Abstract
Background
Autism spectrum disorder (ASD) is a neurodevelopmental disorder (NDD) characterized by significant impairments in social, communicative, and behavioral abilities. However, only a limited number of studies address the genetic basis of ASD in the African population. This study aims to document the genes associated with ASD in Africa and the techniques used to identify them. Additionally, genes identified elsewhere but not yet in Africa are also noted.
Methods
Online databases such as Wiley Online Library, PubMed, and Africa Journal Online were used. The review was conducted using the keyword related to genetic and genomic ASD study in the African population.
Result
In this scoping review, 40 genetic studies on ASD in Africa were reviewed. The Egyptian and South African populations were the most studied, with 25 and 5 studies, respectively. Countries with fewer studies included Tunisia (4), East African countries (3), Libya (1), Nigeria (1), and Morocco (1). Some 61 genes responsible for ASD were identified in the African population: 26 were identified using a polymerase chain reaction (PCR)-based method, 22 were identified using sequencing technologies, and 12 genes and one de novo chromosomal aberration were identified through other techniques. No African study identified any ASD gene with genome-wide association studies (GWAS). Notably, at least 20 ASD risk genes reported in non-African countries were yet to be confirmed in Africa’s population.
Conclusion
There are insufficient genetic studies on ASD in the African population, with sample size being a major limitation in most genetic association studies, leading to inconclusive results. Thus, there is a need to conduct more studies with large sample sizes to identify other genes associated with ASD in Africa’s population using high-throughput sequencing technology.
Keywords: austism spectrum disorders, ASD, gene, genetics, genome, sequencing, africa
1 Introduction
Autism spectrum disorder (ASD), a widespread and clinically heterogeneous neurodevelopmental disease (NDD), is defined by irregularities in social interactions and repetitive or limited behavioral patterns (Hodges et al., 2020) that manifest in early postnatal life. ASD shows various symptoms and severities which significantly burden affected individuals, caregivers, and families (van Heijst and Geurts, 2015). The World Health Organization (WHO) reported ASD as a major worldwide public health issue in underdeveloped nations (World Health Organization, 2013), being among the leading causes of disability in children (Salari et al., 2022). Globally, one in a hundred children have ASD, indicating an increase in the disorder’s prevalence (Zeidan et al., 2022). Although the prevalence of ASD in low- or middle-income countries, including in Africa, is controversial, this disease burden has recently been found to be underestimated (Fombonne et al., 2021; Aderinto et al., 2023). According to a recent global systematic review and meta-analysis, the prevalence of ASD was 0.4% (95% CI: 0.1–1) in Asia, 1% (95% CI: 0.8–1.1) in America, 0.5% (95% CI: 0.2–1) in Europe, 1% (95% CI: 0.3–3.1) in Africa, and 1.7% (95% CI: 0.5–6.1) in Australia (Salari et al., 2022).
With a 50%–90% heritability estimate, ASD is a neuropsychiatric condition that is complicated and genetically heterogeneous (Kainer et al., 2023), although de novo gene variations also have an outstanding contribution. To date, over 200 susceptible genes have been identified as being linked to ASD. Many genes have been found to be the most prevalent risk factors for the development of ASD, including MTHFR, RELN, CACNA1C, SHANK, and VDR (Wiśniowiecka-Kowalnik and Nowakowska, 2019; Wei et al., 2021). Study findings have shown several chromosome regions, including one or more with susceptible genes for autism (Monaco and Bailey, 2001). Moreover, several studies have shown that prenatal, perinatal, and postnatal environmental factors are associated with ASD (Gardener et al., 2011). According to some reports, factors such as medications, chemical exposures, parental age, nutrition, and prenatal environment can account for up to 40%–50% of the variation in ASD liability (Gaugler et al., 2014; Deng et al., 2015). ASD development, however, may also be influenced by the interplay of genetic and environmental factors, as suggested by Tordjman et al. (2014) and Santos et al. (2022). For instance, oxidative stress may be a potential mechanism in genetic disorders linked to ASD that connect genetic and environmental factors. Fragile X messenger ribonucleoprotein (FMRP), primarily involved in mRNA binding, is absent in FXS due to a loss of FMR1 expression. As demonstrated in FMR1-knockout mice, the lack of FMRP increases oxidative stress (De Diego-Otero et al., 2009). Any chromosome or gene changes that lead to an increase in oxidative stress could potentially be a factor in the manifestation of a behavioral autistic phenotype. In addition, the association of particulate matter with an aerodynamic diameter of 10 μm or less (PM10) and cognitive neurodevelopment has been found to be significantly mediated by DNAm; CpG sites mapped several genes, including DYRK1A, which has shown lower expression in South African ASD patients (Feil et al., 2023).
African populations are the most genetically varied in the world, having three times more uncommon variants than in Europe and East Asia (1,000 Genomes Project Consortium et al., 2012); they are exploited to find genes linked to diseases (Gomez et al., 2014). African genomes offer a unique resource for identifying new genetic loci and for very effective genetic fine-mapping due to their extensive genetic diversity and low linkage disequilibrium (Tishkoff and Verrelli, 2003; Campbell and Tishkoff, 2008). Despite Africa’s high human genome diversity, few genes or single nucleotide polymorphisms (SNPs) of potential ASD risk genes have been discovered in that population. This results from (i) no validated tools available for the diagnosis of ASD in the African population, (ii) genetic studies previously conducted in African countries used a small sample size, or (iii) most studies not using high throughput sequence technology for analysis like in the two Egyptian studies (Salem et al., 2013; Abdelrahman et al., 2015).
The present scoping review aims to identify risk genes associated with ASD in the African population, while other ASD-associated genes reported from other populations but not yet identified in Africa will also be discussed.
2 Methodology
The Preferred Reporting Items for Systematic Reviews and Meta-Analyses extension for Scoping Reviews (PRISMA-ScR) checklist was followed in this scoping review (Tricco et al., 2018). The selection included studies conducted in Africa that reported gene(s) or genetic variants associated with ASD, performed only on human beings or human cells, published from 1 January 2000 to 29 February 2024 and published in English. In addition, we excluded papers published as letters, in books, in gray literature, and reviews, as well as papers for research conducted using data from online databases. This scoping review followed Arksey and O'Malley’s scoping framework that proposed six stages of conduct: 1) specify the research question, 2) identify relevant literature, 3) select studies, 4) map out the data, 5) summarize, synthesize, and report the results, and 6) include expert consultation (Arksey and O’Malley, 2005). Electronic research was performed to identify relevant peer-reviewed articles mainly using the Wiley Online Library and PubMed databases. In order to identify genetic variants linked to ASD in Africa, the search approach included multiple sets of broad search terms with “AND” in each database. The search terms: “autism OR autistic OR Asperger Syndrome OR pervasive developmental disorder” were first used. Second, a group of search terms that combined important key terms, including “genetic study OR sequencing OR genomic study OR case/control study OR family-based study”, were employed. Lastly, a search specified the region as “Africa” and the names of individual African countries. We downloaded the details of all articles in the data collection sheet and categorized studies into research themes (genomic association studies, sequencing technology, and PCR-based methodology) and countries where the studies were conducted.
2.1 Article selection
Every research paper identified through the search strategy was exported into collections. Two authors independently examined the titles and abstracts of every article and then sequentially reviewed the full texts of all the articles identified for pertinent publications. The data-charting involved two authors: using a standardized form, the first author charted data from the selected publications, and the second verified the charted data. Study features such as first author, year of publication, country of study, population, sample size, age, genes or genetic variants identified, study design, body tissue type for sampling, and main findings were carefully extracted from every selected paper. Consistent with the scoping review process, the authors did not assess the selected publications for methodological quality or risk of bias. However, some excluded studies from the African continent were project theses. The abstracted data are presented in Supplementary File S1.
3 Result
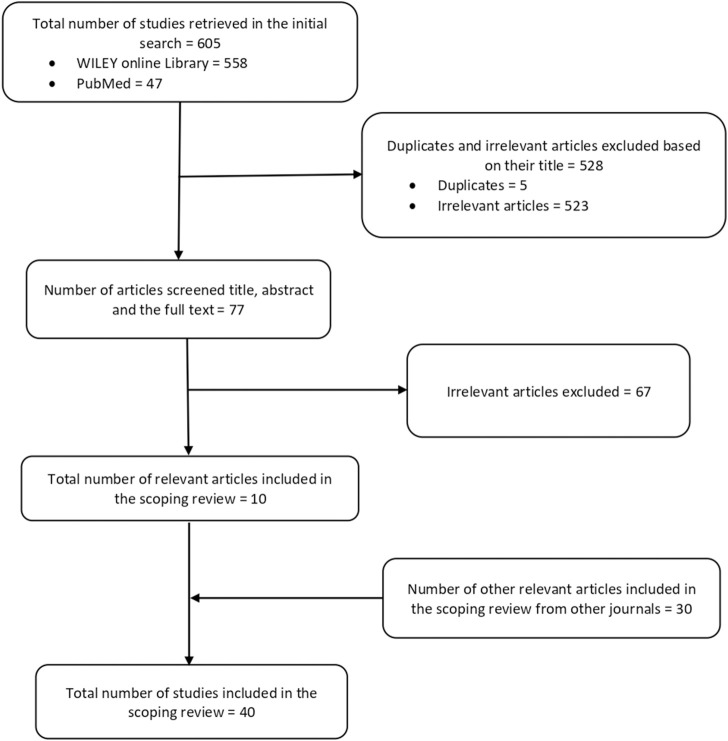
We initially found 605 papers, including 558 articles from the Wiley Online Library and 47 articles from PubMed. Subsequently, we removed 528 articles: five were duplicates, and the remaining 523 lacked the necessary information for being a genetic study conducted in Africa, which is a key variable for this scoping review. Among the 77 articles screened, 67 were deemed irrelevant after screening their titles, abstracts, and full texts; they thus were excluded from the study. Ten relevant articles from the Wiley Online Library and PubMed were eventually included in this scoping review.
Additionally, 30 relevant articles identified by searching other journals in the Africa Journal Online (AJOL) were included in the study. There were no methodological issues that necessitated excluding any related ASD genetic study from Africa in this review. The 40 most pertinent articles (Supplementary File S1) used as source documents for this review were selected through these meticulous selection procedures, summarized in an adapted PRISMA-ScR model shown in Figure 1.
FIGURE 1.
Diagram of the study selection process.
A total of 61 genes were identified in the African population as being responsible for ASD (Table 1). Among the 40 relevant studies, Egyptian and South African ethnicities were the most studied populations, appearing in 25 and 5 articles which represented 62.5% and 12.5%, respectively. Additionally, Tunisia (four articles; 10%), the East African population (Ethiopia, Eritrea, Kenya, and Somalia) (three articles; 7.5%), and the populations of Libya, Nigeria, and Morocco (one article each; 2.5%) (Table 2) were also studied. No other suitable publications were found dealing with the remaining African countries. Furthermore, 20 ASD risk genes identified in non-African populations have not been confirmed/reported in Africa (Table 3).
TABLE 1.
List of ASD genes identified so far in Africa’s population.
| NO | Gene | Country | Study design | Author | Genetic implication |
|---|---|---|---|---|---|
| 1 | AADAT | Egypt | PCR method | Higazi et al., 2021 | Graves’ disease and thyroid disease |
| 2 | CACNA1C | East Africa (Ethiopian, Eritrea, and Kenya) |
WGS | Tuncay et al., 2023 | Timothy syndrome features |
| 3 | CHD7 | CHARGE syndrome | |||
| 4 | COMT | Egypt | PCR method | Karam et al., 2013 | Schizophrenia and ADHD |
| 5 | C4B | Egypt | PCR method | Mostafa and Shehab (2010) | Chronic hypertension, thromboses, and schizophrenia |
| 6 | DRD1 | Egypt | PCR method | Azzam et al., 2018 | Psychosis and schizophrenia |
| 7 | DRD2 A1+ | Egypt | PCR method | Salem et al., 2013 | Type 2 diabetes |
| 8 | DRD4 | Egypt | PCR-RFLP | Kamal et al., 2015 | ADHD |
| 9 | FEZF2 | Morocco | WES | Bensaid et al., 2019 | Uterine inversion and thymic dysplasia |
| 10 | FMR1 | East Africa (Ethiopia, Eritrea, and Kenya) | WEG | Tuncay et al., 2023 | ID and fragile X syndrome |
| 11 | GABA | Egypt | PCR method | El-Ansary et al., 2021 | Neurodevelopmental disorders and epilepsy |
| 12 | GSTM1 | Egypt | PCR method | Said et al., 2021 | Urinary system cancer and metastatic breast cancer |
| 13 | GSTT1 | Egypt | PCR method | Said et al., 2021 | Metastatic breast cancer and sickle cell disease |
| 14 | HAAO | Egypt | PCR method | Higazi et al., 2021 | Hypospadias |
| 15 | HLA-DRB1 | Egypt | PCR method | Mostafa et al., 2012 | Rheumatoid arthritis (RA) |
| 16 | HLA-DR4 | Egypt | PCR method | El-Hossiny et al. (2023) | Rheumatoid arthritis (RA) |
| 17 | HTR1A | Egypt | PCR method | Yahya et al., 2019 | Bipolar disorder |
| 18 | 5-HTTLPR | South Africa | PCR method | Arieff et al., 2010 | Anxiety and depression |
| 19 | 5-HTTLPRS | Egypt | PCR method | Meguid et al., 2015 | Anxiety and depression |
| 20 | 5-HT2A | Egypt | PCR-RFLP | Abdelrahman et al. (2015) | Anxiety, depression, drug addiction, and schizophrenia |
| 21 | IL-1β-511 and IL-1RA | Egypt | PCR method | Saad et al., 2020 | Gastric cancer and osteoporotic fractures |
| 22 | IL-12 | Egypt | PCR method | Ibrahim et al., 2015 | Atherosclerosis and coronary artery disease |
| 23 | MAOA | Egypt | PCR method | Higazi et al., 2021 | Bipolar disorder, depression, and paranoid schizophrenia |
| 24 | PCDHAC1 | East Africa (Ethiopia, Eritrea, and Kenya) | WES | Tuncay et al., 2023 | Higher birth weight percentiles and larger newborn head circumferences |
| 25 | Q6NUR6 | Tunisia | PCR method | Bayou et al., 2010 | ASD |
| 26 | RELN | South Africa and Egypt | PCR method |
Sharma et al., 2013, Abdelhady et al., 2022 |
Bipolar disease, schizophrenia, and autism |
| 27 | SATB2 | Morocco | WES | Mounia et al., 2019 | SAS |
| 28 | SBCADD | East Africa (Somalia, Eritrea, and Ethiopia | Sanger sequencing | Oivind et al., 2007 | Developmental delay |
| 29 | SHANK2 | Ethiopia | NGS | Lu et al., 2018 | Mild-to-moderate ID |
| 30 | SHANK3 | Egypt | PCR-RFLP | Meguid et al., 2020 | Moderate-to-severe ID, severely impaired speech, schizophrenia, and mild dysmorphic features |
| 31 | TAMRTKs | Egypt | PCR method | Mostafa et al., 2022 | Glioblastoma multiforme |
| 32 | TBC1D8 | Libya | WES | Zeglam and Alhmadi, 2020 | Independent prognosis factor for ovarian cancer |
| 33 | TBR1 | Morocco | WES | Mounia et al., 2019 | NDD combining features of ASD, ID, and speech delay |
| 34 | TCF7L2 | East Africa (Ethiopia, Eritrea, and Kenya) | WGS | Tuncay et al., 2023 | Type 2 diabetes |
| 35 | TSHZ3 | Morocco | WES | Mounia et al., 2019 | Autism, cognitive disabilities, and language disturbance |
| 36 | VDR | Egypt | PCR-RFLP | Arafa et al., 2021 | Severity of autoimmune thyroid diseases; HT and GD |
| 37 | ERICH1 | Tunisia | aCGH technology | Fethia et al., 2022> | Adenoma risk |
| 38 | CELF4 | Seizures and neuroticism | |||
| 39 | CHRFAM7A | Schizophrenia and bipolar disease | |||
| 40 | FTHL17 | Tumorigenesis | |||
| 41 | NEXMIF | X-linked intellectual disability, ASD, and epilepsy | |||
| 42 | NLGN4X | Intellectual disability, ASD, anxiety, and ADHD | |||
| 43 | PRKN | Parkinson’s disease | |||
| 44 | SPN | Tumor progression | |||
| 45 | SYCE3 | Human infertility | |||
| 46 | UQCRC2 | Lactic acidosis | |||
| Differential methylated genes | |||||
| 47 | PGC-1α | South Africa | NGS | Bam et al., 2021 | Obesity, diabetes, neurodegeneration, and cardiomyopathy |
| 48 | STOML2 | Pancreatic cancer | |||
| 49 | MFN2 | Charcot–Marie–Tooth disease type 2A | |||
| 50 | FIS1 | Diabetes | |||
| 51 | OPA1 | Optic atrophy | |||
| 52 | GABPA | Hepatocellular carcinoma and bladder cancer | |||
| 53 | AHI1 | South Africa | WG-Methylation | Stathopoulos et al. (2018) | Joubert syndrome and schizophrenia |
| 54 | GLRA2 | X-linked neurodevelopmental disorders and high myopia | |||
| 55 | SETD5 | ID, ASD, and KBG syndrome | |||
| 56 | MTR | Cardiovascular diseases, breast and prostate cancer, birth defects, and congenital anomalies | |||
| 57 | RTL | Egypt | PCR method | Salem et al., 2023 | KOS14 and TS14 |
| 58 | LINE-1 | Schizophrenia, bipolar disorder, and major depressive disorder | |||
| 59 | PCCB | South Africa | Methylation analysis | Stathopoulos et al. (2020) | Propionic acidemia and biotinidase deficiency |
| 60 | PCDHA12 | Developmental delay, movement disorder, epilepsy, microcephaly, and visual impairment | |||
| 61 | De novo balanced (7; 16) (p22.1; p16.2) translocation | Tunisia | Cytogenetics and FISH | Bayou et al. (2008) | Numerous genomic tests for autism have found chromosome 16p, most likely home to an autism-susceptibility variation |
aCGH, micro-array-based comparative genomic hybridization; NGS, next-generation sequencing; WG, whole genome; WES, whole-exome sequencing; ADHD; attention-deficit hyperactivity disorder; ID, intellectual disability; KOS14, Kagami–Ogata syndrome; TS14, temple syndrome; HT, Hashimoto’s thyroiditis; GD, Graves’ disease; SAS, SATB2-associated syndrome.
