Abstract
Nuclear export of incompletely spliced RNAs is a prerequisite for retroviral replication. Complex retroviruses like human immunodeficiency virus (HIV) encode a viral transport factor (Rev), which binds to its target sequence on the RNA genome and directs it into the Crm-1-mediated export pathway. Other retroviruses, like Mason-Pfizer monkey virus, contain cis-acting constitutive RNA transport elements (CTE) which achieve nuclear export of intron-containing RNA via cellular transport factors. Here, we describe the identification and characterization of a novel cis-acting orientation-dependent RNA expression element in the coding region of the murine intracisternal A-type particle (IAP) MIA14. This IAP expression element (IAPE) can functionally replace the Rev system in the expression of HIV-1 Gag proteins but functions independently of Crm-1. The presence of this element is needed for the expression of the IAP Gag proteins, indicating its biological significance. The IAPE can be functionally replaced by placing a CTE on the MIA14 RNA, further supporting its role in mRNA export. Northern blot analysis revealed that total RNA, as well as cytoplasmic RNA, was increased when the element was present. The element was mapped to a predicted stem-loop structure in the 3′ part of the pol open reading frame. There was no overall homology between the IAPE and the CTE, but there was complete sequence identity between short putative single-stranded loops. Deletion of these loops from the IAPE severely reduced Rev-independent Gag expression.
Gene expression is a highly ordered process which is controlled at both the transcriptional and posttranscriptional levels. Prior to their export to the cytoplasm, most pre-mRNAs are modified by removal of intronic sequences, addition of a cap structure to their 5′ends, and cleavage and polyadenylation of their 3′ends (19). Completion of posttranscriptional modifications is a prerequisite for nuclear export, and intron-containing RNAs are generally retained and degraded in the nucleus (24, 25, 53). Retroviruses, on the other hand, require nuclear export of intron-containing RNAs and therefore have devised several strategies to overcome this export block. Most retroviruses produce a single primary transcript which corresponds to the viral RNA genome. This primary transcript serves as mRNA for the inner structural proteins (Gag) and replication enzymes (Pol) but also undergoes one or several splicing events leading to distinct classes of mRNAs. This mode of gene expression fulfils the need for genetic economy but requires the presence of inefficient splice sites and a mechanism for nuclear export of incompletely spliced RNAs (6, 24, 46, 53, 59).
According to their genome organizations, retroviruses are divided into simple and complex viruses. Simple retroviruses encode Gag and Pol proteins from the unspliced RNA and encode the glycoproteins of the viral envelope (Env) from a singly spliced mRNA. Complex retroviruses, like human immunodeficiency virus type 1 (HIV-1), contain several additional open reading frames encoding regulatory proteins that are synthesized from multiply spliced RNAs. Multiply spliced RNAs lack introns and are normally exported from the nucleus, while other viral mRNAs, and in particular the genomic RNA, would normally be retained in the nucleus due to the continued presence of introns (6, 46, 59). To overcome this problem, HIV-1 and other complex retroviruses make use of a posttranscriptional regulatory system consisting of the viral Rev protein and its target RNA structure, the Rev-responsive element (RRE). The RRE is located in the env coding region and is present on all incompletely spliced transcripts (11, 36). Complete removal of intronic sequences removes the RRE, thus discriminating between Rev-dependent and Rev-independent transcripts (46). Nuclear export of RRE-containing RNAs requires Rev binding and multimerization. To achieve its function, Rev contains two distinct domains: an RNA binding domain, which recognizes the RRE and serves as a nuclear localization signal (NLS) (2, 35, 36), and a leucine-rich nuclear export signal (NES) promoting binding to the cellular export receptor Crm-1, a member of the importin-β superfamily of proteins (12, 13, 58).
The export problem is similar in the case of simple retroviruses, but the solutions are different: the genomic RNAs of these viruses also contain introns and have to be exported to the cytoplasm, but there is no Rev-like factor. cis-acting RNA elements that promote efficient nuclear export of viral RNAs in the absence of viral trans-acting factors were first defined in the genomes of viruses from the D-type subgroup of retroviruses (Mason-Pfizer monkey virus [M-PMV] and simian retrovirus types 1 and 2) (4, 65). These elements are located in the intergenic region between env and the 3′ long terminal repeat and have been termed constitutive transport elements (CTE). The CTE are necessary for viral replication and enable cytoplasmic accumulation of the unspliced RNA, while the spliced env mRNA is CTE independent (9). The CTE can functionally replace the export system of complex retroviruses, leading to infectious virus lacking Rev and the RRE, although replication is impaired compared to that of wild-type virus (4, 65). The CTE corresponds to an imperfect inverted repeat folding into a stable secondary structure with two internal loops (8, 9, 55). Mutation of loop sequences abrogated CTE function and inhibited nuclear export of CTE-containing RNAs upon microinjection into Xenopus oocytes, thus defining these regions as potential docking sites for cellular export factors (44, 50). A cellular protein that specifically interacts with the CTE is the Tip-associated protein Tap (15, 63). Tap is a nucleocytoplasmic shuttling protein and contains an RNA binding domain that specifically interacts only with the functional CTE, indicating that Tap is indeed the cellular CTE export factor (3, 28, 29).
A functional CTE has also been observed in the case of the avian retrovirus Rous sarcoma virus (RSV). The RSV genome contains two direct repeats (DR) flanking the src oncogene which mediate cytoplasmic accumulation of unspliced RSV transcripts and weakly substitute for Rev function, as shown by analysis in avian cells (41, 42, 52). Recently, the DR sequences have been shown to mediate nucleocytoplasmic transport of a heterologous RNA in a Crm-1-independent manner, confirming their intrinsic RNA export function (43). CTE-homologous sequences have been suggested for other retroviruses on the basis of computer predictions, but the majority of simple retroviruses do not appear to contain CTE-homologous sequences (8, 54).
Posttranscriptional control elements have also been detected in endogenous retroviruses and pararetroviruses (7, 8, 20, 54). Pararetroviruses, like hepatitis B virus (HBV) replicate via reverse transcription but without chromosomal integration. They contain posttranscriptional regulatory RNA elements (PRE), which substitute for Rev/RRE but appear to function by a different mechanism exerting pleiotropic effects on RNA stability, modification, and export (7, 20, 22). Endogenous retroviruses are stably integrated into the cellular genome, mostly defective, and vertically transmitted (31). A Rev/RRE-analogous system has been observed for the HERV-K subgroup of human endogenous retroviruses (34, 62), while sequences homologous to the CTE have been described for a few members of the endogenous retrovirus group of intracisternal A-type particles (IAPs) (54). Although IAPs are present in rodent cells at 1,000 to 2,000 copies per haploid genome (31), only 10 CTE-like elements were identified by sequence comparison in IAPs and related sequences (8, 54). Only one of these elements led to enhanced gene expression when introduced into a heterologous context, indicating that most IAPs are not expressed via CTE-mediated pathways.
Here, we report the identification and characterization of a new cis-acting RNA element located in the pol region of the murine IAP MIA14 (39). This element was termed the IAP expression element (IAPE). It is required for expression of IAP Gag proteins but can be functionally replaced by a CTE. It efficiently substitutes for Rev/RRE in the expression of HIV-1 structural proteins but shows no overall homology to the CTE. Enhanced expression appears to be due to pleiotropic effects on RNA stability and export. We have mapped the IAPE to a putative stem-loop structure and identified functional domains by mutational analysis. We also provide evidence for the presence of functionally similar cis-acting elements in the pol region of primate foamy virus, suggesting a general role in retroviral gene expression which may be partly conserved in pararetroviruses.
MATERIALS AND METHODS
Plasmids.
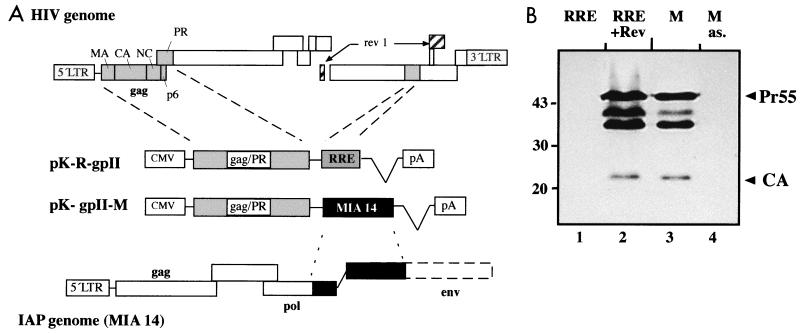
The eukaryotic expression plasmids pK-R-gpII (with the RRE) and pK-gpII (without the RRE) have been described elsewhere (38, 60). Both contain part of the 5′ untranslated region and the complete gag and protease (PR) region of HIV-1 under control of the cytomegalovirus (CMV) promoter-enhancer with simian virus 40 (SV40) splice and polyadenylation sequences. The HIV-1-specific sequences were excised using flanking EcoRI sites and cloned into pBluescript SK(+) to give pBSG. A 1.6-kb fragment corresponding to nucleotides (nt) 4683 to 6291 of the MIA14 sequence (39) was excised with NotI and SmaI and cloned into pK-gpII (pK-gpII-M [Fig. 1]). A similar expression plasmid containing the same 1.6-kb fragment in antisense orientation (pK-gpII-Mas) was cloned by inserting the SmaI-NotI fragment into pBSG and subsequent cloning of an XhoI-NotI fragment into pKex (47).
