Abstract
Different subgroups of feline leukemia virus (FeLV) use different host cell receptors for entry. Subgroup A FeLV (FeLV-A) is the virus that is transmitted from cat to cat, suggesting that cells expressing the FeLV-A receptor are important targets at the earliest stages of infection. FeLV-B evolves from FeLV-A in the infected cat through acquisition of cellular sequences that are related to the FeLV envelope gene. FeLV-Bs have been shown to infect cells using the Pit1 receptor, and some variants can infect cells at a lower efficiency using Pit2. Because these observations were made using receptor proteins of human or rodent origin, the role that Pit1 and Pit2 may play in FeLV-B replication in the cat is unclear. In this study, the feline Pit receptors were cloned and tested for their ability to act as receptors for different FeLV-Bs. Some FeLV-Bs infected cells expressing feline Pit2 and feline Pit1 with equal high efficiency. Variable region A (VRA) in the putative receptor-binding domain (RBD) was a critical determinant for both feline Pit1 and feline Pit2 binding, although other domains in the RBD appear to influence how efficiently the FeLV-B surface unit can bind to feline Pit2 and promote entry via this receptor. An arginine residue at position 73 in VRA was found to be important for envelope binding to feline Pit2 but not feline Pit1. Interestingly, this arginine is not found in endogenous FeLV sequences or in recombinant viruses recovered from feline cells infected with FeLV-A. Thus, while FeLV-Bs that are able to use feline Pit2 can evolve by recombination with endogenous sequences, a subsequent point mutation during reverse transcription may be needed to generate a virus that can efficiently enter the cells using the feline Pit2 as its receptor. These studies suggest that cells expressing the feline Pit2 protein are likely to be targets for FeLV-B infection in the cat.
Feline leukemia virus (FeLV) was originally categorized into three subgroups (FeLV-A, -B, and -C) on the basis of superinfection interference analyses (46, 47). FeLV-A infects most feline cell types, but it is poorly infectious for cells from other species; FeLV-B and -C infect a wide range of nonfeline cell types, although the host cell specificities of FeLV-B and FeLV-C are distinct (16, 46, 48). In addition, T-cell-tropic FeLVs (FeLV-T) have been identified, and these viruses have unique interference properties (30). From these interference and tropism studies, it has been inferred that FeLV-A, -B, -C, and -T each use distinct cellular receptor molecules to initiate infection of the host cell. FeLV-Bs have been shown to use Pit1 (59), which encodes a multiple membrane-spanning phosphate transporter, as a receptor (19, 20, 33, 34, 59). FeLV-T also uses Pit1 for infection, but this class of FeLVs require a cofactor, termed FeLIX, in addition to Pit1 (2). The receptor for FeLV-C has recently been identified, and it is also predicted to encode a multiple membrane-spanning protein with homology to the major facilitator superfamily of proteins (41, 58). The receptor for FeLV-A has not yet been defined.
FeLV-A is found in all infected cats, and it is thought to be the transmissible form of FeLV (14, 15, 17). In contrast, FeLV-B is found with FeLV-A in some but not all chronically infected cats, and it is very poorly transmitted even at high doses (17, 18, 40). Both FeLV-B and FeLV-T have been shown to evolve directly from FeLV-A in infected cats (5, 8, 37, 39, 42, 44, 50). It is possible that these viruses may have a selective advantage for replication once they emerge in an FeLV-A-infected cat because they can circumvent viral interference against the progenitor FeLV-A. In addition, such viruses may also infect an entirely new population of cells that do not express the FeLV-A receptor but do express Pit1.
Subgroup B FeLVs evolve by recombination with portions of endogenous FeLV-like envelope sequences, which have a high degree (≈80%) of homology to FeLV-A (5, 11, 36, 52–54). There are multiple copies of FeLV-related endogenous sequences (enFeLV) in the feline genome, which are transcribed and translated but do not generate infectious virus (3, 22, 27). Because RNA is expressed from enFeLV sequences, enFeLV sequences can recombine with the related infectious viral genome when the two are copackaged. When feline cells are infected with FeLV-A, two major recombinant forms evolve: one in the which the surface unit (SU) of the extracellular envelope glycoprotein is encoded almost entirely by endogenous FeLV sequences, and one in which the N-terminal half encompassing the putative receptor-binding domain (RBD) is encoded by endogenous FeLV sequences, but portions of the C-terminal half are derived from the original FeLV-A parent virus (36).
An FeLV-B in which almost all of the SU coding sequences were acquired from enFeLV was derived from a cat infected with FeLV-A (5). This variant (FeLV-B-90Z) was shown to enter cells using human Pit1 (HuPit1) but not human Pit2 (HuPit2 [4]). Pit2, which is a related phosphate transporter protein that has ≈60% homology with Pit1, functions as the receptor for amphotropic murine leukemia virus (MuLV) (29, 61). When chimeric envelope genes were engineered between FeLV-B-90Z and FeLV-A sequences, some chimeric envelope protein had acquired the ability to recognize HuPit2 as well as HuPit1 as a receptor (4). This suggested that some recombination events that lead to the genesis of FeLV-Bs might facilitate the evolution of viruses with dual Pit1 and Pit2 receptor specificity.
Studies of FeLV-B receptor specificity performed to date have taken advantage of clones of the Pit molecules isolated from nonfeline species. Thus, it is unknown whether any FeLV-Bs can infect cells using feline Pit2. In this study, we cloned the feline Pit cDNAs and examined their ability to function as receptors for a variety of FeLV-B chimeras and mutants. We found that an FeLV-B that had evolved in an FeLV-A-infected cat had acquired dual receptor specificity for both feline Pit1 (FePit1) and feline Pit2 (FePit2). Although this virus could not enter cells using HuPit2, it efficiently infected cells expressing FePit2. A virus that evolved from FeLV-A in feline cells in culture was also shown to use FePit2. This virus recognized FePit2 less efficiently than FePit1. A mutation in the first variable region (VRA) of the envelope was shown to be important in determining whether FeLV-Bs can infect cells as efficiently using FePit2 as using FePit1.
MATERIALS AND METHODS
Cloning of feline Pit proteins.
The FePit2 cDNA was amplified by reverse transcription (RT)-PCR from total RNA that was prepared from a feline fibroblast cell line (AH927) using the Qiagen RNeasy kit. First-strand cDNA synthesis was performed using SuperScriptII RT and RNase H as recommended in a protocol supplied by the manufacturer (Gibco Life Technologies). The cDNA was then used as the template for PCR using various combinations of primers designed to known Pit2 sequences, including those from rat (29), hamster (E36 [62] and CHO-K1 [7]), and human (12). The primer set that resulted in product of the predicted size (≈2 kb) included a 5′ primer that was designed to correspond to the rat Pit2 start codon (5′-ATGGCCATCGATGGGTATCTGTGGATGGTC, where the start codon is italic) and a 3′ primer that corresponds to sequences spanning the stop codon of human Pit 2 (5′-TCACACATATGGAAGGATCCCATAC, where the stop codon is italic). PCR was performed using 0.5 μl of the TaqPlus Precision PCR System (Stratagene), along with 0.125 mM each of the deoxynucleoside triphosphates and 2 ng of each primer in a 50-μl reaction. The ≈2-kb PCR product was separated by gel electrophoresis through agarose, purified from the gel matrix using standard methods (Qiaex II; Qiagen), and introduced into the pCR2.1-TOPO vector (Invitrogen). The FePit2 cDNA was then subcloned into the pLXSN retroviral vector (28) following digestion of both plasmids with EcoRI, resulting in the construct pL(FePit2)SN.