TABLE 2.
National distribution of identified genes associated with ASD in Africa.
| Countries | No. of studies | % | N0 of genes associated with ASD |
| Egypt | 25 | 62.5% | 24 |
| South Africa | 5 | 12.5% | 14 |
| Tunisia | 4 | 10% | 12 |
| East Africa (Ethiopia, Eritrea, Kenya, and Somalia) | 3 | 7.5% | 6 |
| Morocco | 1 | 2.5% | 4 |
| Libya | 1 | 2.5% | 1 |
| Nigeria | 1 | 2.5% | 0 |
TABLE 3.
List of ASD genes identified in non-African populations yet to be confirmed/reported in Africa.
| No. | Gene | Country | Study design | Author |
|---|---|---|---|---|
| 1 | ASMT | China and Sweden | WES and PCR method | Wang et al., 2013; Jonson et al., 2014 |
| 2 | CHD8 | United States | Sanger sequencing | Bernier et al. (2014) |
| 3 | CNTNAP2 | China and Brazil | PCR method | Li et al., 2010; Nascimento et al., 2015 |
| 4 | DRD3 | Netherlands and United Kingdom | SNP genotyping | De Krom et al. (2009) |
| 5 | EN2 | China and India | PCR method | Wang et al., 2008, Sen et al., 2010 |
| 6 | FOXP1 | Taiwan and Canada | PCR method and Sanger sequencing | Chien et al., 2013; Hamdan et al., 2010 |
| 7 | GABRB3 | United States, United Kingdom, and Taiwan | PCR method and Sanger sequencing | Buxbaum et al., 2002; Warrier et al., 2013; Chen et al., 2014 |
| 8 | GRIN2B | China and Korea | Sanger sequencing, PCR method | Pan et al., 2014; Hee Jeong et al., 2012 |
| 9 | HoxA1 | United States and Italy | Sanger sequencing and PCR method | Ingram et al., 2000; Conciatori et al., 2004 |
| 10 | SRRM4 | Italy and Japan | RNA sequencing and GWAS | Irimia et al., 2014; Narita et al., 2020 |
| 11 | MECP2 | United States and China | Sanger sequencing and WES | Nagarajan et al., 2006; Wen et al., 2017 |
| 12 | MET | United States and Italy | PCR method | Campbell et al., 2009; Jackson et al., 2009 |
| 13 | NLGN3 | China and Italy | Sanger sequencing and WES | Xu et al., 2014; Oleari et al., 2023 |
| 14 | NLGN4 | France, Bulgaria, and Japan | PCR and sequencing | Laumonnier et al., 2004; Avdjieva-Tzavella et al., 2012; Toya et al., 2023 |
| 15 | NRXN1 | United States and China | GWAS and array CGH sequencing | Liu et al., 2012; Liu et al., 2012; Griswold et al., 2015 |
| 16 | OXTR | China and United Kingdom | SNP genotyping | Wu et al., 2005; Di Napoli et al., 2014 |
| 17 | PTCHD1 | European, Caucasian | Sequencing | Torrico et al., 2014 |
| 18 | PTEN | France, and United States | WES | Buxbaum et al., 2007, O’Roak et al., 2012 |
| 19 | SLC25A12 | United States and Finland | SNP genotyping | Ramoz et al., 2004, Joni et al., 2008 |
| 20 | SYNGAP1 | Canada | Sanger sequencing | Hamdan et al., 2011, Berryer et al., 2013 |
3.1 Genetic techniques adopted for identifying genetic variants linked to ASD in Africa
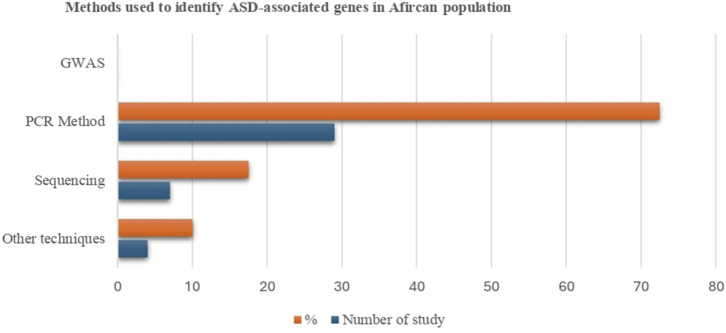
We categorized the research into three main themes based on study design: GWAS (n = 0), sequencing-based technology (n = 7), and PCR-based methods (n = 29) (Figure 2. However, the remaining studies used other molecular biology techniques such as methylation analysis (n = 1), hydride generation technique (n = 1), microarray-based comparative genomic hybridization (aCGH) technology (n = 1), and cytogenetics and fluorescent in situ hybridization (FISH) analysis (n = 1). It was shown that different genetic techniques were performed for various studies, and numerous ASD-associated genes were identified. In the African population, no gene was found that utilized GWAS; 26 genes were identified using a PCR-based method, 22 genes, including 10 differentially methylated, were discovered using sequencing technologies, and 12 were found through other techniques. Except for GWAS, most research methods used globally for genetic studies on ASD have been applied in Africa. Our findings demonstrate that numerous genetic variations could contribute to the underlying genetic basis of ASD in African communities.
FIGURE 2.
Selected articles on the association of genetic factors and ASD by study design.
3.1.1 Genome-wide association studies
Several developed countries have employed the GWAS technique to discover common and rare variants linked to ASD. However, no single GWAS research study was performed for the African population among our retrieved data.
3.1.2 Sequencing technologies
Seven studies used sequencing technologies and identified 12 genes associated with ASD in the African population: CACNA1C, CHD7, FEZF2, FMR1, PCDHAC1, SATB2, SBCADD, SHANK2, TBC1D8, TBR1, TCF7L2, and TSHZ3. In addition, 10 differentially methylated genes—AHI1, FIS1, GABPA, GLRA2, MFN2, MTR, OPA1, PGC-1α, SETD5, and STOML2—were identified as linked to ASD by these technologies. A number of potentially pathogenic variations in known ASD genes—CACNA1C, CHD7, FMR1, TCF7L2, and PCDHAC1—were found to be novel autism susceptibility genes using whole genome sequencing (WGS) on an East African cohort to investigate the genetics of ASD (Tuncay et al., 2023). The results from this study suggested a higher prevalence of ASD in East African children and demonstrated the value of admixture analysis and African genetic diversity in understanding the etiology of complex disorders.
Several studies using next-generation sequencing (NGS) and identifying DNA methylation as causing ASD have been conducted in South Africa. Whole-genome DNA methylation screening showed that GLRA2 and AHI were differentially methylated in children with ASD from South Africa (Stathopoulos et al., 2020). Moreover, it was shown that the DNA methylation-related genes SETD5 and MTR were differentially methylated, linking them to ASD (Stathopoulos et al., 2018). Several genes including, PGC-1α, STOML2, MFN2, FIS1, OPA1 and GABPA, have different levels of methylation related to mitochondrial biogenesis, fission, and fusion in ASD, according to next-generation bisulfite sequencing technology (p < 0.05) (Bam et al., 2021). Furthermore, this South African study showed the correlation between methylation at PGC-1α and metabolomic evidence of mitochondrial malfunction. The results of another South African ASD cohort showed that methylation levels of case and control for PCCB and PCDHA12 varied widely from 9%–49% and 0%–54%, respectively. Further evidence that DNA methylation is a crucial epigenetic component in the pathogenesis of ASD comes from the hypothesis that the differentially methylated genes linked to ASD in this cohort have a functional role in mitochondrial homeostasis and dysregulation.
A previous study showed a mutation in the SBCADD gene among people originating from Somalia and Eritrea. This finding was repeated in a four-year-old autistic Somali boy (Kanavin et al., 2007), suggesting that this mutation is relatively common in this demographic.
3.1.3 PCR-based methods
A total of 29 ASD genetic studies in Africa were conducted using PCR-based approaches such as polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) and real-time PCR. However, these methods identified 26 genes: AADAT, C4B, COMT Val58Met, DRD1, DRD2A1+, DRD4, GABA, GSTM1, GSTT1, HAAO, HLA-DRB1, HLA-DR4, HTR1A, 5-HTTLPR, 5-HTTLPRS, 5-HT2A, IL-1β-511 and IL-1RA, IL-12, LINE-1, MAOA, Q6NUR6, RELN (rs2229864), RTL, SHANK3, TAMRTKS, and VDR. The first study that used sequencing in Africa was conducted in the Egyptian population, linking the DRD1 (rs453) variant to autism (Azzam et al., 2018). The COMT Val58Met polymorphism and ASD in Egyptian young people were also linked (Karam et al., 2013). A case-control study performed by Egyptian researchers revealed strong evidence that the DRD4 7/7 allele could be associated with an increased risk of autism (Kamal et al., 2015).
Additionally, three genes involved in the tryptophan metabolic pathways—MAOA, HAAO, and AADAT—were found to have lower expression levels in patients with ASD, which was correlated with the severity of autism in a comparison between children with ASD, learning-disabled children, and healthy controls (Higazi et al., 2021). The DRD2 A1+ genotype enhanced autism risk in Egypt’s autistic community according to a study on the relationship between autism and genetic variants in several neurotransmitter-related genes such as MAOA, MAOB rs1799836, and DRD2. SNPs and/or mutations in miR-21 or miR-431 were not found in the patients enrolled in the study (Salem et al., 2013). A genetic study conducted on Tunisian people with ASD reported an increase of Q6NUR6 gene expression in blood (Bayou et al., 2010), which might be linked to the cause of ASD in those populations.
The results of numerous investigations point to a correlation between complex diseases brought on by genetic variants and variations in serum protein levels in solid tissues and biofluids. Genetic effects on serum proteins may provide new perspectives on the mechanisms behind the genetics of common diseases and significant characteristics. These new perspectives could help us understand the genetics of prevalent diseases and their key characteristics. Based on the analysis of TAMRTKs blood levels in a sample of Egyptian children with autism, the findings revealed that their levels were upregulated and had strong positive associations with the severity of the condition (Mostafa et al., 2022). In addition, the results of the study supported the immunological etiology of ASD by demonstrating that Egyptian children with ASD had an immunological impairment in the form of an elevated serum level of IL-12, which was positively linked with the severity of autistic symptoms (Ibrahim et al., 2015).
ASD in Egyptian children was found to be associated with abnormally elevated serum levels of IL-1β and IL-1RA, and polymorphisms in the IL-1β-511 and IL-1RA genotype variants may influence the risk of ASD and be used as potential biomarkers of the disorder. These findings were strongly linked to the severity of autism and behavioral impairments (Saad et al., 2020). Evaluation of the possible involvement of RTL and LINE-1 methylation as biomarkers for autism revealed a substantial lower level of methylation than controls in Egyptian autistic patients with p < 0.001 (Salem and Ashaat, 2023). RTL and LINE-1 methylation percentages can both be used as biomarkers for autism. Mostafa and Shehab (2010) suggested that C4B null allele autoimmunity in Egyptian autistic people may be related to the C4B null allele. In the Egyptian ASD population, GABAerigic malfunction was shown to increase apoptosis. As a result, neuronal excitement and an imbalance in the inhibitory system can be connected to GABA synaptopathies and their linkage to apoptosis, which can be used as trustworthy potential biomarkers for autism (El-Ansary et al., 2021). It is interesting to note that all South African autistic ethnic groupings have shown a highly meaningful correlation with the *S/*S variant of 5-HTTLPR (Arieff et al., 2010). Furthermore, the 5-HTTLPRS allele S allele significantly increased in Egyptian autistic children (Meguid et al., 2015). The expression of the HTR1A gene was shown as a potential candidate gene for ASD-related pathways in an explored sample of the Egyptian population (Yahya et al., 2019). Moreover, the 5-HT2A receptor may be involved in the development of ASD, according to a study on Egyptian children with autism (Abdelrahman et al., 2015).
