Abstract
The molecular mechanisms responsible for the heightened reactivity of quiescent T cells in human early life remain largely elusive. Our previous research identified that quiescent adult naïve CD4+ T cells express LINE1 (long interspersed nuclear elements 1) spliced in previously unknown isoforms, and their down-regulation marks the transition to activation. Here, we unveil that neonatal naïve T cell quiescence is characterized by enhanced energy production and protein synthesis. This phenotype is associated with the absence of LINE1 expression attributed to tonic T cell receptor/mTOR complex 1 (mTORC1) signaling and (polypyrimidine tract-binding protein 1 (PTBP1)–mediated LINE1 splicing suppression. The absence of LINE1 expression primes these cells for rapid execution of the activation program by directly regulating protein synthesis. LINE1 expression progressively increases in childhood and adults, peaking in elderly individuals, and, by decreasing protein synthesis, contributes to immune senescence in aging. Our study proposes LINE1 as a critical player of human T cell function across the human life span.
The levels of LINE1 expression regulate the rate of protein synthesis throughout the human life span.
INTRODUCTION
The composition and function of the T cell pool change throughout human life to adapt to various environments and challenges (1). Despite being traditionally viewed as immature or immune deviant, neonatal T cells are now recognized as a distinct developmental layer consisting of fast-reacting naïve T cells (1, 2) without memory lymphocytes (3, 4). This population is predominantly composed of recent thymic emigrants (5), characterized by a limited T cell receptor (TCR) repertoire that displays a higher sensitivity, even to self-peptides (6–8). In addition, studies have indicated that in mice, upon activation, neonatal naïve CD4+ T cells exhibit an early entry into the cell cycle and enhanced proliferation compared to the adult counterparts (9). Overall, these features have been interpreted as favorable for promptly eliciting responses against a diverse array of pathogens in the absence of immunological memory (10, 11). In contrast, in elderly individuals, the T cell pool is predominantly composed of memory T cells, which exhibit reduced reactivity, signs of exhaustion, and immune senescence (12). Nevertheless, the fundamental molecular mechanisms driving the rapid activation in response to antigen threats of naïve T cells in early life and the reduced reactivity in elderly remain obscure.
T lymphocytes must carefully avoid inappropriate activation. In the absence of stimuli, they are actively maintained in a quiescent state characterized by low proliferative rate and energy consumption, yet they remain poised for rapid proliferation and differentiation (13). We have recently discovered a mechanism of transcriptional control governing the quiescence of human naïve CD4+ T cells, mediated by the expression of LINE1 (long interspersed nuclear elements 1), retrotransposable elements that cover about the 17% of the human genome (14). We found that LINE1 undergo splicing to generate previously unknown transcript variants (i.e., LINE1-transcripts), which serve as epigenetic regulators for a whole class of genes associated with T cell activation and effector functions. Notably, T cell activation triggers the down-regulation of LINE1-transcripts under the control of mammalian target of rapamycin complex 1 (mTORC1) signaling, via the splicing suppressor polypyrimidine tract-binding protein 1 (PTBP1). This, in turn, promotes the expression of the genes associated with T cell activation (15).
Here, we sought to investigate the mechanisms responsible for the more rapid activation of quiescent neonatal T cells compared to their adult counterparts. Specifically, we aimed to explore the potential involvement of LINE1 in governing this unique phenotype. Our findings reveal that neonatal naïve CD4+ T cells exhibit a poised quiescent state characterized by increased cellular size/granularity, energy production, and rate of protein synthesis, priming them for rapid activation. We discovered that this unique quiescent state is marked by the absence of LINE1 expression, due to tonic TCR activation induced by self-antigen presentation. This tonic TCR stimulation supports basal mTORC1 signaling and the suppression of LINE1 splicing via PTBP1. We demonstrated that the absence of LINE1 expression enhances the readiness of neonatal T cells for rapid activation by regulating protein synthesis and cell cycle progression. In addition, we explored LINE1 expression across the life span and found that it increases with T cell maturation, becoming overexpressed in T cells during aging, where it leads to reduced protein synthesis, a sign of immune senescence. This study introduces mechanistic insights for T cell adaptation during early life and aging and indicates LINE1 expression as a potential quantitative marker for T cell functionality across the life span.
RESULTS
Naïve CD4+ T cells in neonates exhibit a quiescent phenotype poised for rapid cellular activation
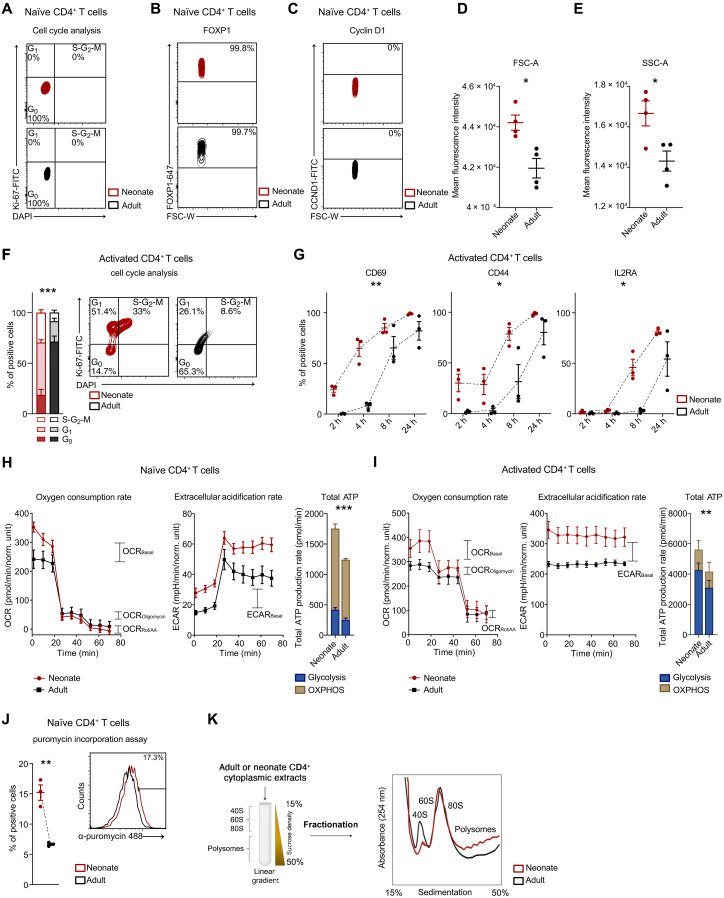
Given the enhanced capacity of neonatal T cells to be rapidly mobilized by antigenic threats compared to adult T cells, we aim to thoroughly investigate their quiescence state and whether they exhibit primed activation compared to adult T cells. We isolated naïve CD4+ T cells from the umbilical cord blood of neonates and the peripheral blood of healthy adult donors (fig. S1A), and examined various phenotypic, functional, and molecular aspects that support T cell quiescence and predispose T cells for activation. Our analysis revealed that, similar to those of adults, neonatal naïve T cells are quiescent. They were in the G0 phase, expressed FOXP1 [a transcription factor known to regulate T cell quiescence (16)], and did not express CyclinD1, which is crucial for entry into the G1 phase (Fig. 1, A to C, and fig. S1, B to D). Despite being quiescent, these cells displayed a substantial increase in cell size [forward scatter (FSC) light] and granularity [side scatter (SSC) light] (Fig. 1, D and E). Upon activation, neonatal T cells progressed more rapidly through the cell cycle (Fig. 1F) and exhibited early and elevated expression of activation markers following TCR stimulation, i.e., CD69, CD44, and IL2RA (Fig. 1G and fig. S1E). Considering that T cell activation relies on energy production and proteome remodeling to which it is intrinsically linked (17), we analyzed these processes both in naïve and activated T cells. Therefore, we examined adenosine triphosphate (ATP) production from glycolysis and mitochondrial respiration in neonatal and adult naïve and activated CD4+ T cells. Our results unveiled that quiescent neonatal naïve T cells were sustained by enhanced ATP production through both oxidative phosphorylation and glycolysis, as measured by the Seahorse ATP rate assay (Fig. 1H). Upon cellular activation, which involves a shift to glycolytic metabolism, we noted that neonatal T cells displayed an increased reliance on glycolysis for ATP production compared to adults (Fig. 1I), suggesting a precocious metabolic switch for these cells. Through puromycin incorporation, we detected a threefold increase in the protein synthesis rate in neonatal quiescent T cells compared to adults (Fig. 1J). Correspondingly, neonatal naïve T cells displayed a higher number of nucleoli, as evidenced by Nucleolin (NCL) immunofluorescence (fig. S1, F and G). This difference persisted in activated T cells, as shown by both puromycin incorporation (fig. S1H) and ribosome profiling, where we observed an increased number of polysomes in neonatal T cells (Fig. 1K).
Fig. 1. Neonatal naïve CD4+ T cells exhibit a quiescent phenotype primed for activation, characterized by enhanced energy production and protein synthesis rate.
(A) Cell cycle analyses were performed using 4′,6-diamidino-2-phenylindole (DAPI) and Ki-67 staining in neonatal or adult naïve CD4+ T cells (n = 4). (B) FOXP1 and (C) CyclinD1 MFI (mean fluorescence intensity) in neonatal or adult naïve CD4+ T cells (n = 3). (D) Cellular size (FSC) and (E) granularity (SSC) MFI in neonatal or adult naïve CD4+ T cells (n = 4). FSC: *P = 0.0103; SSC: *P = 0.0250, unpaired one-tailed t test. (F) Cell cycle analyses in neonatal (n = 4) or adult (n = 5) CD4+ T cells activated for 36 hours. ***P ≤ 0.001, F = 61, two-way ANOVA. (G) Percentage of CD69-, CD44-, and IL2RA-positive naïve CD4+ T cells isolated from neonates or adults and activated for 2, 4, 8, or 24 hours (n = 3). CD69: **P = 0.003, F = 41.9; CD44: *P = 0.01, F = 20; IL2RA: *P = 0.01, F = 17.1, two-way ANOVA. (H and I) Representative kinetic profile of oxygen consumption rate (OCR) (left) and of extracellular acidification rate (ECAR) (middle) measured in neonatal or adult (H) naïve CD4+ T cells or (I) activated for 24 hours. Total ATP production rate (right) is represented as mitochondrial and glycolytic ATP (n = 3). ***P < 0.001, F = 303, two-way ANOVA. **P = 0.002, F = 20.4, two-way ANOVA. (J) Percentage of puromycin-positive neonatal or adult naïve CD4+ T cells (n = 3). **P = 0.003, unpaired two-tailed t test. (K) Schematic representation of polysome profiling technique (left). Polysome profile of neonatal or adult CD4+ T cells activated for 36 hours (right). All data represent mean ± SEM; n refers to individuals.
These findings highlight that neonatal naïve CD4+ T cells are in a quiescent state but are also poised in terms of metabolic processes and protein remodeling, ready for a rapid activation.
