Abstract
Optobiochemical control of protein activities allows investigation of protein functions in living cells with high spatiotemporal resolution. Over the last two decades, numerous natural photosensory domains have been characterized and synthetic domains engineered, then assembled into photoregulatory systmes to control protein function with light. Here we review the field of optobiochemistry, categorizing photosensory domains by chromophore, describing photoregulatory systems by mechanisms of action, and discussing protein classes frequently investigated using optical methods. Finally, we present examples of how spatial or temporal control of proteins in living cells has provided new insights not possible with traditional biochemical or cell biological techniques.
Keywords: optobiochemistry, photosensory domain, signal transduction, optogenetics
INTRODUCTION
Traditional methods for investigating the role of proteins in cell biology have been either disruptive or imprecise. Biochemical assays can quantify catalytic and protein-binding functions of proteins of interest (POI), but do so by breaking the cell and are therefore disruptive. Genetic methods involving the suppression or the overexpression of proteins of interest, or the expression of mutant forms, allow assessment of perturbations on the form or function of the cell, but do not allow the separation of primary and secondary effects and are therefore imprecise.
A revolution in cell biology was initiated by the introduction of fluorescent proteins (FPs) in 1995 (1, 2). The tagging of POIs with FP domains, along with the development of FP-based indicators of biochemical activity, allows real-time observations of how protein localization or enzymatic activity responds to environmental stimuli in living cells (3, 4). However, observations of protein and pathway responses within cells enables only half of the scientific method. Perturbation to test hypotheses about mechanisms and functions represents the other, arguably more important, component of scientific experimentation. Here, to understand protein function in living cells, methods for real-time manipulation of protein expression, localization, or activity are required. In recent years, just as optical methods had previously enabled real-time observation of protein function, optical methods have been developed for real-time control of protein function with high spatiotemporal precision (5–8).
Light-sensitive proteins have been used as optobiochemical tools to modulate diverse protein functions since 2002, beginning with the use of a light-induced interaction by plant phytochromes to regulate transcription in yeast (9). Since then, a variety of photosensory domains have been used for optical regulation of cellular biochemistry (5–8). These domains have been used to regulate fused protein domains via light-induced oligomerization, heterodimerization, dissociation, and conformational change, thereby mimicking many of the mechanisms by which upstream signals naturally regulate proteins. Optical control of protein activities is now advancing our understanding of biochemistry in living cells in two ways that were previously impossible. First, rapid activation of proteins in living cells enables observations of downstream effects with high temporal resolution. This enables more precise quantitation of signaling kinetics and feedback regulation, which is especially useful for building and validating mathematical models of signaling (10). Second, local activation of proteins in living cells enables observations of subcellular effects with high spatial resolution (11). This is especially useful in understanding the role of specific proteins in directional behaviors such as cell migration.
We note that optical control of protein functions is often referred to as optogenetics, but we believe that this is confusing. The term optogenetics was created to describe optical control of the activity of genetically defined neurons as demonstrated in 2005 (12), whereas the use of photosensory domains to control biochemical processes dates to 2002 (9, 17). Thus expanding the term optogenetics from its origins in neuroscience for retroactive continuity with earlier non-neuronal work contradicts a large body of literature recognizing the origin of optogenetics in 2005. Furthermore, describing optical control of biochemistry as optogenetics is a bit of a misnomer, as photons are not modulating genes but rather the biochemical functions of proteins (8). For these reasons, we believe the term optobiochemistry more accurately describes the use of light to control biochemical processes in the cell.
In this review, we will categorize currently available photosensory domains, describe the organizational structure of optobiochemical systems, provide examples of biochemical pathways that are commonly subjected to optical control, and present questions in cell biology that especially benefit from the high spatiotemporal precision provided by optobiochemical control.
PHOTOSENSORY DOMAINS
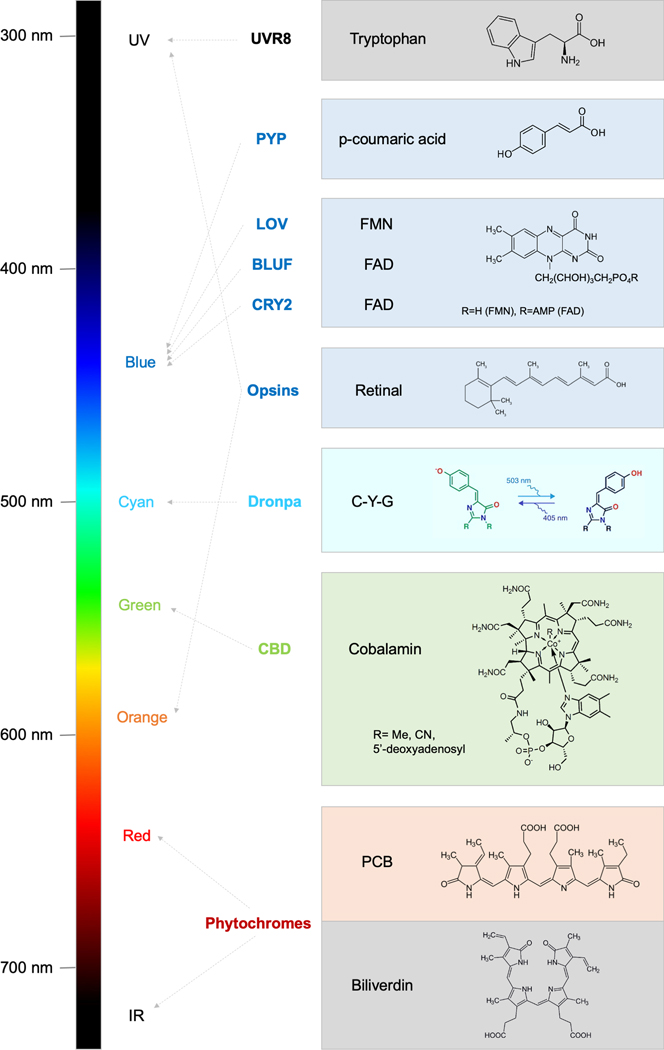
Most known photosensory domains derive from proteins of photosynthetic organisms that transduce photonic stimuli to biochemical responses, but a family of synthetic photosensory domains has also been engineered from non-photosensory domains for the purpose of optobiochemical control. Natural domains include UV-responsive UVR8; photoactive yellow protein (PYP); the flavin-binding blue-light-utilizing FAD-containing (BLUF), light-oxygen-voltage-sensing (LOV), and cryptochrome domains; the cobalamin-binding domain (CBD); and phytochromes (5, 6, 18, 19). The synthetic photoregulatory domains are variants of the green fluorescent protein Dronpa (20, 21). The chromophores of the above domains can be encoded within the protein itself (UVR8 or Dronpa) or supplied in trans as cofactors. The chromophore determines the wavelengths of light used by each photoregulatory domain, and undergoes structural changes that alter the conformation of the protein domain, enabling light absorption to be converted to a change in protein function (Figure 1). Here, we will first describe photosensory domains used for optobiochemical control, categorizing them by chromophore.
Figure 1. Photosensory domains enable control of biology with light from ultraviolet to infrared wavelengths.
Photosensory domains, organized by chromophore structure at right, are connected to their peak absorption wavelengths by arrows. Abbreviations: UV, ultraviolet; IR, infrared; UVR8, UV resistance 8; PYP, photoactive yellow protein; LOV, light-oxygen-voltage-sensing; BLUF, blue-light-utilizing FAD-containing; CRY2, cryptochrome 2; CBD, cobalamin-binding domain; FMN, flavin mononucleotide; FAD, flavin adenine dinucleotide; C-Y-G, cysteine-tyrosine-glycine; PCB, phycocyanobilin.
UVR8
The protein encoded by UV resistance locus 8 (UVR8) in plants and green algae is responsible for inducing photoprotective genes in response to UV-B light (280–310 nm) (22). UVR8 is homodimeric at baseline but monomerizes upon UV-B absorption (23). In Arabidopsis UVR8, a cluster of three tryptophan residues (Trp233, 285, and 337) situated at the dimeric interface functions as the chromophore. Absorption of UV-B light by this tryptophan triad disrupts dimerization-promoting salt bridges by adjacent amino acids (24, 25). The monomerized UVR8 interacts with the E3 ubiquitin ligase COP1, leading to the stabilization of transcription factors HY5 and activation of UV-protection genes. Reversion of UVR8 to its ground state requires an additional plant protein, either RUP1 or RUP2, and occurs over the time course of hours (26). As an optobiochemical tool, UVR8 is unique in using UV light. While UV light is relatively toxic, it does allow for regulation orthogonal to other photosensory domains that use visible wavelengths of light (27).
Photoactive Yellow Protein
Photoactive yellow protein (PYP) is a 14-kD protein from Halorhodospira bacteria containing a Per-Arnt-Sim (PAS) domain bound to a p-hydroxycinnamic acid chromophore (28). PYP functions to mediate negative phototaxis as a photoprotective mechanism, although the signaling pathway is still unknown (29). Absorption of blue light (peak wavelength 450 nm) causes trans-cis isomerization of the chromophore, leading to partial unfolding of the protein and an alteration in its oligomeric state (30). The photoisomerization of the chromophore reverses spontaneously with a half-life of ~1 minute (30).
LOV Domains
The light-oxygen-voltage-sensing (LOV) domain is a subfamily of PAS domains that utilize flavin mononucleotide (FMN) as a chromophore (31). Blue light absorption induces a reversible covalent reaction between a cysteine residue and FMN (32), which then leads to conformational changes in the LOV domain. In Avena phototropin, perhaps the most extensively studied LOV-containing protein, light-induced adduct formation causes the exposure of the C-terminal helix of the LOV2 domain (the Jα helix) (33). This causes the activation of a C-terminal kinase domain and subsequent phototropic gene activation (34). Reversion occurs with a half-life of ~1 minute, but LOV2 mutants with activated-state half-lives of 6 seconds to 20 minutes have been engineered (35).
Another LOV domain, Neurospora Vivid (VVD), contains an N-terminal helix which is exposed by blue light illumination to create a homodimerization interface (36). The activated state of VVD has a relatively long half-life of 5 hours, and this can be further tuned by mutagenesis to 30 seconds to 50 hours (37). Other LOV domains have been found to mediate homodimerization in response to light (38).
LOV domains have also naturally evolved to mediate heterodimerization in response to light. The Arabidopsis LOV-domain protein flavin-binding kelch repeat f-box 1 (FKF1) and its binding partner GIGANTEA (GI) interact in response to blue light. While the isolated FKF1 LOV domain undergoes rapid (~milliseconds) conformational changes in response to light absorption (39), the interaction with GI also requires a portion of FKF1 N-terminal to the LOV domain and occurs relatively slowly (over many minutes) after illumination in cells (40). It also demonstrates negligible reversal after 2 hours, suggesting off-kinetics even slower than VVD (40).
BLUF Domains
Another photosensory domain utilizing a flavin cofactor, in this case flavin adenine dinucleotide (FAD), is the blue-light-utilizing FAD-containing (BLUF) domain. BLUF domains are found in proteobacteria, cyanobacteria, and algae, where they regulate the activity of linked protein domains such as enzymatic function in photoactivated adenylate cyclases and protein-protein interactions (41). BLUF domains noncovalently bind to FAD both in ground and excited states, and blue light induces electron transfer from a tyrosine residue to FAD, i.e. FAD reduction, followed by hydrogen bond rearrangement (42). These subtle conformational changes in the BLUF domain are transduced allosterically to activate linked protein domains (43). Recovery occurs over seconds to minutes (44). For example, in Beggiatoa photoactivated adenylate cyclase (bPAC), light absorption by the BLUF domain mediates the activation of a connected catalytic adenylate cyclase domain. More recently, cyanobacteriochrome-based photoactivated adenylate cyclase (cPAC) was discovered from Microcoleus sp. PCC 7113 (45). cPAC features a bistable photocycle, being activated by blue light and inactivated by green light.