The role of the RELN gene in autism has been elucidated by many discoveries of reduced levels of the reelin protein in the brain and plasma in patients with autism. Evidence first indicated that RELN rs736707 may play a role in autism in South African populations (Sharma et al., 2013). Recently, RELN gene polymorphism (rs2229864) has been reported as a possible contributing factor for ASD genetic susceptibility and severity in Egyptians (Abdelhady et al., 2022). Two different studies were conducted in Egyptian children with autism on HLA allele. One study found that there was a considerable probability that the HLA-DRB1 allele would be linked to a family history of autoimmunity (Mostafa et al., 2013). The HLA-DR4 alleles have been also linked to autism, according to another study conducted in Egypt (El-Hossiny et al., 2023). Children with ASD in Egypt have been shown to have SHANK3 copy number variations, and the variation in SHANK3 copy number revealed the function of SHANK3 in the ASD etiology (Meguid et al., 2020). One potential mechanism underpinning the etiology of ASD is gene–environment interaction. The most common type of null GSTM1 and GSTT1 genotypes in ASD were reported in a gene-environment interaction Egyptian study. That genotype may make autistic children more susceptible to reduced antioxidant status (GST enzyme activity), which could result in improper aluminum detoxification (Said et al., 2021).
Even though most of the results from genetics studies showed a positive association with ASD, some studies have a negative association with autism in the African population. A PCR-amplified DNA MTHFR gene assay revealed that the homozygous 677TT genotype was found in 23% of the children with ASD, whereas the heterozygous 677CT genotype was found in 56% of the ASD group in an Egyptian study on the MTHFR C677T polymorphism. Despite the fact that the 677CT variant alleles were increased in autistic patients, it is doubtful that these alleles cause the wide range of symptoms associated with autism (Shawky et al., 2014). Another Egyptian study analyzed a functional polymorphism of the HFE gene and SLC40A1, and the results highlighted the rarity of this functional polymorphism (Gebril and Meguid, 2011; Na and Oh, 2014). The study of two genes, HoxA1 and HoxB1, appeared to be implicated in the development of autism in various groups. Nevertheless, a study could not find a statistically significant link between the prevalence of autistic children in Egypt and the variations of these two genes (Sheikh et al., 2007). ASD patients have considerably lower levels of reduced glutathione, according to an evaluation of the serum levels of oxidative stress indicators and an analysis of genetic variants of glutathione S-transferase related with autism for children with autism in Nigeria. However, in this study, the distribution of the three polymorphisms GST1, GSTM1, and GSTP1 was not linked to autism (Oshodi et al., 2017).
3.1.4 Other molecular biology techniques
A total of 12 genes was identified through methylation analysis and microarray-based comparative genomic hybridization (aCGH): CELF4, CHRFAM7A, ERICH1, FTHL17, NEXMIF, NLGN4X, PCCB, PCDHA12, PRKN, SPN, SYCE3, UQCRC2 and one de novo balanced (7; 16) (p22.1; p16.2) chromosomal aberration by cytogenetics and fluorescent in situ hybridization (FISH). Through methylation analysis, the results from the South African ASD cohort, both case and control PCCB and PCDHA12, had methylation levels that ranged widely: 9%–49% and 0%–54%, respectively. The PCCB revealed three CpG loci that were differently methylated with p ≤ 0.05; however, PCDHA12 revealed two CpG sites with a significant difference between ASD and control (p ≤ 0.001) (Stathopoulos et al., 2020). In addition, lead, aluminum, and mercury were found at a higher concentrations in the hair of autistic children than in control children. Autism may be caused by exposure to these harmful heavy metals in the environment at critical developmental stages, as shown in the Egyptian study that used the hydride generation technique (Mohamed et al., 2015). The aCGH technology presented high pathogenicity scores in numerous genes, such as PRKN, CHRFAM7A, SYCE3, UQCRC2, SPN, FTHL17, NLGN4X, ERICH1, NEXMIF, and CELF4, in Tunisia children with ASD (Chehbani et al., 2022), and were pathogenic/likely pathogenic to ASD, as showed in this cohort study. A case report in Tunisia showed particular chromosomal abnormality in a person with autism, with de novo balanced (7; 16) (p22.1; p16.2) translocation (Bayou et al., 2008).
4 Discussion
ASD can be caused by both inherited and de novo gene mutations due to its extreme genetic heterogeneity. A staggering number of newly emerging mutations linked to ASD have been discovered due to advances in human genetics and sequencing technologies. The genetic causes of ASD are now better understood because of recent large-scale multinational research projects. To date, there are more than 100 genes linked to autism (Sanders et al., 2015; Grove et al., 2019; Forrest and Penzes, 2020). However, this study has found 61 genes associated with ASD in the African population. ASD-causing genes were found in nine African countries.
The majority of the studies were conducted in Egypt and South-Africa, as their governments acknowledge the importance of science and research in improving people’s standard of living and generating income for their citizens (Hardy et al., 2008; Bond et al., 2012). This has enabled them to establish genetic and genomic facilities while other countries lack an enabling environment for genomic studies, have scarce or no funding, and have complicating political contexts (Omotoso et al., 2022).
Except for GWAS, most research methods used globally for genetic studies on ASD have been applied in Africa. Our findings demonstrate that numerous genetic variations may play a role in the genetic basis of ASD in the African population. Study findings have shown that several chromosomal regions include one or more susceptibility genes for autism (Monaco and Bailey, 2001).
4.1 Advances in genomics for ASD analysis
The quest for the genes underlying prevalent human diseases and associated quantitative features is being revolutionized by GWAS. This method combines a thorough and objective examination of the genome with the capability of finding common alleles with few phenotypic effects (Hirschhorn and Daly, 2005). The effectiveness of this technique depends on the underlying impact sizes carried by the genuine risk variations as well as the statistical power to detect these effects given the sample size and study design. Common genetic variation accounts for around half of ASD genetic risk (Glasson et al., 2004), making GWAS an effective method for locating risk variants. Many loci linked to ASD have been discovered in non-African population using GWAS. Increased number of SNPs (iPSYCH-SSI-Broad Autism Group et al., 2016; Grove et al., 2019) and other types of variation (such as copy number variations and rare structural variants) (De La Torre-Ubieta et al., 2016) that are linked to ASD have been found during the past 10 years through GWAS. According to a meta-analysis of GWAS, there is a strong genetic correlation between ASD and a number of genes relevant to neurodevelopment, including EXT1, ASTN2, MACROD2, and HDAC4 (Anney et al., 2017). In addition, it has been shown through GWAS that traits resembling autism are associated with numerous immune-related genes such as RNF114, CDKN2A, KAZN, SPATA2, and ZNF816A (Arenella et al., 2022). However, GWAS must have a sample size with enough statistical power to identify the genes responsible for human complex disorders (Baranger et al., 2023). Thus, care must be taken in study design, execution, analysis, and interpretation to minimize the drawbacks of this strategy.
Most studies have focused on populations from the United States, Europe, and Asia, and there is little information regarding the genetic composition of ASD across Africa. This scoping review notes that no gene has been found in the African population using GWAS. According to the African Genome Variation Project, the population of Ethiopia has the highest percentage of unique and private genomic diversity in all of Africa, accounting for about 24% of genomic variants (Gurdasani et al., 2015). It represents the highest prevalence of ASD for an East African population (Barnevik-Olsson et al., 2008; Magnusson et al., 2012). Although the prevalence rate of ASD is higher in East African populations than the rest of the globe, no prevalence study has been carried out in East Africa’s nations. Numerous studies carried out in Europe for immigrant populations highlight a higher frequency of ASD among offspring born to parents from East Africa (Barnevik-Olsson et al., 2008; Magnusson et al., 2012). Thus, large-scale GWAS studies are required in Africa to uncover many genetic variants linked to ASD because the continent still retains much of the diversity found in the human genome. Understanding common biological processes that influence health and disease in all groups will result from identifying genetic variations in African populations.
The use of sequencing technology for disease-causing gene mutation discovery has the advantage of emphasizing the importance of this discovery for proper patient management and family counseling (Lohmann and Klein, 2014). The patient’s entire exome or genome may be precisely analyzed using next-generation sequencing and can be found in a single experiment (Gupta and Verma, 2019). Sequencing technology is especially well-suited for studying the genetics of heterogeneous traits like ASD; it has discovered a meaningful number of arising mutations linked to ASD. The ability of next-generation sequencing to find risk-mediating variations in single families or small cohorts of ASD patients was demonstrated in a number of early studies (O’Roak et al., 2012; Sanders et al., 2012). To discover ASD risk genes in the coding regions of human DNA and search for de novo mutations from various populations, several researchers have utilized a variety of sequencing methodologies, such as whole genome sequencing (WGS) and whole exome sequencing (WES) (Michaelson et al., 2012; Neale et al., 2012). Although sequencing technology has only recently been adopted in several African nations, it has allowed researchers to identify numerous genes associated with ASD. Common and rare variants of the voltage-gated calcium channel gene CACNA1C have been associated with autism and neurodevelopmental disorders, including schizophrenia, bipolar disorder, and attention deficit hyperactivity disorder (ADHD) (Buddell et al., 2019). FMR1, the gene linked to fragile X syndrome (FXS), is located on chromosome X, and an unstable CGG repeat in the 5' untranslated region of the gene prevents FMR1 from functioning properly, resulting in FXS. One of the primary gene regulators in the central nervous system (CNS) is FMRP, the protein that results from FMR1. It has been discovered that individuals with ASD who do not have FXS have defective multiple FMRP-regulated pathways (Fyke and Velinov, 2021). The two most commonly identified mutations in ASD patients, CACNA1C and FMR1 (Tuncay et al., 2023), were also found in the East African population. A CHD7 intronic variant causes some important genes linked to neural developmental disorders to become dysregulated; this effect is comparable to that brought on by gene deletion (Zhang et al., 2021). The CHD7 intronic variation is potentially an autism susceptibility locus within the Chinese ASD population (Zhang et al., 2021) and has also been identified in the East African cohort (Tuncay et al., 2023).
In addition, a mediator of canonical Wnt signaling, TCF7L2, is recognized to be crucial for the development of the central nervous system (CNS). Heterozygous de novo mutations in TCF7L2 exhibit neurodevelopmental symptoms (Dias et al., 2021). Previously, TCF7L2 reported in non-African populations (Dias et al., 2021) was also found in East Africa (Tuncay et al., 2023). Protocadherin α (PCDHA) is involved in serotonergic innervation of the brain and synaptic specificity (Katori et al., 2009), making it a good candidate gene for autism. Strong genetic evidence has been reported of the possible role of PCDHA as an autism candidate gene in the Japanese population (Anitha et al., 2013), and the mutation in one PCDHA family gene, PCDHAC1, was also found in East Africa (Tuncay et al., 2023). A 2-methylbutyryl-CoA dehydrogenase deficit, sometimes referred to as “short/branched chain acyl-CoA dehydrogenase deficiency” (SBCADD), is caused by a malfunction in the breakdown process of the amino acid L-isoleucine. Isoleucine is oxidized and used as fuel in the brain. Two cases from Somalia and Eritrea displayed mutated SBCADD gene (Kanavin et al., 2007). Major transcription factors encoded by FEZF2 modulate neuron location and identity. FEZF2 and ASD have been genetically linked in two sizable European ancestry cohorts, and this relationship has been confirmed in two more cohorts (Wang et al., 2009). Interestingly, the FEZF2 gene was also found in the Moroccan population (Bensaid et al., 2019).
The scaffolding proteins found in glutamatergic synapses’ postsynaptic density (PSD) encoded by the SHANK2 and SHANK3 genes, and de novo SHANK2 deletions confirmed in Caucasian ancestry ASD patients (Leblond et al., 2012), were also reported in a patient of Ethiopian ancestry (Lu et al., 2018). Furthermore, SHANK3 deletion was found in Chinese people with ASD through a genome-wide copy number variation analysis (Guo et al., 2017); SHANK3 dose contributes to the pathogenesis of ASD for Egyptian autistic children (Meguid et al., 2020). There is mounting proof that DNA methylation influences ASD characteristics. Retinal degeneration and progressive renal failure are potential outcomes of AHI1 mutations (Parisi et al., 2006), and GLRA2 was found to be a new gene that causes high myopia (Tian et al., 2023). Through whole-genome DNA methylation screening, AHI1 and GLRA2 genes were differentially methylated in a South African cohort with ASD (Stathopoulos et al., 2018), and a United States study provided evidence for an associated haplotype in AHI1 with ASD (Alvarez Retuerto et al., 2008). Moreover, GLRA2 was reported in autistic people from France (Pilorge et al., 2016). The ubiquitin-proteasome pathway that may be responsible for the downregulation of SET domain-containing 5 (SETD5) is often overactive and mutated in human NDDs as well as cancer (Li et al., 2023). The ASD-linked genes SETD5 (Grozeva et al., 2014) and MTR (Muratore et al., 2013), which are directly involved in DNA methylation, were differentially methylated in the ASD South African population (Stathopoulos et al., 2018). Peroxisome proliferator-activated receptor-γcoactivator (PGC-1α) is involved in remodeling muscle tissue into a fiber-like composition, and it increases mitochondrial biogenesis (Liang and Ward, 2006). The differential methylation of the PGC-1α gene was reported in the ASD South-African population (Bam et al., 2021), and a Chinese Han study also suggested that the SIRT1/PGC-1α signaling pathway has a crucial function in patients with ASD (Bu et al., 2017). In addition, the optic atrophy 1 (OPA1) gene, involved in mitochondrial fusion, works in conjunction with stomatin-like protein 2 (STOML2) (Tondera et al., 2009). Study results for those two genes in a South African population with ASD showed that the OPA1 and STOML2 genes were significantly differentially methylated, which may implicate their contribution in ASD etiology (Bam et al., 2021). Furthermore, in a Chilean study, autistic children had significantly higher levels of mtDNA in their oral mucosa, and there was an expression of MFN2, FIS1, and OPA1 involved in the fission/fusion processes of mitochondria (Carrasco et al., 2019). Several co-occurring symptoms linked to SATB2 gene variations have led to identifying a single clinically recognized syndrome known as SATB2-associated syndrome (SAS) (Döcker et al., 2014). There was a decreased metabolic response to tryptophan for SAS patients from United States, and that has been associated with ASD (Boccuto et al., 2013). The SATB2 gene has also been found in Moroccan people (Bensaid et al., 2019). Furthermore, TSHZ3 encoding the zinc-finger homeodomain transcription factor TSHZ3 that was found in networks of human neocortical genes is strongly expressed throughout late fetal development and is implicated in neurodevelopmental and neuropsychiatric diseases (Caubit et al., 2016; Li M. et al., 2018). This gene linked to ASD in a Moroccan study (Bensaid et al., 2019) was identified as one of the ASD-related genes differently expressed in the brain in a Chinese study (Li et al., 2020). Additionally, TBR1, a T-box transcription factor (TF) essential for controlling cortical development (Han et al., 2011), previously reported as ASD risk contributing mutation (O’Roak et al., 2012) in a United States study, was also identified in Moroccan ASD families (Bensaid et al., 2019). Protein coding gene TBC1 domain family member 8 (TBC1D8) is a new ASD gene found in Libyan siblings with ASD (Zeglam and Alhmadi, 2020) but not reported in any other populations.
