Abstract
Apoptosis is the main form of regulated cell death in metazoans. Apoptotic pathways are well characterized in nematodes, flies, and mammals, leading to a vision of the conservation of apoptotic pathways in metazoans. However, we recently showed that intrinsic apoptosis is in fact divergent among metazoans. In addition, extrinsic apoptosis is poorly studied in non-mammalian animals, making its evolution unclear. Consequently, our understanding of apoptotic signaling pathways evolution is a black box which must be illuminated by extending research to new biological systems. Lophotrochozoans are a major clade of metazoans which, despite their considerable biological diversity and key phylogenetic position as sister group of ecdysozoans (i.e. flies and nematodes), are poorly explored, especially regarding apoptosis mechanisms. Traditionally, each apoptotic signaling pathway was considered to rely on a specific initiator caspase, associated with an activator. To shed light on apoptosis evolution in animals, we explored the evolutionary history of initiator caspases, caspase activators, and the BCL-2 family (which control mitochondrial apoptotic pathway) in lophotrochozoans using phylogenetic analysis and protein interaction predictions. We discovered a diversification of initiator caspases in molluscs, annelids, and brachiopods, and the loss of key extrinsic apoptosis components in platyhelminths, along with the emergence of a clade-specific caspase with an ankyrin pro-domain. Taken together, our data show a specific history of apoptotic actors’ evolution in lophotrochozoans, further demonstrating the appearance of distinct apoptotic signaling pathways during metazoan evolution.
Keywords: apoptosis, caspases, BCL-2 family, lophotrochozoans, phylogeny, evolution
Significance.
Apoptosis, a form of programmed cell death, has been long studied in model organisms such as flies and mice and in humans. The restricted focus on these models has led to an overall view that the evolution of genes involved in apoptosis is highly conserved across all animals. The advent of next-generation sequencing has led to a boom in the omics data available across the tree of life. Thanks to this, we explored the evolution of key genes involved in apoptosis in the clade Lophotrochozoa (i.e. molluscs, annelids, flatworms, and brachiopods), one of the three large clades that make up bilaterian animals. We found a complex evolutionary history of apoptosis genes, with multiple losses, gains, divergences, and redundancies, highlighting the value of exploring gene evolution and apoptotic mechanisms in lophotrochozoans.
Introduction
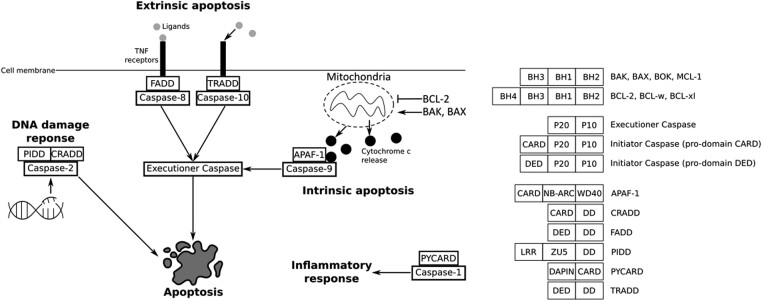
Apoptosis is a form of regulated cell death that sculpts the animal’s body during embryonic development and allows the removal of obsolete tissues or supernumerary cells (Jacobson et al. 1997). Present at the metazoan scale (Vega Thurber and Epel 2007; Ballarin et al. 2010; Kiss 2010; AnvariFar et al. 2017; Krasovec et al. 2019, 2021; Jeffery and Gorički 2021; Krasovec et al. 2022), apoptosis is defined by a conserved set of morphological features that depend on a multigenic family, the caspases (Cohen 1997; Hengartner 2000). While caspases are found in all animals, they have mainly been studied in nematodes, flies, and vertebrates, in which apoptotic signaling pathways are well described (Hengartner 2000; Fan et al. 2005; Steller 2008). Historically, two main apoptotic signaling pathways were described, currently known as extrinsic (formerly the death receptor pathway) and intrinsic (formerly the mitochondrial pathway) apoptosis (Galluzzi et al. 2018). In both cases, caspases play a central role upstream (as initiator caspases) and downstream (as executioner caspases) of the apoptotic signaling pathways (Hengartner 2000; Fan et al. 2005; Kumar 2007). Importantly, each apoptotic pathway is characterized by a specific initiator caspase, itself activated by a protein complex in which a specific activator is involved (Fig. 1). While all caspases are composed of the common P10 and P20 domains, initiator caspases also contain an additional long pro-domain, either a CARD or DED domain, specific for intrinsic and extrinsic apoptoses, respectively.
Fig. 1.
Main apoptotic pathways described in mammals. Initiator caspases rely on a specific activator and are specific to each pathway. Extrinsic apoptosis depends on caspase-8 or caspase-10 and their activators, FADD and TRADD, respectively. Intrinsic apoptosis is controlled by the BCL-2 family (i.e. antiapoptotic BCL-2, proapoptotic BAK or BAX) which can induce mitochondrial outer membrane permeabilization, leading to cytochrome c release. Cytochrome c binds to initiator caspase-9 and APAF-1 forming an activation complex, the apoptosome. Initiator caspases induce activation of the common executioner caspases downstream of the regulation pathways. Caspase-2, activated by CRADD and PIDD, is involved in apoptosis induced by DNA damage. Caspase-1 is activated by PYCARD, leading to the inflammatory response. BCL-2 are characterized by the BH domains, with especially the BH4 for antiapoptotic members. All caspases have the P20 and P10 domains. In addition, initiator caspases possess a long pro-domain, either a CARD or a DED. Activators APAF-1, CRADD, FADD, TRADD, PIDD, and PYCARD have a long domain which can be a CARD or a DED, in addition to a short death-domain.
In mammals, intrinsic apoptosis is activated through mitochondrial outer membrane permeabilization (MOMP), a process controlled by BCL-2 family proteins such as BAX or BAK (Fig. 1) (Hengartner 2000). MOMP leads to the release of cytotoxic molecules such as cytochrome c into the cytosol. Cytochrome c forms the apoptosome complex with the specific adaptor protein APAF-1 (apoptotic protease activating factor 1) and initiator caspase-9, leading to the activation of executioner caspase-3 (Hengartner 2000; Galluzzi et al. 2018; Kalkavan and Green 2018). Key components of the intrinsic pathway are present in flies and nematodes as well (Driscoll 1996; Hengartner 2000; Lettre and Hengartner 2006; Steller 2008; Bender et al. 2012). However, there are several fundamental mechanistic differences between these models and mammals. For instance, cytochrome c is crucial to trigger apoptosis in mammals, whereas it is dispensable in flies and nematodes. Indeed, since BCL-2 members BAX and BAK have not been identified in Drosophila melanogaster and Caenorhabditis elegans genomes, MOMP does not occur. In D. melanogaster, the APAF-1 homologue, DARK, oligomerizes into an eightfold apoptosome complex that does not require cytochrome c for assembly (Hengartner 2000; Dorstyn et al. 2002; Steller 2008). In addition, D. melanogaster apoptotic pathway depends on the removal of inhibition controlled by Grim, Reaper, and Hind (Steller 2008). These three proteins do not have homologues in other species. In C. elegans, the unique BCL-2 homolog CED-9 interacts with the APAF-1 homologue CED-4 (Hengartner 2000). When EGL-1 binds to CED-9, CED-4 is released, resulting in the formation of a homotetrameric complex that activates the caspase CED-3 (Shi 2006). More recently, molecular characterization of the BCL-2 family in the placozoan Trichoplax adhaerens identified a bona fide BAX-like protein which performs MOMP in vitro (Popgeorgiev et al. 2020). Altogether, these findings suggest that although MOMP is ancient in metazoans, intrinsic apoptotic pathways are divergent and have evolved independently in most metazoan phyla. Interestingly, we have recently discovered that Dronc and Ced-3, the initiator caspases of intrinsic apoptosis in flies and nematodes, respectively, are homologues (here orthologous, resulting from speciation) to the vertebrate caspase-2 but not to caspase-9 (Krasovec et al. 2023). Caspase-9 is deuterostome specific, while caspase-2 is ancestral and conserved among bilaterian animals. Interestingly, caspase-2 in mammals is implicated in apoptosis induced by DNA damage, but not in the major intrinsic and extrinsic pathways (Fig. 1) (Krumschnabel et al. 2009a; Krumschnabel et al. 2009b). Caspase-2 is activated by PIDD (P53-induced protein with a death domain) into the PIDDosome platform comprising CRADD (caspase and RIP adapter with death domain) (also named RAIDD—RIP-associated protein with a death domain) (Zhivotovsky and Orrenius 2005). Despite caspase-2 being ancestral to bilaterians (Krasovec et al. 2023), the PIDD activator has not been investigated so far in non-mammals. In addition to caspase-2 and caspase-9, mammals possess other caspases with a CARD pro-domain, the inflammatory caspases, especially caspase-1, which is able to cleave cytokines and thus trigger the inflammatory response (Sollberger et al. 2014). The activator PYCARD (PYD and CARD domain-containing protein) forms the inflammasome complex with caspase-1, allowing its activation. Similar to PIDD, the presence of PYCARD in non-mammals has not yet been investigated (Fig. 1).
Our knowledge on extrinsic apoptosis relies mainly on mammalian data due to its absence in flies and nematodes. This pathway is activated by death receptors (i.e. TNFr—tumor necrosis factor receptors) on cell membranes, themselves able to recruit activator proteins FADD (Fas-associated protein with death domain) or TRADD (tumor necrosis factor receptor type 1–associated death domain) into a DISC (death-inducing signaling complex), which activates caspase-8 or caspase-10, respectively (Fig. 1) (Fan et al. 2005; Galluzzi et al. 2018). Despite this clear molecular cascade, extrinsic apoptosis is poorly described outside of mammals, except for some anecdotal studies, such as in cnidarians or amphioxus in which FADD can induce apoptosis in cultured cells (Yuan et al. 2010; Zhao et al. 2019; Steichele et al. 2021), or in the tunicate Ciona, where caspase-8 is known to be responsible for apoptosis (Krasovec et al. 2024). Consequently, the evolution of extrinsic apoptosis is relatively unknown.
Taken together, it clearly appears that apoptotic signaling pathways’ evolution is a black box that needs to be revisited. Indeed, the lack of knowledge outside three key model species (mammals, deuterostome; flies and nematodes, ecdysozoans) artificially creates biased inferences on apoptotic regulation pathways’ evolutionary history. Thus, the other major group of bilaterians, the lophotrochozoans, remains largely unexplored despite its tremendous diversity. This gap of knowledge creates a critical void, and a comprehensive and global understanding of apoptotic signaling pathway evolution is definitively impossible to reach without exploring lophotrochozoan animals (i.e. molluscs, annelids, brachiopods, and platyhelminths).
In this study, we explored though comparative genomics the apoptotic network of four lineages of lophotrochozoans: molluscs, annelids, platyhelminths, and brachiopods. We conducted exhaustive reciprocal BLAST searches followed by phylogenetic analyses to identify apoptotic actors, especially caspases, BCL-2 family proteins, and activators such as FADD or APAF-1. We then analyzed sequences evolution based on mutation rate and predicted initiator caspase/activator interactions. Our results highlight an unexpected diversity and complexity of the apoptotic network within lophotrochozoans, characterized by the loss of the key intrinsic apoptosis activator APAF-1 in molluscs, annelids, and brachiopods, in parallel with the loss of the extrinsic apoptosis actors in the platyhelminths, notably FADD and DED caspases. Brachiopods lost caspase-2 despite being the conserved and ancestral initiator in bilaterians. Surprisingly, platyhelminths present caspases with an ankyrin pro-domain, a unique feature never observed in vertebrates, flies, or nematodes. Annelids present several species-specific diversifications of initiator caspases, characterized by unusual pro-domains, suggesting a variety of novel functions for these caspases. Instead of a DED or a CARD pro-domain, we identified, among others, caspases with a Z-binding, a kinase, or a PARP catalytic domain. The differential evolution between platyhelminths, molluscs, annelids, and brachiopods suggests specific apoptotic pathways that have not yet been functionally described. Our work highlights the nonlinear, divergent evolution of apoptotic pathways in animals and argues in favor of the study of lophotrochozoans as emerging biological models.