FIG. 1.
Schematic representation of expression plasmids and analysis of HIV-1 Gag expression. (A) All expression plasmids contain part of the 5′ untranslated region of HIV-1 as well as the gag and PR coding regions. RNA elements derived from HIV-1 (RRE; pK-R-gpII) or from the IAP MIA14 (pK-M-gpII) were inserted 3′ of the coding region and are depicted as shaded (RRE) or solid (MIA14) boxes. At the top, the relative positions of the gag-PR region and of the RRE, as well as the coding exons for Rev, are highlighted in the HIV-1 genome. At the bottom, the relative position of the murine element is highlighted in the IAP genome. (B) Immunoblot analysis of transfected HeLa cells using antiserum against HIV-1 CA. Cells were transfected with plasmid pK-R-gpII (in the absence or presence of Rev [lanes 1 and 2]) or pK-gpII-M (containing the IAPE in sense or antisense [as.] orientation [lanes 3 and 4]). The cell lysates were normalized for transfection efficiency. The Gag polyprotein (Pr55) and CA are marked on the right; molecular mass standards (in kilodaltons) are given on the left. The additional proteins migrating between Pr55 and CA correspond to the Gag intermediate cleavage products MA-CA-NC and MA-CA.
In order to map the expression element, various subfragments of the 1.6-kb sequence were excised using restriction enzymes (see Fig. 3A) and inserted into pK-gpII. The plasmids pK-gpII-IAPE sense and antisense were constructed by PCR amplification of nt 5443 to 5811 of the MIA14 sequence. The primers were 5′ MIA (GCTCTAGAGCCAGATAGGCCC), introducing an XbaI site (underlined), and 3′ MIA (GGACTAGTAGAGCCAAGGCTGTCTG), introducing an SpeI site (underlined). The PCR product was digested with XbaI and SpeI and cloned into the XbaI site of pBSG, resulting in inserts in both orientations. Subsequently, XhoI-NotI fragments were cloned into pKex which had been cleaved wih SalI and NotI. The mutated variants were cloned using the same strategy, except that the initial PCR was performed as an overlap PCR to introduce point mutations or deletions. The DI variant was constructed by initially performing two separate reactions using primers 5′MIA (see above) and 3′MIA-DI (CTGGTATCCAAAAAGCTTTCTGTGG) or 5′MIA-DI (CCACAGAAAGCTTTTTGGATACCAG) and 3′MIA. Both PCR fragments were mixed and reamplified using the external primer pair 5′and 3′ MIA. Primers 5′MIA-DII (GTCCAGGGCGTCCGGAAGCCAGCG) and 3′ MIA-DII (CGCTGGCTTCCGGACGCCCT GGAC) and primers 5′MIA-MI (GCTCTAGAGCCAGATAGGCCCCCCGAGATGGTTAAATGG) and 3′MIA-MI (GGACTAGTAGAGCCAAGGCCCCCCGAATTATGGC) served as internal primers for construction of the variants DII and MI, respectively. The presence of the mutations and deletions was verified by sequencing in all cases.
FIG. 3.
Mapping of the MIA14 expression element. (A) Schematic representation depicting the results of a functional analysis of various subfragments derived from the MIA14 element. The relevant restriction sites used for cloning of the subfragments are indicated. All subfragments were inserted into pK-gpII, and transiently transfected cells were analyzed for HIV-1 Gag expression. At least three independent experiments were performed for each construct. The relative (rel.) fluorescence intensity of each fragment is scored on the right. Subregions mediating Rev-independent Gag expression are shown as a solid line; those yielding negative results are depicted as a dashed line. Insertion of a subfragment in reverse orientation is termed antisense (a.s.). The central darkly shaded region corresponds to the region defined by structure prediction analysis (Fig. 4) and termed the IAPE. The lightly shaded region corresponds to a nonoverlapping sequence mediating weak Rev-independent Gag expression. (B) Western blot analysis of HeLa cells transfected with pK-R-gpII, with (+) or without Rev (lanes 1 and 2), pK-gpII-M (lane 3), or pK-gpII-IAPE (lane 4). HIV-1-specific products and marker protein were detected as described in the legend to Fig. 1. (C) Northern blot analysis of nuclear (nuc.; lanes 1 to 3) and cytoplasmic (cyt.; lanes 4 to 6) RNA preparations from HeLa cells transfected with plasmids pK-R-gpII, without and with (+) Rev (lanes 1 and 2 and 4 and 5), or pK-gpII-IAPE (lanes 3 and 6). The blot was hybridized with a radiolabeled probe directed against the gag-PR region present in all constructs. The arrowheads point to specific gag RNAs. The difference in migration results from size differences of the corresponding elements. The additional RNAs of lower mobility correspond to products derived from transcriptional readthrough into the hygromycin resistance transcription unit located 3′ of the polyadenylation signal. The same blot was subsequently stripped and reanalyzed with a probe specific for the cellular housekeeping GAPDH gene (bottom).
The pcDNA3-derived plasmids 3-RRE, 3-CTE, and 3-IAPE were all made from the respective pK-gpII expression vectors by inserting the whole expression cassette, including the CMV promoter, the Gag-PR coding sequence, and the respective RNA element, as ScaI-NotI fragments into pcDNA3, which had been opened with the same enzymes. This replaced the splice and polyadenylation signal from SV40 (present in pKex) by the polyadenylation signal from the bovine growth hormone (BGHpA). Plasmid pL-MIA4 has been described before (57). It contains the MIA14-specific Gag and Pol coding sequence with a replacement of the 5′ 29 codons of gag by the heterologous membrane-targeting signal of Src, which directs the resulting Gag polyprotein to the plasma membrane (57). The derivatives pL-MIA4 ΔSal and ΔNot were constructed by excising the respective coding region, including the CMV promoter, from pL-MIA4 as an SpeI-SalI or SpeI-NotI fragment and inserting it into pKex. For control plasmid 3-MIA4 ΔNot (Fig. 2B), the same SpeI-Not fragment was inserted into pcDNA3.1 (Invitrogen), replacing the CMV promoter, and subsequently, the CTE sequence was inserted into the 3′ untranslated region in order to generate 3-MIA4 ΔNot-CTE. Plasmids pK-gpII-Fo sense and antisense were constructed by PCR amplification of the region from nt 6128 to 6683 from the human foamy virus genome (GenBank accession number U21247.1) with primers 5′ Fo (GGTCTAGAAACAACACCTATAGCCC), introducing an XbaI site (underlined), and 3′ Fo (GGGCTAGCACAACAAGTATAAAGC), introducing an NheI site (underlined). The resulting PCR fragment was cleaved with XbaI and NheI and cloned into the XbaI site (giving both orientations) of pBSG, and SalI-NotI fragments from the resulting plasmids were transferred into pKex.
FIG. 2.
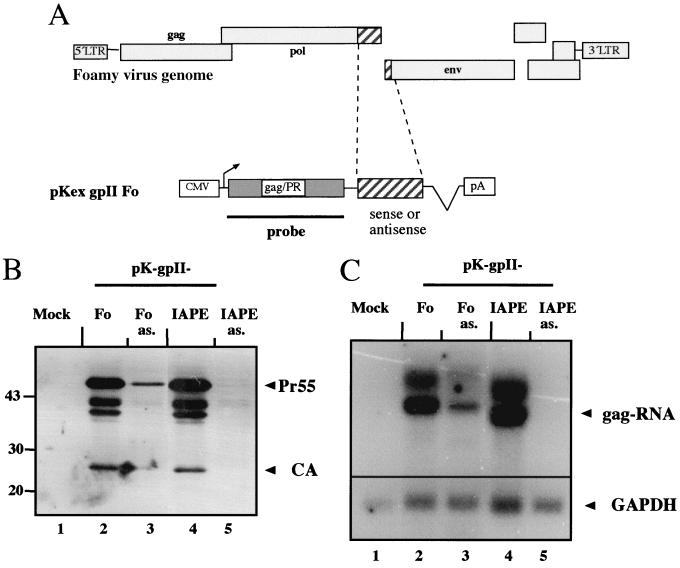
Analysis of IAP Gag expression. (A) Schematic diagram of plasmid pL-MIA4 encoding the gag and pol open reading frames of MIA14 (57) and plasmids 3-MIA4ΔN without and 3-MIA4ΔN-CTE with CTE. The relative positions of the restriction sites used for the deletion constructs pL-MIA4ΔS and pL-MIA4ΔN, as well as the positions of the RNA elements, are indicated. (B) Immunoblot analysis of Cos-7 cells transiently transfected with plasmids pL-MIA4, pL-MIA4ΔS, and pL-MIA4ΔN (lanes 2, 3, and 4) or mock transfected (lane 1) (left blot) and with plasmids 3-MIA4ΔN (lane 5)and 3-MIA4ΔN-CTE (lane 6) (right blot). The cell lysates were analyzed with a polyclonal rabbit anti-IAP Gag serum, and the IAP Gag polyprotein is marked with arrowheads. The additional proteins migrating with higher mobility than the Gag polyprotein correspond to Gag cleavage products (lane 2) (57). (C) Northern blot analysis of Cos cells mock transfected (lane 1) or transfected with pL-MIA4 or pL-MIA4ΔN (lanes 2 and 3). Total RNA was subjected to Northern blot analysis, and IAP Gag-specific RNA was detected with a 32P-labeled probe directed against the gag coding region, as depicted in panel A. The gag RNA is indicated on the right, and RNA standards (in nucleotides) are given on the left.