LINE1 expression is absent in neonatal naïve T cells due to tonic TCR signaling
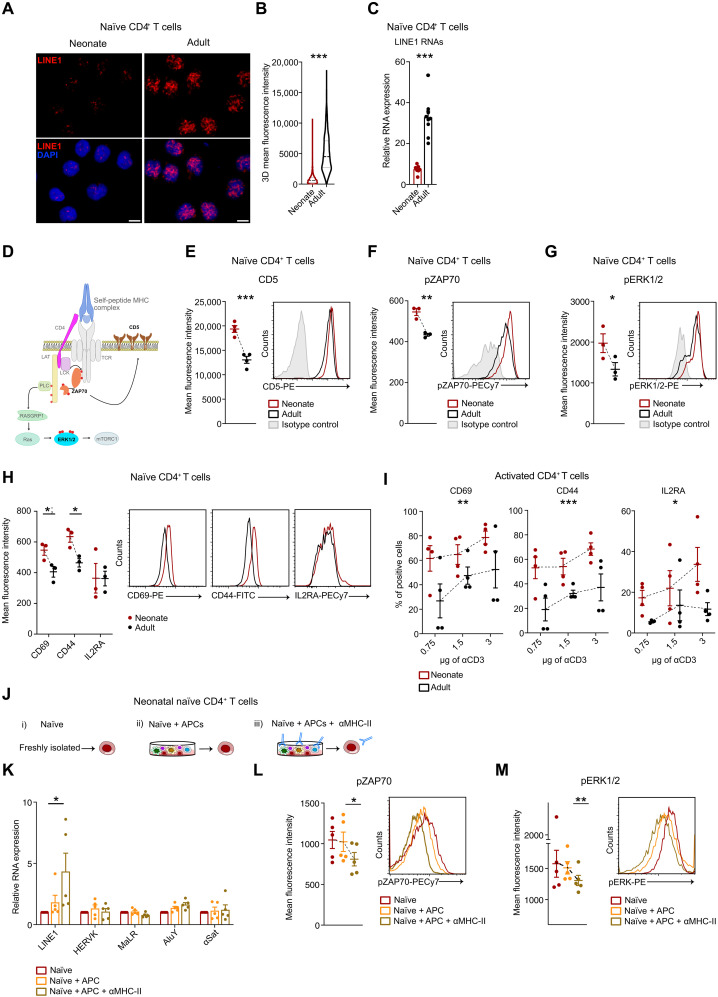
Given that LINE1 expression is known to enforce the quiescence of adult naïve T cells (15), we examined whether LINE1 is similarly expressed in quiescent neonatal naïve CD4+ T cells. We used RNA-FISH and RT-qPCR to assess LINE1 expression, revealing that LINE1 was not expressed in neonatal naïve T cells compared to adult counterparts (Fig. 2, A to C). Specifically, the L1M families, which are mainly expressed in adult naïve T cells, were found to be notably less abundant in neonatal naïve T cells, as revealed by RNA sequencing (RNA-seq) analysis (fig. S2, A and B).
Fig. 2. The expression of LINE1 is absent in neonatal naïve CD4+ T cells due to tonic TCR stimulation.
(A) LINE1 RNA-FISH in naïve CD4+ T cells from neonates or adults, magnification 63×; scale bar, 5 μm, and (B) relative quantification (at least 500 nuclei, n = 3). ***P < 0.001, unpaired two-tailed t test. (C) LINE1 expression by RT-qPCR in naïve CD4+ T cells from neonates (n = 7) or adults (n = 9). ***P < 0.001, unpaired two-tailed t test. (D) Schematic representation of tonic TCR signaling. (E) CD5, (F) phospho-ZAP70 (Tyr319), and (G) phospho-ERK1/2 (Thr202/Tyr204) MFI in neonatal or adult naïve CD4+ T cells. CD5 (n = 4): ***P < 0.001, unpaired two-tailed t test; phospho-ZAP70 (n = 3): **P = 0.0039, unpaired two-tailed t test; phospho-ERK1/2 (n = 3): *P = 0.04, unpaired one-tailed t test. (H) CD69, CD44, and IL2RA MFI in neonatal or adult naïve CD4+ T cells (n = 3). *P = 0.0432; *P = 0.02, unpaired two-tailed t test. (I) Percentage of CD69-, CD44-, and IL2RA-positive naïve CD4+ T cells activated with different anti-CD3 concentrations (n = 4). CD69: **P = 0.0074, F = 9.111; CD44: ***P = 0.0002, F = 20.89; IL2RA: *P = 0.0117, F = 7.870, two-way ANOVA. (J) Schematic representation of the experiments in (K) to (M). Neonatal naïve CD4+ T cells are either freshly processed or cultured in complete medium with autologous APC, with or without α-MHC-II blocking antibodies. (K) LINE1, HERVK, MaLR, AluY, and α-Satellite expression by RT-qPCR (n = 5). *P = 0.05, paired one-tailed t test. (L) Phospo-ZAP70 (Tyr319) and (M) phospo-ERK1/2 (Thr202/Tyr204) MFI (n = 5). *P = 0.0102; **P = 0.0041, paired two-tailed t test. All data represent mean ± SEM; n refers to individuals.
Thymic selection is developmentally regulated and facilitates the production of naïve CD4+ T cells bearing TCR with a higher affinity for self-peptides during early life stages (7, 18). This self-recognition arises from transient interactions between TCR and self-major histocompatibility complex (MHC) peptides presented by antigen-presenting cells (APC), generating subthreshold signaling events in naïve T cells, known as tonic TCR stimulation (19, 20). In our previous work, we reported that TCR stimulation regulates the down-regulation of LINE1 expression in adult T cells (15). As further evidence, by inhibiting the first event of the TCR signaling cascade with PP2 that blocks the lymphocyte-specific protein tyrosine kinase (Lck), the down-regulation of LINE1 expression following TCR stimulation was no longer observed (fig. S2C). Therefore, we investigated the possibility of tonic TCR stimulation occurring in neonatal naïve CD4+ T cells and its potential role in the down-regulation of LINE1 expression. Initially, we examined the expression of CD5, a known reporter for TCR self-reactivity and tonic stimulation (20, 21), and observed that neonatal naïve T cells were predominantly CD5 positive (Fig. 2, D and E). In line with this, ZAP70 was observed to be phosphorylated on Tyr319, and ERK1/2 on Thr202/Tyr204 (Fig. 2, F and G, and fig. S2D). To substantiate these findings, we observed that neonatal naïve T cells already exhibited the expression of early activation markers, namely, CD69 and CD44 (Fig. 2H). The late activation marker IL2RA, in contrast, does not show differences in expression, consistent with a quiescent phenotype (Fig. 2H). Furthermore, neonatal T cells maintained an elevated expression of all these markers under low-dosage TCR stimulation (Fig. 2I and fig. S2E). These data provided evidence of the activation of tonic TCR signal transduction in neonatal naïve T cells. To evaluate the dependency of LINE1 expression on tonic TCR signaling, we analyzed LINE1 expression in freshly isolated neonatal naïve T cells and those cultured with autologous APC, as well as autologous APC blocked with α-MHC-II antibodies (inhibiting tonic TCR signaling). Blocking MHC class II on APC increased LINE1 expression and reduced phosphorylation of ZAP70 and ERK1/2 in neonatal naïve T cells (Fig. 2, J to M). In contrast, T cells cultured with APC showed no significant changes in LINE1 expression and maintained phosphorylation of ZAP70 and ERK1/2 (Fig. 2, J to M). Similar trends were observed in neonatal naïve T cells cultured in medium without APC (fig. S2, F to H). As a control, we tested the expression of other repetitive elements (i.e., HERVK, MaLR, AluY, and α-Satellite) and found no significant changes (Fig. 2K). This suggests that a global transcriptional derepression is not occurring upon tonic TCR stimulation, indicating a specific effect on LINE1 expression. In contrast, in adult naïve CD4+ T cells, no significant differences in LINE1 expression were observed under the same conditions (fig. S2I), supporting the hypothesis of intrinsic developmental differences in TCR sensitivity across different life stages.
Overall, these results demonstrate that LINE1 expression is down-regulated in neonatal naïve CD4+ T cells because of tonic TCR stimulation and suggest that LINE1 expression may be differentially regulated in naïve CD4+ T cells across various developmental ages.
In neonatal naïve CD4+ T cells, LINE1 expression is suppressed by mTORC1 activity via PTBP1
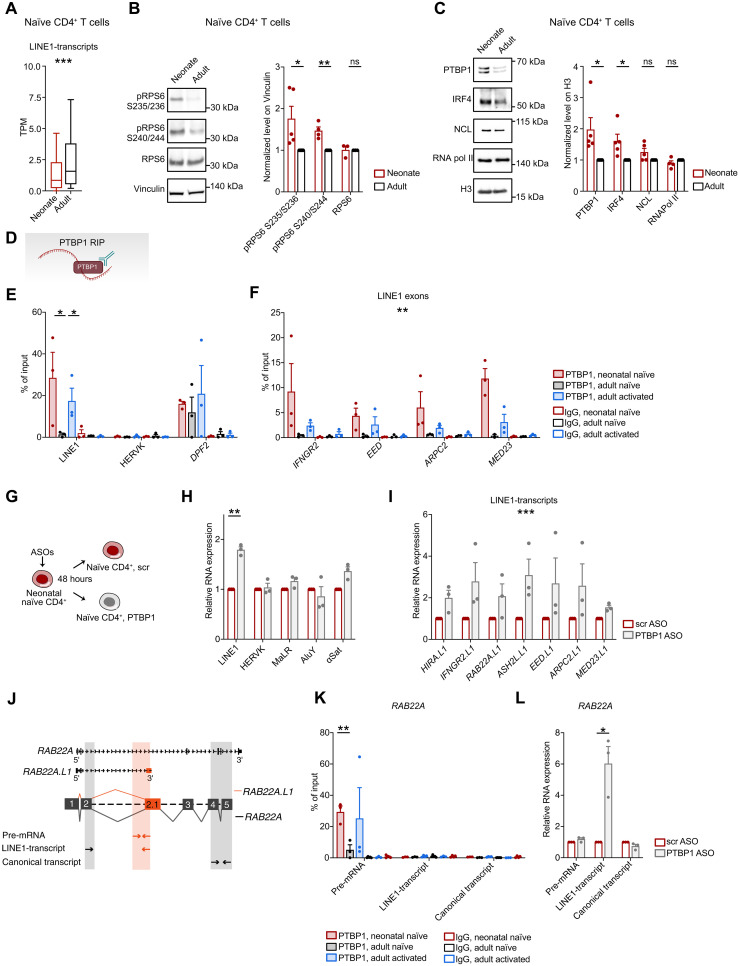
We sought to investigate whether the mechanism responsible for the observed decrease in LINE1 expression in neonatal naïve T cells resembles that documented for adult T cells (15). Specifically, in quiescent adult naïve CD4+ T cells, intronic LINE1 are spliced into exons of previously unkown transcript variants regulated by interferon regulatory factor 4 (IRF4) and maintained at chromatin by NCL. Upon TCR stimulation, mTORC1 is activated and promotes LINE1-transcript down-regulation through the splicing suppressor PTBP1. Initially, we quantified the expression of the 461 LINE1-transcripts identified in our previous study (15) in RNA-seq data and RT-qPCRs from neonatal and adult naïve CD4+ T cells, demonstrating that they are specifically down-regulated in neonates (Fig. 3A, fig. S3A, and table S1). Then, we proved that the mTORC1 pathway was activated in neonatal naïve T cells, as indicated by increased phosphorylation of the ribosomal protein S6 (RPS6) at both Ser235/236 and Ser240/244 (Fig. 3B). To further investigate the role of mTORC1 in down-regulating LINE1 in neonatal naïve T cells, we inhibited its activity with rapamycin and observed a subsequent increase in LINE1 expression (fig. S3B). Considering that robust and tonic mTORC1 signals resulting from self-antigen interactions may influence cell fate decisions in adult naïve T cells (20), we inquired whether a similar scenario occurs in neonatal naïve T cells. In line with a tonic activation pattern, we observed that mTORC1 signaling in neonates is below the threshold when compared to the levels experienced during a short TCR stimulation (fig. S3C). In addition, we observed the elimination of mTORC1 activity in neonatal naïve CD4+ T cells that were cultured without self-antigen presentation by autologous APC (fig. S3D). Next, we analyzed the potential changes in protein levels of regulators of LINE1-transcripts in neonatal naïve CD4+ T cells. We determined that PTBP1 and, to a lesser extent, IRF4 were up-regulated in neonates (Fig. 3C and fig. S3E).
