Keywords: Oral Squamous Cell Carcinoma, Saliva, Non-invasive diagnostics, Tobacco consumers, Biomarkers, Oral Cancer
Abstract
Objective
This review aims to provide a comprehensive analysis of the effectiveness of saliva as a non-invasive diagnostic marker for oral cancer. Despite progress in oral cancer diagnosis and prognosis, the 5-year survival rate remains low due to the resistance to treatment and delayed diagnosis, which can be attributed to various factors including tobacco and alcohol consumption, genetic damage, and human papillomavirus (HPV). The potential use of saliva as an easily accessible non-invasive screening and diagnostic method arises from its direct contact with the lesion site.
Methodology
Data for this study were gathered via a comprehensive literature evaluation using search engines such as the PubMed, Web of Science, Google Scholar, and SciFinder.
Results
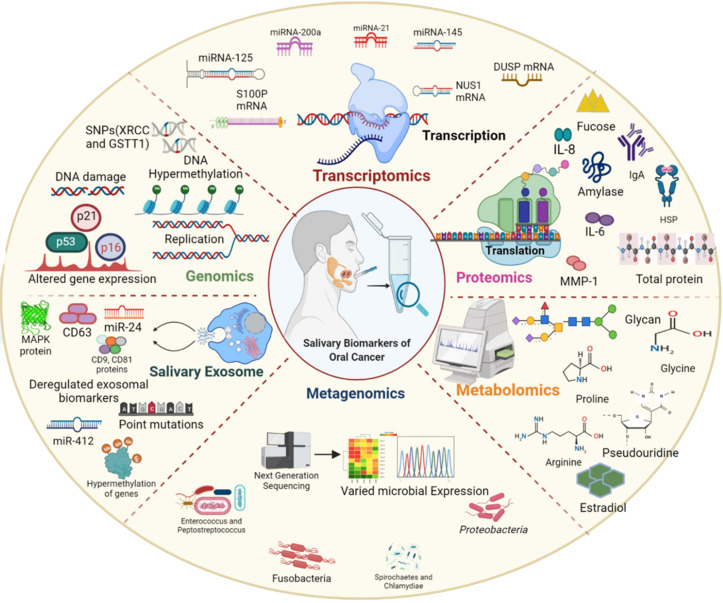
Identifying salivary biomarkers shows potential to transform oral cancer diagnostics by offering a reliable alternative to the traditional invasive methods. Saliva is an abundant reservoir for both cell-bound and cell-free organic and inorganic constituents. Thus, saliva is an appropriate field for research in proteomics, genomics, metagenomics, and metabolomics.
Conclusion
This review provides a comprehensive elucidation of salivary biomarkers and their function in non-invasive oral cancer diagnosis, demonstrating their potential to enhance patient outcomes and reduce the impact of this devastating disease.
Introduction
Oral cancer is characterized by the presence of malignant cells in the oral cavity, which comprises the sublingual space, tongue, lip, soft and hard palates, and buccal mucosa. Oral Squamous Cell Carcinoma (OSCC) originates from squamous cells in the oral cavity and accounts for nearly 90% of all oral malignancies.1 According to a publication issued by the World Health Organization, 377,713 newly diagnosed cases and 177,757 deaths were reported worldwide by 2020, indicating the prevalence of this type of cancer.2 Oral cancer significantly burdens public health and economic resources, primarily due to its high risk of mortality and morbidity, low five-year survival rate, delayed diagnosis, and the continuous emergence of numerous new cases each year.3 Oral cancer is primarily attributed to genetic modifications that occur at both minor and significant levels, such as point mutations involving base insertions, deletions, and substitutions as well as DNA rearrangements encompassing translocation, inversion, deletion, and duplication. Various risk factors, such as human papillomavirus (HPV) infection, tobacco use, smoking, betel quid chewing, and alcohol consumption significantly contributes to the onset and progression of oral cancer.4 These carcinogens can induce mutations and changes in DNA methylation, acetylation, ubiquitination, and histone modifications at the promoter sites of tumor suppressor genes and oncogenes, thereby initiating various malignancies.
Despite numerous advancements in therapeutics and preventive measures, the diagnostic approach to oral cancer still needs improvement, as traditional techniques are non-invasive and evaluate cancer at advanced stages.5 The existing diagnostic methods for oral cancer include visual examination, toluidine dye staining, exfoliative cytology, biopsy, and fluorescence imaging. Tissue biopsy is the gold standard for diagnosing oral cancer, but its invasive nature makes patients apprehensive, which can lead to delays in diagnosis. The uncomfortable nature of the extraction process and delayed diagnosis, which contribute to a high mortality rate, are the primary drawbacks of these screening methods.6 Therefore, it is crucial to identify early predictive biomarkers to enhance the survival of patients living with oral cancer. Biomarkers are molecular entities produced from endogenous genes and exhibit a substantial increase in expression in neoplastic cells. In contrast, these entities may manifest as novel gene products, typically inactive in healthy cells, but capable of regulating the expression of oncogenes and tumor suppressor genes. Biological markers can aid in predicting disease incidence rates, facilitating diagnosis and prognosis, and enabling the monitoring of treatment responses in individuals affected by the disease. Based on the preceding alterations, biomarkers are categorized into several groups: genetic biomarkers, which involve alterations at the gene level; protein biomarkers, which include the discovery of cancer-specific antigens; metabolic biomarkers, which involve the identification of altered metabolites; and metagenomic biomarkers, which detect changes in the microbial genome of the organ and may lead to uncontrolled proliferation, increased vascularization, and altered metabolism.7 Biomarkers for oral cancer can detect changes in DNA, RNA, and protein synthesis as well as in microRNAs, extracellular vesicles, circulating tumor cells, metabolites, and oral microbiota. Oral cancer biomarkers can be used to measure risk, diagnosis, prognostic stage, therapy response, and disease recurrence.
Oral cancer biomarkers can be studied in various body fluids. However, saliva, a susceptible and non-invasive diagnostic fluid that directly interacts with oral cancer lesions, is being considered for screening and diagnosis.3 The capacity of saliva to convey health information reflects that of the human body. Significant developments have been made in technologies designed to measure and analyze small amounts of analytes, including proteins and mRNA. Thus, saliva can be regarded as the circulatory system in the oral cavity. The heterogeneous saliva has been found to be slightly acidic, containing water, proteins, lipids, electrolytes, inorganic elements, DNA and RNA.8 Nucleic acids in saliva may originate locally within the oral cavity or be derived from serum. The examination of salivary biomarkers has been shown as capable of detecting changes in organs located distally from the oral cavity, extending beyond the realm of oral illnesses. This information includes many molecular details that could link the disease prognosis and diagnosis across various body systems.9
Examining saliva provides a wide range of opportunities for scholarly investigation and practical application in both elementary and clinical contexts. The discovery of salivary biomarkers has demonstrated the potential to enhance survival rate by offering a dependable indicator of both the initial and advanced stages of the disease at different intervals. This facilitates the evaluation of drug resistance and the probability of tumor recurrence.10 Moreover, owing to its ease of collection, transportation, storage, and sample volume, saliva shows several advantages over serum and tissue.11 Due to this accessibility and wide range, salivary biomarkers play a more favorable role than invasive procedures in the screening and detection of oral cancer. This review provides a detailed overview of salivary biomarkers and their roles in non-invasive oral cancer diagnostics.
Data for this review was extracted from the PubMed, Science Direct, NIH, and Google Scholar databases using search terms, including “saliva,” “non-invasive,” “biomarkers,” and “oral cancer.” Studies included in the review focused on detecting oral cancer biomarkers present in saliva. Additionally, the research emphasized the interpretation of common oral cancer biomarkers from non-invasive and minimally invasive sample sources. Studies that solely employed invasive methods of oral cancer diagnostics were excluded from the review.
Salivaomics: Salivary biomarkers in oral cancer
Salivaomics, a sophisticated approach within the field of “omics” encompasses five diagnostic components: genetics, transcriptomics, proteomics, metabolomics, and microbiomics. The submandibular, parotid, and sublingual glands produce whole saliva, a unique blend of water, organic compounds, and inorganic molecules. Additionally, the buccal, labial, lingual, and palatal glands contribute to salivary production. The tubarial gland, a newly discovered salivary gland, may reduce the side effects of radiotherapy as revealed using prostate-specific membrane antigen (PSMA) positron emission tomography/computed tomography (PET/CT) imaging.12 Water, organic compounds, and inorganic components are absorbed by salivary glands from the bloodstream and integrated into their secretions. Therefore, saliva is referred to as a “window of health status,” as substances identified in plasma can also be detected in whole saliva.13 Saliva enables mastication, oral hygiene, phonation, digestion, homeostasis, pathogen management, enzymatic digestion, and growth suppression.14 Saliva has garnered significant attention as a diagnostic fluid that is non-invasive and easily collectible. Saliva can be classified into two distinct categories, stimulated and unstimulated. Several methods are available for stimulating salivary production, including masticatory activity and chewing gum. Unstimulated saliva can be obtained without mechanical, masticatory, or external gustatory stimuli.15 Without stimulation, the salivary flow rate and quality are affected by medications and therapy, disease, and physiological and psychological factors.
Salivary biomarkers can either originate locally or be derived from serum, both of which contribute to their presence in the saliva.16 Serum-derived compounds are generated via both transcellular and paracellular pathways. The transcellular pathway utilizes active and passive transport, whereas the paracellular route involves salivary gland-blood capillary ultrafiltration. The salivary glands are surrounded by highly permeable blood capillaries. This characteristic facilitates the transfer of molecules between the bloodstream and saliva.17 Thus, saliva contains indicators that can convey health status, potentially serving as a non-invasive alternative to blood and other invasive methods. Moreover, saliva can serve as a viable substitute for serum in the diagnosis of specific conditions. The importance of saliva in diagnostics has significantly increased due to recent technological advancements. Salivary biomarkers have been employed for screening and detecting numerous malignancies, including oral cancer, periodontal disease, dental caries, viral infections such as HIV, infectious diseases, autoimmune disorders, and drug abuse monitoring.11
Being non-invasive and in close proximity to lesions related to oral cancer, saliva can serve as a diagnostic tool for the identification of oral cancer. Studies have found cell-free DNA, altered DNA, RNA, proteins, metabolites, and microbial communities in the saliva.18 There are significant disparities between the physiological characteristics of the normal and cancerous tissues. Cell-free DNA, RNA, and other waste from apoptotic and necrotic cells in healthy settings are phagocytosed. Under pathological conditions, this mechanism is compromised, causing cellular accumulation in physiological fluids and in tissue microenvironments. Oral cancer tumors contain cell-free DNA and RNA in saliva. Cancerous cells and necrotic bodies present a larger cell-free DNA size range (100–400 base pairs) than apoptotic cells (180–200 base pairs). This phenomenon can be attributed to the enhanced metabolic activity of the malignant cells, which subsequently leads to necrosis. Together with serum-derived findings, there is growing evidence for the release of proteins and nucleic acids from apoptotic, necrotic, and malignant cells.19 Additionally, exosomes contain these compounds. Vesicles released by live cells and exosomes are membrane-bound and range in diameter from 30 nm to 150 nm. Cell-free molecules are crucial for the interplay between cells because they include proteins, messenger RNAs (mRNAs), and microRNAs (miRNAs).20Given the presence of many biomarkers in saliva, it has the potential to be used as a diagnostic medium for the detection of oral cancer. A comprehensive discussion of the numerous components of salivaomics and the specific applications of these components as oral cancer diagnostic biomarkers is presented in subsequent paragraphs.
Salivaomics provides a complex and non-invasive diagnostic method by analyzing genetics, transcriptomics, proteomics, metabolomics, and microbiomics. This method demonstrates substantial promise for the early identification of oral cancer and other disorders, as saliva contains a multitude of indicators. The subsequent paragraphs delve into a comprehensive examination of these elements and their utilization in diagnosing various conditions.
