Abstract
Isolates of bovine viral diarrhea virus (BVDV), the prototype pestivirus, are divided into cytopathic (cp) and noncytopathic (ncp) biotypes according to their effect on cultured cells. The cp viruses also differ from ncp viruses by the production of viral nonstructural protein NS3. However, the mechanism by which cp viruses induce cytopathic effect in cell culture remains unknown. Here we used a genetic approach to isolate ncp variants that arose from a cp virus at low frequency. A bicistronic BVDV (cp strain NADL) was created that expressed puromycin acetyltransferase as a dominant selectable marker. This bicistronic virus exhibited slightly slower growth kinetics and smaller plaques than NADL but remained cp. A number of independent ncp variants were isolated by puromycin selection. Remarkably, these ncp variants produced NS3 and viral RNA at levels comparable to those of the cp parent. Sequence analyses uncovered no change in NS3, but for all ncp variants a Y2441C substitution at residue 15 of NS4B was found. Introduction of the Y2441C substitution into the NADL or bicistronic cp viruses reconstituted the ncp phenotype. Y2441 is highly conserved among pestiviruses and is located in a region of NS4B predicted to be on the cytosolic side of the endoplasmic reticulum membrane. Other engineered substitutions for Y2441 also affected viral cytopathogenicity and viability, with Y2441V being cp, Y2441A being ncp, and Y2441D rendering the virus unable to replicate. The ncp substitutions for Y2441 resulted in slightly increased levels of NS2-3 relative to NS3. We also showed that NS3, NS4B, and NS5A could be chemically cross-linked in NADL-infected cells, indicating that they are associated as components of a multiprotein complex. Although the mechanism remains to be elucidated, these results demonstrate that mutations in NS4B can attenuate BVDV cytopathogenicity despite NS3 production.
Bovine viral diarrhea virus (BVDV), classical swine fever virus (CSFV), and border disease virus (BDV) are economically important pathogens of domestic livestock that also infect wild ruminants (34, 48). They are classified in the Pestivirus genus of the family Flaviviridae that includes two additional genera, the classical flaviviruses like yellow fever virus (YF), dengue virus, and West Nile virus and the hepaciviruses of which hepatitis C virus (HCV) is the sole member. All members of this family are enveloped positive-strand RNA viruses that share similarities in genome organization, protein processing, and RNA replication (reviewed in reference 38). HCV is an important human pathogen affecting an estimated 3% of the world's population (53); however, studies of this virus have been hampered by inefficient replication in cell culture and lack of a small animal model. Given their close resemblance to HCV in terms of translation strategy and processing scheme, BVDV and other pestiviruses are being intensively studied, not only to unravel details of RNA replication that are of intrinsic interest but also to inform future studies of HCV.
The genome of BVDV is about 12.5 kb in length and consists of a single long open reading frame flanked by 5′ and 3′ nontranslated regions (NTRs) (10). Translation is cap independent and mediated by a type III internal ribosome entry site (IRES) (8, 35, 55). The resulting polyprotein is processed co- and posttranslationally by host and virus-encoded proteases to give rise to at least 12 individual proteins in the order NH2-Npro-C-Erns-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A- NS5B-COOH (9, 11, 12) (Fig. 1). Npro is a nonstructural autoprotease that cleaves at its C terminus to liberate the capsid (C) protein (52). C is followed by three virion glycoproteins Erns, E1, and E2, with Erns encoding an RNase of unknown function that is also secreted in nonvirion forms (40, 42). NS3 through NS5B, but not the structural proteins p7 or NS2, are required for pestivirus RNA replication (2, 47). The N-terminal one-third of NS3 encodes a serine protease that functions in concert with NS4A to mediate processing at all downstream cleavage sites (46, 54). The C-terminal NS3 domain encodes an RNA helicase that is required for an unknown step(s) in RNA replication (16, 17, 51). NS5B, the viral RNA-dependent RNA polymerase, has been expressed, purified, and extensively compared to the corresponding enzyme of HCV (1, 24, 57). Essentially nothing is known about the functions of p7, NS2, NS4B, or NS5A, although NS5A has been shown to be a serine phosphoprotein that is tightly associated with one or more cellular kinases (37).
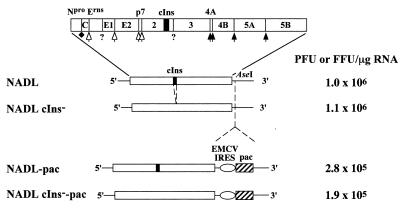
FIG. 1.
Bicistronic BVDV constructs. Shown are diagrams of parental monocistronic (NADL and NADL cIns−) and bicistronic (NADL-Pac and NADL cIns−-Pac) viruses with the indicated 5′ and 3′ NTRs and single open reading frame (ORF) (open box). The NADL polyprotein is illustrated by a more-detailed diagram at the top, with the mature cleavage products and the locations of cleavage sites indicated. Nonstructural proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B are designated as 2, 3, 4A, 4B, 5A and 5B, respectively. The enzymes responsible for polyprotein processing are indicated as follows: Npro autoprotease (solid diamond), ER lumen signal peptidase (open arrow), NS3 serine protease (solid arrow), and unidentified activities (?). The 90-amino-acid cellular insert sequence present in NS2 of NADL and NADL-pac is indicated as a black box. The AseI restriction site downstream of the polyprotein ORF of NADL and NADL cIns− was used to insert the expression cassette containing the EMCV IRES (open oval) and the pac gene (hatched box). On the right are representative RNA specific infectivities determined by an infectious center assay as described in Materials and Methods.
BVDV isolates are divided into two biotypes that are distinguished by their effect on cultured cells. Noncytopathic (ncp) isolates infect permissive host cells without causing cell death; cytopathic (cp) isolates produce rapid cytopathic effects (CPE) and kill cells (34). At least some evidence suggests that cell death is mediated by apoptosis (18, 56). Remarkable clinical correlates exist with these biotypes. BVDV causes a wide variety of clinical symptoms that range from asymptomatic infections to a fatal disease called mucosal disease. In pregnant animals, BVDV can be efficiently transmitted across the placenta, where it can infect the fetus and cause fetal death or congenital abnormalities. In addition, animals can be infected in utero, resulting in the birth of persistently infected BVDV-immunotolerant animals that are a major reservoir for the virus. Such animals sporadically develop mucosal disease and both ncp and cp biotypes (or virus pairs) can be isolated from these animals (reviewed in references 34 and 48). The molecular feature that distinguishes cp from ncp biotypes is the production of a discrete NS3 protein (reviewed in reference 33). Both uncleaved NS2-NS3 (NS2-3) and NS3 are present in cp BVDV-infected cells, whereas only uncleaved NS2-3 is found in ncp BVDV-infected cells. By sequence analysis of multiple pairs of ncp/cp isolates, it has become apparent that deletions, genome duplications and rearrangements, insertions of cellular sequences, and even point mutations in NS2 (22, 23) correlate with NS3 production and cytopathogenicity (reviewed in reference 33). In the case of the prototype cp BVDV strain NADL (National Animal Disease Laboratory), a 270-base cellular sequence encoding 90 amino acids (referred to as the cellular insert or cIns) is found inserted in frame in the NS2 C-terminal region upstream of the NS2-NS3 junction (30) (Fig. 1). By an unknown mechanism, this insertion leads to partial cleavage at the NS2-NS3 site and production of both NS2-3 and NS3. We (29) and others (49) previously showed that deletion of cIns abolished processing at the NS2-NS3 site and converted the virus from cp to ncp. Similar correlates have been obtained for a number of other cp/ncp pairs (22, 31, 32, 47).
The fact that distinct molecular events in independent cp BVDV isolates all lead to NS3 production implicates a central role for this protein in cytopathogenicity. Indeed, NS3 is considered the hallmark of BVDV cytopathogenicity, yet how (and if) NS3 triggers cell death remains unclear. Several hypotheses have been advanced, including the involvement of the NS3 serine protease activity in the apoptotic proteolytic cascade (18, 56) or NS3-dependent upregulation of viral RNA replication leading to higher levels of double-stranded RNA and a consequent trigger of apoptosis (29, 49). The latter idea is based on the observation that cp BVDV NADL RNAs accumulate in infected cells at 5- to 10-fold higher levels than those observed in cells infected with an isogenic ncp derivative lacking cIns (29, 49).
In this study, we used a genetic approach to probe the viral determinants of BVDV cytopathogenicity. A dominant selectable marker placed into BVDV genome allowed us to isolate ncp variants of the cp BVDV strain NADL. Our results revealed an unexpected role for NS4B in attenuating BVDV cytopathogenicity that appears to be independent of NS3 production and enhanced RNA accumulation.
MATERIALS AND METHODS
Cells and viruses.
MDBK cells were obtained from the American Type Culture Collection. Cell monolayers were propagated in Dulbecco's modified essential medium (DMEM) supplemented with sodium pyruvate, penicillin, streptomycin, and 10% heat-inactivated horse serum (HS). Cells were maintained at 37°C with 5% CO2. Seventy to 80% subconfluent cell monolayers grown in 35-mm-diameter wells (about 5 × 105 to 1 × 106 cells per well) were used for the following assays unless otherwise indicated.