Fig. 3. mTORC1/PTBP1 suppress LINE1 expression in neonatal naïve CD4+ T cells.
(A) Expression levels of the 461 LINE1-transcripts identified in (15) in RNA-seq datasets (n = 4). ***P = 2.2 × 10−16, unpaired two-tailed Wilcoxon rank sum test. (B and C) Western blot analysis and quantification of (B) RPS6, phospho-RPS6 (Ser235/236 and Ser240/244) and (C) PTBP1, IRF4, NCL, and RNA Pol II in neonatal or adult naïve CD4+ T cells (Ser235/236, PTBP1, and IRF4, n = 5; Ser240/244, NCL, and RNA pol II, n = 4; RPS6, n = 3). Data were normalized on Vinculin or H3. Ser235/236: *P = 0.0356; Ser240/244: **P = 0.0022; PTBP1: *P = 0.0352; IRF4: *P = 0.0205, unpaired two-tailed t test. (D) Schema of PTBP1 RIP assay. (E and F) RIP for PTBP1 in neonatal or adult naïve and adult activated CD4+ T cells (n = 3). LINE1: neonatal versus adult naïve: *P = 0.05; adult naïve versus activated: *P = 0.03, unpaired one-tailed t test. LINE1 exons: neonatal versus adult naïve: **P = 0.001, F = 16, two-way ANOVA. (G) Schematic representation of PTBP1 knockdown with ASOs. (H) LINE1, HERVK, MaLR, AluY, α-Satellite, and (I) LINE1-transcripts expression by RT-qPCR in neonatal naïve CD4+ T cells treated with PTBP1 or control (scr) ASOs (n = 3). **P = 0.006, paired two-tailed t test; ***P < 0.001, F = 20.8, two-way ANOVA. (J) Scheme of RT-qPCR for RAB22A transcripts to inspect PTBP1 regulation on pre-mRNA, LINE1-transcripts, or canonical transcripts. (K) RIP for PTBP1 in neonatal or adult naïve and adult activated CD4+ T cells (n = 3). Neonatal versus adult naïve: **P = 0.009, unpaired two-tailed t test. (L) RAB22A isoform expression by RT-qPCR in neonatal naïve CD4+ T cells treated with PTBP1 or scr ASOs (n = 3). *P = 0.0445, paired two-tailed t test. All data represent mean ± SEM; n refers to individuals.
PTBP1 has been reported to bind preferentially intronic LINE1 sequences (22). To validate the role of PTBP1 as a LINE1-transcript splicing suppressor in neonatal naïve CD4+ T cells, we first performed RNA immunoprecipitation (RIP) experiments, which showed that PTBP1 specifically binds LINE1 exons (Fig. 3, D to F). Controls with adult naïve and activated T cells confirmed PTBP1 binding to LINE1 exons only upon activation, when the splicing suppression of LINE1 exons has occurred, aligning with our earlier findings (15). No enrichment was observed on HERVK, suggesting that PTBP1 does not indiscriminately bind other transposable elements (Fig. 3E). Furthermore, we found no differences in PTBP1 binding to the DPF2 splicing variant, known to be regulated by PTBP1, across various cell types (23) (Fig. 3E). Second, we used antisense oligonucleotides (ASO) to silence PTBP1 and observed the re-expression of the previously identified LINE1-transcripts as well as of LINE1, but not of other repetitive elements (i.e., HERVK, MaLR, AluY, and α-Satellite) or splicing variants reported to be regulated by PTBP1 (i.e., DPF2 and RPN2) (23) (Fig. 3, G to I, and fig. S3, F to J). Last, consistent with its role as a splicing suppressor of LINE1 exons, we demonstrated that PTBP1 binds the RAB22A pre-mRNA in neonatal naïve T cells and adult activated T cells, but not the spliced mature LINE1-transcript or the canonical protein-coding transcript variant (Fig. 3, J and K). Depleting PTBP1 with ASOs in neonatal naïve CD4+ T cells specifically increased the expression of the LINE1-transcript isoforms (Fig. 3L).
Overall, these results suggest that in neonatal naïve CD4+ T cells, the inhibition of LINE1 expression may be due to tonic mTORC1 activity and increased splicing suppression mediated by PTBP1, which emerges to be primarily specific to LINE1 in agreement to what is already reported by Attig and colleagues (22).
LINE1 expression regulates the rate of protein synthesis and cell cycle progression in CD4+ T cells
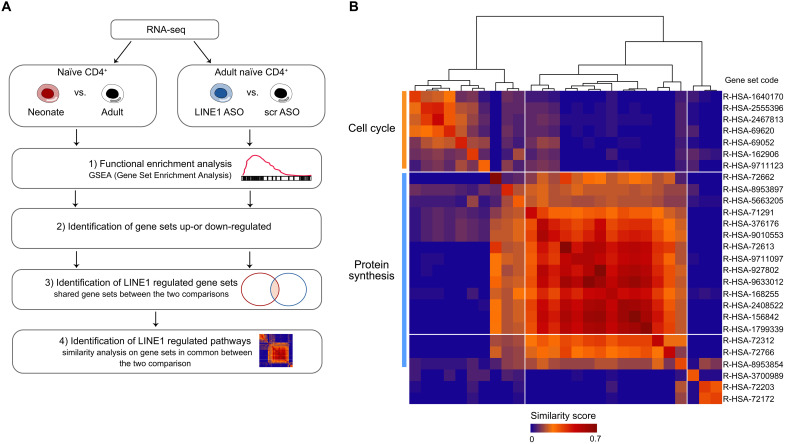
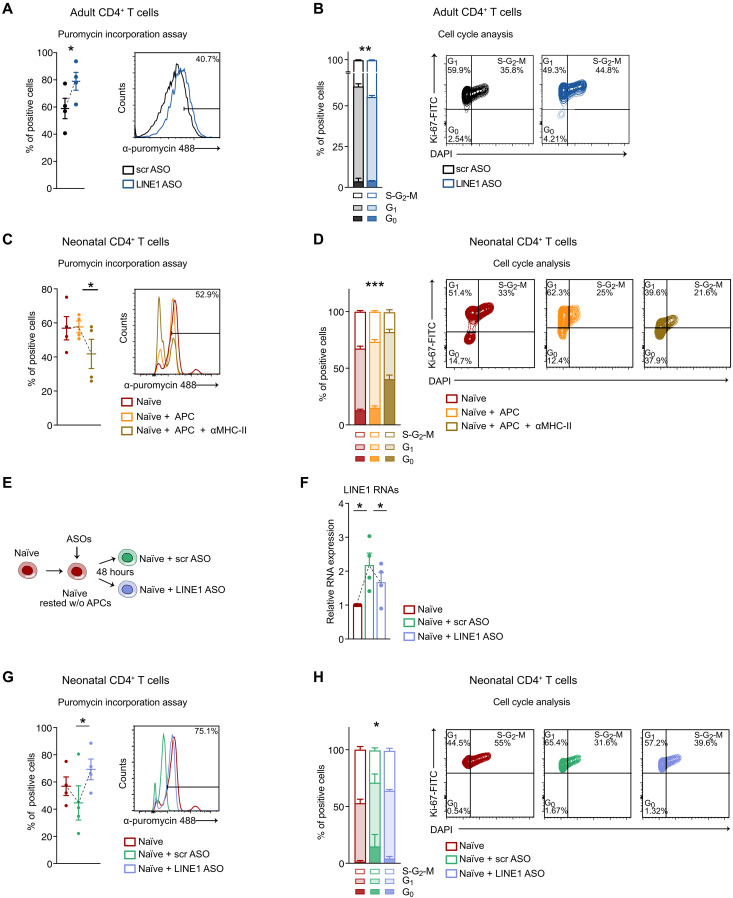
In our previous work, we established that LINE1-transcripts are a crucial regulator of T cell functions playing a role in pausing the expression of genes associated with cellular activation to maintain the quiescent state of naïve CD4+ T cells (15). These findings led us to investigate how the absence of LINE1-transcripts in neonatal naïve T cells might contribute to their quiescent state poised for rapid activation. To distinguish the biological effects attributed to the tonic activation of TCR/mTORC1 from those resulting from the absence of LINE1, we identified classes of genes and biological processes that were consistently altered due to the absence of LINE1 in (i) neonatal versus adult naïve CD4+ T cells and in (ii) adult naïve CD4+ T cells where LINE1 were down-regulated using LINE1 ASOs compared to control cells. We confirmed that the mTORC1 pathway is not active in adult naïve T cells treated with LINE1 ASOs by measuring the level of the target phospho-RPS6 (fig. S4, A and B). Therefore, we conducted RNA-seq under the aforementioned conditions and identified, by gene set enrichment analysis (GSEA), gene sets that were commonly up-regulated or down-regulated in the two comparisons (Fig. 4A). In the neonatal versus adult comparison, we identified 101 up-regulated and 4 down-regulated gene sets, while in the LINE1 ASOs versus control comparison, we found 43 up-regulated and 10 down-regulated gene sets (table S2). Notably, there were 27 shared up-regulated gene sets, while no overlap was observed among the down-regulated ones (fig. S4C and table S2). Hence, we extracted significantly core-enriched genes from the commonly up-regulated gene sets and performed similarity analysis. This analysis revealed a primary cluster of gene sets associated with protein synthesis and a secondary one associated with cell cycle progression (Fig. 4B and table S2). These processes are pivotal for initiating a rapid execution of the T cell activation program (24). Conversely, gene sets showing specific up-regulation in either of the comparisons were associated with different terms, such as metabolism, signal transduction, development, and differentiation (fig. S4, D and E). As discussed earlier, protein synthesis and cell cycle progression are both enhanced in neonatal T cells compared to adults (Fig. 1, F, J and K, and fig. S1H). On the basis of these results, we further investigated the dependence of these two processes on LINE1 expression. Through puromycin incorporation, we confirmed an increase in the rate of protein synthesis in adult T cells where LINE1-transcripts were down-regulated compared to control cells (Fig. 5A). In addition, we observed a higher percentage of cells in the S, G2, or M phases (Fig. 5B), indicating an elevated rate of progression through the cell cycle in the absence of LINE1 expression. To ensure specificity, we knocked down HERVK and found no effects on protein synthesis and cell cycle progression (fig. S5, A and B). To validate the dependency of these pathways on LINE1 expression in neonatal T cells, we analyzed the rate of protein synthesis and cell cycle progression in neonatal CD4+ T cells cultured with autologous APC, comparing conditions with and without MHC-II complex blockade, which modulated LINE1 expression (Fig. 2K). We observed significant down-regulation of protein synthesis and cell cycle progression when blocking tonic TCR stimulation, concomitantly with LINE1 re-expression (Fig. 5, C and D). To formally confirm our hypothesis, we promoted LINE1 expression in neonatal T cells by culturing them without tonic TCR stimulation and then proceeded to knock down LINE1 with ASOs (Fig. 5, E and F). We observed a decrease in protein synthesis and cell cycle progression in neonatal T cells cultured without stimulation (Fig. 5, G and H). This effect was reversed by LINE1 depletion (Fig. 5, G and H). Last, treating neonatal T cells with PTBP1 ASOs to re-express LINE1-transcripts (Fig. 3I) resulted in a consistent reduction of protein synthesis and cell cycle progression (fig. S5, C and D).