Salivary genomics
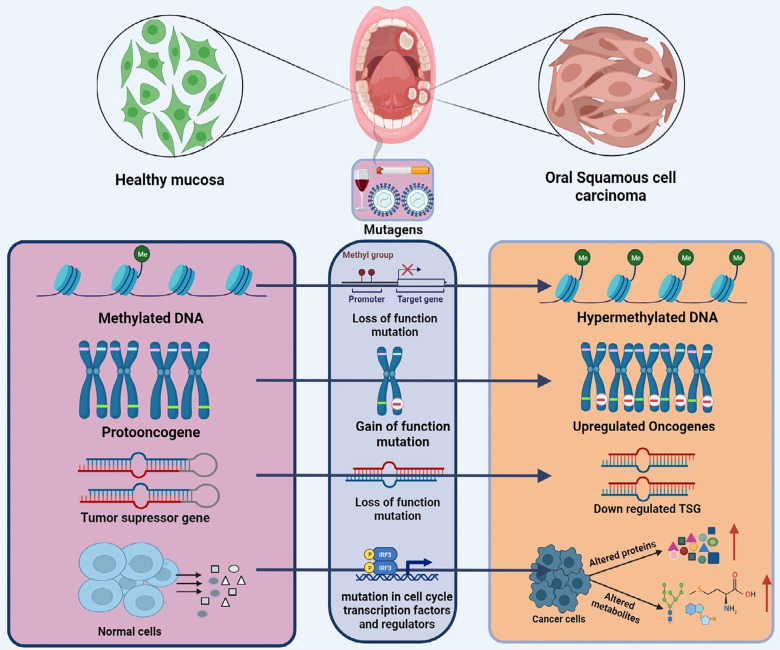
The development of oral cancer is attributed to numerous mutations, including DNA damage, loss of chromosomal segments, modified methylation, genetic instability, circulating tumor DNA, gene polymorphisms, and epigenetic alterations (Figure 1). DNA can be modified by oxidative stress, hydrolysis, base-pair mismatch, chemicals, ionizing radiation, and ultraviolet (UV) radiation. DNA damage response system disruption may initiate cancer progression. This process involves DNA damage recognition and cell cycle regulation by ATM and ATR enzymes. Impaired ATM/ATR kinase is unable to regulate p53, which inhibits p16 and p52 and regulates the cell cycle, DNA repair, and apoptosis signaling. Mutations in p53, which regulate DNA repair, the cell cycle, and apoptosis, may induce cancer. Early oral cancer is associated with mutant p53 levels. Antibodies against p53 have been detected in the saliva of patients with oral cancer, suggesting that saliva can be a non-invasive method for detecting p53 mutations.21
Figure 1. Association of molecular dysbiosis in cancer development-Oral cancer onset stems from various mutations induced by mutagens present in tobacco and alcohol. Numerous molecular dysbiosis, including hypermethylation, gain of function mutation in oncogenes, loss of function mutation in tumor suppressor genes, and several alterations in cell cycle regulators. The cumulative impact of these mutations transforms normal mucosal tissue into oral squamous cell carcinoma.
Cancer development is associated with changes in CpG island methylation at gene promoters. Methylation patterns in OSCC and control participants were examined using non-invasive oral rinse and methylation-specific polymerase chain reaction (PCR). OSCC and control participants expressed eight hypermethylated genes differently.22 The methylation patterns of salivary MGMT, DCC, CCNA1, TIMP-3, p16, and MINT-31 in healthy individuals and patients with Head and Neck Squamous Cell Carcinoma (HNSCC) were measured using quantitative polymerase chain reaction (Q-PCR). Targeted gene hypermethylation was identified in 54.1% of individuals. Epigenetic changes in oral cancer can be detected by salivary methylation.23 Healthy and cancerous cells contain genetic material that differ and can be used as biomarkers. DNA chromosomal abnormalities can cause the spread of cancer. Therefore, examining the abnormalities in ploidy status can aid in the prediction of tumor aggressiveness. Loss of heterozygosity (LOH) occurs when a chromosomal pair loses genetic material and frequently signals early stages of cancer. According to several studies, LOH in tumor suppressor gene regions can cause malignancies.24 OSCC often shows recurrent LOH on chromosomes 3, 17, 9, and 13, as shown by multiple studies.25
Oral cancer detection is possible using circulating tumor DNA (ctDNA) and gene polymorphisms. Tumor DNA was detected in blood, saliva, and urine samples. Somatic mutations make ctDNAs more selective than cell-free DNA. Both non-metastatic and metastatic tumor cells produce ctDNA and exhibit tumor genetics.26 Anticancer treatment steadily reduced ctDNA levels. Low dilution and contamination make salivary ctDNA analyses beneficial. Because saliva touches the tumor, patients with oral cancer have increased levels of ctDNA in their saliva, and previous studies have explored the relationship between salivary TP53 and the dimensions and progression of oral cancer. The prevalence of TP53 mutations in oral rinse is notably higher in cases of oral and oropharyngeal cancers, indicating a clear correlation between saliva and tumor development.27 CDKN2A, HRAS, PIK3CA, and TP53 genes were tested in the saliva of patients with HNSCC for somatic mutations using multiplex PCR. A significantly mutated TP53 mutant was also identified.28 Salivary DNA from patients with HNSCC and healthy controls were examined for 82 mutations. Of the 11 deregulated genes, PMAIP1 and PTPN1 correlated with the study groups. This finding suggest that the salivary biomarkers examined could be used as prognostic indicators for detecting HNSCC.29 Multi-allele gene polymorphisms diversify populations, whereas single-nucleotide polymorphisms (SNPs) control the cell cycle and DNA mismatch repair. Cancer progresses with deregulated SNPs. Researchers have found that oral cancer progression is linked to GSTT1 gene genotype absence.30 Yen, et al.31 (2008) examined salivary SNPs for the DNA-repairing RAD51 family XRCC gene 14q32.3, which affects cancer signaling. PCR restriction fragment length polymorphisms were used to study XRCC1 (rs1799782), XRCC2 (rs2040639), XRCC3 (rs861539), and XRCC4 (rs2075685) polymorphisms. In XRCC2, only rs2040639 was linked to cancer development. XRCC2 (rs2040639) and XRCC3 (rs861539) presented the most significant differences between cancer and control groups. Various studies have demonstrated that alterations in the DNA present in saliva play a crucial role in the early identification and prognosis of oral cancer due to its direct and close interaction with malignant lesions. Table 1 provides a comprehensive list of the diverse salivary DNA biomarkers associated with mutations, aneuploidy, and LOH.
Table 1. Differential expression summary of salivary DNA biomarkers in Oral Cancer.
| Biomarker | Study population | Saliva partition | Methods | Expression | Sensitivity | Function | References |
|---|---|---|---|---|---|---|---|
| 8-OHdG | 90 (30HC, 30 OSCC, 30 OSMF) | Saliva Supernatant | Sandwich ELISA | Upregulated | p<0.0001 | Oxidative DNA damage | 91 |
| P16INK4A, RASSF1A | 83 (40 HC, 43 OSCC) | Salivary rinse | Quantitative methylation specific PCR | Hyper methylation | p<0.001 | Tumor suppressor gene | 92 |
| TIMP3, PCQAP/MED15 | 148 (60 HC, 54 OC, 34 OP) | Saliva Supernatant | Quantitative methylation-specific PCR | Hyper methylation | p<0.05 p<0.0001 | Tumor suppressor gene | 93 |
| Telomerase | 300 (100 HC, 100 PML, 100 OSCC) | Salivary rinse | TRAP assay | Upregulated | p<0.001 | Reverse Transcriptase enzyme | 94 |
| KIF1A, EDNRB | 132 (61 HC, 71 HNSCC) | Salivary rinse | Quantitative methylation-specific PCR | Hyper methylation | p<0.0001 | Intracellular transport and cell cycle | 95 |
| p53 gene codon 63 of exon 4 | 37 (27 HC, 10 OC) | Saliva Supernatant | DNA sequencing and PCR | Loss of p53 gene codon 63 | p<0.05 | Tumor suppressor gene | 96 |
| DAPK1 | 174 (31 HC, 143 HNSCC) | Salivary cell pellet | Nested MSP | Hyper methylation | p<0.0001 | Cell cycle regulator | 97 |
| p16 | 60 (30 HC, 30 OSMF) | Salivary rinse | RT-QMSP | Hyper methylation | - | Tumor suppressor gene | 98 |
| MED15, PCQAP | 184 (94 HC, 90 HNSCC) | Saliva Supernatant | Methylation specific PCR | Hyper methylation | p<0.01 | Transcription factors | 99 |
| Maspin Phospho-Src | 38 (19 HC, 19 TC) | Saliva Supernatant | Immunoreactivity assay | Upregulated | p=0.001 p<0.00001 | Tumor suppressor, cell-cell adhesion, and proliferation | 100 |
| D3S1234, D9S156, and D17S799 | 50 (10 HC, 40HNSCC) | Saliva Supernatant | Microsatellite analysis | Loss of heterozygosity | p<0.001 | Cause Genetic instability | 101 |
| MGMT | 60 (30 HC, 30 HNSCC) | Saliva Supernatant | Methylation-specific PCR | Hyper methylation | p<0.001 | DNA repair pathway | 102 |
HC= Healthy Control; OC= Oral Cancer ;OSCC= Oral Squamous Cell Carcinoma; OSMF= Oral Submucous Fibrosis; OP= Oropharynx; HNSCC= Head and Neck Squamous Cell Carcinoma; PML= Premalignant lesion; 8-OHdG= 8-Hydroxydeoxyguanosine; KIF1A= Kinesin family member 1A; EDNRB= Endothelin receptor type B; TIMP3= Tissue Inhibitor of Metalloproteinases 3; PCQAP = PC2 glutamine/Q-rich-associated protein; PCR= Polymerase Chain Reaction; DAPK1= Death-associated protein kinase 1; OSMF= oral submucous fibrosis; RT-QMSP= Real-Time quantitative methylation-specific PCR; Phospho-Src = Phosphorylated-Src; TC= Tongue Carcinoma; MSP= Methylation specific PCR; Maspin= mammary serine protease inhibitor TRAP= Telomerase Repeated Amplification Protocol; MGMT= methylguanine-DNA-methyltransferase; RAASF1A= Ras association domain-containing protein; MED15= Mediator Complex Subunit 15
The formation of oral cancer involves a wide range of mutations that include DNA damage, changes in chromosomes, patterns of methylation, and variations in genes. Saliva is a valuable diagnostic tool since it can identify genetic abnormalities, such as p53 mutations and LOH, using non-invasive techniques. Moreover, the analysis of DNA in saliva enables the detection of genetic changes and variations linked to the advancement of oral cancer, emphasizing its potential as a diagnostic tool for the early diagnosis and monitoring of the disease.
Salivary transcriptomics
For decades, RNA was thought to facilitate gene expression by converting DNA sequences into proteins. However, further research has shown that certain RNA molecules, known as non-coding RNA (ncRNA), regulate gene expression by controlling the activation and deactivation of genes. These RNA molecules do not encode proteins and include various types, such as short hairpin RNAs, PIWI-interacting RNAs, short interfering RNAs, small nuclear RNAs, small temporal RNAs, and small nuclear RNAs.32 ncRNAs are emerging as key regulators of cancer progression and tumor growth. Their small size and stability in bodily fluids, including blood, urine, and saliva, make them potential candidates for non-invasive diagnostic tests. These tests could revolutionize disease detection, leading to more effective clinical applications and potentially reducing health care costs.
microRNA in oral cancer
miRNAs are non-coding molecules composed of 19–25 nucleotides, which are generated from pre-miRNAs with the aid of drosha in the nucleus and Dicer in the cytoplasm. These molecules play various roles in cell functions, such as proliferation, division, apoptosis, and immunological responses, as well as in inter-pathogen and host communication. Moreover, miRNA transcripts can act as either tumor suppressors or oncogenes due to their diverse effects. Oral cancer initiation and progression is affected by the abnormal regulation of tumor suppressor and oncogene genes.33 It became apparent that circulating miRNAs in saliva were stable, based on their structural assets. MicroRNA (miRNA) analysis was performed on five patients with oral cancer and five study controls using microarray technology to identify differentially expressed miRNAs. The most prominent dysregulated miRNAs were validated using quantitative real-time PCR (qPCR). miR-517b-3p and miR-303b-3p were found to be downregulated in oral cancer, whereas miR-412-3p and miR-512-3p were found to be considerably increased in qPCR analysis. Consequently, a panel of salivary EVs miRNAs can be used as biomarker to identify oral cancer.34 The most prevalent salivary miRNAs, miR-146a-5p, miR-205-5p, miR-92a-3p, and miR-124-3p, set oral cancer apart from control participants in a different study with 216 participants, 108 of whom had head and neck squamous cell carcinoma and 108 were healthy controls.35
Salivary miRNAs were used to screen for 60 individuals, 30 of whom had OSCC. According to a previous study, patients with oral cancer showed considerable downregulation of let-7c, miR-99a, and miR-100.36 Salivary microRNAs (miRNAs) may be promising indicators for oral cancer identification in high-risk populations such as tobacco smokers. Real-time PCR was used to compare salivary miRNA expression between smokeless tobacco users and non-smokers. miR-146a, miR-199a, and miR-155 are considerably upregulated in smokeless tobacco users. These findings indicate that tobacco use may cause epigenetic alterations that lead to oral cancer.37 Quantitative real-time polymerase chain reaction, microarray, and next-generation sequencing indicated that salivary miRNA patterns were distinct between patients living with cancer and healthy controls, as listed in Table 2.