Cryptochromes
Cryptochromes are a family of flavoproteins found in bacteria, fungi, animals and plants. The cryptochrome photolyase homology region (PHR) binds noncovalently to a FAD molecule as the chromophore (46). Blue light (450–500 nm) causes reduction of FAD followed by conformational changes that alter protein-protein interactions. For example, Arabidopsis cryptochrome 2 (CRY2) interacts with the crytochrome-interacting basic helix-loop-helix transcription factor (CIB1) to modulate gene expression upon illumination (47). CRY2 also homo-oligomerizes upon light absorption (48). Reversion occurs spontaneously in the dark with a half-life of ~5 minutes (49), but mutants have been identified with half-lives of 2.5 to 24 min (50). Interestingly the lesser-studied Arabidopsis cryptochrome 1 (CRY1) also undergoes a light-induced heterodimerization, in this case with phytochrome B (PhyB), in a manner dependent on CRY1 sequences outside the PHR (51).
Opsins
Opsins are 7-transmembrane proteins that bind covalently to a retinal cofactor and that transport ions or transduce signals inside cells in response to absorption of light from UV to orange wavelengths (52). The complex of opsin and its cofactor is termed rhodopsin. In microbial rhodopsins, photon absorption triggers photoisomerization of the chromophore retinal from all-trans to 13-cis, which then leads to a series of conformational changes that drive engagement with signaling proteins, ionic pumping, or passive ionic flow (53). Microbial rhodopsins that passively pass or actively pump ions have functions in phototaxis and generating transmembrane ion gradients (54). Other microbial rhodopsins such as sensory rhodopsin couple light reception to bacterial signal transduction (55). A wide variety of microbial ion-conducting rhodopsins are used in neuroscience to alter the membrane potential and thus control neuronal firing in genetically defined neurons in response to light, in a technique termed optogenetics (12). As optogenetics is extensively reviewed elsewhere (56), and its implementation and application are mostly distinct from optobiochemical control in non-neuronal cells, we will not cover it further here.
In metazoan rhodopsins, such as those in mammalian eyes, photon absorption causes isomerization of 11-cis to all-trans retinal and subsequent activation of transducin-class G (Gt) proteins (18). Interestingly, the reverse process is not spontaneous for metazoan rhodopsins, but requires additional proteins to replace all-trans retinal with 11-cis retinal (57). The mechanisms for G protein coupling are conserved between metazoan opsins and ligand-activated G-protein-coupled receptors (GPCRs), so that the intracellular segments of opsins can be swapped with those of ligand-activated G protein-coupled receptors to couple to other classes of G proteins (58–61). These engineered opsins have been further modified to mediate only G-protein or β-arrestin signaling by introducing mutations on β-arrestin binding site (Ser-Ser in the C-terminal tail) or G-protein interacting motif (DRY in the intracellular loop regions) (62). The main use for engineered G-protein-coupled opsins is in neuroscience, where they are used in a true optogenetic way to activate specific G-protein-mediated pathways that modulate neuronal activity positively or negatively in genetically defined neuronal populations.
Dronpa
Dronpa is a reversibly photoswitchable green fluorescent protein whose fluorescence is switched off by 500 nm cyan light and turned on by 400 nm violet light (63). The chromophore of Dronpa, formed by an autocatalytic reaction of Cys62, Tyr63, and Gly64, is in a cis conformation in the fluorescent on state (64). When the chromophore absorbs cyan light, it both fluorescences and, with lower efficiency, is isomerized into the trans conformation (64). This isomerization causes protonation of the chromophore to shift its absorbance away from cyan light and toward violet light, so that over time cyan illumination drives the entire population of chromophores to the trans state. Interestingly, the trans state is associated with partial unfolding of the side of the beta barrel that forms a major dimerization interface (65). A tetrameric fluorescent mutant, DronpaN, was shown to be monomerized when switched off (20), and was further engineered into a photodissociable dimeric Dronpa (pdDronpa) (21). The affinity of the DronpaN tetramer and pdDronpa dimer is relatively low, with dissociation constants in the low-micromolar range, but this low affinity is well suited for their use for steric occlusion when multiple copies are attached to a single polypeptide chain (21). Violet light causes both reactivation of green fluorescence and allows redimerization of the pdDronpa. pdDronpa reverts to the fluorescent dimeric state with a half-life of ~30 minutes (21). DronpaN and pdDronpa are able to report their own oligomeric state through the intrinsic green fluorescence of the baseline bound state. They are also the only photosensory domains regulated by visible light that do not require chemical cofactors.
Cobalamin-binding Domains
The cobalamin-binding domain (CBD) is a photosensory domain from bacterial proteins that regulate gene expression in response to green light and use cobalamin (Vitamin B12) as the chromophore (66). A CBD was first characterized within Myxococcus CarH, a regulator of carotenoid gene transcription (67). In dark, CBD mediates CarH tetramerization, allowing it to bind to carotenoid promoters and repress transcription. Upon absorption of green light (peak 545 nm), CBD monomerizes, inducing release of CarH from DNA and thereby gene activation. Myxococcus CBD apparently relaxes to its tetrameric state with a half-life of ~30 minutes (68). Interestingly, the CBD of Thermus CarH forms a covalent adduct with cobalamin via a histidine residue in response to green light absorption, and thus appears to be irreversible (69). The green absorption of CBD allows it to be used orthogonally from photosensory domains that use blue light.
Phytochromes
Phytochromes, found in plants, fungi, and bacteria, are proteins that are covalently conjugated to tetrapyrrole cofactors through one or two cysteine side chains. Phytochromes absorb green to infrared light, depending on the particular site of cysteine conjugation. Light absorption induces isomerization between the C and D rings of the tetrapyrrole, which causes a shift in absorption wavelength and a change in protein conformation. This conformational change can modulate protein-protein interactions or modulate the activity of a linked enzymatic domain.
For example, the plant phytochromes such as PhyA and PhyB naturally utilize phytochromobilin, a tetrapyrrole synthesized exclusively in plants, or can use phycocyanobilin (PCB), a tetrapyrrole synthesized by cyanobacteria and photosynthetic algae. PhyA and PhyB absorb red light (peak 660 nm) to be activated, and reversion occurs upon illumination of infrared light (peak 720 nm) or spontaneously with a half-life of ~3 seconds (70). In the activated state, PhyA and PhyB bind to phytochrome interacting factors (PIFs). In comparison, the bacterial phytochrome BphP1 uses biliverdin as a chromophore and absorbs 760-nm infrared light at baseline and 640-nm red light after activation. Once activated, BphP1 binds to PpsR2, and this interaction reverses upon 640-nm illumination or spontaneously in ~15 minutes (71).
Some phytochromes transduce light-induced conformational changes allosterically to activate a linked enzyme domain. A family of cyanobacterial phytochromes (cyanobacteriochromes) use two cysteine residues to conjugate to the PCB chromophore, causing absorption at violet wavelengths (peak ~400 nm) (72). Photoisomerization shifts peak absorption to green–orange wavelengths (520–600 nm). These changes can be coupled to enzymatic regulation, e.g. the Fremyella cyanobacteriochrome-based photoswitchable adenylate cyclase (cPAC) is activated by blue light and inactivated by green light (45).
The red or far-red-absorbing phytochromes have the advantage of using wavelengths of light orthogonal to other optobiochemical systems. Red and far-red light is also less phototoxic than blue and green light at equivalent powers. The major disadvantage is that efficacy outside of their native environments may be limited by cofactor availability and toxicity, especially for the plant phytochromes. However, it may be possible to express cyanobacterial enzymes to produce phycocyanobilin in mammalian cells, where the best results are achieved by short bouts of regulated expression (73).
GENERALIZABLE PHOTOREGULATORY MECHANISMS
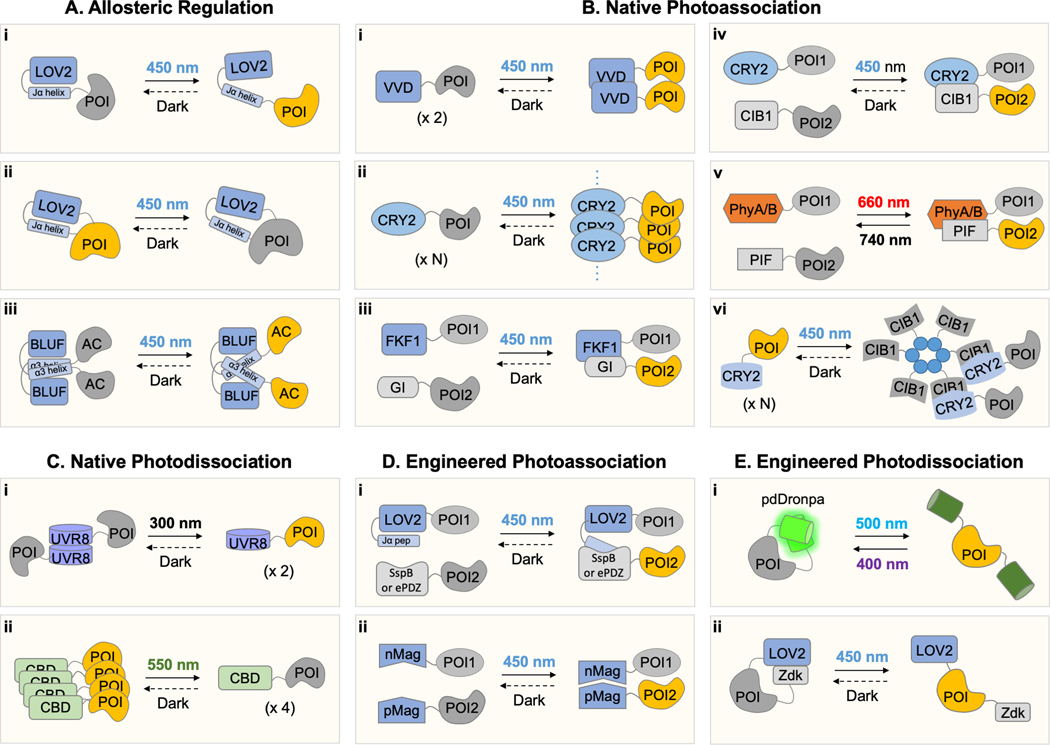
In this section, we review how basic mechanisms of protein regulation can be recapitulated with optical regulation by photosensory domains (Figure 2). The conformational changes in photosensory domains induced by light can be directly transduced allosterically to change the activity of a protein domain of interest. Alternatively, these conformational changes can lead to domain oligomerization, heterodimerization, or monomerization, either naturally or via engineered interactions. Photoregulation of protein-protein interaction can then be used to translocate a target protein to a specific subcellular location, expose the functional domain of a caged protein, or sequester a protein of interest. We will present the first demonstrations for each combination of photosensory domain and mechanism as examples.
Figure 2. Photoregulatory mechanisms.
Strategies for optical control proteins of interest (POIs) categorized by mechanism: (A) allosteric regulation, (B) native photoassociation, (C) native photodissociation, (D) engineered photoasssociation, and (E) engineered photodissociation. An active POI is indicated by yellow coloring. In strategies involving heterodimerization of two POIs, one POI could be a targeting signal and the second POI the activity to be regulated, or the two POIs can be fragments of a protein which will assemble upon heterodimerization. Abbreviation: LOV, light-oxygen-voltage-sensing; POI, protein of interest; BLUF domain, blue-light-utilizing FAD-containing domain; AC, adenylate cyclase; FKF1, flavin-binding kelch repeat f-box 1; GI, GIGANTEA; CRY2, cryptochrome 2; CIB1, crytochrome-interacting basic helix-loop-helix 1; Phy, phytochrome; PIF, phytochrome interacting factor; VVD, vivid; ePDZ, engineered PDZ domain; nMag, negative Magnet domain; pMag, positive Magnet domain; UVR8, UV resistance 8; CBD, cobalamin-binding domain; pdDronpa, photodissociable dimer Dronpa; Zdk, Zdark.
Allosteric Regulation
Allosteric transduction of light-induced conformational changes from a photosensory domain to a linked enzymatic domain is a common natural mechanism for photoregulation, as observed in LOV, BLUF, and phytochrome domains. The partial unfolding or release of the Jα helix of the Avena phototropin LOV2 domain is particularly well understood, and has been used in two ways, either to activate proteins linked to the C-terminus by light-induced exposure, or to change the activity of a protein in which the entire LOV2 domain is inserted into a loop.
