, and the upregulation of miR‐190 could promote gastric cancer progression by targeting FOXP2. 19 In HCC, Xiong et al. found that miR‐190 overexpression leads to the promotion of cancer proliferation and metastasis by downregulating PHLPP1. 20 However, the role and mechanism of miR‐190 in lung cancer are still unknown. Thus, further investigation of miR‐190 is necessary to better understand its function and evaluate its clinical application.
METHODS
Cells, miRNAs and plasmids
Normal lung bronchial epithelial cells BEAS‐2B and lung cancer cells, including A549, H1299, H1944 and HCC827 cells, were purchased from Shanghai Academy of Sciences (Shanghai, China). All cells were cultured in RPMI‐1640 medium (Corning, China) with 10% fetal bovine serum (FBS: Gibco, USA) and 1% streptomycin (Solarbio, China). The cells were placed in a 37°C incubator with 5% CO2, and the culture medium was changed every other 2 days.
The miR‐190 mimic, inhibitor, miR‐190‐3p/5p mimics and miR‐190 overexpression plasmid were purchased from Thermo Fisher Scientific (USA). The siRNA for CBX4 and CBX4 overexpression plasmid were purchased from Ribobio (Guangzhou, China).
Real‐time quantitative polymerase chain reaction (RT‐qPCR)
The RNA from cells were collected using TriZol reagent (Invitrogen, USA), and the concentration was detected by nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). Then, reverse transcription was conducted using a reverse transcription kit (Transgene, China). After that, the Green qPCR SuperMix (Transgene, China) and miRNA RT‐qPCR Starter kit (RiboBio, China) were used for qPCR for mRNA and miRNA, respectively. The primers were obtained from Genewiz (Tianjin, China), and probes for miR‐190 were purchased from Thermo Fisher Scientific (USA).
Colony formation assay
The transfected cells were seeded in six‐well plates at a total number of 1*103 and incubated for an appropriate time. When the colonies reached certain size (one colony contained more than 50 cells), then the cells were fixed and stained with crystal violet (Beyotime, China) for 30 min. The number of colonies were counted and analyzed.
Cell counting kit‐8 (CCK‐8) assay
The transfected cells were seeded in 96‐well plates at a total number of 2*103 per well. Cell counting kit‐8 (Transgen, China) was used for cell proliferation ability detection according to the recommendations of the manufacturer. CCK‐8 reagent was added into each well at a concentration of 10%; after that, the cells were incubated for another 4 h. Then, the absorbance was detected using a microplate reader (Bio‐Rad, USA) at the 450 nm wave. This operation was repeated at the same time every day for 5 days continuously, and the proliferation curves were drawn and analyzed according to the data.
Transwell assay
The transfected cells were seeded in 8‐μm chambers (Millipore, USA) at a total number of 1*105 per chamber with FBS‐free RPMI‐1640 medium. The chambers with cells were then placed into the 24‐well plates with 600 μL 10% FBS RPMI‐1640 medium. Then the cells were incubated for nearly 16–48 h, which mainly depended on the migration speed of different lung cancer cells. When 2–3 cells could be observed in the lower chamber by microscope, the incubation was able to pause. Three‐step stain kit (Thermo Fisher Scientific, USA) was used for cell fixation and staining. The images were captured by a microscope (Olympus, USA) and the number of migrated cells were counted and analyzed.
Animal experiments
The stable miR‐190 overexpressed H1299 cells were used for intravenous injection (total number of 1*105 cells per mouse). The severe immunodeficiency NSG mice of 4–5 weeks old were purchased from SPF Biotech (China). Each group contained three mice. After injection, the living imaging was conducted every week, and the fluorescence intensity value was recorded. After 35 days of growth, the mice were killed by hyperanesthesia. All animal experiments performed were ethically compliant.
Protein extraction and western blot
The cells were washed with cold phosphate‐buffered saline (PBS) and split by radioimmunoprecipitation assay (RIPA) lysis buffer (Solarbio, China) with 1% phenylmethylsulfonyl fluoride (PMSF: Solarbio, China). A bicinchoninic acid (BCA) protein assay kit (Thermo Scientific, USA) was used for protein concentrations detection. Then the proteins were separated by sodium dodecyl sulphate‐polyacrylamide gel electrophoresis (SDS‐PAGE) synthesized by epizyme (Shanghai, China) and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, USA). After being blocked by 5% bovine serum albumin (BSA) and treated with the primary antibodies and secondary antibodies, the separated proteins were visualized by ECL reagent (Millipore, USA).