Results
Evolutionary History of Caspases in Lophotrochozoans
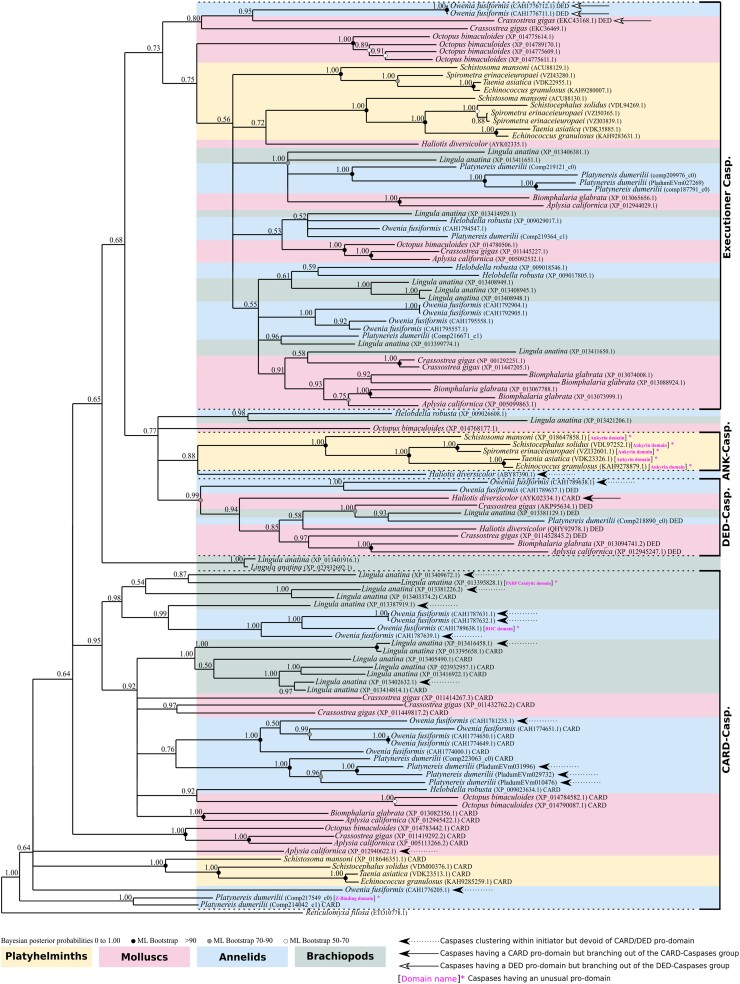
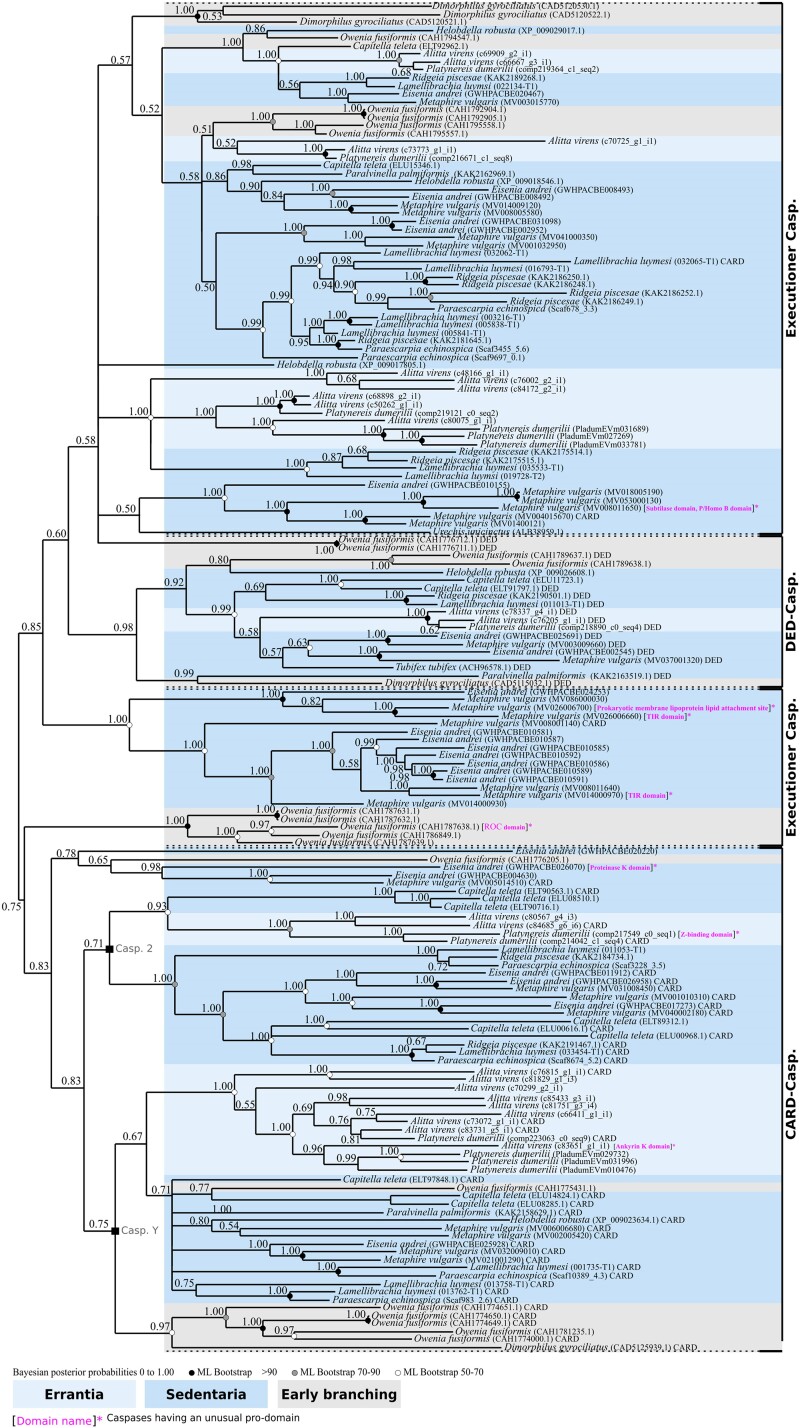
We investigated 60 genomes of lophotrochozoan species including 23 molluscs, 17 annelids, 1 brachiopod, and 19 platyhelminths, to identify their caspase (both initiators and executioners) arsenals (supplementary table S1, Supplementary Material online). We detected sequences of CARD caspases and DED caspases (initiators) as well as executioner caspases. We also identified several caspases presenting an ankyrin pro-domain (ANK caspases), an unexpected feature which has not previously been observed in either deuterostomes or ecdysozoans. To assess the main relationships between the different caspases, we first conducted a phylogenetic analysis on all caspases (both initiator and executioner caspases) from a selection of 13 species representative of lophotrochozoan diversity (Fig. 2; supplementary table S2, Supplementary Material online). We chose this strategy of species sub-selection (while covering the whole diversity of caspase types), as we know from our previous study on this gene family (Krasovec et al. 2023) that their short lengths combined with a high number of sequences leads to poorly resolved topologies. Consequently, this strategy allowed inclusion of all types of caspases at the lophotrochozoan scale without over-saturating the sequence number. The tree was rooted using a Reticulomyxa filosa (foraminifera) caspase-like sequence, already successfully used for metazoan caspase phylogenies (Kaushal et al. 2023; Krasovec et al. 2023). We first noticed a broad separation of the executioner caspases versus all others. Indeed, they form a coherent group (PP = 0.73), along with a clade composed of DED caspases and ANK caspases (plus a few additional unassigned sequences) as sister group. Most caspases with DED pro-domains (DED caspases) form a strongly supported group (PP = 0.99) composed of mollusc, annelid, and brachiopod sequences, but devoid of platyhelminths. ANK caspases form a maximally supported clade (PP = 1.00) specifically restricted to platyhelminths, branching as a sister group to the Haliotis diversicolor sequence and close to DED caspases. The topology is characterized by a polyphyletic distribution of caspases with a CARD pro-domain (CARD caspases), most of them being close to the root.
Fig. 2.
Topology of phylogenetic analysis of caspases at the lophotrochozoan scale made by Bayesian inference method. Caspases with a CARD pro-domain (CARD caspases) present a paraphyletic distribution at the base of the tree. Among them, five sequences do not have a CARD domain. Caspases with a DED pro-domain (DED caspases) are restricted to molluscs and annelids, devoid of platyhelminth representatives. DED caspases form a monophyletic group, with only one from C. gigas out of the cluster. Caspases with an ankyrin pro-domain (ANK caspases) form a monophyletic group specific to platyhelminths. Three CARD caspases are close to DED caspases and ANK caspases. Executioner caspases are grouped into a maximally supported clade composed of sequences from molluscs, annelids, and flatworms. Outgroup is a caspase-like of the foraminifera R. filosa. Node robustness corresponds to posterior probabilities. Bootstrap corresponds to robustness from ML phylogeny which gave similar topology. The analysis includes one brachiopod, five platyhelminths, three annelids, and four molluscs. All caspases analyzed have the common P20 and P10 domains. The presence of a classical CARD or DED pro-domain is indicated next to the sequence name. Caspases with a pro-domain other than CARD or DED are indicated in brackets and in magenta. Phylogeny was conducted on the full length of amino acid sequences containing the pro-domain and the common P20 plus P10 domains.
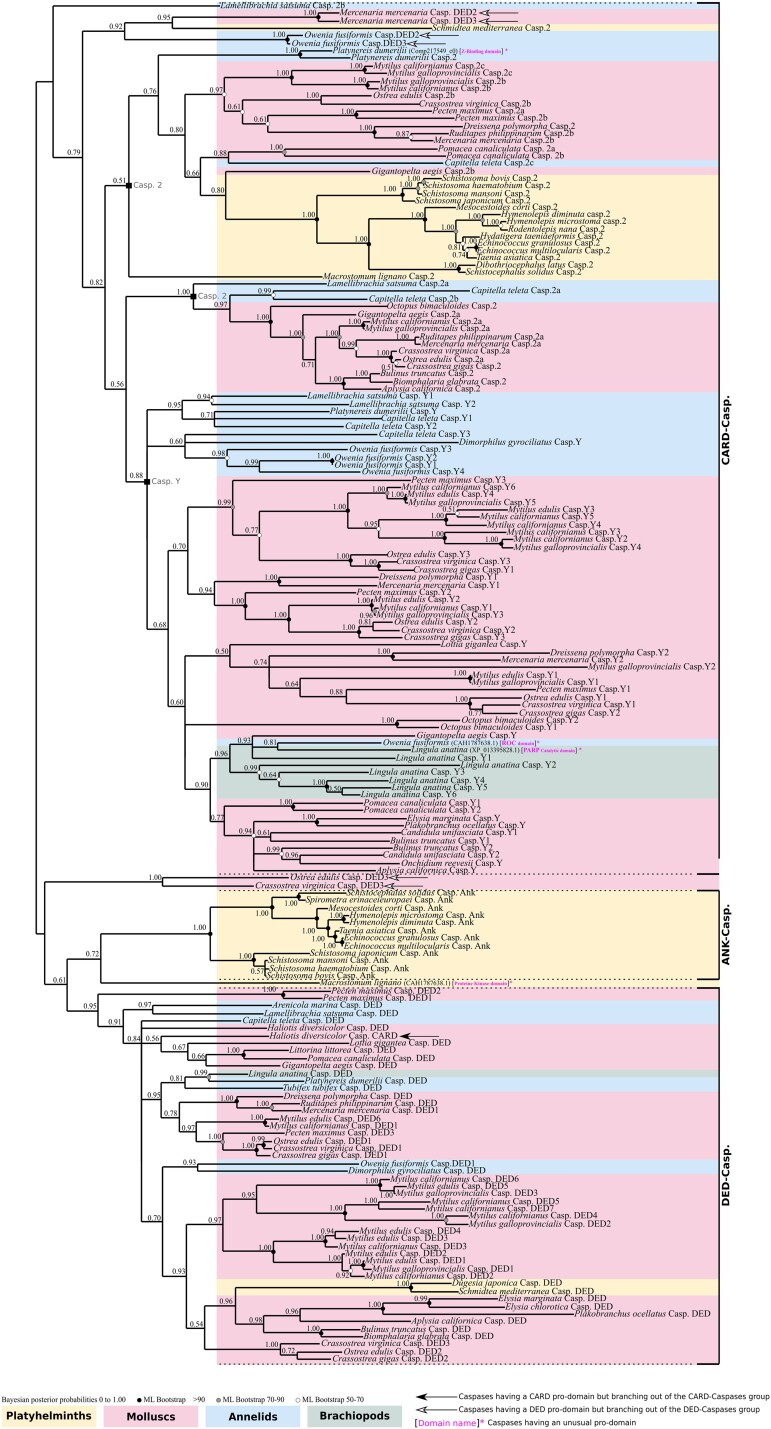
Due to their pivotal role, we next decided to focus more specifically on initiator caspases (supplementary table S3, Supplementary Material online). Due to the short length of caspase sequences, this strategy allowed us to reduce the number of sequences per species and secondarily to increase the sampling to 50 species of lophotrochozoans to conduct a more in-depth analysis of this lineage. We reconstructed their phylogenetic relationships and identified three main groups, consistent with our previous analysis: ANK caspases, DED caspases, and CARD caspases (caspase-2 and caspase-Y) (Fig. 3). ANK caspases and DED caspases are sister groups and together form a well-defined (PP = 0.61) clade. The ANK caspases exhibit maximum support (PP = 1.00) and are specifically restricted to parasitic platyhelminths. Indeed, both the free-living Schmidtea mediterranea and Dugesia japonica do not have ANK caspases. In contrast, the DED caspase group (PP = 0.95) is composed of mollusc, annelid, and brachiopod sequences, together with two nonparasitic platyhelminths, D. japonica and S. mediterranea. Consequently, platyhelminths which have DED caspases do not have ANK caspases and vice versa. This suggests a specific caspase evolution for parasitic platyhelminths.