In the first example of the light-induced exposure of POIs by allosteric regulation (Figure 2A, i), the Avena LOV2 Jα helix was fused to the N-terminal α-helix of the trp transcriptional repressor, with the removal of some initial sequences in the trp helix to create a single chimeric Jα-trp α-helix. As the N-terminal α-helix is the DNA-binding element of trp, it might be expected that spatial overlap of the LOV2 domain with the trp segment of the α-helix would block it from binding DNA. Light-induced release of the α-helix would then enable the trp segment to bind to DNA. Indeed this was observed (74). In a second example, a photoactivatable Rac1 (PA-Rac1) was designed on a similar principle generated by fusing Rac1 to the C-terminus of LOV2 Jα helix (75). Rac1 would be expected to be sterically blocked in dark state by the proximity to the LOV2 domain, whereas blue-light-induced unwinding of Jα helix would allow release of Rac1. Indeed, after screening a large set of fusions, one fusion was found which effectively induced the Rac1 output of membrane ruffling when stimulated with blue light. Interestingly, a crystal structure revealed an interface between Rac1 and the LOV2 domain that contributes to Rac1 docking and inhibition in the dark (75).
The light-induced partial unfolding of the Jα helix has proven to be a general method to functionally release small peptides which can then exert biological effects by binding to other proteins. For example, a light-inducible inhibitor for CaMKII was developed based on conformational change of LOV2-Jα helix (76). This photoactivatable autocamtide inhibitory peptide 2 (PaAIP2) includes AIP2 in the C-terminus of LOV2-Jα helix. As blue light induces conformational change of Jα helix, sterically hindered AIP2 in the C-terminus can be exposed to function as an inhibitor of CaMKII with high spatiotemporal resolutions. In fact, PaAIP2 under blue light inhibited CaMKII-mediated neuronal functions, such as glutamate-induced spine enlargement, field LTP in acute hippocampal slices, and memory formation in inhibitory avoidance learning. As we will discuss later, the general mechanism of light-induced peptide exposure at the end of the Jα helix can be used to engineer light-induced heterodimerizing interactions.
Another mechanism for allosteric regulation involves inserting a photosensory domain into a POI to control its enzymatic activity. The first demonstration of this method was the insertion of Avena phototropin LOV2 into a loop of dihydrofolate reductase (DHFR) that showed co-evolution with the active site. LOV2 insertion resulted in enzymes that could be activated by illumination (77). In a second example, a light-activated caspase-3 was also developed by inserting LOV2 domains to constrain the linker between subunits of caspase-3 (78). Light-induced release of the C-terminal Jα helix provides additional linker flexibility to allow caspase-3 autoactivation. In a third demonstration, insertion of LOV2 into a small loop of the Src kinase domain allowed illumination to inhibit kinase activity (79). This could be explained by the Jα dissociation from the remainder of the LOV2 domain causing the distance between the attachment points to increase, distorting the kinase into a non-catalytic conformation (Figure 2A, ii).
In an unique example of allosteric regulation, the LOV2 domain was inserted between the body and lever arm of kinesin and myosin motor proteins to create a light-actuated hinge. Light-mediated opening of this hinge alters the angle of the lever arm relative to the motor body, causing a displacement of the center of gravity of the molecule and a reversal in the direction of movement (80).
Light-induced changes in LOV and phytochrome domains can also be transduced allosterically to activate enzymes linked at the C-terminus. This is most commonly performed using microbial enzyme domains that are natively regulated by N-terminal sensory domains. For example, in the bacterial EnvZ protein, an N-terminal sensory domain activates a C-terminal histidine kinase domain in response to high osmolality. When the sensory domain is replaced by a bacteriophytochrome domain, the EnvZ histidine kinase domain becomes inhibited by red light (81). Another example is the oxygen-responsive histidine kinase FixL. Replacement of its oxygen-sensing PAS domain with a LOV domain from Bacillus YtvA generated a blue light-responsive histidine kinase (82). As described above, BLUF domains are often found as regulatory elements N-terminal to adenylate cyclase domains (Figure 2A, iii). These PACs are activated when the illuminated BLUF domains induce conformational changes in the linked adenylate cyclase catalytic domains. The BLUF domain could be substituted with the PAS-GAF-PHY domains of bacterial phytochrome BphG1 to create an infrared light-activated adenylate cyclase (83). Likewise the PAS-GAF-PHY domain of another bacterial phytochrome, Deinococcus DrBphP, was used to create a light-activated human phosphodiesterase 2A (84). These examples of allosteric control of enzymatic domains are further discussed in a recent review (85).
Native Photoassociation
Photosensory domains that naturally form homo-oligomers in response to light can be adapted to control proteins whose activity are regulated by oligomerization. For example, the LightON system was developed to control transcription by fusing the VVD LOV domain to the Gal4 DNA binding domain (DBD) and p65 transcription activation domain (TAD) (86) (Figure 2B, i). As Gal4 requires dimerization through a separate domain for DNA binding, it is expected that this protein would only activate gene expression in response to light. Indeed, blue light-induced VVD homodimerization mediates strong dimerization of DBD and subsequent gene expression in mammalian cells. Another example is the use of homodimerizing LOV domains to activate receptor tyrosine kinases (RTKs), many of which are natively activated by binding to dimeric ligands (38).
The light-induced oligomerization of the Arabidopsis thaliana CRY2 PHR domain (Figure 2B, ii) has also been applied to control various signaling proteins including RhoA (48), RTKs (87), and STIM1 (88). OptoTrkB is composed of CRY2 PHR domain and the intracellular region of TrkB, a tyrosine receptor kinase, which can be activated by light-induced receptor dimerization. Clustering can be enhanced by introduction of short extended sequences to the C- terminus of PHR domain to create CRYclust (89), or addition of positive charges to the C-terminus to create CRYhigh (90). Conversely, clustering can be minimized by introduction of negative charges to the C-terminus to create CRYlow, which may be especially useful for creating optically regulated systems based on CRY2-CIB1 heterodimerzation without unwanted clustering.
Light-induced heterodimerizing photosensory domains are readily adaptable to bringing together two functional domains to create a multifunctional protein activity. Following the original example of PhyA/B-PIF3 (9), the FKF1-GI, CRY2-CIB1, and PhyB-PIF6 (Figure 2B, iii–v) heterodimerizing systems have also been used to activate transcription, by fusing one partner to a DNA-binding domain such as GAL4 and the other to a transcriptional activation domain such as VP16 (40, 49, 70). A related use of light-induced heterodimerization is to assemble a functional protein from two nonfunctional fragments, demonstrated initially using CRY2-CIB1 to reconstitute Cre recombinase from split fragments (49).
Light-induced heterodimerization can also be used to regulate proteins whose activity is enhanced by localization to a particular subcellular compartment, or whose activity is required only in specific locations within the cell. For example, the FKF1-GI, CRY2-CIB1, and PhyB-PIF6 photoassociation reactions have been used to bring signaling proteins to the membrane where they are activated by endogenous membrane-bound regulators (40, 70, 91). In addition, the CRY2-CIB1 heterodimerization system was applied to recruit AMPA-type glutamate receptors to excitatory postsynaptic membrane by blue light (92). In this system, postsynaptic density scaffold molecules and AMPA-type glutamate receptors (GluA1) were fused to CRY2 and CIB1, respectively. The light-controlled addition of GluA1 molecules to the postsynaptic density was shown to be sufficient to potentiate postsynaptic currents, enabling spatiotemporally precise control of synaptic strength by blue light.
Interestingly, light-induced heterodimerization can also be used to drive the formation of multivalent protein assemblies for the purpose of sequestering proteins (Figure 2B, vi). LARIAT is a system for light-activated reversible inhibition by assembled trap based on CRY2-CIB1 heterodimerization (93). In the system, the multimerizing domain of CaMKIIa is connected to CIB1, and CRY2 was fused to POI. As the light-induced CRY2-CIB1 interaction mediates the clustering of these proteins, LARIAT can be applied to sequester and inactivate the POI by light.
Native Photodissociation
Light-induced monomerization is a natural mechanism of action of photosensory domains that can be adapted for experimental purposes. Two copies of UVR8 were fused in tandem to a model transmembrane protein, vesicular stomatitis virus glycoprotein (VSVG), to form higher-order oligomers that were retained in the endoplasmic reticulum (ER) (23). UV-B light induced release of the UVR8-UVR8-VSVG fusion proteins from the ER and subsequent expression on the cell surface (Figure 2C, i). This approach was used to control local secretion of a POI from ER membranes in neuronal dendrites.
Another example of the light-induced dissociation to control protein activity is based on the CBD from Myxococcus CarH (68) (Figure 2C, ii). The CBD was fused to intracellular domain of fibroblast growth factor receptor 1 (FGFR1) to tetramerize it in dark. As expected, FGFR1 and downstream MAPK/ERK signaling were activated at baseline. Illumination by green light for 5 minutes decreased the phosphorylation of FGFR1 and ERK, which was then reversed when placed in the dark for 60 minutes.
Finally, the dimeric BLUF-containing protein PixD from Synechocystis creates clusters with PixE in the dark which are dissociated by blue light. This approach was used to create membraneless proteinaceous droplets in mammalian cells which dissolve upon illumination (94).
Engineered Photoassociation
Natural photosensory domains can also be engineered to exhibit light-induced protein-protein interactions. This can be done by linking the light-induced conformational change of the photosensory domain to exposure of protein regions that can then engage in homomeric or heteromeric interactions. For example, PYP was fused to transcription factor GCN4 in a way that was expected to sterically occlude GCN4 dimerization, while light-induced partial unfolding of PYP was expected to release GCN4 for dimerization. This mechanism would resemble that of native VVD if successfully implemented. Indeed, the resulting GCN4Δ25PYP-v2 chimera showed increased binding to DNA upon illumination, although the degree of regulation was modest at only 2-fold induction (95). The inducibility of this system may be constrained by the requirement of a certain minimum length of the GCN4 leucine zipper for homodimerization; a peptide of this minimum length may be difficult to occlude by fusion to the PYP domain.
Two engineered light-induced heterodimerization systems have been based on the Avena LOV2 domain (Figure 2D, i). The improved light-induced dimer (iLID) system is composed of LOV2 fused to the bacterial peptide ssrA. In the iLID system, ssrA is embedded in the C-terminal helix of LOV2 in the dark, and the light-induced conformational change in the C-terminus of LOV2 exposes the ssrA which can then form a heterodimer with sspB (96). Conceptually similar to iLID is the tunable light-controlled interaction protein (TULIP) system. In TULIP, illumination of a fusion protein of LOV2 and a PDZ-recognizing peptide (LOVpep) allows the peptide to bind to an engineered PDZ domain (ePDZ) (97). The iLID and TULIP systems take advantage of the fact that small peptides can be efficiently occluded by embedding in the Arabidopsis LOV2 C-terminus, and that several high-affinity heterodimeric interactions between small peptides and protein domains are well characterized.
Finally, natural light-induced homodimerization can also be converted to engineered heterodimerization. As described abobe, the FAD-dependent small photoreceptor VVD can form a homodimer through its N-terminal α-helix, termed Ncap, upon blue light illumination. The key residues of Ncap, Ile47 and Asn56, at the interface of the VVD homodimer were further engineered to be positively or negatively charged, creating positive Magnet (pMag) or negative Magnet (nMag) (98). Thus, upon blue light illumination, pMag and nMag can recognize each other through electrostatic interactions and selectively generate pMag-nMag heterodimer in this Magnet system (Figure 2D, ii In addition, Magnet variants of different photo-switching kinetics ranging from seconds to hours were created. Light-induced heterodimerization of Magnets was used to reconstitute Cre recombinase from split fragments (99), similar to the earlier demonstration using CRY2-CIB1.