Re-sequencing the complete genome in patients and controls to look for a variant or group of variants enriched or depleted in disease cases yields the most thorough study of candidate genes. However, because such research is still time-consuming and expensive, it has mostly focused on the coding areas of a small number of potential genes. In addition to revealing complex inheritance patterns, such as the inheritance of two autosomal recessive gene mutations in the same patient and family, the use of sequencing technology for disease-causing gene mutation discovery has the advantage of emphasizing the importance of this discovery for proper patient management and family counseling (Lohmann and Klein, 2014). The patient’s entire exome or genome may be precisely analyzed using next-generation sequencing, and single nucleotide variations can be found in a single experiment (Gupta and Verma, 2019). As the most advanced molecular approach available today, PCR technology has a wide range of present and future therapeutic uses, including pathogen detection, assessment of emerging novel infections, surveillance, early detection of biothreat agents, and antibiotic resistance profiling (Yang and Rothman, 2004). PCR is used by a variety of researchers to analyze mutations that arise in many genetic disorders (Reiss and Cooper, 1990). PCR-based methods were mostly adopted in Africa for studying the genetic etiology of ASD. The reelin gene (RELN) plays a key role in the brain’s regulation of neuronal migration, synaptogenesis, and synaptic plasticity. Therefore, the RELN signaling pathway has been linked to a number of neurological conditions in humans, including mental retardation, ASD, and schizophrenia (Jossin, 2020). Two different RELN SNPs previously linked to ASD in non-African population were also identified in Africa. One RELN SNP, rs736707, discovered to be significantly linked to ASD in Asian groups (Chen et al., 2017), was reported as an ASD risk factor for the South African population (Sharma et al., 2013). Another RELN SNP, rs2229864, associated with the genetic predisposition for ASD in the Chinese Han population (Wang et al., 2018), was also found to be related to ASD in Egyptian children (Abdelhady et al., 2022). Brain serotonin (5-HT) is widely recognized to be involved in the regulation of both normal and abnormal behavior (Blier and Ward, 2003; Ögren et al., 2008). Numerous studies have suggested that the neurobiological process underlying ASD is likely to involve function-impairing polymorphisms of the 5-HT system, including 5-HTTLPR, 5-HTR1A, and 5-HTR2A (Chugani, 2002; Richardson-Jones et al., 2010). Strong associations have been found between the 5-HTTLPR polymorphism and various autistic populations worldwide, according to numerous case-control studies. In a United States study, a significant relationship was found between ASD and the serotonin transporter gene promotor polymorphism (5-HTTLPR) (Brune et al., 2006), and was also found in all ASD patients in South Africa (Arieff et al., 2010). Moreover, the 5-HTTLPRS allele was significantly increased in Egyptian autistic children (Meguid et al., 2015). Nevertheless, an association between the 5-HTTLPR polymorphism and autism has not been found by a meta-analysis (Wang et al., 2019). Moreover, the HT2A gene reported in the Croatian (Hranilovic et al., 2010) and Korean (Cho et al., 2007) populations was also shown in Egyptian autistic children (Abdelrahman et al., 2015). The steroid hormone vitamin D is well known for its function in neuronal growth and neuroprotection. Consequently, deficiencies in the vitamin D pathway might contribute to the development of ASD (Zhang et al., 2018). It has been demonstrated that some VDR gene polymorphisms, particularly rs731276, are a risk factor for childhood ASD in the Chinese Han community (Zhang et al., 2018) and are linked to ASD in the Egyptian population (Arafa et al., 2021).
Autoimmunity and various autoimmune disorders may be influenced by complement (C) 4B null alleles, which result in low levels of C4B protein. The C4B null allele had a high risk of association with autism, and there is a connection between the C4B null allele and a family history of autoimmunity (Mostafa and Shehab, 2010). Therefore, autoimmunity in Egyptian patients with ASD may be significantly influenced by the C4B null allele (Mostafa and Shehab, 2010), and the US study’s findings indicate that this allele is significantly linked to an increased risk of autism (Odell et al., 2005). The tryptophan metabolism-related genes such as aminoadipate aminotransferase (AADAT), 3-hydroxyanthranilate oxygenase (HAAO), and monoamine oxygenase A (MAOA) were found in children with ASD in Egypt (Higazi et al., 2021). Interestingly, de novo missense mutation in AADAT found in Chinese families with ASD (Li S.-J. et al., 2018) was suggested as the basic etiology of ASD for those families. ASD patients in the United States have shown decreased expression levels of the genes AADAT, HAAO, and MAOA (Boccuto et al., 2013). In addition, the enzyme catechol-O-methyltransferase (COMT) has been linked to aberrant dopaminergic activity, suggesting that this gene may be involved in the etiology of ASD (Esmaiel et al., 2020). The COMT variant is linked to hyperactivity symptoms in Egyptian children with ASD (Karam et al., 2013), and findings from a United States study suggested that the ASD phenotype may be influenced by the COMT gene (Radoeva et al., 2014).
Dopamine is crucial for maintaining appropriate attention. Hence, research has examined the dopaminergic system’s genes, particularly concerning attention problems. The DRD1 and DRD2 genes were identified in Egyptian ASD autistic patients (Salem et al., 2013; Azzam et al., 2018), and according to the results from an Italian study, both DRD1 and DRD2 receptor SNPs may be regarded as possible risk factors for ASD (Mariggiò et al., 2021). The human dopamine receptor D4 (DRD4) gene, found on chromosome 11p close to the telomere, has an unusually high level of expressed polymorphism. A clinically higher risk for autistic symptoms may be associated with the DRD4 7-repeat allele (Reiersen and Todorov, 2011) according to US findings from Missouri-born twins study, which are similar to the findings of another Egyptian study (Kamal et al., 2015). In the adult human brain, gamma-aminobutyric acid (GABA) is the main inhibitory neurotransmitter. Several neuropsychiatric disorders, including, anxiety, epilepsy, and learning disabilities, have all been linked to GABAergic dysfunction (Coghlan et al., 2012). In a US study, autistic children exhibited considerably lower levels of sensorimotor GABA than healthy controls (Puts et al., 2017), and that was also found in an Egyptian study (El-Ansary et al., 2021). HLA proteins are involved in the development of the central nervous system (CNS), cerebral hemisphere specialization, synaptic function, and neural cell interactions (Torres et al., 2012), and several HLA alleles, including HLA-DRB1 alleles, are linked to ASD (Polleux and Lauder, 2004). Findings from a Han Chinese study suggest that the HLA-DRB1 gene is linked to ASD (Chien et al., 2012), which has been reported in Egypt (Mostafa et al., 2013). Furthermore, a US study suggested that HLA-DR4 is linked to ASD (Torres et al., 2002), where autistic patients demonstrated high HLA DR4 allele frequency, and an Egyptian study has also reported such results (El-Hossiny et al., 2023). In addition, long interspersed nucleotide element-1 (LINE-1) is a type of non-long terminal repeat retrotransposon that has been linked to disorders including ASD due to aberrant DNAm (Saeliw et al., 2018). LINE-1 methylation was shown to be substantially lower in lymphoblastoid cell lines from ASD patients in Italy (Tangsuwansri et al., 2018), and a recent study in Egypt has reported similar results (Salem and Ashaat, 2023). Furthermore, the gene retrotransposon Gag-like 1 (RTL1), a paternally expressed gene, is crucial for placenta maintenance (Sekita et al., 2008). The RTL gene also reported in this study was found in the Iranian population, and the findings showed a sexually dimorphic pattern of RTL in children with ASD (Panahi et al., 2023).
Microarray-based comparative genomic hybridization aCGH technology performed on Tunisian children diagnosed with ASD has reported numerous pathogenic or likely pathogenic variants such as CELF4, CHRFAM7A, ERICH1, FTHL17, NEXMIF, NLGN4X, PRKN, SPN, SYCE3, and UQCRC2 (Chehbani et al., 2022). Several of these were reported to be associated with ASD worldwide, such as both CELF4 and CHRFAM7A in Italy (Bacchelli et al., 2015; Barone et al., 2017), NEXMIF in Switzerland (Lambert et al., 2018), and both NLGN4X and UQCRC2 in China (Yu et al., 2011; Shen et al., 2019). The ASD etiology is thought to be caused by epigenetic processes like DNA methylation, RNA interference, and chromatin alteration (Tremblay and Jiang, 2019). The methylation-dependent regulation of transcription has become an intriguing theory for the etiology of ASD because of next-generation sequencing and the identification of DNA alterations. The PCCB gene mutations found in ASD patients in Belgium (Witters et al., 2016) were differentially hypomethylated in South-Africa patients affected with ASD (Stathopoulos et al., 2020). Another gene, PCDHA12, reported in that South African study was also studied in Japan, providing strong genetic evidence that PCDHA12 may be a viable candidate gene for autism (Anitha et al., 2013). Moreover, genetic and environment interactions have also been identified as potential factors contributing to ASD. Many hazardous metals must be detoxified, and glutathione-S-transferase (GST) genes and associated enzymes are essential. The study findings imply that among Jamaican children with ASD, the existence of active versions of the GSTT1 and GSTM1 genes may be linked to a higher ability for lead (Pb) detoxification (Rahbar et al., 2022), and an Egyptian study also found that the most prevalent genotype associated with ASD, null GSTM1 and GSTT1, may predispose ASD children to a lower antioxidant status (Said et al., 2021), which can ultimately result in improper aluminum detoxification. There is significantly more aluminum in the hair of children diagnosed with ASD, and the development of autism in these children may be significantly influenced by oxidative indicators that result in oxidative damage (Said et al., 2021). However, a Nigerian study reported opposite results on GST genes by discovering that although ASD patients had significantly lower levels of reduced glutathione, there was no correlation between ASD and the distribution of the GSTT1, GSTM1, or GSTP1 polymorphisms (Oshodi et al., 2017). Furthermore, the limited sample size made it difficult to generalize these findings to the whole population. Large sample sizes are typically required in case-control studies to detect such effects because common diseases are caused by complicated interactions among several genetic variations and environmental risk factors (Moonesinghe et al., 2008). In addition, several other genes such as DISC1, GABRB3, GSTP1, HFE, HOXA1/HOXB1, SLC40A1, and MTHFR 677C>T were unrelated to risk of ASD in Africa. Numerous chromosomal abnormalities, both balanced and unbalanced, have been linked to autism (Wassink et al., 2001; The Autism Genome Project Consortium, 2007), and de novo balanced (7; 16) (p22.1; p16.2) translocation was reported in a Tunisian male patient with autism (Bayou et al., 2008). According to our findings, a total of 12 genes associated with ASD in Africa—ERICH1, FTHL17, GABPA, IL-1β-511 and IL-1RA, IL-12, PRKN, Q6NUR6, SPN, STOML2, SYCE3, TAMRTKS, and TBC1D8—were not identified in non-African populations.
On other hand, 20 ASD genes, each reported at least in two population studies, were identified in non-African populations that are yet to be confirmed/reported in Africa, perhaps because molecular research on ASD has continually underrepresented African populations, and the majority of current research on ASD is carried out in high-income countries (Frickel et al., 2023). Acetylserotonin methyltransferase (ASMT), a component of the final stage of melatonin production, was reported in China (Wang et al., 2013; Jonsson et al., 2014). The chromodomain helicase DNA binding protein 8 (CHD8) interacting with beta-catenin for chromatin remodeling (Bernier et al., 2014) was reported in the United States. Contactin-associated protein-like 2 (CNTNAP2), a member of the neurexin superfamily that plays a crucial role in brain development (Li et al., 2010; Nascimento et al., 2016), was reported in China and Brazil. The dopamine-3-receptor (DRD3) gene linked to posttraumatic stress disorder-related impairments in emotion reactivity, executive functioning, and stress-responding (De Krom et al., 2009) was reported in the Netherlands and United Kingdom. Engrailed 2 (EN2), a homeobox transcription factor that plays a role in the cerebellum’s patterning during brain development (Wang et al., 2008; Sen et al., 2010) was reported in China and India. The forkhead-box protein P1(FOXP1) gene encoding a transcription factor crucial for the early development of numerous organ systems, including the brain (Hamdan et al., 2010; Chien et al., 2013), was reported in Taiwan and Canada. Gamma-aminobutyric acid (GABRB3), the primary inhibitory neurotransmitter in the brain (Buxbaum et al., 2002; Warrier et al., 2013; Chen et al., 2014), was reported in the United States, United Kingdom, and Taiwan. The glutamate-binding GluN2 (GRIN2B) involved in circuit construction, brain development, and potential cell migration and differentiation (Yoo et al., 2012; Pan et al., 2015) was reported in China and Korea. HoxA, a key player in the formation of hindbrain neural structures (Ingram et al., 2000; Conciatori et al., 2004), was reported in the United States and Italy. The methyl CpG binding protein 2 (MECP2) gene (Nagarajan et al., 2006; Wen et al., 2017) was reported in the United States and China. The MET mediating hepatocyte growth factor signaling (Campbell et al., 2009; Jackson et al., 2009) was reported in Italy and the United States. Neuroligin 3 (NLGN3) regulating synapse organization (Yu et al., 2011; Oleari et al., 2023) was reported in China and Italy, while Neuroligin4 (NLGN4) was reported in France, Bulgaria, and Japan (Laumonnier et al., 2004; Avdjieva-Tzavella et al., 2012; Toya et al., 2023). In addition, Neurexin-1 (NRXN1) was reported in China and the United States (Liu et al., 2012; Griswold et al., 2015). The oxytocin receptor gene (OXTR) which plays a role in social-emotional behaviors (Wu et al., 2005; Di Napoli et al., 2014) was reported in the United Kingdom and China. The patched domain containing protein 1(PTCHD1) was reported in Spain, the Netherlands, Germany and Italy (Torrico et al., 2015). PTEN, a PI3K/AKT pathway negative regulator, was reported in the United States (Buxbaum et al., 2007). The SLC25A12 gene involved in catalysis of the mitochondrial carrier of aspartate-glutamate exchange (Turunen et al., 2008; Aoki and Cortese, 2016) was reported in Finland and the United Kingdom. SRRM4, a member of an RNA splicing factor family that controls the incorporation of specific genetic material (Irimia et al., 2014; Narita et al., 2020), was reported in Italy and Japan. Lastly, SYNGAP1 that regulates developmental excitatory synapse formation and function was reported in Canada (Berryer et al., 2013).
