Abstract
Virus-cell surface receptor interactions are of major interest. Hepadnaviruses are a family of partially double-stranded DNA viruses with liver tropism and a narrow host range of susceptibility to infection. At least in the case of duck hepatitis B virus (DHBV), host specificity seems controlled partly at the receptor level. The middle portion in the pre-S region of the viral large envelope protein binds specifically to duck carboxypeptidase D (DCPD) but not to its human or chicken homologue. Although domain C of DCPD is implicated in ligand binding, the exact pre-S contact site remains to be determined. We prepared and tested a panel of chimeric constructs consisting of DCPD and human carboxypeptidase D (HCPD). Our results indicate that a short region at the N terminus of domain C (residues 920 to 949) is critical to DHBV binding and is a major determinant for the host specificity of DHBV infection. Replacing this region of the DCPD molecule with its human homologue abolished the DHBV interaction, whereas introducing this DCPD sequence into HCPD conferred efficient DHBV binding. Extensive analysis of site-directed mutants revealed that both conserved and nonconserved residues were important for the pre-S interaction. There were primary sequence variations and secondary structural differences that contributed to the inability of HCPD to bind the DHBV pre-S domain.
A clear definition of attachment and entry within the hepadnavirus infectious life cycle is of major interest. Hepatitis B virus (HBV) is the prototype member of this family of enveloped DNA viruses with hepatotropism and a narrow host range; however, there is no cell culture model system to allow for receptor identification. Duck hepatitis B virus (DHBV), a related avian hepadnavirus, is a suitable model in which to characterize the early events of hepadnavirus infection due to the availability of hepatocytes for infection studies. Duck carboxypeptidase D (DCPD) has been independently identified as a viral binding partner in experiments using DHBV particles and pre-S tagged glutathione S-transferase (GST) fusion proteins (17, 24). Recent studies provide compelling evidence that DCPD is an avian HBV receptor. For example, DCPD-reconstituted kidney and liver cell lines support DHBV attachment and entry (23), and polyclonal antibodies against DCPD block DHBV infection of primary duck hepatocytes (25). Furthermore, DHBV large envelope protein expression downregulates DCPD levels in infected ducks (2).
The mouse, rat, and human homologues of DCPD have been cloned (16, 22, 26). Carboxypeptidase D (CPD) is a type I transmembrane glycoprotein containing three 50-kDa repeats (domains A, B, and C), and each is similar to the carboxypeptidase N/E subfamily of metallocarboxypeptidases (22, 26). The bulk of the protein is extracellular, comprised of domains A and B and most of domain C, followed by a hydrophobic transmembrane anchor and a short (58-residue) cytosolic C-terminal tail (18). Enzymatic activity has been ascribed to domain A and B (5); the physiological role of domain C is unknown. The crystal structure of domain B has been established and used to model the structures of domains A and C, and it has a highly conserved overall topology (1, 12).
An initial mapping study has defined the DCPD binding region within a 66-amino-acid (aa) segment spanning pre-S residues 43 to 108 (15). The contact site was further narrowed to residues 87 to 102 using a large panel of N-terminal, C-terminal, and double-deletion constructs (24). A short pre-S peptide, composed of residues 80 to 104, could bind DCPD, though at reduced efficiency compared with the full-length molecule (24). Breiner et al., using internal deletion mutants, established that residues 85 to 115 formed the major binding domain, while a sequence N-terminal to this domain contributed to full binding affinity (3). Combining the results of surface plasmon resonance spectroscopy with two-dimensional nuclear magnetic resonance analysis, Urban et al. have proposed a two-step receptor-ligand interaction (25). In this model, low-affinity primary complexes form between DCPD and the essential binding sequence comprising residues 85 to 115; these complexes are stabilized via a 60-aa sequence toward the N terminus of the pre-S domain.
Despite extensive ligand scrutiny, the receptor sequences responsible for pre-S interaction remain indistinct. Characterization of the structural basis of DCPD–pre-S interaction may permit the design of antiviral agents to block hepadnavirus infection at the receptor level and may further clarify the species specificity of DHBV infection. Chicken and human CPD molecules do not possess affinity for DHBV pre-S protein (18). Thus, receptor-ligand interaction must be a major determinant of the host specificity of DHBV infection. Our study was designed to identify host-specific sequences that are critical for DHBV binding, and we focused on DCPD domain C, since this region had previously been shown to be responsible for interaction with the DHBV pre-S protein (5).
A 30-aa sequence at the N terminus of DCPD domain C (positions 920 to 949) was found to be a major determinant for a host-specific receptor-ligand interaction, since inserting this 30-aa sequence into human CPD (HCPD) restored DHBV binding capacity. Conversely, replacing a 49-aa region containing this sequence with the human homologue abolished the DCPD-DHBV interaction. Site-directed mutagenesis of this region revealed that replacement of as few as 4 aa with the human sequence, or a 3-aa substitution of conserved residues, abolished DCPD binding to DHBV. Some of the mutants that failed to mediate DHBV binding also displayed low steady-state levels of DCPD protein, suggesting that reduced protein stability was partly responsible. With respect to HCPD, we found that a P-to-LAL conversion promoted a local secondary structural change and reduction of DHBV binding capacity. Other substitutions found in HCPD can also abolish viral binding when introduced into a DCPD background, although these changes do not affect protein secondary structure.
MATERIALS AND METHODS
CPD DNA clones.
The DCPD deletion mutant (delA/B) was similar to the pG180DS33 construct described by Eng et al. (5). To generate this construct, DCPD cDNA cloned into the pcDNA3.1zeo(−) vector (23) was digested with ClaI and KpnI. The 6.5-kb fragment was gel purified and ligated with a 0.2-kb fragment generated by ClaI/KpnI digestion and gel purification of the PCR product between sense primer 5′-ATCAGGTACCGCAAAGGTGGGGTACAGG-3′ (KpnI site underlined) and antisense primer 5′-ACGTAACACTGAAATCCTTC-3′. This construct expresses the N-terminal 48 residues of domain A (containing the signal peptide to allow proper targeting), 37 C-terminal residues of domain B (residues 868 to 904), and the entire 483 residues of domain C (residues 905 to 1387) (Fig. 1A, top). For easy exchange of domain C fragments between DCPD and HCPD, two unique restriction sites were introduced into DCPD domain C by an overlap extension PCR (14). The HindIII site was generated by a G-to-T substitution at nucleotide position 3075, while the BamHI site was created through C3486A and T3489C changes. None of the substitutions altered the encoded amino acid sequence. The HCPD cDNA sequence has been reported previously (22); a full-length clone was inserted into the EcoRI-HindIII sites of the pcDNA3.1zeo(−) vector in two steps. To construct its deletion (delA/B) mutant, HCPD-pcDNA3.1/zeo(−) was double digested with SacII and ClaI. The 7-kb fragment was gel purified and ligated to a 0.2-kb SacII/ClaI fragment of the PCR product of primers 5′-ATCACCGCGGGCGAAGGCGCTATTCAGGTCAACTTCAC-3′ (sense; SacII site underlined) and 5′-GTGGATAGTTCATTACAAGTCCTCTC-3′ (antisense). The resultant protein would lack residues 74 to 857 comprising most of domains A and B (see Fig. 4, construct C). In the course of the sequencing of the del A/B construct of HCPD, we noticed errors in our previous publication (22) as a result of high GC content. In fact, the sequence from the initiation codon to the SacII site is identical to that reported subsequently by Ishikawa et al. (16). Chimeric constructs between HCPD and DCPD domain C were generated either by restriction enzyme digestion or via PCR. Single or multiple point mutations were created by overlap extension PCR and mutants were inserted into the desired backbone using the appropriate restriction enzymes. All mutant constructs were verified by sequence analysis. The primers and detailed procedures used to make these constructs can be obtained from the authors upon request.