The cp NADL strain of BVDV and its isogenic ncp derivative, NADL cIns−, were generated from infectious cDNA clones as previously described (29). Viruses were amplified by growth in MDBK cells as previously described (27). Virus titers were determined by using a plaque (for cp viruses) or focus-forming (for ncp viruses) assay as described below.
BVDV plaque, focus, and puromycin-resistant focus assays.
For the plaque assay, MDBK monolayers were infected with a series of 10-fold dilutions of virus (0.2 ml per well). Following 1 h of adsorption at 37°C, the inoculum was removed, and the cells were overlaid with 0.5% agarose in minimal essential medium (MEM)–5% HS (3 ml per well) and incubated at 37°C. After 3 days, the cells were fixed with 7% formaldehyde for 2 h at room temperature, the agarose overlays were removed, and the monolayers were stained with 1% crystal violet (wf/vol in 50% ethanol) to visualize plaques.
For the focus assay, fixation of the cells and removal of the agarose overlay was followed by monolayer permeabilization with 0.25% Triton X-100 in phosphate-buffered saline (PBS) for 10 min. Monolayers were then washed once with PBS and incubated with a bovine polyclonal anti-BVDV serum (α49; 1/1,000 dilution in PBS) (36) for 2 h. After three additional washes with PBS, monolayers were incubated with peroxidase-conjugated rabbit anti-bovine antibody (Sigma; 1/1,000 dilution in PBS) for 2 h. Monolayers were again washed three times with PBS, and the immunostained foci were visualized by staining with a peroxidase detection kit (Vector Laboratories).
Puromycin-resistant foci were visualized as follows. Infection, overlay, and incubation of cell monolayers were as described above for the plaque assay. After 3 days, an equal volume of MEM–5% HS containing 10 μg of puromycin (Sigma) per ml was added on top of the agarose overlay. After an additional 2 to 3 days of incubation, the top medium and agarose overlay were removed and the monolayers were washed twice with PBS, fixed with 7% formaldehyde, and stained with crystal violet to visualize the foci formed by cells surviving puromycin selection.
Plasmid constructs.
The full-length infectious cDNA clone pACNR/NADL, and its isogenic ncp derivative, pACNR/NADL cIns−, have been described previously (29). The bicistronic full-length clone pACNR/NADL-pac was constructed as follows. The EcoRI-NcoI fragment (containing the encephalomyocarditis virus [EMCV] IRES-puromycin acetyltransferase [pac] cassette) from pRS2/IRES-pac (E. Agapov and C. M. Rice, unpublished data) was filled in and inserted into the unique AseI site (filled in) in the BVDV 3′ NTR of pTET/BVD5′3′ (29). The resulting clone, pTET/BVD5′pac3′, was partially digested with EagI and SdaI, and the large EagI-SdaI fragment was cloned into EagI- and SdaI-digested pACNR/NADL to generate pACNR/NADL-pac. The same EagI-SdaI fragment was similarly cloned into pACNR/NADL cIns− to create pACNR/NADL cIns−-pac.
To engineer the Y2441C mutation identified in the ncp NADL-pac variants into the parental cDNA clones pACNR/NADL and pACNR/NADL-pac, the DrdI (nucleotide [nt] 7462; NADL numbering)-NcoI (nt 8316) portion of the two clones was replaced with the corresponding reverse transcription PCR (RT-PCR) fragment from ncp mutant NADL-pac #8, which contained the Y2441C mutation. The resulting clones were called pACNR/NADL Y2441C and pACNR/NADL Y2441C-pac, respectively.
To construct the additional mutants Y2441A and Y2441D, synthetic oligonucleotides and PCR were used to engineer the following changes (underlined): Y2441A (TAT to GCT) and Y2441D (TAT to GAT). For both mutants two silent mutations, C to G at nt 7696 and T to A at nt 7699, created a novel EcoRV (GATATC) site that served as a marker to distinguish Y2441A and Y2441D from the wild-type sequence. The nucleotide changes for Y2441A also created a new PstI site (CTGCAG) that was used to distinguish Y2441A from Y2441D. These mutations were introduced into pACNR/NADL by replacing the DrdI-NcoI portion with the corresponding PCR fragments, resulting in plasmids pACNR/NADL Y2441A and pACNR/NADL Y2441D, respectively. The Y2441V mutant was constructed by replacing the same DrdI-NcoI portion of pACNR/NADL with the corresponding RT-PCR fragment from the cp revertant of Y2441D, which contains the D to V (GAT to GTT) substitution. The resulting clone was named pACNR/NADL Y2441V.
Standard recombinant DNA technology (41) was used to construct and purify all plasmids, with special care for the full-length clones. All regions amplified by PCR were confirmed by automated nucleotide sequencing. All full-length clones were propagated in Escherichia coli SURE cells (Stratagene) (29). Potentially correct clones from small-scale preparations were first identified by supercoiled plasmid size, checked by PCR and restriction digestions, and finally verified by sequencing of any regions created by PCR. Plasmid DNAs for in vitro transcription were prepared from large-scale bacterial cultures, purified by CsCI-ethidium bromide gradient centrifugation, and verified again by restriction and sequence analyses. For each full-length construct, three independent plasmid templates were prepared, transcribed, and analyzed (see below).
In vitro transcription reaction.
SdaI (MBI Fermentas)-linearized plasmids of full-length BVDV clones were used as DNA templates to generate in vitro transcripts using the T7-MEGAscript kit (Ambion) in the absence of cap analog, as described previously (29). Five-tenths microcurie of [3H]UTP (Dupont) tracer was added to a 20-μl reaction mixture to quantify RNA yield on the basis of [3H]UTP incorporation. The integrity of the full-length RNA transcripts was checked by 1% agarose gel electrophoresis and ethidium bromide staining.
Transfection of MDBK cells and infectious center assay.
Electroporation was used to transfect MDBK cells with in vitro-transcribed RNA. Briefly, confluent MDBK cells were trypsinized and collected, washed three times with ice-cold RNase-free PBS, and resuspended at 2 × 107 cells/ml in PBS. One microgram of in vitro-transcribed RNA was mixed with 0.4 ml of the cell suspension (8 × 106 cells) in a 2-mm-gap cuvette (BTX) and immediately pulsed with a BTX ElectroSquarePorator (0.9 kV; 99-μs pulse length; 10 pulses). Following 10 min of recovery at room temperature, the electroporated mixture was diluted to 10 ml with DMEM–10% HS. A small portion (0.1 ml) was taken to determine RNA-specific infectivity as described below, and the remainder was plated in a 75-cm2 flask and incubated at 37°C for 48 h prior to harvest of the virus as described above. We found that virus yield was enhanced by replacing the medium at 4 h posttransfection, perhaps due to the toxicity of dead cells or cell debris resulting from electroporation.
RNA specific infectivity was measured by an infectious center assay (14, 29). One-tenth milliliter of the 10-ml electroporation mixture was serially diluted by 10-fold with DMEM–10% HS and plated (0.1 ml per well) in 35-mm-diameter wells containing MDBK cell monolayers grown to 50 to 60% confluence. To allow recovery and attachment of the electroporated cells, plates were incubated at 37°C for 4 h, after which the medium was removed and replaced with a 0.5% agarose overlay as described for the plaque assay. Plates were incubated at 37°C for 3 days, and the infectious centers were visualized and counted as described above in plaque (for cp viruses) or focus (for ncp viruses)-forming assay.
Isolation and purification of ncp NADL-pac variants.
A combined infectious center and puromycin-resistant focus-forming assay was used to isolate ncp NADL-pac mutants. MDBK cells were transfected with 1 μg of NADL-pac RNA and plated as described above. After 3 days of incubation at 37°C, puromycin was added to the agarose overlay and the plates were incubated for an additional 2 to 3 days to allow selection for puromycin-resistant cell foci produced by ncp NADL-pac variants. Puromycin-resistant foci were readily visualized by inspection of the bottom of the well against natural light, and the foci were picked with a sterile glass pipette and resuspended in 1 ml of DMEM–10% HS. To assure that each focus was independently derived from the original transfection, only one focus was isolated per well. Virus was eluted from the agarose plug by rocking at 4°C for 12 h. The eluate was used to inoculate a fresh monolayer followed by an additional round of puromycin selection and isolation of a single focus per isolate. The final eluate was amplified in MDBK cells to generate a high-titer virus stock.
Isolation and purification of cp revertants from the lethal Y2441D mutant.
MDBK cells were transfected with 1 μg of pACNR/NADL Y2441D RNA and plated as described above. After 3 days of incubation at 37°C, unstained plaques formed by cp revertants were visualized, picked, plaque purified, and amplified for further analysis.
Protein analysis.