Cells and transfection.
Cos-7 cells and HeLa P4 cells were maintained in Dulbecco modified Eagle medium supplemented with 10% heat-inactivated fetal calf serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 2 mM glutamine. For transient transfection, 5 × 105 cells were seeded in a 10-cm-diameter dish and grown overnight. The medium was exchanged prior to transfection, and 10 μg of the respective plasmid and 1 μg of a reporter construct encoding enhanced green fluorescent protein (eGFP) were transfected using the modified calcium phosphate coprecipitation technique (5). To assess the transfection efficiency, coverslips were fixed for 10 min with 4% paraformaldehyde in phosphate-buffered saline (PBS) at 48 h posttransfection, and GFP-positive cells were counted.
Western blot analysis and enzyme-linked immunosorbent assay.
Cells were harvested at 48 h posttransfection. The cell lysates were normalized for transfection efficiency according to the number of GFP-positive cells and analyzed on 17.5% sodium dodecyl sulfate-polyacrylamide gels (200:1 ratio of acrylamide to N,N-methylenebisacrylamide). The proteins were transferred to nitrocellulose (0.45 μm; Schleicher and Schuell) for 2 h at room temperature, and the membranes were blocked with 10% dry milk in PBS for 1 h and stained overnight with antiserum against HIV-1 capsid protein (CA; 1:2,500 in 5% dry milk in PBS containing 0.5% Triton X-100) or IAP Gag (diluted 1:2,000; kindly provided by K. Lueders). Following an additional blocking step, the blots were incubated for 2 h with peroxidase-conjugated anti-rabbit secondary antibody (diluted 1:10,000; Dianova). Enhanced chemiluminescence (Amersham) was used for detection of specific signals according to the manufacturer's protocol. The blots were exposed to Kodak X-Omat-AR films.
RNA preparation and analysis.
Preparation of total or fractionated RNA was performed as described previously (60). The nuclear and cytoplasmic fractions were subjected to RNA extraction by the RNAzol B method (WAK chemicals) according to the manufacturer's protocol. The RNA content was determined photometrically or by gel analysis. Five micrograms of each RNA was loaded on denaturing formaldehyde gels containing 1% agarose and transferred to Biodyne B membrane (0.45 μm; Pall) by capillary blotting. The transfer efficiency was determined by staining the blot with methylene blue (0.04% in 0.5 M NaOAc, pH 5.2). The blots were prehybridized for >1 h at 42°C in 50% formamide–6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–5× Denhardt solution–0.5% sodium dodecyl sulfate containing 100 μg of denatured salmon sperm DNA/ml and hybridized overnight at 42°C with the specific probes (activity, >107 cpm), which had been labeled by random priming using the Prime It II kit (Stratagene). The blots were washed, sealed, and exposed to X-ray films (Kodak X-Omat-AR) or quantified by phosphorimage analysis.
RESULTS
A sequence from the IAP MIA14 promotes Rev-independent production of the HIV-1 Gag polyprotein.
Expression of HIV-1 structural (Gag) proteins requires binding of the viral Rev protein to the RRE, leading to nuclear export of the mRNA. In our analysis of HIV-1 assembly, we constructed chimeric expression plasmids fusing HIV-1 sequences to segments of the genome of the mouse IAP MIA14 (39) and observed that some of these constructs allowed Rev-independent expression of HIV-1 Gag. For a more detailed analysis, we inserted a 1.6 kb-fragment corresponding to the 3′ part of the MIA14 pol open reading frame and part of the defective env gene in sense or antisense orientation downstream of the HIV-1 Gag-PR coding region into the eukaryotic expression plasmid pK-gpII (Fig. 1A). Cos-7 cells were transfected with these plasmids or with the RRE-containing plasmid pK-R-gpII (in the presence or absence of Rev), and cell lysates normalized for GFP were analyzed by immunoblotting them using antiserum specific for the HIV-1 CA. Cells transfected with pK-R-gpII exhibited a strong Rev-dependent signal corresponding to the Pr55gag polyprotein, the proteolytically cleaved CA protein, and two intermediate cleavage products (Fig. 1B, lanes 1 and 2). A comparable but slightly weaker signal was seen independent of Rev when the plasmid containing MIA14 sequences in sense orientation was transfected (Fig. 1B, lane 3). In contrast, no HIV-1-specific product was observed when the same sequences were present in antisense orientation (Fig. 1B, lane 4), indicating that MIA14 contains an orientation-dependent RNA element that can functionally substitute for the HIV-1 Rev/RRE system.
The MIA14 element is necessary for expression of the structural proteins of the IAP and can be functionally replaced by a CTE.
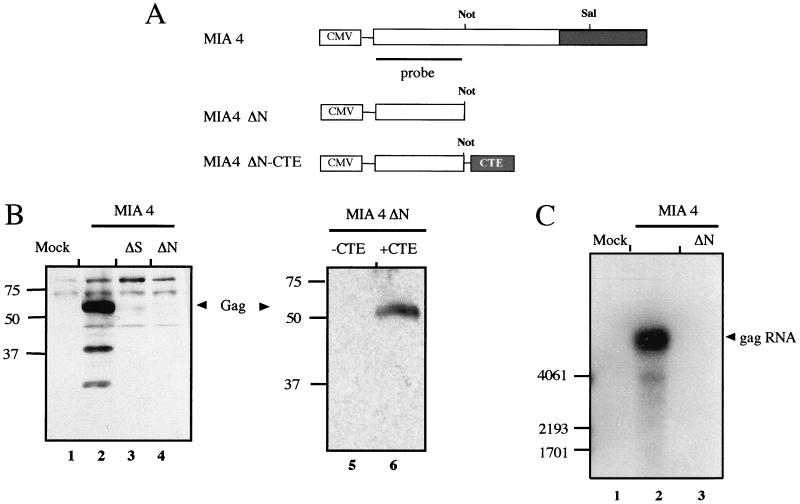
In order to test whether the sequences from MIA14 are important for production of the virus' own structural proteins, we constructed expression vectors for MIA14 Gag proteins. Plasmid pL-MIA4 contains the entire gag open reading frame with a modification in the 5′ matrix region and the complete pol open reading frame, including the 1.6-kb region (57) (Fig. 2A). Derivatives truncated at SalI and NotI sites and partially or completely lacking this 1.6-kb region were termed ΔS and ΔN, respectively (Fig. 2A). Plasmids were transfected into Cos-7 cells, and cell lysates normalized for transfection rate were analyzed by immunoblotting them using a polyclonal rabbit serum against purified IAP Gag. The MIA14 Gag polyprotein and two cleavage products were readily observed in cells transfected with pL-MIA4 (Fig. 2B, lane 2), as described by Welker et al. (57). Cells transfected with pL-MIA4ΔS gave a weak but detectable signal corresponding to the Gag polyprotein (Fig. 2B, lane 3), while no specific signal was observed in the case of pL-MIA4ΔN-transfected cells (Fig. 2B, lane 4). Similar observations were made when transfected cells were analyzed by immunofluorescence, where strong Gag-specific signals were seen for pL-MIA4 while pL-MIA4ΔS gave a weak signal and pL-MIA4ΔN produced no Gag-specific signal (data not shown). Importantly, the MIA14 Gag polyprotein is efficiently translated in vitro from an RNA truncated at the NotI site and thus corresponding to the ΔN construct (reference 10 and data not shown). IAP Gag expression in transfected cells, therefore, appears to require an RNA element which is not needed in vitro.
To analyze directly whether the deleted fragment could be functionally replaced by an export-promoting sequence, we introduced the CTE of M-PMV, which is known to act as an RNA export element (9), into the ΔN construct (Fig. 2A). Cells were transfected with the plasmids 3-IAPΔN and 3-IAPΔN-CTE and analyzed for IAP Gag expression by immunoblot analysis. Figure 2B shows that there was no IAP Gag expression in cells transfected with the construct lacking the murine element and not containing a CTE (Fig. 2B, lane 5), while IAP Gag expression was efficiently restored in the presence of the CTE (Fig. 2B, lane 6).
