Abstract
The RNA polymerase of giardiavirus (GLV) is synthesized as a fusion protein through a −1 ribosomal frameshift in a region where gag and pol open reading frames (ORFs) overlap. A heptamer, CCCUUUA, and a potential pseudoknot found in the overlap were predicted to be required for the frameshift. A 68-nucleotide (nt) cDNA fragment containing these elements was inserted between the GLV 5′ 631-nt cDNA and the out-of-frame luciferase gene that required a −1 frameshift within the 68-nt fragment for expression. Giardia lamblia trophozoites transfected with the transcript of this construct showed a frameshift frequency at 1.7%, coinciding with the polymerase-to-capsid protein ratio in GLV. The heptamer is required for the frameshift but can be replaced with other sequences of the same motif. Mutations placing stop codons in the 0 or −1 frame, located directly before or after the heptamer, implicated the latter as the site for the −1 frameshift. Shortening or destroying the putative stem decreased the frameshift efficiency threefold; the efficiency was fully recovered by mutations to restore the stem. Deleting 18 nt from the 3′ end of the 68-nt fragment, which formed the second stem in the putative pseudoknot, had no effect on the frequency of the frameshift. Chemical probing of the RNA secondary structure in the frameshift region showed that bases resistant to chemical modification were clustered in the putative stem structures, thus confirming the presence of the postulated stem-loop, while all the bases in the loop were chemically modified, thus ruling out their capability of forming a pseudoknot. These results confirmed the conclusion based on data from the mutation study that there is but a simple stem-loop downstream from the heptamer. Together, they constitute the structural elements for a −1 ribosomal frameshift in the GLV transcript.
Although faithful reading of open reading frames (ORFs) in mRNA is most critical for the production of functional proteins, programmed ribosomal frameshifts have been increasingly reported as the means of regulating gene expression (2, 11, 13). An efficient −1 ribosomal frameshift is one of such examples of a programmed posttranscriptional regulation of gene expression. In response to certain specific structural signals in the mRNA, the ribosomes are induced to slip back 1 nucleotide (nt) at a fixed frequency, move into the −1 reading frame at a specific site in the mRNA, and continue translating the rest of the mRNA in the −1 frame (19, 20).
Many viruses are known to depend on this mechanism of −1 ribosomal frameshift to generate the RNA polymerase gene (pol) product in the form of a fusion protein (Gag-Pol) with the capsid protein (Gag) at its N terminus and polymerase (Pol) at its C terminus (2). The production of Gag and Gag-Pol at a fixed ratio during translation enables the inclusion of RNA polymerase in the assembled virus particles at a constant level (42). This inclusion, in turn, makes it possible for continuous replication and transcription of the viral genome inside viral particles within the infected cells.
The −1 ribosomal frameshift was first observed in the retrovirus Rous sarcoma virus, in which the viral polymerase was translated from two overlapping gag and pol ORFs requiring a −1 ribosomal frameshift within the overlapping region (20). This phenomenon of the −1 ribosomal frameshift has since been observed among translations of gene transcripts from a large number of viruses (2), certain Escherichia coli genetic insertion elements (11), and a conventional cellular dnaX gene from E. coli (35, 36). The structural motifs in mRNA that are important for an efficient −1 ribosomal frameshift have been characterized in several viral systems primarily by in vitro translation assays (2, 7, 13). Two structural components have been confirmed to induce such activity. A homopolymeric “slippery” heptamer sequence (X XXY YYZ) is required, where XXX can be any three identical nucleotides, YYY can be either AAA or UUU, and Z can be A, U, or C (4, 8, 9). The second component consists of a stem-loop or a pseudoknot, which is defined as two intertwined stem-loops where a region in the first loop forms base pairs with a downstream sequence to produce a second stem (32). A pseudoknot is essential for the −1 ribosomal frameshift in infectious bronchitis virus (IBV) (3, 5), human coronavirus (16), and yeast killer virus (ScV/L-A) (7). However, among other viruses including human immunodeficiency virus (HIV) (29), human T-cell leukemia virus type 2 (10), human astrovirus serotype 1 (23), potato leaf roll virus (30), and red clover necrotic mosaic virus (21), a pseudoknot is apparently not essential for the −1 ribosomal frameshift. All that is required is a slippery heptamer and a stem-loop located a few nucleotides downstream from it.
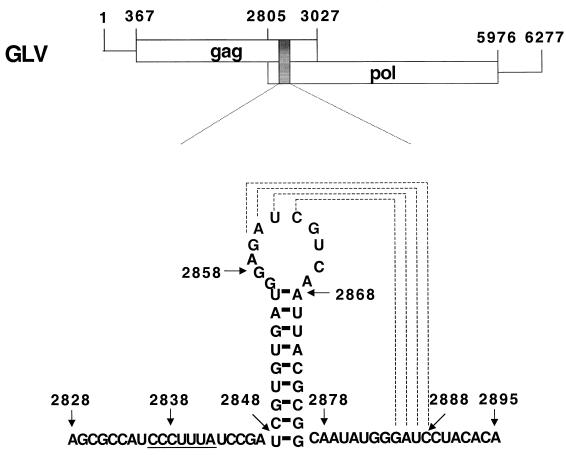
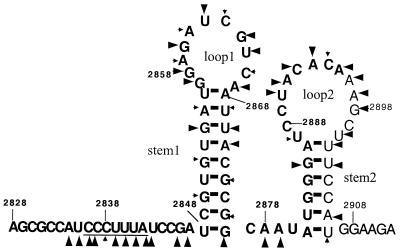
Giardiavirus (GLV) is a small (36-nm diameter) icosahedral virus of the Totiviridae family that specifically infects the trophozoites of Giardia lamblia, an anaerobic protozoan that causes diarrhea and malnutrition in human (26, 38, 39). Its 6,277-nt double-stranded RNA genome contains gag and pol-like ORFs that overlap by 220 nt and are separated by a −1 frameshift (40). Immunostudies with antipeptide sera targeted to regions in the respective ORFs indicated that the 100-kDa capsid protein is encoded by ORF1. They also showed that the N terminus of the 190-kDa GLV minor protein is encoded by ORF1 while its C terminus has all the consensus sequence motifs of the RNA-dependent RNA polymerase (RDRP) family. The 190-kDa protein is therefore most probably a Gag-Pol fusion protein produced by a −1 ribosomal frameshift that is predicted to occur in the 220-nt overlapping region (40). Within this region, we found a putative slippery heptamer, CCCUUUA, at nt 2836 to 2842 and a downstream stem-loop structure at nt 2848 to 2876 as predicted by MFOLD (25). A potential second stem could be also formed between the GAUC at nt 2860 to 2863 in the loop and the downstream GAUC at nt 2885 to 2888 (see Fig. 1), resulting in a pseudoknot (32). Together, they were predicted to constitute the structural requirements for the ribosomal −1 frameshift that lead to the formation of the GLV Gag-Pol fusion protein (40).
FIG. 1.
