Abstract
Brown adipose tissue (BAT) represents an evolutionary innovation enabling placental mammals to regulate body temperature through adaptive thermogenesis. Brown adipocytes are surrounded by a dense network of blood vessels and sympathetic nerves that support their development and thermogenic function. Cold exposure stimulates BAT thermogenesis through the coordinated induction of brown adipogenesis, angiogenesis, and sympathetic innervation. However, how these distinct processes are coordinated remains unclear. Here, we identify Slit guidance ligand 3 (Slit3) as a new niche factor that mediates the crosstalk among adipocyte progenitors, endothelial cells, and sympathetic nerves. We show that adipocyte progenitors secrete Slit3 which regulates both angiogenesis and sympathetic innervation in BAT and is essential for BAT thermogenesis in vivo. Proteolytic cleavage of Slit3 generates secreted Slit3-N and Slit3-C fragments, which activate distinct receptors to stimulate angiogenesis and sympathetic innervation, respectively. Moreover, we introduce bone morphogenetic protein-1 (Bmp1) as the first Slit protease identified in vertebrates. In summary, this study underscores the essential role of Slit3-mediated neurovascular network expansion in enabling cold-induced BAT adaptation. The co-regulation of neurovascular expansion by Slit3 fragments provides a bifurcated yet harmonized approach to ensure a synchronized response of BAT to environmental challenges. This study presents the first evidence that adipocyte progenitors regulate tissue innervation, revealing a previously unrecognized dimension of cellular interaction within adipose tissue.
The regulation of body temperature, thermoregulation, is a fundamental homeostatic process in endothermic animals. Preserving a stable internal temperature ensures the efficiency and fidelity of all cellular reactions and is essential for survival. Brown adipose tissue (BAT) is a specialized type of adipose tissue that is primarily responsible for regulating body temperature through adaptive thermogenesis. Brown adipocytes oxidize substrates and generate heat to maintain euthermia in a cold environment1.
Brown adipocytes are intricately embedded in a dense network of capillaries and sympathetic nerves. The high thermogenic activity of BAT requires a high rate of blood perfusion to supply O2 and substrates. The sympathetic nerves also play a key role in stimulating BAT thermogenesis by releasing norepinephrine in the tissue. Norepinephrine activates adrenergic signaling in thermogenic adipocytes, resulting in enhanced thermogenesis and lipolysis2. Chronic cold exposure increases BAT mass by de novo recruitment of brown adipocytes, as well as by expanding the network of blood vessels and sympathetic nerves in the tissue. This coordinated expansion of the BAT ensures its continuous responsiveness to hormonal and neuronal stimuli and is essential for enhanced thermogenesis in cold3,4. However, how these distinct processes are spatiotemporally coordinated remains unclear. Furthermore, while significant progress has been made in understanding the molecular mechanisms of thermogenic activation in adipocytes, the equally crucial process of remodeling the thermogenic adipose niche remains less understood.
Cell–cell communication is vital for organismal development and function. Ligand-receptor interactions allow cells in complex tissues to coordinate their functions during development, homeostasis, and remodeling. To understand the mechanisms through which different cell types in BAT communicate and synchronize their response to cold to collectively enhance thermogenesis, we recently used single-cell transcriptomic data5 to construct a network of ligand-receptor interactions in BAT6. That study highlighted the central role of adipocyte progenitors, not merely as the source of adipocytes, but also as key communicators and versatile players in the adipose microenvironment. These progenitors have been shown to contribute to a variety of processes, including extracellular matrix remodeling, immune modulation, and angiogenesis7.
Here, we introduce Slit guidance ligand 3 (Slit3) as a crucial factor secreted from brown adipocyte progenitors that mediates crosstalk among adipocyte progenitors, endothelial cells, and sympathetic nerves. Using BAT- and adipocyte progenitor-specific loss of function models, we show that the loss of Slit3 disrupts both angiogenesis and sympathetic innervation, ultimately blunting cold-induced BAT thermogenesis. Furthermore, we show that the proteolytic processing of Slit3 creates two ligands with distinct receptor binding properties and target cells and thus stimulates the coordinated expansion of the BAT neurovascular network. This pathway demonstrates a sophisticated level of intercellular coordination whereby a single factor simultaneously drives these two distinct processes essential for thermogenic activation.
Results
Identification of Slit3-Robo4 signaling axis in BAT
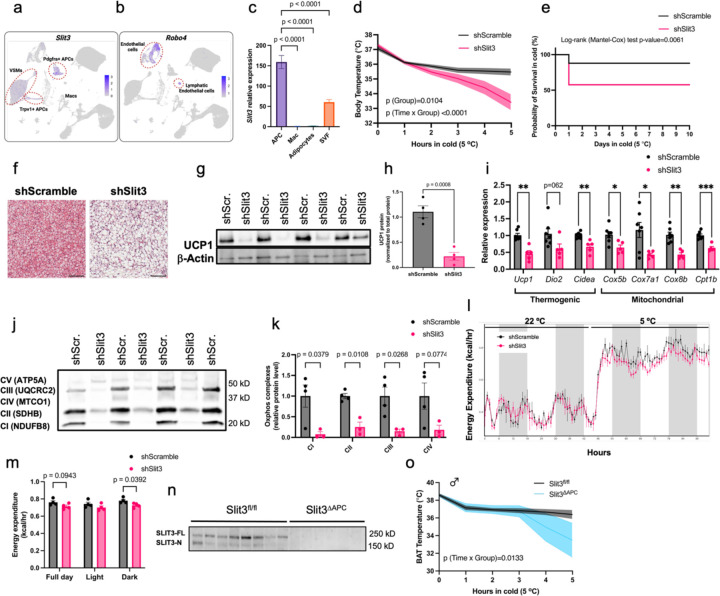
Intercellular communication plays a pivotal role in coordinating tissue adaptation to external challenges. To define the role of intercellular crosstalk in cold-induced BAT thermogenesis, we recently used single-cell transcriptomic data to build a network of ligand-receptor interactions involved in the cold-induced remodeling of BAT6. This analysis revealed the significance of adipocyte progenitors as the major communication “hub” in the adipose niche6. This prompted us to search for secreted ligands expressed in adipocyte progenitors that might mediate the crosstalk between adipocyte progenitors and their niche. We identified a new crosstalk axis in BAT involving the axon guidance molecule Slit3 and its cognate receptor, Robo4. Slit3 is predominantly expressed in Pdgfra+ adipocyte progenitors, Trpv1+ adipocyte progenitors, and vascular smooth muscle cells (Figure 1a), while its receptor Robo4 is exclusively expressed in vascular and lymphatic endothelial cells (Figure 1b). To confirm these results, we assessed Slit3 expression in sorted adipocyte progenitors, macrophages, adipocytes, and total stromal vascular fraction isolated from mouse BAT. Our analysis verified that adipocyte progenitors are the primary source of Slit3 in BAT (Figure 1c and Supplementary Figure 1a-d), whereas Robo4 is exclusively expressed in endothelial cells (Supplementary Figure 1e).
Figure 1. Axon Guidance Molecule Slit3 is essential for BAT thermogenesis.
(a) Slit3 and (b) Robo4 expression in scRNA-seq data of mouse BAT. (c) Slit3 expression in isolated adipocyte progenitors, macrophages, adipocytes, and the total stromal vascular fraction isolated from mouse BAT. (d) Body temperature during cold tolerance test, (e) Kaplan Meier survival curve (N=38 per group), (f) Hematoxylin and eosin (H&E) staining, (g-h) UCP1 protein, (i) expression of thermogenic and mitochondrial genes, and (j-k) Oxphos complexes protein level in BAT of mice receiving local administration of AAV-shSlit3 or scramble shRNA. Scale bar=100 µm. N= 4–7 per group. A representative of 3 independent experiments is shown. (l) hourly energy expenditure and (m) cumulative energy expenditure in mice injected with AAV-shSlit3 or shScramble. N= 4 per group. (n) SLIT3 protein and (o) BAT temperature during cold tolerance test in male Slit3fl/fl and Slit3ΔAPC mice. N= 7–10 per group. Data are presented as means ± SEM and analyzed by one-way ANOVA with Dunnett’s multiple comparison test (c), repeated measures ANOVA (d and o), Log-rank (Mantel-Cox) test (e), unpaired two-sided Student’s t-tests (h, i, k, m).
A previous study reported that Slit3 is secreted from macrophages in white adipose tissue (WAT)8. However, our unbiased scRNA-seq data as well as targeted expression analysis in isolated macrophages did not detect any Slit3 expression in BAT macrophages (Figure 1a and Figure 1c). Analysis of scRNA-seq datasets of mouse inguinal white adipose tissue (ingWAT) and perigonadal white adipose tissue (pgWAT) and human WAT9 also revealed the highest expression of Slit3 in adipocyte progenitors, mesothelial cells, mural cells (vascular smooth muscles and pericytes), and to a lesser degree in adipocytes (Supplementary Figure 2a-b). Consistent with our findings in BAT, Slit3 expression is not detected in macrophages in mouse and human WAT (Supplementary Figure 2a-b). These findings collectively indicate that adipocyte progenitors, rather than macrophages, are the primary source of Slit3 in adipose tissue.