The production of NS3 in virus-infected cells was detected by Western blot using rabbit polyclonal antiserum specific for BVDV NS3 (G40) (9). MDBK cells were infected at a multiplicity of infection (MOI) of 5. At desired time points postinfection, cell monolayers were washed with PBS and lysed in 0.2 ml of standard SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer. Cell lysates were sheared by repeated passage through a 27-gauge needle, boiled for 10 min in the presence of 5% β-mercaptoethanol (β-ME), and clarified by centrifugation at 12,000 × g for 10 min. Proteins from approximately 2 × 104 cells were separated by SDS–8% PAGE and transferred onto nitrocellulose membranes (Amersham), using a semidry Multiphor II Nova blot system (LKB). Membranes were blocked overnight at 4°C in blocking buffer (0.2% Triton X-100, 0.1% Tween 20, and 5% non-fat milk in PBS). After being blocked, the membrane was incubated at room temperature for 3 h with primary antiserum G40 (diluted 1:2,000 in blocking buffer) and then for 2 h with secondary horseradish peroxidase-conjugated goat anti-rabbit serum (Sigma; diluted 1:10,000 in blocking buffer), followed by enhanced chemiluminescence detection (ECL; Pierce) and autoradiography. Repeated wash steps using blocking buffer without milk were performed after each incubation with antiserum. Equal amounts of protein samples were separated on an SDS–10% polyacrylamide gel. Western blotting of β-actin, which served as a loading and transfer control, was conducted essentially as described above except using a mouse anti-β-actin monoclonal antibody (Sigma; 1:5,000 dilution) as the primary antibody and a peroxidase-conjugated goat anti-mouse immunoglobulin G (Boehringer-Mannheim; 1:1,000 dilution) as the secondary antibody.
Metabolic labeling of viral RNA.
MDBK cells were seeded and infected as described above. At 15 h postinfection, cells were treated with actinomycin D (1 μg/ml) for 1 h prior to the addition of [3H]uridine (50 μCi/ml; ICN). After 2 h (18 h postinfection) of labeling, cells were washed once with PBS and total RNA was extracted with Trizol (Gibco-BRL) according to the manufacturer's instructions. RNA from approximately 105 cells was denatured with 1 M glyoxal and 50% dimethyl sulfoxide (DMSO) at 50°C for 1 h and separated by sodium phosphate-buffered electrophoresis on 1% agarose gels. Total RNA was visualized by staining with ethidium bromide, and gels were equilibrated in methanol, processed for fluorography using 3% 2,5-diphenyloxazole in methanol-water, and then dried. Metabolically labeled viral RNA was visualized by fluorography.
RT-PCR.
Total RNA was extracted from virus-infected cells using Trizol (Gibco-BRL). RNA from approximately 104 cells was mixed with a BVDV-specific minus-sense oligonucleotide (DNAgency) and SuperScript II reverse transcriptase (Gibco-BRL). First-strand synthesis reaction mixtures under the manufacturer's conditions were incubated at 44°C for 1 h. The resulting cDNA was then amplified by PCR using high-fidelity KlenTaq LA (Wayne Barnes, Washington University, St. Louis, Mo.) and pairs of BVDV-specific oligonucleotides (DNAgency). PCR products were gel purified and used directly for sequence analysis.
Sequence and computational analyses.
Sequencing reaction mixtures of DNA plasmids and PCR products were prepared using the BigDye cycle sequencing kit (Perkin-Elmer) and separated on an automated ABI sequenator (Perkin-Elmer). Overlapping sequences were assembled and compared to the BVDV NADL sequence (10) using the SeqMan software (DNAStar). NS4B amino acid sequences from 26 pestiviruses were downloaded from GenBank and aligned using the MegAlign program (DNAStar). Membrane protein topology models of the NS4B proteins of BVDV (strain NADL), HCV (type 1b), and YF (strain 17D) were predicted using software on the EMBL-Heidelberg PHD server (39).
Protein labeling and cross-linking.
MDBK cells were seeded and infected as described above. At 18 h postinfection, cells were washed once and incubated in methionine and cysteine-free MEM for 30 min and then labeled with the same medium supplemented with 100 μCi of 35S-protein labeling mixture (NEN) per ml and 2% HS for 3 to 4 h. The cell-permeable and thiol-cleavable cross-linker 3,3′-dithiobis(succinimidylpropionate) (DSP; Pierce) was prepared at 1 mM in 10% DMSO (vol/vol in PBS) according to the manufacturer's instruction. After being labeled, the cells were washed with PBS and incubated with 1 mM DSP at room temperature for 1 h. Non-DSP samples were incubated with 10% DMSO (vol/vol in PBS) instead. The reaction was quenched with 50 mM Tris-Cl pH 7.5 for an additional 15 min. Cells were lysed in SDS lysis buffer without reducing reagent and immunoprecipitated with BVDV protein-specific antiserum as described below. Cross-linked proteins in the precipitates were dissociated when DSP was cleaved with 5% β-ME in standard SDS-PAGE sample buffer at 100°C for 5 min.
Cell lysis and immunoprecipitation.
After being labeled, cell monolayers were washed with PBS and lysed in 0.2 ml of 0.5% SDS lysis buffer containing 20 μg of phenylmethylsulfonyl fluoride/ml. Cell lysates were sheared, heated to 70°C for 10 min, and clarified by centrifugation. Immunoprecipitation reactions using rabbit polyclonal antiserum (usually 5 μl for a 100-μl initial lysate) and cross-linked Staphylococcus aureus cells (Calbiochem) were conducted essentially as previously described (11). The BVDV-specific antisera and their recognition regions are as follows: G40 (9) and WU165 (anti-NS3), WU168 (anti-NS4B), and WU170 (anti-NS5A). The precipitated samples were analyzed by reducing SDS–10% PAGE followed by autoradiography.
RESULTS
Bicistronic BVDV expressing puromycin resistance.
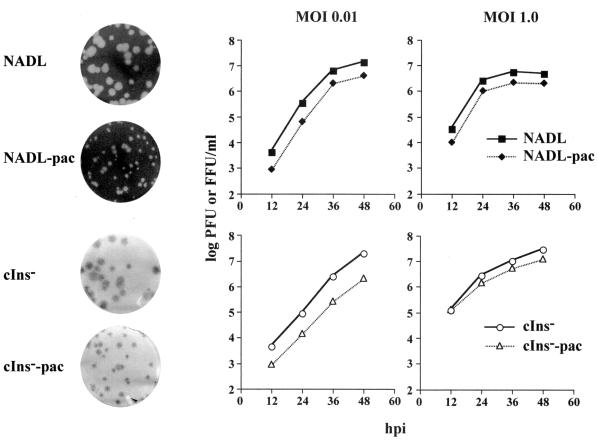
In previous work, we constructed a functional full-length cDNA clone for the prototype cp NADL strain of BVDV (29). Deletion of the 270-base cellular insert in NS2 created an isogenic ncp derivative, NADL cIns− (29). These parental clones were used to create several bicistronic derivatives. Although several strategies can be envisioned, the simplest involved insertion of expression cassettes into the poorly conserved 3′ NTR sequence immediately following the BVDV open reading frame. We employed a convenient unique restriction site (AseI) to introduce the EMCV IRES followed by the puromycin resistance (pac) gene that encodes puromycin N-acetyltransferase (PAC) (Fig. 1). As measured in infectious center assays, RNA transcripts from both pACNR/NADL-pac and pACNR/NADL cIns−-pac yielded specific infectivities within an order of magnitude of the corresponding parental RNAs (Fig. 1: ∼105 versus 106 PFU or focus-forming units [FFU]/μg of RNA). Furthermore, virus yields were similar to those of the parents after 48 h (106 to 107 PFU or FFU/ml), and the cp and ncp phenotypes were maintained with only a slight (4 h) delay of nearly complete CPE in the NADL-pac-infected cells relative to that for NADL. Shown in Fig. 2 are the plaque (NADL and NADL-pac) and focus-forming (NADL cIns− and NADL cIns−-pac) phenotypes of the parental and bicistronic viruses. Both bicistronic derivatives displayed somewhat smaller plaques or foci than their respective parents but produced similar overall virus yields at both low and high MOIs, albeit with slightly slower growth kinetics (Fig. 2).
FIG. 2.
Comparison of bicistronic viruses and their parents. On the left, representative plaque (NADL and NADL-pac) and focus (NADL cIns− and NADL cIns−-pac)-forming assays (see Materials and Methods) are shown. On the right, growth kinetics in MDBK cells of bicistronic and parental viruses infected at an MOI of 0.01 or 1.0 are shown. Viruses were harvested at the indicated hours postinfection (hpi), and titers were determined by plaque (NADL and NADL-pac) or immunostaining (NADL cIns− and NADL cIns−-pac) assays. A set of representative data was plotted by curves on a semilogarithmic scale.
Puromycin selection to isolate ncp variants.