Engineered Photodissociation
DronpaN and pdDronpa are tetrameric and dimeric in their basal state, respectively, but can be dissociated by 500-nm cyan light and reassociated by 400-nm violet light. DronpaN was initially used to release a protein from the cell membrane (20). Subsequently a general design for single-chain photoactivatable proteins was created by fusing two copies of DronpaN on opposite site of an active site of interest (Figure 2E, i). Two chains then interact to assemble a DronpaN tetramer to sterically block the active sites. Cyan light dissociates DronpaN, “uncaging” the active site. This fluorescent light-inducible protein (FLiP) design was successfully demonstrated for GEFs and proteases. The photodissociable dimer pdDronpa was used to create photoswitchable single-chain variants of the kinases Raf1, MEK1/2, and CDK5 (21). In these photoswitchable kinases, one pdDronpa domain was fused to the N-terminus of a constitutively active kinase domain and the second one was inserted in the F-G loop located across the active site from the N-terminus, thus occluding the active site in the dark. Cyan light dissociates the pdDronpa dimer and allows the kinase to phosphorylate its substrates.
LOVTRAP is an optochemical strategy to trap and release a POI through the light-induced conformational change of LOV2 (100) (Figure 2E, ii). In the system, LOV2 domain can be anchored away from the places where a POI functions, for example mitochondria, and the POI is connected to Zdark (Zdk), which binds to LOV2 selectively in dark. Thus, the POI can be trapped by LOV2-Zdk interaction, which can be released by the dissociation of Zdk from LOV2 under blue light. As a demonstration, Rho-family small GTPases were sequestered at mitochondria by LOVTRAP and then released by blue light. In a strategy called Z-lock, the LOV2-Zdk interaction was applied to build an intramolecular block over the functional site of a POI, similar to the strategy employed with DronpaN and pdDronpa (101). Z-lock was successfully used to control the actin-disassembly protein cofilin and α-tubulin acetyl transferase (α-TAT).
Finally, PYP fusions to interacting peptides can exhibit light-induced loss of interaction. A different fusion of PYP to transcription factor GCN4 named GCN4(S)Δ25PYP was unexpectedly found to demonstrate decreased DNA binding upon illumination, the opposite behavior of GCN4Δ25PYP-v2 (102). This was attributed to a light-induced conformational change causing decreased exposure of the homodimerizing interface of GCN4, but the degree of regulation was again modest, with only 3-fold decrease in downstream transcription. Likewise, a fusion of PYP to A-CREB, a negative regulator of the transcription factor CREB, exhibited decreased A-CREB function (a ~3-fold increase in CREB-mediated transcription) upon illumination (103). It was proposed that the blue light-induced conformational change of PYP exposes a hydrophobic patch that then traps A-CREB to prevent its interaction with CREB. Of note, both GCN4 and A-CREB are leucine zippers, so PYP may be exerting a common mechanism of engaging in a weak intramolecular interaction with leucine zippers after illumination.
EXAMPLES OF OPTICALLY CONTROLLED PROTEINS
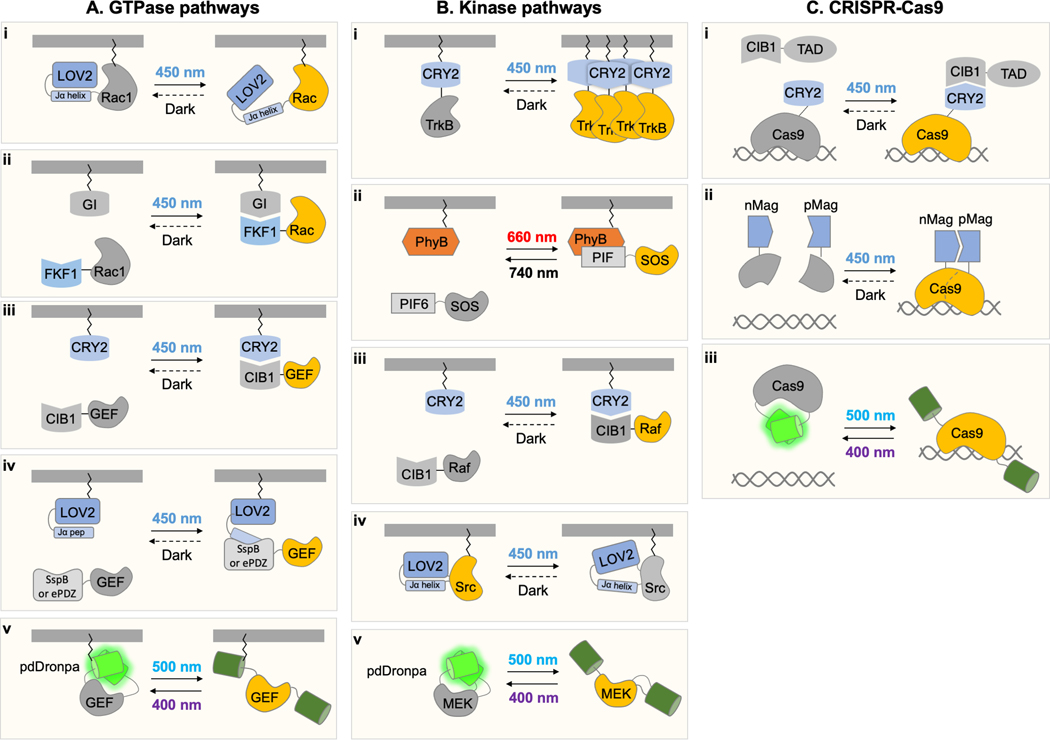
Certain classes of proteins are especially useful to regulate with temporal or spatial specificity, and thus have been controlled by various photosensory domains. Here we provide some examples of proteins that are commonly optically regulated, starting at the membrane and working our way to the nucleus (Figure 3). Given the rapidly expanding use of optobiochemical tools, this list is by no means exhaustive, but serves to showcase the wide variety of biological processes that can be subject to optical control.
Figure 3. Examples of biochemical pathways controlled using multiple optobiochemical strategies.
Strategies are organized from the membrane to the interior of the cell. (A) GTPase pathways have been controlled by (i) allostery between Avena phototropin LOV2 and the GTPase, (ii) membrane recruitment of the GTPase, (iii, iv) membrane recruitment of a guanine nucleotide exchange factor (GEF), (v) pdDronpa-mediated caging and uncaging of a GEF. (B) Kinase pathways have been controlled by (i) CRY2-mediated aggregation of receptor tyrosine kinases, (ii) membrane recruitment of the Ras exchange factor SOS, (iii) membrane recruitment of Raf, (iv) negative allostery between Avena phototropin LOV2 and Src, and (v) pdDronpa-mediated caging and uncaging of the active site. (C) Cas9 proteins have been controlled by (i) recruitment of a transcriptional activation domain (TAD), (ii) reconstitution of split fragments, and (iii) pdDronpa-mediated caging and uncaging of DNA-binding.
Ion Channels
Transmembrane ion flux is the one class of biochemical event that can be regulated both directly using ion-conductive microbial opsins (as commonly done in optogenetics) and indirectly at longer timescales with other photosensory domains. Here we will discuss uses of non-opsin photosensory domains to control ion flux.
Calcium is an important second messenger involved in many cellular processes such as contraction, vesicular release, kinase activation, and gene expression. To control voltage-gated calcium (Cav) channels, the negative regulator RGK was fused to sspB, and LOV2-ssrA was tethered to the plasma membrane (104). Blue light then recruited RGK to the plasma membrane to inhibit Cav channel opening. Another calcium-regulating light-activated channel is OptoSTIM1, a fusion of STIM1 to the CRY2 PHR domain (88). Light-induced oligomerization of STIM1 causes it to bind to and activate the plasma membrane calcium channel CRAC, inducing calcium entry into cells. OptoSTIM1 stimulation in the hippocampus during memory task training was sufficient to improve learning by mice, demonstrating the value of controlling calcium levels in neurons.
The permeability of potassium across the cell membrane is tightly regulated to control the transmembrane potential. Opening of potassium channels hyperpolarizes the resting membrane potential and hinders the generating of action potentials in neurons and cardiomyocytes. An optically controlled potassium channel, blue-light-induced K+ channel 1 (BLINK1), was constructed by fusing LOV2 to the viral potassium channel Kcv channel (105). BLINK was applied to inhibit the escape response of zebrafish larvae by shining blue light.
Rho GTPase Pathways
GTPases are a large family of enzymes that hydrolyze guanosine triphosphate (GTP) to guanosine diphosphate (GDP). In their GTP-bound state, GTPases bind to effector proteins to mediate downstream signaling events, while the GDP-bound form is functionally inactive. The function of GTPases is positively regulated by guanine nucleotide exchange factors (GEFs) that cause the dissociation of GDP and association of GTP, and negatively controlled by GTPase-activating proteins (GAPs) that accelerate its intrinsic GTPase activity to hydrolyze GTP to GDP. Rho subfamily of GTPases, which include RhoA, Rac1 and Cdc42, regulate the formation of actin-based structures and actinomyosin contractility. Thus, various optobiochemical strategies have been applied to control Rho GTPases and thereby cell shape and movement.
One general approach to control Rho GTPase activity involves expressing them in a photoregulatable form. One example is PA-Rac1, whose design was described above (75) (Figure 3A, i). In another example, constitutively active Rac1 mutant can be expressed as a FKF1 fusion for light-induced recruitment to GI-CAAX at the membrane (Figure 3A, ii). The local recruitment of Rac1 effectors then leads to the local formation of lamellipodia (40). Constitutively active mutants of RhoA and Rac1 lacking membrane targeting signals have also been regulated by sequestration at the mitochondria by LOVTRAP followed by release into the cytosol (100). Here, even though cytosolic expression should not be as efficient as membrane expression in activating downstream effects, morphological effects of RhoA and Rac1 release were still observed.
Another general method is to localize GEFs, positive regulators for Rho GTPases, to the membrane in a light-inducible manner, where they can activate endogenous Rho GTPases (Figure 3A, iii). For example, GEFs for Cdc42 and Rac1 were localized to the membrane using the PhyB-PIF6 light-inducible interaction (70). GEFs for RhoA have been recruited to the plasma membrane using the LOVpep-ePDZ and CRY2-CIB1 light-inducible interactions (106, 107) (Figure 3A, iv). Finally, the Rac1 GEF Vav2 can also be released from LOVTRAP sequestration at mitochondria to activate membrane ruffling without specific enrichment at the plasma membrane (100).
A third method is to use a photodissociable Dronpa mutant to create a single-chain GEF that can be pre-localized to the plasma membrane and then activated by light (Figure 3A, v). A single-chain photoswitchable Cdc42 GEF created in this manner allowed repeated cycles of local Cdc42 activation (20).
Kinase Pathways
Kinases are enzymes that catalyze the transfer of a phosphate group to specific substrates. In the case of eukaryotic protein kinases, the substrates are tyrosine, serine, and threonine residues of proteins. As kinase activity is often dynamically regulated, and kinases control multiple adaptive responses by cells including proliferation, survival, metabolism, and differentiation, there is great interest in developing methods to control kinases with high spatiotemporal resolution.
Receptor tyrosine kinases (RTKs) are transmembrane proteins containing an extracellular ligand binding domain and an intracellular kinase domain. RTKs are typically homodimerized by ligand binding to induce autophosphorylation on cytosolic residues which can then recruit additional signaling proteins to activate cytosolic proteins such as GTPases, PI3K, ERK, and Akt. Fusion of light-dependent dimeric or oligomeric domains recapitulates ligand-induced dimerization to enable light-induced RTK activation (Figure 3B, i). For example, optoTrkB, composed of either full-length TrkB or a myristylated TrkB cytosolic domain fused to the CRY2 PHR domain, mediates light-induced local filopodia formation in mature neurons (87), and light-specified axonal differentiation in immature neurons (108). A similarly constructed OptoFGFR enabled light-mediated control of cell polarity and migration (109). A recent study compared activation of downstream kinase pathways by different designs for optically oligomerized TrkB cytosolic domains (110). Photodimerization by the LOV domain from Vaucheria aureochrome or photo-oligomerization by the CRY2 PHR domain, applied to the TrkB cytosolic domain in either a soluble or a membrane-tethered form, was partially effective compared to stimulation by the natural ligand BDNF in activating ERK and Akt. In contrast, simultaneous oligomerization mediated by CRY2 aggregation and membrane recruitment mediated by CRY2-CIB1 heterodimerization was as effective as BDNF. This could be due to the basal cytosolic distribution preventing pathway activation and negative feedback in the dark, while membrane recruitment and oligomerization after illumination allows maximal TrkB phosphorylation and effector recruitment.
