Abstract
The C-type lectins DC-SIGN and DC-SIGNR capture and transfer human immunodeficiency virus (HIV) to susceptible cells, although the underlying mechanism is unclear. Here we show that DC-SIGN/DC-SIGNR-mediated HIV transmission involves dissociable binding and transfer steps, indicating that efficient virus transmission is not simply due to tethering of virus to the cell surface.
Dendritic cells (DCs) are believed to play an important role in HIV infection even though DCs themselves are not readily infectable (2, 4, 13). However, virus bound to DCs can be efficiently transmitted to cocultivated T cells, resulting in efficient virus infection (2). DC-SIGN, a C-type (i.e., calcium-dependent) lectin expressed by DCs, largely accounts for this process since it efficiently captures and transfers primate lentiviruses to receptor-positive cells (3, 9). A closely related lectin expressed on some types of endothelial cells, termed DC-SIGNR (for DC-SIGNRelated), shares 83% amino acid identity with DC-SIGN and also binds and transmits human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus strains (1, 10, 12). Both proteins are type II membrane proteins, with a C-terminal lectin domain, a repeat region most often containing 7.5 repeats of a 23-residue motif, a transmembrane domain, and a short cytoplasmic domain (Fig. 1A) (1, 12). The ability of DC-SIGN and DC-SIGNR to efficiently capture HIV coupled with their expression patterns raises the possibility that they play a role in dissemination of virus between and within hosts. However, it is unknown if DC-SIGN and DC-SIGNR enhance HIV infectivity mainly by concentrating virus particles on the cell surface or if they play a more active role in virus transmission.
FIG. 1.
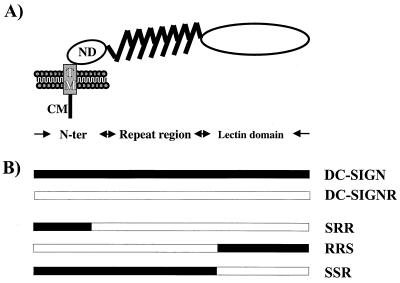
Schematic representation of DC-SIGN and the DC-SIGN/DC-SIGNR chimeras analyzed. (A) Domain structure of DC-SIGN as identified by sequence analysis. DC-SIGNR exhibits a comparable domain organization. N-ter, N terminus; CM, cytoplasmic domain; TM, transmembrane domain; ND, N-terminal domain. (B) Schematic structure of the DC-SIGN/DC-SIGNR chimeras. DC-SIGN/DC-SIGNR chimeras were generated by fusing the N terminus of DC-SIGN to the DC-SIGNR backbone (chimera SRR) and by exchanging the lectin domains of both proteins (chimeras RRS and SSR). For detection of protein expression, all constructs were engineered to contain a C-terminal, antigenic AU-1 tag.
Interactions between DC-SIGN/DC-SIGNR and virus are carbohydrate dependent, involving the C-terminal lectin domain and as-yet-unidentified carbohydrate structures on the HIV envelope (Env) protein (3, 9). Direct protein-protein interactions may also occur. In addition, the repeat region of DC-SIGN is also important for normal function (9). In our previous study, we noted that while DC-SIGN and DC-SIGNR bound multiple virus strains equally well, there were surprising differences in transmission efficiency (10). Some viruses were transmitted by DC-SIGNR more efficiently than by DC-SIGN, while HIV-1 ADA bound to DC-SIGNR but was transmitted to receptor-positive cells very inefficiently. To investigate the structural basis for this variability in virus transmission, we produced several DC-SIGN/DC-SIGNR chimeras (Fig. 1B). An overlap extension PCR technique employing 5′-GGACATTCTTCCAAGGAAACTG and 5′-CAGTTTCCTTGGAAGAATGTCC) as inner primers was used to introduce the lectin domain of DC-SIGN in the DC-SIGNR backbone (chimera RRS) and vice versa (chimera SSR). Using the same technique, we also attached the N terminus of DC-SIGN to the DC-SIGNR backbone (chimera SRR; inner primers 5′-GTCCAAGTGTCCAAGGTCCCCAG and 5′-CTGGGGACCTTGGACACTTGGAC) due to the fact that DC-SIGN contains LL and YXXL motifs which may mediate endocytosis, whereas the YXXL motif is absent in DC-SIGNR. The integrity of the molecular clones was confirmed by sequence analysis.