Fig. 3.
Unrooted phylogenetic analysis of initiator caspases made by Bayesian inference method. Three distinct clades are present. The CARD caspase comprises the caspase-Y specific to molluscs, annelids, and brachiopods, devoid of platyhelminth sequences. The second major group of CARD caspases is the caspase-2 which is split into two subgroups and composed of representatives for each lophotrochozoan clade (molluscs, annelids, brachiopods, platyhelminths). DED caspases are monophyletic and composed of molluscs, annelids, brachiopod, and two nonparasitic platyhelminths. ANK caspases are monophyletic, forming a clade sister to the DED caspases and are restricted to parasitic platyhelminths. Several caspases present unusual pro-domains (brackets, domain names in magenta). The sister group of P. dumerilii caspase-2 is one of them, characterized by a Z-repeat–binding pro-domain. A M. lignano caspase with a protein kinase domain branches as sister to ANK caspases. Inside the caspase-Y group, brachiopod L. anatina has a caspase with a PARP catalytic domain, in addition to O. fusiformis which encodes one with a ROC pro-domain. Node robustness corresponds to posterior probabilities. Bootstrap corresponds to robustness from ML phylogeny which gave a similar topology. The analysis includes 1 brachiopod, 18 platyhelminths, 7 annelids, and 24 molluscs. Phylogeny was conducted on the full length of amino acid sequences containing the pro-domain and the common P20 plus P10 domains.
Both caspase-2 and caspase-Y possess a CARD pro-domain and consequently belong to the CARD caspase type. Caspase-Y is a supported (PP = 0.88) group which we already identified in a previous study and is composed of sequences from molluscs and annelids (Krasovec et al. 2023). Here, we show that caspase-Y is present in brachiopods as well and, interestingly, that there are several duplications in this species harboring six caspase-Y paralogs. Caspase-2 group encompasses sequences from molluscs and annelids, as well as a strongly supported clade of platyhelminth sequences (PP = 1.00). Most lophotrochozoan animals possess caspase-2 (with the exception of the brachiopod Lingula anatina due to a secondary loss), which is consistent with our previous work showing that this caspase is conserved and ancestral in bilaterians.
In addition to the expected caspase types, we discovered four specific caspases with unexpected long pro-domains, which have never been identified before (supplementary fig. S1, Supplementary Material online). The platyhelminth Macrostomum lignano presents a caspase with a protein kinase–like pro-domain, positioned as sister to the ANK caspase group (Fig. 3). We did not detect any other caspases with an unexpected pro-domain in platyhelminths. In addition, we identified a sequence from the brachiopod L. anatina within the caspase-Y clade that has a PARP catalytic and alpha-helical pro-domain instead of the CARD one. Similarly, the annelid Platynereis dumerilii has a caspase with a Z-binding pro-domain which is a paralogue to its caspase-2 (Fig. 3), and Owenia fusiformis encodes a caspase with a ROC domain as a pro-domain.
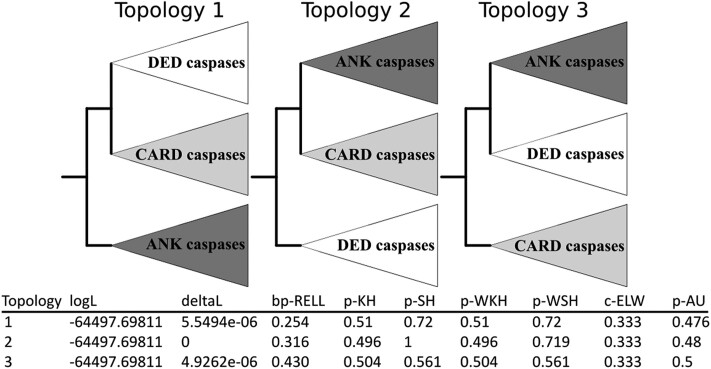
To explore the history of caspase pro-domains acquisition and potentially discover early emerging initiator caspase types, we artificially constructed three topologies corresponding to three potential scenarios of evolution, firstly with ANK caspases as sister to DED caspases + CARD caspases, secondly DED caspases as sister to ANK caspases + CARD caspases, and thirdly CARD caspases as sister to DED caspases + ANK caspases (Fig. 4). Next, we conducted topology tests to evaluate the most likely scenario (Kishino and Hasegawa 1989; Shimodaira and Hasegawa 1999; Shimodaira 2002; Nguyen et al. 2015). Interestingly, regardless of the statistical test used, there is no topology that appears to be the most likely, making it difficult to hypothesize on an ancestral type of caspase, arguing instead for a complex evolutionary model, but also confirming the ancestral presence of both DED caspases and CARD caspases among lophotrochozoans (Fig. 4).
Fig. 4.
Topology tests conducted with IQ-TREE between initiator caspase evolution models. Three artificial topologies were built composed of three clades containing all DED caspases, CARD caspases, and ANK caspases, respectively. Topology tests do not allow identification of a single most-likely scenario, preventing the formation of a hypothesis on the emergence of an ancestral pro-domain within the caspase family. Tests are noted as p-KH for Kishino–Hasegawa test P-value (likelihood method), p-SH for Shimodaira–Hasegawa test P-value, and p-AU for approximately unbiased test P-value. deltaL, logL difference from the maximal logL in the set; bp-RELL, bootstrap proportion using RELL method; c-ELW, expected likelihood weight; p-WKH, P-value of weighted KH test; p-WSH, P-value of weighted SH test. A tree is rejected if its P < 0.05. Tests P-values do not allow to discriminate a more likelihood topology between the three scenarios.
Taken together, our topology indicates that platyhelminths do not possess caspase-Y, and suggests that this caspase is specific to molluscs + annelids + brachiopods. Brachiopods lost the ancestral caspase-2. ANK caspases are specific to parasitic platyhelminths which have lost DED caspases. Conversely, free-living platyhelminths do not have an ANK caspase and retain the DED caspase. The close relationship between ANK caspases and DED caspases in the topology could result in domain shuffling or, in a less likely scenario, could suggest accumulation of divergence of the DED domain of ancestral platyhelminths during evolution.
Diversity of Caspases in Annelids
The presence of unusual caspase pro-domains in some annelid species caught our attention. Therefore, we decided to perform an in-depth investigation of caspases in this lineage, thanks to an increase in species sampling (supplementary table S4, Supplementary Material online). First, we conducted a phylogenetic analysis on a representative set of annelids comprising of two early-branching species, one Errantia and 11 Sedentaria, according to recent annelid phylogenies (Martín-Durán et al. 2021; Lewin et al. 2024). The topology resulting from this analysis showed unresolved nodes, especially regarding CARD caspases, indicating that we were close to sequence saturation in terms of available phylogenetic information, as we previously experienced with caspases (supplementary fig. S2, Supplementary Material online). This could be exacerbated by the overrepresentation of Sedentaria, a group from which most of the annelid-sequenced genomes originate and encompassing species with a large number of caspases. Thus, we conducted a second phylogenetic analysis with modified sampling by removing the Sedentaria Lamellibrachia satsuma which encodes a large set of caspases, and we added the Errantia Alitta virens (Fig. 5). Globally, caspases cluster in a coherent manner, similarly to our previous studies. The majority of executioner caspases cluster together (PP = 0.58). Most DED caspases are in a strongly supported cluster (PP = 0.98), with the exception of two O. fusiformis DED caspases. CARD caspases are closely related, clustering in a strongly supported group (PP = 0.83). CARD caspases are divided into the two types we identified previously: caspase-2 and caspase-Y.
Fig. 5.
Topology of phylogenetic analysis of caspases of annelids made by Bayesian inference method. Most executioner caspases cluster together. Caspases with a DED pro-domain (DED caspases), except for two O. fusiformis sequences, form a monophyletic group. Caspases with a CARD pro-domain (CARD caspases) are split between caspase-Y and caspase-2. A few caspases without long pro-domains branch inside executioner caspases. Annelids cluster in a coherent way according to species phylogeny: the Errantia sequences form several paralogous groups, similarly to Sedentatia sequences. Early-branching annelids are represented by the paleoannelid O. fusiformis, and Dinophiliformia D. gyrociliatus. Globally, each species encodes DED caspases, CARD caspases, and executioner caspases. In addition, several caspases present unusual pro-domains which are a caspase with a protein kinase domain (E. andrei), with a prokaryotic membrane lipoprotein lipid attachment site profile (M. vulgaris), a subtilase domain (M. vulgaris), two caspases having a TIR domain (M. vulgaris), one with a Z-binding domain (M. vulgaris), one with a ROC domain profile (O. fusiformis), and one with a ankyrin domain (A. virens). The presence of a classical CARD or DED pro-domain is indicated next to the sequence name. Caspases with a pro-domain other than CARD or DED are indicated in brackets and in magenta. Bootstrap values correspond to robustness from maximum likelihood phylogeny which gave a similar topology. The analysis includes 2 Errantia, 10 Sedentaria, and 2 early-branching annelids. Phylogeny was conducted on the full length of amino acid sequences containing the pro-domain and the common P20 plus P10 domains.
Early-branching annelid sequences (O. fusiformis and Dimorphillus gyrocilliatus) are distributed along the topology without branching at the base of groups composed of Errantia + Sedentaria, possibly illustrating their uncertain phylogenetic position. Sequences from Errantia (P. dumerilii and A. virens) are clustered within each caspase group, indicating their orthologous relationship. Regarding Sedentaria species, most of them cluster together, especially Clitellata (such as Lamellibrachia luymesi or Ridgeia piscesae) which form several monophyletic sub-clades. Our sampling and the resulting phylogeny indicate significant variation in the caspase family among annelid species. Some annelids have few caspase members, such as Tubifex tubifex or Helobdella robusta with one and four caspases, respectively. Conversely, other species present an expansion of the family with numerous paralogs such as Metaphire vulgaris which encodes 27 caspases. This disparity could be due to genome duplication in this species, which has resulted in numerous paralogs for different genes (Jin et al. 2020).
As hypothesized, we discovered species-specific diversification of caspases with peculiar pro-domains (Fig. 5; supplementary fig. S1, Supplementary Material online). In addition to caspases with a Z-binding and ROC pro-domains in P. dumerilii and O. fusiformis, respectively (see above), we detected a protein kinase domain caspase in Eisenia andrei. In M. vulgaris, we detected two TIR domain caspases, a caspase with a prokaryotic membrane lipoprotein lipid attachment site profile as the pro-domain, and a caspase with a particularly long pro-domain comprising both a subtilase domain and a P/Homo B domain (Fig. 5). A. virens encodes an ANK caspase, similar to those found in platyhelminths, which is likely to be a convergent feature. Although it cannot be ruled out that these particularities are due to errors in gene prediction (artificial chimeric genes), their detection in several species of the same clade (annelids) suggest a specific evolution of caspases in this group. Most of these particular caspases are paralogous to sequences from the same species having a classical domain architecture, suggesting species-specific duplications. After the duplication event, the appearance of unexpected pro-domains may be the result of gene-specific evolution. Taken globally and according to current knowledge, our results indicate that annelids may be the animal group with the greatest diversity and inter-species variation in the caspase family.