Chromatin immunoprecipitation analysis (ChIP)
A chromatin immunoprecipitation assay kit (Millipore, USA) was employed to fulfill the ChIP assay according to the instructions. In brief, the DNA from cells were extracted and broken into 200–500 bp fragments by ultrasound. Then, the specific antibodies or anti‐IgG antibodies (negative control) were added into the lysate and incubated overnight. Then, the immunoprecipitated DNAs were isolated and followed by RT‐qPCR analysis. The results are presented as fold change of % input.
Dual luciferase‐reporter assay
Dual‐luciferase reporter assay system kit (Transgene, China) was utilized to verify the regulation of CBX4 on miR‐190 promoter and the regulation of miR‐190‐3p on CBX4 3′UTR region. The potential combining sites were inserted into PGL3‐basic plasmid (Promega, USA), followed by cotransfection with PRL‐TK plasmid (Promega, USA). The transfection and luciferase detection were conducted according to the manufacturer's recommendations. The experiment was repeated three times.
Statistical analysis
The Service Solutions (SPSS) software of 20.0 version (IBM, USA) was used for statistical calculation. An independent samples t‐test was employed for pair comparison. All data are shown as mean ± standard deviation (SD). GraphPad prism 8.0 software (La Jolla, USA) was utilized for date visualization. The statistical significance was set as p < 0.05.
RESULTS
Downregulation of miR‐190 promoted lung cancer cell invasion
Our previous study demonstrated that miR‐190 inhibited breast cancer metastasis. 21 In this study, we first determined the expression level of miR‐190 in lung cancer cells. RT‐qPCR was performed in normal lung bronchial epithelial cell BEAS‐2B, and lung cancer cells A549, H1299, H1944 and HCC827, and showed that miR‐190 was downregulated in lung cancer cells, especially in H1299 cells (Figure 1a). Then we selected two lung cancer cell lines, A549 and HCC827, in which miR‐190 showed a relatively higher expression. The miR‐190 inhibitor (anti‐190) was transfected into these two cell lines and the inhibition efficiency was verified by RT‐qPCR (Figure 1b). To investigate the role of miR‐190, cell functional experiments were performed. The result of colony formation assay showed that after inhibiting the expression of miR‐190, the number of cell colonies did not change significantly (Figure 1c). Consistent with this, the results of the CCK‐8 assay also showed that downregulation of miR‐190 had no effect on the proliferation curve in both A549 and HCC827 cells (Figure 1d). However, a positive result was obtained by the transwell assay, which showed that the inhibition of miR‐190 increased the migrated cell numbers in both A549 and HCC827 cells (Figure 1e). These results indicated that downregulation of miR‐190 might not affect cell proliferation but significantly promoted cell invasion in lung cancer cells.
FIGURE 1.
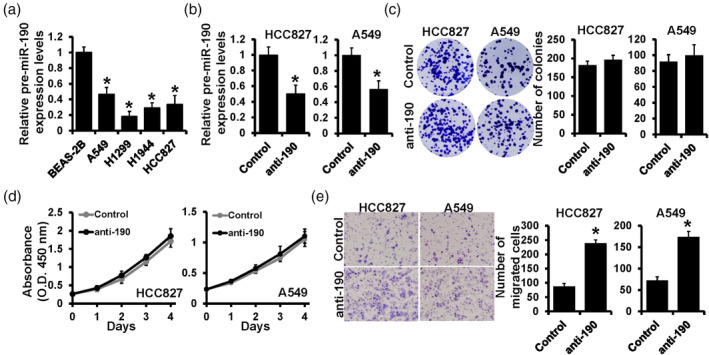
Downregulation of miR‐190 promoted lung cancer cell invasion. (a) The expression level of miR‐190 in normal lung bronchial epithelial cell BEAS‐2B, lung cancer cells A549, H1299, H1944 and HCC827, detected by real‐time quantitative polymerase chain reaction (RT‐qPCR). (b) The detection of miR‐190 expression by RT‐qPCR after anti‐190 transfection in HCC827 cells and A549 cells. (c) The colony formation assay, (d) Cell counting kit‐8 (CCK‐8 assay) and (e) transwell assay were performed in miR‐190 deleted HCC827 cells and A549 cells. *p < 0.05.