Table 2. Differential expression of various salivary microRNA biomarkers.
| Biomarkers | Study population | Saliva partition | Methods | Expression | Function | Sensitivity | References |
|---|---|---|---|---|---|---|---|
| miR-423-5p | 147 (58 HC, 89 OSCC) | Saliva Supernatant (centrifuged at 2600g, 15 minutes) | Microarray and qRT-PCR | Upregulated | Oncogene | p<0.001 | 103 |
| miRNA-let-7a-5p and miRNA- 3928 | 230 (80 HC and 150 HNSCC) | Whole saliva | qRT-PCR | Downregulated | Tumor suppressor gene | p<0.0001 p<0.01 | 104 |
| miR-26a and miR-26b | 28 (14 HC, 14 OLP) | Whole saliva | Real-time PCR | Downregulated | Tumor suppressor gene | p<0.001 | 105 |
| miRNA 31 | 72 (HC and OPMD) | Saliva Supernatant | qRT-PCR | Upregulated | Oncogene | p=0.01 | 106 |
| miRNA-124-3p | 216 (108 HC, 108 HNSCC) | Whole saliva | miRNA PCR Array and qRT-PCR | Downregulated | Tumor suppressor gene | p=0.002 | 107 |
| miRNA-191 miRNA-146a | 78 (24 HC, 14 OSCC, 14 hOSCC, 13 OSCCr, 13hOSCCr) | Oral brush | Real time PCR | Upregulated | Oncogene | p<0.001 | 108 |
| miRNA-125a miRNA-200a | 60 (15 HC, 15 OSCC, 30 OLP) | Whole saliva | qRT-PCR | Downregulated | Tumor suppressor gene | p<0.0001 | 109 |
| miRNA-320a | 62 (15 HC, 15 OSCC and 32 OLP) | Saliva Supernatant (centrifuged at 3000g, 15 minutes) | RT-qPCR | Downregulated | Tumor suppressor gene | p=0.004 | 110 |
| miRNA-139-5p | 50 (25 HC and 25 TSCC) | Saliva Supernatant (centrifuged at 2600g, 15 minutes) | Microarray and qRT-PCR | Downregulated | Tumor suppressor gene | p=0.006 | 111 |
| miRNA-21, miRNA-184 | 100 (20 HC, 40 PMDs, 20 OSCC and 20 RAS) | Saliva Supernatant (centrifuged at 2500g, 10 minutes) | qRT-PCR | Upregulated | Oncogenes | p<0.001 | 112 |
| miRNA-145 | 100 (20 HC, 40 PMDs, 20 OSCC and 20 RAS) | Saliva Supernatant (centrifuged at 2500g, 10 minutes) | qRT-PCR | Downregulated | Tumor suppressor gene | p<0.001 | 112 |
| miRNA-24, miRNA-27b | 34 (9 HC, 9 OSCC, 8 OSCC-R and 8 OLP) | Saliva Supernatant (centrifuged at 2600g, 15 minutes) | Nanostring miRNA assay and RT-qPCR | Upregulated | Oncogenes | p<0.05 | 113 |
| miRNA-136 | 34 (9 HC, 9 OSCC, 8 OSCC-R, and 8 OLP) | Saliva Supernatant (centrifuged at 2600g, 15 minutes) | Nanostring miRNA assay and RT-qPCR | Downregulated | Tumor suppressor gene | p<0.05 | 113 |
| miRNA-9 | 112 (56 HC, 56 HNSCC) | Saliva supernatant | Microarray and qRT-PCR | Upregulated | Oncogenes | p<0.0001 | 114 |
| miRNA-134 | 112 (56 HC, 56 HNSCC) | Saliva supernatant | Microarray and qRT-PCR | Downregulated | Tumor suppressor gene | p<0.0001 | 114 |
| miRNA-148a, miRNA-1250, miRNA-632, miRNA-503, miRNA-323-5p | 34 (9 HC, 9 OSCC, 8 OSCC-R and 8 OLP) | Saliva Supernatant (centrifuged at 2600g, 15 minutes) | Nanostring miRNA assay and RT-qPCR | Downregulated | - | p=0.027 | 113 |
| miRNA-145-5p, miRNA-99b-5p, miRNA-181c, miRNA-197-3p | 14 (7 HC and 7 OLP) | Whole saliva | Microarray and RT-qPCR | Downregulated | Tumor suppressor genes | p=0.034 p=0.011 p=0.028 p=0.057 | 115 |
| miRNA-10b, miRNA-30e | 14 (7 HC and 7 OLP) | Whole saliva | Microarray and RT-qPCR | Upregulated | Oncogenes | p=0.008 p=0.089 | 115 |
p-value < 0.05 was considered statistically significant.
Salivary miRNAs show potential to act as reliable biomarkers for detecting oral cancer. They can indicate abnormalities in genes that regulate tumor growth and genes that promote the development of cancer. Their unique expression patterns, as evidenced by different molecular approaches, indicate their usefulness in detecting oral cancer, especially in high-risk individuals such as tobacco smokers, emphasizing their involvement in the development of cancer.
mRNA in oral cancer
Messenger RNAs (mRNAs) are crucial in cells and act as molecular messengers for protein synthesis. They also play a role in ribosome remodeling and utilize approximately 20% of the cellular energy. Several diseases, including cancer, heart disease, and blood disorders can disrupt mRNA translation. Therefore, regulation of mRNA translation is vital for gene expression. The presence of mRNA in body fluids, particularly saliva, which is a non-invasive liquid biopsy, aids in gene expression analysis in both healthy and diseased states.38 Salivary mRNA serves as a biomarker for a range of malignancies, including oral, breast, lung, and ovarian cancers. The studies on salivary deregulated mRNA levels in oral cancer are listed in Table 3.
Table 3. Expression profile of salivary mRNA in Oral Cancer.
| Biomarkers | Study population | Saliva partition | Methods | Expression | Sensitivity | Function | References |
|---|---|---|---|---|---|---|---|
| NAB2, COL3A1 | 67 (34 HC, 33 OSCC) | Saliva Supernatant | Real-time PCR | Downregulated | p=0.0023 | Growth signal response and cell division | 116 |
| NUS1, RCN1 | 51 (10 HC, 41 OSCC) | Whole saliva | qRT-PCR | Upregulated. | p=0.037 p=0.011 | Protein modification and Ca binding cell signaling | 117 |
| CYP27A1, NPIPB4, MAOB, SIAE | 67 (34 HC, 33 OSCC) | Saliva Supernatant | Real time PCR | Downregulated | p=0.0016 p=0.0059 p=0.0009 p=0.0370 | Protein modification | 116 |
| IL-8 | 85 (31 HC, 34 OSCC, 20 OLP) | Saliva Supernatant | q-PCR | Upregulated. | p=0.013 | Angiogenesis, cell signaling, and replication | 118 |
| OAZ 1 | 125 (25 HC, 25 OSCC, 25 OSCC-R, 25 OLP, 25 I-OLP) | Saliva Supernatant | q-PCR | Upregulated. | p=0.003 | Biosynthesis of polyamine | 119 |
| SAT | 395 (226 HC, 169 OSCC) | Saliva Supernatant | q-PCR | Upregulated. | p<0.002 | Transferase enzyme | 120 |
| S100P | 125 (25 HC, 25 OSCC, 25 OSCC-R, 25 OLP, 25 I-OLP) | Saliva Supernatant | q-PCR | Upregulated. | p=0.003 | Calcium-binding protein | 121 |
| DUSP1 | 125 (25 HC, 25 OSCC, 25 OSCC-R, 25 OLP, 25 I-OLP) | Saliva Supernatant | q-PCR | Upregulated. | p<0.001 | Cell signaling and protein modification | 121 |
| IL1B H3F3A | 64 (32 HC, 32 OSCC) | Saliva Supernatant | Microarray and qPCR | Upregulated. | p<0.001 | Inflammation, cell proliferation, cell signaling, and H3F3A in the DNA binding process | 122 |
| MAP2K3, B2M | 64 (32 HC, 32 OSCC) | Saliva Supernatant | Microarray and qPCR | Upregulated. | p<0.001 | Protein modification and antiapoptotic | 122 |
| Endothelin-1 | 16 (HC, OSCC) | Saliva Supernatant | RT-PCR | Upregulated. | p<0.001 | Vasoactive peptide | 123 |
p-value < 0.05 was considered statistically significant.
HC= Healthy Control; OSCC= oral squamous cell carcinoma; qRT-PCR= quantitative real time polymerase chain reaction; OSCC-R= OSCC recurrence; OLP= oral leukoplakia; NAB2= NGFI-A binding protein 2; NPIPB4= nuclear pore complex interacting protein family, member B4; COL3A1= collagen, type III, alpha 1; MAOB = monoamine oxidase B; qPCR= Quantitative real-time PCR; MAP2K3= Mitogen-activated protein kinase 3; B2M=beta2microglobulin; IL1B= Interleukin 1 beta; H3F3A= H3 histone, family 3A; SIAE= sialic acid acetyltransferase; IL-8= Interleukin 8; S100P= S100 calcium binding protein P; DUSP1= Dual specificity phosphatase 1; OAZ 1= Ornithine decarboxylase antizyme 1; NUS1= nuclear undecaprenyl pyrophosphate synthase 1; RCN1= reticulocalbin 1; SAT= Spermidine/spermine N1-acetyltransferase.
Salivary proteomics
Analysis of the salivary proteomes of individuals with oral cancer is expected to be a promising method for identifying new biomarkers for the disease because oral cancer cells are deeply involved in the salivary environment. Proteins play a crucial role in the survival, development, and division of cells in various biological processes. The disruption of proteins can impair the fundamental functioning of cells, leading to various forms of cancer. The identification of changes in protein expression allows for early cancer detection and prediction.39 Numerous studies have investigated protein levels in the saliva of patients with oral cancer.40Table 4 shows data on the notable alterations in crucial salivary proteins in oral cancer.
Table 4. List of oral cancer protein biomarkers and their expression levels.
| Biomarker | Study population | Saliva partition | Method | Expression | Sensitivity | Reference |
|---|---|---|---|---|---|---|
| LDH | 100 (HC, OLP, OLRs, and OSCC) | Saliva Supernatant (centrifuged at 2600g, 15 minutes) | Spectrophotometry | Upregulated | p<0.02 | 81 |
| IL-8 IL-1β LGALS3BP | 117 (HC, OSCC, PMOD, and under treatment) | Saliva Supernatant | ELISA | Upregulated | p<0.0006 p<0.0001 | 124 |
| IL-6 | 44 (HC and OC) | Saliva Supernatant | Immuno-fluorescence | Upregulated | p<0.05 | 125 |
| MMP-1 | 1160 (HC, OPMD-I, OPMD-II, and OSCC) | Saliva Supernatant | ELISA | Upregulated | p<0.001 | 126 |
| MMP-9 | 88 (HC, OSCC and OPMD) | Saliva Supernatant | ELISA | Upregulated | p<0.001 | 127 |
| IL-17A IL-17F TNF-α | 71 (HC, Differentiated site and grade tumor) | Saliva Supernatant | ELISA | Upregulated | p<0.001 p<0.01 p<0.01 | 128 |
| CYFRA21-1 | 35 (OPMD, OSMF and OSCC) | Saliva Supernatant | ELISA | Upregulated | p=0.01 | 129 |
| TNF- α IL-6 | 60 (HC, OBFOL) | Saliva Supernatant | ELISA | Upregulated | p=0.039 p<0.0001 | 130 |
| L-Fucose | 85 (HC, OPMD and OC) | Saliva Supernatant | Spectrophotometry | Upregulated | p<0.001 | 131 |
| MUC1 | 30 (HC, oral premalignant and OSCC) | Saliva Supernatant | ELISA | Upregulated | p<0.001 | 132 |
| Hsp27 | 45 (HC and OLP) | Saliva Supernatant | ELISA | Upregulated | p<0.001 | 133 |
| Hsp90 L-Fucose | 90 (HC, PMOD, and OC) | Saliva Supernatant | ELISA | Upregulated | p<0.001 | 134 |
| Sialic acid (FSA and PBSA) | 96 (HC, tobacco chewers and OSCC) | Saliva Supernatant | Spectrophotometry | Upregulated | p<0.001 | 135 |
p-value < 0.05 was considered statistically significant.
LDH= Lactate Dehydrogenase; IL= Interleukin; LGALS3BP = galectin-binding protein; MMP= Matrix metallopeptidase; TNF= Tumor Necrosis Factor; CYFRA= Cytokeratin fragment; MUC= Mucin; Hsp= Heat shock protein; FSA= Free sialic acid; PBSA= Protein-bound sialic acid; HC= Healthy Control; OC= Oral Cancer; OLP= Oral Leukoplakia; OLRs= Oral lichen planus; OSCC= Oral Squamous Cell Carcinoma; PMOD=potentially malignant oral disorders; OPMD= Oculopharyngeal muscular dystrophy; OBFOL= oral benign fibro-osseous tumors; IL= Interleukin; MMP= Matrix metalloproteinase; PBSA=Protein-bound sialic acid; FSA= Free sialic acid.