Losses and Gains of Activators is Consistent with Initiator Caspase Inventory
Initiator caspases are activated by specific platforms that depend on an activator which possesses a protein domain similar to their corresponding initiator caspases pro-domain (Fig. 1). Thus, the CARD domain of APAF-1 binds to the CARD pro-domain of caspase-9 (mammals) or caspase-2 (fruit flies and nematode) (Zou et al. 1997, Hengartner 2000; Bratton and Salvesen 2010). Mammalian caspase-2 is activated by CRADD (also known as RAIDD) (Duan and Dixit 1997; Lin et al. 2000; Tinel and Tschopp 2004) and caspase-1 by the PYD and CARD domain-containing protein activator (PYCARD) (Sutterwala et al. 2006). Similarly, FADD and TRADD (containing DED domains) activate initiator caspase-8 and caspase-10 in the DISC complex, respectively (Fan et al. 2005; Pennarun et al. 2010).
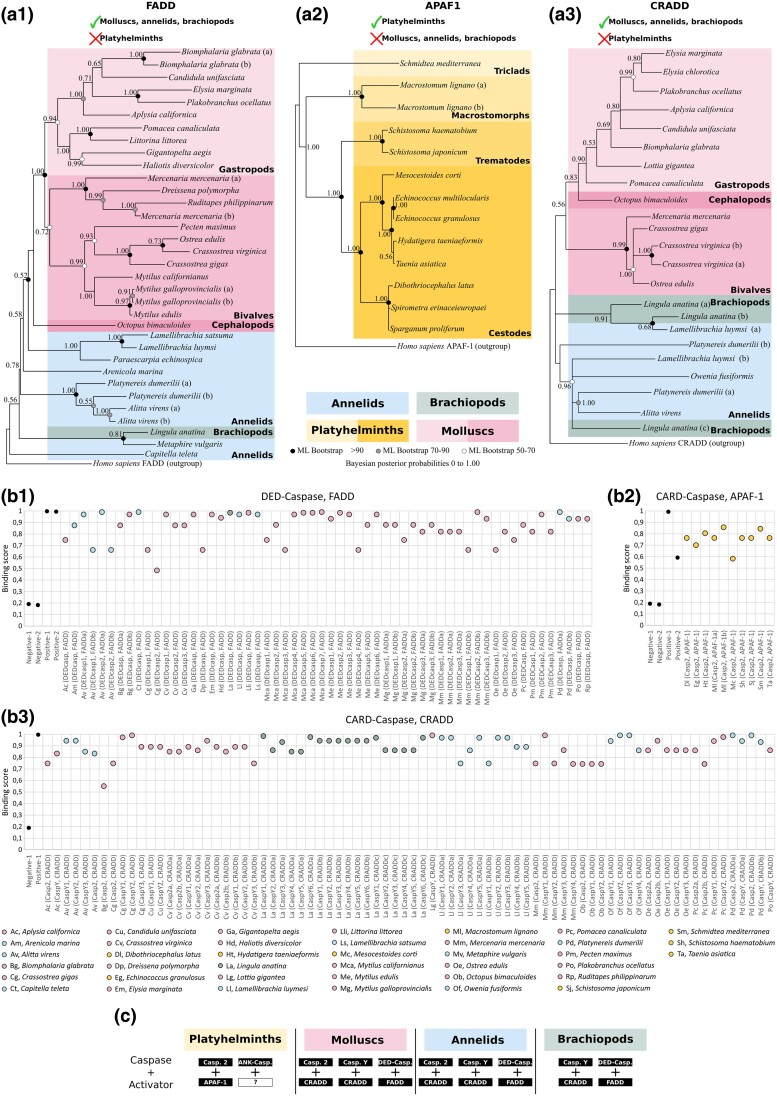
We conducted an exhaustive inventory of caspase activators within lophotrochozoan lineages (Fig. 6a). Interestingly, we found a FADD homologue in the molluscs + annelids + brachiopods group, which is consistent with the presence of DED caspases in these taxa (Fig. 6A1; supplementary table S5, Supplementary Material online). However, FADD was not detected in platyhelminths. Our FADD phylogenetic analysis gave rise to a topology that is globally consistent with the species tree with molluscs grouped in a maximally supported monophyletic clade (PP = 1.00) and split into gastropods and bivalves, in addition to the cephalopod. Annelid and brachiopod sequences branch first in a paraphyletic distribution and appear as sister group to molluscs. The relative topology consistency between sequences and species suggests gene conservation in these groups. Conversely, APAF-1 was only detected in platyhelminths, but, consistently with our previous work (Krasovec et al. 2023), we did not identify any APAF-1 in molluscs + annelids which confirms the loss of this activator in these taxa (Fig. 6A2; supplementary table S6, Supplementary Material online). Here, we did not detect APAF-1 in the brachiopod L. anatina as well. The APAF-1 topology is relatively congruent with the species tree, and all clades are maximally supported (PP = 1.00), with the monophyletic cestode platyhelminths branching with the sister group of trematodes. At the basis of the topology are the triclad platyhelminths. Next, due to the absence of APAF-1, we investigated the potential activators for molluscs + annelids + brachiopods CARD caspases and discovered that these species possess a CRADD homologue, which is absent in platyhelminths (Fig. 6A3; supplementary table S7, Supplementary Material online). Phylogenetic analysis of the CRADD genes suggests a conservation of the gene, as the topology is in accordance with species relationships: molluscs are monophyletic (PP = 0.56) and encompass gastropods (PP = 0.83) and bivalves (PP = 0.99), with the cephalopod as sister of the gastropods. Brachiopod and annelid sequences are distributed between the root of the tree and molluscs. Lastly, we were unable to detect any PIDD, PYCARD, or TRADD (which activate caspase-2, caspase-1, and caspase-10 in mammals, respectively) despite an exhaustive search in all available genomes/transcriptomes/proteomes of lophotrochozoan species from NCBI database. This indicates the potential vertebrate specificity of the PIDD and PYCARD activators, as it was proposed for TRADD (Zmasek and Godzik 2013).
Fig. 6.
Analysis of initiator caspase activators. a) Phylogenetic analysis of initiator Caspase activators at the lophotrochozoan scale made by Bayesian inference method with human sequences as outgroup for FADD (a1), APAF-1 (a2), and CRADD (a3). FADD (containing a DED domain) presents a topology relatively consistent with species relationship. molluscs are monophyletic and split between gastropods and bivalves, with also the presence of a cephalopod. Annelids and brachiopods branch early. The FADD analysis includes 1 brachiopod, 8 annelids, and 20 molluscs and was conducted on the full length of amino acid sequences containing the DED and DD domains. No FADD was detected in platyhelminths. APAF-1 is absent in molluscs, annelids, and brachiopods but was detected in platyhelminths. The analysis includes 12 species and was conducted on the full length of amino acid sequences containing the CARD and the NB-ARC domains. The sequence topology is relatively similar to platyhelminth species relationships, indicating a conservation of this gene within the group. CRADD in absent in platyhelminths but present in molluscs, annelids, and brachiopods, and shows a topology relatively consistent with the species tree, similarly to FADD. The CRADD analysis includes 1 brachiopod, 4 annelids, and 13 molluscs and was conducted on the full length of amino acid sequences containing the CARD and the DD domains. Node robustness corresponds to posterior probabilities. Bootstrap corresponds to robustness from maximum likelihood phylogeny which gave similar topology. b) In silico test of binding affinities made with PSOPIA software between DED caspases with FADD (b1), CARD caspases with APAF-1 (b2), and CARD caspases with CRADD (b3). DED domains from FADD and DED caspases are predicted to bind, suggesting an activation of extrinsic apoptosis. CRADD can theoretically interact with all CARD caspases of these species including caspase-2 and caspase-Y. Predictions indicate that APAF-1 interacts with platyhelminths caspase-2. c) Theoretical couples of initiator Caspase/activator identified in lophotrochozoans resulting from analysis conducted in a and b).
Next, we explored the theoretical initiator caspase/activator binding interactions that could trigger their activation (Fig. 6b). To do so, we evaluated interaction scores (S-score) using PSOPIA software, which is specifically designed to predict protein–protein interactions. First, we calculated the S-scores between DED caspases and FADD for all species where both proteins are present. Consistent with the literature, our results suggest that mollusc, brachiopod, and annelid FADD could activate the corresponding DED caspases (Fig. 6B1). We found the same result with the APAF-1 activator and CARD caspases from platyhelminth species (Fig. 6B2). Interestingly, the S-scores for CRADD and CARD caspases of molluscs + annelids + brachiopods also indicate a high binding probability, with a global preference for caspase-2 rather than caspase-Y (Fig. 6B3), except for a few species such as O. fusiformis.
Our results indicate a consistent relationship between the inventory of initiator caspases and potential interactors, allowing the proposition of activation pairs (Fig. 6c), according to specific loss within each group of lophotrochozoans that we focused on.
BCL-2 Family is Conserved With Nonlinear Evolution Within Lophotrochozoans
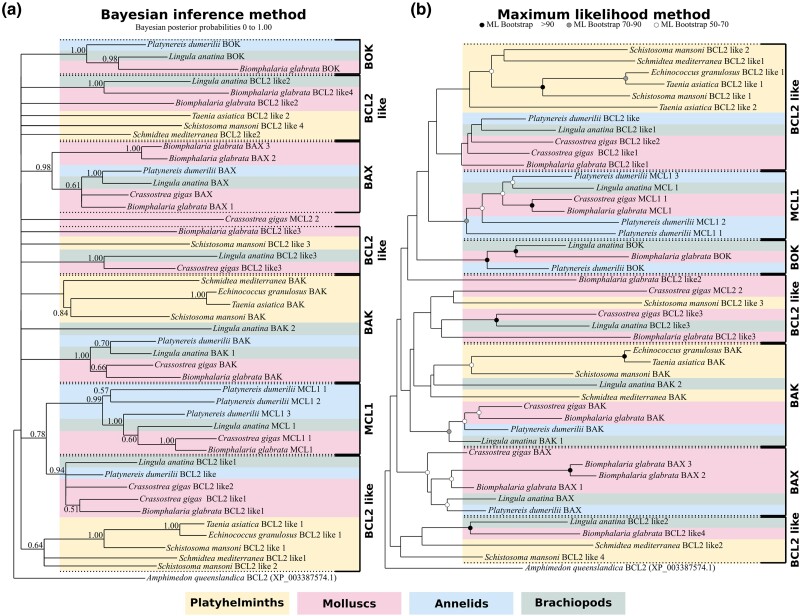
Finally, we focused on the BCL-2 family which is a crucial regulator of MOMP (Chipuk et al. 2010). Within their primary structure, BCL-2 proteins possess one or more conserved sequences, called BCL-2 homology or BH motifs. According to protein architecture, they are divided into multi-motif and “BH3-only.” Multi-motif BCL-2 can be either proapoptotic including BAX, BAK, and BOK which allow cytochrome c release following MOMP in mammals (Kalkavan and Green 2018; Banjara et al. 2020; Moldoveanu and Czabotar 2020) or antiapoptotic such as BCL-2, BCL-XL, and MCL-1. Notably, multi-motif pro- and antiapoptotic BCL-2 members have conserved 3D structure, suggesting a common evolutionary origin. On the other hand, BH3-only proteins represent a polyphyletic group with proapoptotic activities (Aouacheria et al. 2013). Thus, BCL-2 family evolution is complex, with phylogenetic analyses usually producing unclear topologies (Krasovec et al. 2023). Importantly, the short length of BCL-2 proteins restricts the number of informative sites for the phylogenetic analyses. To circumvent this, we conducted phylogenetic analysis on the BCL-2 family by incorporating only two molluscs (Biomphalaria glabrata and Crassostrea gigas), 1 annelid (P. dumerilii), 1 brachiopod (L. anatina), and 4 platyhelminths (Schistosoma mansoni, Taenia asiatica, Echinococcus granulosus, and S. mediterranea) (Fig. 7; supplementary table S8, Supplementary Material online), to represent the diversity of lophotrochozoans without increasing the sequence number. Despite these precautions, the topology obtained by Bayesian inference is of low resolution with numerous unresolved nodes, in line with what we previously observed (Krasovec et al. 2023). Conversely, the topology arising from maximum likelihood (ML) method allows the identification of BCL-2 family relationships (Fig. 7) but with low node robustness. The topology suggests three main proapoptotic clades corresponding to BAK, BAX, and BOK. molluscs, annelids, and brachiopods possess all of them, in addition to numerous specific duplications (i.e. C. gigas BAX or L. anatina BAK). Conversely, platyhelminths only have the BAK proapoptotic, suggesting a loss of both BAX and BOK. Four antiapoptotic clades are identified, one composed of the MCL1 restricted to molluscs and close relatives and three BCL-2–like groups. We named them BCL-2like as evolutionary relationships with BCL-2 are variable and difficult to establish. The relatively clear separation between pro- and antiapoptotic genes clarified which species have a complete BCL-2 inventory (both functions represented). All species from our sampling have both pro- and antiapoptotic BCL-2, but their number is variable. In particular, the three parasitic platyhelminths have only three BCL-2 representatives, while the diversity is high in molluscs and close relatives. Interestingly, there are no differences in BCL-2 family composition between parasitic and nonparasitic platyhelminths, whereas this is the case for the caspase repertoire. Thus, the composition of the ancestral BCL-2 repertoire remains elusive.