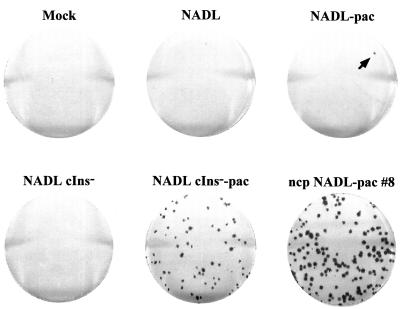
The rationale for using puromycin to select for ncp variants requires that the bicistronic virus replicate and express sufficient levels of PAC to confer resistance, spread from cell to cell, and be ncp, resulting in a focus of viable puromycin-resistant cells. Such foci can then be visualized by staining with crystal violet. The assay to detect ncp variants was optimized using the ncp bicistronic virus, NADL cIns−-pac, which should satisfy all of the conditions above. Cells were infected and overlaid with agarose as for a normal plaque or focus-forming assay, and puromycin was layered over the agarose and allowed to equilibrate as described in Materials and Methods. Under these conditions, uninfected, NADL−, or NADL cIns−-infected cells were killed but foci of cells infected with NADL cIns−-pac were viable, resistant to puromycin, and could be readily visualized by staining with crystal violet (Fig. 3). In the case of cp NADL-pac, most cells in the monolayer were killed, either by the virus or by puromycin, and virus plaques actually showed a halo-like appearance in which the halo consisted of puromycin-resistant virus-infected cells that had not yet succumbed to NADL-induced CPE and death (data not shown). At low frequency, solid foci produced by ncp variants similar to those seen for NADL cIns−-pac appeared in the monolayers infected with NADL-pac (a single focus is visible in the upper right plate shown in Fig. 3, arrow). Relative to the number of PFU per μg of RNA these foci appeared at a frequency of about 10−3 to 10−4. This is about the frequency expected for single base substitutions introduced by bacteriophage T7 RNA polymerase during transcription (10−4) (3, 43) or by RNA-dependent RNA virus replication (10−3 to 10−4) (44, 50). From independent wells, individual ncp variants were isolated, purified by two rounds of focus selection, and expanded to produce small-scale virus stocks. A sample of one of these, ncp NADL-pac #8, is shown in Fig. 3.
FIG. 3.
Puromycin selection of ncp mutants. The combination of infectious center and puromycin-resistant focus assays was performed as described in Materials and Methods. An ncp mutant appeared from NADL-pac-transfected cells (arrow). Shown at the bottom right is an example (isolate #8) of the ncp NADL-pac mutants isolated and purified as described in Materials and Methods.
ncp variants produce NS3 and viral RNA at levels comparable to those of cp parents.
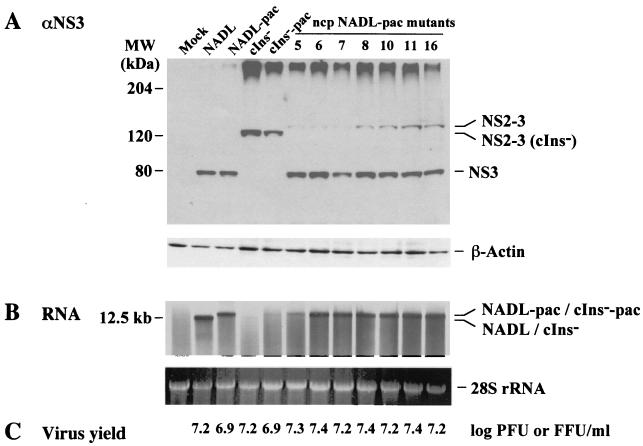
NS3 production has long been considered the molecular signature of cp BVDV, and it has been hypothesized that NS3 itself may be responsible for CPE and the triggering of cell death. In studies comparing NADL with the isogenic derivative NADL cIns−, this correlation held since deletion of the cellular insert (cIns) in NS2 abolished NS3 production and cytopathogenicity (29, 49). We also noticed that despite similar levels of infectious virus production, viral RNA levels in NADL-infected MDBK cells were 5 to 10 times higher than those found in NADL cIns−-infected cells (29, 49). Thus, the accumulation of viral RNA or dsRNA might itself be an effector of apoptosis as seen in other systems (20, 45). This can occur via double-stranded RNA-dependent activation of PKR or perhaps other pathways (reviewed in reference 19). As shown in Fig. 4, levels of NS2-3, NS3, and viral RNA were examined for seven independent ncp NADL-pac variants after infection of MDBK cells at a high MOI (5 PFU or FFU/cell). Surprisingly, NS2-3 and NS3 were detected for all of the variants, with NS3 being present at levels similar to those found for NADL and NADL-pac. Uncleaved NS2-3 levels were somewhat higher for the ncp variants than for NADL or the NADL-pac parent (NS2-3 was also visible in these samples in longer exposures; see below). Viral RNA synthesis for the variants was generally high (except variant #5), characteristic of the cp viruses NADL and NADL-pac rather than the ncp NADL cIns− and NADL cIns−-pac derivatives (Fig. 4B). Virus titers determined at 18 h postinfection were comparable for all of the viruses, regardless of their biotype (Fig. 4C). These results show that the ncp NADL-pac variants retained all the hallmarks of cp NADL and NADL-pac, including NS3 production and RNA accumulation. They did not, however, induce CPE and produced slightly elevated accumulation of NS2-3 relative to that of NS3.
FIG. 4.
Comparison of ncp NADL-pac mutants and parental viruses. Parallel MDBK cells were infected at an MOI of 5 with the viruses shown at the top and analyzed as follows. (A) Protein samples were prepared by lysis of the cells with standard SDS sample buffer at 18 h postinfection, and proteins from approximately 2 × 104 cells were subjected to SDS–8% PAGE. NS3 was detected by Western blotting using NS3-specific antiserum G40 (9) as described in Materials and Methods. In parallel, equal amounts of protein samples were separated by SDS–10% PAGE and β-actin was detected as a loading and transfer control for the Western blot. Only the relevant portions of the gels are shown. The positions of NS2–3, NS3, and β-actin are indicated on the right, and the mobilities of the molecular mass standards are indicated on the left. (B) At 15 h postinfection, virus-infected cells were treated with actinomycin D for 1 h prior to the addition of [3H]uridine. Cells were labeled for an additional 2 h. Total RNA was extracted as described in Materials and Methods. RNA from approximately 105 cells was subjected to 1% agarose gel electrophoresis followed by ethidium bromide staining. Viral RNAs were visualized by fluorography (top panel). Also shown are the ethidium bromide-stained 28S rRNA bands (bottom panel) as a loading control. Only the relevant portions of the gel are shown. (C) Viruses were harvested at 18 h postinfection. Virus titers were determined by plaque assay (cp) or immunostaining (ncp) and are presented as logarithmic values.
A single substitution (Y2441C) in NS4B abolishes cytopathogenicity.
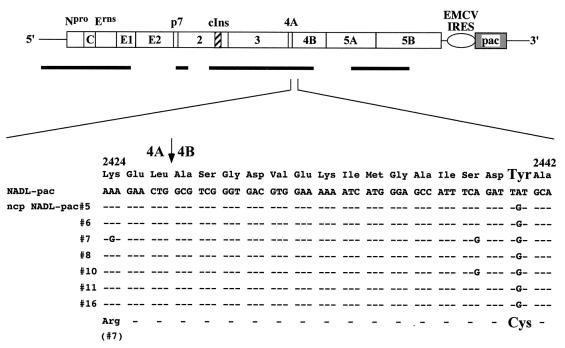
We had expected to find mutations in the ncp variants that abolished NS3 production, either by deletion of cIns or by incorporation of other mutations that block cleavage at the NS2-NS3 site. Clearly this was not the case given that all ncp NADL-pac variants produced NS3 and an NS2-3 species that comigrated with those produced by NADL. This suggested that cIns was present and that other mutations were responsible for the ncp phenotype. We began by sequencing selected regions of ncp variant NADL-pac #8. Using RNA isolated from infected cells, RT-PCR-amplified segments were sequenced directly, without cloning, to determine a population sequence. Surprisingly, sequencing of the NS2-3 region failed to identify any mutation. We then extended the analysis to other regions of the genome, as summarized in Fig. 5. A single A-to-G transition was found at nt 7707 (NADL numbering from reference 10) that changed a Tyr codon (UAU) to a Cys codon (UGU). This corresponds to amino acid residue 2441 of the NADL polyprotein or NS4B residue 15. Strikingly, the same mutation was found in all seven independently isolated ncp variants (Fig. 5). Some variants contained additional mutations, confirming that they were indeed independent isolates. These included a silent mutation in ncp NADL-pac #7 and #10 (UCA to UCG at Ser2439 or NS4B residue 13) and an additional coding change in ncp NADL-pac #7 that resulted in the change of Lys to Arg (AAA to AGA) at position 2424, corresponding to NS4A residue 62.
FIG. 5.
Summary of sequence analysis of the ncp NADL-pac mutants. Shown at the top is the BVDV NADL-pac genome, with the cellular insert sequence (hatched box) and EMCV IRES (open oval)-pac (shaded box) cassette indicated. Thick bars shown below the genome and drawn to scale indicate the regions of the viral cDNA that were sequenced. A small region (amino acids 2424 through 2442) is magnified to show the cDNA nucleotide and amino acid sequences from the parent and seven ncp NADL-pac variants. Identical nucleotide or amino acid sequences are indicated by -. An arrow indicates the 4A/4B cleavage site. A single A-to-G nucleotide mutation leading to replacement of Tyr (Y) with Cys (C) at polyprotein amino acid 2441 (NS4B residue 15) is indicated, with enlarged fonts for Tyr and Cys. Also shown for ncp variant #7 is an A-to-G nucleotide change encoding the replacement of Lys with Arg at polyprotein amino acid 2424 (NS4A residue 62).