Upregulation of miR‐190 inhibited lung cancer metastasis in vitro and in vivo
To further clarify the role of miR‐190 in different lung cancer cells, miR‐190 mimic was transfected into H1299 cells and the expression level of miR‐190 was verified by RT‐qPCR (Figure 2a). Consistent with previous results, the colony formation assay and CCK‐8 assay showed that the overexpression of miR‐190 did not influence colony numbers (Figure 2b) or proliferation curves (Figure 2c). However, the transwell assay showed that overexpression of miR‐190 dramatically decreased the migrated cells of H1299 cells (Figure 2d). For further verification in vivo, the miR‐190 overexpression plasmid was used for constructing miR‐190 stable overexpressed cell lines in H1299, and tumor metastasis models were constructed in mice by intravenous injection. The living imaging result and fluorescence intensity curve showed that upregulation of miR‐190 significantly inhibited the metastasis in vivo (Figure 2e). Thus, we proved that miR‐190 had no effect on cell proliferation but remarkably inhibited cell migration. More convincingly, miR‐190 significantly inhibited tumor metastasis in mice models.
FIGURE 2.
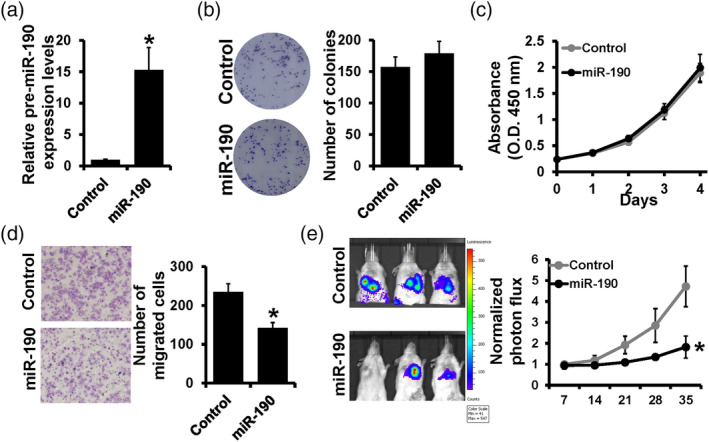
Upregulation of miR‐190 inhibited lung cancer metastasis in vitro and in vivo. (a) The detection of miR‐190 by real‐time quantitative polymerase chain reaction (RT‐qPCR) after miR‐190 mimic transfection in H1299 cells. (b) Colony formation assay, (c) Cell counting kit‐8 (CCK‐8) assay and (d) transwell assay were performed in miR‐190 overexpressed H1299 cells. (e) The living imaging and fluorescence intensity curve of xenograft. *p < 0.05.
MiR‐190 expression was inhibited by CBX4 transcriptionally
MiR‐190 was found to be dramatically downregulated in lung cancer cells and influenced metastasis both in vitro and in vivo; however, the mechanism by which miR‐190 is regulated was still unclear. Herein, we analyzed the expression of genes, which correlated with the miR‐190 expression and found that CBX4 was closely related to the miR‐190 expression (Figure 3a). Then, western blot was performed to detect CBX4 expression in different lung cancer cells. The results showed that CBX4 was significantly upregulated in lung cancer cells compared to normal lung bronchial epithelial cell BEAS‐2B, especially upregulated in H1299 cells, which is completely opposite to miR‐190 expression (Figure 3b). For further investigation of the specific binding site of CBX4 and miR‐190 promoter, four potential binding sites were predicted by the JASPAR website and are shown as a schematic diagram (Figure 3c). ChIP assay was conducted to verify the combination of CBX4 and these four predicted sites, and the results showed that CBX4 had significant binding with site 1 and site 2 (especially site 2), but almost no combination with site 3 or site 4 (Figure 3d). Then, four potential binding sites were inserted into luciferase reporter plasmids, individually or separately, and dual‐luciferase reporter assay showed that CBX4 overexpression notably decreased the luciferase activities of site 1 and site 2 (Figure 3e). Next, the correlation between CBX4 and miR‐190 was analyzed. CBX4 was knocked down by siRNAs transfection in H1299 cells, and the pre‐miR‐190 expression was then detected by RT‐qPCR. The results showed that the expression level of pre‐miR‐190 was increased after CBX4 downregulation (Figure 3f). At the same time, the overexpression of CBX4 resulted in a decrease of pre‐miR‐190 in A549 cells (Figure 3g). These results indicated that CBX4 could inhibit miR‐190 expression by binding to its promoter region.
FIGURE 3.