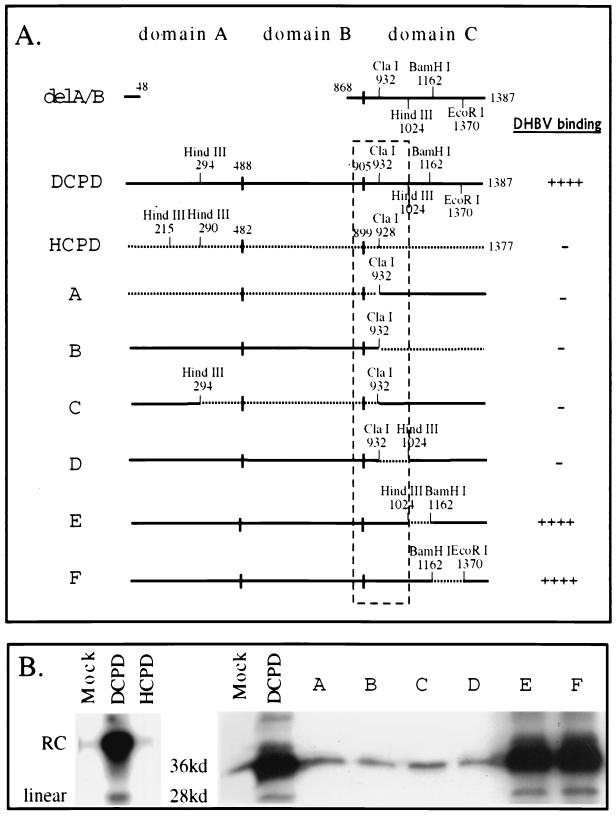
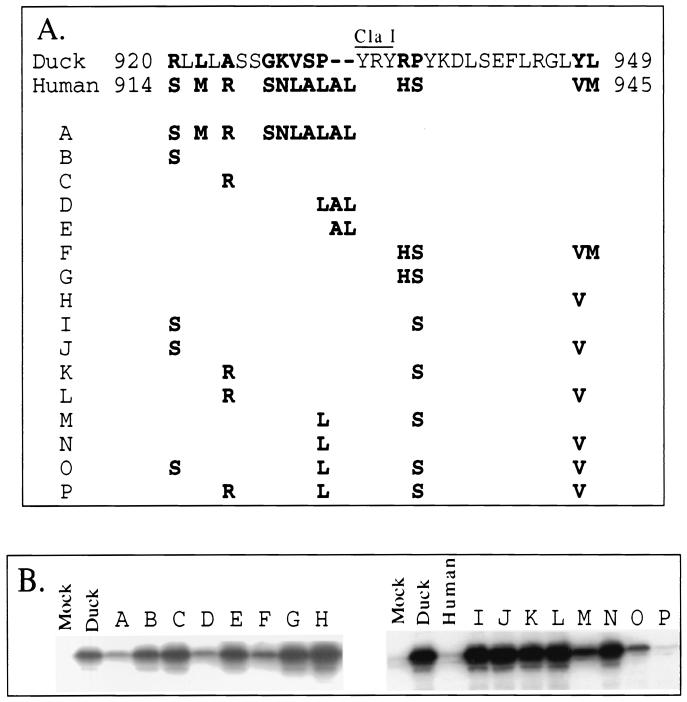
FIG. 1.
Mapping of a host determinant for DHBV binding using full-length CPD constructs. (A) Diagrams of CPD constructs and summary of DHBV binding capacity. Solid lines represent DCPD sequence; dotted lines represent HCPD sequence. Junctions between domains A, B, and C are indicated by vertical bars, and residues at the junctions are shown for the full-length DCPD and HCPD molecules. The number below each restriction site indicates the position of the first amino acid residue encoded by the restriction site. The HindIII and BamHI sites within domain C are absent in the original DCPD cDNA and chimeric constructs A, B, and C. Constructs D, E, and F were obtained by exchanging the 1.3-kb ClaI-EcoRI fragment of full-length DCPD with the corresponding fragment of constructs H, I, and J in Fig. 3, respectively. For the chimeric constructs, residues occupying the junctions are based on the numbering of DCPD rather than HCPD. The boxed area is a region important for DCPD binding of DHBV, where replacement either upstream or downstream of the ClaI site with the human homologue destroys DHBV binding. (B) (Left panel) Lack of affinity between DHBV and HCPD. Bosc cells were transiently transfected with vector DNA (Mock), DCPD cDNA, or HCPD cDNA. After the binding experiment, cell-associated DHBV DNA was revealed by Southern blot analysis. Positions of relaxed circular (RC) and linear DHBV DNA are indicated on the left. (Right panel) Binding of DHBV particles to Bosc cells transfected with DCPD-HCPD chimeric constructs. Bound DHBV was revealed by Western blot analysis of viral large envelope protein. Positions of full-length 36-kDa large envelope protein and the 28-kDa processed form are indicated.