Axon Guidance Molecule Slit3 is essential for BAT thermogenesis.
Slit guidance ligands are large extracellular glycoproteins that bind to transmembrane Robo receptors (Robo1–4) and regulate axon guidance and outgrowth, vascularization and sprouting angiogenesis, inflammatory cell chemotaxis, and tumor metastasis10. To determine the role of Slit3 in BAT, we used AAV-mediated shRNA delivery to knock down Slit3 expression specifically in BAT. We confirmed the significant reduction of Slit3 transcript and protein in BAT, but not WAT of mice receiving Slit3 shRNAs (Supplementary Figure 3a-c). Mice lacking Slit3 expression in BAT exhibited severe impairment in cold-induced BAT thermogenesis and maintained lower core body (Figure 1d) and BAT temperatures (Supplementary Figure 3d) in cold. Severe cold intolerance significantly decreased the survival rate of Slit3-deficient mice. Across three experiments, 42% of Slit3 knockdown mice experienced hypothermia, with their rectal temperature dropping to 30°C or lower within 24 hours. In contrast, only 12% of control mice showed similar symptoms (Log-rank (Mantel-Cox) test p-value=0.0061) (Figure 1e). Detailed analysis of BAT after seven days of cold exposure revealed increased lipid accumulation and whitening (Figure 1f), as well as reduced expression of uncoupling protein 1 (Ucp1) (Figure 1g-h) and other thermogenic and mitochondrial genes in Slit3 knockdown mice (Figure 1i-k).
Loss of Slit3 in BAT significantly reduced energy expenditure, oxygen consumption, and CO2 production in mice housed at 5 °C but not at room temperature (Figure 1l-o and Supplementary Figure 4), without changes in body weight, food intake, locomotor activity, and Respiratory Exchange Ratio (Supplementary Figure 4). A comparable decrease in both Ucp1 transcript and protein expression was noted in mice lacking Slit3 expression in BAT and housed at room temperature. However, other genes associated with thermogenesis and mitochondrial function remained unchanged under this condition (Supplementary Figure 5a-c). Together, these findings demonstrated that Slit3 is essential for cold-induced BAT thermogenesis.
To further confirm that adipocyte progenitor-derived Slit3 is essential for BAT thermogenesis, we specifically deleted Slit3 in Pdgfra-expressing cells by crossing Slit3 floxed mice11 with the Pdgfra-cre strain (Pdgfra-cre;Slit3flox/flox or Slit3ΔAPC). Western blotting revealed a complete loss of Slit3 expression in BAT and WAT of Slit3ΔAPC mice in males and females (Figure 1n and Supplementary Figure 6a-b). Consistent with the phenotypes observed in BAT-specific Slit3 knockdown mice, male Slit3ΔAPC mice, but not females, exhibited impaired BAT thermogenesis and were severely cold-intolerant (Figure 1o and Supplementary Figure 6c). The severe cold sensitivity in male Slit3ΔAPC mice led to hypothermia and required the removal of all animals from cold within 6 hours, preventing the analysis of BAT in response to chronic cold exposure.
Loss of Slit3 impairs cold-induced angiogenesis and sympathetic innervation in BAT.
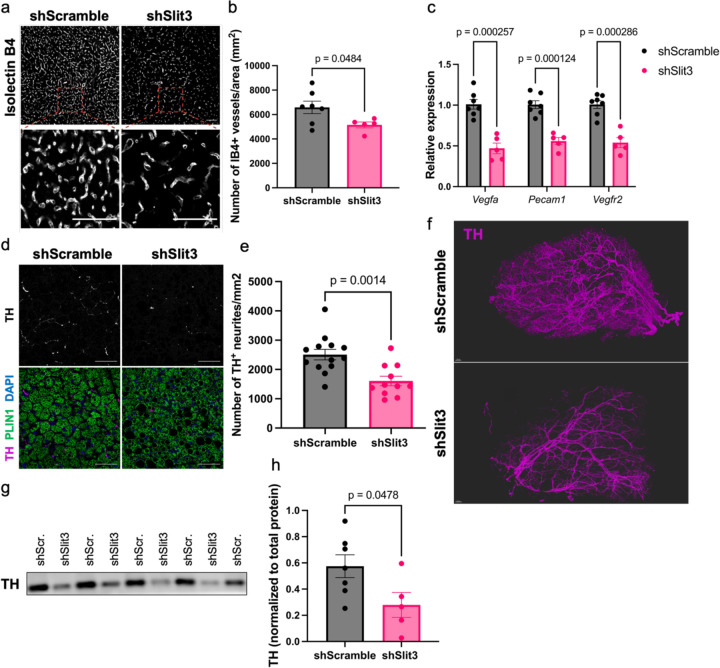
The expansion of BAT neurovasculature is essential for enhanced thermogenesis in the cold3,4. To understand the underlying mechanism of impaired BAT thermogenesis in mice lacking Slit3, we assessed vascularization and sympathetic innervation in BAT of mice receiving Slit3 or scramble following seven days of cold exposure. Analysis of BAT vasculature through immunostaining and molecular analyses revealed that Slit3 knockdown markedly reduced angiogenesis in BAT. This was evidenced by a lower capillary density (Figure 2a-b) and a decrease in the expression of endothelial-specific transcripts (Figure 2c).
Figure 2. Loss of Slit3 impairs cold-induced angiogenesis and sympathetic innervation in BAT.
(a) Representative images of Isolectin B4 staining, (b) quantification of the number of capillaries per area, (c) the expression of endothelial cell markers, (d) representative images of TH and Plin1 staining, (e) quantification of the number of TH+ neurites per area, (f) 3-D reconstruction of TH staining, and (g-h) TH protein level in BAT of mice receiving AAV-shSlit3 or scramble shRNA. Scale bar=50 µm. N= 5–7 per group. Data are presented as means ± SEM and analyzed by unpaired two-sided Student’s t-tests.
Furthermore, the loss of Slit3 resulted in a significant reduction in the density of Tyrosine Hydroxylase (TH)-expressing neurites in BAT (Figure 2d-e). To visualize the architecture and density of sympathetic innervation, we used the Adipo-Clear protocol12 for staining TH-expressing sympathetic nerves in BAT, followed by Lightsheet microscopy. The 3D volumetric images of TH-expressing nerves revealed a severe reduction in parenchymal innervation in BAT of Slit3-deficient mice (Figure 2f). Total TH protein in BAT was also significantly reduced in the absence of Slit3 (Figure 2g-h).
Notably, even when animals were housed at room temperature, loss of Slit3 resulted in a decrease in both capillary density and sympathetic innervation in BAT (Supplementary Figure 7a-d). However, housing at cold temperatures exacerbated these defects. These findings demonstrated the crucial role of Slit3 in the development and expansion of the neurovascular network in BAT.
Bmp1-mediated proteolytic cleavage of Slit3 generates two secreted ligands.
Members of the Slit family are proteolytically cleaved into a large N-terminal and a shorter C-terminal fragment. The cleavage site in Slit proteins has a conserved TSP motif (threonine, serine, proline) in the linker region between the fifth and sixth EGF domains13. The functional implications of Slit cleavage and the distinct roles played by the full-length protein and its fragments remain unclear.