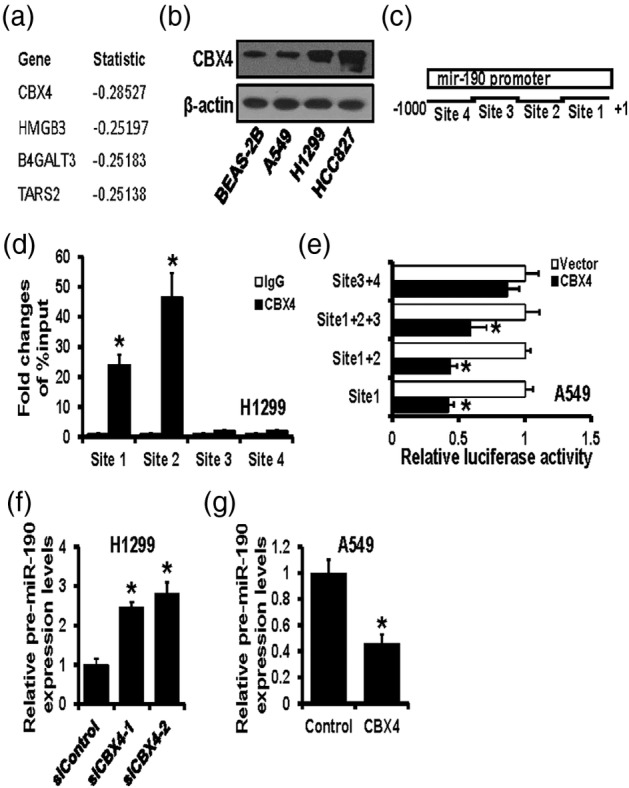
MiR‐190 expression was inhibited by CBX4 transcriptionally. (a) The top four genes closely related to the miR‐190 expression from the Cancer Genome Atlas (TCGA) database. (b) The protein expression level of CBX4 in normal lung bronchial epithelial cell BEAS‐2B, lung cancer cells A549, HCC827 and H1299, detected by western blot. (c) The potential binding sites of CBX4 on miR‐190 promoter predicted by the JASPAR website. (d) ChIP assay validated the binding of CBX4 on miR‐190 in H1299 cells. (e) Dual‐luciferase reporter assay verified the regulation of CBX4 on potential binding sites of miR‐190 promoter. (f) The expression level of pre‐miR‐190 in CBX4 downregulated H1299 cells detected by real‐time quantitative polymerase chain reaction (RT‐qPCR). (g) The expression level of pre‐miR‐190 in CBX4 upregulated A549 cells detected by RT‐qPCR. *p < 0.05.
CBX4 was a direct target of miR‐190‐3p
Interestingly, we observed that in miR‐190 overexpressed H1299 cells, the protein level of CBX4 was also significantly decreased (Figure 4a). MiR‐190 mainly manifested in two mature forms, miR‐190‐3p and miR‐190‐5p. Thus, we transfected miR‐190‐3p and miR‐190‐5p into H1299 cells, respectively, and verified by RT‐qPCR (Figure 4b). Then, the expression level of CBX4 was determined by western blot, and the results showed that the upregulation of miR‐190‐3p led to the downregulation of CBX4, while overexpression of miR‐190‐5p had no effect on CBX4 expression, indicating miR‐190‐3p was able to regulate CBX4 (Figure 4c). Next, the StarBase website was utilized to predict the potential binding site of miR‐190‐3p on the CBX4 3′UTR region (Figure 4d). After inserting wild‐type or mutant CBX4 3′UTR region into luciferase reporter plasmids, a dual‐luciferase reporter assay was performed, and the results showed that miR‐190‐3p dramatically decreased the luciferase activity of wild‐type CBX4 3′UTR (Figure 4e) but had no effects on the mutated group (Figure 4f). Finally, the correlation between miR‐190 and CBX4 was further verified in A549 cells by western blot, and the results showed that downregulation of miR‐190 increased CBX4 expression (Figure 4g). These findings indicated that miR‐190‐3p, not miR‐190‐5p, could bind to the CBX4 3′UTR region and inhibit its expression.
FIGURE 4.