Fig. 7.
Topologies of the BCL2 family at the lophotrochozoan scale obtained by Bayesian inference (BI) and ML methods. Node robustness corresponds to 500 bootstraps and posterior probabilities in ML and BI analyses, respectively. a) The BI tree exhibits numerous unresolved nodes, but the topology allows confirmation of the evolutionary relationships of several groups including BAX or MCL1, among others. b) The ML tree is well resolved and sequences group into several monophyletic groups composed of either proapoptotic or antiapoptotic BCL-2 members. Interestingly, the platyhelminth network is very limited, composed of a unique proapoptotic (BAK), and a small number of BCL-2 sequences, likely antiapoptotic. Conversely, molluscs, brachiopods, and annelids have the three classical proapoptotic BAK, BAX, and BOK, and a large number of antiapoptotic genes including MCL1 and several BCL-2 like genes. The analysis includes 1 brachiopod, 4 platyhelminths, 1 annelid, and 2 molluscs and was conducted on the full length of amino acid sequences containing the BH3, BH1, and BH2 domains.
Discussion
Molluscs, Annelids, and Brachiopods Present Specific Apoptotic Network Architecture
Molluscs and their close relatives, annelids and brachiopods, have an apoptotic network that is distinct from mammals and ecdysozoans. molluscs possess a large diversity of BCL-2 which likely leads to MOMP (conversely to ecdysozoans), as cytochrome c release from mitochondria to cytoplasm has been observed in Helix pomatia and C. gigas (Pirger et al. 2009; Li et al. 2017). The loss of APAF-1, which is the shared component of intrinsic apoptosis, suggests a specific modality of CARD caspase activation. In contrast to ecdysozoans, caspase-2 (and also caspase-Y) in molluscs may be activated by CRADD similarly to what occurs in mammals, despite being more phylogenetically distant (Duan and Dixit 1997; Tinel and Tschopp 2004). This makes these animals the only ones currently known to have a potential intrinsic apoptosis which is not based on an apoptosome, but resulting potentially from caspase-2 activation by CRADD and cytochrome c. The presence of only one CARD domain containing activator in these animals certainly induced an evolutionary constraint leading to the conservation of CRADD. The loss of this unique activator may lead to the inability to trigger apoptosis and could be deleterious for development and homeostasis. The dN/dS is a ratio usually used to evaluate selection pressure on genes during evolution (Nei and Gojobori 1986; Ota and Nei 1994). A ratio <1 indicates a prevalence of synonymous mutations (without impact on protein sequence and likely protein functions) resulting from a purifying selection. Conversely, a ratio >1 corresponds to an accumulation of non-synonymous mutations which likely altered the amino acid composition of proteins, which may indicate positive selection. The ratio we found for lophotrochozoan CRADD < 1 (dN/dS = 0.91) confirms purifying selection on these genes (supplementary table S9, Supplementary Material online).
Caspase-Y is restricted to the molluscs + annelids + brachiopods, indicating a diversification of CARD caspases in this clade. Interestingly, CARD caspase diversification in mammals led to a functional specialization between apoptosis and immunity. Caspase-1 is involved in the inflammatory response, while apoptosis is regulated by other CARD caspases such as caspase-2 or caspase-9 (Sollberger et al. 2014). In molluscs, it has been shown that caspase-2 of Crassostrea angulata is expressed during the apoptotic event of larvae metamorphosis, suggesting a primordial role of this caspase in apoptosis (Yang et al. 2015). Importantly, caspases have been reported to be fundamental in immune response to various stress and inflammation in several mollusc species (Sokolova et al. 2004; Sokolova 2009; Kiss 2010; Romero et al. 2015). Therefore, we may hypothesize that the diversification of CARD caspase in molluscs and their close relatives, annelids and brachiopods, may have induced separate functions as well. Caspase-2 could be involved in apoptosis and caspase-Y in immunity. Interestingly, we did not detect caspase-2 in the brachiopod L. anatina, although this caspase is known to be ancestral and highly conserved in bilaterians. This rather unique feature is only shared with the ambulacraria (hemichordates + echinoderms). This loss could be due to the diversification of caspase-Y (6 paralogues in Lingula) which are consequently the only remaining CARD caspase in brachiopods. Indeed, the loss of one type of CARD caspase is often accompanied by the duplication of the second one (Krasovec et al. 2023). Echinoderms are characterized by the loss of caspase-2 and the duplication of caspase-9, while it is the opposite for urochordates (Krasovec et al. 2023).
molluscs, along with annelids and brachiopods, possess both FADD and DED caspases, two actors known to trigger extrinsic apoptosis in mammals (Fig. 8). The theoretical binding affinity between both strongly suggests that extrinsic apoptosis could be activated with similar modality as that which is already known in mammals. However, the evolutionary relationships of DED caspases at the metazoan scale are still unclear, as the phylogenies performed so far are difficult to analyze and interpret (Sakamaki et al. 2014, 2015). Indeed, the topology of DED caspase sequences are significantly different to the species tree topology, and the branching of animal clades is not consistent with the current knowledge on metazoan relationships. Importantly, the presence of a potential extrinsic apoptosis in molluscs indicates that its absence in flies and nematodes could be a secondary loss that does not reflect the commonality of non-deuterostome animals. Finally, DED caspases and FADD are the only two apoptotic actors identified here to present a positive selection pressure (dN/dS > 1), a selection relaxation that could be due to the presence of both intrinsic and extrinsic apoptosis–like pathways, leading to functional redundancy (supplementary table S9, Supplementary Material online).
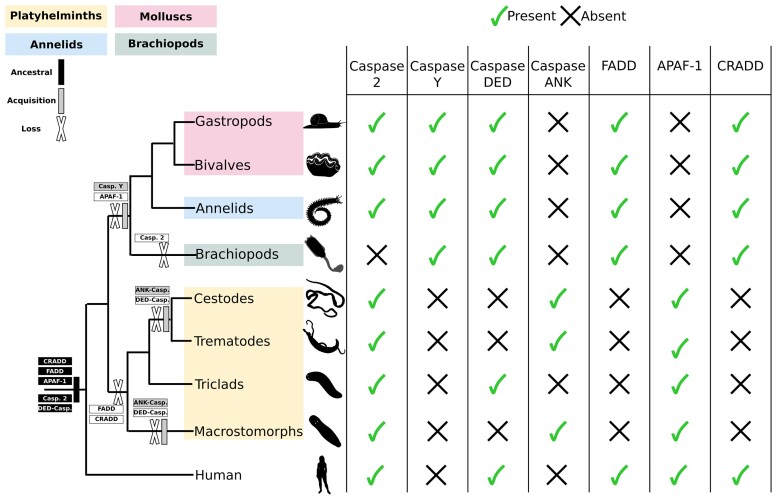
Fig. 8.
Summary of presence and absence of initiator caspases and their potential activators in lophotrochozoans. Caspase-2, which is the shared and ancestral common initiator in animals, is detected in most lophotrochozoans but is lost in brachiopods. Caspase-Y is a specific acquisition of molluscs, annelids, and brachiopods. DED caspases are lost in parasitic platyhelminths, which also acquired the specific ANK caspases. CARD caspases are likely to be activated by APAF-1 in platyhelminths and by CRADD in molluscs, annelids, and brachiopods. FADD, activator of DED caspase, is lost in all platyhelminths. This apoptotic network suggests a loss of extrinsic apoptosis in platyhelminths and a divergence in CARD caspase activation modality within lophotrochozoans.
Taken together, pioneering experimental works on lophotrochozoans (Pirger et al. 2004; Sokolova et al. 2004; Yang et al. 2015; Li et al. 2017) coupled with our study suggest that molluscs and close relatives present original apoptotic pathways which are different to what is known in mammals, flies, and nematodes.
Annelids Present a Variety of Species-Specific Initiator Caspases
Our study identifies annelids as the group with the most diverse caspase repertoire known to date. We discovered a high diversity in this group, both in terms of caspase number variation between species, but also regarding the presence of caspases with unusual pro-domains. Among them is a Z-binding domain caspase encoded by P. dumerilii. Interestingly, Z-binding proteins are known to activate necroptosis in mammals, suggesting a rich diversity of cell death in this annelid (Maelfait et al. 2017). The ROC pro-domain, present in a caspase from O. fusiformis, is known to be involved in protein GTP-binding, especially with kinase (Carlessi et al. 2011). This could allow the interaction with kinase proteins, such as death-associated protein kinase (known as DAPk) which is a mediator of cell death processes, cellular stress or autophagy regulation (Lin et al. 2010; Fujita and Yamashita 2014). The subtilase and P/Homo B domains discovered in a M. vulgaris caspase are both normally found in proteins with protease activity (Rawlings and Barrett 1994; Siezen and Leunissen 1997; Zhou et al. 1998; Anand et al. 2022). Interestingly, subtilisin-like serine proteases are able to cleave various substrates: aspartic acid, serine, and histidine. Consequently, this M. vulgaris caspase seems able to cleave not only aspartate residues, which would make it a very unique caspase in terms of catalytic activity.
Platyhelminths Conserve Intrinsic Apoptotic Actors but Have Lost Extrinsic Apoptosis Network
The history of DED caspase in platyhelminths is particularly interesting, as non-parasitic triclad platyhelminths possess one DED caspase while the parasitic species appear to have lost this gene. In addition, the DED caspase activator FADD has not been detected in platyhelminth genomes despite our investigations (Fig. 8). The loss of one of these actors may lead to a lack of mobilization of the other one, inducing a mutual relaxation of selection pressure leading to their parallel loss during evolution. Gene losses are common in animals with parasitic lifestyles due to genome and morphological simplification (Fairbairn 1970; Keeling 2004; Zarowiecki and Berriman 2015), which could explain the absence of some apoptotic actors in platyhelminths. This absence of extrinsic apoptosis in platyhelminths is shared with ecdysozoans but is likely the result of convergent evolution, as extrinsic apoptotic actors are documented at a large evolutionary scale and are notably present in molluscs, annelids, and brachiopods (this study; Lee et al. 2011; Sakamaki et al. 2014; Li et al. 2015).
However, all parasitic species display an ANK caspase, a type of caspase firstly identified in Schistosoma (Lee et al. 2014), which is unique to parasitic platyhelminths (with the exception of the A. virens annelid ANK caspase). Interestingly, ankyrin domains are known to be involved in protein–protein interaction and recognition (Mosavi et al. 2004). Furthermore, proteins with ankyrin domains are able to interact with caspases (Schweizer et al. 2007; Flütsch et al. 2014). Consequently, ANK caspases may be initiators leading to the activation of upstream executioner caspases. As ANK caspases are close to DED caspases, we may hypothesize that the acquisition of ankyrin pro-domains could be the result of a significant modification of the DED pro-domain of the ancestral DED caspases during evolution. Indeed, the recruitment of a new, but not yet identified, activator with an ankyrin domain was likely positively selected during the establishment of parasitism in these species. These specific caspases suggest a different pathway to the one currently known, which is of primary interest to investigate.
Platyhelminths present an apoptotic network composed of a unique CARD caspase (homologue to caspase-2) and the activator APAF-1, similarly to flies and nematodes. In contrast to ecdysozoans, platyhelminth BCL-2 members seem able to induce MOMP and consequently the subsequent cytochrome c release, at least in S. mediterranea (Bender et al. 2012). The intrinsic apoptosis of platyhelminths appears to be one underlain by CARD domain interactions, implicating the common APAF-1 activator, caspase-2 (like fruit flies and nematodes) and cytochrome c release (as in mammals). The resulting hypothetical apoptosome may share similarities with, or diverge from, apoptosomes from mammals and ecdysozoans and may represent a new type of activation platform (APAF-1 + caspase-2 + cytochrome c). The presence of a single pathway type likely induced purifying selection for conservation of these actors, since a deleterious modification or loss would lead to dysregulation of apoptosis (dN/dS < 1) (supplementary table S9, Supplementary Material online).