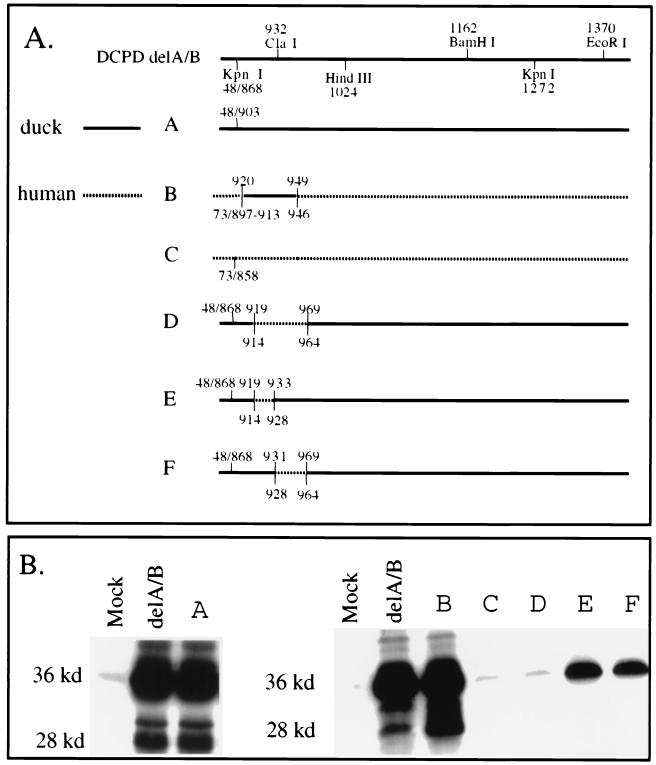
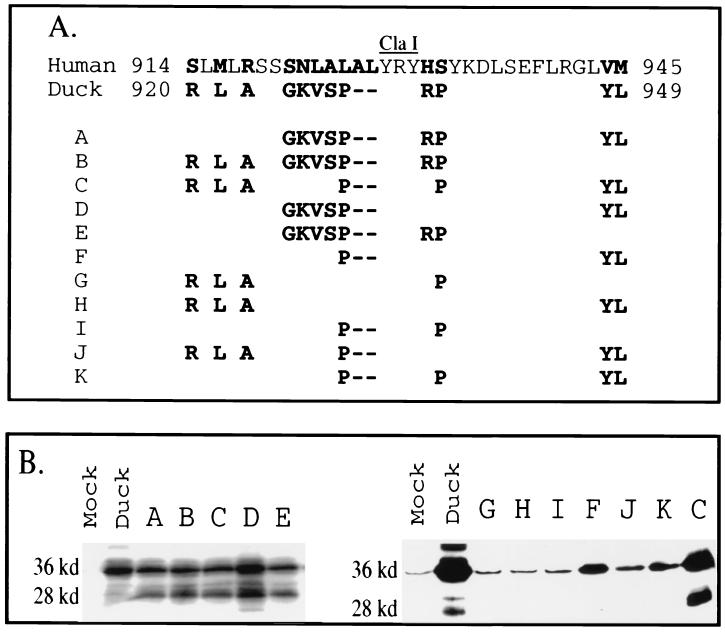
FIG. 4.
Localization of the host determinant to a 30-aa sequence in the N terminus of DCPD domain C. (A) Diagram of chimeric domain C constructs. Solid lines, DCPD; dotted lines, HCPD. Slashes, junctions as a result of deletion. Duck sequences are numbered above the line, while human sequences are numbered below the line. Construct A differs from delA/B of DCPD by removal of an additional 35-aa sequence in domain B (868 to 902). Construct C is the delA/B construct of HCPD. Constructs D, E, and F were based on the delA/B construct of DCPD and had residues 920 to 968, 920 to 932, and 932 to 968 replaced with corresponding human sequences, respectively. Construct B is a deletion mutant of HCPD without domain B for which residues 914 to 945 were replaced with the duck homologue (residues 920 to 949). (B) Binding of DHBV as established by Western blot analysis of viral large envelope protein. Positions of full-length 36-kDa large envelope protein and the 28-kDa truncated form are indicated. Mock, vector DNA-transfected Bosc cells.
Expression of CPD constructs.
We monitored expression of various constructs using an in vitro translation assay. In brief, plasmid DNA (0.5 μg) was transcribed with T7 polymerase and translated in rabbit reticulocyte lysate using TNT Quick coupled Transcription/Translation systems (Promega). Incorporation of [35S]methionine allowed detection of the translation product by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and fluorography. To monitor CPD protein expression in cells, Bosc cells grown in 6-well plates were first transfected with 6 μg of DNA by the calcium-phosphate precipitation method and then lysed 2 days later in 100 μl of lysis buffer containing 10 mM HEPES (pH 7.5), 100 mM NaCl, 1 mM EDTA, and 1% NP-40. Duplicate samples (10 μl each) were separated by SDS–10% PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. After being blocked at room temperature with 3% bovine serum albumin in phosphate-buffered saline (PBS) containing Tween 20 (0.05%; PBST) for 2 h, the blots were incubated at 4°C overnight with a 1:1,000 dilution of rabbit polyclonal antibodies against liver-derived DCPD (23) or the C-terminal 179 aa of HCPD expressed in Escherichia coli (19). After extensive washing, the blots were incubated at room temperature with a 1: 800 dilution of 125I-labeled protein A (low specific activity; New England Nuclear) for 4 h, followed by a wash. Bound protein A was revealed by autoradiography.
Binding of DHBV to CPD-transfected Bosc cells.
The binding assay was performed as described previously (23), and each construct set was tested several times to ensure reproducibility. Bosc cells grown in 60-mm-diameter dishes were transfected with 8 μg of various constructs. Two days later, cells were incubated with 40 μl of prespun viremic duck serum diluted 1:30 in culture medium for 12 h or longer (for full-length constructs, viremic duck serum was diluted 1:10). After a thorough washing step, cells were transferred to 15-ml Falcon tubes in 10 ml of medium. Cells were pelleted down and stored at −80°C before lysis or were lysed immediately with 100 μl of lysis buffer as described above.
Southern blot analysis of DHBV DNA.
Cell lysates were diluted with TEN buffer (10 mM Tris, 1 mM EDTA, 150 mM NaCl), and treated with proteinase K (0.5 mg/ml) in the presence of SDS (0.5%) at 37°C for several hours. The DNA was extracted with phenol-chloroform, precipitated with ethanol, and dissolved in Tris-EDTA (pH 8.0). Following electrophoresis in a 1% agarose gel, and staining with ethidium bromide, DNA was transferred to nylon membranes and hybridized with a randomly primed probe of highly purified PCR-derived DHBV DNA. After thorough washing, hybridization signals were detected by exposing the membranes to Kodak films.
Western blot analysis of large envelope protein.
Cell lysates were electrophoresed by SDS–12% PAGE and transferred to PVDF membranes. Blots were blocked at room temperature with 3% bovine serum albumin in PBST for 2 h and then incubated overnight with a 1:4,000 dilution of rabbit pre-S antibody (23) at 4°C in PBST. After thorough washing, blots were incubated in a 1:20,000 dilution of donkey anti-rabbit antibodies conjugated to horseradish peroxidase (Amersham) for 1 h, followed by a wash. The enhanced chemiluminescence (EC; Pierce) detection system was employed according to the manufacturer's instructions.
RESULTS
Involvement of DCPD residues 868 to 1024 in host-specific interaction with DHBV.
While DCPD has binding affinity for DHBV via domain C (5), HCPD does not interact with the virus (Fig. 1B, left panel). We exchanged various parts of domain C between the two proteins to ascertain why HCPD failed to associate with DHBV. Within domain C there is 82.5% sequence identity between these two proteins (22) (Fig. 2A). A schematic representation of the duck delA/B construct used is shown in Fig. 1A and 3A (top). A large portion of coding sequence for domain A and domain B (nucleotide positions 146 to 2599, corresponding to protein residues 49 to 867) was deleted. The presence of two KpnI sites and of unique ClaI, HindIII, BamHI, and EcoRI sites facilitated the construction of 10 chimeric molecules with HCPD (Fig. 3A, lanes A to J). In vitro translation experiments confirmed production of proteins of the expected size for all constructs (Fig. 3B). These constructs were transiently transfected into Bosc cells (a derivative of human embryonic kidney cell line 293), which are known for high transfection efficiency (23). Transfected cells were incubated at 37°C with viremic duck serum and washed extensively before lysis. Bound DHBV particles were determined by Southern and Western blot analyses.