Fig. 4. The expression of LINE1 in T cells plays a regulatory role in processes related to protein synthesis and cell cycle.
(A) Diagram illustrating the workflow used for identifying commonly altered pathways in RNA-seq datasets, comparing neonatal versus adult naïve CD4+ T cells and adult naïve CD4+ T cells depleted of LINE1 (LINE1 ASOs) versus control cells (scr ASOs): (1) GSEA was conducted on “Curated Reactome.” (2) Identification of gene sets that were either up-regulated or down-regulated. (3) Determination of LINE1-regulated gene sets as those shared between the two comparisons. (4) Identification of shared enriched genes within each gene set identified in (3) and relative similarity analysis. (B) Jaccard coefficients were calculated for core-enriched genes within commonly up-regulated gene sets from the two GSEA comparisons. These coefficients were clustered to assess cross-similarity among gene sets, highlighting that most of these gene sets are associated with molecular pathways related to mRNA translation and cell cycle progression.
Fig. 5. LINE1 expression regulates protein synthesis and cell cycle progression.
(A) Percentage of puromycin-positive adult CD4+ T cells treated with LINE1 or scr ASOs and activated for 24 hours (n = 4). *P = 0.04, paired two-tailed t test. (B) Cell cycle analyses in adult CD4+ T cells treated with LINE1 or scr ASOs and activated for 72 hours (n = 3). **P = 0.009, F = 11.2, two-way ANOVA. (C) Percentage of puromycin-positive neonatal CD4+ T cells either freshly processed or cultured in complete medium with autologous APC, with or without α-MHC-II, and activated for 24 hours (n = 4). *P = 0.03, paired one-tailed t test. (D) Cell cycle analyses in neonatal CD4+ T cells either freshly processed or cultured in complete medium with APC, with or without α-MHC-II, and activated for 72 hours (n = 3). ***P < 0.001, F = 38.7, two-way ANOVA. (E) Schematic representation of the strategy used to induce LINE1 re-expression followed by subsequent depletion in neonatal naïve CD4+ T cells [experiments in (F) to (H)]. Neonatal naïve CD4+ T cells were either freshly isolated or cultured in complete medium to re-express LINE1 and subsequently treated with LINE1 or scr ASOs (F) LINE1 expression by RT-qPCR in neonatal naïve CD4+ T cells (n = 4). Naïve versus naïve + scr ASOs: *P = 0.02; naïve + scr ASOs versus naïve + LINE1 ASOs: *P = 0.05, paired two-tailed t test. (G) Percentage of puromycin-positive neonatal naïve CD4+ T activated for 24 hours (n = 4). *P = 0.03, paired one-tailed t test. (H) Cell cycle analyses in neonatal naïve CD4+ T cells activated for 72 hours (n = 3). *P = 0.04, F = 3.26, two-way ANOVA. All data represent mean ± SEM; n refers to individuals.
Therefore, our findings establish that LINE1 expression controls protein synthesis and cell cycle progression in T cells, contributing to neonatal naïve T cells’ readiness to activation.
LINE1 are overexpressed in T cells during aging, leading to a reduced rate of protein synthesis
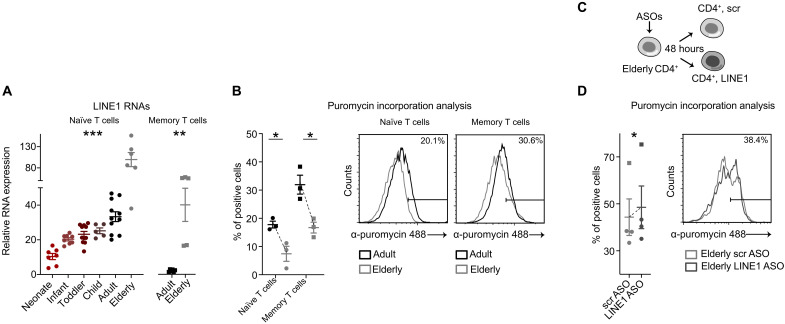
T cells exhibit variation in composition and function across different stages of life (1). In newborns, naïve T cells originate from fetal liver progenitors and mature in the thymus. As individuals grow, progenitor cells shift to the bone marrow, and the thymus begins to regress. In adults, the peripheral pool of naïve T cells is mainly replenished through homeostatic proliferation, with memory T cells comprising a substantial portion (1) (fig. S1A). Thymic involution culminates in individuals above 70 years of age, with only 5% of the initial thymic output of naive T cells remaining (25). Hence, the T cell repertoire is mostly composed of memory T cells and is characterized by immunological senescence and exhaustion (26). We sought to understand the expression of LINE1 elements across the life span from early life to aging, and its impact on T cell functions. To explore this, we analyzed LINE1 expression in naïve CD4+ T cells from various age groups: newborns, infants (0 to 2 years), toddlers (3 to 6 years), children (7 to 13 years), adults (>20 and <65 years), and elderly (>80 years) (table S3). For adults and elderly individuals, we assessed LINE1 expression also for memory T cells. Our findings showed a gradual rise in LINE1 expression in naïve T cells during maturation, starting at minimal levels in neonates, increasing to intermediate levels in toddlers and children, further rising in adults, and peaking in elderly individuals (Fig. 6A, fig. S6A, and table S4). While adult memory CD4+ T cells displayed no LINE1 expression as we have already reported (15), we observed a re-accumulation of LINE1 RNAs in memory CD4+ T cells of elderly individuals (Fig. 6A). RNA-seq analysis on adult and elderly naïve T cells, followed by GSEA, revealed that gene sets associated with protein translation, but not with the cell cycle, are specifically down-regulated in aging (fig. S6B and table S5). Of note, protein synthesis is a pivotal process for T cell activation that supports cell proliferation and function (24). Puromycin incorporation revealed that both naïve and memory CD4+ T cells isolated from elderly individuals showed diminished protein synthesis compared to adults (Fig. 6B). To further validate the association between LINE1 expression and protein synthesis levels, we down-regulated LINE1 with ASOs in naïve and memory CD4+ T cells derived from elderly individuals (Fig. 6C). We observed that LINE1 down-regulation increased protein synthesis (Fig. 6D). This set of data suggests that LINE1 expression could be considered a quantitative parameter that reflects T cell functionality across the life span.
Fig. 6. LINE1 are overexpressed in T cells during aging, reducing the rate of protein synthesis.
(A) Left, LINE1 expression by RT-qPCR in naïve CD4+ T cells isolated from individuals across various age groups: neonates, 0 to 2 years old, 3 to 6 years old, 7 to 13 years old, adults (<65 years old), and elderly (>80 years old). Each data point corresponds to an individual (n = 48). ***P < 0.001, F = 25.6, one-way ANOVA. Right, LINE1 expression by RT-qPCR in memory CD4+ T cells isolated from adult or elderly individuals (n = 11). **P = 0.002, unpaired two-tailed t test. (B) Percentage of puromycin-positive naïve or memory CD4+ T cells isolated from adult or elderly individuals (n = 3). Naïve adult versus elderly: *P = 0.02; memory adult versus elderly: *P = 0.02, unpaired two-tailed t test. (C) Schematic representation of the strategy of LINE1 knockdown with ASOs in elderly naïve and memory CD4+ T cells. (D) Percentage of puromycin-positive naïve or memory CD4+ T cells treated with LINE1 or scr ASOs and activated for 24 hours (naïve, n = 3; memory, n = 1). *P = 0.03, paired one-tailed t test. All data represent mean ± SEM; n refers to individuals.
DISCUSSION
Here, we delve deeper into elucidating the functional implications of expressed LINE1 in shaping the immunological response of CD4+ T cells. Recent findings from our previous work indicate that the expression of evolutionarily old LINE1 elements, particularly L1M, incorporated into exons of transcript variants resulting from alternative splicing, plays a pivotal role in maintaining T cell quiescence in adult T cells (15). These LINE1-transcripts undergo down-regulation upon TCR activation, highlighting a regulatory mechanism that governs the transition from quiescence to activation (15). On the basis of these results, we have now investigated the dynamics of LINE1 expression in neonatal naïve CD4+ T cells to uncover potential mechanisms that could account for the rapid mobilization of neonatal T cells in response to antigen treats. We discovered that in neonatal naïve CD4+ T cells, the expression of LINE1 is maintained at a low level due to tonic TCR signaling mediated by autologous APC. This signaling is adequate to initiate and sustain basal mTORC1 activity in neonates, leading to the suppression of LINE1-transcript splicing. This suppression is mediated by PTBP1, similar to the mechanism described in adults (15). Our findings show that the absence of LINE1 expression enhances the protein synthesis rate and cell cycle progression in neonatal naïve T cells, contributing to their quiescent state poised for rapid activation.