FePit1 was isolated from a feline T-cell (3201) cDNA Lambda ZAP-express library using a Pit-specific probe. The sequence of the FePit1 clone was found to be identical to that of the FePit1 clone described previously (45). Excision of the FePit1 lambda clone created a phagemid in the pBK vector. FePit1 was then introduced into the retroviral vector pLXSN by first linearizing FePit1-pBK using a SacI site in the pBK vector and then modifying the digested DNA with the Klenow fragment to create blunt ends. The linear DNA was digested to use an XhoI site in pBK, and a 3.4-kb fragment was purified from an agarose gel. This FePit1 fragment was ligated to pLXSN that had been digested with both XhoI and HpaI to produce pL(FePit1)SN.
Cell lines.
All cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 100 U of penicillin per ml, 100 μg of streptomycin per ml, 0.25 mg of amphotericin fungicide per ml, and 2 mM l-glutamine (complete DMEM). MDTF (24), MDTF-HuPit1 (62), and MDTF-HuPit2 (10) cells have been described previously.
FePit1 and FePit2 were introduced into MDTF cells by transduction. For this purpose, viral particles packaging L(FePit1)SN or L(FePit2)SN were generated in 293T cells using transient transfection (CaPO4; Stratagene). Three plasmids were transfected into 293T cells: 10 μg of amphotropic MuLV envelope (SV-A-MLV-env [23]), 10 μg of a construct encoding FeLV gag and pol (61E-LTR-ΔΨgag-pol [55]), and 10 μg of either pL(FePit1)SN or pL(FePit2)SN. The following day the cells were washed three times in 5 ml of phosphate-buffered saline (PBS) and fed with 7 ml of fresh complete DMEM. Two days posttransfection, supernatant was collected and filtered through a 0.22-μm filter, and 1 to 2 ml was used to infect MDTF cells that had been seeded the day before at 2 × 105 cells per 10-cm dish. At the time of infection, 4 μg of Polybrene per ml was included, and the final volume was adjusted to 10 ml of medium. The following day the cultures were placed under G418 selection by the addition of 0.6 mg of geneticin per ml (Gibco-BRL) to the medium. Selection continued for approximately 10 days, during which time the medium was replaced every 2 to 3 days. The MDTF-FePit cell lines used here represent pools of G418-resistant cell clones.
Viruses.
Viruses bearing different envelope proteins were generated as described previously, as were several of the FeLV-B envelope expression constructs and packaging system used in this study, which include plasmids expressing FeLV-B: 90Z, 90ZRBD, GA, GARBD, and GARBD-73Q→R envelopes (55). The FeLV-B-90ZVRA and envelope construct was made by the same method using the XhoI and AocI cloning sites in EE(Z2)E (4), such that they contain the complete envelope-open reading frame (ORF) downstream of the cytomegalovirus (CMV) promoter. A plasmid expressing envelope of the SEATO molecular clone of the gibbon ape leukemia virus (GALV-SEATO) has been described previously by Eiden et al. (CIGASenv [60]). Viral particles were made by cotransfecting 293T cells with 10 μg of 61E-LTR-ΔΨgag-pol and 10 μg of FeLV-B or GALV envelope expression plasmids, all of which lack packaging sequences. An MuLV-derived retroviral vector genome that contains a packaging signal and expresses β-galactosidase (pRT43.2Tnlsβgal-1 [60]) was contransfected with the gag-pol and envelope constructs to produce FeLV particles carrying this reporter viral vector genome. The day following transfection, the cells were gently washed three times in PBS, and then 7 ml of complete DMEM was added. The next day, viral supernatant was collected and filtered though a 0.22-μm filter, aliquoted, and stored at −70°C. To examine the infectivity of these particles in a single cycle of infection, recipient cells were seeded onto 24-well dishes at 2 × 104 cells per well. The following day, dilutions of viral supernatants were applied in a 1-ml total volume in the presence of 4 μg of Polybrene per ml.. Approximately 48 h following infection, cells were stained for β-galactosidase activity, and foci of infected cells were scored by visual inspection as described previously (21).
Analyses of FeLV-B variants that arise in infected cells in culture.
The cells from which recombinant FeLV-B variants were isolated were described previously (36). These cells represent feline fibroblast cells that were transfected with a plasmid encoding the FeLV-A-61E proviral genome. In multiple independent transfections, recombinant FeLV-B-like genomes had been detected in feline fibroblast cells (36). Cell-free viral supernatants were collected from a confluent culture of these cell cultures and filtered through a 0.22-μm filter, and then 2 ml was used to infect either MDTF-FePit1, MDTF-FePit2, or MDTF cells that had been seeded the previous day at 2 × 105 per 6-cm dish. These cells were passaged for 10 days and tested for production of FeLV p27gag by enzyme-linked immunosorbent assay (Synbiotics). Cell lines that were positive for FeLV p27gag were lysed (31), and FeLV envelope sequences were amplified with TaqPlus Precision PCR System (Stratagene) using primers and methods described elsewhere (FeLV-pol-1, FeLV-U32-B [43]). A 2.4-kb band was gel purified and cloned into the pcDNA3.1/V5/His vector (Invitrogen), which includes a CMV promoter upstream of the cloning site. Viruses containing these recombinant envelopes were generated as described above by contransfecting 293T cells with the envelope clone, 61E-LTR-ΔΨgag-pol, and the MuLV-based vector genome encoding β-galactosidase.
RESULTS
Comparison of feline and human Pit sequences.
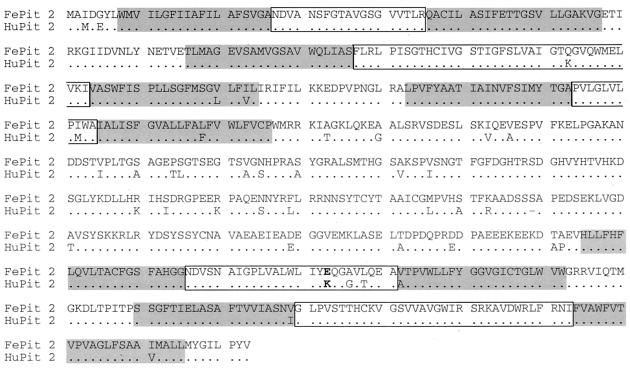
The sequence of FePit1 was first reported by Rudra-Ganguly et al. (45), and this sequence was shown to be 93% identical to HuPit1 at the amino acid level. The clone used in the studies described here was identical in sequence to this previously published FePit1 cDNA. FePit2 (GenBank AF394194), which has not been described previously, showed at the amino acid level approximately 53% identity with FePit1 (data not shown) and 93.7% identity with the HuPit2 protein (Fig. 1). Many of the differences between FePit2 and HuPit2 were within the region that is thought to form a large intracellular domain between the third and fourth extracellular loops (9, 19, 61). Sequences predicted to form the first, second, and fifth extracellular domains were identical between the feline and human proteins. There was a conservative amino acid difference in the predicted extracellular loop three. In loop four, which has been shown to be a critical domain for GALV and FeLV-B interactions (10, 38, 57), there were three differences between the feline and human Pit2 proteins. The most notable difference in FePit2 is a glutamic acid at position 522, where there is a lysine in HuPit2, because this residue has been shown be important for GALV-Pit interactions (6, 10). Specifically, the presence of an uncharged or negatively charged residue at this position has been shown to be important for GALV infections.