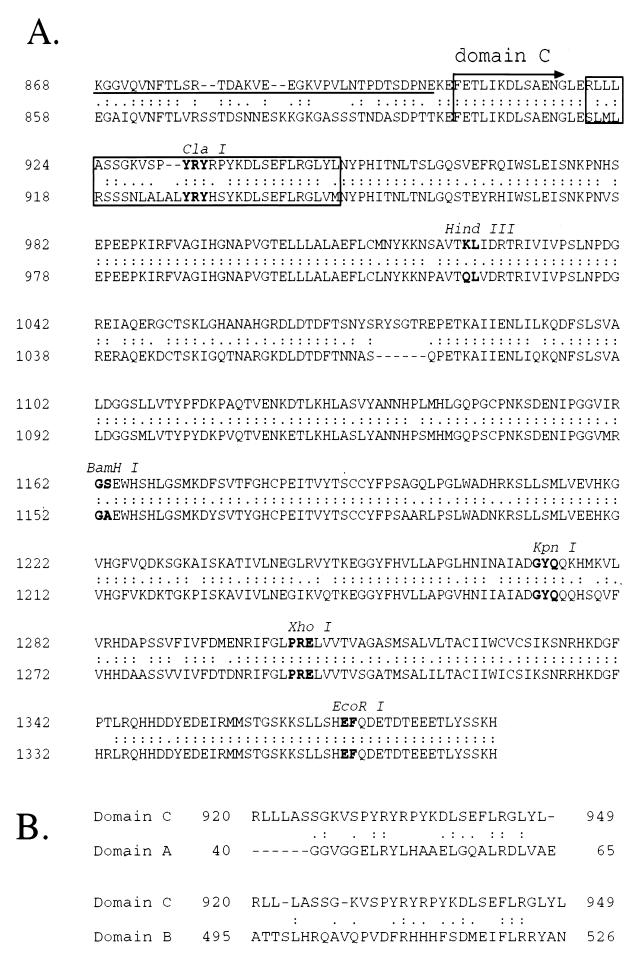
FIG. 2.
(A) Comparison of amino acid sequences of DCPD (top) and HCPD (bottom) in the C-terminal end of domain B and the entire domain C. The start of domain C is indicated. Double dots between the two sequences indicate conserved residues, while single dots denote conservative changes. Dashes within a sequence represent deleted residues. Restriction sites at the cDNA level are shown, and residues encoded are boldfaced. The underlined DCPD sequence (top row) indicates the C-terminal 35 aa of domain B that is dispensable for DHBV binding (see Fig. 4, construct A). The 30 aa residues (32 aa in HCPD) that determine a species barrier against DHBV infection of humans are boxed. (B) Comparison of the 30-aa host determinant in DCPD domain C with its counterparts in domain A and domain B.
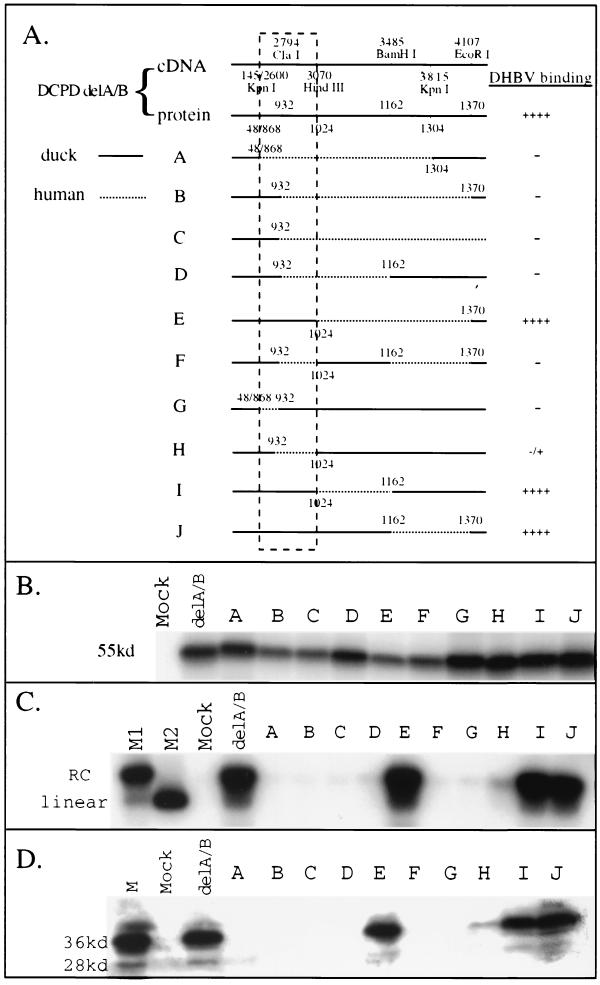
FIG. 3.
Mapping of the host determinant of DHBV binding by restriction fragment exchange of DCPD delA/B construct. (A) Schematic illustration of the constructs and summary of DHBV binding results. For DCPD delA/B, positions of the restriction sites at both the nucleotide (cDNA) and amino acid (protein) levels are indicated. For the chimeric constructs A to J, amino acid positions at the junction between DCPD and HCPD are indicated (numbering based on DCPD). Solid lines indicate duck sequence, while dashed lines denote the human homologue. Slashes between numbers indicate junctions as a result of deletion. Boxed area (KpnI to HindIII) is the host determinant for DHBV binding. (B) In vitro translation of the chimeric constructs. The expected size of the translation product is about 55 kDa. Mock, vector DNA. (C and D) Binding of DHBV to CPD-transfected Bosc cells as determined by Southern blot analysis of viral DNA (C) and Western blot analysis of viral large envelope protein (D). For panel C, lanes M1 and M2 denote virion-derived DHBV DNA and linearized 3-kb DHBV DNA, respectively. Mock, vector DNA-transfected cells. Positions of relaxed circular (RC) virion DNA and linear DNA are indicated. In panel D, lane M contains 0.5 μl of viremic duck serum. Positions of full-length 36-kDa large envelope protein and the 28-kDa truncated form are indicated.
The results of these two assays were highly concordant (Fig. 3C and D). Replacement of large portions of DCPD domain C with its human homologue often led to loss of viral binding capacity (Fig. 3, constructs A, B, and C). However, substitution of a large fragment between HindIII and EcoRI sites did not affect binding capacity (Fig. 3, construct E). In contrast, exchange of as few as 65 residues between the first KpnI site and ClaI site (positions 868 to 932) abolished viral binding (Fig. 3, construct G). Similarly, substitution of the neighboring ClaI-HindIII fragment (encoding 93 residues at positions 932 to 1024) also nearly eliminated DHBV binding (Fig. 3, construct H). These results implicated a 158-aa sequence bracketed by the first KpnI site and the HindIII site (residues 868 to 1024) (Fig. 3A, boxed areas) as a host determinant of the viral receptor-ligand interaction. Constructs in which part or all of this sequence was replaced with HCPD (constructs A, B, C, D, F, G, and H) were inactive in mediating DHBV binding (Fig. 3A).