Tetranectin, a molecule that binds to plasminogen and is implicated in tumor metastasis, was found to show notably reduced levels in a study of 15 participants, including healthy individuals and those with primary and advanced OSCC. This reduction suggests tumor metastasis or advanced tumors.41 Salivary albumin protein, an ultrafiltrate of serum albumin, is increased in various diseases, and researchers have shown that patients with OSCC have higher salivary albumin levels than healthy individuals.42 Survivin, an inhibitor of cell death, is a prospective biomarker for various cancers including OSCC. Salivary survivin levels were assessed in patients with cancer and healthy individuals; those with OSCC had considerably higher levels, suggesting that salivary survivin may be used as a diagnostic biomarker.43 Salivary proteases play a crucial role in the development of oral cancer. Feng et al. found differences in protease levels between healthy individuals and those with OSCC, identifying several upregulated proteases, including MMP 1–3, 10, and 12, ADAMST13, ADAM9, cathepsin V, Kallikrein5, and Kallikrein7.44
Amylase in saliva breaks down starch secreted by the parotid glands. Salivary amylase levels vary among healthy individuals, patients with oral cancer, and those undergoing treatment, with lower levels occurring during cancer therapy.45 According to a previous study, the OSCC group had lower salivary amylase levels than the healthy control group (p=0.12). Awasthi et al. evaluated CYFRA21-1, CA19-9, lactate dehydrogenase (LDH), total protein, and amylase levels in 64 healthy, premalignant, and patients with OSCC. The levels of all biomarkers, except amylase, increased considerably.46 In conclusion, OSCC and premalignant lesions mostly showed low amylase levels. Conversely, the salivary amylase concentration was considerably lower (p<0.001) in healthy individuals than in patients with oral cancer and those undergoing treatment. Amylase levels decreased until week 3, showing increases from weeks 3 to 6.47
Cytokines are proteins that regulate the immune response, cell growth, and blood vessel formation. They can be pro-inflammatory, including IL-1β, IL-6, and TNF-α, or anti-inflammatory, including IL-1, IL-4, and IL-10. Salivary cytokine levels increase in patients with oral cancer, affecting the oral lesions.18,48 A study of 90 patients found higher salivary TNF-α levels in oral leukoplakia and OSCC groups than in healthy controls, suggesting that TNF-α is a biomarker for oral dysplasia.49 Another study compared IL-8 levels between healthy controls and OSCC patients. Patients with OSCC show higher IL-8 levels, indicating their potential as biomarkers of OSCC.50 Apart from these altered salivary biomarkers in oral cancer, other biomarkers, such as cyclin D1, Ki 67, defensin-1, profiling-1, catalase, annexin-1, calcyclin, and SCCA-2 have also been reported in oral cancer.51
Therefore, the examination of salivary proteomes shows potential for identifying new biomarkers for oral cancer, given the complex interaction between oral cancer cells and the salivary environment. Patients living with oral cancer have shown significant changes in important proteins, including tetranectin, albumin, survivin, proteases, and amylase, suggesting that these proteins could be useful for diagnosing the disease. Moreover, alterations in the levels of cytokines in saliva, including TNF-α and IL-8, emphasize the importance of salivary biomarkers in the identification and prediction of oral cancer.
Salivary metagenomics
Metagenomics studies the genomes of multiple microorganisms in complex communities. The oral microbiome comprises various bacteria that form symbiotic relationships in the mouth. It is the second largest component of the gut microbiota, with over 700 bacterial species belonging to 12 phyla and 185 genera, which vary among individuals based on lifestyle and physiological conditions. The 12 phyla were designated as Firmicutes, Proteobacteria, Fusobacteria, Chlamydiae, Bacteroidetes, Actinobacteria, Spirochaetes, SR1, Chloroflexi, Synergistetes, Gracilibacteria, and Saccharibacteria. These microbial communities belong to 70% culturable and 30% non-culturable species.52 In addition to the core microbial community, the oral microbial community also exhibited a spatially and temporally differential pattern. The altered dynamics are influenced by factors such as the disease progression of oral cancer, genetic mutations, and changes in pH levels, smoking, alcohol consumption, pathogen infection, and tooth decay. The examination of altered oral microbiota reflects disease progression in the oral cavity.53
Recent technologies have been developed to study the oral microbiota, including culture and microscopy, DNA microarray, PCR, 16S rRNA, and next-generation sequencing. A correlation was found between oral cancer progression and five microbial genera, namely Enterococcus, Peptostreptococcus, Bacillus, Slackia, and Parvimonas, suggesting their potential as predictive and diagnostic indicators for OSCC detection.54 Significant variations in Prevotella melaninogenica, Streptococcus mitis, and Capnocytophaga gingivalis were identified in another study that compared the oral microbiota of patients with OSCC to that of healthy controls.55 Microbial diversity in the oral cancer, oral leukoplakia, and healthy groups were analyzed using Illumina sequencing. Significant differences were found between the healthy and oral cancer groups but not between the oral lichen planus (OLP) and oral cancer groups. The most differential pattern between the oral cancer and the control group included Prevotella, Streptococcus, and Salmonella. Fusobacterium nucleatum. and Porphyromonas gingivalis has been suggested to play a role in releasing inflammatory cytokines, promoting cell proliferation and invasion in oral cancer, thus it is thought to be involved in cancer metastasis.56 P. gingivalis contributes to TNF-α and metalloproteinase production and inhibits p53, whereas F. nucleatum secretes lipopolysaccharides and cytokines that are linked to cancer development.57
Yang, et al.58 (2018) found increased Fusobacterium and decreased Streptococcus, Actinomyces, Porphyromonas, and Haemophilus in saliva as cancer progressed from stages 1 to 4. Several other studies have shown that Firmicutes (Streptococcus) and Actinobacteria (Rothia) are significantly decreased in patients with cancer compared to those in the non-cancerous group.59 Firmicutes, specifically Streptococcus, contributes to DNA damage via acetaldehyde production, leading to lipid peroxidation and reduced Lactobacillus spp, creating a tumor-friendly environment. Actinobacteria play a role in periodontal diseases, plaque formation, and caries. Alcohol and tobacco use affect the oral microbial diversity. Smokers and alcoholics were compared to the oral microbiome of nonsmokers. Capnocytophaga and Prevotella were upregulated, whereas Staphylococcus, Peptostreptococcus, and Granulicatella were downregulated in smokers compared to those in the control group.60
Metagenomic studies show the oral microbiome’s complex composition and dynamics, which may affect oral cancer detection and progression. Emerging technologies can identify oral cancer-associated microbial genera, demonstrating their predictive and diagnostic potential. Microbial variety underscores their role in inflammation and cancer metastasis, emphasizing the oral microbiota’s role in mouth cancer formation.
Salivary metabolomics
Metabolomics, an advanced omics technique, is associated with abnormal gene expression, drug revelation, and exposure to carcinogens. Different metabolite patterns have been examined using modern technology to detect oral, periodontal, pancreatic, and breast cancers.61 These have included metabolomic analyses of human body fluids, cells, and tissues for metabolic intermediates. Modern metabolomics can quantify disease progression biomarkers using several sophisticated technologies including time-of-flight mass spectrometry (TOF-MS), capillary electrophoresis, and Nuclear Magnetic Resonance (NMR).38 In addition to its core function, saliva contains metabolites that can detect oral cancer. Wei, et al.62 (2011) compared salivary metabolites from OSCC, OLK, and controls using ultra-performance liquid chromatography and TOF-MS. Compared to the OLK and control groups, the OSCC group had significantly higher salivary levels of n-eicosanoic acid and lactic acid, and lower salivary levels of GABA, phenylalanine, and valine. Enhanced glycolysis and anaerobic respiration produce lactic acid, which is a cancer marker. Increased energy utilization causes cancer cells to undergo anaerobic glycolysis, creating hypoxia. Valine provides energy, whereas phenylalanine signals and generates proteins. Lohavanichbutr, et al.63 (2018) found that four salivary metabolites, namely proline, glycine, citrulline, and ornithine, were significantly altered in the healthy and OSCC groups.
Sugimoto, et al.64 (2010) performed capillary electrophoresis-mass spectrometry to compare the levels of OSCC and control salivary metabolites. Study groups discovered 28 metabolites. Phyroline, hydroxycarboxylic acid, choline, tryptophan, threonine, carnitine alpha-aminobutyric acid, and phenylalanine showed significant p-values. Combined biomarkers can differentiate between oral cancer and healthy individuals. Another study examined the salivary metabolites of patients with OSCC and healthy controls, namely L-carnitine, betaine, pipecolinic acid, and choline, using ultraperformance LC-MS. Patients with OSCC showed highly elevated betaine, pipecolinic acid, and choline levels, while under expressing L-carnitine (96.7% specificity, 100% sensitivity).65 Another study used hydrophilic interaction chromatography and performed metabolomic analysis of OSCC to detect salivary metabolites, including propionylcholine, S-carboxymethyl-L-cysteine, phytosphingosine, sphinganine, and N-acetyl-L-phenylalanine.66 Ohshima et al. compared salivary metabolites in OSCC and control groups and found 25 differentially expressed metabolites including hydroxyphenyl acetic acid, choline, and 2-hydroxy-4-methylvaleric acid (p<0.001).67 In tumor cells, choline is metabolized to phosphocholine, followed by its oxidation to betaine.
Sridharan, et al.68 (2019) used LC-MS and mass Hunter profiles to examine salivary metabolites in OSCC, OLP, and healthy controls. The results demonstrated significant (p<0.05) upregulation of inositol 1,3,4-triphosphate, d-glycerate-2-phosphate, 1-methylhistidine, 2-oxoarginine, 4-nitroquinoline-1-oxide, pseudouridine, sphinganine-1-phosphate, and norcocaine nitroxide, as well as downregulation of ubiquinone, estradiol valerate, neuraminic acid, and l-homocysteine acid metabolites. Metabolomic studies have shown that various salivary metabolites exhibit variable expressions. These metabolites may serve as early markers for diagnosing oral cancer either independently or in combination with other metabolites.
Salivary exosomes
Exosomes are nanoparticles with double-layered membranes that originate from the endosomal pathways and bodily fluids. Salivary exosomes contain molecular cargo, support physiological functions, and serve as biomarkers of physiological changes.69 Salivary exosomal materials containing tumor-derived nucleic acids, proteins, and metabolites are stable and suitable for investigating oral cancer. Exosomal content controls immunomodulation and tumor enhancement and indicates microenvironmental fluctuations.70 Studies have shown that salivary exosomes in patients with oral cancer are larger, more numerous, and more diverse.71 In addition, Fourier-transform infrared spectroscopy indicated that salivary exosomes and their contents are protected from cellular nucleases.72 MicroRNAs (miRNAs), proteins, and DNA in salivary exosomes are linked to cancer progression. Salivary exosomal miRNAs are non-coding RNAs that function as tumor suppressors and oncogenes under normal physiological conditions. miR-412-3p, miR-512-3p,73 miR-1246, miR-342-3p, and miR-24-3p74 showed differential patterns and could be used as diagnostic biomarkers. Exosomal miRNAs promote cancer cell migration via the MAPK cell signaling system, upregulate oncogenic genes, and downregulate tumor suppressor genes.
Salivary exosomal proteins demonstrated that patients with oral cancer and healthy individuals have varied expression of MUC5B, HPA, LGALS3BP, A2M, IGHA1, GAPDH, and PKM1/M2. This robust proteomic analysis can distinguish OSCC from the controls with 90% accuracy.75 Oral cancer exosomes have higher levels of CD63, a 25kDa salivary protein that regulates cell invasion and migration, whereas in an exosomal salivary protein study, oral cancer and healthy groups exhibited elevated CD63, as well as downregulated CD9 and CD81.76 As described by the biological role of exosomes in oral cancer progression, the salivary exosomal content can be used as a prognostic and diagnostic biomarker. These nanoparticles are incredibly informative as they are safely packed and delivered to the extracellular environment and show advantages over salivary complexity. Therefore, salivary exosomes have emerged as new tools for the diagnosis of oral cancer.
Common biomarkers from invasive and non-invasive diagnostic tools
The human body consists of several tissues and fluids, including saliva, blood, cerebrospinal fluid (CSF), tears, perspiration, and urine. They show distinct purposes and consist of proteins, electrolytes, hormones, and metabolites that are crucial for health. Blood, saliva, and urine are important for the diagnosis of diseases, and the blood is particularly useful for detecting changes in the body. Tissue biopsy, blood biopsy, exfoliated cell cytology, PET, and CT-SCAN are the key cancer diagnostic methods; however, they have limitations in late-stage diagnosis with painful procedures. Saliva is a non-invasive tool and holds potential to be an alternative to invasive procedures. This fluid is widely available, simply collectible, and storable, especially beneficial in oral cancer, as it is in direct contact with the lesion.77 Several earlier studies have shown that saliva carries the filtrate of plasma and serum and alters cell-free and cell-based tumor cells undergoing necrosis and apoptosis. Hence, saliva can rise above the invasive methods of diagnosis. Several studies have compared and found various significantly altered common biomarkers in the same study cohort using non-invasive and invasive methods of oral cancer diagnosis.
