Abstract
CRISPR gene editing offers unprecedented genomic and transcriptomic control for precise regulation of cell function and phenotype. However, delivering the necessary CRISPR components to therapeutically relevant cell types without cytotoxicity or unexpected side effects remains challenging. Viral vectors risk genomic integration and immunogenicity while non-viral delivery systems are challenging to adapt to different CRISPR cargos, and many are highly cytotoxic. The arginine-alanine-leucine-alanine (RALA) cell penetrating peptide is an amphiphilic peptide that self-assembles into nanoparticles through electrostatic interactions with negatively charged molecules before delivering them across the cell membrane. This system has been used to deliver DNAs, RNAs, and small anionic molecules to primary cells with lower cytotoxicity compared to alternative non-viral approaches. Given the low cytotoxicity, versatility, and competitive transfection rates of RALA, we aimed to establish this peptide as a new CRISPR delivery system in a wide range of molecular formats across different editing modalities. We report that RALA was able to effectively encapsulate and deliver CRISPR in DNA, RNA, and ribonucleic protein (RNP) formats to primary mesenchymal stem cells (MSCs). Comparisons between RALA and commercially available reagents revealed superior cell viability leading to higher numbers of transfected cells and the maintenance of cell proliferative capacity. We then used the RALA peptide for the knock-in and knock-out of reporter genes into the MSC genome as well as for the transcriptional activation of therapeutically relevant genes. In summary, we establish RALA as a powerful tool for safer and effective delivery of CRISPR machinery in multiple cargo formats for a wide range of gene editing strategies.
Keywords: Cell-penetrating peptide, CRISPR gene editing, regenerative medicine, non-viral CRISPR delivery, mesenchymal stem cells
1. Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR) is a precise and tunable tool for reliable genomic and transcriptomic regulation [1]. This technology is based on the use of CRISPR associated (Cas) endonucleases which are directed by guide RNAs (gRNAs) to specific genomic loci to facilitate the knock-out, knock-in, activation, or inhibition, of specific genes [2, 3, 4]. Compared to previous gene editing systems such as zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENS), CRISPR adds ease of design, increased specificity and efficiency, and multiplexed targeting [5]. Despite these advantages, the use of CRISPR for the editing of primary cells is limited by the inability to deliver the necessary machinery in an efficient manner while preserving cell viability and function [6]. Specifically, this is key for applications involving the editing of clinically relevant stem and progenitor cells such as mesenchymal stromal cells (MSCs). These cells must maintain multipotency, proliferation, and immunomodulation post-editing to ensure the production of a large number of healthy cells with the desired therapeutic properties [7].
Among CRISPR delivery strategies to MSCs, viral vectors are often preferred due to strong, long-lasting expression. However, viral vectors are inherently immunogenic, activating antigen presenting cells and innate immune cells causing prolonged inflammation [8, 9, 10, 11]. They are also susceptible to random genomic integration, risking constitutive Cas9 expression which induces significant toxicity in primary stem cells and increases off-target genomic mutations [12, 13]. Viral vectors can also dysregulate natural stem cell pathways [14, 15]. Adenoviral and lentiviral transduction of MSCs was reported to reduce proliferation, induce senescence, and impair differentiation capacity, limiting overall therapeutic potential [16, 17]. In contrast, non-viral vectors reduce the risk of random genomic insertions and can deliver larger genetic payloads with a simpler production process [18]. However, these are not exempt of negative side effects. Physical methods such as electroporation or microinjection enable high editing efficiencies but lead to high cell death [19]. Moreover, physical methods require costly laboratory equipment and are most effective for ex vivo cell modification [20]. In contrast, non-viral nanoparticles composed of polymers, lipids, or inorganic reagents have less drastic effects on cell viability and can be delivered in vivo through systemic or local injection, or via incorporation into 3D biomaterials [21, 22]. Lipid-based strategies have attracted much attention due to their use in mRNA vaccines and have been employed across multiple CRISPR applications [23, 24, 25, 26]. These strategies can achieve high transfection rates allowing for transient gene expression. Additionally, lipids are biodegradable, which prevents severe cytotoxicity and inflammation. However, many commercial lipid vectors still have cytotoxic effects on transfected cells, leading to transcriptomic dysregulation that results in cell death and the disruption of stem cell’s differentiation and proliferation capacity [27, 28, 29]. Jacobsen et al. demonstrated that Lipofectamine transfection alone resulted in transcriptomic changes affecting over 1,000 genes [30]. These unexpected transcriptomic effects undermine the precision of CRISPR gene editing and reduce the number of healthy cells with the desired therapeutic properties.
An additional limitation of current non-viral strategies is the lack of versatility to deliver CRISPR machinery in different molecular formats. Different CRISPR applications require CRISPR components to be delivered in specific cargo types, such as plasmid DNA (pDNA), messenger RNA (mRNA), guide RNA (gRNA), or ribonucleoprotein complexes (RNPs). The choice of CRISPR format depends on factors like the target gene, cell type, and editing goals. However, existing systems lack the capability to efficiently deliver CRISPR machinery in multiple cargo formats, hindering the scalability, cost-efficiency, and standardization of biomanufacturing processes. Each CRISPR component has distinct physiochemical properties that limit their delivery through a broad range of non-viral reagents [31]. For example, Walther et al. attempted to deliver CRISPR components in both mRNA and RNP formats using an ionizable lipid nanoparticle [32]. While this method effectively delivered mRNA, RNP delivery resulted in inefficient nuclear localization and low editing rates (<10%) [32]. Similarly, while Lipofectamine 3000 efficiently delivers CRISPR in pDNA format, it falls short for mRNA and RNP delivery, requiring the Lipofectamine MessengerMAX and CRISPRMAX reagents respectively. This reliance on multiple reagents burdens cell biomanufacturing processes; therefore, new systems are needed to safely deliver CRISPR machinery in diverse molecular formats.
Cell penetrating peptides (CPPs) are a tunable alternative for CRISPR delivery. They can be easily designed and produced and enable competitive transfection rates with reduced cytotoxicity. CPPs have been used to deliver CRISPR components through incorporation into composite lipid or polymeric nanoparticles, direct fusion to Cas proteins, or as self-assembling nanoparticles with CRISPR cargos. Incorporation of CPPs into lipid or cationic polymer nanoparticles can increase transfection rates and add cell-specificity by targeting specific cell surface receptors [33, 34, 35, 36]. However, this method does not address lipid-associated cytotoxicity and transcriptomic dysregulation [34, 35, 36]. Alternatively, CPPs can be fused to Cas endonucleases to allow cellular uptake with minimal effects on cell viability [36]. This method requires careful design since initial attempts achieved low editing efficiencies due to interference between the covalently bound CPP and the endonuclease active site or gRNA binding domains [36]. More recently, Zhang et al. (2024) fused Cas9 or Cas12a to the transactivator of transcription (TAT) peptide sequence [37]. This allowed for effective CRISPR RNP delivery, resulting in ~70% editing efficiency in human CAR-T cells [37]. However, the need of a secondary peptide for endosomal escape limits the applicability of this approach in vivo due to challenges with ensuring that both components reach all target cells [37]. Self-assembling CPP nanoparticles can facilitate the cytoplasmic delivery of CRISPR components without these limitations. Recently, Foss et al. (2023) identified the A5K peptide for delivering Cas9 or Cas12a RNPs to primary human T and B cells [33]. This approach achieved 67–68% knock-out efficiency with no cytotoxicity and minimal transcriptomic perturbation [33]. However, A5K could not deliver the DNA donor sequences required for homology directed repair (HDR) and gene knock-in [33]. Adeno-associated virus (AAV) vectors were required to supplement peptide delivery, which introduces the risk of unexpected side effects such as immunogenicity and complicates manufacturing processes [33]. While existing CPP-CRISPR strategies demonstrate high knock-out efficiencies through RNP delivery, they are unable to deliver diverse CRISPR cargos. This highlights the need of new CPPs capable to effectively facilitate all CRISPR strategies without the need of additional peptides or supporting viral vectors.
The arginine-alanine-leucine-alanine (RALA) peptide is a helical, amphiphilic CPP that self-assembles into nanoparticles in the presence of negatively charged cargos due to electrostatic interactions. This simple assembly mechanism allows for complexing with DNAs, RNAs, and small anionic molecules such as tricalcium phosphate and the chemotherapy drug 5-fluorouracil [38, 39, 40, 41]. These nanoparticles primarily cross the cell membrane through clathrin-mediated endocytosis, but can also use other endocytosis pathways or directly traverse the membrane without forming an endosome [39]. This capacity for multiple transfection pathways expands RALAs applicability to hard-to-transfect cell types including primary MSCs, dendritic cells, dermal fibroblasts, and nucleus pulposus cells [7, 41, 42, 43, 44, 45, 46, 47]. Furthermore, RALA achieves this with minimal cytotoxicity, consistently preserving cell viability compared to lipid-based methods for DNA and RNA delivery [39, 48]. However, although similar arginine rich amphiphiles have been used to encapsulate proteins and CRISPR in RNP format [49], the capacity of RALA to deliver proteins and CRISPR machinery remains unexplored.
Here, we establish the RALA peptide as an effective novel tool for delivering CRISPR machinery into therapeutically relevant primary cells for multiple editing applications. We first characterized RALA’s capacity to complex CRISPR components in pDNA, RNP, and mRNA molecular formats into stable nanoparticles. We then used these nanoparticles to transfect primary MSCs and assess transfection efficiency, cytotoxicity, and proliferation, showing that RALA outperforms commercially available non-viral vectors while maintaining cell proliferation. Next, we effectively employed RALA to deliver pDNA and RNP to knock-in and knock-out red fluorescent protein (RFP) and luciferase (LUC) reporter genes. Finally, we used RALA-mRNA delivery to activate the expression of endogenous genes that could modulate stem cell differentiation towards different lineages. These results demonstrate that RALA enables versatile CRISPR delivery in diverse molecular formats, facilitating a wide range of gene editing approaches for effective stem cell therapies.
2. Materials and Methods
2.1. Materials:
Gene synthesis and cloning were performed by Genscript, USA. pOTTC763 - pX458 with rat ROSA26 gRNA A was a gift from Brandon Harvey (Addgene plasmid # 113161; http://n2t.net/addgene:113161; RRID:Addgene_113161) [50]. The RFP-LUC knock-in donor sequence was designed by first selecting miRFP670nano3 (RFP) and nanoluc (LUC) as small proteins with strong fluorescent and bioluminescent signals respectively [51, 52]. These sequences were separated by a Thosea asigna virus 2A (T2A) linker, placed under the cytomegalovirus (CMV) enhancer fused to chicken beta-actin promoter (CAG), and flanked by 1000 bp homology arms taken from the mRatBN7.2 genome on either side of the gRNA sequence. This entire sequence was constructed via de novo gene synthesis and cloned into pcDNA3-mRFP, a gift from Doug Golenbock (Addgene plasmid # 13032; http://n2t.net/addgene:13032; RRID:Addgene_13032), replacing the RFP sequence.
Plasmids for mRNA production were purchased from addgene. dSpCas9-VPR was a gift from Rasmus Bak (Addgene plasmid # 205247; http://n2t.net/addgene:205247; RRID:Addgene_205247) [53]. pGEM4Z-T7-5’UTR-EGFP-3’UTR-A64 was a gift from Christopher Grigsby & Molly Stevens (Addgene plasmid # 203348 http://n2t.net/addgene:203348; RRID:Addgene_203348). dCas9-VPR and eGFP plasmids were linearized using the Fast Digest Lgui and BcuI restriction enzymes respectively (Fisher scientific). Digested pDNA was gel purified using the QIAquick Gel Extraction Kit (Qiagen) and transcribed using the HiScribe® T7 Quick High Yield RNA Synthesis Kit (NEB). mRNA was purified using the Monarch® RNA Cleanup Kit (NEB). Yield and quality were assessed using a NanoDrop™ 2000 spectrophotometer (Thermo Scientific) and gel electrophoresis respectively.