The above experiments were performed using the artificial mutants lacking most of the A and B domains and thus may have limitations. Therefore, a panel of six chimeras of full-length protein was tested to confirm our observations (Fig. 1). Again, substitution around the ClaI site (Fig. 1A, boxed areas) invariably abolished viral binding (Fig. 1, constructs A, B, C, and D), while replacement of the HindIII-BamHI fragment and BamHI-EcoRI fragment in domain C was tolerated (Fig. 1, constructs E and F).
Transfer of a 30- to 51-aa sequence between DCPD and HCPD alters DHBV binding capacity.
Domain C of DCPD starts at residue 907 of the cDNA characterized by Kuroki and colleagues (18), corresponding to residue 905 in our cDNA clone. The delA/B construct still contained 37 C-terminal residues of domain B. To verify whether the remaining domain B sequence is dispensable for pre-S binding, we removed 35 residues (868 to 902) from the delA/B construct (Fig. 2A, top row). The resulting construct, with the new junction at position 903 (Fig. 4A, construct A), retained full viral binding capability (Fig. 4B, left panel). Comparing the duck sequence 903 to 1024 (HindIII site of the DCPD cDNA) with its human homologue revealed clustering of variant residues within residues 920 to 949 (Fig. 2A, boxed region). Interestingly, the ClaI site, either side of which is important for pre-S interaction (Fig. 1 and 3), sits in the middle of the coding sequence for this domain (Fig. 2A). We therefore introduced duck residues 920 to 949 into the human equivalent of construct A (Fig. 4A, construct B) to test the importance of this region in determining host-specific pre-S binding affinity. Whereas no viral binding occurred in cells transfected with the human delA/B construct (Fig. 4, construct C), construct B acquired binding capacity similar to that of the delA/B construct of DCPD (Fig. 4B). Conversely, replacement of a 49-aa DCPD sequence containing the entire 30-aa stretch and 19 additional C-terminal residues (920 to 968) with the corresponding 51-aa sequence from HCPD abolished DHBV binding capacity (Fig. 4, construct D). These results demonstrate that sequence variations within this 30-aa domain account for the failure of HCPD to interact with DHBV.
Experiments with the deletion construct A and exchange construct B (Fig. 4) appeared to exclude the importance of upstream (868 to 919) and downstream (950 to 1024) sequences in determining the specificity of the receptor–ligand interaction. However, experiments with two additional constructs suggest that these sequences derived from DCPD could maintain low binding capacity when part of the critical sequence 920 to 949 (either side of the ClaI site) was replaced with its human counterpart. For example, construct E had residues 920 to 932 replaced with human sequence and maintained a low viral binding capacity, whereas construct G in Fig. 3 (simplified as construct 3G; with residues 868 to 932 replaced) did not, thus illustrating the contribution of the C-terminal part of domain B to pre-S binding (Fig. 4). Sequence alignment of DCPD and HCPD revealed numerous substitutions between residues 868 and 902 (Fig. 2A). Similarly, construct 4F maintained low binding capacity when residues 932 to 968 were replaced with HCPD, while construct 3H manifested negligible binding with residues 932 to 1024 substituted.
Limited amino acid substitutions within residues 920 to 949 of DCPD abolish DHBV binding.
Comparison of residues 920 to 949 of DCPD with its human homologue reveals seven conservative and five nonconservative amino acid changes, and insertion of an AL dipeptide (Fig. 5A). Of these, eight amino acid substitutions and the dipeptide insertion were upstream, and the remainder were downstream, of the ClaI site. Thus, the corresponding human sequence (924 to 955) is 32 aa in length. We introduced single or multiple variant human residues into the delA/B construct of DCPD to dissect the relative contribution of these individual sequence changes with respect to pre-S binding. None of the single substitutions at position 920, 924, 931, or 948 significantly reduced DHBV binding (Fig. 5, constructs B, C, and H; also data not shown). Similarly, double amino acid substitution failed to modify pre-S binding capacity (Fig. 5, constructs G, I, J, K, L, and N) with the exception of construct M (P931L P936S), for which we observed decreased binding activity. It is noteworthy that these two changes, when combined with two additional changes, substantially reduced or abolished binding capacity (Fig. 5, constructs O and P). Similarly, two nonconservative changes coupled with two conservative changes greatly reduced DHBV binding (Fig. 5, construct F). These results argue against one particular residue as being the sole determinant for pre-S binding; rather, they suggest the importance of many residues (conservative and nonconservative) in viral receptor–pre-S protein interaction. Conversion of 931P of DCPD into the LAL sequence also reduced DHBV binding activity (Fig. 5, construct D). Both the P-to-L substitution and AL insertion are required, since AL insertion or P931L substitution alone failed to modify DHBV binding capacity (Fig. 5, constructs E and N).
FIG. 5.
Effects of DCPD-to-HCPD changes within residues 920 to 949 on DHBV binding capacity. (A) Site-directed mutant constructs. The complete duck sequence between positions 920 and 949 is shown on the top (roman type, conserved residues; boldface, nonconserved residues; hyphen, deletion), and variant human sequences are given below. The position of the ClaI site at the cDNA level is indicated. The parental construct was HCPD delA/B, in which the SacII-KpnI fragment had been replaced with a duck sequence encoding residues 868 to 1304. Mutations were introduced into this construct by overlap extension PCR and cloning. Construct A combined all the mutations upstream of the ClaI site, while construct F had all the mutations downstream. Other constructs had single, double, or multiple mutations on both sides of the ClaI site, or contained a dipeptide insertion. (B) Binding of DHBV as determined by Western blot analysis. Mock, vector DNA-transfected cells. Duck, cells transfected with the delA/B construct of DCPD. Human, cells reconstituted with the delA/B construct of HCPD.
Sequence change in the HCPD 914-to-945 aa sequence is required to confer DHBV binding.
In the reverse experiment, we determined how many human-to-duck sequence changes were required for acquisition of DHBV binding in the delA/B construct of HCPD. Many nonconservative and conservative changes were introduced into residues 911 to 947 of the HCPD delA/B construct. As expected, multiple substitutions introduced into either side of the ClaI site failed to generate viral binding capacity (data not shown). Minimal DHBV binding affinity was established by LAL-to-P conversion followed by two additional substitutions (Fig. 6, construct F). The LAL-to-P change was necessary because, when it was absent, substitution of four or five residues with the duck sequence failed to confer binding capability (Fig. 6, constructs G and H). This finding is consistent with results demonstrated by construct 5D, where the reverse change (from P to LAL) in DCPD was sufficient to reduce DHBV binding. However, the LAL-to-P change by itself was insufficient to induce binding activity (Fig. 5, construct I).
FIG. 6.
Substitution of residues within the host determinant of HCPD with duck sequence confers DHBV binding. (A) Site-directed mutant constructs. The human sequence from position 914 to 945 is given in full (roman type, conserved residues; boldface, nonconserved residues). For DCPD only the variant residues are shown. Hyphens indicate deleted residues. The ClaI site at the cDNA level is indicated. Mutations as listed were introduced into the delA/B construct of HCPD. (B) Binding of DHBV to transiently transfected Bosc cells as analyzed by Western blotting. Positions of the full-length 36-kDa large envelope protein and the 28-kDa truncated form are indicated. Mock, vector DNA-transfected cells; Duck, cells transfected with the delA/B construct of DCPD.