4.2 Recommendations for genomic ASD study
Given the genetic diversity found in Africa, more extensive sequencing of samples from various African populations is lacking. There is a need to conduct sequencing research in various African countries, which could identify new genetic variants linked to ASD in Africa. The use of sequencing techniques to identify ASD risk genes and unreported mutations in well-known sites has shown potential. The potential genes that have not yet been linked to ASD will need to undergo sequencing in larger cohorts and additional experimental validation to prove causation or the genes’ role in the disorder. Most genetic studies for ASD in Africa adopted PCR-based methods; however, fewer ASD risk genes were identified through this study design than in other techniques. The PRC-RFLP method has been primarily performed in Africa, as it is faster and cheaper than sequencing methods. Sequencing is a common technique for the identification of mutations. Additionally, there is a need for many genetic studies that use sequencing technology or GWAS across various African countries, thus helping to identify a significant number of de novo genetic variants associated with ASD due to using different ethnic groups. Several identified studies in this paper have shown that findings can be limited by some technical difficulties and relatively small sample size due to the cost of the sample for analysis. To better understand the hundreds or thousands of common and unusual genetic variants associated with ASD in the African population, sample sizes must be significantly increased. A greater sample size should be used in subsequent efforts to enable additional studies, such as better cataloging of structural variation and improved disease prediction.
4.3 Clinical and therapeutic implications
ASD risk is believed to be influenced by numerous common gene variants. ASD risk genes found in Africa are linked to various human disorders (Table 1). The primary symptoms of ASD, including difficulties with social communication or repetitive activities, are not currently treated with pharmaceuticals; instead, behavioral therapy and the use of tightly regulated learning environments are used as treatments for ASD. Up until now, the US Food and Drug Administration (FDA) has only licensed and approved two medications for the treatment of irritability associated with ASD: the antipsychotics risperidone and aripiprazole (Lamy and Erickson, 2018). However, many additional pharmacological treatments have shown successful therapeutic management of ASD symptoms, including: atypical antipsychotics (risperidone, olanzapine, clozapine) for temper tantrums, aggression, or self-destructive behavior; selective serotonin reuptake inhibitors (sertraline, citalopram, fluoxetine) for anxiety and repetitive behaviors; psychostimulants (methylphenidate) for hyperactivity; opioid antagonists (naltrexone) for hyperactivity (Aman, 2004; Kumar et al., 2012). Additionally, a few of the most effective medications are currently undergoing various stages of clinical trials for the treatment of ASD’s behavioral and neurological symptoms (Eissa et al., 2018).
Precision medicine is a future medical practice based on algorithms that consider the patient’s features, such as their genome, epigenetics, and lifestyle. The main objective of ASD research is to find effective treatments for people with ASD (Fischbach et al., 2016), and the development of a customized medicinal strategy for these individuals depends on the identification of a genetic etiology (Schaefer, 2016). According to data from a Norwegian sample study, many parents of children with ASD obtain positive results from clinical genetic testing (CGT) since there may be an etiological cause (Johannessen et al., 2017). However, investigations are failing if African people and other underrepresented ethnicities are not included in genetic studies. Many unidentified genetic variants probably contribute to medically significant traits in African and non-African populations. Thus, the identification of genes associated with ASD in Africa’s population would help in the pharmaceutical production of therapeutics that would meet the genetic etiology of ASD in Africa.
4.4 Study limitations
The exclusion of gray studies, such as thesis projects, is a limitation given the findings of such excluded studies. However, as genetic research on ASD is not a very developed research field in Africa, theses may contribute significantly to the overall research evidence; hence, their exclusion may have resulted in an incomplete picture of the existing literature. Furthermore, our findings reveal a limitation of small sample sizes in the identified studies, which could account for the discrepancies in the results; thus, future research should focus on using larger samples from multiple sites to generalize its findings more broadly.
5 Conclusion
Our scoping review examined research conducted in Africa on genetic factors potentially associated with ASD, categorized into groups based on study methodology: GWAS, sequencing, PCR-based methods, and other techniques. We found that PCR-based study design is the most performed in Africa. However, only a limited number of ASD risk genes and polymorphisms were reported using that method. Even though sequencing performed in various African countries is limited, many genetic variants associated with ASD were identified through this approach. However, no study identified any ASD gene with GWAS, even though it has been used in non-African countries. Furthermore, larger samples should be used in future studies to enable additional analysis, such as better cataloging of structural variation. As these are developing countries, grants for research collaboration should strengthen research capacity in Africa. African ASD could be highly attributable to genetic factors, as shown in the results of the few studies that have been conducted. Large-scale research on the genetics and prevalence of ASD is required in Africa, which will support increasing efforts to understand the genetic causes of ASD and other NDDs in the continent’s population and thus assist researchers and pharmaceutical industries to propose therapies that will meet the needs of African populations.
Acknowledgments
This document has been produced with the financial assistance of the European Union (Grant no. DCI-PANAF/2020/420-028) through the African Research Initiative for Scientific Excellence (ARISE) pilot program. ARISE is implemented by the African Academy of Sciences with support from the European Commission and the African Union Commission. The contents of this document are the sole responsibility of the authors and can, under no circumstances, be regarded as reflecting the position of the European Union, the African Academy of Sciences, and the African Union Commission.
Funding Statement
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This study was sponsored by the African Research Initiative for Scientific Excellence (ARISE), pilot program (grant number ARISE-PP-40).
Author contributions
OH: conceptualization, investigation, methodology, resources, writing–original draft, writing–review and editing, and formal analysis. JH: methodology, supervision, and writing–review and editing. JU: formal analysis, methodology, supervision, writing–original draft, and writing–review and editing. HO: formal analysis, methodology, and writing–review and editing. LP: methodology, supervision, and writing–review and editing. VB: methodology, supervision, and writing–review and editing. AA: conceptualization, formal analysis, methodology, resources, supervision, validation, writing–original draft, and writing–review and editing. AU: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, software, supervision, validation, visualization, writing–original draft, and writing–review and editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2024.1431093/full#supplementary-material
References
- Abdelhady N. M. S., El-Hefnawy S. M., Elfetoh D. E. A., Abd El Naby S. A. (2022). RELN gene (rs 2229864) polymorphismas genetic risk factor in Egyptian children with autism spectrum disorders. Menoufia Med. J. 35, 1294. [Google Scholar]
- Abdelrahman H. M., Sherief L. M., Alghobashy A. A., Abdel Salam S. M., Hashim H. M., Abdel Fattah N. R., et al. (2015). Association of 5-HT2A receptor gene polymorphisms with gastrointestinal disorders in Egyptian children with autistic disorder. Res. Dev. Disabil. 36C, 485–490. 10.1016/j.ridd.2014.10.023 [DOI] [PubMed] [Google Scholar]
- Aderinto N., Olatunji D., Idowu O. (2023). Autism in Africa: prevalence, diagnosis, treatment and the impact of social and cultural factors on families and caregivers: a review. Ann. Med. Surg. 85, 4410–4416. 10.1097/MS9.0000000000001107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez Retuerto A. I., Cantor R. M., Gleeson J. G., Ustaszewska A., Schackwitz W. S., Pennacchio L. A., et al. (2008). Association of common variants in the Joubert syndrome gene (AHI1) with autism. Hum. Mol. Genet. 17, 3887–3896. 10.1093/hmg/ddn291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman M. G. (2004). Management of hyperactivity and other acting-out problems in patients with autism spectrum disorder. Semin. Pediatr. Neurol. 11, 225–228. 10.1016/j.spen.2004.07.006 [DOI] [PubMed] [Google Scholar]
- Anitha A., Thanseem I., Nakamura K., Yamada K., Iwayama Y., Toyota T., et al. (2013). Protocadherin α (PCDHA) as a novel susceptibility gene for autism. J. Psychiatry Neurosci. Jpn. 38, 192–198. 10.1503/jpn.120058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anney R. J. L., Ripke S., Anttila V., Grove J., Holmans P., Huang H., et al. (2017). Meta-analysis of GWAS of over 16,000 individuals with autism spectrum disorder highlights a novel locus at 10q24.32 and a significant overlap with schizophrenia. Mol. Autism 8, 21. 10.1186/s13229-017-0137-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y., Cortese S. (2016). Mitochondrial aspartate/glutamate carrier SLC25A12 and autism spectrum disorder: a meta-analysis. Mol. Neurobiol. 53, 1579–1588. 10.1007/s12035-015-9116-3 [DOI] [PubMed] [Google Scholar]
- Arafa M., Hasan T., Alghobashy A., Raafat N., Gohary M., Amer M., et al. (2021). Vitamin D receptor gene rs 731276 (taq1) polymorphism and autism susceptibility in children: a single center study. Türk Fiz. Ve Rehabil. DergisiTurkish J. Physiother. Rehabil. 32, 8318–8324. [Google Scholar]
- Arenella M., Cadby G., De Witte W., Jones R. M., Whitehouse A. J., Moses E. K., et al. (2022). Potential role for immune-related genes in autism spectrum disorders: evidence from genome-wide association meta-analysis of autistic traits. Autism 26, 361–372. 10.1177/13623613211019547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arieff Z., Kaur M., Gameeldien H., van der Merwe L., Bajic V. B. (2010). 5-HTTLPR polymorphism: analysis in South African autistic individuals. Hum. Biol. 82, 291–300. 10.3378/027.082.0303 [DOI] [PubMed] [Google Scholar]
- Arksey H., O’Malley L. (2005). Scoping studies: towards a methodological framework. Int. J. Soc. Res. Methodol. 8, 19–32. 10.1080/1364557032000119616 [DOI] [Google Scholar]
- Avdjieva-Tzavella D. M., Todorov T. P., Todorova A. P., Kirov A. V., Hadjidekova S. P., Rukova B. B., et al. (2012). Analysis of the genes encoding neuroligins NLGN3 and NLGN4 in Bulgarian patients with autism. Genet. Couns. Geneva Switz. 23, 505–511. [PubMed] [Google Scholar]
- Azzam A. A., Bahgat D. R., Shahin R. H., Nasralla R. A. (2018). Association study between polymorphisms of dopamine transporter gene (SLC6A3), dopamine D1 receptor gene (DRD1), and autism. J. Med. Sci. Res. 1, 59. 10.4103/jmisr.jmisr_8_18 [DOI] [Google Scholar]
- Bacchelli E., Battaglia A., Cameli C., Lomartire S., Tancredi R., Thomson S., et al. (2015). Analysis of CHRNA7 rare variants in autism spectrum disorder susceptibility. Am. J. Med. Genet. A 167, 715–723. 10.1002/ajmg.a.36847 [DOI] [PubMed] [Google Scholar]
- Bam S., Buchanan E., Mahony C., O’Ryan C. (2021). DNA methylation of PGC-1α is associated with elevated mtDNA copy number and altered urinary metabolites in autism spectrum disorder. Front. Cell Dev. Biol. 9, 696428. 10.3389/fcell.2021.696428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranger D. A. A., Hatoum A. S., Polimanti R., Gelernter J., Edenberg H. J., Bogdan R., et al. (2023). Multi‐omics cannot replace sample size in genome‐wide association studies. Genes Brain Behav. 22, e12846. 10.1111/gbb.12846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnevik-Olsson M., Gillberg C., Fernell E. (2008). Prevalence of autism in children born to Somali parents living in Sweden: a brief report. Dev. Med. Child. Neurol. 50, 598–601. 10.1111/j.1469-8749.2008.03036.x [DOI] [PubMed] [Google Scholar]
- Barone R., Fichera M., De Grandi M., Battaglia M., Lo Faro V., Mattina T., et al. (2017). Familial 18q12.2 deletion supports the role of RNA‐binding protein CELF4 in autism spectrum disorders. Am. J. Med. Genet. A 173, 1649–1655. 10.1002/ajmg.a.38205 [DOI] [PubMed] [Google Scholar]
- Bayou N., Belhadj A., Daoud H., Briault S., Helayem M. B., Chaabouni H., et al. (2010). Exploring the 7p22.1 chromosome as a candidate region for autism. J. Biomed. Biotechnol. 2010, 423894–4. 10.1155/2010/423894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayou N., M’rad R., Belhaj A., Daoud H., Ben Jemaa L., Zemni R., et al. (2008). De novo balanced translocation t (7;16) (p22.1; p11.2) associated with autistic disorder. J. Biomed. Biotechnol. 2008, 231904–231905. 10.1155/2008/231904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaid M., Loe-Mie Y., Lepagnol-Bestel A.-M., Han W., Santpere G., Klarić T., et al. (2019). Multi-hit autism genomic architecture evidenced from consanguineous families with involvement of FEZF2 and mutations in high-risk genes. Neuroscience. 10.1101/759480 [DOI] [Google Scholar]
- Bernier R., Golzio C., Xiong B., Stessman H. A., Coe B. P., Penn O., et al. (2014). Disruptive CHD8 mutations define a subtype of autism early in development. Cell 158, 263–276. 10.1016/j.cell.2014.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berryer M. H., Hamdan F. F., Klitten L. L., Møller R. S., Carmant L., Schwartzentruber J., et al. (2013). Mutations in SYNGAP1 cause intellectual disability, autism, and a specific form of epilepsy by inducing haploinsufficiency. Hum. Mutat. 34, 385–394. 10.1002/humu.22248 [DOI] [PubMed] [Google Scholar]
- Blier P., Ward N. M. (2003). Is there a role for 5-HT1A agonists in the treatment of depression? Biol. Psychiatry 53, 193–203. 10.1016/S0006-3223(02)01643-8 [DOI] [PubMed] [Google Scholar]