The two overlapping ORFs in GLV mRNA. The shaded box represents the 68-nt region within the 220-nt overlap containing the putative structures required for the −1 ribosomal frameshift, which is detailed below. The underlined heptamer is the putative slippery sequence. The stem-loop secondary structure is predicted by MFOLD. The two GAUC regions (linked by dashed lines) may be involved in the formation of an RNA pseudoknot structure.
To verify this assumption, a 68-bp cDNA fragment from nt 2828 to 2895 of the GLV genome containing the two postulated frameshift structural motifs was inserted in front of an out-of-frame luciferase gene in a GLV-based viral vector, pC631-luc (44). This construct requires a −1 frameshift within the 68-nt region for luciferase expression, and the efficiency of the −1 ribosomal frameshift can then be determined by monitoring the luciferase activity in transfected, GLV-infected Giardia trophozoites. We made a large number of mutants with site-directed mutations in the 68-nt region to examine the function of the postulated heptamer and the putative downstream pseudoknot. RNA bases in this putative frameshift region were also probed by chemical modifications to reveal the secondary structures in this region. Results from these two lines of studies helped to delineate the structural requirements for inducing a −1 ribosomal frameshift in the GLV transcript.
MATERIALS AND METHODS
Construction of the recombinant cDNA plasmids.
Based on the GLV genome sequence (GenBank accession number L13218), three primers, each having a HindIII site (underlined) at the 5′ end, were synthesized; fs1, TGGCAAGCTTTGGTACTCAGACAC; fs2, AAAGCTTTGTGTAGGATCCC; and fs3, AAAGCTTCTGTGTAGGATCCC. Using pGEM-GLV, which contains the full-length GLV cDNA (44), as the template, fs1 and fs2 were included in PCR for synthesis of the 68-nt cDNA fragment (nt 2828 to 2895), whereas fs1 and fs3 were used for synthesizing the same cDNA fragment with an extra G added to the 3′ end (a 69-nt cDNA fragment). The 68- and 69-nt PCR fragments were then each inserted into the HindIII-restricted pC631-luc vector (44) between the 5′ 631-nt GLV cDNA and the full-length luciferase gene to constitute pC631(68)luc or pC631(69)luc. The orientation and the fidelity of each fragment were determined by DNA sequencing. The ORFs in both fragments are fused with that of the 5′ 631-nt GLV cDNA. However, the luciferase gene in pC631(68)luc is out of frame and requires a −1 frameshift for its expression, whereas the pC631(69)luc is an in-frame construct.
In vitro site-directed mutagenesis of the recombinant plasmid.
Site-directed mutagenesis was performed with QuickChange as directed by the manufacturer (Life Technologies BRL). For each mutation, two complementary oligonucleotide primers were synthesized with the intended mutation introduced in the midportion of each primer. The PCR-synthesized mutant DNA fragment was amplified in E. coli DH5α cells and purified. Each specific mutation was verified by directly sequencing the cloned mutant plasmid.
In vitro synthesis of chimeric RNA.
Wild-type and mutant plasmids were each restricted with NruI at the 3′ end of the GLV cDNA. In vitro transcription of each linearized plasmid with T7 RNA polymerase was performed in a 20-μl reaction mixture containing 0.5 μg of linearized plasmid DNA as described by the manufacturer (Ambion). The RNA thus synthesized was purified by LiCl precipitation and examined by electrophoresis in a 1.0% agarose–formaldehyde gel for integrity. The concentration of each RNA sample was estimated by measuring its absorption at 260 nm in a Beckman DU7 spectrophotometer.
Transfection of G. lamblia trophozoites and luciferase assay.
In vitro culture of GLV-infected G. lamblia WB trophozoites (WBI) was maintained as described previously (38). Serial passages of the in vitro culture were performed at an inoculation ratio of 1:13 every 3 days into fresh medium to maintain a continuous logarithmic cell growth. Transfection of Giardia trophozoites with RNA was performed by electroporation, and assay of the luciferase activity in the lysate of transfectants was performed as previously described (45).
Probing the RNA structure by chemical modification and primer extension.
The RNA molecule used for chemical probing was the in vitro transcript of pC631(68)luc. It was prepared using a T7 RNA polymerase kit (MegaScript from Ambion) and the NruI-restricted pC631(68)luc as template. The RNA was quantified by spectrophotometry, and its integrity was checked by electrophoresis on a 1% agarose–formaldehyde gel. Approximately 5 μg of the in vitro-synthesized RNA was used in each reaction mixture containing either 0.5% dimethyl sulfate (DMS), 21 mg of 1-cyclohexyl-3-(2-morpholinoethyl)-carbodiimide (CMCT) per ml, or 3.5 mg of kethoxal (KE) per ml as previously described (12). Prior to the probing analysis, the RNA was heated at 70°C for 15 min and cooled slowly to room temperature over a 45-min period in a 200-μl nondenaturing probing buffer, HMK (80 mM HEPES-KOH [pH 7.8], 10 mM MgCl2, 270 mM KCl). The reaction mixtures were incubated at 37°C for 0, 10, and 20 min. The reactions with the DMS and CMCT modifications were terminated by adding 75 μl of the stop buffer (1 M Tris-acetate [pH 7.5], 1 M β-mercaptoethanol, 1.5 M sodium acetate, 0.1 mM EDTA). The KE modification was stabilized by adding 0.5 volume of 250 mM potassium borate (pH 7.0). Chemically modified RNA was precipitated by adding 3 volumes of 100% ethanol.
Sites of RNA modification were mapped by primer extension using the reverse transcriptase Superscript (Life Technologies BRL) with radiolabeled oligonucleotides targeted to the region either 22 nt downstream from stem-loop 1 or 31 nt downstream from stem-loop 2 (see Fig. 4). Briefly, 32P-end-labeled primers were annealed to approximately 1 μg of the chemically modified RNA by incubating the mixture for 15 min at 75°C followed by 10 min on ice. Primer extension was carried out at 50°C for 1 h as specified by the manufacturer. Reaction products were analyzed on a 10% denaturing acrylamide gel along with sequencing ladders prepared by the fmol cycle-sequencing system (Promega).
FIG. 4.
RNA secondary structure predicted by MFOLD on deletion of the first stem-loop structure between nt 2848 and 2876 (stem-loop 1). A 30-bp fragment from nt 2848 to 2877 in the 68-nt region was deleted by site-directed mutagenesis, thus removing stem-loop 1 predicted by MFOLD. The downstream second stem-loop structure predicted between nt 2880 and 2907 (stem-loop 2) in the deletion mutant including the first luciferase codon AUG at the base of the stem contains a 7-bp stem and is 7 nt downstream from the slippery site. The relative frequency of the −1 ribosomal frameshift is not significantly changed from that of the wild type in the mutant transcript transfectant (see Table 1 for actual data). Bold type represents the sequence from GLV mRNA, whereas normal type represents the sequence from the luciferase mRNA.
RESULTS
In vivo efficiency of the −1 ribosomal frameshift mediated by the 68-nt fragment in the ORF overlapping region of GLV mRNA.