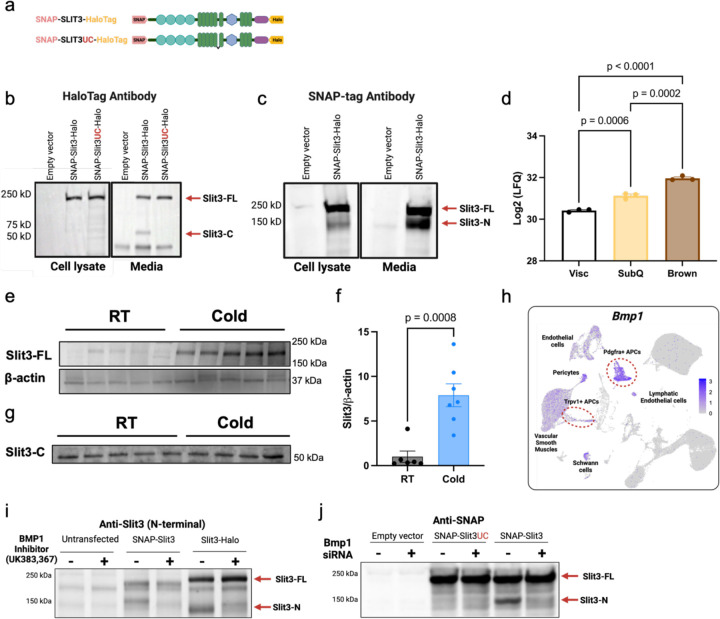
To investigate whether Slit3 is cleaved in adipocyte progenitors, we overexpressed an N- and C-terminal tagged wild-type Slit3 (SNAP-Slit3-HaloTag) as well as an uncleavable Slit3 variant (SNAP-Slit3UC-HaloTag) in an immortalized brown adipocyte progenitor cell line (Figure 3a). Using antibodies against HaloTag and SNAP-tag, we detected the full-length Slit3 protein (Slit3-FL) in the total cell lysates of cells overexpressing either the wild-type or uncleavable Slit3 constructs (Figure 3b-c). The HaloTag antibody detected two bands in the conditioned media of cells overexpressing SNAP-Slit3-HaloTag: a ~200 kDa band corresponding to the tagged full-length Slit3 and a ~60 kDa band matching the expected size of the tagged Slit3C-terminal fragment (Slit3-C). In contrast, the media from cells expressing the uncleavable Slit3 only contained the Slit3-FL (Figure 3b). Similarly, the SNAP-tag antibody detected both the Slit3-FL and a ~150 kDa band corresponding to the tagged Slit3 N-terminal fragment (Slit3-N) in the media of cells overexpressing SNAP-Slit3-HaloTag (Figure 3c). A minor amount of Slit3-N, but not Slit3-C, was also detected in the total cell lysates which contains membrane and membrane-associated proteins (Figure 3c). This finding is consistent with previous reports suggesting that the N-terminal fragment of Slits may remain associated with the plasma membrane, while the C-terminal fragment is more diffusible13. Collectively, these data indicate that Slit3 cleavage in adipocyte progenitors generates Slit3-N and Slit3-C fragments, and that Slit3-FL, Slit3-N, and Slit3-C fragments are secreted factors.
Figure 3. BMP1-mediated proteolytic cleavage of SLIT3 generates two secreted ligands.
(a) Schematic of the tagged Slit3 transgenes. Western blots using (b) HaloTag and (c) SNAP-tag antibodies to visualize SLIT3 fragments in the cell lysate and conditioned media from adipocyte progenitors expressing the indicated plasmids. (d) Log2 Label-Free Quantification (LFQ) intensity of SLIT3 peptides in conditioned media from in vitro differentiated primary Visc, SubQ, and Brown adipocytes quantified by mass spectrometry. N=3 per group. Western blots of (e-f) SLIT3-FL and (g) SLIT3-C in BAT of mice housed at Room temperature (RT) or cold (5°C) for 2 days. N=6–7 per group. (h) Expression of Bmp1 in adipocyte progenitors in scRNA-seq data of mouse BAT. (i) Western blot using Slit3 antibody in adipocyte progenitors transfected with the indicated single-tagged constructs and treated with Bmp1 inhibitor UK383,367 or vehicle. (j) Western blot using SNAP-tag antibody in adipocyte progenitors transfected with siBMP1 or scramble siRNA in cells overexpressing the indicated single-tagged constructs. Data are presented as means ± SEM and analyzed by one-way ANOVA and corrected for multiple comparisons using statistical hypothesis testing with Tukey (d) and unpaired two-sided Student’s t-test (f).
Moreover, analysis of a proteomics dataset of the adipocyte secretome revealed the presence of Slit3 in the conditioned media of in vitro differentiated adipocytes derived from mouse visceral (Visc), subcutaneous inguinal white (SubQ), and interscapular BAT14 (Figure 3d). Notably, Slit3 was significantly more abundant in the conditioned media of brown and SubQ white adipocytes than Visc white adipocytes. These results indicated that Slit3 is secreted by various adipocyte types in vitro, with the highest concentrations observed in the secretome of brown adipocytes.
To examine the presence of Slit3-FL and its fragments in BAT, we used different antibodies raised against different regions of the Slit3 protein. Using the antibody that preferentially detects Slit3-FL, we observed elevated levels of Slit3-FL in BAT of mice housed in cold for 2 days relative to the control group housed at room temperature (Figure 3e-f). Furthermore, an antibody raised against C-terminal epitopes of Slit3 identified a ~55 kDa fragment in BAT, consistent with the size of the Slit3-C fragment (Figure 3g). These findings demonstrate the presence of Slit3 and its proteolytic processing in BAT.
The cleavage of Slit proteins is conserved among Drosophila and vertebrate Slits and is observed in multiple tissues13,15–18. A recent study suggested a role for the metalloprotease Tolkin (Tok) in Slit proteolysis in Drosophila melanogaster15. However, the Slit proteases in vertebrates have not been identified. Tok is a member of the Bmp1/Tolloid family of metalloproteases. In mammals, the Bmp-1/tolloid-like family includes bone morphogenetic protein-1 (Bmp1), tolloid (Tld), and tolloid-like 1 and 2 (Tll1 and Tll2). Given the similarity in the cleavage sites among the members of the Slit family, we hypothesized that the Bmp1/Tolloid proteases might be involved in Slit3 cleavage. Among the members of the Bmp1/Tolloid proteases family, only Bmp1 and Tll1 are expressed in BAT (Figure 3h and Supplementary Figure 8). Notably, Bmp1 is abundantly expressed in Pdgfra- and Trpv1-expressing adipocyte progenitors, the major cell types expressing Slit3 in BAT (Figure 3h). Bmp1 lacks growth factor activity found in other BMP family members and instead acts as a metalloprotease, cleaving precursor proteins involved in extracellular matrix formation, including collagen maturation. Thus, we hypothesized that Bmp1 is the protease responsible for the proteolytic cleavage of Slit3. To discern the role of Bmp1 in Slit3 proteolysis in adipocyte progenitors, we used a combination of pharmacological and genetic loss of function studies. We showed that the inhibition of Bmp1 activity using a small molecule inhibitor, UK383,36719, blocked Slit3 cleavage (Figure 3i). Similarly, knocking down Bmp1 using siRNAs prevented Slit3 cleavage (Figure 3j). Thus, we concluded that Bmp1 is the enzyme responsible for the proteolytic processing of Slit3, establishing Bmp1 as the first Slit protease in vertebrates.
Slit3 fragments promote angiogenesis and sympathetic innervation in BAT.
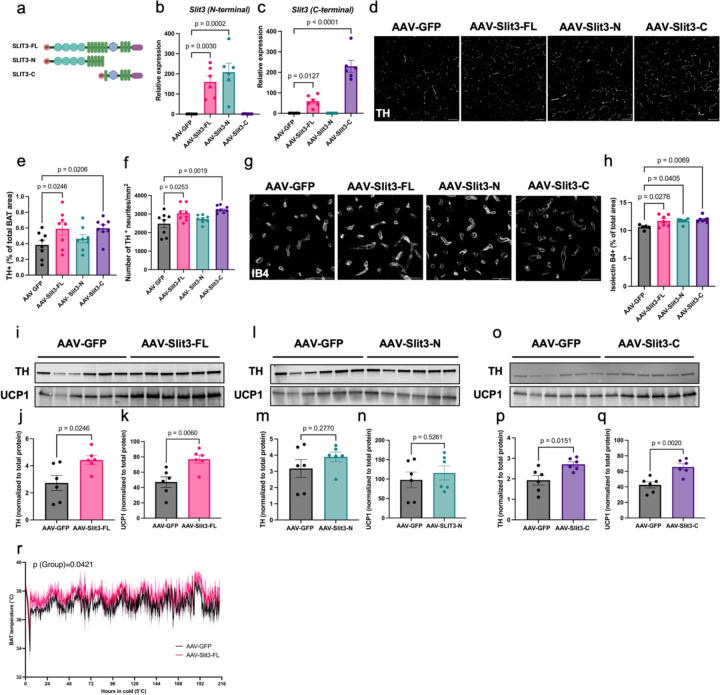
To determine the functions of the individual Slit3 fragments in BAT, we generated AAV constructs for adipocyte-specific overexpression of Slit3-FL, Slit3-N (aa 1–1116), and Slit3-C (aa 1117–1523) (Figure 4a). We confirmed the specific expression of Slit3-FL, Slit3-N, and Slit3-C in interscapular BAT four weeks following local AAV administration (Figure 4b-c). Overexpression of Slit3-FL and Slit3-C, but not Slit3-N, promoted sympathetic innervation in BAT, as indicated by an increased number and relative area of TH-expressing sympathetic neurites (Figure 4d-f). Additionally, overexpression of Slit3-FL, Slit3-N, and Slit3-C led to increased capillary density in BAT (Figure 4g-h). Consistent with the imaging data, mice overexpressing Slit3-FL and Slit3-C, but not SLIT3-N, had higher levels of TH in BAT (Figure 4i-q). Collectively, these effects resulted in increased adaptive thermogenesis, as shown by elevated Ucp1 expression and higher BAT temperatures in mice overexpressing Slit3-FL and Slit3-C, but not Slit3-N, when housed in cold conditions (Figure 4k, 4n, 4q-r). These results demonstrate that Slit3 fragments directly promote the expansion of the neurovascular network in BAT, with Slit3-C driving sympathetic innervation and Slit3-N promoting angiogenesis, collectively enhancing thermogenic function.