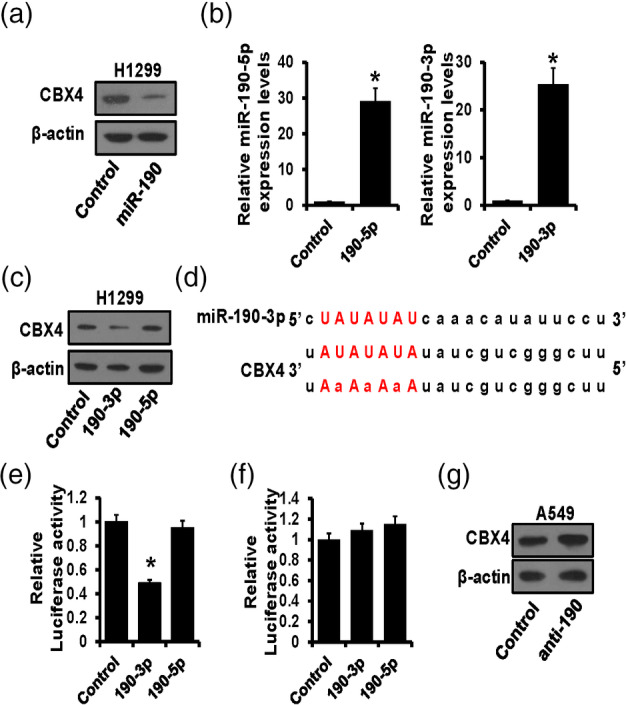
CBX4 was a direct target of miR‐190‐3p. (a) The expression level of CBX4 in miR‐190 overexpressed H1299 cells detected by western blot. (b) The expression of miR‐190‐3p/5p verified by real‐time quantitative polymerase chain reaction (RT‐qPCR) after miR‐190‐3p/5p transfection. (c) The expression level of CBX4 in miR‐190‐3p/5p overexpressed H1299 cells detected by western blot. (d) The potential binding site of miR‐190‐3p on the CBX4 3′UTR region predicted by StarBase. (e) Dual‐luciferase reporter assay verified the regulation of miR‐190‐3p/5p on wild CBX4 3′UTR region. (f) Dual‐luciferase reporter assay verified the regulation of miR‐190‐3p/5p on the mutated CBX4 3′UTR region. (g) The expression level of CBX4 in miR‐190 downregulated A549 cells detected by western blot. *p < 0.05.
DISCUSSION
Lung cancer is a high‐grade malignancy with complex processes during occurrence and metastasis, which involves multiple genes and RNAs. 22 Recent studies have illustrated that miRNAs perform very essential roles in the progression of lung cancer from its initial stages to metastasis. 23 Xue et al. reported that the deficiency of miR‐200 in lung cancer cells promoted the activation and proliferation of adjacent cancer‐associated fibroblasts (CAFs), which resulted in the metastasis cancer cells. 24 Tyagi et al. found that miR‐4466 from nicotine‐activated neutrophils could promote cancer cell stemness and metabolism in lung cancer metastasis. 25 In addition, Song et al. demonstrated that miR‐26a‐5p potentiated lung cancer cell metastasis via JAK2/STAT3 signaling by regulating ITGβ8. 26 Herein, we detected miR‐190 expression level in different lung cancer cells and investigated miR‐190 function in vitro and in vivo, and the results showed that miR‐190 was dramatically downregulated in lung cancer cells compared with normal lung cells, and miR‐190 could inhibit lung cancer cell metastasis both in vitro and in vivo. It is the first study to focus on the role of miR‐190 in lung cancer, which refined its multiple roles in different cancer types. However, it is interesting that miR‐190 could significantly inhibit cancer metastasis but had no effect on cell proliferation. It appears that the inhibitory effect of miR‐190 on lung cancer progression is one‐sided. The same mechanism has been previously reported in multiple studies; for example, one of our previous studies indicated that overexpression of ZNF384 did not influence breast cancer proliferation but promoted breast cancer migration and invasion. 27 This phenomenon might be due to its downstream target genes; however, the underlying mechanisms still needs further exploration.
In this study, we further explored the upstream regulator of miR‐190. The UCSC data set and JASPAR website were utilized to predict the transcription factor that could bind to miR‐190 promoter region. Chromobox4 (CBX4), which is also known as E3 SUMO‐protein ligase, is a key component of polycomb‐repressive complexes 1 (PRC1), 28 which has been reported as a transcriptional factor and regulates a variety of targets implicated in cancer growth, metastasis, and angiogenesis. 29 , 30 , 31 Zeng et al. demonstrated that CBX4 could promote breast cancer progression by regulating the miR‐137‐mediated Notch1 pathway. 32 Wang et al. found that CBX4 inhibited colorectal carcinoma metastasis through recruitment of HDAC3 to the promoter region of Runx2. 33 While, in lung cancer, Hu et al. reported that CBX4 downregulation obviously suppressed the cell growth and migration of human lung cancer cells in vitro and in vivo via regulating BMI‐1. 34 Herein, we predicted that CBX4 was an upstream regulator of miR‐190, which had been verified by ChIP and dual‐luciferase reporter assays. Our findings indicated that CBX4 inhibited miR‐190 expression and involved in lung cancer metastasis, which provides insights into the mechanism underlying miRNAs regulation on lung cancer metastasis.