The RNA transcript of pC631(68)luc or pC631(69)luc was introduced into Giardia WBI trophozoites by electroporation, and the luciferase activity in the cell lysates of the two transfectants was monitored. Since the in vitro transcript from pC631(69)luc contained only one continuous ORF, the luciferase activity found in the lysate of its transfectant, which demonstrated a specific activity of 408,890 ± 71,250 relative light units (RLU)/25 μg of protein in repeated independent experiments, is defined as 100% luciferase expression (Table 1). In comparison, the luciferase activity in the lysate of Giardia trophozoites transfected with the in vitro transcript of pC631(68)luc was 6,768 ± 608 RLU/25 μg of protein and constituted 1.7% of that of the in-frame construct pC631(69)luc (Table 1). Since luciferase expression in the pC631(68)luc transcript transfectant relies on the −1 ribosomal frameshift in the 68-nt overlapping region, the results indicate that this 68-nt fragment in mRNA is indeed capable of causing −1 ribosomal frameshift at a frequency of 1.7%.
TABLE 1.
Luciferase activities of Giardia cells transfected with various mutant transcripts
| Mutation | Luciferase activity (RLU/25 μg of protein)a | % of wild-type activity | Remarks |
|---|---|---|---|
| Ab | |||
| pC631(69)luc | 408,890 ± 71,250 | 100 | Positive control |
| pC631(68)luc | 6,768 ± 608 | 1.7 ± 0.2 | Wild type |
| pC631(69R)luc | 555 ± 270 | 0.13 ± 0.06 | Negative control |
| Bc | |||
| pC631(68)luc | 6,768 ± 608 | 100 | Wild type |
| pC631(69R)luc | 555 ± 270 | 8.2 ± 4.0 | Negative control |
| Stop codons before the heptamer | |||
| GCC/uaa (2831–2833) | 582 ± 115 | 8.6 ± 1.7 | 0 frame stop codon |
| CAU/uaa (2833–2835) | 3,641 ± 677 | 53.8 ± 1.0 | −1 frame stop codon |
| Stop codons after the heptamer | |||
| UCC/Uaa (2843–2845) | 2,558 ± 569 | 37.8 ± 8.4 | 0 frame stop codon |
| GAU/uaa (2846–2848) | 3,526 ± 406 | 52.1 ± 6.0 | 0 frame stop codon |
| CGA/uGA (2845–2847) | 1,211 ± 420 | 17.9 ± 6.2 | −1 frame stop codon |
| UCC/Uaa (2887–2889) | 650 ± 338 | 9.6 ± 5.0 | −1 frame stop codon |
| AUA/uaA (2879–2881) | 3,878 ± 213 | 57.3 ± 3.1 | 0 frame stop codon |
| ACA/uaA (2891–2893) | 4,893 ± 271 | 72.3 ± 4.0 | 0 frame stop codon |
| Other heptamers | |||
| CCC/uuu (2836–2838) | 8,890 ± 724 | 131.2 ± 10.7 | Change to another heptamer |
| CCCUUUA/uuuuuuu (2836–2842) | 29,366 ± 2,748 | 433.9 ± 40.6 | Change to another heptamer |
| Putative pseudoknot | |||
| GAUC/ctag (2885–2888) | 4,792 ± 169 | 70.8 ± 2.5 | Disrupted putative pseudoknot |
| 18-nt deletion (2878–2895) | 6,348 ± 1,002 | 93.8 ± 14.8 | Disrupted putative pseudoknot |
| Stem-loop region | |||
| AUUA/tagt (2868–2871) | 2,829 ± 399 | 41.8 ± 5.9 | Shortened stem |
| CGUG/gcgc (2849–2852) | 2,585 ± 521 | 38.2 ± 7.7 | Shortened stem |
| CGCG/gugc (2872–2875) | 2,186 ± 271 | 32.3 ± 4.0 | Shortened stem |
| CGUG/gcgc (2849–2852)–CGCG/gugc (2872–2875) | 6,937 ± 596 | 102.5 ± 8.8 | Restored stem |
| 30-nt deletion (2848–2877) | 5,787 ± 582 | 85.5 ± 8.6 | Deleted stem |
Each value represents an average of luciferase activities obtained from at least three independent transfection experiments. Wild type and mutants were each transfected in triplicate in each experiment.
Luciferase expression caused by the wild-type 68-nt fragment is compared with that of the in-frame construct pC631(69)luc, which is defined as 100% expression.
Luciferase expression from various mutants is compared with that of pC631(68)luc, which represents 100% of wild-type frameshift frequency.
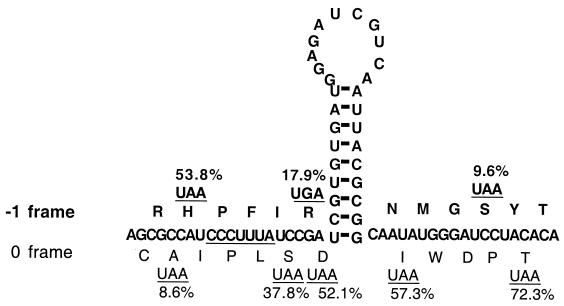
Identification of the site for the −1 ribosomal frameshift.
If the postulated heptamer CCCUUUA is indeed one of the structural elements causing the frameshift, then it would also be the site of the frameshift as predicted by the widely accepted simultaneous-slippage model (19). By site-directed mutagenesis, we placed stop codon UAA in 0 frame at nt 2831 to 2833, 2843 to 2845, 2846 to 2848, 2879 to 2881, or 2891 to 2893 in pC631(68)luc (Fig. 1). If the −1 ribosomal frameshift indeed occurs on top of the heptamer, the first UAA upstream from it in the 0 frame is expected to disrupt luciferase expression whereas the other four downstream stop codons in the 0 frame will no longer be read as stop codons and should not significantly affect luciferase expression. Our data derived from corresponding transfectants of the mutant transcripts (Fig. 2) indicate that the first UAA caused a precipitous drop in luciferase expression to 8.6% ± 1.7% of the wild-type frequency of the frameshift or 0.15% of the expression by the in-frame construct pC631(69)luc, while the other four 0 frame stop codons downstream of the heptamer showed luciferase expression at 37.8% ± 8.4%, 52.1% ± 6.0%, 57.3% ± 3.1%, and 72.3% ± 4.0% of the wild-type frameshift frequency (Fig. 2; Table 1). A negative-control pC631(69R)luc, which has the 69-nt fragment reversed and thus lacks the structural features of the 69-nt fragment, showed 8.2% ± 4.0% of the wild-type frameshift frequency or 0.13% ± 0.06% of the in-frame expression. This value is thus regarded as the noise or background in our assay system.
FIG. 2.
Mutational analysis via generation of stop codons in different reading frames at different locations to identify the slippery site of −1 ribosomal frameshift. The putative slippery heptamer CCCUUUA sequence is underlined. The predicted peptide sequences in the 0 and −1 frames are shown in the one-letter amino acid code. Individual termination codons in either the 0 frame or the −1 frame are created by site-directed mutagenesis. The luciferase activities expressed in the mutant transcript transfectants were determined from three independent experiments, with data within 10% of experimental error. The frequencies of the −1 ribosomal frameshift relative to the wild-type frequency (100%) in the mutant transfectants are each shown at the corresponding position of the termination codon (see Table 1 for actual data).