Figure 4. Slit3 fragments promote angiogenesis and sympathetic innervation in BAT.
(a) Schematic of the Slit3-FL, Slit3-N, and Slit3-C constructs. The expression of Slit3 fragments measured using primers detecting the (b) N-terminal or (c) C-terminal regions of Slit3 transcript in BAT, (d) representative images of TH staining (scale bar=50µm), quantification of (e) percentage of TH+ area and (f) the number of TH+ neurites per area, (g) representative images of Isolectin B4 staining (scale bar=50µm), (h) quantification of the percentage of IB+ capillary area in BAT, TH and UCP1 western blots and quantifications in BAT of mice expressing AAV-GFP, AAV-Slit3-FL (i-k), AAV-Slit3-N (l-n), and AAV-Slit3-C (o-q), and (i) BAT temperature in mice expressing AAV-GFP, AAV-Slit3-FL, AAV-Slit3-N, and AAV-Slit3-C. N= 6–8 per group. Data are presented as means ± SEM and analyzed by one-way ANOVA with Dunnett’s multiple comparison test (b-c, e-f, h), unpaired two-sided Student’s t-test (j-k, m-n, and p-q), and mixed-effects model with the Geisser-Greenhouse correction (r).
Plxna1 is responsible for Slit3-mediated sympathetic innervation in BAT
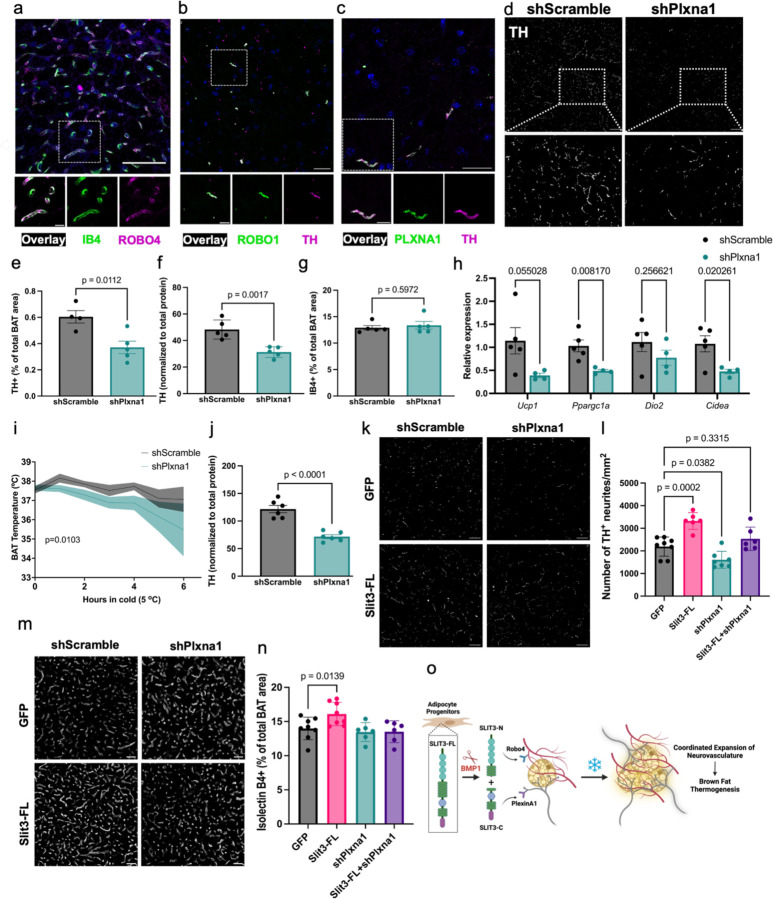
Full-length and N-terminal Slit fragments bind to transmembrane Robo receptors (Robo1–4)36. The receptor binding and bioactivity of the C-terminal fragment (Slit-C) remain less clear. The C-terminal of Slit2 was shown to bind Plexin A1 (Plxna1)20, but the receptor(s) for Slit3-C has not been identified. To identify the receptors that mediate the effects of Slit3 fragments in BAT, we assessed the expression of the putative Slit receptors. Analysis of BAT scRNAseq data showed that among the four members of the Robo family, only Robo4 is expressed in BAT. Robo4 is exclusively expressed in vascular and lymphatic endothelial cells (Figure 1b and Supplementary Figure 1e). To validate this, we co-stained BAT sections with a Robo4 antibody and Isolectin B4. Consistent with the scRNA-seq data, Robo4 was specifically localized to the Isolectin B4-labeled vessels (Figure 5a). Additionally, double staining of BAT with Robo1 and TH antibodies showed Robo1 expression in TH-expressing sympathetic neurites (Figure 5b). Similarly, Plxna1 and TH were co-localized in sympathetic nerves in BAT (Figure 5c).
Figure 5. Plxna1 is responsible for SLIT3-mediated sympathetic innervation in BAT.
co-staining of (a) ROBO4 and Isolectin B4, (b) ROBO1 and TH, and (c) PLXNA1 and TH in BAT. Scale bars= (a) 50µm and (b-c) 25µm. (d) representative images of TH staining (scale bar=50µm), (e) percentage of TH+ area, (f) total TH protein level, (g) the percentage of IB+ capillary area, and (h) expression of thermogenic gene program in BAT of mice receiving AAV-shPlxna1 or scramble shRNA and housed at room temperature. N= 5 per group. (i) BAT temperature during cold tolerance test and (j) total TH protein level in BAT of mice receiving AAV-shPlxna1 or scramble shRNA and housed at cold for 7 days. N= 6–8 per group. (k) representative images of TH staining (scale bar=50µm), (l) number of TH+ neurites per area, (m) representative images of Isolectin B4 staining (scale bar=50µm), (n) quantification of the percentage of IB+ capillary area in BAT of mice expressing Slit3-FL with or without shPlxna1. (o) Proposed model for the mechanisms of Slit3-mediated neurovascular network expansion in BAT. Data are presented as means ± SEM and analyzed by unpaired two-sided Student’s t-test (e-h, j), paired two-sided Student’s t-test (i), and one-way ANOVA with Dunnett’s multiple comparison test (l and n).
Based on our finding that Slit3-C enhances sympathetic innervation (Figure 4d-f) and that Plxna1 is expressed in sympathetic neurites, we hypothesized that Plxna1 might be the receptor mediating Slit3-C signaling in these neurites. To address this, we first injected AAVs delivering Plxna1 shRNAs (shPlxna1) into BAT. qPCR and Western blotting confirmed a partial loss of Plxna1 in the BAT of mice receiving shPlxna1 (Supplementary Figure 9a-b). Intriguingly, even this partial reduction of Plxna1 drastically decreased sympathetic nerve density (Figure 5d-e) and total TH protein (Figure 5f and Supplementary Figure 9c) in BAT, without affecting blood vessel density (Figure 5g). These findings highlight the critical role of Plxna1 in the development and maintenance of sympathetic neurites in BAT. Additionally, knocking down Plxna1 in BAT led to decreased expression of thermogenic genes, impaired cold tolerance, and lower TH protein levels in mice exposed to cold (Figure 5h-j).
To test whether Plxna1 is responsible for Slit3-mediated sympathetic innervation, we overexpressed Slit3-FL in BAT with and without simultaneous Plxna1 knockdown. Loss of Plxna1 expression attenuated the Slit3-induced increase in sympathetic innervation, suggesting that Plxna1 is likely the receptor mediating the effects of Slit3 signaling in sympathetic neurites (Figure 5k-l). While Plxna1 knockdown alone had no effect on capillary density, the pro-angiogenic effects of Slit3-FL were also abolished in Plxna1 knockdown BAT (Figure 5m-n). Considering that Plxna1 is not expressed in endothelial cells, we concluded that the pro-angiogenic effects of Slit3-FL are at least partially due to increased sympathetic innervation. Supporting this notion, sympathetic innervation has been shown to activate angiogenesis via endothelial β-adrenergic receptor signaling in the tumor microenvironment21. In summary, these findings reveal the complex and multifaceted role of Slit3 fragments in orchestrating the expansion of BAT neurovasculature, a crucial step in the activation of BAT thermogenesis (Figure 5o).