Naïve T cells are known for being exceptional long-living cells in humans and follow distinct trajectories during development and throughout life. In early life and childhood, naïve T cells originate in the thymus, where TCR pools expand and diversify in a developmentally regulated manner (1). Following puberty, the thymus experiences progressive involution, and naïve T cells are generated through homeostatic replication (1). Thymic involution culminates in elderly individuals (26). Our findings suggest that LINE1 expression is subjected to developmental regulation, showing a progressive rise from neonates to toddlers, children, and adults. This gradual increase may contribute to enforcing T cell quiescent state through development and the life span by specifically regulating protein synthesis rate. Moreover, T cells undergo notable changes over the course of a lifetime, particularly in elderly individuals, where T cells frequently display senescent and exhausted features (27, 28), contributing to heightened susceptibility to infectious and/or chronic diseases and suboptimal responses to vaccines (29). Recent studies have illustrated the tight regulation of transposable element (TE) expression in the tumor microenvironment (30, 31) during the development of T cell exhaustion (15, 32). In cases of exhausted T lymphocytes infiltrating tumors, LINE1 elements have been shown to be re-expressed (15), and the use of ASOs to pharmacologically target LINE1 has been shown to restore proper immunological responses. In line with this evidence, in the current study, we have demonstrated that LINE1 are overexpressed in aged T cells, inducing a concomitant decrease in the protein synthesis rate and contributing to the immunosenescent phenotype. These findings highlight the potential of expressed LINE1 as previously unidentified inhibitors of immune responses (15, 33). Hence, investigating the impact of LINE1 on T cell functions offers the potential to design innovative immunomodulatory molecules aimed at safeguarding large segments of the population. Our study has proved that LINE1 expression plays a pivotal role in regulating protein synthesis and cell cycle progression, thereby arming T cells for an effective immunological response; however, additional investigations are required to fully clarify the underlying mechanisms. Protein synthesis stands as a fundamental aspect of T cell activation, proliferation, and differentiation, and different studies propose that its regulation in T cells can occur through diverse mechanisms. These mechanisms include the priming of the mRNA translational machinery (17), oversight of energy production and metabolism (34), modulation of amino acid content (35), availability of transcription factors (i.e., cMyc) (36), and the activity of mTORC1 (37). Our study identifies LINE1 as emergent contributors to the regulation of protein synthesis. It is worth highlighting that in mouse embryonic stem cells, full-length, evolutionarily young LINE1 have been reported to positively regulate ribosome biogenesis (38). However, the types of LINE1 sequences and their copy numbers and localization within genomes are species specific (39). Therefore, their regulatory function may depend on the species and the biological context in which they are expressed. In the immune system, the level of LINE1 expression seems to represent an important hallmark of T cell functionality and ability to respond to antigen treats. While the absence of LINE1 expression may be a crucial element contributing to neonatal immune system’s rapid adaptability and effectiveness in defending against novel infections, its overexpression promotes immune senescence and a dysfunctional phenotype. We propose that this is an adaptive characteristic in the initial years of life, tailored to cope with a dynamically changing environment containing a diverse array of pathogens previously unencountered. In contrast, the aberrant accumulation of these transcripts in aging may result from impairments in the mechanisms of LINE1-transcript biogenesis, which requires further investigation.
In conclusion, this study underscores the regulatory role of expressed LINE1 in shaping T cell biology and function across the human life span. Moreover, it sets the stage for future investigation into the potential regulatory roles of LINE1 across diverse contexts, including intrauterine life, health, vaccination, infections, and diseases, spanning both adaptive and innate immunity and other tissues and cell types.
MATERIALS AND METHODS
Human blood samples
Umbilical cord blood samples were collected from healthy full-term neonates (≥37 weeks of gestational age) born by cesarean section and provided by the Neonatal Intensive Care Unit at Mangiagalli Center at Fondazione Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) Cà Granda Ospedale Maggiore Policlinico in Milan. Peripheral blood from healthy adult donors was provided by the Department of Transfusion Medicine and Hematology at IRCCS Cà Granda Ospedale Maggiore Policlinico in Milan. Peripheral blood from child donors was provided by the Nephrology, Dialysis, and Transplant Unit at IRCCS Cà Granda Ospedale Maggiore Policlinico in Milan. All the samples were completely anonymized at the source. Ethics committees of the hospital approved the use of human samples for research purposes, and informed consent was obtained from all participants (authorization nos. 708_2020 and 8263773).
T cell purification and sorting
Human peripheral blood mononuclear cells (PBMCs) and umbilical cord blood mononuclear cells (UCB-MCs) were purified from blood samples by density gradient centrifugation with Ficoll-Paque PLUS. PBMCs and UCB-MCs were stained with antibodies specific for surface markers and sorted by flow cytometry. Naïve CD4+ T cells were sorted as CD4+ CD45RA+ CD45RO−, and APC were sorted as CD4dim, CD8−, CD14+, and CD1c+ with FACSAria SORP (BD Biosciences) to isolate monocytes, B cells, and dendritic cells. The following antibodies were used for flow cytometry–based sorting: αCD4-VioGreen (Miltenyi Biotec, catalog no. 130-113-221; AB_2726032) or αCD4-APC-Cy7 (BD Bioscience, catalog no. 557871; AB_396913); αCD45RA-PECy5 (BD Bioscience, catalog no. 552888; AB_394517); αCD45RO-APC (Miltenyi Biotec, catalog no. 130-113-546; AB_2733383) or αCD45RO-BV605 (BioLegend, catalog no. 304237; AB_2562143); αCD8-VioBlue (Miltenyi Biotec, catalog no. 130-110-683; AB_2659239); αCD14-APC (BD Bioscience, catalog no. 555399; AB_398596); and αCD1c-FITC (Miltenyi Biotec, catalog no. 130-113-301; AB_2726080).
T cell culturing, in vitro activation, and TCR titration
For T cell activation, sorted naïve CD4+ T cells were cultured at a concentration of 1.5 × 106 cells/ml in T-helper 1 (TH1) medium and activated with Dynabeads Human T-activator anti-CD3/anti-CD28 beads (Gibco, catalog no. 1131D). TH1 medium is composed of complete RPMI [RPMI with GlutaMAX-I (Gibco), 10% (v/v) fetal bovine serum (Gibco), 1% (v/v) nonessential amino acids, 1 mM sodium pyruvate, penicillin (50 U/ml), and streptomycin (50 μg/ml)] supplemented with TH1-specific cytokines [recombinant IL-2 (20 IU/ml; Miltenyi Biotec; catalog no. 130-097-744), recombinant IL-12 (10 ng/ml; Miltenyi Biotec; catalog no. 130-096-704), and neutralizing anti-IL-4 (2 μg/ml; Miltenyi Biotec; catalog no. 130-095-753)]. For the TCR titration assay, neonatal and adult naïve CD4+ T cells were cultured for 16 hours at a concentration of 1.5 × 106 cells/ml in complete RPMI supplemented with IL-2 (20 IU/ml) in MaxiSorp Nunc-Immuno 96-well plates precoated with 2 μg of anti-CD28 (BioLegend, catalog no. 302933, RRID:AB_11150591) and different concentrations of anti-CD3 [i.e., 0.75, 1.5, or 3 μg of anti-CD3 (BioLegend, catalog no. 317325, RRID:AB_11147370)]. Cells were then evaluated by FACS (fluorescence-activated cell sorting) analysis for the expression of CD69, CD44, and IL2RA activation markers. For the assay involving naïve CD4+ T cells maintained in culture with autologous APC, with or without an antibody against MHC-II molecules, the procedures were as follows: (i) naïve T cells alone: FACS-sorted naïve T cells were used for subsequent analysis as freshly isolated naïve CD4+ T cells. (ii) Naïve T cells cultured with autologous APC: FACS-sorted naïve CD4+ T cells were cultured at a 1:3 ratio with FACS-sorted APC at a final concentration of 5.5 × 106 cells/ml in complete RPMI. (iii) Naïve T cells cultured with autologous APCs plus anti-MHC-II antibodies: 2.5 × 106 APC were incubated with 30 μg of α-MHC-II antibody in 300 μl of complete RPMI for 40 min at 37°C. After incubation, the excess antibody was washed out, and the APC were incubated with naïve T cells as described in point (ii). After coculture, naïve T cells were magnetic FACS-isolated to be subjected to subsequent analysis: for intracellular staining of phospho-Zap70 and phospho-ERK1/2, cells were cultured for 1 hour; for puromycin and cell cycle analysis, cells were cocultured for 1 hour, FACS isolated, and activated for 24 hours; last, for the RT-qPCR assay, cells were cocultured for 24 hours. T cells were maintained at 37°C in a 5% CO2 humidified incubator.
T cell treatments
To inhibit TCR signaling cascade, adult naïve CD4+ T cells were activated with Dynabeads Human T-activator anti-CD3/anti-CD28 beads (Gibco, catalog no. 1131D) in TH1 medium and treated with or without 10 μM PP2 (Merck; catalog no. P0042-5MG). T cells were treated for 8 hours and then analyzed by RT-qPCR analysis.
To inhibit tonic mTORC1 activity in quiescent neonatal naïve CD4+ T cells, neonatal UCB-MCs (15 × 106 cells/ml) were cultured in complete RPMI with the addition of 125 nM rapamycin (Merck; catalog no. R8781) for 24 hours as done in (15). Subsequently, quiescent neonatal naïve CD4+ T cells were magnetic FACS isolated from the rapamycin-treated UCB-MCs and subjected to RT-qPCR analysis. FANA-ASOs (2′-deoxy-2′-fluoro-β-d-arabinonucleic acid) were used to down-regulate LINE1, PTBP1, and HERVK RNAs, following the sequences and usage protocol outlined in (15). An unrelated scr ASO was used as a control. ASOs were administered without any transfection reagent, following the manufacturer’s instruction, at a final concentration of 10 μM. Quiescent neonatal, adult, or elderly naïve or memory CD4+ T cells were cultured for 48 hours in complete RPMI supplemented with recombinant IL-2 (200 IU/ml) and 10 μM of the specific ASOs. RNA or protein knockdown efficiency was assessed in naïve CD4+ T cells through RT-qPCR and/or flow cytometry analysis. After 48 hours of ASO treatment, naive CD4+ T cells treated with ASOs were activated (in the presence of 10 μM ASOs) and collected after 24 hours (for puromycin incorporation assay, see below) or 36 or 72 hours (for cell cycle analysis, see below). T cells were maintained at 37°C in a 5% CO2 humidified incubator.