We further examined the question in an opposite way. When a stop codon is introduced into the −1 frame, one would expect that codons placed upstream from the site of frameshift will cause little disruption whereas those placed downstream from the shift will have a significant effect. Figure 2 indicates that UAA at nt 2833 to 2835 upstream from the heptamer has a frequency 53.8% ± 1.0% of that of the wild-type frameshift whereas a UGA at nt 2845 to 2847 and a UAA at nt 2887 to 2889, both downstream from the heptamer, lead to 17.9% ± 6.2% and 9.6% ± 5.0% frequencies, i.e., nearly the background level of the frameshift (see Table 1).
Data from Fig. 2 and Table 1 thus provide a strong indication that a −1 ribosomal frameshift may indeed occur at the putative slippery heptamer site CCCUUUA between nt 2836 and 2842 in GLV mRNA. To find if the sequence of this particular heptamer is unique in causing a −1 translation frameshift in Giardia, we replaced it with a slippery heptamer UUUUUUA identified previously in HIV (19) and an engineered UUUUUUU which showed excellent frameshifting activity in vitro in a previous study (4). Giardia trophozoites transfected with each of the mutant transcripts indicated that UUUUUA and UUUUUUU led to 131.2% ± 10.7% and 433.9% ± 40.6% of the wild-type frequency of frameshift, respectively, both of which significantly exceeded the effect from the original heptamer in GLV mRNA. Thus, heptamers functioning well in causing −1 ribosomal frameshift in mammalian cells also worked well in Giardia trophozoites. The advantage in retaining a less efficient slippery heptamer in the mRNA by GLV is not entirely clear. One possible explanation is the need for a rigid molar ratio between the RDRP and the capsid protein in the viral particles for stable maintenance of the virus inside Giardia cells, such as that observed in yeast killer virus (9).
While the CCCUUUA sequence identified in GLV mRNA conforms to the generally accepted structural rule for a slippery heptamer (4, 8, 9), alteration of individual nucleotides in the heptamer invariably reduces the frequency of the frameshift but never abolishes it. For instance, CCCUAUA led to 60% of the wild-type frameshift frequency whereas UCCUUUA, CCCAUUA, and CCCUUUG led to 78, 85, and 98% of the frameshift frequency, respectively (data not shown). Thus, although some of the heptamers may be more slippery for the ribosome than are the others, depending on the nucleotide sequences, there may not be an absolutely non-slippery heptamer. Initiation of a ribosomal frameshift may be dependent primarily on the presence of a ribosome-blocking secondary structure downstream from the heptamer.
A simple stem-loop structure, but not a pseudoknot, constitutes the second structural element for the −1 ribosomal frameshift on GLV mRNA.
The heptamer and a putative pseudoknot-like structure were previously identified within the overlapping region between gag and pol ORFs in GLV mRNA (Fig. 1) (40). To verify whether the putative second stem formation between the two GAUC tetranucleotides at nt 2860 to 2863 and 2885 to 2888 in the 68-nt region is essential for the frameshift (Fig. 1), we altered the sequence at nt 2860 to 2863 from GAUC to CUAG to eliminate the possible pseudoknot formation from the transcript. The resulting mutant retained 70.8% ± 2.5% of the wild-type frameshift frequency, suggesting that the putative pseudoknot structure is not essential for the frameshift. To further confirm this conclusion, an 18-nt deletion from the 3′ end of the 68-nt fragment was performed on the encoding cDNA, which also removed the downstream GAUC tetranucleotide involved in the putative pseudoknot formation. The transfectant containing such a mutant mRNA demonstrated 93.8% ± 14.8% of the wild-type frameshift frequency (Table 1). It is thus clear that neither the pseudoknot structure nor the 3′-end 18 nt of the 68-nt fragment is needed for the occurrence of the −1 ribosomal frameshift. The 50-nt fragment from nt 2828 to 2877 in GLV mRNA thus contains both the essential and sufficient structural elements for the frameshift.
To examine if the loop in the putative stem-loop between nt 2848 and 2876 (Fig. 1) can form a second stem with the downstream sequences elsewhere within the 220-nt ORF overlap outside of the 68-nt fragment that were replaced by the luciferase sequence in our construct, we replaced the 68-nt insert in pC631(68)luc with a fragment including a 215-nt fragment from this 220-nt overlap. The frameshift frequency was the same as in pC631(68)luc (data not shown). It is therefore most likely that all the structural elements required for inducing the −1 ribosomal frameshift are contained within the 50-nt fragment.
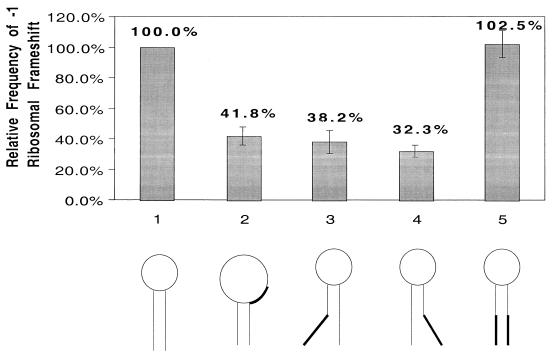
Mutations that shorten the stem in the putative stem-loop in the 68-nt overlapping region decrease the efficiency of the −1 ribosomal frameshift.
A putative stem-loop at nt 2848 to 2876, consisting of a 9-bp stem and a loop of 11 nt, is the only secondary structure predicted by MFOLD (25) in the 68-nt RNA fragment (Fig. 1). To investigate the functional importance of this structure, mutations to shorten the stem were performed on the encoding cDNA. Firstly, AUUA at nt 2868 to 2871 from the top part of the stem was replaced with UAGU, resulting in a shortened stem from 9 to 5 bp and an enlarged loop from 11 to 19 nt (Fig. 3, structure 2). The in vivo luciferase activity derived from this mutant mRNA amounted to 41.8% ± 5.9% of the wild-type frequency of the frameshift. When the tetranucleotide CGUG (nt 2849 to 2852) at the bottom of the stem was changed to GCGC, resulting in reducing the stem to 4 bp and lengthening its distance to the slippery heptamer from 5 to 10 nt, the frameshift frequency was reduced to 38.2% ± 7.7% of the wild-type frequency (Fig. 3, structure 3). Similarly, a mutation from CGCG (nt 2872 to 2875) at the bottom of the stem to GUGC, resulting in a 4-bp stem 10 nt away from the heptamer, changed the frameshift efficiency to 32.3% ± 4.0% of the wild-type frequency (Fig. 3, structure 4). The length of the stem is thus playing an important role in maintaining the efficiency of frameshift. A decrease from 9 to 5 bp results in a two-third loss of the efficiency. However, a comparison of the results in Fig. 3 indicates that the distance between the stem-loop and the slippery heptamer is of somewhat less importance. There is only a moderate effect on the efficiency of frameshift when it is increased from 5 to 10 nt. Restoration of the stem back to 9 bp through a combination of the two previous mutations resulted in a wild-type frameshift frequency at 102.5% ± 8.8% (Fig. 3, structure 5). These data demonstrate the important role of the stem-loop structure in causing the frameshift whereas the actual nucleotide sequences in the stem structure are apparently less important. Further, our earlier alteration of the loop sequence at nt 2860 to 2863 from GAUC to CUAG, which resulted in a 70.8% ± 2.5% frameshift efficiency compared to the wild type, suggested a lack of importance of the actual nucleotide sequence of the loop as well. We thus conclude that it is the stem-loop secondary structure per se that plays an important function in inducing the frameshift (Table 1).