Guide RNAs (gRNAs) for knocking-out the RFP-LUC reporter genes were designed by inputting the miRFP670nano3 sequence into CHOPCHOP with mRATBN7.2, CRISPR/Cas9 and knock-out specified [54]. The top scoring sequences starting with the G codon were selected for optimal compatibility with the precision gRNA synthesis kit (Thermo Fisher A29377). For gRNA targeting TGFB3, BMP2, or TNMD the transcriptional start site for each gene was identified through literature and 400 base pair (bp) sequences immediately upstream of the transcriptional start site (TSS) were downloaded from the National Center for Biotechnology Information (NCBI) genome data viewer [55, 56, 57, 58]. These sequences were then input into CHOPCHOP with mRATBN7.2, CRISPR/Cas9 and activation specified [54]. When possible, the top scoring sequences starting with a G were selected for better compatibility with the T7 promoter. Primers for each gRNA were designed (Table 1) and gRNAs were synthesized and purified according to the precision gRNA synthesis kit protocol.
Table 1.
List of gRNA sequences used in this study.
| Gene | gRNA Sequence | PAM |
|---|---|---|
| ROSA26 | GCAGATCACGAGGGAAGAAG | GGG |
| RFP(A) | GAACTTGCGCACCTCGGTCA | CGG |
| RFP(B) | GGCTTCGTGCGAGACGGTGC | CGG |
| TGFB3 (A) | GAGTCATTCCTGCTACACGT | TGG |
| TGFB3 (B) | GATCGCCCCCGGTCGGACAG | GGG |
| BMP2 (A) | GCGAGGACACCGGCGCGAAG | TGG |
| BMP2 (B) | GCGGCAAGACTGGATCCGCC | GGG |
| BMP2 (C) | GAGGACACCGGCGCGAAGTG | GGG |
| TNMD (A) | GCTGGAAGGGGATCAAGCCG | AGG |
| TNMD (B) | TGTGCAGCATGTATTGCCAC | TGG |
TrueCut™ HiFi Cas9 Protein was purchased from Thermo Fisher. RNPs were formed by mixing Cas9 protein with gRNA at a 1:1 molar ratio for 15 minutes at room temperature. Fluorescently labeled RNPs were produced by staining gRNAs with Ulysis™ Alexa Fluor™ 594 (Invitrogen) according to the manufacturer’s protocols. Labelled gRNA was purified with P-30 gel spin columns (Biorad) which were buffer-exchanged leaving the purified gRNA in pure water.
The RALA peptide (WEARLARALARALARHLARALARALRACEA) was produced by chemical peptide synthesis at Genscript (USA) and provided as a lyophilized powder at 96.6% purity via high-performance liquid chromatography (HPLC). Lipofectamine 3000 was purchased from Thermo Fisher. Branched 25 kD polyethyleneimine (PEI) was purchased from Millipore Sigma.
2.2. Nanoparticle formulation and characterization.
RALA nanoparticles were formed by mixing pDNA, mRNA, gRNA or RNP with the RALA peptide at the specified nitrogen:phosphate (N:P) ratios and incubating for 15 minutes. For RNP nanoparticles Cas9 and gRNA were mixed at a 1:1 molar ratio of Cas9:gRNA and incubated at room temperature for 15 minutes before adding the RALA peptide. RALA nanoparticles were characterized using dynamic light scattering (DLS) (Zetasizer nano ZS, Malvern Panalytical, UK) to measure the mean hydrodynamic diameter (size), zeta potential (charge), and polydispersity index (PDI) [59]. After complexing, 50 μL of nanoparticle mixture was diluted 1:20 vol/vol in ultrapure water and loaded into polystyrene cuvettes or folded capillary zeta cells for size and charge using automated settings.
2.3. MSC Isolation
Rat MSCs were harvested from male Sprague Daley rats (Charles River Laboratories) using protocols approved by the Lehigh University Institutional Animal Care and Use Committee (IACUC). Rats were euthanized and the hind leg tibia and femurs were removed and cleaned. The bone marrow from each extract was collected by flushing with warm Dulbecco’s Modified Eagle Medium (DMEM, low glucose; Gibco) via a 25-gauge needle. The resulting suspension was filtered using a 70 μm filter and seeded at 1 × 106 cells/cm2 onto tissue culture plastic treated for cell adhesion. Cells were grown in expansion media (DMEM supplemented with 10% fetal bovine serum (FBS) (Gemini Bio) 1% penicillin/streptomycin (Thermo Fisher) for 7 days with media changes every 2–3 days. To passage cells, they were first treated with trypsin (0.25% trypsin, 0.1% disodium ethylene diaminetetraacetic acid (EDTA) in Hanks’ balanced salt solution (HBSS) (Corning) for 5 minutes. Trypsin was then inactivated with expansion media and removed by centrifugation for 5 minutes at 200x g. Cells were resuspended in expansion media and reseeded at 5.7 × 103 cells/cm2 on tissue culture plastic treated for cell adhesion.
2.4. Cell culture and transfection protocol.
For RALA transfections, confluent MSCs were first treated with trypsin for 5 minutes before trypsin inactivation using expansion media. Cells were pelleted by centrifugation at 200x g for 5 minutes, resuspended in phosphate buffered saline (PBS), re-pelleted, and finally resuspended in OptiMEM (Gibco). Aliquots of 1 × 104 cells per cm2 of culture area were mixed with complexed RALA-CRISPR nanoparticles. For DNA delivery, 2 × 102 ng/cm2 of pDNA was complexed with RALA at N:P ratios of 5, 7, or 10. For RNP delivery, 2 × 102 ng/cm2 of Cas9 complexed to gRNA was complexed at 100x, 200x, or 500x molar ratios to RALA. Finally, for RNA delivery, 1.3 × 102 ng/cm2 of mRNA at a 1:2 molar ratio of mRNA:gRNA was complexed with RALA at N:P ratios 10, 15, and 20. Nanoparticles were incubated with the cells for 10 minutes at 37°C before cells were moved to 6-well plates and allowed to adhere. After 4 hours, OptiMEM was aspirated, and cells were washed with PBS then cultured in 2 mL of expansion media.
Lipofectamine was used as control according to manufacturer protocols. Briefly, rat MSCs were seeded at 2.6 × 103 cells/cm2 in 6 well plates 24 hours before transfection. For pDNA transfection, 2.08 × 102 ng/cm2 of pDNA was mixed with the 2 μL p3000 reagent per μg DNA then combined with 3 μL/μg Lipofectamine 3000 (Invitrogen) and incubated for 15 minutes before adding over cells. For mRNA delivery, 1.3 × 102 ng/cm2 of mRNA was directly mixed with the 3 μL/μg Lipofectamine MessengerMAX reagent (Invitrogen) and added to cells. For co-transfection with gRNA the mRNA and gRNA were mixed together before adding Lipofectamine. These nanoparticles were added directly to cells which were cultured until the necessary time points.
PEI transfections were performed as previously reported [7]. Briefly, cells were seeded at 2.6 × 103 cells/cm2 in 6-well plates 24 hours before transfection then washed with PBS and incubated in OptiMEM 2 hours before transfection. Nanoparticles were prepared by mixing pDNA or mRNA with PEI and incubating for 30 minutes. For DNA delivery, 2.08 × 102 ng/cm2 of pDNA was complexed with PEI at N:P of 7 [60]. For RNA delivery, 1.3 × 102 ng/cm2 of mRNA at a 1:2 molar ratio of mRNA:gRNA was complexed with PEI at N:P 15. These particles were diluted 10:1 vol/vol in OptiMEM and 500 μL of this mixture was added to each well then diluted with 1 mL of OptiMEM after 15 minutes. Finally, OptiMEM was removed, and cells were washed with PBS and 2 mL of expansion media was added to each well 4 hours after transfection.
After transfection, all fluorescent images were taken with the BZX-800 microscope (Keyence Corporation, USA) in phase contrast mode or with the fluorescein isothiocyanate (FITC), cyanine5 (Cy5), or Texas red (TxRed) filter cubes. Samples were prepared for flow cytometry by detaching cells with 0.25% trypsin, 0.1% EDTA in HBSS for 5 minutes and resuspending in PBS. Samples were assessed using a cytoflex flow cytometer (Beckman Coulter, USA). Samples were recorded at a variable flow rate to maintain the event rate at 400 events/s. The forward scatter (FSC), side scatter (SSC), FITC, and Allophycocyanin (APC) channel gains were set to 15, 20, 1, and 25 respectively. The resulting data was gating on FSC-A and SSC-A to remove debris then gated for FSC-A and FSC-H to select the linear cluster of single cells. GFP, RFP, or Ulysis fluorophore 594 positive thresholds were defined by non-transfected rMSCs for each experiment. The transfection rate is defined as the percentage of GFP, RFP, or Ulysis Fluorophore 594 positive cells compared to the total number of single cells. The cell viability was defined as the percentage of total single cells in transfected samples versus a non-transfected control. The transfection yield is defined as the percentage of GFP, RFP, or Ulysis Fluorophore 594 positive cells compared to the total single cell count of the non-transfected control.
| (eq. 1) |
where: = transfection yield for sample = fluorophore positive cell count, and = total cell count for the non-transfected control sample.
Additionally, the cell doubling time was calculated between days one and 3 using equation 2.
| (eq. 2) |
where: = cell doubling time, = time in days elapsed between cell counts, = cell count at time , and = cell count at initial timepoint.
2.5. RFP Knock-in
The knock-in of RFP was completed by co-transfecting equal amounts of the synthesized RFP-LUC donor plasmid with Addgene plasmid # 113161 containing the Cas9-T2A-GFP and ROSA26 gRNA sequences. Cell media was treated with 250 μg/mL Geneticin (Gibco) from days 2–10 to select for cells expressing the Neomycin/Kanamycin resistance cassette within the RFP donor template. Flow cytometry was performed on days 1, 3, and 10 and fluorescent imaging was performed on days 3 and 14. On day 21 knock-in cells were purified using flow assisted cell sorting (FACS) on a SH800/MA900 cell sorter (Sony).
Genomic DNA was extracted from the RFP knock-in population and purified using the Monarch® Genomic DNA Purification Kit (New England Biotech). Whole genome sequencing (WGS) was performed by Novogene with 350 bp paired-end reads and 30x coverage (90 G of data). The Rattus norvegicus reference genome was downloaded from the University of California at Santa Cruz (UCSC) Genome Browser for the rnor7 [61]. This genome was then modified using reform to insert the RFP-LUC sequence between the homology arm sequences in the fourth chromosome of the genome [62]. The reformed annotation file was then used to build an index then align fastq files from WGS using the bowtie2 algorithm [63]. The read aligned binary alignment map (BAM) files will then be merged to combine all reads into one file, sorted, and indexed using SAMTOOLS [64]. Finally, CRISPResso2 is utilized to visualize the sequences and editing occurrences around the Cas9 cleavage location and confirming the knock-in sequence [65].
2.6. RFP knock-out
To knock-out expression of the RFP-LUC sequence, RNP was formed with Cas9 and gRNA targeting RFP. Two gRNAs were identified for RFP knock-out and are listed in Table 1. RNPs containing both sequences were complexed into nanoparticles with RALA at a molar ratio of 100x and transfected into purified RFP knock-in cells. As a negative control RNP targeting outside of the ROSA26 locus (non-RFP) (gRNA sequence in Table 1) was transfected to ensure that loss of RFP signal was not a result of transfection. Cells were cultured for 7 days to allow for the proliferation of knock-out cells. Flow cytometry was performed on day 7 to assess the RFP signal through the APC channel. Additionally, 6.25 × 103 cells/cm2 of each sample were passage into a 24-well plate for fluorescent imaging and quantification of luciferase expression signal. Bioluminescence was quantified by replacing media with 500 μL of 100x dilute Nano-Glo live cell substrate (Promega) in expansion media. After 30 minutes the average radiance (p/s/cm2/sr) of each well was quantified using a Revvity Lumina III optical in vitro imaging system (IVIS).