However, the role of miR‐190 in cancers has been controversial in previous studies. Some researchers reported that miR‐190 was related to more aggressive phenotypes in cancer progression, including neuroblastoma, 35 bladder cancer, 36 gastric cancer 19 and liver cancer, 20 while in some cancers, miR‐190 also served as a cancer inhibitor, such as in breast cancer 21 , 37 , 38 and colorectal cancer. 39 It is well known that miRNAs mainly work by directly targeting the 3′UTR region of their target genes. Thus, the diverse roles of miR‐190 in different cancers might be due to the different downstream targets. While, in lung cancer, the role of miR‐190 has not previously been defined. Our study is the first to raise the metastasis inhibiting role of miR‐190 in lung cancer. We demonstrated that CBX4 was the upstream regulator of miR‐190, which inhibited its expression transcriptionally. At the same time, CBX4 also served as a direct target of miR‐190‐3p, thus forming a circular regulatory loop. Despite further investigation being needed in order to understand the deep regulatory mechanism, the findings in this study have introduced new strategies for lung cancer metastasis therapy.
In conclusion, we found that miR‐190 was downregulated in lung cancer cells. The overexpression of miR‐190 had no effect on cell proliferation, but significantly inhibited lung cancer metastasis both in vitro and in vivo. In addition, CBX4 could inhibit miR‐190 expression transcriptionally, and CBX4 was a direct target of miR‐190‐3p. Our findings might provide emerging targets and therapy for lung cancer metastasis.
AUTHOR CONTRIBUTIONS
Wen Zhou and Wengui Xu designed the study; Jian Wang, Xiang Zhu, Yue Yu, Jie Ge and Wei Chen performed the experiments and statistical analysis; Jian Wang, Zhu Xiang and Wen Zhou wrote and revised the manuscript. All authors read and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no competing interests.
ACKNOWLEDGMENTS
This study was supported by funds from Tianjin Municipal Health Commission (ZC20013).
Wang J, Zhu X, Yu Y, Ge J, Chen W, Xu W, et al. CBX4/miR‐190 regulatory loop inhibits lung cancer metastasis. Thorac Cancer. 2024;15(26):1889–1896. 10.1111/1759-7714.15415
Jian Wang and Xiang Zhu contributed equally to this study.
Contributor Information
Wengui Xu, Email: wxu06@tmu.edu.cn.
Wen Zhou, Email: zhouwen@tmu.edu.cn.
DATA AVAILABILITY STATEMENT
All data generated or analyzed during this study are included in this article and its supplementary information files.
REFERENCES
- 1. Bade BC, Dela CC. Lung cancer 2020: epidemiology, etiology, and prevention. Clin Chest Med. 2020;41(1):1–24. 10.1016/j.ccm.2019.10.001 [DOI] [PubMed] [Google Scholar]
- 2. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 3. de Sousa V, Carvalho L. Heterogeneity in lung cancer. Pathobiology. 2018;85(1–2):96–107. 10.1159/000487440 [DOI] [PubMed] [Google Scholar]
- 4. Brody H. Lung cancer. Nature. 2020;587(7834):S7. 10.1038/d41586-020-03152-0 [DOI] [PubMed] [Google Scholar]
- 5. Pallis AG, Syrigos KN. Lung cancer in never smokers: disease characteristics and risk factors. Crit Rev Oncol Hematol. 2013;88(3):494–503. 10.1016/j.critrevonc.2013.06.011 [DOI] [PubMed] [Google Scholar]
- 6. Abu RF, Singhi EK, Sridhar A, Faisal MS, Desai A. Lung cancer treatment advances in 2022. Cancer Invest. 2023;41(1):12–24. 10.1080/07357907.2022.2119479 [DOI] [PubMed] [Google Scholar]
- 7. Altorki NK, Markowitz GJ, Gao D, Port JL, Saxena A, Stiles B, et al. The lung microenvironment: an important regulator of tumour growth and metastasis. Nat Rev Cancer. 2019;19(1):9–31. 10.1038/s41568-018-0081-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yin L, Liu X, Shao X, Feng T, Xu J, Wang Q, et al. The role of exosomes in lung cancer metastasis and clinical applications: an updated review. J Transl Med. 2021;19(1):312. 10.1186/s12967-021-02985-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang X, Adjei AA. Lung cancer and metastasis: new opportunities and challenges. Cancer Metastasis Rev. 2015;34(2):169–171. 10.1007/s10555-015-9562-4 [DOI] [PubMed] [Google Scholar]