Slit3 expression is associated with adipose tissue health and inflammation in humans
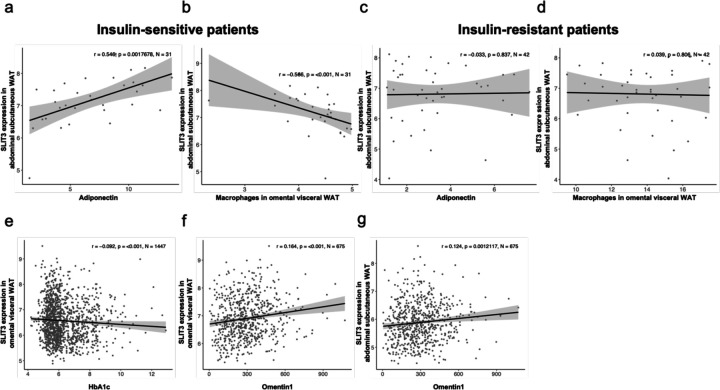
Lastly, to assess the significance of SLIT3 in human obesity, we measured its expression in human adipose tissue biopsies in abdominal subcutaneous WAT and omental visceral WAT collected from two independent cohorts of the Leipzig Obesity BioBank (LOBB). In the metabolically healthy versus unhealthy obese cohort (MHUO), we found that SLIT3 transcript levels in human abdominal subcutaneous WAT were positively correlated with serum adiponectin concentrations (r=546, p=0.00176, N=31) and negatively regulated with the relative number of macrophages in omental visceral WAT (r=566, p<0.001, N=31) in insulin-sensitive patients (Figure 6a-b), but not in insulin-resistant patients (Figure 6c-d).
Figure 6. Slit3 expression is associated with adipose tissue health and inflammation in humans.
SLIT3 expression in abdominal subcutaneous WAT of male and female insulin-sensitive individuals correlated with (a) adiponectin and (b) macrophages in omental visceral WAT. SLIT3 expression in abdominal subcutaneous WAT of male and female insulin-resistant individuals correlated with (c) adiponectin and (d) macrophages in omental visceral WAT. SLIT3 expression in omental visceral WAT of males and females correlated with (e) HbA1c and (f) Omentin1 levels. (g) SLIT3 expression in abdominal subcutaneous WAT of males and females correlated with Omentin1 levels. The Spearman correlation coefficient (r), p-value (p), and number of individuals (N) are denoted on each plot. Gene expression is given as weighted trimmed mean (TMM) values of the log expression ratios.
Additionally, we examined SLIT3 expression in the human cross-sectional cohort comprised of paired samples of omental visceral and abdominal subcutaneous WAT from 1,480 individuals. In this cohort, we found SLIT3 expression in omental visceral WAT negatively correlated with Hemoglobin A1C (HbA1c) (r=0.092, p<0.001, N=1447) (Figure 6e). Additionally, SLIT3 expression in omental visceral and abdominal subcutaneous WAT was positively associated with circulating level of the anti-inflammatory adipokine Omentin1 (r=0.164, p<0.001, N=675 and r=0.124, p=0.00121, N=675, respectively) (Figure 6f-g). Omentin1 has been shown to play important roles in glucose homeostasis, lipid metabolism, insulin resistance, and diabetes22,23. Collectively, these data suggest that SLIT3 may regulate adipose tissue health and inflammation in humans, potentially impacting insulin sensitivity.
Discussion
Recent advances in technical and computational methods for studying complex tissues are beginning to provide a systematic understanding of how cellular crosstalk within the tissue microenvironment drives tissue development, function, and disease mechanisms. Using scRNA-seq data of BAT, this study identified Slit3 as a novel niche factor that mediates crosstalk among adipocyte progenitors, sympathetic nerves, and blood vessels. Our findings show that the coordinated expansion of the BAT neurovasculature stimulated by Slit3 fragments is essential for cold-stimulated BAT thermogenesis. Loss of Slit3 in BAT significantly impairs adaptive thermogenesis and compromises the survival of mice in cold conditions. Our work highlights the critical and dynamic role of the adipose tissue microenvironment and provides new mechanistic insights into the regulation of angiogenesis and sympathetic innervation, two essential processes for maintaining adipose tissue health.
Originally identified as axon guidance factors, Slits regulate the migration and positioning of cells in various tissues. Despite the critical roles of Slit proteins in many biological processes, the biological activities of Slit fragments and the mechanisms controlling the context-dependent outcomes of Slit signaling are still largely unclear. This is in part due to the unknown identity of the Slit proteases, which prevented the separation of Slit fragment activities from that of the full-length protein. Identification of Slit proteases and the specific receptors for the Slit fragments are critical for understanding the mechanisms that control Slit signaling during tissue development, remodeling, and regeneration. In this work, we identified the metalloprotease Bmp1 as the first known vertebrate Slit protease. Through a combination of pharmacological inhibition and genetic knockdown, we found that Bmp1 is responsible for Slit3 cleavage. This finding highlights Bmp1’s role in regulating the biological activities of Slit3, and potentially other Slit family members, in various contexts beyond BAT. Thus, this work advances our understanding of Slit signaling and its regulation in axon guidance and growth, angiogenesis, inflammatory cell chemotaxis, and tumor metastasis.
Plexins are a family of single-pass transmembrane proteins that act as receptors for Semaphorin axon guidance proteins. Semaphorin-Plexin signaling regulate processes such as neuronal axon guidance, angiogenesis, and immunity. Class A plexins (PlxnAs) selectively bind to Class 1 semaphorins and, in conjunction with Neuropilin co-receptors, also interact with Class 3 semaphorins24. Recent studies have challenged the long-held notion that Slit signaling is mediated solely by Robo receptors, revealing that Plxna1 serves as a receptor for the Slit2 C-terminal fragment, functioning independently of both Robo and Neuropilin receptors20. Here, we show that Plxna1 is essential for sympathetic innervation of BAT, and that the loss of Plxna1 diminishes the effects of Slit3 on this process. However, because Plxna1 mediates both Semaphorin and Slit signaling, we cannot exclude the possibility that the reduced sympathetic innervation observed in the absence of Plxna1 is partly due to impaired semaphorin signaling. Further studies are needed to quantitatively assess the contributions of Slit and semaphorin signaling through Plxna1 to BAT innervation.
Adipose tissues are extensively innervated by a network of sympathetic and sensory nerve projections, which facilitate the transmission of information between the adipose tissue and the central nervous system. It is well-established that the sympathetic innervation of adipose tissue is critical for homeostatic control of its function and whole-body metabolism25–27. The plasticity of sympathetic nerves allows for altered neuronal control and adaptation to metabolic challenges. However, the mechanisms underlying the local expansion and remodeling of sympathetic neurites are not well understood. Obesity disrupts norepinephrine-mediated adipocyte lipolysis28, mitochondrial biogenesis29, and adipose tissue remodeling30. These impairments are attributed to dysfunctional local sympathetic innervation resulting from adipose inflammation31. In turn, the impaired sympathetic activity in obese adipose tissue may contribute to more inflammation and further exacerbate adipose dysfunction32. Therefore, preserving the density and functionality of sympathetic nerves may help safeguard adipose tissue health in the context of obesity. To the best of our knowledge, this study is the first to demonstrate that adipocyte progenitors play a regulatory role in sympathetic innervation, introducing a previously unexplored regulator of adipose tissue innervation.
Genome-wide association studies have found genetic variants near the SLIT3 gene to be associated with an increased risk of insulin resistance33 and higher BMI34. Additionally, a differentially methylated CpG in the SLIT3 gene in visceral adipose tissue was found to be associated with the development of type 2 diabetes in obese women35. Genetic variants in PLXNA1 gene are also associated with severe early onset obesity36. These independent findings suggest that SLIT3 and its receptor PLXNA1 may have a role in regulating metabolism in humans. Our analysis of human adipose tissue in two obesity cohorts suggest that SLIT3 signaling may contribute to improved adipose tissue health, reduced inflammation, and insulin sensitivity in human obesity. These findings underscore the need for future research to explore the role of SLIT3 signaling and other axon guidance molecules in obesity and metabolic diseases.
Methods
Animals.
All experimental procedures involving animals were performed in compliance with all relevant ethical regulations applied to the use of small rodents and with approval by the Institutional Animal Care and Use Committees at New York University. C57BL/6J mice (stock no. 000664) were purchased from Jackson Laboratory. Pdgfra-cre;Slit3flox/flox or Slit3ΔAPC were generated by crossing Slit3 floxed mice11 with Pdgfra-cre strain37 (JAX strains 013148). Mice were maintained on a 12-h light-dark cycle at 22 °C and 50% relative humidity, with food and water provided ad libitum. For experiments involving cold acclimation, mice were housed at 5°C and 50% relative humidity in a controlled environmental diurnal chamber (Caron Products & Services) with free access to food and water.
Cold tolerance test.
For cold tolerance tests, mice were single-housed and placed at 5°C in a controlled environmental diurnal chamber (Caron Products & Services). Body temperature was measured with a rectal probe (Physitemp, RET3) and a reader (Physitemp, BAT-12) or using RFID transponder temperature microchips implanted under the interscapular BAT area (Unified Information Devices).
AAV injection.
6–8-week-old male mice were anesthetized with isoflurane and an incision was made above the interscapular area to expose the underlying adipose tissue. AAV particles were injected into each BAT lobe and the incision was closed with suture. Mice were allowed to recover for 3 weeks before analysis.
Slit3 and Plxna1 shRNAs.