2.7. Gene Activation
mRNA encoding the nuclease null Cas9 fused to the tripartite transcriptional activator VPR (dCas9-VPR) was mixed with gRNA targeting either TGFB3, BMP2, or TNMD at a 1:2 mRNA:gRNA molar ratio and complexed with RALA at N:P ratio of 15. Total RNA was extracted 24 hours later using the Trizol reagent (Invitrogen), purified using PureLink™ RNA Mini Kit (Invitrogen), and reverse transcribed using SuperScript™ IV VILO™ Master Mix (Invitrogen). Quantitative PCR (qPCR) was performed using TaqMan™ Fast Advanced Master Mix for qPCR (Applied Biosystems) and specific gene expression assays (Applied Biosystems). TaqMan gene expression assay primers were used to monitor the expression TGFB3, BMP2, and TNMD in comparison to the RPL13A housekeeping control (Table 2). Reactions were prepared according to the manufacturers protocol in a 0.2 mL MicroAmp™ optical 96-well reaction plate with barcode (Applied Biosystems) and PCR was performed in a QuantStudio3 real time PCR system (Applied Biosystems, USA) with the ΔΔCt method. All fold changes were quantified with respect to a control transfected with no gRNA.
Table 2.
List of TaqMan Primers used for qPCR.
| Gene Symbol | TaqMan Assay ID |
|---|---|
| TGFB3 | Rn00565937_m1 |
| BMP2 | Rn01449953_m1 |
| TNMD | Rn00574164_m1 |
| RPL13A | Rn00821946_g1 |
2.8. Statistics
Statistical analyses were performed using GraphPad Prism (version 10.2.1) software. All data was tested for normality using the Shapiro-Wilk test. For normal data with three or more groups, a one-way analysis of variance (ANOVA), or two-way ANOVA when collected on multiple days, was used with a Brown–Forsythe test and a Tukey post-test. Normal data with two groups was analyzed using a student’s t-test. Non-normal data was analyzed using the Kruskal-Wallis test. Numerical and graphical results are displayed as mean ± standard deviation, with the exception of qPCR fold-change data which is displayed as geometric mean ± geometric standard deviation. Significance was accepted at a level of p < 0.05. Sample size (n) is indicated within the corresponding figure legends.
3. Results
3.1. RALA effectively encapsulates and delivers CRISPR machinery in pDNA, mRNA, and RNP molecular cargo formats.
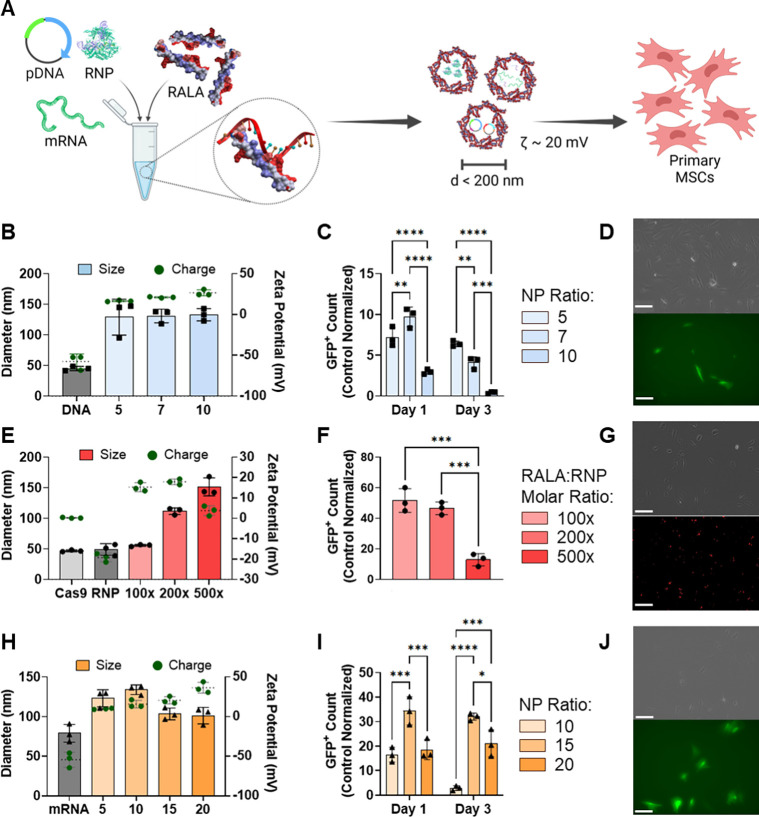
RALA nanoparticles were formed with pDNA, mRNA, and RNP cargos encoding for CRISPR machinery at a range of relevant N:P or molar ratios. These nanoparticles were then characterized by size, charge, and transfection rate (Fig. 1A) [33, 39, 40]. RALA-pDNA nanoparticles at N:P ratios of 5, 7, and 10 formed stable nanoparticles with diameters consistently below 150 nm (Fig. 1B). Alternatively, RALA-pDNA charges positively correlated with the N:P ratio, giving zeta potentials of 16.6 ± 0.67, 21.1 ± 0.57, and 26.6 ± 3.02 mV respectively (Fig. 1B). This trend was also matched in PDI (Supp. Fig. 1), suggesting that higher N:P ratios of 7 and 10 form the most stable nanoparticles. Transfection of pDNA encoding for Cas9-T2A-GFP was assessed by flow cytometry and fluorescent imaging. The transfection rate (percent GFP+) also increased with higher N:P ratios with rates of 11.82 ± 1.53%, 18.92 ± 1.66%, and 22.52 ± 0.07% on day 1 for N:P ratios of 5, 7, and 10 respectively (Fig. 1C–1D, Supp. Fig. 2). However, cell viability was adversely affected at higher N:P ratios, giving N:P = 5 the highest transfection yield (percent GFP+ relative to non-transfected control) (Fig. 1C, Supp. Fig. 3).
Figure 1. RALA encapsulates and delivers CRISPR machinery in pDNA, mRNA and RNP molecular formats into primary MSCs.
A) Schematic of RALA-CRISPR nanoparticle formation. Individual CRISPR cargos (pDNA, mRNA, or RNP) are mixed at specific ratios with the RALA peptide resulting in positively charged nanoparticles with diameters less than 200 nm. B) RALA-pDNA nanoparticle size (left y axis) and zeta potential (right y axis) at different N:P and molar ratios. C) Transfection yield (percent transfected relative to control count) after transfection with N:P ratios of 5, 7 and 10 on days 1 and 3 after transfection, and D) fluorescent images of MSCs 24 hours after transfection with RALA-pDNA using N:P ratio of 5. Scale bar = 100 μm. E) RALA-RNP nanoparticle size (left y axis) and zeta potential (right y axis) at 100x, 200x, 500x RALA: RNP molar ratios, F) Transfection yield (percent transfected relative to control count) after transfection with each ratio 8 hours after transfection, and G) fluorescent images of 100x RALA:RNP transfection with fluorescently labelled gRNA using a Cy5 filter. Scale bar = 100 μm. H) RALA-mRNA nanoparticle size (left y axis) and zeta potential (right y axis) at N:P ratios of 5, 10, 15 and 20, I) Transfection yield (percent transfected relative to control count) after transfection with each ratio on days 1 and 3, and J) fluorescent images of N:P = 15 transfection one day after transfection with FITC filter. Scale bar = 100 μm. * denotes significance p<0.05 (n = 3), **p<0.01 (n = 3), ***p<0.001 (n = 3), ****p<0.0001 (n = 3).
Next, RALA-RNP nanoparticles were characterized for size, charge, and transfection capacity. Cas9 RNPs maintained a similar diameter to Cas9 alone but exhibited a negative charge demonstrating suitability for RALA complexing (Fig. 1E). Nanoparticle formation at RALA:RNP molar ratios of 100x and 200x resulted in diameters below 125 nm and positive charges of 15.3 ± 1.82 and 17.9 ± 1.14 mV respectively, suggesting stable nanoparticle formation. However, at the 500x molar ratio there was an increase in diameter to 151.4 ± 11.6 nm and charge reduction to 3.75 ± 1.98 mV, indicating that the nanoparticles were less stable (Fig. 1E). Transfection of RNPs formed with fluorescently labeled gRNA resulted in nearly 100% transfection rate regardless of molar ratio (Supp. Fig. 5). However, the 100x molar ratio group exhibited the highest cell viability giving it the highest overall transfection yield (Fig. 1F, Supp. Fig. 6). These results were confirmed by localized fluorescent signal of labelled gRNAs within transfected cells (Fig. 1G).
Finally, RALA was complexed to mRNA encoding for dCas9-VPR forming nanoparticles displaying a significant decrease (p<0.05) in particle diameter and increase in charge at N:P ratios above 15. This suggests that N:P ratios of 15 or 20 are required for forming stable complexes. (Fig. 1H). This trend was confirmed through transfection of eGFP mRNA, where an N:P ratio of 15 resulted in a transfection rate of 53.1 ± 1.56% compared to 23.2 ± 4.48% for N:P = 10 (Supp. Fig. 8). At an N:P ratio of 20 the transfection rate was further increased to 71.5 ± 1.42%, but a significant loss in cell viability reduced the total number of edited cells, giving N:P = 15 the highest overall transfection yield (Fig. 1I–1J, Supp. Fig. 8–9).
3.2. RALA outperforms commercial vectors for the delivery of CRISPR pDNA.
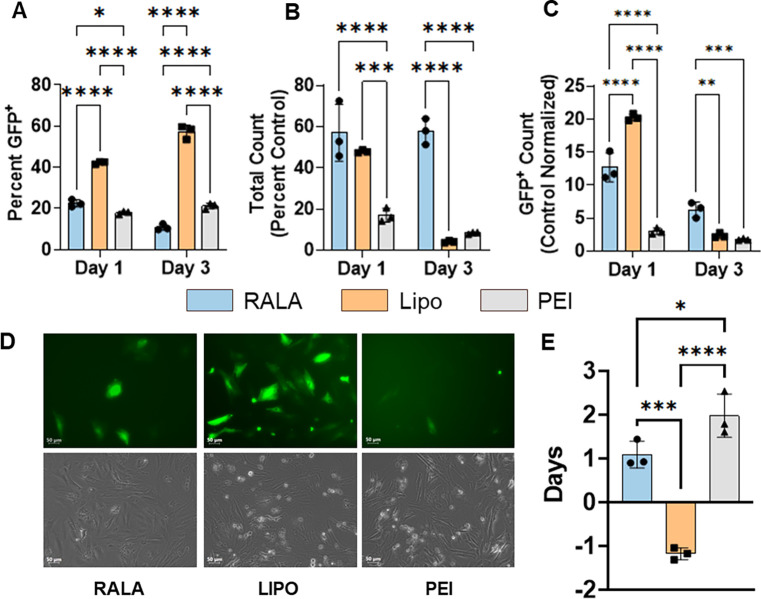
We characterized RALA’s transfection efficiency of pDNA in comparison to commercial vectors Lipofectamine 3000 and polyethyleneimine (PEI). PEI was used at N:P = 7 as optimized in previous reports [7, 60], and Lipofectamine was used according to manufacturer’s protocol at a ratio of 3 μL of Lipofectamine per 1 μg of pDNA. Each of these vectors were used for Cas9-T2A-GFP pDNA transfection and flow cytometry supported by fluorescent imaging were used to assess the transfection rates and cell viability. One day post-transfection RALA achieved a transfection rate of 22.48 ± 1.41% compared to 42.26 ± 0.53% for Lipofectamine and 17.98 ± 0.61% for PEI (Fig. 2A). By day 3, the transfection rate decreased to 10.87 ± 1.23% for RALA whereas the Lipofectamine and PEI rates increased to 57.23 ± 2.68% and 21.21 ± 1.13% respectively (Fig. 2A, Fig. 2D). This trend was inversely correlated with cell viability in which RALA significantly outperformed both Lipofectamine and PEI by day 3 with 57.7 ± 5.2 % of cells remaining viable in comparison to 4.1 ± 0.4 % for Lipofectamine and 8.3 ± 0.4 % for PEI (Fig. 2B, Fig. 2D). This superior viability resulted in higher transfection yields of 6.3 ± 1.0 % in comparison to 2.4 ± 0.3 % and 1.8 ± 0.1 % for Lipofectamine and PEI respectively (Fig. 2C, Fig. 2D). The self-renewal capacity of transfected cells was further analyzed through quantifying cell doubling time. Cell counts from days 1 to 3 revealed that the RALA group maintained a high proliferation rate which was lost in both Lipofectamine and PEI groups (Fig. 2E).