FIG. 3.
Alteration of the putative stem-loop structure downstream from the slippery site by site-directed mutagenesis and assay of the effect from the alteration on the −1 ribosomal frameshift. The five transcripts used for transfecting Giardia trophozoites are as follows: 1, wild type; 2, the AUUA/uagu mutant at nt 2868 to 2871 that shortens the stem from its top by 4 bp and enlarges the loop from 11 to 19 nt without altering its distance from the slippery heptamer; 3, the CGUG/gcgc mutant at nt 2849 to 2852 that shortens the stem from its bottom by 5 bp and increases its distance from the slippery heptamer from 5 to 10 nt without altering the loop; 4, the CGUG/gcgc mutant at nt 2872 to 2875 that shortens the stem from its bottom by 5 bp and increases its distance from the slippery heptamer to 10 nt without altering its loop structure; and 5, the double mutant of CGUC/gcgc (nt 2849 to 2852) and CGCG/gugc (nt 2872 to 2875) that restored the wild-type stem structure with altered nucleotide sequence. The upper panel presents relative frequencies of the −1 ribosomal frameshift derived from luciferase expression in mutant transcript-transfected Giardia trophozoites in triplicate experiments (see Table 1 for actual data).
In a final experiment to verify the essential role played by the stem-loop in causing the frameshift, we deleted the sequence from nt 2848 to 2877 that contains the entire stem-loop structure. Surprisingly, the expression of luciferase amounted to 85.5% ± 8.6% of the wild-type frameshift frequency. Subsequent analysis by the MFOLD program revealed that, serendipitously, a new stem-loop is formed between nucleotides at the 3′ end of the 68-nt fragment and the 5′ terminus of the luciferase mRNA. It has a 6-bp stem and a 14-nt loop located 4 nt downstream from the first stem-loop (Fig. 4). On deletion of the wild-type stem-loop, the downstream stem-loop acquires an additional 1 bp at the bottom of the stem to form a 7-bp stem that is 7 nt downstream from the slippery heptamer (Fig. 4). This distance is within the functional range that we found with our mutants in Fig. 3 (structures 3 and 4). The functional competence of this serendipitous stem-loop, which was an artifact in the chimeric mRNA from pC631(68)luc, nevertheless confirms our conclusion that it is the structure of the stem-loop, and not the sequence therein, that is important in inducing the frameshift.
For the sake of clarity, the actual luciferase activities expressed by transfectants from all the in vivo mutational analysis discussed above are summarized in Table 1.
Probing the secondary structures in the 68-nt region by chemical modifications.
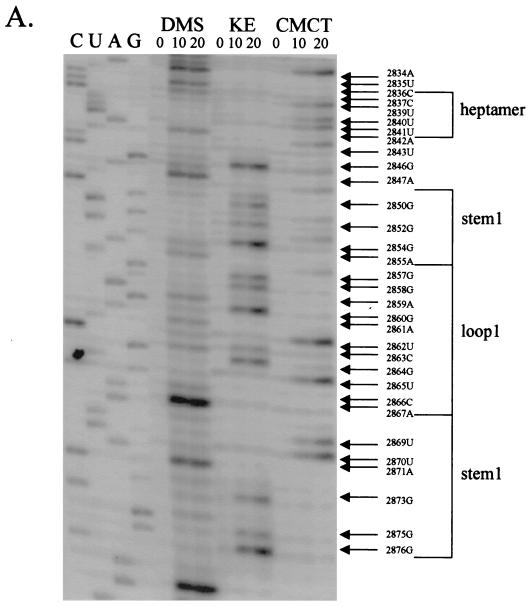
In an attempt to verify the actual secondary structures in the 68-nt RNA fragment under native conditions, we made a 4.6-kb T7 RNA polymerase transcript from pC631(68)luc linearized by NruI. We treated this RNA, containing the GLV 5′ 631-nt RNA, the 68-nt frameshift region, and the entire coding region of the luciferase, with KE, DMS, or CMCT to modify the unpaired bases that are accessible to alkylation by these agents. KE modifies unpaired G residues at the N-1 position. DMS methylates unpaired A residues at N-1 and, much more weakly, the unpaired C residues at N-3, whereas CMCT modifies the N-3 group of unpaired U residues and the N-1 group of unpaired G residues (12). The bases thus modified can no longer be recognized by reverse transcriptase in a primer extension reaction, and each can be identified as a reverse transcriptase stop in subsequent gel electrophoresis (27, 33). A modified base in RNA can be thus readily identified. The oligonucleotide terminating at the modified base moves 1 nt ahead of the corresponding DNA ladder, because primer extension stops immediately before the modified base. Thus, we can expect relatively few stops in stems where the bases are aligned in Watson-Crick pairs and much more stops in loops and unpaired regions free of base-base interaction. The radiolabeled stops often differ in their intensities, presumably due to a partial dissociation among some of the Watson-Crick pairings during the incubation period of chemical probing. The relatively poor efficiency in DMS modification of unpaired C residues also makes unlabeled C's less meaningful in data interpretation.
Results from a chemical probing and primer extension experiment on the in vitro transcript of pC631(68)luc are shown in Fig. 5. When the chemically modified region involving the heptamer and the first putative stem-loop (stem-loop 1) was examined by primer extension (Fig. 5A), bases in the heptamer and its immediate surrounding areas all showed up as transcriptional stops, suggesting that they were all chemically modified. In the region of loop 1, essentially all of the nucleotides were also modified (Fig. 5A). This finding, suggesting that all the bases in the heptamer and loop 1 are unpaired, provides the most direct evidence verifying the secondary structure predicted by MFOLD (Fig. 1). It also rules out the potential involvement of GAUC (nt 2860 to 2863) in loop 1 in forming a pseudoknot as originally postulated in Fig. 1. Regions predicted to form stem 1 (nt 2848 to 2856 and 2868 to 2876) were found largely free of chemical modification except for G-2854, A-2855, and UUA (nt 2869 to 2871), which all form the weaker G-U or A-U pairs in the postulated stem structure, raising the possibility that these base pairs may become partially dissociated during the chemical probing reactions (24). G-2850, G-2852, and G-2875 in stem 1 became lightly labeled after prolonged treatment with KE, which could be attributed to partial denaturation of the stem structure on incubation. Taken together, the results lend strong support to our conclusion from the MFOLD program and the mutational analysis of the predicted secondary structure of stem-loop 1.
FIG. 5.