Adeno-associated viruses (AAV8) harboring three targeting shRNA and one scramble control were purchased from VectorBuilder. The shRNA sequences are listed in Supplementary Table 1. The three shRNAs were mixed and injected into BAT at the dose of 5e+11 genomic copies per lobe.
AAV constructs for the overexpression of Slit3 fragments in BAT.
Mouse Slit3-FL, Slit3-N, and Slit3-C sequences were cloned from a cDNA clone (Origene MR225499L4) and cloned into the pAAV-hAdiponectin-W backbone38 using NEBuilder HiFi DNA Assembly kit using the following primers:
Slit3-FL_fwd (AACTACTCGAGgccaccATGGCCCTCGGCCGGAC), Slit3-FL_rev (tattcaGCGGCCGCTTAAACCTTATCGTCGTCATCCTTGTAATCgctgccGGAACACGCGCGGCA), Slit3-N_fwd (AACTACTCGAGgccaccATGGCCCTCGGCCGGAC), Slit3-N_rev (tattcaGCGGCCGCTTAAACCTTATCGTCGTCATCCTTGTAATCgctgccAACCATGGGTGGGGG), Slit3-C_fwd (tgctgccgccCTGCTACAAACCAGCCCC), Slit3-C_rev (ggttgattatcttctagagcTTACTTGTCGTCATCGTCTTTG), Signal peptide_fwd (tgattccataccagagggtcGCCACCATGGCCCTCGGC), and Signal peptide_rev (tttgtagcagGGCGGCAGCAGGGGGTCC).
pAAV-Adipoq-GFP construct38 was used as the control. AAV8-adipoq-Slit3FL, AAV8-adipoq-Slit3N, AAV8-adipoq-Slit3C, and AAV-Adipoq-GFP were packaged at VectorBuilder. 2e+10 AAV particles were injected into each BAT lobe.
Fluorescence-activated Cell Sorting (FACS).
Adipocytes and the Stromal Vascular Fraction (SVF) were isolated from mouse BAT following the procedure described previously5. The interscapular BAT was dissected, finely minced, and digested for 45 minutes using a cocktail containing type 1 collagenase (1.5 mg ml−1; Worthington Biochemical), dispase II (2.5 U ml−1; Stemcell Technologies), and fatty acid-free BSA (2%; Gemini Bio-Products) in Hanks’ balanced salt solution (Corning Hanks’ Balanced Salt Solution, with calcium and magnesium). The resulting dissociated tissue was subsequently centrifuged at 500g and 4°C for 10 minutes. Adipocytes, located in the uppermost layer, were gently collected using a wide-mouthed transfer pipette and filtered through a 100 µm cell strainer. Brown adipocytes were allowed to float for 5 min at room temperature before they were centrifuged at 30g for 5 minutes at room temperature. This cycle was repeated three times, after which the adipocytes were immediately lysed in Trizol. For the SVF isolation, the pellet was resuspended in 10 ml of 10% FBS in DMEM, filtered through a 100 µm cell strainer into a fresh 50-ml tube, and subsequently centrifuged at 500g for 7 minutes. Red blood cells were lysed in 2 ml of sterile ammonium–chloride–potassium lysis buffer (ACK Lysing Buffer, Lonza) for 5 minutes on ice. The cells were then filtered once more through a 40-μm cell strainer, washed with 20 ml of a solution containing 10% FBS in DMEM, and centrifuged at 500g for 7 minutes. The cells were resuspended in 1 ml of Cell Staining Buffer (BioLegend) before proceeding with staining.
Cells were stained using the fluorescently conjugated antibodies as outlined in Supplementary Table 2. The cells were then incubated with the antibodies at the specified dilutions from Supplementary Table 2 for a duration of 30 minutes, followed by two rounds of washing in Cell Staining Buffer (BioLegend). Cells were sorted using an SH800 sorter using a 100 µm sorting chip (Sony Biotechnology). Debris and doublets were excluded based on forward and side scatter gating, and 7-AAD was used to exclude dead cells. Following sorting, cells were centrifuged at 300 g for 5 minutes and lysed in Trizol for subsequent RNA isolation and gene expression analysis.
RNA isolation and quantitative reverse transcription-PCR (qRT-PCR).
RNA was isolated from cells or tissues using phenol–chloroform extraction and isopropanol precipitation. qRT PCR assays were conducted using a QuantStudio™ 5 Real-Time PCR instrument (Life Technologies Corporation) and SYBR Green (Invitrogen). Relative mRNA expression was determined using the ΔCt method and normalized to the expression of housekeeping genes Rplp0 or Tbp. Primer sequences are available in Supplementary Table 3.
Western blotting.
Cells or tissues were lysed in RIPA buffer supplemented with a protease inhibitor cocktail (cOmplete™, Sigma-Aldrich, Dallas, TX). The primary antibodies are listed in Supplementary Table 2. Primary antibodies were incubated overnight at 4 °C. HRP-coupled secondary antibodies were used at 1:3000 dilution for 1 hour at room temperature. Proteins were detected using the Amersham enhanced chemiluminescence (ECL) prime (GE healthcare, Pittsburgh, PA) using an Odyssey M Imaging System. All the original uncropped and unprocessed scans are provided in the Source Data file. For protein detection in the conditioned media, serum-free and Phenol Red-free media were collected and centrifuged at 300 g to remove cell debris. The supernatant was concentrated using Amicon™ Ultra-4 Centrifugal Filter Units (UFC801024, MilliporeSigma™).
Immunohistochemistry.
Adipose tissues were fixed in 10% formalin and embedded in paraffin. 5 µm sections were prepared and stained with the primary antibodies listed in Supplementary Table 2. Sections were deparaffinized and rehydrated, followed by an antigen retrieval step in Rodent Decloaker buffer using the Decloaking Chamber™ NxGen (BIOC Medical) in 1X rodent antigen retrieval reagent (95 °C for 45 minutes). Sections were then incubated in Sudan Black (0.3% in 70% ethanol) to reduce the autofluorescence signal. Blocking was performed in 5% BSA in PBST (0.1% Tween-20 in PBS) for 30 minutes at room temperature, followed by incubating the section in primary antibodies diluted in 1% BSA in PBST overnight at 4 °C (Supplementary Table 2). The biotinylated isolectin B4 was diluted in 5 ug/ml in a staining buffer containing 0.1mM CaCl2, 0.1mM MgCl2, and 0.1mM MnCl2. The next day, slides were washed with PBST and were incubated with appropriate fluorescently labeled secondary antibodies at a 1:200 dilution (Supplementary Table 2). Slides were stained with DAPI and mounted in a mounting medium. Images were collected on a Leica SP8 confocal microscope and processed with ImageJ.
H&E staining.
Adipose tissues were fixed in 10% formalin and embedded in paraffin. 5 µm sections were prepared and stained with hematoxylin and eosin (H&E) as previously described39. Slides were imaged using an Aperio slide scanner (Leica Biosystems).
Indirect calorimetry.
Mice were individually housed in metabolic cages of a TSE Phenomaster system to obtain measurements for the volume of oxygen consumption (VO2), the volume of carbon dioxide production (VCO2), respiratory exchange ratio (RER), energy expenditure, food intake, and locomotor activity. Mice had ad libitum access to drinking water and standard rodent chow. Measurements were collected first for 48 hours at room temperature, followed by the second 48 hours at 5 °C. Data were analyzed using CalR40.
Dual Energy X-ray Absorptiometry.
Lean and mass were quantified using a Dual Energy X-ray Absorptiometry (DEXA) scanner (Insight, Osteosys).
Tissue clearing and imaging.
Whole BAT sympathetic innervation visualization was achieved using the Adipo-Clear protocol41 with some modifications. One lobe of BAT from each animal was fixed in 3% glyoxal at 4°C overnight. The fixed samples were washed in PBS three times, each time for one hour. Next, the samples were subjected to a series of methanol washes: 20%, 40%, 60%, 80% methanol in H2O/0.1% Triton X-100/0.3 M glycine (B1N buffer, pH 7), and finally 100% methanol, each for 30 minutes. Subsequently, the samples underwent a triple 30-minute delipidation process with 100% dichloromethane (DCM; Sigma-Aldrich) followed by two 30-minute washes in 100% methanol. After delipidation, the tissues were bleached overnight at 4°C with 5% H2O2 in methanol (1 volume of 30% H2O2 to 5 volumes of methanol). The rehydration process followed a reversed methanol/B1N buffer series: 80%, 60%, 40%, 20% methanol in B1N buffer, each step lasting 30 minutes. All the above steps were conducted at 4°C with continuous shaking. Subsequently, samples were washed in B1N buffer twice, each time for 30 minutes, followed by two 1-hour wash in PBS/0.1% Triton X-100/0.05% Tween 20/2 µg/ml heparin (PTwH buffer), before initiating the staining procedure. The samples were incubated with anti-tyrosine hydroxylase antibody (1:200, AB1542, Millipore Sigma) in PTxwH for 4 days. After the primary antibody incubation, the samples underwent a series of PTxwH washes lasting 5 min, 10 min, 15 min, 30 min, 1 hr, 2 hr, 4 hr, and overnight, and then were incubated in donkey anti-sheep IgG Alexa Fluor 647 (1:200, A-21099, Thermo Fisher) in PTxwH for 4 days. Finally, the samples were washed in PTwH for 5 min, 10 min, 15 min, 30 min, 1 hr, 2 hr, 4 hr, and overnight. Finally, the samples were dehydrated using a methanol/H2O series (25%, 50%, 75%, 100%, 100%) for 30 minutes at each step at room temperature. Following dehydration, samples were incubated with 100% DCM for 30 minutes twice, followed by an overnight clearing step in dibenzyl ether (DBE; Sigma-Aldrich). The samples were then stored at room temperature in the dark until imaging. Representative samples were imaged by a blinded experimenter using a Zeiss Lightsheet Z.1 Microscope with two 1920 × 1920-pixel sCMOS cameras. Three-dimensional reconstructions were generated using Imaris (Bitplane).