Dadhich, et al.78 (2014) discovered that individuals with oral cancer and premalignant lesions had significantly upregulated (p<0.0005) sialic acid in their serum and saliva samples as compared to healthy controls. The glycoprotein sialic acid can easily be detected in saliva and has been associated with carcinogenesis. Sartini, et al.79 (2012) reported that patients with OSCC had considerably higher nicotinamide N-methyltransferase levels (p<0.0001) in both saliva and tissue than the healthy group. The study concluded that selected enzymes could be used as early biomarkers for OSCC using a non-invasive sample type. In another study, Bhat, et al.80 (2017) found a significant (p<0.001) decrease in the antioxidant ascorbic acid in the saliva and serum samples of healthy individuals, those with potentially malignant conditions, and patients living with oral cancer. Salivary antioxidant levels can reliably and non-invasively detect OSCC changes reliably and non-invasively.
Gholizadeh, et al.81 (2020) explored the patterns of salivary and plasma lactate dehydrogenase (LDH) levels in healthy controls, OSCC, oral lichen planus, and oral lichenoid eruption. LDH concentrations were considerably higher in patients with OSCC, which is crucial for the maintenance of a healthy oral mucosa. Thus, salivary LDH levels indicate mucosal epithelial damage and metastasis. Dineshkumar, et al.82 (2016) observed that the malignant and OSCC groups had higher saliva, serum IL-6, and pro- and anti-inflammatory cytokine levels than the groups composed of healthy patients and those with premalignant lesions. The identified biomarkers did not differ significantly across non-invasive and invasive sample types. Additional research has compared salivary and serum levels of IL-10, VEGF, TNF alpha, and TNF-β cytokines in healthy individuals. In the OSCC group, saliva and serum showed significantly higher levels of all the potential biomarkers. Thus, salivary cytokines may serve as prognostic biomarkers for oral cancer detection and prognosis biomarkers.83 Other cytokines found in the saliva and serum include IL-8, IL-1β, and eotaxin.84 Saliva and serum tetranectin levels were investigated in healthy controls and patients with OSCC at early and late metastatic stages. Saliva and serum tetranectin levels were considerably lower (p=0.007) in patients with OSCC and subsequent metastases. Matrix metalloproteinases (MMPs) are enzymes responsible for extracellular matrix degradation. MMP levels have been studied in the saliva and serum of OSCC and healthy groups, and they were found to be significantly upregulated in both saliva and serum samples of the OSCC group.85
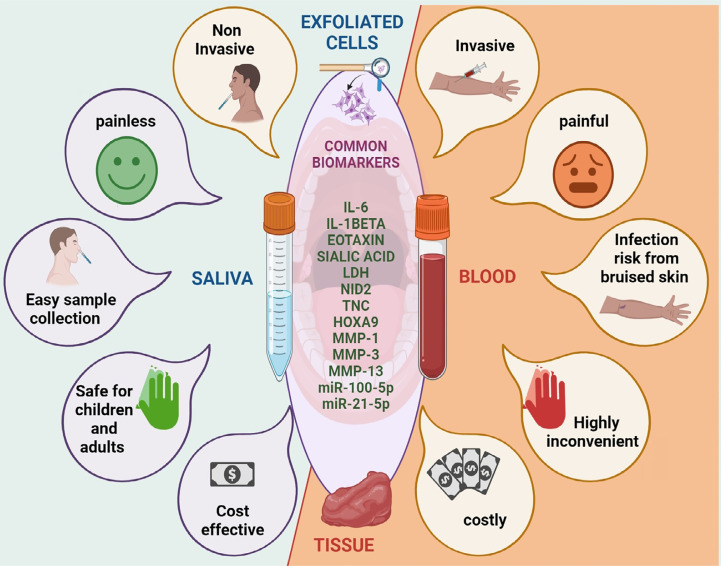
Saliva miRNA profiling is a promising non-invasive oral cancer diagnosis approach. Cao, et al.86 (2018) conducted methylation-specific PCR to compare miRNA promoter methylation in patients with HNSCC and controls. They identified seven methylated genomic loci encoding miRNAs (mgmiRNA) that were altered in saliva and tissues, including mgmiR9-1, mgmiR124-3, mgmiR124-2, mgmiR124-1, mgmiR137, mgmiR129-2, and mgmiR148a. According to this study, a panel of salivary mgmiRNAs may help diagnose HNSCC.86 Systematic studies have evaluated and compared diverse oral cancer biomarkers using invasive and non-invasive sample collection since they carry common biomarkers (Figure 2).
Figure 2. Comparison of non-invasive and invasive biomarkers-Comparison of non-invasive and invasive biomarkers: The utilization of non-invasive approaches such as saliva and oral exfoliated cells conquer the drawbacks associated with invasive methods such as tissue biopsy and fine-needle aspiration cytology (FNAC). The painless, convenient, and safe nature of non-invasive sample collection methods provides an advantage over the painful, inconvenient, and risky aspects associated with invasive methods.
Overall, the various tissues and fluids found in the human body provide essential information about health and disease. The diagnostic efficacy of saliva can be considered remarkable within the realm of non-invasive diagnostic procedures. Saliva shows promise as a non-invasive diagnostic tool for detecting oral cancer. Saliva analysis demonstrates shared biomarkers with invasive techniques, including sialic acid, LDH, cytokines, and miRNAs, highlighting its diagnostic effectiveness and promise as a substitute for invasive treatments.
Validation and Demerits to be taken care of for saliva use
Saliva is a valuable medium for investigating functional indicators associated with disease development because of its non-invasive nature, accessible sample collection, processing, and portability. However, validating small amounts of molecular biomarkers in saliva requires more sophisticated technologies than in other bodily fluids. Various methods have been established for this purpose, but saliva shows limitations such as potential cross-contamination and variability in composition. This section describes the validation and drawbacks of using saliva as the diagnostic medium.
Salivary validation points to be concerned
For saliva sample collection and processing to yield accurate and reliable results, the following guidelines should be followed:
To prevent contamination, patients should refrain from eating or drinking for at least two hours before to sample collection.
Sample collection should be taken from 8 a.m. to 10 a.m. to maintain the circadian rhythm, which can affect biomarker levels.
Saliva samples should avoid mucous contamination, which can affect the analysis.
Saliva samples should be processed and stored as soon as possible to avoid biomarker degradation.
When processing saliva samples, they should be stored at room temperature for immediate processing, 4°C for processing samples within 3–4 h, and −80°C for processing samples after several days to a month.
The number of freeze-thaw cycles should be reduced in the sample because they can cause deterioration of organic contents.
The sample should not be exposed to air, aiming to prevent RNA degradation caused by RNAse activity.
Validation of salivary biomarkers using advanced technology should employ mass spectrometry, polyacrylamide gel electrophoresis, and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) for conventional protein analysis, as well as advanced polymerase chain reaction and microarray for DNA and RNA elucidation. Gas chromatography-mass spectrometry (GC-MS) and microbiomes should be analyzed with next-generation sequencing.87
Demerits of Saliva
Salaried biomarkers are less concentrated than those in other biofluids, making detection difficult, even with advanced technologies.
Accurate saliva collection requires sensitive devices.
The process of biomolecule transfer between blood and saliva is poorly understood, complicating salivary biomarker studies.3
Conclusion
Oral cancer includes cancers that affect the oral cavity, with OSCC being the most common subtype. OSCC is an epithelial neoplasia with high morbidity and mortality rates and low five-year survival rate. Several factors, including late-stage detection and lack of adequate knowledge regarding early alterations, are responsible for most of the health burden associated with oral cancer. These factors, in turn, lead to the progression of oral cancer. Alterations in genetic and epigenetic material are among the early processes that can contribute to the establishment of metastatic cancer, which is both complicated and subtle. These changes are caused by several factors, including mutations, radiation, dental caries, alcohol consumption, and tobacco consumption.88 The incidence of oral cancer is ten times more prevalent among individuals who smoke tobacco products. According to the findings of several scientific studies, tobacco consumption is directly linked to the development of mouth cancer. This is due to the fact that tobacco contains 60 known carcinogenic compounds, which contribute to the development of oral cancer. Screening for altered biomarkers is essential for detecting oral cancer. The current diagnostic methods for oral cancer, including tissue biopsy and fine needle aspiration cytology, show limitations such as invasiveness, cost, and the need for trained personnel. Saliva has been used for the screening and diagnosis of various diseases, including drug abuse, viral infections, autoimmune disorders, oral cancer, and periodontal diseases. The use of saliva, which is non-invasive, holds potential to serve as an alternative to invasive treatments. This body fluid is easily collected, accessible, and stored, in addition to being in immediate contact with oral cancer lesions. Saliva shows a molecular profile that can be used to detect and screen for cancer-associated mutations, as it contains all biomarkers. Thus, it allows the reading of diagnostic areas, including genomics, transcriptomics, proteomics, metabolomics, and metagenomics. The early diagnosis approach assists in treating cancer effectively and on time, helping combat the critical features of cancer. Screening tobacco consumers for early biomarkers might play a significant role. The application of salivaomics, which offers a holistic approach, enables the use of salivary biomarkers in the diagnosis and treatment of oral cancer and other disorders. Non-invasive saliva-based biosensors offer a promising diagnostic approach. These biosensors demonstrate high sensitivity, affordability, and painless detection, making them advantageous for early oral cancer detection. Studies have used various biomarkers and materials, such as salivary CYFRA-21-1 and L-cysteine capped lanthanum hydroxide nanostructures, for biosensor development, with promising results.89,90 Additionally, saliva-based biosensors show potential for detecting other health conditions, including kidney disorders and periodontal health issues, further highlighting their versatility and potential impact in clinical practice.
There are few limitations associated with saliva, such as the possibility of cross-contamination and fluctuations in composition; nonetheless, the benefits of saliva as a non-invasive diagnostic source for oral cancer outweigh these drawbacks. More research needs to be conducted using non-invasive methods for oral cancer detection at an early stage. In this context, saliva may represent a key diagnostic medium and our knowledge on it needs to be expanded. Further research is required to establish protocols for the diagnosis and treatment of oral cancer, including metastatic progression, to ensure its success. This will pave the way for the development of saliva-based biosensors, which will enhance the early identification of oral cancer in vulnerable populations, such as tobacco smokers.
Acknowledgements
We would like to thank the Centre for Medical Biotechnology, Maharshi Dayanand University, Rohtak, for providing support for this work.
Footnotes
Data availability statement
All data generated or analyzed during this study are included in this published article.
Abbreviations: ATM: Ataxia-telangiectasia, mutated.