Apoptotic Signaling Pathways Evolve in a Divergent Way Between Metazoan Clades
The current understanding of apoptotic signaling pathways evolution has benefited from decades of studies conducted mainly on nematodes, flies, and mammals (Ellis and Horvitz 1986; Hengartner 2000; Lettre and Hengartner 2006; Steller 2008). Ecdysozoans are characterized by the loss of numerous genes and the absence of extrinsic apoptosis, whereas mammals have an abundance of different apoptotic players; this has led to the false conclusion that apoptotic pathways are simple in “invertebrates.” In this study, we discovered a large diversity of apoptotic network components in lophotrochozoans, along with clade-specific differences, indicating a complex evolution marked by multiple losses, acquisitions, and diversifications (Fig. 8).
Phylogenetic evidence suggests that the central initiator of intrinsic apoptosis, caspase-9, is deuterostome specific, while caspase-2 is conserved among metazoans and triggers apoptosis in flies and nematodes (Krasovec et al. 2023). Consequently, it is not surprising that the only shared initiator caspase that we have identified between all lophotrochozoan groups studied here is caspase-2. Functional evidence shows that caspase-2 in mammals and its orthologues Dronc and Ced-3 in flies and nematodes, respectively, are involved in a wide range of processes, including cell cycle regulation, tumor suppression, oxidative stress, ageing and cell death, and DNA repair (Tinel and Tschopp 2004; Krumschnabel et al. 2009b, 2; Krumschnabel et al. 2009a; Braga et al. 2008; Olsson et al. 2015). In addition, caspase-2 has the ability to activate both intrinsic and extrinsic apoptoses, in addition to DNA damage pathway apoptosis in mammals (Zhivotovsky and Orrenius 2005; Lavrik et al. 2006; Braga et al. 2008; Olsson et al. 2009). This employment capacity is highlighted by the large number of proteins able to interact with caspase-2, such as APAF-1 in ecdysozoans and CRADD or PIDD in mammals (Duan and Dixit 1997; Lin et al. 2000). This ancestral multifunctionality suggests that a mutation/loss of caspase-2 would lead to destabilization of various signaling cascades, increasing the likelihood of deleterious outcomes.
Our data suggest the existence of a possible apoptotic pathway in molluscs based on the caspase-2/CRADD interaction, and another one in platyhelminths triggered by the caspase-2/APAF-1 complex, in both cases relying on cytochrome c release. Cytochrome c release is likely an ancient trait of apoptosis regulation which is conserved in metazoans (Chipuk et al. 2010; Bender et al. 2012; Banjara et al. 2020), suggesting that intrinsic apoptosis in flies and nematodes reflects an accumulation of derivative features concerning the implication of BCL-2 and mitochondria. The conservation of both anti- and proapoptotic BCL-2 in lophotrochozoans certainly results in their ability to play on mitochondria and apoptosis, but also on their multiple, ancestral, and pleiotropic functions making them crucial in various molecular pathways (Bonneau et al. 2013; Prudent et al. 2013; Popgeorgiev et al. 2019; Popgeorgiev et al. 2020).
Finally, our data indicate that apoptotic signaling pathways have accumulated taxon-specific features during evolution, making these pathways divergent among metazoans and potentially convergent to the ultimate conserved trait: cell death executed by apoptosis regardless of the molecular pathways involved.
Materials and Methods
Sequence Dataset Construction
We conducted BLAST searches for caspases in a maximum of available lophotrochozoan genomes representing molluscs (highlighted in red along the figures), annelids (highlighted in blue), brachiopods (highlighted in dark green) and platyhelminths (highlighted in orange).
Putative lophotrochozoan genes were identified using tBLASTn and BLASTp searches using genes from humans, flies, nematodes, cnidarians, and oysters as queries. We extended the query in accordance to the results (i.e. newly identified caspases were added to the query). Reciprocal BLAST was performed on all genes, including newly identified lophotrochozoan sequences. BLAST searching was conducted using NCBI databases for most species, and on PdumBase in addition to our recent transcriptome for annelid P. dumerilii (Chou et al. 2018; Paré et al. 2023). After identification of genes in target species, proteins were analyzed with ScanProsite (ExPaSy) (Gattiker et al. 2002) to verify the presence of specific domains which are the P20 and P10 for caspases, CARD, and death domains for CRADD, CARD, and NB-ARC for APAF-1, DED, and death domains for FADD (see protein domains architecture, Fig. 1). Sequences arising from BLAST search but devoid of the usual domains were not conserved to prevent false positives. Sampling was conducted on available lophotrochozoan genomes/transcriptomes, and for each species, all caspases and caspase activators were explored.
Caspase family proteins are short (containing the large common P20 and the small P10 domains) with a high number of genes per species, which rapidly limits the relevance of the phylogenetic analyses (see supplementary fig. S2, Supplementary Material online, as example) (Krasovec et al. 2023). To reduce artifact branching and unreadable topologies, and to maximize phylogenetic diversity across lophotrochozoans, the dataset constructed for phylogenetic analysis with all types of caspases was built with restricted number of representative lophotrochozoan species which recapitulate lophotrochozoan diversity. Secondly, analysis of caspases with long pro-domains only was conducted with all species possible (for which unambiguous caspase identifications were made). We provide here Supplementary Files 1 to 4 containing all caspase sequences we found (including the executioner ones) for annelids, brachiopod, molluscs, and platyhelminths, respectively, which are present in the long pro-domain–only phylogeny (topology in Fig. 3). In case of redundancy (several accession numbers for one protein), we kept only one of them. The BCL-2 phylogeny was also conducted on a representative species subset to overcome the same aforementioned artifacts which we already experienced.
FADD, CRADD, and APAF-1 analyses were conducted with all sequences identified at the lophotrochozoans scale which had the corresponding specific domains.
Multiple alignments of protein sequences were generated using MAFFT version 7 (Katoh and Standley 2013) with default parameters, and Clustal Omega (Sievers et al. 2011), to verify the congruence of the different alignments. All sequences were then manually checked in BioEdit 7.2 software to verify the presence of specific domains previously identified. Gblocks version 0.91b (Castresana 2000) was used to remove vacancies and blur sites. Final alignments comprise 424, 218, 344, 337, 556, 106, and 178 amino acids for the following alignments: all caspase alignment, caspases with long pro-domain alignment, annelid caspase alignment, BCL-2 alignment, Apaf-1, CRADD alignment, and FADD alignment, respectively.
Sampling Strategy
Species sampling was conducted to include the highest diversity of lophotrochozoans possible in our analysis. When sampling, we looked for caspases and potential activators in a maximum range of genomes of lophotrochozoan species. Regarding the APAF-1, FADD, CRADD, and initiator caspase sampling, we collected all sequences we detected. The binding score was calculated using PSOPIA for all species were CARD caspases plus APAF-1 and/or CRADD were identified, and same for DED caspases plus FADD. Species sampled in one of our caspase activator phylogenies but which is not included in the PSOPIA score analysis are species for which we did not detect initiator caspases candidates. Similarly, species represented in initiator caspase analysis but not in APAF-1, CRADD, or FADD analyses are species for which we did not detect caspase activator candidates. We maintained a maximum consistency between our analyses by keeping the same species. However, there are some differences resulting in species specificity and likely heterogeneity in genome properties. For example, we fund molluscs in which we did not detect any CRADD (i.e. Gigantopelta aegis), platyhelminths without APAF-1 (i.e. Hymenolepis diminuta), annelids and molluscs devoid of DED caspases (i.e. H. robusta and Candidula unifasciata, respectively), or annelids without CARD caspases (T. tubifex). Further details of this variability are available in supplementary table S1, Supplementary Material online. This variability explains why some species are present in the Caspases analyses but not activator analyses, and conversely. Consequently, the differences in species sampling between our analyses are only the result of the diversity of lophotrochozoans in terms of the repertoire of apoptotic actors.
Regarding the analysis on all type of caspases and BCL-2, we had to change the strategy to run the phylogenetic analysis in accordance with alignment length, in other words, the quantity of phylogenetic information. Caspase and BCL-2 proteins are short, which makes it challenging to conduct phylogenetic analyses with a large number of species, and consequently a large number of sequences (especially when including all of the multigene family), without sacrificing reliability. For these reasons, model animals and taxa were restricted, but animals with functional or well-studied genomic data were prioritized, in addition to be representative of the lophotrochozoans diversity.
Phylogenetic Analysis and Sequence Analysis
Phylogenetic analyses were carried out from the amino acid alignment with ML method using PhyML 3.1 (Guindon et al. 2010). The best amino acid evolution models to conduct analyses were determined using MEGA11 (Tamura et al. 2021) and appeared to be WAG for the caspase alignments, Apaf-1 alignment, CRADD alignment, FADD alignment, and cpREV for the BCL-2 alignment. Node robustness was evaluated by 500 bootstrap replicate sampling.
Bayesian analyses were performed using MrBayes (v3.2.6) (Ronquist and Huelsenbeck 2003) under mixed model. Consensus trees were made using the half-compact parameter. For each analysis, one-fourth of the topologies were discarded as burn-in values, while the remaining ones were used to calculate posterior probability. The run for the caspase alignment was carried out for 3,000,000 generations with 15 randomly started simultaneous Markov chains (1 cold chain, 14 heated chains) and sampled every 100 generations. The runs for APAF-1, CRADD, and FADD alignments were carried out for 500,000 generations with 15 randomly started simultaneous Markov chains (1 cold chain, 14 heated chains) and sampled every 100 generations. For the BCL-2, 3,000,000 generations with 15 randomly started simultaneous Markov chains (1 cold chain, 14 heated chains) were conducted and sampled every 100 generations. ML bootstrap values >50% and Bayesian posterior probabilities are indicated on the Bayesian tree. The outgroup for the all caspase phylogeny is R. filosa (ETO10778.1), a sequence recognized to be sister of metazoan caspases (Klim et al. 2018). For the BCL-2 phylogeny, the outgroup used is a BCL-2–like from Porifera Amphimedon queenslandica (XP_003387574.1). Outgroups for APAF-1, CRADD, and FADD phylogenies are their human relatives.
Topology tests were evaluated using IQ-TREE (Nguyen et al. 2015). We conducted Kishino–Hasegawa test (Kishino and Hasegawa 1989), Shimodaira–Hasegawa test (Shimodaira and Hasegawa 1999), and approximately unbiased test (Shimodaira 2002).
The dN/dS ratio was calculated using SNAP v2.1.1 with default parameters on the ORFs previously codon-aligned by HIVAlign tool. SNAP v2.1.1 sort out raw Dn and Ds allowing next calculation of the ratio as presented in supplementary table S9, Supplementary Material online.
Protein–Protein Binding Score Prediction
Theoretical protein–protein binding scores were calculated using PSOPIA (Murakami and Mizuguchi 2014). Binding scores range from 0 (no affinity) to 1 (maximum binding probability estimation). For each category of measures (CARD caspase/APAF-1; CARD caspase/CRADD; FADD/DED caspase) we included positive controls (the relevant paired vertebrate proteins according to the literature) and negative controls (a caspase plus a randomly selected protein which should not interact). Negative controls for DED caspase/FADD binding (Fig. 6B1) are Negative-1, C. gigas FADD (NP_001295786.1) plus Phodopus campbelli insulin-like growth factor 2 (AFQ62592.1); Negative-2, Mus musculus caspase-8 (AAH49955.1) plus Hydra vulgaris rhamnose-binding lectin (XP_002166914.1). Positive controls for DED caspase/FADD binding (Fig. 6B1) are Positive-1, Homo sapiens caspase-8 (AAD24962.1) plus FADD (CAG33019.1); Positive-2, M. musculus Caspase-8 (AAH49955.1) plus FADD (AAA97876.1). Negative controls for CARD caspase/APAF-1 binding (Fig. 6B2) are Negative-1, M. lignano CARD caspase (PAA59014.1) plus Columba livia Xrcc4 (PKK31261.1), and Negative-2, D. melanogaster Dronc (NP_524017.1) plus H. vulgaris rhamnose-binding lectin (XP_002166914.1). Positive controls for CARD caspase/APAF-1 binding (Fig. 6B2) are Positive-1, H. sapiens caspase-9 (BAA82697.1) plus APAF-1 (NP_863651.1), and Positive-2, C. elegans Ced-3 (AAG42045.1) plus Ced-4 (CAA48781.1). Negative control for CARD caspase/CRADD binding (Fig. 6B3) is Negative-1, M. lignano CARD caspase (PAA59014.1) plus Columba livia Xrcc4 (PKK31261.1). Positive control for CARD caspase/CRADD binding (Fig. 6B3) is Positive-1, H. sapiens caspase-2 (KAI4016188.1) plus CRADD (CAG28571.1).