The Ras-Raf-MEK-ERK signaling pathway has been the subject of multiple optobiochemical studies. This pathway can be initiated by SOS recruitment to the membrane by RTKs to activate the small GTPase Ras, which then recruits the kinase Raf1 to activate MEK and ERK. The Phy-PIF interaction was applied to bring SOS to the plasma membrane, initiating Ras-Raf-MEK-ERK pathway activity (10) (Figure 3B, ii). The light-induced CRY2-CIB1 heterodimerization was also applied to recruit Raf1 to the plasma membrane where it becomes activated by homo-phosphorylation and then in turn activates the ERK signaling pathway (91) (Figure 3B, iii). Light-induced Raf1 activation was used to demonstrate that the pathways driving neurite extension in PC12 cells required activation at least once each 45 minutes for maximal neurite growth.
As described above, single-chain photoswitchable kinases can be engineered by the insertion of pdDronpa domains (21). These photoswitchable kinases can be activated by cyan light and re-inhibited by violet light (Figure 3B, iv). This design was readily generalizable to MEK1, MEK2, Raf, and CDK5. The ability to both turn on and off the kinase activity was used in photoswitchable Raf (psRaf) to create a brief pulse of Raf activation. This allowed the detection of a negative feedback loop from Raf to ERK to a phosphatase (21). In addition, these photoswitchable kinases could be optically controlled in living animals. Finally, single-chain photoinhibitable kinases have been generated (Figure 3B, v). More information on the specific topic of optical control of kinases can be found in a recent review (111).
Transcription Factors
Transcription is a dynamic process during development, with the location and timing of gene expression tightly regulated by genetic programs and environmental cues. While drugs can be used to activate gene expression, the kinetics of drug administration and washout are relatively slow, and no spatial resolution is possible. Transcriptional control by light allows the function of regulated gene expression in development to be assessed at single-cell spatial resolution and with finer temporal resolution.
Light-induced heterodimerization is the most common method for activating transcription in response to light, being used as the initial applications of Phy-PIF (9), FKF1-GI (40), and CRY2-CIB1 interactions (49), and also used to demonstrate iLIDs and TULIPs (112). These systems are essentially light-regulated two-hybrid systems, comprising the fusion of a DNA-binding domain (DBD) to one heterodimerizing domain and a transcriptional activation domain (TAD) to the other. Indeed some natural light-dependent interactions were discovered in two-hybrid screens for light-dependent transcription (47, 51).
Homodimerization can also be used to regulate transcription, although this is restricted to transcription factors that demonstrate a strict dependence of DNA binding on homodimerization. In LightON, homodimerizing VVD was fused to the Gal4 DBD and the p65 TAD to mediate transcription of promoters with Gal4 recognition sites (86). Another LOV-domain protein, EL222 from Erythrobacter litoralis, natively functions as a light-induced transcription factor. Light causes homodimerization and exposure of a DNA-binding motif (113). Thus, EL222 fused to TAD was used to induce activation of promoters with EL222 recognition sites.
Given the large number of systems that have been used for transcriptional control, comparisons between systems have been carried out. For example, when natural and engineered blue-light-inducible dimer systems were compared, the iLID/SspB system shows stronger binding affinity as well as faster reversion kinetics than LOVpep/ePDZ (TULIP) or CRY2/CIB1 system (112). Accordingly, the iLID system, when applied to optically induce the interaction of Gal4 DBD and TAD, shows stronger transcription efficiency than other systems, correlating in vitro and in vivo kinetics of blue-light-inducible dimer systems. Another comparison between FKF1/GI and CRY2/CIB1 revealed that the CRY2/CIB1 system induces stronger light-dependent transcription as well as leakiness than FKF1/GI system (114). In addition, the FKF1/GI is a non-reversible system while CRY2/CIB1 dimerization is reversible. These reports suggest that optical controllers with different biochemical and biophysical properties can be chosen depending on the experimental purposes.
As an example of the utility of precise transcriptional control, the VVD system was used to investigate the function of oscillatory expression of the Asc1 protein that had been observed in mouse neuronal progenitor cells (115). Light-entrained oscillatory Asc1 expression was found to drive progenitor cell proliferation, while sustained expression induced neuronal differentiation, demonstrating that different cell fate outcomes can result from distinct temporal profiles of protein expression.
CRISPR/Cas Proteins
Various classes of CRISPR-associated proteins such as Cas9 and Cas13 are now used widely in biology for RNA-directed gene editing, transcriptional regulation, RNA binding, and RNA editing. Given their ease of use to alter or activate endogenous genes, CRISPR-associated proteins have immense potential as basic research tools for manipulating genetic programs in vitro and in vivo. There is thus intense interest in developing methods for exogenous control of CRISPR-associated protein activities.
Multiple photoregulatory strategies have been developed for optical regulation of CRISPR-associated proteins. For example, fusion to CIB1 and CRY2 enables light-dependent recruitment of the transcriptional activation domains to catalytically inactive Cas9 proteins, which in turn can be directed to a gene of interest by a sequence-specific small guide RNA (116) (Figure 3C, i). Alternatively, the Cas9 protein can be split into two fragments which can be assembled by light-induced heterodimerization of fused nMag and pMag domains (117) (Figure 3C, ii). A single-chain photoswitchable Cas9 was designed by insertion of pdDronpa domains on loops on opposite sides of the DNA-binding site, preventing DNA interaction in the dark and enabling it upon cyan ligth illumination (118) (Figure 3C, iii). This method was generalized from Streptococcus Cas9 to Staphylococcus Cas9. These and other optical approaches to control Cas9-based proteins can be used to perform gene editing or transcriptional activation in only certain cells within a sample, or can be used to limit the function of catalytically active Cas9 to avoid off-target effects. Optical control has also recently been extended to RNA editing with Cas13, by using CRY2 and CIB1 to recruit an editing enzyme to Cas13 (119). More details about the various methods for optical control of CRISPR proteins are available in a recent review (120).
Recombinases
Cre and flp recombinases are enzymes that catalyze site-specific recombination between repeats of a specific recognition sequence, which happens to be 34 base pairs in each case. These recombinases excise DNA sequences between sites in the same orientation, which can be used to remove a gene or a transcriptional block. They invert DNA sequences between sites in opposite orientations, which can also be used to activate gene expression. These recombinases have been extensively used to permanently abolish or induce gene expression in genetically defined cells.
To optically control genome engineering with high spatiotemporal precision, photoactivatable versions of Cre were created in which split Cre fragments were reassembled using light-induced heterodimerizing domains. These were developed based on the VVD-domain Magnets and the CRY2-CIB1 interaction, with both systems having undergone multiple rounds of improvement to achieve ratios of illuminated vs. dark activity of >40 (121, 122).
Flp enables a second set of genetic controls orthogonal to Cre, and can be used together to enable more sophisticated genetic manipulations. Recently, a photoactivatable Flp (PA-Flp) recombinase with a 30-fold increase in PA-Flp activity was developed based on Magnet-driven fragment complementation (123). PA-Flp was successfully used with noninvasive illumination to demonstrate Flp-dependent and Cre-mediated Cav3.1 gene silencing in deep mouse brain regions.
UNIQUE APPLICATIONS OF OPTICAL CONTROL
An inherent characteristic of all optically controlled proteins is the ability to confine activation to a particular place or time. For many processes, such as transcription or gene editing or control of neuronal function, a defined time or location of activation is desired and the optobiochemical tool is used in a simple manner to achieve the desired restriction. For other processes such as signal transduction, spatiotemporal control is only useful when coupled to the testing of hypotheses regarding how signals produce responses depending on space and time. These questions are common in understanding how multiple signals are integrated or restricted to govern complex cellular processes. Thus, optobiochemical methods have been able to provide unique insights in the fields of cell biology, systems biology, and developmental biology. We will thus cover in this final section how complex cellular processes can be explored by optobiochemical tools.
Cellular Computation
Optobiochemical methods allow control of signaling with high temporal resolution and dose precision, which can be used to investigate the role of kinetics and amplitude in the output of specific signaling pathways. Comparing to growth factors, light can selectively stimulate particular signaling networks with high spatiotemporal resolution, which allows the decoding of distinct physiological outcomes from the same signaling node with different temporal dynamics.
Toettcher et al. used optobiochemical activation of the Ras-Raf-ERK pathway to perform fine characterization of essential pathway attributes such as kinetics, feedback, robustness, and cross-activation (10). They found that the Ras-Raf-ERK pathway converts a SOS pulse to ERK translocation with a delay of 3 minutes. Response amplitude was flat for SOS pulse durations of 10 to 120 min, which demonstrates that the pathway can be driven without negative feedback as long as SOS activity is maintained. This observation cannot be made with natural stimuli such as EGF or FGF application, as receptor downregulation causes termination of SOS activation within 60 min in these cases. Another interesting observation is that within each individual cell, ERK nuclear localization responds to Ras activation amplitude in a reliable monotonic fashion. However, the slope of this response differs in different cells. Thus, variability between cells observed with single-point measurements, often termed “noise”, may be due to stochasticity in the sensitivity of different cells, rather than stochasticity within cells. Finally, the ability to control Ras-Raf-ERK signaling onset time and duration enabled unbiased proteomic characterization of responses that occurred at certain intervals after activation or selectively after sustained signaling. Cross-activation of the PKCb pathway occurred with even transient SOS activation whereas cross-activation of mTOR output required sustained SOS signaling. Additional information on the use of optical control to investigate cellular computation can be found in a recent review (124).
Cellular Organization
The internal architecture of eukaryotic cells is both complex and dynamic. A static snapshot of a cell will reveal membrane-lined organelles such as mitochondria, the endoplasmic reticulum, the Golgi apparatus, and various vesicular structures. However, organelles can also be transported long distances via cytoskeleton-associated motors, and stream of vesicles deliver proteins and membranes between other organelles and the plasma membrane (125). In addition, a concept of membraneless organelles has emerged recently with the elucidation of structures such as the nucleolus, P bodies, and stress granules as RNA storage and processing centers with the physical characteristics of liquid droplets (126).
The light-induced heterodimerization of CRY2-CIB1 was used to control organelle locations within cells. Fusion of CIB1 to kinesin or to a dynein adapter protein, along with fusion of CRY2 to organelle-specific membrane proteins, allowed mitochondria, peroxisomes, and lysosomes to be transported along microtubules in either centripetal or centrifugal directions in living cells (127). Similarly, the TULIP system of light-induced heterodimerization between LOVpep and ePDZ was used to control organelle locations within cells (128). Endosome trafficking along microtubules could be regulated by fusion of ePDZ to kinesin or to a dynein-binding protein, and fusing LOVpep to the endosomal protein RAB11. Light-induced coupling of RAB11 to kinesin in axons resulted in an increased rate of axon extension, while coupling to dynein caused a decreased rate of extension. These results demonstrated that cell shape could be regulated by changes in the distribution of membranous organelles.
The LARIAT system was applied to aggregate intracellular membranes (IM-LARIAT) by light-induced heterodimerization of CRY2 and CIB1-Rab5 or CIB1-Rab11, which is specifically tethered to early or late endosomes (94). Blue-light-induced aggregation of these intracellular membranes revealed different roles of Rab5 and Rab11-mediated trafficking in growth cones.
Light-induced oligomerization of CRY2 has also been used to initiate liquid phase transitions and form proteinaceous droplets that can function as artificial membraneless organelles. Specifically, a fusion of CRY2 to the instrinsically disordered region from the protein FUS is soluble in the dark, but assembles into micron-scale droplets upon illumination. By expressing individual enzymes of a multi-enzyme pathway as CRY2-FUS fusions, metabolic flux through the pathway can be enhanced by droplet formation, demonstrating one of the theorized functions of intracellular droplets (129).