To analyze whether differences are also observed on the RNA level, we performed Northern blot analysis of Cos-7 cells transfected with pL-MIA4 or pL-MIA4ΔN. Total RNA from transfected cells normalized for transfection rate was analyzed using a radiolabeled probe detecting the Gag coding region present in both constructs (Fig. 2A). A strong and specific RNA signal was observed for pL-MIA4-transfected cells (Fig. 2C, lane 2), while pL-MIA4ΔN-transfected cells gave a weak RNA signal which was only detected on long-term exposure of the blot (Fig. 2C, lane 3, and data not shown). Taken together, these results suggest that the MIA14 element is not directly needed for protein synthesis but rather for stable intracellular RNA expression and export.
The MIA14 element maps to the pol region and corresponds to a putative stable RNA structure.
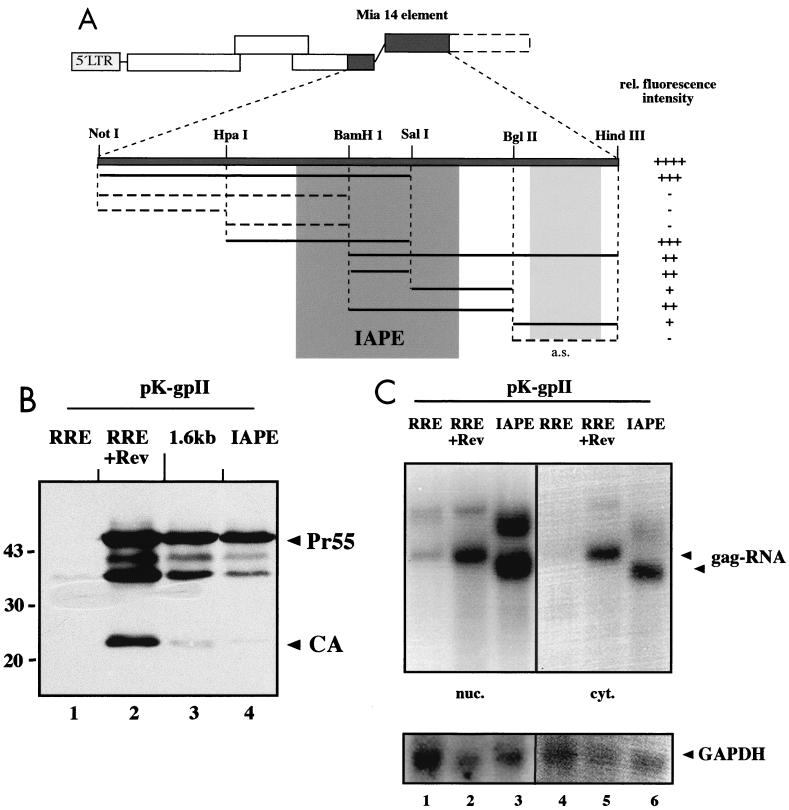
In order to map the newly discovered element more precisely, we constructed expression vectors containing various subfragments of the MIA14 1.6-kb region downstream of the HIV-1 gag gene (Fig. 3A) and analyzed their capacity to promote Rev-independent Gag expression after transient transfection. Cos-7 cells were transfected with the different pK-gpII derivatives, and HIV-1 Gag expression was determined by immunoblotting using CA-specific antiserum or by indirect immunofluorescence. At least three independent experiments were performed for each construct. The results of this analysis are summarized in Fig. 3A, and the relative activity of each fragment is indicated as fluorescence intensity. In general, all subfragments containing the central part of the 1.6-kb element led to Rev-independent expression of HIV-1 Gag, albeit at different efficiencies (Fig. 3A). In contrast, insertion of the 835-nt 5′ NotI-BamHI fragment into pK-gpII did not permit any Gag expression above background. Based on the analysis presented in Fig. 3A, it can be concluded that the main activity resides in the central BamHI-SalI fragment (nt 5501 to 5700 of MIA14), which promoted Rev-independent HIV-1 Gag expression by itself but was enhanced when flanking sequences were present in addition (data not shown). Interestingly, the nonoverlapping 3′ BglII-HindIII fragment also promoted weak, orientation-dependent Gag expression, but this phenotype was not analyzed in more detail.
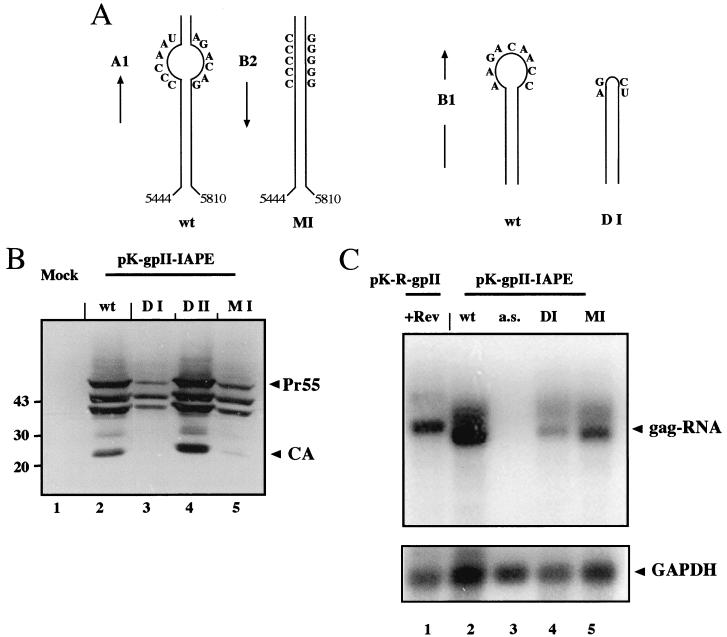
RNA elements commonly correspond to stable secondary structures containing short single-stranded regions with defined specificity for RNA binding proteins. In an independent attempt to map a functional subregion of the MIA14 element, we performed a computer analysis of the folding potential of the 1.6-kb region using the program Mulfold for RNA secondary-structure prediction (26, 27). Initially, a prediction of folding potential was made for the entire 1.6-kb region. Comparison of the 10 most stable predictions revealed a putative stable stem-loop structure in the region from nt 5444 to 5810 of the MIA14 genome. This structure remained stable in folding predictions upon systematic shortening of either the 5′part (nt 4683 to 5443) or the 3′part (nt 5811 to 6291) or both. Comparison of the most stable predictions for the 366-nt region from nt 5444 to 5810 showed a consistent structure with minor differences in loop regions. Sequence analysis of this region revealed several differences from the sequence published by Mietz et al. (39), and the corrected sequence contained only 360 nt. These changes did not alter the folding potential of this region, and its predicted structure is depicted in Fig. 4. Importantly, the region identified by folding prediction (the IAPE [Fig. 3A]) completely overlapped with the experimentally determined active region of the 1.6-kb element (Fig. 3A). To test whether the IAPE promoted Rev-independent HIV-1 Gag expression, pK-gpII derivatives containing this region in sense or antisense orientation were constructed and transfected into HeLa P4 cells. Subsequent immunoblot (Fig. 3B) and immunofluorescence (data not shown) analyses indicated that the IAPE promoted Gag expression almost as efficiently as the entire 1.6-kb fragment (Fig. 3B, lanes 3 and 4), while the same sequence in antisense orientation was completely inactive (data not shown). This result indicates that the IAPE corresponds to the main functional region of the MIA14 element
FIG. 4.
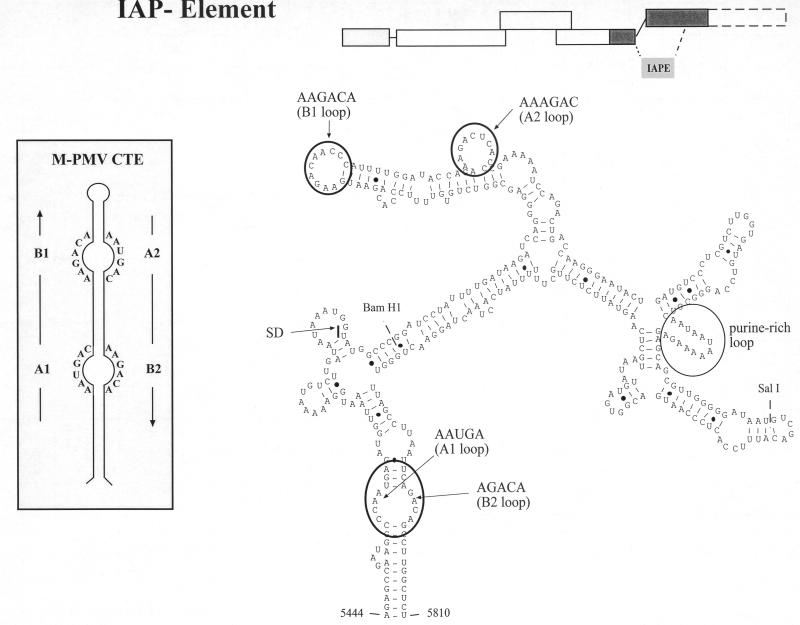
Sequence and predicted secondary structure of the IAPE. The sequence mediating Rev-independent HIV-1 Gag expression was subjected to computer prediction of folding potential using the Mulfold program. This analysis yielded a stable secondary structure corresponding to nt 5444 to 5810 of the MIA14 sequence (shown schematically at the upper right) originally published by Mietz et al. (39). Resequencing of this region showed seven deletions, one insertion, and one nucleotide exchange compared to the original sequence. The corrected sequence was subsequently subjected to folding analysis, yielding the depicted secondary-structure element of 360 nt (IAPE). The calculated free energy value for the proposed RNA structure of the IAPE is −95.8 kcal. A predicted splice donor (SD) within the element is marked. In addition, the BamHI and the SalI site within the element are shown (compare to Fig. 3A). Putative single-stranded regions that have sequence identity with the CTE of D-type retroviruses are circled (compare to inset), as well as a purine-rich single-stranded region. The inset depicts a schematic representation of the mapped secondary structure of the M-PMV CTE and the sequences of its single-stranded regions.