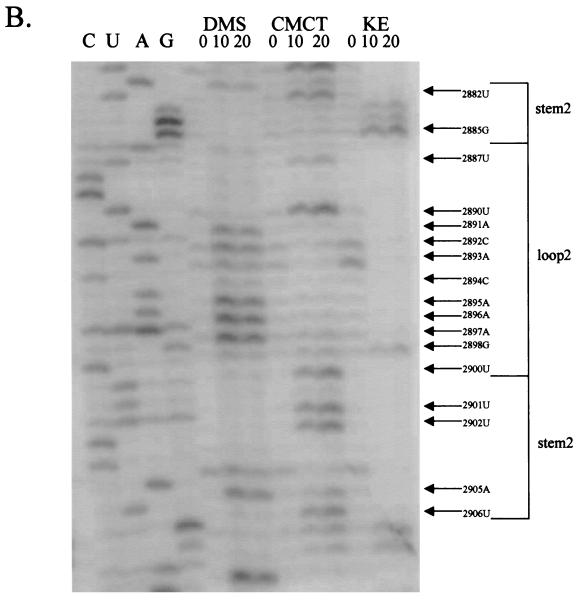
Chemical probings of the pC631(68)luc transcript and structural analysis of the 68-nt frameshift region by primer extension. Chemical modifications of A and C (by DMS), U (by CMCT), and G (by KE) were monitored by reverse transcription using end-labeled primers complementary to the nucleotides either 22 nt downstream of the last residue of stem-loop 1 (A) or 31 nt downstream of the last U residue of stem-loop 2 (B). The durations of incubation in each chemical reaction are indicated in minutes above each lane of gel electrophoresis. The left side of the gel labeled CUAG at the top represents the corresponding sequencing ladder of the cDNA. Arrows indicate transcriptional stops from primer extension representing the chemically modified bases in the treated RNA molecule, which migrate at a distance 1 nt short of that in the corresponding DNA ladder.
We also examined the region in the transcript which encompasses the 3′ end of the 68-nt fragment and the 5′ end of the luciferase mRNA in search of the serendipitous stem-loop structure (stem-loop 2) predicted from MFOLD and the mutational analysis (Fig. 4). Results from primer extension presented in Fig. 5B show that all the bases in the predicted loop 2 region were modified except for the three C's at nt 2888 to 2889 and 2899. However, bases in the postulated stem 2 structure remained largely unmodified, except for G-2985, 2 U's at nt 2901 to 2902, A-2905, and U-2906, all of which are involved in A-U or G-U pairings (Fig. 6). The tetranucleotide GAUC (nt 2885 to 2888), which was originally assumed to be a part of the pseudoknot structure (Fig. 1), has the G and U residues chemically modified. However, since the first two nucleotides, GA, in the tetranucleotide are actually included in the stem 2 structure and unpaired C is usually poorly modified by DMS, the present results are more consistent with the inclusion of this tetranucleotide in the stem-loop 2 structure rather than in a pseudoknot formation (see Discussion). Overall, data from chemical probing have verified the presence of the artifact stem-loop 2 in the chimeric mRNA transcribed from pC631(68)luc.
FIG. 6.
Summary of the results of primer extension on chemically modified RNA presented in Fig. 5. For the 68-nt frameshift region in GLV mRNA and the downstream 17-nt sequence of the luciferase mRNA, bases clearly modified by various chemicals are shown by large arrowheads. Chemical modifications becoming only gradually detectable with incubation time are indicated by small arrowheads. The numbers indicate the nucleotide positions relative to the 5′ end of GLV RNA. Bold type represents the sequence in GLV mRNA, whereas normal type indicates the sequence in luciferase mRNA.
DISCUSSION
A programmed −1 ribosomal frameshift is adopted by many small RNA viruses to generate viral RNA polymerase (Gag-Pol) as fusion protein with the capsid protein (Gag) at its N terminus and RDRP at the C terminus (reviewed in reference 2). Not only does this strategy ensure that Gag and Gag-Pol proteins are synthesized at a fixed ratio during translation, but also the presence of the Gag domain in the fusion protein provides a recognition signal for the RNA polymerase to be incorporated into the newly assembled virus at a fixed ratio (31). GLV apparently also uses the same strategy, which has an estimated ratio of about 1 in 60 between the Gag-Pol fusion protein and the Gag protein in sodium dodecyl sulfate-polyacylanide gel electophoresis of purified GLV particles (40), which is in close agreement with the 1.7% frameshift frequency estimated from the present study. Among the viruses that depend on the −1 ribosomal frameshift for a well-balanced viral protein synthesis, their transcripts have the common structural elements of the slippery heptamer but differ in the structure of a second requirement. Some viruses, such as IBV (3, 5) and ScV/L-A (7), need the structure of a pseudoknot, while others, such as HIV (29, 43), require only a simple stem-loop. Our studies showed that the −1 ribosomal frameshift in GLV transcript translation also requires a slippery heptamer where the actual frameshift takes place but that only a simple downstream stem-loop, rather than a pseudoknot, is required for inducing the shift. This conclusion has been supported by two original observations from the mutational studies. First, a mutation changing nt 2860 to 2863 in loop 1, the tetranucleotide postulated to be involved in a pseudoknot formation (Fig. 1), from GAUC to CUAG did not significantly affect the frequency of the frameshift (Table 1). This suggests that the tetranucleotide is not involved in forming a pseudoknot that plays an important role in inducing ribosomal frameshift. Second, there is support from the MFOLD identification of stem-loop 2 in the chimeric mRNA (Fig. 4), which is formed serendipitously by joining the 5′ luciferase sequence with the 3′ end of the 68-nt fragment. This postulated artifact stem-loop can function at 86% of the wild-type efficiency when brought into the proximity of the heptamer by deleting stem-loop 1 (Fig. 4; Table 1). Since stem-loop 2 contains a loop sequence that cannot form a predicted pseudoknot with the rest of the sequences of the mRNA, a pseudoknot is again ruled out as a requirement for the frameshift. The presence of stem-loops 1 and 2 in the chimeric mRNA was subsequently confirmed by the data from chemical probing and primer extension experiments (Fig. 5). The extensive chemical modifications of essentially all the bases in the two loop regions also indicate that neither loop forms a pseudoknot with another sequence in the mRNA under the native in vitro conditions. There is thus little doubt that no pseudoknot is present in the region where frameshift takes place and that there is no need of for pseudoknot to cause the frameshift.
There could be always some doubt about whether observations made on a chimeric mRNA can fully represent the −1 ribosomal frameshift occurring during translation of GLV mRNA in GLV-infected Giardia. The downstream sequences from the region of frameshift differ widely between the chimeric and GLV mRNA. There is no stem-loop 2 structure predicted within this proximal region in GLV mRNA (data not shown). The presence of an extra stem-loop 2 in the chimeric mRNA may amount to a frameshift enhancer element forcing a translating ribosome to pause and the following ribosome to stack behind it at the frameshift signal, resulting in an increased amount of time required for ribosome slipping. This ribosome stacking has been shown to have an effect on programmed −1 frameshift frequencies (22). However, we do not think that there is a discrepancy in frameshift frequency between the chimeric and GLV mRNA because of the estimated ratio of 2% between the Gag-Pol fusion protein and the Gag protein in sodium dodecyl sulfate-polyacrylanide gel electrophoresis analysis of purified GLV particles (40), which is quite close to the 1.7% frequency derived from the investigation on chimeric mRNA. The stoichiometry of Gag and Gag-Pol maintained by a 1.7% frameshift frequency also closely agrees with the prediction that in a small icosahedral virion such as GLV, RDRP is expected to be packaged with identical capsid subunits at a ratio of 1:60 (15).