- 10. Lu TX, Rothenberg ME. Microrna. J Allergy Clin Immunol. 2018;141(4):1202–1207. 10.1016/j.jaci.2017.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu S, Lu J, Zhu H, Wu F, Mo Y, Xie L, et al. A novel axis of circkif4a‐mir‐637‐stat3 promotes brain metastasis in triple‐negative breast cancer. Cancer Lett. 2024;581:216508. 10.1016/j.canlet.2023.216508 [DOI] [PubMed] [Google Scholar]
- 12. Wang Z, Yang L, Wu P, Li X, Tang Y, Ou X, et al. The circrobo1/klf5/fus feedback loop regulates the liver metastasis of breast cancer by inhibiting the selective autophagy of afadin. Mol Cancer. 2022;21(1):29. 10.1186/s12943-022-01498-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shah V, Shah J. Recent trends in targeting mirnas for cancer therapy. J Pharm Pharmacol. 2020;72(12):1732–1749. 10.1111/jphp.13351 [DOI] [PubMed] [Google Scholar]
- 14. Rupaimoole R, Slack FJ. Microrna therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discovery. 2017;16(3):203–222. 10.1038/nrd.2016.246 [DOI] [PubMed] [Google Scholar]
- 15. Li J, Jin H, Yu H, Wang B, Tang J. Mirna‐1284 inhibits cell growth and induces apoptosis of lung cancer cells. Mol Med Rep. 2017;16(3):3049–3054. 10.3892/mmr.2017.6949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Y, Min L, Ren C, Xu X, Yang J, Sun X, et al. Mirna‐148a serves as a prognostic factor and suppresses migration and invasion through wnt1 in non‐small cell lung cancer. PLoS One. 2017;12(2):e0171751. 10.1371/journal.pone.0171751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen ZH, Tian Y, Zhou GL, Yue HR, Zhou XJ, Ma HY, et al. Cmtm7 inhibits breast cancer progression by regulating wnt/beta‐catenin signaling. Breast Cancer Res. 2023;25(1):22. 10.1186/s13058-023-01620-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hao Y, Yang J, Yin S, Zhang H, Fan Y, Sun C, et al. The synergistic regulation of vegf‐mediated angiogenesis through mir‐190 and target genes. RNA. 2014;20(8):1328–1336. 10.1261/rna.044651.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jia WZ, Yu T, An Q, Yang H, Zhang Z, Liu X, et al. Microrna‐190 regulates foxp2 genes in human gastric cancer. Onco Targets Ther. 2016;9:3643–3651. 10.2147/OTT.S103682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xiong Y, Wu S, Yu H, Wu J, Wang Y, Li H, et al. Mir‐190 promotes hcc proliferation and metastasis by targeting phlpp1. Exp Cell Res. 2018;371(1):185–195. 10.1016/j.yexcr.2018.08.008 [DOI] [PubMed] [Google Scholar]
- 21. Yu Y, Luo W, Yang ZJ, Chi JR, Li YR, Ding Y, et al. Mir‐190 suppresses breast cancer metastasis by regulation of tgf‐beta‐induced epithelial‐mesenchymal transition. Mol Cancer. 2018;17(1):70. 10.1186/s12943-018-0818-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. The L. Lung cancer: some progress, but still a lot more to do. Lancet. 2019;394(10212):1880. 10.1016/S0140-6736(19)32795-3 [DOI] [PubMed] [Google Scholar]
- 23. Hashemi ZS, Khalili S, Forouzandeh MM, Sadroddiny E. Lung cancer and mirnas: a possible remedy for anti‐metastatic, therapeutic and diagnostic applications. Expert Rev Respir Med. 2017;11(2):147–157. 10.1080/17476348.2017.1279403 [DOI] [PubMed] [Google Scholar]
- 24. Xue B, Chuang CH, Prosser HM, Fuziwara CS, Chan C, Sahasrabudhe N, et al. Mir‐200 deficiency promotes lung cancer metastasis by activating notch signaling in cancer‐associated fibroblasts. Genes Dev. 2021;35(15–16):1109–1122. 10.1101/gad.347344.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tyagi A, Wu SY, Sharma S, Wu K, Zhao D, Deshpande R, et al. Exosomal mir‐4466 from nicotine‐activated neutrophils promotes tumor cell stemness and metabolism in lung cancer metastasis. Oncogene. 2022;41(22):3079–3092. 10.1038/s41388-022-02322-w [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 26. Song Q, Liu B, Li X, Zhang Q, Cao L, Xu M, et al. Mir‐26a‐5p potentiates metastasis of human lung cancer cells by regulating itgbeta8‐ jak2/stat3 axis. Biochem Biophys Res Commun. 2018;501(2):494–500. 10.1016/j.bbrc.2018.05.020 [DOI] [PubMed] [Google Scholar]