Figure 2. RALA outperforms commercial vectors for the delivery of pDNA encoding for CRISPR components.
The transfection of pDNA encoding for Cas9-T2A-GFP using RALA, Lipofectamine 3000, or PEI was characterized via flow cytometry. A) Transfection rate defined as the percentage of GFP+ cells in the transfected population. B) Cell viability defined as the total cell counts as a percentage of the non-transfected control count. C) Transfection yield defined as the percentage of GFP+ cells compared to the number of non-transfected control cells. The transfection was further characterized through D) representative fluorescent images of each transfection and E) the calculation of the cell doubling time from days 1 to 3. * denotes significance p<0.05 (n = 3), **p<0.01 (n = 3), ***p<0.001 (n = 3), ****p<0.0001 (n = 3).
3.3. CRISPR knock-in through RALA-mediated pDNA co-delivery.
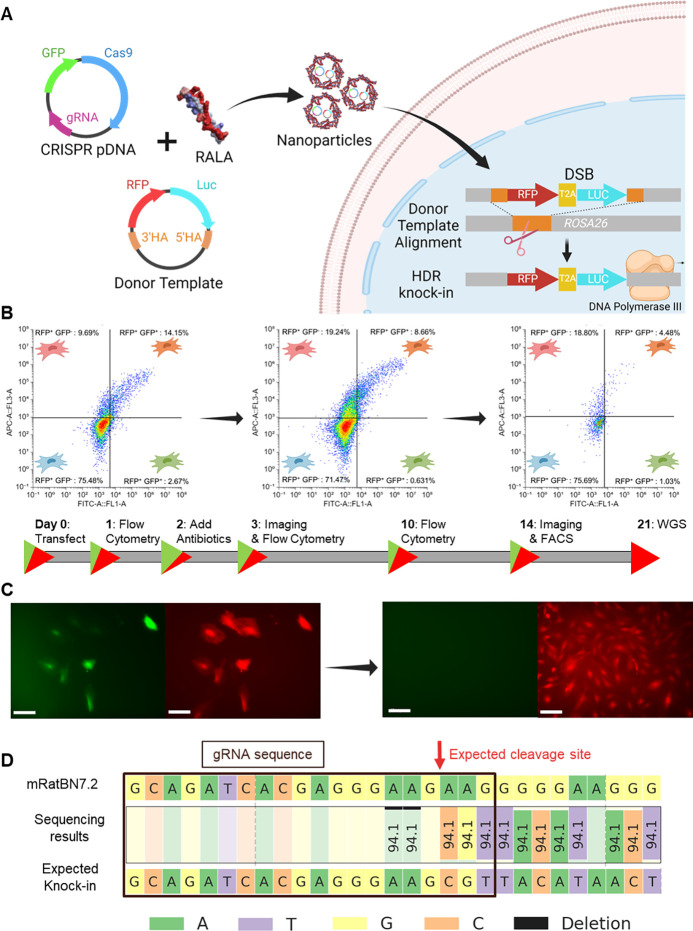
After confirming RALA’s superior properties in comparison to commercial vectors, we used RALA to knock-in an RFP-LUC reporter gene into the genome of primary MSCs. We followed a strategy based on the co-delivery of two pDNAs: (1) encoding for CRISPR machinery (Cas9-T2A-GFP and gRNA) targeting the ROSA26 locus, and (2) DNA donor sequence containing the RFP-LUC genes flanked by homology arms (Fig. 3A). Here, Cas9-T2A-GFP expression makes a double stranded break (DSB) within the ROSA26 safe-harbor locus while homology arms in the donor plasmid align the RFP-LUC sequence over this break for genomic insertion (Fig. 3A). Flow cytometry was performed on days 1, 3, and 10 post-transfection to track the expression of each plasmid. Simultaneous expression of both plasmids was obtained in 12.15 ± 0.86% of transfected cells on day 1 (Fig. 3B). In the remaining cells, 9.70 ± 0.43% expressed RFP from the donor template (RFP+GFP−) compared to only 2.67 ± 0.59% expressing GFP from Cas9-T2A-GFP (RFP−GFP+) (Fig. 3B). As cells proliferated by day 3, the rate of RFP+GFP+ and RFP+GFP− cells significantly decreased whereas a significant increase in RFP+GFP− cells was recorded (Fig. 3B, Supp. Fig. 10). After 10 days of culture, GFP signal was not observed in both RFP+GFP+ and RFP−GFP+ groups while the RFP+GFP− percentage stayed consistent, suggesting knock-in cell proliferation (Fig. 3B). Fluorescent imaging at day 14 after transfection further confirmed constitutive transgene expression with the presence of RFP+ colonies absent of GFP expression (Fig. 3C). These colonies were FACS purified to a homogenous RFP-LUC population. These cells were characterized through IVIS imaging to further confirm constitutive LUC expression (Supp. Fig. 11). Finally, the genomic DNA of RFP-LUC MSCs was sequenced, confirming the presence of the RFP-LUC transgene within the genome (Fig. 3D).
Figure 3. CRISPR knock-in through RALA-mediated pDNA co-delivery.
A) Schematic of RFP-LUC knock-in strategy through pDNA co-delivery. B) Flow cytometry profiling of RFP knock-in cells on days 1, 3, and 10 respectively showing gating representing RFP+GFP−, RFP+GFP+, RFP−GFP+, and RFP−GFP− cells moving clockwise from the top right. Percentages of the total population are listed. C) Representative fluorescent images of RFP knock-in cells taken on days 3 and 14 after transfection. Left image is FITC channel for GFP signal, right image is Cy5 channel for miRFP670nano3 imaging. Scale bar = 100 μm. D) WGS results at the gene knock-in location. The Rattus norvegicus genome assembly (top, mRatBN7.2) is compared to the expected gene knock-in sequence. The gRNA sequence used for knock-in is shown with a black box and the expected cleavage site is marked with a red arrow.
3.4. CRISPR knock-out through RALA-based delivery of Cas9-gRNA RNP.
Next, the capacity for RALA for CRISPR knock-out (CRISPRko) through RNP delivery was assessed through the deletion of RFP expression in the previously edited RFP-LUC MSCs (Fig. 3). RNPs were formed with gRNA A or gRNA B targeting distinct regions within the RFP sequence. Then, the gRNAs were mixed with RALA at a 100x molar ratio as identified in section 3.1 (Fig. 4A). Flow cytometry revealed a clear loss of RFP signal after one week of culture following knock-out (Fig. 4B). Further quantification revealed a 57.3 ± 5.5% and 14.0 ± 1.9% reduction in RFP+ cells after knock-out with gRNAs A and B respectively when compared to a non-RFP targeting control (Fig. 4C). These findings were also supported through fluorescent imaging which demonstrated a clear reduction in RFP signal following knock-out with gRNA A (Fig. 4D). Next, the knock-out was also characterized using an IVIS to capture the bioluminescence of each group resulting from the LUC transgene inserted downstream of RFP (Fig. 4E). All transfected groups experienced a significant loss in bioluminescence compared to a non-transfected control; however, no significant difference was observed between experimental and control groups (Fig. 4F). This result indicates that while knock-out causes a loss of RFP function, the presence of a second start codon at the LUC sequence allows continued expression.
Figure 4. RFP knock-out using RALA-RNP nanoparticles.
A) Schematic of gene knock-out strategy. Cas9 and gRNA targeting the RFP sequence were incubated for 15 minutes to form RNP complexes and then mixed with the RALA peptide to form nanoparticles. These nanoparticles are transfected and the RNPs are transported to the nucleus where they target the RFP gene leading to a loss of fluorescent signal. B) The RFP signal of each population was analyzed one week after transfection using flow cytometry to determine the rate of knock-out. C) The editing efficiency was defined as the percentage of RFP− cells as quantified from flow cytometry results. D) Fluorescent images taken after passaging knock-out cells taken with Cy5 filter. Scale bar 100 μm. E) Average radiance (p/s/cm2/sr) images to visualize the bioluminescence of knock-out cells after passaging and F) quantification of average radiance. * Denotes significance p<0.05 (n = 3), **p<0.01 (n = 3), ***p<0.001 (n = 3), ****p<0.0001 (n = 3).
3.5. RALA-mRNA delivery outperforms commercial transfection reagents.
In comparison to CRISPRki and CRISPRko, transcriptional regulation via CRISPR allows for precise control over gene expression without altering genomic sequences. dCas9-VPR allows activation of specific target genes depending on the chosen gRNA. However, this CRISPR tool requires efficient cellular delivery to achieve strong therapeutic effects [3]. To best accomplish this, we chose to deliver dCas9-VPR as mRNA. This allows for rapid translation in the cytoplasm in a greater number of cells in comparison to pDNA delivery [39, 42]. The capacity for RALA to effectively deliver mRNA for CRISPR transcriptional activation was determined through direct comparison to commercially available Lipofectamine MessengerMAX or PEI for the transfection of mRNA encoding for GFP. On day 1, the RALA group was outperformed by Lipofectamine in transfection rate with GFP+ rates of 53.13 ± 1.55% and 97.18 ± 0.22% respectively (Fig. 5A, Fig. 5 D). However, RALA achieved higher cell viability of 64.81 ± 8.50% compared to 26.0 ± 2.9 % for Lipofectamine (Fig. 5B, Fig. 5 D). This elevated viability led to the RALA group significantly outperforming Lipofectamine with a transfection yield of 34.4 ± 4.7% compared to 25.8 ± 2.9% respectively (Fig. 5C, Fig. 5 D). PEI transfected cells saw a significantly lower transfection rate of 11.3 ± 1.7 % on day 1, resulting in a lower yield of 7.3 ± 1.6 % (Fig. 5A, Fig. 5C). These trends continued on day 3 when the viability of RALA-transfected cells rose to 79.73 ± 1.02% as transfected cells proliferated faster than non-transfected cells. In contrast, the viability of Lipofectamine transfected cells fell to 10.60 ± 1.14% by day 3 (Fig. 5B). Lipofectamine’s negative effects on cell viability were also reflected in the negative doubling time, whereas there was no difference between RALA and PEI groups (Fig. 5E). Overall, RALA transfections resulted in a significantly greater transfection yield of 32.0 ± 1.2 % on day 3 compared to 5.5 ± 0.7 % and 3.5 ± 0.5 % for Lipofectamine and PEI respectively.
Figure 5. RALA mRNA delivery outperforms commercial transfection reagents Lipofectamine MessengerMAX and PEI.