FIG. 1.
Feline Pit2 sequence. The predicted amino acid sequence of FePit2 is shown in single amino acid code. The sequence of HuPit2 is shown below for comparison, with a dot to indicate conserved residues. The putative transmembrane regions are indicated with a gray box, and an open box surrounds the proposed extracellular domains (9, 33, 61). Position 522, which has been shown to be important in determining HuPit2 receptor activity, is indicated in bold (10).
FeLV-B can efficiently infect cells using FePit2.
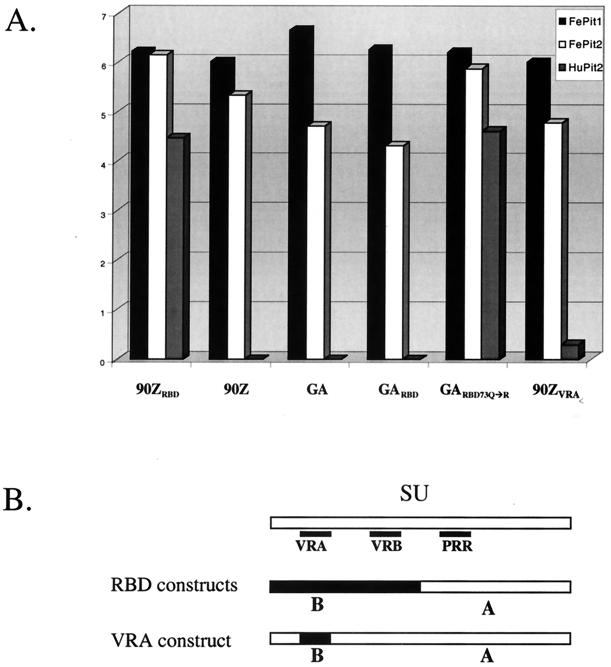
We previously showed that FeLV-B variants differ in their ability to use the HuPit2 protein as a receptor (4, 55). Using a panel of chimeric envelope proteins, we showed that HuPit2 receptor specificity is determined by sequences in the RBD as well as by C-terminal sequences in SU (4, 55). This collection of constructs, which encode SU proteins that differ in the RBD and/or the C-terminal domain, were used to generate viruses that were otherwise identical in containing FeLV-A Gag, polymerase, and envelope transmembrane proteins. We used FeLV-A as the source for the structural and enzymatic proteins because FeLV-B evolves from FeLV-A and differs from it primarily in the sequences encoding SU (36). Each of the viruses with the FeLV-B SU infected cells using FePit1, similar to what has been observed for HuPit1 (4, 55) (Fig. 2 and data not shown). Infectious titers of ≈106/ml were observed when the FeLV-Bs were used to infect MDTF cells engineered to express FePit1 (see Fig. 2, 4, and 6), whereas these viruses did not infect the parental MDTF cell line (data not shown). Surprisingly, we found that the FePit2 receptor allowed at least some infection by all of the FeLV-B variants and chimeras tested, including viruses that could not infect cells using HuPit2. For example, the FeLV-B variant 90Z cannot infect cells using HuPit2 (4) (Fig. 2), yet this virus can infect cells expressing FePit2 and FePit1 with high efficiency. The Gardner-Arnstein (FeLV-B-GA [32]) variant also cannot use HuPit2 as a receptor (55) (Fig. 2), but it could infect MDTF cells expressing FePit2, albeit at ≈100-fold-reduced efficiency relative to FePit1. We also examined an engineered mutant of FeLV-B-GA SU, which encodes a glutamine-to-arginine change at position 73 in VRA. This change confers on the virus the ability to bind to HuPit2 cells, although with low efficiency, and also infect cells using HuPit2 at a level about 20-fold lower than infection levels of HuPit1-expressing cells (55). This virus, FeLV-B-GARBD-73Q→R, was able to efficiently utilize the FePit2 receptor for entry (Fig. 2).
FIG. 2.
Infection of cells expressing Pit1 or Pit2 by viruses pseudotyped with various FeLV-B SUs. (A) Single-cycle infection assay that results in the transduction of a vector genome encoding β-galactosidase. The x axis indicates the envelope-SU of the infecting pseudotyped virus, and the y axis indicates the focus-forming units (FFU) per milliliter of viral supernatant as determined by β-galactosidase activity in a log scale. Within the graph, the shading of the bar depicts the target cells for infection. As shown in the key at the top of the graph, solid black bars, white bars, and gray bars indicate MDTF-FePit1s, MDTF-FePit2s, and MDTF-HuPit2s, respectively. Naive MDTF cells were also exposed to virus in parallel, and no background was observed (55) (data not shown), except in the case of FeLV-B-90ZRBD, which gave an average of 20 FFU/ml in five replicate experiments using the same virus stocks. MDTFs expressing HuPit1 were also infected in parallel, and the results were always similar to those obtained with MDTF-FePit1 cells (55) (data not shown). (B) Schematic of the chimeric FeLV envelope-SUs tested in panel A. The small black boxes show the relative positions of the VRA, VRB, and PRR regions in the SU. The schematic depicting the RBD constructs (GARBD, GARBD-73Q→R and 90ZRBD) illustrates that these pseudotyped viral SUs contain regions derived from FeLV-B (in black) that include the VRA and VRB regions, while the rest of the SU, including the PRR, is supplied by FeLV-A (in white). In the VRA construct, only the VRA region is derived from FeLV-B, while the remainder of the SU is FeLV-A derived.
FIG. 4.
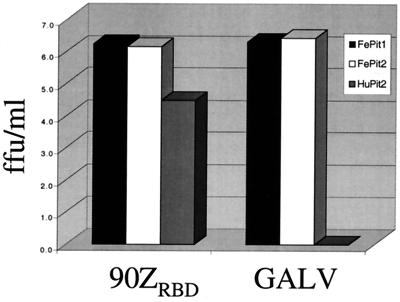
Infection of MDTF-FePit2 cells by GALV. The layout for infection studies is as described in the legend for Fig. 2A, where the targets cells are indicated in the upper right-hand corner and the viral pseudotypes used for infection are indicated on the x axis.
FIG. 6.
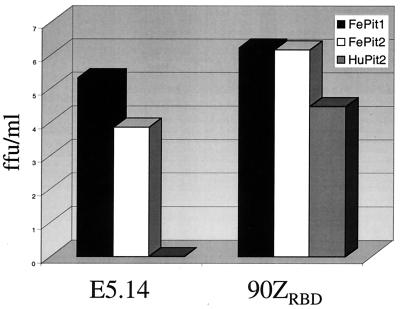
Single-cycle infection assay with FeLV-B-E5.14. The layout for this figure is as described in the legend for Fig. 2A. The x axis indicates the envelope-SU of the virus used in the infection, and the y axis indicates the FFU per milliliter of viral supernatant. Viruses pseudotyped with either FeLV-B-90ZRBD or FeLV-B-E5.14, a representative cloned virus, were used to infect the cells indicated in the right corner of the figure, which shows the corresponding shading used in the bar graph.