The CTE of D-type retroviruses have been shown to mediate Rev-independent expression of HIV-1-derived RNAs by enhancing their nucleocytoplasmic transport (4, 65). One may predict, therefore, that other elements performing the same function work in a similar way, although alternative pathways must be considered. To analyze this for the IAPE, HeLa P4 cells were transiently transfected with pK-R-gpII in the presence or absence of Rev or with pK-gpII-IAPE. The cells were harvested and normalized for GFP expression, and nuclear and cytoplasmic RNA fractions were prepared and subjected to Northern blot analysis. The specific RNA was detected with a labeled antisense probe corresponding to the HIV-1 coding region present in all constructs. This analysis revealed a strong signal for the gag RNA in both the nuclear and cytoplasmic fractions of cells transfected with pK-R-gpII in the presence of Rev (Fig. 3C, lanes 2 and 5). A much weaker signal was observed in the nuclear fraction in the absence of Rev (Fig. 3C, lane 1), while virtually no specific RNA was detected in the cytoplasm in this case (lane 4). In contrast, the nuclear fraction of pK-gpII-IAPE-transfected cells revealed a gag-specific signal significantly stronger than that observed for pK-R-gpII plus Rev (ca. two- to fourfold more specific RNA as determined by phosphorimage analysis [Fig. 3C, lanes 2 and 3]), while comparable amounts of specific RNA were detected in the cytoplasmic fraction in both cases (Fig. 3C, lanes 5 and 6). The additional bands with higher mobilities in the nuclear fractions correspond to read-through products running into the downstream hygromycin expression unit, as determined by rehybridizing the blot with a hygromycin-specific probe (data not shown). Similar read-through products had been observed previously (60), and they are therefore not specific for the elements tested here. Taken together, these results indicate that the IAPE causes a significant increase in gag-specific RNA both in the nucleus and in the cytoplasm of transfected cells, but the relative enhancement is significantly stronger for the cytoplasmic fraction. Interestingly, levels of HIV-1 Gag expression were comparable for the 1.6-kb MIA14 sequence and for the IAPE (Fig. 3B, lanes 3 and 4), while the longer fragment led to much higher steady-state levels of nuclear gag RNA in transfected cells (Fig. 2C and data not shown).
The IAPE shares functionally important sequence motifs with the CTE.
Since the newly recognized element promotes Rev-independent expression of HIV-1 Gag, similar to the CTE of D-type retroviruses, we searched for sequence homologies between these elements. This was of particular importance, since a CTE-like element had previously been identified in the genome of the IAP osteocalcin-related gene (ORG) by sequence homology search (54). This analysis revealed no overall homology between the 360-base sequence from MIA14 and the CTE from D-type retroviruses. However, many RNA elements function through short single-stranded regions containing sequence-specific factor binding sites which are embedded in a structured region with no need for sequence conservation. We therefore analyzed the folding potential of the IAPE in more detail, with particular attention to predicted single-stranded regions. This analysis revealed several short regions of complete sequence identity in the single-stranded loops of the CTE and in the predicted single-stranded regions of the IAPE (Fig. 4). These single-stranded regions have been termed loops A1, B1, A2, and B2 (5′-3′) in the case of the CTE (Fig. 4, inset), and the IAPE contains all four motifs in or very close to predicted single-stranded loops (Fig. 4). Furthermore, the orders of these four putative loop regions, as well as the localization of a predicted splice donor (Fig. 4), are also identical in the IAPE and the CTE. These observations suggest that the elements may share binding motifs for the same or related cellular factors, while their overall sequences are highly divergent.
In order to analyze whether the predicted loop motifs are functionally relevant, we introduced mutations into the IAPE. In construct DI, the sequence corresponding to loop B1 was deleted (Fig. 5A, right), which should also affect the folding potential of loop A2 (not shown). A predicted purine-rich single-stranded region without homology to the CTE (Fig. 4) was deleted in construct DII, while the predicted single-stranded region corresponding to the A1 and B2 loops was converted into a stable stem structure in construct MI by introducing several point mutations (Fig. 5A, left). The variant IAPEs were inserted into pK-gpII, and the resulting plasmids were transiently transfected into HeLa P4 cells. Cell lysates normalized for GFP were subjected to Western and Northern blot analysis. Deletion of the purine-rich loop without homology to the CTE had no effect on IAPE-dependent HIV-1 Gag expression (Fig. 5B, lanes 2 and 4). In contrast, deletion of the B1 sequence in construct DI dramatically reduced HIV-1 Gag expression (Fig. 5B, lane 3), and conversion of the A1-B2 region into a predicted stable stem structure in construct MI led to a severe reduction as well (Fig. 5B, lane 5). Quantitative Western blot analysis of the respective cell lysates revealed ca. 20-fold-reduced Gag levels in the case of the DI construct and ca. 6-fold-reduced Gag levels in the case of the MI construct compared to the wild type (data not shown). Northern blot analysis of total RNA preparations from the same experiment showed a strong signal for the gag-specific RNA in the case of pK-gpII-IAPE (Fig. 5C, lane 2), while no specific RNA was detected in cells transfected with the antisense control (Fig. 5C, lane 3). Cells transfected with constructs DI (Fig. 5C, lane 4) and MI (lane 5) also revealed significantly smaller amounts of gag-RNA, but the observed differences (5-fold reduction for DI and 2.5-fold reduction for MI compared to the wild type; normalized for GAPDH [glyceraldehyde-3-phosphate dehydrogenase] levels) were less than those observed at the protein level.
FIG. 5.
Mutational analysis of IAPE function. (A) Schematic representation of the mutations introduced into the IAPE. The diagram on the left delineates the mutations introduced in order to generate mutant MI, which converts the predicted loops A1 and B2 into a (predicted) stable stem structure. The diagram on the right shows the mutations introduced to delete loop B1, which generated mutant DI. Folding prediction of mutant DI also displayed the disruption of loop A2 (data not shown). Compare to Fig. 4 for localization of the loops. wt, wild type. (B) HeLa cells were transiently transfected with pK-gpII derivatives containing the wild-type IAPE (lane 2) or deletion or substitution variants thereof. Variant DI (lane 3) is deleted in the B1 loop (compare Fig. 4), variant DII (lane 4) is deleted in the purine-rich loop (compare Fig. 4), and variant MI (lane 5) contains substitutions predicted to transform loops A1 and B2 (compare Fig. 4) into a stable RNA helix. Cell lysates normalized for transfection were analyzed by immunoblotting, as described in the legend to Fig. 1. (C) Northern blot analysis of RNA preparations derived from the same experiment shown in panel A. Total RNA was isolated from HeLa cells transiently transfected as indicated above each lane (+Rev, with Rev; a.s., antisense). HeLa cells transfected with pK-R-gpII and a Rev expression vector (lane 1) were analyzed as a control. The gag-specific RNA is marked on the right. Hybridization and detection were performed as described in the legend to Fig. 3C. The same blot was subsequently stripped and reanalyzed with a probe specific for GAPDH (bottom).
The IAPE functions independently of the export receptor Crm-1.