A) Transfection rate defined as the percentage of GFP+ cells in the transfected population at days 1 and 3 post-transfection. B) Cell viability defined as the total cell counts as a percentage of the non-transfected control count. C) Transfection yield defined as the percentage of GFP+ cells compared to the number of non-transfected control cells. The transfection was further characterized through D) representative images of each transfection and E) the cell doubling time from days 1 to 3. * denotes significance p<0.05 (n = 3), **p<0.01 (n = 3), ***p<0.001 (n = 3), ****p<0.0001 (n = 3).
3.6. RALA-mRNA delivery for CRISPR-mediated activation of gene expression.
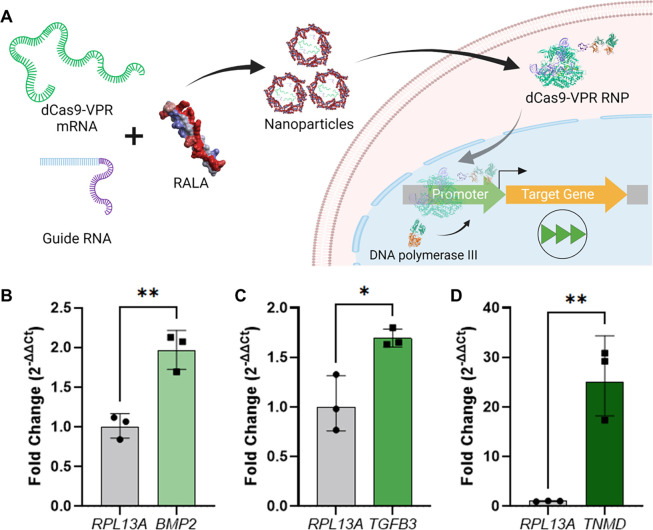
Upon confirming the suitability of RALA for mRNA delivery, mRNA encoding for dCas9-VPR was co-delivered with gRNA for CRISPR-mediated activation of gene expression (CRISPRa) (Fig. 6A). gRNAs were designed to target the therapeutic genes transforming growth factor β3 (TGFB3), bone morphogenic protein 2 (BMP2), and tenomodulin (TNMD). These are well characterized proteins which overexpression drives MSC chondrogenic, osteogenic, and tenogenic differentiation respectively (Table 1) [66, 67, 68]. TGFB3 and BMP2 were both moderately upregulated with fold changes of 1.69 ± 0.075 and 1.95 ± 0.19 respectively, while TNMD was highly upregulated at 25 ± 6.0-fold (Fig. 6B–6D). These activation fold-changes inversely correlated to the basal expression of each gene as indicated by the normalized Ct values (Supp. Fig. 12).
Figure 6. Activation of therapeutic gene expression using dCas9-VPR mRNA.
A) Schematic of CRISPR-mediated activation of gene expression (CRISPRa) strategy followed in this study. dCas9-VPR mRNA is co-delivered with gRNA and immediately forms an RNP upon cell delivery and mRNA translation. This RNP translocates to the nucleus where it binds upstream of the target gene TSS and recruits RNA polymerase III to the promoter region to upregulate transcription. Fold changes resultant of RALA gene activation for B) BMP2, C) TGFB3, and D) TNMD. All genes are compared to a control transfected with no gRNA and the RPL13A housekeeping gene. * denotes significance p<0.05 (n = 3), **p<0.01 (n = 3), ***p<0.001 (n = 3), ****p<0.0001 (n = 3).
4. Discussion
In this study, we establish for the first time the RALA peptide as an efficient delivery vector for a wide range of CRISPR gene editing techniques in therapeutically relevant primary MSCs. Analysis of the size and charge of RALA-CRISPR nanoparticles confirmed the encapsulation of CRISPR machinery in pDNA, mRNA, and RNP formats. Flow cytometry and fluorescent imaging validated effective cellular delivery with low cytotoxicity. Each of these CRISPR formats were delivered to primary MSCs to best suit specific gene editing techniques, demonstrating the flexibility and reliability of RALA-based gene editing. The co-delivery of pDNA encoding for CRISPR machinery and the donor template induced effective knock-in and constitutive expression of an RFP-LUC transgene to generate reporter MSCs. Subsequent delivery of Cas9 RNP targeting the RFP sequence in these reporter MSCs demonstrated robust gene knock-out and loss of RFP signal. Finally, delivery of mRNA encoding for dCas9-VPR upregulated key therapeutic gene targets for MSC chondrogenesis, osteogenesis, and tenogenesis. Taken together, these results establish RALA for the efficiently delivery of CRISPR machinery in different molecular formats to perform a wide range of gene editing modalities.
The RALA cell-penetrating peptide has been used to encapsulate and deliver multiple cargos including pDNA, mRNA, micro RNAs (miRNAs), and small molecules [69, 70, 71]. However, there are no previous reports using RALA for CRISPR gene editing. In this study, RALA serves as delivery vector for diverse gene editing strategies by delivering all CRISPR molecular formats in a cell-friendly manner. RALA’s versatility is accomplished through simple electrostatic interactions between its own positively charged arginine repeats and negatively charged molecules within its cargo. However, optimizing the charge ratios between RALA and CRISPR components is essential for producing stable nanoparticles with suitable transfection capabilities. Hence, the N:P ratio between RALA and pDNA or mRNA, and the molar ratio of RALA to RNP were assessed to optimize the delivery of CRISPR machinery [33, 39, 72]. Size and charge are key indicators of nanoparticle performance, with diameters less than 200 nm and charges of 20–30 mV being optimal for cellular uptake through endocytosis [73]. We observed that all nanoparticles fit these descriptions, but the smallest diameter and highest zeta potential were achieved with N:P = 7 for pDNA, N:P = 20 for mRNA, and 100x molar ratio for RNP. To characterize the cellular delivery of these nanoparticles, pDNA and mRNA encoding for GFP or fluorescently labelled RNPs were transfected and evaluated using fluorescent imaging and flow cytometry. We analyzed the transfection rate and cell viability of each N:P ratio. Combining these two parameters we also defined a transfection yield metric consisting of the percentage of fluorescent cells compared to the number of non-transfected control cells. This metric better describes the potential of each transfection for CRISPR approaches since effective gene editing requires high numbers of edited cells without cytotoxicity [33]. We determined that lower N:P ratios of 5 and 15 for pDNA and mRNA respectively and a 100x RALA:RNP molar ratio resulted in the highest number of edited cells. This result is similar to previous reports establishing an optimal N:P ratio of 6 for pDNA delivery in MSCs [7, 39]. However, we found that a higher N:P ratio of 15 was required for mRNA compared to previous reports using an N:P ratio of 10 [42]. Additionally, we established 100x RALA:RNP as the optimal molar ratio for RNP delivery [42, 74]. These results demonstrate that RALA can effectively encapsulate and deliver all CRISPR molecular formats. This capability enables the optimization of gene editing techniques by directly comparing the editing effects of different CRISPR cargos, without using multiple delivery vectors. Additionally, due to its low cytotoxicity, RALA shows potential for complex gene editing approaches requiring the delivery of multiple components and repeated transfections [75].
CRISPRki is a powerful tool allowing the insertion of customizable transgenes via HDR for gain of function studies and the engineering of advanced cell therapies. For example, Brunger et al. used CRISPR to knock-in IL1RA and TNFR1 genes under the cytokine-responsive CCL2 promoter to engineer auto-regulated anti-inflammatory stem cells [76]. This work demonstrates the advantages of CRISPRki for generating synthetic gene circuits and stimuli-responsive cells. However, HDR is an inefficient process, with knock-in rates typically falling below 5% in mammalian cells [77, 78]. These rates can be improved through chemical treatments and the delivery of additional pDNAs, but these increase the risk of off-target effects, reduce cell viability, and extend ex vivo cell culture times [77, 78]. To increase HDR probability, CRISPR machinery can be delivered as a self-replicating pDNA, resulting in multiple gene copies over a longer time-frame [79]. Still, pDNA must be delivered to a large number of healthy and proliferative cells to allow for effective knock-in [80]. Therefore, we compared the capacity of RALA for CRISPR pDNA transfection to commercial non-viral vectors Lipofectamine 3000 and PEI. RALA outperformed PEI in transfection rate, viability, and yield, and while it had lower transfection rates than Lipofectamine, its higher cell viability resulted in better overall yields. Furthermore, cell viability was constant from days 1 to 3 after RALA transfection whereas it decreased with both Lipofectamine and PEI, demonstrating that RALA maintains the proliferative capacity of transfected cells. Enhanced cell proliferation improves HDR knock-in rates by increasing the number of cell cycles during gene editing [81]. Further, the maintenance of proliferative capacity allows for the rapid growth of edited cells which could otherwise be outcompeted by non-edited cells, limiting the overall therapeutic effect. Therefore, RALA not only delivers CRISPR machinery to a higher number of cells but also increases the likelihood that knock-in will occur during cell division.
While RALA significantly outperformed commercial vectors in this study, we observed lower cell viability post-transfection in comparison to previous reports [39, 82]. This reduced viability on day 1 is likely due to reduced cell adhesion using our suspension transfection method in comparison to previous 2D monolayer transfection [39]. Interestingly, the octa-arginine (R8) cell-penetrating peptide, containing similar arginine content to RALA, has been shown to induce strong cell-adhesion when immobilized on cell culture substrates due to charge interactions [83]. This suggests that the presence of free RALA molecules could interact with integrins in the cell membrane and decrease cell adhesion to tissue culture plastic. Despite this initial lack of cell adhesion, RALA’s capacity to transfect cells in suspension demonstrates the potential of this approach for the editing non-adherent cell types such as hematopoietic stem cells lymphocytes, and monocytes [84]. Future studies will investigate the effect of RALA on cell adhesion and the efficacy of RALA-based CRISPR delivery to non-adherent cells.
After establishing the efficacy of RALA-pDNA transfection, RALA was used for the knock-in of an RFP-LUC reporter into the ROSA26 safe-harbor locus of primary MSCs. The knock-in and constitutive expression of these transgenes were confirmed by flow cytometry, fluorescent imaging, and WGS. The ROSA26 locus was selected due to its constitutive expression in mammalian cells without encoding any essential genes [85]. Therefore, double stranded breaks (DSBs) can be introduced for transgene insertion without risking insertion/deletion (indel) mutations due to non-homologous end joining (NHEJ), the primary CRISPR repair pathway [86]. The continued proliferation and RFP-LUC signal from these knock-in cells demonstrates that RALA-induced gene knock-in at the ROSA26 locus is a promising strategy for CRISPRki. Additionally, future work will target therapeutic locations to enable the quantification of gene expression [87], and the production of stimuli-responsive cells [76].
In addition to gain-of-function therapies through CRISPRki, the knock-out of specific genes also has important therapeutic implications. FDA approved CASGEVY uses CRISPRko to repress the BCL11A enhancer and as a result activate fetal hemoglobin for healthy red blood cell production in sickle cell anemia [88]. Such strategies benefit from the efficient delivery and transient action of CRISPR RNPs for robust editing with minimal off-target cleavage [33]. To evaluate the editing rates following RALA-RNP delivery, the reporter MSCs were treated with two RNPs targeting different locations of the RFP sequence and knock-out was assessed through flow cytometry, fluorescent imaging, and LUC quantification. A large discrepancy was observed in the efficacy of the two gRNA sequences used for this study. While gRNA A achieved a 57% reduction in RFP+ cells, gRNA B offered a 14% reduction. The difference in knock-out efficiency between these two gRNAs demonstrates the importance of gRNA selection and in vitro validation before in vivo studies. Yet, RALA delivery of RNPs containing gRNA A represents a reliable gene editing approach when compared to benchmark non-viral strategies. Although the previously established A5K peptide was able to achieve a higher knock-out rates of 68%, this system is designed specifically for RNP delivery and cannot deliver pDNA or other molecular formats [33]. In contrast, RALA enables a diverse range of CRISPR strategies while still accomplishing high editing rates.