ATR: Ataxia-Telangiectasia Rad3-related
ctDNA: circulating tumor DNA
DNA: Deoxyribonucleic acid
GABA: Gamma-aminobutyric acid
HNSCC: Head and neck squamous cell carcinoma
HPV: human papillomavirus
HIV: Human Immunodeficiency Virus
IL: Interleukins
LDH: Lactate dehydrogenase
LOH: Loss of heterozygosity
LC-MS: Liquid chromatography- mass spectrometry
mRNA: messenger Ribonucleic acid
miRNA: micro Ribonucleic acid
MMP: Matrix Metalloproteinases
MAPK: Mitogen-activated protein kinase
NMR: Nuclear Magnetic Resonance
OLK: Oral leukoplakia
OLP: Oral lichen planus
OSCC: Oral Squamous Cell Carcinoma
PSMA: prostate-specific membrane antigen
PET/CT: Positron emission tomography–computed tomography
PCR: Polymerase chain reaction
RNA: Ribonucleic acid
SNPs: Single Nucleotide Polymorphism
TNF: Tumor necrosis factor
References
- 1.Yang Z, Liang X, Fu Y, Liu Y, Zheng L, Liu F, et al. Identification of AUNIP as a candidate diagnostic and prognostic biomarker for oral squamous cell carcinoma. EBioMedicine. 2019;47:44–57. doi: 10.1016/j.ebiom.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization . Oral health. Geneva: WHO; 2023. [cited 2023 Dec 29]. https://www.who.int/news-room/fact-sheets/detail/oral-health Internet. [Google Scholar]
- 3.Radhika T, Jeddy N, Nithya S, Muthumeenakshi RM. Salivary biomarkers in oral squamous cell carcinoma: an insight. J Oral Biol Craniofac Res. 2016;6(Suppl 1):S51–S54. doi: 10.1016/j.jobcr.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haghshenas MR, Moosazadeh M, Taghiloo S, Sattari S, Valadan R, Mousavi T. Association between human papillomavirus and oral cancer in iranian clinical samples: a meta-analysis review. Iran J Public Health. 2022;51(12):2688–2696. doi: 10.18502/ijph.v51i12.11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mascitti M, Orsini G, Tosco V, Monterubbianesi R, Balercia A, Putignano A, et al. An overview on current non-invasive diagnostic devices in oral oncology. 1510Front Physiol. 2018;9 doi: 10.3389/fphys.2018.01510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khong B, Ferlito S, Quek S, Conte G, Ingrassia A, Lechien JR, et al. Past, present, and future diagnostic methods for the early noninvasive detection of oral premalignant lesions: a state of the art and systematic review. 1455613241245204Ear Nose Throat J. 2024 doi: 10.1177/01455613241245204. [DOI] [PubMed] [Google Scholar]
- 7.Bano S, David MP, Indira AP. Salivary biomarkers for oral squamous cell carcinoma: an overview. IJSS Case Rep Rev. 2015;1(8):39–45. doi: 10.17354/cr/2015/13. [DOI] [Google Scholar]
- 8.Majem B, Rigau M, Reventós J, Wong DT. Non-coding RNAs in saliva: emerging biomarkers for molecular diagnostics. Int J Mol Sci. 2015;16(4):8676–8698. doi: 10.3390/ijms16048676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schafer CA, Schafer JJ, Yakob M, Lima P, Camargo P, Wong DT. Saliva diagnostics: utilizing oral fluids to determine health status. Monogr Oral Sci. 2014;24:88–98. doi: 10.1159/000358791. [DOI] [PubMed] [Google Scholar]
- 10.Yoshizawa JM, Schafer CA, Schafer JJ, Farrell JJ, Paster BJ, Wong DT. Salivary biomarkers: toward future clinical and diagnostic utilities. Clin Microbiol Rev. 2013;26(4):781–791. doi: 10.1128/CMR.00021-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saxena S, Sankhla B, Sundaragiri KS, Bhargava A. A review of salivary biomarker: a tool for early oral cancer diagnosis. 90Adv Biomed Res. 2017;6 doi: 10.4103/2277-9175.211801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valstar MH, Bakker BS, Steenbakkers RJ, Jong KH, Smit LA, Klein Nulent, et al. et al. The tubarial salivary glands: a potential new organ at risk for radiotherapy. Radiother Oncol. 2021;154:292–298. doi: 10.1016/j.radonc.2020.09.034. [DOI] [PubMed] [Google Scholar]
- 13.Cui Y, Yang M, Zhu J, Zhang H, Duan Z, Wang S, et al. Developments in diagnostic applications of saliva in human organ diseases. 100115Med Nov Technol Devices. 2022;13 doi: 10.1016/j.medntd.2022.100115. [DOI] [Google Scholar]
- 14.Carpenter GH. The secretion, components, and properties of saliva. Annu Rev Food Sci Technol. 2013;4:267–276. doi: 10.1146/annurev-food-030212-182700. [DOI] [PubMed] [Google Scholar]
- 15.Mata AD, Marques DN, Silveira JM, Marques JR, Felino ET, Guilherme NF. Effects of gustatory stimulants of salivary secretion on salivary pH and flow: a randomized controlled trial. Oral Dis. 2009;15(3):220–228. doi: 10.1111/j.1601-0825.2009.01513.x. [DOI] [PubMed] [Google Scholar]
- 16.Kaufman E, Lamster IB. The diagnostic applications of saliva: a review. Crit Rev Oral Biol Med. 2002;13(2):197–212. doi: 10.1177/154411130201300209. [DOI] [PubMed] [Google Scholar]
- 17.Kashyap B, Kullaa A. Salivary metabolites produced by oral microbes in oral diseases and oral squamous cell carcinoma: a review. 277Metabolites. 2024;14(5) doi: 10.3390/metabo14050277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bastías D, Maturana A, Marín C, Martínez R, Niklander SE. Salivary biomarkers for oral cancer detection: an exploratory systematic review. 2634Int J Mol Sci. 2024;25(5) doi: 10.3390/ijms25052634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halicka HD, Bedner E, Darzynkiewicz Z. Segregation of RNA and separate packaging of DNA and RNA in apoptotic bodies during apoptosis. Exp Cell Res. 2000;260(2):248–256. doi: 10.1006/excr.2000.5027. [DOI] [PubMed] [Google Scholar]
- 20.Simpson RJ, Lim JW, Moritz RL, Mathivanan S. Exosomes: proteomic insights and diagnostic potential. Expert Rev Proteomics. 2009;6(3):267–283. doi: 10.1586/epr.09.17. [DOI] [PubMed] [Google Scholar]
- 21.Sreelatha S V, Shetty S, Karnaker VK, Jacob AM, Chowdhury CR. Assaying of p53 autoantibodies in saliva for the detection of oral squamous cell carcinoma: a road not taken. [cited 2024 May 14];Indian J Cancer. 2023 Forthcoming; doi: 10.4103/ijc.ijc_870_20. Internet. [DOI] [PubMed] [Google Scholar]
- 22.Nagata S, Hamada T, Yamada N, Yokoyama S, Kitamoto S, Kanmura Y, et al. Aberrant DNA methylation of tumor-related genes in oral rinse: a noninvasive method for detection of oral squamous cell carcinoma. Cancer. 2012;118(17):4298–4308. doi: 10.1002/cncr.27417. [DOI] [PubMed] [Google Scholar]
- 23.Carvalho AL, Henrique R, Jeronimo C, Nayak CS, Reddy AN, Hoque MO, et al. Detection of promoter hypermethylation in salivary rinses as a biomarker for head and neck squamous cell carcinoma surveillance. Clin Cancer Res. 2011;17(14):4782–4789. doi: 10.1158/1078-0432.CCR-11-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Sjöblom T. Targeting loss of heterozygosity: a novel paradigm for cancer therapy. 57Pharmaceuticals (Basel) 2021;14(1) doi: 10.3390/ph14010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambele MA, van Zyl A, Pepper MS, van Heerden MB, van Heerden WF. Amplification of 3q26.2, 5q14.3, 8q24.3, 8q22.3, and 14q32.33 are possible common genetic alterations in oral cancer patients. 683Front Oncol. 2020;10 doi: 10.3389/fonc.2020.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barlebo Ahlborn L, Østrup O. Toward liquid biopsies in cancer treatment: application of circulating tumor DNA. APMIS. 2019;127(5):329–336. doi: 10.1111/apm.12912. [DOI] [PubMed] [Google Scholar]
- 27.Perdomo S, Avogbe PH, Foll M, Abedi-Ardekani B, Facciolla VL, Anantharaman D, et al. Circulating tumor DNA detection in head and neck cancer: evaluation of two different detection approaches. Oncotarget. 2017;8(42):72621–72632. doi: 10.18632/oncotarget.20004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Springer S, Mulvey CL, Silliman N, Schaefer J, Sausen M, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. 293ra104Sci Transl Med. 2015;7(293) doi: 10.1126/scitranslmed.aaa8507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sethi S, Benninger MS, Lu M, Havard S, Worsham MJ. Noninvasive molecular detection of head and neck squamous cell carcinoma: an exploratory analysis. Diagn Mol Pathol. 2009;18(2):81–87. doi: 10.1097/PDM.0b013e3181804b82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shah FD, Begum R, Vajaria BN, Patel KR, Patel JB, Shukla SN, et al. A review on salivary genomics and proteomics biomarkers in oral cancer. Indian J Clin Biochem. 2011;26(4):326–334. doi: 10.1007/s12291-011-0149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yen CY, Liu SY, Chen CH, Tseng HF, Chuang LY, Yang CH, et al. Combinational polymorphisms of four DNA repair genes XRCC1, XRCC2, XRCC3, and XRCC4 and their association with oral cancer in Taiwan. J Oral Pathol Med. 2008;37(5):271–277. doi: 10.1111/j.1600-0714.2007.00608.x. [DOI] [PubMed] [Google Scholar]
- 32.Ranganathan K, Sivasankar V. MicroRNAs: biology and clinical applications. J Oral Maxillofac Pathol. 2014;18(2):229–234. doi: 10.4103/0973-029X.140762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghizoni JS, Nichele R, de Oliveira MT, Pamato S, Pereira JR. The utilization of saliva as an early diagnostic tool for oral cancer: microRNA as a biomarker. Clin Transl Oncol. 2020;22(6):804–812. doi: 10.1007/s12094-019-02210-y. [DOI] [PubMed] [Google Scholar]
- 34.Gai C, Camussi F, Broccoletti R, Gambino A, Cabras M, Molinaro L, et al. Salivary extracellular vesicle-associated miRNAs as potential biomarkers in oral squamous cell carcinoma. 439BMC Cancer. 2018;18(1) doi: 10.1186/s12885-018-4364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salazar-Ruales C, Arguello JV, López-Cortés A, Cabrera-Andrade A, García-Cárdenas JM, Guevara-Ramírez P, et al. Salivary MicroRNAs for early detection of head and neck squamous cell carcinoma: a case-control study in the high altitude mestizo ecuadorian population. 9792730Biomed Res Int. 2018;2018 doi: 10.1155/2018/9792730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yap T, Koo K, Cheng L, Vella LJ, Hill AF, Reynolds E, et al. Predicting the presence of oral squamous cell carcinoma using commonly dysregulated MicroRNA in oral swirls. Cancer Prev Res (Phila) 2018;11(8):491–502. doi: 10.1158/1940-6207.CAPR-17-0409. [DOI] [PubMed] [Google Scholar]
- 37.Al-Rawi NH, Rizvi Z, Mkadmi S, Abu Kou R, Elmabrouk N, Alrashdan MS, et al. Differential expression profile of salivary oncomirnas among smokeless tobacco users. Eur J Dent. 2023;17(4):1215–1220. doi: 10.1055/s-0043-1761191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonne NJ, Wong DT. Salivary biomarker development using genomic, proteomic and metabolomic approaches. 82Genome Med. 2012;4(10) doi: 10.1186/gm383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barros O, D'Agostino VG, Santos LL, Vitorino R, Ferreira R. Shaping the future of oral cancer diagnosis: advances in salivary proteomics. Expert Rev Proteomics. 2024;21(4):149–168. doi: 10.1080/14789450.2024.2343585. [DOI] [PubMed] [Google Scholar]
- 40.Riccardi G, Bellizzi MG, Fatuzzo I, Zoccali F, Cavalcanti L, Greco A, et al. Salivary biomarkers in oral squamous cell carcinoma: a proteomic overview. 37Proteomes. 2022;10(4) doi: 10.3390/proteomes10040037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sajan T, Murthy S, Krishnankutty R, Mitra J. A rapid, early detection of oral squamous cell carcinoma: Real time PCR based detection of tetranectin. Mol Biol Res Commun. 2019;8(1):33–40. doi: 10.22099/mbrc.2019.31544.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rao M, Ramesh A, Adapa S, Thomas B, Shetty J. Correlation of salivary levels of interleukin-6 and albumin with oral squamous cell carcinoma. J Health Res Rev. 2016;3(1):11–14. doi: 10.4103/2394-2010.17749. [DOI] [Google Scholar]
- 43.Li SX, Yang YQ, Jin LJ, Cai ZG, Sun Z. Detection of survivin, carcinoembryonic antigen and ErbB2 level in oral squamous cell carcinoma patients. Cancer Biomark. 2016;17(4):377–382. doi: 10.3233/CBM-160651. [DOI] [PubMed] [Google Scholar]
- 44.Feng Y, Li Q, Chen J, Yi P, Xu X, Fan Y, et al. Salivary protease spectrum biomarkers of oral cancer. 7Int J Oral Sci. 2019;11(1) doi: 10.1038/s41368-018-0032-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramya AS, Uppala D, Majumdar S, Ch Surekha, Deepak KG. Are salivary amylase and pH - prognostic indicators of cancers? J Oral Biol Craniofac Res. 2015;5(2):81–85. doi: 10.1016/j.jobcr.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Awasthi N. Role of salivary biomarkers in early detection of oral squamous cell carcinoma. Indian J Pathol Microbiol. 2017;60(4):464–468. doi: 10.4103/IJPM.IJPM_140_16. [DOI] [PubMed] [Google Scholar]