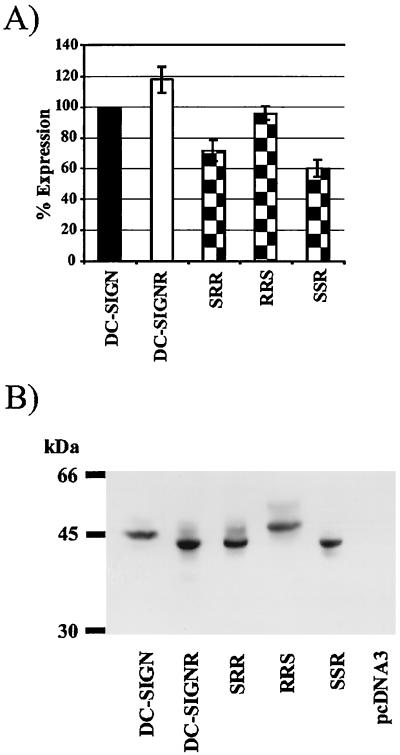
Surface expression levels of DC-SIGN can strongly impact the efficiency of virus binding and transfer (9). Therefore, we introduced an AU-1 tag at the C terminus of all constructs so that surface expression levels could be determined by fluorescence-activated cell sorter (FACS) analysis. Upon transient transfection into 293T cells, the chimeras were expressed efficiently, ranging from 61 to 96% of wild-type DC-SIGN levels (Fig. 2A). Western blot analysis also indicated that expression of the chimeras was comparable to the expression of wild-type DC-SIGN (Fig. 2B). Efficient expression of the chimeras at the cell surface argues that they are correctly folded. In addition, the chimeras retained the ability to bind HIV (see below) as well as ICAM-3 (data not shown).
FIG. 2.
Expression of the DC-SIGN/DC-SIGNR chimeras. (A) Equal amounts of the indicated constructs were transiently transfected into 293T cells. Two days after transfection the cells were stained with anti-AU-1 antibody and analyzed by FACS. The values represent the mean ± standard error of the mean of a single experiment that was carried out in triplicates. Comparable results were obtained in two independent experiments, though the absolute values varied. (B) Analysis of expression by Western blot. The chimeras were transiently expressed in 293T cells, and 2 days after transfection, the cells were lysed and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting with anti-AU-1 antibody. kDa, kilodaltons.
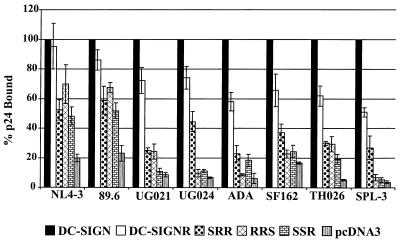
We next investigated the capacity of the DC-SIGN/DC-SIGNR chimeras to bind primary HIV-1 isolates as well as the laboratory-adapted NL4-3 strain. The DC-SIGN/DC-SIGNR chimeras were expressed in 293T cells, and the cells were seeded in 96-well dishes and incubated with 5 ng of p24 antigen of the indicated viral isolates for 3 h at 37°C. We have found that under these conditions, virus bound to either DC-SIGN or to DC-SIGNR remains at the surface of this cell type (9, 10). The cells were vigorously washed and lysed in 0.5% Triton X-100, and the concentration of viral antigen in the lysates was assessed by a commercially available p24 enzyme-linked immunosorbent assay (ELISA). All viruses bound to cells expressing DC-SIGN more efficiently than they bound to cells expressing vector alone by at least fivefold and often by more than 10-fold (Fig. 3). In general, binding to DC-SIGNR was somewhat less efficient, ranging from approximately 51 to 95% of the levels obtained with DC-SIGN. HIV-1 NL4-3 and 89.6 also bound to each of the DC-SIGN/DC-SIGNR chimeras, though at somewhat reduced levels (Fig. 3). Unexpectedly, however, the remaining six virus strains tested either bound poorly or not at all to cells expressing the various chimeras. Binding of these viruses to the RRS and SSR chimeras was particularly weak, in some cases being close to background levels. Thus, despite the high sequence homology between DC-SIGN and DC-SIGNR (83%), the exchange of functional domains between these proteins can dramatically impact their capacity to bind to HIV.
FIG. 3.