Following knock-out, the bioluminescence of the edited cells was also measured. Interestingly, all transfected samples, including the negative control, had reduced bioluminescence compared to the knock-in reporter cells. These results suggest that the knock-out strategy did not enable the suppression of LUC expression. This could be due to the presence of a second start codon before LUC gene, allowing for ribosomal binding downstream of the RFP mutation for sustained LUC expression. Knock-out of both, RFP and LUC sequences could be achieved by co-delivering RNPs targeting both sequences individually. RALA has been previously established as a safe vector for repeated transfections without generating anti-RALA antibodies or over-stimulating the innate immune system [39]. Therefore, this RALA knock-in/knock-out system could serve as a powerful approach for delivering multiple transgenes into cells then selectively turning off genes once the function is no longer required.
CRISPR can also be used to transiently upregulate or inhibit target genes to induce cellular changes without permanent modification of genomic DNA. CRISPRa and CRISPR inhibition (CRISPRi) employ a Cas protein void of nuclease activity, dCas9, to deliver transcriptional machinery to specific sequences and up- or downregulate target gene expression [3]. These methods regulate the transcription of endogenous gene isoforms, thereby ensuring protein stability and reducing immunogenicity risks from foreign proteins [89]. The lack of DNA cleavage also removes indel mutation risks and improves the safety of gene editing [89]. Further, CRISPRa/i systems can be designed for multiplexed gene editing for fine control over multiple gene targets. For example, Truong et al. simultaneously activated SOX9 and repressed PPARG to stimulate MSC chondrogenesis while preventing adipogenesis [90]. However, the viral delivery system used in this study caused inflammation, cell death, and elevated immune cell infiltration upon implantation, which limits its clinical application [90]. Conversely, RALA has been reported to have reduced immune responses in vivo due to transient gene delivery with reduced cytotoxicity [39, 48]. This cell-friendly delivery of CRISPRa/i components serves as a promising approach for precise transcriptomic regulation. Still, CRISPRa/i action is required in a large portion of the target cell population to achieve a significant therapeutic impact. This can be accomplished by delivering CRISPRa/i machinery as mRNA since RALA has demonstrated higher transfection rates compared to pDNA delivery [42]. Additionally, mRNA production through in vitro transcription is more scalable and tunable than RNP due to ease of mRNA modification, synthesis and purification [91].
The capacity of RALA to deliver mRNA encoding for eGFP was characterized in comparison to commercially available Lipofectamine MessengerMAX and PEI. RALA again outperformed PEI in all categories. In comparison to Lipofectamine, RALA resulted in a higher transfection yield due to superior cell viability. This non-cytotoxic delivery of CRISPRa machinery is essential to prevent broad transcriptomic changes that counteract or alter the effects of the desired transcriptional regulation [92]. Having established RALA for mRNA transfection, this system was then used to deliver mRNA encoding dCas9-VPR to activate therapeutically relevant genes. TGFB3, BMP2, and TNMD were selected as well-established targets for the differentiation of MSCs into chondrocytes, osteoblasts, and tenocytes respectively [66, 67, 68]. We found that the fold changes of TGFB3 and BMP2 were lower than TNMD after activation. This follows an inverse correlation to the basal expression of each gene in bone marrow MSCs [93, 94, 95]. This observation fits with previous literature suggesting that activation fold changes depend on basal gene expression [96, 97]. Konermann et al. demonstrated a Pearson’s correlation (r) of 0.94 (p<0.0001) between gene activation and the inverse of basal transcript levels across multiple genes [97]. Importantly, this inverse correlation suggests that activation increases the number of transcripts produced at relatively consistent rates, so the therapeutic effect is independent of the final fold change. For example, a recent study used CRISPRa to upregulate the highly expressed VEGFA gene in endothelial cells, only achieving ~2-fold activation in vitro. Despite this low fold change, the large increase in VEGFA transcripts resulted in profound impacts on healing speed and quality in vivo as determined by wound closure, GAG deposition, and vascularization [98]. Therefore, although RALA-mediated gene activation did not achieve high fold changes for TGFB3 and BMP2, it could result in greater overall therapeutic effects.
5. Conclusion
In conclusion, the RALA peptide is the first non-viral vector able to safely deliver all CRISPR molecular formats to primary stem cells, facilitating a wide range of CRISPR gene editing modalities. Although RALA consistently resulted in lower overall percentages of transfected cells in comparison to Lipofectamine, enhanced cell viability improved transfection yields. This higher number of healthy edited cells improves the precision and efficacy of gene editing for a more profound therapeutic impact. However, the delivery capacity of RALA could be further improved. The transient nature of RALA transfections is beneficial for the safety of gene editing as it avoids constitutive Cas9 nuclease activity but limits the duration of transcriptional regulation techniques such as CRISPRa. Future work will investigate the sustained release of RALA nanoparticles for prolonged gene activation or inhibition. Additionally, RALA lacks cell-specificity. Future work will harness the adaptability and tunability of the RALA peptide to target individual cell types for advanced gene editing approaches. Lastly, RALA transfections are performed without the presence of serum proteins or high salt content [39]. The stability and efficiency of these nanoparticles for in vivo conditions warrants further study. Despite these shortcomings, RALA enables the safe and effective delivery of CRISPR cargos to primary stem cells for reliable gene editing. Therefore, it is a promising delivery approach for regenerative medicine applications and the treatment of stem cell genetic disorders.
Supplementary Material
Funding:
This material is based upon work supported by the National Science Foundation under Grant No. 2347637 to Tomas Gonzalez-Fernandez. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. Additionally, Tomas Gonzalez-Fernandez would like to acknowledge the start-up funds provided by the Department of Bioengineering and the P.C. Rossin College of Engineering & Applied Science at Lehigh University, the Orthoregeneration Network (ON) Kick Starter grant, and the Career Development Award from the American Society of Gene & Cell Therapy. The content is solely the responsibility of the authors and does not necessarily represent the official views of the American Society of Gene & Cell Therapy. Joshua P. Graham would like to acknowledge the support provided through the National Science Foundation Graduate Research Fellowship under Grant No 2234658. Michael Layden would like to acknowledge funds from National Institutes of Health R01GM127615 and the National Science Foundation 1942777.
Footnotes
Competing Interests: The authors have no competing interests to share.
References
- [1].Graham J., Werba L., Federico I., and Gonzalez-Fernandez T., “CRISPR Strategies for Stem Cell Engineering: A New Frontier in Musculoskeletal Regeneration,” Eur. Cell. Mater., vol. 46, pp. 91–118, Nov. 2023, doi: 10.22203/eCM.v046a05. [DOI] [Google Scholar]
- [2].Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., and Charpentier E., “A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity,” Science, vol. 337, no. 6096, pp. 816–821, Aug. 2012, doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Qi L. S. et al. , “Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression,” Cell, vol. 184, no. 3, p. 844, Feb. 2021, doi: 10.1016/j.cell.2021.01.019. [DOI] [PubMed] [Google Scholar]
- [4].Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F., “Genome engineering using the CRISPR-Cas9 system,” Nat. Protoc., vol. 8, no. 11, pp. 2281–2308, Nov. 2013, doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ikmi A., McKinney S. A., Delventhal K. M., and Gibson M. C., “TALEN and CRISPR/Cas9-mediated genome editing in the early-branching metazoan Nematostella vectensis,” Nat. Commun., vol. 5, no. 1, p. 5486, Nov. 2014, doi: 10.1038/ncomms6486. [DOI] [PubMed] [Google Scholar]
- [6].Sinclair F., Begum A. A., Dai C. C., Toth I., and Moyle P. M., “Recent advances in the delivery and applications of nonviral CRISPR/Cas9 gene editing,” Drug Deliv. Transl. Res., vol. 13, no. 5, pp. 1500–1519, May 2023, doi: 10.1007/s13346-023-01320-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gonzalez-Fernandez T. et al. , “Mesenchymal stem cell fate following non-viral gene transfection strongly depends on the choice of delivery vector,” Acta Biomater., vol. 55, pp. 226–238, Jun. 2017, doi: 10.1016/j.actbio.2017.03.044. [DOI] [PubMed] [Google Scholar]
- [8].Chew W. L. et al. , “A multifunctional AAV–CRISPR–Cas9 and its host response,” Nat. Methods, vol. 13, no. 10, pp. 868–874, Oct. 2016, doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mefferd A. L., Kornepati A. V. R., Bogerd H. P., Kennedy E. M., and Cullen B. R., “Expression of CRISPR/Cas single guide RNAs using small tRNA promoters,” RNA, vol. 21, no. 9, pp. 1683–1689, Sep. 2015, doi: 10.1261/rna.051631.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Manjón A. G. et al. , “Unexpected gene activation following CRISPR-Cas9-mediated genome editing,” EMBO Rep., vol. 23, no. 2, p. e53902, Feb. 2022, doi: 10.15252/embr.202153902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Lek A. et al. , “Death after High-Dose rAAV9 Gene Therapy in a Patient with Duchenne’s Muscular Dystrophy,” N. Engl. J. Med., vol. 389, no. 13, pp. 1203–1210, Sep. 2023, doi: 10.1056/NEJMoa2307798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yu K.-R., Natanson H., and Dunbar C. E., “Gene Editing of Human Hematopoietic Stem and Progenitor Cells: Promise and Potential Hurdles,” Hum. Gene Ther., vol. 27, no. 10, pp. 729–740, Oct. 2016, doi: 10.1089/hum.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hanlon K. S. et al. , “High levels of AAV vector integration into CRISPR-induced DNA breaks,” Nat. Commun., vol. 10, no. 1, p. 4439, Sep. 2019, doi: 10.1038/s41467-019-12449-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hacein-Bey-Abina S. et al. , “Efficacy of Gene Therapy for X-Linked Severe Combined Immunodeficiency,” N. Engl. J. Med., vol. 363, no. 4, pp. 355–364, Jul. 2010, doi: 10.1056/NEJMoa1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Nayak S. and Herzog R. W., “Progress and prospects: immune responses to viral vectors,” Gene Ther., vol. 17, no. 3, pp. 295–304, Mar. 2010, doi: 10.1038/gt.2009.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Marasini S. et al. , “Effects of Adenoviral Gene Transduction on the Stemness of Human Bone Marrow Mesenchymal Stem Cells,” Mol. Cells, vol. 40, no. 8, pp. 598–605, Aug. 2017, doi: 10.14348/molcells.2017.0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Guo H., Zhao N., Gao H., and He X., “Mesenchymal Stem Cells Overexpressing Interleukin-35 Propagate Immunosuppressive Effects in Mice,” Scand. J. Immunol., vol. 86, no. 5, pp. 389–395, 2017, doi: 10.1111/sji.12613. [DOI] [PubMed] [Google Scholar]