- 47.Vedam VKV, Boaz K, Natarajan S, Ganapathy S. Salivary amylase as a marker of salivary gland function in patients undergoing radiotherapy for oral cancer. J Clin Lab Anal. 2017;31(3):e22048. doi: 10.1002/jcla.22048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prasad G, McCullough M. Chemokines and cytokines as salivary biomarkers for the early diagnosis of oral cancer. 813756Int J Dent. 2013;2013 doi: 10.1155/2013/813756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deepthi G, Nandan SR, Kulkarni PG. Salivary tumour necrosis factor-a as a biomarker in oral leukoplakia and oral squamous cell carcinoma. Asian Pac J Cancer Prev. 2019;20(7):2087–2093. doi: 10.31557/APJCP.2019.20.7.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Azizi A, Dabirmoghadam P, Keykha F. Comparison of the Concentration of Salivary IL-8 in patients with oral squamous cell carcinoma and healthy subjects. J Res Dent Maxillofac Sci. 2016;1(3):28–32. doi: 10.18869/acadpub.jrdms.1.3.28. [DOI] [Google Scholar]
- 51.Mihara M, Shintani S, Nakahara Y, Kiyota A, Ueyama Y, Matsumura T, et al. Overexpression of CDK2 is a prognostic indicator of oral cancer progression. Jpn J Cancer Res. 2001;92(3):352–360. doi: 10.1111/j.1349-7006.2001.tb01102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verma D, Garg PK, Dubey AK. Insights into the human oral microbiome. Arch Microbiol. 2018;200(4):525–540. doi: 10.1007/s00203-018-1505-3. [DOI] [PubMed] [Google Scholar]
- 53.McLean JS. Advancements toward a systems level understanding of the human oral microbiome. 98Front Cell Infect Microbiol. 2014;4 doi: 10.3389/fcimb.2014.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee WH, Chen HM, Yang SF, Liang C, Peng CY, Lin FM, et al. Bacterial alterations in salivary microbiota and their association in oral cancer. 16540Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-16418-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mager DL, Haffajee AD, Delvin PM, Norris CM, Posner MR, Goodson JM. The salivary microbiota as a diagnostic indicator of oral cancer: A descriptive, non-randomized study of cancer-free and oral squamous cell carcinoma subjects. 27J Transl Med. 2005;3 doi: 10.1186/1479-5876-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belibasakis GN, Senevirantne CJ, Jayasinghe RD, Vo PT, Bostanci N, Choi Y. Bacteriome and mycobiome dysbiosis in oral mucosal dysplasia and oral cancer. [cited 2024 May 10];Periodontol 2000. 2024 Forthcoming; doi: 10.1111/prd.12558. Internet. [DOI] [PubMed] [Google Scholar]
- 57.Gopinath D, Kunnath Menon R, Chun Wie C, Banerjee M, Panda S, Mandal D, et al. Salivary bacterial shifts in oral leukoplakia resemble the dysbiotic oral cancer bacteriome. 1857998J Oral Microbiol. 2020;13(1) doi: 10.1080/20002297.2020.1857998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang CY, Yeh YM, Yu HY, Chin CY, Hsu CW, Liu H, et al. Oral microbiota community dynamics associated with oral squamous cell carcinoma staging. 862Front Microbiol. 2018;9 doi: 10.3389/fmicb.2018.00862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmidt BL, Kuczynski J, Bhattacharya A, Huey B, Corby PM, Queiroz ELS, et al. Changes in abundance of oral microbiota associated with oral cancer. PLoS One. 2014;9(6):e98741. doi: 10.1371/journal.pone.0098741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thomas AM, Gleber-Netto FO, Fernandes GR, Amorim M, Barbosa LF, Francisco AL, et al. Alcohol and tobacco consumption affects bacterial richness in oral cavity mucosa biofilms. 250BMC Microbiol. 2014;14 doi: 10.1186/s12866-014-0250-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mikkonen JJ, Singh SP, Herrala M, Lappalainen R, Myllymaa S, Kullaa AM. Salivary metabolomics in the diagnosis of oral cancer and periodontal diseases. J Periodontal Res. 2016;51(4):431–437. doi: 10.1111/jre.12327. [DOI] [PubMed] [Google Scholar]
- 62.Wei J, Xie G, Zhou Z, Shi P, Qiu Y, Zheng X, et al. Salivary metabolite signatures of oral cancer and leukoplakia. Int J Cancer. 2011;129(9):2207–2217. doi: 10.1002/ijc.25881. [DOI] [PubMed] [Google Scholar]
- 63.Lohavanichbutr P, Zhang Y, Wang P, Gu H, Nagana Gowda GA, Djukovic D, et al. Salivary metabolite profiling distinguishes patients with oral cavity squamous cell carcinoma from normal controls. PLoS One. 2018;13(9):e0204249. doi: 10.1371/journal.pone.0204249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010;6(1):78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang Q, Gao P, Wang X, Duan Y. Investigation and identification of potential biomarkers in human saliva for the early diagnosis of oral squamous cell carcinoma. Clin Chim Acta. 2014;427:79–85. doi: 10.1016/j.cca.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 66.Wang Q, Gao P, Wang X, Duan Y. The early diagnosis and monitoring of squamous cell carcinoma via saliva metabolomics. 6802Sci Rep. 2014;4 doi: 10.1038/srep06802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ohshima M, Sugahara K, Kasahara K, Katakura A. Metabolomic analysis of the saliva of Japanese patients with oral squamous cell carcinoma. Oncol Rep. 2017;37(5):2727–2734. doi: 10.3892/or.2017.5561. [DOI] [PubMed] [Google Scholar]
- 68.Sridharan G, Ramani P, Patankar S, Vijayaraghavan R. Evaluation of salivary metabolomics in oral leukoplakia and oral squamous cell carcinoma. J Oral Pathol Med. 2019;48(4):299–306. doi: 10.1111/jop.12835. [DOI] [PubMed] [Google Scholar]
- 69.Xiao C, Song F, Zheng YL, Lv J, Wang QF, Xu N. Exosomes in head and neck squamous cell carcinoma. 894Front Oncol. 2019;9 doi: 10.3389/fonc.2019.00894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adeola HA, Holmes H, Temilola DO. Sridharan G.editor . Oral cancer: current concepts and future perspectives. London: IntechOpen; 2022. [cited 2024 July 10]. Diagnostic potential of salivary exosomes in oral cancer. Internet. [DOI] [Google Scholar]
- 71.Wang J, Jing J, Zhou C, Fan Y. Emerging roles of exosomes in oral diseases progression. 4Int J Oral Sci. 2024;16(1) doi: 10.1038/s41368-023-00274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zlotogorski-Hurvitz A, Dekel BZ, Malonek D, Yahalom R, Vered M. FTIR-based spectrum of salivary exosomes coupled with computational-aided discriminating analysis in the diagnosis of oral cancer. J Cancer Res Clin Oncol. 2019;145(3):685–694. doi: 10.1007/s00432-018-02827-6. [DOI] [PubMed] [Google Scholar]
- 73.Gai C, Camussi F, Broccoletti R, Gambino A, Cabras M, Molinaro L, et al. Salivary extracellular vesicle-associated miRNAs as potential biomarkers in oral squamous cell carcinoma. 439BMC Cancer. 2018;18(1) doi: 10.1186/s12885-018-4364-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He L, Ping F, Fan Z, Zhang C, Deng M, Cheng B, et al. Salivary exosomal miR-24-3p serves as a potential detective biomarker for oral squamous cell carcinoma screening. 109553Biomed Pharmacother. 2020;121 doi: 10.1016/j.biopha.2019.109553. [DOI] [PubMed] [Google Scholar]
- 75.Winck FV, Ribeiro AC, Domingues RR, Ling LY, Riaño-Pachón DM, Rivera C, et al. Insights into immune responses in oral cancer through proteomic analysis of saliva and salivary extracellular vesicles. 16305Sci Rep. 2015;5 doi: 10.1038/srep16305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zlotogorski-Hurvitz A, Dayan D, Chaushu G, Salo T, Vered M. Morphological and molecular features of oral fluid-derived exosomes: oral cancer patients versus healthy individuals. J Cancer Res Clin Oncol. 2016;142(1):101–110. doi: 10.1007/s00432-015-2005-3. [DOI] [PubMed] [Google Scholar]
- 77.Arantes LM, Carvalho AC, Melendez ME, Carvalho AL. Serum, plasma and saliva biomarkers for head and neck cancer. Expert Rev Mol Diagn. 2018;18(1):85–112. doi: 10.1080/14737159.2017.1404906. [DOI] [PubMed] [Google Scholar]
- 78.Dadhich M, Prabhu V, Pai VR, D'Souza J, Harish S, Jose M. Serum and salivary sialic acid as a biomarker in oral potentially malignant disorders and oral cancer. Indian J Cancer. 2014;51(3):214–218. doi: 10.4103/0019-509X. [DOI] [PubMed] [Google Scholar]
- 79.Sartini D, Pozzi V, Renzi E, Morganti S, Rocchetti R, Rubini C, et al. Analysis of tissue and salivary nicotinamide N-methyltransferase in oral squamous cell carcinoma: basis for the development of a noninvasive diagnostic test for early-stage disease. Biol Chem. 2012;393(6):505–511. doi: 10.1515/hsz-2012-0112. [DOI] [PubMed] [Google Scholar]
- 80.Bhat S, Babu S, Bhat S, Castelino R, Rao K, Madi M. Status of serum and salivary ascorbic acid in oral potentially malignant disorders and oral cancer. Indian J Med Paediatr Oncol. 2017;38(3):306–310. doi: 10.4103/ijmpo.ijmpo_67_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gholizadeh N, Alipanahi Ramandi M, Motiee-Langroudi M, Jafari M, Sharouny H, Sheykhbahaei N. Serum and salivary levels of lactate dehydrogenase in oral squamous cell carcinoma, oral lichen planus and oral lichenoid reaction. 314BMC Oral Health. 2020;20(1) doi: 10.1186/s12903-020-01306-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dineshkumar T, Ashwini BK, Rameshkumar A, Rajashree P, Ramya R, Rajkumar K. Salivary and serum Interleukin-6 levels in Oral premalignant disorders and squamous cell carcinoma: diagnostic value and clinicopathologic correlations. Asian Pac J Cancer Prev. 2016;17(11):4899–4906. doi: 10.22034/APJCP.2016.17.11.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Polz-Dacewicz M, Strycharz-Dudziak M, Dworzanski J, Stec A, Kocot J. Salivary and serum IL-10, TNF-a, TGF-ß, VEGF levels in oropharyngeal squamous cell carcinoma and correlation with HPV and EBV infections. 45Infect Agent Cancer. 2016;11 doi: 10.1186/s13027-016-0093-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee LT, Wong YK, Hsiao HY, Wang YW, Chan MY, Chang KW. Evaluation of saliva and plasma cytokine biomarkers in patients with oral squamous cell carcinoma. Int J Oral Maxillofac Surg. 2018;47(6):699–707. doi: 10.1016/j.ijom.2017.09.016. [DOI] [PubMed] [Google Scholar]
- 85.Ghanbarnia T, Seyedmajidi M, Mehryari M, Bijani A, Pooramir M, Foroughi R. Diagnostic value of serum and saliva matrix metalloproteinase13 (MMP13) in oral squamous cell carcinoma. J Babol Univ Med Sci. 2020;22:39–44. [Google Scholar]
- 86.Cao Y, Green K, Quattlebaum S, Milam B, Lu L, Gao D, et al. Methylated genomic loci encoding microRNA as a biomarker panel in tissue and saliva for head and neck squamous cell carcinoma. 43Clin Epigenetics. 2018;10 doi: 10.1186/s13148-018-0470-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pathiyil V, Udayasankar R. Gokul S.editor . Saliva and salivary diagnostics. London: IntechOpen; 2019. [cited 2024 July 10]. Salivary diagnostics. Internet. [DOI] [Google Scholar]
- 88.Borse V, Konwar AN, Buragohain P. Oral cancer diagnosis and perspectives in India. 100046Sens Int. 2020;1 doi: 10.1016/j.sintl.2020.100046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kumar S, Panwar S, Kumar S, Augustine S, Malhotra BD. Biofunctionalized nanostructured yttria modified non-invasive impedometric biosensor for efficient detection of oral cancer. 1190Nanomaterials (Basel) 2019;9(9) doi: 10.3390/nano9091190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tiwari S, Gupta PK, Bagbi Y, Sarkar T, Solanki PR. L-cysteine capped lanthanum hydroxide nanostructures for non-invasive detection of oral cancer biomarker. Biosens Bioelectron. 2017;89:1042–1052. doi: 10.1016/j.bios.2016.10.020. Pt 2. [DOI] [PubMed] [Google Scholar]