Cloning of SNAP- and Halo-tagged Slit3 Plasmids.
pRP[Exp]-CMV>[SNAP-Slit3-Halo], pRP[Exp]-CMV>[SNAP-Slit3-FL], and pRP[Exp]-CMV>[Slit3-FL-Halo] constructs were generated by VectorBuilder. The N-terminal tag was added immediately downstream of the signal peptide (aa 2–33, GCCCTCGGCCGGACCGGGGCCGGCGCCGCTGTGCGCGCCCGCCTGGCGCTGGGCTTGGCGCTTGCGAGCATCCTGAGCGGACCCCCTGCTGCCGCC).
The ORF in the pRP[Exp]-CMV>[SNAP-Slit3UC-Halo] was generated by the deletion of a 27 bp sequence encoding the Slit cleavage site (CCCACCCATGGTTCTGCTACAAACCAG) from Slit3-FL cDNA.
Transient transfection.
Immortalized brown preadipocytes were transfected with Slit3 overexpression plasmids, Bmp1 or scramble siRNA (Dharmacon GE) using Lipofectamine™ 3000 or Lipofectamine™ RNAiMAX Transfection Reagents (Invitrogen™) following the manufacturer’s instructions.
Bmp1 inhibitor treatment.
Cells were treated with the Bmp1 inhibitor, UK-383,367 (PZ0156–5MG, Sigma Aldrich) at the final concentration of 2.5 µM.
Human data.
The human data used in this research were sourced from the Leipzig Obesity Biobank (LOBB, https://www.helmholtz-munich.de/en/hi-mag/cohort/leipzig-obesity-bio-bank-lobb), which comprises paired samples of abdominal subcutaneous and omental visceral adipose tissue. The metabolically healthy versus unhealthy obese cohort (MHO/MUO) comprises paired samples of omental visceral and abdominal subcutaneous tissues from 31 insulin-sensitive patients (71% female; age: 38.8 ± 11.1 years old; BMI: 45.9 ± 6.9 kg/m²; fasting plasma glucose: 5.2 ± 0.2 mmol/l; fasting plasma insulin: 27.9 ± 13.5 pmol/l) and 42 insulin-resistant patients (71.43% female; age: 47.2 ± 7.7 years old; BMI: 47.3 ± 8.1 kg/m2; fasting plasma glucose: 5.7 ± 0.3 mmol/l; fasting plasma insulin: 113.7 ± 45.7 pmol/l). The cross-sectional cohort (CSC) comprises 1,479 individuals, categorized as either normal/overweight (N = 31; 52% women; age: 55.8 ± 13.4 years; BMI: 25.7 ± 2.7 kg/m2) or obese (N = 1,448; 71% women; age: 46.9 ± 11.7 years; BMI: 49.2 ± 8.3 kg/m²). Adipose tissue samples were collected during elective laparoscopic abdominal surgeries, following established protocols42,43. Body composition and metabolic parameters were assessed using standardized techniques as described previously44,45. The study was approved by the Ethics Committee of the University of Leipzig (approval number: 159–12-21052012) and adhered to the principles outlined in the Declaration of Helsinki. All participants provided written informed consent before being included in the study. Exclusion criteria included individuals under 18 years of age, those with chronic substance or alcohol abuse, smoking within the 12 months prior to surgery, acute inflammatory conditions, concurrent use of glitazones, end-stage malignancies, weight loss greater than 3% in the three months leading up to surgery, uncontrolled thyroid disorders, and Cushing’s disease.
Ribosomal RNA-depleted RNA sequencing data we generated following the SMARTseq protocol46. The libraries were sequenced as single-end reads on a Novaseq 6000 (Illumina, San Diego, CA, USA) at the Functional Genomics Center Zurich, Switzerland. The preprocessing procedures were conducted as previously described47. In brief, adapter and quality-trimmed reads were aligned to the human reference genome (assembly GRCh38.p13, GENCODE release 32), and gene-level expression quantification was performed using Kallisto48 v0.48. For samples with read counts exceeding 20 million, we downsampled them to 20 million reads utilizing the R package ezRun v3.14.1 (https://github.com/uzh/ezRun, accessed on April 27, 2023). The data normalization was carried out using a weighted trimmed mean (TMM) of the log expression ratios, with adjustments made for age, sex, and transcript integrity numbers (TINs). Analyses were conducted in R v4.3.1 (www.R-project.org). Spearman coefficient was used to assess the correlation between SLIT3 expression and metabolic parameters.
Data reporting.
No statistical methods were used to predetermine the sample size. Experiments were not randomized. All imaging and quantifications were performed in a blinded manner. Statistical analyses were performed using GraphPad Prism.
Supplementary Material
Acknowledgment
This work was supported in part by the US National Institutes of Health (grants K01DK125608, R03DK135786, R01DK136724), an award from The G. Harold and Leila Y. Mathers Charitable Foundation, and a grant from Einstein-Mount Sinai Diabetes Center (to F.S.) and by National Institutes of Health grant RC2DK129961 (to P.C.). The Albert Einstein Animal Physiology Core is supported by the NIH grant DK20541. M.B. received funding from grants from the DFG project number 209933838 – SFB 1052 (project B1) and by Deutsches Zentrum für Diabetesforschung (DZD, Grant: 82DZD00601). The Slit3 floxed strain was kindly provided by Dr. Matthew Greenblatt. We thank Adam Mar and Begona Gamallo-Lana at the NYU Langone Rodent Behavior Laboratory (RRID: SCR_017942) for providing technical assistance, Experimental Pathology Research Laboratory (RRID:SCR_017928), Preclinical Imaging Laboratory (RRID:SCR_017937), and Microscopy Laboratory (RRID: SCR_017934). The microscopy shared resource is partially supported by Cancer Center Support Grant P30CA016087. We thank Christian Wolfrum’s group for contributing to the LOBB human adipose tissue RNA sequencing.
Footnotes
Competing interests
M.B. received honoraria as a consultant and speaker from Amgen, AstraZeneca, Bayer, Boehringer-Ingelheim, Lilly, Novo Nordisk, Novartis, and Sanofi. All other authors declare no conflict of interest.
Data availability
The data supporting the findings of this study are available within the paper and its supplementary information files. The human RNA-seq data from the LOBB have not been deposited in a public repository due to restrictions imposed by patient consent but can be obtained from Matthias Blüher upon request.