T cell staining and flow cytometry analysis
For surface marker staining, 1 × 105 neonatal or adult naïve or activated CD4+ T cells were incubated with the antibody of interest (1:50) in MACS buffer (Miltenyi Biotec) for 30 min at 37°C. T cells underwent phosphate-buffered saline (PBS) washing and were subjected to FACS analysis. The following antibodies were used: CD5-PE (BD Bioscience, catalog no. 555353; AB_395757), CD69-PE (BD Bioscience, catalog no. 560968; AB_10565972), CD44-FITC (BD Bioscience, catalog no. 555478; AB_395870), or IL2RA-PECy7 (Invitrogen, catalog no. 25-0259-42; AB_1257140). For CD5 analysis, we used the matched isotype anti-mouse IgG1-PE as control (BD Bioscience, catalog no. 562027; AB_10894761). For PTBP1, NCL, and FOXP1 intracellular staining, 1 × 105 neonatal or adult naive CD4+ T cells were fixed for 30 min at 4°C with the Foxp3 Transcription Factor Fixation/Permeabilization kit (Invitrogen, catalog no. 00-5521-00) according to the manufacturer’s instructions. Cells were incubated with primary antibody in permeabilization buffer 1× (Invitrogen) for 1 hour at room temperature (RT), washed in PBS, incubated with secondary antibody in permeabilization buffer 1×, washed in PBS, and FACS analyzed. The following antibodies were used: PTBP1 (Abcam, catalog no. ab133734; AB_2814646, 1:500), NCL (Abcam, catalog no. ab22758; AB_776878, 1:500), FOXP1 (Cell Signaling, catalog no. 2005, AB_2106979; 1:300), and secondary antibodies conjugated with Alexa Fluor 488 (Invitrogen, catalog no. A32723 or A11034, 1:1000). For CyclinD1 intracellular staining, 1 × 105 neonatal or adult naive CD4+ T cells were fixed for 30 min at 4°C with the Foxp3 Transcription Factor Fixation/Permeabilization kit according to the manufacturer’s instructions, Cells were washed in PBS and stained with CyclinD1-FITC antibody (BD Bioscience, catalog no. 554109; AB_395246, 1:100) in permeabilization buffer 1× for 30 min at RT. Cells were washed with PBS and then analyzed at FACS. For phospho-ZAP70 and phospho-ERK1/2 intracellular staining, 1 × 105 neonatal or adult naive CD4+ T cells were fixed with Phosflow fix buffer I (BD Biosciences, 1:2 in PBS) for 10 min at 37°C, washed in PBS, and incubated for 30 min at RT in Phosflow perm buffer III (BD Biosciences) with anti-phosphoZAP70 (Tyr319)-PECy7 (BioLegend, catalog no. 683702; AB_2572026. 1:100) and anti-phospho-ERK1/2 (Thr202/Tyr204)-PE (BD Bioscience, catalog no. 561991; AB_10895978, 1:100). For phosphoZAP70 (Tyr319) analysis, we used the isotype anti-mouse IgG1-PECy7 as control (BioLegend, catalog no. 400125; AB_2861433); for phospho-ERK1/2 analysis, we used the isotype anti-mouse IgG1-PE as control (BD Bioscience, catalog no. 562027; AB_10894761). T cells underwent PBS washing and were subjected to FACS analysis. For cell cycle analysis, neonatal or adult, naive or activated (for 36 or 72 hours) CD4+ T cells were fixed for 30 min at 4°C with the Foxp3 Transcription Factor Fixation/Permeabilization kit according to the manufacturer’s instructions. Cells were washed in PBS and stained with Ki-67-FITC antibody (BD Bioscience, catalog no. 556026; AB_396302, 1:100) in permeabilization buffer 1× for 30 min at RT. Cells were washed in PBS and incubated with DAPI (4′,6-diamidino-2-phenylindole) (1:5000) in permeabilization buffer 1× for 10 min at RT and analyzed. An average of 104 cells were captured for each staining using FACSCANTO II (BD Biosciences), and data were subsequently analyzed with FlowJo v.10 software (SCR_008520).
Puromycin incorporation assay
Puromycin incorporation was assessed through flow cytometry, following a protocol similar to (17). Briefly, neonatal, adult or elderly CD4+ T cells were incubated with puromycin (5 μg/ml; Gibco, catalog no. A1113803) for 10 min at 37°C in a 5% CO2 humidified incubator. CD4+ T cells were fixed with the Foxp3 Transcription Factor Fixation buffer for 30 min at 4°C, followed by incubation with anti-puromycin–Alexa Fluor 488 antibody (Millipore, catalog no. MABE343-AF488; AB_2736875, 1:500) in permeabilization buffer 1×, for 30 min at RT. T cells were washed in PBS and analyzed, with untreated cells serving as controls for autofluorescence. An average of 104 cells were captured for each staining using FACSCANTO II (BD Biosciences) and data were subsequently analyzed with FlowJo v.10 software.
Polysome profiling
At least 25 × 106 adult or neonatal naïve CD4+ T cells were activated for 36 hours and then treated for 10 min with cycloheximide (100 μg/ml) before harvesting T cells at 500g for 10 min. T cells were lysed in 30 mM tris-HCl, pH 7.5, 100 mM NaCl, 30 mM MgCl2, 0.1% NP-40, cycloheximide (100 μg/ml), and RNasin (30 U/ml; Promega). Lysates were clarified at 16,000g for 10 min at 4°C and cytoplasmic extracts with equal amounts of RNA were loaded on a 15 to 50% sucrose gradient and centrifuged at 4°C in a SW41Ti Beckman rotor for 3 hours and 30 min at 273,000g. The sucrose gradient was analyzed by continuous-flow absorbance at 254 nm and recorded by BioLogic LP software (Bio-Rad).
Seahorse assay
Seahorse assay was performed as in (40) using the Seahorse XF Real-Time ATP Rate Assay Kit (Agilent Technologies, catalog no. 103592-100) following the manufacturer’s instruction. Briefly, 5 × 105 neonatal or adult naïve or 24-hour activated T cells were seeded in poly-l-lysine–coated plates (V7 PS cell culture microplates, Agilent Technologies) and maintained in assay medium (Seahorse XF DMEM Medium, pH 7.4, supplemented with 10 mM XF glucose, 1 mM XF pyruvate, and 2 mM XF glutamine) for 60 min at 37°C. Assay medium was then replaced and 1.5 μM oligomycin and 0.5 μM Rotenone plus Antimycin A were added to measure oxygen consumption rate, extracellular acidification rate, and total ATP production through an XFe24 Analyzer. Mitochondrial ATP production rate was calculated as follows: (OCRBasal − OCROligomycin) × 2 × 2.75. Glycolysis ATP production rate was calculated as follows: (ECARBasal × K) − (OCRBasal − OCRRot/AA) × 0.67. Data were analyzed with WAVE software (SCR_014526).
RNA-FISH
RNA-FISH was carried out as in (15, 41, 42). In brief, T cells were seeded onto poly-lysine microscope slides, fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100 in PBS, and maintained in 20% glycerol-PBS overnight. Subsequently, T cell slides were freeze-thawed on dry ice, deproteinized with 0.1 N HCl, and hybridized with riboprobes at 52.5°C for 3.5 min overnight at 37°C in a water bath. Slides were washed, incubated with Streptavidin HRP (Akoya Biosciences, catalog no. NEL750001EA, 1:1000), and subjected to TSA amplification (Akoya Biosciences, catalog no. NEL763001KT, 1:150). T cell nuclei were counterstained with DAPI 1:5000, and the slides were mounted with ProLong Glass mounting media. RNA-FISH images were acquired using a Leica TCS SP5 Confocal microscope with a Leica HCX PL APO 63, 1.40-NA oil-immersion objective with Z-stacks of 0.29 μm. An additional 2× zoom was used. To ensure comparability between T cells isolated from neonates and adults, RNA-FISH was performed simultaneously. To achieve comparability between different sets of experiments, images were acquired and processed with comparable settings, with at least ∼50 nuclei per individual acquired. RNA-FISH images were analyzed as described in (15).
Nucleolin immunofluorescence
T cells were seeded onto poly-lysine microscope slides, fixed with 3% paraformaldehyde, permeabilized with 0.5% Triton X-100 in PBS, and maintained in 20% glycerol-PBS overnight. T cells were incubated with primary antibodies specific for NCL (Abcam, catalog no. ab22758; AB_776878, 1:500) in a solution of 2% BSA, 10% goat serum, 0.1% Tween, and PBS overnight at 4°C. A secondary antibody conjugated with Alexa Fluor 647 was used (1:1000). T cell nuclei were counterstained with DAPI 1:5000 and glasses were mounted in ProLong Glass antifade mounting medium (Thermo Fisher Scientific).
Protein extraction and Western blotting analysis
To evaluate phospho-ZAP70, phospho-RPS6, and RPS6 levels, total protein extraction was performed following the protocol outlined in (15). A total of 1 × 106 of 2 × 106 neonatal or adult naïve or activated CD4+ T cells were incubated for 10 min at 4°C in 100 μl of RIPA buffer [10 mM tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1 mM EDTA, pH 8, 0.1% SDS supplemented with 1× PIC (Merck), 1 mM NaF, and 10 mM NaVO4]. Cellular lysate was sonicated with Branson digital sonifier for 20 s at 20% of duty (0.2 s ON and 0.5 s OFF). Afterward, debris was sedimented through centrifugation at a maximum speed for 10 min at 4°C. To evaluate PTBP1, IRF4, NCL, and RNA pol II levels, nuclear protein extraction was performed as in (43). Briefly, 1 × 106 to 2 × 106 neonatal or adult naïve CD4+ T cells were resuspended in 100 μl of cytosol buffer (10 mM Hepes KOH, pH 8, 1.5 mM MgCl2, 10 mM NaCl, 1 mM DTT, 10% glycerol, and 1× protease inhibitor), nuclei were collected at 1200g and 4°C, and washed three times with equal volume of cytosolic extraction buffer. Nuclei were resuspended in 50 μl of nuclear buffer (10 mM Hepes KOH, pH 8, 1.5 mM MgCl2, 300 mM NaCl, 1 mM DTT, 0.2% NP-40, 10% glycerol, and 1× protease inhibitor) complemented with 2 mM CaCl2 and 20 U MNase and kept at 37°C for 30 min. Afterward, debris was sedimented through centrifugation at maximum speed for 10 min at 4°C. Proteins were quantified with a Qubit (Invitrogen) fluorometer and used for subsequent Western blotting analysis. Total or nuclear extract (10 to 20 μg) was used for Western blot. Proteins were resolved on 4 to 12% Bolt bis tris gel and transferred into a nitrocellulose membrane. Immunoblotting was performed with the following antibodies: phospho-RPS6 (Ser235/236) (Cell Signaling, catalog no. 2211s; AB_331679, 1:1000), phospho-RPS6 (Ser240/244) (Cell Signaling, catalog no. 5364; AB_10694233, 1:1000), RPS6 (Cell Signaling, catalog no. 2217; AB_331355, 1:1000), vinculin (Sigma-Aldrich, catalog no. V9131; AB_477629, 1:5000), IRF4 (BioLegend, catalog no. 646412; AB_2728478, 1:250), PTBP1 (Abcam, catalog no. ab133734; AB_2814646, 1:2000), NCL (Abcam, catalog no. ab22758; AB_776878, 1:5000), RNA pol II (Abcam, catalog no. ab5408; AB_304868 1:2000), phospo-ZAP70 (Tyr319)/Syk (Tyr352) (Cell Signaling, catalog no. 2701T; AB_331600, 1;1000), and H3 (Abcam, catalog no. ab1791; AB_302613, 1:5000). HRP-conjugated secondary antibodies and West Dura Extended Duration Substrate ECL was used for Western blot detection. Images were acquired using iBright and analyzed with ImageJ software (SCR_003070).
RNA immunoprecipitation
RNA immunoprecipitation assay was performed as done in (15) with minor adaptation in the RNA purification step. In particular, RNA was purified using TRI Reagent, and DNase treatment was performed during a second round of RNA extraction performed with Maxwell using RCS simplyRNA Cells kit (Promega, catalog no. AS1390).