Coupling the LAL-to-P conversion with six or more amino acid changes generated efficient DHBV binding in the HCPD molecule (Fig. 6, constructs A to E). The exact residues substituted did not appear critical, since constructs C and E have different replacement combinations. Thus, more-extensive sequence changes were needed to confer efficient binding of DHBV to HCPD (Fig. 6) than to inactivate DHBV binding in the DCPD background (Fig. 5). This is consistent with the concept that the native HCPD contains multiple amino acid changes, a fraction of which are sufficient to prevent interaction with DHBV.
Role of conserved residues within 920 to 949 of DCPD in pre-S binding.
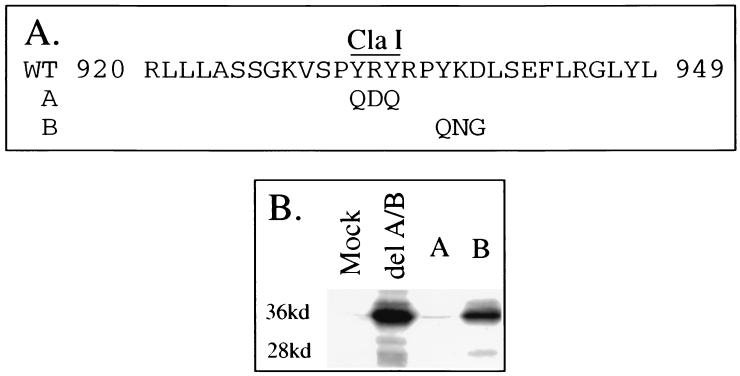
In addition to variant residues, we also evaluated the importance of conserved residues within the 920-to-949 sequence of DCPD in mediating pre-S binding. To do this, two mutant constructs, Y932Q R933D Y934Q (Fig. 7A, construct A) and Y937Q K938N D939G (construct B) were prepared. While the YKD-to-QNG change had no effect, replacement of YRY sequence by QDQ abolished viral binding activity (Fig. 7B).
FIG. 7.
Contribution of conserved residues within the host determinant of DCPD to DHBV binding. (A) Wild-type (WT) sequence between positions 920 and 949 of DCPD domain C and mutated residues in constructs A and B. Mutations as indicated were introduced into the delA/B construct of HCPD in which the SacII–KpnI site was replaced by DCPD sequence encoding residues 868 to 1304. (B) Binding of DHBV particles as revealed by Western blot analysis. Mock, vector DNA-transfected Bosc cells; delA/B, cells transfected with delA/B of DCPD.
CPD protein levels in transfected cells.
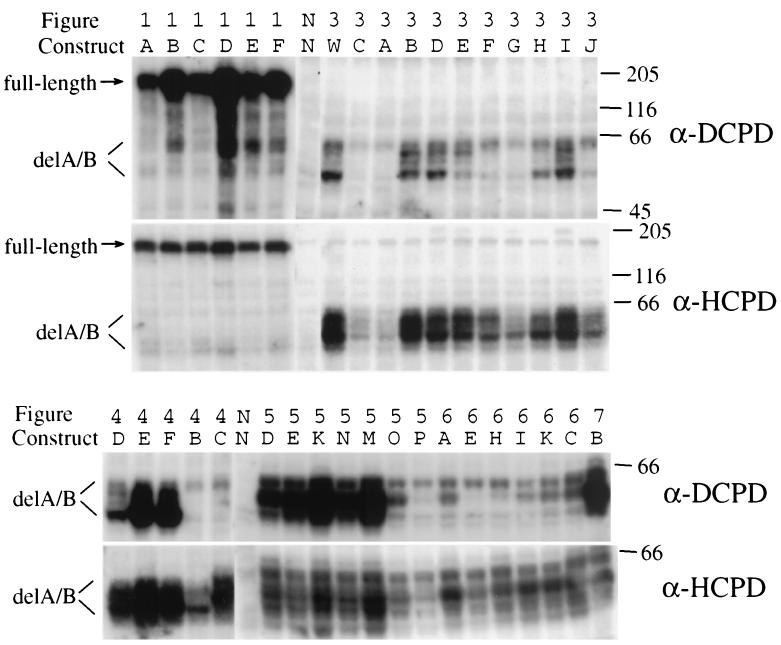
While the negative binding of chimeric constructs and site-directed mutants tested here is consistent with residues 920 to 949 being a direct contact site, it could be explained alternatively by poor expression of protein, its rapid degradation, or failure to reach the cell surface. To evaluate the steady-state levels of CPD protein, we performed Western blot analysis using polyclonal DCPD antibodies (23). Since the DCPD antiserum did not recognize the HCPD sequence efficiently, a rabbit serum against the C-terminal 179 aa of HCPD (19) was employed as well. As shown in Fig. 8, full-length DCPD–HCPD chimeric constructs were all well expressed (stronger signals were obtained with DCPD antibodies, which were raised against the full-length DCPD protein). With regard to the deletion constructs, most ex-pressed at high levels as revealed by the DCPD and HCPD antibodies (DCPD antibodies recognized primarily DCPD-based constructs, while HCPD antibodies could detect both types of constructs). In addition to a band of the predicted size (around 55 kDa), several slower-migrating species up to 62 kDa could be detected as well (Fig. 8). These bands, which differ in their intensity among different constructs, may represent phosphorylated or glycosylated forms of the CPD protein. Several constructs were found to express lower levels of CPD protein, including constructs 3A, 3C, 3G, 3J, 4B, 5O, and 5P. Of these, construct 3J and 4B were known to mediate efficient DHBV binding. Thus, failure of several constructs to mediate DHBV binding may be attributable to protein instability, but a high level of protein expression or stability is not always required for efficient DHBV binding.
FIG. 8.
Detection of CPD proteins in transiently transfected Bosc cells. Samples were analyzed by both DCPD antibodies (against the entire protein) and HCPD antibodies (against the C-terminal 179 residues). The 36 constructs tested here have been described in the figures above (Fig. 1 and 3 to 7) and labeled alphabetically. Samples labeled “NN” are negative controls (nontransfected Bosc cell lysate). Construct labeled “3 W” is the wild-type DCPD delA/B construct. Molecular size markers (in kilodaltons) are shown to the right. CPD proteins expressed by the human and duck delA/B constructs presented as multiple bands, possibly as a result of posttranslational modifications. Note that the two antibodies recognized these samples differently. The DCPD antibody was more sensitive in detecting full-length constructs (1A to 1F) than delA/B constructs. Of the deletion mutants, it recognized DCPD-based constructs (such as 5D, 5E, 5K, 5N, 5 M, and 7B) more efficiently than HCPD-based constructs (4C, 6A, 6E, 6H, 6I, 6K, and 6C). These constructs have been tested in additional transfection and Western blot analysis, and similar results were obtained.
DISCUSSION
The N terminus of CPD domain C as a host-specific determinant for DHBV interaction.