- 27. Meng QX, Wang KN, Li JH, Zhang H, Chen ZH, Zhou XJ, et al. Znf384‐zeb1 feedback loop regulates breast cancer metastasis. Mol Med. 2022;28(1):111. 10.1186/s10020-022-00541-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ren L, Li Z, Zhou Y, Zhang J, Zhao Z, Wu Z, et al. Cbx4 promotes antitumor immunity by suppressing pdcd1 expression in t cells. Mol Oncol. 2023;17(12):2694–2708. 10.1002/1878-0261.13516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen F, Hou W, Yu X, Wu J, Li Z, Xu J, et al. Cbx4 deletion promotes tumorigenesis under kras(g12d) background by inducing genomic instability. Signal Transduct Target Ther. 2023;8(1):343. 10.1038/s41392-023-01623-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fisher ML, Balinth S, Hwangbo Y, Wu C, Ballon C, Goldberg GL, et al. Cancer‐associated fibroblasts promote cancer stemness by inducing expression of the chromatin‐modifying protein cbx4 in squamous cell carcinoma. Carcinogenesis. 2023;44(6):485–496. 10.1093/carcin/bgad048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meng R, Fang J, Yu Y, Hou LK, Chi JR, Chen AX, et al. Mir‐129‐5p suppresses breast cancer proliferation by targeting cbx4. Neoplasma. 2018;65(4):572–578. 10.4149/neo_2018_170814N530 [DOI] [PubMed] [Google Scholar]
- 32. Zeng JS, Zhang ZD, Pei L, Bai ZZ, Yang Y, Yang H, et al. Corrigendum to cbx4 exhibits oncogenic activities in breast cancer via notch1 signaling. Int J Biochem Cell Biol. 2018;105:145. 10.1016/j.biocel.2018.10.001 [DOI] [PubMed] [Google Scholar]
- 33. Wang X, Li L, Wu Y, Zhang R, Zhang M, Liao D, et al. Cbx4 suppresses metastasis via recruitment of hdac3 to the runx2 promoter in colorectal carcinoma. Cancer Res. 2016;76(24):7277–7289. 10.1158/0008-5472.CAN-16-2100 [DOI] [PubMed] [Google Scholar]
- 34. Hu C, Zhang Q, Tang Q, Zhou H, Liu W, Huang J, et al. Cbx4 promotes the proliferation and metastasis via regulating bmi‐1 in lung cancer. J Cell Mol Med. 2020;24(1):618–631. 10.1111/jcmm.14771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Slaby O. Mir‐190 leads to aggressive phenotype of neuroblastoma through indirect activation of trkb pathway. Med Hypotheses. 2013;80(3):325–326. 10.1016/j.mehy.2012.11.033 [DOI] [PubMed] [Google Scholar]
- 36. Huang S, Hua X, Kuang M, Zhu J, Mu H, Tian Z, et al. Mir‐190 promotes malignant transformation and progression of human urothelial cells through cdkn1b/p27 inhibition. Cancer Cell Int. 2021;21(1):241. 10.1186/s12935-021-01937-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu Y, Yin W, Yu ZH, Zhou YJ, Chi JR, Ge J, et al. Mir‐190 enhances endocrine therapy sensitivity by regulating sox9 expression in breast cancer. J Exp Clin Cancer Res. 2019;38(1):22. 10.1186/s13046-019-1039-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sun G, Liu M, Han H. Overexpression of microrna‐190 inhibits migration, invasion, epithelial‐mesenchymal transition, and angiogenesis through suppression of protein kinase b‐extracellular signal‐regulated kinase signaling pathway via binding to stanniocalicin 2 in breast cancer. J Cell Physiol. 2019;234(10):17824–17838. 10.1002/jcp.28409 [DOI] [PubMed] [Google Scholar]
- 39. Zhu S, Li Z, Zheng D, Yu Y, Xiang J, Ma X, et al. A cancer cell membrane coated, doxorubicin and microrna co‐encapsulated nanoplatform for colorectal cancer theranostics. Mol Ther Oncolytics. 2023;28:182–196. 10.1016/j.omto.2022.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article and its supplementary information files.