Binding of HIV-1 to DC-SIGN/DC-SIGNR chimeras. The chimeras were transiently transfected into 293T cells, and the cells were incubated with equal amounts of p24 of the indicated viruses, vigorously washed, and lysed in 0.5% Triton X-100. The amount of p24 antigen in the lysates was quantified by ELISA. The values were normalized to p24 binding to wild-type-DC-SIGN-transfected cells and represent the mean ± Standard error of the mean of three independent experiments.
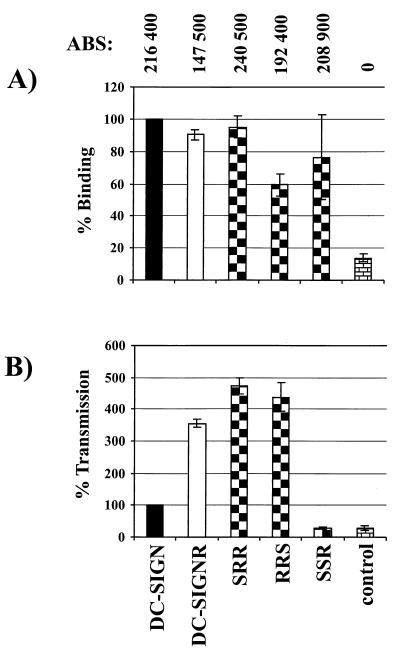
Since all chimeras bound efficiently to NL4-3, we further tested their capacity to transmit this virus strain. In order to limit variability associated with transient transfections, stable T-REX cell lines were engineered to express these proteins upon induction with doxycycline. After induction, surface expression and virus binding and transfer were examined in parallel (Fig. 4A). Surface expression of the chimeras was determined by a quantitative FACS technique that makes it possible to convert mean channel fluorescence into antibody binding sites (ABS). With saturating levels of antibody, there is a direct correlation between ABS with receptor expression levels (7, 9). We found that all of the DC-SIGN/DC-SIGNR chimeras were expressed at levels comparable to those for DC SIGN and higher than those for DC-SIGNR (Fig. 4A). HIV-1 NL4-3 bound to SRR and SSR as efficiently as it did to DC-SIGN and DC-SIGNR and somewhat less well to RRS (Fig. 4A). To test the ability of each chimera to mediate transmission, cells were pulsed with virus, extensively washed, and then cocultivated with C8166 T cells. Three days later, the cells were lysed and luciferase activity was determined. Under these conditions, cells expressing DC-SIGNR transmitted NL4-3 more efficiently than did cells expressing DC-SIGN (Fig. 4B), consistent with an earlier study (9). Virus transmission by chimeras SRR and RRS was comparable to transmission by DC-SIGNR, while chimera SSR did not transfer virus despite binding virus efficiently (Fig. 4B). While it is possible that chimera SSR may transmit virus when expressed in different cellular contexts, our results clearly show that binding and transmission are dissociable functions.
FIG. 4.
Chimera SSR binds but does not transmit HIV-1 NL4-3. Surface expression of the chimeras and binding and transmission of the NL4-3 virus were analyzed in parallel. T-REX cells were induced to express the indicated constructs by overnight incubation with doxycycline. (A) The expression of the chimeras was assessed via quantitative FACS, employing the AU-1 antigenic tag (9). The number of ABS is shown for each cell type at the top of the figure. NL4-3 binding was determined as described in the legend to Fig. 3. (B) Transmission of the NL4-3 virus to C8166 T cells. The T-REX cells were incubated with equal amounts of NL4-3 luciferase reportervirus, washed, and cocultivated with C8166 cells. The luciferase activity in the cultures was determined 3 days after the start of the cocultivation. We set the amount of virus transmitted by DC-SIGN-expressing cells to 100%. Error bars indicate the range of values obtained in quadruplicate samples within a single experiment. Similar results were obtained in an independent experiment.
Virus attached to the cell surface can be more infectious than cell-free virus, indicating that adhesion to cell surface molecules can maintain or even enhance viral infectivity (5, 6, 8). However, DC-SIGN and DC-SIGNR appear to be particularly adept virus binding and transmission factors. DC-SIGN can mediate infection of peripheral blood mononuclear cells even when the amount of virus used is below that needed to establish infection when applied directly to peripheral blood mononuclear cells (3). In addition, virus bound to DCs, presumably via DC-SIGN, can remain infectious for a number of days, a surprising result given the labile nature of HIV-1 in vitro (3). These observations suggest that DC-SIGN may do something other than simply attach virus to the cell surface. Our results are consistent with this in that we have identified a DC-SIGN/DC-SIGNR chimera that binds virus efficiently but fails to transmit it to receptor-positive cells under the conditions that we have examined.