- [18].Gonzalez-Fernandez T., Kelly D. J., and O’Brien F. J., “Controlled Non-Viral Gene Delivery in Cartilage and Bone Repair: Current Strategies and Future Directions,” Adv. Ther., vol. 1, no. 7, p. 1800038, 2018, doi: 10.1002/adtp.201800038. [DOI] [Google Scholar]
- [19].Chastagnier L., Marquette C., and Petiot E., “In situ transient transfection of 3D cell cultures and tissues, a promising tool for tissue engineering and gene therapy,” Biotechnol. Adv., vol. 68, p. 108211, Nov. 2023, doi: 10.1016/j.biotechadv.2023.108211. [DOI] [PubMed] [Google Scholar]
- [20].Geng T., Zhan Y., Wang J., and Lu C., “Transfection of cells using flow-through electroporation based on constant voltage,” Nat. Protoc., vol. 6, no. 8, pp. 1192–1208, Aug. 2011, doi: 10.1038/nprot.2011.360. [DOI] [PubMed] [Google Scholar]
- [21].Liang P. et al. , “CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes,” Protein Cell, vol. 6, no. 5, pp. 363–372, May 2015, doi: 10.1007/s13238-015-0153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hendriks D., Artegiani B., Hu H., de Sousa Lopes S. Chuva, and Clevers H., “Establishment of human fetal hepatocyte organoids and CRISPR–Cas9-based gene knockin and knockout in organoid cultures from human liver,” Nat. Protoc., vol. 16, no. 1, pp. 182–217, Jan. 2021, doi: 10.1038/s41596-020-00411-2. [DOI] [PubMed] [Google Scholar]
- [23].Brunger J. M., Zutshi A., Willard V. P., Gersbach C. A., and Guilak F., “CRISPR/Cas9 Editing of Murine Induced Pluripotent Stem Cells for Engineering Inflammation-Resistant Tissues,” Arthritis Rheumatol., vol. 69, no. 5, pp. 1111–1121, 2017, doi: 10.1002/art.39982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Baden L. R. et al. , “Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine,” N. Engl. J. Med., vol. 384, no. 5, pp. 403–416, Feb. 2021, doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gillmore J. D. et al. , “CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis,” N. Engl. J. Med., vol. 385, no. 6, pp. 493–502, Aug. 2021, doi: 10.1056/NEJMoa2107454. [DOI] [PubMed] [Google Scholar]
- [26].Farhang N., Davis B., Weston J., Ginley-Hidinger M., Gertz J., and Bowles R. D., “Synergistic CRISPRa-Regulated Chondrogenic Extracellular Matrix Deposition Without Exogenous Growth Factors,” Tissue Eng. Part A, vol. 26, no. 21–22, pp. 1169–1179, Nov. 2020, doi: 10.1089/ten.tea.2020.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kon E., Ad-El N., Hazan-Halevy I., Stotsky-Oterin L., and Peer D., “Targeting cancer with mRNA–lipid nanoparticles: key considerations and future prospects,” Nat. Rev. Clin. Oncol., vol. 20, no. 11, pp. 739–754, Nov. 2023, doi: 10.1038/s41571-023-00811-9. [DOI] [PubMed] [Google Scholar]
- [28].Kedmi R., Ben-Arie N., and Peer D., “The systemic toxicity of positively charged lipid nanoparticles and the role of Toll-like receptor 4 in immune activation,” Biomaterials, vol. 31, no. 26, pp. 6867–6875, Sep. 2010, doi: 10.1016/j.biomaterials.2010.05.027. [DOI] [PubMed] [Google Scholar]
- [29].Lv H., Zhang S., Wang B., Cui S., and Yan J., “Toxicity of cationic lipids and cationic polymers in gene delivery,” J. Controlled Release, vol. 114, no. 1, pp. 100–109, Aug. 2006, doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- [30].Jacobsen L. B., Calvin S. A., and Lobenhofer E. K., “Transcriptional effects of transfection: the potential for misinterpretation of gene expression data generated from transiently transfected cells,” BioTechniques, vol. 47, no. 1, pp. 617–624, Jul. 2009, doi: 10.2144/000113132. [DOI] [PubMed] [Google Scholar]
- [31].Hald Albertsen C., Kulkarni J. A., Witzigmann D., Lind M., Petersson K., and Simonsen J. B., “The role of lipid components in lipid nanoparticles for vaccines and gene therapy,” Adv. Drug Deliv. Rev., vol. 188, p. 114416, Sep. 2022, doi: 10.1016/j.addr.2022.114416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Walther J. et al. , “Comparative analysis of lipid Nanoparticle-Mediated delivery of CRISPR-Cas9 RNP versus mRNA/sgRNA for gene editing in vitro and in vivo,” Eur. J. Pharm. Biopharm., vol. 196, p. 114207, Mar. 2024, doi: 10.1016/j.ejpb.2024.114207. [DOI] [PubMed] [Google Scholar]
- [33].Foss D. V. et al. , “Peptide-mediated delivery of CRISPR enzymes for the efficient editing of primary human lymphocytes,” Nat. Biomed. Eng., vol. 7, no. 5, pp. 647–660, May 2023, doi: 10.1038/s41551-023-01032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wang P. et al. , “Thermo-triggered Release of CRISPR-Cas9 System by Lipid-Encapsulated Gold Nanoparticles for Tumor Therapy,” Angew. Chem. Int. Ed., vol. 57, no. 6, pp. 1491–1496, 2018, doi: 10.1002/anie.201708689. [DOI] [PubMed] [Google Scholar]
- [35].Li M. et al. , “Knockdown of hypoxia-inducible factor-1 alpha by tumor targeted delivery of CRISPR/Cas9 system suppressed the metastasis of pancreatic cancer,” J. Controlled Release, vol. 304, pp. 204–215, Jun. 2019, doi: 10.1016/j.jconrel.2019.05.019. [DOI] [PubMed] [Google Scholar]
- [36].Mout R. et al. , “Direct Cytosolic Delivery of CRISPR/Cas9-Ribonucleoprotein for Efficient Gene Editing,” ACS Nano, vol. 11, no. 3, pp. 2452–2458, Mar. 2017, doi: 10.1021/acsnano.6b07600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhang Z. et al. , “Efficient engineering of human and mouse primary cells using peptide-assisted genome editing,” Nat. Biotechnol., vol. 42, no. 2, pp. 305–315, Feb. 2024, doi: 10.1038/s41587-023-01756-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang F. et al. , “Amphiphilic Cationic Peptide-Coated PHA Nanosphere as an Efficient Vector for Multiple-Drug Delivery,” Nanomaterials, vol. 12, no. 17, Art. no. 17, Jan. 2022, doi: 10.3390/nano12173024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].McCarthy H. O. et al. , “Development and characterization of self-assembling nanoparticles using a bio-inspired amphipathic peptide for gene delivery,” J. Controlled Release, vol. 189, pp. 141–149, Sep. 2014, doi: 10.1016/j.jconrel.2014.06.048. [DOI] [PubMed] [Google Scholar]
- [40].McCrudden C. M. et al. , “Peptide delivery of a multivalent mRNA SARS-CoV-2 vaccine,” J. Controlled Release, vol. 362, pp. 536–547, Oct. 2023, doi: 10.1016/j.jconrel.2023.08.053. [DOI] [PubMed] [Google Scholar]
- [41].Sathy B. N. et al. , “RALA complexed α-TCP nanoparticle delivery to mesenchymal stem cells induces bone formation in tissue engineered constructs in vitro and in vivo,” J. Mater. Chem. B, vol. 5, no. 9, pp. 1753–1764, Mar. 2017, doi: 10.1039/C6TB02881K. [DOI] [PubMed] [Google Scholar]
- [42].Udhayakumar V. K. et al. , “Arginine-Rich Peptide-Based mRNA Nanocomplexes Efficiently Instigate Cytotoxic T Cell Immunity Dependent on the Amphipathic Organization of the Peptide,” Adv. Healthc. Mater., vol. 6, no. 13, p. 1601412, 2017, doi: 10.1002/adhm.201601412. [DOI] [PubMed] [Google Scholar]
- [43].Coolen A.-L. et al. , “Poly(lactic acid) nanoparticles and cell-penetrating peptide potentiate mRNA-based vaccine expression in dendritic cells triggering their activation,” Biomaterials, vol. 195, pp. 23–37, Mar. 2019, doi: 10.1016/j.biomaterials.2018.12.019. [DOI] [PubMed] [Google Scholar]
- [44].Yan L.-P. et al. , “Collagen/GAG scaffolds activated by RALA-siMMP-9 complexes with potential for improved diabetic foot ulcer healing,” Mater. Sci. Eng. C, vol. 114, p. 111022, Sep. 2020, doi: 10.1016/j.msec.2020.111022. [DOI] [PubMed] [Google Scholar]
- [45].Chambers P. et al. , “The osteogenic and angiogenic potential of microRNA-26a delivered via a non-viral delivery peptide for bone repair,” J. Controlled Release, vol. 362, pp. 489–501, Oct. 2023, doi: 10.1016/j.jconrel.2023.09.006. [DOI] [PubMed] [Google Scholar]
- [46].Sadowska J. M. et al. , “Development of miR-26a-activated scaffold to promote healing of critical-sized bone defects through angiogenic and osteogenic mechanisms,” Biomaterials, vol. 303, p. 122398, Dec. 2023, doi: 10.1016/j.biomaterials.2023.122398. [DOI] [PubMed] [Google Scholar]
- [47].Barcellona M. N., Ní Néill T., O’Brien F. J., Dixon J. E., Curtin C. M., and Buckley C. T., “Modulation of Inflammation and Regeneration in the Intervertebral Disc Using Enhanced Cell-Penetrating Peptides for MicroRNA Delivery,” Adv. NanoBiomed Res., vol. 4, no. 7, p. 2300112, 2024, doi: 10.1002/anbr.202300112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Bennett R. et al. , “RALA-mediated delivery of FKBPL nucleic acid therapeutics,” Nanomed., vol. 10, no. 19, pp. 2989–3001, Oct. 2015, doi: 10.2217/nnm.15.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Krishnamurthy S. et al. , “Engineered amphiphilic peptides enable delivery of proteins and CRISPR-associated nucleases to airway epithelia,” Nat. Commun., vol. 10, no. 1, p. 4906, Oct. 2019, doi: 10.1038/s41467-019-12922-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bäck S. et al. , “Neuron-Specific Genome Modification in the Adult Rat Brain Using CRISPR-Cas9 Transgenic Rats,” Neuron, vol. 102, no. 1, pp. 105–119.e8, Apr. 2019, doi: 10.1016/j.neuron.2019.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Stacer A. C. et al. , “NanoLuc Reporter for Dual Luciferase Imaging in Living Animals,” Mol. Imaging, vol. 12, no. 7, p. 7290.2013.00062, Oct. 2013, doi: 10.2310/7290.2013.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Oliinyk O. S. et al. , “Single-domain near-infrared protein provides a scaffold for antigen-dependent fluorescent nanobodies,” Nat. Methods, vol. 19, no. 6, pp. 740–750, Jun. 2022, doi: 10.1038/s41592-022-01467-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Jensen T. I. et al. , “Targeted regulation of transcription in primary cells using CRISPRa and CRISPRi,” Genome Res., vol. 31, no. 11, pp. 2120–2130, Nov. 2021, doi: 10.1101/gr.275607.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Labun K., Montague T. G., Krause M., Torres Cleuren Y. N., Tjeldnes H., and Valen E., “CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing,” Nucleic Acids Res., vol. 47, no. W1, pp. W171–W174, Jul. 2019, doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Shukunami C. et al. , “Scleraxis is a transcriptional activator that regulates the expression of Tenomodulin, a marker of mature tenocytes and ligamentocytes,” Sci. Rep., vol. 8, no. 1, p. 3155, Feb. 2018, doi: 10.1038/s41598-018-21194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Schäffer L. et al. , “Oxygen-regulated Expression of TGF-β3, a Growth Factor Involved in Trophoblast Differentiation,” Placenta, vol. 24, no. 10, pp. 941–950, Nov. 2003, doi: 10.1016/S0143-4004(03)00166-8. [DOI] [PubMed] [Google Scholar]