3D Protein Models
3D structures were predicated using Phyre (Kelley et al. 2015) in normal mode, and the best model was selected based on percentage alignment coverage and confidence score. The resultant PDB files were imported to Chimera X (Goddard et al. 2018) for model construction. Functional domains were detected using ScanProsite (ExPaSy).
Supplementary Material
Acknowledgments
The authors acknowledge members of the Uri Frank lab (University of Galway, Ireland) and Eve Gazave lab (Institut Jacques Monod, France).
Contributor Information
Helen R Horkan, Centre for Chromosome Biology, School of Biological and Chemical Sciences, University of Galway, Galway, Ireland.
Nikolay Popgeorgiev, Centre de Recherche en Cancérologie de Lyon, U1052 INSERM, UMR CNRS 5286, Centre Léon Bérard, Université Claude Bernard Lyon 1, Lyon, France; Institut Universitaire de France (IUF), Paris, France.
Michel Vervoort, Université Paris Cité, CNRS, Institut Jacques Monod, F-75013 Paris, France.
Eve Gazave, Université Paris Cité, CNRS, Institut Jacques Monod, F-75013 Paris, France.
Gabriel Krasovec, Centre for Chromosome Biology, School of Biological and Chemical Sciences, University of Galway, Galway, Ireland; Université Paris Cité, CNRS, Institut Jacques Monod, F-75013 Paris, France.
Supplementary Material
Supplementary material is available at Genome Biology and Evolution online.
Author Contributions
G.K. managed the project. G.K. and H.R.H. wrote the manuscript. H.R.H., G.K., M.V., N.P., and E.G. produced the data. All authors contributed to the comments and discussion and approved the manuscript.
Funding
G.K. was a postdoctoral fellow funded by the Irish Research Council (project GOIPD/2020/149) and is currently funded by the Fondation ARC pour la recherche sur le cancer (project ARCPOST-DOC2022070005318). H.R.H. was a doctoral student in the Science Foundation Ireland (Centre for Research Training in Genomic Data Science, grant no. 18/CRT/6214). E.G. is funded by Association pour la Recherche sur le Cancer (grant PJA 20191209482), and La Ligue Nationale Contre le Cancer (grant RS20/75-20). N.P. is funded by Institut Universitaire de France (IUF), Paris, France.
Data Availability
All data needed to evaluate the conclusions in this study are present in the paper and the Supplementary Materials. Alignments used to run the phylogenies are available on Figshare (https://doi.org/10.6084/m9.figshare.26984449.v1.). Any requests can be addressed to the corresponding author G.K. and H.R.H.
Literature Cited
- Anand D, Hummler E, Rickman OJ. ENaC activation by proteases. Acta Physiol (Oxf). 2022:235(1):e13811. 10.1111/apha.13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AnvariFar H, Amirkolaie AK, Miandare HK, Ouraji H, Jalali MA, Üçüncü Sİ. Apoptosis in fish: environmental factors and programmed cell death. Cell Tissue Res. 2017:368(3):425–439. 10.1007/s00441-016-2548-x. [DOI] [PubMed] [Google Scholar]
- Aouacheria A, Rech de Laval V, Combet C, Hardwick JM. Evolution of Bcl-2 homology motifs: homology versus homoplasy. Trends Cell Biol. 2013:23(3):103–111. 10.1016/j.tcb.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballarin L, Schiavon F, Manni L. Natural apoptosis during the blastogenetic cycle of the colonial ascidian Botryllus schlosseri: a morphological analysis. Zool Sci. 2010:27(2):96–102. 10.2108/zsj.27.96. [DOI] [PubMed] [Google Scholar]
- Banjara S, Suraweera CD, Hinds MG, Kvansakul M. The Bcl-2 family: ancient origins, conserved structures, and divergent mechanisms. Biomolecules. 2020:10(1):128. 10.3390/biom10010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender CE, Fitzgerald P, Tait SWG, Llambi F, McStay GP, Tupper DO, Pellettieri J, Sánchez Alvarado A, Salvesen GS, Green DR. Mitochondrial pathway of apoptosis is ancestral in metazoans. Proc Natl Acad Sci U S A. 2012:109(13):4904–4909. 10.1073/pnas.1120680109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau B, Prudent J, Popgeorgiev N, Gillet G. Non-apoptotic roles of Bcl-2 family: the calcium connection. Biochim Biophys Acta. 2013:1833(7):1755–1765. 10.1016/j.bbamcr.2013.01.021. [DOI] [PubMed] [Google Scholar]
- Braga M, Sinha Hikim AP, Datta S, Ferrini MG, Brown D, Kovacheva EL, Gonzalez-Cadavid NF, Sinha-Hikim I. Involvement of oxidative stress and caspase 2-mediated intrinsic pathway signaling in age-related increase in muscle cell apoptosis in mice. Apoptosis. 2008:13(6):822–832. 10.1007/s10495-008-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratton SB, Salvesen GS. Regulation of the apaf-1–caspase-9 apoptosome. J Cell Sci. 2010:123(Pt 19):3209–3214. 10.1242/jcs.073643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlessi R, Levin-Salomon V, Ciprut S, Bialik S, Berissi H, Albeck S, Peleg Y, Kimchi A. GTP binding to the ROC domain of DAP-kinase regulates its function through intramolecular signalling. EMBO Rep. 2011:12(9):917–923. 10.1038/embor.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000:17(4):540–552. 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010:37(3):299–310. 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H-C, Acevedo-Luna N, Kuhlman JA, Schneider SQ. PdumBase: a transcriptome database and research tool for Platynereis dumerilii and early development of other metazoans. BMC Genomics. 2018:19(1):618. 10.1186/s12864-018-4987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen GM. Caspases: the executioners of apoptosis. Biochem J. 1997:326(1):1–16. 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorstyn L, Read S, Cakouros D, Huh JR, Hay BA, Kumar S. The role of cytochrome c in caspase activation in Drosophila melanogaster cells. J Cell Biol. 2002:156(6):1089–1098. 10.1083/jcb.200111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll M. Cell death in C. elegans: molecular insights into mechanisms conserved between nematodes and mammals. Brain Pathol. 1996:6(4):411–425. 10.1111/j.1750-3639.1996.tb00873.x. [DOI] [PubMed] [Google Scholar]
- Duan H, Dixit VM. RAIDD is a new ‘death’ adaptor molecule. Nature. 1997:385(6611):86–89. 10.1038/385086a0. [DOI] [PubMed] [Google Scholar]
- Ellis HM, Horvitz HR. Genetic control of programmed cell death in the nematode C. elegans. Cell. 1986:44(6):817–829. 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- Fairbairn D. Biochemical adaptation and loss of genetic capacity in helminth parasites. Biol Rev Camb Philos Soc. 1970:45(1):29–72. 10.1111/j.1469-185x.1970.tb01075.x. [DOI] [PubMed] [Google Scholar]
- Fan T-J, Han L-H, Cong R-S, Liang J. Caspase family proteases and apoptosis. Acta Biochim Biophys Sin (Shanghai). 2005:37(11):719–727. 10.1111/j.1745-7270.2005.00108.x. [DOI] [PubMed] [Google Scholar]
- Flütsch A, Schroeder T, Barandun J, Ackermann R, Bühlmann M, Grütter MG. Specific targeting of human caspases using designed ankyrin repeat proteins. Biol Chem. 2014:395(10):1243–1252. 10.1515/hsz-2014-0173. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Yamashita T. Role of DAPK in neuronal cell death. Apoptosis. 2014:19(2):339–345. 10.1007/s10495-013-0917-4. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Vitale I, Aaronson SA, Abrams JM, Adam D, Agostinis P, Alnemri ES, Altucci L, Amelio I, Andrews DW, et al. Molecular mechanisms of cell death: recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018:25(3):486–541. 10.1038/s41418-017-0012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattiker A, Gasteiger E, Bairoch A. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl Bioinformatics. 2002:1(2):107–108. [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. UCSF chimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 2018:27(1):14–25. 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010:59(3):307–321. 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hengartner MO. The biochemistry of apoptosis. Nature. 2000:407(6805):770–776. 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- Jacobson MD, Weil M, Raff MC. Programmed cell death in animal development. Cell. 1997:88(3):347–354. 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- Jeffery WR, Gorički Š. Apoptosis is a generator of Wnt-dependent regeneration and homeostatic cell renewal in the ascidian Ciona. Biol Open. 2021:10(4):bio058526. 10.1242/bio.058526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F, Zhou Z, Guo Q, Liang Z, Yang R, Jiang J, He Y, Zhao Q, Zhao Q. High-quality genome assembly of Metaphire vulgaris. PeerJ. 2020:8:e10313. 10.7717/peerj.10313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkavan H, Green DR. MOMP, cell suicide as a BCL-2 family business. Cell Death Differ. 2018:25(1):46–55. 10.1038/cdd.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013:30(4):772–780. 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal V, Klim J, Skoneczna A, Kurlandzka A, Enkhbaatar T, Kaczanowski S, Zielenkiewicz U. Apoptotic factors are evolutionarily conserved since mitochondrial domestication. Genome Biol Evol. 2023:15(10):evad154. 10.1093/gbe/evad154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ. Reduction and compaction in the genome of the apicomplexan parasite cryptosporidium parvum. Dev Cell. 2004:6(5):614–616. 10.1016/s1534-5807(04)00135-2. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015:10(6):845–858. 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J Mol Evol. 1989:29(2):170–179. 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- Kiss T. Apoptosis and its functional significance in molluscs. Apoptosis. 2010:15(3):313–321. 10.1007/s10495-009-0446-3. [DOI] [PubMed] [Google Scholar]
- Klim J, Gładki A, Kucharczyk R, Zielenkiewicz U, Kaczanowski S. Ancestral state reconstruction of the apoptosis machinery in the common ancestor of eukaryotes. G3 (Bethesda). 2018:8(6):2121–2134. 10.1534/g3.118.200295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasovec G, Horkan HR, Quéinnec É, Chambon J-P. The constructive function of apoptosis: more than a dead-end job. Front Cell Dev Biol. 2022:10:1033645. 10.3389/fcell.2022.1033645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasovec G, Horkan HR, Quéinnec É, Chambon J-P. Intrinsic apoptosis is evolutionarily divergent among metazoans. Evol Lett. 2023:8(2):267–282. 10.1093/evlett/qrad057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasovec G, Pottin K, Rosello M, Quéinnec É, Chambon J-P. Apoptosis and cell proliferation during metamorphosis of the planula larva of Clytia hemisphaerica (Hydrozoa, Cnidaria). Dev Dyn. 2021:250(12):1739–1758. 10.1002/dvdy.376. [DOI] [PubMed] [Google Scholar]
- Krasovec G, Renaud C, Quéinnec É, Sasakura Y, Chambon J-P. Extrinsic apoptosis participates to tail regression during the metamorphosis of the chordate Ciona. Sci Rep. 2024:14(1):5729. 10.1038/s41598-023-48411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasovec G, Robine K, Quéinnec E, Karaiskou A, Chambon JP. Ci-hox12 tail gradient precedes and participates in the control of the apoptotic-dependent tail regression during Ciona larva metamorphosis. Dev Biol. 2019:448(2):237–246. 10.1016/j.ydbio.2018.12.010. [DOI] [PubMed] [Google Scholar]
- Krumschnabel G, Manzl C, Villunger A. Caspase-2: killer, savior and safeguard–emerging versatile roles for an ill-defined caspase. Oncogene. 2009a:28(35):3093–3096. 10.1038/onc.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumschnabel G, Sohm B, Bock F, Manzl C, Villunger A. The enigma of caspase-2: the laymen's view. Cell Death Differ. 2009b:16(2):195–207. 10.1038/cdd.2008.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. Caspase function in programmed cell death. Cell Death Differ. 2007:14(1):32–43. 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- Lavrik IN, Golks A, Baumann S, Krammer PH. Caspase-2 is activated at the CD95 death-inducing signaling complex in the course of CD95-induced apoptosis. Blood. 2006:108(2):559–565. 10.1182/blood-2005-07-007096. [DOI] [PubMed] [Google Scholar]