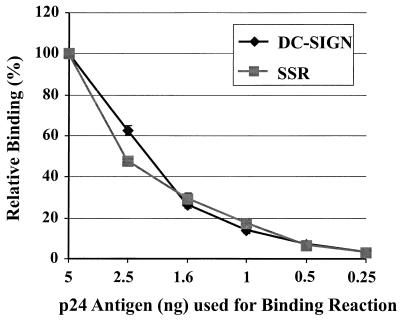
There are a number of general mechanisms that could account for our observation that HIV transmission by DC-SIGN/DC-SIGNR involves separable virus binding and transfer steps. It is possible, for example, that chimera SSR binds virus with reduced affinity. If so, virus may dissociate from this chimera before it engages receptors on adjoining cells. To address this, we added serial dilutions of HIV-1 NL4-3 to cells expressing DC-SIGN or the SSR chimera for 3 h, after which the cells were vigorously washed and the amount of bound p24 antigen was quantified. As shown in Fig. 5, chimera SSR bound virus as well as wild-type DC-SIGN did under all conditions tested. Furthermore, it takes approximately 3 h to wash away unbound virus, since multiple wash and centrifugation steps are involved. Thus, significant differences in off rates should manifest themselves under these conditions. Therefore, we conclude that chimera SSR binds HIV-1 NL4-3 as well as wild-type DC-SIGN.
FIG. 5.
Chimera SSR binds HIV-1 NL4-3 as well as wild-type DC-SIGN. Cells expressing either DC-SIGN or the SSR chimera were incubated with serial dilutions of HIV-1 NL4-3 as indicated for 3 h at 37°C, vigorously washed to remove unbound virus, and lysed, and the amount of bound p24 antigen was quantified via p24 antigen ELISA. Error bars indicate the range of values obtained in quadruplicate samples within a single, representative experiment.
Another possible explanation to account for separable binding and transmission activities involves endocytosis of virus–DC-SIGN complexes. DC-SIGN contains two potential internalization motifs in its cytoplasmic domain, while DC-SIGNR contains one. Thus, chimeras between these two molecules could be endocytosed at different rates. However, we have found that virus bound to either DC-SIGN or DC-SIGNR, when expressed on 293T cells, remains at the cell surface (9, 10). Whether endocytosis of bound virus occurs on DCs remains to be determined. It is also possible that binding of virus to DC-SIGN/DC-SIGNR might modify Env structure or conformation in a manner that impacts receptor binding or Env triggering—events that must necessarily play an important role in virus transmission. Finally, association of DC-SIGN with specific microdomains in the plasma membrane or with molecules on the surface of the target cell could impact transmission. CD4 and the viral coreceptors are asymmetrically distributed on the cell surface (11). Therefore, if the localization of DC-SIGN on the cell surface or if interactions between it and the target cell result in virus presentation to regions with high concentrations of virus receptors, infection efficiency and virus transmission could be enhanced. Recently, it has been found that DC-SIGN colocalizes with CD4 and CCR5 on the surface of alveolar macrophages (B. Lee, unpublished results). Therefore, it will be important to study the distribution of DC-SIGN and DC-SIGNR on relevant primary cell types. While the mechanism that accounts for dissociable virus binding and transmission is not known at present, we can conclude that DC-SIGN/DC-SIGNR-mediated enhancement of virus infectivity is a complex process that cannot be reduced to simple tethering of HIV on the cell surface. Characterizing how DC-SIGN interacts with Env and why this binding event results in marked enhancement of virus transmission could help elucidate the role of DCs in virus transmission and dissemination in vivo and may reveal new targets for antiviral approaches.
Acknowledgments
We thank Victor Holubowsky and Farida Shaheen for generation and quantification of virus stocks.
R.W.D. was supported by NIH grants AI 35383 and 40880, a Burroughs Wellcome Fund Translational Research Award, and an Elizabeth Glaser Scientist Award from the Pediatric AIDS Foundation. S.P. was supported by a fellowship from the Deutsche Forschungsgemeinschaft. F.B. was supported by a fellowship from the Swiss National Science Foundation (grant number 823A-611772). This work was also supported by P30-AI45008 of the Viral/Cell/Molecular Core of the Penn Center for AIDS Research, the Wilhelm-Sander Foundation, and SFB466.