FeLV-B VRA has been shown to be both necessary and sufficient to determine infection of cells using Pit1 when placed in a background of FeLV-A (4), although not when it is placed in the context of murine leukemia virus (56). A virus with an SU that includes just VRA of FeLV-B-90Z, including an arginine at position 73 [FeLV-B-90ZVRA; called EE(Z2)E in reference 4] with the remainder being of FeLV-A origin, infected cells using FePit2 ≈20 less efficiently than it infected cells using FePit1 (Fig. 2). This virus infected cells expressing FePit2 about 10,000 times better than cells expressing HuPit2. Collectively, our data suggest that the VRA region, particularly an arginine at position 73 in VRA, of the RBD is important for FeLV binding to FePit2. Other domains in the RBD of SU are also important for interaction with the FePit2 receptor, but sequences in the C terminus of SU that affect HuPit2 receptor specificity do not impair FePit2 recognition. This is demonstrated by FeLV-B-90Z, which can readily infect cells via FePit2 but not HuPit2 (Fig. 2). Thus, the FePit2 receptor may serve as a receptor for many FeLV-B variants, albeit with variable efficiency.
Differences in FeLV-B infection are determined at the level of virus binding to feline Pit receptors.
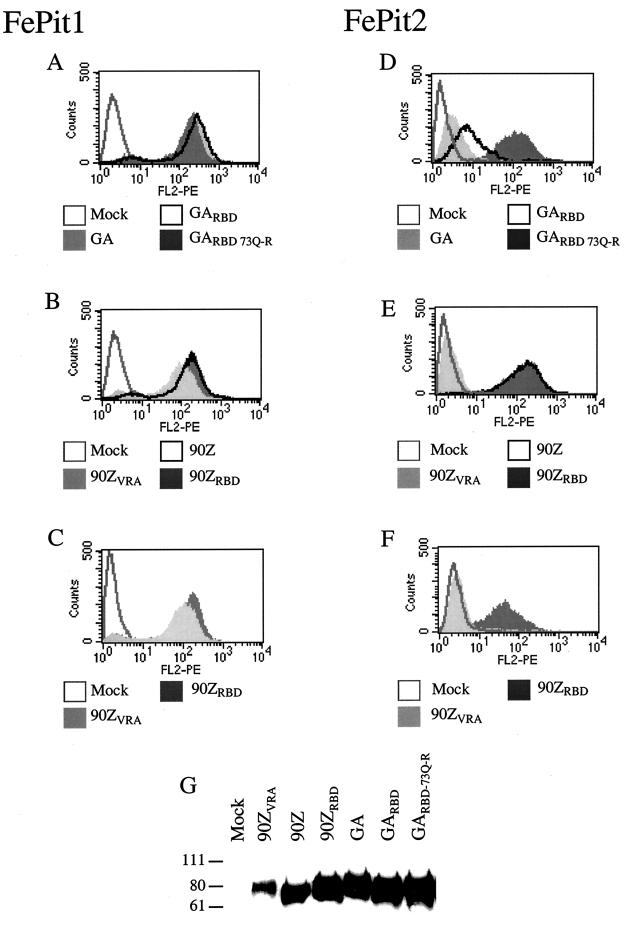
Because there were dramatic differences in the ability of some FeLV-B envelopes to infect cells using FePit1 and FePit2, we asked whether these variations reflected differences in viral binding to these receptors. In order to generate supernatants containing FeLV SUs for use in binding studies, we used constructs to express the FeLV SUs that encoded the signal peptide but lacked the last 10 C-terminal amino acids of SU and the entire TM domain. These SUs, which had a C-terminal hemagglutinin (HA) epitope tag (25) were secreted from the cell. All of the FeLV-B SUs studied here, including the chimera with the FeLV-B VRA in a background of FeLV-A SU, bound to MDTF-FePit1 cells with relatively the same efficiency (Fig. 3A and B). The levels of SU in the cell supernatants used for binding were approximately 10-fold lower for the FeLV-B-90ZVRA SU (Fig. 3G), and so we also performed binding studies with 10-fold less FeLV-B-90ZRBD supernatant than FeLV-B-90ZVRA SUs. We observed a similar shift in fluorescence for these two proteins (Fig. 3C), suggesting that the VRA from FeLV-B SU is sufficient for binding to FePit1 in a background of FeLV-A SU. FeLV-A SU does not bind to either MDTF-FePit1 or MDTF-HuPit1 cells (data not shown).
FIG. 3.
Binding of FeLV-B SU to MDTF cells expressing either FePit1 or FePit2. Panels A to F show overlays of flow cytometry data of FeLV SU-HA supernatants that had been bound to either MDTF-FePit1 cells (panels A, B, and C) or MDTF-FePit2 cells (D, E, and F). Binding was detected using an HA monoclonal antibody. The x axis is fluorescence intensity (log scale), and the y axis is cell number. In panels A-F, the legend below each histogram indicates which FeLV SU-HA was used in the binding assay. In panels A to F, mock represents cells incubated with medium only. In panels A, B, D, and E, 1 ml of cell supernatant was used in the binding experiment. In panels C and F, 1 ml of FeLV-B-90ZVRA and 0.1 ml of FeLV-B-90ZRBD supernatant were used in order to compare more similar levels of protein (see panel G). Panel G is a Western blot that was performed with the supernatants used in the binding assays shown above. Equal amounts of supernatant were used. The sizes of markers (in kilodaltons) are indicated to the left of the blot. The methods used in the immunoprecipitation of the supernatants and Western blot procedure have been described previously (25).
We could also detect binding of the FeLV-B SUs to MDTF-FePit2 cells. However, the binding, as measured by a shift in fluorescence intensity, varied for the different SU proteins. In general, the relative shifts in fluorescence corresponded with the relative infectivity of viruses pseudotyped with these envelope proteins. For example, FeLV-B-90Z and FeLV-B-90ZRBD, which both efficiently infect cells expressing FePit2, bound efficiently to these cells (Fig. 3E). Consistent with the infectivity data, all of the SUs that bound with the highest efficiency encoded an arginine at position 73 (FeLV-B-90Z, FeLV-B-90ZRBD, and FeLV-B-GARBD-73Q→R). In particular, there was better binding with FeLV-B-GARBD-73Q→R SU than with FeLV-B-GARBD or FeLV-B-GA SU (Fig. 3D). This indicates that the residue at position 73 plays a key role in FeLV-B binding to FePit2. However, binding of the chimera encoding the FeLV-B-90ZVRA to cells expressing FePit2 was undetectable using flow cytometry, despite the inclusion of the key arginine at position 73. This may be due in part to the lower levels of FeLV-B-90ZVRA SU present in the supernatant, as shown by Western blot analysis (Fig. 3G). In addition, the level of infection of MDTF-FePit2 cells with viruses pseudotyped with FeLV-B-90ZVRA was reduced about 20-fold in comparison to FeLV-B-90ZRBD. Because we are able to detect infection but not binding, we presume that the methods used for binding are not as sensitive as infection assays, where single events can be quantified.
GALV can infect cells expressing FePit2.
GALV, like FeLV-B-90Z, can infect cells using HuPit1 but not HuPit2 (10, 33). Because FeLV-B-90Z can use FePit2 as a receptor, we asked whether GALV could also use the feline ortholog of Pit2 as its receptor. Viral particles of FeLV-gag-pol were pseudotyped with the GALV-SEATO envelope (60) and used to infect MDTF-FePit2 cells. The infectivity of GALV was indistinguishable on cells expressing FePit2 and FePit1 (Fig. 4) as well as HuPit1 (data not shown). This was in contrast to the results of exposing GALV to MDTF-HuPit2 cells, which were nonpermissive to GALV infection, as expected. Thus, FePit2 functions as a receptor for GALV.