Cellular Motility
Cell motility is a complex and dynamic cellular process, regulated by the continuously changing activity of a network of proteins, including Rho GTPases and their effectors. As the spatiotemporal distribution and activity of these molecules is critical for concerted outcomes such as cell motility, optical controllers have been applied to decipher the signaling pathways involved.
One longstanding question in cell migration is how the Rho GTPases regulate each other. Previous work showed both positive and negative correlations between the activities of RhoA and Rac1, e.g. both were active at the leading edge of randomly migrating cells, but PDGF stimulation activates Rac1 while inhibiting RhoA (130). Expression of constitutively active Rac1 results in inhibition of RhoA, but such long-term manipulations can produce compensatory or secondary changes. Thus whether Rac1 can inhibit RhoA or vice versa was still unclear. Wu et al. found that activation of Rac1 in the PA-Rac1 construct led to a local inhibition of RhoA activity as assessed by a RhoA FRET reporter (75), verifying that acute Rac1 activity can lead to RhoA inhibition. This serves as a reminder that observed activity distributions need not reflect required functions. That is, the visualization of RhoA activity at the leading edge of migrating cells may not reflect a particular requirement for leading edge extension, but rather for an event that closely follows, such as establishment of focal adhesions behind the leading edge.
As cell motility is a dynamic process that requires the tight regulation of various signaling events, only a few of which have been investigated, optobiochemical methods and optical reporters will continue to be powerful tools for understanding the molecular mechanisms underlying random and regulated cell motility.
Tissue Development
Development occurs through the concerted action of multiple cells. Cells can regulate each other across long distances through the secretion and reception of signals, but can also regulate adjacent cells through signals triggered by changes in force or adhesion. Unlike knockouts or drug inhibition of specific protein signals that function at defined steps, the role of cell-cell contacts in tissue organization is not easily investigated by purely chemical or genetic means. Confining chemical and genetic methods to perturb cell force or cell adhesion to a specific developmental time window or to only a few cells requires complex experimental manipulations (e.g. drug-regulated expression in a sparse subset of cells). Active modulation of cell morphology or movement may provide more direct insight into how concerted tissue movements are coordinated.
Along these lines, Wang et al. (11) used PA-Rac to investigate the migratory plasticity of border cells within the Drosophila ovary. Stimulation of PA-Rac on one edge of a border cell cluster was sufficient to move the cluster in the direction of stimulation. Stimulating in the direction opposite from normal migration caused reversed migration, but at slower rates than stimulation in the normal direction. This bias could be due to either environmental conditions somehow inhibiting reverse migration, or competition from a directional endogenous Rac activity in border cells. Removal of the guidance cue receptors PVR and EGFR in just the border cells eliminated this bias, indicating that it did arise from signals in the border cells. Interestingly, the entire cluster of 6 to 8 cells would move together even though only one edge of cell was illuminated, demonstrating collective cell migration. Inhibition of JNK, which promotes cell adhesion, suppressed the collective directional response of the cluster. Thus spatiotemporal control of Rac activation by light revealed the sufficiency of directional Rac signaling in controlling border cell migration, and the role of cell-cell adhesion in collective migration.
Another use of light-activated proteins is to determine temporal requirements for specific proteins in development. For example, Raf1 was known to promote mesoderm formation during germ layer specification during frog embryogenesis, but it was unknown if this effect of Raf1 was confined to the natural time window for germ layer specification. Activation of Raf1 by CRY2-CIB1-mediated membrane recruitment at later stages revealed that Raf1 indeed maintained the ability to directly transform ectoderm to mesoderm (131). In another example, loss of cdk5-family kinases in nematode worms causes a neurite differentiation phenotype in which neurotransmitter vesicles are mislocalized from axons to dendrites, but whether cdk5 was required only in development or was also required for maintenance of axonal specification was unclear. Using a pdDronpa-regulated photoswitchable cdk5, post-developmental cdk5 activation was shown to be sufficient for rescue of a axonal differentiation phenotype in worms, demonstrating the adult axonal phenotype of cdk5 loss was not due to a developmental requirement of cdk5, but reflected a persistent requirement of cdk5 (21).
CONCLUSION
Living cells are remarkable not just for thousands of distinct protein activities they contain, but also for the interconnected and dynamic nature of those protein activities. Proteins involved in signaling, differentiation, or morphology are especially tightly regulated in space or time. Among the critical questions that need to be answered before we can claim an understanding of protein function in living systems are the following: What effects do specific protein activities exert in individual cells? How exactly do patterns of protein activities propagate in space and time to regulate other proteins? How does protein function in one cell influence the phenotypes of neighboring cells in a tissue?
The beginning of the 21st century saw major steps toward a deeper and more dynamic understanding of protein function. With the human genome completely sequenced in 2001, attention turned away from basic protein characterization and categorization to their roles as dynamic signaling entities within computational networks in the cell (132, 133). It was in this setting in 2002 that photosensory domains, specifically Arabidopsis phytochromes and their interacting factors, were first used to control function of a heterologous protein with light. This demonstration opened up the possibility of utilizing light to turn on protein functions at specific times and places in cells, which would allow the systematic investigation of the questions raised above. Light is ideal as a heterologous stimulus for regulating proteins in mammalian cells, as it lacks effects on most mammalian cells, is inexpensive, and can be applied easily at specific locations and times.
In the 18 years since, multiple photosensory domains have been characterized or engineered, and then adopted to control protein activities in living cells with light. In this review, we categorized photosensory domains by chromophore and presented generalizable mechanisms for controlling proteins of interest using these domains. We described protein classes that have been commonly investigated using optical controllers, and for which optical control has been valuable for understanding their functions in living cells. Finally, we discussed specific types of cell biological questions for which optical protein control is already producing significant conceptual insights.
In summary, genetically encoded methods for optical control of protein activity in living cells have rapidly evolved since the first example nearly two decades ago. Researchers now have a variety of photosensory domains to select from, and validated generalizable strategies they can use, to confer optical control on their proteins of interest. Undoubtedly the field will continue to evolve, and we will see additional photosensory domains and creative ways to use them. As a unique research approach that enables nearly real-time four-dimensional control of protein function, optobiochemistry promises to transform the fields of cell biology, systems biology, and developmental biology.
ACKNOWLEDGMENTS
This work was supported by grant CRC-15-04-KIST from the National Research Council of Science & Technology of Korea, Brain Research Program grant 2017M3C7A1043842 through the National Research Foundation of Korea, and Samsung Research Funding & Incubation Center of Samsung Electronics under Project Number SRFC-TC2003-02 (JS), and NIH grant 1R21GM132687 (MZL).
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
ABBREVIATION (PAGE ORDER)
- POI
protein of interest
- FP
fluorescent protein
- UV
ultraviolet
- UVR8
UV resistance 8
- PYP
photoactive yellow protein
- BLUF domain
blue-light-utilizing FAD-containing domain
- LOV domain
light-oxygen-voltage-sensing domain
- CBD
cobalamin-binding domain
- COP1
constitutively photomorphogenic 1
- HY5
hypocotyl 5
- RUP1
repressor of UV-B photomorphogensis 1
- PAS domain
Per-Arnt-Sim domain
- FMN
flavin mononucleotide
- VVD
vivid
- FKF1
flavin-binding kelch repeat f-box 1
- GI
GIGANTEA
- FAD
flavin adenine dinucleotide
- PAC
photoactivated adenylate cyclase
- PHR
photolyase homology region
- CRY2
cryptochrome 2
- CIB1
crytochrome-interacting basic helix-loop-helix 1
- Phy
phytochrome
- Cys
Cysteine
- Tyr
Tyrosine
- Gly
Glysine
- pdDronpa
photodissociable dimer Dronpa
- CarH
repressor of carotenoid gene expression, Histidine
- PIF
phytochrome interacting factor
- BphP1
bacterial phytochrome P1
- PpsR2
regulator of photosynthetic gene expression
- PCB
phycocyanobilin
- PA
photoactivable
- CaMKII
Ca2+/calmodulin-dependent protein kinase II
- PaAIP2
photoactivatable autocamtide inhibitory peptide 2
- LTP
Long-term potentiation
- DHFR
dihydrofolate reductase
- GAF
cGMP-specific phosphodiesterase, adenylyl cyclase, and FhlA
- DrBphP
bacterial phytochrome from Deinococcus radiodurans
- DBD
DNA binding domain
- TAD
transcription activation domain
- RTK
receptor tyrosine kinase
- TrkB
tyrosine receptor kinase B
- STIM1
Stromal interaction molecule 1
- CRAC channel
Ca2+ release-activated Ca2+ channel
- PI3K
Phosphoinositide 3-kinases
- AMPA
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- PSD
postsynaptic density
- GluA1
glutamate receptor A1
- LARIAT
light-activated reversible inhibition by assembled trap
- iLID
improved light-induced dimer
- RGK
Ras-like GTPases RAD/Rem/Gem/Kir
- TULIP
tunable, light-controlled interaction protein tags
- LOVpep
a peptide at the LOV2 C-terminus
- ePDZ
engineered PDZ domain
- pMag
positive Magnet domain
- nMag
negative Magnet domain
- VSVG
vesicular stomatitis virus glycoprotein
- ER
endoplasmic reticulum
- FGFR1
fibroblast growth factor receptor 1
- MAPK
Mitogen-activated protein kinase
- ERK
Extracellular signal-regulated kinase
- FLiP
fluorescent light-inducible protein
- CDK5
cyclin-dependent kinase 5
- LOVTRAP
LOV2 trap and release of protein
- Zdk
Zdark
- α-TAT
alpha-tubulin acetyl transferase
- CREB
cAMP response element-binding protein
- Cav channel
voltage-gated calcium channel
- PDGF-R
platelet-derived growth factor receptor
- K+v channel
voltage-gated potassium channel
- BLINK1
blue-light-induced K+ channel 1
- Ser
serine
- DRY
aspartic acid-arginine-tyrosine
- GTP
guanosine triphosphate
- GDP
guanosine diphosphate
- GEF
guanine nucleotide exchange factor
- GAP
GTPase-activating proteins
- SOS
son of sevenless
- AC
adenylate cyclase
- cAMP
cyclic adenosine monophosphate
- IlaC
infrared-light-activated adenylate cyclase
- NIR
near infrared
- PDE
phosphodiesterase
- cGMP
cyclic guanosine monophosphate
- LAPD
Light-activated phosphodiesterase
- TF
Transcription factor
- CRISPR
clustered regularly interspaced short palindromic repeat
- Cas
CRISPR-associated nuclease protein
- loxP
locus of X-over P1
- Flp
flippase
- FRT
flippase recognition target
- NLS
nucleus localization signal
- LED
light emitting diode
- FRET
fluorescence resonance energy transfer
- PVR
PDGF- and VEGF-related receptor
- EGFR
epidermal growth factor receptor
- JNK
c-jun N-terminal kinase
LITERATURE CITED
- 1.Heim R, Cubitt AB, Tsien RY. 1995. Improved green fluorescence. Nature 373: 663–4 [DOI] [PubMed] [Google Scholar]
- 2.Tsien RY. 1998. The green fluorescent protein. Annu Rev Biochem 67: 509–44 [DOI] [PubMed] [Google Scholar]