REFERENCES
- 1.Bashirova A A, Geijtenbeek T B H, van Duijnhoven G C F, van Vliet S J, Eilering J B, Martin M P, Wu L, Martin T D, Viebig N, Knolle P A, KewalRamani V N, van Kooyk Y, Carrington M. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV-1 infection. J Exp Med. 2001;193:671–678. doi: 10.1084/jem.193.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cameron P U, Freudenthal P S, Barker J M, Gezelter S, Inaba K, Steinman R M. Dendritic cells exposed to human immunodeficiency virus type-1 transmit a vigorous cytopathic infection to CD4+ T cells. Science. 1992;257:383–387. doi: 10.1126/science.1352913. [DOI] [PubMed] [Google Scholar]
- 3.Geijtenbeek T B H, Kwon D S, Torensma R, van Vliet S J, van Duijnhoven G C F, Middel J, Cornelissen I L M H A, Nottet H S L M, KewalRamani V N, Littman D R, Figdor C G, van Kooyk Y. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 4.Granelli-Piperno A, Delgado E, Finkel V, Paxton W, Steinman R M. Immature dendritic cells selectively replicate macrophagetropic (M-tropic) human immunodeficiency virus type 1, while mature cells efficiently transmit both M- and T-tropic virus to T cells. J Virol. 1998;72:2733–2737. doi: 10.1128/jvi.72.4.2733-2737.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jakubik J J, Saifuddin M, Takefman D M, Spear G T. B lymphocytes in lymph nodes and peripheral blood are important for binding immune complexes containing HIV-1. Immunology. 1999;96:612–619. doi: 10.1046/j.1365-2567.1999.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jakubik J J, Saifuddin M, Takefman D M, Spear G T. Immune complexes containing human immunodeficiency virus type 1 primary isolates bind to lymphoid tissue B lymphocytes and are infectious for T lymphocytes. J Virol. 2000;74:552–555. doi: 10.1128/jvi.74.1.552-555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee B, Sharron M, Montaner L J, Weissman D, Doms R W. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Nat Acad Sci USA. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olinger G G, Saifuddin M, Spear G T. CD4-negative cells bind human immunodeficiency virus type 1 and efficiently transfer virus to T cells. J Virol. 2000;74:8550–8557. doi: 10.1128/jvi.74.18.8550-8557.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pöhlmann S, Baribaud F, Lee B, Leslie G J, Sanchez M D, Hiebenthal-Millow K, Münch J, Kirchhoff F, Doms R W. DC-SIGN interactions with human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus. J Virol. 2001;75:4664–4672. doi: 10.1128/JVI.75.10.4664-4672.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pöhlmann S, Soilleux E J, Baribaud F, Leslie G, Morris L S, Trowsdale J, Lee B, Coleman N, Doms R W. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and immunodeficiency viruses and activates infection in trans. Proc Natl Acad Sci USA. 2001;98:2670–2675. doi: 10.1073/pnas.051631398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singer I I, Scott S, Kawka D W, Chin J, Daugherty B L, DeMartino J A, DiSalvo J, Gould S L, Lineberger J E, Malkowitz L, Miller M D, Mitnaul L, Siciliano S J, Staruch M J, Williams H R, Zweerink H J, Springer M S. CCR5, CXCR4, and CD4 are clustered and closely apposed on microvilli of human macrophages and T cells. J Virol. 2001;75:3779–3790. doi: 10.1128/JVI.75.8.3779-3790.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soilleux E J, Barten R, Trowsdale J. DC-SIGN; a related gene, DC-SIGNR; and CD23 form a cluster on 19p13. J Immunol. 2000;165:2937–2942. doi: 10.4049/jimmunol.165.6.2937. [DOI] [PubMed] [Google Scholar]
- 13.Weissman D, Li Y, Ananworanich J, Zhou L J, Adelsberger J, Tedder T F, Baseler M, Fauci A S. Three populations of cells with dendritic morphology exist in peripheral blood, only one of which is infectable with human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1995;92:826–830. doi: 10.1073/pnas.92.3.826. [DOI] [PMC free article] [PubMed] [Google Scholar]