There is also the possibility that some downstream sequence in GLV mRNA, which is absent from the chimeric mRNA, could form a pseudoknot with loop 1 resulting in enhancement of the frameshift frequency. This enhancing effect could compensate for the enhancement attributed to the artifact stem-loop 2 in the chimeric mRNA that is absent from GLV mRNA. Existing evidence does, however, argue against such a possibility; an extension of the 68-nt fragment to a 215-nt portion of the 220-nt overlapping region between the two ORFs in GLV mRNA failed to alter the frequency of frameshift in the chimeric mRNA transfected Giardia. Formation of a pseudoknot between loop 1 and its immediate downstream viral sequence that could enhance the frameshift is therefore unlikely. Although pseudoknot formation between loop 1 and the sequences further downstream in the viral mRNA is still possible, the probability decreases with increasing distance. We think that our current results obtained from studying the chimeric mRNA in Giardia reflect accurately the −1 ribosomal frameshift on GLV mRNA during its translation.
We also showed in this study that all the necessary structural features for a −1 ribosomal frameshift in GLV transcript could be confined within a 50-nt fragment (from nt 2828 to 2877) (Fig. 1). This short fragment of mRNA contains a CCCUUUA heptamer, where the actual −1 frameshift takes place, and a stem-loop 1 5 nt downstream from the heptamer, which presumably retards the movement of the ribosome during mRNA translation and allows a −1-nt slippage of ribosome on the slippery heptamer site of mRNA (14, 20). The heptamer CCCUUUA can be replaced with other heptamers, resulting in changed frameshift frequencies in the deceasing order of UUUUUUU, UUUUUUC, and CCCUUUA. A similar order of decreasing frameshift efficiencies was also observed by Brierley et al. in the IBV system (4), suggesting a hierarchy of efficiency that is innate to the sequences of these heptamers regardless of the viruses or the host cells involved.
Studies of the sites of frameshift in plant viruses and retroviruses have shown that stop codons located within the frameshift region can influence the frameshift efficiency (1, 14, 19, 21, 28). In this work, a termination codon placed before the heptamer but in the −1 frame showed 53.8% of the wild-type frameshift frequency. This was consistent with the results obtained by Honda and Nishimura (17), where an upstream stop codon in the −1 frame before the slippery site suppressed the frameshift by about 50%. Three stop codons in the 0 frame downstream from the heptamer also have lower framshift efficiencies, at 52.1, 57.3, and 72.3% of the wild-type efficiency respectively (Fig. 2). A plausible explanation for the decreased frameshift could be attributed to the nonsense-mediated mRNA decay pathway (6, 34), even though it is not yet known whether such a pathway exists in a lineage as ancient as Giardia. The repressive effect also appeared to be decreasing as the stop codons were placed farther away from the heptamer (41). The stop codon placed at nt 2843 to 2845 in the 0 frame at the immediate 3′ end of the heptamer (Fig. 2) reduced the ribosomal frameshifting to 37.8%. A stop codon placed at the same location in HIV (18) repressed the frameshift by the prokaryote ribosome to a similar extent. Most interestingly, the recoding event was further depressed when the level of the peptide release factors (RF) in vivo was increased, directly linking the decrease in frequency of frameshift with the recruitment of RF by the termination codon (37). It remains to be determined whether repression of the frameshift is also mediated by RF in our case.
In summary, we have succeeded in using the experimental approaches of site-directed mutagenesis and chemical modifications of mRNA to dissect the structural basis of the programmed −1 ribosomal frameshift during translation of GLV mRNA in Giardia. The conclusion, indicating the mere requirement of a heptamer and a downstream stem-loop in the ORF overlapping region, provides to our knowledge the first example of such a mechanism of frameshifting among totiviruses.
ACKNOWLEDGMENTS
We thank Srinivas Garlapati for instructions on how to perform chemical probings of RNA.
This work was supported by grant AI-30475 from the National Institutes of Health.
REFERENCES
- 1.Brault V, Miller W A. Translational frameshifting mediated by a viral sequence in plant cells. Proc Natl Acad Sci USA. 1992;89:2262–2266. doi: 10.1073/pnas.89.6.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brierley I. Ribosomal frameshifting viral RNAs. J Gen Virol. 1995;76:1885–1892. doi: 10.1099/0022-1317-76-8-1885. [DOI] [PubMed] [Google Scholar]
- 3.Brierley I, Digard P, Inglis S C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brierley I, Jenner A J, Inglis S C. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brierley I, Rolley N J, Jenner A J, Inglis S C. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J Mol Biol. 1991;220:889–902. doi: 10.1016/0022-2836(91)90361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Culbertson M R. RNA surveillance. Unforeseen consequences for gene expression, inherited genetic disorders and cancer. Trends Genet. 1999;15:74–80. doi: 10.1016/s0168-9525(98)01658-8. [DOI] [PubMed] [Google Scholar]
- 7.Dinman J D. Ribosomal frameshifting in yeast viruses. Yeast. 1995;11:1115–1127. doi: 10.1002/yea.320111202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinman J D, Icho T, Wickner R B. A −1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc Natl Acad Sci USA. 1991;88:174–178. doi: 10.1073/pnas.88.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dinman J D, Wickner R B. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J Virol. 1992;66:3669–3676. doi: 10.1128/jvi.66.6.3669-3676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falk H, Mador N, Udi R, Panet A, Honigman A. Two cis-acting signals control ribosomal frameshift between human T-cell leukemia virus type II gag and pro genes. J Virol. 1993;67:6273–6277. doi: 10.1128/jvi.67.10.6273-6277.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farabaugh P J. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garlapati S, Chou J, Wang C C. Specific secondary structures in the capsid-coding region of Giardiavirus transcript are required for its translation in Giardia lamblia. J Mol Biol. 2001;308:623–638. doi: 10.1006/jmbi.2001.4568. [DOI] [PubMed] [Google Scholar]
- 13.Gesteland R F, Atkins J F. Recoding: dynamic reprogramming of translation. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 14.Gramstat A, Prufer D, Rohde W. The nucleic acid-binding zinc finger protein of potato virus M is translated by internal initiation as well as by ribosomal frameshifting involving a shifty stop codon and a novel mechanism of P-site slippage. Nucleic Acids Res. 1994;22:3911–3917. doi: 10.1093/nar/22.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harrison S C. Multiple modes of subunit associationin the structures of simple spherical viruses. Trends Biochem Sci. 1984;9:345–351. [Google Scholar]