- 3.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. 2006. The fluorescent toolbox for assessing protein location and function. Science 312: 217–24 [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez EA, Campbell RE, Lin JY, Lin MZ, Miyawaki A, et al. 2017. The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins. Trends Biochem Sci 42: 111–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Repina NA, Rosenbloom A, Mukherjee A, Schaffer DV, Kane RS. 2017. At Light Speed: Advances in Optogenetic Systems for Regulating Cell Signaling and Behavior. Annu Rev Chem Biomol Eng 8: 13–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shcherbakova DM, Shemetov AA, Kaberniuk AA, Verkhusha VV. 2015. Natural photoreceptors as a source of fluorescent proteins, biosensors, and optogenetic tools. Annu Rev Biochem 84: 519–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou XX, Pan M, Lin MZ. 2015. Investigating neuronal function with optically controllable proteins. Front Mol Neurosci 8: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim B, Lin MZ. 2013. Optobiology: optical control of biological processes via protein engineering. Biochem Soc Trans 41: 1183–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. 2002. A light-switchable gene promoter system. Nat Biotechnol 20: 1041–4 [DOI] [PubMed] [Google Scholar]
- 10.Toettcher JE, Weiner OD, Lim WA. 2013. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155: 1422–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, He L, Wu YI, Hahn KM, Montell DJ. 2010. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat Cell Biol 12: 591–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deisseroth K, Feng G, Majewska AK, Miesenbock G, Ting A, Schnitzer MJ. 2006. Next-generation optical technologies for illuminating genetically targeted brain circuits. J Neurosci 26: 10380–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenno L, Yizhar O, Deisseroth K. 2011. The development and application of optogenetics. Annu Rev Neurosci 34: 389–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu X, Mee T, Jia X. 2020. New era of optogenetics: from the central to peripheral nervous system. Crit Rev Biochem Mol Biol 55: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang CL, Ferenczi EA, Lei M. 2020. Editorial: Optogenetics: An Emerging Approach in Cardiac Electrophysiology. Front Physiol 11: 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Josselyn SA. 2018. The past, present and future of light-gated ion channels and optogenetics. Elife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boyden ES. 2015. Optogenetics and the future of neuroscience. Nat Neurosci 18: 1200–1 [DOI] [PubMed] [Google Scholar]
- 18.Spangler SM, Bruchas MR. 2017. Optogenetic approaches for dissecting neuromodulation and GPCR signaling in neural circuits. Curr Opin Pharmacol 32: 56–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar A, Ali AM, Woolley GA. 2015. Photo-control of DNA binding by an engrailed homeodomain-photoactive yellow protein hybrid. Photochem Photobiol Sci 14: 1729–36 [DOI] [PubMed] [Google Scholar]
- 20.Zhou XX, Chung HK, Lam AJ, Lin MZ. 2012. Optical control of protein activity by fluorescent protein domains. Science 338: 810–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou XX, Fan LZ, Li P, Shen K, Lin MZ. 2017. Optical control of cell signaling by single-chain photoswitchable kinases. Science 355: 836–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkins GI. 2014. The UV-B photoreceptor UVR8: from structure to physiology. Plant Cell 26: 21–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen D, Gibson ES, Kennedy MJ. 2013. A light-triggered protein secretion system. J Cell Biol 201: 631–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Christie JM, Arvai AS, Baxter KJ, Heilmann M, Pratt AJ, et al. 2012. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science 335: 1492–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu D, Hu Q, Yan Z, Chen W, Yan C, et al. 2012. Structural basis of ultraviolet-B perception by UVR8. Nature 484: 214–9 [DOI] [PubMed] [Google Scholar]
- 26.Tissot N, Ulm R. 2020. Cryptochrome-mediated blue-light signalling modulates UVR8 photoreceptor activity and contributes to UV-B tolerance in Arabidopsis. Nat Commun 11: 1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller K, Engesser R, Timmer J, Zurbriggen MD, Weber W. 2014. Orthogonal optogenetic triple-gene control in Mammalian cells. ACS Synth Biol 3: 796–801 [DOI] [PubMed] [Google Scholar]
- 28.Kyndt JA, Vanrobaeys F, Fitch JC, Devreese BV, Meyer TE, et al. 2003. Heterologous production of Halorhodospira halophila holo-photoactive yellow protein through tandem expression of the postulated biosynthetic genes. Biochemistry 42: 965–70 [DOI] [PubMed] [Google Scholar]
- 29.Hoff WD, van der Horst MA, Nudel CB, Hellingwerf KJ. 2009. Prokaryotic phototaxis. Methods Mol Biol 571: 25–49 [DOI] [PubMed] [Google Scholar]
- 30.van der Horst MA, Laan W, Yeremenko S, Wende A, Palm P, et al. 2005. From primary photochemistry to biological function in the blue-light photoreceptors PYP and AppA. Photochem Photobiol Sci 4: 688–93 [DOI] [PubMed] [Google Scholar]
- 31.Glantz ST, Carpenter EJ, Melkonian M, Gardner KH, Boyden ES, et al. 2016. Functional and topological diversity of LOV domain photoreceptors. Proc Natl Acad Sci U S A 113: E1442–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. 2000. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry 39: 9401–10 [DOI] [PubMed] [Google Scholar]
- 33.Harper SM, Neil LC, Gardner KH. 2003. Structural basis of a phototropin light switch. Science 301: 1541–4 [DOI] [PubMed] [Google Scholar]
- 34.Briggs WR, Beck CF, Cashmore AR, Christie JM, Hughes J, et al. 2001. The phototropin family of photoreceptors. Plant Cell 13: 993–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hart JE, Sullivan S, Hermanowicz P, Petersen J, Diaz-Ramos LA, et al. 2019. Engineering the phototropin photocycle improves photoreceptor performance and plant biomass production. Proc Natl Acad Sci U S A 116: 12550–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zoltowski BD, Schwerdtfeger C, Widom J, Loros JJ, Bilwes AM, et al. 2007. Conformational switching in the fungal light sensor Vivid. Science 316: 1054–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zoltowski BD, Vaccaro B, Crane BR. 2009. Mechanism-based tuning of a LOV domain photoreceptor. Nat Chem Biol 5: 827–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grusch M, Schelch K, Riedler R, Reichhart E, Differ C, et al. 2014. Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. EMBO J 33: 1713–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakasone Y, Zikihara K, Tokutomi S, Terazima M. 2010. Kinetics of conformational changes of the FKF1-LOV domain upon photoexcitation. Biophys J 99: 3831–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. 2009. Induction of protein-protein interactions in live cells using light. Nat Biotechnol 27: 941–5 [DOI] [PubMed] [Google Scholar]
- 41.Park SY, Tame JRH. 2017. Seeing the light with BLUF proteins. Biophys Rev 9: 169–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonetti C, Mathes T, van Stokkum IH, Mullen KM, Groot ML, et al. 2008. Hydrogen bond switching among flavin and amino acid side chains in the BLUF photoreceptor observed by ultrafast infrared spectroscopy. Biophys J 95: 4790–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohki M, Sato-Tomita A, Matsunaga S, Iseki M, Tame JRH, et al. 2017. Molecular mechanism of photoactivation of a light-regulated adenylate cyclase. Proc Natl Acad Sci U S A 114: 8562–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujisawa T, Masuda S. 2018. Light-induced chromophore and protein responses and mechanical signal transduction of BLUF proteins. Biophys Rev 10: 327–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blain-Hartung M, Rockwell NC, Moreno MV, Martin SS, Gan F, et al. 2018. Cyanobacteriochrome-based photoswitchable adenylyl cyclases (cPACs) for broad spectrum light regulation of cAMP levels in cells. J Biol Chem 293: 8473–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chaves I, Pokorny R, Byrdin M, Hoang N, Ritz T, et al. 2011. The cryptochromes: blue light photoreceptors in plants and animals. Annu Rev Plant Biol 62: 335–64 [DOI] [PubMed] [Google Scholar]
- 47.Liu H, Yu X, Li K, Klejnot J, Yang H, et al. 2008. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 322: 1535–9 [DOI] [PubMed] [Google Scholar]
- 48.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. 2013. Optogenetic protein clustering and signaling activation in mammalian cells. Nat Methods 10: 249–52 [DOI] [PubMed] [Google Scholar]
- 49.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. 2010. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods 7: 973–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taslimi A, Zoltowski B, Miranda JG, Pathak GP, Hughes RM, Tucker CL. 2016. Optimized second-generation CRY2-CIB dimerizers and photoactivatable Cre recombinase. Nat Chem Biol 12: 425–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hughes RM, Vrana JD, Song J, Tucker CL. 2012. Light-dependent, dark-promoted interaction between Arabidopsis cryptochrome 1 and phytochrome B proteins. J Biol Chem 287: 22165–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Briscoe AD. 2008. Reconstructing the ancestral butterfly eye: focus on the opsins. J Exp Biol 211: 1805–13 [DOI] [PubMed] [Google Scholar]
- 53.Zhang F, Vierock J, Yizhar O, Fenno LE, Tsunoda S, et al. 2011. The microbial opsin family of optogenetic tools. Cell 147: 1446–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Spudich JL. 2006. The multitalented microbial sensory rhodopsins. Trends Microbiol 14: 480–7 [DOI] [PubMed] [Google Scholar]
- 55.Inoue K, Tsukamoto T, Sudo Y. 2014. Molecular and evolutionary aspects of microbial sensory rhodopsins. Biochim Biophys Acta 1837: 562–77 [DOI] [PubMed] [Google Scholar]
- 56.Deisseroth K. 2015. Optogenetics: 10 years of microbial opsins in neuroscience. Nat Neurosci 18: 1213–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.von Lintig J, Kiser PD, Golczak M, Palczewski K. 2010. The biochemical and structural basis for trans-to-cis isomerization of retinoids in the chemistry of vision. Trends Biochem Sci 35: 400–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. 2009. Temporally precise in vivo control of intracellular signalling. Nature 458: 1025–9 [DOI] [PubMed] [Google Scholar]
- 59.Masseck OA, Spoida K, Dalkara D, Maejima T, Rubelowski JM, et al. 2014. Vertebrate cone opsins enable sustained and highly sensitive rapid control of Gi/o signaling in anxiety circuitry. Neuron 81: 1263–73 [DOI] [PubMed] [Google Scholar]
- 60.Spoida K, Masseck OA, Deneris ES, Herlitze S. 2014. Gq/5-HT2c receptor signals activate a local GABAergic inhibitory feedback circuit to modulate serotonergic firing and anxiety in mice. Proc Natl Acad Sci U S A 111: 6479–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Wyk M, Pielecka-Fortuna J, Lowel S, Kleinlogel S. 2015. Restoring the ON Switch in Blind Retinas: Opto-mGluR6, a Next-Generation, Cell-Tailored Optogenetic Tool. PLoS Biol 13: e1002143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Siuda ER, McCall JG, Al-Hasani R, Shin G, Il Park S, et al. 2015. Optodynamic simulation of beta-adrenergic receptor signalling. Nat Commun 6: 8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ando R, Mizuno H, Miyawaki A. 2004. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science 306: 1370–3 [DOI] [PubMed] [Google Scholar]
- 64.Andresen M, Stiel AC, Trowitzsch S, Weber G, Eggeling C, et al. 2007. Structural basis for reversible photoswitching in Dronpa. Proc Natl Acad Sci U S A 104: 13005–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mizuno H, Mal TK, Walchli M, Kikuchi A, Fukano T, et al. 2008. Light-dependent regulation of structural flexibility in a photochromic fluorescent protein. Proc Natl Acad Sci U S A 105: 9227–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Padmanabhan S, Jost M, Drennan CL, Elias-Arnanz M. 2017. A New Facet of Vitamin B12: Gene Regulation by Cobalamin-Based Photoreceptors. Annu Rev Biochem 86: 485–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ortiz-Guerrero JM, Polanco MC, Murillo FJ, Padmanabhan S, Elias-Arnanz M. 2011. Light-dependent gene regulation by a coenzyme B12-based photoreceptor. Proc Natl Acad Sci U S A 108: 7565–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kainrath S, Stadler M, Reichhart E, Distel M, Janovjak H. 2017. Green-Light-Induced Inactivation of Receptor Signaling Using Cobalamin-Binding Domains. Angew Chem Int Ed Engl 56: 4608–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jost M, Fernandez-Zapata J, Polanco MC, Ortiz-Guerrero JM, Chen PY, et al. 2015. Structural basis for gene regulation by a B12-dependent photoreceptor. Nature 526: 536–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levskaya A, Weiner OD, Lim WA, Voigt CA. 2009. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461: 997–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kaberniuk AA, Shemetov AA, Verkhusha VV. 2016. A bacterial phytochrome-based optogenetic system controllable with near-infrared light. Nat Methods 13: 591–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rockwell NC, Martin SS, Feoktistova K, Lagarias JC. 2011. Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc Natl Acad Sci U S A 108: 11854–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Uda Y, Miura H, Goto Y, Aoki K. 2020. Improvement of phycocyanobilin synthesis for genetically encoded phytochrome-based optogenetics. bioRxiv [DOI] [PubMed] [Google Scholar]