Virus evolution studies.
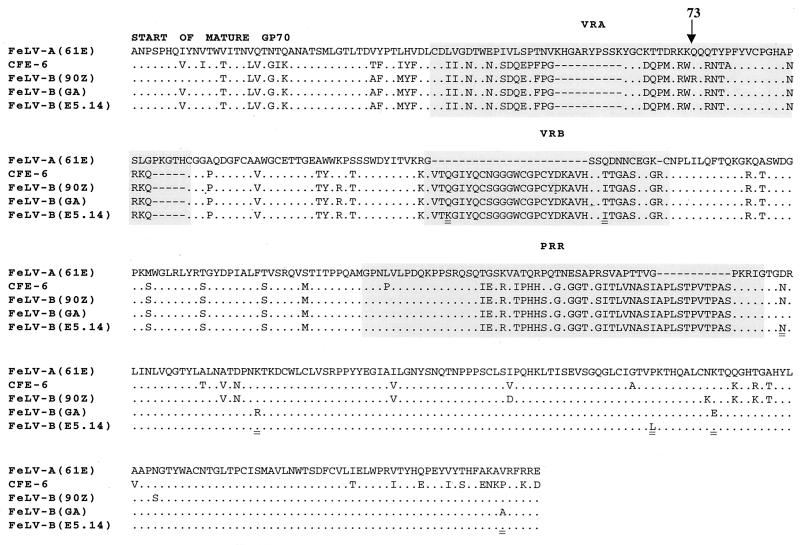
FeLV-B evolves from FeLV-A by recombination with endogenous FeLV-like sequences, but little is known about the receptor specificity of these evolved variants. As described above, all the FeLV-B variants studied efficiently infect cells using FePit1, but they vary in the efficiency with which they infect cells using FePit2. We previously demonstrated the evolution of FeLV-B variants directly from FeLV-A in feline fibroblast cells transfected with an FeLV-A molecular clone, 61E (36). To examine whether these recombinant envelopes could recognize FePit2 as a receptor, viruses from five cell cultures transfected with 61E were used to infect MDTF cells expressing FePit2 or FePit1. Four of the viruses established a productive infection in MDTF-FePit1 cells, but we were only able to transfer detectable FeLV to MDTF-FePit2 cells from one of five feline fibroblast cultures. We analyzed the sequence of the envelope variants that infected MDTF-FePit2 cells and found that all of the variants were very similar in recombinant structure and sequence, suggesting that they likely represent progeny of the same recombinant virus (Fig. 5). Interestingly, these viruses encoded enFeLV-like sequences in the N-terminal two thirds of their SU, including the VRA and VRB RBDs as well as sequences encompassing the proline-rich region. The putative recombination junction in the SU coding region of these culture-derived variants is much like the Gardner-Arnstein FeLV-B clone, which was derived from an isolate from a naturally infected cat (13). The predicted amino acid sequence of this tissue culture-adapted virus differs from that of FeLV-B-GA at seven positions in SU, five of which are found in the FeLV-B-90Z clone (Fig. 5).
FIG. 5.
Amino acid alignment of envelope-SU of FeLV. The predicted amino acid sequences of the mature SU of FeLV-A-61E (35), FeLV-B-90Z (5), FeLV-B-GA (63), and the endogenous FeLV clone CFE-6 (22) are shown. Included is an FeLV-B E5.14 clone that was obtained from MDTF-FePit2 cells infected with supernatant from feline fibroblasts that had been transfected with FeLV-A-61E. FeLV-A-61E is shown as the reference sequence, and for the other SU proteins only amino acids that differ are given. Amino acid residues are shown in single-letter code, and conserved residues are indicated with a dot. A gray box surrounds the VRA, the VRB, and the PRR. Residues of the representative cloned virus FeLV-B-E5.14 (GenBank accession no. AF403716) are double underlined at positions that are divergent between this clone and FeLV-B-GA.
The envelope sequence analyzed above was cloned into an expression plasmid with a CMV promoter 5′ of the inserted envelope clones. To test the receptor specificity of this clone, we produced virus by cotransfection of 293T cells with an FeLV gag-pol construct and a retroviral vector genome encoding β-galactosidase, as described above. We found that the virus expressing the envelope isolated from MDTF-FePit2 cells, FeLV-B-E5.14, could infect cells using either FePit1 or FePit2 (Fig. 6). For viruses containing this envelope, infectivity was 20-fold lower in cells expressing FePit2 than in cells expressing FePit1, which is similar to what we observed for the closely related FeLV-B-GA SU. This shows that FeLV-Bs that use FePit2 as a receptor may evolve from FeLV-A. Additional studies in a variety of cell lines will be needed to determine how commonly such variants arise.
DISCUSSION
FeLV-A is the form of the virus that is spread from cat to cat, suggesting that cells expressing the FeLV-A receptor are important in transmission and/or early virus amplification in the host. FeLV-B evolves from FeLV-A in the infected cat by recombination with endogenous FeLV sequences (5, 11, 36, 52–54). The structure and sequence of variants that emerge from FeLV-A vary both among natural FeLV-B isolates and in viruses that evolve during replication in feline cells in culture (5, 8, 36, 40, 44, 50, 51). This raises the question of whether the specific sequence and structure of some of the FeLV-B variants that emerge over the course of infection confer a particular selective advantage for their replication. Here we show that FeLV-B can infect cells using both FePit1 and FePit2 receptors. A specific arginine residue at position 73 appears to play a key role in determining the efficiency with which particular FeLV-B variants infect cells using FePit2. Together these data suggest that cells expressing FePit2 may be target cells for virus replication in the cat.
We found that the FeLV-B-90Z variant, which was obtained directly from a cat infected with FeLV-A, efficiently infected cells expressing either FePit1 or FePit2. In contrast, infection with an independently derived FeLV-B-GA was approximately100-fold lower in cells expressing FePit2 than in cells expressing FePit1. This allowed us to identify domains of FeLV-B SU that are important for FePit2 specificity. The differences in infectivity were determined by a single amino acid difference, arginine versus glutamine, at position 73 within VRA. As was shown previously for infection with HuPit2 (4), VRA is a minimal determinant for FePit2 specificity. However, VRA alone, even if it encodes arginine at position 73, is not sufficient for high-level infection using FePit2. In fact, we could not detect binding by flow cytometry with FeLV-B-90ZVRA despite the ability of virus pseudotyped with this envelope SU to infect cells expressing FePit2. The decreased sensitivity of the binding assay relative to the infection assay is similar to results obtained in studies using an ecotropic MuLV SU in which measurable binding to the cognate receptor was not observed, although infection was detected (1). Thus, the infection assay is a more sensitive method than flow cytometry for detection of envelope-receptor interactions. Sequences in VRB also appear to influence the interactions between FeLV-B-90Z and FePit2, because a virus encoding FeLV-B-90Z sequences from both VRA and VRB (FeLV-B-90ZRBD) binds and infects cells expressing FePit2 more efficiently than a virus encoding only VRA (FeLV-B-90ZVRA). These data are qualitatively similar to what we observed for infection using HuPit2 (4). However, our findings suggest that viruses encoding VRA and VRB from FeLV-Bs enter cells using the FePit2 receptor more efficiently than they can enter cells using the HuPit2 receptor.