- Boccuto L., Chen C.-F., Pittman A. R., Skinner C. D., McCartney H. J., Jones K., et al. (2013). Decreased tryptophan metabolism in patients with autism spectrum disorders. Mol. Autism 4, 16. 10.1186/2040-2392-4-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond M., Maram H., Soliman A., Khattab R. (2012). Science and innovation in Egypt. London: Lond. R. Soc. [Google Scholar]
- Brune C. W., Kim S.-J., Salt J., Leventhal B. L., Lord C., Cook E. H. (2006). 5-HTTLPR genotype-specific phenotype in children and adolescents with autism. Am. J. Psychiatry 163, 2148–2156. 10.1176/ajp.2006.163.12.2148 [DOI] [PubMed] [Google Scholar]
- Bu X., Wu D., Lu X., Yang L., Xu X., Wang J., et al. (2017). Role of SIRT1/PGC-1α in mitochondrial oxidative stress in autistic spectrum disorder. Neuropsychiatr. Dis. Treat. 13, 1633–1645. 10.2147/NDT.S129081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buddell T., Friedman V., Drozd C. J., Quinn C. C. (2019). An autism-causing calcium channel variant functions with selective autophagy to alter axon targeting and behavior. PLOS Genet. 15, e1008488. 10.1371/journal.pgen.1008488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum J. D., Cai G., Chaste P., Nygren G., Goldsmith J., Reichert J., et al. (2007). Mutation screening of the PTEN gene in patients with autism spectrum disorders and macrocephaly. Am. J. Med. Genet. B Neuropsychiatr. Genet. 144B, 484–491. 10.1002/ajmg.b.30493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum J. D., Silverman J. M., Smith C. J., Greenberg D. A., Kilifarski M., Reichert J., et al. (2002). Association between a GABRB3 polymorphism and autism. Mol. Psychiatry 7, 311–316. 10.1038/sj.mp.4001011 [DOI] [PubMed] [Google Scholar]
- Campbell D. B., Buie T. M., Winter H., Bauman M., Sutcliffe J. S., Perrin J. M., et al. (2009). Distinct genetic risk based on association of MET in families with Co-occurring autism and gastrointestinal conditions. Pediatrics 123, 1018–1024. 10.1542/peds.2008-0819 [DOI] [PubMed] [Google Scholar]
- Campbell M. C., Tishkoff S. A. (2008). African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu. Rev. Genomics Hum. Genet. 9, 403–433. 10.1146/annurev.genom.9.081307.164258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrasco M., Salazar C., Tiznado W., Ruiz L. M. (2019). Alterations of mitochondrial biology in the oral mucosa of Chilean children with autism spectrum disorder (ASD). Cells 8, 367. 10.3390/cells8040367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caubit X., Gubellini P., Andrieux J., Roubertoux P. L., Metwaly M., Jacq B., et al. (2016). TSHZ3 deletion causes an autism syndrome and defects in cortical projection neurons. Nat. Genet. 48, 1359–1369. 10.1038/ng.3681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehbani F., Tomaiuolo P., Picinelli C., Baccarin M., Castronovo P., Scattoni M. L., et al. (2022). Yield of array-CGH analysis in Tunisian children with autism spectrum disorder. Mol. Genet. Genomic Med. 10, e1939. 10.1002/mgg3.1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.-H., Huang C.-C., Cheng M.-C., Chiu Y.-N., Tsai W.-C., Wu Y.-Y., et al. (2014). Genetic analysis of GABRB3 as a candidate gene of autism spectrum disorders. Mol. Autism 5, 36. 10.1186/2040-2392-5-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Bao Y., Xue Y., Sun Y., Hu D., Meng S., et al. (2017). Meta-analyses of RELN variants in neuropsychiatric disorders. Behav. Brain Res. 332, 110–119. 10.1016/j.bbr.2017.05.028 [DOI] [PubMed] [Google Scholar]
- Chien W.-H., Gau S.-F., Chen C.-H., Tsai W.-C., Wu Y.-Y., Chen P.-H., et al. (2013). Increased gene expression of FOXP1 in patients with autism spectrum disorders. Mol. Autism 4, 23. 10.1186/2040-2392-4-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien Y.-L., Wu Y.-Y., Chen C.-H., Gau S. S.-F., Huang Y.-S., Chien W.-H., et al. (2012). Association of HLA-DRB1 alleles and neuropsychological function in autism. Psychiatr. Genet. 22, 46–49. 10.1097/YPG.0b013e32834915ae [DOI] [PubMed] [Google Scholar]
- Cho I. H., Yoo H. J., Park M., Lee Y. S., Kim S. A. (2007). Family-based association study of 5-HTTLPR and the 5-HT2A receptor gene polymorphisms with autism spectrum disorder in Korean trios. Brain Res. 1139, 34–41. 10.1016/j.brainres.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Chugani D. C. (2002). Role of altered brain serotonin mechanisms in autism. Mol. Psychiatry 7 (Suppl. 2), S16–S17. 10.1038/sj.mp.4001167 [DOI] [PubMed] [Google Scholar]
- Coghlan S., Horder J., Inkster B., Mendez M. A., Murphy D. G., Nutt D. J. (2012). GABA system dysfunction in autism and related disorders: from synapse to symptoms. Neurosci. Biobehav. Rev. 36, 2044–2055. 10.1016/j.neubiorev.2012.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conciatori M., Stodgell C. J., Hyman S. L., O’Bara M., Militerni R., Bravaccio C., et al. (2004). Association between the HOXA1 A218G polymorphism and increased head circumference in patients with autism. Biol. Psychiatry 55, 413–419. 10.1016/j.biopsych.2003.10.005 [DOI] [PubMed] [Google Scholar]
- De Diego-Otero Y., Romero-Zerbo Y., Bekay R. E., Decara J., Sanchez L., Fonseca F. R., et al. (2009). α-Tocopherol protects against oxidative stress in the fragile X knockout mouse: an experimental therapeutic approach for the Fmr1 deficiency. Neuropsychopharmacology 34, 1011–1026. 10.1038/npp.2008.152 [DOI] [PubMed] [Google Scholar]
- De Krom M., Staal W. G., Ophoff R. A., Hendriks J., Buitelaar J., Franke B., et al. (2009). A common variant in DRD3 receptor is associated with autism spectrum disorder. Biol. Psychiatry 65, 625–630. 10.1016/j.biopsych.2008.09.035 [DOI] [PubMed] [Google Scholar]
- De La Torre-Ubieta L., Won H., Stein J. L., Geschwind D. H. (2016). Advancing the understanding of autism disease mechanisms through genetics. Nat. Med. 22, 345–361. 10.1038/nm.4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Zou X., Deng H., Li J., Tang C., Wang X., et al. (2015). The relationship among genetic heritability, environmental effects, and autism spectrum disorders: 37 pairs of ascertained twin study. J. Child. Neurol. 30, 1794–1799. 10.1177/0883073815580645 [DOI] [PubMed] [Google Scholar]
- Dias C., Pfundt R., Kleefstra T., Shuurs-Hoeijmakers J., Boon E. M. J., van Hagen J. M., et al. (2021). De novo variants in TCF7L2 are associated with a syndromic neurodevelopmental disorder. Am. J. Med. Genet. A 185, 2384–2390. 10.1002/ajmg.a.62254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Napoli A., Warrier V., Baron-Cohen S., Chakrabarti B. (2014). Genetic variation in the oxytocin receptor (OXTR) gene is associated with Asperger Syndrome. Mol. Autism 5, 48. 10.1186/2040-2392-5-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döcker D., Schubach M., Menzel M., Munz M., Spaich C., Biskup S., et al. (2014). Further delineation of the SATB2 phenotype. Eur. J. Hum. Genet. EJHG 22, 1034–1039. 10.1038/ejhg.2013.280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissa N., Al-Houqani M., Sadeq A., Ojha S. K., Sasse A., Sadek B. (2018). Current enlightenment about etiology and pharmacological treatment of autism spectrum disorder. Front. Neurosci. 12, 304. 10.3389/fnins.2018.00304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Ansary A., Zayed N., Al-Ayadhi L., Qasem H., Anwar M., Meguid N. A., et al. (2021). GABA synaptopathy promotes the elevation of caspases 3 and 9 as pro-apoptotic markers in Egyptian patients with autism spectrum disorder. Acta Neurol. belg. 121, 489–501. 10.1007/s13760-019-01226-z [DOI] [PubMed] [Google Scholar]
- El-Hossiny R. M., El-Baz F., Abdel-Aziz E., Abbass A., Abdel Mageed R., Abdel Raouf B. M. (2023). HLA-DR4 gene expression in a sample of Egyptian autistic children and their mothers: is it a risk factor? Egypt. J. Pediatr. Allergy Immunol. 21, 18–26. 10.21608/ejpa.2023.294364 [DOI] [Google Scholar]
- Esmaiel N. N., Ashaat E. A., Mosaad R., Fayez A., Ibrahim M., Abdallah Z. Y., et al. (2020). The potential impact of COMT gene variants on dopamine regulation and phenotypic traits of ASD patients. Behav. Brain Res. 378, 112272. 10.1016/j.bbr.2019.112272 [DOI] [PubMed] [Google Scholar]
- Feil D., Abrishamcar S., Christensen G. M., Vanker A., Koen N., Kilanowski A., et al. (2023). DNA methylation as a potential mediator of the association between indoor air pollution and neurodevelopmental delay in a South African birth cohort. Clin. Epigenetics 15, 31. 10.1186/s13148-023-01444-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach R. L., Harris M. J., Ballan M. S., Fischbach G. D., Link B. G. (2016). Is there concordance in attitudes and beliefs between parents and scientists about autism spectrum disorder? Autism 20, 353–363. 10.1177/1362361315585310 [DOI] [PubMed] [Google Scholar]
- Fombonne E., MacFarlane H., Salem A. C. (2021). Epidemiological surveys of ASD: advances and remaining challenges. J. Autism Dev. Disord. 51, 4271–4290. 10.1007/s10803-021-05005-9 [DOI] [PubMed] [Google Scholar]
- Forrest M. P., Penzes P. (2020). Autism genetics: over 100 risk genes and counting. Pediatr. Neurol. Briefs 34, 13. 10.15844/pedneurbriefs-34-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickel E., Mahony C., Bam S., Buchanan E., van der Watt M., O’Ryan C. (2023). Molecular autism research in Africa: emerging themes and prevailing disparities. Rev. J. Autism Dev. Disord. 10.1007/s40489-023-00415-0 [DOI] [Google Scholar]
- Fyke W., Velinov M. (2021). FMR1 and autism, an intriguing connection revisited. Genes 12, 1218. 10.3390/genes12081218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardener H., Spiegelman D., Buka S. L. (2011). Perinatal and neonatal risk factors for autism: a comprehensive meta-analysis. Pediatrics 128, 344–355. 10.1542/peds.2010-1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaugler T., Klei L., Sanders S. J., Bodea C. A., Goldberg A. P., Lee A. B., et al. (2014). Most genetic risk for autism resides with common variation. Nat. Genet. 46, 881–885. 10.1038/ng.3039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebril O. H., Meguid N. A. (2011). HFE gene polymorphisms and the risk for autism in Egyptian children and impact on the effect of oxidative stress. Dis. Markers 31, 289–294. 10.3233/DMA-2011-0830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project Consortium Abecasis G. R., Auton A., Brooks L. D., DePristo M. A., Durbin R. M., et al. (2012). An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasson E. J., Bower C., Petterson B., de Klerk N., Chaney G., Hallmayer J. F. (2004). Perinatal factors and the development of autism: a population study. Arch. Gen. Psychiatry 61, 618–627. 10.1001/archpsyc.61.6.618 [DOI] [PubMed] [Google Scholar]
- Gomez F., Hirbo J., Tishkoff S. A. (2014). Genetic variation and adaptation in Africa: implications for human evolution and disease. Cold Spring Harb. Perspect. Biol. 6, a008524. 10.1101/cshperspect.a008524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griswold A. J., Dueker N. D., Van Booven D., Rantus J. A., Jaworski J. M., Slifer S. H., et al. (2015). Targeted massively parallel sequencing of autism spectrum disorder-associated genes in a case control cohort reveals rare loss-of-function risk variants. Mol. Autism 6, 43. 10.1186/s13229-015-0034-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J., Ripke S., Als T. D., Mattheisen M., Walters R. K., Won H., et al. (2019). Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444. 10.1038/s41588-019-0344-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozeva D., Carss K., Spasic-Boskovic O., Parker M. J., Archer H., Firth H. V., et al. (2014). De novo loss-of-function mutations in SETD5, encoding a methyltransferase in a 3p25 microdeletion syndrome critical region, cause intellectual disability. Am. J. Hum. Genet. 94, 618–624. 10.1016/j.ajhg.2014.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Peng Y., Hu Z., Li Y., Xun G., Ou J., et al. (2017). Genome-wide copy number variation analysis in a Chinese autism spectrum disorder cohort. Prim. Care Companion CNS Disord. 7, 44155. 10.1038/srep44155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Verma V. K. (2019). “Next-generation sequencing and its application: empowering in public health beyond reality,” in Microbial technology for the welfare of society. Editor Arora P. K. (Singapore: Springer Singapore; ), 313–341. 10.1007/978-981-13-8844-6_15 [DOI] [Google Scholar]
- Gurdasani D., Carstensen T., Tekola-Ayele F., Pagani L., Tachmazidou I., Hatzikotoulas K., et al. (2015). The african genome variation project shapes medical genetics in Africa. Nature 517, 327–332. 10.1038/nature13997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan F. F., Daoud H., Rochefort D., Piton A., Gauthier J., Langlois M., et al. (2010). De novo mutations in FOXP1 in cases with intellectual disability, autism, and language impairment. Am. J. Hum. Genet. 87, 671–678. 10.1016/j.ajhg.2010.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W., Kwan K. Y., Shim S., Lam M. M. S., Shin Y., Xu X., et al. (2011). TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc. Natl. Acad. Sci. U. S. A. 108, 3041–3046. 10.1073/pnas.1016723108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy B.-J., Séguin B., Ramesar R., Singer P. A., Daar A. S. (2008). South Africa: from species cradle to genomic applications. Nat. Rev. Genet. 9, S19–S23. 10.1038/nrg2441 [DOI] [PubMed] [Google Scholar]
- Higazi A. M., Kamel H. M., Abdel-Naeem E. A., Abdullah N. M., Mahrous D. M., Osman A. M. (2021). Expression analysis of selected genes involved in tryptophan metabolic pathways in Egyptian children with Autism Spectrum Disorder and learning disabilities. Sci. Rep. 11, 6931. 10.1038/s41598-021-86162-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn J. N., Daly M. J. (2005). Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6, 95–108. 10.1038/nrg1521 [DOI] [PubMed] [Google Scholar]
- Hodges H., Fealko C., Soares N. (2020). Autism spectrum disorder: definition, epidemiology, causes, and clinical evaluation. Transl. Pediatr. 9, S55-S65–s65. 10.21037/tp.2019.09.09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hranilovic D., Blazevic S., Babic M., Smurinic M., Bujas-Petkovic Z., Jernej B. (2010). 5-HT2A receptor gene polymorphisms in Croatian subjects with autistic disorder. Psychiatry Res. 178, 556–558. 10.1016/j.psychres.2010.04.007 [DOI] [PubMed] [Google Scholar]
- Ibrahim S., El-Waleely T., Zakaria N., Ismail R. (2015). A study of serum interleukin-12 in a sample of autistic children in Egypt. Egypt. J. Psychiatry 36, 81–87. 10.4103/1110-1105.158115 [DOI] [Google Scholar]
- Ingram J. L., Stodgell C. J., Hyman S. L., Figlewicz D. A., Weitkamp L. R., Rodier P. M. (2000). Discovery of allelic variants of HOXA1 and HOXB1: genetic susceptibility to autism spectrum disorders. Teratology 62, 393–405. [DOI] [PubMed] [Google Scholar]