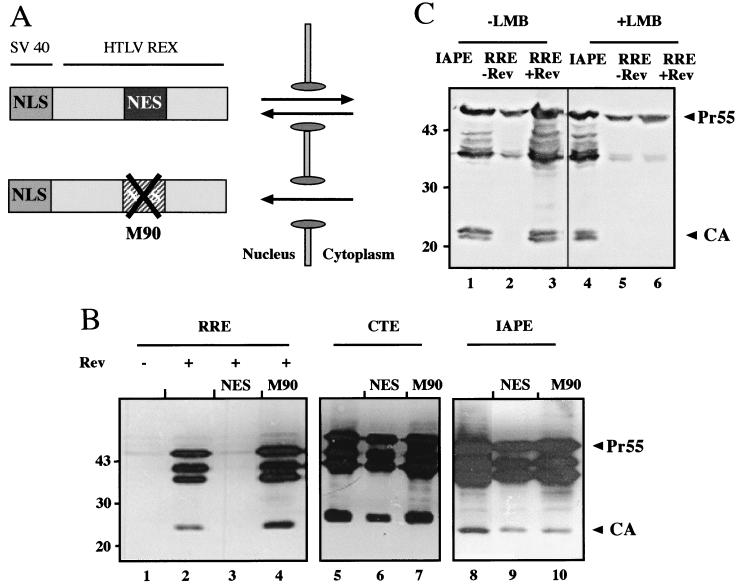
Nuclear RNA export by the Rev/RRE system uses the cellular export receptor Crm-1, which binds to the NES of Rev (13, 14, 37). CTE-dependent nuclear export, on the other hand, occurs independently of Crm-1. To test whether the IAPE uses the Crm-1 pathway, we performed competition experiments using an artificial fusion protein as a Crm-1 competitor. To this end, HIV-1 Gag expression vectors containing the RRE (with or without Rev), the M-PMV CTE, or the IAPE were cotransfected with an expression plasmid encoding a fusion protein carrying the NLS of SV40 and the NES of the human T-cell leukemia virus Rex protein (Fig. 6A) (30, 49). The resulting NLS-NES protein is expected to continuously shuttle between nucleus and cytoplasm and thereby titrate factors of the Crm-1-dependent export pathway (49, 56). A vector encoding the same fusion protein with an inactivating mutation in the NES sequence (M90) was used as a control. As expected, cotransfection of the NLS-NES competitor completely abrogated Rev-dependent Gag expression (Fig. 6B, lane 3), and this effect was dependent on a functional NES (lane 4). In contrast, CTE-mediated HIV-1 Gag expression was not affected by the NLS-NES competitor (Fig. 6B, lanes 5 to 7), confirming that the CTE functions independently of Crm-1. Similarly, HIV-1 Gag expression promoted by the IAPE was not inhibited by the NLS-NES competitor (Fig. 6B, lanes 8 to 10). Thus, the IAPE functions independently of Crm-1, similar to the CTE. This result was confirmed in an independent experiment using a truncated variant of the nucleoporin CAN (also called Nup 214), which also blocks Crm-1-dependent RNA export (1, 48). Cotransfection of the corresponding expression plasmid also abrogated Rev-dependent Gag expression but had no effect on CTE- or IAPE-dependent Gag expression (data not shown). In addition to genetic competition experiments, we also analyzed inhibition of Crm-1-dependent nuclear export by leptomycin B, a drug that specifically binds and inactivates Crm-1 (13). Similar to the results of genetic competition, Rev-dependent HIV-1 Gag expression was abolished in the presence of leptomycin B (Fig. 6C, lanes 3 and 6) while IAPE-dependent Gag expression remained completely unaffected (Fig. 6C, lanes 1 and 4).
FIG. 6.
Analysis of Crm-1 dependence of Gag expression. (A) Schematic representation of a competitor protein for NES-dependent nuclear export (30, 49). The competitor corresponds to a fusion protein containing the NLS of the SV40 large T antigen and the NES of the human T-cell leukemia virus Rex protein lacking the RNA binding domain. This molecule is expected to continuously shuttle between the nucleus and cytoplasm and titrates limiting factors of the NES export pathway. A variant containing an inactivating mutation in the NES domain of the fusion protein (M90) was used as a control. (B) HeLa cells were transiently transfected with plasmid 3-RRE in the absence (−; lane 1) or in the presence (+; lanes 2 to 4) of Rev, with plasmid 3-CTE (lanes 5 to 7), or with plasmid 3-IAPE (lanes 8 to 10). Transfections were performed with or without cotransfected expression plasmids for the NLS-NES competitor (NES) or the M90 variant thereof, as indicated above each lane. Gag expression was determined by immunoblot analysis as described in the legend to Fig. 1. (C) Analysis of Crm-1 dependence of Gag expression using leptomycin B. Cos-7 cells were transiently transfected with plasmid 3-RRE in the absence (lanes 2 and 5) or in the presence (lanes 3 and 6) of Rev or with plasmid 3-IAPE (lanes 1 and 4). Transfection solution was left on the cells for 3 h and then replaced with fresh medium (lanes 1 to 3) or with medium containing 100 nM leptomycin B (lanes 4 to 6). The cells were harvested 16 h after addition of the drug and subsequently analyzed for HIV-1 Gag expression as described in the legend to Fig. 1.
A functionally similar element is present in the same region of the primate foamy virus genome.
A database search for homologous sequences in other retroviral pol genes was not informative, because the pol genes are generally rather conserved due to selection pressure on the coding sequence. Considering that RNA elements may correspond to stable stem-loop structures, we performed a computer-based analysis of the folding potential of different retroviral pol sequences using the Mulfold program as described above. A stable RNA structure in the 3′ region of the pol gene was predicted for the genome of the primate foamy virus (data not shown). The sequence corresponding to the predicted structured region was PCR amplified and cloned into pK-gpII in sense or antisense orientation (Fig. 7A). The corresponding plasmids were transiently transfected into HeLa P4 cells, and HIV-1 Gag expression was determined by Western blot analysis of cell lysates normalized for GFP. Efficient expression of the Pr55gag polyprotein, the cleaved CA, and the two intermediate products was observed when the IAPE (Fig. 7B, lane 4) or the corresponding region from the foamy virus genome (lane 2) was present in sense, but not in antisense, orientation (Fig. 7B, lanes 3 and 5).
FIG. 7.
An expression element is present in the 3′ part of the pol open reading frame of primate foamy virus. (A) Schematic representation of the genome of foamy virus (top) showing the relative position of the region amplified by PCR (hatched box) and cloned in sense and antisense orientations into pK-gpII (bottom). LTR, long terminal repeat. (B) HeLa cells were mock transfected (lane 1) or transiently transfected with pK-gpII derivatives containing sequences derived from foamy virus (Fo; lanes 2 and 3) or the IAPE (lanes 4 and 5) either in sense (even lanes) or in antisense (as.) (odd lanes) orientation. HIV-1 Gag expression was determined by immunoblotting, as described in the legend to Fig. 1. (C) Northern blot analysis of RNA preparations derived from the same experiment shown in panel B. The numbering of lanes is identical to that in panel B. Total RNA was prepared from transfected cells, and gag-specific RNAs were detected using a radiolabeled probe hybridizing to the gag-PR region present in all constructs (depicted in panel A). The same blot was subsequently stripped and reanalyzed with a probe specific for GAPDH (bottom).
Northern blot analysis of total RNA preparations from the same transfection experiment revealed that protein expression levels directly correlated with steady-state RNA accumulation. Strong specific RNA signals were observed for the IAPE and the foamy virus sequence in sense orientation (Fig. 7C, lanes 2 and 4), while all sequences in antisense orientation gave low or undetectable RNA signals (Fig. 7C, lanes 3 and 5). Thus, the 3′ pol region from primate foamy virus apparently contains an orientation-specific RNA element with the capacity to overcome the nuclear restriction of HIV-1-derived RNAs.
DISCUSSION
In this report, we describe the identification and characterization of a new posttranscriptional expression element (IAPE) in the 3′ part of the pol open reading frame of the IAP MIA14 which is required for expression of the virus' own structural proteins. In addition, this element can functionally substitute for the Rev/RRE system of HIV and permits Rev-independent expression of HIV-1 structural proteins. The IAPE functions independently of the Rev export factor Crm-1 but appears to share binding sites with the CTE of D-type viruses, suggesting a partial overlap in function. Accordingly, the IAPE can be functionally replaced by the CTE in the expression of the virus' own structural proteins. However, the IAPE appears to exert additional effects on RNA stability in addition to a presumed role in nuclear RNA export, making it more akin to the PRE of pararetroviruses.
To date, three different mechanisms facilitating RNA export and expression have been characterized for different retro- and pararetroviruses: (i) the Rev/RRE system of primate lentiviruses and the homologous system of the HERV-K family of human endogenous retroviruses; (ii) the CTE of D-type viruses, binding to the cellular export factor Tap, and the functionally homologous DR element from RSV; and (iii) the PRE of HBV and woodchuck HBV, which appear to make use of a different but presently insufficiently characterized cellular pathway. It appears likely that each of these systems is adapted to the particular requirements of the respective virus or virus group, and related posttranscriptional elements are observed in other viruses, as in the thymidine kinase gene of herpes simplex virus (33), and possibly also in cellular gene expression (21). Consistent with their functional differences, insertion of the various elements into heterologous expression vectors leads to different effects on production of the respective proteins, and the relative enhancement of expression mediated by each element may depend on the specific gene of interest as well as the vector context (22, 51).