Previous studies suggested that only some FeLV-Bs can infect cells expressing HuPit2 (4, 38). While FeLV-B 90Z cannot enter cells expressing HuPit2, a chimera encoding the N-terminal half of 90Z SU and the C-terminal half of FeLV-A SU could (FeLV-B-90ZRBD). This suggested that sequences in the C-terminal portion of FeLV SU participate in some aspect of infection via HuPit2. Our studies indicate that the C-terminal sequences of the SU domain that differ between these viruses play a role in post-binding events in cells expressing HuPit2, because FeLV-B-90Z SU binds with low but equal efficiency to HuPit2, as does FeLV-B-90ZRBD (55). In contrast, these C-terminal sequences that distinguish FeLV-B-90Z and FeLV-B-90ZRBD SU did not play a role in determining FePit2 binding and specificity. Thus, it may be possible to define the region of HuPit2 that interacts with the C terminus of SU by examining chimeric feline and human Pit2 receptors. Such studies may provide insights into the domains of SU and the receptor that participate in postbinding stages of entry, such as fusion. In this regard, it is interesting that the C-terminal domain of MuLV SU has sequences that are thought to play a role in fusion activation in a manner that is dependent on their interaction with the N terminus of SU (26). This would imply that there are complex interactions between the N and C termini of SU and specific receptor residues that lead to fusion-activation and that these may be distinct from those that permit binding.
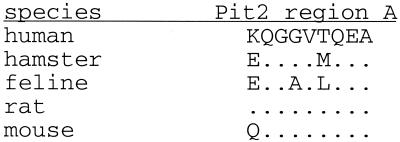
The feline and human Pit2 proteins differ at the amino acid level by 6.3%, with the majority of differences occurring in what is predicted to be a large intracellular domain. One critical difference that may determine the ability of FePit2 to function as an FeLV-B receptor is a glutamic acid in the feline protein that corresponds to a lysine at position 522 in HuPit2. This position, which is predicted to fall in the fourth extracellular loop of the Pit2 protein, has been shown to be a critical determinant for GALV infection (10). GALV, like FeLV-B-90Z, can infect cells using HuPit1 but not HuPit2 (33, 61). When the positively charged lysine at position 522 in HuPit2 is replaced by an uncharged amino acid, this protein functions as a receptor for GALV (10). Moreover, the hamster-derived Pit2 protein can function as a receptor for GALV and for FeLV-B-90Z, and this allele encodes a glutamic acid at position 522 (62) (Fig. 7). This suggests that either a neutral or negatively charged residue at position 522 may be required for infection by GALV and FeLV-B (10). Indeed, we found that GALV is able to infect cells expressing FePit2 as efficiently as it infects cells expressing either FePit1 or HuPit1 (Fig. 4 and data not shown). On the basis of this, we predict that the glutamic acid at position 522 in FePit2 protein plays a critical role in allowing efficient GALV and FeLV-B SU-receptor interactions. As discussed, we have shown that arginine at position 73 in FeLV-B SU plays a critical role in virus entry via FePit2. One model to explain this is that this positively charged residue in SU directly interacts with the negatively charged glutamic acid residue in the receptor. Our data suggest that residues in VRB could act to increase the affinity of the SU-Pit2 binding or provide additional crucial contact residues.
FIG. 7.
Alignment of a domain of the predicted fourth extracellular loop of known Pit2 receptors. Residues 522 through 530, which constitute a domain called region A, are shown for human (61), hamster (62), feline (Fig. 1), rat (29), and mouse (49) homologues of Pit2. Conserved residues are indicated with a dot.
We demonstrated that a recombinant FeLV-B that uses the FePit2 receptor evolved directly from FeLV-A in feline cells in culture. Interestingly, the recombinant that we obtained by this selection was very similar in structure to the natural isolate FeLV-B-GA (32). This may suggest that there is some selection for viruses bearing this recombinant structure, which derives VRA, VRB, and the proline-rich region from enFeLV sequences. This is consistent with our analyses of chimeric viruses, which suggest that the presence of both VRA and VRB may increase infectivity via FePit2. It is interesting that the crucial arginine at residue 73 was not found in the viruses selected from cell culture. This most likely reflects the fact that all endogenous FeLV sequences examined to date encode a glutamine at this position (22, 27). Thus, the evolution of a virus that is able to use FePit2 with high efficiency, such as the FeLV-B-90Z variant found in an infected cat, may arise by both recombination and subsequent point mutation during reverse transcription. In this study we examined only two FeLV-B clones from infected cats, one naturally infected and the other experimentally infected with FeLV-A, and one representative FeLV-B clone that arose in cells transfected with FeLV-A; all three used FePit2 as a receptor, but with variable efficiencies. Now that the FePit2 receptor has been cloned, it will be of interest to analyze a larger panel of natural FeLV-B isolates to determine the prevalence of viruses with dual Pit1 and Pit2 receptor specificity in the infected cat.
While both Pit1 and Pit2 are expressed in many tissues, there are clear differences in the level of expression of these two receptors (19, 20), suggesting that some cell types may express only one or the other of these proteins. Thus, a virus that can enter cells with high efficiency using either receptor would maximize its opportunities for successful propagation and amplification. Moreover, a virus that can use Pit2 as a receptor can superinfect a cell that was previously infected by a Pit1-specific virus. Thus, FeLV-Bs with dual receptor specificity for Pit1 and Pit2 would be predicted to be favored for amplification and selection in a persistently infected cat.
ACKNOWLEDGMENTS
We thank Maribeth Eiden for providing cell lines expressing human Pit receptors and for helpful discussions. We also thank Jenny Riddell, Cara Burns, Jim Sugai, and Sarah Boomer for technical assistance and helpful discussions.
This work was supported by Public Health Service grant CA51080 from the National Cancer Institute. Adam Lauring was supported in part by NIH training grant 2T32 CA09229.