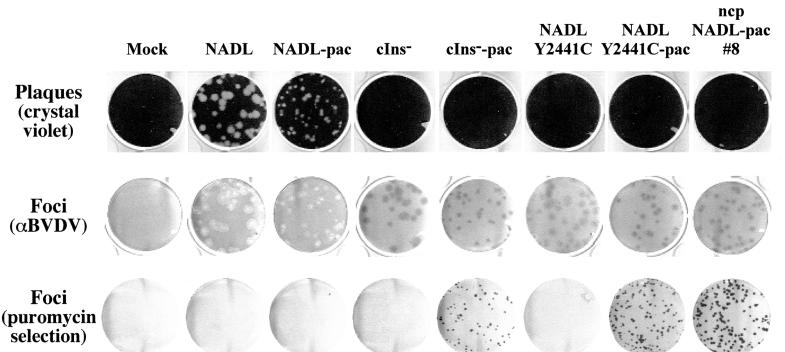
The fact that the Y2441C substitution was found in all ncp variants indicated that this change might be responsible for the ncp phenotype. To test this hypothesis, the Y2441C mutation was introduced into the parental infectious clones pACNR/NADL and pACNR/NADL-pac. Transfection of MDBK cells with the resulting NADL Y2441C and NADL Y2441C-pac RNAs confirmed this hypothesis. As determined by the infectious center assay, both RNAs were highly infectious, forming immunostained foci after 3 days at >105 FFU/μg RNA. Infection of MDBK cells with these viruses did not induce CPE or plaque formation, but infection with NADL Y2441C-pac led to the formation of puromycin-resistant foci that were indistinguishable from those of the original isolates (Fig. 6). These results confirmed that a single amino acid substitution in NS4B was sufficient to confer an ncp phenotype on this otherwise cp genetic background.
FIG. 6.
Introduction of the Y2441C mutation into NADL and NADL-pac reconstitutes the ncp phenotype. MDBK cell monolayers were infected with the indicated viruses at 50 to 100 PFU or FFU per 35-mm-diameter well. The monolayers were analyzed for plaque (top row) or focus (middle row) formation or for puromycin resistance (bottom row) using assays described in Materials and Methods.
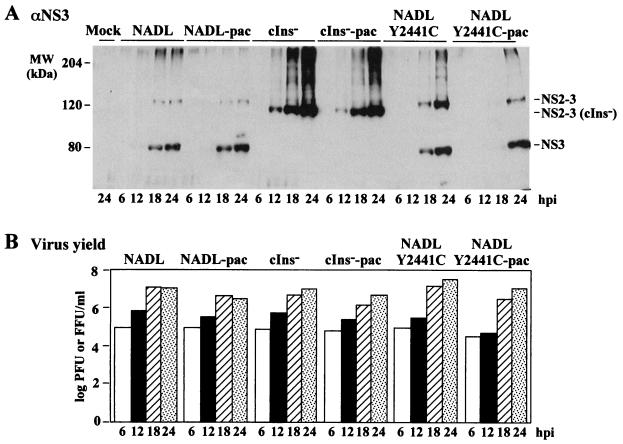
The reconstructed Y2441C variants were further characterized in single-step growth experiments (Fig. 7). Accumulation of NS3-specific products (Fig. 7A) and viral titers (Fig. 7B) were examined at 6, 12, 18, and 24 h postinfection. As shown for the cp viruses NADL and NADL-pac, levels of NS3 and NS2-3 increased over this time period, with virus production peaking at 18 h and declining slightly by 24 h, presumably due to CPE and cell death. In contrast, ncp viral titers for NADL cIns− and NADL cIns−-pac continued to increase even at 24 h and only uncleaved NS2-3 was produced, as expected. Virus production for NADL Y2441C and NADL Y2441C-pac resembled that of the ncp cIns− derivatives and was increasing at 24 h. Although NS3 levels were similar for the cp NADL parents and the reconstructed ncp variants over the time course, the level of NS2-3 relative to NS3 was consistently higher for the ncp variants.
FIG. 7.
Single-step analysis of the reconstructed Y2441C mutants. MDBK cells were infected at an MOI of 5 with the viruses indicated at the top. At the indicated time points postinfection (hpi), protein and virus samples were prepared as described in Materials and Methods. (A) Proteins from approximately 2 × 104 cells were separated by SDS–8% PAGE and analyzed as described in the legend to Fig. 4. Only the relevant portions of the gel are shown. (B) Virus titers were measured by plaque assay (cp) or immunostaining (ncp) and plotted by columns on a semilogarithmic scale.
Other substitutions for Y2441 affect viral cytopathogenicity and viability.
As shown in an alignment of 26 pestivirus sequences (Fig. 8), the Y2441 residue is invariant, as are several other residues in the highly conserved N-terminal region of NS4B. This evolutionary constraint indicates an important role for this domain in pestivirus replication. In particular, the presence of a conserved Tyr residue raised the possibility that this might be a phosphate acceptor site that was required for cytopathogenicity and was abolished by the Cys substitution. In pilot experiments using metabolic labeling with [32P]orthophosphate or immunoblotting of immunoprecipitated BVDV NS4B with phospho-Tyr-specific antibodies, we failed to obtain direct evidence for NS4B phosphorylation at this site (data not shown). Although a comprehensive mutagenesis study of NS4B has not been undertaken, we have examined the effect of two additional substitutions for Y2441 in the NADL background. As shown in Fig. 9, a Y2441A substitution resulted in a viable ncp virus with a focus-forming phenotype similar to that of the the Y2441C substitution. As determined by plaque and focus-forming assays, a Y2441D substitution was nonviable. This was confirmed by unsuccessful attempts to metabolically label viral RNAs and proteins in transfected cells. However, a small number of plaques (<50 PFU/μg of RNA, corresponding to a frequency of 10−4 when compared to the usual 105 to 106 PFU or FFU/μg of RNA for viable constructs) could be detected. These data suggest that viable cp revertants had arisen from the lethal Y2441D mutant. Sequence analysis of five independently isolated cp revertants showed that in all, the engineered Asp codon (GAU) now encoded Val (GUU). Introduction of the Y2441V substitution into the NADL background resulted in a viable cp virus that was similar to NADL in RNA specific infectivity and plaque phenotype (Fig. 9). The fact that the Val substitution is tolerated and gives rise to a cp virus proves that Y2441 (and hence possible Y2441 phosphorylation) is not essential for BVDV cytopathogenicity.
FIG. 8.
Y2441 of NS4B is highly conserved among pestiviruses. Sequences surrounding polyprotein amino acid Y2441 (residue 15 of NS4B) from 26 pestiviruses were aligned as described in Materials and Methods. Numbering corresponds to the N terminus of NS4B. The consensus sequence is shown above the alignment, and amino acid residues that are 100% conserved are shown below. The invariant Y2441 residue is boxed. The GenBank accession numbers of the virus strains/isolates are: BVDV 2–890 (U18059), CP1 (M62430), CP13 (Z73248), CP7 (U63512), DI9 (U03912), ILLC (U86599), ILLNC (U86600), JaCP (U80885), NADL (P19711), Oregon (AF041040), Osloss (M96687), Rit 4350 (AF058699), SD-1 (A44217); CSFV Alfort (P19712), ATCC VR531 (U21328), Brescia (P21530), C (Z46258), D49532, Glentorf (U45478), HCLV (AF091507), J04358, Shimen (AF092448), Steiermark (U21329); BDV BD31 (U70263), C413 (AF002227), and X818 (AF037405). Unnamed isolates are designated by their GenBank accession numbers.
FIG. 9.
Effects of substitutions for Y2441 on viral cytopathogenicity and viability. MDBK cells were transfected with RNA transcripts from the indicated full-length cDNA clones as described in Materials and Methods. The RNA-specific infectivity data from one experiment are presented here. Shown are representative plates of a 10−3 dilution of the transfected cells showing crystal violet-stained plaques (top) or immunostained foci (bottom).
Interactions between NS4B and other BVDV nonstructural proteins.
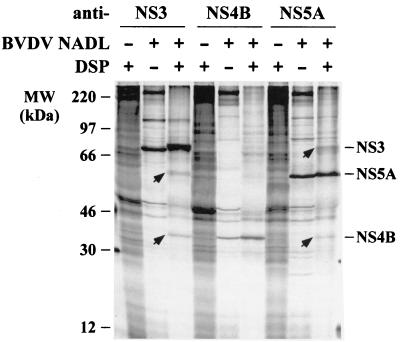
The fact that an ncp Y2441 substitution in NS4B abrogates CPE and alters the NS2-3/NS3 ratio strongly suggests a direct or indirect interaction between these proteins. However, our attempts to demonstrate a direct NS3-NS4B interaction by coimmunoprecipitation were unsuccessful. We then tried to address this question by cross-linking in vivo. NADL-infected cells were metabolically labeled and cross-linked with the cell-permeable and thiol-cleavable cross-linker DSP followed by immunoprecipitation with BVDV protein-specific antiserum. As shown in Fig. 10, detectable levels of NS4B and NS5A were recovered with anti-NS3 antiserum after cross-linking, indicating an association with NS3. Reciprocally, NS3 and NS4B were also recovered with anti-NS5A antiserum. Much lower levels of NS3 and NS5A were also found in the precipitate with anti-NS4B antiserum, yet due to the low level their identities need to be further confirmed. These data, although not sufficient to establish direct protein-protein interactions, strongly suggest associations between NS3, NS4B, and NS5A. Combining these results and the predicted membrane topology of NS4B (see Discussion), we propose that NS3, NS4B, and NS5A are components of a multiprotein complex that is associated with the ER membrane via the membrane-spanning NS4B. NS4B may serve as a membrane anchor and recruit these two otherwise cytoplasmically located proteins to the ER membrane where virus replication is thought to occur.