- 74.Strickland D, Moffat K, Sosnick TR. 2008. Light-activated DNA binding in a designed allosteric protein. Proc Natl Acad Sci U S A 105: 10709–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, et al. 2009. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461: 104–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murakoshi H, Shin ME, Parra-Bueno P, Szatmari EM, Shibata AC, Yasuda R. 2017. Kinetics of Endogenous CaMKII Required for Synaptic Plasticity Revealed by Optogenetic Kinase Inhibitor. Neuron 94: 37–47 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lee J, Natarajan M, Nashine VC, Socolich M, Vo T, et al. 2008. Surface sites for engineering allosteric control in proteins. Science 322: 438–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smart AD, Pache RA, Thomsen ND, Kortemme T, Davis GW, Wells JA. 2017. Engineering a light-activated caspase-3 for precise ablation of neurons in vivo. Proc Natl Acad Sci U S A 114: E8174–E83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dagliyan O, Tarnawski M, Chu PH, Shirvanyants D, Schlichting I, et al. 2016. Engineering extrinsic disorder to control protein activity in living cells. Science 354: 1441–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nakamura M, Chen L, Howes SC, Schindler TD, Nogales E, Bryant Z. 2014. Remote control of myosin and kinesin motors using light-activated gearshifting. Nat Nanotechnol 9: 693–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, et al. 2005. Synthetic biology: engineering Escherichia coli to see light. Nature 438: 441–2 [DOI] [PubMed] [Google Scholar]
- 82.Moglich A, Ayers RA, Moffat K. 2009. Design and signaling mechanism of light-regulated histidine kinases. J Mol Biol 385: 1433–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ryu MH, Kang IH, Nelson MD, Jensen TM, Lyuksyutova AI, et al. 2014. Engineering adenylate cyclases regulated by near-infrared window light. Proc Natl Acad Sci U S A 111: 10167–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gasser C, Taiber S, Yeh CM, Wittig CH, Hegemann P, et al. 2014. Engineering of a red-light-activated human cAMP/cGMP-specific phosphodiesterase. Proc Natl Acad Sci U S A 111: 8803–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ziegler T, Moglich A. 2015. Photoreceptor engineering. Front Mol Biosci 2: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang X, Chen X, Yang Y. 2012. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods 9: 266–9 [DOI] [PubMed] [Google Scholar]
- 87.Chang KY, Woo D, Jung H, Lee S, Kim S, et al. 2014. Light-inducible receptor tyrosine kinases that regulate neurotrophin signalling. Nat Commun 5: 4057. [DOI] [PubMed] [Google Scholar]
- 88.Kyung T, Lee S, Kim JE, Cho T, Park H, et al. 2015. Optogenetic control of endogenous Ca(2+) channels in vivo. Nat Biotechnol 33: 1092–6 [DOI] [PubMed] [Google Scholar]
- 89.Park H, Kim NY, Lee S, Kim N, Kim J, Heo WD. 2017. Optogenetic protein clustering through fluorescent protein tagging and extension of CRY2. Nat Commun 8: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Duan L, Hope J, Ong Q, Lou HY, Kim N, et al. 2017. Understanding CRY2 interactions for optical control of intracellular signaling. Nat Commun 8: 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang K, Duan L, Ong Q, Lin Z, Varman PM, et al. 2014. Light-mediated kinetic control reveals the temporal effect of the Raf/MEK/ERK pathway in PC12 cell neurite outgrowth. PLoS One 9: e92917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sinnen BL, Bowen AB, Forte JS, Hiester BG, Crosby KC, et al. 2017. Optogenetic Control of Synaptic Composition and Function. Neuron 93: 646–60 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee S, Park H, Kyung T, Kim NY, Kim S, et al. 2014. Reversible protein inactivation by optogenetic trapping in cells. Nat Methods 11: 633–6 [DOI] [PubMed] [Google Scholar]
- 94.Dine E, Gil AA, Uribe G, Brangwynne CP, Toettcher JE. 2018. Protein Phase Separation Provides Long-Term Memory of Transient Spatial Stimuli. Cell Syst 6: 655–63 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Morgan SA, Al-Abdul-Wahid S, Woolley GA. 2010. Structure-based design of a photocontrolled DNA binding protein. J Mol Biol 399: 94–112 [DOI] [PubMed] [Google Scholar]
- 96.Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, et al. 2015. Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc Natl Acad Sci U S A 112: 112–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, et al. 2012. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat Methods 9: 379–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kawano F, Suzuki H, Furuya A, Sato M. 2015. Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nat Commun 6: 6256. [DOI] [PubMed] [Google Scholar]
- 99.Kawano F, Okazaki R, Yazawa M, Sato M. 2016. A photoactivatable Cre-loxP recombination system for optogenetic genome engineering. Nat Chem Biol 12: 1059–64 [DOI] [PubMed] [Google Scholar]
- 100.Wang H, Vilela M, Winkler A, Tarnawski M, Schlichting I, et al. 2016. LOVTRAP: an optogenetic system for photoinduced protein dissociation. Nat Methods 13: 755–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Stone OJ, Pankow N, Liu B, Sharma VP, Eddy RJ, et al. 2019. Optogenetic control of cofilin and alphaTAT in living cells using Z-lock. Nat Chem Biol 15: 1183–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Morgan SA, Woolley GA. 2010. A photoswitchable DNA-binding protein based on a truncated GCN4-photoactive yellow protein chimera. Photochem Photobiol Sci 9: 1320–6 [DOI] [PubMed] [Google Scholar]
- 103.Ali AM, Reis JM, Xia Y, Rashid AJ, Mercaldo V, et al. 2015. Optogenetic Inhibitor of the Transcription Factor CREB. Chem Biol 22: 1531–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ma G, Liu J, Ke Y, Liu X, Li M, et al. 2018. Optogenetic Control of Voltage-Gated Calcium Channels. Angew Chem Int Ed Engl 57: 7019–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cosentino C, Alberio L, Gazzarrini S, Aquila M, Romano E, et al. 2015. Optogenetics. Engineering of a light-gated potassium channel. Science 348: 707–10 [DOI] [PubMed] [Google Scholar]
- 106.Valon L, Marin-Llaurado A, Wyatt T, Charras G, Trepat X. 2017. Optogenetic control of cellular forces and mechanotransduction. Nat Commun 8: 14396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oakes PW, Wagner E, Brand CA, Probst D, Linke M, et al. 2017. Optogenetic control of RhoA reveals zyxin-mediated elasticity of stress fibres. Nat Commun 8: 15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Woo D, Seo Y, Jung H, Kim S, Kim N, et al. 2019. Locally Activating TrkB Receptor Generates Actin Waves and Specifies Axonal Fate. Cell Chem Biol 26: 1652–63 e4 [DOI] [PubMed] [Google Scholar]
- 109.Kim N, Kim JM, Lee M, Kim CY, Chang KY, Heo WD. 2014. Spatiotemporal control of fibroblast growth factor receptor signals by blue light. Chem Biol 21: 903–12 [DOI] [PubMed] [Google Scholar]
- 110.Huang P, Liu A, Song Y, Hope JM, Cui B, Duan L. 2020. Optical Activation of TrkB Signaling. J Mol Biol 432: 3761–70 [DOI] [PubMed] [Google Scholar]
- 111.Leopold AV, Chernov KG, Verkhusha VV. 2018. Optogenetically controlled protein kinases for regulation of cellular signaling. Chem Soc Rev 47: 2454–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hallett RA, Zimmerman SP, Yumerefendi H, Bear JE, Kuhlman B. 2016. Correlating in Vitro and in Vivo Activities of Light-Inducible Dimers: A Cellular Optogenetics Guide. ACS Synth Biol 5: 53–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Motta-Mena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, et al. 2014. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol 10: 196–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Quejada JR, Park SE, Awari DW, Shi F, Yamamoto HE, et al. 2017. Optimized light-inducible transcription in mammalian cells using Flavin Kelch-repeat F-box1/GIGANTEA and CRY2/CIB1. Nucleic Acids Res 45: e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Imayoshi I, Isomura A, Harima Y, Kawaguchi K, Kori H, et al. 2013. Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science 342: 1203–8 [DOI] [PubMed] [Google Scholar]
- 116.Polstein LR, Gersbach CA. 2015. A light-inducible CRISPR-Cas9 system for control of endogenous gene activation. Nat Chem Biol 11: 198–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nihongaki Y, Kawano F, Nakajima T, Sato M. 2015. Photoactivatable CRISPR-Cas9 for optogenetic genome editing. Nat Biotechnol 33: 755–60 [DOI] [PubMed] [Google Scholar]
- 118.Zhou XX, Zou X, Chung HK, Gao Y, Liu Y, et al. 2017. A Single-Chain Photoswitchable CRISPR-Cas9 Architecture for Light-Inducible Gene Editing and Transcription. ACS Chem Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhao J, Li B, Ma J, Jin W, Ma X. 2020. Photoactivatable RNA N(6) -Methyladenosine Editing with CRISPR-Cas13. Small: e1907301 [DOI] [PubMed] [Google Scholar]
- 120.Mathony J, Hoffmann MD, Niopek D. 2020. Optogenetics and CRISPR: A New Relationship Built to Last. Methods Mol Biol 2173: 261–81 [DOI] [PubMed] [Google Scholar]
- 121.Morikawa K, Furuhashi K, de Sena-Tomas C, Garcia-Garcia AL, Bekdash R, et al. 2020. Photoactivatable Cre recombinase 3.0 for in vivo mouse applications. Nat Commun 11: 2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Meador K, Wysoczynski CL, Norris AJ, Aoto J, Bruchas MR, Tucker CL. 2019. Achieving tight control of a photoactivatable Cre recombinase gene switch: new design strategies and functional characterization in mammalian cells and rodent. Nucleic Acids Res 47: e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jung H, Kim SW, Kim M, Hong J, Yu D, et al. 2019. Noninvasive optical activation of Flp recombinase for genetic manipulation in deep mouse brain regions. Nat Commun 10: 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Goglia AG, Toettcher JE. 2019. A bright future: optogenetics to dissect the spatiotemporal control of cell behavior. Curr Opin Chem Biol 48: 106–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.van Bergeijk P, Hoogenraad CC, Kapitein LC. 2016. Right Time, Right Place: Probing the Functions of Organelle Positioning. Trends Cell Biol 26: 121–34 [DOI] [PubMed] [Google Scholar]
- 126.Shin Y, Brangwynne CP. 2017. Liquid phase condensation in cell physiology and disease. Science 357 [DOI] [PubMed] [Google Scholar]
- 127.Duan L, Che D, Zhang K, Ong Q, Guo S, Cui B. 2015. Optogenetic control of molecular motors and organelle distributions in cells. Chem Biol 22: 671–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.van Bergeijk P, Adrian M, Hoogenraad CC, Kapitein LC. 2015. Optogenetic control of organelle transport and positioning. Nature 518: 111–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gil AA, Zhao EM, Wilson MZ, Goglia AG, Carrasco-Lopez C, et al. 2019. Optogenetic control of protein binding using light-switchable nanobodies. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pertz O, Hodgson L, Klemke RL, Hahn KM. 2006. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature 440: 1069–72 [DOI] [PubMed] [Google Scholar]
- 131.Krishnamurthy VV, Khamo JS, Mei W, Turgeon AJ, Ashraf HM, et al. 2016. Reversible optogenetic control of kinase activity during differentiation and embryonic development. Development 143: 4085–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Regev A, Shapiro E. 2002. Cells as computation. Nature 419: 343. [DOI] [PubMed] [Google Scholar]
- 133.Eungdamrong NJ, Iyengar R. 2004. Computational approaches for modeling regulatory cellular networks. Trends Cell Biol 14: 661–9 [DOI] [PMC free article] [PubMed] [Google Scholar]