REFERENCES
- 1.Albritton L M, Kim J W, Tseng L, Cunningham J M. Envelope-binding domain in the cationic amino acid transporter determines the host range of ecotropic murine retroviruses. J Virol. 1993;67:2091–2096. doi: 10.1128/jvi.67.4.2091-2096.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson M M, Lauring A S, Burns C C, Overbaugh J. Identification of a cellular cofactor required for infection by feline leukemia virus. Science. 2000;287:1828–1830. doi: 10.1126/science.287.5459.1828. [DOI] [PubMed] [Google Scholar]
- 3.Benveniste R E, Sherr C J, Todaro G J. Evolution of type C viral genes: origin of feline leukemia virus. Science. 1975;190:886–888. doi: 10.1126/science.52892. [DOI] [PubMed] [Google Scholar]
- 4.Boomer S, Eiden M, Burns C C, Overbaugh J. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J Virol. 1997;71:8116–8123. doi: 10.1128/jvi.71.11.8116-8123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boomer S, Gasper P, Whalen L R, Overbaugh J. Isolation of a novel subgroup B feline leukemia virus from a cat infected with FeLV-A. Virology. 1994;204:805–810. doi: 10.1006/viro.1994.1597. [DOI] [PubMed] [Google Scholar]
- 6.Chaudry G J, Eiden M V. Mutational analysis of the proposed gibbon ape leukemia virus binding site in Pit1 suggests that other regions are important for infection. J Virol. 1997;71:8078–8081. doi: 10.1128/jvi.71.10.8078-8081.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaudry G J, Farrell K B, Ting Y T, Schmitz C, Y. S L, Petropoulos C J, Eiden M V. Gibbon ape leukemia virus receptor functions of type III phosphate transporters from CHOK1 cells are disrupted by two distinct mechanisms. J Virol. 1999;73:2916–2920. doi: 10.1128/jvi.73.4.2916-2920.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen H, Bechtel M K, Shi Y, Phipps A, Mathes L E, Hayes K A, Roy-Burman P. Pathogenicity induced by feline leukemia virus, Rickard strain, subgroup A plasmid DNA (pFRA) J Virol. 1998;72:7048–7056. doi: 10.1128/jvi.72.9.7048-7056.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chien M L, Foster J L, Douglas J L, Garcia J V. The amphotropic murine leukemia virus receptor gene encodes a 71-kilodalton protein that is induced by phosphate depletion. J Virol. 1997;71:4564–4570. doi: 10.1128/jvi.71.6.4564-4570.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eiden M V, Farrell K B, Wilson C A. Substitution of a single amino acid residue is sufficient to allow the human amphotropic murine leukemia virus receptor to also function as a gibbon ape leukemia virus receptor. J Virol. 1996;70:1080–1085. doi: 10.1128/jvi.70.2.1080-1085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elder J H, Mullins J I. Nucleotide sequence of the envelope gene of Gardner-Arnstein feline leukemia virus B reveals unique sequence homologies with a murine mink cell focus-forming virus. J Virol. 1983;46:871–880. doi: 10.1128/jvi.46.3.871-880.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia J V, Jones C, Miller A D. Localization of the amphotropic murine leukemia virus receptor gene to the pericentromeric region of human chromosome 8. J Virol. 1991;65:6316–6319. doi: 10.1128/jvi.65.11.6316-6319.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner M B, Rongey R W, Arnstein P, Estes J D, Sarma P, Huebner R J, Rickard C G. Experimental transmission of feline sarcoma to cats and dogs. Nature. 1970;226:807–809. doi: 10.1038/226807a0. [DOI] [PubMed] [Google Scholar]
- 14.Hardy W D, Jr, Hess P W, MacEwen E G, McClelland A J, Zuckerman E E, Essex M, Cotter S M, Jarrett O. Biology of feline leukemia virus in the natural environment. Cancer Res. 1976;36:582–588. [PubMed] [Google Scholar]
- 15.Jarrett O, Hardy W D, Jr, Golder M C, Hay D. The frequency of occurrence of feline leukemia virus subgroups in cats. Int J Cancer. 1978;21:334–337. doi: 10.1002/ijc.2910210314. [DOI] [PubMed] [Google Scholar]
- 16.Jarrett O, Laird H M, Hay D. Determinants of the host range of feline leukemia viruses. J Gen Virol. 1973;20:169–175. doi: 10.1099/0022-1317-20-2-169. [DOI] [PubMed] [Google Scholar]
- 17.Jarrett O, Russell P H. Differential growth and transmission in cats of feline leukemia viruses of subgroups A and B. Int J Cancer. 1978;21:466–472. doi: 10.1002/ijc.2910210411. [DOI] [PubMed] [Google Scholar]
- 18.Jarrett O, Russell P H, Hardy W D. The influence of virus subgroup on the epidemiology of feline leukemia virus. In: Bentveltzen P, editor. Advances in comparative leukemia research. Amsterdam, The Netherlands: Elsevier; 1978. pp. 25–28. [Google Scholar]
- 19.Johann S V, Gibbons J J, O'Hara B. GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J Virol. 1992;66:1635–1640. doi: 10.1128/jvi.66.3.1635-1640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kavanaugh M P, Miller D G, Zhang W, Law W, Kozak S L, Kabat D, Miller A D. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc Natl Acad Sci USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar D V, Berry B T, Roy-Burman P. Nucleotide sequence and distinctive characteristics of the env gene of endogenous feline leukemia provirus. J Virol. 1989;63:2379–2384. doi: 10.1128/jvi.63.5.2379-2384.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landau N R, Page K A, Littman D R. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lander M R, Chattopadhyay S K. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ecotropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J Virol. 1984;52:695–698. doi: 10.1128/jvi.52.2.695-698.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lauring A S, Anderson M M, Overbaugh J. Specificity in receptor usage by T-cell-tropic feline leukemia viruses: implications for in vivo tropism of immunodeficiency-inducing variants. J Virol. 2001;75:8888–8898. doi: 10.1128/JVI.75.19.8888-8898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lavillette D, Boson B, Russell S J, Cosset F L. Activation of membrane fusion by murine leukemia viruses is controlled in cis or in trans by interactions between the receptor-binding domain and a conserved disulfide loop of the carboxy terminus of the surface glycoprotein. J Virol. 2001;75:3685–3695. doi: 10.1128/JVI.75.8.3685-3695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDougall A S, Terry A, Tzavaras T, Cheney C, Rojko J, Neil J C. Defective endogenous proviruses are expressed in feline lymphoid cells: evidence for a role in natural resistance to subgroup B feline leukemia viruses. J Virol. 1994;68:2151–2160. doi: 10.1128/jvi.68.4.2151-2160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–982. [PMC free article] [PubMed] [Google Scholar]
- 29.Miller D G, Edwards R H, Miller A D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moser M, Burns C, Boomer S, Overbaugh J. The host range and interfence properties of two closely related feline leukemia virus variants suggest that they use distinct receptors. Virology. 1998;242:366–377. doi: 10.1006/viro.1997.9008. [DOI] [PubMed] [Google Scholar]
- 31.Moss G B, Overbaugh J, Welch M, Reilly M, Bwayo J, Plummer F A, Ndinya-Achola J O, Malisa M A, Kreiss J K. Human immunodeficiency virus DNA in urethral secretions in men: association with gonococcal urethritis and CD4 cell depletion. J Infect Dis. 1995;172:1469–1474. doi: 10.1093/infdis/172.6.1469. [DOI] [PubMed] [Google Scholar]
- 32.Mullins J I, Casey J W, Nicolson M O, Burck K B, Davidson N. Sequence arrangement and biological activity of cloned feline leukemia virus proviruses from a virus-productive human cell line. J Virol. 1981;38:688–703. doi: 10.1128/jvi.38.2.688-703.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Hara B, Johann S V, Klinger H P, Blair D G, Rubinson H, Dunn K J, Saas P, Vitek S M, Robins T. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1990;1:119–127. [PubMed] [Google Scholar]