Our results clearly show that the newly identified IAPE does not belong to the Rev/RRE family of export systems. There is no evidence for a trans-acting export factor encoded by the MIA14 genome, and the IAPE functions in a heterologous context independent of IAP gene expression. Furthermore, IAPE function cannot be blocked by titrate factors of the Crm-1 export pathway or by biochemical inactivation of Crm-1 with the drug leptomycin B, which both completely and specifically abolished Rev-dependent protein production. There is also no obvious overall homology between the IAPE and the CTE of D-type viruses or of RSV. This element could not be predicted by homology searches based on the CTE sequence, which had been possible for the CTE-like element identified in a different IAP genome (IAP ORG) (54). Furthermore, the predicted structure of the IAPE is quite different from the determined structure of the CTE (Fig. 4), although both regions, as well as the RRE and the PRE, correspond to areas with secondary structure significantly more stable than that of the remainder of the genome (7, 11, 36, 45). However, the observation that there is almost complete sequence identity between predicted single-stranded regions of the IAPE and the four single-stranded loops of the M-PMV CTE, as well as the fact that the CTE could efficiently replace the IAPE in expression of the IAP structural proteins (Fig. 2B), suggests that both elements are likely to share binding sites for common cellular factors, which may include the Tap protein. The loop sequences have been shown to be essential for CTE function in nuclear export (15, 44, 50), and accordingly, deletion of one of these sequences in the IAPE abolished its function as well.
In addition to these analogies, there are also significant differences between the IAPE and the CTE. Most importantly, the presence of the IAPE leads to significantly increased total and nuclear RNA levels as well as increasing cytoplasmic RNA. Considering that enhanced transcription has not been sufficient to overcome Rev dependence in previous experiments (23), it appears unlikely that this increase in total RNA is the sole cause of Rev-independent HIV-1 Gag expression, and an export function is needed in addition to overcome nuclear restriction of gag RNA. Importantly, however, no effect on total or nuclear RNA levels was observed for the CTE, and even multimeric versions of the CTE, which provided multiple factor binding sites and significantly enhanced gene expression, did not cause any increase in total RNA (60). An additional difference between the CTE and the IAPE concerns the observation that the IAPE is fully functional in a vector harboring an intron in the 3′ untranslated region (pK-gpII-IAPE), while the presence of a CTE in an identical context did not yield Rev-independent HIV-1 Gag expression (60). The presence of an intron in the 3′ untranslated region of an mRNA has been shown to direct this RNA into the pathway of nonsense-mediated decay, and virtually all cellular RNAs therefore lack introns in this position (18, 40). It appears likely, therefore, that the IAPE can overcome the restriction of nonsense-mediated decay, a function which is not shared by the CTE.
An increase in steady-state levels of RNA, as observed for the IAPE, has previously been shown for other posttranscriptional control elements. The PRE of the pararetroviruses HBV and woodchuck HBV direct efficient cytoplasmic accumulation of intron-containing transcripts (7, 20) by using a pathway that differs, at least in part, from those of Rev/RRE and CTE (64). In addition, the presence of the PRE significantly increases the half-life of the viral RNA, and this effect is also observed for heterologous (spliced and unspliced) RNAs harboring this element (22, 51, 66). Thus, the PRE appears to function by a dual effect on RNA stability and on RNA nuclear export, and this may be similar to the observed effects of the IAPE. A dual effect on RNA export and stability has also been reported for the RSV DR element (41, 42), which functions as a CTE (43) but in addition appears to increase the stability of the unspliced viral transcript (16, 42).
Intriguingly, the IAPE function of mediating Rev-independent HIV-1 Gag expression is conserved in the same position of the genome of a primate foamy virus and may also be shared by a murine leukemia virus (unpublished observation). Insertion of a predicted structured region from the 3′ end of the pol gene of foamy virus also provided orientation-dependent enhancement of Gag expression and increase of gag RNA levels. The functional significance of this finding and the importance of this region for replication of the respective virus are supported by previously published experiments. Retroviral vectors based on foamy viruses have been constructed by several groups, and transduction by these vectors is absolutely dependent on sequences corresponding to the 3′ part of the pol gene (17, 61), overlapping the region used in our study. In fact, this is the only cis-acting RNA sequence required for transduction besides the viral long terminal repeats and a short stretch in the 5′ gag region (17). Furthermore, insertion of the foamy virus-derived RNA element into murine leukemia virus-based retroviral vectors led to significantly enhanced expression of the transduced genes (61). This effect was also observed for marker genes (e.g., the GFP gene), which were unaffected by the presence of a CTE but could be enhanced by the PRE of pararetroviruses. It can be speculated, therefore, that this functional element may be evolutionarily linked to the PRE of pararetroviruses. Foamy viruses are classified as retroviruses because their genomes are integrated into a host cell chromosome, but foamy virus replication shares many features with pararetrovirus replication, and foamy viruses have been considered an evolutionary link between retro- and pararetroviruses (32). Intriguingly, the PRE, as well as the IAPE and the Fo element, are all localized in the same region of the respective genomes, overlapping the 3′ part of the pol reading frame.
ACKNOWLEDGMENTS
We thank M. Dobbelstein for plasmids and helpful suggestions and K. Lueders for antisera. We also thank B. Henkel for help with tissue culture and M. Dittmar for critically reading the manuscript.
This work was supported in part by grants from the Deutsche Forschungsgemeinschaft to H.-G.K. (SFB545 and Ri192/21-1-1). J. Bohne was supported by the Boehringer Ingelheim Fonds
REFERENCES
- 1.Bogerd H P, Echarri A, Ross T M, Cullen B R. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bohnlein E, Berger J, Hauber J. Functional mapping of the human immunodeficiency virus type 1 Rev RNA binding domain: new insights into the domain structure of Rev and Rex. J Virol. 1991;65:7051–7055. doi: 10.1128/jvi.65.12.7051-7055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun I C, Rohrbach E, Schmitt C, Izaurralde E. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 1999;18:1953–1965. doi: 10.1093/emboj/18.7.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bray M, Prasad S, Dubay J W, Hunter E, Jeang K T, Rekosh D, Hammarskjold M L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc Natl Acad Sci USA. 1994;91:1256–1260. doi: 10.1073/pnas.91.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C A, Okayama H. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. BioTechniques. 1988;6:632–638. [PubMed] [Google Scholar]
- 6.Cullen B R. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology. 1998;249:203–210. doi: 10.1006/viro.1998.9331. [DOI] [PubMed] [Google Scholar]
- 7.Donello J E, Loeb J E, Hope T J. Woodchuck hepatitis virus contains a tripartite posttranscriptional regulatory element. J Virol. 1998;72:5085–5092. doi: 10.1128/jvi.72.6.5085-5092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ernst R K, Bray M, Rekosh D, Hammarskjold M L. Secondary structure and mutational analysis of the Mason-Pfizer monkey virus RNA constitutive transport element. RNA. 1997;3:210–222. [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst R K, Bray M, Rekosh D, Hammarskjold M L. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol Cell Biol. 1997;17:135–144. doi: 10.1128/mcb.17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fehrmann F, Welker R, Krausslich H G. Intracisternal A-type particles express their proteinase in a separate reading frame by translational frameshifting, similar to D-type retroviruses. Virology. 1997;235:352–359. doi: 10.1006/viro.1997.8708. [DOI] [PubMed] [Google Scholar]
- 11.Felber B K, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis G N. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc Natl Acad Sci USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer U, Huber J, Boelens W C, Mattaj I W, Luhrmann R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 13.Fornerod M, Ohno M, Yoshida M, Mattaj I W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 14.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 15.Gruter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber B K, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 16.Guo W, Winistorfer S C, Stoltzfus C M. Selective inhibition of splicing at the avian sarcoma virus src 3′ splice site by direct-repeat posttranscriptional cis elements. J Virol. 2000;74:8513–8523. doi: 10.1128/jvi.74.18.8513-8523.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinkelein M, Schmidt M, Fischer N, Moebes A, Lindemann D, Enssle J, Rethwilm A. Characterization of a cis-acting sequence in the Pol region required to transfer human foamy virus vectors. J Virol. 1998;72:6307–6314. doi: 10.1128/jvi.72.8.6307-6314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hentze M W, Kulozik A E. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 19.Hirose Y, Manley J L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 20.Huang J, Liang T J. A novel hepatitis B virus (HBV) genetic element with Rev response element-like properties that is essential for expression of HBV gene products. Mol Cell Biol. 1993;13:7476–7486. doi: 10.1128/mcb.13.12.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, Carmichael G G. The mouse histone H2a gene contains a small element that facilitates cytoplasmic accumulation of intronless gene transcripts and of unspliced HIV-1-related mRNAs. Proc Natl Acad Sci USA. 1997;94:10104–10109. doi: 10.1073/pnas.94.19.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Y, Wimler K M, Carmichael G G. Intronless mRNA transport elements may affect multiple steps of pre-mRNA processing. EMBO J. 1999;18:1642–1652. doi: 10.1093/emboj/18.6.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iacampo S, Cochrane A. Human immunodeficiency virus type 1 Rev function requires continued synthesis of its target mRNA. J Virol. 1996;70:8332–8339. doi: 10.1128/jvi.70.12.8332-8339.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Izaurralde E, Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- 25.Izaurralde E, Mattaj I W. RNA export. Cell. 1995;81:153–159. doi: 10.1016/0092-8674(95)90323-2. [DOI] [PubMed] [Google Scholar]