- 91.Nandakumar A, Nataraj P, James A, Krishnan R, Mahesh KM. Estimation of salivary 8-Hydroxydeoxyguanosine (8-OHdG) as a potential biomarker in assessing progression towards malignancy: a case-control study. Asian Pac J Cancer Prev. 2020;21(8):2325–2329. doi: 10.31557/APJCP.2020.21.8.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.González-Pérez L, Isaza-Guzmán D, Arango-Pérez E, Tobón-Arroyave S. Analysis of salivary detection of P16INK4A and RASSF1A promoter gene methylation and its association with oral squamous cell carcinoma in a Colombian population. J Clin Exp Dent. 2020;12(5):e452–e460. doi: 10.4317/jced.56647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liyanage C, Wathupola A, Muraleetharan S, Perera K, Punyadeera C, Udagama P. Promoter hypermethylation of tumor-suppressor genes p16 ink4a, RASSF1A,TIMP3, and PCQAP/MED15 in salivary dna as a quadruple biomarker panel for early detection of oral and oropharyngeal cancers. 148Biomolecules. 2019;9(4) doi: 10.3390/biom9040148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Samadi FM, Suhail S, Sonam M, Ahmad MK, Chandra S, Saleem M. Telomerase in saliva: an assistant marker for oral squamous cell carcinoma. J Oral Maxillofac Pathol. 2019;23(2):187–191. doi: 10.4103/jomfp.JOMFP_83_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Demokan S, Chang X, Chuang A, Mydlarz WK, Kaur J, Huang P, et al. KIF1A and EDNRB are differentially methylated in primary HNSCC and salivary rinses. Int J Cancer. 2010;127(10):2351–2359. doi: 10.1002/ijc.25248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liao PH, Chang YC, Huang MF, Tai KW, Chou MY. Mutation of p53 gene codon 63 in saliva as a molecular marker for oral squamous cell carcinomas. Oral Oncol. 2000;36(3):272–276. doi: 10.1016/s1368-8375(00)00005-1. [DOI] [PubMed] [Google Scholar]
- 97.Ovchinnikov DA, Cooper MA, Pandit P, Coman WB, Cooper-White JJ, Keith P, et al. Tumor-suppressor gene promoter hypermethylation in saliva of head and neck cancer patients. Transl Oncol. 2012;5(5):321–326. doi: 10.1593/tlo.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kaliyaperumal S, Sankarapandian S. Evaluation of p16 hypermethylation in oral submucous fibrosis: a quantitative and comparative analysis in buccal cells and saliva using real-time methylation-specific polymerase chain reaction. South Asian J Cancer. 2016;5(2):73–79. doi: 10.4103/2278-330X.181645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ovchinnikov DA, Wan Y, Coman WB, Pandit P, Cooper-White JJ, Herman JG, et al. DNA methylation at the novel CpG sites in the promoter of MED15/PCQAP gene as a biomarker for head and neck cancers. Biomark Insights. 2014;9:53–60. doi: 10.4137/BMI.S16199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Shpitzer T, Hamzany Y, Bahar G, Feinmesser R, Savulescu D, Borovoi I, et al. Salivary analysis of oral cancer biomarkers. Br J Cancer. 2009;101(7):1194–1198. doi: 10.1038/sj.bjc.6605290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.El-Naggar AK, Mao L, Staerkel G, Coombes MM, Tucker SL, Luna MA, et al. Genetic heterogeneity in saliva from patients with oral squamous carcinomas: implications in molecular diagnosis and screening. J Mol Diagn. 2001;4(3):164–170. doi: 10.1016/S1525-1578(10)60668-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Don KR, Ramani P, Ramshankar V, Sherlin HJ, Premkumar P, Natesan A. Promoter hypermethylation patterns of P16, DAPK and MGMT in oral squamous cell carcinoma: a systematic review and meta-analysis. Indian J Dent Res. 2014;25(6):797–805. doi: 10.4103/0970-9290.152208. [DOI] [PubMed] [Google Scholar]
- 103.Romani C, Salviato E, Paderno A, Zanotti L, Ravaggi A, Deganello A, et al. Genome-wide study of salivary miRNAs identifies miR-423-5p as promising diagnostic and prognostic biomarker in oral squamous cell carcinoma. Theranostics. 2021;11(6):2987–2999. doi: 10.7150/thno.45157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fadhil RS, Wei MQ, Nikolarakos D, Good D, Nair RG. Salivary microRNA miR-let-7a-5p and miR-3928 could be used as potential diagnostic bio-markers for head and neck squamous cell carcinoma. PLoS One. 2020;15(3):e0221779. doi: 10.1371/journal.pone.0221779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Du J, Gao R, Wang Y, Nguyen T, Yang F, Shi Y, et al. MicroRNA-26a/b have protective roles in oral lichen planus. 15Cell Death Dis. 2020;11(1) doi: 10.1038/s41419-019-2207-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Uma Maheswari TN, Nivedhitha MS, Ramani P. Expression profile of salivary micro RNA-21 and 31 in oral potentially malignant disorders. Braz Oral Res. 2020;34:e002. doi: 10.1590/1807-3107bor-2020.vol34.0002. [DOI] [PubMed] [Google Scholar]
- 107.Salazar-Ruales C, Arguello JV, López-Cortés A, Cabrera-Andrade A, García-Cárdenas JM, Guevara-Ramírez P, et al. Salivary MicroRNAs for early detection of head and neck squamous cell carcinoma: a case-control study in the high altitude mestizo ecuadorian population. 9792730Biomed Res Int. 2018;2018 doi: 10.1155/2018/9792730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gissi DB, Morandi L, Gabusi A, Tarsitano A, Marchetti C, Cura F, et al. A noninvasive test for MicroRNA expression in oral squamous cell carcinoma. Int J Mol Sci. 2018;19(6):1789. doi: 10.3390/ijms19061789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mehdipour M, Shahidi M, Manifar S, Jafari S, Mashhadi Abbas F, Barati M, et al. Diagnostic and prognostic relevance of salivary microRNA-21, -125a, -31 and -200a levels in patients with oral lichen planus: a short report. Cell Oncol (Dordr) 2018;41(3):329–334. doi: 10.1007/s13402-018-0372-x. [DOI] [PubMed] [Google Scholar]
- 110.Shahidi M, Jafari S, Barati M, Mahdipour M, Gholami MS. Predictive value of salivary microRNA-320a, vascular endothelial growth factor receptor 2, CRP and IL-6 in Oral lichen planus progression. Inflammopharmacology. 2017;25(5):577–583. doi: 10.1007/s10787-017-0352-1. [DOI] [PubMed] [Google Scholar]
- 111.Duz MB, Karatas OF, Guzel E, Turgut NF, Yilmaz M, Creighton CJ, et al. Identification of miR-139-5p as a saliva biomarker for tongue squamous cell carcinoma: a pilot study. Cell Oncol (Dordr) 2016;39(2):187–193. doi: 10.1007/s13402-015-0259-z. [DOI] [PubMed] [Google Scholar]
- 112.Zahran F, Ghalwash D, Shaker O, Al-Johani K, Scully C. Salivary microRNAs in oral cancer. Oral Dis. 2015;21(6):739–747. doi: 10.1111/odi.12340. [DOI] [PubMed] [Google Scholar]
- 113.Momen-Heravi F, Trachtenberg AJ, Kuo WP, Cheng YS. Genomewide study of salivary MicroRNAs for detection of oral cancer. J Dent Res. 2014;93(7) Suppl:86S–93S. doi: 10.1177/0022034514531018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Salazar C, Nagadia R, Pandit P, Cooper-White J, Banerjee N, Dimitrova N, et al. A novel saliva-based microRNA biomarker panel to detect head and neck cancers. Cell Oncol (Dordr) 2014;37(5):331–338. doi: 10.1007/s13402-014-0188-2. [DOI] [PubMed] [Google Scholar]
- 115.Yang Y, Li YX, Yang X, Jiang L, Zhou ZJ, Zhu YQ. Progress risk assessment of oral premalignant lesions with saliva miRNA analysis. 129BMC Cancer. 2013;13 doi: 10.1186/1471-2407-13-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Oh SY, Kang SM, Kang SH, Lee HJ, Kwon TG, Kim JW, et al. Potential salivary mRNA biomarkers for early detection of oral cancer. 243J Clin Med. 2020;9(1) doi: 10.3390/jcm9010243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ueda S, Hashimoto K, Miyabe S, Hasegawa S, Goto M, Shimizu D, et al. Salivary NUS1 and RCN1 levels as biomarkers for oral squamous cell carcinoma diagnosis. In Vivo. 2020;34(5):2353–2361. doi: 10.21873/invivo.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Michailidou E, Tzimagiorgis G, Chatzopoulou F, Vahtsevanos K, Antoniadis K, Kouidou S, et al. Salivary mRNA markers having the potential to detect oral squamous cell carcinoma segregated from oral leukoplakia with dysplasia. Cancer Epidemiol. 2016;43:112–118. doi: 10.1016/j.canep.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 119.Cheng YSL, Jordan L, Rees T, Chen HS, Oxford L, Brinkmann O, et al. Levels of potential oral cancer salivary mRNA biomarkers in oral cancer patients in remission and oral lichen planus patients. Clin Oral Investig. 2014;18(3):985–993. doi: 10.1007/s00784-013-1041-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Elashoff D, Zhou H, Reiss J, Wang J, Xiao H, Henson B, et al. Prevalidation of salivary biomarkers for oral cancer detection. Cancer Epidemiol Biomarkers Prev. 2012;21(4):664–672. doi: 10.1158/1055-9965.EPI-11-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Cheng YS, Rees T, Wright J. A review of research on salivary biomarkers for oral cancer detection. 3Clin Transl Med. 2014;3(1) doi: 10.1186/2001-1326-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li Y, St John MA, Zhou X, Kim Y, Sinha U, Jordan RC, et al. Salivary transcriptome diagnostics for oral cancer detection. Clin Cancer Res. 2004;10(24):8442–8450. doi: 10.1158/1078-0432.CCR-04-1167. [DOI] [PubMed] [Google Scholar]
- 123.Pickering V, Jordan RC, Schmidt BL. Elevated salivary endothelin levels in oral cancer patients: a pilot study. Oral Oncol. 2007;43(1):37–41. doi: 10.1016/j.oraloncology.2005.12.027. [DOI] [PubMed] [Google Scholar]
- 124.Singh P, Verma JK, Singh JK. Validation of salivary markers, IL-1ß, IL-8 and Lgals3bp for detection of oral squamous cell carcinoma in an Indian population. 7365Sci Rep. 2020;10(1) doi: 10.1038/s41598-020-64494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Radulescu R, Greabu M, Totan A, Melescanu Imre M, Miricescu D, Didilescu A, et al. Diagnostic Role of Saliva in Oral Cancers. Rev Chim. 2020;71(7):474–480. doi: 10.37358/RC.20.7.8266. [DOI] [Google Scholar]
- 126.Chang YT, Chu LJ, Liu YC, Chen CJ, Wu SF, Chen CH, et al. Verification of saliva matrix metalloproteinase-1 as a strong diagnostic marker of oral cavity cancer. 2273Cancers (Basel) 2020;12(8) doi: 10.3390/cancers12082273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Smriti K, Ray M, Chatterjee T, Shenoy RP, Gadicherla S, Pentapati KC, et al. Salivary MMP-9 as a biomarker for the diagnosis of oral potentially malignant disorders and oral squamous cell carcinoma. Asian Pac J Cancer Prev. 2020;21(1):233–238. doi: 10.31557/APJCP.2020.21.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zielinska K, Karczmarek-Borowska B, Kwasniak K, Czarnik-Kwasniak J, Ludwin A, Lewandowski B, et al. Salivary IL-17A, IL-17F, and TNF- a Are associated with disease advancement in patients with oral and oropharyngeal cancer. 3928504J Immunol Res. 2020 Aug 13;2020 doi: 10.1155/2020/3928504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rathore A, Katyal S, Jain A, Shetty D. Biochemical analysis of cytokeratin fragment 21-1 concentration and expression of cytokeratin 19 in oral potentially malignant disorders. J Cancer Res Ther. 2020;16(3):452–457. doi: 10.4103/jcrt.JCRT_893_17. [DOI] [PubMed] [Google Scholar]
- 130.Sami SM. Evaluation of cytokine expression tumor necrosis alpha and interleukin-6 in the saliva of oral maxillofacial benign fibro-osseous tumors and health control. SP231338Ann Trop Med Public Health. 2020;23(S13A) doi: 10.36295/ASRO.2020.231338. [DOI] [Google Scholar]
- 131.Sharma M, Sharma E, Prabhu V, Pai VR, D'souza JM, Harish S, et al. Salivary L-fucose as a biomarker for oral potentially malignant disorders and oral cancer. J Cancer Res Ther. 2020;16(3):546–550. doi: 10.4103/jcrt.JCRT_552_17. [DOI] [PubMed] [Google Scholar]
- 132.Gulzar RA, Ajitha P, Subbaiyan H. Comparative evaluation on the levels of salivary Mucin MUC1 in precancerous and cancerous conditions. Int J Dentistry Oral Sci. 2021;8(8):3734–3737. doi: 10.19070/2377-8075-21000765. [DOI] [Google Scholar]
- 133.Bhavana VS, Madhura MG, Kumar BV, Suma S, Sarita Y. Detection of salivary heat shock protein 27 by enzyme-linked immunosorbent assay and its correlation with histopathology of oral leukoplakia. J Oral Maxillofac Pathol. 2018;22(3):307–313. doi: 10.4103/jomfp.JOMFP_86_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bhat S, Babu GS, N SK, Madi M, Buch S, Ullal H. Analysis and comparison of salivary L-Fucose and HSP 70 in oral potentially malignant disorders and oral cancer. Indian J Public Health Res Dev. 2020;11(2):29–33. doi: 10.37506/v11/i2/2020/ijphrd/194746. [DOI] [Google Scholar]
- 135.Azeem MS, Yesupatham ST, Mohiyuddin SM, Sumanth V, Ravishankar S. Usefulness of salivary sialic acid as a tumor marker in tobacco chewers with oral cancer. J Cancer Res Ther. 2020;16(3):605–611. doi: 10.4103/jcrt.JCRT_337_1. [DOI] [PubMed] [Google Scholar]