- 34.Olah Z, Lehel C, Anderson W B, Eiden M V, Wilson C A. The cellular receptor for gibbon ape leukemia virus is a novel high affinity sodium-dependent phosphate transporter. J Biol Chem. 1994;269:25426–25431. [PubMed] [Google Scholar]
- 35.Overbaugh J, Donahue P R, Quackenbush S L, Hoover E A, Mullins J I. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science. 1988;239:906–910. doi: 10.1126/science.2893454. [DOI] [PubMed] [Google Scholar]
- 36.Overbaugh J, Riedel N, Hoover E A, Mullins J I. Transduction of endogenous envelope genes by feline leukaemia virus in vitro. Nature. 1988;332:731–734. doi: 10.1038/332731a0. [DOI] [PubMed] [Google Scholar]
- 37.Pandey R, Bechtel M K, Su Y, Ghosh A K, Hayes K A, Mathes L E, Roy-Burman P. Feline leukemia virus variants in experimentally induced thymic lymphosarcomas. Virology. 1995;214:584–592. doi: 10.1006/viro.1995.0069. [DOI] [PubMed] [Google Scholar]
- 38.Pedersen L, Johann S V, van-Zeijl M, Pedersen F S, O'Hara B. Chimeras of receptors for gibbon ape leukemia virus/feline leukemia virus B and amphotropic murine leukemia virus reveal different modes of receptor recognition by retrovirus. J Virol. 1995;69:2401–2405. doi: 10.1128/jvi.69.4.2401-2405.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phipps A J, Chen H, Hayes K A, Roy-Burman P, Mathes L E. Differential pathogenicity of two feline leukemia virus subgroup A molecular clones, pFRA and pF6A. J Virol. 2000;74:5796–5801. doi: 10.1128/jvi.74.13.5796-5801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phipps A J, Hayes K A, Al-Dubaib M, Roy-Burman P, Mathes L E. Inhibition of feline leukemia virus subgroup A infection by coinoculation with subgroup B. Virology. 2000;277:40–47. doi: 10.1006/viro.2000.0606. [DOI] [PubMed] [Google Scholar]
- 41.Quigley J G, Burns C C, Anderson M M, Sabo K M, Lynch E D, Overbaugh J, Abkowitz J L. Cloning of the cellular receptor for feline leukemia virus subgroup C (FeLV-C), a retrovirus that induces red cell aplasia. Blood. 2000;95:1093–1099. [PubMed] [Google Scholar]
- 42.Rohn J L, Linenberger M L, Hoover E A, Overbaugh J. Evolution of feline leukemia virus variant genomes with insertions, deletions, and defective envelope genes in infected cats with tumors. J Virol. 1994;68:2458–2467. doi: 10.1128/jvi.68.4.2458-2467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohn J L, Moser M S, Gwynn S R, Baldwin D N, Overbaugh J. In vivo evolution of a novel, syncytium-inducing and cytopathic feline leukemia virus variant. J Virol. 1998;72:2686–2696. doi: 10.1128/jvi.72.4.2686-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roy-Burman P. Endogenous env elements: partners in generation of pathogenic feline leukemia viruses. Virus Genes. 1996;11:147–161. doi: 10.1007/BF01728655. [DOI] [PubMed] [Google Scholar]
- 45.Rudra-Ganguly N, Ghosh A K, Roy-Burman P. Retrovirus receptor PiT-1 of the Felis catus. Biochim Biophys Acta. 1998;1443:407–413. doi: 10.1016/s0167-4781(98)00241-3. [DOI] [PubMed] [Google Scholar]
- 46.Sarma P S, Log T. Subgroup classification of feline leukemia and sarcoma viruses by viral interference and neutralization tests. Virology. 1973;54:160–169. doi: 10.1016/0042-6822(73)90125-6. [DOI] [PubMed] [Google Scholar]
- 47.Sarma P S, Log T. Viral interference in feline leukemia-sarcoma complex. Virology. 1971;44:352–358. [PubMed] [Google Scholar]
- 48.Sarma P S, Log T, Jain D, Hill P R, Huebner R J. Differential host range of viruses of feline leukemia-sarcoma complex. Virology. 1975;64:438–446. doi: 10.1016/0042-6822(75)90121-x. [DOI] [PubMed] [Google Scholar]
- 49.Schneiderman R D, Farrell K B, Wilson C A, Eiden M V. The Japanese feral mouse Pit1 and Pit2 homologs lack an acidic residue at position 550 but still function as gibbon ape leukemia virus receptors: implications for virus binding motif. J Virol. 1996;70:6982–6986. doi: 10.1128/jvi.70.10.6982-6986.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheets R L, Pandey R, Jen W C, Roy-Burman P. Recombinant feline leukemia virus genes detected in naturally occurring feline lymphosarcomas. J Virol. 1993;67:3118–3125. doi: 10.1128/jvi.67.6.3118-3125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheets R L, Pandey R, Klement V, Grant C K, Roy-Burman P. Biologically selected recombinants between feline leukemia virus (FeLV) subgroup A and endogenous FeLV element. Virology. 1992;190:849–855. doi: 10.1016/0042-6822(92)90924-e. [DOI] [PubMed] [Google Scholar]
- 52.Soe L H, Shimizu R W, Landolph J, Roy-Burman P. Molecular analysis of several classes of endogenous feline leukemia virus elements. J Virol. 1985;56:701–710. doi: 10.1128/jvi.56.3.701-710.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soe L H, Devi B G, Mullins J I, Roy-Burman P. Molecular cloning and characterization of endogenous feline leukemia virus sequences from a cat genomic library. J Virol. 1983;46:829–840. doi: 10.1128/jvi.46.3.829-840.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stewart M A, Warnock M, Wheeler A, Wilkie N, Mullins J I, Onions D E, Neil J C. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J Virol. 1986;58:825–834. doi: 10.1128/jvi.58.3.825-834.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sugai J, Eiden M, Anderson M, Van Hoeven N, Meiering C D, Overbaugh J. Identification of envelope determinants of feline leukemia virus subgroup B that permit infection and gene transfer to cells expressing human Pit1 or human Pit2. J Virol. 2001;75:6841–6849. doi: 10.1128/JVI.75.15.6841-6849.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tailor C S, Nouri A, Kabat D. A comprehensive approach to mapping the interacting surfaces of murine amphotropic and feline subgroup B leukemia viruses with their cell surface receptors. J Virol. 2000;74:237–244. doi: 10.1128/jvi.74.1.237-244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tailor C S, Takeuchi Y, O'Hara B, Johann S V, Weiss R A, Collins M K. Mutation of amino acids within the gibbon ape leukemia virus (GALV) receptor differentially affects feline leukemia virus subgroup B, simian sarcoma-associated virus, and GALV infections. J Virol. 1993;67:6737–6741. doi: 10.1128/jvi.67.11.6737-6741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tailor C S, Willet B J, Kabat D. A putative cell surface receptor for anemia-inducing feline leukemia virus subgroup C is a member of a transporter family. J Virol. 1999;73:6500–6505. doi: 10.1128/jvi.73.8.6500-6505.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takeuchi Y, Vile R G, Simpson G, O'Hara B, Collins M K, Weiss R A. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol. 1992;66:1219–1222. doi: 10.1128/jvi.66.2.1219-1222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ting Y T, Wilson C A, Farrell K B, Chaudry G J, Eiden M V. Simian sarcoma-associated virus fails to infect Chinese hamster cells despite the presence of functional gibbon ape leukemia virus receptors. J Virol. 1998;72:9453–9458. doi: 10.1128/jvi.72.12.9453-9458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Zeijl M, Johann S V, Closs E, Cunningham J, Eddy R, Shows T B, O'Hara B. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc Natl Acad Sci USA. 1994;91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson C A, Farrell K B, Eiden M V. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cells. J Virol. 1994;68:7697–7703. doi: 10.1128/jvi.68.12.7697-7703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wunsch M, Schulz A S, Koch W, Friedrich R, Hunsmann G. Sequence analysis of Gardner-Arnstein feline leukaemia virus envelope gene reveals common structural properties of mammalian retroviral envelope genes. EMBO J. 1983;2:2239–2246. doi: 10.1002/j.1460-2075.1983.tb01729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]