References
- 1.Cannon B. & Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev 84, 277–359 (2004). 10.1152/physrev.00015.2003 [DOI] [PubMed] [Google Scholar]
- 2.Shamsi F., Wang C. H. & Tseng Y. H. The evolving view of thermogenic adipocytes - ontogeny, niche and function. Nat Rev Endocrinol 17, 726–744 (2021). 10.1038/s41574-021-00562-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xue Y. et al. Hypoxia-independent angiogenesis in adipose tissues during cold acclimation. Cell Metab 9, 99–109 (2009). 10.1016/j.cmet.2008.11.009 [DOI] [PubMed] [Google Scholar]
- 4.Takahashi A., Shimazu T. & Maruyama Y. Importance of sympathetic nerves for the stimulatory effect of cold exposure on glucose utilization in brown adipose tissue. Jpn J Physiol 42, 653–664 (1992). 10.2170/jjphysiol.42.653 [DOI] [PubMed] [Google Scholar]
- 5.Shamsi F. et al. Vascular smooth muscle-derived Trpv1(+) progenitors are a source of cold-induced thermogenic adipocytes. Nat Metab 3, 485–495 (2021). 10.1038/s42255-021-00373-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shamsi F., Zheng R., Ho L. L., Chen K. & Tseng Y. H. Comprehensive analysis of intercellular communication in the thermogenic adipose niche. Commun Biol 6, 761 (2023). 10.1038/s42003-023-05140-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cannavino J. & Gupta R. K. Mesenchymal stromal cells as conductors of adipose tissue remodeling. Genes Dev 37, 781–800 (2023). 10.1101/gad.351069.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y. N. et al. Slit3 secreted from M2-like macrophages increases sympathetic activity and thermogenesis in adipose tissue. Nat Metab 3, 1536–1551 (2021). 10.1038/s42255-021-00482-9 [DOI] [PubMed] [Google Scholar]
- 9.Emont M. P. et al. A single-cell atlas of human and mouse white adipose tissue. Nature 603, 926–933 (2022). 10.1038/s41586-022-04518-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blockus H. & Chedotal A. Slit-Robo signaling. Development 143, 3037–3044 (2016). 10.1242/dev.132829 [DOI] [PubMed] [Google Scholar]
- 11.Xu R. et al. Targeting skeletal endothelium to ameliorate bone loss. Nat Med 24, 823–833 (2018). 10.1038/s41591-018-0020-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin Z., Chi J. & Cohen P. A Clearing Method for Three-Dimensional Imaging of Adipose Tissue. Methods Mol Biol 2448, 73–82 (2022). 10.1007/978-1-0716-2087-8_4 [DOI] [PubMed] [Google Scholar]
- 13.Brose K. et al. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell 96, 795–806 (1999). 10.1016/s0092-8674(00)80590-5 [DOI] [PubMed] [Google Scholar]
- 14.Choi C. H. J. et al. LRG1 is an adipokine that promotes insulin sensitivity and suppresses inflammation. Elife 11 (2022). 10.7554/eLife.81559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kellermeyer R. et al. Proteolytic cleavage of Slit by the Tolkin protease converts an axon repulsion cue to an axon growth cue in vivo. Development 147 (2020). 10.1242/dev.196055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ordan E., Brankatschk M., Dickson B., Schnorrer F. & Volk T. Slit cleavage is essential for producing an active, stable, non-diffusible short-range signal that guides muscle migration. Development 142, 1431–1436 (2015). 10.1242/dev.119131 [DOI] [PubMed] [Google Scholar]
- 17.Nguyen Ba-Charvet K. T. et al. Diversity and specificity of actions of Slit2 proteolytic fragments in axon guidance. J Neurosci 21, 4281–4289 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svensson K. J. et al. A Secreted Slit2 Fragment Regulates Adipose Tissue Thermogenesis and Metabolic Function. Cell Metab 23, 454–466 (2016). 10.1016/j.cmet.2016.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Talantikite M. et al. Inhibitors of BMP-1/tolloid-like proteinases: efficacy, selectivity and cellular toxicity. FEBS Open Bio 8, 2011–2021 (2018). 10.1002/2211-5463.12540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delloye-Bourgeois C. et al. PlexinA1 is a new Slit receptor and mediates axon guidance function of Slit C-terminal fragments. Nat Neurosci 18, 36–45 (2015). 10.1038/nn.3893 [DOI] [PubMed] [Google Scholar]
- 21.Zahalka A. H. et al. Adrenergic nerves activate an angio-metabolic switch in prostate cancer. Science 358, 321–326 (2017). 10.1126/science.aah5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watanabe T., Watanabe-Kominato K., Takahashi Y., Kojima M. & Watanabe R. Adipose Tissue-Derived Omentin-1 Function and Regulation. Compr Physiol 7, 765–781 (2017). 10.1002/cphy.c160043 [DOI] [PubMed] [Google Scholar]
- 23.Yang R. Z. et al. Identification of omentin as a novel depot-specific adipokine in human adipose tissue: possible role in modulating insulin action. Am J Physiol Endocrinol Metab 290, E1253–1261 (2006). 10.1152/ajpendo.00572.2004 [DOI] [PubMed] [Google Scholar]
- 24.Alto L. T. & Terman J. R. Semaphorins and their Signaling Mechanisms. Methods Mol Biol 1493, 1–25 (2017). 10.1007/978-1-4939-6448-2_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guilherme A., Henriques F., Bedard A. H. & Czech M. P. Molecular pathways linking adipose innervation to insulin action in obesity and diabetes mellitus. Nat Rev Endocrinol 15, 207–225 (2019). 10.1038/s41574-019-0165-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang H., Ding X., Cao Y., Wang H. & Zeng W. Dense Intra-adipose Sympathetic Arborizations Are Essential for Cold-Induced Beiging of Mouse White Adipose Tissue. Cell Metab 26, 686–692 e683 (2017). 10.1016/j.cmet.2017.08.016 [DOI] [PubMed] [Google Scholar]
- 27.Murano I., Barbatelli G., Giordano A. & Cinti S. Noradrenergic parenchymal nerve fiber branching after cold acclimatisation correlates with brown adipocyte density in mouse adipose organ. J Anat 214, 171–178 (2009). 10.1111/j.1469-7580.2008.01001.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bougneres P. et al. In vivo resistance of lipolysis to epinephrine. A new feature of childhood onset obesity. J Clin Invest 99, 2568–2573 (1997). 10.1172/JCI119444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heinonen S. et al. Impaired Mitochondrial Biogenesis in Adipose Tissue in Acquired Obesity. Diabetes 64, 3135–3145 (2015). 10.2337/db14-1937 [DOI] [PubMed] [Google Scholar]
- 30.Guo T. et al. Adipocyte ALK7 links nutrient overload to catecholamine resistance in obesity. Elife 3, e03245 (2014). 10.7554/eLife.03245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pirzgalska R. M. et al. Sympathetic neuron-associated macrophages contribute to obesity by importing and metabolizing norepinephrine. Nat Med 23, 1309–1318 (2017). 10.1038/nm.4422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saxton S. N., Withers S. B. & Heagerty A. M. Emerging Roles of Sympathetic Nerves and Inflammation in Perivascular Adipose Tissue. Cardiovasc Drugs Ther 33, 245–259 (2019). 10.1007/s10557-019-06862-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daily J. W., Liu M. & Park S. High genetic risk scores of SLIT3, PLEKHA5 and PPP2R2C variants increased insulin resistance and interacted with coffee and caffeine consumption in middle-aged adults. Nutr Metab Cardiovasc Dis 29, 79–89 (2019). 10.1016/j.numecd.2018.09.009 [DOI] [PubMed] [Google Scholar]
- 34.Liu Y. J. et al. Genome-wide association scans identified CTNNBL1 as a novel gene for obesity. Hum Mol Genet 17, 1803–1813 (2008). 10.1093/hmg/ddn072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baca P. et al. DNA methylation and gene expression analysis in adipose tissue to identify new loci associated with T2D development in obesity. Nutr Diabetes 12, 50 (2022). 10.1038/s41387-022-00228-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Klaauw A. A. et al. Human Semaphorin 3 Variants Link Melanocortin Circuit Development and Energy Balance. Cell 176, 729–742 e718 (2019). 10.1016/j.cell.2018.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roesch K. et al. The transcriptome of retinal Muller glial cells. J Comp Neurol 509, 225–238 (2008). 10.1002/cne.21730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shamsi F. et al. FGF6 and FGF9 regulate UCP1 expression independent of brown adipogenesis. Nat Commun 11, 1421 (2020). 10.1038/s41467-020-15055-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berry R. et al. Imaging of adipose tissue. Methods Enzymol 537, 47–73 (2014). 10.1016/b978-0-12-411619-1.00004-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mina A. I. et al. CalR: A Web-Based Analysis Tool for Indirect Calorimetry Experiments. Cell Metab 28, 656–666.e651 (2018). 10.1016/j.cmet.2018.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chi J., Crane A., Wu Z. & Cohen P. Adipo-Clear: A Tissue Clearing Method for Three-Dimensional Imaging of Adipose Tissue. J Vis Exp (2018). 10.3791/58271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langhardt J. et al. Effects of Weight Loss on Glutathione Peroxidase 3 Serum Concentrations and Adipose Tissue Expression in Human Obesity. Obes Facts 11, 475–490 (2018). 10.1159/000494295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mardinoglu A. et al. Extensive weight loss reveals distinct gene expression changes in human subcutaneous and visceral adipose tissue. Scientific Reports 5, 14841 (2015). 10.1038/srep14841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blüher M. Metabolically Healthy Obesity. Endocr Rev 41 (2020). 10.1210/endrev/bnaa004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klöting N. et al. Insulin-sensitive obesity. Am J Physiol Endocrinol Metab 299, E506–515 (2010). 10.1152/ajpendo.00586.2009 [DOI] [PubMed] [Google Scholar]
- 46.Picelli S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9, 171–181 (2014). 10.1038/nprot.2014.006 [DOI] [PubMed] [Google Scholar]
- 47.Hagemann T. et al. Laminin α4 Expression in Human Adipose Tissue Depots and Its Association with Obesity and Obesity Related Traits. Biomedicines 11 (2023). 10.3390/biomedicines11102806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bray N. L., Pimentel H., Melsted P. & Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–527 (2016). 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the paper and its supplementary information files. The human RNA-seq data from the LOBB have not been deposited in a public repository due to restrictions imposed by patient consent but can be obtained from Matthias Blüher upon request.