- iPSYCH-SSI-Broad Autism Group Robinson E. B., St Pourcain B., Anttila V., Kosmicki J. A., Bulik-Sullivan B., et al. (2016). Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat. Genet. 48, 552–555. 10.1038/ng.3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M., Weatheritt R. J., Ellis J. D., Parikshak N. N., Gonatopoulos-Pournatzis T., Babor M., et al. (2014). A highly conserved program of neuronal microexons is misregulated in autistic brains. Cell 159, 1511–1523. 10.1016/j.cell.2014.11.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson P. B., Boccuto L., Skinner C., Collins J. S., Neri G., Gurrieri F., et al. (2009). Further evidence that the rs1858830 C variant in the promoter region of the MET gene is associated with autistic disorder. Autism Res. 2, 232–236. 10.1002/aur.87 [DOI] [PubMed] [Google Scholar]
- Johannessen J., Nærland T., Hope S., Torske T., Høyland A., Strohmaier J., et al. (2017). Parents’ attitudes toward clinical genetic testing for autism spectrum disorder—data from a Norwegian sample. Int. J. Mol. Sci. 18, 1078. 10.3390/ijms18051078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson L., Anckarsäter H., Zettergren A., Westberg L., Walum H., Lundström S., et al. (2014). Association between ASMT and autistic-like traits in children from a Swedish nationwide cohort. Psychiatr. Genet. 24, 21–27. 10.1097/YPG.0000000000000010 [DOI] [PubMed] [Google Scholar]
- Jossin Y. (2020). Reelin functions, mechanisms of action and signaling pathways during brain development and maturation. Biomolecules 10, 964. 10.3390/biom10060964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainer D., Templeton A. R., Prates E. T., Jacboson D., Allan E. R. O., Climer S., et al. (2023). Structural variants identified using non-Mendelian inheritance patterns advance the mechanistic understanding of autism spectrum disorder. Hum. Genet. Genomics Adv. 4, 100150. 10.1016/j.xhgg.2022.100150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal M., Nady G., Abushady A., Khalil M. (2015). Association of dopamine D4 receptor gene variants with autism. Int. J. Res. Med. Sci., 2658–2663. 10.18203/2320-6012.ijrms20150809 [DOI] [Google Scholar]
- Kanavin O. J., Woldseth B., Jellum E., Tvedt B., Andresen B. S., Stromme P. (2007). 2-methylbutyryl-CoA dehydrogenase deficiency associated with autism and mental retardation: a case report. J. Med. Case Rep. 1, 98. 10.1186/1752-1947-1-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam R. A., Rezk N. A., Abdelrahman H. M., Hassan T. H., Mohammad D., Hashim H. M., et al. (2013). Catechol-O-methyltransferase Val158Met polymorphism and hyperactivity symptoms in Egyptian children with autism spectrum disorder. Res. Dev. Disabil. 34, 2092–2097. 10.1016/j.ridd.2013.04.002 [DOI] [PubMed] [Google Scholar]
- Katori S., Hamada S., Noguchi Y., Fukuda E., Yamamoto T., Yamamoto H., et al. (2009). Protocadherin-alpha family is required for serotonergic projections to appropriately innervate target brain areas. J. Neurosci. Off. J. Soc. Neurosci. 29, 9137–9147. 10.1523/JNEUROSCI.5478-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B., Prakash A., Sewal R. K., Medhi B., Modi M. (2012). Drug therapy in autism: a present and future perspective. Pharmacol. Rep. 64, 1291–1304. 10.1016/S1734-1140(12)70927-1 [DOI] [PubMed] [Google Scholar]
- Lambert N., Dauve C., Ranza E., Makrythanasis P., Santoni F., Sloan-Béna F., et al. (2018). Novel NEXMIF pathogenic variant in a boy with severe autistic features, intellectual disability, and epilepsy, and his mildly affected mother. J. Hum. Genet. 63, 847–850. 10.1038/s10038-018-0459-2 [DOI] [PubMed] [Google Scholar]
- Lamy M., Erickson C. A. (2018). Pharmacological management of behavioral disturbances in children and adolescents with autism spectrum disorders. Curr. Probl. Pediatr. Adolesc. Health Care 48, 250–264. 10.1016/j.cppeds.2018.08.015 [DOI] [PubMed] [Google Scholar]
- Laumonnier F., Bonnet-Brilhault F., Gomot M., Blanc R., David A., Moizard M.-P., et al. (2004). X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am. J. Hum. Genet. 74, 552–557. 10.1086/382137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond C. S., Heinrich J., Delorme R., Proepper C., Betancur C., Huguet G., et al. (2012). Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet. 8, e1002521. 10.1371/journal.pgen.1002521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lin X., Wang M., Hu Y., Xue K., Gu S., et al. (2020). Potential role of genomic imprinted genes and brain developmental related genes in autism. BMC Med. Genomics 13, 54. 10.1186/s12920-020-0693-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Hou Y., Zhang Z., Zhang B., Huang T., Sun A., et al. (2023). Structure, activity and function of the lysine methyltransferase SETD5. Front. Endocrinol. 14, 1089527. 10.3389/fendo.2023.1089527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Santpere G., Imamura Kawasawa Y., Evgrafov O. V., Gulden F. O., Pochareddy S., et al. (2018a). Integrative functional genomic analysis of human brain development and neuropsychiatric risks. Science 362, eaat7615. 10.1126/science.aat7615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.-J., Yu S., Luo H., Li X., Rao B., Wang Y., et al. (2018b). Two de novo variations identified by massively parallel sequencing in 13 Chinese families with children diagnosed with autism spectrum disorder. Clin. Chim. Acta 479, 144–147. 10.1016/j.cca.2018.01.025 [DOI] [PubMed] [Google Scholar]
- Li X., Hu Z., He Y., Xiong Z., Long Z., Peng Y., et al. (2010). Association analysis of CNTNAP2 polymorphisms with autism in the Chinese Han population. Psychiatr. Genet. 20, 113–117. 10.1097/YPG.0b013e32833a216f [DOI] [PubMed] [Google Scholar]
- Liang H., Ward W. F. (2006). PGC-1alpha: a key regulator of energy metabolism. Adv. Physiol. Educ. 30, 145–151. 10.1152/advan.00052.2006 [DOI] [PubMed] [Google Scholar]
- Liu Y., Hu Z., Xun G., Peng Y., Lu L., Xu X., et al. (2012). Mutation analysis of the NRXN1 gene in a Chinese autism cohort. J. Psychiatr. Res. 46, 630–634. 10.1016/j.jpsychires.2011.10.015 [DOI] [PubMed] [Google Scholar]
- Lohmann K., Klein C. (2014). Next generation sequencing and the future of genetic diagnosis. Neurotherapeutics 11, 699–707. 10.1007/s13311-014-0288-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z. A., Mu W., Osborne L. M., Cordner Z. A. (2018). Eighteen-year-old man with autism, obsessive compulsive disorder and a SHANK2 variant presents with severe anorexia that responds to high-dose fluoxetine. BMJ Case Rep. 2018, bcr2018225119. bcr-2018-225119. 10.1136/bcr-2018-225119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson C., Rai D., Goodman A., Lundberg M., Idring S., Svensson A., et al. (2012). Migration and autism spectrum disorder: population-based study. Br. J. Psychiatry 201, 109–115. 10.1192/bjp.bp.111.095125 [DOI] [PubMed] [Google Scholar]
- Mariggiò M. A., Palumbi R., Vinella A., Laterza R., Petruzzelli M. G., Peschechera A., et al. (2021). DRD1 and DRD2 receptor polymorphisms: genetic neuromodulation of the dopaminergic system as a risk factor for ASD, ADHD and ASD/ADHD overlap. Front. Neurosci. 15, 705890. 10.3389/fnins.2021.705890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meguid N. A., Eid O. M., Reda M., Elalfy D. Y., Hussein F. (2020). Copy number variations of SHANK3 and related sensory profiles in Egyptian children with autism spectrum disorder. Res. Autism Spectr. Disord. 75, 101558. 10.1016/j.rasd.2020.101558 [DOI] [Google Scholar]
- Meguid N. A., Gebril O. H., Khalil R. O. (2015). A study of blood serotonin and serotonin transporter promoter variant (5-HTTLPR) polymorphism in Egyptian autistic children. Adv. Biomed. Res. 4, 94. 10.4103/2277-9175.156658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson J. J., Shi Y., Gujral M., Zheng H., Malhotra D., Jin X., et al. (2012). Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 151, 1431–1442. 10.1016/j.cell.2012.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed F. E. B., Zaky E. A., El-Sayed A. B., Elhossieny R. M., Zahra S. S., Salah Eldin W., et al. (2015). Assessment of hair aluminum, lead, and mercury in a sample of autistic Egyptian children: environmental risk factors of heavy metals in autism. Behav. Neurol. 2015, 545674. 10.1155/2015/545674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco A. P., Bailey A. J. (2001). The search for susceptibility genes. Lancet 358, S3. 10.1016/S0140-6736(01)07016-7 [DOI] [PubMed] [Google Scholar]
- Moonesinghe R., Yang Q., Khoury M. J. (2008). Sample size requirements to detect the effect of a group of genetic variants in case-control studies. Emerg. Themes Epidemiol. 5, 24. 10.1186/1742-7622-5-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafa G. A., ElGebaly H. H., Shehab A. A. S., Mohamed M. A. E. H. (2022). Up-regulated serum levels of TAM receptor tyrosine kinases in a group of Egyptian autistic children. J. Neuroimmunol. 364, 577811. 10.1016/j.jneuroim.2022.577811 [DOI] [PubMed] [Google Scholar]
- Mostafa G. A., Shehab A. A. (2010). The link of C4B null allele to autism and to a family history of autoimmunity in Egyptian autistic children. J. Neuroimmunol. 223, 115–119. 10.1016/j.jneuroim.2010.03.025 [DOI] [PubMed] [Google Scholar]
- Mostafa G. A., Shehab A. A., Al-Ayadhi L. Y. (2013). The link between some alleles on human leukocyte antigen system and autism in children. J. Neuroimmunol. 255, 70–74. 10.1016/j.jneuroim.2012.10.002 [DOI] [PubMed] [Google Scholar]
- Muratore C. R., Hodgson N. W., Trivedi M. S., Abdolmaleky H. M., Persico A. M., Lintas C., et al. (2013). Age-dependent decrease and alternative splicing of methionine synthase mRNA in human cerebral cortex and an accelerated decrease in autism. PLoS ONE 8, e56927. 10.1371/journal.pone.0056927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na M., Oh G. (2014). Iron-regulated transporter SLC40A1 gene polymorphism and autism: a pilot study. [Google Scholar]
- Nagarajan R., Hogart A., Gwye Y., Martin M. R., LaSalle J. M. (2006). Reduced MeCP2 expression is frequent in autism frontal cortex and correlates with aberrant MECP2 promoter methylation. Epigenetics 1, e1–e11. 10.4161/epi.1.4.3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita A., Nagai M., Mizuno S., Ogishima S., Tamiya G., Ueki M., et al. (2020). Clustering by phenotype and genome-wide association study in autism. Transl. Psychiatry 10, 290. 10.1038/s41398-020-00951-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento P. P., Bossolani-Martins A. L., Rosan D. B. A., Mattos L. C., Brandão-Mattos C., Fett-Conte A. C. (2016). Single nucleotide polymorphisms in the CNTNAP2 gene in Brazilian patients with autistic spectrum disorder. Genet. Mol. Res. 15. 10.4238/gmr.15017422 [DOI] [PubMed] [Google Scholar]
- Neale B. M., Kou Y., Liu L., Ma’ayan A., Samocha K. E., Sabo A., et al. (2012). Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485, 242–245. 10.1038/nature11011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell D., Maciulis A., Cutler A., Warren L., McMahon W. M., Coon H., et al. (2005). Confirmation of the association of the C4B null allelle in autism. Hum. Immunol. 66, 140–145. 10.1016/j.humimm.2004.11.002 [DOI] [PubMed] [Google Scholar]
- Ögren S. O., Eriksson T. M., Elvander-Tottie E., D’Addario C., Ekström J. C., Svenningsson P., et al. (2008). The role of 5-HT1A receptors in learning and memory. Serot. Cogn. Mech. Appl. 195, 54–77. 10.1016/j.bbr.2008.02.023 [DOI] [PubMed] [Google Scholar]
- Oleari R., Lettieri A., Manzini S., Paganoni A., André V., Grazioli P., et al. (2023). Autism-linked NLGN3 is a key regulator of gonadotropin-releasing hormone deficiency. Dis. Model. Mech. 16, dmm049996. 10.1242/dmm.049996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omotoso O. E., Teibo J. O., Atiba F. A., Oladimeji T., Adebesin A. O., Babalghith A. O. (2022). Bridging the genomic data gap in Africa: implications for global disease burdens. Glob. Health 18, 103. 10.1186/s12992-022-00898-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak B. J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B. P., et al. (2012). Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature 485, 246–250. 10.1038/nature10989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshodi Y., Ojewunmi O., Oshodi T. A., Ijarogbe G. T., Ogun O. C., Aina O. F., et al. (2017). Oxidative stress markers and genetic polymorphisms of glutathione S-transferase T1, M1, and P1 in a subset of children with autism spectrum disorder in Lagos, Nigeria. Niger. J. Clin. P. R. 20, 1161–1167. 10.4103/njcp.njcp_282_16 [DOI] [PubMed] [Google Scholar]
- Pan Y., Chen J., Guo H., Ou J., Peng Y., Liu Q., et al. (2015). Association of genetic variants of GRIN2B with autism. Sci. Rep. 5, 8296. 10.1038/srep08296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panahi Y., Salasar Moghaddam F., Babaei K., Eftekhar M., Shervin Badv R., Eskandari M. R., et al. (2023). Sexual dimorphism in telomere length in childhood autism. J. Autism Dev. Disord. 53, 2050–2061. 10.1007/s10803-022-05486-2 [DOI] [PubMed] [Google Scholar]
- Parisi M. A., Doherty D., Eckert M. L., Shaw D. W. W., Ozyurek H., Aysun S., et al. (2006). AHI1 mutations cause both retinal dystrophy and renal cystic disease in Joubert syndrome. J. Med. Genet. 43, 334–339. 10.1136/jmg.2005.036608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilorge M., Fassier C., Le Corronc H., Potey A., Bai J., De Gois S., et al. (2016). Genetic and functional analyses demonstrate a role for abnormal glycinergic signaling in autism. Mol. Psychiatry 21, 936–945. 10.1038/mp.2015.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polleux F., Lauder J. M. (2004). Toward a developmental neurobiology of autism. Ment. Retard. Dev. Disabil. Res. Rev. 10, 303–317. 10.1002/mrdd.20044 [DOI] [PubMed] [Google Scholar]
- Puts N. A. J., Wodka E. L., Harris A. D., Crocetti D., Tommerdahl M., Mostofsky S. H., et al. (2017). Reduced GABA and altered somatosensory function in children with autism spectrum disorder. Autism Res. 10, 608–619. 10.1002/aur.1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radoeva P. D., Coman I. L., Salazar C. A., Gentile K. L., Higgins A. M., Middleton F. A., et al. (2014). Association between autism spectrum disorder in individuals with velocardiofacial (22q11.2 deletion) syndrome and PRODH and COMT genotypes. Psychiatr. Genet. 24, 269–272. 10.1097/YPG.0000000000000062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahbar M. H., Samms-Vaughan M., Kim S., Saroukhani S., Bressler J., Hessabi M., et al. (2022). Detoxification role of metabolic glutathione S-transferase (GST) genes in blood lead concentrations of Jamaican children with and without autism spectrum disorder. Genes 13, 975. 10.3390/genes13060975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiersen A. M., Todorov A. A. (2011). Association between DRD4 genotype and autistic symptoms in DSM-IV ADHD. J. Can. Acad. Child. Adolesc. Psychiatry J. Acad. Can. Psychiatr. Enfant Adolesc. 20, 15–21. [PMC free article] [PubMed] [Google Scholar]