The mechanisms responsible for hepadnaviral host specificity are not fully understood, although circumstantial evidence indicates that they may lie at the receptor level. For example, HBV transgenic mice support HBV replication (4). Furthermore, DHBV DNA will replicate, although at reduced efficiency, in human HCC cell lines (20). Previous investigations suggest that DCPD lacked affinity for the HBV pre-S protein (17, 24), and the DHBV pre-S domain does not react with CPD molecules of chicken or human origin (18). Thus, the specificity of the receptor-ligand interaction is a major factor that determines DHBV noninfectivity in chickens and humans and prevents HBV infectivity in ducks. There is, however, an exception in heron HBV, where the viral pre-S domain binds DCPD but the virus does not infect duck hepatocytes, indicating the involvement of other components (15). The present study attempted to determine a host-specific element(s) of DCPD binding to DHBV and to initiate an effort at understanding the structural basis of DCPD interaction with the pre-S domain of DHBV large envelope protein. We mapped the host (duck versus human) determinant of pre-S binding to the contiguous KpnI-ClaI and ClaI-HindIII restriction enzyme fragments using chimeric delA/B constructs based on available and artificially created restriction sites (Fig. 3). Experiments with full-length constructs corroborated the importance of the sequence around the ClaI site, thus explaining the initial observation that replacement of DCPD sequence with that of HCPD, either upstream or downstream of the ClaI site, invariably abolished DHBV binding (Fig. 1, constructs A and B). Further analysis revealed a 30-aa sequence (32 aa in HCPD) surrounding the ClaI site as the key determinant of host-specific virus-receptor interaction, as reciprocal fragment exchange reversed the binding properties (Fig. 4). Sequence immediately upstream of this element (the C terminus of domain B) is not required, as indicated by the deletion construct A (Fig. 4). Within the host determinant there are 12 amino acid changes and a dipeptide insertion in HCPD relative to DCPD (Fig. 5). Mutational analysis revealed that P-to-LAL conversion at position 931 reduced DHBV binding (Fig. 5, construct D). Alternatively, four amino acid substitutions, two upstream and two downstream of the ClaI site, were equally effective in eliminating or substantially reducing DHBV binding (Fig. 5, constructs O and P). Thus, a fraction of the sequence changes that have accumulated in this HCPD region are sufficient to render the protein inactive with respect to DHBV binding. However, Western blot analysis revealed reduced levels of DCPD protein in cells transfected with construct 3A, 3C, 3G, 5O, or 5P (Fig. 8), suggesting poor expression or instability associated with these chimeras and mutants. Besides, we do not know whether some of the mutants tested here, despite normal protein expression and stability, could reach the cell surface to mediate virus binding.
DCPD and mammalian CPD homologues have different local secondary structures within the host determinant of DHBV interaction.
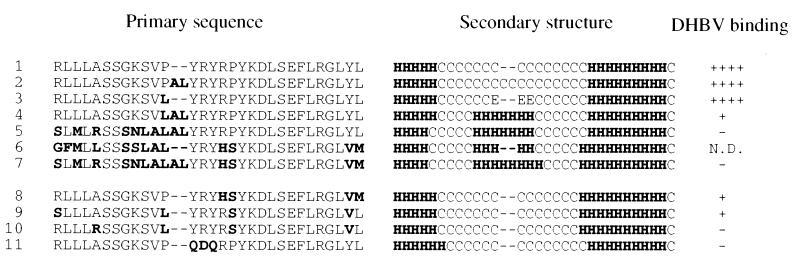
As determined by eight prediction programs for protein secondary structure, domain C of HCPD exhibits a secondary structure different from that of DCPD within this 30- or 32-aa stretch (Fig. 9). An additional helix is formed, or the preexisting helix is extended, in a region corresponding to random coil in DCPD. Inspection of the primary sequence (Fig. 9) reveals that the most conspicuous difference is the 929VSP931 tripeptide sequence of DCPD compared to the 913LALAL917 pentapeptide sequence in HCPD (one copy of AL is derived from substitutions, another copy from insertion). The following observations suggest that the LAL sequence (not the LALAL sequence) is required and sufficient for this structural change. (i) Replacement of 931P in DCPD with the LAL sequence results in helix formation and a reduction in DHBV binding (Fig. 10, row 4). (ii) Mouse and rat CPD molecules, which have the LAL but not LALAL sequence (for lack of AL insertion), adopt the helix structure (Fig. 10, row 6). (iii) Insertion of the AL dipeptide sequence alone into DCPD (resulting in a PAL tripeptide sequence) did not alter the secondary structure or affect DHBV binding (Fig. 10, row 2). Thus, the presence of local randomly coiled protein structure, rather than an α-helix, may be a prerequisite for effective DHBV binding. Other sequence changes that have accumulated in HCPD do not change the predicted secondary structure but still abolish DHBV binding (Fig. 10, rows 8 to 10). Thus, HCPD may contain both secondary structure-dependent and secondary structure-independent mechanisms to abolish the DHBV interaction.
FIG. 9.
Predicted secondary structures of the host determinant of DHBV interaction in DCPD and HCPD. The top two rows show the primary sequences (roman type, conserved; bolface, nonconserved). Rows 1 to 8 show the predicted secondary structure of DCPD (upper line) and HCPD (lower line) according to the methods of Predator (6), SOPM (9), SOPMA (10), GORI (8), GORIII (11), GORIV (7), PHD (21), and the multivariate regression combination (13), respectively. H, helix; E, extended; C, random coil; T, turn. A consensus was assumed when eight of the eight methods gave the same structure prediction for a given amino acid.
FIG. 10.
Predicted secondary structures of the host determinant of CPD molecules and mutants. The secondary structure prediction was based on the Predator program (6). Duck residues (920 to 949) are in roman type, while residues different from wild-type DCPD are boldfaced. Hyphen: deleted sequence. For secondary structure H, E, and C indicate helix, extended, and random coil, respectively. Rows: 1, DCPD; 6, rat and mouse CPD; 7, HCPD; 3, P931L; 2, 4, 5, 8, 9, and 10, constructs E, D, A, F, O, and P in Fig. 5, respectively; 11, construct A in Fig. 7. N.D., not determined.
Does the 920 to 949 sequence element in DCPD constitute a direct contact site for the pre-S domain?
Mutational analysis of residues 920 to 949 suggests that many conserved and variant residues are required for DHBV binding (Fig. 5 to 7). Since most of the mutations that abolish DHBV binding do not alter local secondary structure, these findings implicate this region as a direct contact site for the DHBV pre-S domain. Consistent with this notion, the corresponding sequences in domain A and domain B, which have no binding affinity for DHBV, diverge extensively from that of domain C (Fig. 2B). However, some of the mutants that fail to bind DHBV display low steady-state levels of DCPD protein (constructs 5O and 5P [see Fig. 8]). In addition, we cannot exclude the possibility that for some other constructs that fail to mediate DHBV binding, the mutant proteins fail to reach the cell surface. Thus, other approaches, such as DCPD deletion mutants, and direct in vitro binding assays, will help establish whether the sequence element defined by the DCPD-HCPD chimeras is necessary and sufficient for pre-S binding. Elucidation of the virus contact site on DCPD will present an opportunity to block viral entry into hepatocytes at the receptor level with specific antibodies or competing peptides.