FIG. 10.
Cross-linking of NS4B with other BVDV nonstructural proteins. MDBK cells were infected with BVDV NADL at an MOI of 5, and at 18 h postinfection they were labeled with 35S-protein-labeling mixture for 4 h. Labeled cells were cross-linked with 1 mM DSP for 1 h, lysed, and immunoprecipitated with the following BVDV protein-specific antisera: WU165 (anti-NS3), WU168 (anti-NS4B), and WU170 (anti-NS5A). Immunoprecipitated proteins were solubilized by boiling them in standard SDS sample buffer in the presence of 5% β-ME and analyzed by SDS–10% PAGE. Only the relevant portions of the gel are shown. The presence (+) or absence (−) of virus or DSP is indicated above each lane. BVDV-specific proteins that coimmunoprecipitated with a given antibody target are indicated (arrows).
DISCUSSION
This work identifies NS4B as an important modulator of BVDV NADL cytopathogenicity and uncouples the cp phenotype from NS3 production. By engineering a dominant selectable marker into a permissive site in the BVDV 3′ NTR, ncp variants of an otherwise cp strain were isolated. Seven independent isolates shared a mutation in NS4B. This single substitution, Y2441C at residue 15 of NS4B, was sufficient to attenuate cytopathogenicity in the face of efficient NS3 synthesis, RNA accumulation, and infectious virus production. Although this residue is absolutely conserved among both cp and ncp pestiviruses, we showed that Y2441A also had an ncp phenotype, whereas Y2441V (a revertant isolated from the lethal Y2441D mutation) mimicked the cp parents. Thus, a Tyr residue at position 2441 is not essential for the cp phenotype, nor is Cys at 2441 required for an ncp phenotype on a cp genetic background.
Y2441 and the predicted membrane topology of NS4B.
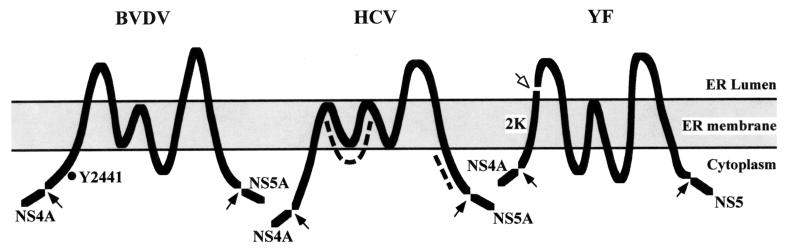
Since little is known about the function of the membrane-associated NS4B protein in replication of members of the Flaviviridae (38), defining a specific role for Y2441 is not possible at this time. To aid in interpreting our results and to facilitate future studies of NS4B, we constructed membrane topology models for BVDV, HCV, and YF NS4B. The results are shown in Fig. 11; specific details are provided in the figure legend. The N and C termini of BVDV NS4B are predicted to be cytoplasmic, consistent with their generation by the viral NS3-NS4A serine protease (46, 54). Six membrane-associated or membrane-spanning segments are predicted, resulting in two ER lumenal loops and one cytoplasmic loop. Polyprotein residue Y2441 (NS4B residue 15) is located in the N-terminal cytoplasmic segment, suggesting that an interaction(s) between NS4B and viral or cellular factors at the cytoplasmic face of the ER membrane is involved in modulating BVDV cytopathogenicity.
FIG. 11.
Membrane topology of NS4B of BVDV, HCV, and YF. Membrane topology of the Flaviviridae NS4B proteins was modeled using the protein prediction server PHD (39), as described in Materials and Methods. The GenBank accession numbers of the virus strains/types are: BVDV strain NADL (P19711), HCV type 1b (CAB46677), and YF vaccine strain 17D (CAA27332). The solid arrows indicate cleavage sites by the viral serine proteases. The open arrow indicates the ER lumen signal peptidase responsible for cleavage at the 2K/4B site of YF. The BVDV Y2441 residue and deletions in HCV NS4B that block NS5A hyperphosphorylation are indicated by the black dot and dashed curves, respectively. Details of the predictions follow. BVDV NS4B: a cytoplasmic N-terminal region (amino acids [aa] 1 to 80; BVDV NS4B numbering), a transmembrane segment (aa 81 to 98), a loop in the ER lumen (aa 99 to 136), three adjacent transmembrane segments (aa 137 to 154, 165 to 180, and 182 to 199), a cytoplasmic loop (aa 200 to 224), a transmembrane segment (aa 225 to 242), a second loop in the ER lumen (aa 243 to 286), a transmembrane segment (aa 287 to 304), and a C-terminal cytoplasmic tail (aa 305 to 347). HCV NS4B: a cytoplasmic N-terminal region (aa 1 to 46; HCV NS4B numbering), five adjacent transmembrane segments (aa 47 to 64, 69 to 86, 91 to 108, 113 to 133, and 138 to 155), a loop in the ER lumen (aa 156 to 172), a transmembrane segment (aa 173 to 190), and a C-terminal region in the cytoplasm (aa 191 to 261). YF NS4B: an N-terminal region translocated into the ER lumen (aa 1 to 36; YF NS4B numbering), a transmembrane segment (aa 37 to 54), a cytoplasmic loop (aa 55 to 81), two adjacent transmembrane segments (aa 82 to 99 and 104 to 122), a second cytoplasmic loop (aa 123 to 173), a transmembrane segment (aa 174 to 191), a loop in the ER lumen (aa 192 to 219), a transmembrane segment (aa 220 to 237), and a C-terminal cytoplasmic tail (aa 238 to 250). Note that for YF, the 2K cleavage product may correspond to the first transmembrane domain of BVDV and HCV NS4B (see Discussion).
HCV NS4B also has cytoplasmic N and C termini that are generated by the viral NS3-NS4A serine protease (15, 26) and six membrane-spanning or associated segments. Only a single large extramembranous loop is predicted, which is found in the ER lumen. This model is consistent with previous data for HCV NS4B. In particular, two deletions (amino acids 69 to 108 and 194 to 222) in HCV NS4B strongly inhibited NS5A hyperphosphorylation in an NS3-NS5A polypeptide but did not affect polyprotein cleavages (21). These data suggest that NS4B containing either of the two deletions can be properly translocated and cleaved. According to the topology model shown in Fig. 11, the amino acid 69 to 108 deletion abolishes two adjacent transmembrane segments with the junction on the cytoplasmic side of the ER membrane; the amino acid 194 to 222 deletion localizes to the C-terminal cytoplasmic region. Removal of either of these two segments should not affect the overall topology of NS4B, thus allowing proper processing. Both deletions affect sequences on the cytoplasmic face of the ER membrane, consistent with their involvement in hyperphosphorylation of NS5A, a hydrophilic protein without any apparent membrane-spanning segments.
The topology model for YF NS4B is distinct. Unlike those of BVDV and HCV NS4B proteins, the N terminus of YF NS4B is predicted to be in the ER lumen (Fig. 11). This is consistent with its generation by host signal peptidase (4, 6, 25). Similar to BVDV and HCV NS4B, the C terminus of YF NS4B is predicted to be cytoplasmic, consistent with its generation by the flavivirus NS2B-NS3 serine protease (5, 7, 13). Between the N and C termini there are five predicted membrane-spanning segments, giving rise to two cytoplasmic loops and one ER lumenal loop. This computer-predicted model is in almost perfect agreement with a previous model predicted from biochemical data (25). The single exception is the location of the last transmembrane segment, which in the previous model was predicted to be at amino acids 191 to 208 (rather than amino acids 220 to 237). In any case, the most striking difference between YF NS4B and the pestivirus and HCV NS4B is that it lacks the cytoplasmic N terminus and the first transmembrane segment. However, it has been found that cleavage at a novel site (called the 4A/2K site) in the YF NS4A region creates a small (23 amino acids) transmembrane peptide preceding NS4B (called the 2K protein) (25). The N terminus of the 2K protein is cytoplasmic and generated by the YF NS2B-NS3 serine protease (25), while its C terminus is produced by signal peptide cleavage in the ER lumen (4, 6, 25). Thus, the 2K peptide may be topologically equivalent to the N-terminal cytoplasmic region and the first transmembrane segment of BVDV and HCV NS4B. It will be interesting to determine if the 2K protein remains functionally associated with YF NS4B after cleavage, perhaps playing a similar role in replication to the covalently linked N-terminal domain predicted for the pestiviruses and HCV.