- Reiss J., Cooper D. N. (1990). Application of the polymerase chain reaction to the diagnosis of human genetic disease. Hum. Genet. 85, 1–8. 10.1007/BF00276316 [DOI] [PubMed] [Google Scholar]
- Richardson-Jones J. W., Craige C. P., Guiard B. P., Stephen A., Metzger K. L., Kung H. F., et al. (2010). 5-HT1A autoreceptor levels determine vulnerability to stress and response to antidepressants. Neuron 65, 40–52. 10.1016/j.neuron.2009.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad K., Abdallah A.-E. M., Abdel-Rahman A. A., Al-Atram A. A., Abdel-Raheem Y. F., Gad E. F., et al. (2020). Polymorphism of interleukin-1β and interleukin-1 receptor antagonist genes in children with autism spectrum disorders. Prog. Neuropsychopharmacol. Biol. Psychiatry 103, 109999. 10.1016/j.pnpbp.2020.109999 [DOI] [PubMed] [Google Scholar]
- Saeliw T., Tangsuwansri C., Thongkorn S., Chonchaiya W., Suphapeetiporn K., Mutirangura A., et al. (2018). Integrated genome-wide Alu methylation and transcriptome profiling analyses reveal novel epigenetic regulatory networks associated with autism spectrum disorder. Mol. Autism 9, 27. 10.1186/s13229-018-0213-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Said S., Moubarz G., Awadalla H., Sharaf N., Hegazy N., Elsaied A., et al. (2021). Role of glutathione-S-transferase M1 (GSTM1) and T1 (GSTT1) genes on aluminum concentration and oxidative markers among autistic children. Egypt. J. Chem. 64, 0–7601. 10.21608/ejchem.2021.94656.4464 [DOI] [Google Scholar]
- Salari N., Rasoulpoor S., Rasoulpoor S., Shohaimi S., Jafarpour S., Abdoli N., et al. (2022). The global prevalence of autism spectrum disorder: a comprehensive systematic review and meta-analysis. Ital. J. Pediatr. 48, 112. 10.1186/s13052-022-01310-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem A. M., Ismail S., Zarouk W. A., Abdul Baky O., Sayed A. A., Abd El-Hamid S., et al. (2013). Genetic variants of neurotransmitter-related genes and miRNAs in Egyptian autistic patients. ScientificWorldJournal 2013, 670621. 10.1155/2013/670621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem S., Ashaat E. (2023). Association of relative telomere length and LINE-1 methylation with autism but not with severity. J. Autism Dev. Disord. 54, 2266–2273. 10.1007/s10803-023-05965-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S. J., He X., Willsey A. J., Ercan-Sencicek A. G., Samocha K. E., Cicek A. E., et al. (2015). Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87, 1215–1233. 10.1016/j.neuron.2015.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S. J., Murtha M. T., Gupta A. R., Murdoch J. D., Raubeson M. J., Willsey A. J., et al. (2012). De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature 485, 237–241. 10.1038/nature10945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos J. X., Rasga C., Marques A. R., Martiniano H., Asif M., Vilela J., et al. (2022). A role for gene-environment interactions in autism spectrum disorder is supported by variants in genes regulating the effects of exposure to xenobiotics. Front. Neurosci. 16, 862315. 10.3389/fnins.2022.862315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer G. (2016). Clinical genetic aspects of ASD spectrum disorders. Int. J. Mol. Sci. 17, 180. 10.3390/ijms17020180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekita Y., Wagatsuma H., Nakamura K., Ono R., Kagami M., Wakisaka N., et al. (2008). Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat. Genet. 40, 243–248. 10.1038/ng.2007.51 [DOI] [PubMed] [Google Scholar]
- Sen B., Surindro Singh A., Sinha S., Chatterjee A., Ahmed S., Ghosh S., et al. (2010). Family‐based studies indicate association of Engrailed 2 gene with autism in an Indian population. Genes Brain Behav. 9, 248–255. 10.1111/j.1601-183X.2009.00556.x [DOI] [PubMed] [Google Scholar]
- Sharma J. R., Arieff Z., Gameeldien H., Davids M., Kaur M., van der Merwe L. (2013). Association analysis of two single-nucleotide polymorphisms of the RELN gene with autism in the South African population. Genet. Test. Mol. Biomark. 17, 93–98. 10.1089/gtmb.2012.0212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shawky R. M., El-baz F., Kamal T. M., Elhossiny R. M., Ahmed M. A., El Nady G. H. (2014). Study of genotype–phenotype correlation of methylene tetrahydrofolate reductase (MTHFR) gene polymorphisms in a sample of Egyptian autistic children. Egypt. J. Med. Hum. Genet. 15, 335–341. 10.1016/j.ejmhg.2014.05.004 [DOI] [Google Scholar]
- Sheikh M., Hamza R., Rashad M., Elhawary N. (2007). The implication of HOXA1 and HOXB1 gene variants in the occurrence and severity of childhood autism in Egypt. Curr. Psychol. 14, 75–88. [Google Scholar]
- Shen L., Feng C., Zhang K., Chen Y., Gao Y., Ke J., et al. (2019). Proteomics study of peripheral blood mononuclear cells (PBMCs) in autistic children. Front. Cell. Neurosci. 13, 105. 10.3389/fncel.2019.00105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos S., Gaujoux R., Lindeque Z., Mahony C., Van Der Colff R., Van Der Westhuizen F., et al. (2020). DNA methylation associated with mitochondrial dysfunction in a South African autism spectrum disorder cohort. Autism Res. 13, 1079–1093. 10.1002/aur.2310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathopoulos S., Gaujoux R., O’Ryan C. (2018). Genome-wide DNA methylation patterns in Autism Spectrum Disorder and mitochondrial function. Genetics. 10.1101/310748 [DOI] [Google Scholar]
- Tangsuwansri C., Saeliw T., Thongkorn S., Chonchaiya W., Suphapeetiporn K., Mutirangura A., et al. (2018). Investigation of epigenetic regulatory networks associated with autism spectrum disorder (ASD) by integrated global LINE-1 methylation and gene expression profiling analyses. PLOS ONE 13, e0201071. 10.1371/journal.pone.0201071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Autism Genome Project Consortium Szatmari P., Paterson A. D., Zwaigenbaum L., Roberts W., Brian J., et al. (2007). Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 39, 319–328. 10.1038/ng1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q., Tong P., Chen G., Deng M., Cai T., Tian R., et al. (2023). GLRA2 gene mutations cause high myopia in humans and mice. J. Med. Genet. 60, 193–203. 10.1136/jmedgenet-2022-108425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff S. A., Verrelli B. C. (2003). Role of evolutionary history on haplotype block structure in the human genome: implications for disease mapping. Curr. Opin. Genet. Dev. 13, 569–575. 10.1016/j.gde.2003.10.010 [DOI] [PubMed] [Google Scholar]
- Tondera D., Grandemange S., Jourdain A., Karbowski M., Mattenberger Y., Herzig S., et al. (2009). SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 28, 1589–1600. 10.1038/emboj.2009.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordjman S., Somogyi E., Coulon N., Kermarrec S., Cohen D., Bronsard G., et al. (2014). Gene × Environment interactions in autism spectrum disorders: role of epigenetic mechanisms. Front. Psychiatry 5, 53. 10.3389/fpsyt.2014.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres A. R., Maciulis A., Stubbs E. G., Cutler A., Odell D. (2002). The transmission disequilibrium test suggests that HLA-DR4 and DR13 are linked to autism spectrum disorder. Hum. Immunol. 63, 311–316. 10.1016/S0198-8859(02)00374-9 [DOI] [PubMed] [Google Scholar]
- Torres A. R., Westover J. B., Rosenspire A. J. (2012). HLA immune function genes in autism. Autism Res. Treat. 2012, 959073. 10.1155/2012/959073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrico B., Fernàndez-Castillo N., Hervás A., Milà M., Salgado M., Rueda I., et al. (2015). Contribution of common and rare variants of the PTCHD1 gene to autism spectrum disorders and intellectual disability. Eur. J. Hum. Genet. 23, 1694–1701. 10.1038/ejhg.2015.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toya A., Fukada M., Aoki E., Matsuki T., Ueda M., Eda S., et al. (2023). The distribution of neuroligin4, an autism-related postsynaptic molecule, in the human brain. Mol. Brain 16, 20. 10.1186/s13041-023-00999-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay M. W., Jiang Y. H. (2019). DNA methylation and susceptibility to autism spectrum disorder. Annu. Rev. Med. 70, 151–166. 10.1146/annurev-med-120417-091431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricco A. C., Lillie E., Zarin W., O’Brien K. K., Colquhoun H., Levac D., et al. (2018). PRISMA extension for scoping reviews (PRISMA-ScR): checklist and explanation. Ann. Intern. Med. 169, 467–473. 10.7326/M18-0850 [DOI] [PubMed] [Google Scholar]
- Tuncay I. O., DeVries D., Gogate A., Kaur K., Kumar A., Xing C., et al. (2023). The genetics of autism spectrum disorder in an East African familial cohort. Cell Genomics 100322, 100322. 10.1016/j.xgen.2023.100322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen J. A., Rehnström K., Kilpinen H., Kuokkanen M., Kempas E., Ylisaukko‐oja T. (2008). Mitochondrial aspartate/glutamate carrier SLC25A12 gene is associated with autism. Autism Res. 1, 189–192. 10.1002/aur.25 [DOI] [PubMed] [Google Scholar]
- van Heijst B. F., Geurts H. M. (2015). Quality of life in autism across the lifespan: a meta-analysis. Autism 19, 158–167. 10.1177/1362361313517053 [DOI] [PubMed] [Google Scholar]
- Wang G., Ye S., Gao L., Han Y., Guo X., Dong X., et al. (2018). Two single-nucleotide polymorphisms of the RELN gene and symptom-based and developmental deficits among children and adolescents with autistic spectrum disorders in the Tianjin, China. Behav. Brain Res. 350, 1–5. 10.1016/j.bbr.2018.04.048 [DOI] [PubMed] [Google Scholar]
- Wang H., Yin F., Gao J., Fan X. (2019). Association between 5-HTTLPR polymorphism and the risk of autism: a meta-analysis based on case-control studies. Front. Psychiatry 10, 51. 10.3389/fpsyt.2019.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Zhang H., Ma D., Bucan M., Glessner J. T., Abrahams B. S., et al. (2009). Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature 459, 528–533. 10.1038/nature07999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Jia M., Yue W., Tang F., Qu M., Ruan Y., et al. (2008). Association of the ENGRAILED 2 (EN2 ) gene with autism in Chinese Han population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 147B, 434–438. 10.1002/ajmg.b.30623 [DOI] [PubMed] [Google Scholar]
- Wang L., Li J., Ruan Y., Lu T., Liu C., Jia M., et al. (2013). Sequencing ASMT identifies rare mutations in Chinese han patients with autism. PLoS ONE 8, e53727. 10.1371/journal.pone.0053727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrier V., Baron-Cohen S., Chakrabarti B. (2013). Genetic variation in GABRB3 is associated with Asperger syndrome and multiple endophenotypes relevant to autism. Mol. Autism 4, 48. 10.1186/2040-2392-4-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassink T. H., Piven J., Patil S. R. (2001). Chromosomal abnormalities in a clinic sample of individuals with autistic disorder. Psychiatr. Genet. 11, 57–63. 10.1097/00041444-200106000-00001 [DOI] [PubMed] [Google Scholar]
- Wei H., Zhu Y., Wang T., Zhang X., Zhang K., Zhang Z. (2021). Genetic risk factors for autism-spectrum disorders: a systematic review based on systematic reviews and meta-analysis. J. Neural Transm. Vienna Austria 1996 128, 717–734. 10.1007/s00702-021-02360-w [DOI] [PubMed] [Google Scholar]
- Wen Z., Cheng T.-L., Li G., Sun S.-B., Yu S.-Y., Zhang Y., et al. (2017). Identification of autism-related MECP2 mutations by whole-exome sequencing and functional validation. Mol. Autism 8, 43. 10.1186/s13229-017-0157-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniowiecka-Kowalnik B., Nowakowska B. A. (2019). Genetics and epigenetics of autism spectrum disorder—current evidence in the field. J. Appl. Genet. 60, 37–47. 10.1007/s13353-018-00480-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witters P., Debbold E., Crivelly K., Vande Kerckhove K., Corthouts K., Debbold B., et al. (2016). Autism in patients with propionic acidemia. Mol. Genet. Metab. 119, 317–321. 10.1016/j.ymgme.2016.10.009 [DOI] [PubMed] [Google Scholar]
- World Health Organization (2013). Meeting report: autism spectrum disorders and other developmental disorders: from raising awareness to building capacity. Geneva, Switzerland: World Health Organization. Available at: https://iris.who.int/handle/10665/103312 (Accessed February 25, 2024). [Google Scholar]
- Wu S., Jia M., Ruan Y., Liu J., Guo Y., Shuang M., et al. (2005). Positive association of the oxytocin receptor gene (OXTR) with autism in the Chinese han population. Biol. Psychiatry 58, 74–77. 10.1016/j.biopsych.2005.03.013 [DOI] [PubMed] [Google Scholar]
- Yahya S., Gebril O. L. A., Raouf E., Elhadidy E. (2019). A PRELIMINARY INVESTIGATION OF HTR1A GENE EXPRESSION LEVELS IN AUTISM SPECTRUM DISORDERS. Int. J. Pharm. Pharm. Sci., 1–3. 10.22159/ijpps.2019v11i8.34141 [DOI] [Google Scholar]
- Yang S., Rothman R. E. (2004). PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 4, 337–348. 10.1016/s1473-3099(04)01044-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. J., Cho I. H., Park M., Yang S. Y., Kim S. A. (2012). Family based association of GRIN2A and GRIN2B with Korean autism spectrum disorders. Neurosci. Lett. 512, 89–93. 10.1016/j.neulet.2012.01.061 [DOI] [PubMed] [Google Scholar]
- Yu J., He X., Yao D., Li Z., Li H., Zhao Z. (2011). A sex-specific association of common variants of neuroligin genes (NLGN3 and NLGN4X) with autism spectrum disorders in a Chinese Han cohort. Behav. Brain Funct. 7, 13. 10.1186/1744-9081-7-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeglam A., Alhmadi S. (2020). Novel homozygous variant of TBC1D8 gene in four Libyan siblings with autistic spectrum disorder and intellectual disability without epilepsy. 9. [Google Scholar]
- Zeidan J., Fombonne E., Scorah J., Ibrahim A., Durkin M. S., Saxena S., et al. (2022). Global prevalence of autism: a systematic review update. Autism Res. 15, 778–790. 10.1002/aur.2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., He H., Yuan B., Wu Z., Wang X., Du Y., et al. (2021). An intronic variant of CHD7 identified in autism patients interferes with neuronal differentiation and development. Neurosci. Bull. 37, 1091–1106. 10.1007/s12264-021-00685-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Li S., Yu L., Liu J. (2018). Polymorphisms in vitamin D receptor genes in association with childhood autism spectrum disorder. Dis. Markers 2018, 7862892. 10.1155/2018/7862892 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.