ACKNOWLEDGMENTS
We thank our colleagues in the Liver Research Center for stimulating discussions, especially Xiaoming Gong for discussions on protein secondary-structure prediction.
This work was supported by NIH grants CA-35711 and AA-08169 and in part by Center of Biomedical Research Excellence grant p20RR. H. C. Spangenberg is supported by a fellowship of the Ernst Jung Stiftung fuer Wissenschaft und Forschung, Germany. J. Li is a Liver Scholar of the American Liver Foundation.
REFERENCES
- 1.Aloy P, Companys V, Vendrell J, Aviles F, Fricker L, Coll M, Gomis-Ruth F. The crystal structure of the inhibitor-complexed carboxypeptidase D domain II and the modelling of regulatory carboxypeptidases. J Biol Chem. 2001;276:16177–16184. doi: 10.1074/jbc.M011457200. [DOI] [PubMed] [Google Scholar]
- 2.Breiner K M, Urban S, Glass B, Schaller H. Envelope protein-mediated down-regulation of hepatitis B virus receptor in infected hepatocytes. J Virol. 2001;75:143–150. doi: 10.1128/JVI.75.1.143-150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breiner K M, Urban S, Schaller H. Carboxypeptidase D (gp180), a Golgi-resident protein, functions in the attachment and entry of avian hepatitis B viruses. J Virol. 1998;72:8098–8104. doi: 10.1128/jvi.72.10.8098-8104.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chisari F V. Hepatitis B virus transgenic mice: models of viral immunobiology and pathogenesis. Curr Top Microbiol Immunol. 1996;206:149–173. doi: 10.1007/978-3-642-85208-4_9. [DOI] [PubMed] [Google Scholar]
- 5.Eng F J, Novikova E G, Kuroki K, Ganem D, Fricker L D. gp180, a protein that binds duck hepatitis B virus particles, has metallocarboxypeptidase D-like enzymatic activity. J Biol Chem. 1998;273:8382–8388. doi: 10.1074/jbc.273.14.8382. [DOI] [PubMed] [Google Scholar]
- 6.Frishman D, Argos P. Incorporation of non-local interactions in protein secondary structure prediction from the amino acid sequence. Protein Eng. 1996;9:133–142. doi: 10.1093/protein/9.2.133. [DOI] [PubMed] [Google Scholar]
- 7.Garnier J, Gibrat J F, Robson B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 1996;266:540–553. doi: 10.1016/s0076-6879(96)66034-0. [DOI] [PubMed] [Google Scholar]
- 8.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 9.Geourjon C, Deleage G. SOPM: a self-optimized method for protein secondary structure prediction. Protein Eng. 1994;7:157–164. doi: 10.1093/protein/7.2.157. [DOI] [PubMed] [Google Scholar]
- 10.Geourjon C, Deleage G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci. 1995;11:681–684. doi: 10.1093/bioinformatics/11.6.681. [DOI] [PubMed] [Google Scholar]
- 11.Gibrat J F, Garnier J, Robson B. Further developments of protein secondary structure prediction using information theory. New parameters and consideration of residue pairs. J Mol Biol. 1987;198:425–443. doi: 10.1016/0022-2836(87)90292-0. [DOI] [PubMed] [Google Scholar]
- 12.Gomis-Ruth F X, Companys V, Qian Y, Fricker L D, Vendrell J, Aviles F X, Coll M. Crystal structure of avian carboxypeptidase D domain II: a prototype for the regulatory metallocarboxypeptidase subfamily. EMBO J. 1999;18:5817–5826. doi: 10.1093/emboj/18.21.5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guermeur Y, Geourjon C, Gallinari P, Deleage G. Improved performance in protein secondary structure prediction by inhomogeneous score combination. Bioinformatics. 1999;15:413–421. doi: 10.1093/bioinformatics/15.5.413. [DOI] [PubMed] [Google Scholar]
- 14.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 15.Ishikawa T, Kuroki K, Lenhoff R, Summers J, Ganem D. Analysis of the binding of a host cell surface glycoprotein to the preS protein of duck hepatitis B virus. Virology. 1994;202:1061–1064. doi: 10.1006/viro.1994.1440. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa T, Murakami K, Kido Y, Ohnishi S, Yazaki Y, Harada F, Kuroki K. Cloning, functional expression, and chromosomal localization of the human and mouse gp180-carboxypeptidase D-like enzyme. Gene. 1998;215:361–370. doi: 10.1016/s0378-1119(98)00270-4. [DOI] [PubMed] [Google Scholar]
- 17.Kuroki K, Cheung R, Marion P L, Ganem D. A cell surface protein that binds avian hepatitis B virus particles. J Virol. 1994;68:2091–2096. doi: 10.1128/jvi.68.4.2091-2096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuroki K, Eng F, Ishikawa T, Turck C, Harada F, Ganem D. gp180, a host cell glycoprotein that binds duck hepatitis B virus particles, is encoded by a member of the carboxypeptidase gene family. J Biol Chem. 1995;270:15022–15028. doi: 10.1074/jbc.270.25.15022. [DOI] [PubMed] [Google Scholar]
- 19.McGwire G B, Tan F, Michel B, Rehli M, Skidgel R A. Identification of a membrane-bound carboxypeptidase as the mammalian homolog of duck gp180, a hepatitis B virus-binding protein. Life Sci. 1997;60:715–724. doi: 10.1016/s0024-3205(96)00642-x. [DOI] [PubMed] [Google Scholar]
- 20.Pugh J C, Yaginuma K, Koike K, Summers J. Duck hepatitis B virus (DHBV) particles produced by transient expression of DHBV DNA in a human hepatoma cell line are infectious in vitro. J Virol. 1988;62:3513–3516. doi: 10.1128/jvi.62.9.3513-3516.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 22.Tan F, Rehli M, Krause S W, Skidgel R A. Sequence of human carboxypeptidase D reveals it to be a member of the regulatory carboxypeptidase family with three tandem active site domains. Biochem J. 1997;327:81–87. doi: 10.1042/bj3270081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong S, Li J, Wands J R. Carboxypeptidase D is an avian hepatitis B virus receptor. J Virol. 1999;73:8696–8702. doi: 10.1128/jvi.73.10.8696-8702.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong S, Li J, Wands J R. Interaction between duck hepatitis B virus and a 170-kilodalton cellular protein is mediated through a neutralizing epitope of the pre-S region and occurs during viral infection. J Virol. 1995;69:7106–7112. doi: 10.1128/jvi.69.11.7106-7112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urban S, Schwarz C, Marx U C, Zentgraf H, Schaller H, Multhaup G. Receptor recognition by a hepatitis B virus reveals a novel mode of high affinity virus-receptor interaction. EMBO J. 2000;19:1217–1227. doi: 10.1093/emboj/19.6.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xin X, Varlamov O, Day R, Dong W, Bridgett M M, Leiter E H, Fricker L D. Cloning and sequence analysis of cDNA encoding rat carboxypeptidase D. DNA Cell Biol. 1997;16:897–909. doi: 10.1089/dna.1997.16.897. [DOI] [PubMed] [Google Scholar]