These NS4B models in the context of the emerging biological functions of NS4B, such as its role in cytopathogenicity and its interactions with other replicase components, suggest that NS4B may be an important scaffold for macromolecular assemblies that affect cell biology as well as viral RNA replication. The presence of NS4B domains on both the cytoplasmic and lumenal faces of the ER membrane makes this protein an ideal candidate for mediating and coordinating interactions in and between these two compartments. Little is known about interactions between viral nonstructural proteins and components of the ER lumen; however, this possibility should certainly not be discounted. Recent work has uncovered a key role for the flavivirus NS1 protein, a glycosylated secretory protein, in early minus-strand RNA synthesis (27, 28). This NS1 function is mediated directly or indirectly through the flavivirus NS4A protein which, like NS4B, is predicted to contain membrane-spanning segments (27).
Role of NS4B in cytopathogenicity.
Until this study, NS3 production invariably correlated with BVDV cytopathogenicity. The fact that a single mutation in NS4B can block this dramatic phenotype suggests several possible models. If NS3 is indeed the trigger for BVDV-induced CPE and cell death, then NS4B might be a cofactor required for this effect. NS4B might directly interact with NS3 or function independently. The cross-linking data, although not sufficient to establish direct protein-protein interactions, suggested that NS4B, NS5A, and NS3 are components of a multiprotein complex. Mutations at Y2441, either to Cys or Ala, give rise to the same ncp phenotype and might do so by disrupting an association between NS4B and NS3 or by abolishing an interaction between NS4B and other host or viral component(s) required for NS3-mediated cytopathogenicity. It is also possible that NS4B itself is the trigger for cell death and that NS3 (or cleavage at the NS2-NS3 site) unmasks this potential. In either scenario, we suggest that the NS4B substitutions are likely loss-of-function mutations, since both the Cys and Ala substitutions for Y2441 have the same ncp phenotype. Since the N-terminal region of NS4B that includes Y2441 is highly conserved, a more comprehensive mutagenesis study with an effort to isolate second-site suppressors may help to clarify the function of this domain and identify interacting viral proteins. It will also be important to see if mutations in NS4B, in particular Y2441, can attenuate cytopathogenicity of cp BVDV isolates other than the NADL prototype.
Another potentially relevant observation concerns the apparent influence of NS4B on the NS2-3/NS3 ratio. Enhanced accumulation of NS2-3 relative to NS3 was observed in both isolated and reconstructed ncp variants (Fig. 4 and 7). The Y2441C mutation could increase the ratio of NS2-3 to NS3 by altering the processing efficiency at the NS2-NS3 site, stabilizing NS2-3 or by destabilizing NS3. This NS4B-mediated effect could occur either via direct or indirect mechanisms. For instance, altered NS2-NS3 processing, enhanced NS2-3 stability, or reduced NS3 stability might be due to a disrupted or weakened interaction with NS4B (or other proteins). If NS3 is indeed the effector of cytopathogenicity, one could argue that destabilization of NS3 might reduce its level to a point where there is insufficient protein to initiate deleterious effects on the host cell. However, we note that NS3 accumulated in cells infected with the BVDV ncp variants to levels that were similar to those of the cp parents at a time when dramatic CPE was observed. Alternatively, it may be the balance between NS2-3 and NS3 that is important, with NS2-3 having an antagonistic or antiapoptotic function. However, this explanation is not supported by the fact that NS2-3 does not exert a dominant antiapoptotic effect in cp BVDV isolates that contain duplicated portions of the NS region and express NS2-3 and NS3 independently (reviewed in reference 33). Since uncleaved NS2-3 is dispensable for BVDV RNA replication, as evidenced by the efficient replication of subgenomic replicons lacking NS2 (2, 47), it may be possible to rigorously exclude a role for NS2-3 if the Y2441C substitution is capable of suppressing cytopathogenicity of subgenomic replicons expressing only NS3-NS5B. Although the isolation of an ncp variant that produces NS3 is unique for BVDV, this is not the case for CSFV or BDV (reviewed in reference 33). In the absence of genome rearrangements or cellular inserts, both NS2-3 and NS3 are produced by CSFV isolates. In spite of NS3 production, most CSFV isolates are ncp in cell culture. It will be of interest to examine the NS2-3/NS3 ratios, NS3 stability, and NS3 levels for ncp CSFV and compare these results to those obtained for the BVDV ncp variants.
In summary, a genetic approach has uncovered an unexpected role for NS4B in pestivirus cytopathogenicity. Mutations in this protein can suppress CPE, regardless of NS3 production and high levels of BVDV RNA accumulation. Attenuating mutations localized to the N-terminal region of NS4B, a domain that is predicted to reside on the cytoplasmic side of intracellular membranes and perhaps functions via interaction with other viral proteins, such as NS3 or cellular factors. Future studies will better define the molecular mechanisms involved in pestivirus cytopathogenicity and the role of NS3 and NS4B in this process.
ACKNOWLEDGMENTS
We thank Rebecca Moran for expert technical assistance. We are also grateful to many colleagues for helpful discussions during the course of this work and to Ilya Frolov, Holly Hanson, Richard Hardy, John Majors, and Tina Myers for critical reading and editing of the manuscript.
This work was supported in part by grants from the Public Health Service (CA57973 and AI24134). L.K.M. was supported by a graduate fellowship from Women & Science.
REFERENCES
- 1.Baginski S G, Pevear D C, Seipel M, Sun S C, Benetatos C A, Chunduru S K, Rice C M, Collett M S. Mechanism of action of a pestivirus antiviral compound. Proc Natl Acad Sci USA. 2000;97:7981–7986. doi: 10.1073/pnas.140220397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behrens S-E, Grassmann C W, Thiel H-J, Meyers G, Tautz N. Characterization of an autonomous subgenomic pestivirus RNA replicon. J Virol. 1998;72:2364–2372. doi: 10.1128/jvi.72.3.2364-2372.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer J C, Bebenek K, Kunkel T A. Unequal human immunodeficiency virus type 1 reverse transcriptase error rates with RNA and DNA templates. Proc Natl Acad Sci USA. 1992;89:6919–6923. doi: 10.1073/pnas.89.15.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cahour A, Falgout B, Lai C-J. Cleavage of the dengue virus polyprotein at the NS3/NS4A and NS4B/NS5 junctions is mediated by viral protease NS2B-NS3, whereas NS4A/NS4B may be processed by a cellular protease. J Virol. 1992;66:1535–1542. doi: 10.1128/jvi.66.3.1535-1542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers T J, Grakoui A, Rice C M. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol. 1991;65:6042–6050. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chambers T J, McCourt D W, Rice C M. Yellow fever virus proteins NS2A, NS2B, and NS4B: identification and partial N-terminal amino acid sequence analysis. Virology. 1989;169:100–109. doi: 10.1016/0042-6822(89)90045-7. [DOI] [PubMed] [Google Scholar]
- 7.Chambers T J, Weir R C, Grakoui A, McCourt D W, Bazan J F, Fletterick R J, Rice C M. Evidence that the N-terminal domain of nonstructural protein NS3 from yellow fever virus is a serine protease responsible for site-specific cleavages in the viral polyprotein. Proc Natl Acad Sci USA. 1990;87:8898–8902. doi: 10.1073/pnas.87.22.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chon S K, Perez D R, Donis R O. Genetic analysis of the internal ribosome entry segment of bovine viral diarrhea virus. Virology. 1998;251:370–382. doi: 10.1006/viro.1998.9425. [DOI] [PubMed] [Google Scholar]
- 9.Collett M S, Larson R, Belzer S K, Retzel E. Proteins encoded by bovine viral diarrhea virus: the genomic organization of a pestivirus. Virology. 1988;165:200–208. doi: 10.1016/0042-6822(88)90673-3. [DOI] [PubMed] [Google Scholar]
- 10.Collett M S, Larson R, Gold C, Strick D, Anderson D K, Purchio A F. Molecular cloning and nucleotide sequence of the pestivirus bovine viral diarrhea virus. Virology. 1988;165:191–199. doi: 10.1016/0042-6822(88)90672-1. [DOI] [PubMed] [Google Scholar]
- 11.Collett M S, Wiskerchen M A, Welniak E, Belzer S K. Bovine viral diarrhea virus genomic organization. Arch Virol. 1991;1991(Suppl 3):19–27. doi: 10.1007/978-3-7091-9153-8_3. [DOI] [PubMed] [Google Scholar]