RNA isolation and RT-qPCR
Total RNA was extracted using the RNeasy Mini kit and the QIAshredder (Qiagen) system following the manufacturer’s instruction. DNase treatment was performed with the RNase-free DNase Set (Qiagen). The SuperScript III First-Strand Synthesis SuperMix kit (Invitrogen) was used to reverse transcribe total RNA in accordance with the manufacturer’s recommendations. Power SYBR Green PCR Master Mix was used to perform real-time quantitative PCR on the StepOnePlus Real-Time PCR System. Gene expression data were normalized on GAPDH as an independent housekeeping gene. Normalized Ct values were calculated as 2−ΔCt or 2−ΔΔCt. A list of primers used is shown in table S6.
RNA-seq and analysis
RNA-seq libraries were generated using RNA extracted from neonatal naïve CD4+ T cells or adult naïve CD4+ T cells that had been treated with either control (Scr) or LINE1 ASOs for 48 hours. RNA extraction was carried out with Maxwell using the RCS simplyRNA Cells kit (catalog no. AS1390). RNA integrity was checked with TapeStation (High Sensitivity RNA Screentape assay, catalog no. 5067-5579). Libraries were prepared using Illumina Stranded mRNA prep kit (catalog no. 20040532). The libraries were sequenced in single end modality with a read length of 150 bp on Illumina NextSeq 500. RNA-seq data were processed with Nextflow version 20.10.1 (44). Ribosomal RNA was filtered out using BBDuk version 38.90 (k = 31 hdist = 1) (https://github.com/BioInfoTools/BBMap/blob/master/sh/bbduk.sh). Adapters were removed with trimmomatic tool version 0.39 (45) and fastQ quality was assessed with multiQC version 1.9 (46). Reads were then aligned using STAR version 2.7.6a against the reference human genome (hg38) using the GENCODE version 40 GTF file including the LINE1-transcripts identified in (15) (STAR command line: --runMode alignReads --genomeLoad NoSharedMemory --outSAMtype BAM Unsorted --outFilterMultimapNmax 100 --outFilterScoreMinOverLread 0 --outFilterMatchNminOverLread 0.3 --outFilterMismatchNmax 2 --outFilterIntronMotifs RemoveNoncanonicalUnannotated --winAnchorMultimapNmax 100 --quantMode GeneCounts TranscriptomeSAM --twopassMode Basic). An average of 20 M reads with ~90% of uniquely mapped reads per sample was obtained. Adult naïve T cells’ RNA-seq (not treated with any ASOs) was retrieved from (47), and fastQ files were downloaded from the European Genome-phenome Archive (EGA) accession number EGAD00001002671 and processed as described above except for the adapters removal step that was achieved using fastp tool version 0.23.2 (default parameters) (48). These libraries were polyA and sequenced in paired-end modality with a read length of 100 bp. An average of 27 M reads with ~93% of uniquely mapped reads per sample was obtained. To mitigate bias due to differences in libraries’ preparation between internally generated and publicly available datasets, paired-end libraries were used as single end (read1) while stranded libraries were used in unstranded modality. Human naive CD4+ T cells of adult and elderly individuals have been obtained from SRP266523 (49), and corresponding fastQ files were processed as described above. On average, 25 M reads with ~94% of uniquely mapped reads were obtained. TE expression was quantified using TEcount version 1.0.1 (https://github.com/bodegalab/tecount); briefly, reads were intersected with the UCSC Repeat Masker version 4.0.3 for human. The reads with a minimum of 10 bp overlap with the TEs were used for counting. Raw TE counts were then normalized using DESeq2 (50) and scaled by mean and SD for each level (e.g., Class). LINE1-transcripts expression was evaluated similarly to (15). In brief, LINE1-transcripts expression was assessed generating transcriptomic reads alignments with STAR (−-quantMode TranscriptomeSAM) and then running Salmon version 1.4.0 (51) (salmon quant --libType “U” --numGibbsSamples 100 --gcBias).
Gene set enrichment analysis
Raw gene counts [STAR --quantMode GeneCounts (52)] were normalized with DESeq2 (50) and used as input for GSEA with GeneTrail version 3.2 (default parameters) (53). The analysis considered the Human Molecular Signatures Database “Curated Reactome” version 7.5.1, which comprises 1615 gene sets. GSEA was performed on two comparisons: (i) neonatal versus adult naïve CD4+ T cells and (ii) adult naïve CD4+ T cells where LINE1 were down-regulated using LINE1 ASOs versus control cells (scr ASOs). For each comparison, up-regulated (or enriched) or down-regulated (ore depleted) gene sets were retrieved. Statistically enriched gene sets (q value ≤0.05) that were in common to both the comparisons were selected, and for each gene within these gene sets, the following information was manually downloaded from GeneTrail web service and imported in R version 4.0.3 for downstream analysis: index in sorted gene list, running sum statistics, position in ranked gene list, and score identifier level statistics. These data were used to extract the genes that contribute to the core enrichment for each gene set. Then, the similarity between the up-regulated common gene sets was measured calculating the Jaccard index of the core enrichment genes for each gene set between the two comparisons. The resulting matrix was clusterized and visualized with heatmap version 0.2.0 (default clustering method) to highlight similarity between common genes and pathways in the two comparisons.
Statistical analysis
Details of statistical analyses, including the number of individuals and nuclei in imaging experiments, as well as the statistical methods used, F values, and P values, are provided in each figure legend, the text, or supplementary tables. The results presented in this article are based on data obtained from a minimum of three individuals, with exceptions specified in figure legends. All statistical analyses were conducted using Prism software (GraphPad Software; SCR_002798). Significance was determined using paired or unpaired t tests (one or two tailed), ordinary two-way analysis of variance (ANOVA), and ordinary one-way ANOVA. Results were considered significant when the P value was <0.05. In box plots, boxes and whiskers represent the 25th to the 75th and the 10th to the 90th percentiles, respectively, with median values indicated. For violin plots, dashed lines indicate the 25th to the 75th percentiles and the median. In bar plots and scatter plots, mean values and SEM are indicated, with each data point representing an individual. For transcriptomic analysis, RNA-seq datasets from four individuals were used for each examined group, and statistical differences in comparisons were assessed using the unpaired Wilcoxon rank-sum test with continuity correction.
Acknowledgments
We are grateful to S. Notarbartolo, J. Geginat, and S. Biffo for critical scientific discussion. We acknowledge the scientific and technical assistance of the INGM Imaging Facility, particularly C. Cordiglieri and A. Fasciani [Istituto Nazionale di Genetica Molecolare “Romeo ed Enrica Invernizzi” (INGM), Milan, Italy].
Funding: This work was funded by the following grants: Ricerca Finalizzata (grant no. GR-2018-12365280), Fondazione AIRC (grant nos. 27066 and 21073), Fondazione Cariplo (grant no. 2019-3416), Fondazione Regionale per la Ricerca Biomedica (FRRB CP2_12/2018), Piano Nazionale di Ripresa e Resilienza (PNRR) (grant no. G43C22002620007), and Progetti di Rilevante Interesse Nazionale (PRIN) (grant no. 2022PKF9S) to B.B. and Fondazione Cariplo (Bando Giovani, grant no. 2018-0321) and Fondazione AIRC (grant no. MFAG 29165) to F.Ma.
Author contributions: Conceptualization: B.B., F.Ma., F.V.B., R.V., V.R., M.P., S.R., and F.Mo. Investigation: F.V.B., R.V., M.P., A.R., F.A.C., C.R., and C.P. Methodology: F.V.B., R.V., M.C., and F.Mo. Formal analysis: F.V.B., R.V., M.P., F.A.C., S.R., and F.Mo. Visualization: B.B., F.Ma., F.V.B., R.V., M.P., A.R., and G.M. Validation: B.B., F.Ma., A.R., T.N., M.P., M.C., and V.R. Data curation: V.R., M.P., F.Mo., and L.P. Software: M.P. Supervision: B.B., F.Ma., V.R., D.P., F.Mo., and L.P. Writing—original draft: B.B., F.Ma., F.V.B., and R.V. Writing—review and editing: B.B., F.Ma., V.R., M.P., A.R., G.M., T.N., S.A., C.P., F.Mo., and L.P. Resources: B.B., A.R., M.C., T.N., G.M., D.P., and E.E. Project administration: B.B. and F.Ma. Funding acquisition: B.B. and F.Ma.
Competing interests: S.A. is a cofounder of the startup CheckMab s.r.l. and T-One Therapeutics s.r.l.; F.Ma. and B.B. are cofounders of the startup T-One Therapeutics s.r.l.; R.V. has a scientific collaboration with the startup T-One Therapeutics s.r.l. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Newly generated RNA-seq data that support the findings of this study have been deposited at ArrayExpress under accession number EMTAB13718 and are available at https://ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-13718. Human RNA-seq of CD4+ naïve T cells of adult and elderly individuals can be downloaded respectively from EGAD00001002671 available at https://ega-archive.org/datasets/EGAD00001002671 and SRP266523 (PRJNA638216) available at https://trace.ncbi.nlm.nih.gov/Traces/?view=study&acc=SRP266523.