- [57].Rogers M. B., Shah T. A., and Shaikh N. N., “Turning Bone Morphogenetic Protein 2 (BMP2) On and Off in Mesenchymal Cells,” J. Cell. Biochem., vol. 116, no. 10, pp. 2127–2138, Oct. 2015, doi: 10.1002/jcb.25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Rangwala S. H. et al. , “Accessing NCBI data using the NCBI Sequence Viewer and Genome Data Viewer (GDV),” Genome Res., vol. 31, no. 1, pp. 159–169, Jan. 2021, doi: 10.1101/gr.266932.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Dahal S., Bastola S., and Ramamurthi A., “JNK2 silencing lipid nanoparticles for elastic matrix repair,” J. Biomed. Mater. Res. A, vol. 112, no. 4, pp. 562–573, 2024, doi: 10.1002/jbm.a.37618. [DOI] [PubMed] [Google Scholar]
- [60].Tierney E. G., Duffy G. P., Hibbitts A. J., Cryan S.-A., and O’Brien F. J., “The development of non-viral gene-activated matrices for bone regeneration using polyethyleneimine (PEI) and collagen-based scaffolds,” J. Controlled Release, vol. 158, no. 2, pp. 304–311, Mar. 2012, doi: 10.1016/j.jconrel.2011.11.026. [DOI] [PubMed] [Google Scholar]
- [61].Nassar L. R. et al. , “The UCSC Genome Browser database: 2023 update,” Nucleic Acids Res., vol. 51, no. D1, pp. D1188–D1195, Jan. 2023, doi: 10.1093/nar/gkac1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Khalfan M. and Gresham D., “Modifying Reference Sequence and AnnotationFiles Quickly and Easily with reform,” Aug. 2022, doi: 10.17605/OSF.IO/5KS7R. [DOI] [Google Scholar]
- [63].Langmead B. and Salzberg S. L., “Fast gapped-read alignment with Bowtie 2,” Nat. Methods, vol. 9, no. 4, pp. 357–359, Mar. 2012, doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Li H. et al. , “The Sequence Alignment/Map format and SAMtools,” Bioinformatics, vol. 25, no. 16, pp. 2078–2079, Aug. 2009, doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Clement K. et al. , “CRISPResso2 provides accurate and rapid genome editing sequence analysis,” Nat. Biotechnol., vol. 37, no. 3, pp. 224–226, Mar. 2019, doi: 10.1038/s41587-019-0032-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Bello A. B. et al. , “Matrilin3/TGFβ3 gelatin microparticles promote chondrogenesis, prevent hypertrophy, and induce paracrine release in MSC spheroid for disc regeneration,” Npj Regen. Med., vol. 6, no. 1, pp. 1–13, Sep. 2021, doi: 10.1038/s41536-021-00160-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Jiang Y. et al. , “Enhanced tenogenic differentiation and tendon-like tissue formation by tenomodulin overexpression in murine mesenchymal stem cells,” J. Tissue Eng. Regen. Med., vol. 11, no. 9, pp. 2525–2536, 2017, doi: 10.1002/term.2150. [DOI] [PubMed] [Google Scholar]
- [68].Na K. et al. , “Osteogenic differentiation of rabbit mesenchymal stem cells in thermo-reversible hydrogel constructs containing hydroxyapatite and bone morphogenic protein-2 (BMP-2),” Biomaterials, vol. 28, no. 16, pp. 2631–2637, Jun. 2007, doi: 10.1016/j.biomaterials.2007.02.008. [DOI] [PubMed] [Google Scholar]
- [69].Murray K. D. et al. , “Enhanced cationic liposome-mediated transfection using the DNA-binding peptide μ (mu) from the adenovirus core,” Gene Ther., vol. 8, no. 6, pp. 453–460, Mar. 2001, doi: 10.1038/sj.gt.3301401. [DOI] [PubMed] [Google Scholar]
- [70].Malim M. H., Böhnlein S., Hauber J., and Cullen B. R., “Functional dissection of the HIV-1 Rev trans-activator--derivation of a trans-dominant repressor of Rev function,” Cell, vol. 58, no. 1, pp. 205–214, Jul. 1989, doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- [71].Tünnemann G., Ter-Avetisyan G., Martin R. M., Stöckl M., Herrmann A., and Cardoso M. C., “Live-cell analysis of cell penetration ability and toxicity of oligo-arginines,” J. Pept. Sci., vol. 14, no. 4, pp. 469–476, 2008, doi: 10.1002/psc.968. [DOI] [PubMed] [Google Scholar]
- [72].Neves A. R., Sousa A., Faria R., Albuquerque T., Queiroz J. A., and Costa D., “Cancer gene therapy mediated by RALA/plasmid DNA vectors: Nitrogen to phosphate groups ratio (N/P) as a tool for tunable transfection efficiency and apoptosis,” Colloids Surf. B Biointerfaces, vol. 185, p. 110610, Jan. 2020, doi: 10.1016/j.colsurfb.2019.110610. [DOI] [PubMed] [Google Scholar]
- [73].El-Sayed A. and Harashima H., “Endocytosis of Gene Delivery Vectors: From Clathrin-dependent to Lipid Raft-mediated Endocytosis,” Mol. Ther., vol. 21, no. 6, pp. 1118–1130, Jun. 2013, doi: 10.1038/mt.2013.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Cole G. et al. , “DNA vaccination via RALA nanoparticles in a microneedle delivery system induces a potent immune response against the endogenous prostate cancer stem cell antigen,” Acta Biomater., vol. 96, pp. 480–490, Sep. 2019, doi: 10.1016/j.actbio.2019.07.003. [DOI] [PubMed] [Google Scholar]
- [75].McCrudden C. M. et al. , “Gene therapy with RALA/iNOS composite nanoparticles significantly enhances survival in a model of metastatic prostate cancer,” Cancer Nanotechnol., vol. 9, no. 1, p. 5, Jun. 2018, doi: 10.1186/s12645-018-0040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Brunger J. M., Zutshi A., Willard V. P., Gersbach C. A., and Guilak F., “Genome Engineering of Stem Cells for Autonomously Regulated, Closed-Loop Delivery of Biologic Drugs,” Stem Cell Rep., vol. 8, no. 5, pp. 1202–1213, May 2017, doi: 10.1016/j.stemcr.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Chu V. T. et al. , “Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells,” Nat. Biotechnol., vol. 33, no. 5, pp. 543–548, May 2015, doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- [78].Shao S. et al. , “Enhancing CRISPR/Cas9-mediated homology-directed repair in mammalian cells by expressing Saccharomyces cerevisiae Rad52,” Int. J. Biochem. Cell Biol., vol. 92, pp. 43–52, Nov. 2017, doi: 10.1016/j.biocel.2017.09.012. [DOI] [PubMed] [Google Scholar]
- [79].Rouches M. V., Xu Y., Cortes L. B. G., and Lambert G., “A plasmid system with tunable copy number,” Nat. Commun., vol. 13, no. 1, p. 3908, Jul. 2022, doi: 10.1038/s41467-022-31422-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Saleh-Gohari N. and Helleday T., “Conservative homologous recombination preferentially repairs DNA double-strand breaks in the S phase of the cell cycle in human cells,” Nucleic Acids Res., vol. 32, no. 12, pp. 3683–3688, Jun. 2004, doi: 10.1093/nar/gkh703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Tran N. T. et al. , “Efficient CRISPR/Cas9-Mediated Gene Knockin in Mouse Hematopoietic Stem and Progenitor Cells,” Cell Rep., vol. 28, no. 13, pp. 3510–3522.e5, Sep. 2019, doi: 10.1016/j.celrep.2019.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].McErlean E. M. et al. , “Rational design and characterisation of an amphipathic cell penetrating peptide for non-viral gene delivery,” Int. J. Pharm., vol. 596, p. 120223, Mar. 2021, doi: 10.1016/j.ijpharm.2021.120223. [DOI] [PubMed] [Google Scholar]
- [83].Yamada Y., Onda T., Hamada K., Kikkawa Y., and Nomizu M., “Effect of Amino Acid Substitution on Cell Adhesion Properties of Octa-arginine,” Biol. Pharm. Bull., vol. 45, no. 10, pp. 1537–1543, 2022, doi: 10.1248/bpb.b22-00430. [DOI] [PubMed] [Google Scholar]
- [84].Diener Y. et al. , “RNA-based, transient modulation of gene expression in human haematopoietic stem and progenitor cells,” Sci. Rep., vol. 5, no. 1, p. 17184, Nov. 2015, doi: 10.1038/srep17184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zambrowicz B. P., Imamoto A., Fiering S., Herzenberg L. A., Kerr W. G., and Soriano P., “Disruption of overlapping transcripts in the ROSA βgeo 26 gene trap strain leads to widespread expression of β-galactosidase in mouse embryos and hematopoietic cells,” Proc. Natl. Acad. Sci., vol. 94, no. 8, pp. 3789–3794, Apr. 1997, doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Peterka M. et al. , “Harnessing DSB repair to promote efficient homology-dependent and -independent prime editing,” Nat. Commun., vol. 13, no. 1, p. 1240, Mar. 2022, doi: 10.1038/s41467-022-28771-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Spisak S. et al. , “Identifying regulators of aberrant stem cell and differentiation activity in colorectal cancer using a dual endogenous reporter system,” Nat. Commun., vol. 15, no. 1, p. 2230, Mar. 2024, doi: 10.1038/s41467-024-46285-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Sheridan C., “The world’s first CRISPR therapy is approved: who will receive it?,” Nat. Biotechnol., vol. 42, no. 1, pp. 3–4, Nov. 2023, doi: 10.1038/d41587-023-00016-6. [DOI] [PubMed] [Google Scholar]
- [89].Chakraborty S., Ji H., Kabadi A. M., Gersbach C. A., Christoforou N., and Leong K. W., “A CRISPR/Cas9-Based System for Reprogramming Cell Lineage Specification,” Stem Cell Rep., vol. 3, no. 6, pp. 940–947, Oct. 2014, doi: 10.1016/j.stemcr.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Truong V. A. et al. , “CRISPRai for Simultaneous Gene Activation and Inhibition to Promote Stem Cell Chondrogenesis and Calvarial Bone Regeneration,” Nucleic Acids Res., vol. 47, no. 13, p. e74, Jul. 2019, doi: 10.1093/nar/gkz267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Leppek K. et al. , “Combinatorial optimization of mRNA structure, stability, and translation for RNA-based therapeutics,” Nat. Commun., vol. 13, no. 1, p. 1536, Mar. 2022, doi: 10.1038/s41467-022-28776-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Gong J., Tang D., and Leong K., “CRISPR/dCas9-mediated cell differentiation,” Curr. Opin. Biomed. Eng., vol. 7, pp. 9–15, Sep. 2018, doi: 10.1016/j.cobme.2018.08.005. [DOI] [Google Scholar]
- [93].Lückgen J., Raqué E., Reiner T., Diederichs S., and Richter W., “NFκB inhibition to lift the mechano-competence of mesenchymal stromal cell-derived neocartilage toward articular chondrocyte levels,” Stem Cell Res. Ther., vol. 13, no. 1, p. 168, Apr. 2022, doi: 10.1186/s13287-022-02843-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Wang Y.-W., Lin W.-Y., Wu F.-J., and Luo C.-W., “Unveiling the transcriptomic landscape and the potential antagonist feedback mechanisms of TGF-β superfamily signaling module in bone and osteoporosis,” Cell Commun. Signal., vol. 20, no. 1, p. 190, Nov. 2022, doi: 10.1186/s12964-022-01002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Dai L. et al. , “Different tenogenic differentiation capacities of different mesenchymal stem cells in the presence of BMP-12,” J. Transl. Med., vol. 13, no. 1, p. 200, Jun. 2015, doi: 10.1186/s12967-015-0560-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Chavez A. et al. , “Highly Efficient Cas9-Mediated Transcriptional Programming,” Nat. Methods, vol. 12, no. 4, pp. 326–328, Apr. 2015, doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Konermann S. et al. , “Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex,” Nature, vol. 517, no. 7536, pp. 583–588, Jan. 2015, doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Zhong C. et al. , “Scaffold-based non-viral CRISPR delivery platform for efficient and prolonged gene activation to accelerate tissue regeneration,” Acta Biomater., vol. 173, pp. 283–297, Jan. 2024, doi: 10.1016/j.actbio.2023.10.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.