- 12.Elbers K, Tautz N, Becher P, Stoll D, Rumenapf T, Thiel H-J. Processing in the pestivirus E2-NS2 region: identification of proteins p7 and E2p7. J Virol. 1996;70:4131–4135. doi: 10.1128/jvi.70.6.4131-4135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falgout B, Pethel M, Zhang Y-M, Lai C-J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991;65:2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frolov I, McBride M S, Rice C M. cis-Acting RNA elements required for replication of bovine viral diarrhea virus-hepatitis C virus 5′ nontranslated region chimeras. RNA. 1998;4:1418–1435. doi: 10.1017/s1355838298981031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grassmann C W, Isken O, Behrens S E. Assignment of the multifunctional NS3 protein of bovine viral diarrhea virus during RNA replication: an in vivo and in vitro study. J Virol. 1999;73:9196–9205. doi: 10.1128/jvi.73.11.9196-9205.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu B, Liu C, Lin-Goerke J, Maley D R, Gutshall L L, Feltenberger C A, Del Vecchio A M. The RNA helicase and nucleotide triphosphatase activities of the bovine viral diarrhea virus NS3 protein are essential for viral replication. J Virol. 2000;74:1794–1800. doi: 10.1128/jvi.74.4.1794-1800.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoff H S, Donis R O. Induction of apoptosis and cleavage of poly(ADPribose) polymerase by cytopathic bovine viral diarrhea virus infection. Virus Res. 1997;49:101–113. doi: 10.1016/s0168-1702(97)01460-3. [DOI] [PubMed] [Google Scholar]
- 19.Kaufman R J. Double-stranded RNA-activated protein kinase mediates virus-induced apoptosis: a new role for an old actor. Proc Natl Acad Sci USA. 1999;96:11693–11695. doi: 10.1073/pnas.96.21.11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kibler K V, Shors T, Perkins K B, Zeman C C, Banaszak M P, Biesterfeldt J, Langland J O, Jacobs B L. Double-stranded RNA is a trigger for apoptosis in vaccinia virus-infected cells. J Virol. 1997;71:1992–2003. doi: 10.1128/jvi.71.3.1992-2003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koch J O, Bartenschlager R. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J Virol. 1999;73:7138–7146. doi: 10.1128/jvi.73.9.7138-7146.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kümmerer B M, Meyers G. Correlation between point mutations in NS2 and the viability and cytopathogenicity of bovine viral diarrhea virus strain Oregon analyzed with an infectious cDNA clone. J Virol. 2000;74:390–400. doi: 10.1128/jvi.74.1.390-400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kümmerer B M, Stoll D, Meyers G. Bovine viral diarrhea virus strain Oregon: a novel mechanism for processing of NS2–3 based on point mutations. J Virol. 1998;72:4127–4138. doi: 10.1128/jvi.72.5.4127-4138.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai V C, Kao C C, Ferrari E, Park J, Uss A S, Wright-Minogue J, Hong Z, Lau J Y. Mutational analysis of bovine viral diarrhea virus RNA-dependent RNA polymerase. J Virol. 1999;73:10129–10136. doi: 10.1128/jvi.73.12.10129-10136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin C, Amberg S M, Chambers T J, Rice C M. Cleavage at a novel site in the NS4A region by the yellow fever virus NS2B-3 proteinase is a prerequisite for processing at the downstream 4A/4B signalase site. J Virol. 1993;67:2327–2335. doi: 10.1128/jvi.67.4.2327-2335.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin C, Prágai B, Grakoui A, Xu J, Rice C M. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J Virol. 1994;68:8147–8157. doi: 10.1128/jvi.68.12.8147-8157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindenbach B D, Rice C M. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol. 1999;73:4611–4621. doi: 10.1128/jvi.73.6.4611-4621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindenbach B D, Rice C M. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71:9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mendez E, Ruggli N, Collett M S, Rice C M. Infectious bovine viral diarrhea virus (strain NADL) RNA from stable cDNA clones: a cellular insert determines NS3 production and viral cytopathogenicity. J Virol. 1998;72:4737–4745. doi: 10.1128/jvi.72.6.4737-4745.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meyers G, Rümenapf T, Thiel H-J. Insertion of ubiguitin-coding sequence identified in the RNA genome of a togavirus. In: Brinton M A, Heinz F X, editors. New aspects of positive-strand RNA viruses. Washington, D.C.: American Society for Microbiology; 1990. pp. 25–29. [Google Scholar]
- 31.Meyers G, Stoll D, Gunn M. Insertion of a sequence encoding light chain 3 of microtubule-associated proteins 1A and 1B in a pestivirus genome: connection with virus cytopathogenicity and induction of lethal disease in cattle. J Virol. 1998;72:4139–4148. doi: 10.1128/jvi.72.5.4139-4148.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyers G, Tautz N, Becher P, Thiel H-J, Kümmerer B M. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J Virol. 1996;70:8606–8613. doi: 10.1128/jvi.70.12.8606-8613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyers G, Thiel H-J. Molecular characterization of pestiviruses. Adv Virus Res. 1996;47:53–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- 34.Moennig V, Plagemann P G. The pestiviruses. Adv Virus Res. 1992;41:53–98. doi: 10.1016/s0065-3527(08)60035-4. [DOI] [PubMed] [Google Scholar]
- 35.Pestova T V, Hellen C U. Internal initiation of translation of bovine viral diarrhea virus RNA. Virology. 1999;258:249–256. doi: 10.1006/viro.1999.9741. [DOI] [PubMed] [Google Scholar]
- 36.Purchio A F, Larson R, Collett M S. Characterization of bovine viral diarrhea virus proteins. J Virol. 1984;50:666–669. doi: 10.1128/jvi.50.2.666-669.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed K E, Gorbalenya A E, Rice C M. The NS5A/NS5 proteins of viruses from three genera of the family Flaviviridaeare phosphorylated by associated serine/threonine kinases. J Virol. 1998;72:6199–6206. doi: 10.1128/jvi.72.7.6199-6206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: LippincottRaven Publishers; 1996. pp. 931–960. [Google Scholar]
- 39.Rost B, Sander C, Schneider R. PHD–an automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 40.Rümenapf T, Unger G, Strauss J H, Thiel H-J. Processing of the envelope glycoproteins of pestiviruses. J Virol. 1993;67:3288–3294. doi: 10.1128/jvi.67.6.3288-3294.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Schneider R, Unger G, Stark R, Schneider-Scherzer E, Thiel H-J. Identification of a structural glycoprotein of an RNA virus as a ribonuclease. Science. 1993;261:1169–1171. doi: 10.1126/science.8356450. [DOI] [PubMed] [Google Scholar]
- 43.Sooknanan R, Howes M, Read L, Malek L T. Fidelity of nucleic acid amplification with an avian myeloblastosis virus reverse transcriptase and T7 RNA polymerase. Biotechniques. 1994;17:1077–1085. [PubMed] [Google Scholar]
- 44.Steinhauer D A, Holland J J. Direct method for quantitation of extreme polymerase error frequencies at selected single base sites in viral RNA. J Virol. 1986;57:219–228. doi: 10.1128/jvi.57.1.219-228.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takizawa T, Ohashi K, Nakanishi Y. Possible involvement of doublestranded RNA-activated protein kinase in cell death by influenza virus infection. J Virol. 1996;70:8128–8132. doi: 10.1128/jvi.70.11.8128-8132.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tautz N, Elbers K, Stoll D, Meyers G, Thiel H-J. Serine protease of pestiviruses: determination of cleavage sites. J Virol. 1997;71:5415–5422. doi: 10.1128/jvi.71.7.5415-5422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tautz N, Harada T, Kaiser A, Rinck G, Behrens S, Thiel H J. Establishment and characterization of cytopathogenic and noncytopathogenic pestivirus replicons. J Virol. 1999;73:9422–432. doi: 10.1128/jvi.73.11.9422-9432.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thiel H-J, Plagemann P G W, Moennig V. . B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. 3rd ed. Vol. 1. New York, N.Y: Lippincott-Raven Publishers; 1996. Pestiviruses pp. 1059–1073. [Google Scholar]
- 49.Vassilev V B, Donis R O. Bovine viral diarrhea virus induced apoptosis correlates with increased intracellular viral RNA accumulation. Virus Res. 2000;69:95–107. doi: 10.1016/s0168-1702(00)00176-3. [DOI] [PubMed] [Google Scholar]
- 50.Ward C D, Flanegan J B. Direct measurement of the poliovirus RNA polymerase error frequency in vitro. J Virol. 1988;62:558–562. doi: 10.1128/jvi.62.2.558-562.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Warrener P, Collett M S. Pestivirus NS3 (p80) protein possesses RNA helicase activity. J Virol. 1995;69:1720–1726. doi: 10.1128/jvi.69.3.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiskerchen M, Belzer S K, Collett M S. Pestivirus gene expression: the first protein product of the bovine viral diarrhea virus large open reading frame, p20, possesses proteolytic activity. J Virol. 1991;65:4508–4514. doi: 10.1128/jvi.65.8.4508-4514.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.World Health Organization. Hepatitis C: global prevalence. Wkly Epidemiol Rec. 1997;72:341–344. [PubMed] [Google Scholar]
- 54.Xu J, Mendez E, Caron P R, Lin C, Murcko M A, Collett M S, Rice C M. Bovine viral diarrhea virus NS3 serine proteinase: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J Virol. 1997;71:5312–5322. doi: 10.1128/jvi.71.7.5312-5322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu H, Grassmann C W, Behrens S E. A stem-loop motif formed by the immediate 5′ terminus of the bovine viral diarrhea virus genome modulates translation as well as replication of the viral genome. J Virol. 2000;74:5825–5835. doi: 10.1128/jvi.74.13.5825-5835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang G, Aldridge S, Clarke M C, McCauley J W. Cell death induced by cytopathic bovine viral diarrhea virus is mediated by apoptosis. J Gen Virol. 1996;77:1677–1681. doi: 10.1099/0022-1317-77-8-1677. [DOI] [PubMed] [Google Scholar]
- 57.Zhong W, Gutshall L L, Del Vecchio A M. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J Virol. 1998;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]