- 16.Herold J, Siddell S G. An ‘elaborated’ pseudoknot is required for high frequency frameshifting during translation of HCV 229E polymerase mRNA. Nucleic Acids Res. 1993;21:5838–5842. doi: 10.1093/nar/21.25.5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda A, Nishimura S. Suppression of translation frameshift by upstream termination codon. Biochem Biophys Res Commun. 1996;221:602–608. doi: 10.1006/bbrc.1996.0642. [DOI] [PubMed] [Google Scholar]
- 18.Horsfield J A, Wilson D N, Mannering S A, Adamski F M, Tate W P. Prokaryotic ribosomes recode the HIV-1 gag-pol-1 frameshift sequence by an E/P site post-translocation simultaneous slippage mechanism. Nucleic Acids Res. 1995;23:1487–1494. doi: 10.1093/nar/23.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacks T, Power M D, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature. 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 20.Jacks T, Varmus H E. Expression of the Rous sarcoma virus pol gene by ribosomal frameshifting. Science. 1985;230:1237–1242. doi: 10.1126/science.2416054. [DOI] [PubMed] [Google Scholar]
- 21.Kim K H, Lommel S A. Identification and analysis of the site of −1 ribosomal frameshifting in red clover necrotic mosaic virus. Virology. 1994;200:574–582. doi: 10.1006/viro.1994.1220. [DOI] [PubMed] [Google Scholar]
- 22.Lopinski J D, Dinman J D, Bruenn J A. Kinetics of ribosomal pausing during programmed −1 translational frameshifting. Mol Cell Biol. 2000;20:1095–1103. doi: 10.1128/mcb.20.4.1095-1103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marczinke B, Bloys A J, Brown T D, Willcocks M M, Carter M J, Brierley I. The human astrovirus RNA-dependent RNA polymerase coding region is expressed by ribosomal frameshifting. J Virol. 1994;68:5588–5595. doi: 10.1128/jvi.68.9.5588-5595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marczinke B, Fisher R, Vidakovic M, Bloys A J, Brierley I. Secondary structure and mutational analysis of the ribosomal frameshift signal of Rous sarcoma virus. J Mol Biol. 1998;284:205–225. doi: 10.1006/jmbi.1998.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 26.Miller R L, Wang A L, Wang C C. Purification and characterization of the Giardia lamblia double-stranded RNA virus. Mol Biochem Parasitol. 1988;28:189–195. doi: 10.1016/0166-6851(88)90003-5. [DOI] [PubMed] [Google Scholar]
- 27.Moazed D, Stern S, Noller H F. Rapid chemical probing of conformation in 16S ribosomal RNA and 30S risosomal subunits using primer extension. J Mol Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 28.Morikawa S, Bishop D H. Identification and analysis of the gag-pol ribosomal frameshift site of feline immunodeficiency virus. Virology. 1992;186:389–397. doi: 10.1016/0042-6822(92)90004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parkin N T, Chamorro M, Varmus H E. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J Virol. 1992;66:5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prufer D, Tacke E, Schmitz J, Kull B, Kaufmann A, Rohde W. Ribosomal frameshifting in plants: a novel signal directs the −1 frameshift in the synthesis of the putative viral replicase of potato leafroll luteovirus. EMBO J. 1992;11:1111–1117. doi: 10.1002/j.1460-2075.1992.tb05151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribas J C, Wickner R B. The Gag domain of the Gag-Pol fusion protein directs incorporation into the L-A double-stranded RNA viral particles in Saccharomyces cerevisiae. J Biol Chem. 1998;273:9306–9311. doi: 10.1074/jbc.273.15.9306. [DOI] [PubMed] [Google Scholar]
- 32.ten Dam E B, Pleij C W, Bosch L. RNA pseudoknots: translational frameshifting and readthrough on viral RNAs. Virus Genes. 1990;4:121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stern S, Moazed D, Noller H F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1998;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- 34.Sun X, Li X, Moriarty P M, Henics T, LaDuca J P, Maquat L E. Nonsense-mediated decay of mRNA for the selenoprotein phospholipid hydroperoxide glutathione peroxidase is detectable in cultured cells but masked or inhibited in rat tissues. Mol Biol Cell. 2001;12:1009–1017. doi: 10.1091/mbc.12.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsuchihashi Z, Brown P O. Sequence requirements for efficient translational frameshifting in the Escherichia coli dnaX gene and the role of an unstable interaction between tRNA(Lys) and an AAG lysine codon. Genes Dev. 1992;6:511–519. doi: 10.1101/gad.6.3.511. [DOI] [PubMed] [Google Scholar]
- 36.Tsuchihashi Z, Kornberg A. Translational frameshifting generates the gamma subunit of DNA polymerase III holoenzyme. Proc Natl Acad Sci USA. 1990;87:2516–2520. doi: 10.1073/pnas.87.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tumer N E, Parikh B A, Li P, Dinman J D. The pokeweed antiviral protein specifically inhibits Ty1-directed +1 ribosomal frameshifting and retrotransposition in Saccharomyces cerevisiae. J Virol. 1998;72:1036–1042. doi: 10.1128/jvi.72.2.1036-1042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang A L, Wang C C. Discovery of a specific double-stranded RNA virus in Giardia lamblia. Mol Biochem Parasitol. 1986;21:269–276. doi: 10.1016/0166-6851(86)90132-5. [DOI] [PubMed] [Google Scholar]
- 39.Wang A L, Wang C C. Viruses of the protozoa. Annu Rev Microbiol. 1991;45:251–63. doi: 10.1146/annurev.mi.45.100191.001343. [DOI] [PubMed] [Google Scholar]
- 40.Wang A L, Yang H M, Shen K A, Wang C C. Giardiavirus double-stranded RNA genome encodes a capsid polypeptide and a gag-pol-like fusion protein by a translation frameshift. Proc Natl Acad Sci USA. 1993;90:8595–8599. doi: 10.1073/pnas.90.18.8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weiss R B, Dunn D M, Shuh M, Atkins J F, Gesteland R F. E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1989;1:159–169. [PubMed] [Google Scholar]
- 42.Wickner R B. Double-stranded RNA viruses of Saccharomyces cerevisiae. Microbiol Rev. 1996;60:250–265. doi: 10.1128/mr.60.1.250-265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yelverton E, Lindsley D, Yamauchi P, Gallant J A. The function of a ribosomal frameshifting signal from human immunodeficiency virus-1 in Escherichia coli. Mol Microbiol. 1994;11:303–313. doi: 10.1111/j.1365-2958.1994.tb00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu D-C, Wang A L, Wang C C. Amplification, expression, and packaging of a foreign gene by giardiavirus in Giardia lamblia. J Virol. 1996;70:8752–8757. doi: 10.1128/jvi.70.12.8752-8757.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu D-C, Wang A L, Wu C-H, Wang C C. Virus-mediated expression of firefly luciferase in the parasitic protozoan Giardia lamblia. Mol Cell Biol. 1995;15:4867–4872. doi: 10.1128/mcb.15.9.4867. [DOI] [PMC free article] [PubMed] [Google Scholar]