- 26.Jaeger J A, Turner D H, Zuker M. Improved predictions of secondary structures for RNA. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaeger J A, Turner D H, Zuker M. Predicting optimal and suboptimal secondary structure for RNA. Methods Enzymol. 1990;183:281–306. doi: 10.1016/0076-6879(90)83019-6. [DOI] [PubMed] [Google Scholar]
- 28.Kang Y, Bogerd H P, Yang J, Cullen B R. Analysis of the RNA binding specificity of the human tap protein, a constitutive transport element-specific nuclear RNA export factor. Virology. 1999;262:200–209. doi: 10.1006/viro.1999.9906. [DOI] [PubMed] [Google Scholar]
- 29.Kang Y, Cullen B R. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katahira J, Ishizaki T, Sakai H, Adachi A, Yamamoto K, Shida H. Effects of translation initiation factor eIF-5A on the functioning of human T-cell leukemia virus type I Rex and human immunodeficiency virus Rev inhibited trans dominantly by a Rex mutant deficient in RNA binding. J Virol. 1995;69:3125–3133. doi: 10.1128/jvi.69.5.3125-3133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuff E L, Lueders K K. The intracisternal A-particle gene family: structure and functional aspects. Adv Cancer Res. 1988;51:183–276. doi: 10.1016/s0065-230x(08)60223-7. [DOI] [PubMed] [Google Scholar]
- 32.Linial M L. Foamy viruses are unconventional retroviruses. J Virol. 1999;73:1747–1755. doi: 10.1128/jvi.73.3.1747-1755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, Mertz J E. HnRNP L binds a cis-acting RNA sequence element that enables intron-dependent gene expression. Genes Dev. 1995;9:1766–1780. doi: 10.1101/gad.9.14.1766. [DOI] [PubMed] [Google Scholar]
- 34.Magin C, Lower R, Lower J. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J Virol. 1999;73:9496–9507. doi: 10.1128/jvi.73.11.9496-9507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malim M H, Bohnlein S, Hauber J, Cullen B R. Functional dissection of the HIV-1 Rev trans-activator—derivation of a trans-dominant repressor of Rev function. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 36.Malim M H, Hauber J, Le S Y, Maizel J V, Cullen B R. The HIV-1 Rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 37.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 38.Mergener K, Facke M, Welker R, Brinkmann V, Gelderblom H R, Krausslich H G. Analysis of HIV particle formation using transient expression of subviral constructs in mammalian cells. Virology. 1992;186:25–39. doi: 10.1016/0042-6822(92)90058-w. [DOI] [PubMed] [Google Scholar]
- 39.Mietz J A, Grossman Z, Lueders K K, Kuff E L. Nucleotide sequence of a complete mouse intracisternal A-particle genome: relationship to known aspects of particle assembly and function. J Virol. 1987;61:3020–3029. doi: 10.1128/jvi.61.10.3020-3029.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nagy E, Maquat L E. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 41.Ogert R A, Beemon K L. Mutational analysis of the Rous sarcoma virus DR posttranscriptional control element. J Virol. 1998;72:3407–3411. doi: 10.1128/jvi.72.4.3407-3411.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogert R A, Lee L H, Beemon K L. Avian retroviral RNA element promotes unspliced RNA accumulation in the cytoplasm. J Virol. 1996;70:3834–3843. doi: 10.1128/jvi.70.6.3834-3843.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paca R E, Ogert R A, Hibbert C S, Izaurralde E, Beemon K L. Rous sarcoma virus DR posttranscriptional elements use a novel RNA export pathway. J Virol. 2000;74:9507–9514. doi: 10.1128/jvi.74.20.9507-9514.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pasquinelli A E, Ernst R K, Lund E, Grimm C, Zapp M L, Rekosh D, Hammarskjold M L, Dahlberg J E. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997;16:7500–7510. doi: 10.1093/emboj/16.24.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patzel V, Sczakiel G. The hepatitis B virus posttranscriptional regulatory element contains a highly stable RNA secondary structure. Biochem Biophys Res Commun. 1997;231:864–867. doi: 10.1006/bbrc.1997.6205. [DOI] [PubMed] [Google Scholar]
- 46.Pollard V W, Malim M H. The HIV-1 Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 47.Rittner K, Sczakiel G. Identification and analysis of antisense RNA target regions of the human immunodeficiency virus type 1. Nucleic Acids Res. 1991;19:1421–1426. doi: 10.1093/nar/19.7.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosorius O, Reichart B, Kratzer F, Heger P, Dabauvalle M C, Hauber J. Nuclear pore localization and nucleocytoplasmic transport of eIF-5A: evidence for direct interaction with the export receptor CRM1. J Cell Sci. 1999;112:2369–2380. doi: 10.1242/jcs.112.14.2369. [DOI] [PubMed] [Google Scholar]
- 49.Roth J, Dobbelstein M. Export of hepatitis B virus RNA on a Rev-like pathway: inhibition by the regenerating liver inhibitory factor IκBα. J Virol. 1997;71:8933–8939. doi: 10.1128/jvi.71.11.8933-8939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saavedra C, Felber B, Izaurralde E. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr Biol. 1997;7:619–628. doi: 10.1016/s0960-9822(06)00288-0. [DOI] [PubMed] [Google Scholar]
- 51.Schambach A, Wodrich H, Hildinger M, Bohne J, Krausslich H G, Baum C. Context dependence of different modules for posttranscriptional enhancement of gene expression from retroviral vectors. Mol Ther. 2000;2:435–445. doi: 10.1006/mthe.2000.0191. [DOI] [PubMed] [Google Scholar]
- 52.Simpson S B, Zhang L, Craven R C, Stoltzfus C M. Rous sarcoma virus direct repeat cis elements exert effects at several points in the virus life cycle. J Virol. 1997;71:9150–9156. doi: 10.1128/jvi.71.12.9150-9156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stutz F, Rosbash M. Nuclear RNA export. Genes Dev. 1998;12:3303–3319. doi: 10.1101/gad.12.21.3303. [DOI] [PubMed] [Google Scholar]
- 54.Tabernero C, Zolotukhin A S, Bear J, Schneider R, Karsenty G, Felber B K. Identification of an RNA sequence within an intracisternal-A particle element able to replace Rev-mediated posttranscriptional regulation of human immunodeficiency virus type 1. J Virol. 1997;71:95–101. doi: 10.1128/jvi.71.1.95-101.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tabernero C, Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. The posttranscriptional control element of the simian retrovirus type 1 forms an extensive RNA secondary structure necessary for its function. J Virol. 1996;70:5998–6011. doi: 10.1128/jvi.70.9.5998-6011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weigel S, Dobbelstein M. The nuclear export signal within the E4orf6 protein of adenovirus type 5 supports virus replication and cytoplasmic accumulation of viral mRNA. J Virol. 2000;74:764–772. doi: 10.1128/jvi.74.2.764-772.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Welker R, Janetzko A, Krausslich H G. Plasma membrane targeting of chimeric intracisternal A-type particle polyproteins leads to particle release and specific activation of the viral proteinase. J Virol. 1997;71:5209–5217. doi: 10.1128/jvi.71.7.5209-5217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 59.Wodrich H, Kraeusslich H-G. Nucleocytoplasmic RNA transport in retroviral replication. Results Probl Cell Differ. 2001;34:197–217. doi: 10.1007/978-3-540-40025-7_12. [DOI] [PubMed] [Google Scholar]
- 60.Wodrich H, Schambach A, Krausslich H G. Multiple copies of the Mason-Pfizer monkey virus constitutive RNA transport element lead to enhanced HIV-1 Gag expression in a context-dependent manner. Nucleic Acids Res. 2000;28:901–910. doi: 10.1093/nar/28.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu M, Chari S, Yanchis T, Mergia A. cis-acting sequences required for simian foamy virus type 1 vectors. J Virol. 1998;72:3451–3454. doi: 10.1128/jvi.72.4.3451-3454.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang J, Bogerd H P, Peng S, Wiegand H, Truant R, Cullen B R. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc Natl Acad Sci USA. 1999;96:13404–13408. doi: 10.1073/pnas.96.23.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yoon D W, Lee H, Seol W, DeMaria M, Rosenzweig M, Jung J U. Tap: a novel cellular protein that interacts with Tip of herpesvirus saimiri and induces lymphocyte aggregation. Immunity. 1997;6:571–582. doi: 10.1016/s1074-7613(00)80345-3. [DOI] [PubMed] [Google Scholar]
- 64.Zang W Q, Yen T S. Distinct export pathway utilized by the hepatitis B virus posttranscriptional regulatory element. Virology. 1999;259:299–304. doi: 10.1006/viro.1999.9777. [DOI] [PubMed] [Google Scholar]
- 65.Zolotukhin A S, Valentin A, Pavlakis G N, Felber B K. Continuous propagation of RRE(−) and Rev(−)RRE(−) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J Virol. 1994;68:7944–7952. doi: 10.1128/jvi.68.12.7944-7952.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zufferey R, Donello J E, Trono D, Hope T J. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]