Supplementary Materials
The PDF file includes:
Figs. S1 to S6
Legends for tables S1 to S6
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S6
REFERENCES AND NOTES
- 1.Davenport M. P., Smith N. L., Rudd B. D., Building a T cell compartment: How immune cell development shapes function. Nat. Rev. Immunol. 20, 499–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudd B. D., Neonatal T cells: A reinterpretation. Annu. Rev. Immunol. 38, 229–247 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucivero G., D’Addario V., Tannoia N., Dell’Osso A., Gambatesa V., Lopalco P. L., Cagnazzo G., Ontogeny of human lymphocytes: Two-color fluorescence analysis of circulating lymphocyte subsets in fetuses in the second trimester of pregnancy. Fetal Diagn. Ther. 6, 101–106 (2009). [DOI] [PubMed] [Google Scholar]
- 4.D'Arena G., Musto P., Cascavilla N., Di Giorgio G., Fusilli S., Zendoli F., Carotenuto M., Flow cytometric characterization of human umbilical cord blood lymphocytes: Immunophenotypic features. Haematologica 83, 197–203 (1998). [PubMed] [Google Scholar]
- 5.Fink P. J., The biology of recent thymic emigrants. Annu. Rev. Immunol. 31, 31–50 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Bogue M., Candéias S., Benoist C., Mathis D., A special repertoire of alpha:beta T cells in neonatal mice. EMBO J. 10, 3647–3654 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong M., Artusa P., Kelly S. A., Fournier M., Baldwin T. A., Mandl J. N., Melichar H. J., Alterations in the thymic selection threshold skew the self-reactivity of the TCR repertoire in neonates. J. Immunol. 199, 965–973 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Thapa P., Guyer R. S., Yang A. Y., Parks C. A., Brusko T. M., Brusko M., Connors T. J., Farber D. L., Infant T cells are developmentally adapted for robust lung immune responses through enhanced T cell receptor signaling. Sci. Immunol. 6, eabj0789 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adkins B., Williamson T., Guevara P., Bu Y., Murine neonatal lymphocytes show rapid early cell cycle entry and cell division. J. Immunol. 170, 4548–4556 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Smith N. L., Wissink E., Wang J., Pinello J. F., Davenport M. P., Grimson A., Rudd B. D., Rapid proliferation and differentiation impairs the development of memory CD8+ T cells in early life. J. Immunol. 193, 177–184 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith N. L., Patel R. K., Reynaldi A., Grenier J. K., Wang J., Watson N. B., Nzingha K., Yee Mon K. J., Peng S. A., Grimson A., Davenport M. P., Rudd B. D., Developmental origin governs CD8+ T cell fate decisions during infection. Cell 174, 117–130.e14 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Mittelbrunn M., Kroemer G., Hallmarks of T cell aging. Nat. Immunol. 22, 687–698 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Chapman N. M., Boothby M. R., Chi H., Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 20, 55–70 (2020). [DOI] [PubMed] [Google Scholar]
- 14.Nurk S., Koren S., Rhie A., Rautiainen M., Bzikadze A. V., Mikheenko A., Vollger M. R., Altemose N., Uralsky L., Gershman A., Aganezov S., Hoyt S. J., Diekhans M., Logsdon G. A., Alonge M., Antonarakis S. E., Borchers M., Bouffard G. G., Brooks S. Y., Caldas G. V., Chen N.-C., Cheng H., Chin C.-S., Chow W., de Lima L. G., Dishuck P. C., Durbin R., Dvorkina T., Fiddes I. T., Formenti G., Fulton R. S., Fungtammasan A., Garrison E., Grady P. G. S., Graves-Lindsay T. A., Hall I. M., Hansen N. F., Hartley G. A., Haukness M., Howe K., Hunkapiller M. W., Jain C., Jain M., Jarvis E. D., Kerpedjiev P., Kirsche M., Kolmogorov M., Korlach J., Kremitzki M., Li H., Maduro V. V., Marschall T., McCartney A. M., McDaniel J., Miller D. E., Mullikin J. C., Myers E. W., Olson N. D., Paten B., Peluso P., Pevzner P. A., Porubsky D., Potapova T., Rogaev E. I., Rosenfeld J. A., Salzberg S. L., Schneider V. A., Sedlazeck F. J., Shafin K., Shew C. J., Shumate A., Sims Y., Smit A. F. A., Soto D. C., Sović I., Storer J. M., Streets A., Sullivan B. A., Thibaud-Nissen F., Torrance J., Wagner J., Walenz B. P., Wenger A., Wood J. M. D., Xiao C., Yan S. M., Young A. C., Zarate S., Surti U., McCoy R. C., Dennis M. Y., Alexandrov I. A., Gerton J. L., O’Neill R. J., Timp W., Zook J. M., Schatz M. C., Eichler E. E., Miga K. H., Phillippy A. M., The complete sequence of a human genome. Science 376, 44–53 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marasca F., Sinha S., Vadala R., Polimeni B., Ranzani V., Paraboschi E. M., Burattin F. V., Ghilotti M., Crosti M., Negri M. L., Campagnoli S., Notarbartolo S., Sartore-Bianchi A., Siena S., Prati D., Montini G., Viale G., Torre O., Harari S., Grifantini R., Solda G., Biffo S., Abrignani S., Bodega B., LINE1 are spliced in non-canonical transcript variants to regulate T cell quiescence and exhaustion. Nat. Genet. 54, 180–193 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Feng X., Wang H., Takata H., Day T. J., Willen J., Hu H., Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nat. Immunol. 12, 544–550 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ricciardi S., Manfrini N., Alfieri R., Calamita P., Crosti M. C., Gallo S., Müller R., Pagani M., Abrignani S., Biffo S., The translational machinery of human CD4+ T cells is poised for activation and controls the switch from quiescence to metabolic remodeling. Cell Metab. 28, 895–906.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandl J. N., Monteiro J. P., Vrisekoop N., Germain R. N., T cell-positive selection uses self-ligand binding strength to optimize repertoire recognition of foreign antigens. Immunity 38, 263–274 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stefanová I., Dorfman J. R., Germain R. N., Self-recognition promotes the foreign antigen sensitivity of naive T lymphocytes. Nature 420, 429–434 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Myers D. R., Norlin E., Vercoulen Y., Roose J. P., Active tonic mTORC1 signals shape baseline translation in naive T cells. Cell Rep. 27, 1858–1874 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Azzam H. S., Grinberg A., Lui K., Shen H., Shores E. W., Love P. E., CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J. Exp. Med. 188, 2301–2311 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Attig J., Agostini F., Gooding C., Chakrabarti A. M., Singh A., Haberman N., Zagalak J. A., Emmett W., Smith C. W. J., Luscombe N. M., Ule J., Heteromeric RNP assembly at LINEs controls lineage-specific RNA processing. Cell 174, 1067–1081.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iannone C., Kainov Y., Zhuravskaya A., Hamid F., Nojima T., Makeyev E. V., PTBP1-activated co-transcriptional splicing controls epigenetic status of pluripotent stem cells. Mol. Cell 83, 203–218.e9 (2023). [DOI] [PubMed] [Google Scholar]
- 24.Wolf T., Jin W., Zoppi G., Vogel I. A., Akhmedov M., Bleck C. K. E., Beltraminelli T., Rieckmann J. C., Ramirez N. J., Benevento M., Notarbartolo S., Bumann D., Meissner F., Grimbacher B., Mann M., Lanzavecchia A., Sallusto F., Kwee I., Geiger R., Dynamics in protein translation sustaining T cell preparedness. Nat. Immunol. 21, 927–937 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Busse P. J., Mathur S. K., Age-related changes in immune function: Effect on airway inflammation. J. Allergy Clin. Immunol. 126, 690–699 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mogilenko D. A., Shchukina I., Artyomov M. N., Immune ageing at single-cell resolution. Nat. Rev. Immunol. 22, 484–498 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikolich-Zugich J., The twilight of immunity: Emerging concepts in aging of the immune system. Nat. Immunol. 19, 10–19 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Goronzy J. J., Weyand C. M., Mechanisms underlying T cell ageing. Nat. Rev. Immunol. 19, 573–583 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar B. V., Connors T. J., Farber D. L., Human T cell development, localization, and function throughout life. Immunity 48, 202–213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grillo G., Lupien M., Cancer-associated chromatin variants uncover the oncogenic role of transposable elements. Curr. Opin. Genet. Dev. 74, 101911 (2022). [DOI] [PubMed] [Google Scholar]
- 31.Grillo G., Keshavarzian T., Linder S., Arlidge C., Mout L., Nand A., Teng M., Qamra A., Zhou S., Kron K. J., Murison A., Hawley J. R., Fraser M., van der Kwast T. H., Raj G. V., He H. H., Zwart W., Lupien M., Transposable elements are co-opted as oncogenic regulatory elements by lineage-specific transcription factors in prostate cancer. Cancer Discov. 13, 2470–2487 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonte P. E., Metoikidou C., Heurtebise-Chretien S., Arribas Y. A., Sutra Del Galy A., Ye M., Niborski L. L., Zueva E., Piaggio E., Seguin-Givelet A., Girard N., Alanio C., Burbage M., Goudot C., Amigorena S., Selective control of transposable element expression during T cell exhaustion and anti-PD-1 treatment. Sci. Immunol. 8, eadf8838 (2023). [DOI] [PubMed] [Google Scholar]
- 33.Marasca F., Gasparotto E., Polimeni B., Vadala R., Ranzani V., Bodega B., The sophisticated transcriptional response governed by transposable elements in human health and disease. Int. J. Mol. Sci. 21, 3201 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Neill L. A., Kishton R. J., Rathmell J., A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Geiger R., Rieckmann J. C., Wolf T., Basso C., Feng Y., Fuhrer T., Kogadeeva M., Picotti P., Meissner F., Mann M., Zamboni N., Sallusto F., Lanzavecchia A., L-Arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell 167, 829–842.e13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Man K., Kallies A., Synchronizing transcriptional control of T cell metabolism and function. Nat. Rev. Immunol. 15, 574–584 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Huang H., Long L., Zhou P., Chapman N. M., Chi H. A.-O., mTOR signaling at the crossroads of environmental signals and T-cell fate decisions. Immunol. Rev. 295, 15–38 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Percharde M., Lin C. J., Yin Y., Guan J., Peixoto G. A., Bulut-Karslioglu A., Biechele S., Huang B., Shen X., Ramalho-Santos M., A LINE1-nucleolin partnership regulates early development and ESC identity. Cell 174, 391–405.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wells J. N., Feschotte C., A field guide to eukaryotic transposable elements. Annu. Rev. Genet. 54, 539–561 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kong B. S., Lee C., Cho Y. M., Protocol for the assessment of human T cell activation by real-time metabolic flux analysis. STAR Protoc. 3, 101084 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marasca F., Cortesi A., Bodega B., 3D COMBO chrRNA-DNA-ImmunoFISH. Methods Mol. Biol. 2157, 281–297 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Marasca F., Cortesi A., Manganaro L., Bodega B., 3D multicolor DNA FISH tool to study nuclear architecture in human primary cells. J. Vis. Exp. 10.3791/60712, (2020). [DOI] [PubMed] [Google Scholar]
- 43.Bodega B., Marasca F., Ranzani V., Cherubini A., Della Valle F., Neguembor M. V., Wassef M., Zippo A., Lanzuolo C., Pagani M., Orlando V., A cytosolic Ezh1 isoform modulates a PRC2-Ezh1 epigenetic adaptive response in postmitotic cells. Nat. Struct. Mol. Biol. 24, 444–452 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Di Tommaso P., Chatzou M., Floden E. W., Barja P. P., Palumbo E., Notredame C., Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35, 316–319 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ewels P., Magnusson M., Lundin S., Käller M., MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L., Yang R., Kwan T., Tang C., Watt S., Zhang Y., Bourque G., Ge B., Downes K., Frontini M., Ouwehand W. H., Lin J.-W., Soranzo N., Pastinen T., Chen L., Paired rRNA-depleted and polyA-selected RNA sequencing data and supporting multi-omics data from human T cells. Sci. Data 7, 376 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen S., Zhou Y., Chen Y., Gu J., fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu B., Jadhav R. R., Gustafson C. E., Le Saux S., Ye Z., Li X., Tian L., Weyand C. M., Goronzy J. J., Distinct age-related epigenetic signatures in CD4 and CD8 T cells. Front. Immunol. 11, 585168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerstner N., Kehl T., Lenhof K., Müller A., Mayer C., Eckhart L., Grammes N. L., Diener C., Hart M., Hahn O., Walter J., Wyss-Coray T., Meese E., Keller A., Lenhof H.-P., GeneTrail 3: Advanced high-throughput enrichment analysis. Nucleic Acids Res. 48, W515–W520 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S6
Legends for tables S1 to S6
Tables S1 to S6