- Lee Y, De Zoysa M, Whang I, Lee S, Kim Y, Oh C, Choi CY, Yeo S-Y, Lee J. Molluscan death effector domain (DED)-containing caspase-8 gene from disk abalone (Haliotis discus discus): molecular characterization and expression analysis. Fish Shellfish Immunol. 2011:30(2):480–487. 10.1016/j.fsi.2010.11.014. [DOI] [PubMed] [Google Scholar]
- Lee EF, Young ND, Lim NTY, Gasser RB, Fairlie WD. Apoptosis in schistosomes: toward novel targets for the treatment of schistosomiasis. Trends Parasitol. 2014:30(2):75–84. 10.1016/j.pt.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Lettre G, Hengartner MO. Developmental apoptosis in C. elegans: a complex CEDnario. Nat Rev Mol Cell Biol. 2006:7(2):97–108. 10.1038/nrm1836. [DOI] [PubMed] [Google Scholar]
- Lewin TD, Liao IJ-Y, Luo Y-J. Annelid comparative genomics and the evolution of massive lineage-specific genome rearrangement in bilaterians. Mol Biol Evol. 2024:41(9):msae172. 10.1093/molbev/msae172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Qu T, Huang B, Ji P, Huang W, Que H, Li L, Zhang G. Cloning and characterization of a novel caspase-8-like gene in Crassostrea gigas. Fish Shellfish Immunol. 2015:46(2):486–492. 10.1016/j.fsi.2015.06.035. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang L, Qu T, Tang X, Li L, Zhang G. Conservation and divergence of mitochondrial apoptosis pathway in the Pacific oyster, Crassostrea gigas. Cell Death Dis. 2017:8(7):e2915. 10.1038/cddis.2017.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Hupp TR, Stevens C. Death-associated protein kinase (DAPK) and signal transduction: additional roles beyond cell death. FEBS J. 2010:277(1):48–57. 10.1111/j.1742-4658.2009.07411.x. [DOI] [PubMed] [Google Scholar]
- Lin Y, Ma W, Benchimol S. Pidd, a new death-domain-containing protein, is induced by p53 and promotes apoptosis. Nat Genet. 2000:26(1):122–127. 10.1038/79102. [DOI] [PubMed] [Google Scholar]
- Maelfait J, Liverpool L, Bridgeman A, Ragan KB, Upton JW, Rehwinkel J. Sensing of viral and endogenous RNA by ZBP1/DAI induces necroptosis. EMBO J. 2017:36(17):2529–2543. 10.15252/embj.201796476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Durán JM, Vellutini BC, Marlétaz F, Cetrangolo V, Cvetesic N, Thiel D, Henriet S, Grau-Bové X, Carrillo-Baltodano AM, Gu W, et al. Conservative route to genome compaction in a miniature annelid. Nat Ecol Evol. 2021:5(2):231–242. 10.1038/s41559-020-01327-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldoveanu T, Czabotar PE. BAX, BAK, and BOK: a coming of age for the BCL-2 family effector proteins. Cold Spring Harb Perspect Biol. 2020:12(4):a036319. 10.1101/cshperspect.a036319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng Z-Y. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004:13(6):1435–1448. 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Mizuguchi K. Homology-based prediction of interactions between proteins using averaged one-dependence estimators. BMC Bioinformatics. 2014:15(1):213. 10.1186/1471-2105-15-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986:3(5):418–426. 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015:32(1):268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M, Forsberg J, Zhivotovsky B. Caspase-2: the reinvented enzyme. Oncogene. 2015:34(15):1877–1882. 10.1038/onc.2014.139. [DOI] [PubMed] [Google Scholar]
- Olsson M, Vakifahmetoglu H, Abruzzo PM, Högstrand K, Grandien A, Zhivotovsky B. DISC-mediated activation of caspase-2 in DNA damage-induced apoptosis. Oncogene. 2009:28(18):1949–1959. 10.1038/onc.2009.36. [DOI] [PubMed] [Google Scholar]
- Ota T, Nei M. Variance and covariances of the numbers of synonymous and nonsynonymous substitutions per site. Mol Biol Evol. 1994:11(4):613–619. 10.1093/oxfordjournals.molbev.a040140. [DOI] [PubMed] [Google Scholar]
- Paré L, Bideau L, Baduel L, Dalle C, Benchouaia M, Schneider SQ, Laplane L, Clément Y, Vervoort M, Gazave E. Transcriptomic landscape of posterior regeneration in the annelid Platynereis dumerilii. BMC Genomics. 2023:24(1):583. 10.1186/s12864-023-09602-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennarun B, Meijer A, de Vries EGE, Kleibeuker JH, Kruyt F, de Jong S. Playing the DISC: turning on TRAIL death receptor-mediated apoptosis in cancer. Biochim Biophys Acta. 2010:1805(2):123–140. 10.1016/j.bbcan.2009.11.004. [DOI] [PubMed] [Google Scholar]
- Pirger Z, Elekes K, Kiss T. Functional morphology of the salivary gland of the snail, Helix pomatia: a histochemical and immunocytochemical study. Acta Biol Hung. 2004:55(1-4):221–232. 10.1556/ABiol.55.2004.1-4.27. [DOI] [PubMed] [Google Scholar]
- Pirger Z, Rácz B, Kiss T. Dopamine-induced programmed cell death is associated with cytochrome c release and caspase-3 activation in snail salivary gland cells. Biol Cell. 2009:101(2):105–116. 10.1042/BC20070168. [DOI] [PubMed] [Google Scholar]
- Popgeorgiev N, Jabbour L, Nguyen TTM, Ralchev N, Gadet R, Manon S, Osigus H-J, Schierwater B, Rimokh R, Gillet G. Pleiotropy of Bcl-2 family proteins is an ancient trait in the metazoan evolution. bioRxiv 816322. 10.1101/816322. 24 October 2019, preprint: not peer reviewed. [DOI]
- Popgeorgiev N, Sa JD, Jabbour L, Banjara S, Nguyen TTM, Akhavan-E-Sabet A, Gadet R, Ralchev N, Manon S, Hinds MG, et al. Ancient and conserved functional interplay between Bcl-2 family proteins in the mitochondrial pathway of apoptosis. Sci Adv. 2020:6(40):eabc4149. 10.1126/sciadv.abc4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudent J, Popgeorgiev N, Bonneau B, Thibaut J, Gadet R, Lopez J, Gonzalo P, Rimokh R, Manon S, Houart C, et al. Bcl-wav and the mitochondrial calcium uniporter drive gastrula morphogenesis in zebrafish. Nat Commun. 2013:4(1):2330. 10.1038/ncomms3330. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ. Families of serine peptidases. Methods Enzymol. 1994:244:19–61. 10.1016/0076-6879(94)44004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero A, Novoa B, Figueras A. The complexity of apoptotic cell death in mollusks: an update. Fish Shellfish Immunol. 2015:46(1):79–87. 10.1016/j.fsi.2015.03.038. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003:19(12):1572–1574. 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sakamaki K, Imai K, Tomii K, Miller DJ. Evolutionary analyses of caspase-8 and its paralogs: deep origins of the apoptotic signaling pathways. Bioessays. 2015:37(7):767–776. 10.1002/bies.201500010. [DOI] [PubMed] [Google Scholar]
- Sakamaki K, Shimizu K, Iwata H, Imai K, Satou Y, Funayama N, Nozaki M, Yajima M, Nishimura O, Higuchi M, et al. The apoptotic initiator caspase-8: its functional ubiquity and genetic diversity during animal evolution. Mol Biol Evol. 2014:31(12):3282–3301. 10.1093/molbev/msu260. [DOI] [PubMed] [Google Scholar]
- Schweizer A, Roschitzki-Voser H, Amstutz P, Briand C, Gulotti-Georgieva M, Prenosil E, Binz HK, Capitani G, Baici A, Plückthun A, et al. Inhibition of caspase-2 by a designed ankyrin repeat protein: specificity, structure, and inhibition mechanism. Structure. 2007:15(5):625–636. 10.1016/j.str.2007.03.014. [DOI] [PubMed] [Google Scholar]
- Shi Y. Mechanical aspects of apoptosome assembly. Curr Opin Cell Biol. 2006:18(6):677–684. 10.1016/j.ceb.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002:51(3):492–508. 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999:16(8):1114–1116. 10.1093/oxfordjournals.molbev.a026201. [DOI] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011:7(1):539. 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen RJ, Leunissen JA. Subtilases: the superfamily of subtilisin-like serine proteases. Protein Sci. 1997:6(3):501–523. 10.1002/pro.5560060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova IM. Apoptosis in molluscan immune defense. Invertebrate Surviv J. 2009:6:49–58. [Google Scholar]
- Sokolova IM, Evans S, Hughes FM. Cadmium-induced apoptosis in oyster hemocytes involves disturbance of cellular energy balance but no mitochondrial permeability transition. J Exp Biol. 2004:207(19):3369–3380. 10.1242/jeb.01152. [DOI] [PubMed] [Google Scholar]
- Sollberger G, Strittmatter GE, Garstkiewicz M, Sand J, Beer H-D. Caspase-1: the inflammasome and beyond. Innate Immun. 2014:20(2):115–125. 10.1177/1753425913484374. [DOI] [PubMed] [Google Scholar]
- Steichele M, Sauermann LS, König A-C, Hauck S, Böttger A. Ancestral role of TNF-R pathway in cell differentiation in the basal metazoan Hydra. J Cell Sci. 2021:134(2):jcs255422. 10.1242/jcs.255422. [DOI] [PubMed] [Google Scholar]
- Steller H. Regulation of apoptosis in Drosophila. Cell Death Differ. 2008:15(7):1132–1138. 10.1038/cdd.2008.50. [DOI] [PubMed] [Google Scholar]
- Sutterwala FS, Ogura Y, Zamboni DS, Roy CR, Flavell RA. NALP3: a key player in caspase-1 activation. J Endotoxin Res. 2006:12(4):251–256. 10.1177/09680519060120040701. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021:38(7):3022–3027. 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinel A, Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science (1979). 2004:304(5672):843–846. 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- Vega Thurber R, Epel D. Apoptosis in early development of the sea urchin, Strongylocentrotus purpuratus. Dev Biol. 2007:303(1):336–346. 10.1016/j.ydbio.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Yang B, Li L, Pu F, You W, Huang H, Ke C. Molecular cloning of two molluscan caspases and gene functional analysis during Crassostrea angulata (Fujian oyster) larval metamorphosis. Mol Biol Rep. 2015:42(5):963–975. 10.1007/s11033-014-3833-y. [DOI] [PubMed] [Google Scholar]
- Yuan S, Liu H, Gu M, Xu L, Huang S, Ren Z, Xu A. Characterization of the extrinsic apoptotic pathway in the basal chordate amphioxus. Sci Signal. 2010:3(139):ra66. 10.1126/scisignal.2000906. [DOI] [PubMed] [Google Scholar]
- Zarowiecki M, Berriman M. What helminth genomes have taught us about parasite evolution. Parasitology. 2015:142(Suppl 1):S85–S97. 10.1017/S0031182014001449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Ren C, Chen T, Sun H, Wu X, Jiang X, Huang W. The first cloned sea cucumber FADD from Holothuria leucospilota: molecular characterization, inducible expression and involvement of apoptosis. Fish Shellfish Immunol. 2019:89:548–554. 10.1016/j.fsi.2019.04.030. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky B, Orrenius S. Caspase-2 function in response to DNA damage. Biochem Biophys Res Commun. 2005:331(3):859–867. 10.1016/j.bbrc.2005.03.191. [DOI] [PubMed] [Google Scholar]
- Zhou A, Martin S, Lipkind G, LaMendola J, Steiner DF. Regulatory roles of the P domain of the subtilisin-like prohormone convertases. J Biol Chem. 1998:273(18):11107–11114. 10.1074/jbc.273.18.11107. [DOI] [PubMed] [Google Scholar]
- Zmasek CM, Godzik A. Evolution of the animal apoptosis network. Cold Spring Harb Perspect Biol. 2013:5(3):a008649. 10.1101/cshperspect.a008649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997:90(3):405–413. 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in this study are present in the paper and the Supplementary Materials. Alignments used to run the phylogenies are available on Figshare (https://doi.org/10.6084/m9.figshare.26984449.v1.). Any requests can be addressed to the corresponding author G.K. and H.R.H.